Abstract
Tamoxifen is successfully used for both treatment and prevention of estrogen-dependent breast cancer, yet side effects and development of resistance remain problematic. Endoxifen is a major active metabolite of tamoxifen that is being investigated for clinical use. We hypothesized that endoxifen and perhaps other major metabolites of tamoxifen may affect the ability of human estrogen sulfotransferase 1E1 (hSULT1E1) and human phenol sulfotransferase 1A1 isoform 1 (hSULT1A1*1) to catalyze the sulfation of estradiol, an important mechanism in termination of estrogen signaling through loss of activity at estrogen receptors. Our results indicated that endoxifen, N-desmethyltamoxifen (N-desTAM), 4-hydroxytamoxifen (4-OHTAM), and tamoxifen-N-oxide were weak inhibitors of hSULT1E1 with Ki values ranging from 10 μM to 38 μM (i.e., over 1000 times higher than the 8.1 nM Km value for estradiol as substrate for the enzyme). In contrast to the results with hSULT1E1, endoxifen and 4-OHTAM were significant inhibitors of the sulfation of 2.0 µM estradiol catalyzed by hSULT1A1*1, with IC50 values (9.9 μM and 1.6 μM, respectively) that were similar to the Km value (1.5 μM) for estradiol as substrate for this enzyme. Additional investigation of the interaction of these metabolites with the two sulfotransferases revealed that endoxifen, 4-OHTAM, and N-desTAM were substrates for hSULT1E1 and hSULT1A1*1, although the relative catalytic efficiencies varied with both the substrate and the enzyme. These results may assist in future elucidation of cell- and tissue-specific effects of tamoxifen and its metabolites.
Introduction
Tamoxifen has been successfully used in the treatment of estrogen-dependent breast cancer for decades; however, its use is limited by a low incidence of endometrial cancer in some patient populations (van Leeuwen et al., 1994; Bernstein et al., 1999). Drug resistance and disease recurrence also occur with tamoxifen therapy. Tamoxifen functions as an anti-estrogen through the formation of 4-hydroxytamoxifen (4-OHTAM) (Jordan et al., 1977) and endoxifen (Wu et al., 2009; Maximov et al., 2014). However, tamoxifen has estrogenic activity in some tissues (MacGregor and Jordan, 1998), and such effects may be mediated through the estrogen receptor (Thompson et al., 1989). Other studies report estrogenic properties of tamoxifen metabolites (Jordan and Gosden, 1982; Jordan, 2007), and these effects may contribute to differential responses to tamoxifen therapy through estrogen receptor-related hormonal stimulation.
Estrogen is important for normal endocrine function (Pasqualini, 2009). Estradiol binds the estrogen receptor to induce cell growth and proliferation (Clemons and Goss, 2001). As one mechanism of inactivation, estradiol is converted into a sulfuric acid ester (sulfate) in a reaction catalyzed by human estrogen sulfotransferase 1E1 (hSULT1E1). Although hSULT1E1 is the primary sulfotransferase involved in the sulfation of estradiol at physiologic substrate concentrations (Zhang et al., 1998), estradiol is also a substrate for human phenol sulfotransferase 1A1 (hSULT1A1) at higher concentrations (Nagar et al., 2006). Sulfation represents a major route for the hormonal inactivation of estrogens, and this mechanism protects surrounding tissues from excessive estrogenic effects and is associated with tumor regression in estrogen-dependent carcinogenesis (Suzuki et al., 2003).
Polymorphisms within hSULT1A1 may affect individual responses to some therapeutic agents that require metabolism by this enzyme (Raftogianis et al., 1997, 1999). Human phenol sulfotransferase 1A1 isoform 1 (hSULT1A1*1) is the dominant variant of human phenol sulfotransferase (Coughtrie et al., 1999) and has been extensively studied in relation to the therapeutic outcome and pharmacogenetics of tamoxifen (Nowell et al., 2002; Wegman et al., 2005; Grabinski et al., 2006; Mercer et al., 2010; Serrano et al., 2011). Although hSULT1A1*1 may enhance the therapeutic effects of tamoxifen in breast cancer cells (Mercer et al., 2010), the complete roles of hSULT1A1*1 or hSULT1E1 in the clinical response to tamoxifen remain to be fully determined.
Major metabolites of tamoxifen, including N-desmethyltamoxifen (N-desTAM), tamoxifen-N-oxide (TAM-NO), 4-OHTAM, and endoxifen, were recently shown to inhibit the sulfation of steroid substrates for hSULT2A1 (Squirewell et al., 2014). Thus, we hypothesized that tamoxifen metabolites were also inhibitors of estradiol sulfation catalyzed by hSULT1E1 and hSULT1A1*1. Decreases in the catalytic activity of either enzyme may increase the physiologic concentrations of unconjugated (active) estradiol as a mechanism of clinical resistance. Moreover, the involvement of estrogen in endometrial carcinogenesis (Rižner, 2013; Hernandez-Ramon et al., 2014) may relate to the endometrial cancer side effect of tamoxifen.
Materials and Methods
Chemicals and Instruments.
Expression plasmids (pReceiver-B02) for hSULT1E1 and hSULT1A1*1 were obtained from GeneCopoeia (Rockville, MD). The Pure Yield Plasmid Mini-Prep System was obtained from Promega (Madison, WI). Antisense (5′-CAG CCT AGG AAC GCC CAA CTT-3′) and sense (5′-GCG TAG AGG ATC GAG ATC GAT-3′) primers for sequencing were obtained from Integrated DNA Technologies (Coralville, IA). Escherichia coli BL21 (DE3) cells were obtained from Life Technologies (Grand Island, NY). DNA grade Hydroxyapatite was purchased from Bio-Rad (Hercules, CA). Bacto tryptone and yeast extract were purchased from BD Biosciences (Sparks, MD). Ampicillin, dithiothreitol (DTT), and granulated LB broth (Miller’s LB Broth) were obtained from Research Products International (Mount Pleasant, IL). Thin-layer chromatography (TLC) sheets (60 Å silica gel without indicator) were obtained from EMD Millipore (Billerica, MA). Adenosine 3′-phosphate 5′-phosphosulfate lithium salt hydrate (PAPS) was obtained from Sigma-Aldrich (St. Louis, MO) and purified upon arrival using a previously described protocol (Sekura, 1981) to a purity greater than 99% as determined by high-pressure liquid chromatography (Sheng and Duffel, 2001). 2-Mercaptothanol, estradiol, estradiol-sulfate, potassium phosphate, (Z)-tamoxifen, (Z)-N-desmethyltamoxifen HCl, (Z)-4-hydroxytamoxifen, and (E/Z)-4-hydroxy-N-desmethyl-tamoxifen hydrochloride hydrate (endoxifen) were purchased from Sigma-Aldrich at the highest available purity (≥98%). Synthesis of tamoxifen-N-oxide (TAM-NO) was performed, as described elsewhere (Foster et al., 1980; Mani and Kupfer, 1991). N-desmethyltamoxifen-sulfamate (N-desTAM-S) and 4-hydroxytamoxifen-sulfate (4-TAM-SO4) were synthesized from sulfuryl imidazolium triflate, according to our previously published method (Squirewell et al., 2014). [3H]-Estradiol (81.0 Ci/mmol) was obtained from Perkin Elmer (Waltham, MA). Radioactive samples were analyzed with a Tri-Carb 2900TR liquid scintillation counter using Econo-Safe liquid scintillation cocktail (Research Products International, Mount Prospect, IL). Data were analyzed using the Enzyme Kinetics Module (version 1.3) of Sigma Plot 11.0 (Systat Software, San Jose, CA).
Expression and Purification of Recombinant hSULT1E1.
A pReceiver-B02 expression clone (2 µl, 108 ng) harboring the gene encoding the native form of hSULT1E1 was transformed into E. coli BL21 (DE3) cells (50 µl). The bacterial cells were incubated on ice for 20 minutes and then heat-shocked at 42°C for 32 seconds. The cells were immediately cooled on ice for 2 minutes, after which was added 180 µl sterile prewarmed Super Optimal broth with Catabolite repression media. The bacterial cells were grown for 1 hour on a reciprocating shaker (250 rpm) at 37°C. Afterward, a 35 µl aliquot of the cell suspension was added to an LB-ampicillin agar plate containing 100 µg/ml ampicillin and incubated at 37°C for 18 hours. A single colony from the LB-ampicillin agar plate was added to 8.0 ml sterile LB containing 100 µg/ml ampicillin and incubated overnight at 37°C on a reciprocating shaker (250 rpm). The bacterial culture was later inoculated into 120 ml sterile LB containing 100 µg/ml ampicillin and incubated overnight at 37°C on a reciprocating shaker (250 rpm). After the incubation, a 20 ml aliquot from the 120 ml bacterial culture was inoculated into 1.0 L sterile Terrific broth containing 100 µg/ml ampicillin and incubated at 37°C on a reciprocating shaker (250 rpm). The bacterial culture was grown to an optical density600 of 1.0 and then induced with 300 µM isopropyl-1-thio-D-galactopyranoside. The culture was then incubated on a reciprocating shaker (250 rpm) overnight at 30°C. Cells were subjected to centrifugation at 10,000g for 1 hour at 4°C, and the supernatant was discarded. The cell pellet was resuspended in 10 ml ice-cold bacterial lysis buffer A [10 mM Tris-HCl, pH 7.5, containing 0.25 M sucrose, 1 mM dithiothreitol, 10% (v/v) glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 µM pepstatin A, 3.3 µM antipain, 10 µM trans-epoxysuccinyl-L-leucylamido-(4-guanidino)-butane, and 100 µM leupeptin] and snap-frozen in liquid nitrogen. The cells were thawed and supplemented with chicken lysozyme such that the final concentration of lysozyme was 0.5 mg/ml. Cells were gently shaken for 15 minutes at 4°C and then snap-frozen in liquid nitrogen to complete the cell lysis. The thawed cells were supplemented with DNase (0.5 mg) and gently shaken for 15 minutes at 4°C. The cytosolic fraction containing hSULT1E1 activity was recovered following centrifugation at 100,000g for 1 hour.
