Abstract
Background
Genome-wide association studies have identified associations of genetic variants at 17q21 near ORMDL3 with childhood asthma.
Objectives
To find out whether associations in this region are specific to particular asthma phenotypes and specific to ORMDL3.
Methods
We examined associations between 244 independent single nucleotide polymorphisms (SNPs) plus 13 previously identified asthma-related SNPs in the region between 34 and 36 Mb on chromosome 17 and early wheezing phenotypes, doctor-diagnosed asthma and atopy at 7½ years, bronchial hyper-responsiveness and lung function at 8½ years in 7,045 children from the Avon Longitudinal Study of Parents and Children (ALSPAC) birth cohort study. With this, cis expression quantitative trait loci (eQTL) signals for the same SNPs were assessed in 875 samples across genes in the same region.
Results
The strongest evidence for phenotypic association was seen for persistent wheezing (rs8076131 near ORMDL3, relative risk ratio (RRR) 1.60 (95% CI 1.40, 1.84), p=1.4×10−11, rs2305480 near GSDML 1.60; 1.39-1.83, p=1.5×10−11 and rs9303277 near IKZF3 1.57; 1.37-1.79, p=4.4×10−11). Similar, but less precisely estimated effects were seen for intermediate-onset wheeze, but there was little evidence of associations with other wheezing phenotypes. There was some evidence of associations with bronchial hyper responsiveness. SNPs across the whole region show strong evidence of association with differential levels of expression at GSDML, IKZF3 and MED24, as well as ORMDL3.
Conclusions
Associations of SNPs in the 17q21 locus are specific to asthma and to specific wheezing phenotypes, and are not explained by associations with intermediate phenotypes, such as atopy or lung function.
Keywords: ALSPAC, wheezing phenotypes, chromosome 17, ORMDL3, gene expression
INTRODUCTION
Asthma is a complex disorder resulting from interactions of genes and environmental factors 1,2. Associations of candidate genes with asthma have been reported, but few were replicated in independent studies 3. Lack of replication may be explained in part by difficulties in defining the asthma phenotype, and by gene-environment interactions leading to variable genetic expression in differing settings 4. Recently, a genome wide association study (GWAS) identified multiple single nucleotide polymorphisms (SNPs) on locus 17q21 that were associated with asthma in several populations of European descent 5. The association of SNPs in this region with asthma has been replicated in similar 6-9 and ethnically diverse 10,11 populations. Most 6-8, but not all 12, studies found that associations were stronger in asthma beginning in early childhood.
Heterogeneity in early childhood wheezing illness and its association with subsequent asthma 13,14 led us to describe 15 and replicate 16 phenotypes of wheezing during the first seven years of childhood, based on data from population-based birth cohort studies. These phenotypes are differentially associated with physician-diagnosed asthma, atopy, lung function and bronchial responsiveness 15. Investigation of the biological origins of well-characterized asthma-related phenotypes is crucial to the better understanding of this often ill-defined clinical entity. Evidence points to early childhood as a critical period in asthma development 17 so we aimed to assess the association of asthma-related phenotypes with a comprehensive set of common, independent variants within the previously identified asthma-associated region of chromosome 17, and to examine the association of these variants with gene expression in the same region.
SUBJECTS AND METHODS
Subjects
The Avon Longitudinal Study of Parents and Children (ALSPAC) is a longitudinal, population-based birth cohort study that recruited pregnant women resident in Avon, UK with expected dates of delivery 1st April 1991 to 31st December 1992. The study methodology has been described in detail previously 18. Ethical approval was obtained from the ALSPAC Law and Ethics Committee and the Local Research Ethics Committees.
Asthma outcomes and wheezing phenotypes
Reports of physician-diagnosed asthma were obtained from questionnaires sent to the mother when the child was aged 7½ years. Skin prick testing (SPT) was performed in all participants at a research clinic at age 7½ years 19. A positive response was defined as a mean weal diameter of >2 mm with no response to negative control solution, and atopy was defined as a positive response to one or more of house dust mite, cat or mixed grass. Spirometry in all participants and bronchial responsiveness to methacholine was measured in a research clinic at age 8½ years 20. Forced vital capacity (FVC), forced expiratory volume in 1 s (FEV1) and forced expiratory flow 25–75% (FEF25_75) were converted to sex-, age- and height-adjusted z-scores. Bronchial hyper-responsiveness (BHR) was defined as being in the highest tertile of the dose-response slope. Responses to the question, “In the past 12 (6 for first questionnaire) months has (your child) had wheezing/wheezing with whistling on the chest?” at 6, 18, 30, 42, 54, 69 and 81 months after birth were used in a longitudinal latent class analysis to define phenotypes of childhood wheezing 15, in children with at least two questionnaire responses. The best fitting model resulted in six phenotypes: 1.”never/infrequent wheeze “ with approximately 10% prevalence of wheezing at 6 months and declining prevalence of sporadic wheeze thereafter, including children who never reported wheeze; 2.”transient early wheeze” with 50–60% prevalence of wheeze up to 18 months, declining to low prevalence from 42 months; 3.”prolonged early wheeze” with peak prevalence of wheeze around 65% at 30 months, declining to low prevalence from 69 months; 4.”intermediate onset wheeze” with low prevalence of wheeze up to 18 months, rising rapidly to high prevalence from age 42 months; 5.”late onset wheeze” with approximately 20% prevalence of wheeze up to 42 months, rising to more than 50% prevalence thereafter; and 6.”persistent wheeze” (7%) with 65% prevalence of wheeze at 6 months and approximately 90% prevalence thereafter. Trajectories of prevalence of wheeze for each phenotype are presented in Figure 1 of Henderson et al.15 and Figure 1 of Savenjie & Granell et al. 16.
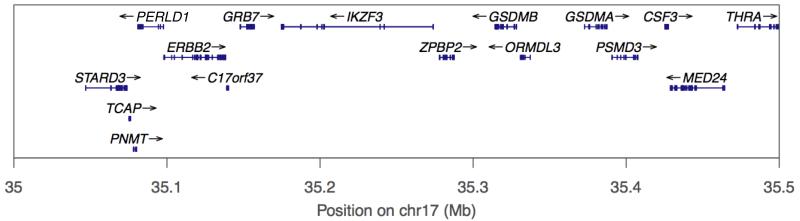
Figure 1.
