Abstract
Molecular genetic research on bacteriophage lambda carried out during its golden age from the mid 1950's to mid 1980's was critically important in the attainment of our current understanding of the sophisticated and complex mechanisms by which the expression of genes is controlled, of DNA virus assembly and of the molecular nature of lysogeny. The development of molecular cloning techniques, ironically instigated largely by phage lambda researchers, allowed many phage workers to switch their efforts to other biological systems. Nonetheless, since that time the ongoing study of lambda and its relatives have continued to give important new insights. In this review we give some relevant early history and describe recent developments in understanding the molecular biology of lambda's life cycle.
Keywords: bacteriophage, lambda, lambdoid phages
Introduction
From the earliest studies on lambda genetics by Jean Weigle (1953), Francois Jacob and Elie Wollman (1953, 1954) and Dale Kaiser (1955 in volume number 1 of VIROLOGY) in the 1950's until it helped usher in the age of genetic engineering in the late 1970’s and early 1980's, phage lambda was in its golden age at the center of the molecular genetic universe. During those decades lambda was one of the few experimental "organisms" that was sufficiently experimentally accessible to be viewed as potentially completely understandable. Its genome was small enough not to be overwhelming, yet it was capable of making a decision of whether to lysogenize or grow lytically and was clearly complex enough to be interesting and informative on many fronts. In particular it was unique in that a genetic and molecular understanding of the control of gene expression seemed to be within almost immediate reach. Those of us lucky enough to be involved in the study of phage lambda during this time were tremendously excited about future possibilities and threw ourselves into the work with all the energy we had.
Lambda was originally discovered in 1951 by Esther Lederberg (1951) at the University of Wisconsin (Madison), when she serendipitously found it was released from the laboratory Escherichia coli strain K-12 after ultraviolet irradiation. Since that time all aspects of lambda's lytic and lysogenic lifestyles have been studied. Lambda became a more important and approachable experimental system when Allan Campbell (1961) isolated a set of suppressible nonsense mutants of lambda (or amber mutations, originally called suppressor sensitive or sus mutations) that identified genes essential for its lytic growth. Additional amber mutations isolated by Sandy Parkinson (1968) and Goldberg and Howe (1969) and prophage mutations isolated by Clarence Fuerst (Mount et al., 1968) that were lethal for lambda lytic growth identified a few more essential genes. Genetic complementation tests of these mutations succeeded in defining all but one of lambda's 29 genes that are essential for plaque formation under laboratory conditions (this includes the frameshifted G and G-T gene pair and the alternate starts of the S105 and S107 genes as well as the C and Nu3 genes as two genes each; see below). We note that two additional genes, Rz and Rz1 should probably also be considered essential genes even though plaques often form in their absence. They are absolutely required for disruption of the outer membrane and lysis unless external physical solution forces that are often present during laboratory growth help to destabilize the outer membrane of infected cells (Berry et al., 2012). Only the small cro gene was not found by the above random mutant hunts. It was discovered during the genetic analysis of the control of lysogeny (Eisen et al., 1970), and its absence leads to the lysogenic state and hence no lytic growth. Recombination frequency and deletion mapping of these and various promoter and operator mutations isolated by others during this time allowed the construction of a detailed genetic map. Analysis of the phage DNA molecule led to the most detailed physical map of any organism's genome at the time. Detailed study of the phenotypes of phage mutants defective in known, mapped genes led to an early overall picture of the lambda lifecycle. Indeed, the careers of both authors here started at about the same time with the analysis of lambda virion morphogenesis by utilizing the various amber mutants to identify the proteins expressed from the lambda morphogenetic genes and determining the nature of the defects in their absence - SRC in the laboratory of Dale Kaiser at Stanford U. and RWH in the laboratory of Jim Watson at Harvard U. (Happily, rather than generating animosity, our early competition in this arena led to mutual respect and a lifelong friendship). We believe that the clever use of forward genetic selections and screens to isolate informative mutations and combinations of mutations and the use of these molecular genetic experiments to deduce the underlying molecular mechanisms reached its zenith in the study of phage lambda during this period. Those were heady days with lambda running at the front of the scientific pack.
We discuss here early experiments that led to our current understanding of phage lambda and molecular biological processes in general. During this period VIROLOGY published many of these important findings. We also mention recent developments to show how far the field has come in the past 60 years (we assume a basic knowledge of the phage lambda life cycle; see Hendrix et al., 1983; Hendrix and Casjens, 2006). We include discussion of phages related to lambda, which are commonly referred to as "lambdoid" phages. This ill-defined (and often incorrectly used) term refers phages with very similar lifestyles to lambda and genomes that are mosaically related to lambda. This definition included the notion that a lambdoid phage is capable of recombination with lambda itself to produce a functional hybrid phage, as was first shown with phage 434 by Kaiser and Jacob (1957). Recent phage genome sequences have caused an expansion of this term to mean a phage with the same functional gene order as lambda and that carries patches of nucleotide sequence homology with lambda or another lambdoid phage. Thus, in theory a single recombination event between lambdoid phages could give rise to a fully functional phage that has all the necessary genes (Hendrix, 2002; Casjens, 2005; Hendrix and Casjens, 2006; Grose and Casjens, 2014). In this discussion we use lambdoid to include for example, the three phages HK97, P22 and N15, which typify three of the best-studied lambdoid groups that have significant differences from lambda. Citations are in general not meant to bestow credit for the original discoveries but to allow the reader access to the literature. Other more detailed historical treatments of some of the topics covered below can be found in the books Bacteriophage lambda (Hershey, 1971) and Lambda II (Hendrix et al., 1983) (and in Gottesman and Weisberg, 2004; Stahl, 2005; Georgopoulos, 2006; Campbell, 2007; Court et al., 2007a).
Historical importance and recent progress
The lambda genome
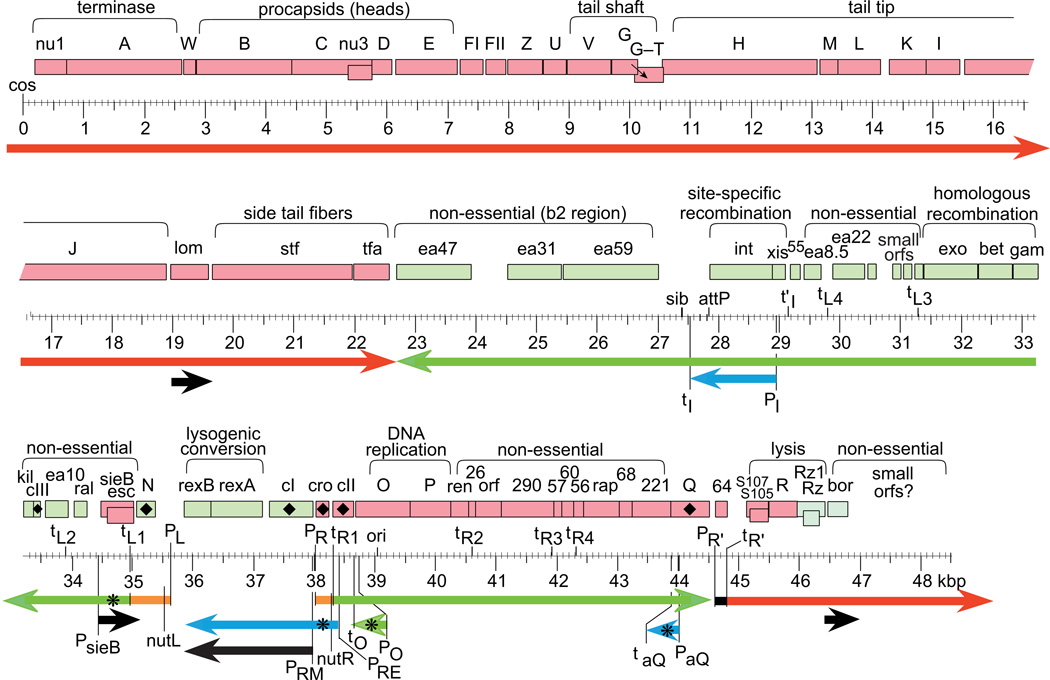
Phage lambda DNA was one of the earliest model systems for studying the physical nature of DNA and genes, and a substantial amount of early research examined the hydrodynamic properties of the lambda chromosome with the goal of, among other things, determining its absolute molecular weight. At the same time methods (now considered to be primitive) were devised for separating the two halves of the lambda chromosome and for mapping genes identified genetically to these halves and to smaller physical intervals (reviewed by Davidson and Szyblaski, 1971). Soon thereafter electron microscopic analysis of absolute length and of lambda DNA in heteroduplex with various altered lambda DNAs gave rise to a major step forward, the construction of a detailed physical map of lambda DNA - a gene map in base pairs (bps) rather than recombination frequency units (Fiandt et al., 1971; Simon et al., 1971). The lambda virion chromosome was also among the first natural linear DNAs whose end structure was understood in detail; it was found not to have blunt ends, but to have complementary 12 bp protruding 5'-ends called cohesive (or sticky) ends, since the two single-strand ends can pair and thus bind to each other (Hershey et al., 1963; Hershey and Burgi, 1965). Meanwhile, lambda contributed greatly to the ultimate mapping of DNA, the complete nucleotide sequence. It is not generally appreciated that the 12 bp lambda cohesive ends were the subject of the first direct nucleotide sequencing of a biological DNA. Ray Wu devised methodology for using DNA polymerase to fill in the cohesive ends with specifically labeled nucleotides, followed by analysis of the product to determine this exact 12 bp nucleotide sequence (Wu and Kaiser, 1968; Onaga, 2014). This sequencing was not a trivial undertaking, as determining this 0.012 kbp of sequence required several years of painstaking work. Fourteen years later, Fred Sanger et al., (1982) used lambda as the subject for the first determination of the complete genome sequence of a dsDNA virus - 48,502 bp - using his dideoxynucleotide chain termination method. This, along with anecdotal sequencing of various lambda genes and mutants of them by the laborious and more error-prone chemical sequencing method (Maxam and Gilbert, 1977) in the previous few years, unleashed a rapid avalanche of studies relating the many extraordinary genetic studies on lambda from the previous decades to the genome sequence. This in turn gave rise to an understanding of phage lambda genes and their expression that was unprecedented at the time (reviewed in Hendrix et al., 1983). Figure 1 shows a map of the lambda chromosome; more detailed annotated maps of lambda, HK97, P22 and N15 can be found in the supplementary material of Hendrix and Casjens (2006).
Figure 1. Map of the bacteriophage lambda chromosome.
The linear virion chromosome is shown with a scale in kbp below. Rectangles indicate known genes with names and functional regions shown above; red genes are transcribed rightward and green leftward; vertically offset gene rectangles are expressed from reading frames that overlap (CNu3, S107–S105 and sieB-esc in the same reading frame, rz-rz1 in different frames) or by programmed frameshifting (G-T, small arrow in figure). Diamonds (♦) mark regulatory genes. Important DNA sites (e.g., P, promoters; t, terminators) are indicated below the genes. Thick horizontal arrows below indicate mRNAs: black, transcripts made in a lysogen; orange, immediate-early transcripts; green, early transcripts; red, late transcripts; blue, transcripts made in response to high CII levels. Asterisks (✳) mark RNAs with regulatory activity (the detailed role of PRE-initiated cro antisense message remains unclear (Spiegelman et al., 1972). The chromosome is circularized in the cell during infection, so the PR'-initiated late transcript is continuous from the lower right to the upper left in the figure.
In a sense the complete nucleotide sequence of the lambda genome and publication of the Lambda II book (Hendrix et al., 1983) heralded the demise of lambda's golden age and its exalted position in molecular genetic research. The complete sequence gave the (false) impression to researchers outside this field that everything important about lambda was known. This impression, along with the approximately simultaneous development of simple molecular cloning techniques that could be used in virtually any molecular biology laboratory, allowed new and powerful molecular genetic access to essentially any biological system a researcher wished to study. Ironically, lambda played a major part in the development of these techniques (below).
Control of transcription during lytic growth
Lambda was one of the first biological entities whose transcriptional regulation was studied and understood in detail. It was found early on, like other phages under study at the time, to have a temporally controlled pattern of transcription and gene expression, called in this case immediate early, early (sometimes called “delayed early”) and late transcription (reviewed by Echols, 1971; Friedman and Gottesman, 1983). Lambda's major transcription units are shown in figure 1. When Jeffrey Roberts (1969), then a student in the Watson/Gilbert lab at Harvard, used purified RNA polymerase to transcribe lambda DNA in vitro, it initiated RNA synthesis at the immediate early promoters PL and PR, but the transcripts made were longer than those observed in vivo. He then used this system to discover a host protein (that he named Rho) that caused shorter transcripts to be made that were the same length as those observed in vivo. Rho was later shown to cause transcription termination at the normal lambda sites in vivo, and to be required for a subset of transcription termination events in E. coli. Thus lambda was directly responsible for the discovery of this first transcription controlling “factor” (if the sigma subunit is considered to be part of the basic RNA polymerase enzyme). Lytic infection by lambda phage carrying a conditional lethal mutation in its N gene drastically lowered protein and transcript synthesis from nearly all lambda genes compared to a wild type infection (e.g., Radding, 1964; Echols, 1971). These findings were rapidly followed by many ingenious molecular genetic studies that converged on the idea that N protein causes termination of the PL and PR immediate early transcripts to fail. The only immediate early genes, N and cro, lie upstream of these terminators, in the PL and PR transcripts, respectively.. Further work showed that transcription is antiterminated at terminators tL1 and tR1 (and other downstream terminators; figure 1) by N protein to allow expression of the downstream (delayed) early genes (summerized in Echols, 1971; Franklin, 1974; Friedman and Gottesman, 1983 and references therein). N protein does not, however, simply inactivate Rho. Instead N somehow makes the transcribing RNA polymerase that has passed over a nut (N utilization) site in its presence insensitive to downstream termination at Rho-dependent and Rho-independent terminators.
Since its identification as a transcription antiterminator, N protein has been the subject of many studies aimed at elucidating the mechanism by which it accomplishes this feat. Jack Greenblatt first succeeded in accomplishing N action in vitro (Greenblatt, 1972), and many subsequent studies have learned much about the detailed roles of the several host proteins that participate in N-mediated antitermination (see Interaction between phage lambda and its host section below for a more detailed discussion of these factors). N protein, a natively unfolded protein (van Gilst and von Hippel, 1997; Goldenberg and Argyle, 2014), binds to the boxB sequence of the nut site in the nascent PL and PR mRNAs, as well as to the host NusA protein (Mogridge et al., 1998; Mishra et al., 2013) and RNA polymerase. It probably binds RNA polymerase on its β subunit near the RNA exit channel (Cheeran et al., 2005). Exactly how N protein blocks termination is not yet known, but the recent observation that it reduces transcriptional slippage may provide a clue if slippage contributes to termination (Parks et al., 2014).
All characterized lambdoid phages possess an N protein-mediated transcription antitermination mechanism, with the sole exception of phage HK022. In lambda antitermination, as outlined above, the nascent PL or PR transcript interacts through its nut sites with N protein, RNA polymerase and other host proteins to create a transcription elongation complex that is insensitive to termination signals. HK022 accomplishes the same end without N and without the known host factors. We now know that nascent PL and PR transcripts of HK022 interact directly with the β’ subunit of the transcribing RNA polymerase through secondary structure formed at the put sites in the nascent RNA (which correspond in position to the nut sites on the lambda transcripts) (Oberto et al., 1993; Sen et al., 2002; Komissarova et al., 2008). The resulting transcription elongation complex, as with lambda, is insensitive to termination signals. Thus, HK022 early transcript antitermination is apparently mediated by RNA alone, rather than by RNA in conjunction with the N and host transcription factors.
HK022 has not finished its dalliance with non-canonical mechanisms of regulating transcription with its unusual mechanism of accomplishing antitermination. It also has a mechanism to promote inappropriate termination in lambda-like phages during co-infection of the same bacterium. While HK022 does not have an N gene, it does have a heterologous gene, called nun, at the same position in the gene order as N in lambda. The Nun protein interacts with the nut sites of lambda as well as with the other host factors and RNA polymerase (Burova et al., 1999; Watnick and Gottesman, 1998; Vitiello et al., 2014). This resembles what happens with N protein, but the resulting transcription elongation complex is biased toward termination. For a phage like lambda such inappropriate termination of transcription would be a lethal event. These stories and others about how HK022 deviates in informative ways from the lambda paradigm, have largely come out of the laboratories of Max Gottesman and the late Bob Weisberg (Weisberg et al., 1999). Ongoing work on these topics provides a good illustration of how phage investigations can still give new information on how RNA polymerase receives and responds to signals.
Following N-mediated antitermination of PR transcription, gene Q is transcribed at the distal end of the early right operon. The Q protein in turn acts as a transcriptional antiterminator that allows read-through of the tR' terminator (figure 1) to allow expression of the late operon (Roberts et al., 1998 and references therein). However, Q protein works by a very different mechanism from that of N protein. Studies with phage lambda and the lambdoid phage 82 have shown that it is a DNA-binding protein that joins the transcription complex at the Q binding element (QBE) between the −35 and −10 parts of the late operon promoter PR' and stays with the transcription elongation complex as it traverses the late operon. In contrast to N-mediated antitermination, there are no known host proteins that participate in its action. The recent structure of the DNAbinding domain of lambda Q protein sheds considerable light on its contacts with the QBE DNA sequence, the R4 region of sigma factor and the tip of the RNA polymerase β flap (Deighan and Hochschild, 2007; Shankar et al., 2007; Muteeb et al., 2012; Vorobiev et al., 2014).
Control of lysogeny
In 1961 Jacob and Monod put forward their operon hypothesis, deduced entirely from their genetic work on the lac operon and phage lambda model systems (Jacob and Monod, 1961). The early observations by Kaiser that CI is required for maintenance of lambda lysogeny and that it is responsible for immunity to superinfecting lambda phages strongly suggested, but did not absolutely prove, that cI encoded a protein that directly turns off all the lambda lytic genes in the prophage state (Kaiser, 1957; Kaiser and Jacob, 1957). The subsequent isolation of suppressible amber nonsense mutants in the cI gene proved that this function is mediated by the protein product of the cI gene (Jacob et al., 1962; Thomas and Lambert, 1962). By the mid 1960s experiments had demonstrated quite clearly that the lack of lytic gene expression from the lambda prophage was explained by blockage of mRNA synthesis by the cI gene product (Jacob and Campbell, 1959; Thomas, 1971; Ptashne, 1971 and references therein). In 1965–66 a competition developed between Mark Ptashne (with the lambda CI repressor) and Wally Gilbert (with the E. coli LacI repressor) to be the first to isolate a transcriptional repressor and thus absolutely prove the existence of repressor proteins that regulate gene expression. They worked in laboratories on different floors of the same building at Harvard, and this race for the repressor caught the attention of ABC television, which filmed a documentary, “The Scientist,” giving CI and LacI a brief moment of popular fame (and somewhat amusing and irritating the other scientists in the building who experienced the filming). Gilbert and Müller-Hill (1966) attained their goal first, but Ptashne (1967) was not far behind, and today both repressors remain important model systems for the study of sequence-specific DNA binding by proteins.
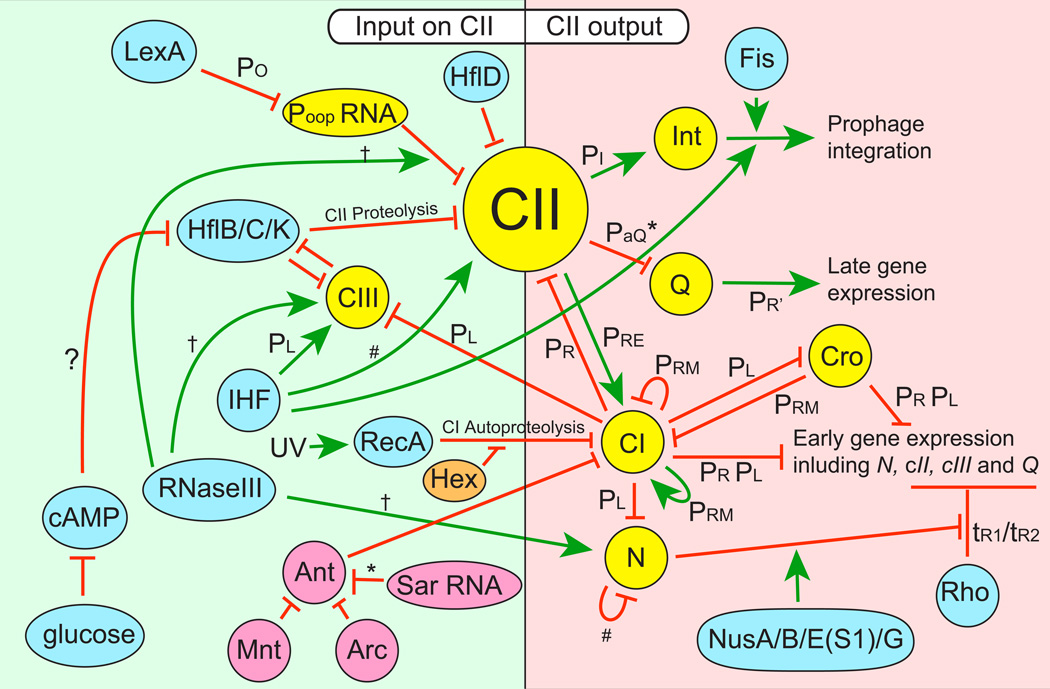
While maintenance of lambda lysogeny only requires one phage protein, the process of establishment of the lysogenic state is more complex. Dale Kaiser's original genetic studies identified two additional genes, cII and cIII, in which defects lower the frequency of establishment by several orders of magnitude (Kaiser, 1957). These two genes lie in the early right and early left operons, respectively. After the discovery of Cro it was first thought that the lysis/lysogeny decision might simply be explained by a direct competition for operator binding between CI and Cro (a bistable genetic switch), but more recent experiments have shown that it is not this simple. We now know that the CII protein is a transcriptional activator that stimulates strong cI gene transcription from the PRE promoter, and that high CII protein levels are critical in ensuring the high CI concentrations required to establish, but not maintain, the lysogenic state (Court et al., 2007a and references therein). The cIII gene encodes a protein that controls the stability of the CII protein by inhibiting the action of the hostencoded HflB(FtsH)-HflC-HflK protease complex (Kobiler et al., 2007; Bandyopadhyay et al., 2010). In addition to this host protease control of CII stability, the host physiological state impacts the lysis/lysogeny decision through the following: (1) cAMP and ppGpp levels may control HflBCK levels (Hong et al., 1971; Rao and Raj, 1973; Joung et al., 1994; Slominska et al., 1999). (2) RNase III is under strong growth rate regulation (Wilson et al., 2002), and its action controls cIII mRNA stability (Altuvia et al., 1991). (3) Integration host factor (IHF) modulates CII and CIII levels (Mahajna et al., 1986; Krinke and Wulff, 1990; Giladi et al., 1998) and N gene translation (Kameyama et al., 1991; Wilson et al., 2002). (4) RecA is activated by DNA damage to cause CI inactivation (see below). (5) HflD may directly inhibit DNA binding by CII protein (Parua et al., 2010). (6) It has also been suggested that host proteins DnaA and SeqA may affect PL transcription and thus the lysis/lysogeny decision (Wegrzyn et al., 2012). Finally, (7) the OOP RNA made from the lambda Po promoter is antisense to the 3' portion of cII gene mRNA and acts to destabilize that message with the help of RNase III, and the OOP promoter is repressed by the host LexA protein (Krinke and Wulff, 1990; Krinke et al., 1991; Lewis et al., 1994). Thus, the current consensus is that CII is the central player in establishing lysogeny, and figure 2 summarizes this complex regulatory circuitry.
