Abstract
The U2AF heterodimer has been well studied for its role in defining functional 3′ splice sites in pre-mRNA splicing, but many fundamental questions still remain unaddressed regarding the function of U2AF in mammalian genomes. Through genome-wide analysis of U2AF-RNA interactions, we report that U2AF has the capacity to directly define ~88% of functional 3′ splice sites in the human genome, but numerous U2AF binding events also occur in intronic locations. Mechanistic dissection reveals that upstream intronic binding events interfere with the immediate downstream 3′ splice site associated either with the alternative exon, to cause exon skipping, or with the competing constitutive exon, to induce exon inclusion. We further demonstrate partial functional impairment with leukemia-associated mutations in U2AF35, but not U2AF65, in regulated splicing. These findings reveal the genomic function and regulatory mechanism of U2AF in both normal and disease states.
Pre-mRNA splicing takes place in the spliceosome, consisting of U1, U2 and U4/U6•U5 small nuclear ribonucleoprotein particles (snRNPs)1. U1 defines functional 5′ splice sites (5′ SSs) whereas U2 recognizes functional 3′ splice sites (3′ SSs) by base-pairing with the branch-point sequence (BPS). Because the BPS is quite degenerate in higher eukaryotic cells, the addition of U2 snRNP requires multiple auxiliary factors, the most important one being the U2AF heterodimer, consisting of a 65-kDa and a 35-kDa subunit2,3. U2AF65 binds the polypyrimidine tract (Py tract) immediately downstream of the BPS, and U2AF35 contacts the AG dinucleotide4,5. After a series of ATP-dependent steps, the U4/U6•U5 tri-snRNP complex joins the initial prespliceosome to convert it into the mature spliceosome1.
Although it has been well established that U2AF defines functional 3′ SSs on model genes, it remains unclear whether U2AF has the capacity to directly bind all functional 3′ SSs in eukaryotic genomes. In budding yeast, Mud2 is the U2AF65 ortholog, but the gene is non-essential, probably because of highly invariant BPS in this organism6,7. In fission yeast, a substantial fraction of intron-containing genes seem to lack typical Py tracts, and multiple introns appear to be insensitive to a temperature-sensitive mutant of U2AF8,9. In mammals, high levels of splicing-enhancer factors, such as SR proteins, are able to bypass U2AF to initiate spliceosome assembly10, and there also exist multiple genes related to both U2AF65 (refs. 11-13) and U2AF35 (refs. 14-16). Therefore, the functional requirement for U2AF might be bypassed by multiple mechanisms, thus raising a general question regarding the degree of the involvement of U2AF in the definition of 3′ SSs in mammalian genomes. This fundamental question has remained unaddressed despite the availability of genome-wide U2AF65-RNA interaction data17.
Computational analysis and experimental studies have also suggested that definition of many noncanonical introns in mammalian genomes may still involve U2AF but not via its direct RNA-binding activity typically seen on canonical introns18,19. Interestingly, introns that contain a strong Py tract can support spliceosome assembly in an AG-independent manner20, and U2AF65 appears to be sufficient to support splicing of such AG-independent introns, at least in vitro21. However, the U2AF35 subunit is responsible for directly contacting the AG dinucleotide on typical functional 3′ SSs22,23, and this partnership appears to be enforced by U2AF65-dependent stability control of U2AF35 (ref. 24). Furthermore, some RNA-binding proteins, such as DEK and hnRNP A1, were previously suggested to prevent U2AF from binding to pyrimidine-rich sequences elsewhere in mammalian genomes25,26, but these mechanisms await genome-wide evidence.
Both U2AF65 and U2AF35 have also been implicated in regulated splicing27,28. In general, alternative splice sites are weak, and suboptimal binding may render them particularly sensitive to levels of U2AF; moreover, U2AF is further subjected to competition with or enhancement by other RNA-binding proteins, such hnRNP C17, YB-1 (ref. 19), TIA-1 and TIAR29,30, and PTB31. Although these mechanisms may readily explain U2AF-dependent exon inclusion, the mechanism for U2AF-regulated exon skipping observed in vivo27 has been largely unknown. One potential mechanism is via skipping caused by U2AF binding on exons, as demonstrated with exon-tethered U2AF32. Last, but not least, multiple mutations in both U2AF65 and U2AF35 have been reported to associate with myelodysplasia (MDS) and related blood disorders32-35. However, it has been unclear how such mutations might affect the function of U2AF in constitutive and regulated splicing, thus underscoring the importance of understanding the regulatory function of U2AF in mammalian cells.
Given such an array of mechanistic and functional issues that remain to be addressed, we set out to perform genome-wide analysis of U2AF-RNA interactions in the human genome. By defining the genomic landscape of U2AF binding and its functional requirement in regulated splicing, we provide critical mechanistic insights into the function of U2AF in normal and disease states.
RESULTS
Genome-wide mapping of U2AF-RNA interactions
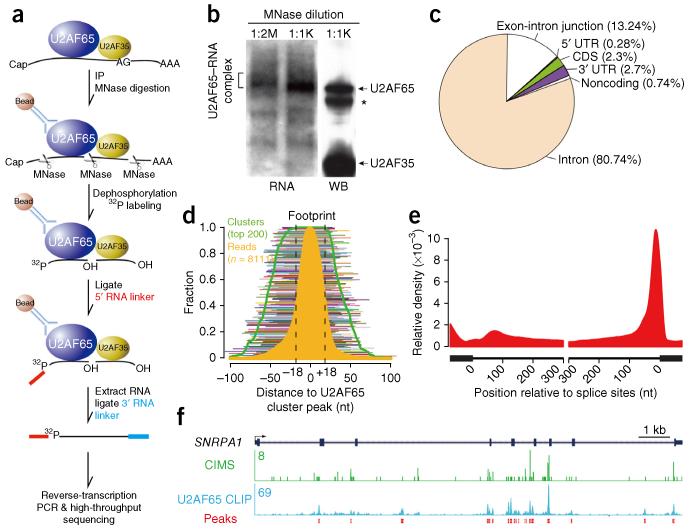
To map U2AF65-RNA interactions, we initially used standard cross-linking and immunoprecipitation coupled with deep sequencing (CLIP-seq) for library construction31. Although the monoclonal anti-U2AF65 antibody (MC3) quantitatively precipitated the U2AF heterodimer from HeLa cells (Supplementary Fig. 1a), we could not efficiently ligate the 3′ RNA linker to immunoprecipitated RNA on the U2AF complex. Reasoning that there was potential steric hindrance at the 3′ end, we first ligated the 5′ linker to 32P-labeled RNA on the complex (Fig. 1a), producing U2AF65–RNA complexes readily detectable by autoradiography (Fig. 1b). We then ligated recovered RNA to the 3′ linker and performed reverse transcription, PCR amplification and deep sequencing. Out of a total of 19.5 million sequenced tags, 9.3 million could be uniquely mapped to the human genome (Supplementary Table 1); these results also showed a reasonable concordance with the previously published data17 on U2AF65-RNA interaction (Supplementary Fig. 1b).
