A common bean microRNA, that targets a trancription factor, positively controls root development and symbiotic rhizobia infection and nodulation.
Abstract
Micro-RNAs are recognized as important posttranscriptional regulators in plants. The relevance of micro-RNAs as regulators of the legume-rhizobia nitrogen-fixing symbiosis is emerging. The objective of this work was to functionally characterize the role of micro-RNA172 (miR172) and its conserved target APETALA2 (AP2) transcription factor in the common bean (Phaseolus vulgaris)-Rhizobium etli symbiosis. Our expression analysis revealed that mature miR172c increased upon rhizobial infection and continued increasing during nodule development, reaching its maximum in mature nodules and decaying in senescent nodules. The expression of AP2-1 target showed a negative correlation with miR172c expression. A drastic decrease in miR172c and high AP2-1 mRNA levels were observed in ineffective nodules. Phenotypic analysis of composite bean plants with transgenic roots overexpressing miR172c or a mutated AP2-1 insensitive to miR172c cleavage demonstrated the pivotal regulatory role of the miR172 node in the common bean-rhizobia symbiosis. Increased miR172 resulted in improved root growth, increased rhizobial infection, increased expression of early nodulation and autoregulation of nodulation genes, and improved nodulation and nitrogen fixation. In addition, these plants showed decreased sensitivity to nitrate inhibition of nodulation. Through transcriptome analysis, we identified 114 common bean genes that coexpressed with AP2-1 and proposed these as being targets for transcriptional activation by AP2-1. Several of these genes are related to nodule senescence, and we propose that they have to be silenced, through miR172c-induced AP2-1 cleavage, in active mature nodules. Our work sets the basis for exploring the miR172-mediated improvement of symbiotic nitrogen fixation in common bean, the most important grain legume for human consumption.
The symbiotic nitrogen fixation (SNF) occurring in the legume-rhizobia symbiosis takes place in root-developed specialized organs called nodules. Nodulation is a complex process that involves communication between rhizobia and legumes through molecular signals, including rhizobial lipochitin-oligosaccharide symbiotic signals known as nodulation factors (NFs), that triggers a root-signaling cascade essential for rhizobia infection (for review, see Crespi and Frugier, 2008; Oldroyd and Downie, 2008; Kouchi et al., 2010; Murray, 2011; Oldroyd, 2013). Nuclear Ca2+ oscillations, or calcium spiking, is one of the earliest NF-induced responses in legume root hairs. Perception and transduction of the calcium-spiking signal involves Ca2+/CALMODULIN-DEPENDENT PROTEIN KINASE (CCaMK), which interacts with the nuclear protein CYCLOPS, and other downstream components, such as the transcriptional regulators NODULATION SIGNALING PATHWAY (NSP1)/NSP2, NUCLEAR FACTOR YA1 (NF-YA1)/YA2, ETHYLENE-RESPONSIVE FACTOR REQUIRED FOR NODULATION1, and NODULE INCEPTION (NIN), which, in turn, control the expression of early nodulation genes.
Legumes strictly regulate the number of developing nodules in response to internal and external cues. An important internal cue is the systemic feedback regulatory mechanism called autoregulation of nodulation (AON), which consists of root-derived and shoot-derived long-distance signals. AON is initiated in response to rhizobial NF during nodule primordium formation by the root production of CLAVATA3/Embryo-Surrounding Region Protein-related (CLE) peptides (Reid et al., 2011a). Some CLE peptides are predicted, although not proven, to act as the ligand for a shoot CLAVATA1-like Leu-rich repeat receptor kinase (Okamoto et al., 2009). Activation of this receptor is proposed to initiate the production of a shoot-derived inhibitor that is transported to the root, where it inhibits further nodule formation (for review, see Magori and Kawaguchi, 2009; Ferguson et al., 2010; Kouchi et al., 2010; Reid et al., 2011b). Soil nitrogen is an important external cue for the control of nodulation (Streeter and Wong, 1988). Recent work indicates that nitrate inhibition of nodulation may function via an up-regulation of a nitrate-induced CLE peptide that is perceived by a Leu-rich repeat receptor kinase in the root (Okamoto et al., 2009; Reid et al., 2011a).
In recent years, microRNAs (miRNAs), a class of noncoding RNA 21 to 24 nucleotides in length, have been identified as central regulators of gene expression in plants, controlling fundamental processes such as stress response, phytohormone regulation, organ morphogenesis, and development (Rogers and Chen, 2013). The plant miRNA precursors, generally transcribed by RNA polymerase II, adopt stem-loop structures that are processed by several enzymes and generate mature miRNAs that are exported to the cytosol. The role of miRNAs in posttranscriptional regulation is mediated by the almost perfect complementarity with their target mRNAs, thereby causing their degradation or their translational inhibition (Zhang et al., 2006; Rogers and Chen, 2013).
Progress in high-throughput sequencing technologies has facilitated the genome-wide identification of large miRNA populations and their target mRNAs in different legumes (for review, see Simon et al., 2009; Bazin et al., 2012; Bustos-Sanmamed et al., 2013). Conserved and legume-specific miRNA families differentially expressed during nodule organogenesis have been reported for Medicago truncatula, soybean (Glycine max), and Lotus japonicus (Subramanian et al., 2008; Lelandais-Brière et al., 2009; De Luis et al., 2012; Turner et al., 2012; Dong et al., 2013). Recently, Formey et al. (2014) identified miRNAs from M. truncatula roots that respond to treatments with purified NF. However, evidence for the functional involvement of miRNAs in rhizobial infection and the functionality of nodules has only been obtained for a small number of candidates. The involvement of M. truncatula microRNA166 (miR166), miR169, and miR164 in nodule development has been reported. miR169 controls nodule meristem maintenance through the repression of NF-YA1 (previously called HAEM ACTIVATOR PROTEIN2-1), a nodule-responsive transcription factor (TF; Combier et al., 2006), while miR166 and its target gene, HOMEODOMAIN-LEUCINE ZIPPER protein of class III TF, regulate meristem activity and vascular differentiation in roots and nodules (Boualem et al., 2008). The overexpression of miR164, a conserved miRNA targeting NAC1 (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF1-2], and cup-shaped cotyledon [CUC2] domain1) TF in roots, affected nodule organogenesis presumably through the deregulation of auxin responses (D’haeseleer et al., 2011). In soybean, the overexpression of miR482, miR1512, and miR1515 results in increased nodule numbers without affecting root development or the number of nodule primordia (Li et al., 2010). Recently, Turner et al. (2013) reported that the overexpression of soybean miR160, which targets a set of repressor auxin response factors, resulted in an enhanced sensitivity to auxin and inhibition of nodule development, apparently through a reduction in cytokinin sensitivity. Likewise, the overexpression of M. truncatula miR160 affected root gravitropism and nodule number (Bustos-Sanmamed et al., 2013). Specific variants of L. japonicus and M. truncatula miR171 target the GRAS-family NSP2 TF, a key regulator of the common symbiotic pathway for rhizobial and arbuscular mychorrizal symbioses (Ariel et al., 2012; De Luis et al., 2012; Lauressergues at al., 2012). M. truncatula roots overexpressing miR171h showed deceased arbuscular mychorrizal colonization (Lauressergues at al., 2012), while in L. japonicus, miR171c regulates the maintenance and establishment of the nodule but not the bacterial infection (De Luis et al., 2012). In addition, the role of L. japonicus miR397 in nodule copper homeostasis, through the regulation of a member of the laccase copper protein family, has been documented (De Luis et al., 2012).
Common bean (Phaseolus vulgaris) is the most important crop legume for human consumption and the main source of proteins for people in African and Central/South American countries (Broughton et al., 2003). Our research is focused on identifying and functionally characterizing common bean miRNAs. High-throughput sequencing of small RNAs generated from different organs of common bean let us identify more than 100 conserved miRNAs and to predict novel miRNAs (Peláez et al., 2012). Common bean miRNAs that respond to drought, salinity, nutrient deficiencies, or metal toxicity stresses have been identified, and their target genes have been predicted or validated (Arenas-Huertero et al., 2009; Valdés-López et al., 2010; Contreras-Cubas et al., 2012). The roles of miR399 in the common bean root response to phosphorus deficiency (Valdés-López et al., 2008) and of miR398 in the regulation of copper homeostasis and response to biotic interactions (Naya et al., 2014) have been demonstrated. In this work, we analyzed the role of miR172 in common bean roots and nodules.
miR172 is conserved in all angiosperm lineages; its conserved targets are TFs from the APETALA2 (AP2) family. The miR172 node that involves the miR156 node is one of the best-understood networks that regulate developmental timing in Arabidopsis (Arabidopsis thaliana) and other plants (Rubio-Somoza and Weigel, 2011). Aukerman and Sakai (2003) first described that miR172 promotes flowering by repressing AP2 genes, primarily through translation inhibition (Chen, 2004) but also through mRNA cleavage (Kasschau et al., 2003; Jung et al., 2007). In addition, the miR172 node regulates the juvenile-to-adult phase transition during shoot development (Wu et al., 2009; Huijser and Schmid, 2011). Such developmental transitions are coordinated by the antagonistic activities of the miR156 and miR172 nodes. miR156 targets a subset of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) TFs that bind to the miR712 promoter and directly promote its transcription, resulting in AP2 silencing (Wu et al., 2009).
miR172 has been identified in the legumes M. truncatula, L. japonicus, soybean, and common bean; it is highly accumulated in mature nodules relative to other plant tissues (Lelandais-Brière et al., 2009; Wang et al., 2009; Valdes-López et al., 2010; De Luis et al., 2012). Yan et al. (2013) reported the regulation of soybean nodulation by miR172 that involves a complex regulatory circuit in which miR156 regulates miR172 expression, which, in turn, controls the level of its AP2 target gene. They propose that AP2, directly or indirectly, controls the expression of nonsymbiotic hemoglobin, which is essential for regulating the levels of nodulation and nitrogenase activity (Yan et al., 2013). Very recently, Wang et al. (2014) demonstrated that soybean miR172c modulates rhizobial infection and nodule organogenesis. They showed that miR172c regulates nodule formation by repressing its target gene NODULE NUMBER CONTROL1 (NNC1), an AP2 TF, which directly targets and represses the early nodulin gene ENOD40 that plays a key role in nodulation. However, it is not clear whether miR172c controls early responses that are critical to establish a functional symbiosis between legumes and rhizobia.
The aim of this work was to analyze the role of the miR172 node in the common bean-rhizobia symbiosis. We determined an increased expression of miR172c upon rhizobial infection and during nodulation, showing a negative correlation with AP2-1 expression. We achieved the overexpression of miR172c and of a mutagenized AP2-1 insensitive to miR172 cleavage in composite common bean plants. Common bean plants with increased miR172c levels showed an improved symbiotic phenotype as well as lower sensitivity to nitrate inhibition of nodulation. We explored the possible role of AP2-1 as a transcriptional activator and/or repressor. Candidate target genes for downstream transcriptional activation by AP2-1 were identified; these could be relevant in the nodule senescence process. Our work extends the knowledge of miR172 function in the nodulation of common bean, an agronomically important legume.
RESULTS
Common Bean miR172 Isoforms and Target Genes
The Arabidopsis genome contains five loci that generate miR172 isoforms miR172a to miR172e, while 12 miR172 isoforms are reported for soybean (www.mirbase.org, version 20). The high-throughput small RNA sequencing analysis by Peláez et al. (2012) led us to identify four isoforms of common bean miR172.
In this work, we analyzed the recently published (Schmutz et al., 2014; www.phytozome.net/commonbean.php, v1.0) common bean genome sequence and identified six MIR172 loci that map in different common bean chromosomes. The most stable secondary structure of the miR172 precursors was predicted, and these showed the expected stem-loop structure. The six isoforms of mature common bean miR172, 20 or 21 nucleotides long, were designated miR172a to miR172f (Supplemental Fig. S1). The nucleotide sequences of miR172a, miR172b, and miR172c isoforms differ. However, although encoded by different loci, the sequences of miR172e and miR172f are identical to miR172a, while miR172d is identical to miR172c (Supplemental Fig. S1).
