Abstract
The paramyxovirus family has a genome consisting of a single strand of negative sense RNA. This genome acts as a template for two distinct processes: transcription to generate subgenomic, capped and polyadenylated mRNAs, and genome replication. These viruses only encode one polymerase. Thus, an intriguing question is, how does the viral polymerase initiate and become committed to either transcription or replication? By answering this we can begin to understand how these two processes are regulated. In this review article, we present recent findings from studies on the paramyxovirus, respiratory syncytial virus, which show how its polymerase is able to initiate transcription and replication from a single promoter. We discuss how these findings apply to other paramyxoviruses. Then, we examine how trans-acting proteins and promoter secondary structure might serve to regulate transcription and replication during different phases of the paramyxovirus replication cycle.
INTRODUCTION
The family Paramyxoviridae is large and diverse. It encompasses viruses that infect reptilian, avian and mammalian hosts, and includes a number of human pathogens, such as respiratory syncytial virus (RSV), mumps (MuV), measles (MeV), parainfluenza viruses (PIV 1 to 5), and the newly emerged Nipah and Hendra viruses. The family is divided into two subfamilies, the Pneumovirinae and Paramyxovirinae, which contain two and seven genera, respectively (1). The paramyxoviruses have a single stranded, negative sense RNA genome, and so are members of the non-segmented, negative sense (NNS) RNA virus order. The paramyxoviruses also share a similar (although not identical) cohort of genes as each other. During their replication cycle, the viral genome is transcribed to produce subgenomic, capped and polyadenylated mRNAs and replicated to produce encapsidated antigenome and genome RNAs (2). Despite the fact that approximately two-thirds of the paramyxovirus genome encodes proteins involved in performing and regulating gene expression and genome replication, paramyxoviruses only encode one polymerase. This raises questions that have puzzled researchers for more than three decades, namely how does the polymerase become committed to either mRNA transcription or genome replication, and how can these processes be differentially regulated? In this review, we attempt to address these questions. The review is divided into three parts. Part 1 presents an overview of paramyxovirus transcription and replication and discusses previously proposed models. In part 2, we describe relatively new findings regarding RSV transcription and replication, propose a revised model that fits these data, and discuss if this revised model can be applied across the paramyxovirus family. With models to describe possible mechanisms of transcription and replication initiation, it is possible to consider how these processes might be regulated during infection and in part 3 we describe information available regarding regulation of transcription and replication, highlighting similarities and differences across the family.
PART 1: PARAMYXOVIRUS TRANSCRIPTION AND REPLICATION
Overview of paramyxovirus transcription and replication
The general strategy of paramyxovirus transcription and replication is similar to that of other NNS RNA viruses (2, 3) and much of what we know has been as a result of studies on another virus in the order, vesicular stomatitis virus (VSV), a member of the family Rhabdoviridae. However, there is a considerable body of research on paramyxoviruses, as reviewed previously (2), and this is described below.
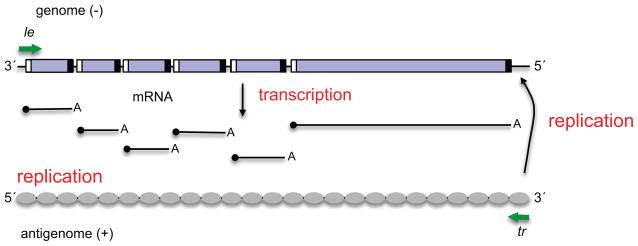
The core polymerase is a complex of two proteins, the large polymerase subunit L, which contains the enzymatic domains involved in RNA synthesis, capping and cap methylation, and the phosphoprotein, P, which is an essential cofactor (4) (5) (6) (7). The polymerase executes transcription and replication by recognizing and responding to a number of essential cis-acting elements within the virus genome (Figure 1). At the beginning and end of each gene are short (~10–13 nt) conserved signals termed gene start (gs) and gene end (ge) signals, respectively, and between each gene is a short, non-transcribed intergenic region. At the 3′ end of the genome, before the first gene, is a leader (le) promoter region, which is ~40–55 nt long, and at the 5′ end is a trailer region which is variable in length, depending on the virus (2). An important feature of the genome template is that it is associated along its length with an interlinking polymer of nucleoprotein (N) to form a helical nucleocapsid, such that cis-acting RNA elements are buried within the N-RNA structure (8–10). Most likely for this reason, when the polymerase transcribes the genome to produce mRNAs, it cannot access individual genes independently. Instead, it first engages the template at or near the 3′ end of the genome, within the le promoter. The polymerase then moves along the genome, presumably with the N subunits of the nucleocapsid being displaced and replaced as the polymerase passes by. As the polymerase proceeds, it responds to the gs and ge signals it encounters to generate the subgenomic mRNAs: at a gs signal, the polymerase initiates mRNA synthesis (opposite the first nucleotide of the gs) and at the ge signal, it releases the RNA (2). The polymerase can then scan the intergenic region to locate the next gs signal and begin mRNA synthesis of the next gene (11). This allows the polymerase to generate subgenomic RNAs. The mRNAs are also modified to contain a 5′ methyl cap and 3′ poly A tail. Work with VSV and the paramyxovirus, Sendai virus (SeV) indicates that the complement of the gs signal, which lies at the 5′ end of the mRNA, contains a signal that directs the capping reaction and methylation of the cap (12) (13, 14). The ge signal contains a poly U tract, and it is thought that stuttering of the polymerase on this U-stretch leads to polyadenylation of the mRNA (3). Similarly to cellular capping, there is evidence that addition of the cap is important to allow the transcribing polymerase to transition into an elongation mode: in the case of RSV, if capping is inhibited, the polymerase aborts RNA synthesis approximately 45–50 nt after initiating at the gs signal (15).
Figure 1.
Schematic diagram illustrating a representative paramyxovirus genome and transcription and RNA replication products. The genes are represented by purple boxes, and gs and ge signals are illustrated with white and black boxes, respectively. The le and tr promoters at the 3′ ends of the genome and antigenome, respectively, are indicated with green arrows. The genome acts as a template for mRNA and antigenome synthesis, and the antigenome as a template for genome RNA synthesis. The mRNA caps are indicated with black circles. The antigenome is shown covered with grey ovals, representing N protein, to indicate that it is encapsidated. The genome is also encapsidated, but this is not shown so that the cis-acting signals can be clearly seen.
To replicate the genome, the polymerase also initiates RNA synthesis at the le promoter. In this case, it must initiate precisely opposite the first nucleotide of the template. During replication, the polymerase does not respond to the gene junction signals, but instead elongates the nascent RNA along the complete length of the genome to produce a positive sense antigenome. The 3′ end of the antigenome contains the complement of the trailer, referred to here as tr promoter. The tr promoter in turn signals the polymerase to initiate and perform genome RNA synthesis. The antigenome and genome RNAs are not capped, but instead are encapsidated with N protein, which is delivered to the elongating RNA in a complex with P (N0P, where N0 is a monomer of N) (5). It is thought that concurrent encapsidation causes the polymerase to enter a super-processive mode, allowing it to disregard the ge signals and extend to the end of the template (16) (17) (18). Encapsidation initiation appears to be dependent on cis-acting elements in the le and tr promoters (18). These elements could function in the context of the promoter within the template strand, to recruit a specific pool of polymerase that is capable of delivering N protein onto the RNA that it is synthesizing. Alternatively, they could function at the 5′ end of the nascent RNA product to signal an initial nucleation event that begins polymerization of N protein onto the growing RNA chain.
