Abstract
Genotoxic agents can cause replication fork stalling in dividing cells because of DNA lesions, eventually leading to replication fork collapse when the damage is not repaired. Small Ubiquitin-like Modifiers (SUMOs) are known to counteract replication stress, nevertheless, only a small number of relevant SUMO target proteins are known. To address this, we have purified and identified SUMO-2 target proteins regulated by replication stress in human cells. The developed methodology enabled single step purification of His10-SUMO-2 conjugates under denaturing conditions with high yield and high purity. Following statistical analysis on five biological replicates, a total of 566 SUMO-2 targets were identified. After 2 h of hydroxyurea treatment, 10 proteins were up-regulated for SUMOylation and two proteins were down-regulated for SUMOylation, whereas after 24 h, 35 proteins were up-regulated for SUMOylation, and 13 proteins were down-regulated for SUMOylation. A site-specific approach was used to map over 1000 SUMO-2 acceptor lysines in target proteins. The methodology is generic and is widely applicable in the ubiquitin field. A large subset of these identified proteins function in one network that consists of interacting replication factors, transcriptional regulators, DNA damage response factors including MDC1, ATR-interacting protein ATRIP, the Bloom syndrome protein and the BLM-binding partner RMI1, the crossover junction endonuclease EME1, BRCA1, and CHAF1A. Furthermore, centromeric proteins and signal transducers were dynamically regulated by SUMOylation upon replication stress. Our results uncover a comprehensive network of SUMO target proteins dealing with replication damage and provide a framework for detailed understanding of the role of SUMOylation to counteract replication stress. Ultimately, our study reveals how a post-translational modification is able to orchestrate a large variety of different proteins to integrate different nuclear processes with the aim of dealing with the induced DNA damage.
All cellular processes are tightly regulated via post-translational modifications (PTMs) including small chemical modifications like phosphorylation and acetylation and including modifications by small proteins belonging to the ubiquitin family (1). These post-translational modifications frequently regulate protein-protein interactions via specific domains, exemplified by the archetypical phosphor-tyrosine-interacting SH2-protein-interaction module (2). The reversible nature of these modifications enables rapid and transient cellular signal transduction. As a result of these post-translational modifications, functional proteomes are extremely complex (3).
Ubiquitination, the process of ubiquitin conjugation to target proteins is best known for its role in targeting proteins for degradation by the proteasome, but importantly also regulates target proteins in a degradation-independent manner (4). The ubiquitin-like (Ubl) family includes small ubiquitin-like modifiers (SUMOs)1, FUBI, HUB1, Nedd8, ISG15, FAT10, URM1, UFM1, Atg12, and Atg8 (5, 6). SUMOs are predominantly located in the nucleus, regulating all nuclear processes, including transcription, splicing, genome stability, and nuclear transport (7).
Similar to the ubiquitin system, SUMO conjugation is mediated by E1, E2, and E3 enzymes (8). The SUMO E1 is a dimer consisting of SAE1 and SAE2. A single E2 enzyme, Ubc9, mediates conjugation of SUMO to all target proteins. SUMO E3 enzymes include PIAS protein family members and the nucleoporin RanBP2. SUMO proteases remove SUMOs from target proteins and mediate the maturation of SUMO precursors to enable SUMO conjugation to the epsilon amino group of lysines situated in target proteins (9). A significant set of SUMO-2 acceptor lysines are situated in the SUMO consensus motif ΨKxE (8, 10). This motif is directly recognized by Ubc9, with coordinated binding of the lysine and the acidic residue of the motif to the catalytic core of the E2 enzyme (11).
The essential role of SUMO to maintain genome stability is particularly well studied (12–14). Organisms deficient for SUMOylation display increased sensitivity for different types of DNA damaging agents including double strand breaks (IR), intrastrand crosslinks (UV), alkylation (MMS), and replication fork blockage (HU) (12–14). Mice deficient for Ubc9 die at the early postimplantation stage showing DNA hypo-condensation and chromosomal aberrancies (15). The trimeric replication clamp PCNA is one of the best studied SUMO target proteins in yeast (16, 17), where SUMOylation enables the interaction with the helicase Srs2 to prevent recombination (18–20). Multiple SUMO target proteins relevant for the DNA Damage Response have been identified in mammalian systems, including DNA topoisomerase I (21), DNA topoisomerase II α and β (22, 23), the BLM helicase (24), 53BP1 (25), BRCA1 (26), HERC2, RNF168 (27), and MDC1 (28–31). In yeast, significant numbers of SUMO target proteins have been identified upon MMS and UV treatment using proteomics approaches (32, 33).
Currently, we are limited in our understanding of the role of SUMOylation in mammalian cells during the DDR because only a limited number of SUMO target proteins are known to be specifically SUMOylated in response to DNA damage. In this study, we are focusing on the role of SUMOylation with respect to replication, because SUMOylation-impaired organisms are particularly sensitive to replication stress (34–36). We have used a proteomics approach to purify and identify SUMO-2 target proteins and acceptor lysines from cells exposed to replication stress. We have uncovered sets of SUMO target proteins specifically up-regulated or down-regulated in response to replication stress, revealing a highly interactive network of proteins that is coordinated by SUMOylation to cope with replication damage. Our results shed light on the target protein network that is coordinated by SUMOylation to maintain genome stability during replication stress.
EXPERIMENTAL PROCEDURES
Antibodies
The primary antibodies used were: Mouse monoclonal anti-polyHistidine, clone HIS-1 (Sigma, H1029), mouse monoclonal anti-SUMO-2/3 (Abcam, ab81371, Cambridge, UK), rabbit polyclonal anti-SUMO-2/3 (developed with Eurogentec, Liege, Belgium) (37), rabbit polyclonal anti gamma-H2AX (Bethyl, A300–081A, Montgomery, TX), rabbit polyclonal anti-53BP1 (Bethyl, A300–272A), mouse monoclonal anti EME1 (ImmuQuest, IQ284, Seamer, UK), rabbit polyclonal anti B-Myb (Mybl2) (Bethyl, A301–654A), rabbit polyclonal anti-FOXM1 (Santa Cruz Biotechnology, sc-502) and rabbit polyclonal anti-MDC1 (Bethyl, A300–052A).
Electrophoresis and Immunoblotting
Whole cell extracts or purified protein samples were separated on Novex Bolt 4–12% Bis-Tris Plus gradient gels (Life Technologies, Carlsbad, CA) using MOPS buffer or via regular SDS-PAGE using a Tris-glycine buffer and transferred onto Hybond-C nitrocellulose membranes (GE Healthcare Life Sciences) using a submarine system (Life Technologies). Membranes were stained with Ponceau S (Sigma) to stain total protein and blocked with PBS containing 8% milk powder and 0.05% Tween-20 before incubating with the primary antibodies as indicated. Gels were stained with Coomassie using the Colloidal Blue Staining kit according to the manufacturer's instructions (Life Technologies).
Flow Cytometry
Cells were harvested by a mild trypsin treatment, subsequently washed two times with phosphate-buffered saline (PBS) and resuspended in 1.3 ml of PBS. Afterward, 3.75 ml of 100% ethanol was added and the cells were fixed at 4 °C overnight. On the day of flow cytometry analysis, the cells were centrifuged at 250 relative centrifugal force for 2 min, the supernatant was removed and the cells were washed with PBS and 2% fetal calf serum. Then, the cells were pelleted again and resuspended in 500 μl of PBS complemented with 2% fetal calf serum, 25 μg/ml propidium iodide (Sigma) and 100 μg/ml RNase A (Sigma) and then incubated for 30 min at 37 °C. FACS analysis was performed on a BD LSRII system and all gathered data was analyzed using BD FACS DIVA Software (BD Biosciences, Franklin Lakes, NJ).
Cell Culture, Cell Line Generation, and Hydroxyurea Treatment
Cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) and 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies). U2OS cells were infected using a bicistronic lentivirus encoding His10-SUMO-2 (His10-S2) and GFP separated by an IRES. Following infection, cells were sorted for low GFP levels using a FACSAria II (BD Biosciences). To induce DNA replication damage, an asynchronously growing cell population was incubated in medium containing 2 mm Hydroxyurea (Sigma) for either 2 h or 24 h. In all cases, the cells were then harvested and subjected to flow cytometry analysis, or Ni-NTA purification to enrich SUMO conjugates. For the proteome-wide identification of SUMO-2 acceptor lysines, U2OS cells were infected with bicistronic lentiviruses encoding His10-SUMO-2-K0-Q87R-IRES-GFP, abbreviated as His10-S2-K0-Q87R.
