Genes of thujone biosynthesis are critical for herbivore resistance in a woody Redcedar.
Abstract
Western redcedar (WRC; Thuja plicata) produces high amounts of oxygenated thujone monoterpenoids associated with resistance against herbivore feeding, particularly ungulate browsing. Thujones and other monoterpenoids accumulate in glandular structures in the foliage of WRC. Thujones are produced from (+)-sabinene by sabinol and sabinone. Using metabolite analysis, enzyme assays with WRC tissue extracts, cloning, and functional characterization of cytochrome P450 monooxygenases, we established that trans-sabin-3-ol but not cis-sabin-3-ol is the intermediate in thujone biosynthesis in WRC. Based on transcriptome analysis, full-length complementary DNA cloning, and characterization of expressed P450 proteins, we identified CYP750B1 and CYP76AA25 as the enzymes that catalyze the hydroxylation of (+)-sabinene to trans-sabin-3-ol. Gene-specific transcript analysis in contrasting WRC genotypes producing high and low amounts of monoterpenoids, including a glandless low-terpenoid clone, as well as assays for substrate specificity supported a biological role of CYP750B1 in α- and β-thujone biosynthesis. This P450 belongs to the apparently gymnosperm-specific CYP750 family and is, to our knowledge, the first member of this family to be functionally characterized. In contrast, CYP76AA25 has a broader substrate spectrum, also converting the sesquiterpene farnesene and the herbicide isoproturon, and its transcript profiles are not well correlated with thujone accumulation.
Western redcedar (WRC; Thuja plicata) is a Cupressaceae species native to the Pacific Northwest of North America. This evergreen gymnosperm tree has been used for thousands of years by First Nations people as a preferred building material for houses and boats and a weaving material for baskets, mats, and clothing as well as for carvings, totem poles, and various other practical and ceremonial purposes (Hebda and Mathewes, 1984). WRC is highly valued in the forest industry for its light, uniform, and durable wood. As a building material, WRC provides high-dimensional stability and good thermal insulation properties. Because of its high durability, WRC wood is widely used for exterior applications, such as roofing and sidings, decks, and exterior furniture. In addition, the essential oil of WRC, which is rich in mono- and sesquiterpenoids, is used in the cosmetics and fragrance industry because of its antimicrobial and scent qualities.
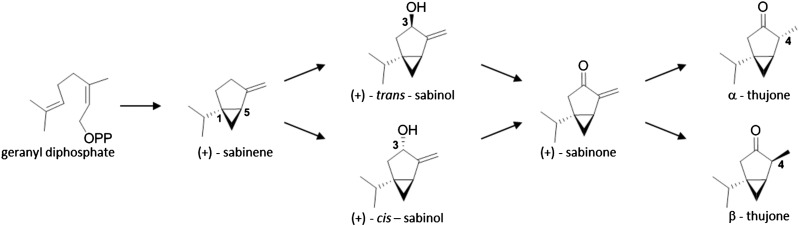
To support sustainable reforestation and harvest of WRC, a breeding program was established (Russell and Ferguson, 2008; Russell and Yanchuk, 2012). Traits of particular interest in WRC breeding include heartwood durability, growth, and environmental adaptation as well as resistance to pathogens and herbivores. Decay resistance of WRC heartwood is attributed to the accumulation of terpenoid- and lignan-specialized (i.e. secondary) metabolites (Morris and Stirling, 2012), some of which are named after this species (e.g. thujaplicin terpenoids and plicatic acid and thujaplicatin lignans; Anderson and Gripenberg, 1948; Gardner et al., 1959; MacLean and Murakami, 1966). The oxidized monoterpenoids α- and β-thujone are characteristic specialized metabolites of the foliage and bark/phloem (Fig. 1).
Figure 1.
Proposed biosynthesis of α- and β-thujone in WRC. The monoterpenenoids α- and β-thujone are proposed to derive from GDP through a four-step biosynthetic pathway starting with (+)-sabinene synthase (Foster et al., 2013). Sabinene may be oxidized to either (+)-cis- or (+)-trans-sabinol followed by oxidation to (+)-sabinone as the proposed direct precursor to α- and β-thujone. Here, we provide evidence for (+)-trans-sabinol but not (+)-cis sabinol as an intermediate in thujone biosynthesis in the gymnosperm WRC. In contrast, in the angiosperm common garden sage, thujone is formed by the (+)-cis-sabinol intermediate (Karp and Croteau, 1982). This article describes (+)-trans-sabinol dehydrogenase enzyme activity in WRC tissue extract and gene discovery and functional characterization of WRC (+)-sabinene-3-hydroxylase. The proposed pathway was redrawn based on Karp and Croteau (1982).
Herbivore feeding, specifically browsing by ungulate deer and elk, is a major ecological problem and economic cost factor for WRC reforestation. High foliage terpenoid content has been associated with reduced herbivore damage (Vourc’h et al., 2001, 2002; Burney and Jacobs, 2011; Kimball et al., 2012). The monoterpenes (+)-sabinene and α- and β-thujone are abundant in WRC foliage and account for 65% to 83% of terpenoids in WRC essential oil (Von Rudloff et al., 1988; Kimball et al., 2005; Tsiri et al., 2009; Tisserand and Young, 2014). These three monoterpenes are also found in other Cupressaceae species, such as the genera Juniperus spp. and Cupressus spp. They are also major monoterpenes in species of other plant families, including Asteraceae (e.g. species of wormwood [Artemisia spp.] or tansy [Tanacetum vulgare]) and Lamiaceae (e.g. common garden sage [Salvia officinalis]; Tisserand and Young, 2014). Based on characterization of purified enzyme preparations from common garden sage, Karp and Croteau (1982) proposed a biosynthetic pathway leading from GDP by (+)-sabinene to thujone (Fig. 1). Karp and Croteau (1982) showed that activity of cytochrome P450 catalyzed hydroxylation of (+)-sabinene synthesis in common garden sage.
In recent work with WRC, we discovered and functionally characterized the monoterpene synthase gene encoding (+)-sabinene synthase (Foster et al., 2013). Terpene synthases are well characterized in gymnosperms and angiosperms (Keeling and Bohlmann, 2006; Chen et al., 2011). In contrast and to the best of our knowledge, a P450 gene involved in monoterpene biosynthesis has not yet been functionally characterized in a gymnosperm. The only functionally characterized terpene-modifying P450 genes of gymnosperms are CYP725A of taxol diterpene biosynthesis in Taxus spp. (Croteau et al., 2006; Rontein et al., 2008), CYP720B1 and CYP720B4 of diterpene resin acid biosynthesis in loblolly pine (Pinus taeda; Ro et al., 2005) and Sitka spruce (Picea sitchensis; Hamberger et al., 2011), and CYP706M1 of nootkatone sesquiterpene biosynthesis in yellow cypress (Callitropsis nootkatensis; Cankar et al., 2014).
Building on a previous WRC transcriptome analysis (Foster et al., 2013), we describe here, to our knowledge, the first gene discovery and functional characterization of monoterpene-oxidizing P450s in a gymnosperm, specifically WRC (+)-sabinene hydroxylases CYP750B1 and CYP76AA25. A biosynthetic function of CYP750B1 in α- and β-thujone biosynthesis is supported by profiles of metabolite and transcript accumulation in contrasting genotypes of WRC with distinct patterns of terpenoid and thujone accumulation. WRC CYP750B1 is, to our knowledge, the first functionally characterized member of the apparently gymnosperm-specific CYP750 family.
RESULTS
Monoterpene Profiles of WRC Foliage and Bark
Metabolite analysis by gas chromatography (GC)-mass spectrometry (MS) revealed similar monoterpenoid profiles in extracts of foliage and green bark of WRC saplings (genotype 5309) as shown with the results from three biological replicates (Table I). Genotype 5309 represents a phenotype with high levels of monoterpenoids and high resistance to deer browsing when tested against lower monoterpenoid genotypes (J. Russell, unpublished data). α-thujone was the major monoterpenoid in foliage and bark, accounting for two-thirds of total monoterpenoid content and 0.2% to 0.4% (w/w) of tissue dry weight. The second most abundant compound in both foliage and bark was (+)-sabinene followed by β-thujone.
Table I. Quantification and identification of monoterpenoids in WRC foliage and bark extracts.
