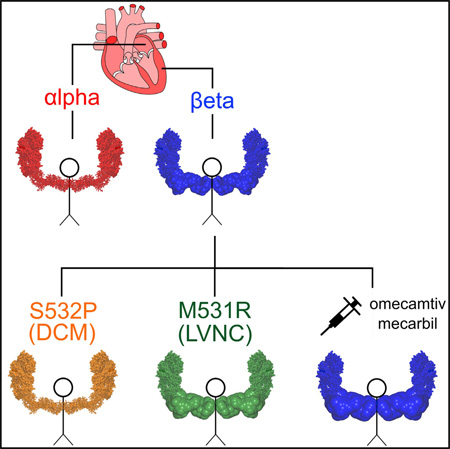
SUMMARY
Cardiomyopathies due to mutations in human β-cardiac myosin are a significant cause of heart failure, sudden death, and arrhythmia. To understand the underlying molecular basis of changes in the contractile system’s force production due to such mutations and search for potential drugs that restore force generation, an in vitro assay is necessary to evaluate cardiac myosin’s ensemble force using purified proteins. Here, we characterize the ensemble force of human α- and β-cardiac myosin isoforms and those of β-cardiac myosins carrying left ventricular non-compaction (M531R) and dilated cardiomyopathy (S532P) mutations using a utrophin-based loaded in vitro motility assay and new filament-tracking software. Our results show that human α- and β-cardiac myosin, as well as the mutants, show opposite mechanical and enzymatic phenotypes with respect to each other. We also show that omecamtiv mecarbil, a previously discovered cardiac-specific myosin activator, increases β-cardiac myosin force generation.
Graphical Abstract
INTRODUCTION
With the advent of technologies to easily obtain the complete sequence of the human genome, an exponential increase will be seen in our understanding of the underlying genetics that lead to human disease. Where we have a deep understanding of the biochemical and biophysical basis of the machineries and pathways involved in those genetic changes, the development of modern therapeutics that specifically target the actual machinery and pathways altered by individual mutations will demand increasing attention. Prime examples of such genetic disease are hypertrophic (HCM), dilated (DCM), and left ventricular non-compaction (LVNC) cardiomyopathies, which result from mutations in one of several of the proteins that make up the cardiac sarcomere. Here, we describe an approach to understand the molecular basis of the alterations in power output that result from such mutations. We also show, studying one example of a myosin small-molecule activator, that small molecules binding to the mutant sarcomeric protein complex will be able to mitigate the effects of hypertrophic and dilated cardiomyopathy mutations at their sources, leading to new therapeutic approaches for these genetic diseases.
Human α- and β-cardiac myosins drive contraction of the heart atria and ventricles, respectively (Lompré et al., 1984; Reiser and Kline, 1998; Reiser et al., 2001). β-Cardiac myosin is a major site for cardiomyopathy mutations; over 250 mutations in the β-cardiac myosin motor domain have been reported as the cause of cardiomyopathy diseases that disrupt the morphology and proper functioning of the left ventricle, which delivers oxygenated blood throughout the body (Bing et al., 2000; Colegrave and Peckham, 2014; Harvey and Leinwand, 2011; Kimura, 2010; Marston, 2011). Although significant progress has been made in understanding the effects of some cardiomyopathy mutations from studies with animal models and patient biopsy samples (Cuda et al., 1997; Keller et al., 2004; Palmiter et al., 2000), a detailed investigation of cardiomyopathy mutations at the molecular level has been hampered by the lack of an expression system with high yields of human cardiac myosin (Sata and Ikebe, 1996).
In recent years, a mammalian expression system has been developed for human striated muscle myosins that has made it possible (1) to determine enzymatic differences between purified, homogeneous human α- and β-cardiac myosins and (2) to study cardiomyopathy-causing mutations in the human β-cardiac myosin backbone (Resnicow et al., 2010; Srikakulam and Winkelmann, 1999). The human α- and β-cardiac motor domains (subfragment 1 of myosin, or S1) can be expressed and purified using this system (Deacon et al., 2012). Purified β-cardiac S1 demonstrates good actin-activated ATPase activity and displaces actin filaments in actin-gliding assays (Sommese et al., 2013). Stopped-flow measurements have been performed to determine the rates of transitions in the actin-myosin ATPase cycle for these human cardiac S1s (Bloemink et al., 2014; Deacon et al., 2012).
Cardiac myosin operates under varying loads in the heart; to recapitulate the effect of cardiomyopathy mutations on cardiac myosin’s function under physiological conditions, it is essential to measure its power output under load. Thus, fundamental to muscle contraction is the force-velocity curve, where the product of the force and velocity at any point along the curve is a measure of the power output. With increasing load, actin-gliding velocity decreases and ultimately reaches zero when load is equal to the maximal ensemble force (Daniels et al., 1984; Sonnenblick, 1962).
The goal of this study is (1) to illustrate the effects of two cardiomyopathy mutations on human cardiac myosin power output with a reconstituted purified protein system and (2) to demonstrate that screening of small compounds that restore mutant cardiac myosin power output to wild-type (WT) levels is a feasible path forward.
One way to measure actin-gliding velocities at different loads is using a loaded in vitro motility assay in which increasing concentrations of an actin-binding protein is utilized as the load-bearing molecule (Bing et al., 2000; Greenberg and Moore, 2010; Haeberle and Hemric, 1995; Warshaw et al., 1990). This assay has been used to compare power output and maximal force generation between myosin variants (Greenberg et al., 2010; Haeberle, 1994; Malmqvist et al., 2004) and between different solution conditions (Greenberg and Moore, 2010; Janson et al., 1992). The assay is highly limited, however, by the time it takes to analyze motility data and by an excessive amount of user intervention and subjectivity in the analysis. Furthermore, the current assay with α-actinin does not yield highly reproducible results with the motor domain S1, presumably due to the small size of this domain compared to two-headed myosin constructs and the non-specific attachment of α-actinin on the surface (Sommese et al., 2013).
Here, we present (1) in vitro motility analysis software that rapidly performs actin filament tracking objectively with no user intervention and (2) a customized loaded motility assay for human cardiac short subfragment 1 (sS1; spans the N-terminal motor domain and the ELC binding region in the lever arm) with utrophin as the load molecule. To achieve proper control of myosin and load molecule ratios, a truncated utrophin construct of similar size to sS1 is engineered to anchor on a glass surface via the same attachment used for sS1. Using these new tools, we compare the WT forms of purified homogeneous human α- and β-cardiac sS1. In addition, we characterize two cardiomyopathy mutations, M531R and S532P, which are adjacent in the primary structure of β-cardiac myosin and are expected to have very different effects on cardiac myosin activity. M531R has been found in patients with LVNC (Kaneda et al., 2008). S532P, on the other hand, has been associated with DCM (Kamisago et al., 2000). To assess whether our assay is sensitive to power output changes due to binding of small molecules to cardiac myosin, we characterize the effect of omecamtiv mecarbil (Malik et al., 2011; Morgan et al., 2010), which is a previously discovered cardiac myosin activator.
