Abstract
The APOBEC family of single-stranded DNA cytosine deaminases comprises a formidable arm of the vertebrate innate immune system. Pre-vertebrates express a single APOBEC, whereas some mammals produce as many as eleven enzymes. The APOBEC3 subfamily displays both copy number variation and polymorphisms, consistent with ongoing pathogenic pressures. These enzymes restrict the replication of many DNA-based parasites, such as exogenous viruses and endogenous transposable elements. APOBEC1 and activation-induced cytosine deaminase (AID) have specialized functions in RNA editing and antibody gene diversification, respectively, whereas APOBEC2 and APOBEC4 appear to have different functions. Nevertheless, the APOBEC family protects against both periodic viral zoonoses as well as exogenous and endogenous parasite replication. This review highlights viral pathogens that are restricted by APOBEC enzymes, but manage to escape through unique mechanisms. The sensitivity of viruses that lack counterdefense measures highlights the need to develop APOBEC-enabling small molecules as a new class of anti-viral drugs.
APOBEC hallmarks
DNA deamination
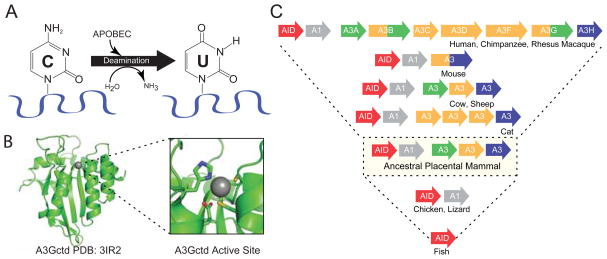
The fundamental biochemical activity of the APOBEC family of enzymes is DNA cytosine deamination (Figure 1A). This activity was originally demonstrated using E. coli-based mutation assays, and subsequently elaborated in a wide variety of biochemical and virological experimental systems [(Harris et al., 2002; Petersen-Mahrt et al., 2002); reviewed by (Aydin et al., 2014; Desimmie et al., 2014; Di Noia and Neuberger, 2007; Feng et al., 2014; Imahashi et al., 2012; Malim and Bieniasz, 2012; Refsland and Harris, 2013; Shandilya et al., 2014; Strebel, 2013)]. APOBEC cytosine to uracil (C-to-U) deaminase activity is largely specific to single-stranded DNA substrates and requires a minimum of five contiguous deoxy-nucleotides (three bases on the 5′ side of the target cytosine and one on the 3′ side) (Harjes et al., 2013; Nabel et al., 2013). DNA C-to-U deamination occurs through a zinc-mediated hydrolytic mechanism, in which a conserved glutamic acid deprotonates water, and the resulting zinc-stabilized hydroxide ion attacks the 4-position of the cytosine nucleobase, with the net replacement of the amine group (NH2) with a carbonyl group (double-bonded oxygen) (Figure 1A).
Figure 1. APOBEC hallmarks.
(A) A schematic of the single-stranded DNA cytosine deamination reaction catalyzed by APOBEC family members. (B) A ribbon model of the A3G catalytic domain showing its globular structure and a blow-up of the zinc-coordinating active site (cysteines are depicted in yellow, histidine in blue, and the catalytic glutamate in red). (C) Schematics of the A3 gene composition of several current mammals depicted above the repertoire of a likely common ancestor and current non-mammalian vertebrates. The color scheme distinguishes phylogenetic subfamilies. See the text for additional details.
A second hallmark property of the APOBEC enzymes, which has been exploited to deduce biological functions, is an intrinsic local dinucleotide preference, with one enzyme preferring the target cytosine to be preceded by a purine (AID), one preferring the target cytosine to be preceded by another cytosine (APOBEC3G), and the remainder preferring the target cytosine to be preceded by a thymine (APOBEC1, APOBEC3A/B/C/D/F/H) [(Carpenter et al., 2010; Kohli et al., 2010; Rathore et al., 2013; Wang et al., 2010) and references therein]. APOBEC2 and APOBEC4 cannot be classified this way because they have yet to elicit activity. Chimeric enzymes constructed by swapping domains between proteins of different specificity have been particularly informative by implicating amino acid residues in a loop adjacent to the active site in governing these minus-one nucleobase preferences (loop 7; described in greater detail below). Although these enzymes may be grouped by dinucleotide preferences, it is important to note that the identities of the single-stranded DNA substrate minus-two and plus-one bases are also influential and that other factors such as DNA integrity and secondary structures may also be key determinants [references above and (Holtz et al., 2013; Nabel et al., 2013; Rausch et al., 2009; Yu et al., 2004)].
Globular protein organization
Considerable high-resolution structural information is now available for several human APOBEC family members including APOBEC3A (A3A), APOBEC3C (A3C), APOBEC3F (A3F), and APOBEC3G (A3G) [most recent structures by (Bohn et al., 2013; Byeon et al., 2013; Kitamura et al., 2012; Li et al., 2012b; Siu et al., 2013) and older work referenced therein; reviewed recently by (Feng et al., 2014; Refsland and Harris, 2013; Salter et al., 2014; Shandilya et al., 2014; Siu et al., 2013)]. Each APOBEC family member is comprised of either one or two conserved zinc-coordinating domains and, in the case of the double-domain enzymes, the two halves are most likely joined by a flexible linker.
The C-terminal catalytic domain of A3G illustrates several of the family’s structural hallmarks (Figure 1B). First, each deaminase domain has an overall globular architecture comprised of five β-strands and six α-helices. The β-strands are organized into a hydrophobic core β-sheet core, and the α-helices are positioned around this core. β2 is split in A3G into β2 and β2′ but this feature is shorter or continuous in A3A, A3C, and A3F. The loops between secondary structure elements vary in length, composition, and conservation and are thought to have key roles in nucleic acid binding, local target selection, and overall function. Second, the catalytic site is characterized by a glutamate and a histidine in an HxE motif located at the end of a conserved α-helix and two cysteines in a CPx2-4C motif at the end of an adjacent conserved α-helix, which serves to coordinate a single zinc ion (α2 and α3 based on N- to C-terminal numbering of secondary structural elements; x represents a less conserved amino acid; Figure 1B). These α-helices are anchored into the globular structure through a conserved β-strand located in the center of a hydrophobic β-sheet that comprises the core of each domain. Each α-β-α catalytic motif is encoded by a single exon that most likely evolved (and continues to evolve) as a contiguous block enabling each of the five distinct APOBEC subgroups to be distinguished (elaborated below). This core α-β-α catalytic motif is most likely derived from the more ancient RNA and/or free-base deaminating enzymes (Chen et al., 2008; Conticello, 2008; Prochnow et al., 2007). Finally, high-resolution information of the non-catalytic domain of A3G and other double-domain enzymes is still lacking, but strong homology with the solved catalytic domain structures, including no significant insertions or deletions, indicates that they will have a similar overall structural organization.
Positive selection and copy number variation
One of the most fascinating hallmarks of the APOBEC family and a probable characteristic of all virus restriction factors is rapid evolution evidenced by elevated rates of amino acid substitution mutations and gene copy number variations (Harris et al., 2012; Johnson and Sawyer, 2009; Meyerson and Sawyer, 2011; Ortiz et al., 2006). A higher ratio of amino acid altering mutations relative to silent mutations is called positive selection. All of the A3 subfamily members show compelling evidence for positive selection (Duggal et al., 2013; Henry et al., 2012; Sawyer et al., 2004; Zhang and Webb, 2004), consistent with ancient and likely ongoing battles with viral pathogens. There is also tremendous variation in A3 gene copy number between branches of the mammalian phylogenetic tree (Conticello et al., 2005; Harris and Liddament, 2004; LaRue et al., 2009; LaRue et al., 2008; Münk et al., 2008) (Figure 1C). For instance, humans, chimpanzees, and rhesus macaques, and most other primates share a similar seven gene A3 locus comprised of three single domain genes (A3A/C/H) and four double domain genes (A3B/D/F/G) (Hultquist et al., 2011; Schmitt et al., 2011; Virgen and Hatziioannou, 2007). In contrast, mice have just one double-domain A3 gene (Harris and Liddament, 2004; Li et al., 2012a; Sanville et al., 2010). Other present day mammals have different copy numbers and overall gene organizations (Conticello et al., 2005; Harris and Liddament, 2004; LaRue et al., 2009; LaRue et al., 2008; Münk et al., 2008).
Genomic sequences have enabled investigators to deduce that the origin of the mammal-specific A3 gene subfamily most likely occurred through duplication of an ancestral AID/APOBEC1 locus (these genes are still located adjacent to one another in most vertebrates, but separated in others, such as primates, by a large chromosomal inversion) (Conticello et al., 2005; Harris and Liddament, 2004; LaRue et al., 2009; LaRue et al., 2008). The tandem head-to-tail organization of the ancestral A3 gene cluster provided the necessary substrate for rapid evolutionary diversification through multiple unequal crossing-over events, with some leading to gene expansions and others to contractions. Selective pressures from diverse viral infections most likely led to the expanded A3 gene repertoire observed in many present day mammals. Nevertheless, deletions are also common as observed by one A3 gene in rodents (due to a relatively ancient deletion early in the rodent lineage) and one in pigs (due to a relatively recent deletion specific to the Suidae lineage). Copy number and amino acid alternations also occur within a single species evidenced by the circulation of a common A3B deletion in humans (Kidd et al., 2007), the existence of seven distinct A3H haplotypes in humans that encode stable or unstable proteins (OhAinle et al., 2008; Ooms et al., 2013; Refsland et al., 2014; Wang et al., 2011), two human A3A translation initiation sites (Henry et al., 2012; Stenglein et al., 2010; Thielen et al., 2010), multiple transcription initiation sites and alternative splicing events (LaRue et al., 2008; Lassen et al., 2010; Münk et al., 2008; Santiago et al., 2008), a polymorphism in mice that affects splicing (exon composition) (Jónsson et al., 2006; Li et al., 2012a; Sanville et al., 2010), and the likelihood that many other variants await discovery and functional investigation.
Human APOBEC3 enzymes and HIV restriction
Deaminase-dependent restriction mechanism
Permissive and non-permissive cell fusion experiments deduced the existence of a dominant cellular factor that blocked the replication of human immunodeficiency virus type 1 (HIV-1) lacking its viral infectivity factor (Vif) (Madani and Kabat, 1998; Simon et al., 1998). In 2002, a subtractive hybridization approach yielded a variety of mRNA species expressed differentially between a permissive T-cell line called CEM-SS and its non-permissive parental line CEM [(Sheehy et al., 2002). One of these mRNAs (CEM15), independently named APOBEC3G and commonly abbreviated A3G (Harris et al., 2002; Jarmuz et al., 2002)], was sufficient to convert a permissive cell to a non-permissive phenotype (Sheehy et al., 2002). After demonstrating its potent DNA cytosine deaminase activity (Harris et al., 2002), a viral cDNA deamination mechanism was quickly unraveled (Harris et al., 2003; Mangeat et al., 2003; Zhang et al., 2003). This work provided a compelling mechanistic explanation for prior reports of strand-biased retroviral G-to-A mutation (Pathak and Temin, 1990; Vartanian et al., 1994; Wain-Hobson et al., 1995).