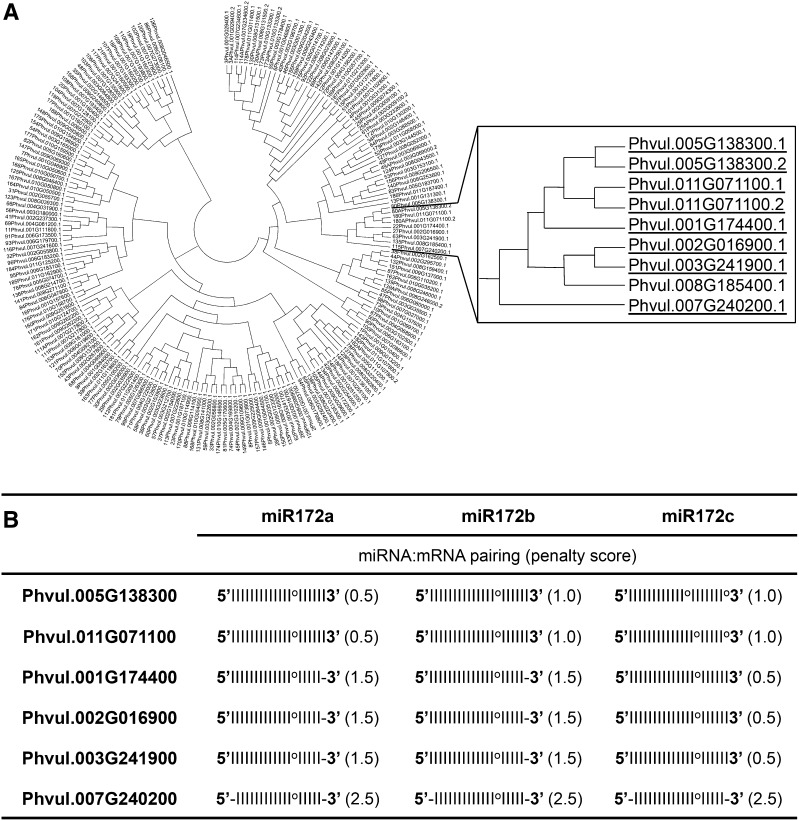
The conserved targets for miR172 in different plants are transcripts that encode TFs from the AP2 superfamily. In Arabidopsis, six AP2 TF genes, AP2, TARGET OF EARLY ACTIVATION TAGGED1 (TOE1), TOE2, TOE3, SCHLAFMUTZE, and SCHNARCHZAPFEN, which act as floral repressors, are targeted by miR172 and silenced through translation inhibition or cleavage (Aukerman and Sakai, 2003; Schmid et al., 2003; Chen, 2004; Jung et al., 2007). In soybean, 10 AP2 TF genes have been proposed as miR172 targets (Song et al., 2011). A recent analysis of the common bean transcriptome by RNA sequencing (RNA-seq) combined with available gene calls (O’Rourke et al., 2014) identified 202 transcripts encoding AP2-type TFs. The phylogenetic tree generated using sequence alignments of the common bean AP2 proteins is depicted in Figure 1A. From the whole set (202), we identified eight AP2 transcripts, encoded by six loci, with putative miR172 binding sites within their coding regions. The six predicted AP2 target genes showed base pairing with the three different miR172 isoforms (Fig. 1B). In each case, the penalty score for miRNA:mRNA (Jones-Rhoades and Bartel, 2004) was low; the highest score was observed in Phvul.007G240200, with the three miR172 isoforms. From the predicted AP2 targets, Phvul.005G138300, hereafter denominated as AP2-1, has been experimentally validated as a target of common bean miR172 (Arenas-Huertero et al., 2009). In addition, Phvul.011G071100 was identified as a target in a common bean degradome analysis (D. Formey, L.P. Íñiguez, P. Peláez, Y.F. Li, R. Sunkar, F. Sánchez, J.L. Reyes, and G. Hernández, unpublished data). Interestingly, the transcripts of AP2 predicted targets were organized in a single clade of the phylogenetic tree (Fig. 1A). However, this clade also includes the Phvul.008G185400.1 transcript, which has an AP2 domain (http://www.phytozome.net/commonbean.php) but lacks a detectable miR172 binding site and, therefore, is not proposed as a target (Fig. 1A). The recently published Phaseolus vulgaris Gene Expression Atlas (Pv GEA; O’Rourke et al., 2014) showed a very low expression of this AP2 gene in all the tissues reported (reads per kilobase per million = 6, highest values in leaves and pods). Therefore, Phvul.008G185400 AP2 is perhaps highly expressed in tissue-, development-, or environment-specific conditions not yet analyzed and its transcript level could be regulated by factors other than miR172. In addition, we could not detect a Phvul.008G185400.1 ortholog in the soybean genome sequence, perhaps indicating that it could be a pseudogene.
Figure 1.
Common bean AP2 transcripts with predicted miR172 binding sites. A, Neighbor-joining tree of AP2 proteins retrieved from the common bean genome sequence (http://www.phytozome.net/commonbean.php, v1.0). The clade including AP2 transcripts with miR172 binding sites (underlined) is shown in the inset. B, Pairing of the three different miR172 isoforms (a–c) with the predicted binding sites of the six different AP2 transcripts highlighted in A. Watson-Crick base pairing is indicated by lines, G:U base pairing is indicated by circles, and dashes indicate mismatches. Penalty scores, shown in parentheses, were calculated as described by Jones-Rhoades and Bartel (2004).
Differential Expression of miR172, Predicted AP2 Target Genes, miR156, and SPL6 in Plant Tissues
The differential expression of miR172 isoforms in plant organs/tissues at different developmental stages has been reported for Arabidopsis and soybean (Aukerman and Sakai, 2003; Wu et al., 2009; Yan et al., 2013; Wang et al., 2014). In this work, we performed expression analyses of the miR172 isoforms and their putative AP2 target genes (Fig. 1) in different tissues of SNF common bean plants 18 d post inoculation (dpi) with Rhizobium etli (Fig. 2).
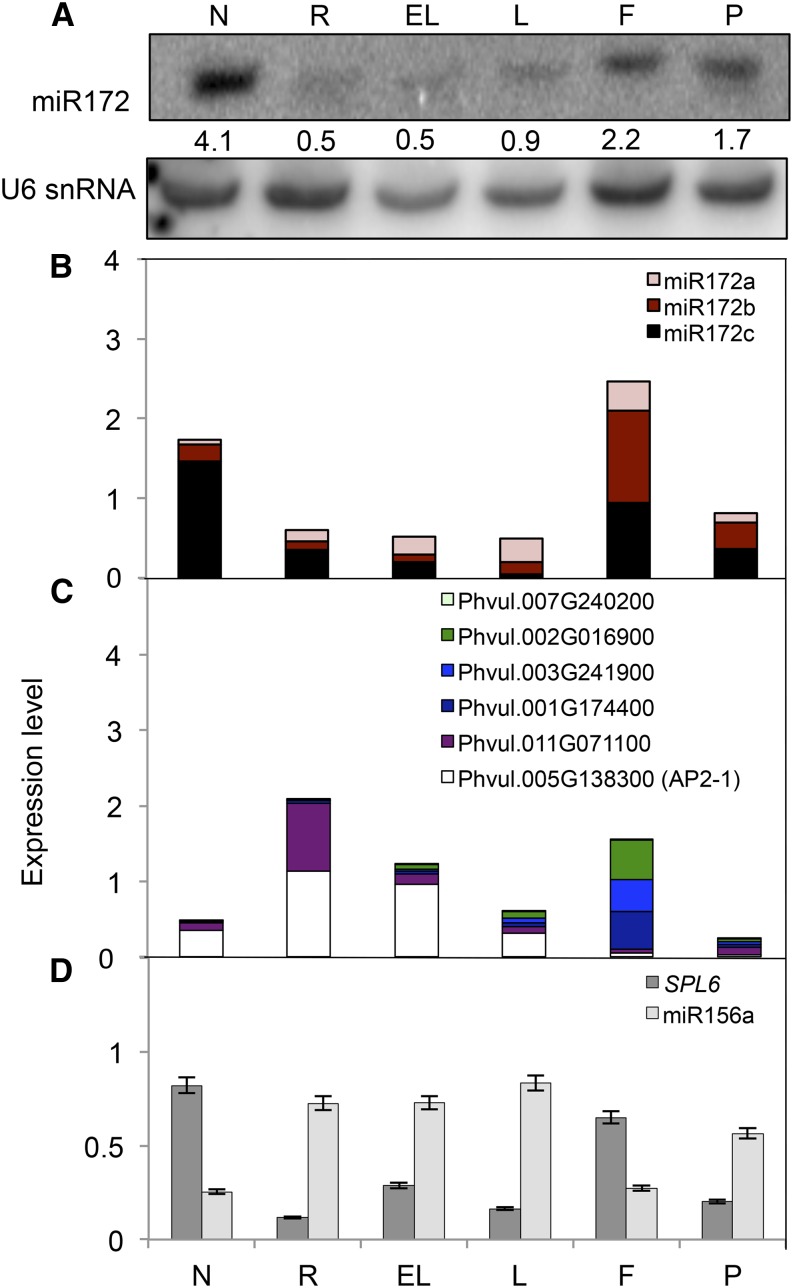
Figure 2.
Expression analysis of miR172, AP2 target genes, miR156a, and its SPL transcription factor target gene in different tissues of R. etli CE3-inoculated common bean plants (18 dpi). A, Mature miR172 levels detected by northern-blot analysis using U6 small nuclear RNA (snRNA) as a loading control for normalization. Signal intensities of the miR172 and U6 hybridization bands for each tissue were determined to calculate normalized expression levels. Numbers in each lane indicate normalized values of miR172 signal intensity. N, Nodules; R, roots; EL, embryonic leaves; L, leaves; F, flowers; P, pods. B to D, Transcript levels of mature miR172 isoforms (B; Supplemental Fig. S1), predicted AP2 target genes (C; Fig. 1), and mature miR156a and its target gene SPL6 (D) determined by qRT-PCR. Expression level refers to gene expression, based on threshold cycle (Ct) value, normalized with the expression of the housekeeping miR159 or UBIQUITIN CONJUGATING ENZYME9 (UBC9) gene.
Northern-blot analysis revealed that mature miR172 transcripts were most abundant in nodules followed by flowers (Fig. 2A). The miR172a probe was used for blot hybridization, but the quantified signals might reflect the combined levels of miR172 isoforms whose sequences differ only in two nucleotides (Supplemental Fig. S1). For real-time quantitative reverse transcription (qRT)-PCR expression analysis, specific primers were synthesized for each of the miR172 isoforms (a, b, and c; Supplemental Fig. S1; Supplemental Table S1). The data obtained by northern-blot and qRT-PCR expression analyses showed similar trends regarding the highest levels in nodules and flowers and the lowest levels in roots and leaves (Fig. 2, A and B). The observed variation in the levels of cumulative miR172 expression for each tissue may be attributable to the different sensitivities of the two methods. In addition, qRT-PCR analysis revealed differential expression of the miR172 isoforms among tissues, especially in those tissues with higher cumulative levels. Nodules showed the highest level of miR172c and very low amounts of miR172a and miR172b, while flowers showed the highest level of miR172b, followed by miR172c and a low amount of miR172a (Fig. 2B).
The transcript levels of each of the AP2 TF genes proposed as miR172 targets (Fig. 1) were determined by qRT-PCR (Fig. 2C) in tissues from SNF bean plants (Fig. 2C). Cumulative AP2 transcript levels were very high in roots and very low in nodules, thus showing a negative correlation with cumulative miR172 levels in these tissues (Fig. 2, B and C). In roots, the most highly expressed of the AP2 genes were Phvul.005G138300 (AP2-1) and Phvul.011G071100; AP2-1 was also highly expressed in embryonic leaves (Fig. 2C). However, a distinct pattern was observed in flowers, where the expression of these two genes was negligible and Phvul.001G174400, Phvul.003G241900, and Phvul.002G16900 were highly expressed.
The miR156 node has been implicated in upstream negative regulation of the miR172 node (Rubio-Somoza and Weigel, 2011). Arabidopsis miR156 represses miR172 expression by targeting members of the SPL family of TFs that directly bind to the MIR172 promoter and positively regulate its expression (Wu et al., 2009). Here, we determined the levels of mature miR156a (Peláez et al., 2012) in common bean tissues. The expression of miR156a was elevated in roots and leaves but low in nodules and flowers, showing an opposite trend of the cumulative expression of miR172 (Fig. 2D). We identified 32 SPL genes in the common bean genome, and 14 of these showed putative miR156 binding sites, including Phvul.009G165100, which was validated as a common bean miR156a target (Arenas-Huertero et al., 2009). Comparative sequence analysis with the Arabidopsis SPL gene family indicated that the common bean Phvul.009G165100 SPL gene is an ortholog to Arabidopsis SPL6. We analyzed the expression of the validated miR156a target SPL6 gene in different common bean organs. As shown in Figure 2D, common bean SPL6 was highly expressed in nodules and flowers while it was decreased in roots and leaves, thus showing a negative correlation with miR156 expression. We also searched for SPL transcription factor binding sites (TFBS) in the 5′ (promoter) region of each of the six MIR172 loci mapped in the genome, but we could not identify any, while 35 other TFBS were present in one or more of these loci (Supplemental Table S2).
Our data (Fig. 2) showed miR172c as the isoform with the highest expression in nodules and low expression in roots. Its expression pattern is opposite that of AP2-1 (Phvul.005G138300), the experimentally validated target (Arenas-Huertero et al., 2009) that showed the highest expression in roots. Therefore, we then focused our analysis on miR172c and AP2-1 in common bean plants interacting with R. etli.
Expression Analysis of miR172 and AP2-1 during Symbiosis
To assess a possible role of miR172c/AP2-1 in the common bean-R. etli symbiosis, we determined their expression in inoculated roots (Fig. 3) and in effective nodules at different developmental stages (Fig. 4A).
