Abstract
Helicases are ubiquitous motor proteins that separate and/or rearrange nucleic acid duplexes in reactions fueled by adenosine triphosphate (ATP) hydrolysis. Helicases encoded by bacteria, viruses, and human cells are widely studied targets for new antiviral, antibiotic, and anticancer drugs. This review summarizes the biochemistry of frequently targeted helicases. These proteins include viral enzymes from herpes simplex virus, papillomaviruses, polyomaviruses, coronaviruses, the hepatitis C virus, and various flaviviruses. Bacterial targets examined include DnaB-like and RecBCD-like helicases. The human DEAD-box protein DDX3 is the cellular antiviral target discussed, and cellular anticancer drug targets discussed are the human RecQ-like helicases and eIF4A. We also review assays used for helicase inhibitor discovery and the most promising and common helicase inhibitor chemotypes, such as nucleotide analogues, polyphenyls, metal ion chelators, flavones, polycyclic aromatic polymers, coumarins, and various DNA binding pharmacophores. Also discussed are common complications encountered while searching for potent helicase inhibitors and possible solutions for these problems.
Keywords: motor protein, ATPase, RNA binding proteins, molecular probes, antivirals, antibiotic, anticancer
Helicases are tiny molecular motors fueled by adenosine triphosphate (ATP) hydrolysis that grab one strand of DNA or RNA and peel it from its complementary strand. In cells, DNA helicases play key roles in DNA replication, recombination, and repair. Cells need RNA helicases for transcription, translation, and RNA splicing. More than 10 years ago, several potent antiviral drugs were discovered that inhibit an essential herpes simplex virus (HSV) helicase complex, and this discovery inspired many others to study helicases as drug targets.1 , 2 Discovering similarly potent and specific inhibitors for other helicases has been quite challenging, but considerable progress has been made in recent years. This review discusses recent progress toward making helicases more tractable drug targets. We discuss below data recently published or deposited in the PubChem BioAssay,3 common chemical scaffolds identified as hits in high-throughput screens, how hits have been optimized, and novel new high-throughput assays. Because numerous other reviews on helicase biochemistry, helicase assays suitable for screening, and the role of helicases in biology are available,4, 5, 6, 7, 8, 9 we will only briefly review key points before discussing inhibitor development in more detail. Throughout this article, helicase inhibitors will be identified by a PubChem Compound Identification (CID) number, which can be used to access a wealth of other data for each compound by searching the CID in the PubChem Compound database.10 A PubChem Assay Identification (AID) number will also be noted for assays used to identify or characterize discussed helicase inhibitors.3
There are many reasons why helicase inhibitor development is challenging. We have encountered two basic problems in our efforts to discover hepatitis C virus (HCV) helicase inhibitors. First, high-throughput screens using assays monitoring helicase-catalyzed nucleic acid duplex separation yield few hits, and second, most of the hits act by binding the nucleic acid substrate. For example, the Scripps Research Institute Molecular Screening Center tested 290,735 compounds in the National Institutes of Health (NIH) small-molecule collection using an assay that monitors the ability of the HCV helicase to separate duplex DNA (PubChem BioAssay AID 1800).11 Only 500 compounds (0.2%) were confirmed as hits (AID 1943), the most potent hits were assay artifacts (AID 485301), and the most potent hits did not inhibit HCV RNA replication in a cell-based assay (AID 463235). Many assay artifacts, or false leads, seen in helicase assays that monitor DNA duplex separation, such as the one used for HCV helicase, result from a compound’s ability to interact with the helicase’s DNA substrate. Such DNA binding compounds are still very difficult to identify in a high-throughput format. Solutions to various helicase inhibitor development problems include extensive counterscreening, innovative assays monitoring helicases in cells or helicase interactions with other proteins, and structure-based design. As discussed below, all these efforts have led to potent, specific, and some drug-like helicase inhibitors.
The goal of this article is to discuss how new helicase inhibitors were discovered and optimized in the past few years. These small molecules target proteins linked to diverse diseases, such as viral infections, bacterial infections, premature aging, and cancer. Due to space limitations, this is not a comprehensive review of these subjects. Rather, we intend to update other helicase articles, such as Xu Guang Xi’s review about helicases as antiviral and anticancer drug targets.12 We have chosen to focus on how new helicase inhibitors were identified and optimized, common screening problems, and the chemistry of common helicase inhibitor chemotypes. Numerous other reviews are available that are focused on either viral helicases7 , 13 or cancer-linked helicases.14, 15, 16 Many excellent resources also cover helicase biochemistry in more detail, but most are focused on specific helicases such as the ones encoded by HSV,17, 18, 19 HCV,20, 21, 22, 23, 24 flaviviruses,25 or humans.16 , 26, 27, 28
Introduction to Helicase Structure and Function
Cells use helicases whenever they need to access DNA or RNA, and all life forms encode helicases.4 , 5 The only exceptions are some viruses that replicate in a cell’s nucleus, where they might hijack cellular helicases to access or copy genetic material.8 DNA helicases separate the two strands of the double helix when it is copied, repaired, or transcribed into RNA. Cells need RNA helicases for messenger RNA (mRNA) transcription, translation, and to assemble or disassemble RNA-protein complexes such as the ribosome. Viruses with RNA genomes also use helicases to resolve RNA duplexes formed after replication. In other words, helicases guard access to our genomes. As genome guardians, helicases are linked to a myriad of disorders caused by abnormal gene expression, cell proliferation, and infectious pathogen replication.19 , 27 , 29
RNA and DNA helicases are complex molecular motors that use ATP to fuel nucleic acid base pair separation and/or rearrangement.6 , 9 On the molecular level, most helicases grab one strand of DNA or RNA and move along it to displace its complement. Some helicases assemble into oligomeric rings (typically hexamers),30 , 31 which encircle one DNA (or RNA) strand,32 whereas non–ring helicases do not form rings but rather function as monomers,33 dimers,34 or higher order oligomers.35 , 36 Ring helicases consist of identical subunits, each of which contains a domain that resembles the Escherichia coli protein RecA.37 ATP binds at the interface of two RecA-like domains such that there are six ATP binding sites on a hexameric ring helicase. Sequential or concerted ATP hydrolysis causes a ring helicase to spin down a nucleic acid strand.30 Non–ring helicases38 consist of two RecA-like domains covalently linked in tandem on the same polypeptide,39 and ATP binds between these “motor domains.”40 ATP binding and hydrolysis cause a non–ring helicase to expand and contract so that the helicase moves along DNA (or RNA) like an inchworm.41, 42, 43 The above characterization likely oversimplifies how helicases function as molecular motors, and exactly how these molecular machines assemble is still a subject of considerable research and debate.
Both ring and non–ring helicases must first load on single-stranded DNA (or RNA) before they can separate a duplex. Once loaded on single-stranded DNA (or RNA), most helicases move in either one of two possible directions. Some move from the 5′-end to the 3′-end of the strand to which they are bound, and others move in a 3′ to 5′ direction.44 , 45
In addition to movement directionality and oligomeric state, helicases are also classified based on their genetic similarities. All helicase genes evolved from the same common ancestor, and helicase proteins share common signature sequences indicative of family relationships. Helicase families are then grouped into superfamilies.46 , 47 Most members of helicase superfamily 1 (SF1)48 and superfamily 2 (SF2)49 are non–ring helicases, and members of superfamily 3 (SF3) and superfamily 4 (SF4) are typically ring helicases.9 HSV and human coronaviruses (CoV)50 encode the SF1 helicases that will be discussed below. SF2 helicase drug targets to be discussed are the NS3 proteins encoded by HCV and related viruses, the cellular DEAD-box proteins,51 and human RecQ-like helicases.52 SF3 helicases discussed below include viral DNA helicases encoded by human papillomaviruses (HPVs)53 , 54 and polyomaviruses (e.g., simian virus 40 [SV40]).31 All SF4 helicases discussed below, as targets for new antibiotics, resemble the E. coli DnaB hexamer, which unwinds DNA and coordinates leading and lagging strand DNA replication.55 Many other helicases in other helicase superfamilies (i.e., Rho-like helicases in superfamily 5 and the MCM proteins in superfamily 6)9 and the related AAA+ superfamily47 could someday be important drug targets, but they will not be further discussed here because specific small molecules that inhibit them have not yet been reported in the literature.
Helicases as Drug Targets
The primary motivation to discover potent and specific helicase inhibitors is to control the ability of an organism to access genetic material. In theory, one could use helicase inhibitors to control any aspect of gene replication or expression, but the goal of most present efforts is to find helicase inhibitors that simply prevent the replication of infectious pathogens or cancer cells. Antibiotics could be developed from potent and specific inhibitors of bacterial helicases, such as the DnaB55 protein that acts at bacterial replication forks, or proteins involved in recombination, such as RecBCD.36 Inhibitors of cellular helicases could function as antivirals or be used to control cancer cells or make them more sensitive to chemotherapy.15
Bacteria-Encoded Helicases
Much of what we know about helicases comes from studies performed with proteins first purified from benign E. coli laboratory strains, such as the E. coli helicase that coordinates DNA replication, called DnaB.55 , 56 Inhibitors of E. coli helicases could be used, however, to treat pathogenic strains of E. coli, which cause more than 100 million gastrointestinal infections each year and about 170,000 deaths.57 Like E. coli, other gram-negative pathogenic bacteria, such as Pseudomonas aeruginosa, encode an SF4 DnaB-like helicase that they use to coordinate leading and lagging strand DNA replication. P. aeruginosa causes pneumonia, urinary tract infections, and sepsis.58 Gram-positive bacteria encode DnaB-like proteins that have been targeted to find treatments for Bacillus anthracis, the causative agent of anthrax, and Staphylococcus aureus. S. aureus causes many natural and hospital-acquired infections, which typically respond to current antibiotics.58 However, new S. aureus drugs are desperately needed because of the evolution of methicillin-resistant S. aureus, which is resistant to penicillin and other beta-lactam antibiotics.59
The other bacterial helicase targets discussed here are the non–ring helicases that form the multifunctional RecBCD complex.60 The RecBCD complex prepares DNA for homologous recombination and/or repair. RecB and RecD are both SF1 helicases, which move in opposite directions, on complementary strands, to drive the translocation of the RecBCD complex along DNA.61 A separate nuclease function of RecB degrades both strands of DNA as the complex translocates. Upon reaching a Chi recognition sequence, RecB stops cleaving the 3′-strand but continues to digest the 5′-strand, leaving an extended 3′ overhang of single-stranded DNA (ssDNA).62 RecA binds the 3′ overhang and helps it invade a homologous duplex to form a DNA crossover to be resolved at a Holliday junction.37 RecBCD also destroys bacteriophage DNA that lacks Chi sites, unless the virus encodes a protective protein.63 Most work with RecBCD has been done with the E. coli complex, but pathogenic bacteria, such as the ulcer causing Helicobacter pylori, encode RecBCD homologues. The H. pylori RecBCD homolog, which will be discussed later, is called AddAB.64
Virus-Encoded DNA Helicases
As noted above, only helicase inhibitor–based drugs target an HSV helicase. HSV is in the family Herpesviridae, members of which cause chickenpox/shingles (Varicella zoster), cold sores (HSV-1), and genital herpes (HSV-2). HSV-2 infects 40 to 60 million people worldwide, with 1 to 2 million cases each year, and typically spreads through infected mucosa.65 All HSV are fully enveloped double-stranded DNA viruses, which encode seven proteins needed for DNA replication: the origin binding protein UL9, the single-stranded DNA binding protein ICP8, a heterodimeric polymerase (UL30 and UL42), and a heterotrimeric helicase-primase complex (UL5, UL8, and UL52). UL52 is the primase, and UL5 is the helicase. UL8 coordinates helicase-primase activity with the help of ICP8 and assists helicase processivity. UL5 is an SF1 helicase, and it unwinds in a 5′ to 3′ direction, but only in the presence of UL52.66 , 67
Papillomaviruses and polyomaviruses encode the other viral DNA helicases that are widely studied as drug targets. Both families have smaller DNA genomes than the viruses in Herpesviridae, and their circular genomes encode few proteins other than an SF3 helicase. Host DNA polymerases and other replication proteins assemble around the viral SF3 helicase to synthesize new viral DNA in the same bidirectional semi-discontinuous manner used by most cellular organisms. The prototype polyomavirus, SV40, was accidentally introduced to humans in the polio vaccine, but it has not yet been clearly linked to any human disease. SV40 is closely related to two human polyamaviruses, which cause debilitating illnesses in immunocompromised patients. One is the BK virus, which is named after the transplant patient from whom it was first isolated, and the other is the John Cunningham virus (JCV).68 The polyomavirus SF3 helicase is the large tumor antigen (TAg), a well-conserved viral protein transcribed soon after infection.69 TAg contains an N-terminal J-domain, which stimulates the ATPase activity of the Hsp70 chaperone, a central origin–binding domain, and a C-terminal helicase domain fueled by ATP hydrolysis. Like other replicative ring helicases, TAg forms two hexamers at the origin of DNA replication that move in opposite directions as the replication bubble opens and DNA is copied.31
Members of the family Papillomaviridae have a circular genome like SV40, but the papillomavirus genome is larger and more complex. More than 120 different strains of HPV infect more than 440 million people, and they cause about 250,000 deaths per year. Most HPV strains cause genital warts, but several pathogenic variants, such as HPV16, cause cervical cancer.70 The HPV helicase is the viral E1 protein, which has both site-specific binding activity and unwinding activity. Like TAg, E1 is an SF3 ring helicase with many functions and domains: a C-terminal helicase/ATPase domain, a central origin DNA binding domain, and an N-terminal regulatory domain. The N-terminal E1 region contains a nuclear localization signal, a nuclear export signal, a conserved cyclin-binding motif, and several phosphorylation sites. E1 hexamers form at each replication fork at either end of a replication bubble. Exactly how the hexamers coordinate their activity either by interacting with each other or other proteins at the replication fork is still a subject of investigation. The internal DNA binding hairpins of each subunit form a spiral staircase, and these hairpins pull the DNA through the center of the ring one nucleotide at a time, such that six ATP molecules are used to move six nucleotides.53
RNA Helicases Needed for Virus Replication
Two classes of RNA helicases have been studied as drug targets: helicases encoded by viruses and cellular helicases needed for virus replication. The most widely studied viral helicase is the nonstructural protein 3 (NS3) encoded by HCV. NS3 is an SF2, non–ring, 3′ to 5′ helicase. HCV is the only species in the genus Hepacivirus, which is part of the family Flaviviridae.71 The genus Flavivirus, as well as its numerous important human pathogens such as yellow fever virus (YFV), Japanese encephalitis (JEV), West Nile virus, and dengue virus, is also part of Flaviviridae.25 All flaviviruses encode NS3 proteins similar in form and function to the HCV helicase.25 , 72, 73, 74 NS3 proteins are multifunctional, with the N-terminal domain functioning as a protease needed for viral polyprotein processing and the C-terminal domain functioning as a helicase. HCV and related viruses encode the only proteins known that are both proteases and helicases. NS3 proteases are active only when combined with another viral peptide. The HCV NS3 protease is active only when combined with HCV nonstructural protein 4A (NS4A),75 , 76 and NS3 from flaviviruses is activated by nonstructural protein 2B (NS2B).77 , 78 Unlike HCV NS3, flavivirus helicases possess an RNA triphosphatase activity, meaning they can cleave the terminal phosphate present at the 5′-end of RNA, to prepare the genome for capping.79 HCV genomes are not capped and HCV translation instead begins at an internal ribosome entry site (IRES).80
HCV infects about 1 in 50 people alive today, but most HCV patients are unaware of their illness because the virus destroys the liver so slowly that it causes few symptoms.81 After years of infection, most HCV patients develop fibrosis, cirrhosis, hepatocellular carcinoma, or liver failure.82 Effective HCV treatments combine pegylated human interferon, ribavirin, and an inhibitor of the NS3/NS4A protease.83 , 84 The flaviviruses are primarily transmitted to humans through mosquitoes, and they cause numerous mild to fatal diseases. Vaccines are available for both YFV and JEV, but currently there are no specific drugs available for the treatment of flavivirus diseases.85
Coronaviruses (viruses in the family Coronaviridae) are also (+)RNA viruses, but they have genomes more than three times the size of HCV. Coronaviruses typically cause upper respiratory infections (i.e., the common cold), but some species, such as severe acute respiratory syndrome coronavirus (SARS-CoV), can cause life-threatening pneumonia. Another deadly coronavirus was recently isolated from patients in Saudi Arabia.86 The SARS-CoV helicase is nonstructural protein nsp13.50 Like NS3 from flaviviruses, the SARS-CoV helicase has RNA 5′ triphosphatase activity.87
Cellular Helicases as Antiviral Targets
Viruses that enter the nucleus often hijack cellular helicases during viral replication. For example, the human cellular helicases DDX1, DDX3, DDX24, MDA-5, RNA helicase A (RHA), and Werner syndrome protein (WRN) have been linked to human immunodeficiency virus (HIV) replication. All six are SF2 helicases. RHA was the first cellular helicase linked to HIV replication88 and is encapsulated in the HIV particle.89 The melanoma differentiation-associated gene 5 (MDA-5) helicase was called “RH116” in the study linking it to HIV replication.90 A dominant negative helicase-defective WRN allele inhibits HIV long terminal repeat transactivation and HIV replication.91 DDX3 facilitates the export of HIV RNA transcripts from the nucleus into the cytoplasm.92 DDX1 restricts HIV-1 Rev function in human astrocytes,93 , 94 and DDX24 helps with HIV packaging.95 Although all the above helicases could serve as antiviral drug targets, small-molecule inhibitors have been found so far only for DDX3, as will be discussed in more detail below.
