Abstract
The role of the BCR–ABL oncogene in the progression of chronic myeloid leukemia (CML) to blast crisis (BC) is unknown. The appearance of chromosomal aberrations in patients with CML BC has led to many attempts to elucidate a mechanism whereby BCR–ABL affects DNA damage and repair. BCR–ABL-expressing cells have been found to accumulate genetic abnormalities, but the mechanism leading to this genomic instability is controversial. In this study, we review the effects of BCR–ABL on DNA repair mechanisms, centrosomes, checkpoint activation and apoptosis. BCR–ABL has diverse effects on these mechanisms, but which of these effects are necessary for the progression of CML to BC is still unresolved.
Keywords: chronic myeloid leukemia, BCR–ABL, DNA damage, blast crisis
Introduction
It is tempting to think of chronic myeloid leukemia (CML) as a solved medical mystery. Thanks to many decades of work, it is clear that CML is a two-stage disease of the bone marrow brought on by the t(9;22) chromosomal translocation, which leads to the production of the p210 BCR–ABL protein. The initial chronic phase is marked by hyperproliferation of white blood cells, which is the direct result of the constitutive kinase activity of BCR–ABL and activation of signal transduction pathways. Inhibition of this kinase with the drug imatinib leads to alleviation of hyperproliferative symptoms.1,2 Physicians are happy with this successful example of targeted therapeutics and patients are living well. However, there is still an incomplete understanding of the pathogenesis of CML blast crisis (BC). The second stage of CML, BC, occurs 3–5 years after diagnosis of untreated CML chronic phase. One of the hallmarks of CML BC is the acquisition of additional chromosomal aberrations beyond the initial t(9;22) chromosomal rearrangement. Although a few abnormalities are more common than others, such as trisomy 8, isochromosome 17q or the presence of an additional Philadelphia chromosome, secondary mutations vary greatly and reflect an overall genomic instability rather than a direct effect on any particular locus.3 This review addresses the issue of whether this genomic instability is a direct consequence of BCR–ABL, making this disease a rare example of a one hit malignancy, or whether there are other, unrelated events driving CML BC. Our intent is to provide a ‘tour’ of the literature with emphasis on critical conclusions and important open questions rather than a complete compilation of the work in this field.
BCR–ABL-expressing cells have increased DNA damage and genetic aberrations
The consequences of BCR–ABL expression on DNA damage and genetic aberrations have been studied for many years. For purposes of this review, we define DNA damage as unresolved DNA breaks or mutations (that is, lesions in need of repair) and genetic aberrations as inappropriately repaired lesions that have led to fixed alterations in the genome. The theme of a growing body of literature is that BCR–ABL expression is associated with a consistent but small increase in both DNA damage and genetic aberrations in cells. Pierre Laneuville4 first suggested that interleukin 3-dependent cell lines transformed to cytokine independence by expression of BCR–ABL showed features of genomic instability. Laneuville later used the Big Blue mouse, in which point mutations in the transgenic lac Z gene lead to loss of lac Z reactivity in tissues, and reported that expression of BCR–ABL led to a small but significant increase in point mutations in BCR–ABL-expressing cells in vivo.5 However, it is still unresolved whether point mutations are a major mechanism of genetic instability associated with CML BC (see below). Several groups, including our own, have pursued more specifically the role of BCR–ABL in the generation of chromosomal instability, one phenotype clearly associated with CML BC. Deutsch et al.6 analyzed sister chromatid exchange and chromosome-specific fluorescence in situ hybridization to show that BCR–ABL-expressing cell lines have an increased incidence of sister chromatid exchange and an increase in chromosomal translocations after DNA damage. This result was confirmed on a more genome-wide basis using spectral karyotyping by the Skorski laboratory,7 and we have recently extended these observations to primary CML cells compared with normal cells.8 Several common themes emerge from these studies. Genetic and chromosomal abnormalities are consistently increased in BCR–ABL-expressing cells, although the increase is modest. Increases are observed both spontaneously after long periods of expression of BCR–ABL, and with increased frequency after induction of DNA damage by genotoxic agents. The alterations appear to be random (the cited experiments were carried out under conditions that did not allow for selections of mutations that provided a growth advantage) consistent with a general ‘mutator phenotype’ rather than induction of a specific genetic lesion. Importantly, most of these assays are laborious and expensive and, as a consequence, few of these papers have in-depth structure-function studies to identify which domain(s) of BCR–ABL is necessary for the alteration in DNA repair that leads to the accumulation of genetic abnormalities. Overall, these data lead to the question, ‘How does BCR–ABL alter genomic stability?’ This question remains incompletely resolved but the available data are summarized here.
DNA damage may arise in BCR–ABL-expressing cells in a number of ways. This topic and the induction of mutations in BCR–ABL itself have been recently reviewed and these studies are only summarized here.9 BCR–ABL has been shown to induce the production of reactive oxygen species, which cause oxidative damage and mutations.10–12 Recently, it was also shown that the B-cell-specific mutator enzyme activation-induced cytidine deaminase is expressed in CML lymphoid BC cells and contributes to mutations within BCR–ABL itself.13 In adults, lymphoid BC represents a minority of BC patients, however, and it is not clear that activation-induced cytidine deaminase contributes to the more common myeloid BC. Alternatively, damage could develop because of the uncontrolled proliferation of cells expressing BCR–ABL. As polymerases themselves cause errors during DNA replication,14 an increase in the number of cells produced could simply lead to an increased chance of aberrancy. Alternatively, it is speculated that activated tyrosine kinase oncogenes decrease the fidelity of the G1/S cell cycle checkpoint although this has not been confirmed. Overall, there are several suggested effects of BCR–ABL that may lead to an increased rate of DNA damage in CML chronic phase cells. None of these are conclusively shown to be necessary for progression to CML BC, and it is rather likely that there are several mechanisms that lead to a modest increase in the rate of DNA damage in BCR–ABL-expressing cells.
