Abstract
Emergence of antibiotic resistant pathogens is changing the way scientists look for new antibiotic compounds. This race against the increased prevalence of multi-resistant strains makes it necessary to expedite the search for new compounds with antibiotic activity and to increase the production of the known. Here, we review a variety of new scientific approaches aiming to enhance antibiotic production in Streptomyces. These include: (i) elucidation of the signals that trigger the antibiotic biosynthetic pathways to improve culture media, (ii) bacterial hormone studies aiming to reproduce intra and interspecific communications resulting in antibiotic burst, (iii) co-cultures to mimic competition-collaboration scenarios in nature, and (iv) the very recent in situ search for antibiotics that might be applied in Streptomyces natural habitats. These new research strategies combined with new analytical and molecular techniques should accelerate the discovery process when the urgency for new compounds is higher than ever.
Keywords: antibiotics, Streptomyces, co-culture, interactions, signals
Introduction
Since pioneering work leading to the isolation of the antibiotic streptomycin in 1943 by Waksman et al. (Jones et al., 1944), huge progress has been made in the elucidation of the molecular basis and mechanism behind the production of these biological weapons by the Streptomyces genus. Following this initial discovery, thousands of compounds produced by these microorganisms have been described and utilized in order to fight infections, and they comprise over two-thirds of all known antibiotic compounds (Omura, 1992; Berdy, 2005; Hopwood, 2007). Nowadays, widespread antibiotic resistance (McArthur et al., 2013; Mak et al., 2014; Lin et al., 2015) has rendered a large number of these compounds ineffective and is currently urging the scientific community to push the boundaries of classical microbiology toward a faster and more efficient secondary metabolite search.
Antibiotic biosynthesis is carried out by a high number of proteins encoded by genomic clusters and is tightly regulated (Bibb and Hesketh, 2009). Normally, there is specific regulation for each product, mediated by Cluster-Situated Regulators and also global or pleiotropic mechanisms of regulation that can control several pathways at the same time (Rokem et al., 2007; Martín and Liras, 2012). Therefore, there are complex regulatory networks that control the onset of production of the secondary metabolites (Liu et al., 2013). These networks respond to multiple signals, many of which are still unknown, and therefore empirical methods are needed to trigger the production of cryptic secondary metabolites.
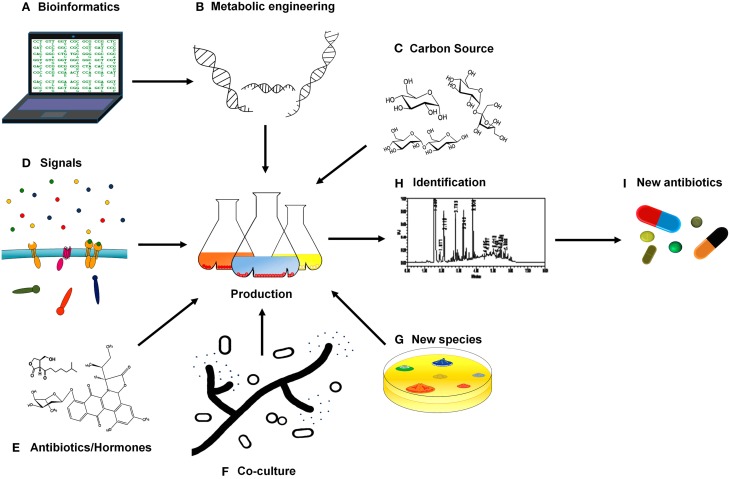
Genome sequencing combined with in silico prediction has revealed that microorganisms of the genus Streptomyces harbor a high number of secondary metabolism clusters (Aigle et al., 2014; Bachmann et al., 2014; Ikeda et al., 2014). Bioinformatics and “omics” based engineering has become a powerful tool in this field, allowing the identification of secondary metabolite gene clusters and their possible products by similarity searching (Chaudhary et al., 2013) (Figure 1A). The use of techniques developed in recent years in metabolic engineering will also be of tremendous value and the perfect complement in this urgent quest for new antibiotic compounds (Aigle and Corre, 2012; Weber et al., 2015). Changing promoters, introducing biosynthetic clusters in others species, or rewiring transcriptional and post-transcriptional regulation are methods for unveiling new antibiotics or modifying products previously discarded to obtain new molecules with antibiotic activity (Figure 1B). Removal of endogenous secondary metabolites gene-clusters, based on the previously described competition for precursors, has been also shown as an alternative for improving antibiotic production (Komatsu et al., 2010; Gómez-Escribano and Bibb, 2011, 2014). Although all these techniques have shown their efficacy, when cultured in axenic conditions most bacteria express a limited number of these clusters. That is the reason that makes the unlocking of Streptomyces cryptic pathways (potentially abundant) one of the most feasible methods for antibiotic(s) discovery in this counter clock race against multi-resistant strains. Moreover, new approaches designed with a view to awake silent or cryptic pathways will also surely result in the discovery of highly valuable secondary metabolites with antifungal, herbicidal, anti-cancer, immunosuppressive, anti-inflammatory, antihelmintic, or antiviral activities, amongst others, widening the interest for the research in this area.
