Abstract
Mononucleotide microsatellites are tandem repeats of a single base pair, abundant within coding exons and frequent sites of mutation in the human genome. Because the repeated unit is one base pair, multiple mechanisms of insertion/deletion (indel) mutagenesis are possible, including strand-slippage, dNTP-stabilized, and misincorportion-misalignment. Here, we examine the effects of polymerase identity (mammalian Pols α, β, κ, and η), template sequence, dNTP pool size, and reaction temperature on indel errors during in vitro synthesis of mononucleotide microsatellites. We utilized the ratio of insertion to deletion errors as a genetic indicator of mechanism. Strikingly, we observed a statistically significant bias towards deletion errors within mononucleotide repeats for the majority of the 28 DNA template and polymerase combinations examined, with notable exceptions based on sequence and polymerase identity. Using mutator forms of Pol β did not substantially alter the error specificity, suggesting that mispairing-misalignment mechanism is not a primary mechanism. Based on our results for mammalian DNA polymerases representing three structurally distinct families, we suggest that dNTP-stabilized mutagenesis may be an alternative mechanism for mononucleotide microsatellite indel mutation. The change from a predominantly dNTP-stabilized mechanism to a strand-slippage mechanism with increasing microsatellite length may account for the differential rates of tandem repeat mutation that are observed genome-wide.
1. INTRODUCTION
Microsatellites are tandem repeats of one to six base pairs per repeat unit that are found in all organisms, at varying abundance [1]. Microsatellites are three-fold more abundant in Saccharomyces cerevisiae than prokaryotes [2]; within eukarya, vertebrates have the longest microsatellite alleles, particularly rodents [3]. Longer and more abundant microsatellite alleles may provide evolutionary advantages by allowing a species to adapt more quickly to a changing environment [4,5]. Evolutionary adaptation through microsatellite mutability has been shown for many different species, including the circadian rhythm of Drosophila, the social behavior of voles, and, dramatically, the skeletal morphology of domestic dogs (for review see [4,6]). Among primates, the most common microsatellites are mononucleotide repeats, especially [A/T] tracts [3,7]. Mononucleotide microsatellite repeats are frequent sites of genetic variation among individual genomes [8]. For example, mononucleotide microsatellites greater than 8 units in length make up 10% of all insertion/deletions (indel) polymorphisms in the European population.
Microsatellites are interspersed throughout genomes, in a nonrandom distribution [9,10]. Numerous microsatellites, including mononucleotide repeats, are present within protein coding and regulatory sequences of all species examined [11–13], and 14% of all proteins contain at least one microsatellite within the coding region [14]. When present within protein coding exons, indels within microsatellites result in frameshifts. In E. coli, abundant microsatellite alleles within stress response genes may be an evolutionary strategy to adapt to stressful conditions [15]. In humans, frameshift mutagenesis is associated with many disease states, including cancers [16–18]. Even when present in intronic regions, microsatellite length variation can affect gene transcription and splicing efficiency. For example, an intronic [T]11 allele within the MRE11 gene, important in DNA repair of DSBs, is frequently deleted to [T]9 in tumors with microsatellite instability. This results in skipping of exon five and a subsequent premature stop codon, which decreases the expression of the MRE11 protein [16].
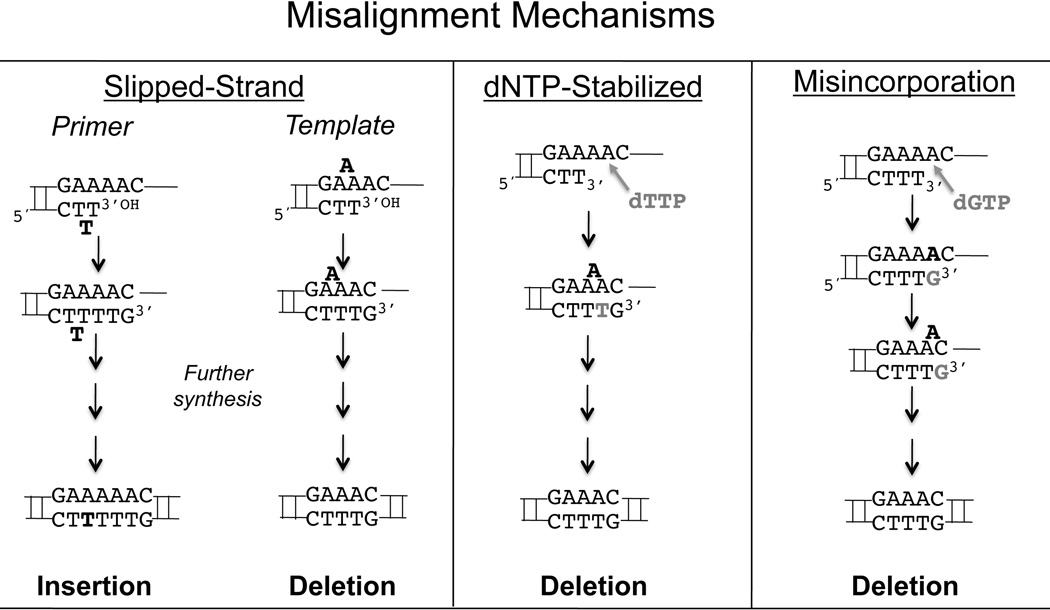
Three potential mechanisms could give rise to frameshift mutagenesis: slipped-strand misalignment, dNTP-mediated misalignment, and misincorporation-misalignment. Only slipped-strand misalignment is unique to repetitive DNA. Less than ten years after Watson and Crick published the model of DNA structure and the rules for base pairing [19,20], Fresco and Alberts reported an explanation of how DNA can break those rules and accommodate unpaired bases that were looped out of the DNA strand, allowing in-register bases to align [21]. In 1966, Streisinger et al. examined frameshift mutagenesis from a genetic and biochemical perspective [22]. Based on analyses of bacteriophage T4 mutants, the authors found that the original wild-type sequence contained repeated bases at the sites where mutations were formed. From these observations, the Streisinger strand-slippage model was formed. This model has been widely applied to explain the mechanism of indel mutations within microsatellites [23–26].
In the slipped-strand misalignment mechanism, repetitive DNA strands separate from each other and rehybridize out of frame, forming an indel loop (IDL) (Figure 1). Because the DNA is repetitive, this out of register duplex can still present a correctly paired 3’-primer terminus to the DNA polymerase. With continued synthesis, the looped base(s) results in an insertion or deletion, depending on whether the IDL is located in primer or template strand, respectively. Slipped-strand misalignment is thermodynamically stabilized by longer tandem repeats in which more correctly aligned bases can form more hydrogen bonds to overcome the destabilizing effects of the IDL. In addition, the more repeat units present in a tandem repeat, the more opportunities there are for IDLs to form, and the more distance there can be between the IDL and the primer terminus in the polymerase active site. Structural evidence of misaligned DNA shows that DNA bases form both extrahelical loops and intrahelical bulges with extra bases stacked within the helix [27]. The first structure of a catalytically competent DNA polymerase co-crystallized with a slipped-strand intermediate was made using Pol lambda, an X-family polymerase [28]. Garcia-Diaz et al. isolated five different deletion intermediates with unpaired nucleotides in the template strand. In all cases, the unpaired bases were extrahelical and the primer terminus contained properly paired bases. These biochemical studies, combined with genetic studies showing that longer microsatellites have higher mutation frequencies than shorter microsatellites [29–31] support the Streisinger model.
Figure 1. Mechanisms of Mononucleotide Indel Mutation.
The three panels demonstrate the three methods of indel error production that are possible for mononucleotide microsatellite alleles. The potential outcome of each mechanism provides a testable hypothesis to determine how mutations are produced in various mononucleotide alleles.
