Abstract
Characterizing the nature of the adaptive process at the genetic level is a central goal for population genetics. In particular, we know little about the sources of adaptive substitution or about the number of adaptive variants currently segregating in nature. Historically, population geneticists have focused attention on the hard-sweep model of adaptation in which a de novo beneficial mutation arises and rapidly fixes in a population. Recently more attention has been given to soft-sweep models, in which alleles that were previously neutral, or nearly so, drift until such a time as the environment shifts and their selection coefficient changes to become beneficial. It remains an active and difficult problem, however, to tease apart the telltale signatures of hard vs. soft sweeps in genomic polymorphism data. Through extensive simulations of hard- and soft-sweep models, here we show that indeed the two might not be separable through the use of simple summary statistics. In particular, it seems that recombination in regions linked to, but distant from, sites of hard sweeps can create patterns of polymorphism that closely mirror what is expected to be found near soft sweeps. We find that a very similar situation arises when using haplotype-based statistics that are aimed at detecting partial or ongoing selective sweeps, such that it is difficult to distinguish the shoulder of a hard sweep from the center of a partial sweep. While knowing the location of the selected site mitigates this problem slightly, we show that stochasticity in signatures of natural selection will frequently cause the signal to reach its zenith far from this site and that this effect is more severe for soft sweeps; thus inferences of the target as well as the mode of positive selection may be inaccurate. In addition, both the time since a sweep ends and biologically realistic levels of allelic gene conversion lead to errors in the classification and identification of selective sweeps. This general problem of “soft shoulders” underscores the difficulty in differentiating soft and partial sweeps from hard-sweep scenarios in molecular population genomics data. The soft-shoulder effect also implies that the more common hard sweeps have been in recent evolutionary history, the more prevalent spurious signatures of soft or partial sweeps may appear in some genome-wide scans.
Keywords: natural selection, population genetics, selective sweeps
THERE is a growing body of evidence that positive directional selection has a pervasive impact on genetic variation within and between species (reviewed in Hahn 2008; Sella et al. 2009; Cutter and Payseur 2013). In stark contrast to predictions from Kimura’s neutral theory of molecular evolution (Kimura 1968), there is strong evidence that in at least some taxa a large fraction of nucleotide differences between species have been fixed by positive selection (Fay et al. 2001, 2002; Smith and Eyre-Walker 2002; Begun et al. 2007; Boyko et al. 2008; Langley et al. 2012). A great deal of emphasis has therefore been placed on identifying the population genetic signatures of adaptation, in hopes of revealing the genetic basis of recent phenotypic innovations. Typically this is done by investigating genetic variation at a locus of interest using one or more summary statistics capturing information about allele frequencies (Tajima 1989; Fu and Li 1993; Fay and Wu 2000; Kim and Stephan 2002; Achaz 2009), linkage disequilibrium (Kelly 1997; Kim and Nielsen 2004), or haplotypic diversity (Hudson et al. 1994; Sabeti et al. 2002; Voight et al. 2006) and asking whether the values of these statistics differ from the expectation under neutrality. More recently, these efforts have taken the form of genome-wide population genetic scans (e.g., Sabeti et al. 2002; Nielsen et al. 2005; Voight et al. 2006; Pickrell et al. 2009). These scans have identified many loci that appear to have experienced recent positive selection.
While adaptation is an important evolutionary force, the precise mode by which beneficial mutations become fixed in populations remains unclear. According to the classic model of a “selective sweep,” a novel beneficial mutation occurs once, quickly rising in frequency until it becomes fixed. Immediately following the fixation, the vicinity around the selected site contains no polymorphism except that introduced by recombination or mutation during the sweep (Maynard Smith and Haigh 1974). This is because in the sweep region all sampled individuals will have coalesced extremely recently, and in the absence of recombination the most recent common ancestor (MRCA) will be no older than the time at which the selected mutation occurred. As genetic (recombination) distance to the selected site increases, so does the expected size of the coalescent tree, and thus polymorphism can approach background genomic levels (Kaplan et al. 1989). Perhaps due to its relative simplicity in comparison to other models of selection, this model has received a great deal of theoretical attention; the expected impact of a selective sweep on patterns of genetic variation at linked sites has thus been well characterized (e.g., Maynard Smith and Haigh 1974; Kaplan et al. 1989; Stephan et al. 1992; Barton 1998; Durrett and Schweinsberg 2004; Mcvean 2007; Messer and Neher 2012).
In contrast to the “hard-sweep” model described above, an alternative model concerns adaptation via a mutation that is initially fitness neutral (or nearly so) and thus evolves by drift until, perhaps due to an environmental shift, it becomes beneficial and sweeps to fixation (Gillespie 1991; Orr and Betancourt 2001). Importantly, the frequency at which the sweep begins may be substantially greater than 1/2N (where N is the population size), the starting frequency of a hard sweep, and as a consequence the beneficial mutation may be linked to numerous backgrounds. Thus, although the effect of this type of adaptation on linked variation is not as well described as Maynard Smith and Haigh’s model (Maynard Smith and Haigh 1974), it is expected to produce a less severe reduction in diversity (Innan and Kim 2004; Przeworski et al. 2005)—hence the term “soft sweeps” for this model (Hermisson and Pennings 2005). Because multiple distinct haplotypes may be sweeping rather than just one, there will be an excess of intermediate alleles relative to the expectation under the hard-sweep model (Przeworski et al. 2005; Barrett and Schluter 2008) and sometimes relative to the expectation under neutrality (Teshima et al. 2006).
Although the soft-sweep model has only recently been considered through the lens of population genetics, it is a compelling alternative to the hard-sweep model because it eliminates the need for a population to wait for an adaptive mutation to occur after an environmental shift: if any segregating polymorphisms have acquired a positive selection coefficient because of the environmental shift, natural selection will act immediately on this standing variation (Gillespie 1991; Barrett and Schluter 2008). Indeed, artificial selection on quantitative traits often results in an immediate phenotypic response, and the strength of this response is correlated with the amount of standing variation in the trait of interest (Falconer and Mackay 1996). Thus, if a population’s selective environment changes frequently enough, soft sweeps may be the primary mode of adaptation. Furthermore, because at the onset of a soft sweep the adaptive allele may be at frequency >>1/2N (Innan and Kim 2004), the probability of fixation once the sweep begins is higher (Kimura 1957), although of course such an allele would have already had to have drifted to this frequency. In support of the soft sweep model, recent population genetic studies have identified candidate adaptive events that appear to have been driven by selection on standing variation (e.g., Hamblin and Di Rienzo 2000; Scheinfeldt et al. 2009; Jones et al. 2012; Peter et al. 2012). An alternative phenomenon resulting in a somewhat similar type of soft sweep is that of recurrent mutation to the adaptive allele, each of which could occur on distinct haplotypes during the sweep. This scenario is plausible when the population mutation rate to the adaptive allele is high enough (Pennings and Hermisson 2006a).
A third scenario of positive selection that has garnered a great deal of attention over the past 2 decades is that of “partial” or “incomplete” sweeps, where a sweeping allele has not reached fixation (Hudson et al. 1994; Voight et al. 2006; Pritchard et al. 2010). Under this model, a new mutation is initially beneficial and rapidly increases in frequency, but does not reach fixation, perhaps because the sweep is still ongoing at the time the population is sampled. Searching for signals of partial sweeps may thus reveal adaptive evolution in action. For this reason the signal of partial sweeps has been the focus of a popular class of haplotype-based tests for selection (Hudson et al. 1994; Sabeti et al. 2002; Voight et al. 2006; Sabeti et al. 2007); these methods have identified numerous candidate loci, apparently supporting the partial sweep model. On the other hand, because the sojourn of an adaptive mutation is expected to be brief (Fisher 1937; Maynard Smith and Haigh 1974), selective sweeps must be commonplace if we are to frequently catch them in the act. Alternatively, partial sweeps may be observed under a scenario that is essentially the opposite of the soft sweep scenario: an initially beneficial allele rapidly increases in frequency until an environmental shift eliminates its selective advantage and the allele begins to drift, or a frequency dependence on fitness renders the beneficial allele less so as it increases in frequency (Gillespie 1991).
While the relative prominence of each of these modes of adaptive evolution is unknown, for some parameter combinations the three models may have distinct effects on patterns of genetic variation. Thus it is appealing to design population genetic tests to distinguish among models and to reveal the extent to which each has been responsible for recent adaptation. In this article we examine population genetic summary statistics that can be used to identify partial or soft sweeps and show that their effectiveness may be compromised in regions linked to recent hard sweeps. Indeed, it appears that the pattern of variation at a site linked to a hard sweep, depending on genetic distance and the strength of selection, can be agonizingly similar to that of a partial or soft sweep—a phenomenon we call the “soft-shoulder effect.” The soft-shoulder effect implies, somewhat counterintuitively, that the more common hard sweeps have been in recent evolutionary history, the more prevalent spurious signatures of soft or partial sweeps may appear in genome-wide scans.
Materials and Methods
Statistics used to detect selection
We used three classes of population genetic summary statistics to differentiate among regions evolving neutrally, regions recently experiencing hard selective sweeps, and regions recently experiencing soft sweeps. These statistics include: (1) those capturing the number of and frequency of derived alleles [nucleotide diversity or π, Nei and Li 1979; Tajima 1983; the number of polymorphisms (S); Tajima’s D, Tajima 1989; and Fay and Wu’s and H statistics, Fay and Wu 2000]; (2) those measuring the number and frequency of distinct haplotypes [the number of distinct haplotypes (Hudson et al. 1994) and haplotype homozygosity—the fraction of pairs of haplotypes that are identical (Depaulis and Veuille 1998)]; (3) those containing linkage disequilibrium (LD) information (Kelly’s ZnS, Kelly 1997, and Kim and Nielsen’s ω statistic, Kim and Nielsen 2004, fixing the focal site to the center of the window being examined). We used support vector machines (SVMs) leveraging these statistics to distinguish among evolutionary models as described below.
