Background: Mutations in NCF2 predispose individuals to lupus.
Results: NCF2 Arg-395 → Trp is associated with lupus; Arg-395 stabilizes the C terminus of NCF4 and NCF2 loop 395–402.
Conclusion: Trp-395 disrupts the NADPH oxidase complex.
Significance: The results support a nuanced rather than strictly proinflammatory role for this pathway in immune regulation with important consequences for autoimmunity.
Keywords: autoimmune disease, autoimmunity, computational biology, human genetics, NADPH oxidase, NCF2, systemic lupus erythematosus
Abstract
In a case-control association study with 3716 North Americans of Hispanic descent and 4867 North Americans of European descent, we show that the associations of rs17849502 (NCF2 His-389 → Gln) and rs13306575 (NCF2 Arg-395 → Trp) with systemic lupus erythematosus are independent. We have shown that His-389 → Gln disrupts the binding of NCF2 to the ZF domain of VAV1, resulting in decreased NADPH oxidase activity. With respect to Arg-395 → Trp, using protein docking and structure analyses, we provide a model for the involvement of this mutation in the structure and function of the NADPH oxidase complex. This model assigns a central role to Arg-395 in the structure and stability of the quaternary NCF2/NCF4/VAV1/RAC1 NADPH oxidase complex. Arg-395 stabilizes the C-terminal tail of NCF4 and the conformation of NCF2 loop 395–402, which in turn stabilize the evolutionarily conserved interactions of NCF2/NCF4 with the DH domain of VAV1 and RAC1 region 120–137. Our findings are consistent with the high levels of conservation of all of the residues involved in these interactions.
Introduction
The superoxide-generating leukocyte NADPH oxidase has long been considered to be of main importance for the generation of reactive oxygen species (ROS) inside the phagosome, where these promote the killing of microbes, or for the release of extracellular ROS mainly thought to contribute to tissue damage.
The NADPH oxidase is composed of multiple subunits, two of which (CYBA and CYBB) are localized in cellular membranes. Upon activation, the three cytosolic subunits (NCF1, NCF2, and NCF4) translocate to the membrane where they, along with RAC-GTP (either RAC1 or RAC2), form the active NOX2 complex (1–3). In resting cells, RAC-GDP is present as a complex with Rho-GDP disassociation inhibitor, but this complex rapidly dissociates, and RAC-GTP forms in stimulated cells (3). This process is facilitated by activation of guanine nucleotide exchange factors (GEFs)3 and is accompanied by translocation of RAC to the membrane (1, 3). The VAV proteins (VAV1, VAV2, and VAV3) are a family of GEFs for Rho family GTPases, and VAV1 and VAV3 are expressed in leukocytes. Upon phosphorylation, VAV acquires GEF activity for RAC, facilitating the transition of RAC to an active GTP-bound state (4). Thus, VAV family GEFs regulate RAC. There is a direct interaction of VAV1 with NCF2 that enhances the GEF activity of VAV1 and causes a positive feedback loop to amplify RAC nucleotide exchange and NADPH oxidase activation (5). NADPH oxidase activity is absent or profoundly decreased in patients with chronic granulomatous disease (CGD) because of mutations in CYBB, CYBA, NCF1, NCF2, or NCF4 resulting in bacterial and fungal infections, aberrant inflammation, and in some patients, autoimmunity (6, 7).
SLE is a multisystem autoimmune disorder with extensive immune dysregulation and chronic inflammation affecting multiple organs. Its development is influenced by the effects of variants in multiple genes associated with immune regulation. We have undertaken a combined genetic, functional, and structural approach to test the candidacy of NCF2, which encodes an essential regulatory subunit of the NADPH oxidase that is activated by RAC-GTP, as a risk factor for SLE. We have shown that genetic variation in NCF2 is strongly associated with increased SLE risk (8). The association between NCF2 and SLE in North Americans of European ancestry could be attributed almost exclusively to a single non-synonymous coding mutation in exon 12, the effect of which is the substitution of histidine (His) 389 with glutamine (Gln) (His-389 → Gln) in the PB1 domain of the NCF2 protein. The association of this mutation in Hispanic Americans was at first inconclusive given the low minor allele frequency of this SNP in North Americans of Hispanic descent (HispNA) (8).
In the present study we replicated the association of NCF2 His-389 → Gln in North Americans of European descent and confirmed that this variant is a risk factor in the North American Hispanic population as well. In addition, a functionally important NCF2 variant, Arg-395 → Trp, was identified in Hispanics but not in Americans of European descent. We present here a combined genetic and structural analysis of the effect of the Arg-395 → Trp mutation on the interactions of the NCF2 PB1 domain with other proteins in the NADPH oxidase complex and propose a mechanism for the functional consequences of this mutation.
MATERIALS AND METHODS
Subjects and Genotyping
All subjects provided written consent using consent forms approved by their respective institutional review boards. This study was approved by the institutional review board of the University of Southern California School of Medicine as well as by those of the institutions providing the samples. Ethnicity was self-reported and verified by parental and grandparental ethnicity when known. Controls were defined as adults with a self-reported absence of SLE or any other autoimmune disease in the subjects or their first-degree relatives. 3716 (1840 unaffected controls, 1876 with SLE) unrelated self-identified HispNA and 4867 (1082 controls, 3785 with SLE) North Americans of European descent were genotyped at multiple SNPs in and surrounding NCF2 using the Illumina ImmunoChip array as part of a larger association study. Individuals with SLE satisfied the revised criterion for SLE from the American College of Rheumatology (9, 10).
Admixture Correction
To address population structure, which can be a source of confounders (11, 12), we performed a principal component analysis on a subset of SNPs that are informative for ancestry (13).
Statistical Analyses
The significance of association was calculated using logistic regression with SLE status as the dependent variable and the additive dosage of the SNP (or SNPs in the case of multiple regression) and the three first principal component analysis axes as independent variables with custom routines written in R (14).
Evolutionary Conservation
Protein sequences for NCF2, NCF4, VAV1, and RAC1 were obtained from NCBI for all organisms where the gene was annotated. These sequences were aligned using ClustalW 2.1 (15) with default settings and plotted using custom routines written in R, which replicate the ClustalW color coding.