DNA polymerases also create indel errors in non-repetitive DNA sequences, observations that led to the dNTP-stabilized and misincorporation-misalignment models of frameshift mutagenesis [32–36]. In the dNTP-stabilized mechanism, substrate misalignment occurs in the polymerase active site, after dNTP substrate binding. In this model, the legitimate templating base is skipped because the incoming dNTP correctly binds to the next 5’ base, creating a bulged template and, after continued synthesis, a deletion mutation (Figure 1). Structural support for this model is provided by Dpo4, an archaebactrial Y-family DNA polymerase. Using undamaged templates, Ling et al. isolated a structure of Dpo4 with a skipped template base and a correctly paired next base in the active site [37]. Possibly, the skipped base was in a conformation that obscured the hydrogen binding surface from the active site of the polymerase, creating a non-instructive base. In that case, when the dNTP complementary to the 5’ base entered the active site, the non-informative base was skipped.
The misincorporation-misalignment mechanism begins as a base misincorporation event (Figure 1). The newly added mismatched nucleotide can relocate to the next template position, if the 5’ templating base is complementary to the misinserted base [38]. Polymerase misincorporation events often create a poor DNA substrate for further polymerase elongation synthesis, slowing the polymerase catalytic rate and allowing substrate realignment with the correctly pairing 5’ neighbor [39]. In support of this model, when DNA polymerase dNTP pools are biased with excess dGTP, mutational hotspots are formed at purines with 5’ C’s, and when dNTP pools are biased with excess dATP, hotspots are formed at 5’ T’s [38]. This mechanism of minus-one deletion errors has been proposed to operate in both repeated and non-repeated sequences [38]. We note that misincorporation followed by substrate realignment to the 3’ (previous) templating base would result in insertion errors by this mechanism. Overall, the rate of this misincorporation-misalignment mechanism must be lower than the other indel mechanisms, as it will be the product of the polymerase misincorporation rate multiplied by the one in four probability that a neighboring base template is complementary to the newly misincoporated base.
For mononucleotide microsatellites, the expected genetic outcomes of the three possible indel mechanisms are distinct: strand-slippage mediated misalignment is expected to result in both insertions and deletions within a mononucleotide microsatellite, whereas the dNTP-stabilized mechanism is expected to result in deletions. The misincorporation-misalignment mechanism can result in either insertion or deletion errors, but is expected to be dependent on both the sequence context and the base misincorporation rate of the polymerase. We recently formalized the relationship between microsatellite mutability and repeat number, using segmented regression models and the human 1000 genomes dataset [29]. Our results showed that short tandem repeats and microsatellites differ in the rate of mutation as a function of repeat number. Paradoxically, tandem repeat mutability grows at a slower rate after crossing a threshold length; for mononucleotides, the change point in mutational behavior from fast to slow occurred between 8 and 9 repeat units (genome-wide average). Our genome analyses also indicated that mononucleotide repeats differ in mutational behavior, as compared to di-, tri-, and tetranucleotide repeats. To understand the mechanisms underlying these observations, we undertook an in-depth examination of mononucleotide microsatellite errors created by several mammalian DNA polymerases. We utilized the ratio of insertion to deletion errors as a genetic indicator of mechanism (Figure 1). We examined polymerase errors within all four microsatellite templates, with biased dNTP pools, and at different reaction temperatures. All polymerases examined favor deletion over insertion errors within most mononucleotide repeats, with notable exceptions based on sequence and polymerase identity. Based on these data, we suggest that dNTP-stabilized misalignment may be an alternative mechanism of indel errors within mononucleotide microsatellites.
2. MATERIAL AND METHODS
2.1. DNA templates
The construction of microsatellite-containing Herpes simplex virus type 1 thymidine kinase (HSV-tk) vectors has been previously described [40–42]. Briefly, the MluI (position 83) to StuI (position 180) HSV-tk target sequence within the gapped duplex of molecule contains 88 bp of HSV-tk gene coding region sequence. Microsatellites are inserted in frame, in the sequence contexts shown in Table 1 and Figure S1. The mononucleotide vectors have been previously characterized [29,43]. For this study, we constructed an additional [G]9 allele. However, a [C]9 allele within the same sequence context failed to produce phenotypically TK+ bacteria, presumably due to the presence of a proline codon at the 3’ end of the repeat. In-frame mononucleotide A and T repeats longer than 8 units also fail to produce phenotypically TK+ bacteria, due to E. coli RNA polymerase frameshifting within long A/T repeats [44].
Table 1.
Sequences and contexts of microsatellite alleles used in this study.
| Allele | Template Sequence (Microsatellite allele in bold) |
|---|---|
| Wild type | 5’- GCG CGT TCT CGA -3’ |
| [T]8 | 5’- GCG CGT TTT TTT TCT CGA -3’ |
| [A]8 | 5’- GCG CGA AAA AAA ACT CGA -3’ |
| [G]9 | 5’- GCG CGG GGG GGG GCT TCT CGA -3’ |
| [G]10 | 5’- GCG CGG GGG GGG GGT TCT CGA -3’ |
| [C]10 | 5’- GCG CCC CCC CCC CGT TCT CGA -3’ |
| [C]10-reverse | 3’- CGC GCC CCC CCC CCA AGA GCT-5’ |
2.2. In vitro HSV-tk mutagenesis assay
Pol κ and Pol η reactions
Purified full-length human pol κ and pol η were purchased from Enzymax (Lexington, KY, USA). Unless otherwise indicated, in vitro gap-filling reactions (100 µl) contained approximately 75 fmol of gapped DNA substrate, 250 µM dNTPs, 25mM potassium phosphate buffer pH 7.2, 5 mM MgCl2, 5 mM DTT, 100 µg/ml non-acetylated BSA, 10% glycerol and 37.5 fmol pol κ or 50 fmol pol η, and were incubated at 37°C for 2 h. Complete gap filling was verified by agarose gel electrophoresis [45]. An aliquot of DNA from complete gap-filling reactions was used to transform E. coli strain FT334.
Pol α and Pol β reactions
Human DNA pol α-primase was purchased from Chimerx (Madison, WI). Rat recombinant DNA pol β (WT, I260M and Y265W) was purified as a hexahistadine fusion protein as previously described [46]. DNA synthesis templates were created by hybridization of GStu2 oligonucleotide to ssDNAs, to initiate synthesis at HSV-tk position 169. The in vitro reactions (100 µl) contained 1 pmol of template DNA. Reaction conditions for pol α were 20mM Tris-HCl (pH 7.5), 10 mM MgCl2, 2.5 mM DTT, 1 mM dNTPs, and 20 units pol α-primase. All reactions were incubated at 37 °C for 2 hours and terminated with 15 mM EDTA and incubation at 68 °C for 3 minutes. Reaction conditions for pol β were 50 mM Tris-HCl (pH 8.5), 50 mM NaCl, 10 mM MgCl2, 250 µM dNTPs, 1 mM dithiothreitol, 200 µg/ml acetylated BSA, and 10–15 pmol WT, 40 pmol Y260Q and 12.5 pmol Y265W enzyme. All reactions were incubated at 37 °C for 60 minutes. To sample the polymerase reactions for mutations, small fragments were prepared by MluI and StuI restriction digestion and hybridized to the corresponding gapped DNA duplex (described above), forming heteroduplex plasmid molecules, as described [40,41]. An aliquot of the heteroduplex DNA was used to electroporate E. coli strain FT334.
2.3. HSV-tk mutant and polymerase error frequencies
Following electroporation, bacteria were plated on VBA + Chloramphenicol (Cm) or VBA + Cm + 5-fluoro-2’-deoxyuridine (FUdR) selective media to determine the HSV-tk mutant frequency [47]. The observed HSV-tk mutant frequency is defined as the number of CmRFudRR/CmR viable cells, and is reported as the mean of two, or mean of three ± standard deviation independent polymerase reactions. The MluI to StuI DNA sequences were determined for independent mutants isolated from two independent polymerase reactions per template, as described [40]. Overall polymerase error frequencies (Pol EFs) for each template, which includes both microsatellite and coding region mutations, were determined by the following equation:
Pol EF = (Observed MF− ssDNA MF) × (# sequenced mutants with an in-target mutation/total number mutants sequenced).
