Abstract
Blockade of P-selectin/PSGL-1 interactions holds significant potential for treatment of disorders of innate immunity, thrombosis, and cancer. Current inhibitors remain limited due to low binding affinity or by the recognized disadvantages inherent to chronic administration of antibody therapeutics. Here we report an efficient approach for generating glycosulfopeptide mimics of N-terminal PSGL-1 through development of a stereoselective route for multi-gram scale synthesis of the C2 O-glycan building block and replacement of hydrolytically labile tyrosine sulfates with isosteric sulfonate analogs. Library screening afforded a compound of exceptional stability, GSnP-6, that binds to human P-selectin with nanomolar affinity (Kd ~ 22 nM). Molecular dynamics simulation defines the origin of this affinity in terms of a number of critical structural contributions. GSnP-6 potently blocks P-selectin/PSGL-1 interactions in vitro and in vivo and represents a promising candidate for the treatment of diseases driven by acute and chronic inflammation.
Introduction
The vascular endothelium forms a dynamic interface between blood elements and peripheral tissues. Characteristically, leukocyte-endothelial interactions are mediated by transient tethering, followed by rapid integrin activation and subsequent transendothelial migration.1,2 The recruitment of leukocytes to sites of inflammation is mediated by selectin adhesion molecules and their ligands.3 P-selectin4,5, found on activated platelets and vascular endothelium, is rapidly translocated to the cell surface within minutes of an inflammatory stimulus, E-selectin6 is expressed on endothelial cells after de novo synthesis within a few hours of activation, while L-selectin is expressed on most leukocytes and functions as a homing receptor to mediate binding of lymphocytes to high endothelial venules of peripheral lymph nodes.7 Excessive trafficking of leukocytes to extravascular locations can lead to tissue injury contributing to the development of inflammatory bowel disease, chronic obstructive pulmonary disease, atherosclerosis, and post-thrombotic syndrome, among a variety of other disorders. Thus, selectins, as a mediator of early adhesion and intracellular signaling events in the inflammatory cascade, represent a promising target for the design of agents that limit adverse inflammatory responses.
While structurally diverse glycoprotein counter-receptors bind selectins with high affinity, the most well characterized ligand is P-selectin-glycoprotein-ligand-1 (PSGL-1).8 PSGL-1 binds all three selectins, but with highest affinity to P-selectin.9 Ligation of P-selectin expressed on endothelial cells by PSGL-1 constitutes the initial ‘capture and rolling’ step in the leukocyte-endothelial cell adhesion cascade.10 Likewise, the interaction of PSGL-1 with P-selectin on activated platelets promotes formation of leukocyte-platelet aggregates that contributes to adhesion and infiltration of inflammatory cells and both activated platelets and soluble P-selectin promote leukocyte infiltration.11-13 Significantly, the engagement of PSGL-1 to P-selectin activates intracellular signaling pathways that induces the β2-integrin LFA-1 to adopt an extended conformation associated with the intermediate affinity state, which supports leukocyte deceleration and cell arrest onto the endothelium.14 PSGL-1 also activates the expression of intracellular protein kinases, such as Rho/Rock kinase, which mediates cell migration, and MAPK kinase that controls expression of pro-inflammatory cytokines.15,16 Blockade of P-selectin/PSGL-1 interactions holds significant potential for the treatment of disorders due to maladaptive acute or chronic inflammatory responses.17-19
The role of P-selectin/PSGL-1 in a number of disease states has led to the design of a variety of biologics, small molecules, and glycopeptide mimics to target these interactions. Although P-selectin and PSGL-1 blocking antibodies are undergoing clinical evaluation for the treatment of sickle cell disease and Crohn's disease, they are expensive to manufacture, limited in shelf-life, and the development of antibodies against monoclonal therapeutics, including chimeric and humanized monoclonal antibodies, continues to limit the effectiveness of antibody therapy especially when there is need for daily or long-term administration.20 Small molecule inhibitors designed through modifications of sialyl Lewis x (sLex) continue to be limited by their low potency and off-target toxicity. For example, GMI-1070 has demonstrated efficacy in treating sickle cell disease, but its low activity to P-selectin (IC50 ~ 423 μM) requires infusion of ~ 2 gram of drug per day.21 Likewise, PSI-697 only weakly inhibits human platelet-monocyte aggregation, which is almost certainly attributable to its low Kd ~ 200 μM.22 Similarly, the glycomimetic, bimosiamose (TBC1269), is a pan-selectin inhibitor with an IC50 of 70 μM against P-selectin and an IC50 of > 500 μM against E- and L-selectin.23 Most existing P-selectin inhibitors have been designed to mimic the core 2 O-glycan bearing sLex moiety, but often fail to account for the crucial contributions of multiple clustered tyrosine sulfates.21,24,25 Indeed, Leppänen et. al.26-28 have shown that high affinity binding of P-selectin to PSGL-1 requires stereospecific interactions with both clustered tyrosine sulfates (Tyr-SO3H) and a nearby core 2 O-glycan bearing a sLex-containing hexasaccharide epitope (C2-O-sLex). To date, attempts to synthesize mimics of the N-terminus of PSGL-1 have been limited by the acid sensitivity of tyrosine sulfates,29,30 poor selectivity in key glycosylation steps,31 and incompatible protecting groups for oligosaccharide synthesis.32
We report an efficient approach for the generation of a diverse set of glycopeptide mimics of PSGL-1. Key features of this synthesis include an efficient stereoselective route that has lead to multi- gram scale synthesis of the C2 O-glycan and replacement of hydrolytically labile tyrosine sulfates with stable, isosteric sulfonate analogs affording compounds with high affinity to P-selectin (Kd 14-22 nM). In the process, we identified a high affinity, chemically stable compound, termed GSnP-6 that blocks PSGL-1/P-selectin interactions in vitro and in vivo.
Results
Compound design and strategy
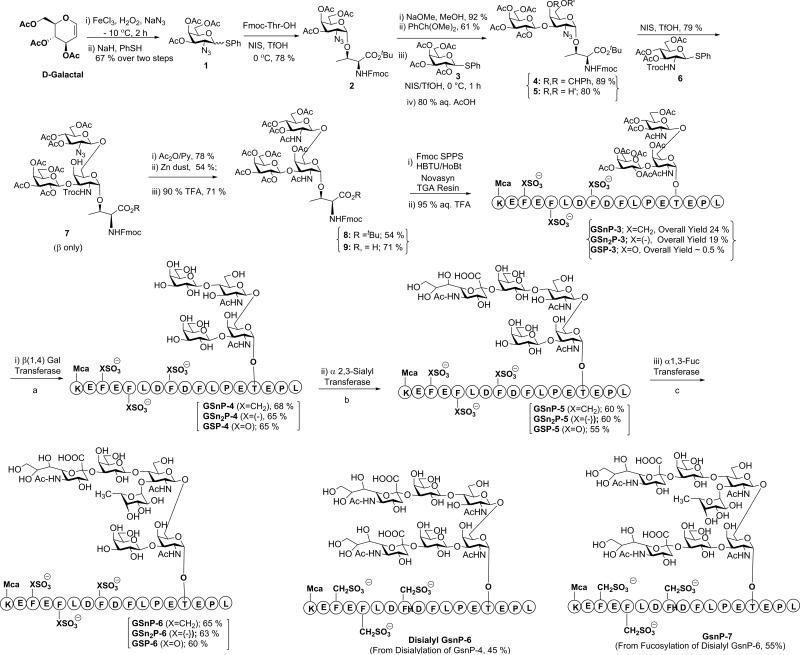
In our approach, we envisaged a building block containing protecting groups fully compatible with peptide synthesis, after which sLex would be incorporated. Appendages were selected so that the resulting compounds could be selectively extended by glycosyltransferases to provide a diverse array of glycopeptide mimetics of PSGL-1. In this regard, we previously identified a threonine-derived core-2 O-glycan as an appropriate building block for such studies.33 However, attempts to synthesize the glycoamino acid building block suffered from suboptimal regioselectivity (α/β : 1:2:6) in the key glycosylation step. Furthermore, use of a glycosyl imidate donor required a triflic azide mediated diazotransfer step, which is potentially explosive, thus, prohibiting deployment of this scheme for preparative scale synthesis (> 50 g). As a result, the core-2 O-glycan was only obtained on milligram scale. In order to circumvent these problems, a more efficient scheme was developed that lead to multi gram scale synthesis of this key compound. Synthesis began from 3,4,6 tri-O-acetyl-D-galactal, which can be readily converted to a halide via a one pot azidochlorination step (Figure 1). Although previously reported,34 direct coupling of the chloride intermediate with a Fmoc-threonine acceptor was unsuccessful due to its rapid decomposition. Therefore, this intermediate was converted in situ to the thioglycoside donor 1. Significantly, this two-step, one pot procedure could be safely carried out on a preparative scale (>50 gram) to provide donor 1 in 67% yield. Azidochlorination does not completely eliminate explosive risk, but it is substantially reduced and greatly facilitates safe, large-scale production. Coupling with the Fmoc-threonine acceptor afforded 2 in 78% yield with high α selectivity (α/β: 95:5). Glycoamino acid 2 was then converted to diol 5 by following previously reported steps.33 Significantly, this new scheme provided acceptor 5 in less than 7 steps with 21% overall yield, which established this route for this key building block as the most efficient and convenient to date.31-33
Figure 1. Synthesis of the Core-2 glycan and subsequent enzymatic steps to afford a family of 17 glycopeptide mimetics of PSGL-1.
Enzymatic steps (a) UDP-Gal, β-1,4-GalT (bovine), alkaline phosphatase, 130 mM HEPES, pH 7.4, 40 mM sodium cacodylate, pH 7.0, 20 mM MnCl2, and 0.02% NaN3; (b) α 2,3-(N)-sialylT CMP-NeuAc 50 mM MOPS, pH 7.4, 0.1% bovine serum albumin, and 0.02% NaN3, 14 h; (c) GDP-Fuc, α1,3-FucT-VI, 50 mM MOPS, pH 7.4, 20 mM MnCl2 and 0.02% NaN3, 16 h. The desialylated GSnP-6 was obtained from GSnP-4 in 45% yield. GSnP-7 was obtained in 55% yield by fucosylation of disialylated GSnP-6. 4-(Sulfomethyl)phenylalanine series (n): GSnP-6(X: CH2; R=H); GSnP-7 (X: CH2; R=Sialyl); 4-(Sulfo)phenylalanine series (n2): GSn2P-6 (X: bond; R=H); Tyrosine O-sulfate series: GSP-6 (X: O; R=H). The last numeral refers to the size of the glycan (e.g. 6 for hexasaccharide).
