SUMMARY
Triple-negative breast cancer is a heterogeneous disease characterized by poor clinical outcomes and a shortage of targeted treatment options. To discover molecular features of triple-negative breast cancer, we performed quantitative proteomics analysis of twenty human-derived breast cell lines and four primary breast tumors to a depth of more than 12,000 distinct proteins. We used this data to identify breast cancer subtypes at the protein level and demonstrate the precise quantification of biomarkers, signaling proteins, and biological pathways by mass spectrometry. We integrated proteomics data with exome sequence resources to identify genomic aberrations that affect protein expression. We performed a high-throughput drug screen to identify protein markers of drug sensitivity and understand the mechanisms of drug resistance. The genome and proteome provide complementary information that, when combined, provide a powerful engine for therapeutic discovery. This resource is available to the cancer research community to catalyze further analysis and investigation.
INTRODUCTION
A key challenge for medicine in the twenty-first century is to harness the predictive power of molecular data to eradicate cancer (Arteaga and Baselga, 2012; Vidal et al., 2012; Weinstein et al., 1997). Like other cancers, breast cancer is caused by a series of inherited and/or acquired genetic aberrations that eventually lead to uncontrolled cell proliferation and metastasis. The diverse genetic “drivers” of breast cancer have been characterized in exquisite detail (Banerji et al., 2012; Curtis et al., 2012; Perou et al., 2000; Prat and Perou, 2011; The Cancer Genome Atlas Network, 2012; Vogelstein et al., 2013). However, characterization of the proteome has lagged behind.
At the functional level, relevant genomic aberrations affect cellular functions by altering the activity and abundance of proteins. These effects are context specific and very much depend on the unique catalog of proteins expressed by different cell types. For example, a mutation in the BRAF kinase might have different functional outcomes in skin cancer than in liver or breast cancer. In addition to driving cellular functions, proteins are the most actionable and druggable cellular components. Therefore, protein measurements are important to understand breast cancer and delineate breast cancer therapies.
In fact, protein measurements are being used today to classify breast cancer types according to their receptor status, in which the presence or absence of three cellular receptors (estrogen receptor ESR1, progesterone receptor PGR, and human epidermal growth factor receptor-2 ERBB2) is assessed via immunohistochemistry. Despite the reduced number of molecular features measured, this classification is the most useful today for chemotherapy selection. Irrespective of genomic aberrations, more than 80% of breast cancers express one or more of these receptors (Howlader et al., 2014) and are treatable by hormone deprivation and/or ERBB2 inhibition (Untch et al., 2014). Targeted therapies are not currently available for tumors that do not express these receptors, which are collectively referred to as triple-negative breast cancer (TNBC). TNBC is an important and unmet clinical problem. It tends to be more aggressive, is correlated with worse prognosis than receptor-positive subtypes (Hudis and Gianni, 2011), and is more common among young and African American women (Howlader et al., 2014). Identifying subtypes within the TNBC type, and proteins within those subtypes that can serve as therapeutic targets will be extremely valuable.
Among protein measurements, reverse-phase protein arrays (RPPA) have been one the most widely adopted tools for integrated genomics and drug sensitivity analysis, but a key limitation of RPPA technology is its lack proteome coverage, generally less than two hundred analytes (Tibes et al., 2006). As such, mRNA expression has been used as a proxy for protein levels, despite mediocre quantitative concordance (Gygi et al., 1999; Maier et al., 2009). Both mRNA and protein expression using RPPA outperform genomic data as predictors of drug sensitivity and clinical outcomes (Costello et al., 2014; Yuan et al., 2014). These results highlight the potential of systematic protein expression analyses for breast cancer research in general and drug discovery in particular.
It is an excellent time to further investigate the triple-negative breast cancer proteome using more comprehensive techniques. Mass spectrometry in the form of “shotgun proteomics” is highly quantitative, and has reached the speed and sensitivity to measure proteomes at a depth comparable to gene expression studies (Kim et al., 2014; Wilhelm et al., 2014). In fact, proteomics is already making an impact in breast cancer research (Geiger et al., 2012a; Gholami et al., 2013; Kennedy et al., 2014), but yet, to show its full potential, proteomics needs to be integrated with other types of big data.
Here we present an integrative approach using quantitative mass spectrometry to characterize TNBC proteomes both as readouts of genetic abnormality and as predictors of drug sensitivity. The goal of this work is to refine our understanding of breast cancer biology as an integrated ‘proteogenomic’ landscape and to identify molecular diagnostic markers to improve drug selection in triple-negative breast cancer.
RESULTS
The triple-negative breast cancer proteome
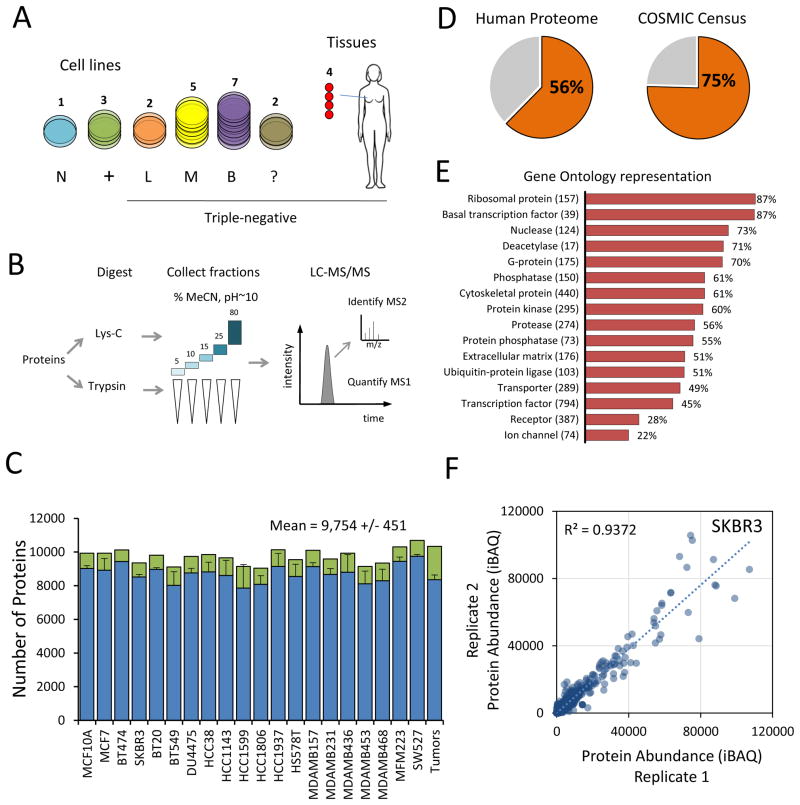
We assembled a panel of twenty human breast cell lines and four clinical tumors to analyze the proteomic landscape of TNBC (Figure 1A). These included 16 triple-negative cell lines covering mesenchymal, luminal, and basal-like subtypes, as well as 3 receptor-positive and 1 non-tumorigenic cell line to serve as a basis for comparison (Lehmann et al., 2011; Neve et al., 2006). Primary tumor tissues were derived from patients with metastatic triple-negative breast cancer (stage II–III). Cell lines were cultured and analyzed in duplicate to assess the precision of protein quantification. Proteins were digested in parallel with either lysyl-endopeptidase (LysC) or trypsin and separated at the peptide-level into five fractions to enhance proteome coverage (Figure 1B). We used liquid chromatography tandem mass spectrometry (LC-MS/MS) on a hybrid quadrupole-orbitrap mass spectrometer to acquire quantitative profiles of the peptides present in each fraction.
Figure 1. Mass spectrometry-based profiling of triple-negative breast cancer.
(A) Overview of samples analyzed. N: normal epithelial, +: ER/PR/ERBB2+, L: luminal-like, M: mesenchymal-like, B: basal-like, ?: not matched. TNBC cell line classifications according to (Lehmann et al., 2011) (B) Workflow of proteomics sample preparation and data collection. (C) Average number of proteins identified in each replicate (blue bars), total number of proteins for each cell line (green bars). Error bars represent S.D. (D) Percent of identified proteins relative to the Uniprot/Swiss-Prot database (left) and the COSMIC census (right). (E) Number and percent representation of indicated gene ontology categories. (F) Representative scatter plot for cell line SKBR3 replicate protein measurements showing quantitative reproducibility of iBAQ protein abundance.