The ssDNA MF is the HSV-tk mutant frequency of the single-stranded DNA used to make the gapped DNA duplex, or as the template for primer-extension reactions. Mutants that do not contain a mutation within the target are due to either background errors in the DNAs used to create the gapped duplex or polymerase strand displacement synthesis. Because multiple mutational events per target sequence are created by some polymerases, error frequencies were also adjusted (Pol EFest) to reflect the number of mutational events that were detectable as single mutational events, as described [42,48]. All frameshift errors and base substitutions that caused an amino acid change were considered detectable. Only detectable events were used for Pol EFest. Within the microsatellite target, a unit-based indel is defined as any mutational event that resulted in an insertion or deletion of a microsatellite base. Interruptions are defined as base substitutions or insertions of nucleotides that are not the same base as the mononucleotide repeat template sequence.
3. RESULTS
Unlike larger motif sizes, unit-based indel mutations within single base mononucleotide repeats can arise by three mechanisms (Figure 1). The strand-slippage mechanism of misalignment mutagenesis is expected to produce pre-mutational IDL intermediates in both the template and primer strands. Because these IDLs can migrate to positions well removed from the 3’OH within longer repeats, DNA polymerases are expected to extend these pre-mutational intermediates efficiently, resulting in both insertion and deletion mutations. The precise proportion of insertions to deletions is expected to vary among polymerases, reflecting each enzyme’s interactions with the DNA and inherent fidelity for extending IDL-containing substrates. In contrast, only deletion mutations are expected to result from dNTP-stabilized misalignment mechanism, while indels produced by the misincorporation-misalignment mechanism are expected to be highly dependent on DNA polymerase misincorporation fidelity and DNA sequence context. Using this theoretical genetic framework, we set out to intensively examine the mechanism by which DNA polymerases create errors within each of the four mononucleotide repeats, of lengths at or near the 8–9 unit change point in mutational behavior [29]. We examined the effects of five variables on mononucleotide mutational specificity: template sequence, sequence context, polymerase identity, reaction temperature, and dNTP substrate pools.
3.1. Template sequence and flanking sequence content
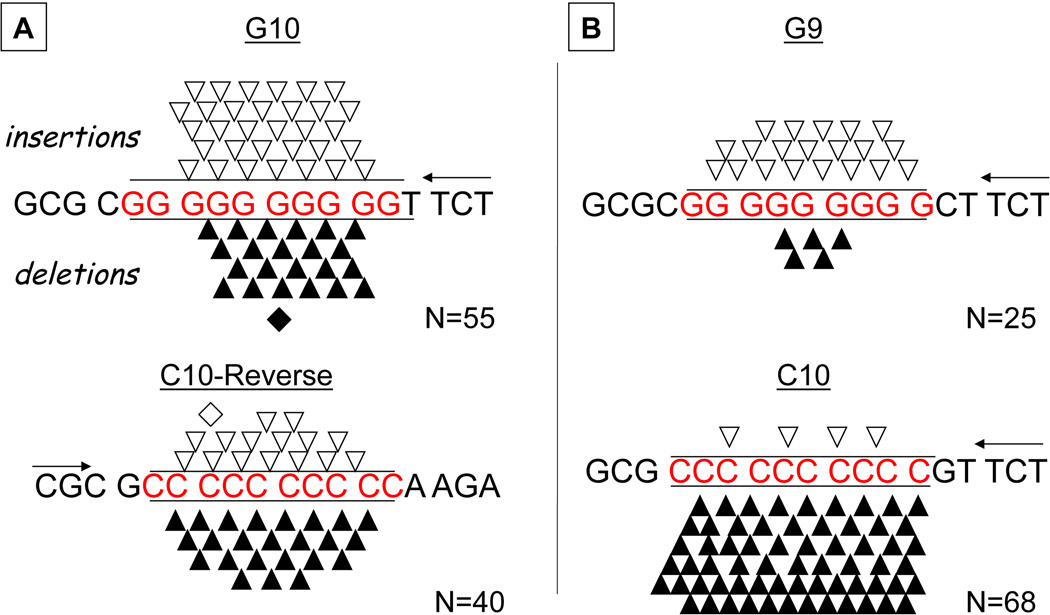
Using our established HSV-tk in vitro assay, we measured DNA Pol β errors within [G]10 and the complementary [C]10-R mononucleotide repeats. The WT Pol β microsatellite error frequencies within the [G]10 and complementary [C]10-R repeats are not different (Table S1), and DNA sequence analyses of independent mutants revealed an approximately equal proportion of unit-based (1 nucleotide) insertion and deletion errors (Figure 2). These results demonstrate that both polydG and polydC sequences can form IDL intermediates within both the template and primer strands during the in vitro DNA polymerase reactions. One of the basic predictions of the strand-slippage model is that as microsatellite alleles increase in length, the mutability of the repeated sequence also increases. Therefore, we examined how changing the length of the G repeat allele affected both the absolute frequency and specificity of errors. As expected, we measured an ~two-fold lower mutation frequency within the [G]9 repeat, compared to the [G]10 repeat (Table S1). Although 80% of Pol β errors within [G]9 were unit insertions (Figure 2), this difference from the [G]10 specificity is not statistically significant (p=0.0776, Fisher’s exact test). These Pol β results are entirely consistent with the traditional strand slippage mechanism.
Figure 2. Specificity of WT Pol β errors is template sequence dependent.
The sequences of the template strands are indicated by the middle line, with the microsatellite sequence in red. Arrow indicates direction of DNA synthesis (5’ to 3’ direction). (A). Error specificity within complementary [G]10 and [C]10-R alleles. (B). Error specificity within [G]9 and [C]10 alleles inserted in the same sequence context as the [G]10 allele. Open triangles, single base insertion; open diamond, two base deletion; closed triangles, single base deletion; closed diamond, two base deletion. N, number of independent microsatellite errors observed. See Table S1 for mutation frequencies.
Next, to test sequence context effects, we examined Pol β errors produced within a [C]10 repeat inserted within the same strand and sequence context as the [G]10 allele, rather than in the complementary strand. For this construct, the WT Pol β mutant frequency was ~two-fold higher than that of the [C]10-R allele (Table S1). Surprisingly, the error specificity for [C]10 in this sequence context was highly skewed towards deletion errors: of the 68 microsatellite mutants recovered from four independent reactions, only 4 were insertion errors at the microsatellite, while 64 were deletions (Figure 2). The difference in Pol β error specificity between the [C]10 repeat in the two sequence contexts is highly statistically significant (p<0.0001, Fisher’s exact test). We previously reported the Pol β error frequencies within [T]8 and [A]8 repeats [43] (see Table S1). The error specificity for both of these alleles was also highly skewed towards unit deletions within the microsatellites (Figure 3A). Therefore, of the six microsatellites examined, Pol β produced an unbiased proportion of insertion and deletion errors only within two alleles: the complementary [G]10/[C]10-R template sequences.