Schematic of chromosome 17 between 35.0 and 35.5 Mb. Vertical red lines delineate region between 35.2 and 35.4 Mb around ORMDL3 (marked in green) containing 13 asthma-related SNPs 5, 22, 23.
Genetic data
Details on genotyping, quality control and exclusions can be found in the online repository.
Previously reported SNPs associated with childhood asthma are located in the region containing ORMDL3 and 14 nearby genes (Online Table A1 & Figure 1), between 35.0 and 35.5Mb on chromosome 17 5. To allow for associations outside this locus, SNPs within the region between 34.0 and 36.0 Mb were extracted in the form of allelic dosages. To avoid redundancy, linkage disequilibrium (LD) based data pruning was undertaken based on a pairwise tagging approach. Using analysis software Haploview v4.2 21 and after taking best genotypes from the imputation process (best guess genotypes were only used for the variant pruning process), this removed SNPs correlated with remaining tags at r2≥0.80. From an initial set of 749 SNPs across the region, pruning yielded a working data set of 257 variants including 13 SNPs at the 17q21 locus (35.2 to 35.4 Mb) previously reported to be associated with asthma. As a sensitivity analysis and to further explore the patterns of association with gene expression, a further pruned SNP set was generated using a more stringent threshold r2≥0.20, yielding a set of 63 variants.
Expression data
mRNA-quantified expression data were collected for 947 unrelated ALSPAC participants from lymphoblastoid cell lines (LCLs) established from blood samples taken when the children were 9 years old. Of these, 875 also had genotypic data available and 815 also had data on asthma status at 7½ years. For more technical details see the online repository.
Children with expression data were selected on the basis of availability of cell lines and completeness of phenotypic data across a wide range of outcomes, neither of which was associated with asthma outcome.
Statistical analyses
Multinomial logistic regression was used to estimate relative risk ratios (RRR), also known as “multinomial odds ratios”, with 95% confidence intervals (CI) and p-values for associations of SNP dosages with wheezing phenotypes (never/infrequent wheeze was used as the reference group). Posterior probabilities of phenotype membership for each child were used as weights in the regression models, in order to account for uncertainty in phenotype membership. Logistic, ordinal logistic and linear regression models were used to estimate associations of SNP dosages with asthma, atopy, BHR and lung function outcomes. We included terms for interaction with atopy in logistic regression models of the association of the top-ranked SNPs with asthma. The direction of association depends on the arbitrary assignment of one allele (that with greatest population frequency) as “wild-type”. Therefore, RRRs and odds ratios (ORs) less than 1 were inverted, and mean differences were coded as absolute values, so all associations are reported as positive and reported risk alleles are swapped accordingly. Conditional analyses were performed by adjusting for the dosages of 13 SNPs previously reported as asthma-related5,22,23 (Online Table A1). Q-Q plots were used to assess overall evidence for associations. The cumulative incidence of wheezing from 6 to 81 months was estimated using life tables in a survival-time data framework. We estimated the population-attributable risk fraction (PARF) to quantify the reduction in prevalence of a wheezing phenotype if the genotype in all subjects was changed to “no risk alleles”:
Normalized and transformed expression levels at each of the 15 genes (STARD3 to MED24) in the region 35.0 to 35.5Mb were regressed on SNP dosages in a linear regression model assuming an additive genetic model as for other analyses. The variance in expression levels explained by genotypic variation (r2) was reported along with p-values adjusted for 3855 independent tests (945 when the reduced SNP set based on a linkage disequilibrium (LD) pruning threshold of r2≥0.8 was used) using Bonferroni-corrections.
All analyses were performed using Stata Version 11.0 (Stata Corp, College Station, Texas).
RESULTS
A total of 7,045 children with central-European ancestry had at least two questionnaire responses on wheezing between 6 and 81 months and data on SNP dosages. Of these children, 3595 (51.0%) were males, 1085 (19.7%) had asthma, 3140 (46.7%) had an asthmatic or allergic mother, 1516 (22.3%) were exposed to maternal smoking during pregnancy and 948 (20.1%) were atopic (Table 1). The estimated numbers (%)with each wheezing phenotype were never/infrequent (4815, 68.4%), transient early (715, 10.2%), prolonged early (526, 7.5%), intermediate-onset (162, 2.3%), late-onset (335, 4.8%) and persistent wheezing (492, 7.0%)
Table 1.