In total, more than 450 peptide fractions were analyzed, yielding approximately 20 million high-resolution mass spectra. Across the entire dataset, we identified 289,819 non-redundant peptide sequences mapping to at least 12,775 distinct proteins encoded by 11,466 genes (protein FDR <1%). To facilitate comparison of specific protein isoforms, we additionally retained in our data truncated protein isoforms having high sequence coverage, bringing the total proteins analyzed to 15,524. The median protein had 15 peptide matches, 4 isoform-specific peptide matches, and shared peptides with only one other protein in the dataset (Figure S1A–C). Median protein sequence coverage was 52%.
The number of proteins identified was consistent across cell lines, tissues, and replicates. On average, 80% of proteins were identified in both replicates. At least 9,000 proteins were found in each cell line (Figure 1C), which agrees well with other recent deep proteome experiments (Beck et al., 2011; Geiger et al., 2012b; Gholami et al., 2013; Nagaraj et al., 2011). These proteins represent 56% of the 20,537 genes annotated in Uniprot/Swiss-Prot and at least 75% of genes included in the catalog of somatic mutations in cancer (COSMIC) (Figure 1D). As expected, we achieved near complete coverage of gene ontology categories involved in core cellular functions such as primary metabolism, protein synthesis, and general transcription, and lower coverage of tissue-specific categories such as transcription factors and receptors (Figure 1E).
To infer protein absolute abundances (as copies/cell) we used the intensity-based approach for absolute quantitation (iBAQ). Quantitative reproducibility between biological replicates was uniformly high across all cell lines, with an average R2 equal to 0.92 (Figure 1F, Figure S1D). Proteins that were highly abundant and identified in all samples were the most reproducibly quantified (median CV = 16%, Figure S1E). By comparison, the average R2 between different cell lines was 0.72, indicating significant differences in global protein expression.
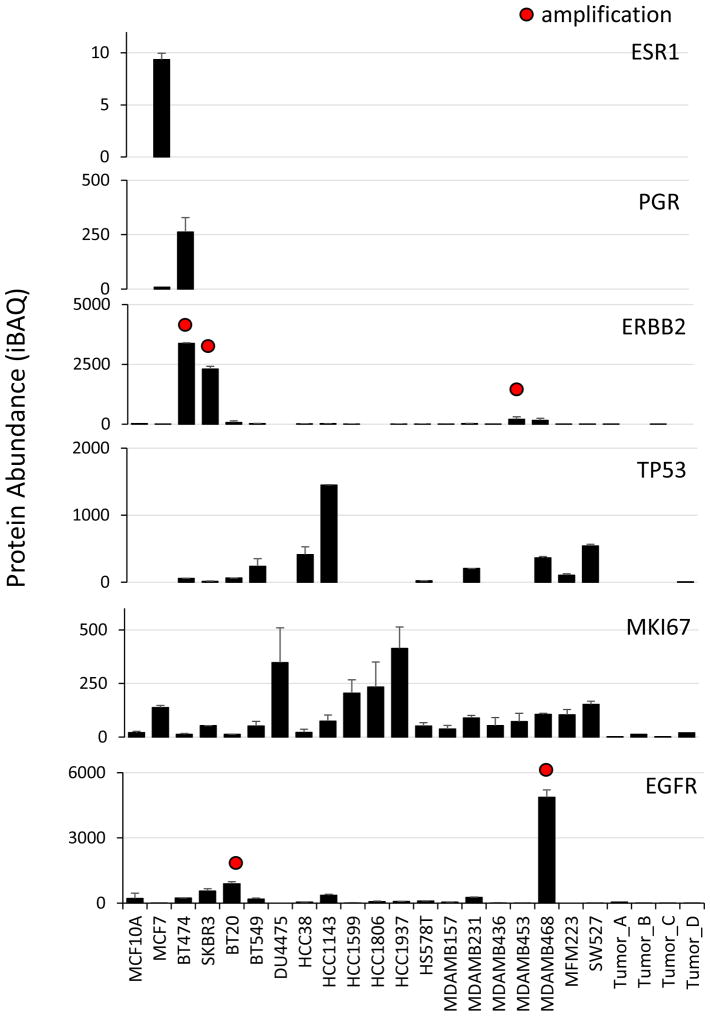
The data presented here comprises more than 200,000 quantitative measurements of absolute protein abundance (Table S1). Innovations in instrumentation and extensive peptide fractionation prior to analysis have greatly increased the sensitivity and reproducibility of “shotgun proteomics” analysis, and our quantitative results compared favorably with a recent targeted proteomics study on many of the same cell lines (Kennedy et al., 2014) completed by the CPTAC (Clinical Proteomic Tumor Analysis Consortium). To facilitate use and dissemination of the data, we have developed a web resource (https://zucchini.gs.washington.edu/BreastCancerProteome/) in which protein abundances can be queried, and correlated to genomic and drug sensitivity data, as presented below. To demonstrate the validity of our data set as a quantitative resource, we examined several clinical breast cancer biomarkers including ESR1, PGR, and ERBB2 (Figure 2). These measurements accurately reproduce the known classification of cell lines based on immunocytochemistry (Subik et al., 2010) and correspond with known copy number amplifications. In contrast to antibody staining, which assesses the presence or absence of expression, mass spectrometry provides sensitive and precise quantitation over a broad range. This is an important consideration for markers such as Ki-67, which are dynamically expressed in all cells. As another example, the cell line MDA-MB-453 stains negative for ERBB2 (Vranic et al., 2011) and was classified as a TNBC cell line (Neve et al., 2006), despite bearing a copy number amplification. However, our results show that MDA-MB-453 expressed ERBB2 at levels 20-fold higher than the median, compared to several hundred-fold overexpression of ERBB2 by cell lines such as BT474 and SKBR3.
Figure 2. Quantification of clinical breast cancer biomarkers.
ESR1: estrogen receptor, PGR: progesterone receptor, ERBB2: human epidermal growth factor receptor-2, TP53: tumor protein p53, MKI67: Ki-67 antigen, EGFR: human epidermal growth factor receptor. Sample labels are shown in the bottom panel. Absolute protein abundance was calculated using intensity-based absolute quantification (iBAQ). Error bars represent S.D. Red dots indicate gene copy number amplification (>7 copies).
