Abstract
Many viruses encode short transmembrane proteins that play vital roles in virus replication or virulence. Because these proteins are often less than 50 amino acids long and not homologous to cellular proteins, their open reading frames were often overlooked during the initial annotation of viral genomes. Some of these proteins oligomerize in membranes and form ion channels. Other miniproteins bind to cellular transmembrane proteins and modulate their activity, whereas still others have an unknown mechanism of action. Based on the underlying principles of transmembrane miniprotein structure, it is possible to build artificial small transmembrane proteins that modulate a variety of biological processes. These findings suggest that short transmembrane proteins provide a versatile mechanism to regulate a wide range of cellular activities, and we speculate that cells also express many similar proteins that have not yet been discovered.
Keywords: transmembrane protein, traptamer, viroporin, hydrophobic, randomized library
Introduction
How to remain small?
Virus has a strategy.
Transmembrane proteins.
Viruses are biological haiku. Like the traditional stylized Japanese poetry, viruses are small, constructed according to strict rules, and lacking extraneous features that detract from the central message. Viruses thus provide an exceptionally clear view into the soul of the poet, natural selection. Over the course of evolution, natural selection has identified and refined the core imperatives of the virus life cycle: genome amplification and transmission to new hosts. Central to rapid genome amplification is small genome size. Viruses have adopted a number of strategies to maintain the small size of their genomes; chief among them is a parasitic lifestyle, in which the host cell provides the biochemical infrastructure that supports virus replication.
Another strategy employed by viruses to maintain small genome size is the use of very small proteins. These proteins, some shorter than 50 amino acids, were often overlooked during sequence analysis of viral genomes or biochemical analysis of infected cells, and they cloud the distinction between proteins and peptides. We regard a polypeptide chain as a protein regardless of its length if it is a primary translation product from viral mRNA. Cleavage products of large precursor proteins are not discussed in this review, even though some of them resemble the proteins discussed here. Many viral miniproteins are extremely hydrophobic and span viral or cellular membranes. These viral transmembrane miniproteins constitute the subject of this review.
Poxviruses, papillomaviruses, polyomaviruses, paramyxoviruses, lentiviruses, influenza viruses, and presumably many other virus families encode short transmembrane proteins that are not homologous to cellular proteins. Thus, these proteins are not restricted to RNA or DNA viruses or to a particular viral genome replication strategy. Why do viruses use short transmembrane proteins instead of soluble globular proteins replete with hydrophilic amino acids? The active site of enzymes is often constructed from only a small number of charged or polar amino acids in a particular conformation for catalysis, so most of the protein can be viewed as a scaffold to steer the catalytic amino acids into the proper three-dimensional position in a suitable environment to carry out their chemical roles. Similarly, for proteins that bind to other macromolecules, most amino acids perform the back-office function of supporting the relatively few amino acids that actually constitute the interacting surface. For an organism that puts a premium on small size, like a virus, there are clear advantages to using proteins that can dispense with a large scaffold. However, because the vast majority of very short proteins consisting of a mixture of hydrophobic and hydrophilic amino acids are presumably unfolded or aggregate, they are unlikely to be biologically active.
In contrast, owing to underlying chemical principles, transmembrane domains form short, folded proteins (94a). First, fewer than 25 amino acids is sufficient to span membranes. Second, the predominance of amino acids with hydrophobic side chains ensures their stable incorporation into membranes. Most important, the energetics of lipid-protein interactions results in the generation of a stable protein structure in the absence of a discrete scaffold, regardless of the amino acid sequence (66, 105). Because the main-chain polypeptide backbone is studded with polar amine and carboxyl groups, a protein with a hydrophobic composition is faced with a dilemma: How can it simultaneously accommodate the energetically favorable insertion of hydrophobic side chains into a membrane with the unfavorable presence of main-chain hydrophilic groups in the hydrophobic lipid bilayer? There is a simple solution: The amine and carboxyl groups form intramolecular hydrogen bonds along the axis of the protein roughly perpendicular to the membrane plane, resulting in the formation of an α-helix (94a). This shields the main-chain polar groups from the hydrophobic core of the membrane and forces the side chains to be radially displayed in an energetically favorable position facing the hydrophobic fatty acid groups in the lipid bilayer, where they can interact specifically with complementary surfaces generated by other transmembrane domains (104). Transmembrane miniproteins may self-associate in the membrane and form homooligomers or associate with the transmembrane segments of other cellular or viral proteins and influence their activity. Thus, the chemistry of hydrophobic proteins and the surrounding membrane imposes a productive, stable fold on the protein.
Some viral miniproteins, known as viroporins, homooligomerize in membranes to form ion channels with an aqueous pore (81). The utility of a transmembrane protein to conduct charged molecules through a membrane is obvious. Other viral miniproteins act by binding to the membrane-spanning domains of much larger cellular transmembrane proteins and modulating their expression or activity. Still other miniproteins have an unknown mechanism of action. In this review, we describe the structure, biological activities, and biochemical properties of representative animal virus transmembrane miniproteins. We then discuss artificial proteins modeled on these viral proteins and close with a consideration of the implications of these findings.
Influenza Virus M2 Proteins
Flu M2 protein.
The amantadine target.
Stop the pandemic.
Influenza viruses, enveloped viruses with segmented RNA genomes, are of great medical importance. Influenza A virus causes most influenza epidemics in humans and was responsible for the 1918 Spanish influenza pandemic, which killed between 50 and 100 million people. Influenza A virus encodes the M2 protein (A/M2), a 97-amino-acid integral membrane phosphoprotein with a single membrane-spanning domain and a 54-amino-acid carboxy terminus cytoplasmic tail (93). A tetramer of M2 exists in cell membranes and the envelope of the virus particle, where it acts as a proton-selective ion channel (Figure 1) (47, 92, 93, 117). Hence, M2 is a viroporin. Numerous RNA and DNA viruses encode viroporins, most of which facilitate the assembly and release of virus particles from infected cells. For example, the nonenveloped polyomaviruses SV40 and JC virus encode viroporins, which facilitate virus release (96, 120). The reader is referred to the excellent review by Nieva et al. (81) for a comprehensive discussion of viroporins.
Figure 1.
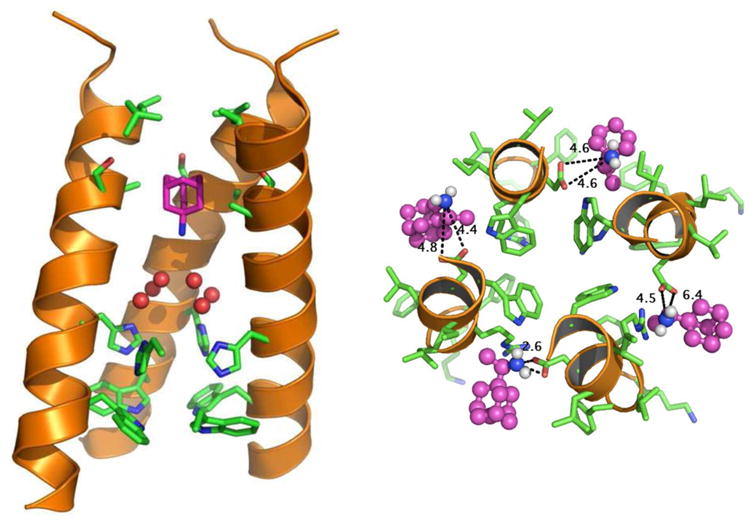
Structure of the M2 proton channel. (a) Lateral view of the transmembrane domains of the influenza A virus M2 protein tetramer (ribbons) bound to the inhibitor amantadine (purple), as determined by solid state nuclear magnetic resonance (NMR) spectrometry. One helix from the foreground of the M2 tetramer has been removed for clarity. Six water molecules (red spheres) from the high-resolution crystal structure are superimposed on the NMR structure. The essential histidine and tryptophan side chains are shown in stick representation (green and blue) in the bottom portion of the structure. (b) An end-on view of the M2 tetramer (main chain, ribbons; amino acid side chains, green and blue sticks), showing the central aqueous pore. Adapted with permission from Reference 132, copyright 2011 American Chemical Society; and Reference 52, copyright 2008 National Academy of Sciences USA.
M2 proton channel activity is essential for influenza virus replication, possibly by acting at several different steps in the virus life cycle (93, 136, 137). Internalization of the influenza virion deposits it in the endosome, but virus uncoating and delivery of the viral genome into the cytoplasm requires acidification of the interior of the virion (44). Virion acidification is mediated by the proton channel activity of the M2 tetramer, which conducts protons from the endosomal lumen into the virion interior (135). The M2 proton channel is not constitutively open, but it requires low pH for maximal activity (92). Detailed genetic, biochemical, and biophysical studies provided insight into the molecular basis of these activities. The transmembrane domain of M2 mediates tetramerization and is sufficient for proton channel activity (25, 57, 65). Mutational analysis and biochemical studies identified the crucial amino acids that mediate M2 channel activity, including two polar amino acids on a single turn of the transmembrane α-helix, histidine 37 and tryptophan 41 (91). Replacing the histidine with any of several amino acids allows M2 to transport Na+ and K+ as well as H+, showing that histidine confers ion selectivity (129, 131). The tryptophan appears to play an important role in activating the channel in response to low pH, by participating in a pH-dependent conformation change. Molecular modeling and structural studies showed that the essential histidine and tryptophan side chains lie in the central pore of the M2 tetramer (Figure 1) (1, 7, 50, 106, 134, 135) and suggest that the M2 tetramer is not a static conduit of protons but rather adopts a series of conformational states during ion passage (49, 62, 113, 135). Influenza B virus M2 (BM2) also forms a proton channel (134). Although the sequence of BM2 diverges greatly from that of A/M2, it contains the essential histidine and tryptophan residues noted above.
