Abstract
Background
The pilidium larva is a novel body plan that arose within a single clade in the phylum Nemertea - the Pilidiophora. While the sister clade of the Pilidiophora and the basal nemerteans develop directly, pilidiophorans have a long-lived planktotrophic larva with a body plan distinctly different from that of the juvenile. Uniquely, the pilidiophoran juvenile develops inside the larva from several discrete rudiments. The orientation of the juvenile with respect to the larval body varies within the Pilidiophora, which suggests that the larval and juvenile anteroposterior (AP) axes are patterned differently. In order to gain insight into the evolutionary origins of the pilidium larva and the mechanisms underlying this implied axial uncoupling, we examined the expression of the Hox genes during development of the pilidiophoran Micrura alaskensis.
Results
We identified sequences of nine Hox genes and the ParaHox gene caudal through a combination of transcriptome analysis and molecular cloning, and determined their expression pattern during development using in situ hybridization in whole-mounted larvae. We found that Hox genes are first expressed long after the pilidium is fully formed and functional. The Hox genes are expressed in apparently overlapping domains along the AP axis of the developing juvenile in a subset of the rudiments that give rise to the juvenile trunk. Hox genes are not expressed in the larval body at any stage of development.
Conclusions
While the Hox genes pattern the juvenile pilidiophoran, the pilidial body, which appears to be an evolutionary novelty, must be patterned by some mechanism other than the Hox genes. Although the pilidiophoran juvenile develops from separate rudiments with no obvious relationship to the embryonic formation of the larva, the Hox genes appear to exhibit canonical expression along the juvenile AP axis. This suggests that the Hox patterning system can maintain conserved function even when widely decoupled from early polarity established in the egg.
Electronic supplementary material
The online version of this article (doi:10.1186/s12915-015-0133-5) contains supplementary material, which is available to authorized users.
Keywords: Biphasic life cycle, Hox, Indirect development, Larval evolution, Nemertea, Pilidium
Background
The life history of many marine invertebrates is strikingly biphasic. In the most extreme cases, often termed maximally indirect development, adults and larvae differ so dramatically that they were originally described as different animals. Such development is exemplified by the pilidiophoran worms of the phylum Nemertea. During embryogenesis, a pilidium larva forms, complete with a domed episphere, apical sensory organ, blind stomach, and ciliated band for feeding and swimming. Days to weeks after the larval body plan is established, a juvenile begins to develop from distinct rudiments, called imaginal discs, that eventually fuse around the larval stomach. Once complete, the juvenile escapes the larval body and devours the larval tissues in a catastrophic metamorphosis.
The maximally indirect development of the pilidiophorans appears to be derived from more-or-less direct development [1-3]. The sister clade to the Pilidiophora (the Hoplonemertea) and the basal Palaeonemertea both have rather direct modes of development (Figure 1). Both have ciliated larvae that gradually grow into juvenile worms rather than passing through a morphologically distinct larval stage. Thus, the pilidium could be viewed as an insertion of a novel body plan into the life cycle of a more directly developing nemertean.
Figure 1.
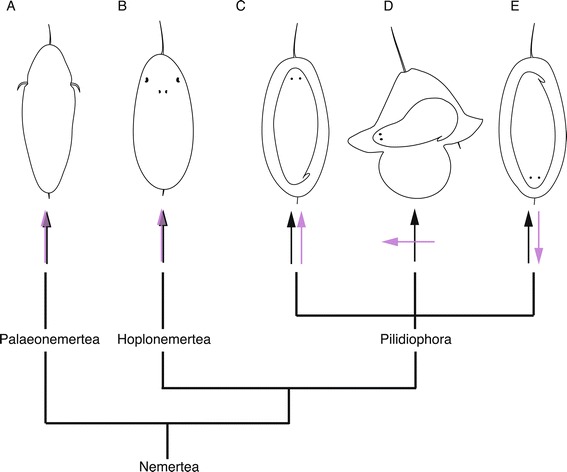
Distribution of larval types and larval-juvenile axial orientations among major Nemertean clades. Phylogeny (bottom) shows relationships among three major nemertean clades. Above: orientation of anteroposterior (AP) axis of larva (black arrows) and juvenile (purple arrows) of (A) a typical palaeonemertean larva, (B) a typical hoplonemertean larva, and (C) the larva of pilidiophoran Micrura rubramaculosa [62], in which the larval and juvenile AP axes coincide; (D) a typical pilidium, in which larval and juvenile axes are roughly perpendicular; and (E) the larva of M. akkeshiensis, in which larval and juvenile AP axes oppose one another.
If the pilidium is a true novelty, one might expect different mechanisms patterning the larval versus juvenile body. Diversity of the orientation of the juvenile with respect to the larva within the Pilidiophora provides indirect evidence of such mechanistic decoupling. While all pilidiophorans build worm-shaped juveniles inside larval bodies, the anteroposterior (AP) axes of the larva and the juvenile appear to be decoupled. In a typical pilidium, the juvenile AP axis is roughly perpendicular to the larval AP axis (Figure 1D). In some pilidia, however, the AP axis of the juvenile coincides with that of the larva, as in the sock-like larvae called pilidium incurvatum [4] and pilidium recurvatum of Riserius sp. [4-7], the non-feeding bullet-shaped larvae of Micrura rubramaculosa and M. verrilli [8], and the trochophore-like pilidium nielseni [9,10] (Figure 1C). However, in the lecithotrophic larvae of M. akkenshiensis [11] and several other species, the AP axis of the larva and juvenile oppose one another [11] (Figure 1E). This diversity suggests that pilidiophoran evolution may have been accompanied by the dissociation of axial patterning mechanisms between life history stages.
In order to understand the mechanics of pilidiophoran development and the origin of a novel larval body plan, we examined the expression of the Hox genes during development of a pilidiophoran nemertean M. alaskensis. Hox genes are highly conserved patterning genes that are expressed in and determine the identity of domains along the AP axis of many animals [12-14]. We also looked at expression of one of the three ParaHox genes, caudal (Cdx), which is a posterior marker in other animals [15].
Very little is known about patterning mechanisms in nemerteans [16-18]. Here, we report for the first time on the patterns of expression of Hox genes in this phylum, specifically during the development of the pilidium larva, and discuss the relevance of these findings to the origins of the pilidium larva.
