Abstract
Background
The endosymbiosis in trypanosomatids is characterized by co-evolution between one bacterium and its host protozoan in a mutualistic relationship, thus constituting an excellent model to study organelle origin in the eukaryotic cell. In this association, an intense metabolic exchange is observed between both partners: the host provides energetic molecules and a stable environment to a reduced wall symbiont, while the bacterium is able to interfere in host metabolism by enhancing phospholipid production and completing essential biosynthesis pathways, such as amino acids and hemin production. The bacterium envelope presents a reduced cell wall which is mainly composed of cardiolipin and phosphatidylcholine, being the latter only common in intracellular prokaryotes. Phosphatidylinositol (PI) is also present in the symbiont and host cell membranes. This phospholipid is usually related to cellular signaling and to anchor surface molecules, which represents important events for cellular interactions.
Methods
In order to investigate the production of PI and its derivatives in symbiont bearing trypanosomatids, aposymbiotic and wild type strains of Angomonas deanei, as well as isolated symbionts, were incubated with [3H]myo-inositol and the incorporation of this tracer was analyzed into inositol-containing molecules, mainly phosphoinositides and lipoproteins. Gene searches and their phylogenies were also performed in order to investigate the PI synthesis in symbiontbearing trypanosomatids.
Results
Our results showed that the bacterium did not incorporate the tracer and that both strains produced similar quantities of PI and its derivatives, indicating that the symbiont does not influence the production of these metabolites. Gene searches related to PI synthesis revealed that the trypanosomatid genome contains an inositol transporter, PI synthase and the myo-inositol synthase. Thus, the host is able to produce PI either from exogenous myo-inositol (inositol transporter) or from myo-inositol synthesized de novo. Phylogenetic analysis using other organisms as references indicated that, in trypanosomatids, the genes involved in PI synthesis have a monophyletic origin. In accordance with experimental data, sequences for myo-inositol transport or for myo-inositol and PI biosynthesis were not found in the symbiont.
Conclusions
Altogether, our results indicate that the bacterium depends on the host to obtain PI.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-015-0854-x) contains supplementary material, which is available to authorized users.
Keywords: Phosphatidylinositol metabolism, Symbiosis, Phylogenetic analysis
Background
The Trypanosomatidae family is known worldwide, since it contains pathogenic species from genera Trypanosoma and Leishmania, which affect human health and the economy [1]. However, most protozoa in this family are non-pathogenic to humans and inhabit only invertebrate hosts during the whole life cycle. Between these monoxenics, six species, including Angomonas deanei, maintain an obligatory symbiotic relationship with an intracellular bacterium [2,3]. The mutualistic co-evolution between these primitive partners makes this relationship an excellent model to study the origin of organelles in the eukaryotic cell [4]. Symbiosis in trypanosomatid protozoa has a monophyletic origin and the bacterium is phylogenetically related to the Alcaligenaceae family of the Betaproteobacteria class, thus belonging to the Gram-negative group [2,3,5]. Treatment of trypanosomatids with chloramphenicol generated an aposymbiotic strain, which can be used as a useful tool to understand how the symbiont influences the host [4]. Previous studies revealed an intense metabolic exchange between the symbiotic bacterium and the protozoan, which is reflected in an enhanced growth rate and low nutritional requirements of symbiont-bearing trypanosomatids when compared to the aposymbiotic ones [6-9]. Actually, it has been demonstrated that the symbiotic bacterium completes several metabolic routes of the host cell, such as those involved in the production of amino acids, vitamins, and heme [10-13]. Furthermore, the presence of the symbiont is related to alterations in trypanosomatid ultrastructure [14], surface charge [15], and carbohydrate plasma membrane composition [16].