RESULTS
FAST Facilitates Unbiased, Automated, and Rapid In Vitro Motility Data Analysis
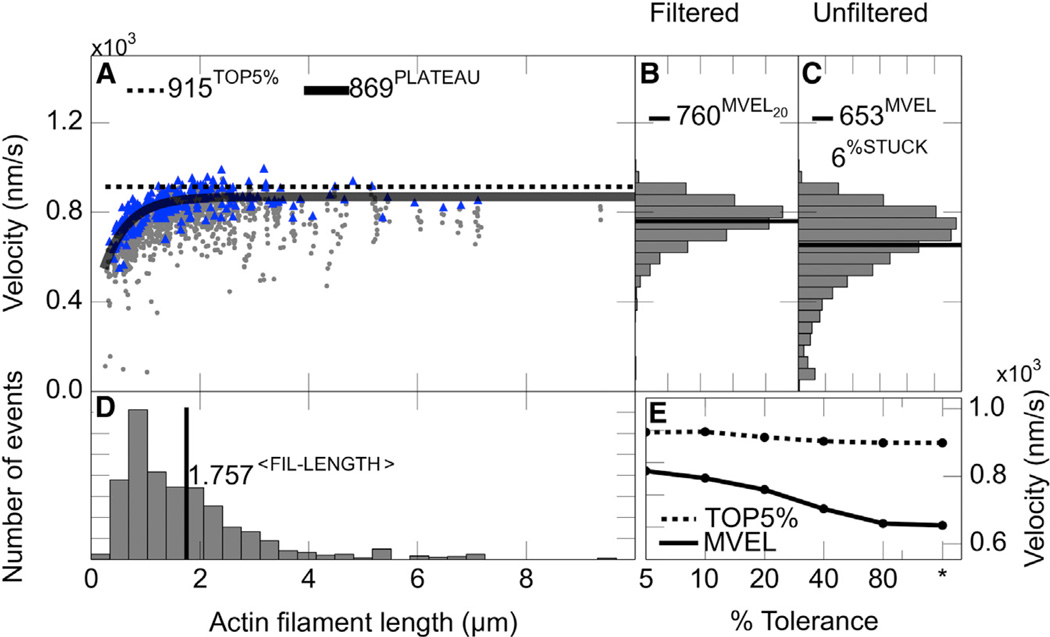
A bottleneck limiting high-throughput data acquisition for in vitro motility assays has been the lack of rapid, unbiased, and automated analysis software. Some automated filament tracking software programs are packaged together with analog image-acquisition systems (Homsher et al., 1992) not readily compatible with state-of-the-art digital images. Also, we found the digital analysis tools that are available (Ruhnow et al., 2011) do not have the high throughput we seek for our studies and are not well optimized for actin filament tracking. Here, we describe new actin filament tracking and in vitro motility data analysis software, FAST (Fast Automated Spud Trekker). FAST completes the analysis of a 30-frame in vitro motility movie in less than 30 s, providing a detailed output. Graphical output of FAST has multiple panels showing different aspects of the analysis (Figure 1; see the Supplemental Experimental Procedures and Figure S1 for the tracking algorithm).
Figure 1. The FAST Output for an Unloaded Human β-Cardiac sS1 In Vitro Motility Experiment.
For this and all unloaded velocity experiments, filaments tracked for at least ten consecutive frames were included in the analysis, velocity and filament lengths were averaged over five consecutive frames, a 20% tolerance filter was applied to eliminate intermittently moving filaments with velocities fluctuating higher than 20% of their mean velocity, and stuck filaments were excluded from the scatterplot analysis.
(A) Scatterplot of filament length and velocities. Maximum velocity of each filament is in blue (triangle), and lower velocities are in gray (circles). The solid line is the fit to single exponential decay function to the maximum velocities (blue) to obtain the PLATEAU velocity. The dashed line marks the mean of the top 5% velocities (TOP5%).
(B) A 20%tolerance-filtered filament velocity distribution excluding stuck filaments. The line marks the mean of the filtered velocity distribution (MVEL20; subscript 20 is for the percent tolerance value).
(C) The unfiltered filament velocity distribution excluding stuck filaments. The line marks the mean of the unfiltered distribution (MVEL). Filaments with a mean velocity less than one pixel length per second (80 nm/s) are classified as stuck. %STUCK represents the time-weighted fraction of stuck filaments.
(D) Filament length distribution. The line marks the mean of the distribution.
(E) Mean of the velocity distribution (solid line) and TOP5% velocity (dashed line) from the data subjected to different levels of tolerance filtering (see the Supplemental Experimental Procedures). Lower percent tolerance values indicate more stringent filtering allowing only filaments with smoother movement. Asterisk (*) is for the unfiltered velocity distribution. See also Figure S1.
Maximum velocity (PLATEAU and TOP5%) is estimated from a distribution that is filtered with respect to filaments with fluctuating velocities (Figures 1A–1C; see the Supplemental Experimental Procedures). Filtered (Figure 1B) and unfiltered (Figure 1C) distributions are displayed in FAST output side by side to show the effect of the filtering.
Another feature of FAST is to estimate the percentage of stuck filaments (Figure 1C). Together with %STUCK, filament length distribution (Figure 1D) is a useful metric in assessing the quality of motility in unloaded conditions (see Experimental Procedures).
FAST output also displays how the mean of the velocity distribution and TOP5% velocities change with decreasing tolerance in filtering (Figure 1E).
Loaded In Vitro Motility Assay with Utrophin as the Load Molecule Provides a Readout for Human Cardiac sS1’s Ability to Overcome Load
For loaded in vitro motility, we designed a utrophin construct that is similar in size to human β-cardiac sS1 (Figure S2) and anchored on the surface via the same attachment system used for human β-cardiac sS1 (Figure 2A). This allows proper maintenance of myosin to load molecule ratios on the surface.
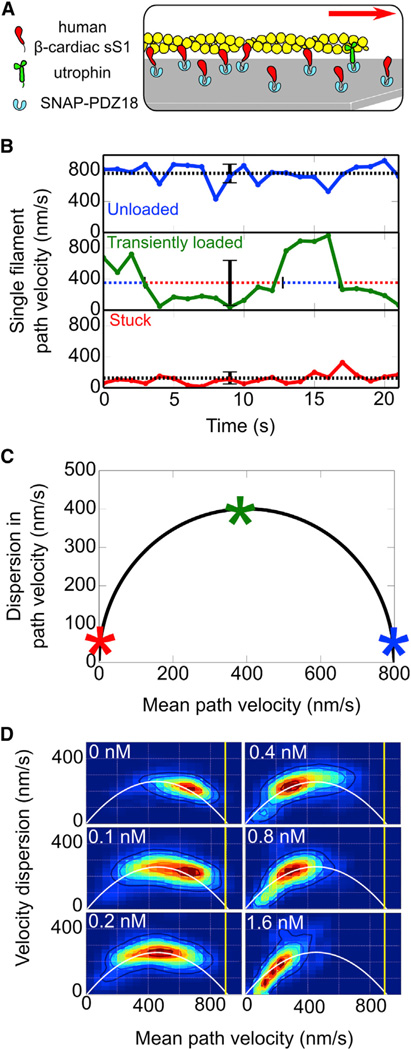
Figure 2. Utrophin Causes Actin Filaments to Glide with Intermittent Stops in the Loaded In Vitro Motility Assay.
(A) Cartoon representation of the loaded in vitro motility setup with human β-cardiac sS1 and utrophin on a surface coated with the anchoring protein SNAP-PDZ18 (light blue). Myosin (red) and utrophin (green) are anchored on the surface via the same attachment system involving binding of a C-terminal eight-residue peptide that specifically binds to the SNAP-PDZ18. Approximately 400-fold less utrophin (1 nM) than myosin (400 nM) is required to inhibit actin filament (yellow) movement. Smoothly moving actin filaments encounter utrophin at their front (minus) end and stop moving temporarily (see Movies S1, S2, S3, and S4).
(B) Wild-type β-cardiac sS1 instantaneous velocity-time trajectories for an unloaded actin filament (blue), a stuck filament (red), and a transiently loaded filament (green) with utrophin. Dashed lines and error bars mark the mean velocity and the SD of instantaneous velocities, respectively, along the trajectories. Unloaded and stuck intervals of the transiently loaded trajectory are marked by blue and red, respectively, over the dashed mean velocity line.
(C) Effect of utrophin on gliding filaments is explained by a model in which utrophin stops filament gliding at the maximal velocity (Supplemental Experimental Procedures). Stars on the curve are for the unloaded (blue), stuck (red), and transiently loaded (green) velocity trajectories in (B).
(D) Heatmaps show the probability density for experimental measurements of filament mean velocity and velocity dispersion at different utrophin concentrations (upper left corner). The white solid line is for the best parabola fit to aggregated probability densities from all utrophin concentrations. The yellow vertical line marks the unloaded TOP5% velocity.
See also Figure S2.
The new utrophin construct is ideal for loaded in vitro motility as (1) it is stable as a monomer and (2) the range of utrophin concentrations required for actin-motility inhibition is low enough not to compete with sS1 binding to SNAP-PDZ18.
Careful observation of movies revealed that with utrophin, “mobile filaments” become stuck at their front (minus) end, remain immobile temporarily, and then continue gliding (see Movies S1, S2, S3, and S4). Thus, filament velocities fluctuate between unloaded velocities and zero, enhancing dispersion in instantaneous velocities and decreasing average velocities (Figure 2B, unloaded versus 0.2 nM utrophin) due to utrophin. As the duration of stops increases with increasing utrophin concentration, filaments remain immobile longer, trapping velocity values around zero, thus causing dispersion in velocities to decrease (Figure 2B, stuck).
