Abstract
The splicing factor SF3B1 is the most commonly mutated gene in the myelodysplastic syndrome (MDS), particularly in patients with refractory anemia with ring sideroblasts (RARS). We investigated the functional effects of SF3B1 disruption in myeloid cell lines: SF3B1 knockdown resulted in growth inhibition, cell cycle arrest and impaired erythroid differentiation and deregulation of many genes and pathways, including cell cycle regulation and RNA processing. MDS is a disorder of the hematopoietic stem cell and we thus studied the transcriptome of CD34+ cells from MDS patients with SF3B1 mutations using RNA sequencing. Genes significantly differentially expressed at the transcript and/or exon level in SF3B1 mutant compared with wild-type cases include genes that are involved in MDS pathogenesis (ASXL1 and CBL), iron homeostasis and mitochondrial metabolism (ALAS2, ABCB7 and SLC25A37) and RNA splicing/processing (PRPF8 and HNRNPD). Many genes regulated by a DNA damage-induced BRCA1–BCLAF1–SF3B1 protein complex showed differential expression/splicing in SF3B1 mutant cases. This is the first study to determine the target genes of SF3B1 mutation in MDS CD34+ cells. Our data indicate that SF3B1 has a critical role in MDS by affecting the expression and splicing of genes involved in specific cellular processes/pathways, many of which are relevant to the known RARS pathophysiology, suggesting a causal link.
Introduction
The myelodysplastic syndromes (MDS) are a heterogeneous group of clonal hematopoietic stem cell (HSC) malignancies characterized by blood cell dysplasia and peripheral blood cytopenia. Approximately 30–40% of MDS patients will develop acute myeloid leukemia (AML).1
The recent discovery of somatic splicesomal mutations in MDS has revealed a new leukemogenic pathway involving spliceosomal dysfunction.2, 3 Somatic mutations in the splicing factor genes SF3B1, U2AF1, SRSF2 and ZRSR2 are frequent in MDS patients.4 Importantly, these genes encode proteins that are all involved in 3′-splice site recognition during pre-messenger RNA (pre-mRNA) processing. Splicing factor gene mutations occur in over 50% of MDS patients, are highly specific to this disorder, and occur in a mutually exclusive manner.5, 6, 7
SF3B1, encoding a core component of the U2 small nuclear ribonucleoprotein, involved in the recognition of the branchpoint sequence, is the most commonly mutated gene found in MDS (20–28% of all patients).2, 8, 9 We and others have shown that mutations of the splicing factor SF3B1 are found in a high percentage (>70%) of MDS patients whose disease is characterised by the presence of ring sideroblasts, including both refractory anemia with ring sideroblasts (RARS) and refractory cytopenia with multilineage dysplasia and ring sideroblasts (RCMD-RS).2, 8 The close association between SF3B1 mutation and the presence of ring sideroblasts is consistent with a causal relationship and makes this the first gene to be strongly associated with a specific morphological feature of MDS. Ring sideroblasts are characterised by an excess accumulation of iron in the mitochondria of erythroblasts,10 and SF3B1 mutant RARS cases show altered iron distribution characterised by coarse iron deposits compared with wild-type RARS cases.11 SF3B1 mutations are generally more prevalent in low-risk MDS and have been shown to be independent predictors of favorable clinical outcome in MDS in most studies.8, 11 The clinical consequences of SF3B1 mutation in MDS are clear, but the functional consequences of these mutations in human cells remain poorly understood. Altered RNA splicing has been suggested as the mechanism underlying the observed phenotypic changes concomitant to splicing factor gene mutations, including SF3B1; 3, 12, 13 however, the target genes in the HSC of MDS cases with SF3B1 mutations are yet to be defined.
SF3B1 mutations in MDS are primarily heterozygous point mutations. The presence of hotspots and the absence of nonsense or frameshift mutations in SF3B1 in MDS patients suggest that SF3B1 mutations are likely to be gain/change-of-function (neomorphic) mutations. A heterozygous Sf3b1+/− knockout mouse model has been shown to develop ring sideroblasts, suggesting that haploinsufficiency of SF3B1 may lead to their formation.11 Recent similar studies have not made this observation, however.14, 15 Thus, it is yet to be determined whether SF3B1 mutations found in MDS are loss-of-function mutations or gain/change-of-function mutations. In this study, we thus evaluated the effects of SF3B1 knockdown on cell growth, gene expression and splicing in a range of myeloid cell lines and performed RNA sequencing (RNA-Seq) on the CD34+ cells of MDS patients harboring SF3B1 mutations.
Materials and methods
Myeloid cell lines culture
K562, HEL, TF1 and SKM1 cells were cultured in Roswell Park Memorial Institute medium 1640 (Sigma-Aldrich, Gillingham, UK) containing 10% fetal bovine serum, at 37 °C and 5% CO2. TF1 and SKM1 cultures were supplemented with 2 and 1 ng/ml of granulocyte-macrophage colony-stimulating factor, respectively.
SF3B1 knockdown
Three non-overlapping small interfering RNAs (siRNAs) targeting SF3B1 and two different scramble sequences with guanine–cytosine content similar to the siRNA sequences (Stealth Select RNAi, Invitrogen) were used to knock down SF3B1 in myeloid cell lines. For each transfection, 30 pmol of siRNA and 2 × 106 cells were electroporated in an Amaxa Nucleofector I, using the Amaxa cell optimization kit V (Amaxa, Gaithersburg, MD, USA). Evaluation of green fluorescent protein-positive cells obtained using the pmaxGFP fluorescent expression plasmid confirmed >80% of successfully transfected cells after 24 h. Readout data are reported as mean±s.e.m. Statistical analysis was performed using Student's t-test.
Cell growth assay
Live and dead cells were assessed by trypan blue viability testing and counted using a hemocytometer.
Cell cycle analysis
Cells were fixed with ice cold methanol, incubated with 40 μg/ml propidium iodide and 10 μg/ml RNaseA, as previously described.16 Data were acquired on a BD LSRII flow cytometer (BD Bioscience; Franklin Lakes, NJ, USA) and analyzed using FLOWJO software 7.6.4 (FlowJo, Ashland, OR, USA).
Erythroid differentiation
To induce erythroid differentiation, K562, TF1 and HEL cells were cultured with hemin 50 μm for 72 h. Erythroid differentiation was studied by analyzing the expression level of γ-globin (HBG1) using quantitative real-time PCR (qRT-PCR) as described previously.17 Expression of the erythroid markers CD36, CD71 and CD235a was evaluated by flow cytometry as described previously.16
Quantitative real-time PCR
Total RNA was reverse transcribed using Retroscript kit (Ambion, Life Technologies, Paisley, UK). The expression levels of SF3B1, ABCB7, FTMT, HBG1, KLF1 and B2M were determined using Assays-on-Demand (Applied Biosystems, Foster City, CA, USA). B2M expression levels were used to normalize for differences in input complementary DNA (cDNA). Triplicate samples were run on a LightCycler 480 Real-time PCR system (Roche Diagnostics, Lewes, UK) and expression ratios were calculated using the ddCT method.18
Gene expression profiling
Total RNA (100 ng for each sample) was amplified and labeled with the 3′ IVT Express Kit (Affymetrix, Santa Clara, CA, USA), and hybridized to Affymetrix Human Genome U133 Plus 2.0 GeneChips, as described previously.19, 20 CEL files were pre-processed using Robust Multi-chip Average and data analysis was performed using GeneSpring 7.3.1 (Agilent, Santa Clara, CA, USA). Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) 7.5 (Qiagen, Manchester, UK), as previously described.19 Analysis of gene set up- or downregulation was performed using Gene Set Enrichment Analysis,21 as previously described (false discovery rate<0.1).2
Splicing analysis using human genome exon-junction microarray
Total RNA was DNAse treated (Invitrogen), purified (Agencourt RNA Clean XP) and amplified (100 ng) using Ambion WT Expression Kit (Affymetrix). The cDNA was subsequently fragmented and labeled using WT terminal labeling kit (Affymetrix). Samples were hybridized onto Affymetrix Human genome exon-junction arrays, which on average contain 119 unique probes per gene spanning every exon and known exon–exon junction. CEL files were analyzed using GenoSplice technology (www.genosplice.com).22, 23 Data were normalized using quantile normalization. Analysis at the exon level was performed taking into account only exon probes. Analysis at the splicing variant level was performed by taking into account exon–exon junction probes using the FAST DB splicing patterns annotation (release fastdb_2012_2). Unpaired Student's t-test was used to determine exon and splicing pattern variation between cells with SF3B1 knockdown and scramble controls. Genes were considered significantly deregulated when fold change was ⩾1.5 and P-value <0.05. Gene Ontology, KEGG and REACTOME analyses of differentially regulated genes were performed using DAVID.24
Splicing analysis using qRT-PCR and Sanger sequencing
Splicing analysis of TP53 was performed using Sanger sequencing of gel-extracted individual bands from PCR-amplified cDNA. Primers and PCR conditions are listed in Supplementary Table S1.
