Abstract
Nucleic acids hold promise as biomolecules for future applications in biomedicine and biotechnology. Their well-defined structures and compositions afford unique chemical properties and biological functions. Moreover, the specificity of hydrogen-bonded Watson-Crick interactions allows the construction of nucleic acid sequences with multiple functions. In particular, the development of nucleic acid probes as essential molecular engineering tools will make a significant contribution to advancements in biosensing, bioimaging and therapy. The molecular beacon (MB), first conceptualized by Tyagi and Kramer in 1996, is an excellent example of a double-stranded nucleic acid (dsDNA) probe. Although inactive in the absence of target, dsDNA probes can report the presence of a specific target through hybridization or specific recognition-triggered change in conformation. MB probes are typically fluorescently labeled oligonucleotides that range from 25 to 35 nucleotides (nt) in length, and their structure can be divided into three components: stem, loop and reporter. The intrinsic merit of MBs depends on predictable design, reproducibility of synthesis, simplicity of modification, and built-in signal transduction. Using resonance energy transfer (RET) for signal transduction, MBs are further endowed with increased sensitivity, rapid response and universality, making them ideal for chemical sensing, environmental monitoring and biological imaging, in contrast to other nucleic acid probes. Furthermore, integrating MBs with targeting ligands or molecular drugs can substantially support their in vivo applications in theranositics.
In this review, we survey advances in bioanalytical and biomedical applications of rationally designed MBs, as they have evolved through the collaborative efforts of many researchers. We first discuss improvements to the three components of MBs: stem, loop and reporter. The current applications of MBs in biosensing, bioimaging and therapy will then be described. In particular, we emphasize recent progress in constructing MB-based biosensors in homogeneous solution or on solid surfaces. We expect that such rationally designed and functionalized MBs will open up new and exciting avenues for biological and medical research and applications.
Graphical abstract
1. Introduction
Nucleic acids, especially DNA bases, make an ideal framework for the molecular engineering of probes for various unique applications in bioanalysis. Simplicity of synthesis, suitability for structural modification, and high selectivity and affinity are the hallmarks of nucleic acids. Specifically, DNA molecules can be engineered to efficiently recognize target nucleic acids and other molecules, such as proteins and small molecules. Advances in molecular biology and the chemical synthesis of nucleic acids have benefited the development of nucleic acid probes, many types of which have been designed and applied in the fields of biology, chemistry, and medicine.1-3 For instance, nucleic acid probes, in particular those based on DNA, are essential tools for exploring the biological processes of nucleic acid duplication, recombination, transcription and expression. In the postgenomic era, there is a continuing demand for highly sensitive and selective DNA probes, and many kinds of DNA probes have been developed in recent years through various molecular engineering strategies.4
First reported by Tyagi and Kramer in 1996, molecular beacons (MBs) are oligonucleotide hybridization probes that can report the presence of specific nucleic acids in homogeneous solutions.5 These single-stranded DNA molecules consist of a stem-and-loop structure doubly labeled with a fluorophore and a quencher group on each end. In the absence of targets, MBs act like switches that are normally closed by the stem part, and in the “off” position, little fluorescence background is observed because of quenching. However, upon binding with their targets, conformational changes open the hairpin, and fluorescence is turned “on”. MBs are characterized by simple operation and high sensitivity and specificity. 6-8 As such, they have become a class of nucleic acid probes widely used in biosensing, bioimaging and therapy.
In this review, we describe efforts to explore and construct rationally designed MBs based on improvements in their three components: loop, stem and reporter. We first show how, for example, random single-stranded DNA (ssDNA), aptamers or DNAzymes are employed in the engineering of the loop region. Then we discuss how the stem is modulated by special DNA structures, metal ions and small molecules, as well as by external physical triggers and nucleotide analogues. As reporters, novel signal transduction strategies based on different regimes endow the rationally designed MB with low background and amplified signals. With this background, the applications of MBs in biosensing, bioimaging and therapy will be described.
2. Fundamentals and Design of Molecular Beacons
MBs contain loop, stem and signal reporter, as shown in Figure 1. (1) Loop: The probe sequence is the real determinant of MB specificity, and the length of the probe sequence should be such that it dissociates itself from the target at a temperature 7-10 °C higher than the annealing temperature of polymerase chain reaction (PCR).9-10 The melting temperature of the probe-target hybrid can be predicted using the percentage of guanine and cytosine (GC) content, which can be calculated with commercial software packages like Oligo 6.0.11-12 In general, the length of the probe sequence should range from 15 to 30 nucleotides and not form any secondary structure.13-14 If, however, this does happen, then the frame of the probe can be moved along the target to reach a nonself-complementary sequence. Increase in probe length results in improved affinity, but also leads to reduced specificity. (2) Stem: As a “lock” to maintain the closed hairpin structure of the MB in the absence of target, the stem portion of MBs usually involves Watson-Crick hydrogen bonding of natural DNA base pairs. The criteria for stem sequence include length, sequence and GC content. The stem should have a melting temperature 7-10 °C higher than the detection temperature.15 Maximum stability of MBs is achieved with 15-25 base sequences together with 5-7 bps in the stem.16-17 (3) Reporter: Reporters generally consist of two elements: fluorophore and quencher. Many different fluorophores have been tested for their efficiency, depending on the quencher group. Flexibility in the use of reporter dye expands the use of MBs in multiplex detection reactions where multiple targets can be distinguished in the same solution. Proper selection of fluorophore is critical for improved signal-to-background ratio. Recently, dyes with special properties, such as pyrene and lanthanide ion complexes, have been employed as effective signal transduction output tools to construct MBs.18-23 Capture and transfer of energy from an excited fluorophore is termed quenching, and the substances involved in it are termed quenchers. A commonly used quencher, 4-(4′-dimethylaminophenylazo) benzoic acid, or Dabcyl, is a nonfluorescent chromophore that can serve as a universal quencher for a variety of fluorophores. It optimally quenches fluorescein, but its quenching efficiency decreases to 93-98 % for dyes emitting longer wavelengths.24 The use of metal ions has made it possible to use varioust types of quenchers with different properties.25 Various nanomaterials, such as gold nanoparticles(AuNPs), single-wall carbon nanotubes (SWNT) and graphene oxide (GO), have higher efficiency as quenchers.26-29
Figure 1.
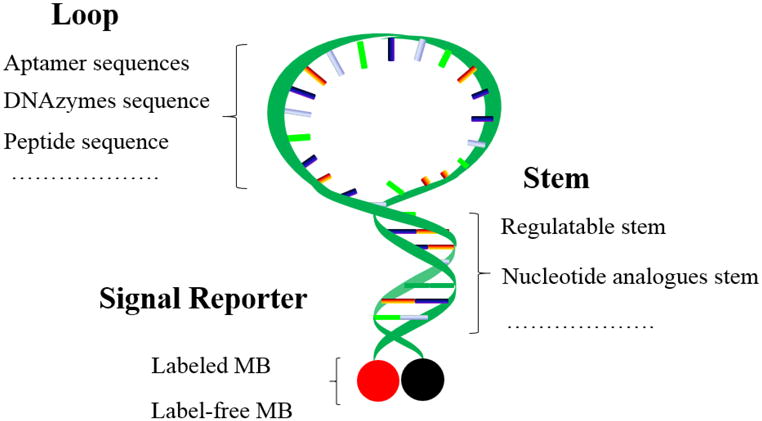
Molecular beacon structure and rationally designed strategies for different parts.
3. Rationally Designed Molecular Beacons
The chemical nature of nucleic acids allows their easy synthesis and modification. As a result, various ways have been proposed to engineer MBs with advanced bioavailability, regulating ability, and multifunctional properties. Herein, we discuss the efforts to explore and construct rationally designed MBs to boost performance, expand targeting repertoire, and incorporate additional functions.
3.1 Loop
MBs contain a target-binding region (loop) flanked by two complementary stem sequences. Accordingly, the thermodynamic and kinetic properties of MBs are, to a large extent, related to the favorability of the loop-target hybridization event, and the loop domain spans an 18-30 single-strand region which is complementary to the target sequence, as shown in Figure 2A. The loop domain is typically used as the target-binding region, but it is limited to the detection of oligonucleotide-binding targets.
Figure 2.
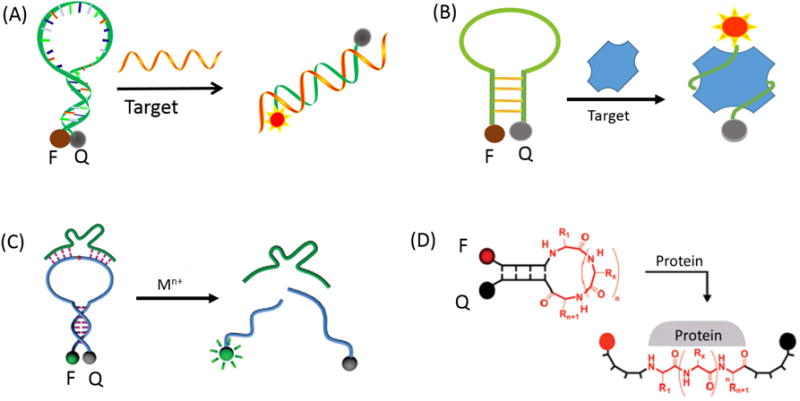
Loop region engineering: (A) Schematic representation of the recognition mechanism for the random DNA; (B) Representation of molecular aptamer beacon; (C) Representation of DNAzyme-based molecular beacon. Reprinted with permission from ref. 36. Copyright (2010) American Chemical Society. (D) Representation of hairpin peptide molecular beacon. Reprinted with permission from ref. 38. Copyright (2007) American Chemical Society.
3.1.1 Aptamers for Functional Loop Portion
Aptamers are single-stranded oligonucleotides that possess high stability and selectivity for specific targets generated through a technology termed “systematic evolution of ligands by exponential enrichment” (SELEX).30-32 Immense combinatorial libraries that contain trillions of different sequences are used to select different aptamers against a variety of targets, including metal ions, metabolites, proteins, and even whole cells. A novel nucleic acid probe, termed molecular aptamer beacon (MAB), has been developed by combining the high binding affinity of aptamers with the sensitive signal transduction of MBs, as shown in Figure 2B. For example, to investigate protein-DNA interaction, our group developed a MAB that targets single-stranded DNA-binding (SSB) protein.30 Apart from intact MABs, split MABs have been constructed by dividing the MAB sequences into two subunits. In this design, the two subunits of split MBs can be induced to assemble a target-MAB complex upon target addition. In addition, our group developed a novel dual-pyrene-labeled MAB, which can bind to target protein platelet-derived growth factor (PDGF)-BB with high affinity.21 When the dual-pyrene-labeled MAB is free in solution without the target protein, the pyrene molecules are spatially separated, and only the monomer emission peaks can be observed. However, binding between MAB and target protein brings the pyrene molecules at the 3′- and 5′-ends together, allowing the formation of an excimer.
3.1.2 DNAzyme for Functional Loop Portion
DNAzymes, also called catalytic DNA or deoxyribozymes, are also functional DNA molecules selected in vitro. 33-35 The active sites of DNAzymes can distinguish substrates at the atomic level by short DNA strands. Meanwhile, their backbones are negatively charged and have a certain flexibility that exposes their bases to the outside, achieving, in turn, highly efficient catalysis. Thus, in many sensing platforms, DNAzymes have been implemented as the loop region of MBs for the construction of catalytic molecular beacons (CAMB), as shown in Figure 2C.36 This CAMB sensing system has lower background fluorescence signal, improved signal-to-noise ratio, and, thus, higher sensitivity compared to linear substrate sensing systems. Most importantly, in this construction, the DNAzyme strand is liberated from its role as quencher and can be readily adapted to sense a broad range of targets, including metal ions and organic molecules. Zhang et al. first reported a general CAMB strategy that combines the highly efficient quenching of MB with amplified sensing through enzymatic turnovers provided by the catalytic beacon, a design which allows detection of metal ions, such as lead ion (Pb2+), with high sensitivity.
3.1.3 Using Special Recognition Sites for Functional Loop Portion
In another class of novel MBs, a special recognition site, such as a disulfide or peptide sequence, is embedded in the loop sequence for the detection of biomolecules.37-38 For instance, Qu et al. proposed a disulfide-bonded MB (SSMB), in which the loop consists of single-strand DNA sequences, including a disulfide bond, and the fluorophore/quencher pair is labeled at the 5′- and 3′-termini of the hairpin structure.37 In this design, the disulfide bond is able to react efficiently via a thiol-disulfide exchange reaction upon the addition of glutathione (GSH). This leads to the dissociation in solution of the two fragments lacking sufficient base pair stability. Under these conditions, quantitative analysis of intracellular GSH was realized in K562 cells. Seitz et al. proposed a hairpin peptide MB (HPMB) consisting of a central, protein-specific peptide sequence flanked by two DNA-analogous self-complementary peptide nucleic acid (PNA) arm segments (Figure 2D).38 The hairpin-like structure enforces interactions between terminally attached chromophores, resulting in structural reorganization upon target binding. The results obtained with two different protein targets and two different combinations of labels suggest a broad applicability. In addition to these recognition sites, more special chemical bonds need to be explored to construct MBs with advanced functionality.