| Monoterpene |
RI DB1 |
RI HP5 |
Foliage | Bark | ||
|---|---|---|---|---|---|---|
| Determined | Reference | Determined | Reference | |||
| mg g−1 dry wt ± sea | ||||||
| α-Thujeneb,c | 932 | 932 | 924 | 924 | 0.007 ± 0.0018 | 0.021 ± 0.0078 |
| α-Pineneb,d | 938 | 936 | 929 | 932 | 0.019 ± 0.0038 | 0.086 ± 0.0287 |
| Camphened,e | 950 | 955 | 943 | 946 | 0.001 ± 0.0001 | 0.001 ± 0.0007 |
| Sabineneb,c,f | 970 | 973 | 970 | 969 | 0.507 ± 0.0320f | 0.481 ± 0.1523f |
| β-Pineneb,d | 973 | 972 | 974 | 974 | 0.001 ± 0.0005 | 0.004 ± 0.0019 |
| β-Myrceneb,c | 986 | 987 | 988 | 988 | 0.056 ± 0.0071 | 0.083 ± 0.0364 |
| α-Phellandreneb,d | 996 | 1,002 | 1,002 | 1,002 | 0.188 ± 0.0211 | 0.039 ± 0.0341 |
| α-Terpineneb,c | 1,011 | 1,013 | 1,014 | 1,014 | 0.053 ± 0.0080 | 0.033 ± 0.0094 |
| p-Cymeneb,d | 1,015 | 1,015 | 1,022 | 1,022 | 0.009 ± 0.0038 | 0.023 ± 0.0133 |
| β-Phellandreneb,d | 1,021 | 1,023 | 1,028 | 1,025 | 0.010 ± 0.0017 | 0.001 ± 0.0210 |
| (S)-Limoneneb,c | 1,023 | 1,025 | 1,026 | 1,024 | 0.030 ± 0.0006 | 0.066 ± 0.0002 |
| γ-Terpineneb,c | 1,051 | 1,051 | 1,058 | 1,054 | 0.054 ± 0.0064 | 0.031 ± 0.0095 |
| trans-Sabinene hydratee,g | 1,054 | 1,053 | 1,064 | 1,065 | 0.018 ± 0.0088 | 0.017 ± 0.0073 |
| p-Cymenened,e | 1,075 | 1,075 | 1,088 | 1,082 | 0.001 ± 0.0004 | 0.003 ± 0.0024 |
| Terpinoleneb,g | 1,081 | 1,086 | 1,086 | 1,086 | 0.023 ± 0.0098 | 0.012 ± 0.0041 |
| cis-Sabinene hydrateb,g | 1,082 | 1,096 | 1,083 | 1,083 | 0.016 ± 0.0135 | 0.006 ± 0.0048 |
| α-Thujoneb,c,f | 1,088 | 1,089 | 1,103 | 1,101 | 3.507 ± 0.6845f | 2.304 ± 0.9221f |
| β-Thujoneb,h,f | 1,098 | 1,103 | 1113 | 1,112 | 0.364 ± 0.0802f | 0.216 ± 0.0745f |
| Thujol/neothujole,g | 1,134 | 1,136 | 1,148 | 1,149 | 0.159 ± 0.0372 | 0.003 ± 0.0022 |
| Cymen-8-ole,g | 1,162 | 1,169 | 1,183 | 1179 | 0.010 ± 0.0047 | |
| Terpinen-4-olb,c | 1,164 | 1,164 | 1,174 | 1,174 | 0.014 ± 0.0026 | 0.019 ± 0.0100 |
| trans-Sabinyl acetateb,g | 1,275 | 1,278 | 1,292 | 1,289 | 0.002 ± 0.0004 | 0.006 ± 0.0026 |
| Carvacrole,g | 1,279 | 1,278 | 1,302 | 1,298 | 0.011 ± 0.0024 | |
se based on three biological replicates.
Identified by comparison with authentic standards.
Quantified using concentration series of authentic standard.
Quantity estimated using sabinene Rf (nonoxygenated compounds).
Identified based on comparison with NIST or Massfinder4 databases.
The three major monoterpene compounds are shown in boldface.
Quantity estimated using terpinene-4-ol Rf (oxygenated compounds).
Quantity estimated using α-thujone Rf.
Detection of (+)-trans-Sabinol, (+)-cis-Sabinol, and (+)-Sabinone
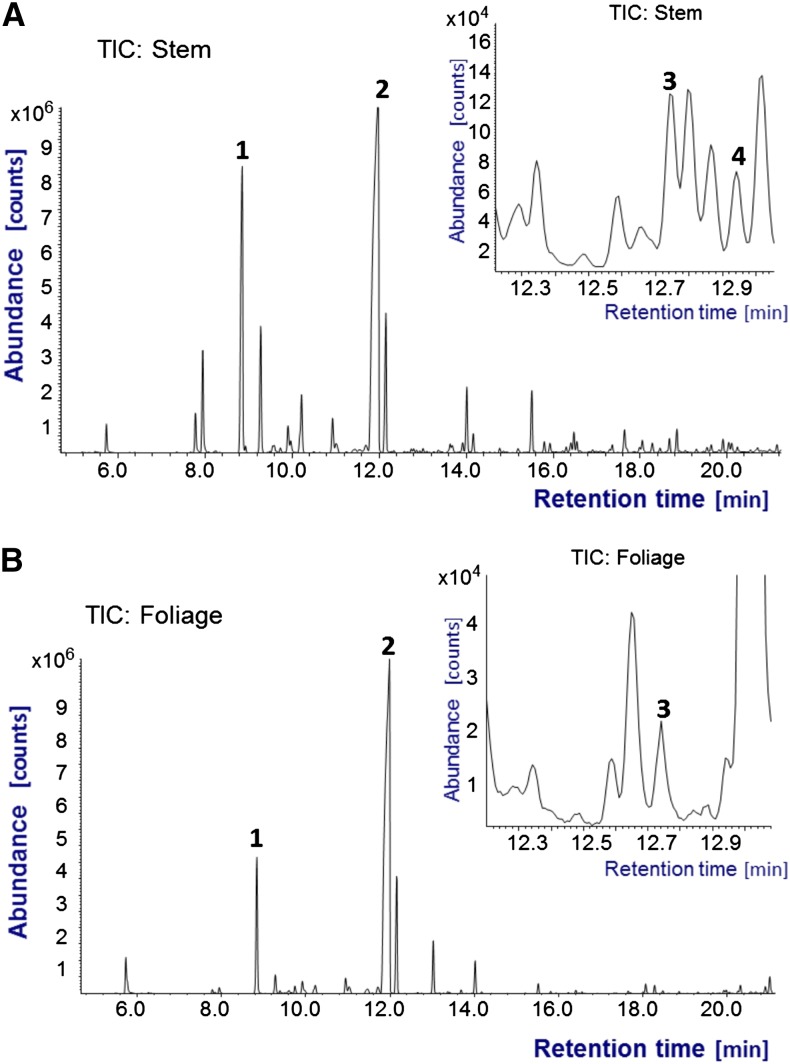
Thujone biosynthesis is proposed to start with (+)-sabinene synthase activity (Foster et al., 2013) and proceed through the intermediates (+)-trans- or (+)-cis-sabinol and (+)-sabinone (Fig. 1). We synthesized authentic standards of (+)-trans- and (+)-cis-sabinol and (+)-sabinone to support detection of these compounds in WRC. NMR spectra were recorded to determine the absolute configuration of the (+)-sabinol 3-hydroxyl group (Supplemental Fig. S1). (+)-trans- and (+)-cis-Sabinol and (+)-sabinone could be separated by GC, and their MS patterns were obtained (Supplemental Fig. S2). We detected (+)-trans-sabinol, but not (+)-cis-sabinol, and (+)-sabinone in WRC tissue extracts (Fig. 2).
Figure 2.
Detection of (+)-trans sabinol and (+)-sabinone in WRC green bark and foliage tissue extracts. Shown are total ion chromatograms (TICs) of green bark (A) and foliage (B) extracts. Insets represent magnifications of TIC traces between 12.2 and 14 min. Extracts were separated on a DB1 column. (+)-Sabinene (peak 1), α-thujone (peak 2), and (+)-trans-sabinol (peak 3) were detected in bark and foliage tissue. In addition, (+)-sabinone (peak 4) was detected in bark tissue. (+)-cis-Sabinol was not detected.
Identification of P450 Candidate Genes for Functional Characterization
In previous work, we identified in the transcriptome of WRC several putative P450 candidates for thujone biosynthesis, including CYP750B1 (Foster et al., 2013). We reanalyzed the WRC transcriptome assembly, with focus on P450s and their annotation according to established clans, families, and nomenclature (Nelson, 2006). Our analysis identified WRC P450 sequences corresponding to most of the plant P450 clans; except, no WRC P450s were found for clans 710, 711, and 746.
All currently known plant P450s with functions as monoterpene oxidases are members of the CYP71 clan (Hamberger and Bak, 2013). We, therefore, targeted WRC P450 contigs falling into families of this clan for additional analysis. We found a total of 26 different contigs belonging to the CYP71 clan (Fig. 3). Within the CYP71 clan, WRC sequences clustered with families representing P450s of well-defined functions in biological processes, such as phenylpropanoid biosynthesis (CYP73 and CYP98 families), flavonoid biosynthesis (CYP75 family), gibberellic acid biosynthesis (CYP701 family), chlorophyll oxidation (CYP89 family), and fatty acid oxidation (CYP77 family). Other WRC P450 sequences fell into families CYP76 (oxidation of terpenoids, fatty acids, lignans, and xenobiotics), CYP81 (oxidation of flavonoids, glucosinolates, and xenbiotics), CYP93 (oxidation of flavonoids and glucosinolates), and CYP736 (glucosinolate metabolism). We did not find WRC representatives of lineage-specific families, such as CYP705, CYP719, and CYP726, members of family CYP84, a family involved in S-lignin biosynthesis, and pollen-specific CYP703. Also, no WRC sequences were identified that cluster within the CYP71 family involved in terpenoid metabolism.
Figure 3.
Phylogeny of WRC P450 contigs and full-length sequences of the CYP71 clan and their association with CYP450 families of the CYP71 clan. Nine different full-length P450 sequences were annotated according to P450 nomenclature standards (Nelson, 2006). Phylogenetic relationship was analyzed using the Parsimony method based on a ClustalW protein sequence alignment in MEGA. Quality of the tree was analyzed by bootstrapping. Branches with bootstrap support lower than 50% are collapsed in a strict consensus tree.
Four WRC P450 sequences, including the full-length CYP750B1, fell into the gymnosperm-specific CYP750 family of unknown functions (Fig. 3). Transcripts abundance of CYP750B1 was previously shown to correlate with transcripts of (+)-sabinene synthase (Foster et al., 2013), which made this sequence an interesting candidate for functional characterization and testing of a possible role in thujone biosynthesis. In addition, we targeted eight WRC full-length CYP76 family members (Fig. 3) for functional characterization that represent the highest sequence similarity with known terpenoid-metabolizing enzymes, such as peppermint (Mentha × piperita) limonene-3-hydroxylase (CYP71D13; Lupien et al., 1999), strawberry (Fragaria vesca) pinene-10-hydroxylase (Aharoni et al., 2004), or Madagascar periwinkle (Catharanthus roseus) geraniol 10-hydroxylase (CYP76B6; Collu et al., 2001). These eight WRC CYP76 sequences belong to subfamily CYP76AA, with the sago palm (Cycas rumphii) expressed sequence tag CYP76AA1 as its first described member (National Center Biotechnology Information [NCBI] gene identification no. 27916177), and subfamily CYP76Z, with Sitka spruce (UniGene CYP76Z1) as its first described member (NCBI gene identification no. 1706841). To the best of knowledge, functions for members of the CYP76AA and CYP76Z subfamilies have not yet been identified in any species.