Overview of DNA damage response and repair mechanisms
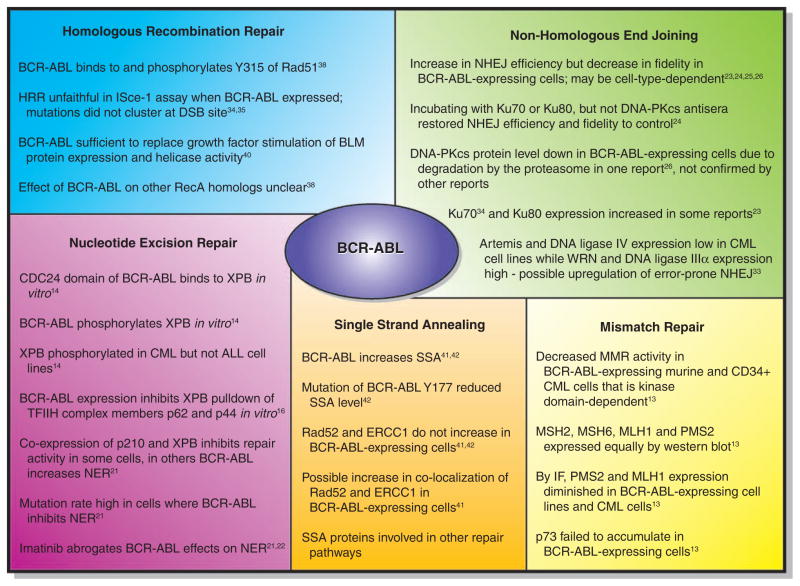
More extensive work has been carried out studying the effects of BCR–ABL on the DNA damage response and this literature will be reviewed in detail here, as well as summarized in Figure 1. The DNA damage response is quite complex but can be shortly summarized: DNA damage may occur either as single-nucleotide alterations, single-strand breaks, or double-strand breaks (DSBs). Single-strand breaks are prone to degrade to DSBs and the two will be considered together here. Single-nucleotide alterations are repaired by mismatch repair (MMR) or by nucleotide excision repair (NER). Strand breaks are repaired by either high-fidelity homologous recombination when a sister chromatid is available as a template (during the S or G2 phase of the cell cycle), or by non-homologous end joining (NHEJ), which may lead to short deletions in the repaired strands. DNA mutations can occur as the result of several situations; when there are mutations in single-nucleotide repair pathways, mutations in proteins necessary for the complex process of DNA DSB recognition and repair or, alternatively, when there is a failure of cell cycle checkpoints that allows subsequent replication of damaged DNA. The latter may occur because of defects in sensing DNA damage or defects in proteins necessary to execute the cell cycle arrest. Effects of BCR–ABL on all of these pathways have been described.
Figure 1.
BCR–ABL affects DNA repair processes.
Single-nucleotide repair
Although point mutations in the kinase domain of BCR–ABL have been detected and are important for imatinib resistance,15,16 it is unclear at this time just how prominent or deleterious point mutations in the rest of the genome are in patients transitioning to CML BC. It is noteworthy that recent work in acute myeloid leukemia with normal cytogenetics has revealed such mutations. 17,18 Given the rapid advances in DNA sequencing approaches, it is likely that more information on point mutations in CML BC will be available in the next few years.
Mismatch repair
MMR corrects DNA mismatches when a base or a few bases are incorrectly incorporated or mutated. In eukaryotes, mismatches in DNA are bound by MutSα (MSH2-MSH6 heterodimer) or MutSβ (MSH2-MSH3 heterodimer), which initiates the repair process along with MutL1 (MLH1-PMS1 heterodimer). A nick is made in one DNA strand, which is then degraded past the mismatch, followed by filling in of the proper bases by DNA polymerase δ. Only one paper has examined the effect of BCR–ABL on MMR. Stoklosa et al.19 used an elegant assay to determine the effect of BCR–ABL on MMR. By incorporating an EGFP gene with a point mutation in the start codon into BCR–ABL-positive or -negative cells, they are able to detect MMR products as GFP+ cells. They determined that in both murine hematopoietic cells expressing BCR–ABL, as well as primary CD34+ CML cells, there was a decrease in MMR activity compared with controls. This was due to BCR–ABL kinase function in examined cell lines, as imatinib treatment returned MMR activity to control level. Mechanistically, there appeared to be no effects of BCR–ABL on MSH2, MSH6, MLH1 or PMS2 expression by western blot, but immunofluorescence data suggest that PMS2 and MLH1 expression may in fact be diminished in BCR–ABL-expressing cell lines as well as CML cells compared with controls.19 Further exploration of the effects on these and other MMR proteins should be revealing.