3.2 Stem
After selecting the probe sequence, two complementary arm sequences are added on either side of the probe sequence to serve as the stem sequence. Since the stem is created by intramolecular hybridization, the melting temperature of the stem cannot be predicted by the percent-GC rule. Stem structure is a key factor in MB design. However, once the stem is formed, its strength cannot be easily changed to meet the needs of specific target binding. To address this issue, various new strategies have been advanced to improve the controllability of stem design, and some of these approaches are described below.
3.2.1 Modulating Stem Sequence by Special DNA Structure
One approach would modulate the stem sequence by incorporating a G-quadruplex motif in the stem region.39 Both monovalent and divalent cations (e.g, K+, Na+ and Mg2+) can be used in this type of MB, as shown in Figure 3A. Mismatch discrimination could be achieved at lower temperatures and over a broader temperature range compared to MBs based solely on duplex formation. A typical application of this type of quadruplex-MB is the recently developed DNAzyme MB.40 Two separate oligonucleotides that self-assemble into a G-quadruplex structure with hemin were employed as the catalytic unit for generating amplified signals. Without terminal dye modification, this new colorimetric DNA detection system could achieve a detection limit of 1 pM, 3-5 orders of magnitude lower than that of normal DNAzyme MB detection methods. Subsequently, Mohanty et al. reported that thioflavin T (ThT) could specifically bind to a DNA quadruplex derived from the human telomeric repeat sequence (HTG), generating a 2100-fold fluorescence enhancement.41 Inspired by these results, Bruchez et al. envisioned using this interaction to generate a label-free MB with the HTG/ThT complex, providing a fluorogenic signal.42 By extending the HTG sequence to promote hairpin formation, folding of HTG into a quadruplex and subsequent binding of ThT are suppressed, while disruption of the stem by target binding liberates the HTG and allows fluorogenic association with ThT in solution. As a label-free sensor, this ThT-based MB (TMB) has been shown to effectively detect DNA, RNA and protein.43
Figure 3.
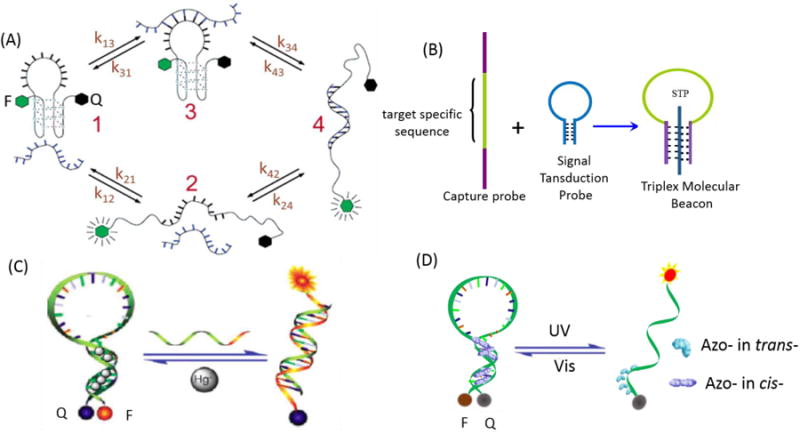
Stem region engineering: (A) G-quadruplex motif-based molecular beacon. Reprinted with permission from ref. 39. Copyright (2006) American Chemical Society. (B) DNA triple helix-based molecular beacon; (C) Metal ion-modulated molecular beacon. Reprinted with permission from ref. 52. Copyright (2009) Royal Society of Chemistry. (D) Photo-regulated molecular beacon.
The DNA triple helix is another useful special structure-regulating motif in the design of MB stems.44 It can be formed in two main ways. First, an oligopyrimidine strand can bind to the major groove of the duplex parallel to the strand carrying the purine tract using Hoogsteen base-pairing. Second, an oligopurine can bind to the purine strand in an antiparallel orientation using reverse Hoogsteen base-pairing.45-47 Extending this design, our group reported a new type of aptamer-based triplex MB which consists of a central, target-specific aptamer sequence flanked by two arm segments and a dual-labeled oligonucleotide serving as a signal transduction probe (STP), as shown in Figure 3B.48 When the aptamer's two arm segments bind with the loop sequence of STP, the STP forms an “open” configuration, while formation of an aptamer/target complex releases the STP to produce new signal readouts.
3.2.2 Modulating Stem Sequence by Metal Ions and Small Molecules
Since the binding of mercury ion (Hg2+) by thymine-thymine (T-T) pairs is strong and highly selective, duplexes that contain a T-T pair are thermally stabilized in the presence of Hg2+.49-51 Taking advantage of metal-dependent pairing, our group developed a MB whose target binding properties could be controlled by Hg2+. As such, Hg2+ could selectively bind between two T bases and promote these T-T mismatches to form stable T-Hg2+-T base pairs, as shown in Figure 3C.52 By adjusting the concentration of Hg2+, MBs were made to work properly at different temperatures with the desired kinetic response and selectivity. Furthermore, to reduce toxicity and shorten analysis time, an alternative MB was developed by our group by replacing the T-Hg2+-T structure with the C-Ag+-C structure.53 Besides these metal ions, Tseng et al. introduced an adenosine-coralyne-adenosine-based MB. In this design, the presence of coralyne promotes A-A mismatches to form stable A2-coralyne-A2 complexes in the stem, leading to the conformational change of A12-MB-A12 from a random-coil structure to a hairpin structure.54-55
Our group also proposed a way to restrict the labeled dyes to the hydrophobic cavity of cyclodextrin (CD).56 This bonding, which acts like extra base pairs to form the Watson-Crick duplex, achieves variation in the level of spatial proximity of the two labels and thus the degree of conformational constraint. This inclusion interaction of γ-CD with stem labels offers unprecedented, new, easy and predictable degrees of freedom to tune stem stability, allowing for the facile introduction of further functionalities of DNA probes to increase signal-to-background ratio and improve target binding specificity for multiplexing.57-58
3.2.3 Modulating the Stem Sequence by External Stimulus
The design of MBs responsive to external physical triggers is another direction for stem engineering. Compared with a chemically triggered response, photoregulation enables remote control of the timing, location, intensity and dosage of light, while, at the same time, minimizing the adverse effects of other chemical components. Our group designed an MB incorporated with azobenzene moieties in the stem to facilitate reversible photocontrollable switching (Figure 3D).59 The results suggest that the azobenzene-incorporated MBs have potential for applications which require highly efficient light-driven molecular motors. Yang et al. have carried forward this idea by developing “caged” MBs.60 In this design, either biotin-avidin interactions, or triazole linkages, were utilized to lock the MB stems via a photocleavable linker bearing an o-nitrobenzyl moiety. Opening of the “cage” by seconds of UV-light irradiation efficiently recovered the target binding affinity of the MBs. Besides photoinduced regulation, the design of other external triggers, even multiple triggers, can be a promising direction for future MB stem engineering.
3.2.4 Combination with Nucleotide Analogues
Special modified nucleotide analogues can also modulate the MB stem sequence, as shown in Figure 4. Our group investigated, designed, and synthesized a new locked nucleic acid (LNA)-based MB which contains one or more LNA nucleotide monomers with a bicyclic furanose unit locked in an RNA mimicking sugar conformation.61-62 The methylene bridge connecting the 2′-oxygen of the ribose and the 4′-carbon endows LNA with many attractive properties, such as high binding affinity, excellent base mismatch discrimination capability, and decreased susceptibility to nuclease digestion, properties which can be extended to MB probes, making them more robust tools in bioapplications. For instance, such LNA-MB not only functioned at room temperature but also hybridized with its target at 90°C, implying its high affinity.
Figure 4.
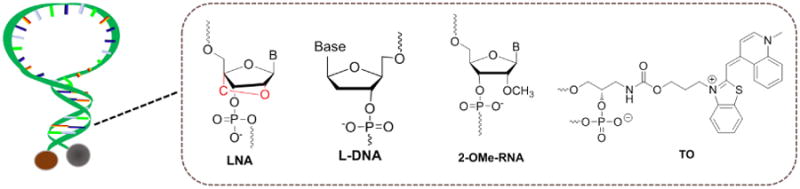
Stem region engineering combined with nucleotide analogues: structures of LNA, L-DNA, 2-OMe-RNA and TO.
Following that, our group also created a modified MB that incorporates unnatural enantiomeric L-DNA or 2′-O-Me-modified RNA in the stem. 63-64 L-DNA has the same physical characteristics as D-DNA, except that L-DNA cannot form stable duplexes with D-DNA. The incorporation of L-DNA into the stem region of MB can reduce intra- and intermolecular stem invasions, thereby increasing the melting temperature, improving selectivity to its target and enhancing biostability. Recently, Yang et al. employed an L-DNA-based MB for the photothermal study of palladium (Pd)-nanosheets in living cells, establishing the nano-thermometer as a useful tool for intracellular temperature measurement.65
Thiazole orange (TO) represents another promising oligonucleotide alternative which can be linked covalently to the phosphodiester or to the 5′-terminus of DNA.66-68 The stem stability of MB could be enhanced when TO dimer was embedded as an artificial DNA base.69 Similarly, an MB that incorporates components of an artificially expanded genetic information system in its stem was proposed by our group, and the result demonstrated that the MB could not be opened by unwanted stem invasion by adventitious standard DNA. This design can improve the “darkness” of the beacon in real-world applications.70
3.3 Reporter
In the presence of the target molecule, the loop region of traditional MBs forms a hybrid helix that is longer and more stable than the stem helix. This interaction forces MBs to undergo a conformational change, which separates the stem helix. Because the quencher is no longer positioned near the fluorophore, fluorescence is restored and signals the binding of MB to its target. Conventional MBs use organic fluorophores and quenchers, such as fluorescein or Dabcyl. However, incomplete quenching and limited sensitivity still hinder the further development of conventional MBs. To solve these problems, rationally designed fluorescent MBs, phosphorescent MBs and label-free MBs, among other approaches, have been proposed.
3.3.1 Labeled-Molecular Beacons
Some of the major drawbacks of conventional MBs based on fluorophore-quencher pairs are residual fluorescence from incomplete quenching and limited sensitivity. To solve these problems, an in-stem molecular beacon (ISMB) has been developed. In this design, threoninol nucleotides attached to a fluorophore and a quencher are incorporated into the stem region as pseudo base pairs (Figure 5A).71 In the absence of target DNA, the fluorophore and quencher are stacked together in the middle of the stem region of the closed beacon, and background fluorescence is greatly suppressed as a result of the close stacking. In contrast, the presence of target generates strong emission from the intercalated fluorophore. As a result, the signal-to-noise ratio is greatly improved relative to that of conventional MBs.
Figure 5.
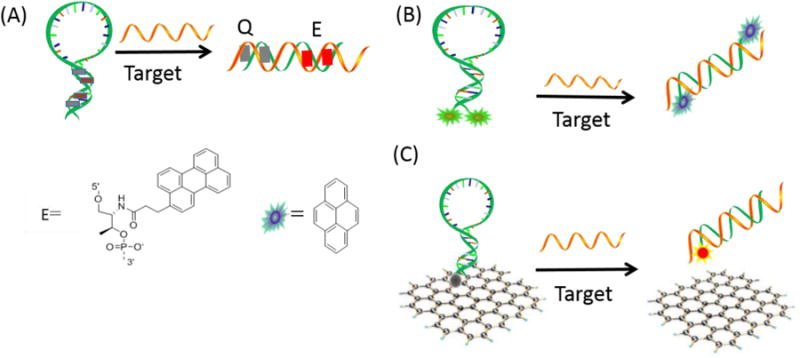
Reporting region engineering of fluorescent molecular beacon. (A) ISMB design and structures of candidate perylene quenchers; (B) EMB; Schematic representation of dual-pyrene-labeled EMB. (C) Schematic representation of target-induced fluorescence change of GO-quenched molecular beacon.