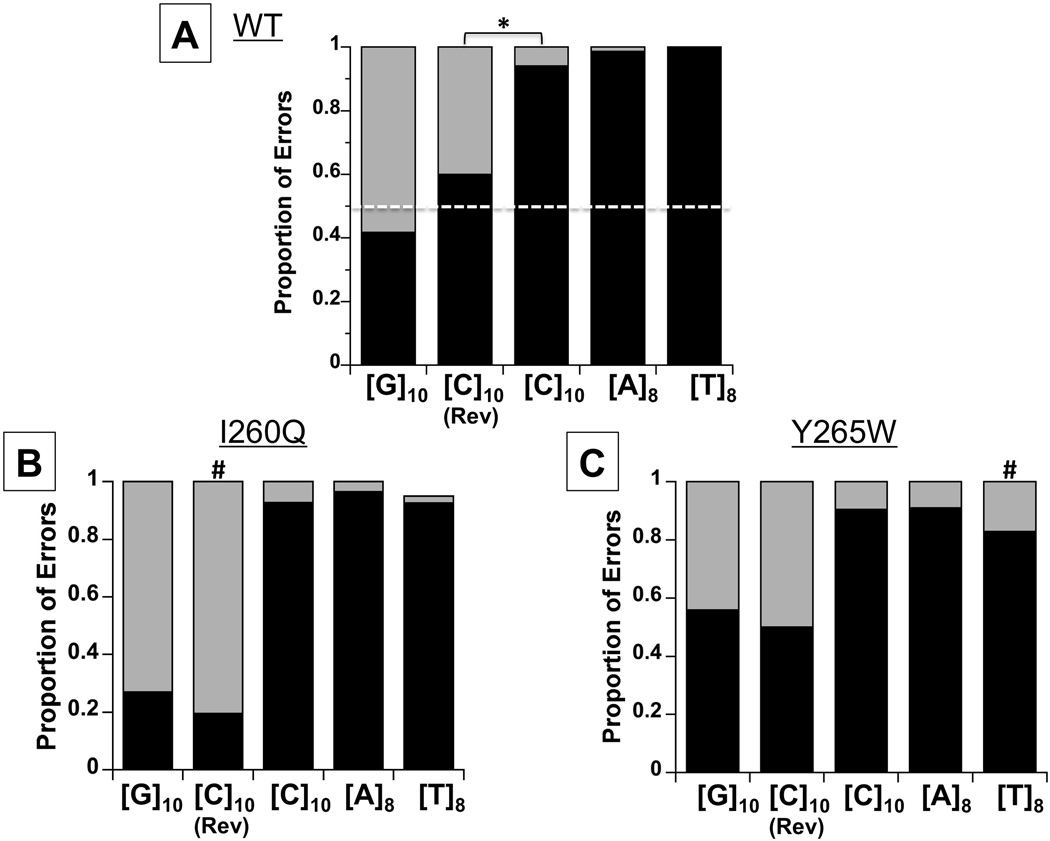
Figure 3. Effect of mutator forms of Pol β on mononucleotide error specificity.
In vitro replication reactions were performed using the mononucleotide microsatellite templates shown (see Tables S1 for frequencies). Bars represent the proportion deletion (black) versus insertion (grey) errors within each microsatellite. (A). WT; (B). I260Q. For the [T]8 template, 2 mutants were interruptions within the microsatellite. (C). Y265W. White dashed line indicates 50% insertion:50% deletion ratio expected for unbiased strand slippage. *, significantly different sequence contexts (p<0.0001, Fisher’s exact test); #, significantly different relative to WT Pol β (p<0.001, Fisher’s exact test).
3.2. DNA polymerase identity
To examine whether the very high proportion of Pol β deletion errors observed for the [C]10, [A]8, and [T]8 templates are produced by the misincorporation-misalignment mechanism, we tested variant Pol β forms with decreased fidelity. For these experiments, we used two Pol β variants, both located in the hinge region and previously described to affect the positioning of dNTP and/or DNA substrates: I260Q [49,50] and Y265W [46,51]. Using the I260Q mutator, we observed a 1.5- to 3.7-fold increase in mutations formed within all microsatellites examined, relative to WT Pol β (Table S1). The error specificity for this mutator polymerase was not significantly different from WT Pol β for the [G]10, [C]10, [A]8, and [T]8 alleles (Figure 3B). However, we did measure an increased proportion of insertion errors within the [C]10-R template, relative to WT Pol β (p=0.0003, Fisher’s exact test; ; Figure 3B). Using the Y265W mutator, we observed no change in microsatellite mutation frequency (Table S1) or error specificity (Figure 3C) for the [G]10, [C]10-R, [C]10 or [A]8 templates, relative to WT Pol β. However, we did observe a statistically significant increase in Y265W insertion errors at the [T]8 microsatellite (17%, p=0.0008, Fisher’s exact test), relative to WT, together with an ~two-fold increased microsatellite mutation frequency (Figure 3C).
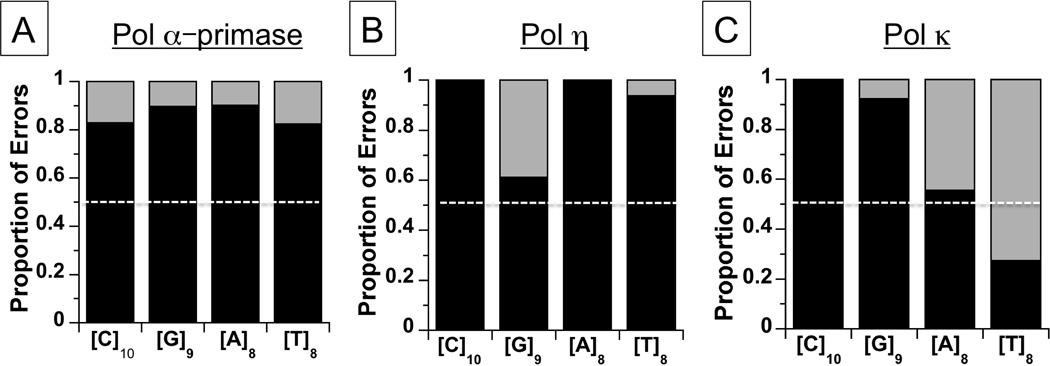
Next, we examined three additional DNA polymerases with varying structures and fidelities (human Pol α-primase, Pol η and Pol κ) for the production of errors within the microsatellite alleles that failed to produce an unbiased insertion:deletion error spectrum by WT Pol β. All enzymes are proofreading deficient, to avoid the confounding effects of exonuclease enzymatic activity on error frequency and specificity. Strikingly, we discovered that over 80% of all indel errors created by all polymerases (including wild-type Pol β) copying the four microsatellites examined ([C]10, [G]9, [A]8, [T]8) were deletions (531/645) (Figures 3 and 4). When comparing to the expected unbiased result of 50% insertions: 50% deletions, the difference is highly statistically significant (p<0.0001, Χ2=262.6, 1 degree of freedom). This bias is most pronounced for the [C]10 template, where only 15 insertions (7%) were observed among the 213 errors produced by six polymerase forms. For Pol α-primase, nearly every indel mutation recovered was a deletion, regardless of the mononucleotide template sequence (Figure 4A). In fact, we observed only two additional instances where any polymerase created an unbiased ratio of insertions to deletions within these templates: Pol η and the [G]9 template (Figure 4B), and Pol κ and the [A]8 template (Figure 4C). We began this study with the hypothesis that strand-slippage misalignment will result in an unbiased proportion of both insertion and deletion errors, since pre-mutational IDL intermediates should be formed in both strands and can migrate to positions well removed from the 3’OH within the long microsatellite repeat. Altogether, however, our data for the microsatellite error specificity of multiple polymerases do not support this hypothesis. Instead, the genetic outcomes we expected from the slipped strand mechanism were the exception, not the rule.
Figure 4. Error specificity of mammalian DNA polymerases within mononucleotide microsatellites.
In vitro replication reactions were performed using the mononucleotide microsatellite templates shown (see Table S2 for frequencies). Bars represent the percentage of deletion (black) versus insertion (grey) errors within each microsatellite. (A). Pol α-primase; (B). Pol η; (C). Pol κ. White dashed line indicates 50% insertion:50% deletion ratio expected for unbiased strand slippage.
3.3. Effects of reaction temperature
The traditional strand-slippage mechanism proposed for microsatellite mutagenesis [52] requires DNA strand denaturation, followed by misaligned renaturation of partially synthesized DNA duplexes. Thus, the relative strength of hydrogen bonding between basepairs and the local sequence context of the repeats may influence this mechanisms of mutagenesis. As an alternative means of probing mechanism, we examined the effects of reaction temperature on Pol κ mutational specificity within A/T and G/C microsatellites. Assuming that more denatured templates will result in the formation of more abundant slipped-strand substrates, we expected that the likelihood of creating pre-mutational misaligned DNA templates via strand slippage will be greater in polymerase reactions conducted nearer to the microsatellite melting temperature (TM), because at the TM, 50% of the duplexed molecules are expected to be partially denatured. The approximate TM of the [T]8 and [C]10 microsatellite duplexes is 16°C and 40°C, respectively. Therefore, at standard 37°C reaction conditions, the equilibrium is shifted towards denaturation (more misalignment probability) for the [T]8 template (TM =16°C) but towards fully hybridized (less misalignment probability) for the [C]10 template (TM =40°C).