Results
Development of M. alaskensis
The development and a staging scheme for M. alaskensis is documented in detail by Maslakova [19], and is briefly summarized here. Fertilized zygotes undergo spiral cleavage and pass through a square-shaped blastula (blastosquare) stage (Figure 2A). By day 1, they have developed into swimming gastrulae with an apical tuft. By day 2, a ciliated band spanning two lateral lobes becomes evident. At about 3 days, the larvae begin to feed on microalgae (Figure 2B). Food particles pass through the funnel-shaped esophagus, which is equipped with two ciliated ridges, and enter the blind gut. As the larva grows, the lappets grow larger and two additional elaborations of the ciliated band appear, called the lobes (anterior and posterior, in reference to the juvenile AP axis). The first rudiments of the juvenile, the cephalic imaginal discs, appear as invaginations of the larval episphere as early as 5 to 7 days, and give rise to the juvenile head. We refer to this stage as the ‘cephalic-discs stage’ (Figure 2C). The timing of development, especially the emergence of the juvenile rudiments, is highly variable, and depends on the water temperature, feeding regime, and, possibly, other factors. As the cephalic discs are growing, a pair of trunk discs invaginates from the hyposphere near the larval stomach. These discs give rise to the majority of the trunk of the juvenile. We refer to this stage as the ‘trunk-discs stage’ (Figure 2D). The third and final pair of imaginal discs, called the cerebral-organ discs, arise as early as 2 weeks, as invaginations of the inner epidermis of the larval lappets at the lower end of the esophageal ciliated ridges. These discs are located anterior to the trunk discs (along the AP axis of the future juvenile), and give rise to the cerebral organs. Thus, this stage is called ‘cerebral-organ-discs stage’ (Figure 2E). At about the same time, an unpaired proboscis rudiment appears as a small cluster of cells near the larval epidermis in-between the two cephalic discs. The proboscis rudiment later fuses with the cephalic discs to form the head rudiment, while the trunk and cerebral organ discs fuse with each other to form the trunk rudiments. An unpaired dorsal disc arises dorsal to the trunk rudiments between the stomach and the larval epidermis, at about the same time as the proboscis rudiment. This characterizes the ‘head-and-trunk stage’ (Figure 2F). As early as 4 weeks after fertilization, the dorsal disc fuses with the trunk rudiments and, subsequently, the head rudiment fuses with the trunk rudiment to form a toroid of juvenile tissue around the stomach. We refer to this stage as the ‘torus stage’ (Figure 2G). The juvenile proboscis continues to grow, first extending beyond the margin of the juvenile head (‘extended-proboscis stage’, Figure 2H), then reaching the dorsal margin of the juvenile trunk epidermis (‘complete-proboscis stage’, Figure 2I). The juvenile tissues grow over the stomach and esophagus (before fusing, ‘hood stage’, Figure 2J), eventually enclosing the gut (after fusing, ‘pre-metamorphosis stage’, Figure 2K). As early as 35 days, the juvenile ruptures the larval enclosure and engulfs the larval tissues in a radical metamorphosis.
Figure 2.
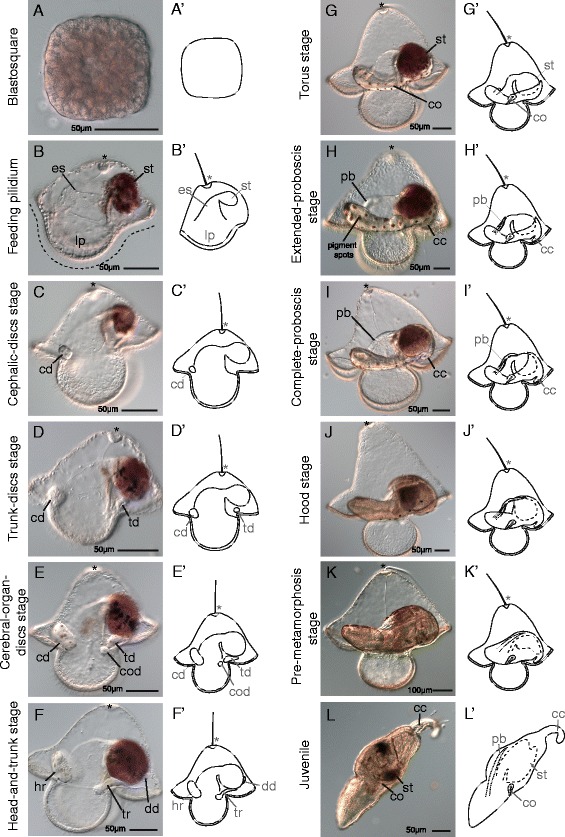
Development of Micrura alaskensis. Differential interference contrast images. Lateral views except in (A). Apical organ (indicated by asterisk) orientated up, juvenile anterior to the left. (A) Blastosquare stage. Polar view (animal or vegetal). (B) Feeding pilidium stage larva. Esophagus (es), leads to blind stomach (st), which is dark red due to algal food. Ciliated band, indicated with dashed line, spans lobes and lappets (lp). (C) Cephalic-discs-stage larva. One of the paired cephalic discs (cd) labeled. (D) Trunk-discs-stage larva. One of the paired trunk discs (td) labeled. (E) Cerebral-organ-discs-stage larva. One of the paired cerebral-organ discs (cod) labeled. (F) Head-and-trunk-stage larva. Cephalic discs have merged with each other to become the head rudiment (hr). Cerebral-organ discs have merged with the trunk discs to become the trunk rudiment (tr). Dorsal disc (dd) is present. (G) Torus-stage larva. All discs have merged together to form toroid of juvenile tissue around larval stomach. Cerebral organ (co) is labeled. (H) Extended-proboscis-stage larva. Proboscis (pb) has begun to grow over the esophagus towards the stomach. Caudal cirrus (cc) is evident. Membrane housing juvenile (the amnion) is decorated with red-brown pigment spots in a polka-dot pattern. Amniotic pigment spots develop in earlier stages, but are quite clear in this image. (I) Complete-proboscis-stage larva. Proboscis has grown to meet stomach. (J) Hood-stage larva. Dorsal tissues of juvenile overgrow proboscis. (K) Pre-metamorphosis-stage larva. Juvenile is complete inside larva. Metamorphosis (not shown) is rapid and radical; juvenile breaks out of larval body and devours all larval tissues in a minute or less. (L) Juvenile after metamorphosis. Larval structures, including amniotic pigment, are inside juvenile stomach.
M. alaskensis Hox gene sequences
Nine contigs containing Hox gene sequences were recovered from the M. alaskensis developmental transcriptome. Full coding sequences were isolated from seven of the nine genes. Characteristic residues and motifs were found in all nine sequences and permitted assignment to known paralog groups [20-23]. See Additional file 1 for alignment of M. alaskensis Hox genes with those of other bilaterians. Bayesian phylogenetic analysis supports assignment to paralog groups (PG) based on the presence of characteristic residues (Additional file 2). M. alaskensis has representatives from PG1 (Labial/Lab), PG2 (Proboscipedia/Pb), PG3 (Hox3), PG4 (Deformed/Dfd), PG5 (Sex combs reduced/Scr), PG6 (Lox5), PG7 (Antennapedia/Antp), PG8 (Lox4), and PG9-PG13 (Post2). Cdx and Six3/6 were also isolated and identified via phylogenetic analysis (see Additional file 2 for Cdx, data for Six3/6 not shown). Additional file 3 lists details of each gene, including paralog group, length of predicted open reading frame, and GenBank accession number.
Six Hox genes and the Cdx gene have been previously cloned from a single nemertean species, the pilidiophoran Ramphogordius (= Lineus) sanguineus [17]; however, their expression had not been characterized. M. alaskensis genes MaLab, MaHox3, MaDfd, MaLox5, MaAntp, and MaPost2 isolated by us are orthologous to R. sanguineus genes LsHox1, LsHox3, LsHox4, LsHox6, LsHox7, and LsHox9, respectively.
