SUMMARY
MYC is an oncoprotein transcription factor that is overexpressed in the majority of malignancies. The oncogenic potential of MYC stems from its ability to bind regulatory sequences in thousands of target genes, which depends on interaction of MYC with its obligate partner, MAX. Here, we show that broad association of MYC with chromatin also depends on interaction with the WD40-repeat protein WDR5. MYC binds WDR5 via an evolutionarily conserved “MYC box IIIb” motif that engages a shallow, hydrophobic, cleft on the surface of WDR5. Structure-guided mutations in MYC that disrupt interaction with WDR5 attenuate binding of MYC to ~80% of its chromosomal locations and disable its ability to promote induced pluripotent stem cell formation and drive tumorigenesis. Our data reveal WDR5 as a key determinant for MYC recruitment to chromatin and uncover a tractable target for the discovery of anti-cancer therapies against MYC-driven tumors.
INTRODUCTION
The MYC oncogenes encode a family of related transcription factors (c-, L-, and N-MYC) that are overexpressed in the majority of malignancies and contribute to ~100,000 cancer-related deaths annually in the USA alone (Vita and Henriksson, 2006). MYC proteins derive their oncogenicity from an aggregate of effects on cell growth, proliferation, metabolism, genome stability, and apoptosis—actions that in turn depend on their function as sequence-specific transcriptional regulators (Tansey, 2014). Although the precise number of MYC target genes is the subject of debate, it is clear that MYC proteins drive tumorigenesis by regulating thousands of genes and, depending on context, can function as both transcriptional activators and repressors.
Key to the actions of MYC as a transcription factor is its ability to bind specific DNA elements within the regulatory regions of its target genes. To bind DNA, MYC must heterodimerize with its obligate partner, MAX (Blackwood and Eisenman, 1991), forming a basic helix-loop-helix (bHLH) DNA-binding domain (DBD) that recognizes the major groove of DNA. MYC–MAX heterodimers preferentially bind the “E-box” motif (CACGTG) that is found in promoters and enhancers controlled by MYC, although they can also to bind to E-box variants and sequences that lack this motif entirely (Tansey, 2014). The importance of MAX to the function of MYC is supported by the widespread overlap of these proteins on chromatin (Lin et al., 2012), the impact of mutations that disrupt MYC–MAX dimerization on MYC’s oncogenicity (Amati et al., 1993), and by the demonstration that genetic inhibition of MYC–MAX association causes tumor regression in multiple model systems of cancer (Annibali et al., 2014; Soucek et al., 2013).
Despite the role of direct DNA interaction in recruiting MYC to its target genes, evidence indicates that binding of MYC to DNA in the cell is influenced by additional factors. There is a particularly strong bias for MYC to bind chromatin that is enriched in ‘active’ histone modifications such as histone H3 lysine 4 (K4) and 79 (K79) methylation (Guccione et al., 2006; Lin et al., 2012; Sabo et al., 2014; Walz et al., 2014; Zeller et al., 2006). Indeed, based on the correlation between MYC binding and these epigenetic marks, it has been proposed that H3K4/79 methylation is strictly required for MYC to engage target gene chromatin (Guccione et al., 2006). Whether this requirement reflects the accessibility of the DNA in modified nucleosomes, recognition of methylated histones by epigenetic ‘readers’, or some other process, is unknown.
Structure-function analyses of MYC have delineated a critical transcriptional activation domain (TAD) in the amino-terminal third of the protein and a bHLH DNA-binding domain in the carboxy-terminal third of the protein (Tansey, 2014). The intervening central portion of MYC, in contrast, is poorly understood, but is likely to have important functions as it contains three highly conserved sequences known as “MYC boxes” (Mb) IIIa, IIIb, and IV (Meyer and Penn, 2008). MbIIIa contributes to transcriptional repression by MYC, likely via association with histone deactylases (Kurland and Tansey, 2008). MbIV is required for MYC to bind naked DNA through an unknown mechanism (Cowling et al., 2006). And MbIIIb has, as yet, no known function. Here, we report that MbIIIb acts by binding directly to WDR5, a WD40-repeat protein present in multiple chromatin regulatory complexes, including H3K4 methyltransferases. We show that interaction with WDR5 is not required for MYC to bind naked DNA, but is required for MYC to broadly associate with target genes in vivo and to drive tumorigenesis. Properties of the MYC–WDR5 interface make it a potentially viable point for the discovery of small molecule inhibitors that disrupt the MYC–WDR5 interaction and interfere with MYC function in cancer cells.
RESULTS
Identification of WDR5 as a direct MYC-interaction partner
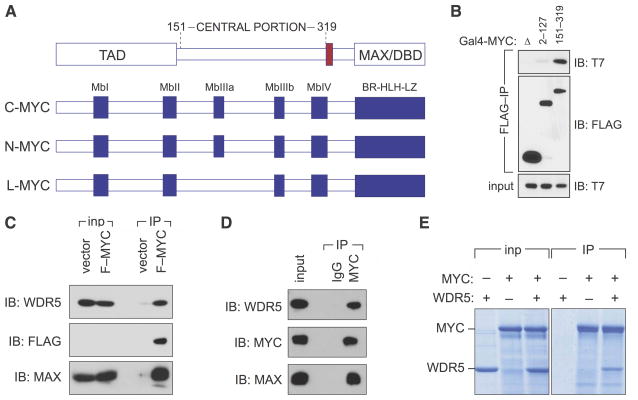
To illuminate the function of the central portion of c-MYC (residues 151–319; Figure 1A), we searched for factors that interact with this region using a two-pronged approach that involved two-hybrid and proteomic screening (Figure S1). One protein identified in both assays was WDR5, a WD40-repeat-containing protein that assembles into a number of chromatin regulatory complexes including the MLL/SET methyltransferases that methylate H3K4 and the MOF/NSL histone acetyltransferases that acetylate histone H4. Using co-immunoprecipitation, we confirmed that T7-tagged WDR5 binds to the central portion of MYC after transient co-expression with a FLAG-Gal4-MYC (151–319) fusion in human cells, but does not associate with the MYC TAD (residues 2–127) under similar conditions (Figure 1B). We observed that retrovirally-expressed FLAG-MYC binds endogenous WDR5 (Figure 1C). And we found that endogenous MYC robustly associates with endogenous WDR5 (Figure 1D). Consistent with identification of WDR5 via a two-hybrid assay, we also determined that interaction between the two proteins is direct, as recombinant full-length MYC and WDR5 interact when expressed and purified from Escherichia coli (Figure 1E). WDR5 has also been identified as a MYC-binding partner by Ullius et al., (2014), and was detected in a recent proteomic survey for MYC-interacting proteins (Dingar et al., 2014). Together, these data establish that WDR5 is a bona-fide and direct interaction partner that associates with the central portion of MYC.
Figure 1. WDR5 binds directly to the central portion of MYC.
