Abstract
Background
Cytochrome P450s (CYPs) encode one of the most diverse enzyme superfamily in nature. They catalyze oxidative reactions of endogenous molecules and exogenous chemicals.
Methods
We identified CYPs genes through in silico analysis using EST, RNA-Seq and genome databases of channel catfish. Phylogenetic analyses and conserved syntenic analyses were conducted to determine their identities and orthologies. Meta-analysis of RNA-Seq databases was conducted to analyze expression profile of CYP genes following bacterial infection.
Results
A full set of 61 CYP genes were identified and characterized in channel catfish. Phylogenetic tree and conserved synteny provided strong evidence of their identities and orthorlogy. Lineage-specific gene duplication was evident in a number of clans in channel catfish. CYP46A1 is missing in the catfish genome as observed with syntenic analysis and RT-PCR analysis. Thirty CYPs were found up- or down-regulated in liver, while seven and eight CYPs were observed regulated in intestine and gill following bacterial infection.
Conclusion
We systematically identified and characterized a full set of 61 CYP genes in channel catfish and studied their expression profiles after bacterial infection. Strikingly large numbers of CYP genes appear to be involved in the bacterial defense processes.
General significance
This work provides an example to systematically study CYP genes in non-model species. Moreover, it provides a basis for further toxicological and physiological studies in channel catfish.
Keywords: Cytochrome P450, CYP, catfish, genome, immunity, bacterial infection
1. Introduction
Cytochrome P450s (CYPs), named for their characteristic spectral property of Soret absorption peak at 450 nm when binding with carbon monoxide, constitute a widespread and highly diverse heme-thiolate enzyme superfamily (1-4). These enzymes catalyze the oxidative reactions, which are not only involved in synthesizing or metabolizing of endogenous molecules, such as arachidonic acid, eicosanoid, retinoic acid, steroid, vitamin D3, bile acid, biogenic amine, prostaglandin and cholesterol, but also engaged in the detoxification processes of exogenous substrates, including pharmaceuticals, and foreign chemicals (5-10). Moreover, it has been demonstrated that the expression level of many CYPs is regulated during the host defense responses in liver, as well as in extrahepatic tissues such as kidney and brain in mice or rats (11-21).
With the application of the next-generation sequencing, rapid progress has been made for expanding CYP family from model species to non-model species. At present, thousands of CYPs have been identified from all domains of life including Animalia, Plantae, Fungi, Protista, Archaea, Bacteria and even virus (22-33). All these CYPs are classified into clans, families and subfamilies based on sequence similarities (40% amino acid sequence identity rule for membership in a family and 55% amino acid sequence identity rule for membership in a subfamily), phylogenetic relationships and syntenic relationships (23, 34-36). According to these rules, vertebrate CYPs could be clustered into 19 families within 10 clans-including CYP clan 2 (include CYP1, CYP2, CYP17 and CYP21 family), CYP clan 3 (include CYP3 and CYP5 family), CYP clan 4 (include CYP4 family), CYP clan 7 (include CYP7, CYP8 and CYP39 family), CYP clan 19 (include CYP19 family), CYP clan 20 (include CYP20 family), CYP clan 26 (include CYP16 and CYP26 family), CYP clan 46 (include CYP46 family), CYP clan 51 (include CYP51 family) and mitochondrial clan (include CYP11, CYP24 and CYP27 family) (22, 23, 35). Generally, families among CYP5-51 are involved in endogenous metabolism, whereas CYP1-3 families and several subfamilies of CYP4 play an important role in detoxification processes.
Currently, all 19 vertebrate CYP families have been identified in teleost fish, including rainbow trout (Oncorhynchus mykiss) (37-44), Japanese pufferfish (Fugu rubripes) (45, 46), zebrafish (Danio rario) (47-56), Atlantic salmon (Salmo salar) (57-60), European seabass (Dicentrarchus labrax) (61-63), largemouth bass (Micropterus salmoides) (64), medaka (Oryzias latipes) (65-71), common carp (Cyprinus carpio) (72-74), mummichog (Fundulus heteroclitus) (75-78), three-spined stickleback (Gasterosteus aculeatus) (70), gilthead seabream (Sparus aurata) (79), fathead minnow (Pimephales promelas) (80, 81), half-smooth tongue sole (Cynoglossus semilaevis) (82, 83). By far, with the accomplishment of whole genome sequence (84, 85), CYP genes in Japanese pufferfish (45) and zebrafish (55) have been analyzed at the whole genome level, revealing 54 CYPs (update to 61 CYPs, Table 3) and 94 CYPs are presented in the Japanese pufferfish and zebrafish, respectively.
Table 3.
Gene number variation of CYPs among human, chicken, frogs, Japanese pufferfish, zebrafish and catfish in diverse subfamilies. Shading indicates lineage-specific CYP subfamilies.
| CYP450 subfamily |
gene # Human |
gene # Mouse |
gene # Chicken |
gene # Western clawed frog |
gene # African clawed frog |
gene # Japanese pufferfish |
gene # zebrafish |
gene # catfish |
|---|---|---|---|---|---|---|---|---|
| 1A | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 1 |
| 1B | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1C | 1 | 1 | 1 | 2 | 2 | 1 | ||
| 1D | 1 | 1 | 1 | |||||
| 2A | 3 | 4 | ||||||
| 2B | 1 | 5 | ||||||
| 2C | 4 | 15 | 1 | |||||
| 2D | 1 | 9 | 1 | 7 | 5 | |||
| 2E | 1 | 1 | ||||||
| 2F | 1 | 1 | 1 | |||||
| 2G | 1* | |||||||
| 2H | 2 | |||||||
| 2I | ||||||||
| 2J | 1 | 7 | 5 | 1 | ||||
| 2K | 3 | 11 | 2 | |||||
| 2M | 1 | |||||||
| 2N | 3 | 1 | ||||||
| 2P | 1 | 6 | ||||||
| 2Q | 8 | 5 | ||||||
| 2R | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 2S | 1 | 1 | ||||||
| 2T | 1 | |||||||
| 2U | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 2V | 1 | |||||||
| 2W | 1 | 1 | 2 | |||||
| 2X | 2 | 7 | 4 | |||||
| 2Y | 2 | 2 | 2 | |||||
| 2Z | 2 | |||||||
| 2AA | 12 | 1 | ||||||
| 2AB | 1 | 5 | 2 | 3 | ||||
| 2AC | 3 | 12 | 6 | |||||
| 2AD | 1 | 3 | 4 | |||||
| 2AE | 2 | |||||||
| 2AM | 6 | 4 | ||||||
| 2AN | 5 | 5 | ||||||
| 2AP | 1 | |||||||
| 2AQ | 2 | 2 | ||||||
| 2AR | 1 | |||||||
| 2AS | 1 | |||||||
| 2AT | 1 | 1 | ||||||
| 3A | 4 | 8 | 2 | 6 | 4 | 3 | 1 | 4 |
| 3B | 2 | |||||||
| 3C | 4 | 1 | ||||||
| 3D | 1 | |||||||
| 4A | 2 | 7 | ||||||
| 4B | 1 | 1 | 1 | |||||
| 4F | 6 | 9 | 1 | 3 | 2 | 1 | 1 | 2 |
| 4T | 4 | 10 | 1 | 1 | 2 | |||
| 4V | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 2 |
| 4X | 1 | 1 | ||||||
| 4Z | 1 | |||||||
| 5A | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| 7A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 7B | 1 | 1 | 1 | 1 | 1 | |||
| 7C | 1 | 1 | 1 | |||||
| 7D | 1 | 1 | ||||||
| 8A | 1 | 1 | 1 | 2 | 1 | 2 | ||
| 8B | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 3 |
| 11A | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 |
| 11B | 2 | 2 | 1 | 1 | ||||
| 11C | 1 | 1 | 1 | |||||
| 16A | 1 | |||||||
| 17A | 1 | 1 | 1 | 1 | 1 | 4 | 2 | 2 |
| 19A | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 |
| 20A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 21A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 24A | 1 | 1 | 1 | 1 | 1 | 1 | ||
| 26A | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| 26B | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 26C | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 27A | 1 | 1 | 1 | 4 | 3 | 5 | 5 | 7 |
| 27B | 1 | 1 | 1 | 1 | 1 | 1 | ||
| 27C | 1 | 1 | 1 | 2 | 1 | 1 | 1 | |
| 39A | 1 | 1 | 1 | 1 | 1 | 1 | ||
| 46A | 1 | 1 | 1 | 6 | 3 | 1 | 4 | |
| 51A | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| Total | 57 | 100 | 48 | 95 | 80 | 61 | 94 | 61 |
CYP2G subfamily is not mammal specific, because it is also found in lizard (belong to tetrapod).
Several channel catfish CYPs have been characterized previously including CYP1B1, CYP2X1, CYP11A1, CYP17A1, CYP19A1 and CYP19A2 (86-89). In recent years, following the development of genomic resources of channel catfish (90), particularly the ESTs (91, 92), transcriptome sequences generated by RNA-Seq (93-95) and the draft whole genome sequence, the systematic analysis of CYP genes in channel catfish genome becomes feasible. Channel catfish (Ictalurus punctatus) is the leading aquaculture species in the United States. Its sustainable production is threatened by the widespread ESC and columnaris disease outbreaks caused by the two pathogens: E. ictaluri and F. columnare. Here we report the identification of a full set of CYP genes, their phylogenetic analysis and syntenic analysis in the channel catfish, and their involvement in response to bacterial infections with E. ictaluri and F. columnare.
2. Materials and Methods
2.1. CYP homologous genes collection and database mining
In order to identify the complete set of CYP genes in channel catfish, we collected all the fish CYPs (Anguilla japonica, Carassius auratus, Carassius carassius, Cyprinus carpio, Cynoglossus semilaevis, Danio rario, Dicentrarchus labrax, Fugu rubripes, Fundulus heteroclitus, Gasterosteus aculeatus, Gobiocypris rarus, Micropterus salmoides, Oncorhynchus mykiss, Oreochromis niloticus, Oryzias latipes, Pimephales promelas, Salmo sala, Sparus aurata, Squalus acanthias Stenotomus chrysops) from the database of NCBI (http://www.ncbi.nlm.nih.gov), Ensembl (http://www.ensembl.org) and CYP homepage (http://drnelson.uthsc.edu/CytochromeP450.html) (22). In addition, other CYP homologous genes from human, mouse, chicken and frog (Xenopus laevis and Xenopus tropicalis) were also collected.
More than 1000 collected CYPs were first used as query sequences to search against channel catfish EST and RNA-Seq databases. In order to pull all potential CYP genes, the cutoff value was set at the level of 1e-5 such that conserved transcripts are captured initially for further analysis. The retrieved sequences were then translated using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Further, the predicted ORFs were verified by BLASTP against NCBI non-redundant (Nr) protein sequence database. All identified channel catfish CYP transcripts and all other query sequences were utilized to search channel catfish draft genome sequence using TBLASTN program. The following steps are the same with above with the exception of applying FGENESH from SoftBerry (http://linux1.softberry.com/berry.phtml) to predict exons and amino acid sequences using genomic sequences.
2.2. Phylogenetic and syntenic analyses
In order to identify channel catfish CYPs, all the amino acids from channel catfish and other species were used to conduct phylogenetic analyses. We constructed separate phylogenetic trees for different CYP clans since the number of sequences under study was too many to fit a page. Sequence alignment was performed using MUSCLE (Multiple Sequence Comparison by Log-Expectation) (96). JTT (Jones-Taylor-Thornton) and gamma distributed rate with invariant sites (G+I) model was proposed by ProtTest3 (97). Maximum likelihood phylogenetic trees were built with MEGA 5.10 with 1000 bootstrap replications (98).
Shared synteny was searched by examining the conserved co-localization of neighboring genes on scaffold (unpublished data) of channel catfish and zebrafish. Generally, in order to obtain the location information of these channel catfish CYPs on scaffolds, all identified CYP transcripts together with query sequences from other species were used as queries to blast against channel catfish draft genome sequence. Neighboring genes of the channel catfish CYPs were predicted by FGENESH (http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind). Neighboring genes of zebrafish were identified from Zv9 database in ENSEMBL.