The cell extract (440 mg protein) was applied to a DE-52 anion exchange column (2.5 × 20 cm) equilibrated with buffer B [50 mM Tris-HCl, pH 7.5, containing 0.25 M sucrose, 1 mM DTT, 10% (v/v) glycerol, and 0.05% (v/v) Tween 20] and then washed with approximately 1 L buffer B to remove proteins that did not bind to this column. Once the absorbance of the eluate at 280 nm had reached a baseline value, the hSULT1E1 was eluted with a linear gradient formed between 200 ml buffer B and 200 ml buffer B containing 0.1 M KCl. The fractions containing hSULT1E1 were then combined and concentrated by ultrafiltration (Amicon stirred cell with a YM10 membrane; Millipore, Bedford, MA). The concentration of potassium chloride was then reduced through successive dilution and concentration by ultrafiltration, with the dilutions carried out using the same buffer to be employed for the subsequent hydroxyapatite chromatography step [i.e., buffer C: 10 mM potassium phosphate, pH 6.8, 0.25 M sucrose, 1 mM DTT, and 0.05% (v/v) Tween 20]. The resulting protein (26 mg) was applied to a column of hydroxyapatite (2.5 × 3.0 cm) that had been equilibrated with buffer C. Buffer C was used to wash the column and remove all nonbinding proteins, and the elution of hSULT1E1 was carried out with a linear gradient formed between 80 ml buffer C and 80 ml buffer C containing 0.4 M potassium phosphate. The fractions containing hSULT1E1 activity were pooled and concentrated by ultrafiltration using an Amicon membrane. Approximately 10 mg purified hSULT1E1 was recovered from the hydroxyapatite column. The subunit molecular mass of hSULT1E1 was found to be approximately 35 kDa by SDS-PAGE, which is consistent with previously reported data for this enzyme (Aksoy et al., 1994). The purity of hSULT1E1 was greater than 94% when analyzed by densitometry on SDS-PAGE. At each purification step, hSULT1E1-containing fractions were identified and quantitated with a previously described methylene blue assay (Duffel et al., 1989) using 25 µM estradiol as substrate. Protein concentration was determined using a standard Bradford assay (Bradford, 1976) with bovine serum albumin as a standard. The complete DNA coding sequence of hSULT1E1 was verified using the sense and antisense sequencing primers described above.
Expression and Purification of Recombinant hSULT1A1*1.
A pReceiver-B02 expression clone (0.6 µl, 114 ng) harboring the gene encoding the native form of hSULT1A1*1 was transformed into E. coli BL21 (DE3) cells (50 µl). The expression and extraction of hSULT1A1*1 from these cells were carried out utilizing a procedure similar to that described above for hSULT1E1. The cell extract (300 mg protein) was applied to a DE-52 anion exchange column (2.5 × 15 cm) equilibrated with buffer B and washed with approximately 1 L buffer B to elute those proteins that did not bind to the column. Once the absorbance of the eluate at 280 nm had reached a baseline value, the hSULT1A1*1 was eluted with a linear gradient formed between 200 ml buffer B and 200 ml buffer B containing 0.1 M KCl. The fractions containing hSULT1A1*1 were then combined, and the concentration of potassium chloride was reduced through successive dilution and concentration by ultrafiltration with buffer C. The protein obtained from the DE-52 anion exchange column (40 mg protein) was applied to a hydroxyapatite column (2.5 × 5.0 cm) that had been equilibrated with buffer C, and the column was then washed with buffer C to remove all nonbinding proteins. Once the absorbance of the eluate at 280 nm returned to baseline, separation was achieved with a linear gradient formed between 100 ml buffer C and 100 ml buffer C containing 0.4 M potassium phosphate. Analysis by SDS-PAGE revealed minor impurities after the column of hydroxyapatite. Thus, to prepare this mixture for the next step in purification, the fractions containing hSULT1A1*1 activity were combined and the concentration of potassium phosphate reduced through successive dilution and concentration by ultrafiltration with buffer C. The hSULT1A1*1 obtained from the hydroxyapatite (16 mg protein) was loaded onto a second hydroxyapatite column (2.5 × 5.0 cm) equilibrated with buffer C. Following the initial removal of nonbinding proteins, the hSULT1A1*1 was eluted with a linear gradient formed between 100 ml buffer C and 100 ml buffer C containing 80 mM potassium phosphate. The fractions containing hSULT1A1*1 with the highest activity were combined and concentrated. Approximately 8 mg purified hSULT1A1*1 was recovered from the second column of hydroxyapatite. The subunit molecular mass of the hSULT1A1*1 was found to be approximately 34 kDa, which is consistent with previously reported data for this enzyme (Wilborn et al., 1993). The purity of hSULT1A1*1 was determined to be greater than 96% when analyzed by densitometry on SDS-PAGE. Chromatography fractions were analyzed for hSULT1A1*1 activity at pH 7.4 using the methylene blue assay and 25 µM 2-naphthol as substrate. Protein concentration was determined at each step of the purification process using the Bradford assay with bovine serum albumin as a standard. The complete DNA coding sequence of hSULT1A1*1 was verified using the antisense and sense sequencing primers described above.
Inhibition of hSULT1E1-Catalyzed Sulfation of Estradiol.
Assays for estradiol sulfation catalyzed by hSULT1E1 were performed utilizing the following procedure. Each 200 μl reaction was performed at pH 7.4 and contained 0.25 M potassium phosphate, 50 µM PAPS, and 8.3 mM 2-mercaptoethanol. [3H]-Estradiol and tamoxifen metabolites were dissolved in absolute ethanol, and they were added to the reaction mixture in volumes such that the final concentration of ethanol in each assay was 2% (v/v). The reactions were initiated by the addition of 1.0 µl purified hSULT1E1 (3.0 ng) and incubated for 4 minutes at 37°C. The reactions were then terminated by the addition of 800 μl 0.25 M Tris-HCl, pH 8.7 (Nishiyama et al., 2002), and 4.0 ml chloroform. Samples were vortexed vigorously for 20 seconds and subjected to centrifugation at 1500 rpm for 5 minutes to separate the phases. A 500 μl aliquot of the upper aqueous phase containing [3H]-estradiol-sulfate was added to 10 ml liquid scintillation cocktail, and the radioactivity was determined using a Perkin Elmer Tri-Carb 2900TR liquid scintillation analyzer.
Inhibition of hSULT1A1*1-Catalyzed Sulfation of Estradiol.
Assays for estradiol sulfation catalyzed by hSULT1A1*1 were performed as described below. Each 200 μl reaction was performed at pH 7.4 and contained 0.25 M potassium phosphate, 50 µM PAPS, and 8.3 mM 2-mercaptoethanol. [3H]-Estradiol and tamoxifen metabolites were dissolved in absolute ethanol, and they were added to the reaction mixture in volumes such that the final concentration of ethanol in each assay was 2% (v/v). The reactions were initiated by the addition of 1.0 µl purified hSULT1A1*1 (0.74 µg) and incubated for 10 minutes at 37°C. The reactions were then terminated by the addition of 800 μl 0.25 M Tris-HCl, pH 8.7 (Nishiyama et al., 2002), and 4.0 ml chloroform. Samples were vortexed vigorously for 20 seconds and subjected to centrifugation at 1500 rpm for 5 minutes to separate the phases. A 500 μl aliquot of the upper aqueous phase containing [3H]-estradiol-sulfate was added to 10 ml liquid scintillation cocktail, and the radioactivity was determined as described above.
Tamoxifen Metabolites as Substrates for hSULT1E1 and hSULT1A1*1.