Cellular Helicases as Targets for Cancer Chemotherapy
As reviewed elsewhere in more detail,12 , 26 , 96 cells need helicases to evade the effect of drugs that kill cancer cells by damaging cancer cell DNA. For example, topoisomerase inhibitors, such as camptothecin, cause DNA strand breaks that need to be repaired by homologous recombination for a cell to survive or for a tumor cell to duplicate. Many of the human proteins needed for recombinational DNA repair resemble the E. coli helicase RecQ protein, and helicase inhibitors that target human RecQ-like helicases could, in theory, make cancer cells more sensitive to chemotherapy.97 , 98
Human RecQ-like helicases were first discovered when some of their genes were found to be linked to various autosomal recessive diseases, such as the premature aging disorder Werner syndrome.28 There are five known human RecQ-like proteins: RECQ1, WRN, BLM, RecQ4, and RecQ5. Mutations in the WRN gene (RECQL2) cause Werner syndrome,99 and mutations in the BLM gene (RECQL3) cause Bloom syndrome.100 Mutations in the RecQ4 gene (RECQL4) have been implicated in Rothmund-Thomson syndrome,101 Rapadilino syndrome,102 and Baller-Gerold syndrome.103 RecQ-like helicases play key roles in DNA replication, recombination, and repair; stabilize the replication fork; and are needed for telomere stability.52 As evidence that targeting RecQ-like helicases might aid chemotherapy, cells exposed to small hairpin RNA (shRNA) targeting WRN are more sensitive to methylselenic acid,104 and cells with mutated WRN respond more slowly to DNA damage from UV light or chemotherapeutic agents, such as the topoisomerase inhibitor camptothecin.105 RecQ5 also aids in the recovery of stalled replication forks after camptothecin treatment.97 The RecQ-like proteins are not the only helicases that could potentially serve as anticancer drug targets. Many other helicases are needed to repair genomes, and they form complex networks with other key proteins, including tumor suppressors105 and oncoproteins.106 Mutations in these networks lead to synthetic lethality, suggesting that small-molecule inhibitors of such helicases might reproduce the same phenotype.16
Another manner in which helicase inhibitors could be used to treat cancers would be to halt the activity of a helicase needed to express a specific oncogene. Protein synthesis dysregulation is associated with many human cancers, and it might result from abnormal activity of RNA helicases needed for translation. The best-studied example is the RNA helicase eIF4A, which prepares mRNA for ribosome binding by unwinding secondary structures in the 5′ untranslated region (UTR). Cells exposed to an eIF4A inhibitor show reduced expression of an oncoprotein that has been linked to breast cancer, called Mucin 1.106
Below we will discuss small molecules that inhibit the above helicases and how they were identified using high-throughput screening (HTS) techniques. Of course, other methods could be used to modulate helicase activity. For example, RNA interference is the main technique used to demonstrate that a helicase is a potential therapeutic target. RNA interference was used to show that HIV replication requires both DDX3107 and RNA helicase A,108 that human liver cells need the RNA helicase p68 to support HCV replication,109 and that suppression of the Werner helicase makes cells more sensitive to cancer chemotherapy.110, 111, 112 Viral helicases encoded as parts of polyproteins are obviously more difficult to selectively knock down using small interfering RNA (siRNA), but they have been inhibited using other biological macromolecules. For example, HCV replication has been repressed using therapeutic antibodies113 , 114 and RNA aptamers directed against the NS3 helicase.115, 116, 117
Typical High-Throughput Assays Used to Identify Helicase Inhibitors
Most high-throughput assays used to identify helicase inhibitors in compound collections either detect the ability of a helicase to separate DNA (or RNA) strands or cleave ATP, which fuels helicase movements. ATP hydrolysis (ATPase) assays are typically easier to design and execute, are less costly, and are simpler to perform in a high-throughput format. In addition, numerous commercial kits available, designed to monitor protein kinases,118 can be modified to detect helicase-catalyzed ATP hydrolysis. Strand separation (i.e., unwinding) assays require more sophisticated reagents, such as modified oligonucleotides, that are not needed in ATPase assays.
Helicase-catalyzed ATP hydrolysis is measured by monitoring either the loss of ATP or the appearance of adenosine diphosphate (ADP) or inorganic phosphate (Pi). Most Pi assays are based on the Fiske-SubbaRow method119 or more sensitive ammonium molybdate reagents that incorporate the dye malachite green.120 , 121 Colorimetric phosphate assays can be challenging to perform as screens because either ATP must be removed or multiple reagents must be added in a precisely timed procedure. Proprietary colorimetric reagents such as Biomol Green reagent (Enzo Life Sciences, Farmingdale, NY) or the CytoPhos reagent (Cytoskeleton, Inc., Denver, CO)122 are more amenable to HTS. Miyata et al.123 recently reported an interesting new variant of these classic phosphate assays that uses the dye quinaldine red. A quinaldine red–phosphate complex absorbs light where many white assay plates emit when excited at 430 nm, so that white plate fluorescence decreases when quinaldine red forms a complex with phosphate and molybdate. As discussed below, Seguin et al.124 used this quinaldine red assay to discover new inhibitors of the SV40 TAg helicase.
The alternatives to detecting Pi in an ATPase assay are to couple ATP hydrolysis to another reaction, detect ATP remaining, or detect ADP. The classic coupled ATPase assays link ATP hydrolysis to either nicotinamide adenine dinucleotide (NADH) reduction via pyruvate kinase and lactate dehydrogenase125 or methylthioguanosine (MESG) hydrolysis via purine nucleoside phosphorylase.122 , 126 Neither coupled assay is particularly useful in HTS because many small molecules absorb in the same wavelengths as NADH and MESG. There are commercial assays, however, that detect ATP and ADP through coupled luminescent reactions (e.g., ADP glo; Promega, Madison, WI) or by using ADP sensors. ADP sensors use antibodies bound to a fluorescent ADP analogue, which can be displaced by native ADP produced in a helicase-catalyzed reaction. ADP sensor assays, commonly referred to as “Transcreener” assays, can be monitored with fluorescence intensity, polarization, or time-resolved fluorescence resonance energy transfer and are available from Bellbrook Labs (Madison, WI) or Cisbio BioAssays (Marcoule, France).
With all these possibilities, and only a few published studies that directly compare various methods, choosing an ATPase-based helicase screen can be challenging. Our laboratory prefers colorimetric ATPase assays for their low cost, precision, and simplicity,127 but other laboratories prefer other techniques. For example, Seguin et al.128 compared a commercial malachite green–based kit (BioAssays System, Hayward, CA) and the ADP Hunter kit (DiscoverRX, Freemont, CA) and reported that ADP Hunter kit was more sensitive and had a higher signal/background in assays with the SV40 helicase.
Helicase unwinding assays are performed as end-point assays or as continuous assays. The prototype helicase unwinding end-point assay measures the conversion of double-stranded DNA to single-stranded DNA using an isotope-labeled oligonucleotide. Such assays can be adapted to HTS using GeneClean Glassmilk (MP Biomedicals, Santa Ana, CA) and filter plates129 or by using a scintillation proximity assay (SPA), where a radiolabeled oligonucleotide is captured with a biotin-labeled oligonucleotide, which then binds to a scintillant bead.130 Radioactive helicase end-point assays have also been done with a FlashPlate (PerkinElmer, Waltham, MA).131 , 132 Two unwinding end-point assays that do not use radioisotopes have been described. One uses electrochemiluminescence (ECL) and a substrate made by attaching a DNA nucleotide to a ruthenium chelate, which is trapped by a biotin-labeled strand and streptavidin-coated magnetic beads that are detected by ECL.133 Another uses an enzyme-linked immunosorbent assay to detect displacement of a digoxigenin (DIG)–labeled strand from a biotin-labeled strand in a streptavidin-coated well plate.134
Continuous helicase unwinding assays typically monitor changes in Förster resonance energy transfer (FRET) between “donor” and “acceptor” chromophores tethered to complementary strands of DNA (or RNA).135, 136, 137, 138 Continuous assays are simpler and often less costly than end-point assays, but they are plagued by compound interference because many library compounds absorb or emit light at wavelengths that overlap those of the fluorophores being monitored. As an attempt to minimize compound interference, similar assays have been developed that monitor either fluorescence polarization139 or time-resolved fluorescence instead of fluorescence intensity.140
Our laboratory relies mainly on FRET-based assays in which one strand of a helicase substrate is made of a molecular beacon (i.e., an oligonucleotide containing both a FRET donor and acceptor that can form a hairpin).141 In such a molecular beacon-based helicase assay (MBHA), substrate fluorescence decreases when ATP activates the helicase. An MBHA has two advantages over other FRET-based assays. First, no oligonucleotide trap is required in the reaction, making it simpler and less costly.11 Second, compounds that bind the helicase substrate can be detected because they decrease substrate fluorescence in the absence of the helicase (or ATP).24 , 142 As discussed below, DNA binding compounds are frequent, nonspecific hits in helicase assays, and identifying them early is critical for efficient helicase inhibitor development.
Proof of Concept: HSV Helicase/Primase Inhibitors
The inspiration for much of the research discussed here comes from the fact that helicase inhibitors already have been demonstrated to be potent antiviral agents that rival many of the drugs used to treat herpes infections.18 , 19 More than 10 years ago, Boehringer Ingelheim1 and Bayer2 discovered anti-HSV drugs that target the UL5/8/52 complex. Boehringer Ingelheim identified their inhibitors, typified by BILS 22BS ( Fig. 1), by screening for compounds that inhibit helicase-catalyzed DNA strand separation. These aminothiazolylphenyls inhibit primase-catalyzed RNA synthesis and helicase-catalyzed ATP hydrolysis in the presence of nucleic acids, but they do not inhibit helicase-catalyzed ATP hydrolysis in the absence of DNA. Interestingly, the BILS series stabilizes a helicase/primase:DNA complex, possibly preventing the primase recycling needed to initiate new Okazaki fragments.1
Figure 1.
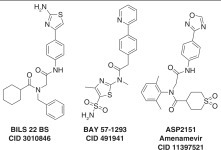
Herpes simplex virus (HSV) helicase/primase inhibitors. BILS 22 BS (CID 3010846), BAY 57-1293 (CID 491941), and ASP2151 (CID 11397521).