Immunofluorescence
Primary antibodies used for immunofluorescence were rabbit polyclonal anti-53BP1, rabbit polyclonal anti-phospho-Histone H2AX (Ser139) (gamma H2AX). Secondary antibody was anti-rabbit IgG AlexaFluor 594 (Life Technologies). Cells were cultured on circular glass slides in 24-well plates. After Hydroxyurea treatment for either 2 h or 24 h, medium was removed, cells were fixed with 4% paraformaldehyde for 20 min at room temperature in PBS and cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min. Next, cells were washed twice with PBS and once with PBS with 0.05% Tween-20 (PBS-T). Cells were then blocked for 10 min with 0.5% blocking reagent (Roche, Basel, Switzerland) in 0.1 m Tris, pH 7.5 and 0.15 m NaCl (TNB) and incubated with primary antibody in TNB for one hour. Coverslips were washed five times with PBS-T and incubated with the secondary antibody in TNB for one hour. Next, coverslips were washed five times with PBS-T and dehydrated by washing once with 70% ethanol, once with 90% ethanol, and once with 100% ethanol. After drying the cells, coverslips were mounted onto a microscopy slide using citifluor/DAPI solution (500 ng/ml) and sealed with nail varnish. Images were recorded on a Leica TCS SP8 confocal microscope system equipped with 405, 488, 552 and 638-nm lasers for excitation, and a 63× lens for magnification, and were analyzed with Leica confocal software.
Purification of His10-SUMO-2 Conjugates
U2OS cells expressing His10-SUMO-2 were washed, scraped, and collected in ice-cold PBS. For total lysates, a small aliquot of cells was kept separately and lysed in 2% SDS, 1% N-P40, 50 mm TRIS pH 7.5, 150 mm NaCl. The remaining part of the cell pellets were lysed in 6 m guanidine-HCl pH 8.0 (6 m guanidine-HCl, 0.1 m Na2HPO4/NaH2PO4, 10 mm TRIS, pH 8.0). The samples were snap frozen using liquid nitrogen, and stored at −80 °C.
For His10-SUMO-2 purification, the cell lysates were first thawed at room temperature and sonicated for 5 s using a sonicator (Misonix Sonicator 3000, Farmingdale, NY) at 30 Watts to homogenize the lysate. Protein concentrations were determined using the bicinchoninic acid (BCA) protein assay reagent (Thermo Scientific) and lysates were equalized. Subsequently, imidazole was added to a final concentration of 50 mm and β-mercaptoethanol was added to a final concentration of 5 mm. His10-SUMO-2 conjugates were enriched on nickel-nitrilotriacetic acid-agarose beads (Ni-NTA) (Qiagen, Venlo, The Netherlands), and subsequently the beads were washed using wash buffers A-D. Wash buffer A: 6 m guanidine-HCl, 0.1 m Na2HPO4/NaH2PO4 pH 8.0, 0.01 m Tris-HCl pH 8.0, 10 mm imidazole pH 8.0, 5 mm β-mercaptoethanol, 0.1% Triton X-100. Wash buffer B: 8 m urea, 0.1 m Na2HPO4/NaH2PO4 pH 8.0, 0.01 m Tris-HCl pH 8.0, 10 mm imidazole pH 8.0, 5 mm β-mercaptoethanol, 0.1% Triton X-100. Wash buffer C: 8 m urea, 0.1 m Na2HPO4/NaH2PO4 pH 6.3, 0.01 m Tris-HCl pH 6.3, 10 mm imidazole pH 7.0, 5 mm β-mercaptoethanol, no Triton X-100. Wash buffer D: 8 m urea, 0.1 m Na2HPO4/NaH2PO4 pH 6.3, 0.01 m Tris-HCl, pH 6.3, no imidazole, 5 mm β-mercaptoethanol, no Triton X-100. Wash buffers employed for immunoblotting experiments contained 0.2% Triton X-100. Samples were eluted in 7 m urea, 0.1 m NaH2PO4/Na2HPO4, 0.01 m Tris/HCl, pH 7.0, 500 mm imidazole pH 7.0. For site-specific purification, we used the strategy developed previously by our group (38, 39).
Sample Preparation and Mass Spectrometry
SUMO-2 enriched samples were supplemented with 1 m Tris-(2-carboxyethyl)-phosphine hydrochloride (TCEP) to a final concentration of 5 mm, and incubated for 20 min at room temperature. Iodoacetamide (IAA) was then added to the samples to a 10 mm final concentration, and samples were incubated in the dark for 15 min at room temperature. Lys-C and Trypsin digestions were performed according to the manufacturer's specifications. Lys-C was added in a 1:50 enzyme-to-protein ratio, samples were incubated at 37 °C for 4 h, and subsequently three volumes of 100 mm Tris-HCl pH 8.5 were added to dilute urea to 2 m. Trypsin (V5111, Promega, Madison, WI) was added in a 1:50 enzyme-to-protein ratio and sample were incubated overnight at 37 °C. For site-specific sample preparation, we used the strategy developed previously by our group (39).
Subsequently, digested samples were desalted and concentrated on STAGE-tips as described previously (40) and eluted with 80% acetonitrile (Sigma, 34998) in 0.1% formic acid (Sigma, 09676). Eluted fractions were vacuum dried employing a SpeedVac RC10.10 (Jouan, Nantes, France) and dissolved in 10 μl 0.1% formic acid before online nanoflow liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS).
All the experiments were performed on an EASY-nLC 1000 system (Proxeon, Odense, Denmark) connected to a Q-Exactive Orbitrap or a Q-Exactive Plus Orbitrap (Thermo Fisher Scientific, Schwerte, Germany) through a nano-electrospray ion source. For the Q-Exactive, peptides were separated in a 13 cm analytical column with an inner-diameter of 75 μm, in-house packed with 1.8 μm C18 beads (Reprospher-DE, Pur, Dr. Manish, Ammerbuch-Entringen, Germany). The Q-Exactive Plus was coupled to 15 cm analytical columns with an inner-diameter of 75 μm, in-house packed with 1.9 μm C18 beads (Reprospher-DE, Pur, Dr. Manish, Ammerbuch-Entringen, Germany), employing a column oven (PRSO-V1, Sonation, Biberach) to heat the column to 50 °C.
The gradient length was 120 min from 2% to 95% acetonitrile in 0.1% formic acid at a flow rate of 200 nL/minute. The mass spectrometers were operated in data-dependent acquisition mode with a top 10 method. Full-scan MS spectra were acquired at a target value of 3 × 106 and a resolution of 70,000, and the higher-collisional dissociation (HCD) tandem mass spectra (MS/MS) were recorded at a target value of 1 × 105 and with a resolution of 17,500 with a normalized collision energy (NCE) of 25%. The maximum MS1 and MS2 injection times were 20 ms and 60 ms, respectively. The precursor ion masses of scanned ions were dynamically excluded (DE) from MS/MS analysis for 60 s. Ions with charge 1, and greater than 6 were excluded from triggering MS2 events.
For samples enriched for identification of SUMO-2 acceptor lysines, a 120 min gradient was used for chromatography. Data dependent acquisition with a top 5 method was used. Maximum MS1 and MS2 injection times were 20 ms and 250 ms, respectively. Resolutions, normalized collision energy and automatic gain control target were set as mentioned previously. Dynamic exclusion was set to 20 s.
Protein Level SUMOylation Data Analysis
For protein-level data analysis, four experimental conditions were performed in biological quintuplicate, and all samples were measured in technical triplicate, resulting in a total of 60 runs. All RAW data was analyzed using MaxQuant (version 1.4.1.2) and its integrated search engine Andromeda. The first search was carried out with 20 ppm, whereas the main search used 6 ppm for precursor ions. Mass tolerance of MS/MS spectra were set to 20 ppm to search against an in silico digested UniProt reference proteome for Homo sapiens (13 Nov 2013, 88704 proteins). Additionally, MS/MS data were searched against a list of 262 common mass spectrometry contaminants by Andromeda.
Database searches were performed with Trypsin/P and Lys-C specificity, allowing two missed cleavages. Carbamidomethylation of cysteine residues was considered as a fixed modification. Oxidation of methionine, phosphorylation and acetylation of protein N termini were considered as variable modifications. Match between runs was performed with 0.7 min match time window and 20 min alignment time window. The minimum peptide length was set to 6. Protein groups considered for quantification required at least two peptides, including unique and razor peptides. Peptides and proteins were identified with a false discovery rate of 1% according to Cox et al. (41).