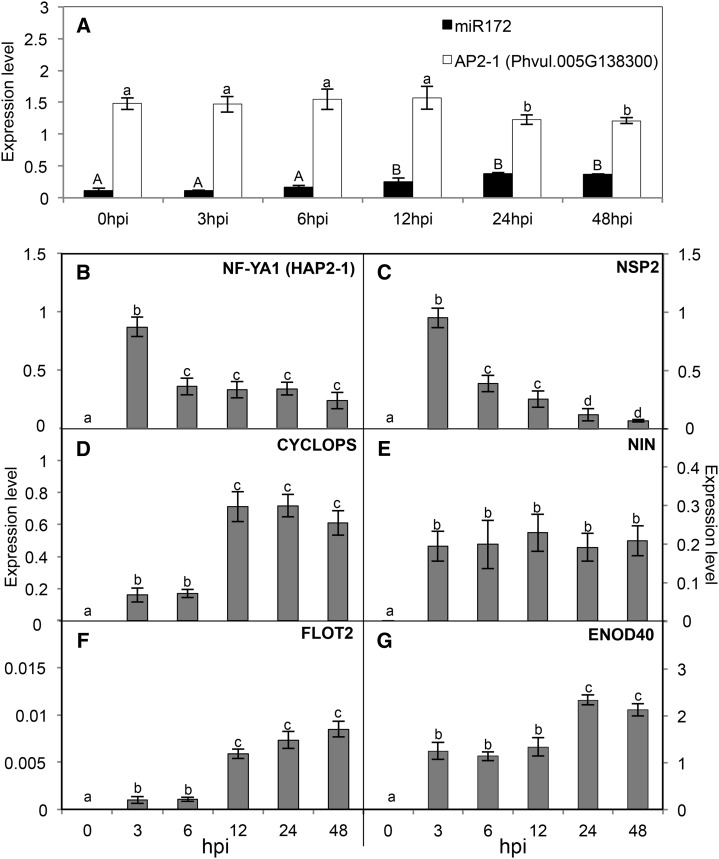
Figure 3.
Increased expression of miR172c and of early nodulation genes upon rhizobial infection. Expression levels of mature miR172c, AP2-1 (Phvul.005G138300; A), and early nodulation genes (B–G) were determined in roots inoculated with R. etli CE3 at the initial time (0) and after the indicated hpi. The common bean early nodulation genes were identified in the Pv GEA (O’Rourke et al., 2014): NF-YA1, Phvul.001G196800 (B); NSP2, Phvul.009G122700 (C); CYCLOPS, Phvul.002G128600 (D); NIN, Phvul.009G115800 (E); FLOT2, Phvul.009G090700 (F); and ENOD40, Phvul.002G064200 (G). Values represent averages ± sd from three biological replicates and two technical replicates each. Expression level refers to gene expression, based on Ct value, normalized with the expression of the housekeeping miR159 or UBC9 gene. Different lowercase letters indicate statistically different groups (ANOVA, P < 0.001); in A, lowercase and uppercase letters were used for AP2-1 and miR172c values, respectively.
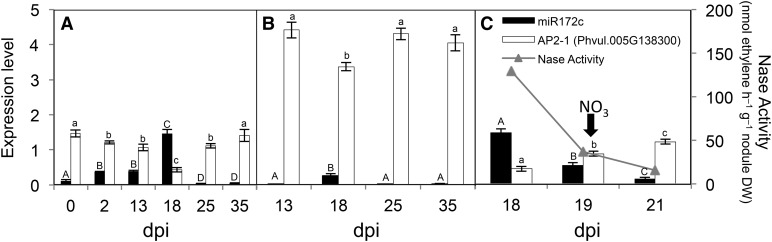
Figure 4.
miR172c and AP2-1 are differentially regulated in effective versus ineffective R. etli symbioses. Transcript levels were determined by qRT-PCR in inoculated roots or nodules harvested at the indicated dpi. A, Plants inoculated with the CE3 wild-type strain; determinations at 0 and 2 dpi were done in inoculated roots. B, Plants inoculated with the fix− R. etli nifA− mutant. C, Plants inoculated with the CE3 wild-type strain were grown for 18 d and watered with nitrogen-free nutrient solution. Subsequently, these plants were watered with nutrient solution supplemented with 10 mm KNO3 (black arrow). Nitrogenase (Nase) activity and transcript levels were determined at 18 dpi and at 1 or 3 d after nitrate addition (19 or 21 dpi). Values represent averages ± sd from three biological replicates and two technical replicates each. Expression level refers to gene expression, based on Ct value, normalized with the expression of the housekeeping miR159 or UBC9 gene. Different letters indicate statistically different groups (ANOVA, P < 0.001); lowercase and uppercase letters were used for AP2-1 and miR172c values, respectively.
To analyze miR172c/AP2-1 regulation at early stages of the symbiosis, common bean plantlets were grown in plastic square bioassay dishes and R. etli inoculum was applied directly to the roots. For gene expression analysis, only the responsive root zone, where initial bacteria-host recognition takes place, was collected at the initial time (0 h) and at 3, 6, 12, 24, and 48 h post inoculation (hpi). The mature miR172c level increased significantly after 6 h, whereas the high expression level of the AP2-1 target gene decreased significantly after 12 h; both transcript levels persisted until 48 h (Fig. 3A).
To investigate if miR172c up-regulation correlated with relevant events in the rhizobial infection process, we determined the expression of early nodulation genes in inoculated roots (Fig. 3, B–G). O’Rourke et al. (2014) identified common bean nodulation genes that were highly expressed in young and/or mature nodules and that are homologous to cognate nodulation genes previously identified in other legume species. From these, we selected six early nodulation genes for expression analysis: the TF genes NF-YA1 (Phvul.001G196800.1), NSP2 (Phvul.009G122700.1), and NIN (Phvul.009G115800); CYCLOPS (Phvul.002G128600.1), coding for a nuclear protein that interacts with CCaMK; FLOTILLIN-LIKE2 (FLOT2; Phvul.009G090700.1), coding for a lipid raft component; and ENOD40 (Phvul.008G291800), which lacks an open reading frame but encodes two small peptides and may function as a cell-cell signaling molecule for nodulation (Crespi and Frugier, 2008; Oldroyd and Downie, 2008; Kouchi et al., 2010; Murray, 2011; Oldroyd, 2013). Figure 3, B to G, shows the expression levels of the early nodulation genes in the responsive zone of R. etli-inoculated roots. The expression of all the genes tested increased significantly after the initial time (3 h). Highest levels of NF-YA1 and NSP2 decreased gradually after 3 h. Increased NIN expression persisted, whereas CYCLOPS and FLOT2 transcripts increased further after 6 h and persisted until 48 h. The ENOD40 transcript level increased significantly after 12 h and persisted until 48 h.
We then analyzed the regulation of the miR172 node during the development of effective nodules elicited by the R. etli CE3 wild-type strain (Fig. 4A). Nodules from inoculated common bean plants were harvested at different developmental stages, as defined by the differential expression of nodule development marker genes (Ramírez et al., 2005; Van de Velde et al., 2006; Supplemental Table S3). Immature, prefixing, 13-dpi nodules showed the highest ENOD55 expression and low (19.6%) nitrogenase activity. At 18 dpi, the nodules were fully developed and showed the highest nitrogenase activity and expression of the PHOSPHOENOLPYRUVATE CARBOXYLASE (PEPc) gene, essential for carbon assimilation in mature nodules (Ramírez et al., 2005). By 35 dpi, nodules had low nitrogenase activity (11%) and high CYSTEINE PROTEINASE (CP) gene expression, described as being specific for nodule senescence (Van de Velde et al., 2006; Supplemental Table S3). As shown in Figure 4A, the increased expression level of miR172c observed at 2 dpi (or 48 h in Fig. 3A) persisted in immature, prefixing nodules (13 dpi). In contrast, miR172c increased significantly to its highest level in mature, fully active nodules (18 dpi). Afterward, a drastic decrease in miR172c level was observed; it remained barely detectable until nodule senescence (35 dpi). Slightly decreased levels of AP2-1 transcripts persisted in immature nodules (13 dpi), a further decrease was observed in mature 18-dpi nodules, and afterward, the level of AP2-1 transcripts gradually increased. The lowest level of AP2-1 correlated with the highest level of miR172c in mature nodules (18 dpi).
Taken together, these data suggest that miR172c is involved in rhizobial infection and nodule development/function.
Altered Expression of miR172c and AP2-1 in Ineffective Symbioses
Because the maximum level of miR172c expression and AP2-1 silencing correlated with the peak of SNF (Fig. 4A; Supplemental Table S2), we assessed the regulation of the miR172 node in ineffective, nonfixing common bean-R. etli symbioses.
The nitrogen fixation genes regulator A (NifA)/RNA polymerase sigma factor complex is a master regulator of the N2 fixation genes in rhizobia. Transcriptional analysis of the R. etli nifA− (CFNX247) mutant strain demonstrated the nifA dependency of symbiotic genes on the symbiotic plasmid (Girard et al., 1996). The symbiotic phenotype of common bean plants inoculated with the R. etli nifA− mutant strain was drastically altered, as evidenced by a diminished amount of early-senescent nodules with few infected cells having bacteroids and devoid of nitrogenase activity and with symptoms characteristic of nitrogen deprivation in the leaves (Supplemental Fig. S2; Supplemental Table S3). As shown in Figure 4B, the ineffective nodules elicited by R. etli nifA− had nearly undetectable levels of miR172c, although a minor, but significant, increase was observed in 18-dpi ineffective nodules. Meanwhile, the AP2-1 target gene was highly induced at the different developmental stages of ineffective nodules; these values were even higher (approximately 2-fold) than those observed during effective symbiosis (Fig. 4, A and B). A slight but significant decrease in AP2-1 transcript was observed in 18-dpi ineffective nodules, when the miR172c level increased (Fig. 4B).
A similar effect was observed when the abolishment of SNF was achieved by adding nitrate to effective R. etli-elicited nodules (Fig. 4C), a well-known phenomenon in the legume-rhizobia symbiosis (Streeter and Wong, 1988). A short time (1 and 3 d) after nitrate addition, nitrogenase activity decreased drastically and nodules senesced (Fig. 4C; Supplemental Table S3). The latter correlated with the drastic decrease in mature miR172c and a concomitant increase of AP2-1 transcript level in the ineffective nodules (Fig. 4C).
Taken together, these data indicate a contrasting regulation of miR172c/AP2-1 expression in effective versus ineffective symbioses.
Effect of miR172c Overexpression on Root Development and Rhizobial SNF
To further investigate the role of miR172c and its target gene AP2-1 in SNF, we aimed to modulate their expression in common bean composite plants with transgenic roots and untransformed aerial organs, generated through Agrobacterium rhizogenes-mediated genetic transformation. This protocol has been used as an alternative method for stable transformation in common bean and other recalcitrant species (Estrada-Navarrete et al., 2007). The construct for miR172 overexpression (OE172) contained the 35S cauliflower mosaic virus promoter fused to the miR172c precursor. The OEAP2m plasmid contained a mutagenized AP2-1 gene that is insensitive to miR172 cleavage due to nucleotide substitutions in the miR172 binding site. Both constructs as well as the control empty vector (EV) contain the tdTomato (red fluorescent protein) reporter gene (Supplemental Fig. S3). We obtained several composite plants and determined the level of transgene expression for each plant (Supplemental Fig. S4). The OE172 composite plants showed very high levels of mature miR172c in both nodules and roots as well as a decreased level of AP2-1 transcript. Roots and nodules of OEAP2m plants showed very high levels of AP2-1. The variation in the degree of overexpression between individual transgenic roots is because each results from an independent transformation event.
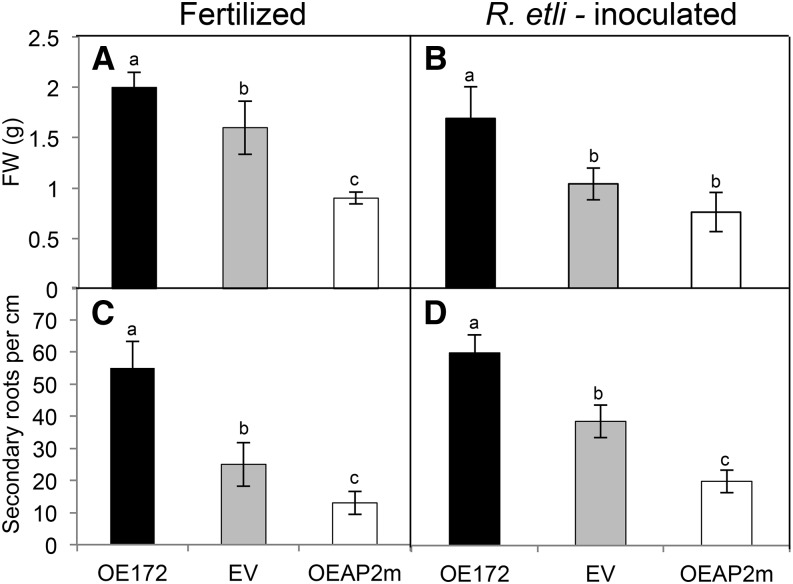
We first assessed if miR172 overexpression affected the root phenotype of fertilized (noninoculated) common bean plants as compared with those inoculated with R. etli. As shown in Figure 5, roots with high miR172c showed increased biomass and density of secondary roots, both in fertilized and SNF composite plants. The opposite phenotype was observed in OEAP2m composite plants. These data indicate that miR172 had a positive effect on root biomass/architecture independent of the presence of rhizobia.