Figure 1.
Mapping of U2AF65-RNA interactions in the human genome. (a) Schematic illustration of U2AF65 CLIP-seq. IP, immunoprecipitation; MNase, micrococcal nuclease. (b) The U2AF65–RNA complexes trimmed by two different concentrations of MNase (1:2M, 1:2,000,000 dilution; 1:1K, 1:1,000 dilution), detected by autoradiography. WB, western blotting. Asterisk indicates IgG heavy chain. Bracketed RNA–protein adducts were recovered for CLIP library construction. (c) Genomic distribution of U2AF65 CLIP-seq peaks. UTR, untranslated region; CDS, coding sequence. (d) U2AF65 footprint on RNA. A set of high-density clusters used to derive the footprint is shown by lines of different colors. Green line, cluster density; orange area, density of all tags in the clusters; black dashed lines, deduced U2AF65 footprint. (e) Metagene analysis of U2AF65-RNA interactions on a composite pre-mRNA. Relative density (normalized tag count) is shown. (f) U2AF65 binding on a gene example, showing raw tags, peaks and identified cross-linking–induced mutation sites (CIMS).
We detected U2AF65 binding mostly in intronic regions (80.74%) and in an additional fraction (13.24%) corresponding to exon-intron boundaries, which together accounted for 94% of mapped U2AF65 binding events in the human genome (Fig. 1c). We also detected U2AF65 binding on exons (2.3%) and 3′ untranslated regions (2.7%), results consistent with the negative impact of exon-bound U2AF65 on splicing32 and with a positive function of U2AF65 in 3′-end formation36. By compiling a set of frequent U2AF65 binding events (8,111 tags on 200 top clusters), we estimated the average U2AF65 footprint to be ~36 nt (Fig. 1d). From cross-linking–induced mutation sites (CIMS), as described earlier37, which display a characteristic distribution of base deletions but not insertions or substitutions (Supplementary Fig. 1c–f), we made a similar estimate of the U2AF footprint (Supplementary Fig. 1g). Metagene analysis demonstrated prevalent U2AF65 binding at 3′ SSs (Fig. 1e), as illustrated with the SNRPA1 gene, on the basis of both mapped tags and identified CIMS (Fig. 1f). These data demonstrated high-fidelity mapping results for U2AF65-RNA interactions in the human genome.
U2AF recognition of ~88% of functional 3′ SSs in the human genome
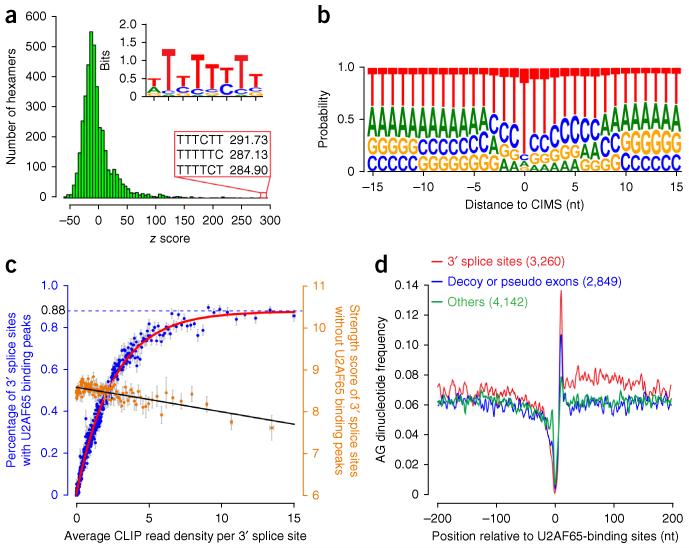
Consistently with the biochemically defined binding specificity of U2AF, motif analysis showed highly pyrimidine-enriched sequences on mapped U2AF65-binding sites (Fig. 2a). The top 50 hexamers alone, which all consist of pyrimidines (top 20 in Supplementary Fig. 2a), account for 80% of all mapped U2AF65-binding sites, in contrast to ~20% for 50 randomly selected hexamers (Supplementary Fig. 2b). Alignment of mapped U2AF65-binding sites according to the centers of CIMS in individual tags generated a Py tract–like sequence typical of those associated with canonical 3′ SSs (Fig. 2b). This high-quality data set allowed us to address two critical rules deduced from previous in vitro studies.
Figure 2.
Specificity of U2AF65-RNA interactions in the human genome. (a) Enriched motifs for U2AF65 binding. The top three motifs are shown. Inset, consensus sequence, deduced from the top 50 motifs. (b) Nucleotide frequency centered on identified CIMS. (c) Maximum-likelihood analysis to determine the capacity of U2AF65-occupied 3′ SSs in the human genome. Each blue dot represents the averaged occupancy of a group of 50 genes, on the basis of CLIP-seq tag density. Individual groups were sorted according to the averaged CLIP-seq tag density at 3′ SSs. Each orange dot represents the average strength score of 3′ SSs among a group of 100 3′ SSs that exhibited no U2AF65 binding peak. Error bars, s.d. (n = 50 genes for blue dot; n = 100 splice sites for orange dot). (d) The frequency of the AG dinucleotide from the mapped U2AF65-binding sites on annotated 3′ SSs (red), deduced decoy and pseudo exons (blue) or other intronic regions (green).
The first rule concerns the U2AF65 coverage of functional 3′ SSs in mammalian genomes. U2AF65 was previously mapped to 58% of active 3′ SSs in HeLa cells17. However, this simple counting method probably missed many U2AF-dependent 3′ SSs, especially among genes that are expressed at modest to low levels. We thus developed a maximum-likelihood approach to estimate the percentage of 3′ SSs that could be directly bound by U2AF65. We first sorted expressed genes according to the average CLIP-tag density per annotated 3′ SS in each gene and then divided these genes into consecutive groups of 50 genes. This allowed us to calculate the coverage of annotated 3′ SSs by U2AF65 with s.d. in all groups. We next determined the percentage of 3′-SS coverage when the CLIP-tag density per 3′ SS was progressively increased. We observed that the coverage reached saturation at ~88%, with increasing levels of U2AF65 binding at annotated 3′ SSs (Fig. 2c).