For Pol κ errors, the deletion bias of [G] and [C] templates vs. [A] and [T] templates (Figure 4C) is statistically significant (p<0.0001, Fisher’s exact test). We hypothesized that if strand slippage were the major mechanism of creating misalignment errors within the microsatellites, then adjusting the reaction temperature might shift the mutational specificity to favor an unbiased ratio of insertion:deletion errors. To test this, we first lowered the reaction temperature for the [T]8 template to 16°C. Although the Pol κ microsatellite error frequency at this temperature was slightly lower than that measured at the 37°C reaction (Table 2), the mutational specificity of the 16°C reactions still significantly favored insertion mutations (92%). Next, we raised the reaction temperature for the [C]10 template to 47°C (above the TM), expecting this to favor DNA melting and increase strand misalignments. Instead, raising the reaction temperature of Pol κ reaction reduced the mutation frequency, to 180 × 10−4, approximately 5-fold lower than at 37°C. Importantly, all mutants recovered from 47°C reaction conditions using the [C]10 template were deletions, similar to results when reactions were performed at 37°C (Table 2). These experiments indicate that the biased Pol κ mutational specificity observed within mononucleotide [T] and [C] sequences is unlikely to reflect differences in the thermodynamic properties of the DNA primer-template.
Table 2.
Effect of varying reaction temperature on Pol κ error frequencies and specificity at mononucleotide microsatellites.
| Template | Reaction Temperature |
Pol Error Frequency × 10−4 | Microsatellite Errors | ||
|---|---|---|---|---|---|
| Observed | Microsatellite | Insertion | Deletion | ||
| [T]8 | 37°C | 480 | 330 | 47 | 9 |
| 16°Ca | 230 | 150 | 36 | 3 | |
| [C]10 | 37°C | 1300 | 960 | 0 | 33 |
| 47°Cb | 210 | 180 | 0 | 30 | |
Reactions incubated for 2–4 hours
Reactions incubated for 30–120 mins
3.4. Effects of dNTP substrate pools
DNA polymerase accuracy is affected by dNTP levels [53,54] and relative dNTP pool sizes. For misalignment-based errors, dNTP levels or pools can affect the rate of polymerase base misincorporation errors or extension of slipped strand IDL intermediates, but should have no effect on the initial creation of slipped strand intermediates. Therefore, if the bias towards deletion errors within mononucleotide repeats is caused by the preferential extension of IDLs within the template strand over IDLs in the primer strand, we hypothesized that we could shift the specificity of microsatellite mutations by altering the concentration of incoming dNTPs. In experiments using the [C]10 template, almost all of the microsatellite mutants recovered from the various polymerase reactions were deletions. Therefore, we hypothesized that increasing the concentration of the incoming correct dNTP, relative to the other three dNTP substrates (250 µM dGTP and 20 µM of the other dNTPs), would force the polymerase to utilize primer slipped strands, thereby expanding the microsatellite. However, creating a dNTP pool bias did not substantially change the mutational specificity of Pol κ for the [C]10 template. Approximately 90% of all microsatellite mutants recovered were deletions with the pool biased conditions as compared to 100% deletions in standard conditions (Table 3). Because this reaction condition dramatically lowers the total concentration of dNTPs, we also performed control reactions with 20 µM of all four dNTPs (no bias). As expected, the overall Pol κ microsatellite error frequency was lowered 15-fold by decreasing the dNTP levels (Table 3), but deletions still accounted for greater than 90% of mutations.
Table 3.
Effect of dNTP pool biases on Pol κ error frequencies and specificity at mononucleotide microsatellites.
| Template/ Correct dNTP |
[dNTP], µM | Microsatellite Pol EFa × 10−4 (N) |
Microsatellite Errors | ||
|---|---|---|---|---|---|
| Correct | Other three | Insertions | Deletions | ||
| [C]10 /dGTPb | 250 | 20 | 200 (75) | 2 | 15 |
| 20 | 20 | 63 (37) | 1 | 24 | |
| [A]8 / dTTP | 20 | 250 | 100 (28) | 3 | 8 |
| 250 | 250 | 100 (38) | 4 | 5 | |
| [T]8 / dATPb | 20 | 250 | 300 (41) | 0 | 10 |
Calculated from the Observed HSV-tk mutant frequencies and the total number of independent mutational events shown in parentheses.
See Table 2 for unbiased [dNTP] conditions results.
We attempted also to change the Pol κ mutational specificity of the [T]8 template, which is 84% insertions using our standard reaction conditions of 250 µM each dNTP. We lowered the concentration of the incoming dNTP (dATP) to 20 µM while keeping the other three substrates at 250 µM. In these conditions, the microsatellite mutation frequency was not altered relative to standard conditions (Table 3). However, the mutational specificity was significantly affected, as all indel mutants sequenced from the 20µM dATP/250µM dG, T, CTP reactions were now deletions (p<0.0001, Fisher’s Exact Test). Also, unlike mutants recovered from reactions using the standard dNTP concentrations, 50% of all microsatellite mutants from 20µM dATP/250µM dG,T,CTP reactions were interruptions, consisting of base substitutions and insertions of non-template bases (p<0.0001, Fisher’s Exact Test) (Figure S2). Therefore, when dATP levels were lowered in the [T]8 reactions, insertions of the templating base were eliminated, and insertions of incorrect bases made up over 25% of all microsatellite mutants recovered. In similar reactions, we lowered the dTTP concentration to 20µM when utilizing the [A]8 template. In this case, we found that lowering the correct dNTP had no effect on either the frequency of errors (Table 3), the specificity of indels (p=0.6424), or the frequency of interruptions (Figure S2).