Characteristics of children included in analyses
| Number of children with data available | Number (%) of children with characteristica | Sub-sample with expression data (n=947) | Number (%) with characteristic in subsamplea | |
|---|---|---|---|---|
| Male sex | 7045 | 3595 (51.0) | 875 | 413 (47.2) |
| Maternal history of asthma or allergy | 6723 | 3140 (46.7) | 855 | 423 (49.5) |
| Paternal history of asthma or allergy | 4963 | 1994 (40.2) | 671 | 267 (39.8) |
| Prenatal maternal smoking | 6786 | 1516 (22.3) | 857 | 136 (15.9) |
| Maternal lower education levelb | 6853 | 4012 (58.5) | 864 | 445 (51.5) |
| Wheezing phenotypec | 7045 | 947 | ||
| Never/Infrequent | 4815 (68.4) | 682 (72.0) | ||
| Transient Early | 715 (10.2) | 81 (8.6) | ||
| Prolonged Early | 526 (7.5) | 58 (6.1) | ||
| Intermediate Onset | 162 (2.3) | 20 (2.1) | ||
| Late Onset | 335 (4.8) | 48 (5.1) | ||
| Persistent | 492 (7.0) | 58 (6.1) | ||
| Doctor-diagnosed asthma ever by 7½ years | 5497 | 1085 (19.7) | 815 | 149 (18.3) |
| Skin prick test sensitivity at 7.5 years | 4710 | 948 (20.1) | 728 | 123 (16.9) |
| Mean [IQR] FVCd at 8.5 years (l) | 4831 | 0.02 [1.26] | 833 | 0.03 [1.23] |
| Mean [IQR] FEV1d at 8.5 years (l) | 4758 | 0.02 [1.29] | 821 | 0.04 [1.20] |
| Mean [IQR] FEF25_75d at 8.5 years (l/s) | 4831 | 0.02 [1.33] | 833 | 0.02 [1.21] |
| BHRd at 8.5 years | 3216 | 875 | ||
| No BHR (LSDRS<0) | 616 (19.2) | 93 (15.5) | ||
| Positive BHR (1st tertile) | 869 (27.0) | 152 (25.3) | ||
| Positive BHR (2nd tertile) | 867 (27.0) | 184 (30.7) | ||
| Positive BHR (3rd tertile) | 864 (27.0) | 171 (28.5) |
Unless labeled as Mean [IQR]
Educated to GCE level (school leaving certificate) or lower
Estimated using posterior probabilities
FVC= forced vital capacity (adjusted z-scores), FEV1= forced expiratory volume in 1 s (adjusted z-scores), FEF25_75= forced 6 expiratory flow 25–75% (adjusted z-scores), BHR=bronchial hyper-responsiveness, LSDRS=least square dose response slope
Associations of ORMDL3 region SNPs with doctor-diagnosed asthma, atopy, BHR and lung function
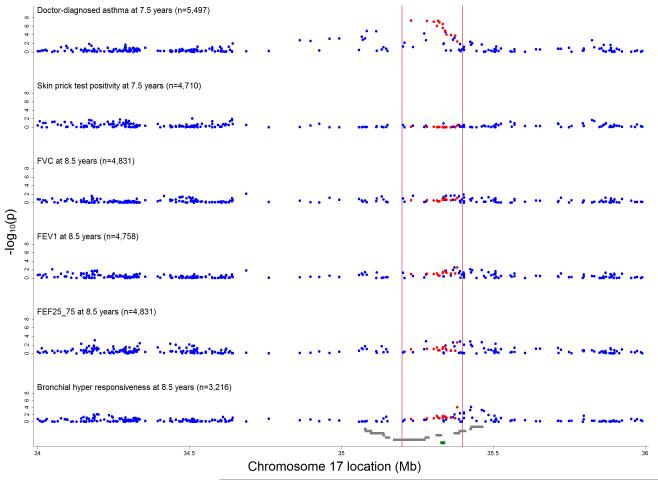
Q-Q plots showed substantial deviation from expected values for doctor-diagnosed asthma ever at 7½ years, and small deviation for BHR but not for the other phenotypes (Online Figure A1). Consistent with previously reported GWAS, the strongest evidence of association with doctor-diagnosed asthma was for variants in the region between 35.0 and 35.5 Mb (Figure 2). Corresponding conditional p-values are plotted in the Online Figure A2 which shows that the 13 previously asthma-related SNPs explain the majority of the observed associations. The strongest evidence for association with doctor-diagnosed asthma was for the previously asthma-related SNPs: rs9303277 near IKZF3 (OR = 1.30; 95% CI 1.19-1.43, p =4.6×10−8) and rs2290400 near GSDML (OR = 1.30; 1.18-1.43, p=4.7×10−8 (Online Table A2 & A3). The strongest evidence of association with BHR was for rs1042658 near CSF3 (1.20; 1.10-1.31, p=7.5×10−5) and for the previously asthma-related SNP rs3859192 near GSDM1 (1.19; 1.09-1.30, p=8.2×10−5) (Online Table A3). Corresponding figures for crude (Online Figure A3) and conditional (Online Figure A4) ORs and mean differences for the various asthma outcomes can be found in the online repository. No evidence of effect modification by atopy was observed in the association between top 10 ranked SNPs and asthma (Online Table A4). Conditioning on each of the top SNPs did not show evidence of independent associations with doctor-diagnosed asthma ever (See Online Repository and Online Table A5).
Figure 2.
Inverse log10 transformed P-values for crude associations of 257 independent SNPs between 34 and 36 Mb on chromosome 17 with doctor-diagnosed asthma ever, skin prick test positivity, bronchial hyper- responsiveness, FVC, FEV1 and FEF25_75. Vertical red lines delineate region between 35.2 and 35.4 Mb around ORMDL3 (marked in green) containing 13 asthma-related SNPs 5, 22, 23 (highlighted in red). Grey gene tracks show loci in the order PNMT, PERLD1, ERBB2, C17orf37, GRB7, IKZF3, ZPBP2, GSDML, GSDM1, PSMD3, CSF3, MED24.
Associations of ORMDL3 region SNPs with wheezing phenotypes
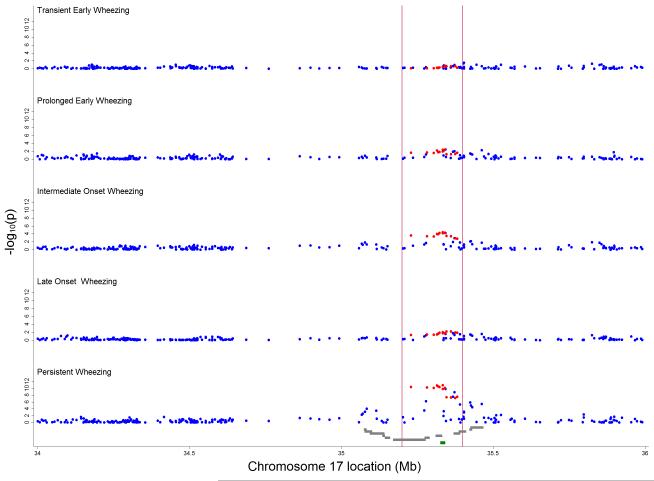
For the intermediate-onset and persistent wheezing phenotypes, Q-Q plots showed substantial deviations from expected values (Online Figure A5). The strongest evidence of association was for SNPs in the region between 35.2 and 35.4 Mb with persistent wheezing, although there was some evidence of association for SNPs in the flanking regions between 35.0 and 35.2 and between 35.4 and 35.6 Mb (Figure 3). For the other phenotypes, there was little evidence of any association.
