The difference in flesh-rind transport of nutrients and water due to the anatomic structural differences among citrus varieties might be an important factor that influences fruit senescence behavior.
Abstract
Citrus (Citrus spp.), a nonclimacteric fruit, is one of the most important fruit crops in global fruit industry. However, the biological behavior of citrus fruit ripening and postharvest senescence remains unclear. To better understand the senescence process of citrus fruit, we analyzed data sets from commercial microarrays, gas chromatography-mass spectrometry, and liquid chromatography-mass spectrometry and validated physiological quality detection of four main varieties in the genus Citrus. Network-based approaches of data mining and modeling were used to investigate complex molecular processes in citrus. The Citrus Metabolic Pathway Network and correlation networks were constructed to explore the modules and relationships of the functional genes/metabolites. We found that the different flesh-rind transport of nutrients and water due to the anatomic structural differences among citrus varieties might be an important factor that influences fruit senescence behavior. We then modeled and verified the citrus senescence process. As fruit rind is exposed directly to the environment, which results in energy expenditure in response to biotic and abiotic stresses, nutrients are exported from flesh to rind to maintain the activity of the whole fruit. The depletion of internal substances causes abiotic stresses, which further induces phytohormone reactions, transcription factor regulation, and a series of physiological and biochemical reactions.
Fruits are usually morphologically classified into different groups, such as silique (e.g. Arabidopsis [Arabidopsis thaliana]), pome (e.g. apple [Malus domestica]), berry (e.g. tomato [Solanum lycopersicum]), and hesperidium (e.g. sweet orange [Citrus sinensis]). Fruit ripening and senescence are inevitable and irreversible processes in the plant life cycle, and the underlying mechanisms are unique among different fruit types. According to the amount of ethylene biosynthesis and its signal transduction, fruits are physiologically classified into climacteric fruit (e.g. tomato, apple, and banana [Musa spp.]) and nonclimacteric fruit (e.g. citrus, strawberry [Fragaria spp.], and grape [Vitis vinifera]).
The complex regulation of senescence is one of the most important topics in fruit research. Experimental evidence has shown that transcription factors (TFs) and phytohormones are involved in controlling the fruit senescence process accompanied by physiological changes in color, texture, aroma, and nutritional components (Prasanna et al., 2007; Vicente et al., 2007; Defilippi et al., 2009; Seymour et al., 2013). Studies on the regulatory role of ethylene in fruit senescence have been mainly focused on the climacteric model fruit tomato, including the ethylene biosynthesis/perception by the target ethylene receptors and the signal transduction cascade that involves several TFs such as ethylene-responsive factors (ERFs), ETHYLENE INSENSITIVE3 (EIN3), EIN3-Like, APETALA2 (AP2), and WRKY TFs (Seymour et al., 2013). Meanwhile, abscisic acid (ABA) also plays an important role in ripening regulation. Studies on strawberry (Jia et al., 2011) and tomato (Sun et al., 2012) showed that reduced expression of the key enzyme 9-cis-epoxycarotenoid dioxygenase (NCED) of ABA biosynthesis can down-regulate the expression of ripening-related genes, suggesting that ABA acts as a promoter of ripening. ABA treatment of mango (Mangifera indica variety Alphonso; Parikh et al., 1990) and tomato (Martínez-Madrid et al., 1996) could stimulate ethylene synthesis. Moreover, analysis of the maturation of a sweet orange ABA-deficient mutant revealed a role for ABA in the regulation of citrus fruit coloration (Rodrigo et al., 2003). Furthermore, a large group of TFs have been characterized to be related to senescence, including families of NAC (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF1-2], and cup-shaped cotyledon [CUC2]), WRKY, C2H2-type zinc finger, AP2/Ethylene-Responsive Element Binding Protein (EREBP), MYB, and so on (Lim et al., 2007; Asif et al., 2014).
High-throughput molecular biological techniques like transcriptomic, proteomic, and metabolomic approaches have been widely used to explore aging-related mechanisms in fruits, as reported in tomato (Kok et al., 2008; Karlova et al., 2011; Osorio et al., 2011), pepper (Capsicum annuum; Osorio et al., 2012), grape (Fasoli et al., 2012), peach (Prunus persica; Jiang et al., 2014), apple (Zheng et al., 2013), melon (Cucumis melo; Bernillon et al., 2013), strawberry (Kang et al., 2013), and cassava (Manihot esculenta; Vanderschuren et al., 2014). The combination analysis of different omics data sets by network construction has been used to unravel the regulatory relationship or changes of metabolic pathways during the ripening and senescence process in strawberry (Fait et al., 2008), tomato (Enfissi et al., 2010; Lee et al., 2012), and peach (Lombardo et al., 2011).
The study of senescence in citrus has important scientific and socioeconomic benefits. As one of the most important edible fruits in the world, citrus has tremendous economic impact (Food And Agriculture Organization of the United Nations, FAO statistic, http://faostat3.fao.org). The anatomic structure of citrus fruit is distinctive from that of model fruits such as silique of Arabidopsis or berry of tomato. Hesperidium is a modified berry developed from syncarpous pistil, with soft leathery rind and flesh containing vascular bundles and a mass of segments with juice vesicles (Bennici and Tani, 2004; Ladaniya, 2008). Citrus fruit contains a large number of nutritional components beneficial to health, such as carbohydrates, fibers, vitamin C, vitamin E, provitamin A, pantothenic acid, carotenoids, flavonoids, limonoids, polysaccharides, and phenolic compounds (Silalahi, 2002). Therefore, citrus fruit can be a particularly advantageous model for studying the senescence of woody perennial fruits. After harvest, citrus fruit remains animate and active. The senescence of nonclimacteric citrus fruit is a gradual physiological change process with dysfunction or malfunction of the fruit tissues in response to water, nutrient, and temperature stresses, which ultimately affects the fruit quality. Citrus can be classified into two classes in commercial postharvest practices: tight-skin (hard-peel) citrus (such as sweet orange and pummelo [Citrus grandis]) and loose-skin (easy-peel) citrus (such as Satsuma mandarin [Citrus unshiu] and Ponkan mandarin [Citrus reticulata]). The two classes have different degrees of tightness in flesh-rind anatomic structure and different storage characteristics, as tight-skin citrus fruit has a longer storage life than loose-skin ones. According to empirical data, the commercial storage life of loose-skin citrus fruit is always shorter than 50 d, and the wilt of its flesh is previous to that of the rind during senescence; however, tight-skin citrus can be stored with good consumption quality for half a year, and the decay of its rind is antecedent to that of flesh. With the implementation of citrus genome projects, the genomes of mandarin and sweet orange have been fully sequenced (Xu et al., 2013; Wu et al., 2014), making it possible to analyze the biological mechanism of citrus fruit senescence at the genomic level. The rapid development of high-throughput techniques enables advances in omics-based research by transcriptomics, proteomics, and metabolomics. Analyses from multiple omics perspectives of citrus senescence have been conducted recently in our research group (Zhu et al., 2011; Yun et al., 2013; Ma et al., 2014). With the development of citrus postharvest research, efforts should be focused on a comprehensive and systemic insight into citrus fruit senescence based on high-throughput data.
In this study, the postharvest senescence process of citrus fruit was comprehensively analyzed based on transcriptomic and metabolomic data sets. We chose four major citrus fruit varieties: Satsuma mandarin, Ponkan mandarin, sweet orange (Citrus sinensis), and Shatian pummelo. They were sampled every 10 d during 50 d after harvest (DAH), almost covering the commercial storage period of loose-skin citrus. Additionally, conventional physiological quality was measured and data were collected during 0 to 200 DAH, which covered the whole senescence period of tight-skin citrus. Furthermore, we also compared the expression of genes related to phytohormones during the senescence process of two model fruits (i.e. tomato and grape) with that of citrus. In general, this study was aimed to further uncover the rind-flesh communication of hesperidium, to characterize the differential storage behaviors of different citrus varieties, to reveal the important physiological changes during storage, and to demonstrate the specific nonclimacteric characteristics of citrus fruit.
RESULTS
Profiling of Transcriptome, Metabolome, and Physiological Quality Data Sets
This study included the most economically important citrus varieties in the Asian market, including Satsuma mandarin, Ponkan mandarin, sweet orange, and pummelo. Transcriptomes of 96 individual samples were obtained by microarrays, including two tissue types (rind and flesh) at six time points (0–50 DAH) from the four citrus varieties (Fig. 1; Supplemental Data Set S1). Clustering analysis of 96 microarray data sets provides a bird’s-eye view of the transcriptome data set (Fig. 2). We dissected all the differences of gene expression at three independent levels: differences between four varieties (variety level), differences between rind and flesh (tissue level), and differences between different time stages (time level). The subsequent research was guided by the overall clustering results and was focused on multilevel omics characterization at the three independent levels.
Figure 1.
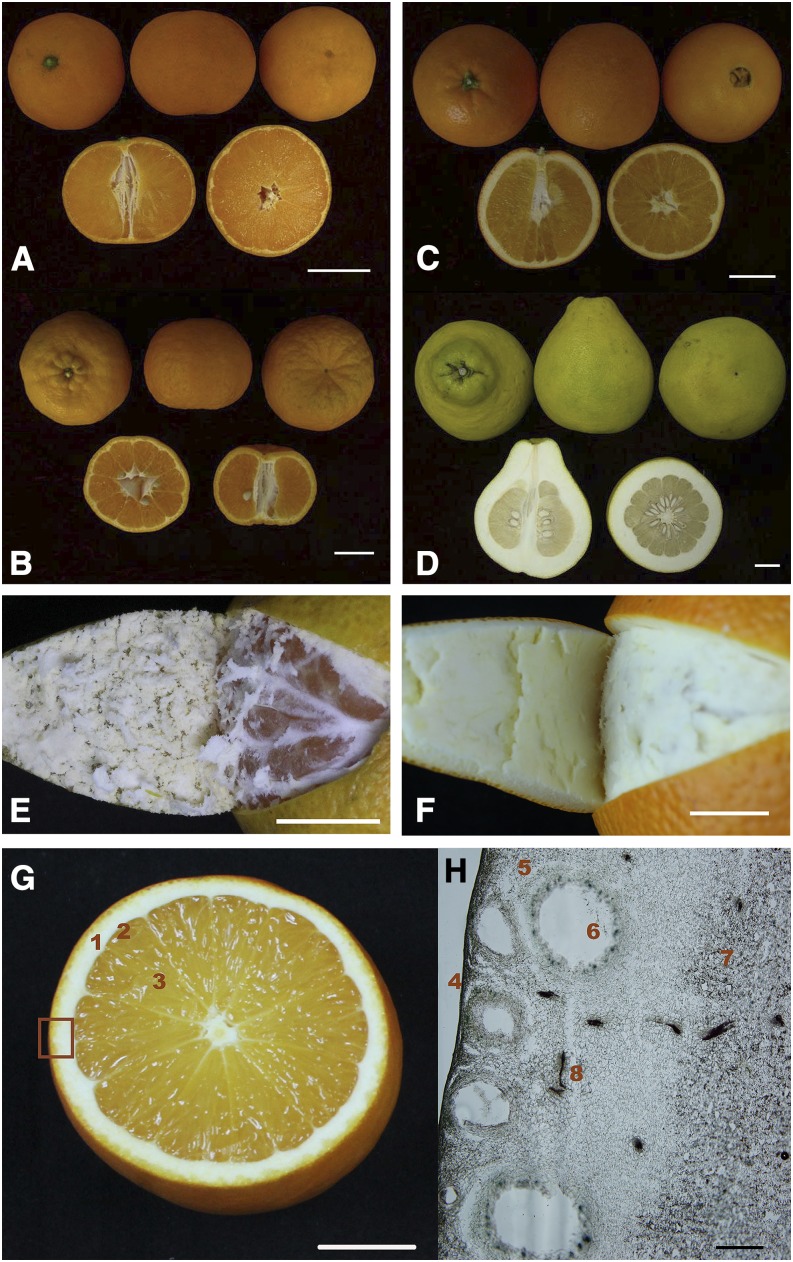
Description of anatomic structures of hesperidium. A to D, Anatomic structures of four citrus varieties: Satsuma mandarin (A), Ponkan mandarin (B), sweet orange (C), and Shatian pummelo (D). Bars = 30 mm. E and F, Appearance of the flesh-rind anatomic structure of loose-skin Satsuma (E) and tight-skin sweet orange (F). Bars = 10 mm. G and H, Detailed anatomical structures of an orange fruit. G, Hesperidium fruit has two major distinct regions, rind (1) and flesh. The flesh comprises segment epidermis (2) and juice vesicles (3). Bar = 20 mm. H, Light microscopy image of orange fruit rind. The rind comprises a series of parts: epidermis (4), flavedo area (5), oil gland (6), albedo area (7), and lignin dye with phloroglucinol-HCl (8). Bar = 0.25 mm. Hesperidium fruits have two distinct tissues: flesh (endocarp or pulp) and rind (pericarp or peel). The flesh is the edible part of most citrus fruits, including juice vesicles with segment membranes and vascular bundles. The rind is further divided into two parts: flavedo (exocarp) is the external chromoplast-rich colored layer, and albedo (mesocarp) is the white layer characterized by numerous intercellular air spaces.