Splicing analysis of cyclins CCNA2 and STK6 was performed using a qRT-PCR on cDNA, using primers specific for splice junctions corresponding to exon inclusion or skipping.25 Primers are listed in Supplementary Table S2. TBP expression levels were used to normalize for differences in input cDNA.
RNA sequencing
Bone marrow samples were obtained from MDS patients and healthy controls and CD34+ cells isolated using MACS magnetic cell separation columns (Miltenyi Biotec, Germany), as described previously.19, 20 RNA extracted from bone marrow CD34+ cells obtained from 12 MDS patients and 5 healthy controls was used for deep RNA-Seq. Eight of the 12 MDS cases (four RARS and four RCMD-RS) had SF3B1 gene mutation, whereas four cases (all RCMD) had no known mutations in splicing factor genes (SF3B1, SRSF2, U2AF1 or ZRSR2) (Supplementary Table S3), as determined by targeted next-generation sequencing data from a previous study.7
Total RNA was DNase treated (Invitrogen), purified using XP beads (Beckman Coulter, High Wycombe, UK) and processed (100 ng) using NEBNext Ultra directional mRNA Library prep (NEB, Hitchin, UK) as per the manufacturer's protocol for Illumina with two alterations: custom indexes were designed in house and an additional purification step was performed using XP beads (Beckman Coulter) rather than size selection. Samples were run on HiSeq2000 Illumina sequencing machine (Illumina, San Diego, CA, USA). Sequencing reads were mapped to the human genome using TopHat,26 which can handle reads spanning exon–exon boundaries. Data analysis was performed using edgeR27 to evaluate the whole transcript expression (false discovery rate<0.05) and with DEXSeq28 to evaluate differential exon usage (false discovery rate<0.05). At the expression level, only genes with at least 10 reads in four or more samples were included in the analysis; at the exon level, exons with fewer than 10 reads were excluded. Gender was included as an additional factor in the DEXSeq model testing for differential exon usage. Selected differentially expressed exons were validated using qRT-PCR (Supplemetary Figure S1, Supplementary Information). Integrative Genomics Viewer (IGV) v2.3 (http://www.broadinstitute.org/igv/) was used for visualization of the sequence reads. Pathway analysis was performed using IPA and Gene Set Enrichment Analysis, as described above. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE63569.
Results
SF3B1 knockdown inhibits cell growth, induces cell cycle arrest and impairs erythroid differentiation
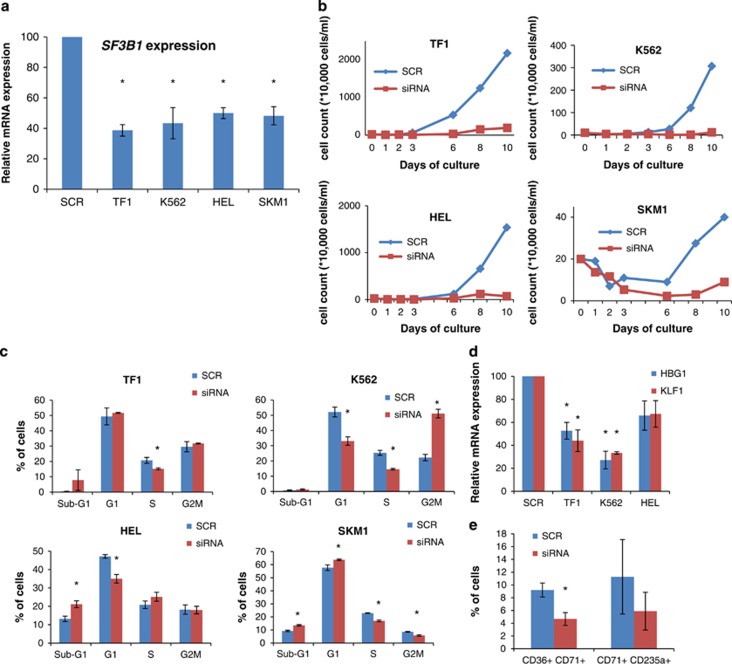
SF3B1 was knocked down using siRNA technology in four myeloid cell lines (TF1, K562, HEL and SKM1) that we found to be wild type for SF3B1 (Supplementary Information), resulting in a significant decrease in expression ranging between 50 and 60% (Figure 1a).
Figure 1.
Effects of SF3B1 knockdown in myeloid cell lines. Each cell line transfected with siRNA targeting SF3B1 was compared with the corresponding cell line transfected with the scramble control. (a) SF3B1 mRNA expression measured 3 days after knockdown. (b) Growth curves of cells with SF3B1 knockdown, compared with cells transfected with the scramble control, as assessed by trypan blue exclusion. (c) Cell cycle analysis of cell lines following SF3B1 knockdown. (d) Erythroid differentiation in myeloid cell lines with SF3B1 knockdown treated with 50 μm hemin, as measured by HBG1 and KLF1 expressions relative to the scramble control. (e) Percentage of CD36+CD71+ and CD71+CD235a+ populations in K562 cells with SF3B1 knockdown compared with the scramble control. Results in subpanels a–d were obtained from scramble n=2 and SF3B1 siRNA as follows: a, n=3; b, n=3; c, n=3 for SKM1 and TF1, n=6 for HEL and n=9 for K562; d, n=3 for TF1 and HEL, n=6 for K562. Results in subpanel e were obtained from scramble n=4 and SF3B1 siRNA n=4. *P<0.05.
Cell growth was inhibited in all four cell lines with SF3B1 knockdown in comparison with the scramble control (Figure 1b). Cell cycle arrest in different phases was detected in different myeloid cell lines with SF3B1 knockdown (Figure 1c). K562 cells showed a significant G2M cell cycle arrest with a concomitant reduction in the percentage of cells in the G1 and S phase. TF1 cells showed a significant decrease in the percentage of cells in the S phase. HEL cells showed a significant decrease in the percentage of cells in the G1 phase and a significant increase in the sub-G1 cell population, indicating increased apoptosis. Similarly, SKM1 cells showed a significant decrease in the percentage of cells in S and G2M phase with a concomitant increase in the percentage of cells in the sub-G1 and G1 phase, indicating cell cycle arrest in the G1 phase and an increase in apoptosis (Figure 1c).
Three cell lines (TF1, K562 and HEL) with SF3B1 knockdown were cultured with hemin to induce erythroid differentiation. We assessed the expression of the erythroid differentiation markers HBG1 and KLF1 using qRT-PCR. A significant reduction in the expression of HBG1 and KLF1 was observed in TF1 and K562 cell lines with SF3B1 knockdown (Figure 1d). In addition, we observed a reduction in the percentage of CD36+CD71+ and CD71+CD235a+ erythroid populations (significant for the CD36+CD71+ population) in K562 cells with SF3B1 knockdown compared with the scramble control (Figure 1e), suggesting that normal SF3B1 function is required for erythroid differentiation.17
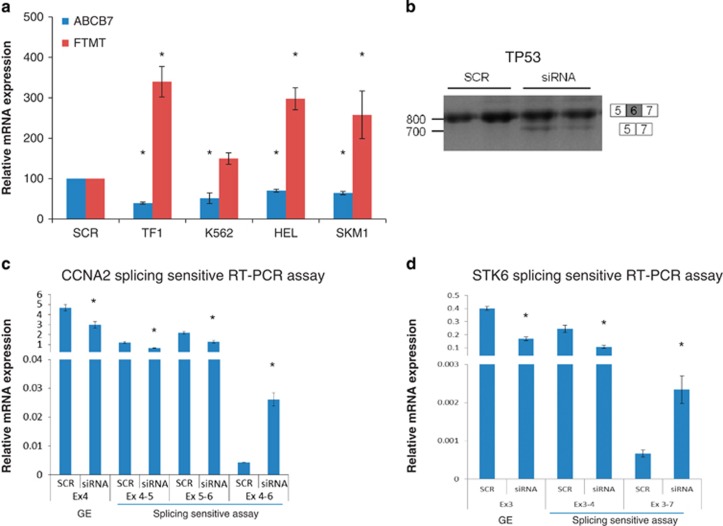
RARS is characterized by FTMT accumulation and low expression levels of the iron transporter ABCB7.17, 29 We have previously shown that SF3B1 knockdown leads to decreased ABCB7 expression and increased FTMT expression in K562 cells.17 In this study, we have extended these observations to the other three myeloid cell lines investigated (Figure 2a). In addition, restoration of SF3B1 expression to normal levels after 10 days of culture was followed by restoration of ABCB7 expression levels to normal (Supplementary Figure S2).