The Bayer compounds, in contrast, were discovered using a cell-based high-throughput cell survival assay, not intended to find helicase inhibitors per se.2 The initial hit from a screen of 420,000 compounds (BAY 38-9489) was optimized to a highly potent (IC50 = 12 nM) thiazole amide derivative called BAY 57-1293 (Fig. 1). Genetic analysis of HSV resistant to BAY 57-1293 revealed that mutations in UL5 or UL52 confer resistance to BAY 57-1293. BAY 57-1293 inhibits purified UL5/52-catalyzed ATP hydrolysis in the presence of DNA (IC50 = 30 nM). In HSV-infected guinea pigs143 and rabbits,144 BAY 57-1293 relieves symptoms and prevents viral relapse, but resistance mutations are common in both clinical and laboratory isolates of HSV-1.145 Some alleles (e.g., K356T in UL5) confer resistance to both BAY 57-1293 and BILS 22 BS. However, some HSV alleles resistant to BAY 57-1293 (e.g., A899T in UL52) are still sensitive to BILS 22 BS.146
The first helicase inhibitor to show success in the clinic is a herpes primase/helicase inhibitor called ASP2151 (amenamevir; Fig. 1). ASP2151 is an oxadiazolylphenyl-containing compound that inhibits purified UL5/8/52-catalyzed ATP hydrolysis (IC50 = 78 nM), primer synthesis (IC50 < 30 nM), and DNA unwinding (IC50 < 100 nM). ASP2151 inhibits HSV in cell culture147 and guinea pig models.148 ASP2151 is also effective against thymidine kinase–deficient HSV strains resistant to acyclovir,149 and resistance to ASP2151 is 1000 times less common than seen for acyclovir.150 When administered to patients with genital herpes, ASP2151 significantly reduces the median time for lesion healing.151 , 152
Inhibitors of Helicase-Catalyzed ATP Hydrolysis
The most obvious inhibitors of helicase-catalyzed ATP hydrolysis are nucleotide and nucleobase analogues. As reviewed previously,8 , 153 nucleotide analogues have been extensively tested as inhibitors of the NS3 helicase, but few inhibit the enzyme with IC50 values less than 50 µM. More recently, synthesized new ring expanded nucleosides (REN) were tested if they inhibited HIV-1 replication by targeting the cellular helicase DDX3. The most potent REN, CID 44586781, inhibits DDX3-catalyzed RNA unwinding, and it suppresses HIV-1 replication in T cells and macrophages.154
Other than nucleotides, the most common phamacophores explored as inhibitors of helicase-catalyzed ATP hydrolysis are polyphenols made of two or three linked phenyl rings. Biphenyls have been studied as inhibitors of SV40 TAg,124 , 128 HPV E1,155 and DDX3,156 and triphenylmethanes have been twice studied as NS3 inhibitors.157 , 158
Biphenyls were first noted as helicase inhibitors by researchers at Boehringer Ingelheim, who optimized this chemotype as a lead to treat HPV. As previously reviewed,54 , 159 Boehringer Ingelheim tested its compound collection for inhibitors of HPV E1-catalyzed ATP hydrolysis, and the most promising screening hit was a biphenysulfonacetic acid (CID 515118; Fig. 2A), which inhibited HPV6 E1-catalyzed ATP hydrolysis with an IC50 value of 2 µM (Fig. 2A).160 Compound optimization guided the discovery of CID 515164 (Fig. 2A), which inhibits HPV6 E1 500 times more potently than CID 515118. These biphenylsulfonacetic acids are reversible, but not linear competitive, HPV helicase inhibitors, suggesting they bind an allosteric site, and they inhibit E1 isolated from some strains dramatically better than E1 isolated from other HPV strains. Compound specificity results from the presence of tyrosine at position 486 in HPV E1. When another residue is present in this position, the compounds bind more weakly. For example, they are less active against HPV11 than they are against HPV6.155
Figure 2.
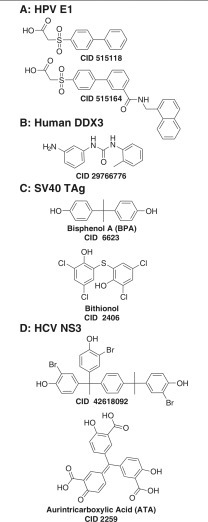
Polyphenyl helicase inhibitors. (A) Inhibitors of the human papillomavirus (HPV) E1-catalyzed adenosine triphosphate (ATP) hydrolysis: CID 515118, IC50 = 2 µM160; CID 515164, IC50 = 0.004 µM.155,160 (B) Human DDX3 inhibitors. CID 29766776, IC50 = 5 µM.156 (C) Inhibitors of simian virus 40 (SV40) TAg-catalyzed ATP hydrolysis: bisphenol A (CID 6623), IC50 = 41 µM128; bithionol (CID 2406), IC50 = 4 µM.124 (D) Hepatitis C virus (HCV) helicase inhibitors: CID 42618092, IC50 = 10 µM157; aurintricarboxylic acid (CID 2259), IC50 = 1.4 µM.158
Another polyphenyl helicase inhibitor was reported in a modeling study designed to find compounds that bind the RNA binding cleft of DDX3. Using docking, virtual screening, and tests of the ability of hits to inhibit DDX3-catalyzed DNA unwinding or ATP hydrolysis, Radi et al.156 found a potent N,N′-diarylurea (CID 29766776; Fig. 2B) DDX3 helicase inhibitor (IC50 = 5 µM), which also inhibits HIV-1 replication in cell-based assays (IC50 = 15 µM) without detectable toxicity at 100 µM. CID 29766776 notably resembles the potent antibacterial and antifungal triclocarban (CID 7547).
Compounds similar to triclocarban (CID 7547) also inhibit SV40 TAg. The Southern Research Specialized Biocontainment Screening Center tested compounds in the NIH collection for their ability to inhibit SV40 TAg-catalyzed ATP hydrolysis (AID 1909). After examining their 2153 hits in dose-response assays (AID 1903), compound interference counterscreens (AID 2501), and cytotoxicity assays (AID 2102), as well as examining the common features of the hits, the team discovered that Bisphenol A (BPA; CID 6623; Fig. 2C) inhibits TAg with an IC50 value of 41 µM. BPA is used in many plastic consumer food containers, and as an estrogen receptor agonist, it might present an environmental hazard. BPA inhibits TAg-dependent DNA replication (EC50 = 6 µM), but it is cytotoxic at similar concentrations.128 Remarkably similar chemotypes were also obtained in screens of other libraries with a different assay monitoring TAg-catalyzed ATP hydrolysis.124 For example, bithionol (CID 2406; Fig. 2C) and hexachlorophene (CID 3598), both of which are Food and Drug Administration–approved drugs, inhibit TAg.124 The bisphenol-like moiety, flexibility of the linker group, and the presence of substituents at positions 2 and 4 on the phenols are all essential features needed for this chemotype to inhibit SV40 TAg. Importantly, both bithionol and hexachlorophene inhibit SV40 and BKV cell culture, and they are less toxic than BPA.124
Unpublished experiments in our laboratory have also noted that some of the above biphenyls also inhibit the HCV helicase, but biphenyls are not as potent as triphenyl methanes known to inhibit NS3. Triphenylmethanes were first noted as NS3 inhibitors when the dye, soluble blue HT, was found to dock in the ATP binding site and inhibit NS3 in assays with an IC50 value of 40 µM. A crystal structure (PDB code 2ZJO) of blue HT bound to NS3 shows blue HT in the ATP binding site, and it has been used to design CID 42618092 (Fig. 2D), a more potent triphenylmethane that inhibits NS3 helicase and the HCV replicon.157 Mukherjee et al.158 found that a similar compound called aurintricarboxylic acid (ATA; CID 2259; Fig. 2D) is an even more effective HCV helicase inhibitor, with an IC50 value of 1.4 µM. ATA also inhibits human RECQ1-catalyzed DNA unwinding (AID 2708) and the BLM helicase (AID 2528).
Like other proteins with P-loop “Walker”-type ATP binding sites,161 magnesium forms a bridge needed for ATP to fuel helicase action.121 In theory, this bridge could be blocked by metal ion chelators, such as aryl diketoacids (ADKs). ADKs inhibit the unwinding activity of SARS-CoV helicase with IC50 values ranging from 5.4 to 13.6 µM. Dihydroxychromones, a class of naturally occurring flavonoids, are bioisosteres of ADKs with better stability and safety. Dihydroxychromones containing arylmethyl groups, catechol groups, or both inhibit SARS-CoV helicase-catalyzed ATP hydrolysis and DNA unwinding. For example, CID 45270979, which contains an arylmethyl and a catechol moiety on either side of a dihydroxychromone, inhibits SARS-CoV helicase-catalyzed DNA unwinding (IC50 = 8.1 µM) but not ATP hydrolysis. When two arylmethyl groups are on either side of the pharmacophore (e.g., CID 56929932), the compound inhibits both Nsp13-catalyzed ATP hydrolysis (IC50 = 4 µM) and DNA unwinding (IC50 = 11 µM). CID 56929932 also inhibits HCV replication in cells (EC50 = 4 µM), but its antiviral effect against SARS-CoV has not yet been reported. Similar compounds with only one arylmethyl or catechol group do not inhibit the SARS-CoV helicase.162 , 163
Inhibitors of Helicase-Catalyzed Nucleic Acid Separation
The main problem with targeting helicases through their ATP binding site is that the motor domains lining the ATP binding cleft are highly conserved.46 , 47 , 161 Helicase DNA (or RNA) binding sites are less similar, so in theory, compounds binding in place of nucleic acids might be less promiscuous. However, small molecules targeting helicase nucleic acid binding sites have been hard to discover. To find compounds that directly target unwinding, most teams have focused on compounds that inhibit helicase-catalyzed unwinding but do not inhibit helicase-catalyzed ATP hydrolysis.23 One problem with this approach is that a vast majority of compounds that inhibit unwinding do so by interacting with the nucleic acid substrate, not the enzyme itself. Examples include ethidium bromide, actinomycin D, 4′,6′-diamidino-2-phenylindole (DAPI), daunorubicin, distamycin, ellipticine, mitoxantrone, nalidixic acid, or netropsin, many of which have been studied as helicase inhibitors since the first studies were done with herpes UL9164 and the human RecQ-like proteins.165
Many DNA binding pharmacophores, such as anthracyclines, acridones, tropolones, and amidinoanthracyclines, have been optimized as HCV helicase inhibitors, and these have been reviewed elsewhere.21 , 23 The inhibitory effects of optimized acridones and tropolones on HCV helicase have been recently confirmed in our laboratory.142 Fluoroquinolone antibiotics, which also bind nucleic acids, have also been studied as inhibitors of SV40 TAg166 and the HCV helicase.167
Flavones comprise another pharmacophore with nucleic acid binding capacity that has been frequently seen in screens for helicase inhibitors. For example, myricetin (CID 5281672) and related flavones, such as luteolin and morin, all inhibit the hexameric replicative helicases, and myricetin inhibits gram-negative bacteria growth, with a minimal inhibitory concentration (MIC) as low as 0.25 mg/mL.168 Myricetin (CID 5281672) and scutellarein (CID 5281697) also inhibit SARS-CoV helicase with IC50 values of 2.7 µM and 0.9 µM, respectively.50 , 169 However, myricetin is also a potent inhibitor of numerous DNA and RNA polymerases and telomerases,170 likely due to nonspecific interactions with DNA or nucleic acid binding proteins.
Although some of the discussion above suggests that helicases function nonspecifically on any duplex structure, many helicases are known to act mainly on specific sequences or secondary structures such as hairpins, G-quadruplexes, or Holliday junctions.171 It might be possible, therefore, to use small molecules that bind certain sequences or mimic DNA structures to target specific helicases needed in a disease pathway. For example, porphyrins that mimic a G-quadruplex inhibit the RecQ helicase,172 and similar bismuth porphyrin complexes inhibit the SARS helicase.173
Optimization of helicase inhibitors that bind nucleic acids is challenging because of the lack of HTS assays capable of detecting small molecule–DNA interactions. Most groups have relied on assays that monitor the ability of a small molecule to decrease the fluorescence of DNA stained with a fluorescent intercalator (e.g., ethidium bromide174 or thiazole orange175). Such fluorescent intercalator displacement (FID) assays, however, do not detect all compounds that interact with DNA. For example, the Scripps Research Institute Molecular Screening Center tested 290,731 compounds in the NIH small-molecule collection and found 487 hits (AID 1845), but later Li et al.142 found that several of the compounds that did not test positive in this ethidium bromide–based FID did, in fact, bind DNA. Li et al. therefore developed a different DNA binding assay using SYBR green I, which can detect the interaction of a wider range of compounds with DNA, but there is still no guarantee that all DNA binding compounds will affect the fluorescence of a SYBR green I–stained DNA. In our laboratory, we therefore use an MBHA11 to simultaneously detect compounds that bind DNA and inhibit helicase action.24
Polycyclic Aromatic Polymers as NS3 Inhibitors
Li et al.142 used the MBHA to design specific NS3 inhibitors from polycyclic aromatic polymers purified from the yellow dye primuline. The basic scaffold of the primuline derivatives is similar to the symmetrical benzothiazole polymers (e.g., (BIP)2B, CID 247520; Fig. 3) that were first noted to inhibit HCV helicase by ViroPharma,176, 177, 178 except that they are made from benzothiazoles (rather than benzimidazoles) that are linked head to tail rather than head to head. The symmetrical benzimidazoles inhibit HCV helicase by binding in place of RNA,177 but many retain an ability to interact with nucleic acids,178 so they are rather promiscuous, inhibiting NS3 from flaviviruses, and human DDX3.177
Figure 3.
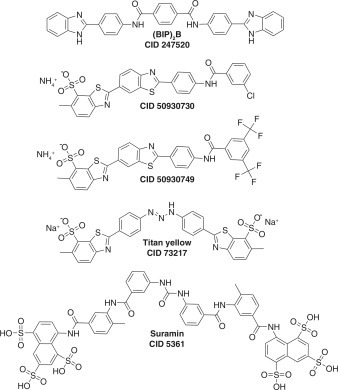
Polcyclic polymers that inhibit the hepatitis C virus (HCV) NS3 helicase. (BIP)2B, CID 247520, IC50 = 5 µM177; CID 50930730, IC50 = 2 µM142; CID 50930749, IC50 = 15 µM127; titan yellow (CID 73217), IC50 = 12 µM; suramin (CID 5361), IC50 = 4 µM.158
Li et al.142 chose to study primuline because Belon and Frick179 found a related dye called thioflavine S to be a hit in a small screen.179 To better understand how the dyes exert their action, Li et al. purified their active components and found that thioflavine S is composed of two major components. The related dye primuline is composed of two major and four minor components, all of which are 1- to 4-unit-long benzothiazole oligomers terminating with a p-aminobenzene group.142 Their potency in helicase assays correlates with the length of the benzothiazole chain. All are reversible helicase inhibitors, and they inhibit NS3h by preventing the protein from binding single-stranded DNA or RNA.158 They also have three undesirable properties. First, they bind nucleic acids, albeit with a lower affinity than with which they inhibit NS3h. Second, they displace unrelated proteins from single-stranded DNA or RNA. Third, they inhibit the NS3 protease even in the absence of the helicase domain, suggesting a nonspecific interaction with NS3.127 These properties were minimized through synthetic diversification. All derivatives were tested for their ability to inhibit HCV helicase, to bind DNA, and to displace E. coli single-stranded DNA binding protein from an oligonucleotide. The most potent and specific compounds were tested for their ability to either inhibit NS3-catalyzed ATP hydrolysis or peptide cleavage.127 , 180 The rationale for this extensive counterscreening effort was that benzothiazoles such as those found in primuline could be promiscuous, acting nonspecifically. The most potent and specific helicase inhibitor synthesized is a 3-Cl benzoyl analogue synthesized from the primuline dimer (CID 50930730; Fig. 3).180 CID 50930730 inhibits NS3 helicase but does not interact with DNA, affect the SSB-DNA interaction, or potently inhibit the NS3 ATPase or NS3 protease.