A/M2 is the target of the antiviral medicines amantadine and rimantadine, which block virus uncoating by inhibiting the proton channel activity of M2 (94). Mutations in M2 can cause amantadine resistance (41), and peptide reconstitution and mutant expression experiments demonstrated that the transmembrane domain was sufficient to reconstitute amantadine-sensitive proton channels in lipid bilayers and cells (25, 65). More recent studies demonstrated that amantadine binds inside the aqueous pore of the M2 tetramer and inhibits proton channel activity by physically obstructing ion flow (Figure 1) (15, 52). The clinical utility of these medicines is limited by the high prevalence of drug-resistant virus strains (119). Thus, more complete understanding of the structure-function relationships of the influenza virus M2 proteins is likely to lead to the rational design of improved therapeutics (8, 133).
The cytoplasmic carboxy terminus tail of A/M2 serves another interesting function. The last step in the release of enveloped viruses is membrane scission, whereby the extruded nascent virion at the plasma membrane is pinched off to separate from the cell. In several virus families, membrane scission is mediated by a cellular protein complex called endosome sorting complex required for transport (ESCRT) (130), but influenza virus release is ESCRT independent (13, 18). Instead, an amphipathic helix in the membrane-proximal segment of the cytoplasmic domain of M2 inserts into the plasma membrane at the neck of the budding virion and alters membrane curvature, leading to membrane scission and virion release (Figure 2a) (99). Viral mutants lacking this segment (or lacking M2 entirely) display a striking beads-on-a-string budding phenotype due to incomplete scission (Figure 2b) (73, 97) and are attenuated in vivo (138). Thus, the transmembrane domain of M2 not only forms the proton channel required for virus entry, but also targets the hydrophilic carboxy terminus of M2 to the cell membrane, where it can participate in virus release.
Figure 2.
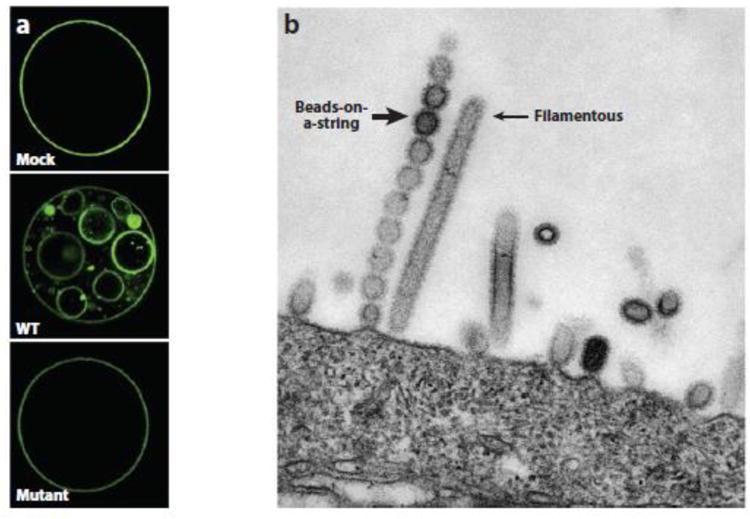
Role of M2 in membrane scission. (a) In an in vitro model of membrane scission, giant unilamellar vesicles containing a low level of cholesterol were mock treated (top) or incubated with wild-type (WT, middle) or mutant (bottom) amphipathic helix peptide from M2. Note the appearance of many pinched-off, small vesicles in the sample treated with the wild-type peptide. Reprinted from Reference 99, copyright 2010, with permission from Elsevier. (b) MDCK-M2Stop70 cells were infected with an influenza A virus mutant lacking the M2 gene. After 12 hours, cells were imaged by transmission electron microscopy. Elongated filamentous budding virion structure is indicated by a thin arrow; beads-on-a-string budding virion structure is indicated by a thick arrow. Host cell is at the bottom. Adapted from Reference 73 with permission from the American Society for Microbiology.
Human Immunodeficiency Virus Vpu
In the cell membrane,
HIV needs Vpu.
Escape host defense.
HIV-1 causes AIDS. In addition to its major structural genes, HIV-1 encodes accessory proteins that assist in virus replication or counter host immune defenses. One of these proteins is Vpu, an approximately 80-amino-acid, multifunctional, single-span transmembrane phosphoprotein that resides primarily in intracellular membranes (116, 123). Notably, Vpu causes downregulation of the immune protein (and HIV receptor) CD4 and antagonizes the antiviral protein known as tetherin or Bst2. Vpu acts as an adaptor to link newly synthesized CD4 in the endoplasmic reticulum (ER) to the βTrCP-1 and βTrCP-2 subunits of the cytosolic SCF E3 ubiquitin ligase complex, resulting in polyubiquitination and proteasomal degradation of CD4 (14, 32, 69, 107). The p97 ATPase plays a role in the extraction of CD4 from the ER membrane prior to degradation (11, 68). In addition, Vpu may cause the ER retention of CD4 as another mechanism to ensure low cell-surface levels of CD4 (68). Although the carboxy-terminal domain of Vpu seems the primary determinant of CD4 downregulation (23), the transmembrane domain may also play a role, possibly by affecting Vpu oligomerization (67).
The mechanism by which Vpu antagonizes tetherin is incompletely understood. Tetherin is an integral membrane protein that inhibits the production of infectious HIV and many other enveloped viruses, an activity that is blocked by Vpu (Figure 3a). Electron microscopy showed that tetherin prevents release of budded virions from the surface of the host cell (Figure 3b). (56, 79, 84, 128). Direct interactions between the transmembrane domains of Vpu and tetherin appear to be required for tetherin inhibition (5, 24, 51, 74, 109). Vpu downregulates cell-surface expression of tetherin (40, 79, 128), but there is considerable debate (summarized in References 6 and 23) whether Vpu prevents newly synthesized tetherin from reaching the cell surface, inhibits recycling of internalized tetherin to the cell surface, or directly removes tetherin from the plasma membrane. Similarly, the relative importance of Vpu-mediated degradation versus tetherin redistribution is unclear, and the intracellular site of Vpu-induced degradation is the subject of debate.
Figure 3.
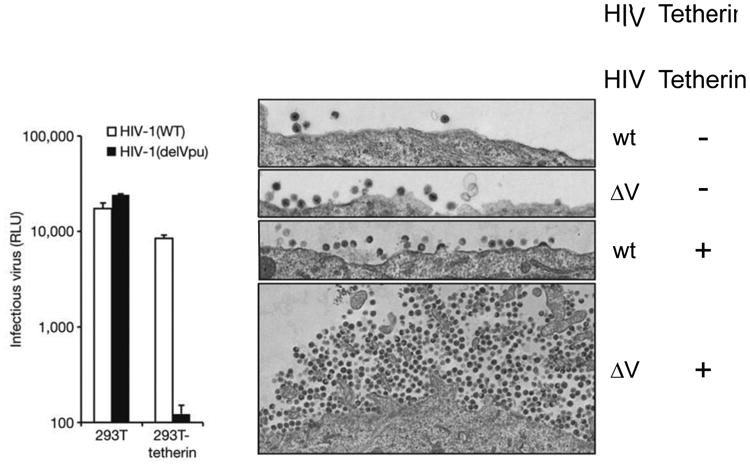
Vpu antagonizes the antiviral effect of tetherin. (a) Unmodified 293T cells or cells expressing tetherin were infected with wild-type HIV or HIV with a deletion in Vpu (delVpu), and after a single-cycle infection, virus production was assessed by infecting HeLa-TZMbl indicator cells with the medium and measuring expression of the integrated HIV-responsive luciferase gene. (b) Unmodified HT1080 cells or cells expressing exogenous tetherin were infected with wild-type HIV or delVpu (ΔV) and examined two days later by transmission electron microscopy. Note the large number of delVpu virions associated with the surface of cells expressing tetherin. Abbreviations: RLU, relative light units; wt, wild type. Reprinted from Reference 79 with permission from Macmillan Publishers, Ltd.
The transmembrane domain of Vpu also oligomerizes to generate an ion channel (108), but viroporin activity does not appear to be essential for its effects on CD4 expression or virus release (12). The three-dimensional structure of the Vpu transmembrane helix has been solved by nuclear magnetic resonance (NMR) spectrometry, but the oligomeric state of the protein in its active form is not known with certainty (83).
Fibropapillomavirus E5 Proteins
The E5 proteins
Regulate cell receptors.
Transmembrane ligands.
