Abstract
In this review, we discuss new emerging medical applications of the rapidly evolving field of mammalian synthetic biology. We start with simple mammalian synthetic biological components and move towards more complex and therapy-oriented gene circuits. A comprehensive list of ON–OFF switches, categorized into transcriptional, post-transcriptional, translational and post-translational, is presented in the first sections. Subsequently, Boolean logic gates, synthetic mammalian oscillators and toggle switches will be described. Several synthetic gene networks are further reviewed in the medical applications section, including cancer therapy gene circuits, immuno-regulatory networks, among others. The final sections focus on the applicability of synthetic gene networks to drug discovery, drug delivery, receptor-activating gene circuits and mammalian biomanufacturing processes.
Keywords: synthetic biology, mammalian cells, gene therapy
1. Introduction
Synthetic Biology is a new, emerging field that aims to enhance functionality of cells by incorporating gene networks into cells through the application of engineering principles to biology. The early, pioneering studies were predominantly focused on standardization of network components to increase transfection efficiency and applications in the prokaryotic cell [1,2], while more recent applications are emerging in yeast and mammalian cell lines [3–5]. Eukaryotic cells, in contrast to prokaryotic cells, have developed sophisticated mechanisms to withstand invasion of foreign RNA/DNA material, such as the major histocompatibility complex-dependent mechanism. Furthermore, eukaryotic cells are highly compartmentalized, which poses obstacles for the application of synthetic gene networks [6,7]. It might also be used to compartmentalize applications of gene networks. In addition, the highly complex nuclear packaging of DNA and its tight regulation might hamper appropriate gene expression regulation and stable transfection. It was therefore surprising that initial synthetic biology studies performed in yeast and mammalian cell lines presented considerably positive results [8–12]. These pioneering studies were soon followed by studies in primary cells [13,14]. The majority of synthetic gene networks are expressed from plasmids, but these genetic constructs can also be inserted into the chromosome of immortalized cell lines, using various recombinase-based techniques, thus creating stable cell lines. Once synthetic biology was proved to be applicable to eukaryotic cells, the focus started to shift towards adding new tools to the reservoir of current technologies, including modification of (disease related) signalling pathways, new interventions at the gene level (known as gene therapy) and therapies involving insertion of modified (immune) cells (known as cell therapy) into patients. In this respect, synthetic biology may be considered a natural extension to the field of systems biology and systems medicine. Therefore, it holds great promise for future treatment of disease.
The emerging applications of synthetic biology to medicine are relatively new and have been the topic of a few, recent reviews [3,15,16].
In the chapter on medical applications, we review synthetic gene networks for cancer therapy, diabetes, enhancing the immune system and treatment of cardiovascular diseases. Towards this end, the main focus is on drug discovery, including gene networks for antibiotic and anti-cancer drug screening and circuits for assaying receptor activation.
2. Mammalian synthetic biology
Over the past years, synthetic biology applied to higher eukaryotes, such as mammalian cells, has rapidly evolved from the development of simple gene switches and gene networks to complex and to therapy-oriented circuits. Currently, mammalian synthetic biology provides strategies for gene- and cell-based therapies with a wide range of applications, such as artificial insemination, personalized medicine and the treatment of cancer, and metabolic and immune disorders. The application to other fields, e.g. cardiovascular disease, will also be briefly outlined. In the following sections, we will review mammalian synthetic biological gene switches, which form the basis to construct more complex gene networks.
3. Switches in mammalian synthetic biology (figures 1 and 2)
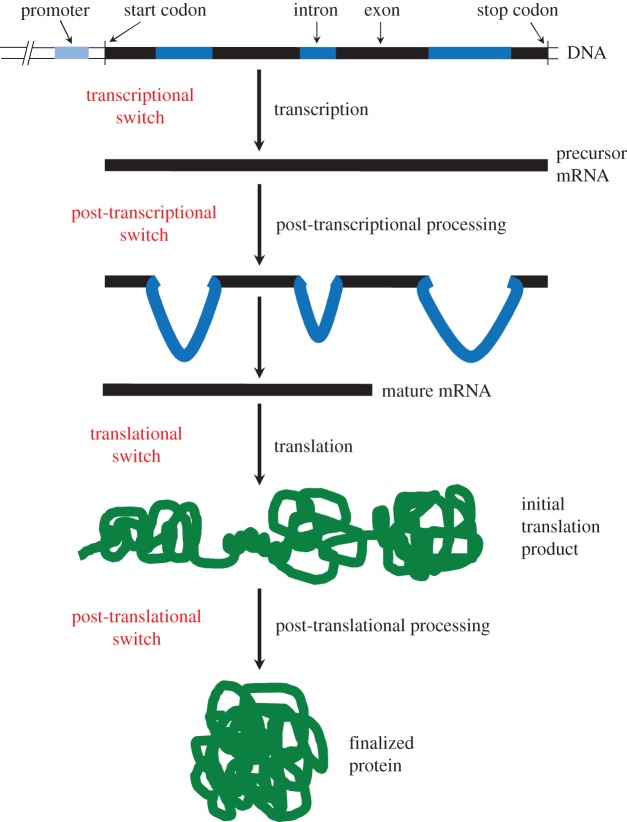
Figure 1.
ON–OFF switches in synthetic biology and their regulatory location in genetic signal processing. ON–OFF switches can control either translation, post-translational, transcriptional or post-transcriptional events. Examples of switches at all four levels are described in the text and a comprehensive list is presented in table 1. (Online version in colour.)
Figure 2.
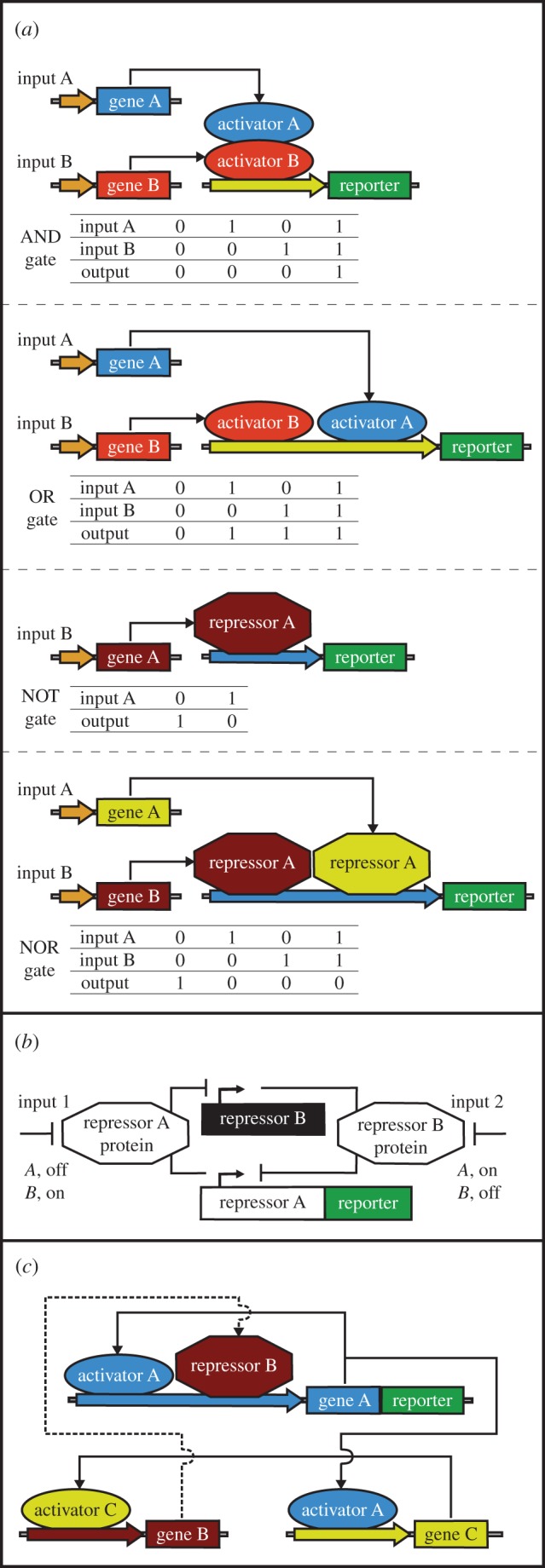
Gene networks in mammalian synthetic biology. (a) Boolean logic gates. The exemplified AND gate consists of two gene expression activators, under the control of two input signals, to enable gene expression (output) only when both activators are switched on by the input signals. The OR gate is formed by two gene expression activators, regulated by two input signals, that output gene expression when either one of the activators is switched ON. The NOT gate is composed of a repressor that shuts down the expression of a target gene in response to an input signal. Two repressors controlled by two input signals can form a NOR gate to inhibit gene expression when either one the two repressors is activated. (b) Toggle switch. The exemplified network is formed by two trans-acting repressors that inhibit each other's expression in response to input signals. An input signal applied for a short period of time sets the toggle switch into one of its pre-determined states. The reporter gene can be fused to either repressors or be under the control of these repressors. (c) Oscillator. The exemplified system is composed of two activators and a repressor. The first activator is cis-acting and is also trans-triggering expression of the second activator. The second activator then switches on the expression of the repressor. The repressor feeds back with a time-delay to inhibit expression of the first activator, generating oscillations in the concentrations of the first activator over time. In the exemplified network, the reporter gene is fused to the first activator, but it could also be regulated by this activator. (Online version in colour.)
In order to harvest the full potential of synthetic biology, biological parts should be re-organized into controllable and well-defined switches, logical operators and other systems with predictable functions. Similar to electronic switches, synthetic biological switches should be able to switch a signal between two discrete states: ON and OFF. In the case of electronic switches, the signal is electricity; in the case of biological switches, a biological signal propagates. A crucial difference between electronic and mammalian molecular switches resides in the complexity, spatial context and mobility of constitutive parts and of interacting elements. Biological switches have been shown to be context sensitive, often unbalanced in strength between individual elements, and sometimes excessively resource demanding for the machinery of cells [6,17]. Despite these difficulties, reproducible ON and OFF switches have been designed and applied in prokaryotic cells with great success [17,18]. A comprehensive list of ON–OFF switches in mammalian synthetic biology is presented in table 1, indicating the function, origin, effector molecule and applications for each switch.
Table 1.