Structural Analyses and Molecular Modeling
Experimental structures were downloaded from the Protein Data Bank (PDB) (16). The construction of the model of the ternary assembly NCF2/NCF4/VAV1 was described previously (8). Briefly, the preferred binding positions of excised His and Gln probes were mapped on the surface of VAV1 using the computational anchoring spots mapping program ANCHORS MAP (17). Docking of NCF2 (chain A, PDB code 1oey (18)) to VAV1 (chain B, PDB code 2vrw (19)) was performed using the geometric-electrostatic-hydrophobic (GEH) version of the program MolFit (20) followed by post-scan filtering in which additional properties of the interface, desolvation energy, and statistical propensity measures were tested (21). Several conformers of VAV1 were used in the docking. We found that a conformer similar to the RAC1-bound structure of VAV1 produced the best model. The model of the quaternary NCF2/NCF4/VAV1/RAC1 assembly was constructed by overlaying the structure of VAV1 from the VAV1/RAC1 (PDB code 2vrw (19)) complex onto VAV1 in the NCF2/NCF4/VAV1 model (PDB found in the supplemental material). The rotamer of only one residue, Lys-132, was modified to highlight the proposed interaction with NCF2 Asp-398. Buried surface areas were calculated with NACCESS (22). Molecular graphics and analyses were performed with the UCSF Chimera package (23).
RESULTS
His-389 → Gln and Arg-395 → Trp Are Independently Associated with SLE in Hispanic Americans
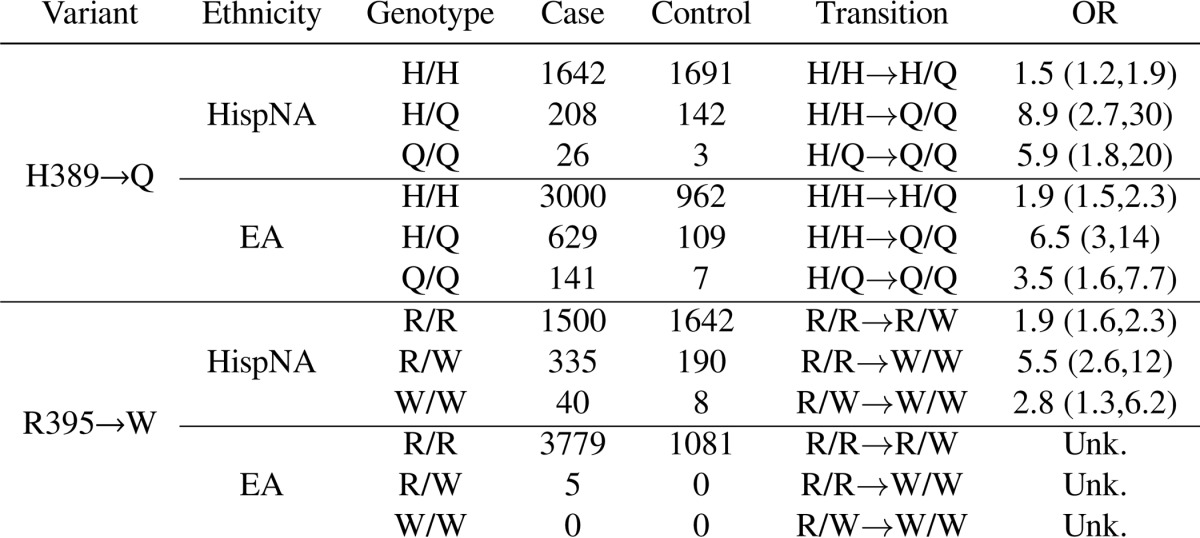
In a follow-up to our previous study (8), we genotyped 140 SNPs in and surrounding NCF2 in a case-control association study of 3716 (1876 SLE cases and 1840 healthy controls) HispNA and 4867 (3785 SLE cases and 1082 healthy controls) North Americans of European descent (EA). We found two significant non-synonymous variants in NCF2 (Table 1). Both rs17849502 (NCF2 His-389 → Gln) and s13306575 (NCF2 Arg-395 → Trp) were significantly associated in HispNA (p = 4.91 × 10−9 and 1.54 × 10−11, respectively). In EA, only rs17849502 (NCF2 His-389 → Gln) was significantly associated (p = 9.47 × 10−14). These two variants in HispNA are independent, as shown by their genome-wide significance (p ≤ 5 × 10−8) in a logistic regression model containing both terms (“rs17849502+rs13306575” row in Table 1). These variants clearly arose in a His-389/Arg-395 background, as there were no individuals who were homozygous for Gln-389 and had any copies of Trp-395 or any individuals who were homozygous for Trp-395 and had any copies of Gln-389 (Table 2). The W395/W genotype is associated with SLE in HispNA with an odds ratio of 5.5 (95% confidence interval: 2.6, 12.0) over R395/R, whereas in EA subjects the W/W genotype is not found (Table 3).
TABLE 1.
NCF2 alleles associated with SLE singly and together in HispNA and EA subjects
| Model | Ethnicity | rs17849502 | rs13306575 |
|---|---|---|---|
| rs17849502 | EA | 9.5 × 10−14 | |
| rs17849502 | HispNA | 4.9 × 10−9 | |
| rs13306575 | HispNA | 1.5 × 10−11 | |
| rs17849502+rs13306575 | HispNA | 3.6 × 10−9 | 1.4 × 10−11 |
TABLE 2.
Coincidence table of North Americans of Hispanic decent diagnosed with SLE with alleles of rs17849502 (columns) and rs13306575 (rows)
The probability of seeing no individuals homozygous for Trp-395 or Gln-389 and heterozygous for the other allele or homozygous for both Trp-395 and Gln-389 (which corresponds to the three zeros in the lower right of the table) is 0.0181.
| His/His | His/Gln | Gln/Gln | |
|---|---|---|---|
| Arg/Arg | 1310 | 164 | 26 |
| Arg/Trp | 291 | 44 | 0 |
| Trp/Trp | 40 | 0 | 0 |
TABLE 3.
Counts of NCF2 variants in cases and controls in HispNA and EA subjects
Odds ratios (OR) are ±95% confidence interval of specific genotype transitions.