Figure 2.
Effects of SF3B1 knockdown on gene expression and splicing. (a) ABCB7 and FTMT expression levels in TF1, K562, HEL and SKM1 cells with SF3B1 knockdown, as measured by qRT-PCR 48 h post transfection. Each cell line transfected with siRNA targeting SF3B1 was compared with the corresponding cell line transfected with the scramble control. (b) Reverse transcription-PCR of TP53 exons 5–7 showing aberrant splicing in K562 cells with SF3B1 knockdown (siRNA) compared with scramble (SCR). (c, d). qRT-PCR analysis using primers that monitor general gene expression (GE) in a constitutive exon (Ex4 of CCNA2 and Ex3 of STK6) or primers specific for splice junctions corresponding to exon inclusion or skipping in the cyclin A2 (CCNA2) and Aurora Kinase A (STK6) genes in K562 cells. Cells with SF3B1 knockdown (siRNA) show alternative splicing events. Results in subpanel a were obtained from scramble n=3 and SF3B1 siRNA n=4. Results in subpanel c–d were obtained from scramble n=2 and SF3B1 siRNA n=3. *P<0.05.
Taken together, these data show that SF3B1 knockdown results in inhibition of cell growth, induction of cell cycle arrest and impairment of erythroid differentiation in myeloid cell lines.
SF3B1 knockdown alters gene expression
To evaluate the effects of SF3B1 knockdown on global gene expression, gene expression profiling was performed in the four myeloid cell lines. For each cell line, we compared the expression profiles of cells treated with two different siRNAs targeting SF3B1 with those of cells treated with a scramble control, 48 h post transfection.
We identified many genes that were up- or downregulated by >2-fold in each cell line treated with SF3B1 siRNAs (Supplementary Figure S3A and S3B). Four genes were upregulated (TFDP1, LOC100505759, MKRN1 and WRNIP1) and five genes were downregulated (ZC3H7A, CREBZF, SGK494, WSB1 and two probesets for SF3B1) in all four cell lines with SF3B1 knockdown.
Pathway analysis was performed on the up- and downregulated genes in each cell line with SF3B1 knockdown using IPA. Significant deregulation of pathways related to cell cycle regulation was observed in all cell lines and of mTOR signaling and AMPK signaling pathways in three cell lines (Table 1). We performed Gene Set Enrichment Analysis to identify pathways and processes showing coordinated up- or downregulation. Upregulated gene sets include p53 signaling in K562 and SKM1 cells, and several gene sets associated with regulation of transcription, spliceosome and splicing in K562 cells (Supplementary Table S4). Downregulated gene sets associated with the mitochondrial function were found in K562 and TF1 cells, and with cell cycle regulation in SKM1 and HEL cells (Supplementary Table S4). These data show that SF3B1 knockdown in the cell lines studied results in deregulation of many genes and pathways including cell cycle and RNA processing.
Table 1. List of significant deregulated pathways in cell lines obtained using IPA.
| TF1 | SKM1 | K562 | HEL |
|---|---|---|---|
| Selenoamino acid metabolism | Estrogen-mediated S-phase entry | AMPK signaling | Estrogen-mediated S-phase entry |
| Mitochondrial dysfunction | Pentose phosphate pathway | HMGB1 signaling | Cyclins and cell cycle regulation |
| AMPK signaling | Cell cycle: G1/S checkpoint regulation | mTOR signaling | PI3K signaling in B lymphocytes |
| Regulation of eIF4 and p70S6K signaling | Small-cell lung cancer signaling | Polyamine regulation in colon cancer | mTOR signaling |
| mTOR signaling | Lysine biosynthesis | Cell cycle regulation by BTG family proteins | Breast cancer regulation by stathmin1 |
| Role of NANOG in mammalian embryonic stem cell pluripotency | Actin cytoskeleton signaling | ILK signaling | Purine metabolism |
| Cyclins and cell cycle regulation | Chronic myeloid leukemia signaling | Cyclins and cell cycle regulation | Glioma signaling |
| Polyamine regulation in colon cancer | Glioma signaling | Ceramide signaling | Aldosterone signaling in epithelial cells |
| Assembly of RNA polymerase II complex | Assembly of RNA polymerase III complex | ErbB signaling | fMLP signaling in neutrophils |
| Signaling by Rho family GTPases | TNFR1 signaling | Cell cycle: G1/S checkpoint regulation | |
| Glucocorticoid receptor signaling | Non-small cell lung cancer signaling | ||
| Glycine, serine and threonine metabolism | Growth hormone signaling | ||
| Role of CHK proteins in cell cycle checkpoint control | Small-cell lung cancer signaling | ||
| VDR/RXR activation | Protein ubiquitination pathway | ||
| Lysine degradation | Nitric oxide signaling in the cardiovascular system | ||
| Tight junction signaling | AMPK signaling | ||
| Ubiquinone biosynthesis | Glioblastoma multiforme signaling | ||
| Mitotic roles of polo-like kinase | |||
| Wnt/β-catenin signaling | |||
| PI3K/AKT signaling | |||
| Renal cell carcinoma signaling | |||
| Regulation of eIF4 and p70S6K signaling | |||
| TNFR2 signaling | |||
| Fructose and mannose metabolism | |||
| Telomerase signaling |
Gene expression profiling was performed in all four cell lines with SF3B1 knockdown, and genes showing >2-fold change were used for the analysis.
SF3B1 knockdown impact on splicing
The genome-wide effects of SF3B1 knockdown on splicing were investigated in two myeloid cell lines (K562 and TF1) using human genome exon-junction arrays. The splicing profile of cells with SF3B1 knockdown (using two different siRNA per cell line) was compared with that of cells transfected with the scramble control.
We observed 2027 differentially expressed exons from 1419 genes, and 507 significant differentially regulated splicing variants (including exon skipping, intron retention and alternative splice sites) of 384 genes in cells with SF3B1 knockdown. For example, we observed differential exon usage of CDC7 and SRSF11 in the data from both the cell lines, and of TP53 in TF1 cells. We found a significant overrepresentation of 3′ acceptor splice sites affected by alternative splicing events compared with 5′ donor splice sites (5:1 ratio, P=0.0027, χ2 test with Yates' correction), consistent with the known role of SF3B1 in the recognition of 3′ splice sites. Gene ontology analysis was performed using DAVID (http://david.abcc.ncifcrf.gov/), and many themes showed significant enrichment of genes affected at the level of exon usage and splice variants (Table 2). Cell cycle and RNA degradation were found to be consistently deregulated at both levels (Table 2). We investigated TP53 differential exon usage by PCR amplification and Sanger sequencing of gel-extracted bands. We observed exon skipping that was present in the SF3B1 knockdown cells only (Figure 2b).
Table 2. Pathway analysis using human genome exon-junction array data at two different levels: differential exon usage and different splicing variant.