Label-free quantification was carried out, as described in Fig. 4, using MaxLFQ without fast LFQ. Proteins identified by the same set of peptides were combined to a single protein group by MaxQuant.
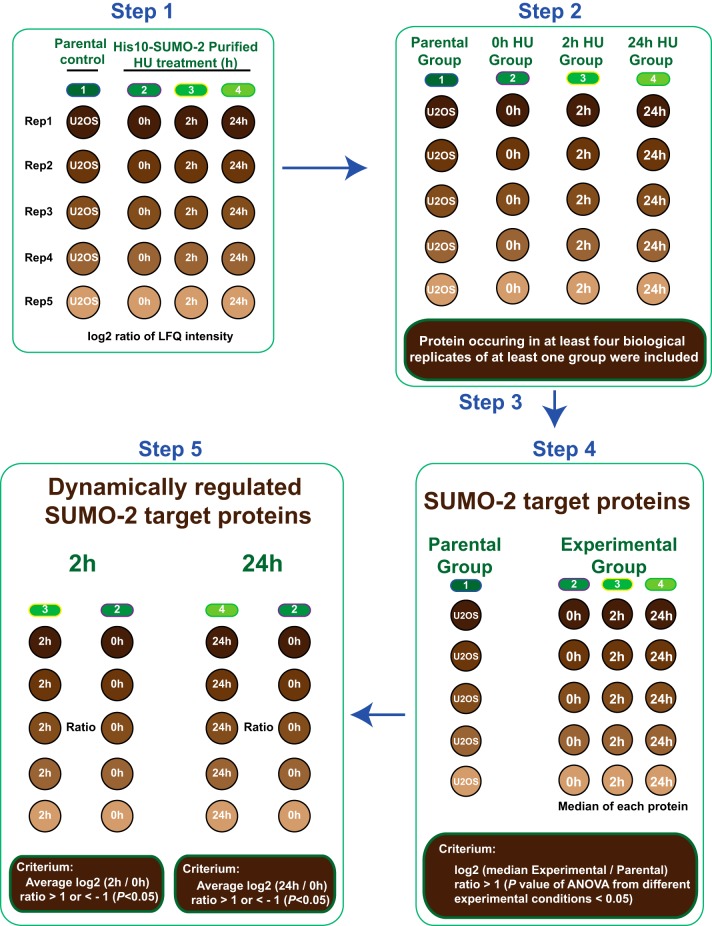
Fig. 4.
Label Free Quantification Strategy. Cartoon depicting our strategy for Label Free Quantification (LFQ) to select SUMO-2 target proteins and to identify significantly up- or down- regulated SUMO-2 target proteins in response to 2 h or 24 h HU treatment. Step 1: Protein lists generated by MaxQuant were further analyzed by Perseus and LFQ intensities were log2 transformed. Step 2: Different experiments were divided into four groups based on experimental conditions: A parental control group for U2OS control samples and three experimental groups for SUMO-2 samples purified from U2OS cells expressing His10-SUMO-2 treated with HU for 2 h or 24 h or mock treated. Inclusion criteria are depicted. Step 3: Imputation of the missing values by normally distributed values with 1.8 downshift (log2) and 0.3 randomized width (log2). Step 4: Proteins were considered as SUMO-2 target proteins using the indicated criteria. Step 5: Significantly up- or down-regulated SUMO-2 target proteins in response to 2 h or 24 h HU treatment were identified as indicated.
Protein lists generated by MaxQuant were further analyzed by Perseus (version 1.5.0.31) (42). Proteins identified as a common contaminant were filtered out, and then all the LFQ intensities were log2 transformed as described in Fig. 4, step 1. Scatter plots were generated for each experimental condition to compare the differences between biological replicates and to derive Pearson correlations. Different experiments were annotated into four groups as described in Fig. 4, step 2: a control group for the parental U2OS cell line, a 0 h HU group (untreated), a 2 h HU group and a 24 h HU group for the different time points of HU treatment of the U2OS cell line expressing His10-SUMO-2. Proteins identified in at least one treatment condition and found in at least four biological replicates were included for further analysis. For each experimental condition individually, missing values were imputed using Perseus software by normally distributed values with a 1.8 downshift (log2) and a randomized 0.3 width (log2) as described in Fig. 4, step 3. For heat maps, the log2 ratios of LFQ intensities were plotted by hierarchical clustering to compare biological replicates. For identification of SUMO-2 target proteins, selection criteria are detailed in Fig. 4 step 4. Proteins are considered to be SUMO-2 target proteins when the median log2 ratio of the LFQ intensity in the experimental group minus the median log2 ratio of the LFQ intensity in the parental control group was greater than 1 and the p value of ANOVA was smaller than 0.05. Term enrichment analysis (Gene Ontology) of SUMOylated proteins by protein annotation was carried out using Perseus. Term enrichment was determined by Fisher Exact testing, and p values were corrected for multiple hypotheses testing using the Benjamini and Hochberg false discovery rate. Final corrected p values were filtered to be less than 0.02.
Subsequently, average log2 ratios for 2 h of HU treatment (LFQ ratio 2 h/0 h HU treatment) and average log2 ratios for 24 h of HU treatment (LFQ ratio 24 h/0 h HU treatment) were calculated, and p values of each protein across all treatment conditions were calculated by ANOVA, using permutation-based FDR. SUMOylated proteins were considered significantly up- or down-regulated when their average log2 ratio (2 h/0 h HU treatment) or average log2 ratio (24 h/0 h HU treatment) was greater than 1 or less than −1, and corresponding ANOVA p values were less than 0.05; selection criteria are detailed in Fig. 4 step 5. Volcano plots to demonstrate significant changes in protein SUMOylation upon HU treatment were created by plotting the Student's t test -log10(P) value of LFQ 2 h/0 h against the average log2(LFQ ratio 2 h/0 h) value, and the Student's t test -log10(P) value of 24h/0h against the average log2(LFQ ratio 24 h/0 h). Significantly regulated SUMOylated proteins after 2 h or 24 h of HU treatment were selected to perform functional protein interaction analysis by STRING (string-db.org, version 9.1) using a confidence score of medium or higher (p > 0.4). STRING analysis results were visualized using Cytoscape (version 3.1.0).
Site level SUMOylation data analysis
Site-specific purifications were performed in biological quadruplicate, and all samples were measured in technical duplicate. All 24 RAW files were analyzed by MaxQuant (version 1.5.1.2). The first search was carried out with a mass accuracy of 20 ppm, whereas the main search used 6 ppm for precursor ions. Database searches were performed with Trypsin/P specificity, allowing two missed cleavages. Carbamidomethylation of cysteine residues was considered as a fixed modification. Mass tolerance of MS/MS spectra was set to 20 ppm to search against an in silico digested UniProt reference proteome for Homo sapiens (29 October 2014, 88812 proteins). Additionally, MS/MS data were searched against a list of 245 common mass spectrometry contaminants by Andromeda. Pyro-QQTGG (K), QQTGG (K) and phosphorylation (STY) were set as variable modifications. In order to increase identification certainty, diagnostic peaks were searched within MS/MS spectra corresponding to SUMOylated peptides. The default minimum Andromeda score for accepting MS/MS spectra corresponding to modified peptides was left at 40 and the minimum delta score was left at 6. Additionally identified Pyro-QQTGG and QQTGG sites without diagnostic peaks, matching reversed sequences, or having a localization probability of less than 0.9 were excluded. Match between runs was used with 0.7 min match time window and 20 min alignment time window. Sequence windows of −15 to +15 with respect to the identified SUMO-2 acceptor lysines were used to generate a SUMOylation motif, employing iceLogo software.
Intensities of Pyro-QQTGG and QQTGG were further analyzed by Perseus (1.5.0.31). Pyro-QQTGG and QQTGG site intensities were normalized by the total site-peptide intensities in each sample, and then all the intensities were log2 transformed. Experiments were annotated into three groups: untreated control, 2 h HU treatment, and 24 h HU treatment. Sites occurring in at least one treatment condition and identified in all four biological repeats, were included for further analysis. Missing values were imputed using Perseus software by normally distributed values with a 1.8 downshift (log2) and a randomized 0.3 width (log2), for each experimental condition individually. Volcano plots to demonstrate significant changes in protein SUMOylation upon HU treatment were created by plotting -log10(P) values of 2 h/0 h HU and 24 h/0 h HU from pairwise comparisons of SUMO-2 target proteins against the average LFQ Ratio 2 h/0 h (log2) and LFQ Ratio 24 h/0 h (log2).