ON–OFF switches in mammalian synthetic biology.
| switch levela | no. | name | ON switchb | OFF switchc | origin | effectord | applications | references |
|---|---|---|---|---|---|---|---|---|
| transcriptional switches | ||||||||
| 1 | ABA | yes | no | Arabidopsis thaliana, yeast | abscisic acid | gene therapy, experimental tool to control diverse cellular activities in vivo | [19] | |
| 2 | AIR | yes | no | Aspergillus nidulans | acetaldehyde | gas-adjustable transgene expression, biopharmaceutical manufacturing | [20] | |
| 3 | ART | yes | no | Chlamydia pneumoniae | l-arginine | biopharmaceutical manufacturing, gene therapy | [21] | |
| 4 | BEARON, BEAROFF | yes | yes | Campylobacter jejuni | bile acid | gene- and cell-based therapies | [22] | |
| 5 | BirA-tTA | no | yes | Escherichia coli | biotin (vitamin H) | biopharmaceutical manufacturing, cell therapy | [23] | |
| 6 | BIT | yes | no | Escherichia coli | biotin (vitamin H) | gene therapy, tissue engineering, biopharmaceutical manufacturing | [24] | |
| 7 | Cry2-CIB1 | yes | no | Arabidopsis thaliana, yeast | blue light | protein translocation, transcription control, Cre-mediated DNA recombination using light | [25] | |
| 8 | CTA, CTS | yes | yes | Comamonas testosteroni, Homo sapiens | food additives (benzoate, vanillate) | gene- and cell-based therapy | [26] | |
| 9 | cTA, rcTA | yes | yes | Pseudomonas putida | cumate | regulation of gene expression level and duration | [27] | |
| 10 | Ecdysone | yes | no | Homo sapiens, Drosophila melanogaster | Ecdysone | transient inducible expression, gene therapy | [28] | |
| 11 | EcR:RXR | yes | no | Homo sapiens, Locusta migratoria | ecdysone | ligand-dependent induction of reporter gene, gene therapy | [29] | |
| 12 | electro-genetic | yes | no | Aspergillus nidulans | electricity, acetaldehyde | electric power-adjustable transcription control; mammalian cell-based control of microelectronic circuits | [30] | |
| 13 | ER-p65-ZF | yes | no | Homo sapiens, yeast | 4,4′-dyhydroxybenzil | generating a wide array of orthogonal gene switches | [31] | |
| 14 | E.REX | yes | yes | Escherichia coli | erythromycin | gene therapy, tissue engineering, in vivo gene-function analyses, drug discovery, biopharmaceutical manufacturing | [32] | |
| 15 | EthR | no | yes | Mycobacterium tuberculosis | 2-phenylethyl-butyrate | drug discovery | [33] | |
| 16 | GAL4-ER | yes | yes | yeast, Homo sapiens | oestrogen, 4-hydroxytamoxifen | regulating heterologous genes, identifying Fos (oncogene) targets | [34] | |
| 17 | GAL4-hPR | yes | yes | yeast, Homo sapiens | mifepristone | positive and negative regulation of gene expression, also in vivo | [35,36] | |
| 18 | GAL4-Raps | yes | yes | yeast, Homo sapiens | rapamycin and rapamycin derivatives | targeted gene expression control | [37] | |
| 19 | GAL4-TR | yes | no | yeast, Homo sapiens | thyroid hormone | study nuclear receptor signalling | [38] | |
| 20 | GyrB | yes | yes | Escherichia coli | coumermycin, novobiocin | rapid regulation of gene expression | [39] | |
| 21 | HEA-3 | yes | no | Homo sapiens | 4-hydroxytamoxifen | gene therapy | [40] | |
| 22 | Intramer | no | yes | synthetic SELEX-derived aptamers | theophylline | controlling transgene expression, construction of complex gene networks | [41] | |
| 23 | LacI | yes | no | Escherichia coli | IPTG | reversible regulation of gene expression, predictable levels of de-repressed gene expression | [42–46] | |
| 24 | LAD | yes | no | Arabidopsis thaliana, yeast | blue light | light-controlled gene expression regulation; protein hetero- and homo-dimerization induced by light | [47] | |
| 25 | LightOn | yes | no | Neurospora crassa, yeast | blue light | light-switchable transcription control | [48] | |
| 26 | NICE | yes | yes | Arthrobacter nicotinovorans | 6-hydroxynicotine | therapeutic cell engineering, biopharmaceutical manufacturing | [49] | |
| 27 | PPAR* | yes | no | Homo sapiens | rosiglitazone | gene therapy | [50] | |
| 28 | PEACE | no | yes | Pseudomonas putida | flavonoids (e.g. phloretin) | biopharmaceutical manufacturing, gene- and cell-based therapies | [51] | |
| 29 | PIT | yes | yes | Streptomyces coelicolor | pristinamycin I, virginiamycin | compatible with the Tet-OFF system, thus two different gene activities can be controlled in the same cell | [12] | |
| 30 | REDOX | no | yes | Streptomyces coelicolor | NADH | process development, biopharmaceutical manufacturing | [52] | |
| 31 | QuoRex | yes | yes | Streptomyces coelicolor, Streptomyces pristinaespiralis | butyrolactones (e.g. SCB1) | small molecule-adjustable gene control systems for clinical application | [53] | |
| 32 | ST-TA | yes | yes | Streptomyces coelicolor, Escherichia coli, Herpes simplex | γ-butyrolactone, tetracycline | Design of therapeutic gene circuits for gene and cell-based therapies | [54] | |
| 33 | TIGR | no | yes | Streptomyces albus | temperature | temperature-inducible gene regulation | [55] | |
| 34 | TraR | yes | no | Agrobacterium tumefaciens | N-(3-oxo-octanoyl)homoserine lactone | versatile gene expression control | [56] | |
| 35 | TET-OFF, TET-ON | yes | yes | Escherichia coli, Herpes simplex | tetracycline, doxycycline | reversible and tight gene expression control | [11,57] | |
| 36 | TRT | yes | no | Chlamydia trachomatis | l-tryptophan | synthetic bidirectional communication between mammalian cells | [58] | |
| 37 | UREX | yes | no | Deinococcus radiodurans | uric acid | self-sufficient control of pathologic metabolites, gene- and cell-based therapies | [59] | |
| 38 | VAC | yes | yes | Caulobacter crescentus | vanillic acid | biopharmaceutical manufacturing, gene- and cell-based therapies | [60] | |
| 39 | ZF-ER, ZF-RXR/EcR | yes | yes | Mus musculus, Homo sapiens, Drosophila melanogaster | 4-hydroxytamoxifen, ponasterone-A | spatio-temporal control of gene expression, genomic and proteomic research, gene therapy | [61] | |
| 40 | ZF-Raps | yes | no | Homo sapiens | rapamycin | gene expression control, gene therapy | [62] | |
| 41 | ZF switches | yes | no | Mus musculus, Homo sapiens, Drosophila melanogaster | 4-hydroxytamoxifen, mifepristone | study of gene function, alteration of phenotypes of cells or organisms | [63] | |
| 42 | ZF(TF)s | yes | no | Xenopus laevis, Homo sapiens | ethyl-4-hydroxybenzoate, propyl-4-hydroxybenzoate | genomic and proteomic research, gene therapy | [64] | |
| post-transcriptional switches | ||||||||
| 1 | aptamer RNAi | yes | no | synthetic SELEX-derived aptamer | theophylline | regulation of gene expression by small molecules without engineered proteins | [65] | |
| 2 | aptamer RNAi | no | yes | synthetic SELEX-derived aptamer | theophylline | construction of conditional RNAi systems | [66] | |
| 3 | aptamer RNAi miRNA | yes | no | synthetic SELEX-derived aptamer | theophylline, tetracycline, hypoxanthine | probing and programming of cellular function | [67] | |
| 4 | aptamer Splicing | yes | yes | Homo sapiens, MS2 bacteriophage | MS2, p65, p50, b-catenin | programmable sensing-actuation devices for autonomous cell behaviour control | [68] | |
| 5 | aptazyme | no | yes | synthetic SELEX-derived aptamer, Schistosoma mansoni | theophylline | regulation of therapeutic gene expression | [69] | |
| 6 | replicon CytTS | yes | no | Sindbis virus | temperature | expression of toxic proteins, bioprocess engineering | [70] | |
| 7 | TET-OFF-shRNA, TET-ON-shRNA | yes | yes | Escherichia coli, Herpes simplex, Homo sapiens | doxycycline | basic or translational research, development of gene-based therapeutics | [71] | |
| 8 | theo aptamer | no | yes | synthetic SELEX-derived aptamer | theophylline | basic, biotechnological, and biomedical research | [72] | |
| 9 | 3′ UTR aptazyme | yes | no | synthetic SELEX-derived aptamers, tobacco ringspot virus | theophylline, tetracycline | functional response regulation, gene and cellular therapy | [73] | |
| 10 | 5′ UTR aptazyme | no | yes | synthetic SELEX-derived aptamer, Schistosoma mansoni | theophylline | control of gene expression, gene therapy | [74] | |
| translational switches | ||||||||
| 1 | Hoechst aptamer | no | yes | synthetic RNA sequence | Hoechst dyes | translational gene regulation, biology and medical research | [75] | |
| 2 | H23 aptamer | no | yes | Archaeoglobus fulgidus | L7Ae, L7KK | gene expression regulation, construction of a NOR gate | [76] | |
| 3 | L7Ae aptamer | yes | yes | Archaeoglobus fulgidus | L7Ae | detection, repair or rewiring of intrinsic cellular defects, characterization of complex circuits | [77] | |
| 4 | MS2 aptamer | no | yes | MS2 bacteriophage | MS2 | study RNA–protein interactions, clone RNA-binding proteins | [78] | |
| post-translational switches | ||||||||
| 1 | AID | no | yes | Arabidopsis thaliana, Oryza sativa, Gossypium hirsutum | auxins (e.g. IAA) | control protein expression and study protein function | [79] | |
| 2 | ER DD | no | yes | Homo sapiens | CMP8, 4-hydroxytamoxifen | regulate the intracellular concentration of any protein | [80] | |
| 3 | FM | yes | no | Homo sapiens | AP21998 | delivery of polypeptides that require rapid and regulated delivery | [81] | |
| 4 | HaloTag | no | yes | Rhodococcus sp. RHA1 | HyT13 | validating potential drug targets in disease models | [82,83] | |
| 5 | HDV-aptazyme | no | yes | hepatitis delta virus | theophylline, guanine | dynamic range of gene regulation | [84] | |
| 6 | PROTAC | no | yes | Homo sapiens | proteolysis targeting chimeric molecules (PROTACS) | chemical knockouts, control of protein function | [85] | |
| 7 | shield DD | yes | no | Homo sapiens | shields (e.g. Shld1) | conditional control of proteins levels | [86] | |
| 8 | shield LID | no | yes | Homo sapiens | shields (e.g. Shld1) | conditional control of proteins levels | [87] | |
| 9 | TMP DD | yes | no | Escherichia coli | trimethoprim (TMP) | tuneable regulation of protein expression in the mammalian central nervous system | [88] | |
aLevel of gene expression where the switching occurs.
bON switchability by an effector; other than removing the effector which confers the OFF state.
cOFF switchability by an effector; other than removing the effector which confers the ON state.
dA ligand or other physical stimuli (e.g. temperature, electromagnetic radiation, electricity) which stabilizes the switch either in its ON or OFF state.
3.1. Transcriptional switches (figures 1 and 2)
The first step towards a mammalian synthetic switch was the implementation of the lactose regulator, discovered in Escherichia coli in the early 1960s by Jacob & Monod [89], in mammalian cells in the late 1980s [42–45]. In this transcriptional control system, gene expression can be switched ON by adding the lactose analogue isopropyl β-d-1-thiogalactopyranoside (IPTG). When IPTG subsequently binds to the Lac repressor, it facilitates its dissociation from the lac operator DNA sequence in a dose-dependent manner, and the expression of targeted genes downstream of the lac operator is switched ON [42].