Figure 2. Lambda regulatory circuits.
The figure shows phage lambda-encoded functions (yellow ovals) and host encoded functions (blue ovals) that impact the lysis-lysogeny decision and control the lytic gene expression cascade. Phage P22 (Liao et al., 1987;Wu et al., 1987) and phage 434 (Shkilnyj et al., 2013) genes are pink and orange ovals respectively. Green arrows denote positive (activation) functions and red lines denote inhibitory (repression) functions. Asterisks (*) indicate antisense RNA interactions, hashes (#) indicate translational repression, and daggers (†) indicate mRNA stability controls; other regulatory events are at the level of transcription except IHF and FIS physical participation in the integrase complex that catalyzes the integration of prophage DNA.
When a decision is made to establish lysogeny, lambda ensures that the proteins that favor the lytic growth pathway disappear quickly, thereby minimizing stuttering between the lytic and lysogenic life cycle directions and ensuring that a decision is definitive. The O, N, CII, CIII and Xis proteins are all extremely metabolically unstable and are destroyed within a few minutes of being made. Thus, when their synthesis stops, their DNA replication initiation, delayed early gene expression and lysogeny promoting activities are quickly lost (Kobiler et al., 2004). The host proteases utilized for this purpose are ClpX/ClpP for O (Gonciarz-Swiatek et al., 1999; Thibault and Houry, 2012), Lon for N (Gottesman et al., 1981; Maurizi, 1987; Mikita et al., 2013) and FtsH for CII and CIII (Shotland et al., 2000; Kobiler et al., 2002; Kobiler et al., 2007), and Lon for Xis (Leffers and Gottesman, 1998).
In addition to preventing the lytic gene transcription cascade, the CI protein has two other known functions, turning down expression of the host pckA gene and autoregulating its own synthesis. Array analysis of mRNAs showed that the E. coli pckA gene (which encodes phosphoenolpuruvate carboxykinase) is the most highly down-regulated host gene in a lambda lysogen (Chen et al., 2005). This gene is required for growth on succinate and some other carbon sources. It is not known why lowering its expression might be beneficial, but the fact that the pckA promoter also overlaps lambdoid phage 21, P22, 434 and H-19B repressor binding targets (all different from lambda CI operator specificity), suggests that this regulation is evolutionarily important to the lambdoid phages. Louis Reichardt, a student in Kaiser's lab in the early 1970's, devised a quantitative assay for CI repressor protein and performed an early set of intricate and elegant experiments that showed that CI activates its own synthesis at low CI levels and represses at high levels (Reichardt and Kaiser, 1971). This was one of the first demonstrations of autoregulation of a gene. Other experiments illuminated the amazing intricacies of CI and Cro binding to the three tandem operators at PL to regulate the divergent PRM and PL promoters (Ptashne, 1992 and references therein). More recent study of the lambda prophage repressor has shown that it forms multimers that bind simultaneously to operators OL and OR at PL and PR, respectively, thus looping the DNA between these two promoters, and that this looping is important in the autoregulation that keeps the CI concentration within narrow limits in lysogens (Lewis et al., 2011; Cui et al., 2013a and 2013b; Hensel et al., 2013; Michalowski and Little, 2013; Priest et al., 2014). Host supercoil density impacts this looping, but its role in vivo is not known (Ding et al., 2014).
The CI-CII-CIII circuitry is nearly universally present in the lambdoid phages, but some of these phages have additional controls on lysogeny. The first of these to be discovered was the P22 antirepressor (Ant), which binds to repressor and causes it to release repression (Susskind and Botstein, 1975). Transcription of the ant gene in a prophage is kept off by two additional phage-encoded repressors, Mnt and Arc, and a small antisense RNA, Sar, that controls the translation of ant mRNA (Prell and Harvey, 1983; Liao et al., 1987; Wu et al., 1987; Schaefer and McClure, 1997). Salmonella lambdoid phage gifsy-1 encodes an antirepressor GfoR that is repressed by the host LexA repressor (Lemire et al., 2011). Other phages such as N15 encode a different type of antirepressor control system. Here, transcription of AntA (an antirepressor whose mode of action is not known) is negatively regulated by the N15 prophage repressor and by premature transcriptional termination that is mediated by processing of a portion of the leader of the antirepressor operon mRNA (Ravin et al., 1999; Mardanov and Ravin, 2007). The exact biological role(s) of antirepressors remains unclear; they could be prophage induction modules (see below) but could also impact the lytic/lysogeny decision.
Integration and excision
An aspect of the establishment of lysogeny not discussed above is the physical integration of the phage chromosome into the host chromosome. The phenomenon of lysogeny had been known but poorly understood for several decades before lambda was discovered (Lwoff, 1953). The quiescent phage genome (prophage) in a lysogen appeared to be attached to the host chromosome in some way that could be mapped in bacterial conjugation experiments (Lederberg and Lederberg, 1953; Wollman, 1953), but the nature of such an attachment was not known. Our current understanding of prophage insertion derives from the idea of site-specific recombination between a circular phage lambda genome and the host chromosome put forward by Allan Campbell (1962). Numerous subsequent genetic and physical tests of this idea by Campbell and others soon validated this model (see Gottesman and Weisbeg, 1971). The existence of a lambda encoded function, integrase (Int), that was necessary for prophage insertion was simultaneously demonstrated by the isolation of int defective mutants by Zissler (1967; Zissler et al. 1971), Gingery and Echols (1967) and Gottesman and Yarmolinsky (1968). Howard Nash and coworkers accomplished in vitro integration (reviewed by Weisberg and Landy, 1983), and many subsequent ingenious genetic and biochemical studies elucidated the complex mechanism of the integration reaction. The host integration host factor (IHF; named for this reaction but now known to have many pleiotropic functions in E. coli and Fis protein were found to be involved (Ball and Johnson, 1991). Genetic studies by Ethan Signer, Robert Weisberg and others combined with biochemical studies from the Art Landy lab have resulted in a marvelously detailed picture of the integration protein DNA complex bound to its target (the attachment site, att) and its actions that result in att site-specific recombination and lambda prophage insertion (Seah et al., 2014; Tong et al., 2014 and references therein).
As Esther Lederberg originally discovered (above), lambda prophages are induced by DNA damaging agents such as UV light or mitomycin C to enter a cycle of lytic phage growth. Release of CI repression causes the lytic gene expression cascade to initiate, and the prophage is excised from the host chromosome to allow circle-to-circle DNA replication (below). Interestingly, the CI repressor contains a cryptic autoprotease activity that is activated by binding to the host RecA recombination protein, but only when RecA is activated by ssDNA (Little, 1984; Kim and Little, 1993; Mustard and Little, 2000). Integration and excision are directional; while Int (with its host factors) is sufficient to integrate lambda DNA, another phage-encoded protein Xis is required for the reverse excision reaction. Control of the relative amounts of Int and Xis are necessary to ensure that integration occurs during establishment of lysogeny and excision occurs during induction of a lysogen. This is accomplished in two ways. First, when lysogeny is being established by high CII levels (above) the Int/Xis ratio needs to be high to ensure that integration occurs. To accomplish this, CII protein activates the promoter PI, and its transcript encodes Int but not Xis. Second, a process dubbed retroregulation lowers the level of Int expression during lytic infection. Here, a site sib in the RNA downstream of int and the PI transcript causes more rapid decay of PL initiated int mRNA early after infection, ensuring that Int is made in large amounts only when a high CII level commits the cell to lysogeny. In addition, the sib site is separated from PL in the integrated prophage, so after induction moderate levels of both Int and Xis made from PL ensure excision (Schindler and Echols, 1981; Gottesman et al., 1982; Guarneros et al., 1982). CII also activates a third promoter, PaQ, whose product is a small antisense RNA of the Q gene, which presumably helps keep the late operon off while a decision is being whether to lysogenize (Ho and Rosenberg, 1985; Hoopes and McClure, 1985). In vivo visualization experiments have recently shown that, after injection, lambda DNA is confined near its point of entry and replication-driven movement of the host DNA allows proper juxtaposition of the host and phage attachment sites. These experiments also demonstrate that even bacterial attachment site location in the chromosome affects the frequency of lysogenization (Tal et al., 2014).
In an interesting variation of lysogeny, lambdoid phages N15, øKO2 and PY54 do not integrate into their host's genome, but exist as linear plasmid prophages (Ravin and Shulga, 1970; Hertwig et al., 2003; Casjens et al., 2004). In these phages the int gene is replaced by a ptl gene that encodes the enzyme, called protelomerase, that creates the ends of these plasmids. These ends are covalently closed hairpins in which one DNA strand turns around and becomes the other strand with no break in the phosphodiester backbone (Rybchin and Svarchevsky, 1999). The mechanism of action of protelomerase has been examined, and it recognizes a specific inverted repeat sequence, makes staggered nicks 6 bp apart in the two DNA strands, separates the strands between the nicks, and after strand swapping ligates the ends to form two hairpins (Huang et al., 2004; Aihara et al., 2007). These prophages encode their own plasmid segregation and replication proteins (Ravin, 2003; Ravin et al., 2003) and so do not turn off their early genes as completely as integrated prophages (Ravin et al., 2000).
DNA replication and homologous recombination
Lambda DNA replication initiates at a single origin and proceeds bidirectionally from that point to generate circle-to-circle replication until several tens of circles accumulate in the infected cell. At that point replication switches (by a still poorly understood mechanism) to rolling circle mode to generate the concatemeric DNA molecules that are the substrate for DNA packaging into virions (below) (Skalka et al., 1972; Wake et al., 1972; Narajczyk et al., 2007). The lambda genome carries two essential DNA replication genes, O and P. Four dimers of O protein bind at four sequence repeats in the origin, which lies inside the O gene, to form an "O-some" complex. The other essential replication protein, P, binds to the host DnaB helicase, and this binary complex binds the O-some to form the ori:O:P:DnaB preprimosomal complex (Zylicz et al., 1998 and references therein). Host chaperones DnaK-DnaJ-GrpE (see below) are involved in activating the preprimosomal complex to recruit the rest of the host replication machinery (Learn et al., 1997). Thus, the early study of lambda replication contributed greatly to an understanding of the detailed steps required to accomplish controlled initiation. In addition, the origin is activated by rightward transcription from PR, but this phenomenon remains poorly understood (Leng and McMacken, 2002; Hayes et al., 2012; Olszewski et al., 2014). In some other lambdoid phages like P22, the P gene is replaced by a gene that encodes an actual DnaB type helicase (Wickner, 1984a and 1984b), and phages APSE-1 and N15 encode a putative DNA polymerase (van der Wilk et al., 1999) or a bifunctional helicase-primase (Ravin et al., 2000; Mardanov and Ravin, 2006), respectively.
Genetic recombination was first demonstrated for lambda by Jacob and Wollman (1954), and mutants of lambda that recombine at reduced frequency were isolated by Franklin (1967), Echols and Gingery (1968) and Signer and Weil (1968). These mutations were found to affect genes exo and bet that lie in the early PL transcript. Exo is an exonuclease (Zhang et al., 2011 and refs therein) and Bet is a strand-annealing protein (Matsubara et al., 2013 and refs therein). These two proteins can substitute for RecA in homologous recombination. In addition, a host RecBCD nuclease inhibitor is encoded by the gam gene, which is located immediately upstream of exo and bet (Court et al., 2007b and references therein). Lambda also carries two genes in its nin region that affect recombination, orf/ ninB and rap/ninG. Orf protein is a recombination mediator that binds to host single-strand binding protein and ssDNA (Maxwell et al., 2005; Curtis et al., 2011 and 2014), and Rap protein is a DNA structure-specific endonuclease that cleaves Holliday junction branch points (Sharples et al., 1998; Poteete et al., 2002). Homologous recombination occurs by several different pathways, and these phage-encoded proteins participate with host proteins in various ways (see for example Poteete, 2013). We will not review these in detail here, but these lambdoid proteins continue to be important in ongoing studies of the systems that catalyze homologous recombination.
Early work on lambda recombination also led directly to our current understanding of host RecBCD action. In about 1969 mutants were isolated by David Henderson in Ken and Noreen Murray's laboratory that increased the growth of lambda that was missing its recombination genes. These were studied further by Frank Stahl, Gerry Smith and coworkers (Stahl, 2005 and references therein; Smith et al. 1981a and 1981b). These mutations turned out to have created a unique asymmetric 8 bp nucleotide sequence (now called the Chi site, for crossover hotspot instigator), and their study gave rise to our current understanding that the RecBCD nuclease enters DNA at double-strand breaks and degrades both strands (differentially) from there until it hits a properly oriented Chi site. Passing over a Chi site "civilizes" the RecBCD exonuclease (to use Stahl's words) so that it continues along the DNA stands as a helicase, separating but not degrading them. Finally the host RecA protein is recruited to initiate a recombination event. Wild type lambda has no native Chi site, but its E. coli host has one every 5 kbp on average, and they are highly preferentially oriented according to the direction replication passes over them. It is now believed that Chi plays a major role in RecBCD-mediated recombinational repair of collapsed E. coli replication forks (Kuzminov, 2001). The lack of Chi sites in most invading foreign DNAs may also allow the RecBCD nuclease to help protect E. coli from such "invaders" (Stahl, 2005).
Other lambdoid phages carry different recombination genes in the exo/bet/gam chromosomal location. The best studied of these are genes abc1, abc2, erf and arf of phage P22. The two Abc proteins inhibit RecBCD nuclease, Erf is a strand annealing protein, and Arf is a poorly understood recombination accessory function (Botstein and Matz, 1970; Poteete and Fenton, 1983; Poteete et al., 1988 and 1991). Other lambdoid phages carry a third type of recombination genes, recE and recT, that encode an exonuclease (ExoVIII) and a strand annealing protein, respectively (Kushner et al., 1974; Hall et al., 1993; Zhang et al., 2009). The latter two genes were originally identified as E. coli genes, but were later found to lie in the defective lambdoid prophage Rac (Clark et al., 1993); various fully functional lambdoid phages such as gifsy-1 carry recE and recT genes (Lemire et al., 2008). In addition, some other lambdoid phage nin regions carry a rusA gene which encodes a Holliday junction resolvase that is different from lambda Rap (Sharples et al., 1994).
Recently the lambda exo and bet recombination functions have been put to valuable technological use in accomplishing very efficient homologous recombination in vivo. Don Court and coworkers devised and optimized systems in which short 30–50 bp homologous tails on a selectable gene (generated by amplification by PCR primers with such tails) are inserted by homologous recombination into any desired site in a bacterial genome according to the sequence of the tails. Then, since the inserted gene is chosen to be counterselectable, homologous recombination can replace the inserted gene with any DNA that has the proper homologies (summarized in Thomason et al., 2014). This now widely used strategy, called "recombineering," is dependent upon high levels of ectopically expressed Exo and Bet and allows rapid, efficient and scarless engineering of bacterial genomes. Very recently, Farzadfard and Lu (2014) have used lambda bet-mediated ssDNA recombineering to develop a genomic memory system that "writes" (recombines) a chosen sequence into a second site in the genome of E. coli in response to endogenous expression of ssDNA containing the sequence. The system thus genomically encodes a memory of past expression of the promoter engineered to express the ssDNA.
Interactions between phage lambda and its host
In the early 1970s Costa Georgopoulos, David Friedman and Nat Sternberg independently devised methods of isolating mutations in the E. coli host that allowed lambda virions to adsorb and DNA to be injected, but which block phage infection at some later stage. These mutations affect lambda's head assembly, DNA replication and early transcription. They alter specific host proteins that are required for successful transcription and for proper protein folding/unfolding dynamics.
The host mutations that affect head assembly identified the host GroEL-GroES protein folding chaperone (also now known as HSP60-HSP10) (Georgopoulos et al., 1973; Sternberg, 1973). The functions defined by these host mutations were named “gro“ for to their inability of grow lambda (Georgopoulos and Herskowitz, 1971), and the “E” was added to reflect the fact that mutations in lambda gene E (major head protein) overcome this gro block (Georgopoulos et al., 1973). However, in actual fact, mutations in either gene E or B (portal protein) have this property, and although this particular reaction has still not been studied in biophysical detail, subsequent work suggests that it is probably folding of the gene B protein that actually requires this host chaperone (Murialdo and Becker, 1978; Kochan and Murialdo, 1983; Georgopoulos, 2006). GroEL was originally known simply as GroE, but it acquired the L when its smaller partner, GroES, was discovered (Tilly et al., 1981). In agreement with the genetic evidence that GroEL-GroES interacts with lambda head proteins, examination of the results of lambda infection of a groEL mutant host showed that head production was seriously aberrant as seen in a B or C minus infection (Georgopoulos et al., 1973). The mechanism of GroEL-GroES protein folding chaperone action has now been studied in detail with other more tractable substrates (Horwich and Fenton, 2009).
The groE gene was one of the first bacterial genes to be cloned. Its cloning relied on the confluence of microbial genetics in the form of the groE mutants isolated by Costa Georgopoulos, and one of the earliest recombinant DNA techniques in the form of a shotgun (random) library of E. coli DNA fragments in a phage lambda vector. One of us (RWH) packed some of Costa’s groE mutants in a suitcase and went to visit Ron Davis at Stanford. Ron had constructed one of the first libraries of E. coli DNA cloned into phage lambda genomic DNA and packaged in vitro into lambda virions (below), and when this phage library was plated on a groE mutant bacterial strain it contained plaque-forming phages at a frequency of 10−4 relative to wild type E. coli. The rare plaque-forming phages carried an 8 kbp insert of E. coli DNA. (As an indication of how new the recombinant DNA technology was, the size of the insert was measured not with restriction enzymes and agarose gels but by the much better established technique of measuring the density of phage carrying the insert in an equilibrium CsCl gradient.) The 8 kbp insert was subsequently shown to contain both the groEL and groES genes and led to the identification of the GroEL protein as a 60 kDa protein with ATPase activity that assembles into a cylindrical 14 subunit structure (two stacked rings of seven subunits) (Hendrix, 1979). Sequencing of this cloned DNA led to the realization, which was very surprising at the time, that GroEL had homologues in the chloroplasts of green plants in the form of “rubisco large subunit binding protein” (Hemmingsen et al., 1988). Rubisco is the photosynthesis enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase, and the plant GroEL homologue has a role in folding its large subunit (Gatenby, 1996). While we were cloning and characterizing GroE, very similar experiments were done by Barbara and Tom Hohn in Basel, Switzerland, using another early lambda library of E. coli DNA that was constructed by Ken and Noreen Murray (Georgopoulos and Hohn, 1978; Hohn et al., 1979).
The host mutants that affect DNA replication identified the DnaK-DnaJ-GrpE protein folding/unfolding chaperone (DnaK is also called HSP70) (Georgopoulos and Herskowitz 1971; Georgopoulos and Welch, 1993 and references therein), which is now known to be required for disassembly/loosening of the protein complex that forms at the origin of lambda (above), thus enabling the replication fork to move away from the origin (Hoffmann et al., 1992; Osipiuk et al., 1993; Wyman et al., 1993). In addition to pointing out that protein folding, assembly and disassembly can be catalyzed by other proteins, it was noticed early that transcription of the genes encoding the GroE and DnaK protein chaperone systems is enhanced at high temperatures. Further examination of this latter phenomenon by many workers has led to our current understanding of the mechanisms of stress control of gene expression (Tilly et al., 1983; Georgopoulos and Welch, 1993). Thus, lambda work was seminal in the nucleation of the idea of catalyzed protein folding and in the discovery and understanding of two important chaperone systems. Before this, all protein folding was thought to be spontaneous.