2.3. Degenerate primers design and Touchdown RT-PCR for CYP46
CYP46 sequences from human, mouse, rat, pig, cow, frog, Japanese pufferfish, green spotted pufferfish, and zebrafish were first aligned using software MUSCLE. The degenerate primers were designed by CODEHOP (Consensus-Degenerate Hybrid Oligonucleotide Primers) (99) using the alignment file generated as the input file. Parameters were selected as maximum core degeneracy: 64, target clamp temperature: 60°C, Genetic code: standard, and Codon usage: Ictalurus punctatus. Two sets of degerate primers were chosen to conduct the reaction with the estimated size of production around 400 bp and 200 bp, respectively (Table 1).
Table 1.
Degenerate primers used for CYP46A1 amplification
| primer name | forward primer | reverse primer |
| primer 1 | agagggcccagacggarrtngayga | cggggtcccagggagaanggrwarta |
| primer 2 | gaccttcttcatcgccggncangarac | agccgcagggtctccttnarnacytg |
One healthy channel catfish (Marion strain) was sacrificed in this study. Following manufacturer’s protocol, total RNA (RNeasy Mini Kit, Qiagen, USA) was extracted from brain. The extracted RNA was quantified using UV-spectrophotometer and then reverse-transcribed using iScript™ cDNA Synthesis Kit (Bio-Rad, USA). Degenerated primers were applied to amplify brain cDNA. PCR amplification was carried out using KAPA HiFi PCR kit (Kapa Biosystems, USA). The 25 μl PCR reaction mixture contained 5.0 μl of 5×KAPA HiFi Fidelity Buffer, 1 μl of KAPA dNTP Mix (10 mM), 0.5 μl of KAPA HiFi DNA Polymerase, 1 μl (10 pmol/μl) of each primers, 1 μl of template, and 15.5 μl PCR-grade water. Touchdown PCR was carried out on a Bio-Rad PCR system using the cycling conditions: denaturation, 95°C/4 min, 10 cycles of 95°C /30s, 65°C /30s (each cycle decrease 1°C), and 72°C/30s, 20 cycles of 95°C/30s, 55°C/30s and 72°C /30s, then, 72°C 10 min. PCR products were resolved on 1.5% agarose gel.
2.4. Meta-analyses of RNA-Seq datasets
Illumina-based RNA-Seq reads were retrieved from three RNA-Seq datasets: liver samples from catfish experimentally challenged with E. ictaluri (3 day and 14 day post infection) (SRA accession number SRP028159) (93), intestine samples from catfish challenged with E. ictaluri (3 h, 24 h and 3 day post infection) (SRA accession number SRP009069) (94) and gill samples from catfish challenged with F. columnare (4 h, 24 h and 48 h post infection) (SRA accession number SRP012586) (95). Trimmed high quality RNA-seq reads were mapped onto the deduced channel catfish CYPs reference assembly using CLC Genomics Workbench software (version 5.5.2; CLC bio, Aarhus, Denmark). Mapping parameters were set as ≥ 95% of the reads in perfect alignment and ≤ 2 mismatches. After the total mapped reads number for each transcript was determined, normalization was conducted in order to determine RPKM (Reads Per Kilobase of exon per Million mapped reads). The proportions-based Kal’s test was performed to identify the differently expressed genes comparing with control sample and fold changes were calculated. Transcripts with absolute fold change value ≥ 2, p-value ≤ 0.05 and total read number ≥ 5 were included in the analyses as differently expressed genes.
3. Results
3.1. Identification of CYP genes in channel catfish
CYP genes collected from various species (as detailed in Materials and Methods) were used as queries to conduct BLAST searches against the channel catfish transcriptome and genome databases. All sequences with significant hits were assembled into unique sequences of 61 CYP genes in channel catfish (Table 2). Among all these genes, 40 sequences were identified in both databases with full-length or nearly full-length CYPs, while the remaining 21 genes were identified in the genome database with partial sequences of transcripts in the transcriptome database. All these sequences have been deposited to GenBank with their accession numbers being summarized in Table 2.
Table 2. Cytochrome P450 genes of channel catfish.
| Catfish CYPs |
Transcripts or genome |
Sequence integrity |
Accession number of NCBI |
Linkage group | Reference |
|---|---|---|---|---|---|
| CYP1A1 | both | complete | JT412024.1 | not available | this study |
| CYP1B1 | both | complete | AAY90143.1 | 6 | [92] |
| CYP1C1 | both | complete | JT408041.1 | 16 | this study |
| CYP2K8 | both | complete | JT406642.1 | 3 | this study |
| CYP2K17 | both | complete | JT415766.1 | 3 | this study |
| CYP2M1 | both | complete | JT414187.1 | 22 | this study |
| CYP2R1 | both | complete | JT416283.1 | 21 | this study |
| CYP2U1 | both | complete | JT416818.1 | 21 | this study |
| CYP2X1 | both | complete | AAG30296.1 | 9 | [93] |
| CYP2X20 | genome | partial | KF531904 | 9 | this study |
| CYP2X21 | genome | complete | KF531901 | 9 | this study |
| CYP2X22 | genome | partial | KF531905 | 14 | this study |
| CYP2Y7 | genome | partial | KF531905 | 3 | this study |
| CYP2Y8 | genome | partial | KF531907 | 3 | this study |
| CYP2AA14 | both | complete | JT411919.1 | 11 | this study |
| CYP2AD6 | genome | partial | KF531902 | 28 | this study |
| CYP2AD8 | genome | partial | KF531903 | 28 | this study |
| CYP2AD9 | both | complete | JT418663.1 | 28 | this study |
| CYP2AD10 | genome | partial | KF531908 | 28 | this study |
| CYP3A125 | genome | partial | KF531909 | 20 | this study |
| CYP3A126 | both | complete | JT418825.1 | 20 | this study |
| CYP3A127 | genome | partial | KF531910 | 20 | this study |
| CYP3A128 | genome | partial | KF531911 | 20 | this study |
| CYP3C5 | both | complete | JT408739.1 | 28 | this study |
| CYP4F60 | both | complete | JT479785.1 | 5 | this study |
| CYP4F66 | genome | Partial | KF531912 | 5 | this study |
| CYP4T16 | genome | partial | KF531913 | 19 | this study |
| CYP4T17 | both | partial | JT320237.1 | 19 | this study |
| CYP4V7 | both | complete | JT408109.1 | 9 | this study |
| CYP4V8 | both | complete | JT414161.1 | 29 | this study |
| CYP5A1 | both | complete | JT316268.1 | 4 | this study |
| CYP7A1 | both | complete | JT408390.1 | 1 | this study |
| CYP7C1 | both | complete | JT411121.1 | 11 | this study |
| CYP7D1 | both | complete | JT415644.1 | 18 | this study |
| CYP8A1a | both | complete | JT411628.1 | 3 | this study |
| CYP8A1b | both | partial | JT470032.1 | 7 | this study |
| CYP8B5 | both | complete | JT323293.1 | 28 | this study |
| CYP8B6 | genome | partial | KF531914 | 8 | this study |
| CYP8B7 | genome | partial | KF531915 | 20 | this study |
| CYP11A1 | both | complete | NP_001187241.1 | 14 | unpublished |
| CYP11C1 | both | complete | JT399700.1 | not available | this study |
| CYP17A1 | both | complete | NP_001187242.1 | 6 | unpublished |
| CYP17A2 | both | partial | JT364637.1 | 11 | this study |
| CYP19A1 | both | complete | Q92111.1 | 6 | [94] |
| CYP19A2 | both | complete | AF417239.1 | 18 | [95] |
| CYP20A1 | both | complete | JT411689.1 | 13 | this study |
| CYP21A1 | both | complete | JT407837.1 | 1 | this study |
| CYP24A1 | genome | partial | KF531916 | 7 | this study |
| CYP26A1 | both | complete | JT413111.1 | 5 | this study |
| CYP26B1 | both | complete | JT416269.1 | 23 | this study |
| CYP26C1 | both | complete | JT411012.1 | 7 | this study |
| CYP27A2 | both | complete | JT244274.1 | 13 | this study |
| CYP27A3 | genome | partial | KF531917 | not available | this study |
| CYP27A14 | both | complete | JT406399.1 | 17 | this study |
| CYP27A15 | genome | partial | KF531918 | 17 | this study |
| CYP27A16 | genome | partial | KF531919 | 17 | this study |
| CYP27A17 | genome | partial | KF531920 | 17 | this study |
| CYP27A18 | genome | partial | KF531921 | 17 | this study |
| CYP27B1 | both | complete | JT483829.1 | 28 | this study |
| CYP27C1 | both | complete | JT408218.1 | 13 | this study |
| CYP51A1 | both | complete | JT418451.1 | 28 | this study |
Based on the rules described previously for CYP genes nomenclature (35), the 61 CYP genes can be classified into 9 CYP clans (Table 2) including CYP clans 2, 3, 4, 7, 19, 20, 26, 51 and mitochondrial clan. These represented all vertebrate CYP clans but clan 46 (CYP46) which is absent in channel catfish.
The 61 channel catfish CYPs fall into 16 families (Table 2) including CYP1, CYP2, CYP3, CYP4, CYP5, CYP7, CYP8, CYP11, CYP17, CYP19, CYP20, CYP21, CYP24, CYP26, CYP27 and CYP51 families. Three families, CYP16, CYP39 and CYP46, of the vertebrate CYPs are missing from channel catfish. Of the 16 CYP families in channel catfish, CYP2, CYP3, CYP4 and CYP27 are the four biggest families contributed more than half (32 CYPs) of the CYP genes in channel catfish.
3.2. Location of CYPs on linkage groups
To better understand the genome distribution of channel catfish CYPs, we examined their locations on channel catfish linkage map (101). Of the 61 channel catfish CYPs, genomic locations of 58 CYPs can be determined, while genomic locations for three CYPs: CYP1A1, CYP11C1, and CYP27A3, cannot yet be determined. The 58 channel catfish CYPs were located on 21 linkage groups of the channel catfish genome (Table 2). Among which, linkage group 28 had the largest number of CYPs including five family of eight different genes, followed by LG17 and LG20 with five CYPs each.
3.3. Specific CYPs and comparative studies
A comparison of CYP gene numbers in various species is shown in Table 3. Mammals (human and mouse) had eight specific subfamilies: CYP2A, CYP2B, CYP2E, CYP2S, CYP2T, CYP4A, CYP4X and CYP4Z; chickens had one specific subfamily, CYP2H that is absent from other taxa analyzed. Amphibians have eight specific subfamilies: CYP2Q, CYP2AM, CYP2AN, CYP2AP, CYP2AQ, CYP2AR, CYP2AS and CYP2AT. A large number of 18 CYP subfamilies appeared to be teleost-specific: CYP2K, CYP2M, CYP2N, CYP2P, CYP2V, CYP2X, CYP2Y, CYP2Z, CYP2AA, CYP2AD, CYP2AE, CYP3B, CYP3C, CYP3D, CYP7C, CYP7D, CYP11C, and CYP16A (Table 3). It is noteworthy that these teleost-specific CYPs belong to five families, and the vast majority (11) of these belongs to CYP2 family. Of the 18 teleost-specific subfamilies, 10 were identified from channel catfish (Table 3).
Within the 37 CYP subfamilies identified in channel catfish, 13 (CYP2K, CYP2X, CYP2Y, CYP2AD, CYP3A, CYP4B, CYP4F, CYP4T, CYP8A, CYP8B, CYP17A, CYP19A and CYP27A) possess multiple copies of CYP genes while 24 possess a single copy for each CYP gene (Table 3). Among these subfamilies, CYP27A subfamily obtained the largest number of CYP gene copies of seven (Table 3).