Following the inclusion of a prokaryotic transcriptional system into mammalian cells, the next step was the construction of the first mammalian synthetic transcription factor, the tetracycline-controlled transactivator (tTA), by Gossen and Bujard in 1992 [11]. This was achieved by fusing the prokaryotic tetracycline (Tet) repressor, TetR, with the C-terminal domain of virion protein 16 (VP16) from herpes simplex virus. Promoters responsive to tTA (Ptet) consist of heptamerized tetracycline operator (tetO) sequences and minimal promoters derived from viral or cellular RNA polymerase II promoters [11]. TetR and tetO sequences originate from the Tn10 transposon, natively present in E. coli [11]. Consequently, genes downstream of Ptet are transcribed only in the presence of tTA. Doxycycline, a tetracycline analogue, binds to tTA with relatively high affinity. This promotes the dissociation of tTA from the heptamerized tetO DNA sequence, inhibiting the expression of the gene downstream of Ptet, setting the switch to the OFF state [90,91]. A reverse tetracycline-controlled transactivator rtTA has also been created by point mutating the tTA gene [57,92]. These point mutations completely reverse the tetracycline responsiveness of rtTA: rtTA requires tetracyclines (or tetracycline derivatives) for binding to tetO sequences to set the switch to the ON state [57,92]. The tTA-dependent control circuit is also referred to as the Tet-Off System and the rtTA system is also known as the Tet-On System.
Based on these developments and using prokaryotic regulators, with DNA-binding capacity controlled allosterically by small molecules, several mammalian transgene control switches have been developed [93,94] (see also figure 1). Switches controlled by antibiotics [12,27,32], hormones and hormone analogues [29,40,95], quorum-sensing substances [53], and immune suppressive and anti-diabetic drugs [50,96] have recently been engineered expanding the arsenal of tools of synthetic biologists.
The second generation of synthetic transcriptional switches are regulated by metabolites such as amino acids [21], vitamins [23], gaseous acetaldehyde [20], food and cosmetics additives [33,51]. These second-generation compounds may be used for regulating gene therapy more effectively than the previously mentioned small molecules, such as hormones, which have known and potent side effects. Moreover, mammalian transcriptional switches that respond to physical factors, such as electricity [30], temperature [55] and blue light [25,47,48] have also been developed (see also figure 1). Thus, synthetic biologists can already choose from a plethora of transcriptional switches to fulfil the requirements for various individual applications.
3.2. Post-transcriptional switches (figures 1 and 2)
The abovementioned transcriptional switches regulate the transcription of DNA into mRNA. Post-transcriptional synthetic biological switches control the function, stability and/or splicing of mRNA molecules. One of these techniques, RNA interference (RNAi), led to the development of short interfering RNAs (siRNA) technology, which allows control over mRNA degradation (cf. [97]). RNAi involves the cleavage of double-stranded RNA molecules (naturally occurring in viruses [98]), such as short hairpin RNAs (shRNA), into 21–23 nucleotide long RNA duplexes by the endogenous enzyme Dicer [99]. The obtained RNA duplexes consist of siRNA or micro RNAs (miRNA). One of the two strands of these RNA duplexes is then incorporated into the RNA-induced silencing complex (RISC), which degrades complementary mRNA sequences to which the siRNA or miRNA of RISC binds to, leading to mRNA degradation and post-transcriptional repression. In principle, post-transcriptional control can be engineered by switch-controlled generation of siRNA and miRNA molecules. A TetR-switch has been developed which regulates the expression of shRNA by doxycycline and the resulting shRNA controls the expression of a transgene in vitro and in vivo [71]. Similarly, a hybrid switch based on both RNAi and Lac and tet repressor proteins has been developed for regulating gene expression both at the transcriptional and post-transcriptional level [100].
Post-transcriptional switches can also be created by employing aptamers. Aptamers are small, single-stranded, highly folded nucleic acids with high affinity and specificity for their target molecules (small molecules or protein ligands), which inhibit their biological functions [101]. By integrating aptamers into siRNA or miRNA, sophisticated switches have been obtained in which the RNAi function is ligand-controlled [65–67]. By integrating a theophylline-binding aptamer into an essential structural loop of a shRNA, the processing of the shRNA by Dicer could be inhibited in a theophylline dose-dependent fashion, making the post-translational control of the shRNA probe inducible [65]. Gene expression has also been regulated by controlling mRNA splicing using an aptamer. For this design, the theophylline-binding aptamer was used to cloak an essential splicing element within the pre-mRNA, inhibiting splicing in the presence of theophylline [41,72] (see also table 1).
An aptamer (sensor) together with a ribozyme (actuator) forms an aptazyme (also called allosteric ribozymes). The ribozyme part is an RNA molecule with enzymatic function, capable of catalysing specific biochemical reactions. In the presence of a ligand that binds to the aptamer, the ribozyme part of the aptazyme self-cleaves and functionality is induced. Thus, aptazymes can also act as post-transcriptional switches. For instance, by incorporating an aptazyme into the 5′ or 3′ untranslated region (UTR) of an mRNA, the 5′ and 3′ UTR structural elements necessary for gene expression can be chopped off, in a ligand-dependent manner, thereby inducing ligand-controlled silencing of gene expression [69,73,74].
3.3. Translational switches (figure 2)
Once the mature mRNA reaches the ribosomes, translation of the mRNAs into amino acid chains or peptides begins. Recently, control switches have been engineered to regulate the expression of genes into proteins; for example, RNA-binding proteins, like the archaeal ribosomal protein L7Ae, can be switched on by an input protein to block translation or by binding to an RNA motif integrated in the 5′ UTR of an mRNA transcript [78,102]. An aptamer controlled by the Hoechst 33258 dye has also been shown to inhibit translation in the presence of Hoechst 33258, thus functioning as a translational switch [75].
3.4 Post-translational switches (figure 2)
Following the translation of mRNA into proteins, at the post-translational level, switches have been developed to control the half-life of proteins or protein trafficking. To degrade a protein, the protein in question is tagged with a degradation signal and it reaches the endogenous ubiquitin–proteasome system, where it is degraded. As an example, an auxin-inducible protein degradation system from plants was shown to degrade proteins of interest in mammalian cells under the control of auxin [79]. Destabilized or ligand-induced degradation protein domains can be fused to a target protein to control the degradation of the entire fusion protein [80,85–87,103]. A noteworthy example is the HaloTag system [82,83] which involves the fusion of a dehalogenase enzyme to the protein of interest. By adding small hydrophobic molecules that covalently bind to the HaloTag, the fusion protein unfolds and is efficiently degraded by the proteasome. Recently, switches have also been developed to control protein secretion [81] (see also figure 1).
Another type of post-translational switch involves inteins. An intein (also known as protein intron) is a segment within a protein that is capable of excising itself, while the remaining protein segments (the exteins) are joined with a peptide bond in the process of protein splicing [104,105]. As inteins are transcribed and translated together with the target protein before they undergo autocatalytic self-excision and splicing, inteins increase the time it takes to transcribe, translate and post-translationally process a target gene. The intein domain can be transcribed and translated by two separate genes (known as split inteins) and the resulting precursor proteins splice each other, process termed trans-splicing, to yield a single functional protein [106,107]. The protein splicing and trans-splicing processes can be triggered by small molecules or protein inputs to obtain intein-based post-translational switches [108–111].
3.5. Rheostat switches
Most synthetic gene circuits developed to date employ digital logic. However, there has been rising interest in the design and implementation of analogue synthetic gene circuits. These circuits allow for the construction of multi-signal integration and are able to execute complex computational functions in living cells, such as ratiometric operations, power-law implementations, logarithmically linear sensing, among others [112]. The majority of synthetic gene circuits developed to date have been implemented in prokaryotes or lower eukaryotes, i.e. yeast. However, few mammalian implementations of synthetic gene circuits are available in the literature. Relevant examples of such implementations include the RNAi-based logic evaluator developed by Rinaudo et al. [113], and the singe-cell half-adder/subtractor constructed by Auslander et al. [114]. The logic evaluator was developed with the objective of sensing endogenous molecular inputs, processing it through Boolean logic and eliciting a physiological response [113]. This system was implemented by targeting siRNAs to endogenous mRNA signals, which allows for logic operations to be executed depending on the presence or absence of these endogenous inputs [113]. The single-cell biocomputers developed by Auslander et al. [114] were based on coupling three logic gates in a combinatorial fashion, thus enabling single cells to execute half-adder and half-subtractor operations. This sophisticated approach relied on the activity of two transactivators, ETI (erythromycin-dependent transactivator) and TtgA1 (phloretin-dependent transactivator), and their corresponding DNA-binding sequences, PETR2 and PTtgR1. The transcriptional activation properties of these transcription factors are disabled by the presence of erythromycin and phloretin, respectively, which function as the inputs of the system [114]. By coupling this layer of transcriptional control to C/D and MS2 boxes, Auslander et al. were able to couple XOR logic gates to N-IMPLY and AND gates, thus creating a half-subtractors and half-adders, respectively.
4. Synthetic gene networks in mammalian cells
Synthetic biological ON–OFF switches and/or a set of genes can be rationally selected and combined, to interact in a predictable and controllable manner, forming a system with a pre-set function, a synthetic gene network, also known as synthetic gene circuit. In order to rationally construct higher-order gene networks for advanced therapeutic applications, a toolbox of well-characterized and well-controllable standardized parts should be available. The following subsections present gene networks that were developed to function as standardized parts in a higher-order gene network. In some cases, these simple gene networks are also used as therapies.
In order to perform logical operations in mammalian cells, programmable Boolean logic gates were constructed in 2004, by combining heterogeneous transcription factors [115]. Moreover, similarly to electronic circuits, several multicomponent circuits with complex trigger-controlled topology have been designed with time-delay [116], bandpass [117] and hysteretic [118] properties. A gene network operated by a Boolean AND gate was applied for targeting cancer cells [119], where the AND gate activity was achieved when both pre-set conditions were met, leading to the expression of apoptotic genes and cancer cell death.
Boolean logic gates have also been engineered based on synthetic transcription factor-containing zinc finger motifs and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 motifs [120]. These are attractive parts for engineering higher-order networks because (i) zinc fingers and CRISPR/Cas9 can be created to recognize virtually any DNA sequence and (ii) they can function without interfering with each other. Indeed, the bacterial CRISPR/Cas system has been shown to be particularly versatile and easy to use. Many bacteria use CRISPR-based immune systems to degrade genetic materials of invading phages [121,122]. In these systems, short RNAs expressed from CRISPR loci are used to guide an endonuclease protein (Cas9) to recognize and cleave invading genetic material. Recently, Cas9 was used as a programmable tool for genome editing in mammalian cells [123–125]. In this context, small customizable guide RNAs (gRNAs) can be designed to programme and target Cas9 endonuclease to specific loci in living cells to induce double (or single)-stranded breaks in DNA. Upon cleavage, error-prone or template-directed repair pathways are triggered, generating variants of the original target loci. Recently, Qi et al. [126] showed that an endonuclease-deficient Cas9 (dCas9, with D10A H841A mutations relative to the wild-type Cas9) can be used as a programmable ‘CRISPRi’ tool for gene silencing in E. coli. Several groups have shown that dCAS9/gRNA can be used to inhibit or partially stimulate synthetic or endogenous mammalian genes, when dCAS9 was fused to an actuator (VP64), depending on the position of gRNA binding with respect to the TATA box [127–129]. Inhibitory circuits in mammalian cells have recently been introduced using dCAS9 systems [124,130,131]. Recently, CRISPR regulatory devices were layered to obtain cascaded circuits [132], and the expression of functional gRNAs from RNA polymerase II promoters (normally expressed from RNA polymerase II promoters) and multiplexed production of proteins and gRNAs from a single transcript in human cells was made possible [133]. In the above discussed switches, the switching molecule should be continuously present in order to maintain the switch in either the ON or OFF state. To reversibly set the switch to ON or OFF positions by transiently applying a trigger molecule, toggle switches have been developed [12,32,134,135]. Examples of how these toggle switches have been employed include monitoring the environment of immune cells in lymph nodes or the presence of hormones or signalling molecules.