|
Pathways affected by diferential exon usage |
Pathways affected by differential splice variant usage |
||
|---|---|---|---|
| Pathway description | P-value | Pathway description | P-value |
| KEGG pathway | |||
| Ubiquitin-mediated proteolysis | 1.82E−05 | RNA degradation | 7.85E−05 |
| Cell cycle | 1.02E−04 | Oocyte meiosis | 4.23E−03 |
| Spliceosome | 3.22E−04 | Cell cycle | 8.45E−03 |
| Oocyte meiosis | 3.88E−04 | Progesterone-mediated oocyte maturation | 2.17E−02 |
| Phosphatidylinositol signaling system | 8.92E−04 | Aminoacyl-tRNA biosynthesis | 4.04E−02 |
| Inositol phosphate metabolism | 1.68E−03 | ||
| RNA degradation | 2.30E−02 | ||
| Ribosome | 2.58E−02 | ||
| One carbon pool by folate | 2.65E−02 | ||
| Aminoacyl-tRNA biosynthesis | 2.91E−02 | ||
| Selenoamino acid metabolism | 3.93E−02 | ||
| Insulin signaling pathway | 4.03E−02 | ||
| REACTOME pathway | |||
| Cell cycle, mitotic | 1.19E−09 | Cell cycle, mitotic | 4.43E−05 |
| Gene expression | 1.47E−04 | Gene expression | 1.29E−02 |
| DNA repair | 1.75E−03 | Cell cycle checkpoints | 2.20E−02 |
| APC-Cdc20-mediated degradation of Nek2A | 9.97E−03 | ||
| Cell cycle checkpoints | 1.13E−02 | ||
| Transcription | 1.22E−02 | ||
| Signaling by Wnt | 2.91E−02 | ||
| Signaling by NGF | 3.63E−02 | ||
| Cdc20:Phospho-APC/C-mediated degradation of cyclin A | 4.70E−02 | ||
We also investigated whether two cell cycle genes (CCNA2 and STK6) previously shown to be aberrantly spliced in HeLa cells with SF3B1 knockdown,25 were also aberrantly spliced in K562 cells with SF3B1 knockdown in our study. Consistent with the finding in HeLa cells, we observed aberrant splicing of these genes using a qRT-PCR strategy as described previously (Figure 2c and d).25
RNA-Seq in HSPC from MDS patients with SF3B1 mutations
To gain insight into the spectrum of genes that are deregulated or aberrantly spliced in association with SF3B1 mutation in the hematopoietic stem and progenitor cells (HSPC) of MDS patients, we used deep RNA-Seq to compare the transcriptome of bone marrow CD34+ cells from eight MDS patients with SF3B1 mutation (SF3B1 mutants), four MDS patients with no known splicing mutation (wild type) and five healthy controls (control) (Supplementary Table S3). Using IGV, we evaluated the expression of the SF3B1 alleles in SF3B1 mutant cases and observed a range of 45–52% of mutant allele frequency, indicating that both wild-type and mutant alleles were equally expressed (Supplementary Table S3, Supplementary Figure S4).
We used edgeR to perform differential gene expression analysis of SF3B1 mutants versus wild type and control. At the whole transcript level, we observed a total of 526 genes (253 upregulated and 273 downregulated) significantly differentially expressed in SF3B1 mutants in comparison with wild type (Supplementary Table S5). In the comparison of SF3B1 mutants with control, we found 1823 significantly differentially expressed genes (646 upregulated and 1177 downregulated) (Supplementary Table S6). Genes linked to the pathogenesis of RARS and RCMD-RS, such as ALAS2 and ABCB7, were deregulated (ALAS2 upregulated and ABCB7 downregulated) in both comparisons of SF3B1 mutant with wild type and control. We also observed upregulation of the mitochondrial genes SLC25A37 when comparing SF3B1 mutant with control and GLRX5 in both comparisons of SF3B1 mutant with wild type and control.
We performed an analysis using 121 genes either known to be expressed in erythroid cells30 or described as erythroid transcription factors in the literature. We found 42 differentially expressed genes (37 upregulated and 5 downregulated) when comparing SF3B1 mutant with control. These included heme biosynthetic enzymes (for example, ALAS2, ALAD, FECH and UROD), globin genes (for example, HBQ1, HBA2, HBB and HBA1) and transcription factors (for example, GATA1, GATA2 and KLF1). In the comparison of SF3B1 mutant with wild type, we found a total of 32 differentially expressed genes (31 upregulated and 1 downregulated) of which 28 were overlapping with the differentially expressed genes found when comparing SF3B1 mutant with control.
We then performed pathway analysis on the significantly differentially expressed genes using IPA. Many pathways, including heme biosynthesis, mitotic roles of polo-like kinase and TNFR2 signaling, were significantly deregulated in the comparison of SF3B1 mutant with wild type (Table 3). When comparing SF3B1 mutant with control, significantly deregulated pathways included apoptosis signaling, p53 signaling, cell cycle regulation and heme degradation (Supplementary Table S7). We next performed Gene Set Enrichment Analysisand many gene sets showing significant enrichment were identified in SF3B1 mutant versus wild type and control. Upregulated gene sets included several that were related to mitochondrial function, cell cycle checkpoints and mRNA splicing. In the comparison of SF3B1 mutant with control cases, downregulated gene sets included several that were related to cell differentiation and apoptosis (Supplementary Table S8). Many of these deregulated pathways and gene sets are relevant to the known pathophysiology of MDS and in particular of RARS and RCMD-RS.
Table 3. Pathway analysis (IPA) of the significant differentially expressed genes between SF3B1 mutant and wild type obtained using edgeR.
| Ingenuity canonical pathways | P-value | Ratio | Molecules |
|---|---|---|---|
| Lymphotoxin β receptor signaling | 0.00074131 | 9.68E−02 | NFKBID, BCL2L1, CASP3, RELB, CXCL1 and TRAF1 |
| Agranulocyte adhesion and diapedesis | 0.001071519 | 5.73E−02 | CXCL3, PODXL2, CCL4,CLDN19, CXCL14, CXCL1, CCL3L1/CCL3L3, MMP2, CXCL2, MYH7B and CCL4L1/CCL4L2 |
| Heme biosynthesis II | 0.001096478 | 1.67E−01 | FECH, ALAS2, CPOX and HMBS |
| Hepatic fibrosis/hepatic stellate cell activation | 0.00162181 | 5.81E−02 | CXCL3, LEPR, IL6R, MMP2, LBP, IL6, MYH7B, AGTR1 and PGF |
| Communication between innate and adaptive immune cells | 0.001995262 | 6.25E−02 | CCL4, TLR7, CCL3L1/CCL3L3, CD83, IGHG1, IGHA1 and IL6 |
| Colorectal cancer metastasis signaling | 0.003467369 | 4.48E−02 | BCL2L1, ADCY9, CDH1, JUN, CASP3, PTGER3, DIRAS3, IL6R, TLR7, MMP2, IL6 and PGF |
| IL-17A signaling in fibroblasts | 0.004570882 | 1E−01 | NFKBID, JUN, IL6 and NFKBIZ |
| Toll-like receptor signaling | 0.004570882 | 7.81E−02 | JUN, TLR7, TNFAIP3, LBP and TRAF1 |