Quantitative analysis of TNBC proteomic subtypes
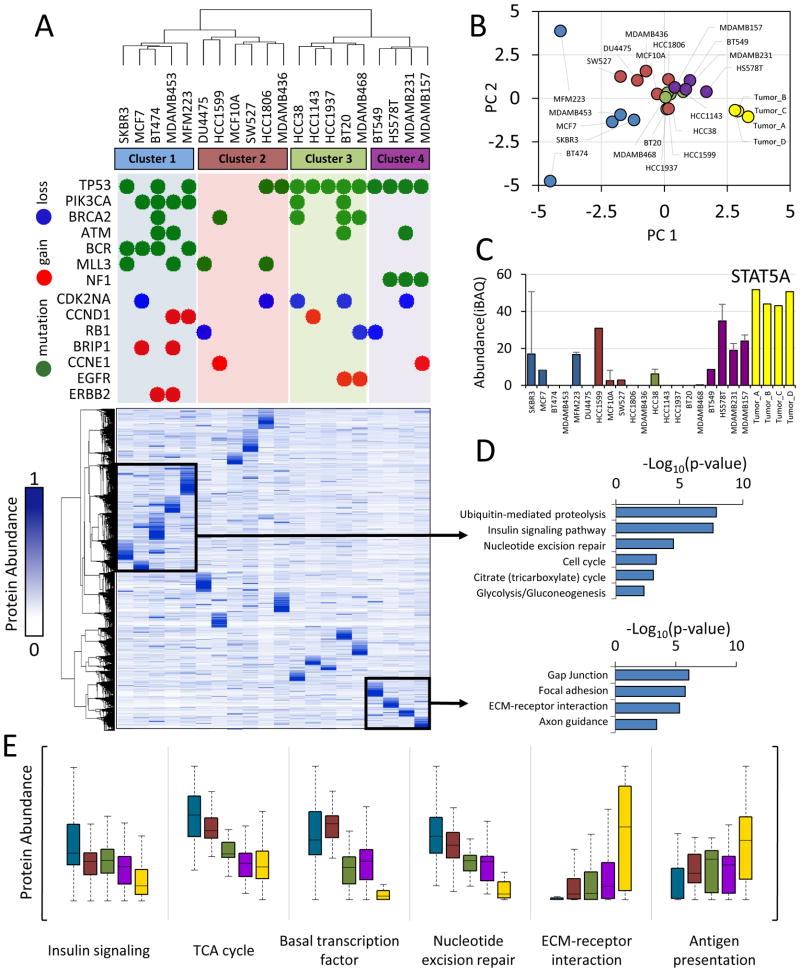
Molecular subtyping using gene expression or copy-number aberration has been used extensively to characterize clinical breast cancer specimens and cell lines (Banerji et al., 2012; Lehmann et al., 2011; Prat and Perou, 2011). We used hierarchical clustering to identify patterns based on correlation of protein expression profiles. This approach classified the panel of cell lines into two overarching groups containing four clusters (Figure 3A). To illustrate the relationship between driver gene alterations and proteome profiles, we show the most frequent census mutations and copy number aberrations for each cell line (Figure 3A, upper). Cell lines with similar genetic abnormalities tended to cluster together. As has been observed previously (The Cancer Genome Atlas Network, 2012), PIK3CA mutations were associated with luminal breast cancer subtypes (80% of the cell lines in cluster 1), whereas TP53 mutations were characteristic of triple-negative breast cancer (100% of the cell lines in clusters 3 and 4). Mutations in the tumor suppressor NF1 were exclusive to the mesenchymal-like subtype (cluster 4) and BCR mutations were exclusive to luminal cells (cluster 1).
Figure 3. The triple-negative breast cancer proteome.
(A) Hierarchical clustering of protein expression profiles computed using centered Pearson’s correlation identified four proteome subtypes as indicated. Protein expression values were normalized to a scale from 0 to 1 prior to clustering. Frequent genetic aberrations are overlaid onto the proteome clustering results. Green circles represent exonic mutations. Red and blue circles represent copy number gain (>7 copies) or loss (0 copies), respectively. Colored background shading corresponds to cluster membership. At the time of writing, exome sequence and copy number data were not available for MCF10A, SW527. (B) Scatter plot of principal component 1 and 2. Principal component analysis was performed using protein expression profiles. Each point represents a sample. Colors represent hierarchical cluster membership from (A). (C) Biological pathways enriched from the indicated proteins clusters. Inverted log10 p-values are shown. (D) Representative example of a protein upregulated in cluster 4 and tumors. STAT5A: signal transducer and activator of transcription 5A. Error bars represent S.D. (E) Distribution of protein abundances within each cluster (colors) for indicated biological processes. For all panels, cluster membership is indicated by the same colors used in (A), with tumor samples indicated in yellow.
Protein expression patterns within subtype clusters were still highly cell-type specific. To better illustrate this, we used principal component analysis (PCA) to project the distances between each proteome onto a two-dimensional coordinate system. Some of the sample proteomes formed tight clusters, while others were more distantly related to those in the same group (Figure 3B). Additional principal component dimensions are necessary to capture the proximity of cell lines such as MFM223, BT474, and HCC1599 to their respective subtypes. Intra-subtype correlation was also modest in earlier classification studies using mRNA expression (Lehmann et al., 2011), and the differences in mRNA may be further amplified at the protein level. The heterogeneity of protein expression underscores the importance of data-driven cell line selection in cancer research.
Accurate analysis of genes, transcripts, or proteins from heterogeneous clinical specimens represents a major challenge for precision medicine. The proteins expressed >10-fold in tumors versus the cell lines were enriched with proteins from blood cells and plasma (p<0.001). These proteins accounted for as much as 20% of the total proteome intensity from the tumors. Since TNBC cell lines should better represent the cellular component of the tumor we correlated tumor samples to the centroids from each cell line cluster to identify which proteomic subtype they belonged to, and found that they were all more similar to clusters 3 and 4, an observation which can also be made based on PCA (Figure 3B).
Nevertheless, many proteins significantly over- or under-expressed within each cluster could be identified. We were particularly interested in potential drug targets and proteins known to be involved in cancer biology. For example, the protein STAT5A, a pro-survival transcription factor, was expressed at high levels in the tumors and mesenchymal-like cell lines (Figure 3C). Using the first cluster as an example, we show how these proteins can be identified using our web-based resource (Figure S2A). The transcription factor FOXA1 was exclusively expressed by luminal-like cells, while TGFB1 was not found (Figure S2B). PPM1A, a protein involved in the suppression of TGF-β signaling pathways (Lin et al., 2006), was decreased in TNBC, while many proteins involved in immunity and metastasis such as POSTN, MYLK, and HLA-A were expressed at higher levels in TNBC (Figure S3A). Some of these proteins are thought to be provided by tumor-infiltrating immune cells and fibroblasts (Quail and Joyce, 2013), but here we show they are also abundant in the homogenous conditions of cell culture.
The composition of each cluster showed striking similarity to subtypes defined by mRNA expression arrays and morphological studies (Kenny et al., 2007; Lehmann et al., 2011; Neve et al., 2006). Cluster 1 contained the luminal breast cancer cell lines SKBR3, MCF7, and BT474 as well as “luminal-androgen-receptor” cell lines MFM-223 which expresses the androgen receptor protein, and MDA-MB-453 which overexpresses ERBB2 as described above. The set of proteins that were highly expressed by these cell lines was enriched for functions typically expected of cancer cells including insulin and ErbB signaling, glycolysis, and nucleotide excision repair (Figure 3D). Cluster 2, most similar to the “basal-like 2” gene expression subtype, contained, DU4475, SW527, HCC1806, MDA-MB-436, and the normal breast epithelial cell line MCF10A. Cluster 3 included all “basal-like 1” cell lines: HCC38, HCC1143, HCC1937, BT20, and MDA-MB-468. Cluster 4, containing BT549, HS578T, MDA-MB-231, and MDA-MB-157, was identical to “mesenchymal-like/claudin-low” subtype (Lehmann et al., 2011), all showing stellate morphology in 3D culture (Kenny et al., 2007), and high invasiveness in chamber assays (Neve et al., 2006) (Figure 3D). To better understand the biology of each subtype, we compared the distribution of protein abundance within gene ontology categories. Interestingly, luminal-like cells expressed higher levels of pathways associated with proliferation such as cell cycle, growth factor signaling, metabolism, and DNA damage repair mechanisms (Figure 3E, Figure S3B). TNBC cell types, particularly the tumors and more invasive cells, expressed higher levels of pathways associated with metastasis such as ECM-receptor interaction, cell adhesion, and angiogenesis (Figure 3E, Figure S3B). The expression of proliferation and metastasis pathways were mutually exclusive, an observation also made in an analysis of mRNA expression profiles from claudin-low tumors (Prat et al., 2010). Thus, therapies targeting immune and metastatic signaling are an exciting avenue for TNBC treatment.