We next asked whether those U2AF65-unbound 3′ SSs contain a strong consensus and thus do not require U2AF65, as in those in budding yeast, or conversely whether they might drift from the consensus, therefore representing noncanonical introns. For this purpose, we sorted expressed genes similarly according to the average CLIP-tag density per 3′ SS and then grouped those splice sites in individual genes without detectable U2AF65 peaks into consecutive groups of 100 3′ SSs. We next calculated the averaged strength score of U2AF65-unbound 3′ SSs in each group according to the method of Yeo and Burge38. We detected progressive decreases in the averaged strength scores with U2AF65-unbound 3′ SSs (Fig. 2c).
Combined, these data indicate that, among genes that show less-efficient U2AF binding in general, the lack of U2AF binding in unoccupied introns is probably due to limited expression, but among genes that show extensive U2AF binding on canonical 3′ SSs, the lack of U2AF binding in remaining introns probably results from poor consensus in their 3′ SSs. Therefore, coupled with the maximum-likelihood analysis, our data suggest that about 12% of functional 3′ SSs may not require U2AF65’s binding activity or even its function for their recognition in the human genome, although to definitively distinguish between these two possibilities represents a major challenge.
Additional U2AF binding events beyond functional 3′ SSs
The second rule concerns the ability of U2AF65 to discriminate between Py tracts with or without a flanking AG dinucleotide in mammalian genomes. In vitro binding studies suggest that U2AF efficiently binds Py tracts followed by AG but binds much less to Py tracts that do not end with an AG dinucleotide22,23. Such specificity may be enhanced by additional RNA-binding factors, such as DEK and hnRNP A1 (refs. 25,26). Because U2AF65 functions as a heterodimer in conjunction with U2AF35, on the basis of their tight interactions in coimmunoprecipitation experiments (Supplementary Fig. 1a), it is likely that the mapped genomic U2AF65 binding events largely reflect the action of the U2AF heterodimer in vivo; this allowed us to directly test whether U2AF indeed prefers Py tracts followed by an AG dinucleotide in the human genome.
Comparing U2AF65 binding events on functional 3′ SSs and on other regions, we found that both U2AF65-bound 3′ SSs and non–3′ SSs exhibited a similar profile of the S65 score, a measure of U2AF65 binding affinity based on previous systematic evolution of ligands by exponential enrichment (SELEX) experiments (Supplementary Fig. 2c)18. We next segregated U2AF65 binding events on non–3′ SSs into two classes. The first contains potential decoy exons (those with flanking sequences that resemble a 3′ or 5′ splice site) or pseudo exons (those with flanking potential 3′ and 5′ splice sites separated by a sequence up to 250 nt)36, and the second has no obvious evidence for any splicing signals. We found that U2AF65 binding at functional 3′ SSs is strongly associated with a downstream AG; U2AF65 binding at decoy and pseudo exons shows less enrichment with a downstream AG; and the remaining U2AF65 binding in other intronic locations exhibits no selective enrichment with a downstream AG (Fig. 2d). These data suggest that, despite the presence of other specificity-enhancing factors, U2AF is still able to bind certain other locations in pre-mRNA besides functional 3′ SSs. These U2AF65 binding events may interfere with functional definition of adjacent bona fide 3′ SSs as a mechanism to modulate alternative splicing (described below) and/or may reflect a role of U2AF65 in other RNA-metabolism steps39,40.
Key roles of U2AF in gene expression and regulated splicing
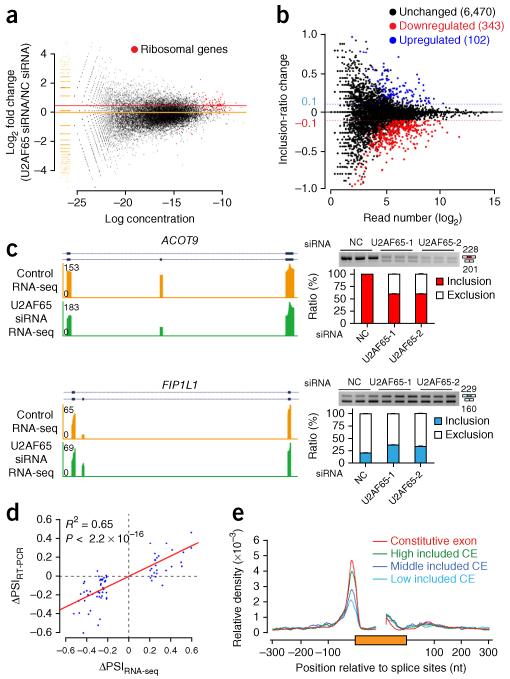
Despite the existence of potential U2AF65-independent introns, it is clear from our genome-wide analysis that U2AF65 occupies the majority of functional 3′ SSs. To determine whether U2AF65 is essential for gene expression, we performed RNA sequencing (RNA-seq), generating 14.1 and 16.8 million uniquely mapped tags before and after U2AF65 RNA interference (RNAi), respectively, and detected both up- and downregulated genes upon U2AF65 knockdown. However, we noted from gene ontology term analysis that most upregulated genes are either intronless histone genes or abundant ribosomal genes, thus indicating that these genes appear upregulated because of their insensitivity to U2AF65 depletion. After normalizing the RNA-seq data set against the ribosomal genes, we found that the majority of genes were downregulated in U2AF65-depleted cells (Fig. 3a). These results suggest that U2AF65 is required for efficient expression of most intron-containing genes, despite the possibility that U2AF65 may not be required for removal of a subset of introns in them.
Figure 3.
Roles of U2AF65 in gene expression and alternative splicing. (a) Global analysis of gene expression by RNA-seq. Ribosomal-gene transcripts (red dots) were used to normalize the levels of the individual genes in U2AF65-knockdown cells. Orange and red lines illustrate the normalization scale. NC, negative control. (b) Altered alternative-splicing events determined by RNA-seq. Significantly induced and repressed splicing events in U2AF65-knockdown cells are indicated by blue and red dots, respectively, which were identified by two-sided Fisher’s exact test with a threshold of inclusion-ratio change ≥0.10 for individual detected cassette exons. (c) Splicing of two representative genes in response to U2AF65 knockdown. RNA-seq data were validated by RT-PCR (gels and quantification at right) in HeLa cells treated independently with two different U2AF65 siRNAs. Error bars, s.d. (n = 3 cell cultures). Uncropped gels are shown in Supplementary Data Set 1. (d) Comparison between the alternative-splicing events detected by RNA-seq and validated by semiquantitative RT-PCR. P < 2.2 × 10−16 by F test (n = 70 randomly selected alternative-splicing events). ΔPSI, difference in percentage spliced in, in HeLa cells treated with U2AF65 siRNA or negative control (NC) siRNA. (e) U2AF65 binding densities (normalized tag count) on genes with different exon inclusion levels. CE, alternative cassette exon.