We also used the iHS (integrated haplotype score) statistic (Voight et al. 2006) to search for signatures of partial sweeps. iHS builds on the extended haplotype homozygosity (EHH) statistic, which is simply the fraction of all pairs of chromosomes sharing the same allele at a focal segregating site that is identical within some range of positions (Sabeti et al. 2002). Briefly, iHS is based on integrated EHH, or iHH, which is calculated by taking the sum of all EHH values computed for a given allele at a given site within increasingly larger regions surrounding the focal polymorphism and terminating the summation when EHH drops to 0.05 (Voight et al. 2006). Unstandardized iHS is then defined as
| (1) |
where iHHA is the iHH score for the ancestral allele at the focal polymorphism and iHHD is the iHH score for the derived allele. Given that iHHD will be elevated for younger (typically lower frequency) alleles, Voight et al. (2006) recommend standardizing each iHS score according to the mean and standard deviation of iHS scores of polymorphisms having a similar derived allele frequency to that of the focal polymorphism.
Coalescent simulations
To obtain the expected distribution of all statistics immediately following a hard sweep, immediately following a soft sweep, and under neutrality, we performed coalescent simulations for each of these scenarios. For these simulations, we set θ = 4Nμ (where μ is the rate of mutation to neutral alleles) to 0.01 per base pair, and sampled 50 individuals. For soft sweeps from standing variation, we drew the initial selected frequency of the sweeping allele uniformly from the range U(0.05, 0.2). For soft sweeps from recurrent mutation, the rate of mutation to the adaptive allele (4NμA, where μA is the per-base-pair adaptive mutation rate) was selected from U(1, 2.5). For hard and soft sweeps the selection coefficient α = 2Ns was set to 1000, 2000, or 5000. Unless stated otherwise, 1000 replicate simulations were performed for each evolutionary scenario. We also simulated hard sweeps within 10-kb chromosomes with gene conversion rates 4Nγ ranging from 0 to 500 in increments of 50 (with α = 1000 and ρ = 100), as well as sweeps completing at various times in the past (number of generations post-sweep: 0.000625 × 2N, 0.00125 × 2N, 0.0025 × 2N, 0.005 × 2N, 0.01 × 2N, 0.02 × 2N, 0.04 × 2N, 0.08 × 2N, 0.16 × 2N, 0.32 × 2N, 0.64 × 2N, and 1.28 × 2N). To examine spatial patterns of variation around selective sweeps, we simulated sets of 210-kb chromosomes experiencing recent hard sweeps in the center (with α = 1000, ρ = 2100, and θ = 2100) and with 4Nγ of 0 and 2100, respectively, as well as sets of these large chromosomes occurring at the same range of ages post-sweep as listed above. Similarly, we also simulated a set of 210-kb chromosomes with a soft sweep occurring in the center (again with the initial selected frequency ranging from 0.05 to 0.2). Note that these larger simulations, which were subdivided into 21 adjacent 10-kb windows in downstream analyses, have the same per-base-pair crossover and mutation rates as the smaller simulated chromosomes.
All simulations were performed using our in-house coalescent simulation software, discoal. Source code in C and compilation instructions for discoal are available for download from GitHub (http://github.com/kern-lab/discoal). Supporting Information, Table S1 describes each set of coalescent simulations we performed for this study.
Testing sites linked to hard sweeps for spurious evidence of soft sweeps
We examined regions flanking simulated hard sweeps for patterns of variation expected under a soft sweep as follows. First, we simulated 1000 chromosomes each 100 kb in length, with a hard sweep occurring at position 5000 along the chromosome (i.e., toward the far left end) and reaching fixation immediately prior to sampling (simulation sets 9–11 from Table S1). We subdivided the chromosome into 10 adjacent windows each 10 kb in length and calculated values of the population genetic summary statistics listed above. We then classified each window for each simulation as evolving neutrally, recently experiencing a soft sweep, or recently experiencing a hard sweep according to the values of various summary statistics as described below. Importantly, the only window directly experiencing any selection was the first window of the chromosome, which experienced a hard sweep at its center. The extent to which linked neutral windows were misclassified as selected therefore reveals how linkage to a hard sweep creates patterns of genetic variation that are misleadingly consistent with hard or soft sweeps. We repeated this process, which is outlined in Figure S1, for three different values of α: 1000, 2000, and 5000. For these simulations the locus-wide value of ρ was set to equal α.
Classifying simulated regions as hard sweeps, soft sweeps, or neutral
We developed a classifier to label each window within each of our larger simulations as a hard sweep, a soft sweep from standing variation, or neutral. We trained this classifier by simulating 1000 chromosomes 10 kb in length (total ρ = 100) either experiencing a hard sweep at its center, experiencing a soft sweep from standing variation at its center, experiencing a soft sweep from recurrent mutation at its center, or evolving neutrally as described above (simulation sets 1–8 from Table S1). For these training simulations the selection coefficient and per-base-pair recombination rate was identical to that used for the corresponding set of larger simulations to which the classifier was to be applied.
Next, we calculated values of various population genetic summary statistics (listed above) for each of these 10-kb simulated windows. We then trained a SVM to classify each of these windows as a hard sweep, a soft sweep, or as evolving neutrally. Briefly, SVMs work by discovering a hyperplane that optimally separates two sets of multidimensional data known to belong to two different classes (training data). Additional data whose classes are unknown are then classified according to the side of the hyperplane on which they lie (Vapnik and Lerner 1963). SVMs can be extended to allow for three or more classes (e.g., Knerr et al. 1990; Platt et al. 2000) and to utilize nonlinear decision surfaces by mapping data to higher-dimensional space using kernel functions (Aizerman et al. 1964; Boser et al. 1992; Cortes and Vapnik 1995). We constructed a training set from 1000 simulated 10-kb windows containing a hard sweep, 1000 soft sweeps, and 1000 neutrally evolving regions (simulations described above). We then trained a multiclass SVM using Knerr et al.’s method (Knerr et al. 1990) of extending binary classification to three or more classes; for this SVM we used a radial basis kernel function. Briefly, we performed a coarse grid search to obtain optimal values of the SVM’s cost hyperparameter and the radial basis function’s γ-hyperparameter, performing 10-fold cross-validation for each hyperparameter combination. The set of values tested for each of these hyperparameters was [10−4, 10−3, 10−2, 10−1, 10°, 101, 102, 103, 104]. The combination of these hyperparameters yielding the highest cross-validation accuracy was then used to classify each 10-kb window from the 100-kb simulated chromosomes, as well as simulated 10-kb chromosomes experiencing hard sweeps of varying ages and with varying gene conversion rates. All training and classification was performed via the scikit-learn python library (http://scikit-learn.org).
Testing sites linked to hard sweeps for spurious evidence of partial sweeps
We performed 5000 coalescent simulations of 50-kb regions with a hard sweep occurring in the center of the chromosome (i.e., position 25,000) with α = ρ = 1000, 5000 such simulations with α = ρ = 2000, and 5000 simulations of neutral evolution (simulation sets 12–15 from Table S1). We then calculated iHS scores for each polymorphism in each simulation using iHS_calc and standardized them based on allele frequency using WHAMM.pl (both available at http://coruscant.itmat.upenn.edu/whamm/index.html). Standardization was performed according to the mean and variance of iHS scores within 20 equally sized allele-frequency bins from the neutral simulations—this step is important for ensuring that iHS scores do not correlate with allele frequency under neutrality. We then calculated the average absolute value of standardized iHS scores in each 1-kb window along the 50-kb region across all simulations with the same selection coefficient. We also calculated one-sided P-values for each polymorphism’s iHS score by comparing it to those of neutral polymorphisms of similar allele frequency (i.e., found in the same of the 20 allele-frequency bins): for a score above the neutral median, the P-value was the fraction of neutral polymorphisms in the frequency bin with a greater or equal iHS score, while for below-median scores the P-value was the fraction of lesser or equal neutral iHS scores. Because these P-value calculations take allele frequency into account, they were performed using unstandardized iHS scores.
Results
Hard sweeps produce signatures of soft sweeps at linked loci
The well-known signature of a soft sweep is that near the selected site, multiple haplotypes carrying the beneficial allele increase in frequency. A fixation caused by a hard sweep will leave only a single haplotype carrying the beneficial allele in the immediate vicinity of the selected site (modulo mutations that occur during the sweep; Messer and Neher 2012). Consider, however, a neutral locus linked to a beneficial allele at some intermediate genetic distance. During a hard sweep, recombination between the selected and neutral loci will lead to multiple haplotypes linked to the beneficial allele during its sojourn through the population. Thus, in linked regions far enough away from the target of selection, there will be several haplotypes at elevated frequencies after a hard sweep (Begun and Aquadro 1994; Hudson et al. 1994), similar to the expectation for the selected locus after a soft sweep. We therefore reasoned that scans for soft sweeps might produce false positives in the “shoulders” of a hard sweep.
To test this hypothesis we used coalescent simulations to generate population samples from 100-kb chromosomes experiencing a hard sweep at one end of the chromosome, partitioning these chromosomes into adjacent 10-kb windows for analysis (simulation sets 9–11 from Table S1). The distances between the centers of these windows and the selective sweep range from 0.1 to 0.9 units of ρ/α, where ρ is the population recombination rate 4NrL between sites L bp apart with per-base-pair crossover rate r, and α is the selection coefficient 2Ns, which we set to 1000. Assuming a population size of N = 1 × 104, these distances correspond to 0.25–2.25 cM (roughly 0.25–2.25 Mb in the human genome; Kong et al. 2002), while with N = 1 × 106 they correspond to 0.0025–0.025 cM (∼1.25–12.5 kb in Drosophila; Comeron et al. 2012); these distances will be greater for larger values of α. We then compared the patterns of variation within each of these windows to those from simulated 10 kb chromosomes evolving neutrally, experiencing a hard sweep, or experiencing a soft sweep.