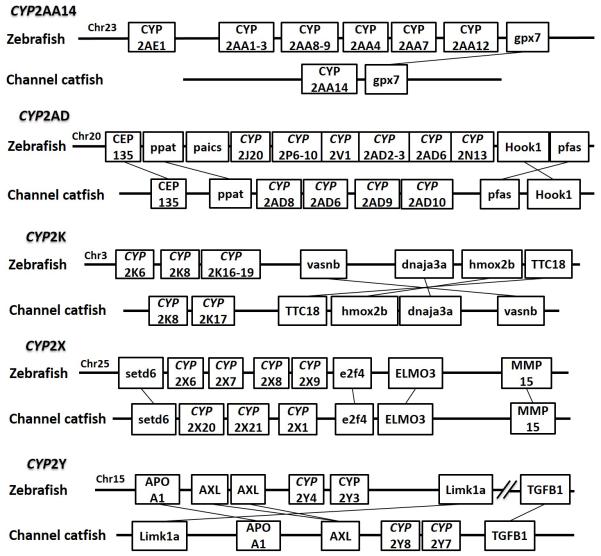
Compared to zebrafish, channel catfish genome appeared to have lost several subfamilies including CYP2N, CYP2P, CYP2V, CYP2AE, CYP39A, and CYP46A. In addition, channel catfish had much fewer copies for CYP2K, CYP2AA. Zebrafish had 11 copies of CYP2K while channel catfish had only 2 copies; similarly, zebrafish had 12 copies of CYP2AA, while channel catfish had just a single copy of CYP2AA (Table 3).
3.4. Phylogenetic analysis and lineage-specific gene duplication
A total of 61 channel catfish CYP genes have been phylogenetically analyzed. First, the 61 genes were categorized into Clans by initial phylogenetic analysis. Each clade (Clan) then was subsequently analyzed separately due to the enormous size of the phylogenetic tree. As shown in Figs 1-9, phylogenetic analyses provided clear evidence for the identities of most channel catfish CYP genes. Among these, CYP1A1, CYP1B1, CYP1C1, CYP2K8, CYP2K17, CYP2M1, CYP2R1, CYP2U1, CYP2AD6, CYP4V7, CYP4V8, CYP5A1, CYP7A1, CYP7C1, CYP7D1, CYP11A1, CYP11C1, CYP17A1, CYP17A2, CYP19A1, CYP19A2, CYP20A1, CYP21A1, CYP24A1, CYP26A1, CYP26B1, CYP26C1, CYP27A2, CYP27A3, CYP27B1, CYP27C1 and CYP51A1 were placed into their corresponding clades containing other teleost equivalents, respectively, with strong bootstrap support.
Figure 1.
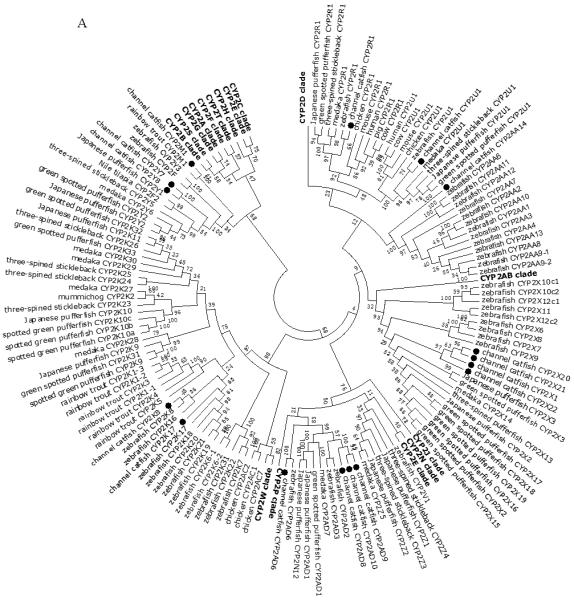
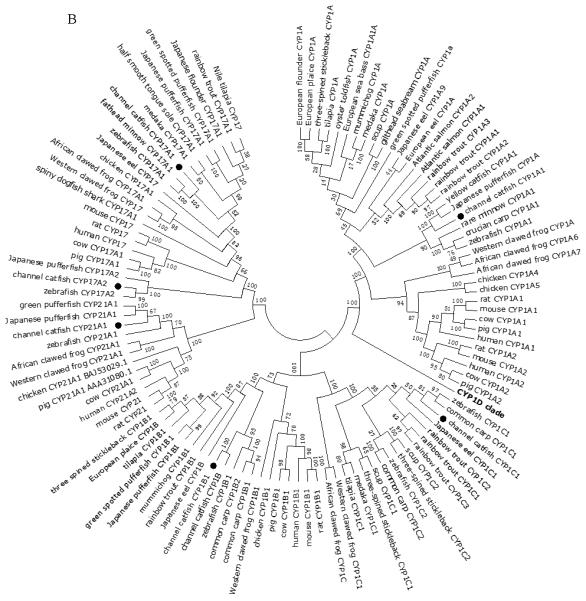
Phylogenetic relationship of channel catfish CYP genes in clan 2. (A) Phylogenetic relationship of channel catfish CYP2 subfamily. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches. (B) Phylogenetic relationship of channel catfish CYP1, CYP7 and CYP21 subfamilies. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Figure 9.
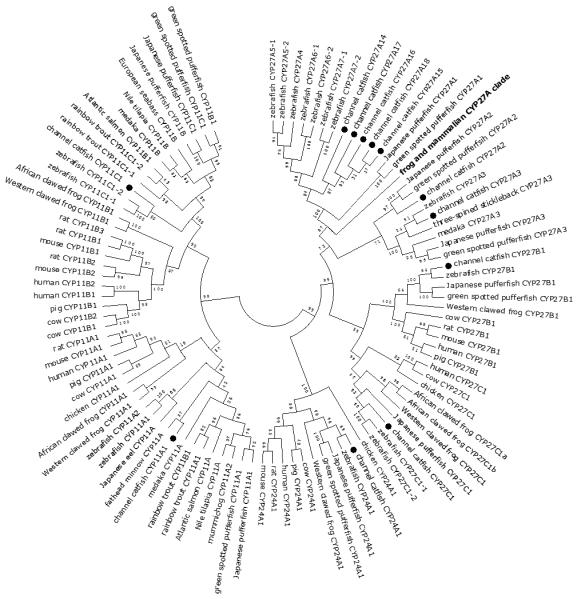
Phylogenetic relationship of channel catfish CYP genes in mitochondria clan. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Lineage-specific gene duplication was evident in a number of clans (Clan 2, Clan 3, Clan 4, Clan 7 and mitochondrial clan). For instance, within Clan 2, two CYP2Y genes (CYP2Y7 and CYP2Y8), four CYP2X genes (CYP2X1, CYP2X20, CYP2X21 and CYP2X22), one CYP2AA gene (CYP2AA14), and three CYP2AD genes (CYP2AD8, CYP2AD9 and CYP2AD10) were clustered together; similarly, four CYP3A genes (CYP3A125, CYP3A126, CYP3A127 and CYP3A128), one CYP3C gene (CYP3C5), two CYP4F genes (CYP4F60 and CYP4F66), two CYP4T genes (CYP4T16 and CYP4T17), two CYP8A genes (CYP8A1a and CYP8A1b), three copies of CYP8B genes (CYP8B5, CYP8B6 and CYP8B7) and five CYP27A genes (CYP27A14, CYP27A15, CYP27A16, CYP27A17 and CYP27A18) clustered together to form their own clade adjacent to zebrafish counterparts in teleost branch, respectively, suggesting channel catfish lineage-specific gene duplication. Similar lineage-specific gene duplication may have also occurred as many of zebrafish genes were clustered together themselves as well (Figs 1-4, Fig9).
Figure 4.
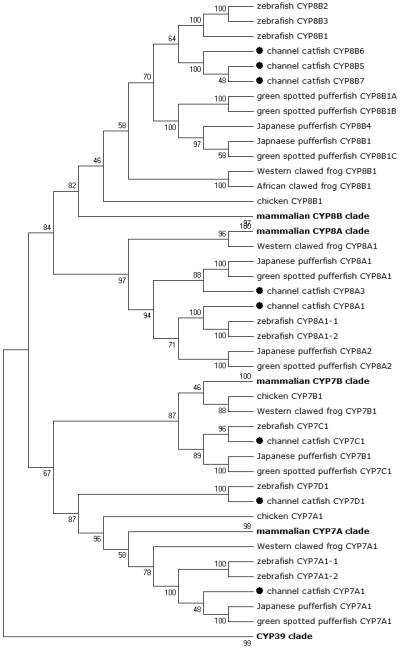
Phylogenetic relationship of channel catfish CYP genes in clan 7. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
3.5. Syntenic analysis and tandem duplication of channel catfish CYPs
Though phylogenetic relationships provide strong support for the identities of most CYP genes, syntenic analyses were required to provide additional evidence for orthologies or otherwise the paralogies. Syntenic analyses for a group of CYP genes were conducted, including subfamily 2K, subfamily 2X, subfamily 2Y, subfamily 2AA, subfamily 2AD, subfamily 3A, subfamily 3C, subfamily 4F, subfamily 4T, subfamily 8A, subfamily 27A, subfamily 27B and subfamily 27C.
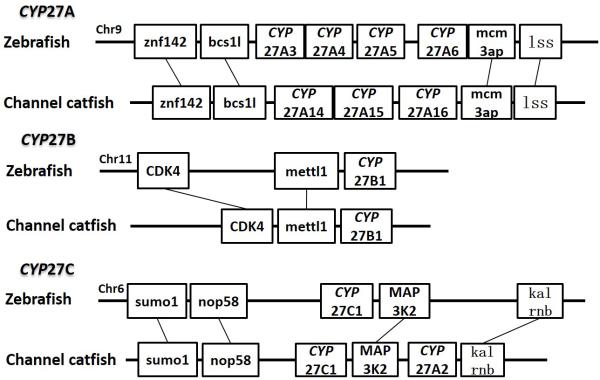
Positions of these channel catfish CYPs and their neighbor genes were identified from the draft genome scaffolds. And the genes were also identified from the zebrafish genome. As shown in Figs. 10-14, conserved syntenic blocks were characterized in these subfamilies between channel catfish and zebrafish counterparts. On the one hand, the conserved syntenies provide the strongest evidence for the orthologies, and on the other hand, a number of CYP genes were found to be located together in tandem, apparently as a result of tandem gene duplications. For instance, channel catfish CYP2AA14 shared the conserved neighbor genes with a cluster of eight zebrafish CYP2AA genes on chromosome 23; channel catfish CYP3C5 shared the conserved neighboring genes with three CYP3C genes on chromosome 3; channel catfish CYP27B1 shared the conserved neighboring genes with zebrafish CYP27B1 on chromosome 11; and channel catfish CYP27C1 shared the conserved neighboring genes with zebrafish CYP27C1 on chromosome 6. Tandem CYP genes were found within the conserved syntenies, suggesting both lineage-specific gene duplication in channel catfish and zebrafish, and the paralogies among the channel catfish genes and among the zebrafish genes within the conserved syntenic blocks. For instance, four channel catfish CYP2AD genes (CYP2AD6, CYP2AD8, CYP2AD9 and CYP2AD10) were present in tandem in the channel catfish genome within the conserved syntenic region in zebrafish genome that included 11 CYP2 genes arranged in tandem arrays (CYP2J, five CYP2Ps, one CYP2V, three CYP2ADs and one CYP2N); Similarly, two channel catfish CYP2K genes (CYP2K8 and CYP2K17) corresponded six CYP2K genes, all present in tandem arrays in both species; three channel catfish CYP2X genes (CYP2X1, CYP2X20 and CYP2X21), two channel catfish CYP2Y genes (CYP2Y7 and CYP2Y8), four channel catfish CYP3A genes (CYP3A125, CYP3A126, CYP3A127 and CYP3A128), two channel catfish CYP4F genes (CYP4F60 and CYP4F66), two channel catfish CYP4T genes (CYP4T16 and CYP4T17) and three channel catfish CYP27 genes (CYP27A14, CYP27A15 and CYP27A16) were all present in tandem in the channel catfish genome, corresponding the conserved syntenies of zebrafish genomic regions containing four CYP2X genes, two CYP2Y genes, CYP3A65, CYP4F3, CYP4T8 and four CYP27A genes of zebrafish, respectively. In addition, two CYP8A genes were identified in the channel catfish genome that had only one counterpart in the zebrafish genome, the CYP8A1 gene. We annotated the one with identical neighboring genes in channel catfish as CYP8A1a gene, and its paralog as CYP8A1b in channel catfish because this paralog shared only KCNB1 neighboring gene with zebrafish CYP8A1 gene, following the nomenclature of duplicated genes in zebrafish.
Figure 10.
Syntenic analysis of subfamilies CYP2AA, CYP2AD, CYP2K, CYP2X and CYP2Y
Figure 14.