Nucleotide excision repair
NER is the main DNA repair pathway for ultraviolet (UV)- induced damage, and also works to correct other bulky helix distortions. UV lesions are detected by the DNA damage binding protein heterodimer, made up of DNA damage binding protein 1 and DNA damage binding protein 2, and helix distortions are recognized by the xeroderma pigmentosum group C-Rad23B complex. These complexes then recruit other NER proteins to carry out repair. The transcription factor TFIIH, consisting of XPB, XPD, p62, p52, p44 and p34, is the NER helicase, and XPG and XPF create incisions in the DNA for proliferating-cell nuclear antigen and replication protein A binding and DNA polymerase δ or ε repair. Several groups have studied the interactions of BCR–ABL with this complex of proteins with conflicting results.20–23
Initial reports have shown that the CDC24 homology domain of BCR–ABL23 as well as BCR22 was found to interact with the XBP protein in yeast cells. In vitro assays have shown that p210 BCR–ABL (but not p185 BCR–ABL) was pulled down by GST-XPB, an interaction that was not dependent on the kinase activity of BCR–ABL. However, a complex of BCR–ABL and XPB was not convincingly shown in human BCR–ABL+ cell lines, suggesting that the interaction may not occur at physiologic levels of BCR–ABL expression. It was later suggested that the mechanism of this NER defect may be a result of BCR–ABL interfering with overall formation of the TFIIH complex formation.24 It was found that the presence of BCR–ABL prevented the pull-down of p62 and p44 with XPB in vitro,24 which may be the result of steric hindrance induced by BCR–ABL binding to XPB. However, this negative interaction may not be physiologically relevant. Although BCR–ABL has been shown to translocate to the nucleus,25,26 subsequent attempts27 have been unsuccessful in replicating this data, and we believe BCR–ABL to be primarily a cytoplasmic protein in the absence of substantial DNA damage. BCR–ABL and TFIIH therefore have different subcellular localizations and limited interaction in vivo, as suggested by the very weak ability to pull-down a BCR–ABL/XPB complex,23 and the absence of p210 from purified TFIIH complexes.24
The functional effects of BCR–ABL expression on NER are also controversial. Several groups have shown that expression of BCR–ABL makes cells sensitive to UV- and cisplatin-induced (NER-type) damage.21–23 The cell line 27-1 has a defect in NER that is corrected by expression of XPB. However, the presence of BCR–ABL leads to the inability of XPB to correct this defect.24 An interesting interpretation comes from Canitrot et al.20, who determined that the effects of BCR–ABL on NER activity are cell-type dependent. In BaF3 cells, BCR–ABL decreased NER activity and sensitized the cells to UV, in accordance with the above studies. However, in all other cell lines tested, the presence of BCR–ABL actually increased NER and decreased UV sensitivity. 20 In all cases, imatinib abrogated the effects, suggesting the necessity of the kinase domain of BCR–ABL.20,28 The results of Laurent et al.21 corroborate the data that suggest that BCR–ABL decreases sensitivity of MO7E and 4A2+ cells to UV. Importantly, their data make clear the large number of variables at play in assessing the rapid and integrated series of events that occur after DNA damage. They show that after UV-induced DNA damage, BCR–ABL-expressing cells showed less DNA damage than parental cells as measured by the number of cyclobutane pyrimidine dimers. However, BCR–ABL cells showed an increase in DNA breaks as assessed by the Comet assay. NER works through induction of DNA strand ‘nicks’ (really single-strand breaks) that will be converted to DSBs in the alkaline comet assay. Thus, the investigators suggest that BCR–ABL effects in the comet assay indicate an effect on the DNA repair process leading to faster DNA repair.21 Their studies leave open the question of the fidelity of this repair. On top of the conflicting results from different cell lines,20–23 the role of BCR–ABL in affecting NER is of questionable importance, as UV-induced damage is not likely to be observed in the hematopoietic system, which is not exposed to UV. At this point, although the effects of BCR–ABL on NER in cell lines are documented, its physiologic significance in the progression of CML chronic phase to CML BC remains unclear.
DSB repair
It is well documented that partial deletions, duplications and translocations are commonly observed in patients with CML BC.3 In addition, Dierov et al.8 have shown that BCR–ABL-expressing cell lines and CML cells acquire more translocations and aneuploid events than control cells after exogenous DNA damage (as assessed by spectral karyotyping analysis). As translocations arise as a result of the mis-repair of DNA DSBs, many labs have analyzed how BCR–ABL may have a role in altering the efficiency or fidelity of DSB repair pathways.
Non-homologous end joining
NHEJ repair of DSBs occurs rapidly after breakage and does not require a homologous template, and thus is the preferred response to DSBs when cells are in G0 or G1. The canonical NHEJ pathway involves the binding to a DSB by the Ku70/Ku80 heterodimer and recruitment of the DNA-PKcs-Artemis nuclease. Ligation steps are carried out by DNA ligase IV along with XRCC4. NHEJ is unfaithful in that most breaks repaired in this manner are marked by short deletions and insertions of 1–4 nucleotides, and often translocations are formed when chromosomes are incorrectly juxtaposed.
The effects of BCR–ABL on NHEJ repair may be cell-type dependent. Slupianek et al.29 used a fluorescent detection assay to look at NHEJ repair products after restriction digest. They found an approximately twofold increase in NHEJ activity in blunt-end repair in BCR–ABL-expressing 32Dcl3 cells compared with parental, and a fourfold increase in the case of 5′ overhang repair. The fidelity of the repair was compromised, with more small additions and larger deletions in the presence of BCR–ABL.29 Importantly, these results have been recently confirmed.30 Gaymes et al.30 present data that agrees with Slupianek, in which BCR–ABL-expressing CML patient samples and K562 cells showed a three- to fivefold increase in end-ligation efficiency compared with normal CD34+ cells. This was accompanied by an increased frequency of misrepair, which included large (30–400 bp) deletions. Mechanistically, they found that Ku70 and Ku80 were involved, as incubation with antisera against these proteins restored the frequency and size of deletions to control levels. The same was not true of DNA-PKcs, as the use of antisera to that protein did not alter deletion size.30 In contrast to these results, Pastwa et al.31 found that K562 myeloid leukemia cells (which express BCR–ABL but not wild-type p53) showed fewer repair products from DNA with 5′ overhangs than normal human lymphocytes, but no difference in blunt-end repair was observed between the two cell types. When imatinib was included in the assay, the same result was obtained, leading the investigators to question which genetic component may actually be responsible for the difference, the expression of BCR–ABL or the lack of wild-type p53 in the K562 cells.31 Overall, these data support the concept that BCR–ABL expression increases repair of DNA DSBs through the error-prone NHEJ mechanism.