Figure 2.
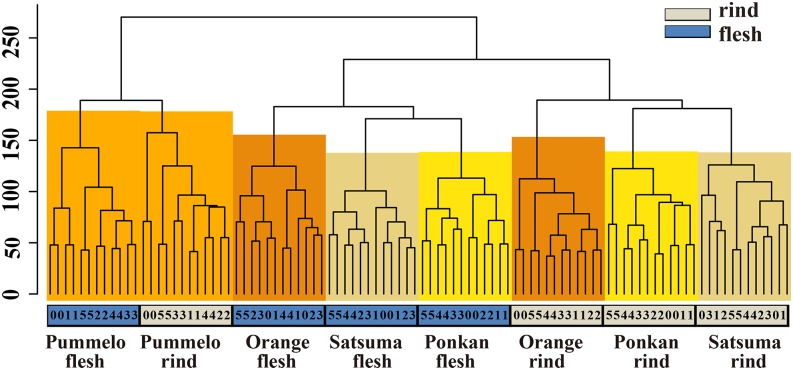
Hierarchical cluster analysis of all microarray expression data sets. The expression values of all probe sets of the 96 microarrays before DEGs identification were analyzed by hierarchical clustering with Euclidean distance. The cluster dendrogram shows the clear distinctions in expression pattern at the tissue, variety, and time levels. The y axis represents the height value of hierarchical clustering. Numbers 0 to 5 represent 0 to 50 DAH.
To characterize the differentially expressed genes (DEGs) during senescence, a total of 11,586 DEGs were identified and analyzed at the variety, tissue, and time levels (Fig. 3; Supplemental Data Set S2). The 96 samples were divided into different groups and subgroups based on the differences at the three levels (Supplemental Table S1). Then, significance analysis of microarrays (Tusher et al., 2001) and biweight value restriction were used to identify the DEGs of each subgroup. First, DEGs at the variety level are shown in Figure 3A: DEGs in Ponkan and Satsuma (rind, 381; flesh, 391) and DEGs in pummelo and sweet orange (rind, 371; flesh, 404) had greater numbers than the other two variety combinations, which is consistent with the commercial classification of tight- and loose-skin citrus fruit. The analysis also showed that pummelo possessed a higher number of unique genes (rind, 1,788; flesh, 2,221) than the other three varieties, which is consistent with the hierarchical dendrogram of the entire transcriptome (Fig. 2). Second, Venn diagrams of DEGs at the tissue level (Fig. 3B) showed that there were similar numbers of DEGs in the rind and flesh. Third, the distribution of DEGs at the time level (Fig. 3C) demonstrated that most varieties showed the maximum numbers of DEGs at the first and last phases except for Ponkan, which had the maximum number of DEGs at 20 DAH. To obtain insights into the functions of DEGs, an enrichment analysis based on Mapman annotation was also conducted at the three levels (Supplemental Data Set S3). Clusters of some significant Mapman gene functions (BINs; hypergeometric P ≤ 0.01) are shown in Figure 3D. Many BINs were identified to be significant at least at one level, including the BINs related to phytohormones, transporters, stresses, and TFs. These genes might play important roles during the postharvest senescence process of citrus fruit. It is noteworthy that, at the tissue level, the numbers of DEGs in the rind and the flesh showed no dramatic difference (Fig. 3B), while the enriched functions were greatly different between the rind and the flesh (Fig. 3D). Many BINs were enriched in the rind, such as AP2/EREBP TFs (BIN 27.3.3), WRKY TFs (BIN 27.3.32), ATP-binding cassette (ABC) transporters (BIN 34.16), amino acid transporters (BIN 34.3), lignin biosynthesis (BIN 16.2.1.1011), dihydroflavonol metabolism (BIN 16.8.3), sulfate transporters (BIN 34.6), ethylene synthesis (BIN 17.5.1.1002), and ABA synthesis (BIN 17.1.1.1002), indicating that the rind is more actively engaged in senescence.
Figure 3.
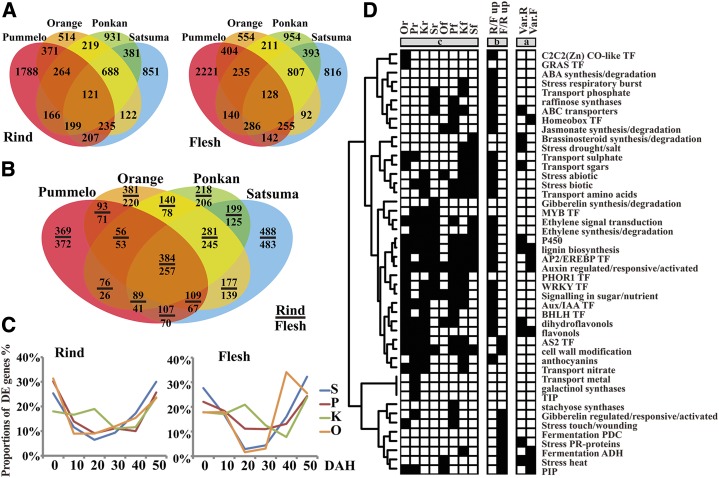
Identification and function enrichment analysis of DEGs at three levels. A and B, Venn diagrams show DEGs at the variety and tissue levels. C, Proportions of DEGs numbers at the time level. D, Gene function enrichment analysis of DEGs based on Mapman annotation at three levels: variety level (a), tissue level (b), and time level (c). Important Mapman gene function BINs with significant DEGs distribution (hypergeometric P ≤ 0.01) are marked in black.
To characterize the metabolism during senescence, the levels of primary and secondary metabolites were detected by gas chromatography (GC)-mass spectrometry (MS) and liquid chromatography (LC)-mass spectrometry in the corresponding citrus samples of microarrays. In total, 64 metabolites were identified (Table I; Supplemental Data Set S4). The primary metabolites included 11 sugars or sugar alcohols, 10 organic acids, six amino acids, and five other primary metabolites. Moreover, 32 secondary metabolites were identified, including flavonoids in particular, such as hesperetin, neohesperidin, and naringin; phenols and terpenoids were also identified, which play important roles in signaling, abiotic stress reaction, and microbial infection (Grassmann et al., 2002). Although the metabolome data set showed complex changes at the time and variety levels, there were distinctive differences in the compositions of metabolites at the tissue level. The levels of Fru, Glc, Suc, citric acid, and almost all secondary metabolites such as flavonoids and terpenoids are higher in the flesh than in the rind, indicating that there is a greater accumulation of nutrients in the flesh.
Table I. List of metabolites detected by GC-MS and LC-MS.
Numbers in parentheses represent isomers detected by GC-MS and LC-MS.
| Primary Metabolites | Secondary Metabolites |
|---|---|
| Sugar and sugar alcohols | |
| Glc | Phe |
| Fru | N-Feruloylputrescine |
| 4-Ketoglucose | Quercetin dihexose-deoxyhexose |
| Gal | Sinapic acid |
| Lyxose | Feruloylquinic acid |
| Man | Naringin |
| Xylofuranose | Naringenin |
| Myoinositol | Naringenin chalcone-hexose |
| Glucopyranose | Quercetin-3-O-rhamnoside |
| Suc | Neohesperidin |
| Glycerol | Hesperetin |
| Organic acids | |
| Citric acid | Naringenin chalcone-dihexose |
| Aconitic acid | Rutin |
| 2-Butenedioic acid | Kaempferol-3-O-rutinoside |
| Malic acid | Phloretin-C-diglycoside |
| Malonic acid | Naringenin chalcone-hexose(2) |
| Oxalic acid | Naringenin(2) |
| Butanedioic acid | Naringin(2) |
| Butanoic acid | Hesperetin(2) |
| Isocitric acid | Hydroxylated naringenin-hexose |
| Pentanoic acid | Hesperetin(3) |
| Amino acids | |
| Asn | Neohesperidin(2) |
| Asp | Isosakuranetin |
| Citrulline | Isosakuranetin-7-rutinoside |
| Gly | Sinensetin |
| Ala | Tetramethyl-o-scutellarein |
| Val | Tetramethyl-o-scutellarein(3) |
| Others | |
| 5-Aminohexanoic acid | Sinensetin(2) |
| 3-Amino-6-methoxypicolinic acid | Tetramethyl-o-scutellarein(2) |
| Cyclohexanone, 2-[(dimethylamino)methyl]- | Tetramethyl-o-scutellarein(4) |
| Hexadecanoic acid | 3-3-4-5-6-7-8-Heptamethoxyflavone |
| Hexadecanoic acid, 2,3-bisoxypropylester | Sinensetin(3) |
Many harvested citrus fruits can be stored for a long period of time. To investigate the physiological behavior of citrus fruit after 50 DAH, the physiological quality data were investigated until 200 DAH. To ensure the reliability of the results, we repeated our data in a total of 12 batches of fruits in 3 years (2009, 2010, and 2012; Supplemental Data Set S5). A principal component analysis individual factor map (Supplemental Figure S1) showed that the time trajectory had a chronological linear distribution in principal component 1, indicating that the loss of physiological quality during long storage is a gradual process. Figure 4A demonstrates the results of the hierarchical cluster analysis of the physiological quality measurement, which divides the physiological quality data set into two independent clusters: one cluster shows downward trends, including acid-related parameters (such as total acid, titratable acid [TA], citric acid, malate, and quinic acid), sugar-related parameters (such as total sugar, total soluble solids [TSS], Fru, Glc, and Suc), and juice yield; the other cluster shows upward trends, including the parameters related to odor components (such as ethanol, methanol, and acetaldehyde), weight loss, total sugar-total acid ratio, and TSS-TA ratio. Furthermore, some important physiological parameters, like respiration rate, sugars, organic acids, and odor components, were investigated in detail. The postharvest senescence process could be roughly divided into three time intervals (TI-1, 0–50 DAH; TI-2, 50–100 DAH; and TI-3,100–200 DAH) based on the fluctuations of the physiological quality data (Fig. 4, B–E). It was found that the physiological change in TI-1 was the most dramatic. The respiration rate increased at 60 to 130 DAH and then decreased after 130 DAH, which might be the result of stress resistance against the rise of environmental temperature at TI-2 and physiological disorders within the fruit at TI-3. The metabolic levels of organic acids and sugars showed sharp increases at TI-1 and TI-2 and decreased gradually thereafter. The odor components generally increased especially at TI-3, which is opposite to the profiles of sugars and acids, indicating that the commercial value of the fruit was greatly lost at the terminal stage. These data provide a foundational description of the storage process of citrus fruit and may guide further experiments on the postharvest physiology of citrus fruit.
Figure 4.
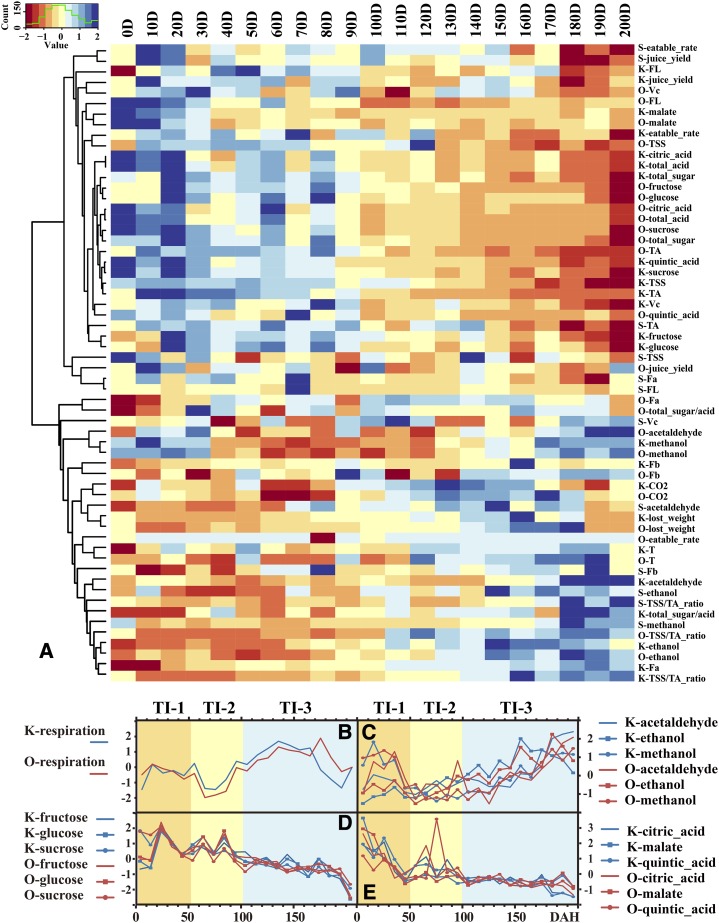
Analysis of conventional physiological quality data. A, Hierarchical cluster analysis of conventional physiological quality data during 200 DAH in sweet orange (O), Ponkan (K), and Satsuma (S). B to E, Important fruit quality indices: respiration rate (B) and levels of odor components (C), reducing sugars (D), and organic acids (E). Data were processed with Z-score normalization, and repeated data were averaged and hierarchically clustered using Spearman distance. CO2, Respiration rate; Fa, Fb, and FL, color parameters; T, laboratory temperature; Vc, ascorbic acid.