Syntenic analysis of subfamilies CYP27A, CYP27B and CYP27C.
3.6. Meta-analyses of CYP expression after bacterial infection using RNA-Seq datasets
Three RNA-Seq datasets, RNA-Seq data from the liver of catfish infected with E. ictaluri (93), RNA-Seq data from the intestine of catfish infected with E. ictaluri (94), and RNA-Seq data from the gill of catfish infected with F. columnare (95), were used in this study. These datasets were analyzed to determine expression profile of all CYP genes after bacterial infection, using the cut-off of 2-fold change, p-value < 0.05, and reads number per gene ≥ 5. There were a total of 36 channel catfish CYPs showed significant differential expression for at least one time point post infection in the three tissues compared with control (Table 4).
Table 4.
Differentially expressed CYPs in channel catfish following Edwardsiella ictaluri and Flavobacterium columnare infection. Bold values indicate time-points where CYP was significantly changed relative to the control and absolute fold change was larger than two. Dash indicates all values of time-points in the experiment were out of threshold (threshold: p-value < 0.05, reads number per gene ≥ 5 and fold change ≥ 2).
| CYP gene | ESC liver | ESC intestine | Columnaris gill | |||||
|---|---|---|---|---|---|---|---|---|
| 3d | 14d | 3h | 24h | 3d | 4h | 24h | 48h | |
| CYP1C1 | 16.84 | 1.81 | - | - | - | - | - | - |
| CYP2K17 | - | - | 2.26 | 1.07 | 1.27 | - | - | - |
| CYP2M1 | −2.96 | 1.20 | - | - | - | - | - | - |
| CYP2R1 | −6.80 | 1.10 | 1.20 | −2.50 | −1.93 | - | - | - |
| CYP2X1 | −1.07 | −2.31 | −5.65 | −3.76 | −4.66 | - | - | - |
| CYP2X20 | −14.78 | −1.85 | - | - | - | −1.43 | −2.73 | −2.57 |
| CYP2X21 | −31.69 | −2.01 | - | - | - | - | - | - |
| CYP2X22 | −52.52 | 1.08 | - | - | - | - | - | - |
| CYP2Y8 | −2.56 | −1.00 | - | - | - | - | - | - |
| CYP2AA14 | - | - | −1.26 | −1.90 | −2.23 | - | - | - |
| CYP2AD6 | - | - | - | - | - | 1.83 | 1.07 | 2.88 |
| CYP2AD8 | 2.32 | −1.02 | - | - | - | - | - | - |
| CYP3A128 | 3.26 | 2.03 | - | - | - | - | - | - |
| CYP3C5 | 5.93 | 1.58 | - | - | - | - | - | - |
| CYP4F60 | 5.29 | 7.98 | - | - | - | - | - | - |
| CYP4F66 | 2.17 | 1.71 | - | - | - | - | - | - |
| CYP4T16 | 2.41 | −1.44 | 2.75 | 2.76 | 2.58 | 1.65 | 1.98 | 3.25 |
| CYP4V7 | −32.48 | −1.06 | - | - | - | - | - | - |
| CYP7A1 | −11.72 | −1.78 | - | - | - | - | - | - |
| CYP7D1 | 10.19 | 2.09 | - | - | - | - | - | - |
| CYP8A1a | 27.60 | 2.29 | - | - | - | - | - | - |
| CYP8A1b | 30.23 | 1.67 | - | - | - | - | - | - |
| CYP8B5 | 2.05 | 1.41 | - | - | - | - | - | - |
| CYP8B6 | 2.14 | 1.39 | - | - | - | - | - | - |
| CYP8B7 | −4.68 | −6.30 | - | - | - | - | - | - |
| CYP11A1 | 2.72 | 1.19 | - | - | - | - | - | - |
| CYP17A1 | - | - | - | - | - | −1.25 | −3.74 | −3.39 |
| CYP19A1 | - | - | - | - | - | 1.88 | 2.06 | 1.86 |
| CYP21A1 | 3.35 | −1.04 | - | - | - | - | - | - |
| CYP24A1 | −6.78 | −2.15 | - | - | - | - | - | - |
| CYP26A1 | −2.87 | −1.09 | 1.16 | −2.67 | −1.24 | −1.29 | −5.92 | −2.90 |
| CYP27A2 | 2.17 | 2.01 | - | - | - | - | - | - |
| CYP27A3 | −6.63 | 1.06 | - | - | - | - | - | - |
| CYP27A14 | −2.59 | −1.00 | - | - | - | - | - | - |
| CYP27A17 | - | - | - | - | - | −3.93 | −1.08 | −1.31 |
| CYP51A1 | 7.28 | 2.96 | −1.21 | −2.33 | −2.27 | 1.22 | −2.18 | −2.13 |
Several channel catfish CYPs exhibited drastic induction or suppression after bacterial infection. As shown in table 4, 3 days after E. ictaluri infection in the liver, drastic up-regulation was found in CYP8A1b, CYP8A1a and CYP1C1. CYP8A1b was up-regulated over 30-fold; CYP8A1a was up-regulated over 27-fold; while CYP1C1 was up-regulated 17-fold. In contrast, dramatic down-regulation was observed with CYP2X22, CYP4V7, CYP2X21, CYP2X20 and CYP7A1 with 52-fold, 32-fold, 31-fold, 14-fold and 11-fold reduction in expression after bacterial infection, respectively.
The involvement of CYP genes in the disease response appeared to be at early stages after infection. As shown in Table 4, numerous CYP genes exhibited differential expression 3 days after infection, but at the time point of 14 days after E. ictaluri infection in the liver, only two CYP genes, CYP4F60 and CYP8B7, were highly up-regulated or down-regulated, with eight-fold up and six-fold down for CYP4F60 and CYP8B7, respectively.
Many CYP genes were regulated by bacterial infection in the liver, but only few were regulated in the intestine and the gill (Table 4). Compared with the liver where 30 CYP genes were significantly regulated by bacterial infection compared with control, only 7 CYP genes were regulated in the intestine, and 8 CYP genes were regulated in the gill. Not only the number of CYP genes under regulation was much fewer in the intestine and gill, the level of induction and suppression was also much less dramatic. The most highly down-regulated CYP gene in the intestine is CYP2X1 (down 5.6X), and the most highly down-regulated CYP gene in the gill is CYP26A1 (down 5.9 X). Apparently, much of this difference was caused by the expression patterns of CYP genes, which are naturally expressed most abundantly in the liver (Table 4). Nonetheless, the use of the high throughput next generation sequencing RNA-Seq allowed detection of CYP gene expression in tissues other than the liver. A total of seven CYP genes were detected in the intestine: CYP2K17, CYP2R1, CYP2X1, CYP2AA14, CYP4T16, CYP26A1 and CYP51A1; and a total of eight CYP genes were detected in the gill: CYP2X20, CYP2AD6, CYP4T16, CYP17A1, CYP19A1, CYP26A1, CYP27A17, and CYP51A1. Three of these genes, CYP4T16, CYP26A1, and CYP51A1 were expressed in all three tested tissues.
4. Discussion
CYP genes play key roles in many crucial biological processes including oxidative transformation of xenobiotics and metabolism of endogenous substrates. They belong to one of the most widespread and diverse gene families that consist more than 18,500 members (24) among various species. In spite of their importance, only a few CYP genes were characterized from channel catfish (86-89). Systematic analysis of channel catfish CYP genes has been lacking. In this study, we identified 61 catfish CYP genes, which may represent the vast majority, if not all, CYP genes in the channel catfish genome. This assessment is based on the resources we used for the identification of these genes: several hundred thousands of ESTs (101-106), RNA-Seq (93-95), and the draft genome sequences that are yet not published, but represent over 200× genome coverage (unpublished data). This repertoire of genomic resources allowed thorough identification of CYPs in channel catfish. However, it is possible that additional CYP genes are yet to be discovered. At any rate, we believe that the vast majority of channel catfish CYPs in channel catfish have been discovered in this study.
Phylogenetic and syntenic analyses allowed annotation of these genes, and revealed that many of the CYP genes had gone through lineage-specific gene duplications, leading to the presence of a large number of paralogues within a species. Meta-analysis was conducted for the analysis of CYP gene expression. To our surprise, a large number of CYP genes, 36 in total, were up- or down-regulated after bacterial infection, suggesting their involvement in disease responses. The transcripts of all 61 P450 genes in the channel catfish not only were detected from the liver, where they are believed to be expressed, a significant number of CYP genes were expressed in tissues other than the liver, 7 in the intestine and 8 in the gill.
4.1. CYP families 1-4
The significance of CYP genes in detoxification of a variety of environmental pollutants, food additives, organic compounds and even drugs in aquatic species has been well established in previous studies (32, 55). This feature is particularly important for channel catfish that Inhabits at the bottom of water column. Among all the CYPs, the genes in families of CYP1, CYP2, CYP3 and CYP4 were reported to be highly involved in metabolism of xenobiotics, drugs and fatty acids (55). Identification of CYPs in these four families is critical for understanding of the detoxification mechanism in channel catfish.
In the present study, we identified 30 CYP genes belong to CYP families 1, 2, 3 and 4 in channel catfish (Table 2). In contrast to those CYPs primarily involved in endogenous metabolism, CYP genes among families 1-4 exhibited a larger degree of divergence across species (Table 3). This is strikingly significant in some fish-specific subfamilies belong to CYP2 family and CYP3 family. For example, gene number differences were observed in CYP2K, CYP2N, CYP2P, CYP2X, CYP2AA, CYP2AD, CYP3A and CYP3C subfamilies among species (Table 3). This observation indicated that these CYP subfamilies exhibited high level of divergence could be result of individual gene duplication or gene loss, or remnants of the genome duplication (55). Among these, CYP2N and CYP2P genes were commonly found in teleost species (45, 55, 64, 108, 109) but absent in catfish. In killifish, these two genes have been shown to catalyze benzphetamine N-demethylation and metabolize arachidonic acid (108, 109). In catfish, however, their function could be replaced by other CYP genes during evolution. For example, CYP2X1 in channel catfish have been proved to possess benzphetamine demethylase activity (110). In addition, only subfamilies CYP1A, CYP1B, CYP2U and CYP2R appear to be evolutionarily conserved across species. Of these four subfamilies, CYP2R1 and CYP2U1 catalyze modifications on the endogenous substrates vitamin D and arachidonic acid (101-112), while CYP1A and CYP1B are induced by a variety of drugs or contaminants (32, 113-115).
Phylogenetic analysis and syntenic analysis provided strong support for the identity of the majority of CYPs in channel catfish. However, for subfamily CYP2Y, CYP2X, CYP2AA, CYP2AD, CYP3A, CYP3C, CYP4F and CYP4T, phylogenetic analysis did not yield informative conclusion concerning their identities because members in these subfamilies formed their own clades in the phylogenetic tree, respectively. Consistent with phylogeny, syntenic analyses showed that genes in these CYP subfamilies existed as tandem duplication arrays which shared synteny with corresponding gene clusters in zebrafish, indicating that members of these subfamilies could be derived from recent lineage-specific gene duplication events. As such, these tandem arranged CYP genes are paralogous one another within a conserved syntenic block. The high level of lineage-specific multiplication of these CYP genes may suggest that the involved organisms were under evolutionary selection for the rapid expansion of such CYP genes, perhaps in the face of heavy environmental pollution.
4.2. CYP families 5-51
CYP genes in families 5-51, including CYP5A1, CYP7A1, CYP7C1, CYP7D1, CYP11A1, CYP11C1, CYP17A1, CYP17A2, CYP19A1, CYP19A2, CYP20A1, CYP21A1, CYP24A1, CYP26A1, CYP26B1, CYP26C1, CYP27A2, CYP27A3, CYP27B1, CYP27C1 and CYP51A1 are mainly involved in the metabolism of endogenous substrates (32, 36). In contrast to CYP gene families 1-4, all CYP subfamilies in families 5-51 have single copy gene in channel catfish with the exception of CYP8A, CYP8B, CYP17A, CYP19A and CYP27A, in which 2, 3, 2, 2 and 7 copies are presented in each of them. Phylogenetic analysis provides clear evidence for majority of CYP genes in these families, where they exhibit 1:1 correspondence with teleost counterparts, indicating conservation of enzyme activities and physiological functions.