What emerges from this description of transcription and replication is that the le and tr promoters and the gs signals are all multifunctional entities, which are not only important for directing initiation of RNA synthesis, but also directing modification of the RNA products. These modifications enable the polymerase to elongate the RNA and also serve to protect the RNA from nucleases. This multifunctional nature of the cis-acting signals complicates analysis of the initial events in RNA synthesis, particularly in cell-based assays in which abortive (i.e. prematurely released), unmodified RNAs might be unstable, and this is the reason why understanding mechanisms underlying transcription and replication initiation and regulation has proven difficult.
NNS RNA virus transcription and replication initiation models
A complexity in understanding mechanisms by which the polymerase is coordinated between transcription and RNA replication is that it is difficult to conclusively define where transcription begins on the viral genome. The mRNA for the first protein-coding gene is initiated at the first gs signal (at ~ nt 40–55), but how the polymerase accesses this signal has been the focus of debate (19–21). Three models have been proposed to explain how this could happen, based largely on studies with paramyxoviruses (mainly SeV, PIV-3 and RSV) and VSV.
Model 1: According to this model, transcription and replication are both initiated in exactly the same way, opposite the first nucleotide of the le promoter (16, 22). In its simplest version, this model postulates that a single pool of polymerase can initiate both processes. The polymerase begins transcription by first synthesizing an RNA transcript complementary to the le region (le+). At or near the end of the le, this RNA is released and the polymerase is able to scan the template to locate the first gs signal and reinitiate RNA synthesis. It is then committed to mRNA transcription. Replication would occur when N protein accumulates to a sufficiently high concentration to initiate encapsidation very quickly after initiation of le+ synthesis. If the le+ RNA becomes encapsidated before being released, this stabilizes the polymerase-template-nascent RNA complex and commits the polymerase to replication. As described in detail in later sections, this model is the most consistent with what is known regarding paramyxoviruses.
Model 2: The second model proposes that there are two pools of polymerase, a transcriptase and a replicase that consists of L-P in complex with different proteins (20, 21, 23). According to this model, the replicase initiates at the 3′ end of the genome, whereas the transcriptase initiates directly at the first gs signal and then proceeds to synthesize capped and polyadenylated mRNAs. This model is consistent with results from a number of studies with the rhabdovirus VSV (24) (25) (23) (26).
Model 3: Finally, a universal model has been proposed which attempts to tie together the two models described above (19). This model postulates that N0P is required for RNA synthesis initiation from position 1 of the template, to aid initiation from the 3′ end of the linear genome. According to this model, when the virus first enters the cell, when N0P would not be present, the polymerase cannot initiate at the 3′ end, and instead relies on cellular factors to help the polymerase access the first gs signal to begin transcription. However, once N0P has been synthesized, the polymerase instead switches to initiating at the 3′ end. When N0P is present at a low level, the polymerase synthesizes a le+ RNA, but because this is not encapsidated, the polymerase aborts RNA synthesis and reinitiates mRNA synthesis at the gs signal. At high levels of N0P, the le+ transcript becomes encapsidated and the polymerase is committed to encapsidation. This model is supported by the finding that in an in vitro SeV experiment, low concentrations of N0P enhanced transcription, whereas high concentrations caused a shift from transcription to replication. It is also based on results from VSV UV mapping experiments, which showed that the polymerase initiated transcription at the 3′ end of the le if the nucleocapsid template was derived from a virus particle, but at the gs signal in infected cells (26). However, arguments have been made against this model (20, 21) and more recent studies with VSV and RSV have shown that the polymerase can initiate opposite nucleotide 1 of the template independently of N0P (6, 27).
One problem with trying to develop a universal transcription/replication model to explain all the data available for VSV and the paramyxoviruses is that there are differences between the viruses and they might not utilize a common mechanism. One difference that is of particular significance is that the organization of transcription signals in the le promoter is different in the paramyxoviruses than in VSV, with the paramyxovirus transcription signals being relatively circumscribed (as described below) and the VSV transcription signals existing throughout almost all of le (25, 28) (Figure 2). This distinction could reflect the presence of a 3′ terminal transcription start site on the paramyxovirus promoter versus an internal start site on the VSV promoter.
Figure 2.
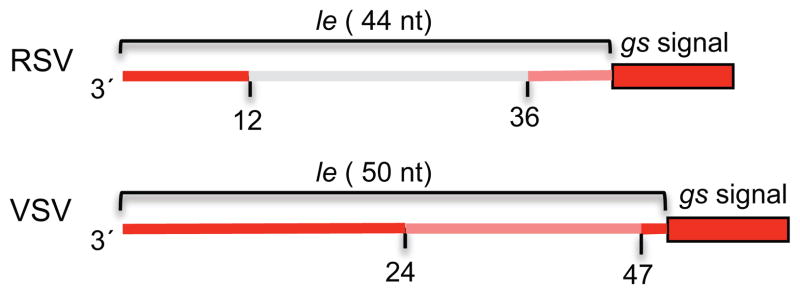
Schematic diagrams of the le and gs regions of RSV (a paramyxovirus) and VSV (a rhabdovirus) showing the positions of the transcription-specific signals in the respective viruses. Regions in which mutations reduced transcription to less than 15% of wt levels are shown in red. Regions in which mutations reduced transcription to 15–40% of wt levels are shown in orange. Substitution of the region shown in gray had no effect on transcription. The numbers underneath indicate nucleotide positions. It should be noted that not all signals have been mapped precisely.
PART 2: A REVISED MODEL FOR PARAMYXOVIRUS TRANSCRIPTION AND REPLICATION INITIATION
If the data regarding VSV are put aside, model 1, described above, is highly consistent with almost all available data regarding the paramyxoviruses. However, based on our recent studies with RSV we propose a slightly revised version of this model, which is extremely simple and is supported with experiments performed in vitro, using a minigenome system and analyzing RNA from RSV infected cells. This model is also consistent with what is known about mechanisms of RNA synthesis initiation by RNA dependent RNA polymerases of other RNA viruses.
Organization of the RSV le promoter
The RSV le region is 44 nt in length and is followed by a 10 nt gs signal for the first gene (29). Mutation analysis of the RSV le region, using a minigenome system, has shown it can be divided into three segments. The 3′ terminal ~12 nt are required for both transcription and RNA replication, nucleotides 13 to ~36 are not required for transcription, but are required for replication, and a U-rich region at the end of le, nucleotides ~37–44, increases transcription efficiency, but is not essential for transcription; this region has no effect on replication (18). The first 13 nt were shown to be sufficient to signal RNA synthesis initiation, indicating this region contains the core promoter (30, 31). This core promoter is capable of recruiting both transcription- and replication-competent polymerase, suggesting that it either recruits a single pool of polymerase that subsequently becomes differentiated, or has the capability to recruit both transcriptase and replicase forms of polymerase (30). Nucleotides located at positions 3, 5, 8, 9, 10 and 11 of the le region are of particular significance: if any of these nucleotides was mutated, both transcription and RNA replication were completely, or almost completely abrogated (32). Thus, the 3′ end of the RSV le region contains a core promoter necessary for signaling transcription and replication initiation (Figure 3).
Figure 3.