Figure 3.
Inverse log10 transformed P-values for crude associations of 257 independent SNPs between 34 and 36 Mb on chromosome 17 with wheezing phenotypes (compared with never/infrequent wheeze). Vertical red lines delineate region between 35.2 and 35.4 Mb around ORMDL3 (marked in green) containing 13 asthma-related SNPs 5, 22, 23 (highlighted in red). Grey gene tracks show loci in the order PNMT, PERLD1, ERBB2, C17orf37, GRB7, IKZF3, ZPBP2, GSDML, GSDM1, PSMD3, CSF3, MED24.
SNPs near the IKZF3, ZPBP2, GSDML, ORMDL3 and GSDM1 genes were associated with intermediate-onset (p-values ≤0.001) and persistent wheezing (p-values ≤1.5×10−9) (Table 2). The magnitude of associations appeared similar for these two phenotypes (RRR in the range 1.46 to 1.60), and were generally larger than the corresponding odds ratios for association with doctor-diagnosed asthma (OR<1.30). The strongest evidence of association was for persistent wheezing and rs8076131 near ORMDL3 (RRR 1.60; 95% CI 1.40-1.84, p=1.4×10−11), rs2305480 near GSDML (1.60; 1.39-1.83, p=1.5×10−11) and rs9303277 near IKZF3 (1.57; 1.37-1.79, p=4.4×10−11) (Table 2). Respective associations for the less frequent (162 children, prevalence 2.3%) intermediate-onset wheezing phenotype were: 1.60; 1.27-2.02, p=7.5×10−5, 1.58; 1.25-1.99, p=1.1×10−4 and 1.52; 1.21-1.91, p=2.8×10−4. In contrast, associations with other wheezing phenotypes were smaller (RRR < 1.20) (Table 2). Similarly, genes in the flanking regions were only associated with intermediate-onset and persistent wheezing, but the magnitude of associations was smaller. A list of the top crude associations for intermediate-onset and persistent wheezing can be found in the Online Table A3.
Table 2.
Association of variants in the region of chromosome 17 between 35.0 and 35.5 Mb (around 17q21) with wheezing phenotypes in 7,045 children
| Nearby gene a | SNP b | Position (Mb) | MAF c | Overall P-value | Ph d | Risk allele± | RRR (95%CI) e | P-value for Ph | Other SNPs (overall p-value) f |
|---|---|---|---|---|---|---|---|---|---|
|
Region between 35.0 and 35.2 Mb
g | |||||||||
| PNMT | rs876493 | 35.078071 | 0.41 | 0.002 | TE | A (common) | 1.02 (0.91, 1.14) | 0.79 | NA |
| PE | G | 1.10 (0.97, 1.25) | 0.13 | ||||||
| IO | G | 1.35 (1.06, 1.73) | 0.02 | ||||||
| LO | G | 1.08 (0.93, 1.26) | 0.33 | ||||||
| P | G | 1.29 (1.11, 1.48) | 6.52×10−4 | ||||||
|
| |||||||||
| PERLD1 | rs2941504 | 35.084426 | 0.31 | 0.001 | TE | G (common) | 1.01 (0.91, 1.13) | 0.81 | NA |
| PE | A | 1.06 (0.94, 1.20) | 0.36 | ||||||
| IO | A | 1.27 (1.00, 1.60) | 0.05 | ||||||
| LO | A | 1.10 (0.95, 1.28) | 0.21 | ||||||
| P | A | 1.31 (1.14, 1.51) | 9.98×10−5 | ||||||
|
| |||||||||
| ERBB2 | rs2952155 | 35.229995 | 0.24 | 0.004 | TE | C (common) | 1.03 (0.91, 1.16) | 0.64 | No signals |
| PE | T | 1.06 (0.93, 1.22) | 0.39 | ||||||
| IO | T | 1.29 (1.00, 1.67) | 0.05 | ||||||
| LO | T | 1.10 (0.93, 1.29) | 0.27 | ||||||
| P | T | 1.31 (1.13, 1.53) | 3.68×10−4 | ||||||
|
| |||||||||
|
Region between 35.2 and 35.4 Mb
| |||||||||
| IKZF3 | rs9303277 | 35.229995 | 0.48 | 2.24×10−11 | TE | T (common) | 1.02 (0.92, 1.12) | 0.77 | rs9635726 (0.004) |
| PE | C | 1.14 (1.02, 1.28) | 0.024 | ||||||
| IO | C | 1.52 (1.21, 1.91) | 2.81×10−4 | ||||||
| LO | C | 1.15 (1.00, 1.32) | 0.05 | ||||||
| P | C | 1.57 (1.37, 1.79) | 4.43×10−11 | ||||||
|
| |||||||||
| ZPBP2 | rs11557467 | 35.282160 | 0.48 | 3.52×10−11 | TE | T (common) | 1.02 (0.92, 1.13) | 0.73 | rs8067378 (6.37×10−11) |
| PE | G | 1.14 (1.02, 1.28) | 0.02 | rs12150079 (3.57×10−6) | |||||
| IO | G | 1.50 (1.20, 1.89) | 4.11×10−4 | ||||||
| LO | G | 1.16 (1.01, 1.33) | 0.04 | ||||||
| P | G | 1.56 (1.37, 1.78) | 5.99×10−11 | ||||||
|
| |||||||||
| GSDML | rs2305480 | 35.323475 | 0.48 | 1.69×10−12 | TE | T (common) | 1.03 (0.93, 1.15) | 0.51 | rs7216389 (4.82×10-12) |