We also used the RNA-seq data to deduce altered splicing events in an unbiased manner. U2AF65 has been implicated as a regulator of alternative splicing12,24,27, but it has been unclear how extensively U2AF65 is involved in regulated splicing and, if so, what mechanism(s) underlie such regulatory function. Taking advantage of 75-nt sequences from both ends of our libraries, we generated sequence contigs that cover alternative splice junctions, thus permitting calculation of the splicing ratio (percentage spliced in, PSI) of individual annotated cassette exons, as described previously41. The data revealed 102 and 343 (out of a total of 6,915) cassette exons that showed significantly increased and decreased inclusion, respectively, in response to U2AF65 knockdown (Fig. 3b).
Most identified alternative-splicing events are evident even from RNA-seq tags mapped on the alternative and flanking exons (examples in Fig. 3c). We validated ~70 randomly selected alternative-splicing events by PCR with reverse transcription (RT-PCR) and found that the induced exon-inclusion (Supplementary Fig. 3a) or skipping (Supplementary Fig. 3b) events detected by RNA-seq were well correlated with RT-PCR results (Fig. 3d). These data demonstrated that U2AF65 is extensively involved in the regulation of alternative splicing. Importantly, although three-quarters of events induced by U2AF65 RNAi showed increased exon skipping, results consistent with a decisive role of U2AF65 binding at the 3′ SS of the alternative exon in dictating the selection of the exon, the remaining one-quarter exhibited increased exon inclusion, thus raising an important mechanistic question of how U2AF65 might be directly involved in repressing such alternative-splicing events (described below).
Multiple mechanisms for U2AF-regulated alternative splicing
We observed that U2AF65 binding levels are generally proportional to the levels of exon inclusion (Fig. 3e), thus suggesting that the 3′ SSs of alternative exons are weaker in general relative to the flanking competing exons and, as a result, that the alternative exons are preferentially affected when U2AF65 is reduced in short interfering RNA (siRNA)-treated cells. Although U2AF65 RNAi-induced exon-skipping events could be comprehended on the basis of existing knowledge, it remains to be determined whether and how some U2AF65 RNAi–induced exon-inclusion events might also result from a direct effect of U2AF65. We noted many examples in which U2AF65 bound on exons (GANAB and ANKRD10 genes, Supplementary Fig. 4a,b); this is actually consistent with a recent report showing splicing inhibition by exon-bound U2AF65 in minigene assays32. However, this mechanism could not explain other U2AF65 RNAi–induced exon-inclusion events, particularly those linked to intronic U2AF65 binding events. We noted that, among 102 U2AF65 depletion–induced exon-inclusion events, ~80% showed U2AF65 binding in intronic regions that flank the alternative exon, but this was not the case with the majority of introns that flank a constitutive exon, thus suggesting that U2AF65 binding around those regulated exons may be directly responsible for their U2AF65-dependent changes in alternative splicing.
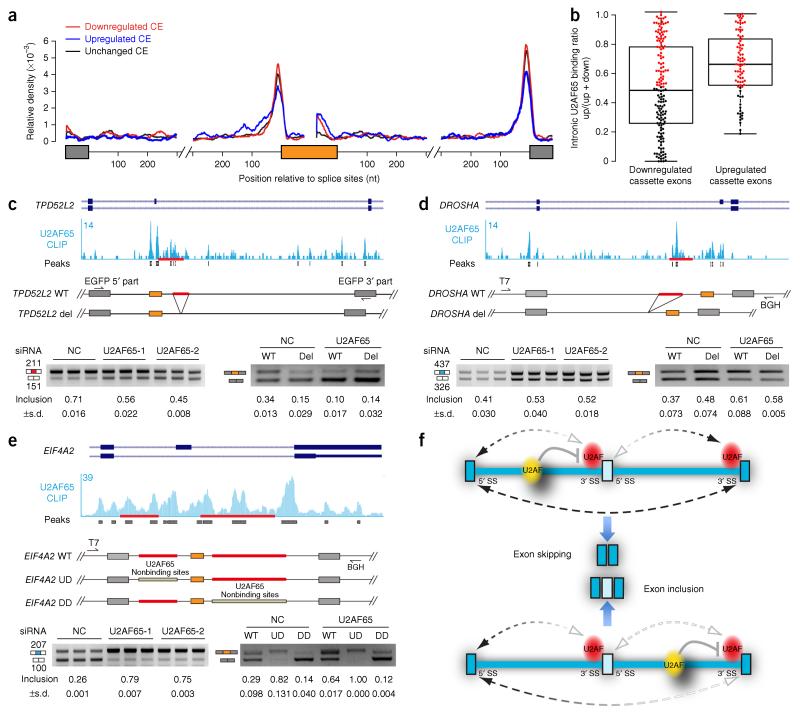
To aid in mechanistic dissection, we constructed a U2AF65 RNA map based on detected exon-inclusion or skipping events in response to U2AF65 RNAi (Fig. 4a). Although we saw no obvious trend for U2AF65-dependent exon inclusion or skipping at first, close inspection of the RNA map revealed higher intronic binding events upstream of U2AF65-repressed alternative exons in the immediate intronic regions of 300 nt that we displayed (Fig. 4a). When we considered all intronic binding events among U2AF65 RNAi–induced alternative splicing events, we found that the percentage of upstream intronic U2AF65 binding events over total intronic (both upstream and downstream) binding events was evenly distributed among induced exon-skipping events. In contrast, higher U2AF65 binding in upstream intronic regions was mostly linked to induced exon-inclusion cases (Fig. 4b). These observations suggest two combinatory mechanisms in operation: one is more sensitive to U2AF65 binding at the 3′ SS of the alternative exon relative to intronic binding events, as associated with induced exon-skipping cases, and the other is more dependent on U2AF65 binding in upstream intronic regions, as predominantly associated with induced exon-inclusion events.
Figure 4.