To understand if primuline derivatives reach their target in cells, Ndjomou et al.127 exploited the fact that most retain fluorescent properties similar to primuline.181 Most of the primuline derivatives absorb light near 360 nm and emit light near 500 nm. The new compounds stain live cells harboring subgenomic HCV replicons, and some derivatives decrease the amount of HCV RNA present in a hepatoma cell line with an enhanced ability to harbor HCV replicons. The primuline derivative that is the most potent HCV antiviral is CID 50930749 (Fig. 3).127
Mukherjee et al.158 used a DNA binding assay to find other compounds that also prevent HCV from loading on DNA. The two most effective compounds they found were titan yellow (CID 73217) and the polysulfonated naphthalene suramin (CID 5361) (Fig. 3), which inhibit HCV helicase with IC50s of 12 µM and 4 µM, respectively. Both suramin and titan yellow are not specific like the optimized primuline derivatives. Suramin and titan yellow also prevent the E. coli single-stranded DNA binding protein from binding to DNA158; suramin inhibits the activity of human eIF4A182 and human RecQ-like proteins (AID 2549), and it prevents the RNA-induced silencing complex from loading on RNA.183
Antibacterial Agents Targeting DnaB
Several groups have searched large compound collections for inhibitors of the bacterial replicative helicase, DnaB. For example, McKay et al.184 tested more than 230,000 compounds and found a series of triaminotriazines that inhibit P. aeruginosa DnaB-catalyzed DNA unwinding (AID 261721). The compounds do not prevent gram-negative bacterial cell growth and are cytotoxic toward HeLa cells, but the most potent, CID 4041506 ( Fig. 4A), inhibits the growth of gram-positive bacteria, such as S. aureus, with an MIC of 4 µg/mL.
Figure 4.
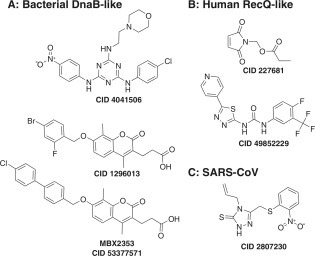
New inhibitors of helicase-catalyzed DNA unwinding. (A) Inhibitors targeting bacterial DnaB-like helicases. CID 4041506, IC50 = 5 µM184; CID 1296013, IC50 = 12 µM185; CID 53377571, IC50 = 1 µM. (B) Inhibitors targeting human RecQ-like helicase. CID 227681, IC50 (WRN) = 20 µM187; CID 49852229, IC50 (BLM) = 5 µM (AID 504662). (C) Inhibitor targeting severe acute respiratory syndrome coronavirus (SARS-CoV) helicase. CID 2807230, IC50 = 5.7 µM.93
In a more recent study, Aiello et al.185 tested 78,588 compounds from the Microbiotix (MBX) library for their ability to inhibit B. anthracis DnaB, as well as 108,026 compounds in the National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infectious Disease (NSRB) collection for the ability to inhibit DNA unwinding catalyzed by S. aureus DnaC. The ICCB-Longwood/NSRB Screening Facility (Harvard Medical School) has deposited results for assays performed with the S. aureus helicase in the PubChem BioAsay (AID 485395). The new DnaB inhibitors discovered in these campaigns include coumarins (5 compounds), benzothiazoles (2 compounds), rhodanines (4 compounds), triazines (2 compounds), N-phenylpyrroles (2 compounds), and three not easily classified compounds. The most promising DnaB inhibitor in this set is an aminocoumarin (CID 1296013; Fig. 4A) that inhibits growth of a variety of gram-positive bacteria (MIC 5 µg/mL).185 This coumarin scaffold has since been further optimized, and the optimized compound incorporates a biphenyl moiety reminiscent of the phamacophore seen in SV40 and HPV inhibitors (CID 53377571; Fig. 4A).186
New Inhibitors of Human RecQ-like Helicases
Several high-throughput screens using unwinding assays have recently focused on finding inhibitors of human RecQ-like proteins, and as with the DnaB assays noted above, much of the data are available on the PubChem BioAssay.
Aggarwal et al.187 first showed that RecQ-like helicase inhibitors could be valuable molecular probes when they characterized the effects of NSC19630 (CID 227681; Fig. 4B), which was identified as a potent WRN inhibitor in a screen of 2000 compounds from the National Cancer Institute Diversity Set. CID 227681 inhibits WRN helicase but does not affect the activity of related RECQ1, E. coli RecQ, and DnaB under the same conditions. CID 227681 does not appear to interact with DNA in FID assays, but when it is administered to cells, it induces double-stranded DNA (dsDNA) breaks, apoptosis, replication fork stalling, and mitotic checkpoint control, and it delays the cells in S-phase. These NSC19630-induced cellular phenotypes have all been shown to be WRN dependent, suggesting a “dominant negative” mechanism of action.
More extensive screens have been performed with the WRN (AID 651767), RECQ1 (AID 2549), and BLM (AID 2528) helicases, and data are available in the PubChem BioAssay. The NIH Chemical Genomics Center performed a quantitative high-throughput screen to measure IC50 values for more than 250,000 compounds in assays with each of the three RecQ-like helicases. Hits in these screens include compounds such as those discussed above, including triphenylmethanes, biphenyls, DNA binding compounds, suramin, and anthracenediones. This project led to the development of a potent, selective BLM inhibitor that became NIH molecular probe ML216 (CID 49852229; Fig. 4B). ML216 inhibits BLM with an IC50 of 0.97 µM and WRN with an IC50 value of 12 µM, but the compound has no effect against the related RECQ1 helicase. ML216 (CID 49852229) treatment sensitizes cells to aphidicolin, and ML216 inhibits the proliferation of only cells that express BLM.188
New Inhibitors of the SARS-CoV Helicase
Another recent example of a new helicase inhibitor discovered using an unwinding assay-based high-throughput screen was identified from the Maybridge Hitfinder chemical library and is an inhibitor of the SARS-CoV helicase, CID 2807230 (Fig. 4C). CID 2807230 blocks the ability of nsp13 to unwind double-stranded RNA (IC50 = 5.7 µM) and dsDNA (IC50 = 5.30 µM) but not the ATPase activity. CID 2807230 inhibits the SARS-CoV replication in cells without apparent toxicity. CID 2807230 also inhibits the WRN helicase (AID 651768), but it does not inhibit HCV helicase, Dengue helicase, Moloney murine leukemia virus reverse transcriptase, or the E. coli DNA polymerase I, Klenow fragment polymerase.189
Disrupting Helicase Interactions with Key Partners
The best example of an antiviral drug that disrupts a critical helicase interaction affects the binding of HPV E1 to the HPV E2 protein, which helps load the E1 helicase on the HPV origin of replication. E2 is a DNA binding protein that regulates viral gene transcription, and E2 helps segregate the HPV genome when host cells divide. The impressive development of E1-E2 interaction inhibitors has been recently reviewed.54 , 159 Briefly, Boehringer Ingelheim first found compounds that prevent inhibitors of E1-E2 binding with an SPA using purified E2, E1, and radiolabeled HPV DNA. Since E2 binds DNA very tightly, most inhibitors reduce the signal in this assay by disrupting the E1-E2 interaction.54 Structure-based design and further chemical optimization led to CHEMBL1207308 ( Fig. 5A), which is still one of the most potent small-molecule helicase inhibitors (IC50 = 6 nM). Lower molecular weight, less complex E1-E2 interaction inhibitors were discovered using a radiolabeled CHEMBL1207308 analogue and an SPA to screen an expanded compound library.54 An intriguing hit in the later screen was a racemic mixture with a structure similar to repaglinide, a type 2 diabetes drug.190 Chemical optimization led to CID 11330698 (Fig. 5A), which is a potent inhibitor of the E1-E2 interaction (IC50 = 20 nM). Unfortunately, CID 11330698 is rapidly metabolized and lowers glucose levels by 6% when administered to rats at 1 mg/kg.54
Figure 5.
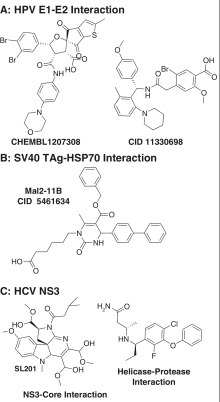
Compounds that disrupt the interactions of helicases with other proteins. (A) Inhibitors for the human papillomavirus (HPV) E1-E2 interaction. CHEMBL1207308, IC50 = 0.006 µM206; CID 11330698, IC50 = 0.02 µM.54 (B) Inhibitor of the simian virus 40 (SV40) TAg-Hsp70 interaction. CID 5461634, IC50 = 20 µM.191 (C) Compounds targeting the interaction of hepatitis C virus (HCV) helicase with the core protein (IC50 = 15 µM)192 and the NS3 protease (IC50 = 0.1 µM).193
The interaction of the similar SF3 helicase from SV40 with a key partner has also been exploited to find antiviral compounds. The J-domain of SV40 TAg stimulates the ATPase of Hsp70, an important heat shock protein that helps protect cells from virus-induced stress. By testing compound ability to inhibit TAg J-domain–stimulated Hsp70-catalyzed ATP hydrolysis, Wright et al.191 found that MAL2-11B (CID 5461634; Fig. 5B) inhibits TAg-stimulated Hsp70 ATPase activity, endogenous TAg ATPase activity, and SV40 replication in plaque assays by 4.5-fold when tested at 100 µM. MAL2-11B also inhibits BK virus DNA replication in human kidney cells by 90% when the cells are treated with 15 µM MAL2-11B.
Small molecules have also been reported that disrupt the interaction of the NS3 helicase with the NS3 protease and an HCV structural protein called “core” (Fig. 5C). HCV core is a highly basic protein that helps pack the viral RNA genome in the virus capsid. Mousseau et al.192 designed an AlphaScreen to detect the interaction between the NS3 helicase and HCV core, and they used it to show that core peptides and an indoline alkaloid-type compound (called SL201; Fig. 5C) disrupt the core–NS3 helicase interaction. SL201 also prevents core from forming dimers, suggesting that core dimers must form in order for core to bind NS3. SL201 inhibits HCV virus production in cell culture.
More recently, new compounds were found that bind the interface between the NS3 helicase and protease, so that they lock the protein in a “closed” conformation where peptides cannot access the NS3 protease active site. The compounds, like the one shown in PDB file 4B75 (Fig. 5C), are potent protease inhibitors (IC50 = 0.1 µM) and inhibit HCV replication (EC50 = 0.4 µM) without apparent toxicity in cell culture.193 The effects of these new allosteric NS3 inhibitors on NS3-catalyzed RNA unwinding or ATP hydrolysis have not been reported. However, the compounds have no effect on NS3 lacking its helicase domain.
Targeting Biological Functions of Helicases
Helicase inhibitors identified using biochemical assays often do not exert biological effects because they fail to enter cells, or they are unstable if they successfully enter cells. To find helicase inhibitors with better pharmacological properties, several groups have designed assays that depend on active helicase. An in vitro translation assay and one example of a cell-based helicase assay suitable for HTS are discussed below. They have been used successfully to find eIF4A and RecBCD inhibitors.
Since eIF4A is needed for cap-dependent translation, eIF4A inhibitors often block translation of capped RNA but not translation initiated from an IRES, like the one used to initiate HCV polyprotein synthesis. Inhibitors of either IRES-mediated or cap-dependent translation can therefore be identified using bicistronic reporter vectors, such as one where the cap-dependent reading frame encodes firefly luciferase, and the IRES-expressed reading frame encodes renilla luciferase. Both enzymes in this system produce light but with different substrates, so they can be monitored simultaneously in the same assay. Using this system, Novac et al.182 screened more than 90,000 compounds, identifying many known translation inhibitors and nucleic acid binding ligands, as well as helicase inhibitors that do not interact with RNA, such as suramin (Fig. 3C). Bordeleau et al.194 used the same assay to screen marine extracts for natural products that inhibit translation and found that hippuristanol (CID 9981822; Fig. 6A) selectively inhibits eIF4A’s ability to bind RNA. Hippuristanol binds to amino acids near two conserved motifs in the C-terminal domain of eIF4A.195 Another translation inhibitor, pateamine A (CID 10053416; Fig. 6A), stimulates the rate at which eIF4A cleaves ATP by enhancing the protein’s affinity for RNA.196 Pateamine A induces eIF4A dimerization, thereby forcing eIF4A to engage in RNA binding and preventing it from participating in ribosome recruitment needed for translation.197 Another eIF4A-dependent translation inhibitor, silvestrol (CID 21301152; Fig. 6A), enhances mouse lymphoma sensitivity to chemotherapy198 and blocks translation of the MUC1-C oncoprotein.106
Figure 6.
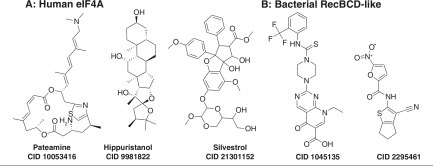
Inhibitors found using assays monitoring the biological function of helicases. (A) Inhibitors of eIF4A-dependent translation: pateamine (CID 10053416),196 hippuristanol (CID 9981822),194 and silvestrol (CID 21301152).198 (B) Inhibitors of RecBCD-dependent bacteriophage defense. CID 1045135, IC50 = 2.5 µM; CID 2295461, IC50 = 16 µM.199
RecBCD also has a biological role that can be exploited to identify inhibitors of the helicase complex. RecBCD prevents phages lacking Chi sites in their DNA from infecting E. coli. Amundsen et al.199 exploited the ability of RecBCD to protect against phage infection in a clever assay to find RecBCD inhibitors in 326,100 compounds in the NIH collection (AID 449731). They used T4 phage lacking the protective gene 2 protein, which caps DNA to prevent RecBCD binding. Compounds inhibiting RecBCD therefore allow phage to lyse E. coli. To find possible drugs to treat H. pylori, the same system was used except that the host lacked the E. coli RecBCD gene and instead contained the H. pylori RecBCD homolog, called AddAB (AID 435030), present. As a counterscreen, they added compounds to E. coli without phage, to identify compounds that simply act by killing cells (AID 449728). The most active and specific new RecBCD inhibitors include nine nitrofurans, one cyanothiophene, one modified pyrimidopyridone, and one nitrothiazole. Two of the most potent of these RecBCD inhibitors are CID 1045135 and CID 2295461 (Fig. 6B). In helicase assays, both compounds specifically inhibit RecBCD helicase, and CID 1045135 also inhibits the RecBCD nuclease.
