Abstract
Activated macrophages are crucial for restriction of microbial infection but may also promote inflammatory pathology in a wide range of both infectious and sterile conditions. The pathways that regulate macrophage activation are therefore of great interest. Recent studies in silico have putatively identified key transcription factors that may control macrophage activation, but experimental validation is lacking. Here, we generated a macrophage regulatory network from publicly available microarray data, employing steps to enrich for physiologically relevant interactions. Our analysis predicted a novel relationship between the AP-1 family transcription factor Junb and the gene Il1b, encoding the pyrogen Interleukin-1β, which macrophages express upon activation by inflammatory stimuli. Previously, Junb has been characterized primarily as a negative regulator of the cell cycle, whereas AP-1 activity in myeloid inflammatory responses has largely been attributed to c-Jun. We confirmed experimentally that Junb is required for full expression of Il1b, and of additional genes involved in classical inflammation, in macrophages treated with LPS and other immunostimulatory molecules. Furthermore, Junb modulates expression of canonical markers of alternative activation in macrophages treated with Interleukin-4. Our results demonstrate that JUNB is a significant modulator of both classical and alternative macrophage activation. Further, this finding provides experimental validation for our network modeling approach, which will facilitate the future use of gene expression data from open databases to reveal novel, physiologically relevant regulatory relationships.
Introduction
Macrophages, tissue-resident phagocytic cells of the innate immune system, are critical sentinels in the detection and containment of infectious microbes and the initiation of inflammatory Type I immune responses. In addition to these functions, collectively referred to as classical activation, macrophages may also undergo alternative activation, resulting in distinct non-inflammatory programs that are important in Type II immune responses, wound healing, and tissue homeostasis (1, 2). Given the central role of macrophages in diverse immune functions, it is important to develop a more systematic understanding of the transcriptional networks that govern their activation and polarization.
One recently developed tool that may yield great insight into mechanisms of macrophage activation is regulatory network analysis, a statistical method for identifying components of a dataset that co-vary across a broad range of samples or conditions (3). A wealth of macrophage transcriptional data is available in public databases, but such data are generally considered unsuitable for network analysis due to the confounding effects of technical variation resulting from the use of diverse nucleic acid amplification procedures and expression profiling platforms. In this study, we present the results of a regulatory network analysis approach that is based on mutual information and data processing inequality procedures (4–8) applied to strictly standardized and normalized public datasets. We further improved the power of this approach to identify physiological relationships by using existing literature to strengthen predictions in a series of steps that we term “knowledge-based enrichment.”
Our network model led us to examine the AP-1 transcription factor JUNB for its role in myeloid immune activation. Although JUNB has historically been studied primarily in the contexts of cell cycle regulation and differentiation, several recent bioinformatic studies, like the one presented here, have predicted a role for JUNB in the regulation of myeloid immune responses (3, 9). However, there is currently little experimental evidence to support this prediction. To directly test the importance of JUNB in macrophage activation, we characterized the transcriptional responses of JUNB-deficient macrophages to diverse stimuli. Confirming our network prediction, we found that JUNB modulates subsets of immune-related genes in macrophages treated with microbial ligands (referred to as classically activated or M(LPS) macrophages) as well as with the cytokine Interleukin-4 (IL-4), which stimulates polarization of alternatively activated M(IL-4) macrophages (10). To our knowledge, this is one of the first reports of a transcription factor that promotes polarization of both M(LPS) and M(IL-4) macrophages. Furthermore, this study provides experimental validation for several recent predictions made in silico (3, 9), demonstrating the power of network analysis to lead to new insights into immune regulation.
Materials and Methods
GEO data preprocessing
All mouse macrophage microarray datasets warehoused in the Gene Expression Omnibus (GEO) or ArrayExpress database as of 2010 were downloaded. Data were log2 transformed and each experimental sample was normalized to a baseline sample (e.g., untreated or time zero) to reduce inter-dataset technical variation. Each microarray dataset was then z-scaled to minimize distribution variation. This dataset consisted of 40 studies and 243 samples, which were subsequently collapsed by averaging technical and biological replicates in order to reduce bias from studies that used larger sample groups. Author-provided gene identifiers were used to map datasets to one another, and studies that failed to map to at least 40% of the total gene set were removed, leaving 87 samples from 18 studies (Supplemental Table 1). Data were further filtered by removal of genes not present in at least 60% of the remaining arrays, resulting in probes for 11,816 unique genes.
ARACNe network prediction
A regulatory network model was generated using a mutual information metric with p-value thresholding and application of the data processing inequality as implemented in ARACNe (4, 5). Briefly, missing data were estimated in R using K-nearest neighbors as implemented in the impute package and pairwise mutual information was calculated using ARACNe. Thresholds for p-values were evaluated from 10−3 to 10−15, and the data processing inequality (DPI) was applied at a tolerance of 0.10. For each p-value threshold, a false discovery rate was estimated by generating a background model based on the average number of edge predictions over 1000 random iterations. Random iterations were based on datasets in which gene expression values were unchanged (to preserve the distribution) but randomly ordered for each array.
A p-value was chosen for thresholding based on the F-score calculated from precision and recall. Calculation of these parameters is limited by the lack of availability of a suitably large database of mouse macrophage transcriptional regulation interactions; the most extensive such database, InnateDB (11), contains fewer than 200 such relationships. To obtain a more robust estimation of precision and recall, we based our calculations on all human and mouse interactions catalogued in BioGRID (12), including physical associations, under the assumption that co-expressed genes would be enriched in these data because a physical association implies some degree of co-expression. The maximal F-score was found to occur at p-value of 10−6, which was chosen for thresholding and subsequent DPI application.
Knowledge enrichment
Knowledge enrichment was performed in two steps. First, triangular regulatory subnets were identified by retrieving triangular subnets (i.e., those with three nodes and three edges) from the ARACNe-predicted full network, followed by removal of triangular subsets without exactly one transcription factor as defined by comparison with the RIKEN transcription factor database (13). Triangular subsets were subjected to knowledge enrichment by searching PubMed for each transcription factor-gene combination and only retaining triangular subnets in which at least two citations were found for one transcription factor-gene combination (a “strong” association), the rationale being that these subnets were further enriched for true biological interactions. Triangular subnets were recombined into a complete network delineating transcription factor-gene relationships, but not the originally predicted gene-gene relationships.