Differential expression of cancer signaling proteins
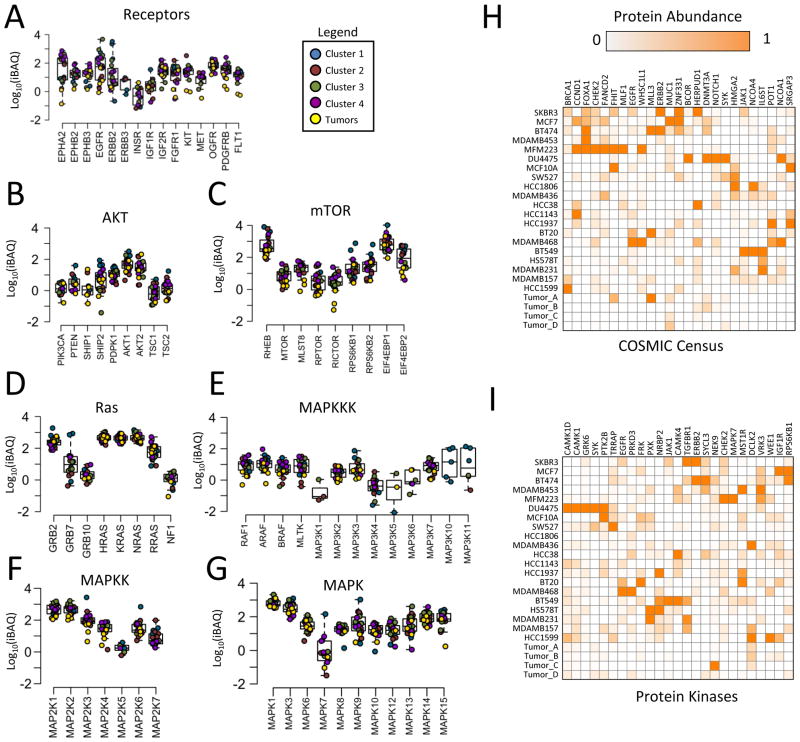
The cancer genome has been studied extensively (Futreal et al., 2004; Vogelstein et al., 2013). We sought to characterize the abundance of proteins derived from cancer census genes and signaling pathways (Figure 4, Figure S4). The abundance of most signaling proteins spanned two to three orders of magnitude, but others were expressed similarly across all cell lines (Figure 4A–G). These proteins included several members of the RAS-MAPK pathway such as GRB2, HRAS/KRAS/NRAS, MEK1/2, and ERK1/2. In certain cases expression of these proteins was associated with proteomic-based breast cancer subtypes. For example, CHEK2, HMGA2, POT1, and IL6STwere highly expressed by members of clusters 1 through 4, respectively (Figure 4H–I). However, protein expression was generally variable and cell-type specific. MLL3 was specifically expressed by BT474, BT20, and tumor A, which were each from different clusters (Figure 4H). HCC1806 and MDA-MB-436 specifically lacked expression of the protein kinase AKT1/2 (Figure 4B). PKCα was expressed at high levels in each of the cell lines from cluster 4, but also was highly expressed in DU4475 (Figure S4J). These results show that despite overall concordance of whole proteome profiles with various cellular phenotypes, in most cases the expression of particular cancer proteins did not uniformly belong to one subtype or another. The identification of proteins with very specific outliers or large dynamic range provides a valuable resource for TNBC drug development efforts. EGFR, ERBB2, ESR1, and PGR exemplify these properties (Figure 4A, Figure S4D) and are already routine clinical targets in breast cancer, but there are many others. For example, ephrin type A receptors, which are involved in embryonic development and not normally present in adult tissues, were overexpressed by several orders of magnitude in many TNBC cell lines compared to luminal-like cells (Figure 4A). With the increasing availability of comprehensive quantitative proteomics datasets, protein expression should continue to be one of the most valuable parameters for drug development and clinical diagnostics.
Figure 4. Expression of cancer signaling proteins.
(A–G) Distribution of absolute abundance for each protein in the signaling network. Chart titles indicate subnetwork membership. Each data point represents a sample, color coded according to cluster membership from Figure 4A. (H) Top 25 most differentially expressed proteins (highest standard deviation between different samples) from the COSMIC gene census or (I) the protein kinase superfamily.
Isoform-specific protein expression
The identification and quantification of protein isoforms resulting from alternative splicing is a significant challenge in proteomics, arising from the reduced number of isoform-specific peptides that are amenable to analysis by mass spectrometry. For this dataset, we first relied on isoform-specific peptides to unambiguously identify proteins mapping to the same gene in the Uniprot sequence database. This led to the identification of 1,860 protein isoforms that corresponded to 844 genes, 52 of which were members of the COSMIC census. Next, we examined the relative quantification of protein isoforms. Protein isoforms share long segments of identical sequence but are missing certain protein domains, resulting in altered signal intensity from those parts of the protein.
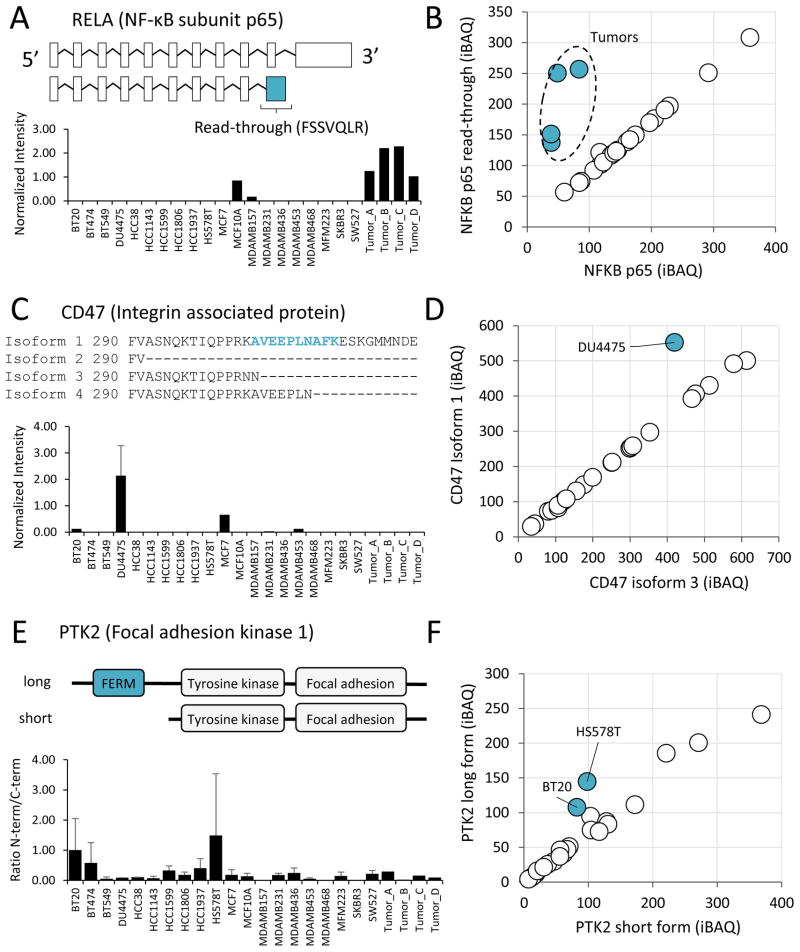
We relied on manual inspection to analyze the expression of isoforms for proteins involved in cancer progression. For most proteins, different isoforms were nearly perfectly correlated, indicating no difference in expression of specific isoforms, but there were notable exceptions. For example, we identified variants in the p65 subunit of the transcription factor NF-κB, the tumor antigen CD47, and focal adhesion kinase PTK2. The protein sequence of the NF-κB p65 variant is identical to the canonical sequence until proline 344, followed by the read-through translation of 33 amino acids and an early stop (Figure 5A). The alternative sequence lacks many important regulatory regions including the residues phosphorylated by IKKB that directly affect its transcriptional activity (Sakurai et al., 1999). The p65 variant was detected in two cell lines and was expressed at higher levels in all four tumor samples (Figure 5B). This result was confirmed by an isoform-specific peptide, FSSVQLR, which matched no other entry in the Uniprot protein sequence database (Figure 5A). This finding was especially interesting since the tumor proteomes were enriched in immuno-modulatory pathways. NF-κB modulates the inflammatory response and plays an important role in cancer by promoting metastasis (Huber et al., 2004; Luo et al., 2004).
Figure 5. Differential expression of protein isoforms.
(A) Schematic of RELA (NF-κB subunit p65) mRNA sequence variants and intensity-based quantification of the isoform-specific peptide FSSVQLR in each sample. Peptide intensity was divided by the total proteome intensity for normalization. The location of an exon read-through event is indicated. (B) Scatterplot of the full length NF-κB protein versus the read-through variant highlighting off-diagonal samples. (C) Four alternative splice variants encode the cytoplasmic tail of integrin associated protein CD47. The sequence of these variants is shown along with the quantification of the peptide specific to isoform 1, AVEEPLNAFK. (D) Scatterplot of CD47 isoform 1 versus isoform 3 highlighting off-diagonal samples. (E) Schematic of N-terminally truncated form of focal adhesion kinase PTK2 and quantification of N-terminal/C-terminal intensity in each sample. (F) Scatterplot of PTK2 long form versus short form highlighting off-diagonal samples.