Future Directions
Despite the recent progress and numerous newly reported helicase inhibitors with promising properties, only a few highly potent and specific helicase inhibitors have been developed, most of which target the HSV and HPV helicases. More work clearly needs to be done before helicase inhibitors become a common drug class. Standard in vitro helicase assays still yield few hits and are confounded by compounds that act nonspecifically or that simply make DNA more difficult to separate. Use of the molecular beacon-based helicase assays is helpful for identifying inhibitors that exert their effects by interacting with nucleic acids. However, improved methods to identify DNA binding agents in a high-throughput format are still needed.
High-resolution structures of the best compounds described above bound to their targets would also speed their development and the design of more potent and specific inhibitors. Co-structures of a helicase-bound inhibitor guided the design of a few of the above compounds, notably those that bind the HCV NS3 ATP binding site (PDB 2ZJO),157 the NS3 protease-helicase interface (PDB 2B75),193 and HPV E2 (PDB 1R6N).200 Similar co-structures with other compounds highlighted above should be possible to obtain, too, because many of the proteins discussed above have already been crystallized and high-resolution models are already available.
Even though few co-structures exist today, there is still a wealth of structural information for most of the targets discussed here, and these data are underused in many drug discovery programs. Available structures could be used for virtual screening, docking, or compound optimization. Some of this work has been done already, but due to space limitations, we have not discussed it here in much detail. Examples of helicase inhibitors discovered through molecular modeling include compounds targeting the nucleic acid binding site,201 flavivirus NS3,202 and the human DDX3 ATP binding site.203 , 204 Similar work with other targets might prove fruitful.
More studies also need to be done to examine the biological implication of helicase inhibition and whether these compounds are reaching their desired targets in cells. Monitoring a helicase in a cell is a difficult problem to solve, but it is not impossible, as has been demonstrated with cell-based RecBCD assays.199 Single-molecule enzymology205 might help in this regard, and it might also open up new frontiers for monitoring helicases either in cells or in cell-free extracts, which could be useful in screening, target identification, or compound optimization. Regardless, a multidisciplinary approach that combines novel in vitro and cell-based screening methods, structural biology, and rational design will be needed to design new antibiotics, antivirals, and anticancer drugs that function by targeting DNA or RNA helicases.
Acknowledgments
We thank Frank Schoenen and Jennifer Golden (University of Kansas) for helpful advice while preparing this review.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the NIH (RO1 AI088001) and a grant from the UWM research foundation (RGI 101X219).
References
- 1.Crute J.J., Grygon C.A., Hargrave K.D., Simoneau B., Faucher A.M., Bolger G., Kibler P., Liuzzi M., Cordingley M.G. Herpes Simplex Virus Helicase-Primase Inhibitors Are Active in Animal Models of Human Disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 2.Kleymann G., Fischer R., Betz U.A., Hendrix M., Bender W., Schneider U., Handke G., Eckenberg P., Hewlett G., Pevzner V., et al. New Helicase-Primase Inhibitors as Drug Candidates for the Treatment of Herpes Simplex Disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Zhou Z., Han L., Karapetyan K., Dracheva S., Shoemaker B.A., et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delagoutte E., von Hippel P.H. Helicase Mechanisms and the Coupling of Helicases within Macromolecular Machines. Part I: Structures and Properties of Isolated Helicases. Q. Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 5.Delagoutte E., von Hippel P.H. Helicase Mechanisms and the Coupling of Helicases within Macromolecular Machines. Part II: Integration of Helicases into Cellular Processes. Q. Rev. Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- 6.Pyle A.M. Translocation and Unwinding Mechanisms of RNA and DNA Helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 7.Kwong A.D., Rao B.G., Jeang K.T. Viral and Cellular RNA Helicases as Antiviral Targets. Nat. Rev. Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frick D.N., Lam A.M. Understanding Helicases as a Means of Virus Control. Curr. Pharm. Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 10.Bolton E.E., Wang Y., Thiessen P.A., Bryant S.H. PubChem: Integrated Platform of Small Molecules and Biological Activities. Annu. Rep. Comp. Chem. 2008;4:217–241. [Google Scholar]
- 11.Belon C.A., Frick D.N. Monitoring Helicase Activity with Molecular Beacons. BioTechniques. 2008;45:433–440. doi: 10.2144/000112834. 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi X.G. Helicases as Antiviral and Anticancer Drug Targets. Curr. Med. Chem. 2007;14:883–915. doi: 10.2174/092986707780362998. [DOI] [PubMed] [Google Scholar]
- 13.Frick D.N. Helicases as Antiviral Drug Targets. Drug News Perspect. 2003;16:355–362. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Brabant A.J., Stan R., Ellis N.A. DNA Helicases, Genomic Instability, and Human Genetic Disease. Annu. Rev. Genomics Hum. Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R., Brosh R.M.J. Helicases as Prospective Targets for Anti-Cancer Therapy. Anticancer Agents Med. Chem. 2008;8:390–401. doi: 10.2174/187152008784220339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal M., Brosh R.M.J. Hitting the Bull’s Eye: Novel Directed Cancer Therapy through Helicase-Targeted Synthetic Lethality. J. Cell. Biochem. 2009;106:758–763. doi: 10.1002/jcb.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleymann G. New Antiviral Drugs That Target Herpesvirus Helicase Primase Enzymes. Herpes. 2003;10:46–52. [PubMed] [Google Scholar]
- 18.Kleymann G. Helicase Primase: Targeting the Achilles Heel of Herpes Simplex Viruses. Antivir. Chem. Chemother. 2004;15:135–140. doi: 10.1177/095632020401500303. [DOI] [PubMed] [Google Scholar]
- 19.Field H.J., Mickleburgh I. The Helicase-Primase Complex as a Target for Effective Herpesvirus Antivirals. Adv. Exp. Med. Biol. 2013;767:145–159. doi: 10.1007/978-1-4614-5037-5_7. [DOI] [PubMed] [Google Scholar]
- 20.Bretner M., Najda A., Podwinska R., Baier A., Paruch K., Lipniacki A., Piasek A., Borowski P., Kulikowski T. Inhibitors of the NTPase/Helicases of Hepatitis C and Related Flaviviridae Viruses. Acta Pol. Pharm. 2004;61(Suppl):26–28. [PubMed] [Google Scholar]
- 21.Frick D.N. The Hepatitis C Virus NS3 Protein: A Model RNA Helicase and Potential Drug Target. Curr. Issues Mol. Biol. 2007;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- 22.Belon C.A., Frick D.N. Helicase Inhibitors as Specifically Targeted Antiviral Therapy for Hepatitis C. Future Virol. 2009;4:277–293. doi: 10.2217/fvl.09.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belon C.A., Frick D.N. In: Hepatitis C: Antiviral Drug Discovery and Development. He Y., Tan S.L., editors. Caister Academic; Norfolk, UK: 2011. NS3 Helicase Inhibitors; pp. 237–256. [Google Scholar]
- 24.Hanson A.M., Hernandez J.J., Shadrick W.R., Frick D.N. Identification and Analysis of Inhibitors Targeting the Hepatitis C Virus NS3 Helicase. Methods Enzymol. 2012;511:463–483. doi: 10.1016/B978-0-12-396546-2.00021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lescar J., Luo D., Xu T., Sampath A., Lim S.P., Canard B., Vasudevan S.G. Towards the Design of Antiviral Inhibitors against Flaviviruses: The Case for the Multifunctional NS3 Protein from Dengue Virus as a Target. Antiviral Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Chu W.K., Hickson I.D. RecQ Helicases: Multifunctional Genome Caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 27.Suhasini A.N., Brosh R.M.J. DNA Helicases Associated with Genetic Instability, Cancer, and Aging. Adv. Exp. Med. Biol. 2013;767:123–144. doi: 10.1007/978-1-4614-5037-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suhasini, A. N., Brosh, R. M. J. Disease-Causing Missense Mutations in Human DNA Helicase Disorders. Mutat. Res., in press. [DOI] [PMC free article] [PubMed]
- 29.Larsen N.B., Hickson I.D. RecQ Helicases: Conserved Guardians of Genomic Integrity. Adv. Exp. Med. Biol. 2013;767:161–184. doi: 10.1007/978-1-4614-5037-5_8. [DOI] [PubMed] [Google Scholar]
- 30.Singleton M.R., Sawaya M.R., Ellenberger T., Wigley D.B. Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 31.Li D., Zhao R., Lilyestrom W., Gai D., Zhang R., DeCaprio J.A., Fanning E., Jochimiak A., Szakonyi G., Chen X.S. Structure of the Replicative Helicase of the Oncoprotein SV40 Large Tumour Antigen. Nature. 2003;423:512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 32.Skordalakes E., Berger J.M. Structural Insights into RNA-Dependent Ring Closure and ATPase Activation by the Rho Termination Factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 33.Eoff R.L., Raney K.D. Intermediates Revealed in the Kinetic Mechanism for DNA Unwinding by a Monomeric Helicase. Nat. Struct. Mol. Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- 34.Wong I., Chao K.L., Bujalowski W., Lohman T.M. DNA-Induced Dimerization of the Escherichia coli rep Helicase: Allosteric Effects of Single-Stranded and Duplex DNA. J. Biol. Chem. 1992;267:7596–7610. [PubMed] [Google Scholar]
- 35.Sikora B., Chen Y., Lichti C.F., Harrison M.K., Jennings T.A., Tang Y., Tackett A.J., Jordan J.B., Sakon J., Cameron C.E., et al. Hepatitis C Virus NS3 Helicase Forms Oligomeric Structures That Exhibit Optimal DNA Unwinding Activity In Vitro. J. Biol. Chem. 2008;283:11516–11525. doi: 10.1074/jbc.M708125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B. Crystal Structure of RecBCD Enzyme Reveals a Machine for Processing DNA Breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 37.Story R.M., Weber I.T., Steitz T.A. The Structure of the E. coli recA Protein Monomer and Polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 38.Lohman T.M., Tomko E.J., Wu C.G. Non-hexameric DNA Helicases and Translocases: Mechanisms and Regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 39.Yao N., Hesson T., Cable M., Hong Z., Kwong A.D., Le H.V., Weber P.C. Structure of the Hepatitis C Virus RNA Helicase Domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 40.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal Structures of Complexes of PcrA DNA Helicase with a DNA Substrate Indicate an Inchworm Mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 41.Soultanas P., Wigley D.B. DNA Helicases: ‘Inching Forward’. Curr. Opin. Struct. Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 42.Gu M., Rice C.M. Three Conformational Snapshots of the Hepatitis C Virus NS3 Helicase Reveal a Ratchet Translocation Mechanism. Proc. Natl. Acad. Sci. U. S. A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Appleby T.C., Anderson R., Fedorova O., Pyle A.M., Wang R., Liu X., Brendza K.M., Somoza J.R. Visualizing ATP-Dependent RNA Translocation by the NS3 Helicase from HCV. J. Mol. Biol. 2011;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris P.D., Raney K.D. DNA Helicases Displace Streptavidin from Biotin-Labeled Oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 45.Fischer C.J., Maluf N.K., Lohman T.M. Mechanism of ATP-Dependent Translocation of E.coli UvrD Monomers along Single-Stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Gorbalenya A.E., Koonin E.V. Helicases: Amino Acid Sequence Comparisons and Structure-Function Relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 47.Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary History and Higher Order Classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Gilhooly N.S., Gwynn E.J., Dillingham M.S. Superfamily 1 Helicases. Front. Biosci. 2013;5:206–216. doi: 10.2741/s367. [DOI] [PubMed] [Google Scholar]
- 49.Byrd A.K., Raney K.D. Superfamily 2 helicases. Front. Biosci. 2012;17:2070–2088. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keum Y.S., Jeong Y.J. Development of Chemical Inhibitors of the SARS Coronavirus: Viral Helicase as a Potential Target. Biochem. Pharmacol. 2012;84:1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henn A., Bradley M.J., De La Cruz E.M. ATP Utilization and RNA Conformational Rearrangement by DEAD-box Proteins. Annu. Rev. Biophys. 2012;41:247–267. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett R.J., Keck J.L. Structure and Function of RecQ DNA Helicases. Crit. Rev. Biochem. Mol. Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 53.Enemark E.J., Joshua-Tor L. Mechanism of DNA Translocation in a Replicative Hexameric Helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 54.White P.W., Faucher A.M., Goudreau N. Small Molecule Inhibitors of the Human Papillomavirus E1-E2 Interaction. Curr. Top. Microbiol. Immunol. 2011;348:61–88. doi: 10.1007/82_2010_92. [DOI] [PubMed] [Google Scholar]