In the key glycosylation step with acceptor 5, we anticipated that the axial 4-OH group would be of low reactivity, especially when carrying a substituent at O-3. However, an undesired tetrasaccharide was identified in approximately 20% yield.33 Both desired and undesired compounds had similar retention factor values. Thus, chromatographic separation was challenging and laborious, particularly at a preparative scale. To address this problem, we performed low temperature activation (-10°C) with glucosamine donor 6 at 0.8 Equiv (as opposed to 1.2 Equiv), which produced only the β-glycoside 7 in 79% yield (Supplementary Figures 1 and 2). The (1→6)-linkage in 7 was confirmed by NOESY spectrum, which displayed a cross-peak between H-1 of the glucosamine residue and H-6 of the galactosamine residue. gHMBC NMR of 7 confirmed that O-6 was glycosylated in revealing cross-peaks from 101.3 ppm (C-1 of the glucosamine residue, A-C1) and 4.02 ppm (H-6 of the galactosamine residue, BH6), as well as from 4.70 ppm (H-1 of the glucosamine residue, A-H1) and 69.5 ppm (C-6 of the galactosamine residue, B-C6) (Supplementary Figures 3-7). Acetylation of 7 with Py/Ac2O, zinc reduction, and TFA mediated tert-butyl ester deprotection provided the core-2 O-glycan 9 (Supplementary Figures 8 and 9). This new route improved not only selectivity and yield, but also enabled the production of gram quantities of intermediate 9 and facilitated the synthesis of a broad range of structural mimics of PSGL-1.
With the availability of this glycoamino acid building block, glycopeptide mimics of the N-terminal domain of PSGL-1 were synthesized using a Fmoc-assisted SPPS strategy (Figure 1). Coupling reactions were performed using 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and 1-hydroxy-benzotriazol (HOBt) and the deprotection step was carried out with 20% piperidine. Addition of the sterically hindered core-2 glycoamino acid 9 proceeded smoothly without formation of any side products. However, the incorporation of Fmoc-Tyr-(OSO3−) amino acid in peptide synthesis is extremely challenging owing to its acid lability.35 As an alternative approach, enzymatic sulfation has proven less than ideal since tyrosine sulfotransferase is not commercially available and while the enzymatic reaction is optimal at 30°C, tyrosine sulfotransferase is unstable above 20°C.36 Chemical sulfation of tyrosine residues after peptide synthesis also suffers from low specificity,30 incomplete reaction with sulfating reagents37, and low yield.38 Together, these factors place significant limitations on the use of Tyr (OSO3−), which has limited their broad application in drug design. Therefore, we elected to replace the acid labile Tyr (OSO3−) moiety with bioiosteric, hydrolytically stabile analogues, namely, Fmoc-Phe (p-CH2SO3H)39,40 and Fmoc-Phe (p-SO3H)41. Such sulfonate mimics can be readily incorporated in solid phase peptide synthesis without the need for any side chain protecting groups using standard HBTU coupling conditions. The chromogenic amino acid, Fmoc-Lys(Mca)-OH was also incorporated at the N-terminus of the PSGL-1 sequence for facile detection and quantification. After peptide synthesis, the deprotection of sulfonate mimics could be readily carried out with 95% aq. TFA at room temperature. Saponification of O-acetate groups in GS(n)P-3 was achieved with catalytic NaOMe to afford the glycopeptide mimics, which were further purified by RP-HPLC. In synthesizing the native tyrosine O-sulfate sequence, GSP-3, deprotection was performed at 0°C to suppress the degradation of tyrosine O-sulfates.42 When tyrosine O-sulfates were employed in peptide synthesis, overall yield of GSP-3 was less than 0.5% indicating the labile nature of tyrosine sulfates.36,43 In contrast, use of hydrolytically stable tyrosine sulfonate analogues resulted in a dramatic improvement in overall yield to afford GSnP-3 and GSn2P-3 in 24% and 19% yields, respectively.
The sLex moiety was incorporated into GS(n)P-3 by using glycosyltransferases in stepwise order to sequentially add galactose, sialic acid, and fucose. The highly efficient β1,4-galactosyltransferase (β1,4-Gal T) is commercially available and was used to initially install β1,4-galactose. Since sialyltransferases have low efficiency towards a fucosylated Lex structure, fucose was installed after sialylation. Thus, galactose was first appended in a β1,4 linkage to GlcNAc by treatment with β1,4- GalT in the presence UDP-Galactose to produce GS(n)P-4. Similarly, addition of sialic acid was achieved by incubation of GSnP-4 with α2,3-sialyltransferase (α,2,3-(N)-sialylT) in the presence of CMP-NeuAc to obtain GSnP-5. The sLex structure was completed by incubation of GS(n)P-5 with human α1,3-fucosyltransferase V (1,3-FucT V) and GDP-Fucose to produce GS(n)P-6. All enzymatic steps proceeded with 60 to 70% yield without observed interference of the sulfonate residues.44 In addition, desialylated glycopeptide mimics were generated through additional sialylation of GSnP-4 to provide disialyl GSnP-6 (45% yield) followed by fucosylation to provide GSnP-7 in 55% yield. Using this approach, a diverse array of 17 glycopeptide mimics of PSGL-1 was synthesized, including GSnP[3-6], disialyl GSnP-6, GSnP-7, GSn2P[3-6], GSP[3-6], and three GSnP core-1 peptides (Figure 1, Supplementary Figures 10-17).
GSnP-6 demonstrates nanomolar affinity to P-selectin
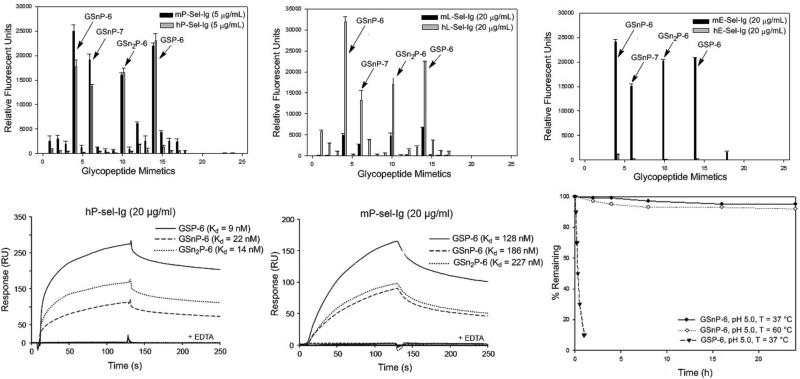
The binding affinity of this set of glycopeptide mimics towards P-, L-, and E-selectin was initially screened using microarray technology. The 17 glycopeptide mimics along with five glycan standards (sialyl Lex, NA2, NA2,3, NA2,6 and LNnT) were printed on a NHS-activated glass slide. The slide was then incubated with recombinant Ig chimeras of P-, L-, or E-selectin (5-20 μg/mL) followed by Alexa-488 labeled anti-human IgG antibody (5 μg/mL). Similar to the native, N-terminal PSGL-1 sequence containing tyrosine sulfate (GSP-6), GSnP-6 bound to P-selectin more strongly than to E- or L-selectin (Figure 2A-C, Supplementary Figure 18). Likewise, GSnP-7, a sialylated extension of GSnP-6, showed higher affinity to P-selectin. However, disialyl GSnP-6, lacking the fucosyl residue displayed lower affinity to P-selectin, consistent with the key contribution of α1,3-fucose. Binding of glycopeptide mimics was Ca2+ dependent and inhibited by EDTA.
Figure 2. Binding of P-, E-, or L-selectin to PSGL-1 glycopeptide mimics and GSnP-6 stability analysis.
Microarray binding studies of glycopeptide mimics toward (A) human and mouse P-selectin (5 μg/mL), (B) human and mouse L-selectin (20 μg/mL), and (C) human and mouse E-selectin (20 μg/mL). The compounds printed on the microarray in the order depicted here in A-C are detailed in Supplementary Figure 18. Reference compounds included sialyl Lewis × (sLex), the biantennary glycans NA2, NA2,3, NA2,6, as well as lacto-N-neo-tetraose (LNnT) and biotin. Bound selectin-Igs were detected using Alexa-488 labeled anti-human IgG antibody (5 μg/mL). Three lectins RCA-1, AAL, and PNA were used to confirm the sequence of enzymatic steps. Monoclonal antibodies CHO131, PSG2 antibody and PL-1 were used to confirm the presence of sLex, tyrosine sulfates, and the peptide sequence, respectively. Biacore binding analysis to human P-selectin with observed rate constants for (D) GSnP-6, kon 3.1 × 105 M−1 s−1, koff 6.9 × 103 M−1s; GSn2P-6, kon 6.4 × 105 M−1s−1, koff 6.8 × 103 M−1s. Biacore binding analysis to mouse P-selectin with observed rate constants for (E) GsnP-6, kon 4.9 × 104 M−1s−1, koff 8.0 × 103 M−1s−; GSn2P-6 kon 5.3 × 104 M−1s−1, koff 9.0 × 103 M−1s. (F) Temperature- and pH-dependent stability studies of GSnP-6.
Dissociation constants (Kd) were determined using a Biacore binding assay after initial capture of biotinylated GS(n)P-6 onto streptavidin-coated sensor chips followed by flow through of P-, L- or E-sel-Ig (2.5 to 60 μg/mL). Dissociation constants for GSnP-6 and GSn2P-6 to human P-selectin were 22 nM and 14 nM (Figure 2D), respectively, compared to the reported Kd of 73 nM for native PSGL-1.9 The Kd of GSnP-6 to murine P-selectin was ~9-fold lower than to human P-selectin (Figure 2E). GSnP-6 bound to E- and L-selectin with lower affinity (Supplementary Figure 19), consistent with reports of PSGL-1 binding to E- and L-selectin at a Kd of 2 to 5 μM.28
Sulfotyrosine exhibits poor stability under acid and near neutral conditions, thereby, limiting its use in drug development. For example, rapid decomposition occurs at 37°C upon exposure to TFA and other scavengers used in peptide synthesis and the hydrolysis rate of sulfotyrosine increases with increasing temperature.42,43 Consistent with these observations, the native N-terminal PSGL-1 sequence, GSP-6, rapidly degraded under low pH conditions (pH ~ 5; T= 37°C). In contrast, the GSnP-6 mimic demonstrated superior stability when subjected to both low pH (pH 5; T = 37°C) and high temperature (pH 7.5; T = 60°C), without observed degradation products as detected by RP-HPLC (Figure 2F).