As shown in Figure 1A, nucleotide diversity (π) is extremely depleted in the window containing the just-completed hard sweep, as expected under the hard-sweep model. As we examine windows further and further away from the sweep, π increases from its trough, gradually recovering toward the neutral expectation. In several of these windows we observe median values of π that appear more consistent with soft sweeps than either hard sweeps or neutrality. Similarly, in windows flanking the hard sweep, the number of distinct haplotypes (K; Figure 1B) and ZnS (which measures linkage disequilibrium; Figure 1C) both often match the expectation of soft sweeps more than hard sweeps or neutrality (additional statistics are shown in Figure S2). The shoulder effect exhibited by ω is less pronounced (Figure S2H), perhaps unsurprisingly as this statistic is designed to capture the patterns of linkage disequilibrium expected flanking either side of a completed hard sweep (Kim and Nielsen 2004). However, according to a wide range of summary statistics capturing allele frequency, haplotype frequency, and linkage disequilibrium information, a large stretch within the shoulders of hard sweeps will more closely resemble soft sweeps than either hard sweeps or neutral evolution. We obtain qualitatively similar results when considering soft selective sweeps from recurrent mutation (Figure S2).
Figure 1.
Distributions of summary statistics in neutral, selected, and linked regions. (A) π-values from simulated smaller windows (genetic distance of ρ = 100) experiencing hard sweeps, soft sweeps from standing variation, or neutral evolution are shown on the right. The remaining box plots on the left show the values of π obtained in 10 adjacent windows from large simulations (total genetic distance of ρ = 1000) with a just-completed hard sweep in the center of the leftmost window. The genetic distances of sites in each of these windows from the selective sweep are shown on the x-axis. (B) Values of haplotype homozygosity are shown for the same simulations and windows as in A. (C) Values of ZnS for the same simulations and windows.
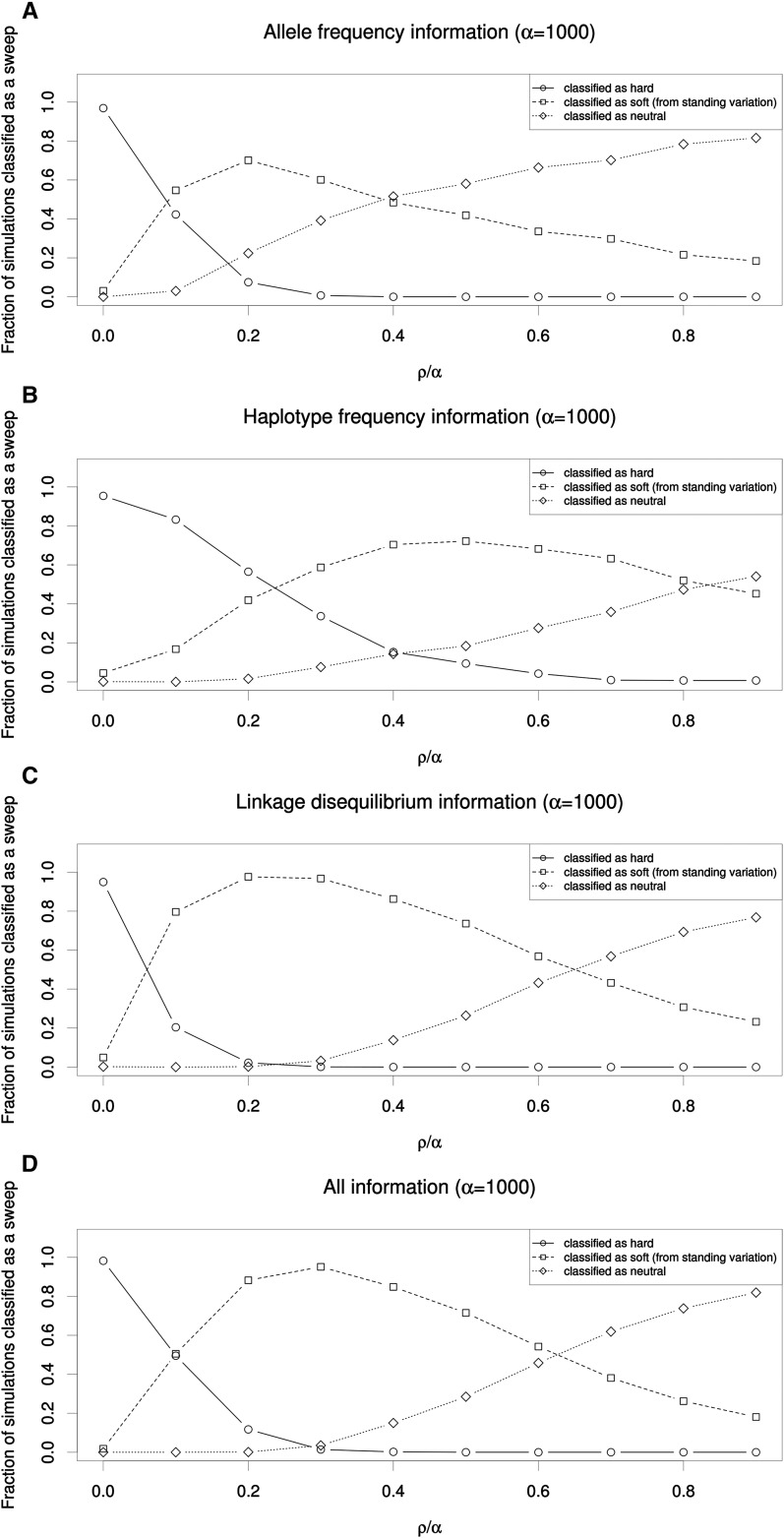
To further demonstrate this effect, we asked what fraction of genomic windows flanking a hard sweep might be mistaken for soft sweeps. We used SVM classifiers that examine the values of several summary statistics to infer whether a 10-kb window is more consistent with those obtained from simulated hard sweeps, soft sweeps from standing variation, or from neutral simulations (Materials and Methods). Because SVMs have been shown to be powerful for detecting selection and can handle multidimensional data (Pavlidis et al. 2010; Ronen et al. 2013), they are ideally suited for assessing the extent to which regions flanking hard sweeps resemble soft sweeps. We trained and applied four such SVMs: (1) one using allele-frequency information (π and S); (2) one using haplotype frequency information (K and haplotype homozygosity); (3) one using LD information (ZnS and ω); and (4) one using all summary statistics we examined (Materials and Methods).
These SVMs were for the most part extremely accurate on test data (e.g., the SVM utilizing all statistics recovers the correct class 98.8% of the time). Perhaps the only exception is that the haplotype-based SVM struggles somewhat to distinguish soft sweeps from neutrality (Table S2). Unsurprisingly, the window containing the simulated hard sweep was nearly always classified correctly by all four SVMs (Figure 2 shows results for ρ = α = 1000). As one moves away from the site of selection, a substantial fraction of linked windows were also classified as having a hard sweep, especially when using haplotype frequency information (Figure 2B), undoubtedly because at shorter genetic distances little or no recombination has occurred between the selected site and the neutral locus being queried.
Figure 2.
Classification of regions flanking hard sweeps reveals the soft-shoulder effect. (A) The relationship between distance from a simulated hard sweep and the fraction of simulated windows classified as experiencing a hard sweep, a soft sweep, or neutral is shown for an SVM classifier that leverages values of π and the number of segregating sites. Each window accounts for one-tenth of the simulated chromosome. The values on the x-axis show the distance (in terms of ρ/α) of the center of the tested window from the simulated hard sweep—i.e., the selected window is shown at position 0.0, and the window furthest from the sweep is shown at 0.9. For these simulations total recombination distance (ρ) across the simulated chromosome was set to 1000, as was α. (B) The same plot as A is shown for an SVM using the number of distinct haplotypes and haplotype homozygosity to classify each window. (C) Classifications by an SVM leveraging ω and ZnS. (D) Classifications by an SVM using our full set of summary statistics.
Strikingly, we observe that a large fraction of flanking windows are incorrectly classified as soft sweeps by each SVM. The relationship between this fraction and distance from the hard sweep is nonmonotonic, initially increasing with distance, peaking at well over 50% for each classifier (at a distance ranging from 0.2 to 0.5 × ρ/α), and then decreasing at further distances where patterns of genetic variation begin to more closely match the neutral expectation. Overall, for each SVM, >99% of simulated hard sweeps yield at least one spurious soft sweep call in one of the nine downstream windows. Thus, patterns of allele frequency, haplotype frequency, and LD flanking hard sweeps will often closely match those expected from soft sweeps. Indeed, combining all of these pieces of information into one classifier does not alleviate this problem (Figure 2D). Together these results suggest that the coalescent histories of regions flanking hard sweeps are very similar to those of soft sweeps.
This soft-shoulder effect can be understood through the lens of recombination: if a neutral locus is far enough from the selected site such that one or more recombination events are likely to occur during the sojourn of the beneficial allele, then two or more haplotypes will be linked to the beneficial allele and thus caught up in the sweep. This will result in an excess of intermediate frequency alleles, multiple high- or intermediate-frequency haplotypes, and increased LD, just as expected under a soft sweep (see Figure 1 and Figure S2). At still greater genetic distances too many recombination events are likely between the selected and neutral loci to yield any detectable signal of selection. We observed a similar trend for stronger values of α (Figure S3 and Figure S4) and also when using SVMs trained to distinguish among soft sweeps from recurrent mutation, hard sweeps, and neutral evolution (Figure S5).
Hard sweeps produce signatures of partial sweeps at linked loci
An allele that rapidly rises from frequency 1/2N to intermediate or high frequency will result in a long, nearly identical haplotype among that class of chromosomes, while chromosomes containing the ancestral haplotype will exhibit much greater diversity as they share a much older MRCA (e.g., Meiklejohn et al. 2004; Saunders et al. 2005; Tishkoff et al. 2007). A class of statistics designed to uncover this phenomenon thus seeks to identify polymorphisms where one allele is associated with far less haplotypic diversity than the other allele (Hudson et al. 1994; Sabeti et al. 2002, 2007; Voight et al. 2006). We reasoned that, because hard sweeps bring linked alleles to intermediate frequencies depending on the recombination distance between the selected and linked sites (Gillespie 2000), the soft shoulder effect might also confound statistics used to detect partial sweeps.