Deutsch et al.32 also searched for a mechanism by which BCR–ABL may be affecting NHEJ. They found that while there was no difference in the protein expression of Ku70 and Ku80, cells expressing BCR–ABL expressed less DNA-PKcs than their wild-type counterparts.32 This protein expression was not due to a difference in the amount of mRNA in the cells, but rather degradation of DNA-PKcs, as inhibition of the proteasome restored protein levels. This was also the case in CD34+ cells from CML patients compared with normal controls.32 However, as with other results, this has not been confirmed. Preliminary work in our laboratory (Dierov, Carroll, unpublished) showed a decrease in expression of a full size DNA-PKcs enzyme but appearance of a smaller protein fragment. Gaymes et al.30 determined that DNA-PKcs was not involved in the large deletions in NHEJ products in CML cells, and Slupianek et al.29 detected similar expression levels of DNA-PKcs in normal and BCR–ABL-expressing cells. They also determined that there was an increase in Ku70 and Ku80 protein expression in BCR–ABL cells after irradiation, while no difference in protein level was observed before irradiation.29 This response could actually be a response to the increase in DNA DSBs, rather than a BCR–ABL-mediated protein upregulation, but corroborates the importance of Ku70 and Ku80 in leading to large deletions in NHEJ products.30,33 Evidence that the response could be due to an increase in native breaks comes from Brady et al.33 who examined normal peripheral blood lymphocytes and CD34+ cells after 6Gy irradiation and determined that they in fact have a similar pattern (although lower overall level) of NHEJ activity and misrepair compared with untreated myeloid leukemia cells. The comparisons are not completely isogenic, but it is interesting to speculate that it is the excessive amount of DNA damage that is driving constitutive expression of NHEJ proteins in untreated myeloid leukemia cells, including the BCR–ABL-expressing K562 line. Certainly, there is increasing evidence that the effects of BCR–ABL may be mediated through alterations in the level of expression or effects on the function of Ku70 and Ku80 although the exact cause–effect relationships remain to be determined.
A second, more error-prone pathway of NHEJ exists, although it is not well-defined.34 DNA ligase IIIα (which usually participates in single-strand break repair and base excision repair35) is thought to function as a backup for DNA ligase IV when it is unavailable or inefficiently recruited.36 In addition, poly ADP-ribose polymerase was shown to have a role in NHEJ in cells deficient in Ku70/Ku80.37 Werner (WRN) protein, which may actually have a role in canonical NHEJ, is also important in preventing genomic instability, as cells lacking WRN generate extensive deletions after NHEJ repair.38 These alternative NHEJ proteins lead to an increased frequency of errors in repair products as a result of microhomology-mediated ligation (low-fidelity repair). The error-prone repair of NHEJ products often observed led Sallmyr et al.39 to examine whether alternative NHEJ proteins were involved in repair of DSBs in CML. CML cell lines expressed reduced steady-state levels of Artemis and DNA ligase IV as compared with normal hematopoietic cells, as well as increased levels of WRN and DNA ligase IIIα as assessed by western blot. Although most comparisons are between nonisogenic cell lines, the same overexpression of WRN and DNA ligase IIIα is observed in P210MO7e cells over parental MO7e, and a CML patient sample with high BCR–ABL expression also has correspondingly high WRN and DNA ligase IIIα compared with a CML patient sample with lower BCR–ABL expression. No changes were observed in the expression of Ku80, DNA-PKcs (in contrast with a previous report32), XRCC4, XRCC1 or poly ADP-ribose polymerase-1.39 Interestingly, small interfering RNA knockdown of either WRN or DNA ligase IIIa resulted in a greater increase in both endogenous and IR-induced DSBs in K562 cells than NC10 cells, as well as a decrease in end joining efficiency. In addition, Artemis overexpression in K562 cells led to a decrease in the size of DNA deletions at misrepaired DSBs, suggesting the downregulation of Artemis in CML cells may be important mechanistically in abnormal DSB repair.39 This will be a compelling mechanism if it is confirmed in isogenic cell lines or CML patient samples, or with the use of imatinib to inhibit BCR–ABL expression.
Homologous recombination repair
Homologous recombination repair (HRR) involves the use of a sister chromatid to repair a DSB with greater fidelity than NHEJ. DNA around the DSB is resected toward the 5′ end on both strands, followed by invasion of the sister chromatid by RAD51 and its accessory proteins. RecQ family helicases (BLM, WRN, RTS, RecQL1 and RecQL5) unwind DNA during HRR both at initiation and resolution of intermediates.
Cells expressing BCR–ABL showed enhanced HRR efficiency as assessed by ISce-I expression assay,29,40,41 but the repair was unfaithful.29,40 Although the number of repair events was similar to that of parental cells, the mutation frequency was 100-fold higher in terms of single base substitutions. Interestingly, these mutations did not cluster near the DSB site.
A few proteins have been examined for their role in BCR–ABL-mediated HRR efficiency and fidelity. RAD51 expression increased in BCR–ABL-expressing cells after irradiation29 and in one report under steady-state conditions.41 Indeed, the addition of a RAD51 antisense construct to the HRR efficiency assay decreased the amount of RAD51 protein as well as the number of HRR events that occurred.41 In addition, RAD51 was found to colocalize with phosphorylated H2AX in immunofluorescence studies,29,40 but there is some concern that this represents an artifact of increased expression of Rad51 after the induction of DNA damage. In addition, BCR–ABL was found to directly associate with RAD51 through IP and to phosphorylate RAD51 on the Y315 site. This phosphorylation could enhance RAD51/RAD52 complex42 formation, stimulating HRR.43 What role BCR–ABL may have in stimulating other RecA homologs is unclear, as it appears the expression level of some of these proteins may have changed, but more work is needed to clarify the overlapping roles of this family of proteins.44
BLM has been shown to be necessary for normal DSB repair.45 Slupianek et al.46 examined the expression of BLM in BCR–ABL-expressing and control cells, and determined that BCR–ABL was sufficient to replace the growth factor stimulation of BLM protein expression and helicase activity. There may also be an effect on the colocalization of BLM and RAD51 in BCR–ABL-expressing cells, which could have an effect on HRR.