Integrated Analysis of Metabolome and Transcriptome Data Sets by the Citrus Metabolic Pathway Network
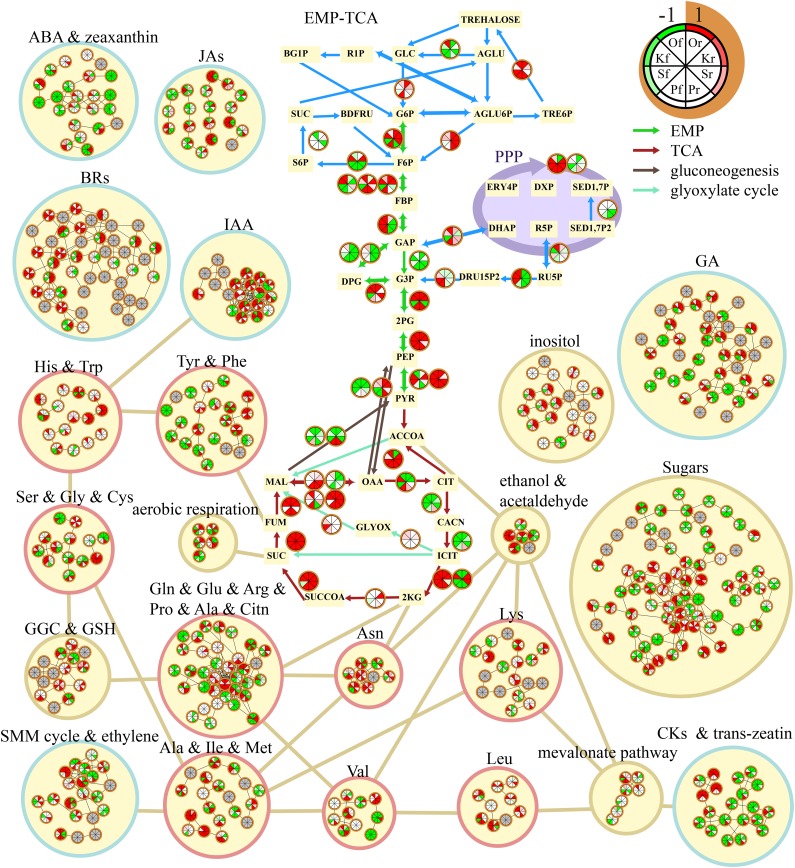
To obtain a global view of citrus postharvest senescence, we constructed a genome-scale Citrus Metabolic Pathway Network (CitrusCyc) using AraCyc, a database containing biochemical pathways of Arabidopsis, developed at The Arabidopsis Information Resource (http://www.arabidopsis.org; Zhang et al., 2005) based on sequence similarities. CitrusCyc integrates the information of 3,490 metabolism-related genes (enzymes) and 2,614 metabolites, aiming to explain the mechanisms of cellular functions in citrus. It contains the major metabolic pathways such as reactions of sugars, acids, lipids, amino acids, and most phytohormones (Supplemental Figure S2). In a metabolic network, a biological reaction can be affected by its adjacent reactions in a pathway. Therefore, in this research, the transcriptome data set of the 96 microarrays was mapped to CitrusCyc by transforming the data to parameters of recalculated expression values of reaction genes (RE-values) using the network-based diffusion method (Allen et al., 2013), considering the influence of neighboring reactions in CitrusCyc (Fig. 5). Three kinds of correlation networks were constructed based on the transcriptome and metabolome information in CitrusCyc: coexpression networks of recalculated expression values (Networks-RR), a correlation network of primary metabolites (Network-MM), and a correlation network between metabolites and recalculated expression values (Network-MR).
Figure 5.
Sketch map of some pathways in CitrusCyc. Colors in each pie represent the RE-values (−1 to 1) of different samples (Or, Of, Kr, Kf, Sr, Sf, Pr, and Pf). 2KG, 2-Ketoglutarate; 2PG, 2-phosphoglycerate; ACCOA, acetyl-CoA; AGLU, α-Glc; AGLU6P, α-Glc-6-P; BDFRU, β-d-Fru; BG1P, β-d-Glc-1-P; CACN, cis-aconitate; CIT, citrate; DHAP, dihydroxy-acetone-phosphate; DRU15P2, d-ribulose-15-bisphosphate; DXP, xylulose-5-phosphate; EMP, glycolysis; ERY4P, erythrose-4-phosphate; F6P, Fru-6-P; FBP, Fru-1,6-bisP; FUM, fumaric acid; G6P, Glc-6-P; GAP, d-glyceraldehyde-3-phosphate; GGC, γ-glutamyl cycle; GLC, Glc; GLYOX, glyoxylate; ICIT, isocitrate; MAL, malate; OAA, oxaloacetate; P3G, 3-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; R1P, Glc-1-P; R5P, Rib-5-P; RU5P, ribulose-5-phosphate; SED1,7-P, d-sedoheptulose-7-phosphate; SED1,7-P2, d-sedoheptulose-1,7-bisphosphate; SMM cycle, S-methyl-Met cycle; SUC, succinic acid; SUCCOA, succinyl-CoA; TCA, tricarboxylic acid cycle; TRE6P, trehalose-6-phosphate.
Networks-RR were constructed (Spearman coefficient ≥ 0.75) and clustered by Markov cluster (MCL) algorithm in the rind and the flesh. The results revealed the modularity characteristics of the networks: the reaction genes belonging to the same pathway always fall into the same subnetwork (Supplemental Data Set S6), including the subnetworks of down-regulated genes (Subr2 in the rind and Subf1 in the flesh) and the subnetworks of up-regulated genes (Subr1 in the rind and Subf2 in the flesh). In the rind, the genes in the pathways related to brassinosteroid biosynthesis, GA biosynthesis, tricarboxylic acid cycle, and glycolysis II were in Subr1, and the genes in the pathways related to chlorophyll, Glu, and starch fell into Subr2. In the flesh, Subf1 contains the genes in the reactions related to starch, ascorbate, plant sterol, and jasmonic acid, and Subf2 comprises the genes for glycolysis, glyoxylate cycle, the tricarboxylic acid cycle, and the biosynthesis of cutin, flavin, and jasmonic acid. The modularity indicates that the metabolic pathways in a subnetwork always cooperate during senescence. Additionally, a Network-MM (Supplemental Data Set S7) was constructed. The network shows frequent negative correlations between organic acids and sugars, and such negative correlations were more frequently observed in Satsuma and Ponkan than in sweet orange and pummelo.
The integrated analysis of metabolome and transcriptome data sets in CitrusCyc was focused on certain important metabolites with significant influences on fruit quality, such as sugars and acids, as well as on the identification of the active reactions of these important metabolites. A Network-MR was constructed by calculating the Jaccard distance (0.4 or greater; Supplemental Data Set S8). A correlation with a high absolute Jaccard distance value means that the metabolite has been catalyzed by a corresponding reaction. The results show that some important organic acids and sugars are involved in different catalytic reactions. There were correlations between malate and malic dehydrogenase (Enzyme Commission [EC] 1.1.1.37), malate and pyruvic-malic carboxylase (EC 1.1.1.40), and citric acid and cleavage enzyme, and the correlations were stronger in the rind than in the flesh. Additionally, correlations were found between Fru and fructokinase (EC 2.7.1.4), Fru and β-fructofuranosidase (EC 3.2.1.26), Suc and Suc fructosyltransferase (EC 2.4.1.99), Suc and β-fructofuranosidase (EC 3.2.1.26), Glc and trehalase (EC 3.2.1.28), Glc and β-d-glucosidase (EC 3.2.1.21), and Glc and 4-α-glucanotransferase (EC 2.4.1.25), while these correlations were stronger in the flesh than in the rind (Supplemental Data Set S8). The statistical data of the numbers of first-level and second-level linkages in Network-MR (Supplemental Table S2) suggest that, for the first-level linkages, organic acids (such as succinate, citric acid, isocitric acid, and malate) had more significantly correlated reactions in the rind, while sugars (such as Glc, Fru, Gal, and Suc) had more significantly correlated reactions in the flesh, and for the second-level linkages, more significant correlated reactions of organic acids were observed in both the rind and the flesh. These results indicate that the reactions of sugars are more active in the flesh, while the reactions of organic acids are more active in the rind and have greater influence on the metabolic network.
Differential Expression Patterns of Transcription Factors among Citrus Varieties
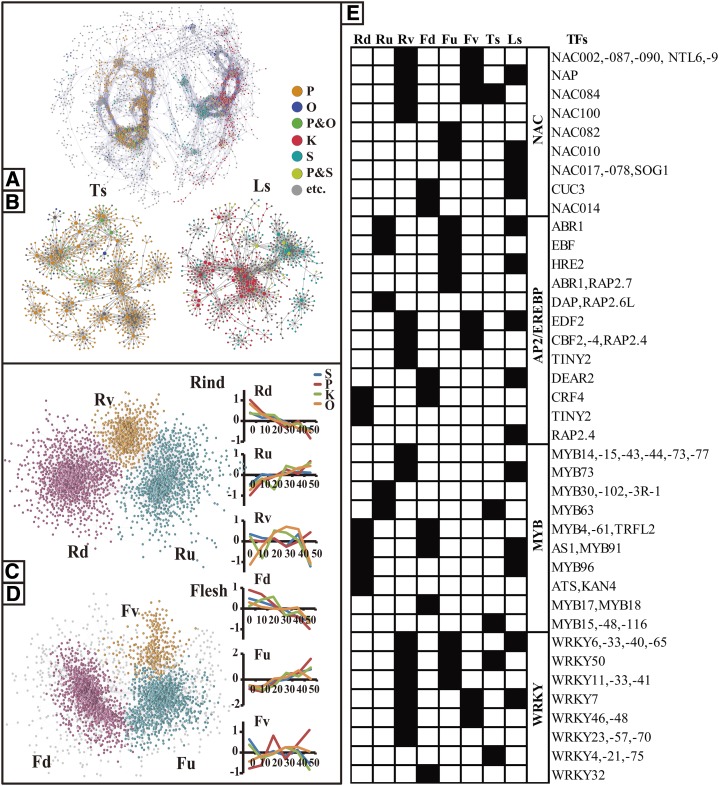
To uncover the differential expression patterns among citrus varieties during the postharvest senescence process, we calculated the Spearman rank correlation coefficient for constructing a coexpression network of DEGs at the variety level (Network-DV; Fig. 6, A and B; Supplemental Data Set S9). The network demonstrates modularity characteristics, since the TF-centered networks fell spontaneously into two clusters: the DEGs highly expressed in pummelo or orange gathered in one subnetwork (tight-skin citrus subnetwork [SubTs]) and the DEGs highly expressed in Ponkan or Satsuma fell into another subnetwork (loose-skin citrus subnetwork [SubLs]). The distinction between these two subnetworks is consistent with the commercial classification of tight- and loose-skin citrus fruits; therefore, it provides an effective way to analyze the gene expression related to variety characteristics and storage properties.
Figure 6.
Overview and TF hubs of Network-DV and coexpression networks of DEGs at the time level (Networks-DT). A and B, Whole Network-DV (A) and its TF-centered subnetworks (B). Network-DV was constructed based on the data set of DEGs at the variety level. The data set was normalized by biweight, resulting in an eight-dimension matrix, and the matrix was calculated by Spearman coefficient (>0.75). Network-DV was unaffectedly divided into two subnetworks: SubTs and SubLs. Bigger nodes represent TF hubs in the subnetworks. Different colors represent the DEGs significant in pummelo (P), orange (O), pummelo and orange (P&O), Ponkan (K), Satsuma (S), Ponkan and Satsuma (P&S), and others (etc.). C and D, Networks-DT of rind and flesh. Networks-DT were constructed based on data sets of DEGs at the variety level in the rind and the flesh. Data sets were normalized by biweight, forming a 48-dimension matrix, and the resulting matrix was calculated by Spearman coefficient (>0.75). Then, the MCL algorithm, a fast and scalable unsupervised cluster algorithm for networks based on the simulation of (stochastic) flow in graphs, was used, which divided each Networks-DT into three major subnetworks (nodes in the same color). Line graphs show the average gene expression values normalized by biweight in each subnetwork. E, Major TF hubs in subnetworks of Networks-DT and Network-DV. TFs from some important TF families that fell into a subnetwork of Networks-DT or Network-DV are marked in black.