Gene expression in the development of M. alaskensis
No Hox gene expression was detected at any stage of development in the larval body, cephalic discs, cerebral organ discs, or the proboscis rudiment. We are confident that the in situ hybridization protocol works on pilidial structures and early stages because we have expression data for other genes at early stages (for example, MaSix3/6, which is expressed at blastosquare and in the pilidial body during feeding-pilidium stage, see Additional file 4). Hox genes were expressed at various later developmental stages in different patterns in the trunk discs and the dorsal disc, and, as a rule, expression became more prominent, and occupied larger domains in more advanced developmental stages.
Labial (MaLab)
No expression was observed in gastrulae or young pilidia (data not shown). Weak expression of MaLab is first noticeable in the trunk discs as soon as they emerge (Figure 3A). At the head-and-trunk stage, expression becomes more prominent and occupies a broader domain in each trunk disc, and is also detectable in the dorsal disc (Figure 3B). In complete-proboscis-stage larvae, expression extends to the anterior margin of the trunk disc, bordering the cerebral organ, and posteriorly throughout most of the juvenile trunk, except the very posterior (Figure 3C).
Figure 3.
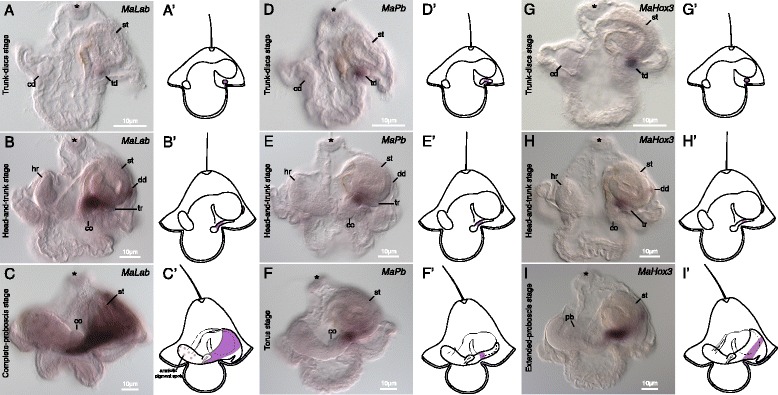
Expression of MaLab, MaPb, and MaHox3 in M. alaskensis larval development. All images and diagrams show lateral views with larval anterior up and future juvenile anterior to the left. Asterisk marks apical organ. (A) MaLab is expressed in the trunk discs (td) at trunk-discs stage. The cephalic disc (cd) and stomach (st) are marked for reference. (B) In head-and-trunk-stage larvae, MaLab is expressed in portion of the trunk rudiment (tr) and dorsal disc (dd). The head rudiment (hr) is marked for reference. (C) In complete-proboscis-stage larvae MaLab is expressed in a large portion of the juvenile posterior, from just up against the cerebral organ (co). (D) MaPb is expressed initially in trunk discs (td) at trunk-discs stage. (E) In head-and-trunk-stage larvae, MaPb is expressed in a central domain of the trunk rudiment (tr). (F) In torus-stage larvae, MaPb is expressed in a small central domain of the juvenile posterior. (G) The onset of MaHox3 expression occurs at the trunk-discs stage in the trunk discs (td). (H) In head-and-trunk-stage larvae, MaHox3 is expressed in a central domain of the trunk rudiment (tr) and in a portion of the dorsal disc (dd). MaHox3 is not expressed in the larval body or head rudiment (hr). (I) In extended-proboscis-stage larvae, MaHox3 is expressed in a narrow stripe of the juvenile posterior. Proboscis rudiment (pb) marked in image. Anterior spots are amniotic pigment spots (see Figure 2) that happen to be in focus in this image. (A’-I’) diagrammatically illustrate expression patterns in the three respective developmental stages.
Proboscipedia (MaPb)
Similar to MaLab, expression of MaPb is first evident in the trunk discs as soon as they emerge (Figure 3D). By head-and-trunk stage, MaPb is expressed clearly in the middle portion of the trunk rudiments, but not in the dorsal disc (Figure 3E). About a fourth of each trunk rudiment is stained, forming a stripe about midway from the anterior portion of the trunk discs to the posterior tip of the trunk (Figure 3E). By torus stage, the expression of MaPb remains localized to a fairly narrow region of the trunk about midway between the cerebral organ and the posterior end (Figure 3F).
Hox3 (MaHox3)
As is the case with MaLab and MaPb, no expression of MaHox3 is detectable until the trunk-discs stage, and expression is first evident in the trunk discs (Figure 3G). At the head-and-trunk stage, the expression domain occupies the anterior portion of the trunk discs (Figure 3H). The size of the expression domain is similar to MaPb. Unlike MaPb, expression of MaHox3 at this stage is also detectable in the dorsal disc (Figure 3H). At extended-proboscis stage, the expression in the dorsal disc and the trunk discs forms a continuous band encircling the juvenile trunk about half way between the cerebral organs and the posterior end (Figure 3I).
Deformed (MaDfd)
MaDfd is not expressed at early stages, including the trunk-discs stage (Figure 4A). By the head-and-trunk stage, MaDfd is expressed in a very small patch in trunk rudiments, similar in location to MaHox3, but slightly more posterior, as well as in the anterior portion of the dorsal disc (Figure 4B). By torus stage, MaDfd is expressed in a very thin band around the juvenile trunk (Figure 4C).
Figure 4.
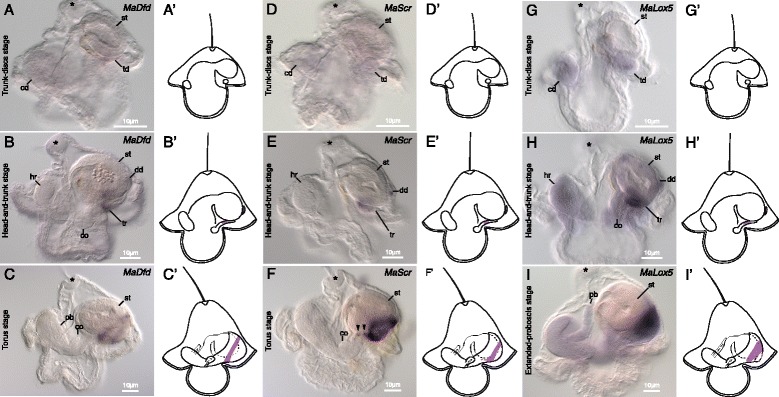
Expression of MaDfd, MaScr, and MaLox5 in M. alaskensis larval development. All images and diagrams show lateral views with larval anterior up and future juvenile anterior to the left. Asterisk marks apical organ. (A) MaDfd expression is not evident during the trunk-discs stage anywhere in the larval or juvenile body, including the trunk discs (td), the cephalic disc (cd), and stomach (st). (B) MaDfd is first clearly expressed during the head-and-trunk stage in small domains of each trunk rudiment (tr) and in a portion of the dorsal disc (dd). The head rudiment (hr) is indicated for reference. (C) In torus-stage larvae, MaDfd is expressed in a very narrow ring of the juvenile posterior. Proboscis rudiment (pb) marked for reference. (D) MaScr expression is not evident during the trunk-discs stage anywhere in the larval or juvenile body. (E) The onset of MaScr expression occurs during the head-and-trunk stage in domains of each trunk rudiment (tr) and in a portion of the dorsal disc (dd). (F) In torus-stage larvae, MaScr is expressed in a ring of the juvenile posterior with an additional few small spots of expression laterally towards the anterior of the juvenile (arrowheads). (G) MaLox5 expression is not evident during the trunk-discs stage anywhere in the larval or juvenile body. Faint background staining is observed in both pairs of discs. (H) The onset of strong MaLox5 expression occurs during the head-and-trunk stage in domains of the each trunk rudiment (tr) and in a portion of the dorsal disc (dd). Faint background staining persists in all juvenile rudiments, including head rudiment (hr) as well as portions of the stomach. (I) MaLox5 is expressed in a patch near the posterior end of the juvenile in extended-proboscis-stage larvae. (A’-I’) diagrammatically illustrate expression patterns in the three respective developmental stages.