CD47 is an atypical G-protein coupled receptor with five membrane spanning domains that participates in integrin signaling and is proposed to have many important roles in cancer (Sick et al., 2012). We detected two of the four known alternative splice variants which differentially encode the cytoplasmic tail. The cell line DU4475 expressed higher levels of the long isoform (Figure 5C–D), which is highly expressed in neurons (Brown and Frazier, 2001). While little is known about the functional differences between the isoforms, it is likely that this tail mediates intracellular signaling downstream of the receptor.
PTK2, or focal adhesion kinase 1, is a tyrosine protein kinase involved in cell migration (McLean et al., 2005). We confirmed the presence an N-terminally truncated form of this protein which lacks the FERM (4.1-Ezrin-Radixin-Moesin) domain (Figure 5E). The FERM domain regulates PTK2 localization and interaction with other proteins to affect its activity (Frame et al., 2010). Interestingly, the full-length form appeared to be expressed higher in HS578T and BT20 cells based on the relative intensity of N-terminal versus C-terminal peptides (Figure 5E–F). The differential expression of structural protein variants, many of which occur post-translationally, could be a significant regulatory mechanism in cancer. Further work will be necessary to systematically identify and accurately quantify these events.
Proteogenomic analysis identifies signatures of driver mutations
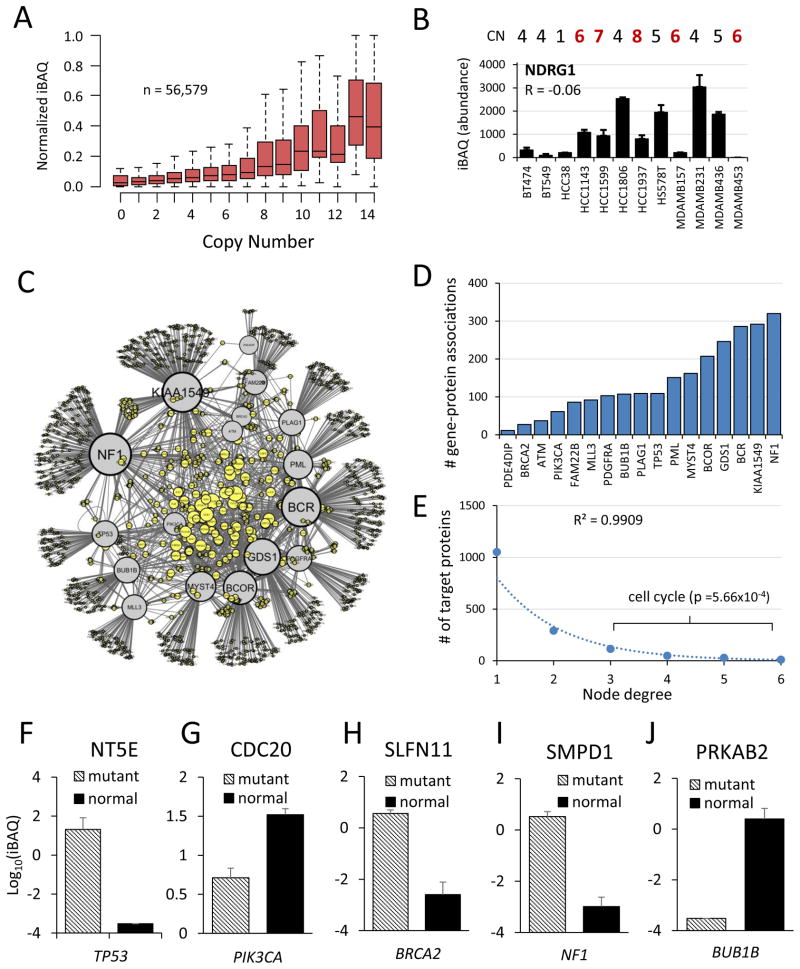
Genetic aberrations such as sequence mutations and amplifications, which typically occur in regulatory proteins, can have pleiotropic downstream effects on other proteins that more directly drive cancer phenotypes. We integrated publicly available exome sequence and gene copy number (CN) data from COSMIC (Forbes et al., 2011) with proteome profiles from 18 cell lines. Protein abundance trended positively with gene CN. The average expression of all proteins in each CN bin correlated strongly with CN (R = 0.96). However, it was more variable and correlated poorly on a pairwise basis (n = 56,579, R = 0.19) (Figure 6A). For example, the cancer census gene NDRG1 was not correlated with CN (R = −0.06) and was not highly expressed even when amplified (Figure 6B). This poor correlation is expected for proteins under high transcriptional, translational or proteasomal control.
Figure 6. Proteogenomic associations.
(A) Boxplot showing relationship of protein abundance versus gene copy number. Protein abundances were row-normalized to a scale of 0 to 1 to account for differences in absolute expression. (B) NDRG1 (N-myc downstream regulated gene 1), a representative protein that was not correlated with copy number. CN: copy number. CN>6 highlighted in red. R represents Pearson’s correlation. Error bars represent S.D. between replicate measurements.
(C) Network of gene-protein associations. Each edge represents an association (p < 0.001) between a mutated census gene (gray nodes) and protein expression (yellow nodes). Only genes from the COSMIC census mutated in at least 3 cell lines were analyzed. Node size represents the number of connections. The network was plotted in Cytoscape using ‘edge-weighted spring embedded’ layout so that genes with common associations cluster together. (D) Number of outgoing associations for each mutated gene in network. (E) Number of incoming associations for each target protein in network (node degree distribution). Cell cycle proteins were enriched among proteins with 3 or more associated genes (p = 5.66 × 10−4). (F–J) Representative gene-protein associations (p < 0.001) for common genetic lesions in breast cancer. Protein is indicated in chart title, and mutated gene shown in italics below plot. Error bars represent S.E.M.
Driver mutations occur frequently in regulatory proteins such as protein kinases, E3 ubiquitin ligases, and transcription factors which alter the physiology of the cell by modulating the abundance or activity of other proteins. For example, our data showed that DU4475, the cell line with an APC mutation, expressed more than 4-fold median levels of β-catenin (P = 3.3×10−4, heteroscedastic t-test) (Table S1), which APC normally targets for degradation. Initially we characterized cellular subtypes according to protein abundance profiles and asked whether frequent genetic mutations were associated with these subtypes (Figure 3). An alternative analysis approach is to group cell lines by their mutational status, and ask whether the abundance of specific proteins are associated with these mutations, as in the β-catenin and APC example.
We reasoned that mutations in certain driver genes, such as those in the same signaling pathway, would likely converge to regulate common effectors. To determine the global effects of driver gene mutations on protein expression, we systematically evaluated gene-protein associations for frequently mutated census genes (n ≥ 3 cell lines) by comparing the abundance of each protein in cell lines with versus without a mutation, and plotted this information as a network. Driver genes and their protein targets formed clusters according to their shared associations (Figure 6C). The number of significant (P<0.001) associations for each gene ranged from 11 to 320 (Figure 6D). The network degree distribution fit an exponential function (R2 = 0.99), revealing 233 ‘hub’ proteins, each associated with 3 or more cancer census genes (Figure 6E). ‘Cell cycle’ was the only significantly enriched gene ontology term among hub proteins (P = 5.66×10−4). While not surprising, it demonstrates that dysregulation of cell cycle protein abundance may be a common effect of diverse genetic mutations.
On an individual basis, proteins regulated downstream of genetic lesions (e.g. TP53 loss-of-function) might represent more suitable therapeutic targets than the gene product itself. Several highly significant (P<0.001) gene-protein associations are shown (Figure 6F–J). In the case of TP53, nearly all of the significantly associated proteins were involved in DNA metabolism and repair. One such protein was ecto-5′-nucleotidase (NT5E or CD73), a GPI-anchored cell surface enzyme involved in the production of membrane-permeable nucleosides which can be used for nucleotide salvage (Zimmerman, 1992). Targeting it by siRNA or small molecule inhibition (using adenosine [(α,β)-methylene] diphosphate) arrested the cell cycle and triggered apoptosis in MDA-MB-231 breast cancer cells (Zhi et al., 2010). Monoclonal antibodies against NT5E were also demonstrated to block breast cancer metastasis in vivo (Stagg et al., 2010). NT5E may be an effective drug target specifically for cancers with TP53 mutations. In addition to the discovery of potential drug targets, these proteins could also be used as markers to infer whether or not a mutation is deleterious.