MBs with two fluorophores, instead of a fluorophore-quencher pair, have also been designed. The excimer molecular beacon (EMB) is a typical class of multiple fluorophore-labeled MB. EMB is a dual-pyrene-labeled hairpin DNA structure with large Stokes shift and long fluorescence lifetime, a design which affords an effective strategy for detection in complex biological environments. Two pyrene derivatives are modified on the 5′- and 3′-end of the typical EMB (Figure 5B). 72 In the absence of target DNA, the hairpin structure brings the two pyrene moieties into close proximity and allows the formation of an excimer that emits fluorescence. Our group engineered EMB probes that contain 1-4 pyrene monomers on the 5′-end and quencher Dabcyl on the 3′-end for real-time probing of DNA sequences. In the presence of target, the EMBs switch to a stem-open conformation, separating the pyrene label from the quencher molecule and generating an excimer emission signal proportional to the target concentration.19 Water soluble metallo-phthalocyanines (MPcs) are near-infrared fluorophores that possess high extinction coefficients, favorable quantum yields, and narrow absorption/emission envelopes and which display extraordinarily high photostabilities.73-74 These qualities of MPc dyes can provide dimer-based MB probes (DBMBs) with unique properties and high signal-to-background ratio.75
Using multiple quenchers to pair with one fluorophore is believed to provide better quenching efficiency because of the improved absorption efficiency and the increased probability of dipole-dipole coupling between the quenchers and the fluorophore as a result of a collective quenching effect. Using this as a working principle, our group designed a molecular assembly of superquenchers (SQs) for molecular interaction studies and for ultrasensitive bioanalysis.76 MBs with SQs are highly efficient in signaling the hybridization of MBs with target nucleic acids. With an increased number of Dabcyl moieties in the molecular assembly, the background fluorescence intensity decreased dramatically, and a 320-fold enhancement of fluorescent signal was achieved upon target addition, compared to about 14-fold from a conventional MB prepared with the same monomer quencher.
Other than multiple fluorophores or superquencher labeling, quencher-free MBs were designed using a monolabeled beacon structure via changes in the microenvironment of the fluorophores. Fluorescence quenching by nucleobases (normally G bases) has been reported previously, mostly via photoinduced electron transfer between the nucleobases and the excited state of the fluorophore.77 However, other factors, such as changes in solvent polarity, hydrophobicity around the fluorophores, and the pH of the media, must also be considered when designing quencher-free MBs.
Nanomaterials, such as metallic nanoparticles, semiconductor nanocrystals (quantum dots), carbon nanomaterials, silica nanoparticles and magnetic nanoparticles, exhibit unique optical, electronic, magnetic, and catalytic properties. These properties make them ideal candidates for signal reporter elements in constructing new biosensors. Recently, some nanomaterials which can serve as “nanoquenchers” have attracted considerable attention. These MB systems contain hairpin-structured, fluorophore-labeled oligonucleotides and special nanomaterials. For instance, our group used SWNTs as the “nanoquencher” for MBs, greatly improving the signal-to- background ratio compared to conventional MBs.27 Yang et al. also investigated the binding of MBs to nanoscale graphene oxide (NGO), and the results showed that NGO can also adsorb MBs and decrease their background fluorescence. This result indicates that the target DNA can hybridize with MB on the NGO surface followed by the release of MB from NGO (Figure 5C).28 AuNPs can also quench fluorescent dyes based on their efficient absorption of internal energy, transferred electrons, as well as long-range resonant energy transfer, otherwise known as FRET. By applying AuNPs to MBs, as a substitution for organic quenchers, and using either covalent or noncovalent modification, the signal-to-background ratio can be greatly improved. Dubertret et al. constructed the first AuNP-based MB by attaching a fluorophore-tagged stem loop probe at the surface of small AuNPs (1.4 nm), leading to high quenching efficiency toward a wide range of organic fluorophores.26 Later on, larger-sized AuNPs (13 nm) were functionalized with MBs to explore higher quenching efficiency and improved signal output.78
Fluorescence detection is quite sensitive and functions as the typical signaling method for MB recognition events. However, applications can be hindered by the background signals from ubiquitous endogenous fluorescent components in the biological environment.79-80 Moreover, such issues as photobleaching and blinking, as well as toxicity, can also restrict applications. Nonfluorescence detection methods have thus begun to gain some attention for MB engineering. For example, phosphorescence with a long emission triplet lifetime could be suitable for reducing background signals, since an elongated delay time can avoid the influences of short-lived background fluorescence and scattered light. However, during detection in complex biological systems, these MBs, similar to other fluorescence probes, suffer severely from background signal interference. To address this issue, our group developed a room temperature phosphorescence (RTP)-based MB. This MB contains a europium(III) (Eu3+) complex as a phosphorescent signaling reporter and black hole quencher 2 (BHQ-2) as a phosphorescent quencher (Figure 6).81 In the MB, the luminescence of the Eu3+ complex was quenched by BHQ-2 as a result of the stem-closed conformation of the MB. In the presence of a target, the MB switched to a stem-open conformation, which separated the luminescent complex from BHQ-2 and generated a Eu3+ emission signal proportional to the target concentration. Combined with RTP assays, this probe can directly and sensitively detect and quantify the target molecule in cell media with no need for sample cleanup.
Figure 6.
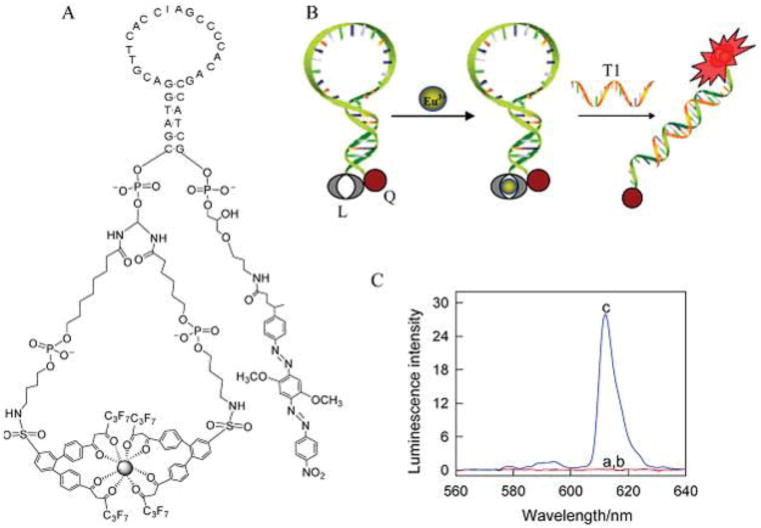
Reporting region engineering of phosphorescent molecular beacon. (A) Structure of the designed molecular beacon. (B) Signaling scheme of molecular beacon hybridization with complementary target DNA. (C) RTP emission spectra of molecular beacon before and after target DNA addition. Reprinted with permission from ref. 81. Copyright (2011) American Chemical Society.
Whereas optical detection methods have historically dominated state-of-the-art real-time or near real-time genosensors, the application of electrochemical methods to the sensing of biologically related species may provide very significant advantages. Specifically, the advantages of bioelectronics approaches include speed, sensitivity, and low cost/mass/power requirements for electrochemical detection; the relatively high stability and environmental insensitivity of electroactive labels; and the availability of electroactive labels with nonoverlapping redox potentials for multicolor labeling and the simultaneous detection of multiple analytes. Plaxco et al. firstly developed an electrochemical biosensor for sensitive DNA detection based on conformation changes in MBs.82 By monitoring the change of electron transfer efficiency, down to 10 pM target DNA could be measured by cyclic voltammetry. Grinstaff et al. constructed a “signal-on” electrochemical DNA sensor by using two short strand DNAs, one is the capture part and the other is the signaling part that is linked with a flexible PEG spacer.83 The hybridization of target DNAs to the triblock DNA probe induces a conformational change that brings the electrochemical reporter close to the surface of the gold electrode. This change of conformation leads to electrochemical “signal on” amplification that can be used for quantitative analysis of target DNAs. Besides target DNA, Fan et al. developed a target-responsive electrochemical aptamer switch (TREAS) approach for the detection of ATP and Plaxco et al. also proposed a “signal-off” electronic MAB sensor for the detection of thrombin.84-85
3.3.2 Label-Free Molecular Beacons
The labeling steps for MBs are sometimes challenging due to the relatively high cost, limited available probes, limited synthesis techniques, and time-consuming operation. Additionally, reduced sensitivity can occur if the affinity of MB for its target is affected by extra chemical modification. Therefore, to ensure high affinity while achieving a low-cost, rapid, simple and noncovalent method of detection, “label-free” strategies, such as colorimetry and surface enhanced Raman scattering (SERS), have begun to attract attention as promising alternatives to traditional MBs. For example, Ren et al. demonstrated a novel concept for a label-free, quadruplex-based functional MB by using the G-quadruplex motif as a substitute for Watson-Crick base pairing in the MB stem and a specific G-quadruplex binder, N-methylmesoporphyrin IX (NMM), as a reporter.86 This MB showed high sensitivity in assays for Uracil-DNA glycosylase (UDG) activity/inhibition and detection of DNA sequences based on the unique fluorescence increase that occurs as a result of the strong interaction between NMM and the folded quadruplex upon removal of uracil by UDG or displacement of block sequence by target DNA. Many groups have combined G-quadruplex DNAzyme with MBs to develop a novel bifunctional colorimetric oligonucleotide probe for DNA and other biomolecule detection.87-91 The oligonucleotide forms double stem-loops and a G-quadruplex structure in the absence of targets, yielding a catalytically active DNAzyme that promotes a process that leads to the generation of a colorimetric signal. In the presence of targets, the loops of the hairpin structures are combined and disrupted, which induces the G-quadruplex DNAzyme to dissociate, causing, in turn, a decrease of catalytic activity and enabling the separate analysis of biomolecules (Figure 7A). In addition, a horseradish peroxidase (HRP)-mimicking DNAzyme sequence was locked by a triplex-based MB. Upon target addition, switching of triplex-helix MB allowed the product strand to self-assemble into a hemin/G-quadruplex-HRP-mimicking DNAzyme able to biocatalyze the formation of a colored product, thus providing a flexible and simple method for analyte detection (Figure 7B).92 Different from covalent labeling of an electronic tag at the end of MABs, Kim et al. developed an electrochemical detection of thrombin by employing MABs with an intercalation tag.93 In the presence of thrombin, due to the conformation change of MAB when binding with thrombin, intercalated methylene blue tags were released, resulting in a decrease of current intensity. The detection limit of this method was 11 nM thrombin.
Figure 7.
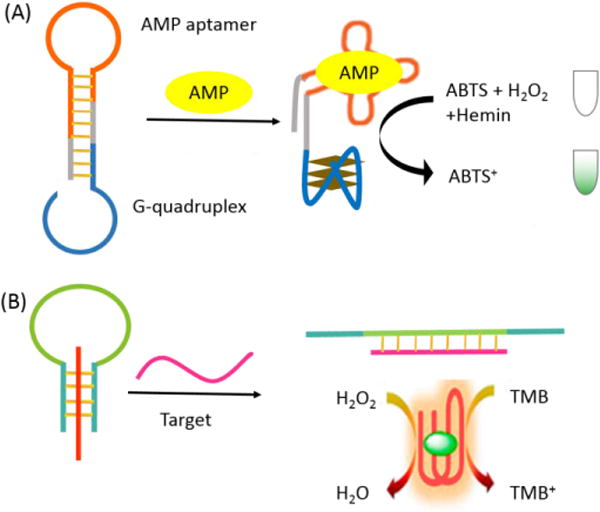
Reporting region engineering of label-free molecular beacon. (A) Analysis of adenosine monophosphate (AMP) by the aptamer-DNAzyme hairpin structure. (B) Construction and operation of the triplex molecular beacon using G-quadruplex sequence as the signal transduction element. The signals are generated from the released G-quadruplex upon target binding.
4. Bioanalytical and Biomedical Applications of Molecular Beacons
Because of their many advantages and unique properties, MBs are attractive systems for a variety of applications in biosensing, bioimaging and therapy. The scope of target molecules in these applications has been expanded from DNA and RNA to small molecules, proteins and even cancer cells.
4.1 Applications of Molecular Beacons in Biosensing
4.1.1 Gene-Detection Assays
Soon after MBs were first introduced, they found important applications in real-time polymerase chain reactions (RT-PCR), which require prompt signal production with high specificity.94-95 To monitor DNA amplification of the target sequence during the PCR process, MBs designed to hybridize with the forward/reverse PCR product are introduced into the PCR solution. With an increasing number of PCR cycles, a growing number of amplified target DNA molecules are produced and hybridize with the MB during the annealing stage (Figure 8A). This process generates increasing fluorescence from the MB, and this fluorescence provides a measure of PCR progress in real time.6-7 Besides PCR, Wang et al. also presented a sensitive platform based on a polymerase-induced isothermal strand-displacement polymerization reaction containing an MB probe for single-strand DNA detection.96 In addition, MBs have also been employed as effective reporters for the detection of rolling circle amplification (RCA) and nucleic acid sequence-based amplification (NASBA) products (Figure 8B).97-98
Figure 8.