Sex combs reduced (MaScr)
Similar to MaDfd, MaScr is not apparently expressed until the head-and-trunk stage (Figure 4E). At the head-and-trunk stage, MaScr is expressed in a region of both trunk rudiments and the posterior region of the dorsal disc (Figure 4F). The expression domain in the trunk rudiment is slightly wider than that of MaPb, MaHox3, and MaDfd. During the torus stage, MaScr is expressed in a thin belt around the juvenile trunk located somewhat more posterior to the domains of MaPb, MaHox3, and MaDfd (compare Figure 4F with Figures 3F,I and 4C). A few small localized spots of expression occur laterally on either side of the juvenile trunk just anterior to the main expression domain (Figure 4F).
Lox5 (MaLox5)
The faint staining observed at early stages throughout the juvenile rudiments (see Figure 4G) likely represents either very weak expression or possibly just background staining (Figure 4G). Clear MaLox5 expression is first detected at the head-and-trunk stage in the trunk rudiments and dorsal disc (Figure 4H). Within the trunk rudiments, the expression domain of MaLox5 appears to be slightly broader than that of MaScr. In extended-proboscis-stage larvae, the expression domain appears as a broad band encircling the juvenile trunk near the posterior end (Figure 4I).
Antennapedia (MaAntp)
Expression of MaAntp is first detectable at the head-and-trunk stage (Figures 5A,B). At this stage, MaAntp is expressed in posterior domains of the trunk rudiments and the posterior part of the dorsal disc (Figure 5B). By complete-proboscis stage, expression is confined to a fairly broad band encircling the juvenile trunk near the posterior end (Figure 5C).
Figure 5.
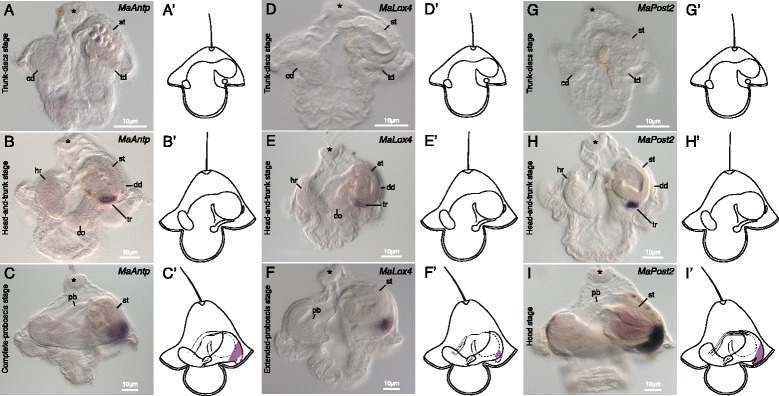
Expression of MaAntp, MaLox4, and MaPost2 in M. alaskensis larval development. All images and diagrams show lateral views with larval anterior up and future juvenile anterior to the left. Asterisk marks apical organ. (A) MaAntp expression is not evident during the cerebral-organ-discs stage anywhere in the larval or juvenile body, including the trunk discs (td), the cephalic disc (cd), cerebral organ discs (cod), or stomach (st). (B) Initial MaAntp expression occurs during the head-and-trunk stage in domains of each trunk rudiment (tr) and in a portion of the dorsal disc (dd). The head rudiment (hr) is indicated for reference. (C) MaAntp is expressed in a patch near the posterior end of the juvenile in complete-proboscis-stage larvae. Proboscis rudiment (pb) marked for reference. (D) MaLox4 is not expressed in trunk-discs-stage larvae. (E) MaLox4 expression first occurs during the head-and-trunk stage in domains of each trunk rudiment (tr), with no evident expression in the dorsal disc (dd). (F) MaLox4 is expressed in a small spot near the posterior end of the juvenile in extended-proboscis-stage larvae. (G) MaPost2 is not expressed in trunk-discs-stage larvae. (H) MaPost2 expression first occurs during the head-and-trunk stage in posterior domains of each trunk rudiment (tr), with no evident expression in the dorsal disc (dd). (I) MaPost2 is expressed in the juvenile caudal cirrus in hood-stage larvae. (A’-I’) diagrammatically illustrate expression patterns in the three respective developmental stages.
Lox4 (MaLox4)
Faint expression of MaLox4 is first evident at the head-and-trunk stage in the trunk rudiments (Figure 5E). By the extended-proboscis stage, expression of MaLox4 appears as a patch near the posterior of the juvenile (Figure 5F).
Post2 (MaPost2)
MaPost2 is not expressed in early stages (Figure 5G), and is first detectable at the head-and-trunk stage at the very posterior of the trunk rudiments (Figure 5H). No expression was observed in the dorsal disc. By the hood stage, MaPost2 is expressed in a broad domain at the very posterior of the juvenile trunk including the juvenile caudal cirrus (Figure 5I).
Caudal (MaCdx)
In addition to the expression of Hox genes, we also examined the expression of MaCdx during pilidial development. MaCdx is first expressed in the trunk-discs stage in the posterior portion of trunk discs. By the head-and-trunk stage, MaCdx is expressed in the posterior portion of the trunk rudiments (Figure 6B). At the extended-proboscis stage, MaCdx is expressed in a ring around the juvenile trunk just anterior to the juvenile caudal cirrus (Figure 6C).
Figure 6.
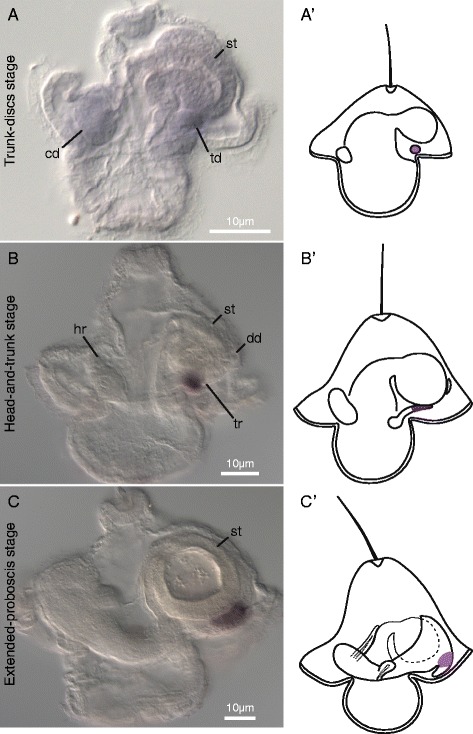
MaCdx expression in M. alaskensis larval development. All images and diagrams show lateral views with larval anterior up and future juvenile anterior to the left. (A) MaCdx is expressed in trunk discs of trunk-discs-stage larvae. A single trunk disc (td), cephalic disc (cd), and the stomach (st) are labeled for reference. (B) MaCdx expression in head-and-trunk stage occurs in posterior domains of each trunk rudiment (tr), with no evident expression in the dorsal disc (dd). The head rudiment (hr) is indicated for reference. (C) MaCdx is expressed just anterior to the juvenile caudal cirrus in extended-proboscis-stage larvae. (A’-C’) diagrammatically illustrate expression patterns in the three respective developmental stages.