Tamoxifen metabolites were investigated as substrates for the enzymes using a previously described protocol that determines the incorporation of a radiolabeled sulfuryl moiety from [35S]-PAPS into products of the reaction (Lyon et al., 1981). Each 50 μl reaction was performed at pH 7.4 and contained 50 µM [35S]-PAPS with 0.25 M potassium phosphate, 8.3 mM 2-mercaptoethanol, and the indicated concentrations of tamoxifen metabolites dissolved in dimethylsulfoxide, with a final dimethylsulfoxide concentration of 2% (v/v). The reactions were initiated by the addition of either purified hSULT1E1 (0.86 µg) or hSULT1A1*1 (0.74 µg) at 37°C, incubated for 20 minutes, and terminated with 50 µl methanol. A 10 μl aliquot of the resulting mixture was applied to Silica Gel 60 TLC sheets (w/o indicator) and developed in chloroform/methanol (3:7) until the solvent migrated approximately 8 cm from the origin. An area of the TLC sheet 5.5 cm below and including the solvent front (i.e., that contained the section of the radiolabeled sulfated products) was excised and placed in 10 ml scintillation cocktail for determination of radioactivity. The location of the sulfated products on TLC was determined prior to the radiolabeled assay using synthesized standards for 4-TAM-SO4 and N-desTAM-S.
Determination of the Kinetic Mechanism of Inhibition.
Endoxifen, 4-OHTAM, N-desTAM, TAM-NO, N-desTAM-S, 4-TAM-SO4, and tamoxifen were used as inhibitors of the enzymes. Data were fit to rate equations for competitive, noncompetitive, uncompetitive, or mixed inhibition using a nonlinear least-squares algorithm in the Enzyme Kinetics Module (version 1.3) of Sigma Plot 11.0, and the model with the highest value for the coefficient of determination, r2, was selected. In cases where r2 was not significantly different, the model with the lowest corrected Akaike Information Criterion was selected.
Identification of Enzyme Reaction Products by Liquid Chromatography and Mass Spectrometry.
Products of sulfation catalyzed by hSULT1E1 and hSULT1A1*1 were identified by liquid chromatography–mass spectrometry (LC-MS) analysis on a Waters Q-TOF Premiere mass spectrometer, as described previously (Squirewell et al., 2014). Briefly, each 50 μl reaction was performed at pH 7.4 and used 50 μM substrate with 50 µM PAPS in the presence of 0.25 M potassium phosphate, 8.3 mM 2-mercaptoethanol, and 2% ethanol (v/v). The reactions were initiated with the addition of either hSULT1E1 (4.6 µg) or hSULT1A1*1 (3.7 µg) at 37°C for 60 minutes and terminated with 50 µl methanol. A 10 μl aliquot of each sample was analyzed using a Waters Aquity (UPLC) BEH C18 column (2.1 mm × 100 mm; 1.7 μm) operated at a flow rate of 0.25 ml/min and a UV wavelength of 213 nm. A linear gradient system was programmed to 40% acetonitrile with 0.1% (v/v) formic acid for 15 minutes and 40–70% (v/v) acetonitrile with 0.1% (v/v) formic acid for 5 minutes, and then sustained at 70% acetonitrile with 0.1% formic acid for 10 minutes. The liquid chromatography-eluate was subjected to mass spectral analysis through interface with an electrospray ionization source operated in negative ion mode.
Results
Metabolites of Tamoxifen Are Weak Inhibitors of the Sulfation of Estradiol Catalyzed by hSULT1E1.
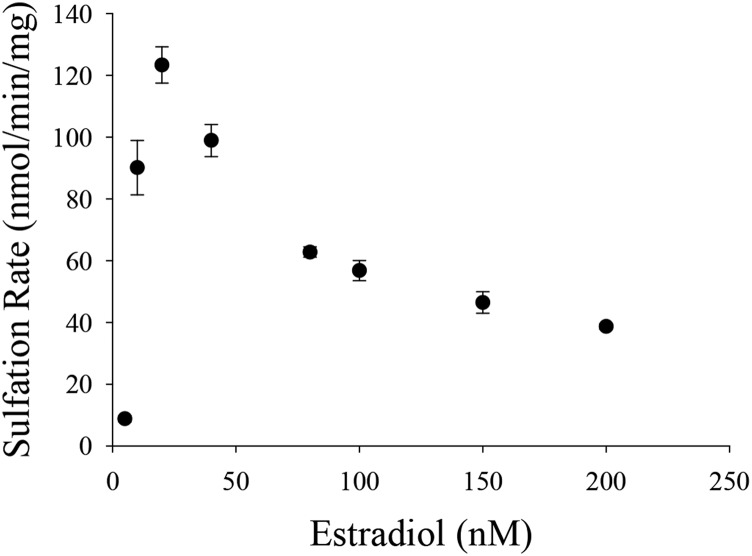
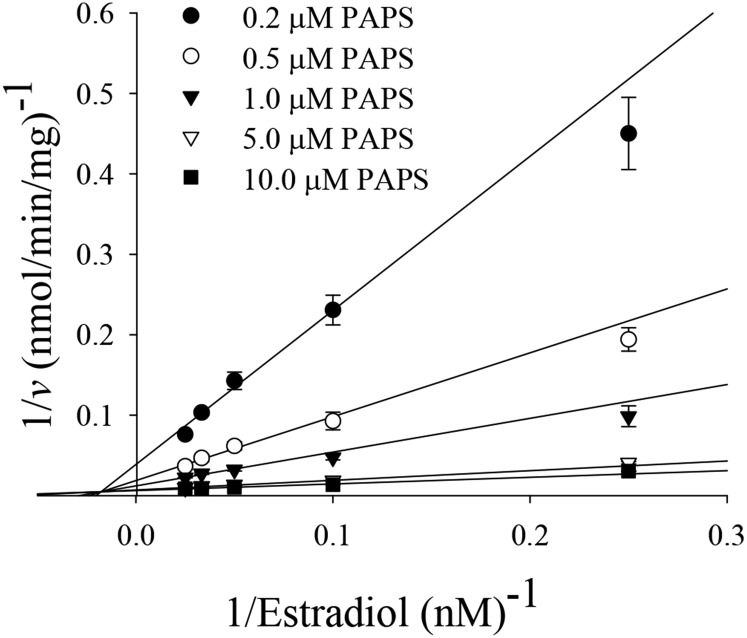
Endoxifen, 4-OHTAM, TAM-NO, and N-desTAM were investigated as inhibitors of hSULT1E1 using estradiol as substrate. The sulfation of estradiol was initially examined with 200 µM PAPS and a substrate concentration range between 5.0 and 200 nM to determine the concentrations of estradiol where minimal substrate inhibition occurred (Fig. 1). The sulfation of estradiol could not be described using a simple substrate inhibition model, nor could the data be described using an equation that assumes partial substrate inhibition as noted in previous studies with hSULT1E1 (Zhang et al., 1998). Due to variations in the methodology and reaction conditions used to determine the sulfation of estradiol, it is possible that changes in the enzyme environment (i.e., pH 7.4 in the current study versus pH 6.3 in previous work) could contribute to the differences in substrate inhibition that were observed. Thus, an equation that accurately represents substrate inhibition during the hSULT1E1-catalyzed sulfation of estradiol at pH 7.4 may be more complex than previously assumed. In efforts to determine the kinetic constants for estradiol sulfation and to verify the kinetic mechanism of hSULT1E1, estradiol (50 µM) was examined with varied PAPS concentrations (50 nM–100 µM) to determine those concentrations of PAPS where minimal substrate inhibition occurred. Substrate inhibition was not observed with PAPS at pH 7.4 (Supplemental Fig. 1), and this has been previously reported elsewhere (Falany et al., 1995). Sulfation rates were then examined with varied concentrations of estradiol (4 nM–40 nM) and varied concentrations of PAPS (0.2 µM–10.0 µM). The data from this study were best described with a sequential rate equation (Fig. 2), which is in agreement with the kinetic mechanism of hSULT1E1 previously determined by Zhang et al. (1998). PAPS displayed a Km value of 1.2 ± 0.3 µM, and the Km, Ki, Vmax, and kcat/Km derived from the hSULT1E1-catalyzed sulfation of estradiol was determined to be 8.1 ± 1.6 nM, 56 ± 4 nM, 179 ± 9 nmol/min/mg, and 1.6 ± 0.3 minute−1nM−1, respectively.
Fig. 1.
Initial velocities of hSULT1E1-catalyzed sulfation of estradiol in the presence of 50 µM PAPS. Data are the means ± S.E. from triplicate determinations.
Fig. 2.
Initial rates for the sulfation of estradiol catalyzed by hSULT1E1 at low concentrations of estradiol and varied concentrations of PAPS. Data were fit to a sequential rate equation, and individual data points are the means ± S.E. from triplicate determinations.