Functional Characterization of CYP750B1 and CYP76AA25 as (+)-Sabinene 3-Hydroxylases
Full-length coding sequences for CYP750B1, CYP76AA20, CYP76AA21, CYP76AA22, CYP76AA23, CYP76AA24, CYP76AA25, CYP76AA26, and CYP76Z2 were retrieved directly from the transcriptome assembly (for six of nine P450s) or obtained by additional 5′RACE and 3′RACE cloning based on partial contig sequences (for three of nine P450s). P450s were individually expressed from the pESC-LEU2d-u plasmid in yeast (Saccharomyces cerevisiae) BY4741 cells containing a genomically integrated plant cytochrome P450 reductase (lodgepole pine [Pinus contorta] cytochrome P450 reductase [LpCPR]). Assays were performed with microsomal membrane fractions of the transformed yeast cells containing 0.55 to 0.75 mg mL−1 of total protein, corresponding to 0.8 to 5.4 μg of P450 protein per assay, which was determined by the CO difference spectra analysis. LpCPR activity of the microsomal membrane preparations was confirmed by cytochrome c reduction assay. For example, LpCPR activity was 1.98 and 1.92 μmol min−1 mg−1 of protein for microsomal membrane preparations containing CYP750B1 and CYP76AA25, respectively.
Each of the nine expressed P450 proteins was tested against a panel of 19 different substrates, including eight different monoterpenes, seven different sesquiterpenes, and four nonterpenoid substrates (Table II). Six of nine P450 proteins (CYP76AA20, CYP76AA21, CYP76AA22, CYP76AA23, CYP76AA25, and CYP750B1) were active with one or several of the substrates tested: CYP76AA20 was active with the herbicide isoproturon, CYP76AA21 and CYP76AA22 were active with the monoterpene (S)-limonene and isoproturon, and CYP76AA23 was active with the sesquiterpenes cadinene and cedrene and isoproturon. None of the terpenoid reaction products could be detected in WRC tissue extracts. Two P450 proteins, CYP76AA25 and CYP750B1, were active with (+)-sabinene (Table II).
Table II. Relative catalytic activity of CYP76AA25 and CYP750B1 with different monoterpene, sesquiterpene, and nonterpenoid substrates.
| Substrate | CYP76AA25 kcat | CYP750B1 kcat |
|---|---|---|
| % | ||
| Monoterpenes | ||
| (+)-Sabinene | 100a | 100b |
| (R)-Limonene | 0 | 0 |
| (S)-Limonene | ndc | ndc |
| 3-Carene | 0 | 0 |
| Geraniol | 0 | 0 |
| Linalool | 0 | 0 |
| Nerol | ndc | 0 |
| Terpinolene | 0 | 0 |
| Sesquiterpenes | ||
| Farnesene | 29 | 0 |
| Cadinene | 0 | 0 |
| Caryophyllene | 0 | 0 |
| Cedrene | 0 | 0 |
| Germacrene | 0 | 0 |
| Humulene | 0 | 0 |
| Longifolene | 0 | 0 |
| Nonterpenoid | ||
| Naringenine | 0 | 0 |
| Isoproturon | 5 | 0 |
| 7-Methoxycoumarine | 0 | 0 |
| Matairesinol | 0 | 0 |
100% is 1.30 min−1.
100% is 0.20 min−1.
Trace activity of <0.1% substrate conversion in standard assays (kcat) could not be determined.
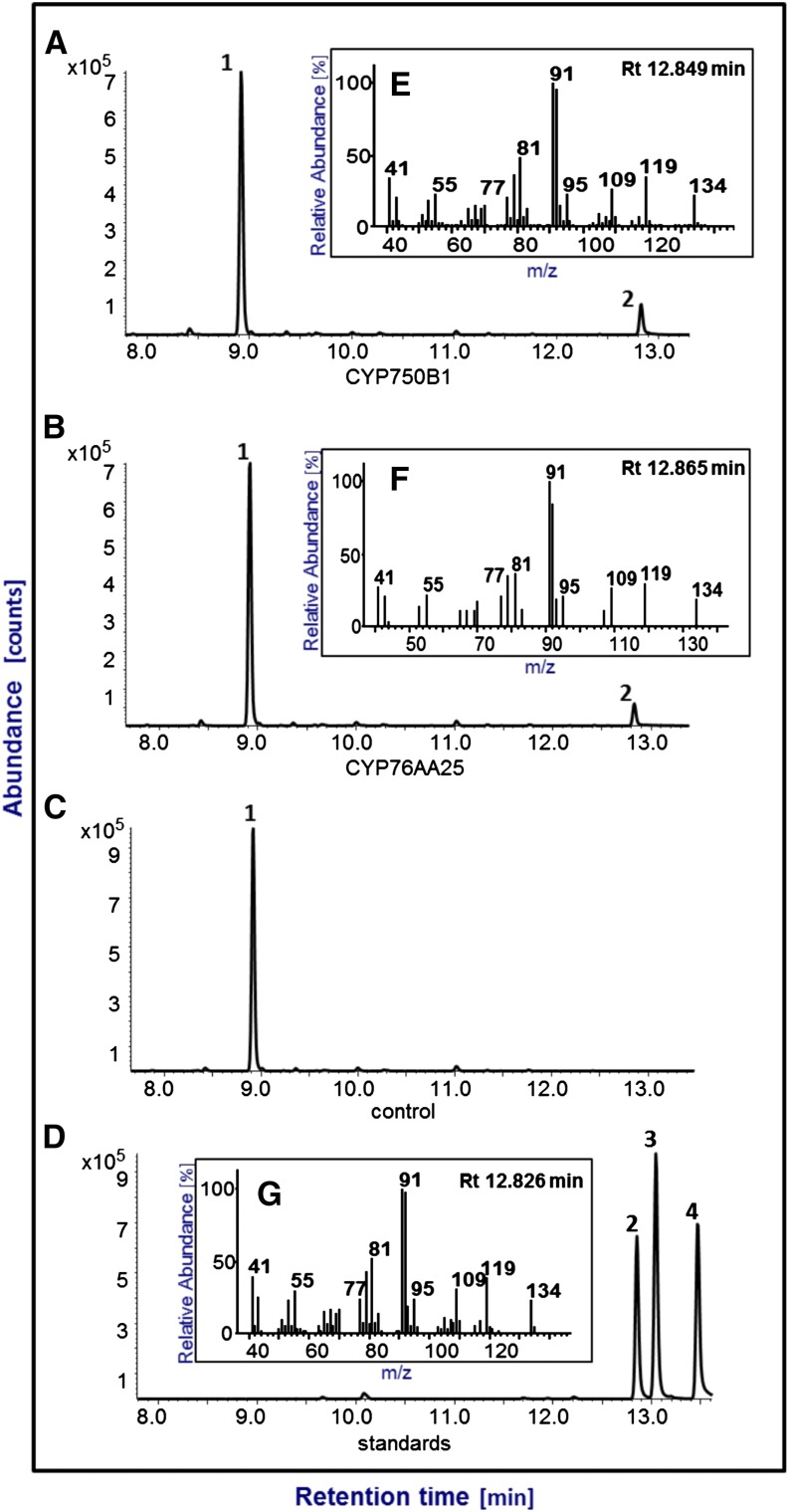
Both CYP76AA25 and CYP750B1 catalyzed the 3-hydroxylation of (+)-sabinene and produced stereoselectively (+)-trans-sabin-3-ol but not (+)-cis-sabinol (Fig. 4). For CYP750B1, (+)-sabinene is the preferred substrate (Km = 110 ± 0.3 μm); a very minor conversion was detected with (S)-limonene. In addition to (+)-sabinene (Km = 299 ± 0.2 μm), CYP76AA25 also converted the sesquiterpene farnesene (Km = 10.3 ± 0.2 μm) and the herbicide isoproturon (Km = 30.5 ± 0.4 μm; Figs. 5 and 6). Minor conversion of nerol and (S)-limonene by CYP76AA25 was also detected. These results revealed CYP750B1 and CYP76AA25 as (+)-sabinene 3-hydroxylases producing stereoselectively (+)-trans-sabinol. In contrast to CYP76AA25, CYP750B1 seems to be more selective for (+)-sabinene as a substrate (Table II).
Figure 4.
Conversion of (+)-sabinene (1) by CYP750B1 and CYP76AA25 producing stereoselectively (+)-trans-sabinol (2). Shown are total ion chromatograms of (+)-sabinene conversion by CYP750B1 (A), (+)-sabinene conversion by CYP76AA25 (B), empty vector control reactions (C), and authentic standards (D). Peaks correspond to (+)-sabinene (1), (+)-trans-sabinol (2), (+)-sabinone (3), and (+)-cis-sabinol (4). Insets show the mass fragmentation pattern of CYP750B1 assay product peak 2 (E) and CYP76AA25 assay product peak 2 (F) coeluting with the (+)-trans-sabinol authentic standard (G). Enzyme and control reactions were incubated for 1 h at 30°C. Pentane extracts of assays and standards were separated on a DB1 column. Rt, Retention time.
Figure 5.