(A) Schematic representation of MYC proteins. Functional domains of MYC are shown at top (transcriptional activation domain, TAD; MAX/DBD, MAX-interaction and DNA binding domain; nuclear localization signal colored red). Below represents conserved sequences in MYC family members. Shown are five “MYC boxes” (Mb) and the basic region-helix-loop-helix-leucine zipper (BR-HLH-LZ). (B) WDR5 binds to the central portion of MYC. FLAG-tagged Gal4 DBD alone (Δ), or fused to the indicated portions of MYC, was transiently expressed in HEK293 cells together with T7-tagged WDR5, immunoprecipitated (IP) with anti-FLAG antibodies, and subject to immunoblotting (IB) with either anti-T7 or -FLAG antibodies. (C) Full-length MYC binds WDR5. Co-immunoprecipitation of retrovirally-expressed FLAG-tagged c-MYC (F–MYC) with endogenous WDR5 in extracts of stably-transduced HEK293 cells. FLAG immunoprecipitates (IP) were immunoblotted with antibodies against WDR5, the FLAG epitope, or MAX. 0.5% of the input (inp) is shown for WDR5; 10% is shown for MAX and FLAG (MYC). Due to low abundance, FLAG-tagged MYC is not detectable in the input under these conditions. (D) Endogenous MYC and WDR5 interact. Co-immunoprecipitation of endogenous MYC, MAX, and WDR5 proteins from HEK293 cells treated with proteasome inhibitor MG132 for 90 minutes. Extracts were subject to IP with antibodies against MYC, or an IgG control, and immunoblotted with the indicated antibodies. 0.04% of the input is shown for WDR5; 1% is shown for MAX and MYC. (E) MYC and WDR5 interact directly. Recombinant FLAG-tagged MYC and recombinant untagged WDR5 were mixed in equimolar ratios, and subject to immunoprecipitation with anti-FLAG resin. Immunoprecipitates, and a sample of the input, were resolved by SDS-PAGE followed by staining with Coomassie Blue.
MYC and WDR5 co-localize extensively on chromatin
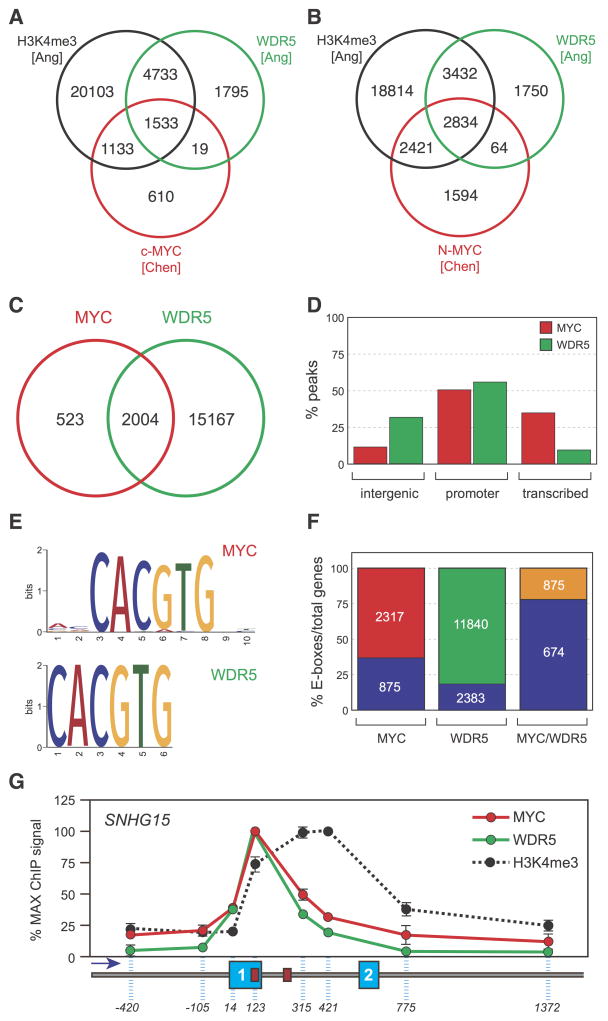
If MYC and WDR5 interact in a meaningful way, we might expect these two proteins to bind to the same or overlapping sites on chromatin. Indeed, we analyzed two distinct published chromatin immunoprecipitation-sequencing (ChIP-seq) datasets from mouse embryonic stem cells (Ang et al., 2011; Chen et al., 2008) and found a widespread overlap between the two proteins, with 1552/3295 (47%) of sites for c-MYC—and 2907/6913 (42%) of sites for N-MYC—overlapping with a site bound by WDR5 (Figure 2A, B). To probe the extent of this overlap further, we directly compared localization of MYC and WDR5 in HEK293 cells engineered to express FLAG-epitope-tagged MYC at an average concentration of ~30,000 molecules per cell (Figure S2); a level sufficient for suppression of endogenous MYC expression (Penn et al., 1990) but well below that where promiscuous binding to all active loci is observed (Lin et al., 2012). Our ChIP-seq analysis identified 2,527 high-confidence binding sites for exogenous MYC (Figure 2C), the majority of which are located either at promoters or within transcription units (Figure 2D). We mapped 17,171 sites for WDR5 (Figure 2C), which also displayed a marked preference for promoters but showed significantly higher enrichment in intergenic regions than MYC (Figure 2D). Importantly, comparison of these profiles revealed that 2004/2527—79%—of the binding sites for MYC overlap with WDR5 (Figure 2C). Motif enrichment analysis showed highly-significant enrichment of E-boxes in both the MYC and WDR5 binding sites (Figure 2E) with 77% (674/875) of the MYC/WDR5 colocalized genes carrying this motif (Figure 2F), compared to the 38% (875/2317) average presence of E-boxes in MYC-bound loci. Finally, we performed high resolution ChIP mapping at the SNHG15 long non-coding RNA gene (Figure 2G), which bound both MYC and WDR5 in our ChIP-seq analyses. We observed that MYC and WDR5 distribute identically across SNHG15, with the signals for both proteins peaking at an E-box within exon 1. Because MYC binding is known to correlate with H3K4 trimethylation, we also monitored distribution of this modification across the SNHG15 gene (Figure 2G). Interestingly, the profiles for MYC and WDR5 are both noticeably different from that for H3K4 trimethylation, showing that—at this level of resolution and at this locus—binding of MYC correlates better with WDR5 than with trimethylation of H3 at lysine 4. We conclude that there is a widespread and intimate overlap of MYC and WDR5 binding sites on chromatin.
Figure 2. MYC and WDR5 co-localize on chromatin.
(A) MYC and WDR5 binding sites overlap in mouse embryonic stem cells. Summary of analysis of published ChIP-seq datasets (Ang et al., 2011; Chen et al., 2008) comparing localization of c-MYC, WDR5, and H3K4 trimethylation (H3K4me3) in the two studies. Numbers inside the Venn diagram report the number of binding sites for each factor/modification mapped, and the overlap between them. (B) As in (A), except comparing N-MYC, WDR5, and H3K4me3. (C) MYC and WDR5 binding sites overlap in HEK293 cells. Venn diagram, showing the number of binding sites detected for virally-expressed FLAG-tagged MYC (red) and endogenous WDR5 (green) in HEK293 cells by ChIP-seq, and the overlap between them. n=3. (D) Percentage distribution of MYC and WDR5 ChIP-seq peaks into intergenic, promoter, and transcription units, as defined by HOMER (Heinz et al., 2010). (E) E-box motifs are enriched in MYC and WDR5 binding sites. Motif enrichment analysis was performed on the ChIP-seq data from HEK293 cells. For both MYC (top) and WDR5 (bottom), E-boxes are centrally located in ChIP-seq peaks and the enrichment is highly significant (WT-MYC, p=2.9×10−109; WDR5, p=2.4×10−134). (F) MYC/WDR5-bound genes are enriched in E-boxes. The figure shows the total distribution of E-boxes in genes (containing one or more peaks) for each protein, together with the subset of genes where MYC and WDR5 both bind (MYC/WDR5). Numbers in the lower blocks represent the number of E-box-containing genes; numbers in the upper blocks represent the total number of E-box-containing genes in each category. (G) MYC distribution at the SNHG15 locus closely parallels that of WDR5. ChIP analysis was performed in HEK293 cells, probing for distribution of MYC, WDR5, or H3K4me3 across the 5′ end of the SNHG15 gene. Dashed blue lines indicate the relative position of the primers used to interrogate ChIP DNA. Exons 1 and 2 are numbered blue boxes, and the direction of transcription is indicated by the arrow. SNHG15 carries two perfect E-box motifs, cartooned as red boxes. Signals are normalized according to the maximum ChIP signal for each protein. Data are presented as mean +/− SEM. n=3.