The dispersion in path filament velocities and their average values fall on a parabola, predicted by a stop model, and the increase in utrophin concentration causes these points to move along the parabola from high to low mean path-velocity values (Figures 2C and 2D; see the Supplemental Experimental Procedures). The velocity dispersion in unloaded conditions (Figure 2C, 0 nM utrophin) is likely due to the small fraction of dead heads that bind actin in rigor and halt gliding temporarily.
Percent Time Mobile Is the Proper Measure for the Assessment of Actin-Gliding Inhibition by Utrophin
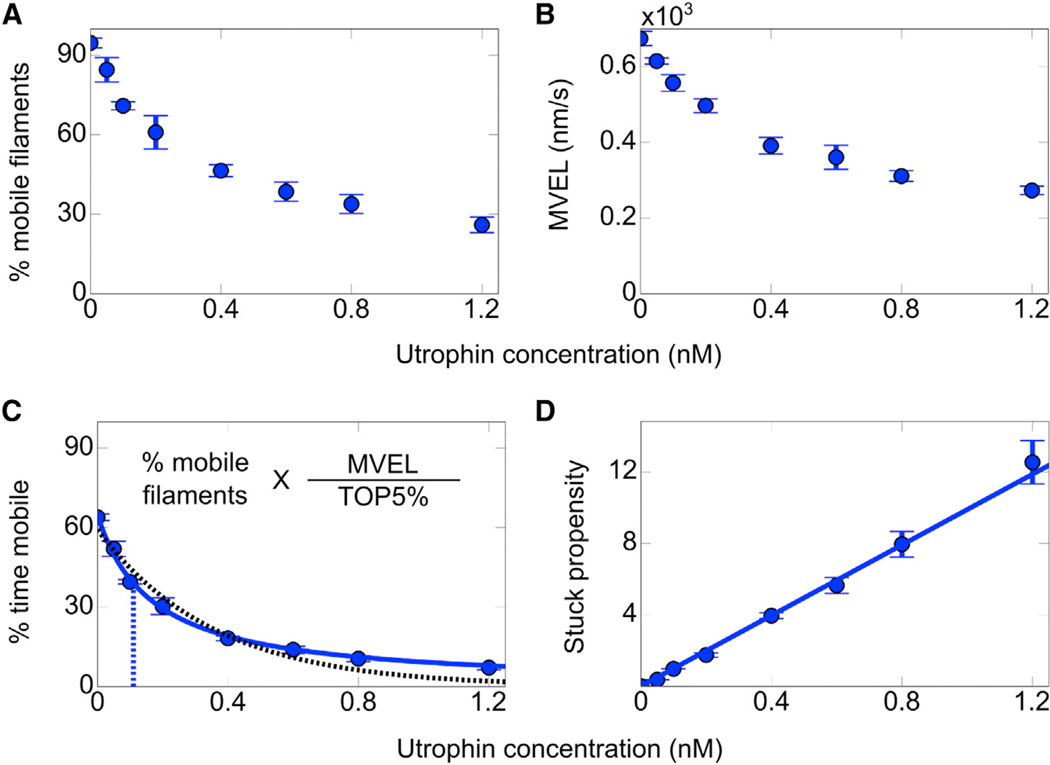
We observe that both the time-weighted fraction of mobile filaments (percent mobile filaments, Figure 3A) and the mean velocity of the unfiltered velocity distribution (MVEL, Figure 3B) decay as a function of utrophin concentration. However, decay in MVEL or percent mobile filaments alone does not capture the full impact of utrophin. Effective utrophin surface density for the inhibition of mobile filaments is reduced due to filaments that are relatively permanently bound to utrophin molecules; these utrophin molecules are no longer accessible to mobile filaments, thus leading to overestimation of MVEL. Similarly, “percent mobile filaments” overestimates the mobility of filaments, since “mobile filaments” do not glide at all times, remaining immobile when bound to utrophin.
Figure 3. Percent Time Mobile Is the Measure of Actin-Gliding Inhibition by Utrophin.
(A and B) Percent mobile filaments (A) and MVEL (B) as a function of utrophin concentration.
(C) The stop model fits well to human β-cardiac sS1 loaded in vitro motility percent time mobile data. Percent time mobile is equal to the product of percent mobile filaments (A) and MVEL (B) normalized by unloaded TOP5% velocity. Blue solid and black dashed lines are for the best stop model and single exponential fits, respectively. KS is indicated by the vertical dashed blue line. Each loaded in vitro motility point is the average from ≥1,000 filament tracks and ≥5,000 instantaneous velocities averaged over five frames. Confidence intervals are the SEM from at least four movie replicates.
(D) Alternative representation of β-cardiac sS1 loaded motility data as stuck propensity, which is equivalent to percent time stuck (100% − percent time mobile) divided by percent time mobile. Unloaded stuck propensity is subtracted from each point. The line shows the best linear equation fit to the data points. See also Figure S3.
For proper assessment of filament mobility, we define “percent time mobile,” which is equal to the percent mobile filaments corrected for the times spent as immobile (Figures 3A and 3B) and normalized by the unloaded TOP5% velocity (Figure 3C; see the Supplemental Experimental Procedures).
The Stop Model Enables Comparison of Ensemble Forces from Percent Mobile Data
To extract cardiac myosin ensemble force from percent mobile data, a mathematical function that relates utrophin concentration to percent time mobile is required. We observe that percent time stuck (100% − percent time mobile) normalized by percent time mobile depends linearly on the utrophin concentration (Figure 3D). Writing normalized percent time stuck (also referred to as stuck propensity) as a linear function of utrophin, we derive an expression (Equation 1) that relates utrophin concentration to percent time mobile with two parameters: (1) M0, which is percent time mobile in the absence of utrophin; and (2) KS, which is analogous to KD (i.e., dissociation constant) used for binding equilibrium relationships (for derivation, see the Supplemental Experimental Procedures):
| (Equation 1) |
KS can be interpreted as the disassociation constant for the utrophin-actin interaction in the presence of cardiac sS1 on the surface. We consider higher KS to indicate lower affinity between utrophin and actin due to higher force generated by the cardiac sS1 to break the utrophin-actin interaction.
This mathematical model (the stop model) fits better than a single exponential fit when applied to β-cardiac sS1 percent time mobile data (Figure 3C; see Figure S3A). As expected, we observe that increases in cardiac sS1 concentration cause Ks to increase, indicative of higher ensemble force due to higher myosin density on the surface (Figure S3B).
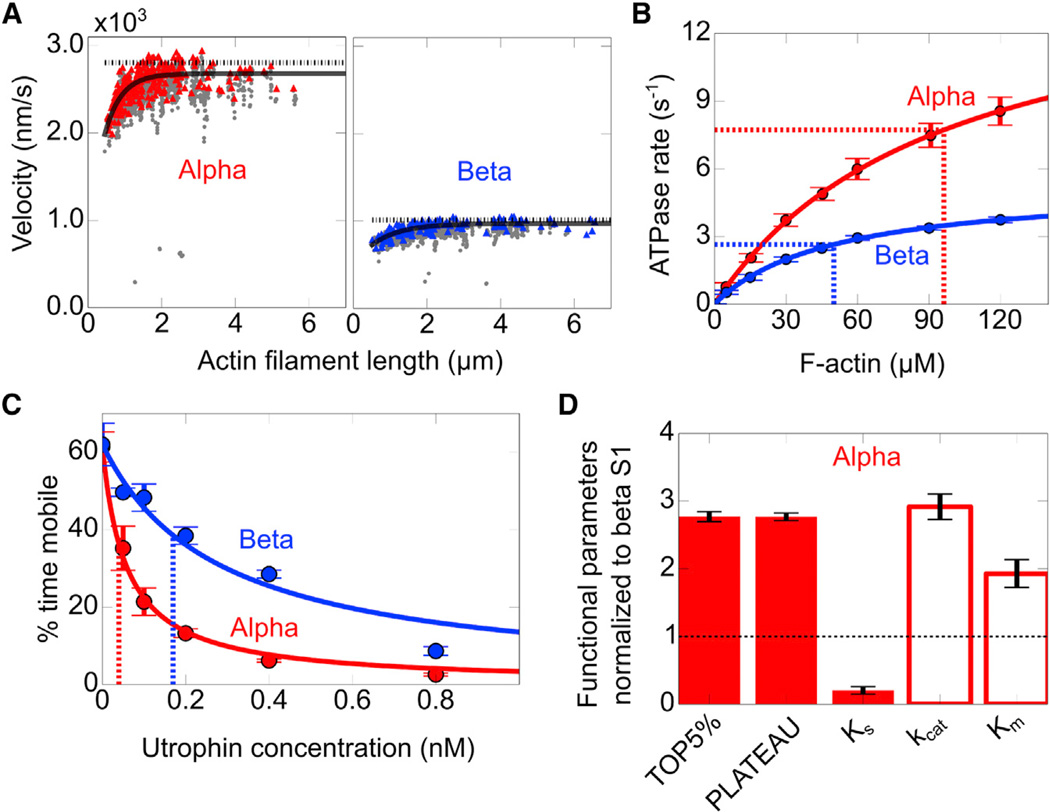
Human α-sS1 Has 3-Fold Higher Maximal Velocity and ATPase Rate but Generates Less Ensemble Force Than β-sS1
Mammalians have two isoforms of cardiac myosin, α and β, which are expressed in different ratios across different species and in different sections of the heart (Lompré et al., 1984; Reiser and Kline, 1998; Reiser et al., 2001). Using FAST and the new loaded motility assay with utrophin, we compared human α- and β-cardiac sS1, which are expected to have large differences in their maximal unloaded velocities and ensemble forces based on previous studies with animal and human α/β isoforms (Alpert et al., 2002; Malmqvist et al., 2004; Noguchi et al., 2003; VanBuren et al., 1995).