Furthermore, we studied the function of TFs in more detail by analyzing the TF-centered subnetworks in Network-DV. SubTs and SubLs include mainly four TF families as hubs (Fig. 6E). AP2/EREBP TFs respond to environmental stresses and inner phytohormone ethylene (Licausi et al., 2013). The AP2 domain TF Abscisic Acid Responsive1 (ABR1) functions as an ABA repressor and is responsive to ABA and stress treatments like cold, high salt concentration, and drought (Pandey et al., 2005). NAC TFs were shown to be senescence related (Balazadeh et al., 2010). NAC Domain-Containing Protein (NAP; NAC029) plays an important role in leaf senescence (Guo and Gan, 2006). NAC Transcription Factor-Like9 (NTL9) is involved in osmotic stress signaling, which results in the regulation of leaf senescence (Yoon et al., 2008). WRKY TFs belong to a superfamily with different functions and are involved in the response to biotic and abiotic stresses (Pandey and Somssich, 2009; Chen et al., 2012). MYB TFs function as stress-responsive factors, as key factors in the signaling of many hormones (Du et al., 2009), and as regulators of anthocyanin biosynthesis in citrus (Butelli et al., 2012). In our results, AP2/EREBP and NAC family TFs were mainly present in the SubLs, including Ethylene Response DNA Binding Factor2 (CL9870), Related to AP2.4 (CL3372), DREB and EAR Motif Protein2 (CL4034), ABR1 (CL11397), ERF018 (CL8491), ERF071 (CL788), ERF013 (CL6697), NAP (CL2421), NAC010 (CL7396), NAC017 (CL13252), NAC078 (CL3268), etc. These AP2/EREBP TFs are involved in the response to abiotic stresses and ABA, and the NAC TFs play an important role in senescence. On the other hand, the SubTs contains many WRKY TFs that are responsive to biotic stresses, such as WRKY4 (CL8232; Lai et al., 2008; Ren et al., 2010), WRKY75 (CL11313; Encinas-Villarejo et al., 2009), and WRKY50 (CL12354; Gao et al., 2011). These results suggest that the TFs related to abiotic and biotic stresses are critical factors that can be used to distinguish tight- and loose-skin citrus and play different roles in the senescence of different citrus varieties.
Transporter-Mediated Nutrient Transportation from Flesh to Rind
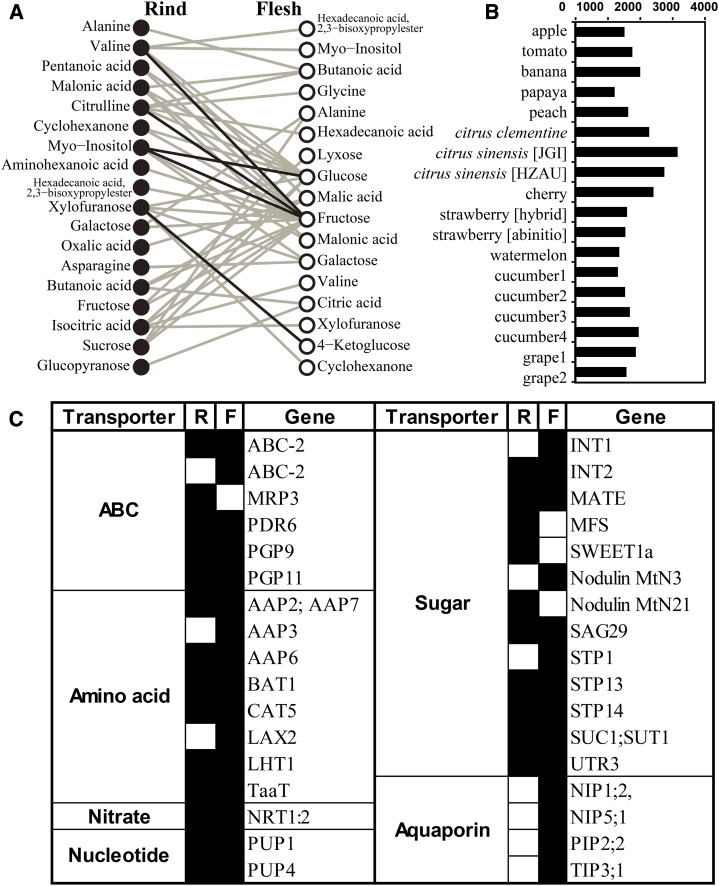
There is a transfer of nutrients and water between the rind and the flesh after harvest in hesperidium. A previous study of labeled CO2 indicated that, in developing citrus fruits, photosynthates are accumulated via dorsal vascular bundles in the rind and are slowly transferred through nonvascular tissue juice stalks (Koch, 1984). It was suggested that there is a carrier-mediated and energy-dependent active sugar transport in the juice sacs before the maturation of Satsuma fruit (Chen et al., 2002). However, no previous study has been focused on the nutrient transport in citrus fruit after harvest. In this experiment, much evidence suggested that nutrients and water are transported from the flesh to the rind in citrus fruit during storage. As a previous study suggested, the sink is much more active than the source in the transport process during fruit development (Roitsch, 1999). Function enrichment analysis of DEGs at the tissue level (Fig. 3D) shows that there are more enriched functions in the rind than in the flesh, mainly including ethylene and ABA synthesis, secondary metabolism, transporter- and TF-related functions, stress responses, etc. This result suggests that the rind could be the sink (energy consumption) tissue in an individual citrus fruit. Additionally, the data for metabolite levels (Supplemental Data Set S4) show that Suc, Fru, and Glc are mainly accumulated in the flesh. The correlation network of metabolites between the rind and the flesh (Network-Mrf; Fig. 7A) illustrates that the Glc and Fru in the flesh are at the center of the network with the highest connectivity, and their coexpression partners include inositol, Fru, Gal, and many other metabolites in the rind. It is commonly recognized that the transfer of nutrients among plant tissues may rely on a minority of specific substances to work efficiently. Therefore, as Network-Mrf indicates, the efficient and economical model of transportation between the tissues of detached citrus fruit is as follows: nutrients (possibly Glc and Fru) are transported from the flesh to the rind, but not from the opposite direction (mass metabolites are transported from the rind to the flesh). In addition, invertase genes (CL13030, CL3976, CL4459, CL10238, CL12157, and CL698), which encode the enzymes that can break Suc down into the monosaccharides Glc and Fru, were highly expressed in the rind (Supplemental Data Set S2). Therefore, it can be speculated that Suc might also be involved in the flesh-rind nutrient transfer.
Figure 7.
Description of communication between flesh and rind. A, Network-Mrf. The Spearman correlation coefficient was calculated for each metabolite between the rind and the flesh in four varieties. Significant correlations (Spearman correlation > 0.75) supported by multiple varieties are represented. The black lines represent correlations occurring in four varieties, and the gray lines represent correlations occurring in three varieties. B, Genome-wide scale comparison of copy numbers of transporter genes between citrus and other model fruits. Genome-wide statistics show copy numbers of all orthologous genes annotated to transporters in 18 genomes of fruit species (genome data were downloaded from the databases listed in Supplemental Table S6). C, List of up-regulated transporter genes in the rind and the flesh. Transporter genes that were up-regulated in the rind or the flesh are marked in black. AAP2, AMINO ACID PERMEASE2; ABC-2, ABC2-TYPE TRANSPORTER FAMILY PROTEIN; BAT1, BIDIRECTIONAL AMINO ACID TRANSPORTER1; CAT5, CATIONIC AMINO ACID TRANSPORTER5; INT1, INOSITOL TRANSPORTER1; LAX2, LIKE AUXIN RESISTANT2; LHT1, LYSINE HISTIDINE TRANSPORTER1; MATE, MULTIDRUG AND TOXIC-COMPOUND EXTRUSION EFFLUX FAMILY PROTEIN; MFS, MAJOR FACILITATOR SUPERFAMILY PROTEIN; MRP3, MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN3; NIP1;2, NOD26-LIKE INTRINSIC PROTEIN1;2; Nodulin MtN3, NODULIN MtN3 FAMILY PROTEIN; NRT1:2, NITRATE TRANSPORTER1:2; PDR6, PLEIOTROPIC DRUG RESISTANCE6; PGP9, P-GLYCOPROTEIN9; PIP2;2, PLASMA MEMBRANE INTRINSIC PROTEIN2;2; PUP1, PURINE PERMEASE1; SAG29, SENESCENCE-ASSOCIATED GENE29; STP1, SUGAR TRANSPORTER1; SUC1, SUCROSE-PROTON SYMPORTER1; TaaT, TRANSMEMBRANE AMINO ACID TRANSPORTER FAMILY PROTEIN; TIP3;1, TONOPLAST INTRINSIC PROTEIN3;1; UTR3, UDP-GAL TRANSPORTER3.
We also focused on the transporters to analyze the transportation process of nutrients and water. Genome-wide statistics of the copy numbers of the orthologous genes in the 18 genomes of fruit species show that, compared with other fruit species, citrus has a larger gene copy number of transporters, such as transporters of ABC, amino acids, calcium, plasma membrane intrinsic proteins (PIP), and sugars (Fig. 7B; Supplemental Data Set S10), indicating that transporters play special roles in the species-specific characteristics of citrus. Although the expression of most transporter genes was suppressed in response to the stresses of the storage environment, some important transporter genes were up-regulated (Fig. 7C; Supplemental Data Set S2). Several genes related to sugar transporters, such as INT1 (CL248), INT2 (CL196), UTR3 (CL2889), STP1 (CL4560), STP13 (CL965), STP14 (CL413), SUT1 (CL2401), Early-Responsive to Dehydration6-like major facilitator superfamily protein (CL6047 and CL8900), and the bidirectional sugar transporter SWEET1a (CL10442), were all up-regulated with time. The STPs are membrane transporters that specifically transfer hexoses from the apoplastic space into the cell and function as high-affinity hexose transporters that can be induced in programmed cell death (Norholm et al., 2006). The SUTs can load Suc to phloem (Weise et al., 2000), and CitSUT1 was reported to be strongly expressed in source and sugar-exporting organs and was repressed in mature leaf discs by exogenous Suc, Glc, and Man in citrus (Li et al., 2003). The up-regulation of these sugar transporters definitely indicates the existence of sugar transport in citrus fruit. Subsequently, we investigated the aquaporin genes, another group of transporters that affect water loss and fruit quality. Aquaporins conduct efficient water transport through cell membranes in plants and belong to a large major intrinsic protein family, which includes tonoplast intrinsic proteins (TIPs), PIPs, NOD26-like intrinsic proteins (NIP), small basic intrinsic proteins, and X intrinsic proteins (Zhang et al., 2013). In our results, although many aquaporins were down-regulated, it is noteworthy that there were still quite a few aquaporin genes, such as PIP1;2 (CL11753), PIP2;2 (CL4793), and NIP1;2 (CL10874), which were up-regulated, especially in the flesh of Ponkan and Satsuma (Supplemental Data Set S2), indicating that the flesh of Ponkan and Satsuma may suffer from more severe water loss. Furthermore, many other kinds of transporters related to amino acids, nitrates, ABC, cations, and anions were also up-regulated (Fig. 7C), suggesting that there are various transport processes during the postharvest senescence of citrus fruit. Taken together, all these results suggest that, during storage, there is a transporter-mediated transportation process in which water and nutrients (such as Glc, Fru, and probably Suc) are transported from the flesh to the rind.
Characteristics of the Citrus Senescence Process
Transcriptome, metabolome, and physiological data show that the postharvest senescence of citrus fruit is a continuous and irreversible process, unlike that of climacteric fruits, which have a dramatic increase in ethylene synthesis and respiration. The data for important physiological qualities show that the greatest volatility appeared at 0 to 50 DAH (Fig. 4, B–E). This time interval is regarded as a key period for storage physiology and, therefore, is the focus of our transcriptomic and metabolomic research. Moreover, Networks-DT were constructed. The networks clustered by MCL algorithm (van Dongen, 2000) showed modularity, as genes in the same module had similar patterns of change at the time level (Fig. 6, C and D; Supplemental Data Set S11). Networks-DT of the rind or the flesh contained three distinct subnetworks: subnetworks containing down-regulated genes (SubRd and SubFd), subnetworks containing up-regulated genes (SubRu and SubFu), and subnetworks containing genes with differential expression patterns among different varieties (SubRv and SubFv). Analysis of Networks-DT and the metabolic and physiological quality data sets at the time level could comprehensively present the senescence process of citrus fruit.