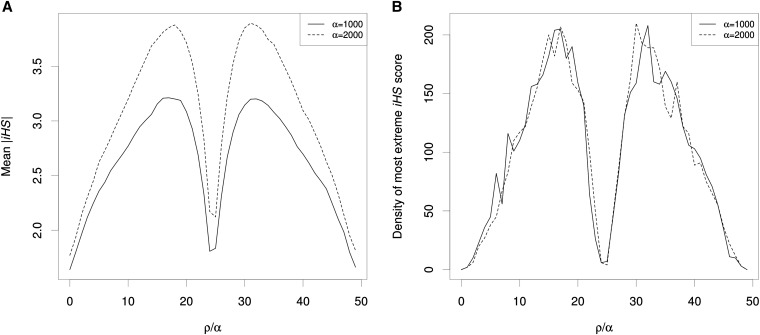
To address this question we simulated 50-kb chromosomes either evolving neutrally or with a hard sweep occurring in the center of the chromosome (simulation sets 12–15 in Table S1; Materials and Methods). For these simulations we fixed ρ at 1000 and α at 1000; all other conditions are similar to those used in the simulations to detect signatures of soft sweeps, except that the selected site is now in the center of all windows. We used the integrated haplotype score statistic, or iHS (Voight et al. 2006), to measure long-range haplotype homozygosity at each segregating site. For each SNP in each simulation we calculated the iHS value and then calculated empirical P-values using the distribution of iHS values from the neutral simulations, while controlling for allele frequency (Materials and Methods). Rather than taking its maximum value at the actual selected site, we found that |iHS| initially increases with increasing distance from the sweep in either direction, before peaking at a distance of ∼0.15 ρ/α and then decreasing back toward the neutral expectation with further distance from the sweep (Figure 3A). The lack of signal at the site of selection is expected, as a fixed sweep does not leave the signature of selection detected by iHS in the close vicinity of the selected site. Next, for each simulated hard sweep we asked where along the chromosome the most extreme iHS value was found; the distribution of these locations is shown in Figure 3B. We found that the density of extreme iHS scores closely mirrored the trend observed for average |iHS|, increasing from a trough at the sweep to a peak near 0.15 ρ/α, and then again decreasing with further distance. The iHS scores found in these peaks are quite extreme: in 98.6% of simulations our empirical P-value estimate was zero (i.e., the iHS score was more extreme than any score observed in any of the neutral simulations). When doubling α, we obtain a similar shoulder effect peaking near 0.15 ρ/α from the hard sweep (Figure 3).
Figure 3.
Elevated iHS scores flanking hard sweeps. (A) Mean absolute-value standardized iHS is shown flanking a just-completed hard sweep, which occurred in the center of the simulated region whose total genetic distance (ρ) was set to equal the selection coefficient (α). (B) The distribution of locations of the most extreme iHS score (i.e., lowest P-value in each simulation with a hard sweep at the center.
These findings strongly support our hypothesis that the shoulders of a completed hard sweep will resemble a partial sweep. Indeed, if we consider P-values <8.47 × 10−7 (the most conservative threshold we can establish given our number of simulated SNPs under neutrality) to be indicative of a partial sweep, almost every completed hard sweep (97.6%) produces a false partial sweep signal at least 0.2 ρ/α away from the true hard sweep—with α = 1000 and N = 10000, this corresponds to 0.2 cM, or roughly 200 kb in the human genome. Because iHS is intended to detect partial sweeps (Voight et al. 2006), it is unsurprising that its values are often not extremely high at the site of a hard sweep (Figure 3, A and B, centers); however, the elevated iHS observed flanking the hard sweeps could be misinterpreted as evidence of partial sweeps. As with soft sweeps above, this spurious signature of partial sweeps is the result of intermediate levels of recombination between the neutral and selected loci during a hard sweep. At genetic distances where a small number of recombination events is expected, a soft shoulder effect should prevail near the sites of hard sweeps in such a way that it may be difficult to differentiate true soft or partial sweeps from the shoulders of completed hard sweeps without prior information as to the location of hard sweeps themselves. If one instead examines very large genomic windows for evidence of partial sweeps, a window centered on a hard sweep may be mistaken for single partial sweep. For example, for the average hard sweep with α = 1000, 48% of all SNPs in the simulated chromosome exhibit significant iHS scores (in the upper or lower 2.5% tail of the distribution from neutral simulations). Such a window would likely be considered as a strong candidate for a partial sweep; indeed such a fraction of significant iHS scores is qualitatively similar to the top candidate partial sweeps highlighted in Voight et al. (2006, Table 1).
Stochasticity during and after the sweep exacerbates the soft shoulder effect
We have demonstrated that the shoulders of hard sweeps often present patterns of variation closely matching those generated at the site of soft sweeps and partial sweeps. However, our analyses thus far have made a number of simplifications: we have considered significant windows only in isolation and not in their genomic context, we have examined patterns of variation only at the moment the sweep ends, and we have not considered the effects of gene conversion. In the following we consider relaxing each of these assumptions in turn and show how patterns of genetic variation are affected.
Examining large genomic regions and misidentifying the target of selection:
Rather than considering smaller windows independently of one another, scans for selective sweeps can examine spatial patterns of diversity across large genomic regions to identify the putative target of selection (e.g., Kim and Stephan 2002; Nielsen et al. 2005). Such strategies may be able to mitigate the soft-shoulder problem by discovering that the signature of a soft sweep occurs in the shoulder of a hard sweep. In other words, if a scan is able to identify the true target of selection within a larger genomic window containing a single recent hard sweep, then it should be correctly classified (see Figure 2) and no spurious soft sweeps will be called.
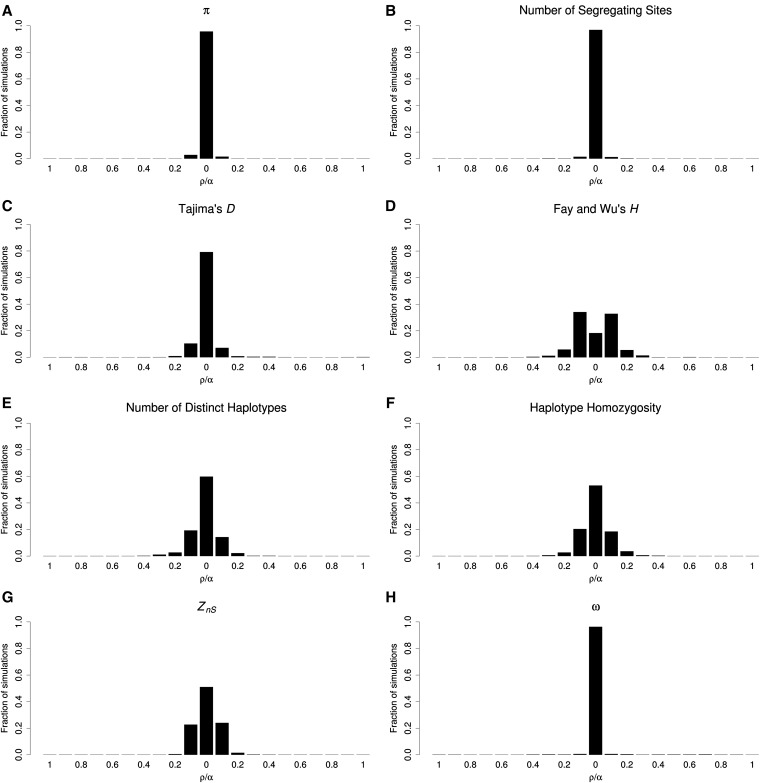
To assess the utility of such a strategy, we simulated large chromosomal regions of 210 kb that we then subdivided into 21 windows, with a hard sweep occurring in the central window (with α = 1000 and ρ = 2100, or ρ = 100 per window as above; simulation set 16 in Table S1). We then asked how often the strongest signal of selection was observed in the central window. We found that for some statistics this technique often misidentifies the target of selection. For example, K, haplotype homozygosity, and ZnS exhibit their strongest signals in the incorrect window in 40.2, 46.9, and 49.0% of simulations, respectively. Thus, if these statistics are used to identify selective sweeps, the inferred target of selection may often be located far from the true selected site, and in turn the mode of selection may be misclassified. Fortunately, some statistics fare much better: π, the number of segregating sites (S), and ω identify the wrong window only 4.3, 3.1, and 3.7% of the time, respectively. The distributions of the strongest sweep signal for these and other statistics are shown in Figure 4. It is important to note that applying this type of approach to iHS does not mitigate any of the problems with detecting partial sweeps. Because the shoulder of the hard sweep has a larger value of iHS than the sweep itself, the shoulder could be identified as a significant partial sweep regardless of the size of the region examined.
Figure 4.
Signals of selection from various summary statistics in windows containing or flanking a hard sweep. For each summary statistic, we examined each individual simulation and located the window exhibiting the most extreme value (in the direction suggestive of a hard sweep). This figure shows the histogram of these locations for each statistic. The total genetic distance of each simulated chromosome (ρ) was 2100. The chromosome was subdivided into 21 equally sized windows (ρ = 100) with a hard selective sweep occurring in the central window (α = 1000). (A) π, (B) number of segregating sites, (C) Tajima’s D, (D) Fay and Wu’s H, (E) number of distinct haplotypes, (F) haplotype homozygosity, (G) Zns, and (H) ω.