Molecular dynamics simulations
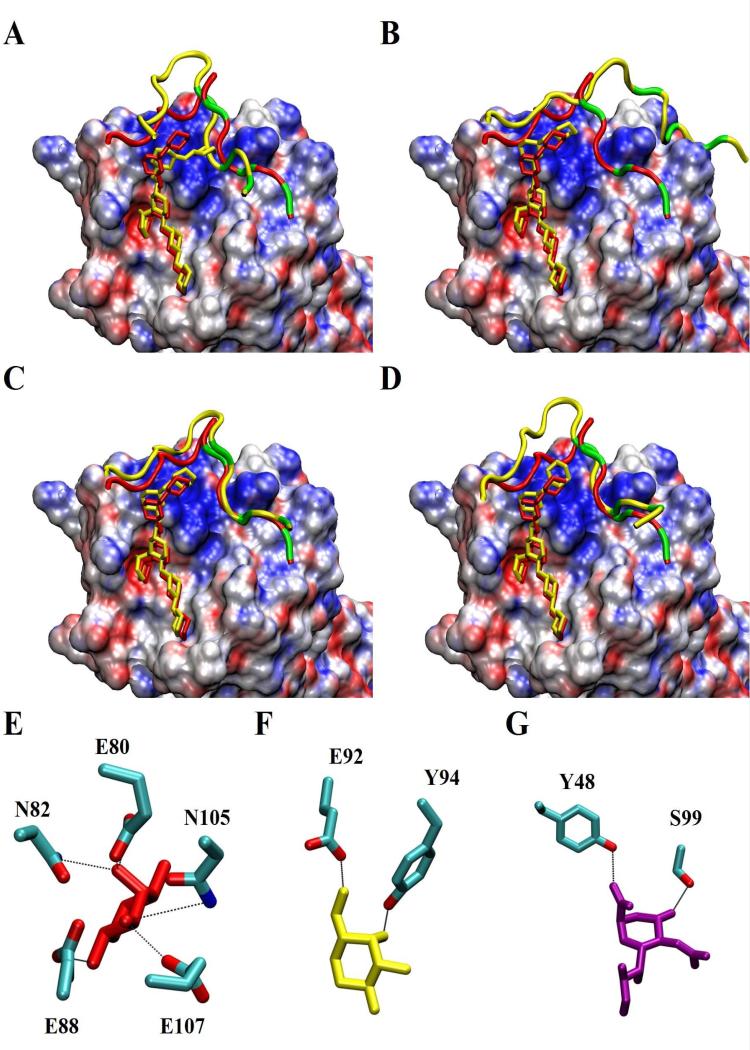
Computational simulations were used to predict key structural residues responsible for P-selectin/GSnP-6 interactions. Molecular dynamics (MD) simulations were first validated by reproducing all structural attributes of the PSGL-1/P-selectin interaction, as defined by crystallographic data and experimental investigations. The positions of the glycan and the peptide components of PSGL-1 were monitored over the course of simulating its interaction with P-selectin (Supplementary Figure 20). Simulations confirmed that the ligand remained stable in the binding site, with the glycan displaying less positional variation than the peptide. In addition, all experimentally observed hydrogen bonds and salt bridges between the ligand and the protein were observed during the MD simulations. Of note, the dynamic motions of the system weakened some of these interactions relative to others (Supplementary Table 2). In particular, while the hydrogen bonds associated with the glycan appeared markedly stable, the interactions between the tyrosine sulfate residues and the P-selectin surface residues were relatively unstable and depended heavily on the protonation state of H114. This is consistent with a recent report of Cao et al.45, who demonstrated that the protonation state of H114 impacts the binding affinity of PSGL-1 to P-selectin. As expected, full protonation of this histidine enhanced the stability of these interactions, particularly those associated with tyrosine sulfate (Y607) and H114. Similarly, the structure (Figure 3) and stability (Supplementary Figure 20) of GSnP-6 in the fully-protonated H114 complex was comparable to that observed for the native PSGL-1 sequence, and retained all of the key non-covalent interactions (Supplementary Table 1). In the case of neutral H114, the interactions between the tyrosine sulfonates and the protein were unstable, leading to orientation disruption of the peptide component in GSnP-6. The frequency of the rebinding interaction was too low to observe statistical convergence in the MD simulation (Supplementary Figure 20). Thus, only simulations with fully-protonated H114 were employed in the subsequent analysis of interaction energies.
Figure 3. Interactions of the N-terminus of PSGL-1 and GSnP-6 bound to P-selectin, as a function of the protonation state of H114.
Conformation of PSGL-1 (A) and GSnP-6 (B) ligands most similar to the average shape acquired from MD simulations performed with neutral H114. The crystal structure of the PSGL-1 ligand is shown in red with a splined representation of the peptide backbone, sulfated amino acid positions in green, and a stick representation for monosaccharide rings. Assuming that H114 is fully protonated leads to optimal reproduction of the crystallographic data for PSGL-1 (C) and leads to similar binding for GSnP-6 (D). The solvent-accessible surface of P-selectin is colored according to the electrostatic potential (acidic region: red; basic region: blue). (E-G) Hydrogen bonds between Fuc (red), Core-2 Gal (yellow), and Neu5Ac (purple) and P-selectin residues.
Combinations of five GB parameterizations and protein dielectric constant values (ε) ranging from 1 to 5 were initially evaluated in order to determine which, if any, set of conditions reproduced the relative contributions of PSGL-1 constituent features to P-selectin binding affinity. Optimal conditions were then employed to assess the interaction energies of GSnP-6 with P-selectin, as well as the contributions of distinct structural features of the analogue. The MM/GBSA calculations using either the GB OBC2 and GB OBC1 model produced comparable results with ε = 4.0, in agreement with relative experimental values (Supplementary Table 2). Since the GB OBC1 parameterization has been successfully employed to compute interaction energies in other carbohydrate-protein systems involving charged moieties, it was selected over the GB OBC2 parameterization (Supplementary Figure 21).46 This combination was able to correctly rank the relative contributions to binding, with Fuc ≈ total sulfate > Neu5Ac, and identified that the second sulfate made a larger contribution (by -1.3 to -1.6 kcal/mol) than the other two.
The experimental interaction energies of P-selectin with PSGL-1 ligand and the analogue GSnP-6 were similar at -9.8 kcal/mol (Kd = 73 nM) and -10.5 kcal/mol (22 nM), respectively. Using the combination of GB OBC1 with ε = 4.0, the absolute interaction energies of P-selectin with PSGL-1 and GSnP-6 were predicted to be -45.1 ± 3.4 kcal/mol and -44.6 ± 3.1 kcal/mol, respectively (Table 1). As anticipated, due to the omission of entropic penalties, predicted interaction energies were almost 4-fold larger than experimental values.47-49 Nonetheless, interaction energies were statistically equivalent, which is in agreement with experimental data. To facilitate detection on binding microarrays, GSnP-6 was chemically derivatized with a 7-amino-4-methylcoumarin (MCA) tag, which alters net charge, as well as local properties of the terminal lysine. The MCA tag was not included in the modeling, which employed only a lysine residue. The remaining residues in the peptide sequence of PSGL-1 and GSnP-6 render essentially identical contributions to the interaction energies. Due to the similarities of the two ligands, the relative contributions of the glycans to binding affinity were also indistinguishable within standard deviations. Likewise, the individual contributions from each sulfate group were comparable for each ligand, despite chemical differences (Table 1). An unexpected and potentially significant observation concerns the second tyrosine sulfate residue (607), whose strong affinity (-8.4 to -9.5 kcal/mol) appears to arise predominantly from van der Waals interactions (-6.6 to -7.0 kcal/mol). In summary, computational analysis reproduced salient structural and energetic features of PSGL-1/P-selectin interactions and predicted that GSnP-6 would behave comparably, consistent with observed affinity data (Figure 3).
Table 1.
Per-residue MM/GBSA interaction energiesa for interactions of P-selectin with residues in PSGL-1 and GSnP-6
| PSGL-1 | GSnP-6 | PSGL-1 | GSnP-6 | ||
|---|---|---|---|---|---|
| Y/YC 605b | −1.4 ± 0.6 | −1.3 ± 0.6 | Neu5Ac | −2.7 ± 0.8 | −2.6 ± 0.8 |
| Y/YC 607b | −5.5 ± 0.7 | −6.0 ± 0.6 | Core-2 Gal | −4.2 ± 0.7 | −4.2 ± 0.7 |
| Y/YC 610b | −0.8 ± 0.4 | −0.6 ± 0.3 | GlcNAc | −3.3 ± 0.6 | −3.2 ± 0.5 |
| SO3– 605c | −1.9 ± 0.8 | −2.3 ± 0.9 | Fuc | −4.8 ± 1.3 | −4.7 ± 1.3 |
| SO3– 607c | −2.9 ± 0.9 | −3.5 ± 0.8 | GalNAc | −0.6 ± 0.3 | −0.4 ± 0.2 |
| SO3– 610c | −0.8 ± 0.6 | −0.8 ± 0.4 | Gal | 0.1 ± 0.1 | 0.1 ± 0.1 |
| Subtotal | −13.3 ± 1.7 | −14.1 ± 1.5 | Subtotal | −15.5 ± 1.8 | −15.0 ± 1.8 |
| Amino Acids | |||||
| K603e | N/A | 1.3 ± 0.1 | L613 | −4.1 ± 1.1 | −3.4 ± 1.2 |
| E604 | 0.1 ± 0.2 | −0.6 ± 0.1 | P614 | −1.9 ± 1.2 | −1.3 ± 1.0 |
| E606 | −1.2 ± 0.4 | −1.2 ± 0.3 | E615 | −0.9 ± 0.2 | −0.9 ± 0.2 |
| L608 | −2.2 ± 1.2 | −3.3 ± 0.8 | T616d | −1.4 ± 0.3 | −1.3 ± 0.4 |
| D609 | −1.8 ± 0.7 | −1.9 ± 0.7 | E617 | −0.6 ± 0.3 | −0.5 ± 0.3 |
| D611 | −0.7 ± 0.1 | −0.7 ± 0.1 | P618 | −0.4 ± 0.1 | −0.4 ± 0.2 |
| F612 | −1.2 ± 0.6 | −0.9 ± 0.5 | |||
| Subtotal | −16.3 ± 2.3 | −15.1 ± 2.1 | |||
| Total Interaction Energy | −45.1 ± 3.4 | −44.6 ± 3.1 | |||
All results are in kcal/mol. The entropy contributions are not included in these results.
Contribution from tyrosine sulfate or tyrosine sulfonate not including the SO3– group.
SO3– is counted as a residue in the energy decomposition, instead of –O-SO3– or –CH2–SO3–.
Glycosylation site.
Numbering based on the crystal structure.
