Abstract
Cranial sensory placodes derive from discrete patches of the head ectoderm, and give rise to numerous sensory structures. During gastrulation, a specialized “neural border zone” forms around the neural plate in response to interactions between the neural and non-neural ectoderm and signals from adjacent mesodermal and/or endodermal tissues. This zone subsequently gives rise to two distinct precursor populations of the peripheral nervous system: the neural crest and the pre-placodal ectoderm (PPE). The PPE is a common field from which all cranial sensory placodes arise (adenohypophyseal, olfactory, lens, trigeminal, epibranchial, otic). Members of the Six family of transcription factors are major regulators of PPE specification, in partnership with co-factor proteins such as Eya. Six gene activity also maintains tissue boundaries between the PPE, neural crest and epidermis by repressing genes that specify the fates of those adjacent ectodermally-derived domains. As the embryo acquires anterior-posterior identity, the PPE becomes transcriptionally regionalized, and it subsequently subdivides into specific placodes with distinct developmental fates in response to signaling from adjacent tissues. Each placode is characterized by a unique transcriptional program that leads to the differentiation of highly specialized cells, such as neurosecretory cells, somatic sensory receptor cells, chemosensory neurons, peripheral glia and supporting cells. In this review, we summarize the transcriptional and signaling factors that regulate key steps of placode development, influence subsequent sensory neuron specification, and discuss what is known about mutations in some of the essential PPE genes that underlie human congenital syndromes.
Keywords: Six1, Six4, Eya, Pax, Branchio-otic syndrome, Branchio-otic-renal syndrome, olfactory, trigeminal, epibranchial, cranial sensory neurons
Introduction
The vertebrate head contains a number of specialized sensory organs that are derived from discrete patches in the embryonic ectoderm called cranial sensory placodes (reviewed by Webb and Noden, 1993; Baker and Bonner-Fraser, 2001; Streit, 2004, 2007; Schlosser 2006, 2010; Patthey et al., 2014; Saint-Jeannet and Moody, 2014; Moody and Saint-Jeannet, 2014). Cranial sensory placodes give rise to the anterior pituitary gland, the olfactory epithelium, the lens, the auditory and vestibular organs and their associated sensory ganglia, and in aquatic species the lateral line and electroreceptive organs and their associated sensory ganglia. In addition, placodes give rise to the large neurons in the sensory ganglia of the trigeminal, facial, glossopharyngeal and vagus cranial nerves. All of these structures are crucial for an animal to successfully interact with other animals and to navigate through its environment. Therefore, it is critically important that these structures develop properly and be replaced, either naturally or by medical intervention, when they are damaged. The focus of this review is to present our current understanding of the transcriptional and signaling pathways that transform a common pre-placodal precursor field into these diverse and highly specialized structures.
Experiments performed in a number of animals demonstrate that placode development is highly conserved across vertebrates (reviewed in Patthey et al., 2014; Saint-Jeannet and Moody, 2014). In chick, frog, fish and mouse, the embryonic ectoderm is divided into neural ectoderm and non-neural ectoderm during gastrulation. As this process is completed, the ectoderm surrounding the nascent neural ectoderm, called the neural border (NB) zone, is specified to give rise to the neural crest and the pre-placodal ectoderm (PPE) (Figure 1). During neural tube closure (called neurulation), signals that establish the anterior-posterior (A–P) axis of the embryo also impose regional identity on the PPE, and subsequent signals from adjacent tissues cause the PPE to separate into many discrete placodes that have distinct developmental fates. These placodes will then undergo morphogenetic movements and cellular differentiation processes that result in the numerous specialized sensory structures that characterize the vertebrate head.
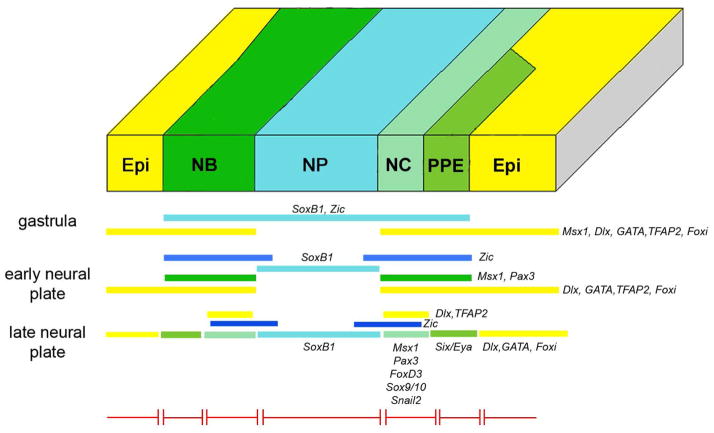
Figure 1.
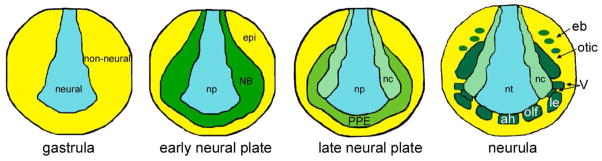
The ectodermal domains depicted at different stages of Xenopus embryonic development. At gastrulation, the early embryonic ectoderm is divided into two domains neural (blue) and non-neural (yellow). Interactions between these two domains and signaling from underlying tissues establish a neural border (NB) zone (green) between the early neural plate (np) and the epidermis (epi). At later neural plate stages the NB zone divides into the medially located neural crest (nc; light green) and the laterally located pre-placodal ectoderm (PPE, dark green). After the the PPE acquires initial anterior-posterior patterning and the neural tube (nt) begins to roll up during neurulation, the PPE breaks up into individual placodes in response to local signaling and differential transcription factor expression. These include: those derived from the anterior PPE field (the adenohypophyseal (ah), olfactory (olf), and lens (le)); those derived from the intermediate field (V; ophthalmic placode is dorsal to lens placode, and maxillomandibular placode is posterior to it); and those derived from the posterior field (otic and epibranchial (eb)).
One of the key outcomes of placode specification is the genesis of cranial sensory neurons. Chemosensory, auditory, proprioceptive, mechanoreceptive, and nociceptive neurons are generated from placode cells with distinct properties based upon transcriptional specification and subsequent programs of gene expression. These properties include: the acquisition of bipolar morphology with a “basal” receptive process (similar to a dendrite) specialized for sensory transduction, and an apical process (an axon) for transmitting information to the central nervous system. In addition, placode derived sensory neurons are capable of generating action potentials and/or vesicular neurotransmitter release, and thus must acquire a full range of molecular regulators of neuronal excitability. We will review data that indicates that these key neuronal properties, and therefore the specific identity of cranial sensory neurons, reflects transcriptional specification of placode precursors, definition of placode signaling centers, and subsequent interactions with adjacent neural crest derived mesenchymal cells. We will evaluate this issue by focusing on chemoreceptive olfactory sensory neurons that emerge from the olfactory placode during embryonic development. Olfactory receptor neurons are paradigmatic cranial sensory receptor neurons: they are found in all animals, with remarkably similar cellular and molecular properties (Axel, 2005; Hallem and Carlson, 2004; Buck, 2000).
Over the past two decades a number of highly conserved transcription and signaling factor genes have been identified that are expressed at these different steps of placode development, allowing us to begin to understand the molecular pathways that induce and specify the fate of these important cells. We will review what is known regarding three steps of placode development: initial induction and specification of the pre-placodal field; subdividing the field into region-specific cranial placodes; and differentiation of some of the specialized cells (olfactory receptor cells and cranial ganglion sensory neurons). We will also review the contribution of some of the key genes involved in placode development to human congenital syndromes that include craniofacial and auditory defects.
Induction and specification of the pre-placodal field
Based on several experimental approaches, it is now widely accepted that the cranial sensory placodes are derived from a common precursor field in the embryonic ectoderm (reviewed in Streit, 2004, 2007; Schlosser 2006, 2010; Patthey et al., 2014; Saint-Jeannet and Moody, 2014; Moody and Saint-Jeannet, 2014). Classic histological descriptions of cranial sensory placode formation identified the origin of all cranial placodes from a common precursor region called the pre-placodal ectoderm (PPE; also called the “pan-placodal region” or “common placodal field”), which is a U-shaped band of ectoderm that surrounds the anterior margin of the neural plate (von Kupffer, 1895; Platt, 1896; Knouff, 1935; reviewed in Schlosser, 2005) (Figure 1). Fate mapping studies in amphibians and chick also showed that all placodes originate from a common PPE field (Couly and LeDouarin, 1987; Couly and LeDouarin, 1990; Streit, 2002; reviewed in Schlosser and Ahrens, 2004; Streit, 2004; Pieper et al., 2011), and transplantation studies in frog and chick showed that the PPE is initially competent to give rise to all the different types of placodes (Jacobson, 1963; Groves and Bronner-Fraser, 2000). This pan-placodal competence appears to be regulated by a common molecular signature. At early stages of chick, frog, fish and mouse development, the entire PPE field expresses Six and Eya genes (Esteve and Bovolenta, 1999; Kobayashi et al., 2000; Pandur and Moody, 2000; David et al., 2001; Ghanbari et al., 2001; McLarren et al., 2003; Schlosser and Ahrens, 2004; Streit, 2004; Litsiou et al., 2005; Christophorou et al., 2009; Sato et al., 2010). In both frog and chick the expression domains of these pan-placodal genes overlap with the fate maps of the placodes (reviewed in Schlosser and Ahrens, 2004; Streit, 2004; Pieper et al., 2011), and these genes are required for the proper development of several placodes and their derivatives (reviewed in Saint-Jeannet and Moody, 2014; Moody and Saint-Jeannet, 2014, and discussed in detail below). Furthermore, studies that explanted the PPE into culture to reveal its developmental potential in the absence of tissue interactions showed that a PPE molecular “ground” state must first be attained before a specific placode identity can be achieved (Martin and Groves, 2005; Bailey et al., 2006). We will first address how the pan-placodal field arises, and discuss the transcriptional program that both specifies the PPE molecular state and maintains its boundaries.
Formation of the neural border zone
During gastrulation, the embryonic ectoderm is divided into two transcriptionally distinct fields: the neural ectoderm and the non-neural ectoderm (Figure 1). Interactions between these two fields and signals originating from the underlying mesodermal and endodermal tissues initiate the formation of an intervening zone of ectoderm with the potential to form the neural crest and the PPE, both of which make significant but distinct contributions to the peripheral nervous system. This neural border (NB) zone was experimentally confirmed by explanting pieces of neural plate into the epidermis; both neural crest-specific and PPE-specific genes were induced at the border between the neural plate explant and the host epidermis (Selleck and Bronner-Fraser, 1995; Selleck and Bronner-Fraser, 2000; Mancilla and Mayor, 1996; Woda et al, 2003; Glavic et al., 2004; Litsiou et al., 2005; Ahrens and Schlosser, 2005). Several studies have shown that the NB zone initially expresses a distinct set of genes, called “NB-specifying” genes, that are required for the later expression of neural crest-specific and/or PPE-specific genes (reviewed in Meulemans and Bronner-Fraser, 2004; Sargent, 2006; Park and Saint-Jeannet, 2010; Grocott et al., 2012). These include members of the Dlx, Msx, Pax and Zic families, as well as TFAP2α, GATA and Foxi genes. In most vertebrates examined, Dlx, Msx1, GATA, Foxi, and TFAP2α are expressed broadly in the non-neural ectoderm at blastula and gastrula stages, but their expression domains also overlap with the border of the neural ectoderm, which expresses SoxB1 and Zic genes (Figure 2) (reviewed in detail in Grocott et al., 2012; Groves and LaBonne, 2014). As the neural ectoderm thickens into an early neural plate, boundaries form between these expression domains: SoxB1 genes are confined to the neural plate, Zic genes span the lateral neural plate and medial NB zone, Msx1 and Pax3 become confined to the NB zone, and Dlx, GATA, Foxi and TFAP2α no longer overlap with neural ectoderm (Figure 2). At late neural plate/neurula stages, Msx1 and Pax3 become confined to the neural crest region, along with neural crest-specifying genes (FoxD3, Sox9/10, Snail2); in addition some Dlx genes and TFAP2α are newly expressed in the neural crest. The PPE expresses Six and Eya genes, and the epidermis expresses Dlx, GATA and Foxi genes (Figure 2). These changing expression patterns indicate that NB-specifying genes have dynamic roles in the transcriptional pathway that leads to PPE formation. What is the experimental evidence for their functions?
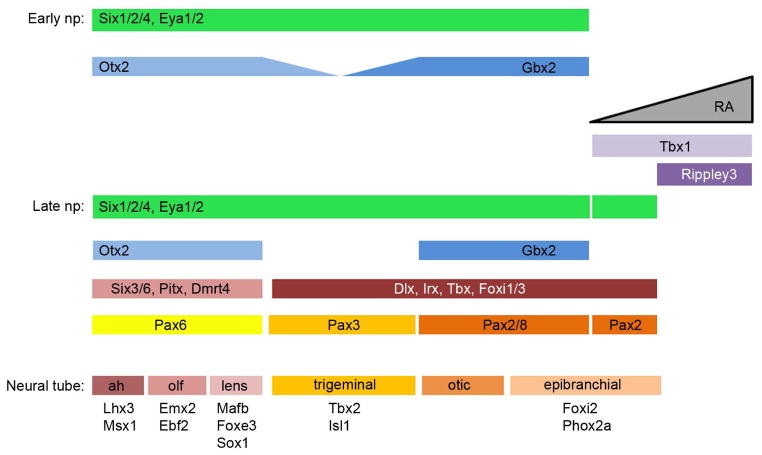
Figure 2.
Several steps are involved in forming the pre-placodal ectoderm.
Top: The embryonic ectoderm of a neurula stage has been flattened into a sheet. On the left is represented the fields apparent at early neural plate stages: epidermis (Epi), neural border zone (NB) and neural plate (NP). On the right the NB has divided into its neural crest (NC) and pre-placodal ectoderm (PPE) derivatives.
Bottom: Different sets of transcription factors are differentially expressed in these ectodermal domains over developmental time. At gastrula stages, the expression domains of neural plate genes (blue bar; SoxB1 and Zic) and epidermal genes (yellow bars; Msx1, Dlx, GATA, TFAP2, Foxi) overlap in a region that will become the neural border zone. At early neural plate stages, SoxB1 gene expression recedes from the NB zone, Zic genes (purple bars) are no longer expressed in the medial region of the neural plate, Msx1 and Pax3 expression (dark green bars) is confined to the NB zone, whereas Dlx, GATA, TFAP2 and are expressed in both the NB zone and epidermis (yellow bars). At late neural plate stages Six and Eya genes are expressed in the PPE domain (medium green bars) and “neural crest specifying” genes (Msx1, Pax3, FoxD3, Sox9/10, Snail2) are expressed in the neural crest domain (light green bars). Dlx3 and TFAP2 also are expressed in the neural crest domain, whereas Dlx5/6, GATA and Foxi1 are expressed in the epidermis (yellow bars), and are now excluded from the PPE. Once these four domains are formed, their boundaries appear to be maintained by mutual repression between domain-specific transcription factors (red bars).
Distal-less related homeobox transcription factors, in particular Dlx3, Dlx5, and Dlx6, have two important functions in the NB zone (Figure 3): they are required for the expression of both neural crest and PPE genes, and they repress neural plate genes (Feledy et al., 1999; Beanan and Sargent, 2000; Luo et al., 2001a; Solomon and Fritz, 2002; Woda et al., 2003; McLarren et al., 2003; Kaji and Artinger, 2004; Esterberg and Fritz, 2009). In the neural plate transplantation experiments described above, both neural crest and PPE markers were induced only when Dlx gene activity was intact (Woda et al., 2003). It has been proposed that Dlx factors promote PPE formation by regulating a BMP antagonist in the NB zone (Esterberg and Fritz, 2009), and/or by interacting with GATA factors (Solomon and Fritz, 2002; McLarren et al., 2003; Phillips et al., 2006; Pieper et al., 2012). However, Dlx and GATA factors are not equivalent in activity; while loss of either Dlx3 or GATA2 significantly reduces PPE gene expression, only Dlx3 can expand PPE gene expression (Pieper et al., 2012). A PPE-specific enhancer in the Six1 gene contains putative binding sites for both GATA factors and homeodomain (HD) containing factors such as Dlx (Sato et al., 2010). Mutating the GATA sites reduced expression of a Six1 enhancer-reporter construct, whereas mutating the HD sites eliminated it.
Figure 3.