| Differential regulation of cytokine production in macrophages and T-helper cells by IL-17A and IL-17F | 0.004786301 | 1.67E−01 | CCL4, CXCL1 and IL6 |
| Granulocyte adhesion and diapedesis | 0.007762471 | 4.95E−02 | CXCL3, CCL4, CLDN19, CXCL14, CXCL1, CCL3L1/CCL3L3, MMP2, CXCL2 and CCL4L1/CCL4L2 |
| Airway pathology in chronic obstructive pulmonary disease | 0.009772372 | 1.82E−01 | CXCL3 and MMP2 |
| TNFR2 signaling | 0.016595869 | 8.82E−02 | JUN, TNFAIP3 and TRAF1 |
| Heme bosynthesis from uroporphyrinogen-III I | 0.018620871 | 1.82E−01 | FECH and CPOX |
| Aryl hydrocarbon receptor signaling | 0.024547089 | 4.09E−02 | TGM2, CCNE1, GSTM2, ALDH1A1, JUN, NQO2 and IL6 |
| NRF2-mediated oxidative stress response | 0.025118864 | 4.1E−02 | GSR, JUN, GSTM2, NQO2, DNAJC6, JUND, FOSL1 and ABCC4 |
| Role of IL-17A in psoriasis | 0.025703958 | 1.43E−01 | CXCL3 and CXCL1 |
| Gα12/13 signaling | 0.028840315 | 4.72E−02 | BTK, CDH7, CDH1, JUN, MEF2D and CDH11 |
| Tetrapyrrole biosynthesis II | 0.029512092 | 1.43E−01 | ALAS2 and HMBS |
| CDK5 signaling | 0.033884416 | 5.15E−02 | FOSB, PPP1CC, ADCY9, PPM1J and EGR1 |
| CD40 signaling | 0.033884416 | 5.63E−02 | JUN, TNFAIP3, MAPKAPK2 and TRAF1 |
| Mitotic roles of polo-like kinase | 0.041686938 | 5.41E−02 | PLK3, PPM1J, ANAPC13 and CCNB1 |
| Adenine and adenosine salvage III | 0.042657952 | 1.18E−01 | PNP and ADAT3 |
| Tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde | 0.046773514 | 1.11E−01 | HAAO and KYNU |
| T-helper cell differentiation | 0.047863009 | 5.56E−02 | STAT4, IL6R, IL6 and RORC |
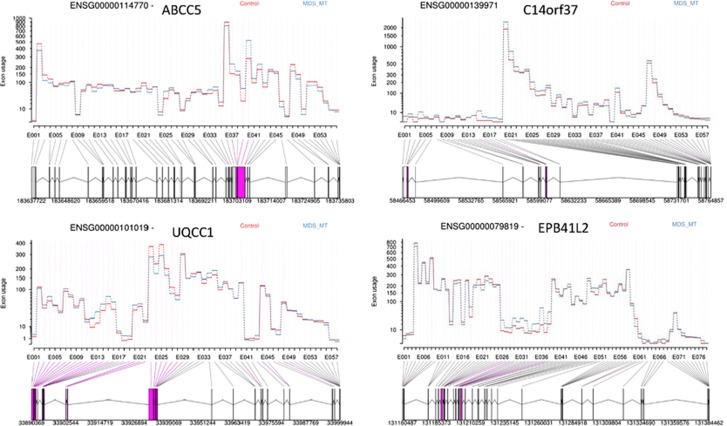
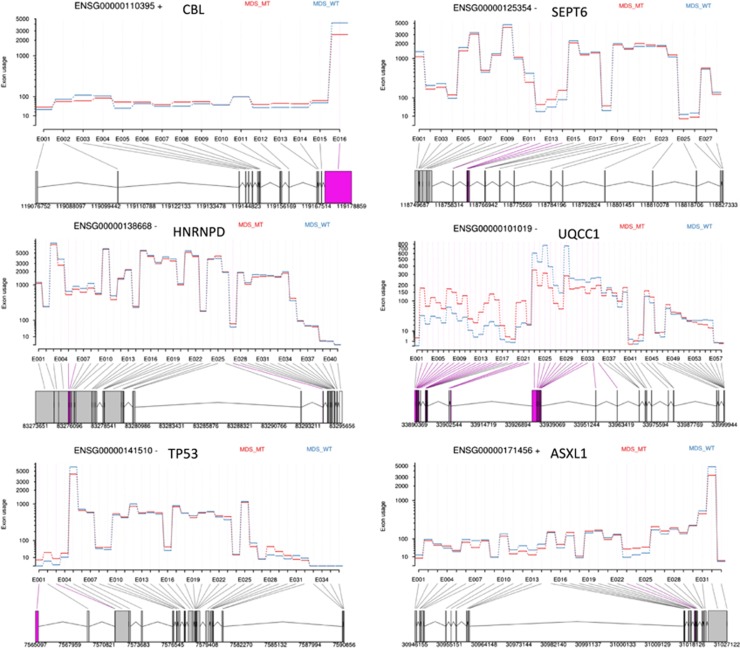
DEXSeq was used to perform differential exon usage analysis of the RNAseq data to evaluate aberrantly spliced genes. At the exon level, we observed a total of 3506 exons (corresponding to 1924 genes) significantly differentially expressed in SF3B1 mutant compared with control (Table 4, Figure 3, Supplementary Table S9). Differential exon usage was observed in at least one exon of genes known to be involved in MDS pathophysiology (TP53 and EZH1), erythroid genes (ALAD, UROD and EPB42) and genes associated with cell cycle (AURKB and CRNDE) and RNA processing (RBM5, RBM25, PRPF40A and HNRNPD). When comparing SF3B1 mutant with wild type cases, we found 3097 significantly differentially expressed exons (corresponding to 2022 genes) (Table 5, Figure 4, Supplementary Table S10). We found differential exon usage in at least one exon of genes involved in MDS pathophysiology (CBL, ASXL1 and DNMT3A), mitochondrial function (ALAS2, NDUFAF6), erythroid differentiation (NFE2L2, PPOX and HMBS) and mRNA processing (HNRNPD, U2AF2 and PRPF8). Interestingly, UQCC1, a gene involved in mitochondrial biogenesis31 and showing abnormal splicing in SF3B1 mutant cases in uveal melanoma,32 showed differential exon usage and upregulation in SF3B1 mutant cases compared with wild type and control in our study.
Table 4. Top ranking genes showing differential exon usage between SF3B1 mutant and control, obtained from RNA sequencing data analysis using DEXSeq.
| Gene ID | Exon ID | Gene name | Chromosome | Strand | P-value | Padj | FC |
|---|---|---|---|---|---|---|---|
| ENSG00000125354 | E011 | SEPT6 | X | −1 | 1.67E−78 | 5.19E−73 | 0.822522658 |
| ENSG00000111843 | E006 | TMEM14C | 6 | 1 | 2.80E−51 | 4.36E−46 | 0.803897136 |
| ENSG00000131669 | E006 | NINJ1 | 9 | −1 | 1.09E−33 | 1.13E−28 | 0.835812796 |
| ENSG00000088986 | E014 | DYNLL1 | 12 | 1 | 1.21E−28 | 9.43E−24 | 0.913566493 |
| ENSG00000223865 | E018 | HLA-DPB1 | 6 | 1 | 3.05E−27 | 1.90E−22 | 1.357267219 |
| ENSG00000101019 | E025 | UQCC1 | 20 | −1 | 1.71E−26 | 8.88E−22 | 0.889537433 |
| ENSG00000132199 | E025 | ENOSF1 | 18 | −1 | 2.57E−24 | 1.14E−19 | 0.905515696 |
| ENSG00000125991 | E049 | ERGIC3 | 20 | 1 | 4.11E−23 | 1.42E−18 | 0.870728412 |
| ENSG00000101019 | E024 | UQCC1 | 20 | −1 | 2.01E−22 | 6.25E−18 | 0.878172451 |
| ENSG00000118495 | E010 | PLAGL1 | 6 | −1 | 2.78E−22 | 7.85E−18 | 1.753766932 |
| ENSG00000266086 | E005 | RP11-159D12.5 | 17 | −1 | 1.43E−18 | 3.43E−14 | 1.051487592 |
| ENSG00000119777 | E023 | TMEM214 | 2 | 1 | 1.35E−18 | 3.43E−14 | 0.724952987 |
| ENSG00000130066 | E011 | SAT1 | X | 1 | 2.63E−17 | 5.83E−13 | 1.224469811 |
| ENSG00000101019 | E029 | UQCC1 | 20 | −1 | 3.49E−17 | 7.24E−13 | 0.987866168 |
| ENSG00000141425 | E015 | RPRD1A | 18 | −1 | 2.14E−16 | 3.91E−12 | 1.151614234 |
| ENSG00000268400 | E058 | CTD-3214H19.4 | 19 | 1 | 2.08E−16 | 3.91E−12 | 0.720165623 |
| ENSG00000127586 | E055 | CHTF18 | 16 | 1 | 2.45E−16 | 4.24E−12 | 0.83461565 |
| ENSG00000130066 | E015 | SAT1 | X | 1 | 4.80E−16 | 7.86E−12 | 1.112053709 |
| ENSG00000160789 | E072 | LMNA | 1 | 1 | 1.00E−15 | 1.56E−11 | 0.74131183 |
| ENSG00000130066 | E010 | SAT1 | X | 1 | 1.22E−15 | 1.81E−11 | 1.175909464 |
| ENSG00000119777 | E024 | TMEM214 | 2 | 1 | 6.13E−15 | 8.66E−11 | 0.769414939 |
| ENSG00000266086 | E004 | RP11-159D12.5 | 17 | −1 | 1.51E−14 | 2.04E−10 | 1.073999383 |
| ENSG00000168675 | E009 | LDLRAD4 | 18 | 1 | 4.10E−14 | 5.10E−10 | 0.882283199 |
| ENSG00000160789 | E071 | LMNA | 1 | 1 | 6.34E−14 | 7.58E−10 | 0.907377148 |
| ENSG00000214021 | E019 | TTLL3 | 3 | 1 | 1.13E−13 | 1.30E−09 | 1.027729333 |
| ENSG00000196576 | E041 | PLXNB2 | 22 | −1 | 1.59E−13 | 1.77E−09 | 0.53703246 |
| ENSG00000166508 | E021 | MCM7 | 7 | −1 | 2.21E−13 | 2.37E−09 | 1.034956343 |
| ENSG00000114770 | E040 | ABCC5 | 3 | −1 | 2.75E−13 | 2.79E−09 | 1.290748901 |
| ENSG00000196365 | E028 | LONP1 | 19 | −1 | 2.78E−13 | 2.79E−09 | 0.876937237 |
| ENSG00000130066 | E002 | SAT1 | X | 1 | 3.59E−13 | 3.49E−09 | 0.967812734 |
| ENSG00000102119 | E017 | EMD | X | 1 | 3.76E−13 | 3.55E−09 | 1.050545479 |
| ENSG00000130066 | E014 | SAT1 | X | 1 | 5.55E−13 | 5.08E−09 | 1.168469304 |
| ENSG00000266086 | E006 | RP11-159D12.5 | 17 | −1 | 5.72E−13 | 5.08E−09 | 1.073091463 |
| ENSG00000055609 | E005 | KMT2C | 7 | −1 | 6.09E−13 | 5.26E−09 | 1.089688525 |
| ENSG00000228315 | E065 | GUSBP11 | 22 | −1 | 6.70E−13 | 5.63E−09 | 1.125912723 |
| ENSG00000156860 | E023 | FBRS | 16 | 1 | 7.27E−13 | 5.95E−09 | 1.10075677 |
| ENSG00000268400 | E071 | CTD-3214H19.4 | 19 | 1 | 8.37E−13 | 6.67E−09 | 1.139015436 |
| ENSG00000214021 | E065 | TTLL3 | 3 | 1 | 1.15E−12 | 8.34E−09 | 1.269221488 |
| ENSG00000122566 | E004 | HNRNPA2B1 | 7 | −1 | 1.10E−12 | 8.34E−09 | 1.160615984 |
| ENSG00000101557 | E035 | USP14 | 18 | 1 | 1.13E−12 | 8.34E−09 | 1.043528763 |
Genes are ranked by adjusted P-value (Padj).