GSnP-6 inhibits P- and L-selectin binding to PSGL-1
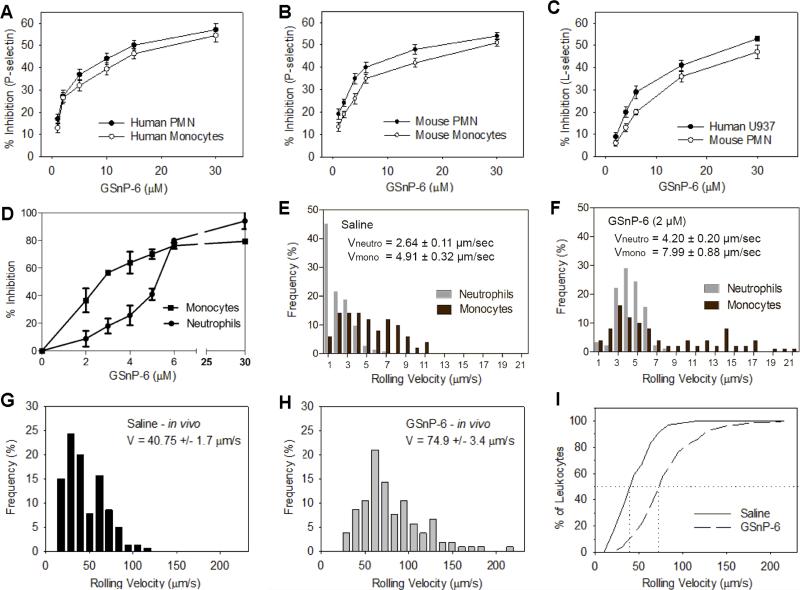
Flow cytometry was initially used to characterize the ability of GSnP-6 to block binding of selectin Fc chimeras to murine and human leukocytes. Recombinant mouse P-, L-, or E-selectin Fc chimera (2 μg/mL) were incubated with murine leukocytes and GSnP-6 (0 - 30 μM). Likewise, recombinant human P-, L-, or E-selectin Fc chimera (2 μg/mL) were incubated with human peripheral blood leukocytes, as well as the human U937 cell line in the presence of GSnP-6 (0 - 30 μM). Binding of selectin chimeras was detected with PE-conjugated anti-Fc antibody, quantified as mean fluorescent intensity, and plotted as percent inhibition. Specificity was confirmed with PSGL-1 blocking antibodies. GSnP-6 inhibited P-selectin dependent interactions in a dose dependent manner in both human and mouse leukocytes, including human neutrophils (IC50 ~ 14 μM), human monocytes (IC50 ~ 20 μM), mouse neutrophils (IC50 ~ 19 μM), and mouse monocytes (IC50 ~ 28 μM) (Figure 4A-B). GSnP-6 also inhibited PSGL-1/L-selectin interactions at a somewhat lower potency (IC50 ~ 30 μM) for human and mouse leukocytes (Figure 4C). Inhibition of E-selectin interactions was not observed over this concentration range (Supplementary Figure 22), which is consistent with the lower binding affinity of PSGL-1 for E-selectin and the notion that leukocytes interact with E-selectin through alternate ligands including CD44 and ESL-1 on mouse, and L-selectin on human leukocytes.50,51
Figure 4. GSnP-6 inhibits selectin adhesive interactions in vitro and in vivo.
(A-B) GSnP-6 (0 - 30 μM) was incubated with (A) human circulating PMN and monocytes or (B) mouse circulating PMN and monocytes and species appropriate P-selectin chimera, analyzed by flow cytometry and plotted as percent inhibition vs. PBS control. GSnP-6 inhibited P-selectin dependent interactions in a dose dependent manner in human and mouse leukocytes, including human neutrophils (IC50 ~ 14 μM), human monocytes (IC50 ~ 20 μM), mouse neutrophils (IC50 ~ 19 μM), and mouse monocytes (IC50 ~ 28 μM). (C) GSnP-6 inhibited PSGL-1/L-selectin interactions to human U937 and mouse neutrophils with a lower potency (IC50 ~ 30 μM). Data are mean ± SEM, n=3. (D-F) GSnP-6 inhibits P-selectin/PSGL-1 adhesion under shear in vitro. (D) GSnP-6 (0-6 μM) exhibits dose dependent inhibition of human neutrophil and human monocyte rolling and arrest on a recombinant ICAM-1/P-selectin substrate under shear; n=3 PBS, n=3/GSnP-6 dose, velocities (μm/s) of 120 cells per condition were measured, data are plotted as mean ± SEM. (E-F) GSnP-6 increases rolling velocity of neutrophils and monocytes, cumulative frequency histograms are shown for PBS vehicle control (E) and 2 μM GSnP-6 (F); Vneutrophil: saline vs. GSnP-6, p < 0.0001 (Student's t-test); Vmonocyte: saline vs. GSnP-6, p < 0.0001 (Student's t-test). (G-I) GSnP-6 (4 μmol/kg, n=4 mice, 5 leukocytes/vessel analyzed in 28 vessels) or saline control (n=3 mice, 5 leukocytes/vessel analyzed in 21 vessels) were delivered IV and leukocyte rolling velocity was recorded 15-25 min post-surgical stimulation of the mouse cremaster to characterize P-selectin dependent responses in vivo; Vmean: saline vs. GSnP-6, p < 0.01 (Student's t-test). (I) Cumulative frequency histogram of leukocyte rolling velocities, median values indicated by vertical lines (median velocity saline = 35.5 μm/s, median velocity GSnP-6 = 64.0 μm/s).
GSnP-6 inhibits leukocyte rolling and arrest
PSGL-1 interactions with P-selectin have been shown to signal activation of CD18 (β2-integrin) to adopt a high affinity extended conformation resulting in integrin dependent deceleration and cell arrest. Inhibition of this interaction leads to an increase in rolling velocity and a reduction in cell arrest.51 Human neutrophils and monocytes were incubated with GSnP-6 (0-30 μM) or PBS vehicle for 10 min and perfused through a microfluidic flow chamber at 2 dynes/cm2 over glass substrates coated with ICAM-1 and P-selectin Fc chimeras.52 Total cell capture, defined as the number of cells that rolled or arrested (velocity ≤ 1 μm/sec), was determined, along with leukocyte rolling velocity. Videomicroscopic analysis demonstrated that GSnP-6 lead to a dose-dependent inhibition of monocyte and neutrophil capture in shear flow, with an IC50 of 3 and 5 μM, respectively (Figure 4D). At a concentration as low as 2 μM, GSnP-6 increased the rolling velocity of human neutrophils and monocytes and resulted in 80% inhibition at 6 μM, a dose that abrogated the fraction achieving arrest (Figure 4D-F). In contrast, incubation with the sulfated polysaccharide, fucoidan, at a concentration of up to 50 μg/mL (~50 μΜ) did not affect the efficiency of capture, rolling, or arrest when compared to PBS control (data not shown). GSnP-6 at concentrations as high as 30 μM did not perturb leukocyte rolling and arrest on substrates coated with ICAM-1 and E- selectin (Supplementary Figure 23). This implies that GSnP-6 specifically inhibits the interaction between PSGL-1 and P-selectin in neutrophils and monocytes over this concentration range.
Intravital microscopy was performed within 30 min of surgical stimulation of the murine cremaster muscle to determine the ability of GsnP-6 to inhibit leukocyte binding to microvascular endothelium in vivo.53 Seven venules per mouse (n = 4, saline control; n = 3, GSnP-6, 4 μmol/kg IV) with a diameter of 30 to 40 μm were analyzed and the velocities of 5 to 10 rolling leukocytes were determined in each venule by individually tracking leukocyte distance/time (μm/s). A significant increase in mean rolling velocity was observed after intravenous administration of GSnP-6 when compared to saline vehicle control (Figure 4G-H, saline: 40.8 ± 1.7 μm/s vs. GSnP-6: 74.9 ± 3.4 μm/s; p ≤ 0.01; Student's t-test). Cumulative frequency histogram analysis demonstrated a 2-fold increase in median rolling velocities (Figure 4I, saline: 35.5 μm/s vs. GSnP-6: 64.0 μm/s; Supplementary Figure 24).
GSnP-6 inhibits early thromboinflammatory events in vivo
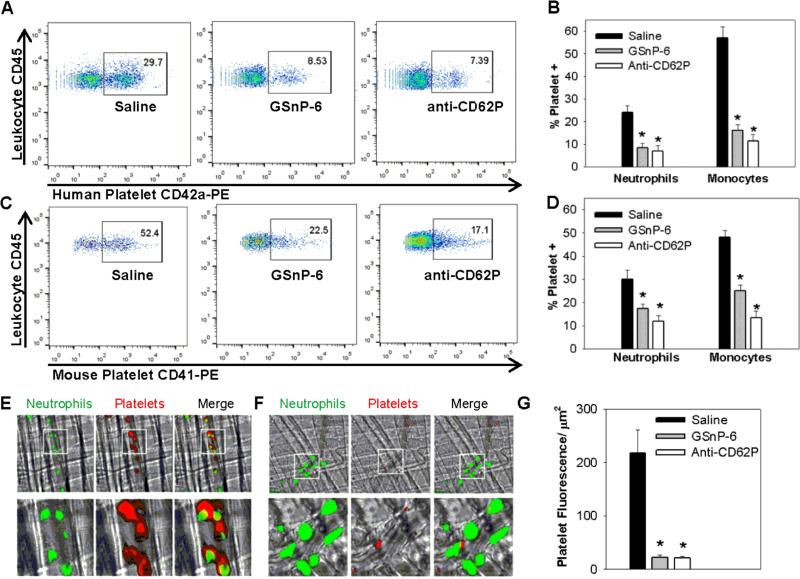
PSGL-1/P-selectin interactions are responsible for the formation of leukocyte-platelet aggregates, which promotes the infiltration of leukocytes, as well as the release of neutrophil extracellular traps (NETs) and other procoagulant factors.54 Human and murine platelet-leukocyte aggregates were quantified in whole blood using dual-label flow cytometry.55 Human or mouse blood was stimulated with a thrombin receptor-activating peptide to induce platelet P-selectin expression and dosed with 40 μM GSnP-6. GSnP-6 inhibited 65% of platelet-neutrophil and 72% of platelet-monocyte aggregate formation in human blood (Figure 5A-B; p < 0.05; Student's t-test) and 42% of platelet-neutrophil and 47% of platelet-monocyte aggregate formation in murine blood (Figure 5C-D; p < 0.05; Student's t-test).
Figure 5. GSnP-6 limits platelet-leukocyte aggregation in vitro and in vivo.