The symbiont presents an envelope composed of two membrane units and a reduced cell wall, which does not allow the symbiont either to survive or to replicate when isolated from the host [17]. Although phosphatidylcholine (PC) is a major component of symbiont membranes, once isolated the bacterium produces very low amounts of this phospholipid. Considering the host’s phospholipid composition, A. deanei has PC as a major constituent, followed by phosphatidylethanolamine (PE) and PI. Interestingly, comparisons between wild type and the aposymbiotic strains indicate that the presence of the symbiont is correlated with an increase in phospholipid production [18]. Accordingly, it has been reported that intracellular bacteria may complement some steps of the PI metabolism from the host, as already observed in plants as well as in animal cells infected by pathogenic prokaryotes [19,20].
In trypanosomatids, phospholipids are not obtained directly from the environment (host or medium), but synthesized using the common headgroups (such as choline, ethanolamine, inositol) and diacylglycerol. In T. brucei, the genes responsible for de novo biosynthesis of all phospholipids were identified [21,22]. The biosynthesis of PI in trypanosomatids occurs by condensation of the headgroup, in this case myo-inositol, with cytidine diphosphate-diacylglycerol (CDP-DAG). In the trypanosomatid Leishmania donovani, the uptake of myo-inositol occurs through an inositol/H+ symporter, which belongs to the glucose transporter superfamily [23,24], while in T. cruzi myo-inositol transport occurs through an inositol/Na+ symporter [25]. Similar to mammals, in trypanosomatids PI synthase (PIS) catalyzes the production of PI from CDP-DAG and myo-inositol, with the reaction taking place in the endoplasmic reticulum (ER) and in the Golgi apparatus [21,22]. While all eukaryotes are able to synthesize PI, most prokaryotes do not present this ability. Few prokaryotes present a PIS, which was probably obtained by lateral gene transfer from Archaea [26]. In these organisms, the production of PI occurs as follows: first, phosphatidylinositol monophosphate (PIP) is produced from inositol phosphate and cytidine monophosphate-phosphatidyl; then, PIP releases an inorganic phosphate (Pi), thus generating PI [27].
PI has an important role in cell signaling in trypanosomatids, as well as in other eukaryotic cells, being the precursor of important molecules such as phosphorylated inositol lipids, also called phosphoinositides [28,29]. Mainly located in the cytosolic layer of membranes, PI can be sequentially phosphorylated by different specific kinases (PI-K). When PI-4,5-bisphosphate is formed, it can be hydrolyzed by the PI-phospholipase C (PI-PLC) yielding inositol triphosphate, which is released into the cytoplasm, and DAG [30]. Some PI-Ks were already identified in the genome of trypanosomatids and also by biochemical activity [21,22].
Phosphatidylinositol and phosphoinositides also participate in the establishment and maintenance of symbiotic relationships, as observed in symbiosis between fungi and plants [31] and between bacteria and plants [19,32]. It was previously demonstrated that the symbiotic bacteria Rhizobium leguminosarum presents a phosphotransferase whose activity increases the PI 4-phosphate content of the plant host, which is essential to the nodulation process [19]. In the symbiosis between Medicago truncatula and Rhizobium bacteria, nodulation is stimulated by the production of bacterial Nod factors, thus promoting the activation of calcium spikes in the host in a phosphoinositide-dependent signaling [32]. Furthermore, it has also been reported that intracellular prokaryotes, such as pathogenic bacteria, can promote alterations in the PI, PIP, and PIP2 content in the host [20].
In this work, we investigated the production of PI, PIP, and phosphatidylinositol biphosphate (PIP2) in A. deanei and its aposymbiotic strain in order to verify whether the bacterium influences the PI metabolism of the protozoan host. Furthermore, the enzymes involved in PI biosynthesis were also investigated, at the genome level and phylogenetically, to provide further information about this metabolic pathway in the trypanosomatid family.
Methods
Cell growth and the obtainment of endosymbiont fraction
Wild type (Wt) and aposymbiotic strains of A. deanei were grown at 28°C in Warren's culture medium [33] supplemented with 10% fetal calf serum. In all assays, both strains were cultivated for 24 hours, which corresponds to the exponential growth phase. The endosymbiont fractions were obtained as described in [18].