RESULTS
A Quantitative Proteomics Approach to Identify SUMO-2 Target Proteins and Acceptor Lysines that are Dynamically SUMOylated in Response to Replication Stress
Strategy to Enrich SUMO-2 Conjugates
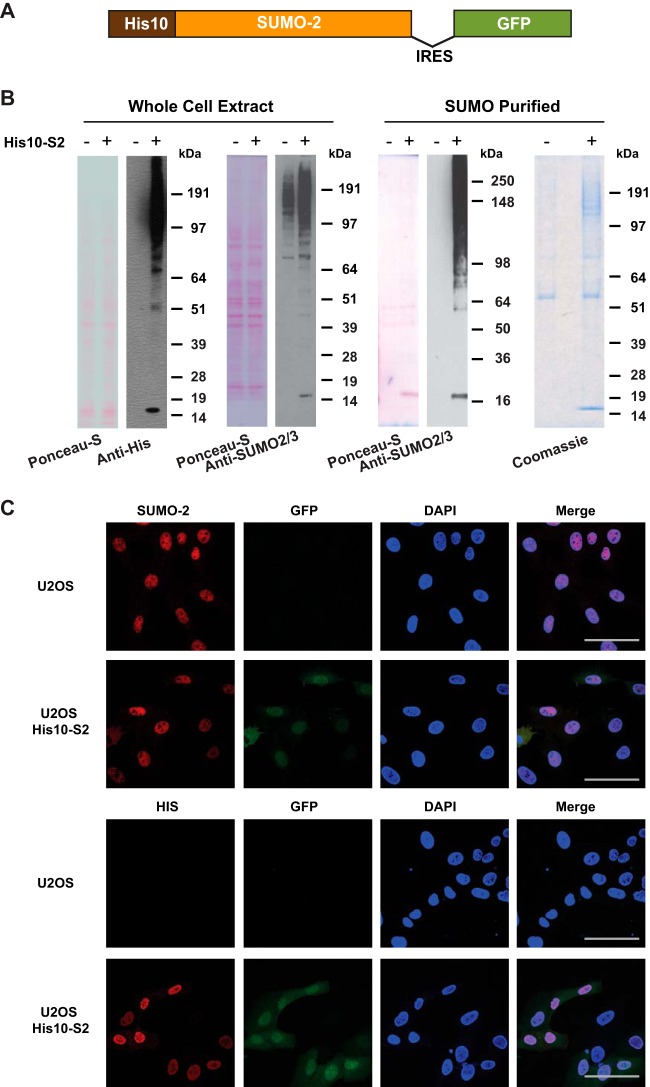
In order to purify SUMO-2 target proteins from cells under replication stress conditions, we first generated a U2OS cell line stably expressing His10-tagged SUMO-2 (abbreviated as His10-S2). To facilitate flow cytometry sorting of cells expressing a low level of His10-S2, GFP was linked to the His10-S2 cDNA via an internal ribosome entry site (IRES) (Fig. 1A). A homogeneous population of low level GFP expressing cells was selected by flow cytometry to avoid overexpression artifacts. Immunoblotting analysis confirmed the relatively low expression level of His10-SUMO-2 compared with endogenous SUMO-2 levels in U2OS cells, and the efficient enrichment of SUMO-2 conjugates by Ni-NTA purification. Ponceau S staining is shown as a loading control. Coomassie staining showed the low level of nonspecific binding of non-SUMOylated proteins (Fig. 1B). Analysis of the cells by confocal microscopy revealed that His10-SUMO-2 was predominantly located in the nucleus (Fig. 1C).
Fig. 1.
Generation and validation of U2OS cells stably expressing His10-SUMO-2. A, Schematic representation of the His10-SUMO-2-IRES-GFP construct used in this project. U2OS cells were infected with a lentivirus encoding His10-SUMO-2 (His10-S2) and GFP separated by an Internal Ribosome Entry Site (IRES) and cells stably expressing low levels of GFP were sorted by flow cytometry. B, Expression levels of SUMO-2 in U2OS cells and His10-SUMO-2 (His10-S2) expressing stable cells were compared by immunoblotting. Whole cell extracts were analyzed by immunoblotting using anti-polyHistidine and anti-SUMO-2/3 antibody to confirm the expression of SUMO-2 in U2OS cells and His10-SUMO-2 (His10-S2) expressing stable cells. Ponceau-S staining is shown as a loading control. Additionally, a His-pulldown was performed using Ni-NTA agarose beads to enrich SUMOylated proteins, and purification of His10-SUMO-2 conjugates was confirmed by immunoblotting using anti-SUMO-2/3 antibody. Ponceau-S staining and Coomassie staining were performed to confirm the purity of the final fraction. The experiment was performed in three biological replicates. C, The predominant nuclear localization of His10-SUMO-2 was visualized via confocal fluorescence microscopy after immunostaining with the indicated antibodies. DAPI staining was used to visualize the nuclei. Scale bars represent 75 μm.
Replication Stress Induction on HU Treatment
To investigate global changes of proteins that are dynamically SUMOylated during early and late replication damage events, we employed a label free quantitative proteomics approach (Fig. 2A). It was reported before that after short replication blocks, replication forks can stay viable and are able to restart after release from the replication block. In contrast, prolonged stalling of replication forks is known to result in the generation of DNA double strand breaks (DSBs) in S phase and requires HR-mediated restart (43). We cultured U2OS and U2OS cells which stably expressed His10-tagged SUMO-2 (His10-S2) in regular medium and then treated these cells with 2 mm of the replication inhibiting agent hydroxyurea (HU) for 2 h or for 24 h in order to induce replication fork stalling.
Fig. 2.
A strategy for discerning SUMOylation dynamics during replication stress. A, Cartoon depicting the strategy to study SUMOylation dynamics during replication stress. U2OS cells expressing His10-SUMO-2 were treated with 2 mm Hydroxyurea (HU) for 2 h or 24 h to induce DNA replication fork stalling and double strand breaks, respectively. Parental U2OS cells and U2OS cells expressing His10-SUMO-2 were mock treated as negative controls. SUMO-2 target proteins were purified by Ni-NTA purification. To study SUMO-2 targets that dynamically respond to replication stress, five biological replicates were performed. B, Purification of His10-SUMO-2 conjugates via NTA purification was confirmed by immunoblotting. Whole cell extracts and SUMO-2 purified proteins of the differently treated cells were run on 4–12% Bis-Tris polyacrylamide gels and levels of His10-SUMO-2 conjugates were compared by immunoblotting using anti-SUMO-2/3 antibody.
We first used Ni-NTA purification to enrich His10-SUMO-2 conjugates from U2OS cells stably expressing His10-SUMO-2, after treatment with HU for either 2 h, 24 h, or after mock treatment. Parental U2OS cells were included as a negative control. Immunoblotting analysis was employed to assess global purified SUMO-2 conjugates. The total level of SUMO-2 conjugation appeared to be equal and the immunoblotting analysis also confirmed our highly efficient enrichment for SUMO-2 conjugates (Fig. 2B).
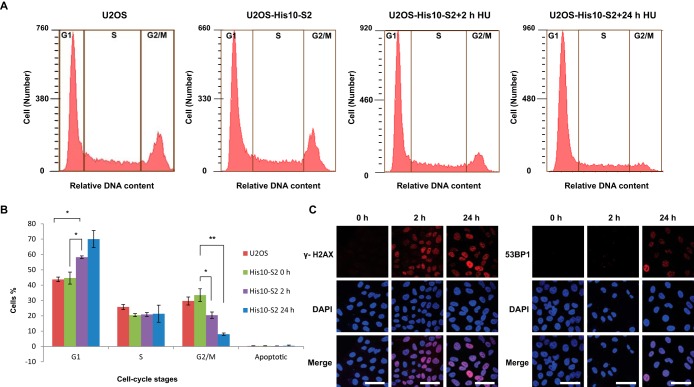
As indicated in Fig. 3A and 3B, flow cytometry analysis from three independent experiments confirmed the enrichment of cells in the G1 phase and a decreased number of G2/M cells after 2 h HU treatment, and a further decrease of G2/M phase cells upon 24 h HU treatment, which confirmed stalling of the replication forks.
Fig. 3.
HU-induced DNA damage in U2OS stably expressing His10-SUMO-2. A, DNA content analysis of Hydroxyurea treated and nontreated cells. Flow cytometry was employed to confirm an increase in G1 phase cells upon HU treatment and a corresponding decrease in G2/M phase cells. B, The percentage of cells in each cell cycle phase is depicted. Error bars indicate the standard deviation from three independent replicates. Asterisks indicate significant differences by two-tailed Student's t testing. * p < 0.05, ** p < 0.001. C, Localization of γH2AX and 53BP1 on HU treatment. Cells were treated with 2 mm HU for 2 h or 24 h or left untreated. Cells were then fixed, permeabilized, and immunostained for γH2AX (red) or 53BP1 (red), and DNA was stained with DAPI (blue). Scale bars represent 75 μm.