A3G-focused studies were followed by additional work demonstrating HIV-1 restriction in model cell-based systems using overexpression of A3F and multiple other family members [reviewed by (Desimmie et al., 2014; Malim and Bieniasz, 2012; Refsland and Harris, 2013)]. However, conflicting results were reported for all human A3 family members over the next decade, with some studies showing HIV-1 restriction and others not (except A3G). Therefore, a variety of experimental approaches clarified the role of APOBEC, including stable A3 expression in permissive T-cell lines, A3 knockdown and knockout studies in non-permissive T-cell lines, and Vif separation-of-function experiments in primary T lymphocytes was used to deduce that the combined activities of A3D, A3F, A3G, and A3H are responsible for HIV-1 restriction and G-to-A mutagenesis [(Hultquist et al., 2011; Ooms et al., 2013; Refsland et al., 2012; Refsland et al., 2014) and references therein].
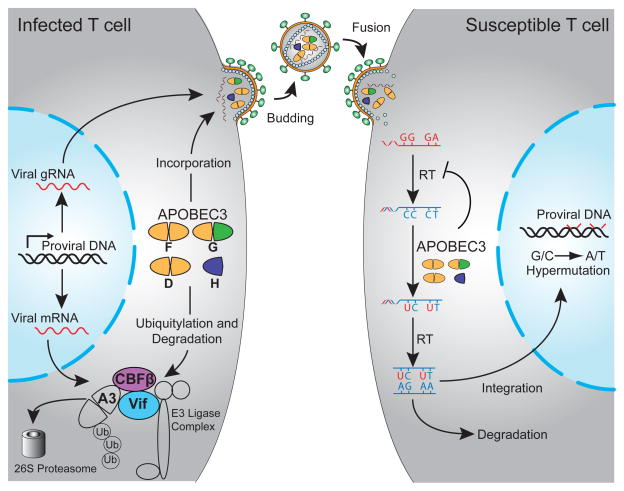
The current model for HIV-1 restriction is shown in Figure 2 [adapted from (Harris et al., 2012)]. In the absence of Vif, A3D, A3F, A3G, and/or A3H form cytoplasmic ribonucleoprotein complexes with HIV-1 Gag and one or more cellular RNA species [7SL, Y1, and viral genomic RNA have been implicated (Apolonia et al., 2015; Bogerd and Cullen, 2008; Strebel and Khan, 2008; Tian et al., 2007; Wang et al., 2007; Wang et al., 2008; Zhen et al., 2012)]. RNA binding requires the nucleocapsid domain of Gag (although heterologous RNA-binding proteins can substitute), and the importance of an RNA bridge is highlighted by several studies showing the sensitivity of Gag-A3 complexes to RNase A treatment (Alce and Popik, 2004; Apolonia et al., 2015; Douaisi et al., 2004; Schafer et al., 2004; Svarovskaia et al., 2004). A3D, A3F, A3G, and A3H have been observed to oligomerize in living cells, and this property correlates with the capacity to restrict HIV-1 infectivity (Li et al., 2014). Although precise mechanistic details will require additional investigation, RNA-protein interactions clearly mediate the packaging of restrictive A3 enzymes into assembling HIV-1 particles.
Figure 2. Model for HIV-1 restriction by APOBEC3 enzymes and virus protection by Vif.
HIV-1 infection of T cells allows expression of Vif, which recruits CBF-β to form an E3 ubiquitin ligase complex that degrades several different A3 proteins (A3D, A3G, A3F, and A3H). In the absence of Vif, these A3 proteins can package into virions. During subsequent infection of susceptible cells, these A3 proteins deaminate viral cDNA cytosines to uracils and directly impede reverse transcription. Viral cDNA uracils template the insertion of genomic strand adenines and result in hallmark G-to-A hypermutations. See the text for details [adapted from (Harris et al., 2012), based upon prior studies].
During or shortly after budding and before the conical capsid becomes fully closed (matures), a significant fraction of packaged A3 enzymes enter the viral core (i.e., become encapsidated). This step of the restriction mechanism is evidenced by chimeric A3 enzymes and amino acid substitution mutants that package, but somehow fail to breach the core and inhibit viral infectivity (Donahue et al., 2015; Haché et al., 2005; Song et al., 2012). Upon receptor binding and fusion, the conical capsid is deposited in the cytosol of a target cells, reverse transcription occurs concomitant with RNase H activity to degrade template viral genomic RNA, and single-stranded viral cDNA becomes susceptible to the mutagenic activity of an encapsidated A3 enzyme. The viral reverse transcriptase enzyme uses the resulting viral cDNA uracils to template the insertion of genomic strand adenines. A single round of virus replication and A3 mutagenesis can suppress viral infectivity by several logs and convert up to 10% of all genome plus-strand guanines into adenines, accounting for the phenomenon of retroviral G-to-A hypermutation (Harris et al., 2003; Liddament et al., 2004; Mangeat et al., 2003; Yu et al., 2004; Zhang et al., 2003). Interestingly, although a single viral genome can be co-mutated by two different A3 enzymes in model single-cycle experiments (Liddament et al., 2004), co-mutated sequences rarely occur in primary HIV-1 isolates, suggesting that the number of A3 molecules per particle may be low during pathogenic infections (Ebrahimi et al., 2012; Sato et al., 2014).
Deaminase-independent mechanism
Multiple studies have noted that significant HIV-1 restriction can still occur upon overexpression of catalytically defective variants of A3G and A3F (Chaurasiya et al., 2014; Holmes et al., 2007a; Holmes et al., 2007b; Iwatani et al., 2007; Newman et al., 2005). This deaminase activity-independent effect appears to be greater for A3F than for A3G (Albin et al., 2014; Browne et al., 2009; Holmes et al., 2007a; Kobayashi et al., 2014; Schumacher et al., 2008). Primary cell studies also suggest a deaminase-independent component (Gillick et al., 2013). A number of models have been proposed for this catalytic activity-independent restriction mechanism including binding genomic RNA to impede reverse transcription, binding tRNA to prevent reverse transcription initiation, binding reverse transcriptase directly, and others [e.g., (Gillick et al., 2013; Holmes et al., 2007a; Wang et al., 2012); reviewed by (Holmes et al., 2007b)]. However, the prevailing model to explain this phenomenon is genomic RNA binding, which causes a steric block to reverse transcription. Because A3G and A3F are capable of binding both RNA and single-stranded DNA, such binding effectively diminishes the overall kinetics of reverse transcription. Interestingly, although a minority of HIV-1 restriction is attributable to this mechanism, deaminase-independent mechanisms appear dominant for A3-mediated restriction of several other parasitic elements (detailed below).
Vif-mediated counterdefense mechanism
Early studies showed that Vif function was required in virus-producing, but not in target cells, and that the absence of Vif in the producer cells somehow resulted in less viral cDNA accumulation in target cells (Gabuzda et al., 1992; von Schwedler et al., 1993). The discovery of A3G led rapidly to unraveling the mechanism of Vif-mediated counterdefense (Conticello et al., 2003; Kao et al., 2003; Marin et al., 2003; Sheehy et al., 2003; Stopak et al., 2003; Yu et al., 2003). A major clue was the observation that fluorescently-tagged A3G appeared brighter in cells without Vif, than in cells co-expressing Vif [rather than, for instance, re-localization; e.g., (Conticello et al., 2003)]. Similar decreases in A3G intensity were observed by immunoblot comparisons of cell extracts with and without co-expressed Vif [e.g., (Conticello et al., 2003)]. In both instances, A3G signal could be recovered by treating cells with proteasome inhibitors such as MG132 [e.g., (Conticello et al., 2003)]. Several groups thereby converged upon a polyubiquitination and degradation mechanism (Conticello et al., 2003; Kao et al., 2003; Marin et al., 2003; Sheehy et al., 2003; Stopak et al., 2003; Yu et al., 2003). Proteomic studies and genetic experiments implicated an E3 ubiquitin ligase consisting of CUL5, ELOB, ELOC, and RBX1 (Yu et al., 2003).
Over the ensuing decade, additional progress on Vif function was made through a wide variety of genetic and virologic studies but broader progress was constrained due to purification issues that prevented biochemical and structural approaches. Many labs invested significant effort on Vif purification in heterologous systems, such as E. coli, with mostly negative results. Speculating that this problem may be due to a missing cellular co-factor, a series of quantitative proteomic experiments revealed the transcription co-factor CBF-β as an abundant Vif-interacting protein (Jäger et al., 2011; Zhang et al., 2011). CBF-β co-precipitated with CUL5 and ELOC, but only in the presence of Vif, suggesting membership in the Vif-ligase complex itself. Indeed, CBF-β enabled Vif expression in E. coli, and the purification of a Vif-CBF-β ubiquitin ligase complex with polyubiquitination specificity for HIV-restrictive (A3G), but not non-restrictive (A3A) enzymes. Knockdown studies demonstrated that CBF-β was required for Vif expression and function against restrictive A3 enzymes. These advances led to a revised model for Vif-mediated A3 counteraction in which Vif hijacks CBF-β to nucleate the formation of an active ubiquitin ligase complex that protects HIV-1 from lethal restriction (Figure 2). Many aspects of this model, including an extensive interface between Vif and CBF-β, have been validated recently through the first X-ray crystal structure of the HIV-1 Vif ligase complex (Guo et al., 2014).
Conservation of the A3 restriction and Vif counteraction mechanisms
As described above, all mammals encode at least one A3 and often multiple A3s. The A3-mediated restriction mechanism is conserved since enzymes from many different mammals elicit retrovirus restriction activity (frequently against HIV-1 or HIV-based vectors). However, lentiviruses are only known to exist in a small subset of mammals. A comprehensive examination of restriction and Vif-mediated counteraction activities using host A3 enzymes and HIV-1, simian immunodeficiency virus (SIV) mac239, feline immunodeficiency virus (FIV), bovine immunodeficiency virus (BIV), and maedi-visna virus (MVV) Vif proteins demonstrated conservation of both mechanisms (LaRue et al., 2010). As expected, each Vif protein selectively degraded the A3 enzyme of its lentiviral host. However, some cross degradation was observed as might also be expected because the A3 enzymes are relatively conserved (especially in comparison to Vif). Surprisingly, recent work has demonstrated that CBF-β is specifically required for function of primate lentiviral Vif proteins, but not for non-primate Vifs (Ai et al., 2014; Han et al., 2014; Yoshikawa et al., 2014; Zhang et al., 2014). These data indicate that either the Vif proteins of FIV, BIV, and MVV do not require a CBF-β-like factor, or these viruses have evolved to use one or more other as-yet-unknown cellular factors. Additional work will be needed to distinguish between these intriguing possibilities.
Evidence for HIV-1 restriction and hypermutation in vivo
Prior to the discovery of APOBEC3 enzymes and the elucidation of HIV-1 Vif counteraction mechanism, many studies reported strand-biased G-to-A mutations in patient-derived viral sequences [e.g., (Janini et al., 2001; Vartanian et al., 1994; Wain-Hobson et al., 1995)]. These and other reports have combined to suggest that multiple A3 enzymes can impact the virus in vivo. This is clearly evidenced by the fact that both 5′-GG-to-AG and 5′-GA-to-AA mutations are observed in patient-derived sequences and in viral sequences from humanized models [i.e., A3G and A3D/F/H attributable mutations (Krisko et al., 2013; Sato et al., 2014)]. However, because these data rely on standard nucleic acid purification and PCR amplification procedures, which survey all available substrates, it is possible that these hypermutations represent dead-end replication intermediates that may never have completed reverse transcription and resulted in a productive infection. In other words, some fraction of these sequences are genetic dead-ends that may never have been propagated regardless of APOBEC (i.e., interesting artifacts recovered through technology). This possibility is supported by far fewer viral G-to-A mutations observed in analyses of viral RNA in sera (from virus or virus-like particles) compared to viral DNA from cells of infected individuals [integrated or non-integrated viral DNA sequences; e.g., (Sato et al., 2014)].