[3H]myo-inositol incorporation and labeling of the PI
A. deanei PI was metabolically labeled as follows: log phase cells were incubated with 1 μCi/mL [3H]-myo-inositol in culture medium for 24 h. Protozoa were then washed 3 times with PBS, with centrifugation (1,800 × g), and the cell pellet was quantified by protein concentration and used for lipid extraction as described below. The isolated symbiont fraction was also incubated with the radiolabeled inositol. The fraction was resuspended in cell growth medium containing 1 μCi/mL [3H]-myo-inositol for 1 or 3 h at 28°C. After incubation, the fraction was washed 3 times with PBS, with centrifugation (1,800 × g), and then homogenized. Protein concentration was quantified for the different assays.
Lipid extraction, separation, and identification of phospholipids
The methodology previously described by our group [18] was adopted for lipid extraction and separation. After identification of the phospholipid by thin layer chromatography (TLC), plates were dried in a fume hood and the lipid spots were visualized by exposure to iodine vapors. Autoradiography of the TLC plates was carried out using X-ray film (Kodak T-Mat) at −70°C for 10 days, and the film was developed following the manufacturer’s specifications. For the identification of radiolabeled PI spots, two methods were adopted: in the first, plates were exposed to a 3H-sensitive screen in a cassette stored at −20°C for 10 days and the radioactivity was then detected in Storm Phosphor Image. In the second method, PI spots identified by exposure to iodine vapor were scraped off the plate and put into vials containing scintillation solution. The radioactivity was measured using a Beckman counter.
Chemicals
The solvents, plates and other reagents used in this work for extraction, separation, and identification of phospholipids by thin layer chromatography were obtained from Merck. [3H]-myo-inositol was purchased from Perkin Elmer. All reagents were high purity grade. Deionized water was used to prepare all media and solutions.
Statistical analysis
The PI quantifications are expressed as mean ± standard error (±SE), using one-way ANOVA followed by post-hoc Tukey's test for multiple comparisons. Data were considered statistically significant when p < 0.05. Although experiments considered a starting concentration of 107 cells/mL, PI measures were normalized by protein quantification.
Genomic analysis
For genomic analysis, data from A. deanei and its corresponding symbiotic bacterium were obtained from our previous publication where DNA extraction and sequencing, followed by gene calling and the functional annotation, were described [5]. The assembly and associated annotations are available in NCBI's GenBank database under accession number ATMG00000000. In this work, sequences of interest are identified according to their protein identifiers. Reference sequences used in the present genomic analyses were selected from phylogenetically related organisms, as follows: Leishmania major and Leishmania donovani for comparisons with A. deanei, and Mycobacterium tuberculosis RGTB327 and Methanothermobacter thermautotrophicus (str. Delta H, all available from GenBank) for comparisons with the symbiotic bacterium.
Phylogenetic analysis
To ensure wide representation of taxa from all branches of life, candidate sequences for phylogenetic analyses were selected by similarity searches of the A. deanei protein sequence against the whole NCBI non-redundant (nr) database (maximum E-value cutoff of 1E-10), as previously described [3]. New trypanosomatid sequences used in phylogenetic analysis were deposited in GenBank (KP689381- KP689400). The resulting datasets were aligned by Muscle v. 3.8.31 [34], ambiguously aligned positions were removed by Gblocks [35] using the “with half” option for gap treatment, and maximum likelihood phylogenetic analyses were performed by RAxML v. 8.0.24 [36], using the WAG substitution model [37], gamma-distributed rate heterogeneity categories, and empirical residue frequencies. Trees were edited and drawn in TreeGraph2 [38] and Dendroscope [39], with cosmetic adjustments done in Inkscape (http://inkscape.org).