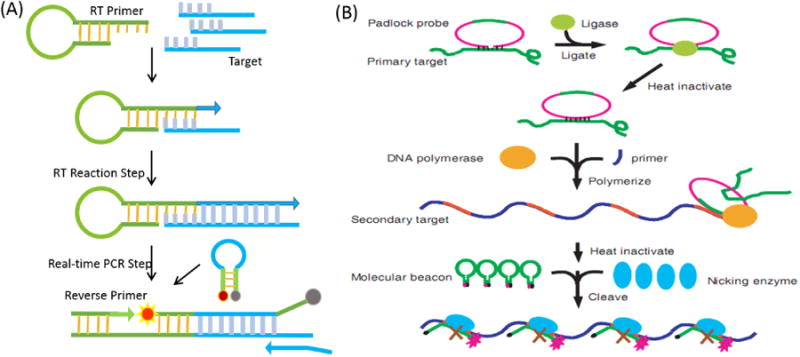
Molecular beacons serving as signal reporter for real-time PCR (A) and RCA-based (B) amplification gene assays. Reprinted with permission from ref. 97. With an increasing number of PCR or RCA cycles, a growing number of amplified target DNA molecules is produced and hybridize with the molecular beacon during the process. Copyright (2002) Oxford University Press.
Single nucleotide polymorphisms (SNPs) are single nucleotide variations in the genome, accounting for 80-90 % of all human genetic variations.99-101 Trace amounts of SNPs can be detected by PCR amplification. Thus, the combination of sensitive PCR methodology with selective MB probe design has generated a powerful method for real-time base-mutation analysis. In one study, MBs were used for point mutation detection in a methylenetetrahydrofolate reductase (MTHFR) gene, which represents an important marker for cardiovascular disease and neural tube defects.102 As the strands synthesized during PCR cycles accumulated, the number of MB probes that bound to the targets increased, leading to more intense fluorescence from the open fraction of MBs. The target specificity of the method was confirmed by the target selective signaling MB design. Li et al. designed a RT-PCR technique using a universal MB (U-MB) and a 5′-universal template primer (5′-UT primer) to achieve SNP detection.103 Alternative methods not requiring PCR amplification have recently used MBs to directly analyze SNP samples. For example, a ligase-based point mutation detection assay uses an allele-specific discriminating primer and a common primer, each having a 10 base-pair complementary arm with fluorescent labels at their 5′- and 3′-ends. A reverse MB (rMB) is formed by the complementary arm sequences of the ligated primers. Thus, real-time single pair fluorescence resonance energy transfer (spFRET) measurements were performed in a poly (methyl methacrylate) (PMMA) microfluidic device to detect rMBs formed in a ligation detection reaction (LDR) assay where point mutations in K-ras codon 12 were detected (Figure 9A).104 Additionally, various types of enzyme-based amplification methods have been employed with MBs for sensitive and specific SNP analysis.105-106 Among these enzymatic methods, DNA endonuclease-based approaches are the most widely used strategies. Zhang et al. recently developed an MB-based “Y”-shaped probe with strong SNP identification ability (Figure 9B).107 The Y-shaped junction consists of three parts: a signal MB probe, an assistant probe and target DNA. The MB probe contains the nicking endonuclease site for a Nt. BbvCI enzyme, which recognizes double strand DNA, but only hydrolyzes one specific strand (MB). By changing the target DNA-bound sequence of the MB and assistant probes, while retaining the enzyme cleavage region, this smart design guarantees universal DNA target detection with the added potential of SNP selectivity.
Figure 9.
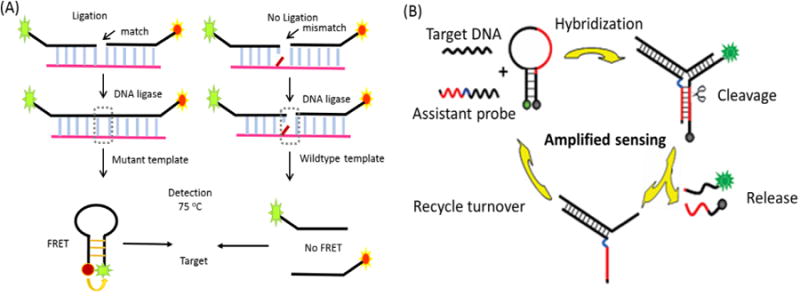
Molecular beacons serving as signal reporter for single nucleotide polymorphism detection. (A) A reverse MB (rMB) is formed by the complementary arm sequences of the ligated primers. Real-time single-pair fluorescence resonance energy transfer (spFRET) measurements were performed in a poly (methyl methacrylate) (PMMA) microfluidic device to detect rMBs formed in a ligation detection reaction (LDR) assay where point mutations in K-ras codon 12 were detected. (B) Design of a molecular beacon-based junction probe system for amplified single nucleotide polymorphism detection. Reprinted with permission from ref. 107. Copyright (2010) American Chemical Society.
4.1.2 Detection of Biomolecules in Solution
The use of MBs to investigate interactions of biomolecules has been achieved in past decades. The single-stranded DNA-binding protein (SSB) from E. coli was the first target biomolecule detected by an MB. Based on increased understanding of the characteristics and functions of MBs, a new molecular scheme was developed for the detection of small molecules, such as nicotinamide adenine dinucleotide (NAD) and ATP.108 The MB was designed to bridge two short oligonucleotides with its loop sequence. Under these conditions, the DNA ligase from E. coli will only catalyze the ligation of two short oligonucleotides in the presence of NAD. The ligation results in the opening of the MB and the recovery of the fluorescence signal (Figure 10A). Following this discovery, MB-based assays have been designed to study more specific and more sophisticated processes involving DNA and proteins, such as DNA ligation and phosphorylation, which are involved in DNA duplication, DNA recombination, and the repair of DNA damage.109
Figure 10.
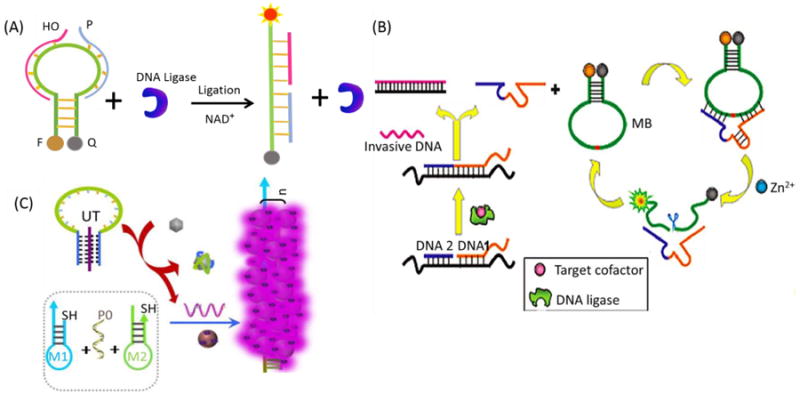
Molecular beacons used for detection of biomolecules in solution. (A) Analysis of NAD+ using molecular beacon and E. coli DNA ligase. (B) Strategy of the catalytic DNAzyme molecular beacon cascade for amplified fluorescence detection of small biological molecules. Reprinted with permission from ref. 116. Copyright (2011) American Chemical Society. (C) Schematic illustration of the structure and working principle of a triplex molecular beacon-based universal SERS detector with amplified signal for the detection of multiple target analytes. Reprinted with permission from ref. 117. Copyright (2014) American Chemical Society.
Since aptamers are selected by SELEX, which can bind any targets ranging from ions, small molecules and peptides to proteins, viruses and even whole cells, MABs have gained increasing attention in the field of molecular recognition to study various biomolecules. Takenaka et al. constructed a K+-sensing MAB for the specific detection of K+ in buffer solution.110 Two different dyes are labeled at the 5′- and 3′-termini of a K+-specific MAB. In the presence of K+, FRET can be observed and used as a quantitative tool for the detection of K+. Another cocaine-targeting MAB was reported by Stojanovic et al.111 In this design, a fluorophore and quencher were labeled on either end of a cocaine aptamer beacon. After addition of cocaine molecules, the MABs were able to fold into a three-way junction conformation to bring fluorophore and quencher into close proximity, resulting in decreased fluorescence intensity. Even in complex serum media, 10 μM of cocaine could be detected. A similar fluorophore-quencher labeling strategy was reported by Ellington et al. who constructed MABs for direct detection of thrombin.112 However, generalizability remains a problem since the recognition element of a MAB is aptamer sequence-specific and, hence, limited by the aptamer-binding target against which it is deployed. For our designed triplex MB, one STP can detect multiple targets by selecting different types of aptamers. This strategy allows for the facile introduction of various targets without increasing the complexity and cost of MB synthesis.56 By virtue of altering the aptamer sequence, the universality of the approach could be achieved, and three biomolecules, including thrombin, adenosine triphosphate (ATP) and L-argininamide (L-Arm), which are important substrates in the biological reaction, were detected.
To improve the sensitivity of biosensing for targets at lower concentration, multiple amplification platforms have been constructed. Willner's group provided an autonomous aptamer-based machine activated by polymerase, a nicking enzyme, and an MB for amplified detection of cocaine.113 Yang et al. have developed a target recycling amplification method for highly sensitive and selective microRNA (miRNA) detection.64 The combination of low fluorescence background from a 2-OMe-RNA-modified MB and nuclease-assisted signal amplification leads to ultra-high assay sensitivity, and the powerful discriminating ability of the MB enables the differentiation of highly similar miRNAs that differ by only one base. Zhang et al. developed a series of ligation-triggered DNAzyme cascades by combining a split DNAzyme-based background reduction strategy with CAMB-based amplification in one system, resulting in ultra-high sensitivity.114-116 Combining zero-background signal and CAMB-based signal amplification significantly improved the sensitivity of the sensing system, resulting in the ultra-sensitive detection of such biomolecules as ATP or NAD+, as shown in Figure 10B. To further achieve ultra-high sensitivity for multianalyte detection, our group described a universal triplex MB-based SERS biodetection approach combined with hybridization chain reaction (HCR) that uses a single-stranded DNA as a universal trigger (UT) to induce SERS-active hot-spot formation, allowing, in turn, detection of a broad range of targets (Figure 10C).117
4.1.3 Detection of Biomolecules on Solid Surfaces
The use of MBs as biosensors on solid surfaces is also expanding rapidly. Compared to MB assays performed in homogeneous phase, surface-immobilized MBs allow simultaneous detection of multiple targets through spatial encoding.118 A glass slide surface was the first solid surface chosen for immobilization of MBs. Our group designed LNA-incorporated MBs and immobilized them on a glass slide surface.119 Around 25-fold fluorescence enhancement was observed for these LNA-incorporated MBs. Taking advantage of the preferential exodeoxyribonuclease activity of exonuclease III in combination with the difference in diffusivity between an oligonucleotide and a mononucleotide toward a negatively charged ITO electrode, a highly sensitive and selective electrochemical MB (eMB)-based DNA sensor has been developed by Hsing and Tang et al. These two sensors can achieve electrochemical detection of DNA in homogeneous solution, with sensing signals amplified by an exonuclease III-based target recycling strategy.120-121
Even though microscope glass slides have been commonly used as a substrate for MB immobilization, they play only a passive role. Therefore, novel substrate materials with desired functions are in great demand. Among all potential substrates, the gold surface has become particularly important since it can be used as a quenching agent through RET or a “contact quenching” process. Quencher-free MBs can be immobilized onto a gold surface. For example, Krauss et al. have developed two prototype MB biosensors by anchoring MBs labeled with only a fluorophore onto a gold surface through standard gold-thiol chemistry.122 The quenching of the gold surface for both MBs was found to be more than 95%, which is comparable to that obtained in solution phase assays by using Dabcyl as the quencher. Based on the excellent quenching of the gold surface, more than 20-fold fluorescence enhancement was achieved, and complementary targets were detected down to 10 nM. Besides fluorescence detection, we fabricated our triplex MB on a gold film surface, and the central sequence of the capture probe was designed as an ATP aptamer, as shown in Figure 11A. 123 Initially, a “triplex-stem” MB structure is formed, and the R6G-functionalized silver nanoparticles (AgNPs) are physically separated from the gold surface, producing a SERS signal in the “off” state (low SERS effect). In the presence of target, however, the dye-functionalized AgNPs are brought close to the gold surface, which creates an intense electromagnetic field, thereby allowing detection of ATP down to 50 nM.
Figure 11.