Considerable effort has therefore been invested in characterizing viral and/or host genetic variations in an attempt to gauge the impact of the A3 restriction mechanism in vivo. Host genetic studies have the potential to be especially informative. However, most studies have shown conflicting effects or a clean negative result. For instance, a deletion of the full A3B gene that is common in some Southeast Asian populations provided an opportunity to unambiguously show that the encoded protein is not a factor in HIV-1 infection rates, viral loads, and CD4-positive T cell counts, and has no measurable effect on virus replication in primary cells ex vivo (Imahashi et al., 2014). In contrast, recent studies comparing stable and unstable A3H proteins have indicated that some naturally occurring viral variants with hypo-functional Vif alleles may be susceptible to restriction by stable A3H enzymes (Ooms et al., 2013; Refsland et al., 2014). Moreover, HIV-1 loads appeared lower and T cell counts higher overall in patients with stable A3H proteins in comparison to those with unstable proteins, suggesting that A3H may help to control viral pathogenesis in vivo (Ooms et al., 2013). Additionally intriguing is that a significant subset of global HIV-1 isolates may have Vif proteins that are specifically hypofunctional toward A3H because a large proportion of the human population is homozygous for unstable A3H haplotypes, therefore exerting little or no selective pressure to maintain full Vif function (Refsland et al., 2014). Given the potential potency of A3-mediated HIV-1 restriction and the plethora of circulating haplotypes, these enzymes should be further investigated as candidate factors in long-term non-progression, elite control, and overt resistance to infection.
HIV-1 drug resistance studies have also provided valuable portals into understanding the potential impact of A3 enzymes in vivo. Several common antiretroviral drug resistance mutations are potential A3-mediated G-to-A events (Berkhout and de Ronde, 2004; Haché et al., 2006; Jern et al., 2009; Kim et al., 2010; Kim et al., 2014; Mulder et al., 2008; Pillai et al., 2008; Sadler et al., 2010). For instance, deep-sequencing studies have revealed that drug resistance mutations are more common in HIV-1 cultures in the presence of A3G than in its absence, and that potential A3-mediated drug resistance mutations pre-exist in clinical HIV-1 populations (Kim et al., 2010; Mulder et al., 2008). In addition, adaptive immune responses exert strong selective pressures on HIV-1, and sequencing studies have shown that many putative immune escape mutants correspond to potential A3-mediated G-to-A editing sites (Kim et al., 2014). Finally, significant proportions of transmission-associated mutations also correspond to potential A3-mediated G-to-A mutation events (Wood et al., 2009). These mutations may have been required for escaping immune responses and/or adapting to differences in host factor compositions (restriction or dependence factors) during or shortly after transmission. Taken together, these studies strongly implicate A3 mutagenesis in providing HIV-1 with mutational fuel and evolutionary diversity upon which natural adaptive immune and pharmacologic pressures may act. As detailed below, many concepts in APOBEC restriction extend far beyond HIV-1 to multiple DNA parasites.
Human APOBEC3 enzymes and HTLV-1 restriction
HTLV-1 restriction
Cell culture over-expression experiments have indicated that multiple A3 enzymes can restrict the infectivity of human T-cell leukemia virus type 1 (HTLV-1) (Derse et al., 2007; Mahieux et al., 2005; Navarro et al., 2005; Ooms et al., 2012; Sasada et al., 2005; Strebel, 2005). However, in head-to-head experiments, HTLV-1 appears considerably more resistant than HIV-1 to restriction by A3D, A3F, and A3G (Derse et al., 2007; Navarro et al., 2005; Ooms et al., 2012). Only stably expressed haplotypes of A3H caused strong restriction of both viruses (Ooms et al., 2012). HTLV-1 also appeared preferentially susceptible to restriction by A3A and A3B (Ooms et al., 2012). Both deaminase-dependent and independent mechanisms have been noted (Derse et al., 2007; Mahieux et al., 2005; Navarro et al., 2005; Ooms et al., 2012; Sasada et al., 2005; Strebel, 2005). The impact of these enzymes on HTLV-1 infectivity in primary lymphocytes has yet to be examined in part due to technical challenges of lower virus infectivity and strong preferences for cell-to-cell, rather than cell-free transmission.
However, in contrast HIV-1 sequences derived from patient samples, characteristic G-to-A mutations are relatively rare in HTLV-1 sequences. An initial study of HTLV-1 in 10 patients found no evidence for hypermutation (Mahieux et al., 2005). A subsequent larger study analyzed the entire sequence of HTLV-1 proviruses from 60 adult T-cell leukemia (ATL) patients and 10 HTLV-1 carriers and found inactivating mutations in nearly 50% of cases with G-to-A changes in an A3G context accounting only for a small subset of nonsense changes (Fan et al., 2010). These observations are consistent with the fact that HTLV-1 provokes strong cytotoxic T cell responses in vivo, which often select for mutational inactivation of dominant viral epitopes encoded by plus-strand genes such as Tax (Bangham et al., 2014). This manifests in ATL as oligoclonally expanded pools of T cells, with each pool characterized by a single replication-defective provirus insertion (Bangham et al., 2014).
A3 counteraction mechanism of HTLV-1
As mentioned above, HTLV-1 is relatively resistant to restriction by A3G, in comparison to Vif-deficient HIV-1 (Derse et al., 2007; Navarro et al., 2005; Ooms et al., 2012). This resistance phenotype correlated with lower levels of encapsidated A3G. An elegant mutational analysis revealed that resistance was not due to a viral accessory protein, rather to a unique C-terminal extension of the viral nucleocapsid (NC) protein (Derse et al., 2007). If 20 residues of this extension were deleted or mutated, then HTLV-1 became exquisitely sensitive to A3G restriction through an encapsidation and hypermutation mechanism analogous to that described above for Vif-deficient HIV-1. This work was significant by its demonstration of a novel mechanism of A3 resistance by exclusion. Thus, other potentially susceptible DNA parasites may use a similar exclusion mechanism or possess a novel strategy for preventing A3 enzymes from attacking single-stranded DNA replication intermediates.
Retrovirus restriction by murine A3 – in vivo insights from animal models
MMTV restriction
The first direct evidence that APOBEC family members protect against retroviral infection in vivo was obtained by studying mouse mammary tumor virus (MMTV) infection of A3-defective mice (Okeoma et al., 2007). A3 mRNA is detectable in a wide variety of wild-type mouse tissues with the highest levels occurring in the thymus and lymph nodes, suggesting preferential activity in lymphoid tissues (Okeoma et al., 2007). C57BL/6 (B6) mice were infected subcutaneously with MMTV and compared to infections of A3-mutant mice created by gene trap technology. The mutant (designated A3−/−) expressed an in-frame fusion of the first four A3 exons to the β-galactosidase gene and was confirmed to lack deaminase activity as well as inhibitory activity after packaging into HIV virions. Since MMTV must infect and activate B and T cells during transmission to target mammary epithelial cells (Golovkina et al., 1992; Golovkina et al., 1998; Held et al., 1993a; Held et al., 1993b), lymphocytes were examined in infected mice in the presence and absence of functional A3. Two-fold higher levels of activated B and T cells were observed in A3−/− mice at 4 to 6 days post-infection compared to those obtained from wild-type or heterozygous mice, consistent with a dominantly active A3 enzyme. Levels of MMTV DNA in lymph nodes also were increased by 10-fold in the homozygous A3-mutant animals (Okeoma et al., 2007). Interestingly, A3 characteristic G-to-A mutations were rarely found in MMTV sequences, implying that this virus restriction mechanism may be largely and possibly exclusively deamination-independent (MacMillan et al., 2013; Nair et al., 2014). Although some mechanistic details remain to be determined, these experiments clearly demonstrated that endogenous A3 functions in vivo to limit MMTV infection.
Interestingly, mice express two different A3 isoforms (Abudu et al., 2006; Jónsson et al., 2006; Li et al., 2012a; Sanville et al., 2010; Takeda et al., 2008). All nine exons combine to encode a longer isoform in BALB/c mice, whereas eight exons encode a shorter isoform in B6 mice due to an exon 5 skip during splicing. Both isoforms have intact N- and C-terminal deaminase domains but, unlike the human double-domain enzymes, the N-terminal domain is responsible for DNA cytosine deamination (Hakata and Landau, 2006; Jónsson et al., 2006; MacMillan et al., 2013). In addition, BALB/c mice express lower levels of A3 compared to B6 mice, suggesting that specific mouse strains, such as BALB/c, may be less restrictive for the replication of murine retroviruses (Li et al., 2012a; Okeoma et al., 2009a; Okeoma et al., 2009b; Sanville et al., 2010; Takeda et al., 2008). A3 mRNA is expressed in many tissues, including lymphocytes, one of the major cell types infected by MMTV (Golovkina et al., 1998), thus providing a primary barrier to the establishment of infection. A3 is also expressed in mammary epithelial cells, providing a secondary barrier to the establishment of infection and helping to prevent milk-borne viral transmission (Okeoma et al., 2010). Therefore, A3 most likely restricts MMTV replication at multiple steps during virus replication and transmission in vivo. Consistent with function against both exogenous viruses and endogenous retroelements (discussed below), A3-knockout mice do not appear to have a defect in development, survival, or fertility (Mikl et al., 2005).
MuLV restriction
Multiple early studies indicated that murine leukemia viruses (MuLV) are considerably more resistant to murine A3 than to enzymes from other species, such as human A3G (Abudu et al., 2006; Bishop et al., 2004; Langlois et al., 2009; Rulli et al., 2008). This observation led to suggestions that MuLVs were naturally resistant to the A3 enzyme of its host species, and that murine A3 was ineffective in controlling these viral infections. However, several studies have used wild-type and A3-null mice (true knockout and gene trap models) to demonstrate that A3 restricts MuLV infection in vivo (Langlois et al., 2009; Low et al., 2009; Mikl et al., 2005; Takeda et al., 2008). A3-null animals showed 10- to 100-fold increases in overall numbers of productively Friend (F)-MuLV infected cells in both the spleen and the bone marrow (Takeda et al., 2008). Similar overall increases in infected cell numbers and proportional increases in viral loads were reported for Moloney (M)-MuLV infection of A3 null animals compared to heterozygous or wild-type littermates (Low et al., 2009). Furthermore, the differences between heterozygous and mutant A3 were only observed within the first 10 days after virus introduction (Low et al., 2009). As anticipated, the presence of two copies of A3 in mice prolonged the latency of M-MuLV-induced T-cell lymphomas and decreased metastasis to the kidneys (Low et al., 2009). These results are consistent with the idea that APOBEC family proteins serve as part of the innate immune system, which is important at early times after infection to induce an adaptive response (Moris et al., 2014). Neither of these MuLV studies, as well as an independent series of experiments (Langlois et al., 2009), reported evidence for MuLV G-to-A hypermutation by endogenous A3. Nevertheless, more sensitive methods such as deep sequencing suggest that both MuLV and MMTV may accumulate low levels of G-to-A mutation (Barrett et al., 2014; MacMillan et al., 2013; Smith et al., 2011).