Figure 3.
Examples of genes showing significant differential exon usage between MDS patients with SF3B1 mutation in comparison with control, obtained from RNA-Seq data analysis using DEXSeq. The graphs show some of the top ranking genes with significant differential exon usage. The exons highlighted in purple represent the significant differential exon usage.
Table 5. Top ranking genes showing differential exon usage between SF3B1 mutant and wild type, obtained from RNA sequencing data analysis using DEXSeq.
| Gene ID | Exon ID | Gene name | Chromosome | Strand | P-value | Padj | FC |
|---|---|---|---|---|---|---|---|
| ENSG00000125354 | E011 | SEPT6 | X | −1 | 9.38E−92 | 3.53E−86 | 0.827252395 |
| ENSG00000132199 | E025 | ENOSF1 | 18 | −1 | 5.10E−33 | 9.59E−28 | 0.911519852 |
| ENSG00000189283 | E004 | FHIT | 3 | −1 | 1.73E−28 | 2.18E−23 | 3.663047029 |
| ENSG00000101019 | E025 | UQCC1 | 20 | −1 | 9.24E−28 | 8.69E−23 | 0.72317914 |
| ENSG00000111843 | E006 | TMEM14C | 6 | 1 | 9.84E−26 | 7.40E−21 | 0.91596329 |
| ENSG00000125991 | E049 | ERGIC3 | 20 | 1 | 9.81E−23 | 6.15E−18 | 0.791616483 |
| ENSG00000071082 | E025 | RPL31 | 2 | 1 | 3.50E−21 | 1.88E−16 | 0.717351893 |
| ENSG00000101019 | E029 | UQCC1 | 20 | −1 | 2.62E−20 | 1.23E−15 | 0.701671242 |
| ENSG00000211644 | E016 | IGLV1-51 | 22 | 1 | 3.99E−20 | 1.67E−15 | 0.441359946 |
| ENSG00000088986 | E014 | DYNLL1 | 12 | 1 | 5.65E−20 | 2.12E−15 | 0.845893878 |
| ENSG00000028310 | E077 | BRD9 | 5 | −1 | 3.97E−18 | 1.36E−13 | 0.767361674 |
| ENSG00000127586 | E055 | CHTF18 | 16 | 1 | 2.12E−17 | 6.64E−13 | 0.878313474 |
| ENSG00000101019 | E007 | UQCC1 | 20 | −1 | 3.20E−17 | 9.25E−13 | 1.47227241 |
| ENSG00000160710 | E024 | ADAR | 1 | −1 | 1.30E−16 | 3.51E−12 | 1.320985378 |
| ENSG00000101019 | E024 | UQCC1 | 20 | −1 | 2.02E−16 | 5.06E−12 | 0.71746253 |
| ENSG00000118495 | E010 | PLAGL1 | 6 | −1 | 4.10E−16 | 9.65E−12 | 2.538692764 |
| ENSG00000255863 | E017 | AC073610.5 | 12 | −1 | 1.20E−15 | 2.65E−11 | 0.882954589 |
| ENSG00000211644 | E017 | IGLV1-51 | 22 | 1 | 1.38E−15 | 2.88E−11 | 0.485819764 |
| ENSG00000075218 | E014 | GTSE1 | 22 | 1 | 2.48E−15 | 4.91E−11 | 1.468605485 |
| ENSG00000137133 | E004 | HINT2 | 9 | −1 | 2.09E−14 | 3.93E−10 | 0.839329231 |
| ENSG00000205593 | E031 | DENND6B | 22 | −1 | 5.28E−14 | 9.46E−10 | 4.609575952 |
| ENSG00000101019 | E013 | UQCC1 | 20 | −1 | 1.05E−13 | 1.74E−09 | 2.895712194 |
| ENSG00000101019 | E026 | UQCC1 | 20 | −1 | 1.11E−13 | 1.74E−09 | 0.732192837 |
| ENSG00000101019 | E028 | UQCC1 | 20 | −1 | 1.10E−13 | 1.74E−09 | 0.698206407 |
| ENSG00000103426 | E028 | CORO7-PAM16 | 16 | −1 | 1.21E−13 | 1.83E−09 | 0.838759667 |
| ENSG00000205593 | E030 | DENND6B | 22 | −1 | 3.43E−13 | 4.97E−09 | 4.61396886 |
| ENSG00000101019 | E010 | UQCC1 | 20 | −1 | 4.54E−13 | 6.33E−09 | 1.74387423 |
| ENSG00000111640 | E017 | GAPDH | 12 | 1 | 8.37E−13 | 1.12E−08 | 1.387332969 |
| ENSG00000100387 | E034 | RBX1 | 22 | 1 | 1.24E−12 | 1.61E−08 | 1.875629945 |
| ENSG00000271270 | E006 | TMCC1-AS1 | 3 | 1 | 2.12E−12 | 2.66E−08 | 2.979442225 |
| ENSG00000028310 | E078 | BRD9 | 5 | −1 | 3.00E−12 | 3.64E−08 | 0.795339192 |
| ENSG00000101019 | E027 | UQCC1 | 20 | −1 | 3.26E−12 | 3.84E−08 | 0.703747197 |
| ENSG00000255103 | E077 | KIAA0754 | 1 | 1 | 3.38E−12 | 3.85E−08 | 1.663519412 |
| ENSG00000268400 | E035 | CTD-3214H19.4 | 19 | 1 | 3.57E−12 | 3.95E−08 | 1.463054031 |
| ENSG00000205593 | E032 | DENND6B | 22 | −1 | 4.28E−12 | 4.60E−08 | 4.764064117 |
| ENSG00000136044 | E042 | APPL2 | 12 | −1 | 5.63E−12 | 5.88E−08 | 0.720698938 |
| ENSG00000101019 | E005 | UQCC1 | 20 | −1 | 1.00E−11 | 9.92E−08 | 1.53637968 |
| ENSG00000167323 | E043 | STIM1 | 11 | 1 | 9.97E−12 | 9.92E−08 | 0.766910234 |
| ENSG00000101019 | E016 | UQCC1 | 20 | −1 | 1.35E−11 | 1.28E−07 | 2.2692562 |
| ENSG00000161013 | E025 | MGAT4B | 5 | −1 | 1.36E−11 | 1.28E−07 | 1.097105348 |
Genes are ranked by adjusted P-value (Padj).
Figure 4.
Examples of genes showing significant differential exon usage in MDS patients with SF3B1 mutation in comparison with wild type, obtained from RNA-Seq data analysis using DEXSeq. The graphs show some of the top ranking genes with significant differential exon usage. The exons highlighted in purple represent the significant differential exon usage.
To identify pathways affected by differential exon usage, we performed pathway analysis on the genes showing significantly differentially expressed exons using IPA. In the comparison of SF3B1 mutant with wild type and control, we observed many pathways to be affected, including cell cycle, heme biosynthesis, DNA damage response, mitochondrial and haematopoietic progenitor cells pathway (Supplementary Table S11 and S12). Using DAVID functional annotation tool, we observed significant enrichment of biological themes including alternative splicing, RNA binding, mitochondrion, spliceosome and cell cycle.