Results
Phosphoinositide formation in A. deanei
Wild type and aposymbiotic strains of A. deanei were grown in culture medium containing [3H]myo-inositol in order to quantify the incorporation of this tracer in PI and in phosphoinositides. There is no significant differences in the incorporation of exogenous [3H]-myo-inositol into PI, PIP and PIP2 (p > 0,05 in all tests) after comparing wild type and aposymbiotic strain of A. deanei (Figure 1). The wild type cells presented a mean of 1382 ± 105 CPM/mg of protein, while the aposymbiotic cells presented 1299 ± 124 CPM/mg. The tracer was mainly incorporated in PI: 1328 ± 302 CPM/mg in wild type and 1536 ± 450 CPM/mg in aposymbiotic strain. PIP and PIP2 labeling were very low when compared with PI: wild type cells presented 70 ± 7 CPM/mg for PIP and 314 ± 91 CPM/mg for PIP2, while aposymbiotic cells presented similar values for PIP (70 ± 9 CPM/mg) and for PIP2 (397 ± 193 CPM/mg) (Figure 1).
Figure 1.
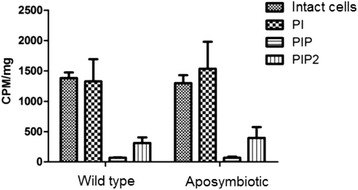
A. deanei synthesis of PI and its derivatives after incorporation of [3H]myo-inositol. Wild type and aposymbiotic cells presented similar quantities of PI, PIP and PIP2. The graph represents the mean CPM/mg of protein ± SE of five independent experiments after 24 hours of incubation with radiolabeled inositol.
To analyze the [3H]myo-inositol incorporation in the symbiont, the A. deanei wt strain was fractionated for obtainment of isolated bacteria which were incubated for one or three hours in the presence of the tracer. After extraction, identification of lipids, and radioactivity measurement, PI, PIP, and PIP2 spots were not labeled in the symbiont fraction (Figure 2). These results indicate that, once isolated from the host, the bacterium is not able to synthesize PI and phosphorylated PI species using exogenous myo-inositol as substrate.
Figure 2.
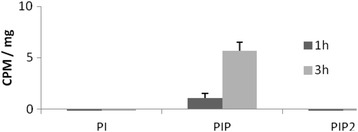
Isolated symbionts were incubated with [3H]myo-inositol in order to verify the synthesis of PI, PIP, and PIP2. The graph represents the mean CPM/mg of protein ± SE of five independent experiments using symbionts incubated for 1 h or 3 h with the radiolabeled tracer. No significant radioactivity values were observed (the highest radioactivity inositide, PIP, presented less than 6 CPM/mg).
PI synthesis pathway
Searches for gene sequences involved in PI biosynthesis were performed in genomes of symbiont-harboring trypanosomatids and their symbionts, as well as in genomes of trypanosomatids that do not contain the bacterium. Results revealed that the host protozoan contains PIS genes (Table 1 ), indicating that PI may be synthesized from CDP-diacylglycerol and myo-inositol, as already reported in other trypanosomatids. Three sequences from the A. deanei genome deposited in GenBank (EPY39094.1, EPY39654.1 and EPY28298.1) were identified as PIS (E. C. 2.7.8.11). On the other hand, searching for PIS in the symbiont genomes using related sequences in Archaea and some bacteria (AIS) did not show any candidate genes (Table 1).PI synthesis requires exogenous myo-inositol or myo-inositol synthesized by a de novo pathway. The de novo synthesis of myo-inositol occurs by action of an isomerase, called L-myo-inositol 1-phosphate synthase (MIPS) (E. C. 5.5.1.4), that uses glucose as substrate to produce myo-inositol. Investigations in genomes of A. deanei and its symbiont revealed the presence of MIPS in the host trypanosomatid, but not in the bacterium.
Table 1.
Gene sequences for PI metabolism in A. deanei and its symbiont
| Protein | Reference (organism) | A. deanei | Symbiont |
|---|---|---|---|
| Inositol transporter | PRF 1583317 (L. donovani) | EPY28738.1, EPY351511, EPY29848.1 | NF |
| PIS (eukaryotic) | XP_001684255.1 (L. major) | EPY39094.1, EPY39654.1, EPY28298.1 | NF |
| PIS (prokaryotic) | AFE17439.1 (M. tuberculosis) | NF | NF |
| AIPS | O27726.1 (M. thermatotrophicus) | NF | NF |
NF : gene not found.