Figure 1.
Schematic overview of new approaches for antibiotic discovery in Streptomyces. (A,B) Biosynthetic clusters and regulatory elements can be predicted with bioinformatic tools, opening up new possibilities to metabolic engineering. (C) Modification of the culture medium that is crucial in antibiotic production such as different carbon sources. (D) Searching for the signals that activate different regulatory systems triggering antibiotic production. (E) Communication molecules like antibiotics and hormones may also boost secondary metabolism. (F) Co-culture of Streptomyces species with fungi and bacteria might simulate interspecies interactions and thus induce antibiotic production. (G) The discovery of new Streptomyces species could also reveal new compounds with antibiotic activities. (H) The compounds produced have to be identified and isolated. (I) All these approaches contribute to the elucidation of the nature and activity of new antibiotics.
Studying bacterial sensors to find key molecules triggering antibiotic burst: the nutritional signals
It is widely known that media composition has a great impact on microbial secondary metabolites, comprising of activators of signaling cascades that trigger their production (Yang et al., 2010). Based on systematic culture modification of easily accessible parameters, some strategies such as “One Strain-Many Compounds” (OSMAC) were proven successful more than a decade ago, leading to the isolation of up to 20 different metabolites from species of Streptomyces (Bode et al., 2002). In particular, chemical compounds present in Streptomyces niches but not found in culture media are thought to play a role in cryptic metabolite activation as signals in sensory mechanisms, triggering regulatory cascades responsible for the tuning of the secondary metabolites synthesis. That is the case of N-acetylglucosamine, the monomer of chitin that has been shown to act as a signal, mediated by the DasR global regulator (Rigali et al., 2008), that controls antibiotic production (Swiatek et al., 2012; Nazari et al., 2013). The recently reported cellobiose-induced production of thaxtomin A by Streptomyces scabies is also of interest (Francis et al., 2015). More generally, the effect of carbon source(s) in antibiotic production is also a subject of study (Figure 1C), since some of the most used carbon sources, in which bacteria are growing more “comfortably,” repress secondary metabolism (Sánchez et al., 2010). We could possibly use less “efficient” carbon sources, which however might induce more antibiotic production in an effort to establish a balance between growth yield and antibiotic yield.
Regarding signal translation into responses, two component systems (TCS) are the main signaling pathways in Streptomyces and their role in the antibiotic production complex regulatory network has just been started to be decoded (Rodríguez et al., 2013). Nevertheless, the signals that activate the different Streptomyces' TCS remain mostly unknown and just a few of them have been described to date (Figure 1D). As an example, in S. coelicolor phosphate is the signal triggering PhoP/R (Sola-Landa et al., 2003), nitrogen balance seems to be related with AfsQ (Shu et al., 2009) and DraR/K (Yu et al., 2012) regulation, the level of iron seems to be the signal activating AbrA1/2 activator (Rico et al., 2014) and the presence of heme-oxidative stress provokes SenS/R reaction (Bogel et al., 2007; Ortiz de Orue Lucana and Groves, 2009). Most of the signals sensed by the TCS present in the genome of the Streptomyces spp. sequenced so far (more than 100 genomes) remain elusive. Phosphate, nitrogen and iron are compounds present in laboratory culture media. However, other compounds found in nature but absent in lab culture might also act as the signals triggering other TCSs responses (or different regulatory mechanisms) and therefore controlling antibiotic production. So, the addition of low concentration of rare earth elements to the culture medium may activate the secondary metabolism in S. coelicolor and in other Streptomyces species. Scandium has been the one studied more in depth but yttrium, lanthanum, cerium, and europium can also provoke antibiotic production boost (Kawai et al., 2007). The molecular mechanism under this induction, however, has not been described yet.
New developments in Streptomyces research, also linked with nutrients, since their depletion is coupled with sporulation, are exploring new solutions in order to wake silent pathways through morphological differentiation, namely sporulation recovery. As has been recently described, physiological differentiation is tightly linked to secondary metabolism and therefore recovering sporulation capacities of some Streptomyces might also lead to the discovery of new compounds (Chater, 2013; Kalan et al., 2013). Another nutrient-related deficiency of axenic cultures is the absence of siderophores. Some Streptomyces species are defective in the production of these iron-chelating compounds and need to utilize those released by other species in order to differentiate, produce secondary metabolites or even grow in lab conditions (Yamanaka et al., 2005; Eto et al., 2013; Lambert et al., 2014). Therefore, the addition of purified siderophores or the co-culture of non-producer species with siderophore producers are strategies also related with nutrient supply that might be used to awake silent pathways.