Discussion
The trochozoan Hox complement typically contains 10 to 11 Hox genes [24-26]. Orthologs of all but two of the genes in the typical trochozoan repertoire were identified in M. alaskensis; we have not found orthologs of the central gene Lox2 or of the posterior gene Post1 in the M. alaskensis developmental transcriptome. Attempts to isolate Lox2 and Post1 from genomic DNA of M. alaskensis using degenerate PCR were not successful. Both genes have been found in annelids, mollusks, and brachiopods, but not in bryozoans [27]. Post1 and Post2 are likely a result of a gene duplication that occurred in the trochozoan lineage [21]. Lox4 and Lox2 are also related by a duplication event that likely took place within the Trochozoa [21]. One possible explanation for the absence of Post1 and Lox2 in M. alaskensis (if, indeed, the absence is real, and not a result of lack of expression, or low level of expression at surveyed developmental stages) is that the duplication events happened after the split between the Nemertea and the other trochozoan phyla. Alternatively, Post1 and Lox2 may have been lost in the nemertean lineage. Post1 is not involved in axial patterning in either of the three annelid species [26,28,29] or in the two mollusk species in which posterior Hox genes have been analyzed. Post1 is genomically disconnected from the rest of the cluster in the annelids Capitella teleta and Platynereis dumerilii [26,30]. This suggests that Post1 may have been removed from participation in vectorial patterning in the trochozoan lineage. Perhaps, its absence in the M. alaskensis developmental transcriptome is related to an evolutionary loss of body patterning function.
The principal and remarkable finding here is that, based on their expression, all nine Hox genes in M. alaskensis likely participate in vectorial regionalization along the AP axis of the developing juvenile, but are not expressed at detectable levels in the pilidial larval body. Furthermore, it appears that Hox genes do not pattern the entire juvenile body, but only the posterior region, which develops from the trunk imaginal discs and dorsal rudiment. The canonical function of Hox genes in other bilaterians, such as mollusks [31-33], polychaetes [26,28,34-38], deuterostomes [39-41], and acoels [42,43], is AP patterning of the juvenile/adult trunk. Thus, with the caveat that we only have data for a single species, the role of Hox genes in axial patterning of the adult body appears to be conserved in nemerteans.
Although we do not have evidence from double labeling, based on the position of expression domains with respect to the morphological landmarks (for example, cerebral organ discs or the caudal cirrus), it is quite clear that these domains partially overlap along the AP axis of the juvenile body. Interestingly, the orthologs of anterior genes show earlier and more anterior patterns of expression compared to the orthologs of the more posterior genes. Because we do not know the genomic organization of Hox cluster in nemerteans (or even whether the genes are arranged in a cluster), it is unclear whether the spatial and temporal pattern of expression of these genes in pilidial development corresponds to the order of arrangement in the genome. Therefore we do not claim that Hox genes in M. alaskensis exhibit co-linearity. Nevertheless, it is noteworthy that the pattern of expression of Hox genes in M. alaskensis development corresponds to the order of arrangement of their orthologs in animals with known genomic organization of the Hox cluster (for example, C. teleta [26], Lottia gigantea [25], and Branchiostoma floridae [44]).
One of the Hox genes, MaScr, has a peculiar pattern of expression: in a few small patches on the lateral surface of the juvenile trunk just anterior to its belt-like domain. The position of these dots roughly corresponds to the position of the nephridial openings in the juvenile trunk of M. alaskensis [19]. We do not claim here that this is what they are, because we do not have an easy way to label nephridial openings in the same preparation as the in situ hybridization, but the pattern is suggestive.
The ParaHox gene Cdx patterns posterior structures in a number of spiralians, such as the gastropods Gibbula varia and Patella vulgate, the polychaete Platynereis dumerilii, and the nemertean Lineus viridis [15,45]. Similarly, in M. alaskensis it is expressed in the posterior end of the developing juvenile.
Because Hox genes are not expressed during M. alaskensis embryogenesis or at any other time in the larval tissues, the pilidial body must be patterned by mechanisms other than Hox genes. At this time it is unclear whether the pilidium larva and juvenile share other patterning mechanisms, or if the pilidial AP axis is patterned by a novel mechanism. Preliminary experiments with inhibitors of the Wnt and fibroblast growth factor pathways suggest that these may be involved in patterning the pilidium (Hiebert, unpublished).
Hox gene expression has been studied in many other animals, but most of those develop more or less directly [41,46,47]. In most direct-developers, the adult axial properties can be traced directly back to initial asymmetries in the egg [48,49]. Even in some indirect-developers, such as some annelids or the fruit fly, a blueprint of the adult is already present at the larval stage [28,49,50]. In the pilidium, there is no clear early blueprint of the adult worm, and we found that Hox patterning is not shared between early pilidial and adult stages. The same is true for another group of maximally indirectly developing animals: the sea urchins. In urchins, the Hox genes are also more or less limited to patterning the juvenile structures as they form inside the larva [51,52]. But, in urchins, the adult body is highly modified and the Hox expression pattern is difficult to relate to that in other bilaterians. Nemertean adult body, however, is clearly comparable to that of other bilaterians. It is remarkable that Hox genes maintain conserved expression in the juvenile body of M. alaskensis even though it develops separately from the larva and expression of Hox genes is evident even before the individual juvenile rudiments form a contiguous domain.
Expression patterns of Hox and Cdx genes in pilidial development suggest that the larval body may represent an ontogenetic ‘insertion’ into a more direct developmental program because these adult patterning mechanisms are not shared by the larva. Thus, less constraint may exist on phase-specific evolution in the pilidiophorans than in animals whose larval stage preforms the adult. This may be relevant to how the pilidium arose, diversified, and how catastrophic metamorphosis evolved in nemerteans.
Conclusions
We find that Hox genes in the pilidiophoran Micrura alaskensis pattern the AP axis of the juvenile trunk as it arises from isolated rudiments inside the pilidium larva. The pilidium, an evolutionarily novel body plan, is patterned without the use of Hox genes. The lack of shared axial patterning mechanisms (Hox and Cdx) across the two life stages may help explain the extreme morphogenetic uncoupling between the larva and the juvenile.