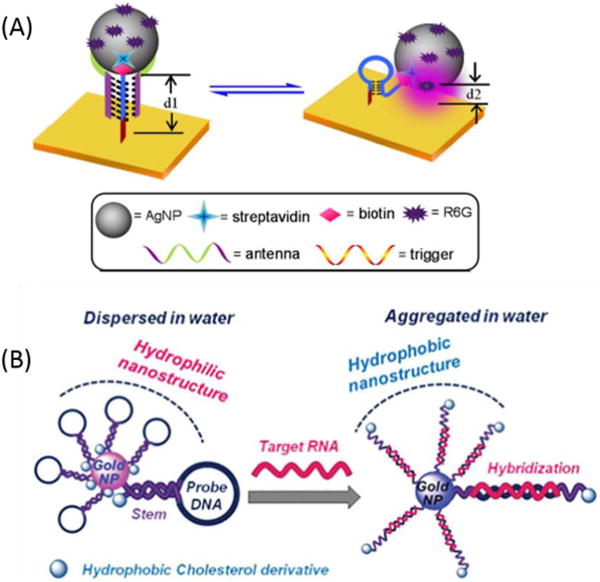
Molecular beacons used for detection of biomolecules on a solid surface. (A) Working principle of the triplex molecular beacon-based DNA nanomachine-directed reversible SERS-active substrate and corresponding reversible SERS “hot-spot” generation through assembly and disassembly of AgNPs on a gold film surface. Reprinted with permission from ref. 123. Copyright (2012) American Chemical Society. (B) Schematic representation of the AuNP sensor with hydrophobic molecular beacons. Reprinted with permission from ref. 125. Copyright (2014) Royal Society of Chemistry.
Nanomaterials are structures with a size of 100 nanometers or smaller in at least one dimension. Based on quantum effects that result from the large surface area-to-volume ratio, nanomaterials possess unique optical, electronic, magnetic and mechanical properties.124 Therefore, MBs have been immobilized onto different nanomaterial surfaces to develop multifunctional probes. Herein we will focus on AuNPs, quantum dots (QDs) and magnetic nanoparticles (MNPs). Somoza et al. reported a gene sensor using AuNPs modified with MB-like structures bearing a cholesterol derivative (Figure 11B).125 In the absence of the target sequence, the cholesterol molecules are buried inside the nanostructure, stabilizing the nanostructure in aqueous media. In the presence of the target sequence, the hairpin unfolds, exposing the cholesterol units to the water molecules. A significant change occurs in the solubility of nanoparticles when the cholesterol moiety is exposed to water molecules. This resulted in the detection of three target genes with specific single-point mutations involved in different diseases. Liu et al. have constructed a dry reagent strip-type nucleic acid biosensor based on an MB-modified AuNP probe and a lateral flow test strip. Using this sensor, target DNA as low as 50 pM was visualized within 15 min with great discrimination against single-base mismatched target.126 Tang et al. developed a multiplex nucleic acid detection assay using an MB-modified AuNP probe labeled with different fluorophores. In this proof-of-principle study, the loop sequences of MBs, which consisted of complementary sequences to three different tumor-suppressor genes, were labeled with FAM, Cy5, and Rox and then bound to 15 nm AuNPs.127 Using different targets on single particles, these MB-modified AuNPs could detect specific messenger RNA (mRNA) sequences and discriminated against single-base mismatched DNA.
Organic dyes have been widely used for MB construction. However, organic dyes are sensitive to the physiological environment and vulnerable to photobleaching under normal imaging conditions. In contrast, inorganic nanomaterials, such as QDs, are usually bright and stable, even under a relatively harsh environment.128-129 Importantly, QDs always emit the same wavelength, no matter what excitation wavelength is used.130 To demonstrate the feasibility of using QDs for MB construction, Zhang et al. demonstrated the excellent performance characteristics of a QD-FRET nanosensor for DNA detection with ultra-high sensitivity and excellent selectivity and simplicity.131 The detection limit of their nanosensing system is 100-fold lower than that of conventional FRET probe-based assays, as monitored by confocal fluorescence spectroscopy. In addition, with strong anti-photobleaching properties, QDs-based MBs are stable in complex environment, which means they will be very good candidates for intracellular imaging.
4.2 Applications of Molecular Beacons in Bioimaging
4.2.1 Intracellular Bioimaging
Development of sensitive, specific molecular probes is one of the central challenges in bioimaging. Combined with aptamers, which are small, polyanionic probes, MBs exhibit faster tissue penetration and uptake, shorter residence in blood and nontarget organs, and higher ratio of target accumulation, thus affording high potential for imaging intracellular biomolecules, such as mRNA and ATP, or even in vivo cancer imaging.
Yang et al developed procedures for detecting the levels of expression of multiple genes in fixed as well as viable cells using MB imaging technology. The results demonstrated that simultaneous delivery of MBs targeting surviving and cyclinD1 mRNAs produced strong fluorescence in breast cancer but not in normal breast cells.132 Because of their unique optical properties, ease of functionalization, size, and efficient cellular internalization, AuNPs are frequently used as an effective vector for MB intracellular bioimaging. The synthesis, transport, and distribution of mRNA in living cells can be monitored with good spatiotemporal resolution to provide important information for functional genomics.133 A new live cell mRNA detection methodology was described by Wright's group.134 The strategy retains the advantages of gold-oligonucleotide constructs, such as intracellular uptake and high specificity, but also exhibits the additional advantage of preserving spatial localization of target mRNA within the cell. Meanwhile, abundant fluorophore-labeled MBs are coupled to the AuNP surface. Since the loop sequence of MBs was designed to target the specific mRNA of respiratory syncytial virus (RSV) and the enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH), these constructs can realize spatial localization of intracellular target mRNA imaging in live cells. Tang et al. also reported simultaneous and multicolor intracellular imaging of tumor-related mRNA based on AuNPs modified with MBs. 127, 135 Ju et al. proposed a nicked MB-functionalized AuNP probe for in situ imaging and detection of intracellular telomerase activity. In the presence of telomerase in the cellular environment, the telomerase signal probe could be elongated from its 3′-end to produce a telomeric repeated sequence that is complementary to the corresponding stem at the 3′-end of MB, leading to substitutional hybridization to open the hairpin (Figure 12A).136-137
Figure 12.
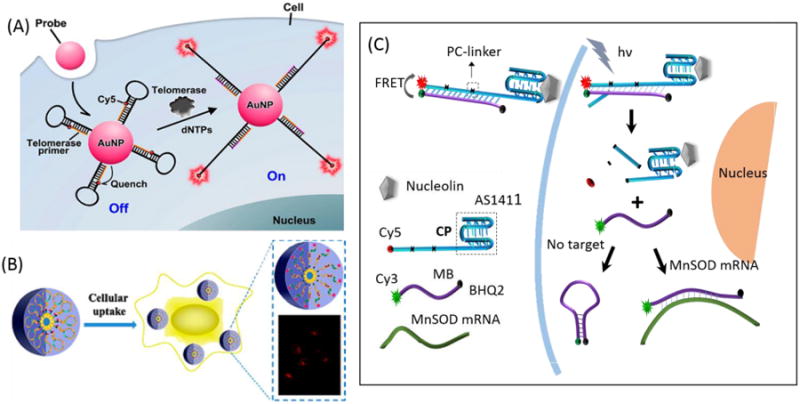
Molecular beacons used for intracellular imaging. (A) Illustration of the nicked molecular beacon-functionalized AuNPs for in situ imaging of intracellular telomerase. Reprinted with permission from ref. 136. Copyright (2014) American Chemical Society. (B) Working principle of switchable molecular aptamer beacon micelle flares for molecular imaging in living cells. Reprinted with permission from ref. 140. Copyright (2013) American Chemical Society. (C) Analysis of the molecular beacon probe for spatiotemporal MnSOD mRNA detection in living cells.
Other nanostructured materials have also been used to fabricate MB delivery systems to achieve bioimaging. Polyethylenimine (PEI)-grafted graphene nanoribbon has been used for cellular delivery of LNA-modified MB for recognition of miRNA.138 Multifunctional SnO2 nanoparticles were proposed by using folic acid for cell-specific delivery and MB conjugated to fluorescence stannic oxide (SnO2) nanoparticles with a disulfide linkage for imaging the intracellular target miRNA-21.139
Additionally, functional DNA-micelles, which contain both hydrophilic and hydrophobic segments that self-assemble under certain conditions, have been employed for intracellular imaging. Recently, we reported a MAB-micelle system for intracellular molecule detection, which we term switchable aptamer micelle flare (SAMF), as shown in Figure 12B. 140-141 This SAMF was modified with a lipid tail to form the nanostructure of micelle flares. Each hydrophobic diacyllipid tail is incorporated into a hydrophilic switchable aptamer head through solid-phase phosphoramidite chemistry on an automatic DNA synthesizer, followed by self-assembly to form a uniform spherical nanostructure. In the presence of ATP or mRNA, the conformation of the SAMF containing the aptamer is altered, leading to the restoration of fluorescence. SAMFs have many advantages over previous approaches, including facile probe modification, capable self-delivery, high signal-to-background ratio, excellent target selectivity, and superior biocompatibility. Therefore, SAMFs can provide efficient delivery and imaging platforms for living cells without using nanomaterials that may cause potential toxic effects.
To further eliminate the background fluorescence of MBs in living cells, an approach was developed with two MBs that targeted adjacent regions on the mRNA. When both MBs were hybridized to the mRNA sequence at adjacent positions, the FRET signal would occur and indicate the presence of the target. This method could be used to both visualize mRNA distribution and track the migration of the mRNA through the cell and even into adjacent cells in the oocyte.142
Considering that the spatiotemporal dynamics of specific biomolecules remain difficult to image inside living cells, our group designed a self-delivered MB for photo-initiated real-time imaging and detection of mRNA in living cells via direct hybridization of an extended internalizing aptamer and an MB, as shown in Figure 12C. 143 With this fluorescent aptamer as an internalizing carrier, the MB could be efficiently delivered into the cytoplasm of targeted cells, and its internalized amount, as well as its intracellular distribution, could be tracked before photoactivation.
4.2.2 In vivo Bioimaging
In 2011, our group was the first to report MAB probes for in vivo cancer cell imaging. These MABs were equipped with a fluorophore and a quencher attached at either terminus for tumor imaging in a mouse.144 This MAB could effectively recognize tumors with high sensitivity and specificity, thus establishing the efficacy of fluorescent MABs for diagnostic applications, as shown in Figure 13. In the absence of a target, the MAB was hairpin-structured, resulting in quenched fluorescence. As expected, the MAB could be activated by target cancer cells with rapid restoration of fluorescence achieved at the tumor site compared to other areas. To further explore novel MABs with ultra-low background and high stability, Wang et al. proposed an activatable fluorescence probing platform based on a self-assembled fluorophore-labeled MAB/SWNT assembly and presented the first attempt to apply SWNT-based activatable probes for in vitro and in vivo cancer cell imaging with high specificity and sensitivity.145 In vitro assays confirmed that Cy5-Sgc8c/SWNT was specifically activated by target cancer cells and that it effectively improved sensitivity for detecting CCRF-CEM cells both in buffer and serum. In comparison with “always on” probes, in vivo applications also demonstrated that Cy5-Sgc8c/SWNT could achieve contrast-enhanced cancer imaging.
Figure 13.
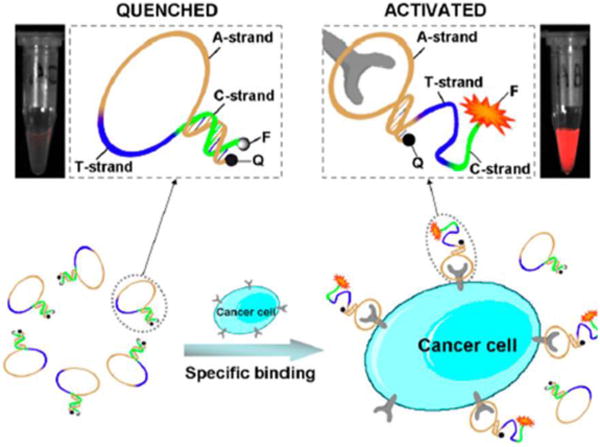
Molecular beacons used for in vivo imaging. Schematic representation of the novel strategy for in vivo cancer imaging using activatable molecular aptamer beacon based on cell membrane protein-triggered conformation alteration. Reprinted with permission from ref. 144. Copyright (2011) National Academy of Sciences, USA.
4.3 Applications of Molecular Beacons in Therapy
The last two decades have witnessed the development and application of MBs in a variety of fields, including target analysis and molecular and cellular imaging. The efficient design, versatility of features and functionalities, reproducible chemical synthesis and modification, generally impressive target binding selectivity and affinity, relatively rapid tissue penetration, low immunogenicity, and rapid systemic clearance all make MBs ideal recognition elements for use as therapeutics.