- 55.Itsathitphaisarn O., Wing R.A., Eliason W.K., Wang J., Steitz T.A. The Hexameric Helicase DnaB Adopts a Nonplanar Conformation during Translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajendran S., Jezewska M.J., Bujalowski W. Multiple-Step Kinetic Mechanism of DNA-Independent ATP Binding and Hydrolysis by Escherichia coli Replicative Helicase DnaB Protein: Quantitative Analysis Using the Rapid Quench-Flow Method. J. Mol. Biol. 2000;303:773–795. doi: 10.1006/jmbi.2000.4124. [DOI] [PubMed] [Google Scholar]
- 57.Kaper J.B., Nataro J.P., Mobley H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 58.Bereket W., Hemalatha K., Getenet B., Wondwossen T., Solomon A., Zeynudin A., Kannan S. Update on Bacterial Nosocomial Infections. Eur. Rev. Med. Pharmacol. Sci. 2012;16:1039–1044. [PubMed] [Google Scholar]
- 59.Enright M.C., Robinson D.A., Randle G., Feil E.J., Grundmann H., Spratt B.G. The Evolutionary History of Methicillin-Resistant Staphylococcus aureus (MRSA) Proc. Natl. Acad. Sci. U. S. A. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eggleston A.K., Rahim N.A., Kowalczykowski S.C. A Helicase Assay Based on the Displacement of Fluorescent, Nucleic Acid–Binding Ligands. Nucleic Acids Res. 1996;24:1179–1186. doi: 10.1093/nar/24.7.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dillingham M.S., Spies M., Kowalczykowski S.C. RecBCD Enzyme Is a Bipolar DNA Helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 62.Anderson D.G., Kowalczykowski S.C. The Translocating RecBCD Enzyme Stimulates Recombination by Directing RecA Protein onto ssDNA in a Chi-Regulated Manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 63.Court R., Cook N., Saikrishnan K., Wigley D. The Crystal Structure of Lambda-Gam Protein Suggests a Model for RecBCD Inhibition. J. Mol. Biol. 2007;371:25–33. doi: 10.1016/j.jmb.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 64.Cromie G.A. Phylogenetic Ubiquity and Shuffling of the Bacterial RecBCD and AddAB Recombination Complexes. J. Bacteriol. 2009;191:5076–5084. doi: 10.1128/JB.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cernik C., Gallina K., Brodell R.T. The Treatment of Herpes Simplex Infections: An Evidence-Based Review. Arch. Intern. Med. 2008;168:1137–1144. doi: 10.1001/archinte.168.11.1137. [DOI] [PubMed] [Google Scholar]
- 66.Marintcheva B., Weller S.K. A Tale of Two HSV-1 Helicases: Roles of Phage and Animal Virus Helicases in DNA Replication and Recombination. Prog. Nucleic Acid Res. Mol. Biol. 2001;70:77–118. doi: 10.1016/s0079-6603(01)70014-1. [DOI] [PubMed] [Google Scholar]
- 67.Muylaert I., Tang K.W., Elias P. Replication and Recombination of Herpes Simplex Virus DNA. J. Biol. Chem. 2011;286:15619–15624. doi: 10.1074/jbc.R111.233981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hartman E.A., Huang D. Update on PML: Lessons from the HIV Uninfected and New Insights in Pathogenesis and Treatment. Curr. HIV/AIDS Rep. 2008;5:112–119. doi: 10.1007/s11904-008-0018-0. [DOI] [PubMed] [Google Scholar]
- 69.Fanning E., Knippers R. Structure and Function of Simian Virus 40 Large Tumor Antigen. Annu. Rev. Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 70.zur Hausen H. Papillomaviruses and Cancer: From Basic Studies to Clinical Application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 71.Chène P. NS3 Helicases as Drug Targets. RNA. 2009;2:2–2.6. [Google Scholar]
- 72.Wu J., Bera A.K., Kuhn R.J., Smith J.L. Structure of the Flavivirus Helicase: Implications for Catalytic Activity, Protein Interactions, and Proteolytic Processing. J. Virol. 2005;79:10268–10277. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mastrangelo E., Milani M., Bollati M., Selisko B., Peyrane F., Pandini V., Sorrentino G., Canard B., Konarev P.V., Svergun D.I., et al. Crystal Structure and Activity of Kunjin Virus NS3 Helicase; Protease and Helicase Domain Assembly in the Full Length NS3 Protein. J. Mol. Biol. 2007;372:444–455. doi: 10.1016/j.jmb.2007.06.055. [DOI] [PubMed] [Google Scholar]
- 74.De Colibus L., Speroni S., Coutard B., Forrester N.L., Gould E., Canard B., Mattevi A. Purification and Crystallization of Kokobera Virus Helicase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:193–195. doi: 10.1107/S1744309107005283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. NS3 is a Serine Protease Required for Processing of Hepatitis C Virus Polyprotein. J. Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao N., Reichert P., Taremi S.S., Prosise W.W., Weber P.C. Molecular Views of Viral Polyprotein Processing Revealed by the Crystal Structure of the Hepatitis C Virus Bifunctional Protease-Helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 77.Luo D., Xu T., Hunke C., Gruber G., Vasudevan S.G., Lescar J. Crystal Structure of the NS3 Protease-Helicase from Dengue Virus. J. Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Assenberg R., Mastrangelo E., Walter T.S., Verma A., Milani M., Owens R.J., Stuart D.I., Grimes J.M., Mancini E.J. Crystal Structure of a Novel Conformational State of the Flavivirus NS3 Protein: Implications for Polyprotein Processing and Viral Replication. J. Virol. 2009;83:12895–12906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wengler G., Wengler G. The NS 3 Nonstructural Protein of Flaviviruses Contains an RNA Triphosphatase Activity. Virology. 1993;197:265–273. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 80.Spahn C.M., Kieft J.S., Grassucci R.A., Penczek P.A., Zhou K., Doudna J.A., Frank J. Hepatitis C Virus IRES RNA-Induced Changes in the Conformation of the 40S Ribosomal Subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 81.Edlin B.R. Perspective: Test and Treat This Silent Killer. Nature. 2011;474:S18–S19. doi: 10.1038/474S18a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McHutchison J.G. Understanding Hepatitis C. Am. J. Manag. Care. 2004;10:S21–S29. [PubMed] [Google Scholar]
- 83.Lange C.M., Sarrazin C., Zeuzem S. Review Article: Specifically Targeted Anti-viral Therapy for Hepatitis C—A New Era in Therapy. Aliment Pharmacol. Ther. 2010;32:14–28. doi: 10.1111/j.1365-2036.2010.04317.x. [DOI] [PubMed] [Google Scholar]
- 84.Garber K. Hepatitis C: Move Over Interferon. Nat. Biotechnol. 2011;29:963–966. doi: 10.1038/nbt.2031. [DOI] [PubMed] [Google Scholar]
- 85.Mackenzie J.S., Gubler D.J., Petersen L.R. Emerging Flaviviruses: The Spread and Resurgence of Japanese Encephalitis, West Nile and Dengue Viruses. Nat. Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 86.Al-Ahdal M.N., Al-Qahtani A.A., Rubino S. Coronavirus Respiratory Illness in Saudi Arabia. J. Infect. Dev. Ctries. 2012;6:692–694. doi: 10.3855/jidc.3084. [DOI] [PubMed] [Google Scholar]
- 87.Ivanov K.A., Ziebuhr J. Human Coronavirus 229E Nonstructural Protein 13: Characterization of Duplex-Unwinding, Nucleoside Triphosphatase, and RNA 5′-Triphosphatase Activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reddy T.R., Tang H., Xu W., Wong-Staal F. Sam68, RNA Helicase A and Tap Cooperate in the Post-transcriptional Regulation of Human Immunodeficiency Virus and Type D Retroviral mRNA. Oncogene. 2000;19:3570–3575. doi: 10.1038/sj.onc.1203676. [DOI] [PubMed] [Google Scholar]
- 89.Roy B.B., Hu J., Guo X., Russell R.S., Guo F., Kleiman L., Liang C. Association of RNA Helicase A with Human Immunodeficiency Virus Type 1 Particles. J. Biol. Chem. 2006;281:12625–12635. doi: 10.1074/jbc.M510596200. [DOI] [PubMed] [Google Scholar]
- 90.Cocude C., Truong M.J., Billaut-Mulot O., Delsart V., Darcissac E., Capron A., Mouton Y., Bahr G.M. A Novel Cellular RNA Helicase, RH116, Differentially Regulates Cell Growth, Programmed Cell Death and Human Immunodeficiency Virus Type 1 Replication. J. Gen. Virol. 2003;84:3215–3225. doi: 10.1099/vir.0.19300-0. [DOI] [PubMed] [Google Scholar]
- 91.Sharma A., Awasthi S., Harrod C.K., Matlock E.F., Khan S., Xu L., Chan S., Yang H., Thammavaram C.K., Rasor R.A., et al. The Werner Syndrome Helicase Is a Cofactor for HIV-1 Long Terminal Repeat Transactivation and Retroviral Replication. J. Biol. Chem. 2007;282:12048–12057. doi: 10.1074/jbc.M608104200. [DOI] [PubMed] [Google Scholar]
- 92.Yedavalli V.S., Neuveut C., Chi Y.H., Kleiman L., Jeang K.T. Requirement of DDX3 DEAD Box RNA Helicase for HIV-1 Rev-RRE Export Function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 93.Fang J., Kubota S., Yang B., Zhou N., Zhang H., Godbout R., Pomerantz R.J. A DEAD Box Protein Facilitates HIV-1 Replication as a Cellular Co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 94.Fang J., Acheampong E., Dave R., Wang F., Mukhtar M., Pomerantz R.J. The RNA Helicase DDX1 Is Involved in Restricted HIV-1 Rev Function in Human Astrocytes. Virology. 2005;336:299–307. doi: 10.1016/j.virol.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 95.Ma J., Rong L., Zhou Y., Roy B.B., Lu J., Abrahamyan L., Mouland A.J., Pan Q., Liang C. The Requirement of the DEAD-Box Protein DDX24 for the Packaging of Human Immunodeficiency Virus Type 1 RNA. Virology. 2008;375:253–264. doi: 10.1016/j.virol.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 96.Sharma S., Doherty K.M., Brosh R.M.J. DNA Helicases as Targets for Anti-Cancer Drugs. Curr. Med. Chem. Anticancer Agents. 2005;5:183–199. doi: 10.2174/1568011053765985. [DOI] [PubMed] [Google Scholar]
- 97.Hu Y., Lu X., Zhou G., Barnes E.L., Luo G. Recql5 Plays an Important Role in DNA Replication and Cell Survival after Camptothecin Treatment. Mol. Biol. Cell. 2009;20:114–123. doi: 10.1091/mbc.E08-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu X., Lou H., Luo G. A Blm-Recql5 Partnership in Replication Stress Response. J. Mol. Cell Biol. 2011;3:31–38. doi: 10.1093/jmcb/mjq056. [DOI] [PubMed] [Google Scholar]
- 99.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional Cloning of the Werner’s Syndrome Gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 100.Gray M.D., Shen J.C., Kamath-Loeb A.S., Blank A., Sopher B.L., Martin G.M., Oshima J., Loeb L.A. The Werner Syndrome Protein Is a DNA Helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 101.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 Cause a Subset of Cases of Rothmund-Thomson Syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 102.Kellermayer R., Siitonen H.A., Hadzsiev K., Kestila M., Kosztolanyi G. A Patient with Rothmund-Thomson Syndrome and All Features of RAPADILINO. Arch. Dermatol. 2005;141:617–620. doi: 10.1001/archderm.141.5.617. [DOI] [PubMed] [Google Scholar]
- 103.Van Maldergem L., Siitonen H.A., Jalkh N., Chouery E., De Roy M., Delague V., Muenke M., Jabs E.W., Cai J., Wang L.L., et al. Revisiting the Craniosynostosis-Radial Ray Hypoplasia Association: Baller-Gerold Syndrome Caused by Mutations in the RECQL4 Gene. J. Med. Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng W.H., Wu R.T., Wu M., Rocourt C.R., Carrillo J.A., Song J., Bohr C.T., Tzeng T.J. Targeting Werner Syndrome Protein Sensitizes U-2 OS Osteosarcoma Cells to Selenium-Induced DNA Damage Response and Necrotic Death. Biochem. Biophys. Res. Commun. 2012;420:24–28. doi: 10.1016/j.bbrc.2012.02.104. [DOI] [PubMed] [Google Scholar]
- 105.Blander G., Zalle N., Leal J.F., Bar-Or R.L., Yu C.E., Oren M. The Werner Syndrome Protein Contributes to Induction of p53 by DNA Damage. FASEB J. 2000;14:2138–2140. doi: 10.1096/fj.00-0171fje. [DOI] [PubMed] [Google Scholar]
- 106.Jin, C., Rajabi, H., Rodrigo, C. M., Porco, J. A. J., Kufe, D. Targeting the eIF4A RNA Helicase Blocks Translation of the MUC1-C Oncoprotein. Oncogene, in press. [DOI] [PMC free article] [PubMed]
- 107.Ishaq M., Hu J., Wu X., Fu Q., Yang Y., Liu Q., Guo D. Knockdown of Cellular RNA Helicase DDX3 by Short Hairpin RNAs Suppresses HIV-1 Viral Replication without Inducing Apoptosis. Mol. Biotechnol. 2008;39:231–238. doi: 10.1007/s12033-008-9040-0. [DOI] [PubMed] [Google Scholar]
- 108.Bolinger C., Sharma A., Singh D., Yu L., Boris-Lawrie K. RNA Helicase A Modulates Translation of HIV-1 and Infectivity of Progeny Virions. Nucleic Acids Res. 2010;38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goh P.Y., Tan Y.J., Lim S.P., Tan Y.H., Lim S.G., Fuller-Pace F., Hong W. Cellular RNA Helicase p68 Relocalization and Interaction with the Hepatitis C Virus (HCV) NS5B Protein and the Potential Role of p68 in HCV RNA Replication. J. Virol. 2004;78:5288–5298. doi: 10.1128/JVI.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grandori C., Wu K.J., Fernandez P., Ngouenet C., Grim J., Clurman B.E., Moser M.J., Oshima J., Russell D.W., Swisshelm K., et al. Werner Syndrome Protein Limits MYC-Induced Cellular Senescence. Genes Dev. 2003;17:1569–1574. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Futami K., Takagi M., Shimamoto A., Sugimoto M., Furuichi Y. Increased Chemotherapeutic Activity of Camptothecin in Cancer Cells by siRNA-Induced Silencing of WRN Helicase. Biol. Pharm. Bull. 2007;30:1958–1961. doi: 10.1248/bpb.30.1958. [DOI] [PubMed] [Google Scholar]
- 112.Masuda K., Banno K., Yanokura M., Tsuji K., Kobayashi Y., Kisu I., Ueki A., Yamagami W., Nomura H., Tominaga E., et al. Association of Epigenetic Inactivation of the WRN Gene with Anticancer Drug Sensitivity in Cervical Cancer Cells. Oncol. Rep. 2012;28:1146–1152. doi: 10.3892/or.2012.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Artsaenko O., Tessmann K., Sack M., Haussinger D., Heintges T. Abrogation of Hepatitis C Virus NS3 Helicase Enzymatic Activity by Recombinant Human Antibodies. J. Gen. Virol. 2003;84:2323–2332. doi: 10.1099/vir.0.19299-0. [DOI] [PubMed] [Google Scholar]