Testing the degree of enrichment is limited by the absence of a robust regulatory interaction reference database. In our case, testing is further limited by the fact that existing databases are based on published literature, which is similarly used to enrich our network predictions. For testing, we therefore eliminated all strong associations and based our enrichment analysis only on weak associations. Using all mouse and human interactions (including physical) in InnateDB, as well as all human regulatory interactions in the Human Transcriptional Regulation Database (14), allowed testing of weak associations against a reference database. Similar comparisons were performed with BioGRID and other subsets of InnateDB (e.g., mouse-specific interactions, or transcriptional regulation interactions), but insufficient associations were discovered to perform the analysis. Transcription factor-specific subnets were parsed using in-house scripts, and network predictions were visualized in Cytoscape (15). All analysis described above was performed using customized Perl scripts.
Mice
Junbfl/fl mice (from E. Passegué, UCSF) and Lyz2Cre/Cre mice were crossed in-house to generate Junbfl/fl x Lyz2Cre/Cre mice. All mouse work was conducted with the approval of the UCSF Institutional Animal Care and Use Committee in strict accordance with the guidelines of the Office of Laboratory Animal Welfare.
Derivation and stimulation of macrophages
Macrophages were derived from bone marrow by culturing for 8 d in RPMI supplemented with 10% serum, 10% supernatant from 3T3-M-CSF cells, and 1 mM sodium pyruvate, with feeding on day 5. Resident peritoneal macrophages were isolated by peritoneal lavage with ice-cold 10 mL PBS containing 1mm EDTA and 3% FBS. Unless otherwise indicated, macrophages were plated in 12-well dishes at a density of 8 x 105 cells per well and treated with LPS (Sigma; 100 ng/mL), CpG (Invivogen ODN1826, 1.5 μg/mL), imiquimod (Invivogen, 5 μg/mL), poly(I:C) (Sigma, 2.5 μg/mL), IL-4 (Peprotech, 20 ng/mL) or transfected cyclic di-GMP (10 μg/mL; kind gift of D. Burdette and R. Vance, UC Berkeley). Lipofectamine 2000 (Life Technologies) was used for transfection.
IL-1β ELISA
Macrophages were plated in 6-well dishes at a density of 106 cells/well in 5 mL media. After 3 h LPS stimulation, ATP (Sigma) was added to a final concentration of 5 mM. 50 μL aliquots of media were removed at indicated timepoints for analysis using the Ready-Set- Go IL-1β ELISA (eBioscience) according to the manufacturer's protocol, with recombinant IL- 1β (eBioscience) as a standard.
Western blot
Cell lysates were harvested in RIPA buffer supplemented with 1 mM PMSF and 1X Complete Protease Inhibitor Cocktail (Roche). Proteins were separated on 10% NuPage bis-tris gels (Life Technologies), transferred to PVDF membrane and immunoblotted with antibodies to JUNB (Cell Signaling) or β-actin (Abcam).
Phagocytosis assay
BMDM were incubated with 3μm FITC-labeled beads (Spherotech ECFP-F1) at a cell: bead ratio of 5:1 or with 1μm Fluosphere beads (Life Technologies) at 10:1 for 24 h. Cells were labeled with anti-F4/80 antibody as described below, and uptake of fluorescent beads was measured on an LSRII (BD Biosciences).
Flow cytometry
BMDM were blocked with anti-CD16/32 (2.4G2; UCSF Monoclonal Antibody Core), stained with antibodies to myeloid surface markers, and analyzed on an LSRII (BD Biosciences). Intracellular staining was performed after 4 h incubation (for BMDM) or 2 h incubation (for peritoneal macrophages) with GolgiPlug (BD) and monensin (eBioscience), using Cytofix/Cytoperm (BD) according to manufacturer protocols. Results were analyzed using FACSDiva (BD) and FlowJo (TreeStar) software. Antibodies to the following were used: Ly6C (clone HK1.4), CD11c (N418), IL-1β (NJTEN3), TNF (TN3-19), rat IgG1 isotype (eBRG1), all eBioscience; Ly6G (1A8), Biolegend; Ly6B.2 (7/4), Cedarlane; CD11b (M1/70), F4/80 (BM8), Gr-1 (RB6-8C5), all UCSF Monoclonal Antibody Core; and CD68 (FA-11), ABD Serotec. Myeloid populations were delineated as previously described (16): granulocytes (CD11b+ Ly6G+), eosinophils (CD11b+ Ly6cint SSChi), monocytes (CD11b+ F4/80int SSClo Ly6c as indicated), red pulp macrophages (F4/80hi CD11b−), conventional DC (CD11chi), and plasmacytoid DC (CD11cint Siglec H+).
Junb microarray analysis
Following a 4 h LPS stimulation, RNA was isolated from BMDM using the RNaqueous Micro kit (Ambion). Purified RNA was amplified (Amino Allyl MessageAmp II kit, Life Technologies) with minor modifications to manufacturer’s instructions to generate amino allyl incorporated amplified RNA (aaRNA). aaRNA was coupled to Cy3 dye (GE Healthcare Life Sciences) and hybridized overnight to a SurePrint G3 Mouse Gene Expression 8x60K microarray, which was washed and scanned per manufacturer’s instructions (Agilent). Raw intensities were extracted using Feature Extraction software (Agilent) and quantile normalized using Limma (17). Differentially expressed genes were identified using Significance Analysis for Microarrays (SAM) (18), and data were visualized as heat maps using custom Perl scripts. Genes that were increased by at least twofold according to SAM (1% FDR) in LPS-treated Junbfl/fl BMDM were considered to be LPS-induced. JUNB-dependent genes were identified using SAM (10% FDR, 1.5-fold change threshold) to compare LPS-stimulated gene expression values, normalized to average baseline expression, between Junbfl/fl and JunbΔLyz2 macrophages. The set of genes regulated by JUNB in response to LPS was defined as the intersection of LPS-induced genes and JUNB-dependent genes. All data are available in GEO under accession number GSE50542.