MYC binds WDR5 via the evolutionarily-conserved MbIIIb motif
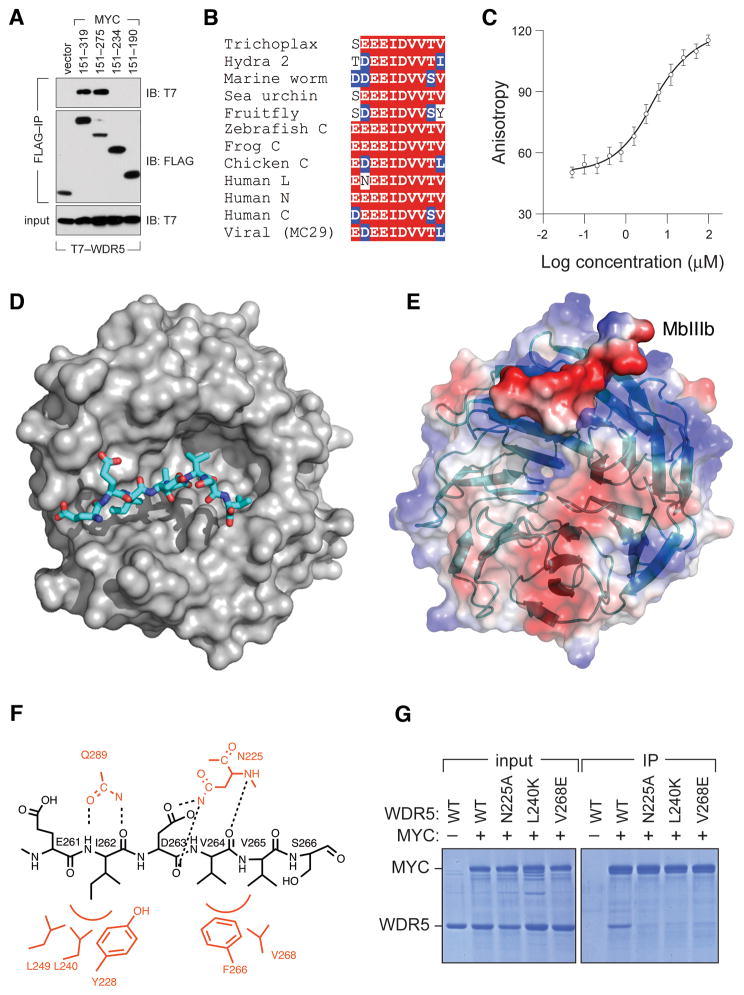
We next asked how MYC binds WDR5. Co-immunoprecipitation analysis of a set of transiently-expressed FLAG-Gal4–MYC fusions showed MYC residues 151–275 of MYC bind T7-WDR5, whereas residues 151–234 do not (Figure 3A). Alignment of the intervening region of c-MYC with evolutionarily diverse MYC proteins (Figure S3A) revealed that this segment contains MbIIIb, a conserved 10 amino acid block featuring an invariant “EEIDVV” core present in all MYC family members from all species (Figure 3B). Interestingly, WDR5 shows a similar pattern of deep conservation, which prompted us to ask whether MbIIIb is sufficient for interaction of MYC with WDR5. Using fluorescence polarization anisotropy (FPA), we showed that a 10-residue MbIIIb peptide (residues 258–267 of human c-MYC) binds to WDR5 with a Kd of 9.3 μM (Figure 3C), confirming that MbIIIb is a self-contained WDR5-interaction module.
Figure 3. MbIIIb is a WDR5-interaction module.
(A) Residues 235–275 of MYC are necessary to bind WDR5. Lysates were prepared from HEK293 cells transiently expressing FLAG-tagged Gal4DBD fused to the indicated portions of MYC, as well as T7-epitope-tagged WDR5, and subject to FLAG IP. Recovered proteins were probed with antibodies against the FLAG or T7 peptides. (B) MbIIIb contains an invariant “EEIDVV” motif. Alignment of MbIIIb sequences from indicated MYC proteins, showing homology to residues 258–267 of human c-MYC. See also Figure S3. (C) MbIIIb is sufficient to bind WDR5 in vitro. Saturation binding curve of FITC-labeled MbIIIb peptide with purified recombinant WDR5. Data are presented as mean +/− SEM. (D) Structure of the MbIIIb–WDR5 complex. The MYC MbIIIb peptide is shown in stick representation (colored by atom type) and WDR5 is shown in surface representation (grey). (E) Surface potential of the MbIIIb–WDR5 complex. WDR5 is shown in transparent surface/ribbon representation. MbIIIb is shown as solid surface representation. Positively-charged features are colored blue; negatively-charged features are colored red. (F) Summary of important interactions between the MbIIIb peptide and WDR5. The MYC peptide is in black, and critical WDR5 residues are in red. Red arcs indicate hydrophobic contacts. Intermolecular hydrogen bonds are shown as dotted lines. (G) Mutation of key residues in WDR5 required for interaction with MbIIIb disrupt interaction with full-length MYC. Recombinant FLAG-tagged WT MYC and recombinant untagged WDR5 (WT or the indicated mutants) were mixed in equimolar ratios, and subject to immunoprecipitation with anti-FLAG resin. Immunoprecipitates, and a sample of the input, were resolved by SDS-PAGE followed by staining with Coomassie Blue.
To understand how MYC interacts with WDR5, we solved the X-ray crystal structure of WDR5 in complex with an MbIIIb peptide at 1.9 Å resolution (Table 1; Figure 3D). The overall structure of WDR5 in this complex is almost identical to the reported apo structure for WDR5 [all backbone atom RMSD 0.21 Å; (Schuetz et al., 2006)] indicating that WDR5 does not undergo significant structural alterations when bound to MbIIIb. In the complex, the MbIIIb peptide binds WDR5 in an extended conformation on one face of the WDR5 β-propeller disc, lying in a cleft formed from loops that link blades of the β-propeller (Figure 3E). The cleft to which MbIIIb binds is hydrophobic in nature but surrounded by positive charge (Figure 3E), consistent with the negative electrostatic potential of the MbIIIb peptide. Within the WDR5 cleft are two hydrophobic pockets that mediate critical interactions with residues in the EEIDVV core: pocket 1 is created by tyrosine 228 (Y228), leucine 240 (L240) and leucine 249 (L249) of WDR5, which accommodate isoleucine 262 (I262) of MYC; pocket 2 is created by phenylalanine 266 (F266) and valine 268 (V268) of WDR5, which accommodate side chains of valines 264 and 265 (V264; V264) of MYC (Figure 3F). The complex is also stabilized by intramolecular hydrogen bonds, including those involving the side chains of asparagine 225 (N225) and glutamine 289 (Q289) of WDR5. Accordingly, mutation of residues N225, L240, or V268 in WDR5 block interaction of recombinant WDR5 with recombinant full-length MYC in vitro (Figure 3G), demonstrating that the MbIIIb interaction surface defined in the structure is relevant to interaction of WDR5 with the entire MYC protein.