Figure 5.
miR172c and AP2-1 control the root development of fertilized and R. etli-inoculated common bean plants. Root fresh weight (FW; A and B) and density of secondary roots (C and D) were determined in composite bean plants with transgenic roots overexpressing miR172c (OE172) or a mutated insensitive AP2-1 target gene (OEAP2m) as compared with control EV transformed roots. A set of composite plants was fertilized with full-nutrient solution for 10 d (A and C), and another set was inoculated with R. etli and watered with nitrogen-free nutrient solution for 21 dpi (B and D). Values represent averages ± sd from roots of eight independent composite plants each. Different lowercase letters indicate statistically different groups (ANOVA, P < 0.001).
To analyze if the positive effect of miR172 on root development (Fig. 5) also affects rhizobial infection and SNF, we investigated the response of composite plants altered in miR172 content to R. etli infection, early symbiotic stages, and nodule development/function.
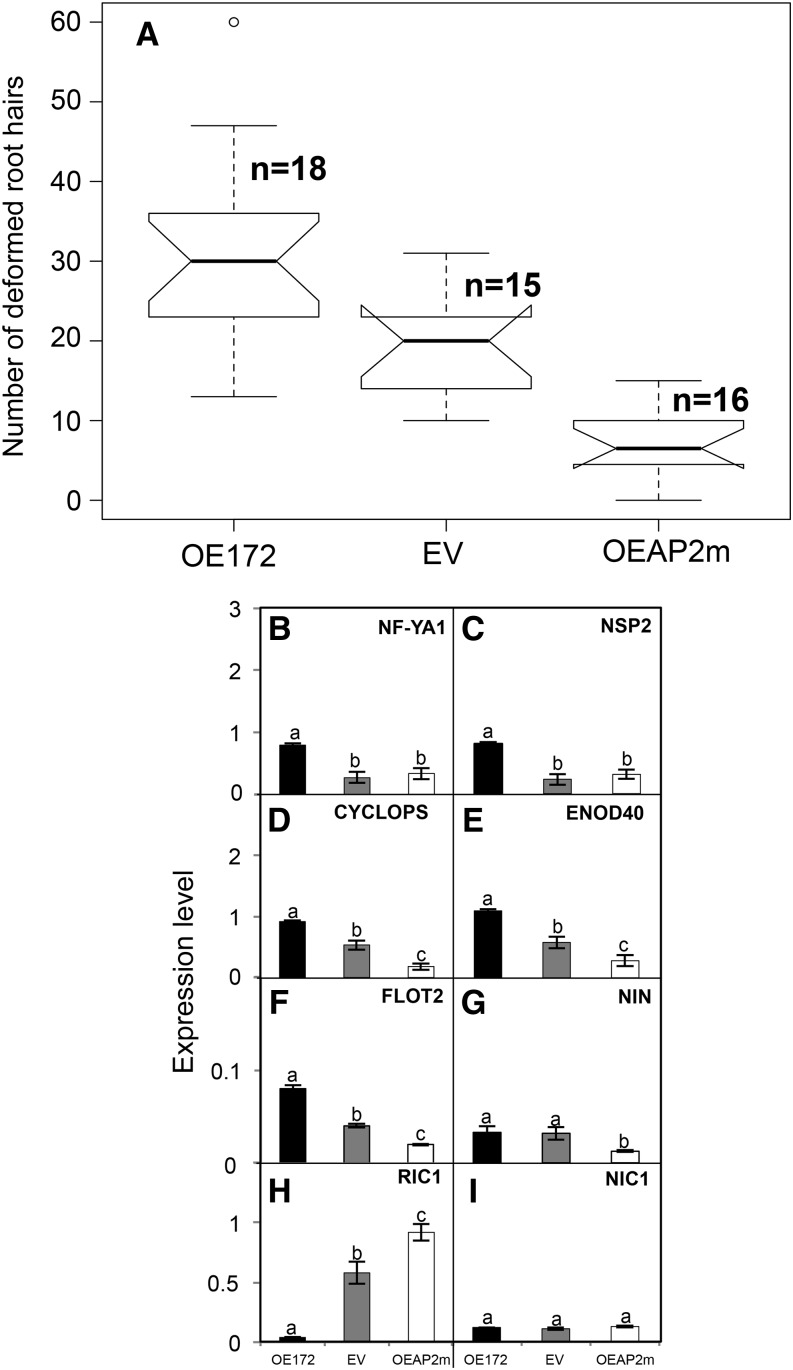
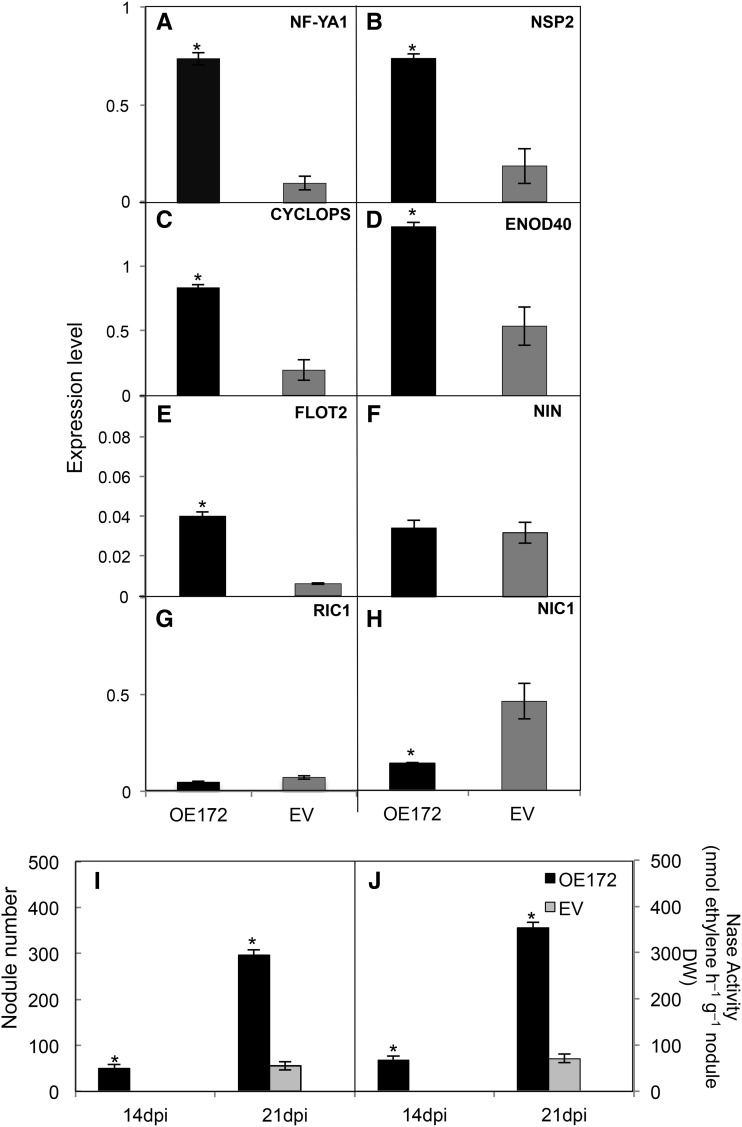
Figure 6 shows data for the analysis of rhizobial infection and early nodulation gene expression. For these experiments, the plastic square bioassay dish system was used for the inoculation and growth of OE172, EV, or OEAP2m composite plants. Notably, the amount of deformed root hairs was significantly higher in 48-hpi inoculated roots that overexpress miR172, while the opposite effect was observed in OEAP2m roots (Fig. 6A; Supplemental Fig. S5). A correlation of altered root hair deformation and the expression of early nodulation genes essential for rhizobial infection was observed after determining the transcript level of selected genes (NF-YA1, NSP2, CYCLOPS, ENOD40, FLOT2, and NIN) in the responsive root zone from 0 to 48 hpi (Fig. 6, B–G). All the genes tested showed increased expression in OE172 inoculated roots; NIN expression was increased only as compared with OEAP2m inoculated roots (Fig. 6, B–G).
Figure 6.
miR172c and AP2-1 control the rhizobial infection of common bean roots. Roots from OE172, EV, or OEAP2m composite plants were inoculated with R. etli CE3, and at 48 hpi, the root responsive zones were harvested for analysis. A, Quantification of the number of deformed root hairs (branched and swollen root hair) per 0.5 cm; each box plot indicates the number of the transgenic roots analyzed for each construct. B to I, Expression analysis of selected early nodulation genes. Most gene identifiers are indicated in the legend to Figure 3; RIC1, Phvul.005G096900; and NIC1, Phvul005G097000. Values represent averages ± sd from three biological replicates and two technical replicates each. Expression level refers to gene expression, based on Ct value, normalized with the expression of the housekeeping UBC9 gene. Different lowercase letters indicate statistically different groups (ANOVA, P < 0.001).
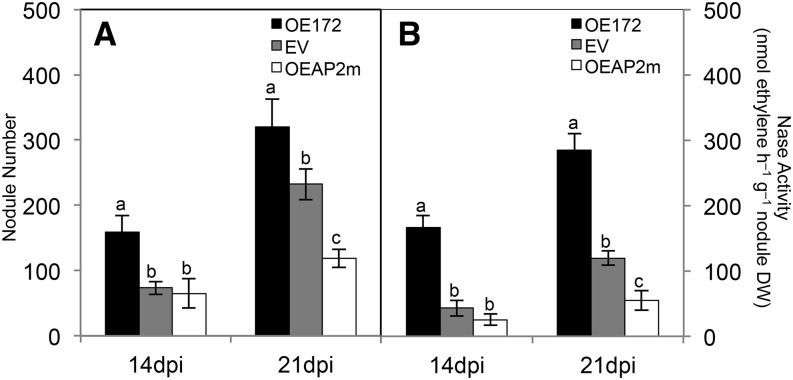
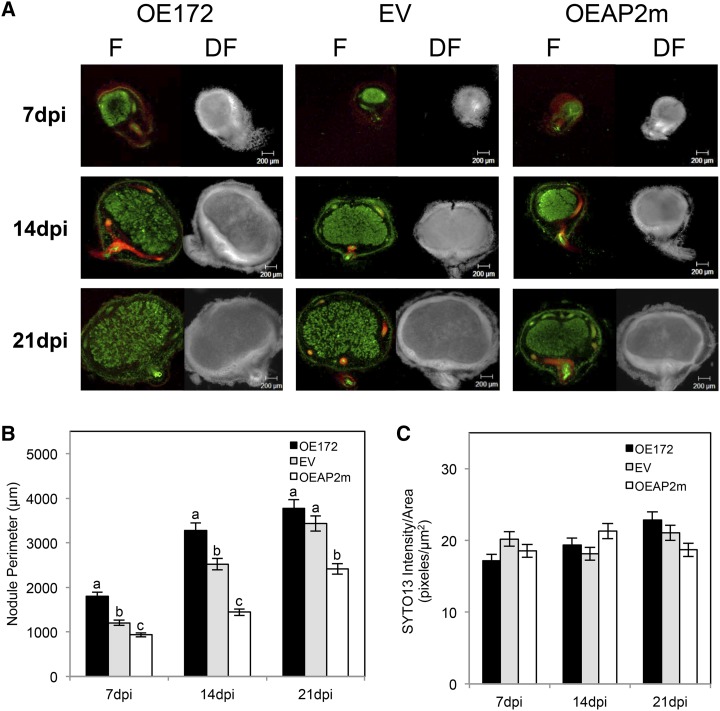
Nodule number and nitrogenase activity as well as histological analysis of nodules stained with SYTO13 (a fluorescent dye binding nucleic acids) from composite plants overexpressing miR172 or AP2m and control (EV) plants are presented in Figures 7 and 8. At 14 and 21 dpi, OE172 plants showed increased nodule number and nitrogenase activity that correlated with higher PEPc and reduced CP expression (Fig. 7; Supplemental Table S4). In addition, OE172 plants showed accelerated nodule development: nodule primordia and well-formed nodules (approximately 50 per root) were easily observed at 5 and 7 dpi, respectively, while only some unorganized primordia and very few tiny nodules were seen in EV plants. In addition, young (7 and 14 dpi) nodules from OE172 plants were larger (increased perimeter; Fig. 8, A and B). In contrast, OEAP2m plants at 21 dpi had fewer nodules with diminished nitrogenase activity and higher CP expression (Fig. 7; Supplemental Table S4). At all time points analyzed, the OEAP2m nodules had significantly reduced perimeters (Fig. 8, A and B). Similar values for SYTO13 intensity per nodule area were obtained for EV, OE172, and OEAP2m nodules, indicating similar bacteroid densities in infected cells (Fig. 8C).
Figure 7.
miR172c and AP2-1 control nodule number (A) and nitrogenase (Nase) activity (B) of SNF common bean. Plants were inoculated with R. etli CE3 for the indicated dpi. Values represent averages ± sd from five replicate samples per time point. Different lowercase letters from each set of values at different dpi indicate statistically different groups (ANOVA, P < 0.001).
Figure 8.
