Abstract
Understanding normal and cancer stem cells should provide insights into the origin of prostate cancer and their mechanisms of resistance to current treatment strategies. In this study, we isolated and characterized stem-like cells present in the immortalized human prostate cell line, RWPE-1. We used a reporter system with green fluorescent protein (GFP) driven by the promoter of s-SHIP (for stem-SH2-domain-containing 5′-inositol phosphatase) whose stem cell-specific expression has been previously shown. We observed that s-SHIP-GFP-expressing RWPE-1 cells showed stem cell characteristics such as increased expression of stem cell surface markers (CD44, CD166, TROP2) and pluripotency transcription factors (Oct4, Sox2), and enhanced sphere-forming capacity and resistance to arsenite-induced cell death. Concomitant increased expression of the long noncoding RNA H19 was observed, which prompted us to investigate a putative role in stemness for this oncofetal gene. Targeted suppression of H19 with siRNA decreased Oct4 and Sox2 gene expression and colony-forming potential in RWPE-1 cells. Conversely, overexpression of H19 significantly increased gene expression of these two transcription factors and the sphere-forming capacity of RWPE-1 cells. Analysis of H19 expression in various prostate and mammary human cell lines revealed similarities with Sox2 expression, suggesting that a functional relationship may exist between H19 and Sox2. Collectively, we provide the first evidence that s-SHIP-GFP promoter reporter offers a unique marker for the enrichment of human stem-like cell populations and highlight a role in stemness for the long noncoding RNA H19.
Introduction
Prostate cancer is the second most frequently diagnosed cancer and the sixth leading cause of cancer death in males worldwide and the second in developing countries [1]. Characterizing the prostate cells that are more susceptible to transformation and capable of cancer maintenance represents an essential step to a better understanding and more efficient treatment of the disease. Conventional therapeutics target and kill proliferating cells, sparing the putative cancer stem cell fraction and allowing for the recurrence of the disease. Successful therapy must not only kill the proliferating tumor cells but also eliminate or differentiate the cancer stem cells. Thus, improvements in isolating purified normal or cancer stem cell fractions will then be crucial for advancing therapeutic potential in this field [2].
In vitro cell culture may provide convenient models for studying normal and cancer prostate stem cells since stem cell properties reside naturally among populations of normal or neoplastic cells propagated in culture. Primary cell cultures not only represent an ideal approach to investigate the nature and functions of normal or cancer stem cells in vitro but also present inconvenient features, such as a short life span. To circumvent this difficulty, several studies used nonmalignant-immortalized or malignant cell lines that contain sub-populations of cells with stem cell properties [3]. Enrichment in stem-like cell subpopulations could be achieved by multiple approaches: by modifying cell culture conditions such as cultivating cells in low-calcium serum-free defined medium [4] or under hypoxia [5]; by using a combination of cell surface markers, such as CD133 [6], CD44 [7], CD166 [8,9], or TROP2 [10]; by developing functional assays on cytoprotective activity such as the side population assay based on the efflux of Hoechst 33342 fluorescent dye by the ATP-binding cassette transporter and the ALDEFLUOR assay based on activity of aldehyde dehydrogenases detoxifying enzyme activity [11–13]; or finally by using reporter vectors containing fluorescent protein driven by stemness gene promoters [14,15].
In this study, we applied this latter approach using the green fluorescent protein (GFP) cDNA under the control of the promoter of s-SHIP. Ship1 gene (SH2-containing Inositol 5′-Phosphatase-1) encodes a 145-kDa signaling protein with 5′ phosphatase activity. From this Ship1 gene, a second protein (∼104 kDa) is encoded but lacking the amino-terminal SH2 domain compared with the SHIP1 protein, it is expressed in embryonic stem cells and bone marrow cells enriched for the stem cell population [16,17]. This protein was termed s-SHIP, suggesting its potential for expression in stem cells. The SHIP1 protein is produced from a full-length mRNA, whereas s-SHIP expression is produced from an internal promoter within intron 5/6 of the full-length ship1 gene [18]. Stem cell-specific expression of s-SHIP promoter was determined by generating a transgenic mouse containing the 11.5 kb s-SHIP promoter driving the expression of GFP [18]. In these mice, s-SHIP promoter expression marks activated stem cells in the developing mammary tissue at puberty and during pregnancy [19]. Expression of the transgene was also observed in embryonic prostatic buds, suggesting that s-SHIP promoter expression may also mark prostate stem/progenitor cells [18]. To test this hypothesis, we used as a model the nontumorigenic human prostate cell line RWPE-1 that was derived from normal human prostate epithelium immortalized by human papillomavirus 18 [20]. RWPE-1 cells and its derivatives contain stem, intermediate, and differentiated cell types and offer valuable models for studies of adult prostate stem cells [21,22].
In this report, we show that s-SHIP-GFP promoter reporter tracks subsets of RWPE-1 cells enriched in stem cell characteristics such as enhanced stem cell marker expression. In this subset population, higher expression of the long noncoding RNA (LncRNA) H19 [23] was observed and further investigations strongly suggested that H19 may play a role in prostate stemness through the expression of key pluripotency transcription factors, especially Sox2. Altogether, these data provide new insights into the genetic network controlling stem cell identity, uncovering a role for the long noncoding RNA H19 as a potential stemness regulator.
Materials and Methods
Prostate and mammary cell lines and cell culture
RWPE-1 cells (a gift of Dr. B.S. Kundsen; Fred Hutchinson Cancer Research Center) were maintained in Keratinocyte Serum-Free Medium (KSFM Gibco; Life Technologies) supplemented with 5 ng/mL epidermal growth factor (EGF, PeproTech), bovine pituitary extract (Gibco; Life Technologies), and Zell Shield (Minerva Biolabs; Biovalley). Normal human prostate epithelial cells (PrEC) were obtained from Lonza and cultured in PrEC basal media containing PrEGM SingleQuot Kit supplements and growth factors (Lonza). Human androgen-dependent (LNCaP) and androgen-independent (PC-3 and DU145) prostate cancer epithelial cells were obtained from American Type Culture Collection (ATCC), and were maintained in RPMI 1640 Medium (Gibco; Life Technologies) supplemented with 10% fetal bovine serum (FBS, Gibco; Life Technologies) and Zell Shield. The highly metastatic M12 subline (a gift of Dr. B.S. Kundsen) was cultured in RPMI 1640 medium supplemented with 10 ng/mL EGF, 0.1 μM dexamethasone (Sigma Aldrich), 5 μg/mL insulin, 5 μg/mL transferin, and 5 ng/mL selenium (ITS medium; Sigma) and Zell Shield. The estrogen-sensitive MCF7 and T47D and the estrogen-insensitive MDA-MB-231 human cancerous mammary epithelial cell lines were obtained from the ATCC and maintained routinely in RPMI 1640 medium containing 10% of FBS and Zell Shield. Normal mammary epithelial cells (hTERT, hMEC) were obtained from ATCC and maintained in MEGM (Lonza) supplemented with gentamycin and 1% penicillin/streptomycin. All cells were maintained at 37°C, in a humidified incubator of 5% CO2.