Recently, a role for SF3B1 in the maintenance of genomic stability has also been reported where it functions in a DNA damage-induced mRNA splicing complex with BRCA1 and BCLAF1.33 Given that deregulation of the DNA damage response pathway was highlighted by the IPA pathway analysis, we performed an analysis using genes regulated by the BRCA1–BCLAF1–SF3B1 complex. Several genes regulated by this complex showed differential exon usage in SF3B1 mutant compared with control (including NUMA1, RB1, CHUK and ABL1) and compared with wild type (NUMA1, PIAS1, SMAD4, BIRC2 and PTK2) (Supplementary Table S13). The overrepresentation of genes regulated by the BRCA1–BCLAF1–SF3B1 complex was significant in SF3B1 mutant compared with control (P<0.001) and compared with wild type (P=0.0498, hypergeometric test). We also found many genes to be affected at the transcript level, including BIRC3, BCL2A1, GYPB, HBB and HBBP1 when comparing SF3B1 mutant with wild type and control (Supplementary Table S13).
Discussion
The identification of frequent somatic mutations of SF3B1 in MDS patients with ring sideroblasts suggests a direct correlation between the presence of mutations and this particular phenotype.2, 11, 13 However, the mechanism by which SF3B1 mutation leads to MDS with ring sideroblasts remains to be elucidated.
To illuminate the role of SF3B1 mutation in MDS pathophysiology, we have determined the effects of SF3B1 disruption on cell growth and gene expression in human hematological cells. First, we investigated the effects of SF3B1 knockdown on cell function and gene expression in myeloid cell lines. Second, we used RNA-Seq to study the global gene expression changes and splicing abnormalities associated with the presence of SF3B1 mutations in the HSPC of MDS patients.
We have shown that SF3B1 knockdown in four myeloid cell lines resulted in inhibition of cell growth and disruption of the cell cycle. It has recently been reported that Sf3b1+/− mice showed reduced numbers of HSC and compromised reconstitution capacity in lethally irradiated mice.14, 15 Thus, reduced expression of SF3B1 appears to result in impaired cell growth in hematopoietic cells. Similar observations have been made following reduced expression of other splicing factor genes, including U2AF1.3, 4, 34
We used two different microarray platforms to identify genes deregulated at the transcriptional and exon level by SF3B1 knockdown in myeloid cell lines. Gene expression profiling identified many deregulated genes, with four genes consistently upregulated, including TFDP1, and five genes downregulated, including CREBZF, in all four cell lines with SF3B1 knockdown, several of which have a role in the control of cell growth. TFDP1 is involved in the control of transcriptional activity of G1/S cell cycle checkpoint genes,35 and CREBZF is a potent suppressor of cell growth, the effects of which are mediated through the tumor suppressor p53.36 Next, we employed exon-junction arrays to study the transcriptome in K562 and TF1 cells. We identified many differentially expressed exons and differentially regulated splicing variants in cells with SF3B1 knockdown. For example, we observed differential exon usage of the CDC7 and SRSF11 genes, involved in cell cycle regulation and splicing, respectively, and of the TP53 gene. Several pathways including cell cycle, RNA processing, mitochondrion and apoptosis/p53 pathway were consistently deregulated in the cell lines with SF3B1 knockdown.
In summary, our data on myeloid cell lines suggest that the phenotype observed in cells with SF3B1 knockdown is mediated by aberrant splicing and expression of target genes involved in key biological processes.
RNA-Seq is the method of choice for a comprehensive analysis of global gene expression and splicing. In all relevant studies published to date, RNA-Seq has been performed on unfractionated bone marrow mononuclear cells from a small number of MDS patients with SF3B1 mutations (n⩽3 per study), revealing interesting data.11, 37, 38 MDS arise in the HSC, and it is thus of critical importance that the effects of SF3B1 mutation on the transcriptome are studied in the cell of origin. In this study, we performed RNA-Seq on purified bone marrow CD34+ cells from 12 MDS patients, eight with ring sideroblasts and SF3B1 mutation and four without mutation in other splicing factor genes, and from five healthy controls.
We identified many significantly differentially expressed genes at the transcript level and the exon level when comparing SF3B1 mutant with wild type and control. CD34+ cells from RCMD-RS and RARS patients display a particular expression profile of mitochondria-related genes;17, 20 we observed upregulation of ALAS2 (heme biosynthesis enzyme) and downregulation of ABCB7 (involved in the transport of iron from the mitochondria to the cytoplasm) in SF3B1 mutants compared with wild type and control, in agreement with our previous reports.17, 20 SLC25A37, encoding a mitochondrial iron importer, and GLRX5, encoding another mitochondrial protein, were both significantly upregulated in CD34+ cells of SF3B1 mutants compared with wild type and control. A recent study showed SLC25A37 mRNA upregulation in SF3B1 mutant bone marrow mononuclear cells of three MDS patients with RARS/-T.38 Evidence is thus mounting to suggest that upregulation of the iron importer SLC25A37 and downregulation of iron exporter ABCB7 may be linked to the increased mitochondrial iron accumulation observed in MDS patients with ring sideroblasts.39 Deregulation of these genes could also be consistent with an impaired ability of mitochondrial pathways to use iron that may result in an attempt to increase iron availability.
Importantly, we observed many genes associated with porphyrin and heme biosynthesis showing differential exon usage in SF3B1 mutant cases, indicating altered splicing. These include ALAD and UROD when comparing SF3B1 mutant with control and ALAS2 and PPOX when comparing with wild type. We suggest that the aberrant expression and exon usage of mitochondrial and heme-related genes in the CD34+ cells of MDS patients with SF3B1 mutation has a role in ring sideroblast formation and abnormal iron homeostasis observed in this patient group. Intriguingly, the most common congenital sideroblastic anemia (X-linked sideroblastic anemia) results from an ALAS2 mutation.40
The relative expression levels of the transcription factors GATA1 and GATA2 differed from normal in the CD34+ cells of MDS patients with SF3B1 mutation. The dynamic and strictly regulated change of expression from GATA2 to GATA1 during erythropoiesis has a crucial role,41 and any alteration of this pattern may be predicted to lead to aberrant erythropoiesis. We suggest that the aberrant expression and splicing of erythroid-related genes observed in the CD34+ cells of MDS patients with SF3B1 mutation may have a role in the ineffective erythropoiesis found in these patients.
In a previous study, RNA-Seq was used to compare the transcriptome of bone marrow mononuclear cells of two MDS patients with SF3B1 mutation to that of one healthy control.11 One hundred and thirty genes showed significant differential expression, 28 of which overlap with the genes differentially expressed between SF3B1 mutant and control in our study of CD34+ cells. These genes include MAP3K8 and CLEC5A, which have been previously shown to be downregulated in MDS.19, 42 Furthermore, 350 genes showed differential exon usage, and 52 of these were in common with the genes showing differential exon usage in our study of CD34+ cells. The overlapping genes include EZH1, ASXL1, CBL1 and SMAD4.
The mutant forms of ASXL1, CBL and TP53 are known to have an important role in the molecular pathogenesis of MDS, and intriguingly all these genes were found to be alternatively spliced in the HSPC of MDS patients harboring SF3B1 mutations.
A recent study identified a DNA damage-induced BRCA1 protein complex containing BCLAF1 and SF3B1 (BRCA1–BCLAF1–SF3B1 complex).33 In response to DNA damage, the complex regulates pre-mRNA splicing of genes involved in DNA damage signaling and repair and hence affects their transcription and pre-mRNA maturation.33 Importantly, we found many of the genes regulated by this complex to be differentially expressed and to show differential exon usage in our study. Genes showing differential transcript level changes included BIRC3 and BCL2A1, whereas NUMA1 showed differential exon usage when comparing SF3B1 mutant with wild type and control. When comparing SF3B1 mutant with wild type, we also found differential exon usage of the PIAS1 gene, a DNA damage response regulator.43, 44 It has been demonstrated that abrogation of members of this complex including BRCA1 and BCLAF1 results in genomic instability,33 a common feature of malignant cells. In MDS patients with mutant SF3B1, the function of this complex may be impaired, with possible downstream effects on the efficiency of DNA damage repair.
Many genes involved in RNA splicing and processing were found to show differential transcript levels, as well as differential exon usage in our study, including HNRNPD, U2AF2, PRPF8 and RBM25. A recent study showed RNA processing genes to be misspliced in the bone marrow mononuclear cells of MDS/AML patients with U2AF1 mutation.45 Intriguingly, there is limited overlap between the genes misspliced as a result of U2AF1 mutation and the RNA processing/splicing genes deregulated/aberrantly spliced in our SF3B1 mutant cases in both comparisons of SF3B1 mutant with wild type and control, suggesting that this process may be affected by different splicing mutations even if their target genes are distinct.