Human α-Cardiac sS1 Is a Faster Motor Than β-Cardiac sS1
To assess whether human α- and β-cardiac sS1 have different maximal velocities and catalytic activities, we measured the unloaded velocities (Figure 4A) and actin-activated ATPase rates (Figure 4B) for the two human isoforms with the same lever arm length (see Figure S4A). α-Cardiac sS1 showed 3-fold higher maximum velocities than β. From the Michaelis-Menten equation fit to actin-activated ATPase rates, α-cardiac sS1 showed a 3-fold higher maximal ATPase rate (kcat) and a 2-fold higher Km than β, suggesting that α-cardiac sS1 has significantly lower affinity for actin (Table S1; Figure 4B).
Figure 4. α-Cardiac sS1 Has Nearly 3-Fold Higher Actin-Activated Enzymatic Activity and Unloaded Gliding Velocity but Generates Less Force Than β-Cardiac sS1.
(A) Representative unloaded velocity scatterplots for α- (left) and β-human cardiac sS1 (right), as described in Figure 1. For description of lines, see Figure 1A (a 20% tolerance filter was applied).
(B) Actin-activated ATPase rates for α (red) and β (blue) human cardiac sS1. Solid lines show the best Michaelis-Menten fits for the ATPase rates (rate = kcat[actin]/(Km+[actin]). Dashed lines mark the Km and half maximal ATPase rates (0.5 kcat). Confidence intervals are the SEM from three replicate measurements for each of three myosin preparations.
(C) Representative loaded in vitro motility percent time mobile data for α (red) and β (blue) human cardiac sS1. Lines show the best stop model fit to percent time mobile data. Dashed lines mark KS values determined from the fit. Confidence intervals are the SEM from at least four movie replicates.
(D) Normalized α-cardiac sS1 motility parameters (TOP5%, PLATEAU, KS; filled bars) and ATPase (kcat, Km; empty bars) values with respect to β-cardiac sS1. For unloaded motility parameters, 95% confidence intervals are the SEM from at least four replicate movies from three different preparations of sS1. For KS, the 95% confidence interval is estimated from 100 iterations of stop model fitting to bootstrapped data (Supplemental Experimental Procedures). For ATPase values, rates are the average of at least three replicates from three different preparations of sS1 (Table S1).
See also Figure S4.
β-Cardiac sS1 Produces More Force Than α
To determine the ensemble force differences between α- and β-cardiac sS1, we measured the utrophin-dependent percent time mobile changes for the two isoforms. With increasing utrophin concentration, α-cardiac sS1 percent time mobile decays more sharply than that of β-cardiac sS1, indicative of lower ensemble force generation (Figure 4C). To determine the decay parameter, KS, we fit the stop model to the percent mobile data. From the fits, the α-cardiac sS1 KS is significantly lower than the β-cardiac sS1 KS (Figure 4D; Table S1), indicating a higher force for β-cardiac sS1.
In ventricles, there is ~5% α-cardiac myosin, together with ~95% β. To determine the effect of low levels of α-cardiac sS1 on β-cardiac sS1 velocities, we measured unloaded velocities for α- and β-cardiac sS1 mixed at different ratios. From 0% to 10% α-cardiac sS1, we did not observe a significant change in velocities. Also, we observed that the α/β-cardiac sS1 mixture velocities deviated from linearity with a downward curvature against the fraction of α-cardiac sS1, indicating that β dominates α-cardiac sS1 (Figure S4B), where downward curvature observed in myosin-mixing experiments is generally attributed to lower force generation by the titrated species (Harris et al., 1994; Warshaw et al., 1990). This observation is in agreement with our results from α- and β-cardiac sS1 loaded motility experiments, indicating that β-cardiac sS1 generates higher force than α.
Two Neighboring Cardiomyopathy Mutations, M531R and S532P, Have Opposite Effects on Human β-Cardiac Myosin Function
One aim for developing FAST and the new loaded in vitro motility assay was for high-throughput maximal velocity and ensemble force characterization of cardiomyopathy causing mutations in the human β-cardiac myosin motor domain.
Maximal velocity and ensemble force directly determine myosin’s maximal power output (Spudich, 2014). It has been hypothesized that different classes of cardiomyopathy mutations may change cardiac myosin’s power output differently leading to different phenotypes (Sivaramakrishnan et al., 2009).
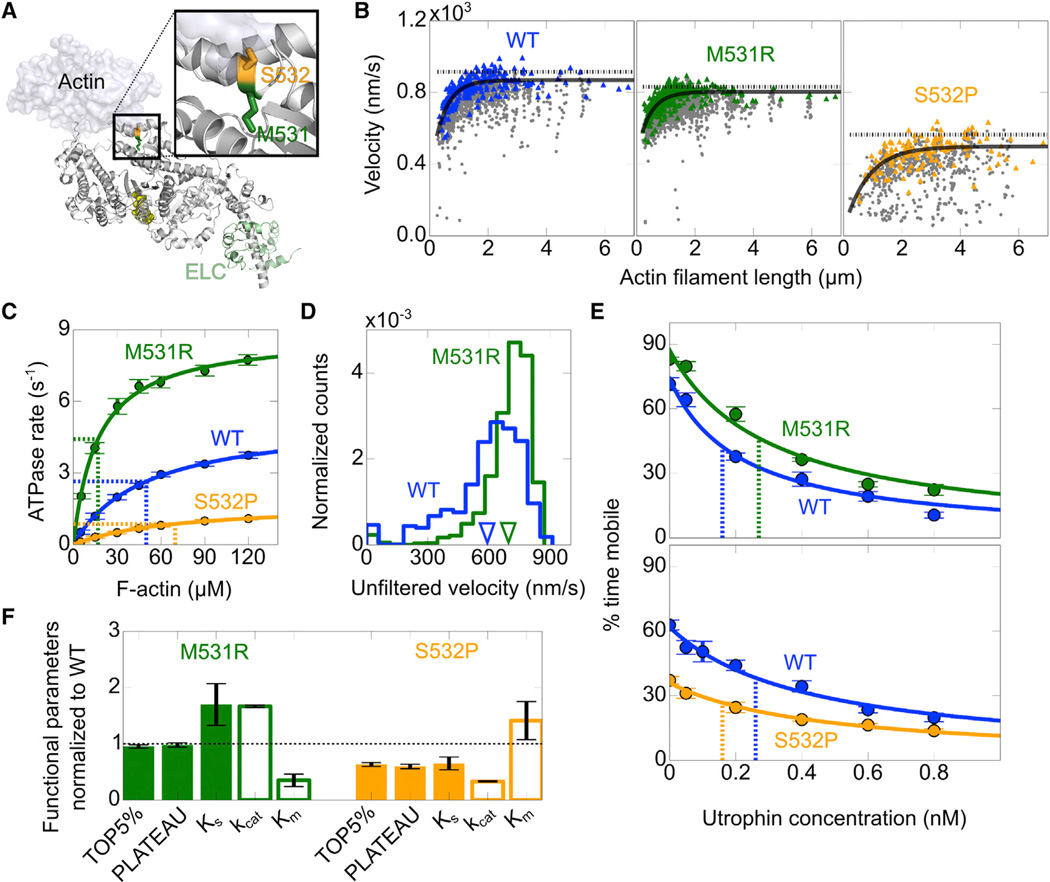
To illustrate the usefulness of the assay, we chose two cardiomyopathy mutations, M531R and S532P, which are known to cause LVNC and DCM, respectively (Kamisago et al., 2000; Kaneda et al., 2008). LVNC is associated with severe trabeculae formation in the left ventricular wall and impaired systolic function (Udeoji et al., 2013), and DCM is associated with dilation of the left ventricular wall and reduced systolic function (Hershberger et al., 2013). The two mutations are located at the actin-binding interface of the β-cardiac motor domain, most likely affecting the actin-myosin interaction (Figure 5A).