Reverse transcription and quantitative PCR
Macrophage RNA was harvested 4 h post stimulation using the RNeasy kit (Qiagen) with on-column Turbo DNase treatment (Ambion). Reverse transcription was performed with Superscript III (Life Technologies) primed with dT(20)V. Quantitative PCR was performed in a Step One Plus RT PCR System (Applied Biosystems) using PerfeCTa 2x qPCR mix (Quanta). Transcript levels were normalized to levels of actin mRNA. The following primer sequences were used: Il1b: F- CAACCAACAAGTGATATTCTCCATG, R- GATCCACACTCTCCAGCTGCA; actin: F- ACCCTAAGGCCAACCGTGAA, R-CCGCTCGTTGCCAATAGTGA; Il12b: F- ACAAGACTTTCCTGAAGTGTGAAG, R-GCTCTTGATGTTGAACTTCAAGTCC, Tnf: F-GACGTGGAACTGGCAGAAGAG, R- TTGGTGGTTTGTGAGTGTGAG, Ifnb: F- CAGCTCCAAGAAAGGACGAAC, R- GGCAGTGTAACTCTTCTGCAT; Junb: F-ATCCTGCTGGGAGCGGGGAACTGAGGGAAG, R- AGAGTCGTCGTGATAGAAAGGC; Arg1: F-TGGGAGGCCTATCTTACAGA, R-CATGTGGCGCATTCAC; Retlna: F-GGATGCCAACTTTGAATAGG, R-GCACACCCAGTAGCAGTCAT; Il4ra: F-CAAGCCCTTTGGGGATGAC, R-CAGGACAGCTTTCCAAGGC; Mrc1: F- CTCTGTTCAGCTATTGGACGC, R- CGGAATTTCTGGGATTCAGCTTC.
Meta-analysis of ChIP-Seq data
Raw reads from chromatin immunoprecipitation and sequencing (ChIP-Seq) of JUNB from bone marrow-derived dendritic cells were downloaded from GEO (accession GSE36104) (9). These paired end reads were mapped to version mm10 of the Mus musculus genome using Bowtie2 (19). Mapped reads were sorted and converted from SAM to BAM format using Samtools (20). Peaks were identified in MACS2 (21) using signal over background. Although we identified all peaks with a q < 0.05, our highest confidence peaks had an adjusted q-value of < 10−5.The closest gene to each JUNB binding peak was identified by calculating the distance to the nearest transcriptional start site. Genes were considered “bound” when this distance was less than or equal to 5 kilobases in any of the four timepoints assayed. Genes bound by JUNB were intersected with JUNB-dependent and JUNB-independent genes (identified by microarray above).
Arginase assay
Macrophages were plated in 96-well plates at a density of 105 cells per well and treated with IL-4 (20 ng/mL) overnight. After a PBS rinse, cells were lysed in 100 μl PBS + 0.4% Triton-X100, and the lysate was processed per manufacturer instructions using the QuantiChrom Arginase Assay Kit (BioAssay Systems). Absorbance was measured at 430 nm.
Cytometric bead array
Cytokine concentrations were measured in cell culture supernatant using the Milliplex MAP Mouse Cytokine/Chemokine Magnetic Bead Panel kit (EMD Millipore) according to the manufacturer's instructions. Measurements were made on a MAGPIX instrument (Luminex).
Results
Knowledge-enriched analysis of macrophage regulatory networks
In order to identify novel transcriptional relationships that regulate macrophage activation, we generated a de novo regulatory network prediction based on microarray expression data obtained from the public GEO database (http://www.ncbi.nlm.nih.gov/geo/). Our initial dataset consisted of 243 microarrays from 40 individual studies, each of which examined the transcriptional responses of mouse macrophages to various stimuli (Supplemental Table 1). The data from each individual study were internally normalized to a baseline condition, reducing variance inherent in technical differences between studies and enriching for physiological changes in gene expression. These data were used to generate a mutual information-based network linking genes whose expression co-varied over a range of conditions (see Methods and Supplemental Fig. 1).
Despite a low false discovery rate, estimated using a background model comprised of randomly ordered expression values (not shown), this approach yielded a large number of predicted regulatory interactions. However, due to the heterogeneity of our dataset, we expected that some proportion of these interactions (also known as “edges”) would reflect technical covariation within subsets of microarrays, rather than biological co-variation. To enrich for biological relationships, we subjected the network to a literature-based refinement process. In this procedure, triangular subnets, comprised of a transcription factor connected to two genes that are likewise connected to one another, were identified as being candidates for improved biological signal based on their multiple routes of connectedness. In a further refinement, we identified “knowledge-enriched” triangular subnets, in which at least one transcription factor-gene relationship was found to have two or more citations in PubMed, lending credibility to at least one of the predictions. These knowledge-enriched subnets were recombined into a new global network (Supplemental Fig. 1). The specificity of the knowledge-enriched predictions was tested by comparison with reference databases that track regulatory, physical, and other types of interactions. The refinements improved specificity for biological interactions by several fold, demonstrating the value of the approach in improving the frequency of biologically relevant predictions (at the expense of sensitivity) (Fig. 1A). The resultant network map is likely to be a rich source of undiscovered regulatory interactions.
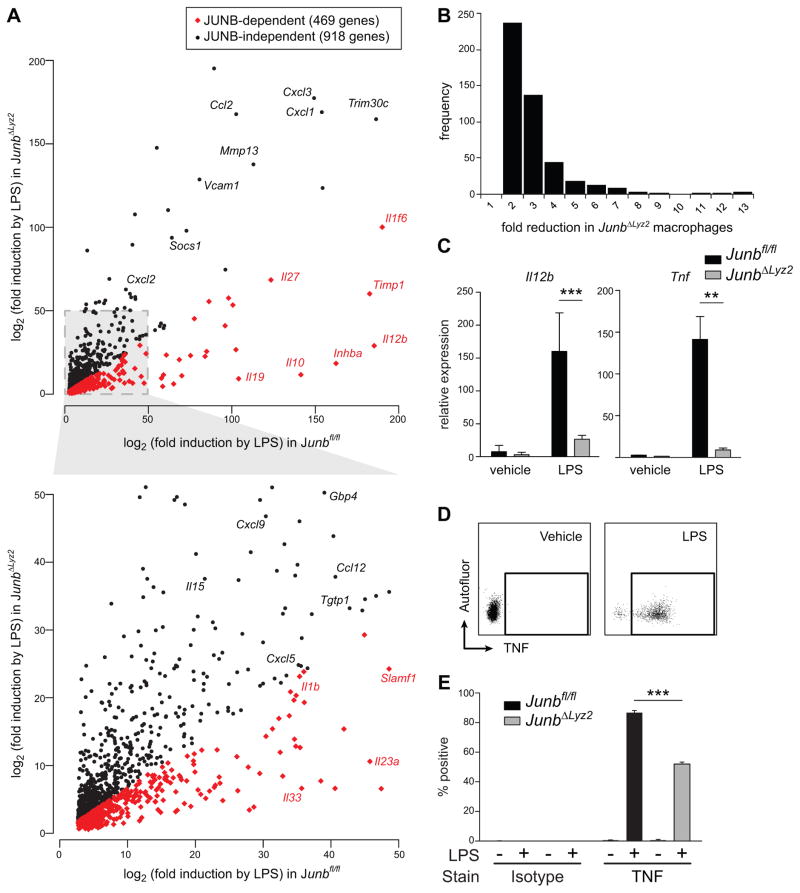
Figure 1. Network analysis predicts a regulatory interaction between Junb and Il1b in macrophages.