Arg-395 → Trp Destabilizes the C-terminal Interaction of NCF4 Segment 330–339 with the NCF2 PB1 Domain and the Conformation of NCF2 Loop 395–402
In a previous study we showed that the mutation NCF2 His-389 → Gln (rs17849502) weakens the interaction of the PB1 domain of NCF2, with VAV1, leading to the reduced production of ROS. His-389 was shown to interact with the zinc finger (ZF) domain of VAV1 in the RAC1-bound conformation (8). In the current study we analyzed the effect of the NCF2 mutation Arg-395 → Trp and the role of loop 395–402 in light of the structure of the complex between the NCF2 and NCF4 PB1 domains (PDB code 1oey (18)) and of our model structures of the NCF2/NCF4/VAV1 (8) and NCF2/NCF4/VAV1/RAC1 assemblies.
The crystal structure of the complex between the NCF2 and NCF4 PB1 domains suggests that the role of NCF2 Arg-395 is to stabilize the conformation of the C-terminal segment of NCF4 (residues 330–339) and NCF2 loop 395–402. NCF4 segment 330–339 is unstructured in free NCF4 (PDB code 2dyb (24)), whereas in the complex with NCF2 it is structured (PDB code 1oey (18)) and makes several highly conserved interactions with NCF2 (Table 4). The contacts of NCF4 segment 330–339 contribute 36% to the total buried surface area in the NCF2/NCF4 complex (510 of 1417 Å2). Of particular importance are the interactions of NCF2 Arg-395. The guanidine group of Arg-395 makes an intermolecular hydrogen bond to one carboxyl oxygen of the C-terminal NCF4 residue Pro-339, and the hydrophobic part of Arg-395 interacts with NCF2 Leu-402, stabilizing the conformation of loop 395–402 (Figs. 1 and 4E). Of note is the dual role of residue NCF4 339, which interacts with both NCF2 residues 395 and 402, thereby stabilizing the structure of NCF2 loop 395–402. The three-way interaction among NCF2 Arg-395 and Leu-402 with NCF4 Pro-339 is disrupted upon the substitution of tryptophan (Trp) for Arg-395, which precludes the electrostatic interaction with the carboxyl oxygen at NCF4 Pro-339 (Fig. 1B).
TABLE 4.
Conserved residues in the NCF2/NCF4/VAV1/RAC1 quaternary assembly
% conservation, the percentage of species containing the indicated residues in the multiply aligned protein. % interaction cons., the percentage of species that maintains the indicated type of interaction between residues in the multiply aligned protein. Less than 1% of the aligned positions were gaps; therefore gaps are ignored for the calculation of conservation. The alignments upon which this table is based can be seen in supplemental Fig. 1.
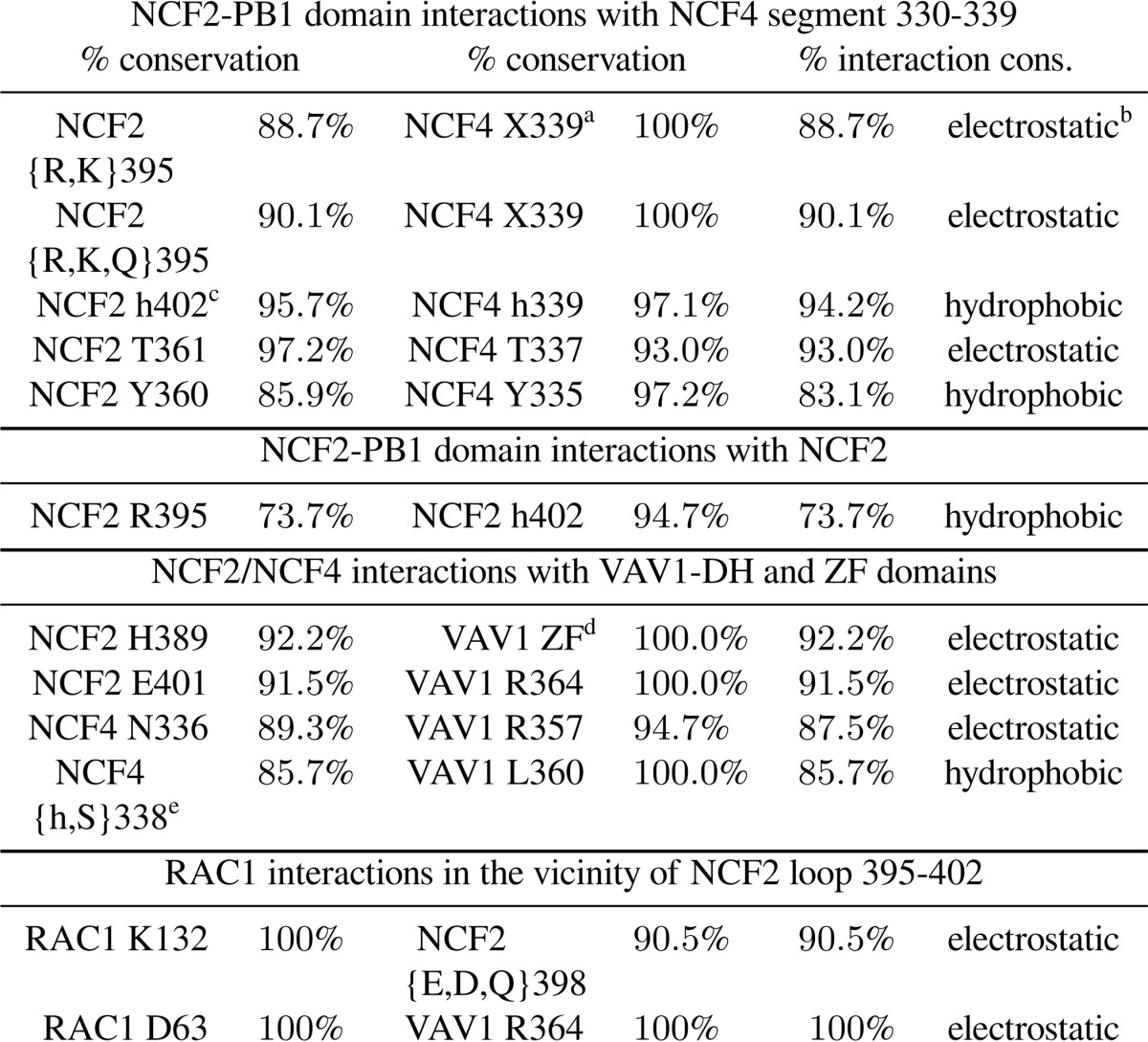
a Interaction with carboxyl or carbonyl oxygen at this location.
b Electrostatic interactions include ion pairing and hydrogen bonding within 3.5 Å of acceptor and donor atoms.
c h, hydrophobic.
d His-389 binds four residues in the ZF domain of VAV1: Glu-509/Asp-509, His-510/Asn-510, Glu-556, and Glu-559.
e In some species Ser or Asp is found in NCF4 position 338; its side chain can form a hydrogen bond to the conserved VAV1 residue Asp-361 or Arg-357, respectively.