Generally, fruit ripening and senescence are associated with a series of changes in color, texture, aroma, and nutritional components caused by the participation of related genes. Color parameters FL and Fa/Fb measured by a Chroma Meter revealed that the fruit surface brightness decreased with time, and the rind color slowly turned to red from orange and then gradually faded during the long period of time in storage (Supplemental Data Set S5). The change of rind color is mainly due to the degradation of chlorophyll and the change of anthocyanins and flavonoids. Flavonoids are a big family that includes over 4,000 unique structures and are recognized as nutritional components and pigments (Middleton et al., 2000; Tripoli et al., 2007). Citrus is a main source of dietary flavonoids. Many flavonoids were identified by LC-MS in this experiment, such as hesperetin, neohesperidin, and naringin, and the data for the metabolite levels showed diverse patterns of change (Supplemental Data Set S4). Networks-DT indicate that most of the genes related to flavonoids (including anthocyanins) and chlorophyll fell into the down-regulated groups (SubRd and SubFd; Table II). Meanwhile, modifications of the cell wall led to changes in the firmness and texture of the fruit. The distribution of related genes in Networks-DT (Supplemental Data Set S11) shows that high expression of cell wall modification genes occurred at the early storage in Ponkan and Satsuma and at the late storage in orange and pummelo. It is noteworthy that the alteration of flavor is attributed to the changes of sugars, organic acids, alcohols, aldehydes, etc. Analysis of Network-MR indicates that sugar reactions might be more active in the flesh and organic acid reactions might be more active in the rind. The reliable physical quality data set among 0 to 200 DAH (Supplemental Data Set S5) demonstrates that although the major sugars and organic acids were decreased, the TSS-TA ratio and levels of odor components were increased with fluctuations. Network-MM shows that there were more frequent negative correlations between organic acids and sugars in loose-skin citrus (Satsuma and Ponkan) than in tight-skin citrus (orange and pummelo; Supplemental Data Set S7). These results suggest that there is a conversion of acids to sugars during senescence. Both the respiratory pathways of glycolysis and tricarboxylic acid cycle have important functions in energy and substance supply. Networks-DT show that most glycolysis and tricarboxylic acid cycle-related genes were up-regulated, especially in the rind (Table II), indicating that the rind of citrus fruit has an activated nutrient consumption. Additionally, most aquaporins and other transporters were down-regulated. SAGs are markers of leaf senescence that are up-regulated during both dark-induced and natural leaf senescence (Yamada et al., 2014). In citrus fruit, most SAGs showed higher expression in the rind than in the flesh and were down-regulated during the senescence process, with the exception of SAG29 (also called SWEET15), which functions as a bidirectional sugar transporter (Chen et al., 2010). Therefore, the senescence process might be connected with the transfer of nutrients between the rind and the flesh.
Table II. Numbers of DEGs in corresponding function classes in subnetworks of Networks-DT.
The subnetwork names SubRd, SubRu, SubRv, SubFd, SubFu, and SubFv represent the subnetworks in Figure 6. Each number presents the number of DEGs in the corresponding function class in the subnetwork.
| Function | SubRd | SubRu | SubRv | SubFd | SubFu | SubFv |
|---|---|---|---|---|---|---|
| Basic metabolism | ||||||
| Tricarboxylic acid | 3 | 7 | 1 | 3 | 5 | 0 |
| Cell wall modification | 2 | 2 | 7 | 4 | 2 | 4 |
| Fermentation | 1 | 3 | 1 | 1 | 6 | 1 |
| Gluconeogenesis | 2 | 1 | 1 | 0 | 0 | 1 |
| Glycolysis | 2 | 13 | 2 | 3 | 9 | 1 |
| Starch degradation | 8 | 2 | 1 | 10 | 1 | 0 |
| Starch synthesis | 2 | 0 | 1 | 4 | 0 | 0 |
| Suc degradation | 2 | 2 | 4 | 2 | 1 | 1 |
| Suc synthesis | 1 | 0 | 1 | 2 | 0 | 0 |
| ATP synthesis | 4 | 13 | 2 | 5 | 10 | 1 |
| Ascorbate and glutathione | 9 | 4 | 1 | 4 | 3 | 2 |
| Flavonoids | 19 | 9 | 4 | 11 | 7 | 0 |
| Carotenoids | 1 | 0 | 0 | 4 | 0 | 0 |
| Wax | 2 | 4 | 0 | 0 | 0 | 0 |
| Tetrapyrrole synthesis | 7 | 2 | 2 | 4 | 2 | 1 |
| Transporter | ||||||
| Aquaporin | 10 | 0 | 0 | 6 | 3 | 0 |
| Transporter, amino acids | 5 | 3 | 3 | 1 | 3 | 2 |
| Transporter, sugars | 9 | 6 | 1 | 5 | 6 | 0 |
| Transporter, Suc | 0 | 0 | 1 | 0 | 0 | 1 |
| Transmembrane transport | 69 | 37 | 17 | 47 | 32 | 5 |
| Inner cell transport | 4 | 11 | 6 | 9 | 7 | 2 |
| Phytohormones | ||||||
| ABA induced | 3 | 0 | 2 | 2 | 0 | 2 |
| ABA signal transduction | 2 | 1 | 1 | 2 | 1 | 1 |
| ABA synthesis/degradation | 2 | 3 | 1 | 4 | 2 | 0 |
| Auxin induced | 8 | 4 | 3 | 13 | 8 | 1 |
| Auxin synthesis/degradation | 0 | 1 | 1 | 1 | 1 | 0 |
| Brassinosteroid induced | 1 | 0 | 0 | 0 | 0 | 0 |
| Brassinosteroid signal transduction | 1 | 1 | 1 | 1 | 0 | 0 |
| Brassinosteroid synthesis/degradation | 3 | 3 | 0 | 2 | 2 | 1 |
| Cytokinin signal transduction | 1 | 0 | 0 | 1 | 1 | 0 |
| Cytokinin synthesis/degradation | 1 | 1 | 0 | 2 | 0 | 0 |
| Ethylene induced | 3 | 0 | 2 | 4 | 1 | 1 |
| Ethylene signal transduction | 1 | 2 | 11 | 1 | 2 | 6 |
| Ethylene synthesis/degradation | 2 | 2 | 2 | 1 | 4 | 0 |
| GA related | 8 | 1 | 0 | 2 | 0 | 0 |
| Jasmonic acid related | 2 | 3 | 4 | 2 | 2 | 0 |
| Salicylic acid related | 1 | 0 | 0 | 0 | 0 | 0 |
Furthermore, senescence regulation was illustrated by the distinct distribution of TFs in Networks-DT (Fig. 6E). All WRKY TFs with functions in pathogen defense and response to water deficit or other stresses were in the SubRv, indicating that the stress response was more active in the rind with differences among varieties. NAC TFs such as NAP, NTL9 and NTL6, which are identified as senescence-related genes, were located in SubRv and SubRv. Additionally, many MYB TFs, which function mainly in abiotic stress responses, were down-regulated with time (in SubRd and SubFd); it remains unknown why these MYB TFs were down-regulated with storage time. Nevertheless, most of the MYB TFs were differentially expressed in the rind, suggesting their potential roles in abiotic stresses during storage.
Expression Analysis of Phytohormone-Related Genes in Citrus and Climacteric/Nonclimacteric Model Fruit
To analyze the nonclimacteric characteristics of citrus fruit, we focused on the phytohormone-related features at the terminal stage of the life cycle of citrus fruit. DEGs were identified by comparing the transcription data of postripening and preripening samples from citrus, climacteric model fruit (tomato), and nonclimacteric model fruit (grape; Table III; Supplemental Fig. S3; Supplemental Data Set S12). Previous studies have shown that, although many regulatory factors are involved in fruit ripening and senescence, the phytohormones ethylene and ABA are the most important regulatory factors in climacteric and nonclimacteric fruits (Lafuente and Sala, 2002; Zhang et al., 2009; Sun et al., 2010; Klee and Giovannoni, 2011). Previous studies also have demonstrated the functional relationship between ethylene and ABA during senescence: ethylene can induce the accumulation of ABA during citrus ripening (Lafuente and Sala, 2002), and ABA can trigger the ripening and ethylene biosynthesis of tomato fruit with induced expression of ACS2, ACS4, and ACO1 (Zhang et al., 2009).
Table III. Expression trends of the genes related to ethylene and ABA synthesis and regulation by comparing the postripening and preripening data in three species.
Up means up-regulation in postripening/preripening, and down means down-regulation in postripening/preripening. Numbers represent the numbers of genes regulated by ethylene/ABA. AAO, ABA-aldehyde oxidase; ACO, 1-aminocyclopropane-1-carboxylic acid oxidase; ACS, 1-aminocyclopropane-1-carboxylic acid synthase; CCD, carotenoid cleavage dioxygenase; CYP707, the ABA 8′-hydroxylase gene.
| Function | Genes | Tomato | Citrus Flesh | Citrus Rind | Grape |
|---|---|---|---|---|---|
| Ethylene synthesis | ACO1 | Up | Up | Up | Down |
| ACO2 | Up | Up | |||
| ACO4 | Up | Up | |||
| ACS1 | Up | Up | Up | Down | |
| ACS2 | Up | ||||
| ACS5 | Up | ||||
| ACS6 | Down | Down | Down | ||
| ACS7 | Down | ||||
| ACS8 | Up | Up/down | |||
| ACS9 | Down | ||||
| ACS10 | Up | Up | Down | ||
| ACS12 | Up | Down | |||
| Ethylene regulated | Up, six; down, one | Up, four; down, two | Up, two; down, five | ||
| ABA synthesis | NCED3 | Down | Up/down | Up | Down |
| NCED4 | Down | Down | Down | Up | |
| NCED5 | Down | Up | Up/down | ||
| NCED6 | Up | ||||
| NCED9 | Up | ||||
| CYP707 | Up | Up | Up/down | Up | |
| CCD8 | Up | ||||
| CCD7 | Up | ||||
| AAO2 | Up/down | ||||
| AAO4 | Down | Up/down | |||
| ABA1 | Down | Down | |||
| ABA2 | Up | ||||
| ABA3 | Up | Down | |||
| ABA regulated | Up, one; down, three | Up, three; down, two | Up, one; down, four | ||
Ethylene is a key regulation factor of climacteric fruit ripening, and tomato fruit is a model for studying ethylene synthesis in climacteric fruits. Previous research in tomato indicated that, during the System-1 stage of the ethylene synthesis pathway, less and autoinhibitory ethylene is synthesized by LeACS1A, LeACS6, LeACO1, LeACO3, and LeACO4. At the transition stage, the ripening regulators were demonstrated to play critical roles, and LeACS4 induces a large increase of autocatalytic ethylene, resulting in a negative feedback on System-1. LeACS2, LeACS4, LeACO1, and LeACO4 are thus responsible for the high ethylene production in System-2 (Cara and Giovannoni, 2008). Our analysis of the postripening and preripening gene expression in System-1 and System-2 in tomato shows that the expression of some genes was up-regulated, such as that of ACS1 (AT3G61510.1), ACS2 (AT1G01480.1), ACS5 (AT5G65800.1), ACS8 (AT4G37770.1), ACS10 (AT1G62960.1), ACS12 (AT5G51690.1), ACO1 (AT2G19590.1), and ACO2 (AT1G62380.1; Table III). On the other hand, although the regulation of nonclimacteric fruit ripening does not strongly depend on the role of ethylene, ethylene-related genes can also play important roles in the senescence process. Grape is a typical nonclimacteric fruit. Data for grape senescence show that ACS1, ACS6, ACS7, ACS10, ACS12, and ACO1 were generally down-regulated (Table III), indicating that the ethylene synthesis pathway is not thoroughly activated in grape berry during ripening. Furthermore, ethylene-related genes also showed differential expression profiles between species. In general, most genes regulated by ethylene signals were up-regulated in tomato but down-regulated in grape. As a nonclimacteric fruit, citrus usually produces a relatively low amount of ethylene throughout maturation. Previous research has shown that exogenous ethylene treatment of citrus fruit could change the pigments and accelerate respiration (Fujii et al., 2007), and similar to climacteric fruits, green citrus fruit on the tree shows a rise of ethylene production accompanied by the up-regulation of ACS1, ACO1, and ERS1 (Katz et al., 2004). Similar to the maturation of citrus fruits on the tree, ethylene also seems to be involved in the senescence of postharvest citrus fruits. The data for citrus senescence show that some ethylene synthesis genes, such as ACO1 (CL2446), ACO4 (CL169, and CL1150), ACS1 (CL7938), and ACS10 (CL13016), were up-regulated (Table III). These ACOs and ACSs were reported to be responsible for the ethylene synthesis in System-1 and System-2, especially ACO4, which can induce a massive production of autocatalytic ethylene in System-2 (Cara and Giovannoni, 2008). Taken together, these results suggest that there is a System-2-like ethylene synthesis during citrus senescence.
Although the mechanism of ABA to affect the ripening physiology of fruits has not been fully elucidated, previous research has demonstrated that ABA is related to fruit ripening in climacteric fruits (Chernys and Zeevaart, 2000; Zhang et al., 2009). Meanwhile, ABA is associated with the regulation of nonclimacteric fruit ripening and facilitates the senescence of nonclimacteric fruits, such as grape (Coombe, 1992), strawberry (Ofosu-Anim et al., 1996), and citrus (Rodrigo et al., 2006). ABA is also considered as a ripening inducer in grape and strawberry (Jia et al., 2011). In our results, down-regulated expression of NCED3 (AT3G14440.1), NCED4 (AT4G19170.1), and NCED5 (AT1G30100.1) was observed in tomato (Table III). Oxidative cleavage of cis-epoxycarotenoids by NCED is a critical step in the regulation of ABA synthesis in higher plants. In addition, more genes in the ABA synthesis pathway are involved in the ripening process of grape. NCED3 (AT3G14440.1), NCED4 (AT4G19170.1), NCED5 (AT1G30100.1), NCED6 (AT3G24220.1), and NCED9 (AT1G78390.1) all showed an up-regulation trend (Table III), indicating that ABA plays an important role in grape ripening. In citrus data from our experiment, NCEDs were more active in the rind than in the flesh. For example, NCED3 (CL6281) and NCED5 (CL2472) had up-regulation profiles in the rind of Ponkan, pummelo, and Satsuma (Supplemental Data Set S2). And carotenoid biosynthesis is connected with the ABA synthesis pathway (Sandmann, 2001). Therefore, it can be suggested that ABA is not only involved in the senescence of citrus fruit but also is involved in the change of the rind color.