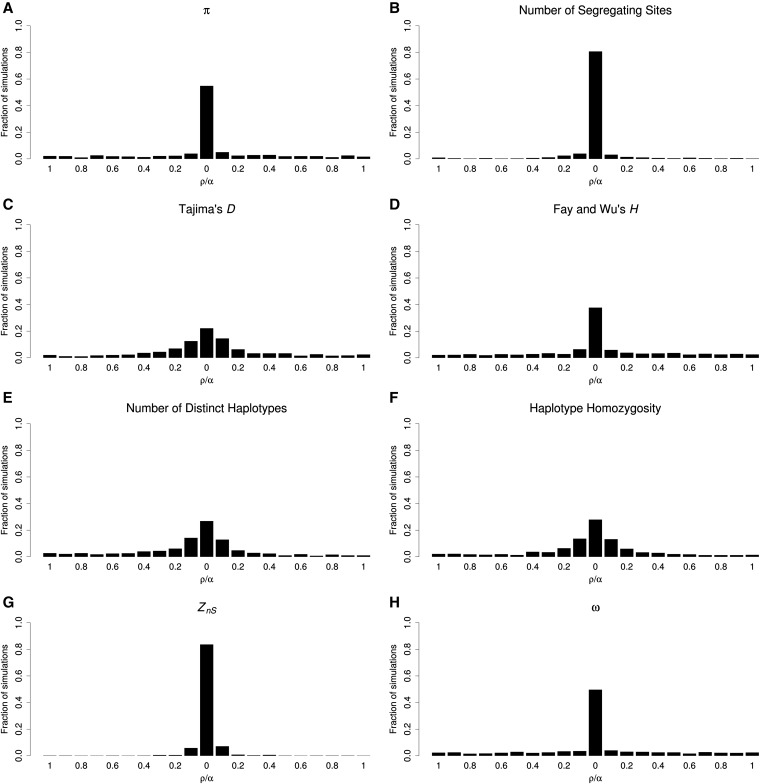
For soft sweeps, where stochastic forces have a greater impact on patterns of diversity, most statistics identify the selected window much less reliably. For example, not only do the error rate of K and haplotype homozygosity increase to 73.2 and 72.1%, respectively, but even π (error rate of 45.2%) and ω (50.4%) perform poorly (Figure 5; data from simulation set 17 in Table S1). Compared to π, the number of segregating sites has a somewhat more modest error rate (19.3%). Interestingly, the error rate of ZnS is lower than that for hard sweeps (16.4%), perhaps because hard sweeps completely eliminate variation in the most immediate vicinity of the selected site, thereby reducing the number of windows with high LD (Kim and Nielsen 2004). We observe a similar dispersal of the signal of soft sweeps arising from recurrent mutation events (Figure S6; data from simulation set 18 in Table S1). Interestingly, the ω statistic performs much more poorly for soft sweeps from recurrent mutations than those from standing variation, recovering the wrong window in nearly 90% of instances (Figure S6H).
Figure 5.
Signals of selection from various summary statistics in windows containing or flanking a soft sweep. For each summary statistic, we examined each individual simulation and located the window exhibiting the most extreme value (in the direction suggestive of a soft sweep). This figure shows the histogram of these locations for each statistic. The total genetic distance of each simulated chromosome (ρ) was 2100. The chromosome was subdivided into 21 equally sized windows (ρ = 100) with a soft selective sweep (with α = 1000; initial selected frequency ranging from 0.05 and 0.2) occurring in the central window. (A) π, (B) number of segregating sites, (C) Tajima’s D, (D) Fay and Wu’s H, (E) number of distinct haplotypes, (F) haplotype homozygosity, (G) Zns, and (H) ω.
Selecting the size of genomic regions to examine:
Our results above demonstrate that, at least in the case of hard sweeps, it may be possible to detect the targets of selection with reasonable precision by examining loci within their larger genomic context, therefore also likely inferring the correct mode of selection. However, a serious limitation of this approach is its inability to uncover multiple sweeps fairly close to one another (i.e., separated by a distance smaller than a predefined threshold). Because the results in the previous section hold only when there is no more than one sweep within each large region examined, it is natural to ask how extensions allowing for more than one sweep might perform.
To identify separate sweeps, we can segment the genome into groups of consecutive windows more consistent with neutrality or with either mode of positive selection. We assessed the robustness of this approach by applying our SVM classifier to each window within our large simulated hard sweeps (α = 1000 and ρ = 2100; simulation set 16 from Table S1) and asking how often in our simulations the signature of selection around the target site appeared to have decayed back to the neutral expectation with increasing distance from the hard sweep, but then at some even greater distance had risen enough to cause our classifier to once again favor positive selection. Of the 979 of 1000 simulated sweeps in which our SVM correctly classifies the target window as a hard sweep (using the full set of summary statistics), 68.1% had at least one window classified as a sweep, but separated from the true hard sweep by at least one window classified as neutral. Remarkably, every one of these spurious secondary sweeps was more consistent with a soft than hard sweep according to our classifier.
While on average a selective sweep should result in a sharp valley of diversity (or peak in linkage disequilibrium) near the selected site with a monotonic recovery toward the neutral expectation with increasing physical distance, patterns of genetic variation in each linked window have a large stochastic component (Kim and Stephan 2002). To demonstrate this point we have generated plots showing the values of various summary statistics in each sub-window from each individual simulation from simulation sets 16–18, 21, and 22 (described in Table S1); the patterns expected on average poorly match those presented by many individual simulations (plots available at https://github.com/kern-lab/softshoulders). Both mutation and recombination events occurring during and after the sweep contribute to noise, and drift plays a greater role as distance from the selected site increases. This stochasticity can often cause violations of the expectation of monotonicity, creating secondary signals of selection far away from the sweep. Because only windows very tightly linked to the hard sweep will more closely resemble hard sweeps than soft sweeps (Figure 1), these secondary peaks are more likely to resemble soft sweeps.
Hard sweeps leave behind the signal of soft sweeps, even at the selected site:
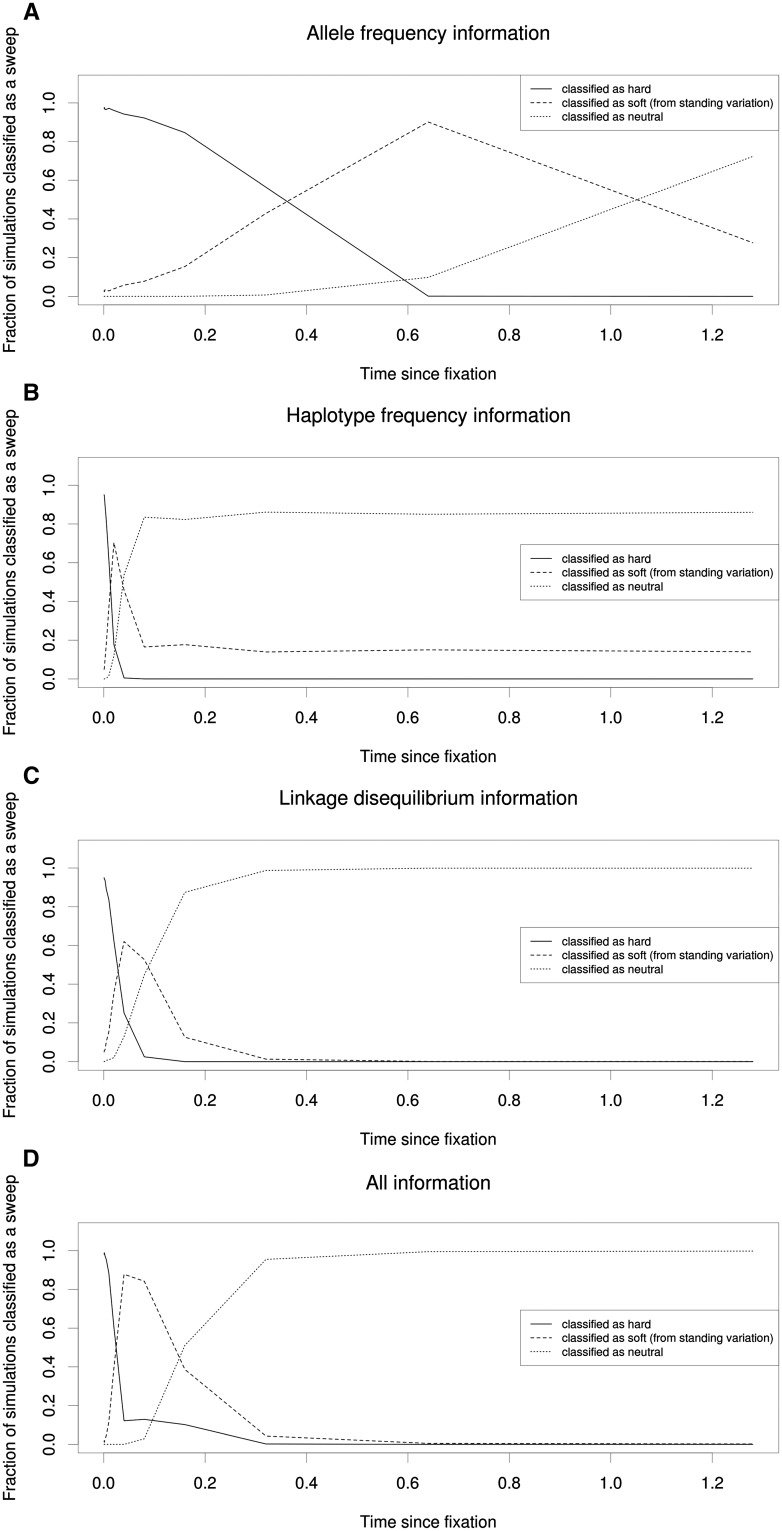
Our results thus far have examined only patterns of diversity at the exact moment that a sweep ends. To examine the effects of sampling a population some time after the sweep ends, we asked whether older hard sweeps are often misclassified as soft sweeps from standing variation at various times post-fixation. For these simulations we focused only on the window containing the selected site. While it may not be immediately obvious why the recovery from a hard sweep would result in false signatures of soft sweeps, note that the values of many of our statistics are at their most extreme after a hard sweep and that their values under a soft sweep typically lie between the extreme of the hard sweep and the neutral expectation (Figure 1); of the statistics we examine, Tajima’s D is the sole exception to this rule. Thus, although much of the power to detect any type of sweep is lost fairly soon after an advantageous allele has fixed (Przeworski 2002), during the recovery phase the values of many statistics may pass through the range normally produced by soft sweeps. For this reason, hard sweeps may possibly leave a “soft shadow.”