| PE | T (common) | 1.17 (1.05, 1.31) | 0.006 | rs2290400 (6.66×10-12) | |||||
| IO | T (common) | 1.58 (1.25, 1.99) | 1.11×10−4 | ||||||
| LO | T (common) | 1.16 (1.01, 1.34) | 0.03 | ||||||
| P | T (common) | 1.60 (1.39, 1.83) | 1.5×10−11 | ||||||
|
| |||||||||
|
Region between 35.2 and 35.4 Mb
| |||||||||
| ORMDL | rs8076131 | 35.334438 | 0.48 | 8.55×10−13 | TE | A (common) | 1.05 (0.95, 1.17) | 0.31 | rs4795405 (3.08×10−12) |
| PE | A (common) | 1.18 (1.06, 1.33) | 0.004 | rs4378650 (6.76×10−12) | |||||
| IO | A (common) | 1.60 (1.27, 2.02) | 7.52×10−5 | rs4794820 (7.04×10−12) | |||||
| LO | A (common) | 1.19 (1.03, 1.37) | 0.02 | rs8079416 (1.06×10−8) | |||||
| P | A (common) | 1.60 (1.40, 1.84) | 1.41×10−11 | rs3744246 (5.57×10−4) | |||||
|
| |||||||||
| GSDM1 | rs3902025 | 35.372780 | 0.44 | 1.20×10−9 | TE | T (common) | 1.09 (0.98, 1.20) | 0.12 | rs4795408 (1.38×10−8) |
| PE | T (common) | 1.17 (1.04, 1.31) | 0.009 | rs3859192 (2.23×10−8) | |||||
| IO | T (common) | 1.46 (1.16, 1.85) | 0.001 | rs7219080 (5.34×10−8) | |||||
| LO | T (common) | 1.15 (1.00, 1.33) | 0.05 | rs3894194 (6.36×10−8) | |||||
| P | T (common) | 1.53 (1.33, 1.75) | 1.46×10−9 | ||||||
|
| |||||||||
|
Region between 35.4 and 35.5 Mb
| |||||||||
| PSMD3 | rs2227321 | 35.424820 | 0.37 | 1.60×10−6 | TE | G (common) | 1.00 (0.90, 1.11) | 0.95 | rs8075668 (6.80×10−6) |
| PE | G (common) | 1.14 (1.01, 1.28) | 0.04 | rs2241245 (0.003) | |||||
| IO | G (common) | 1.38 (1.09, 1.76) | 0.009 | ||||||
| LO | G (common) | 1.17 (1.01, 1.35) | 0.04 | ||||||
| P | G (common) | 1.42 (1.23, 1.64) | 1.36×10−6 | ||||||
|
| |||||||||
| CSF3 | rs1042658 | 35.427428 | 0.38 | 3.45×10−5 | TE | T | 1.05 (0.95, 1.16) | 0.36 | NA |
| PE | T | 1.12 (1.00, 1.26) | 0.05 | ||||||
| IO | T | 1.36 (1.08, 1.70) | 0.008 | ||||||
| LO | T | 1.15 (1.00, 1.32) | 0.05 | ||||||
| P | T | 1.34 (1.18, 1.53) | 1.13×10−5 | ||||||
|
| |||||||||
| MED24 | rs8065443 | 35.462466 | 0.41 | 5.84×10−6 | TE | A | 1.07 (0.96, 1.18) | 0.22 | rs2302776 (1.77×10−4) |
| PE | A | 1.18 (1.05, 1.32) | 0.006 | ||||||
| IO | A | 1.31 (1.05, 1.65) | 0.02 | ||||||
| LO | A | 1.17 (1.02, 1.35) | 0.03 | ||||||
| P | A | 1.36 (1.2, 1.56) | 4.15×10−6 | ||||||
From online database SNP Function Prediction (FuncPred): http://manticore.niehs.nih.gov/snpfunc.htm
SNP with the smallest overall p-value, only for genes for which there is evidence of association for at least one variant (p<0.001). A complete list of results for all SNPs is available from the authors on request. SNPs highlighted in bold are the 13 previously reported asthma-related SNPs5 22 23.
MAF=Minor allele frequency, RRR=Relative risk ratio
Ph= Phenotype: TE=Transient early, PE=Prolonged early, IO=Intermediate onset, LO=Late onset, P=Persistent
Risk allele is rare allele unless indicated otherwise
Overall p-values for other SNPs within the same gene which are also associated with intermediate onset and persistent wheezing only.
Conditional analyses including the 13 previously asthma-related SNPs essentially removed any evidence for association (Online Figure A6) and demonstrated that these original signals explain the majority of observed signal across the region. Corresponding figures for crude (Online Figure A7) and conditional (Online Figure A8) RRRs for the wheezing phenotypes can be found in the online repository.
Online Figure A9 shows a low cumulative incidence of wheezing up to approximately 6 months with a large increment between 6 and 18 months. Stratification by top SNPs rs8076131 near ORMDL3 and rs2305480 near GSDML shows the lack of association with wheeze at 6 months, and emergence of association from age 18 months, together with possible departures from a per-allele genetic model (particularly for rs2305480). The estimated PARF for the top SNP rs8076131 near ORMDL3 was 35% for intermediate-onset and 40% for persistent wheezing.