Another well-studied miniprotein is the bovine papillomavirus (BPV) E5 protein. The papillomaviruses are small DNA viruses that infect humans and a wide variety of animals, with human papillomaviruses (HPV) being responsible for ∼5% of all human cancers. BPV type 1 and related fibropapillomaviruses from other ungulate species (e.g., pigs and sheep) encode a small, hydrophobic E5 protein and cause skin warts or papillomas with a prominent fibroblastic component (in contrast to the HPV, which induce purely epithelial papillomas) (20, 103a). Only 43 or 44 amino acids long, all fibropapillomavirus E5 proteins share an extremely hydrophobic central portion that serves as a transmembrane domain, a glutamine at position 17, an aspartic acid or glutamic acid at position 33, and two cysteines near the carboxy terminus that stabilize homodimer formation between two E5 monomers (21). Although their hydrophobic nature is a defining feature of the E5 proteins, the sequence of hydrophobic amino acids is quite divergent among different E5 proteins. Because ∼22 amino acids are required to span the membrane and the transmembrane domain of E5 is thought to be in the middle of the molecule, only about 10 amino acids from each end of E5 are likely to protrude from either surface of the membrane. Because E5 thus lacks a large soluble domain, it can be regarded in essence as a free-standing transmembrane domain (140a) (Figure 4).
Figure 4.
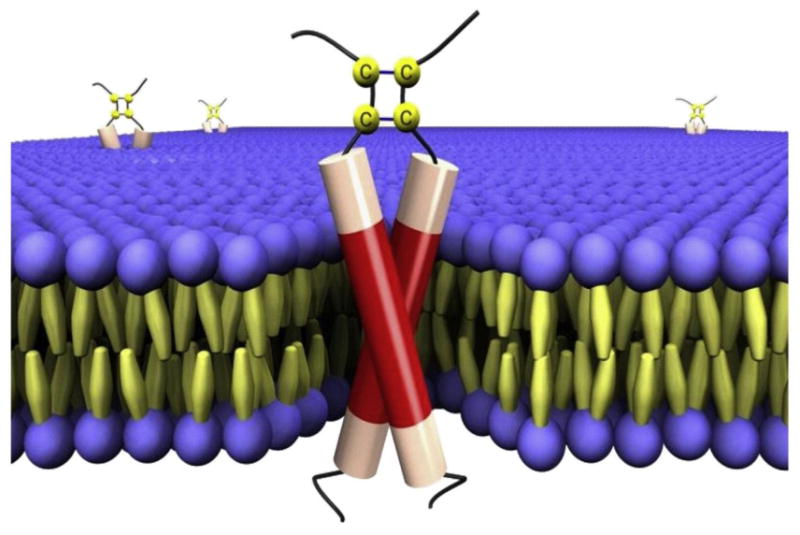
BPV1 E5 protein. A schematic representation of BPV1 E5 dimers embedded in a lipid bilayer. The transmembrane domain of the E5 protein is shown in red, with the disulfide-bonded carboxy terminus at the top (in the extracellular/luminal space). The membrane phospholipids are represented as spheres and the fatty acids as spindles. Reprinted from Reference 140a, copyright 2010, with permission from Elsevier.
The 44-amino-acid BPV E5 protein is the major oncogene product of BPV, inducing stable transformation of cultured fibroblasts, and is thought to be responsible for the fibrotropism of this virus in vivo (20, 21). Extensive genetic and biochemical studies demonstrated that the E5 protein stably transforms cells by causing sustained activation of the cellular platelet-derived growth factor (PDGF) β receptor (82, 87). This receptor is normally activated by PDGF binding, which results in dimerization of the receptor, transautophosphorylation of the intracellular domain of the receptor at specific tyrosine residues, and initiation of intracellular signaling cascades. Unlike PDGF, which binds to the extracellular domain of the PDGF β receptor, the E5 dimer acts by forming a complex with the transmembrane domain of two molecules of the receptor, resulting in receptor dimerization and activation (Figure 5) (22, 37, 58, 85, 86, 111). Indeed, the ligand-binding domain of the receptor is not required for E5 action, demonstrating that E5 activates the receptor in a ligand-independent fashion. Thus the normal ligand and the E5 protein use different biochemical mechanisms to activate the same receptor.
Figure 5.
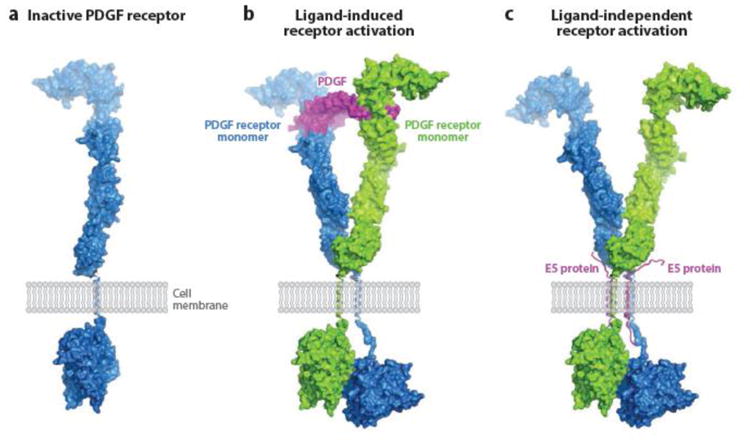
Model for E5-induced PDGF receptor activation. (a) Surface representation of monomeric inactive PDGF β receptor, showing the extracellular ligand-binding domain, the single transmembrane domain (shown as a single helical ribbon), and the intracellular kinase domain in an inactive, closed conformation. (b) PDGF receptor activation induced by PDGF binding. Pre formed PDGF dimer (center) binds to the extracellular domains of two PDGF receptor monomers and thereby facilitates dimerization via tight contacts, which allow one of the intracellular kinase domains to maintain an active, open conformation, ready to catalyze transphosphorylation. (c) PDGF receptor activation induced by BPV E5 binding. Instead of imposing PDGF receptor dimerization through ligand binding as seen in (b), BPV E5 (two transmembrane helixes) promotes receptor dimerization through direct contacts with the receptor transmembrane segments. The following structures were used to generate these models: crystal structure of PDGF-BB in complex with the ligand-binding domains of PDGF β receptor (108a), crystal structures of the ectodomain of c-Kit before and after SCF binding (140c), and 3-D cryoEM reconstruction of the entire c-Kit transmembrane receptor in complex with SCF (82a). Abbreviations: PDGF, platelet-derived growth factor; SCF, stem cell factor. Figure prepared by Yarden Opatowsky (Bar-Ilan University) and adapted from Reference 108 (copyright 2010 National Academy of Sciences, USA) and Reference 140c (copyright 2007 Elsevier), with permission.
The isolation of compensatory mutations that allow a mutant E5 protein to bind and activate a PDGF β receptor transmembrane mutant unable to recognize the wild-type viral protein demonstrated that E5 and the PDGF β receptor transmembrane domain contact one another directly (26), and models of the complex between the E5 protein and the PDGF β receptor have been proposed based on extensive mutational analysis and molecular modeling (2, 53, 118). This analysis also demonstrated that the cysteines are required for E5 homodimerization and can be replaced with a heterologous dimerization domain that forces the E5 monomers into the correct rotational orientation within the dimer (48, 72). Glutamine 17 and aspartic acid 33 of E5 are thought to make a hydrogen bond with an essential threonine in the transmembrane domain of the receptor and a salt bridge with an essential lysine in the extracellular juxtamembrane domain of the receptor, respectively (54, 55, 76).
The interaction between the E5 protein and the PDGF β receptor is highly specific. The PDGF β receptor appears to be the only receptor tyrosine kinase activated by the E5 protein in transformed fibroblasts, the interaction is disrupted by conservative mutations in the PDGF β receptor transmembrane domain, and the E5 protein cannot transform cells by interacting with the closely related PDGF α receptor (39, 77, 78, 86--88, 111). The E5 protein also engages the transmembrane subunit of the vacuolar ATPase (38), but the biochemical basis for this interaction and its biological consequences are unclear (21).
These experiments demonstrated that small proteins unrelated to the normal ligand can activate receptor tyrosine kinases. They also demonstrated that a freestanding transmembrane domain not linked to a large soluble domain is sufficient for biological activity and that it acts by binding specifically to the transmembrane domain of a much larger cellular protein and modulating its activity. Similarly, the p12 transmembrane protein of HTLV-1 and its 66-amino-acid p8 cleavage product bind and modulate cellular transmembrane proteins that regulate several aspects of T lymphocyte function (27).
Some HPV also express hydrophobic E5 proteins, which play a role in virus replication and have weak transforming activity (28, 34, 59, 61). The 83-amino-acid HPV16 E5 protein modulates the activity of the epidermal growth factor receptor by enhancing or prolonging ligand-induced receptor activation, possibly by affecting receptor trafficking (21, 35, 90, 114, 115). Recent studies suggest that the HPV16 E5 protein may hexamerize to form an ion channel required for its mitogenic activity (139). Thus, HPV16 E5 may be a viroporin.
Poxviruses are A Rich Source of Small Membrane Proteins
The poxviruses
Are very large and complex.
Lots of small proteins.
Poxviruses are large, enveloped, double-stranded DNA viruses that include smallpox virus, which is thought to have caused more human deaths than any other infectious disease in history prior to its eradication. Poxviruses replicate in the cytoplasm, where they generate a variety of membrane structures (19) (Figure 6). For vaccinia virus, the prototype mammalian poxvirus, numerous mutants interfere with the biogenesis or function of viral membranes. Studies of these mutants identified viral genes required for viral membrane formation, including those encoding some of the smallest known naturally occurring transmembrane proteins.
Figure 6.