(A-D) Anti-coagulated human or mouse blood was dosed with 40 μM GSnP-6 at room temperature and stimulated for platelet P-selectin expression with thrombin receptor-activating peptide (40 μM human PAR1-activating peptide, 200 μM mouse PAR4-activating peptide). Platelet-leukocyte aggregates were quantified by two-color flow cytometry, CD45+ monocyte and neutrophil populations were discerned through characteristic side scatter as quantified as % platelet positive in saline control, 40 μM GSnP-6, and anti-CD62P (5 μg/mL human KPL-1, 5 μg/mL mouse RB40.34) treated samples. (A) Representative scatter plot of human samples incubated with anti-CD42a-PE and anti-CD45-APC, (B) % platelet positive neutrophils and monocytes in human samples, GSnP-6 inhibited 65% of platelet-neutrophil and 72% of platelet-monocyte aggregate formation in human blood. (C) Representative scatter plot of mouse samples incubated with anti-CD41-PE and anti-CD45-APC, (D) % platelet positive neutrophils and monocytes in mouse samples, GSnP-6 inhibited 42% of platelet-neutrophil and 47% of platelet-monocyte aggregate formation in mouse blood. Data representative of triplicate sample mean ± SEM, *p<0.05 vs. saline control (Student's t-test). (E-G) In vivo platelet-leukocyte aggregation in a TNF-α model of venular inflammation. (E) Intravital microscopy of venular inflammation 3 hr after administration of TNF-α demonstrates platelet aggregation and platelet-PMN binding after administration of saline control (PMN green; platelet red). (F) Platelet aggregation and platelet-PMN binding are not observed after administration of GSnP-6 (4 μmol/kg IV). (G) The platelet inhibitory effect of GSnP-6 was equivalent to that observed for anti-CD62P, a P-selectin blocking antibody (75 μg/mouse IV) and was significantly less than saline control (*p < 0.05 vs. saline (Student's t-test), n=5 mice/treatment, 8-10 venules/mouse analyzed; error bars are SEM).
The capacity of GSnP-6 to inhibit early thromboinflammatory events in vivo was characterized by platelet adhesion to leukocytes and platelet aggregation in a cremaster model of TNF-α-induced venular inflammation.56 TNF-α was administered intrascrotally (0.5 μg) to male mice (6-8 wks old, C57BL/6) 3 hr prior to imaging. P-selectin blocking antibody (RB40.34), GSnP-6, or saline was subsequently administered at the time of imaging and platelet microaggregate formation and platelet-neutrophil binding was monitored in venules (d 30-45 μm, 8-10 venules/mouse, n = 5 mice/test group) over 2 min intervals through visualization of platelets and neutrophils labeled with Dylight 649-anti-CD42b and Alexa Fluor 488-anti-Gr-1 antibodies, respectively. Three hours after intrascrotal administration of TNF-α, venule endothelial cells display ~700/mm2 adherent neutrophils and limited rolling flux (~1 WBC/min) (Supplementary Figure 25). Although administration of anti-CD62P or GSnP-6 at this stage would not be expected to alter established populations of adherent leukocytes, leukocyte-platelet interactions and platelet aggregate formation can be examined. Platelet accumulation was quantified by calculating median integrated fluorescence intensity, normalized to vessel surface area. GSnP-6 significantly reduced platelet-neutrophil interactions and platelet thrombi formation, similar to that observed after administration of a P-selectin blocking antibody (*p < 0.05; Student's t-test, Figure 5C, Supplementary Figure 25).
Discussion
The PSGL1/P-selectin pathway plays a critical role in disorders of innate immunity12,17,57,58, thrombosis19,59, and cancer60,61 and, as a consequence, the pharmacological blockade of this pathway represents an important therapeutic target. While the PSGL-1 glycan is a critical component of a diverse set of cell-cell recognition processes, as a complex glycoconjugate its recognition occurs in the context of other aglycone components that support leukocyte recruitment under shear stress of blood flow. Specifically, PSGL-1 is representative of a novel class of “glycosulfopeptide” (GSP) that includes GPIbα62, and endoglycan63, among many other important biomacromolecules. Detailed structural and mechanistic studies of PSGL-1 have revealed that high affinity recognition requires stereospecific interactions with both clustered tyrosine sulfates and a core 2 O-glycan that bears a sLex-containing hexasaccharide epitope.26-28 Thus, while first generation small molecule inhibitors of P-selectin/PSGL-1 interactions designed to mimic the sLex moiety have proven valuable in validating targets, they have yet to achieve clinical benefit, which is likely related to their low binding affinity to selectins.
Despite the potential of sLex-GSPs, such as PSGL-1, to serve as tools for biological studies and the promise of PSGL-1 glycopeptide mimetics as therapeutic agents, their utility has been limited by low yield and limited stability. For example, synthesis of the C2 O-glycan typically suffers from suboptimal regioselectivity and may involve a triflic azide mediated diazotransfer step, which is potentially explosive; thus, prohibiting preparative scale synthesis (> 50 g). Likewise, the desire to generate clustered tyrosine sulfates by direct incorporation of Fmoc-Tyr-(OSO3-) amino acids during peptide synthesis is extremely challenging, owing to its acid lability. Alternatively, the sulfation of tyrosine residues by enzymatic sulfation is hindered by the instability of tyrosine sulfotransferase above 20°C and chemical sulfation is limited by low specificity, incomplete reaction, and low yield.
This report highlights three important milestones that effectively circumvent these problems. First, we have developed an efficient stereoselective route that has lead to multi-gram scale synthesis of the C2 O-glycan. This scheme is the shortest and most convenient reported to date, which has facilitated the synthesis of a broad range of glycosulfopeptide mimics. Second, we have determined that the acid labile Tyr (OSO3-) moiety can be readily replaced with bioisosteric, hydrolytically stable Phe (p-CH2SO3H) and Phe (p-SO3H), increasing overall yield from < 0.5% to 24%. The significance of these new synthetic strategies was demonstrated by the identification and production of a PSGL-1 glycopeptide mimic, GSnP-6, from a GSP library, which is very stable and has the highest affinity (Kd 22 nM) to P-selectin reported to date. Our studies have confirmed that GSnP-6 potently blocks in a dose-dependent manner, the adhesion of platelet and leukocyte subsets implicated in the pathogenesis of variety of diseases. Finally, we have used GSnP-6 to establish a molecular dynamics model as an important tool for providing new structural insights for the design and evaluation of additional novel PSGL-1 glycomimetics.
In conclusion, a combination of chemical and enzymatic synthesis provided a diverse array of glycopeptide mimetics of PSGL-1. In the process, GSnP-6 was identified as a chemically stable compound that potently inhibits P-selectin/PSGL-1 interactions both in the microvasculature and in model systems that reveal its antagonist specificity in blocking platelet, monocyte, and neutrophil thromboinflammatory adhesion. A key discovery was the incorporation of stable sulfonated isosters, which permits facile peptide synthesis and provides a greater therapeutic window than the parent physiological ligand with considerable potential for application in disease states characterized by both acute and chronic inflammation.
Methods
General
All reagents were purchased from commercial sources and used as received, unless otherwise indicated. All solvents were dried and distilled by standard protocols. All reactions were performed under an inert atmosphere of argon or nitrogen, unless otherwise indicated. For peptide synthesis, Fmoc-Leu Novasyn TGA resin (resin loading: 0.3 mmol/g), DMF (Biotech grade) and piperidine were purchased from Novabiochem. Fmoc-Phe(CH2SO3H)-OH and Fmoc-Phe(SO3H)-OH amino acids were purchased from RSP Amino Acids LLC. Coupling agents HBTU, HOBt and all other Fmoc protected amino acids were purchased from EMD chemicals. Free reducing glycans, used as standards, were obtained from V-LABS, Inc. (Covington, LA). EZ-link NHS-Biotin was purchased from Thermo Scientific (Rockford, IL). Biotinylated lectins were from Vector Labs (Burlingame, CA). Cyanine 5- and Alexa488-labeled Streptavidin, Alexa488- and Alexa633-label secondary antibodies were purchased from Life Technologies (Woburn, MA). Recombinant human ST3Gal IV and ST3Gal 1 were kind gifts from Dr. Kelley Moremen at University of Georgia. Studies with the anti-sulfotyrosine monoclonal antibody PSG2 were kindly performed with help from Dr. Kevin Moore, Oklahoma Medical Research Foundation. Recombinant selectin Fc chimeras were purchased from R&D systems (Minneapolis, MN). Protein A/G was purchased from Fischer Scientific. Fucoidan control was obtained from Sigma-Aldrich. The NHS-activated NEXTERION® Slide H slides were purchased from Schott North America (Louisville, KY). 1H and 13C were recorded with Inova 600 MHz spectrometer. High Resolution MALDI spectra were recorded by Harvard University Mass Spectrometry Center Waters Micro MX MALDI-TOF instrument. Optical rotation values were recorded using Perkin Elmer Polarimeter. All organic extracts were dried over sodium sulfate and concentrated under aspirator vacuum.
All experimental protocols for compound synthesis, characterization data sets for all compounds, including NMR, HPLC, and MALDI profiles, as well as details of computational and biological studies are provided in Supplementary Information.
Microarray printing, binding assay, and scanning
For a multi-panel experiment on a single slide, the array layout was designed using Piezoarray software according to the dimension of a standard 14-chamber adaptor. The adaptor was applied on the slide to separate a single slide to 14 chambers sealed from each other during the experiment. All glycopeptides and control samples were printed on a NHS-activated glass slide (Schott, North America) in phosphate buffer (300 mM sodium phosphates, pH 8.5). The average spot volume was within 10% variation (intra-tip) of 0.33 nL. The average spot size was 100 μm. The following glycans were derivatized with 2-amino-N-(2-aminoethyl)-benzamide (AEAB)64: asialobiantennary N-glycan NA2 (Galβ1-4GlcNAcβ1-2Manα1-6(Galβ1-4GlcNAcβ1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAc), the 2,3-disiaylated biantennary N-glycan NA2,3 (Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-6(Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAc), the 2,6-disialylated biantennary N-glycan NA2,6 (Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAc), and lacto-N-neo-tetraose (LNnT) (Galβ1-4GlcNAcβ1-3Galβ1-4Glc). These AEAB-derivatives and biotin-NHNH2, all at 50 μM concentration were printed on the microarray as controls and reference standards. After printing, the slide was placed in a high moisture chamber at 50°C and incubated for 1 h. The slide was subsequently washed and blocked with 50 mM ethanolamine in 0.1 M Tris buffer (pH 8.0) for 1 h, dried by centrifugation, and stored desiccated at -20°C prior to use. The binding assay was performed by incubating the slide with human or mouse P- E- or L- selectin (5 μg/mL in calcium buffer (pH 8.0). Binding is Ca2+-dependent and was quantitatively inhibited by addition of EDTA. Detection was accomplished using Alexa 488 goat anti Human IgG (5 μg/mL). The slide was subsequently washed (TSM Buffer 5 times), centrifuged, and analyzed with a Perkin Elmer ProScanArray microarray scanner equipped with 4 lasers covering an excitation range from 488 to 637 nm. The scanned images were analyzed using ScanArray Express software (Supplementary Figure 18).