Figure 5. M531R and S532P Have Opposite Impact on WT β-Cardiac sS1 Enzymatic and Mechanical Activity.
(A) M531 (green) and S532 (orange) are side by side on the actin-binding interface. The inset shows a close-up of the mutation positions (for details of the structural model, rendered using Pymol, see the Supplemental Experimental Procedures).
(B) Representative unloaded velocity scatter plots comparing PLATEAU and TOP5% velocities for WT (blue), M531R (green), and S532P (orange). For description of lines, see Figure 1A (a 20% tolerance filter was applied).
(C) Actin-activated ATPase rates for WT β-cardiac sS1 (blue), M531R (green), and S532P (orange). For description of lines and estimation of confidence intervals, see Figure 4B and Table S1 legend.
(D) Representative unfiltered velocity distributions comparing M531R β-cardiac sS1 (green) and WT (blue). MVEL values for M531R and WT are marked on the velocity axis (inverted triangles). Velocity distributions are from four movies for WT and M531R collected on the same slide.
(E) Lines show the best stop model fit to unfiltered percent time mobile loaded motility data. Dashed lines indicate the KS determined from the fit. Confidence intervals are the SEM from at least four movie replicates.
(F) M531R (green) and S532 (orange) motility (TOP5%, PLATEAU, KS; filled bars) and ATPase (kcat, Km; empty bars) values normalized to WT values. For estimation of confidence intervals, see Figure 4D (legend) and Table S1.
The Human β-Cardiac Myosin Cardiomyopathy Mutations M531R and S532P Are Gain- and Loss-of-Function Mutations, Respectively
To determine whether the two mutations have a strong impact on human β-cardiac sS1, we performed in vitro motility (Figure 5B) and actin-activated ATPase measurements (Figure 5C). M531R has slightly lower maximum velocities than WT. On the other hand, S532P has significantly depressed maximum velocities as compared to WT (Table S1).
M531R and S532P have significantly higher and lower maximal ATPase rates, respectively, than WT (Figure 5C; Table S1). M531R and S532P have lower and higher Km values, respectively, than WT, suggesting that M531R and S532P have higher and lower affinity, respectively, for actin than WT.
M531R and S532P Generate Higher and Lower Ensemble Force, Respectively, Than WT
We performed loaded in vitro motility measurements for M531R and S532P with the utrophin-based assay to estimate their ensemble force as compared to WT. Although M531R has slightly lower maximum 20% filtered velocities than WT (Figure 5B), it has a higher unloaded mean velocity than WT, since M531R has a tighter velocity distribution concentrated closer to maximum velocity values (Figure 5D). Having a tight velocity distribution with significantly fewer filaments gliding at lower velocities indicates that M531R is more efficient in overcoming the internal load due to dead myosin heads in unloaded conditions than WT. From stop model fits, M531R has higher KS values than WT, suggesting that M531R generates higher ensemble force than WT, whereas lower KS values for S532P indicate that S532P generates lower ensemble force than WT (Figures 5E and 5F; Table S1). These loaded in vitro motility results with human β-cardiac myosin support the conclusion that M531R and S532P are gain- and loss-of-function mutations, respectively.
Omecamtiv Mecarbil Increases β-Cardiac sS1 Force Generation but Decreases Unloaded Velocity
A major goal of cardiomyopathy research is to find therapies that reverse or prevent disease progression and restore normal cardiac function. We and others have shown that cardiomyopathy mutations in cardiac myosin alter its power output, and it is thought that the change in cardiac output triggers signaling cascades leading to the disease phenotype (Sivaramakrishnan et al., 2009; Spudich, 2014). It has been suggested that small molecules that specifically interact with a cardiac sarcomeric protein, including myosin itself, may restore cardiac function in patients with cardiomyopathy mutations and reverse the effect of those mutations (Sivaramakrishnan et al., 2009; Spudich, 2014).
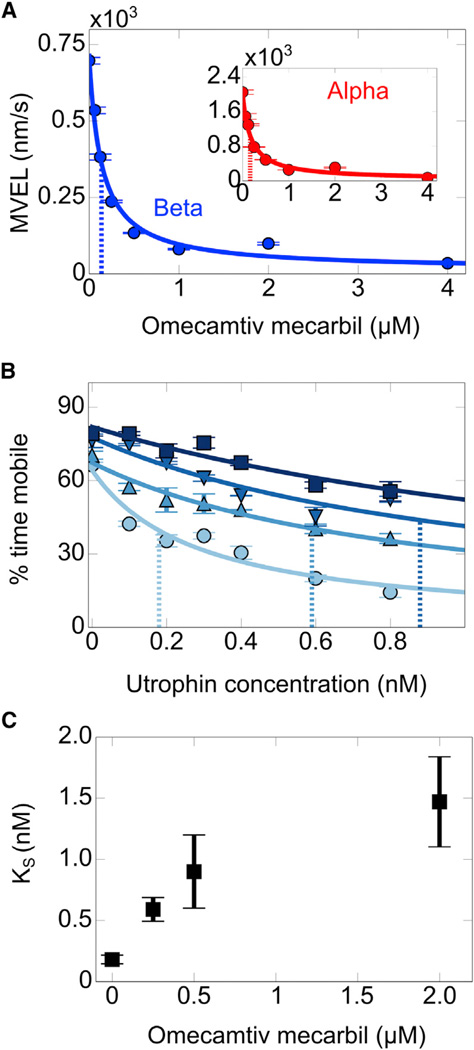
Omecamtiv mecarbil (OM) is a small compound that was discovered to bind specifically to cardiac myosin and enhance cardiac output in animal models (Malik et al., 2011) and in humans (Teerlink et al., 2011); however, demonstration of increased force production with OM has not been achieved with a purified reconstituted contractile system. Here, we use the utrophin-based loaded in vitro motility assay to show that OM does indeed increase human β-cardiac sS1 ensemble force production.
First, to test whether OM affects human α- and β-cardiac sS1 similarly, we measured their unloaded velocities as a function of OM concentration. Both α- and β-cardiac sS1 unloaded velocities drop sharply with OM (Figure 6A). It has previously been reported that at saturating concentrations of OM, human (Liu et al., 2013) and porcine β-cardiac myosin (Liu et al., 2015) unloaded velocities drop several-fold. KD values determined from fitting a binding isotherm to α- and β-cardiac sS1 unloaded velocity data are within similar range (0.14–0.17 µM; see Table S1), suggesting that OM binds to human α- and β-cardiac sS1 with similar affinities.
Figure 6. Omecamtiv Mecarbil Binding to Cardiac sS1 Increases Ensemble Force Generation but Decreases Its Unloaded Velocity.
(A) MVEL decreases with increasing concentration of omecamtiv mecarbil for both α- (red, inset) and β-cardiac (blue) sS1. Lines show the best binding isotherm fit to data (Supplemental Experimental Procedures). Dashed lines mark the KD determined from the fits. Confidence intervals are the SEM from at least four movie replicates.
(B) Representative loaded in vitro motility data for β-cardiac sS1 at different concentrations of omecamtiv mecarbil (●, 0 µM; :▲, 0.25 µM; ;▼, 0.5 µM; ■, 2 µM). Increase in color intensity indicates increase in omecamtiv mecarbil concentration. Solid lines are for the best stop model fits to percent time mobile data. Dashed lines mark KS determined from the fits. Confidence intervals are the SEM from at least four movie replicates.
(C) KS determined from stop model fits to β-cardiac sS1 loaded motility data. 95% confidence intervals are estimated from 100 iterations of stop model fitting to bootstrapped data (Supplemental Experimental Procedures).
To determine the ensemble force changes due to OM binding to β-cardiac sS1, we performed loaded in vitro motility measurements as a function of OM concentration. OM shifts the percent time mobile data to higher values (Figure 6B), thus causing KS to increase with OM (Figure 6C), which demonstrates that binding of OM increases β-cardiac sS1 ensemble force.