Syntenic analysis of CYP8A genes indicated that there were two conserved syntenic blocks in channel catfish in comparison with just one in zebrafish. One of them contains five conserved genes and the other one possesses only two conserved genes in the block. We simply named the first one as CYP8A1a and the second one as CYP8A1b, following the nomenclature rule of zebrafish duplicated genes. Apparently, segmental duplication in the catfish genome accounted for the observed additional copy of the CYP8A1 gene. It is likely that several genes in the conserved syntenic block of the catfish genome were lost after segmental duplication.
For CYP27A subfamily, syntenic analyses were only available for CYP27A14, CYP27A15 and CYP27A16. These three genes showed a conserved syntenic block with zebrafish CYP27A cluster. Syntenic analysis of CYP8B genes, CYP27A17 gene and CYP27A18 gene were not available at this stage because missing information of their neighboring genes. Gap filling in the genomic sequence is required to provide a better resource for syntenic analysis in the future.
CYP46A1 gene in mammals is a cholesterol 24-hydroxylase enzyme and only present in the brain. It has been widely identified among species including teleost fish and plays an essential role in the majority of cholesterol turnover in vertebrate central nervous system (116). Though the function of CYP46 gene has not been studied in teleost, mutation of CYP46A1 gene could lead to serious neurodegenerative disease including Multiple Sclerosis, Alzheimer and Huntington Diseases in human (117). Four CYP46A genes have been identified in zebrafish (55). However, in channel catfish, no CYP46 homolog was found in any databases (EST, RNA-seq and genome sequence) in this study. Syntenic analyses with zebrafish indicated a potential gene loss in the conserved block between ak7a gene and SPTLC2 gene (Fig 15). We had also tried to amplify this gene in the catfish brain tissue using degenerate primers, in case that the gene may have existed, but had not been found. We were unable to amplify the CYP46 gene transcripts from the brain (data not shown). Although the negative PCR results still does not exclude the possibility of CYP46 presence in channel catfish, it is highly possible that channel catfish has lost the CYP46 gene set in its genome. Further analyses are required to validate this speculation. In addition to CYP46, the catfish appeared not to possess CYP16 and CYP39. CYP16 was also lost in the zebrafish genome (55), and CYP39 was lost in the fugu genome (45).
Figure 15.
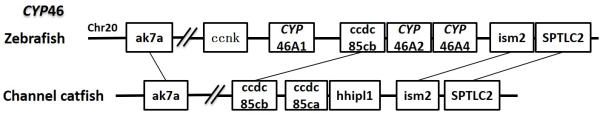
Syntenic analysis of CYP46.
4.3. CYP gene expression in disease defense response
RNAseq-based expression analysis has become a robust method to assess transcriptional profile to different challenge experiment (118). As described in our recent studies (93-95), using the 100 bp paired-end reads, we successfully captured comprehensive transcriptome from catfish intestine and gill after E. ictaluri and Columnare infection, respectively. The expression patterns of differentially expressed genes from these two studies were validated by quantitative real-time RT-PCR with average correlation coefficient around 0.9 (p < 0.001). In the present work, these two datasets from RNA-Seq of the intestine and gill after E. ictaluri and Columnare infection, along with dataset from RNA-Seq of the liver after ESC challenge, were utilized to analyze expression profiles of CYP genes in channel catfish after infection.
The expression patterns of CYP genes following E. ictaluri infection in the liver exhibited drastic differences as compared to the control. Up-regulation was observed 3 days after infection for a large number of CYPs including CYP1C1, CYP2AD8, CYP3A128, CYP3C5, CYP4F60, CYP4F66, CYP4T16, CYP7D1, CYP8A1a, CYP8A1b, CYP8B5, CYP8B6, CYP11A1, CYP21A1, CYP27A2 and CYP51A1. This is the first report involving induction of this large number of CYP genes after infection. In a study conducted by Sewer et al. (14), mRNAs of all three of the CYP4A subfamily members (CYP4A1, CYP4A2 and CYP4A3) were found to be induced 2- to 6-fold in the F344 rat livers after LPS administration. Later (15), the same group observed that this induction was not unique to LPS, CYP4A mRNA expression levels were also induced by irritants such as SiO2 and BaSO4. Though the mechanism behind this is not clear, it is possible that these CYPs are involved in the synthesis of mediators of inflammation cascade. Although the detailed mechanism for the involvement of such a large number of CYP genes in defense responses after infection is unknown at present, the disease induced expression suggested that they are important mediators in defense. Additional research is warranted to explore how the CYP genes are involved.
Down-regulation of CYP genes were observed 3 days after infection in liver with CYP2M1, CYP2R1, CYP2X1, CYP2X20, CYP2X21, CYP2X22, CYP2Y8, CYP4V7, CYP7A1, CYP8B7, CYP24A1, CYP26A1, CYP27A3 and CYP27A14. Similar observation has been reported by Chaluvadi et al., who demonstrated that infection of mice with enteropathogenic bacterium C. rodentium could cause selective down-regulation of hepatic cytochrome P450 mRNA and protein levels (119). Cui et al. also found that CYP4F4 and CYP4F5 were suppressed at early stage but induced after 24h of LPS treatment, and reached the highest levels at 2 weeks post-injury (18). In addition, Renton and Nicholson reported that CYPs were down-regulated after LPS treatment in the brain of rats (16). In all these cases, it is hypothesized that the pattern of down-regulation CYPs is a pathophysiological consequence of inflammatory process, and regulated by inflammatory mediators, such as cytokines. Similar study has been reported by Morgan (11), who found that down-regulation in enzyme activities of CYP1A1, CYP2C11, CYP2C12, CYP2E1 and CYP3A2 were triggered by inflammation mediators including cytokines IL-1, IL-6, or tumor necrosis factor (TNF) in liver of rats. Though the mechanisms under this down-regulation of CYP genes have not been clearly established, several hypotheses have been proposed by Morgan (12): firstly, the down-regulation of CYP genes may be associated with oxidative damage in liver; secondly, CYP genes down-regulation during an inflammatory response could be related to their function in metabolism of arachidonic acid; Lastly, the reason for down-regulation of CYP enzymes in the liver could be related to their ability to form nitric oxide. Further work is needed to elucidate the role of CYPs in the inflammation response, and particularly after the bacterial infection.
5. Conclusion
In summary, we systematically identified and characterized a repertoire of 61 CYP genes in channel catfish and studied their expression profile after bacterial infection. Strikingly large numbers of CYP genes appear to be involved in the bacterial defense processes.
Figure 2.
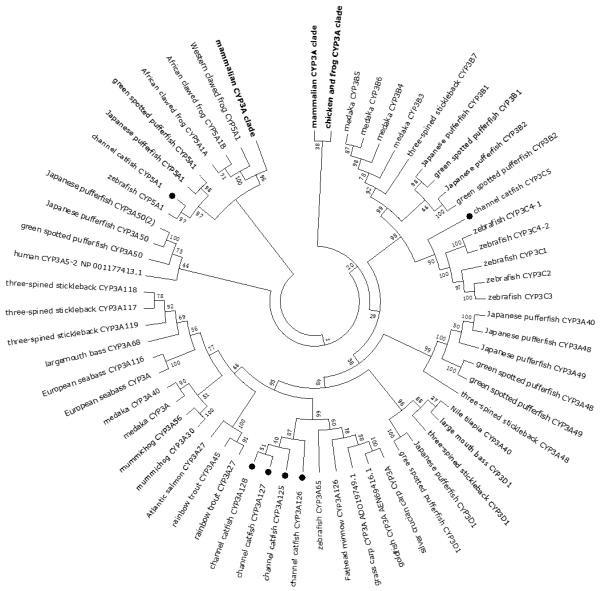
Phylogenetic relationship of channel catfish CYP genes in clan 3. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Figure 3.
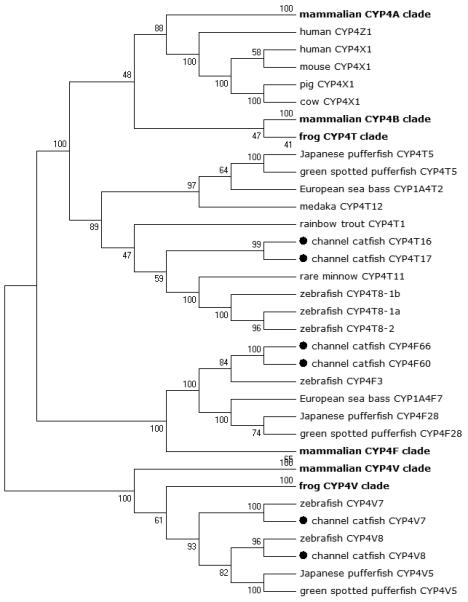
Phylogenetic relationship of channel catfish CYP genes in clan 4. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Figure 5.
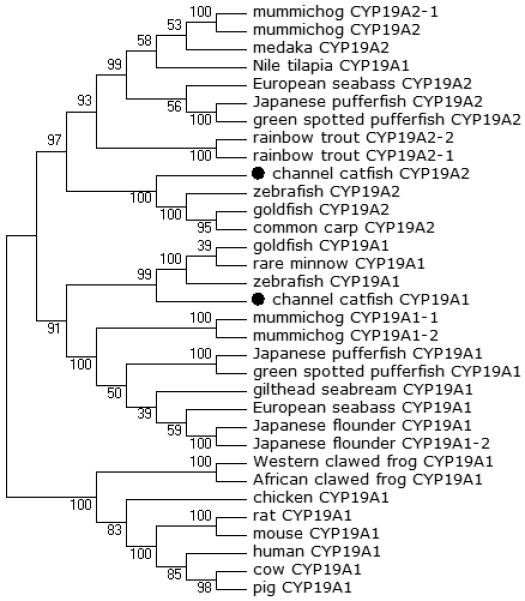
Phylogenetic relationship of channel catfish CYP genes in clan 19. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Figure 6.
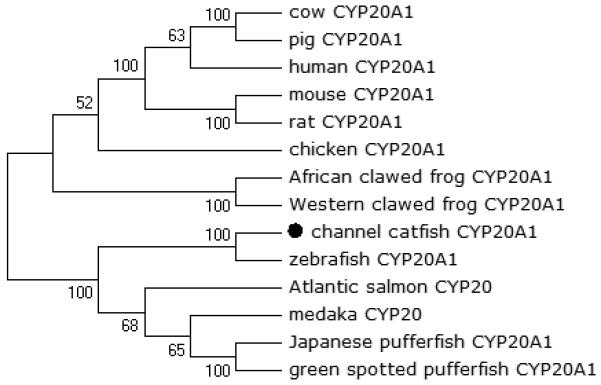
Phylogenetic relationship of channel catfish CYP genes in clan 20. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Figure 7.
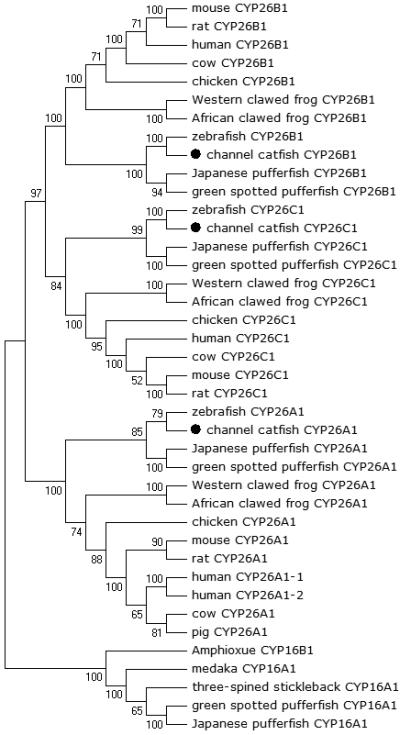
Phylogenetic relationship of channel catfish CYP genes in clan 26. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Figure 8.