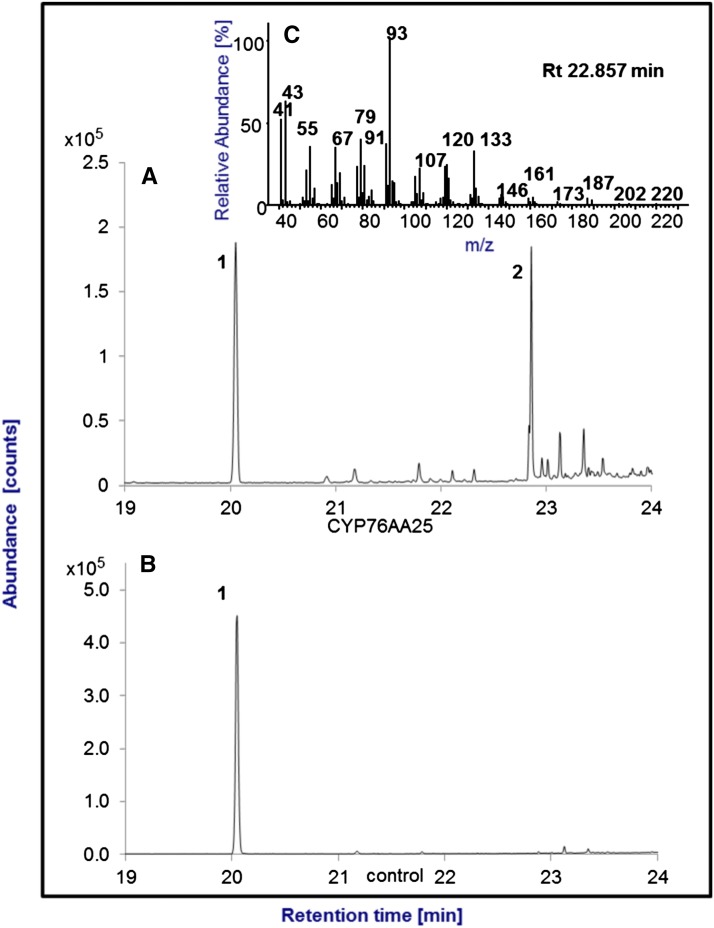
Farnesene conversion by CYP76AA25. Shown are total ion chromatograms of conversion of farnesene (1) in assays with CYP76AA25 (A) and lack of conversion of farnesene in negative control assays without CYP76AA25 after 1 h of incubation at 30°C and separation of the pentane extract on a DB1 column (B). The reaction product (2) formed could not be identified with any of our available authentic standards. Based on mass fragmentation pattern (C), this product is similar or identical to 2,6,10-trimethyldodeca-2,6,10-trienal. Rt, Retention time.
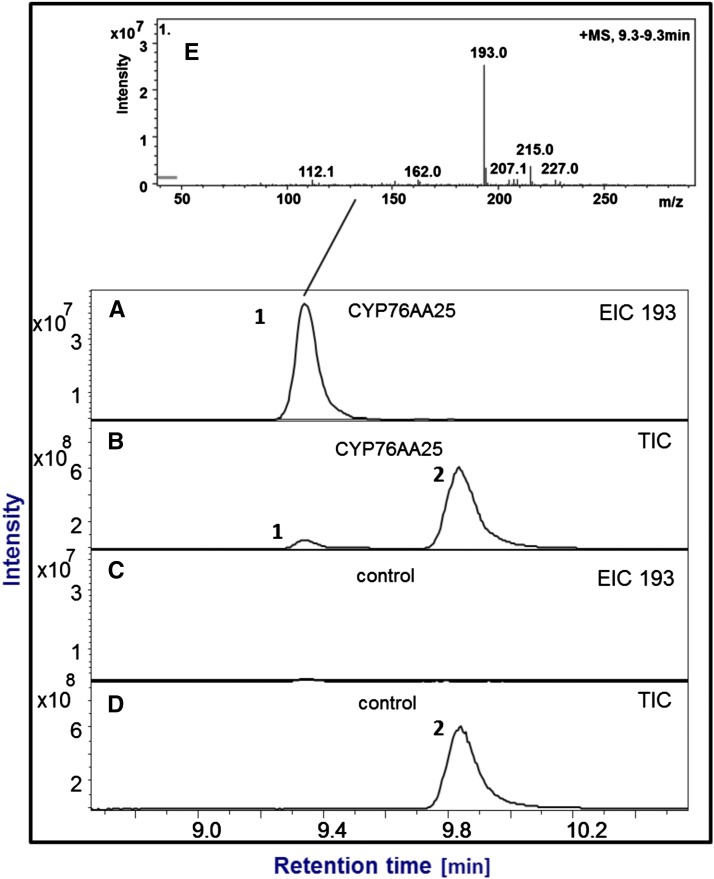
Figure 6.
Isoproturon conversion by CYP76AA25. Shown are total ion chromatograms (TICs) and extracted ion chromatograms (EICs; 193) from liquid chromatography-MS analyses. A and B, Product (1) formed during conversion of isoproturon (2) by CYP76AA25. C and D, Negative control without CYP76AA25. E, Mass spectrum of the product with the major mass ion adducts 193 (M + H) and 215 (M + Na). Assays were performed with 1 h of incubation at 30°C. Reaction product (retention time = 9.32 min) of molecular mass 192 is in agreement with demethylation to N-(4-isopropylphenyl)-N'-methylurea, a known product for CYP450-catalyzed isoproturon conversion (Robineau et al., 1998).
Transcript Profiles of CYP750B1 But Not CYP76AA25 Correlate with Thujone Accumulation
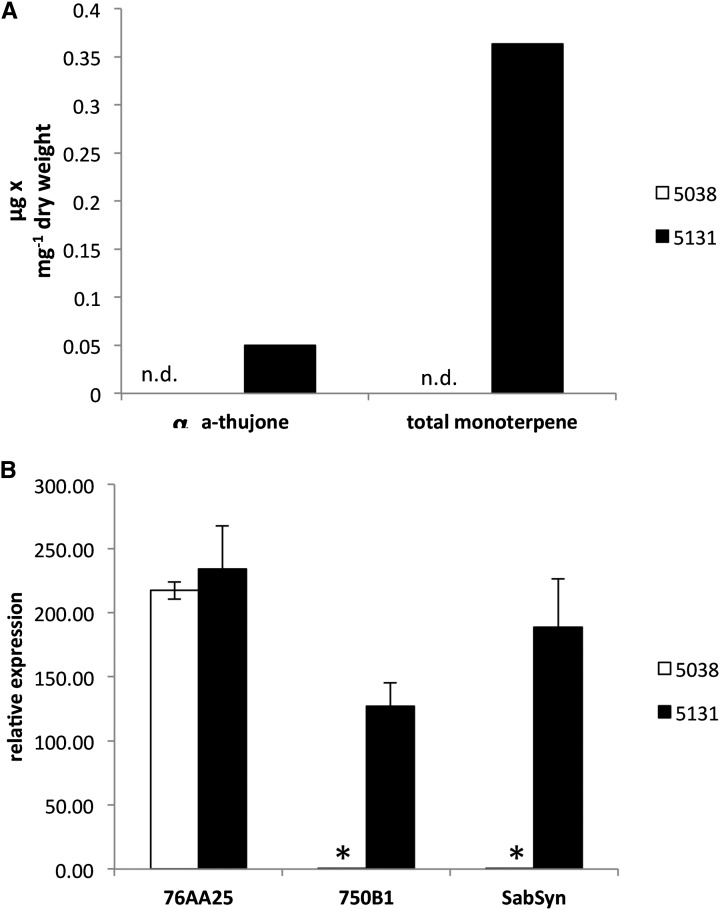
Transcript abundance of CYP750B1 and CYP76AA25 was measured first in two contrasting WRC genotypes, 5038 and 5313. Genotype 5038 represents a glandless, low-thujone phenotype that does not accumulate monoterpenes as shown in Foster et al. (2013). For comparison, genotype 5131 represents a typical phenotype with foliar glands for accumulation of thujone and other monoterpenes. Extreme differences in monoterpene content of foliage of 5038 and 5131 are shown in Figure 7. Patterns of high transcript abundance of (+)-sabinene synthase and CYP750B1 in 5131 and their very low levels in 5038 correlate well with the presence and the absence of thujone in 5131 and 5038, respectively (Fig. 7), confirming previous observations (Foster et al., 2013). In addition, we found that transcript abundance of CYP76AA25 did not correlate with differences in thujone content in 5131 and 5038 (Fig. 7).
Figure 7.
Contrasting patterns of α-thujone accumulation and transcript accumulation in WRC glandless genotype 5038 foliage, which does not accumulate monoterpenes, and monoterpenoid-accumulating genotype 5131 foliage. A, Total monoterpene and α-thujone content in foliage of genotypes 5038 and 5131. Monoterpenes, including thujones, were not detectable (n.d.) in glandless 5038. B, Transcript accumulation for CYP76AA25, CYP750B1, and (+)-sabinene synthase quantified by qRT-PCR. Data are shown as normalized expression against EF-α expression. Error bars show se based on three biological replicates. *, Significant differences of transcript abundance between 5038 and 5131 as determined by t test (P < 0.05).
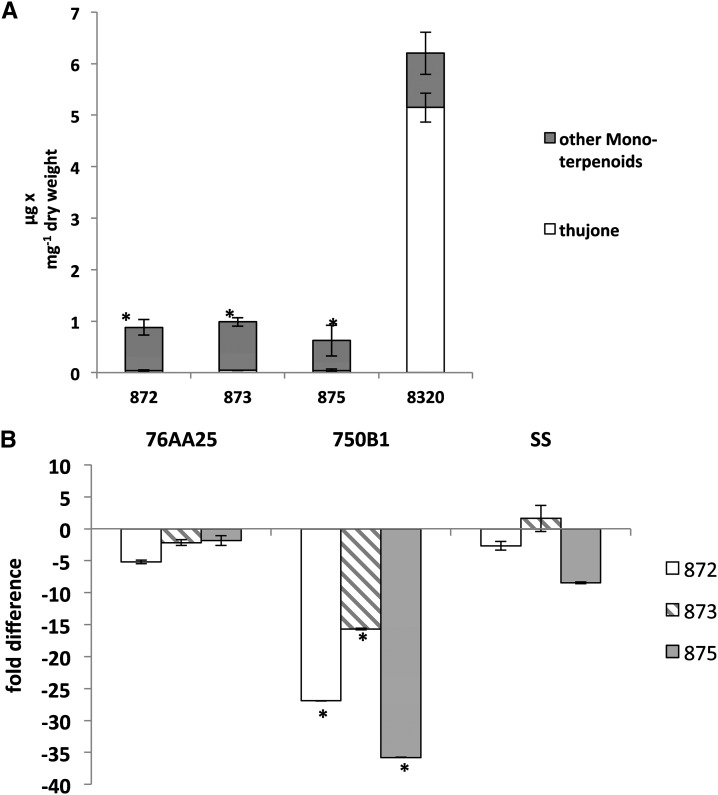
To extend this analysis, we compared thujone levels and transcript abundance in four different second generation inbred lines (S2), three of which (872, 873, and 875) had drastically reduced α-thujone levels compared with a reference line 8320 (Fig. 8). Comparison of transcript abundance in lines 872, 873, and 875 relative to line 8320 showed significantly reduced levels of CYP750B1 transcripts but not CYP76AA25 transcripts in the low α-thujone lines (Fig. 8). These results together with results from the characterization of P450 enzyme functions support a specific role of CYP750B1 in thujone biosynthesis in WRC. A function of CYP76AA25 in thujone biosynthesis is ambiguous.