To further ratify that HU treatments induced the anticipated DNA damage response, we measured the formation of the phosphorylated histone variant H2AX (γH2AX) foci after 2 h and 24 h HU treatment and mock treatment was used as negative control (Fig. 3C). As reported before (44), γH2AX accumulated during 2 h HU treatment and further increased numbers of foci were observed after 24 h HU treatment. Furthermore, we checked the formation of Double Strand Break (DSB)-associated 53BP1 foci, and confirmed a large increase in foci upon 24 h HU treatment (Fig. 3C).
Identification of SUMOylated Proteins Using Label Free Quantification
His10-SUMO-2 purification coupled to mass spectrometry and label free quantification as described in the experimental procedures were used to study the abundance of SUMOylated proteins in response to replication stress (Fig. 4). In total, five biological replicates with three technical repeats for each condition were analyzed in this study. Label free quantification was performed using the MaxQuant software suite which quantifies proteins by MS1 peak intensities (41). Peak intensities measured during individual runs were matched to all other runs.
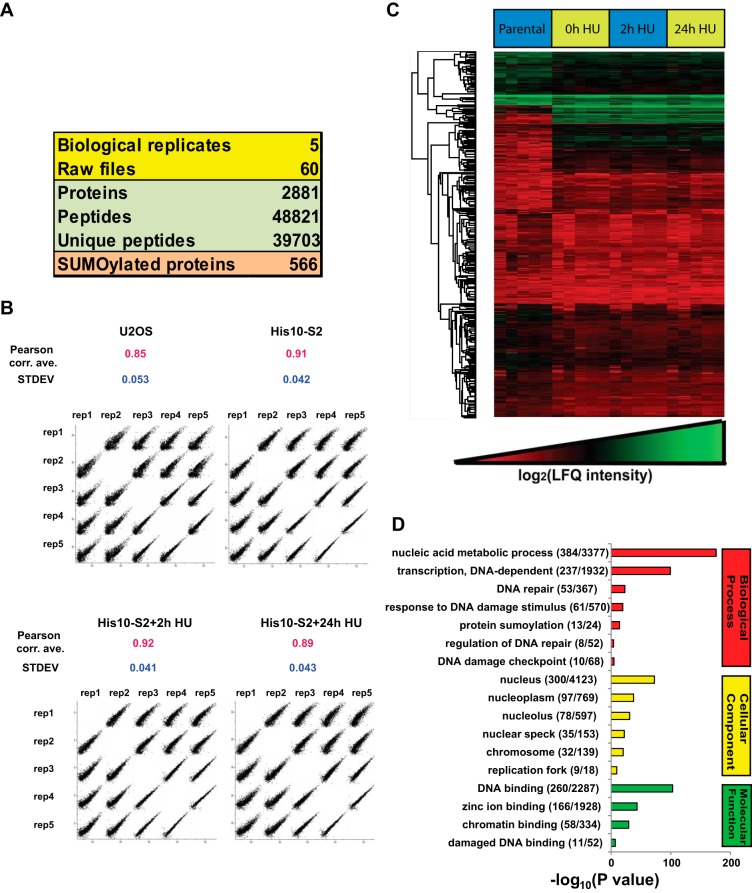
A total of 2,881 proteins were identified from 48,821 peptides and 566 of them were considered as SUMO-2 target proteins in response to DNA replication stress (Fig. 5A and supplemental Table S1). After filtering out contaminants and putative false positives, all the LFQ intensities were transformed by log2. The multiple scatter plot in Fig. 5B shows high correlation within each condition throughout different biological replicates. SUMO-2 purified fractions showed more correlation than parental control because of specific enrichment of SUMOylated proteins by the affinity purification. Next, log2 ratios of all the LFQ intensities were used to generate a heat map by hierarchical clustering of all proteins. The heat map also visualized a high correlation between the biological replicates (Fig. 5C).
Fig. 5.
Overview of the SUMO proteomics results. A, Overview of the proteomic experiments. Out of 2,881 proteins identified with 48,821 peptides, 566 proteins were considered as SUMO-2 target proteins after filtering by LFQ intensities as described in Fig. 4. B, LFQ intensity scatter plot. Each condition of each biological replicate was plotted together to visualize the correlation between the experiments. Pearson correlation averages were calculated for each condition and standard deviations (S.D.) are indicated. C, Heat map of log2 LFQ intensities. Hierarchical clustering was performed for all identified proteins. Within each biological replicate, the sample order from left to right was U2OS, U2OS His10-SUMO-2 (mock treated), U2OS His10-SUMO-2 (2 h HU), and U2OS His10-SUMO-2 (24 h HU). D, GO term enrichment analysis of the SUMOylated proteins identified. The bar chart shows GO terms for biological processes, cellular components and molecular functions.
Subsequently, LFQ ratios corresponding to proteins derived from SUMO-2 enriched fractions purified from either the parental U2OS cell line or U2OS cells stably expressing His10-SUMO-2 were compared, in order to filter out non-specifically binding proteins (Fig. 4). After selecting proteins that were found in at least four biological replicates in at least one experimental condition in SUMO-2 purified samples, missing LFQ ratios were imputed as described in the experimental procedures. Proteins were considered as SUMO-2 target proteins when they were enriched at least twofold from His10-SUMO-2 expressing cells compared with U2OS parental control cells. The SUMOylated protein list is provided in supplemental Table S1.
To assess the biological function of SUMOylated proteins identified in this study, we performed Gene Ontology (GO) term enrichment analysis using Perseus (Fig. 5D, supplemental Table S2 and supplemental Table S3). For GO Biological Processes, proteins involved in the DNA damage response were found to be significantly enriched. Fifty-three proteins were related to DNA repair, 61 proteins were related to the DNA damage response. For GO Cellular Compartments, 300 proteins were found to be located in the nucleus. For GO Molecular Functions, 260 proteins were involved in DNA binding and 11 proteins were involved in damaged DNA binding.
Analysis of SUMOylated Protein Dynamics During Replication Stress
SUMOylation dynamics in response to replication stress was explored using bioinformatics analysis as described under “Experimental Procedures” and in Fig. 4. Ten proteins were significantly increased in SUMOylation and 2 proteins were significantly decreased in SUMOylation after 2 h HU treatment. After 24 h HU treatment, 35 proteins were significantly increased in SUMOylation and 13 proteins were significantly decreased in SUMOylation (Table S4).
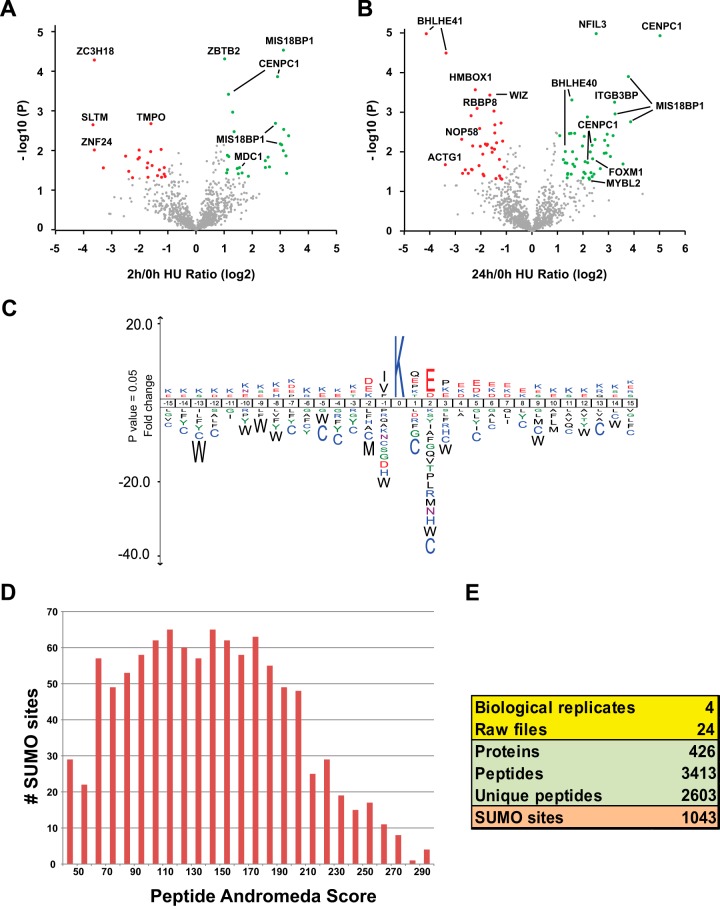
Volcano plots shown in Fig. 6A and 6B indicate the significance and magnitude of SUMO-2 target protein changes after 2h and 24h HU treatment. p values less than 0.05 were considered significant. STRING analysis of significantly regulated SUMOylated proteins was performed. Fig. 6C shows the interaction of proteins significantly increased or decreased in SUMOylation after 2 h of HU treatment. SUMOylation of CHAF1A and PCNA was significantly decreased. On the other hand, SUMOylation of MCM4, MYBL2 and FOXM1 was found to be increased. Similar to the finding of Li et al. (45), PCNA is a hub connecting several other SUMOylated proteins, including CHAF1A, FOXM1, MYBL2, ATRIP, and MCM4. After 24 h of HU treatment (Fig. 6D), BHLHE41, CHAF1A, and DNMT1 were significantly decreased in SUMOylation. Additionally, BHLHE40, BARD1, MDC1, RMI1, and BRCA1 were greatly increased in SUMOylation. EME1 was found to be modestly down-regulated and additionally high-lighted in Fig. 6B. Most of the SUMOylated proteins were significantly interacting to each other throughout the STRING network. In conclusion, DNA replication stress caused by HU treatment changed the SUMOylation of a distinct subset of proteins with key functions in the DNA damage response.