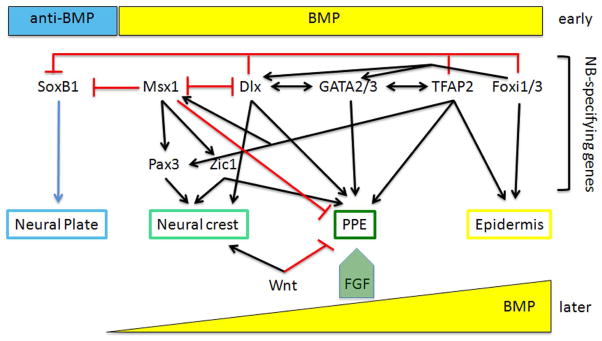
During gastrulation, early, high BMP levels activate several NB-specifying genes and low BMP levels, controlled by the secretion of BMP antagonists, allow the expression of neural genes (e.g., SoxB1 transcription factors). The NB-specifying genes interact with each other to promote (black arrows) the formation of the NB zone, and they repress (red bars) the expression of neural genes. Indicated are the various interactions, described in the text, that result in the separation of the NB zone into the neural crest and the pre-placodal ectoderm (PPE). Contributing to this separation are several signaling pathways. Later, high levels of BMP promote an epidermal fate, but this pathway must be attenuated to produce neural crest and PPE. An FGF pulse is required for PPE formation. Wnt postitively regulates neural crest formation and represses PPE formation.
Other experiments indicate that Dlx also functionally interacts with Msx1 (Figure 3). Dlx and Msx expression domains partially overlap in the NB zone (Arkell and Beddington, 1997; Feledy et al., 1999; McLarren et al., 2003; Schlosser and Ahrens, 2004; Monsoro-Burq et al., 2005), and these proteins are known to inhibit each other through the formation of heterodimers (Zhang et al., 1997; Givens et al., 2005). Differential knock-down of these genes bias NB zone cells towards either a neural crest fate (Msx-high, Dlx-low) or a PPE fate (Msx-low, Dlx-high) (Phillips et al., 2006). GST assays with the PPE-specific Six1 enhancer construct indicates that both Dlx5 and Msx1 can bind to the HD sites, and reporter assays indicate that Dlx5 activates Six1 whereas Msx1 represses it (Sato et al., 2010). Msx1 activates two other NB-specifying genes: Pax3 and Zic1 (Figure 3). Interactions between Pax3 and Zic1 are well known to initiate neural crest gene expression (Tribulo et al., 2003; Monsoro-Burq et al., 2005; Sato et al., 2005). By manipulating the timing of Pax3 expression with inducible constructs, Hong and Saint-Jeannet (2007) showed that Pax3 has an early role in forming the NB zone and a later role in neural crest specification when co-expressed with Zic1; alternatively, expression of Zic1 in the absence of Pax3 leads to PPE gene expression (Figure 3).
Foxi1 and TFAP2α also are required for both neural crest and PPE formation (Figure 3). Loss of Foxi1 in Xenopus embryos expands neural plate (Sox2) and reduces NB-specifying (Dlx), neural crest (FoxD3), PPE (Six1) and epidermis (keratin) genes; in gain-of-function experiments Sox2, FoxD3 and Six1 were reduced whereas Dlx and keratin were up-regulated (Matsuo-Takasaki et al., 2005). By utilizing a hormone-inducible version of Foxi1, these authors showed that it is the gastrula stage expression of Foxi1 that regulates NB zone formation. TFAP2α is a key regulator of epidermis genes (Nguyen et al., 1998; Luo et al., 2002; Luo et al., 2003; Zhang et al., 2006). Loss of TFAP2α down-regulates epidermis (keratin), NB-specifying (Msx1, Pax3), neural crest (FoxD3, Slug, Sox9), and PPE (Six1, Eya1) genes, and up-regulates neural plate genes; the opposite phenotypes are observed with gain-of-function experiments (Luo et al., 2002; Hoffman et al., 2007; Li and Cornell, 2007; de Croze et al., 2011). Epistasis analyses show that TFAP2α acts upstream of other NB-specifying genes, with Pax3 being a direct transcriptional target (Luo et al., 2002; de Croze et al., 2011).
These studies indicate that transcriptional interactions between the NB-specifying genes lead to a NB transcriptional state from which either neural crest or PPE cells can arise, depending upon the timing and combinations of genes that are expressed (Figures 2, 3). A recent study showed that a maintained expression of TFAP2α, Foxi1 and GATA in the NB zone is required for PPE formation, and this is accomplished by cross-regulation between these factors after local BMP signaling is attenuated (Bhat et al., 2013). Likewise, in human embryonic stem cell cultures, the expression of TFAP2α, GATA and DLX genes must precede the expression of PPE genes for placode cells to differentiate (Leung et al., 2013). Thus, the NB zone transcriptional state appears to set the stage for PPE-specific gene expression.
Induction of the PPE genes by signaling factors
After the NB zone is established, signals from the local environment induce a distinct set of PPE-specific transcriptional regulators, in particular members of the Six and Eya gene families (reviewed in Bhattacharyya and Bronner-Fraser, 2004; Schlosser and Ahrens, 2004; Streit, 2004; Grocott et al., 2012; Saint-Jeannet and Moody, 2014; Moody and Saint-Jeannet, 2014). Although the neural plate grafting experiments discussed above indicate that interactions between the neural and non-neural ectoderm are necessary for the induction of PPE genes, ablation and transplantation experiments also show that they are not sufficient. Experiments in chick and Xenopus showed that head mesoderm provides an additional, required, PPE-inducing signal (Litsiou et al., 2005; Ahrens and Schlosser, 2005). Both studies identified fibroblast growth factor (FGF) as the likely additional signal (Figure 3); FGF also promotes PPE gene expression in fish (Esterberg and Fritz, 2009; Kwon et al., 2010). Consistent with these findings, endogenous FGFs are expressed in the adjacent head mesoderm and anterior neural ridge (Shamim and Mason, 1999; Ohuchi et al., 2000; Eagleson and Dempewolf, 2002; Ahrens and Schlosser, 2005; Shim et al., 2005). However, several studies also showed that FGF signaling alone is not sufficient to induce PPE genes (Phillips et al., 2001; Maroon et al., 2002; Leger and Brand, 2002; Liu et al., 2003; Solomon et al., 2004; Litsiou et al., 2005; Ahrens and Schlosser, 2005). It appears that the level of FGF signaling is important: while low levels of FGF signaling promote PPE fate, high levels inhibit it (Brugmann et al., 2004; Hong and Saint-Jeannet, 2007). Grocott et al. (2012) propose that FGF promotes PPE formation by both inhibiting PPE-repressing factors and up-regulating PPE-specific gene expression. However, this transcriptional control may not be direct because two recently characterized PPE-specific enhancers of Six1, which are conserved across humans, mouse, chick and frog, do not contain any obvious FGF pathway binding sites (Sato et al., 2010; Sato et al., 2012). Study of other enhancers is warranted, therefore, to elucidate how the FGF pathway may regulate the expression of the Six1 gene, as well as other PPE-specific genes.
Bone morphogenetic protein (BMP) signaling plays important roles in both NB zone and PPE gene induction (Figure 3). Most NB-specifying genes are induced by BMP (Suzuki et al., 1997; Luo et al., 2001a; Luo et al., 2001b; Luo et al., 2003; Tribulo et al., 2003; Bhat et al., 2013). Maintained high BMP levels promotes an epidermal fate, whereas subsequent lower levels of BMP signaling, experimentally accomplished by expressing either dominant-negative BMP receptors or BMP antagonists, preferentially expand PPE gene expression domains (Brugmann et al., 2004; Glavic et al., 2004; Ahrens and Schlosser, 2004; Litsiou et al., 2005; Kwon et al., 2010). In fact, in order for FGF8 to induce PPE genes in the non-neural ectoderm the level of BMP signaling in the epidermis that activates the NB-specifying and epidermal genes, must be reduced (Ahrens and Schlosser, 2004). BMP levels also can distinguish between induction of neural crest versus PPE fate in Xenopus ectodermal explants (Brugmann et al., 2004), and BMP levels that induce a neural crest fate activate TFAP2α alone, whereas BMP levels that induce a PPE fate activate TFAP2α plus Foxi1 and Gata3 (Bhat et al., 2013). The timing of the BMP signaling also is critical (Figure 3). In fish, high levels of BMP at gastrula stages activate NB-specifying genes, whereas at neural plate stages lower levels of BMP are necessary to activate PPE genes (Kwon et al., 2010; Bhat et al., 2013). Similarly, human embryonic stem cells directed to a placode fate require an early pulse of BMP to activate NB-specifying genes, and a later attenuation of BMP signaling to activate PPE genes (Leung et al., 2013; Dincer et al., 2013).
Wnt signaling appears to antagonize PPE induction (Figure 3). In both chick and Xenopus embryos, elevated Wnt signaling represses PPE genes, whereas reducing it expands the PPE (Brugmann et al., 2004; Litsiou et al., 2005; Matsuo-Takasaki et al., 2005; Hong and Saint-Jeannet, 2007). Secreted anti-Wnt factors are highly expressed in the anterior neural plate and the underlying chordomesoderm (Bradley et al., 2000; Pera and De Robertis, 2000; Carmona-Fontaine et al., 2007; Takai et al., 2010), which may account for the restriction of PPE gene expression to the ectoderm surrounding the anterior neural plate (Figure 1). In chick NB zone explants, if Wnt signaling is attenuated in the presence of low BMP, placode markers are expressed, whereas if Wnt signaling persists with low BMP, neural crest markers are expressed (Patthey et al., 2008, 2009). In fact, it appears that a pulse of FGF in a BMP low/Wnt Low environment is the most effective means of inducing PPE genes (Litsiou et al., 2005). These authors propose that the FGF pulse confers a “neural border” state on the ectoderm, and that those cells within the NB zone that are protected from Wnt signaling become PPE whereas those that are exposed to Wnt signaling become neural crest. It is interesting to note that one of the conserved PPE-specific enhancers of Six1 contains a Wnt/Lef1 site (Sato et al., 2012); this suggests that Wnt regulation of Six1 may be direct but this site needs to be functionally tested.
Another important signal in PPE formation is retinoic acid (RA). Raldh2, the RA-synthesizing enzyme, is expressed in a discrete U-shaped ectodermal domain around the anterior neural plate that appears coincident with the PPE (Chen et al., 2001). Decreasing RA signaling during PPE formation expands the posterior limit of a similar U-shaped FGF8 expression domain in the cranial mesoderm (Shiotsugu et al., 2004), suggesting that endogenous RA contributes to limiting the PPE to the head (Figures 1, 4). This may be accomplished via the differential expression of two RA-regulated genes: Tbx1, a T-box transcription factor, and Ripply3/Dscr6, a Groucho-associated co-repressor (Arima et al., 2005). Ripply3 and Tbx1 expression domains in the PPE partially overlap; in regions where only Tbx1 is expressed, PPE genes are induced, whereas in regions where Tbx1 and Ripply3 overlap, PPE genes are repressed (Janesick et al., 2012). Thus, RA signaling appears to be required for the formation of the posterior, Tbx1-positive part of the PPE, and for restricting the posterior boundary of the PPE by co-inducing Ripply3 (Figure 4).
Figure 4.
A summary of the anterior-posterior (A-P) patterning of the PPE into anterior, intermediate and posterior domains. Anterior is to the left and posterior is to the right. At early neural plate (np) stages the anterior-specific expression of Otx2 and posterior-specific expression of Gbx2, and their mutual repression in the intermediate zone set the stage for A-P patterning of the PPE, which uniformly expresses Six and Eya genes. Subsequently, RA signaling that locally up-regulates Tbx1 and Ripply3 likely causes new Six/Eya expression in a more posterior domain that will give rise to the most posterior epibranchial (eb) placodes. At late neural plate stages, other transcription factors become restricted to either the anterior domain or the intermediate/posterior domains. Presumably these transcriptional combinations, together with further local signaling, results in region-specific Pax gene expression, leading to the specification of distinct placode fates. Subsequently each of these placodes (ah, adenohypophyseal; olf, olfactory; lens; V, trigeminal; otic; eb, epibranchial) expresses unique combinations of additional genes.
PPE transcriptional regulators
Six and Eya genes were among the first transcriptional regulators to be implicated in placode development because they are expressed in the classically described U-shaped PPE domain that surrounds the anterior neural plate (Figure 1) (Esteve and Bovolenta, 1999; Kobayashi et al., 2000; Pandur and Moody, 2000; David et al., 2001; Ghanbari et al., 2001; McLarren et al., 2003; Bessarab et al., 2004; Schlosser and Ahrens, 2004; Sato et al., 2010). Functional studies in several animals have since demonstrated that several members of these two gene families have important roles in the initial specification of the PPE and the formation and differentiation of many of its derivatives.
Drosophila Sine oculis (SO), which is essential for fly visual system formation (Cheyette et al., 1994; Serikaku and O’Tousa, 1994), is the founding member of the highly conserved Six (Sine oculis homeobox) gene family. There are 6 vertebrate Six genes that are clustered into two groups (Six1/Six4/Six6 and Six2/Six3/Six5) on two separate chromosomes; these are closely syntenic in frog, mouse and human and slightly rearranged in chick and zebrafish (Moody and Saint-Jeannet, 2014). The proteins are functionally grouped into three sub-families (Six1/Six2; Six3/Six4; Six5/Six6) based on sequence variations in both the homeodomain (HD), which binds to DNA, and the Six domain (SD), which binds to co-factors that regulate DNA binding specificity and protein activity (Pignoni et al., 1997; Kawakami et al., 2000; Kobayashi et al., 2001). Six genes play major roles in eye, muscle, kidney, genital, limb and craniofacial development (Kawakami et al., 1996; Brodbeck and Englert, 2004). Although Six3 plays an important role in lens differentiation (e.g., Oliver et al., 1996), we will not consider it further because it is not expressed in the PPE or in other placodes, and its function in lens development is discussed in detail elsewhere (Ogino et al., 2012). Instead, we will focus on the three members of the family (Six1, Six2, Six4) that are strongly expressed in the PPE, placodes, and numerous placode derivatives (reviewed by Brugmann and Moody, 2005).
Several experiments indicate that Six1 plays a central role in PPE and placode development. Six1 loss-of-function in Xenopus results in reduced expression of other early PPE markers (Eya1, Sox11, Sox2/3) (Brugmann et al., 2004; Schlosser et al., 2008). In another study that reduced Six1 activity by repressing Six1 targets, placode-specific gene expression likewise was lost, and the otic placode was malformed (Christophorou et al., 2009); in fish and mouse, Six1 knock-down also resulted in inner ear defects (Bricaud and Collazo, 2006; Bricaud and Collazo, 2011; Oliver et al., 1995; Laclef et al., 2003; Zheng et al., 2003; Ozaki et al., 2004; Zou et al., 2004; Konishi et al., 2006). Although Six1 is expressed in the mouse PPE (Sato et al., 2010), its loss only appears to affect later stages of placode development (e.g., Ikeda et al., 2007, 2010; Chen et al., 2009). This lack of an early phenotype may be due to gene redundancy because in contrast, the olfactory placode does not form in Six1/Six4 compound null mice (Chen et al., 2009). Gain-of-function experiments in both chick and frog demonstrate that Six1 up-regulates early-expressed PPE genes and represses epidermal, neural crest and neural plate genes (Brugmann et al., 2004; Christophorou et al., 2009). However, ectopic expression of Six1 outside the NB zone is not sufficient to induce other PPE markers (Brugmann et al., 2004; Schlosser et al., 2008; Christophorou et al., 2009), confirming that the appropriate signaling environment of the NB zone (Figure 3) is necessary to induce PPE gene expression.
Although Six2 and Six4 are frequently used as PPE and placode markers in a number of animals, their functional roles have not yet been described in any detail. Placode deficiencies have not been reported in either Six2-null or Six4-null mice (Ozaki et al., 2001; Self et al., 2006), but this may be due to redundant functions between the family members. It will be important to elucidate the cooperative roles of Six1, Six2 and Six4 in PPE and placode development, particularly because they are known to have distinct roles in muscle differentiation and kidney development (Ohto et al., 1998; Spitz et al., 1998; Fougerousse et al., 2002; Xu et al., 2003; Brodbeck and Englert, 2004; Himeda et al., 2004).