Figure 8.
Thujone, monoterpene, and transcript accumulation in WRC foliage of four different second generation inbred lines, including three lines with low-thujone phenotypes. A, Content of α-thujone and total content of other monoterpenes in three low-thujone lines (872, 873, and 875) and line 8,320. B, Transcript accumulation for CYP76AA25, CYP750B1, and sabinene synthase quantified by qRT-PCR and normalized to EF-α expression (shown as x fold difference in low-α-thujone lines [872, 873, and 875] relative to line 8,320). Error bars show se based on three biological replicates per line. SS, Sabinene synthase; *, significant differences (t test, P < 0.05) between low-α-thujone lines 872, 873, and 875 and line 8320.
In Vitro Conversion of (+)-trans-Sabinol to (+)-Sabinone by WRC Tissue Extracts
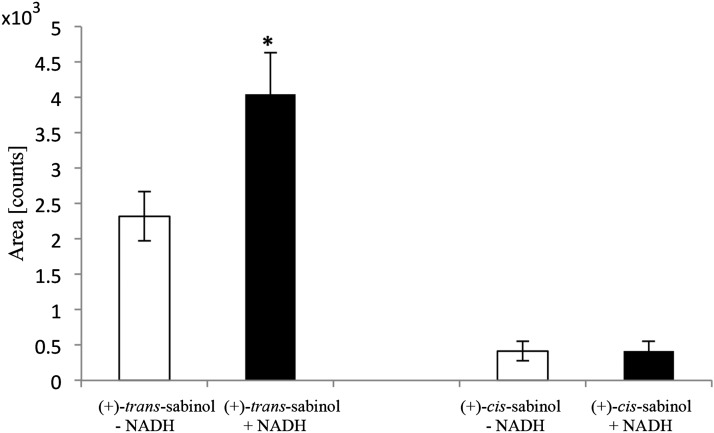
Given that (+)-trans-sabinol, but not (+)-cis-sabinol, was present in WRC tissue extracts (Fig. 2) and after the identification of (+)-trans-sabinol, but not (+)-cis-sabinol, as the product of CYP750B1 (Fig. 4), we tested if, indeed, (+)-trans-sabinol could serve as the substrate in the next step of thujone biosynthesis, presumably involving (+)-sabinol dehydrogenase activity to yield (+)-sabinone (Fig. 1). Using cell-free tissue extracts of soluble proteins, we tested the NADH-dependent conversion of both (+)-trans- and (+)-cis-sabinol. We detected NADH-dependent conversion of (+)-trans-sabinol but not (+)-cis-sabinol to sabinone (Fig. 9). This result together with lines of evidence from tissue metabolite analysis [presence of (+)-trans-sabinol but not (+)-cis-sabinol] and stereoselective formation of (+)-trans-sabinol by CYP750B1 strongly support a role of (+)-trans-sabinol as the biological intermediate in thujone biosynthesis in WRC.
Figure 9.
NADH-dependent conversion of (+)-trans-sabinol but not (+)-cis-sabinol by cell-free protein extracts of WRC tissue. (+)-trans-Sabinol is converted to (+)-sabinone by cell-free protein extracts. Assays containing soluble protein extracts were incubated for 90 min at 30°C with either (+)-trans-sabinol or (+)-cis-sabinol with and without NADH. Assays products were extracted, separated on a DB1 column, and analyzed by GC-MS. Sabinone production was confirmed by comparing product peak retention time and mass fragmentation with authentic standards, and areas were calculated for ion 108 (mass to charge ratio). Shown are average values for quadruplicate experiments. *, Significant differences with and without NADH added in assays with (+)-trans-sabinol as determined by t test (P < 0.05).
DISCUSSION
Monoterpenoids and in particular, thujones have been shown to influence deer-browsing resistance in WRC (Vourc’h et al., 2001, 2002; Kimball et al., 2012). We recently identified a monoterpene synthase gene, (+)-sabinene synthase, encoding the first step in thujone biosynthesis in WRC (Foster et al., 2013). Here, we report the cloning and functional characterization of a gymnosperm-specific P450 enzyme CYP750B1, which functions as a (+)-sabinene 3-hydroxylase and catalyzes the second of four steps in the thujone pathway (Fig. 1). To the best of our knowledge, this is the first report of a functionally characterized P450 gene of monoterpene biosynthesis in any gymnosperm. CYP750B1 is also, to our knowledge, the first functionally characterized member of the apparently gymnosperm-specific CYP750 family. CYP750B1 produces stereoselectively (+)-trans-sabinol but not (+)-cis-sabinol, which was shown with a set of newly synthesized authentic standards. A role of (+)-trans-sabinol as a relevant intermediate in thujone biosynthesis was substantiated with enzyme assays using WRC tissue extracts, which selectively converted (+)-trans-sabinol but not (+)-cis-sabinol to (+)-sabinone.
A P450 enzyme in thujone biosynthesis was originally shown by Karp and Croteau (1982) in common garden sage. Thujone biosynthesis in common garden sage, an angiosperm, and WRC, a gymnosperm, seems to involve different stereoisomers: (+)-cis- and (+)-trans-sabinol, respectively. A sabinene hydroxylase has not yet been cloned and characterized from common garden sage or any other angiosperm species, and it is not known to which P450 family such a gene would belong. Given that WRC CYP750B1 is a member of the gymnosperm-specific CYP750 family, it is quite possible that P450s of different stereospecificities in the formation (+)-trans- and (+)-cis-sabinol evolved independently in WRC and common garden sage.
Our initial functional characterization of a set of nine P450 candidates revealed two P450s, CYP750B1 and CYP76AA25, that were able to convert (+)-sabinene to (+)-trans-sabinol. Several lines of evidence supported a role of CYP750B1 in thujone biosynthesis, whereas such a role was not supported for CYP76AA25. On the biochemical level, CYP750B1 seems to have narrow substrate specificity, with (+)-sabinene being the only substrate identified in a panel of 19 different compounds. On the molecular level, our work made use of two highly unique phenotype resources with regard to conifer monoterpene chemistry: (1) a clonally propagated glandless genotype (line 5038) and (2) a set of three different low-thujone S2 lines (872, 873, and 875). Foliar glands are the major site of monoterpene biosynthesis and accumulation in WRC. The glandless clone does not accumulate monoterpenes, consistent with a lack of expression of monoterpene biosynthesis (Russell and Ferguson, 2008; Russell and Yanchuk, 2012; Foster et al., 2013). The low-thujone S2 lines 872, 873, and 875 seem to be blocked more specifically in thujone biosynthesis. The phenotypes of these materials contrast with the high-monoterpene and high-thujone chemistry that is typical for WRC. Using these materials, we found a strong correlation of CYP750B1 transcript abundance with thujone accumulation in all of the different genotypes investigated. Transcript accumulation of both CYP750B1 and sabinene synthase was extremely low in the glandless, thujone-nonaccumulating line (Fig. 7). In all three low-thujone S2 lines, CYP750B1 transcript levels were significantly reduced compared with a reference line, whereas in this phenotype, sabinene synthase transcripts were not significantly reduced, which is in agreement with the presence of sabinene (Fig. 8). In contrast to CYP750B1, transcript abundance of CYP76AA25 did not correlate with thujone accumulation or transcript accumulation of sabinene synthase, and CYP76AA25 transcript abundance was unaffected in the glandless genotype and the low-thujone S2 lines (Figs. 7 and 8). Hence, in contrast to CYP750B1, there was no supporting molecular evidence for a role of CYP76AA25 in thujone biosynthesis.
In the context of naturally occurring variation of monoterpenoid and thujone accumulation in WRC, this work and the work by Foster et al. (2013) suggest that differences in transcript abundance of sabinene synthase and CYP750B1 is an important and potentially causative component for this chemophenotypic variation. Transcript abundance of both the sabinene synthase and CYP750B1 genes correlates with thujone accumulation and the presence and absence of glands, and both of these traits contribute to deer-browsing resistance. These genes may be used as transcript biomarkers to screen large numbers of WRC accessions in a high-throughput and automated fashion in addition to using existing methods of screening for chemical and histology phenotype. In future work, it will be important to identify the factors that control expression of these two genes and possibly other genes in the thujone pathway as well. Using the glandless WRC phenotype, it might be possible to identify genes or mechanisms that control development of secretory structures for monoterpene biosynthesis and accumulation as well as gland-specific gene expression of thujone biosynthesis.
Our results conclusively showed that the gymnosperm-specific cytochrome P450 enzyme CYP750B1 catalyzes the stereospecific monoterpene hydroxylation of (+)-sabinene, which is a critical step in the biosynthesis of α- and β-thujone, major defense compounds for deer-browsing resistance in WRC. To date, only a few gymnosperm draft genomes have been published (Birol et al., 2013; Nystedt et al., 2013; De La Torre et al., 2014; Neale et al., 2014), and a genome sequence for WRC is not yet available. Therefore, in the absence of RNA interference lines, it is not possible to conclude if other genes exist in WRC with similar or overlapping functions to those of CYP750B1. It is important to note that, in other gymnosperm trees, it has been shown that specific biochemical functions in terpenoid defense pathways are covered with several gene copies, such as multiple functionally similar monoterpene synthases (Hall et al., 2011; Roach et al., 2014) or multiple P450s of diterpene resin acid biosynthesis (Hamberger et al., 2011).