Polar effects of U2AF65 binding on recognition of downstream 3′ SSs. (a) Normalized U2AF65 binding events (normalized tag count) on unaffected cassette exons (black) or upregulated (blue) or downregulated (red) cassette exons in U2AF65-knockdown cells. The composite gene model contains flanking intronic sequences of 300 nt. (b) Box-plot analysis of relative distribution of intronic binding events flanking U2AF65 RNAi–repressed or enhanced exons. ‘Up’ and ‘down’ represent upstream and downstream intronic U2AF65 binding events, respectively. Center line, median; bottom and top box limits, first and third quartiles, respectively; whiskers, minimum and maximum. The events were selected with at least one binding peak in intronic regions, with 81 upregulated cases and 153 downregulated cases. (c–e) Minigene constructs and induced alternative splicing in response to U2AF65 RNAi, analyzed by RT-PCR. Top, CLIP analysis of U2AF65 binding to intronic regions downstream and upstream of the alternative exon, respectively (c,d) and U2AF65 binding to both introns flanking the alternative exon (e), with schematics of constructs shown below. Bottom left, RT-PCR analysis showing splicing response of these genes to U2AF65 RNAi. Bottom right, RT-PCR for specific deletion mutants (c,d) and the replacement mutant (e) at the prevalent U2AF65 binding site(s), tested in both control siRNA–treated and U2AF65 siRNA–treated HeLa cells. Error bars, s.d. (n = 3 cell cultures). Uncropped images of gels are in Supplementary Data Set 1. WT, wild type; del, deletion. (f) A proposed model for the polar effects of intronically bound U2AF65 interfering with the recognition of the immediate downstream 3′ SS in regulated splicing.
Polar effects of U2AF binding on downstream 3′-SS recognition
The prevalent intronic U2AF65 binding events could be more directly appreciated in specific examples (Fig. 4c–e). To explore the regulatory mechanism(s), we chose these representative genes and performed mutational analysis on their minigenes to avoid potential indirect effects of U2AF65 depletion. On the TPD52L2 gene, U2AF65 binding predominantly took place within the downstream intron, and independent knockdown of U2AF65 by two different siRNAs caused increased skipping of the alternative exon (Fig. 4c). Deletion of the major U2AF65-binding site near the 5′ SS of the alternative exon induced exon skipping in the same way as in U2AF65-depleted cells, and this mutant minigene no longer responded to U2AF65 knockdown (Fig. 4c). In comparison, on the DROSHA gene the major U2AF65 binding event occurred in the intron upstream of the alternative exon, and U2AF65 RNAi increased the inclusion of the alternative exon (Fig. 4d). Mutational analysis showed that deletion of the intronic U2AF65-binding site induced exon inclusion, and the mutant minigene became largely insensitive to U2AF65 RNAi (Fig. 4d). (The remaining U2AF-dependent effect was probably due to other minor binding events in the upstream intron.) These data suggest that the intronic U2AF65 binding events on these two tested cases are directly responsible for U2AF65-dependent splicing responses.
We next dissected a more complex case on the EIF4A2 gene, where U2AF65 bound extensively on both upstream and downstream introns, and U2AF65 depletion induced a net increase in exon inclusion (Fig. 4e). In this case, instead of constructing simple deletion mutants (because deletion of the U2AF-binding sequence would remove most of the upstream or downstream intron), we replaced the U2AF65-binding sequence with a non–U2AF65-binding sequence of similar length in either the upstream or the downstream intron. Interestingly, we detected enhanced exon inclusion when the upstream U2AF65-binding site was substituted but reduced exon inclusion when the downstream U2AF65-binding site was replaced, and on both mutant minigenes U2AF65 knockdown resulted in no further effects (Fig. 4e). Finally, we took a non–U2AF65-responsive gene (C1orf43), inserted the U2AF65-binding site from DROSHA in either the upstream or the downstream intron and found that the extra upstream U2AF65-binding site increased exon skipping, whereas the extra downstream site repressed exon skipping, both in a U2AF65-dependent manner (Supplementary Fig. 4c).
The simplest interpretation of the above combined results is that intronic U2AF65 binding events interfere with the recognition of the immediate downstream functional 3′ SS according to rules for context-dependent splicing42. In the case of TPD52L2, release of such inhibition increases the competitiveness of the flanking 3′ SS, thereby suppressing the selection of the upstream 3′ SS associated with the alternative exon. This is also the case with U2AF65 binding in the downstream intron of the EIF4A2 gene. However, the removal of U2AF65 competition from the upstream intron in both DROSHA and EIF4A2 genes enhances the competitiveness of the 3′ SS of the alternative exon, thus allowing more efficient exon inclusion in each case. When both competing events operate in the same alternative-splicing unit, a stronger one wins, as in the case of the EIF4A2 gene, thus generating a net effect of exon inclusion in U2AF65-depleted cells. From these findings, we propose a polar mechanism for intronic U2AF65 binding to interfere with the recognition of the downstream 3′ SS (Fig. 4f).
Coordinated action of U2AF65 and U2AF35 in regulated splicing
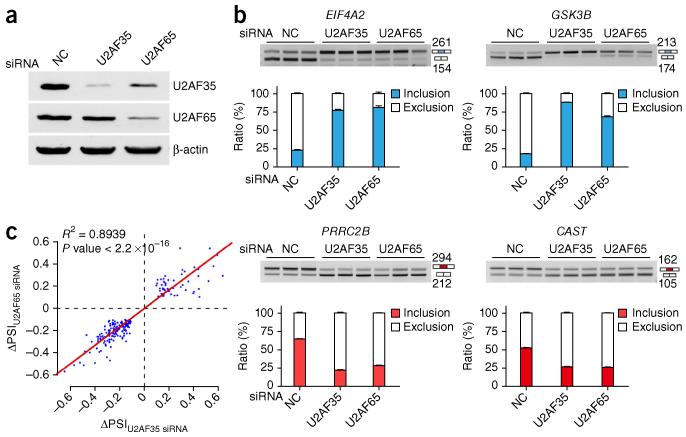
It has been unclear thus far whether U2AF65 acts predominantly alone or in conjunction with U2AF35 or other U2AF35-related molecules in regulated splicing. Because the majority of U2AF65 appears to exist as a heterodimer with U2AF35 in the cell, it is likely that the heterodimer may have a dominant role in both constitutive and regulated splicing in the human genome. To directly test this hypothesis, we used alternative splicing as a functional readout to compare the cellular response to U2AF65 and U2AF35 RNAi. As previously reported24, U2AF35 RNAi reduced the expression of only U2AF35, whereas U2AF65 RNAi reduced the levels of both subunits (Fig. 5a).
Figure 5.
Coordinated action of U2AF65 and U2AF35 in regulated splicing. (a) Western blotting analysis of RNAi-mediated U2AF65 and U2AF35 knockdown, showing reduced U2AF35 in U2AF65 siRNA–treated cells. β-actin is a loading control. (b) Splicing response of representative genes in response to RNAi against U2AF65 or U2AF35. Error bars, s.d. (n = 3 cell cultures). Uncropped gels are provided in Supplementary Data Set 1. (c) Global concordance of U2AF65- and U2AF35-dependent splicing revealed by RASL-seq. P < 2.2 × 10−16 by F test (n = 208 splicing-changed events). ΔPSI, difference in percentage spliced in, in HeLa cells treated with U2AF65 siRNA, U2AF35 siRNA or negative control (NC) siRNA.