4. DISCUSSION
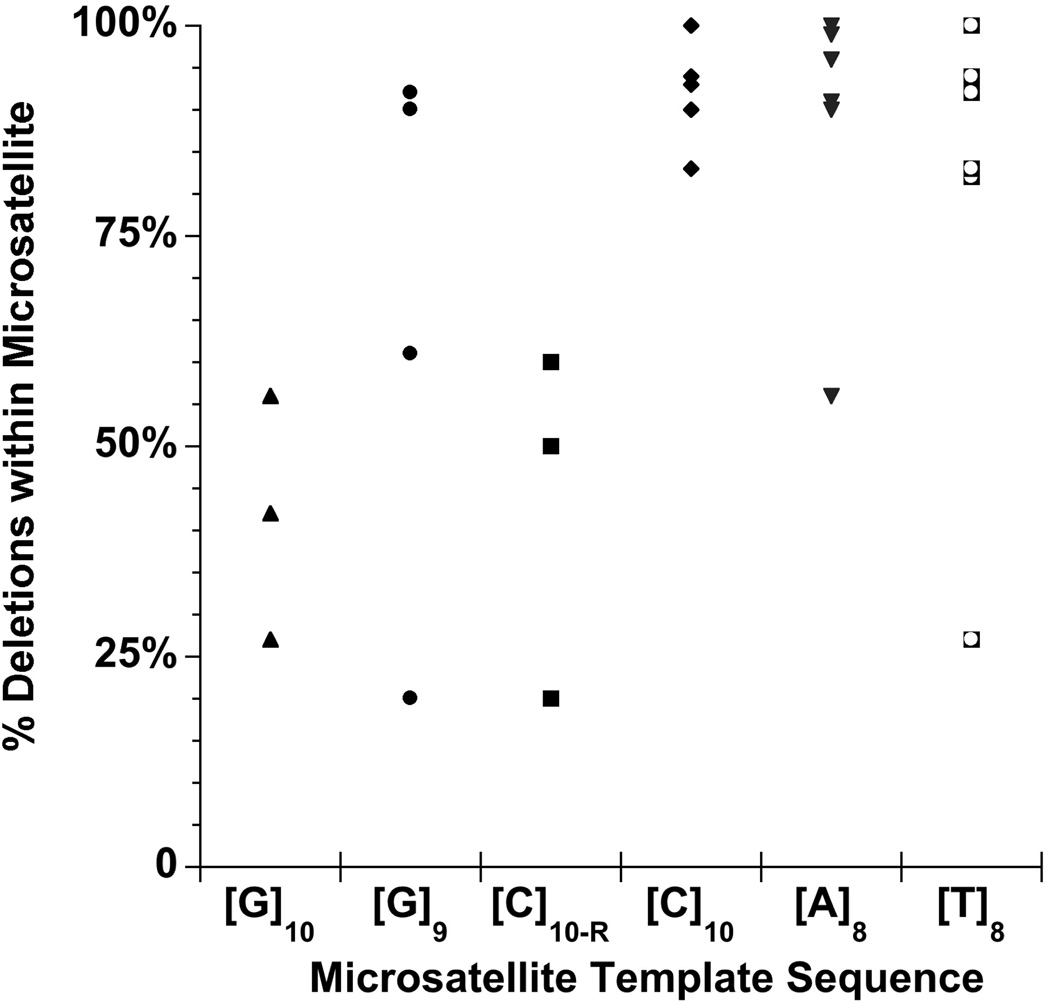
Repetitive DNA has the propensity to form slipped-strand intermediates. This characteristic led to the generally accepted view that microsatellites mutate via slipped-strand misalignment [23–26]. Our recent genome-wide analyses indicated that mononucleotide repeats differ in mutational behavior, as compared to di-, tri-, and tetranucleotide repeats, and that a change point in mutational behavior occurs between 8 and 9 repeat units [29]. In this study, we compared the error spectra of several mammalian DNA polymerases under various reaction conditions to test mechanisms of mononucleotide mutagenesis. Within mononucleotide microsatellites, the insertion or deletion of a single base unit can occur by any of three mechanisms (Figure 1). We began this study with the simple assumption that formation of the slipped-strand premutational intermediates in the traditional Streisinger model shoud occur with approximately equal probability on either the primer or template strand. Thus, if no other factors contribute to creating mutational biases, we hypothesized that both deletion and insertion errors should be produced with similar frequency within mononucleotide microsatellites. For three mononucleotide sequences (poly dC, dG, dA), we did observe the unbiased production of both insertion and deletion errors for at least one polymerase (Figure 5), supporting our assumption that IDLs can initially form on both strands in nearly equal proportions. Strikingly, however, none of the six DNA polymerases examined, representing three structurally distinct DNA polymerases, produced an unbiased proportion of both insertion and deletion errors within all four of the mononucleotide microsatellites. Indeed, for the 28 polymerase/template combinations tested, we observed a statistically significant bias towards deletion errors within mononucleotide repeats for the majority of DNA template and polymerase combinations examined (Figure 5). The unbiased production of insertions and deletions was the exception, not the rule, being observed in only a few instances: Pol β and the [G]10 and [C]10-R alleles (Figure 3), Pol η and the [G]9 template (Figure 4B), and Pol κ and the [A]8 template (Figure 4C).
Figure 5. DNA polymerases display a pronounced bias towards deletion errors within all four mononucleotide microsatellites.
The proportion of deletion errors created within each indicated mononucleotide template sequence is plotted for the six polymerases tested: Pol α-primase, Pol β (WT, I260Q, Y265W), Pol η, and Pol κ.
What mechanisms can account for this pronounced deletion bias for DNA polymerase errors within mononucleotide microsatellites? Possibly, the creation of the IDL substrates occurs during DNA polymerase binding, in a manner that is selective for only template strand IDL formation. For this mechanism to be consistent with our data, the polymerase-mediated template IDLs must be formed only within specific mononucleotide sequences and sequence contexts, differing for each polymerase. For example, one would need to propose that Pol β DNA binding creates template IDLs within the [C]10 sequence context, but not within the [C]10-R context (Figure 2), or that Pol η binding creates template IDLs within [A]8, [T]8, and [C]10 mononucleotides, but not within the [G]9 microsatellite. Another possibility is that the IDL structures formed within the microsatellite template strands may be differentially stabilized by direct polymerase-DNA interactions. While we cannot formally rule out these possibilities, we consider these mechanisms to be unlikely, given the extensive differences in protein-DNA interactions observed among Pol α-primase [55], Pol β [56], Pol η [57], and Pol κ [58] ternary structures.
Alternatively, the predominance of polymerase deletion errors could be explained by a traditional strand slippage mechanism in which IDL premutational intermediates are either more efficiently extended when present in the template strand, or less efficiently extended when in the primer strand. We do not expect that differential extension of IDLs located in the template versus primer strand would be a major factor influencing specificity, because of the length of the repeated tracts examined. To more directly probe this mechanism, we examined Pol κ errors within poly dG and poly dC template microsatellites under different reaction conditions. Pol κ predominantly makes deletion errors in both of these repeats (Figure 4C). For reactions using the [C]10 template, lowering the three non-complementary dNTPs to 20 µM, while keeping the correct dGTP at 250 µM, reduced the microsatellite error frequency but did not affect the deletion bias within this template (Table 3). This indicates that even with a >10-fold excess of the correct incoming dNTP, Pol κ is unlikely to extend the primer strand IDL and make insertion errors on the [C]10 template. Furthermore, if differential utilization of premutational IDLs is the mechanism, then one would have to postulate that Pol κ can efficiently extend both a guanine-IDL and a cytosine-IDL when present in the template strand, but cannot efficiently extend the same IDL substrates when present in the primer strand. Finally, the most pronounced deletion bias we observed is within the [C]10 template sequence, where all of the polymerases examined created 80–100% deletion errors when copying this microsatellite (Figure 5). For the differential extension mechanism to be correct, one would have to assume that the extension biases for IDLs are similar among B, X, and Y family polymerases, which have very distinct active site geometries.
In addition to strand slippage, the misincorporation-misalignment mechanism (Figure 1C) could produce indels within microsatellites. In this case, the indel error rate is dependent on both the rate of polymerase misincorporation and the rate of relocating the newly synthesized mispair to form a correctly hybridized base pair [59]. Although this mechanism can result in either deletions or insertions, we do not believe this is a major mechanism underlying polymerase errors within mononucleotide microsatellites, for the following reasons. First, we did not observe major changes in error specificity for the two mutator Pol β forms tested, relative to WT Pol β. The I260Q variant affects DNA template positioning within the Pol β active site, and cannot discriminate correct from incorrect dNTP substrates [50]. However, we only observed a significantly altered spectrum of I260Q indel errors when copying the [C]10-R microsatellite template (Figure 3B). This may be an isolated example of misincorporation-misalignment, in which dG-A mispairs are formed at the base 5’ to the microsatellite, followed by primer strand misalignment and extension of a correct dG-C basepair. Consistent with this, we previously reported that the I260M Pol β variant also creates an increased proportion of insertion over deletion errors within a [TC]11 microsatellite allele [60]. The Y265W mutator displayed a 15-fold increased base substitution and frameshift error frequency within sequences of the HSV-tk coding region [46], but this variant did not create an increased microsatellite frequency or altered mutational specificity within four of the five microsatellites examined (Figure 3C). Second, the rate of base mispair formation varies greatly among the four polymerases we tested. Yet, deletions dominated all of the mononucleotide mutational spectra. At the [C]10 template, for example, the only C that could participate in the misincorporation-misalignment mechanism and form deletions is the last one, because only the mispairs formed at the last C could relocate to the following 5’ base to form a correct base pair. Because the base 5’ to the [C]10 microsatellite is a G (Table 1), misincorporation-misalignment would require formation of a C·dCMP mispair, which is a rare misinsertion event for most DNA polymerases. For example, Ohashi et al. determined the C-C mispair frequency for full-length human Pol κ to be 2.0×10−4 [61]. In contrast, the rate of Pol κ single base deletions at the [C]10 template is 960 × 10−4, a difference of nearly 500 fold, suggesting that misincorporation misalignment is responsible for few, if any, of the Pol κ errors on this template. Altogether, these data argue against misincorporation-misalignment being a major mechanism of mononucleotide microsatellite mutagenesis.