Moreover, a membrane inositol transporter that would allow the entrance of exogenous myo-inositol into the cytoplasm was identified in the A. deanei genome. Represented by 3 sequences in GenBank (GenBank ID: EPY28738.1, EPY351511, and EPY29848.1), such genes have high similarity to those described in L. donovani (PRF 1583317). The symbiotic bacterium lacks sequences that codify for this protein. It is worth mentioning that mRNAs for PIS and inositol membrane transporter were identified in the transcriptome of A. deanei (submitted to publication).
Our phylogenetic analyses showed that A. deanei genes involved in PI production cluster with those described in other species of this genus, such as A. desouzai. The Angomonas group is inserted in the Trypanosomatidae family cluster, which also contains the regular trypanosomatids, and is placed among eukaryotes (Figure 3 and Additional file 1: Figure S1, Additional file 2: Figure S2 and Additional file 3: Figure S3). These results suggest that PI synthesis genes present a monophyletic origin in this family.
Figure 3.
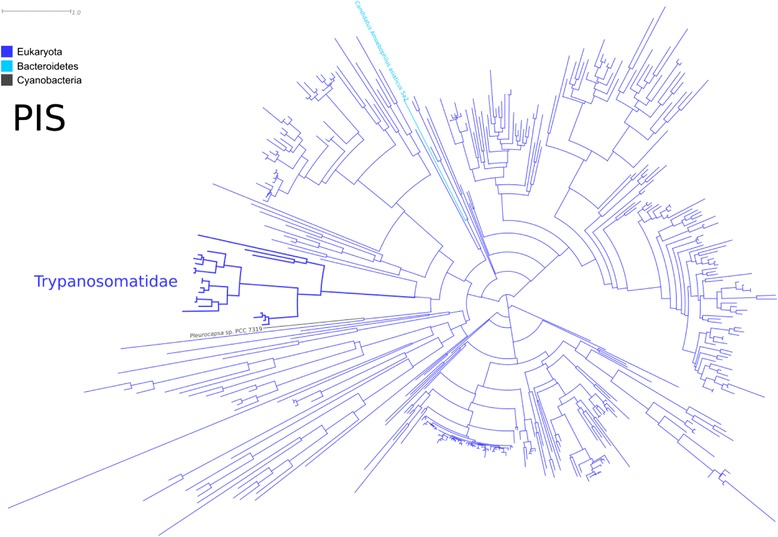
Molecular phylogenetic analysis of PIS by the maximum likelihood method. Branches are colored based on taxonomic affiliation, according to the legend on the left. The clade comprising the Trypanosomatidae family is represented by thicker branches and labeled accordingly. The only two prokaryotic sequences present in this tree are indicated by name. See Additional file 1: Figure S1 for a full version of this tree showing all names and including bootstrap support values.
Discussion
Our previous studies revealed that the symbiotic bacterium enhances the host phospholipid biosynthesis, including PI [19]. Here, we have shown that the bacterium genome does not contain genes for PI biosynthesis, including PI synthase, nor for inositol transporters, which were previously described in few prokaryotes, such as in Mycobacterium sp. and in the archaea M. thermautotrophicus [26]. Despite the absence of these genes in the symbiont, the host trypanosomatid presents both PIS and inositol membrane transporter genes, as supported by the transcriptome data of A. deanei (submitted to publication), indicating that the bacterium obtains its PI directly from the host. Taking these data together, we can suggest that the symbiotic association maintains the optimal lipid content: the prokaryote stimulates the glycerophospholipid metabolism in the protozoan, while the host supplies the symbiont with key phospholipids.