Fig. 6.
Volcano plots and STRING protein interaction network of dynamically regulated SUMO-2 target proteins. A, B, Volcano plots to show significantly altered SUMO-2 targets in response to 2 h HU treatment (A) or 24 h HU treatment (B). The -log10(P) value of 2 h/0 h and 24 h/0 h from pairwise comparisons of SUMO-2 target proteins purified from mock treated cells and HU-treated cells were plotted against the average LFQ ratio 2 h/0 h (log2) and LFQ ratio 24 h/0 h (log2). The red dots represent proteins decreased for SUMOylation in response to HU with an average log2 ratio smaller than −1. The green dots represent proteins increased for SUMOylation in response to HU with an average log2 ratio greater than 1. C, STRING analysis of dynamically regulated SUMO-2 target proteins after 2 h Hydroxyurea treatment. p value: 1.42*10−7. Up-regulated SUMOylated proteins are colored in green and down-regulated SUMOylated proteins are colored in red. D, STRING analysis of dynamically regulated SUMO-2 target proteins after 24 h Hydroxyurea treatment. p value: 6.94*10−14. Up-regulated SUMOylated proteins are colored in green and down-regulated SUMOylated proteins are colored in red.
Site-specific SUMOylation Dynamics During Replication Stress
Previously we have developed methodology to study SUMOylation at the site-specific level (supplemental Fig. S1) (38, 39). Here we used similar methodology to map SUMO-2 acceptor lysines in U2OS target proteins. First, we generated U2OS cells stably expressing lysine-deficient His10-SUMO-2 with a conserved mutation at its C-terminal part, Q87R, which mimics the localization of an arginine in yeast SUMO, Smt3. Characterization of the cell line is shown in supplemental Fig. S2. We used this cell line to enrich SUMOylation sites from mock treated cells, or cells treated with HU for 2 h or 24 h to induce replication stress. In total, 1,043 SUMOylation sites were identified in 426 proteins (supplemental Tables S5 and S6), at a false discovery rate (FDR) below 1%. Mass accuracy was within 3 p.p.m. for 98.8% of all identified sites and within 6 p.p.m. for all sites, with an average absolute mass error of 0.78 p.p.m. We identified 83 peptides comodified by SUMOylation and phosphorylation, of which 24 SUMOylated peptides were not found in the nonphosphorylated form (supplemental Table S9).
In order to study the dynamics of SUMO-2 acceptor sites in response to HU treatment, SUMO-2 site MS1 peak intensities were normalized and compared between samples. Volcano plots were used to visualize SUMOylation site dynamics in response to 2 h or 24 h HU treatment. (Fig. 7A and 7B). Sites displaying a significant change of at least 2-fold in response to HU treatment are highlighted. After 2 h of HU treatment, 27 sites were significantly up-regulated and 23 sites were significantly down-regulated. After 24 h of HU treatment, 49 sites were up-regulated and 38 sites were down-regulated (Table S7). Overall, the dynamic SUMO-2 sites correspond well with the results from our site-independent approach (supplemental Table S8).
Fig. 7.
Volcano plots of dynamically regulated SUMOylation sites and SUMOylation motif analysis. A, B, Volcano plots showing dynamically regulated SUMO-2 acceptor sites in response to 2 h HU treatment (A) or 24 h HU treatment (B). The -log10(P) value from pairwise comparisons of SUMO-2 acceptor lysines purified from mock treated cells and HU-treated cells were plotted against the average LFQ Ratio 2 h/0 h (log2) and LFQ Ratio 24 h/0 h (log2). The red dots represent sites decreased for SUMOylation in response to HU with an average log2 ratio smaller than −1.0 and with p < 0.05. The green dots represent sites increased for SUMOylation in response to HU with an average log2 ratio greater than 1.0 and with p < 0.05. C, All SUMO-2 acceptor lysines identified in this study (1043 sites) were used to generate a SUMOylation motif employing IceLogo software. The height of the amino acid letters represents the fold change as compared with amino acid background frequency. All amino acid changes were significant with p < 0.05 by two-tailed Student's t test. D and E, Summary of the SUMO-2 acceptor lysines identified (E) with their peptide Andromeda scores (Median = 141.36) (D).
Verification of Dynamic SUMO Targets on DNA Replication Stress
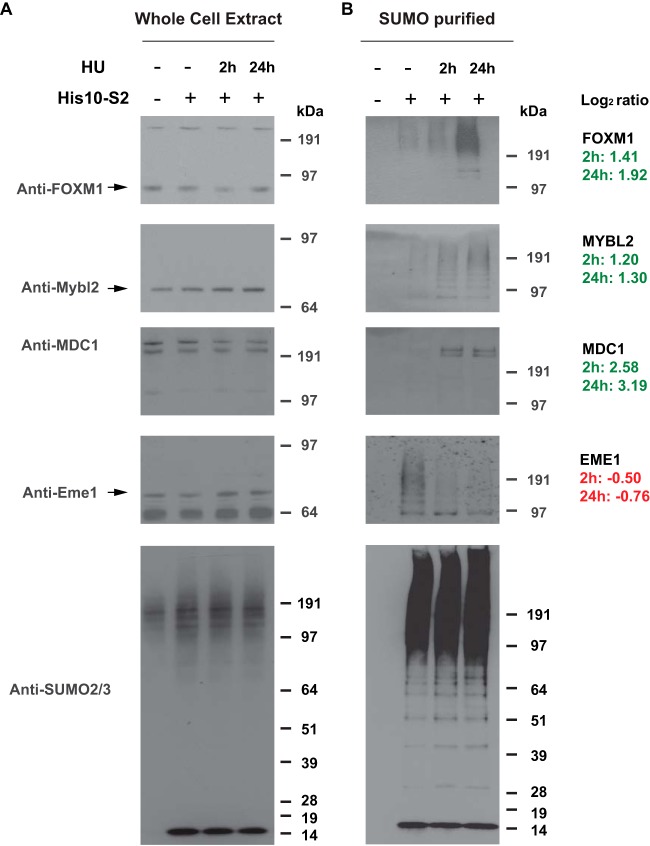
We verified SUMOylation dynamics upon DNA replication stress by immunoblotting analysis for a subset of the identified dynamic SUMO-2 targets, including FOXM1, MYBL2, MDC1, and EME1 (Fig. 8). To study whether the HU treatment was efficient, flow cytometry was used to confirm the expected effects on cell cycle progression (Fig. 3A and 3B). All four of these SUMO-2 target proteins were found to be dynamically SUMOylated in accordance with the LFQ data derived from the mass spectrometry analysis, whereas the total amount of SUMO remained stable. As such, we demonstrated the feasibility of our approach, providing a powerful tool for analysis of SUMOylation dynamics in general, as well as a reliable resource of SUMO-2 target proteins dynamically regulated in response to replication stress.
Fig. 8.