Six proteins can bind to co-factor proteins that do not bind to DNA on their own, but nonetheless modulate Six transcriptional activity (Zhu et al., 2002; Tessmar et al., 2002; Giot et al., 2003; Brugmann et al., 2004; Bricaud and Collazo, 2011). The best studied vertebrate Six co-factors are the four members of the Eya family; they are homologous to Drosophila Eyes absent (Eya), which plays an essential role in fly eye development as a SO co-factor (Bonini et al., 1993). Like the Six genes, vertebrate Eya genes are expressed in numerous embryonic tissues, including the eyes, muscles, kidneys, the PPE and placodes (Duncan et al., 1997; Xu et al., 1997; Sahly et al., 1999; David et al., 2001; Ishihara et al., 2008; Neilson et al., 2010; Modrell and Baker, 2012). In Xenopus, chick and fish, the expression patterns of Eya1 and Eya2 are nearly identical to that of Six1 (Sahly et al., 1999; David et al., 2001; McLarren et al., 2003; Bane et al., 2005; Neilson et al., 2010), suggesting that they have important roles in PPE and placode development. Eya proteins contain a highly conserved protein-binding domain called the Eya domain (ED) located at the C-terminus. In Drosophila, the ED interacts with the SD of the SO protein (Pignoni et al., 1997). In vertebrates, the interaction between the Six1 SD and the Eya1 ED is essential for Eya1 nuclear translocation and for exerting the transcriptional function of the complex (Ohto et al., 1999; Ikeda et al., 2002; Patrick et al., 2013). It should be kept in mind, however, that Eya1 can bind to several other proteins, so its function is likely not restricted to the Six1 transcriptional pathway (Heanue et al., 1999; Ohto et al., 1999; Giot et al., 2003; Kenyon et al., 2005). Eya also functions as a phosphatase (Rayapureddi et al., 2003; Tootle et al., 2003), which impacts whether the Six1 transcriptional complex activates or represses targets (Li et al., 2003), and as a substrate for mitogen-activated protein kinase (Hsiao et al., 2001). There have been several Eya1 loss- and gain-of function studies in Xenopus, fish and mouse (Johnson et al., 1999; Xu et al., 1999; Xu et al., 2002; Zou et al., 2004; Nica et al., 2006; Zou et al., 2006b; Li et al., 2010). Although defects are seen in various placode derivatives, to our knowledge no study has addressed the role of this protein in PPE formation. To date, Eya2-null mouse phenotypes have not been reported and while Eya3-null mice have behavioral abnormalities consistent with its regulation of neural plate cell survival in Xenopus (Kriebel et al., 2007), craniofacial deficits were not noted (Soker et al., 2008). Loss of Eya4 in fish and mouse results in inner ear defects (Schonberger et al., 2005; Depreux et al., 2008; Wang et al., 2008). Sorting out the distinctive and/or overlapping roles of the different Eya proteins will be important for understanding how these proteins contribute to the transcriptional regulations of placode development.
Another well-described interactor with Six proteins is Groucho. Six1/Six2 can act as both transcriptional activators and repressors, depending on the presence of Eya or Groucho co-factors (Silver et al., 2003). Using activating (Six1VP16) and repressing (Six1EnR) Six1 constructs, it was shown that Six1 transcriptionally activates PPE genes in cooperation with Eya1, and transcriptionally represses epidermal and neural crest genes in cooperation with Groucho (Brugmann et al., 2004; Christophorou et al., 2009). In the fish otocyst, Groucho is required (Bajoghli et al., 2005). Further, combined Six1/Eya1 activation of target genes is required for hair cell fate, whereas combined Six1/Groucho repression of target genes is required for neural cell fate (Bricaud and Collazo, 2011). Because both Eya and Groucho genes are endogenously expressed in the PPE, these data indicate that Six1 functions in PPE development as both a transcriptional activator and a repressor, depending on the co-factor with which it interacts.
Several lines of evidence indicate that there are likely to be additional modifiers of Six transcriptional activity that are developmentally relevant to PPE and placode development. We embarked on a screen of vertebrate homologues of reported novel fly SO-interactors (Giot et al., 2003; Kenyon et al., 2005) because the SD of fly SO differs by only four non-conserved amino acids from the SD of Xenopus and human Six1/Six2. We identified Xenopus genes with amino acid sequences homologous to 20/25 of the fly SO-interactors. We analyzed the expression patterns of the homologues of 11 of the fly genes, and found that most overlap extensively in the PPE and placodes with Six1 (Neilson et al., 2010). To date, functional data are only available for one of these: Sine oculis binding protein (Sobp). In fly, this protein is co-expressed with SO in the eye imaginal disc where its misexpression interferes with normal eye neurogenesis (Kenyon et al., 2005). In mouse, a recessive mutation in Sobp caused abnormal cochlear development (Chen et al., 2008), and in humans, a truncating mutation of SOBP caused craniofacial abnormalities in a family in which one member had hearing loss (Basel-Vanagaite et al., 2007; Birk et al., 2010). There also are likely to be protein regulators that modify Six1 function by binding to or modifying the activity of Eya (or other putative co-factors). For example, Dachshund (Dac), which has an important role in Drosophila eye development in cooperation with Eya and SO (Chen et al., 1997), binds to both Eya and DNA, but does not have a direct interaction with SO. Vertebrate Dac is expressed widely in embryonic tissues, including placodes (McLarren et al., 2003; Schlosser, 2006; Grocott et al., 2012), and can regulate the transcriptional effectiveness of Six/Eya complexes (Heanue et al., 1999; Ikeda et al., 2002; Li et al., 2003). However, a specific role for Dac or for other potential members of the Six/Eya transcriptional complex in PPE and placode development remains to be revealed.
Based on studies showing that kidney development is regulated by a transcriptional network that includes Six, Eya, Pax and Fox genes (reviewed in Brodbeck and Englert, 2004), it was proposed that a similar network might regulate sensory placode development (Bhattacharyya and Bronner-Fraser, 2004). At the time there was only sufficient evidence for the lens placode, so an update is warranted. As discussed above, Pax3 and Foxi1 are important for NB zone formation, and Six and Eya are critical for PPE and placode development. Explant studies suggest that a PPE-wide molecular ground state includes expression of Pax6 (Bailey et al., 2006), and, as we will discuss below, Pax genes are critical in conferring individual placode identity. Foxi1 and Foxi3 are important for otic placode development (Nissen et al., 2003; Solomon et al., 2003; Ohyama and Groves, 2004; Khatri and Groves, 2013), Foxe3 for lens placode development (Kenyon et al., 1999; Brownell et al., 2000; Ogino et al., 2012) and Foxg1 is expressed in all placodes in the mouse and plays a role in olfactory and auditory development (Hatini et al., 1999; Pauley et al., 2006; Duggan et al., 2008; Hwang et al., 2009; Kawauchi et al., 2009). Thus, there is little doubt that Six/Eya/Pax/Fox genes are important regulators in PPE and placode development. However, their epistatic and transcriptional relationships have not yet been clearly delineated. Furthermore, there are certain to be numerous additional members of the PPE/placode regulatory network. For example, Sox, Irx, and Tbx genes have been experimentally placed in the pathway (Brugmann et al., 2004; Schlosser and Ahrens, 2004; reviewed in Streit, 2004; Grocott et al., 2012). In Drosophila, SO is known to directly regulate several genes required for eye formation (reviewed in Jusiak et al., 2014) that also are involved in placode development. These include: eyeless (vertebrate Pax6; involved in PPE ground state and anterior placodes), dac (see above), atonal (vertebrate Atoh1-8; involved in placode neurogenesis), prospero (Prox1/2; involved in vertebrate lens development; Mizuno et al., 1999; Wigle et al., 1999), and hedgehog (vertebrate Shh, involved in adenohypophyseal placode development; reviewed in Saint-Jeannet and Moody, 2014). A ChIP-Seq analysis of SO binding to DNA isolated from developing fly eye-antennal imaginal discs identified nearly 6,000 putative SO target genes, over half of which do not have a described function in eye development (Jusiak et al., 2014). Although flies do not have the equivalent of cranial sensory placodes, we speculate that some of these genes may prove important in the transcriptional regulation of vertebrate placode development because of the significant conservation of many gene regulatory networks between flies and vertebrates. Using a different approach, we recently conducted an expression screen to identify potential new targets of Six1 in Xenopus ectodermal explants, and identified 72 up-regulated and 58 down-regulated genes (Yan et al., 2014). Like the fly study, most candidates are of unknown function, but over 30 of these genes are expressed in the PPE and placodes, suggesting they may be part of the placode transcriptional network. Functionally analyzing these new putative SO/Six1 targets in both flies and vertebrates is likely to significantly advance our understanding of the PPE regulatory state.
The PPE transcriptional regulators have two important roles. They set the transcriptional landscape for subsequent development into specific placodes with distinct fates (Figure 4; to be discussed below). Additionally, they maintain the boundaries between the PPE, neural crest and epidermis (Figure 2). Several studies have shown that the expression domains of transcription factors that are initially broad and overlapping become discrete stripes with distinct boundaries (reviewed in Schlosser and Ahrens, 2004; Streit, 2004; Schlosser, 2006; Groves and LaBonne, 2014; Moody and Saint-Jeannet, 2014). The formation of expression domains with sharp boundaries suggests that these factors are mutually repressive. There is a lot of experimental support for this idea. For example, in embryo gain-of-function assays Dlx5/6 and Zic2 both repress Six1, and Six1 represses Dlx5/6 and Zic2 (Woda et al., 2003; Brugmann et al., 2004). Epidermal, neural crest, and neural plate genes have mutually repressive interactions with Six1 (Brugmann et al., 2004; Matsuo-Takasaki et al., 2005; Christophorou et al., 2009). Thus, after the fates of the four major ectodermal sub-domains are specified and express unique sets of transcription factors, these same factors appear to continue as maintenance factors to preserve the boundaries between the sub-domains (Figures 2 and 3). However, experiments that control the timing and spatial localization of gene expression are needed to fully understand the molecular interactions that establish and maintain the boundaries between the different ectodermal sub-domains.
Breaking the PPE into individual placodes with different developmental fates
After the PPE is established as a separate sub-domain in the embryonic ectoderm with distinct boundaries from the other sub-domains (epidermis, neural crest, neural plate), the tissue undergoes several steps of regionalization (reviewed in Bailey and Streit, 2006; Grocott et al., 2012; Groves and LaBonne, 2014; Saint-Jeannet and Moody, 2014). Classical work in amphibians demonstrated that when pieces of PPE were transplanted to novel sites at early stages, they gave rise to placodes that were appropriate for their new A-P position, whereas when transplanted later they gave rise to placodes that were appropriate for their original A-P position (Jacobson, 1963). These results indicate that cells within the PPE are initially competent to form any placode, and only after interactions with adjacent tissues at specific A-P addresses do they become specified to particular placode fates (Jacobson, 1966). This idea has since been supported in chick, frog and fish by similar transplantation studies using molecular markers (Streit, 2002; Bhattacharyya et al., 2004; Toro and Varga, 2007; Xu et al., 2008; Ladher et al., 2010; Pieper et al., 2011). Although the early PPE is competent to form all types of placodes and has a unified molecular ground state, a few hours later it begins to acquire separate anterior (adenohypophyseal, olfactory, lens), intermediate (trigeminal) and posterior (otic, lateral line, epibranchial) domains (Figure 4). Fate mapping studies show that at late PPE stages precursors of anterior and posterior regions begin to sort out (Streit, 2002; Bhattacharyya et al., 2004; Dutta et al., 2005; Toro and Varga, 2007; Xu et al., 2008; Ladher et al., 2010; Pieper et al., 2011), concomitant with the differential expression of several sets of transcription factors that are placode-specific (Figure 4). What are the transcriptional and signaling factors that regulate this process? Similar to the program that regulates developmental patterning of the neural tube and the neural crest, the PPE appears to first undergo A-P patterning and then subsequent regionalization due to interactions with nearby signaling centers that maintain or repress specific sets of transcription factors.
One potential mechanism that accomplishes A-P regionalization of the PPE is the differential regulation of Six gene expression. An in-depth analysis of Six1 enhancers identified a genomic region that is conserved across tetrapods and activates reporter expression only in the anterior PPE (Sato et al., 2010). Although a posterior-specific enhancer was not identified, there are other conserved enhancers that are active in combinations of the PPE sub-domains: intermediate + posterior; adenohypophyseal + posterior; anterior + posterior (Sato et al., 2012). It will be very important to identify the factors that bind to these enhancers to determine the direct regulators of region-specific expression of Six1. Evidence from Xenopus implicates RA signaling for posterior Six1 expression. First, the most posterior lateral line and epibranchial placodes form posterior to the initial Six1/Eya1 PPE expression domain, and they only express Six1/Eya1 during neurulation (Schlosser and Ahrens, 2004). This later, posterior expression is likely to occur in response to RA regulation of Rippley3/Tbx1 expression described above (Janesick et al., 2012) (Figure 4). Another potential mechanism for A-P regionalization of the PPE is the differential expression of Otx2 and Gbx2 (Steventon et al., 2012). Otx2 is expressed in the anterior PPE domain, Gbx2 in the posterior PPE domain, and where their expression overlaps in the intermediate PPE domain both genes are down-regulated by their mutual inhibition (Figure 4). How this patterned expression is regulated is not yet understood, but there is evidence that differential responsiveness to BMP signaling may be involved (Sjödal et al., 2007). Also, there is evidence for differential responsiveness to Wnt signaling. For example, in axin1 mutant fish that harbor increased Wnt activity, the most anterior placodes are lost and the posterior placodes are expanded (Heisenberg et al., 1996). In Xenopus ectodermal explants, an anterior placode gene (Dmrt4) can be induced by co-expression of FGF8 and Noggin (an anti-BMP factor), but adding Wnt to the cocktail only induces a posterior placode gene (Pax8) (Park and Saint-Jeannet, 2008).
Other genes that are initially expressed broadly in the PPE also gradually become restricted to mostly anterior (e.g., Six3, Six6, Pitx3) versus mostly posterior (e.g., Dlx, Irx, Tbx, Foxi3) placodes (Figure 4) (Khatri and Groves, 2003; Schlosser and Ahrens, 2004; Schlosser, 2006; Sjödal and Gunhaga, 2008; reviewed in Grocott et al., 2012). This may be in direct response to differential expression of Otx2 and Gbx2. But, it is generally accepted that local signals from underlying mesodermal and endodermal tissues, including specific combinations of BMP, FGF, Wnt, and RA signaling, significantly contribute to placode-specific transcriptional identity (reviewed in detail in Baker and Bronner-Fraser, 2001; Schlosser and Ahrens, 2004; Streit, 2004; Schlosser, 2006; Saint-Jeannet and Moody, 2014). Some signaling pathways appear to be unique for certain placodes, including: Sonic Hedgehog (adenohypophyseal; Lewis et al., 1999; Treier et al., 2001; Herzog et al., 2003); Somatostatin/Nociceptin (olfactory, lens; Llera-Forero et al., 2013); Platelet Derived Growth Factor (trigeminal; McCabe and Bronner-Fraser, 2008) and Notch (otic and epibranchial; Abello et al., 2007; Jayasena et al., 2008).
One result of local signaling is region-specific induction of different members of the Pax gene family (Baker and Bronner-Fraser, 2001; Schlosser and Ahrens, 2004). As shown in Figure 4, Pax genes are differentially expressed: Pax6 in anterior placodes, Pax3 in the ophthalmic part of the trigeminal placode, and Pax2 and Pax8 in posterior placodes. In fact, the appropriate Pax genes are expressed in the PPE domains that fate map to these specific placodes prior to placode separation (Pieper et al., 2012; Grocott et al., 2012), suggesting that they are involved in initiating placode identity. Experimental results support this conclusion: transplantation experiments indicate that the onset of Pax expression correlates with the acquisition of placode identity (Baker et al., 1999; Baker and Bronner-Fraser, 2000); numerous loss-of-function studies show that the Pax genes are required for the development of these respective placodes (e.g., Hans et al., 2004; Mackereth et al., 2005; Dude et al., 2009; Shaham et al., 2012); and placode domains segregate by mutual repression between the Pax genes (Wakamatsu, 2011; reviewed in Grocott et al., 2012).
Regulation of cellular differentiation
Some genes that are broadly expressed in the PPE (e.g., Six, Eya, Dlx, Foxi, Irx), or are critical for setting up individual placode identity (Pax) have maintained expression in placodes as they differentiate. For example, Six and Eya genes are expressed in each neurogenic placode but repressed in the non-neural lens placode. This suggests that PPE genes have additional, later roles in regulating the differentiation of placode precursor cells into the distinct cell types found in the mature placode-derived organs. The transcriptional networks for some placodes are dealt with in detail elsewhere (adenohypophysis, Pogoda and Hammerschmidt, 2007; lens, Charlton-Perkins et al., 2011, Ogino et al., 2012; lateral line, Chitnis and Nogare, 2014). Herein, we focus on two programs of differentiation: the olfactory receptor neurons and the sensory neurons in the ganglia of the trigeminal and epibranchial cranial nerves.
Specifying Olfactory Receptor Neurons
The transcriptional and signaling mechanisms that establish the PPE, the NB zone and resulting placode subdomains result in a remarkable dual end-point: the definition of organizer regions that will drive subsequent sensory neuron differentiation (Richman and Tickle, 1989; 1992; LaMantia et al., 1993; 2000; Shou et al., 2000; Kawauchi et al., 2005), as well as the precursor populations whose fate will be determined by local signaling from those organizers. This key outcome for placode development is demonstrated dramatically by the olfactory placode, which emerges as a distinct ectodermal domain as initial generation of placodes draws to a close—approximately E9.0 in the mouse (Figure 5). Once the olfactory placodal ectoderm is established, a series of inductive events follows that drives olfactory epithelial (OE) and olfactory receptor neuron (ORN) differentiation, via signaling within the ectoderm as well as signaling between ectodermal and mesenchymal compartments.