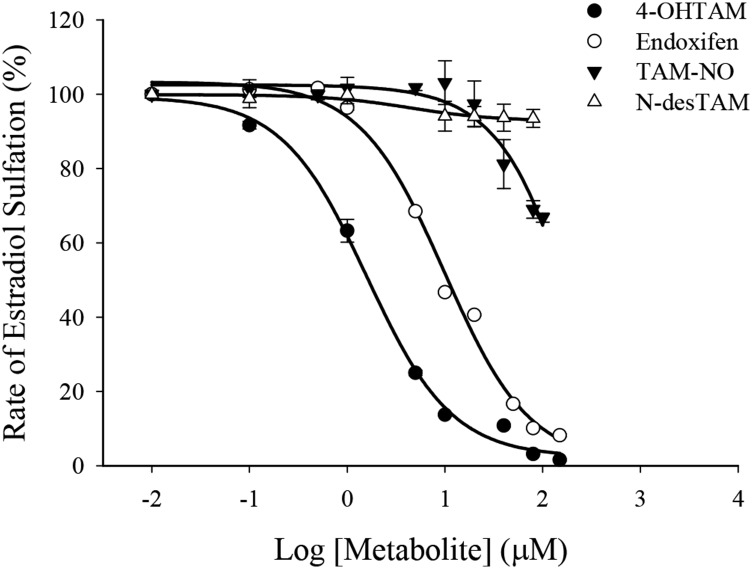
Endoxifen, 4-OHTAM, N-desTAM, and TAM-NO were all weak inhibitors of estradiol sulfation catalyzed by hSULT1E1 (Fig. 3). Tamoxifen did not exhibit significant inhibition of hSULT1E1 up to the limits of its solubility in the assay (data not shown). Endoxifen, 4-OHTAM, TAM-NO, and N-desTAM displayed greater than 95% inhibition of the enzyme within their solubility limits. The calculated IC50 values ranged from 7.0 μM to 21.0 μM for the inhibition of the sulfation of 7.0 nM estradiol, with 4-OHTAM being the most potent inhibitor. The kinetic mechanism of inhibition, apparent (app) maximum velocity (Vmax), Michaelis-Menten constant (Km), inhibitor dissociation constant (Ki), and catalytic efficiency constant (kcat/Km) for inhibitors of the hSULT1E1-catalyzed sulfation of estradiol are reported in Table 1, with the initial velocity data in Supplemental Fig. 2. N-desTAM was a mixed inhibitor of hSULT1E1 with a Ki value of 10 μM, whereas endoxifen and 4-OHTAM were noncompetitive inhibitors with Ki values of 30 μM and 38 μM, respectively. Initial velocity data for TAM-NO showed a significant deviation from a noncompetitive inhibition model at 80 μM inhibitor concentration (Supplemental Fig. 2D). Other standard inhibition models (e.g., competitive and mixed inhibition) also failed to describe this behavior at higher inhibitor concentration. This observation was reproducible in later studies with TAM-NO, and its cause remains unclear. Also of note, the estradiol concentrations used to determine the inhibitor dissociation constant for each metabolite (0.5–1.3 × Km) were lower than the estradiol concentrations used in initial velocity studies with hSULT1E1 (0.5–5 × Km). Thus, Vmax and Km values for the hSULT1E1-catalyzed sulfation of estradiol in the presence of metabolites (Table 1) are higher because they do not account for the substrate inhibition reflected in the Vmax and Km values (179 nmol/min/mg and 8.1 nM, respectively) for estradiol when determined at higher substrate concentrations in the absence of tamoxifen metabolites.
Fig. 3.
Inhibition of the hSULT1E1-catalyzed sulfation of 7.0 nM estradiol by major metabolites of tamoxifen. Rates of sulfation of estradiol for uninhibited controls were 62, 67, 58, and 69 nmol/min/mg protein for studies with endoxifen, N-desTAM, 4-OHTAM, and TAM-NO, respectively. Data are the means ± S.E. from triplicate determinations.
TABLE 1.
Inhibition of hSULT1E1-catalyed sulfation of estradiol by metabolites of tamoxifen
The sulfation of estradiol was determined using 3.0 ng purified hSULT1E1 in the presence of varied concentrations of inhibitor and either 7.0 nM estradiol (for IC50 values) or 3 nM–10 nM estradiol for determination of the best fit to a kinetic model of inhibition and the kinetic constants for that fit to the model. In the case of TAM-NO, the inconclusive fit to a kinetic model is denoted as N.A. The data are expressed as the means ± S.E. from three independent experiments. Calculation of kcat values was based on 70,252 as the dimeric molecular mass of hSULT1E1.
| Metabolite | IC50 | Inhibition Model | Vmax(app) | Km(app) | kcat/Km | Ki |
|---|---|---|---|---|---|---|
| μM | nmol/min/mg | nM | min−1nM−1 | μM | ||
| Endoxifen | 21 ± 1 | Noncompetitive | 680 ± 237 | 63 ± 25 | 0.76 ± 0.4 | 30 ± 1 |
| N-desTAM | 8.2 ± 0.9 | Mixed | 435 ± 66 | 38 ± 7 | 0.81 ± 0.2 | 10 ± 1 |
| 4-OHTAM | 7.0 ± 1.1 | Noncompetitive | 553 ± 149 | 57 ± 17 | 0.68 ± 0.3 | 38 ± 1 |
| TAM-NO | 18 ± 1 | N.A. |
Endoxifen and 4-OHTAM Are Potent Inhibitors of the Sulfation of Estradiol Catalyzed by hSULT1A1*1.
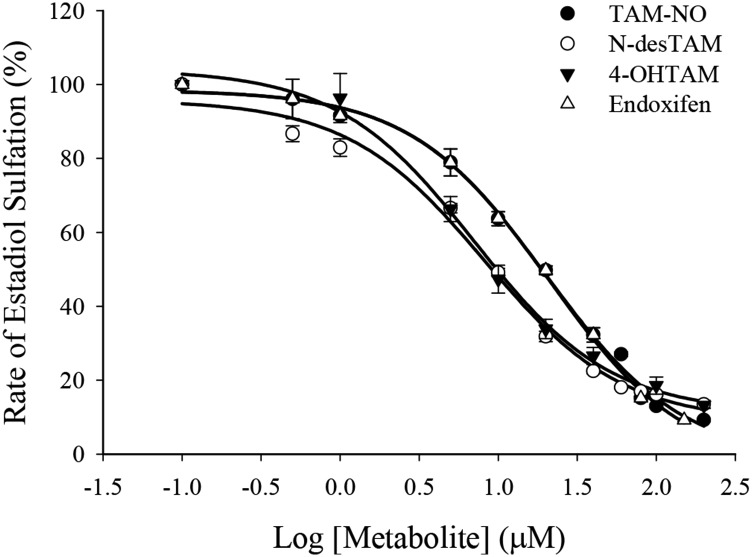
Endoxifen, 4-OHTAM, TAM-NO, and N-desTAM were investigated as inhibitors of hSULT1A1*1 at pH 7.4 using estradiol as substrate. The sulfation of estradiol was initially examined with PAPS (50 µM) and varied concentrations of estradiol (0.1–25.0 μM) to determine the concentrations of estradiol where minimal substrate inhibition occurred (Fig. 4). The Km, Ki, Vmax, and kcat/Km derived from the hSULT1A1*1-catalyzed sulfation of estradiol were 1.5 ± 0.2 µM, 14 ± 2 µM, 11 ± 1 nmol/min/mg, and 0.5 ± 0.1 minute−1µM−1, respectively. Estradiol sulfation was later examined using a single concentration of estradiol (5 µM) with varied concentrations of PAPS (1.0–100 µM) to determine the concentrations of PAPS that were saturating for the enzyme (Supplemental Fig. 3). Subsequent inhibition studies used 50 µM PAPS as cosubstrate. Of the metabolites studied, only endoxifen and 4-OHTAM were significant inhibitors of estradiol sulfation catalyzed by hSULT1A1*1 (Fig. 5). These metabolites displayed greater than 95% inhibition of the enzyme within their solubility limits, with IC50 values of 1.6 ± 0.9 μM for 4-OHTAM and 9.9 ± 0.9 μM for endoxifen. TAM-NO was also an inhibitor of hSULT1A1*1; however, the calculated IC50 value for this metabolite was greater than 100 µM when examined with 2 µM estradiol as substrate. N-desTAM and tamoxifen (data not shown) were not significant inhibitors of estradiol sulfation within their solubility limits. Initial velocity data on the inhibition of hSULT1A1*1 are shown in Supplemental Fig. 4. At estradiol concentrations of 0.5–2.5 µM, the data for 4-OHTAM–mediated inhibition of the enzyme were described well by a competitive inhibition model with a Ki value of 1.6 ± 0.1 μM (apparent Km and Vmax values under these reaction conditions were 3.9 ± 0.6 μM and 18 ± 2 nmol/min/mg, respectively). Although the data fit a competitive inhibition model for 4-OHTAM as inhibitor at these low concentrations of estradiol, there was a significant deviation of endoxifen from any simple inhibition models, as was especially apparent at 40 μM endoxifen (Supplemental Fig. 4A). This observation was reproducible in later studies with endoxifen, and its cause remains unclear.