Proteomics of drug sensitivity
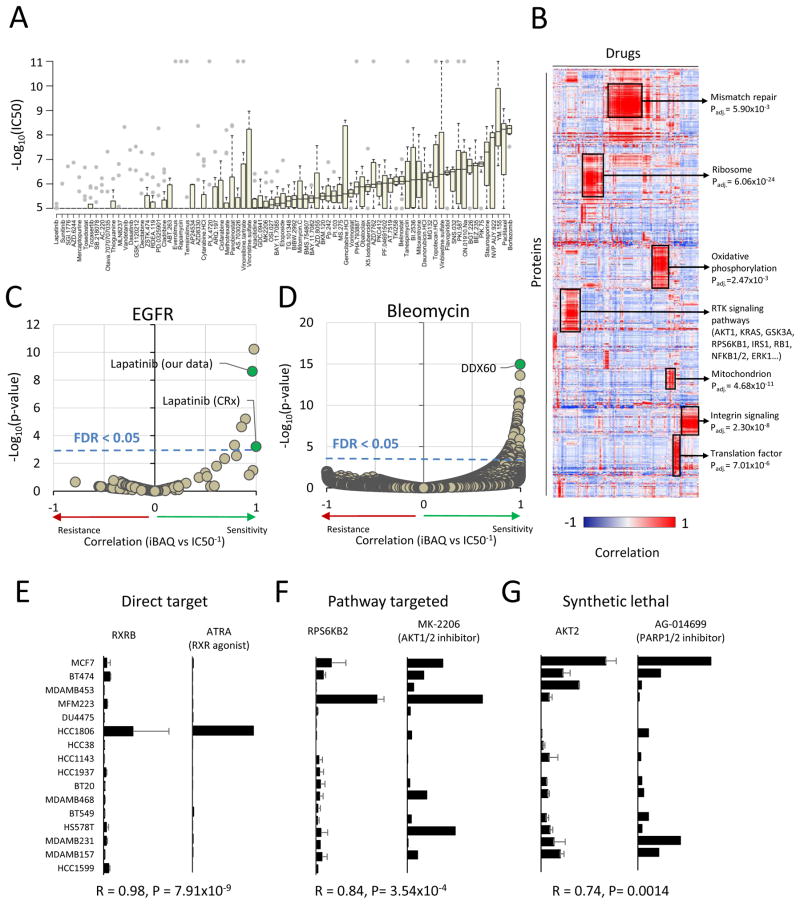
To generate a resource for drug sensitivity prediction, we screened the sixteen TNBC cell lines from our panel against a library of 160 compounds at eight different concentrations spanning four orders of magnitude. We used this data to determine the IC50, defined as the dose required to reach a 50% reduction in cell viability, for each drug in each cell line (Table S2). Approximately three quarters (123/160) of the compounds elicited a measurable response in at least one cell line, and each cell line was sensitive to at least 5 compounds at sub-micromolar doses. The distribution of responses for each drug was diverse (Figure 7A). The IC50 distribution for most drugs spanned a wide range, 790-fold on average. Some drugs were very specific with few sensitive cell lines (e.g. everolimus, methotrexate, lapatinib), while other drugs were indiscriminate with few resistant cell lines (e.g. bortezomib, paclitaxel, MG132).
Figure 7. Protein expression and drug sensitivity.
(A) Distribution of drug sensitivity (−log10IC50) values across 16 TNBC cell lines for each drug in order of increasing median sensitivity. Drugs with sub-micromolar IC50 in at least one cell line are shown. Grey dots represent outlier values (>1.5× interquartile range). (B) Hierarchical clustering of drug-protein associations. Pairwise Pearson’s correlation was calculated systematically between drug sensitivity (inverted IC50) and protein abundance (iBAQ) values and clustered in both dimensions. Enriched gene ontology terms are shown for several clusters with Benjamini-Hochberg adjusted p-value. (C) Association of drug sensitivity with EGFR expression. The EGFR inhibitor lapatinib was significantly associated in both drug screen datasets (CRx: P = 6.2×10−4, our data: P = 2.4×10−9, FDR < 0.05). (D) Association of protein expression with bleomycin sensitivity. The protein DDX60 was significantly associated bleomycin sensitivity (P = 1.1×10−15, FDR < 0.05). (E–G) Pairwise comparison of protein expression and drug sensitivity for three examples. Direct target: expression of the target protein indicates sensitivity to the drug. Pathway target: expression of a protein in the pathway of the drug target, but not the target itself, indicates sensitivity. Synthetic lethal: expression of a protein in an independent pathway from the drug target indicates sensitivity. Left panel: protein abundance (iBAQ) across cell lines. Right panel: drug sensitivity (inverse IC50, M−1) across the same cell lines. RXRB: retinoid X receptor beta, RPS6KB2: ribosomal protein S6 kinase-2, AKT1: RAC-alpha serine/threonine-protein kinase. ATRA: RXR agonist all-trans retinoic acid, MK-2206: pan-isoform AKT inhibitor, AG-014699: poly-ADP ribose polymerase 1/2 inhibitor. Pearson’s correlation and p-value is indicated below the plots. CRx: Data from (Yang et al., 2013). Panel A includes only data generated in this study. For panels B–G, data from the CRx was included. Missing IC50 values were not imputed.
Next, we combined our pharmacological data set with publicly accessible data from the Genomics of Drug Sensitivity in Cancer (CRx) resource (Yang et al., 2013) and performed regression analysis against mass spectrometry-derived protein abundances to discover proteomic markers of drug sensitivity or resistance. We used hierarchical clustering to analyze global patterns among drug sensitivity-protein expression relationships, revealing many distinct clusters (Figure 7B). Drugs targeting proteins in the same pathway (e.g. BRAF and MEK inhibitors) showed similar correlation profiles. Interestingly, proteins that were part of the same pathways or complexes also clustered together, which did not occur using protein expression data alone (Figure 3A). The cluster that was highly enriched with mitochondrial proteins was associated with sensitivity to drugs that might depend on mitochondrial protein expression (belinostat, vorinostat, obatoclax). For example, since protein acetylation is known to be enriched within the mitochondrial space, cells with more mitochondria might be more sensitive to deacetylase inhibition. In a similar vein, the cluster that was enriched with translation factors was associated with increased sensitivity to proteasome inhibitors MG132 and bortezomib. These results show that integration of proteomics and drug sensitivity data using regression analysis provides a rich resource to identify unexpected modes-of-action and to discover new features of target pathways.
We used the regression analysis to select the most effective and robust drugs for known targets. For example, EGFR expression was, as expected, strongly associated with sensitivity to the EGFR inhibitor lapatinib in both drug screens (our data: R = 0.96, P = 2.36 × 10−9; CRx: R = 0.99, P = 6.2 × 10−4) (Figure 7C). Proteomics data can also be used to uncover mechanisms of drug sensitivity. For example, several cell lines were hypersensitive to the drug bleomycin, an antibiotic used to treat plantar warts as well as many forms of cancer by inducing DNA damage. Expression of DDX60, an antiviral RNA/DNA helicase that binds cytosolic DNA (Miyashita et al., 2011), was most significantly associated with sensitivity to bleomycin (R = 0.99, P = 1.1 × 10−15) (Figure 7D).
We curated these drug sensitivity results to ask whether drug sensitivity associated with (1) genetic mutations or protein expression of the drug target itself, (2) proteins in the same pathway as the target, or (3) other literature-supported ‘synthetic lethal’ interactions. Drug sensitivity associated strongly with both genomic and proteomic features of known targets. For example, we found that sensitivity to all-trans retinoic acid (ATRA) was correlated with the expression of its target protein RXRB (R = 0.98, P = 7.91 × 10−9). HCC1806 cells, which expressed the highest level of RXRB, were >200-fold more sensitive than the median cell line (Figure 7E). The cell line DU4475, which harbors the hyperactive BRAF-V600E mutation, was hypersensitive to both BRAF and MEK inhibitors (6,000-fold and 100,000-fold versus median, respectively) despite similar expression of the target proteins.