Alterations in the nodule development of OE172 and OEAP2m R. etli-inoculated composite bean plants. A, Fluorescent (F; left) and corresponding dark-field (DF; right) micrographs of central sections of nodules harvested at the indicated dpi. Red fluorescence from the tdTomato reporter gene expressed in A. rhizogenes transformed roots and green fluorescence from SYTO13 staining were observed. Magnification = 5×. B and C, Nodule perimeter (B) and SYTO13 fluorescence intensity per infection area (C) were calculated using the ImageJ program. Values represent averages ± sd from 10 replicate nodule images per condition. Different lowercase letters in B indicate statistically different groups (ANOVA, P < 0.001); values from C were not statistically different.
We assessed if increased nodulation in OE172 common bean plants could be related to alterations in the AON. In soybean, AON involves long-distance signaling requiring the interaction of RHIZOBIA-INDUCED CLE peptides (RIC1/RIC2), with NODULE AUTOREGULATION RECEPTOR KINASE (NARK) in the leaf and the subsequent inhibition of nodulation via the production of a shoot-derived inhibitor. For local nitrate inhibition, the nitrate-induced CLE peptide (NIC1) interacts with NARK in the root, leading to a nitrate-induced inhibitor (Reid et al., 2011a). The homologous common bean RIC and NIC genes RIC1 (Phvul.005G096900), RIC2 (Phvul.011G135900), and NIC1 (Phvul.005G097000) were identified from the Pv GEA (O’Rourke et al., 2014). As in soybean (Reid et al., 2011a), the common bean RIC1 genes were expressed in inoculated common roots at early stages of rhizobial infection, while RIC2 was expressed at later time points in prefixing and mature nodules. Figure 6, H and I, shows the expression levels of RIC1 and NIC1 genes in 48-hpi inoculated roots from OE172, EV, and OEAP2m plants. RIC1 expression in control (EV) transgenic roots indicates the rhizobial induction of CLE-derived peptides for AON. Interestingly, the level of RIC1 was decreased significantly in OE172 inoculated roots that showed increased nodulation, while it was increased significantly in OEAP2m roots with diminished nodulation (Fig. 6H). As expected, the expression of NIC1 was low in the transgenic inoculated roots (Fig. 6I) under nitrogen-free conditions. These data point to the involvement of a common bean AON mechanism in the miR172 control of nodulation.
miR172c Overexpression Decreases the Sensitivity to Nitrate Inhibition of Rhizobial Symbiosis
Nitrogen (nitrate or ammonia) in the soil perceived by legume plants is an important external stimulus that inhibits nodulation as part of the AON mechanism. Considering the improved rhizobial infection and nodule development/function in common bean plants with increased miR172c (Figs. 6–8), we assessed if these alterations could be related to a decreased sensitivity to external nitrate inhibition of R. etli nodulation (Fig. 9). For this experiment, we applied a low nitrate concentration (1 mm) to R. etli-inoculated OE172, EV, and OEAP2m composite plants. The nodulation of inoculated OEAP2m plants, with low miR172c, was completely abolished when low nitrate was added. For this reason, we analyzed plants overexpressing miR172c as compared with control (EV) plants (Fig. 9). As expected for nitrate inhibition of the rhizobial infection process, the expression level of most early nodulation genes was reduced in EV-inoculated plants in the presence of nitrate (Fig. 9, A–F) as compared with the nitrogen-free condition (Fig. 6, B–G). Notably, in the presence of nitrate, the early nodulation genes NF-YA1, NSP2, CYCLOPS, ENOD40, and FLOT2 showed significantly increased expression in OE172 inoculated roots (Fig. 9, A–F). In fact, the expression level of early nodulation genes in OE172 inoculated roots was similar in the absence (Fig. 6, B–G) or presence (Fig. 9, A–F) of nitrate. As expected, while the RIC1 gene was barely detectable, NIC1 was expressed in the responsive root zone of EV plants inoculated in the presence of nitrate, and it was increased in OE172 roots (Fig. 9, G–H).
Figure 9.
miR172c controls the sensitivity to nitrate inhibition of rhizobial symbiosis. To analyze the effect of nitrate in the symbiosis of R. etli-inoculated OE172 or EV composite plants, 1 mm KNO3 was added to the nutrient solution used to water each set of plants daily. A to H, Expression analysis of selected early nodulation genes determined at 48 hpi in the root responsive zone. Gene identifiers are indicated in the legends to Figures 3 and 6. Values represent averages ± sd from three biological replicates and two technical replicates each. Expression level refers to gene expression, based on Ct value, normalized with the expression of the housekeeping UBC9 gene. I and J, Nodules were counted (I) and nitrogenase (Nase) activity (J) was determined at the indicated dpi. Values represent averages ± sd from five replicate samples per time point. Asterisks indicate the level of statistically significant difference, if any, among values from OE172 and EV roots (Student’s t test, P < 0.01).
Nitrate inhibition of nodulation was evident in control (EV) plants that presented delayed and diminished nodulation (Fig. 9I). These plants also showed decreased nitrogenase activity and PEPc expression and increased ENOD55 (Fig. 9J; Supplemental Table S4) as compared with nitrogen-free inoculated plants (Fig. 7B). Notably, inoculated OE172 plants in the presence of nitrate showed few active nodules at 14 dpi, while at 21 dpi they had a similar number of mature nodules with slightly higher nitrogenase activity as compared with plants inoculated without nitrogen (Figs. 7 and 9, I and J; Supplemental Table S4). In addition, we observed that a higher nitrate concentration (3 mm) totally blocked the nodulation of EV plants, while OE172 plants were able to form active nodules (approximately 100 per root) at 21 dpi.
Exploring the Downstream AP2-1 Regulation in SNF Plants
TFs from the AP2 superfamily are widespread in plants and control diverse developmental programs and stress responses. Different AP2 family members have been classified as activators or repressors of specific target genes (Licausi et al., 2013). In this work, we aimed to predict target genes for AP2-1 transcriptional activation or repression by analyzing data from the root and nodule libraries reported in the Pv GEA (O’Rourke et al., 2014), especially those from young roots and mature effective nodules that were derived from common bean tissues similar to those analyzed in this work. However, a caveat of this analysis is that the Pv GEA does not include libraries from roots inoculated with rhizobia for short periods, so we could not predict AP2-1 targets that would be regulated during the rhizobial infection process.
As shown in Figures 2 and 4, AP2-1 showed high expression in common bean roots as opposed to mature nodules. We hypothesized that genes with an expression pattern similar to AP2-1 are likely to be involved in root function/development and to be positively regulated by AP2-1, thus providing information on a possible mechanism of action of miR172/AP2-1. We identified 114 genes that had an expression pattern similar to AP2-1, designated AP2-1 coexpressed genes (Supplemental Fig. S6). In order to support the latter contention, we searched for TFBS in the 5′ promoter region of AP2-1 coexpressed genes. Besides WRKY, the most statistically overrepresented (P = 0 and 0.001) TFBS were ETHYLENE-RESPONSIVE FACTOR2 (ERF2) and DEHYDRATION-RESPONSIVE ELEMENT BINDING 1B (DREB1B), which belong to the AP2 superfamily. These TFBS were identified in 82% of AP2-1 coexpressed genes (Supplemental Table S5).
Sixty-seven of the 114 AP2-1 coexpressed genes could be assigned to a Gene Ontology (GO) category (Table I; Supplemental Table S5). The most statistically significant (P = 0.006) assigned GO category, which included 19 coexpressed genes, is GO:0004672, associated with protein kinase activity (Table I). We validated by qRT-PCR the expression of protein kinase activity genes in roots and nodules of control (EV) plants and also of plants overexpressing AP2-1. We hypothesized that those genes positively targeted by AP2-1, with high expression in roots and low expression in nodules from wild-type or control EV plants (Supplemental Fig. S6), would show a different expression pattern, higher and/or similar in roots and nodules, in OEAP2m plants that have constitutively enhanced expression of the AP2-1 transcriptional regulator. Table II shows six AP2-1 coexpressed genes assigned to the protein kinase activity category (GO:0004672) whose expression levels agree with our hypothesis. In control (EV) plants, these genes were highly expressed in roots as compared with nodules, similar to AP2-1, thus validating the Pv GEA data (O’Rourke et al., 2014; Supplemental Fig. S6), while they showed an increased and/or similar expression in both tissues from OEAP2m plants (Table II). Interestingly, these genes shared the DREB, ERF, or both TFBS in their promoter regions (Table II; Supplemental Table S5). Transcriptomic analyses of M. truncatula nodule senescence have shown that protein kinases genes are one of the highly induced gene classes at different stages of this process (Van de Velde et al., 2006; Pérez Guerra et al., 2010). On this basis, we asked if the some of the AP2-1 coexpressed genes from the protein kinase activity GO category might be related to nodule senescence in common bean. As shown in Table II, four of the protein kinase activity genes analyzed (Phvul.002G326600, Phvul.007G049500, Phvul.007G049000, and Phvul.006G174500) were induced in senescent (35 dpi) as compared with mature (18 dpi) nodules, thus indicating their possible relation to common bean nodule senescence.
Table I. GO categories statistically overrepresented for genes coexpressed with AP2-1 in roots.
| GO Identifier | Description (Molecular Function) | P |
|---|---|---|
| GO:0004672 | Protein kinase activity | 0.006 |
| GO:0008703 | 5-Amino-6-(5-phosphoribosylamino)uracil reductase activity | 0.016 |
| GO:0005516 | Calmodulin binding | 0.018 |
| GO:0005471 | ATP:ADP antiporter activity | 0.024 |
| GO:0004674 | Protein Ser/Thr kinase activity | 0.036 |
| GO:0004351 | Glu decarboxylase activity | 0.055 |
| GO:0004435 | Phosphatidylinositol phospholipase C activity | 0.055 |
Table II. Selected genes coexpressed with AP2-1 from the statistically overrepresented GO:0004672 category: protein kinase activity.
Expression level was determined by qRT-PCR from 21-dpi mature nodules (N) and roots (R) of EV and OEAP2m R. etli-inoculated composite plants and from 18 dpi mature or 35 dpi senescent nodules (N) from R. etli-inoculated wild-type plants. Values represent averages ± sd from three independent biological replicates and two technical replicates. TFBS for ERF and DREB (subfamilies of the AP2 TF family) were identified as statistically overrepresented in the promoter sequence of each gene as indicated.
| Gene Identifiera | Annotationa | TFBS | Expression Level |
|||||
|---|---|---|---|---|---|---|---|---|
| 21 dpi |
Wild Type |
|||||||
| EV |
OEAP2m |
18 dpi |
35 dpi |
|||||
| R | N | R | N | N | N | |||
| Phvul.002G326600 | Aminocyclopropane carboxylate oxidase | DREB | 0.76 ± 0.08 | 0.35 ± 0.13 | 1.2 ± 0.06 | 1.03 ± 0.07 | 0.11 ± 0.01 | 1.03 ± 0.07 |
| Phvul.007G049500 | Ser/Thr protein kinase | ERF | 0.61 ± 0.09 | 0.25 ± 0.03 | 0.93 ± 0.08 | 0.97 ± 0.09 | 0.10 ± 0.02 | 0.53 ± 0.05 |
| Phvul.007G049000 | Ser/Thr protein kinase | DREB | 0.10 ± 0.007 | 0.043 ± 0.01 | 0.02 ± 0.003 | 0.03 ± 0.004 | 0.07 ± 0.009 | 0.21± 0.03 |
| Phvul.006G174500 | Glycogen synthase kinase-3α | DREB | 0.45 ± 0.06 | 0.24 ± 0.03 | 0.71 ± 0.07 | 0.57 ± 0.06 | 0.16 ± 0.02 | 0.37 ± 0.03 |
| Phvul.011G169600 | Ser/Thr protein kinase | DREB | 0.35 ± 0.02 | 0.18 ± 0.02 | 0.52 ± 0.03 | 0.47 ± 0.03 | 0.33 ± 0.03 | 0.36 ± 0.02 |
| Phvul.008G263900 | Ser/Thr protein kinase | ERF and DREB | 1.2 ± 0.06 | 0.15 ± 0.04 | 0.57 ± 0.02 | 0.50 ± 0.09 | 0.0023 ± 0.0003 | 0.0019 ± 0.0002 |
The root/nodule expression pattern of AP2-1 was opposite that of miR172c, which showed highest expression in mature effective nodules and very low expression in young roots (Figs. 2 and 4). O’Rourke et al. (2014) have described a set of 402 common bean genes highly expressed in mature effective nodules as compared with all other tissues analyzed in the Pv GEA. These genes are likely involved in the establishment of symbiosis and SNF as supported by their assigned GO categories (O’Rourke et al., 2014). Here, we hypothesized that these nodule-enhanced genes, lowly expressed in young roots with increased AP2-1 (Figs. 2 and 4), are candidates for AP2-1 negative regulation. To this end, we searched for TFBS in the 5′ promoter region of nodule-enhanced genes, but we did not find a statistical overrepresentation of AP2 TFBS.