Fig. 4.
Initial velocities of hSULT1A1*1-catalyzed sulfation of estradiol in the presence of 50 µM PAPS.
Fig. 5.
Inhibition of the hSULT1A1*1-catalyzed sulfation of 2.0 µM estradiol by major metabolites of tamoxifen. Reactions were conducted with 0.74 µg purified enzyme in the presence of the indicated concentrations of inhibitor. Rates of sulfation of estradiol for uninhibited controls were 5.1, 5.8, 5.7, and 5.7 nmol/min/mg protein for studies with endoxifen, N-desTAM, 4-OHTAM, and TAM-NO, respectively. The calculated IC50 values for 4-OHTAM and endoxifen were 1.6 ± 0.9 µM and 9.9 ± 0.9 µM, respectively. Data points are the means ± S.E. from triplicate determinations.
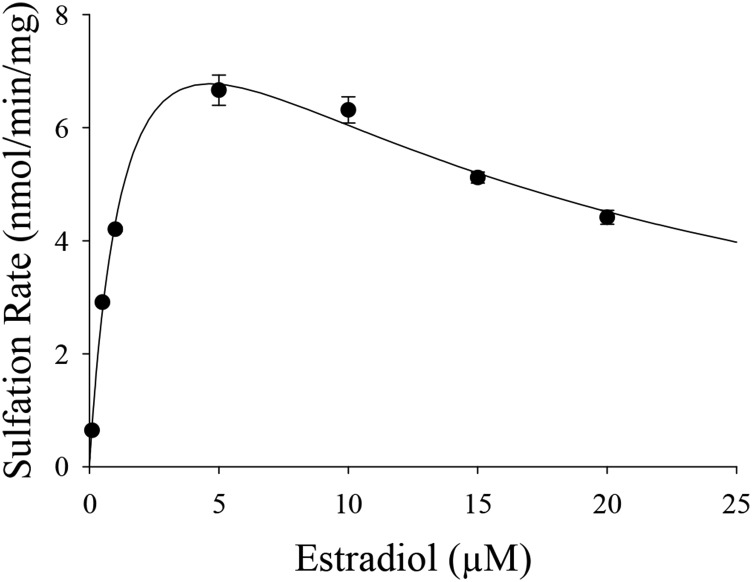
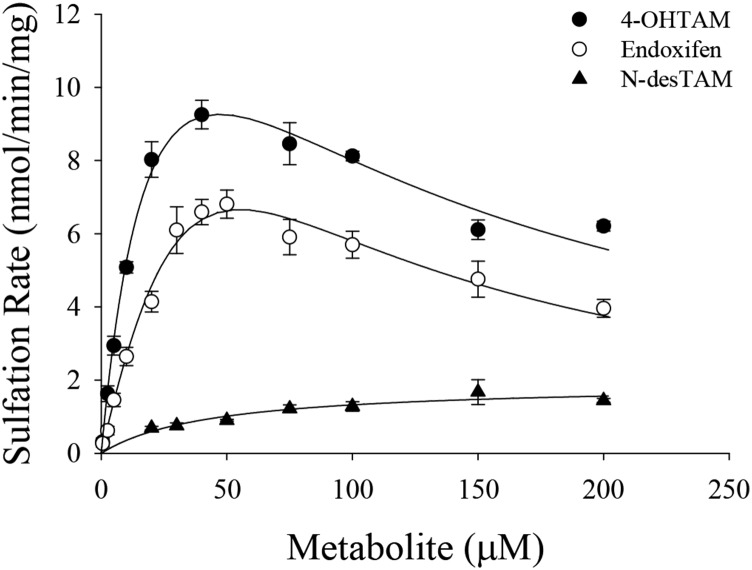
Characterization of 4-OHTAM, N-desTAM, and Endoxifen as Substrates for hSULT1E1.
Previous studies have shown that 4-OHTAM is a substrate for hSULT1E1 (Falany et al., 2006). However, the sulfation kinetics of either endoxifen or N-desTAM with hSULT1E1 have never been fully examined. In efforts to ascertain the metabolic fate of these metabolites, endoxifen, 4-OHTAM, and N-desTAM were examined as substrates for hSULT1E1 (Fig. 6). Sulfation kinetics for endoxifen, 4-OHTAM, and N-desTAM were best described using a substrate inhibition model, and the kinetic constants obtained for the hSULT1E1-catalyzed sulfation of these metabolites are reported in Table 2. Relative catalytic efficiencies for the metabolites were endoxifen > 4-OHTAM > N-desTAM. The enzymatic reactions were analyzed by LC-MS, and the negative ion electrospray ionization-mass spectrometry (ESI-MS) of products formed by the hSULT1E1-catalyzed sulfation of endoxifen, 4-OHTAM, and N-desTAM (Supplemental Figs. 5–7, respectively) were similar to those of products catalyzed by hSULT2A1 in our previous study (Squirewell et al., 2014). The retention times of endoxifen-sulfate, 4-TAM-SO4, and N-desTAM-S on LC-MS chromatograpy (data not shown) were 16.06, 16.30, and 21.91 minutes, respectively.
Fig. 6.
Sulfation of 4-OHTAM, N-desTAM, and endoxifen catalyzed by hSULT1E1. Data are the means ± S.E. from triplicate determinations. Curves represent the fit of the data to an equation for uncompetitive substrate inhibition.
TABLE 2.
Kinetic constants for the hSULT1E1-catalyzed sulfation of 4-OHTAM, N-desTAM, and endoxifen
The sulfation of each metabolite was determined using 0.86 µg purified hSULT1E1. Data were fit to a standard uncompetitive substrate inhibition equation and are the means ± S.E. from three independent experiments. Calculation of kcat values was based on 70,252 as the dimeric molecular mass of hSULT1E1.
| Metabolite | Km | Vmax | kcat/Km | Ki |
|---|---|---|---|---|
| µM | nmol/min/mg | min−1µM−1 | µM | |
| 4-OHTAM | 24 ± 5 | 12 ± 1 | 0.036 ± 0.008 | 387 ± 133 |
| Endoxifen | 24 ± 5 | 19 ± 2 | 0.057 ± 0.013 | 283 ± 86 |
| N-desTAM | 96 ± 52 | 26 ± 11 | 0.019 ± 0.013 | 144 ± 105 |
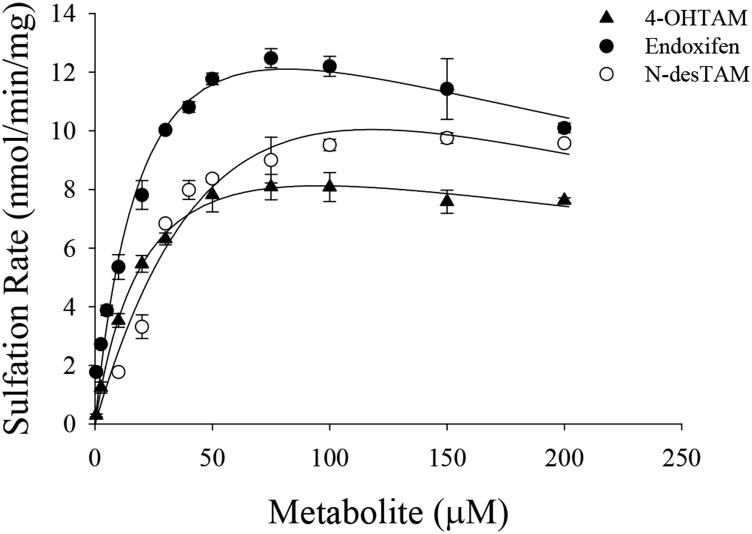
Characterization of 4-OHTAM, N-desTAM, and Endoxifen as Substrates for hSULT1A1*1.
Although 4-OHTAM is a known substrate for hSULT1A1*1 (Nagar et al., 2006), endoxifen and N-desTAM have never been formally examined as substrates for this enzyme. The results from the current study indicated that 4-OHTAM, N-desTAM, and endoxifen were all substrates for hSULT1A1*1. The kinetics of N-desTAM sulfation was best described using a Michaelis-Menten equation, whereas the data for the sulfation of 4-OHTAM and endoxifen were best described using a substrate inhibition model (Fig. 7). Kinetic constants obtained from the hSULT1A1*1-catalyzed sulfation of 4-OHTAM, N-desTAM, and endoxifen are shown in Table 3. In this study, hSULT1A1*1 displayed higher catalytic activity with endoxifen than with N-desTAM, as indicated by a ninefold higher kcat/Km. Additionally, the enzyme displayed a much higher catalytic activity with 4-OHTAM than N-desTAM with a 22-fold higher kcat/Km. The enzymatic reactions were analyzed by LC-MS, and the negative ion ESI-MS of the products formed by the hSULT1A1*1-catalyzed sulfation of 4-OHTAM, endoxifen, and N-desTAM (Supplemental Figs. 8–10, respectively) were similar to previous ESI-MS data from reactions catalyzed by hSULT2A1 (Squirewell et al., 2014). The retention times of 4-TAM-SO4, endoxifen-sulfate, and N-desTAM-S on LC-MS chromatography (data not shown) were 16.30, 16.07, and 21.81 minutes, respectively.