MATERIALS AND METHODS
Plant Material and Terpenes
All WRC (Thuja plicata) plant material was from the breeding program of the British Columbia Ministry of Forests, Lands and Natural Resource Operations at Cowichan Lake Research Station (CLRS). Four second generation inbred seedling lines (S2; 872, 873, 875, and 8320) and three clones (genotypes 5309, 5131, and 5038) were used for this study. S2 reference line 8320 has normal accumulation of thujone and other monoterpenes, whereas S2 lines 872, 873, and 875 accumulate low amounts of thujone and normal amounts of sabinine and other monoterepenes. Reference genotypes 5309 and 5131 have above-normal thujone accumulation, whereas 5038, a glandless genotype, has no detectable accumulation of monoterpenoids, including thujone. S2 lines were sown in March of 2013 in a greenhouse, and foliage was harvested from three individual seedlings per line in November of 2014 at CLRS for metabolite and transcript analyses. The clones were propagated as rooted cuttings in 2009 in a greenhouse and transferred from CLRS to the University of British Columbia greenhouse facility in 2011. Sapling trees were moved outside in October of 2013, maintained under outside conditions in 2-gallon pots, and harvested in September of 2014 for metabolite and transcript analyses. Monoterpene standards were purchased from Chromadex and Sigma-Aldrich. Sabinyl acetate was from Extrasynthese. (+)-cis- and (+)-trans-sabinol and (+)-sabinone were synthesized as described below.
Synthesis and Confirmation of (+)-cis-Sabinol, (+)-trans-Sabinol, and (+)-Sabinone
(+)-cis-Sabinol, (1S,3S,5S)-1-isopropyl-4-methylidenebicyclo[3.1.0]hexan-3-ol, was synthesized following Umbreit and Sharpless (1977) and Sirisoma et al. (2001). In detail, (+)-sabinene (70 µL; 0.43 mmol) was dissolved in dichloromethane (5 mL); SeO2 (0.0027 mmol) and t-butylhydroperoxide (240 µL) were added, and the reaction was stirred at room temperature overnight. After the addition of more SeO2 (0.009 mmol), stirring was continued for another 2 h. Benzene (1 mL) was added, and stirring continued for 2 h; then, the reaction was evaporated to near dryness, diluted with diethyl ether (10 mL), washed with 10% KOH, saturated with NaCl, dried (MgSO4), filtered, and evaporated to near dryness again. The remaining liquid was applied to a silica gel column (10 g), the flask was rinsed with petroleum ether (500 μL), and the petroleum ether was loaded onto the same column. The column was washed with 8 × 8 mL of petroleum ether, 8 × 5 mL of 4% (v/v) ether/ethyl acetate, and 1 × 5 mL of 5% (v/v) petroleum ether/ethyl acetate. (+)-cis-Sabinol was eluted in 7 × 5 mL of 5% (v/v) petroleum ether/ethyl acetate. Eluates were pooled, and one-half of the eluate was evaporated to dryness to give 15 mg (0.1 mmol) of the target compound as one single isomer as determined by 1H and 13C NMR. The 1H (CDCl3, 400 MHz, reference 7.26 ppm) data are: δ 4.98 (d, 1H, J = 2.3 Hz, H-10a), 4.87 (d, 1H, J = 2 Hz, H-10b), 4.25 to 4.16 (bs, 1H, H-3), 2.26 (dd, 1H, J = 12.2 Hz, J = 7.6 Hz, H-2a), 1.70 (dd, 1H, J = 8.3 Hz, J = 3.4 Hz, H-5), 1.57 (ddd, J = 12.2 Hz, J = 8.3 Hz, J = 1.5 Hz, H-2b), 1.40 (m, 1H, H-7), 0.95 (d, 1H, J = 6.8 Hz, H-8), 0.89 (d, 1H, J = 6.8 Hz, H-9), 0.65 (ddd, J = 8.3 Hz, J = 4.9 Hz, J = 1 Hz, H6a), and 0.57 (dd, 1H, J = 4.8 Hz, J = 3.4 Hz, H-6b). 13C (CDCl3, 100 MHz, reference 77.0 ppm): δ 156.11 (C-4), 101.96 (C-10), 71.42 (C-3), 37.63 (C-2), 33.43 (C-1), 32.9 (C-7), 27.87 (C-5), 19.61 (C-8 or C-9), 19.45 (C-9 or C8), and 17.79 (C-6).
(+)-trans-Sabinol, (1S,3R,5S)-1-isopropyl-4-methylidenebicyclo[3.1.0]hexan-3-ol, was obtained by basic hydrolysis of sabinyl acetate (40 mg) in ethanolic KOH (10%; Garside et al., 1969). After a 10-min incubation at room temperature, 3 parts of water was added to the solution, and the compound was extracted in an equal volume of pentene. The pentane phase was directly used or further purified by silica gel column chromatography (1.5 g). (+)-trans-Sabinol was purified by washing the column using pentene methyl tertiary-butyl ether (MTBE) gradient (3 mL of pentene, 1 mL of 90% pentene/10% MTBE, and 1 mL of 80% pentene/20% MTBE). (+)-trans-Sabinol was eluted in 70% pentene/30% MTBE, and the solvent was removed to give 27 mg (0.177 mmol; yield of 68%) of (+)-trans-sabinol, which was identified by 1H and 13C NMR. The 1H NMR (CDCl3, 400 MHz, reference 7.26 ppm) data are: δ 4.99 (s, 1H, H-10a), 4.93 (s, 1H, H-10b), 4.43 (d, 1H, J = 7.4 Hz, H-3), 2.05 (ddd, 1H, J = 13.9 Hz, J = 7.4 Hz, J = 2.1 Hz, H-2a), 1.72 (d, 1H, J = 13.8 Hz, H-2b), 1.64 (dd, 1H, J = 8.7 Hz, J = 3.4 Hz, H-5), 1.43 (hep, 1H, J = 6.8 Hz, H-7), 1.04 (t, 1H, J = 3.7 Hz, H-6a), 0.92 (d, 3 H, J = 6.8 Hz, H-8 or -9), 0.87 (d, J = 6.8 Hz, H-9 or -8), and 0.80 (ddd, 1H, J = 8.5 Hz, J = 4.2 Hz, J = 2.1 Hz, H-6b). 13C (CDCl3, 100 MHz, reference 77.0 ppm): 157.20 (C-4), 106.69 (C-10), 75.08 (C-3), 37.67 (C-1), 37.20 (C-2), 32.56 (C-7), 28.93 (C-5), 19.99 (C-6), 19.72 (C-8 or C-9), and 19.55 (C-9 or C-8).
(+)-Sabinone, (1S,5S)-1-isopropyl-4 methylidenebicyclo[3.1.0]hexan-3-on, was obtained by oxidation of both (+)-cis- and (+)-trans-sabinol following Dess and Martin (1983). In detail, (+)-sabinol (15 mg; 0.1 mmol) was dissolved in dichloromethane (2 mL), a slight excess of Dess-Martin periodinane reagent (47 mg; 0.11 mmol) was added, and the reaction was stirred for 6 h at room temperature. The reaction was diluted with pentane (1 mL) and applied directly to a pentane prewashed silica gel column (3 g). The compound was eluted with a gradient from 0% to 4% acetone in pentane. Product-containing fractions were pooled, concentrated in volume, and then diluted with pentane or acetone for additional analysis. (+)-Sabinone was confirmed by NMR. The 1H (600 MHz, acetone-d6, reference 2.05 ppm) data are: δ 5.60 (s, 1H, H-10a), 5.24 (s, 1H, H-10b), 2.50 (dd, 1H, J = 18.9, J = 2.7, H-2a), 2.23 (d, 1H, J = 19, H-2b), 2.14 (dd, 1H, J = 8.5, J = 3.6, H-5), 1.52 (q, 1H, J = 7 Hz, H-6), 1.11 (ddd, 1H, J = 8.3, J = 5.2, J = 2.9, H-6a), 0.99 (d, 3H, J = 6.7 Hz, H-8), 0.95 (d, 3H, J = 6.8 Hz, H-9), and 0.42 (dd, 1H, J = 5 Hz, J = 3.7 Hz, H-6b). 13C (150.9 MHz, acetone-d6, reference 29.84 ppm): δ 205.1 (C-3), 149.4 (C-10), 112.6 (C-4), 41.5 (C-2), 33.1 (C-5), 30.1 (C-1), 26.8 (C-7), 22.0 (C-6), 19.6 (C-9), and 19.5 (C-8).
Terpenoid Extraction from WRC Tissue
Foliage tips (approximately 2 cm in length) were collected into GC-MS vials containing 1 mL of pentane spiked with isobutyl benzene (5 µg mL−1) as the internal standard. After overnight incubation, samples were centrifuged for 15 min at 1,000g, and the pentane phase was transferred into new vials for GC-MS analysis.
GC-MS and Liquid Chromatography-MS Analysis
Monoterpenoid analysis was done by GC-MS (Agilent 6890A/5975C and 7890A/7000A). GC conditions were as follows. Injections were done in pulsed splitless mode with inlet temperature at 250°C. Helium was used as a carrier gas. Terpenoid metabolites from enzyme assay extracts were separated on a DB-WAX Column (122-7032; J&W; 30 m × 250 µm; 0.25-µm film thickness) with oven temperature at 40°C for 1 min; temperature was increased by 15°C min−1 to 150°C and 30°C min−1 to 250°C and held for 8 min. Average carrier gas velocity was 33 cm s−1. Metabolites from tissue extracts were separated on a DB1 Capillary Column (122-0132; J&W; 30 m × 250 µm; 0.25-µm film thickness). The oven temperature started at 40°C followed by a 6°C min−1 increase to 160°C and a 40°C min−1 increase to 300°C and ended with a hold for 5 min; the average carrier gas velocity was set to 33 cm s−1. For validation of metabolite identification in tissue extracts, additional analysis was performed on a HP5 Capillary Column (19091S-433; Agilent; 30 m × 250 µm; 0.25-µm film thickness), with oven temperature starting at 40°, increasing by 3°C min−1 to 120°C and 15°C min−1 to 280°C, and finally holding for 5 min; the average carrier gas velocity was set to 38 cm s−1. Terpenoids were identified by (1) comparison with authentic standards, (2) comparison with retention indices (RIs) published for DB1 and HP5 capillary columns searching MassFinder4’s internal databases (massfinder.com/wiki/MassFinder_4) and the National Institute of Standards and Technology (NIST) data collection incorporated into W9N08L database (Wiley), or (3) accessing the NIST compound information at http://webbook.nist.gov/chemistry. For quantification, response factors (Rfs) were established with concentration series of authentic standards. Isobutylbenzene (Sigma-Aldrich) was used as an internal standard for all experiments.