The occurrence of gene transfer has been previously described in A. deanei. Analysis of biosynthetic routes revealed that many horizontal gene transfers occurred from diverse bacterial lineages to the nucleus, thus indicating previous symbiotic associations in trypanosomatids. Some of these transferred sequences originated from the present endosymbiont [3]. However, in the present work, gene searches and phylogenetic analyses have never grouped trypanosomatid sequences with any bacterial ortholog, revealing that PIS, MIPS, and the inositol membrane transporter sequences found in A. deanei and other trypanosomatids have eukaryotic origin and are not derived from bacterial gene transfers.
Conclusions
In this work, we showed that A. deanei is able to synthesize PI and phosphoinositides from [3H]myo-inositol incorporation, which was corroborated by genomic and phylogenetic data. The presence of the symbiotic bacterium does not influence the [3H]myo-inositol incorporation by the host trypanosomatid. The isolated symbiont is not able to produce its own PI, which is evidenced by the lack of sequences coding enzymes and proteins for PI synthesis in its genome. The overall data provide strong evidence for the view that PI is produced by the host and further incorporated, processed, and metabolically employed by the bacterium in its biological processes. We also suggest that the above mentioned PI metabolism route, shared in this mutualistic association, would represent a mechanism used by the host to control the bacterium size and division.
Acknowledgements
We thank Drs. Erney Camargo and Marta Teixeira for scientific discussion and FAPERJ, FAPESP (grant #2013/14622-3, São Paulo Research Foundation), CAPES and CNPq for financial support.
Abbreviations
- AIS
Archaeal inositol synthase
- CDP-DAG
Cytidine diphosphate-diacylglycerol
- ER
Endoplasmic reticulum
- MIPS
L-myo-inositol 1-phosphate synthase
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- Pi
Inorganic phosphate
- PI
Phosphatidylinositol
- PI-K
Phosphatidylinositol kinase
- PI-PLC
Phosphatidylinositol phospholipase C
- PIP
Phosphatidylinositol monophosphate
- PIP2
Phosphatidylinositol biphosphate
- PIP3
Phosphatidylinositol triphosphate
- PIS
Phosphatidylinositol synthase
Additional files
Full version of the molecular phylogenetic analysis of PIS by the maximum likelihood method. Branches were colored based on taxonomic affiliation, according to the legend on the right. Numbers on nodes represent bootstrap support values. The clade comprising the Trypanosomatidae family was represented by thicker branches.
Molecular phylogenetic analysis of the inositol transporter by the maximum likelihood method. Branches were colored based on taxonomic affiliation, according to the legend on the right. Numbers on nodes represent bootstrap support values. The clade comprising the Trypanosomatidae family was represented by thicker branches.
Molecular phylogenetic analysis of MIPS by the maximum likelihood method. Branches were colored based on taxonomic affiliation, according to the legend on the right. Numbers on nodes represent bootstrap support values. The clade comprising the Trypanosomatidae family were represented by thicker branches.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ACAM performed all biochemical assays, ACAM and JMPA performed phylogenetic analyses, MCMM, MEL, FGM conceived and designed the experiments, JMPA and ATRV conceived the phylogenetic analyses, MCMM, MEL, FGM, WS, ATRV contributed with reagents, materials and analysis tools, ACAM, MEL, JMPA, MCMM wrote the paper. All authors read and approved the final version of the manuscript.
Contributor Information
Allan C de Azevedo-Martins, Email: allan@biof.ufrj.br.
João MP Alves, Email: alvesjmp@gmail.com.
Fernando Garcia de Mello, Email: fgmello@biof.ufrj.br.
Ana Tereza R Vasconcelos, Email: atrv@lncc.br.
Wanderley de Souza, Email: wsouza@biof.ufrj.br.
Marcelo Einicker-Lamas, Email: einicker@biof.ufrj.br.
Maria Cristina M Motta, Email: motta@biof.ufrj.br.