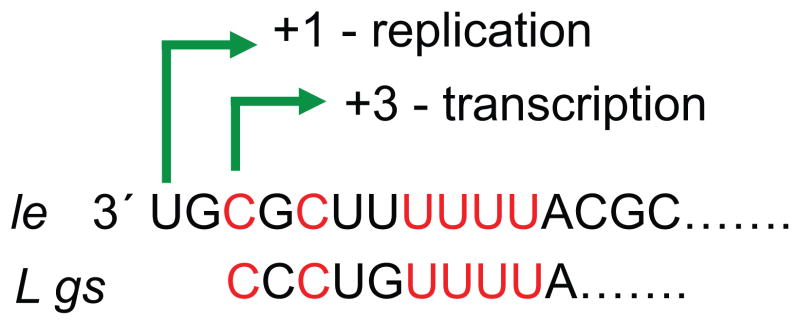
Diagram showing the alignment of the RSV le and L gs sequences. The figure shows the 3′ terminal 15 nt of the le and the first 10 nt of the L gene. The nucleotides shown in red in the le region were shown to be essential for both transcription and replication. These nucleotides align with the L gs signal. The green arrows show the experimentally determined initiation sites at positions 1 and 3 of le, which we propose are the replication and transcription initiation sites, respectively.
Transcription initiation in RSV
A hint as to how transcription is initiated came serendipitously from minigenome studies examining how the RSV polymerase is able to initiate antigenome synthesis opposite nt 1 of the le (described in more detail below). In these studies, it was noticed that some RNA appeared to be initiated at position 3 of the le promoter, in addition to the expected initiation site at position 1 (33). Primer extension analysis of RNA isolated from RSV infected cells confirmed this finding, clearly showing that RNA is initiated from both the position 1 and 3 sites in the le promoter. Indeed in infected cells, RNA initiated from position 3 was significantly more abundant than RNA initiated from position 1, suggesting that initiation from position 3 is the dominant initiation event (31). Inspection of the le core promoter sequence showed that it bears very strong resemblance to one of the RSV gs sequences, with the essential nucleotides from positions 3–11 of le aligning perfectly (Figure 3). These findings suggest that nt 3–11 of the le promoter element function similarly to a gs signal and position the polymerase to initiate opposite position 3. Further analysis of the RNAs generated from the position 1 and 3 initiation sites indicated that the RNA initiated from position 1 was elongated relatively efficiently, consistent with it being a replication product, as expected. In contrast, RNA initiated from position 3 was short and heterogeneous in length, with the majority of transcripts varying from approximately 20–25 nucleotides long (31). Why these transcripts were released after such a short distance, whereas mRNA transcripts are elongated, is not completely clear.
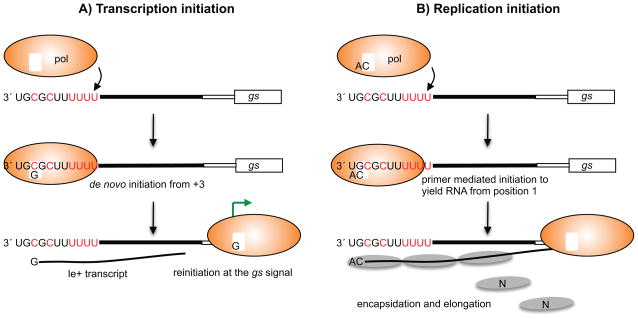
These findings suggest a very simple and logical model for transcription initiation, illustrated in Figure 4A. According to this model, the polymerase interacts with nucleotides 3–11 of the le region. This core promoter positions the polymerase so that its active site is opposite nucleotide 3, similarly to the way in which a gs signal positions the polymerase to reinitiate mRNA synthesis at a gene junction. The polymerase initiates RNA synthesis at position 3, but is unable to enter a stable elongation mode and releases the RNA after approximately 25 nucleotides. Synthesis of this abortive RNA allows the polymerase to break contacts with the promoter. Having released the abortive RNA, the polymerase can scan the template (as it does at the gene junctions) and locate the gs signal of the first gene, aided by the U-rich region at the end of le. Here it can reinitiate RNA synthesis, extend the RNA beyond 25 nucleotides, and cap the 5′ end. This commits the polymerase to transcription.
Figure 4.
Diagrams showing the models for initiation of transcription (A) and replication (B) in RSV infection. The figure shows the relationship between the polymerase, NTPs, the le promoter region and gs signal in each case. The le region is shown as three sections, reflecting the distribution of cis-acting signals, as determined by mapping analysis. The first eleven nucleotides of the le region are written, with the core promoter element shown in red type. The black line in the middle of le indicates sequence required specifically for replication, and the white box at the end of le indicates a U-rich sequence that enhances transcription. The gs signal is shown as a large white box. In the replication initiation model (B), N protein binding to the nascent le+ RNA allows encapsidation. N is likely delivered to the RNA as a soluble N0P complex, but the P protein is not shown for simplicity.
Replication initiation in RSV
While the results described above provide a compelling explanation for how RSV transcription is initiated, they raise the question: if the gs-like element in the le core promoter positions the polymerase to initiate opposite nucleotide 3 of the genome, how does the polymerase initiate replication from position 1? In considering this, it is helpful to appreciate that replication of a linear viral genome presents a challenge to a polymerase. The reason being that RNA synthesis initiation is a complex enzymatic process, in which the polymerase and template must stabilize and position the two incoming NTPs sufficiently well to allow formation of the first phosphodiester bond. This is even more challenging at the end of a linear template because there is limited RNA template for the polymerase to associate with. For this reason, viruses with linear genomes have evolved complex mechanisms of replication initiation that help provide stability to the initiating complex (34). An initiation processes that has been particularly well characterized is that utilized by phi6 bacteriophage. In this case, the polymerase enters internally on the template and then ratchets in a 5′ to 3′ direction to reposition the active site opposite nucleotide 1 (35). Based on this model and what is known regarding other RNA viruses, a logical mechanism for RSV replication initiation would be internal entry at the promoter element at nt 3–11 followed by events to enable initiation at position 1. Evidence as to how this might happen during RSV replication came from studies probing the role of the first nucleotide in determining the replication start site (36). It was found that if the first nucleotide of the tr promoter was mutated, the replication product was restored to wild type sequence in a single round of replication. These findings indicate that when the polymerase initiates RNA replication, it is able to select the initiating ATP independently of the template. Similar results were obtained for the RSV le promoter, and in this case there was evidence that the second nucleotide, a cytidine, was also selected in a template independent manner (33). Studies with rotavirus and Dengue virus, have shown that their viral polymerases can self generate a dinucleotide primer (37, 38). Taken together, these findings suggest a replication initiation model in which the polymerase becomes loaded with the first two nucleotides of the replication product, ATP and CTP, independently of the template and self-generates a primer. It could then bind the core promoter element, ratchet backwards by one nucleotide to position the 5′ AC primer opposite nucleotides 1 and 2, and then use the primer to initiate RNA synthesis. This would give the appearance that replication is initiated from position 1, although the first templated nucleotide insertion would be opposite position 3 (Figure 4B).