Associations of ORMDL3 region SNPs with expression
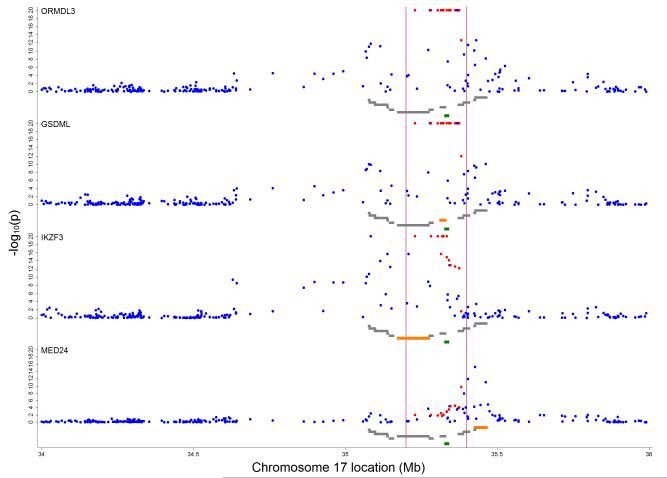
Expression of ORMDL3, GSDML, IKZF3 and MED24 genes showed evidence for association with genetic variation in the region between 35.0 and 35.5 Mb (Figure 4). Expression of other genes showed little evidence for association (Online Figure A10). Of those SNPs showing strongest evidence for association with the wheezing phenotypes (as reported in Table 2), greatest evidence for association was exhibited by the SNPs: rs9303277 (in IKZF3) for ORMDL3, IKZF3 and GSDML expression (r2=0.20, Bonferroni corrected p-value =1.39×10−44, r2=0.13, p=2.53×10−27 and r2=0.19, p=6.09×10−44 respectively), rs11557467 (in ZPBP2) for ORMDL3, IKZF3 and GSDML expression (r2=0.20, p=6.75×10−45, r2=0.13, p=1.78×10−26 and r2=0.20, p=3.64×10−45 respectively), rs2290400 (in GSDML) for ORMDL3, IKZF3 and GSDML expression (r2=0.20, p=3.96×10−45, r2=0.12, p=2.43×10−25 and r2=0.19, p=4.17×10−43 respectively) and rs4378650 (in ORMDL3) for ORMDL3, IKZF3 and GSDML expression (r2=0.18, p=8.00×10−41 , r2= 0.12, p=3.39×10−23 and r2=0.16, p=8.39×10−36 respectively) amongst other associations (Table 3 and online Table A6). The strongest evidence for variable expression at MED24 was seen for rs1042658 (in CSF3) (r2=0.07, p=2.62×10−12) which was the SNP showing the strongest association with BHR. A list of the top 10 crude associations for expression of ORMDL3, GSDML, IKZF3 and MED24 genes can be found in the online Table A7. In a reduced set of 63 SNPs based on a more stringent LD pruning threshold of r2≥0.2, associations with expression were still concentrated on the loci IKZF3, GSDML, ORMDL3, MED24 and ZPBP2 (Online Table A8). We found no evidence of associations between expression and wheezing phenotypes (See online repository and online Table A9).
Figure 4.
Inverse log10 transformed P-values for crude associations of 257 independent SNPs between 34 and 36 Mb on chromosome 17 with gene specific expression levels. Vertical red lines delineate region between 35.2 and 35.4 Mb around ORMDL3 (marked in green) containing 13 asthma-related SNPs 5, 22, 23 (highlighted in red). Grey gene tracks show loci in the order PNMT, PERLD1, ERBB2, C17orf37, GRB7, IKZF3, ZPBP2, GSDML, GSDM1, PSMD3, CSF3, MED24 with the reported marker in orange. Figure shows results for ORMDL3, GSDML, IKZF3 and MED24.
Table 3.
Association of variants in the region of chromosome 17 between 35.0 and 35.5 Mb (around 17q21) with expression quantitative traits for local genes in 875 children
| Nearby gene | SNP a | ORMDL3 expression (r2) b | pc | Other eQTLd | expression (r2) b | pc |
|---|---|---|---|---|---|---|
|
Region between 35.0 and 35.2 Mb
| ||||||
| PNMT | rs876493 | 0.05 | 3.38×10−8 | IKZF3 | 0.05 | 6.28×10−8 |
| GSDML | 0.05 | 4.50×10−7 | ||||
|
| ||||||
| PERLD1 | rs2941504 | 0.05 | 6.10×10−9 | IKZF3 | 0.08 | 1.47×10−14 |
| GSDML | 0.05 | 5.37×10−7 | ||||
|
| ||||||
| ERBB2 | rs2952155 | 0.05 | 2.48×10−8 | IKZF3 | 0.43 | 4.08×10−11 |
| GSDML | −0.33 | 1.93×10−5 | ||||
|
| ||||||
|
Region between 35.2 and 35.4 Mb
| ||||||
| IKZF3 | rs9303277 | 0.20 | 1.39×10−44 | IKZF3 | 0.13 | 2.53×10−27 |
| ZPBP2 | 0.02 | 0.04 | ||||
| GSDML | 0.19 | 6.09×10−44 | ||||
|
| ||||||
| ZPBP2 | rs11557467 | 0.20 | 6.75×10−45 | IKZF3 | 0.13 | 1.78×10−26 |
| ZPBP2 | 0.02 | 0.09 | ||||
| GSDML | 0.20 | 3.64×10−45 | ||||
|
| ||||||
| GSDML | rs2305480 | 0.16 | 1.78×10−35 | IKZF3 | 0.07 | 1.15×10−12 |
| GSDML | 0.17 | 6.33×10−36 | ||||
|
| ||||||
| ORMDL3 | rs8076131 | 0.16 | 4.05×10−34 | IKZF3 | 0.07 | 4.86×10−12 |
| GSDML | 0.16 | 4.03×10−33 | ||||
|
| ||||||
| GSDM1 | rs3902025 | 0.10 | 1.40×10−19 | IKZF3 | 0.02 | 0.02 |
| GSDML | 0.10 | 1.83×10−20 | ||||
| MED24 | 0.02 | 0.46 | ||||
|
| ||||||
|
Region between 35.4 and 35.5 Mb
| ||||||
| PSMD3 | rs2227321 | 0.05 | 2.79×10−7 | GSDML | 0.04 | 0.001 |
| MED24 | 0.02 | 0.20 | ||||
|
| ||||||
| CSF3 | rs1042658 | 0.03 | 0.001 | GSDML | 0.03 | 0.001 |
| MED24 | 0.07 | 2.62×10−12 | ||||
|
| ||||||
| MED24 | rs8065443 | 0.04 | 2.59×10−5 | GSDML | 0.05 | 3.77×10−7 |
| MED24 | 0.05 | 3.84×10−8 | ||||
SNP near each gene with the smallest overall p-value from primary analyses with wheeze phenotypes (Table 2) and their relationships with expression quantitative traits for genes in the ORMDL3 region of chromosome 17. Supplemental Table A6 shows the complete list of expression quantitative traits for the SNPs reported in this table. A complete list of SNPs and their relationship with expression quantitative traits for all loci in this region is available from the authors.
r2 refers to variance in expression explained in expression levels by SNP. For all results, 875 samples were available with both genotype and expression data.