We performed additional biochemical experiments using Pol κ as a means of deciphering misalignment mechanisms. The Pol κ insertion:deletion ratio for the [A]8 template (45%:55%) demonstrates that slipped-strand intermediates with loops in both the primer and template strand are formed within the [A/T] microsatellite, and both can be substrates for the enzyme. Within the [T]8 template, however, Pol κ creates both indel errors, but is significantly biased towards insertions. The insertion bias may be caused by a higher Pol κ extension efficiency from IDL intermediates containing looped A nucleotides (primer) versus looped T nucleotides (template). To alter polymerase extension efficiency during synthesis of the [T]8 template, we lowered the incoming dATP levels relative to the other three dNTPs. This pool bias resulted in the elimination of T insertion errors within the microsatellite (Table 3) while increasing the number of interruptions within the microsatellite. The interruptions are created, in many cases, by the insertion of non-T bases within the microsatellite repeat (Figure S2). We interpret these results to indicate that under pool biased conditions, the slipped primer strand intermediates continue to arise, concomitant with an increased rate of base misinsertions, resulting in interruptions by the misincorporation-misalignment mechanism (Figure 1). We also performed temperature change experiments as an attempt to shift the equilibrium of the DNA duplexes towards more fully hybridized molecules. The Pol κ [T]8 microsatellite error frequency was lowered by decreasing the reaction temperature to 16°C, but the insertion bias was still prominent (Table 2). One interpretation of our results is that at low temperatures, the DNA duplex is more stable, allowing for fewer slipped-strand intermediates. Notably, this insertion bias within the [T]8 sequence is unique to Pol κ, as the other five polymerases examined displayed very strong biases towards only deletion errors (Figure 5).
Our results for polymerase errors produced within mononucleotide microsatellites differ dramatically from our initial expectations of the genetic consequences from simple strand-slippage mutagenesis. At this time, we cannot formally rule out the possibility that differential formation and/or utilization of IDL premutational intermediates, as a function of repeat sequence and polymerase, causes the pronounced deletion biases we observed. A direct test of this mechanism will require single molecule studies in which varying microsatellite DNA substrates are created with differentially labelled strands to allow visualization of IDL intermediates in the absence and presence of purified polymerases. Never-the-less, a much more simple explanation of all our results is that dNTP-stabilized mutagenesis within mononucleotide microsatellites is more common than previously believed. Interestingly, we recently reported that Pol κ frameshift errors within the HSV-tk gene coding region arise in a sequence context where C was significantly favoured as the 5’ neighbour of the deleted base [62]. In the case of the [C]10 template, there are 9 independent 5’ C’s that could participate in the deletion event. The dNTP-stabilized misalignment model postulates that the templating base assumes conformation that is non-instructive to the polymerase [37], resulting in that base being skipped during incorporation of the next incoming dNTP. Perhaps, dCMPs within the microsatellite interact with Pol κ amino acid residues, resulting in an active site geometry that is slightly altered compared to other bases. This effect may be enhanced by the 10-base repeat of the [C]10 mononucleotide template. Despite the mechanistic basis, our data indicate that there is a highly conserved bias towards mononucleotide deletion bias by polymerases from at least three different evolutionary families.
5. CONCLUSIONS
Based on the data and arguments presented here, we support a model in which polymerase errors within mononucleotide microsatellites can result from a dNTP-mediated misalignment mechanism, depending on DNA sequence and DNA polymerase identity. The change from a predominantly dNTP-stabilized mechanism to a strand-slippage mechanism with increasing microsatellite length may account for the differential rates of tandem repeat mutation that we observed genome-wide [29]. This is an important consideration, not only because mononucleotide microsatellites are present within genes, but also because intracellular dNTP ratios and levels are variable in cells. In both nontumorigenic and cancer cells, concentrations are not equal among the four dNTPs, and the relative abundance of each dNTP varies with cell type [63,64]. The natural asymmetry of dNTPs is hypothesized to be regulated to maximize replication accuracy [65]. Indeed, perturbations in the balance of dNTP pools have been shown to increase mutagenesis in human cells extracts [66] and in yeast, in which the mechanism is through misinsertion and altered proofreading activity [54]. Regulation of dNTP pools is altered in response to endogenous or exogenous factors [53], utilizing cell cycle control proteins p53 [67] and pRb [68]. Both dNTP concentration and pool balance can affect cell-cycle checkpoints, polymerase activity, and the identity of the DNA polymerases present at the replication fork [53,69].
Supplementary Material
ACKNOWLEDGEMENTS
We thank Suzanne Hile for critical reading of the manuscript and Amanda Breski for technical assistance.
FUNDING
This work was supported by the National Institutes of Health [grant R01-GM087472 to K.A.E.]. Funding for open access charge: The Jake Gittlen Memorial Cancer Research Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement: The authors declare no conflict of interest.
REFERENCES
- 1.Galindo CL, McIver LJ, McCormick JF, Skinner MA, Xie Y, Gelhausen RA, Ng K, Kumar NM, Garner HR. Global microsatellite content distinguishes humans, primates, animals, and plants. Mol Biol Evol. 2009;26:2809–2819. doi: 10.1093/molbev/msp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Field D, Wills C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proceedings of the National Academy of Sciences. 1998;95:1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Research. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 5.Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Molecular Biology and Evolution. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 6.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montgomery SB, Goode DL, Kvikstad E, Albers CA, Zhang ZD, Mu XJ, Ananda G, Howie B, Karczewski KJ, Smith KS, Anaya V, Richardson R, Davis J, MacArthur DG, Sidow A, Duret L, Gerstein M, Makova KD, Marchini J, McVean G, Lunter G. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome research. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katti MV, Ranjekar PK, Gupta VS. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Molecular Biology and Evolution. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- 10.Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 11.Loire E, Higuet D, Netter P, Achaz G. Evolution of coding microsatellites in primate genomes. Genome Biol Evol. 2013;5:283–295. doi: 10.1093/gbe/evt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome research. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 14.Marcotte EM, Pellegrini M, Yeates TO, Eisenberg D. A census of protein repeats. Journal of Molecular Biology. 1999;293:151–160. doi: 10.1006/jmbi.1999.3136. [DOI] [PubMed] [Google Scholar]
- 15.Rocha EPC, Matic I, Taddei Fo. Over-representation of repeats in stress response genes: a strategy to increase versatility under stressful conditions? Nucleic Acids Research. 2002;30:1886–1894. doi: 10.1093/nar/30.9.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannini G, Rinaldi C, Ristori E, Ambrosini MI, Cerignoli F, Viel A, Bidoli E, Berni S, D'Amati G, Scambia G, Frati L, Screpanti I, Gulino A. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004;23:2640–2647. doi: 10.1038/sj.onc.1207409. [DOI] [PubMed] [Google Scholar]