Enzymatic conversion of nonterpenoid compounds (isoproturon, naringenine, 7-methoxycoumarine, and matairesinol) was monitored by liquid chromatography-MS. For assays with isoproturon, extracts were separated on a C18 Column (Atlantis T3; Waters; 2.1 [i.d.] × 100 mm; 5-µm pore size) at a flow rate of 0.5 mL min−1 using a water-acetonitrile (ACN) gradient (0–1 min with 5% [v/v] ACN, 1–14 min with 5%–90% ACN, and 14–15 min with 90% ACN) containing 0.2% (v/v) formic acid in the mobile phase. Negative ion electrospray mass spectra of enzyme product and isoproturon were recorded by a coupled MSD-Trap-XCT_Plus (AgilentTechnologies Inc.).
Assignment of (+)-Sabinol Stereochemistry
Assignment of cis- and trans-configuration was based on the distance of C-6 methylene and C-3 hydroxyl groups (Supplemental Fig. S1). Because the 3-hydroxyl only in the cis isomer is close through space proximity to the C-6 CH2 group, the corresponding protons are shifted significantly to low field in the 1H NMR spectrum (cis, 0.65 ppm for 6a and 0.57 ppm for 6b; trans, 1.08 ppm for 6a and 0.85 ppm for 6b). The assignment agrees with data published by Ohloff et al. (1966) and Mamane et al. (2004). (+)-cis-Sabinol structure was determined by 1H and 13C NMR. A one-dimensional Nuclear Overhauser Effect difference spectroscopy revealed a correlation between H-3 and the syn proton at C-6 for the cis isomer, whereas no correlation between H-3 and any of the C-6 protons was observed for the trans isomer, confirming the trans and cis assignments.
RNA Isolation and Measurement of Transcript Abundance
RNA was isolated from WRC using Concert Plant Reagent (Invitrogen) following the manufacturer’s protocol for small-scale RNA isolation using 30 mg fresh weight of WRC tissue material. Sugar and other impurities as well as genomic DNA were removed using the Plant RNeasy RNA Mini Kit (Qiagen) and the RNAse Free DNAse Set (Qiagen); 1 µg of RNA was transcribed with Superscript III (Invitrogen) and Oligo(dT)20VN for 60 min at 42°C. The resulting complementary DNA (cDNA) was diluted to 3 ng μL−1. Quantitative PCR analysis was performed in a BioRad CFX96 Real-Time PCR Detection System following the SsoFast EvaGreen protocol (BioRad) and using primers as shown in Supplemental Table S1. Data were analyzed using the LinRegPCR Program (Ruijter et al., 2009) as described in Zifkin et al. (2012). Representative PCR products were purified and sequenced to confirm product specificity. We assessed four reference genes for quantitative reverse transcription (qRT)-PCR analysis in foliage of different WRC genotypes: actin, elongation factor-α (EF-α), glyceraldehyde 3-phosphate dehydrogenase, and RNA-Polymerase III. No variation was observed for EF-α and actin in the different genotypes. Relative transcription abundance of target genes was calculated by normalizing data against actin and EF-α expression.
Reassembly of WRC Transcriptome Sequences
Generation of 42 m pairs of 75-bp WRC transcriptome sequences using the Illumina Genome Analyzer IIx Platform was described by Foster et al. (2013). For quality control before assembly, we used FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequence trimming was performed with Trimmomatic (Lohse et al., 2012); 12 bp were trimmed from the 5′ end of reads. Additional trimming was done at the 3′ end of reads if they fell below a quality score of Q20. A minimum read length of 50 bp was used as a threshold. De novo transcriptomic assembly was performed on 25 m paired trimmed sequences using Trinity assembler (Grabherr et al., 2011). The assembly generated 75,507 contigs with an average length of 907 bp.
Identification of P450 Candidate Sequences
For the identification of candidate P450 sequences in the dataset of 75,507 transcriptome contigs, we used a set of 38 bait sequences (Supplemental Table S2) extracted from the NCBI Protein Database representing P450 families of all 11 plant P450 clades for a tBlastN search. The identified WRC contigs were consolidated and tested for redundancies and possible short overlapping regions, which were not recognized by the Trinity assembler, using the CLC Contig Assembler Tool. The obtained unique contigs were translated into protein sequences and analyzed for their phylogenetic relation to the bait sequences and each other. All sequence analyses were done using CLC Workbench, with the exception of phylogenetic reconstruction (see below)
P450 Full-Length cDNA Cloning
RNA was isolated from WRC foliage and reverse transcribed as described above. Amplicons were derived by PCR using gene-specific primer pairs (Supplemental Table S1) and Phusion Polymerase (NEB). Gel-purified (QIAquick; Qiagen) fragments were ligated into pJET2.1 vector (Fermentas), and the resulting plasmids were transformed into Alpha-Select Gold Efficiency Escherichia coli cells (Bioline). Plasmids were purified (QIAprep; Qiagen), and the nucleotide sequences were verified (3730 DNA Analyzer; Applied Biosystems) using Big Dye Reagent (Life Technologies) and vector-specific primer.
Phylogenetic Sequence Analysis
All phylogenetic analyses were conducted in MEGA5 (Nei and Kumar, 2000; Tamura et al., 2007). Protein sequences were aligned using ClustalW (Thompson et al., 1994). Phylogenetic relationships were reconstructed using maximum likelihood. Bootstrap values above 50% are shown as a percentage next to the branches.
Expression of CYP76AA25 and CYP750B1 in Yeast
A cassette for Uracil-Specific Excision Reagent (USER)-based cloning was inserted into multicloning site1 (MSC1) of the pESC-LEU2d vector (Ro et al., 2006) using the BamHI and HindIII site based on Hamann and Møller (2007). A PacI restriction site close to MCS2 was silenced by a 2-nucleotide exchange to enable USER cloning and obtain pESC-LEU2d-u (Supplemental Table S1). CYP76AA25 and CYP750B1 cDNAs were cloned into PacI, and Nb.BbvCI (NEB) digested pESC-LEU2d-u after amplification with T7 polymerase using USER-compatible primers (Supplemental Table S1) and treatment with USER enzyme (NEB). The resulting constructs were transformed into yeast (Saccharomyces cerevisiae) strain BY4741. The yeast BY4741 also contained a conifer (lodgepole pine [Pinus contorta]) CPR (accession no. KJ914574) integrated in the genome (BY4741:LpCPR). Proteins were expressed, and microsomal fractions containing the recombinant protein were isolated as described previously (Pompon et al., 1996; Ro et al., 2005; Hamberger et al., 2011). The amount of expressed P450 was calculated following the method by Omura and Sato (1964).
P450 Enzyme Assays
Microsomal preparations (30 µL) were added on ice to 270 µL of reaction mixture containing 50 mm potassium phosphate (pH 7.5), 0.8 mm NADPH, and 100 µm substrate in a GC-MS glass vial. Assays were incubated under constant shaking at 30°C for 1 h for qualitative analysis or variable times of 1 to 180 min for kinetic analysis. Reactions were stopped by adding 300 µL of pentane, rigorous mixing for 1 min, and subsequent freezing at −80°C. After phase separation by centrifugation at 500g for 10 min, the pentane layer was analyzed by GC-MS.
Sabinol Dehydrogenase Assays
Soluble protein extracts from WRC foliage were obtained using a modified protocol described for common garden sage (Salvia officinalis) monoterpene dehydrogenases (Dehal and Croteau, 1987). In detail, 1 g of fresh frozen foliage tissue (line 5309) was ground into a fine powder under liquid nitrogen and resuspended on ice in extraction buffer (pH 6.5; 250 mm Suc, 20 mm sodium pyrosulfite, 100 mm sodium phosphate, 10 mm sodium ascorbate, 5 mm dithiothreitol [DTT], 1 mm EDTA, and 0.0275 g mL−1 insoluble polyvinylpolypyrrolidone). After centrifugation for 20 min at 27,000g at 4°C, 2.5 mL of supernatant was buffer exchanged into assay buffer (Tris-HCl, pH 7.5 and 2 mm DTT) and cleared of low-Mr metabolites on PD-10 Desalting Columns (GE-Healthcare). Enzyme assays were performed with 100 µL of desalted protein extract mixed with 200 µL of assay buffer (Tris-HCl, pH 7.5, and 2 mm DTT), 1 mm NADH, and 100 µm either (+)-trans- or (+)-cis-sabinol. Assays were incubated for 90 min at 30°C. Negative controls were done without NADH. Reactions were stopped by adding 300 µL of pentane, rigorous mixing for 1 min, and subsequent freezing at −80°C. After phase separation by centrifugation at 500g for 10 min, the pentane layer was analyzed by GC-MS.
Sequence data from this article can be found in the National Center Biotechnology Information GenBank under accession numbers CYP750B1 (KP004988), CYP76AA20 (KP015848), CYP76AA21 (KP015849), CYP76AA22 (KP015850), CYP76AA23 (KP015851), CYP76AA24 (KP015852), CYP76AA25 (KP015853), CYP76AA26 (KP015854), and CYP76Z2 (KP015855).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Determination of (+)-sabinol stereochemistry.
Supplemental Figure S2. GC-MS separation and detection of (+)-trans- and (+)-cis-sabinol and (+)-sabinone.
Supplemental Table S1. Primers used for full-length cDNA isolation, qRT-PCR, and pESC-LEU2d modification.
Supplemental Table S2. P450 sequences with accession numbers and P450 family and clan associations used as search baits against the WRC transcriptome.