Table 1.
Crystallographic Data and Refinement Statistics
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 80.76, 87.18, 44.91 |
| Wavelength (Å) | 0.98 |
| Resolutiona (Å) | 50.0–1.9 (1.93–1.90) |
| Total reflections | 248,946 |
| Unique reflections | 25,784 |
| Completeness (%) | 99.9 (100.0) |
| Redundancy | 9.7 (9.4) |
| Overall (I/σI) | 24.8 (4.3) |
| Rsymb (%) | 7.4 (67.3) |
| Structure refinement | |
| No. reflections | 25,709 |
| Rwork/Rfree (%) | 15.9/18.6 |
| No. atoms | |
| Protein | 2,466 |
| Ligand | 62 |
| Water | 179 |
| B-factors | |
| Protein | 36.6 |
| Ligand | 64.1 |
| Water | 44.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.9 |
| Ramachandran favoredc (%) | 97 |
| Ramachandran allowed (%) | 3.2 |
| PDB ID code | 4Y7R |
The numbers in parentheses refer to the highest-resolution shell
Rsym = Σhkl Σi |Ihkl,i - <Ihkl>|/Σhkl Σ<Ihk>
Validation was done by MolProbity as implemented in Phenix
MYC binds WDR5 via an interaction surface that is shared with RBBP5 and KANSL2
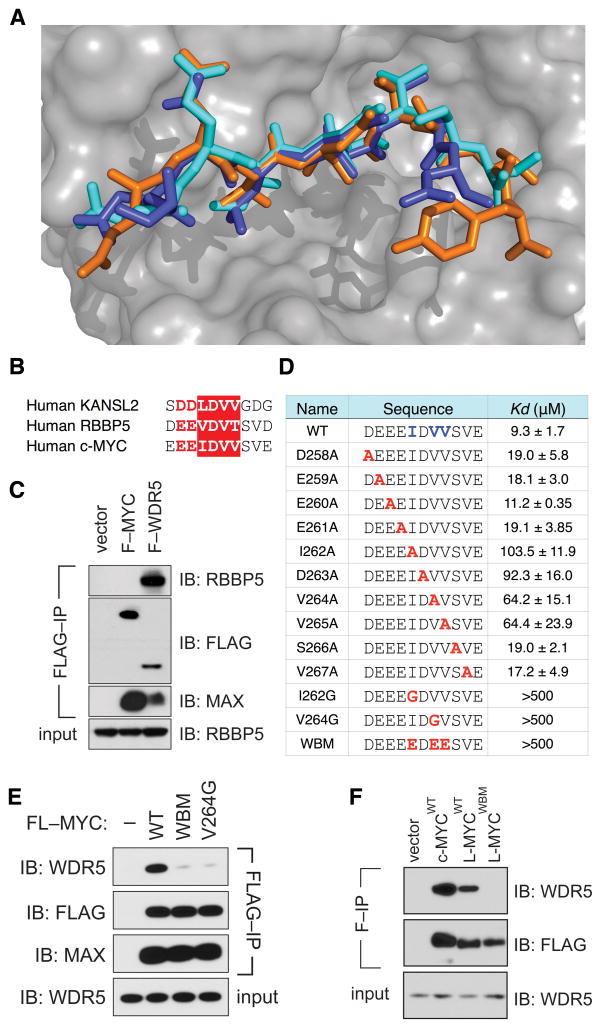
WDR5 interacts with a number of proteins, including MLL/SET methyltransferases (Patel et al., 2008), histone H3 tails (Migliori et al., 2012), and the RBBP5 (Avdic et al., 2011; Odho et al., 2010) and KANSL2 (Dias et al., 2014) components of the MLL/SET and NSL complexes, respectively. The MLL/SET and H3 binding sites are located on the opposite side of WDR5 that binds MbIIIb, but RBBP5 and KANSL2 bind to the same site on WDR5 that interacts with MYC. Indeed, comparative analysis (Figure 4A, Figure S4) revealed that MYC binds WDR5 in a manner virtually identical to RBBP5 (all backbone atom RMSD 0.35 Å) and KANSL2 (all backbone atom RMSD 0.25 Å), with all three proteins making key contacts with the same two hydrophobic pockets in WDR5. These structural similarities are reflected at the sequence level, as the invariant core motif of MbIIIb is highly similar to the corresponding regions of RBBP5 and KANSL2 (Figure 4B). Consistent with the identical patterns of WDR5-binding shared by these three proteins, binding of MYC and RBBP5 to WDR5 appears mutually exclusive, as full-length FLAG-MYC recovered from cells is devoid of co-associated endogenous RBBP5, even under conditions where FLAG-WDR5 and endogenous RBBP5 associate strongly (Figure 4C). Based on these observations, we conclude that MYC binds WDR5 via a modality that is shared with core components of the MLL/SET and NSL histone modifiers. Furthermore, because of the overlapping nature of these interactions, we conclude that MYC must interact with WDR5 in a way that does not involve (or disrupts) MLL/SET or NSL2 complexes.
Figure 4. MYC binds WDR5 via an interaction surface that is shared with RBBP5 and KANSL2.
(A) MbIIIb interacts with WDR5 in a manner similar to that of RBBP5 and KANSL2. The figure shows a superimposition of the MbIIIb peptide (cyan, this study), the RBBP5 peptide (orange, PDB ID: 3P4F) and the KANSL2 peptide (purple, PDB ID: 4CY2) in complex with WDR5 (semi-transparent surface). See also Figure S4. (B) Conservation of the MbIIIb core in KANSL2 and RBBP5. Sequence alignment of the regions of KANSL2, RBBP5, and MYC shown to bind to WDR5. The IDVV/LDVV/VDVT core is boxed in red; conserved negatively-charged residues preceding the core are in red. (C) MYC associates with WDR5 but not RBBP5. Lysates were prepared from HEK293 cells transiently expressing FLAG-tagged MYC (F–MYC) or WDR5 (F–WDR5), subject to FLAG immunoprecipitation, and immunoblotted with antibodies against endogenous RBBP5, endogenous MAX, or the FLAG epitope. (D) Conserved hydrophobic residues in the MbIIIb core mediate interaction with WDR5. FPA was used to determine the equilibrium dissociation constants for each of the indicated peptides with recombinant WDR5. (E) MbIIIb is an essential point of contact between MYC and WDR5. Co-immunoprecipitation of stably expressed FLAG-tagged c-MYC (WT, WBM, or the V264G mutant) with endogenous WDR5 in extracts of retrovirally-transduced HEK293 cells. FLAG immunoprecipitates (IP) were immunoblotted (IB) with antibodies against WDR5, the FLAG peptide, or MAX. 2% of input is shown for WDR5. (F) L-MYC associates with WDR5 in an MbIIIb-dependent manner. FLAG-tagged variants of the indicated MYC proteins were transiently-expressed in HEK293 cells, subject to FLAG immunoprecipitation (F–IP) and immunoblotted (IB) with antibodies against the FLAG peptide or endogenous WDR5.