Plasmids and siRNA transfections
The 11.5-kb s-SHIP promoter GFP construct (a gift of the late Dr. LR Rohrschneider, FHCRC) has been previously described [18]. RWPE-1 cells were grown to 70%–80% confluence and transfected with 1 μg of DNA using PEI/ExGen 500 (Euromedex) according to the manufacturer's instructions; 24 h after transfection, the culture medium was replaced with fresh medium. Between 5 and 7 days after transfection, GFP-positive RWPE-1 cells were sorted by using an FACSAria cell sorter [Becton Dickinson (BD)], then grown for 1 week, sorted to a purity of ≥95%, and analyzed. Human long noncoding RNA H19 cDNA was amplified by standard polymerase chain reaction (PCR) with Go Taq Hot Start DNA Polymerase (Promega) and cDNA obtained from MDA-MB-231 cells as the template. The primers were forward, 5′-AGCAGGGTGAGGGAGGG GGT-3′ and reverse, 5′-GTAACAGTGTTTATTGATG-3′. The amplicon was cloned into pcDNA3.1 (−) vector using Not1 and BamH1 sites. RWPE-1 cells were transfected with pcDNA3-H19 or empty pcDNA3 as a control, using PEI/ExGen 500, and transfected cells were allowed to recover for 48 h before selection with G418 (0.5 mg/mL; Sigma) for 4 weeks and beyond this time when all the control (nontransfected) cells had expired. For siRNA, transfections were performed using DharmaFect according to the manufacturer's transfection protocol (Dharmacon; GE Healthcare), and total RNA was extracted 48 h after transfection for analysis. The siRNA sequences are presented in Supplementary Table S1 (Supplementary materials are available online at http://www.liebertpub.com/scd).
Antibodies and flow cytometry
All primary antibodies were from eBiosciences: anti-CD44 (APC) (Cat. No. 17-0441), rat IgG2b κ isotype control (APC) (Cat. No. 17-4031), anti-Human CD166 (ALCAM) PerCP-eFluor®710 (Cat. No. 46-1668), mouse IgG1 κ isotype control PerCP-eFluor®710 (Cat. No. 46-4714), anti-Human TROP2 (EGP-1) (Cat. No. 14-6024), mouse IgG2a κ isotype control purified (Cat. No. 14-472), anti-CD49f APC (Cat. No. 17-0495), anti-Human CD29 (APC) (Cat. No. 17-0299), and rat IgG2a κ isotype control (APC) (Cat. No. 17-4321). APC-goat anti-mouse Ig multiple adsorption (Cat. No. 550826) secondary antibody was from BD Pharmingen. For immunofluorescence staining, 2.5×105 cells were incubated in the dark with primary antibody at a recommended concentration for 30 min on ice; cells were washed with complete medium and resuspended with secondary antibody at a recommended concentration for 30 min, if necessary, or directly analyzed on a flow cytometer (Canto II; BD).
RNA isolation and quantitative RT-PCR
5×105 Cells were plated into a 100-mm-diameter dish, a cell density that allow the cells to continue exponential growth for an additional 24 h. Total RNA was then isolated using the RNeasy Plus Extraction kit (Qiagen). Reverse transcription (RT) was performed using QuantiTect Reverse Transcription kit (Qiagen) on 1 μg of RNA, according to the manufacturer's protocol. Quantitative RT-PCR (qRT-PCR) was achieved on Stratagene Mx3005P (Agilent Technologies) using KAPA SYBR® Fast Universal qPCR kit (Kapa Biosystems; CliniSciences): 2 μL of DNase-treated cDNA (from a 50 μL RT reaction) was added to 10 μL Master Mix, 2 μL of oligo mix (1 μM). Experiments were performed in triplicate, and the comparative threshold cycle method was used for the calculation of amplification fold. The expression level of each gene was normalized by the expression level of the “housekeeping” RPLP0 gene (ΔCt method). Primer sequences are presented in Supplementary Table S1.
Growth studies
Cell proliferation was evaluated by plating 2×104 cells per plate in a six-well plate in 2 mL of culture medium. After 24, 48, 72, and 96 h, cells were isolated after trypsinization using trypsin-versene (EDTA) solution and cell counting was done under the microscope using a Malassez counting chamber. For colony-forming unit (CFU) assay, single cells (50, 100, and 200 cells per well) were plated in six-well plates and cultured for 2 weeks in K-SFM medium. Cells were fixed with 4% paraformaldehyde made in phosphate-buffered saline (PBS) for 10 min at room temperature, rinsed with PBS, and stained with Giemsa solution (Sigma). The plates were gently washed with PBS and dried. Macroscopic colonies visible by naked eyes were enumerated. Colony formation efficiency was evaluated as follows: CFU (%)=colonies/input cells×100.
Sphere formation assay
Cells (100, 250, 500 cells) were plated in 0.5 mL of PrEC basal media containing PrEGM SingleQuot Kit supplements and growth factors, in ultra-low attachment 24-well plates (Costar) and grown for 2 weeks. To not disturb sphere formation and to avoid sphere aggregation, fresh medium was gently added to each well after 5 and 10 days and no medium was aspirated. Total number of spheres was counted under a microscope and photographed. Sphere formation efficiency was evaluated as follows: sphere-forming unit (SFU) (%)=spheres/input cells×100.
Chemoresistance
RWPE-1 and RW-GFP cells (105 cells per well in six-well plates) were plated and allowed to attach for 24 h; then, they were treated with doxorubicin (D1515; Sigma) at 0.5 μg/mL for 48 h and photographed. P53 activation was determined by western blot: Cells were lysed in RIPA buffer containing protease and phosphatase inhibitors (protease inhibitor P8340; phosphatase inhibitor cocktail 2 P5726; Sigma), and proteins were quantified with BCA protein assay (Pierce). Proteins were reduced in NuPAGE LDS Sample buffer with NuPAGE Reducing Agent (Invitrogen; Life Technologies) at 70°C for 10 min. Proteins were separated on SDS-PAGE 4%–12% (Invitrogen; Life Technologies) and transferred onto PolyVinylidene Fluoride (PVDF) membrane (Millipore). After saturation in PBS 0.2% of casein, membranes were incubated with primary antibodies overnight at 4°C (p53 Do-1 sc-126, Santa Cruz; β-actin sc-47778, Santa Cruz). Membranes were washed with PBS 0.5% Tween for 30 min and incubated with secondary antibodies conjugated with Horse Radish Peroxidase (HRP) for 30 min at room temperature. Membranes were analyzed with SuperSignal west Dura Chemiluminescence Substrate (Pierce). For arsenite-induced cytolethality, cells (105 cells per well in six-well plates) were plated and allowed to attach for 24 h; fresh medium was added containing sodium arsenite (NaAsO2, No. 35000; Sigma) at different concentrations (between 5 and 40 μM); after 48 h, viable cells were photographed and counted with a Malassez hemocytometer (trypan blue method).
Statistical analysis
Data are expressed as mean values±standard error of the mean of at least three independent experiments. The statistical analysis was done by using Student's t-test, and P value <0.05 was considered significant.