- 114.Prabhu R., Khalap N., Burioni R., Clementi M., Garry R.F., Dash S. Inhibition of Hepatitis C Virus Nonstructural Protein, Helicase Activity, and Viral Replication by a Recombinant Human Antibody Clone. Am. J. Pathol. 2004;165:1163–1173. doi: 10.1016/S0002-9440(10)63377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hwang B., Cho J.S., Yeo H.J., Kim J.H., Chung K.M., Han K., Jang S.K., Lee S.W. Isolation of Specific and High-Affinity RNA Aptamers against NS3 Helicase Domain of Hepatitis C Virus. RNA. 2004;10:1277–1290. doi: 10.1261/rna.7100904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Umehara T., Fukuda K., Nishikawa F., Kohara M., Hasegawa T., Nishikawa S. Rational Design of Dual-Functional Aptamers That Inhibit the Protease and Helicase Activities of HCV NS3. J. Biochem. 2005;137:339–347. doi: 10.1093/jb/mvi042. [DOI] [PubMed] [Google Scholar]
- 117.Fukuda K., Umehara T., Sekiya S., Kunio K., Hasegawa T., Nishikawa S. An RNA Ligand Inhibits Hepatitis C Virus NS3 Protease and Helicase Activities. Biochem. Biophys. Res. Commun. 2004;325:670–675. doi: 10.1016/j.bbrc.2004.10.089. [DOI] [PubMed] [Google Scholar]
- 118.Koresawa M., Okabe T. High-Throughput Screening with Quantitation of ATP Consumption: A Universal Non-Radioisotope, Homogeneous Assay for Protein Kinase. Assay Drug Dev. Technol. 2004;2:153–160. doi: 10.1089/154065804323056495. [DOI] [PubMed] [Google Scholar]
- 119.Simoni R.D., Hill R.L., Vaughan M. The Determination of Phosphorus and the Discovery of Phosphocreatine and ATP: The Work of Fiske and SubbaRow. J. Biol. Chem. 2002;277:21e. [PubMed] [Google Scholar]
- 120.Lanzetta P.A., Alvarez L.J., Reinach P.S., Candia O.A. An Improved Assay for Nanomole Amounts of Inorganic Phosphate. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 121.Frick D.N., Banik S., Rypma R.S. Role of Divalent Metal Cations in ATP Hydrolysis Catalyzed by the Hepatitis C Virus NS3 Helicase: Magnesium Provides a Bridge for ATP to Fuel Unwinding. J. Mol. Biol. 2007;365:1017–1032. doi: 10.1016/j.jmb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Funk C.J., Davis A.S., Hopkins J.A., Middleton K.M. Development of High-Throughput Screens for Discovery of Kinesin Adenosine Triphosphatase Modulators. Anal. Biochem. 2004;329:68–76. doi: 10.1016/j.ab.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 123.Miyata Y., Chang L., Bainor A., McQuade T.J., Walczak C.P., Zhang Y., Larsen M.J., Kirchhoff P., Gestwicki J.E. High-Throughput Screen for Escherichia coli Heat Shock Protein 70 (Hsp70/DnaK): ATPase Assay in Low Volume by Exploiting Energy Transfer. J. Biomol. Screen. 2010;15:1211–1219. doi: 10.1177/1087057110380571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Seguin S.P., Ireland A.W., Gupta T., Wright C.M., Miyata Y., Wipf P., Pipas J.M., Gestwicki J.E., Brodsky J.L. A Screen for Modulators of Large T Antigen’s ATPase Activity Uncovers Novel Inhibitors of Simian Virus 40 and BK Virus Replication. Antiviral Res. 2012;96:70–81. doi: 10.1016/j.antiviral.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suzich J.A., Tamura J.K., Palmer-Hill F., Warrener P., Grakoui A., Rice C.M., Feinstone S.M., Collett M.S. Hepatitis C Virus NS3 Protein Polynucleotide-Stimulated Nucleoside Triphosphatase and Comparison with the Related Pestivirus and Flavivirus Enzymes. J. Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rieger C.E., Lee J., Turnbull J.L. A Continuous Spectrophotometric Assay for Aspartate Transcarbamylase and ATPases. Anal. Biochem. 1997;246:86–95. doi: 10.1006/abio.1996.9962. [DOI] [PubMed] [Google Scholar]
- 127.Ndjomou J., Kolli R., Mukherjee S., Shadrick W.R., Hanson A.M., Sweeney N.L., Bartczak D., Li K., Frankowski K.J., Schoenen F.J., et al. Fluorescent Primuline Derivatives Inhibit Hepatitis C Virus NS3-Catalyzed RNA Unwinding, Peptide Hydrolysis and Viral Replicase Formation. Antiviral Res. 2012;96:245–255. doi: 10.1016/j.antiviral.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Seguin S.P., Evans C.W., Nebane-Akah M., McKellip S., Ananthan S., Tower N.A., Sosa M., Rasmussen L., White E.L., Maki B.E., et al. High-Throughput Screening Identifies a Bisphenol Inhibitor of SV40 Large T Antigen ATPase Activity. J. Biomol. Screen. 2012;17:194–203. doi: 10.1177/1087057111421630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sivaraja M., Giordano H., Peterson M.G. High-Throughput Screening Assay for Helicase Enzymes. Anal. Biochem. 1998;265:22–27. doi: 10.1006/abio.1998.2875. [DOI] [PubMed] [Google Scholar]
- 130.Kyono K., Miyashiro M., Taguchi I. Detection of Hepatitis C Virus Helicase Activity Using the Scintillation Proximity Assay System. Anal. Biochem. 1998;257:120–126. doi: 10.1006/abio.1998.2560. [DOI] [PubMed] [Google Scholar]
- 131.Earnshaw D.L., Pope A.J. FlashPlate Scintillation Proximity Assays for Characterization and Screening of DNA Polymerase, Primase, and Helicase Activities. J Biomol. Screen. 2001;6:39–46. doi: 10.1177/108705710100600106. [DOI] [PubMed] [Google Scholar]
- 132.Hicham Alaoui-Ismaili M., Gervais C., Brunette S., Gouin G., Hamel M., Rando R.F., Bedard J. A Novel High Throughput Screening Assay for HCV NS3 Helicase Activity. Antiviral Res. 2000;46:181–193. doi: 10.1016/s0166-3542(00)00085-1. [DOI] [PubMed] [Google Scholar]
- 133.Zhang L., Schwartz G., O’Donnell M., Harrison R.K. Development of a Novel Helicase Assay Using Electrochemiluminescence. Anal. Biochem. 2001;293:31–37. doi: 10.1006/abio.2001.5093. [DOI] [PubMed] [Google Scholar]
- 134.Hsu C.C., Hwang L.H., Huang Y.W., Chi W.K., Chu Y.D., Chen D.S. An ELISA for RNA Helicase Activity: Application as an Assay of the NS3 Helicase of Hepatitis C Virus. Biochem. Biophys. Res. Commun. 1998;253:594–599. doi: 10.1006/bbrc.1998.9813. [DOI] [PubMed] [Google Scholar]
- 135.Houston P., Kodadek T. Spectrophotometric Assay for Enzyme-Mediated Unwinding of Double-Stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5471–5474. doi: 10.1073/pnas.91.12.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bjornson K.P., Amaratunga M., Moore K.J., Lohman T.M. Single-Turnover Kinetics of Helicase-Catalyzed DNA Unwinding Monitored Continuously by Fluorescence Energy Transfer. Biochemistry. 1994;33:14306–14316. doi: 10.1021/bi00251a044. [DOI] [PubMed] [Google Scholar]
- 137.Boguszewska-Chachulska A.M., Krawczyk M., Stankiewicz A., Gozdek A., Haenni A.L., Strokovskaya L. Direct Fluorometric Measurement of Hepatitis C Virus Helicase Activity. FEBS Lett. 2004;567:253–258. doi: 10.1016/j.febslet.2004.04.072. [DOI] [PubMed] [Google Scholar]
- 138.Tani H., Fujita O., Furuta A., Matsuda Y., Miyata R., Akimitsu N., Tanaka J., Tsuneda S., Sekiguchi Y., Noda N. Real-Time Monitoring of RNA Helicase Activity Using Fluorescence Resonance Energy Transfer In Vitro. Biochem. Biophys. Res. Commun. 2010;393:131–136. doi: 10.1016/j.bbrc.2010.01.100. [DOI] [PubMed] [Google Scholar]
- 139.Xu H.Q., Zhang A.H., Auclair C., Xi X.G. Simultaneously Monitoring DNA Binding and Helicase-Catalyzed DNA Unwinding by Fluorescence Polarization. Nucleic Acids Res. 2003;31:e70. doi: 10.1093/nar/gng070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Earnshaw D.L., Moore K.J., Greenwood C.J., Djaballah H., Jurewicz A.J., Murray K.J., Pope A.J. Time-Resolved Fluorescence Energy Transfer DNA Helicase Assays for High Throughput Screening. J. Biomol. Screen. 1999;4:239–248. doi: 10.1177/108705719900400505. [DOI] [PubMed] [Google Scholar]
- 141.Tyagi S., Kramer F.R. Molecular Beacons: Probes That Fluoresce upon Hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 142.Li K., Frankowski K.J., Belon C.A., Neuenswander B., Ndjomou J., Hanson A.M., Shanahan M.A., Schoenen F.J., Blagg B.S., Aube J., et al. Optimization of Potent Hepatitis C Virus NS3 Helicase Inhibitors Isolated from the Yellow Dyes Thioflavine S and Primuline. J. Med. Chem. 2012;55:3319–3330. doi: 10.1021/jm300021v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Baumeister J., Fischer R., Eckenberg P., Henninger K., Ruebsamen-Waigmann H., Kleymann G. Superior Efficacy of Helicase-Primase Inhibitor BAY 57-1293 for Herpes Infection and Latency in the Guinea Pig Model of Human Genital Herpes Disease. Antivir. Chem. Chemother. 2007;18:35–48. doi: 10.1177/095632020701800104. [DOI] [PubMed] [Google Scholar]
- 144.Kaufman H.E., Varnell E.D., Gebhardt B.M., Thompson H.W., Atwal E., Rubsamen-Waigmann H., Kleymann G. Efficacy of a Helicase-Primase Inhibitor in Animal Models of Ocular Herpes Simplex Virus Type 1 Infection. J. Ocul. Pharmacol. Ther. 2008;24:34–42. doi: 10.1089/jop.2007.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Biswas S., Smith C., Field H.J. Detection of HSV-1 Variants Highly Resistant to the Helicase-Primase Inhibitor BAY 57-1293 at High Frequency in 2 of 10 Recent Clinical Isolates of HSV-1. J. Antimicrob. Chemother. 2007;60:274–279. doi: 10.1093/jac/dkm182. [DOI] [PubMed] [Google Scholar]
- 146.Biswas S., Kleymann G., Swift M., Tiley L.S., Lyall J., Aguirre-Hernandez J., Field H.J. A Single Drug-Resistance Mutation in HSV-1 UL52 Primase Points to a Difference between Two Helicase-Primase Inhibitors in Their Mode of Interaction with the Antiviral Target. J. Antimicrob. Chemother. 2008;61:1044–1047. doi: 10.1093/jac/dkn057. [DOI] [PubMed] [Google Scholar]
- 147.Chono K., Katsumata K., Kontani T., Kobayashi M., Sudo K., Yokota T., Konno K., Shimizu Y., Suzuki H. ASP2151, a Novel Helicase-Primase Inhibitor, Possesses Antiviral Activity against Varicella-Zoster Virus and Herpes Simplex Virus Types 1 and 2. J. Antimicrob. Chemother. 2010;65:1733–1741. doi: 10.1093/jac/dkq198. [DOI] [PubMed] [Google Scholar]
- 148.Katsumata K., Chono K., Sudo K., Shimizu Y., Kontani T., Suzuki H. Effect of ASP2151, a Herpesvirus Helicase-Primase Inhibitor, in a Guinea Pig Model of Genital Herpes. Molecules. 2011;16:7210–7223. doi: 10.3390/molecules16097210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Himaki T., Masui Y., Chono K., Daikoku T., Takemoto M., Haixia B., Okuda T., Suzuki H., Shiraki K. Efficacy of ASP2151, a Helicase-Primase Inhibitor, against Thymidine Kinase-Deficient Herpes Simplex Virus Type 2 Infection In Vitro and In Vivo. Antiviral Res. 2012;93:301–304. doi: 10.1016/j.antiviral.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 150.Chono K., Katsumata K., Kontani T., Shiraki K., Suzuki H. Characterization of Virus Strains Resistant to the Herpes Virus Helicase-Primase Inhibitor ASP2151 (Amenamevir) Biochem. Pharmacol. 2012;84:459–467. doi: 10.1016/j.bcp.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 151.Tyring S., Wald A., Zadeikis N., Dhadda S., Takenouchi K., Rorig R. ASP2151 for the Treatment of Genital Herpes: A Randomized, Double-Blind, Placebo- and Valacyclovir-Controlled, Dose-Finding Study. J. Infect. Dis. 2012;205:1100–1110. doi: 10.1093/infdis/jis019. [DOI] [PubMed] [Google Scholar]
- 152.Katsumata K., Weinberg A., Chono K., Takakura S., Kontani T., Suzuki H. Susceptibility of Herpes Simplex Virus Isolated from Genital Herpes Lesions to ASP2151, a Novel Helicase-Primase Inhibitor. Antimicrob. Agents Chemother. 2012;56:3587–3591. doi: 10.1128/AAC.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Borowski P., Mueller O., Niebuhr A., Kalitzky M., Hwang L.H., Schmitz H., Siwecka M.A., Kulikowsk T. ATP-Binding Domain of NTPase/Helicase as a Target for Hepatitis C Antiviral Therapy. Acta Biochim. Pol. 2000;47:173–180. [PubMed] [Google Scholar]
- 154.Yedavalli V.S., Zhang N., Cai H., Zhang P., Starost M.F., Hosmane R.S., Jeang K.T. Ring Expanded Nucleoside Analogues Inhibit RNA Helicase and Intracellular Human Immunodeficiency Virus Type 1 Replication. J. Med. Chem. 2008;51:5043–5051. doi: 10.1021/jm800332m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.White P.W., Faucher A.M., Massariol M.J., Welchner E., Rancourt J., Cartier M., Archambault J. Biphenylsulfonacetic Acid Inhibitors of the Human Papillomavirus Type 6 E1 Helicase Inhibit ATP Hydrolysis by an Allosteric Mechanism Involving Tyrosine 486. Antimicrob. Agents Chemother. 2005;49:4834–4842. doi: 10.1128/AAC.49.12.4834-4842.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Radi M., Falchi F., Garbelli A., Samuele A., Bernardo V., Paolucci S., Baldanti F., Schenone S., Manetti F., Maga G., et al. Discovery of the First Small Molecule Inhibitor of Human DDX3 Specifically Designed to Target the RNA Binding Site: Towards the Next Generation HIV-1 Inhibitors. Bioorg. Med. Chem. Lett. 2012;22:2094–2098. doi: 10.1016/j.bmcl.2011.12.135. [DOI] [PubMed] [Google Scholar]
- 157.Chen C.S., Chiou C.T., Chen G.S., Chen S.C., Hu C.Y., Chi W.K., Chu Y.D., Hwang L.H., Chen P.J., Chen D.S., et al. Structure-Based Discovery of Triphenylmethane Derivatives as Inhibitors of Hepatitis C Virus Helicase. J. Med. Chem. 2009;52:2716–2723. doi: 10.1021/jm8011905. [DOI] [PubMed] [Google Scholar]