The host mutants in which lambda fails to express (delayed) early genes affect the ability of the lambda N protein to cause RNA polymerase to bypass terminators and thus transcribe the downstream essential early genes (above). These mutations alter several transcription factors now called NusA, NusB, NusE and NusG (originally for N under supplied but more recently N utilization substance) that are all required for successful N antitermination (Georgopoulos, 1971; Friedman, 1971; Friedman and Baron, 1974; Friedman and Gottesman, 1983; Friedman and Court, 1995). NusA is an RNA-binding protein that interacts with the α subunit of RNA polymerase and modulates transcriptional pausing (Prasch et al., 2009; Yang et al., 2009; Yang and Lewis, 2010; Schweimer et al., 2011; Zhou et al., 2011; Muteeb et al., 2012; Kolb et al., 2014), as well as interacting with lambda N protein (Mishra et al., 2013). The NusB-NusE complex binds the BoxA RNA sequence of the nut sites and binds RNA polymerase (Burmann et al., 2010; Stagno et al., 2011). NusG protein homologues are present in all three domains of life, and in E. coli NusG is a transcriptional pause-suppressing factor that might also be involved in suppressing aberrant anti-sense transcription in E. coli (Herbert et al., 2010; Peters et al., 2012; Yakhnin and Babitzke, 2014). It has two domains, an N-terminal domain that binds RNA polymerase (Belogurov et al., 2009) and a C-terminal domain that binds NusE and Rho (Burmann et al., 2010). NusE, which is now known to be ribosomal small subunit protein S10, binds NusG and as such is thought to be involved in ensuring that a ribosome closely follows the transcribing RNA polymerase, thus coupling translation to transcription (Burmann et al., 2010; McGary and Nudler, 2013). In addition, NusA interacts with translesion DNA polymerases to affect DNA repair (Cohen et al., 2009), and with NusB it may even be involved in intracellular RNA folding and processing (Bubunenko et al., 2013). Thus, these early lambda-host interaction genetic studies revealed previously unsuspected complexities of transcription and led directly to the discovery and characterization of factors that control RNA polymerase elongation and termination. The study of the mechanisms of action of these factors continues to the present.
Virion assembly
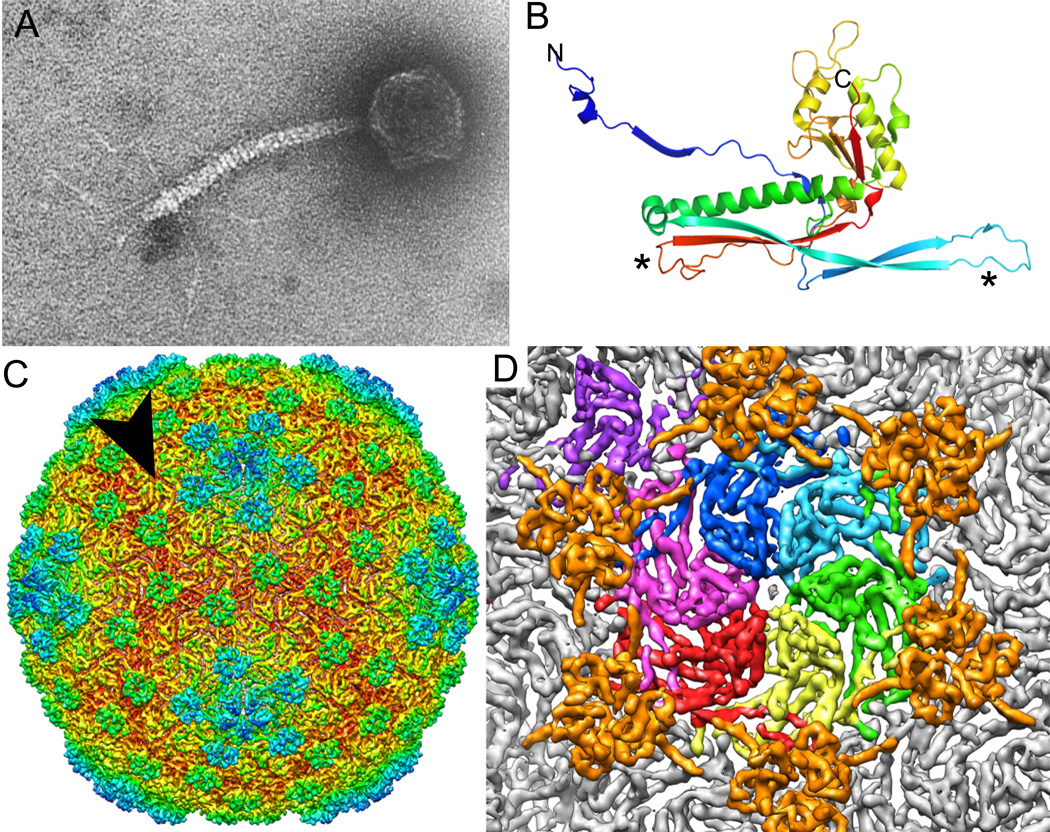
The study of the assembly of phage lambda virions began in the 1960s when Jean Weigle (1966 and 1968) showed that lambda heads and tails assemble independently and spontaneously join in vitro to form infectious virions and with the deduction in the Eduard Kellenbeger, Louis Siminovitch and Dale Kaiser laboratories that gene E encodes its major capsid protein (MCP—also called coat protein) (Karamata et al., 1962; Kemp et al., 1968; Casjens et al., 1970). Figure 3A shows an electron micrograph of the phage lambda virion (with the side fibers that are not present in laboratory strains, see below).
Figure 3. The phage lambda virion.
A. Negatively stained electron micrograph of the Ur-lambda virion. Micrograph is modified from Hendrix and Duda (1992) and reproduced with permission of publisher.
B. Ribbon diagram of the phage HK97 virion MCP subunit. The N- and C-termini of the protein are indicated, and the asterisks mark the locations of K169 and N356 whose side chains are crosslinked to adjacent subunits in the virion (from Helgestrand et al., 2003; protein database code 1OHG with rainbow color depiction by MacPyMOL copyright 2009–2010 Schrodinger, LLC).
C. Subnanometer-resolution 3-dimensional cyroelectron microscopic reconstruction of the phage lambda head. Coloration ranges from red to blue with increasing radius, and the black arrowhead indicates the center of a coat protein hexamer as shown in panel D.
D. Close up view centered on a major capsid protein hexamer of the head shown in panel C. Six gene D protein trimers are indicated in orange and seven major coat protein (gene E protein) subunits are shown in other colors. These colored subunits form one shell asymmetric unit. Panels C and D are modified from Lander et al. (2008) and reproduced with permission of publisher.
At that time, the study of viral structural proteins and their assembly was a non-trivial enterprise. For example, one of us (SRC) began his Ph.D. thesis project in 1968 by simply attempting to determine how many different proteins are present in the lambda virion. I had been toiling over various strategies for separating lambda virion proteins solubilized with urea, acetic acid or guanidine hydrochloride for nearly a year without noticeable success, when Bill Studier (who was studying phage T7 at the Brookhaven Laboratory) gave a seminar at Stanford describing the sodium dodecyl-sulfate (SDS)-polyacrylamide gel electrophoresis technique that he and Jake Maizel had developed (Studier and Maizel, 1969; Maizel, 2000). The combined facts that boiling in 0.1% SDS disassembles virtually all protein complexes and that electrophoresis could be performed in SDS solutions instantly made the prospects for my thesis very bright. This, along with what was probably the first published use of a slab gel apparatus that allowed accurate comparison of adjacent lanes, allowed me to get my thesis started with a definitive proof that gene E encodes the major head protein of lambda (Casjens et al., 1970). I note here that it was the ability to do the rigorous controls necessary to really prove (rather than surmise or strongly suggest) our conclusions that first attracted me to phage work and has kept me here for nearly 50 years. Uli Laemmli (a phage T4 worker) and Jake Maizel soon modified this technique to give considerably higher resolution (called discontinuous SDS gel electrophoresis). In a curious twist of fate this now universally used and extremely highly cited method was published only in a figure legend in Uli's paper on T4 morphogenetic protein cleavage (Laemmli, 1970; cited 215,930 times at the time of this writing).
With the advent of discontinuous SDS gel electrophoresis and methods to tamp down host protein synthesis relative to lambda protein synthesis in infected cells (Ptashne, 1967; Hendrix, 1971) came the rapid identification of the rest of the morphogenetic gene products as specific bands in such gels (Buchwald et al., 1970; Hendrix, 1971; Georgopoulos et al., 1973). This in turn led to analysis of the structures made by infections with various mutant phages and the gene products in them. These studies also illuminated the common protein cleavages that accompany lambda and many other virus assembly processes and make them irreversible and/or allow access to final conformations not available to the full-length proteins (Murialdo and Siminovitch, 1972; Georgopoulos et al., 1973; Hendrix and Casjens, 1974 and 1975). This information allowed the deduction of the basic assembly pathway of lambda virions (the order of action and general role of each of the morphogenetic proteins; summarized in Georgopoulos et al., 1983; Hendrix and Casjens, 2006), but it did not elucidate the detailed functions of each of the twenty-one phage-encoded proteins required for virion assembly. Work continues in this direction to this day.
The fact that lambda procapsids (precursor head structures) are assembled first and DNA is subsequently pumped into them by a DNA translocase was initially suggested by analysis of head-like structures made in vivo (Hendrix and Casjens, 1975) and was rigorously proven by Kaiser et al. (1975). Two proteins, the products of the Nu1 and A genes are required for inserting DNA into the procapsid. The Nu1 protein recognizes the DNA to be packaged (Shinder and Gold, 1988; de Beer et al., 2002). The A protein cleaves lambda DNA at the cos site during packaging to create the cohesive ends (Wang and Kaiser, 1973; Feiss et al., 1983; Davidson et al., 1991) and is also the ATP cleavage-driven translocase that moves the DNA into the procapsid (Duffy and Feiss, 2002; Tsay et al., 2010). Neither of these proteins is present in the completed virion. Mike Feiss and coworkers performed extensive and informative genetic analyses of lambda DNA packaging (Sippy et al., 2014; Feiss and Rao, 2012 and references therein), and Carlos Catalano's laboratory has studied its biochemistry (e.g., Andrews and Catalano, 2013; Singh et al., 2013). Most viruses do not depend on the host to help build their virions (except for chaperones, above), but lambda DNA packaging initiation requires the host IHF protein, which binds to a sequence near the Nu1 protein recognition sites and bends the DNA. This bend allows formation of the DNA:Nu1:A nucleoprotein complex that initiates cos site cleavage and DNA packaging (Xin et al., 1993; Sanyal et al., 2014). Laser tweezer measurements of the force generated by this molecular motor in vitro by Douglas Smith and collaborators have shown it to be among the most powerful nanomotors known (Tsay et al., 2009 and 2010).
Lambda tail assembly work has focused largely on how its shaft is assembled. Isao Katsura and Roger Hendrix found that the length of the gene H-encoded tape measure protein determines the length of the shaft. It acts as a measuring device, most likely by serving as a template for shaft assembly (Katsura and Hendrix, 1984; Katsura, 1987). Curiously, the H protein is proteolytically cleaved before tail attachment to heads, but the role of this cleavage and the protease that causes it remain unknown (Hendrix and Casjens, 1974; Tsui and Hendrix, 1983). In addition, Levin et al. (1993) found that a programmed translational frameshift is required for expression of the essential T reading frame as a C-terminal extension of the gene G protein (called G-T protein). The frameshift ensures the proper ratio (~30:1) of G and G-T proteins. This was one of the first demonstrations of programmed frameshifting in prokaryotes, and it is now known to be an almost universal feature of the assembly of long phage tails, with the curious apparent exception of phage T4 and its allies (Xu et al., 2004a). This phenomenon has only been studied in more detail in lambda, where both G and G-T proteins are required for tail assembly, but neither is present in completed tails. The G and G-T proteins act as a chaperone in which the G domain binds the tape measure protein, and the T domain of G-T binds the major tail subunit (the gene V protein) (Xu et al., 2013 and 2014). An understanding of the detailed dynamics of this assembly process awaits future study.
The different lambdoid phages have quite diverse virion structures (below) and as such have proven to be very fertile ground in studies of virion assembly - in particular studies of phages P22 and HK97 have been very informative. Salmonella phage P22 was of interest early because it was the first phage found to carry out generalized transduction (Zinder and Lederberg, 1952). Horst Schmieger isolated P22 mutants with greatly increased frequencies of transduction (Schmieger, 1972; Casjens et al., 1992b), which made P22 the easiest transducing phage to use in the laboratory. As such it contributed significantly to the rise of Salmonella enterica as one of the important bacterial model organisms (Neidhardt, 1996). Myron Levine, David Botstein and Jonathan King, studied P22 on its own merits with the (prescient) view that phages related to lambda would benefit from the early lambda studies but would be different enough from lambda to be interesting and informative. Transduction is the result of errors in phage DNA packaging, and the study of P22 has led to a much better understanding of headful DNA packaging (Tye et al., 1974; Jackson et al., 1978; Casjens and Hayden, 1988; Casjens et al., 1992c; Wu et al., 2002; Leavitt et al., 2013). P22 was also the first virus found to require a scaffolding protein for proper assembly of its MCP into a capsid shell. Scaffolding protein is present in large numbers inside of procapsids, but it exits the structure before DNA enters to form complete virions. This was one of the first discoveries of a protein that catalyzes the assembly of other proteins (King and Casjens, 1974; Earnshaw et al., 1976; Weigele et al., 2005; Chen et al., 2011; Padilla-Meier et al., 2012), and scaffolding proteins have since been found to be a general feature of the assembly of large virus particles.
The study of lambdoid phage HK97 began in the Hendrix lab because its tail has a slightly different length from that of lambda, but it was soon noticed that its MCP behaved as a smear near the top of an SDS electrophoresis gel of virion proteins (Popa et al., 1991). Analysis of this phenomenon showed that its head proteins are very different from those of lambda and that its virion MCP is autocatalytically cross-linked into inter-locked rings as "molecular chain mail" (Duda, 1998; Wikoff et al., 2000; Helgstrand et al., 2003; Dierkes et al., 2009). We now know that such a chain mail arrangement is not unique to HK97 and may not be uncommon in phage capsids (Duda, 1998; Gilakjan and Kropinski, 1999). In addition, structural studies in the Alasdair Steven and Jack Johnson laboratories led to a uniquely detailed picture of HK97 procapsid maturation (MCP shell expansion) into virion heads (Duda et al., 2006; Hendrix and Johnson, 2012; Cardone et al., 2014). The study of P22 and epsilon15 (a remote lambda relative that is not quite lambdoid) gave rise to the first asymmetric cryo-electron microscopic (EM) reconstructions of virions that show the tail as well as the MCP at subnanometer resolution (Jiang et al., 2006 and 2008; Lander et al., 2006; Chang et al., 2006; Tang et al., 2011). Finally, and perhaps most importantly, the HK97 MCP shell crystallizes and remains today as the only large phage MCP that has been solved to atomic resolution by x-ray crystallography. The fold of the HK97 MCP was novel at the time of its determination (Wikoff et al., 2000) (figure 3B). Subsequent high resolution cryoEM structures have shown that this protein fold (with minor variations) is present in all tailed phage MCPs examined, including those of lambdoid phages lambda, P22, Sf6 and CUS-3 (Lander et al., 2008; Parent et al., 2012a and 2014a; Singh et al., 2013; Rizzo et al., 2014), as well as herpesvirus MCP (Baker et al., 2005), the MCP of an archaea virus with tailed phage morphology (Pietila et al., 2013), and a cellular nanocompartment (McHugh et al., 2014). In addition, Pell et al. (2009) determined the structure of the lambda tail tube subunit (gene V protein). This fold is also present in the protein that makes up the tail tubes of both noncontractile and contractile tails of other phages, as well as in the shaft of the bacterial type VI secretion apparatus (reviewed by Davidson et al., 2012). Thus, the lambdoid phages gave us the structures - now called the "HK97 fold" and the "tail tube fold" - of what are surely two of the most abundant proteins on Earth.
Cell lysis
Five lambda genes have been identified that are important in lysis of infected cells, S105, S107, R, Rz and Rz1. The enzyme responsible for cleaving the cell wall peptidoglycan is encoded by gene R (Campbell, 1961; Harris et al., 1967). This endolysin is a true lysozyme in some phages like P22 and a transglycosidase in lambda (Bienkowska-Szewczyk et al., 1981; Weaver et al., 1985; Evrard et al., 1998; Mooers and Matthews, 2006). Ryland Young and colleagues have described the mechanism(s) by which the lambda R protein is released into the periplasm. Release is controlled by the S105 and S107 holin proteins, which are expressed from different start codons of the same reading frame, resulting in one protein, S107 that is two amino acids longer than the other, S105. The shorter one, S105, causes lysis and S107 inhibits lysis; thus, the ratio of the two determines the exact timing of lysis (Wang et al., 2000a). Lambda S105 protein is a small membrane protein that forms large physical holes in the membrane that allow endolysin escape across the cytoplasmic membrane. These holes are irregular, average >300 nm in diameter and involve hundreds of S105 molecules (Savva et al., 2008; To and Young, 2014). Exactly how S105 molecules aggregate to form such large holes after accumulating in the membrane in a non-hole-forming state remains mysterious. More recent study of the lambdoid phage 21 has shown that its mechanism of endolysin release is different from that of lambda. Its endolysin is secreted by the host Sec translocon machinery into the periplasm in a catalytically inactive form, tethered to the inner membrane by an N-terminal transmembrane domain. When the phage 21 "pinholin" forms small holes that depolarize the inner membrane, the endolysin is released from the membrane and is activated to cleave the peptidoglycan (Pang et al., 2009 and 2013; Xu et al., 2004b). In addition to holin disruption of the inner membrane and cell wall breakdown, the outer membrane must also be disrupted for complete cell lysis to occur. The two lambda proteins that perform this still poorly understood process are Rz and Rz1, which are an inner membrane protein and an outer membrane lipoprotein, respectively. Together they form the "spanin" complex (so named because it spans the entire periplasm) (Berry et al., 2008 and 2012; Young, 2014). Spanins are not always essential in laboratory phage infections because buffeting of cells suspended in solution can physically disrupt the outer membrane in their absence; however, their nearly universal presence in tailed phages suggests a critical evolutionary importance. Curiously, in lambdoid and many other phages the Rz1 gene lies wholly within the Rz gene in its +1 reading frame, where it is a spectacular example of complete gene overlap. These three features of cell lysis, a peptidoglycan degrading enzyme, a holin or pinholin and a spanin, are essentially universal among the tailed phages, and their actions were unraveled through study of the lambdoid phages.
DNA delivery from the virion into target cells
Most mutations that protect bacteria from killing by bacteriophage infection are due to the loss of the primary surface receptors that virions bind to on the cell surface, and studies with lambda identified the LamB protein (an outer membrane porin involved in maltose uptake) as its primary receptor (Thirion and Hofnung, 1972; Randall-Hazelbauer and Schwartz, 1973; Roessner et al., 1983). Other lambdoid phages utilize different surface receptors. For example ø80 binds the FhuA outer membrane protein (Vostrov et al., 1996; Endriss and Braun, 2004), P22 binds (and cleaves) the O-antigen polysaccharide (Wright and Kanegasaki, 1971), and Sf6 utilizes Oantigen and OmpA protein (Parent et al., 2014b). In all phages where it has been studied in detail, fibers or spikes at the distal tip of the tail make the first interaction with the host. In lambda the J protein that resides at the tip of the lambda tail (R. Duda and R. Hendrix, unpublished) and its C-terminal 149 AAs (of 1132 AAs) are sufficient to interact with LamB (Charbit et al., 1994; Wang et al., 2000b; Berkane et al., 2006; Meyer et al., 2012). In addition to receptor alterations, mutations in the host inner membrane protein ManY (mannose phosphotransferase) were found that block injection of lambda DNA. Mutations in the phage V or H genes overcome this block, and this role of ManY does not depend on its enzymatic activity (Emmons et al., 1975; Scandella and Arber, 1976; Elliott and Arber, 1978; Esquinas-Rychen and Emi, 2001). Phage HK97, which has a tail that is quite similar to that of lambda, requires the host's inner membrane glucose transporter protein PstG and periplasmic chaperone FkpA for successful DNA entry, and these requirements are determined by the tape measure protein (Cumby et al., 2014). The actual role of host proteins in DNA injection remains a mystery.
A reconstruction of the early laboratory history of lambda revealed some unexpected additional information about lambda's tail fibers. When Dale Kaiser began his work on lambda as a graduate student at Caltech he saw that the phage made very small plaques, and he isolated a mutant that made larger, more robust plaques. This large plaque size made his classic experiments defining the cI, cII and cIII genes (above) possible, since plaque turbidity is much more visible in larger plaques. Subsequently, as a postdoc at the Pasteur Institute in Paris, he crossed the large plaque mutant he had brought from Pasadena with the version of lambda in use in Paris at that time to make what came to be called lambda Pasadena/Paris or lambda PaPa. This version of lambda is the one used in virtually all laboratory experiments thereafter, and it soon came to be thought of as lambda wild type. Finally, although lambda PaPa has only one short J protein fiber at its tail tip, and does not have long tail fibers, its genome sequence contains a side tail fiber gene stf and a tail fiber addition gene tfa downstream of J. The STF protein has substantial sequence similarity to the phage T4 gene 37 tail fiber protein but contains a frameshift mutation (Hendrix and Duda, 1992). When it was examined, the original lambda prophage in E. coli K-12 (Lederberg, 1951), which has been called Ur-lambda, was found to lack the frameshift mutation in the stf gene and to produce virions that have long thin side tail fibers (figure 3A); the two halves of stf generated by the frameshift in lambda PaPa were originally called orf401 and orf314 by Sanger et al. (1982). These long fibers cause lambda to make smaller plaques (they probably splay out and slow diffusion in agar), so the stf frameshift mutation was present in the plaque picked by Kaiser. The side fibers apparently bind weakly to E. coli outer membrane protein OmpC, and, although they are not essential, they speed up adsorption (Hendrix and Duda, 1992).