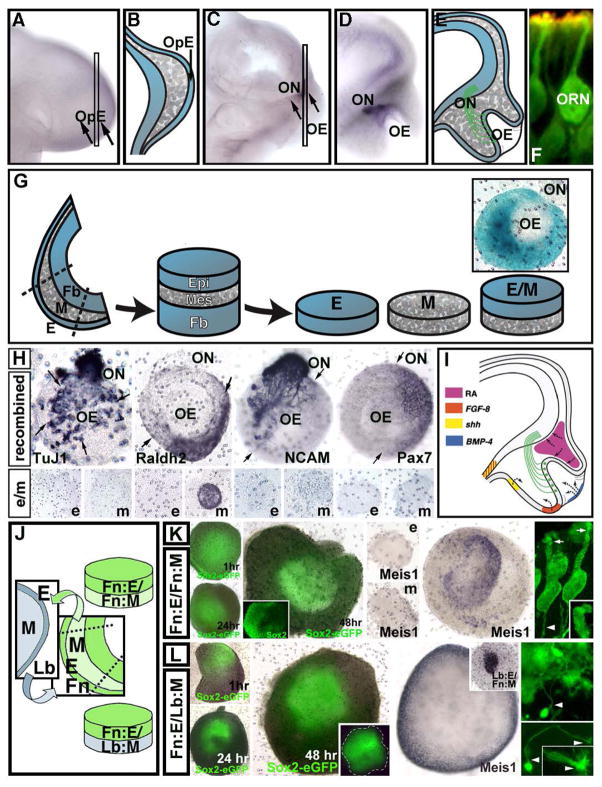
Figure 5.
The genesis of olfactory receptor neurons (ORNs) from the olfactory placodal ectoderm (OpE). Top row: Whole mouse embryos stained for the neuronal marker NCAM (which labels ORNs and axons in the olfactory nerve—ON), and schematics showing major structures that differentiate from the olfactory placode. A, B) The undifferentiated OpE at embryonic day (E)9.0 is a slightly thickened epithelial domain on the lateral/ventral surface of the head. C, D, E) Within 1.5 days, at E10.5, the OpE invaginates to form the olfactory epithelium (OE). ORNs are generated primarily in the medial OE, and their axons (ON, olfactory nerve) extend through the frontonasal mesenchyme (gray hatched regions in E) and enter the forebrain. F) An ORN at E10.5, labeled for the olfactory marker protein (green, a relatively specific marker), and the ORN selective adenylcyclase, ACIII (red), which labels the dendritic knob and associated cilia where odor receptor molecules are concentrated, and sensory transduction occurs. G) A schematic of the approach to isolate the olfactory placode at E9.0, separate the OpE (E), the frontonasal mesenchyme (Mes), and the forebrain neuroepithelium (Fb), and recombine them in vitro to assess OpE/ORN differentiation. The inset (right) shows a recombined OpE/frontonasal mesenchyme explant with mesenchyme from a ROSA-26 gene trap mouse that labels all cells with β-galactosidase (blue), and OpE from a WT embryo. There is no movement of cells from the mesenchyme into the OpE over 48 hours in vitro; however, there is growth of olfactory axons (ON) and cells that migrate with them. H) The top row shows recombined explants (OpE and fronto-nasal mesenchyme) labeled for molecular markers for differentiating ORNs (TuJ1 and NCAM), a synthetic enzyme for retinoic acid (Raldh2) and Pax7, which is restricted to the lateral mesenchyme. The expression patterns and intensity of each marker is parallel to that seen in the differentiation of the olfactory placode/OE in vivo at E11. The bottom row shows that the differentiation of the OE, and expression of relevant markers, with the exception of Raldh2, depends upon mesenchymal/epithelial interactions—epithelium (e) nor mesenchyme (m) expresses any of the other markers when isolated. I) A schematic showing the role of signaling sources of Fgf8, Bmp4, Shh and RA, established via earlier placodal transcriptional mechanisms, in establishing OE neurogenesis and ON trajectory. J) Schematic of mesenchyme “swapping” experiments in vitro to assess specificity of OpE/frontonasal mesenchyme interactions. K) Isolated E9.0 frontonasal OpE (Fn:E) recombined with frontonasal mesenchyme (Fn:M) leads to the restriction and patterned expression of Sox2 in the OpE/OE over 48 hours. The same explant is shown here, imaged at the times indicated. The inset shows the medial (high) to lateral (low) pattern of Sox2 expression facilitated by Fn:E/Fn:M interactions. This pattern is key for specifying ORN stem cells. In addition, Fn:E/Fn:M interactions induce Meis1 in a population of cells that additional evidence (Tucker et al., 2010; Murdoch et al., 2010) shows are likely OE neural stem cells. At far right, these patterning events yield cells in Fn:E/Fn:M explants that have all of the morphological hallmarks of ORNs. In addition, these cells have physiological properties seen in ORNs (Rawson et al., 2010). L) When OpE (Fn:E) is recombined with limb bud mesenchyme (Lb:M), Sox2 is not patterned, and Meis1 expression is seen in the mesenchyme rather than OpE. The OpE still generates neurons (far right) in this heterologous pairing of E and M; however, these neurons lack apical dendrites, their axons cannot enter the limb bud mesenchyme and instead end in elaborate growth cones (arrow, lower right panel), and they lack the physiological hallmarks of ORNs (Rawson et al., 2010).
Placode specification alone—establishment of the PPE and subsequent definition of distinct placodes—does not yield a population of neural stem cells capable of generating ORNs and related neuronal classes (including the GnRH neurons that are generated in the olfactory placode and migrate to the hypothalamus during early embryogenesis; Wray, 2010). Separating the olfactory placodal ectoderm from the underlying neural crest derived mesenchyme at E9.0 in the mouse prior to initial morphogenesis and neurogenesis in the olfactory placode, results in a failure of OE and ORN differentiation in vitro (LaMantia et al., 2000; Bhasin et al., 2003; Rawson et al., 2010). Similarly, mutation of Pax6 disrupts the migration of neural crest derived mesenchyme as well as the capacity of placodal ectoderm to generate ORNs (Grindley et al., 1994; Anchan et al., 1996; LaMantia et al., 2000). In contrast, when olfactory placodal ectoderm and neural crest-derived frontonasal mesenchyme are apposed to one another, a fully patterned OE with a coherent olfactory nerve emerges (Figure 5). This patterning includes, critically, the restriction and up-regulation of Sox2 in the placodal ectoderm, as well as the induction of Meis1, which identifies a population of slowly dividing precursors in the nascent OE (Figure 5). Over the next 48 hours, the transcriptional profile of precursor cells in the OE diversifies further. Key factors that influence specification of the PPE, including Six1 and Sox2 remain in subsets of OE progenitors. The putative stem cells, found primarily in the lateral portion of the OE, express high levels of Meis1, Pbx1/2/3 and Pax7, low levels of Sox2 and high levels of Six1 (Ikeda et al., 2007, 2010; Chen et al., 2009; Tucker, 2010). In contrast, the presumed transit amplifying cells, found in the medial portion of the OE, express reversed levels of Sox2 (high) and Six1 (low). Pax6 is expressed throughout the medial region, and subsets of cells express bHLH neurogenic genes (Ascl1, Ngn1, NeuroD1). Differentiating ORNs and GnRH neurons also are found in the medial portion of the OE. Thus, the placodal ectoderm established by E9.0 in the mouse as a fairly uniform transcriptional field acquires within two days a stunning degree of transcriptional diversity that parallels precursor classes that have the capacity to generate ORNs and other OE neuronal classes.
It seemed likely that the key facilitator of this rapid and dramatic acquisition of cellular diversity and identity were signals exchanged between the placodal ectoderm—especially the “organizer” domains established as the placodal ectoderm is defined—and the adjacent neural crest-derived mesenchyme that accumulates between E8.0 and E9.0 in the mouse. FGF and BMP domains that emerge during early placode formation are potential sources of signals for further cellular differentiation. Their action on the underlying mesenchyme likely drives the capacity of the mesenchyme to support further signaling to the OE. Accordingly, we asked whether the epithelial and mesenchymal tissues that constitute the nascent olfactory primordium (which will form not only the OE and ORNs, but cartilaginous structures of the nose itself) during early gestation in the mouse (E9.0) interact via cardinal morphogenetic signals, particularly FGFs (LaMantia et al., 2000; Kawauchi et al., 2005; Balmer et al., 2005; Tucker et al., 2010; Lassiter et al., 2014), BMPs (Shou et al., 2000; LaMantia et al., 2000), and RA (LaMantia et al., 1993; LaMantia et al., 2000; Bhasin et al., 2003) to facilitate ORN differentiation. Our results are quite clear: these signals, available from distinct zones in the olfactory placodal ectoderm and frontonasal mesenchyme, influence axial patterning and neuronal differentiation in the OE (Figure 5).
The identity of OE stem cells that generate the initial embryonic OE neuronal lineage remained uncertain until recently. For several decades, the location and cellular characteristics of embryonic OE stem cells—slowly, symmetrically dividing stem cells in a specific location or niche within the OE that have the capacity to generate all differentiated cell types of the mature tissue—were undefined. Initial localization studies indicated that progenitors expressed neuronal bHLH transcription factors, including Ascl1, NeuroG1, and NeuroD1, which were important for OE neurogenesis. From earliest observations (Guillemot et al., 1993) through several studies of loss-of-function mutants (Cau et al., 1997; 2000; 2002; Murray et al., 2003; Tucker et al., 2010; Krolewski et al., 2012) it was clear that Ascl1 and the other bHLH genes were important for the expansion of the numbers of ORNs and other OE cell classes. Nevertheless, in the absence of Ascl1, ORNs are still produced. Indeed, every neuronal and non-neuronal cell type found in the OE can be identified in these mutants (Guillemot et al., 1993; Tucker et al., 2010) although their numbers appear to be dramatically diminished.
Finally, it was not clear how the specificity of sensory neuron identity was established from distinct placodal domains. For example, trigeminal proprioceptive and mechanoreceptive neurons are also derived from placodal ectodermal precursors that express Six1 and Sox2; however, these cells delaminate from the placode, and coalesce with adjacent neural crest cells that become nociceptive sensory neurons, rather than retaining a mesenchymal identity (as is the case for those adjacent to the OE). We found that the anterior-posterior identity of the mesenchyme is essential for establishing specific sensory neuron identity. When the limb bud mesenchyme, which contains several of the same molecular constituents found in frontonasal mesenchyme, is apposed to the nascent OE, neurogenesis occurs. These neurons, however, lack the functional properties of ORNs, and their axons cannot penetrate the basal lamina between the anomalously induced OE and the underlying heterologous mesenchyme (Figure 5; Rawson et al., 2010). Similar frontonasal mesenchymal cues also specify the generation of GnRH neurons (Schwarting et al,. 2001; Gamble et al., 2005; Messina et al., 2011; Cariboni et al., 2011; Parkash et al., 2012); these cells do not differentiate when olfactory placodal ectoderm is induced by limb mesenchyme.
Together, these observations on the initial differentiation of ORNs from the olfactory placodal ectoderm establish two key points. First, through regulated expression of Six1, Sox2 and other Six/Sox genes, the placodal ectoderm acquires the capacity to generate neurons; however, that capacity is only realized, and neuronal identity is specified by, interactions with adjacent neural crest-derived mesenchymal cells. Second, the distinct zones of FGF, BMP and other signaling factors established at placodal ectoderm boundary regions are not only essential for defining placodal limits; they are also critical for providing signals that drive cranial sensory neuron differentiation, including for special sensory receptor neurons like ORNs. The relationship between the signaling molecules, which are shared by many placodal domains, and the distinctive sensory neuron classes that emerge from each placodal region remain unknown. Nevertheless, it is clear that the transcriptional and signaling mechanisms that define the cranial placodes specify local neural ectodermal progenitors whose subsequent fate depends upon interactions with the underlying neural crest-derived mesenchyme, orchestrated by signaling domains established as placodal identity emerges.
Cranial ganglion sensory neurons
The trigeminal and epibranchial placodes are specialized niches that produce the large neurons in the sensory ganglia of the trigeminal (V), facial (VII), glossopharyngeal (IX) and vagus (X) cranial nerves. Unlike the neural crest cells that contribute the small neurons and glia to these ganglia, the placode-derived neurons are specified within the placode, delaminate from it and migrate to the coalescing ganglion as post-mitotic neurons (Graham et al., 2007). The signaling pathways involved in the neurogenesis in these placodes have recently been reviewed in detail (Lassiter et al., 2014). In general, the Notch pathway plays a critical role in selecting which cells in the neurogenic placode field will become neurons, the FGF pathway regulates delamination, the Wnt pathway contributes to delamination, Pax and bHLH gene expression, and the BMP pathway regulates bHLH and other neural differentiation genes.
Prior to delamination of these neurons, however, PPE factors (e.g., Six1) and SoxB1 neural stem cell factors (e.g., Sox2, Sox3) play important roles in regulating neurogenesis. Six1 expression is maintained in the outer, proliferative layer of the placode and is down-regulated as cells delaminate from the epithelium and coalesce into ganglia (Pandur and Moody, 2000; Schlosser et al., 2008). SoxB1 genes, which are important in maintaining neural stem cells in the central nervous system (reviewed in Bergsland et al., 2011; Wegner, 2013; Moody et al., 2013; Thiel, 2013), are expressed in localized regions of the PPE after Six and Eya genes but before individual placodes segregate (Schlosser and Ahrens, 2004). In the cranial nerve placodes, SoxB1 genes are expressed at high levels in the inner layer of the placode ectoderm (Schlosser et al., 2008) (Figure 6). bHLH transcription factors (e.g., NeuroG, NeuroD) promote the generation of neural progenitors, cause them to exit the cell cycle, and promote neuronal differentiation in both the central nervous system and in the neurogenic placodes (reviewed in Schlosser, 2006; Castro and Guillemot, 2011). In Xenopus cranial nerve placodes, the expression of NeuroG and NeuroD is complimentary to that of Six1 (Schlosser and Northcutt, 2000; Schlosser and Ahrens, 2004; Schlosser et al., 2008). NeuroG is first expressed in the inner placode layer and later in the delaminating neural progenitor cells; NeuroG expression is lost as the coalescing progenitor cells differentiate into neurons. NeuroD is expressed later than NeuroG in scattered cells within the inner placode layer, and it remains expressed in most of the placode-derived ganglion cells. These expression patterns suggest that Six1 maintains placode cells in a proliferative, undifferentiated “precursor” state, SoxB1 factors transition these cells to a neural stem cell state, and bHLH factors specify neural progenitors (with Notch pathway input; Lassiter et al., 2014) and regulate their differentiation into the sensory neurons of the cranial ganglia (Figure 6).
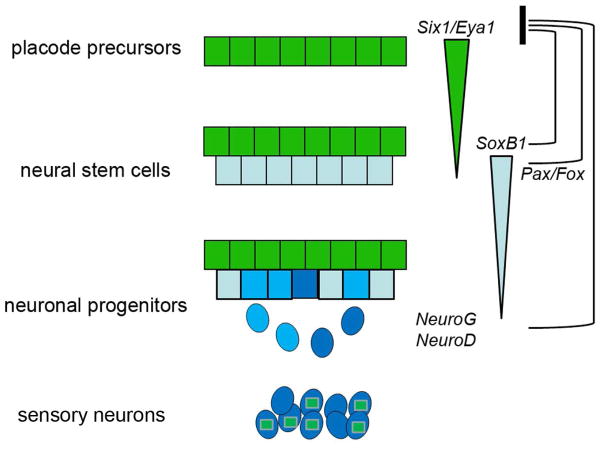
Figure 6.
A model of the gene regulatory pathway proposed to control the onset of neurogenesis in neurogenic placodes that form sensory ganglia of cranial nerves V, VII, IX and X. Six/Eya maintain proliferative, undifferentiated placode precursors (dark green). Six/Eya are required for the expression of SoxB1 genes in neural stem cells (light blue), which are only detected in the deeper-positioned cells in the placode once Six/Eya protein levels decrease. SoxB1 genes are required for the expression of bHLH neural progenitor genes (darker blue), which are only detected once the levels of SoxB1 protein decrease. NeuroG (medium blue) is detected in the deeper layer of the placode and in delaminating cells. Later, NeuroD (dark blue) is detected in these same cells and in the sensory neurons in the coalescing ganglion. These neurons re-express Six/Eya genes (green squares), which are required for cell survival. Several studies indicate that the genes expressed at each step of the process, including Pax and Fox genes that are involved in placode identity, feedback (black bars) to negatively regulate Six/Eya genes, thus promoting differentiation.