Ripening and senescence are a continuous and indivisible process in the plant life cycle (Watada et al., 1984). The major differences between climacteric and nonclimacteric fruit appear in the terminal stage (i.e. ripening and senescence) of development. Although postharvest handling and storage could affect the physiology of citrus fruit, it could not stop the process of ripening and senescence in the fruit life cycle (we only focused on phytohormone-related differences between postripening and preripening; physiological differences of fruit while attached and removal from a plant have not been studied). In this section, we intended to reveal the differences in fruit physiological activity during ripening and senescence among different species, aiming to uncover the general biological significance of the process. Comparison of the postripening and preripening transcription data among the three species reveals an interesting phenomenon: although there were no significant differences in the copy numbers of the homologous genes related to the phytohormones ethylene and ABA (Supplemental Data Set S10), the relationship of the numbers of up-regulated ethylene biosynthesis genes is tomato > citrus > grape, while that of ABA is tomato < citrus < grape. This reveals that citrus has specific nonclimacteric characteristics, as indicated by the functions of ethylene and ABA in senescence. As a nonclimacteric fruit, citrus is different from the climacteric fruit tomato in that its respiration and ethylene production rates do not exhibit climacteric characteristics (dramatic increases in ethylene synthesis and respiration during ripening; Fig. 4B). Moreover, citrus is also different from the nonclimacteric fruit grape, since citrus has a System-2-like pathway of ethylene production but a smaller number of up-regulated ABA synthesis genes compared with grape.
DISCUSSION
This study introduced data sets from commercial microarrays, GC-MS, LC-MS, and validated physiological quality detection. We focused on the analyses of TFs, phytohormones, transporters, and pathways related to physical qualities and characterized the citrus senescence process based on data features and relationships at three independent levels: differences among four varieties (variety level), differences between tissues (tissue level), and differences between different time stages (time level). In addition, we also compared the expression patterns of phytohormone-related genes in tomato, grape, and citrus to uncover the nonclimacteric characteristics of citrus.
In recent years, next-generation sequencing has been widely used to assess the copy number of transcripts. Although microarray technology is still reliable and can provide accurate and sensitive measurement (Willenbrock et al., 2009), some limitations of the Affymetrix genechip Citrus Genome Arrays used in this study are apparent: (1) this citrus array was designed in 2006 based on citrus EST collections without the support of genome annotations; and (2) as the senescence characteristics of different citrus varieties were to be investigated, the coverage rates of array probe sets for the genomes of different citrus varieties should be revaluated. To address these problems, we first improved the annotation of the array probes based on citrus genomic resources (Xu et al., 2013; Wu et al., 2014) and Arabidopsis functional annotations (Lamesch et al., 2012) to obtain an accurate and comprehensive view of the gene function information. We also calculated the coverage rates of the array probe sets for the genes in citrus genomes. The results show that the citrus array could cover about 64% of ESTs of Citrus paradisi, about 56% of genes in the genome of clementine mandarin (Citrus clementina), about 51% to 56% of genes in the whole genome of sweet orange, and 78% of genes expressed in the fruit tissues of sweet orange (BLAST E value ≤ 1e-5; Supplemental Table S3).
Fruit Senescence Is Less Affected by Genetic Background
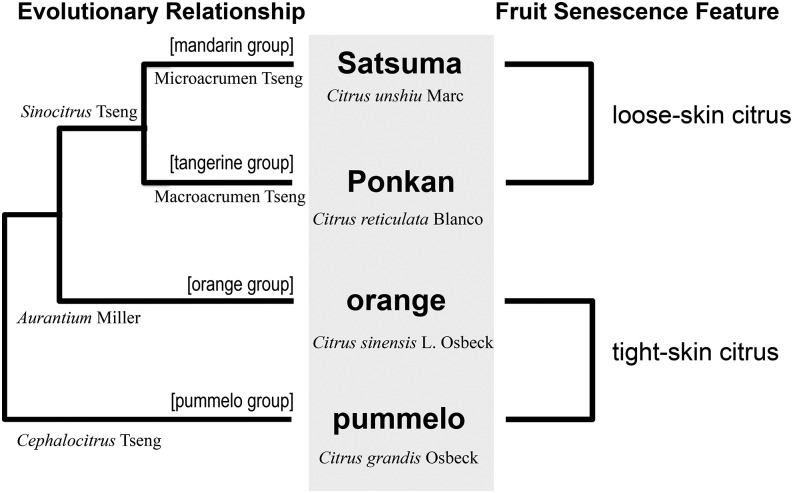
One of our major concerns in this study is the specific distinctions in postharvest senescence between different citrus varieties. The chosen citrus varieties (Satsuma, Ponkan, sweet orange, and pummelo) represent four groups in the Citrus genus, namely mandarin, tangerine, orange, and pummelo (Fig. 8). Based on the system classification in genus Citrus (Swingle and Reece, 1967; Tanaka, 1977) supported by single-nucleotide polymorphism evidence (Abkenar et al., 2004; Garcia-Lor et al., 2013), the evolutionary relationship of citrus varieties is also demonstrated by the hierarchical dendrogram of microarray expression data in Figure 2 (pummelo has the maximum differentiation, while Satsuma and Ponkan have the most similar transcriptome patterns during storage). On the other hand, commercial citrus fruits are divided into tight-skin citrus (e.g. orange and pummelo) and loose-skin citrus (e.g. Satsuma and Ponkan), and the greatest differences between these two classes are the degree of tightness of the flesh-rind anatomic structure (Fig. 1, E and F) and the different storage characteristics, as tight-skin citrus has a longer storage life than loose-skin citrus (based on empirical data and Supplemental Fig. S4). A Venn diagram of DEGs numbers at the variety level (Fig. 3A) shows that DEGs in Ponkan and Satsuma, and DEGs in pummelo and orange, have greater numbers than the other two-variety combination. Additionally, the differentiation between the two modules of coexpression network of DEGs at the variety level (Fig. 6A) is consistent with that between the tight-skin and loose-skin citrus. These results indicate that the major difference of the transcriptome at the variety level during senescence is the difference between tight-skin and loose-skin fruit. All of these analyses suggest that fruit senescence is less affected by genetic background and more influenced by other factors, which also can be the major factors responsible for the fundamental differences between tight-skin and loose-skin citrus fruit (Fig. 8).
Figure 8.
Evolutionary relationships and storing features of the four citrus varieties. Satsuma, Ponkan, sweet orange, and pummelo represent four groups (i.e. mandarin, tangerine, orange, and pummelo) in the genus Citrus. The evolutionary relationships of citrus varieties were produced based on the system of classification in the genus Citrus (Swingle and Reece, 1967; Tanaka, 1977).
Differential Flesh-Rind Communications Are Important Factors Affecting the Postharvest Properties of Different Citrus Varieties
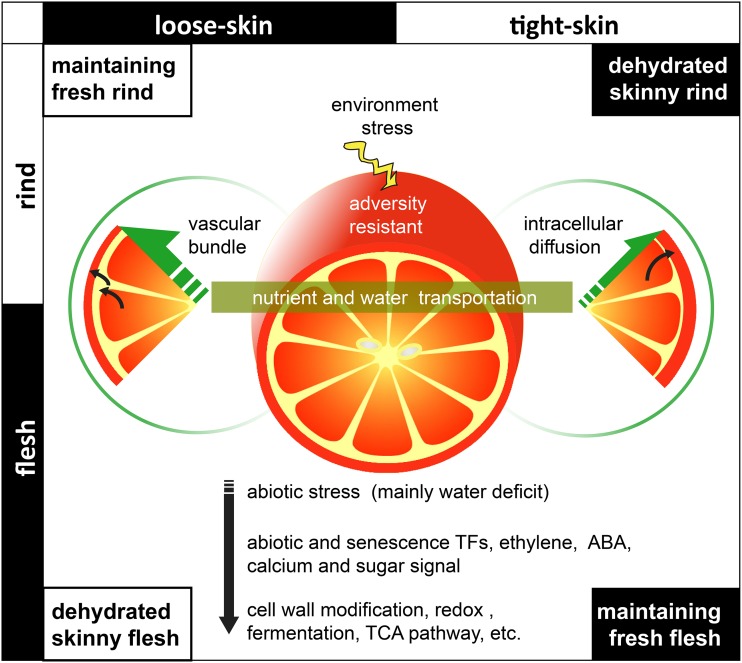
Tight-skin and loose-skin citrus fruits have greatly different flesh-to-rind nutrient transport. When the fruit loses steady nutrient supply from the tree and a new sink-source relationship is established after harvest, its physiological activity during senescence relies on the internal flesh-to-rind transport of nutrients and water. The previous results of our research have suggested that the hesperidium flesh-rind connection during senescence is a transporter-mediated transportation process that transfers water and nutrients from the flesh to the rind. Long-term observation of citrus storage suggests that, during the storage of most tight-skin citrus fruits, the stress of water deficit results in desiccation, leading to wilted rind and a shriveled appearance; on the other hand, most loose-skin citrus fruits can maintain the natural quality of rind for a period of storage, but their flesh tissues are dehydrated and skinny as a consequence of losing nutrients and water. These phenomena indicate that the two classes of citrus fruits have distinctly different flesh-rind transfer of nutrients and water after harvest. The different flesh-rind anatomic structures might be responsible for the different ways of nutrient and water transportation (Fig. 1, E and F): as the albedo of tight-skin citrus fruit closely covers the segment membranes, the transportation mainly depends on the intercellular pipelines and vascular bundles between the flesh and the rind. Since there is a space between the flesh and the rind as well as a highly developed vascular bundle system in loose-skin citrus fruit, the transportation is accomplished mainly through vascular bundles.
We hypothesized that the distinction of flesh-rind communication due to anatomic differences between citrus varieties is an important factor that influences the senescence process. Based on this hypothesis, the citrus senescence process was modeled in detail. As fruit rind is directly exposed to the environment, which results in energy expenditure because of the responses to biotic and abiotic stresses, nutrients are transported from the flesh to the rind to maintain the bioactivity of the whole fruit. The depletion of internal substances causes abiotic stresses (mainly water deficit stress), which further induces phytohormone reactions, transcription factor regulation, and a series of physiological and biochemical reactions (Fig. 9).
Figure 9.
Sketch map model of the citrus senescence process. TCA, Tricarboxylic acid.
The hypothesis and model are supported by sufficient evidence of postharvest phenomena and the results in this research. Taking loose-skin citrus fruits as an example, during storage, nutrients and water are transported from the flesh to the rind in response to environmental stresses. The flesh-rind transportation of loose-skin citrus fruits relies mainly on the powerful vascular bundle system between the flesh and the rind, which can efficiently transport materials, especially water, to maintain the bioactivity of the rind. The unimpeded material supply channel to the rind not only results in the phenomenon that loose-skin fruits always have fresh rind with dehydrated and skinny flesh in storage but also accelerates the senescence (phenomena from observation for years; Y. Cheng, unpublished data). Gene expression analysis shows that, during storage, although many transporters were down-regulated in response to stresses, some important transporters of sugars, ABC, cations, and anions were up-regulated with time and had higher expression at the early stage in loose-skin citrus than in tight-skin citrus (Supplemental Table S4). It is notable that some aquaporin genes were up-regulated. For example, PIP2;2, TIP3;1, NIP1;2, and NIP5;1 were up-regulated in loose-skin citrus (Ponkan and Satsuma; Supplemental Data Set S2). The powerful vascular bundle system and the active aquaporins of loose-skin citrus lead to a rapid water loss as well as accelerate the abiotic stress-induced senescence. The internal abiotic stresses (possibly mainly water deficit stress) induce a lot of responses of hormones like ethylene and ABA and the regulation of abiotic stress-related TFs, and they also substantially impact the overall energy-related transcripts and metabolism. Network-DV (Fig. 6E) shows that AP2/EREBP family TFs (involved in the response to abiotic stresses and ABA) and NAC family TFs (involved in senescence) mainly fall into the SubLs and act as critical distinguishing factors between tight-skin and loose-skin citrus. The enormous nutrient consumption and the activated responses to abiotic stresses influence a range of metabolic pathways. We found that large numbers of genes with important functions showed higher expression in fruit tissues at the early stage in loose-skin citrus than in tight-skin citrus, including the genes of cell wall modification, fermentation pathway, ethylene synthesis and signaling, mitochondrial electron transport, tricarboxylic acid pathway, and genes related to stress, redox, calcium, and sugar signaling (Supplemental Table S4). This result implies that the short postharvest life of loose-skin citrus might be the result of the overconsumption of nutrients at the earlier stages in response to abiotic stresses (possibly water deficit stress). Conversely, tight-skin citrus fruits have a less developed vascular bundle system, and nutrients and water are transported from the flesh to the rind mainly through an intracellular diffusion pathway. This inefficient transportation results in the phenomenon that the quality of the flesh could be maintained for a longer time than that of the rind during senescence.