Spying microbial conversations: bacterial hormones and antibiotics as signals
The presence of antibiotics is an important piece of information for the microorganism in order to respond to threats (as a signal of the competitors presence) or even to coordinate efforts with other antibiotic producing neighbors (combining strategies in order to repel a common menace). Therefore, the addition of certain antibiotics or bacterial hormones in low concentrations to the culture medium might also be an alternative for antibiotic production stimulation (Figure 1E). Over the last 5 years there have been several reports on the role of antibiotic compounds as auto-inducers of antibiotic production (Romero et al., 2011). Even more, the importance of molecules previously described as antimicrobials, in inter-specific communication between Streptomyces species and, as a consequence, in the regulation of antibiotic production has been recently described (Nodwell, 2014). For example, the antibiotic jadomycin B, an angucycline, produced by Streptomyces venezuelae, triggers different antibiotic production levels in S. coelicolor depending on the concentration (Wang et al., 2014a). Additionally, hormones also play a role in communication between bacteria. Among them, gamma-butyrolactones have been demonstrated to promote antibiotic production in many streptomycetes (Sidda and Corre, 2012). These molecules are involved in cell to cell communication processes (quorum sensing) in which bacteria use the production and detection of autoinducers in order to synchronize gene expression and population growth (Garg et al., 2014). For example, A-factor, a gamma-butyrolactone, autoinduces morphological differentiation and secondary metabolite production in S. griseus (Horinouchi and Beppu, 2007). Recently, an exogenous butyrolactone has also been showed to increase validamicyn antibiotic production in Streptomyces hygroscopicus 5008 (Tan et al., 2013). Although, many Streptomyces species are apparently capable of synthesizing gamma-butyrolactones (Takano, 2006) a recent research has also made clear that antibiotic production can also be triggered by different hormones like avenolide in Streptomyces avermitilis or methylenomycin furans in S. coelicolor (Corre et al., 2008; Kitani et al., 2011). Regulation of antibiotic production by microbial signaling molecules such as hormones and foreign antibiotics is widespread. A better understanding of the nature and functions of these signals could drive us to their potential use as activators of silent pathways.
Exploiting microbial communal living: co-cultures
Routine laboratory work with Streptomyces, as with other microorganisms, has been basically done in axenic cultures. However, antibiotic functions can only be understood in the context of Streptomyces' habitat. Traditionally, antibiotics have been considered biological weapons that allow the bacteria to compete with others microorganisms, either by killing them or inhibiting their growth. Nevertheless, antibiotics are also signals that trigger adaptive responses (Yim et al., 2007; Fajardo and Martínez, 2008; Aminov, 2009). Antibiotics have evolved as a result of interactions (mainly competitive but also cooperative) with other organisms and their natural role has just started to be elucidated (Davies, 2009, 2013). It is, therefore, not unreasonable to think that the presence of either foreign neighboring species in its environmental niches or its buddy's signals could trigger different patterns of secondary metabolites production (Vetsigian et al., 2011).
Although streptomycetes have been considered as normal inhabitants of soil, recent studies show that Streptomyces species are also frequent in different habitats in the underwater world, mainly in sediments from shallow and deep water habitats and marine dwelling animals, and as symbionts of plants and invertebrates (Seipke et al., 2012; Raveh et al., 2013). Besides, the variety of organisms that share these different niches with Streptomyces is huge. In order to achieve a laboratory scenario that resembles more closely the environmental conditions, co-culture of two or three species has emerged as a powerful tool (Figure 1F). This aims to mimic real simple situations in nature that will facilitate the discovery of new secondary metabolites. Although just a limited number of experiments have been carried out to date using this new approach, results are promising. One of the seminal experiments in this area used different combinations of streptomycetes in co-culture to produce the stimulation of antibiotic production and differentiation (Ueda et al., 2000). More recently, co-cultures of several Streptomyces species with different fungi and bacteria have reported an induction of new molecules or the stimulation of previously known compounds either in Streptomyces, in the other partner or even in both partners (Seyedsayamdost et al., 2012; Watrous et al., 2013; Moody, 2014). An example shows that pairwise co-culture of Streptomyces coelicolor with five different actinomycetes produces a range of compounds of unknown identity, among them, at least 12 different desferrioxamines, that were not produced when S. coelicolor was grown under pure culture (Traxler et al., 2013). Similarly the co-culture of the predator bacteria Myxococcus xanthus with S. coelicolor showed that S. coelicolor increases actinorhodin production in order to repel the invader when it senses the presence of the predator (Pérez et al., 2011). In some cases substance-mediated induction has been discarded, with cell-to-cell interaction being the causative agent of antibiotic biosynthetic pathways “decryption.” Thus, the interaction of Streptomyces with mycolic acid-containing bacteria such as Tsukumurella pulmonis in co-cultures provokes the synthesis of new natural antibiotic products (i.e., alchivemycin A by S. endus) although not mediated via any chemical substance (Onaka et al., 2011). Presence of plant pathogens has also been shown to trigger the production of secondary metabolites able to suppress Verticillium dahlia, such as prodiginines, by S. lividans (Meschke et al., 2012). Streptomyces products obtained in the presence of plant invaders are becoming an interesting tool for biocontrol initiatives that are being developed in order to fight plant plagues (Taechowisan et al., 2005; de Oliveira et al., 2010; Meschke and Schrempf, 2010; Cuesta et al., 2012; Meschke et al., 2012; Palaniyandi et al., 2013a,b). One step further of “natural co-culture” lies in the culture of Streptomyces in the presence of human pathogens pushing the evolutionary mechanisms of Streptomyces toward the biosynthesis of natural compounds able to outcompete the pathogen. So, Streptomyces clavuligerus co-cultured with methicillin resistant Staphylococcus aureus was able to synthesize holomycin, a S. aureus chemical inhibitor not detected in axenic cultures (Charusanti et al., 2012). Other interesting interactions are shown in Table 1. In this way, co-culture allows the induction of secondary metabolism even when signals that trigger the response remain unknown or the induction is due to a combination of factors that is hardly reproducible in axenic conditions.