There are also data to suggest a mechanism for how the RNA initiated at position 1 can become encapsidated. Studies performed in vitro, using purified, recombinant RSV L-P complexes, have shown that they initiate RNA synthesis at both the position 1 and 3 sites of the promoter (39). Thus, in the case of RSV, N protein is not required for replication initiation at position 1, indicating that in infected cells N protein becomes recruited after initiation of RNA synthesis. This suggests that encapsidation begins by N binding to a signal at the 5′ end of the nascent RNA. In a minigenome experiment, in which a position 3C-to-U substitution was introduced into the le promoter to create a promoter sequence: 3′ UGUGCUUUU (the 3C-to-U substitution is underlined), a high level of full-length encapsidated replication product was generated from position 3 (33). This suggests that 5′ AC at the end of the RNA correlates with encapsidation and replication elongation. Based on these findings, we propose that when the polymerase initiates antigenome synthesis at position 1, the nascent RNA that is synthesized contains cis-acting elements that signal initiation of encapsidation, with 5′ AC playing a key role.
Together, these proposed mechanisms for transcription and replication initiation integrate to form a cohesive model. The binding site for the RSV polymerase is located at nt 3–11 of the le region and RSV L-P complex alone, with no other viral proteins, is able to interact with this signal to initiate RNA synthesis. Most frequently, the polymerase initiates directly at position 3. Because the RNA initiated at this site lacks a complete encapsidation signal, the polymerase is unable to enter an efficient elongation mode and generates abortive transcripts. Having released the le+ transcript, the polymerase can scan forward and locate the gs signal at position 45 to reinitiate RNA synthesis and become committed to transcription. Less frequently, the polymerase is able to become loaded with ATP and CTP and self-generate a primer. When this happens the polymerase can generate an RNA that is apparently initiated at position 1, and engage in RNA replication.
Can the RSV transcription and replication initiation model be applied to other paramyxoviruses?
As described above, paramyxoviruses are divided into the paramyxovirinae and pneumovirinae. These two sub-families clearly share many similarities in transcription and replication mechanisms, but they differ significantly in the organization of their promoters. Therefore, is it reasonable to think that the model proposed for RSV, a pneumovirus, will apply to other paramyxoviruses?
There are three differences in the organization of the pneumovirus and paramyxovirus promoters. First, whereas the RSV promoters are contained entirely within the le or tr regions the promoters of the paramyxovirinae are bipartite. One promoter element (promoter element I) lies within approximately the first 12 nt of the template (40–42) and is sensitive to mutation at almost all positions (40), similarly to the core promoter of RSV. The other (promoter element II) lies further downstream, between nucleotides 79 and 96 of the respiro- and morbilliviruses and between 77 and 94 of the rubulaviruses, and is a more simple motif repeated three times, either (CNNNNN)3 or (NNNNGC)3 (40–46). Second, the paramyxovirinae promoter elements need to be positioned in the appropriate phase relative to N protein and aligned adjacent to each other on the helical nucleocapsid to be functional (43, 44, 47)(see Figure 8 in reference 43 for an illustration of the promoter alignment on the nucleocapsid helix). In contrast N phasing has no effect on the efficiency of the RSV promoters (48). Third, whereas the RSV promoter must lie near the 3′ end of the template to be functional, this is not the case in the paramyxovirinae (30, 47, 49). Together, these data suggest that the structure that the polymerase recognizes to form its initial contacts with the promoter is distinct between the pneumo- and paramyxovirinae, with the pneumovirus polymerase relying, at least in part, on the unique structure that would be present at the 3′ end of the nucleocapsid, and the polymerase of the paramyxovirinae recognizing promoter bases by virtue of their positioning within the N-RNA helix. However, having bound the promoter in the nucleocapsid, it would be expected that the N-RNA structure would become relaxed at the initiation site to allow the RNA into the polymerase active site. Thus, in both the pneumo- and paramyxovirinae the next step would be expected to involve a direct polymerase-RNA interaction.
Aside from this difference in initial promoter recognition, other aspects of transcription initiation are conserved between pneumo and paramyxovirinae. Aside from the necessity for a core promoter, the le sequence prior to the first gs signal is not essential for transcription (50–52). In the paramyxovirinae, the first gs signal lies on the opposite face of the helical nucleocapsid from the promoter elements, suggesting that it is not seen in conjunction with them (53), and like the RSV gs signal, it can function if it is placed at varying distances relative to the promoter, although its natural position is optimal (51, 52, 54, 55). Finally, it has been known for a long time that the SeV and MeV polymerases synthesize a heterogeneous population of abortive transcripts from a site at, or near, the 3′ end of le (16, 56, 57). These data are very consistent with what has been found for RSV, and with a model that the polymerase initiates transcription from the 3′ end of the promoter.
The question that remains is: does the polymerase of the paramyxovirinae use a gs-like sequence at the 3′ end of the le promoter for initiation, and are transcription and replication initiated from two different start sites? Figure 5 shows the 3′ terminal promoter elements (promoter element I) and gs sequences for a representative virus species of each paramyxovirus genus. The most striking observation is that while the promoter sequences do not obviously align, all of them begin with 3′ UG, meaning that the replication product is initiated 5′ AC. This suggests that the mechanism that RSV uses to select initiating NTPs to begin replication is probably conserved throughout the family. On the other hand, identity between the promoter and gs signals is not that obvious in the paramyxovirus subfamily. However, as noted above, these signals are multifunctional and differences between them might be important for determining the fate of the RNA products (e.g. whether they will be capped, encapsidated, or aborted). Indeed, we only noticed the identity between the RSV promoter and L gs signal because the 3′ terminal le nucleotides required specifically for transcription (rather than replication) had been identified, but this information is not available for the paramyxovirinae. Thus, the promoters and gs signals of the paramyxovirinae might contain a conserved polymerase binding and initiation signal, even if not readily apparent. With this in mind, a closer inspection of the sequence alignments suggests that there might be a gs-like signal in the paramyxovirus promoters. By color coding the pyrimidine and purine nucleotides, it is apparent that the promoters and gs signals both contain a motif consisting of a pyrimidine stretch, followed by 1–3 nt of variable sequence that typically contains a purine, followed by a second pyrimidine stretch. This motif typically aligns slightly internally on the promoter, although the optimal alignment varies between viruses. This observation, coupled with the fact that apparently all paramyxoviruses begin replication with 5′ AC suggests that the members of the paramyxovirus family share a common mechanism of replication initiation, in which the polymerase binds to a pyrimidine-rich motif, slightly internally on the template, and perhaps self-generates a primer to initiate opposite position 1.
Figure 5.
Promoter and gs sequences for one virus species from each of the paramyxovirus genera (indicated in parentheses). The gs signals for each of the viral genes are shown. The promoter sequences shown were defined as those nucleotides that were identical or almost identical between the le and tr promoters; the minimal core promoters might be more constrained than what is shown. Pyrimidine and purine residues are shown in orange and green type, respectively.
Although the sequence alignments in Figure 5 suggest that there may be a gs-like sequence within the promoters of the paramyxovirinae, it is difficult to conclude that their polymerase initiates transcription internally, in a similar fashion to RSV. A method to identify initiation sites experimentally is to perform primer extension analysis on RNA from virus-infected cells. However, to detect RNA initiated from position 3 of the RSV le promoter, we needed to use a primer that hybridized very close to the 5′ end of the RNA to detect the short abortive transcripts. We could find no evidence in the literature of a similar analysis for the paramyxovirinae. Sequence alignment also does not give a clear indication of an internal initiation site, particularly given that RNA polymerases typically initiate synthesis with a purine residue and so initiation opposite position 2 or 3 of the promoter would be unlikely. In the absence of any data indicating the presence of an internal initiation site for transcription, it is probably prudent to assume that the paramyxovirinae differ from the pneumovirinae in terms of the exact transcription initiation site, and instead initiate both transcription and replication in the same way, opposite the first nucleotide of the le. As described below, the model that the polymerase of the paramyxovirinae initiates transcription opposite position 1 is supported by data regarding transcription and replication regulation, which appears to be slightly different in RSV versus SeV and measles virus, as described below.