Results
Isolation of RWPE-1 cells with s-SHIP promoter activity
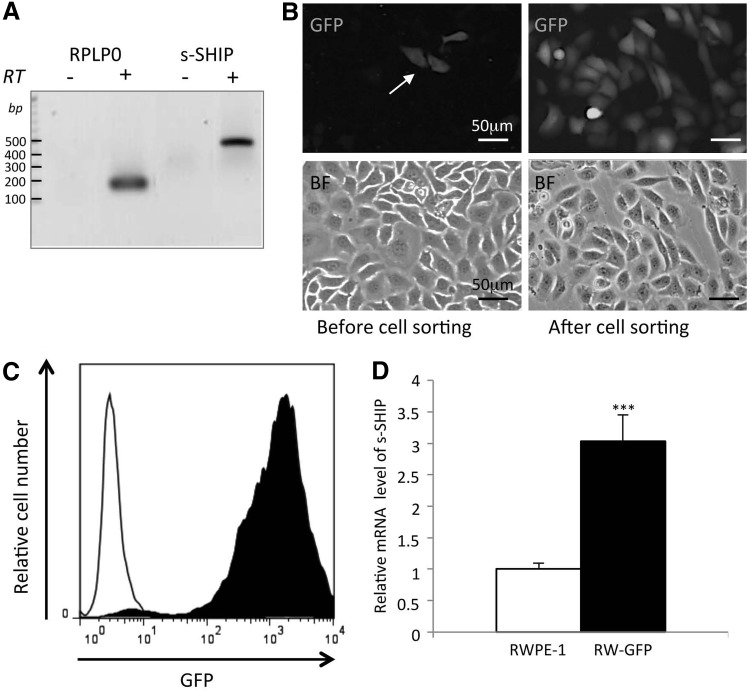
Previous studies showed that the intron 5/6 promoter region of the Ship1 gene regulates expression of a shorter isoform of the protein, called s-SHIP, in various stem/progenitor cell populations [18,19]. We tested whether high s-SHIP promoter expression could enrich stem-like cells within a human cell line. In this attempt, we used as a model the nontumorigenic prostate cell line RWPE-1 in which s-SHIP transcript expression could be detected (Fig. 1A). Since s-SHIP promoter region shows high conservation between human and mouse (Supplementary Fig. S1), we hypothesized that a DNA construct incorporating the 11.5 kb s-SHIP promoter from the mouse upstream of the GFP substituted at the normal translation start site for s-SHIP [18] could be used to detect the live human cells with higher s-SHIP promoter activity. Among RWPE-1 cells that were transfected with 11.5 kb-GFP, a small number of GFP-positive (GFP+) cells were observed (Fig. 1B) and then isolated by FACS. After two rounds of cell sorting, a population of GFP+ cells were obtained (Fig. 1C) and then tested for native s-SHIP transcript expression. We observed a four-fold increased expression as compared with the parental population (Fig. 1D), thus demonstrating that 11.5 kb murine s-SHIP promoter region is functional in human prostate cells and allows enrichment in cells expressing endogenous s-SHIP mRNA. This population of RWPE-1 cells expressing a higher level of s-SHIP transcript was named RW-GFP cells and was then analyzed for its stemness characteristics.
FIG. 1.
Isolation of RW-GFP cells, a subset of RWPE-1 cells with higher levels of s-SHIP promoter expression. (A) RT-PCR analysis of s-SHIP transcript in parental RWPE-1 cell line. (B) RWPE-1 cells were transfected with s-SHIP-GFP promoter reporter (11.5 kb-GFP) and few GFP+ cells were obtained (arrow, left panel), then isolated by FACS, and amplified in culture to create RW-GFP cells (right panels). (Scale bar=50 μm) (C) Flow cytometry analysis of green fluorescent protein (GFP) expression in RW-GFP cells (filled graph) after two rounds of cell sorting and cell amplification as compared with parental cells (empty graph). (D) Quantitative RT-PCR analysis of endogenous s-SHIP transcript expression in RW-GFP cells as compared with parental RWPE-1 cells. Data represent mean values±standard error of the mean of three independent experiments, P values was determined by Student's test ***P<0.001.
s-SHIP-GFP promoter reporter tracks subsets of RWPE-1 cells enriched in clonogenic properties and chemoresistance
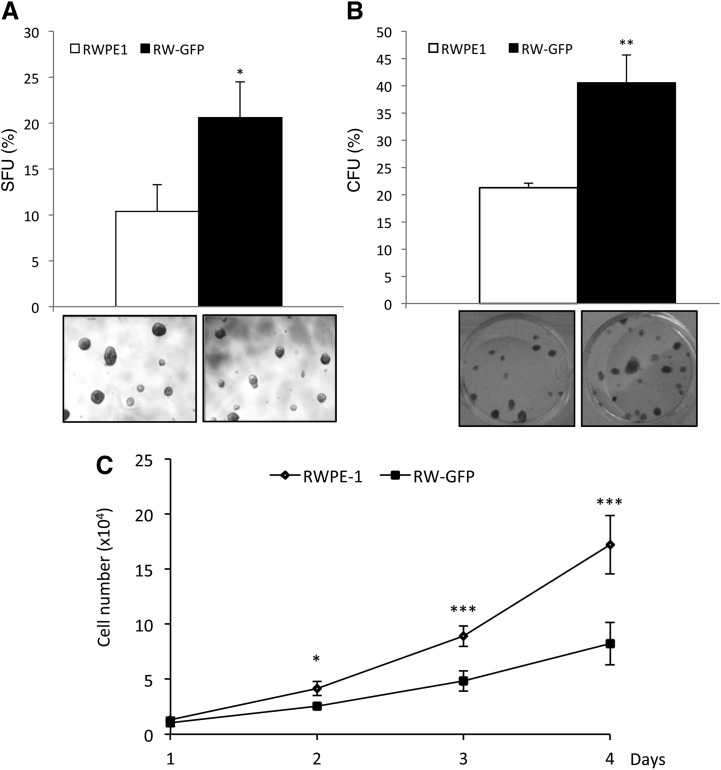
When progenitor/stem cells are cultivated in defined culture conditions, they have the ability to grow in an anchorage-independent manner and to form suspension spheres [24,25]. We showed that cells with higher s-SHIP expression (RW-GFP cells) were able to form more spheres compared with the parental RWPE-1 cells when cultivated in defined PrEGM serum-free medium and onto ultra-low attachment plates (Fig. 2A). RW-GFP-derived spheres appeared to be smaller with a homogeneous distribution of size as compared with RWPE-1-derived spheres (Supplementary Fig. S2). Similarly, the colony-forming efficiency of RW-GFP cells was significantly higher, compared with parental RWPE-1 cells (Fig. 2B). In contrast, RW-GFP cells grew slower than parental RWPE-1 cells (Fig. 2C).
FIG. 2.
Clonogenicity and growth curve of RW-GFP cells. (A) When cultivated at a low density in liquid PrEGM culture medium in ultra-low attachment 24-well plates in nonadherent conditions, RW-GFP cells formed more spheres than parental RWPE-1 cells; lower panels represent spheres formed after 15 days of culture. (B) When 50–200 cells per well were plated in six-well plates in K-SFM medium, RW-GFP cells formed more colonies than parental RWPE-1 parental cells; lower panels are pictures of 2-weeks-old colonies after fixation and Giemsa staining. (C) RW-GFP cells grew slower than parental RWPE-1 cells; cell growth curves were obtained after plating 2×104 cells per well in six-well plates, and viable cell numbers were determined daily after trypsinization. (A, B, C) Data represent mean values±standard error of the mean of three independent experiments, P values was determined by Student's test ***P<0.001, **P<0.01, and *P<0.05.
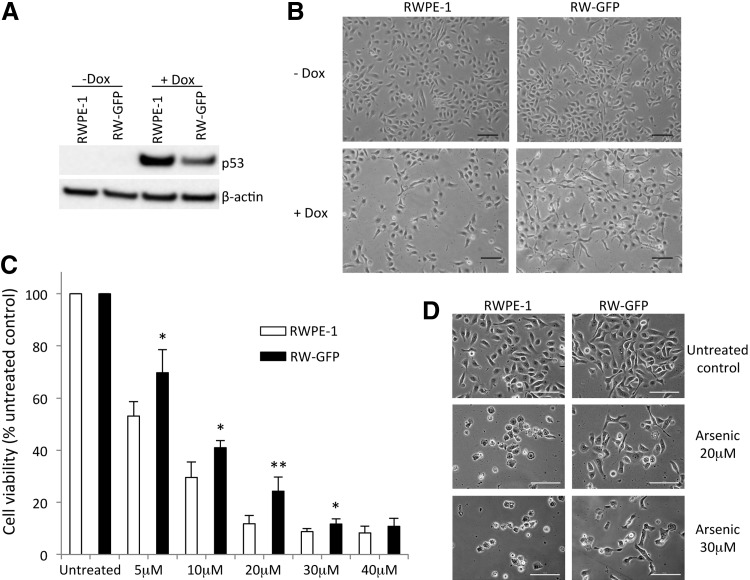
Stemness is associated to chemoresistance [26]; both RWPE-1 and RW-GFP cells responded to chemotherapeutic drug doxorubicin by inducing p53 protein expression and cytolethality (Fig. 3A, B); interestingly, p53 induction was lower in RW-GFP cells together with better cell survival as compared with parental RWPE-1 cells (Fig. 3 A, B). This prompted us to investigate innate resistance to arsenite-induced cytolethality since this was a trait associated with stemness in RWPE-1 cells as previously described [22]. After short-term (ie, 48 h) arsenite exposure (Fig. 3C, D), a clear difference in arsenite-induced cytotoxicity occurred between RW-GFP cells and the parental RWPE-1 cells at all concentrations tested, especially at 20 μM with a two-fold increase in cell survival for the RW-GFP cells as compared with parental RWPE-1 cells (24.3%±5.4% vs. 11.8%±3.2%).