Biacore binding assay of PSGL-1 mimics
Biotinylated GS(n)P-6 was first captured quantitatively on a streptavidin-coated sensor chip (GE Healthcare). Different amounts of P-, L- or E -sel-Ig were incubated in the wells (2.5 - 60 μg/mL) and bound selectins were detected using fluorescently labeled anti-human IgG. PL1 (Santa Cruz, CA) and CHO131 (Santa Cruz, CA) are mAbs that recognize the peptide sequence and C2 sLex respectively, which were used to confirm the immobilization step and to compare the relative amount of glycopeptides on the chip (Supplementary Figure 19). The bound mAbs and selectins were removed from the sensor chip by injecting 10 mM glycine-HCl, pH 2.0 and 50 mM EDTA, respectively, at 20 μL/min for 30 sec. The kinetic parameters were obtained by plotting the curves using a 1:1 binding model provided by the Biacore evaluation software.
At low GSnP-6 density (< 5 RU), no plateau was observed even at highest concentrations of L- or E- Sel-Ig (60 μg/mL). By contrast P-Selectin (P-Sel-Ig), generated a saturated binding curve irrespective of the GsnP-6 coated densities used (< 5-60 RU), indicating that affinity of P-Sel-Ig for GSnP-6 is less dependent on the density of the immobilized ligand. The comparison of GSnP's densities (< 5 RU vs 60 RU) indicate that binding of L- or E-Sel-Ig has ~ 25 fold lower affinity for the immobilized GSnP-6 than P-Sel-Ig.
Molecular dynamics (MD) simulation studies
Force field parameters for the oligosaccharide and SO3− moiety in the YS and YCS residues were taken from the GLYCAM06 (version h)65 parameter set, while those for the protein came from AMBER12 (ff99sb).66 Parameters for the linkage between the SO3− group and the amino acid side chains were approximated from existing terms in two parameter sets (Supplementary Table 3). Ensemble-averaged partial atomic charges for the YS and YCS residues were developed according to the standard GLYCAM protocol65, from a collection of 300 snapshots extracted at 0.1 ns intervals from MD simulations (30 ns) performed in explicit solvent (TIP3P) for the zwitterionic forms of each amino acid. The initial coordinates for the charge calculations were based on the crystal structure of a monomer of the P-selectin/PSGL-1 complex obtained from the Protein Data Bank (PDB entry code: 1G1S). The coordinates for the tyrosine sulfonate (YCS) residues were generated by replacing the phenolic oxygen atom in the YS residue with a methylene group. Molecular electrostatic potentials were computed at the HF/6-31G*//HF/6-31G* level with the Gaussian03 software package67, and restrained electrostatic potential charge fitting was performed using the RESP procedure with a restraint weight of 0.01. During charge fitting, the amino acid backbone charges were constrained to the standard values employed in ff99sb. The MD simulations were initiated with RESP charge sets computed for single conformations of each residue. The MD simulations were performed under nPT conditions (12 Å TIP3P water buffer in a cubic box, covalent bonds involving hydrogen atoms constrained using the SHAKE algorithm, a time step of 2 fs with heating from 5 to 300K over a period of 50 ps controlled by the Berendsen thermostat). Prior to MD simulation, the systems were subjected to energy minimization under nVT conditions (500 steps steepest descent (SD), followed by 24500 steps of conjugate-gradient (CG) minimization).
Prior to simulation, water molecules and sodium ions in the crystal structure were removed, and the strontium ion substituted by magnesium, because parameters for strontium ion are not available in AMBER. Missing residue E604 was added to Y605 at the N-terminus of the ligand using the LEaP module in AMBER12 with the backbone conformation copied from the E154-Y155 sequence. Sodium counter ions were added to each protein-glycopeptide complex to achieve neutrality using LEaP, prior to solvation with TIP3P water (8 Å buffer in an octahedral box). Energy minimization of the solvated complexes was performed in two-steps under nVT conditions. Initially, the positions of water molecules and counter ions were minimized (500 steps SD followed by 24500 steps CG), during which all other solute atoms were restrained (100 kcal/mol·Å2). In the second step, all restraints were removed with the exception of those on the protein backbone, and the minimization cycle was repeated. Subsequently, heating to 300°K was performed over 50 ps (nVT) with a weak restraint (10 kcal/mol·Å2) on the backbone atoms of P-Selectin only. Systems were then equilibrated at 300°K for 0.5 ns (nPT ensemble, with the Berendsen thermostat) prior to production MD, under the same conditions (covalent bonds involving hydrogen atoms constrained using SHAKE, 2 fs time step). Production MD simulations were for 250 ns performed with the GPU implementation of PMEMD from AMBER12. In all MD simulations, a non-bonded cut-off of 8 Å was applied to van der Waals interactions, with long-range electrostatics treated with the particle mesh Ewald approximation, and mixed 1-4 non-bonded scale factors applied, as recommended for systems containing both carbohydrates and proteins (SCEE=SCNB=1.0 for the oligosaccharide and SCEE=1.2 and SCNB=2.0 for the protein).65
Molecular Mechanics-Generalized Born Solvent Accessible Surface Area (MM/GBSA) calculations were carried out on 10,000 snapshots extracted evenly from the entire simulation trajectory using the single-trajectory method68 with the MMPBSA.py.MPI module. The ability to correctly predict the relative per-residue contributions to affinity is essential if the MM-GBSA calculations are to be employed in the rational design of inhibitors based on the PSGL-1 structure. There are several parameterizations of the GBSA model, none of which has been established as optimal for use in analyzing glycopeptide-protein binding. Further, estimating the affinity of PSGL-1/P-selectin interactions faces the additional challenge of quantifying the potentially highly polarizing influence of multiple charge-charge interactions. In a non-polarizable classical force field, one approach to addressing this limitation is to employ an internal dielectric constant (ε) greater than unity in the MMGBSA analysis. Again, as in the case of the GB approximation, no optimal value for ε has been reported for this type of interaction, although typical values are below 4.0 D. Lastly, entropic effects, arising from changes in conformational flexibility may be estimated separately, but may require very long MD simulations in order to achieve convergence, and are frequently omitted when computing estimates of relative affinity.69 Because of the novel nature of interactions involving YS and YCS, the suitability of five different GB implementations was examined, specifically: 1) the Hawkins, Cramer, Truhlar pairwise generalized Born model with parameters described by Tsui and Case (GBHCT model, igb=1) ; the modified GB model developed by Onufriev, Bashford and Case, with the following values for α, β and γ: GB OBC1 (α=0.8, β=0.0, γ=2.909125, igb=2) and GB OBC2 (α=1.0, β=0.8, γ=4.8, igb=5) ; the GBn models described by Mongan, Simmerling, McCammon, Case and Onufriev: GBn1, igb=7; GBn2, igb=8. In addition, six different internal dielectric (ε) values (ε = 1.0, 1.5, 2.0, 3.0, 4.0 and 5.0) were considered in the MM/GBSA calculations.
Flow cytometry
Flow cytometry was used to quantify binding inhibition of (P-, L-, E-) selectin chimeras to human and mouse leukocytes.58 Heparinized whole mouse blood was collected via cardiac puncture under the approval of the Animal Care and Use Committee of BIDMC. Normal human blood draws were collected in citrate under approval of the BIDMC Institutional Review Board. The human monocyte cell line U937 was obtained from ATCC and cultured according to supplied protocols. Cells were incubated with increasing concentrations of GSnP-6 (0 - 30 μM) and Fc chimeras of human or mouse P-, L-, and E-selectin (R&D Systems, 2 μg/mL) followed by PE-conjugated anti-Fc (1:100). The interaction of the selectins with mouse leukocytes, human leukocytes, or human U937 was analyzed by flow cytometry (BD LSR II) and quantified (FlowJo) as mean fluorescent intensity and plotted as percent inhibition. PSGL-1 specificity was examined in the presence of blocking antibodies KPL1 (anti-human PSGL-1) and 4RA10 (anti-mouse PSGL-1). Inhibition experiments were performed in triplicate. GSnP-6 inhibited binding of P- and L-selectin-IgG chimeras in a dose dependent manner. The P-selectin inhibitor KF38789 was included as a reference compound. Inhibition of E-selectin-IgG binding was not observed over the examined concentration range (Supplementary Figure 22).
In vitro flow assay
Recombinant human ICAM-1-Fc, P-selectin-Fc, and E-selectin-Fc were purchased from R&D Systems (Lot #: DLA0914011, FDA0913111, and BGU0413091 respectively). Protein A/G was purchased from Fischer Scientific (PI-21186). Neutrophils and monocytes were isolated from freshly collected human blood from healthy donors consented through an approved UC Davis institutional review board protocol. Whole blood was layered over neutrophil or mononuclear cell separation media, Polymophoprep, an Axis Shield formulation purchased from Cosmo Bio USA. After centrifugation myeloid cells were extracted and washed with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffered salt solution. Coverslips were prepared as follows: 25 mm diameter, #1.5 glass coverslips were Piranha etched to remove organic molecules and to deposit hydroxyl group molecules on the surface. The etched coverslips were submerged in acetone with 1% 3-aminopropyltriethoxysilane (APTES) to add aminosilane groups. Recombinant human ICAM-1-Fc along with either P-selectin-Fc or E-selectin-Fc were absorbed at 5 μg/mL for 1 hour prior to the experiment. Human neutrophils were suspended at a concentration of 106 cells/mL in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffered salt solution and allowed to incubate for 10 minutes with PBS (vehicle control), 2 μM, 3 μM, 4 μM, 5 μM, 6 μM and 30 μM concentrations of GSnP-6. Treated cells were perfused through a microfluidic flow chamber at a calculated shear stress of 2 dynes/cm2 and cell motion recorded at three to five fields of view at 7.5 frames/second for 2 minutes along the centerline of the channel using a Matrox Meteor II and ImagePro Plus software to analyze cell kinetics. To further observe neutrophil behavior, buffer was infused and images were collected at two additional locations along the channel. Rolling and arrested neutrophils were determined by tracking the cell centroid over 10 second increments utilizing MTrack2 software plugin for ImageJ. Neutrophils moving at a velocity of less than 1 μm/s were considered to be arrested.