PART 3: REGULATION OF PARAMYXOVIRUS POLYMERASE ACTIVITY
Kinetics of paramyxovirus RNA synthesis
Although paramyxoviruses are relatively simple viruses, it would be expected that they would have evolved mechanisms to regulate RNA synthesis to maximize use of available templates and avoid production of non-functional and potentially detrimental RNAs (e.g. unencapsidated negative sense RNA could hybridize to mRNAs). Consistent with this idea, studies in which RSV, measles and SeV RNAs were examined over a single cycle of infection have provided evidence for temporal control of RNA synthesis (58–60). In the case of measles virus, where quantitative analysis of each RNA species was performed, it was found that mRNA accumulated in an exponential fashion from 2–24 hours post infection, at which point it reached a plateau. In contrast, genome and antigenome levels remained at a low level until 12 hours post infection, then increased exponentially at an equivalent rate until 24 hours post infection; thereafter, the rate of replicative RNA accumulation decreased, but the rate of genome accumulation exceeded that of antigenome (59). These findings are essentially similar to those with RSV (58). The experiments with SeV measured antigenome and genome RNAs, specifically. In this case, it was found that in the initial stages of infection, the ratio of antigenome to genome RNA increased significantly compared to input virus, reflecting very high use of the le promoter and synthesis of positive sense antigenome RNA, but at later stages, the ratio reversed due to very active production of genome RNA from the tr promoter (60). These studies indicate that there are at least two points in which paramyxovirus gene expression and genome replication are regulated: first, between transcription and replication, and second, between positive and negative sense RNA synthesis from the le and tr promoters, respectively. The mechanisms by which these two transitions might be regulated are described below. In addition, other regulatory mechanisms might come into play to temper RNA synthesis, to avoid over-stimulation of the innate immune response, and/or to ready nucleocapsids for packaging (61–67).
Transition from mRNA to antigenome synthesis
The factor responsible for “switching on” replication during paramyxovirus infection is the viral N protein, which is required to encapsidate the replication product as the RNA is being synthesized (5, 68). Although the N protein is required to enable replication of all paramyxoviruses, the extent to which it regulates a switch between transcription and replication appears to differ depending on the virus. In experiments using the RSV minigenome system it was shown that while increasing the level of N (or N and P) resulted in an increase in antigenome synthesis, there was no apparent inhibition of transcription, even at very high levels of N protein (69). The reason why RSV transcription is not affected by N protein concentration is now clear: the dominant initiation event from the le promoter is from position 3, not position 1 (31). There is no evidence that any RNA initiated at position 3 can be elongated into a replication product. This means that transcription would always be the dominant initiation event, regardless of how much N protein is available for encapsidation. However, a corollary of the RSV transcription/replication model is that if the polymerase initiates at position 1, but N levels are low, then this polymerase could abort RNA synthesis after ~25 nt and engage in transcription. This would mean that N protein levels would control a switch between transcription and replication, but only for the relatively small proportion of polymerase that happens to initiate at position 1, not polymerase that initiates at position 3. Thus, in the case of RSV, rather than thinking of transcription switching to replication, it is more appropriate to think in terms of transcription being a constitutive event, and replication being switched on once N has accumulated, with just a small cost to transcription.
If transcription and replication were both initiated from position 1 of the le promoter by a single pool of polymerase, then these would be expected to be interchangeable events. This indeed seems to be the case for at least two of the paramyxovirinae. In experiments performed in vitro with SeV and in cellulo with measles virus, increasing the level of N protein led to an increase in replication and a discernable decrease in transcription (19, 59). This difference in the effect of N protein on transcription between RSV, MeV and SeV provides evidence that different paramyxoviruses might initiate transcription using different initiation sites.
Transition from positive to negative sense RNA synthesis
As described above, at a late stage of infection there is a transition from synthesis of positive to negative sense RNA (58–60). This transition ensures accumulation of a high proportion of genome sense nucleocapsids for packaging into virus particles (70). One plausible mechanism by which this transition could occur is by action of a trans-acting protein that is either absent, or at only a low level in virus particles, but which accumulates over the course of infection. There does not appear to be a common protein to control this transition in the paramyxoviruses and consideration of this provides further evidence of how divergent the paramyxoviruses are.
The protein most likely responsible for regulating a shift from positive to negative sense RNA synthesis in the pneumovirinae is the M2-2 protein. M2-2 is expressed from an alternative and downstream open reading frame in the M2 gene (71, 72). If M2-2 is over-expressed, it inhibits all RNA synthesis (72–74). However, examination of the kinetics of RNA accumulation in a recombinant RSV containing a deletion of M2-2 suggests the protein specifically inhibits positive sense RNA synthesis (58). How M2-2 functions to accomplish this is not known, but it binds to the L protein (74), and in this and other respects, shares similarities with a protein encoded by some of the paramyxovirinae, the C protein.
A number of viruses within the paramyxovirinae have the capability to express C protein from an alternative open reading frame within the P gene (2, 75). The SeV C protein has been studied extensively. It is a multifunctional protein, which plays a role in allowing the virus to evade the host immune response and inhibit apoptosis. However, it also plays an important role in modulating viral RNA synthesis. If the C protein is over-expressed it inhibits all RNA synthesis (76, 77). However, at normal expression levels, the protein functions in a promoter specific manner, inhibiting RNA synthesis from the le promoter, but not from the tr promoter (49, 76–78). Indeed, in recent work using recombinant viruses lacking C protein, it was shown that the le promoter is a stronger promoter than the tr, contrary to what had been thought previously (60), and that C protein is the key factor that ensures that genome RNA is more abundant in cells than antigenome (60, 70). C protein binds to the L subunit of the polymerase and the strength of this interaction correlates with the ability of C protein to exert its inhibitory effect (79, 80). Thus, C protein acts directly to modify the polymerase complex and affect its behavior at the le promoter.
The ferla, avula and rubulavirus genera of the paramyxovirinae do not express a C protein (75) and it is not clear if they have a functional homolog. However, one possible candidate is their V protein. The V proteins are multi-functional proteins, expressed from the same gene that expresses P, by almost all paramyxovirinae. The V proteins can inhibit RNA replication by binding N protein (64), but the V proteins of some viruses have also been shown to bind to L (65, 81). In the case of PIV-2, a rubula virus, L-V interaction was responsible for inhibiting RNA synthesis in a minigenome system (65). Thus, it is possible that V might bind L to fulfill a similar function as C protein in some of the paramyxovirinae.
Finally, another factor that could play a role in regulating the polymerase between different RNA synthesis activities is P protein phosphorylation. The P proteins are heavily phosphorylated (as indicated by their name), but in most cases in which the effect of phosphorylation status has been examined, there has been either inconsistent results between in vitro assays and recombinant virus, or no detectable effect (82–87). However, in experiments with the rubulaviruses, PIV5 and mumps virus, regulatory effects of phosphorylation have been clearly demonstrated (88–90). In addition, sumoylation been shown to impact RNA synthesis (91). Given these findings, it would be interesting to determine to what extent phosphorylation and other post-translational modifications can play a role in temporal regulation.