Although more complex in network topology, functionally, synthetic mammalian oscillators constitute synthetic biological parts that can be integrated into higher-order circuits or applied alone to reprogramme circadian clocks [136,137] or to govern metabolic [138], repair [139] and signalling pathways [140] in mammalian cells. Such a synthetic mammalian oscillator has been developed using a time-delayed negative feedback loop, but these systems have been shown to dampen their oscillations due to noise and/or epigenetic silencing [141]. The addition of a positive feedback loop may overcome these limitations and generate autonomous, self-sustained and tuneable oscillatory expression of reporter genes [141]. A low-frequency mammalian oscillator has also been developed, by silencing of the tetracycline-controlled transactivator using siRNA encoded in the introns of the mRNA, in order to facilitate robust and autonomous expression of a fluorescent reporter protein with periods of 26 h [142]. In order to generate transcriptional and translational time-delay for tuning oscillators, inteins (described in the Post-translational switches section) could also be employed [143–145]. All of these synthetic biological control circuits described in this subsection contribute to the development of mammalian cell biocomputers [114] and gene networks with advanced functions, described in the following subsections.
5. Synthetic gene networks for advanced medical applications (figure 3)
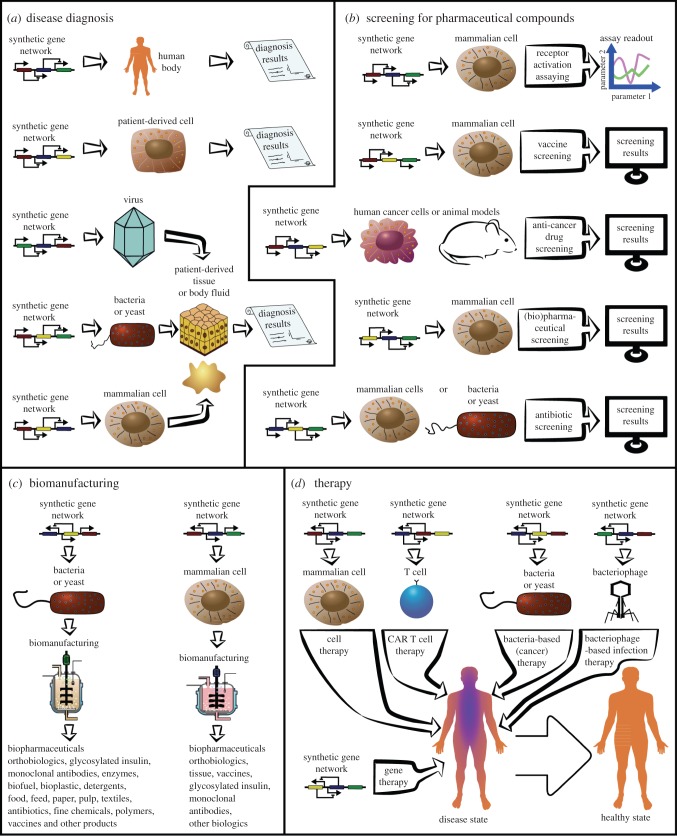
Figure 3.
Synthetic biology enlightens the healthcare spectrum from diagnosis of diseases, to drug screening, biomanufacturing and to therapy. (a) Synthetic biology can be applied to diagnose diseases either by inserting gene circuits into the human body or into patient-derived cells. Alternatively, viruses, bacteria or mammalian cells can be used as synthetic network carriers and added to patient-derived tissue or body fluids. In either case, the diagnostic synthetic network should recognize diseased stated and generate a measurable readout in response to the disease. (b) Following disease diagnosis, synthetic biology can also be applied to screen for pharmaceutical compounds to combat diseases. In this case, synthetic circuits inserted into various organisms should mimic disease-related states and the alteration of these states by added compounds should yield a measurable readout. Synthetic gene networks can also be applied to monitor the activation or inactivation of trans-membrane receptor, which are potential drug targets. Additionally, synthetic biology holds great promise to screen for vaccines [146]. (c) After pharmaceuticals are identified, synthetic biology can significantly enhance the manufacturing of these pharmaceuticals and other biologics. (d) Last but not least, synthetic biology enhances state-of-the-art treatments such as gene therapy, cell therapy, CAR T cell immunotherapy, bacterial-based therapy for cancer and other diseases and bacteriophage-based therapy for infectious diseases. (Online version in colour.)
One of the ultimate goals of mammalian synthetic biology is to improve human health. For this purpose, gene networks that contribute to the diagnosis and/or treatment of diseases are emerging and/or have been already developed.
Imaging of cellular behaviour has been greatly improved by enriching target cells with new functions. A gene network has been created that enables the tracking of human cell fate by retaining memory of exposure to stimuli [147]. In these memory circuits, a brief doxycycline, UV or hypoxic stimulus upregulates the expression of a synthetic transcription factor which retains its presence over longer periods of time through a positive feedback loop. This memory circuit can change gene expression, growth rates and cell viability for several generations after the initial stimulus, offering a new diagnostic and therapeutic window. Implementing these networks with toggle switches (see above) in migrating immune cells would enable monitoring of the location of hypoxic regions in non-dividing tumour or atherosclerotic tissue.
A slightly more elaborate circuit has been designed to detect cell line lineages [148]. This multi-input RNAi-based Boolean logic circuit identified the presence of HeLa cells by the expression levels of a customizable set of endogenous miRNAs. When these pre-determined conditions are met, a fluorescent signal or an apoptosis signal is triggered, causing the diagnosis or death of the targeted cells. This offers unique capabilities to monitor complex miRNA dynamics in cells under a wide variety of conditions. Similarly, in order to target cancer cells, a tunable dual-promoter integrator has been developed, which expresses an effector gene when two internal input promoters show high activity levels [119].
In order to control cellular behaviour in a more sophisticated way, several synthetic gene networks have been engineered that interface with native cellular pathways [68]. For instance, a class of RNA controllers have been constructed, which recognize innate signalling through the nuclear factor κB and Wnt signalling pathways in human cells, and rewire these signalling cascades to generate new behaviour by regulating alternative RNA splicing. Furthermore, several groups have engineered the MAPK pathway either through modifying its scaffolding proteins [149] or by dynamically controlling regulators in the pathway [150].
A therapeutic gene network that is capable of sensing and regulating host metabolic signalling pathways to normal levels has been developed as well. In one study, a synthetic signalling cascade was interfaced as a sensing-controller device to regulate blood glucose homeostasis after illumination with blue light [151], while in another study, insulin deficiency was corrected for by an insulin expression mammalian gene network under the control of radio waves [152]. A slightly more sophisticated, self-sufficient, sensor-effector gene network was designed which senses uric acid concentrations in blood and expresses urate oxidase in a uric acid-dependent manner. Urate oxidase then restores urate homeostasis [59].
While these therapeutic networks described above were supplied through external devices, endogenous cells may also be used as a therapeutic agent. Thymus-derived (T) lymphocytes are a type of white blood cell that plays a crucial role in cell-mediated immunity, especially in the response to invading pathogens. Human T cells are important targets for synthetic biology because they can be extracted from patients, genetically engineered and inserted back into patients to treat chronic infection or cancer [153]. A synthetic, drug-responsive, RNA-based switch has been implemented to control cytokine expression in mice, in turn governing T cell proliferation [73]. In this system, the input ligands and regulatory targets can be modified to meet various therapeutic needs. Recently, T cells were reprogrammed for treating acute lymphoid leukaemia with considerable success by modifying their chimeric antigen receptors (CARs) [154].
In addition to the above described RNA-based switch, a synthetic signal cascade has been developed in order to confront multiple risk factors of cardiovascular disease simultaneously [155]. In this circuit, the chimeric trace-amine-associated receptor 1, activated by the clinically licensed antihypertensive drug guanabenz (Wytensin®), is coupled to cAMP and cAMP-dependent phosphokinase A (PKA)-mediated activation of the cAMP-response element binding protein (CREB1). After activation, CREB1 drives the expression of bi-functional therapeutic peptide hormone GLP-1-Fc-leptin which attenuates hyperglycaemia and dyslipidaemia. Thus, this gene network adds to the antihypertensive effect of Wytensin® two other functions to create a three-in-one treatment strategy for cardiovascular diseases [155].
5.1. Synthetic gene networks in bacterial antibiotic resistance (figure 3)
Recent advances in molecular biology and genetic engineering have refined the ability to synthesize, design and modify bacteriophages [156]. Indeed, this has enabled innovative strategies and technologies for bacteriophage-based tools to be applied to the treatment of infectious diseases, within a synthetic biology context. The main phage-enabled technologies used to this end are phage display, bio-part development and genomic recombineering [156]. Phage display, a methodology involving fusing random peptide libraries to phage coat proteins, has yielded significant advances in the areas of vaccine development and drug delivery [156–159], with particular applications to management of Pseudomonas aeruginosa, Staphylococcus aureus and Bacillus anthracis [156]. Phage-based bio-part development represents a core strategy of synthetic biology. Phage bio-parts are orthogonal in both prokaryotic and eukaryotic cells and their robustness has been demonstrated in the past decades through their ubiquitous presence in common laboratory protocols. Specifically, the T7 promoter has been extensively used in synthetic biology due to its ability to drive high-level gene expression in an orthogonal, low-toxicity fashion [160–162]. Additionally, phage-occurring recombinases such as Cre, PhiC31 and Bxb1 have been widely used in the construction of several gene networks, including Boolean logic gates and counters [163,164]. Phage-based genomic recombineering also represents a powerful tool for the development of synthetic biology applications that tackle infectious diseases. An example of this is the phage λ Red recombination system used in conjunction with flanking homology target sequences in transformed DNA [156]. This methodology has been used to induce genomic modifications to several bacterial species which are known to represent high disease burdens, i.e. E. coli, Salmonella enterica, P. aeruginosa, Yersinia pestis, Shigella flexneri and Vibrio cholerae [165–170].
In addition to the referred phage-enabled technologies, bacteriophage-based synthetic gene networks with therapeutic and diagnostic orientations have recently been developed. These networks can be categorized into antimicrobial and antibiotic-sensitizing. Antimicrobial synthetic gene networks are based on engineering existing phages through the addition or improvement of biological functions. Examples of this include the biofilm-degrading network designed by Lu & Collins [171] and the lysis-inducing network constructed by Westwater et al. [172]. These networks were based on the T7 and M13 phages, respectively. Rather than delivering toxic compounds to target infectious cells, antibiotic-sensitizing networks use phages as the chassis for generating biochemical responses to antibiotics. This concept was elegantly illustrated by Edgar et al. [173]: phage λ was used to generate antibiotic re-sensitizing particles through the delivery of wild-type copies of gyrA and rpsL [156]. Phage performance was assayed by transducing these genes in streptomycin and fluoroquinolone-resistant target cells, i.e. caused by point mutations in rpsL and gyrA, respectively. Expression of the wild-type copies resulted in the biosynthesis of enzymes that showed sensitivity to these antibiotics [173].
6. Synthetic gene networks for drug discovery (figure 3)
While synthetic gene networks may be used for tackling disease by modifying or reprogramming genetic material, mammalian synthetic gene networks can also be developed to screen for novel pharmaceutical compounds. This therapeutic approach has the advantage of keeping the genetic material of the treatment subject intact, thus reducing the severity of controversial ethical issues [174–176].