Bone marrow-derived cells previously were shown to be required for efficient M-MuLV infection (Brightman et al., 1990; Davis et al., 1987; Li and Fan, 1990). Because increased virus levels also were observed in bone marrow after infection of A3-mutant mice, primary bone-marrow-derived dendritic cells (BMDCs) were infected in culture with Moloney virus that had packaged HA-tagged A3 Δexon5 (the isoform expressed in B6 mice) (Low et al., 2009). As anticipated, the presence of B6 A3 reduced the infectivity of M-MuLV by ~2-fold in BMDC lacking functional A3 expression. More surprising was the observation that infectivity of M-MuLV with packaged functional A3 (Δexon5) could be reduced further by infection of BMDCs expressing A3 (Δexon5) (Low et al., 2009). These data suggested that A3 in the recipient cells could also suppress the infectivity of murine retroviruses.
Another interesting story emerged from studies of an endogenous MuLV (AKV). Similar to the work described above for MuLV, splenocytes and thymocytes purified from A3-null animals were >10-fold more susceptible to infection by AKV (Langlois et al., 2009). However, here restriction correlated with a significant increase in viral G-to-A mutations (Langlois et al., 2009). Moreover, since this experiment was performed ex vivo, this study provides a second clear example of endogenous A3 restricting the incoming viral particles in target cells (a still contentious issue discussed further below). Nevertheless, these studies demonstrate that endogenous A3 controls MuLV infection and pathogenesis in vivo. These conclusions alone are important but additionally interesting by implying an evolutionary advantage for retroviruses to evolve partial resistance, rather than complete resistance, to A3 restriction and mutagenesis.
The murine A3 locus also has been identified as the Resistance to Friend Virus (Rfv3) gene, shown previously to regulate the neutralizing antibody response to Friend virus infection (Santiago et al., 2008). The underlying mechanism is complex and not yet fully understood. An indirect possibility is that the increased antigenic diversity of hypermutated and even non-infectious viruses may provoke stronger AID-dependent adaptive immune responses (Smith et al., 2011). A direct explanation is that murine A3 may contribute directly to somatic mutation of expressed antibody gene DNA sequences and, together with AID, create more robust antibody responses (Halemano et al., 2014). This is supported by observations of increased levels of antibody gene G-to-A and C-to-T mutations in wild-type compared to A3-null animals infected in parallel with F-MuLV (Halemano et al., 2014). However, this single report contrasts with many prior studies indicating total ablation of antibody gene somatic hypermutation in AID-null animals that presumably still expressed endogenous A3 [original work by (Muramatsu et al., 2000); reviewed in (Di Noia and Neuberger, 2007)]. In any event, these studies are significant because they highlight potential synergy and crosstalk between the innate A3 restriction system and the adaptive antibody response to retrovirus infection.
The contribution of murine APOBEC1 and AID to retrovirus restriction is less clear. One study implicated APOBEC1 in F-MuLV restriction, as both G-to-A and C-to-T hypermutations were detected in 5′-TC motifs using differential DNA denaturation PCR (3D-PCR) of genomic DNA samples at multiple time points after infection of newborn OF-1/Swiss mice (Petit et al., 2009). In contrast, a more recent study infected B6 animals with Friend virus complex, evaluated acute infection levels, and found no discernable difference between APOBEC1 wild-type and null animals (Barrett et al., 2014). APOBEC1-null animals were also analyzed in parallel with A3-null animals in the AKV study described above, and G-to-A mutation levels were not above background by deep sequencing (Langlois et al., 2009). Together, these results suggest that A3, but not APOBEC1, restricts F-MuLV infectivity and causes hypermutations during viral replication in mice. Strain-specific differences between Swiss versus B6 mice may explain these observed differences. In addition, Abelson MuLV infection of mice induced AID in non-germinal center B cells, which then triggered the DNA-damage response and restricted proliferation of infected cells (Gourzi et al., 2006, 2007). Since no viral G-to-A mutations were observed, these data suggest that APOBEC family members may use multiple mechanisms to activate innate immunity to viruses.
A3 counteraction mechanisms of murine retroviruses
As mentioned above, several studies have indicated that murine retroviruses are more resistant to murine A3 than to enzymes from other species, such as human A3G (Abudu et al., 2006; Bishop et al., 2004; Langlois et al., 2009; Rulli et al., 2008). Analogous to the A3 counteraction mechanism of HTLV-1, some work has indicated a virion exclusion mechanism in which cytoplasmic A3 is simply not packaged into assembling particles (Abudu et al., 2006; Doehle et al., 2005). In support of this idea, cell culture studies with epitope-tagged proteins have indicated that murine A3 packages into MuLV particles less efficiently than human A3G. In addition, MuLV protease may cleave packaged A3 and provide a second layer of defense against restriction (Abudu et al., 2006).
Recent data indicate that glyco-Gag affords protection from the anti-viral effects of murine A3 (Boi et al., 2014; Kolokithas et al., 2010; Nitta et al., 2012; Stavrou et al., 2013). Almost all MuLVs encode a longer glycosylated form of the capsid precursor protein Gag (glyco-Gag), which originates from translation initiation at a CUG start codon upstream of the normal cytoplasmic Gag start codon (Berlioz and Darlix, 1995). This glyco-Gag protein has an N-terminal 88 amino acid extension with a signal peptide that directs synthesis of the protein across the ER membrane, allowing glycosylation and transport to the cell surface. Subsequently, the glycosylated Gag is cleaved into two proteins of 55 and 40 kDa. The latter is maintained as a type II transmembrane protein, which is necessary for a late step of viral assembly as well as neurovirulence, whereas the C-terminal 40 kDa protein is released from cells (Fujisawa et al., 1997; Low et al., 2007). Glycosylation and post-translational processing may differ according to the cell type infected (Fujisawa et al., 1997).
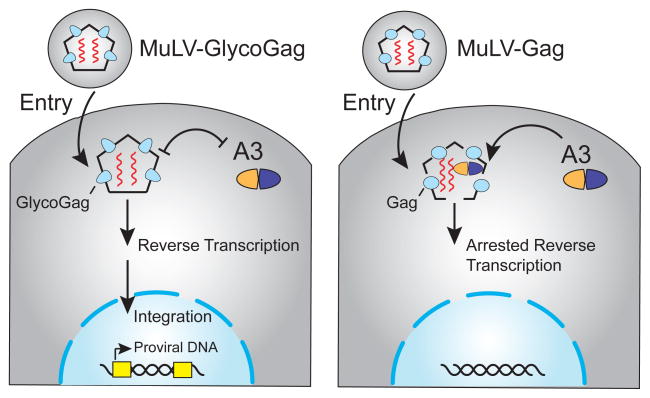
Several studies have shown that glyco-Gag defective particles are less infectious than wild-type MuLV particles (Boi et al., 2014; Kolokithas et al., 2010; Nitta et al., 2012; Stavrou et al., 2013). This restriction phenotype is largely alleviated in A3-deficient cells and animals (Boi et al., 2014; Kolokithas et al., 2010; Stavrou et al., 2013). Moreover, glyco-Gag-defective viruses reverted to wild-type function during infections of A3-expressing animals, but not A3-null animals, demonstrating the importance of glyco-Gag in antagonizing A3-dependent restriction (Stavrou et al., 2013). Recent data have also indicated that loss of N-linked glycosylation sites in glyco-Gag result in increased hypermutation by A3 (Rosales Gerpe et al., 2015). Interestingly, glyco-Gag-mutant virions are less stable than wild-type particles during ultracentrifugation with detergent (Stavrou et al., 2013). Further, A3 incorporation during cell culture and in vivo replication caused defects in reverse transcription when glyco-Gag was absent (Boi et al., 2014; Stavrou et al., 2013). These studies combined to suggest a mechanism in which glyco-Gag stabilizes the viral core and shields viral reverse transcription complexes from the restrictive activities of A3, as well as affording protection from other innate immune effector proteins such as the DNA nuclease Trex1 (Stavrou et al., 2013) (Figure 3).
Figure 3.
Model for glyco-Gag protection from restriction by murine A3. The left image depicts glyco-Gag as an oblong blue shape that prevents A3 from accessing reverse transcription complexes. Capsids are primarily composed of the classical Gag (circles not depicted on the left panel). The right panel depicts Gag as a blue oval that causes the capsid to be more loosely formed and susceptible to the A3-mediated block in reverse transcription. Although low levels of G-to-A mutation have been reported, these changes are a minor outcome of A3 activity and are not depicted for clarity. See the text for additional details.
A3 counteraction mechanisms of other retroviruses
The foamy viruses (FVs) use the Bet protein to antagonize APOBEC. Bet, like Vif, is encoded at the 3′ end of the retroviral genome and is not required for virus replication in cell lines (Baunach et al., 1993). Mutations in the feline FV bet open reading frame lead to reduced viral titers in CRFK (feline) cells expressing feline A3s and increased G-to-A hypermutations (Lochelt et al., 2005). Nevertheless, Bet has no sequence homology to Vif and appears to act by a different mechanism than either Vif or glyco-Gag (Chareza et al., 2012; Lochelt et al., 2005; Russell et al., 2005). Unlike Vif, which acts as an adapter between APOBEC and an E3 ligase, Bet does not induce A3 degradation, but prevents packaging of particular A3s into foamy virus particles. Feline FV Bet has been shown to bind to feline A3 (Lochelt et al., 2005), and prototype FV Bet can prevent human A3G dimerization and function (Jaguva Vasudevan et al., 2013; Perkovic et al., 2009; Russell et al., 2005). Bioinformatic analysis has identified six conserved motifs encoded within the bel2 portion of the bet mRNA, and these motifs appear to be required for creating high affinity complexes between Bet and A3 (Lukic et al., 2013). Interestingly, Bet is expressed at high levels in infected cells, both in culture and in animals, consistent with inactivation of A3 by Bet binding or sequestration (Alke et al., 2001; Lukic et al., 2013). In addition, A3s may be able to inhibit FV replication in both producer as well as target cells (Lochelt et al., 2005), which may be linked to the fact that spumaviruses can initiate reverse transcription in producer cells (Moebes et al., 1997). Therefore, FVs antagonize A3-induced hypermutation using a mechanism distinct from those described above.