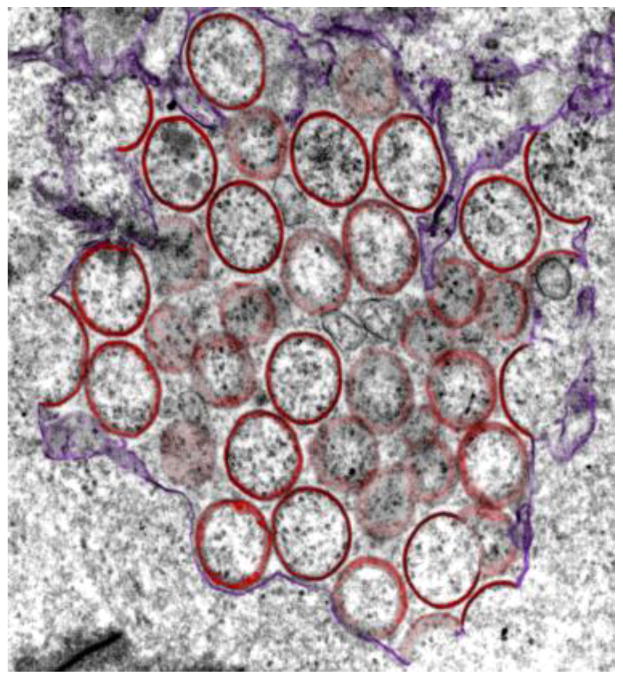
Poxvirus membrane formation. Numerous viral transmembrane miniproteins are required for proper poxvirus membrane biogenesis, infectivity, and/or virulence. This transmission electron micrograph shows incomplete viral membranes (pseudocolored red) forming from endoplasmic reticulum (ER) membranes (purple) in cells infected with vaccinia virus lacking the A30.5 miniprotein. Adapted from Reference 71 with permission from the American Society for Microbiology.
The initial annotation of the vaccinia virus genome had a threshold of 65 amino acids for the identification of viral open reading frames (ORFs), resulting in the identification of ∼200 viral genes (36). However, this analysis overlooked several small genes, including the O3L gene, which encodes a 35-amino-acid membrane-associated virion protein with a single transmembrane domain (101, 102). O3L mutants display a marked defect in infectivity and plaque formation because of impaired virus entry. O3L is nearly absolutely conserved in all orthopoxviruses, and a small ORF with the potential to encode a similarly sized hydrophobic protein is located in the same genomic position in more distantly related poxviruses. Even though these distantly related proteins share little sequence similarity, O3L from divergent poxviruses can replace the vaccinia virus version in transcomplementation experiments and in recombinant viral genomes, demonstrating their functional equivalence (102). Analysis of chimeric O3L proteins and truncation mutants revealed that the transmembrane domain of O3L was sufficient for stable complex formation with other viral proteins involved in poxvirus entry and for the restoration of infectivity of O3L mutants (100, 102). Despite its importance, the transmembrane domain of O3L can tolerate many mutations without loss of activity, implying that it has flexible sequence requirements (100). The I2L protein is another short transmembrane vaccinia virion protein required for virus entry (80).
Additional small transmembrane proteins are involved in vaccinia virus biogenesis. The 87-amino-acid L2R virion membrane protein has two putative membrane-spanning domains, which may insert into membranes as a helix-turn-helix hairpin. The absence of L2R impairs proteolytic processing of other virion proteins and results in abnormal production of crescent membranes during the formation of virions (70). L2R binds to another small transmembrane virion protein required for virion biogenesis, the 42-amino-acid A30.5 protein, which resides in the ER membrane (Figure 6) (71). There is an ORF with the potential to encode a small hydrophobic protein at the same genomic position as A30.5 in all vertebrate poxviruses, although the amino acid sequences vary. Three additional membrane proteins required for proper virion biogenesis and maturation are A14L, its binding partner A17L, and A13L (75, 98, 124, 125, 127). A14L is a dimeric 90-amino-acid glycosylated virion phosphoprotein with two well-conserved transmembrane domains (75), and A13L is a monomeric 70-amino-acid virion membrane phosphoprotein with a single predicted transmembrane domain required for virion maturation (127). The 79-amino-acid I5L protein also contains two putative transmembrane domains and is exposed on the surface of the intact virion. Although it is dispensable for virus growth in cultured cells, I5L enhances virus replication and virulence (110, 126). Similarly, the A14.5L gene encodes a 53-amino-acid transmembrane protein dispensable for replication in cell culture but required for virulence in mice (10). It is not clear why poxviruses devote so many different miniproteins to virion production, but this strategy undoubtedly provides opportunities for regulation of this complex process.
Paramyxovirus SH Proteins
The SH protein.
Short hydrophobic protein.
The name says it all.
The paramyxoviruses are enveloped RNA viruses, which include important human pathogens such as mumps virus and respiratory syncytial virus. Many paramyxoviruses encode a short hydrophobic (SH) integral membrane virion protein between 44 and 60 amino acids long. These proteins display little amino acid sequence homology, but they are encoded in the same region of the viral genomes, suggesting they serve similar functions. SH proteins are not required for virus replication in cultured cells (Figure 7), but viruses lacking the SH gene are attenuated in vivo (42, 121, 140). Notably, the SH proteins of respiratory syncytial virus and parainfluenza virus 5 are interchangeable in vivo, despite marked sequence differences (31). Similarly, the mumps virus SH protein can functionally complement the simian virus 5 SH deletion mutant (Figure 7) (140). This latter finding is particularly striking because mumps virus SH is a type I transmembrane protein, i.e., with its carboxy-terminus in the cytoplasm, whereas SV5 SH displays the opposite orientation. Clearly, the overall hydrophobic composition rather than the precise sequence of SH was selected, perhaps in combination with a few key residues.
Figure 7.
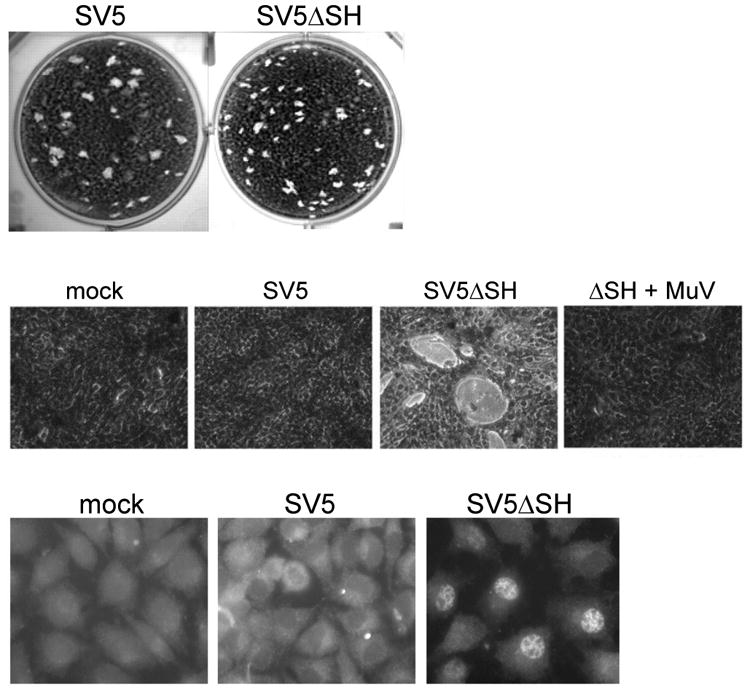
Effects on SH proteins on virus replication and apoptosis.(a) Permissive BSR-T7 cells were infected with wild-type recombinant simian virus 5 (SV5) or an SV5 mutant lacking the SH gene (SV5ΔSH), and plaque formation was assessed. (b) MBDK cells were mock infected, infected with wild-type SV5, infected with SV5ΔSH, or coinfected with SV5ΔSH and wild-type mumps virus (MuV), which express MuV SH protein (ΔSH + MuV). Photomicrographs were taken five days later. Note the marked cytopathic effect in cells infected with SV5ΔSH but not in the coinfected cells. (c) L929 cells were mock infected or infected with wild-type SV5 or SV5ΔSH. One day later, localization of NF-κB p65 was determined by immunofluorescence in permeabilized cells. Note nuclear translocation of NF-κB in cells infected with SV5ΔSH. Adapted from Reference 140 with permission from the American Society for Microbiology.
Paramyxoviruses lacking the SH protein display enhanced cytopathic effect and induction of apoptosis (Figure 7), suggesting that a major physiologic role of the SH protein is the inhibition of apoptosis induced by viral replication (31, 43, 64, 140). The premature cell death induced by these mutants presumably reduces virus yield and contributes to virus attenuation. Human metapneumoviruses lacking the SH protein induce enhanced secretion of proinflammatory mediators in the airway of infected mice as a consequence of increased activation of the transcription factor NF-κB (9). Further studies implied that the SH proteins of divergent paramyxovirus types block tumor necrosis factor-α-mediated activation of NF-κB (Figure 7) (63, 140). The direct molecular targets of SH proteins remain to be identified, and perhaps the miniproteins themselves can be used as affinity reagents to isolate cell proteins that regulate apoptosis.
Recent structural studies indicate that the single transmembrane domain of respiratory syncytial virus SH protein homooligomerizes to form a pentameric ion channel in detergent micelles and lipid bilayers (33). Moreover, human cells expressing SH showed pH-dependent ion channel activity, but it is not clear if this biochemical property is responsible for the activities in cells and animals described above.
Artificial Small Transmembrane Proteins Modeled on Viral Miniproteins
Traptamer proteins
Can alter cell behavior.