FIGURE 1.
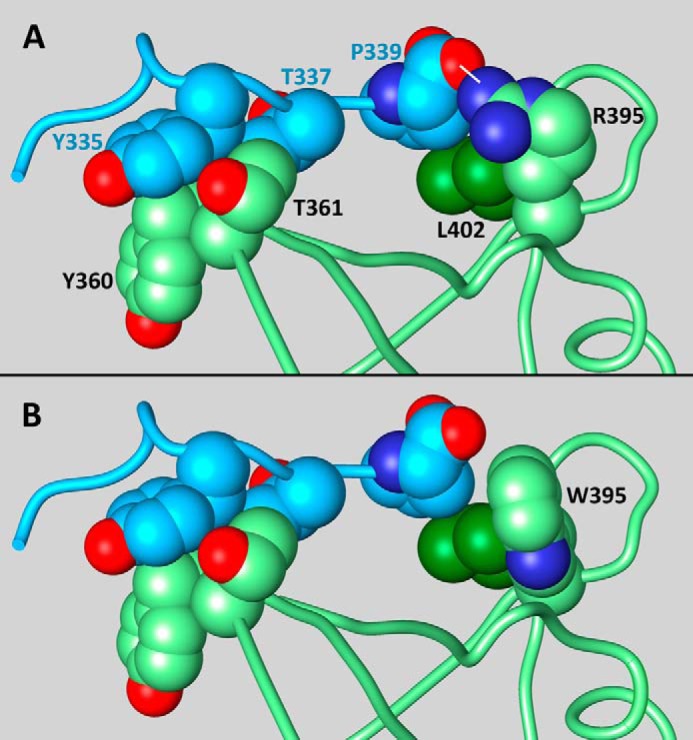
Conserved interactions that stabilize the conformation of NCF4 segment 330–339 and NCF2 loop 395–402 and the effect of mutation Arg-395 → Trp. The atoms of the interacting residues are shown as spheres of van der Waals radii. A, shows the interaction between NCF4 (blue) and NCF2 (green) in the experimental structure of the complex (18). Oxygen atoms are colored red, and nitrogen atoms are in dark blue. Note the cluster on the right formed by NCF2 Arg-395 (green), Leu-402 (dark green), and NCF4 Pro-339 (blue), which is stabilized by electrostatic and hydrophobic interactions. The hydrogen bond between the guanidine nitrogen of Arg-395 and the C-terminal carboxyl of Pro-339 is indicated by a white dash. B, a model of the Arg-395 → Trp mutation. Arg was replaced with Trp without changing the backbone conformation, selecting a side-chain rotamer that does not clash with the rest of the domain (χ1 = 66.0°, χ2 = −6.3° (26, 27)). The hydrogen bond between Arg-395 and NCF4 Pro-339 cannot be formed with Trp in position NCF2 395, and the hydrophobic interactions with NCF2 Leu-402 are weakened (see “Results”).
FIGURE 4.
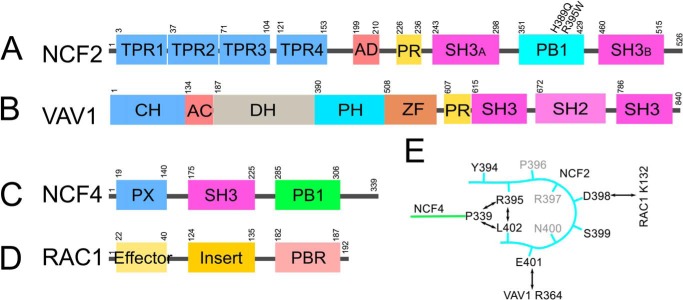
Schematic of the domain structures of NCF2 (A), VAV1 (B), NCF4 (C), and RAC1 (D) and quaternary interactions among NCF2, VAV1, NCF4, and RAC1 involving NCF2 loop 395–402 (E).
It has been suggested previously that the side chain of Trp-395 introduces significant clashes within the PB1 domain of NCF2 (25). We tested all of the common rotamers of Trp (26, 27) and found that most of them would clash with the rest of NCF2 and therefore distort loop 395–402. One low probability rotamer could be introduced in position 395 without intermolecular clashes (χ1 = 66.0°, χ2 = −6.3° (Fig. 1B)). This rotamer cannot make a hydrogen bond to the carbonyl oxygen of NCF4 P339. Furthermore, although the interaction of Arg-395 with Leu-402 buries 103 Å2 of their surfaces, the interaction of this Trp rotamer with Leu-402 buries only 85 Å2 of their surfaces. We concluded that the mutation Arg-395 → Trp weakens the interaction of NCF2 with the C-terminal tail of NCF4 and distorts or destabilizes NCF2 loop 395–402.