Interestingly, the betaretroviruses lack a common mechanism to avoid APOBEC-mediated restriction. For example, the Mason-Pfizer monkey virus (MPMV) has been reported to be resistant to expression rhesus monkey A3G by excluding this enzyme from virions (Doehle et al., 2006). The mechanism for A3G exclusion is unclear. Nevertheless, mouse A3, but not rhesus A3G, is bound by MPMV Gag and packaged into viral particles where it inhibits viral infectivity (Doehle et al., 2006). In contrast, the betaretrovirus MMTV packages A3, which then blocks subsequent reverse transcription (MacMillan et al., 2013). Like many MuLVs, the packaged A3 caused only low-level hypermutation of the proviruses that escaped A3 inhibition (MacMillan et al., 2013). Effects of A3 on MMTV replication were most apparent in mouse strains that express high levels of this deaminase (Okeoma et al., 2009b), whereas the related TBLV, which has an altered LTR and induces T-cell lymphomas, replicates well in mouse strains that express either high or low levels of A3 (Bhadra et al., 2009; Meyers et al., 1989; Mustafa et al., 2003). Furthermore, unlike MPMV, MMTV, and TBLV, complex retroviruses express a doubly spliced mRNA and the Rem precursor protein (Indik et al., 2005; Mertz et al., 2005). The Rem precursor is cleaved into an N-terminal signal peptide (Rem-SP) that serves a Rev-like function, whereas the function of the C-terminal 203 amino acid protein has not been determined (Byun et al., 2012; Byun et al., 2010). One possibility is that the activity of the Rem precursor or the C-terminus provides the role of the glycosylated Gag protein of MuLVs.
APOBEC3 involvement in endogenous virus and transposon restriction
Although the role of APOBECs as anti-viral factors was initially shown with exogenous retroviruses, including HIV-1, subsequent studies demonstrated fundamental roles for these enzymes in suppressing the mobilization of endogenous retroviruses and retrotransposons. These parasitic elements occupy a large fraction of the human genome and, although mostly defective, the remaining functional elements must be exquisitely controlled to prevent excessive genome damage and potential genetic catastrophe.
One major family of endogenous parasites that is controlled by APOBEC proteins is comprised of autonomous LINE-1 (L1) transposons and related non-autonomous Alu transposons, which require L1 gene products for transposition. These elements rely on integration-primed reverse transcription for copying from one location of the genome and inserting in another (i.e., copy and paste mechanism). Initial studies demonstrated L1 restriction by overexpressing various A3 and AID members in cell culture experiments (Bogerd et al., 2006b; Chiu et al., 2006; Kinomoto et al., 2007; MacDuff et al., 2009; Muckenfuss et al., 2006; Stenglein and Harris, 2006). Restriction did not correlate with A3 localization to the nuclear compartment, where L1 reverse transcription occurs (Stenglein and Harris, 2006). In all of these instances, the inhibition of transposition occurred without detectable G-to-A mutation, suggesting that the major mechanism of inhibition may be linked to the strong RNA-binding activity of these enzymes. Consistent with this idea, AID overexpression inhibited production of L1 ORF1 [equivalent to Gag capsid proteins (Metzner et al., 2012)]. However, a recent study blocked uracil DNA repair and observed some L1 G-to-A mutation (Richardson et al., 2014). Thus, similar to other examples discussed earlier, the mechanism of L1 and Alu restriction by A3 family members may involve both deaminase-dependent and -independent activities.
However, a major drawback to the aforementioned studies is a dependence on A3/AID overexpression and L1/Alu transposition from a reporter plasmid inserted into chromosomal DNA. Only two studies have attempted to address the impact of endogenous A3 enzymes on transposition. One study depleted endogenous A3B in both HeLa and human embryonic stem cell lines and observed a significant 3 to 5-fold increase in L1 transposition from a transfected reporter plasmid (Wissing et al., 2011). The second study reported an inverse correlation between L1 mobility in primates and expression levels of endogenous A3B and PiWi proteins (Marchetto et al., 2013). Thus, more work will be necessary to establish the precise mechanisms and the identities of the A3 family members that are most relevant to suppressing the transposition of L1 and Alu elements.
Endogenous retroviruses are also substrates for restriction and hypermutation by A3 family members. Like exogenous viruses, but unlike L1/Alu elements, these parasites require terminal long-terminal repeats (LTRs) for reverse transcription and gene expression, most of which are inactive (Bannert and Kurth, 2004). Initial studies demonstrated that mouse intracisternal A particles (IAPs) and MusD elements are susceptible to restriction and hypermutation by overexpressed A3 enzymes (Bogerd et al., 2006a; Esnault et al., 2005; Esnault et al., 2006). Interestingly, the inhibition and hypermutation of LTR-dependent elements is also observed by overexpressing A3 enzymes in heterologous systems, as evidenced by suppression of Ty1 element replication in yeast (Dutko et al., 2005; Schumacher et al., 2005). These studies suggest that at least one aspect of the restriction mechanism does not require additional mammalian proteins as cofactors. Although most mechanistic studies have been performed in model systems, bioinformatics approaches have revealed that significant fractions of some, but not all, endogenous retroviruses have been rendered inactive by a G-to-A hypermutation mechanism, most likely mediated by A3 enzymes based on hallmark signatures (Anwar et al., 2013; Jern and Coffin, 2008; Jern et al., 2007; Lee et al., 2008).
DNA viruses and the APOBEC family
Although the vast majority of information about APOBEC inhibition of viruses pertains to retroviruses and retroelements, APOBEC has been reported to be a restriction factor for multiple DNA-containing viruses [reviewed by (Moris et al., 2014)]. Hepatitis B virus (HBV) is one of the most studied instances of APOBEC-mediated inhibition of a DNA virus. HBV is a pararetrovirus that is a major cause of liver cirrhosis and cancer (Beggel et al., 2013; Bonvin and Greeve, 2008). Similar to foamy virus, HBV has a reverse transcriptase that copies packaged pregenomic RNA into DNA within the nascent capsid of the producer cells (Jones and Hu, 2013). Unlike retroviruses, the reverse trascriptase is covalently attached to the 5′ end of the minus-strand DNA and does not fully complete plus strand synthesis within producer cells. The remaining single-stranded DNA region represents a natural target for APOBEC family enzymes (Beggel et al., 2013). An initial report using Huh7 hepatoma cells suggested that HBV DNA does not exhibit G-to-A hypermutation after transfection of A3G, but that pregenomic RNA was inefficiently packaged (Seppen, 2004; Turelli et al., 2004). A3G appeared associated with viral cores in the cytoplasm, and similar observations were made for A3B, A3C, A3F, and A3G in another hepatoma cell line (Suspène et al., 2005; Turelli et al., 2004). Further investigation revealed that G-to-A hypermutations were observed at low frequencies (< 1 in 10 genomes) using transfection of HBV and A3G in another hepatoma cell line (Rosler et al., 2004). Both G-to-A and C-to-T substitutions were observed with A3B, A3F, and A3G, but not A3C, suggesting that both strands of HBV DNA may be susceptible to deamination (Suspène et al., 2005). AID also has been reported to be associated with an HBV ribonucleoprotein complex and to deaminate viral RNA in tissue culture experiments (Liang et al., 2013). Recent experiments have interrogated endogenous APOBEC3 proteins in multiple cell culture models. Treatment of hepatocyte cells with interferon α or an antibody to crosslink the lymphotoxin β receptor results in induction of A3A and A3B, respectively, and in G-to-A hypermutations and clearance of HBV covalently closed circular DNA (cccDNA) replication intermediates (Lucifora et al., 2014). Thus, analysis of cell culture models of HBV infection have indicated roles for multiple APOBEC family proteins in virus restriction.
Analysis of patients chronically infected with HBV paints a somewhat different picture of APOBEC restriction. A3G levels appear to be low in primary hepatocytes, but can be induced by interferon α (Bonvin et al., 2006). In addition, human A3B, A3C, A3G, A3H, and AID mRNAs are upregulated by inflammation, which often accompanies viral infection (Endo et al., 2007; Vartanian et al., 2010). HBV may replicate in non-hepatic cells, although replication in hematopoietic cells appears to be extremely low (Rosler et al., 2004; Untergasser et al., 2006). HBV DNA sequences from the livers of four patients with high levels of viremia were enriched by 3D-PCR (Suspène et al., 2005). Two of these patient samples gave PCR products at a denaturation temperature of 90°C, and sequencing of these products revealed that a small number had G-to-A mutations. The context of these mutations was consistent with the preference of A3G (Suspène et al., 2005). In another study, DNA samples were obtained from patients with liver cirrhosis and analyzed by 3D-PCR at 88.7°C. Fifteen of 17 DNAs were amplified under this condition, and five were cloned and sequenced (Vartanian et al., 2010). G-to-A mutations were observed in HBV minus strands with a sequence context consistent with A3G activity. The remainder of the mutations showed a sequence context more typical of A3C, rather than AID (Vartanian et al., 2010). Deep sequencing also was performed on PCR products obtained in reactions with a 95°C denaturation. Four of five samples showed G-to-A hypermutation varying from 10 to 35% in the X gene, which is single-stranded in virions (Vartanian et al., 2010). Taken together with the fact that mutated sequences are rarely recovered by normal high denaturation temperature PCR, the level of HBV hypermutation appears to be significantly lower than that reported for a number of retroviruses.
Data from several laboratories have indicated that loss of the pre-capsid (core) antigen may be due to APOBEC-mediated editing (Noguchi et al., 2007; Turelli et al., 2004; Vartanian et al., 2010). Serum samples from 47 HBeAg-positive and 33 HBeAg-negative treatment-naïve patients were subjected to deep sequencing (Beggel et al., 2013). Hypermutation rates were ca. 15-fold greater in HBeAg-negative patients and were preferentially in the HBV virion-associated single-stranded region. Similar to other studies, the context of the G-to-A mutations suggested editing by A3G (Beggel et al., 2013; Vartanian et al., 2010). Because pre-core antigen expression is associated with high viremia and the coding region contains several optimal sites for APOBEC-mediated mutation, seroconversion to HBeAg negativity may well represent innate immune selection for particular HBV variants (Beggel et al., 2013; Vartanian et al., 2010).
Interestingly, transfusion-transmitted virus (TTV) found in the blood of healthy patients and in HBV carriers contains G-to-A hypermutations, indicating that viruses that lack reverse transcriptase can be subjected to APOBEC family restriction (Tsuge et al., 2010). TTV is a single-stranded negative-sense DNA virus, which is predicted to be a good target for A3 enzymes (Irshad et al., 2006). In addition, single-stranded DNA viruses belonging to the parvovirus family can be restricted by APOBEC, particularly A3A (Chen et al., 2006; Narvaiza et al., 2009). Curiously, two different parvoviruses are inhibited by A3A, suggesting a conserved mechanism, yet inhibition appears independent of catalytic activity (Narvaiza et al., 2009).
Double-stranded DNA viruses also may also be substrates for APOBEC family members. Double-stranded DNA viruses may yield exposed single-stranded DNA during transcription or genome replication. Human papilloma viruses (HPVs) for example are sexually transmitted, and about 24 of the 300 genotypes are associated with human cancers (zur Hausen, 2008). HPV types 1a and 16 in plantar skin warts and precancerous cervical lesions were shown to have hypermutations (Vartanian et al., 2008). A3A, A3B, and A3H are expressed in keratinocytes, where HPV has been shown to replicate. Further, co-transfection experiments have shown that overexpression of these enzymes could induce HPV hyperediting (Vartanian et al., 2008).