DISCUSSION
Our results provide important insights into the functioning of human cardiac myosin, actin-binding proteins, and OM. Furthermore, we demonstrate that the utrophin-based loaded in vitro motility assay more generally provides a quantitative platform to diagnose cardiomyopathy mutations and to screen for potential drugs that restore cardiac myosin function. This assay and FAST analysis software are well designed for high-throughput screening purposes.
Human α- and β-Cardiac sS1 Evolved to Function at Different Force Ranges and Timescales
We show here that human α-cardiac sS1 is faster but generates lower force than β. In the adult human heart, the cardiac myosin composition is ~95% β and ~5% α in the ventricles and ~5% β and ~95% α in the atria (Reiser et al., 2001). Ventricles are responsible for pumping blood out of the heart and work against higher loads than atria, which only aid the largely passive flow of blood into the ventricles and contract over a shorter duration of time than the ventricles. Our findings with human α- and β-cardiac myosin are consistent with the idea that β-cardiac myosin generates the high force required for ventricular contraction, while the α isoform provides lower force but faster contraction required for atrial function, and they are consistent with isometric force measurements of human atrial and ventricular myocardium (Noguchi et al., 2003).
M531R and S532P May Alter the Structural Organization in the Human Cardiac sS1 Actin-Binding Interface, Resulting in Opposite Phenotypes
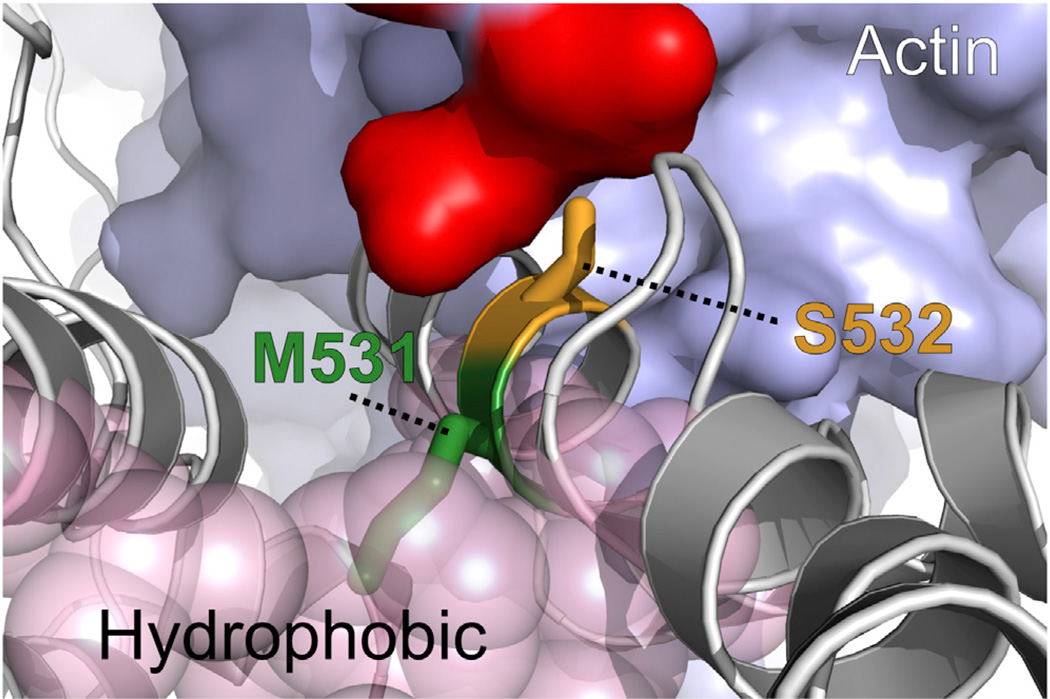
Residues M531 and S532 are located on an α helix that is at the actin-binding interface of human β-cardiac myosin (Figure 5A). Both residues are highly conserved in the cardiac myosin family (Figure S5). We showed that M531R and S532P have opposite impacts on human cardiac myosin ATPase activity and mechanical output, indicating that the two mutations may perturb the structural arrangement in the actin-binding interface differently. The lower Km and higher maximal ATPase rate for M531R suggest that M531R leads to a conformational change favoring the transition from the free to the actin-bound state of the motor; oppositely, the higher Km and lower maximal ATPase rate for S532P suggest that S532P disrupts the transition from the free to the actin-bound state.
The M531 side chain is buried in the hydrophobic core of the motor, while the S532 side chain is positioned near the motor surface at the actin-binding site (Figure 7). Thermodynamically, the positively charged mutant arginine side chain at position 531 is not likely to be accommodated in the hydrophobic environment that the M531 residue normally resides in (Harms et al., 2011). Sequence analysis of a representative myosin universe indicates that only hydrophobic residues are favored at position 531 (Figure S5). We expect the M531R mutation to cause a structural perturbation in the α-helical region exposing the arginine side chain in solution, in a conformation favoring arginine interaction with the negatively charged actin interface, promoting actin binding. On the other hand, we expect the S532P mutation to deform the actin-binding interface by destabilizing the α helix, since proline is an α-helix breaker (Pace and Scholtz, 1998). From the sequence analysis, we also found proline not to be favored at position 532 (Figure S5).
Figure 7. The Mutations M531R and S532P Are Likely to Cause Local Structural Changes with a Major Impact on the Actin-Myosin Interaction.
Local environments of M531 (green) and S532 (orange) correlate with the sequence conservation profiles in Figure S6. Both positions are on an α helix in close proximity to a patch of negatively charged residues (red) on actin. M531 (green) is buried in the hydrophobic core, and S532 (orange) is exposed to solvent. See also Figure S5.
Increased Force Generation by OM May Offset Its Effect in Unloaded Velocity Reduction
It was previously shown that OM binds to the myosin catalytic domain and operates by an allosteric mechanism to increase the transition rate of myosin into the strongly actin-bound force-generating state and enhances systolic force generation in animal models, thus increasing cardiac power output (Malik et al., 2011). Recent studies show that additional effects at the molecular level include a nearly 20-fold decrease in actin-gliding velocity and a 2-fold decrease in the actin-activated ATPase of porcine β-cardiac myosin at saturating OM (Liu et al., 2015). Strongly bound (to actin) state time (referred as ts) and total ATPase cycle time (referred as tc) are inversely proportional to unloaded velocity and the actin activated ATPase rate, respectively. Therefore, such changes in velocity and ATPase rate would indicate a ~20-fold increase in ts and a 2-fold increase in tc, resulting in a 10-fold increase in the duty ratio ts/tc, which would be predicted to result in increased ensemble force (Spudich, 2014). Our results showing increased force generation with OM at the molecular level are consistent with those observations. The reduction in unloaded velocity by OM seen here and previously reported (Liu et al., 2013, 2015) is unexpected and perhaps could be explained by an OM-based decrease on a load-dependent ADP release rate. Such an effect could lengthen the strongly bound state time of the myosin head, further increasing the duty ratio, leading to a further increase in force production.
Perhaps related to this putative decrease in load-dependent ADP release rate is the finding that OM prolongs the systolic phase of the contraction, which is thought to be the primary cause of myocardial ischemia observed in some human subjects (Teerlink et al., 2011). This side effect was more pronounced with human subjects carrying 3 µM (1,200 ng/ml) OM, which was the highest concentration tested. Our results indicate that OM leads to a dose-dependent increase in ensemble force from 0 to 2 µM. This is in the range of the therapeutic window for OM function in vivo.
The increase in ensemble force by OM apparently offsets the decrease in velocity, resulting in an overall increase in power output (p = F × v) seen in treated patients (Teerlink et al., 2011).
EXPERIMENTAL PROCEDURES
Expression and Purification of Human Cardiac Myosin in C2C12 Cells
For steady-state ATPase measurements and in vitro motility gliding assays, two different human cardiac short S1 (sS1) constructs, one with a C-terminal EGFP tag and the other with C tag (-RSTIDVW) were used, respectively. For both constructs, a flexible GSG linker separates sS1 from the C-terminal tag (see Figure S2). The human α-cardiac sS1 constructs span residues 1–810. The human β-cardiac sS1 constructs span residues 1–808. The cloning, expression, and purification of the EGFP-tagged construct is described elsewhere (Sommese et al., 2013). C-tagged constructs were cloned, expressed, and purified similarly. The only difference is that 10% sucrose was added to the human cardiac sS1 preparations after elution from the Q-column to keep the myosin more stable. The human cardiac sS1 retains its motility properties, our most sensitive test of stability, when flash frozen in liquid nitrogen and stored at −80°C in 10% sucrose. After purification, EGFP-tagged constructs were exchanged into ATPase assay buffer conditions using Amicon centrifugal filter units (Millipore).