FIG. 3.
Chemoresistance of RW-GFP cells. (A, B) RWPE-1 and RW-GFP cells were untreated (−Dox) or treated (+Dox) with doxorubicin (Dox, 0.5 μg/mL) for 48 h, and (A) immunoblot analysis was then performed on whole cell lysate for p53 expression, using anti-β-actin antibodies as a loading control; (B) cells were photographed (scale bar=100 μm), (C, D) RWPE-1 and RW-GFP cells were untreated or treated with arsenite (5, 10, 20, 30, 40 μM) for 48 h and (C) viable cells were counted using the trypan blue dye exclusion method, and (D) cells were photographed (scale bar=100 μm); (C) Data represent mean values±standard error of the mean of five independent experiments; P values were determined by Student's test **P<0.01, *P<0.05.
Taken together, these results suggested that higher s-SHIP expression was associated to lower proliferation and higher clonogenicity, a property of stem-like cells. We then investigated whether this phenotype was also associated to the expression of stemness markers.
RW-GFP cells expressed higher levels of stem cell markers and pluripotency transcription factors
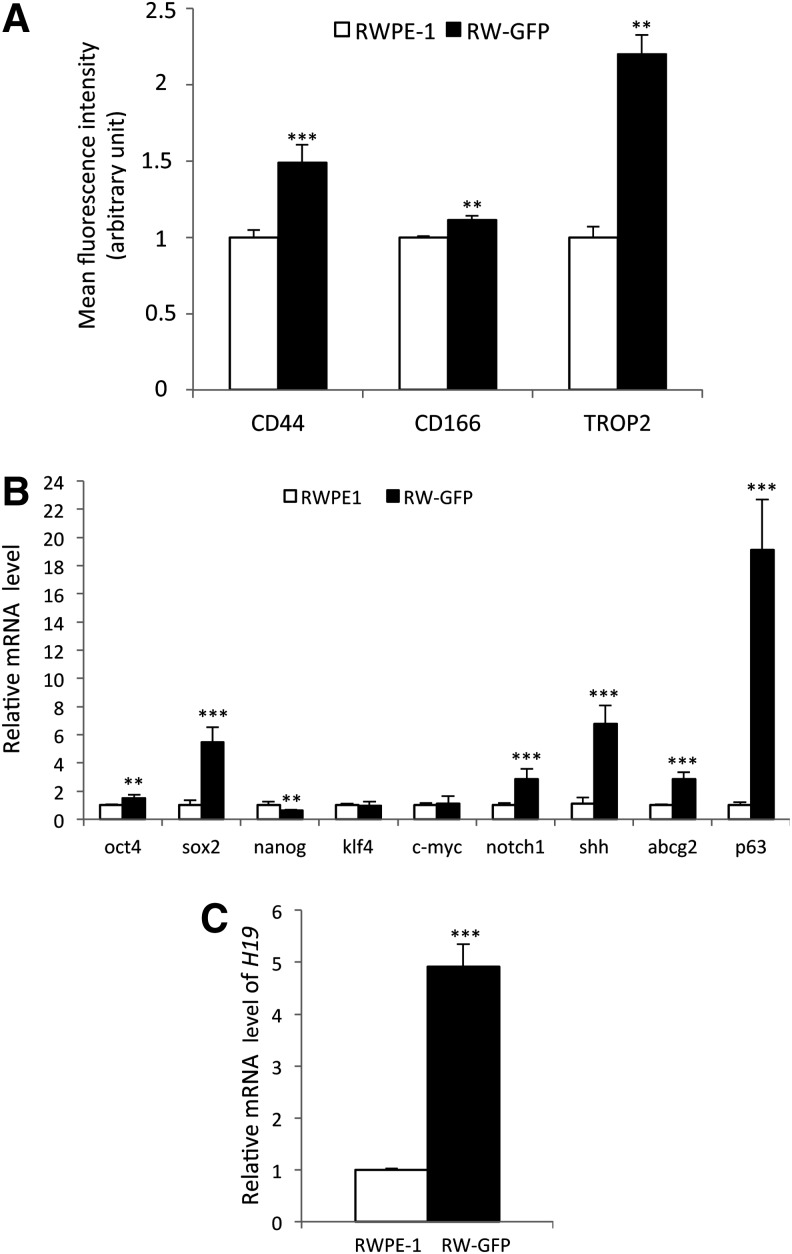
RW-GFP cell stemness characteristics were further analyzed for cell surface marker expression and mRNA profile of stemness-related genes. Although CD44 expression level was already high in parental RWPE-1 cells, RW-GFP subpopulation shifted significantly to a higher CD44 expression level (Fig. 4A and Supplementary Fig. S3A). Next, we examined whether cell surface expression of CD166 and TROP2, two other prostate stem cell markers [9,10], was modified in RW-GFP cells. As expected, CD166 and TROP2 expression levels were also significantly increased as compared with parental RWPE-1 cells (Fig. 4A and Supplementary Fig. S3A). Similarly, integrin α6 (CD49f) and β1 (CD29) expression showed a homogeneous expression of both markers with a slightly increased expression as compared with parental RWPE-1 cells (Supplementary Fig. S3B).
FIG. 4.
RW-GFP cells exhibit a higher expression level of stem cell markers and a significant increase of long noncoding RNA H19 expression. (A) Fold-enrichment over the parental RWPE-1 cells for CD44, CD166, and TROP2 cell surface marker expression analyzed by flow cytometry; data are expressed as mean fluorescence intensity (MFI) of RW-GFP cells (black bars) as compared with parental RWPE-1 cells (open bars). (B) Expression of stemness genes and (C) long noncoding RNA H19 was analyzed by qRT-PCR using total RNA from parental RWPE-1 cells (open bars) or RW-GFP cells (closed bars). Levels were normalized to those of RPLP0 internal control. Graphs show fold enrichment in RW-GFP cells over the parental RWPE-1 cells for each gene. (A, B, C) Data represent mean values±standard error of the mean of three independent experiments; P values were determined by Student's test ***P<0.001, **P<0.01, and *P<0.05.
Transcription regulators such as octamer-binding transcription factor 4 (oct4), sex determining region Y-box 2 (sox2), nanog, kruppel-like factor 4 (klf4), and c-myc are associated with induction or maintenance of stemness in different tissues [27]. We observed a significantly higher expression of oct4 and sox2, but not nanog, klf4, or c-myc, in cells with high s-SHIP promoter activity (RW-GFP) as compared with parental RWPE-1 cells (Fig. 4B). Examination of mRNA expression profile was extended to different stemness-related genes, such as the ATP-binding cassette transporter G2 (abcg2), or genes implicated in signaling pathways regulating stem cells [sonic hedgehog (shh) and notch1], or in prostate development (p63 transcription factor) [28]. We observed significant increased expression in RW-GFP cells for all of them, with p63 expression being the most important (∼18-fold). The long noncoding (lnc) RNA H19 is involved in prostate carcinogenesis [29] and plays a role in early development [23]. These results led us to examine the expression of the noncoding RNA H19 in RW-GFP cells. As shown in Figure 4C, a marked increased expression (approximately five-fold) was observed in RW-GFP cells as compared with parental cells; this prompted us to further investigate the relationship between H19 expression and stemness characteristics in RWPE-1 cells.