4.3.1 Gene Therapy
Most human diseases, including cancer, can be treated with the introduction of genetic materials, including plasmid DNAs, antisense oligonucleotides or small interfering RNAs, into somatic tissues.146-150 Hybridization between the loop portion of the MB and its complementary sequence has greatly facilitated the development of disease diagnosis and gene therapy; therefore, functional MBs have recently become an effective molecular tool able to enhance gene expression or inhibit the production of deleterious proteins, thus serving as excellent candidates for gene therapy.151-152 For instance, our group introduced a sensitive and selective approach for combined mRNA detection and gene therapy using SAMFs (Figure 14A).141 Just like pyrotechnic flares, which produce brilliant light when activated, SAMFs undergo a significant burst of fluorescence enhancement upon target binding. This hybridization event subsequently induces gene silencing, leading to apoptosis of cancer cells. In addition, Huh et al. constructed hyaluronic acid-coated nanocontainers which could be endocytosed by cluster determinant 44 (CD44)-over-expressing cancer cells for the efficient intracellular delivery of miR-34a MBs, and this technique is expected to be further developed as a cancer diagnostic, as well as a miRNA-based MB therapy against metastatic cancer (Figure 14B).153
Figure 14.
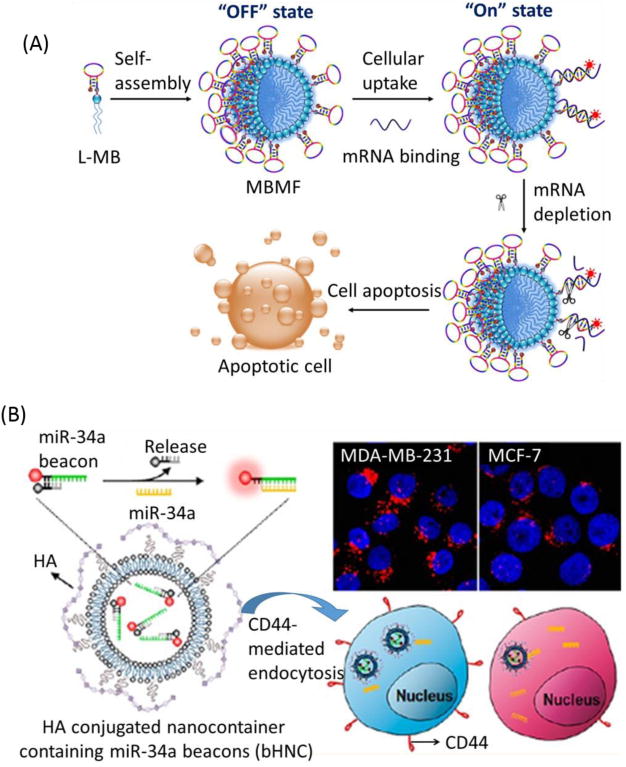
Molecular beacons used for gene therapy. (A) Analysis of molecular beacon micelle flares for intracellular mRNA gene therapy. Diacyllipid-molecular beacon conjugates (L-MBs) self-assemble into MBMFs and enter living cells. (B) Illustration of miR-34a beacon delivery system for targeted intracellular recognition of miR-34a based on HA-coated nanocontainers that encapsulate the miR-34a beacons (bHNCs). Reprinted with permission from ref. 154. Copyright (2012) American Chemical Society.
4.3.2 Drug Delivery
Based on easy synthesis and suitability for structural modification, covalent crosslinking can be used for MB drug delivery. Conventional photosensitizers, such as chlorin e6 (Ce 6), have been successfully conjugated to the terminus of MBs.154 Tang et al. coupled two photosensitizer (PS) molecules, respectively, onto opposite ends of a single MB, termed bi-PS MB.155 This MB was sequence-specific to surviving breast cancer mRNA. Thus, singlet oxygen could be effectively used to kill breast cancer cells (SK-BR-3, high surviving mRNA expression), but not normal cells (Figure 15A). Saito et al reported a drug molecule-releasing system controllable by intramolecular quenching based on a MB strategy by using photoactive probe oligodeoxynucleotides (ODNs).The photo reaction of the probe ODN containing a photoactive group and a triplet quencher at the ends of the strand was very inefficient when it was in the closed state, whereas irradiation of the open-form ODN hybridized with the complementary DNA resulted in a rapid release of the functional drug molecule from the ODN. This MB-based drug-release system will facilitate the rational design of a controllable prodrug for cancer therapy.156 In our group, taking advantage of the strong absorption in the near-infrared region, gold nanorods (AuNRs) are emerging as an efficient photothermal therapy (PTT) nanomaterials. In order to manipulate the quenching and recovery of PS fluorescence, a photosensitizer-conjugated MAB was designed for the probes, which were assembled on AuNRs, thus achieving controlled singlet oxygen generation (SOG) for photodynamic therapy (PDT).157 The strategy of utilizing a highly selective MAB combined with the synergistic effect of PTT and PDT promises to be a more efficient therapeutic regimen against cancer cells than nonspecific methods using either PTT or PDT alone (Figure 15B).
Figure 15.
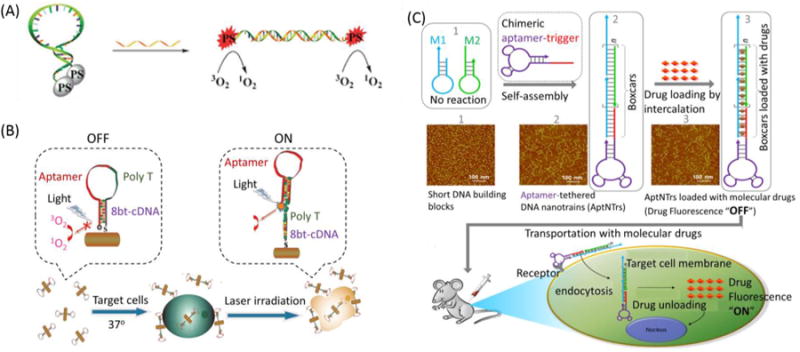
Molecular beacons used for drug delivery. (A) Concept of mRNA-triggered bi-PS molecular beacon for therapy. Reprinted with permission from ref. 155. Copyright (2011) Royal Society of Chemistry. (B) Investigation of MAB-conjugated photosensitizer and AuNRs for photothermal therapy (PTT) and photodynamic therapy (PDT). Reprinted with permission from ref. 157. Copyright (2012) American Chemical Society. (C) Design of the self-assembly of aptamer-tethered DNA nanotrains (aptNTrs) for transport of molecular drugs in theranostic applications. Reprinted with permission from ref. 161. Copyright (2013) National Academy of Sciences, USA
Besides covalent crosslinking, intercalation is another important drug-loading mode for MBs. The anticancer drug doxorubicin (Dox) can intercalate into the double-stranded GC or CG stem sequences, leading to fluorescence quenching and decreased drug cytotoxicity.158-159 Tang et al. proposed MB-based AuNPs associated with intracellular tumor-related mRNA, in which MBs were utilized as the drug carriers for activated release upon recognition of specific mRNA.160 MB-based DNA self-assembly has allowed the construction of various nanostructures for applications in therapy. Recently, our group reported aptamer-tethered DNA nanotrains (aptNTrs) as carriers for targeted drug transport in cancer therapy.161 In this design, tandem “boxcars” formed after the self-assembly of two MB-structured DNA probes served as carriers transporting anticancer drugs to target cells, followed by the induction of selective cytotoxicity, as shown in Figure 15C. Combined with nanomaterials, DNA polymers formed by self-assembly on AuNPs can be variously designed, for example, by labeling with imaging fluorescent tags or the simultaneous loading of recognition elements and anticancer drugs. AuNP-DNA conjugates show high stability and good biocompatibility, and the size of the complex can be controlled by changing the length of the self-assembled DNA biopolymer shell, which may provide a new and highly effective means for cancer therapy. 162
To further improve the limited drug payload capacity of the MB itself, Schafer et al. fabricated a MAB on the surface of mesoporous nanoparticles which served as nanovalves for drug delivery. 163 Upon target/MAB complex formation, the single-stranded neck region would allow the release of the loaded drug by their reduced size. This design was therefore shown to be a suitable drug transport system for high-performance delivery of anticancer drugs, including camptothecin (CPT), paclitaxel (PAX) and doxorubicin (DXR).
5. Conclusions
Since their first appearance in the literature about two decades ago, molecular beacons have assumed an increasingly prominent role in many important bioanalytical fields. This review mainly focuses on rationally designed MBs and the various efficient and practical strategies which have been successfully developed to engineer them with stem tunability, high sensitivity and high specificity. These properties make MBs ideal tools for biosensing and enzyme monitoring, as well as ATP and mRNA tracing in living cells. In combination with other platforms and technologies, MBs can now be used for bioimaging and even therapy. In spite of the many still unresolved issues and challenges, multidisciplinary approaches have converged for the further evolution of innovative concepts in developing the properties of MBs for bioanalytical and biomedical applications.
Acknowledgments
We sincerely appreciate Dr. Liang Cui, Dr. Jianbo Liu, Ms. Pinting Tang and Ms. Huiming Liu for their kind help with manuscript preparation. This work is supported by grants awarded from the National Natural Science Foundation of China (21135001, 21405038), the Foundation for Innovative Research Groups of NSFC (21221003), the “973” National Key Basic Research Program (2011CB91100-0), and the Fundamental Research Funds for the Central Universities. C. W. acknowledges support from the American Chemical Society, Division of Analytical Chemistry Fellowship, sponsored by the Society for Analytical Chemists of Pittsburgh.
References
- 1.Hall BD, Spiegelman S. Proc Natl Acad Sci USA. 1961;47:137–163. doi: 10.1073/pnas.47.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolton ET, McCarthy BJ. Proc Natl Acad Sci USA. 1962;48:1390–1397. doi: 10.1073/pnas.48.8.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nygaard AP, Hall BD. Biochem Biophys Res Commun. 1963;12:98–104. doi: 10.1016/0006-291x(63)90242-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang KM, Tang ZW, Yang CY, Kim YM, Fang XH, Li W, Wu YR, Medley CD, Cao ZH, Li J, Colon P, Tan WH. Angew Chem Int Ed. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 6.Tyagi S, Marras SAE, Kramer FR. Nat Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- 7.Tyagi S, Bratu DP, Kramer FR. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 8.Li JW, Fang XH, Schuster SM, Tan WH. Angew Chem Int Ed. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Broude NE. Trends Biotechnol. 2002;20:249–256. doi: 10.1016/s0167-7799(02)01942-x. [DOI] [PubMed] [Google Scholar]
- 10.Ortiz E, Lizardi PM, Estrada G. Mol Cell Probe. 1998;12:219–226. doi: 10.1006/mcpr.1998.0175. [DOI] [PubMed] [Google Scholar]
- 11.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 13.Kostrikis LG, Tyagi S, Mhlanga MM, Ho DD, Kramer FR. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 14.Ortiz E, Estrada G, Lizardi PM. Mol Cell Probes. 1998;12:219–226. doi: 10.1006/mcpr.1998.0175. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S, Bratu DP, Kramer FR. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 16.J. A. M. Vet and S. A. E. Marras, Humana Press Inc, Totowa. 2004, 288, 273-289.