Recent studies also have documented APOBEC-mediated 5′-TC-to-TT hypermutation signatures in numerous cancers, including HPV-associated cervical and head-and-neck squamous cell carcinomas (HNSCCs) (Alexandrov et al., 2013; Burns et al., 2013; Roberts et al., 2013). HNSCCs are particularly informative because some are HPV positive and others are HPV negative. Higher overall levels of APOBEC signature mutations are observed in HPV positive cancers (Henderson et al., 2014; Vieira et al., 2014). In addition, hotspots for APOBEC mutagenesis and oncogene activation occur in PIK3CA, which encodes the catalytic subunit of a phosphoinositide-3-kinase that is activated in large proportions of cervical and HNSCCs (Henderson et al., 2014). These hypermutations are most likely due to A3B, and possibly A3A, because these are the only family members induced by HPV infection (Vieira et al., 2014; Warren et al., 2015). Thus, it is tempting to speculate that cancer mutagenesis may be the result of collateral DNA damage of an antiviral response to HPV infection.
DNA viruses that infect B cells also might be expected to be inhibited by AID since this family member is activated in the germinal centers to trigger adaptive responses to infection. In addition, AID can be activated in non-B-cells that normally lack AID expression by pathogen expression (Gourzi et al., 2006, 2007). Such viruses have been shown to inhibit AID function through mechanisms distinct from those described for retroviruses. Epstein-Barr virus (EBV) is a herpes virus (double-stranded DNA) that replicates in B cells. This virus antagonizes the effects of AID by upregulation of a host regulatory microRNA (miR-155) and the latency-associated protein EBNA2 (Tobollik et al., 2006). Kaposi’s sarcoma herpes virus (KSHV) also infects B cells (Mesri et al., 1996), and infection of tonsillar B cells upregulates AID (Bekerman et al., 2013). KSHV often maintains its genome in a latent state, which requires the expression of four proteins and 12 micro RNAs (Cai et al., 2005; Dittmer et al., 1998; Pfeffer et al., 2005; Samols et al., 2005). Latency prevents viral DNA replication and associated AID damage, but AID expression activates KSHV lytic replication and loss of infectivity (Bekerman et al., 2013). To antagonize AID, KSHV encodes two different miRs that bind to the 3′ UTR of AID mRNA. Binding of viral miRs at several different sites is believed to block translation of AID and triggering of the innate immune response to KSHV (Bekerman et al., 2013).
Not all DNA viruses have been shown to be susceptible to APOBEC-mediated restriction. Vaccinia virus does not appear to be inhibited by APOBEC family members, perhaps due to the sequestration of its replication complex in cytoplasmic bodies (Kremer et al., 2006). Therefore, DNA viruses may avoid APOBEC-mediated restriction by encoding an inhibitor, preventing incorporation into virions, avoiding induction of inflammation and APOBEC enzymes, replication in cells with low levels of APOBEC, or replicating in privileged subcellular locations.
Conclusions
In this review, we have summarized the extraordinary functions of the APOBEC family of proteins, including participating in antibody diversification, editing of mRNA, and acting as retrovirus and retrotransposon restriction factors. Moreover, mechanisms for inhibition of parasites are diverse, including both deamination-dependent and -independent processes. For their part, viruses have responded with a variety of strategies to circumvent restriction from poly-ubiquitination and degradation mechanisms to exclusion and protection mechanisms. It is likely that many more A3 antagonizing measures exist and await discovery. An overall picture is emerging in which the A3 family members, including AID and APOBEC1, provide an overlapping defense against a wide variety of parasitic elements. This is important for controlling endogenous elements, but also for protecting against exogenous viral infections as well as from zoonotic transmissions. Nearly every DNA-based parasite may be susceptible and, if so, invariably evolved at least one protective measure. Many protective measures have been described thus far and undoubtedly several more await discovery. Clearly, there is still much work to be done, and this story has no finite ending.
Acknowledgments
We thank N. Shaban for assistance with figures. This work was supported by NIH grants R21 AI105710 and R01 CA167053 to J.P.D. and R01 AI064046 and P01 GM091743 to R.S.H. We apologize to colleagues whose work could not be cited because of space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Reuben S. Harris, Email: rsh@umn.edu.
Jaquelin P. Dudley, Email: jdudley@austin.utexas.edu.
References
- Abudu A, Takaori-Kondo A, Izumi T, Shirakawa K, Kobayashi M, Sasada A, Fukunaga K, Uchiyama T. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr Biol. 2006;16:1565–1570. doi: 10.1016/j.cub.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Ai Y, Zhu D, Wang C, Su C, Ma J, Ma J, Wang X. Core-binding factor subunit beta is not required for non-primate lentiviral Vif-mediated APOBEC3 degradation. J Virol. 2014;88:12112–12122. doi: 10.1128/JVI.01924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, Brown WL, Harris RS. Catalytic activity of APOBEC3F is required for efficient restriction of Vif-deficient human immunodeficiency virus. Virology. 2014;450–451:49–54. doi: 10.1016/j.virol.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinsk M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alke A, Schwantes A, Kido K, Flotenmeyer M, Flugel RM, Lochelt M. The bet gene of feline foamy virus is required for virus replication. Virology. 2001;287:310–320. doi: 10.1006/viro.2001.1065. [DOI] [PubMed] [Google Scholar]
- Anwar F, Davenport MP, Ebrahimi D. Footprint of APOBEC3 on the genome of human retroelements. J Virol. 2013;87:8195–8204. doi: 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 2015;11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin H, Taylor MW, Lee JE. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure. 2014;22:668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Bangham CR, Cook LB, Melamed A. HTLV-1 clonality in adult T-cell leukaemia and non-malignant HTLV-1 infection. Semin Cancer Biol. 2014;26:89–98. doi: 10.1016/j.semcancer.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett BS, Guo K, Harper MS, Li SX, Heilman KJ, Davidson NO, Santiago ML. Reassessment of murine APOBEC1 as a retrovirus restriction factor in vivo. Virology. 2014;468–470:601–608. doi: 10.1016/j.virol.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baunach G, Maurer B, Hahn H, Kranz M, Rethwilm A. Functional analysis of human foamy virus accessory reading frames. J Virol. 1993;67:5411–5418. doi: 10.1128/jvi.67.9.5411-5418.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggel B, Munk C, Daumer M, Hauck K, Haussinger D, Lengauer T, Erhardt A. Full genome ultra-deep pyrosequencing associates G-to-A hypermutation of the hepatitis B virus genome with the natural progression of hepatitis B. Journal of viral hepatitis. 2013;20:882–889. doi: 10.1111/jvh.12110. [DOI] [PubMed] [Google Scholar]
- Bekerman E, Jeon D, Ardolino M, Coscoy L. A role for host activation-induced cytidine deaminase in innate immune defense against KSHV. PLoS Pathog. 2013;9:e1003748. doi: 10.1371/journal.ppat.1003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, de Ronde A. APOBEC3G versus reverse transcriptase in the generation of HIV-1 drug-resistance mutations. AIDS. 2004;18:1861–1863. doi: 10.1097/00002030-200409030-00022. [DOI] [PubMed] [Google Scholar]
- Berlioz C, Darlix JL. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S, Lozano MM, Dudley JP. BALB/Mtv-null mice responding to strong mouse mammary tumor virus superantigens restrict mammary tumorigenesis. J Virol. 2009;83:484–488. doi: 10.1128/JVI.01374-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Cullen BR. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA. 2008;14:1228–1236. doi: 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006a;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006b;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boi S, Kolokithas A, Shepard J, Linwood R, Rosenke K, Van Dis E, Malik F, Evans LH. Incorporation of mouse APOBEC3 into murine leukemia virus virions decreases the activity and fidelity of reverse transcriptase. J Virol. 2014;88:7659–7662. doi: 10.1128/JVI.00967-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- Bonvin M, Greeve J. Hepatitis B: modern concepts in pathogenesis--APOBEC3 cytidine deaminases as effectors in innate immunity against the hepatitis B virus. Current opinion in infectious diseases. 2008;21:298–303. doi: 10.1097/QCO.0b013e3282fe1bb2. [DOI] [PubMed] [Google Scholar]
- Brightman BK, Davis BR, Fan H. Preleukemic hematopoietic hyperplasia induced by Moloney murine leukemia virus is an indirect consequence of viral infection. J Virol. 1990;64:4582–4584. doi: 10.1128/jvi.64.9.4582-4584.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun H, Halani N, Gou Y, Nash AK, Lozano MM, Dudley JP. Requirements for mouse mammary tumor virus Rem signal peptide processing and function. J Virol. 2012;86:214–225. doi: 10.1128/JVI.06197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun H, Halani N, Mertz JA, Ali AF, Lozano MM, Dudley JP. Retroviral Rem protein requires processing by signal peptidase and retrotranslocation for nuclear function. Proc Natl Acad Sci U S A. 2010;107:12287–12292. doi: 10.1073/pnas.1004303107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amst) 2010;9:579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chareza S, Slavkovic Lukic D, Liu Y, Rathe AM, Munk C, Zabogli E, Pistello M, Lochelt M. Molecular and functional interactions of cat APOBEC3 and feline foamy and immunodeficiency virus proteins: different ways to counteract host-encoded restriction. Virology. 2012;424:138–146. doi: 10.1016/j.virol.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nature chemistry. 2014;6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt S, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Davis BR, Brightman BK, Chandy KG, Fan H. Characterization of a preleukemic state induced by Moloney murine leukemia virus: evidence for two infection events during leukemogenesis. Proc Natl Acad Sci U S A. 1987;84:4875–4879. doi: 10.1073/pnas.84.14.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D, Hill SA, Princler G, Lloyd P, Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc Natl Acad Sci U S A. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle BP, Bogerd HP, Wiegand HL, Jouvenet N, Bieniasz PD, Hunter E, Cullen BR. The betaretrovirus Mason-Pfizer monkey virus selectively excludes simian APOBEC3G from virion particles. J Virol. 2006;80:12102–12108. doi: 10.1128/JVI.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue JP, Levinson RT, Sheehan JH, Sutton L, Taylor HE, Meiler J, D’Aquila RT, Song C. Genetic analysis of the localization of APOBEC3F to human immunodeficiency virus type 1 virion cores. J Virol. 2015;89:2415–2424. doi: 10.1128/JVI.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douaisi M, Dussart S, Courcoul M, Bessou G, Vigne R, Decroly E. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem Biophys Res Commun. 2004;321:566–573. doi: 10.1016/j.bbrc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Duggal NK, Fu W, Akey JM, Emerman M. Identification and antiviral activity of common polymorphisms in the APOBEC3 locus in human populations. Virology. 2013;443:329–337. doi: 10.1016/j.virol.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko JA, Schafer A, Kenny AE, Cullen BR, Curcio MJ. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr Biol. 2005;15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi D, Anwar F, Davenport MP. APOBEC3G and APOBEC3F rarely co-mutate the same HIV genome. Retrovirology. 2012;9:113. doi: 10.1186/1742-4690-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Marusawa H, Kinoshita K, Morisawa T, Sakurai T, Okazaki IM, Watashi K, Shimotohno K, Honjo T, Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 2007;26:5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ma G, Nosaka K, Tanabe J, Satou Y, Koito A, Wain-Hobson S, Vartanian JP, Matsuoka M. APOBEC3G generates nonsense mutations in human T-cell leukemia virus type 1 proviral genomes in vivo. J Virol. 2010;84:7278–7287. doi: 10.1128/JVI.02239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Baig TT, Love RP, Chelico L. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol. 2014;5:450. doi: 10.3389/fmicb.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa R, McAtee FJ, Zirbel JH, Portis JL. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: identification of differences in processing in vitro and in vivo. J Virol. 1997;71:5355–5360. doi: 10.1128/jvi.71.7.5355-5360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. Role of Vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillick K, Pollpeter D, Phalora P, Kim EY, Wolinsky SM, Malim MH. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. J Virol. 2013;87:1508–1517. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkina TV, Chervonsky A, Dudley JP, Ross SR. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- Golovkina TV, Dudley JP, Ross SR. B and T cells are required for mouse mammary tumor virus spread within the mammary gland. J Immunol. 1998;161:2375–2382. [PubMed] [Google Scholar]