An early example for this approach is the employment of a streptogramin-controlled transcriptional switch [12] for the identification of bioavailable, non-cytotoxic streptogramin antibiotics [177]. This streptogramin-controlled transcriptional switch contains a protein domain that is involved in conferring the antibiotic resistance, which is activated by adding libraries of metabolic compounds to cells that contained the streptogramin-controlled switch. This method clearly identified new streptogramin antibiotics that modified the transcription activity of the switch.
Another example is the development of a drug discovery circuit that led to the identification of anti-tuberculosis drugs [33]. In Mycobacterium tuberculosis, ethionamide activator (EthA) converts the pro-drug ethionamide into an antimycobacterial nicotinamide adenine dinucleotide derivative which is effective against M. tuberculosis [178]. The expression of the EthA is inhibited by the ethionamide repressor (EthR), increasing the resistance of this pathogen to ethionamide, which is the last-line-of-defence in treatment of TBC [178]. In the anti-tuberculosis drug discovery gene network, the activity of EthR is monitored and a screening for compounds that inhibit EthR was carried out to identify the food additive ‘strawberry flavour’ (2-phenyl ethyl butyrate) as a new line of defence against multidrug-resistant M. tuberculosis [33].
Besides gene networks for antibiotic discovery, a gene network that allows screening for anti-cancer drugs has also been developed [179,180]. Cancer is characterized by cells that uncontrollably proliferate in healthy tissues where cells are arrested in the resting phase (G0) or growth 1 (also known as gap 1, G1) phases of their cell cycle. In order to treat cancer, proliferating cancer cells should be targeted and destroyed without adversely affecting healthy tissue. For this purpose, a mammalian synthetic gene network was developed, which expressed the cyclin-dependent kinase inhibitor p27Kip1 under the control of tTA to arrest mammalian cells in their cell cycle in a tetracycline-dependent manner. Chinese hamster ovary cells containing this gene circuit grow normally in the presence of tetracycline, which inhibits tTA and thus, p27Kip1 expression is switched OFF. When tetracycline is removed, tTA promotes the expression of p27Kip1 and cells are arrested in the G1 phase. By maintaining p27Kip1 expression for prolonged periods of time, a fraction of cells escape the arrest in their G1 phase and resumed growth. This way, similarly to cancer development, a mixed cell population is obtained which contains proliferating and proliferation-arrested cells. Approved cancer therapeutics killed proliferating cells, without adversely affecting growth-arrested cells. Therefore, cells with this gene network that are led into this mixed population could be used to screen compounds that selectively kill neoplastic cells [179,180].
In addition to screening for anti-cancer drugs and antibiotics, gene circuits for identifying compounds that modify the activity of cell membrane receptors are also engineered. Cell membrane receptors are extremely important drug targets. In fact, one family of plasma membrane receptors, the G protein-coupled receptors (GPCRs), represent the core drug target in modern medicine [181]; over 50% of all prescription drugs currently on the market act on GPCRs [182]. Despite this large number, only 50 members out of the approximately 1000 members of the large GPCR family are currently targeted by drugs [183]. Therefore, developing drugs that target GPCRs represents an important goal in pharmacology. A common approach for finding GPCR targeting drugs is to screen for compounds that modify the activity of GPCRs. Although receptor activation assays have already been developed [184], synthetic biology offers possibilities to improve the efficiency and controllability of receptor activation monitoring. We have recently developed an activity assay for a subset of GPCRs that are sensitive to blood flow. The assay detects GPCR activity by GFP and produces a dose–response curve enabling testing of new drugs for their effect on this subset of receptors (data yet not published)
Recently, synthetic gene networks have been established that record the activation of plasma membrane receptors such as GPCRs, receptor tyrosine kinases and steroid hormone receptors [185]. The activation of these plasma membrane receptors is coupled to secondary messengers, which then promote the transcription of a firefly luciferase reporter gene. This circuit was used to identify a ligand for the orphan G protein-coupled receptor 1 (GPR1), suggesting that this receptor is involved in the regulation of inflammation [185]. Recently, a synthetic GPCR was designed where the C-terminus was replaced by Gα1234 to test the role of this receptor and their drugs in tumour metastasis [186]. In addition, we have modified a GPCR system to monitor the activity of a mechanosensitive GPCR. We have shown that this GPCR becomes activated at higher shear stress values and it was not responsive to para-/autocrine effects. By seeding cells containing this gene network in parallel-plate flow chambers, we can screen compound libraries for new drugs that modify the shear stress-sensing ability of this GPCR.
Another human health improving application of synthetic biology that does not modify the genetic material of patients is the manufacturing of biotherapeutics. Although a large proportion of biomanufacturing is carried out in bacteria, some products need to be manufactured in mammalian cells, predominantly those from the area of biopharmaceuticals. A common example for biomanufacturing in mammalian cells is the production of monoclonal antibodies [187,188]. In general, mammalian proteins that require mammalian-specific post-translational modifications, such as glycosylation, carboxylation, hydroxylation, sulfation and amidation are needed to be expressed in mammalian cells. When expressing proteins in mammalian cells, the rate-limiting step seems to be the folding and secretion of the protein of interest. Mammalian synthetic biology was also employed to increase the rate of protein folding and secretion for the manufacturing of recombinant protein biopharmaceuticals [189,190].
In conclusion, synthetic biology has been pioneered in prokaryotic cells, but it is now emerging as a novel therapy-oriented field in mammalian cells and organisms. Initially, basic mammalian synthetic biological parts and components have been created to switch biological signals between two discrete states (ON and OFF) to gate biological signals and to oscillate parameters of interest. Using these elementary components, more complex and therapy-focused gene networks have been engineered. Mammalian synthetic biology aims to cure severe health disorders from their root cause, to treat symptoms of diseases, to help identify pharmaceuticals and to enhance the manufacturing of biopharmaceuticals. Moreover, synthetic biology has the potential to revolutionize state-of-the-art therapeutic approaches, such as gene therapies, CAR T cell therapies and other cell therapies. For this, it is also necessary to engineer new modules and components and to characterize and generate component libraries for mammalian synthetic biology. Such a toolbox would help the field to prevent, diagnose and treat disease in general. In the near future, the emergence of more and more therapy-oriented gene networks is expected and existing and future synthetic circuits might be subjected to clinical trials. A key challenge of synthetic biology enhanced gene therapies remains the development of vectors that can efficiently deliver the gene networks to the cells and tissues of interest. For synthetic biology to live up to its clinical potential, it should also be better integrated with clinicians. Additionally, clinical trials will be required to evaluate the potency of the treatments and their side effects, and arising ethical issues have to be taken into account. Further work is required to clinically implement synthetic biology, but this field remains one of the most promising and exciting areas of science and technology of our times.
References
- 1.Elowitz MB, Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338. ( 10.1038/35002125) [DOI] [PubMed] [Google Scholar]
- 2.Gardner TS, Cantor CR, Collins JJ. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342. ( 10.1038/35002131) [DOI] [PubMed] [Google Scholar]
- 3.Lienert F, Lohmueller JJ, Garg A, Silver PA. 2014. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 15, 95–107. ( 10.1038/nrm3738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber W, Fussenegger M. 2009. Engineering of synthetic mammalian gene networks. Chem. Biol. 16, 287–297. ( 10.1016/j.chembiol.2009.02.005) [DOI] [PubMed] [Google Scholar]
- 5.Tigges M, Fussenegger M. 2009. Recent advances in mammalian synthetic biology-design of synthetic transgene control networks. Curr. Opin. Biotechnol. 20, 449–460. ( 10.1016/j.copbio.2009.07.009) [DOI] [PubMed] [Google Scholar]
- 6.Kwok R. 2010. Five hard truths for synthetic biology. Nature 463, 288–290. ( 10.1038/463288a) [DOI] [PubMed] [Google Scholar]
- 7.May T, Eccleston L, Herrmann S, Hauser H, Goncalves J, Wirth D. 2008. Bimodal and hysteretic expression in mammalian cells from a synthetic gene circuit. PloS ONE 3, e2372 ( 10.1371/journal.pone.0002372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. 2004. Noise minimization in eukaryotic gene expression. PLoS Biol. 2, e137 ( 10.1371/journal.pbio.0020137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blake WJ, KÆrn M, Cantor CR, Collins JJ. 2003. Noise in eukaryotic gene expression. Nature 422, 633–637. ( 10.1038/nature01546) [DOI] [PubMed] [Google Scholar]
- 10.Bashor CJ, Helman NC, Yan SD, Lim WA. 2008. Using engineered scaffold interactions to reshape map kinase pathway signaling dynamics. Science 319, 1539–1543. ( 10.1126/science.1151153) [DOI] [PubMed] [Google Scholar]
- 11.Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA 89, 5547–5551. ( 10.1073/pnas.89.12.5547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. 2000. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 18, 1203–1208. ( 10.1038/81208) [DOI] [PubMed] [Google Scholar]
- 13.Kalos M, June CH. 2013. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity 39, 49–60. ( 10.1016/j.immuni.2013.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.June CH, Maus MV, Plesa G, Johnson LA, Zhao Y, Levine BL, Grupp SA, Porter DL. 2014. Engineered T cells for cancer therapy. Cancer Immunol. Immunother. 63, 969–975. ( 10.1007/s00262-014-1568-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auslander S, Fussenegger M. 2013. From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol. 31, 155–168. ( 10.1016/j.tibtech.2012.11.006) [DOI] [PubMed] [Google Scholar]
- 16.Ruder WC, Lu T, Collins JJ. 2011. Synthetic biology moving into the clinic. Science 333, 1248–1252. ( 10.1126/science.1206843) [DOI] [PubMed] [Google Scholar]
- 17.Cameron DE, Bashor CJ, Collins JJ. 2014. A brief history of synthetic biology. Nat. Rev. Microbiol. 12, 381–390. ( 10.1038/nrmicro3239) [DOI] [PubMed] [Google Scholar]