- 158.Mukherjee S., Hanson A.M., Shadrick W.R., Ndjomou J., Sweeney N.L., Hernandez J.J., Bartczak D., Li K., Frankowski K.J., Heck J.A., et al. Identification and Analysis of Hepatitis C Virus NS3 Helicase Inhibitors Using Nucleic Acid Binding Assays. Nucleic Acids Res. 2012;40:8607–8621. doi: 10.1093/nar/gks623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.D’Abramo C.M., Archambault J. Small Molecule Inhibitors of Human Papillomavirus Protein-Protein Interactions. Open Virol. J. 2011;5:80–95. doi: 10.2174/1874357901105010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Faucher A.M., White P.W., Brochu C., Grand-Maitre C., Rancourt J., Fazal G. Discovery of Small-Molecule Inhibitors of the ATPase Activity of Human Papillomavirus E1 Helicase. J. Med. Chem. 2004;47:18–21. doi: 10.1021/jm034206x. [DOI] [PubMed] [Google Scholar]
- 161.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly Related Sequences in the Alpha- and Beta-Subunits of ATP Synthase, Myosin, Kinases and Other ATP-Requiring Enzymes and a Common Nucleotide Binding Fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee C., Lee J.M., Lee N.R., Kim D.E., Jeong Y.J., Chong Y. Investigation of the Pharmacophore Space of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) NTPase/Helicase by Dihydroxychromone Derivatives. Bioorg. Med. Chem. Lett. 2009;19:4538–4541. doi: 10.1016/j.bmcl.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kim M.K., Yu M.S., Park H.R., Kim K.B., Lee C., Cho S.Y., Kang J., Yoon H., Kim D.E., Choo H., et al. 2,6-Bis-arylmethyloxy-5-hydroxychromones with Antiviral Activity against Both Hepatitis C Virus (HCV) and SARS-Associated Coronavirus (SCV) Eur. J. Med. Chem. 2011;46:5698–5704. doi: 10.1016/j.ejmech.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kwok Y., Zhang W., Schroth G.P., Liang C.H., Alexi N., Bruice T.W. Allosteric Interaction of Minor Groove Binding Ligands with UL9-DNA Complexes. Biochemistry. 2001;40:12628–12638. doi: 10.1021/bi0109865. [DOI] [PubMed] [Google Scholar]
- 165.Brosh R.M.J., Karow J.K., White E.J., Shaw N.D., Hickson I.D., Bohr V.A. Potent Inhibition of Werner and Bloom Helicases by DNA Minor Groove Binding Drugs. Nucleic Acids Res. 2000;28:2420–2430. doi: 10.1093/nar/28.12.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ali S.H., Chandraker A., DeCaprio J.A. Inhibition of Simian Virus 40 Large T Antigen Helicase Activity by Fluoroquinolones. Antivir. Ther. 2007;12:1–6. [PubMed] [Google Scholar]
- 167.Khan I.A., Siddiqui S., Rehmani S., Kazmi S.U., Ali S.H. Fluoroquinolones Inhibit HCV by Targeting Its Helicase. Antivir. Ther. 2012;17:467–476. doi: 10.3851/IMP1937. [DOI] [PubMed] [Google Scholar]
- 168.Xu H., Ziegelin G., Schroder W., Frank J., Ayora S., Alonso J.C., Lanka E., Saenger W. Flavones Inhibit the Hexameric Replicative Helicase RepA. Nucleic Acids Res. 2001;29:5058–5066. doi: 10.1093/nar/29.24.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Keum Y.S., Jeong Y.J. Identification of Myricetin and Scutellarein as Novel Chemical Inhibitors of the SARS Coronavirus Helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Griep M.A., Blood S., Larson M.A., Koepsell S.A., Hinrichs S.H. Myricetin Inhibits Escherichia coli DnaB Helicase but Not Primase. Bioorg. Med. Chem. 2007;15:7203–7208. doi: 10.1016/j.bmc.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 171.Sharma S. Non-B DNA Secondary Structures and Their Resolution by RecQ Helicases. J. Nucleic Acids. 2011;2011:724215. doi: 10.4061/2011/724215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wu X., Maizels N. Substrate-Specific Inhibition of RecQ Helicase. Nucleic Acids Res. 2001;29:1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yang N., Tanner J.A., Wang Z., Huang J.D., Zheng B.J., Zhu N., Sun H. Inhibition of SARS Coronavirus Helicase by Bismuth Complexes. Chem. Commun. (Camb). 2007;42:4413–4415. doi: 10.1039/b709515e. [DOI] [PubMed] [Google Scholar]
- 174.Boger D.L., Fink B.E., Brunette S.R., Tse W.C., Hedrick M.P. A Simple, High-Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 175.Boger D.L., Tse W.C. Thiazole Orange as the Fluorescent Intercalator in a High Resolution Fid Assay for Determining DNA Binding Affinity and Sequence Selectivity of Small Molecules. Bioorg. Med. Chem. 2001;9:2511–2518. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- 176.Phoon C.W., Ng P.Y., Ting A.E., Yeo S.L., Sim M.M. Biological Evaluation of Hepatitis C Virus Helicase Inhibitors. Bioorg. Med. Chem. Lett. 2001;11:1647–1650. doi: 10.1016/s0960-894x(01)00263-3. [DOI] [PubMed] [Google Scholar]
- 177.Belon C.A., High Y.D., Lin T.I., Pauwels F., Frick D.N. Mechanism and Specificity of a Symmetrical Benzimidazolephenylcarboxamide Helicase Inhibitor. Biochemistry. 2010;49:1822–1832. doi: 10.1021/bi901974a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tunitskaya V.L., Mukovnya A.V., Ivanov A.A., Gromyko A.V., Ivanov A.V., Streltsov S.A., Zhuze A.L., Kochetkov S.N. Inhibition of the Helicase Activity of the HCV NS3 Protein by Symmetrical Dimeric Bis-benzimidazoles. Bioorg. Med. Chem. Lett. 2011;21:5331–5335. doi: 10.1016/j.bmcl.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 179.Belon C., Frick D.N. Thioflavin S Inhibits Hepatitis C Virus RNA Replication and the Viral Helicase with a Novel Mechanism. FASEB J. 2010;24:lb202. [Google Scholar]
- 180.Li, K., Frankowski, K. J., Hanson, A. M., Ndjomou, J., Shanahan, M. A., Mukherjee, S., Shadrick, W. R., Sweeney, N. L., Belon, C. A., Neuenswander, B., et al. Hepatitis C Virus NS3 Helicase Inhibitor Discovery. In Probe Reports from the NIH Molecular Libraries Program, in press. [PubMed]
- 181.Graham R.K., Caiger P. Fluorescence Staining for the Determination of Cell Viability. Appl Microbiol. 1969;17:489–490. doi: 10.1128/am.17.3.489-490.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Novac O., Guenier A.S., Pelletier J. Inhibitors of Protein Synthesis Identified by a High Throughput Multiplexed Translation Screen. Nucleic Acids Res. 2004;32:902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tan G.S., Chiu C.H., Garchow B.G., Metzler D., Diamond S.L., Kiriakidou M. Small Molecule Inhibition of RISC Loading. ACS Chem. Biol. 2012;7:403–410. doi: 10.1021/cb200253h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McKay G.A., Reddy R., Arhin F., Belley A., Lehoux D., Moeck G., Sarmiento I., Parr T.R., Gros P., Pelletier J., et al. Triaminotriazine DNA Helicase Inhibitors with Antibacterial Activity. Bioorg. Med. Chem. Lett. 2006;16:1286–1290. doi: 10.1016/j.bmcl.2005.11.076. [DOI] [PubMed] [Google Scholar]
- 185.Aiello D., Barnes M.H., Biswas E.E., Biswas S.B., Gu S., Williams J.D., Bowlin T.L., Moir D.T. Discovery, Characterization and Comparison of Inhibitors of Bacillus anthracis and Staphylococcus aureus Replicative DNA Helicases. Bioorg. Med. Chem. 2009;17:4466–4476. doi: 10.1016/j.bmc.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li B., Pai R., Di M., Aiello D., Barnes M.H., Butler M.M., Tashjian T.F., Peet N.P., Bowlin T.L., Moir D.T. Coumarin-Based Inhibitors of Bacillus anthracis and Staphylococcus aureus Replicative DNA Helicase: Chemical Optimization, Biological Evaluation, and Antibacterial Activities. J. Med. Chem. 2012;55:10896–10908. doi: 10.1021/jm300922h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Aggarwal M., Sommers J.A., Shoemaker R.H., Brosh R.M.J. Inhibition of Helicase Activity by a Small Molecule Impairs Werner Syndrome Helicase (WRN) Function in the Cellular Response to DNA Damage or Replication Stress. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nguyen G.H., Dexheimer T.S., Rosenthal A.S., Chu W.K., Singh D.K., Mosedale G., Bachrati C.Z., Schultz L., Sakurai M., Savitsky P., et al. A Small Molecule Inhibitor of the BLM Helicase Modulates Chromosome Stability in Human Cells. Chem. Biol. 2013;20:55–62. doi: 10.1016/j.chembiol.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Adedeji A.O., Singh K., Calcaterra N.E., Dediego M.L., Enjuanes L., Weiss S., Sarafianos S.G. Severe Acute Respiratory Syndrome Coronavirus Replication Inhibitor That Interferes with the Nucleic Acid Unwinding of the Viral Helicase. Antimicrob. Agents Chemother. 2012;56:4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Grell W., Hurnaus R., Griss G., Sauter R., Rupprecht E., Mark M., Luger P., Nar H., Wittneben H., Muller P. Repaglinide and Related Hypoglycemic Benzoic Acid Derivatives. J. Med. Chem. 1998;41:5219–5246. doi: 10.1021/jm9810349. [DOI] [PubMed] [Google Scholar]
- 191.Wright C.M., Seguin S.P., Fewell S.W., Zhang H., Ishwad C., Vats A., Lingwood C.A., Wipf P., Fanning E., Pipas J.M., et al. Inhibition of Simian Virus 40 Replication by Targeting the Molecular Chaperone Function and ATPase Activity of T Antigen. Virus Res. 2009;141:71–80. doi: 10.1016/j.virusres.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mousseau G., Kota S., Takahashi V., Frick D.N., Strosberg A.D. Dimerization-Driven Interaction of Hepatitis C Virus Core Protein with NS3 Helicase. J. Gen. Virol. 2011;92:101–111. doi: 10.1099/vir.0.023325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Saalau-Bethell S.M., Woodhead A.J., Chessari G., Carr M.G., Coyle J., Graham B., Hiscock S.D., Murray C.W., Pathuri P., Rich S.J., et al. Discovery of an Allosteric Mechanism for the Regulation of HCV NS3 Protein Function. Nat. Chem. Biol. 2012;8:920–925. doi: 10.1038/nchembio.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional Characterization of IRESes by an Inhibitor of the RNA Helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 195.Lindqvist L., Oberer M., Reibarkh M., Cencic R., Bordeleau M.E., Vogt E., Marintchev A., Tanaka J., Fagotto F., Altmann M., et al. Selective Pharmacological Targeting of a DEAD Box RNA Helicase. PLoS One. 2008;3:e1583. doi: 10.1371/journal.pone.0001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bordeleau M.E., Matthews J., Wojnar J.M., Lindqvist L., Novac O., Jankowsky E., Sonenberg N., Northcote P., Teesdale-Spittle P., Pelletier J. Stimulation of Mammalian Translation Initiation Factor eIF4A Activity by a Small Molecule Inhibitor of Eukaryotic Translation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bordeleau M.E., Cencic R., Lindqvist L., Oberer M., Northcote P., Wagner G., Pelletier J. RNA-Mediated Sequestration of the RNA Helicase eIF4A by Pateamine A Inhibits Translation Initiation. Chem. Biol. 2006;13:1287–1295. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 198.Bordeleau M.E., Robert F., Gerard B., Lindqvist L., Chen S.M., Wendel H.G., Brem B., Greger H., Lowe S.W., Porco J.A.J., et al. Therapeutic Suppression of Translation Initiation Modulates Chemosensitivity in a Mouse Lymphoma Model. J. Clin. Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Amundsen S.K., Spicer T., Karabulut A.C., Londono L.M., Eberhart C., Fernandez Vega V., Bannister T.D., Hodder P., Smith G.R. Small-Molecule Inhibitors of Bacterial AddAB and RecBCD Helicase-Nuclease DNA Repair Enzymes. ACS Chem. Biol. 2012;7:879–891. doi: 10.1021/cb300018x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang Y., Coulombe R., Cameron D.R., Thauvette L., Massariol M.J., Amon L.M., Fink D., Titolo S., Welchner E., Yoakim C., et al. Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor. J. Biol. Chem. 2004;279:6976–6985. doi: 10.1074/jbc.M311376200. [DOI] [PubMed] [Google Scholar]
- 201.Kandil S., Biondaro S., Vlachakis D., Cummins A.C., Coluccia A., Berry C., Leyssen P., Neyts J., Brancale A. Discovery of a Novel HCV Helicase Inhibitor by a De Novo Drug Design Approach. Bioorg. Med. Chem. Lett. 2009;19:2935–2937. doi: 10.1016/j.bmcl.2009.04.074. [DOI] [PubMed] [Google Scholar]
- 202.Mastrangelo E., Pezzullo M., De Burghgraeve T., Kaptein S., Pastorino B., Dallmeier K., de Lamballerie X., Neyts J., Hanson A.M., Frick D.N., et al. Ivermectin is a Potent Inhibitor of Flavivirus Replication Specifically Targeting NS3 Helicase Activity: New Prospects for an Old Drug. J. Antimicrob. Chemother. 2012;67:1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hogbom M., Collins R., van den Berg S., Jenvert R.M., Karlberg T., Kotenyova T., Flores A., Karlsson Hedestam G.B., Schiavone L.H. Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP. J. Mol. Biol. 2007;372:150–159. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 204.Maga G., Falchi F., Garbelli A., Belfiore A., Witvrouw M., Manetti F., Botta M. Pharmacophore Modeling and Molecular Docking Led to the Discovery of Inhibitors of Human Immunodeficiency Virus–1 Replication Targeting the Human Cellular Aspartic Acid–Glutamic Acid–Alanine–Aspartic Acid Box Polypeptide 3. J. Med. Chem. 2008;51:6635–6638. doi: 10.1021/jm8008844. [DOI] [PubMed] [Google Scholar]
- 205.Ha T., Kozlov A.G., Lohman T.M. Single-Molecule Views of Protein Movement on Single-Stranded DNA. Annu. Rev. Biophys. 2012;41:295–319. doi: 10.1146/annurev-biophys-042910-155351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Goudreau N., Cameron D.R., Deziel R., Hache B., Jakalian A., Malenfant E., Naud J., Ogilvie W.W., O’Meara J., White P.W., et al. Optimization and Determination of the Absolute Configuration of a Series of Potent Inhibitors of Human Papillomavirus Type-11 E1-E2 Protein-Protein Interaction: A Combined Medicinal Chemistry, NMR and Computational Chemistry Approach. Bioorg. Med. Chem. 2007;15:2690–2700. doi: 10.1016/j.bmc.2007.01.036. [DOI] [PubMed] [Google Scholar]