Verification of SUMO targets showing SUMOylation dynamics in response to replication stress. U2OS cells and U2OS cells expressing His10-tagged SUMO-2 were either mock treated or treated with Hydroxyurea (2 mm) for 2 h or 24 h as described, and His10-SUMO-2 conjugates were purified by Ni-NTA purification. SUMOylation dynamics induced by DNA replication stress was analyzed for four different SUMO-2 targets identified in the mass spectrometry screen using the indicated antibodies, and equal levels of SUMO conjugates in all samples were verified via immunoblotting using anti-SUMO-2/3 antibody. The fold changes in SUMOylation (log2) of these proteins as found in our proteomics screen are indicated on the right.
DISCUSSION
Our knowledge on the role of SUMOylation to maintain genome stability during replication is limited, because of limited insight into all involved SUMO target proteins. To address this, we have optimized a purification procedure to enrich SUMO target proteins, reaching a depth of 566 proteins. The optimized methodology employs the His10 tag, enabling the use of denaturing buffers to inactivate proteases and combining a high yield with a high purity; a major improvement over the His6 tag as a result of the usage of a much higher concentration of competing imidazole during the purification procedure to reduce the binding of contaminating proteins.
We have used the optimized methodology to study SUMOylation dynamics in response to HU-induced replication stress, resulting from the depletion of dNTPs required for DNA replication. A group of 12 dynamic SUMO-2 targets was identified when cells were treated for 2 h with HU, including 10 up-regulated and 2 down-regulated proteins. When treating U2OS cells for 24 h with HU, 48 dynamic SUMO-2 targets were identified including 35 up-regulated and 13 down-regulated proteins. As a cautionary note, we cannot exclude the possibility that changes in total levels of some proteins could underlie some of the observed changes in SUMOylation. More than half (2h: 70%, 24h: 52%) of these targets are functionally connected, indicating tight interactions between the SUMO-orchestrated proteins. The identified SUMO-regulated functional groups include key replication factors, DDR-components, a transcription-factor network, centromeric proteins, and signal transducers.
Identification of sites of modification is the most reliable manner to study post-translational modification of proteins (5). Powerful site-specific methodology is available to study phosphorylation and ubiquitination. In contrast, this has remained a major challenge in the SUMOylation field (5). Several years ago, we have developed a novel approach, enabling the identification of a limited set of SUMO-2 acceptor lysines in endogenous target proteins (38). Recently, we have further optimized this methodology by employing the His10-tag, enabling large-scale identification of SUMOylation sites (39). The use of a lysine-deficient version of SUMO-2 enabled the enrichment of SUMOylated peptides after Lys-C digestion. This is a key step to reduce the complexity of samples prior to mass spectrometry analysis. Using this methodology, we have identified 1043 SUMOylation sites in this project, including 382 sites not previously identified (39). Similar methodology could be employed to study other ubiquitin-like modifications.
Proliferating Cell Nuclear Antigen (PCNA) is one of the most strongly down-regulated SUMO target proteins after two hours of HU treatment and is also one of the most highly connected components of the SUMO-target protein interaction network as depicted in Fig. 6C. PCNA is a trimeric replication clamp that serves as platform for replicating polymerases. SUMOylated PCNA is known to interact with the helicase Srs2 to counteract recombination (18–20). Down-regulation of PCNA SUMOylation at early time points could represent a mechanism to explain increased recombination as a result of HU treatment, to counteract replication fork stalling (46).
MCM4 is a second key replication factor that we identified in our screen. MCM4 is a component of the DNA replication licensing factor Mini Chromosome Maintenance (MCM), which consists of MCM2–7 hexamers. The MCM complex plays an essential role in replication licensing and ensures that the entire genome is replicated exactly once during S-phase, avoiding reduplication or leaving genomic regions unreplicated (47). It acts as a helicase to unwind DNA, enabling access for the replication machinery to duplicate DNA. Interestingly, we previously found that MCM4 SUMOylation preferentially occurs during G1, a time point when MCM complexes are loaded on DNA for replication-licensing (48). Our data indicate a small increase in MCM4 SUMOylation in response to HU-treatment, which potentially could be involved in a cellular attempt to complete replication by firing of dormant origins in response to replication fork stalling (49). It is currently unclear how SUMOylation precisely regulates MCM4.
Furthermore, we identified a set of known SUMOylated DNA damage response factors, including MDC1, BRCA1, and BLM. MDC1 was previously found to be SUMOylated in response to ionizing radiation (28–31), and BRCA1 was found to be SUMOylated in response to cisplatin and HU (26). SUMOylation regulates the interaction of BLM and RAD51 at damaged replication forks (50) and the accumulation of ssDNA at stalled replication forks. The identification of SUMOylated BRCA1 and BLM is thus in agreement with the existing literature, and underlines the validity of our approach.
After prolonged exposure of cells to HU, SUMOylation of a cluster of centromeric proteins was induced, including CENPC1, CENPH, and MIS18A (51) which is required for regulating CENPA deposition. Our data thus indicate co-regulation of centromeric proteins by SUMOylation in response to replication stress. The functional significance of these findings could further be explored, because other studies have demonstrated that SUMOylation at centromeres plays a key role to regulate cell cycle progression (23, 52–54).
Finally, it is interesting to note that SUMOylation regulates a number of factors involved in other modifications, demonstrating extensive signaling crosstalk. These proteins include the ATR-interacting protein ATRIP, ubiquitin E3 ligases RAD18 and BRCA1, the lysine-specific demethylases KDM5D, KDM5C, and KDM4A. SUMOylation of HDAC1 and BRCA1 was previously demonstrated to promote enzymatic activity (26, 55), and it would be interesting to determine the functional relevance of SUMOylation for these other SUMO target proteins. Concerning crosstalk, we also obtained evidence for mixed SUMO-ubiquitin chains in our study, including linkages of SUMO-2 to lysines 11, 48, and 63 of ubiquitin. Furthermore, we found 83 peptides simultaneously modified by both SUMO-2 and phosphorylation (supplemental Table S9).
In summary, our study provides a critical framework to understand the role of SUMOylation to maintain genome stability during replication stress. The identified target proteins can be studied in detail at the functional level. Replication problems frequently lead to genome instability, one of the enabling characteristics of cancer cells (56). A detailed understanding of the role of SUMOylation in genome instability could potentially be employed to counteract oncogenesis, similarly to recent successful efforts to disrupt signal transduction by the ubiquitin-like protein Nedd8 (57, 58). Moreover, the developed methodology could be widely employed to study SUMOylation, but also ubiquitination and signal transduction by other ubiquitin-like proteins.
Ultimately, our study reveals how SUMO regulates a network of target proteins in response to replication stress to coordinate the cellular DNA damage response. This network not only consists of known DNA damage response factors, but also includes replication factors, transcriptional regulators, chromatin modifiers and centromeric proteins, revealing how a post-translational modification is able to orchestrate a large variety of different proteins to integrate different nuclear processes with the aim of dealing with the induced DNA damage.
Supplementary Material
Footnotes
Author contributions: A.C.V. designed research; Z.X. and J.C. performed research; I.A.H., J.S., and J.V.O. contributed new reagents or analytic tools; Z.X., J.C., I.A.H., and A.C.V. analyzed data; Z.X., J.C., I.A.H., and A.C.V. wrote the paper.
* This work was supported by the Marie Curie Initial Training Network UPStream of the European Union (A.C.O.V. and J.V.O.). This work was furthermore supported by the European Research Council (A.C.O.V.) and the Netherlands Organization for Scientific Research (NWO) (A.C.O.V.). The NNF Center for Protein Research is supported by a generous donation from the Novo Nordisk Foundation. None of the authors have a financial interest related to this work. The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001736.
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S9.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S9.