NCF2 Arg-395 Is Evolutionarily Conserved, as Are Its Interaction Partners, NCF2 Leu-402 and NCF4 Pro-339
Evolutionarily, the most common amino acid at NCF2 position 395 is arginine (Arg). However, NCF2 395 is Lys or Gln in a few species and, rarely, Leu (supplemental Fig. 1). The hydrogen bond interaction with the backbone carboxyl oxygen (or carbonyl oxygen in the few species with longer NCF4 chains) is therefore evolutionarily conserved (90%) (Table 4 and Fig. 2A). The substitution of lysine (Lys) for Arg-395 in a few species preserves the charged interaction, as Lys is also a positive basic residue. Glutamine (Glu) in Rattus norvegicus retains the ability to hydrogen bond with the C-terminal carboxyl oxygen of NCF4, but it may slightly affect the position of NCF4 339 because the side chain of Gln is shorter than that of Lys or Arg. The importance of this interaction is further supported by the rarely occurring small hydrophobic residue Leu at NCF2 395, which is still able to interact with NCF2 Leu-402 and maintain the stability of loop 395–402 and its interactions with VAV1 and RAC1 (see below). The length of NCF4 (Fig. 2C) is conserved at position 339, but even longer NCF4 molecules can interact with a positive or polar residue at NCF2 395 via their carbonyl group at position 339. Furthermore, the C-terminal tail itself is strongly conserved in mammals. NCF2 402 is conserved as Leu, except in one case where it is Pro, and hence the hydrophobic contact with the hydrophobic part of Lys/Arg/Gln/Leu in NCF2 position 395 and with Pro/Thr in NCF4 position 339 is conserved.
FIGURE 2.
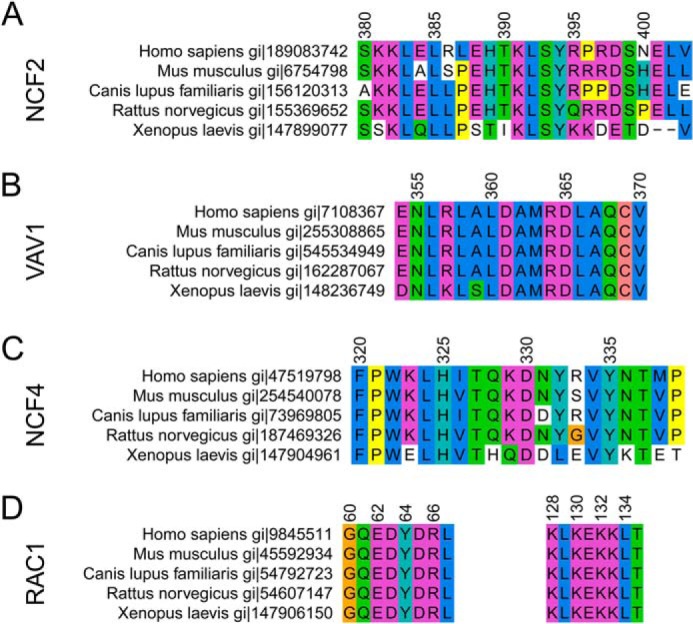
Evolutionary conservation of relevant regions of NCF2 (A), VAV1 (B), NCF4 (C), and RAC1 (D) in selected species. Colors are coded according to the ClustalW (15) color scheme. Numbering corresponds to the human alignment.
Mutations NCF2 His-389 → Gln and NCF2 Arg-395 → Trp Affect the Stability and Structure of the NCF2/NCF4/VAV1 Complex and Its Interactions with RAC1
Another role of the C-terminal segment of NCF4 is revealed in our model structure of the ternary NCF2/NCF4/VAV1 complex. Once bound to NCF2, NCF4 segment 330–339 is stabilized and participates in the binding of the complex of the NCF2/NCF4 PB1 domains to VAV1 (8). The model of the ternary NCF2/NCF4/VAV1 complex was constructed by computational docking of the NCF2 PB1 domain to free VAV1 or to normal modes, variants of VAV1 extracted from the VAV1/RAC1 complex, using the geometric-electrostatic-hydrophobic version of MolFit (20). In an independent computation, the preferred binding positions (anchoring spots) of excised His and Gln probes were mapped on the surface of VAV1 (17). The docking and anchoring spots mapping results were analyzed, searching for binding locations with high geometric-electrostatic-hydrophobic complementarity scores. Binding locations also must utilize anchoring spots that clearly prefer His over Gln and allow binding of the NCF4 PB1 domain without clashes. Our model suggested that NCF2/NCF4 is more likely to bind to the RAC1-bound conformation of VAV1 than to free VAV1. The binding occurs through conserved interactions of NCF2 His-389 with the ZF domain of VAV1 and the NCF2/NCF4 complex with the VAV1 DH domain as summarized in Table 4.
Based on our computational model, the surface area buried upon formation of the complex between NCF2/NCF4 and VAV1 is 1109 Å2. This is well within the range (800–1400 Å2) obtained from statistical studies of biologically relevant heterocomplexes (28). Excluding NCF4 from the complex leaves only 636 Å2 of buried surface area, a reduction of 43%. This reduction is due to loss of contacts with NCF4 segments 330–339, emphasizing the importance of the stabilization of this segment to which NCF2 Arg-395 contributes significantly. The interactions between NCF4 and VAV1 are highly conserved (see Table 4) and include a hydrogen bond interaction between NCF4 Asn-336/Ser-336 and VAV1 Lys-357/Arg-357 and a hydrophobic interaction between NCF4 Met-338/Val-338/Thr-338/Ala-338 and VAV1 Leu-360. In several species position NCF4 338 is occupied by hydrophilic residues Ser/Glu, which can interact with the conserved VAV1 residues Asp-361 and Arg-357, respectively. The conservation of these predicted interactions emphasizes their importance in maintaining the structure of the NCF2/NCF4/VAV1 complex.
The importance of NCF2 loop 395–402, which is stabilized by inter- and intramolecular interactions as outlined above, is revealed when RAC1 is added to the assembly. A model of the quaternary assembly NCF2/NCF4/VAV1/RAC1 (Fig. 3) was obtained by overlaying VAV1 from the complex VAV1/RAC1 (PDB code 2vrw (19)) onto VAV1 in our ternary complex model. It shows that RAC1 loops 60–66 and 131–137 interact with NCF2 loop 395–402 (Fig. 3) utilizing highly conserved residues (Table 4). Loop 60–66 interacts with NCF2 indirectly through Asp-63, which forms a hydrogen bond with conserved VAV1 residue Arg-364 that is also hydrogen-bonded to NCF2 residue Glu-401. RAC1 loop 60–66 is part of switch II, which changes conformation between the GTP-bound RAC1 (PDB code 1mh1 (29)) and the nucleotide-free RAC1 bound to VAV1 (19). Region 120–137 is an insertion domain found to play a role in RAC1 activity in the NADPH oxidase pathway (29), in line with our predicted interaction between NCF2 residue Asp-398 and RAC1 residue Lys-132.