Fig. 7.
Sulfation of 4-OHTAM, N-desTAM, and endoxifen catalyzed by hSULT1A1*1. Data are the means ± S.E. from triplicate determinations. Curves represent fit of the data to a simple Michaelis Menten equation (for N-desTAM) and to an equation for uncompetitive substrate inhibition (for 4-OHTAM and endoxifen).
TABLE 3.
Kinetic constants for the hSULT1A1*1-catalyzed sulfation of 4-OHTAM, N-desTAM, and endoxifen
The sulfation of each metabolite was determined using 0.74 µg purified hSULT1A1*1. Data are the means ± S.E. from three independent experiments and were fit to either a standard uncompetitive substrate inhibition equation (for 4-OHTAM and endoxifen) or a Michaelis Menten equation (for N-desTAM). Calculation of kcat values was based on 68,312 as the dimeric molecular mass of hSULT1A1*1.
| Metabolite | Km | Vmax | kcat/Km | Ki |
|---|---|---|---|---|
| µM | nmol/min/mg | min−1µM−1 | µM | |
| 4-OHTAM | 26 ± 5 | 20 ± 3 | 0.050 ± 0.012 | 84 ± 19 |
| Endoxifen | 118 ± 82 | 35 ± 20 | 0.020 ± 0.018 | 26 ± 18 |
| N-desTAM | 44 ± 14 | 1.9 ± 0.2 | 0.003 ± 0.001 |
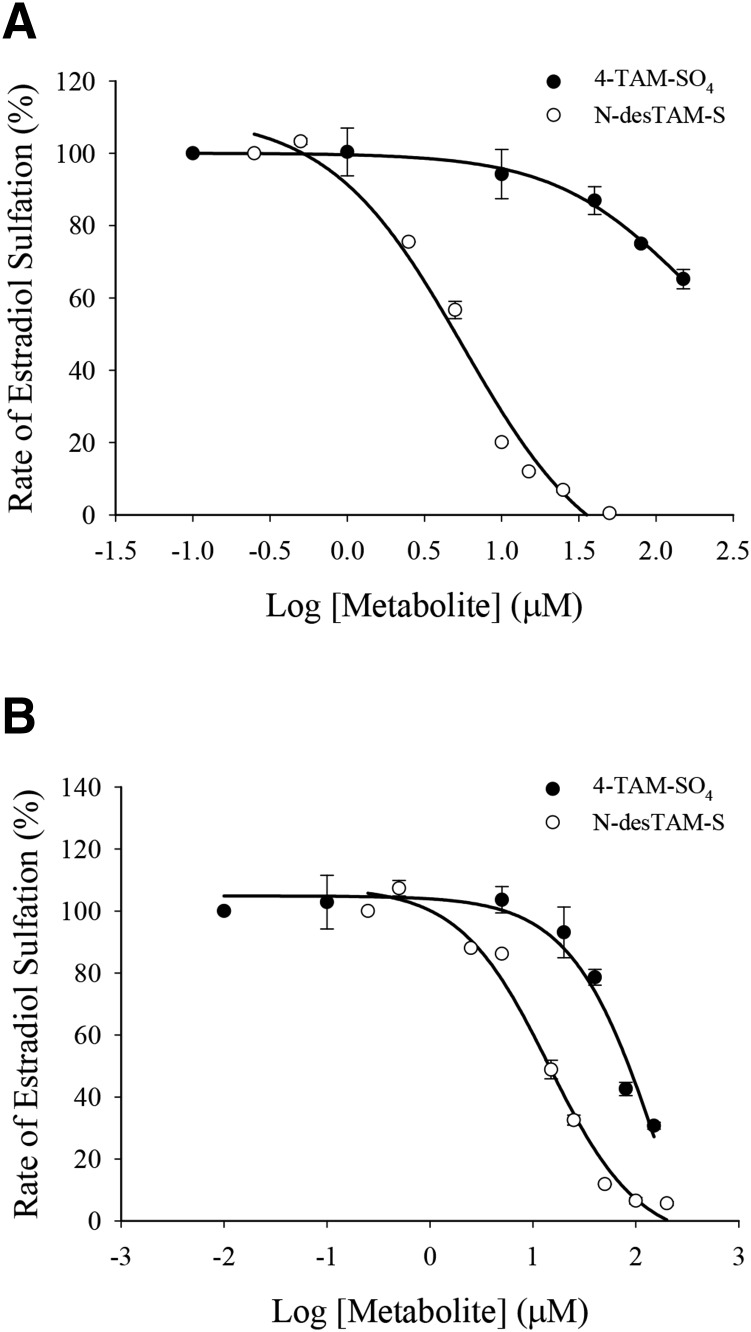
N-desTAM-S and 4-TAM-SO4 Are Weak Inhibitors of Estradiol Sulfation Catalyzed by hSULT1E1 and hSULT1A1*1.
As seen in Fig. 8A, N-desTAM-S was a weak inhibitor of hSULT1E1 with an IC50 value of 5.6 ± 0.9 µM, and 4-TAM-SO4 was a very weak inhibitor of the enzyme with an IC50 value greater than 100 µM. These metabolites were also weak inhibitors of hSULT1A1*1 with an IC50 value of 14 ± 1 µM for N-desTAM-S and an IC50 value greater than 70 µM for 4-TAM-SO4 (Fig. 8B).
Fig. 8.
(A) Inhibition of the hSULT1E1-catalyzed sulfation of 7.0 nM estradiol by N-desTAM-S with a calculated IC50 value of 5.6 ± 0.9 µM, and inhibition by 4-TAM-SO4 with an IC50 value greater than 100 µM. (B) Inhibition of the hSULT1A1*1-catalyzed sulfation of 2.0 µM estradiol by N-desTAM-S with a calculated IC50 value of 14 ± 1 µM, and inhibition by 4-TAM-SO4 with an IC50 value greater than 70 µM.
Discussion
Human SULT1E1 catalyzes the sulfation of estrogens and various endogenous and exogenous molecules that contain phenol functional groups. Although known to catalyze the sulfation of hydroxysteroids such as dehydroepiandrosterone and pregnenolone, hSULT1E1 functions primarily in the sulfation of estradiol. Estradiol promotes cellular growth and proliferation when bound to the estrogen receptor (Clemons and Goss, 2001), and it is also conjugated in a reaction catalyzed by hSULT1E1 as one mechanism for inactivation of its role in cell signaling via the estrogen receptor. Although hSULT1E1 is the principal enzyme responsible for the sulfation of estradiol at physiologic substrate concentrations (Zhang et al., 1998), hSULT1A1*1 is also capable of catalyzing the sulfation of estradiol, albeit at micromolar concentrations (Falany, 1997; Shatalova et al., 2005).
Due to the roles of hSULT1E1 and hSULT1A1*1 in estrogen metabolism, we were interested in determining the interactions of tamoxifen and its major metabolites with these enzymes. We hypothesized that major metabolites of tamoxifen could inhibit the catalytic function of hSULT1E1 and/or hSULT1A1*1 and thus potentially serve as modulators of estrogen metabolism. Changes in the concentration of hormonally active estradiol might then play a role in the endometrial effects of tamoxifen as well as in the observed differential responses to tamoxifen therapy. Each metabolite was a weak inhibitor of hSULT1E1 when examined with estradiol (7.0 nM) as substrate, with IC50 values ranging from 7.0 µM to 21.0 µM (Table 1). Furthermore, the inhibition constant (Ki) for inhibitors of hSULT1E1 ranged from 10 µM to 38 µM (Table 1), and these values were orders of magnitude higher than the Km value (8.1 nM) determined for estradiol sulfation. The weak inhibition of hSULT1E1 by 4-OHTAM, N-desTAM, and endoxifen suggests that these metabolites are unlikely to interfere in the inactivation of estradiol in tissues that express hSULT1E1. Also of note in this regard, previous studies have shown that hSULT1E1 is poorly expressed in breast cancer cells (Falany and Falany, 1996; Suzuki et al., 2003). Nonetheless, even if other tumor tissues were to express hSULT1E1, the weak interactions with this isoform of sulfotransferase relative to those of estradiol suggest that inhibition of hSULT1E1 by endoxifen and the other tamoxifen metabolites examined is unlikely to play a role in altering estradiol concentrations within tumor tissues.