Expression and Purification of Utrophin and Other Actin-Binding Proteins
The pDEST17 plasmid encoding the first 416 residues of mouse utrophin was a gift from David Thomas’ laboratory (University of Minnesota). Using this plasmid, the utrophin construct with C tag (see Figure S2) was designed and cloned into a pet47-b vector. The new construct has an N-terminal 6×-his tag for nickel affinity purification. The final plasmid was transformed into the Rosetta (DE3) bacterial cell line for protein expression (see the Supplemental Experimental Procedures).
Expression and Purification of SNAP-PDZ18
The SNAP-PDZ18 fusion construct has an N-terminal 10×-his tag and FLAG tag and a FLAG-tag linker between SNAP and the PDZ18, which is at the C terminus. SNAP-PDZ18 was cloned into a pHFT2 plasmid for expression. SNAP-PDZ18 was expressed and purified using nickel affinity chromatography following the same protocol as for the purification of PDZ18 (Huang et al., 2009) (see the Supplemental Experimental Procedures).
Preparation of Other Proteins and Materials
Expression and purification of full-length human gelsolin is described elsewhere (Sommese et al., 2013). Bovine actin, which is identical in protein sequence to human actin, was a gift from MyoKardia. OM, the structure of which was confirmed by nuclear magnetic resonance and mass spectrometry, was a gift from Leslie Leinwand.
Unloaded and Loaded In Vitro Motility Measurements
The motility assay was as described previously (Kron et al., 1991), with modifications described in the Supplemental Experimental Procedures. Reagents were loaded in the following order: (1) SNAP-PDZ18, incubated 2 min; (2) BSA to block the surface from nonspecific subsequent attachments, incubated 2 min; (3) a mixture of C-tagged human cardiac sS1 and utrophin, incubated for 3 min; (4) ABBSA (defined in Sommese et al., 2013) to wash the channels; and (5) tetramethylrhodamine (TMR)-phalloidin-labeled bovine actin, ATP, an oxygen-scavenging system, and an ATP-regeneration system. Movies were obtained as described in the Supplemental Experimental Procedures. Assays for each cardiac sS1 variant were repeated with at least three fresh protein preparations. At each condition, at least four movies of 30 s duration were recorded. While there was some prep-to-prep variability in absolute velocities, the relative differences between WT β-cardiac sS1 and the α-isoform or mutants were preserved. To minimize any systematic errors due to batch-to-batch differences in C2C12 cells, when the α-cardiac sS1 or the mutant β-cardiac sS1s were prepared, a fresh WT β-cardiac sS1 was prepared in parallel. For measurements with OM, OM is diluted in AB and incubated with cardiac sS1 for 10 min on ice before loading, then included in all buffers such that the final concentrations of the OM in the motility assay buffer is as indicated in the figures.
Actin-Activated Steady-State ATPase Measurements
Actin-activated steady-state ATPase measurements were performed as described previously (Sommese et al., 2013) but using bovine cardiac actin. Human cardiac sS1 that was EGFP tagged was used. The absorbance of EGFP allowed accurate measurement of the concentration of human cardiac sS1. Actin-activated ATPase measurements for each human cardiac sS1 variant were repeated with at least three fresh myosin preparations.
Supplementary Material
Highlights.
In vitro assay and analysis software measures human cardiac myosin power output
α-Cardiac myosin has higher velocity but generates less force than β-cardiac myosin
M531R and S532P respectively increase and decrease β-cardiac myosin force output
Omecamtiv mecarbil increases β-cardiac myosin force but decreases its velocity
ACKNOWLEDGMENTS
We thank Ruth Sommese for designing and cloning the SNAP-PDZ18 affinity clamp construct. We thank Arjun Adhikari for expressing and purifying SNAP-PDZ18, which was initially designed for a separate project. We thank Ron Rock for providing the plasmid for the original PDZ18 (also called as ePDZ-b1) affinity clamp construct. We thank David Thomas for providing the original plasmids for Utrophin. We thank Henrik Flyvbjerg, Carol Cho, Masataka Kawana, and Suman Nag for useful discussions. Omecamtiv mecarbil was a kind gift from Leslie Leinwand. This work was funded by NIH grants GM33289 and HL117138 (to J.A.S.) and by NIH Clinical and Translational Science Award grant KL2 RR025743 and the Advanced Residency Training at Stanford Fellowship (to E.C.Y.). J.A.S. is a founder of MyoKardia and a member of its scientific advisory board.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, one table, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.04.006.
AUTHOR CONTRIBUTIONS
T.A., E.C.Y., K.M.R., and J.A.S. designed the research. T.A., E.C.Y., K.M.R., and S.S. performed the research. T.A. contributed new reagents and analytic tools. T.A. analyzed data. T.A., K.M.R., and J.A.S. wrote the manuscript.
REFERENCES
- Alpert NR, Brosseau C, Federico A, Krenz M, Robbins J, Warshaw DM. Molecular mechanics of mouse cardiac myosin isoforms. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H1446–H1454. doi: 10.1152/ajpheart.00274.2002. [DOI] [PubMed] [Google Scholar]
- Bing W, Knott A, Marston SB. A simple method for measuring the relative force exerted by myosin on actin filaments in the in vitro motility assay: evidence that tropomyosin and troponin increase force in single thin filaments. Biochem. J. 2000;350:693–699. [PMC free article] [PubMed] [Google Scholar]
- Bloemink M, Deacon J, Langer S, Vera C, Combs A, Leinwand L, Geeves MA. The hypertrophic cardiomyopathy myosin mutation R453C alters ATP binding and hydrolysis of human cardiac β-myosin. J. Biol. Chem. 2014;289:5158–5167. doi: 10.1074/jbc.M113.511204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrave M, Peckham M. Structural implications of β-cardiac myosin heavy chain mutations in human disease. Anat. Rec. (Hoboken) 2014;297:1670–1680. doi: 10.1002/ar.22973. [DOI] [PubMed] [Google Scholar]
- Cuda G, Fananapazir L, Epstein ND, Sellers JR. The in vitro motility activity of beta-cardiac myosin depends on the nature of the beta-myosin heavy chain gene mutation in hypertrophic cardiomyopathy. J. Muscle Res. Cell Motil. 1997;18:275–283. doi: 10.1023/a:1018613907574. [DOI] [PubMed] [Google Scholar]
- Daniels M, Noble MI, ter Keurs HE, Wohlfart B. Velocity of sarcomere shortening in rat cardiac muscle: relationship to force, sarcomere length, calcium and time. J. Physiol. 1984;355:367–381. doi: 10.1113/jphysiol.1984.sp015424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon JC, Bloemink MJ, Rezavandi H, Geeves MA, Leinwand LA. Erratum to: Identification of functional differences between recombinant human α and β cardiac myosin motors. Cell. Mol. Life Sci. 2012;69:4239–4255. doi: 10.1007/s00018-012-1111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, Moore JR. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton (Hoboken) 2010;67:273–285. doi: 10.1002/cm.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, Kazmierczak K, Szczesna-Cordary D, Moore JR. Cardiomyopathy-linked myosin regulatory light chain mutations disrupt myosin strain-dependent biochemistry. Proc. Natl. Acad. Sci. USA. 2010;107:17403–17408. doi: 10.1073/pnas.1009619107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeberle JR. Calponin decreases the rate of cross-bridge cycling and increases maximum force production by smooth muscle myosin in an in vitro motility assay. J. Biol. Chem. 1994;269:12424–12431. [PubMed] [Google Scholar]