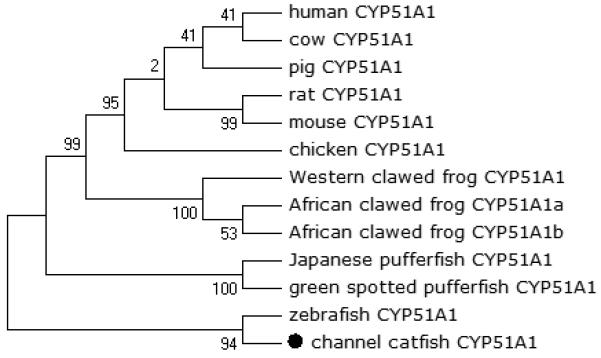
Phylogenetic relationship of channel catfish CYP genes in clan 51. MUSCLE alignments of all the amino acid sequences were used to generate a phylogenetic tree using Maximum likelihood method. The tree was shown with 10,000 replicates from the bootstrap test and the percentage of bootstrap values were given next to the branches.
Figure 11.
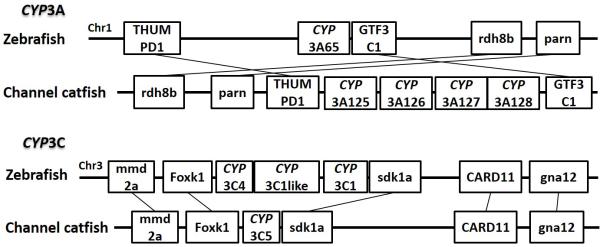
Syntenic analysis of subfamilies CYP3A and CYP3C
Figure 12.
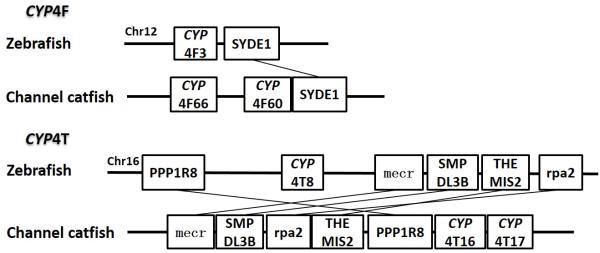
Syntenic analysis of subfamilies CYP4F and CYP4T
Figure 13.
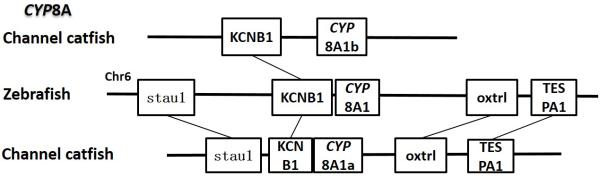
Syntenic analysis of subfamily CYP8A
Acknowledgements
We thank Alabama Supercomputer Center for providing the computer capacity for the bioinformatic analysis. This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2009-2009-35205-05101, 2010-65205-20356 and 2012-67015-19410 from the USDA National Institute of Food and Agriculture (NIFA). D. Nelson contribution supported by NIH U41-HG003345.
Reference
- 1.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes I. evidence for its hemoprotein nature. The Journal of biological chemistry. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 2.Werck-reichhart D, Feyereisen R. Protein family review Cytochromes P450 : a success story. Genome biology. 2000;1:3003.1–3003.9. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhardt R. Cytochromes P450 as versatile biocatalysts. Journal of biotechnology. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 4.Hrycay EG, Bandiera SM. The monooxygenase, peroxidase, and peroxygenase properties of cytochrome P450. Archives of biochemistry and biophysics. 2012;522:71–89. doi: 10.1016/j.abb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Nehert DW, Gonzalez F. P450 Genes: Structure, Evolution, and Regulation. Biochemistry. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- 6.Degtyarenko KN, Archakov AI. Molecular evolution of P450 superfamily and P450-containing monooxygenase systems. FEBS letters. 1993;332:1–8. doi: 10.1016/0014-5793(93)80470-f. [DOI] [PubMed] [Google Scholar]
- 7.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chemical reviews. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 8.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems--biological variations of electron transport chains. Biochimica et biophysica acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 10.Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philosophical Transactions of The Royal Society B. 2013;368 doi: 10.1098/rstb.2012.0431. 20120431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan ET. Regulation of cytochromes p450 during inflammation and infection. Drug Metabolism Reviews. 1997;29:1129–1188. doi: 10.3109/03602539709002246. [DOI] [PubMed] [Google Scholar]
- 12.Morgan ET. Regulation of cytochrome p450 by inflammatory mediators: why and how? Drug metabolism and disposition. 2001;29:207–212. [PubMed] [Google Scholar]
- 13.Renton KW. Cytochrome P450 regulation and drug biotransformation during inflammation and infection. Current Drug Metabolism. 2004;5:235–243. doi: 10.2174/1389200043335559. [DOI] [PubMed] [Google Scholar]
- 14.Sewer MB, Koop DR, Morgan ET. Endotoxemia in rats is associated with induction of the P450 4A subfamily and suppression of several other forms of cytochrome P450. Drug Metabolism and Disposition. 1996;4:401–407. [PubMed] [Google Scholar]
- 15.Sewer MB, Koop DR, Morgan ET. Differential inductive and suppressive effects of endotoxin and particulate irritants on hepatic and renal cytochrome P-450 expression. The Journal of pharmacology and experimental therapeutics. 1997;280:1445–1454. [PubMed] [Google Scholar]
- 16.Renton KW, Nicholson TE. Hepatic and central nervous system cytochrome P450 are down-regulated during lipopolysaccharide-evoked localized inflammation in brain. The Journal of pharmacology and experimental therapeutics. 2000;294:524–530. [PubMed] [Google Scholar]
- 17.Monshouwer M, Witkamp RF, Nujmeijer SM, Van Amsterdam JG, Van Miert a S. Suppression of cytochrome P450- and UDP glucuronosyl transferase-dependent enzyme activities by proinflammatory cytokines and possible role of nitric oxide in primary cultures of pig hepatocytes. Toxicology and applied pharmacology. 1996;137:237–244. doi: 10.1006/taap.1996.0077. [DOI] [PubMed] [Google Scholar]
- 18.Cui X, Kalsotra a, Robida a M., Matzilevich D, Moore a N., Boehme CL, Morgan ET, Dash PK, Strobel HW. Expression of cytochromes P450 4F4 and 4F5 in infection and injury models of inflammation. Biochimica et biophysica acta. 2003;1619:325–331. doi: 10.1016/s0304-4165(02)00491-9. [DOI] [PubMed] [Google Scholar]
- 19.Richardson TA, Sherman M, Antonovic L, Kardar SS, Henry W, Kalman D, Morgan ET. Hepatic and renal cytochrome P450 gene regulation during Citrobacter rodentium infection in wildtype and Toll-like Receptor 4 mutant mice. Drug Metabolism and Disposition. 2007;34:354–360. doi: 10.1124/dmd.105.007393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson TA, Morgan ET. Hepatic cytochrome p450 gene regulation during endotoxin induced inflammation in nuclear receptor knockout mice. The journal of pharmacology and experimental therapeutics. 2005;314:703–709. doi: 10.1124/jpet.105.085456. [DOI] [PubMed] [Google Scholar]
- 21.Satarug S, Lang MA, Yongvanit P, Lang MA. Induction of cytochrome P450 2A6 expression in humans by the carcinogenic parasite infection, opisthorchiasis viverrini. Cancer Epidemiol Biomarkers Prevention. 1996;5:795–800. [PubMed] [Google Scholar]
- 22.Nelson DR. The cytochrome p450 homepage. Human genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson DR. Progress in tracing the evolutionary paths of cytochrome P450. Biochimica et biophysica acta. 2011;1814:14–18. doi: 10.1016/j.bbapap.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Nelson DR. A world of cytochrome P450s. Philosophical Transactions of The Royal Society B. 2013;368 doi: 10.1098/rstb.2012.0430. 2013. 20120430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2003;450:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D. Cytochromes p450. The Arabidopsis book / American Society of Plant Biologists. 2011;9:e0144. doi: 10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldwin WS, Marko PB, Nelson DR. The cytochrome P450 (CYP) gene superfamily in Daphnia pulex. BMC genomics. 2009;10:169. doi: 10.1186/1471-2164-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo H, Bao Z, Du H, Zhang L, Wang S, Sun L, Mou X, Hu X. Identification of Cytochrome P450 (CYP) genes in Zhikong scallop (Chlamys farreri) Journal of Ocean University of China. 2012;12:97–102. [Google Scholar]
- 29.Ai J, Zhu Y, Duan J, Yu Q, Zhang G, Wan F, Xiang Z. Genome-wide analysis of cytochrome P450 monooxygenase genes in the silkworm, Bombyx mori. Gene. 2011;480:42–50. doi: 10.1016/j.gene.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Diaz GJ, Murcia HW, Cepeda SM. Cytochrome P450 enzymes involved in the metabolism of aflatoxin B1 in chickens and quail. Poultry science. 2010;89:2461–2469. doi: 10.3382/ps.2010-00864. [DOI] [PubMed] [Google Scholar]
- 31.Nelson DR, Ming R, Alam M, Schuler M. a. Comparison of Cytochrome P450 Genes from Six Plant Genomes. Tropical Plant Biology. 2008;1:216–235. [Google Scholar]
- 32.Uno T, Ishizuka M, Itakura T. Cytochrome P450 (CYP) in fish. Environmental toxicology and pharmacology. 2012;34:1–13. doi: 10.1016/j.etap.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Lee DS, Nioche P, Hamberg M, Raman CS. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature. 2008;455:363–368. doi: 10.1038/nature07307. [DOI] [PubMed] [Google Scholar]
- 34.Nebert DW, Adesnik M, Coon MJ, Estabrook RW, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, Kemper B, Levin W, et al. The P450 gene superfamily: recommended nomenclature. DNA. 1987;6:1–11. doi: 10.1089/dna.1987.6.1. [DOI] [PubMed] [Google Scholar]
- 35.Nelson DR. Cytochrome P450 nomenclature, Methods in molecular biology (Clifton, N.J.) 2004;320:1–10. doi: 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- 36.Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus : the origin and evolution of animal cytochrome P450s. Philosophical Transactions of the Royal Society. 2013;368 doi: 10.1098/rstb.2012.0474. 20120474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai N, Tanaka M, Adachi S, Millerh WL, Nagahama Y. Rainbow trout cytochrome P-450c17 (17 alpha-hydroxylase/17, 20-lyase). cDNA cloning, enzymatic properties and temporal pattern of ovarian P450c17 mRNA expression during oogenesis. FEBS letters. 1992;301:60–64. doi: 10.1016/0014-5793(92)80210-8. [DOI] [PubMed] [Google Scholar]
- 38.Buhler DR, Yang YH, Dreher TW, Miranda CL, Wang JL. Cloning and sequencing of the major rainbow trout constitutive cytochrome P450 (CYP2K1): identification of a new cytochrome P450 gene subfamily and its expression in mature rainbow trout liver and trunk kidney. Archives of biochemistry and biophysics. 1994;312:45–51. doi: 10.1006/abbi.1994.1278. [DOI] [PubMed] [Google Scholar]
- 39.Activity A, Yang Y, Wang J, Miranda CL, Buhler DR. CYP2M1 : cloning, sequencing, and expression of a new cytochrome P450 from rainbow trout liver with fatty acid (omega-6)-hydroxylation activity. Archives of biochemistry and biophysics. 1998;352:271–280. doi: 10.1006/abbi.1998.0607. [DOI] [PubMed] [Google Scholar]
- 40.Carvan M, Ponomareva L, Solis W, Matlib R, Puga A, Nebert D. Trout CYP1A3 Gene: Recognition of Fish DNA Motifs by Mouse Regulatory Proteins. Marine biotechnology (New York, N.Y.) 1999;1:155–166. doi: 10.1007/pl00011763. [DOI] [PubMed] [Google Scholar]
- 41.Råbergh CM, Vrolijk NH, Lipsky MM, Chen TT. Differential expression of two CYP1A genes in rainbow trout (Oncorhynchys mykiss) Toxicology and applied pharmacology. 2000;165:195–205. doi: 10.1006/taap.2000.8941. [DOI] [PubMed] [Google Scholar]
- 42.Kusakabe M, Kobayashi T, Todo T, Mark Lokman P, Nagahama Y, Young G. Molecular cloning and expression during spermatogenesis of a cDNA encoding testicular 11beta-hydroxylase (P45011beta) in rainbow trout (Oncorhynchus mykiss) Molecular reproduction and development. 2002;62:456–469. doi: 10.1002/mrd.10145. [DOI] [PubMed] [Google Scholar]
- 43.Lee SJ, Buhler DR. Cloning, tissue distribution, and functional studies of a new cytochrome P450 3A subfamily member, CYP3A45, from rainbow trout (Oncorhynchus mykiss) intestinal ceca. Archives of Biochemistry and Biophysics. 2003;412:77–89. doi: 10.1016/s0003-9861(03)00029-8. [DOI] [PubMed] [Google Scholar]
- 44.Gomez CF, Constantine L, Moen M, Vaz A, Wang W, Huggett DB. Ibuprofen metabolism in the liver and gill of rainbow trout, Oncorhynchus mykiss. Bulletin of environmental contamination and toxicology. 2011;86:247–51. doi: 10.1007/s00128-011-0200-8. [DOI] [PubMed] [Google Scholar]