We used our recently developed technology43, based on RNA-mediated oligonucleotide annealing, selection and ligation coupled with sequencing (RASL-seq), to conduct a cost-effective survey of alternative-splicing events in HeLa cells subjected to U2AF65 and U2AF35 RNAi. Using this oligonucleotide ligation–based approach, which was designed to specifically interrogate a large set of annotated splicing events (~5,000), we detected 1,892 alternative-splicing events in control siRNA–treated HeLa cells, among which 271 and 334 events showed significant changes (statistics described in Online Methods) in response to U2AF65 and U2AF35 depletion, respectively, which we extensively validated by RT-PCR (Fig. 5b and Supplementary Fig. 5a,b). Notably, we detected nearly identical sets of alternative-splicing events induced by RNAi against U2AF65 and U2AF35 with either RASL-seq (Fig. 5c and Supplementary Table 1) or RT-PCR (Supplementary Fig. 5c).
Because U2AF35 was reduced in U2AF65 siRNA–treated cells, and this might account for the similar effects of U2AF35 and U2AF65 on alternative splicing, we further tested splicing responses in U2AF65 siRNA–treated cells in which expression of U2AF35 was restored. We found largely identical induced splicing events with or without exogenously expressed U2AF35 (Supplementary Fig. 5d), and we further confirmed this by RT-PCR (Supplementary Fig. 5e). Altogether, these data demonstrated that U2AF35 functions largely in conjunction with U2AF65 in the regulation of alternative splicing in mammalian cells.
Causal effects of MDS mutations in U2AF35 but not U2AF65
Recent advances in disease studies have revealed the association of prevalent mutations in splicing factors, including both U2AF65 and U2AF35, with MDS and related blood disorders35. A published analysis suggested that the MDS-associated mutations in U2AF35 caused widespread defects in pre-mRNA processing in HeLa cells33; this is reminiscent of global downregulation of gene expression in U2AF65-depleted cells (Fig. 3a). Because such dramatic defects are expected to cause cell lethality, it has remained unclear how such null-like mutations in U2AF35 would permit the transition of initial MDS to eventual acute myeloid leukemia in patients with MDS. We decided to use U2AF-regulated alternative-splicing events as functional measurements to determine the nature of MDS-associated mutations in U2AF35 and to extend the analysis to mutations in U2AF65.
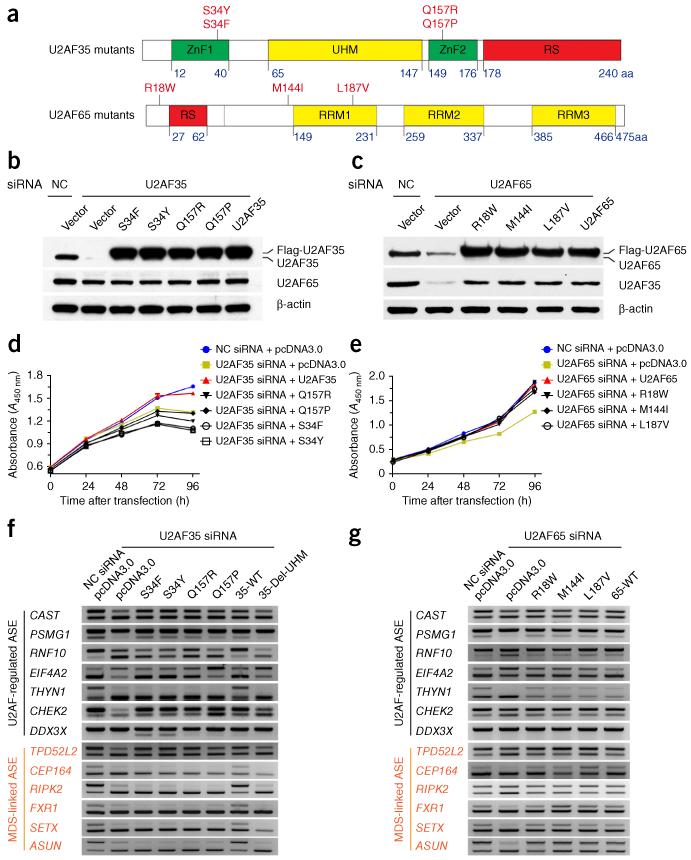
We constructed expression units for mutant U2AF65 (official symbol U2AF2) and U2AF35 (official symbol U2AF1) that carry individual mutations identified in patients with MDS (Fig. 6a) and expressed them in individual U2AF35- and U2AF65-knockdown cells (Fig. 6b,c). We observed that all U2AF35 mutations retarded cell growth upon introduction into HeLa cells (Fig. 6d), but surprisingly none of the U2AF65 mutations that we tested displayed the same growth phenotype (Fig. 6e). These observations imply that, in contrast to those in U2AF35, the mutations in U2AF65 might be neutral; this is consistent with their low frequency among patients with MDS.
Figure 6.
Splicing defects induced by MDS-associated mutations in U2AF35 but not in U2AF65. (a) Schematic illustration of mutations in U2AF65 and U2AF35 detected in leukemia. (b,c) Western blot analysis of U2AF35 and U2AF65 expression in knockdown of U2AF35 (b) or U2AF65 (c) and reconstitution of knockdown cells with wild-type and mutant genes. β-actin is a loading control. (d,e) Growth of HeLa cells expressing the indicated mutants of U2AF35 (d) or U2AF65 (e) and the indicated siRNAs. Error bars, s.d. (n = 3 cell cultures). (f,g) RT-PCR analysis of functional rescue of RNAi-induced alternative splicing events (ASE) with individual U2AF mutants. Genes in black, representative genes initially scored by RASL-seq in U2AF65 siRNA–treated HeLa cells. Genes in red, genes expressed in HeLa cells and shown to exhibit induced exon skipping in blood cells of patients with MDS carrying mutations in U2AF35. 35-WT, wild-type U2AF35; 35-Del-UHM, U2AF35 lacking the U2AF65 interaction domain; 65-WT, wild-type U2AF65.