The mechanism of DNA release from tailed phage virions and passage into cells remains poorly understood. The right end of the DNA in lambda virions extends from the head about one-third of the way into the tail, so it is primed for directional release during injection (Thomas, 1974). The great majority of the DNA is packed very tightly into phage heads with no DNA-binding proteins holding it in place (Casjens and Hendrix, 1974; Earnshaw et al., 1979; Chang et al., 2006; Lander et al., 2006 and 2008). Phosphate charge repulsion, DNA bending, partial dehydration and low entropy state must be overcome to package DNA, and its compression in the phage head creates a kind of DNA pressure inside the capsid. DNA is spontaneously released from lambda virions when they bind purified LamB receptor (Roessner et al., 1983; Roessner and Ihler, 1987); however, Molineux (2006 and 2013) and Panja and Molineux (2010) have argued that cellular turgor pressure acts against phage DNA injection into cells and have reviewed the ongoing discussion of the exact nature of the forces that drive DNA delivery into cells from virions. Walter Gelbart, Alex Evilevitch and co-workers have studied the energetics of lambda DNA condensation and release (e.g., Grayson et al., 2006; Qiu et al., 2011; Liu et al., 2014). Rob Phillips and co-workers have found that lambda DNA release on pure LamB in vitro is complete in about 10 sec, while injection into cells appears to average about 5 min (Van Valen et al., 2012). Unless the DNA-bound dye confounds the latter experiments, this suggests that forces inside the cell do affect the rate of DNA entry into cells from lambda virions (Van Valen et al., 2012).
Very little is known about the role of the other lambda tail proteins in DNA injection beyond the fact that the H protein is injected into the host, probably into the inner membrane and ahead of the DNA (Roessner et al., 1983; Roessner and Ihler, 1987), and that there must be an as yet uncharacterized rearrangement of the tail tip that allows release of the DNA and H protein from the virion. Curiously, one of the minor tail tip proteins, the L protein, contains a (4Fe-4S)+2 iron-sulfur cluster (Tam et al., 2013). While the function of the cluster is not known, the ability of the L protein to form the complex with this cluster is essential for both tail assembly and DNA injection (Dai, 2009). While lambda appears to release only a few molecules of one protein from the virion during DNA injection, the short-tailed lambdoid phage P22 releases 6–20 molecules each of three different proteins with its DNA (Botstein et al., 1973; Casjens and King, 1974; Israel, 1977). The roles of these proteins also remain unclear. It is not known how the DNA traverses the periplasm and penetrates the inner membrane DNA. At least in the short tailed phages the injected proteins may form a temporary channel through the periplasm (Perez et al., 2009; reviewed by Casjens and Molineux, 2012; see also Hu et al., 2013). Indeed one of the earliest observations in this area, that bacteria infected with lambda can take up exogenously added lambda DNA only if it has at least one cohesive end is still not understood (Kaiser and Hogness, 1960; Kaiser, 1962).
Non-essential accessory genes
In addition to lambda's 29 essential lytic genes, and cI, cII, cIII and int, which are essential for efficient lysogeny, 38 additional open reading frames (ORFs) whose protein products are not absolutely essential to lambda lytic growth or lysogeny in the laboratory were identified in the original genome sequence (as delineated in Hendrix et al., 1983; Hendrix and Casjens, 2006). Many of these have been shown to be functional genes that produce proteins. Recent ribosome profiling analysis of lambda by Jeff Roberts and co-workers (Liu et al., 2013) confirmed that essentially all of these ORFs are translated. This observation, like the determination of the lambda genome sequence before it, provides striking confirmation of the accuracy of the catalog of lambda genes with identifiable functions, which was determined largely by genetic methods, decades before development of many of the sophisticated techniques that contemporary students may assume have always been available. Surprisingly, the ribosome profiling also identified 55 possibly translated ORFs in addition to the 71 previously known genes. These "new" ORFs are all quite small (all but one are ≤76 codons in length), and none completely overlaps a previously identified gene. It is not known if the short putative proteins they might encode (or perhaps the act of their translation?) have functions. The profiling technique used is relatively new, and the authors do not rule out the possibility that some or all of the putative new genes could be the result of artifacts of the method. On the other hand, if ongoing work confirms the reality of even a fraction these new potential genes, lambda will once again have shown that the descriptions found in textbooks fall short, even for a “simple” little virus like lambda.
Each of the major lambda transcription units contains accessory genes whose functions are known. The late region includes the lom and bor genes that are expressed from the prophage and appear to help the host bind to eukaryotic cells and confer blood serum resistance on the host, respectively (Barondess and Beckwith, 1995; Vaca Pacheco et al., 1997). Ea47, Ea31 and Ea59 (the latter encodes an ATP-dependent endonuclease; Benchimol et al., 1982) are nonessential genes that lie between the tail region and int whose biological roles are not known. The early left operon contains a number of useful but nonessential genes. For example, the exo, bet and gam proteins catalyze homologous recombination (see DNA replication and homologous recombination above), the kil gene product disrupts host division septum formation (Greer, 1975; Haeusser et al., 2014), and the less well studied bin and ral functions block host replication initiation (Sergueev et al., 2001) and modulate host restriction-modification systems (Loenen and Murray, 1986), respectively. Nonessential genes rexA and rexB are co-transcribed with the cI repressor gene and are discussed below with lysogenic conversion genes. Near the 3'-end of lambda's early right operon lie ten contiguous nonessential nin genes, so named because they are absent in large deletions that remove tR2 and make transcription of the essential gene Q (and thus the late genes) high enough to allow the phage to be N independent (Court and Sato, 1969). The nin genes include the ren, orf (ninB), rap (ninG) and orf221, genes that encode protection from rexAB exclusion (Toothman and Herskowitz, 1980b), a homologous recombination function (Tarkowski et al., 2002; Curtis et al., 2014), a Holliday junction resolvase active in homologous recombination (Barik, 1993; Sharples et al., 2004) and a serine/threonine/tyrosine protein phosphatase (Voegtli et al., 2000) whose role is unknown, respectively.
Lysogenic conversion
Accessory genes that are expressed from the prophage and alter the host in some way are called lysogenic conversion genes, e.g. the lambda bor and lom genes (above). Many temperate phages carry genes, called superinfection exclusion genes, that protect the prophage-carrying host from attack by other phages, and lambda is no exception. Its rexAB (above) and sieB genes block infection by certain other phages (e. g., Parma et al., 1992; Engelberg-Kulka et al., 1998; Ranade and Poteete, 1993). In both these cases the detailed mechanism of exclusion remains unclear, but it occurs after DNA from the superinfecting phage has entered the cell and in both cases a second protein, ren and esc, respectively, protect lambda from self-exclusion (Toothman and Herskowitz, 1980a; Ranade and Poteete, 1993). Other lambdoid phages encode exclusion proteins that operate through other mechanisms. Several examples are as follows: The P22 sieA gene blocks injection of DNA by some phages (Susskind et al., 1974), phage 933W encodes a tyrosine kinase that excludes HK97 (Friedman et al., 2011)., and the phage ø80 and N15 cor genes block adsorption of superinfecting phages to the outer membrane FhuA protein (Vostrov et al., 1996; Uc-Mass et al., 2004).
Many lambdoid phages encode proteins that modify the bacterial O-antigen surface polysaccharide. These modifications change the spectrum of phages that can adsorb to the O-antigens of such lysogens and also alter the antigenicity and thus the pathogenicity of the host bacterium (Allison and Verma, 2000; Broadbent et al., 2010; Kintz et al., 2015). O-antigen modifying genes are present in two common forms in lambdoid phages. Some phages, for example Shigella phages Sf6 and Sf101, encode O-acetyl transferases that are expressed from the prophage and acetylate sugar moieties in the O-antigen repeat (Verma et al., 1991; Allison and Verma, 2000; Jakhetia et al., 2014). Other lambdoid phages, for example P22, SfX and epsilon34, carry a three gene gtrABC operon; the first two genes in such a cluster, gtrA and gtrB, are rather highly conserved and are thought to encode an undecaprenol phosphate-glucose flippase that moves the sugar part of its substrate from the cytoplasmic to periplasmic side of the inner membrane (Liu et al., 1996) and a bactoprenol transferase that transfers the glucose from UDP-glucose to form undecaprenol phosphate-glucose (Guan et al., 1999), respectively. The third gene, gtrC, is much more variable and different gtrC's encode different glycosyltransferases that add various sugars as side chains to the O-antigen backbone (Markine-Goriaynoff et al., 2004; Thanweer et al., 2008; Villafane et al., 2008; Davies et al., 2013). Interestingly, the phage P22 prophage gtr operon is subject to on/off phase variation that is epigenetically controlled by the host Dam methylase and OxyR regulatory protein (Broadbent et al., 2010).
A variation on lysogenic conversion in the lambdoid phages is the presence of genes that affect the host during lytic infection. It has been known for over half a century that prophages often encode virulence factors that are important in bacterial diseases, e.g., diphtheria toxin (Freeman, 1951; Uchida et al., 1971; Pappenheimer and Murphy, 1983), but it was assumed that these are expressed from uninduced prophages. A surprising more recent observation is that the Shiga-like toxins do not follow this paradigm. The Shiga-like toxins are important in the human disease E. coli hemorrhagic colitis, and the stxA and stxB genes that encode the toxin are carried mostly by lambdoid phage 933W and its close relatives (Plunkett et al., 1999; Smith et al., 2012), where they lie within the late operon between the promoter and lysis genes (Neely and Friedman, 1998). David Friedman, Matthew Waldor and colleagues have studied two of these phages, 933W and H-19B, and they find that high level expression of stxAB is largely dependent on prophage induction and replication of the prophage to increase the DNA copy number. In addition, phage mediated cell lysis is required to release the toxin from the cell (Wagner et al., 2002; Tyler et al., 2004). Thus, it appears that “spontaneous” induction, lysis and death of a small fraction of these bacteria produces the toxin that is presumably advantageous to the bacterium during disease. This seems to be an example of sib selection and/or altruism in which lysis of a few members of a bacterial clone give an advantage to their clonal siblings (Livny and Friedman, 2004).
Molecular cloning of DNA
In 1968 Peter Lobban, a Ph.D. student in Dale Kaiser's laboratory in the Biochemistry Department of Stanford University was the first person to realize that the tools to clone DNA existed and that it would be a useful thing to do. He presented this proposal at a Kaiser lab group meeting, defended the idea in his Ph.D. preliminary exam, and proceeded to do his thesis work on cloning methodology (Lobban, 1972; Lobban and Kaiser, 1973). This idea, which in part sprang from the fact that Kaiser's laboratory was very near those of Arthur Kornberg and Robert Lehman where DNA polymerase and DNA ligase were being characterized at that time (e.g., Olivera and Lehman, 1967; Englund et al., 1968), was so powerful that others devised more rapidly attainable cloning goals and were able to publish before Lobban (Jackson et al., 1972; Cohen et al., 1973). Nonetheless, the idea of molecular cloning originated with him (Kornberg, 1991; Berg, 1993; Casjens, 1994; Yi, 2008; for more extensive historical treatments of molecular cloning and genetic engineering see Lear, 1978; Judson, 1979; and McElheny, 2014).
In addition, other important parts of cloning technology were also developed with phage lambda. Werner Arber's discovery and characterization of bacterial restriction-modification systems was done using phage lambda (Arber, 1971 and references therein). Ron Davis at Stanford developed phage lambda into a powerful cloning vector by showing that some restriction enzymes leave sticky ends and then utilizing these enzymes for DNA cloning (Mertz and Davis, 1972; Cameron et al., 1975), Kaiser and Masuda (1973) devised a procedure for packaging exogenously added lambda DNA into lambda virions, and lambda-based cosmid vectors were developed for cloning relatively large DNA sections up to about 40 kbp (Hohn and Murray, 1977; Collins and Hohn, 1978).
Thus, lambda contributed greatly to the development of DNA cloning technology, but these advances contributed to the demise of the golden age of lambda, since molecular cloning became the first step in being able to understand cellular (both prokaryotic and eukaryotic) DNA function at a molecular level. Because lambda was no longer uniquely experimentally accessible, and because the basic aspects of its lifecycle were by then fairly well understood, the 1980s and 1990s saw fewer and fewer laboratories dedicated solely to the study of phage lambda. Nonetheless, a handful of laboratories persisted in delving deeper into the details of the life cycles of lambda and its relatives. In addition, the well-understood lambda system has often been used for proof-of-principle demonstrations for new technologies. Thus, even though they are the sole objects of study in relatively few laboratories at present, the lambdoid phages continue to give new insights into the amazing details of gene expression, gene function and virus life cycles to this day.
Bacteriophage diversity and the evolution of modular viral genomes
The early decision to study a small number of chosen bacteriophages (including the most highly studied phages lambda, T4 and T7) for highly focused hypothesisdriven research certainly sped up the attainment of our current overall knowledge of gene expression mechanisms and numerous viral functions immensely. However, at the same time it restricted our view of the diversity of life on Earth and in particular the diversity and natural abundance of bacteriophages. Even in the early days of the study of phages several lambda relatives (e.g., phages 21, 434, P22 and ø80) had been isolated and were known to form viable hybrids with lambda (Kaiser and Jacob, 1957). Because of their different repressor-operator specificities, these other phages were of seminal use in developing an understanding of the control of early gene expression and the reasons why phage lambda cannot infect a lambda lysogen (e.g., Bode and Kaiser (1965) showed that the lambda cII and cIII genes can't complement from a heteroimmune repressed hybrid prophage). Analysis of heteroduplex DNA pairs with lambda showed early on that these phages have mosaically related genomes in which patches of highly related sequence are interspersed with patches of less related or unrelated sequences (Fiandt et al., 1971; Simon et al., 1971; Botstein, 1980; Campbell and Botstein, 1983; Casjens et al., 1992a; Campbell, 1994). Phage genome sequences have greatly increased our understanding of this genome mosaicism in recent years (Hendrix et al., 2000; Juhala et al., 2000; Hendrix, 2002; Casjens, 2005; Casjens and Thuman-Commike, 2011; Grose and Casjens, 2014). Figure 4 shows some of the better-characterized variations among lambdoid phages.
Figure 4. The lambdoid phage diversity menu.
A schematic map of the lambda virion chromosome and its major transcripts are shown above (colored as in figure 1). The "menus" of various functional sections are labeled above and listed below the map in colored rectangles. In each section's menu a few lambdoid phages that have very different genes in this region and are best understood in that particular regard are indicated; however, the extant diversity of most sections is larger than shown and remains poorly defined in many cases. In some cases the different phages indicated within one menu section have clearly nonhomologous genes that encode parallel functions (e.g., SfV long contractile, lambda long non-contractile and P22 short tails; lambda Exo, P22 Erf and gifsy-2 RecE type homologous recombination systems), and in some cases the genes are extremely divergent homologues (e.g., portal and MCPs in the procapsid assembly section and probably all the different prophage repressors with different operator binding specificities). Each of the functional sections shown is populated by at least one gene in most lambdoid phages, although there are a few exceptions (e.g., Fels-1 has no nin region genes and HK022 has no N gene). In addition, any given lambdoid phage may have additional scattered accessory genes (e.g., in lambda the lom and rexAB genes between the tail and tail fiber genes and immediately downstream of cI, respectively, and in P22 the rha and ant genes between the lysis and DNA packaging genes and between tail and tailspike genes in P22, respectively).
Many relatives of phage lambda have now been isolated, and at present over 80 such phage genomes have been completely sequenced (compiled in Grose and Casjens, 2014), along with hundreds of related prophages in bacterial genome sequences (see, for example, Casjens and Thuman-Commike (2011) for the 45 such P22-like prophages known at that time). All these phages infect members of the Enterobacteriaceae bacterial family, but their diversity is nonetheless huge. These 80 phages fall into seventeen different "types" or "clusters" that all have lambda-like gene (functional module) orders and apparently similar gene expression cascades (Grose and Casjens, 2014). (We note that there are a very small number of notable exceptions to this synteny; some phage N15 and ASPE-1 early genes have an order that is different from that of lambda; van der Wilk et al., 1999; Ravin et al., 2000.) The seventeen lambdoid phage clusters were defined in terms of overall sequence similarity, in which a dot matrix plot comparison of members of different clusters at relatively low stringency shows recognizable nucleic acid similarity over more than 50% of the genome within but not between clusters (Grose and Casjens, 2014). A very preliminary survey of prophages present in the hundreds of reported enterobacterial genome sequences has revealed at least four additional lambdoid phage clusters (S. Casjens, unpublished). Clearly, the lambda life-style has been extremely successful and has diverged to fill many presumed niches.
All these lambdoid phages are transitively related by patches of similar sequence that can potentially undergo homologous recombination. The "unrelated" patches that lie in parallel chromosomal locations can be either homologous sequence that has diverged to the point that it is no longer significantly related or entirely unrelated sequence form a different source that performs the same function. An example of the former is the lambdoid prophage repressor. The repressors all perform the same role, shutting off the lytic genes in the prophage, yet they are extremely variable and often barely recognizable or even not recognizable as being homologous. The lambdoid repressors that have been examined have similarities in their DNA-binding domain folds, and so are almost certainly ancient homologues (Pabo et al., 1990; Watkins et al., 2008). We are not aware of a recent analysis, but in 1992 eleven different repressor operator-binding specificities were known (Casjens et al., 1992a), and the currently known sequence diversity of these proteins suggests a very much larger repertoire. Another example of very divergent homologues is the procapsid assembly gene cluster (including the MCP and portal protein) of which there are fourteen very different types among the lambdoid phages (Grose and Casjens, 2014). Examples of apparently nonhomologous proteins that perform analogous functions in various lambdoid phages are the three largely unrelated tail types - short, long contractile and long noncontractile (e.g., P22, SfV and lambda, respectively), the integrases of integrating phages like lambda and P22 and the protelomerases of phages N15, PY54 and øKO2 (above), and the multiple types of homologous recombination modules (above). We have previously discussed the differences and similarities among the four best-studied lambdoid phages, lambda, HK97, P22 and N15 in more depth, and the reader is referred to Hendrix and Casjens (2006) for more examples of such mosaic relationships.
We believe that extant lambdoid phage diversity has been attained in several ways. These phages are likely extremely old, and natural divergence has had sufficient time to make even some of their homologous protein sequences no longer significantly related. New mosaic boundaries/novel sequence joints can form in two ways. First, highly divergent lambdoid phages could recombine through illegitimate recombination events to generate a new boundary. Second, illegitimate recombination could incorporate novel sequences into a phage by insertion between or replacement of resident genes. These foreign inserts between pre-existing lambdoid phage genes have been called "morons" since there is more DNA when they are present (Hendrix et al., 2000; Juhala et al., 2000), and the known morons usually carry their own promoters for independent expression. The sources of such inserts and replacements may be divergent lambdoid phages, other phages or genes with no phage relationship, since their sources are currently unknown. Thus, new mosaic boundaries likely form very rarely, but the probability of such a fortuitous event between two phages giving rise to a functional progeny phage is greatly increased if the two participating phages have the same gene order (as all lambdoid phages have). Given the enormous numbers of phages on Earth (Whitman et al., 1998; Wommack and Colwell, 2000; Hendrix, 2002), we imagine that this type of novel joint formation may well be random, and that all possible single-recombination novel joints between divergent lambdoid phage genomes might be formed. However, only the extremely small fraction of such recombinants that has a functional, advantageous phage genome would be found as the phages present today. Once a new functional mosaic boundary is formed in a lambdoid phage genome, homologous recombination between similar sequences in other mosaically related lambdoid phages could rapidly cause reassortment to form new combinations of such boundaries (and sequence patches). Interestingly, although both of these processes, formation and reassortment of mosaic patches, will tend to homogenize the genetic makeup of the participating phages, neither process is fast enough to mask the presence of clear clustering of different types within the lambdoid phage group (Grose and Casjens, 2014).
The future
Lambdoid phage research was seminal in the attainment of our current understanding on such varied fronts as gene and operon structure and function, the mechanisms of control of gene expression by regulatory proteins and small RNAs, protein chaperones, prophage integration, protein and RNA turnover, virion assembly and modular virus genome evolution. Although from outside lambdoid phages may be seen as "fully understood," we believe that there is still much that these phages can teach us. In addition to their roles as model systems, the amazing abundance of tailed phages on our planet means that they are extremely important in all ecological processes that involve bacteria such as global carbon and nitrogen cycling (Suttle, 2007; Ankrah et al., 2014). Phages are involved through both their ability to kill bacteria and so affect their population size as well as modification of bacteria through lysogenic conversion and transduction. Thus, it is important to understand phages in their own right.