- Gourzi P, Leonova T, Papavasiliou FN. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 2006;24:779–786. doi: 10.1016/j.immuni.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Gourzi P, Leonova T, Papavasiliou FN. Viral induction of AID is independent of the interferon and the Toll-like receptor signaling pathways but requires NF-kappaB. J Exp Med. 2007;204:259–265. doi: 10.1084/jem.20061801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- Haché G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Haché G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8:148–157. [PubMed] [Google Scholar]
- Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281:36624–36631. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- Halemano K, Guo K, Heilman KJ, Barrett BS, Smith DS, Hasenkrug KJ, Santiago ML. Immunoglobulin somatic hypermutation by APOBEC3/Rfv3 during retroviral infection. Proc Natl Acad Sci U S A. 2014;111:7759–7764. doi: 10.1073/pnas.1403361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Liang W, Hua D, Zhou X, Du J, Evans SL, Gao Q, Wang H, Viqueira R, Wei W, Zhang W, Yu XF. Evolutionarily conserved requirement for core binding factor beta in the assembly of the human immunodeficiency virus/simian immunodeficiency virus Vif-cullin 5-RING E3 ubiquitin ligase. J Virol. 2014;88:3320–3328. doi: 10.1128/JVI.03833-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes S, Solomon WC, Li M, Chen KM, Harjes E, Harris RS, Matsuo H. Impact of H216 on the DNA binding and catalytic activities of the HIV restriction factor APOBEC3G. J Virol. 2013;87:7008–7014. doi: 10.1128/JVI.03173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- Held W, Shakhov AN, Izui S, Waanders GA, Scarpellino L, MacDonald HR, Acha-Orbea H. Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J Exp Med. 1993a;177:359–366. doi: 10.1084/jem.177.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W, Waanders GA, Shakhov AN, Scarpellino L, Acha-Orbea H, MacDonald HR. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993b;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Henry M, Terzian C, Peeters M, Wain-Hobson S, Vartanian JP. Evolution of the primate APOBEC3A cytidine deaminase gene and identification of related coding regions. PloS one. 2012;7:e30036. doi: 10.1371/journal.pone.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007a;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci. 2007b;32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Holtz CM, Sadler HA, Mansky LM. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 2013;41:6139–6148. doi: 10.1093/nar/gkt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imahashi M, Izumi T, Watanabe D, Imamura J, Matsuoka K, Ode H, Masaoka T, Sato K, Kaneko N, Ichikawa S, Koyanagi Y, Takaori-Kondo A, Utsumi M, Yokomaku Y, Shirasaka T, Sugiura W, Iwatani Y, Naoe T. Lack of association between intact/deletion polymorphisms of the APOBEC3B gene and HIV-1 risk. PloS one. 2014;9:e92861. doi: 10.1371/journal.pone.0092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imahashi M, Nakashima M, Iwatani Y. Antiviral mechanism and biochemical basis of the human APOBEC3 family. Front Microbiol. 2012;3:250. doi: 10.3389/fmicb.2012.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indik S, Gunzburg WH, Salmons B, Rouault F. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology. 2005;337:1–6. doi: 10.1016/j.virol.2005.03.040. [DOI] [PubMed] [Google Scholar]
- Irshad M, Joshi YK, Sharma Y, Dhar I. Transfusion transmitted virus: A review on its molecular characteristics and role in medicine. World journal of gastroenterology : WJG. 2006;12:5122–5134. doi: 10.3748/wjg.v12.i32.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan DSB, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Research. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2011;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaguva Vasudevan AA, Perkovic M, Bulliard Y, Cichutek K, Trono D, Haussinger D, Munk C. Prototype foamy virus Bet impairs the dimerization and cytosolic solubility of human APOBEC3G. J Virol. 2013;87:9030–9040. doi: 10.1128/JVI.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janini M, Rogers M, Birx DR, McCutchan FE. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J Virol. 2001;75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Stoye JP, Coffin JM. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007;3:2014–2022. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Sawyer SL. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics. 2009;61:163–176. doi: 10.1007/s00251-009-0358-y. [DOI] [PubMed] [Google Scholar]
- Jones SA, Hu J. Protein-primed terminal transferase activity of hepatitis B virus polymerase. J Virol. 2013;87:2563–2576. doi: 10.1128/JVI.02786-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson SR, Haché G, Stenglein MD, Fahrenkrug SC, Andrésdóttir V, Harris RS. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006;34:5683–5694. doi: 10.1093/nar/gkl721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, Wolinsky SM. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol. 2010;84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Lorenzo-Redondo R, Little SJ, Chung YS, Phalora PK, Maljkovic Berry I, Archer J, Penugonda S, Fischer W, Richman DD, Bhattacharya T, Malim MH, Wolinsky SM. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014;10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Koizumi Y, Takeuchi JS, Misawa N, Kimura Y, Morita S, Aihara K, Koyanagi Y, Iwami S, Sato K. Quantification of deaminase activity-dependent and -independent restriction of HIV-1 replication mediated by APOBEC3F and APOBEC3G through experimental-mathematical investigation. J Virol. 2014;88:5881–5887. doi: 10.1128/JVI.00062-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Maul RW, Guminski AF, McClure RL, Gajula KS, Saribasak H, McMahon MA, Siliciano RF, Gearhart PJ, Stivers JT. Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. J Biol Chem. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolokithas A, Rosenke K, Malik F, Hendrick D, Swanson L, Santiago ML, Portis JL, Hasenkrug KJ, Evans LH. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J Virol. 2010;84:10933–10936. doi: 10.1128/JVI.01023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer M, Suezer Y, Martinez-Fernandez Y, Munk C, Sutter G, Schnierle BS. Vaccinia virus replication is not affected by APOBEC3 family members. Virol J. 2006;3:86. doi: 10.1186/1743-422X-3-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko JF, Martinez-Torres F, Foster JL, Garcia JV. HIV restriction by APOBEC3 in humanized mice. PLoS Pathog. 2013;9:e1003242. doi: 10.1371/journal.ppat.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois MA, Kemmerich K, Rada C, Neuberger MS. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J Virol. 2009;83:11550–11559. doi: 10.1128/JVI.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Andrésdóttir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O’Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Jónsson SR, Silverstein KAT, Lajoie M, Bertrand D, El-Mabrouk N, Hötzel I, Andrésdóttir V, Smith TPL, Harris RS. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104, 120. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Lengyel J, Jónsson SR, Andrésdóttir V, Harris RS. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J Virol. 2010;84:8193–8201. doi: 10.1128/JVI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Wissing S, Lobritz MA, Santiago M, Greene WC. Identification of two APOBEC3F splice variants displaying HIV-1 antiviral activity and contrasting sensitivity to Vif. J Biol Chem. 2010 doi: 10.1074/jbc.M110.154054. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YN, Malim MH, Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol. 2008;82:8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hakata Y, Takeda E, Liu Q, Iwatani Y, Kozak CA, Miyazawa M. Two genetic determinants acquired late in mus evolution regulate the inclusion of exon 5, which alters mouse APOBEC3 translation efficiency. PLoS Pathog. 2012a;8:e1002478. doi: 10.1371/journal.ppat.1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem Biol. 2012b;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QX, Fan H. Combined infection by Moloney murine leukemia virus and a mink cell focus-forming virus recombinant induces cytopathic effects in fibroblasts or in long-term bone marrow cultures from preleukemic mice. J Virol. 1990;64:3701–3711. doi: 10.1128/jvi.64.8.3701-3711.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Kitamura K, Wang Z, Liu G, Chowdhury S, Fu W, Koura M, Wakae K, Honjo T, Muramatsu M. RNA editing of hepatitis B virus transcripts by activation-induced cytidine deaminase. Proc Natl Acad Sci U S A. 2013;110:2246–2251. doi: 10.1073/pnas.1221921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Lochelt M, Romen F, Bastone P, Muckenfuss H, Kirchner N, Kim YB, Truyen U, Rosler U, Battenberg M, Saib A, Flory E, Cichutek K, Munk C. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc Natl Acad Sci U S A. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low A, Datta S, Kuznetsov Y, Jahid S, Kothari N, McPherson A, Fan H. Mutation in the glycosylated gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J Virol. 2007;81:3685–3692. doi: 10.1128/JVI.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low A, Okeoma CM, Lovsin N, de las Heras M, Taylor TH, Peterlin BM, Ross SR, Fan H. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology. 2009;385:455–463. doi: 10.1016/j.virol.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukic DS, Hotz-Wagenblatt A, Lei J, Rathe AM, Muhle M, Denner J, Munk C, Lochelt M. Identification of the feline foamy virus Bet domain essential for APOBEC3 counteraction. Retrovirology. 2013;10:76. doi: 10.1186/1742-4690-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDuff DA, Demorest ZL, Harris RS. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 2009;37:1854–1867. doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan AL, Kohli RM, Ross SR. APOBEC3 inhibition of mouse mammary tumor virus infection: the role of cytidine deamination versus inhibition of reverse transcription. J Virol. 2013;87:4808–4817. doi: 10.1128/JVI.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral vif protein. Journal of Virology. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieux R, Suspene R, Delebecque F, Henry M, Schwartz O, Wain-Hobson S, Vartanian JP. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J Gen Virol. 2005;86:2489–2494. doi: 10.1099/vir.0.80973-0. [DOI] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harbor perspectives in medicine. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD, Yeo GW, Muotri AR, Gage FH. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J Virol. 2005;79:14737–14747. doi: 10.1128/JVI.79.23.14737-14747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Arvanitakis L, Rafii S, Moore MA, Posnett DN, Knowles DM, Asch AS. Human herpesvirus-8/Kaposi’s sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2390. [Google Scholar]
- Metzner M, Jack HM, Wabl M. LINE-1 retroelements complexed and inhibited by activation induced cytidine deaminase. PloS one. 2012;7:e49358. doi: 10.1371/journal.pone.0049358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S, Gottlieb PD, Dudley JP. Lymphomas with acquired mouse mammary tumor virus proviruses resemble distinct prethymic and intrathymic phenotypes defined in vivo. J Immunol. 1989;142:3342–3350. [PubMed] [Google Scholar]