Expression of the long noncoding RNA H19 favors stemness phenotype of RWPE-1 cells
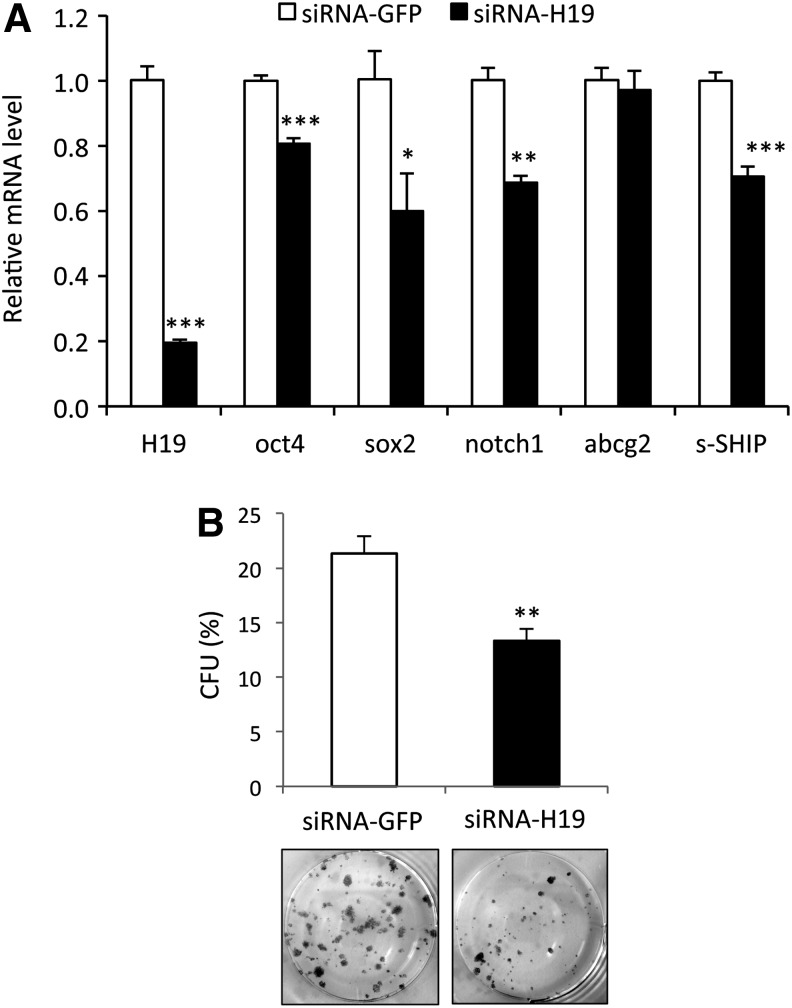
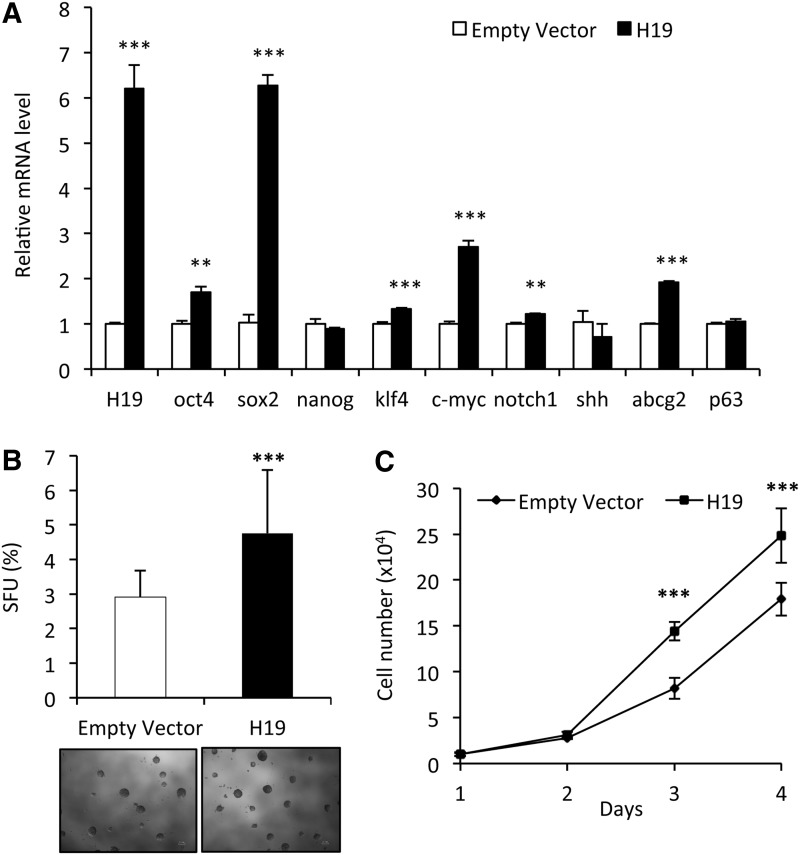
To evaluate the role of H19 in RWPE-1 stemness, we analyzed whether the expression level of H19 correlated with stemness marker expression and sphere formation. First, we delivered siRNA to knock down H19 expression in RWPE-1 cells. As shown in Figure 5A, 2 days after transfection of RWPE-1 cells with H19 siRNA, H19 RNA level was repressed till 80% of the negative control. Expression of oct4, sox2, notch1, and s-SHIP was significantly reduced, with sox2 showing the most dramatic reduction (Fig. 5A). Furthermore, colony-forming efficiency of these transfected cells was examined, and we observed a significant decrease of CFU potential for cells transfected with H19 siRNA, compared with GFP siRNA-transfected control cells (Fig. 5B). Next, we determined the impact of H19 overexpression on stem cell characteristics in RWPE-1 cells. Cells were transfected and selected by G418, and H19 RNA level was analyzed by qRT-PCR. H19 RNA level was significantly increased approximately six-fold in RW-pcDNA-H19 cells relative to that in RW-pcDNA3 control cells (Fig. 6A). Then, expression of several stemness genes was examined. A significantly higher expression of oct4, sox2, klf4, c-myc, and abcg2 but not nanog or p63 was observed in RW-pcDNA-H19 cells as compared with RW-pcDNA3 control cells (Fig. 6A). Thus, RWPE-1 cells showed a global increase in stemness gene expression when H19 gene is overexpressed, but with some difference with RWPE-1 cells expressing a higher level of s-SHIP transcript (Fig. 4B). Since H19 gene overexpression increased the expression of stemness factors, we next investigated whether lnc RNA H19 influenced the clonogenicity and the proliferation of RWPE-1 cells. When RW-pcDNA-H19 and RW-pcDNA3 control cells were analyzed for their sphere formation capabilities in ultra-low attachment plates, we observed that control RW-pcDNA3 exhibited a lower sphere-forming potential than the parental RWPE-1 cells, suggesting that long-term culture in the presence of G418 affected their clonogenicity (Fig. 6B). Nevertheless, RW-pcDNA-H19 cells that have been cultivated in identical culture conditions exhibited a significant increase of SFU potential (Fig. 6B), suggesting that H19 overexpression promotes a stem-like cell phenotype similar to the RW-GFP cells. However, regarding the difference of RW-GFP cells, when proliferation in two-dimensional culture conditions was analyzed, RW-pcDNA-H19 grew faster than RW-pcDNA3 control cells (Fig. 6C).
FIG. 5.