Recent work has also found the lambdoid phages to have technological applications and human health importance. Lambda and P22 have been developed into powerful and versatile protein display agents, where both the head decoration proteins lambda D (Sternberg and Hoess, 1995; Mikawa et al., 1996; Nicastro et al., 2013; Chang et al., 2014) and P22 Dec (Parent et al., 2012b), as well as the lambda major tail V protein (Maruyama et al., 1994; Zanghi et al., 2007) and P22 MCP head protein (Kang et al., 2008; Servid et al., 2013) have been used successfully to display foreign peptides and proteins. Lambda virions have also been used as DNA or protein vaccine delivery vehicles in mammals (Jepson and March, 2004; Lankes et al., 2007; Clark et al., 2011; Mattiacio et al., 2011; Saeedi et al., 2014). Trevor Douglas and Peter Prevelige are developing P22 into a flexible multi-use nanoparticle platform, where foreign molecular cargo can be placed on the surface or within the interior of the particle (O'Neil et al., 2011 and 2102; Lucon et al., 2012; Uchida et al., 2012) to make, for example, magnetic resonance contrast enhancing agents (Qazi et al., 2013 and 2014) or enzymecontaining nanoreactors (Patterson et al., 2012 and 2014). Finally, the stability of cross-linked HK97 capsids may give them stability advantages, and HK97 nanoparticles are under development (Huang et al., 2011). Human health relevance comes from the fact that these phages carry bacterial disease virulence factor genes such as the lom and bor genes of lambda and the Shiga-like toxin stx genes (above).
It is now nearly 65 years since Esther Lederberg discovered and named bacteriophage lambda, but we still haven’t reached the mythical goal of a complete understanding of its molecular life cycle. If nothing else, work on these phages should have taught us that there are many levels of intricacy and sophistication in all aspects of phage life cycles, and each time a level is "understood" it opens doors to yet deeper levels of understanding. Each time we think we have learned something new and important about lambda, we have really learned something new and important about biology more generally, and the lambdoid phages haven’t yet run out of interesting biology to illuminate. Despite the changing fashions in research, the vicissitudes of NIH grant funding, and the changing cast of characters studying them, these phages have been a remarkably consistent and productive source of new biological insight that remains an important experimental model system in the vanguard of biological science. This seems to us unlikely to change any time soon.
Highlights.
Phage lambda research was seminal to our current understanding of molecular biology.
We describe some of the events that occurred during early lambda research.
We also describe recent findings in phage lambda research.
Acknowledgements
The authors' research is supported by NIH grants GM47795 and GM114817 from the U.S. Public Health Service to RWH and SRC, respectively. We thank our colleagues in the phage research field for many years of gratifyingly open and stimulating discussions. We especially thank Costa Georgopoulos, Peter Lobban and two anonymous reviewers for their welcome suggestions and corrections of our less than perfectly accurate memories.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aihara H, Huang WM, Ellenberger T. An interlocked dimer of the protelomerase TelK distorts DNA structure for the formation of hairpin telomeres. Mol. Cell. 2007;27:901–913. doi: 10.1016/j.molcel.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Kornitzer D, Kobi S, Oppenheim AB. Functional and structural elements of the mRNA of the cIII gene of bacteriophage lambda. J. Mol. Biol. 1991;218:723–733. doi: 10.1016/0022-2836(91)90261-4. [DOI] [PubMed] [Google Scholar]
- Andrews BT, Catalano CE. Strong subunit coordination drives a powerful viral DNA packaging motor. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5909–5914. doi: 10.1073/pnas.1222820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankrah NY, May AL, Middleton JL, Jones DR, Hadden MK, Gooding JR, LeCleir GR, Wilhelm SW, Campagna SR, Buchan A. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 2014;8:1089–1100. doi: 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber W. Host-controlled variation. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 83–96. [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–16970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CA, Johnson RC. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J. Bacteriol. 1991;173:4032–4038. doi: 10.1128/jb.173.13.4032-4038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay K, Parua PK, Datta AB, Parrack P. Escherichia coli HflK and HflC can individually inhibit the HflB (FtsH)-mediated proteolysis of lambda CII in vitro. Arch. Biochem. Biophys. 2010;501:239–243. doi: 10.1016/j.abb.2010.06.030. [DOI] [PubMed] [Google Scholar]
- Barik S. Expression and biochemical properties of a protein serine/threonine phosphatase encoded by bacteriophage lambda. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10633–10637. doi: 10.1073/pnas.90.22.10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barondess JJ, Beckwith J. bor gene of phage lambda, involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J. Bacteriol. 1995;177:1247–1253. doi: 10.1128/jb.177.5.1247-1253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28:112–122. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchimol S, Lucko H, Becker A. A novel endonuclease specified by bacteriophage lambda. Purification and properties of the enzyme. J. Biol. Chem. 1982;257:5211–5219. [PubMed] [Google Scholar]
- Berg P. Dissections and reconstructions of genes and chromosomes. In: Frängsmyr T, editor. Nobel Lectures, Chemistry 1971–1980. Singapore: World Scientific Publishing Co.; 1993. pp. 385–402. [Google Scholar]
- Berkane E, Orlik F, Stegmeier JF, Charbit A, Winterhalter M, Benz R. Interaction of bacteriophage lambda with its cell surface receptor: an in vitro study of binding of the viral tail protein gpJ to LamB (Maltoporin) Biochemistry. 2006;45:2708–2720. doi: 10.1021/bi051800v. [DOI] [PubMed] [Google Scholar]
- Berry J, Summer EJ, Struck DK, Young R. The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Molec. Microbiol. 2008;70:341–351. doi: 10.1111/j.1365-2958.2008.06408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Rajaure M, Pang T, Young R. The spanin complex is essential for lambda lysis. J. Bacteriol. 2012;194:5667–5674. doi: 10.1128/JB.01245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Szewczyk K, Lipinska B, Taylor A. The R gene product of bacteriophage lambda is the murein transglycosylase. Mol. Gen. Genet. 1981;184:111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- Bode V, Kaiser AD. Repression of the c2 and c3 cistrons of phage lambda in a lysogenic bacterium. Virology. 1965;25:111–121. doi: 10.1016/0042-6822(65)90258-8. [DOI] [PubMed] [Google Scholar]
- Botstein D. A theory of modular evolution in bacteriophages. Ann. N. Y. Acad. Sci. 1980;354:484–491. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- Botstein D, Matz MJ. A recombination function essential to the growth of bacteriophage P22. J. Mol. Biol. 1970;54:417–440. doi: 10.1016/0022-2836(70)90119-1. [DOI] [PubMed] [Google Scholar]
- Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Molec. Microbiol. 2010;77:337–353. doi: 10.1111/j.1365-2958.2010.07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubunenko M, Court DL, Al Refaii A, Saxena S, Korepanov A, Friedman DI, Gottesman ME, Alix JH. Nus transcription elongation factors and RNase III modulate small ribosome subunit biogenesis in Escherichia coli. Molec. Microbiol. 2013;87:382–393. doi: 10.1111/mmi.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald M, Murialdo H, Siminovitch L. The morphogenesis of bacteriophage lambda. II. Identification of the principal structural proteins. Virology. 1970;42:390–400. doi: 10.1016/0042-6822(70)90282-5. [DOI] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- Cameron JR, Panasenko SM, Lehman IR, Davis RW. In vitro construction of bacteriophage lambda carrying segments of the Escherichia coli chromosome: selection of hybrids containing the gene for DNA ligase. Proc. Natl. Acad. Sci. U. S. A. 1975;72:3416–3420. doi: 10.1073/pnas.72.9.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. Sensitive mutants of bacteriophage lambda. Virology. 1961;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Campbell A. The episomes. Advan. Genet. 1962;11:101–118. [Google Scholar]
- Campbell A. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- Campbell A. Phage integration and chromosome structure. A personal history. Annu. Rev. Genet. 2007;41:1–11. doi: 10.1146/annurev.genet.41.110306.130240. [DOI] [PubMed] [Google Scholar]
- Campbell A, Botstein D. Evolution of the lambdoid phages. In: Hendrix R, Roberts JW, Stahl FW, Weisberg R, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1983. pp. 365–380. [Google Scholar]
- Cardone G, Duda RL, Cheng N, You L, Conway JF, Hendrix RW, Steven AC. Metastable intermediates as stepping stones on the maturation pathways of viral capsids. MBio. 2014;5:e02067-14. doi: 10.1128/mBio.02067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. DNA packaging by the lambdoid phages - from pure beginnings to applications in genetic engineering. Bioesssays. 1994;16:847–851. [Google Scholar]
- Casjens S. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 2005;8:451–458. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hendrix RW. Locations and amounts of major structural proteins in bacteriophage lambda. J. Mol. Biol. 1974;88:535–545. doi: 10.1016/0022-2836(74)90500-2. [DOI] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hayden M. Analysis in vivo of the bacteriophage P22 headful nuclease. J. Mol. Biol. 1988;199:467–474. doi: 10.1016/0022-2836(88)90618-3. [DOI] [PubMed] [Google Scholar]
- Casjens S, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Casjens S, Molineux IJ. Short noncontractile tail machines: adsorption and DNA delivery by podoviruses. Adv. Exp. Med. Biol. 2012;726:143–179. doi: 10.1007/978-1-4614-0980-9_7. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hohn T, Kaiser AD. Morphological proteins of phage lambda: identification of the major head protein as the product of gene E. Virology. 1970;42:496–507. doi: 10.1016/0042-6822(70)90293-x. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hatfull G, Hendrix R. Evolution of dsDNA tailed-bacteriophage genomes. Sem. Virol. 1992a;3:383–397. [Google Scholar]
- Casjens S, Sampson L, Randall S, Eppler K, Wu H, Petri JB, Schmieger H. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 1992b;227:1086–1099. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno E, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 1992c;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- Casjens S, Gilcrease EB, Huang WM, Bunny KL, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 2004;186:1818–1832. doi: 10.1128/JB.186.6.1818-1832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chang JR, Song EH, Nakatani-Webster E, Monkkonen L, Ratner DM, Catalano CE. Phage lambda capsids as tunable display nanoparticles. Biomacromolecules. 2014;15:4410–4419. doi: 10.1021/bm5011646. [DOI] [PubMed] [Google Scholar]
- Charbit A, Werts C, Michel V, Klebba PE, Quillardet P, Hofnung M. A role for residue 151 of LamB in bacteriophage lambda adsorption: possible steric effect of amino acid substitutions. J. Bacteriol. 1994;176:3204–3209. doi: 10.1128/jb.176.11.3204-3209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeran A, Babu Suganthan R, Swapna G, Bandey I, Achary MS, Nagarajaram HA, Sen R. Escherichia coli RNA polymerase mutations located near the upstream edge of an RNA:DNA hybrid and the beginning of the RNA-exit channel are defective for transcription antitermination by the N protein from lambdoid phage H-19B. J. Mol. Biol. 2005;352:28–43. doi: 10.1016/j.jmb.2005.06.052. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Golding I, Sawai S, Guo L, Cox EC. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005;3:e229. doi: 10.1371/journal.pbio.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Sharma V, Brenowitz S, Chu CC, Sandler S, Satin L, Templin A, Berger I, Cohen A. Genetic and molecular analyses of the C-terminal region of the recE gene from the Rac prophage of Escherichia coli K-12 reveal the recT gene. J. Bacteriol. 1993;175:7673–7682. doi: 10.1128/jb.175.23.7673-7682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JR, Bartley K, Jepson CD, Craik V, March JB. Comparison of a bacteriophage-delivered DNA vaccine and a commercially available recombinant protein vaccine against hepatitis B. FEMS Immunol. Med. Microbiol. 2011;61:197–204. doi: 10.1111/j.1574-695X.2010.00763.x. [DOI] [PubMed] [Google Scholar]
- Cohen SE, Godoy VG, Walker GC. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J. Bacteriol. 2009;191:665–672. doi: 10.1128/JB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. U. S. A. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J, Hohn B. Cosmids: a type of plasmid gene-cloning vector that is packageable in vitro in bacteriophage lambda heads. Proc. Natl. Acad. Sci. U. S. A. 1978;75:4242–4246. doi: 10.1073/pnas.75.9.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D, Sato K. Studies of novel transducing variants of lambda: dispensability of genes N and Q. Virology. 1969;39:348–352. doi: 10.1016/0042-6822(69)90060-9. [DOI] [PubMed] [Google Scholar]
- Court DL, Oppenheim AB, Adhya SL. A new look at bacteriophage lambda genetic networks. J. Bacteriol. 2007a;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court R, Cook N, Saikrishnan K, Wigley D. The crystal structure of lambda-Gam protein suggests a model for RecBCD inhibition. J. Mol. Biol. 2007b;371:25–33. doi: 10.1016/j.jmb.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Cui L, Murchland I, Dodd IB, Shearwin KE. Bacteriophage lambda repressor mediates the formation of a complex enhancer-like structure. Transcription. 2013a;4:201–205. doi: 10.4161/trns.26101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Murchland I, Shearwin KE, Dodd IB. Enhancer-like long-range transcriptional activation by lambda CI-mediated DNA looping. Proc. Natl. Acad. Sci. U. S. A. 2013b;110:2922–2927. doi: 10.1073/pnas.1221322110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumby N, Reimer K, Mengin-Lecreulx D, Davidson AR, Maxwell KL. The phage tail tape measure protein, an inner membrane protein, and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Molec. Microbiol. 2015 doi: 10.1111/mmi.12918. in press. [DOI] [PubMed] [Google Scholar]
- Curtis FA, Reed P, Wilson LA, Bowers LY, Yeo RP, Sanderson JM, Walmsley AR, Sharples GJ. The C-terminus of the phage lambda Orf recombinase is involved in DNA binding. J. Mol. Recognit. 2011;24:333–340. doi: 10.1002/jmr.1079. [DOI] [PubMed] [Google Scholar]
- Curtis FA, Malay AD, Trotter AJ, Wilson LA, Barradell-Black MM, Bowers LY, Reed P, Hillyar CR, Yeo RP, Sanderson JM, Heddle JG, Sharples GJ. Phage ORF family recombinases: conservation of activities and involvement of the central channel in DNA binding. PLoS One. 2014;9:e102454. doi: 10.1371/journal.pone.0102454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XX. Ph. D. Dissertation. University of Pittsburgh; 2009. Expression, purification and characterization of bacteriophage lambda tail tip proteins. [Google Scholar]
- Davidson A, Yau P, Murialdo H, Gold M. Isolation and characterization of mutations in the bacteriophage lambda terminase genes. J. Bacteriol. 1991;173:5086–5096. doi: 10.1128/jb.173.16.5086-5096.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. Long noncontractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 2012;726:115–142. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- Davidson N, Szyblaski W. Physical and chemical characteristics of lambda DNA. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 45–82. [Google Scholar]
- Davies MR, Broadbent SE, Harris SR, Thomson NR, van der Woude MW. Horizontally acquired glycosyltransferase operons drive salmonellae lipopolysaccharide diversity. PLoS Genet. 2013;9:e1003568. doi: 10.1371/journal.pgen.1003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- Deighan P, Hochschild A. The bacteriophage lambdaQ anti-terminator protein regulates late gene expression as a stable component of the transcription elongation complex. Molec. Microbiol. 2007;63:911–920. doi: 10.1111/j.1365-2958.2006.05563.x. [DOI] [PubMed] [Google Scholar]
- Dierkes LE, Peebles CL, Firek BA, Hendrix RW, Duda RL. Mutational analysis of a conserved glutamic acid required for self-catalyzed cross-linking of bacteriophage HK97 capsids. J. Virol. 2009;83:2088–2098. doi: 10.1128/JVI.02000-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Manzo C, Fulcrand G, Leng F, Dunlap D, Finzi L. DNA supercoiling: A regulatory signal for the lambda repressor. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15402–15407. doi: 10.1073/pnas.1320644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94:55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hendrix RW, Huang WM, Conway JF. Shared architecture of bacteriophage SPO1 and herpesvirus capsids. Curr. Biol. 2006;16:R11–R13. doi: 10.1016/j.cub.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Duffy C, Feiss M. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J. Mol. Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Casjens S, Harrison S. Assembly of the head of bacteriophage P22, X-ray diffraction from heads, proheads and related structures. J. Mol. Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Hendrix RW, King J. Structural studies of bacteriophage lambda heads and proheads by small angle X-ray diffraction. J. Mol. Biol. 1979;134:575–594. doi: 10.1016/0022-2836(79)90368-1. [DOI] [PubMed] [Google Scholar]
- Echols H. Regulation of lytic development. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 247–270. [Google Scholar]
- Echols H, Gingery R. Mutants of bacteriophage lambda defective in vegetative genetic recombination. J. Mol. Biol. 1968;34:239–249. doi: 10.1016/0022-2836(68)90249-0. [DOI] [PubMed] [Google Scholar]
- Eisen H, Brachet P, Pereira da Silva L, Jacob F. Regulation of repressor expression in lambda. Proc. Natl. Acad. Sci. U. S. A. 1970;66:855–862. doi: 10.1073/pnas.66.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J, Arber W. E. coli K-12 pel mutants, which block phage lambda DNA injection, coincide with ptsM, which determines a component of a sugar transport system. Mol. Gen. Genet. 1978;161:1–8. doi: 10.1007/BF00266608. [DOI] [PubMed] [Google Scholar]
- Emmons S, MacCosham V, Baldwin R. Tandem genetic duplications in phage lambda. III. The frequency of duplication mutants in two derivatives of phage lambda is independent of known recombination systems. J. Mol. Biol. 1975;91:133–146. doi: 10.1016/0022-2836(75)90154-0. [DOI] [PubMed] [Google Scholar]
- Endriss F, Braun V. Loop deletions indicate regions important for FhuA transport and receptor functions in Escherichia coli. J. Bacteriol. 2004;186:4818–4823. doi: 10.1128/JB.186.14.4818-4823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund PT, Deutscher MP, Jovin TM, Kelly RB, Cozzarelli NR, Kornberg A. Structural and functional properties of Escherichia coli DNA polymerase. Cold Spring Harb Symp. Quant. Biol. 1968;33:1–9. doi: 10.1101/sqb.1968.033.01.005. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Reches M, Narasimhan S, Schoulaker-Schwarz R, Klemes Y, Aizenman E, Glaser G. RexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15481–15486. doi: 10.1073/pnas.95.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquinas-Rychen M, Emi B. Facilitation of bacteriophage lambda DNA injection by inner membrane proteins of the bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS) J. Molec. Microbiol. Biotechnol. 2001;3:361–370. [PubMed] [Google Scholar]
- Evrard C, Fastrez J, Declercq JP. Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes. J. Mol. Biol. 1998;276:151–164. doi: 10.1006/jmbi.1997.1499. [DOI] [PubMed] [Google Scholar]
- Farzadfard F, Lu TK. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science. 2014;346:1256272. doi: 10.1126/science.1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss M, Rao VB. The bacteriophage DNA packaging machine. Adv. Exp. Med. Biol. 2012;726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- Feiss M, Kobayashi I, Widner W. Separate sites for binding and nicking of bacteriophage lambda DNA by terminase. Proc. Natl. Acad. Sci. U. S. A. 1983;80:955–959. doi: 10.1073/pnas.80.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiandt M, Hradecna S, Lozeron H, Szybalski W. Electron micrographic mapping of eletions, insertions, inversions, and homologies in the DNAs of coliphges lambda and ø80. In: Hershey AD, editor. Bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 329–354. [Google Scholar]
- Franklin NC. Deletions and functions of the center of the ø80-lambda phage genome. Evidence for a phage function promoting genetic recombination. Genetics. 1967;57:301–318. doi: 10.1093/genetics/57.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin NC. Altered reading of genetic signals fused to the N operon of bacteriophage lambda: genetic evidence for modification of polymerase by the protein product of the N gene. J. Mol. Biol. 1974;89:33–48. doi: 10.1016/0022-2836(74)90161-2. [DOI] [PubMed] [Google Scholar]
- Freeman V. Studies on the virulence of bacteriophge-infected strains of Corynebacterium diphtheriae. J. Bacteriol. 1951;61:675–688. doi: 10.1128/jb.61.6.675-688.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. A bacterial mutant affecting lambda development. In: Hershey ADE, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 733–738. [Google Scholar]
- Friedman DI, Baron LS. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974;58:141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- Friedman DI, Court DL. Transcription antitermination: the lambda paradigm updated. Molec. Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- Friedman D, Gottesman M. Lytic mode of lambda development. In: Hendrix R, Roberts J, Stahl FW, Weisberg R, editors. Lambda II. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1983. pp. 21–52. [Google Scholar]