Transmembrane Ninjas!
The studies summarized above demonstrate that viral miniproteins carry out diverse functions that support viral replication, some by forming complexes with native cellular transmembrane proteins. We reasoned that in a large collection of artificial proteins composed of random hydrophobic sequences, many of them would insert into the membrane and adopt a stable α-helical conformation. Although the vast majority of such proteins are presumably inert, by chance some of them would bind to the transmembrane domain of a cellular protein and display biological activity. To test this idea, we constructed retroviral libraries expressing hundreds of thousands of small proteins containing totally randomized hydrophobic segments, expressed these proteins in cells, and imposed biological selection to identify the rare proteins in the libraries that displayed a desired activity. Genes encoding active proteins were then recovered from genomic DNA from the selected cells and characterized. We named these artificial transmembrane proteins traptamers, for transmembrane aptamers.
Using this approach, we isolated traptamers that activate the PDGF β receptor and transform cells (17, 29, 30, 95, 122), others that activate the erythropoietin (EPO) receptor and induce erythroid differentiation (16), and still others that downregulate expression of the HIV coreceptor CCR5 and inhibit HIV infection (103). The transmembrane domain sequences of these proteins are different and not homologous to known proteins (Figure 8). Some active traptamers are only 29 amino acids long, contain no sequences whatsoever from naturally occurring proteins, and have markedly reduced chemical complexity compared with natural proteins (17). These studies revealed that the sequence of the transmembrane domain is sufficient to determine the identity of its target. Furthermore, traptamers can exert a variety of effects on both single-pass and multipass transmembrane proteins. These interactions are specific: Traptamers that activate the PDGF β receptor do not activate the EPO receptor and vice versa, traptamers that downregulate CCR5 do not downregulate CXCR4, and a traptamer selected to activate the human EPO receptor does not activate the mouse EPO receptor. Finally, multiple traptamers with diverse sequences can productively interact with the same target transmembrane domain (Figure 8), but traptamers that recognize the same target can engage the target in slightly different ways (89, 95).
Figure 8.

Divergent sequences of traptamers. The sequences of the wild-type BPV1 E5 protein and the indicated traptamers are shown, together with their cellular targets. The randomized segment of each traptamer is shown in blue. In TC2-3, the transmembrane domain of E5 was replaced with a randomized transmembrane domain. TM36-4 contains an N-terminal poly-His tag, and BY1PC2 contains an N-terminal HA tag. Note the divergent sequences of the traptamers that recognize the same target. Abbreviations: EPO, erythropoietin; PDGF, platelet-derived growth factor.
Naturally occurring proteins and traptamers are subject to different evolutionary pressures. For example, viral proteins surviving natural selection are constrained by the evolutionary demands of virus replication and were thus presumably selected to recognize their targets within a certain range of affinities, possibly to recognize more than one target, or to exert a certain influence on their targets. In contrast, a traptamer, which can be selected for a single activity, can concentrate on that particular activity: bind its target with highest affinity, bind a single target, activate a single signaling pathway. Thus, traptamers can be specialists, unlike natural proteins, which may need to serve many masters.
Because of the intimate association of viruses with their hosts, viruses are powerful probes to identify and characterize important cellular regulatory nodes, thereby providing fundamental insights into numerous cellular processes. Traptamers can also be used to dissect cellular pathways. However, unlike intrinsic viral genes, which sample only those activities that benefit the virus, traptamers can be selected to perturb a wide variety of cellular processes. Traptamers are thus analogous to cellular proto-oncogenes transduced by acutely transforming retroviruses. Even though these oncogenes do not contribute to virus replication in most cases, study of them has illuminated many aspects of biology. The genes acquired by retroviruses in nature are restricted to the existing repertoire of cellular genes, and selection is restricted to tumor formation. In principle, the use of artificial proteins and different selection schemes eliminate both of these limitations and will result in the isolation of proteins to probe many aspects of cell behavior.
Conclusions and Implications
Natural and artificial short transmembrane proteins display numerous biochemical and biological activities. Most viral miniproteins were discovered by viral genome sequencing. Proteomic analysis of infected cells or purified virions may be useful in future discovery or validation efforts. Ribosome profiling, which has identified novel translated ORFs in cytomegalovirus, will also be useful for identifying new miniproteins (45, 112).
A striking feature of viral miniproteins is the relatively flexible sequence requirements in the transmembrane domain. This flexibility is revealed by the ability of these proteins to tolerate many hydrophobic substitution mutations and by the finding that related viruses often encode sequence-divergent but interchangeable miniproteins. For example, influenza virus A/M2 and BM2 proteins share little sequence similarity, yet both act as proton channels, and SH proteins with opposite transmembrane orientations can replace one another. In some cases, a few key conserved residues are embedded in a hydrophobic context whose sequence can drift, as illustrated by the histidine and tryptophan in M2 and the essential hydrophilic residues in fibropapillomavirus E5 proteins. Indeed, an inactive traptamer with a monotonous polyleucine transmembrane domain can be converted into an oncoprotein by introducing two specific amino acids into the polyleucine segment (95). This sequence flexibility suggests that sequence conservation is not the best criterion to assess the likelihood that miniproteins carry out similar functions.
Another striking property of these miniproteins is their high specificity. Transmembrane domains are not inert protein segments whose sole biological role is to anchor a protein in a membrane and separate its extracellular and intracellular domains so they can exist on opposite sides of the membrane. Rather, transmembrane domains can exert specific effects on cells. This is perhaps seen most clearly with the BPV E5 protein, a freestanding transmembrane domain that activates a particular cellular receptor. Such specificity could arise because natural selection weeded out nonspecific activities of the miniprotein that interfered with virus replication. However, the isolation of traptamers that are specific for the selected target and that ignore closely related proteins not encountered during in vitro selection suggests that small transmembrane proteins are intrinsically specific. Nevertheless, it is possible that some miniproteins recognize multiple targets or exert off-target effects.
Influenza A virus M2 is an antiviral drug target, and other viral miniproteins may provide novel therapeutic opportunities. Potential targets include the SH proteins of pathogenic paramyxoviruses, numerous miniproteins required for poxvirus replication, HIV Vpu, and small transmembrane proteins such as hepatitis C virus p7 processed from larger precursor proteins. Proteins residing in virus or cell membranes may be particularly accessible to hydrophobic molecules acting at the cell surface. The size and relatively simple structure of these proteins should facilitate rational drug design, and screening for molecules that inhibit the ion channel activity of viroporins may be a useful surrogate assay to identify compounds with antiviral activity (132).
Because up to 30% of cell proteins and many viral proteins span membranes (60), it may be possible to build small transmembrane proteins that regulate many cellular and viral processes. Artificial miniproteins may have activities that never arose during evolution or were lost during natural selection. Can traptamers instigate a constellation of signaling pathways that is distinct from that initiated by natural ligands? Traptamers have been isolated that inhibit the PDGF β receptor by altering which tyrosine residues are phosphorylated in response to PDGF, demonstrating that traptamers can indeed fine-tune receptor activity (89). Can traptamers be selected that form membrane channels with novel specificity or endow existing transmembrane proteins with activities that cells never found useful?
Given the versatility of transmembrane miniproteins, their repeated emergence during virus evolution, and the relative ease of constructing similar proteins in the laboratory, it seems likely that cells also adopted this biochemical strategy. In fact, bacteria make wide use of small regulatory transmembrane proteins such as the 30-amino-acid MgtR protein of Salmonella enterica, which binds to the inner membrane protein MgtC and stimulates its degradation (3, 4, 28a, 46). We predict that many similar proteins will be discovered in various cells by using the approaches pioneered to identify viral miniproteins. Indeed, increasingly sophisticated bioinformatics and biochemical approaches are being used to identify short, expressed ORFs in various organisms, but the biological activity of the vast majority of them has not been assessed (e.g., 28a, 140b). It is also possible that somatic mutations may confer oncogenic activity on small transmembrane proteins and thus contribute to the development of cancer in humans. Thus, as has frequently been the case, studies of viruses are providing unexpected insights into how proteins work and how cells function.
Summary Points.
Many viruses encode small transmembrane proteins, some shorter than 50 amino acids.
Viral miniproteins regulate many aspects of viral replication and cell physiology.
Viroporins, one class of miniproteins, oligomerize and form ion channels.
The BPV E5 oncoprotein and other miniproteins bind to viral or cellular transmembrane proteins and regulate their activity.
Artificial small transmembrane proteins that display a variety of biological activities can be constructed and selected.
Cells are likely to express similar proteins that have eluded detection.
Acknowledgments
I apologize to my colleagues whose work could not be cited here because of space limitations. I thank Yarden Opatowsky for preparing Figure 5 and Jan Zulkeski for assistance in preparing this manuscript. Work in the author's laboratory on small transmembrane proteins is supported by a grant from the National Cancer Institute (R01 CA037157) and by generous gifts from Mrs. Laurel Schwartz.