References
- 1.World Health Organization . Update of american trypanosomiasis and leishmania control and research: final report. Rio de Janeiro: PAHO/WHO; 2007. [Google Scholar]
- 2.Teixeira MMG, Borghesan TC, Ferreira RC, Santos M, Takata CS, Campaner M, et al. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 2011;162(3):503–24. doi: 10.1016/j.protis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Alves JM, Serrano MG, Maia Da Silva F, Voegtly LJ, Matveyev AV, Teixeira MM, et al. Genome evolution and phylogenomic analysis of candidatus kinetoplastibacterium, the betaproteobacterial endosymbionts of Strigomonas and Angomonas. Genome Biol Evol. 2013;5(2):338–50. doi: 10.1093/gbe/evt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motta MCM. Endosymbiosis in trypanosomatids as a model to study cell evolution. O Parasitol J. 2010;4:139–47. doi: 10.2174/1874421401004010139. [DOI] [Google Scholar]
- 5.Motta MC, Martins AC, de Souza SS, Catta-Preta CM, Silva R, Klein CC, et al. Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0060209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang KP, Chang CS, Sassa S. Heme biosynthesis in bacterium-protozoon symbioses: enzymic defects in host hemofagellates and complemental role of their intracellular symbiotes. Proc Natl Acad Sci U S A. 1975;72:2979–83. doi: 10.1073/pnas.72.8.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mundim MH, Roitman I. Extra nutritional requirements of artificially aposymbiotic Crithidia deanei. J Protozool. 1977;24:329–31. doi: 10.1111/j.1550-7408.1977.tb00988.x. [DOI] [Google Scholar]
- 8.Fiorini JE, Silva PM F e, Soares MJ, Brasil RP. Três novas espécies de tripanosomatídeos de insetos isolados em Alfenas, Minas Gerais, Brasil. Mem Inst O C. 1989;84:69–74. doi: 10.1590/S0074-02761989000100013. [DOI] [Google Scholar]
- 9.Frossard ML, Seabra SH, DaMatta RA, de Souza W, de Mello FG, Motta MC. An endosymbiont positively modulates ornithine decarboxylase in host trypanoso-matids. Biochem Biophys Res Com. 2006;343:443–9. doi: 10.1016/j.bbrc.2006.02.168. [DOI] [PubMed] [Google Scholar]
- 10.Alves JM, Klein CC, da Silva FM, Costa-Martins AG, Serrano MG, Buck GA, et al. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol. 2013;13:190. doi: 10.1186/1471-2148-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein CC, Alves JM, Serrano MG, Buck GA, Vasconcelos AT, Sagot MF, et al. Biosynthesis of vitamins and cofactors in bacterium-harbouring trypanosomatids depends on the symbiotic association as revealed by genomic analyses. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0079786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alves JMP, Voegtly L, Matveyev AV, Lara AM, Silva FM, Serrano MG, et al. Identification and phylogenetic analysis of heme synthesis genes in trypanosomatids and their bacterial endosymbionts. PLoS One. 2011;6:e23518. doi: 10.1371/journal.pone.0023518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camargo EP, Freymüller E. Endosymbiont as supplier of ornithine carbamoyltransferase in a trypanosomatid. Nature. 1977;270:52–3. doi: 10.1038/270052a0. [DOI] [PubMed] [Google Scholar]
- 14.Freymuller JE, Camargo EP. Ultrastructural differences between species of trypanosomatids with and without endosymbionts. J Protozool. 1981;28:175–82. doi: 10.1111/j.1550-7408.1981.tb02829.x. [DOI] [PubMed] [Google Scholar]
- 15.Oda LM, Alviano CS, Costa e Silva Filho F, Angluster J, Roitman I, De Souza W. Surface anionic group in symbiont-bearing and symbiont-free strains of Crithidia deanei. J Protozool. 1984;31:131–4. doi: 10.1111/j.1550-7408.1984.tb04301.x. [DOI] [Google Scholar]
- 16.McLaughlin GL, Cain GD. Characterization of whole-cell and organelle protein synthesis in normal and aposymbiotic strains of Crithidia oncopelti and Blastocrithidia culicis. Comparative Biochem Physiol B, 82(3):479–86.