- Friedman DI, Mozola CC, Beeri K, Ko CC, Reynolds JL. Activation of a prophage-encoded tyrosine kinase by a heterologous infecting phage results in a self-inflicted abortive infection. Molec. Microbiol. 2011;82:567–577. doi: 10.1111/j.1365-2958.2011.07847.x. [DOI] [PubMed] [Google Scholar]
- Gatenby A. Chaperonins of photosynthetic organisms. In: Ellis RJ, editor. The Chaperonins. San Diego, CA: Academic Press; 1996. pp. 91–106. [Google Scholar]
- Georgopoulos CP. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc. Natl Acad Sci U S A. 1971;68:2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. Toothpicks, serendipity and the emergence of the Escherichia coli DnaK (Hsp70) and GroEL (Hsp60) chaperone machines. Genetics. 2006;174:1699–1707. doi: 10.1534/genetics.104.68262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C, Herskowitz I. Escherichia coli mutants blocked in lambda DNA synthesis. In: Hershey ADE, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 553–564. [Google Scholar]
- Georgopoulos CP, Hohn B. Identification of a host protein necessary for bacteriophage morphogenesis (the groE gene product) Proc. Natl. Acad. Sci U S A. 1978;75:131–135. doi: 10.1073/pnas.75.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C, Welch WJ. Role of the major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- Georgopoulos CP, Hendrix RW, Casjens SR, Kaiser AD. Host participation in bacteriophage lambda head assembly. J. Mol. Biol. 1973;76:45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Tilly K, Casjens S. Lambdoid phage head assembly. In: Hendrix RW, Roberts J, Stahl F, Weisberg R, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1983. pp. 279–304. [Google Scholar]
- Giladi H, Koby S, Prag G, Engelhorn M, Geiselmann J, Oppenheim AB. Participation of IHF and a distant UP element in the stimulation of the phage lambda PL promoter. Molec. Microbiol. 1998;30:443–451. doi: 10.1046/j.1365-2958.1998.01079.x. [DOI] [PubMed] [Google Scholar]
- Gilakjan ZA, Kropinski AM. Cloning and analysis of the capsid morphogenesis genes of Pseudomonas aeruginosa bacteriophage D3: another example of protein chain mail? J. Bacteriol. 1999;181:7221–7227. doi: 10.1128/jb.181.23.7221-7227.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Muller-Hill B. Isolation of the lac repressor. Proc. Natl. Acad. Sci. U. S. A. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingery R, Echols H. Mutants of bacteriophage lambda unable to integrate into the host chromosome. Proc. Natl. Acad. Sci. U. S. A. 1967;58:1507–1514. doi: 10.1073/pnas.58.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AR, Howe M. New mutations in the S cistron of bacteriophage lambda affecting host cell lysis. Virology. 1969;38:200–202. doi: 10.1016/0042-6822(69)90148-2. [DOI] [PubMed] [Google Scholar]
- Goldenberg DP, Argyle B. Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study. Biophys. J. 2014;106:905–914. doi: 10.1016/j.bpj.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonciarz-Swiatek M, Wawrzynow A, Um SJ, Learn BA, McMacken R, Kelley WL, Georgopoulos C, Sliekers O, Zylicz M. Recognition, targeting, and hydrolysis of the lambda O replication protein by the ClpP/ClpX protease. J. Biol. Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- Gottesman ME, Weisbeg R. Prophage insertion and excision. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 113–138. [Google Scholar]
- Gottesman ME, Yarmolinsky MB. Integration-negative mutants of bacteriophage lambda. J. Mol. Biol. 1968;31:487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Gottesman ME, Weisberg RA. Little lambda, who made thee? Microbiol. Mol. Biol. Rev. 2004;68:796–813. doi: 10.1128/MMBR.68.4.796-813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman ME, Oppenheim A, Court D. Retroregulation: control of gene expression from sites distal to the gene. Cell. 1982;29:727–728. doi: 10.1016/0092-8674(82)90434-2. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Gottesman M, Shaw JE, Pearson ML. Protein degradation in E. coli: the lon mutation and bacteriophage lambda N and CII protein stability. Cell. 1981;24:225–233. doi: 10.1016/0092-8674(81)90518-3. [DOI] [PubMed] [Google Scholar]
- Grayson P, Evilevitch A, Inamdar MM, Purohit PK, Gelbart WM, Knobler CM, Phillips R. The effect of genome length on ejection forces in bacteriophage lambda. Virology. 2006;348:430–436. doi: 10.1016/j.virol.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J. Positive control of endolysin synthesis in vitro by the gene N protein of phage lambda. Proc. Natl. Acad. Sci. U. S. A. 1972;69:3606–3610. doi: 10.1073/pnas.69.12.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer H. The kil gene of bacteriophage lambda. Virology. 1975;66:589–604. doi: 10.1016/0042-6822(75)90231-7. [DOI] [PubMed] [Google Scholar]
- Grose JH, Casjens S. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468–470:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S, Bastin DA, Verma NK. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology. 1999;145(Pt 5):1263–1273. doi: 10.1099/13500872-145-5-1263. [DOI] [PubMed] [Google Scholar]
- Guarneros G, Montanez C, Hernandez T, Court D. Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc. Natl. Acad. Sci. U. S. A. 1982;79:238–242. doi: 10.1073/pnas.79.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Hoashi M, Weaver A, Brown N, Pan J, Sawitzke JA, Thomason LC, Court DL, Margolin W. The Kil peptide of bacteriophage lambda blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet. 2014;10:e1004217. doi: 10.1371/journal.pgen.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SD, Kane MF, Kolodner RD. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J. Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AW, Mount DW, Fuerst CR, Siminovitch L. Mutations in bacteriophage lambda affecting host cell lysis. Virology. 1967;32:553–569. doi: 10.1016/0042-6822(67)90032-3. [DOI] [PubMed] [Google Scholar]
- Hayes S, Horbay MA, Hayes C. A CI-independent form of replicative inhibition: turn off of early replication of bacteriophage lambda. PLoS One. 2012;7:e36498. doi: 10.1371/journal.pone.0036498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 Å resolution. J. Mol. Biol. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Hemmingsen SM, Woolford C, van der Vies SM, Tilly K, Dennis DT, Georgopoulos CP, Hendrix RW, Ellis RJ. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Identification of proteins coded by phage lambda. In: Hershey ADE, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 355–370. [Google Scholar]
- Hendrix RW. Purification and properties of GroE, a host protein involved in bacteriophage assembly. J. Mol. Biol. 1979;129:375–392. doi: 10.1016/0022-2836(79)90502-3. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Casjens SR. Protein cleavage in bacteriophage lambda tail assembly. Virology. 1974;61:156–159. doi: 10.1016/0042-6822(74)90250-5. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Casjens SR. Assembly of bacteriophage lambda heads: protein processing and its genetic control in petit lambda assembly. J. Mol. Biol. 1975;91:187–199. doi: 10.1016/0022-2836(75)90159-x. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Casjens S. Bacteriophage Z and its genetic neighborhood. In: Calendar R, editor. The Bacteriophages. 2nd Edition. New York City NY: Oxford Press; 2006. pp. 409–447. [Google Scholar]
- Hendrix RW, Duda RL. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science. 1992;258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Johnson JE. Bacteriophage HK97 capsid assembly and maturation. Adv. Exp. Med. Biol. 2012;726:351–363. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Roberts J, Stahl F, Weisberg RE. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- Hensel Z, Weng X, Lagda AC, Xiao J. Transcription-factor-mediated DNA looping probed by high-resolution, single-molecule imaging in live E. coli cells. PLoS Biol. 2013;11:e1001591. doi: 10.1371/journal.pbio.1001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SM. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J. Mol. Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey AD. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. [Google Scholar]
- Hershey AD, Burgi E, Ingraham L. Cohesion of DNA molecules isolated from phage lambda. Proc. Natl. Acad. Sci. U. S. A. 1963;49:748–755. doi: 10.1073/pnas.49.5.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey AD, Burgi E. Complementary structure of interacting sites at the ends of lambda DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 1965;53:325–330. doi: 10.1073/pnas.53.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig S, Klein I, Lurz R, Lanka E, Appel B. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Molec. Microbiol. 2003;48:989–1003. doi: 10.1046/j.1365-2958.2003.03458.x. [DOI] [PubMed] [Google Scholar]
- Ho YS, Rosenberg M. Characterization of a third, cII-dependent, coordinately activated promoter on phage lambda involved in lysogenic development. J. Biol. Chem. 1985;260:11838–11844. [PubMed] [Google Scholar]
- Hoffmann HJ, Lyman SK, Lu C, Petit MA, Echols H. Activity of the Hsp70 chaperone complex-DnaK, DnaJ, and GrpE-in initiating phage lambda DNA replication by sequestering and releasing lambda P protein. Proc. Natl. Acad. Sci. U. S. A. 1992;89:12108–12111. doi: 10.1073/pnas.89.24.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B, Murray K. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3259–3263. doi: 10.1073/pnas.74.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T, Hohn B, Engel A, Wurtz M, Smith PR. Isolation and characterization of the host protein GroE involved in bacteriophage lambda assembly. J. Mol. Biol. 1979;129:359–373. doi: 10.1016/0022-2836(79)90501-1. [DOI] [PubMed] [Google Scholar]
- Hong JS, Smith GR, Ames BN. Adenosine 3':5'-cyclic monophosphate concentration in the bacterial host regulates the viral decision between lysogeny and lysis. Proc. Natl. Acad. Sci. U. S. A. 1971;68:2258–2262. doi: 10.1073/pnas.68.9.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes BC, McClure WR. A CII-dependent promoter is located within the Q gene of bacteriophage lambda. Proc. Natl. Acad. Sci. U. S. A. 1985;82:3134–3138. doi: 10.1073/pnas.82.10.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich A, Fenton W. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide folding. Q. Rev. Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RK, Steinmetz NF, Fu CY, Manchester M, Johnson JE. Transferrin-mediated targeting of bacteriophage HK97 nanoparticles into tumor cells. Nanomedicine (Lond) 2011;6:55–68. doi: 10.2217/nnm.10.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WM, Joss L, Hsieh T, Casjens S. Protelomerase uses a topoisomerase IB/Y-recombinase type mechanism to generate DNA hairpin ends. J. Mol. Biol. 2004;337:77–92. doi: 10.1016/j.jmb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Israel V. E proteins of bacteriophage P22. I. Identification and ejection from wild-type and defective particles. J. Virol. 1977;23:91–97. doi: 10.1128/jvi.23.1.91-97.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Symons RH, Berg P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1972;69:2904–2909. doi: 10.1073/pnas.69.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 1978;118:365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Jacob F, Wollman E. Induction of phage development in lysogenic bacteria. Cold Spr. Harbor Symp. Quant. Biol. 1953;18:101–108. doi: 10.1101/sqb.1953.018.01.019. [DOI] [PubMed] [Google Scholar]
- Jacob F, Wollman E. Etude genetique d'un bacteriophage tempere d'Escherichia coli. I. Le systeme genetique du bacteriophage. Ann. Inst. Pasteur. 1954;87:653–673. [PubMed] [Google Scholar]
- Jacob F, Campbell A. Sur le systeme de repression assurant l'immunite chez les bacteries lysogenes. C. R. Hebd. Seances Acad. Sci. 1959;248:3219–3221. [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jacob F, Sussman R, Monod J. On the nature of the repressor ensuring the immunity of lysogenic bacteria. C. R. Hebd. Seances Acad. Sci. 1962;254:4214–4216. [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hryc C, Zhang Q, Jakana J, Haase-Petingeill C, Afonine P, Adams P, King J, Jaing W, Chiu W. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proc. Natl. Acad. Sci. U. S. A. 2008;110:12301–12306. doi: 10.1073/pnas.1309947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakhetia R, Marri A, Stahle J, Widmalm G, Verma NK. Serotype-conversion in Shigella flexneri: identification of a novel bacteriophage, Sf101, from a serotype 7a strain. BMC Genomics. 2014;15:742. doi: 10.1186/1471-2164-15-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson CD, March JB. Bacteriophage lambda is a highly stable DNA vaccine delivery vehicle. Vaccine. 2004;22:2413–2419. doi: 10.1016/j.vaccine.2003.11.065. [DOI] [PubMed] [Google Scholar]
- Joung J, Koepp D, Hochschild A. Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science. 1994;265:1863–1866. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- Judson HF. The eighth day of creation. New York, NY: Simon and Schuster; 1979. [Google Scholar]
- Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- Kaiser AD. A genetic study of the temperate coliphage. Virology. 1955;1:424–443. doi: 10.1016/0042-6822(55)90036-2. [DOI] [PubMed] [Google Scholar]
- Kaiser AD. Mutations in a temperate bacteriophage affecting its ability to lysogenize Escherichia coli. Virology. 1957;3:42–49. doi: 10.1016/0042-6822(57)90022-3. [DOI] [PubMed] [Google Scholar]
- Kaiser AD. The production of phage chromosome fragments and their capacity for genetic transfer. J. Mol. Biol. 1962;4:275–287. doi: 10.1016/s0022-2836(62)80005-9. [DOI] [PubMed] [Google Scholar]
- Kaiser AD, Jacob F. Recombination between related bacteriophages and the genetic control of immunity and prophage localization. Virology. 1957;4:509–517. doi: 10.1016/0042-6822(57)90083-1. [DOI] [PubMed] [Google Scholar]
- Kaiser AD, Hogness DS. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J. Mol. Biol. 1960;2:392–415. doi: 10.1016/s0022-2836(60)80050-2. [DOI] [PubMed] [Google Scholar]
- Kaiser D, Masuda T. In vitro assembly of bacteriophage Lambda heads. Proc. Natl. Acad. Sci. U. S. A. 1973;70:260–264. doi: 10.1073/pnas.70.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D, Syvanen M, Masuda T. DNA packaging steps in bacteriophage lambda head assembly. J. Mol. Biol. 1975;91:175–186. doi: 10.1016/0022-2836(75)90158-8. [DOI] [PubMed] [Google Scholar]
- Kameyama L, Fernandez L, Court DL, Guarneros G. RNaselll activation of bacteriophage lambda N synthesis. Molec. Microbiol. 1991;5:2953–2963. doi: 10.1111/j.1365-2958.1991.tb01855.x. [DOI] [PubMed] [Google Scholar]
- Kang S, Lander GC, Johnson JE, Prevelige PE. Development of bacteriophage P22 as a platform for molecular display: genetic and chemical modifications of the procapsid exterior surface. Chembiochem. 2008;9:514–518. doi: 10.1002/cbic.200700555. [DOI] [PubMed] [Google Scholar]
- Karamata D, Kellenberger E, Kellenberger G, Terzi M. On a particle accompanying the development of lambda coliphage. Pathol. Microbiol. (Basel) 1962;25:575–585. [PubMed] [Google Scholar]
- Katsura I. Determination of bacteriophage lambda tail length by a protein ruler. Nature. 1987;327:73–75. doi: 10.1038/327073a0. [DOI] [PubMed] [Google Scholar]
- Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–698. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- Kemp CL, Howatson AF, Siminovitch L. Electron microscopy studies of mutants of lambda bacteriophage. I. General description and quantitation of viral products. Virology. 1968;36:490–502. doi: 10.1016/0042-6822(68)90174-8. [DOI] [PubMed] [Google Scholar]
- Kim B, Little JW. LexA and lambda Cl repressors as enzymes: specific cleavage in an intermolecular reaction. Cell. 1993;73:1165–1173. doi: 10.1016/0092-8674(93)90645-7. [DOI] [PubMed] [Google Scholar]
- King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- Kintz E, Davies M, Hammarlöf D, Canals R, Hinton J, van der Woude M. A BTP1 prophage gene present in invasive non-typhoidal Salmonella determines composition and length of the O-antigen of the LPS. Molec. Micro. 2015 doi: 10.1111/mmi.12933. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiler O, Koby S, Teff D, Court D, Oppenheim AB. The phage lambda CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14964–14969. doi: 10.1073/pnas.222172499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiler O, Oppenheim AB, Herman C. Recruitment of host ATP-dependent proteases by bacteriophage lambda. J. Struct. Biol. 2004;146:72–78. doi: 10.1016/j.jsb.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Kobiler O, Rokney A, Oppenheim AB. Phage lambda CIII: a protease inhibitor regulating the lysis-lysogeny decision. PLoS One. 2007;2:e363. doi: 10.1371/journal.pone.0000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan J, Murialdo H. Early intermediates in bacteriophage lambda prohead assembly. II. Identification of biologically active intermediates. Virology. 1983;131:100–115. doi: 10.1016/0042-6822(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Kolb KE, Hein PP, Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J. Biol. Chem. 2014;289:1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Velikodvorskaya T, Sen R, King RA, Banik-Maiti S, Weisberg RA. Inhibition of a transcriptional pause by RNA anchoring to RNA polymerase. Mol. Cell. 2008;31:683–694. doi: 10.1016/j.molcel.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. For the love of enzymes: the odyssey of a biochemist. Cambridge, MA: Harvard University Press; 1991. pp. 275–281. [Google Scholar]
- Krinke L, Wulff DL. RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA. Genes Dev. 1990;4:2223–2233. doi: 10.1101/gad.4.12a.2223. [DOI] [PubMed] [Google Scholar]
- Krinke L, Mahoney M, Wulff DL. The role of the OOP antisense RNA in coliphage lambda development. Molec. Microbiol. 1991;5:1265–1272. doi: 10.1111/j.1365-2958.1991.tb01900.x. [DOI] [PubMed] [Google Scholar]
- Kushner SR, Nagaishi H, Clark AJ. Isolation of exonuclease VIII: the enzyme associated with sbcA indirect suppressor. Proc. Natl. Acad. Sci. U. S. A. 1974;71:3593–3597. doi: 10.1073/pnas.71.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U S A. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankes HA, Zanghi CN, Santos K, Capella C, Duke CM, Dewhurst S. In vivo gene delivery and expression by bacteriophage lambda vectors. J. Appl. Microbiol. 2007;102:1337–1349. doi: 10.1111/j.1365-2672.2006.03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear J. Recombinant DNA: the untold story. New York, NY: Crown Publishers; 1978. [Google Scholar]
- Learn BA, Um SJ, Huang L, McMacken R. Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt JC, Gilcrease EB, Wilson K, Casjens SR. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology. 2013;440:117–133. doi: 10.1016/j.virol.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. Lysogenicity in E. coli K-12. Genetics. 1951;36:560. [Google Scholar]
- Lederberg EM, Lederberg J. Genetic Studies of Lysogenicity in Escherichia Coli. Genetics. 1953;38:51–64. doi: 10.1093/genetics/38.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers GG, Jr, Gottesman S. Lambda Xis degradation in vivo by Lon and FtsH. J. Bacteriol. 1998;180:1573–1577. doi: 10.1128/jb.180.6.1573-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire S, Figueroa-Bossi N, Bossi L. A singular case of prophage complementation in mutational activation of recET orthologs in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2008;190:6857–6866. doi: 10.1128/JB.00769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire S, Figueroa-Bossi N, Bossi L. Bacteriophage crosstalk: coordination of prophage induction by trans-acting antirepressors. PLoS Genet. 2011;7:e1002149. doi: 10.1371/journal.pgen.1002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng F, McMacken R. Potent stimulation of transcription-coupled DNA supercoiling by sequence-specific DNA-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9139–9144. doi: 10.1073/pnas.142002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin ME, Hendrix RW, Casjens SR. A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J. Mol. Biol. 1993;234:124–139. doi: 10.1006/jmbi.1993.1568. [DOI] [PubMed] [Google Scholar]
- Lewis D, Le P, Zurla C, Finzi L, Adhya S. Multilevel autoregulation of lambda repressor protein CI by DNA looping in vitro. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14807–14812. doi: 10.1073/pnas.1111221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Liao SM, Wu TH, Chiang CH, Susskind MM, McClure WR. Control of gene expression in bacteriophage P22 by a small antisense RNA. I. Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev. 1987;1:197–203. doi: 10.1101/gad.1.2.197. [DOI] [PubMed] [Google Scholar]
- Little JW. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Cole RA, Reeves PR. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J. Bacteriol. 1996;178:2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Sae-Ueng U, Li D, Lander GC, Zuo X, Jonsson B, Rau D, Shefer I, Evilevitch A. Solid-to-fluid-like DNA transition in viruses facilitates infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14675–14680. doi: 10.1073/pnas.1321637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang H, Gu Z, Roberts JW. High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11928–11933. doi: 10.1073/pnas.1309739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Friedman DI. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Molec. Microbiol. 2004;51:1691–1704. doi: 10.1111/j.1365-2958.2003.03934.x. [DOI] [PubMed] [Google Scholar]
- Lobban PE. Ph. D. Dissertation. Stanford University; 1972. An enzymatic method for the end-to-end joining of DNA molecules. [Google Scholar]
- Lobban PE, Kaiser AD. Enzymatic end-to end joining of DNA molecules. J. Mol. Biol. 1973;78:453–471. doi: 10.1016/0022-2836(73)90468-3. [DOI] [PubMed] [Google Scholar]