Endoxifen was shown to be a relatively good substrate for hSULT1E1 with a calculated kcat/Km of 0.057 ± 0.013 minute−1µM−1, which suggests that hSULT1E1 may contribute to the in vivo formation of endoxifen-O-sulfate (Supplemental Fig. 5). This information may be useful when evaluating the pharmacokinetic properties of endoxifen. The properties of sulfated metabolites of tamoxifen have been largely overlooked, although our recent findings show that the product sulfamate of N-desTAM, N-desTAM-S, is a potent inhibitor of the sulfation of endogenous steroid substrates catalyzed by hSULT2A1 (Squirewell et al., 2014). Thus, the pharmacokinetic properties of sulfated tamoxifen metabolites as well as their effects on surrounding tissues are subjects for future investigations.
N-desTAM was a notably good substrate for hSULT1E1 with a calculated kcat/Km higher than the catalytic efficiency constant determined for the sulfation of this metabolite either by hSULT1A1*1 in the current work or by hSULT2A1 in our previous findings (Squirewell et al., 2014). These results suggest that hSULT1E1 might potentially generate sufficient concentrations of N-desTAM-S to inhibit the genotoxic effects of tamoxifen due to the action of hSULT2A1, which is possible given the coexpression of hSULT1E1 and hSULT2A1 in tissues such as the liver (Radominska et al., 1990; Miki et al., 2002) and endometrium (Falany et al., 1998; Rubin et al., 1999; Singh et al., 2008; Andersson et al., 2010).
As with hSULT1E1, we determined that some metabolites of tamoxifen were inhibitors of estradiol sulfation catalyzed by hSULT1A1*1. This was of particular interest to our studies because the expression of hSULT1A1*1 in breast cancer is associated with an increased patient survival in tamoxifen-treated women (Nowell et al., 2002; Wegman et al., 2005). Endoxifen and 4-OHTAM were potent inhibitors of the enzyme with IC50 values of 9.9 μM and 1.6 μM, respectively (Fig. 5). Furthermore, the interactions of either endoxifen or 4-OHTAM with hSULT1A1*1 were of similar magnitude to the Michaelis constant determined for estradiol sulfation catalyzed by this enzyme (Km = 1.5 µM). Rižner (2013) reports a physiologic estradiol concentration in postmenopausal women of only 30 pM, whereas the mean plasma concentrations of major tamoxifen metabolites are reported to be in the nanomolar range (Brauch et al., 2009). Other studies report that the concentrations of tamoxifen metabolites in tissues are 6- to 60-fold higher than those in serum (Lien et al., 1991; Decensi et al., 2003). Given the high expression of hSULT1A1 in breast cancer cells (Falany and Falany, 1996) as well as the abundance of the metabolites in relation to the physiologic estradiol concentrations, endoxifen or 4-OHTAM might potentially inhibit the catalytic function of hSULT1A1 in breast tumor tissue. Such inhibition might increase the localized concentrations of hormonally active estradiol, thus decreasing the therapeutic efficacy of tamoxifen as one mechanism of clinical resistance. This may have implications in the ongoing clinical trials of endoxifen (NCT01327781 and NCT01273168; ClinicalTrials.gov), because the steady-state tissue concentrations of endoxifen might increase from its direct use.
Of the metabolites studied, 4-OHTAM was the best substrate for the hSULT1A1*1 (Table 3). Moreover, the rate of sulfation for 4-OHTAM was higher with hSULT1A1*1 than for either hSULT1E1 (Table 2) or hSULT2A1 (Squirewell et al., 2014). These studies suggest that hSULT1A1*1 could be important for the in vivo formation of 4-TAM-SO4, a metabolite of interest due to its reported apoptotic effects in breast cancer cells and potential role(s) in the therapeutic efficacy of tamoxifen (Mercer et al., 2010). Thus, changes in the catalytic activity or expression of hSULT1A1*1 could significantly alter the formation of 4-TAM-SO4 in breast cancer tissue. This might be a concern for the population of patients who are homozygous for the thermally labile and low-activity hSULT1A1*2 allele, as the formation of 4-TAM-SO4 may be limited by this genetic polymorphism. It is important to note that 4-TAM-SO4 is also a product of sulfation catalyzed by hSULT2A1 and hSULT1E1. However, there is no known evidence that significant expression of either of these enzymes occurs in breast cancer cells.
In summary, this study examined the interactions of major tamoxifen metabolites with purified hSULT1E1 and hSULT1A1*1. Endoxifen, 4-OHTAM, TAM-NO, and N-desTAM were weak inhibitors of estradiol sulfation catalyzed by hSULT1E1, which suggests that these metabolites of tamoxifen are unlikely to interfere with estrogen inactivation catalyzed by this particular isoform of sulfotransferase. However, because 4-OHTAM and endoxifen were relatively potent inhibitors of estradiol sulfation catalyzed by hSULT1A1*1, there is a potential for these metabolites to alter the concentrations of hormonally active estrogen in tissues where hSULT1A1*1 is expressed and hSULT1E1 is not. Additional roles of hSULT1E1 and hSULT1A1*1 in the variable response to tamoxifen therapy will be the subject of future investigations.
Supplementary Material
Acknowledgments
The authors thank Dr. Duncan I. Mackie for helpful advice and assistance in the bacterial expression of hSULT1E1 and hSULT1A1*1.
Abbreviations
- DTT
dithiothreitol
- ESI-MS
electrospray ionization-mass spectrometry
- hSULT2A1
human hydroxysteroid sulfotransferase 2A1
- hSULT1A1
human phenol sulfotransferase 1A1
- hSULT1A1*1
human phenol sulfotransferase 1A1 isoform 1
- hSULT1E1
human estrogen sulfotransferase 1E1
- LC-MS
liquid chromatography-mass spectrometry
- N-desTAM
N-desmethyltamoxifen
- N-desTAM-S
N-desTAM sulfamate
- 4-OHTAM
4-hydroxytamoxifen
- PAPS
adenosine 3′-phosphate 5′-phosphosulfate lithium salt hydrate
- TAM-NO
tamoxifen N-oxide
- 4-TAM-SO4
4-hydroxytamoxifen sulfate
- TLC
thin-layer chromatography
Authorship Contributions
Participated in research design: Squirewell, Duffel.
Conducted experiments: Squirewell.
Performed data analysis: Squirewell, Duffel.
Wrote or contributed to writing of the manuscript: Squirewell, Duffel.
Footnotes
This work was supported by the National Institutes of Health National Cancer Institute [Grant R01 CA038683].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aksoy IA, Wood TC, Weinshilboum R. (1994) Human liver estrogen sulfotransferase: identification by cDNA cloning and expression. Biochem Biophys Res Commun 200:1621–1629. [DOI] [PubMed] [Google Scholar]
- Andersson H, Helmestam M, Zebrowska A, Olovsson M, Brittebo E. (2010) Tamoxifen-induced adduct formation and cell stress in human endometrial glands. Drug Metab Dispos 38:200–207. [DOI] [PubMed] [Google Scholar]
- Bernstein L, Deapen D, Cerhan JR, Schwartz SM, Liff J, McGann-Maloney E, Perlman JA, Ford L. (1999) Tamoxifen therapy for breast cancer and endometrial cancer risk. J Natl Cancer Inst 91:1654–1662. [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. [DOI] [PubMed] [Google Scholar]
- Brauch H, Mürdter TE, Eichelbaum M, Schwab M. (2009) Pharmacogenomics of tamoxifen therapy. Clin Chem 55:1770–1782. [DOI] [PubMed] [Google Scholar]
- Clemons M, Goss P. (2001) Estrogen and the risk of breast cancer. N Engl J Med 344:276–285. [DOI] [PubMed] [Google Scholar]
- Coughtrie MW, Gilissen RA, Shek B, Strange RC, Fryer AA, Jones PW, Bamber DE. (1999) Phenol sulphotransferase SULT1A1 polymorphism: molecular diagnosis and allele frequencies in Caucasian and African populations. Biochem J 337:45–49. [PMC free article] [PubMed] [Google Scholar]
- Decensi A, Robertson C, Viale G, Pigatto F, Johansson H, Kisanga ER, Veronesi P, Torrisi R, Cazzaniga M, Mora S, et al. (2003) A randomized trial of low-dose tamoxifen on breast cancer proliferation and blood estrogenic biomarkers. J Natl Cancer Inst 95:779–790. [DOI] [PubMed] [Google Scholar]
- Duffel MW, Binder TP, Rao SI. (1989) Assay of purified aryl sulfotransferase suitable for reactions yielding unstable sulfuric acid esters. Anal Biochem 183:320–324. [DOI] [PubMed] [Google Scholar]
- Falany CN. (1997) Enzymology of human cytosolic sulfotransferases. FASEB J 11:206–216. [DOI] [PubMed] [Google Scholar]
- Falany CN, Krasnykh V, Falany JL. (1995) Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol 52:529–539. [DOI] [PubMed] [Google Scholar]