- 17.Motta MCM, Monteiro-Leal LH, de Souza W, Almeida DF, Ferreira LCS. Detection of penicilin-binding proteins in endosymbiosis of the trypanosomatid Crithidia deanei. J Euk Microbiol. 1997;44:492–6. doi: 10.1111/j.1550-7408.1997.tb05729.x. [DOI] [Google Scholar]
- 18.Azevedo-Martins AC, Frossard ML, de Souza W, Einicker-Lamas M, Motta MCM. Phosphatidylcholine synthesis in Crithidia deanei: the influence of the endosymbiont. FEMS Microbiol Lett. 2007;275:229–36. doi: 10.1111/j.1574-6968.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- 19.Basu SS, York JD, Raetz CRH. A Phosphotransferase that generates Phosphatidylinositol 4-Phosphate (PtdIns-4-P) from Phosphatidylinositol and Lipid A in Rhizobium leguminosarum: a membrane-bound enzyme linking lipid A and PtdIns-4-P biosynthesis. J Biol Chem. 1999;274(16):11139–49. doi: 10.1074/jbc.274.16.11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pizarro-Cerdá J, Cossart P. Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat Cell Biol. 2004;6(11):1026–33. doi: 10.1038/ncb1104-1026. [DOI] [PubMed] [Google Scholar]
- 21.Van Hellemond JJ, Tielens AGM. Adaptations in the lipid metabolism of the protozoan parasite Trypanosoma brucei. FEBS Lett. 2006;580(23):5552–8. doi: 10.1016/j.febslet.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 22.Smith TK, Bütikofer P. Lipid metabolism in Trypanosoma brucei. Mol Biochem Parasitol. 2010;172(2):66–79. doi: 10.1016/j.molbiopara.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langford CK, Ewbank SA, Hanson SS, Ullman B, Landfear SM. Molecular characterization of two genes encoding members of the glucose transporter superfamily in the parasitic protozoan Leishmania donovani. Mol Chem Parasitol. 1992;55(1–2):51–64. doi: 10.1016/0166-6851(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 24.Drew ME, Langford CK, Klamo EM, Russell DG, Kavanaugh MP, Landfear SM. Functional expression of a myo-inositol/H+ symporter from Leishmania donovani. Mol Cell Biol. 1995;15:5508–15. doi: 10.1128/mcb.15.10.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira MM, Einicker-Lamas M. Inositol Metabolism in Trypanosoma cruzi: Potential Target for Chemotherapy Against Chagas’ Disease. An Acad Bras Ci. 2000;72:3. [DOI] [PubMed]
- 26.Michell RH. Inositol lipids: from an archaeal origin to phosphatidylinositol 3,5-bisphosphate faults in human disease. FEBS J. 2013;280:6281–94. doi: 10.1111/febs.12452. [DOI] [PubMed] [Google Scholar]
- 27.Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol. 2008;9(2):151–61. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 28.Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–35. doi: 10.1016/S0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- 29.Asaki TS, Asaki JS, Akai TS, Akasuga ST, Uzuki AS. The physiology of phosphoinositides, metabolism and functions of phosphoinositides. Bio Pharm Bull. 2007;30:1599–604. doi: 10.1248/bpb.30.1599. [DOI] [PubMed] [Google Scholar]
- 30.Gardocki ME, Jani N, Lopes JM. Phosphatidylinositol biosynthesis: biochemistry and regulation. Biochim Biophys Acta. 2005;1735(2):89–100. doi: 10.1016/j.bbalip.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Plett JM, Kemppainen M, Kale SD, Kohler A, Legué V, Brun A, et al. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol. 2011;21:1197–203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 32.Engstrom EM, Ehrhardt DW, Mitra RM, Long SR. Pharmacological analysis of nod factor-induced calcium spiking in Medicago truncatula, Evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol. 2002;128(4):1390–401. doi: 10.1104/pp.010691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren LG. Metabolism of Schizotrypanum cruzi, Chagas, I. Effect of culture age and substrate concentration on respiratory rate. J Parasitol. 1960;46:529–39. doi: 10.2307/3274932. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:4540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 36.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 37.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–9. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 38.Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]