What experimental observations place Six1 upstream in this pathway? First, knockdown experiments in Xenopus show that Six1 (and Eya1) are required for the expression of SoxB1 genes, bHLH genes and differentiated neuron markers (Schlosser et al., 2008). In mouse, bHLH factors also appear to be regulated by Six1 and Eya1 in neurogenic placodes. For example, In Eya1-null neurogenic placodes, NeuroG- and NeuroD-positive cells are depleted, delamination is blocked, and sensory neuron differentiation markers (Phox2a/b, SCG10) are not expressed (Zou et al., 2004). Similar, but weaker phenotypes are seen in Six1-null embryos (Zou et al., 2004), perhaps due to the remaining activity of Six2 and Six4 (Konishi et al., 2006). Conversely, gain-of-function assays in Xenopus indicate that the level of Six1/Eya1 expression is critical for their function in the neural differentiation pathway. Increasing the levels of Six1/Eya1 in the neurogenic placodes increases proliferation, resulting in a larger domain of SoxB1-expressing neural stem cells, but fewer bHLH-expressing neural progenitors (Schlosser et al., 2008). Lowering the levels of Six1/Eya1 promotes the transition of SoxB1-positive cells to bHLH-positive cells. These observations indicate that Six/Eya genes maintain a proliferative precursor cell, but for neural differentiation to occur they need to be down-regulated.
In other systems Six genes also keep precursor cells in a proliferative state prior to cell type differentiation. The loss of Six1 in mice appears to decrease proliferation, which results in apoptosis (Li et al., 2003; Ozaki et al., 2004). In humans, SIX1 overexpression was identified in hyper-proliferating cell populations (e.g., primary breast cancers and metastatic lesions) (Ford et al., 1998). Six1 overexpression also influences cell proliferation by directly activating the transcription of CyclinA1 (Coletta et al., 2004), indicating that Six1 may maintain cells in an immature state by influencing cell cycle regulation. In fact, reactivating Six1 in adult tissues leads to mis-regulated cell proliferation and a number of human cancers (Li et al., 2013; Patrick et al., 2009; Patrick et al., 2013).
Six genes have an even later role in neurogenesis in the cranial nerve placodes. Six1 and Six4 are secondarily expressed in subsets of differentiating sensory neurons in the cranial ganglia (Konishi et al., 2006), and gene knockout studies show that they are required to prevent apoptosis. Six1-null, Eya1-null and Six1/Six4-null mice have small trigeminal, vestibulocochlear and epibranchial sensory ganglia (Xu et al., 1999; Zheng et al., 2003; Zou et al., 2004; Konishi et al., 2006). During embryonic stages these ganglia begin to form, but the loss of Six1 and Six4 leads to cell autonomous apoptosis of newly formed cranial ganglion neurons. It is proposed that Six genes contribute to sensory neuron survival by regulating the expression of the anti-apoptotic gene Bcl-x (Konishi et al., 2006).
The description of the transcriptional path that leads to cranial neuron formation is far from complete. First, there are likely to be other neural stem cell genes involved in this process. For example, neither Sox2 nor Sox3 is expressed in Xenopus trigeminal placodes (Schlosser and Ahrens, 2004), whereas Sox11 is expressed in the PPE and is up-regulated by Six1 (Brugmann et al., 2004). Second, the critical levels of transcription factors for varying phenotypes has not yet been explored rigorously in ganglion neuron differentiation; the use of null mutants may not be clinically relevant for human syndromes in which only one allele of Six1 or Eya1 is affected. Third, studies in mouse indicate that the Six1-21 enhancer functions to integrate input from Sox, Pax, Fox, and bHLH factors (Sato et al., 2012), suggesting a transcriptional mechanism by which Six1 expression can be down-regulated to permit differentiation to proceed (Figure 6). This now needs to be experimentally tested. Finally, not all cranial ganglia are equally affected by the loss of Six/Eya genes (Zou et al., 2004), suggesting A-P positional input into the regulatory program. In the future it will be important to identify all the factors involved in this process and determine precisely how they interact to regulate cranial nerve placode neurogenesis. We expect that the late steps in this process will be very similar to neural plate and neural crest neurogenesis, but the regulatory inputs will obviously be placode specific (e.g., Grocott et al., 2012).
NB zone, PPE and placode genes involved in human congenital syndromes
In humans, craniofacial anomalies and congenital hearing loss are among the most common developmental defects. As expected from their roles in placode specification and differentiation, mutations in NB-specifying, PPE and placode genes often are associated with congenital syndromes characterized by craniofacial and auditory phenotypes (Table 1). The craniofacial defects are likely caused by perturbations in the neural crest derivations of the NB zone, which give rise to the skeleton and connective tissue of the face; the hearing deficits are likely to be caused by perturbations in the otic placode derivative of the NB zone. At first glance it might be surprising that the phenotypes associated with each syndrome often extend beyond craniofacial and auditory tissues (Table 1), but it is well established that these genes are also expressed in other tissues later during development. Conversely, it is remarkable that the phenotypes are not more severe considering that work from experimental model systems show that these genes have critical roles in placode development. However, it must be kept in mind that many of the mutations identified in the human patients are only on one allele of the involved gene, whereas experimental gene knockdowns are often complete nulls. It is interesting that hearing loss is the dominant placode deficit identified in humans. This is a phenotype that can easily be clinically tested, whereas the consequences of disruptions in olfactory and cranial sensory neurons might be subtle. Nonetheless, correlating the anatomical phenotypes in humans carrying mutations with the experimental animals helps confirm the predicted roles of these genes in patients. Furthermore, it is very useful to identify the causative genes for hearing loss, because genetic screening might identify affected infants early enough for intervention. For a full description of the syndromes summarized below, please refer to the Online Mendelian Inheritance in Man website (OMIM.org).
Table 1.
Human congenital syndromes associated with NB-specifying and PPE genes and craniofacial defects
| Name of syndrome | Location | ||
|---|---|---|---|
| MSX1 | Ectodermal dysplasia Orofacial cleft Tooth agenesis |
4p16.2 | Defects in hair, nails, teeth and/or sweat glands Cleft lip/palate Lack of a tooth |
| PAX3 | Cranio-facial-deafness-hand syndrome Waardenburg syndrome |
2q36.1 | Flat facial profile, craniofacial abnormalities, sensorineural hearing loss Pigment abnormalities, sensorineural hearing loss, craniofacial abnormalities |
| DLX3 | Ameliogenesis imperfect Trichodontoosseous syndrome |
17q21.33 | Hair, tooth, and bone defects |
| DLX5 | Split-hand/foot malformation 1 | 7q21.3 | Defects in hands and feet; sensorineural hearing loss |
| GATA2 | Emberger syndrome | 3q21.3 | Congenital deafness, lower limb lymphedema, leukemias |
| GATA3 | Hypoparathyroidism with sensorineural deafness syndrome (HDRS) | 10p14 | Hypoparathryoidism, sensorineural deafness, renal dysplagis |
| FOXI1 | Enlarged vestibular aqueduct | 5q35.1 | Sensorineural and mixed hearing loss |
| TFAP2-α | Branchio-oculo-facial (BOFS) | 6p24.3 | Branchial clefts, dysmorphic face, external and middle ear anomalies w/ conductive deafness |
| SIX1 | Branchio-otic syndrome 3 (BOS3) Deafness, autosomal dominant 23 (DFNA23) |
14q23.1 | Branchial arch defects, deafness, lacrimal duct stenosis. Hearing loss |
| EYA1 | Oro-facial-cervical syndrome Branchio-otic Syndrome 1(BOS1) Branchio-oto-renal syndrome 1 (BOR1) |
8q13.3 | Facial anomalies, low-set ears, preauricular fistulas, hearing loss, skeletal defects. Hearing loss, structural defects in outer, middle and inner ear, brachial fistulas. BOS with renal defects |
| EYA4 | Deafness, autosomal dominant 10 Cardiomyopathy, dilated, 1J |
6q23.2 | Progressive hearing loss Dilated cardiopathy, sensorineural hearing loss |
| PAX6 | Aniridia Various ocular syndromes Peters anomaly |
11p13 | Defects in the iris of the eye Defects in the neural retina Defects in cornea, lens, and iris |
| OTX2 | Micropthalmia, syndromic 5 Pituitary hormone deficiency, combined, 6 |
14q22.3 | Small, dysmorphic retinas Hypoplasia of anterior and/or posterior pituitary |
| TBX1 | Conotruncal anomaly face syndrome DiGeorge syndrome Tetralogy of Fallot Velocardiofacial syndrome |
22q11.21 | Cardiac anomalies, facial anomalies Defects in thymus, cardiac outflow tracts, facial anomalies Cardiac anomalies Cardiac anomalies, cleft palate, facial anomalies |
| PAX2 | Papillorenal syndrome Renal hypoplasia |
10q24.31 | Ocular and renal anomalies; high frequency hearing loss Renal agenesis |
| PAX8 | Congenital hypothyroidism | 2q13 | Thyroid dysgenesis |
Information from the Online Mendelian Inheritance in Man website (OMIM.org).
Mutations in NB-specifying genes often result in defects that likely are caused by perturbations in both neural crest and placode development. Defects in MSX1 cause ectodermal dysplasia, tooth agenesis and orofacial clefting, phenotypes that are associated with its dominant role in neural crest development. Mutations in PAX3 are associated with Waardenburg syndrome and Craniofacial-deafness-hand syndrome. Waardenburg patients have pigment cell defects (neural crest) and both syndromes are characterized by craniofacial defects (neural crest) and hearing loss (placode). Mutations in DLX3 cause two syndromes that have hair, tooth (neural crest) and bone defects, whereas mutations in DLX5 result in split-hand/foot malformation 1 syndrome, patients of which have hearing loss (placode). Mutations in GATA2 are mostly associated with leukemias, as this gene plays a prominent role in hematopoiesis. But one form, called Emberger syndrome, also has hearing loss (placode); mutations in GATA3 also can cause deafness (placode). Mutations in FOXI1 cause Enlarged Vestibular Aqueduct syndrome, characterized by hearing loss (placode). Mutations in TFAP2α cause Branchio-oculo-facial syndrome; these patients have craniofacial dysmorphologies (neural crest) and hearing deficits (placode).
PPE gene mutations also are causative of a few human congenital syndromes (Table 1). SIX1 mutations underlie one form of Branchio-otic syndrome (BOS3), whose phenotypes include craniofacial defects and hearing loss (Ruf et al., 2004). Nine mutations in BOS3 patients from 16 unrelated families have been reported to date; seven are missense mutations in the SD and two are missense or deletion mutations in the HD (Ruf et al., 2004; Ito et al., 2006; Sanggaard et al., 2007; Kochhar et al., 2008; Noguchi et al., 2011). The mutations either disrupt the Six1-Eya1 interactions or the ability of Six1 to bind to DNA (Patrick et al., 2009). In zebrafish, expression of mutant Six1 mRNA that carries a BOS patient mutation (R110W) interferes with the Six1/Eya1 interaction that promotes cell proliferation and hair cell formation (Bricaud and Collazo, 2011). The Catweasel (Cwe) mouse mutant is thought to be a good model for the related Brachio-otic-renal syndrome (BOR), which is similar to BOS but also includes kidney defects, because it harbors a missense mutation in the SD that is similar to at least one BOR family (Bosman et al., 2009; Mosrati et al., 2011). Heterozygous-Cwe mice have an ectopic row of hair cells in the cochlea, and homozygous-Cwe mice do not have hair cells in either the cochlea, semicircular canals or utricle. To our knowledge, no human syndromes have been assigned to mutations in SIX2 or SIX4, but one affected locus in BOS patients contains the Six1, Six4, and Six6 genes (Ruf et al., 2004).
Patients diagnosed with BOS1 and BOR1 harbor EYA1 mutations (Abdelhak et al., 1997; Kumar et al., 1997; Rodriguez-Soriano, 2003; Spruijt et al., 2006); cataracts of the lens also have been associated with EYA1 mutations (Azuma et al., 2000). Often the defects in the protein lie in the ED, where they interfere with the interaction between Eya1 and Six proteins (Buller et al., 2001; Ozaki et al., 2002). Partial deletions of EYA1 that include other genes cause Oto-facio-cervical syndrome, which includes both craniofacial and hearing defects (Rickard et al., 2001; Estefania et al., 2006). To date, no mutations in EYA2 or EYA3 have been reported in humans. However, mutations in EYA4 are involved in Autosomal Dominant Sensorineural Deafness 10 (DFNA10; Wayne et al., 2001; Makishima et al., 2007) and Dilated Cardiomyopathy with Sensorineural Hearing Loss, Autosomal Dominant (CMD1J (Schonberger et al., 2005), implicating defects in the otic placode. The described mutations, which include small insertions, amino acid substitutions and large deletions, are all predicted to truncate the EYA4 protein within the ED, which is required for interactions with SIX proteins.
Mutations in genes that lie downstream of SIX/EYA in the placode development pathway have fewer identifiable craniofacial and hearing phenotypes. While Irx and Gbx2 have important roles in placode development (Figure 4), no human congenital syndromes have been associated with these genes so far. Although Pax6 is an important early marker of the PPR “ground” state (Bailey et al., 2006), mutations in PAX6 are associated primarily with ocular defects, and a small number of patients have lens defects. Likewise, although patients with PAX2 mutations have high frequency hearing deficits, the major abnormalities are found in the kidneys. PAX8 mutations are associated with thyroid dysgenesis without any reported hearing loss. Mutations of TBX1 cause a large number of cardiac defects and patients have craniofacial anomalies (neural crest), but no obvious placode-derived defects. OTX2 mutations cause eye defects as well as defects in both the anterior (placode) and posterior (neural) pituitary. Perhaps because these genes play overlapping roles in placode differentiation, mutation in one is compensated by the activities of the other genes in the regulatory network.
Conclusions
Placodes contribute to important secretory cells and cranial sensory organs that are critical for the normal behavior of an animal. Many of the genes involved in placode development are highly conserved from invertebrates to vertebrates, indicating that the underlying transcriptional pathways are evolutionarily very old (Bassham and Postlthwait, 2005; Schlosser, 2005; Schlosser, 2007; Gasparini et al., 2013; Graham and Shimeld, 2013; Luttrell and Swalla, 2014). Mutation of genes involved in placode development lead to a variety of congenital syndromes in humans that share craniofacial dysmorphologies and hearing loss. Future work is needed to identify the specific molecular functions of the Six (and other) genes that have key roles in PPE and placode development, and identify all of the potential co-factors; this will reveal further genetic causes of craniofacial syndromes. In addition, identifying and understanding the function of all of the genes involved in PPE specification and placode differentiation pathways will have a major impact on craniofacial tissue repair efforts. Elucidating the basic molecular mechanisms by which PPE cells are induced and transformed from the embryonic ectoderm into numerous differentiated cell types and how the process differs from that described for the closely related neural crest will be critical for designing techniques for sensory organ replacement from various stem and progenitor cell sources. In one case, that of olfactory sensory neurons, the embryonic mechanisms are likely maintained and adapted in the adult to facilitate olfactory sensory neuron replacement throughout life seen in most vertebrates, including mammals (Schwob, 2002; Leung et al, 2007). It is exciting that human embryonic stem cells have been successfully differentiated into placode cells (Chen et al., 2012; Leung et al., 2013), in some instances generating trigeminal sensory neurons capable of in vivo engraftment in chick and mouse embryos, mature lens fibers, and anterior pituitary cells capable of producing human growth hormone and adrenocorticotropic hormone in vivo (Dincer et al., 2013). Understanding the placode gene regulatory network in the embryo will surely enhance in vitro differentiation steps for regenerating placode derivatives, making it possible to repair and regenerate cranial sensory organs.
Acknowledgments
We apologize to those authors whose work was not cited due to space limitations. Work from the authors’ laboratories is supported by NIH R01 grants DE022065 (SAM), DC011534 (A-SL) and HD29178 (A-SL).