Single-strand annealing
Single-strand annealing (SSA) is a non-conservative HRR process that repairs DSBs that occur between identical repeats. This process is very unfaithful, as it results in the deletion of sequences between repeats, as well as one of the repeats. SSA is not completely independent from other DSB repair mechanisms, as many of the proteins involved overlap with other repair pathways. Specifically, a number of HRR proteins and processes are common to both pathways, with the exception of Rad51, which does not participate in SSA.
Cramer et al.47 analyzed the role of BCR–ABL in promoting SSA, and determined there was an increase in SSA activity with increasing BCR–ABL expression. Fernandes, et al.48 also determined that there was an increase in SSA with BCR–ABL expression, and that imatinib abrogated the effect. Control cells were not supplied with growth factors that would match the effect of BCR–ABL, and thus the ability of the cells to grow and proliferate may affect the results. In fact, adding stromal cell conditioned media at the same time as imatinib inhibited the abrogation. Like HRR, SSA is regulated by Rad proteins (particularly Rad52) and it is not clear that the effects described are isolated to SSA. Interestingly, no effects were observed on the protein levels of ERCC1 or Rad52 in BCR–ABL-expressing cells. At this point, further work needs to be carried out to clarify the effects of BCR–ABL on SSA.
Checkpoint regulation
An alternative hypothesis to a direct effect of BCR–ABL on DNA repair is that BCR–ABL expression alters the cellular ability to efficiently recognize and respond to DNA damage. Ataxia telangiectasia mutated protein (ATM) and its homolog ataxia telangiectasia and Rad3-related protein (ATR) are phosphatidylinositol 3-kinase family members that sense and respond to DSBs. Signaling commences through the checkpoint kinases, as ATM phosphorylates Chk2 and ATR phosphorylates Chk1. ATM can also phosphorylate BRCA1, which has a role in DSB repair. ATM has been shown to interact with and phosphorylate c-Abl after irradiation.49
When CML BC cell lines and patient samples were analyzed for mutations in ATM in the Abl-binding region and the region implicated in sporadic hematological malignancy, no nucleotide changes were detected.50 In addition, ATM has been shown to associate with BCR–ABL under immunoprecipitation conditions, however, no difference was observed in the kinase activity of ATM in terms of its ability to phosphorylate either p53 or Chk2.25 It is therefore unlikely that ATM has a significant role in the transition to CML BC. As an ATM homolog that regulates chromosome stability, ATR was also analyzed as a potential instigator in the transition to CML BC. As with ATM, ATR was found to associate with BCR–ABL after DNA damage and Dierov et al.25 initially reported that BCR–ABL expression appeared to inhibit ATR function. This was not confirmed, however, in recent studies by Niebrowska-Skorska et al.27 although the concept remains intriguing, there is not conclusive evidence at this point that BCR–ABL alters the function of the ATR/Chk1 axis.
Centrosomal hypertrophy
Centrosomes are vital members of a cell’s mitotic machinery as they organize the mitotic spindle to ensure bipolar separation of chromosomes. Consequently, centrosome aberrations are a possible cause of aneuploidy, which is a feature of CML BC. In addition, centrosomes may have a role not only structurally, but also as regulators of mitotic entry through their association with p53.51 The role of centrosome defects on causing genetic instability is under examination,52,53 and one group has examined these defects in the context of CML.54,55
Giehl et al.55 determined that centrosome abnormalities correlated with CML disease stage and preceded chromosomal aberrations. Importantly, in the first study, primary patient samples were used, giving added weight to the data. The investigators suggest that the acquisition of centrosome defects may be one of the driving forces of progression to CML BC and contribute to the chromosomal instability.55 A causative relationship with BCR–ABL was later elucidated by this same group,54 in which they used U937 cells with a tetracycline-inducible BCR–ABL in long-term culture experiments. They determined that constant expression of BCR–ABL led to increasing centrosome aberrations over time. Further, turning BCR–ABL off halfway through the experiment through doxycycline withdrawal led to a reversion toward baseline.54 Overall, these data suggest that BCR–ABL introduces centrosome abnormalities, which may contribute to the chromosomal aberrancies found in CML BC.