DISCUSSION
The key role of the miR172 node in Arabidopsis flowering time and phase transition is well known; similar roles have also been documented in maize (Zea mays), rice (Oryza sativa), and barley (Hordeum vulgare; Zhu and Helliwell, 2011). Besides conserved roles, specialized/particular species-specific functions of miR172, such as the induction of tuberization in potato (Solanum tuberosum), have been reported (Martin et al., 2009). In legumes, conserved roles of the miR172 node have been documented for L. japonicus (control of flowering time; Yamashino et al., 2013) and soybean (control of juvenile-to-adult phase transition; Yoshikawa et al., 2013). In addition, the control of nodulation during the rhizobia symbiosis has been proposed as a family-specific acquired function of miR172 in different legumes and has been demonstrated for soybean (Yan et al., 2013; Wang et al., 2014). In this work, we identified the miR172 node as a relevant regulator of rhizobial infection and nodulation in common bean.
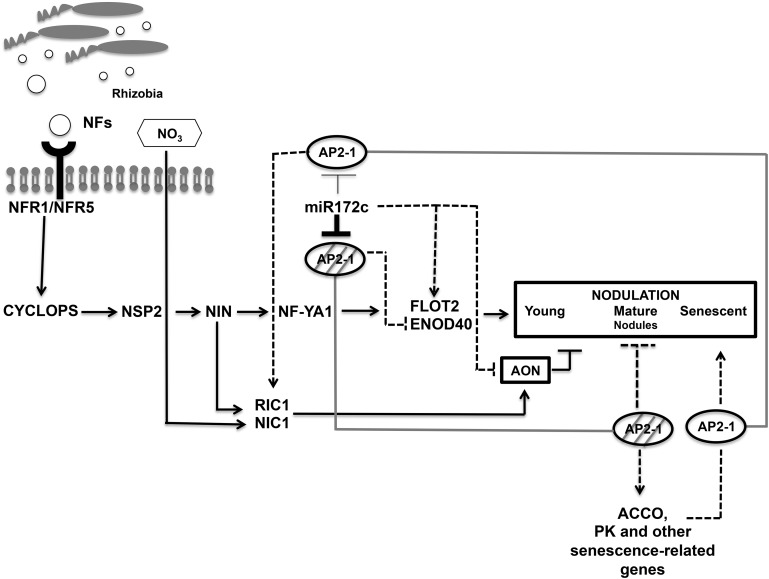
We propose that different miR172 isoforms regulate different processes: miR172b is involved in flowering, while miR172c mainly regulates nodulation. Our data indicate that these miR172 isoforms exert their effects by silencing different target genes from the AP2 TF superfamily. Transcripts from two AP2 genes (Phvul.005G138300 and Phvul.011G071100) are likely to function in roots and are cleaved by miR172c in nodules. Three other AP2 genes (Phvul.003G241900, Phvul.002G16900, and Phvul.001G174400) are likely to function in young flowers, which showed a high level of these transcripts as well as of miR172, thus suggesting that, in flowers, the AP2 target genes are silenced by miR172-induced translation inhibition, similar to Arabidopsis (Chen, 2004). Our work focused on the analysis of the miR172c/AP2-1 (Phvul.005G138300) node in the common bean-rhizobia nitrogen-fixing symbiosis, and our proposed regulatory model is summarized in Figure 10.
Figure 10.
Model of miR172 node regulation in common bean-rhizobia symbiosis. Positive regulation is represented with arrows and negative regulation with lines. The root signaling cascade, triggered by rhizobial NF, is essential for rhizobia infection and nodule development in different legumes. Regulation of common bean rhizobial infection and nodulation by miR172c and AP2-1 are represented by dashed arrows or lines. A high level of miR172c (thick line) induces AP2-1 degradation (hatched circle), while active AP2-1 (circle) is present when the miR172c level is very low (thin line). miR172c positively regulates early nodulation gene expression and rhizobial infection, silencing AP2-1 that may repress ENOD40 expression. Nodule number is positively regulated by miR172c; AON decreases through low RIC1/NIC1 expression, perhaps regulated by AP2-1. In mature nodules, abundant miR172c silences AP2-1, an activator of senescence-related genes that are further required during nodule senescence when AP2-1 levels are recovered. ACCO, Aminocyclopropane carboxylase oxidase.
In Arabidopsis, miR156 represses miR172 expression by targeting SPL TFs that directly bind to the MIR172 promoter and positively regulate its expression (Wu et al., 2009). Transgenic soybean roots overexpressing miR156 showed a reduction in nodulation, decreased miR172 level, and decreased expression of two SPL genes proposed as miR156 targets, although evidence for the binding of SPL to MIR172 promoters for transcription activation was not provided (Yan et al., 2013). Recently, Wang et al. (2015) reported that the overexpression of miR156 affects several aspects of plant architecture in L. japonicus, including underdeveloped roots and reduced nodulation, which correlate with the repression of several early symbiotic genes. However, the authors did not analyze a possible regulation of miR172 by miR156, which may be related to the miR156 effects in nodulation that they showed (Wang et al., 2015). In common bean, we observed opposite levels of mature miR156a as compared with miR172 and also a negative correlation between miR156a and its validated target SPL6 gene (Phvul.009G165100). However, we could not identify SPL TFBS in any of the promoter regions of the six MIR172 loci from the common bean genome, so it is difficult to propose SPL as a direct transcriptional regulator of miR172. However, the binding of SPL proteins to yet unknown sequence motifs present in MIR172 promoters cannot be ruled out. Alternatively, miR156a may exert its negative regulation over common bean miR172 through other target genes not yet identified. For example, transcripts coding for tryptophan-aspartic acid repeats protein domain proteins, which may be involved in microtubule organization, protein-protein and protein-DNA interactions, or chromatin conformation, have been validated as miR156 targets in M. truncatula and L. japonicus, but their specific regulatory function has not been analyzed (Naya et al., 2010; Wang et al., 2015).
In this work, we showed that miR172c has a positive effect on root development independent of rhizobium infection. In addition, miR172c is relevant for the control of rhizobial infection. This miRNA increased after 6 h in R. etli-inoculated roots, when infection threads are formed, and this is related to the increase in root hair deformation observed in plants that overexpress miR172c. Preliminary data indicate that the roots overexpressing AP2m induce irregular infection threads (B. Nova-Franco, O. Valdés-López, and G. Hernández, unpublished data). Together, these data indicate a regulatory/signaling role of miR172c in the rhizobial infection of common bean (Fig. 10). Up-regulation of miR172c was concomitant with that of early nodulation genes, mainly expressed in the cortical cells, that are involved in infection thread initiation/progression (i.e. FLOT2 and ENOD40) and act downstream of NSP2, NIN, and NF-YA1 (Murray, 2011; Oldroyd, 2013), whose expression was highest after 3 h of rhizobial inoculation of common bean roots. Therefore, we propose that miR172c-mediated control of rhizobial infection is exerted at the level of cortical cell division downstream of NF perception, Ca2+ spiking, CCaMK, NSP2, and NIN (Fig. 10). In addition, our data on the repression of RIC1 in roots overexpressing miR172c indicate the involvement of this miRNA in the AON at early stages of the common bean symbiosis. Soyano et al. (2014) reported that the AON L. japonicus CLE root signal genes CLE-RS1 and CLE-RS2, which are orthologous to soybean RIC1 and RIC2, are directly transcribed by NIN, the essential inducer for nodule primordium formation. This constitutes a complex regulatory circuit with a long-distance feedback loop involved in the homeostatic regulation of nodule organ production in L. japonicus (Soyano et al., 2014). In soybean, Wang et al. (2014a) recently reported that NARK negatively regulates miR172 transcription during nodule primordium formation to prevent excess nodulation. In this work, we showed that both NIN and RIC1 are expressed at early stages of rhizobial infection in common bean roots and that OEAP2 roots with decreased nodulation showed increased RIC1 levels. Taken together, these data would indicate a positive regulation of NIN and AP2-1 to RIC1, thus leading to reduced nodulation through AON in common bean (Fig. 10). Sequence analysis of the RIC1 promoter region led us to identify NIN- and DREB/ERF-enriched regions, something that supports the latter contention. However, further work is required to fully demonstrate which TFs activate RIC1 expression in common bean.
The regulation of nodulation through AON signaling is also relevant for the inhibition of nodulation that occurs when nitrate is present in the rhizosphere. For local nitrate inhibition, the nitrate-induced CLE peptide in soybean (NIC) interacts with NARK in the root, leading to a nitrate-induced inhibition of nodulation in soybean (Reid et al., 2011a). Our data indicate that common bean miR172c is a signaling component of the nitrate-induced AON (Fig. 10). In the presence of nitrate, rhizobia-inoculated roots that overexpress miR172c developed more active nodules and showed very low expression of NIC1 that correlates with the up-regulation of NF-YA1, NSP2, CYCLOPS, ENOD40, and FLOT2 early symbiotic genes. The expression of NIN was similar in EV and OE172 roots inoculated in the absence or presence of nitrate, which is in agreement with data from L. japonicus reported by Soyano et al. (2015). The legume-rhizobia symbiosis with increased resistance to soil nitrate is relevant for improving plant growth and crop production. Our better understanding of the elements involved in the control of this phenomenon, such as miR172c in common bean, opens the possibility to exploit it for the future improvement of symbiosis.
The effect of miR172c in rhizobial infection and nodulation of common bean is likely to be directly exerted by its target gene, the AP2-1 transcriptional regulator. TFs from the AP2 superfamily may function as repressors or as activators of transcription (Licausi et al., 2013). Work recently published by Yan et al. (2013) and Wang et al. (2014) about the mechanism of action of AP2 in soybean nodulation points to the repressor role of AP2. In this work, we explored the possible role of AP2-1 as a transcriptional activator and/or repressor of genes relevant for common bean rhizobium infection and nodulation. Recently, Soyano et al. (2015) reported that in L. japonicus, the NIN TF could repress or activate transcription in different scenarios of rhizobial infection in the presence or absence of nitrate. They postulated that such dual regulatory functions might depend on specific coactivator or corepressor molecules that would interact with the same NIN TF in different scenarios.
We identified genes that significantly coexpress with AP2-1 and are candidates for transcriptional activation by this TF (Fig. 10). Several of these genes were assigned to the protein kinase activity GO category. The best candidates are four protein kinases that are repressed in mature nodules and induced in roots and in senescent nodules of control plants, whereas they show high and/or constitutive expression in roots and nodules of AP2m overexpressing plants. This group includes the Phvul.007G049500 gene annotated (www.phytozome.net/commonbean.php) as a Ser/Thr protein kinase with a Cys-rich receptor-like protein kinase, Domain of Unknown Function26 (transmembrane), and Ser/Thr protein kinase domains. We found that this common bean kinase gene is similar (48%) to the Arabidopsis CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE29 (CRK29) gene and to M. truncatula SymCRK (52% similarity) and has domains characteristic of the Cys-rich kinase family. Several members of this family are induced during M. truncatula nodule senescence (Van de Velde et al., 2006; Pérez Guerra et al., 2010). Specifically, SymCRK is involved in senescence and defense-like reactions during the M. truncatula-Sinorhizobium meliloti symbiosis (Berrabah et al., 2014). Another gene from this group encodes the aminocyclopropane carboxylase oxidase, the ethylene-forming enzyme. This and other genes encoding enzymes from the ethylene biosynthetic pathway are up-regulated during M. truncatula nodule senescence (Van de Velde et al., 2006). Ethylene plays a positive role in nodule senescence and also a significant inhibitory role in rhizobial infection and nodule formation (Van de Velde et al., 2006; Murray, 2011). Our interpretation of these results is that AP2-1 transcriptional regulation is important in common bean roots but that this TF needs to be silenced for an adequate nodule function (SNF), something that is achieved by posttranscriptional target cleavage mediated by miR172c. AP2-1 silencing in effective mature nodules would maintain functionality by avoiding senescence through the down-regulation of nodule senescence genes activated by this TF (Fig. 10). Ineffective nodules where miR172c is not induced and AP2-1 remains elevated, as well as OEAP2 nodules, showed early senescence. Alternatively, other protein kinases, proposed as AP2-1 targets, may participate in signaling pathways important for root development or nodule senescence. Nodule-specific protein kinases essential for signaling pathways during initial stages of nodulation are known (Oldroyd and Downie, 2008; Kouchi et al., 2010; Murray, 2011; Oldroyd, 2013).
Regarding AP2 transcriptional repression, Wang et al. (2014) recently reported that soybean NNC1, a miR172 target, represses ENOD40 expression, which results in negative regulation of early stages of rhizobial symbiosis. In this work, we showed that the expression of common bean ENOD40 is decreased in rhizobia-inoculated roots that overexpress AP2m. In addition, we identified ERF- and DREB-enriched regions in the ENOD40 5′ promoter region. So it is conceivable that, as in soybean, common bean ENOD40 expression is repressed by AP2-1 (Fig. 10). However, it is important to consider that soybean NNC1 is not the ortholog of common bean AP2-1. We identified NNC1 (glyma12g07800) as the ortholog (84% similarity) of the Phvul.011G071100 AP2 gene, identified as a miR172 target in a common bean degradome analysis (D. Formey, L.P. Íñiguez, P. Peláez, Y.F. Li, R. Sunkar, F. Sánchez, J.L. Reyes, and G. Hernández, unpublished data) but not further analyzed in this work; while common bean AP2-1 was identified as the ortholog (93% similarity) of the soybean AP2 gene glyma15g04930 that was predicted but not validated as the miR172 target (Wang et al., 2014). Therefore, different miR172 target genes from the AP2 family analyzed in soybean and in common bean may have different mechanisms for transcriptional regulation.