- 45.Nelson DR. Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Archives of biochemistry and biophysics. 2003;409:18–24. doi: 10.1016/s0003-9861(02)00553-2. [DOI] [PubMed] [Google Scholar]
- 46.Kim JH, Raisuddin S, Ki JS, Lee JS, Han KN. Molecular cloning and beta-naphthoflavone-induced expression of a cytochrome P450 1A (CYP1A) gene from an anadromous river pufferfish, Takifugu obscurus. Marine pollution bulletin. 2008;57:433–440. doi: 10.1016/j.marpolbul.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Chiang EF, Yan YL, Guiguen Y, Postlethwait J. Chung B, Two Cyp19 (P450 aromatase) genes on duplicated zebrafish chromosomes are expressed in ovary or brain. Molecular biology and evolution. 2001;18:542–550. doi: 10.1093/oxfordjournals.molbev.a003833. [DOI] [PubMed] [Google Scholar]
- 48.Goto-Kazeto R, Kight KE, Zohar Y, Place AR, Trant JM. Localization and expression of aromatase mRNA in adult zebrafish. General and comparative endocrinology. 2004;139:72–84. doi: 10.1016/j.ygcen.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Ge W. Cloning of zebrafish ovarian P450c17 (CYP17, 17α-hydroxylase/17, 20-lyase) and characterization of its expression in gonadal and extra-gonadal tissues. General and Comparative Endocrinology. 2004;135:241–249. doi: 10.1016/j.ygcen.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Gu X, Xu F, Wang X, Gao X, Zhao Q. Molecular cloning and expression of a novel CYP26 gene (cyp26d1) during zebrafish early development. Gene expression patterns. 2005;5:733–739. doi: 10.1016/j.modgep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Tseng H, Hseu T, Buhler DR, Wang W, Hu C. Constitutive and xenobiotics-induced expression of a novel CYP3A gene from zebrafish larva. Toxicology and applied pharmacology. 2005;205:247–258. doi: 10.1016/j.taap.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 52.Wang-Buhler JL, Lee SJ, Chung WG, Stevens JF, Tseng HP, Hseu TH, Hu CH, Westerfield M, Yang YH, Miranda CL, Buhler DR. CYP2K6 from zebrafish (Danio rerio): cloning, mapping, developmental/tissue expression, and aflatoxin B1 activation by baculovirus expressed enzyme. Comparative biochemistry and physiology. Toxicology & pharmacology. 2005;140:207–219. doi: 10.1016/j.cca.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Corley-Smith GE, Su H, Wang-Buhler J, Tseng H, Hu C, Hoang T, Chung W, Buhler DR. CYP3C1, the first member of a new cytochrome P450 subfamily found in zebrafish (Danio rerio) Biochemical and biophysical research communications. 2006;340:1039–1046. doi: 10.1016/j.bbrc.2005.12.110. [DOI] [PubMed] [Google Scholar]
- 54.Jönsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Basal and 3, 3′, 4, 4′, 5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicology and applied pharmacology. 2007;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jönsson ME, Nelson DR, Stegeman JJ. Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish. BMC genomics. 2010;11:643. doi: 10.1186/1471-2164-11-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scornaienchi ML, Thornton C, Willett KL, Wilson JY. Cytochrome P450-mediated 17beta-estradiol metabolism in zebrafish (Danio rerio) The Journal of endocrinology. 2010;206:317–325. doi: 10.1677/JOE-10-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arukwe A. Complementary DNA cloning, sequence analysis and differential organ expression of beta-naphthoflavone-inducible cytochrome P4501A in Atlantic salmon (Salmo salar) Comparative biochemistry and physiology. Toxicology & pharmacology. 2002;133:613–624. doi: 10.1016/s1532-0456(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 58.Lyssimachou A, Jenssen BM, Arukwe A. Brain cytochrome P450 aromatase gene isoforms and activity levels in Atlantic salmon after waterborne exposure to nominal environmental concentrations of the pharmaceutical ethynylestradiol and antifoulant tributyltin. Toxicological sciences : an official journal of the Society of Toxicology. 2006;91:82–92. doi: 10.1093/toxsci/kfj136. [DOI] [PubMed] [Google Scholar]
- 59.Vang S-H, Kortner TM, Arukwe A. Steroidogenic acute regulatory (StAR) protein and cholesterol side-chain cleavage (P450scc) as molecular and cellular targets for 17alpha-ethynylestradiol in salmon previtellogenic oocytes. Chemical research in toxicology. 2007;20:1811–1819. doi: 10.1021/tx700228g. [DOI] [PubMed] [Google Scholar]
- 60.Sanden M, Olsvik PA. Intestinal cellular localization of PCNA protein and CYP1A mRNA in Atlantic salmon Salmo salar L. exposed to a model toxicant. BMC physiology. 2009;9:3. doi: 10.1186/1472-6793-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stien X, Amichot M, Berge JB, Lafaurie M. Molecular cloning of a CYP1A cDNA from the teleost fish Dicentrarchus labrax. Comparative biochemistry and physiology Toxicology & pharmacology. 1998;121:241–248. doi: 10.1016/s0742-8413(98)10045-2. 1998. [DOI] [PubMed] [Google Scholar]
- 62.Sabourault C, Bergé J, Lafaurie M, Girard JP, Amichot M. Molecular cloning of a phthalate-inducible CYP4 gene (CYP4T2) in kidney from the sea bass, Dicentrarchus labrax. Biochemical and biophysical research communications. 1998;251:213–219. doi: 10.1006/bbrc.1998.9429. [DOI] [PubMed] [Google Scholar]
- 63.Vaccaro E, Salvetti A, Del Carratore R, Nencioni S, Longo V, Gervasi PG. Cloning, tissue expression, and inducibility of CYP 3A79 from sea bass (Dicentrarchus labrax) Journal of biochemical and molecular toxicology. 2007;21:32–40. doi: 10.1002/jbt.20153. [DOI] [PubMed] [Google Scholar]
- 64.Barber DS, McNally AJ, Garcia-Reyero N, Denslow ND. Exposure to p, p’ -DDE or dieldrin during the reproductive season alters hepatic CYP expression in largemouth bass (Micropterus salmoides) Aquatic Toxicology. 2007;81:27–35. doi: 10.1016/j.aquatox.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Melo AC, Ramsdell JS. Sexual dimorphism of brain aromatase activity in medaka: induction of a female phenotype by estradiol. Environmental health perspectives. 2001;109:257–264. doi: 10.1289/ehp.01109257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim I, Kim YJ, Yoon Y, Kawamura S, Lee Y, Lee J. Cloning of cytochrome P450 1A (CYP1A) genes from the hermaphrodite fish Rivulus marmoratus and the Japanese medaka Oryzias latipes. Marine environmental research. 2004;58:125–129. doi: 10.1016/j.marenvres.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 67.Kullman SW, Kashiwada S, Hinton DE. Analysis of medaka cytochrome P450 3A homotropic and heterotropic cooperativity. Marine environmental research. 2004;58:469–473. doi: 10.1016/j.marenvres.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 68.Kashiwada S, Hinton DE, Kullman SW. Functional characterization of medaka CYP3A38 and CYP3A40: kinetics and catalysis by expression in a recombinant baculovirus system. Comparative biochemistry and physiology Toxicology & pharmacology. 2005;141:338–348. doi: 10.1016/j.cca.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 69.Yokota H, Abe T, Nakai M, Murakami H, Eto C, Yakabe Y. Effects of 4-tert-pentylphenol on the gene expression of P450 11beta-hydroxylase in the gonad of medaka (Oryzias latipes) Aquatic toxicology (Amsterdam, Netherlands) 2005;71:121–132. doi: 10.1016/j.aquatox.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 70.Goldstone JV, Stegeman JJ. Gene structure of the novel cytochrome P4501D1 genes in stickleback (Gasterosteus aculeatus) and medaka (Oryzias latipes) Marine environmental research. 2008;66:19–20. doi: 10.1016/j.marenvres.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamauchi R, Ishibashi H, Hirano M, Mori T, Kim J, Arizono K. Effects of synthetic polycyclic musks on estrogen receptor, vitellogenin, pregnane X receptor, and cytochrome P450 3A gene expression in the livers of male medaka (Oryzias latipes) Aquatic toxicology (Amsterdam, Netherlands) 2008;90:261–268. doi: 10.1016/j.aquatox.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 72.El-kady MA, Mitsuo R, Kaminishi Y, Itakura T. cDNA cloning, sequence analysis and expression of 3-methylcholanthrene-inducible cytochrome P450 1B1 in carp (Cyprinus carpio) Environmental Science. 2004;11:231–240. 2004. [PubMed] [Google Scholar]
- 73.Itakura T, El-Kady M, Mitsuo R, Kaminishi Y. Complementary DNA cloning and constitutive expression of cytochrome P450 1C1 in the gills of carp (Cyprinus carpio) Environmental Science. 2005;12:111–120. [PubMed] [Google Scholar]
- 74.Barney ML, Patil JG, Gunasekera RM, Carter CG. Distinct cytochrome P450 aromatase isoforms in the common carp (Cyprinus carpio): sexual dimorphism and onset of ontogenic expression. General and comparative endocrinology. 2008;156:499–508. doi: 10.1016/j.ygcen.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 75.Morrison HG, Weil EJ, Karchner SI, Sogin ML, Stegeman JJ. Molecular cloning of CYP1A from the estuarine fish Fundulus heteroclitus and phylogenetic analysis of CYP1 genes: update with new sequences. Comparative biochemistry and physiology Part C, Pharmacology, toxicology & endocrinology. 1998;121:231–240. doi: 10.1016/s0742-8413(98)10044-0. [DOI] [PubMed] [Google Scholar]
- 76.Hegelund T, Celander MC. Hepatic versus extrahepatic expression of CYP3A30 and CYP3A56 in adult killifish (Fundulus heteroclitus) Aquatic Toxicology. 2003;64:277–291. doi: 10.1016/s0166-445x(03)00057-2. [DOI] [PubMed] [Google Scholar]
- 77.Wang L, Scheffler BE, Willett KL. CYP1C1 messenger RNA expression is inducible by benzo[α]pyrene in Fundulus heteroclitus embryos and adults. Toxicological sciences. 2006;93:331–340. doi: 10.1093/toxsci/kfl072. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zanette J, Jenny MJ, Goldstone JV, Woodin BR, Watka L. a, Bainy ACD, Stegeman JJ. New cytochrome P450 1B1, 1C2 and 1D1 genes in the killifish Fundulus heteroclitus: Basal expression and response of five killifish CYP1s to the AHR agonist PCB126. Aquatic toxicology (Amsterdam, Netherlands) 2009;93:234–243. doi: 10.1016/j.aquatox.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calò M, Bitto a, Lo Cascio P, Polito F, Lauriano ER, Minutoli L, Altavilla D, Squadrito F. Cytochrome P450 (CYP1A) induction in sea bream (Sparus Aurata) gills and liver following exposure to polychlorobiphenyls (PCBs) Veterinary research communications. 2009;33:181–184. doi: 10.1007/s11259-009-9279-3. [DOI] [PubMed] [Google Scholar]
- 80.Halm S, Kwon JY, Rand-Weaver M, Sumpter JP, Pounds N, Hutchinson TH, Tyler CR. Cloning and gene expression of P450 17alpha-hydroxylase, 17, 20-lyase cDNA in the gonads and brain of the fathead minnow Pimephales promelas. General and comparative endocrinology. 2003;130:256–266. doi: 10.1016/s0016-6480(02)00592-0. [DOI] [PubMed] [Google Scholar]
- 81.Christen V, Caminada D, Arand M, Fent K. Identification of a CYP3A form (CYP3A126) in fathead minnow (Pimephales promelas) and characterisation of putative CYP3A enzyme activity. Analytical and bioanalytical chemistry. 2010;396:585–595. doi: 10.1007/s00216-009-3251-5. [DOI] [PubMed] [Google Scholar]