Footnotes
Disclosure Statement: The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Acharya R, Carnevale V, Fiorin G, Levine BG, Polishchuk AL, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc Natl Acad Sci USA. 2010;107:15075–80. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adduci AJ, Schlegel R. The transmembrane domain of the E5 oncoprotein contains functionally discrete helical faces. J Biol Chem. 1999;274:10249–58. doi: 10.1074/jbc.274.15.10249. [DOI] [PubMed] [Google Scholar]
- 3.Alix E, Blanc-Potard AB. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008;27:546–57. doi: 10.1038/sj.emboj.7601983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alix E, Blanc-Potard AB. Hydrophobic peptides: novel regulators within bacterial membrane. Mol Microbiol. 2009;72:5–11. doi: 10.1111/j.1365-2958.2009.06626.x. [DOI] [PubMed] [Google Scholar]
- 5.Arias JF, Iwabu Y, Tokunaga K. Structural basis for the antiviral activity of BST-2/tetherin and its viral antagonism. Front Microbiol. 2011;2:250. doi: 10.3389/fmicb.2011.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arias JF, Iwabu Y, Tokunaga K. Sites of action of HIV-1 Vpu in BST-2/tetherin downregulation. Curr HIV Res. 2012;10:283–91. doi: 10.2174/157016212800792423. [DOI] [PubMed] [Google Scholar]
- 7.Balannik V, Carnevale V, Fiorin G, Levine BG, Lamb RA, et al. Functional studies and modeling of pore-lining residue mutants of the influenza A virus M2 ion channel. Biochemistry. 2010;49:696–708. doi: 10.1021/bi901799k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balannik V, Wang J, Ohigashi Y, Jing X, Magavern E, et al. Design and pharmacological characterization of inhibitors of amantadine-resistant mutants of the M2 ion channel of influenza A virus. Biochemistry. 2009;48:11872–82. doi: 10.1021/bi9014488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao X, Kolli D, Liu T, Shan Y, Garofalo RP, Casola A. Human metapneumovirus small hydrophobic protein inhibits NF-κB transcriptional activity. J Virol. 2008;82:8224–29. doi: 10.1128/JVI.02584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betakova T, Wolffe EJ, Moss B. The vaccinia virus A14.5L gene encodes a hydrophobic 53-amino-acid virion membrane protein that enhances virulence in mice and is conserved among vertebrate poxviruses. J Virol. 2000;74:4085–92. doi: 10.1128/jvi.74.9.4085-4092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binette J, Dube M, Mercier J, Halawani D, Latterich M, Cohen EA. Requirements for the selective degradation of CD4 receptor molecules by the human immunodeficiency virus type 1 Vpu protein in the endoplasmic reticulum. Retrovirology. 2007;4:75. doi: 10.1186/1742-4690-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolduan S, Votteler J, Lodermeyer V, Greiner T, Koppensteiner H, et al. Ion channel activity of HIV-1 Vpu is dispensable for counteraction of CD317. Virology. 2011;416:75–85. doi: 10.1016/j.virol.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, et al. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–78. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Butticaz C, Michielin O, Wyniger J, Telenti A, Rothenberger S. Silencing of both β-TrCP1 and HOS (β-TrCP2) is required to suppress human immunodeficiency virus type 1 Vpu-mediated CD4 down-modulation. J Virol. 2007;81:1502–5. doi: 10.1128/JVI.01711-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–92. doi: 10.1038/nature08722. Amantadine binds inside the influenza A virus M2 channel, inhibiting proton transport and blocking infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cammett TJ, Jun SJ, Cohen EB, Barrera FN, Engelman DM, DiMaio D. Construction and genetic selection of small transmembrane proteins that activate the human erythropoietin receptor. Proc Natl Acad Sci USA. 2010;107:3447–52. doi: 10.1073/pnas.0915057107. The transmembrane domain of a traptamer targets it to a novel target, the erythropoietin receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chacón KM, Petti LM, Scheideman EH, Pirazzoli V, Politi K, DiMaio D. De novo selection of oncogenes. Proc Natl Acad Sci USA. 2014;111:E6–14. doi: 10.1073/pnas.1315298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372:221–32. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chichón FJ, Rodríguez MJ, Risco C, Fraile-Ramos A, Fernández JJ, et al. Membrane remodelling during vaccinia virus morphogenesis. Biol Cell. 2009;101:401–14. doi: 10.1042/BC20080176. [DOI] [PubMed] [Google Scholar]
- 20.Corteggio A, Altamura G, Roperto F, Borzacchiello G. Bovine papillomavirus E5 and E7 oncoproteins in naturally occurring tumors: Are two better than one? Infect Agent Cancer. 2013;8:1. doi: 10.1186/1750-9378-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiMaio D, Petti LM. The E5 proteins. Virology. 2013;445:99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummond-Barbosa DA, Vaillancourt RR, Kazlauskas A, DiMaio D. Ligand-independent activation of the platelet-derived growth factor β receptor: requirements for bovine papillomavirus E5-induced mitogenic signaling. Mol Cell Biol. 1995;15:2570–81. doi: 10.1128/mcb.15.5.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dube M, Bego MG, Paquay C, Cohen EA. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology. 2010;7:114. doi: 10.1186/1742-4690-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dube M, Paquay C, Roy BB, Bego MG, Mercier J, Cohen EA. HIV-1 Vpu antagonizes BST-2 by interfering mainly with the trafficking of newly synthesized BST-2 to the cell surface. Traffic. 2011;12:1714–29. doi: 10.1111/j.1600-0854.2011.01277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duff KC, Ashley RH. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992;190:485–89. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 26.Edwards AP, Xie Y, Bowers L, DiMaio D. Compensatory mutants of the bovine papillomavirus E5 protein and the platelet-derived growth factor β receptor reveal a complex direct transmembrane interaction. J Virol. 2013;87:10936–45. doi: 10.1128/JVI.01475-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards D, Fenizia C, Gold H, Fernanda de Castro-Amarante M, Buchmann C, et al. Orf-I and Orf-II-encoded proteins in HTLV-1 infection and persistence. Viruses. 2011;3:861–85. doi: 10.3390/v3060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fehrmann F, Klumpp DJ, Laimins LA. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J Virol. 2003;77:2819–31. doi: 10.1128/JVI.77.5.2819-2831.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Fontaine F, Fuchs RT, Storz G. Membrane localization of small proteins in Escherichia coli. J Biol Chem. 2011;86:32464–74. doi: 10.1074/jbc.M111.245696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeman-Cook L, Dixon AM, Frank JB, Xia Y, Ely L, et al. Selection and characterization of small random transmembrane proteins that bind and activate the platelet-derived growth factor β receptor. J Mol Biol. 2004;338:907–20. doi: 10.1016/j.jmb.2004.03.044. [DOI] [PubMed] [Google Scholar]
- 30.Freeman-Cook LL, Edwards APB, Dixon AM, Yates KE, Ely L, et al. Specific locations of hydrophilic amino acids in constructed transmembrane ligands of the platelet-derived growth factor β receptor. J Mol Biol. 2005;345:907–21. doi: 10.1016/j.jmb.2004.10.072. [DOI] [PubMed] [Google Scholar]
- 31.Fuentes S, Tran KC, Luthra P, Teng MN, He B. Function of the respiratory syncytial virus small hydrophobic protein. J Virol. 2007;81:8361–66. doi: 10.1128/JVI.02717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J Gen Virol. 1997;78(Pt. 3):619–25. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- 33.Gan SW, Tan E, Lin X, Yu D, Wang J, et al. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J Biol Chem. 2012;287:24671–89. doi: 10.1074/jbc.M111.332791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genther SM, Sterling S, Duensing S, Munger K, Sattler C, Lambert PF. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J Virol. 2003;77:2832–42. doi: 10.1128/JVI.77.5.2832-2842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genther-Williams SM, Disbrow GL, Schlegel R, Lee D, Threadgill DW, Lambert PF. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Res. 2005;65:6534–42. doi: 10.1158/0008-5472.CAN-05-0083. [DOI] [PubMed] [Google Scholar]
- 36.Goebel SJ, Johnson GP, Perkus ME, Davis SW, Winslow JP, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–66. 517–63. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein DJ, Andresson T, Sparkowski JJ, Schlegel R. The BPV-1 E5 protein, the 16 kDa membrane pore-forming protein and the PDGF receptor exist in a complex that is dependent on hydrophobic transmembrane interactions. EMBO J. 1992;11:4851–59. doi: 10.1002/j.1460-2075.1992.tb05591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein DJ, Finbow ME, Andresson T, McLean P, Smith K, et al. Bovine papillomavirus E5 oncoprotein binds to the 16K component of vacuolar H+-ATPases. Nature. 1991;352:347–49. doi: 10.1038/352347a0. [DOI] [PubMed] [Google Scholar]