We began by examining relatively recently completed sweeps—fixing 0.000625 × 2N generations ago (with α = 1000)—and then repeatedly doubled the time since fixation in order to search for a fixation time where the sweep was more often classified as soft than hard according to the same test used above (simulation set 19 from Table S1; Materials and Methods). Again, we performed this analysis using four SVMs (allele-frequency information, haplotype frequency information, LD information, and all summary statistics). As expected, we found that the misclassification rate increased with time since fixation, with the fraction of simulations classified as soft dramatically overtaking that of hard sweeps between 0.01 and 0.02 × 2N generations ago when using haplotype frequency information (Figure 6B). When using LD information, this shift occurs between 0.02 and 0.04 × 2N generations ago (Figure 6C). The SVM using allele-frequency information maintains accuracy for a much longer period, with the fraction of spurious soft sweep calls overtaking correct hard sweep calls after between 0.32 and 0.64 × 2N generations (Figure 6A). The SVM incorporating all summary statistics exhibits this shift faster than that using allele-frequency information, but not as quickly as those using statistics measuring haplotype frequencies and LD (Figure 6D). Eventually these classifiers cannot detect the sweep at all, classifying the majority of simulated sweeps as neutral rather than hard or soft. Again, this occurs soonest when using haplotype frequency information. This is perhaps because a single mutation or recombination event can create a new distinct haplotype while hardly affecting summaries of allele frequency across the entire window, although it is important to note that in regions with lower mutation and/or recombination rates this decline may be far less steep. The classifier using LD information also loses power to detect sweeps well before that using allele-frequency information, which has power to detect >90% of sweeps with α = 1000 even after 0.64 × 2N generations, although it misclassifies nearly all of these as soft. Note that the classifier using haplotype information continues to classify a small but consistent minority (∼15%) of simulations as soft sweeps as time since the sweep increases to 1.28 × 2N; this is an artifact of the relative lack of power this SVM has to distinguish soft sweeps from neutrality (i.e., even truly neutral windows are classified as soft at this rate; Table S2). We observe a similar decay in the ability to properly classify hard sweeps of increasing ages when the soft sweep model we consider is that of recurrent mutation rather than selection on standing variation (Figure S7).
Figure 6.
The relationship between age of the sweep and misclassification rate. (A) Allele-frequency information: the fraction of simulated windows containing a hard sweep (α = 1000) classified as hard, soft, or neutral by an SVM leveraging allele-frequency information is shown according to the time in the past at which the sweep completed (in units of 2N generations). The most recent sweep examined in this plot completed 0.000625 × 2N generations ago, and we examined older sweeps by continually doubling the time since fixation, stopping at a sweep time of 1.28 × 2N generations in the past. (B) Same as A, but using a classifier leveraging haplotype information. (C) Results from an SVM leveraging LD information. (D) An SVM leveraging our full set of summary statistics.
Because neutral evolution following a selective sweep should introduce noise into spatial patterns of variation, we also asked whether increasing the time since the completion of the selective sweep affected the location of the strongest signal of selection. We again simulated sweeps occurring in the center of a larger chromosome (ρ = 2100) with varying fixation times and with α = 1000 and subdivided these chromosomes into 21 windows (simulation set 21 in Table S1). Perhaps unsurprisingly, we found that the passage of time since the completion of the sweep obscured the location of the target of selection. For example, while the highest value of haplotype homozygosity occurred in the incorrect window 44.9% of the time for a sweep occurring 0.000625 × 2N generations ago, this fraction increases to 51.0% after 0.01 × 2N generations, and to 91.4% after 0.08 × 2N generations. ω fares better, with only 3% of sweeps incorrectly located after 0.000625 × 2N generations, an error rate of 49.6% after 0.08 × 2N generations, and 95.6% after 0.32 × 2N generations. π performs far better than both of these statistics, with an error rate of 4.2% after 0.000625 × 2N generations, which increases to 38.1% after 0.64 × 2N generations—lower than the error rates exhibited by haplotype homozygosity and ω after much shorter periods (0.01 and 0.08 × 2N generations, respectively). While this error rate increases to a considerable 72.4% after 1.28 × 2N generations, this is substantially lower than the error rates of haplotype homozygosity after 0.08 and 0.32 × 2N generations, respectively. Thus it appears that haplotype frequency-based signatures of selection decay more rapidly following the completion of a selective sweep than LD-based signatures, which in turn decay far more rapidly than nucleotide diversity (for examples, see plots of individual simulations available at https://github.com/kern-lab/softshoulders).
The effect of gene conversion:
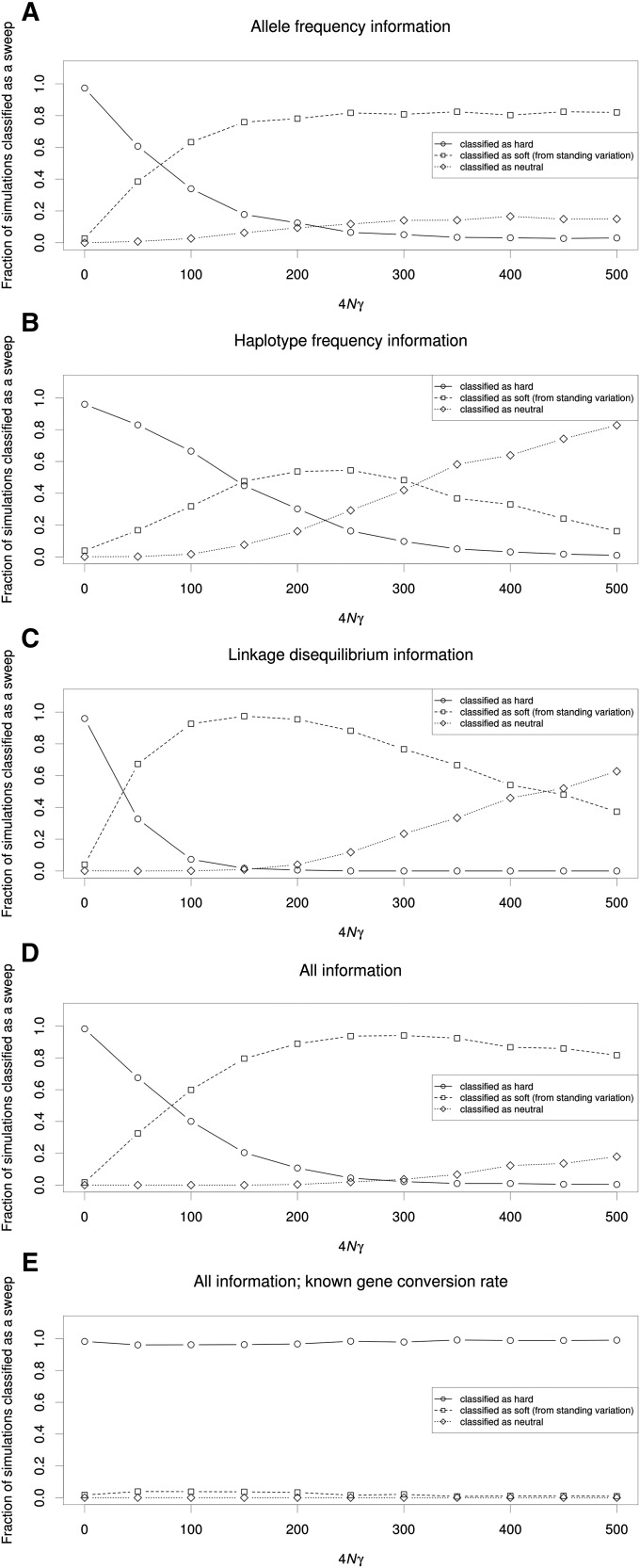
Allelic gene conversion can result in lineages escaping a selective sweep by acquiring the adaptive mutation after the sweep has begun, similar to a recurrent adaptive mutation (Begun and Aquadro 1994; Hamblin and Di Rienzo 2000; Hamblin et al. 2002). We therefore simulated hard selective sweeps with various rates of gene conversion using a mean tract length of 518 bp (simulation set 20 from Table S1), as estimated in Drosophila (Comeron et al. 2012), and asked how often these simulations would be misclassified as soft by our classifiers. We found that as the total gene conversion rate for our 10-kb locus increased from 4Nγ = 0 to 4Nγ = 500, where γ is the gene conversion tract initiation rate per base pair, the fraction of simulated windows misclassified as soft increases at first, suggesting that the greater amounts of variation introduced to the class of sweeping chromosomes causes them to resemble soft sweeps; indeed at 4Nγ = 250 the majority of simulated hard sweeps are misclassified as soft by each classifier (Figure 7A-D). Even greater rates of gene conversion begin to erode the sweep signal further so that most simulations are misclassified as neutral. This problem is especially troublesome when using haplotypic information: at 4Nγ = 500, our haplotype-frequency-based SVM classifies only 17.2% of simulations as sweeps (nearly all of which are misclassified as soft), whereas the allele-frequency-based SVM recovers 85.0% of sweeps in this case (although again nearly all are misclassified as soft). This is likely because a single gene conversion event can create a new haplotype, but has a very small impact on nucleotide variation. Note that when 4Nγ = 500, our ratio of gene conversion to crossover rates is similar to that observed in Drosophila (5 conversion events for every crossover vs. ∼4.9 from Comeron et al. 2012). Thus, even when examining the true target of selection, gene conversion can cause hard sweeps to frequently be misclassified as soft or to be missed altogether (Jones and Wakeley 2008). However, if the true gene conversion rate at the target of selection is known, then it is possible to accurately distinguish hard sweeps from soft sweeps and neutrality (Figure 7E) in the context of our SVM. Again, we obtain similar results when using recurrent adaptive mutation as our SVM’s model of soft sweeps (Figure S8).
Figure 7.
The relationship between gene conversion rate and sweep misclassification rate. (A) Allele-frequency information: the relationship between the locus-wide gene-conversion rate and the fraction of simulated windows containing a hard sweep (α = 1000) classified as hard, soft, or neutral. (B) Same as A, but using a classifier leveraging haplotype information. (C) Results from an SVM leveraging LD information. (D) An SVM leveraging our full set of summary statistics. (E) An SVM leveraging our full set of summary statistics and trained from simulated regions experiencing the correct gene conversion rate.