In addition, Yan et al. (2013) postulated that the regulation of soybean nodulation by miR172 is explained by the AP2 repression of nonsymbiotic hemoglobin (Hb) gene expression that is essential for regulating the level of nodulation; however, the authors did not provide evidence demonstrating AP2 binding and direct transcriptional repression of Hb genes. To explore if this circuit is functioning in common bean, we first identified Hb genes encoded by the common bean genome: five symbiotic leg-hemoglobin (Lb) genes having greatly increased expression in effective nodules and four nonsymbiotic Hb genes (O’Rourke et al., 2014; Supplemental Fig. S7). From the latter, Phvul011G048600 and Phvul.011G048700 were identified as orthologs of the nonsymbiotic Hb-1 and Hb-2 genes in soybean, respectively. In common bean, these Hb genes showed similar low expression in roots and nodules of wild-type plants and also in composite plants that overexpress AP2-1 or that have very low AP2-1 resulting from miR172 overexpression (Supplemental Fig. S7). Therefore, our data differ from those of Yan et al. (2013) and lead us to conclude that AP2-1 repression of Hb-1 genes is not relevant for common bean nodulation. Our exploration of other common bean symbiotic genes repressed by AP2-1 included the identification of TFBS statistically overrepresented in the promoter regions of 402 common bean genes reported by O’Rourke et al. (2014) as nodule-enhanced genes. The expression pattern of these genes is similar to that of miR172c and opposite to that of AP2-1, which shows low expression in mature nodules and high expression in roots, suggesting these as candidates for transcriptional repression by AP2-1. However, AP2 (ERF and DREB) TFBS were not overrepresented in these genes. The latter is different from our data from AP2-1 coexpressed genes proposed as being activated by this TF; these genes did show overrepresentation of ERF/DREB TFBS in their promoter regions. Further work is required to demonstrate the direct transcription repression, if any, of the miR172c target gene AP2-1 in common bean.
Legume crops with increased nodulation/decreased nitrate inhibition of nodulation would be relevant for sustainable agriculture. This work sets the basis for further exploration, through genetic/genomic approaches, for common bean cultivars with improved traits resulting from increased miR172 in roots and nodules.
MATERIALS AND METHODS
Identification and Analysis of miR172 Precursor Genes, Isoforms, and Target Genes
The common bean (Phaseolus vulgaris) genome sequence recently published (Schmutz et al., 2014; http://www.phytozome.net/commonbean.php, v1.0) was analyzed, and six miR172 isoforms (a–f) were identified. Of these, four isoforms were described previously through RNA-seq analysis of common bean small RNAs by Peláez et al. (2012). The secondary RNA structure of each miR172 isoform was predicted using mfold software (Zuker, 2003) available at http://mfold.ma.albany.edu, and only the lowest energy structure generated for each sequence was chosen (Supplemental Fig. S1).
Since the only targets identified for miR172 from different plant species belong to the AP2-type TF family, we focused our analysis on identifying common bean miR172 targets within this gene family. We performed target prediction analysis for all the common bean AP2 gene transcripts identified in the Pv GEA (O’Rourke, et al., 2014) using the Web server psRNATarget (http://plantgrn.noble.org/psRNATarget/; Dai and Zhao, 2011). Stringent criteria were used to predict targets; that is, an alignment spanning at least 18 bp with maximum penalty score of 3. Score calculation considered 0.5 points for each G:U wobble, one point for each non-G:U mismatch, and two points for each bulged nucleotide in either RNA strand (Jones-Rhoades and Bartel., 2004). In addition, we constructed a phylogenetic cladogram from the amino acid sequences of common bean AP2 genes; these were aligned using ClustalX version 2.1 (Larkin et al., 2007). The sequence-aligned file was used to construct the bootstrapped neighbor-joining tree using the NJ clustering algorithm and Phylip output format (.phb). The reliability of the phylogenetic analysis was estimated from 1,000 bootstrap resamplings, and the tree was viewed using the program MEGA version 5.2.1 (Tamura et al., 2011).
Plant Material and Growth Conditions
Common bean seeds from the Mesoamerican cv Negro Jamapa 81 were surface sterilized and germinated for 2 d at 26°C to 28°C in darkness. Plants were grown in a hydroponic system under controlled environmental conditions as described previously (Valdés-López et al., 2010). The hydroponic trays contained 8 L of Franco and Munns (1982) nutrient solution. The volume and pH (6.5) of the trays were controlled throughout the experiment. For SNF conditions, plantlets adapted by growth for 7 d in the hydroponic system with nitrogen-free nutrient solution were inoculated with 200 mL of a saturated liquid culture of the Rhizobium etli CE3 wild-type strain or the R. etli fix− mutant strain CFNX247 (ΔnifA::ΩSp/Sm; Girard et al., 1996). Plants were harvested at different times (dpi) for analysis; tissues for RNA isolation were collected directly into liquid nitrogen and stored at −80°C.
To analyze the initial events of rhizobial infection, 2-d-old seedlings were placed in plastic square bioassay dishes (24 × 24 cm; Corning) with solidified nitrogen-free Fähreus medium (Vincent, 1970). Plates containing common bean seedlings were incubated in a growth chamber at 25°C with a 16-h photoperiod. After 2 d, seedlings were inoculated by applying 1 mL of R. etli CE3 saturated liquid culture directly to the root and were further incubated at various times (3–48 h). After specific incubation times, the root responsive zones were detached, frozen in liquid nitrogen, and stored at −80°C until used.
Plasmid Construction, Plant Transformation, and Production of Composite Plants
To generate a plasmid to overexpress the pre-miR172 in common bean transgenic roots, a 217-bp PCR product was obtained using common bean nodule complementary DNA (cDNA) as template and the specific primers Fw-pre172 (5′-CACCCCAGTCACTGTTTGCCGGTGGAG-3′) and R-pre172 (5′-AAAAACCTCCTTTGCTCTGAGCGT-3′), based on the Phvul.001G233200 sequence. The PCR product was cloned by T-A annealing into pCR 2.1-TOPO (Invitrogen) and sequenced. To construct the OE172 plasmid, the pre-miR172 region was excised using the XhoI and BamHI sites of the vector and cloned into the pTDTO plasmid that carries the reporter tdTomato (red fluorescent protein) gene (Aparicio-Fabre et al., 2013; Supplemental Fig. S3).
The complete cDNA clone of common bean AP2-1 was obtained by PCR amplification using cDNA from roots and the specific primers Fw-AP2 (5′-CAGCTACCTTTCCGCCAAATGC-3′) and Rv-AP2 (5′-TAGGCTGGGATGGTGTCTGCAG-3′), based on the Phvul.005G138300 sequence. The 1,127-bp product was cloned by T-A annealing into pCR 2.1 TOPO (Invitrogen) and analyzed by sequencing. Mutations of the putative miR172 cleavage site of AP2-1 were introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The PstI site present in the wild-type miR172 cleavage site (5′-CTGCAGCATCATCAGGATTCT-3′) was eliminated by changing the A to a C at the 3′ position of the recognition site for this enzyme. Additionally, the nucleotides at positions 9, 10, and 11 of the miR172 cleavage site were modified by introducing a BglII site. The primers used were forward (5′-TTCTCTACTGCCGCCAGATCTGGATTCTCAATT-3′) and reverse (5′-AATTGAGAATCCAGATCTGGCGGCAGTAGAGAA-3′). The changes were checked by sequencing, and the modified AP2 was cloned into plasmid pTDTO to obtain plasmid OEAP2m. The nucleotide changes in AP2m introduced an amino acid substitution (Arg for Ser), but this does not seem to affect AP2-1 function (Supplemental Fig. S3).
Common bean composite plants with transformed root system and untransformed aerial system were generated as described (Estrada-Navarrete et al., 2007; Aparicio-Fabre et al., 2013). For plant transformation, Agrobacterium rhizogenes K599 strains bearing the EV, OE172, or OEAP2m plasmids were used. Selected composite plants were grown under controlled environmental conditions in pots with vermiculite and watered daily with B&D nutrient solution (Broughton and Dilworth, 1971), either nitrogen free for the symbiotic condition or with 10 mm potassium nitrate for the full-nutrient condition. SNF plants were adapted by growing in pots for 7 d and then inoculated with R. etli CE3. For experiments designed to analyze the effect of nitrate on symbiosis, B&D nutrient solution supplemented with 1 or 3 mm KNO3 was used to water the inoculated plants daily from 1 dpi. Composite plants were analyzed phenotypically at different dpi; transgenic roots and nodules were collected in liquid nitrogen and stored at −80°C.
To analyze the initial events of rhizobial infection in transgenic roots, plastic square bioassay dishes were used to grow selected composite plants under the same conditions described above. Plates containing composite plants were sealed with Parafilm, and the root zone was covered with aluminum foil. After 2 d, composite plants were inoculated by applying 1 mL of R. etli CE3 saturated liquid culture directly to the root. At 48 hpi, the root responsive zones were detached and stored at −80°C until used or collected into phosphate-buffered saline (PBS) buffer for microscopic analyses.
Phenotypic Analysis
Nitrogenase activity was determined in detached nodulated roots by the acetylene reduction assay essentially as described by Hardy et al. (1968). Specific activity is expressed as nmol ethylene h–1 g–1 nodule dry weight.
The root fresh weight and the number of secondary roots per plant were determined in composite plants grown under full-nutrient (10 d) or symbiotic (21 dpi) conditions.
Microscopic analysis was performed on transgenic nodules at different developmental stages from EV, OE172, and OEAP2m composite plants. The protocol described by Haynes et al. (2004) was used for tissue staining with the nucleic acid-binding dye SYTO13. Nodule sections were stained with SYTO13 (1 µL mL–1) in 80 mm PIPES, pH 7, for 5 min, then mounted in 1% (v/v) PBS/50% (v/v) glycerol and analyzed. Images were obtained using the Zeiss LSM 510 laser scanning microscope attached to an Axiovert 200 M. SYTO13 excitation was obtained at 488 nm using an argon laser and an HFT UV 488/543/633-nm dual dichroic excitation mirror with an LP 560 emission filter for detection. Sequentially, red fluorescence from the reporter gene was observed by exciting at 543 nm with a helium/neon laser, with the same dual dichroic excitation mirror and a BP 500-530 IR emission filter. Images were processed using the LSM 510 version 4.2 5P1 software (Carl Zeiss MicroImaging). For the determination of nodule perimeter and SYTO13 intensity per infected area, 10 images from individual nodule replicates from each condition were analyzed using the ImageJ program.
Statistical analyses of symbiotic parameters (root biomass/architecture, nodulation, and nitrogenase activity) were performed using one-way ANOVA and multiple paired Student’s t tests (P < 0.001).
For analyses of root hair deformation and infection thread induction by rhizobial infection, the root responsive zones from inoculated composite plants grown in plastic square bioassay dishes, as described above, were collected at 48 hpi into PBS buffer. Responsive zone root samples were stained with 0.01% (w/v) Methylene Blue for 1 h and washed three times with double-distilled water; infection events were observed in an optical microscope.
RNA Isolation and Analysis
Total RNA was isolated from 100 mg of frozen nodules, 250 mg of frozen roots, or 200 mg of other frozen tissues from wild-type or composite plants grown under similar conditions, using Trizol reagent (Life Technologies) following the manufacturer’s instructions. These samples were preserved at −80°C until tested. Genomic DNA removal, cDNA synthesis, and quality verification for qRT-PCR were performed as reported (Hernández et al., 2007).
RNA preparations were used to detect mature miR172 in different plant tissues by low-molecular-weight RNA-gel hybridization using 15 μg of total RNA, as reported (Naya et al., 2014). Synthetic DNA oligonucleotides with antisense sequence corresponding to miR172 (5′-ATGCAGCATCATCAAGATTCT-3′) and to U6 snRNA (5′-CCAATTTTATCGGATGTCCCCG-3′) were used as probes after radioactive labeling. Hybridization of U6 snRNA was used as a loading control. The signal intensities of miR172 and U6 hybridization bands were determined using ImageQuant 5.2 software (Molecular Dynamics). Normalized miR172 expression levels were calculated related to U6 snRNA.
For the quantification of transcript levels of mature miRNAs, cDNA was synthesized from 1 μg of total RNA using the NCode miRNA First-Strand cDNA Synthesis Kit (Invitrogen) or the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) for transcripts of selected genes. Resulting cDNAs were then diluted and used to perform qRT-PCR assays using SYBR Green PCR Master Mix (Applied Biosystems), following the manufacturer’s instructions. The sequences of oligonucleotide primers used for qRT-PCR amplification of each gene are provided in Supplemental Table S1. Reactions were analyzed in a real-time thermocycler (Eco Illumina Real-Time PCR System; Illumina) with settings of 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 57°C for 60 s. Relative expression for each sample was calculated with the comparative Ct method. The Ct value obtained after each reaction was normalized with the Ct value of miR159 for miRNA levels or with the Ct value of UBC (Phvul.006G110100) for expression levels of miRNAs and transcripts, respectively.