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies Branchio–Oto–Renal (BOR) syndrome and identifies a novel gene family. Nature Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Abello G, Khatri S, Giraldez F, Alsina B. Early regionalization of the otic placode and its regulation bu the Notch signaling pathway. Mech Dev. 2007;124:631–645. doi: 10.1016/j.mod.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev Biol. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Anchan RM, Drake DP, Gerwe EA, Haines CFn, LaMantia AS. A failure of retinoid-mediated induction accompanies the loss of the olfactory pathway during mammalian forebrain development. J Comp Neurol. 1997;379:171–184. [PubMed] [Google Scholar]
- Arima K, Shiotsugu J, Niu R, Khandpur R, Martinez M, Shin Y, Koide T, Cho KW, Kitayama A, Ueno N. Global analysis of RAR-responsive genes in the Xenopus neurula using cDNA microarrays. Dev Dyn. 2005;232:414–431. doi: 10.1002/dvdy.20231. [DOI] [PubMed] [Google Scholar]
- Arkell R, Beddington RS. BMP-7 influences pattern and growth of the developing hindbrain of mouse embryos. Development. 1997;124:1–12. doi: 10.1242/dev.124.1.1. [DOI] [PubMed] [Google Scholar]
- Axel R. Scents and sensibility: a molecular logic of olfactory perception (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:6110–6127. doi: 10.1002/anie.200501726. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Molec Genet. 2000;9:363–366. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- Bailey AP, Streit A. Sensory organs: making and breaking the pre-placodal region. Curr Top Dev Biol. 2006;72:167–204. doi: 10.1016/S0070-2153(05)72003-2. [DOI] [PubMed] [Google Scholar]
- Bailey AP, Bhattacharyya S, Bronner-Fraser M, Streit A. Lens specification is the ground state of all sensory placodes, from which FGF promotes olfactory identity. Dev Cell. 2006;11:505–517. doi: 10.1016/j.devcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Bajoghli B, Aghaallaei N, Czerby T. Groucho corepressor proteins regulate otic vesicle outgrowth. Dev Dyn. 2005;233:760–771. doi: 10.1002/dvdy.20398. [DOI] [PubMed] [Google Scholar]
- Baker CVH, Bronner-Fraser M. Establishing neuronal identity in vertebrate neurogenic placodes. Development. 2000;127:3045–3056. doi: 10.1242/dev.127.14.3045. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Baker CV, Stark MR, Marcelle C, Bronner-Fraser M. Competence, specification and induction of Pax-3 in the trigeminal placode. Development. 1999;126:147–156. doi: 10.1242/dev.126.1.147. [DOI] [PubMed] [Google Scholar]
- Balmer CW, LaMantia AS. Noses and neurons: morphogenesis and neural induction in the olfactory pathway. Dev Dyn. 2005;234:464–481. doi: 10.1002/dvdy.20582. [DOI] [PubMed] [Google Scholar]
- Bane BC, Van Rybroek JM, Kolker SJ, Weeks DL, Manaligod JM. EYA1 expression in the developing inner ear. Ann Otol Rhinol Laryngol. 2005;114:853–858. doi: 10.1177/000348940511401108. [DOI] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Rainschtein L, Inbar D, Gothelf D, Hennekam R, Straussberg R. Autosomal recessive mental retardation syndrome with anterior maxillary protrusion and strabismus: MRAMS syndrome. Am J Med Genet. 2007;143A:1687–1691. doi: 10.1002/ajmg.a.31810. [DOI] [PubMed] [Google Scholar]
- Bassham S, Postlthwait JH. The evolutionary history of placodes: a molecular genetic investigation of the larvacean urochordate Oikoleura dioica. Development. 2005;132:4259–4272. doi: 10.1242/dev.01973. [DOI] [PubMed] [Google Scholar]
- Beanan MJ, Sargent TD. Regulation and function of Dlx3 in vertebrate development. Dev Dyn. 2000;218:545–5. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1026>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessarab DA, Chong SW, Korzh V. Expression of zebrafish six1 during sensory organ development and myogenesis. Dev Dyn. 2004;230:781–786. doi: 10.1002/dvdy.20093. [DOI] [PubMed] [Google Scholar]
- Bhasin N, Maynard TM, Gallagher P, LaMantia AS. Mesenchymal/epithelial interactions regulate retinoid signaling in the olfactory placode. Dev Biol. 2003;261:82–98. doi: 10.1016/s0012-1606(03)00295-1. [DOI] [PubMed] [Google Scholar]
- Bhat N, Kwon HJ, Riley BB. A gene network that coordinates preplacodal competence and neural crest specification in zebrafish. Dev Biol. 2013;373:107–117. doi: 10.1016/j.ydbio.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Bronner-Fraser M. Hierarchy of regulatory events in sensory placode development. Curr Opin Genet Dev. 2004;14:520–526. doi: 10.1016/j.gde.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Bailey AP, Bronner-Fraser M, Streit A. Segregation of lens and olfactory precursors from a common territory: cell sorting and reciprocity of Dlx5 and Pax6 expression. Dev Biol. 2004;271:403–414. doi: 10.1016/j.ydbio.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Birk E, Har-Zahav A, Manzini CM, Pasmanik-Chor M, Kornreich L, Walsh CA, Noben-Trauth K, Albin A, Simon AJ, Colleaux L, Morad Y, Rainshtein L, Tischfield DJ, Wang P, Magai N, Shoshani N, Rechavi G, Gothelf D, Maydan G, Shohat M, Basel-Vanagaite L. SOBP is mutated in syndromic and non-syndromic intellectual disability and is highly expressed in the brain limbic system. Am J Hum Genet. 2010;87:694–700. doi: 10.1016/j.ajhg.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bosman EA, Quint E, Fuchs E, Hrabe de Angelis M, Steel KP. Catweasel mice: a novel role for Six1 in sensory patch development and a model for branchio-otic-renal syndrome. Dev Biol. 2009;328:285–296. doi: 10.1016/j.ydbio.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley L, Sun B, Collins-Racie L, LaVallie E, McCoy J, Sive H. Different activities of the frizzled-related proteins frzb2 and sizzled2 during Xenopus anteroposterior patterning. Dev Biol. 2000;227:118–132. doi: 10.1006/dbio.2000.9873. [DOI] [PubMed] [Google Scholar]
- Bricaud O, Collazo A. The transcription factor six1 inhibits neuronal and promotes hair cell fates in the developing zebrafish (Danio rerio) inner ear. J Neurosci. 2006;26:10438–10451. doi: 10.1523/JNEUROSCI.1025-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricaud O, Collazo A. Balancing cell numbers during organogenesis: Six1a differentially affects neurons and sensory hair cells in the inner ear. Dev Biol. 2011;357:191–201. doi: 10.1016/j.ydbio.2011.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck S, Englert C. Genetic determination of nephrogenesis: the Pax/Eya/Six gene network. Pediatr Nephrol. 2004;19:249–255. doi: 10.1007/s00467-003-1374-z. [DOI] [PubMed] [Google Scholar]
- Brownell I, Dirksen M, Jamrich M. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. genesis. 2000;27:81–93. doi: 10.1002/1526-968x(200006)27:2<81::aid-gene50>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Moody SA. Induction and specification of the vertebrate ectodermal placodes: precursors of the cranial sensory organs. Biol Cell. 2005;97:303–319. doi: 10.1042/BC20040515. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- Buck LB. The molecular architecture of odor and pheromone sensing in mammals. Cell. 2000;100:611–618. doi: 10.1016/s0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- Buller C, Xu X, Marquis V, Schwanke R, Xu PX. Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum Mol Genet. 2001;10:2775–2781. doi: 10.1093/hmg/10.24.2775. [DOI] [PubMed] [Google Scholar]
- Cariboni A, Davidson K, Rakic S, Maggi R, Parnavelas JG, Ruhrberg C. Defective gonadotropin-releasing hormone neuron migration in mice lacking SEMA3A signalling through NRP1 and NRP2: implications for the aetiology of hypogonadotropic hypogonadism. Hum Mol Genet. 2011;20:336–344. doi: 10.1093/hmg/ddq468. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Acuna G, Ellwanger K, Niehrs C, Mayor R. Neural crests are actively precluded from the anterior neural fold by a novel inhibitory mechanism dependent on Dickkopf1 secreted by the prechordal mesoderm. Dev Biol. 2007;309:208–221. doi: 10.1016/j.ydbio.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Castro DS, Guillemot F. Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle. 2011;10:4026–4031. doi: 10.4161/cc.10.23.18578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127:2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Charlton-Perkins M, Brown NL, Cook TA. The lens in focus: a comparison of lens development in Drosophila and vertebrates. Mol Genet Genomics. 2011;286:798–811. doi: 10.1007/s00438-011-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Kim EH, Xu PX. Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev Biol. 2009;326:75–85. doi: 10.1016/j.ydbio.2008.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Chen W, Jongkamonwiwat N, Abbas L, Eshtan SJ, Johnson SL, Kuhn S, Milo M, Thurlow JK, Andrews PW, Marcotti W, Moore HD, Rivolta MN. Restoration of auditory evoked response by human ES-derived otic progenitors. Nature. 2012;490:278–282. doi: 10.1038/nature11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pollet N, Niehrs C, Pieler T. Increased XRALDH2 activity has a posteriorizing effect on the central nervous system of Xenopus embryos. Mech Dev. 2001;101:91–103. doi: 10.1016/s0925-4773(00)00558-x. [DOI] [PubMed] [Google Scholar]
- Chen Z, Montcouquioi M, Calderon R, Jenkins NA, Copeland NG, Kelley MW, NobenTrauth K. Jac1/Sobp, encoding a nuclear zinc finger protein, is critical for cochlear growth, cell fate and patterning of the organ of Corti. J Neurosci. 2008;28:6633–6641. doi: 10.1523/JNEUROSCI.1280-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Chitnis AB, Nogare DD. Lessons from the zebrafish lateral line system. In: Moody SA, editor. Principles of Developmental Genetics. 2. Elsevier Inc; 2014. in press. [Google Scholar]
- Christophorou NA, Bailey AP, Hanson S, Streit A. Activation of Six1 target genes is required for sensory placode formation. Dev Biol. 2009;336:327–336. doi: 10.1016/j.ydbio.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Muller-Tidow C, Golub TR, Kawakami K, Ford HL. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci USA. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couly GF, LeDouarin NM. Mapping of the early neural primordium in quail-chick chimeras. II The prosencephalic neural plate and neural folds: implications for the genesis of cephalic human congenital abnormalities. Dev Biol. 1987;120:198–214. doi: 10.1016/0012-1606(87)90118-7. [DOI] [PubMed] [Google Scholar]
- Couly GF, LeDouarin NM. Head morphogenesis in embryonic avian chimeras: evidence for a segmental pattern in the ectoderm corresponding to the neuromeres. Development. 1990;108:543–558. doi: 10.1242/dev.108.4.543. [DOI] [PubMed] [Google Scholar]
- de Croze N, Maczkowiak F, Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci USA. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Ahrens K, Wedlich D, Schlosser G. Xenopus Eya1 demarcates all neurogenic placodes as well as migrating hypaxial muscle precursors. Mech Dev. 2001;103:189–192. doi: 10.1016/s0925-4773(01)00355-0. [DOI] [PubMed] [Google Scholar]
- Depreux FF, Darrow K, Conner DA, Eavey RD, Liberman MC, Seidman CE, Seidman JG. Eya4-deficient mice are a model for heritable otitis media. J Clin Invest. 2008;118:651–658. doi: 10.1172/JCI32899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dincer Z, Piao J, Niu L, Ganat Y, Kriks S, Zimmer B, Shi S-H, Tabar V, Studer L. Specification of functional cranial placode derivatives from human pluripotent stem cells. Cell Reports. 2013;5:1–16. doi: 10.1016/j.celrep.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dude CM, Kuan CY, Bradshaw JR, Greene ND, Relaix F, Stark MR, Baker CV. Activation of Pax3 target genes is necessary but not sufficient for neurogenesis in the ophthalmic trigeminal placode. Dev Biol. 2009;326:314–326. doi: 10.1016/j.ydbio.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J. Foxg1 is required for development of the vertebrate olfactory system. J Neurosci. 2008;28:5229–5239. doi: 10.1523/JNEUROSCI.1134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MK, Kos L, Jenkins NA, Gilbert DJ, Copeland NG, Tomarev SI. Eyes absent, a gene family found in several metazoan phyla. Mammal Genome. 1997;8:479–485. doi: 10.1007/s003359900480. [DOI] [PubMed] [Google Scholar]
- Dutta S, Dietrich JE, Aspock G, Burdine RD, Schier A, Westerfield M, Varga ZM. Pitx3 defines an equivalence domain for lens and anterior pituitary placode. Development. 2005;132:1579–1590. doi: 10.1242/dev.01723. [DOI] [PubMed] [Google Scholar]
- Eagleson GW, Dempewolf RD. The role of the anterior neural ridge and Fgf-8 in early forebrain patterning and regionalization in Xenopus laevis. Comp Biochem Physiol B Biochem Mol Biol. 2002;132:179–189. doi: 10.1016/s1096-4959(01)00521-8. [DOI] [PubMed] [Google Scholar]
- Estefania E, Ramirez-Camacho R, Gomar M, Trinidad A, Arellano B, Garcia-Berrocal JR, Verdaguer JM, Vilches C. Point mutation of an EYA1-gene splice site in a patient with oto-facial-cervical syndrome. Ann Hum Genet. 2006;70:140–144. doi: 10.1111/j.1529-8817.2005.00204.x. [DOI] [PubMed] [Google Scholar]
- Esterberg R, Fritz A. Dlx3b/4b are required for the formation of the preplacodal region and otic placode through local modulation of BMP activity. Dev Biol. 2009;325:189–199. doi: 10.1016/j.ydbio.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P, Bovolenta P. cSix4, a member of the six gene family of transcription factors, is expressed during placode and somite development. Mech Dev. 1999;85:161–165. doi: 10.1016/s0925-4773(99)00079-9. [DOI] [PubMed] [Google Scholar]
- Feledy JA, Beanan MJ, Sandoval JJ, Goodrich JS, Lim JH, Matsuo-Takasaki M, Sato SM, Sargent TD. Inhibitory patterning of the anterior neural plate in Xenopus by homeodomain factors Dlx3 and Msx1. Dev Biol. 1999;212:455–464. doi: 10.1006/dbio.1999.9374. [DOI] [PubMed] [Google Scholar]
- Ford HL, Kabingu EN, Bump EA, Mutter GL, Pardee AB. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc Natl Acad Sci USA. 1998;95:12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougerousse F, Durand M, Lopez S, Suel L, Demignon J, Thornton C, Ozaki H, Kawakami K, Barbet P, Beckmann JS, Maire P. Six and Eya expression during human somitogenesis and MyoD family activation. J Muscle Res Cell Motil. 2002;23:255–264. doi: 10.1023/a:1020990825644. [DOI] [PubMed] [Google Scholar]
- Gamble JA, Karunadasa DK, Pape JR, Skynner MJ, Todman MG, Bicknell RJ, Allen JP, Herbison AE. Disruption of ephrin signaling associates with disordered axophilic migration of the gonadotropin-releasing hormone neurons. J Neurosci. 2005;25:3142–3150. doi: 10.1523/JNEUROSCI.4759-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini F, Degasperi V, Shimeld SM, Burighel P, Manni L. Evolutionary conservation of the placodal transcriptional network during sexual and asexual development of chordates. Dev Dyn. 2013;242:752–766. doi: 10.1002/dvdy.23957. [DOI] [PubMed] [Google Scholar]
- Ghanbari H, Seo HC, Fjose A, Brändli AW. Molecular cloning and embryonic expression of Xenopus Six homeobox genes. Mech Dev. 2001;101:271–277. doi: 10.1016/s0925-4773(00)00572-4. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, Rubenstein JL, Robert B, Mellon PL. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem. 2005;280:19156–19165. doi: 10.1074/jbc.M502004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavic A, Maris Honoré S, Gloria Feijóo C, Bastidas F, Allende ML, Mayor R. Role of BMP signaling and the homeoprotein Iroquois in the specification of the cranial placodal field. Dev Biol. 2004;272:89–103. doi: 10.1016/j.ydbio.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Graham A, Blentic A, Dusque S, Begbie J. Delamination of cells from neurogenic placodes does not involve an epithelial-to-mesenchymal transition. Development. 2007;134:4141–4145. doi: 10.1242/dev.02886. [DOI] [PubMed] [Google Scholar]
- Graham A, Shimeld SM. The origin and evolution of the ectodermal placodes. J Anat. 2013;222:32–40. doi: 10.1111/j.1469-7580.2012.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Grocott T, Tambalo M, Streit A. The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective. Dev Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Groves AK, Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–3499. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
- Groves AK, LaBonne C. Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev Biol. 2014;389:2–12. doi: 10.1016/j.ydbio.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. The odor coding system of Drosophila. Trends Genet. 2004;20:453–459. doi: 10.1016/j.tig.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Hans S, Liu D, Westerfield M. Pax8 and Pax2a function synergistically in otic specification, downstream of the Foxi1 and Dlx3b transcription factors. Development. 2004;131:5091–5102. doi: 10.1242/dev.01346. [DOI] [PubMed] [Google Scholar]
- Hatini V, Xin Y, Balas G, Lai E. Dynamics of placodal lineage development revealed by targeted transgene expression. Dev Dyn. 1999;215:332–343. doi: 10.1002/(SICI)1097-0177(199908)215:4<332::AID-AJA5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;15:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Houart C, Take-Uchi M, Rauch GJ, Young N, Coutinho P, Masai I, Caneparo L, Concha ML, Geisler R, Dale TC, Wilson SW, Stemple DL. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes Dev. 2001;15:1427–1434. doi: 10.1101/gad.194301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog W, Zeng X, Lele Z, Sonntag C, Ting JW, Chang CY, Hammerschmidt M. Adenohypophysis formation in the zebrafish and its dependence on sonic hedgehog. Dev Biol. 2003;245:36–49. doi: 10.1016/s0012-1606(02)00124-0. [DOI] [PubMed] [Google Scholar]