- Loenen WA, Murray NE. Modification enhancement by the restriction alleviation protein (Ral) of bacteriophage lambda. J. Mol. Biol. 1986;190:11–22. doi: 10.1016/0022-2836(86)90071-9. [DOI] [PubMed] [Google Scholar]
- Lucon J, Qazi S, Uchida M, Bedwell GJ, LaFrance B, Prevelige PE, Jr, Douglas T. Use of the interior cavity of the P22 capsid for site-specific initiation of atom-transfer radical polymerization with high-density cargo loading. Nat. Chem. 2012;4:781–788. doi: 10.1038/nchem.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lwoff A. Lysogeny. Bacteriol. Rev. 1953;17:269–337. doi: 10.1128/br.17.4.269-337.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajna J, Oppenheim AB, Rattray A, Gottesman M. Translation initiation of bacteriophage lambda gene cII requires integration host factor. J. Bacteriol. 1986;165:167–174. doi: 10.1128/jb.165.1.167-174.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel JV. SDS polyacrylamide gel electrophoresis. Trends. Biochem. Sci. 2000;25:590–592. doi: 10.1016/s0968-0004(00)01693-5. [DOI] [PubMed] [Google Scholar]
- Mardanov AV, Ravin NV. Functional characterization of the repA replication gene of linear plasmid prophage N15. Res. Microbiol. 2006;157:176–183. doi: 10.1016/j.resmic.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Mardanov AV, Ravin NV. The antirepressor needed for induction of linear plasmid-prophage N15 belongs to the SOS regulon. J. Bacteriol. 2007;189:6333–6338. doi: 10.1128/JB.00599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markine-Goriaynoff N, Gillet L, Van Etten JL, Korres H, Verma N, Vanderplasschen A. Glycosyltransferases encoded by viruses. J. Gen. Virol. 2004;85:2741–2754. doi: 10.1099/vir.0.80320-0. [DOI] [PubMed] [Google Scholar]
- Maruyama IN, Maruyama HI, Brenner S. Lambda foo: a lambda phage vector for the expression of foreign proteins. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8273–8277. doi: 10.1073/pnas.91.17.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Malay AD, Curtis FA, Sharples GJ, Heddle JG. Structural and functional characterization of the Red beta recombinase from bacteriophage lambda. PLoS One. 2013;8:e78869. doi: 10.1371/journal.pone.0078869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiacio J, Walter S, Brewer M, Domm W, Friedman AE, Dewhurst S. Dense display of HIV-1 envelope spikes on the lambda phage scaffold does not result in the generation of improved antibody responses to HIV-1 Env. Vaccine. 2011;29:2637–2647. doi: 10.1016/j.vaccine.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi MR. Degradation in vitro of bacteriophage lambda N protein by Lon protease from Escherichia coli. J. Biol. Chem. 1987;262:2696–2703. [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL, Reed P, Zhang RG, Beasley S, Walmsley AR, Curtis FA, Joachimiak A, Edwards AM, Sharples GJ. Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11260–11265. doi: 10.1073/pnas.0503399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElheny VK. Watson and DNA. Lexington, KY: Basic Books; 2014. [Google Scholar]
- McGary K, Nudler E. RNA polymerase and the ribosome: the close relationship. Curr. Opin. Microbiol. 2013;16:112–117. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Fontana J, Nemecek D, Cheng N, Aksyuk AA, Heymann JB, Winkler DC, Lam AS, Wall JS, Steven AC, Hoiczyk E. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 2014;33:1896–1911. doi: 10.15252/embj.201488566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JE, Davis RW. Cleavage of DNA by R1 restriction endonuclease generates cohesive ends. Proc. Natl. Acad. Sci. U. S. A. 1972;69:3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335:428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski CB, Little JW. Role of cis-acting sites in stimulation of the phage lambda P(RM) promoter by CI-mediated looping. J. Bacteriol. 2013;195:3401–3411. doi: 10.1128/JB.02148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa Y, Maruyama I, Brenner S. Surface display of proteins on bacteriophage lambda heads. J. Mol. Biol. 1996;262:21–30. doi: 10.1006/jmbi.1996.0495. [DOI] [PubMed] [Google Scholar]
- Mikita N, Cheng I, Fishovitz J, Huang J, Lee I. Processive degradation of unstructured protein by Escherichia coli Lon occurs via the slow, sequential delivery of multiple scissile sites followed by rapid and synchronized peptide bond cleavage events. Biochemistry. 2013;52:5629–5644. doi: 10.1021/bi4008319. [DOI] [PubMed] [Google Scholar]
- Mishra S, Mohan S, Godavarthi S, Sen R. The interaction surface of a bacterial transcription elongation factor required for complex formation with an antiterminator during transcription antitermination. J. Biol. Chem. 2013;288:28089–28103. doi: 10.1074/jbc.M113.472209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogridge J, Legault P, Li J, Van Oene MD, Kay LE, Greenblatt J. Independent ligand-induced folding of the RNA-binding domain and two functionally distinct antitermination regions in the phage lambda N protein. Mol. Cell. 1998;1:265–275. doi: 10.1016/s1097-2765(00)80027-1. [DOI] [PubMed] [Google Scholar]
- Molineux IJ. Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it? Virology. 2006;344:221–229. doi: 10.1016/j.virol.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Molineux IJ, Panja D. Popping the cork: mechanisms of phage genome ejection. Nat. Rev. Microbiol. 2013;11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- Mooers BH, Matthews BW. Extension to 2268 atoms of direct methods in the ab initio determination of the unknown structure of bacteriophage P22 lysozyme. Acta Crystallogr. D Biol. Crystallogr. 2006;62:165–176. doi: 10.1107/S0907444905037212. [DOI] [PubMed] [Google Scholar]
- Mount D, Harris A, Fuerst C, Siminovitch L. Mutation in bacteriophage lambda affecting particle morphogenesis. Virology. 1968;35:134–145. doi: 10.1016/0042-6822(68)90313-9. [DOI] [PubMed] [Google Scholar]
- Murialdo H, Becker A. A genetic analysis of bacteriophage lambda prohead assembly in vitro. J. Mol. Biol. 1978;125:57–74. doi: 10.1016/0022-2836(78)90254-1. [DOI] [PubMed] [Google Scholar]
- Murialdo H, Siminovitch L. The morphogenesis of phage lambda. V. Form-determining function of the genes required for the assembly of the head. Virology. 1972;48:824–835. doi: 10.1016/0042-6822(72)90163-8. [DOI] [PubMed] [Google Scholar]
- Mustard JA, Little JW. Analysis of Escherichia coli RecA interactions with LexA, lambda CI, UmuD by site-directed mutagenesis of recA. J. Bacteriol. 2000;182:1659–1670. doi: 10.1128/jb.182.6.1659-1670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muteeb G, Dey D, Mishra S, Sen R. A multipronged strategy of an antiterminator protein to overcome Rho-dependent transcription termination. Nucleic Acids Res. 2012;40:11213–11228. doi: 10.1093/nar/gks872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narajczyk M, Baranska S, Wegrzyn A, Wegrzyn G. Switch from theta to sigma replication of bacteriophage lambda DNA: factors involved in the process and a model for its regulation. Mol. Genet. Genomics. 2007;278:65–74. doi: 10.1007/s00438-007-0228-y. [DOI] [PubMed] [Google Scholar]
- Neely MN, Friedman DI. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene. 1998;223:105–113. doi: 10.1016/s0378-1119(98)00236-4. [DOI] [PubMed] [Google Scholar]
- Neidhardt F, editor. Escherichia coli and Salmonella cellular and molecular biology. Washington, D.C: ASM Press; 1996. [Google Scholar]
- Nicastro J, Sheldon K, El-Zarkout FA, Sokolenko S, Aucoin MG, Slavcev R. Construction and analysis of a genetically tuneable lytic phage display system. Appl. Microbiol. Biotechnol. 2013;97:7791–7804. doi: 10.1007/s00253-013-4898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil A, Prevelige PE, Basu G, Douglas T. Coconfinement of fluorescent proteins: spatially enforced communication of GFP and mCherry encapsulated within the P22 capsid. Biomacromolecules. 2012;13:3902–3907. doi: 10.1021/bm301347x. [DOI] [PubMed] [Google Scholar]
- O'Neil A, Reichhardt C, Johnson B, Prevelige PE, Douglas T. Genetically programmed in vivo packaging of protein cargo and its controlled release from bacteriophage P22. Angew Chem. Int. Ed. Engl. 2011;50:7425–7428. doi: 10.1002/anie.201102036. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Lehman IR. Linkage of polynucleotides through phosphodiester bonds by an enzyme from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1967;57:1426–1433. doi: 10.1073/pnas.57.5.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski P, Szambowska A, Baralska S, Narajczyk M, Wegrzyn G, Glinkowska M. A dual promoter system regulating lambda DNA replication initiation. Nucleic Acids Res. 2014;42:4450–4462. doi: 10.1093/nar/gku103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaga LA. Ray Wu as Fifth Business: Deconstructing collective memory in the history of DNA sequencing. Stud. Hist. Philos. Biol. Biomed. Sci. 2014;46:1–14. doi: 10.1016/j.shpsc.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Osipiuk J, Georgopoulos C, Zylicz M. Initiation of lambda DNA replication. The Escherichia coli small heat shock proteins, DnaJ and GrpE, increase DnaK's affinity for the lambda P protein. J. Biol. Chem. 1993;268:4821–4827. [PubMed] [Google Scholar]
- Pabo CO, Aggarwal AK, Jordan SR, Beamer LJ, Obeysekare UR, Harrison SC. Conserved residues make similar contacts in two repressor-operator complexes. Science. 1990;247:1210–1213. doi: 10.1126/science.2315694. [DOI] [PubMed] [Google Scholar]
- Padilla-Meier GP, Gilcrease EB, Weigele PR, Cortines JR, Siegel M, Leavitt JC, Teschke CM, Casjens SR. Unraveling the role of the C-terminal helix turn helix of the coat-binding domain of bacteriophage P22 scaffolding protein. J. Biol. Chem. 2012;287:33766–33780. doi: 10.1074/jbc.M112.393132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang T, Fleming TC, Pogliano K, Young R. Visualization of pinholin lesions in vivo. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2054–E2063. doi: 10.1073/pnas.1222283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang T, Savva CG, Fleming KG, Struck DK, Young R. Structure of the lethal phage pinhole. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18966–18971. doi: 10.1073/pnas.0907941106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Molineux IJ. Dynamics of bacteriophage genome ejection in vitro and in vivo. Phys. Biol. 2010;7:045006. doi: 10.1088/1478-3975/7/4/045006. [DOI] [PubMed] [Google Scholar]
- Pappenheimer AM, Jr, Murphy JR. Studies on the molecular epidemiology of diphtheria. Lancet. 1983;2:923–926. doi: 10.1016/s0140-6736(83)90449-x. [DOI] [PubMed] [Google Scholar]
- Parent KN, Gilcrease EB, Casjens SR, Baker TS. Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology. 2012a;427:177–188. doi: 10.1016/j.virol.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Deedas CT, Egelman EH, Casjens SR, Baker TS, Teschke CM. Stepwise molecular display utilizing icosahedral and helical complexes of phage coat and decoration proteins in the development of robust nanoscale display vehicles. Biomaterials. 2012b;33:5628–5637. doi: 10.1016/j.biomaterials.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Tang J, Cardone G, Gilcrease EB, Janssen ME, Olson NH, Casjens SR, Baker TS. Three-dimensional reconstructions of the bacteriophage CUS-3 virion reveal a conserved co. Virology. 2014a:464–465. 55–66. doi: 10.1016/j.virol.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Erb ML, Cardone G, Nguyen K, Gilcrease EB, Porcek NB, Pogliano J, Baker TS, Casjens SR. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Molec. Microbiol. 2014b;92:47–60. doi: 10.1111/mmi.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Genetics of the left arm of the chromosome of bacteriophage lambda. Genetics. 1968;59:311–325. doi: 10.1093/genetics/59.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AR, Court C, Lubkowska L, Jin DJ, Kashlev M, Court DL. Bacteriophage lambda N protein inhibits transcription slippage by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014;42:5823–5829. doi: 10.1093/nar/gku203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parma D, Snyder M, Sobolevski S, Nawroz M, Brody E, Gold L. The Rex system of bacteriophage lambda: tolerance and altruistic cell death. Genes Dev. 1992;6:497–510. doi: 10.1101/gad.6.3.497. [DOI] [PubMed] [Google Scholar]
- Parua PK, Mondal A, Parrack P. HflD, an Escherichia coli protein involved in the lambda lysis-lysogeny switch, impairs transcription activation by lambda CII. Arch. Biochem. Biophys. 2010;493:175–183. doi: 10.1016/j.abb.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Patterson DP, Prevelige PE, Douglas T. Nanoreactors by programmed enzyme encapsulation inside the capsid of the bacteriophage P22. ACS Nano. 2012;6:5000–5009. doi: 10.1021/nn300545z. [DOI] [PubMed] [Google Scholar]
- Patterson DP, Schwarz B, Waters RS, Gedeon T, Douglas T. Encapsulation of an enzyme cascade within the bacteriophage P22 virus-like particle. ACS Chem. Biol. 2014;9:359–365. doi: 10.1021/cb4006529. [DOI] [PubMed] [Google Scholar]
- Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U S A. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez GL, Huynh B, Slater M, Maloy S. Transport of phage P22 DNA across the cytoplasmic membrane. J. Bacteriol. 2009;191:135–140. doi: 10.1128/JB.00778-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietila MK, Laurinmaki P, Russell DA, Ko CC, Jacobs-Sera D, Hendrix RW, Bamford DH, Butcher SJ. Structure of the archaeal headtailed virus HSTV-1 completes the HK97 fold story. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10604–10609. doi: 10.1073/pnas.1303047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett G, 3rd, Rose DJ, Durfee TJ, Blattner FR. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 1999;181:1767–1778. doi: 10.1128/jb.181.6.1767-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa MP, McKelvey TA, Hempel J, Hendrix RW. Bacteriophage HK97 structure: wholesale covalent cross-linking between the major head shell subunits. J. Virol. 1991;65:3227–3237. doi: 10.1128/jvi.65.6.3227-3237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR. Involvement of DNA replication proteins in phage lambda Red-mediated homologous recombination. PLoS One. 2013;8:e67440. doi: 10.1371/journal.pone.0067440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC. DNA-binding properties of the Erf protein of bacteriophage P22. J. Mol. Biol. 1983;163:257–275. doi: 10.1016/0022-2836(83)90006-2. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC, Murphy KC. Modulation of Escherichia coli RecBCD activity by the bacteriophage lambda Gam and P22 Abc functions. J. Bacteriol. 1988;170:2012–2021. doi: 10.1128/jb.170.5.2012-2021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC, Semerjian AV. Bacteriophage P22 accessory recombination function. Virology. 1991;182:316–323. doi: 10.1016/0042-6822(91)90675-2. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC, Wang HR. Recombination-promoting activity of the bacteriophage lambda Rap protein in Escherichia coli K-12. J. Bacteriol. 2002;184:4626–4629. doi: 10.1128/JB.184.16.4626-4629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch S, Jurk M, Washburn RS, Gottesman ME, Wohrl BM, Rosch P. RNA-binding specificity of E. coli NusA. Nucleic Acids Res. 2009;37:4736–4742. doi: 10.1093/nar/gkp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell HH, Harvey AM. P22 antirepressor protein prevents in vivo recA-dependent proteolysis of P22 repressor. Mol. Gen. Genet. 1983;190:427–431. doi: 10.1007/BF00331072. [DOI] [PubMed] [Google Scholar]
- Priest DG, Cui L, Kumar S, Dunlap DD, Dodd IB, Shearwin KE. Quantitation of the DNA tethering effect in long-range DNA looping in vivo and in vitro using the Lac and lambda repressors. Proc. Natl. Acad. Sci. U. S. A. 2014;111:349–354. doi: 10.1073/pnas.1317817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Isolation of the lambda phage repressor. Proc. Natl. Acad. Sci. U. S. A. 1967;57:306–313. doi: 10.1073/pnas.57.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Repressor and its action. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 221–237. [Google Scholar]
- Ptashne M. A genetic switch - phage λ and higher organisms. 2nd ed. Cambridge, MA: Cell Press and Blackwell Scientific Publications; 1992. [Google Scholar]
- Qazi S, Liepold LO, Abedin J, Johnson B, Prevelige PE, Frank JA, Douglas T. P22 viral capsids as nanocomposite high-relaxivity MRI contrast agents. Mol. Pharm. 2013;4:781–784. doi: 10.1021/mp300208g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi S, Uchida M, Usselman R, Shearer R, Edwards E, Douglas T. Manganese(III) porphyrins complexed with P22 virus-like particles as T1-enhanced contrast agents for magnetic resonance imaging. J. Biol. Inorg. Chem. 2014;19:237–246. doi: 10.1007/s00775-013-1075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Rau DC, Parsegian VA, Fang LT, Knobler CM, Gelbart WM. Salt-dependent DNA-DNA spacings in intact bacteriophage lambda reflect relative importance of DNA self-repulsion and bending energies. Phys Rev Lett. 2011;106:028102. doi: 10.1103/PhysRevLett.106.028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding CM. Nuclease activity in defective lysogens of phage lambda. Biochem. Biophys. Res. Commun. 1964;15:8–12. doi: 10.1016/0006-291x(64)90093-2. [DOI] [PubMed] [Google Scholar]
- Ranade K, Poteete AR. Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. J. Bacteriol. 1993;175:4712–4718. doi: 10.1128/jb.175.15.4712-4718.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall-Hazelbauer L, Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J. Bacteriol. 1973;116:1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RN, Raj CV. Salmonella typhimurium mutants affecting establishment of lysogeny. Mol. Gen. Genet. 1973;125:119–123. doi: 10.1007/BF00268864. [DOI] [PubMed] [Google Scholar]
- Ravin NV. Mechanisms of replication and telomere resolution of the linear plasmid prophage N15. FEMS Microbiol. Lett. 2003;221:1–6. doi: 10.1016/S0378-1097(03)00125-3. [DOI] [PubMed] [Google Scholar]
- Ravin V, Shulga MG. The evidence of extrachromosomal location phage prophage N15. Virology. 1970;40:800–805. doi: 10.1016/0042-6822(70)90125-x. [DOI] [PubMed] [Google Scholar]
- Ravin NV, Svarchevsky A, Deho G. The anti-immunity system of phage-plasmid N15: identification of the antirepressor gene and its control by a small processed RNA. Molec. Microbiol. 1999;34:980–984. doi: 10.1046/j.1365-2958.1999.01658.x. [DOI] [PubMed] [Google Scholar]
- Ravin NV, Rech J, Lane D. Mapping of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J. Mol. Biol. 2003;329:875–889. doi: 10.1016/s0022-2836(03)00525-4. [DOI] [PubMed] [Google Scholar]
- Ravin V, Ravin N, Casjens S, Ford ME, Hatfull GF, Hendrix RW. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 2000;299:53–73. doi: 10.1006/jmbi.2000.3731. [DOI] [PubMed] [Google Scholar]
- Reichardt L, Kaiser AD. Control of lambda repressor synthesis. Proc. Natl. Acad. Sci. U. S. A. 1971;68:2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo AA, Suhanovsky MM, Baker ML, Fraser LC, Jones LM, Rempel DL, Gross ML, Chiu W, Alexandrescu AT, Teschke CM. Multiple functional roles of the accessory I-domain of bacteriophage P22 coat protein revealed by NMR structure and CryoEM modeling. Structure. 2014;22:830–841. doi: 10.1016/j.str.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb. Symp. Quant. Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Roessner CA, Ihler GM. Sequence of amino acids in lamB responsible for spontaneous ejection of bacteriophage lambda DNA. J. Mol. Biol. 1987;195:963–966. doi: 10.1016/0022-2836(87)90502-x. [DOI] [PubMed] [Google Scholar]
- Roessner CA, Struck DK, Ihler GM. Injection of DNA into liposomes by bacteriophage lambda. J. Biol. Chem. 1983;258:643–648. [PubMed] [Google Scholar]
- Rybchin VN, Svarchevsky AN. The plasmid prophage N15: a linear DNA with covalently closed ends. Molec. Microbiol. 1999;33:895–903. doi: 10.1046/j.1365-2958.1999.01533.x. [DOI] [PubMed] [Google Scholar]
- Saeedi A, Ghaemi A, Tabarraei A, Moradi A, Gorji A, Semnani S, Soleimanjahi H, Adli AH, Hosseini SY, Vakili MA. Enhanced cell immune responses to hepatitis C virus core by novel heterologous DNA prime/lambda nanoparticles boost in mice. Virus Genes. 2014;49:11–21. doi: 10.1007/s11262-014-1070-z. [DOI] [PubMed] [Google Scholar]
- Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J. Mol. Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Sanyal SJ, Yang TC, Catalano CE. Integration host factor assembly at the cohesive end site of the bacteriophage lambda genome: Implications for viral DNA packaging and bacterial gene regulation. Biochemistry. 2014 doi: 10.1021/bi501025s. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva CG, Dewey JS, Deaton J, White RL, Struck DK, Holzenburg A, Young R. The holin of bacteriophage lambda forms rings with large diameter. Molec. Microbiol. 2008;69:784–793. doi: 10.1111/j.1365-2958.2008.06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandella D, Arber W. Phage lambda DNA injection into Escherichia coli pel− mutants is restored by mutations in phage genes V or H. Virology. 1976;69:206–215. doi: 10.1016/0042-6822(76)90207-5. [DOI] [PubMed] [Google Scholar]
- Schaefer KL, McClure WR. Antisense RNA control of gene expression in bacteriophage P22. II. Kinetic mechanism and cation specificity of the pairing reaction. RNA. 1997;3:157–174. [PMC free article] [PubMed] [Google Scholar]
- Schindler D, Echols H. Retroregulation of the int gene of bacteriophage lambda: control of translation completion. Proc. Natl. Acad. Sci. U. S. A. 1981;78:4475–4479. doi: 10.1073/pnas.78.7.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Schweimer K, Prasch S, Sujatha PS, Bubunenko M, Gottesman ME, Rosch P. NusA interaction with the alpha subunit of E. coli RNA polymerase is via the UP element site and releases autoinhibition. Structure. 2011;19:945–954. doi: 10.1016/j.str.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah NE, Warren D, Tong W, Laxmikanthan G, Van Duyne GD, Landy A. Nucleoprotein architectures regulating the directionality of viral integration and excision. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12372–12377. doi: 10.1073/pnas.1413019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergueev K, Yu D, Austin S, Court D. Cell toxicity caused by products of the p(L) operon of bacteriophage lambda. Gene. 2001;272:227–235. doi: 10.1016/s0378-1119(01)00535-2. [DOI] [PubMed] [Google Scholar]