Inhibition of long noncoding RNA H19 expression decreases stemness gene expression and clonogenicity. (A) Knockdown of H19 by siRNA reduced expression of several stemness genes; expression was analyzed by qRT-PCR using total RNA from control (GFP) siRNA-transfected RWPE-1 cells (open bars) or from H19 siRNA-transfected RWPE-1 cells (closed bars) and levels were normalized to those of RPLP0 internal control. Graphs show fold decrease over control siRNA-transfected RWPE-1 cells for each gene. (B) When 50–200 cells per well were plated in six-well plates in K-SFM medium, H19 siRNA-transfected RWPE-1 cells formed fewer colonies than control siRNA-transfected RWPE-1 cells; lower panels are pictures of 2-week-old colonies after fixation and Giemsa staining. (A, B) Data represent mean values±standard error of the mean of three independent experiments; P values were determined by Student's test ***P<0.001, **P<0.01, and *P<0.05.
FIG. 6.
Overexpression of long noncoding RNA H19 favors stemness gene expression and clonogenicity. RWPE-1 cells were transfected with either pcDNA-H19 or pcDNA-control and selected with G418 to obtain stable neor cell lines. (A) Overexpression of H19 in RWPE-1 cells increased expression of several stemness genes; expression was analyzed by qRT-PCR using total RNA from empty vector-expressing G418-resistant RWPE-1 cells (open bars) or from H19-expressing G418-resistant RWPE-1 cells (closed bars), and levels were normalized to those of RPLP0 internal control. Graphs show fold change over empty vector-expressing G418-resistant RWPE-1 cells for each gene. (B) Cells were cultivated at a low density in PrEGM culture medium in ultra-low attachment 24-well plates in liquid cultures, and RW-pcDNA-H19 formed more spheres than RW-pcDNA3 control cells; lower panels represent spheres formed after 15 days of culture. (C) RW-pcDNA-H19 cells grew faster than RW-pcDNA3 control cells; cell growth curves were obtained after plating 2×104 cells per well in six-well plates, and viable cell numbers were determined daily after trypsinization. (A, B, C) Data represent mean values±standard error of the mean of three independent experiments; P values were determined by Student's test ***P<0.001, **P<0.01.
Correlation between H19 and sox2 expression in various prostate and breast cell lines
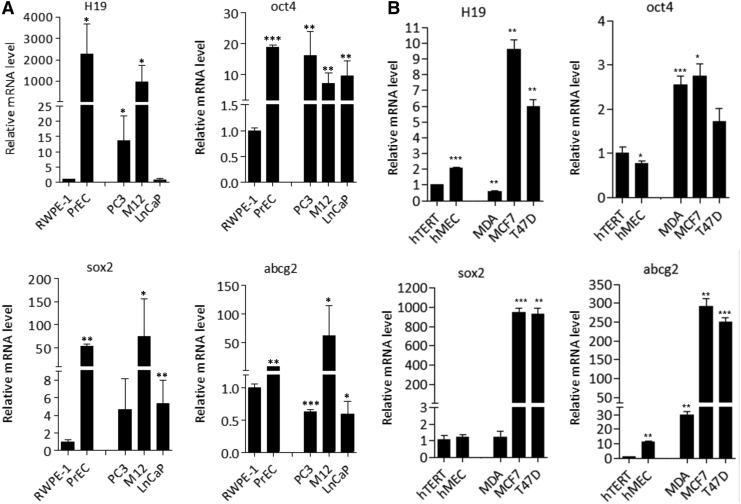
H19 loss- or gain-of-function experiments in RWPE-1 cells (Figs. 5 and 6) suggested a positive correlation between H19 expression and sox2, oct4, and abcg2 stemness gene expression. To assess this correlation, we determined the level of expression of these four genes in another population of normal human PrEC and in three different prostate cancer cell lines: two androgen-independent (PC3 and M12) and one androgen-dependent (LNCaP) cell lines. In the PrEC normal cell line, high H19 RNA expression was detected as compared with RWPE-1 cells (Fig. 7A). Similarly, PrEC cells exhibited high increased expression for sox2 (54-fold), oct4 (19-fold), and abcg2 (8.4-fold) as compared with RWPE-1 cells (Fig. 7A). In cancerous cells, the M12 cell line expressed the highest H19, sox2, and abcg2 expression levels (Fig. 7A). To extend these observations and further investigate a putative co-expression between H19 and stemness genes in epithelial cells, we used a set of mammary cell populations with two nonmalignant cell lines (hTERT and hMEC) and three mammary cancer cell lines (MDA-MB-231, MCF7, T47D). Again, a strong positive correlation was observed between H19, sox2, and abcg2 expression and to a lesser extent with oct4 expression (Fig. 7B).
FIG. 7.
Long noncoding RNA H19 expression is associated with sox2 expression in various prostate and mammary cell lines. H19, oct4, sox2, and abcg2 expression was analyzed by qRT-PCR using total RNA from (A) prostate nonmalignant RWPE-1 and prostate epithelial cells (PrEC) and from PC3, M12, LNCaP prostate cancer cell lines, and (B) mammary hTERT and hMEC nonmalignant immortalized cell lines and breast tumor cell lines (MDA-MB-231, MCF7, T47D). Levels were normalized to those of RPLP0 internal control. Graphs show fold enrichment over (A) RWPE-1 cells and (B) hTERT cells for each gene. (A, B) Data represent mean values±standard error of the mean of three independent experiments, P values was determined by Student's test ***P<0.001, **P<0.01, and *P<0.05.
Discussion
Understanding of the prostate stem cell biology is essential since there is a growing body of evidence suggesting that benign prostate hyperplasia and prostate cancer may arise from the stem or stem-like cell compartments [30]. In this study, we took advantage of the s-SHIP promoter-driven reporter to detect and isolate s-SHIP-expressing human prostate cells. First, we demonstrated that s-SHIP promoter activity was well correlated with expression of different stemness markers and sphere-formation capabilities in human prostate RWPE-1 cell line. These results are in good agreement with studies performed in mice models and identify s-SHIP as a stem cell-specific isoform of SHIP1, which is expressed in both pluripotent ES cells and adult tissue-specific multipotent cells such as hematopoietic stem cells [16,17]. Rohrschneider et al. confirmed that s-SHIP promoter, located within the intron 5/6 of the SHIP1 gene, promotes stem cell-specific transcription of the GFP reporter gene in a transgenic mouse model (Tg11.5kb-GFP) [18]. In these mice, GFP expression from the s-SHIP promoter marks activated mammary stem cells that specifically localize to the cap cell region of terminal end buds in developing mammary glands and alveolar units [19,31,32]. Regarding the prostate tissue, s-SHIP promoter-GFP reporter is expressed in a specific subset of cells within the embryonic prostate [18] and we have recently extended this result to prostate postnatal development and demonstrated that in developing prostatic buds of newborn mice, GFP-expressing cells correspond to a population of basal cells with stem cell characteristics (manuscript in preparation).
All of these experiments were previously performed in mouse tissues. In this work, we provide the first evidence that s-SHIP promoter expression could also be used for the enrichment of human stem-like cells. SIP-110, the human homolog of s-SHIP, is expressed in various human cell lines, including hematopoietic [33], mammary, and melanoma cell lines (our unpublished data), suggesting that the s-SHIP-GFP promoter reporter should be functional in these cells. More experiments are now needed to determine whether our approach could be useful in other cell types, including cancer cell lines with the ultimate goal to isolate and characterize human cancer stem cells. The exact role of s-SHIP as a signaling molecule remains to be determined in these stem cell populations. Even though this protein lacks the SH2 domain as compared with SHIP1, s-SHIP contains cell signaling domains, including the 5′-inositol phosphatase domain [16], which might play a key role in controlling phosphatidylinositol-3-kinase (PI3K)/AKT activation. From this point of view, an interesting study has recently demonstrated the importance of the enzymatic activity of SHIP2, another 5′-inositol phosphatase, for breast cancer stem cells [34]. Altogether, these studies support the idea that s-SHIP promoter expression offers a valuable and unique marker of mammary and prostate stem cell populations that could lead to the identification of molecules involved in stem cell biology.