- 17.Owczarzy R, Vallone PM, Gallo FJ, Paner TM, Lane MJ, Benight AS. Biopolymers. 1997;44:217–239. doi: 10.1002/(SICI)1097-0282(1997)44:3<217::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Yamana K, Ohshita Y, Fukunaga Y, Nakamura M, Maruyama A. Bioorg Med Chem. 2008;16:78–83. doi: 10.1016/j.bmc.2007.04.053. [DOI] [PubMed] [Google Scholar]
- 19.Conlon P, Yang CJ, Wu Y, Chen Y, Martinez K, Kim Y, Stevens N, Marti AA, Jockusch S, Turro NJ, Tan W. J Am Chem Soc. 2007;130:336–342. doi: 10.1021/ja076411y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamana K, Iwase R, Furutani S, Tsuchida H, Zako H, Yamaoka T, Murakami A. Nuc Acids Res. 1999;27:2387–2392. doi: 10.1093/nar/27.11.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang CJ, Jockusch S, Vicens M, Turro NJ, Tan WH. Proc Natl Acad Sci USA. 2005;102:17278–17283. doi: 10.1073/pnas.0508821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Häner R, Biner SM, Langenegger SM, Meng T, Malinovskii VL. Angew Chem Int Ed. 2010;49:1227–1230. doi: 10.1002/anie.200905829. [DOI] [PubMed] [Google Scholar]
- 23.Nagatoishi S, Nojima T, Juskowiak B, Takenaka S. Angew Chem Int Ed. 2005;44:5067–5070. doi: 10.1002/anie.200501506. [DOI] [PubMed] [Google Scholar]
- 24.Tsourkas A, Behlke MA, Bao G. Nucleic Acids Res. 2002;30:4208–4215. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunner J, Kraemer R. J Am Chem Soc. 2004;126:13626–13627. doi: 10.1021/ja047252a. [DOI] [PubMed] [Google Scholar]
- 26.Dubertret B, Calame M, Libchaber AJ. Nat Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 27.Yang RH, Jin J, Chen Y, Shao N, Kang H, Xiao ZY, Tang ZW, Wu YR, Zhu Z, Tan WH. J Am Chem Soc. 2008;130:8351–8358. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 28.Lu CH, Li J, Liu JJ, Yang HH, Chen X, Chen GN. Chem Eur J. 2010;16:4889–4894. doi: 10.1002/chem.200903071. [DOI] [PubMed] [Google Scholar]
- 29.Yeh HY, Yates MV, Mulchandani A, Chen W. Chem Commun. 2010;46:3914–3916. doi: 10.1039/c001553a. [DOI] [PubMed] [Google Scholar]
- 30.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 31.Robertson DL, Joyce GF. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 32.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 33.Breaker RR. Nat Biotechnol. 1997;15:427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 34.Breaker RR. Science. 2000;290:2095–2096. doi: 10.1126/science.290.5499.2095. [DOI] [PubMed] [Google Scholar]
- 35.Joyce GF. Annu ReV Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 36.Zhang XB, Wang ZD, Xing H, Xiang Y, Lu Y. Anal Chem. 2010;82:5005–5011. doi: 10.1021/ac1009047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo YS, Wang H, Sun YS, Qu B. Chem Commun. 2012;48:3221–3223. doi: 10.1039/c2cc17552e. [DOI] [PubMed] [Google Scholar]
- 38.Thurley S, Röglin L, Seitz O. J Am Chem Soc. 2007;129:12693–12695. doi: 10.1021/ja075487r. [DOI] [PubMed] [Google Scholar]
- 39.Bourdoncle A, Estévez Torres A, Gosse C, Lacroix L, Vekhoff P, Le Saux T, Jullien L, Mergny JL. J Am Chem Soc. 2006;128:11094–11105. doi: 10.1021/ja0608040. [DOI] [PubMed] [Google Scholar]
- 40.Fu R, Li T, Lee SS, Park HG. Anal Chem. 2011;83:494–500. doi: 10.1021/ac102719x. [DOI] [PubMed] [Google Scholar]
- 41.Mohanty J, Barooah N, Dhamodharan V, Harikrishna S, Pradeepkumar PI, Bhasikuttan AC. J Am Chem Soc. 2013;135:367–376. doi: 10.1021/ja309588h. [DOI] [PubMed] [Google Scholar]
- 42.Faverie AR, Guédin A, Bedrat A, Yatsunyk LA, Mergny JL. Nucleic Acids Res. 2014;42:e65. doi: 10.1093/nar/gku111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan XH, Wang Y, Armitage BA, Bruchez MP. Anal Chem. 2014;86:10864–10869. doi: 10.1021/ac502986g. [DOI] [PubMed] [Google Scholar]
- 44.Grossmann TN, Röglin L, Seitz O. Angew Chem Int Ed. 2007;46:5223–5225. doi: 10.1002/anie.200700289. [DOI] [PubMed] [Google Scholar]
- 45.Salunkhe M, Wu TF, Letsinger RL. J Am Chem Soc. 1992;14:8768–8772. [Google Scholar]
- 46.Chen Y, Lee SH, Mao CD. Angew Chem Int Ed. 2004;43:5335–5338. doi: 10.1002/anie.200460789. [DOI] [PubMed] [Google Scholar]
- 47.Grossmann TN, Roglin L, Seitz O. Angew Chem Int Ed. 2007;46:5223–5225. doi: 10.1002/anie.200700289. [DOI] [PubMed] [Google Scholar]
- 48.Zheng J, Li JS, Jiang Y, Jin JY, Wang KM, Yang RH, Tan WH. Anal Chem. 2011;83:6586–6592. doi: 10.1021/ac201314y. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka Y, Oda S, Yamaguchi H, Kondo Y, Kojima C, Ono A. J Am Chem Soc. 2007;129:244–245. doi: 10.1021/ja065552h. [DOI] [PubMed] [Google Scholar]
- 50.Ono A, Togashi H. Angew Chem Int Ed. 2004;43:4300–4302. doi: 10.1002/anie.200454172. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Lu Y. Angew Chem Int Ed. 2007;46:7587–7590. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]
- 52.Yang RH, Jin JY, Long LP, Wang Y, Wang H, Tan WH. Chem Commun. 2009;3:322–324. doi: 10.1039/b816638b. [DOI] [PubMed] [Google Scholar]
- 53.Wang YX, Li JS, Wang H, Jin JY, Liu JH, Wang KM, Tan WH, Yang RH. Anal Chem. 2010;82:6607–6612. doi: 10.1021/ac101114w. [DOI] [PubMed] [Google Scholar]
- 54.Lin YH, Tseng W. Chem Commun. 2012;48:6262–6264. doi: 10.1039/c2cc31382k. [DOI] [PubMed] [Google Scholar]
- 55.Kuo CY, Tseng WL. Chem Commun. 2013;49:4607–4609. doi: 10.1039/c3cc40976g. [DOI] [PubMed] [Google Scholar]
- 56.Zheng J, Li JS, Gao XX, Jin JY, Wang KM, Tan WH, Yang RH. Anal Chem. 2010;82:3914–3921. doi: 10.1021/ac1004713. [DOI] [PubMed] [Google Scholar]
- 57.Jin F, Lian Y, Li J, Zheng J, Hu Y, Liu J, Huang J, Yang R. Anal Chim Acta. 2013;799:44–50. doi: 10.1016/j.aca.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 58.Gao X, Deng T, Li J, Yang R, Shen G, Yu RQ. Analyst. 2013;138:2755–2760. doi: 10.1039/c3an00122a. [DOI] [PubMed] [Google Scholar]
- 59.Kang HZ, Liu HP, Phillips JA, Cao ZH, Kim YM, Chen Y, Yang ZY, Li JW, Tan WH. Nano Lett. 2009;9:2690–2696. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C, Zhu Z, Song Y, Lin H, Yang CJ, Tan WH. Chem Commun. 2011;47:5708–5710. doi: 10.1039/c1cc10481k. [DOI] [PubMed] [Google Scholar]
- 61.Vester B, Wengel J. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Yang CYJ, Medley CD, Benner SA, Tan WH. J Am Chem Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 63.Kim Y, Yang CJ, Tan WH. Nucleic Acids Res. 2007;5:7279–7287. doi: 10.1093/nar/gkm771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin X, Zhang C, Huang Y, Zhu Z, Chen X, Yang CJ. Chem Commun. 2013;49:7243–7245. doi: 10.1039/c3cc43224f. [DOI] [PubMed] [Google Scholar]
- 65.Ke GL, Wang CM, Ge Y, Zheng NF, Zhu Z, Yang CYJ. J Am Chem Soc. 2012;134:18908–18911. doi: 10.1021/ja3082439. [DOI] [PubMed] [Google Scholar]
- 66.Wittenhagen LM, Carreon JR, Prestwich EG, Kelley SO. Angew Chem Int Ed. 2005;44:2542–2546. doi: 10.1002/anie.200462836. [DOI] [PubMed] [Google Scholar]
- 67.Brunner J, Barton JK. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 68.Svanvik N, Nygren J, Westman G, Kubista M. J Am Chem Soc. 2001;123:803–809. doi: 10.1021/ja002294u. [DOI] [PubMed] [Google Scholar]
- 69.Berndl S, Wagenknecht HA. Angew Chem Int Ed. 2009;48:2418–2421. doi: 10.1002/anie.200805981. [DOI] [PubMed] [Google Scholar]
- 70.Sheng P, Yang Z, Kim Y, Wu Y, Tan W, Benner SA. Chem Commun. 2008:5128–5130. doi: 10.1039/b811159f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kashida H, Takatsu T, Fujii T, Sekiguchi K, Liang X, Niwa K, Takase T, Yoshida Y, Asanuma H. Angew Chem Int Ed. 2009;48:7044–7047. doi: 10.1002/anie.200902367. [DOI] [PubMed] [Google Scholar]
- 72.Fujimoto K, Shimizu H, Inouye M. J Org Chem. 2004;69:3271–3275. doi: 10.1021/jo049824f. [DOI] [PubMed] [Google Scholar]
- 73.Nesterova I, Verdree V, Pakhomov S, Strickler K, Allen M, Hammer R, Soper S. Bioconjugate Chem. 2007;18:2159–2168. doi: 10.1021/bc700233w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verdree V, Pakhomov S, Su G, Allen M, Countryman A, Hammerand R, Soper S. J Fluoresc. 2007;17:547–563. doi: 10.1007/s10895-007-0210-4. [DOI] [PubMed] [Google Scholar]
- 75.Nesterova IV, Erdem SS, Pakhomov S, Hammer RP, Soper SA. J Am Chem Soc. 2009;131:2432–2433. doi: 10.1021/ja8088247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang CJ, Lin H, Tan W. J Am Chem Soc. 2005;127:12772–12773. doi: 10.1021/ja053482t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saito Y, Mizuno E, Bag SS, Suzuka I, Saito I. Chem Commun. 2007:4492–4494. doi: 10.1039/b709715h. [DOI] [PubMed] [Google Scholar]
- 78.Song S, Liang Z, Zhang J, Wang L, Li G, Fan C. Angew Chem Int Ed. 2009;48:8670–8674. doi: 10.1002/anie.200901887. [DOI] [PubMed] [Google Scholar]
- 79.Billinton N, Knight AW. Anal Biochem. 2001;291:175–197. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- 80.Mansfield JR, Gossage KW, Hoyt CC, Levenson RM. J Biomed Opt. 2005;10:04120–041209. doi: 10.1117/1.2032458. [DOI] [PubMed] [Google Scholar]
- 81.Li J, Zhou W, Ouyang X, Yu H, Yang R, Tan W, Yuan J. Anal Chem. 2011;83:1356–1362. doi: 10.1021/ac102710w. [DOI] [PubMed] [Google Scholar]
- 82.Fan C, Plaxco KW, Heeger AJ. Proc Natl Acad Sci USA. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Immoos CE, Lee SJ, Grinstaff MW. J Am Chem Soc. 2004;126:10814–10815. doi: 10.1021/ja046634d. [DOI] [PubMed] [Google Scholar]
- 84.Zuo X, Song S, Zhang J, Pan D, Fan C. J Am Chem Soc. 129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 85.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew Chem Int Ed. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 86.Hu D, Huang Z, Pu F, Ren J, Qu X. Chem Eur J. 2011;17:1635–1641. doi: 10.1002/chem.201001331. [DOI] [PubMed] [Google Scholar]
- 87.Zhang L, Zhu J, Li T, Wang E. Anal Chem. 2011;83:8871–8876. doi: 10.1021/ac2006763. [DOI] [PubMed] [Google Scholar]
- 88.Fu R, Li T, Lee SS, Park HG. Anal Chem. 2011;83:494–500. doi: 10.1021/ac102719x. [DOI] [PubMed] [Google Scholar]
- 89.Xiao Y, Pavlov V, Niazov T, Dishon A, Kotler M, Willner I. J Am Chem Soc. 2004;126:7430–7431. doi: 10.1021/ja031875r. [DOI] [PubMed] [Google Scholar]
- 90.Huang Y, Chen J, Zhao S, Shi M, Chen ZF, Liang H. Anal Chem. 2013;85:4423–4430. doi: 10.1021/ac3037443. [DOI] [PubMed] [Google Scholar]
- 91.Teller C, Shimron S, Willner I. Anal Chem. 2009;81:9114–9119. doi: 10.1021/ac901773b. [DOI] [PubMed] [Google Scholar]
- 92.Gao W, Zhang L, Zhang YM, Liang RP, Qiu JD. J Phys Chem C. 2014;118:14410–14417. [Google Scholar]
- 93.Xiao Y, Lubin AA, Baker BR, Plaxco KW, Heeger AJ. Proc Natl Acad Sci USA. 2006;103:16677–16680. doi: 10.1073/pnas.0607693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.NCBI dbSNP. [Accessed 21 July 2011]; http://www.ncbi.nlm.nih.gov/snp/index.html.