- 18.Khalil AS, Collins JJ. 2010. Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379. ( 10.1038/nrg2775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang FS, Ho WQ, Crabtree GR. 2011. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal. 4, rs2 ( 10.1126/scisignal.2001449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, Aubel D, Weber CC, Fussenegger M. 2004. Gas-inducible transgene expression in mammalian cells and mice. Nat. Biotechnol. 22, 1440–1444. ( 10.1038/nbt1021) [DOI] [PubMed] [Google Scholar]
- 21.Hartenbach S, Daoud-El Baba M, Weber W, Fussenegger M. 2007. An engineered l-arginine sensor of Chlamydia pneumoniae enables arginine-adjustable transcription control in mammalian cells and mice. Nucleic Acids Res. 35, e136 ( 10.1093/nar/gkm652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossger K, Charpin-El-Hamri G, Fussenegger M. 2014. Bile acid-controlled transgene expression in mammalian cells and mice. Metab. Eng. 21, 81–90. ( 10.1016/j.ymben.2013.11.003) [DOI] [PubMed] [Google Scholar]
- 23.Weber W, Bacchus W, Daoud-El Baba M, Fussenegger M. 2007. Vitamin H-regulated transgene expression in mammalian cells. Nucleic Acids Res. 35, e116 ( 10.1093/nar/gkm466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber W, Lienhart C, Baba MD, Fussenegger M. 2009. A biotin-triggered genetic switch in mammalian cells and mice. Metab. Eng. 11, 117–124. ( 10.1016/j.ymben.2008.12.001) [DOI] [PubMed] [Google Scholar]
- 25.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975. ( 10.1038/nmeth.1524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie M, Ye H, Hamri GC, Fussenegger M. 2014. Antagonistic control of a dual-input mammalian gene switch by food additives. Nucleic Acids Res. 42, e116 ( 10.1093/nar/gku545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullick A, et al. 2006. The cumate gene-switch: a system for regulated expression in mammalian cells. BMC Biotechnol. 6, 43 ( 10.1186/1472-6750-6-43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.No D, Yao TP, Evans RM. 1996. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA 93, 3346–3351. ( 10.1073/pnas.93.8.3346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palli SR, Kapitskaya MZ, Potter DW. 2005. The influence of heterodimer partner ultraspiracle/retinoid X receptor on the function of ecdysone receptor. FEBS J. 272, 5979–5990. ( 10.1111/j.1742-4658.2005.05003.x) [DOI] [PubMed] [Google Scholar]
- 30.Weber W, Luzi S, Karlsson M, Sanchez-Bustamante CD, Frey U, Hierlemann A, Fussenegger M. 2009. A synthetic mammalian electro-genetic transcription circuit. Nucleic Acids Res. 37, e33 ( 10.1093/nar/gkp014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang J, McLachlan MJ, Zhao H. 2013. Orthogonal control of endogenous gene expression in mammalian cells using synthetic ligands. Biotechnol. Bioeng. 110, 1419–1429. ( 10.1002/bit.24807) [DOI] [PubMed] [Google Scholar]
- 32.Weber W, et al. 2002. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 20, 901–907. ( 10.1038/nbt731) [DOI] [PubMed] [Google Scholar]
- 33.Weber W, Schoenmakers R, Keller B, Gitzinger M, Grau T, Daoud-El Baba M, Sander P, Fussenegger M. 2008. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc. Natl Acad. Sci. USA 105, 9994–9998. ( 10.1073/pnas.0800663105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braselmann S, Graninger P, Busslinger M. 1993. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proc. Natl Acad. Sci. USA 90, 1657–1661. ( 10.1073/pnas.90.5.1657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Xu J, Pierson T, O′Malley BW, Tsai SY. 1997. Positive and negative regulation of gene expression in eukaryotic cells with an inducible transcriptional regulator. Gene Therapy 4, 432–441. ( 10.1038/sj.gt.3300402) [DOI] [PubMed] [Google Scholar]
- 36.Wang YL, Omalley BW, Tsai SY, Omalley BW. 1994. A regulatory system for use in gene-transfer. Proc. Natl Acad. Sci. USA 91, 8180–8184. ( 10.1073/pnas.91.17.8180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liberles SD, Diver ST, Austin DJ, Schreiber SL. 1997. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc. Natl Acad. Sci. USA 94, 7825–7830. ( 10.1073/pnas.94.15.7825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collingwood TN, Butler A, Tone Y, Clifton-Bligh RJ, Parker MG, Chatterjee VK. 1997. Thyroid hormone-mediated enhancement of heterodimer formation between thyroid hormone receptor beta and retinoid X receptor. J. Biol. Chem. 272, 13 060–13 065. ( 10.1074/jbc.272.20.13060) [DOI] [PubMed] [Google Scholar]
- 39.Zhao HF, Boyd J, Jolicoeur N, Shen SH. 2003. A coumermycin/novobiocin-regulated gene expression system. Hum. Gene Therapy 14, 1619–1629. ( 10.1089/104303403322542266) [DOI] [PubMed] [Google Scholar]
- 40.Roscilli G, Rinaudo CD, Cimino M, Sporeno E, Lamartina S, Ciliberto G, Toniatti C. 2002. Long-term and tight control of gene expression in mouse skeletal muscle by a new hybrid human transcription factor. Mol. Therapy J. Am. Soc. Gene Therapy 6, 653–663. ( 10.1016/S1525-0016(02)90717-3) [DOI] [PubMed] [Google Scholar]
- 41.Auslander D, Wieland M, Auslander S, Tigges M, Fussenegger M. 2011. Rational design of a small molecule-responsive intramer controlling transgene expression in mammalian cells. Nucleic Acids Res. 39, e155 ( 10.1093/nar/gkr829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyborski DL, Short JM. 1991. Analysis of inducers of the E. coli lac repressor system in mammalian cells and whole animals. Nucleic acids Res. 19, 4647–4653. ( 10.1093/nar/19.17.4647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu MC, Davidson N. 1987. The inducible lac operator–repressor system is functional in mammalian cells. Cell 48, 555–566. ( 10.1016/0092-8674(87)90234-0) [DOI] [PubMed] [Google Scholar]
- 44.Hu MC, Davidson NA. 1990. Combination of derepression of the lac operator–repressor system with positive induction by glucocorticoid and metal ions provides a high-level-inducible gene expression system based on the human metallothionein-iia promoter. Mol. Cell Biol. 10, 6141–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown M, Figge J, Hansen U, Wright C, Jeang KT, Khoury G, Livingston DM, Roberts TM. 1987. Lac repressor can regulate expression from a hybrid sv40 early promoter containing a lac operator in animal cells. Cell 49, 603–612. ( 10.1016/0092-8674(87)90536-8) [DOI] [PubMed] [Google Scholar]
- 46.Cronin CA, Gluba W, Scrable H. 2001. The lac operator–repressor system is functional in the mouse. Genes Dev. 15, 1506–1517. ( 10.1101/gad.892001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. 2009. Induction of protein–protein interactions in live cells using light. Nat. Biotechnol. 27, 941–945. ( 10.1038/nbt.1569) [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Chen X, Yang Y. 2012. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9, 266–269. ( 10.1038/nmeth.1892) [DOI] [PubMed] [Google Scholar]
- 49.Malphettes L, Weber CC, El-Baba MD, Schoenmakers RG, Aubel D, Weber W, Fussenegger M. 2005. A novel mammalian expression system derived from components coordinating nicotine degradation in Arthrobacter nicotinovorans pAO1. Nucleic Acids Res. 33, e107 ( 10.1093/nar/gni107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tascou S, Sorensen TK, Glenat V, Wang M, Lakich MM, Darteil R, Vigne E, Thuillier V. 2004. Stringent rosiglitazone-dependent gene switch in muscle cells without effect on myogenic differentiation. Mol. Therapy 9, 637–649. ( 10.1016/j.ymthe.2004.02.013) [DOI] [PubMed] [Google Scholar]
- 51.Gitzinger M, Kemmer C, El-Baba MD, Weber W, Fussenegger M. 2009. Controlling transgene expression in subcutaneous implants using a skin lotion containing the apple metabolite phloretin. Proc. Natl Acad. Sci. USA 106, 10 638–10 643. ( 10.1073/pnas.0901501106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber W, Link N, Fussenegger M. 2006. A genetic redox sensor for mammalian cells. Metab. Eng. 8, 273–280. ( 10.1016/j.ymben.2005.12.004) [DOI] [PubMed] [Google Scholar]
- 53.Weber W, et al. 2003. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic acids Res. 31, e71 ( 10.1093/nar/gng071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Folcher M, Xie M, Spinnler A, Fussenegger M. 2013. Synthetic mammalian trigger-controlled bipartite transcription factors. Nucleic Acids Res. 41, e134 ( 10.1093/nar/gkt405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber W, et al. 2003. Conditional human VEGF-mediated vascularization in chicken embryos using a novel temperature-inducible gene regulation (TIGR) system. Nucleic Acids Res. 31, e69 ( 10.1093/nar/gng069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli F, De Francesco R, Cortese R. 2003. A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep. 4, 159–165. ( 10.1038/sj.embor.embor734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. 2000. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA 97, 7963–7968. ( 10.1073/pnas.130192197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bacchus W, Lang M, El-Baba MD, Weber W, Stelling J, Fussenegger M. 2012. Synthetic two-way communication between mammalian cells. Nat. Biotechnol. 30, 991–996. ( 10.1038/nbt.2351) [DOI] [PubMed] [Google Scholar]
- 59.Kemmer C, Gitzinger M, Daoud-El Baba M, Djonov V, Stelling J, Fussenegger M. 2010. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol. 28, 355–360. ( 10.1038/nbt.1617) [DOI] [PubMed] [Google Scholar]
- 60.Gitzinger M, Kemmer C, Fluri DA, El-Baba MD, Weber W, Fussenegger M. 2012. The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 40, e37 ( 10.1093/nar/gkr1251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magnenat L, Schwimmer LJ, Barbas CF, III, 2008. Drug-inducible and simultaneous regulation of endogenous genes by single-chain nuclear receptor-based zinc-finger transcription factor gene switches. Gene Therapy 15, 1223–1232. ( 10.1038/gt.2008.96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivera VM, et al. 1996. A humanized system for pharmacologic control of gene expression. Nature Med. 2, 1028–1032. ( 10.1038/nm0996-1028) [DOI] [PubMed] [Google Scholar]
- 63.Beerli RR, Schopfer U, Dreier B, Barbas CF., III 2000. Chemically regulated zinc finger transcription factors. J. Biol. Chem. 275, 32 617–32 627. ( 10.1074/jbc.M005108200) [DOI] [PubMed] [Google Scholar]
- 64.Schwimmer LJ, Gonzalez B, Barbas CF., III 2012. Benzoate X receptor zinc-finger gene switches for drug-inducible regulation of transcription. Gene Therapy 19, 458–462. ( 10.1038/gt.2011.112) [DOI] [PubMed] [Google Scholar]
- 65.An CI, Trinh VB, Yokobayashi Y. 2006. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer–small molecule interaction. RNA 12, 710–716. ( 10.1261/rna.2299306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar D, An CI, Yokobayashi Y. 2009. Conditional RNA interference mediated by allosteric ribozyme. J. Am. Chem. Soc. 131, 13 906–13 907. ( 10.1021/ja905596t) [DOI] [PubMed] [Google Scholar]