FIGURE 3.
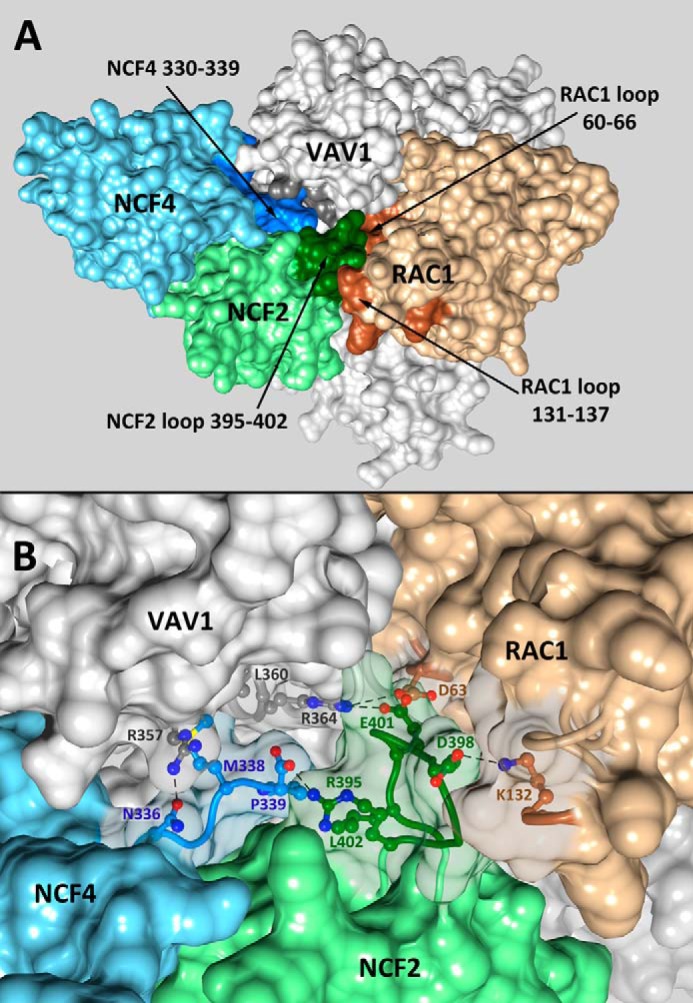
Overall view (A) and details (B) of the quaternary complex NCF2/NCF4/VAV1/RAC1 showing the centrality of NCF2 loop 395–402. The solvent-accessible surfaces of NCF2, NCF4, VAV1, and RAC1 are shown in green, blue, gray, and beige, respectively, with the interacting regions in darker shades. B, details of the interactions of the NCF2/NCF4 PB1 domains with VAV1 and RAC1. The surfaces of the interacting residues were made transparent. The interacting residues are indicated, and hydrogen bonds are denoted by dashed lines.
DISCUSSION
In the present study we replicated and confirmed the strong association of the NCF2 His-389 → Gln mutation in North American SLE subjects of European descent and further demonstrated the association of two causal mutations within the PB1 domain of NCF2 (Arg-395 → Trp and His-389 → Gln) in North American SLE subjects of Hispanic descent. These two mutations, identified within a short stretch of the PB1 domain of NCF2, arose independently (Tables 1 and 2) and are very important for the structure and function of the NADPH oxidase complex.
We have shown by computational modeling that the His-389 → Gln mutation in NCF2 reduces its binding efficiency to the guanine nucleotide exchange factor VAV1 (8). The model predicted that the NCF2 His-389 residue resides in a pocket on the surface of the VAV1 ZF domain (Fig. 4). The effect of the NCF2 His-389 → Gln mutation on NADPH oxidase function was tested with site-specific mutations in NCF2. These showed a 2-fold decrease in ROS production downstream of VAV1-dependent FcγR signaling but had no effect on VAV1-independent NADPH oxidase activity elicited by phorbol myristate acetate (8). Our assessment that this variant has reduced function is further supported by the fact that the NCF2 His-389 → Gln variant was reported in patients with very early onset inflammatory bowel disease. At least in one such subject, impaired ROS production and function was reported in primary leukocytes (30). The occurrence of this NCF2 variant in two different autoimmune diseases strongly supports its functional importance.
As shown here, the Arg-395 → Trp mutation is present in North American SLE subjects of Hispanic descent but not in North American SLE subjects of European descent. NCF2 Arg-395 is highly conserved evolutionarily, suggesting structural and functional importance. Indeed, our model locates Arg-395 at the critical junction of NCF2, NCF4, and VAV1 and close to RAC1 (Fig. 3A), suggesting a mechanism for the functional consequences of the Arg-395 → Trp mutation. One role of Arg-395 is to stabilize the interaction between NCF2 and the C-terminal tail of NCF4 via a hydrogen bond to the carboxyl oxygen of NCF4 residue 339. Thus, whereas in free NCF4 the C-terminal tail (residues 330–339) is disordered (PDB code 2dyb (24)), this segment is ordered in the crystal structure of the NCF2/NCF4 complex (PDB code 1oey (18)). Another role of Arg-395 is to stabilize the conformation of NCF2 loop 395–402 by interacting with Leu-402. This loop makes highly conserved interactions with NCF4, VAV1, and RAC1 (Table 4) and is therefore central to the formation of the quaternary NCF2/NCF4/VAV1/RAC1 complex (Fig. 3). The mutation Arg-395 → Trp precludes formation of the hydrogen bond to NCF4, thus destabilizing the NCF2/NCF4 interaction. It also weakens the hydrophobic interaction with NCF2 Leu-402, thereby affecting the structure of NCF2 loop 395–402 and its interactions with the other proteins in the NCF2/NCF4/VAV1/RAC1 complex.