- Meyerson NR, Sawyer SL. Two-stepping through time: mammals and viruses. Trends Microbiol. 2011;19:286–294. doi: 10.1016/j.tim.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikl MC, Watt IN, Lu M, Reik W, Davies SL, Neuberger MS, Rada C. Mice deficient in APOBEC2 and APOBEC3. Mol Cell Biol. 2005;25:7270–7277. doi: 10.1128/MCB.25.16.7270-7277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, McClure MO, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris A, Murray S, Cardinaud S. AID and APOBECs span the gap between innate and adaptive immunity. Front Microbiol. 2014;5:534. doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O’Brien SJ, Löchelt M, Yuhki N. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Mustafa F, Bhadra S, Johnston D, Lozano M, Dudley JP. The type B leukemogenic virus truncated superantigen is dispensable for T-cell lymphomagenesis. J Virol. 2003;77:3866–3870. doi: 10.1128/JVI.77.6.3866-3870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel CS, Lee JW, Wang LC, Kohli RM. Nucleic acid determinants for selective deamination of DNA over RNA by activation-induced deaminase. Proc Natl Acad Sci U S A. 2013;110:14225–14230. doi: 10.1073/pnas.1306345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Sanchez-Martinez S, Ji X, Rein A. Biochemical and biological studies of mouse APOBEC3. J Virol. 2014;88:3850–3860. doi: 10.1128/JVI.03456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Nitta T, Lee S, Ha D, Arias M, Kozak CA, Fan H. Moloney murine leukemia virus glyco-gag facilitates xenotropic murine leukemia virus-related virus replication through human APOBEC3-independent mechanisms. Retrovirology. 2012;9:58. doi: 10.1186/1742-4690-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Fujimoto Y, Ochi H, Abe H, Maekawa T, Yatsuji H, Shirakawa K, Takaori-Kondo A, Chayama K. Dual effect of APOBEC3G on Hepatitis B virus. J Gen Virol. 2007;88:432–440. doi: 10.1099/vir.0.82319-0. [DOI] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM, Huegel AL, Lingappa J, Feldman MD, Ross SR. APOBEC3 proteins expressed in mammary epithelial cells are packaged into retroviruses and can restrict transmission of milk-borne virions. Cell Host Microbe. 2010;8:534–543. doi: 10.1016/j.chom.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- Okeoma CM, Low A, Bailis W, Fan HY, Peterlin BM, Ross SR. Induction of APOBEC3 in vivo causes increased restriction of retrovirus infection. J Virol. 2009a;83:3486–3495. doi: 10.1128/JVI.02347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM, Petersen J, Ross SR. Expression of murine APOBEC3 alleles in different mouse strains and their effect on mouse mammary tumor virus infection. J Virol. 2009b;83:3029–3038. doi: 10.1128/JVI.02536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, Barbour JD, Simon V. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Ooms M, Krikoni A, Kress AK, Simon V, Munk C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J Virol. 2012;86:6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M, Bleiber G, Martinez R, Kaessmann H, Telenti A. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology. 2006;3:11. doi: 10.1186/1742-4690-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkovic M, Schmidt S, Marino D, Russell RA, Stauch B, Hofmann H, Kopietz F, Kloke BP, Zielonka J, Strover H, Hermle J, Lindemann D, Pathak VK, Schneider G, Lochelt M, Cichutek K, Munk C. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein bet. J Biol Chem. 2009;284:5819–5826. doi: 10.1074/jbc.M808853200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Petit V, Guetard D, Renard M, Keriel A, Sitbon M, Wain-Hobson S, Vartanian JP. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol. 2009;385:65–78. doi: 10.1016/j.jmb.2008.10.043. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pillai SK, Wong JK, Barbour JD. Turning up the volume on mutational pressure: is more of a good thing always better? (A case study of HIV-1 Vif and APOBEC3) Retrovirology. 2008;5:26. doi: 10.1186/1742-4690-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- Rathore A, Carpenter MA, Demir O, Ikeda T, Li M, Shaban NM, Law EK, Anokhin D, Brown WL, Amaro RE, Harris RS. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J Mol Biol. 2013;425:4442–4454. doi: 10.1016/j.jmb.2013.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch JW, Chelico L, Goodman MF, Le Grice SF. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J Biol Chem. 2009;284:7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, Brown WL, Reilly C, Emerman M, Harris RS. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife. 2014;3:e02008. doi: 10.7554/eLife.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales Gerpe MC, Renner TM, Belanger K, Lam C, Aydin H, Langlois MA. N-Linked glycosylation protects gammaretroviruses against deamination by APOBEC3 proteins. J Virol. 2015;89:2342–2357. doi: 10.1128/JVI.03330-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosler C, Kock J, Malim MH, Blum HE, von Weizsacker F. Comment on “Inhibition of hepatitis B virus replication by APOBEC3G”. Science. 2004;305:1403. doi: 10.1126/science.1101974. author reply 1403. [DOI] [PubMed] [Google Scholar]
- Rulli SJ, Jr, Mirro J, Hill SA, Lloyd P, Gorelick RJ, Coffin JM, Derse D, Rein A. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J Virol. 2008;82:6566–6575. doi: 10.1128/JVI.01357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RA, Wiegand HL, Moore MD, Schafer A, McClure MO, Cullen BR. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter JD, Morales GA, Smith HC. Structural insights for HIV-1 therapeutic strategies targeting Vif. Trends Biochem Sci. 2014;39:373–380. doi: 10.1016/j.tibs.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Montano M, Benitez R, Messer RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, Greene WC. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanville B, Dolan MA, Wollenberg K, Yan Y, Martin C, Yeung ML, Strebel K, Buckler-White A, Kozak CA. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog. 2010;6:e1000974. doi: 10.1371/journal.ppat.1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasada A, Takaori-Kondo A, Shirakawa K, Kobayashi M, Abudu A, Hishizawa M, Imada K, Tanaka Y, Uchiyama T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005;2:32. doi: 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Takeuchi JS, Misawa N, Izumi T, Kobayashi T, Kimura Y, Iwami S, Takaori-Kondo A, Hu WS, Aihara K, Ito M, An DS, Pathak VK, Koyanagi Y. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 2014;10:e1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schmitt K, Guo K, Algaier M, Ruiz A, Cheng F, Qiu J, Wissing S, Santiago ML, Stephens EB. Differential virus restriction patterns of rhesus macaque and human APOBEC3A: implications for lentivirus evolution. Virology. 2011;419:24–42. doi: 10.1016/j.virol.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Haché G, MacDuff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc Natl Acad Sci U S A. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppen J. Unedited inhibition of HBV replication by APOBEC3G. J Hepatol. 2004;41:1068–1069. doi: 10.1016/j.jhep.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Shandilya SM, Bohn MF, Schiffer CA. A computational analysis of the structural determinants of APOBEC3’s catalytic activity and vulnerability to HIV-1 Vif. Virology. 2014;471–473:105–116. doi: 10.1016/j.virol.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Simon JH, Gaddis NC, Fouchier RA, Malim MH. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- Siu KK, Sultana A, Azimi FC, Lee JE. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Guo K, Barrett BS, Heilman KJ, Evans LH, Hasenkrug KJ, Greene WC, Santiago ML. Noninfectious retrovirus particles drive the APOBEC3/Rfv3 dependent neutralizing antibody response. PLoS Pathog. 2011;7:e1002284. doi: 10.1371/journal.ppat.1002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Sutton L, Johnson ME, D’Aquila RT, Donahue JP. Signals in APOBEC3F N-terminal and C-terminal deaminase domains each contribute to encapsidation in HIV-1 virions and are both required for HIV-1 restriction. J Biol Chem. 2012;287:16965–16974. doi: 10.1074/jbc.M111.310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou S, Nitta T, Kotla S, Ha D, Nagashima K, Rein AR, Fan H, Ross SR. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc Natl Acad Sci U S A. 2013;110:9078–9083. doi: 10.1073/pnas.1217399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Strebel K. APOBEC3G & HTLV-1: inhibition without deamination. Retrovirology. 2005;2:37. doi: 10.1186/1742-4690-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K. HIV accessory proteins versus host restriction factors. Current opinion in virology. 2013;3:692–699. doi: 10.1016/j.coviro.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Khan MA. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology. 2008;5:55. doi: 10.1186/1742-4690-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois MA, Neuberger MS, Rada C, Miyazawa M. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J Virol. 2008;82:10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen BK, McNevin JP, McElrath MJ, Hunt BV, Klein KC, Lingappa JR. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J Biol Chem. 2010;285:27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wang T, Zhang W, Yu XF. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 2007;35:7288–7302. doi: 10.1093/nar/gkm816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobollik S, Meyer L, Buettner M, Klemmer S, Kempkes B, Kremmer E, Niedobitek G, Jungnickel B. Epstein-Barr virus nuclear antigen 2 inhibits AID expression during EBV-driven B-cell growth. Blood. 2006;108:3859–3864. doi: 10.1182/blood-2006-05-021303. [DOI] [PubMed] [Google Scholar]
- Tsuge M, Noguchi C, Akiyama R, Matsushita M, Kunihiro K, Tanaka S, Abe H, Mitsui F, Kitamura S, Hatakeyama T, Kimura T, Miki D, Hiraga N, Imamura M, Takahashi S, Hayses CN, Chayama K. G to A hypermutation of TT virus. Virus Res. 2010;149:211–216. doi: 10.1016/j.virusres.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Zedler U, Langenkamp A, Hosel M, Quasdorff M, Esser K, Dienes HP, Tappertzhofen B, Kolanus W, Protzer U. Dendritic cells take up viral antigens but do not support the early steps of hepatitis B virus infection. Hepatology. 2006;43:539–547. doi: 10.1002/hep.21048. [DOI] [PubMed] [Google Scholar]
- Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- Vartanian JP, Henry M, Marchio A, Suspene R, Aynaud MM, Guetard D, Cervantes-Gonzalez M, Battiston C, Mazzaferro V, Pineau P, Dejean A, Wain-Hobson S. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010;6:e1000928. doi: 10.1371/journal.ppat.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Meyerhans A, Sala M, Wain-Hobson S. G-->A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci U S A. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira VC, Leonard B, White EA, Starrett GJ, Temiz NA, Lorenz LD, Lee D, Soares MA, Lambert PF, Howley PM, Harris RS. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio. 2014:5. doi: 10.1128/mBio.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA, Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J Virol. 2007;81:13932–13937. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain-Hobson S, Sonigo P, Guyader M, Gazit A, Henry M. Erratic G-->A hypermutation within a complete caprine arthritis-encephalitis virus (CAEV) provirus. Virology. 1995;209:297–303. doi: 10.1006/viro.1995.1261. [DOI] [PubMed] [Google Scholar]
- Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J Exp Med. 2010;207:141–153. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J Mol Biol. 2008;375:1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ao Z, Chen L, Kobinger G, Peng J, Yao X. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J Virol. 2012;86:3777–3786. doi: 10.1128/JVI.06594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol. 2015;89:688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem. 2011;286:36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa R, Takeuchi JS, Yamada E, Nakano Y, Ren F, Tanaka H, Munk C, Harris RS, Miyazawa T, Koyanagi Y, Sato K. Vif determines the requirement for CBF-beta in APOBEC3 degradation. J Gen Virol. 2014 doi: 10.1099/jgv.0.000027. Online Dec 16. pii: jgv.0.000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum Mol Genet. 2004;13:1785–1791. doi: 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]
- Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2011;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang H, Li Z, Liu X, Liu G, Harris RS, Yu XF. Cellular requirements for bovine immunodeficiency virus Vif-mediated inactivation of bovine APOBEC3 proteins. J Virol. 2014;88:12528–12540. doi: 10.1128/JVI.02072-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A, Du J, Zhou X, Xiong Y, Yu XF. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PloS one. 2012;7:e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]