Can RNA secondary structure in the promoters play a regulatory role?
As described above, it is well established that the genome and antigenome RNAs of the paramyxoviruses are encapsidated along their length with N protein at all times of infection. Thus, these RNAs are not expected to form secondary structures, and cis-acting signals are comprised entirely of primary sequence. However, recent work with RSV indicates that the tr promoter might not be completely encapsidated at all times, suggesting the possibility that promoter RNA secondary structure could play a regulatory role. It was found that antigenome sense RNA isolated from RSV infected cells is heterogeneous, containing one to three nucleotide additions at the 3′ end (6). Nucleotide addition was shown to occur because the tr RNA could adopt a secondary structure that allowed limited templated nucleotide addition. A similar phenomenon has been described for Borna disease virus, another NNS RNA virus (92). In the case of Borna disease virus, the 3′ nucleotide addition enables the virus to generate replicative RNA lacking a 5′ triphosphate, allowing it to escape immune surveillance (93), but this is not the case in RSV and so the significance of the nucleotide addition is not clear. However, it was found that in the context of a naked RNA template, the three-nucleotide addition significantly inhibited RSV tr promoter activity (6). Thus, one possible explanation for this finding is that the capacity for forming a secondary structure and adding nucleotides onto the tr promoter serves as a mechanism for sensing available N protein. According to this hypothesis, if N protein levels are low as the polymerase completes antigenome synthesis, encapsidation of the RNA lags behind and there is the opportunity for the RNA to form a secondary structure and additional nucleotides to be added onto the 3′ end of the tr promoter. In this case, the newly synthesized antigenome RNA cannot act as a template to produce genome RNA, and polymerase remains available to reinitiate RNA synthesis on available genome templates, resulting in increased N protein expression. On the other hand, if N protein is abundant, the newly synthesized antigenome RNA is encapsidated as it is synthesized and does not have the opportunity to form a secondary structure to modify the 3′ end of the tr promoter. In this case, the polymerase can efficiently initiate genome RNA synthesis from the tr promoter. While considerable work is required to test this hypothesis, the idea that limited RNA secondary structure might play a role in regulating transcription and replication is intriguing.
The possibility that promoter secondary structure might play a role in its regulation has also been suggested for SeV (60). This idea emerged from studies to define the sequence within the le promoter that is responsible for inhibition by C protein. Swaps between le and tr promoter sequences failed to identify a specific primary sequence that could be responsible (60, 77). However, there was a correlation between lack of predicted promoter secondary structure and C protein inhibition (60). It is difficult to reconcile the idea that RNA secondary structure plays a role in SeV given that the bipartite promoter appears to function by being positioned on the same face of a helical nucleocapsid. Furthermore, mutation analysis of the PIV-3 promoters using the minigenome system failed to identify a role for RNA secondary structure (94). However, promoter secondary structures might form as the N-RNA template is relaxed during polymerase binding and such secondary structure effects might not be detectable in a minigenome assay due to the excess of N protein in this system. Therefore, while the data to support the idea are limited, it is interesting to speculate that perhaps the genome and antigenome 3′ termini are not entirely encapsidated at all times during infection, and that under certain circumstances, secondary structures may play a role in governing promoter activity.
FUTURE DIRECTIONS
In our view, the main gaps in knowledge in paramyxovirus transcription and replication research fall in two main areas. First, it will be valuable to have an understanding of how the RNA synthesis machinery functions, at a molecular level. While there are structures for many paramyxovirus proteins, there is relatively little structural information available regarding the L protein. An exciting development in NNS RNA virology in recent years includes establishment of enzymatic assays using recombinant SeV, VSV, and RSV polymerase (6, 12, 27, 95–97). This breakthrough has opened up the opportunity to perform detailed mechanistic and structural studies of the polymerase complex (4, 98). The second main area to pursue involves developing a better appreciation of temporal and perhaps spatial regulation of polymerase and nucleocapsid function. There is relatively little known about what happens to the paramyxovirus nucleocapsid and polymerase once they have been delivered into a cell, or how cellular proteins might alter their structures and functions. The ability to generate recombinant paramyxoviruses, coupled with developments in high-throughput “omics” approaches and high-resolution microscopy, has the potential to provide enormous insight into the interplay between the virus and the host cell. Finally, as we think this review conveys, a true understanding of paramyxovirus transcription and replication mechanisms will come from continued research on a variety of viruses so that we can appreciate the similarities and differences between them.
Acknowledgments
The authors would like to thank Drs. Laurent Roux and Michael Hoffman for valuable discussions regarding their data and Michael Mawhorter for helpful comments on the manuscript. RF would like to thank Dr. Sean Whelan for inviting us to write this review, and for many stimulating conversations regarding NNS RNA virus transcription and replication mechanisms. Research in the Fearns laboratory is currently funded by NIH R01AI113321, AstraZeneca and Alios Biopharma. The opinions expressed in this article are those of the authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sarah L. Noton, Email: slnoton@bu.edu.
Rachel Fearns, Email: rfearns@bu.edu.