It is noteworthy that NCF2 interacts with VAV1 at two points, via His-389 (8) and via Glu-401 (within loop 395–402), which is hydrogen-bonded to VAV1 Arg-364. These interactions and the interactions of NCF4 with VAV1 are highly conserved (Table 4). The binding of NCF2/NCF4 to VAV1 appears to restrict the relative positions of the VAV1 DH and ZF domains to the conformation adequate for RAC1 binding. NCF2 loop 395–402 is predicted to also interact with RAC1 through the highly conserved (90.5%) contact NCF2-Asp-398 ↓ RAC1 Lys-132. Lys-132 of RAC1 is perfectly evolutionarily conserved across all species that have an annotated RAC1 gene (Fig. 2D). It is located within the RAC1 insertion domain, region 120–137, that has been found to play a role in its activity in the NADPH oxidase pathway (29), in line with our predicted interaction between NCF2 residue Asp-398 and RAC1 residue Lys-132 (8). Another region of RAC1, loop 60–66, interacts with NCF2 loop 395–402 indirectly as both RAC1 Asp-63 and NCF2 Glu-401 are hydrogen-bonded to VAV1 Arg-364. RAC1 loop 60–66 is part of switch II, which changes conformation between the GTP-bound RAC1 (PDB code 1mh1 (29)) and the nucleotide-free RAC1 bound to VAV1 (19). The change in switch II is necessary for the release of GDP, and its proximity to NCF2 loop 395–402 suggests that the latter contributes to the stabilization of the nucleotide-free RAC1, thus enhancing the nucleotide exchange process.
The mutation Arg-395 → Trp was found previously in a CGD patient from a Mexican kindred (31). The maternal allele of the CGD patient had an Arg-395 → Trp mutation in NCF2 in addition to an in-frame deletion of nine nucleotides in the middle of exon 2 causing the deletion of three amino acids, 19KKD21. Although this allele produced a nearly complete NCF2 mRNA transcript, the protein was not detected by immunoblot. Patiño et al. (31) speculated that the absence of the three amino acids produces an unstable protein that is degraded soon after its synthesis. Furthermore, the N terminus of NCF2 (TPR domain) binds to RAC-GTP; the structure of this complex is available (32). From this structure, we can deduce that NCF2 19KKD21 does not interact directly with RAC, but the deletion is likely to distort the N-terminal end of the TPR domain, possibly affecting the NCF2-TPR ↓ RAC-GTP complex, which is important for NADPH assembly. Therefore, given the complexity of this allele in the CGD patient, the effect of Arg-395 → Trp by itself cannot be assessed.
CGD develops when NADPH oxidase activity is absent or profoundly decreased. It is either X-linked (when caused by null mutations in CYBB) or autosomal recessive (when caused by mutations in all of the other subunit genes). However, only ≈5% of CGD are homozygous (or compound heterozygotes) for null mutations in NCF2 (31).
It is noteworthy that patients who have CGD, as well as CGD carriers, have a higher risk of developing autoimmune diseases (33). Moreover, patients who have CGD and mothers carrying X-linked CYBB mutations have a 5–10% incidence of cutaneous and/or mucosal lesions, characteristic of those seen in SLE (34–36). However, a CGD phenotype is not seen in our SLE subjects even in the presence of a homozygous Gln-389 or Trp-395 NCF2 genotype. This may indicate that the mutations in NCF2 seen in SLE subjects do not cause the catastrophic reduction in NADPH oxidase function seen in CGD patients. The phagocytes of SLE patients remain capable of generating a reduced respiratory burst that is sufficient for antimicrobial activity. Conversely, in immune cells (e.g. antigen-presenting cells and B lymphocytes), ROS production is much more limited (37–39) and the role of ROS is not antimicrobial. Instead, NADPH oxidase activity regulates phagosomal pH, and derivatives of ROS can function as signaling molecules within and between neighboring immune cells. Thus, reduced levels of ROS in these cells may influence antigen processing, immunoregulation, control of cell activation, and differentiation (39, 40), functions that are essential to the development of autoimmune disease.
Increased oxidative stress has been amply documented in SLE (41). To a large degree it is secondary to the chronic inflammation that occurs during the course of disease progression and/or during active disease. It may originate from many non-NADPH oxidase sources such as dysfunctional mitochondria in lupus T-lymphocytes linked to enhanced mitochondrial oxygen consumption (42) or polymorphisms in mitochondrial DNA complex I (43).
Paradoxically, NADPH oxidase deficiency has been shown to have a proinflammatory effect that likely drives oxidative stress. Thus, diminished ROS production by phagocytic cells due to NADPH oxidase deficiency can cause impaired digestion of microbes or debris followed by increased Toll-like receptor stimulation (41) and/or altered redox status of cellular proteins resulting in increased proinflammatory cytokine production via the NFκB pathway.
The NCF2 variants we have described increase the risk of developing SLE but may have no discernible effects on disease progression. Indeed, so far we have not detected any striking differences in the clinical spectrum of disease in SLE patients with these NCF2 variants from patients without them. It is therefore likely that the effects of the NCF2 variants occur in the original process of disease establishment, prior to the “oxidative stress” that appears later in active lupus.
Finally, our findings provide additional support for the emerging paradigm shift that ROS produced by the NADPH oxidase complex are not exclusively proinflammatory byproducts of cellular responses to infectious or inflammatory stimuli. Instead, they have a nuanced function in regulating immune responses with important consequences for the development of autoimmunity.
Supplementary Material
Acknowledgments
We thank the members of the Systemic Lupus Erythematosus Genetics Consortium (John B. Harley, Marta E Alarcón-Riquelme, Lindsey A. Criswell, Patrick M. Gaffney, Chaim O. Jacob, Robert P. Kimberly, Kathy L. M. Sivils, Betty P. Tsao, Timothy J. Vyse, and Carl D. Langefeld) for their involvement in the collection of samples and genotyping of these variants. We thank Mary C. Dinauer invaluable insights into the function of the NADPH oxidase complex. Chimera was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of health Grant P41-GM103311 from NIGMS).
Note Added in Proof
The supplemental file that contains the model of the NCF2/NCF4/VAV1/RAC1 NADPH oxidase complex was missing in the version of this article that was published as a Paper in Press on March 20, 2015. The model structure is now available as a supplement.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1AR0571720 (to C. O. J.). This work was also supported by a grant from the Alliance for Lupus Research (to C. O. J.).

This article contains supplemental Fig. 1 and PDB model of the NCF2/NCF4/VAV1/RAC1 NADPH oxidase complex.
- GEF
- guanine exchange factor
- CGD
- chronic granulomatous disease
- SLE
- systemic lupus erythematosus
- HispNA
- North Americans of Hispanic descent
- EA
- North Americans of European descent
- PDB
- Protein Data Bank
- ROS
- reactive oxygen species
- ZF
- zinc finger.