- Himeda CL, Ranish JA, Angello JC, Maire P, Aebersold R, Hauschka SD. Quantitative proteomic identification of six4 as the trex-binding factor in the muscle creatine kinase enhancer. Mol Cell Biol. 2004;24:2132–2143. doi: 10.1128/MCB.24.5.2132-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol. 2007;308:679–691. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao FC, Williams A, Davies EL, Rebay I. Eyes absent mediates cross-talk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Dev Cell. 2001;1:51–61. doi: 10.1016/s1534-5807(01)00011-9. [DOI] [PubMed] [Google Scholar]
- Hwang CH, Simeone A, Lai E, Wu DK. Foxg1 is required for proper separation and formation of sensory cristae during inner ear development. Dev Dyn. 2009;238:2725–2734. doi: 10.1002/dvdy.22111. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ookawara S, Sato S, Ando Z, Kageyama R, Kawakami K. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev Biol. 2007;311:53–68. doi: 10.1016/j.ydbio.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Kageyama R, Suzuki Y, Kawakami K. Six1 is indispensable for production of functional progenitor cells during olfactory epithelial development. Int J Dev Biol. 2010;54:1453–1464. doi: 10.1387/ijdb.093041ki. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Ikeda K, Sato S, Yajia H, Kawakami K. Differential expression of Eya1 and Eya2 during chick early embryonic development. Gene Expr Patterns. 2008;8:357–367. doi: 10.1016/j.gep.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Ito T, Noguchi Y, Yashima T, Kitamura K. SIX1 mutation associated with enlargement of the vestibular aqueduct in a patient with branchio–oto syndrome. Laryngoscope. 2006;116:797–799. doi: 10.1097/01.mlg.0000209096.40400.96. [DOI] [PubMed] [Google Scholar]
- Jacobson AG. The determination and positioning of the nose, lens and ear. III Effects of reversing the antero–posterior axis of epidermis, neural plate and neural fold. J Exp Zool. 1963;154:293–303. doi: 10.1002/jez.1401540305. [DOI] [PubMed] [Google Scholar]
- Jacobson AG. Inductive processes in embryonic development. Science. 1966;152:25–34. doi: 10.1126/science.152.3718.25. [DOI] [PubMed] [Google Scholar]
- Janesick A, Shiotsugu J, Taketani M, Blumberg B. RIPPLY3 is a retinoic acid-inducible repressor required for setting the borders of the pre-placodal ectoderm. Development. 2012;139:1213–1224. doi: 10.1242/dev.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena CS, Ohyama T, Segli N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Cook SA, Erway LC, Matthews AN, Sanford LP, Paradies NE, Friedman RA. Inner ear and kidney anomalies caused by IAP insertion in an intron of the Eya1 gene in a mouse model of BOR syndrome. Hum Mol Genet. 1999;8:645–653. doi: 10.1093/hmg/8.4.645. [DOI] [PubMed] [Google Scholar]
- Jusiak B, Karandikar UC, Kwak SJ, Wang F, Wang H, Chen R, Mardon G. Regulation of Drosophila eye development by the transcription factor Sine oculis. PLoS One. 2014;9:e89695. doi: 10.1371/journal.pone.0089695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji T, Artinger KB. dlx3b and dlx4b function in the development of Rohon-Beard sensory neurons and trigeminal placode in the zebrafish neurula. Dev Biol. 2004;276:523–540. doi: 10.1016/j.ydbio.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Ohto H, Ikeda K, Roeder RG. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 1996;24:303–310. doi: 10.1093/nar/24.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes-structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hébert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Kim J, Santos R, Wu HH, Lander AD, Calof AL. Foxg1 promotes olfactory neurogenesis by antagonizing Gdf11. Development. 2009;136:1453–1464. doi: 10.1242/dev.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KL, Li DJ, Clouser C, Tran S, Pignoni F. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev Dyn. 2005;234:497–504. doi: 10.1002/dvdy.20442. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Moody SA, Jamrich M. A novel fork head gene mediates early steps during Xenopus lens formation. Development. 1999;126:5107–5116. doi: 10.1242/dev.126.22.5107. [DOI] [PubMed] [Google Scholar]
- Khatri SB, Groves AK. Expression of the Foxi2 and Foxi3 transcription factors during development of chicken sensory placodes and pharyngeal arches. Gene Expr Patterns. 2013;13:38–42. doi: 10.1016/j.gep.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouff RA. The developmental pattern of ectodermal placodes in Rana pipiens. J Comp Neurol. 1935;62:17–71. [Google Scholar]
- Kobayashi M, Osanai H, Kawakami K, Yamamoto M. Expression of three zebrafish Six4 genes in the cranial sensory placodes and the developing somites. Mech Dev. 2000;98:151–155. doi: 10.1016/s0925-4773(00)00451-2. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nishikawa K, Suzuki T, Yamamoto M. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev Biol. 2001;232:315–326. doi: 10.1006/dbio.2001.0185. [DOI] [PubMed] [Google Scholar]
- Kochhar A, Orten DJ, Sorensen JL, Fischer SM, Cremers CW, Kimberling WJ, Smith RJ. SIX1 mutation screening in 247 branchio-otic-renal syndrome families: a recurrent missense mutation associated with BOR. Hum Mutat. 2008;29:565. doi: 10.1002/humu.20714. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Ikeda K, Iwakura Y, Kawakami K. Six1 and Six4 promote survival of sensory neurons during early trigeminal gangliogenesis. Brain Res. 2006;1116:93–102. doi: 10.1016/j.brainres.2006.07.103. [DOI] [PubMed] [Google Scholar]
- Kriebel M, Muller F, Hollemann T. Xeya3 regulates survival and proliferation of neural progenitor cells within the anterior neural plate of Xenopus embryos. Dev Dyn. 2007;236:1526–1534. doi: 10.1002/dvdy.21170. [DOI] [PubMed] [Google Scholar]
- Krolewski RC, Packard A, Jang W, Wildner H, Schwob JE. Ascl1 (Mash1) knockout perturbs differentiation of nonneuronal cells in olfactory epithelium. PloS One. 2012;7:e51737. doi: 10.1371/journal.pone.0051737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Deffenbacher K, Cremers CW, Van Camp G, Kimberling WJ. Brachio-oto-renal syndrome: identification of novel mutations, molecular characterization, mutation distribution and prospects for genetic testing. Genet Test. 1997;1:243–251. doi: 10.1089/gte.1997.1.243. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Bhat N, Sweet EM, Cornell RA, Riley BB. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010;6:e1001133. doi: 10.1371/journal.pgen.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech Dev. 2003;120:669–679. doi: 10.1016/s0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Ladher RK, O’Neill P, Begbie J. From shared lineage to distinct functions: development of the inner ear and epibranchial placodes. Development. 2010;137:1777–1785. doi: 10.1242/dev.040055. [DOI] [PubMed] [Google Scholar]
- LaMantia A-S, Colbert MC, Linney E. Retinoic acid induction and regional differentiation prefigure olfactory pathway formation in the mammalian forebrain. Neuron. 1993;10:1035–1048. doi: 10.1016/0896-6273(93)90052-s. [DOI] [PubMed] [Google Scholar]
- LaMantia AS, Bhasin N, Rhodes K, Heemskerk J. Mesenchymal/epithelial induction mediates olfactory pathway formation. Neuron. 2000;28:411–425. doi: 10.1016/s0896-6273(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Lassiter RNT, Stark MR, Zhao T, Zhou CJ. Signaling mechanisms controlling cranial placode neurogenesis and delamination. Dev Biol. 2014;389:39–49. doi: 10.1016/j.ydbio.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger S, Brand M. Fgf8 and Fgf3 are required for zebrafish ear placode induction, maintenance and inner ear patterning. Mech Dev. 2002;119:91–108. doi: 10.1016/s0925-4773(02)00343-x. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- Leung AW, Morest KD, Li JY. Differential BMP signaling controls formation and differentiation of multipotent preplacodal progenitors from human embryonic stem cells. Dev Biol. 2013;379:208–220. doi: 10.1016/j.ydbio.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, Drossopoulou G, Paton IR, Morrice DR, Robertson KE, Burt DW, Ingham PW, Tickle C. Expression of ptc and gli genes in talpid3 suggests bifurcation in Shh pathway. Development. 1999;126:32397–2407. doi: 10.1242/dev.126.11.2397. [DOI] [PubMed] [Google Scholar]
- Li W, Cornell RA. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol. 2007;304:338–354. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Li Y, Manaligod JM, Weeks DL. EYA1 mutations associated with the branchio-otic renal syndrome result in defective otic development in Xenopus laevis. Biol Cell. 2010;102:277–292. doi: 10.1042/BC20090098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang L, Li X, Li L, Ma W, Wu J, Zhang M. Six1 promotes proliferation of pancreatic cancer cells via upregulation of Cyclin D1 expression. PLoS One. 2013;8:e59203. doi: 10.1371/journal.pone.0059203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–4062. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- Liu D, Chu H, Maves L, Yan YL, Morcos PA, Postlethwait JH, Westerfield M. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development. 2003;130:2213–2224. doi: 10.1242/dev.00445. [DOI] [PubMed] [Google Scholar]
- Llleras-Forero L, Tambalo M, Christophorou N, Chambers D, Houart C, Streit A. Neuropeptides: developmental signals in placode progenitor formation. Dev Cell. 2013;26:195–203. doi: 10.1016/j.devcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Sargent TD. Distinct roles for Distal-less genes Dlx3 and Dlx5 in regulating ectodermal development in Xenopus. Mol Reprod Dev. 2001a;60:331–337. doi: 10.1002/mrd.1095. [DOI] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Lim JH, Sargent TD. Differential regulation of Dlx gene expression by a BMP morphogenetic gradient. Int J, Dev Biol. 2001b;45:681–684. [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Thomas ML, Weeks DL, Sargent TD. Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev Biol. 2002;245:136–144. doi: 10.1006/dbio.2002.0621. [DOI] [PubMed] [Google Scholar]
- Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2a. Proc Natl Acad Sci USA. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell SM, Swalla BJ. Genomic and evolutionary insights into chordate development. In: Moody SA, editor. Principles of Developmental Genetics. 2. Elsevier Inc; 2014. in press. [Google Scholar]
- Mackereth MD, Kwak SJ, Fritz A, Riley BB. Zebrafish pax8 is required for otic placode induction and plays a redundant role with Pax2 genes in the maintenance of the otic placode. Development. 2005;132:371–382. doi: 10.1242/dev.01587. [DOI] [PubMed] [Google Scholar]
- Makishima T, Madeo AC, Brewer CC, Zalewski CK, Butman JA, Sachdev V, Arai AE, Holbrook BM, Rosing DR, Griffith AJ. Nonsyndromic hearing loss DFNA10 and a novel mutation of EYA4: evidence for correlation of normal cardiac phenotype with truncating mutations of the Eya domain. Am J Med Genet. 2007;143A:1592–1598. doi: 10.1002/ajmg.a.31793. [DOI] [PubMed] [Google Scholar]
- Mancilla A, Mayor R. Neural crest formation in Xenopus laevis: mechanism of Xslug induction. Dev Biol. 1996;177:580–589. doi: 10.1006/dbio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Mann ZF, Kelley MW. Development of the inner ear. In: Moody SA, editor. Principles of Developmental Genetics. 2. Elsevier Inc; 2014. in press. [Google Scholar]
- Maroon H, Walshe J, Mahmood R, Kiefer P, Dickson C, Mason I. Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development. 2002;129:2099–2108. doi: 10.1242/dev.129.9.2099. [DOI] [PubMed] [Google Scholar]
- Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2005;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- Matsuo-Takasaki M, Matsumura M, Sasai Y. As essential role of Foxi1a for ventral specification of the cephalic ectoderm during gastrulation. Development. 2005;132:3885–3894. doi: 10.1242/dev.01959. [DOI] [PubMed] [Google Scholar]
- McCabe KL, Bronner-Fraser M. Essential role for PDGF signaling in ophthalmic trigeminal placode induction. Development. 2008;135:1863–1874. doi: 10.1242/dev.017954. [DOI] [PubMed] [Google Scholar]
- McLarren KW, Litsiou A, Streit A. DLX5 positions the neural crest and preplacode region at the border of the neural plate. Dev Biol. 2003;259:34–47. doi: 10.1016/s0012-1606(03)00177-5. [DOI] [PubMed] [Google Scholar]
- Messina A, Ferraris N, Wray S, Cagnoni G, Donohue DE, Casoni F, Kramer PR, Derijck AA, Adolfs Y, Fasolo A, Pasterkamp RJ, Giacobini P. Dysregulation of Semaphorin7A/beta1-integrin signaling leads to defective GnRH-1 cell migration, abnormal gonadal development and altered fertility. Hum Mol Gen. 2011;20:4759–4774. doi: 10.1093/hmg/ddr403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Mochii M, Yamamoto TS, Takahashi TC, Eguchi G, Okada TS. Pax-6 and Prox-1 expression during lens regeneration from Cynops iris and Xenopus cornea: evidence for a genetic program common to embryonic lens development. Differentiation. 1999;65:141–149. doi: 10.1046/j.1432-0436.1999.6530141.x. [DOI] [PubMed] [Google Scholar]
- Modrell MS, Baker CV. Evolution of electrosensory ampullary organs: conservation of Eya4 expression during lateral line development in jawed vertebrates. Evol Dev. 2012;14:277–285. doi: 10.1111/j.1525-142X.2012.00544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SA, Klein SL, Karpinski BA, Maynard TM, LaMantia AS. On becoming neural: what the embryo can tell us about differentiating neural stem cells. Amer J Stem Cells. 2013;2:74–94. [PMC free article] [PubMed] [Google Scholar]
- Moody SA, Saint-Jeannet J-P. Development of the Pre-placodal ectoderm and cranial sensory placodes. In: Moody SA, editor. Principles of Developmental Genetics. 2. Elsevier Inc; 2014. in press. [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and Wnt signals during Xenopus neural crest induction. Dev Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Mosrati MA, Hammami B, Rebeh IB, Ayadi L, Dhouib L, Ben Mahfoudh K, Hakim B, Charfeddine I, Mnif J, Ghorbel A, Masmoudi S. A novel dominant mutation in SIX1, affecting a highly conserved residue, results in only auditory defects in humans. Eur J Med Genet. 2011;54:e484–e488. doi: 10.1016/j.ejmg.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Murray RC, Navi D, Fesenko J, Lander AD, Calof AL. Widespread defects in the primary olfactory pathway caused by loss of Mash1 function. J Neurosci. 2003;23:1769–1780. doi: 10.1523/JNEUROSCI.23-05-01769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson KM, Pignoni F, Yan B, Moody SA. Developmental expression patterns of candidate co-factors for vertebrate Six family transcription factors. Dev Dyn. 2010;239:3446–3466. doi: 10.1002/dvdy.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VH, Schmid B, Trout J, Connors SA, Ekker M, Mullins MC. Ventral and lateral regions of the zebrafish gastrula, including the neural crest progenitors, are established by a bmp2b/swirl pathway of genes. Dev Biol. 1998;199:93–110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- Nica G, Herzog W, Sonntag C, Nowak M, Schwarz H, Zapata AG, Hammerschmidt M. Eya1 is required for lineage-specific differentiation, but not for cell survival in the zebrafish adenohypophysis. Dev Biol. 2006;292:189–204. doi: 10.1016/j.ydbio.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Nissen RM, Yan J, Amsterdam A, Hopkins N, Burgess SM. Zebrafish foxi modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–2554. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- Noguchi Y, Ito T, Nishio A, Honda K, Kitamura K. Audiovestibular findings in a branchio-oto syndrome patient with a SIX1 mutation. Acta Otolaryngol. 2011;131:413–418. doi: 10.3109/00016489.2010.543146. [DOI] [PubMed] [Google Scholar]
- Ogino H, Ochi H, Reza HM, Yasuda K. Transcription factors involved in lens development from the preplacodal ectoderm. Dev Biol. 2012;363:333–347. doi: 10.1016/j.ydbio.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Ohto H, Takizawa T, Saito T, Kobayashi M, Ikeda K, Kawakami K. Tissue and developmental distribution of Six family gene products. Int J Dev Biol. 1998;42:141–148. [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga S, Ozaki H, Sato S, Kawakami K. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Kimura S, Watamoto M, Itoh N. Involvement of fibroblast growth factor (FGF)18-FGF8 signaling in the specification of left-right asymmetry and brain and limb development of the chick embryo. Mech Dev. 2000;95:55–66. doi: 10.1016/s0925-4773(00)00331-2. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–646. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Oliver G, Loosli F, Koster R, Wittbrodt J, Gruss P. Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- Ozaki H, Watanabe Y, Takahashi K, Kitamura K, Tanaka A, Urase K, Momoi T, Sudo K, Sakagami J, Asano M, Iwakura Y, Kawakami K. Six4, a putative myogenin gene regulator, is not essential for mouse embryonic development. Mol Cell Biol. 2001;21:3343–3350. doi: 10.1128/MCB.21.10.3343-3350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki H, Watanabe Y, Ikeda K, Kawakami K. Impaired interactions between mouse Eya1 harboring mutations found in patients with branchio-oto-renal syndrome and Six, Dach, and G proteins. J Hum Genet. 2002;47:107–116. doi: 10.1007/s100380200011. [DOI] [PubMed] [Google Scholar]