To validate our structure, we measured the interaction of MbIIIb mutant peptides with recombinant WDR5 using a fluorescent polarization anisotropy assay (Figure 4D). Alanine substitutions at I262, V264, and V265 reduced the affinity of interaction by six- to 10-fold, while mutation of V264 to glycine (V264G), or simultaneous mutation of I262/V264/V265 to glutamic acid (“WBM”), disrupted interaction with WDR5 (Kd >500 μM). Thus, as predicted by the structure, hydrophobic resides in the MbIIIb core are critical to binding WDR5. Importantly, both the WBM and V264G mutations also prevented the interaction of full-length FLAG-MYC with endogenous WDR5 after retroviral-mediated expression in human cells, while leaving MYC expression, and association with endogenous MAX, unaffected (Figure 4E). The WBM mutation also disrupts the interaction of FLAG-tagged L-MYC with endogenous WDR5 (Figure 4F). These data demonstrate that MbIIIb is the sole essential element in MYC required for binding to WDR5.
Association with WDR5 is broadly required for MYC to bind target gene chromatin
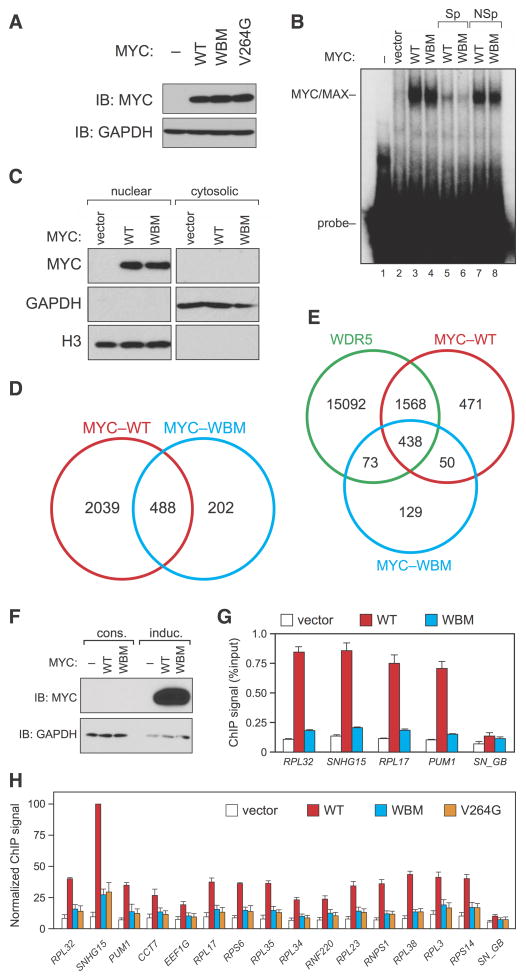
We next characterized how mutations in MYC that disrupt interaction with WDR5 impact the ability of MYC to bind target genes. The WBM mutation does not affect MYC expression (Figure 5A) or association with MAX (Figure 4E), demonstrating that this is a true separation of function mutation that is selective for WDR5. Consistent with the ability of the WBM MYC mutant to bind MAX, this mutation does not alter binding to a canonical E-box in naked DNA (Figure 5B). Moreover, the WBM mutation has no effect on nuclear localization of MYC—as measured by biochemical fractionation (Figure 5C) and immunohistochemistry (Figure S5A), demonstrating that the mutant protein is localized in the nucleus and retains its DNA binding characteristics. When we performed ChIP-seq to examine binding of the WBM mutant to chromatin, however, we found that 80% (2039/2527) of WT MYC binding sites are lost upon introduction of the WBM mutation (Figure 5D), 78% (1568/2004) of which are at sites where MYC and WDR5 co-localize (Figure 5E). Imperceptible differences in the expression of the two forms of MYC are not responsible for these differences in chromatin association, as vast overexpression of MYC under the control of a potent inducible promoter (Figure 5F) fails to rescue chromatin binding of the WBM mutant (Figure 5G). Moreover, the failure of the WBM MYC mutant to bind chromatin is not due to changes in WDR5 localization, as this mutant has little if any impact on recruitment of WDR5 to a set of representative MYC/WDR5-bound loci (Figure S5B). Finally, the effect of the WBM mutation on MYC binding is recapitulated by the more conservative V264G single point mutation (Figure 5H), supporting the idea that loss of WDR5 interaction underlies the defect in chromatin binding we observe in the WBM mutant. We conclude that interaction with WDR5 is required for MYC to effectively recognize its target genes in the context of chromatin.
Figure 5. Interaction of MYC with WDR5 is broadly required for MYC to bind target genes in the context of chromatin.
(A) The WT, WBM, and V264G forms of MYC are expressed equivalently in HEK293 cells. Immunoblot, showing relative levels of the MYC proteins in HEK293 cells used in the ChIP experiments presented in this figure. (B) The WBM mutation does not affect the ability of MYC/MAX heterodimers to bind naked DNA. Extracts were prepared from HEK293 cells expressing the indicated MYC proteins and a gel shift assay performed using a radio-labeled double-stranded DNA probe carrying an E-box. To confirm the specificity of the interaction, reactions also contained an excess of unlabeled specific competitor DNA (“Sp”, lanes 5–6) or a non-specific competitor with a mutant E-box (“NSp”, lanes 7–8). (C) The WBM mutation does not alter nuclear localization of MYC. Stably-transduced HEK293 cells expressing FLAG-tagged WT or WBM MYC were subjected to biochemical fractionation into nuclear and cytosolic extracts, which were then resolved by SDS-PAGE and MYC proteins detected by FLAG immunoblot (MYC). Extracts were also probed with antibodies against GAPDH (cytosolic) or histone H3 (nuclear). (D) Interaction with WDR5 is required for MYC to bind a majority of target sites in chromatin. Venn diagram, showing the number of binding sites mapped for WT-MYC (red) and WBM-MYC (blue) by ChIP-seq in HEK293 cells, and the overlap between them. n=3. (E) Summary of 293 ChIP-seq data for WDR5, WT-MYC, and WBM-MYC. The Venn diagram summarizes the number of binding sites mapped for each protein, and the overlap between each of them. (F) Inducible overexpression of WT or WBM MYC. Immunoblot, comparing expression levels of FLAG-MYC via constitutive lentiviral overexpression in HEK293 cells (“cons”; used in ChIP experiments presented in panels D, E, and H) versus doxycycline-inducible overexpression in HEK293 cells (“induc”; used in panel G). At this exposure, the constitutively-overexpressed MYC is undetectable. (G) Overexpression does not rescue chromatin binding by the WBM MYC mutant. ChIP, performed on inducible MYC-expressing cells presented in panel (F). Binding to four MYC/WDR5-bound genes was determined via Q-PCR. “SN_GB” corresponds to a primer set within the SNHG15 gene body that binds neither MYC nor WDR5. Data are presented as mean +/− SEM. n=3. (H) The WBM and V264G MYC mutants are defective in chromatin binding. ChIP was performed on HEK293 cells stably expressing the indicated FLAG-tagged MYC proteins (panel A). Binding to 15 MYC target genes was determined via Q-PCR. ChIP signals are presented as the percentage of the maximum signal for WT MYC at SNHG15. Data are presented as mean +/− SEM. n=3. See also Figure S5.