Methods
Collection of adults, fertilization of gametes, and larval culture
Adult M. alaskensis Coe, 1901 (Heteronemertea; Lineidae) were collected from mudflats in Coos Bay near Charleston, OR, USA during May to August of 2009 to 2013 on negative tides. Adult worms were transported to the Oregon Institute of Marine Biology and kept in a flow-through seawater system in 150 mL glass custard dishes until dissection. Adults were either collected bearing ripe gametes or allowed to ripen in the laboratory over a few weeks. Primary oocytes and sperm were dissected from live females and males and transferred to 0.45 μm filtered seawater (FSW). After around 30 minutes, oocytes underwent germinal vesicle break down and were fertilized with a dilute suspension of sperm. Larval culture followed methods described by Maslakova [19]. In short, embryos were kept in custard dishes until reaching a swimming stage, at which point the larvae were transferred to gallon jars kept at ambient sea temperature (12 to 16°C) with constant stirring using acrylic paddles [53]. Larval concentrations during the first few weeks of development were approximately one larval per milliliter. Subsequently, larvae were maintained at close to one larva per 10 mL. FSW was exchanged every 2 to 3 days via reverse-filtration. The larvae were fed Rhodomonas lens (104 cells/mL) after each water change.
Isolation of Hox genes
Hox-containing contigs were retrieved from a M. alaskensis developmental transcriptome containing transcripts from seven developmental stages (gastrula, young feeding pilidium, cephalic-discs stage, cerebral-organ-discs stage, head-and-trunk stage, and hood to pre-metamorphosis stages). The assembly contained nine unique Hox gene transcripts, seven of which included a full open reading frame. Long fragments of each Hox gene were amplified from cDNA libraries from torus stage and extended-proboscis- to hood-stage larvae. Primers were designed from the transcriptome contigs using Primer3 [54,55]. Additional sequence fragments were amplified by rapid amplification of cDNA ends (RACE [56]) from cDNA libraries derived from head-and-trunk-stage, torus-stage, and extended-proboscis-stage to hood-stage larvae. PCR and RACE products were subcloned into PGEM-t (Promega, Madison, WI, USA) vectors and then transformed into One Shot Top10 chemically competent Escherichia coli cells (Invitrogen, Carlsbad, CA, USA). Plasmid DNA was isolated using QIAprep Spin Miniprep Kit (Qiagen Valencia, CA, USA) and sequenced in both forward and reverse directions on a ABI 3730xl DNA Analyzer platform (Sequetech, Mountain View, CA, USA) using T7 and SP6 primers.
Orthology assessment and phylogenetic analysis
Orthology was determined by the presence of characteristic residues in the homeodomain and flanking regions and by phylogenetic analysis. An amino acid sequence alignment was made by eye using Geneious version 7.0.4 [57] and included the complete homeodomain, the 12 amino acids just 3′ of the homeodomain, and the eight amino acids just 5′ of the homeodomain. Hox and Cdx fragments from M. alaskensis were aligned with Cdx and Hox complements of a deuterostome Branchiostoma florida, two ecdysozoans (Tribolium castaneum, Drosophila melanogaster), and five lophotrochozoans (an annelid C. teleta, a bryozoan Bugula turrita, a nemertean Ramphogordius (=Lineus) sanguineus, a brachiopod Lingula anatina, and a mollusk Euprymna scolopes). Sequence data from representative metazoans were retrieved using the National Center for Biotechnology ([58]; accession numbers listed in Additional file 5).
Phylogenetic analysis (Bayesian inference) was conducted using MrBayes version 3.2.1 [59,60]. Hox fragments were aligned using the homeodomain and the twelve 3′ amino acids. The analysis was done with the Rtrev amino acid model with a gamma-shaped distribution of rates across sites. Drosophila eve was selected as an out-group. The analysis was done with five heated chains with 5,000,000 generations and was sampled every 500 generations. Four independent runs were conducted. The first 25% samples from the cold chain was discarded as burn-in. Trees were visualized and manipulated using Figtree version 1.4.0.
Whole mount in situ hybridization
Gastrulae and larvae were relaxed in 1:1 mixture of 0.37 M MgCl2 in FSW for 10 minutes, then fixed in 4% paraformaldehyde in FSW overnight at 4°C. Fixed larvae were washed three times in 1X phosphate-buffered saline (PBS), dehydrated in methanol, and stored at −20°C. A protocol for in situ hybridization was modified from Seaver and Kaneshige [61]. Larvae were rehydrated in PBS, followed by three washes in PBS with 0.1% Tween-20 (PTw). All wash and incubation steps were 5 to 10 minutes, unless otherwise indicated. Fixed tissues were acetylated in 1% triethanolamine in PTw with 0.3% acetic anhydride, and then incubated in 1% triethanolamine in PTw with 0.6% acetic anhydride. After two quick rinses with PTw, larvae were re-fixed with 4% paraformaldehyde for 30 to 60 minutes. The fixative was removed with four washes of PTw. Larvae were transferred to hybridization buffer (50% formamide, 5X saline-sodium citrate (SSC)pH7, 50 μg/mL heparin, 0.1% Tween-20, 1% SDS, 50 μg/mL boiled salmon sperm DNA made up in diethyl dicarbonatewater) at hybridization temperature (63°C) for 4 hours to overnight. Digoxygenin-labeled riboprobes were synthesized using linearized template DNA using DIG RNA Labeling Mix and following standard protocol (Roche, Penzberg, Germany). Probes were quantified using a Qubit fluorometer (Invitrogen) then diluted to 1 ng/μL in hybridization buffer and either used right away or stored at −20°C. Diluted probes were saved after hybridization and reused up to five times. Before hybridization, probes were denatured at 80°C to 90°C for 10 minutes. Larvae were hybridized with the probe in hybridization buffer at 63°C for 2 to 3 days. After hybridization, the probe was washed out with hybridization buffer. Then larvae were washed through a graded series of hybridization buffer/2X SSC (75/25, 50/50, 25/75, 100/0). Larvae were washed of excess probe by incubating in 0.05X SSC for 30 minutes at 63°C. This step was repeated once, followed by transfer through a series (70/30, 30/70, 0/100) to Tris-buffered saline and Tween (TBST; 0.15 M NaCl; 0.2 M Tris buffer, pH 7.5). Larvae were washed four times in TBST, then blocked in TBST with 0.1% Tween-20, 5% normal goat serum, and 2 ng/μL bovine serum albumin). The color reaction took place in 1X Detection Buffer (Roche) with 4.4 μL of 75 mg/mL Nitro Blue Tetrazolium (Sigma, St. Louis, MO, USA) and 3.3 μL of 50 mg/mL 5-bromo-4-chloro-3-indolyl phosphate (Sigma). Staining was carried out in the dark from 1 hour to 2 days, depending on the probe. The staining reaction was terminated by washing larvae in PTw. Stained larvae were mounted on slides in 80% glycerol in PBS. Photomicrographs were obtained using a Leica DFC400 digital color camera mounted on an Olympus BX51 microscope equipped with differential interference contrast optics. Ten to twenty specimens were examined for each gene and stage.
Acknowledgements
We thank Elaine Seaver, Néva Meyer, and Emi Yamaguchi, who all helped immensely with the development of the in situ hybridization protocol. For help with transcriptomics, we thank William Cresko, Susie Basham, Pete Batzel, Andrew Nishida, Mikhail Matz, Galina Aglyamova, and Eli Meyer. We are especially grateful to George von Dassow for many thoughtful suggestions and insightful comments. This work was supported by the National Science Foundation Graduate Research Fellowship DGE-0829517 to LH and the NSF grant IOS-1120537 to SM.