(A) Gene-gene relationships (edges) identified in the initial expression analysis (ARACNe) were enriched for biological relevance based on known interactions; the graph depicts the percent of edges remaining after each of two sequential refinements (triangular subnets and knowledge-enriched) that also appear in two databases of known biological interactions (InnateDB and HTRI). Numbers above bars indicate fold enrichment from the initial to the final datasets. (B) Regulatory network analysis identified significant covariance between Il1b and 7 transcription factors, including Rel and Junb. (C) Splenocytes from 4 week old mice (n = 4 of each genotype) were analyzed by flow cytometry to quantify frequencies of the indicated myeloid cells. RPMs, red pulp macrophages. cDC, conventional DC. pDC, plasmacytoid DC. (D-F) Macrophages were differentiated from the bone marrow of Junbfl/fl or Junbfl/fl x Lyz2 Cre/Cre (JunbΔLyz2) mice. (D) Deletion of Junb was assessed by quantitative PCR (qPCR) on BMDM genomic DNA. Means + SD for three technical replicates are shown. (E) BMDM from the indicated genotypes were stimulated with LPS for 4 h. Expression of JUNB (top panel) and β-actin (bottom panel) was measured in lysates by Western blot. (F) BMDM were labeled with antibodies to the indicated myeloid surface markers and analyzed by flow cytometry. Data shown are means + SD averaged from three independent experiments. (G) BMDM were incubated for 24 h with fluorescent beads of the indicated diameter, and phagocytosis was measured by flow cytometry. **, p < 0.01 by t-test.
A role for the JUNB transcription factor in macrophage transcriptional responses to LPS
Because we were interested in pathways affecting macrophage activation, we focused on network predictions surrounding the gene Il1b, which encodes the pro-inflammatory cytokine IL-1β. IL-1β is a potent pyrogen produced by macrophages in response to a variety of pro-inflammatory stimuli. As an internal control, our network analysis correctly predicted a co-regulatory relationship between Il1b and the gene Rel, which encodes a subunit of NF-κB, a transcription factor known to be required for Il1b induction in macrophages (22) (Fig. 1B).
The network analysis also predicted a relationship between Il1b and Junb, a member of the AP-1 family of transcription factors, which are activated downstream of MAP kinase signaling and regulate a wide variety of cell processes, including differentiation, proliferation, activation, and apoptosis (23). The dimeric AP-1 is assembled from subunits of the JUN (c-JUN, JUNB, or JUND), FOS (c-FOS, FOSB, FRA-1, FRA-2), or ATF protein families. Interestingly, JUNB was recently reported to bind extensively to the chromatin of LPS-treated dendritic cells (DCs) (9), and was also identified as one of the most highly expressed and extensively connected transcription factors in a regulatory network model from human monocyte-derived macrophages (3). However, despite these observations, no role has been demonstrated for JUNB in the regulation of Il1b, or indeed of any gene expressed in activated macrophages. In fact, in myeloid cells, AP-1 activity in response to inflammatory stimuli has until recently been attributed almost exclusively to c-JUN:c-FOS heterodimers (24–26), which represent AP-1's canonical and most well-characterized composition.
Systemic deletion of Junb results in embryonic lethality (27). Therefore, to test whether JUNB does indeed regulate Il1b in macrophages, we crossed mice bearing floxed alleles of Junb to mice expressing Cre recombinase under control of the Lyz2 promoter, resulting in excision of Junb in a subset of myeloid lineages (monocytes, macrophages, granulocytes, and a small fraction of DCs) (28). The resulting mice are healthy, breed as expected, and exhibit no gross pathology. Previously, conditional deletion of Junb in hematopoietic stem cells was shown to result in a marked expansion of myelomonocytic progenitors (29); importantly, the myeloid compartment of Junbfl/fl x Lyz2Cre/Cre mice (referred to henceforth as JunbΔLyz2) mice is indistinguishable from Junbfl/fl controls (Fig. 1C). We confirmed loss of Junb expression in bone marrow-derived macrophages (BMDM) produced from JunbΔLyz2 mice by both quantitative RT-PCR (Fig. 1D) and Western blot (Fig. 1E). JunbΔLyz2 BMDM had typical morphology (unpublished data) and normal levels of macrophage-specific surface markers (Fig. 1F), and they phagocytosed fluorescent beads as efficiently as JUNB-sufficient macrophages (Fig. 1G). Together, these data indicate that myeloid-restricted deletion of Junb does not disrupt macrophage proliferation, differentiation, or phagocytic function.
In validation of our network model prediction, we observed significant defects in induction of Il1b mRNA (Fig. 2A) and secretion of IL-1β protein (Fig. 2B) in JunbΔLyz2 BMDM following stimulation with LPS or LPS + ATP, respectively. The defects were partial, indicating that JUNB is not absolutely required for Il1b induction but rather augments its expression. Decreased protein in supernatant was not merely due to a secretion defect, as we also observed decreased expression of pro-IL-1β in intact JunbΔLyz2 BMDM (Fig. 2C,D). IL-1β production was also reduced in peritoneal macrophages from JunbΔLyz2 mice stimulated with LPS ex vivo, confirming that JUNB deficiency affects cytokine production in a distinct type of macrophage (Fig. 2E).
Figure 2. JUNB regulates IL-1β production in macrophages treated with LPS.
(A) Following 4 h treatment with LPS, Il1b mRNA levels were assessed by reverse transcription (RT) and qPCR. (B) BMDM were pre-treated for 3 h with LPS followed by addition of 5 mM ATP. IL-1β was measured in supernatant by ELISA. (C, D) BMDM were treated for 4 h with LPS, and intracellular pro-IL-1β was detected by flow cytometry. (C) Isotype staining and mock-treated controls were used to establish gates for F4/80+ BMDM. (D) The graph depicts the percent of BMDM positive for intracellular pro-IL-1β. (E) Peritoneal macrophages were stimulated for 2 h with LPS and stained for intracellular pro-IL-1β as in C, D. Data shown are means + SE (D, E) or SD (A, B) and are representative of three (A-D) or two (E) independent experiments. *, p < 0.05. **, p < 0.01. ***, p < 0.001 by t-test.