Thus, characterization of our stem-like enriched RWPE-1 cell population uncovered a potential role for lncRNA H19 in stemness. The H19 gene, located in human in 11p15.5 locus, is submitted to genomic imprinting, being expressed only from the maternal allele [35]. No protein associated to this transcript has been discovered and it has been identified initially as a riboregulator [36], and, more recently, as an miRNA precursor [37]. A status of oncofetal gene has been ascribed to H19: During embryogenesis, H19 is highly expressed both in extraembryonic tissues and in the embryo; after birth, its expression is repressed to a basal activity that subsists in several tissues [38–41]. In cancer, H19 gene is reexpressed and acts as an oncogene [42–44].
In recent years, lncRNAs have been emerging as important components of gene regulation that may play key roles in regulating quiescence, survival, and self-renewal of pluripotent stem cells [45]. Till date, few mechanisms of action of lncRNAs in stem cell biology have been investigated. Among them, lncRNA-RoR (Regulator Of Reprogramming or lincRNA-ST8SIA3) is a suppressor of p53 response [46] whereas p53 suppression promotes stem cells expansion by activation of self-renewing divisions, symmetric division, and reprogramming of somatic/progenitor cells in stem cells [47,48]. LncRNA-RoR can also act as an endogenous miRNA sponge (impairing miR-145 repression) to positively regulate Oct4, Nanog, and Sox2 and promote embryonic stem cell self-renewal [49]. Similarly, H19 interacts with p53, impairs its signaling [50], and acts as an miRNA sponge to antagonize let-7 miRNAs [51]. In breast cancer stem cells, Let-7 miRNAs down-regulate self-renewal and tumorigenicity [52], and in prostate cancer cells, loss of Let-7 up-regulates EZH2 (a component of PRC2, polycomb repressive complex) and induces a stem cell signature [53]. Interestingly, H19 by squelching Let-7 miRNAs could enhance EZH2 expression and activity to promote epithelial-mesenchymal transition in bladder cancer [54]. In RWPE-1 cell line, H19 could confer stemness phenotype by these multiple pathways. In addition, knockdown of H19 in parthenogenetic embryonic stem cells (pES) promotes differentiation of pES to epidermis, showing that H19 RNA contributes to stem cell integrity [55].
Finally, H19 loss- or gain-of-function experiments showed coregulation of H19, sox2, and abcg2 and to a lesser extent oct4. Even if H19 could act by modifying chromatin conformation at these loci, our results suggest a direct role of H19 in the regulation of these genes (Figs. 5A and 6A). Conversely, oct4 and sox2 positively control H19 expression by impairing the methylation of imprinting control region and promoter [56], and a defect of oct4 and sox2 binding sites is associated with loss of expression of H19 in Beckwith–Wiedemann syndrome [57], suggesting an amplification loop of H19/Sox2 regulation.
In conclusion, our study is the first to demonstrate that s-SHIP-GFP promoter reporter offers a unique marker for the enrichment of human stem-like cell populations and this strategy enabled us to unveil lncRNA H19 as a potential factor for inducing and maintaining the biological nature of stem cells. Further analysis should be performed to determine the exact role and the mechanisms of action of H19 RNA.
Supplementary Material
Acknowledgments
This study was supported by grants from the Centre National de la Recherche Scientifique (CNRS), the Région Nord-Pas de Calais (plan de lutte contre le cancer), and the SIRIC ONCOLille (INCa-DGOS-Inserm 6041). R.P.B. was supported by the Philippe Foundation. E.A. was supported by INCA (PLBio 2010-180). H.B.L.R. and G.B. were supported by postdoctoral fellowships from the Région Nord-Pas de Calais and the Institut Pasteur de Lille. The authors thank the flow cytometry platform of the BioImaging Center of Lille (BICeL) and Lucie Deschamps for their support, Cateline Guerardel for technical assistance, and Dr. Jérôme Vicogne for his revision of this article. R.P.B. dedicates this article to the memory of his mentor and friend, the late Dr. Larry R. Rohrschneider, who was the initiator of the s-SHIP project.
Author Disclosure Statement
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E. and Forman D. (2011). Global cancer statistics. CA Cancer J Clin 61:69–90 [DOI] [PubMed] [Google Scholar]
- 2.Goldstein AS, Stoyanova T. and Witte ON. (2010). Primitive origins of prostate cancer: in vivo evidence for prostate-regenerating cells and prostate cancer-initiating cells. Mol Oncol 4:385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miki J. and Rhim JS. (2008). Prostate cell cultures as in vitro models for the study of normal stem cells and cancer stem cells. Prostate Cancer Prostatic Dis 11:32–39 [DOI] [PubMed] [Google Scholar]
- 4.Litvinov IV, Vander Griend AJ, Xu Y, Antony L, Dalrymple SL. and Isaacs JT. (2006). Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res 66:8598–8607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Liang D, Liu J, Axcrona K, Kvalheim G, Stokke T, Nesland JM. and Suo Z. (2011). Prostate cancer cell lines under hypoxia exhibit greater stem-like properties. PLoS One 6:e29170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ. and Collins AT. (2004). CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci 117(Pt16):3539–3545 [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Liu H, Zhang BH, Cadaneanu RM, Mayle AM. and Garraway IP. (2012). Epcam, CD44, and CD49f distinguish sphere-forming human prostate basal cells from a subpopulation with predominant tubule initiation capability. PLoS One 7:e34219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajasekhar VK, Studer L, Gerald W, Socci ND. and Scher HI. (2011). Tumor-initiating stem-like cells in human prostate cancer exhibit increased NF-κB signalling. Nat Commun 2:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, Lawson D, Chen D, Li Y, Guo C, et al. (2012). Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One 7:e42564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP. and Witte ON. (2008). Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci U S A 105:20882–20887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, et al. (2010). High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res 70:5163–5173 [DOI] [PubMed] [Google Scholar]
- 12.Foster BA, Gangavarapu KJ, Mathew G, Azabdaftari G, Morrison CD, Miller A. and Huss WJ. (2013). Human prostate side population cells demonstrate stem cell properties in recombination with urogenital sinus mesenchyme. PLoS One 8:e55062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangavarapu KJ, Azabdaftari G, Morrison CD, Miller A, Foster BA. and Huss WJ. (2013). Aldhehyde dehydrogenase and ATP binding cassette transporter G2 (ABCG2) functional assays isolate different populations of prostate stem cells where ABCG2 function selects for cells with increased stem cell activity. Stem Cell Res Ther 4:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, Repass J, Zaehres H, Shen JJ. and Tang DG. (2011). NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene 30:3833–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang S, Furuhashi M, Nakane R, Nakazawa S, Goudarzi H, Hamada J. and Iizasa H. (2013). Isolation and characterization of human breast cancer cells with SOX2 promoter activity. Biochem Biophys Res Commun 437:205–211 [DOI] [PubMed] [Google Scholar]
- 16.Tu Z, Ninos JM, Ma Z, Wang JW, Lemos MP, Desponts C, Ghansah T, Howson JM. and Kerr WG. (2001). Embryonic and hematopoietic stem cells express a novel SH2-containing inositol 5′-phosphatase isoform that partners with the Grb2 adapter protein. Blood 98:2028–2038 [DOI] [PubMed] [Google Scholar]
- 17.Desponts C, Ninos JM. and Kerr WG. (2006). s-SHIP associates with receptor complexes essential for pluripotent stem cell growth and survival. Stem Cells Dev 15:641–646 [DOI] [PubMed] [Google Scholar]
- 18.Rohrschneider LR, Custodio JM, Anderson TA, Miller CP. and Gu H. (2005). The intron-5/6 promoter region of the ship1 gene regulates expression in stem/progenitor cells of the mouse embryo. Dev Biol 283:503–521 [DOI] [PubMed] [Google Scholar]