Additional files
Alignment of Hox and Cdx genes and flanking sequences. Alignment includes the Hox homeodomain, twelve residues 3′ of the homeodomain, and eight residues 5′ of the homeodomain for representative species. Sequences are aligned against Drosophila melanogaster Antp. Yellow highlighting indicates conserved residues that differ from other paralog groups. Green highlighting indicates positions of conserved peptide motifs outside the homeodomain.
Bayesian phylogenetic analysis of M. alaskensis Hox genes. Bf, Branchiostoma floridae; Tc, Tribolium castaneum; Dm, Drosophila melanogaster; Ct, Capitella teleta; Bt, Bugula turrita; Ls, Lineus sanguineus; La, Lingula anatine; Es, Euprymna scolopes. Numbers at branch points indicate Bayesian posterior probabilities. Values lower than 50% are not included. Colors denote paralog groups (PG). Arrowheads indicate location of M. alaskensis Hox genes.
Table of Hox, Cdx, and Six3/6 genes isolated from M. alaskensis .
MaSix3/6 expression in M. alaskensis development. (A) Polar view (animal or vegetal) of blastosquare. (B-D) Feeding pilidium, lateral view. Stomach (st), cephalic disc (cd), and trunk disc (td) labeled. Asterisk marks apical organ. (A’-D’) diagrammatically illustrate expression patterns in the respective developmental stages.
Table of sequences used for alignment and phylogenetic analysis.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LH designed the study, carried out the experiments, and drafted the manuscript. SM conceived of the study; obtained funding; and participated in data analysis, writing, and critical revisions of the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Laurel S Hiebert, Email: lhiebert@uoregon.edu.
Svetlana A Maslakova, Email: svetlana@uoregon.edu.
References
- 1.Thollesson M, Norenburg JL. Ribbon worm relationships: a phylogeny of the phylum Nemertea. Proc Biol Sci. 2003;270:407–15. doi: 10.1098/rspb.2002.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maslakova SA. The invention of the pilidium larva in an otherwise perfectly good spiralian phylum Nemertea. Integr Comp Biol. 2010;50:734–43. doi: 10.1093/icb/icq096. [DOI] [PubMed] [Google Scholar]
- 3.Andrade SCS, Montenegro H, Strand M, Schwartz ML, Kajihara H, Norenburg JL, et al. A transcriptomic approach to ribbon worm systematics (Nemertea): resolving the Pilidiophora problem. Mol Biol Evol. 2014;31:3206–15. doi: 10.1093/molbev/msu253. [DOI] [PubMed] [Google Scholar]
- 4.Dawydoff C. Les formes larvaires de polyclades et de nemertes du plancton indochinois. Bull Biol Fr Belg. 1940;74:443–96. [Google Scholar]
- 5.Fewkes JW. On the developmental stages of certain worm larvae. Bull Mus Comp Zool. 1883;11:167–208. [Google Scholar]
- 6.Cantell CE. Some developmental stages of peculiar nemertean larva Pilidium recurvatum Fewkes from Gullmarfjord (Sweden) Arkiv Fur Zoologi. 1966;19:143–7. [Google Scholar]
- 7.Hiebert TC, von Dassow G, Hiebert LS, Maslakova SA. The peculiar nemertean larva pilidium recurvatum belongs to Riserius sp. a basal heteronemertean that eats Carcinonemertes errans, a hoplonemertean parasite of Dungeness crab. Invertebr Biol. 2013;132:207–25. doi: 10.1111/ivb.12023. [DOI] [Google Scholar]
- 8.Schwartz ML. Untying a Gordian Knot of Worms: Systematics and Taxonomy of the Pilidiophora (phylum Nemertea) from Multiple Data Sets. Washington, DC: The George Washington University; 2009. [Google Scholar]
- 9.Maslakova SA, von Dassow G. A non-feeding pilidium with apparent prototroch and telotroch. J Exp Zool B Mol Dev Evol. 2012;318:586–90. doi: 10.1002/jezb.22467. [DOI] [PubMed] [Google Scholar]
- 10.Maslakova SA, Hiebert TC. From trochophore to pilidium and back again - a larva’s journey. Int J Dev Biol. 2014;58:585–91. doi: 10.1387/ijdb.140090sm. [DOI] [PubMed] [Google Scholar]
- 11.Iwata F. On the development of the nemertean Micrura akkeshiensis. Embryologia. 1958;4:103–31. doi: 10.1111/j.1440-169X.1958.tb00150.x. [DOI] [Google Scholar]
- 12.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–85. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 13.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-N. [DOI] [PubMed] [Google Scholar]
- 14.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 15.de Rosa R, Prud’homme B, Balavoine G. Caudal and even-skipped in the annelid Platynereis dumerilii and the ancestry of posterior growth. Evol Dev. 2005;7:574–87. doi: 10.1111/j.1525-142X.2005.05061.x. [DOI] [PubMed] [Google Scholar]
- 16.Henry JQ, Perry KJ, Wever J, Seaver E, Martindale MQ. β-Catenin is required for the establishment of vegetal embryonic fates in the nemertean, Cerebratulus lacteus. Dev Biol. 2008;317:368–79. doi: 10.1016/j.ydbio.2008.02.042. [DOI] [PubMed] [Google Scholar]
- 17.Kmita-Cunisse M, Loosli F, Bierne J, Gehring WJ. Homeobox genes in the ribbon worm Lineus sanguineus: evolutionary implications. Proc Natl Acad Sci U S A. 1998;95:3030–5. doi: 10.1073/pnas.95.6.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loosli F, Kmita-Cunisse M, Gehring WJ. Isolation of a Pax-6 homolog from the ribbon worm Lineus sanguineus. Proc Natl Acad Sci U S A. 1996;93:2658–63. doi: 10.1073/pnas.93.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maslakova SA. Development to metamorphosis of the nemertean pilidium larva. Front Zool. 2010;7:30. doi: 10.1186/1742-9994-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharkey M, Graba Y, Scott MP. Hox genes in evolution: protein surfaces and paralog groups. Trends Genet. 1997;13:145–51. doi: 10.1016/S0168-9525(97)01096-2. [DOI] [PubMed] [Google Scholar]
- 21.de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, et al. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–6. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 22.Kourakis MJ, Martindale MQ. Combined-method phylogenetic analysis of Hox and ParaHox genes of the metazoa. J Exp Zool. 2000;288:175–91. doi: 10.1002/1097-010X(20000815)288:2<175::AID-JEZ8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.Balavoine G, de Rosa R, Adoutte A. Hox clusters and bilaterian phylogeny. Mol Phylogenet Evol. 2002;24:366–73. doi: 10.1016/S1055-7903(02)00237-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang GF, Fang XD, Guo XM, Li L, Luo RB, Xu F, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- 25.Simakov O, Marletaz F, Cho SJ, Edsinger-Gonzales E, Havlak P, Hellsten U, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493:526–31. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fröbius AC, Matus DQ, Seaver EC. Genomic organization and expression demonstrate spatial and temporal Hox gene colinearity in the lophotrochozoan Capitella sp. I PLoS One. 2008;3 doi: 10.1371/journal.pone.0004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Passamaneck YJ, Halanych KM. Evidence from Hox genes that bryozoans are lophotrochozoans. Evol Dev. 2004;6:275–81. doi: 10.1111/j.1525-142X.2004.04032.x. [DOI] [PubMed] [Google Scholar]