Next, we used whole-transcriptome microarrays to assess the global impact of Junb on macrophage responses to LPS. We found that approximately 34% of LPS-induced genes exhibited significantly diminished upregulation in JUNB-deficient macrophages relative to their expression in JUNB-sufficient macrophages (Fig. 3A; 469 JUNB-dependent genes [Supplemental Table 2 A] versus 918 JUNB-independent genes [Supplemental Table 2 B]). JUNB-deficient macrophages were not merely poorly responsive to LPS in general, as the majority of genes were induced equally in both JUNB-deficient and JUNB-sufficient macrophages (Fig. 3A and Supplemental Table 2). The JUNB-dependent genes included cytokines, chemokines, interferon-associated genes, and other genes with established immune functions, several of which were previously shown to be decreased following knockdown of Junb in DCs treated with LPS (30). Expression of a small number of genes, such as Il12b, which encodes a subunit of the cytokine IL-12, was drastically reduced in the absence of JUNB, consistent with the previous observation that JUNB binds to an enhancer in the Il12b promoter (31). However, nearly 70% of affected genes, like Il1b, were reduced by only 2–3 fold (Fig. 3B). Together, these data suggest that JUNB is not absolutely required for activation of gene expression, but that it fine-tunes gene expression and, by inference, modulates the output of other transcription factors. Quantitative RT-PCR was used to validate additional predicted targets (Il12b, Tnf), confirming their JUNB dependence (Fig. 3C). Additionally, intracellular cytokine staining revealed a defect in production of TNF protein in JUNB-deficient macrophages following LPS stimulation (Fig. 3D,E). Thus, JUNB regulates production of multiple cytokines and other immune-related genes in macrophages treated with LPS, affecting levels of both transcript and protein.
Figure 3. JUNB modulates expression of a cluster of immune-related genes in LPS-treated macrophages.
(A-C) Junbfl/fl or JunbΔLyz2 BMDM were treated for 4 h with LPS. RNA was harvested, amplified, and analyzed by microarray (A, B) or reverse transcribed into cDNA (C). (A) Genes exhibiting < 2-fold difference (black dots) or > 2-fold difference (red dots) in Junbfl/fl versus JunbΔLyz2 macrophages. Results shown are averages from four technical replicates. (B) Histogram of fold-reduction in gene expression for JUNB-dependent genes. (C) Levels of the indicated transcripts were measured by RT-qPCR. Means + SD are shown. Representative results from one of three experiments are shown. **, p < 0.01. ***, p < 0.001 by t-tests. (D) Gating strategy for measurement of intracellular TNF. Isotype controls (not shown) were used as described for pro-IL-1β. (E) Induction of intracellular TNF after 4 h of LPS treatment. Bars represent means + SE for % of BMDM positive for TNF and are representative of three independent experiments.
JUNB likely regulates transcription both directly and indirectly
To gauge whether JUNB regulates transcription through direct binding of regulatory elements in target genes (9, 30–32), we cross-referenced our microarray data with a previous study that performed ChIP-Seq on LPS-treated DCs, which are ontologically and functionally closely related to macrophages (9). We identified 178 genes, including Il1b and Il12b, that exhibited JUNB-dependent expression in macrophages and that were also directly bound by JUNB in DCs (Supplemental Table 3 A). These bound and regulated genes accounted for 38% of all JUNB-dependent genes examined. JUNB binding was not observed proximal to the remaining 291 JUNB-dependent genes (Supplemental Table 3 B), consistent with indirect regulation of these targets by JUNB. Interestingly, 41% of JUNB-independent genes were also bound by JUNB (376 of 919 examined; Supplemental Tables 3 C and D), suggesting that binding of this transcription factor is not sufficient to alter gene expression. This observation is consistent with the previous report's suggestion that JUNB likely cooperates with several other factors to activate transcription (9). Alternatively, these binding sites could represent sites at which JUNB binds without impacting gene expression, as has been observed for other transcription factors (33). Although we have not assessed binding of JUNB to target genes in macrophages here, our analysis of ChIP-Seq data from a closely related cell type, combined with a previous report of JUNB binding to the Il12b enhancer in macrophages (31), provides strong evidence that JUNB modulates transcription at least in part through direct activation.
In addition to binding directly to regulatory elements, JUNB may affect transcription indirectly by altering the levels of other transcription factors, such as other AP-1 family members. However, we observed no defect in mRNA expression of the AP-1 transcription factors c-Jun, Jund, and Fos in JUNB-deficient macrophages either at baseline or upon stimulation with LPS (Fig. 4A); in fact, expression of some of these factors was slightly increased in the absence of JUNB. Nevertheless, these data do not rule out the possibility that JUNB affects transcription indirectly; for example, although Jun and Fos transcript levels are intact in JUNB-deficient macrophages, it is possible that protein levels or activities are altered in the absence of JUNB, or that other AP-1 family members are affected. Furthermore, we identified several other transcription factors, including Aff1, Mxd1, Crem, and Stat3, whose expression is defective in the absence of JUNB in macrophages (Supplemental Table 2 A) and whose transcriptional initiation regions are directly bound by JUNB in DCs (9). Altered transcription downstream of these factors may therefore account for the JUNB-dependent regulation of genes we observed in the absence of direct JUNB binding. Interestingly, Junb itself is bound by JUNB (9), suggesting that its own expression may be subject to feedback. Altogether, our analyses indicate that JUNB is likely to affect transcription both directly, through binding of target gene regulatory regions, and indirectly, by altering levels of other transcription factors.
Figure 4. JUNB affects late transcription of primary and secondary genes. (A).
Expression of AP-1 family members was assessed by microarray in BMDM with or without LPS stimulation as described in Fig. 3A. SD from 4 technical replicates is shown. (B) A subset of JUNB-dependent and JUNB-independent genes was identified by microarray and further classified into primary, secondary, tolerizable and non-tolerizable categories. +, Junbfl/fl BMDM. Δ, JunbΔLyz2 BMDM. (C) Junbfl/fl or JunbΔLyz2 BMDM were treated with LPS, and RNA was harvested at the indicated timepoints. Transcript of Il1b (left panel) or Il12b (right panel) was measured by RT-qPCR. Graphs depict means + SD and are representative of two independent experiments, each with three technical replicates.