- 17.Huang H, Papadopoulos N, McKinley AJ, Farrington SM, Curtis LJ, Wyllie AH, Zheng S, Willson JKV, Markowitz SD, Morin P, Kinzler KW, Vogelstein B, Dunlop MG. APC Mutations in colorectal tumors with mismatch repair deficiency. Proceedings of the National Academy of Sciences, U.S.A. 1996;93:9049–9054. doi: 10.1073/pnas.93.17.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lea IA, Jackson MA, Li X, Bailey S, Peddada SD, Dunnick JK. Genetic pathways and mutation profiles of human cancers: site- and exposure-specific patterns. Carcinogenesis. 2007;28:1851–1858. doi: 10.1093/carcin/bgm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson JD, Crick FH. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 20.Watson JD, Crick FH. General Implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 21.Fresco JR, Alberts BM. The accommodation of noncomplementary bases in helical polyribonucleotides and deoxyribonucleic acids. Proceedings of the National Academy of Sciences of the United States of America. 1960;46:311. doi: 10.1073/pnas.46.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symposia on Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Levinson G, Gutman GA. Slipped strand mispairing: a major mechanism for DNA sequence evolution. Molecular Biology Evolution. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 24.Henderson ST, Petes TD. Instability of simple sequence DNA in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 27.Joshua-Tor L, Frolow F, Appella E, Hope H, Rabinovich D, Sussman JL. Three-dimensional structures of bulge-containing DNA fragments. J Mol Biol. 1992;225:397–431. doi: 10.1016/0022-2836(92)90929-e. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 29.Ananda G, Walsh E, Jacob KD, Krasilnikova M, Eckert KA, Chiaromonte F, Makova KD. Distinct mutational behaviors differentiate short tandem repeats from microsatellites in the human genome. Genome Biol Evol. 2013;5:606–620. doi: 10.1093/gbe/evs116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelkar YD, Strubczewski N, Hile SE, Chiaromonte F, Eckert KA, Makova KD. What is a microsatellite: a computational and experimental definition based upon repeat mutational behavior at A/T and GT/AC repeats. Genome Biol Evol. 2010;2:620–635. doi: 10.1093/gbe/evq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weirdl M, Dominska M, Petes TD. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efrati E, Tocco G, Eritja R, Wilson S, Goodman M. Abasic translesion synthesis by DNA polymerase β violates the "A-rule". Journal of Biological Chemistry. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 33.Kunkel TA. The mutational specificity of DNA polymerase β during in vitro DNA synthesis. Journal of Biological Chemistry. 1985;260:5787–5796. [PubMed] [Google Scholar]
- 34.Kunkel TA. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. The Journal of Biological Chemistry. 1986;261:13581–13587. [PubMed] [Google Scholar]
- 35.Kunkel TA. Misalignment-Mediated DNA Synthesis Errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 36.Tippin B, Kobayashi S, Bertram JG, Goodman MF. To Slip or Skip, Visualizing Frameshift Mutation Dynamics for Error-prone DNA Polymerases. Journal of Biological Chemistry. 2004;279:45360–45368. doi: 10.1074/jbc.M408600200. [DOI] [PubMed] [Google Scholar]
- 37.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 38.Bebenek K, Kunkel TA. Frameshift errors initiated by nucleotide misincorporation. Proc Natl Acad Sci U S A. 1990;87:4946–4950. doi: 10.1073/pnas.87.13.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- 40.Eckert KA, Mowery A, Hile SE. Misalignment-mediated DNA polymerase beta mutations: comparison of microsatellite and frame-shift error rates using a forward mutation assay. Biochemistry. 2002;41:10490–10498. doi: 10.1021/bi025918c. [DOI] [PubMed] [Google Scholar]
- 41.Hile SE, Eckert KA. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 2008;36:688–696. doi: 10.1093/nar/gkm1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hile SE, Wang X, Lee MY, Eckert KA. Beyond translesion synthesis: polymerase kappa fidelity as a potential determinant of microsatellite stability. Nucleic acids research. 2012;40:1636–1647. doi: 10.1093/nar/gkr889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ananda G, Hile SE, Breski A, Wang Y, Kelkar Y, Makova KD, Eckert KA. Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes. PLoS Genet. 2014;10(7):e1004498. doi: 10.1371/journal.pgen.1004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18:3529–3535. doi: 10.1093/nar/18.12.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdulovic AL, Hile SE, Kunkel TA, Eckert KA. The in vitro fidelity of yeast DNA polymerase delta and polymerase varepsilon holoenzymes during dinucleotide microsatellite DNA synthesis. DNA Repair (Amst) 2011;10:497–505. doi: 10.1016/j.dnarep.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opresko PL, Shiman R, Eckert KA. Hydrophobic interactions in the hinge domain of DNA polymerase beta are important but not sufficient for maintaining fidelity of DNA synthesis. Biochemistry. 2000;39:11399–11407. doi: 10.1021/bi000698t. [DOI] [PubMed] [Google Scholar]
- 47.Eckert KA, Hile SE, Vargo PL. Development and use of an in vitro HSV-tk forward mutation assay to study eukaryotic DNA polymerase processing of DNA alkyl lesions. Nucleic Acids Research. 1997;25:1450–1457. doi: 10.1093/nar/25.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opresko PL, Sweasy JB, Eckert KA. The mutator form of polymerase beta with amino acid substitution at tyrosine 265 in the hinge region displays an increase in both base substitution and frame shift errors. Biochemistry. 1998;37:2111–2119. doi: 10.1021/bi9722711. [DOI] [PubMed] [Google Scholar]
- 49.Starcevic D, Dalal S, Jaeger J, Sweasy JB. The hydrophobic hinge region of rat DNA polymerase beta is critical for substrate binding pocket geometry. J Biol Chem. 2005;280:28388–28393. doi: 10.1074/jbc.M502178200. [DOI] [PubMed] [Google Scholar]
- 50.Starcevic D, Dalal S, Sweasy J. Hinge residue Ile260 of DNA polymerase beta is important for enzyme activity and fidelity. Biochemistry. 2005;44:3775–3784. doi: 10.1021/bi047956x. [DOI] [PubMed] [Google Scholar]
- 51.Shah AM, Maitra M, Sweasy JB. Variants of DNA polymerase Beta extend mispaired DNA due to increased affinity for nucleotide substrate. Biochemistry. 2003;42:10709–10717. doi: 10.1021/bi034885d. [DOI] [PubMed] [Google Scholar]
- 52.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 53.Davidson MB, Katou Y, Keszthelyi A, Sing TL, Xia T, Ou J, Vaisica JA, Thevakumaran N, Marjavaara L, Myers CL, Chabes A, Shirahige K, Brown GW. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31:895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Research. 2011;39:1360. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baranovskiy AG, Babayeva ND, Suwa Y, Gu J, Pavlov YI, Tahirov TH. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic acids research. 2014;42:14013–14021. doi: 10.1093/nar/gku1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase beta. Biochemistry. 2014;53:2768–2780. doi: 10.1021/bi500139h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 59.Kunkel TA, Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 60.Dalal S, Hile S, Eckert KA, Sun KW, Starcevic D, Sweasy JB. Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry. 2005;44:15664–15673. doi: 10.1021/bi051179z. [DOI] [PubMed] [Google Scholar]
- 61.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J Biol Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 62.Baptiste BA, Eckert KA. DNA polymerase kappa microsatellite synthesis: two distinct mechanisms of slippage-mediated errors. Environ Mol Mutagen. 2012;53:787–796. doi: 10.1002/em.21721. [DOI] [PubMed] [Google Scholar]
- 63.Leeds JM, Slabaugh MB, Mathews CK. DNA precursor pools and ribonucleotide reductase activity: distribution between the nucleus and cytoplasm of mammalian cells. Molecular and Cellular Biology. 1985;5:3443–3450. doi: 10.1128/mcb.5.12.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W, Tan S, Paintsil E, Dutschman GE, Gullen EA, Chu E, Cheng YC. Analysis of deoxyribonucleotide pools in human cancer cell lines using a liquid chromatography coupled with tandem mass spectrometry technique. Biochem Pharmacol. 2011;82:411–417. doi: 10.1016/j.bcp.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mathews CK. DNA precursor metabolism and genomic stability. The FASEB Journal. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 66.Martomo SA, Mathews CA. Effects of biological DNA precursor pool assymetry upon accuracy of DNA replication in vitro. Mutation Research. 2002;499:197–211. doi: 10.1016/s0027-5107(01)00283-4. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 68.Angus SP, Wheeler LJ, Ranmal SA, Zhang X, Markey MP, Mathews CK, Knudsen ES. Retinoblastoma Tumor Suppressor Targets dNTP Metabolism to Regulate DNA Replication. Journal of Biological Chemistry. 2002;277:44376–44384. doi: 10.1074/jbc.M205911200. [DOI] [PubMed] [Google Scholar]
- 69.Godoy VG, Jarosz DF, Walker FL, Simmons LA, Walker GC. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 2006;25:868–879. doi: 10.1038/sj.emboj.7600986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.