Apoptosis inhibition
When cells acquire DNA damage, the damage-sensing proteins recognize the error and stimulate repair, but when the damage is too great the cell may instead signal to undergo apoptosis. This prevents the replication of potentially harmful mutations, and evading apoptosis is one of the known hallmarks of cancer. Another of the known cellular roles of BCR–ABL is the inhibition of apoptosis. McGahon et al.57 determined that the resistance of K562 cells to apoptosis56 was a consequence of BCR–ABL expression, as antisense oligonucleotide treatment against BCR–ABL rendered the cells susceptible to apoptosis. Leukemic HL-60 cells expressing p185 BCR–ABL were also found to be more resistant to apoptosis after cytotoxic agents than normal HL-60 cells. Expression of BCR–ABL in these cells led to the downregulation of Bcl-2 and increase in Bcl-xL levels, although Bax levels did not change. The investigators determined that Bcl-xL was important in BCR–ABL-mediated resistance to apoptosis in HL-60 cells, as knockdown of Bcl-xL increased susceptibility of HL-60.BCR–ABL cells to staurosporine.58
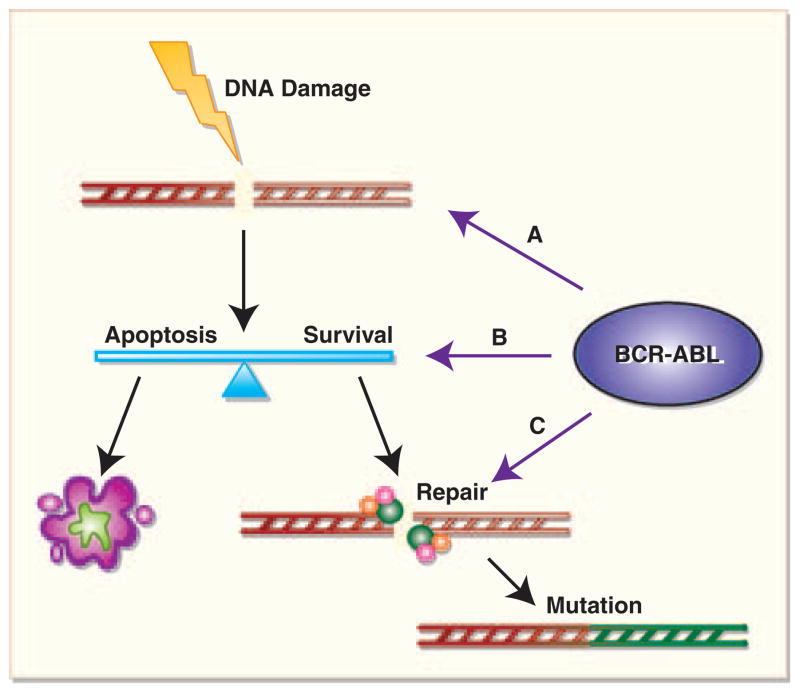
Despite confirming that BCR–ABL-expressing cells were resistant to apoptosis induced by drug treatment, Keeshan et al.59 determined that BCR–ABL cells actually had a proapoptotic expression profile that is determined by the level of expression of the protein. Normal 32D cells and cells expressing a low level of BCR–ABL were found to upregulate p53, p21 and Bax, and downregulate Bcl-2 in response to cytotoxic insult. In contrast, cells expressing a high level of BCR–ABL expressed constitutively high levels of p53, p21 and Bax, and low levels of Bcl-2, and cytotoxic insult did not alter these levels. An explanation for this expression profile was elucidated in a later paper, in which it was determined that the translocation of pro-apoptotic proteins Bax and Bad to the mitochondria was blocked in cells with high BCR–ABL expression. In addition, BCR–ABL was found to prevent late mitochondrial depolarization and prevent caspase 9 and caspase 3 processing.60 Therefore, despite the unexpected pro-apoptotic protein expression in cells expressing high levels of BCR–ABL, the proteins do not function normally to carry out apoptosis after cytotoxic insult. A critical question is whether BCR–ABL expression allows cells with damaged DNA to survive long enough to recruit error-prone DNA repair pathways leading to survival of CML cells with mis-repaired DNA. Such a mechanism (c.f. Figure 2) may provide a model to integrate the multiple DNA repair abnormalities described. After DNA damage, cells either repair DNA rapidly and continue to grow (if damage is minimal); undergo apoptosis (after large amounts of damage); or arrest, repair DNA and then resume growth (after intermediate damage). It may be that BCR–ABL alters the apoptotic threshold of cells, increasing the pool of cells undergoing active DNA repair, and the alterations in DNA repair processes described reflect the increase in the size of the pool of cells undergoing DNA repair. It will be important for future experiments that the degree of apoptosis induction is correlated with alterations in the activity of DNA repair enzymes.
Figure 2.
BCR–ABL affects DNA mutation through the induction of DNA damage, alteration of the apoptotic threshold and mediation of DNA repair pathways. (A) Represents DNA damage caused by BCR–ABL. Once damage occurs, BCR–ABL may inhibit apoptosis, thus allowing cells to survive with more damage than can be effectively repaired (B). BCR–ABL also interacts directly and indirectly with repair proteins and can contribute to rapid but low-fidelity repair (C). The end result is an accumulation of mutations in CML BC.
Conclusions
The bulk of data convincingly shows that BCR–ABL-expressing cells are subject to the accumulation of mutated DNA (Figure 2). Our opinion is that this is a consequence of effects of BCR–ABL expression on the fidelity of DNA repair processes, although we cannot, with current data, eliminate the possibility that BCR–ABL alters the accumulation of DNA damage. Which DNA repair process is critical in preventing the development of CML BC is unclear, and resolution of this vexing problem is probably dependent on development of a robust model of CML BC in mice. Using such a model, DNA repair processes can be altered to allow for definition of the critical DNA repair process. Finally, whether or not these effects of BCR–ABL on DNA damage accumulation are dependent on BCR–ABL kinase activity and will be abrogated in patients on ABL kinase inhibitor therapy is unclear. Clinical data clearly suggest that ABL kinase inhibitors delay the development of CML BC. Whether it is permanently prevented or only forestalled remains to be observed.