JUNB affects transcription of both primary and secondary transcriptional targets
In order to understand whether JUNB preferentially regulates a specific category of genes, we attempted to classify the JUNB-dependent genes identified by microarray. Gene ontology analysis did not reveal enrichment of specific gene categories among JUNB-dependent genes; instead, JUNB modulated expression of a representative subset of LPS-induced gene ontology categories (unpublished data). Next, we examined 48 LPS-induced genes that were previously classified as primary or secondary response genes on the basis of their activation kinetics, as well as whether they require new protein synthesis for induction (discussed further below) (34, 35). Nine of these targets, including eight primary response genes and a single secondary response gene (Il12b), were determined to be JUNB-dependent by microarray (Fig. 4B). Although the pool of primary gene targets examined was larger (41 primary genes versus 7 secondary genes), the percentage of JUNB-dependent genes was similar in both categories (16% of primary and 10.5% of secondary genes examined) (Fig. 4B). Finally, we examined whether these 48 genes were tolerizable or non-tolerizable according to the classification scheme proposed by Foster et al., in which macrophages previously exposed to LPS will selectively upregulate non-tolerizable genes, but not tolerizable genes, upon secondary LPS stimulation (36). Among the 48 primary/secondary response genes examined in (35), 19 genes were further classifiable as tolerizable or non-tolerizable, including three JUNB-dependent genes (Fig. 4B). Thus, JUNB-dependent genes include both primary and secondary response genes as well as tolerizable genes. Although our analysis did not reveal any non-tolerizable genes that are JUNB-dependent, this is likely due to the small size of the gene sets that were searched for overlap (i.e., secondary genes versus non-tolerizable genes versus JUNB-dependent genes). On the whole, we conclude that JUNB modulates the transcription of a broad range of genes from several ontological and regulatory categories without obvious bias toward any given category.
In order to further characterize JUNB activity, we examined the kinetics of transcription of Il1b, a primary response gene, and Il12b, a secondary response gene, in macrophages treated with LPS. Primary response genes do not require new protein synthesis for induction, whereas secondary response genes do; furthermore, primary genes tend to be upregulated more swiftly than secondary genes, though exceptions exist (35). Consistent with previously reported activation kinetics, Il1b was markedly induced in wild-type macrophages as early as 1 h post stimulation, whereas robust upregulation of Il12b was not observed until 2 h post stimulation (Fig. 4C). Interestingly, Il1b induction was intact in JUNB-deficient macrophages at early timepoints, but by 4 h post-treatment it lagged behind JUNB-sufficient controls (Fig. 4C). In contrast, upregulation of Il12b was almost completely abrogated in JUNB-deficient macrophages at all timepoints assayed (Fig. 4C). Thus, JUNB modulates expression of both primary and secondary genes with delayed kinetics, and appears to be most important from 2 to 4 h post stimulation, perhaps reflecting the time needed to upregulate JUNB itself. In the case of the rapidly induced Il1b, this delayed involvement results in an initially intact response followed by a leveling off in JUNB-deficient macrophages, whereas with the more gradually induced Il12b, dramatic JUNB dependency is observed from the time that transcript levels first begin to increase.
JUNB modulates immune-related gene expression downstream of multiple pattern recognition receptors
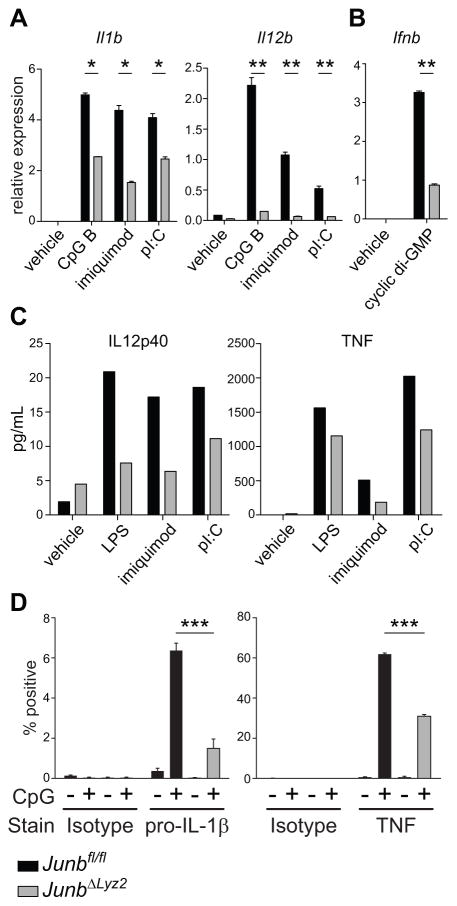
Having established that JUNB contributes to transcriptional responses to LPS, which signals through Toll-like receptor (TLR) 4, we wished to test whether JUNB also modulates the transcriptional output of other pattern recognition receptors. Stimulation of JUNB-deficient BMDM with the synthetic ligands poly(I:C), imiquimod, or CpG DNA resulted in defective induction of the target genes Il1b and Il12b (Fig. 5A), implicating JUNB in transcriptional responses downstream of TLRs 3, 7, and 9, respectively. JUNB-deficient macrophages were also defective in their response to transfected cyclic-di-GMP (Fig. 5B), which elicits robust expression of Interferon-β (Ifnb) in wild-type macrophages through the cytosolic sensor STING (37). JUNB-deficient macrophages treated with a variety of TLR ligands produced less IL-12, IL-1β, and TNF protein than JUNB-sufficient macrophages, as measured by ELISA (Fig. 5C) and intracellular cytokine staining (Fig. 5D), indicating defects in macrophage responses at the level of both transcript and protein. Taken together, these results demonstrate the broad involvement of JUNB in generating macrophage responses to multiple immunostimulatory ligands.
Figure 5. JUNB regulates cytokine expression downstream of multiple pattern recognition receptors.
(A-D) Junbfl/fl or JunbΔLyz2 BMDM were treated for 4 h with the indicated ligands. Transcripts of the indicated cytokines were measured by RT-qPCR (A, B), and secreted protein was measured in the supernatant by cytometric bead array (C). (D) Production of pro-IL-1β and TNF by intracellular cytokine staining. Data are depicted as means + SD and are representative of two (C, D) or at least three (A, B) experiments. *, p < 0.05. **, p < 0.01. ***, p < 0.001 by t-tests.