Table 1.
Streptomyces co-cultures involved in antibiotic production.
| Co-cultured species | Effects | References |
|---|---|---|
| Streptomyces coelicolor–five actinomycetes | Production of multiple cryptic compounds and antibiotics (i.e., prodiginines and actinorhodines) | Traxler et al., 2013 |
| Combinations of 76 Streptomyces spp. | Stimulation of various antibiotics | Ueda et al., 2000 |
| Streptomyces coelicolor–Bacillus subtilis | Increase of undecylprodigiosin production. Earlier onset of production | Luti and Mavituna, 2011 |
| Streptomyces spp.–Tsukamurella pulmonis | Production of novel antibiotics (i.e., alchivemycin A by S. endus) | Onaka et al., 2011 |
| Streptomyces coelicolor–Myxococcus xanthus | Increase of actinorhodin production in S. coelicolor | Pérez et al., 2011 |
| Streptomyces clavuligerus–Staphylococcus aureus | Production of holomycin | Charusanti et al., 2012 |
| Streptomyces cinnabarinus–Alteromonas sp. | Induction of lobocompactol production | Cho and Kim, 2012 |
| Streptomyces sp. Mg1–Bacillus subtilis | Production of chalcomycin A | Barger et al., 2012 |
| Streptomyces sp.–Proteobacteria | Production of the antibiotic resistomycin | Carlson et al., 2015 |
| Streptomyces coelicolor–Corallococcus coralloides | Increase of antibiotic production of undecylprodigiosin and earlier onset | Schäberle et al., 2014 |
| Streptomyces fradiae 007–Penicillium sp. WC-29-5 | Production of four aromatic polyketides | Wang et al., 2014b |
| Streptomyces lividans–Bacillus subtilis | Induction of prodiginine production | Vargas-Bautista et al., 2014 |
| Streptomyces lividans–Verticillium dahliae | Increase of antibiotic production of prodiginines | Meschke et al., 2012 |
Taking the lab to the field: in situ culture for antibiotic discovery
Uncultured bacteria make up approximately 99% of all species. These “undomesticated” microorganisms are potentially a huge unexplored source of antibiotic compounds (Lewis, 2013). The recent publication by Ling et al. of new methods for in situ cultivation of previously uncultivable microbial species opens up a new world of possibilities enabling the search for natural products in previously inaccessible sources (Ling et al., 2015). These new methods are, respectively, based on cultivation of the microorganism in their natural environment using a multichannel device that allows diffusion of nutrients and growth factors (Nichols et al., 2010) and on the use of siderophores as growth factors, to microorganisms out of their environment (D'onofrio et al., 2010). As a proof of concept of the in situ culture approach, teixobactin, a new antibiotic produced by the Gram-negative bacteria Elephteria terrae with excellent activity against Gram-positive pathogens, was discovered in an extract obtained using iChip devices (Ling et al., 2015). Many new species are being described every day and it is thought that most of Streptomyces species remain undiscovered to date, foreseeing unlimited possibilities for future antimicrobial discovery (Figure 1G).
Identifying compounds: technical advances in secondary metabolite detection
Although production of cryptic secondary metabolites is the main goal, it is important to consider other aspects, such as the identification of the compounds produced in each condition. This is the first step for the purification and elucidation of their structures and activity. Previous technical problems have been solved by emerging analytical techniques like nanospray desorption electrospray ionization (NanoDESI) and matrix-assisted laser desorption ionization–time of flight (MALDI–TOF) imaging mass spectrometry that allow researchers to gain an in situ global chemical view of bacterial secretions (Watrous et al., 2013; Fang and Dorrestein, 2014; Hsu and Dorrestein, 2015). Therefore, the use of new advanced techniques applied in addition to classical detection/identification methods such as High Performance Liquid Chromatography (HPLC) or Mass spectrophotometry (MS) (Figure 1H) will also be crucial in this crusade against resistant pathogens.
Concluding remarks
A new universe of possibilities for antibiotic discovery (Figure 1I) is opening up through multiple strategies whereby the aforementioned genetic techniques may be complemented by deeper studies on bacterial relationships and elucidation of the compounds serving as signals for regulator systems. As a consequence of both, these novel techniques and the need for new and more effective products, the following years might therefore present a new golden age for antibiotic discoveries after 70 years of the pioneering discoveries in Streptomyces.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our laboratory is funded by: The Spanish Ministerio de Ciencia e Innovación (MICINN) [BFU2010-17551] and the Junta de Castilla y León (JCyL) CSI099A12-1. We specially thank Dr. Alexia Hapeshi for her valuable comments and English proofreading. We also thank the reviewers of the manuscript for their valuable suggestions.