- 82.Deng S, Chen S, Xu J, Liu B. Molecular cloning, characterization and expression analysis of gonadal P450 aromatase in the half-smooth tongue-sole, Cynoglossus semilaevis. Aquaculture. 2009;287:211–218. [Google Scholar]
- 83.Chen CF, Wen HS, Wang ZP, He F, Zhang JR, Chen XY, Jin GX, Shi B, Shi D, Yang YP, Li JF, Qi BX, Li N. Cloning and expression of P450c17-I (17α-hydroxylase/17, 20-lyase) in brain and ovary during gonad development in Cynoglossus semilaevis. Fish physiology and biochemistry. 2010;36:1001–1012. doi: 10.1007/s10695-009-9378-7. [DOI] [PubMed] [Google Scholar]
- 84.Aparicio S, Chapman J, Stupka E, Putnam N, Chia J-M, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MDS, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJK, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science (New York, N.Y.) 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 85.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch G-J, White S, Chow W, Kilian B, Quintais LT, Guerra-Assunção J. a., Zhou Y, Gu Y, Yen J, Vogel J-H, Eyre T, Redmond S, Banerjee R, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013 doi: 10.1038/nature12111. doi: 10.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Willett KL, Ganesan S, Patel M, Metzger C, Quiniou S, Waldbieser G, Scheffler B. In vivo and in vitro CYP1B mRNA expression in channel catfish. Marine environmental research. 2006;62(Suppl):332–336. doi: 10.1016/j.marenvres.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 87.Schlenk D, Furnes B, Zhou X, Debusk BC. Cloning and sequencing of cytochrome P450 2X1 from channel catfish (Ictalurus punctatus) Marine environmental research. 2002;54:391–394. doi: 10.1016/s0141-1136(02)00202-7. [DOI] [PubMed] [Google Scholar]
- 88.Trant JM. Isolation and characterization of the cDNA encoding the channel catfish (Ictalurus punctatus) form of cytochrome P450arom. General and comparative endocrinology. 1994;95:155–168. doi: 10.1006/gcen.1994.1113. [DOI] [PubMed] [Google Scholar]
- 89.Kazeto Y, Trant JM. Molecular biology of channel catfish brain cytochrome P450 aromatase (CYP19A2): cloning, preovulatory induction of gene expression, hormonal gene regulation and analysis of promoter region. Journal of molecular endocrinology. 2005;35:571–583. doi: 10.1677/jme.1.01805. [DOI] [PubMed] [Google Scholar]
- 90.Liu Z. Development of genomic resources in support of sequencing, assembly, and annotation of catfish genome. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. 2011;6:11–17. doi: 10.1016/j.cbd.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 91.Li P, Peatman E, Wang S, Feng J, He C, Baoprasertkul P, Xu P, Kucuktas H, Nandi S, Somridhivej B, Serapion J, Simmons M, Turan C, Liu L, Muir W, Dunham R, Brady Y, Grizzle J, Liu Z. Towards the ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs. BMC Genomics. 2007;8:177. doi: 10.1186/1471-2164-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang S, Abernathy J, Waldbieser G, Lindquist E, Richardson P, Lucas S, Wang M, Li P, Thimmapuram J, Liu L, Vullaganti D, Kucuktas H, Murdock C, Small B, Wilson M, Liu H, Jiang Y, Lee Y, Chen F, Lu J, Wang W, Peatman E, Xu P, Somridhivej B, Baoprasertkul P, Quilang J, Sha Z, Bao B, Wang Y, Wang Q, Takano T, Nandi S, Liu S, Wong L, Kaltenboeck L, Quiniou S, Bengten E, Miller N, Trant J, Rokhsar D, Liu Z. Catfish Genome Consortium: Assembly of 500,000 inter-specific catfish expressed sequence tags and large scale gene-associated marker development for whole genome association studies. Genome Biology. 2010;11:R8. doi: 10.1186/gb-2010-11-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang R, Sun L, Bao L, Zhang J, Jiang Y, Yao J, Song L, Feng J, Liu S, Liu Z. Bulk segregant RNA-seq reveals expression and positional candidate genes and allele-specific expression for disease resistance against enteric septicemia of catfish. BMC Genomics. 2013;14:929. doi: 10.1186/1471-2164-14-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li C, Zhang Y, Wang R, Lu J, Nandi S, Mohanty S, Terhune J, Liu Z, Peatman E. RNA-Seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish & Shellfish Immunology. 2012;32:816–827. doi: 10.1016/j.fsi.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 95.Sun F, Peatman E, Li C, Liu S, Jiang Y, Zhou Z, Liu Z. Transcriptomic signatures of attachment, NF-κB suppression and IFN stimulation in the catfish gill following columnaris bacterial infection. Developmental and Comparative Immunology. 2012;38:169–180. doi: 10.1016/j.dci.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 96.Liu S, Zhang Y, Zhou Z, Waldbieser G, Sun F, Lu J, Zhang J, Jiang Y, Zhang H, Wang X, Rajendran KV, Kucuktas H, Peatman E, Liu Z. Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote. BMC Genomics. 2012;13:595. doi: 10.1186/1471-2164-13-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics (Oxford, England) 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic acids research. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ninwichian P, Peatman E, Liu H, Kucuktas H, Somridhivej B, Liu S, Li P, Jiang Y, Sha Z, Kaltenboeck L, Abernathy JW, Wang W, Chen F, Lee Y, Wong L, Wang S, Lu J, Liu Z. Second-generation genetic linkage map of catfish and its integration with the BAC-based physical map. G3 (Bethesda, Md) 2012;2:1233–1241. doi: 10.1534/g3.112.003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ju Z, Karsi A, Kocabas A, Patterson A, Li P, Cao D, Dunham R, Liu Z. Transcriptome analysis of channel catfish (Ictalurus punctatus): genes and expression profile from the brain. Gene. 2000;261:373–382. doi: 10.1016/s0378-1119(00)00491-1. [DOI] [PubMed] [Google Scholar]
- 103.Cao D, Kocabas A, Ju Z, Karsi A, Li P, Patterson A, Liu Z. Transcriptome of channel catfish (Ictalurus punctatus): initial analysis of genes and expression profiles of the head kidney. Animal genetics. 2001;32:169–188. doi: 10.1046/j.1365-2052.2001.00753.x. [DOI] [PubMed] [Google Scholar]
- 104.Karsi A, Patterson A, Feng J, Liu Z. Translational machinery of channel catfish: I. A transcriptomic approach to the analysis of 32 40S ribosomal protein genes and their expression. Gene. 2002;291:177–186. doi: 10.1016/s0378-1119(02)00595-4. [DOI] [PubMed] [Google Scholar]
- 105.Kocabas AM, Li P, Cao D, Karsi A, He C, Patterson A, Ju Z, Dunham R, Liu Z. Expression profile of the channel catfish spleen: analysis of genes involved in immune functions. Marine biotechnology (New York, N.Y.) 2002;4:526–536. doi: 10.1007/s10126-002-0067-0. [DOI] [PubMed] [Google Scholar]
- 106.Li P, Peatman E, Wang S, Feng J, He C, Baoprasertkul P, Xu P, Kucuktas H, Nandi S, Somridhivej B, Serapion J, Simmons M, Turan C, Liu L, Muir W, Dunham R, Brady Y, Grizzle J, Liu Z. Towards the ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs. BMC genomics. 2007;8:177. doi: 10.1186/1471-2164-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang S, Peatman E, Abernathy J, Waldbieser G, Lindquist E, Richardson P, Lucas S, Wang M, Li P, Thimmapuram J, Liu L, Vullaganti D, Kucuktas H, Murdock C, Small BC, Wilson M, Liu H, Jiang Y, Lee Y, Chen F, Lu J, Wang W, Xu P, Somridhivej B, Baoprasertkul P, Quilang J, Sha Z, Bao B, Wang Y, Wang Q, Takano T, Nandi S, Liu S, Wong L, Kaltenboeck L, Quiniou S, Bengten E, Miller N, Trant J, Rokhsar D, Liu Z. Assembly of 500,000 inter-specific catfish expressed sequence tags and large scale gene-associated marker development for whole genome association studies. Genome biology. 2010;11:R8. doi: 10.1186/gb-2010-11-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oleksiak MF, Wu S, Parker C, Karchner SI, Stegeman JJ, Zeldin DC. Identification, functional characterization, and regulation of a new cytochrome P450 subfamily, the CYP2Ns. J. Biol. Chem. 2000;275:2312–2321. doi: 10.1074/jbc.275.4.2312. [DOI] [PubMed] [Google Scholar]
- 109.Oleksiak MF, Wu S, Parker C, Qu W, Cox R, Zeldin DC, Stegeman JJ. Identification and regulation of a new vertebrate cytochrome P450 subfamily, the CYP2Ps, and functional characterization of CYP2P3, a conserved arachidonic acid epoxygenase/19-hydroxylase. Arch. Biochem. Biophys. 2003;411:223–234. doi: 10.1016/s0003-9861(02)00734-8. [DOI] [PubMed] [Google Scholar]
- 110.Mosadeghi S, Furnes B, Matsuo AY, Schlenk D. Expression and characterization of cytochrome P450 2X1 in channel catfish (Ictalurus punctatus) Biochim. Biophys. Acta. 2007;1770:1045–1052. doi: 10.1016/j.bbagen.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 111.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc. Natl. Acad. Sci. USA. 2004;101:7711–7715. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chuang SS, Helvig C, Taimi M, Ramshaw H. a, Collop AH, Amad M, White J. a, Petkovich M, Jones G, Korczak B. CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes omega- and (omega-1)-hydroxylation of fatty acids. The Journal of biological chemistry. 2004;279:6305–6314. doi: 10.1074/jbc.M311830200. [DOI] [PubMed] [Google Scholar]
- 113.Jung J-H, Kim M, Yim UH, Ha SY, An JG, Won JH, Han GM, Kim NS, Addison RF, Shim WJ. Biomarker responses in pelagic and benthic fish over 1 year following the Hebei Spirit oil spill (Taean, Korea) Marine pollution bulletin. 2011;62:1859–1866. doi: 10.1016/j.marpolbul.2011.04.045. [DOI] [PubMed] [Google Scholar]
- 114.Brammell BF, McClain JS, Oris JT, Price DJ, Birge WJ, Elskus A. CYP1A expression in caged rainbow trout discriminates among sites with various degrees of polychlorinated biphenyl contamination. Archives of environmental contamination and toxicology. 2010;58:772–782. doi: 10.1007/s00244-009-9368-x. [DOI] [PubMed] [Google Scholar]
- 115.Bugiak B, Weber LP. Hepatic and vascular mRNA expression in adult zebrafish (Danio rerio) following exposure to benzo-a-pyrene and 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Aquatic toxicology (Amsterdam, Netherlands) 2009;95:299–306. doi: 10.1016/j.aquatox.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 116.Russell DW, Halford RW, Ramirez DMO, Shah R, Kotti T. Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annual review of biochemistry. 2009;78:1017–1040. doi: 10.1146/annurev.biochem.78.072407.103859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leoni V, Caccia C. 24S-hydroxycholesterol in plasma: a marker of cholesterol turnover in neurodegenerative diseases. Biochimie. 2013;95:595–612. doi: 10.1016/j.biochi.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 118.Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biology. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chaluvadi MR, Kinloch RD, Nyagode BA, Richardson TA, Raynor MJ, Sherman M, Antonovic L, Strobel HW, Dillehay DL, Morgan ET. Regulation of Hepatic Cytochrome P450 Expression in Mice with Intestinal or Systemic Infections of Citrobacter rodentium. Drug Metabolism and Diposition. 2009;37:366–374. doi: 10.1124/dmd.108.024240. [DOI] [PMC free article] [PubMed] [Google Scholar]