- Falany JL, Azziz R, Falany CN. (1998) Identification and characterization of cytosolic sulfotransferases in normal human endometrium. Chem Biol Interact 109:329–339. [DOI] [PubMed] [Google Scholar]
- Falany JL, Falany CN. (1996) Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res 56:1551–1555. [PubMed] [Google Scholar]
- Falany JL, Pilloff DE, Leyh TS, Falany CN. (2006) Sulfation of raloxifene and 4-hydroxytamoxifen by human cytosolic sulfotransferases. Drug Metab Dispos 34:361–368. [DOI] [PubMed] [Google Scholar]
- Foster AB, Griggs LJ, Jarman M, van Maanen JMS, Schulten HR. (1980) Metabolism of tamoxifen by rat liver microsomes: formation of the N-oxide, a new metabolite. Biochem Pharmacol 29:1977–1979. [DOI] [PubMed] [Google Scholar]
- Grabinski JL, Smith LS, Chisholm GB, Drengler R, Rodriguez GI, Lang AS, Kalter SP, Garner AM, Fichtel LM, Hollsten J, et al. (2006) Genotypic and allelic frequencies of SULT1A1 polymorphisms in women receiving adjuvant tamoxifen therapy. Breast Cancer Res Treat 95:13–16. [DOI] [PubMed] [Google Scholar]
- Hernandez-Ramon EE, Sandoval NA, John K, Cline JM, Wood CE, Woodward RA, Poirier MC. (2014) Tamoxifen-DNA adduct formation in monkey and human reproductive organs. Carcinogenesis 35:1172–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC. (2007) New insights into the metabolism of tamoxifen and its role in the treatment and prevention of breast cancer. Steroids 72:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC, Collins MM, Rowsby L, Prestwich G. (1977) A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol 75:305–316. [DOI] [PubMed] [Google Scholar]
- Jordan VC, Gosden B. (1982) Importance of the alkylaminoethoxy side-chain for the estrogenic and antiestrogenic actions of tamoxifen and trioxifene in the immature rat uterus. Mol Cell Endocrinol 27:291–306. [DOI] [PubMed] [Google Scholar]
- Lien EA, Solheim E, Ueland PM. (1991) Distribution of tamoxifen and its metabolites in rat and human tissues during steady-state treatment. Cancer Res 51:4837–4844. [PubMed] [Google Scholar]
- Lyon ES, Marcus CJ, Wang JL, Jakoby WB. (1981) Hydroxysteroid sulfotransferase. Methods Enzymol 77:206–213. [DOI] [PubMed] [Google Scholar]
- MacGregor JI, Jordan VC. (1998) Basic guide to the mechanisms of antiestrogen action. Pharmacol Rev 50:151–196. [PubMed] [Google Scholar]
- Mani C, Kupfer D. (1991) Cytochrome P-450-mediated activation and irreversible binding of the antiestrogen tamoxifen to proteins in rat and human liver: possible involvement of flavin-containing monooxygenases in tamoxifen activation. Cancer Res 51:6052–6058. [PubMed] [Google Scholar]
- Maximov PY, McDaniel RE, Fernandes DJ, Bhatta P, Korostyshevskiy VR, Curpan RF, Jordan VC. (2014) Pharmacological relevance of endoxifen in a laboratory simulation of breast cancer in postmenopausal patients. J Natl Cancer Inst 106:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KE, Apostolov EO, da Costa GG, Yu X, Lang P, Roberts DW, Davis W, Basnakian AG, Kadlubar FF, Kadlubar SA. (2010) Expression of sulfotransferase isoform 1A1 (SULT1A1) in breast cancer cells significantly increases 4-hydroxytamoxifen-induced apoptosis. Int J Mol Epidemiol Genet 1:92–103. [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Nakata T, Suzuki T, Darnel AD, Moriya T, Kaneko C, Hidaka K, Shiotsu Y, Kusaka H, Sasano H. (2002) Systemic distribution of steroid sulfatase and estrogen sulfotransferase in human adult and fetal tissues. J Clin Endocrinol Metab 87:5760–5768. [DOI] [PubMed] [Google Scholar]
- Nagar S, Walther S, Blanchard RL. (2006) Sulfotransferase (SULT) 1A1 polymorphic variants *1, *2, and *3 are associated with altered enzymatic activity, cellular phenotype, and protein degradation. Mol Pharmacol 69:2084–2092. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Ogura K, Nakano H, Kaku T, Takahashi E, Ohkubo Y, Sekine K, Hiratsuka A, Kadota S, Watabe T. (2002) Sulfation of environmental estrogens by cytosolic human sulfotransferases. Drug Metab Pharmacokinet 17:221–228. [DOI] [PubMed] [Google Scholar]
- Nowell S, Sweeney C, Winters M, Stone A, Lang NP, Hutchins LF, Kadlubar FF, Ambrosone CB. (2002) Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst 94:1635–1640. [DOI] [PubMed] [Google Scholar]
- Pasqualini JR. (2009) Estrogen sulfotransferases in breast and endometrial cancers. Ann N Y Acad Sci 1155:88–98. [DOI] [PubMed] [Google Scholar]
- Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. (1990) Human liver steroid sulphotransferase sulphates bile acids. Biochem J 272:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. (1997) Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun 239:298–304. [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Weinshilboum RM. (1999) Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem Pharmacol 58:605–616. [DOI] [PubMed] [Google Scholar]
- Rižner TL. (2013) Estrogen biosynthesis, phase I and phase II metabolism, and action in endometrial cancer. Mol Cell Endocrinol 381:124–139. [DOI] [PubMed] [Google Scholar]
- Rubin GL, Harrold AJ, Mills JA, Falany CN, Coughtrie MW. (1999) Regulation of sulphotransferase expression in the endometrium during the menstrual cycle, by oral contraceptives and during early pregnancy. Mol Hum Reprod 5:995–1002. [DOI] [PubMed] [Google Scholar]
- Sekura RD. (1981) Adenosine 3′-phosphate 5′-phosphosulfate. Methods Enzymol 77:413–415. [Google Scholar]
- Serrano D, Lazzeroni M, Zambon CF, Macis D, Maisonneuve P, Johansson H, Guerrieri-Gonzaga A, Plebani M, Basso D, Gjerde J, et al. (2011) Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian Tamoxifen Prevention Trial. Pharmacogenomics J 11:100–107. [DOI] [PubMed] [Google Scholar]
- Shatalova EG, Walther SE, Favorova OO, Rebbeck TR, Blanchard RL. (2005) Genetic polymorphisms in human SULT1A1 and UGT1A1 genes associate with breast tumor characteristics: a case-series study. Breast Cancer Res 7:R909–R921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng JJ, Duffel MW. (2001) Bacterial expression, purification, and characterization of rat hydroxysteroid sulfotransferase STa. Protein Expr Purif 21:235–242. [DOI] [PubMed] [Google Scholar]
- Singh MN, Stringfellow HF, Walsh MJ, Ashton KM, Paraskevaidis E, Abdo KR, Martin-Hirsch PL, Phillips DH, Martin FL. (2008) Quantifiable mRNA transcripts for tamoxifen-metabolising enzymes in human endometrium. Toxicology 249:85–90. [DOI] [PubMed] [Google Scholar]
- Squirewell EJ, Qin X, Duffel MW. (2014) Endoxifen and other metabolites of tamoxifen inhibit human hydroxysteroid sulfotransferase 2A1 (hSULT2A1). Drug Metab Dispos 42:1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nakata T, Miki Y, Kaneko C, Moriya T, Ishida T, Akinaga S, Hirakawa H, Kimura M, Sasano H. (2003) Estrogen sulfotransferase and steroid sulfatase in human breast carcinoma. Cancer Res 63:2762–2770. [PubMed] [Google Scholar]
- Thompson EW, Katz D, Shima TB, Wakeling AE, Lippman ME, Dickson RB. (1989) ICI 164,384, a pure antagonist of estrogen-stimulated MCF-7 cell proliferation and invasiveness. Cancer Res 49:6929–6934. [PubMed] [Google Scholar]
- van Leeuwen FE, Benraadt J, Coebergh JWW, Kiemeney LALM, Gimbrère CH, Otter R, Schouten LJ, Damhuis RA, Bontenbal M, Diepenhorst FW, et al. (1994) Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet 343:448–452. [DOI] [PubMed] [Google Scholar]
- Wegman P, Vainikka L, Stål O, Nordenskjöld B, Skoog L, Rutqvist LE, Wingren S. (2005) Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res 7:R284–R290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilborn TW, Comer KA, Dooley TP, Reardon IM, Heinrikson RL, Falany CN. (1993) Sequence analysis and expression of the cDNA for the phenol-sulfating form of human liver phenol sulfotransferase. Mol Pharmacol 43:70–77. [PubMed] [Google Scholar]
- Wu X, Hawse JR, Subramaniam M, Goetz MP, Ingle JN, Spelsberg TC. (2009) The tamoxifen metabolite, endoxifen, is a potent antiestrogen that targets estrogen receptor alpha for degradation in breast cancer cells. Cancer Res 69:1722–1727. [DOI] [PubMed] [Google Scholar]
- Zhang H, Varlamova O, Vargas FM, Falany CN, Leyh TS. (1998) Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem 273:10888–10892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.