- 95.Ding C. Trends Biotechnol. 2007;25:279–283. doi: 10.1016/j.tibtech.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 96.Guo QP, Yang XH, Wang KM, Tan WH, Li W, Tang HX, Li HM. Nucleic Acids Res. 2009;37:e20. doi: 10.1093/nar/gkn1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nilsson M, Gullberg M, Dahl F, Szuhai K, Raap AK. Nucleic Acids Res. 2002;30:e66. doi: 10.1093/nar/gnf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ayele W, Baar MPd, Goudsmit J, Kliphuis A, Tilahun T, Wolday D, Abebe A, Mengistu Y, Pollakis G. J Virol Methods. 2005;130:22–29. doi: 10.1016/j.jviromet.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 99.Risch NJ. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 100.Gut IG. Hum Mutat. 2001;17:475–492. doi: 10.1002/humu.1131. [DOI] [PubMed] [Google Scholar]
- 101.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 102.Giesendorf BAJ, Vet JAM, Tyagi S, Mensink EJMG, Trijbels FJM, Blom HJ. Clin Chem. 1998;44:482–486. [PubMed] [Google Scholar]
- 103.Li X, Huang Y, Guan Y, Zhao M, Li Y. Anal Chem. 2006;78:7886–7890. doi: 10.1021/ac061518+. [DOI] [PubMed] [Google Scholar]
- 104.Wabuyele MB, Farquar H, Stryjewski W, Hammer RP, Soper SA, Cheng YW, Barany F. J Am Chem Soc. 2003;125:6937–6945. doi: 10.1021/ja034716g. [DOI] [PubMed] [Google Scholar]
- 105.Kiesling T, Cox K, Davidson EA, Dretchen K, Grater G, Hibbard S, Lasken RS, Leshin J, Skowronski E, Danielsen M. Nucleic Acids Res. 2007;35:e117. doi: 10.1093/nar/gkm654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li JJ, Chu Y, Lee BYH, Xie XS. Nucleic Acids Res. 2008;36:e36. doi: 10.1093/nar/gkn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kong RM, Zhang XB, Zhang LL, Huang Y, Lu DQ, Tan W, Shen GL, Yu RQ. Anal Chem. 2010;83:14–17. doi: 10.1021/ac1025072. [DOI] [PubMed] [Google Scholar]
- 108.Tang ZW, Wang KM, Tan WH, Ma CB, Li J, Liu LF, Guo QP, Meng XX. Nucleic Acids Res. 2005;33:e97. doi: 10.1093/nar/gni096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tang ZW, Liu P, Ma CB, Yang XH, Wang KM, Tan WH. Anal Chem. 2011;83:2505–2510. doi: 10.1021/ac102742k. [DOI] [PubMed] [Google Scholar]
- 110.Ueyama H, Takagi M, Takenaka S. J Am Chem Soc. 2002;124:14286–14287. doi: 10.1021/ja026892f. [DOI] [PubMed] [Google Scholar]
- 111.Stojanovic MN, Prada P, Landry DW. J Am Chem Soc. 2000;122:11547–11548. doi: 10.1021/ja0022223. [DOI] [PubMed] [Google Scholar]
- 112.Hamaguchi N, Ellington A, Stanton M. Anal Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 113.Shlyahovsky B, Li D, Weizmann Y, Nowarski R, Kotler M, Willner I. J Am Chem Soc. 2007;129:3814–3815. doi: 10.1021/ja069291n. [DOI] [PubMed] [Google Scholar]
- 114.Zhao XH, Kong RM, Zhang XB, Meng HM, Liu WN, Tan WH, Shen GL, Yu RQ. Anal Chem. 2011;83:5062–5066. doi: 10.1021/ac200843x. [DOI] [PubMed] [Google Scholar]
- 115.Zhao XH, Gong L, Zhang XB, Yang B, Fu T, Hu R, Tan WH, Yu RQ. Anal Chem. 2013;85:3614–3620. doi: 10.1021/ac303457u. [DOI] [PubMed] [Google Scholar]
- 116.Lu LM, Zhang XB, Kong RM, Yang B, Tan WH. J Am Chem Soc. 2011;133:11686–11691. doi: 10.1021/ja203693b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng J, Hu YP, Bai JH, Ma C, Tan WH, Yang RH. Anal Chem. 2014;86:2205–2212. doi: 10.1021/ac404004m. [DOI] [PubMed] [Google Scholar]
- 118.Situma C, Moehring AJ, Noor MAF, Soper SA. Anal Biochem. 2007;363:35–45. doi: 10.1016/j.ab.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zuo X, Yang X, Wang KM, Tan WH. Anal Chim Acta. 2006;567:173–178. [Google Scholar]
- 120.Xuan F, Luo XT, Hsing IM. Anal Chem. 2012;84:5216–5220. doi: 10.1021/ac301033w. [DOI] [PubMed] [Google Scholar]
- 121.Liu SF, Lin Y, Wang L, Liu T, Cheng CB, Wei WJ, Tang B. Anal Chem. 2014;86:4008–4015. doi: 10.1021/ac500426b. [DOI] [PubMed] [Google Scholar]
- 122.Du H, Disney MD, Miller BL, Krauss TD. J Am Chem Soc. 2003;125:4012–4013. doi: 10.1021/ja0290781. [DOI] [PubMed] [Google Scholar]
- 123.Zheng J, Jiao AL, Yang RH, Li HM, Tan WH. J Am Chem Soc. 2012;134:19957–19960. doi: 10.1021/ja308875r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen T, Shukoor MI, Chen Y, Yuan Q, Zhu Z, Zhao Z, Gulbakan B, Tan WH. Nanoscale. 2011;3:546–556. doi: 10.1039/c0nr00646g. [DOI] [PubMed] [Google Scholar]
- 125.Latorre A, Posch C, Garcimartin Y, Ortiz–Urda S, Somoza A. Chem Commun. 2014;50:3018–3020. doi: 10.1039/c3cc47862a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mao X, Xu H, Zeng Q, Zeng L, Liu G. Chem Commun. 2009:3065–3067. doi: 10.1039/b822582f. [DOI] [PubMed] [Google Scholar]
- 127.Qiao G, Gao Y, Li N, Yu Z, Zhuo L, Tang B. Chem Eur J. 2011;17:11210–11215. doi: 10.1002/chem.201100658. [DOI] [PubMed] [Google Scholar]
- 128.Beer D, Weber J. Opt Commun. 1972;5:307–309. [Google Scholar]
- 129.Aldana J, Wang YA, Peng X. J Am Chem Soc. 2001;123:8844–8850. doi: 10.1021/ja016424q. [DOI] [PubMed] [Google Scholar]
- 130.Guo W, Li JJ, Wang YA, Peng X. Chem Mater. 2003;15:3125–3133. [Google Scholar]
- 131.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Nat Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 132.Peng XH, Cao ZH, Xia JT, Carlson GW, Lewis MM, Wood WC, Yang Y. Cancer Res. 2005;65:1909–1917. doi: 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]
- 133.Medley CD, Drake TJ, Tomasini JM, Rogers RJ, Tan WH. Anal Chem. 2005;77:4713–4718. doi: 10.1021/ac050881y. [DOI] [PubMed] [Google Scholar]
- 134.Jayagopal A, Halfpenny KC, Perez JW, Wright DW. J Am Chem Soc. 2010;132:9789–9796. doi: 10.1021/ja102585v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li N, Chang C, Pan W, Tang B. Angew Chem Int Ed. 2012;51:7426–7430. doi: 10.1002/anie.201203767. [DOI] [PubMed] [Google Scholar]
- 136.Qian R, Ding L, Yan L, Lin M, Ju HX. J Am Chem Soc. 2014;136:8205–8208. doi: 10.1021/ja5042995. [DOI] [PubMed] [Google Scholar]
- 137.Qian R, Ding L, Yan L, Lin M, Ju HX. Anal Chem. 2014;86:8642–8648. doi: 10.1021/ac502538w. [DOI] [PubMed] [Google Scholar]
- 138.Dong H, Ding L, Yan F, Ji H, Ju HX. Biomaterials. 2011;32:3875–3882. doi: 10.1016/j.biomaterials.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 139.Dong H, Lei J, Ju H, Zhi F, Wang H, Guo W, Zhu Z, Yan F. Angew Chem Int Ed. 2012;51:4607–4612. doi: 10.1002/anie.201108302. [DOI] [PubMed] [Google Scholar]
- 140.C. C. Wu, T. Chen, D. Han, M. You, L. Peng, S. Cansiz, G. Z. Zhu, C. Li, X. Xiong, E. Jimenez, C. J. Yang and W. H. Tan, ACS Nano, 2013, 7, 5724-5731.
- 141.Chen T, Wu CS, Jimenez E, Zhu Z, Dajac JG, You M, Han D, Zhang X, Tan WH. Angew Chem Int Ed. 2013;52:2012–2016. doi: 10.1002/anie.201209440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Colin Donnell M. Ph D Dissertation. University of Florida; USA: 2007. Method and Material Development for the Detection and Analysis of Cancer Cells. [Google Scholar]
- 143.Qiu LP, Wu C, You M, Han D, Chen T, Zhu G, Jiang J, Yu R, Tan WH. J Am Chem Soc. 2013;135:12952–12955. doi: 10.1021/ja406252w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shi H, He X, Wang K, Wu X, Ye X, Guo Q, Tan W, Qing Z, Yang X, Zhou B. Proc Natl Acad Sci USA. 2011;108:3900–3905. doi: 10.1073/pnas.1016197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yan LA, Shi H, He X, Wang K, Tang J, Chen M, Ye X, Xu F, Lei Y. Anal Chem. 2014;86:9271–9277. doi: 10.1021/ac5024149. [DOI] [PubMed] [Google Scholar]
- 146.Horton HM, Anderson D, Hernandez P, Barnhart KM, Norman JA, Parker SE. Proc Natl Acad Sci USA. 1999;96:1553–1558. doi: 10.1073/pnas.96.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hao L, Patel PC, Alhasan AH, Giljohann DA, Mirkin CA. Small. 2011;7:3158–3162. doi: 10.1002/smll.201101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang J, Zhang PC, Mao HQ, Leong KW. Gene Ther. 2002;9:1254–1261. doi: 10.1038/sj.gt.3301794. [DOI] [PubMed] [Google Scholar]
- 152.Okamoto A, Tanabe K, Inasaki T, Saito I. Angew Chem Int Ed. 2003;115:2606–2608. doi: 10.1002/anie.200250832. [DOI] [PubMed] [Google Scholar]
- 153.Monia BP, Johnston JF, Geiger T, Muller M, Fabbro D. Nat Med. 1996;2:668–675. doi: 10.1038/nm0696-668. [DOI] [PubMed] [Google Scholar]
- 154.E. Kim, J. Yang, J. Park, S. Kim, N. H. Kim, J. I. Yook, J. S. Suh, S. Haam and Y. M. Huh, ACS Nano, 2012, 6, 8525-8535.
- 155.Zhu Z, Tang ZW, Phillips JA, Yang R, Wang H, Tan WH. J Am Chem Soc. 2008;130:10856–10857. doi: 10.1021/ja802913f. [DOI] [PubMed] [Google Scholar]
- 156.Gao Y, Qiao G, Zhuo L, Li N, Liu Y, Tang B. Chem Commun. 2011;47:5316–5318. doi: 10.1039/c1cc10557d. [DOI] [PubMed] [Google Scholar]
- 157.J. Wang, G. Zhu, M. You, E. Song, M. I. Shukoor, K. Zhang, M. B. Altman, Y. Chen, Z. Zhu, C. Z. Huang and W. H. Tan, ACS Nano, 2012, 6, 5070-5077.
- 158.Fan P, Suri AK, Fiala R, Live D, Patel DJ. J Mol Biol. 1996;258:480–500. doi: 10.1006/jmbi.1996.0263. [DOI] [PubMed] [Google Scholar]
- 159.Qiao G, Zhuo L, Gao Y, Yu L, Li N, Tang B. Chem Commun. 2011;47:7458–7460. doi: 10.1039/c1cc11490e. [DOI] [PubMed] [Google Scholar]
- 160.Pan W, Yang H, Zhang T, Li Y, Li N, Tang B. Anal Chem. 2013;85:6930–6935. doi: 10.1021/ac401405n. [DOI] [PubMed] [Google Scholar]
- 161.Zhu GZ, Zheng J, Song EQ, Donovan M, Zhang K, Liu C, Tan WH. Proc Natl Acad Sci USA. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.J. Zheng, G. Z. Zhu, Y. H. Li, C. Li, M. You, T. Chen, E. Song, R. H. Yang and W. H. Tan, ACS Nano, 2013, 7, 6545-6554.
- 163.Özalp VC, Schäfer T. Chem Eur J. 2011;17:9893–9896. doi: 10.1002/chem.201101403. [DOI] [PubMed] [Google Scholar]