Intravital microscopy
Surgical preparation of the mouse cremaster was performed as previously described. All experiments were performed in the BIDMC Center for Hemostasis and Thrombosis Research Core and were approved by the Animal Care and Use Committee of the Beth Israel Deaconess Medical Center. Mice were anesthetized with an intraperitoneal injection of ketamine HCl (125 mg/kg), xylazine (12.5 mg/kg), and atropine (0.25 mg/ml) and placed on a 37°C surgical blanket. The jugular vein was cannulated with PE 10 tubing to allow introduction of reagents, including GSnP-6 (4 μmol/kg in saline), saline vehicle control, or 0.5 μm Dragon Green microspheres (Bangs Laboratories, Inc) for the measurement of centerline velocity. The trachea was intubated with PE90 to facilitate breathing. The cremaster muscle was exteriorized, pinned to the stage, and superfused with thermocontrolled bicarbonate buffered saline equilibrated with 5% CO2 in N2. The surgical procedure was accomplished within 10 minutes. Microvessel data were obtained using an Olympus AX microscope with 60x or 40x water immersion objectives recorded with a Hamamatsu C9300-201/Gen III videoscope image intensifier interface. Coordinated image acquisition and offline data analyses were carried out using SlideBook software (Intelligent Imaging Innovations). At the termination of the experiment, blood was drawn via cardiac puncture and analyzed for complete blood count. Venules (30 - 40 μm in diameter) were observed 15 to 25 minutes after surgical stimulation of tissue to study P-selectin-dependent rolling. Vessel diameter was measured using SlideBook ruler. Centerline blood flow velocity (Vcl) was determined by measuring frame-to-frame displacement of single fluorescent microspheres in the center of the vessel wall. Volumetric blood flow rate (Q) was calculated using the equation Q = (Vcl)(0.625)(Acs), where Acs is the cross-sectional area of the vessel (πr2) and 0.625 is an empirical correction factor. Wall shear rate (γw) was calculated as γw = 2.12 × [(8 × 0.625 × Vcl)/Dv)], where Dv is the vessel diameter. Hemodynamic parameters are reported in Supplementary Table 4. Recordings of each vessel were analyzed in brightfield for 30 to 60 s and leukocyte rolling flux was characterized as the number of leukocytes passing a plane perpendicular to the vessel axis (Supplementary Figure 24). Total leukocyte flux was determined as a product of systemic leukocyte concentration (cells/μL) and volumetric blood flow rate (μL/s). Leukocyte rolling was characterized as the rolling flux fraction, which is the number of rolling leukocytes reported as a percentage of total leukocyte flux. Rolling velocity (μm/s) was measured by tracking individual leukocyte translation over 2 sec.
Assessment of platelet-leukocyte aggregation in vitro
Platelet-leukocyte aggregates were quantified in whole blood using dual-label flow cytometry.55 Anti-coagulated human or mouse blood was incubated with 40 μM GSnP-6 at room temperature and stimulated with thrombin receptor-activating peptide (human PAR1-activating peptide 40 μM; mouse PAR4-activating peptide 200 μM) to induce platelet P-selectin expression. Platelet-leukocyte aggregates were quantified by two-color flow cytometry by incubating human samples with anti-CD42a-PE and anti-CD45-APC and mouse samples with anti-CD41-PE and anti-CD45-APC. CD45+ monocyte and neutrophil populations were discerned through characteristic side scatter and quantified as % platelet positive in saline control, GSnP-6 (40 μM), or anti-CD62P (human KPL-1 (5 μg/mL); mouse RB40.34 (5 μg/mL) treated samples.
Assessment of platelet-leukocyte interactions and platelet aggregate formation in vivo
TNF-α was administered intrascrotally (0.5 μg) to male mice (6-8 wks old, C57BL/6) 3 hr prior to surgical preparation and imaging. Surgical preparation of the cremaster was performed, as described above. Agents were administered through the jugular cannula just prior to cremaster exposure and imaging, including saline vehicle control, GSnP-4 (4 μmol/kg), anti-CD62P blocking antibody (clone RB40.34, 3 mg/kg), anti-platelet Dylight 649 anti-CD42b (Emfret, 1 μL/gm), Alexa Fluor 488 anti-Gr-1 (0.5 μg/gm). Microvessel data were obtained using an Olympus AX microscope with a 60x water immersion objective recorded with a Hamamatsu C9300-201/Gen III videoscope image intensifier interface. Coordinated image acquisition and offline data analyses were carried out using SlideBook software (Intelligent Imaging Innovations). For each treatment condition, platelet accumulation was characterized as median integrated fluorescence plotted during a 2 minute time interval from 5 mice (8-10 venules/mouse). The platelet signal was quantified as area under the curve for each individual capture, plotted against time, and normalized to vessel surface area. Platelet aggregation and platelet-PMN binding is observed in saline control vessels while limited platelet signal is captured in GSnP-6 and anti-CD62P treatment animals (Supplementary Figure 25A-D). Characterization of leukocyteendothelial interactions at 3 hours post-TNFα activation revealed minimal rolling flux and similar levels of adherent leukocytes between all groups (Supplementary Figure 25E-F).
Supplementary Material
Acknowledgements
The authors acknowledge support from National Institutes of Health (DK069275, HL106018, GM103694, HL60963, AI047294, HL085607, GM094919, GM103390), the Science Foundation of Ireland (08/IN.1/B2070) and the European Research Development Fund. VRK would like to thank Dr. Sunia Trauger (Harvard University, FAS-Center for Systems Biology) and Dr. Jim Lee (DFCI, Molecular Biology Core Facility) for help with mass spectrometry. CH would like to thank Glenn Merrill-Skoloff (BIDMC Center for Hemostasis and Thrombosis Research Core) for assistance with intravital microscopy.
Footnotes
Author contributions
VRK, RDC and ELC jointly conceived the project, and directed the chemistry and biology, respectively. MYR was responsible for execution of the chemistry. YY and XLS are responsible for the execution of enzymatic steps and binding affinity studies. Molecular modeling was directed by RJW and carried out by XW. CH, ED, and DHP carried out cellular and in vivo studies. VM and SIS carried out in vitro shear studies. All authors contributed to the preparation of the manuscript.
Additional information
Supplementary information and chemical compound characterization data sets are available in the online version of the paper. Reprints and permissions information is available online at www.nature.com/reprints. Correspondence and requests for materials should be addressed to EC.
Competing financial interests
A patent based on these investigations has been filed by VRK, MYRS, RDC, and ELC. The remaining authors declare no conflict of interest.
References
- 1.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nature reviews. Immunology. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 2.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 3.Zarbock A, Ley K, McEver RP, Hidalgo A. Leukocyte ligands for endothelial selectins: specialized glycoconjugates that mediate rolling and signaling under flow. Blood. 2011;118:6743–6751. doi: 10.1182/blood-2011-07-343566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palabrica T, et al. Leukocyte accumulation promoting fibrin deposition is mediated in vivo by P-selectin on adherent platelets. Nature. 1992;359:848–851. doi: 10.1038/359848a0. [DOI] [PubMed] [Google Scholar]
- 5.Mayadas TN, Johnson RC, Rayburn H, Hynes RO, Wagner DD. Leukocyte rolling and extravasation are severely compromised in P selectin-deficient mice. Cell. 1993;74:541–554. doi: 10.1016/0092-8674(93)80055-j. [DOI] [PubMed] [Google Scholar]
- 6.Weller A, Isenmann S, Vestweber D. Cloning of the mouse endothelial selectins. Expression of both E- and P-selectin is inducible by tumor necrosis factor alpha. J Biol Chem. 1992;267:15176–15183. [PubMed] [Google Scholar]
- 7.Girard JP, Moussion C, Forster R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol. 2012;12:762–773. doi: 10.1038/nri3298. [DOI] [PubMed] [Google Scholar]
- 8.Carlow DA, et al. PSGL-1 function in immunity and steady state homeostasis. Immunol Rev. 2009;230:75–96. doi: 10.1111/j.1600-065X.2009.00797.x. [DOI] [PubMed] [Google Scholar]
- 9.Mehta P, Cummings RD, McEver RP. Affinity and kinetic analysis of P-selectin binding to P-selectin glycoprotein ligand-1. J Biol Chem. 1998;273:32506–32513. doi: 10.1074/jbc.273.49.32506. [DOI] [PubMed] [Google Scholar]
- 10.McEver RP, Cummings RD. Role of PSGL-1 binding to selectins in leukocyte recruitment. J Clin Invest. 1997;100:485–491. doi: 10.1172/JCI119556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HB, et al. P-selectin primes leukocyte integrin activation during inflammation. Nat Immunol. 2007;8:882–892. doi: 10.1038/ni1491. [DOI] [PubMed] [Google Scholar]
- 12.Sreeramkumar V, et al. Neutrophils scan for activated platelets to initiate inflammation. Science. 2014;346:1234–1238. doi: 10.1126/science.1256478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Costa Martins P, et al. Platelet-monocyte complexes support monocyte adhesion to endothelium by enhancing secondary tethering and cluster formation. Arterioscler Thromb Vasc Biol. 2004;24:193–199. doi: 10.1161/01.ATV.0000106320.40933.E5. [DOI] [PubMed] [Google Scholar]
- 14.Ma YQ, Plow EF, Geng JG. P-selectin binding to P-selectin glycoprotein ligand-1 induces an intermediate state of alphaMbeta2 activation and acts cooperatively with extracellular stimuli to support maximal adhesion of human neutrophils. Blood. 2004;104:2549–2556. doi: 10.1182/blood-2004-03-1108. [DOI] [PubMed] [Google Scholar]
- 15.Fox R, et al. PSGL-1 and mTOR regulate translation of ROCK-1 and physiological functions of macrophages. EMBO J. 2007;26:505–515. doi: 10.1038/sj.emboj.7601522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidari KI, Weyrich AS, Zimmerman GA, McEver RP. Engagement of P-selectin glycoprotein ligand-1 enhances tyrosine phosphorylation and activates mitogen-activated protein kinases in human neutrophils. J Biol Chem. 1997;272:28750–28756. doi: 10.1074/jbc.272.45.28750. [DOI] [PubMed] [Google Scholar]
- 17.Dong ZM, Brown AA, Wagner DD. Prominent role of P-selectin in the development of advanced atherosclerosis in ApoE-deficient mice. Circulation. 2000;101:2290–2295. doi: 10.1161/01.cir.101.19.2290. [DOI] [PubMed] [Google Scholar]
- 18.Sato C, et al. P-selectin glycoprotein ligand-1 deficiency is protective against obesity-related insulin resistance. Diabetes. 2011;60:189–199. doi: 10.2337/db09-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Bruhl ML, et al. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J Exp Med. 2012;209:819–835. doi: 10.1084/jem.20112322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baert F, et al. Influence of immunogenicity on the long-term efficacy of infliximab in Crohn's disease. New Engl J Med. 2003;348:601–608. doi: 10.1056/NEJMoa020888. [DOI] [PubMed] [Google Scholar]