Supplementary Material
Acknowledgments
We thank Craig Ferguson (CLRS) for generous access to plant materials; Melina Biron, David Kaplan, Alfonso Lara Quesada, and Elizabeth Steves (University of British Columbia) for plant maintenance; David Nelson (University of Tennessee) for naming of P450 genes; Katrin Geissler (University of British Columbia) for the yeast expression cell line; and Karen Reid (University of British Columbia) for outstanding laboratory management.
Glossary
- ACN
acetonitrile
- cDNA
complementary DNA
- CLRS
Cowichan Lake Research Station
- DTT
dithiothreitol
- GC
gas chromatography
- MS
mass spectrometry
- MTBE
methyl tertiary-butyl ether
- NIST
National Institute of Standards and Technology
- qRT
quantitative reverse transcription
- Rf
response factor
- RI
retention index
- WRC
Western red cedar
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Strategic Grant to J.M. and J.B. and Discovery Grant to J.B.), Genome British Columbia (User Partnership Program Project funds for bioinformatics analysis to J.M., J.H.R., and J.B.), the Austrian Science Fund (Erwin Schroedinger Fellowship to M.B.), Michael Smith Laboratories and the University of British Columbia (funds for analytical work of metabolite profiling), and the University of British Columbia (Distinguished Scholar Award to J.B.).
Articles can be viewed without a subscription.
References
- Aharoni A, Giri AP, Verstappen FWA, Bertea CM, Sevenier R, Sun Z, Jongsma MA, Schwab W, Bouwmeester HJ (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16: 3110–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AB, Gripenberg J (1948) Antibiotic substances from the heart wood of Thuja plicata D. Don; the constitution of β-thujaplicin. Acta Chem Scand 2: 644–650 [DOI] [PubMed] [Google Scholar]
- Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Yuen MMS, Keeling CI, Brand D, Vandervalk BP, et al. (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29: 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney OT, Jacobs DF (2011) Ungulate herbivory of regenerating conifers in relation to foliar nutrition and terpenoid production. For Ecol Manage 262: 1834–1845 [Google Scholar]
- Cankar K, van Houwelingen A, Goedbloed M, Renirie R, de Jong RM, Bouwmeester H, Bosch D, Sonke T, Beekwilder J (2014) Valencene oxidase CYP706M1 from Alaska cedar (Callitropsis nootkatensis). FEBS Lett 588: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Collu G, Unver N, Peltenburg-Looman AMG, van der Heijden R, Verpoorte R, Memelink J (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508: 215–220 [DOI] [PubMed] [Google Scholar]
- Croteau R, Ketchum REB, Long RM, Kaspera R, Wildung MR (2006) Taxol biosynthesis and molecular genetics. Phytochem Rev 5: 75–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Birol I, Bousquet J, Ingvarsson PK, Jansson S, Jones SJN, Keeling CI, MacKay J, Nilsson O, Ritland K, et al. (2014) Insights into conifer giga-genomes. Plant Physiol 166: 1724–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal SS, Croteau R (1987) Metabolism of monoterpenes: specificity of the dehydrogenases responsible for the biosynthesis of camphor, 3-thujone, and 3-isothujone. Arch Biochem Biophys 258: 287–291 [DOI] [PubMed] [Google Scholar]
- Dess DB, Martin JC (1983) Readily accessible 12-I-5 oxidant for the conversion of primary and secondary alcohols to aldehydes and ketones. J Org Chem 48: 4155–4156 [Google Scholar]
- Foster AJ, Hall DE, Mortimer L, Abercromby S, Gries R, Gries G, Bohlmann J, Russell J, Mattsson J (2013) Identification of genes in Thuja plicata foliar terpenoid defenses. Plant Physiol 161: 1993–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JAF, Barton GM, MacLean H (1959) The polyoxyphenols of western red cedar (thuja plicata donn.). Can J Chem 37: 1703–1709 [Google Scholar]
- Garside P, Halsall TG, Hornby GM (1969) Action of peracetic acid on (+)-sabinol. J Chem Soc C 5: 716–721 [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DE, Robert JA, Keeling CI, Domanski D, Quesada AL, Jancsik S, Kuzyk MA, Hamberger B, Borchers CH, Bohlmann J (2011) An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J 65: 936–948 [DOI] [PubMed] [Google Scholar]
- Hamann T, Møller BL (2007) Improved cloning and expression of cytochrome P450s and cytochrome P450 reductase in yeast. Protein Expr Purif 56: 121–127 [DOI] [PubMed] [Google Scholar]
- Hamberger B, Bak S (2013) Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci 368: 20120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Ohnishi T, Hamberger B, Séguin A, Bohlmann J (2011) Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol 157: 1677–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebda RJ, Mathewes RW (1984) Holocene history of cedar and native Indian cultures of the north american pacific coast. Science 225: 711–713 [DOI] [PubMed] [Google Scholar]
- Karp F, Croteau R (1982) Evidence that sabinene is an essential precursor of C(3)-oxygenated thujane monoterpenes. Arch Biochem Biophys 216: 616–624 [DOI] [PubMed] [Google Scholar]
- Keeling CI, Bohlmann J (2006) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol 170: 657–675 [DOI] [PubMed] [Google Scholar]
- Kimball BA, Russell JH, Griffin DL, Johnston JJ (2005) Response factor considerations for the quantitative analysis of western redcedar (Thuja plicata) foliar monoterpenes. J Chromatogr Sci 43: 253–258 [DOI] [PubMed] [Google Scholar]
- Kimball BA, Russell JH, Ott P (2012) Phytochemical variation within a single plant species influences foraging behavior of deer. Oikos 5: 743–751 [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40: W622–W627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien S, Karp F, Wildung M, Croteau R (1999) Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species: cDNA isolation, characterization, and functional expression of (-)-4S-limonene-3-hydroxylase and (-)-4S-limonene-6-hydroxylase. Arch Biochem Biophys 368: 181–192 [DOI] [PubMed] [Google Scholar]
- MacLean H, Murakami K (1966) Lignans of western red cedar (Thuja plicata Donn). V. Hydroxythujaplicatin methyl ether. Can J Chem 44: 1827–1830 [Google Scholar]
- Mamane V, Gress T, Krause H, Fürstner A (2004) Platinum- and gold-catalyzed cycloisomerization reactions of hydroxylated enynes. J Am Chem Soc. 126: 8654–8655 [DOI] [PubMed] [Google Scholar]
- Morris PI, Stirling R (2012) Western red cedar extractives associated with durability in ground contact. Wood Sci Technol 46: 991–1002 [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, et al. (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S (2000) Molecular Evolution and Phylogenetics, Ed 1 Oxford University Press, New York, pp 1–333 [Google Scholar]
- Nelson DR. (2006) Cytochrome P450 nomenclature, 2004. Methods Mol Biol 320: 1–10 [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Ohloff G, Uhde G, Thomas AF, Kováts Esz (1966) The absolute configuration of thujane. Tetrahedron 22: 309–320 [Google Scholar]
- Omura T, Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239: 2370–2378 [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Ro DK, Arimura G, Lau SYW, Piers E, Bohlmann J (2005) Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102: 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943 [DOI] [PubMed] [Google Scholar]
- Roach CR, Hall DE, Zerbe P, Bohlmann J (2014) Plasticity and evolution of (+)-3-carene synthase and (-)-sabinene synthase functions of a sitka spruce monoterpene synthase gene family associated with weevil resistance. J Biol Chem 289: 23859–23869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robineau T, Batard Y, Nedelkina S, Cabello-Hurtado F, LeRet M, Sorokine O, Didierjean L, Werck-Reichhart D (1998) The chemically inducible plant cytochrome P450 CYP76B1 actively metabolizes phenylureas and other xenobiotics. Plant Physiol 118: 1049–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein D, Onillon S, Herbette G, Lesot A, Werck-Reichhart D, Sallaud C, Tissier A (2008) CYP725A4 from yew catalyzes complex structural rearrangement of taxa-4(5),11(12)-diene into the cyclic ether 5(12)-oxa-3(11)-cyclotaxane. J Biol Chem 283: 6067–6075 [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JH, Ferguson CF (2008) Preliminary results from five generations of a western redcedar (Thuja plicata) selection study with self mating. Tree Genet Genomes 4: 509–518 [Google Scholar]
- Russell JH, Yanchuk AD (2012) Breeding for growth improvement and resistance to multiple pests in Thuja plicata. Gen Tech Rep 240: 40–44 [Google Scholar]
- Sirisoma NS, Höld KM, Casida JE (2001) alpha- and beta-Thujones (herbal medicines and food additives): synthesis and analysis of hydroxy and dehydro metabolites. J Agric Food Chem 49: 1915–1921 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserand R, Young R (2014) Essential Oil Safety, Ed 2 Churchill Livingstone, London, pp 1–784 [Google Scholar]
- Tsiri D, Graikou K, Pobłocka-Olech L, Krauze-Baranowska M, Spyropoulos C, Chinou I (2009) Chemosystematic value of the essential oil composition of Thuja species cultivated in Poland-antimicrobial activity. Molecules 14: 4707–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit MA, Sharpless KB (1977) Allylic oxidation of olefins by catalytic and stoichiometric Selenium dioxide with tert-Butyl hydroperoxide. J Am Chem Soc 99: 5526–5528 [Google Scholar]
- Von Rudloff E, Lapp MS, Yeh FC (1988) Chemosystematic study of Thuja plicata: multivariate analysis of leaf oil terpene composition. Biochem Syst Ecol 16: 119–125 [Google Scholar]
- Vourc’h G, Russell J, Martin JL (2002) Linking deer browsing and terpene production among genetic identities in Chamaecyparis nootkatensis and Thuja plicata (Cupressaceae). J Hered 93: 370–376 [DOI] [PubMed] [Google Scholar]
- Vourc’h G, Martin JL, Duncan P, Escarré J, Clausen TP (2001) Defensive adaptations of Thuja plicata to ungulate browsing: a comparative study between mainland and island populations. Oecologia 126: 84–93 [DOI] [PubMed] [Google Scholar]
- Zifkin M, Jin A, Ozga JA, Zaharia LI, Schernthaner JP, Gesell A, Abrams SR, Kennedy JA, Constabel CP (2012) Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol 158: 200–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.