References
- 1.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy: classification and nomenclature of viruses: Ninthe Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; 2012. [Google Scholar]
- 2.Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams and Wilkins; pp. 1449–1496. [Google Scholar]
- 3.Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 4.Morin B, Kranzusch PJ, Rahmeh AA, Whelan SP. The polymerase of negative-stranded RNA viruses. Current opinion in virology. 2013;3:103–110. doi: 10.1016/j.coviro.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horikami SM, Curran J, Kolakofsky D, Moyer SA. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noton SL, Deflube LR, Tremaglio CZ, Fearns R. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS pathogens. 2012;8:e1002980. doi: 10.1371/journal.ppat.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazumder B, Adhikary G, Barik S. Bacterial expression of human respiratory syncytial viral phosphoprotein P and identification of Ser237 as the site of phosphorylation by cellular casein kinase II. Virology. 1994;205:93–103. doi: 10.1006/viro.1994.1623. [DOI] [PubMed] [Google Scholar]
- 8.Cox R, Pickar A, Qiu S, Tsao J, Rodenburg C, Dokland T, Elson A, He B, Luo M. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc Natl Acad Sci U S A. 2014;111:15208–15213. doi: 10.1073/pnas.1413268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruigrok RW, Crepin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Current opinion in microbiology. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 11.Fearns R, Collins PL. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J Virol. 1999;73:388–397. doi: 10.1128/jvi.73.1.388-397.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino T, Kobayashi M, Iwama M, Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J Biol Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- 13.Wang JT, McElvain LE, Whelan SP. Vesicular Stomatitis Virus mRNA Capping Machinery Requires Specific cis-Acting Signals in the RNA. J Virol. 2007;81:11499–11506. doi: 10.1128/JVI.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stillman EA, Whitt MA. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J Virol. 1999;73:7199–7209. doi: 10.1128/jvi.73.9.7199-7209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagace L, Massariol MJ, Do F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal S, Kolakofsky D. Modified model for the switch from Sendai virus transcription to replication. J Virol. 1989;63:1951–1958. doi: 10.1128/jvi.63.5.1951-1958.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gubbay O, Curran J, Kolakofsky D. Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J Gen Virol. 2001;82:2895–2903. doi: 10.1099/0022-1317-82-12-2895. [DOI] [PubMed] [Google Scholar]
- 18.McGivern DR, Collins PL, Fearns R. Identification of internal sequences in the 3′ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J Virol. 2005;79:2449–2460. doi: 10.1128/JVI.79.4.2449-2460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curran J, Kolakofsky D. Nonsegmented negative-strand RNA virus RNA synthesis in vivo. Virology. 2008;371:227–230. doi: 10.1016/j.virol.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Whelan SP. Response to “Non-segmented negative-strand RNA virus RNA synthesis in vivo”. Virology. 2008;371:234–237. doi: 10.1016/j.virol.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 21.Banerjee AK. Response to “Non-segmented negative-strand RNA virus RNA synthesis in vivo”. Virology. 2008;371:231–233. doi: 10.1016/j.virol.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Kolakofsky D, Le Mercier P, Iseni F, Garcin D. Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology. 2004;318:463–473. doi: 10.1016/j.virol.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Qanungo KR, Shaji D, Mathur M, Banerjee AK. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc Natl Acad Sci U S A. 2004;101:5952–5957. doi: 10.1073/pnas.0401449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keene JD, Thornton BJ, Emerson SU. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc Natl Acad Sci U S A. 1981;78:6191–6195. doi: 10.1073/pnas.78.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whelan SP, Wertz GW. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol. 1999;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whelan SP, Wertz GW. Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc Natl Acad Sci U S A. 2002;99:9178–9183. doi: 10.1073/pnas.152155599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morin B, Rahmeh AA, Whelan SP. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. The EMBO journal. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Pattnaik AK. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J Virol. 1999;73:444–452. doi: 10.1128/jvi.73.1.444-452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol. 2013;372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowton VM, Fearns R. Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3′ end of the nucleocapsid and can scan to access internal signals. J Virol. 2005;79:11311–11322. doi: 10.1128/JVI.79.17.11311-11322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tremaglio CZ, Noton SL, Deflube LR, Fearns R. Respiratory syncytial virus polymerase can initiate transcription from position 3 of the leader promoter. J Virol. 2013;87:3196–3207. doi: 10.1128/JVI.02862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fearns R, Peeples ME, Collins PL. Mapping the transcription and replication promoters of respiratory syncytial virus. J Virol. 2002;76:1663–1672. doi: 10.1128/JVI.76.4.1663-1672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noton SL, Fearns R. The first two nucleotides of the respiratory syncytial virus antigenome RNA replication product can be selected independently of the promoter terminus. RNA. 2011;17:1895–1906. doi: 10.1261/rna.2813411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol. 2004;85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 35.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 36.Noton SL, Cowton VM, Zack CR, McGivern DR, Fearns R. Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion. Proc Natl Acad Sci U S A. 2010;107:10226–10231. doi: 10.1073/pnas.0913065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, Patton JT. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. Rna. 2000;6:1455–1467. doi: 10.1017/s1355838200001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, Sallamand C, Debart F, Vasseur JJ, Canard B. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS pathogens. 2012;8:e1002912. doi: 10.1371/journal.ppat.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noton SL, Aljabr W, Hiscox JA, Matthews DA, Fearns R. Factors affecting de novo RNA synthesis and back-priming by the respiratory syncytial virus polymerase. Virology. 2014;462–463:318–327. doi: 10.1016/j.virol.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman MA, Banerjee AK. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology. 2000;269:201–211. doi: 10.1006/viro.2000.0223. [DOI] [PubMed] [Google Scholar]
- 41.Marcos F, Ferreira L, Cros J, Park MS, Nakaya T, Garcia-Sastre A, Villar E. Mapping of the RNA promoter of Newcastle disease virus. Virology. 2005;331:396–406. doi: 10.1016/j.virol.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 42.Mioulet V, Barrett T, Baron MD. Scanning mutagenesis identifies critical residues in the rinderpest virus genome promoter. J Gen Virol. 2001;82:2905–2911. doi: 10.1099/0022-1317-82-12-2905. [DOI] [PubMed] [Google Scholar]
- 43.Murphy SK, Ito Y, Parks GD. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J Virol. 1998;72:10–19. doi: 10.1128/jvi.72.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walpita P. An internal element of the measles virus antigenome promoter modulates replication efficiency. Virus research. 2004;100:199–211. doi: 10.1016/j.virusres.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 46.Walpita P, Peters CJ. Cis-acting elements in the antigenomic promoter of Nipah virus. J Gen Virol. 2007;88:2542–2551. doi: 10.1099/vir.0.83035-0. [DOI] [PubMed] [Google Scholar]
- 47.Vulliemoz D, Roux L. “Rule of six”: how does the Sendai virus RNA polymerase keep count? J Virol. 2001;75:4506–4518. doi: 10.1128/JVI.75.10.4506-4518.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samal SK, Collins PL. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vulliemoz D, Roux L. Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally. J Virol. 2002;76:7987–7995. doi: 10.1128/JVI.76.16.7987-7995.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cordey S, Roux L. Further characterization of a paramyxovirus transcription initiation signal: search for required nucleotides upstream and importance of the N phase context. J Gen Virol. 2007;88:1555–1564. doi: 10.1099/vir.0.82701-0. [DOI] [PubMed] [Google Scholar]
- 51.Hoffman MA, Thorson LM, Vickman JE, Anderson JS, May NA, Schweitzer MN. Roles of human parainfluenza virus type 3 bases 13 to 78 in replication and transcription: identification of an additional replication promoter element and evidence for internal transcription initiation. J Virol. 2006;80:5388–5396. doi: 10.1128/JVI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vulliemoz D, Cordey S, Mottet-Osman G, Roux L. Nature of a paramyxovirus replication promoter influences a nearby transcription signal. J Gen Virol. 2005;86:171–180. doi: 10.1099/vir.0.80435-0. [DOI] [PubMed] [Google Scholar]