- 21.Chang J, et al. GMI-1070, a novel pan-selectin antagonist, reverses acute vascular occlusions in sickle cell mice. Blood. 2010;116:1779–1786. doi: 10.1182/blood-2009-12-260513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Japp AG CR, Tattersall L, Lang NN, Meng X, Weisel K, Katz A, Burt D, Fox KA, Feuerstein GZ, Connolly TM, Newby DE. Effect of PSI-697, a novel P-selectin inhibitor, on platelet-monocyte aggregate formation in humans. J Am Heart Assoc. 2013;2:1–6. doi: 10.1161/JAHA.112.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beeh KM, et al. Bimosiamose, an inhaled small-molecule pan-selectin antagonist, attenuates late asthmatic reactions following allergen challenge in mild asthmatics: A randomized, double-blind, placebo-controlled clinical cross-over-trial. Pulm Pharmacol Ther. 2006;19:233–241. doi: 10.1016/j.pupt.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Kranich R, et al. Rational design of novel, potent small molecule pan-selectin antagonists. J Med Chem. 2007;50:1101–1115. doi: 10.1021/jm060536g. [DOI] [PubMed] [Google Scholar]
- 25.Huang A, et al. Discovery of 2-[1-(4-Chlorophenyl)cyclopropyl]-3-hydroxy-8-(trifluoromethyl)quinoline-4-carboxylic acid (PSI-421), a P-selectin Inhibitor with improved pharmacokinetic properties and oral efficacy in models of vascular injury. J Med Chem. 2010;53:6003–6017. doi: 10.1021/jm9013696. [DOI] [PubMed] [Google Scholar]
- 26.Leppänen A, et al. A novel glycosulfopeptide binds to P-selectin and inhibits leukocyte adhesion to P-selectin. J Biol Chem. 1999;274:24838–24848. doi: 10.1074/jbc.274.35.24838. [DOI] [PubMed] [Google Scholar]
- 27.Leppänen A, White SP, Helin J, McEver RP, Cummings RD. Binding of glycosulfopeptides to P-selectin requires stereospecific contributions of individual tyrosine sulfate and sugar residues. J Biol Chem. 2000;275:39569–39578. doi: 10.1074/jbc.M005005200. [DOI] [PubMed] [Google Scholar]
- 28.Leppänen A, Yago T, I. OV, McEver RP, Cummings RD. Model glycosulfopeptides from P-selectin glycoprotein ligand-1 require tyrosine sulfation and a core 2-branched O-glycan to bind to L-selectin. J Biol Chem. 2003;278:26391–26400. doi: 10.1074/jbc.M303551200. [DOI] [PubMed] [Google Scholar]
- 29.Koeller KM, Smith MEB, Huang R-F, Wong C-H. Chemoenzymatic synthesis of a PSGL-1 N-terminal glycopeptide containing tyrosine sulfate and α-O-linked sialyl Lewis X. J Am Chem Soc. 2000;122:4241–4242. [Google Scholar]
- 30.Huang K-T, et al. Multi-enzyme one-pot strategy for the synthesis of sialyl Lewis X-containing PSGL-1 glycopeptide. Carbohydr Res. 2006;341:2151–2155. doi: 10.1016/j.carres.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 31.Baumann K, Kowalczyk D, Kunz H. Total synthesis of the glycopeptide recognition domain of the P-selectin glycoprotein ligand 1. Angew Chem Int Ed. 2008;47:3445–3449. doi: 10.1002/anie.200705762. [DOI] [PubMed] [Google Scholar]
- 32.Vohra Y, Buskas T, Boons G-J. Rapid assembly of oligosaccharides: A highly convergent strategy for the assembly of a glycosylated amino acid derived from PSGL-1. J Org Chem. 2009;74:6064–6071. doi: 10.1021/jo901135k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnamurthy VR, et al. Synthesis of an Fmoc-threonine bearing core-2 glycan: A building block for PSGL-1 via Fmoc-assisted solid-phase peptide synthesis. Carbohydr Res. 2010;345:1541–1547. doi: 10.1016/j.carres.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plattner C, Höfener M, Sewald N. One-pot azidochlorination of glycals. Org Lett. 2011;13:545–547. doi: 10.1021/ol102750h. [DOI] [PubMed] [Google Scholar]
- 35.Yagami T, Kitagawa K, Aida C, Fujiwara H, Futaki S. Stabilization of a tyrosine O-sulfate residue by a cationic functional group: Formation of a conjugate acid-base pair. J Pept Res. 2000;56:239–249. doi: 10.1034/j.1399-3011.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- 36.Baumann K, et al. Sulfated and non-sulfated glycopeptide recognition domains of P-selectin glycoprotein ligand 1 and their binding to P- and E-selectin. Angew Chem Int Ed. 2009;48:3174–3178. doi: 10.1002/anie.200805999. [DOI] [PubMed] [Google Scholar]
- 37.Bunschoten A, et al. A general sequence independent solid phase method for the site specific synthesis of multiple sulfated-tyrosine containing peptides. Chem Commun. 2009:2999–3001. doi: 10.1039/b823425f. [DOI] [PubMed] [Google Scholar]
- 38.Simpson LS, Widlanski TS. A comprehensive approach to the synthesis of sulfate esters. J Am Chem Soc. 2006;128:1605–1610. doi: 10.1021/ja056086j. [DOI] [PubMed] [Google Scholar]
- 39.Roosenburg S, et al. Stabilized (111)in-labeled sCCK8 analogues for targeting CCK2-receptor positive tumors: Synthesis and evaluation. Bioconjug Chem. 2010;21:663–670. doi: 10.1021/bc900465y. [DOI] [PubMed] [Google Scholar]
- 40.Lam SN, Acharya P, Wyatt R, Kwong PD, Bewley CA. Tyrosine-sulfate isosteres of CCR5 N-terminus as tools for studying HIV-1 entry. Bioorg Med Chem. 2008;16:10113–10120. doi: 10.1016/j.bmc.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acharya P, et al. Structure-based identification and neutralization mechanism of tyrosine sulfate mimetics that inhibit HIV-1 entry. ACS Chem Biol. 2011;6:1069–1077. doi: 10.1021/cb200068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yagami T, Shiwa S, Futaki S, Kitagawa K. Evaluation of the final deprotection system for the solid-phase synthesis of Tyr(SO3H)-containing peptides with 9-fluorenylmethyloxycarbonyl (Fmoc)-strategy and its application to the synthesis of cholecystokinin (CCK)-12. Chem Pharm Bull. 1993;41:376–380. doi: 10.1248/cpb.41.376. [DOI] [PubMed] [Google Scholar]
- 43.Balsved D, Bundgaard JR, Sen JW. Stability of tyrosine sulfate in acidic solutions. Anal Biochem. 2007;363:70–76. doi: 10.1016/j.ab.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Koeller KM, Smith MEB, Wong C-H. Tyrosine sulfation on a PSGL-1 glycopeptide influences the reactivity of glycosyltransferases responsible for synthesis of the attached O-glycan. J Am Chem Soc. 2000;122:742–743. [Google Scholar]
- 45.Cao TM, Takatani T, King MR. Effect of extracellular pH on selectin adhesion: Theory and experiment. Biophys J. 2013;104:292–299. doi: 10.1016/j.bpj.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massova I, Kollman PA. Computational alanine scanning to probe protein–protein interactions: A novel approach to evaluate binding free energies. J Am Chem Soc. 1999;121:8133–8143. [Google Scholar]
- 48.Wang JM, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 49.Wong S, Amaro RE, McCammon JA. MM-PBSA captures key role of intercalating water molecules at a protein-protein interface. J Chem Theory Comput. 2009;5:422–429. doi: 10.1021/ct8003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon SI, Hu Y, Vestweber D, Smith CW. Neutrophil tethering on E-selectin activates beta 2 integrin binding to ICAM-1 through a mitogen-activated protein kinase signal transduction pathway. J Immunol. 2000;164:4348–4358. doi: 10.4049/jimmunol.164.8.4348. [DOI] [PubMed] [Google Scholar]
- 51.Yago T, et al. E-selectin engages PSGL-1 and CD44 through a common signaling pathway to induce integrin alphaLbeta2-mediated slow leukocyte rolling. Blood. 2010;116:485–494. doi: 10.1182/blood-2009-12-259556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altman SM, Dixit N, Simon SI. Detection of bidirectional signaling during integrin activation and neutrophil adhesion. Methods Mol Biol. 2014;1124:235–248. doi: 10.1007/978-1-62703-845-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridger VC, Hellewell PG, Norman KE. L- and P-selectins collaborate to support leukocyte rolling in vivo when high-affinity P-selectin-P-selectin glycoprotein ligand-1 interaction is inhibited. Am J Pathol. 2005;166:945–952. doi: 10.1016/S0002-9440(10)62314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark SR, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13:463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 55.Kling D, et al. Pharmacological control of platelet-leukocyte interactions by the human anti-P-selectin antibody inclacumab - Preclinical and clinical studies. Thromb Res. 2013;131:401–410. doi: 10.1016/j.thromres.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 56.Li J, et al. Neutrophil AKT2 regulates heterotypic cell-cell interactions during vascular inflammation. J Clin Invest. 2014;124:1483–1496. doi: 10.1172/JCI72305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulligan MS, et al. Neutrophil-dependent acute lung injury. Requirement for P-selectin (GMP-140). J Clin Invest. 1992;90:1600–1607. doi: 10.1172/JCI116029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.An G, et al. P-selectin glycoprotein ligand-1 is highly expressed on Ly-6Chi monocytes and a major determinant for Ly-6Chi monocyte recruitment to sites of atherosclerosis in mice. Circulation. 2008;117:3227–3237. doi: 10.1161/CIRCULATIONAHA.108.771048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falati S, et al. Accumulation of tissue factor into developing thrombi in vivo is dependent upon microparticle P-selectin glycoprotein ligand 1 and platelet P-selectin. J Exp Med. 2003;197:1585–1598. doi: 10.1084/jem.20021868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laubli H, Borsig L. Selectins promote tumor metastasis. Sem Cancer Biol. 2010;20:169–177. doi: 10.1016/j.semcancer.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 61.Labelle M, Beguma S, Hynes RO. Platelets guide the formation of early metastatic niches. Proc Natl Acad Sci U S A. 2014:E3053–E3061. doi: 10.1073/pnas.1411082111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez JA, et al. The alpha and beta chains of human platelet glycoprotein Ib are both transmembrane proteins containing a leucine-rich amino acid sequence. Proc Natl Acad Sci U S A. 1988;85:2135–2139. doi: 10.1073/pnas.85.7.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fieger CB, Sassetti CM, Rosen SD. Endoglycan, a member of the CD34 family, functions as an L-selectin ligand through modification with tyrosine sulfation and sialyl Lewis x. J Biol Chem. 2003;278:27390–27398. doi: 10.1074/jbc.M304204200. [DOI] [PubMed] [Google Scholar]
- 64.Song X, et al. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;30:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirschner KN, et al. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frisch MJ, et al. Gaussian 03. Gaussian, Inc.; Wallingford CT: 2004. [Google Scholar]
- 68.Kollman PA, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 69.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem. 2011;32:866–877. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.