The MYC–WDR5 interaction is required for MYC-driven tumorigenesis
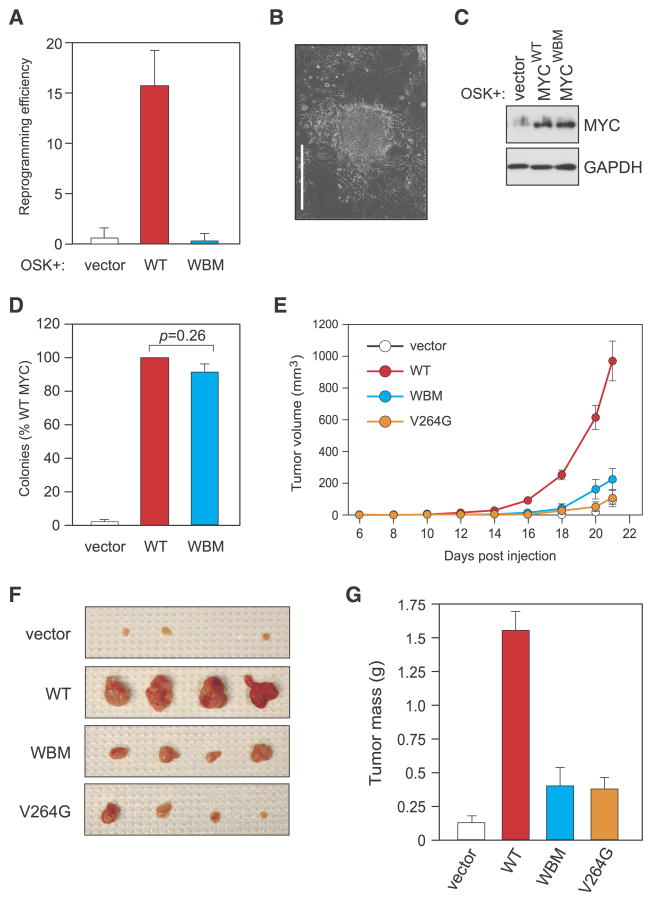
To determine whether the defects in chromatin binding we observe upon loss of the MYC–WDR5 interaction impacts the biological activity of MYC, we tested WDR5-binding-deficient MYC mutants in three assays of MYC function. First, we tested the ability of MYC to cooperate with OCT3/4, SOX2, and KLF4 (OSK) to reprogram mouse embryo fibroblasts to induced pluripotent stem cells (iPSCs; (Nakagawa et al., 2008)). We selected this assay because it requires a DNA-binding competent form of MYC (Nakagawa et al., 2010), and because WDR5 is essential for the initial phases of somatic cell reprogramming and iPSc generation (Ang et al., 2011). In this context, WT MYC efficiently stimulated iPSC colony formation but the WBM MYC protein did not (Figure 6A, B). Indeed, the combination of WBM MYC with OSK was comparable in reprogramming efficiency to that seen with OSK alone, despite the fact that both forms of MYC were expressed at equivalent levels (Figure 6C). We conclude that interaction with WDR5 is required for MYC to stimulate efficient somatic cell reprogramming.
Figure 6. Interaction with WDR5 is required for MYC to stimulate iPSC formation and to drive tumorigenesis.
(A) The MYC–WDR5 interaction is required for MYC to stimulate iPSC formation. Mouse embryo fibroblasts (MEFs) were retrovirally-transduced with viruses expressing OCT3/4, SOX2, and KLF4 (OSK), with or without virus expressing WT- or WBM-MYC. Transduced cells were grown under conditions that promote iPS formation and iPS colonies scored as described in Experimental Procedures. Results are presented in terms of reprogramming efficiency (number of iPSc colonies per 100,000 transduced MEFs). Data are presented as mean +/− SEM. n=5. (B) Photomicrograph of representative iPS colony formed by WT MYC. Scale bar represents 400 μM. (C) Immunoblotting of MYC protein expression in MEFs used for iPSC reprogramming two-days post transduction. (D) The MYC–WDR5 interaction is dispensable for MYC-driven transformation in vitro. NIH3T3 cells were engineered to express vector control, or FLAG-tagged WT or WBM mutant MYC, and tested for ability to promote colony formation when plated in soft agar. After 30 days, colonies were blind-counted and colony numbers expressed relative to WT MYC (100%). Student’s t-test showed no significant difference between WT and WBM MYC in this assay (p=0.26). Data are presented as mean +/− SEM. n=6. (E) The MYC–WDR5 interaction is required for the tumorigenicity of MYC in vivo. The same engineered NIH3T3 cells used in panel D were injected into the flanks of athymic nude mice (vector, n=10; WT, n=11; WBM, n=11; V264G, n=12). The graph shows tumor volumes at the indicated times post-injection. Data are presented as mean +/− SEM. (F) Representative tumors from 4 injection sites for vector control and each of the MYC variants. (G) Tumor masses from mice in panel E. Tumors were excised 21 days after injection and weighed. For Student’s t-test: vector versus WT, p=0.00000010; WT versus WBM, p=0.0000025; WT versus V264G, p=0.00000044; vector versus WBM, p=0.038; vector versus V264G, p=0.0091; WBM versus V264G, p=0.45. Data are presented as mean +/− SEM. See also Figure S6.
Next, we asked whether the WBM mutation impacts the ability of MYC to promote “transformation”, as measured by anchorage-independent growth of engineered NIH3T3 fibroblasts in soft agar. Remarkably, in this assay, loss of interaction with WDR5 had no detectable effect on MYC function, as both the WT and WBM forms of MYC stimulated equivalent levels of colony formation above the ‘vector only’ control (Figure 6D, Figure S6A). Moreover, we detected no notable differences in either the size or morphology of colonies formed with either MYC protein (Figure S6B), demonstrating that the MYC–WDR5 interaction is dispensable for in vitro transformation by MYC under our assay conditions.
Others have reported that MYC mutants incapable of DNA binding can drive transformation in vitro (Conacci-Sorrell et al., 2014), but it is clear that in vivo tumorigenesis by MYC depends on its chromatin-binding abilities (Tansey, 2014). To follow our MYC mutants in a more demanding and tumor-relevant context, therefore, we injected the same fibroblasts as in Figure 6D into the flanks of athymic nude mice and monitored tumor formation (Figure 6E–G, Figure S6C,D). Compared to WT MYC, which produced large, aggressively growing, tumors during the 21 day course of this experiment, both the WBM and V264G mutants of MYC were significantly less tumorigenic, promoting slower-growing tumors that were, on average, 80% smaller than WT MYC-expressing tumors after resection (WT vs WBM p<0.001; WT vs V264G p<0.001). The dramatic deficit in the behavior of WDR5-binding-deficient MYC mutants in mice, compared to in vitro transformation assays, illustrates that these assays measure fundamentally different aspects of MYC function. Importantly, this deficit also reveals a critical role for the MYC–WDR5 interaction in the tumorigenicity of MYC in vivo.