References
- Aigle B., Corre C. (2012). Waking up Streptomyces secondary metabolism by constitutive expression of activators or genetic disruption of repressors. Methods Enzymol. 517, 343–366. 10.1016/B978-0-12-404634-4.00017-6 [DOI] [PubMed] [Google Scholar]
- Aigle B., Lautru S., Spiteller D., Dickschat J. S., Challis G. L., Leblond P., et al. (2014). Genome mining of Streptomyces ambofaciens. J. Ind. Microbiol. Biotechnol. 41, 251–263. 10.1007/s10295-013-1379-y [DOI] [PubMed] [Google Scholar]
- Aminov R. I. (2009). The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11, 2970–2988. 10.1111/j.1462-2920.2009.01972.x [DOI] [PubMed] [Google Scholar]
- Bachmann B. O., van Lanen S. G., Baltz R. H. (2014). Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 41, 175–184. 10.1007/s10295-013-1389-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger S. R., Hoefler B. C., Cubillos-Ruiz A., Russell W. K., Russell D. H., Straight P. D. (2012). Imaging secondary metabolism of Streptomyces sp. Mg1 during cellular lysis and colony degradation of competing Bacillus subtilis. Antonie Van Leeuwenhoek 102, 435–445. 10.1007/s10482-012-9769-0 [DOI] [PubMed] [Google Scholar]
- Berdy J. (2005). Bioactive microbial metabolites. J. Antibiot. 58, 1–26. 10.1038/ja.2005.1 [DOI] [PubMed] [Google Scholar]
- Bibb M., Hesketh A. (2009). Analyzing the regulation of antibiotic production in streptomycetes. Methods Enzymol. 458, 93–116. 10.1016/S0076-6879(09)04804-6 [DOI] [PubMed] [Google Scholar]
- Bode H. B., Bethe B., Hofs R., Zeeck A. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3, 619–627. [DOI] [PubMed] [Google Scholar]
- Bogel G., Schrempf H., Ortiz de Orue Lucana D. (2007). DNA-binding characteristics of the regulator SenR in response to phosphorylation by the sensor histidine autokinase SenS from Streptomyces reticuli. FEBS J. 274, 3900–3913. 10.1111/j.1742-4658.2007.05923.x [DOI] [PubMed] [Google Scholar]
- Carlson S., Tanouye U., Omarsdottir S., Murphy B. T. (2015). Phylum-specific regulation of resistomycin production in a Streptomyces sp. via microbial coculture. J. Nat. Prod. 78, 381–387. 10.1021/np500767u [DOI] [PubMed] [Google Scholar]
- Charusanti P., Fong N. L., Nagarajan H., Pereira A. R., Li H. J., Abate E. A., et al. (2012). Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction. PLoS ONE 7:e33727. 10.1371/journal.pone.0033727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F. (2013). Curing baldness activates antibiotic production. Chem. Biol. 20, 1199–1200. 10.1016/j.chembiol.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Chaudhary A. K., Dhakal D., Sohng J. K. (2013). An insight into the “-omics” based engineering of streptomycetes for secondary metabolite overproduction. Biomed Res. Int. 2013:968518. 10.1155/2013/968518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. Y., Kim M. S. (2012). Induction of antifouling diterpene production by Streptomyces cinnabarinus PK209 in co-culture with marine-derived Alteromonas sp. KNS-16. Biosci. Biotechnol. Biochem. 76, 1849–1854. 10.1271/bbb.120221 [DOI] [PubMed] [Google Scholar]
- Corre C., Song L., O'rourke S., Chater K. F., Challis G. L. (2008). 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc. Natl. Acad. Sci. U.S.A. 105, 17510–17515. 10.1073/pnas.0805530105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta G., García-de-la-Fuente R., Abad M., Fornes F. (2012). Isolation and identification of actinomycetes from a compost-amended soil with potential as biocontrol agents. J. Environ. Manage. 95(Suppl.), S280–S284. 10.1016/j.jenvman.2010.11.023 [DOI] [PubMed] [Google Scholar]
- Davies J. (2009). Everything depends on everything else. Clin. Microbiol. Infect. 15(Suppl. 1), 1–4. 10.1111/j.1469-0691.2008.02682.x [DOI] [PubMed] [Google Scholar]
- Davies J. (2013). Specialized microbial metabolites: functions and origins. J. Antibiot. 66, 361–364. 10.1038/ja.2013.61 [DOI] [PubMed] [Google Scholar]
- de Oliveira M. F., da Silva M. G., van der Sand S. T. (2010). Anti-phytopathogen potential of endophytic actinobacteria isolated from tomato plants (Lycopersicon esculentum) in southern Brazil, and characterization of Streptomyces sp. R18(6), a potential biocontrol agent. Res. Microbiol. 161, 565–572. 10.1016/j.resmic.2010.05.008 [DOI] [PubMed] [Google Scholar]
- D'onofrio A., Crawford J. M., Stewart E. J., Witt K., Gavrish E., Epstein S., et al. (2010). Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 17, 254–264. 10.1016/j.chembiol.2010.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D., Watanabe K., Saeki H., Oinuma K., Otani K., Nobukuni M., et al. (2013). Divergent effects of desferrioxamine on bacterial growth and characteristics. J. Antibiot. 66, 199–203. 10.1038/ja.2012.111 [DOI] [PubMed] [Google Scholar]