- 67.Beisel CL, Chen YY, Culler SJ, Hoff KG, Smolke CD. 2011. Design of small molecule-responsive microRNAs based on structural requirements for drosha processing. Nucleic Acids Res. 39, 2981–2994. ( 10.1093/nar/gkq954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Culler SJ, Hoff KG, Smolke CD. 2010. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255. ( 10.1126/science.1192128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ketzer P, Haas SF, Engelhardt S, Hartig JS, Nettelbeck DM. 2012. Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res. 40, e167 ( 10.1093/nar/gks734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boorsma M, Nieba L, Koller D, Bachmann MF, Bailey JE, Renner WA. 2000. A temperature-regulated replicon-based DNA expression system. Nat. Biotechnol. 18, 429–432. ( 10.1038/74493) [DOI] [PubMed] [Google Scholar]
- 71.Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. 2006. A versatile tool for conditional gene expression and knockdown. Nat. Methods 3, 109–116. ( 10.1038/nmeth846) [DOI] [PubMed] [Google Scholar]
- 72.Kim DS, Gusti V, Dery KJ, Gaur RK. 2008. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol. Biol. 9, 23 ( 10.1186/1471-2199-9-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen YY, Jensen MC, Smolke CD. 2010. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl Acad. Sci. USA 107, 8531–8536. ( 10.1073/pnas.1001721107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Auslander S, Ketzer P, Hartig JS. 2010. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 6, 807–814. ( 10.1039/b923076a) [DOI] [PubMed] [Google Scholar]
- 75.Werstuck G, Green MR. 1998. Controlling gene expression in living cells through small molecule–RNA interactions. Science 282, 296–298. ( 10.1126/science.282.5387.296) [DOI] [PubMed] [Google Scholar]
- 76.Hara T, Saito H, Inoue T. 2013. Directed evolution of a synthetic RNA–protein module to create a new translational switch. Chem. Commun. 49, 3833–3835. ( 10.1039/c3cc38688k) [DOI] [PubMed] [Google Scholar]
- 77.Kodama K, Komatsu S, Ueda Y, Takayama T, Yajima J, Nanto S, Matsuoka H, Saito S, Hirayama A. 2010. Stabilization and regression of coronary plaques treated with pitavastatin proven by angioscopy and intravascular ultrasound: the together trial. Circ. J. 74, 1922–1928. ( 10.1253/circj.CJ-10-0038) [DOI] [PubMed] [Google Scholar]
- 78.Stripecke R, Oliveira CC, McCarthy JE, Hentze MW. 1994. Proteins binding to 5′ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol. Cell. Biol. 14, 5898–5909. ( 10.1128/MCB.14.9.5898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922. ( 10.1038/nmeth.1401) [DOI] [PubMed] [Google Scholar]
- 80.Miyazaki Y, Imoto H, Chen LC, Wandless TJ. 2012. Destabilizing domains derived from the human estrogen receptor. J. Am. Chem. Soc. 134, 3942–3945. ( 10.1021/ja209933r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rivera VM, et al. 2000. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science 287, 826–830. ( 10.1126/science.287.5454.826) [DOI] [PubMed] [Google Scholar]
- 82.Los GV, et al. 2008. Halotag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. Chem. Biol. 3, 373–382. ( 10.1021/cb800025k) [DOI] [PubMed] [Google Scholar]
- 83.Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM. 2011. Small-molecule hydrophobic tagging-induced degradation of halotag fusion proteins. Nat. Chem. Biol. 7, 538–543. ( 10.1038/nchembio.597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nomura Y, Zhou LL, Miu A, Yokobayashi Y. 2013. Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth. Biol. 2, 684–689. ( 10.1021/sb400037a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneekloth JS, Jr, Fonseca FN, Koldobskiy M, Mandal A, Deshaies R, Sakamoto K, Crews CM. 2004. Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 126, 3748–3754. ( 10.1021/ja039025z) [DOI] [PubMed] [Google Scholar]
- 86.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. 2006. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004. ( 10.1016/j.cell.2006.07.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonger KM, Chen LC, Liu CW, Wandless TJ. 2011. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat. Chem. Biol. 7, 531–537. ( 10.1038/nchembio.598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iwamoto K, Hamada H, Eguchi Y, Okamoto M. 2011. Mathematical modeling of cell cycle regulation in response to DNA damage: exploring mechanisms of cell-fate determination. Biosystems 103, 384–391. ( 10.1016/j.biosystems.2010.11.011) [DOI] [PubMed] [Google Scholar]
- 89.Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356. ( 10.1016/S0022-2836(61)80072-7) [DOI] [PubMed] [Google Scholar]
- 90.Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. 1999. Identification and classification of p53-regulated genes. Proc. Natl Acad. Sci. USA 96, 14 517–14 522. ( 10.1073/pnas.96.25.14517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. 1996. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl Acad. Sci. USA 93, 10 933–10 938. ( 10.1073/pnas.93.20.10933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766–1769. ( 10.1126/science.7792603) [DOI] [PubMed] [Google Scholar]
- 93.Weber W, Fussenegger M. 2004. Approaches for trigger-inducible viral transgene regulation in gene-based tissue engineering. Curr. Opin. Biotechnol. 15, 383–391. ( 10.1016/j.copbio.2004.07.003) [DOI] [PubMed] [Google Scholar]
- 94.Weber W, Fussenegger M. 2006. Pharmacologic transgene control systems for gene therapy. J. Gene Med. 8, 535–556. ( 10.1002/jgm.903) [DOI] [PubMed] [Google Scholar]
- 95.Nordstrom JL. 2002. Antiprogestin-controllable transgene regulation in vivo. Curr. Opin. Biotechnol. 13, 453–458. ( 10.1016/S0958-1669(02)00356-7) [DOI] [PubMed] [Google Scholar]
- 96.Chong H, Ruchatz A, Clackson T, Rivera VM, Vile RG. 2002. A system for small-molecule control of conditionally replication-competent adenoviral vectors. Mol. Therapy J. Am. Soc. Gene Therapy 5, 195–203. ( 10.1006/mthe.2002.0531) [DOI] [PubMed] [Google Scholar]
- 97.Mittal V. 2004. Improving the efficiency of RNA interference in mammals. Nat. Rev. Genet. 5, 355–365. ( 10.1038/nrg1323) [DOI] [PubMed] [Google Scholar]
- 98.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80, 5059–5064. ( 10.1128/JVI.80.10.5059-5064.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. 2002. Human dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21, 5875–5885. ( 10.1093/emboj/cdf582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deans TL, Cantor CR, Collins JJ. 2007. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell 130, 363–372. ( 10.1016/j.cell.2007.05.045) [DOI] [PubMed] [Google Scholar]
- 101.Mayer G. 2009. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 48, 2672–2689. ( 10.1002/anie.200804643) [DOI] [PubMed] [Google Scholar]
- 102.Saito H, Kobayashi T, Hara T, Fujita Y, Hayashi K, Furushima R, Inoue T. 2010. Synthetic translational regulation by an L7Ae–kink-turn RNP switch. Nat. Chem. Biol. 6, 71–78. ( 10.1038/nchembio.273) [DOI] [PubMed] [Google Scholar]
- 103.Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. 2010. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 17, 981–988. ( 10.1016/j.chembiol.2010.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. 2002. Inteins: structure, function, and evolution. Annu. Rev. Microbiol. 56, 263–287. ( 10.1146/annurev.micro.56.012302.160741) [DOI] [PubMed] [Google Scholar]
- 105.Anraku Y, Mizutani R, Satow Y. 2005. Protein splicing: its discovery and structural insight into novel chemical mechanisms. IUBMB Life 57, 563–574. ( 10.1080/15216540500215499) [DOI] [PubMed] [Google Scholar]
- 106.Lockless SW, Muir TW. 2009. Traceless protein splicing utilizing evolved split inteins. Proc. Natl Acad. Sci. USA 106, 10 999–11 004. ( 10.1073/pnas.0902964106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elleuche S, Poggeler S. 2010. Inteins, valuable genetic elements in molecular biology and biotechnology. Appl. Microbiol. Biotechnol. 87, 479–489. ( 10.1007/s00253-010-2628-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mootz HD, Muir TW. 2002. Protein splicing triggered by a small molecule. J. Am. Chem. Soc. 124, 9044–9045. ( 10.1021/ja026769o) [DOI] [PubMed] [Google Scholar]
- 109.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. 2003. Conditional protein splicing: a new tool to control protein structure and function in vitro and in vivo. J. Am. Chem. Soc. 125, 10 561–10 569. ( 10.1021/ja0362813) [DOI] [PubMed] [Google Scholar]
- 110.Shi J, Muir TW. 2005. Development of a tandem protein trans-splicing system based on native and engineered split inteins. J. Am. Chem. Soc. 127, 6198–6206. ( 10.1021/ja042287w) [DOI] [PubMed] [Google Scholar]
- 111.Selgrade DF, Lohmueller JJ, Lienert F, Silver PA. 2013. Protein scaffold-activated protein trans-splicing in mammalian cells. J. Am. Chem. Soc. 135, 7713–7719. ( 10.1021/ja401689b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. 2013. Synthetic analog computation in living cells. Nature 497, 619–623. ( 10.1038/nature12148) [DOI] [PubMed] [Google Scholar]
- 113.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. 2007. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 25, 795–801. ( 10.1038/nbt1307) [DOI] [PubMed] [Google Scholar]
- 114.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. 2012. Programmable single-cell mammalian biocomputers. Nature 487, 123–127. [DOI] [PubMed] [Google Scholar]
- 115.Kramer BP, Fischer C, Fussenegger M. 2004. Biologic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 87, 478–484. ( 10.1002/bit.20142) [DOI] [PubMed] [Google Scholar]
- 116.Weber W, Stelling J, Rimann M, Keller B, Daoud-El Baba M, Weber CC, Aubel D, Fussenegger M. 2007. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl Acad. Sci. USA 104, 2643–2648. ( 10.1073/pnas.0606398104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Greber D, Fussenegger M. 2010. An engineered mammalian band-pass network. Nucleic Acids Res. 38, e174 ( 10.1093/nar/gkq671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kramer BP, Fussenegger M. 2005. Hysteresis in a synthetic mammalian gene network. Proc. Natl Acad. Sci. USA 102, 9517–9522. ( 10.1073/pnas.0500345102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nissim L, Bar-Ziv RH. 2010. A tunable dual-promoter integrator for targeting of cancer cells. Mol. Syst. Biol. 6, 444 ( 10.1038/msb.2010.99) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lohmueller JJ, Armel TZ, Silver PA. 2012. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 40, 5180–5187. ( 10.1093/nar/gks142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhaya D, Davison M, Barrangou R. 2011. CRISPR–Cas systems in Bacteria and Archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. ( 10.1146/annurev-genet-110410-132430) [DOI] [PubMed] [Google Scholar]
- 122.Westra ER, Buckling A, Fineran PC. 2014. CRISPR–Cas systems: beyond adaptive immunity. Nat. Rev. Microbiol. 12, 317–326. ( 10.1038/nrmicro3241) [DOI] [PubMed] [Google Scholar]
- 123.Cho SW, Kim S, Kim JM, Kim JS. 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232. ( 10.1038/nbt.2507) [DOI] [PubMed] [Google Scholar]
- 124.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339, 823–826. ( 10.1126/science.1232033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cong L, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. ( 10.1126/science.1231143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. ( 10.1016/j.cell.2013.02.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jinek M, Charpentier E, Chylinski K, Doudna CJH, Lim W, Qi L. 2013. Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription. U.S. Patent 20140068797 A1, March 6, 2014.