- Servid A, Jordan P, O'Neil A, Prevelige P, Douglas T. Location of the bacteriophage P22 coat protein C-terminus provides opportunities for the design of capsid-based materials. Biomacromolecules. 2013;14:2989–2995. doi: 10.1021/bm400796c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar S, Hatoum A, Roberts JW. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mo. Cell. 2007;27:914–927. doi: 10.1016/j.molcel.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Chan SN, Mahdi AA, Whitby MC, Lloyd RG. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Corbett LM, Graham IR. lambda Rap protein is a structure-specific endonuclease involved in phage recombination. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13507–13512. doi: 10.1073/pnas.95.23.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Curtis FA, McGlynn P, Bolt EL. Holliday junction binding and resolution by the Rap structure-specific endonuclease of phage lambda. J. Mol. Biol. 2004;340:739–751. doi: 10.1016/j.jmb.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Shinder G, Gold M. The Nul subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J. Virol. 1988;62:387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkilnyj P, Colon MP, Koudelka GB. Bacteriophage 434 Hex protein prevents RecA-mediated repressor autocleavage. Viruses. 2013;5:111–126. doi: 10.3390/v5010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotland Y, Shifrin A, Ziv T, Teff D, Koby S, Kobiler O, Oppenheim AB. Proteolysis of bacteriophage lambda CII by Escherichia coli FtsH (HflB) J. Bacteriol. 2000;182:3111–3116. doi: 10.1128/jb.182.11.3111-3116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer ER, Weil J. Recombination in bacteriophage lambda. I. Mutants deficient in general recombination. J. Mol. Biol. 1968;34:261–271. doi: 10.1016/0022-2836(68)90251-9. [DOI] [PubMed] [Google Scholar]
- Simon M, Davis RW, Davidson N. Heteroduplexs of DNA moleculaes of lambdoid phages: physical mapping of their base sequence relationships by electron microscopy. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 313–328. [Google Scholar]
- Singh P, Nakatani E, Goodlett DR, Catalano CE. A pseudo-atomic model for the capsid shell of bacteriophage lambda using chemical cross-linking/ mass spectrometry and molecular modeling. J. Mol. Biol. 2013;425:3378–3388. doi: 10.1016/j.jmb.2013.06.026. [DOI] [PubMed] [Google Scholar]
- Sippy J, Patel P, Vahanian N, Sippy R, Feiss M. Genetics of critical contacts and clashes in the DNA packaging specificities of bacteriophages λ and 21. Virology. 2014;467C:115–123. doi: 10.1016/j.virol.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A, Poonian M, Bartl P. Concatemers in DNA replication: electron microscopic studies of partially denatured intracellular lambda DNA. J. Mol. Biol. 1972;64:541–550. doi: 10.1016/0022-2836(72)90081-2. [DOI] [PubMed] [Google Scholar]
- Slominska M, Neubauer P, Wegrzyn G. Regulation of bacteriophage lambda development by guanosine 5'-diphosphate-3'-diphosphate. Virology. 1999;262:431–441. doi: 10.1006/viro.1999.9907. [DOI] [PubMed] [Google Scholar]
- Smith DL, Rooks DJ, Fogg PC, Darby AC, Thomson NR, McCarthy AJ, Allison HE. Comparative genomics of Shiga toxin encoding bacteriophages. BMC Genomics. 2012;13:311. doi: 10.1186/1471-2164-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR, Kunes SM, Schultz DW, Taylor A, Triman KL. Structure of chi hotspots of generalized recombination. Cell. 1981a;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- Smith GR, Comb M, Schultz DW, Daniels DL, Blattner FR. Nucleotide sequence of the chi recombinational hot spot chi+D in bacteriophage lambda. J. Virol. 1981b;37:336–342. doi: 10.1128/jvi.37.1.336-342.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman WG, Reichardt LF, Yaniv M, Heinemann SF, Kaiser AD, Eisen H. Bidirectional transcription and the regulation of phage lambda repressor synthesis. Proc. Natl. Acad. Sci. U. S. A. 1972;69:3156–3160. doi: 10.1073/pnas.69.11.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno JR, Altieri AS, Bubunenko M, Tarasov SG, Li J, Court DL, Byrd RA, Ji X. Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination. Nucleic Acids Res. 2011;39:7803–7815. doi: 10.1093/nar/gkr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW. Chi: a little sequence controls a big enzyme. Genetics. 2005;170:487–493. doi: 10.1093/genetics/170.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. Properties of a mutant of Escherichia coli defective in bacteriophage lambda head formation (groE). II. The propagation of phage lambda. J. Mol. Biol. 1973;76:25–44. doi: 10.1016/0022-2836(73)90079-x. [DOI] [PubMed] [Google Scholar]
- Sternberg N, Hoess RH. Display of peptides and proteins on the surface of bacteriophage lambda. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1609–1613. doi: 10.1073/pnas.92.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Maizel JV. T7-directed protein synthesis. Virology. 1969;39:575–586. doi: 10.1016/0042-6822(69)90105-6. [DOI] [PubMed] [Google Scholar]
- Susskind MM, Botstein D. Mechanism of action of Salmonella phage P22 antirepressor. J. Mol. Biol. 1975;98:413–424. doi: 10.1016/s0022-2836(75)80127-6. [DOI] [PubMed] [Google Scholar]
- Susskind MM, Botstein D, Wright A. Superinfection exclusion by P22 prophage in lysogens of Salmonella typhimurium. III. Failure of superinfecting phage DNA to enter sieA+ lysogens. Virology. 1974;62:350–366. doi: 10.1016/0042-6822(74)90398-5. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Tal A, Arbel-Goren R, Costantino N, Court DL, Stavans J. Location of the unique integration site on an Escherichia coli chromosome by bacteriophage lambda DNA in vivo. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7308–7312. doi: 10.1073/pnas.1324066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W, Pell LG, Bona D, Tsai A, Dai XX, Edwards AM, Hendrix RW, Maxwell KL, Davidson AR. Tail tip proteins related to bacteriophage lambda gpL coordinate an iron-sulfur cluster. J. Mol. Biol. 2013;425:2450–2462. doi: 10.1016/j.jmb.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Lander G, Olia A, Li R, Casjens S, Prevelige P, Jr, Cingolani G, Baker T, Johnson J. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in P22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski TA, Mooney D, Thomason LC, Stahl FW. Gene products encoded in the ninR region of phage lambda participate in Red-mediated recombination. Genes Cells. 2002;7:351–363. doi: 10.1046/j.1365-2443.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- Thanweer F, Tahiliani V, Korres H, Verma NK. Topology and identification of critical residues of the O-acetyltransferase of serotype-converting bacteriophage, SF6, of Shigella flexneri. Biochem. Biophys. Res. Commun. 2008;375:581–585. doi: 10.1016/j.bbrc.2008.08.069. [DOI] [PubMed] [Google Scholar]
- Thibault G, Houry WA. Role of the N-terminal domain of the chaperone ClpX in the recognition and degradation of lambda phage protein O. J. Phys. Chem. B. 2012;116:6717–6724. doi: 10.1021/jp212024b. [DOI] [PubMed] [Google Scholar]
- Thirion JP, Hofnung M. On some genetic aspects of phage lambda resistance in E. coli K12. Genetics. 1972;71:207–216. doi: 10.1093/genetics/71.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO. Chemical linkage of the tail to the right-hand end of bacteriophage lambda DNA. J. Mol. Biol. 1974;87:1–9. doi: 10.1016/0022-2836(74)90555-5. [DOI] [PubMed] [Google Scholar]
- Thomas R. Cpontrol circuits. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1971. pp. 211–220. [Google Scholar]
- Thomas R, Lambert L. On the occurrence of bacterial mutations permitting lysogenization by clear variants of temperate bacteriophages. J. Mol. Biol. 1962;5:373–374. doi: 10.1016/s0022-2836(62)80079-5. [DOI] [PubMed] [Google Scholar]
- Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 2014;106:1 16 1–1 16 39. doi: 10.1002/0471142727.mb0116s106. [DOI] [PubMed] [Google Scholar]
- Tilly K, Murialdo H, Georgopoulos C. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc. Natl. Acad. Sci. U S A. 1981;78:1629–1633. doi: 10.1073/pnas.78.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, VanBogelen RA, Georgopoulos C, Neidhardt FC. Identification of the heat-inducible protein C15.4 as the groES gene product in Escherichia coli. J. Bacteriol. 1983;154:1505–1507. doi: 10.1128/jb.154.3.1505-1507.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KH, Young R. Probing the structure of the S105 hole. J. Bacteriol. 2014;196:3683–3689. doi: 10.1128/JB.01673-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Warren D, Seah NE, Laxmikanthan G, Van Duyne GD, Landy A. Mapping the lambda Integrase bridges in the nucleoprotein Holliday junction intermediates of viral integrative and excisive recombination. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12366–12371. doi: 10.1073/pnas.1413007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toothman P, Herskowitz I. Rex-dependent exclusion of lambdoid phages. II. Determinants of sensitivity to exclusion. Virology. 1980a;102:147–160. doi: 10.1016/0042-6822(80)90077-x. [DOI] [PubMed] [Google Scholar]
- Toothman P, Herskowitz I. Rex-dependent exclusion of lambdoid phages. III. Physiology of the abortive infection. Virology. 1980b;102:161–171. doi: 10.1016/0042-6822(80)90078-1. [DOI] [PubMed] [Google Scholar]
- Tsay JM, Sippy J, Feiss M, Smith DE. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14355–14360. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay JM, Sippy J, DelToro D, Andrews BT, Draper B, Rao V, Catalano CE, Feiss M, Smith DE. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J. Biol. Chem. 2010;285:24282–24289. doi: 10.1074/jbc.M110.129395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui LC, Hendrix RW. Proteolytic processing of phage lambda tail protein gpH: timing of the cleavage. Virology. 1983;125:257–264. doi: 10.1016/0042-6822(83)90199-x. [DOI] [PubMed] [Google Scholar]
- Tye BK, Huberman JA, Botstein D. Non-random circular permutation of phage P22 DNA. J. Mol. Biol. 1974;85:501–528. doi: 10.1016/0022-2836(74)90312-x. [DOI] [PubMed] [Google Scholar]
- Tyler JS, Mills MJ, Friedman DI. The operator and early promoter region of the Shiga toxin type 2-encoding bacteriophage 933W and control of toxin expression. J. Bacteriol. 2004;186:7670–7679. doi: 10.1128/JB.186.22.7670-7679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uc-Mass A, Loeza EJ, de la Garza M, Guarneros G, Hernandez-Sanchez J, Kameyama L. An orthologue of the cor gene is involved in the exclusion of temperate lambdoid phages. Evidence that Cor inactivates FhuA receptor functions. Virology. 2004;329:425–433. doi: 10.1016/j.virol.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Uchida M, Morris DS, Kang S, Jolley CC, Lucon J, Liepold LO, LaFrance B, Prevelige PE, Jr, Douglas T. Site-directed coordination chemistry with P22 virus-like particles. Langmuir. 2012;28:1998–2006. doi: 10.1021/la203866c. [DOI] [PubMed] [Google Scholar]
- Uchida T, Gill DM, Pappenheimer AM., Jr Mutation in the structural gene for diphtheria toxin carried by temperate phage. Nat. New Biol. 1971;233:8–11. doi: 10.1038/newbio233008a0. [DOI] [PubMed] [Google Scholar]
- Vaca Pacheco S, Garcia Gonzalez O, Paniagua Contreras GL. The lom gene of bacteriophage lambda is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol. Lett. 1997;156:129–132. doi: 10.1111/j.1574-6968.1997.tb12717.x. [DOI] [PubMed] [Google Scholar]
- van der Wilk F, Dullemans AM, Verbeek M, van den Heuvel JF. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology. 1999;262:104–113. doi: 10.1006/viro.1999.9902. [DOI] [PubMed] [Google Scholar]
- Van Gilst MR, von Hippel PH. Assembly of the N-dependent antitermination complex of phage λ: NusA and RNA bind independently to different unfolded domains of the N protein. J. Mol. Biol. 1997;274:160–173. doi: 10.1006/jmbi.1997.1389. [DOI] [PubMed] [Google Scholar]
- Van Valen D, Wu D, Chen YJ, Tuson H, Wiggins P, Phillips R. A single-molecule Hershey-Chase experiment. Curr. Biol. 2012;22:1339–1343. doi: 10.1016/j.cub.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma NK, Brandt JM, Verma DJ, Lindberg AA. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage Sf6 that adds group antigen 6 to Shigella flexneri. Molec. Microbiol. 1991;5:71–75. doi: 10.1111/j.1365-2958.1991.tb01827.x. [DOI] [PubMed] [Google Scholar]
- Villafane R, Zayas M, Gilcrease EB, Kropinski AM, Casjens SR. Genomic analysis of bacteriophage epsilon 34 of Salmonella enterica serovar Anatum (15+) BMC Microbiol. 2008;8:227. doi: 10.1186/1471-2180-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello CL, Kireeva ML, Lubkowska L, Kashlev M, Gottesman M. Coliphage HK022 Nun protein inhibits RNA polymerase translocation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2368–E2375. doi: 10.1073/pnas.1319740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegtli WC, White DJ, Reiter NJ, Rusnak F, Rosenzweig AC. Structure of the bacteriophage lambda Ser/Thr protein phosphatase with sulfate ion bound in two coordination modes. Biochemistry. 2000;39:15365–15374. doi: 10.1021/bi0021030. [DOI] [PubMed] [Google Scholar]
- Vorobiev SM, Gensler Y, Vahedian-Movahed H, Seetharaman J, Su M, Huang JY, Xiao R, Kornhaber G, Montelione GT, Tong L, Ebright RH, Nickels BE. Structure of the DNA-binding and RNA-polymerase-binding region of transcription antitermination factor lambdaQ. Structure. 2014;22:488–495. doi: 10.1016/j.str.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrov A, Vostrukhina O, Svarchevsky A, Rybchin V. Proteins responsible for lysogenic conversion caused by coliphages N15 and ø80 are highly homologous. J. Bacteriol. 1996;178:1484–1486. doi: 10.1128/jb.178.5.1484-1486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner PL, Livny J, Neely MN, Acheson DW, Friedman DI, Waldor MK. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Molec. Microbiol. 2002;44:957–970. doi: 10.1046/j.1365-2958.2002.02950.x. [DOI] [PubMed] [Google Scholar]
- Wake RG, Kaiser AD, Inman RB. Isolation and structure of phage lambda head-mutant DNA. J. Mol. Biol. 1972;64:519–540. doi: 10.1016/0022-2836(72)90080-0. [DOI] [PubMed] [Google Scholar]
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000a;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- Wang J, Hofnung M, Charbit A. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 2000b;182:508–512. doi: 10.1128/jb.182.2.508-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Kaiser AD. Evidence that the cohesive ends of mature lambda DNA are generated by the gene A product. Nat. New Biol. 1973;241:16–17. doi: 10.1038/newbio241016a0. [DOI] [PubMed] [Google Scholar]
- Watkins D, Hsiao C, Woods KK, Koudelka GB, Williams LD. P22 c2 repressor-operator complex: mechanisms of direct and indirect readout. Biochemistry. 2008;47:2325–2338. doi: 10.1021/bi701826f. [DOI] [PubMed] [Google Scholar]
- Weaver LH, Rennell D, Poteete AR, Mathews BW. Structure of phage P22 gene 19 lysozyme inferred from its homology with phage T4 lysozyme. Implications for lysozyme evolution. J. Mol. Biol. 1985;184:739–741. doi: 10.1016/0022-2836(85)90318-3. [DOI] [PubMed] [Google Scholar]
- Wegrzyn G, Licznerska K, Wegrzyn A. Phage lambda-new insights into regulatory circuits. Adv. Virus Res. 2012;82:155–178. doi: 10.1016/B978-0-12-394621-8.00016-9. [DOI] [PubMed] [Google Scholar]
- Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. Molecular genetics of bacteriophage P22 scaffolding protein's functional domains. J. Mol. Biol. 2005;348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Weigle J. Induction of Mutations in a Bacterial Virus. Proc. Natl. Acad. Sci. U. S. A. 1953;39:628–636. doi: 10.1073/pnas.39.7.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle J. Assembly of phage lambda in vitro. Proc. Natl. Acad. Sci. U. S. A. 1966;55:1462–1466. doi: 10.1073/pnas.55.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle J. Studies on head-tail union in bacteriophage lambda. J. Mol. Biol. 1968;33:483–489. doi: 10.1016/0022-2836(68)90204-0. [DOI] [PubMed] [Google Scholar]
- Weisberg R, Landy A. Site-specific recombination. In: Hendrix R, Roberts J, Stahl FW, Weisberg R, editors. Lambda II. Cold Spring Harbor NY: Cold Spring Harbor Laboratory; 1983. pp. 211–250. [Google Scholar]
- Weisberg RA, Gottesmann ME, Hendrix RW, Little JW. Family values in the age of genomics: comparative analyses of temperate bacteriophage HK022. Annu. Rev. Genet. 1999;33:565–602. doi: 10.1146/annurev.genet.33.1.565. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. Oligonucleotide synthesis byEscherichia coli dnaG primase in conjunction with phage P22 gene 12 protein. J. Biol. Chem. 1984a;259:14044–14047. [PubMed] [Google Scholar]
- Wickner S. DNA-dependent ATPase activity associated with phage P22 gene 12 protein. J. Biol. Chem. 1984b;259:14038–14043. [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wilson HR, Yu D, Peters HK, 3rd, Zhou JG, Court DL. The global regulator RNase III modulates translation repression by the transcription elongation factor. N. Embo J. 2002;21:4154–4161. doi: 10.1093/emboj/cdf395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman E. Sur le déterminisme génétique de la lysogénie. Ann. Inst. Pasteur. 1953;84:281. [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Kanegasaki S. Molecular aspects of lipopolysaccharides. Physiol. Rev. 1971;51:748–784. doi: 10.1152/physrev.1971.51.4.748. [DOI] [PubMed] [Google Scholar]
- Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Molec. Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- Wu R, Kaiser AD. Structure and base sequence in the cohesive ends of phage lambda. J. Mol. Biol. 1968;57:491–499. doi: 10.1016/s0022-2836(68)80012-9. [DOI] [PubMed] [Google Scholar]
- Wu TH, Liao SM, McClure WR, Susskind MM. Control of gene expression in bacteriophage P22 by a small antisense RNA. II. Characterization of mutants defective in repression. Genes Dev. 1987;1:204–212. doi: 10.1101/gad.1.2.204. [DOI] [PubMed] [Google Scholar]
- Wyman C, Vasilikiotis C, Ang D, Georgopoulos C, Echols H. Function of the GrpE heat shock protein in bidirectional unwinding and replication from the origin of phage lambda. J. Biol. Chem. 1993;268:25192–25196. [PubMed] [Google Scholar]
- Xin W, Cai ZH, Feiss M. Function of IHF in lambda DNA packaging. II. Effects of mutations altering the IHF binding site and the intrinsic bend in cosB on lambda development. J. Mol. Biol. 1993;230:505–515. doi: 10.1006/jmbi.1993.1167. [DOI] [PubMed] [Google Scholar]
- Xu J, Hendrix RW, Duda RL. A conserved frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell. 2004a;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Xu J, Hendrix RW, Duda RL. A balanced ratio of proteins from gene G and frameshift-extended gene GT is required for phage lambda tail assembly. J. Mol. Biol. 2013;425:3476–3487. doi: 10.1016/j.jmb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hendrix RW, Duda RL. Chaperone-protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length. J. Mol. Biol. 2014;426:1004–1018. doi: 10.1016/j.jmb.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Struck DK, Deaton J, Wang IN, Young R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. U S A. 2004b;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Babitzke P. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr. Opin. Microbio. 2014;18:68–71. doi: 10.1016/j.mib.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lewis PJ. The interaction between RNA polymerase and the elongation factor NusA. RNA Biol. 2010;7:272–275. doi: 10.4161/rna.7.3.12021. [DOI] [PubMed] [Google Scholar]
- Yang X, Molimau S, Doherty GP, Johnston EB, Marles-Wright J, Rothnagel R, Hankamer B, Lewis RJ, Lewis PJ. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 2009;10:997–1002. doi: 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D. Cancer, viruses, and mass migration: Paul Berg's venture into eukaryotic biology and the advent of recombinant DNA research and technology, 1967–1980. J. Hist. Biols. 2008;41:589–636. doi: 10.1007/s10739-008-9149-9. [DOI] [PubMed] [Google Scholar]
- Young R. Phage lysis: three steps, three choices, one outcome. J. Microbiol. 2014;52:243–258. doi: 10.1007/s12275-014-4087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghi CN, Sapinoro R, Bradel-Tretheway B, Dewhurst S. A tractable method for simultaneous modifications to the head and tail of bacteriophage lambda and its application to enhancing phage-mediated gene delivery. Nucleic Acids Res. 2007;35:e59. doi: 10.1093/nar/gkm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, McCabe KA, Bell CE. Crystal structures of lambda exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11872–11877. doi: 10.1073/pnas.1103467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xing X, Herr AB, Bell CE. Crystal structure ofE. coliRecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks. Structure. 2009;17:690–702. doi: 10.1016/j.str.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ha KS, La Porta A, Landick R, Block SM. Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA. Mol. Cell. 2011;44:635–646. doi: 10.1016/j.molcel.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N, Lederberg J. Genetic exchange in Salmonella. J. Bacteriol. 1952;64:679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zissler J. Integration negative (int) mutations of phage lambda. 1967, Virology. 1967;31:189. doi: 10.1016/0042-6822(67)90030-x. [DOI] [PubMed] [Google Scholar]
- Zissler J, Signer E, Sachaefer F. The role of recombination in growth of bacteriophage lambda. In: Hershey AD, editor. Bacteriophage Lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories; 1971. pp. 455–475. [Google Scholar]
- Zylicz M, Liberek K, Wawrzynow A, Georgopoulos C. Formation of the preprimosome protects lambda O from RNA transcription-dependent proteolysis by ClpP/ClpX. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15259–15363. doi: 10.1073/pnas.95.26.15259. [DOI] [PMC free article] [PubMed] [Google Scholar]