We next selected seven alternative-splicing events to test the ability of individual U2AF mutations to rescue the splicing responses induced by U2AF RNAi (Fig. 6f,g). We found that wild-type U2AF35 was able to fully rescue the splicing changes, but the mutant U2AF35 that lacks the U2AF65 interaction domain failed to rescue them, and individual U2AF35 mutations displayed variable rescuing effects. On the CAST, TPD52L2 and PSMG1 genes, for example, all mutant U2AF35 expression units fully rescued the splicing defects. On the CHEK2 and DDX3X genes, however, the mutants showed either full or partial rescuing effects, but on the THYN1 gene, none of the mutants was able to rescue. Among all U2AF35 mutants tested, the Q157P mutation appears most interesting, showing full effects on four genes, a partial effect on one and no effect on the other two (Fig. 6f). In contrast to U2AF35 mutations and consistently with the lack of growth phenotype, all U2AF65 mutants functioned like the wild-type protein (Fig. 6g). These data suggest that the MDS-associated mutations in U2AF35 are partially compromised at the functional level, but are not null, whereas the mutations in U2AF65 are all likely to be neutral.
To further examine critically altered splicing events detected by RNA-seq in patients with MDS who carry mutations in U2AF35 (ref. 44), we noted that six genes (labeled in red in Fig. 6f,g) from the reported 35 alternatively spliced genes in blood cells also showed expression in HeLa cells. Because the alternative-splicing events in these genes were not covered by our RASL-seq pool, we designed specific PCR primers to detect individual U2AF35 mutation–associated alternative-splicing events and found that, although wild-type U2AF35 was able to rescue U2AF35 RNAi-induced splicing, none of the U2AF35 mutants showed the ability to rescue (Fig. 6g). Importantly, the induced exon-skipping events on all six genes were identical to those observed in blood cells44. In contrast, all U2AF65 mutants behaved identically to the wild-type gene in rescuing U2AF65 RNAi–induced alternative splicing (Fig. 6g). The ability to detect the same effects of U2AF35 mutations in HeLa cells as those in patient blood cells suggests that the induced alternative-splicing events directly result from mutations in the U2AF35 gene, rather than being due to selection of different cell types, as part of MDS etiology.
DISCUSSION
Our data demonstrate a predominant role of U2AF in defining functional 3′ SSs in the human genome. Interestingly, our analysis also suggests that U2AF65 may not be able to directly bind ~12% of introns, whose recognition may be aided by other intronic splicing–enhancer factors, such as YB1 (ref. 19). However, the existence of a fraction of introns that lack U2AF65 binding is consistent with previous observations in fission yeast8,9, thus raising the question of which specific splicing factors might fulfill such a role in defining those noncanonical 3′ SSs. Although several RNA-binding splicing factors have structures related to those of U2AF65 or U2AF35 (ref. 16), the available functional evidence suggests that most of them function in synergy with, rather than independently of, U2AF13-15,45. Therefore, it remains unclear how those potential U2AF-independent introns might be recognized in mammalian genomes.
The preferential binding of U2AF65 to functional 3′ SSs over other pyrimidine-rich sequences in the genome is enforced by the U2AF35 subunit, and other factors may provide additional proofreading functions25,26. However, our genome-wide binding data clearly show that U2AF65 can also bind to various locations that are not part of annotated 3′ SSs, and these binding events do not seem to depend on a downstream AG dinucleotide. This is consistent with the proposed function of U2AF65 in promoting nuclear export of intronless transcripts in Drosophila39 and with U2AF65 binding on some spliced mRNAs40. A more recent study showed that hnRNP C is able to prevent U2AF65 from binding to many Alu-containing transcripts to suppress exonization of those Alu elements17. Therefore, U2AF binding appears to be a highly regulated process in mammalian genomes.
U2AF has been implicated in the regulation of alternative splicing. Our data suggest U2AF binding on the 3′ SS of alternative exons as a predominant mechanism for U2AF-regulated splicing. In addition, U2AF65 also binds in other locations, such as those on exonic regions, to interfere with the selection of the exon32. We found that U2AF65 also binds various intronic locations, which appear to selectively interfere with the recognition of immediate downstream 3′ SSs to dictate the splicing outcome. This splice-site competition model provides a universal mechanism for the regulation of alternative splicing by both sequence-specific RNA-binding proteins and core components of the splicing machinery42.
A major advance in the field is the identification of specific mutations in multiple splicing factors, including U2AF65 and U2AF35, in specific types of myeloid leukemia. The mutations have been generally considered to drive the disease, although this actually remains to be functionally determined. A HeLa cell–based study indicated that U2AF35 mutations cause a slow-growth phenotype and more-severe defects in gene expression, thus implying that those mutations might be functionally null33. However, RNA-seq detected only limited splicing defects in patient blood cells44. This agrees with our observation that mutant U2AF35 is still functional in rescuing many, but not all, U2AF35 RNAi–induced changes in alternative splicing. Thus, the mutations in U2AF35 are not functionally null. In contrast, a parallel analysis indicated that the disease-associated mutations in U2AF65 might be neural mutations, and this is consistent with the low frequency of those mutations in MDS patients35. These findings thus provide critical insights into the nature of specific mutations in the splicing regulators. The challenge ahead will be to link specific molecular defects to the etiology of the disease.
ONLINE METHODS
Cell culture and transfection
HeLa cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% FBS at 37 °C and 5% CO2. Lipofectamine 2000 or Lipofectamine RNAi Max (Life Technology) was used to transfect plasmid or siRNA. For double transfection, cells were transfected with the reporter minigene 48 h after siRNA transfection and harvested 24 h later for RNA and protein analysis. Sense sequences of siRNAs are as follows: U2AF65-1 siRNA, 5′-GCACGGTGGACTGATTCGTdT dT-3′; U2AF65-2 siRNA, 5′-GCAAGTACGGGCTTGTCAAdTdT-3′; U2AF35 siRNA, 5′-GAAAGTGTTGTAGTTGATTGAdTdT-3′.
Plasmids and antibodies
To construct the expression plasmids for U2AF65 and U2AF35, Flag-tagged primer sets were designed to amplify the coding region of individual genes, and the PCR products were inserted into pcDNA3.0 between the HindIII and BamHI sites. U2AF35-Del-UHM was made by PCR to delete amino acids 65 to 147 from the U2AF35 coding region. All other site-specific mutations, including those for the U2AF35 mutants S34Y, S34F, Q157R and Q157P, and the U2AF65 mutants R18W, M144I and L187V, were generated by a PCR-based method (KOD Plus, TOYOBO). Supplementary Table 2 lists the primers used to construct U2AF-expression plasmids described above, and Supplementary Table 3 lists the primers used to construct and analyze individual minigenes in transfected cells. All constructs were verified by DNA sequencing.
The following primary antibodies were used: anti-U2AF65 (U4758, Clone MC3, Sigma-Aldrich, 1:10,000), anti-U2AF35 (ab86305, Abcam, 1:2,000), anti–β-actin (A1978, Clone AC-15, Sigma-Aldrich, 1:20,000), anti-GAPDH (10494-1-AP Proteintech, 1:10,000) and anti-Flag (F1804, Clone M2, Sigma-Aldrich, 1:3,000). Primary antibodies were detected with horseradish peroxidase (HRP)-labeled secondary antibodies (anti-rabbit IgG, A8275, Sigma-Aldrich, 1:10,000; anti-mouse IgG, A9044, Sigma-Aldrich, 1:10,000) for western blotting.