Acknowledgments
We would like to thank our colleagues and the anonymous reviewers for helpful discussions. We apologize to colleagues whose work we are not able to discuss because of space limitations.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 2.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein R. Cytogenetics of chronic myelogenous leukemia. Semin Hematol. 1988;25:20–34. [PubMed] [Google Scholar]
- 4.Laneuville P, Sun G, Timm M, Vekemans M. Clonal evolution in a myeloid cell line transformed to interleukin-3 independent growth by retroviral transduction and expression of p210bcr/abl. Blood. 1992;80:1788–1797. [PubMed] [Google Scholar]
- 5.Salloukh HF, Laneuville P. Increase in mutant frequencies in mice expressing the BCR-ABL activated tyrosine kinase. Leukemia. 2000;14:1401–1404. doi: 10.1038/sj.leu.2401855. [DOI] [PubMed] [Google Scholar]
- 6.Deutsch E, Jarrousse S, Buet D, Dugray A, Bonnet ML, Vozenin-Brotons MC, et al. Down-regulation of BRCA1 in BCR-ABL-expressing hematopoietic cells. Blood. 2003;101:4583–4588. doi: 10.1182/blood-2002-10-3011. [DOI] [PubMed] [Google Scholar]
- 7.Koptyra M, Cramer K, Slupianek A, Richardson C, Skorski T. BCR// ABL promotes accumulation of chromosomal aberrations induced by oxidative and genotoxic stress. Leukemia. 2008;22:1969–1972. doi: 10.1038/leu.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dierov J, Sanchez PV, Burke BA, Padilla-Nash H, Putt ME, Ried T, et al. BCR/ABL induces chromosomal instability after genotoxic stress and alters the cell death threshold. Leukemia. 2009;23:279–286. doi: 10.1038/leu.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skorski T. BCR/ABL, DNA damage and DNA repair: implications for new treatment concepts. Leuk Lymphoma. 2008;49:610–614. doi: 10.1080/03093640701859089. [DOI] [PubMed] [Google Scholar]
- 10.Kim JH, Chu SC, Gramlich JL, Pride YB, Babendreier E, Chauhan D, et al. Activation of the PI3K/mTOR pathway by BCR-ABL contributes to increased production of reactive oxygen species. Blood. 2005;105:1717–1723. doi: 10.1182/blood-2004-03-0849. [DOI] [PubMed] [Google Scholar]
- 11.Koptyra M, Falinski R, Nowicki MO, Stoklosa T, Majsterek I, Nieborowska-Skorska M, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108:319–327. doi: 10.1182/blood-2005-07-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sattler M, Verma S, Shrikhande G, Byrne CH, Pride YB, Winkler T, et al. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. J Biol Chem. 2000;275:24273–24278. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- 13.Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 15.Willis SG, Lange T, Demehri S, Otto S, Crossman L, Niederwieser D, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106:2128–2137. doi: 10.1182/blood-2005-03-1036. [DOI] [PubMed] [Google Scholar]
- 16.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCRABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 17.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoklosa T, Poplawski T, Koptyra M, Nieborowska-Skorska M, Basak G, Slupianek A, et al. BCR/ABL inhibits mismatch repair to protect from apoptosis and induce point mutations. Cancer Res. 2008;68:2576–2580. doi: 10.1158/0008-5472.CAN-07-6858. [DOI] [PubMed] [Google Scholar]
- 20.Canitrot Y, Falinski R, Louat T, Laurent G, Cazaux C, Hoffmann JS, et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood. 2003;102:2632–2637. doi: 10.1182/blood-2002-10-3207. [DOI] [PubMed] [Google Scholar]
- 21.Laurent E, Mitchell DL, Estrov Z, Lowery M, Tucker SL, Talpaz M, et al. Impact of p210(Bcr-Abl) on ultraviolet C wavelength-induced DNA damage and repair. Clin Cancer Res. 2003;9:3722–3730. [PubMed] [Google Scholar]
- 22.Maru Y, Kobayashi T, Tanaka K, Shibuya M. BCR binds to the xeroderma pigmentosum group B protein. Biochem Biophys Res Commun. 1999;260:309–312. doi: 10.1006/bbrc.1999.0822. [DOI] [PubMed] [Google Scholar]
- 23.Takeda N, Shibuya M, Maru Y. The BCR-ABL oncoprotein potentially interacts with the xeroderma pigmentosum group B protein. Proc Natl Acad Sci USA. 1999;96:203–207. doi: 10.1073/pnas.96.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maru Y, Bergmann E, Coin F, Egly JM, Shibuya M. TFIIH functions are altered by the P210BCR-ABL oncoprotein produced on the Philadelphia chromosome. Mutat Res. 2001;483:83–88. doi: 10.1016/s0027-5107(01)00229-9. [DOI] [PubMed] [Google Scholar]
- 25.Dierov J, Dierova R, Carroll M. BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell. 2004;5:275–285. doi: 10.1016/s1535-6108(04)00056-x. [DOI] [PubMed] [Google Scholar]
- 26.Vigneri P, Wang JY. Induction of apoptosis in chronic myelogenous leukemia cells through nuclear entrapment of BCR-ABL tyrosine kinase. Nat Med. 2001;7:228–234. doi: 10.1038/84683. [DOI] [PubMed] [Google Scholar]
- 27.Nieborowska-Skorska M, Stoklosa T, Datta M, Czechowska A, Rink L, Slupianek A, et al. ATR-Chk1 axis protects BCR/ABL leukemia cells from the lethal effect of DNA double-strand breaks. Cell Cycle. 2006;5:994–1000. doi: 10.4161/cc.5.9.2722. [DOI] [PubMed] [Google Scholar]
- 28.Sliwinski T, Czechowska A, Szemraj J, Morawiec Z, Skorski T, Blasiak J. STI571 reduces NER activity in BCR/ABL-expressing cells. Mutat Res. 2008;654:162–167. doi: 10.1016/j.mrgentox.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Slupianek A, Nowicki MO, Koptyra M, Skorski T. BCR/ABL modifies the kinetics and fidelity of DNA double-strand breaks repair in hematopoietic cells. DNA Repair. 2006;5:243–250. doi: 10.1016/j.dnarep.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaymes TJ, Mufti GJ, Rassool FV. Myeloid leukemias have increased activity of the nonhomologous end-joining pathway and concomitant DNA misrepair that is dependent on the Ku70/86 heterodimer. Cancer Res. 2002;62:2791–2797. [PubMed] [Google Scholar]