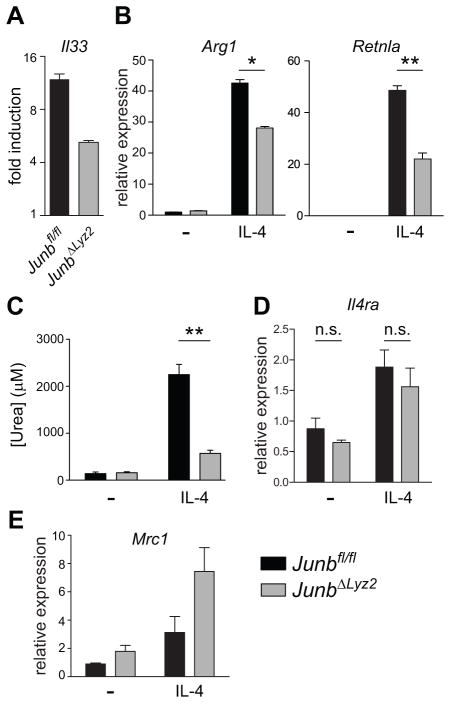
JUNB regulates alternative macrophage activation
Finally, we sought to determine whether JUNB modulated M(IL-4) macrophage responses in addition to M(LPS) responses. Our microarray analysis had revealed that LPS-treated macrophages upregulated Il33, a Type II cytokine, in a JUNB-dependent manner (Fig. 3A, 6A). Since IL-4-driven alternative activation is a central feature of Type II immunity, we evaluated the induction of M(IL-4) responses in JUNB-sufficient and JUNB-deficient BMDM treated with IL-4. JUNB-deficient BMDMs exhibited marked defects both in the transcriptional induction of the M(IL-4) target genes Arg1 and Retnla (Fig. 6B) and in the upregulation of arginase activity (Fig. 6C). Importantly, expression of the IL-4 receptor was intact in JUNB-deficient macrophages both at baseline and following treatment with IL-4, as was expression of another M(IL-4) target, Mrc1 (Fig. 6D,E). These data indicate that JUNB-deficient macrophages are not generally unresponsive to IL-4 signaling; rather, JUNB appears to act downstream of the IL-4 receptor to positively regulate some aspects of M(IL-4) polarization.
Figure 6. JUNB modulates alternative activation markers in macrophages treated with IL- 4.
(A) BMDM were treated with LPS for 4 h and gene expression was measured on microarrays, as described in Figure 3A. Statistical significance was as described in the methods. (B-E) BMDM were treated for 4 h (B, D, E) or 18 h (C) with IL-4. Levels of the indicated mRNA targets were measured by RT-qPCR (B, D, E) and arginase enzyme activity in cell lysates was assessed (C). Results shown are means + SD and are representative of three (B) or two (C, D, E) experiments. *, p < 0.05. **, p < 0.01. ***, p < 0.001 by t-tests.
Discussion
In this study, regulatory network analysis on community-generated gene expression data led us to predict a role for the AP-1 transcription factor Junb in macrophage activation. Whereas previous studies in silico have suggested such a role without providing experimental validation, we have demonstrated here that JUNB regulates macrophage responses to a variety of immunostimulatory ligands. The defects we observed in JUNB-deficient macrophages included altered transcription of a subset of immune-related genes, decreased secretion of M(LPS)-associated cytokines in response to TLR ligands, and weakened M(IL-4) polarization following treatment with IL-4. It is notable that JUNB positively regulates both M(LPS) and M(IL-4) activation, since to our knowledge, virtually all previously described transcription factors with demonstrated roles in macrophage polarization specifically promote either M(LPS) or M(IL-4) activation, with the exception of C/EBPα (38). Importantly, at baseline, JunbΔLyz2 mice exhibit no gross defects in survival or development, suggesting that deletion of JUNB does not disrupt the homeostatic functions of monocytes and tissue macrophages but specifically affects activation. This selectivity raises the possibility that therapeutic targeting of JUNB in macrophages, for example through use of liposomes to deliver drugs selectively to phagocytic cells, might be an effective way to dampen harmful inflammatory responses while minimizing the toxicity associated with systemic inhibition of the canonical c-JUN:c-FOS AP-1 pathway (39).
Our results deepen our understanding of the contributions of individual AP-1 subunits to innate immune responses. Previous studies have provided conflicting data as to whether JUNB is a negative regulator of gene expression, a competitive inhibitor of JNK-mediated phosphorylation of c-JUN, or a transcriptional activator in its own right. Although c-JUN and JUNB appear to play antagonistic roles in processes such as cell cycle progression, JUNB can also compensate to transcribe c-JUN target genes in the absence of c-JUN (23, 40). Furthermore, during CD4+ T cell and macrophage activation, JUNB appears to act as a transcriptional activator even in the presence of c-JUN ((32) and this work). Although previous studies of AP-1 signaling in macrophages have largely focused on c-JUN and c-FOS, our data demonstrate that JUNB also shapes macrophage transcriptional responses to LPS and other stimuli. Together with previous studies, our results suggest that c-JUN and JUNB both serve as positive and non-redundant transcription factors in the induction of immune-related genes in macrophages.
The kinetics of JUNB-dependent transcription may offer a clue to how this transcription factor is regulated. Whereas expression of Il12b and Il1b was unaffected by the absence of JUNB in the first two hours after LPS treatment, defects in transcription emerged in JUNB-deficient macrophages between two and four hours after stimulation. The time frame of the transcriptional defect tracks with the expression pattern of JUNB itself: we were not able to detect JUNB in resting macrophages by Western blot, but observed robust accumulation of Junb mRNA and JUNB protein by four hours after LPS stimulation. It is possible, therefore, that JUNB activity is regulated primarily at the level of expression, although phosphorylation by the upstream MAP kinase JNK has been reported in T cells (32).
Unlike an amplifier that promotes bistability, our data are consistent with a model in which JUNB serves to fine-tune the output of other transcription factors, including c-JUN. Several lines of evidence support this model: first, most (~70%) of the LPS-induced genes whose expression was defective in the absence of JUNB experienced only a modest (two- to three-fold) decrease in expression. Second, in contrast to c-JUN, JUNB appears to activate only a subset of promoters containing AP-1 sites (32). Finally, a previous study in LPS-stimulated DCs proposed that JUNB be considered one of several “Primer” factors that act in concert with a variety of other lineage- and environment-specific transcription factors to regulate the transcriptional output of the cell (9).
Importantly, the bioinformatic analysis that initially led us to examine JUNB in macrophage activation was performed on publicly available microarray data from a wide range of studies, each with its own set of protocols and sample treatments. Our work thus serves to demonstrate that despite the technical challenges inherent in starting with heterogeneous data, meaningful biological interactions can be identified with proper data processing. As increasingly extensive and diverse datasets become available, many of which introduce the additional variable of genetic heterogeneity across humans, future studies should not be deterred by these limitations, which can be overcome with sufficiently large datasets and appropriate analysis approaches.