- Fajardo A., Martínez J. L. (2008). Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11, 161–167. 10.1016/j.mib.2008.02.006 [DOI] [PubMed] [Google Scholar]
- Fang J., Dorrestein P. C. (2014). Emerging mass spectrometry techniques for the direct analysis of microbial colonies. Curr. Opin. Microbiol. 19, 120–129. 10.1016/j.mib.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis I. M., Jourdan S., Fanara S., Loria R., Rigali S. (2015). The cellobiose sensor CebR is the gatekeeper of Streptomyces scabies pathogenicity. MBio 6:e02018-14. 10.1128/mBio.02018-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N., Manchanda G., Kumar A. (2014). Bacterial quorum sensing: circuits and applications. Antonie Van Leeuwenhoek 105, 289–305. 10.1007/s10482-013-0082-3 [DOI] [PubMed] [Google Scholar]
- Gómez-Escribano J. P., Bibb M. J. (2011). Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 4, 207–215. 10.1111/j.1751-7915.2010.00219.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Escribano J. P., Bibb M. J. (2014). Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 41, 425–431. 10.1007/s10295-013-1348-5 [DOI] [PubMed] [Google Scholar]
- Hopwood D. A. (2007). Streptomyces in Nature and Medicine. The Antibiotic Makers. New York, NY: Oxford University Press Inc. [Google Scholar]
- Horinouchi S., Beppu T. (2007). Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 83, 277–295. 10.2183/pjab.83.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. C., Dorrestein P. C. (2015). Visualizing life with ambient mass spectrometry. Curr. Opin. Biotechnol. 31C, 24–34. 10.1016/j.copbio.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Kazuo S. Y., Omura S. (2014). Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 41, 233–250. 10.1007/s10295-013-1327-x [DOI] [PubMed] [Google Scholar]
- Jones D., Metzger H. J., Schatz A., Waksman S. A. (1944). Control of Gram-negative bacteria in experimental animals by Streptomycin. Science 100, 103–105. 10.1126/science.100.2588.103 [DOI] [PubMed] [Google Scholar]
- Kalan L., Gessner A., Thaker M. N., Waglechner N., Zhu X., Szawiola A., et al. (2013). A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem. Biol. 20, 1214–1224. 10.1016/j.chembiol.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Kawai K., Wang G., Okamoto S., Ochi K. (2007). The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 274, 311–315. 10.1111/j.1574-6968.2007.00846.x [DOI] [PubMed] [Google Scholar]
- Kitani S., Miyamoto K. T., Takamatsu S., Herawati E., Iguchi H., Nishitomi K., et al. (2011). Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U.S.A. 108, 16410–16415. 10.1073/pnas.1113908108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Uchiyama T., Omura S., Cane D. E., Ikeda H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651. 10.1073/pnas.0914833107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S., Traxler M. F., Craig M., Maciejewska M., Ongena M., van Wezel G. P., et al. (2014). Altered desferrioxamine-mediated iron utilization is a common trait of bald mutants of Streptomyces coelicolor. Metallomics 6, 1390–1399. 10.1039/C4MT00068D [DOI] [PubMed] [Google Scholar]
- Lewis K. (2013). Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–387. 10.1038/nrd3975 [DOI] [PubMed] [Google Scholar]
- Lin J., Nishino K., Roberts M. C., Tolmasky M., Aminov R. I., Zhang L. (2015). Mechanisms of antibiotic resistance. Front. Microbiol. 6:34. 10.3389/fmicb.2015.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L. L., Schneider T., Peoples A. J., Spoering A. L., Engels I., Conlon B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. 10.1038/nature14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Chater K. F., Chandra G., Niu G., Tan H. (2013). Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. 10.1128/MMBR.00054-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luti K. J., Mavituna F. (2011). Elicitation of Streptomyces coelicolor with dead cells of Bacillus subtilis and Staphylococcus aureus in a bioreactor increases production of undecylprodigiosin. Appl. Microbiol. Biotechnol. 90, 461–466. 10.1007/s00253-010-3032-2 [DOI] [PubMed] [Google Scholar]
- Mak S., Xu Y., Nodwell J. R. (2014). The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 93, 391–402. 10.1111/mmi.12689 [DOI] [PubMed] [Google Scholar]
- Martín J. F., Liras P. (2012). Cascades and networks of regulatory genes that control antibiotic biosynthesis, in Reprogramming Microbial Metabolic Pathways, ed Link S. (Dordrecht: Springer; ), 115–138. [DOI] [PubMed] [Google Scholar]