DISCUSSION
The role of MYC proteins as transcription factors means that their ability to engage specific gene sequences within the context of chromatin is paramount to their activity. It is well-established that binding of MYC to target genes requires interaction with MAX. However, it is also clear that precisely where MYC binds to chromatin is influenced by additional factors. The work presented here argues that one of these additional factors is interaction of MYC with WDR5. Based on our findings, we hypothesize that, in order to stably associate with a majority of target genes, MYC–MAX complexes make at least two important contacts—one with DNA, and the other with pre-bound and proximal WDR5. We further propose that the avidity provided by this mechanism not only stabilizes MYC on chromatin, but provides flexibility in terms of where MYC binds by integrating both genetic—DNA sequence—and epigenetic—WDR5—determinants, the latter of which may be positioned in a context-dependent manner. Finally, because the MYC–WDR5 interface is small and structurally well-defined, we posit that small molecules capable of interacting with the MbIIIb-binding pocket on WDR5 could disrupt MYC function and provide a novel approach for targeting MYC-driven tumors.
MbIIIb and the central portion of MYC
By comparison to other regions of the protein, the central portion of MYC is understudied. Yet this portion contains three highly-conserved Myc boxes, implying some important role for these elements in MYC function. Studies of MYC boxes I and II within the MYC TAD have been instrumental in defining how MYC functions (Tansey, 2014), but to our knowledge MbIIIb has not been systematically studied in any mammalian MYC protein. Early work using large internal deletion mutants failed to reach a consensus on the importance of the central portion of MYC, with deletion mutants encompassing MbIIIb active in some in vitro transformation assays, but not others (Biegalke et al., 1987; Heaney et al., 1986; Stone et al., 1987). As reported here, however, behavior in such in vitro assays does not necessarily recapitulate what occurs in tumorigenesis in vivo—a discrepancy that has also been reported for other variant MYC proteins, such as “MYC-nick” which lacks the entire DNA-binding domain of MYC and yet efficiently promotes transformation in vitro (Conacci-Sorrell et al., 2014). We suggest that the non-essential nature of MbIIIb for in vitro transformation, and a failure to analyze appropriate mutants in more stringent contexts, is the reason MbIIIb has not been the focus of more attention in the past.
MYC box IIIb is extraordinarily conserved. The EEIDVV core of MbIIIb is present in all MYC family members from all animals, including Trichoplax adhaerens, the simplest known metazoan. This EEIDVV core is also present in MYC from the unicellular eukaryote Capsaspora owczarzaki (Young et al., 2011), which possesses a WDR5 homolog that is 75% identical to its human counterpart (Genbank accession: EFW41757.1). The level of conservation of the MbIIIb core highlights the significance of this motif in MYC proteins throughout evolution. Importantly, our analyses reveal the role of both the acidic and the hydrophobic residues in the EEIDVV core for interaction with WDR5. Our ability to account for all residues in the core in terms of WDR5 binding provides a molecular rationalization for the conservation of these residues in MYC and leads to the inexorable conclusion that MbIIIb is conserved because it is a docking site for WDR5. Moreover, because MYC binds WDR5 and not the WD40-protein RBBP5, these data also indicate that MbIIIb is sufficient to discriminate between different types of WD40-repeat containing proteins. Based on the universal conservation of the MbIIIb core motif, and how it engages WDR5, we predict that all MYC proteins associate with WDR5 via this element.
WDR5 as a critical co-factor for MYC proteins
Four observations point to a fundamental role for WDR5 for MYC activity. First, the WDR5-interaction motif is strictly preserved in all metazoan MYC proteins, as discussed above. Second, MYC and WDR5 extensively co-localize on chromatin, as documented in HEK293 and mouse embryonic stem cells. Third, structure-guided point mutations in MYC that disrupt interaction with WDR5 block interaction of MYC with the majority of its target genes, but do not impact MYC expression, localization, interaction with MAX, or binding to naked DNA. Finally, these same mutations attenuate the ability of MYC to promote somatic cell reprogramming and to drive tumorigenesis in mice. Although it is formally possible that these mutations perturb some unknown aspect of MYC behavior, the most parsimonious way to reconcile the evolutionary, structural, genomic, and genetic data presented here is to conclude that direct interaction with WDR5 facilitates MYC recruitment to its target genes, which in turn is required for its functions in iPSc formation and tumorigenesis.
One important question raised by this work centers on the biochemical context in which MYC and WDR5 associate. Recent work by Ullius et al., (2014) may shed light on this issue. Specifically, they reported that MYC binds directly to two components of MLL/SET methyltransferases, WDR5 and ASH2L. Although they did not investigate how MYC associates with WDR5 nor interrogate the significance of the interaction, they examined published ChIP-seq data sets from normal human epidermal keratinocytes and similarly found a widespread overlap of MYC and WDR5 on chromatin and an enrichment of E-boxes in motifs bound by WDR5 (Ullius et al., 2014). In contrast to our study, however, Ullius et al., concluded that these interactions allow MYC to bind functional MLL/SET complexes. This may be possible via interaction of MYC with ASH2L, but cannot occur via the interaction of MYC with WDR5 we describe here. Because MYC binds WDR5 via an interaction surface that is identical to RBBP5, and because RBBP5 is required for the full enzymatic activity of MLL/SET complexes, MYC cannot associate with active and complete MLL/SET complexes via its interaction with WDR5. A similar argument can be made for KANSL2 and the NSL histone acetyltransferase complexes. The observation that WDR5 binding to chromatin precedes that of MYC (Ullius et al., 2014) and is not impacted by the WBM mutation (Figure S5B) strongly implies that WDR5 must be brought to chromatin through a mechanism that is MYC-independent, and likely as part of a chromatin regulatory complex. One possibility is that MYC associates separately with pre-bound but biochemically-distinct forms of ASH2L and WDR5, the latter of which is part of a complex that contains neither RBBP5 nor KANSL2, such as the ATAC histone acetyltransferase (Suganuma et al., 2008). Alternatively, MYC may initially recognize a chromatin-bound MLL/SET (or KANSL2) complex—perhaps by interaction with proteins such as ASH2L—and then selectively disengage RBBP5 (or KANSL2) from the complex, leaving in its wake a ‘ghost’ complex that is enzymatically inactive but anchored to its original location on chromatin. In this way, MYC binding would be linked to active histone modifications, as many studies have shown (Tansey, 2014), but by direct recognition of core enzymatic components, rather than through a dedicated set of ‘reader’ proteins. Further work will be required to decipher the biochemical form of WDR5 that binds MYC.
Therapeutic Possibilities
MYC is one of the most important oncoproteins in human malignancy, suggesting that effective therapies to treat cancer could stem from the discovery of drug-like molecules that interfere with MYC in cancer cells. Consistent with this notion, MYC is a highly-validated anti-cancer target, as genetic inhibition of MYC consistently causes tumor regression in mouse animal models, including in those cancers where MYC is not the primary oncogenic lesion (Soucek et al., 2013). Although recent success has been achieved with small-molecules that inhibit MYC gene transcription via inhibition of the bromodomain-containing protein Brd4 (Delmore et al., 2011), these inhibitors will likely only work in the subset of tumors where MYC transcription is Brd4-dependent. Targeting the MYC protein itself might be expected to more globally inhibit MYC-driven tumors, but the amino-terminal transactivation and carboxyl-terminal DNA-binding domain of MYC are unfolded in solution and difficult to target with small molecules, and there is as yet no small, soluble, and potent inhibitors of MYC function. Our work, however, suggests that the MYC–WDR5 interface may be a viable point for development of MYC inhibitors. Association of MYC with WDR5 appears to be almost entirely mediated by MbIIIb, which binds weakly to WDR5 and in a shallow hydrophobic cleft that buries just ~1000 Å2 of protein surface. The tractable and structured properties of this interface, together with its universal conservation in MYC proteins, make it well-suited for the discovery of small molecules that attenuate MYC function by preventing stable recruitment of MYC to chromatin. This could be a promising strategy for targeting the many and diverse tumors that depend on MYC for their survival.