The network method described herein is also notable in that it permits enrichment for physiologically relevant predictions, as we have demonstrated through extensive experimental validation of one of these predictions. Furthermore, in revealing a novel cell-type-specific function for JUNB in innate immune responses, this study lays the groundwork for future therapeutic targeting of the inflammatory response modulated by this key regulator.
Supplementary Material
Acknowledgments
We thank E. Passegué (UCSF) for Junbfl/fl mice; D. Burdette and R. Vance (UC Berkeley) for cyclic di-GMP; and J. DeRisi and D. Poscablo for technical assistance.
Abbreviations used in this article
- BMDM
bone marrow derived macrophage
- DC
dendritic cell
- cDC
conventional dendritic cell
- pDC
plasmacytoid dendritic cell
- RPM
red pulp macrophage
- pI:C
poly-inositol:cytosine
- GEO
Gene Expression Omnibus
- ChIP-Seq
chromatin immunoprecipitation and sequencing
- TLR
Toll-like receptor
References
- 1.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margolin AA, Wang K, Lim WK, Kustagi M, Nemenman I, Califano A. Reverse engineering cellular networks. Nat Protoc. 2006;1:662–671. doi: 10.1038/nprot.2006.106. [DOI] [PubMed] [Google Scholar]
- 6.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 7.Nemenman I, Escola GS, Hlavacek WS, Unkefer PJ, Unkefer CJ, Wall ME. Reconstruction of metabolic networks from high-throughput metabolite profiling data: in silico analysis of red blood cell metabolism. Ann N Y Acad Sci. 2007;1115:102–115. doi: 10.1196/annals.1407.013. [DOI] [PubMed] [Google Scholar]
- 8.Taylor RC, Acquaah-Mensah G, Singhal M, Malhotra D, Biswal S. Network inference algorithms elucidate Nrf2 regulation of mouse lung oxidative stress. PLoS Comput Biol. 2008;4:e1000166. doi: 10.1371/journal.pcbi.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, Blecher-Gonen R, Bornstein C, Amann-Zalcenstein D, Weiner A, Friedrich D, Meldrim J, Ram O, Cheng C, Gnirke A, Fisher S, Friedman N, Wong B, Bernstein BE, Nusbaum C, Hacohen N, Regev A, Amit I. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege J-L, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL, Lynn DJ. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatr-Aryamontri A, Breitkreutz B-J, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O’Donnell L, Reguly T, Breitkreutz A, Sellam A, Chen D, Chang C, Rust J, Livstone M, Oughtred R, Dolinski K, Tyers M. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanamori M, Konno H, Osato N, Kawai J, Hayashizaki Y, Suzuki H. A genome-wide and nonredundant mouse transcription factor database. Biochem Biophys Res Commun. 2004;322:787–793. doi: 10.1016/j.bbrc.2004.07.179. [DOI] [PubMed] [Google Scholar]
- 14.Bovolenta LA, Acencio ML, Lemke N. HTRIdb: an open-access database for experimentally verified human transcriptional regulation interactions. BMC Genomics. 2012;13:405. doi: 10.1186/1471-2164-13-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinforma Oxf Engl. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim CC, Nelson CS, Wilson EB, Hou B, DeFranco AL, DeRisi JL. Splenic Red Pulp Macrophages Produce Type I Interferons as Early Sentinels of Malaria Infection but are Dispensable for Control. PloS One. 2012;7:e48126. doi: 10.1371/journal.pone.0048126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods San Diego Calif. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheibel M, Klein B, Merkle H, Schulz M, Fritsch R, Greten FR, Arkan MC, Schneider G, Schmid RM. IκBβ is an essential co-activator for LPS-induced IL-1β transcription in vivo. J Exp Med. 2010;207:2621–2630. doi: 10.1084/jem.20100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jochum W, Passegué E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 24.Yao J, Mackman N, Edgington TS, Fan ST. Lipopolysaccharide induction of the tumor necrosis factor-alpha promoter in human monocytic cells. Regulation by Egr-1, c-Jun, and NF-kappaB transcription factors. J Biol Chem. 1997;272:17795–17801. doi: 10.1074/jbc.272.28.17795. [DOI] [PubMed] [Google Scholar]
- 25.Napolitani G, Bortoletto N, Racioppi L, Lanzavecchia A, D’Oro U. Activation of src-family tyrosine kinases by LPS regulates cytokine production in dendritic cells by controlling AP-1 formation. Eur J Immunol. 2003;33:2832–2841. doi: 10.1002/eji.200324073. [DOI] [PubMed] [Google Scholar]
- 26.Nakahara T, Uchi H, Urabe K, Chen Q, Furue M, Moroi Y. Role of c-Jun N-terminal kinase on lipopolysaccharide induced maturation of human monocyte-derived dendritic cells. Int Immunol. 2004;16:1701–1709. doi: 10.1093/intimm/dxh171. [DOI] [PubMed] [Google Scholar]
- 27.Schorpp-Kistner M, Wang Z-Q, Angel P, Wagner EF. JunB is essential for mammalian placentation. EMBO J. 1999;18:934–948. doi: 10.1093/emboj/18.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 29.Passegué E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Gomard T, Michaud H-A, Tempé D, Thiolon K, Pelegrin M, Piechaczyk M. An NF-kappaB-dependent role for JunB in the induction of proinflammatory cytokines in LPS-activated bone marrow-derived dendritic cells. PloS One. 2010;5:e9585. doi: 10.1371/journal.pone.0009585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu C, Gagnidze K, Gemberling JH, Plevy SE. Characterization of an activation protein-1-binding site in the murine interleukin-12 p40 promoter. Demonstration of novel functional elements by a reductionist approach. J Biol Chem. 2001;276:18519–18528. doi: 10.1074/jbc.M100440200. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.So AY-L, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 37.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee B, Qiao L, Lu M, Yoo HS, Cheung WW, Mak R, Schaack J, Feng G-S, Chi N-W, Olefsky JM, Shao J. C/EBPα Regulates Macrophage Activation and Systemic Metabolism. Am J Physiol Endocrinol Metab. 2014 doi: 10.1152/ajpendo.00002.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 40.Passegué E, Jochum W, Behrens A, Ricci R, Wagner EF. JunB can substitute for Jun in mouse development and cell proliferation. Nat Genet. 2002;30:158–166. doi: 10.1038/ng790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.