- McArthur A. G., Waglechner N., Nizam F., Yan A., Azad M. A., Baylay A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. 10.1128/AAC.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meschke H., Schrempf H. (2010). Streptomyces lividans inhibits the proliferation of the fungus Verticillium dahliae on seeds and roots of Arabidopsis thaliana. Microb. Biotechnol. 3, 428–443. 10.1111/j.1751-7915.2010.00165.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meschke H., Walter S., Schrempf H. (2012). Characterization and localization of prodiginines from Streptomyces lividans suppressing Verticillium dahliae in the absence or presence of Arabidopsis thaliana. Environ. Microbiol. 14, 940–952. 10.1111/j.1462-2920.2011.02665.x [DOI] [PubMed] [Google Scholar]
- Moody S. C. (2014). Microbial co-culture: harnessing intermicrobial signaling for the production of novel antimicrobials. Future Microbiol. 9, 575–578. 10.2217/fmb.14.25 [DOI] [PubMed] [Google Scholar]
- Nazari B., Kobayashi M., Saito A., Hassaninasab A., Miyashita K., Fujii T. (2013). Chitin-induced gene expression in secondary metabolic pathways of Streptomyces coelicolor A3(2) grown in soil. Appl. Environ. Microbiol. 79, 707–713. 10.1128/AEM.02217-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols D., Cahoon N., Trakhtenberg E. M., Pham L., Mehta A., Belanger A., et al. (2010). Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 76, 2445–2450. 10.1128/AEM.01754-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodwell J. R. (2014). Are you talking to me? A possible role for γ-butyrolactones in interspecies signalling. Mol. Microbiol. 94, 483–485. 10.1111/mmi.12787 [DOI] [PubMed] [Google Scholar]
- Omura S. (1992). The expanded horizon for microbial metabolites–a review. Gene 115, 141–149. 10.1016/0378-1119(92)90552-Z [DOI] [PubMed] [Google Scholar]
- Onaka H., Mori Y., Igarashi Y., Furumai T. (2011). Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 77, 400–406. 10.1128/AEM.01337-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz de Orue Lucana D., Groves M. R. (2009). The three-component signalling system HbpS-SenS-SenR as an example of a redox sensing pathway in bacteria. Amino Acids 37, 479–486. 10.1007/s00726-009-0260-9 [DOI] [PubMed] [Google Scholar]
- Palaniyandi S. A., Yang S. H., Suh J. W. (2013a). Extracellular proteases from Streptomyces phaeopurpureus ExPro138 inhibit spore adhesion, germination and appressorium formation in Colletotrichum coccodes. J. Appl. Microbiol. 115, 207–217. 10.1111/jam.12212 [DOI] [PubMed] [Google Scholar]
- Palaniyandi S. A., Yang S. H., Zhang L., Suh J. W. (2013b). Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 97, 9621–9636. 10.1007/s00253-013-5206-1 [DOI] [PubMed] [Google Scholar]
- Pérez J., Muñoz-Dorado J., Braña A. F., Shimkets L. J., Sevillano L., Santamaría R. I. (2011). Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microb. Biotechnol. 4, 175–183. 10.1111/j.1751-7915.2010.00208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh A., Delekta P. C., Dobry C. J., Peng W., Schultz P. J., Blakely P. K., et al. (2013). Discovery of potent broad spectrum antivirals derived from marine actinobacteria. PLoS ONE 8:e82318. 10.1371/journal.pone.0082318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico S., Yepes A., Rodríguez H., Santamaría J., Antoraz S., Krause E. M., et al. (2014). Regulation of the AbrA1/A2 two-component system in Streptomyces coelicolor and the potential of its deletion strain as a heterologous host for antibiotic production. PLoS ONE 9:e109844. 10.1371/journal.pone.0109844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigali S., Titgemeyer F., Barends S., Mulder S., Thomae A. W., Hopwood D. A., et al. (2008). Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9, 670–675. 10.1038/embor.2008.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez H., Rico S., Díaz M., Santamaría R. I. (2013). Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microb. Cell Fact. 12:127. 10.1186/1475-2859-12-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokem J. S., Lantz A. E., Nielsen J. (2007). Systems biology of antibiotic production by microorganisms. Nat. Prod. Rep. 24, 1262–1287. 10.1039/b617765b [DOI] [PubMed] [Google Scholar]
- Romero D., Traxler M. F., López D., Kolter R. (2011). Antibiotics as signal molecules. Chem. Rev. 111, 5492–5505. 10.1021/cr2000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez S., Chávez A., Forero A., Garcia-Huante Y., Romero A., Sánchez M., et al. (2010). Carbon source regulation of antibiotic production. J. Antibiot. 63, 442–459. 10.1038/ja.2010.78 [DOI] [PubMed] [Google Scholar]
- Schäberle T. F., Orland A., König G. M. (2014). Enhanced production of undecylprodigiosin in Streptomyces coelicolor by co-cultivation with the corallopyronin A-producing myxobacterium, Corallococcus coralloides. Biotechnol. Lett. 36, 641–648. 10.1007/s10529-013-1406-0 [DOI] [PubMed] [Google Scholar]