1 The abbreviations used are:
- 53BP1
- Tumor suppressor p53-binding protein 1
- Atg8
- Ubiquitin-like protein ATG8
- Atg12
- Ubiquitin-like protein ATG12
- BHLHE40/41
- Class E basic helix-loop-helix protein 40/41
- BLM
- Bloom syndrome protein, RecQ helicase-like
- BRCA1
- Breast cancer type 1 susceptibility protein
- CENPC1
- CENPC, Centromere protein C
- CENPH
- Centromere protein H
- CHAF1A
- Chromatin assembly factor 1, subunit A
- DDR
- DNA damage response
- dNTPs
- deoxynucleotide triphosphates
- DSBs
- Double strand breaks
- EME1
- Crossover junction endonuclease EME1
- FAT10
- UBD or ubiquitin D
- FOXM1
- Forkhead box protein M1
- FUBI
- Ubiquitin-like protein FUBI
- GO
- Gene Ontology
- HERC2
- HECT domain and RCC1-like domain-containing protein 2, E3 ligase
- His10-S2
- His10-SUMO-2-IRES-GFP
- His10-S2-K0-Q87R
- His10-SUMO-2-K0-Q87R-IRES-GFP
- HU
- Hydroxyurea
- HUB1
- Ubiquitin-like modifier HUB1
- IAA
- Iodoacetamide
- IR
- Ionizing radiation
- IRES
- Internal ribosome entry site
- ISG15
- Ubiquitin-like modifier ISG15
- K0
- Lysine-deficient
- LFQ
- Label free quantification
- MAFF
- Transcription factor MafF
- MCM4
- Minichromosome maintenance complex component 4
- MDC1
- Mediator of DNA-damage checkpoint 1
- MIS18A
- Protein Mis18-alpha
- MMS
- Methyl methanesulfonate
- MYBL2
- Myb-related protein B
- NACC1
- Nucleus accumbens-associated protein 1
- Nedd8
- Ubiquitin-like protein NEDD8
- PCNA
- Proliferating Cell Nuclear Antigen
- PIAS
- E3 SUMO-protein ligase
- RANBP2
- RAN binding protein 2
- RMI1
- RecQ-mediated genome instability protein 1
- RNF168
- E3 ubiquitin-protein ligase RNF168
- SAE1
- SUMO activating enzyme subunit 1
- SAE2
- SUMO-activating enzyme subunit 2, UBA2
- Srs2
- ATP-dependent DNA helicase SRS2
- STRING
- Search Tool for the Retrieval of Interacting Genes and Proteins
- SUMO
- Small Ubiquitin-like Modifier
- TCEP
- Tris(2-carboxyethyl)phosphine hydrochloride
- TOP2A
- DNA topoisomerase II α
- UBC9
- SUMO-conjugating enzyme UBC9
- Ubl
- Ubiquitin like
- UFM1
- Ubiquitin-fold modifier 1
- URM1
- Ubiquitin related modifier 1
- UV
- Ultraviolet.
REFERENCES
- 1. Deribe Y. L., Pawson T., Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 [DOI] [PubMed] [Google Scholar]
- 2. Scott J. D., Pawson T. (2009) Cell Signaling in Space and Time: Where Proteins Come Together and When They're Apart. Science 326, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 4. Komander D., Rape M. (2012) The Ubiquitin Code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 5. Vertegaal A. C. O. (2011) Uncovering Ubiquitin and Ubiquitin-like Signaling Networks. Chem. Rev. 111, 7923–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 7. Flotho A., Melchior F. (2013) Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 [DOI] [PubMed] [Google Scholar]
- 8. Gareau J. R., Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hickey C. M., Wilson N. R., Hochstrasser M. (2012) Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodriguez M. S., Dargemont C., Hay R. T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 11. Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 12. Ulrich H. D. (2012) Ubiquitin and SUMO in DNA repair at a glance. J. Cell Sci. 125, 249–254 [DOI] [PubMed] [Google Scholar]
- 13. Jackson S. P., Durocher D. (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 [DOI] [PubMed] [Google Scholar]
- 14. Bergink S., Jentsch S. (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
- 15. Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., Dejean A. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 [DOI] [PubMed] [Google Scholar]
- 16. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 17. Stelter P., Ulrich H. D. (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 [DOI] [PubMed] [Google Scholar]
- 18. Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 [DOI] [PubMed] [Google Scholar]
- 19. Armstrong A. A., Mohideen F., Lima C. D. (2012) Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483, 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., Sung P., Ulrich H. D. (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 [DOI] [PubMed] [Google Scholar]
- 21. Mao Y., Sun M., Desai S. D., Liu L. F. (2000) SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc. Natl. Acad. Sci. U.S.A. 97, 4046–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mao Y., Desai S. D., Liu L. F. (2000) SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem. 275, 26066–26073 [DOI] [PubMed] [Google Scholar]
- 23. Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J. M. (2008) Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 133, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eladad S., Ye T. Z., Hu P., Leversha M., Beresten S., Matunis M. J., Ellis N. A. (2005) Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14, 1351–1365 [DOI] [PubMed] [Google Scholar]
- 25. Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009) Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., Ng T., Solomon E. (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 [DOI] [PubMed] [Google Scholar]
- 27. Danielsen J. R., Povlsen L. K., Villumsen B. H., Streicher W., Nilsson J., Wikstrom M., Bekker-Jensen S., Mailand N. (2012) DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 197, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo K., Zhang H., Wang L., Yuan J., Lou Z. (2012) Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 31, 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galanty Y., Belotserkovskaya R., Coates J., Jackson S. P. (2012) RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yin Y., Seifert A., Chua J. S., Maure J. F., Golebiowski F., Hay R. T. (2012) SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vyas R., Kumar R., Clermont F., Helfricht A., Kalev P., Sotiropoulou P., Hendriks I. A., Radaelli E., Hochepied T., Blanpain C., Sablina A., van A. H., Olsen J. V., Jochemsen A. G., Vertegaal A. C., Marine J. C. (2013) RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 20, 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Psakhye I., Jentsch S. (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 [DOI] [PubMed] [Google Scholar]
- 33. Cremona C. A., Sarangi P., Yang Y., Hang L. E., Rahman S., Zhao X. (2012) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. (2006) Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127, 509–522 [DOI] [PubMed] [Google Scholar]
- 35. Branzei D., Vanoli F., Foiani M. (2008) SUMOylation regulates Rad18-mediated template switch. Nature 456, 915–920 [DOI] [PubMed] [Google Scholar]
- 36. Branzei D., Foiani M. (2010) Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 [DOI] [PubMed] [Google Scholar]
- 37. Vertegaal A. C., Ogg S. C., Jaffray E., Rodriguez M. S., Hay R. T., Andersen J. S., Mann M., Lamond A. I. (2004) A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 279, 33791–33798 [DOI] [PubMed] [Google Scholar]
- 38. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van D. H., Gnad F., Mann M., Vertegaal A. C. (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 39. Hendriks I. A., D'Souza R. C., Yang B., Verlaan-de V. M., Mann M., Vertegaal A. C. (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 41. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 42. Cox J., Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13 Suppl 16, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones R. M., Petermann E. (2012) Replication fork dynamics and the DNA damage response. Biochem. J. 443, 13–26 [DOI] [PubMed] [Google Scholar]
- 44. Petermann E., Orta M. L., Issaeva N., Schultz N., Helleday T. (2010) Hydroxyurea-Stalled Replication Forks Become Progressively Inactivated and Require Two Different RAD51-Mediated Pathways for Restart and Repair. Mol. Cell 37, 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma L., Aslanian A., Sun H. Y., Jin M. J., Shi Y., Yates J. R., Hunter T. (2014) Identification of Small Ubiquitin-like Modifier Substrates with Diverse Functions Using the Xenopus Egg Extract System. Mol. Cell. Proteomics 13, 1659–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galli A., Schiestl R. H. (1996) Hydroxyurea induces recombination in dividing but not in G1 or G2 cell cycle arrested yeast cells. Mutat. Res. 354, 69–75 [DOI] [PubMed] [Google Scholar]
- 47. Blow J. J., Dutta A. (2005) Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schimmel J., Eifler K., Sigurdsson J. O., Cuijpers S. A. G., Hendriks I. A., Verlaan-de Vries M., Kelstrup C. D., Francavilla C., Medema R. H., Olsen J. V., Vertegaal A. C. O. (2014) Uncovering SUMOylation Dynamics during Cell-Cycle Progression Reveals FoxM1 as a Key Mitotic SUMO Target Protein. Mol. Cell 53, 1053–1066 [DOI] [PubMed] [Google Scholar]
- 49. Ge X. Q., Jackson D. A., Blow J. J. (2007) Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 21, 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ouyang K. J., Woo L. L., Zhu J., Huo D., Matunis M. J., Ellis N. A. (2009) SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 7, e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. (2007) Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell 12, 17–30 [DOI] [PubMed] [Google Scholar]
- 52. Zhang X. D., Goeres J., Zhang H., Yen T. J., Porter A. C., Matunis M. J. (2008) SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S. J. (2002) The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 54. Mukhopadhyay D., Arnaoutov A., Dasso M. (2010) The SUMO protease SENP6 is essential for inner kinetochore assembly. J. Cell Biol. 188, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. David G., Neptune M. A., DePinho R. A. (2002) SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277, 23658–23663 [DOI] [PubMed] [Google Scholar]
- 56. Hanahan D., Weinberg R. A. (2011) Hallmarks of Cancer: The Next Generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 57. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 58. Nawrocki S. T., Griffin P., Kelly K. R., Carew J. S. (2012) MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin. Investig. Drugs 21, 1563–1573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.