When examining the larger genomic neighborhood of a selective sweep, gene conversion also may obscure the location of the target of selection. We performed large simulations (ρ = 2100, subdivided into 21 windows) with a hard sweep occurring in the middle of the central window, and a gene conversion rate (4Nγ) of either 0 or 2100 (simulation set 22 in Table S1). When 4Nγ = 0 the most extreme value of π is found in the central window all but 4.2% of the time, but when 4Nγ = 2100 this error rate increases to 86.4%. At this gene conversion rate, haplotype homozygosity performs similarly (85.7%; up from 47.5% when 4Nγ = 0), as does ω (90.5%; up from 3.4% when 4Nγ = 0). Thus, accurately locating selective sweeps in the presence of gene conversion may be particularly difficult, especially given that in these simulations the ratio of gene conversion to crossover rates was 1:1, probably far lower than the true ratio in many species. Again, this insight can be gleaned by examining the patterns of variation flanking individual simulated hard selective sweeps experiencing allelic gene conversion events (available online at https://github.com/kern-lab/softshoulders).
Discussion
The question of whether adaptation more commonly proceeds from a single, new advantageous mutation (hard sweeps) or from either standing variation or multiple new mutations (soft sweeps) has recently attracted much attention in the field of molecular population genetics. We have shown that the shoulders of completed hard selective sweeps produce a pattern of diversity that is difficult to distinguish from that generated by completed soft sweeps or by partial sweeps, a pattern that we call the soft-shoulder effect. The soft-shoulder effect results from recombination events between the site of a hard sweep and a neutral locus during the sojourn of the beneficial mutation; this recombination creates multiple sweeping flanking haplotypes, strong linkage disequilibrium, and an excess of intermediate-frequency alleles, just as would be observed at the site of a soft sweep. Our results show that this phenomenon confounds efforts to differentiate neutral loci flanking hard sweeps from selection on standing variation or from sweeping advantageous mutations that have not yet fixed. We also find evidence that the soft shoulders of hard sweeps would likely be mistaken for selection on recurrent de novo mutations occurring during the sweep phase. This should not be surprising, as this alternative form of soft sweep creates patterns of variation that are in many ways qualitatively similar to the effect of sweeps acting on standing variation (Pennings and Hermisson 2006 a,b).
Our results imply that the soft shoulder effect may be problematic for genome-wide scans for selection where the selected sites are not known a priori—even if hard sweeps were the sole mode of adaptation, such scans might identify a large number of putative soft sweeps. Perhaps more worryingly, these spurious soft sweeps may be identified far from the true target of selection, potentially resulting in considerable wasted effort characterizing loci that are in fact evolving neutrally. Thus, efforts to uncover the sites and modes of positive selection across the genome should be interpreted with care if they are unable to confidently discriminate true targets of selection from linked sites (cf. Begun and Aquadro 1994; Hudson et al. 1994). Such efforts must also be able to distinguish between recent soft sweeps and older hard sweeps, as we have shown that such older events may often be misclassified as soft sweeps by methods that consider only recently completed sweeps (as our SVM did).
Just as with soft sweeps, we have shown that partial sweeps are difficult to distinguish from the shoulders of completed hard sweeps. Statistical tests for evidence of partial sweeps have been applied to genome-wide variation data in humans to detect candidate adaptive mutations currently sweeping through human populations (e.g., Sabeti et al. 2002; Redon et al. 2006; Voight et al. 2006; Conrad et al. 2010; Ferrer-Admetlla et al. 2014). Again, because such genome-wide scans have no prior knowledge of which sites may have been selected, regions linked to, but some genetic distance away from, completed hard sweeps could be misclassified as partial sweeps due to the soft shoulder effect. Thus, although these scans have power to detect partial sweeps, they may often misidentify both the target locus and the mode of selection if hard sweeps are common. Tests that attempt to recover the positively selected polymorphism or a small window containing several candidate polymorphisms (e.g., Grossman et al. 2010) may be misled in this case, as the true target of selection will have already reached fixation. Instead, the test may incorrectly single out hitchhiking alleles that escaped the sweep via a recombination event; these mutations are most likely fitness neutral or nearly so and may be located far from the positively selected locus. For example, for moderately strong selection, the soft shoulder could appear kilobases away from the selected site in Drosophila, and multiple megabases away in humans, and for stronger selection coefficients these distances could be greater. Therefore, caution should be taken in attempting to identify the sites directly under selection, or even the Gene Ontology categories of genes associated with apparent partial sweeps—these may be nothing more than innocent bystanders (cf. Pavlidis et al. 2012).
It is worth noting that many signals of selection used in molecular population genetics are focused on the regions flanking selected sites. Indeed, modern-day studies of the effect of hitchhiking on variation have often explicitly focused on linked neutral loci (e.g., Kaplan et al. 1989; Przeworski 2002), and several commonly used tests for selection—e.g., Fay and Wu’s H (Fay and Wu 2000)—are often significant only in flanking regions (i.e., in the “shoulders”; Figure 4D). However, the development of many methods for detecting selection has been accompanied by a focus on the region directly surrounding the selected site itself, rather than the flanking regions (for exceptions, see the next paragraph). Despite the fact that previous studies have found the same patterns as shown here for the shoulders of completed sweeps (e.g., Figure 6 in Pennings and Hermisson 2006b), these studies have generally not discussed the implications of these results. For instance, Peter et al. (2012) sought to classify previously identified sweeps as hard or soft (from standing variation); they observed that misidentifying the target of selection increases the fraction of sweeps falsely classified as soft, but did not discuss the implications of this for genome-wide scans of selection. The results presented here suggest that this misidentification is itself a problem when conducting whole-genome studies.
Although the problem of distinguishing soft or partial sweeps from the shoulders of hard sweeps is difficult, it may not be intractable. Hard sweeps leave an expected spatial pattern of variation around the target of selection, with a large region of low diversity flanked by regions with skewed allele frequencies and high linkage disequilibrium within each shoulder but not across the selected site (Stephan et al. 2006). These patterns are in fact the basis of tests for selection that use the spatial arrangement of allele-frequency skews (e.g., Kim and Stephan 2002; Nielsen et al. 2005) and LD (Kim and Nielsen 2004; Pavlidis et al. 2010). Based on the results laid out above, we expect that these methods used to detect completed hard sweeps should find regions directly abutting the loci discovered by methods employed to detect soft or partial sweeps. Indeed, there is a significant overlap between loci identified as sweeps in the human genome using iHS (Voight et al. 2006) and the composite likelihood-ratio test of Nielsen et al. (2005) that is intended to detect completed sweeps (Williamson et al. 2007). The extent of the overlap between significant loci may be determined by a number of factors, including the statistical power of each test and the frequency of recurrent hitchhiking events, which may cause much of the genome to resemble a shoulder. Nevertheless, a simple heuristic could prove effective: whenever a putative soft or partial sweep is found, one can attempt to rule out the possibility that it is simply the shoulder of another event by searching for evidence of a nearby hard sweep. Here we tested a similar strategy of examining only the strongest signature of a sweep within a large genomic window, effectively imposing a distance cutoff such that any signature of positive selection found within a prespecified distance of another putative sweep with stronger support would be ignored. For soft sweeps, we found that this approach could dramatically reduce the false-positive rate; however, one could miss many true sweeps, especially when positive selection repeatedly acts on the same locus or cluster of neighboring loci or if the region examined is large. These are important limitations when examining populations where recent positive selection is thought to be widespread (e.g., Drosophila: Begun et al. 2007; Langley et al. 2012).
We also employed a more sensitive approach of requiring at least one putatively neutral window to separate distinct putative sweeps, but found that this approach was not robust to soft shoulders. The failure of our latter approach seems to be driven by stochasticity in the signal of selection surrounding a sweep: while on average this signal dissipates monotonically with increasing distance from the target of selection, many individual windows do not closely resemble this average case and instead exhibit secondary signal peaks due to noise and indeed often present the strongest peak outside of the selected window (see Figure 4, Figure 5, and plots of individual simulations available: https://github.com/kern-lab/softshoulders). Thus, noise in the signal of selection may result in not only false-positive soft sweep calls, but also in the mislocalization of true sweeps—especially when using haplotypic information. This may be especially troublesome near soft sweeps, where the location of the peak signal of selection is particularly unpredictable.
When examining the spatial distribution of various summary statistics in our large-scale simulations containing selective sweeps, we observed that some statistics (e.g., π, ω) more reliably identify the target of selection than others. This suggests that it may be possible to devise a method, perhaps one combining multiple allele-frequency-, haplotype-, and LD-based statistics, that more accurately identifies sweep locations (although gene conversion and the passage of time since the sweep may complicate this; see below). This would in turn allow for the characterization of targets of recent positive selection and inferences about the phenotypic changes they underlie, as well as more accurate inferences of the type of positive selection responsible for these sweeps.
Alternatively, it may prove difficult to disentangle the different modes of adaptive evolution from one another, even if one can correctly identify the target of selection. This more pessimistic view is supported by multiple considerations. First, the distinction between hard and soft sweeps may not be so distinct (Jensen 2014). While new mutations always begin at frequency 1/2N, soft sweeps from standing variation may be selecting on variants from frequency 2/2N to (2N − 1)/2N. For selection that acts on polymorphisms at low frequencies, the patterns of variation at both the selected site and in the flanking regions may be indistinguishable from a true hard sweep. Second, as shown here, the shadow of a hard sweep may leave the apparent signature of a soft sweep even at the site of selection. In addition, allelic gene conversion at the selected site during the sweep can place the advantageous allele onto new backgrounds. This pattern is exactly the same as that expected by soft sweeps via recurrent adaptive evolution (Begun and Aquadro 1994; Hamblin and Di Rienzo 2000; Hamblin et al. 2002). Although this may not seem to be a likely event, gene conversion occurs at a high rate (Comeron et al. 2012), and even with short conversion tract lengths it will affect a large proportion of all sweeps (Figure 7; also see Jones and Wakeley 2008).