- 128.Farzadfard F, Perli SD, Lu TK. 2013. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/CAS. ACS Synth. Biol. 2, 604–613. ( 10.1021/sb400081r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. ( 10.1126/science.1225829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. 2013. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979. ( 10.1038/nmeth.2598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Perez-Pinera P, et al. 2013. RNA-guided gene activation by CRISPR-CAS9-based transcription factors. Nat. Methods 10, 973–976. ( 10.1038/nmeth.2600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kiani S, Beal J, Ebrahimkhani MR, Huh J, Hall RN, Xie Z, Li Y, Weiss R. 2014. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–726. ( 10.1038/nmeth.2969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. 2014. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/CAS toolkit in human cells. Mol. Cell 54, 698–710. ( 10.1016/j.molcel.2014.04.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M. 2004. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 22, 867–870. ( 10.1038/nbt980) [DOI] [PubMed] [Google Scholar]
- 135.Greber D, El-Baba MD, Fussenegger M. 2008. Intronically encoded siRNAs improve dynamic range of mammalian gene regulation systems and toggle switch. Nucleic Acids Res. 36, e101 ( 10.1093/nar/gkn443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gillette MU, Sejnowski TJ. 2005. Physiology: biological clocks coordinately keep life on time. Science 309, 1196–1198. ( 10.1126/science.1111420) [DOI] [PubMed] [Google Scholar]
- 137.Schibler U, Sassone-Corsi P. 2002. A web of circadian pacemakers. Cell 111, 919–922. ( 10.1016/S0092-8674(02)01225-4) [DOI] [PubMed] [Google Scholar]
- 138.Kaasik K, Lee CC. 2004. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430, 467–471. ( 10.1038/nature02724) [DOI] [PubMed] [Google Scholar]
- 139.Lahav G. 2004. The strength of indecisiveness: oscillatory behavior for better cell fate determination. Sci. STKE 2004, pe55. [DOI] [PubMed] [Google Scholar]
- 140.Covert MW, Leung TH, Gaston JE, Baltimore D. 2005. Achieving stability of lipopolysaccharide-induced NF-kappa B activation. Science 309, 1854–1857. ( 10.1126/science.1112304) [DOI] [PubMed] [Google Scholar]
- 141.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. 2009. A tunable synthetic mammalian oscillator. Nature 457, 309–312. ( 10.1038/nature07616) [DOI] [PubMed] [Google Scholar]
- 142.Tigges M, Denervaud N, Greber D, Stelling J, Fussenegger M. 2010. A synthetic low-frequency mammalian oscillator. Nucleic Acids Res. 38, 2702–2711. ( 10.1093/nar/gkq121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mather W, Bennett MR, Hasty J, Tsimring LS. 2009. Delay-induced degrade-and-fire oscillations in small genetic circuits. Phys. Rev. Lett. 102, 068105 ( 10.1103/PhysRevLett.102.068105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Purcell O, Savery NJ, Grierson CS, di Bernardo M. 2010. A comparative analysis of synthetic genetic oscillators. J. R. Soc. Interface R. Soc. 7, 1503–1524. ( 10.1098/rsif.2010.0183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. 2008. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519. ( 10.1038/nature07389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.IOM (Institute of Medicine) 2011. The science and applications of synthetic and systems biology. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- 147.Burrill DR, Inniss MC, Boyle PM, Silver PA. 2012. Synthetic memory circuits for tracking human cell fate. Genes Dev. 26, 1486–1497. ( 10.1101/gad.189035.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. 2011. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311. ( 10.1126/science.1205527) [DOI] [PubMed] [Google Scholar]
- 149.O′Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. 2011. Tunable signal processing in synthetic map kinase cascades. Cell 144, 119–131. ( 10.1016/j.cell.2010.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bardwell L. 2011. Synthetic biology: modulating the map kinase module. Curr. Biol. 21, R249–R251. ( 10.1016/j.cub.2011.02.042) [DOI] [PubMed] [Google Scholar]
- 151.Ye H, Daoud-El Baba M, Peng RW, Fussenegger MA. 2011. Synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332, 1565–1568. ( 10.1126/science.1203535) [DOI] [PubMed] [Google Scholar]
- 152.Stanley SA, Gagner JE, Damanpour S, Yoshida M, Dordick JS, Friedman JM. 2012. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 336, 604–608. ( 10.1126/science.1216753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ye H, Aubel D, Fussenegger M. 2013. Synthetic mammalian gene circuits for biomedical applications. Curr. Opin. Chem. Biol. 17, 910–917. ( 10.1016/j.cbpa.2013.10.006) [DOI] [PubMed] [Google Scholar]
- 154.Grupp SA, et al. 2013. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 368, 1509–1518. ( 10.1056/NEJMoa1215134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ye HF, Charpin-El Hamri G, Zwicky K, Christen M, Folcher M, Fussenegger M. 2013. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc. Natl Acad. Sci. USA 110, 141–146. ( 10.1073/pnas.1216801110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Citorik RJ, Mimee M, Lu TK. 2014. Bacteriophage-based synthetic biology for the study of infectious diseases. Curr. Opin. Microbiol. 19C, 59–69. ( 10.1016/j.mib.2014.05.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yacoby I, Shamis M, Bar H, Shabat D, Benhar I. 2006. Targeting antibacterial agents by using drug-carrying filamentous bacteriophages. Antimicrob. Agents Chemother. 50, 2087–2097. ( 10.1128/AAC.00169-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jacobsson K, Frykberg L. 1996. Phage display shot-gun cloning of ligand-binding domains of prokaryotic receptors approaches 100% correct clones. BioTechniques 20, 1070–1081. [DOI] [PubMed] [Google Scholar]
- 159.Jönsson K, Guo BP, Monstein HJ, Mekalanos JJ, Kronvall G. 2004. Molecular cloning and characterization of two Helicobacter pylori genes coding for plasminogen-binding proteins. Proc. Natl Acad. Sci. USA 101, 1852–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Temme K, et al. 2012. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 40, 8773–8781. ( 10.1093/nar/gks597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Noireaux V, Bar-Ziv R, Libchaber A. 2003. Principles of cell-free genetic circuit assembly. Proc. Natl Acad. Sci. USA 100, 12 672–12 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim J, White KS, Winfree E. 2006. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2, 68 ( 10.1038/msb4100099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. 2009. Synthetic gene networks that count. Science, 324, 1199–1202. ( 10.1126/science.1172005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. 2013. Amplifying genetic logic gates. Science 340, 599–603. ( 10.1126/science.1232758) [DOI] [PubMed] [Google Scholar]
- 165.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli k-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645. ( 10.1073/pnas.120163297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Husseiny MI, Hensel M. 2005. Rapid method for the construction of Salmonella enterica serovar Typhimurium vaccine carrier strains. Infect. Immun. 73, 1598–1605. ( 10.1128/IAI.73.3.1598-1605.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Beloin C, Deighan P, Doyle M, Dorman CJ. 2003. Shigella flexneri 2a strain 2457T expresses three members of the H-NS-like protein family: characterization of the Sfh protein. Mol. Genet. Genomics 270, 66–77. ( 10.1007/s00438-003-0897-0) [DOI] [PubMed] [Google Scholar]
- 168.Yamamoto S, Izumiya H, Morita M, Arakawa E, Watanabe H. 2009. Application of λ Red recombination system to Vibrio cholerae genetics: simple methods for inactivation and modification of chromosomal genes. Gene 438, 57–64. ( 10.1016/j.gene.2009.02.015) [DOI] [PubMed] [Google Scholar]
- 169.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38, 113–116. ( 10.1016/S0928-8244(03)00181-0) [DOI] [PubMed] [Google Scholar]
- 170.Lesic B, Rahme LG. 2008. Use of the λ Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol. Biol. 9, 20 ( 10.1186/1471-2199-9-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lu TK, Collins JJ. 2007. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl Acad. Sci. USA 104, 11 197–11 202. ( 10.1073/pnas.0704624104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Westwater C, Kasman LM, Schofield DA, Werner PA, Dolan JW, Schmidt MG, Norris JS. 2003. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob. Agents Chemother. 47, 1301–1307. ( 10.1128/AAC.47.4.1301-1307.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Edgar R, Friedman N, Molshanski-Mor S, Qimron U. 2012. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl. Environ. Microbiol. 78, 744–751. ( 10.1128/AEM.05741-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schmidt M, Ganguli-Mitra A, Torgersen H, Kelle A, Deplazes A, Biller-Andorno N. 2009. A priority paper for the societal and ethical aspects of synthetic biology. Syst. Synth. Biol. 3, 3–7. ( 10.1007/s11693-009-9034-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bedau MA, Parke EC, Tangen U, Hantsche-Tangen B. 2009. Social and ethical checkpoints for bottom-up synthetic biology, or protocells. Syst. Synth. Biol. 3, 65–75. ( 10.1007/s11693-009-9039-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rabinow P, Bennett G. 2009. Synthetic biology: ethical ramifications 2009. Syst. Synth. Biol. 3, 99–108. ( 10.1007/s11693-009-9042-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aubel D, Morris R, Lennon B, Rimann M, Kaufmann H, Folcher M, Bailey JE, Thompson CJ, Fussenegger M. 2001. Design of a novel mammalian screening system for the detection of bioavailable, non-cytotoxic streptogramin antibiotics. J. Antibiot. 54, 44–55. ( 10.7164/antibiotics.54.44) [DOI] [PubMed] [Google Scholar]
- 178.Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, Locht C, Besra GS. 2000. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 275, 28 326–28 331. [DOI] [PubMed] [Google Scholar]
- 179.Gonzalez-Nicolini V, Fux C, Fussenegger M. 2004. A novel mammalian cell-based approach for the discovery of anticancer drugs with reduced cytotoxicity on non-dividing cells. Invest. New Drugs 22, 253–262. ( 10.1023/B:DRUG.0000026251.00854.77) [DOI] [PubMed] [Google Scholar]
- 180.Gonzalez-Nicolini V, Fussenegger M. 2005. In vitro assays for anticancer drug discovery: a novel approach based on engineered mammalian cell lines. Anticancer Drugs 16, 223–228. ( 10.1097/00001813-200503000-00001) [DOI] [PubMed] [Google Scholar]
- 181.Filmore D. 2004. It's a GPCR world. Cell-based screening assays and structural studies are fueling G-protein coupled receptors as one of the most popular classes of investigational drug targets. Mod. Drug Discov. 7, 24–28. [Google Scholar]
- 182.Xiao X, Min JL, Wang P, Chou KC. 2013. Predict drug–protein interaction in cellular networking. Curr. Top. Med. Chem. 13, 1707–1712. ( 10.2174/15680266113139990121) [DOI] [PubMed] [Google Scholar]
- 183.Esbenshade TA. 2005. G protein-coupled receptors as targets for drug discovery. In G protein-coupled receptors in drug discovery (eds Lundstrom KH, Chiu ML.). Boca Raton, FL: Taylor and Francis. [Google Scholar]
- 184.PerkinElmer Inc. Receptor activation. See http://www.perkinelmer.co.uk/pages/020/cellularimaging/assays/receptoractivation.xhtml (accessed 16 March 2015).
- 185.Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, Lee KJ. 2008. The genetic design of signaling cascades to record receptor activation. Proc. Natl Acad. Sci. USA 105, 64–69. ( 10.1073/pnas.0710487105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Seifert R, Wenzel-Seifert K, Kobilka BK. 1999. GPCR-G alpha fusion proteins: molecular analysis of receptor–G-protein coupling. Trends Pharmacol. Sci. 20, 383–389. ( 10.1016/S0165-6147(99)01368-1) [DOI] [PubMed] [Google Scholar]
- 187.Li F, Vijayasankaran N, Shen AY, Kiss R, Amanullah A. 2010. Cell culture processes for monoclonal antibody production. mAbs 2, 466–479. ( 10.4161/mabs.2.5.12720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Marx U, et al. 1997. Monoclonal antibody production—the report and recommendations of ECVAM workshop 23. Atla-Altern. Lab. Anim. 25, 121–137. [Google Scholar]
- 189.McLeod J, James DC. 2011. Engineering protein folding and secretion in eukaryotic cell factories. In Comprehensive biotechnology (ed. Moo-Young M.). Amsterdam, The Netherlands: Elsevier Science and Technology Books. [Google Scholar]
- 190.Dinnis DM, James DC. 2005. Engineering mammalian cell factories for improved recombinant monoclonal antibody production: lessons from nature? Biotechnol. Bioeng. 91, 180–189. ( 10.1002/bit.20499) [DOI] [PubMed] [Google Scholar]