- 19.Bai L. and Rohrschneider LR. (2010). s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev 24:1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bello DB, Webber MM, Kleinman HK, Wartinger DD. and Rhim JS. (1997). Androgen responsive adult human prostatic epithelial cell lines immortalysed by human papillomavirus 18. Carcinogenesis 18:1215–1223 [DOI] [PubMed] [Google Scholar]
- 21.Tokar EJ, Ancrile BB, Cunha GR. and Webber MM. (2005). Stem/progenitor and intermediate cells and the origin of human prostate cancer. Differentiation 73:463–473 [DOI] [PubMed] [Google Scholar]
- 22.Tokar EJ, Qu W, Liu J, Liu W, Webber MM, Phang JM. and Waalkes MP. (2010). Arsenic-specific stem cell selection during malignant transformation. J Natl Cancer Inst 102:638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vennin C, Dahmani F, Spruyt N. and Adriaenssens E. (2013). Role of non-coding RNA in cells: example of the H19/IGF2 locus. Adv Biosci Biotechnol 4:34–44 [Google Scholar]
- 24.Shi X, Gipp J. and Bushman W. (2007). Anchorage-independent culture maintains prostate stem cells. Dev Biol 312:396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin L, Lukacs RU, Lawson DA, Cheng D. and Witte ON. (2007). Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells 25:2760–2769 [DOI] [PubMed] [Google Scholar]
- 26.Ni J, Cozzi P, Hao J, Duan W, Graham P, Kearsley J. and Li Y. (2014). Cancer stem cells in prostate cancer chemoresistance. Curr Cancer Drug Targets 14:225–240 [DOI] [PubMed] [Google Scholar]
- 27.Calloni R, Cordero EA, Henriques JA. and Bonatto D. (2013). Reviewing and updating the major molecular markers for stem cells. Stem Cells Dev 22:1455–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pignon JC, Grisanzio C, Geng Y, Song J, Shivdasani RA. and Signoretti S. (2013). P63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc Natl Acad Sci U S A 110:8105–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berteaux N, Lottin S, Adriaenssens E, Van Coppenolle F, Leroy X, Coll J, Dugimont T. and Curgy JJ. (2004). Hormonal regulation of H19 gene expression in prostate epithelial cells. J Endocrinol 183:69–78 [DOI] [PubMed] [Google Scholar]
- 30.Prajapati A, Gupta S, Mistry B. and Gupta S. (2013). Prostate stem cells in the development of benign prostate hyperplasia and prostate cancer: emerging role and concepts. Biomed Res Int 2013:107954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huo Y. and Macara IG. (2014). The Par3-like polarity protein Par3L is essential for mammary stem cell maintenance. Nat Cell Biol 16:526–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kogata N, Oliemuller E, Wansbury O. and Howard BA. (2014). Neuregulin-3 regulates epithelial progenitor cell positioning and specifies mammary phenotype. Stem Cells Dev 23:2758–2770 [DOI] [PubMed] [Google Scholar]
- 33.Kavanaugh WM, Pot DA, Chin SM, Deuter-Reinhart M, Jefferson AB, Norris FA, Masiarz FR, Cousens LS, Majerus PW. and Williams LT. (1996). Multiple forms of an inositol polyphosphate 5-phosphatase form signaling complexes with Shc and Grb2. Curr Biol 6:438–445 [DOI] [PubMed] [Google Scholar]
- 34.Fu C-H, Lin R-J, Yu J, Chang W-W, Liao G-S, Chang W-Y, Tseng L-M, Tsai Y-F, Yu J-C. and Yu AL. (2014). A novel oncogenic role of inositol phosphatase SHIP2 in ER-negative breast cancer stem cells: involvement of JNK/vimentin activation. Stem Cells 32:2048–2060 [DOI] [PubMed] [Google Scholar]
- 35.Bartolomei MS, Zemel S. and Tilghman SM. (1991). Parental imprinting of the mouse H19 gene. Nature 351:153–155 [DOI] [PubMed] [Google Scholar]
- 36.Brannan CI, Dees EC, Ingram RS. and Tilghman SM. (1990). The product of the H19 gene may function as an RNA. Mol Cell Biol 10:28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai X. and Cullen BR. (2007). The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13:313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Kahri AI, Heikkilä P, Ilvesmäki V. and Voutilainen R. (1995). H19 and insulin-like growth factor-II gene expression in adrenal tumors and cultured adrenal cells. J Clin Endocrinol Metab 80:492–496 [DOI] [PubMed] [Google Scholar]
- 39.Ariel I, Weinstein D, Voutilainen R, Schneider T, Lustig-Yariv O, de Groot N. and Hochberg A. (1997). Genomic imprinting and the endometrial cycle. The expression of the imprinted gene H19 in the human female reproductive organs. Diagn Mol Pathol 6:17–25 [DOI] [PubMed] [Google Scholar]
- 40.Adriaenssens E, Dumont L, Lottin S, Bolle D, Leprêtre A, Delobelle A, Bouali F, Dugimont T, Coll J. and Curgy JJ. (1998). H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am J Pathol 153:1597–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adriaenssens E, Lottin S, Dugimont T, Fauquette W, Coll J, Dupouy JP, Boilly B. and Curgy JJ. (1999). Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene 18:4460–4473 [DOI] [PubMed] [Google Scholar]
- 42.Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, Dugimont T. and Curgy JJ. (2002). Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23:1885–1895 [DOI] [PubMed] [Google Scholar]
- 43.Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T. and Adriaenssens E. (2005). H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 280:29625–29636 [DOI] [PubMed] [Google Scholar]
- 44.Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, et al. (2014). Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta 1843:1414–1426 [DOI] [PubMed] [Google Scholar]
- 45.Ng JH. and Ng HH. (2010). LincRNAs join the pluripotency alliance. Nat Genet 42:1035–1036 [DOI] [PubMed] [Google Scholar]
- 46.Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, Lu Z, Bai C, Watabe K. and Mo YY. (2013). The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res 23:340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP. and Pelicci PG. (2009). The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138:1083–1095 [DOI] [PubMed] [Google Scholar]
- 48.Bonizzi G, Cicalese A, Insinga A. and Pelicci PG. (2012). The emerging role of p53 in stem cells. Trends Mol Med 18:6–12 [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X. and Liu H. (2013). Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 25:69–80 [DOI] [PubMed] [Google Scholar]
- 50.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J. and Fang G. (2012). Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 279:3159–3165 [DOI] [PubMed] [Google Scholar]
- 51.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, et al. (2013). The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 52:101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J. and Song E. (2007). let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131:1109–1123 [DOI] [PubMed] [Google Scholar]
- 53.Kong D, Heath E, Chen W, Cher ML, Powell I, Heilbrun L, Li Y, Ali S, Sethi S, et al. (2012). Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One 7:e33729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo M, Li Z, Wang W, Zeng Y, Liu Z. and Qiu J. (2013). Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett 333:213–221 [DOI] [PubMed] [Google Scholar]
- 55.Yin Y, Wang H, Liu K, Wang F, Ye X, Liu M, Xiang R, Liu N. and Liu L. (2014). Knockdown of H19 enhances differentiation capacity to epidermis of parthenogenetic embryonic stem cells. Curr Mol Med 14:737–748 [DOI] [PubMed] [Google Scholar]
- 56.Zimmerman DL, Boddy CS. and Schoenherr CS. (2013). Oct4/Sox2 binding sites contribute to maintaining hypomethylation of the maternal igf2/h19 imprinting control region. PLoS One 8:e81962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abi Habib W, Azzi S, Brioude F, Steunou V, Thibaud N, Neves CD, Le Jule M, Chantot-Bastaraud S, Keren B, et al. (2014). Extensive investigation of the IGF2/H19 imprinting control region reveals novel OCT4/SOX2 binding site defects associated with specific methylation patterns in Beckwith-Wiedemann syndrome. Hum Mol Genet 23:5763–5773 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.