Perspectives of Future Research on Citrus Fruit Senescence
Plant organisms all experience senescence. Leaf senescence is the most extensively studied senescence process in higher plants. At the end of life, a leaf destroys itself through the active degeneration of cellular structures and makes its last contribution to the plant by recycling nutrients to the actively growing part. It is a highly complex and a genetically controlled process, which involves a lot of regulatory factors, including transcription regulators, receptors and signaling components for hormones and stress responses, and regulators of metabolism. Leaf senescence is the result of programmed cell death (Lim et al., 2007) accompanied by decreased expression of genes related to photosynthesis and protein synthesis. The SAGs are up-regulated during leaf senescence and are often regarded as molecular markers of leaf senescence. Our analysis of citrus fruit shows that most SAGs are down-regulated in fruit tissues, and the physiological and biochemical reactions and substance transport are highly active. Compared with that of the leaf, the senescence of citrus fruit may not rely on the genetically controlled voluntary cell death process but tends to be the result of environmental stresses, which cause the exhaustion and imbalance of endogenous substances due to the need to maintain the physiological activity of the fruit. This postulation needs to be further validated. Furthermore, we also found some similarities in senescence between citrus fruit and leaf. Previous research on leaf senescence indicated that leaf senescence is controlled by a series of external and internal factors, including age, levels of plant hormones, environmental stresses, and pathogens. The plant hormones ethylene, abscisic acid, jasmonic acid, salicylic acid, auxin, and brassinosteroids are believed to be inducers/promoters, while cytokinins and polyamines are antagonists of senescence (Guo and Gan, 2012). It was also found that TFs are involved in leaf senescence (Guo and Gan, 2006). In our research, we found that similar plant hormones (such as ABA and ethylene) and TFs (such as WRKY TFs, AP2 TFs, and NAC TFs) are involved in citrus fruit senescence. However, the common senescence signaling pathways of the fruit and the leaf need further verification.
MATERIALS AND METHODS
Material Preparation
To ensure a comprehensive data collection of citrus fruits, commercially mature fruits of four citrus varieties, Satsuma mandarin (Citrus unshiu), Ponkan mandarin (Citrus reticulata), sweet orange (Citrus sinensis; var Newhall navel), and Shatian pummelo (Citrus grandis), were selected for this study (Fig. 1). Fruits were collected in Yichang, Hubei Province, China, in 2008 at harvest season. Randomly selected healthy fruits with uniform color and size were stored at ambient temperature (16°C–20°C, with relative humidity of 85%–90%). Fruits were mainly divided into two parts, rind and flesh tissues, and were sampled every 10 d during 50 d after harvest (0, 10, 20, 30, 40, and 50 d) for the microarray and metabolic analyses. Each sample was mixed from 10 fruits and was frozen immediately in liquid nitrogen and stored at −80°C until use.
Transcriptome Analysis
RNA Hybridization and Microarray Expression Analysis
Two total RNA samples were independently isolated according to the method described by Tao et al. (2004) and were hybridized with the commercial genechip Citrus Genome Arrays (Affymetrix) in double repeats. The obtained raw CEL files were analyzed using Bioconductor software for R (http://www.r-project.org/). Quality control and MAS5 present/absent call values were obtained using the XPS package, and the expression values were obtained by the robust multiarray average method using the Affy package. Transcriptome data sets on the genechip Citrus Genome Arrays platform in this article can be found in the Gene Expression Omnibus (GEO) with accession number GSE63706.
Identification of DEGs
The DEGs were identified at three levels: variety level, tissue level, and time level. To analyze the DEGs at each level, we first divided the samples into different groups and then subgrouped the samples into small groups to identify DEGs based on Supplemental Table S1. The significance analysis of microarrays method (Tusher et al., 2001) was used to detect DEGs in each group with restriction of P < 0.01 and fold change greater than 2. DEGs of each subgroup were defined with biweight value greater than 0.5. Taking the identification of DEGs at the variety level as an example, samples were first divided into two groups (rind and flesh); then, each group, such as the rind group, was further divided into four subgroups, one for each species. Significantly expressed genes in each subgroup were identified by significance analysis of microarrays and restriction of biweight value and defined as DEGs of orange in the rind at the variety level.
Annotation of Microarray Probe Sets
We annotated the genechip Citrus Genome Arrays probe sets using GMAP (Wu and Watanabe, 2005) and BLAST, aiming to find the sequence similarities of the probe sets to the genes in the genomes of sweet orange and clementine mandarin (Citrus clementina; Xu et al., 2013; Wu et al., 2014), and the Mapman annotations of Arabidopsis (Arabidopsis thaliana) were homologously mapped to the annotations of the probe sets. Finally, the probe sets with similar consensus sequences were grouped by their mapping results to the two citrus genomes and the cluster names were customized as CL numbers. The subsequent gene function analysis was based on CL genes. CL gene expression was defined as the average value of probe sets in a cluster. Unless otherwise specified, DEGs in this article refer to differentially expressed CL genes (Supplemental Data Set S1).
Verification of Microarray Expression Data with Quantitative Real-Time PCR
Total RNA extraction, complementary DNA synthesis, and quantitative real-time PCR of sweet orange were performed as described previously (Zhu et al., 2011). Primers were designed by Primer Express 3.0 (Supplemental Table S5). The average Pearson correlation coefficient for quantitative real-time PCR and microarray expression is 0.76.
Metabolic Analysis
Profiling of Primary Metabolites
A 150-mg sample was extracted with methanol, and the extracts were analyzed by GC-MS (Thermo Fisher; ISQII) with four replicates as described by Yun et al. (2013). Customized reference spectrum databases, including the National Institute of Standards and Technology and the Wiley Registry, were utilized for the identification and annotation of the metabolites recorded by GC-MS based on retention indices and mass spectral similarities. The raw output data of GC-MS were processed by MetAlign (Lommen, 2009) for baseline correction and retention time alignment. The data processed by MetAlign were corrected by the unique mass-to-charge ratio value of internal standard metabolites, and then the sum value of retention time was normalized by Z-score transformation for further analysis.
Profiling of Secondary Metabolites
Secondary metabolite analysis was performed using the QTOF 6520 mass spectrometer (Agilent Technologies) coupled to a 1200 series Rapid Resolution HPLC system and the Zorbax Eclipse Plus C18 1.8-μm, 2.1- × 100-mm reverse-phase analytical column (Agilent Technologies). Metabolites were identified based on accurate masses and tandem mass spectrometry by searching against the MassBank metabolite database. Four replicates were performed for each sample. The metabolites were putatively assigned and analyzed by ultra-performance liquid chromatography-quadrupole time of flight-mass spectrometry as described by Yun et al. (2013).
Coexpression Analysis
In this experiment, we analyzed the coexpression relationships using transcriptome and metabolome data. All coexpression networks were constructed from a Spearman coefficient matrix computed by R, and the threshold was set to 0.75 for samples with a number larger than 20 and 0.9 for samples with a number smaller than 20. The data used for gene coexpression analysis were preprocessed by biweight transformation, and the metabolomic data used for coexpression analysis were preprocessed by Z-score transformation.
Metabolic Network Analysis
We introduced the parameter RE-value to represent the expression of the reaction genes in the citrus metabolic network. CitrusCyc was constructed by mapping citrus genes to AraCyc (Zhang et al., 2005) based on the sequence similarities with Arabidopsis. Then, the diffusion method (Allen et al., 2013) was used to process the biweight-normalized expression data for neutralizing the influence of adjoining reactions in the metabolic network, resulting in the RE-value. Coexpression networks of reaction genes in CitrusCyc were constructed using RE-values (Spearman coefficient ≥ 0.75), and the networks were clustered by MCL algorithm in the rind and the flesh. Additionally, the correlation network between metabolite levels and RE-values was constructed based on Jaccard distance (0.4 or greater; Jaccard distance has two values: positive and negative).
Genome-Wide Comparison of Gene Copy Numbers and Gene Expression between Citrus and Other Model Fruits
To investigate the copy numbers of citrus functional genes, genome-wide protein sequences of sequenced fruit species were downloaded from genome project Web sites (Supplemental Table S6). Homologous genes were identified by BLAST, clustered by MCL, and annotated by Arabidopsis gene identifiers, and their copy numbers were counted.
To investigate the nonclimacteric characteristics of citrus fruits, tomato (Solanum lycopersicum) and grape (Vitis vinifera) were selected to be compared with the four citrus varieties at the transcription level. We collected a total of 247 array samples in 10 independent experiments on fruit senescence (from fruits before or after harvest) of the two model fruits from the GEO database. The ratio of postripening to preripening expression values with a fold change threshold of 2 or greater was used for the identification of DEGs (Supplemental Table S7).
Determination of Fruit Physiological Quality
To study the physiological changes of citrus fruits during a long period of storage, another group of fruits was sampled, and the sampling was repeated in a total of 12 batches within 3 years (2009, 2010, and 2012). The fruits were harvested from the same orchard in Yichang as the samples for the microarray and metabolic analyses, including sweet oranges, Satsuma mandarin (in 2012), and Ponkan mandarin. The postharvest handling and storage conditions imitated the commercial standards of the Chinese citrus industry (temperature, 6°C–25°C; humidity, 35%–100%). They were evaluated by measurements of fruit weight loss (%), three color parameters (Fa, Fb, and FL), fruit flesh TSS, TA, ascorbic acid, organic acids, reducing sugars, and odor components about every 10 d during 200 d after harvest. The respiration rate was measured with an infrared gas analyzer (model GXH-305H; Junfang Science & Technology Institute of Physical and Chemical Research). TSS were determined with a refractometer (model Pocket PAL-1; Atago) according to the manufacturer’s instructions. Color measurements were performed on the surface of the fruits around the equatorial region with a Konica Minolta Chroma Meter CM-5. TA and ascorbic acid contents were also determined. An Agilent 7890A gas chromatograph (Agilent Technologies) was used to analyze the organic acids (malic acid, citric acid, and quinic acid), reducing sugars (Fru, Glc, and Suc), and odor components (ethanol, methanol, and acetaldehyde) in the flesh using the methods described by Sun et al. (2013).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Principal component analysis using conventional physiological quality data during 0 to 200 DAH.
Supplemental Figure S2. General map of CitrusCyc constructed based on AraCyc.
Supplemental Figure S3. Venn diagrams of numbers of DEGs identified by comparing the transcription data of postripening and preripening samples of tomato, grape, and citrus.
Supplemental Figure S4. Rot rate of four varieties during 70-DAH storage.
Supplemental Table S1. Sample classification for the identification of DEGs at three levels.
Supplemental Table S2. Proportions of the first and second level linkages to the total linkages in the Network-MR.
Supplemental Table S3. Ratio of array-covered genes from different citrus gene resources.
Supplemental Table S4. Numbers of DEGs at early stage (0–10 DAH) by comparing samples of loose- and tight-skin citrus.
Supplemental Table S5. Quantitative real-time PCR verification of selective probe sets in orange rind and flesh.
Supplemental Table S6. Database resources for the investigation of homologous gene copies.
Supplemental Table S7. List of collected samples of tomato and grape from the GEO database.
Supplemental Data Set S1. Annotation of clustered citrus microarray probe sets (CL genes).
Supplemental Data Set S2. Expression of all DEGs at the tissue, variety, and time levels.
Supplemental Data Set S3. Gene function enrichment analysis of DEGs at the tissue, variety, and time levels.
Supplemental Data Set S4. Data set of all metabolite levels after Z-score normalization.
Supplemental Data Set S5. Data set of physiological quality during 0 to 200 DAH.
Supplemental Data Set S6. Major clusters in Networks-RR.
Supplemental Data Set S7. Spearman correlation coefficient values between primary metabolites (in Network-MM).
Supplemental Data Set S8. Jaccard distances between metabolite levels and RE-values (Network-MR).
Supplemental Data Set S9. Gene list of the Network-DV.
Supplemental Data Set S10. Genome-wide scale comparison of gene copy numbers between citrus and other model fruits.
Supplemental Data Set S11. Gene list of subnetworks in Networks-DT.
Supplemental Data Set S12. Expression data of DEGs during the senescence process of grape and tomato.
Supplementary Material
Acknowledgments
We thank Hanhui Kuang, Chunying Kang, and Wenwu Guo (Key Laboratory of Horticultural Plant Biology [Ministry of Education], Huazhong Agricultural University) as well as Zhongchi Liu (Department of Cell Biology and Molecular Genetics, University of Maryland) for advice on this work.