CLIP-seq, RNA-seq and RASL-seq
CLIP-seq for U2AF65 was performed as previously described31, with additional modifications. Briefly, we first ligated the 5′ linker to RNA on the immunoprecipitated U2AF complex. T4 Polynucleotide Kinase (3′ phosphatase minus) was next used to 32P-label the 5′ end of RNA so that both ends of the RNA contained a phosphate group, thus preventing self-ligation of immunoprecipitated RNA. After blotting onto nitrocellulose, recovery of U2AF65-RNA adducts, and removal of proteins by proteinase K digestion, a 3′ linker containing 3′ puromycin was ligated to isolated RNA. The remaining steps were identical to the CLIP-seq procedure described previously31.
CLIP-seq reads were processed as previously described31. Briefly, reads were trimmed to remove sequencing adaptors and homopolymeric runs of ≥20 nt and mapped to the human genome (hg18) with Bowtie2 (version 0.12.7 with parameters -q -l 25 -m 2 –best). Significant peaks, footprint, and CIMS were determined as previously described31,37. De novo motif finding was implemented with the RSA tools oligonucleotide analysis algorithm (http://rsat.ulb.ac.be/) with input U2AF65 peaks46.
Paired-end RNA-seq was performed with RNA isolated from control siRNA– or U2AF65 siRNA–treated HeLa cells. Supplementary Table 4 lists PCR primers used for validating U2AF-regulated splicing events.
Both ends of RNA-seq libraries were sequenced for 75 nt, and these were separately mapped (allowing two mismatches) into a custom Bowtie index built from UCSC Known Genes (hg18) not including introns and then joined together to reconstruct into full contigs of ~200 nt. The splicing ratio was calculated with reads mapped on specific splice junctions. Significant changes in alternative splicing in response to U2AF depletion were identified by two-sided Fisher’s exact test with a threshold of inclusion-ratio change ≥0.10. In the current study, we mainly focused our analysis on cassette exons.
The library construction and data analysis for RASL-seq were performed as previously described43.
Maximum-likelihood analysis of U2AF binding percentage on 3′ SSs
We first sorted all genes by the averaged CLIP-seq read density at annotated 3′ SSs, from low to high. For smoothing, we grouped 50 genes as one data point. As the averaged density increases, an increased percentage of U2AF65 occupancy on annotated 3′ SSs is expected, as we observed. A logarithmic regression model leads to the estimate of ~88% functional 3′ SSs recognizable by U2AF65, thus implying the existence of ~12% of functional 3′ SSs that may not be bound directly by U2AF65 in the human genome. We next asked whether those potential U2AF65-independent sites tend to have low splice-site strength, which is calculated as previously described38.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to members of the Fu laboratory for numerous stimulating discussions during the course of this investigation. This work was supported by China 973 program grants (2011CB811300 and 2012CB910800), a Chinese 111 program grant (B06018) and US National Institutes of Health grants (HG004659 and GM049369) to X.-D.F. and a US National Institutes of Health grant (DK098808) to D.-E.Z. and X.-D.F.
Footnotes
Accession codes. The CLIP-seq, RASL-seq and RNA-seq data for U2AF65 have been deposited in the Gene Expressing Omnibus database under accession number GSE61603.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
T.W., Y. Zhang, D.-E.Z. and X.-D.F. designed the experiments; C.S. and T.W. performed the biochemical experiments; B.Y., J.H., Y. Zhou, A.D. and J.Z. were responsible for bioinformatics analysis of genomics data; J.Q. and H.L. performed RASL-seq and RNA-seq; P.T., L.J., G.C., H.S. and D.-E.Z. contributed to additional data analysis; and T.W., C.S., B.Y. and X.-D.F. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 5.Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 6.Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 7.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 8.Sridharan V, Heimiller J, Singh R. Genomic mRNA profiling reveals compensatory mechanisms for the requirement of the essential splicing factor U2AF. Mol. Cell. Biol. 2011;31:652–661. doi: 10.1128/MCB.01000-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sridharan V, Singh R. A conditional role of U2AF in splicing of introns with unconventional polypyrimidine tracts. Mol. Cell. Biol. 2007;27:7334–7344. doi: 10.1128/MCB.00627-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacMillan AM, McCaw PS, Crispino JD, Sharp PA. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl. Acad. Sci. USA. 1997;94:133–136. doi: 10.1073/pnas.94.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai H, Chan EK, Kiyosawa K, Fu XD, Tan EM. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J. Clin. Invest. 1993;92:2419–2426. doi: 10.1172/JCI116848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS ONE. 2007;2:e538. doi: 10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page-McCaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65-related splicing activity. RNA. 1999;5:1548–1560. doi: 10.1017/s1355838299991938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tronchère H, Wang J, Fu XD. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- 15.Shepard J, Reick M, Olson S, Graveley BR. Characterization of U2AF6, a splicing factor related to U2AF35. Mol. Cell. Biol. 2002;22:221–230. doi: 10.1128/MCB.22.1.221-230.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mollet I, Barbosa-Morais NL, Andrade J, Carmo-Fonseca M. Diversity of human U2AF splicing factors. FEBS J. 2006;273:4807–4816. doi: 10.1111/j.1742-4658.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 17.Zarnack K, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray JI, Voelker RB, Henscheid KL, Warf MB, Berglund JA. Identification of motifs that function in the splicing of non-cannonical introns. Genome Biol. 2008;9:R97. doi: 10.1186/gb-2008-9-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei WJ, et al. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 2012;40:8622–8636. doi: 10.1093/nar/gks579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 21.Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 23.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 24.Pacheco TR, Coelho MB, Desterro JM, Mollet I, Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell. Biol. 2006;26:8183–8190. doi: 10.1128/MCB.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 26.Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol. Cell. 2012;45:314–329. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 27.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Guiner C, et al. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, et al. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. USA. 2011;108:11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 34.Thol F, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 35.Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danckwardt S, et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 39.Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 2004;14:775–786. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 40.Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao R, et al. Nuclear matrix factor hnRNP U/SAF-A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Mol. Cell. 2012;45:656–668. doi: 10.1016/j.molcel.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu X-D, Ares M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Przychodzen B, et al. Patterns of missplicing due to somati U2AF1 mutations in myoloid neoplasms. Blood. 2013;122:999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010;24:2389–2394. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Helden J, André B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.