- 31.Pastwa E, Poplawski T, Czechowska A, Malinowski M, Blasiak J. Non-homologous DNA end joining repair in normal and leukemic cells depends on the substrate ends. Z Naturforsch (C) 2005;60:493–500. doi: 10.1515/znc-2005-5-619. [DOI] [PubMed] [Google Scholar]
- 32.Deutsch E, Dugray A, AbdulKarim B, Marangoni E, Maggiorella L, Vaganay S, et al. BCR-ABL down-regulates the DNA repair protein DNA-PKcs. Blood. 2001;97:2084–2090. doi: 10.1182/blood.v97.7.2084. [DOI] [PubMed] [Google Scholar]
- 33.Brady N, Gaymes TJ, Cheung M, Mufti GJ, Rassool FV. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res. 2003;63:1798–1805. [PubMed] [Google Scholar]
- 34.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 35.Gottlich B, Reichenberger S, Feldmann E, Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur J Biochem/FEBS. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 37.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshima J, Huang S, Pae C, Campisi J, Schiestl RH. Lack of WRN results in extensive deletion at nonhomologous joining ends. Cancer Res. 2002;62:547–551. [PubMed] [Google Scholar]
- 39.Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowicki MO, Falinski R, Koptyra M, Slupianek A, Stoklosa T, Gloc E, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104:3746–3753. doi: 10.1182/blood-2004-05-1941. [DOI] [PubMed] [Google Scholar]
- 41.Slupianek A, Hoser G, Majsterek I, Bronisz A, Malecki M, Blasiak J, et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol. 2002;22:4189–4201. doi: 10.1128/MCB.22.12.4189-4201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen G, Yuan SS, Liu W, Xu Y, Trujillo K, Song B, et al. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 43.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 44.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 45.Langland G, Elliott J, Li Y, Creaney J, Dixon K, Groden J. The BLM helicase is necessary for normal DNA double-strand break repair. Cancer Res. 2002;62:2766–2770. [PubMed] [Google Scholar]
- 46.Slupianek A, Gurdek E, Koptyra M, Nowicki MO, Siddiqui KM, Groden J, et al. BLM helicase is activated in BCR/ABL leukemia cells to modulate responses to cisplatin. Oncogene. 2005;24:3914–3922. doi: 10.1038/sj.onc.1208545. [DOI] [PubMed] [Google Scholar]
- 47.Cramer K, Nieborowska-Skorska M, Koptyra M, Slupianek A, Penserga ET, Eaves CJ, et al. BCR/ABL and other kinases from chronic myeloproliferative disorders stimulate single-strand annealing, an unfaithful DNA double-strand break repair. Cancer Res. 2008;68:6884–6888. doi: 10.1158/0008-5472.CAN-08-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandes MS, Reddy MM, Gonneville JR, DeRoo SC, Podar K, Griffin JD, et al. BCR-ABL promotes the frequency of mutagenic single-strand annealing DNA repair. Blood. 2009;114:1813–1819. doi: 10.1182/blood-2008-07-172148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, et al. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 50.Melo JV, Kumberova A, van Dijk AG, Goldman JM, Yuille MR. Investigation on the role of the ATM gene in chronic myeloid leukaemia. Leukemia. 2001;15:1448–1450. doi: 10.1038/sj.leu.2402223. [DOI] [PubMed] [Google Scholar]
- 51.Tritarelli A, Oricchio E, Ciciarello M, Mangiacasale R, Palena A, Lavia P, et al. p53 localization at centrosomes during mitosis and postmitotic checkpoint are ATM-dependent and require serine 15 phosphorylation. Mol Biol Cell. 2004;15:3751–3757. doi: 10.1091/mbc.E03-12-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marx J. Cell biology. Do centrosome abnormalities lead to cancer? Science (New York) 2001;292:426–429. doi: 10.1126/science.292.5516.426. [DOI] [PubMed] [Google Scholar]
- 53.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 54.Giehl M, Fabarius A, Frank O, Erben P, Zheng C, Hafner M, et al. Expression of the p210BCR-ABL oncoprotein drives centrosomal hypertrophy and clonal evolution in human U937 cells. Leukemia. 2007;21:1971–1976. doi: 10.1038/sj.leu.2404834. [DOI] [PubMed] [Google Scholar]
- 55.Giehl M, Fabarius A, Frank O, Hochhaus A, Hafner M, Hehlmann R, et al. Centrosome aberrations in chronic myeloid leukemia correlate with stage of disease and chromosomal instability. Leukemia. 2005;19:1192–1197. doi: 10.1038/sj.leu.2403779. [DOI] [PubMed] [Google Scholar]
- 56.Martin SJ, Lennon SV, Bonham AM, Cotter TG. Induction of apoptosis (programmed cell death) in human leukemic HL-60 cells by inhibition of RNA or protein synthesis. J Immunol. 1990;145:1859–1867. [PubMed] [Google Scholar]
- 57.McGahon A, Bissonnette R, Schmitt M, Cotter KM, Green DR, Cotter TG. BCR-ABL maintains resistance of chronic myelogenous leukemia cells to apoptotic cell death. Blood. 1994;83:1179–1187. [PubMed] [Google Scholar]
- 58.Amarante-Mendes GP, McGahon AJ, Nishioka WK, Afar DE, Witte ON, Green DR. Bcl-2-independent Bcr-Abl-mediated resistance to apoptosis: protection is correlated with up regulation of Bcl-xL. Oncogene. 1998;16:1383–1390. doi: 10.1038/sj.onc.1201664. [DOI] [PubMed] [Google Scholar]
- 59.Keeshan K, Mills KI, Cotter TG, McKenna SL. Elevated Bcr-Abl expression levels are sufficient for a haematopoietic cell line to acquire a drug-resistant phenotype. Leukemia. 2001;15:1823–1833. doi: 10.1038/sj.leu.2402309. [DOI] [PubMed] [Google Scholar]
- 60.Keeshan K, Cotter TG, McKenna SL. High Bcr-Abl expression prevents the translocation of Bax and Bad to the mitochondrion. Leukemia. 2002;16:1725–1734. doi: 10.1038/sj.leu.2402576. [DOI] [PubMed] [Google Scholar]