- Ozaki H, Nakamura K, Funahashi J, Ikeda K, Yamada G, Tokano H, Okamura HO, Kitamura K, Muto S, Kotaki H, Sudo K, Horai R, Iwakura Y, Kawakami K. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev. 2000;96:253–257. doi: 10.1016/s0925-4773(00)00396-8. [DOI] [PubMed] [Google Scholar]
- Park BY, Saint-Jeannet JP. Hindbrain-derived Wnt and Fgf signals cooperate to specify the otic placode in Xenopus. Dev Biol. 2008;324:108–121. doi: 10.1016/j.ydbio.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BY, Saint-Jeannet J-P. Colloquium Series on Developmental Biology. 3. Vol. 1. Morgan & Claypool Publishers; 2010. Induction and segregation of the vertebrate cranial placodes. [PubMed] [Google Scholar]
- Parkash J, Cimino I, Ferraris N, Casoni F, Wray S, Cappy H, Prevot V, Giacobini P. Suppression of beta1-integrin in gonadotropin-releasing hormone cells disrupts migration and axonal extension resulting in severe reproductive alterations. J Neurosci. 2012;32:16992–17002. doi: 10.1523/JNEUROSCI.3057-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C, Gunhaga L, Edlund T. Early development of the central and peripheral nervous systems is coordinated by Wnt and BMP signals. PLoS One. 2008;3:e1625. doi: 10.1371/journal.pone.0001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C, Edlund T, Gunhaga L. Wnt-regulated temporal control of BMP exposure directs the choice between neural plate border and epidermal fate. Development. 2009;136:73–83. doi: 10.1242/dev.025890. [DOI] [PubMed] [Google Scholar]
- Patthey C, Schlosser G, Shimeld SM. The evolutionary history of vertebrate cranial placodes –I: cell type evolution. Dev Biol. 2014;389:82–97. doi: 10.1016/j.ydbio.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Patrick AN, Schiemann BJ, Yang K, Zhao R, Ford HL. Biochemical and functional characterization for six SIX1 Branchio-otic-renal syndrome mutations. J Biol Chem. 2009;284:20781–20790. doi: 10.1074/jbc.M109.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick AN, Cabrera JH, Smaith AL, Chen XS, Ford HL, Zhao R. Structure function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR Syndrome. Nature Struct Mol Biol. 2013;20:447–453. doi: 10.1038/nsmb.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera EM, De Robertis EM. A direct screen for secreted proteins in Xenopus embryos identifies distinct activities for the Wnt antagonists Crescent and Frzb-1. Mech Dev. 2000;96:183–195. doi: 10.1016/s0925-4773(00)00394-4. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–365. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Kwon HJ, Melton C, Houghtaling P, Fritz A, Riley BB. Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev Biol. 2006;294:376–390. doi: 10.1016/j.ydbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Pieper M, Eagleson GW, Wosniok W, Schlosser G. Origin and segregation of cranial placodes in Xenopus laevis. Dev Biol. 2011;360:257–275. doi: 10.1016/j.ydbio.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Pieper M, Ahrens K, Rink E, Peter A, Schlosser G. Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development. 2012;139:1175–1187. doi: 10.1242/dev.074468. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Platt JB. Ontogenetic differentiation of the ectoderm in necturus. II On the development of the peripheral nervous system. Quart J Mic Sci. 1896;38:485–547. [Google Scholar]
- Pogoda HM, Hammerschmidt M. Molecular genetics of pituitary development in zebrafish. Semin Cell Dev Biol. 2007;18:543–558. doi: 10.1016/j.semcdb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Rawson NE, Lischka F, Yee KK, Peters AZ, Tucker ES, Meechan DW, Zirlinger M, Maynard TM, Burd GB, Dulac C, Pevny LH, LaMantia AS. Specific mesenchymal/epithelial induction of olfactory receptor, vomeronasal, and gonadotropin-releasing hormone (GnRH) neurons. Dev Dyn. 2010;239:1723–1738. doi: 10.1002/dvdy.22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Richman JM, Tickle C. Epithelia are interchangeable between facial primordia of chick embryos and morphogenesis is controlled by the mesenchyme. Dev Biol. 1989;136:201–210. doi: 10.1016/0012-1606(89)90142-5. [DOI] [PubMed] [Google Scholar]
- Richman JM, Tickle C. Epithelial-mesenchymal interactions in the outgrowth of limb buds and facial primordia in chick embryos. Dev Biol. 1992;154:299–308. doi: 10.1016/0012-1606(92)90069-s. [DOI] [PubMed] [Google Scholar]
- Rickard S, Parker M, van’t Hoff W, Barnicoat A, Russell-Eggitt I, Winter RM, Bitner-Glindzicz M. Oto-facial-cervical (OFC) syndrome is a contiguous gene deletion syndrome involving EYA1: molecular analysis confirms allelism with BOR syndrome and further narrows the Duane syndrome critical regions to 1cM. Hum Genet. 2001;108:398–403. doi: 10.1007/s004390100495. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Soriano J. Brachio-oto-renal syndrome. J Nephrol. 2003;16:603–605. [PubMed] [Google Scholar]
- Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RMJ, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahly I, Andermann P, Petit C. The zebrafish eya1 gene and its expression pattern during embryogenesis. Dev Genes Evol. 1999;209:399–410. doi: 10.1007/s004270050270. [DOI] [PubMed] [Google Scholar]
- Saint-Jeannet JP, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014;389:13–27. doi: 10.1016/j.ydbio.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanggaard KM, Rendtorff ND, Kjaer KW, Eiberg H, Johnsen T, Gimsing S, Dyrmose J, Nielsen KO, Lage K, Tranebjaerg L. Branchio-otic-renal syndrome: detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses. Eur J Hum Genet. 2007;15:1121–1131. doi: 10.1038/sj.ejhg.5201900. [DOI] [PubMed] [Google Scholar]
- Sargent TD. Transcriptional regulation at the neural plate border. Adv Exp Med Biol. 2006;589:32–44. doi: 10.1007/978-0-387-46954-6_3. [DOI] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Sato S, Ikeda K, Ochi H, Ogino H, Yajima H, Kawakami K. Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev Biol. 2010;344:158–171. doi: 10.1016/j.ydbio.2010.04.029. [DOI] [PubMed] [Google Scholar]
- Sato S, Ikeda K, Shioi G, Nakao K, Yajima H, Kawakami K. Regulation of Six1 expression by evolutionarily conserved enhancers in tetrapods. Dev Biol. 2012;368:95–108. doi: 10.1016/j.ydbio.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Evolutionary origins of vertebrate placodes: insights from developmental studies and from comparisons with other deuterostomes. J Exp Zool B Dev Evol. 2005;304:347–399. doi: 10.1002/jez.b.21055. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Schlosser G. How old genes make a new head: redeployment of Six and Eya genes during the evolution of vertebrate cranial placodes. Integr Comp Biol. 2007;47:343–359. doi: 10.1093/icb/icm031. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Making senses development of vertebrate cranial placodes. Int Rev Cell Mol Biol. 2010;283:129–234. doi: 10.1016/S1937-6448(10)83004-7. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G, Northcutt RG. Development of neurogenic placodes in Xenopus laevis. J Comp Neurol. 2000;418:121–146. [PubMed] [Google Scholar]
- Schonberger J, Wang L, Shin JT, Kim SD, Depreux FFS, Zhu H, Zon L, Pizard A, Kim JB, MacRae CA, Mungall AJ, Seidman JG, Seidman CE. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nature Genetics. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- Schwarting GA, Kostek C, Bless EP, Ahmad N, Tobet SA. Deleted in colorectal cancer (DCC) regulates the migration of luteinizing hormone-releasing hormone neurons to the basal forebrain. J Neurosci. 2001;21:911–919. doi: 10.1523/JNEUROSCI.21-03-00911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec. 2002;269:33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- Self M, Lsagutin OV, Bowling B, Hendrix J, Cal Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. Origins of the avian neural crest: The role of neural plate-epidermal interactions. Development. 1995;121:525–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. Avian neural crest cell fate decisions: a diffusible signal mediates induction of neural crest by the ectoderm. Int J Dev Neurosci. 2000;18:621–627. doi: 10.1016/s0736-5748(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multilevel regulator of ocular development. Prog Retin Eye Res. 2012;31:351–376. doi: 10.1016/j.preteyeres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Shamim H, Mason I. Expression of Fgf4 during development of the chick embryo. Mech Dev. 1999;85:189–192. doi: 10.1016/s0925-4773(99)00093-3. [DOI] [PubMed] [Google Scholar]
- Shim S, Bae N, Park Sy, Kim WS, Han JK. Isolation of Xenopus FGF-8b and comparison with FGF-8a. Molec Cells. 2005;19:310–317. [PubMed] [Google Scholar]
- Shiotsugu J, Katsuyama Y, Arima K, Baxter A, Koide T, Song J, Chandraratna RA, Blumberg B. Multiple points of interaction between retinoic acid and FGF signaling during embryonic axis formation. Development. 2004;131:2653–2667. doi: 10.1242/dev.01129. [DOI] [PubMed] [Google Scholar]
- Shou J, Murray RC, Rim PC, Calof AL. Opposing effects of bone morphogenetic proteins on neuron production and survival in the olfactory receptor neuron lineage. Development. 2000;127:5403–5413. doi: 10.1242/dev.127.24.5403. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Davies EL, Doyon L, Rebay I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol Cell Biol. 2003;23:5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödal M, Gunhaga L. Expresion patterns of Shh, Ptc2, Raldh3, Pitx2, Isl1, Lim3 and Pax6 in the developing chick hypophyseal placode and Rathke’s pouch. Gene Expr Patterns. 2008;8:481–485. doi: 10.1016/j.gep.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Sjödal M, Edlund T, Gunhaga L. Time of exposure to BMP signals plays a key role in the specification of the olfactory and lens placodes ex vivo. Dev Cell. 2007;13:141–149. doi: 10.1016/j.devcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Kwak SJ, Fritz A. Genetic interactions underlying otic placode induction and formation. Dev Dyn. 2004;230:419–433. doi: 10.1002/dvdy.20067. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Logsdo JM, Fritz A. Expression and phylogenetic analyses of three zebrafish Foxi class genes. Dev Dyn. 2003;228:301–307. doi: 10.1002/dvdy.10373. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Fritz A. Concerted action of two dlx paralogs in sensory placode formation. Development. 2002;129:3127–3136. doi: 10.1242/dev.129.13.3127. [DOI] [PubMed] [Google Scholar]
- Soker T, Dalke C, Puk O, Floss T, Becker L, Bolle I, Favor J, Hans W, Holter SM, Horsch M, Kallnik M, Kling E, Moerth C, Schrewe A, Stigloher C, Topp S, Gailus-Durner V, Naton B, Beckers J, Fuchs H, Ivandic B, Klopstock T, Schulz H, Wolf E, Wurst W, Bally-Cuif L, de Angelis MH, Graw J. Pleio-tropic effects in Eya3 knockout mice. BMC Dev Biol. 2008;8:118. doi: 10.1186/1471-213X-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci U S A. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt L, Hoefsloot LH, van Schaijk GH, van Waardenburg D, Kremer B, Brackel HJ, de Die-Smulders CE. Identification of a novel EYA1 mutation presenting in a newborn with laryngomalacia, glossoptosis, retrognathia, and pectus excavatum. Am J Med Genet A. 2006;140:1343–1345. doi: 10.1002/ajmg.a.31285. [DOI] [PubMed] [Google Scholar]
- Streit A. Extensive cell movements accompany formation of the otic placode. Dev Biol. 2002;249:237–254. doi: 10.1006/dbio.2002.0739. [DOI] [PubMed] [Google Scholar]
- Streit A. Early development of the cranial sensory nervous system: from a common field to individual placodes. Dev Biol. 2004;276:1–15. doi: 10.1016/j.ydbio.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Steventon B, Mayor R, Streit A. Mutual repression between Gbx2 and Otx2 in sensory placodes reveals a general mechanism for ectodermal patterning. Dev Biol. 2012;367:55–65. doi: 10.1016/j.ydbio.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Ueno N, Hemmati-Brivanlou A. Xenopus msx1 medicates epidermal induction and neural inhibition by BMP4. Development. 1997;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- Takai A, Inomata H, Arakawa A, Yakura R, Matsuo-Takasaki M, Sasai Y. Anterior neural development requires Del1, a matrix-associated protein that attenuates canonical Wnt signaling via the Ror2 pathway. Development. 2010;137:3293–3302. doi: 10.1242/dev.051136. [DOI] [PubMed] [Google Scholar]
- Tessmar K, Loosli F, Wittbrodt J. A screen for co-factors of Six3. Mech Dev. 2002;117:103–113. doi: 10.1016/s0925-4773(02)00185-5. [DOI] [PubMed] [Google Scholar]
- Thiel G. How Sox2 maintains neural stem cell identity. Biochem J. 2013;450:e1–2. doi: 10.1042/BJ20130176. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Toro S, Varga ZM. Equivalent progenitor cells in the zebrafish anterior preplacodal field give rise to adenohypophysis, lens, and olfactory placodes. Semin Cell Dev Biol. 2007;18:534–542. doi: 10.1016/j.semcdb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Tribulo C, Ayba MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by Bmp gradient is essential for neural crest specification. Development. 2003;130:6441–6452. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- Treier M, O’Connell S, Gleiberman A, Price J, Szeto DP, Burgess R, Chuang PT, McMahom AP, Rosenfeld MG. Hedgehog signaling is required for pituitary gland development. Development. 2001;128:377–386. doi: 10.1242/dev.128.3.377. [DOI] [PubMed] [Google Scholar]
- Tucker ES, Lehtinen MK, Maynard TM, Zirlinger M, Dulac C, Rawson NE, Pevny LH, LaMantia AS. Proliferative and transcriptional identity of two distinct classes of neural precursors in the mammalian olfactory epithelium. Development. 2010;137:2471–81. doi: 10.1242/dev.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kupffer C. Studien zur vergleichenden Entwicklungsgeschichte des Kpofes des Cranioten 3. Heft: Die Entwicklung der Kopfnerven von Ammocoetes planeri; Munchen: 1895. [Google Scholar]
- Wakamatsu Y. Mutual repression between Pax3 and Pax6 is involved in the positioning of ophthalmic trigeminal placode in avian embryo. Dev Growth Differ. 2011;53:994–1003. doi: 10.1111/j.1440-169X.2011.01311.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Sewell WF, Kim SD, Shin JT, MacRae CA, Zon LI, Seidman JG, Seidman CE. Eya4 regulation of Na+/K+-ATPase is required for sensory system development in zebrafish. Development. 2008;135:3425–3434. doi: 10.1242/dev.012237. [DOI] [PubMed] [Google Scholar]
- Wayne S, Robertson NG, DeClau F, Chen N, Verhoeven K, Prasad S, Tranebjarg L, Morton CC, Ryan AF, Van Camp G, Smith RJ. Mutations in the transcriptional activator Eya4 cause late-onset deafness at the DFN10 locus. Hum Mol Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- Webb JF, Noden DM. Ectodermal placodes: Contributions to the development of the vertebrate head. Am Zool. 1993;33:434–447. [Google Scholar]
- Wegner M. SOX after SOX: SOXession regulates neurogenesis. Genes Dev. 2013;25:2423–2428. doi: 10.1101/gad.181487.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens fibre elongation. Nature Genetics. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Woda JM, Pastagia J, Mercola M, Artinger KB. Dlx proteins position the neural plate border and determine adjacent cell fates. Development. 2003;130:331–342. doi: 10.1242/dev.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurons. J Neuroendocrinol. 2010;22:743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Cheng J, Epstein JA, Maas RL. Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc Natl Acad Sci USA. 1997;94:11974–11979. doi: 10.1073/pnas.94.22.11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Laclef C, Maire P, Maas RL, Peters H, Xu X. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development. 2002;129:3033–3044. doi: 10.1242/dev.129.13.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Dude CM, Baker CV. Fine-grained fate maps for the ophthalmic and maxillomandibular trigeminal placodes in the chick embryo. Dev Biol. 2008;317:174–186. doi: 10.1016/j.ydbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Ranganathan R, Streit A, Moody SA. Microarray identification of novel genes downstream of Six1, a critical factor in cranial placode, somite and kidney development. Dev Dyn. 2014 doi: 10.1002/dvdy.24229. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Luo T, Sargent TD. Expression of TFAP2beta and TFAP2gamma genes in Xenopus laevis. Gene Expr Patterns. 2006;6:589–595. doi: 10.1016/j.modgep.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- Zou D, Silvius D, Fritzsch B, Xu PX. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Rodrigo-Blomqvist S, Enerback S, Xu PX. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas of the inner ear. Dev Biol. 2006;298:430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]