Bonferroni corrected p values (based on 3855 tests)
Other eQLT, results are shown where evidence for association exceeds a threshold when taking a Bonferroni corrected p value for 3855 tests (257 independent loci*15 expression quantitative traits). All raw p values are <0.0002.
DISCUSSION
Common SNPs in the 17q21 locus are associated with childhood asthma. Based on a birth cohort study with detailed information on phenotypes of asthma and allergic disease we found that these SNPs were strongly associated with persistent and intermediate-onset childhood wheezing phenotypes, characterized by onset before age 30 months 15. These SNPs were also strongly associated with a diagnosis of asthma in mid-childhood as previously reported5,22,23, however we found that the magnitude of these associations was smaller and the p-values were weaker than for persistent wheeze. Little evidence of associations with early wheezing that resolved, or with late-onset wheezing were found. We did not find evidence that SNPs in the 17q21 locus were associated with atopy or lung function in mid-childhood, but found some evidence of associations with BHR. SNPs in the 17q21 locus were associated with expression of the ORMDL3, GSDML, IKZF3 and MED24 genes but no association between asthma phenotypes and expression data was found. Our analyses were well-powered to detect associations of modest strength within phenotype groups. We have previously shown that these phenotypes have face validity in their associations with other markers of asthma and its intermediate phenotypes and can be replicated in independent data 16. The phenotypes reported here have some consonance with those described by Stein & Martinez in the Tucson study 13,14. The transient early phenotype is similar in both studies, the ALSPAC prolonged early wheezing phenotype corresponds approximately to the Tucson “non-atopic wheezers”, and our intermediate-onset, late-onset and persistent wheezing phenotypes, all of which were associated with atopy and bronchial hyper-responsiveness, correspond approximately to the Tucson “IgE-associated wheeze/asthma” phenotype, although with earlier age of onset. Our study provides further evidence for the distinct nature of these phenotypes, based on their differential associations with genetic variants in the 17q21 region.
Studies reporting associations of SNPs in the 17q21 locus with early-onset asthma have used varying definitions of this outcome. Bouzigon et al. used ordered subset regression to classify early-onset asthma (<4 years), which was strongly associated with four SNPS in the 17q21 locus 7. Flory et al. reported that associations did not differ according to age of onset 12, while Bisgaard et al. reported evidence of a stronger association of SNPs in the 17q12-21 region with asthma that began below the age of 3 years 6 and, consistent with our results, a lack of association with allergic sensitization. They suggested that these associations reflected increased susceptibility to non-atopic asthma. However, the situation may be more complicated than a simple dichotomy between atopic and non-atopic disease. Galanter et al. reported stronger associations between ORMDL3 SNPs and asthma when the latter was associated with raised serum IgE concentrations 10.
In the first report of the association of the 17q21 locus with asthma, Moffatt et al. suggested ORMDL3 as a promising candidate on the basis of gene expression studies in EBV-transformed lymphoblastoid cell lines 5. However, the function of ORMDL3 remains to be fully elucidated and it is possible that other genes in this region, or more distant genes, contain the true causal variants 24. We confirmed that SNPs at the 17q21 locus were associated with expression of ORMDL3 but, additionally, identified associations with GSDML, IKZF3 and MED24. ORMDL3 encodes a member of a family of transmembrane proteins that are anchored in the endoplasmic reticulum, ubiquitously expressed in adult and foetal tissues and show evidence of functional conservation between species 25. It has recently been reported to bind and inhibit sarco-endoplasmic reticulum Ca2+ ATPase (SERCA) activity, which may contribute to airway remodelling in asthma 26 and provides a plausible mechanisms whereby ORMDL3 may contribute to non-allergic mechanisms of airway inflammatory responses 27.
The loci, other than ORMDL3, implicated in this study have a number of possible connections to the persistent wheeze phenotype and to measureable changes in gene expression. Firstly, GSDML, IKZF3 and ZPBP2 (and MED24/THRAP4 by way of regional association) have all been directly highlighted in GWAS for conditions that have an aetiological contribution from autoimmunity, including ulcerative colitis 28, rheumatoid arthritis 29, biliary cirrhosis 30 and type 1 diabetes 31. Whilst not providing direct evidence that observed associations are underpinned by autoimmunity, this does confirm the likely importance of immunological processes in the development of wheezing phenotypes. Secondly, these loci have been shown to play roles in the action and control of transcription and gene expression. For example, IKZF3 encodes a member of the IKAROS family of zinc finger proteins which are haemopoietic-specific transcription factors involved in the regulation of lymphocyte development 32. Lastly, observed associations between genetic variation here and expression patterns may be the result of more global patterns of transcriptional activity. Variation at MED24/THRAP4 encodes a component of the mediator complex which is thought to be required for the expression of almost all genes 33,34, which may be related to non-specific differential expression at multiple loci but which has downstream effects that contribute to the overall risk of persistent wheeze. Taken together, these aspects make it entirely plausible that variation at multiple loci contribute to observed phenotypes but considerable effort will be required to elucidate precise mechanisms. Although associations of wheezing phenotype related SNPs with expression data were found in this study, the absence of a direct association of expression profiles with asthma phenotypes is likely to be due to measurement error, confounding and sample size limitation.
The results of this study suggest that associations of SNPs in the 17q21 locus with susceptibility to asthma in early and mid childhood are specific to asthma and to specific wheezing phenotypes, and are not explained by associations with intermediate phenotypes, such as atopy or lung function. Elucidation of causal mechanisms has the potential to facilitate disease prediction in children with wheezing in preschool years, contribute to separation of discrete phenotypes of childhood asthma, and hence identify risk factors that may be targets for primary or secondary disease prevention.
Supplementary Material
KEY MESSAGES.
Asthma-related genetic variants are associated with intermediate-onset and persistent wheezing but not with early-onset wheezing
These associations are not explained by associations with intermediate phenotypes, such as atopy or lung function
Elucidation of causal mechanisms has the potential to identify risk factors that may be targets for primary or secondary disease prevention
ACKNOWLEDGEMENTS
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. This publication is the work of the authors and AJH & JACS will serve as guarantors for the contents of this paper.