Statistical analyses of gene expression (miR172c, AP2-1, and early symbiotic genes) from wild-type and composite SNF plants were performed using one-way ANOVA and multiple paired Student’s t tests (P < 0.001).
Identification of Root-Enhanced Genes Coexpressed with AP2-1
To identify common bean genes with an expression pattern similar to that of the AP2-1 target gene, the Euclidian distance between Z scores for each gene was determined in RNA-seq samples from the Pv GEA (O’Rourke et al., 2014). The tissue samples analyzed were as follows: young roots, prefixing effective (5 dpi) nodules, effective (21 dpi) nodules, ineffective (21 dpi) nodules, roots from nonsymbiotic plants grown in full-nutrient solution, nodule-detached roots (5 dpi), effective nodule-detached roots (21 dpi), and ineffective nodule-detached roots (21 dpi). A threshold Euclidian distance of 0.9 was established as significant. A total of 114 genes within the threshold were identified as genes coexpressed with AP2-1 (Supplemental Table S5).
Gene Sequence Analysis for the Identification of TFBS
The CLOVER program (Frith et al., 2004) was used to identify TFBS in 5′ promoter regions of AP2-1 coexpressed genes (Supplemental Table S5) and of genes highly expressed in mature effective nodules described previously by O’Rourke et al. (2014). For this analysis, a 2,000-bp sequence from the region immediately upstream of the transcription start site of each gene was retrieved from the common bean genome sequence (Schmutz et al., 2014; http://www.phytozome.net/commonbean.php, v1.0).
Promoter regions (2,000-bp sequence) of selected early nodulation genes were tested for DREB/ERF or NIN binding sites using http://plants.rsat.eu/. A Markov order of 2 was used for predicting cis-regulatory element-enriched regions with default parameters. The cis-regulatory element-enriched regions were also searched in the promoter region of each locus encoding an miR172 isoform; 1,000 bp upstream of the transcription start site of isoforms a and c and 1,500 bp upstream of the precursors of isoforms b, d, e, and f were analyzed (Supplemental Table S2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. miR172 isoforms encoded in the common bean genome and the most stable secondary structures predicted for their precursors.
Supplemental Figure S2. Symbiotic phenotypes of common bean plants inoculated with the R. etli nifA− mutant strain as compared with the CE3 wild-type strain.
Supplemental Figure S3. Schematic representation of plasmids used for miR172c or AP2-1m overexpression.
Supplemental Figure S4. Overexpression of miR172c and AP2m in transgenic roots and nodules of composite bean plants.
Supplemental Figure S5. miR172 and AP2-1 control rhizobial infection in common bean roots.
Supplemental Figure S6. Expression pattern of AP2-1 coexpressed genes.
Supplemental Figure S7. Expression analysis of common bean symbiotic (Lb) and nonsymbiotic (Hb) hemoglobin genes.
Supplemental Table S1. Primer sequences for qRT-PCR.
Supplemental Table S2. TFBS identified in the 5′ promoter region of each MIR172 gene.
Supplemental Table S3. Nitrogenase activity and expression analysis of marker genes for nodule development.
Supplemental Table S4. Expression analysis of marker genes for nodule development in transgenic nodules of OE172, EV, or OEAP2m plants.
Supplemental Table S5. AP2-1 coexpressed genes: assigned GO categories and identified TFBS.
Supplementary Material
Acknowledgments
We thank Dr. Jesús Arellano (Universidad Autónoma del Estado de Morelos [UAEM]) and María del Socorro Sánchez Correa (Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México) for technical assistance in plant transformation, Dr. Ramón Suárez (UAEM) for cooperation in qRT-PCR determinations, Darla Boydston (Noble Foundation) for helping in the graphic design of the figures, and Dr. Michael Dunn (Centro de Ciencias Genómicas-Universidad Nacional Autónoma de México) for critically reviewing the article.
Glossary
- SNF
symbiotic nitrogen fixation
- NF
nodulation factor
- TF
transcription factor
- AON
autoregulation of nodulation
- miRNA
microRNA
- RNA-seq
RNA sequencing
- Pv GEA
Phaseolus vulgaris Gene Expression Atlas
- dpi
days post inoculation
- qRT
quantitative reverse transcription
- TFBS
transcription factor binding sites
- hpi
hours post inoculation
- EV
empty vector
- GO
Gene Ontology
- cDNA
complementary DNA
- PBS
phosphate-buffered saline
- Ct
threshold cycle
Footnotes
This work was supported by the Dirección General de Asuntos del Personal Académico/Universidad Nacional Autónoma de México (grant nos. PAPIIT: IN209710 and IN210814) and the Consejo Nacional de Ciencia y Tecnología, México (studentship nos. 351615 and 340334 to B.N.-F. and L.P.I., respectively).
Articles can be viewed without a subscription.
References
- Aparicio-Fabre R, Guillén G, Loredo M, Arellano J, Valdés-López O, Ramírez M, Iñiguez LP, Panzeri D, Castiglioni B, Cremonesi P, et al. (2013) Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol 13: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero C, Pérez B, Rabanal F, Blanco-Melo D, De la Rosa C, Estrada-Navarrete G, Sánchez F, Covarrubias AA, Reyes JL (2009) Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol 70: 385–401 [DOI] [PubMed] [Google Scholar]
- Ariel F, Brault-Hernandez M, Laffont C, Huault E, Brault M, Plet J, Moison M, Blanchet S, Ichanté JL, Chabaud M, et al. (2012) Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula. Plant Cell 24: 3838–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Bustos-Sanmamed P, Hartmann C, Lelandais-Brière C, Crespi M (2012) Complexity of miRNA-dependent regulation in root symbiosis. Philos Trans R Soc Lond B Biol Sci 367: 1570–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, et al. (2014) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54: 876–887 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Hernández G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.): model food legumes. Plant Soil 252: 55–128 [Google Scholar]
- Bustos-Sanmamed P, Mao G, Deng Y, Elouet M, Khan GA, Bazin J, Lelandais-Brière C (2013) Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct Plant Biol 40: 1208–1220 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Cubas C, Rabanal FA, Arenas-Huertero C, Ortiz MA, Covarrubias AA, Reyes JL (2012) The Phaseolus vulgaris miR159a precursor encodes a second differentially expressed microRNA. Plant Mol Biol 80: 103–115 [DOI] [PubMed] [Google Scholar]
- Crespi M, Frugier F (2008) De novo organ formation from differentiated cells: root nodule organogenesis. Sci Signal 1: re11. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luis A, Markmann K, Cognat V, Holt DB, Charpentier M, Parniske M, Stougaard J, Voinnet O (2012) Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol 160: 2137–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’haeseleer K, Den Herder G, Laffont C, Plet J, Mortier V, Lelandais-Brière C, De Bodt S, De Keyser A, Crespi M, Holsters M, et al. (2011) Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol 191: 647–661 [DOI] [PubMed] [Google Scholar]
- Dong Z, Shi L, Wang Y, Chen L, Cai Z, Wang Y, Jin J, Li X (2013) Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean nodules by high-throughput sequencing. Int J Mol Sci 14: 2717–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Navarrete G, Alvarado-Affantranger X, Olivares JE, Guillén G, Díaz-Camino C, Campos F, Quinto C, Gresshoff PM, Sanchez F (2007) Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat Protoc 2: 1819–1824 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52: 61–76 [DOI] [PubMed] [Google Scholar]
- Formey D, Sallet E, Lelandais-Brière C, Ben C, Bustos-Sanmamed P, Niebel A, Frugier F, Combier JP, Debellé F, Hartmann C, et al. (2014) The small RNA diversity from Medicago truncatula roots under biotic interactions evidences the environmental plasticity of the miRNAome. Genome Biol 15: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AA, Munns DN (1982) Nodulation and growth of Phaseolus vulgaris in solution culture. Plant Soil 66: 149–160 [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z (2004) Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res 32: 1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard L, Valderrama B, Palacios R, Romero D, Dávila G (1996) Transcriptional activity of the symbiotic plasmid of Rhizobium etli is affected by different environmental conditions. Microbiology 142: 2847–2856 [Google Scholar]
- Hardy RW, Holsten RD, Jackson EK, Burns RC (1968) The acetylene-ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiol 43: 1185–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes JG, Czymmek KJ, Carlson CA, Veereshlingam H, Dickstein R, Sherrier DJ (2004) Rapid analysis of legume root nodule development using confocal microscopy. New Phytol 163: 661–668 [DOI] [PubMed] [Google Scholar]
- Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, et al. (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4: 205–217 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP (2012) The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, Gouzy J, Crespi M (2009) Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng Y, Wu T, Subramanian S, Yu O (2010) Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol 153: 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Magori S, Kawaguchi M (2009) Long-distance control of nodulation: molecules and models. Mol Cells 27: 129–134 [DOI] [PubMed] [Google Scholar]
- Martin A, Adam H, Díaz-Mendoza M, Zurczak M, González-Schain ND, Suárez-López P (2009) Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136: 2873–2881 [DOI] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Naya L, Khan GA, Sorin C, Hartmann C, Crespi M, Lelandais-Brière C (2010) Cleavage of a non-conserved target by a specific miR156 isoform in root apexes of Medicago truncatula. Plant Signal Behav 5: 328–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya L, Paul S, Valdés-López O, Mendoza-Soto AB, Nova-Franco B, Sosa-Valencia G, Reyes JL, Hernández G (2014) Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS ONE 9: e84416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M (2009) Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol 50: 67–77 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- O’Rourke JA, Iniguez LP, Fu F, Bucciarelli B, Miller SS, Jackson SA, McClean PE, Li J, Dai X, Zhao PX, et al. (2014) An RNA-Seq based gene expression atlas of the common bean. BMC Genomics 15: 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez P, Trejo MS, Iñiguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes JL, Sánchez F (2012) Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genomics 13: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Guerra JC, Coussens G, De Keyser A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M (2010) Comparison of developmental and stress-induced nodule senescence in Medicago truncatula. Plant Physiol 152: 1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez M, Graham MA, Blanco-López L, Silvente S, Medrano-Soto A, Blair MW, Hernández G, Vance CP, Lara M (2005) Sequencing and analysis of common bean ESTs: building a foundation for functional genomics. Plant Physiol 137: 1211–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM (2011a) Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact 24: 606–618 [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Hayashi S, Lin YH, Gresshoff PM (2011b) Molecular mechanisms controlling legume autoregulation of nodulation. Ann Bot (Lond) 108: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16: 258–264 [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, et al (2014) A reference genome for common bean and genome-wide analysis of dual domestication. Nat Genet 46: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SA, Meyers BC, Sherrier DJ (2009) MicroRNAs in the rhizobia legume symbiosis. Plant Physiol 151: 1002–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M (2014) Nodule inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc Natl Acad Sci USA 111: 14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Hayashi M (2015) NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus. Plant Cell Physiol 56: 368–376 [DOI] [PubMed] [Google Scholar]
- Streeter J, Wong PP (1988) Inhibition of legume nodule formation and N2 fixation by nitrate. Crit Rev Plant Sci 7: 1–23 [Google Scholar]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, Arunachalam SP, Yu O, Subramanian S (2013) Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol 162: 2042–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Yu O, Subramanian S (2012) Genome organization and characteristics of soybean microRNAs. BMC Genomics 13: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ 31: 1834–1843 [DOI] [PubMed] [Google Scholar]
- Valdés-López O, Yang SS, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernández G (2010) MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol 187: 805–818 [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JCP, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S (2006) Aging in legume symbiosis: a molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JMA (1970) Manual for the Practical Study of Root Nodule Bacteria. IBP Handbook No. 15, Blackwell Scientific Publications, Edinburgh
- Wang Y, Li P, Cao X, Wang X, Zhang A, Li X (2009) Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem Biophys Res Commun 378: 799–803 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang L, Zou Y, Chen L, Cai Z, Zhang S, Zhao F, Tian Y, Jiang Q, Ferguson BJ, et al. (2014) Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26: 4782–4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z, Amyot L, Tian L, Xu Z, Gruber MY, Hannoufa A (2015) Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus. Mol Genet Genomics 290: 471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Yamawaki S, Hagui E, Ueoka-Nakanishi H, Nakamichi N, Ito S, Mizuno T (2013) Clock-controlled and FLOWERING LOCUS T (FT)-dependent photoperiodic pathway in Lotus japonicus. I. verification of the flowering-associated function of an FT homolog. Biosci Biotechnol Biochem 77: 747–753 [DOI] [PubMed] [Google Scholar]
- Yan Z, Hossain MS, Wang J, Valdés-López O, Liang Y, Libault M, Qiu L, Stacey G (2013) miR172 regulates soybean nodulation. Mol Plant Microbe Interact 26: 1371–1377 [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Ozawa S, Sentoku N, Itoh J, Nagato Y, Yokoi S (2013) Change of shoot architecture during juvenile-to-adult phase transition in soybean. Planta 238: 229–237 [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289: 3–16 [DOI] [PubMed] [Google Scholar]
- Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62: 487–495 [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.