Another potential mechanism of drug sensitivity is synthetic lethality, in which the right combination of genetic, proteomic, or pharmacologic perturbations leads to cell death. Synthetic lethality tends to occur between proteins in the same pathway. For example, the AKT1/2 inhibitor MK-2206 was not associated with expression of AKT isoforms, but was significantly associated with expression of RPS6KB2 (R = 0.84, P = 3.54×10−4) (Figure 7F), which lies downstream in the signaling pathway (Shaw and Cantley, 2006). Other drugs correlated with proteins that are not known to be in the same pathway, but have been previously proposed to be synthetic lethal relationships in genetic datasets. For example, poly-ADP ribose polymerase (PARP) inhibition disrupts DNA repair leading to genotoxic stress and cellular senescence, a process shown to be accelerated in overactive AKT signaling mutants (Chatterjee et al., 2013; Mendes-Pereira et al., 2009). In our data, AKT protein expression was also significantly correlated with sensitivity to PARP inhibition using AG-014699 (R = 0.74, P = 0.0014) (Figure 7G).
Finally, we explored how the differences in drug sensitivity and target expression between members of a signaling pathway relate to pathway structure. In the Akt-mTOR-S6K signaling pathway, ribosomal protein S6 kinases (RPS6KB1/2) are activated by mTOR. Curiously, despite its association with MK-2206 sensitivity, expression of either RPS6KB1 or RPS6KB2 was inversely correlated with the S6K inhibitor PF-4708671 in luminal breast cancer cells (R = −0.96, P = 0.04) (Figure S5A). This is consistent with the suggestion that S6K inhibition may amplify upstream cancer signaling due to the chronic ablation of a negative feedback loop (Carracedo et al., 2008; Manning, 2004). Thus, the tumorigenic action of this protein may be best targeted indirectly (Figure S5B). Unlike RPS6KB2, RPS6KB1 expression did not correlate with AKT1/2 inhibitor MK-2206 sensitivity but instead was most highly correlated with the p21-activated kinase (PAK) inhibitor IPA-3 (R = 0.99, P = 1.91×10−12). Based on images from the Human Protein Atlas (Figure S5C), RPS6KB1 and PAK2 are localized to the nucleus whereas RPS6KB2 and PAK1 are cytoplasmic (Uhlen et al., 2010). Thus, the reported activation of PAK1 downstream of S6K (Ishida et al., 2007) might be localized and isoform-specific. Together, these results demonstrate that integrated analysis of drug sensitivity and protein expression provides a useful strategy for drug selection, finding diagnostic markers, and identifying potential mechanisms of cellular signaling. Further experimentation will be required to confirm these findings.
Finally, to demonstrate the potential clinical utility of these results, we asked how many proteins from the drug association analysis could be identified in primary tumors. We found that 73% (6,798/9,292) were quantifiable in the four clinical specimens we analyzed (Figure S5D). Of these, 494 were at least 5-fold more abundant than the average sample in at least 1 tumor. For example, the abundance of the protein kinase AKT2 was higher in one of the tumor samples than in any cell line analyzed in this study (Figure S5E).
DISCUSSION
Despite the success of large-scale ‘omic’ studies in providing molecular targets for therapeutic intervention, these studies have been limited by the lack of comprehensive protein data. Mass spectrometry-based proteomics has advanced rapidly and it has become a routine to reproducibly quantify near-complete proteomes using this technology. Here we used mass spectrometry to interrogate the proteomes of TNBC. We then integrated proteomics, genomics and drug sensitivity data to study the effects of genomic aberrations in the proteome and build prediction models of drug response using proteomics.
This dataset is a useful resource to further explore the biology of TNBC. For example, many of the recently described metastatic stem cell pathways were highly expressed at the protein level in TNBC compared to luminal breast cells. The most invasive TNBC cells and solid tumors expressed low levels of proteins involved in cell proliferation and high levels of proteins involved in the epithelial-to-mesenchymal transition. Thus, the highly specialized nature of metastatic TNBC cells may be one reason they are so difficult to treat using conventional cytotoxic agents that target highly proliferative cells. Precise knowledge of the proteomes of these cells can guide the development of new drugs to target the metastatic transition.
Machine learning has become a useful tool to capture the molecular features responsible for differences in drug sensitivity (Barretina et al., 2012; Costello et al., 2014; Weinstein et al., 1997; Yang et al., 2013). Statistically significant differences in drug sensitivity based on cellular subtype have been observed (Lehmann et al., 2011), but the effect sizes are small compared to treatment strategies directed towards precise molecular insults. Examples include ERBB2 amplification (trastuzumab), BCR-ABL fusion (imatinib), or BRAF-V600E mutation (vemurafenib), all of which result in orders-of-magnitude increases in drug sensitivity. In reality, large effect sizes are needed to make an impact in the clinic. In this study, drug sensitivity and the expression of cancer-related proteins was not generally attributable to subtypes derived by clustering global protein profiles. Considering these cells were all derived from the same tissue type (breast) and were cultured in the same conditions, the dynamic range and specificity of protein expression for established regulatory proteins and drug targets was surprising. Using regression and prior knowledge to interrogate mechanisms of protein expression in drug sensitivity, we found that in many cases, drug sensitivity was strongly correlated with the expression of the drug target itself (e.g. retinoic acid receptors, EGFR) or proteins in the same biological pathway (e.g. S6K expression as a marker for sensitivity to AKT inhibitors).
With the exception of drugs targeting amplified genes, the importance of protein expression in drug efficacy might be underestimated. While it is evident that the target of a drug must be expressed at some level in order for the drug to take effect, many drugs are developed with the assumption that the target is expressed at similar levels in all cells. Even in the case of gene amplification, copy number does not fully account for differences in protein expression between specimens. In any case, quantitative analysis of drug targets and genetic abnormalities at the protein level might represent a useful addition to the current adjuvant therapy selection algorithm. Indeed, this is already routine for estrogen, progesterone, and epidermal growth factor receptor-2. Larger panels of cell lines will be necessary to capture rare genetic events and to enable more robust machine learning approaches. This will facilitate the discovery of less obvious markers of drug sensitivity, such as synthetic lethal interactions. Proteomics could also provide an indispensable tool to rescue clinical trial results which do not improve patient outcomes in aggregate, but have many exceptional responses that might be due to underlying molecular features.
This study builds upon other deep proteomic characterizations of cancer (Geiger et al., 2012b; Gholami et al., 2013; Nagaraj et al., 2011; Zhang et al., 2014) and represents the first deep proteome characterization targeting triple-negative breast cancer. With the development of large “omics” approaches, personalized, predictive medicine is the prevailing direction of next-generation healthcare technology (Tian et al., 2012). Systematic, data-driven approaches are necessary to meet this goal. We anticipate that genome-scale nucleic acid sequencing and protein analysis will provide the basic molecular diagnostics toolbox for precision cancer medicine. Triple-negative breast cancer is one of many unmet clinical needs that will benefit from future research in this area.
EXPERIMENTAL PROCEDURES
Sample preparation
Samples were lysed in denaturing buffer and centrifuged at 12,000 g for 10 min to pellet insoluble material. Protein extracts were reduced with 5mM DTT at 55°C and alkylated with 15mM iodoacetamide at room temperature in the dark. Extracts from each sample (25μg) were diluted and digested in solution overnight with either lysyl-endopeptidase (Lys-C) (Wako) or sequencing grade trypsin (Promega). Peptides were desalted and fractionated on StageTips (Rappsilber et al., 2007) by basic reverse-phase using a step-wise gradient of increasing acetonitrile (5%, 10%, 15%, 25%, 80%) in 0.1% NH4OH. The resulting fractions were analyzed by LC-MS/MS.