The UK Medical Research Council and the Wellcome Trust (Grant ref: 092731), and the University of Bristol provide core support for ALSPAC study. R.G. was supported by the UK Medical Research Council (Grant no. 0401540). JPK is funded by a Wellcome Trust 4-year PhD studentship in molecular, genetic, and life course epidemiology (WT083431MA).
Abbreviations used
- ALSPAC
Avon longitudinal study of parents and children
- BHR
Bronchial hyper-responsiveness
- GWAS
genomewide association study
- FEV1
force expiratory volume per second
- FEF25-75
forced expiratory flow 25–75%
- FVC
forced vital capacity
- LSDRS
least square dose response slope
- Mb
Megabase
- OR
odd ratio
- RRR
relative risk ratio
- SNP
single nucleotide polymorphism
Footnotes
The Online Repository contains additional methods and results, 10 figures including all conditional analysis and 9 tables.
WEB RESOURCES
SNP Function Prediction (FuncPred): http://manticore.niehs.nih.gov/snpfunc.htm
References
- 1.von Mutius E. Gene-environment interactions in asthma. J Allergy Clin Immunol. 2009;123(1):3–11. doi: 10.1016/j.jaci.2008.10.046. quiz 12-3. [DOI] [PubMed] [Google Scholar]
- 2.Martinez FD. Gene-environment interactions in asthma: with apologies to William of Ockham. Proc Am Thorac Soc. 2007;4(1):26–31. doi: 10.1513/pats.200607-144JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8(3):169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 4.von Mutius E. Influences in allergy: epidemiology and the environment. J Allergy Clin Immunol. 2004;113(3):373–9. doi: 10.1016/j.jaci.2003.12.040. quiz 80. [DOI] [PubMed] [Google Scholar]
- 5.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–73. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 6.Bisgaard H, Bonnelykke K, Sleiman PM, Brasholt M, Chawes B, Kreiner-Moller E, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179(3):179–85. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 7.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. New England Journal of Medicine. 2008;359(19):1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 8.Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18(8):902–8. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64(4):629–35. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galanter J, Choudhry S, Eng C, Nazario S, Rodriguez-Santana JR, Casal J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177(11):1194–200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung TF, Sy HY, Ng MC, Chan IH, Wong GW, Tang NL, et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64(4):621–8. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 12.Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124(3):605–7. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 13.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. New England Journal of Medicine. 1995;332:133–38. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 14.Stein RT, Martinez FD. Asthma phenotypes in childhood: lessons from an epidemiological approach. Paediatr.Respir.Rev. 2004;5(2):155–61. doi: 10.1016/j.prrv.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Henderson J, Granell R, Heron J, Sherriff A, Simpson A, Woodcock AA, et al. Associations of wheezing phenotypes in the first six years of life with atopy, lung function and airway responsiveness in mid childhood. Thorax. 2008 doi: 10.1136/thx.2007.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savenije OEGR, Caudri D, Koppelman GH, Smit HA, Wijga A, de Jongste JC, Brunekreef B, Sterne JA, Postma DS, Henderson J, Kerkhof M. Comparison of childhood wheezing phenotypes in 2 birth cohorts: ALSPAC and PIAMA. J. Allergy Clin Immunol. 2011;127(6):1505–12. doi: 10.1016/j.jaci.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Martinez FD. Toward asthma prevention--does all that really matters happen before we learn to read? N Engl J Med. 2003;349(15):1473–75. doi: 10.1056/NEJMe030041. [DOI] [PubMed] [Google Scholar]
- 18.Boyd AGJ, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Davey Smith G. Cohort Profile: The ‘Children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2012 doi: 10.1093/ije/dys064. First published online: April 16, 2012. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts G, Peckitt C, Northstone K, Strachan D, Lack G, Henderson J, et al. Relationship between aeroallergen and food allergen sensitization in childhood. Clin.Exp.Allergy. 2005;35(7):933–40. doi: 10.1111/j.1365-2222.2005.02280.x. [DOI] [PubMed] [Google Scholar]
- 20.Kotecha SJ, Watkins WJ, Heron J, Henderson J, Dunstan FD, Kotecha S. Spirometric lung function in school-age children: effect of intrauterine growth retardation and catch-up growth. Am J Respir Crit Care Med. 2010;181(9):969–74. doi: 10.1164/rccm.200906-0897OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JCFB, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Moffatt MFGI, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO, GABRIEL Consortium A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers AJRB, Lasky-Su JA, Murphy A, Lazarus R, Klanderman BJ, Sylvia JS, Ziniti JP, Lange C, Celedón JC, Silverman EK, Weiss ST. Assessing the reproducibility of asthma candidate gene associations, using genome-wide data. Am J Respir Crit Care Med. 2009;179(12):1084–90. doi: 10.1164/rccm.200812-1860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wjst M. ORMDL3--guilt by association? Clin Exp Allergy. 2008;38(10):1579–81. doi: 10.1111/j.1365-2222.2008.03086.x. [DOI] [PubMed] [Google Scholar]
- 25.Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Duarte R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3(6):RESEARCH0027. doi: 10.1186/gb-2002-3-6-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahn K, Hirst SJ, Ying S, Holt MR, Lavender P, Ojo OO, et al. Diminished sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) expression contributes to airway remodelling in bronchial asthma. Proc Natl Acad Sci U S A. 2009;106(26):10775–80. doi: 10.1073/pnas.0902295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19(1):111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 28.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Invernizzi P, Lu Y, Kosoy R, Bianchi I, Podda M, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42(8):658–60. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt C, Tonnelle C, Dalloul A, Chabannon C, Debre P, Rebollo A. Aiolos and Ikaros: regulators of lymphocyte development, homeostasis and lymphoproliferation. Apoptosis. 2002;7(3):277–84. doi: 10.1023/a:1015372322419. [DOI] [PubMed] [Google Scholar]
- 33.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16(1):55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30(5):256–63. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.