- 53.Le Mercier P, Garcin D, Garcia E, Kolakofsky D. Competition between the Sendai virus N mRNA start site and the genome 3′-end promoter for viral RNA polymerase. J Virol. 2003;77:9147–9155. doi: 10.1128/JVI.77.17.9147-9155.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fearns R, Collins PL, Peeples ME. Functional analysis of the genomic and antigenomic promoters of human respiratory syncytial virus. J Virol. 2000;74:6006–6014. doi: 10.1128/jvi.74.13.6006-6014.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cordey S, Roux L. Transcribing paramyxovirus RNA polymerase engages the template at its 3′ extremity. J Gen Virol. 2006;87:665–672. doi: 10.1099/vir.0.81353-0. [DOI] [PubMed] [Google Scholar]
- 56.Leppert M, Rittenhouse L, Perrault J, Summers DF, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 57.Horikami SM, Moyer SA. Synthesis of leader RNA and editing of the P mRNA during transcription by purified measles virus. J Virol. 1991;65:5342–5347. doi: 10.1128/jvi.65.10.5342-5347.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bermingham A, Collins PL. The M2–2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc Natl Acad Sci U S A. 1999;96:11259–11264. doi: 10.1073/pnas.96.20.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plumet S, Duprex WP, Gerlier D. Dynamics of viral RNA synthesis during measles virus infection. J Virol. 2005;79:6900–6908. doi: 10.1128/JVI.79.11.6900-6908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irie T, Okamoto I, Yoshida A, Nagai Y, Sakaguchi T. Sendai virus C proteins regulate viral genome and antigenome synthesis to dictate the negative genome polarity. J Virol. 2014;88:690–698. doi: 10.1128/JVI.02798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gander JR, Schwan LM, Hoffman MA. Analysis of nucleotides 13–96 of the human parainfluenza virus type 3 antigenomic promoter reveals positive- and negative-acting replication elements. Virology. 2011;419:90–96. doi: 10.1016/j.virol.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keller MA, Parks GD. Positive- and negative-acting signals combine to determine differential RNA replication from the paramyxovirus simian virus 5 genomic and antigenomic promoters. Virology. 2003;306:347–358. doi: 10.1016/s0042-6822(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 63.Curran J, Boeck R, Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. The EMBO journal. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horikami SM, Smallwood S, Moyer SA. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222:383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- 65.Nishio M, Ohtsuka J, Tsurudome M, Nosaka T, Kolakofsky D. Human parainfluenza virus type 2 V protein inhibits genome replication by binding to the L protein: possible role in promoting viral fitness. J Virol. 2008;82:6130–6138. doi: 10.1128/JVI.02635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sleeman K, Bankamp B, Hummel KB, Lo MK, Bellini WJ, Rota PA. The C, V and W proteins of Nipah virus inhibit minigenome replication. J Gen Virol. 2008;89:1300–1308. doi: 10.1099/vir.0.83582-0. [DOI] [PubMed] [Google Scholar]
- 67.Witko SE, Kotash C, Sidhu MS, Udem SA, Parks CL. Inhibition of measles virus minireplicon-encoded reporter gene expression by V protein. Virology. 2006;348:107–119. doi: 10.1016/j.virol.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 68.Baker SC, Moyer SA. Encapsidation of Sendai virus genome RNAs by purified NP protein during in vitro replication. J Virol. 1988;62:834–838. doi: 10.1128/jvi.62.3.834-838.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fearns R, Peeples ME, Collins PL. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 70.Irie T, Nagata N, Yoshida T, Sakaguchi T. Paramyxovirus Sendai virus C proteins are essential for maintenance of negative-sense RNA genome in virus particles. Virology. 2008;374:495–505. doi: 10.1016/j.virol.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 71.Ahmadian G, Randhawa JS, Easton AJ. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. The EMBO journal. 2000;19:2681–2689. doi: 10.1093/emboj/19.11.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collins PL, Hill MG, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci U S A. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng X, Park H, Zhou H, Jin H. Overexpression of the M2–2 protein of respiratory syncytial virus inhibits viral replication. J Virol. 2005;79:13943–13952. doi: 10.1128/JVI.79.22.13943-13952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kitagawa Y, Zhou M, Yamaguchi M, Komatsu T, Takeuchi K, Itoh M, Gotoh B. Human metapneumovirus M2–2 protein inhibits viral transcription and replication. Microbes and infection/Institut Pasteur. 2010;12:135–145. doi: 10.1016/j.micinf.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 75.Lo MK, Sogaard TM, Karlin DG. Evolution and structural organization of the C proteins of paramyxovirinae. PloS one. 2014;9:e90003. doi: 10.1371/journal.pone.0090003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Curran J, Marq JB, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 79.Grogan CC, Moyer SA. Sendai virus wild-type and mutant C proteins show a direct correlation between L polymerase binding and inhibition of viral RNA synthesis. Virology. 2001;288:96–108. doi: 10.1006/viro.2001.1068. [DOI] [PubMed] [Google Scholar]
- 80.Horikami SM, Hector RE, Smallwood S, Moyer SA. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 81.Sweetman DA, Miskin J, Baron MD. Rinderpest virus C and V proteins interact with the major (L) component of the viral polymerase. Virology. 2001;281:193–204. doi: 10.1006/viro.2000.0805. [DOI] [PubMed] [Google Scholar]
- 82.Barik S, McLean T, Dupuy LC. Phosphorylation of Ser232 directly regulates the transcriptional activity of the P protein of human respiratory syncytial virus: phosphorylation of Ser237 may play an accessory role. Virology. 1995;213:405–412. doi: 10.1006/viro.1995.0013. [DOI] [PubMed] [Google Scholar]
- 83.Dupuy LC, Dobson S, Bitko V, Barik S. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase; phosphorylation by casein kinase 1 occurs mainly at Ser(215) and is without effect. J Virol. 1999;73:8384–8392. doi: 10.1128/jvi.73.10.8384-8392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu C, Gupta KC. Functional significance of alternate phosphorylation in Sendai virus P protein. Virology. 2000;268:517–532. doi: 10.1006/viro.1999.0176. [DOI] [PubMed] [Google Scholar]
- 85.Hu CJ, Kato A, Bowman MC, Kiyotani K, Yoshida T, Moyer SA, Nagai Y, Gupta KC. Role of primary constitutive phosphorylation of Sendai virus P and V proteins in viral replication and pathogenesis. Virology. 1999;263:195–208. doi: 10.1006/viro.1999.9953. [DOI] [PubMed] [Google Scholar]
- 86.Lu B, Ma CH, Brazas R, Jin H. The major phosphorylation sites of the respiratory syncytial virus phosphoprotein are dispensable for virus replication in vitro. J Virol. 2002;76:10776–10784. doi: 10.1128/JVI.76.21.10776-10784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mazumder B, Barik S. Requirement of casein kinase II-mediated phosphorylation for the transcriptional activity of human respiratory syncytial viral phosphoprotein P: transdominant negative phenotype of phosphorylation-defective P mutants. Virology. 1994;205:104–111. doi: 10.1006/viro.1994.1624. [DOI] [PubMed] [Google Scholar]
- 88.Pickar A, Xu P, Elson A, Li Z, Zengel J, He B. Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication. J Virol. 2014;88:4414–4422. doi: 10.1128/JVI.03673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun D, Luthra P, Li Z, He B. PLK1 down-regulates parainfluenza virus 5 gene expression. PLoS pathogens. 2009;5:e1000525. doi: 10.1371/journal.ppat.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Timani KA, Sun D, Sun M, Keim C, Lin Y, Schmitt PT, Schmitt AP, He B. A single amino acid residue change in the P protein of parainfluenza virus 5 elevates viral gene expression. J Virol. 2008;82:9123–9133. doi: 10.1128/JVI.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun D, Xu P, He B. Sumoylation of the P protein at K254 plays an important role in growth of parainfluenza virus 5. J Virol. 2011;85:10261–10268. doi: 10.1128/JVI.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martin A, Hoefs N, Tadewaldt J, Staeheli P, Schneider U. Genomic RNAs of Borna disease virus are elongated on internal template motifs after realignment of the 3′ termini. Proc Natl Acad Sci U S A. 2011;108:7206–7211. doi: 10.1073/pnas.1016759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider U, Schwemmle M, Staeheli P. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc Natl Acad Sci U S A. 2005;102:3441–3446. doi: 10.1073/pnas.0405965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoffman MA, Banerjee AK. Analysis of RNA secondary structure in replication of human parainfluenza virus type 3. Virology. 2000;272:151–158. doi: 10.1006/viro.2000.0369. [DOI] [PubMed] [Google Scholar]
- 95.Li J, Rahmeh A, Morelli M, Whelan SP. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Molecular cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 97.Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SP. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci U S A. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]