In a scenario involving realistic rates of gene conversion, even with prior knowledge of the selected site—and even sampling the data at the time of fixation—it becomes very difficult to disentangle soft from hard sweeps, or distinguish either from neutrality. That gene conversion may cause hard sweeps to go undetected underscores the need for tests for selection that are more sensitive than those designed with completed hard sweeps in mind: tests designed to uncover soft sweeps may be well suited for detecting hard sweeps undergoing gene conversion, for example. Moreover, our results suggest that if gene-conversion rate estimates across the genome are available, selection scans may be able to leverage this information to construct a more accurate expectation of the impact of selection on linked polymorphism, and thus more accurately detect sweeps and classify the mode of selection (Figure 7E). Unfortunately, gene conversion also obscures the spatial signal of a selective sweep, making it more difficult to accurately locate the target of selection. This effect may be just as severe on allele-frequency information as on haplotype frequencies. The passage of time following fixation also obscures the sweep location, although nucleotide diversity is more robust to this problem. Thus it appears that gene conversion could be the biggest obstacle to accurate sweep detection and classification.
Despite the pessimism engendered by these results, it should be remembered that scans for selection have successfully identified the targets of adaptive evolution in multiple cases (e.g., Rockman et al. 2004; Schlenke and Begun 2004; Tishkoff et al. 2007; Yi et al. 2010). While such examples may represent the patterns produced only by the strongest selection coefficients, and may be skewed toward only certain modes of selection, they represent the promise of population genetic methods for identifying targets of natural selection. The continued deployment of an array of approaches designed to detect different signals produced by selection remains one of the best ways to identify the genes underlying adaptive phenotypes (cf. Li et al. 2008).
Whatever the approach taken, future searches for recent adaptive events must be robust to altered patterns of nucleotide and haplotype diversity and LD observed near classic hitchhiking events, including those experiencing gene conversion, especially when these searches are conducted on a genome-wide scale. Otherwise, such efforts may fail to deliver their promise of identifying either the genomic loci undergoing recent adaptation or the types of positive selection they have experienced.
Supplementary Material
Acknowledgments
We thank Jody Hey and David Begun for helpful discussion as well as Rasmus Nielsen and two reviewers for constructive comments. D.R.S. was supported by the National Institutes of Health (NIH) under Ruth L. Kirschstein National Research Service Award F32 GM105231. A.D.K. was supported by Rutgers University, by National Science Foundation Award MCB-1161367, and by the National Institute of General Medical Sciences of the NIH under award no. R01GM078204.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.174912/-/DC1.
Communicating editor: R. Nielsen
Literature Cited
- Achaz G., 2009. Frequency spectrum neutrality tests: one for all and all for one. Genetics 183: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizerman A., Braverman E. M., Rozoner L., 1964. Theoretical foundations of the potential function method in pattern recognition learning. Autom. Remote Control 25: 821–837. [Google Scholar]
- Barrett R. D., Schluter D., 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23: 38–44. [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Begun D. J., Aquadro C. F., 1994. Evolutionary inferences from DNA variation at the 6-phosphogluconate dehydrogenase locus in natural populations of Drosophila: selection and geographic differentiation. Genetics 136: 155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Holloway A. K., Stevens K., Hillier L. W., Poh Y.-P., et al. , 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5: e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boser, B. E., I. M. Guyon, and V. N. Vapnik, 1992 A training algorithm for optimal margin classifiers. Proceedings of the fifth annual workshop on Computational learning theory, pp. 144–152. Pittsburgh, PA. [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Pinto D., Redon R., Feuk L., Gokcumen O., et al. , 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C., Vapnik V., 1995. Support-vector networks. Mach. Learn. 20: 273–297. [Google Scholar]
- Cutter A. D., Payseur B. A., 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaulis F., Veuille M., 1998. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15: 1788–1790. [DOI] [PubMed] [Google Scholar]
- Durrett R., Schweinsberg J., 2004. Approximating selective sweeps. Theor. Popul. Biol. 66: 129–138. [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F., 1996. Introduction to Quantitative Genetics, Ed. 4 Longmans Green, Harlow, Essex, United Kingdom. [Google Scholar]
- Fay J. C., Wu C.-I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wyckoff G. J., Wu C.-I., 2001. Positive and negative selection on the human genome. Genetics 158: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wyckoff G. J., Wu C.-I., 2002. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature 415: 1024–1026. [DOI] [PubMed] [Google Scholar]
- Ferrer-Admetlla A., Liang M., Korneliussen T., Nielsen R., 2014. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 31: 1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1937. The wave of advance of advantageous genes. Ann. Eugen. 7: 355–369. [Google Scholar]
- Fu Y.-X., Li W.-H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1991. The Causes of Molecular Evolution. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- Gillespie J. H., 2000. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman S. R., Shylakhter I., Karlsson E. K., Byrne E. H., Morales S., et al. , 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327: 883–886. [DOI] [PubMed] [Google Scholar]
- Hahn M. W., 2008. Toward a selection theory of molecular evolution. Evolution 62: 255–265. [DOI] [PubMed] [Google Scholar]
- Hamblin M. T., Di Rienzo A., 2000. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 66: 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., Thompson E. E., Di Rienzo A., 2002. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 70: 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Bailey K., Skarecky D., Kwiatowski J., Ayala F. J., 1994. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kim Y., 2004. Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Natl. Acad. Sci. USA 101: 10667–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. D., 2014. On the unfounded enthusiasm for soft selective sweeps. Nat. Commun. 5: 5281. [DOI] [PubMed] [Google Scholar]
- Jones D. A., Wakeley J., 2008. The influence of gene conversion on linkage disequilibrium around a selective sweep. Genetics 180: 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R., Langley C., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Nielsen R., 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167: 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1957. Some problems of stochastic processes in genetics. Ann. Math. Stat., 882–901. [Google Scholar]
- Kimura M., 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. [DOI] [PubMed] [Google Scholar]
- Knerr S., Personnaz L., Dreyfus G., 1990. Single-layer learning revisited: a stepwise procedure for building and training a neural network, pp. 41–50 in Neurocomputing, edited by Fogelman Soulié F., Hérault J. Springer-Verlag, Berlin. [Google Scholar]
- Kong A., Gudbjartsson D. F., Sainz J., Jonsdottir G. M., Gudjonsson S. A., et al. , 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C. G., Schrider D. R., et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Costello J. C., Holloway A. K., Hahn M. W., 2008. “Reverse ecology” and the power of population genomics. Evolution 62: 2984–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McVean G., 2007. The structure of linkage disequilibrium around a selective sweep. Genetics 175: 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Kim Y., Hartl D. L., Parsch J., 2004. Identification of a locus under complex positive selection in Drosophila simulans by haplotype mapping and composite-likelihood estimation. Genetics 168: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., Neher R. A., 2012. Estimating the strength of selective sweeps from deep population diversity data. Genetics 191: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W.-H., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M. J., Clark A. G., et al. , 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Betancourt A. J., 2001. Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Jensen J. D., Stephan W., 2010. Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations. Genetics 185: 907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Jensen J. D., Stephan W., Stamatakis A., 2012. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol. Biol. Evol. 29: 3237–3248. [DOI] [PubMed] [Google Scholar]
- Pennings P. S., Hermisson J., 2006a Soft sweeps II: molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 23: 1076–1084. [DOI] [PubMed] [Google Scholar]
- Pennings P. S., Hermisson J., 2006b Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2: e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B. M., Huerta-Sanchez E., Nielsen R., 2012. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 8: e1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Coop G., Novembre J., Kudaravalli S., Li J. Z., et al. , 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19: 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt J. C., Cristianini N., Shawe-Taylor J., 2000. Large margin DAGs for multiclass classification, pp. 547–553 in Advances in Neural Information Processing Systems, edited by Solla S. A., Leen T. K., Müller K.-R. MIT Press, Cambridge, MA. [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M., 2002. The signature of positive selection at randomly chosen loci. Genetics 160: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M., Coop G., Wall J. D., 2005. The signature of positive selection on standing genetic variation. Evolution 59: 2312–2323. [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K. R., Feuk L., Perry G. H., et al. , 2006. Global variation in copy number in the human genome. Nature 444: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Hahn M. W., Soranzo N., Loisel D. A., Goldstein D. B., et al. , 2004. Positive selection on MMP3 regulation has shaped heart disease risk. Curr. Biol. 14: 1531–1539. [DOI] [PubMed] [Google Scholar]
- Ronen R., Udpa N., Halperin E., Bafna V., 2013. Learning natural selection from the site frequency spectrum. Genetics 195: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., Reich D. E., Higgins J. M., Levine H. Z., Richter D. J., et al. , 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., Varilly P., Fry B., Lohmueller J., Hostetter E., et al. , 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M. A., Slatkin M., Garner C., Hammer M. F., Nachman M. W., 2005. The extent of linkage disequilibrium caused by selection on G6PD in humans. Genetics 171: 1219–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt L. B., Biswas S., Madeoy J., Connelly C. F., Schadt E. E., et al. , 2009. Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history. Mol. Biol. Evol. 26: 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke T. A., Begun D. J., 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., Petrov D. A., Przeworski M., Andolfatto P., 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5: e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. G., Eyre-Walker A., 2002. Adaptive protein evolution in Drosophila. Nature 415: 1022–1024. [DOI] [PubMed] [Google Scholar]
- Stephan W., Wiehe T. H., Lenz M. W., 1992. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor. Popul. Biol. 41: 237–254. [Google Scholar]
- Stephan W., Song Y. S., Langley C. H., 2006. The hitchhiking effect on linkage disequilibrium between linked neutral loci. Genetics 172: 2647–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima K. M., Coop G., Przeworski M., 2006. How reliable are empirical genomic scans for selective sweeps? Genome Res. 16: 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S. A., Reed F. A., Ranciaro A., Voight B. F., Babbitt C. C., et al. , 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik V., Lerner A., 1963. Pattern recognition using generalized portrait method. Autom. Remote Control 24: 774–780. [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S. H., Hubisz M. J., Clark A. G., Payseur B. A., Bustamante C. D., et al. , 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z. X. P., et al. , 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.