A recent study showed that defects in the splicing factor gene PRPF8 result in missplicing in myeloid malignancies and are associated with the presence of ring sideroblasts in advanced MDS and AML.46 Interestingly, we observed differential exon usage of PRPF8 in SF3B1 mutant cases compared with control in our study, indicating a link between SF3B1 and PRPF8 and the ring sideroblast phenotype. Our finding of differential exon usage of multiple RNA processing/splicing genes in the HSPC of cases with SF3B1 mutation points towards an exacerbation of aberrant splicing, with a wider number of downstream target genes affected. Emerging evidence from our study on SF3B1 and the study on U2AF145 thus supports the hypothesis that in MDS patients harboring splicing factor mutations, there is widespread disruption of the splicing machinery as a consequence of downstream effects of the mutant protein. Loss-of-function mutations in zebrafish Prpf8 have been shown to result in missplicing of TP53.47 We found TP53 to be aberrantly spliced in MDS patients with SF3B1 mutation compared with wild type and control.
Splicing factor mutations, including SF3B1, are considered to be founder mutations and must confer a selective growth advantage to the HSC, but how this occurs remains a mystery. We have identified differential splicing in several genes known to have a role in MDS pathogenesis, including TP53 and ASXL1, as well as genes involved in DNA damage repair, in the HSPC of MDS patients harboring SF3B1 mutations. We suggest that the aberrant splicing of such genes may confer oncogenic properties that help drive the malignant process. In this study, we have determined the target genes of SF3B1 mutations in the HSPC of MDS patients with ring sideroblasts, shedding light on the mechanisms underlying this phenotype.
Acknowledgments
This work was supported by Leukaemia and Lymphoma Research of the United Kingdom. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust Grant reference 090532/Z/09/Z and MRC Hub Grant G0900747 91070) for the generation of the RNA-Seq data. PV is a senior clinical investigator of Research Foundation—Flanders. We are also grateful to the patients who participated in this study and to all physicians for providing samples and clinical data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Heaney ML, Golde DW. Myelodysplasia. N Engl J Med. 1999;340:1649–1660. doi: 10.1056/NEJM199905273402107. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Boultwood J, Dolatshad H, Varanasi SS, Yip BH, Pellagatti A. The role of splicing factor mutations in the pathogenesis of the myelodysplastic syndromes. Adv Biol Regul. 2014;54:153–161. doi: 10.1016/j.jbior.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je EM, Yoo NJ, Kim YJ, Kim MS, Lee SH. Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int J Cancer. 2013;133:260–265. doi: 10.1002/ijc.28011. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, Della Porta MG, Pascutto C, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- Cazzola M, Invernizzi R, Bergamaschi G, Levi S, Corsi B, Travaglino E, et al. Mitochondrial ferritin expression in erythroid cells from patients with sideroblastic anemia. Blood. 2003;101:1996–2000. doi: 10.1182/blood-2002-07-2006. [DOI] [PubMed] [Google Scholar]
- Visconte V, Rogers HJ, Singh J, Barnard J, Bupathi M, Traina F, et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120:3173–3186. doi: 10.1182/blood-2012-05-430876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Thol F, Kosmider O, Kade S, Loffeld P, Dreyfus F, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26:1137–1140. doi: 10.1038/leu.2011.321. [DOI] [PubMed] [Google Scholar]
- Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26:542–545. doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]
- Matsunawa M, Yamamoto R, Sanada M, Sato-Otsubo A, Shiozawa Y, Yoshida K, et al. Haploinsufficiency of Sf3b1 leads to compromised stem cell function but not to myelodysplasia. Leukemia. 2014;28:1844–1850. doi: 10.1038/leu.2014.73. [DOI] [PubMed] [Google Scholar]
- Wang C, Sashida G, Saraya A, Ishiga R, Koide S, Oshima M, et al. Depletion of Sf3b1 impairs proliferative capacity of hematopoietic stem cells but is not sufficient to induce myelodysplasia. Blood. 2014;123:3336–3343. doi: 10.1182/blood-2013-12-544544. [DOI] [PubMed] [Google Scholar]
- Yip BH, Pellagatti A, Vuppusetty C, Giagounidis A, Germing U, Lamikanra AA, et al. Effects of L-leucine in 5q- syndrome and other RPS14-deficient erythroblasts. Leukemia. 2012;26:2154–2158. doi: 10.1038/leu.2012.82. [DOI] [PubMed] [Google Scholar]
- Nikpour M, Scharenberg C, Liu A, Conte S, Karimi M, Mortera-Blanco T, et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia. 2013;27:889–896. doi: 10.1038/leu.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Pellagatti A, Cazzola M, Giagounidis A, Perry J, Malcovati L, Della Porta MG, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24:756–764. doi: 10.1038/leu.2010.31. [DOI] [PubMed] [Google Scholar]
- Pellagatti A, Cazzola M, Giagounidis AA, Malcovati L, Porta MG, Killick S, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006;108:337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandoura S, Weiss E, Rautou PE, Fasseu M, Gustot T, Lemoine F, et al. Gene- and exon-expression profiling reveals an extensive LPS-induced response in immune cells in patients with cirrhosis. J Hepatol. 2013;58:936–948. doi: 10.1016/j.jhep.2012.12.025. [DOI] [PubMed] [Google Scholar]
- Wang E, Aslanzadeh V, Papa F, Zhu H, de la Grange P, Cambi F. Global profiling of alternative splicing events and gene expression regulated by hnRNPH/F. PLoS One. 2012;7:e51266. doi: 10.1371/journal.pone.0051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boultwood J, Pellagatti A, Nikpour M, Pushkaran B, Fidler C, Cattan H, et al. The role of the iron transporter ABCB7 in refractory anemia with ring sideroblasts. PLoS One. 2008;3:e1970. doi: 10.1371/journal.pone.0001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JS, Mohandas N, Conboy JG. High frequency of alternative first exons in erythroid genes suggests a critical role in regulating gene function. Blood. 2006;107:2557–2561. doi: 10.1182/blood-2005-07-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker EJ, Wanschers BF, Szklarczyk R, Mountford HS, Wijeyeratne XW, van den Brand MA, et al. Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet. 2013;9:e1004034. doi: 10.1371/journal.pgen.1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ, et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell. 2014;54:445–459. doi: 10.1016/j.molcel.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol Biol Cell. 2006;17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargou RC, Wagener C, Bommert K, Arnold W, Daniel PT, Mapara MY, et al. Blocking the transcription factor E2F/DP by dominant-negative mutants in a normal breast epithelial cell line efficiently inhibits apoptosis and induces tumor growth in SCID mice. J Exp Med. 1996;183:1205–1213. doi: 10.1084/jem.183.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Misra V. Effects of cyclic AMP response element binding protein-Zhangfei (CREBZF) on the unfolded protein response and cell growth are exerted through the tumor suppressor p53. Cell Cycle. 2014;13:279–292. doi: 10.4161/cc.27053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconte V, Avishai N, Mahfouz R, Tabarroki A, Cowen J, Sharghi-Moshtaghin R, et al. Distinct iron architecture in SF3B1-mutant myelodysplastic syndrome patients is linked to an SLC25A37 splice variant with a retained intron Leukemiae-pub ahead of print 23 May 2014; doi: 10.1038/leu.2014.170 [DOI] [PubMed]
- Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- Cazzola M, Invernizzi R. Ring sideroblasts and sideroblastic anemias. Haematologica. 2011;96:789–792. doi: 10.3324/haematol.2011.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T, Yamamoto M. A regulatory network governing Gata1 and Gata2 gene transcription orchestrates erythroid lineage differentiation. Int J Hematol. 2014;100:417–424. doi: 10.1007/s12185-014-1568-0. [DOI] [PubMed] [Google Scholar]
- Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J Clin Invest. 2013;123:4627–4640. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- Przychodzen B, Jerez A, Guinta K, Sekeres MA, Padgett R, Maciejewski JP, et al. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122:999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic-Kozaric A, Przychodzen B, Singh J, Konarska MM, Clemente MJ, Otrock ZK, et al. PRPF8 defects cause missplicing in myeloid malignancies Leukemiae-pub ahead of print 30 April 2014; doi: 10.1038/leu.2014.144 [DOI] [PMC free article] [PubMed]
- Keightley MC, Crowhurst MO, Layton JE, Beilharz T, Markmiller S, Varma S, et al. In vivo mutation of pre-mRNA processing factor 8 (Prpf8) affects transcript splicing, cell survival and myeloid differentiation. FEBS Lett. 2013;587:2150–2157. doi: 10.1016/j.febslet.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.