- Seipke R. F., Kaltenpoth M., Hutchings M. I. (2012). Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol. Rev. 36, 862–876. 10.1111/j.1574-6976.2011.00313.x [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost M. R., Traxler M. F., Clardy J., Kolter R. (2012). Old meets new: using interspecies interactions to detect secondary metabolite production in actinomycetes. Methods Enzymol. 517, 89–109. 10.1016/B978-0-12-404634-4.00005-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D., Chen L., Wang W., Yu Z., Ren C., Zhang W., et al. (2009). afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 81, 1149–1160. 10.1007/s00253-008-1738-1 [DOI] [PubMed] [Google Scholar]
- Sidda J. D., Corre C. (2012). Gamma-butyrolactone and furan signaling systems in Streptomyces. Methods Enzymol. 517, 71–87. 10.1016/B978-0-12-404634-4.00004-8 [DOI] [PubMed] [Google Scholar]
- Sola-Landa A., Moura R. S., Martín J. F. (2003). The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U.S.A. 100, 6133–6138. 10.1073/pnas.0931429100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek M. A., Urem M., Tenconi E., Rigali S., van Wezel G. P. (2012). Engineering of N-acetylglucosamine metabolism for improved antibiotic production in Streptomyces coelicolor A3(2) and an unsuspected role of NagA in glucosamine metabolism. Bioengineered 3, 280–285. 10.4161/bioe.21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taechowisan T., Lu C., Shen Y., Lumyong S. (2005). Secondary metabolites from endophytic Streptomyces aureofaciens CMUAc130 and their antifungal activity. Microbiology 151, 1691–1695. 10.1099/mic.0.27758-0 [DOI] [PubMed] [Google Scholar]
- Takano E. (2006). γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9, 287–294. 10.1016/j.mib.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Tan G. Y., Bai L., Zhong J. J. (2013). Exogenous 1,4-butyrolactone stimulates A-factor-like cascade and validamycin biosynthesis in Streptomyces hygroscopicus 5008. Biotechnol. Bioeng. 110, 2984–2993. 10.1002/bit.24965 [DOI] [PubMed] [Google Scholar]
- Traxler M. F., Watrous J. D., Alexandrov T., Dorrestein P. C., Kolter R. (2013). Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio 4:e00459-13. 10.1128/mBio.00459-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Kawai S., Ogawa H., Kiyama A., Kubota T., Kawanobe H., et al. (2000). Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J. Antibiot. 53, 979–982. 10.7164/antibiotics.53.979 [DOI] [PubMed] [Google Scholar]
- Vargas-Bautista C., Rahlwes K., Straight P. (2014). Bacterial competition reveals differential regulation of the pks genes by Bacillus subtilis. J. Bacteriol. 196, 717–728. 10.1128/JB.01022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsigian K., Jajoo R., Kishony R. (2011). Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 9:e1001184. 10.1371/annotation/1d584443-c6b8-423b-8027-5f9034d4599f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ji J., Li X., Wang J., Li S., Pan G., et al. (2014a). Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 111, 5688–5693. 10.1073/pnas.1324253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang L., Zhuang Y., Kong F., Zhang C., Zhu W. (2014b). Phenolic polyketides from the co-cultivation of marine-derived Penicillium sp. WC-29-5 and Streptomyces fradiae 007. Mar Drugs 12, 2079–2088. 10.3390/md12042079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J. D., Phelan V. V., Hsu C. C., Moree W. J., Duggan B. M., Alexandrov T., et al. (2013). Microbial metabolic exchange in 3D. ISME J. 7, 770–780. 10.1038/ismej.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Charusanti P., Musiol-Kroll E. M., Jiang X., Tong Y., Kim H. U., et al. (2015). Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol. 33, 15–26. 10.1016/j.tibtech.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Oikawa H., Ogawa H. O., Hosono K., Shinmachi F., Takano H., et al. (2005). Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 151, 2899–2905. 10.1099/mic.0.28139-0 [DOI] [PubMed] [Google Scholar]
- Yang Y. H., Song E., Lee B. R., Kim E. J., Park S. H., Kim Y. G., et al. (2010). Rapid functional screening of Streptomyces coelicolor regulators by use of a pH indicator and application to the MarR-like regulator AbsC. Appl. Environ. Microbiol. 76, 3645–3656. 10.1128/AEM.02617-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G., Wang H. H., Davies J. (2007). Antibiotics as signalling molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 1195–1200. 10.1098/rstb.2007.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Zhu H., Dang F., Zhang W., Qin Z., Yang S., et al. (2012). Differential regulation of antibiotic biosynthesis by DraR-K, a novel two-component system in Streptomyces coelicolor. Mol. Microbiol. 85, 535–556. 10.1111/j.1365-2958.2012.08126.x [DOI] [PubMed] [Google Scholar]