- Haeberle JR, Hemric ME. Are actin filaments moving under unloaded conditions in the in vitro motility assay? Biophys. J. 1995;68:306S–310S. discussion 310S–311S. [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Schlessman JL, Sue GR, García-Moreno B. Arginine residues at internal positions in a protein are always charged. Proc. Natl. Acad. Sci. USA. 2011;108:18954–18959. doi: 10.1073/pnas.1104808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DE, Work SS, Wright RK, Alpert NR, Warshaw DM. Smooth, cardiac and skeletal muscle myosin force and motion generation assessed by cross-bridge mechanical interactions in vitro. J. Muscle Res. Cell Motil. 1994;15:11–19. doi: 10.1007/BF00123828. [DOI] [PubMed] [Google Scholar]
- Harvey PA, Leinwand LA. The cell biology of disease: cellular mechanisms of cardiomyopathy. J. Cell Biol. 2011;194:355–365. doi: 10.1083/jcb.201101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- Homsher E, Wang F, Sellers JR. Factors affecting movement of F-actin filaments propelled by skeletal muscle heavy meromyosin. Am. J. Physiol. 1992;262:C714–C723. doi: 10.1152/ajpcell.1992.262.3.C714. [DOI] [PubMed] [Google Scholar]
- Huang J, Nagy SS, Koide A, Rock RS, Koide S. A peptide tag system for facile purification and single-molecule immobilization. Biochemistry. 2009;48:11834–11836. doi: 10.1021/bi901756n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson LW, Sellers JR, Taylor DL. Actin-binding proteins regulate the work performed by myosin II motors on single actin filaments. Cell Motil. Cytoskeleton. 1992;22:274–280. doi: 10.1002/cm.970220407. [DOI] [PubMed] [Google Scholar]
- Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- Kaneda T, Naruse C, Kawashima A, Fujino N, Oshima T, Namura M, Nunoda S, Mori S, Konno T, Ino H, et al. A novel beta-myosin heavy chain gene mutation, p.Met531Arg, identified in isolated left ventricular non-compaction in humans, results in left ventricular hypertrophy that progresses to dilation in a mouse model. Clin. Sci. 2008;114:431–440. doi: 10.1042/CS20070179. [DOI] [PubMed] [Google Scholar]
- Keller DI, Coirault C, Rau T, Cheav T, Weyand M, Amann K, Lecarpentier Y, Richard P, Eschenhagen T, Carrier L. Human homozygous R403W mutant cardiac myosin presents disproportionate enhancement of mechanical and enzymatic properties. J. Mol. Cell. Cardiol. 2004;36:355–362. doi: 10.1016/j.yjmcc.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Kimura A. Molecular basis of hereditary cardiomyopathy: abnormalities in calcium sensitivity, stretch response, stress response and beyond. J. Hum. Genet. 2010;55:81–90. doi: 10.1038/jhg.2009.138. [DOI] [PubMed] [Google Scholar]
- Kron SJ, Toyoshima YY, Uyeda TQ, Spudich JA. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 1991;196:399–416. doi: 10.1016/0076-6879(91)96035-p. [DOI] [PubMed] [Google Scholar]
- Liu Y, White HD, Winkelmann DA, Forgacs E. The affect of omecamtiv mecarbilon the phosphate dissociation and motile properties of the recombinant human β-cardiac heavymeromyosin. Biophys. J. 2013;104:153a. [Google Scholar]
- Liu Y, White HD, Belknap B, Winkelmann DA, Forgacs E. Omecamtiv mecarbil modulates the kinetic and motile properties of porcine β-cardiac myosin. Biochemistry. 2015;54:1963–1975. doi: 10.1021/bi5015166. [DOI] [PubMed] [Google Scholar]
- Lompré AM, Nadal-Ginard B, Mahdavi V. Expression of the cardiac ventricular alpha- and beta-myosin heavy chain genes is developmentally and hormonally regulated. J. Biol. Chem. 1984;259:6437–6446. [PubMed] [Google Scholar]
- Malik FI, Hartman JJ, Elias KA, Morgan BP, Rodriguez H, Brejc K, Anderson RL, Sueoka SH, Lee KH, Finer JT, et al. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 2011;331:1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmqvist UP, Aronshtam A, Lowey S. Cardiac myosin isoforms from different species have unique enzymatic and mechanical properties. Biochemistry. 2004;43:15058–15065. doi: 10.1021/bi0495329. [DOI] [PubMed] [Google Scholar]
- Marston SB. How do mutations in contractile proteins cause the primary familial cardiomyopathies? J. Cardiovasc. Transl. Res. 2011;4:245–255. doi: 10.1007/s12265-011-9266-2. [DOI] [PubMed] [Google Scholar]
- Morgan BP, Muci A, Lu PP, Qian X, Tochimoto T, Smith WW, Garard M, Kraynack E, Collibee S, Suehiro I, et al. Discovery of omecamtiv mecarbil the first, selective, small molecule activator of cardiac Myosin. ACS Med Chem Lett. 2010;1:472–477. doi: 10.1021/ml100138q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Camp P, Jr, Alix SL, Gorga JA, Begin KJ, Leavitt BJ, Ittleman FP, Alpert NR, LeWinter MM, VanBuren P. Myosin from failing and non-failing human ventricles exhibit similar contractile properties. J. Mol. Cell. Cardiol. 2003;35:91–97. doi: 10.1016/s0022-2828(02)00282-1. [DOI] [PubMed] [Google Scholar]
- Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter KA, Tyska MJ, Haeberle JR, Alpert NR, Fananapazir L, Warshaw DM. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J. Muscle Res. Cell Motil. 2000;21:609–620. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
- Reiser PJ, Kline WO. Electrophoretic separation and quantitation of cardiac myosin heavy chain isoforms in eight mammalian species. Am. J. Physiol. 1998;274:H1048–H1053. doi: 10.1152/ajpheart.1998.274.3.H1048. [DOI] [PubMed] [Google Scholar]
- Reiser PJ, Portman MA, Ning XH, Schomisch Moravec C. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H1814–H1820. doi: 10.1152/ajpheart.2001.280.4.H1814. [DOI] [PubMed] [Google Scholar]
- Resnicow DI, Deacon JC, Warrick HM, Spudich JA, Leinwand LA. Functional diversity among a family of human skeletal muscle myosin motors. Proc. Natl. Acad. Sci. USA. 2010;107:1053–1058. doi: 10.1073/pnas.0913527107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhnow F, Zwicker D, Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 2011;100:2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata M, Ikebe M. Functional analysis of the mutations in the human cardiac beta-myosin that are responsible for familial hypertrophic cardiomyopathy. Implication for the clinical outcome. J. Clin. Invest. 1996;98:2866–2873. doi: 10.1172/JCI119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaramakrishnan S, Ashley E, Leinwand L, Spudich JA. Insights into human beta-cardiac myosin function from single molecule and single cell studies. J. Cardiovasc. Transl. Res. 2009;2:426–440. doi: 10.1007/s12265-009-9129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommese RF, Sung J, Nag S, Sutton S, Deacon JC, Choe E, Leinwand LA, Ruppel K, Spudich JA. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc. Natl. Acad. Sci. USA. 2013;110:12607–12612. doi: 10.1073/pnas.1309493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenblick EH. Force-velocity relations in mammalian heart muscle. Am. J. Physiol. 1962;202:931–939. doi: 10.1152/ajplegacy.1962.202.5.931. [DOI] [PubMed] [Google Scholar]
- Spudich JA. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 2014;106:1236–1249. doi: 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam R, Winkelmann DA. Myosin II folding is mediated by a molecular chaperonin. J. Biol. Chem. 1999;274:27265–27273. doi: 10.1074/jbc.274.38.27265. [DOI] [PubMed] [Google Scholar]
- Teerlink JR, Clarke CP, Saikali KG, Lee JH, Chen MM, Escandon RD, Elliott L, Bee R, Habibzadeh MR, Goldman JH, et al. Dose-dependent augmentation of cardiac systolic function with the selective cardiac myosin activator, omecamtiv mecarbil: a first-in-man study. Lancet. 2011;378:667–675. doi: 10.1016/S0140-6736(11)61219-1. [DOI] [PubMed] [Google Scholar]
- Udeoji DU, Philip KJ, Morrissey RP, Phan A, Schwarz ER. Left ventricular noncompaction cardiomyopathy: updated review. Ther. Adv. Cardiovasc. Dis. 2013;7:260–273. doi: 10.1177/1753944713504639. [DOI] [PubMed] [Google Scholar]
- VanBuren P, Harris DE, Alpert NR, Warshaw DM. Cardiac V1 and V3 myosins differ in their hydrolytic and mechanical activities in vitro. Circ. Res. 1995;77:439–444. doi: 10.1161/01.res.77.2.439. [DOI] [PubMed] [Google Scholar]
- Warshaw DM, Desrosiers JM, Work SS, Trybus KM. Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J. Cell Biol. 1990;111:453–463. doi: 10.1083/jcb.111.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.