EXPERIMENTAL PROCEDURES
Plasmids and Recombinant DNA manipulations
Details of plasmids used in this analysis are presented in Extended Experimental Procedures.
Two-hybrid and proteomic screening
Yeast two-hybrid Screening was performed by Hybrigenics Services, S.A.S, France using a Gal4-DBD-MYC (220–270) fusion as bait. For proteomic screening details, see Extended Experimental Procedures.
Production of recombinant proteins
For production of recombinant MYC and WDR5, appropriate pSumo plasmids were transformed into BL21 DE3 Rosetta 2 cells (EMD Millipore) and induced and purified on Ni NTA Agarose (Qiagen). For FPA assays and structure determination, large-scale protein production was carried out by fermentation at 30°C overnight. Additional details are provided in Extended Experimental Procedures.
Structural determination
Amino-terminally truncated WDR5 (24-334) was crystallized in the presence of a five-fold molar excess of the MbIIIb peptide. A single flash-cooled crystal diffracted to 1.9 Å, and data were collected on the Life Sciences Collaborative Access Team (LS-CAT) 21-ID-D beamline at the Advanced Photon Source (APS), Argonne National Laboratory. The WDR5–MbIIIb structure was determined by molecular replacement method using the WDR5-RBBP5 peptide complex (PDB ID: 2XL2) as the search molecule in Phaser (McCoy et al., 2007). The model was refined to a final R and R-free values of 17% and 21%, respectively.
Chromatin immunoprecipitation and analyses
ChIP experiments were performed as described in Extended Experimental Procedures. ChIP-seq data sets have been deposited at the Gene Expression Omnibus (GEO).
Anchorage-independent growth
Soft agar assays, in retrovirally-transduced NIH3T3 cells, were performed as described (Overholtzer et al., 2006). Cultures were grown for 30 days before colonies were counted.
iPSC reprogramming
These experiments were performed using MEFs that carry a GFP–IRES–puro cassette integrated in the Nanog locus (Nanog-GFP) (Maherali et al., 2007). Retroviral transduction was used to express OSK and MYC proteins. Transduced MEFs were plated onto Mitomycin-C treated SNL feeder cells. Three weeks after plating, iPS cell colonies were visualized for activation of the Nanog-GFP reporter and staining for alkaline phosphatase activity. Reprogramming efficiency was calculated as total number of alkaline phosphatase-positive colonies per 100,000 transduced MEFs.
In vivo tumorigenesis assays
6-week-old athymic nude mice (strain Foxn1nu/nu; Harlan Laboratories) were subcutaneously injected with NIH3T3 cells stably expressing MYC proteins. Tumor size was measured every two days and ellipsoidal tumor volume calculated. Tumor mass was determined by resection of tumors from surrounding tissue and weighing. Experiments with mice were pre-approved by the Vanderbilt Institutional Animal Care and Use Committee and followed all state and federal rules and regulations.
Supplementary Material
Acknowledgments
We thank Y. Dou, S. Hiebert, R. Ohi, H. McDonald, A. Reynolds, E. Sturgill, and K. Zaret for reagents, advice, and insightful discussions. This work was supported by the Edward P. Evans Foundation (to W.P.T.), the Melanoma Research Foundation (to W.P.T.), the Vanderbilt Ingram Cancer Center Support grant (NIH: CA68485), the NCI SPORE in Breast Cancer (P50 CA098131), the Cellular, Biochemical, and Molecular Sciences Training Program (T32 GM008554), the National Center for Advancing Translational Sciences (UL1 TR000445), and grants F30AG039164 (to B.C.G), R01NS078289 (to K.C.E.), R01LM011177 (to Z.Z), R01CA148950 (to C.M.E) and 5DP1OD006933 (to S.W.F) from the National Institutes of Health.
Footnotes
Accession numbers. ChIP-seq data are deposited at GEO with accession number GSE60897. Structural data are deposited at the PDB with accession code 4Y7R.
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annibali D, Whitfield JR, Favuzzi E, Jauset T, Serrano E, Cuartas I, Redondo-Campos S, Folch G, Gonzalez-Junca A, Sodir NM, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdic V, Zhang P, Lanouette S, Groulx A, Tremblay V, Brunzelle J, Couture JF. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19:101–108. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- Biegalke BJ, Heaney ML, Bouton A, Parsons JT, Linial M. MC29 deletion mutants which fail to transform chicken macrophages are competent for transformation of quail macrophages. J Virol. 1987;61:2138–2142. doi: 10.1128/jvi.61.7.2138-2142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Ngouenet C, Anderson S, Brabletz T, Eisenman RN. Stress-induced cleavage of Myc promotes cancer cell survival. Genes Dev. 2014;28:689–707. doi: 10.1101/gad.231894.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, Chandriani S, Whitfield ML, Cole MD. A conserved Myc protein domain, MBIV, regulates DNA binding, apoptosis, transformation, and G2 arrest. Mol Cell Biol. 2006;26:4226–4239. doi: 10.1128/MCB.01959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J, Van Nguyen N, Georgiev P, Gaub A, Brettschneider J, Cusack S, Kadlec J, Akhtar A. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 2014;28:929–942. doi: 10.1101/gad.240200.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingar D, Kalkat M, Chan PK, Srikumar T, Bailey SD, Tu WB, Coyaud E, Ponzielli R, Kolyar M, Jurisica I, et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. Journal of proteomics. 2014 doi: 10.1016/j.jprot.2014.09.029. [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Heaney ML, Pierce J, Parsons JT. Site-directed mutagenesis of the gag-myc gene of avian myelocytomatosis virus 29: biological activity and intracellular localization of structurally altered proteins. J Virol. 1986;60:167–176. doi: 10.1128/jvi.60.1.167-176.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nature reviews Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem. 2010;285:32967–32976. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem. 2008;283:32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- Penn LJ, Brooks MW, Laufer EM, Land H. Negative autoregulation of c-myc transcription. Embo J. 1990;9:1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. Structural basis for molecular recognition and presentation of histone H3 by WDR5. Embo J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, de Lange T, Ramsay G, Jakobovits E, Bishop JM, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- Tansey WP. Mammalian MYC proteins and cancer. New Journal of Science. 2014;2103:1–27. [Google Scholar]
- Ullius A, Luscher-Firzlaff J, Costa IG, Walsemann G, Forst AH, Gusmao EG, Kapelle K, Kleine H, Kremmer E, Vervoorts J, et al. The interaction of MYC with the trithorax protein ASH2L promotes gene transcription by regulating H3K27 modification. Nucleic Acids Res. 2014;42:6901–6920. doi: 10.1093/nar/gku312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Seminars in cancer biology. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SL, Diolaiti D, Conacci-Sorrell M, Ruiz-Trillo I, Eisenman RN, King N. Premetazoan ancestry of the Myc-Max network. Molecular biology and evolution. 2011;28:2961–2971. doi: 10.1093/molbev/msr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.