- 39.Goldstein DJ, Li W, Wang LM, Heidaran MA, Aaronson SA, et al. The bovine papillomavirus type 1 E5 transforming protein specifically binds and activates the β-type receptor for platelet-derived growth factor but not other tyrosine kinase-containing receptors to induce cellular transformation. J Virol. 1994;68:4432–41. doi: 10.1128/jvi.68.7.4432-4441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Habermann A, Krijnse-Locker J, Oberwinkler H, Eckhardt M, Homann S, et al. CD317/tetherin is enriched in the HIV-1 envelope and downregulated from the plasma membrane upon virus infection. J Virol. 2010;84:4646–58. doi: 10.1128/JVI.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–24. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He B, Leser GP, Paterson RG, Lamb RA. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250:30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- 43.He B, Lin GY, Durbin JE, Durbin RK, Lamb RA. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J Virol. 2001;75:4068–79. doi: 10.1128/JVI.75.9.4068-4079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–78. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 45.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol. 2008;70:1487–501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hobbs EC, Fontaine F, Yin X, Storz G. An expanding universe of small proteins. Curr Opin Microbiol. 2011;14:167–73. doi: 10.1016/j.mib.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 48.Horwitz BH, Burkhardt AL, Schlegel R, DiMaio D. 44-amino-acid E5 transforming protein of bovine papillomavirus requires a hydrophobic core and specific carboxyl-terminal amino acids. Mol Cell Biol. 1988;8:4071–78. doi: 10.1128/mcb.8.10.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu F, Luo W, Cady SD, Hong M. Conformational plasticity of the influenza A M2 transmembrane helix in lipid bilayers under varying pH, drug binding, and membrane thickness. Biochim Biophys Acta. 2011;1808:415–23. doi: 10.1016/j.bbamem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–8. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, et al. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J Biol Chem. 2009;284:35060–72. doi: 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, et al. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2008;105:10967–72. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King G, Oates J, Patel D, van den Berg HA, Dixon AM. Towards a structural understanding of the smallest known oncoprotein: investigation of the bovine papillomavirus E5 protein using solution-state NMR. Biochim Biophys Acta. 2011;1808:1493–501. doi: 10.1016/j.bbamem.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Klein O, Kegler-Ebo D, Su J, Smith S, DiMaio D. The bovine papillomavirus E5 protein requires a juxtamembrane negative charge for activation of the platelet-derived growth factor β receptor and transformation of C127 cells. J Virol. 1999;73:3264–72. doi: 10.1128/jvi.73.4.3264-3272.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein O, Polack GW, Surti T, Kegler-Ebo D, Smith SO, DiMaio D. Role of glutamine 17 of the bovine papillomavirus E5 protein in platelet-derived growth factor β receptor activation and cell transformation. J Virol. 1998;72:8921–32. doi: 10.1128/jvi.72.11.8921-8932.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–29. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kochendoerfer GG, Salom D, Lear JD, Wilk-Orescan R, Kent SB, DeGrado WF. Total chemical synthesis of the integral membrane protein influenza A virus M2: role of its C-terminal domain in tetramer assembly. Biochemistry. 1999;38:11905–13. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 58.Lai CC, Henningson C, DiMaio D. Bovine papillomavirus E5 protein induces oligomerization and trans-phosphorylation of the platelet-derived growth factor β receptor. Proc Natl Acad Sci USA. 1998;95:15241–46. doi: 10.1073/pnas.95.26.15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leechanachai P, Banks L, Moreau F, Matlashewski G. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene. 1992;7:19–25. [PubMed] [Google Scholar]
- 60.Lehnert U, Xia Y, Royce TE, Goh CS, Liu Y, et al. Computational analysis of membrane proteins: genomic occurrence, structure prediction and helix interactions. Q Rev Biophys. 2004;37:121–46. doi: 10.1017/s003358350400397x. [DOI] [PubMed] [Google Scholar]
- 61.Leptak C, Ramon Y, Cajal S, Kulke R, Horwitz BH, et al. Tumorigenic transformation of murine keratinocytes by the E5 genes of bovine papillomavirus type 1 and human papillomavirus type 16. J Virol. 1991;65:7078–83. doi: 10.1128/jvi.65.12.7078-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–70. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z, Xu J, Patel J, Fuentes S, Lin Y, et al. Function of the small hydrophobic protein of J paramyxovirus. J Virol. 2011;85:32–42. doi: 10.1128/JVI.01673-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Y, Bright AC, Rothermel TA, He B. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J Virol. 2003;77:3371–83. doi: 10.1128/JVI.77.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2009;106:12283–88. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mackenzie KR. Folding and stability of α-helical integral membrane proteins. Chem Rev. 2006;106:1931–77. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 67.Magadán JG, Bonifacino JS. Transmembrane domain determinants of CD4 downregulation by HIV-1 Vpu. J Virol. 2012;86:757–72. doi: 10.1128/JVI.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Magadán JG, Pérez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010;6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Margottin F, Bour SP, Durand H, Selig L, Benichou S, et al. A novel human WD protein, h-β TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–74. doi: 10.1016/s1097-2765(00)80056-8. HIV Vpu targets CD4 for degradation by targeting it to the SCF ubiquitin ligase complex. [DOI] [PubMed] [Google Scholar]
- 70.Maruri-Avidal L, Domi A, Weisberg AS, Moss B. Participation of vaccinia virus l2 protein in the formation of crescent membranes and immature virions. J Virol. 2011;85:2504–11. doi: 10.1128/JVI.02505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maruri-Avidal L, Weisberg AS, Moss B. Direct formation of vaccinia virus membranes from the endoplasmic reticulum in the absence of the newly characterized L2-interacting protein A30.5. J Virol. 2013;87:12313–26. doi: 10.1128/JVI.02137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mattoon D, Gupta K, Doyon J, Loll PJ, DiMaio D. Identification of the transmembrane dimer interface of the bovine papillomavirus E5 protein. Oncogene. 2001;20:3824–34. doi: 10.1038/sj.onc.1204523. [DOI] [PubMed] [Google Scholar]
- 73.McCown MF, Pekosz A. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006;80:8178–89. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, et al. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mercer J, Traktman P. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J Virol. 2003;77:8857–71. doi: 10.1128/JVI.77.16.8857-8871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer AN, Xu YF, Webster MK, Smith AS, Donoghue DJ. Cellular transformation by a transmembrane peptide: structural requirements for the bovine papillomavirus E5 oncoprotein. Proc Natl Acad Sci USA. 1994;91:4634–38. doi: 10.1073/pnas.91.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nappi VM, Petti LM. Multiple transmembrane amino acid requirements suggest a highly specific interaction between the bovine papillomavirus E5 oncoprotein and the platelet-derived growth factor β receptor. J Virol. 2002;76:7976–86. doi: 10.1128/JVI.76.16.7976-7986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nappi VM, Schaefer JA, Petti LM. Molecular examination of the transmembrane requirements of the platelet-derived growth factor β receptor for a productive interaction with the bovine papillomavirus E5 oncoprotein. J Biol Chem. 2002;277:47149–59. doi: 10.1074/jbc.M209582200. [DOI] [PubMed] [Google Scholar]
- 79.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–30. doi: 10.1038/nature06553. HIV Vpu antagonizes the ability of tetherin to inhibit HIV release. [DOI] [PubMed] [Google Scholar]
- 80.Nichols RJ, Stanitsa E, Unger B, Traktman P. The vaccinia virus gene I2L encodes a membrane protein with an essential role in virion entry. J Virol. 2008;82:10247–61. doi: 10.1128/JVI.01035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat Rev Microbiol. 2012;10:563–74. doi: 10.1038/nrmicro2820. An overview of viroporins, an important class of viral miniproteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nilson LA, DiMaio D. Platelet-derived growth factor receptor can mediate tumorigenic transformation by the bovine papillomavirus E5 protein. Mol Cell Biol. 1993;13:4137–45. doi: 10.1128/mcb.13.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82a.Opatowsky Y, Lax I, Tomé F, Bleichert F, Unger VM, Schlessinger J. Structure, domain organization, and different conformational states of stem cell factor-induced intact KIT dimers. Proc Natl Acad Sci USA. 2014;111:1772–77. doi: 10.1073/pnas.1323254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, et al. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J Mol Biol. 2003;333:409–24. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- 84.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petti L, DiMaio D. Stable association between the bovine papillomavirus E5 transforming protein and activated platelet-derived growth factor receptor in transformed mouse cells. Proc Natl Acad Sci USA. 1992;89:6736–40. doi: 10.1073/pnas.89.15.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petti L, DiMaio D. Specific interaction between the bovine papillomavirus E5 transforming protein and the β receptor for platelet-derived growth factor in stably transformed and acutely transfected cells. J Virol. 1994;68:3582–92. doi: 10.1128/jvi.68.6.3582-3592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petti L, Nilson LA, DiMaio D. Activation of the platelet-derived growth factor receptor by the bovine papillomavirus E5 transforming protein. EMBO J. 1991;10:845–55. doi: 10.1002/j.1460-2075.1991.tb08017.x. The 44-amino-acid bovine papillomavirus E5 oncoprotein activates the PDGF β receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Petti LM, Reddy V, Smith SO, DiMaio D. Identification of amino acids in the transmembrane and juxtamembrane domains of the platelet-derived growth factor receptor required for productive interaction with the bovine papillomavirus E5 protein. J Virol. 1997;71:7318–27. doi: 10.1128/jvi.71.10.7318-7327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petti LM, Talbert-Slagle K, Hochstrasser ML, DiMaio D. A single amino acid substitution converts a transmembrane protein activator of the platelet-derived growth factor β receptor into an inhibitor. J Biol Chem. 2013;288:27273–86. doi: 10.1074/jbc.M113.470054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pim D, Collins M, Banks L. Human papillomavirus type 16 E5 gene stimulates the transforming activity of the epidermal growth factor receptor. Oncogene. 1992;7:27–32. [PubMed] [Google Scholar]
- 91.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–6. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–28. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 93.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 94.Pinto LH, Lamb RA. Controlling influenza virus replication by inhibiting its proton channel. Mol Biosyst. 2007;3:18–23. doi: 10.1039/b611613m. [DOI] [PubMed] [Google Scholar]