LC-MS/MS
Peptide fractions were analyzed on an EASY-nLC-1000 (Thermo) coupled to a hybrid quadrupole-orbitrap Q-Exactive mass spectrometer (Thermo) configured for data dependent acquisition. Raw mass spectra were searched using Sequest (release 2012.01.0 of UW Sequest) against a concatenated forward and reverse version of the Uniprot human protein sequence database (v11/29/2012). Peptide spectral matches for all fractions corresponding to the same sample were filtered to reach a protein identification false discovery rate of less than 1%, resulting in an aggregate peptide-level FDR of less than 0.1% for the entire dataset. Protein quantifications were calculated using the intensity-based absolute quantitation (iBAQ) approach (Schwanhäusser et al., 2011).
Drug screen and curve fitting
Compounds were added to cells using the CyBi-Well Vario Workstation (CyBio) and incubated at 37°C, 5% CO2 for 96 hours. Cell viability was measured by luminescence using quantitation of ATP as an indicator of metabolically active cells. Measurements were corrected for background luminescence and percentage cell viability is reported as relative to the DMSO solvent control. Non-linear curve fitting was performed using MATLAB’s ‘nlinfit’ function. External drug sensitivity data (IC50) was downloaded from the “Genomics of Drug Sensitivity in Cancer” resource (Yang et al., 2013), release 2.0 (http://www.cancerrxgene.org).
Statistical analysis
Significance tests and correlation analysis were performed using built-in functions within Microsoft Office Excel 2013 or R statistical computing environment version 3.1.0. Gene enrichment significance testing was performed in DAVID version 6.7 using the EASE metric, a modified Fisher’s exact test(Huang et al., 2009). All error bars represent standard deviation unless otherwise noted.
Supplementary Material
Acknowledgments
We thank Elisabeth Mahen and Chaozhong Song for excellent technical support and members of the Villén laboratory as well as Elizabeth O’Day for critical reading of this manuscript. This work was supported by a Howard Temin Pathway to Independence Award K99/R00 from NIH/NCI (R00CA140789) to J.V., a National Science Foundation grant (DBI-1355899) to S.-I.L, and funds from the South Sound CARE Foundation, the Washington Research Foundation, the Gary E. Milgard Family Foundation (to C.A.B.).
Footnotes
ACCESSION NUMBERS
The raw mass spectrometry files associated with this work are available for download at https://www.proteomicsdb.org/#projects/4167?accessCode=ecf333f8f2323901987aefd9f2982fb38a95acc11538b62af14884df25f553ca
AUTHOR CONTRIBUTIONS
R.T.L. and J.V. designed research. R.T.L., E.M.P., and K.M.H. performed proteomics experiments under J.V.’s supervision. C.P.M. performed drug sensitivity assays under C.A.B.’s supervision. H.Y.I. provided reagents. R.T.L. performed proteomics data analysis, and integrative analysis. S.-I.L. analyzed drug sensitivity data, and supervised statistical analysis. D.H. developed the web-based resource. R.T.L. and J.V. wrote the paper, and all authors edited it.
Authors declare no financial conflicts of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arteaga CL, Baselga J. Impact of Genomics on Personalized Cancer Medicine. Clin Cancer Res. 2012;18:612–618. doi: 10.1158/1078-0432.CCR-11-2019. [DOI] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Frazier WA. Integrin-associated protein (CD47) and its ligands. Trends Cell Biol. 2001;11:130–135. doi: 10.1016/s0962-8924(00)01906-1. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P, Choudhary GS, Sharma A, Singh K, Heston WD, Ciezki J, Klein EA, Almasan A. PARP Inhibition Sensitizes to Low Dose-Rate Radiation TMPRSS2-ERG Fusion Gene-Expressing and PTEN-Deficient Prostate Cancer Cells. PLoS ONE. 2013;8:e60408. doi: 10.1371/journal.pone.0060408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JC, Heiser LM, Georgii E, Gönen M, Menden MP, Wang NJ, Bansal M, Ammad-ud-din M, Hintsanen P, Khan SA, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol. 2014;32:1202–1212. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol. 2010;11:802–814. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Madden SF, Gallagher WM, Cox J, Mann M. Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 2012a;72:2428–2439. doi: 10.1158/0008-5472.CAN-11-3711. [DOI] [PubMed] [Google Scholar]
- Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative Proteomic Analysis of Eleven Common Cell Lines Reveals Ubiquitous but Varying Expression of Most Proteins. Mol Cell Proteomics. 2012b;11:M111014050. doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholami AM, Hahne H, Wu Z, Auer FJ, Meng C, Wilhelm M, Kuster B. Global Proteome Analysis of the NCI-60 Cell Line Panel. Cell Rep. 2013;4:609–620. doi: 10.1016/j.celrep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LAG, Cronin KA. US Incidence of Breast Cancer Subtypes Defined by Joint Hormone Receptor and HER2 Status. J Natl Cancer Inst. 2014;106:dju055. doi: 10.1093/jnci/dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huber MA, Azoitei N, Baumann B, Grünert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudis CA, Gianni L. Triple-Negative Breast Cancer: An Unmet Medical Need. The Oncologist. 2011;16:1–11. doi: 10.1634/theoncologist.2011-S1-01. [DOI] [PubMed] [Google Scholar]
- Ishida H, Li K, Yi M, Lemon SM. p21-activated kinase 1 is activated through the mammalian target of rapamycin/p70 S6 kinase pathway and regulates the replication of hepatitis C virus in human hepatoma cells. J Biol Chem. 2007;282:11836–11848. doi: 10.1074/jbc.M610106200. [DOI] [PubMed] [Google Scholar]
- Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, Kim JS, Zhang Y, Wang X, Ivey RG, et al. Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Nat Methods. 2014;11:149–155. doi: 10.1038/nmeth.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, et al. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Inhibition of NF-κB in cancer cells converts inflammation- induced tumor growth mediated by TNFα to TRAIL-mediated tumor regression. Cancer Cell. 2004;6:297–305. doi: 10.1016/j.ccr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat Rev Cancer. 2005;5:505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Sick E, Jeanne A, Schneider C, Dedieu S, Takeda K, Martiny L. CD47 update: a multifaceted actor in the tumour microenvironment of potential therapeutic interest. Br J Pharmacol. 2012;167:1415–1430. doi: 10.1111/j.1476-5381.2012.02099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg J, Divisekera U, McLaughlin N, Sharkey J, Pommey S, Denoyer D, Dwyer KM, Smyth MJ. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci USA. 2010;107:1547–1552. doi: 10.1073/pnas.0908801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG, et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer Basic Clin Res. 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Price ND, Hood L. Systems cancer medicine: towards realization of predictive, preventive, personalized and participatory (P4) medicine. J Intern Med. 2012;271:111–121. doi: 10.1111/j.1365-2796.2011.02498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Untch M, Konecny GE, Paepke S, von Minckwitz G. Current and future role of neoadjuvant therapy for breast cancer. Breast. 2014;23:526–537. doi: 10.1016/j.breast.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Vidal M, Chan DW, Gerstein M, Mann M, Omenn GS, Tagle D, Sechi S, Workshop Participants. The human proteome - a scientific opportunity for transforming diagnostics, therapeutics, and healthcare. Clin Proteomics. 2012;9:6. doi: 10.1186/1559-0275-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranic S, Gatalica Z, Wang ZY. Update on the molecular profile of the MDA-MB-453 cell line as a model for apocrine breast carcinoma studies. Oncol Lett. 2011;2:1131–1137. doi: 10.3892/ol.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. An Information-Intensive Approach to the Molecular Pharmacology of Cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Van Allen EM, Omberg L, Wagle N, Amin-Mansour A, Sokolov A, Byers LA, Xu Y, Hess KR, Diao L, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol. 2014;32:644–652. doi: 10.1038/nbt.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X, Wang Y, Zhou X, Yu J, Jian R, Tang S, Yin L, Zhou P. RNAi-mediated CD73 suppression induces apoptosis and cell-cycle arrest in human breast cancer cells. Cancer Sci. 2010;101:2561–2569. doi: 10.1111/j.1349-7006.2010.01733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H. 5′-Nucleotidase: molecular structure and functional aspects. Biochem J. 1992;285(Pt 2):345–365. doi: 10.1042/bj2850345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.