- 28.Kulakova M, Bakalenko N, Novikova E, Cook CE, Eliseeva E, Steinmetz PRH, et al. Hox gene expression in larval development of the polychaetes Nereis virens and Platynereis dumerilii (Annelida, Lophotrochozoa) Dev Genes Evol. 2007;217:39–54. doi: 10.1007/s00427-006-0119-y. [DOI] [PubMed] [Google Scholar]
- 29.Bakalenko NI, Novikova EL, Nesterenko AY, Kulakova MA. Hox gene expression during postlarval development of the polychaete Alitta virens. EvoDevo. 2013;4:13. doi: 10.1186/2041-9139-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui JHL, McDougall C, Monteiro AS, Holland PW, Arendt D, Balavoine G, et al. Extensive chordate and annelid macrosynteny reveals ancestral Homeobox gene organization. Mol Biol Evol. 2012;29:157–65. doi: 10.1093/molbev/msr175. [DOI] [PubMed] [Google Scholar]
- 31.Hinman VF, O’Brien EK, Richards GS, Degnan BM. Expression of anterior Hox genes during larval development of the gastropod Haliotis asinina. Evol Dev. 2003;5:508–21. doi: 10.1046/j.1525-142X.2003.03056.x. [DOI] [PubMed] [Google Scholar]
- 32.Lee PN, Callaerts P, de Couet HG, Martindale MQ. Cephalopod Hox genes and the origin of morphological novelties. Nature. 2003;424:1061–5. doi: 10.1038/nature01872. [DOI] [PubMed] [Google Scholar]
- 33.Samadi L, Steiner G. Expression of Hox genes during the larval development of the snail, Gibbula varia (L.) - further evidence of non-colinearity in molluscs. Dev Genes Evol. 2010;220:161–72. doi: 10.1007/s00427-010-0338-0. [DOI] [PubMed] [Google Scholar]
- 34.Wysocka-Diller JW, Aisemberg GO, Baumgarten M, Levine M, Macagno ER. Characterization of a homologue of bithorax-complex genes in the leech Hirudo medicinalis. Nature. 1989;341:760–3. doi: 10.1038/341760a0. [DOI] [PubMed] [Google Scholar]
- 35.Aisemberg GO, Macagno ER. Lox1, an antennapedia-class homeobox gene, is expressed during leech gangliogenesis in both transient and stable central neurons. Dev Biol. 1994;161:455–65. doi: 10.1006/dbio.1994.1044. [DOI] [PubMed] [Google Scholar]
- 36.Nardelli-Haefliger D, Bruce AEE, Shankland M. An axial domain of HOM/Hox gene expression is formed by morphogenetic alignment of independently specified cell lineages in the leech Helobdella. Development. 1994;120:1839–49. doi: 10.1242/dev.120.7.1839. [DOI] [PubMed] [Google Scholar]
- 37.Kourakis MJ, Master VA, Lokhorst DK, Nardelli-Haefliger D, Wedeen CJ, Martindale MQ, et al. Conserved anterior boundaries of Hox gene expression in the central nervous system of the leech Helobdella. Dev Biol. 1997;190:284–300. doi: 10.1006/dbio.1997.8689. [DOI] [PubMed] [Google Scholar]
- 38.Irvine SQ, Martindale MQ. Expression patterns of anterior Hox genes in the polychaete Chaetopterus: correlation with morphological boundaries. Dev Biol. 2000;217:333–51. doi: 10.1006/dbio.1999.9541. [DOI] [PubMed] [Google Scholar]
- 39.Wada H, Garcia-Fernandez J, Holland PWH. Colinear and segmental expression of amphioxus Hox genes. Dev Biol. 1999;213:131–41. doi: 10.1006/dbio.1999.9369. [DOI] [PubMed] [Google Scholar]
- 40.Seo HC, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- 41.Aronowicz J, Lowe CJ. Hox gene expression in the hemichordate Saccoglossus kowalevskii and the evolution of deuterostome nervous systems. Integr Comp Biol. 2006;46:890–901. doi: 10.1093/icb/icl045. [DOI] [PubMed] [Google Scholar]
- 42.Hejnol A, Martindale MQ. Coordinated spatial and temporal expression of Hox genes during embryogenesis in the acoel Convolutriloba longifissura. BMC Biol. 2009;7:65. doi: 10.1186/1741-7007-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreno E, Nadal M, Baguna J, Martinez P. Tracking the origins of the bilaterian Hox patterning system: insights from the acoel flatworm Symsagittifera roscoffensis. Evol Dev. 2009;11:574–81. doi: 10.1111/j.1525-142X.2009.00363.x. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Fernandez J, Holland PWH. Archetypal organization of the amphioxus Hox gene-cluster. Nature. 1994;370:563–6. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- 45.Charpignon V. Homeobox-containing genes in the nemertean Lineus: key players in the antero-posterior body patterning and in the specification of the visual structures. PhD Thesis. Faculty of Science, University of Basel. 2007.
- 46.Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121:333–46. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- 47.Wang BB, Muller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-X. [DOI] [PubMed] [Google Scholar]
- 48.Martindale MQ. The evolution of metazoan axial properties. Nat Rev Genet. 2005;6:917–27. doi: 10.1038/nrg1725. [DOI] [PubMed] [Google Scholar]
- 49.Goldstein B, Freeman G. Axis specification in animal development. Bioessays. 1997;19:105–16. doi: 10.1002/bies.950190205. [DOI] [PubMed] [Google Scholar]
- 50.Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–99. doi: 10.1046/j.1525-142X.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- 51.Arenas-Mena C, Martinez P, Cameron RA, Davidson EH. Expression of the Hox gene complex in the indirect development of a sea urchin. Proc Natl Acad Sci U S A. 1998;95:13062–7. doi: 10.1073/pnas.95.22.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arenas-Mena C, Cameron AR, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000;127:4631–43. doi: 10.1242/dev.127.21.4631. [DOI] [PubMed] [Google Scholar]
- 53.Strathmann MF. Reproduction and Development of Marine Invertebrates of the Northern Pacific Coast. Seattle, WA: University of Washington Press; 1987. [Google Scholar]
- 54.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–91. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 55.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:12. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matz MV, Alieva NO, Chenchik A, Lukyanov S. Amplification of cDNA ends using PCR suppression effect and step-out PCR. Methods Mol Biol. 2003;221:41–9. doi: 10.1385/1-59259-359-3:41. [DOI] [PubMed] [Google Scholar]
- 57.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, et al. GenBank. Nucleic Acids Res. 2013;41:D36–42. Available: html://http://ncbi.nlm.nih.gov. [DOI] [PMC free article] [PubMed]
- 59.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 60.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 61.Seaver EC, Kaneshige LM. Expression of ‘segmentation’ genes during larval and juvenile development in the polychaetes Capitella sp I and H. elegans. Dev Biol. 2006;289:179–94. doi: 10.1016/j.ydbio.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 62.Schwartz ML, Norenburg JL. Three new species of Micrura (Nemertea: Heteronemertea) and a new type of heteronemertean larva from the Caribbean Sea. Caribb J Sci. 2005;41:528–43. [Google Scholar]