- 94a.Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 95.Ptacek JB, Edwards APB, Freeman-Cook LL, DiMaio D. Packing contacts can mediate highly specific interactions between artificial transmembrane proteins and the PDGFβ receptor. Proc Natl Acad Sci USA. 2007;104:11945–50. doi: 10.1073/pnas.0704348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raghava S, Giorda KM, Romano FB, Heuck AP, Hebert DN. The SV40 late protein VP4 is a viroporin that forms pores to disrupt membranes for viral release. PLoS Pathog. 2011;7:e1002116. doi: 10.1371/journal.ppat.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roberts KL, Leser GP, Ma C, Lamb RA. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J Virol. 2013;87:9973–82. doi: 10.1128/JVI.01363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rodríguez JR, Risco C, Carrascosa JL, Esteban M, Rodríguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J Virol. 1998;72:1287–96. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–13. doi: 10.1016/j.cell.2010.08.029. The influenza A virus M2 protein inserts into cell membranes to facilitate virus release. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Satheshkumar PS, Chavre J, Moss B. Role of the vaccinia virus O3 protein in cell entry can be fulfilled by its Sequence flexible transmembrane domain. Virology. 2013;444:148–57. doi: 10.1016/j.virol.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Satheshkumar PS, Moss B. Characterization of a newly identified 35-amino-acid component of the vaccinia virus entry/fusion complex conserved in all chordopoxviruses. J Virol. 2009;83:12822–32. doi: 10.1128/JVI.01744-09. The 35-amino-acid O3L virion protein is required for poxvirus entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Satheshkumar PS, Moss B. Sequence-divergent chordopoxvirus homologs of the O3 protein maintain functional interactions with components of the vaccinia virus entry-fusion complex. J Virol. 2012;86:1696–705. doi: 10.1128/JVI.06069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scheideman EH, Marlatt SA, Xie Y, Hu Y, Sutton RE, DiMaio D. Transmembrane protein aptamers that inhibit CCR5 expression and HIV coreceptor function. J Virol. 2012;86:10281–92. doi: 10.1128/JVI.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103a.Schlegel R, Wade-Glass M, Rabson MS, Yang YC. The E5 transforming gene of bovine papillomavirus encodes a small, hydrophobic polypeptide. Science. 1986;233:464–67. doi: 10.1126/science.3014660. [DOI] [PubMed] [Google Scholar]
- 104.Schneider D. Rendezvous in a membrane: close packing, hydrogen bonding, and the formation of transmembrane helix oligomers. FEBS Lett. 2004;577:5–8. doi: 10.1016/j.febslet.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 105.Schneider D, Finger C, Prodohl A, Volkmer T. From interactions of single transmembrane helices to folding of α-helical membrane proteins: analyzing transmembrane helix-helix interactions in bacteria. Curr Protein Pept Sci. 2007;8:45–61. doi: 10.2174/138920307779941578. [DOI] [PubMed] [Google Scholar]
- 106.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–95. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schubert U, Anton LC, Bacik I, Cox JH, Bour S, et al. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J Virol. 1998;72:2280–88. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schubert U, Ferrer-Montiel AV, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 108a.Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci USA. 2010;107:11307–12. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Skasko M, Wang Y, Tian Y, Tokarev A, Munguia J, et al. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem. 2012;287:58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sood CL, Ward JM, Moss B. Vaccinia virus encodes I5, a small hydrophobic virion membrane protein that enhances replication and virulence in mice. J Virol. 2008;82:10071–78. doi: 10.1128/JVI.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Staebler A, Pierce JH, Brazinski S, Heidaran MA, Li W, et al. Mutational analysis of the β-type platelet-derived growth factor receptor defines the site of interaction with the bovine papillomavirus type 1 E5 transforming protein. J Virol. 1995;69:6507–17. doi: 10.1128/jvi.69.10.6507-6517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–93. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stouffer AL, Ma C, Cristian L, Ohigashi Y, Lamb RA, et al. The interplay of functional tuning, drug resistance, and thermodynamic stability in the evolution of the M2 proton channel from the influenza A virus. Structure. 2008;16:1067–76. doi: 10.1016/j.str.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Straight SW, Herman B, McCance DJ. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J Virol. 1995;69:3185–92. doi: 10.1128/jvi.69.5.3185-3192.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Straight SW, Hinkle PM, Jewers RJ, McCance DJ. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J Virol. 1993;67:4521–32. doi: 10.1128/jvi.67.8.4521-4532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Strebel K, Klimkait T, Martin MA. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–23. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- 117.Sugrue RJ, Hay AJ. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–24. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Surti T, Klein O, Aschheim K, DiMaio D, Smith SO. Structural models of the bovine papillomavirus E5 protein. Proteins. 1998;33:601–12. [PubMed] [Google Scholar]
- 119.Suzuki H, Saito R, Masuda H, Oshitani H, Sato M, Sato I. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J Infect Chemother. 2003;9:195–200. doi: 10.1007/s10156-003-0262-6. [DOI] [PubMed] [Google Scholar]
- 120.Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, et al. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 2010;6:e1000801. doi: 10.1371/journal.ppat.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Takeuchi K, Tanabayashi K, Hishiyama M, Yamada A. The mumps virus SH protein is a membrane protein and not essential for virus growth. Virology. 1996;225:156–62. doi: 10.1006/viro.1996.0583. [DOI] [PubMed] [Google Scholar]
- 122.Talbert-Slagle K, Marlatt S, Barrera FN, Khurana E, Oates J, et al. Artificial transmembrane oncoproteins smaller than the bovine papillomavirus E5 protein redefine the sequence requirements for activation of the platelet-derived growth factor β receptor. J Virol. 2009;83:9773–85. doi: 10.1128/JVI.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Terwilliger EF, Cohen EA, Lu YC, Sodroski JG, Haseltine WA. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–67. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Traktman P, Liu K, DeMasi J, Rollins R, Jesty S, Unger B. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J Virol. 2000;74:3682–95. doi: 10.1128/jvi.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Unger B, Mercer J, Boyle KA, Traktman P. Biogenesis of the vaccinia virus membrane: genetic and ultrastructural analysis of the contributions of the A14 and A17 proteins. J Virol. 2013;87:1083–97. doi: 10.1128/JVI.02529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Unger B, Nichols RJ, Stanitsa ES, Traktman P. Functional characterization of the vaccinia virus I5 protein. Virol J. 2008;5:148. doi: 10.1186/1743-422X-5-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Unger B, Traktman P. Vaccinia virus morphogenesis: A13 phosphoprotein is required for assembly of mature virions. J Virol. 2004;78:8885–901. doi: 10.1128/JVI.78.16.8885-8901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–52. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Venkataraman P, Lamb RA, Pinto LH. Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J Biol Chem. 2005;280:21463–72. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- 130.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–41. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–71. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang J, Ma C, Balannik V, Pinto LH, Lamb RA, Degrado WF. Exploring the requirements for the hydrophobic scaffold and polar amine in inhibitors of M2 from influenza A virus. ACS Med Chem Lett. 2011;2:307–12. doi: 10.1021/ml100297w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang J, Ma C, Fiorin G, Carnevale V, Wang T, et al. Molecular dynamics simulation directed rational design of inhibitors targeting drug-resistant mutants of influenza A virus M2. J Am Chem Soc. 2011;133:12834–41. doi: 10.1021/ja204969m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang J, Pielak RM, McClintock MA, Chou JJ. Solution structure and functional analysis of the influenza B proton channel. Nat Struct Mol Biol. 2009;16:1267–71. doi: 10.1038/nsmb.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang J, Qiu JX, Soto C, DeGrado WF. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza A virus. Curr Opin Struct Biol. 2011;21:68–80. doi: 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Watanabe S, Watanabe T, Kawaoka Y. Influenza A virus lacking M2 protein as a live attenuated vaccine. J Virol. 2009;83:5947–50. doi: 10.1128/JVI.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Watanabe T, Watanabe S, Ito H, Kida H, Kawaoka Y. Influenza A virus can undergo multiple cycles of replication without M2 ion channel activity. J Virol. 2001;75:5656–62. doi: 10.1128/JVI.75.12.5656-5662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Watanabe T, Watanabe S, Kim JH, Hatta M, Kawaoka Y. Novel approach to the development of effective H5N1 influenza A virus vaccines: use of M2 cytoplasmic tail mutants. J Virol. 2008;82:2486–92. doi: 10.1128/JVI.01899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wetherill LF, Holmes KK, Verow M, Muller M, Howell G, et al. High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors. J Virol. 2012;86:5341–51. doi: 10.1128/JVI.06243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wilson RL, Fuentes SM, Wang P, Taddeo EC, Klatt A, et al. Function of small hydrophobic proteins of paramyxovirus. J Virol. 2006;80:1700–9. doi: 10.1128/JVI.80.4.1700-1709.2006. The simian virus 5 SH protein can be replaced by the divergent mumps virus SH protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140a.Windisch D, Hoffmann S, Afonin S, Vollmer S, Benamira S, et al. Structural role of the conserved cysteines in the dimerization of the viral transmembrane oncoprotein E5. Biophys J. 2010;99:1764–72. doi: 10.1016/j.bpj.2010.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140b.Yang X, Tschaplinski TJ, Hurst GB, Jawdy S, Abraham PE, et al. Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome Res. 2011;21:634–41. doi: 10.1101/gr.109280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140c.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–34. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]