Glossary
- TF
transcription factor
- ABA
abscisic acid
- DAH
days after harvest
- DEGs
differentially expressed genes
- ABC
ATP-binding cassette
- GC-MS
gas chromatography-mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
- TA
titratable acid
- TSS
total soluble solids
- CitrusCyc
Citrus Metabolic Pathway Network
- RE-value
recalculated expression value of reaction genes
- Networks-RR
coexpression networks of recalculated expression values
- Network-MM
correlation network of primary metabolites
- Network-MR
correlation network between metabolites and recalculated expression values
- MCL
Markov cluster
- EC
Enzyme Commission
- Network-DV
network of differentially expressed genes at the variety level
- SubTs
tight-skin citrus subnetwork
- SubLs
loose-skin citrus subnetwork
- Network-Mrf
correlation network of metabolites between the rind and the flesh
- GEO
Gene Expression Omnibus
- Networks-DT
coexpression networks of differentially expressed genes at the time level
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31221062 and 31271968), the Program for New Century Excellent Talents in University (grant no. NCET–12–0859), the National Modern Agriculture (Citrus) Technology Systems of China (grant no. CARS–27), and the Huazhong Agricultural University Scientific and Technological Self-Innovation Foundation.
Articles can be viewed without a subscription.
References
- Abkenar AA, Isshiki S, Tashiro Y (2004) Phylogenetic relationships in the “true citrus fruit trees” revealed by PCR-RFLP analysis of cpDNA. Sci Hortic (Amsterdam) 102: 233–242 [Google Scholar]
- Allen J, Weinrich M, Hoppitt W, Rendell L (2013) Network-based diffusion analysis reveals cultural transmission of lobtail feeding in humpback whales. Science 340: 485–488 [DOI] [PubMed] [Google Scholar]
- Asif MH, Lakhwani D, Pathak S, Gupta P, Bag SK, Nath P, Trivedi PK (2014) Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC Plant Biol 14: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Bennici A, Tani C (2004) Anatomical and ultrastructural study of the secretory cavity development of Citrus sinensis and Citrus limon: evaluation of schizolysigenous ontogeny. Flora 199: 464–475 [Google Scholar]
- Bernillon S, Biais B, Deborde C, Maucourt M, Cabasson C, Gibon Y, Hansen TH, Husted S, de Vos RCH, Mumm R, et al. (2013) Metabolomic and elemental profiling of melon fruit quality as affected by genotype and environment. Metabolomics 9: 57–77 [Google Scholar]
- Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24: 1242–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cara B, Giovannoni JJ (2008) Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci 175: 106–113 [Google Scholar]
- Chen J, Zhang S, Zhang L, Xie M, Tao J, Wu J (2002) Physiological mechanism of postphloem sugar transport in citrus fruits. Zhongguo Nong Ye Ke Xue 36: 1530–1534 [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JA (2000) Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol 124: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe B. (1992) Research on development and ripening of the grape berry. Am J Enol Vitic 43: 101–110 [Google Scholar]
- Defilippi BG, Manriquez D, Luengwilai K, Gonzalez-Aguero M (2009) Aroma volatiles: biosynthesis and mechanisms of modulation during fruit ripening. Adv Bot Res 50: 1–37 [Google Scholar]
- Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX (2009) Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (Mosc) 74: 1–11 [DOI] [PubMed] [Google Scholar]
- Encinas-Villarejo S, Maldonado AM, Amil-Ruiz F, de los Santos B, Romero F, Pliego-Alfaro F, Muñoz-Blanco J, Caballero JL (2009) Evidence for a positive regulatory role of strawberry (Fragaria × ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. J Exp Bot 60: 3043–3065 [DOI] [PubMed] [Google Scholar]
- Enfissi EM, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, et al. (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A (2008) Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol 148: 730–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, Porceddu A, Venturini L, Bicego M, Murino V, et al. (2012) The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24: 3489–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Shimada T, Sugiyama A, Nishikawa F, Endo T, Nakano M, Ikoma Y, Shimizu T, Omura M (2007) Profiling ethylene-responsive genes in mature mandarin fruit using a citrus 22K oligoarray. Plant Sci 173: 340–348 [Google Scholar]
- Gao QM, Venugopal S, Navarre D, Kachroo A (2011) Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 155: 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lor A, Curk F, Snoussi-Trifa H, Morillon R, Ancillo G, Luro F, Navarro L, Ollitrault P (2013) A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group (Citrinae, Rutaceae) and the origin of cultivated species. Ann Bot (Lond) 111: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann J, Hippeli S, Elstner EF (2002) Plant’s defence and its benefits for animals and medicine: role of phenolics and terpenoids in avoiding oxygen stress. Plant Physiol Biochem 40: 471–478 [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan SS (2012) Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ 35: 644–655 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang L, Shi Y, Lu Z, Yu Z (2014) Proteomic analysis of peach fruit during ripening upon post-harvest heat combined with 1-MCP treatment. J Proteomics 98: 31–43 [DOI] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z (2013) Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E, Lagunes PM, Riov J, Weiss D, Goldschmidt EE (2004) Molecular and physiological evidence suggests the existence of a system II-like pathway of ethylene production in non-climacteric Citrus fruit. Planta 219: 243–252 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Koch KE. (1984) The path of photosynthate translocation into citrus fruit. Plant Cell Environ 7: 647–653 [Google Scholar]
- Kok EJ, Lehesranta SJ, van Dijk JP, Helsdingen JR, Dijksma WTP, Van Hoef AMA, Koistinen KM, Karenlampi SO, Kuiper HA, Keijer J (2008) Changes in gene and protein expression during tomato ripening: consequences for the safety assessment of new crop plant varieties. Food Sci Technol Int 14: 503–518 [Google Scholar]
- Ladaniya M. (2008) Citrus Fruit: Biology, Technology and Evaluation. Academic Press,Elsevier, San Diego [Google Scholar]
- Lafuente MT, Sala JM (2002) Abscisic acid levels and the influence of ethylene, humidity and storage temperature on the incidence of postharvest rindstaning of ‘Navelina’ orange (Citrus sinensis L. Osbeck) fruit. Postharvest Biol Technol 25: 49–57 [Google Scholar]
- Lai Z, Vinod K, Zheng Z, Fan B, Chen Z (2008) Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Li CY, Shi JX, Weiss D, Goldschmidt EE (2003) Sugars regulate sucrose transporter gene expression in citrus. Biochem Biophys Res Commun 306: 402–407 [DOI] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lombardo VA, Osorio S, Borsani J, Lauxmann MA, Bustamante CA, Budde CO, Andreo CS, Lara MV, Fernie AR, Drincovich MF (2011) Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiol 157: 1696–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A. (2009) MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem 81: 3079–3086 [DOI] [PubMed] [Google Scholar]
- Ma Q, Ding Y, Chang J, Sun X, Zhang L, Wei Q, Cheng Y, Chen L, Xu J, Deng X (2014) Comprehensive insights on how 2,4-dichlorophenoxyacetic acid retards senescence in post-harvest citrus fruits using transcriptomic and proteomic approaches. J Exp Bot 65: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Madrid MC, Serrano M, Riquelme F, Romojaro F (1996) Polyamines, abscisic acid and ethylene production in tomato fruit. Phytochemistry 43: 323–326 [Google Scholar]
- Middleton E Jr, Kandaswami C, Theoharides TC (2000) The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol Rev 52: 673–751 [PubMed] [Google Scholar]
- Norholm MHH, Nour-Eldin HH, Brodersen P, Mundy J, Halkier BA (2006) Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS Lett 580: 2381–2387 [DOI] [PubMed] [Google Scholar]
- Ofosu-Anim J, Kanayama Y, Yamaki S (1996) Sugar uptake into strawberry fruit is stimulated by abscisic acid and indoleacetic acid. Physiol Plant 97: 169–174 [Google Scholar]
- Osorio S, Alba R, Damasceno CM, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JK, Fei Z, et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Nikoloski Z, Kochevenko A, Fernie AR, Giovannoni JJ (2012) Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol 159: 1713–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H, Nair G, Modi V (1990) Some structural changes during ripening of mangoes (Mangifera indica var. Alphonso) by abscisic acid treatment. Ann Bot (Lond) 65: 121–127 [Google Scholar]
- Prasanna V, Prabha TN, Tharanathan RN (2007) Fruit ripening phenomena: an overview. Crit Rev Food Sci Nutr 47: 1–19 [DOI] [PubMed] [Google Scholar]
- Ren XJ, Huang WD, Li WZ, Yu DQ (2010) Tobacco transcription factor WRKY4 is a modulator of leaf development and disease resistance. Biol Plant 54: 684–690 [Google Scholar]
- Rodrigo MJ, Alquezar B, Zacarías L (2006) Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J Exp Bot 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Marcos JF, Alférez F, Mallent MD, Zacarías L (2003) Characterization of Pinalate, a novel Citrus sinensis mutant with a fruit-specific alteration that results in yellow pigmentation and decreased ABA content. J Exp Bot 54: 727–738 [DOI] [PubMed] [Google Scholar]
- Roitsch T. (1999) Source-sink regulation by sugar and stress. Curr Opin Plant Biol 2: 198–206 [DOI] [PubMed] [Google Scholar]
- Sandmann G. (2001) Carotenoid biosynthesis and biotechnological application. Arch Biochem Biophys 385: 4–12 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C (2013) Fruit development and ripening. Annu Rev Plant Biol 64: 219–241 [DOI] [PubMed] [Google Scholar]
- Silalahi J. (2002) Anticancer and health protective properties of citrus fruit components. Asia Pac J Clin Nutr 11: 79–84 [DOI] [PubMed] [Google Scholar]
- Sun L, Sun Y, Zhang M, Wang L, Ren J, Cui M, Wang Y, Ji K, Li P, Li Q, et al. (2012) Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol 158: 283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang M, Ren J, Qi J, Zhang G, Leng P (2010) Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biol 10: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhu A, Liu S, Sheng L, Ma Q, Zhang L, Nishawy EM, Zeng Y, Xu J, Ma Z, et al. (2013) Integration of metabolomics and subcellular organelle expression microarray to increase understanding the organic acid changes in post-harvest citrus fruit. J Integr Plant Biol 55: 1038–1053 [DOI] [PubMed] [Google Scholar]
- Swingle WT, Reece PC (1967) The botany of citrus and its wild relatives. InReuther W, Webber HJ, Batchelor LD, eds, The Citrus Industry, Vol 1 University of California, Riverside, pp 190–430 [Google Scholar]
- Tanaka T. (1977) Fundamental discussion of Citrus classification. Studia Citrogia 14: 1–6 [Google Scholar]
- Tao N, Cheng Y, Xu J, Xu Q, Deng X (2004) An effective protocol for the isolation of RNA from the pulp of ripening citrus fruits. Plant Mol Biol Rep 22: 305 [Google Scholar]
- Tripoli E, La Guardia M, Giammanco S, Di Majo D, Giammanco M (2007) Citrus flavonoids: molecular structure, biological activity and nutritional properties. A review. Food Chem 104: 466–479 [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderschuren H, Nyaboga E, Poon JS, Baerenfaller K, Grossmann J, Hirsch-Hoffmann M, Kirchgessner N, Nanni P, Gruissem W (2014) Large-scale proteomics of the cassava storage root and identification of a target gene to reduce postharvest deterioration. Plant Cell 26: 1913–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen SM (2000) Graph clustering by flow simulation. PhD thesis. University of Utrecht, The Netherlands [Google Scholar]
- Vicente AR, Saladie M, Rose JKC, Labavitch JM (2007) The linkage between cell wall metabolism and fruit softening: looking to the future. J Sci Food Agric 87: 1435–1448 [Google Scholar]
- Watada A, Herner R, Kader A, Romani R, Staby G (1984) Terminology for the description of developmental stages of horticultural crops. HortScience 19: 20–21 [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock H, Salomon J, Søkilde R, Barken KB, Hansen TN, Nielsen FC, Møller S, Litman T (2009) Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA 15: 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, et al. (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol 32: 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, et al. (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45: 59–66 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240: 399–408 [DOI] [PubMed] [Google Scholar]
- Yoon HK, Kim SG, Kim SY, Park CM (2008) Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells 25: 438–445 [PubMed] [Google Scholar]
- Yun Z, Gao H, Liu P, Liu S, Luo T, Jin S, Xu Q, Xu J, Cheng Y, Deng X (2013) Comparative proteomic and metabolomic profiling of citrus fruit with enhancement of disease resistance by postharvest heat treatment. BMC Plant Biol 13: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DY, Ali Z, Wang CB, Xu L, Yi JX, Xu ZL, Liu XQ, He XL, Huang YH, Khan IA, et al. (2013) Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 8: e56312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P (2009) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, Rhee SY (2005) MetaCyc and AraCyc: metabolic pathway databases for plant research. Plant Physiol 138: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Song J, Campbell-Palmer L, Thompson K, Li L, Walker B, Cui Y, Li X (2013) A proteomic investigation of apple fruit during ripening and in response to ethylene treatment. J Proteomics 93: 276–294 [DOI] [PubMed] [Google Scholar]
- Zhu A, Li W, Ye J, Sun X, Ding Y, Cheng Y, Deng X (2011) Microarray expression profiling of postharvest Ponkan mandarin (Citrus reticulata) fruit under cold storage reveals regulatory gene candidates and implications on soluble sugars metabolism. J Integr Plant Biol 53: 358–374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.