REFERENCES
- 1. Nauseef W. M. (2004) Assembly of the phagocyte NADPH oxidase. Histochem. Cell Biol. 122, 277–291 [DOI] [PubMed] [Google Scholar]
- 2. Durand D., Vivès C., Cannella D., Pérez J., Pebay-Peyroula E., Vachette P., Fieschi F. (2010) NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 169, 45–53 [DOI] [PubMed] [Google Scholar]
- 3. Groemping Y., Rittinger K. (2005) Activation and assembly of the NADPH oxidase: a structural perspective. Biochem. J. 386, 401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katzav S. (2009) Vav1: a hematopoietic signal transduction molecule involved in human malignancies. Int. J. Biochem. Cell Biol. 41, 1245–1248 [DOI] [PubMed] [Google Scholar]
- 5. Ming W., Li S., Billadeau D. D., Quilliam L. A., Dinauer M. C. (2007) The Rac effector p67phox regulates phagocyte NADPH oxidase by stimulating Vav1 guanine nucleotide exchange activity. Mol. Cell. Biol. 27, 312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schäppi M. G., Jaquet V., Belli D. C., Krause K. H. (2008) Hyperinflammation in chronic granulomatous disease and anti-inflammatory role of the phagocyte NADPH oxidase. Semin. Immunopathol. 30, 255–271 [DOI] [PubMed] [Google Scholar]
- 7. Matute J. D., Arias A. A., Wright N. A., Wrobel I., Waterhouse C. C., Li X. J., Marchal C. C., Stull N. D., Lewis D. B., Steele M., Kellner J. D., Yu W., Meroueh S. O., Nauseef W. M., Dinauer M. C. (2009) A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood 114, 3309–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacob C. O., Eisenstein M., Dinauer M. C., Ming W., Liu Q., John S., Quismorio F. P., Jr., Reiff A., Myones B. L., Kaufman K. M., McCurdy D., Harley J. B., Silverman E., Kimberly R. P., Vyse T. J., Gaffney P. M., Moser K. L., Klein-Gitelman M., Wagner-Weiner L., Langefeld C. D., Armstrong D. L., Zidovetzki R. (2012) Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. Proc. Natl. Acad. Sci. U.S.A. 109, E59–E67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hochberg M. C. (1997) Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 40, 1725. [DOI] [PubMed] [Google Scholar]
- 10. Tan E. M., Cohen A. S., Fries J. F., Masi A. T., McShane D. J., Rothfield N. F., Schaller J. G., Talal N., Winchester R. J. (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 25, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 11. Bouaziz M., Ambroise C., Guedj M. (2011) Accounting for population stratification in practice: a comparison of the main strategies dedicated to genome-wide association studies. PLoS One 6, e28845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng H. W. (2001) Population admixture may appear to mask, change or reverse genetic effects of genes underlying complex traits. Genetics 159, 1319–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baye T. M., Tiwari H. K., Allison D. B., Go R. C. (2009) Database mining for selection of SNP markers useful in admixture mapping. BioData Min. 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. R Core Team (2012) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 15. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 16. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ben-Shimon A., Eisenstein M. (2010) Computational mapping of anchoring spots on protein surfaces. J. Mol. Biol. 402, 259–277 [DOI] [PubMed] [Google Scholar]
- 18. Wilson M. I., Gill D. J., Perisic O., Quinn M. T., Williams R. L. (2003) PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 12, 39–50 [DOI] [PubMed] [Google Scholar]
- 19. Rapley J., Tybulewicz V. L., Rittinger K. (2008) Crucial structural role for the PH and C1 domains of the Vav1 exchange factor. EMBO Rep. 9, 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berchanski A., Shapira B., Eisenstein M. (2004) Hydrophobic complementarity in protein-protein docking. Proteins 56, 130–142 [DOI] [PubMed] [Google Scholar]
- 21. Kowalsman N., Eisenstein M. (2009) Combining interface core and whole interface descriptors in postscan processing of protein-protein docking models. Proteins 77, 297–318 [DOI] [PubMed] [Google Scholar]
- 22. Hubbard S. J., Thorton J. M. (1993) NACCESS Computer Program, Department of Biochemistry and Molecular Biology, University College London, London [Google Scholar]
- 23. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 24. Honbou K., Minakami R., Yuzawa S., Takeya R., Suzuki N. N., Kamakura S., Sumimoto H., Inagaki F. (2007) Full-length p40phox structure suggests a basis for regulation mechanism of its membrane binding. EMBO J. 26, 1176–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim-Howard X., Sun C., Molineros J. E., Maiti A. K., Chandru H., Adler A., Wiley G. B., Kaufman K. M., Kottyan L., Guthridge J. M., Rasmussen A., Kelly J., Sánchez E., Raj P., Li Q. Z., Bang S. Y., Lee H. S., Kim T. H., Kang Y. M., Suh C. H., Chung W. T., Park Y. B., Choe J. Y., Shim S. C., Lee S. S., Han B.G., Olsen N. J., Karp D. R., Moser K., Pons-Estel B. A., Wakeland E. K., James J. A., Harley J. B., Bae S.C., Gaffney P. M., Alarcón-Riquelme M., GENLES. Looger L. L., Nath S. K. (2014) Allelic heterogeneity in NCF2 associated with systemic lupus erythematosus (SLE) susceptibility across four ethnic populations. Hum. Mol. Genet. 23, 1656–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dunbrack R. L., Jr., Cohen F. E. (1997) Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 6, 1661–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dunbrack R. L. (2002) Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 12, 431–440 [DOI] [PubMed] [Google Scholar]
- 28. Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2004) A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 336, 943–955 [DOI] [PubMed] [Google Scholar]
- 29. Hirshberg M., Stockley R. W., Dodson G., Webb M. R. (1997) The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nat. Struct. Biol. 4, 147–152 [DOI] [PubMed] [Google Scholar]
- 30. Dhillon S. S., Fattouh R., Elkadri A., Xu W., Murchie R., Walters T., Guo C., Mack D., Huynh H. Q., Baksh S., Silverberg M. S., Griffiths A. M., Snapper S. B., Brumell J. H., Muise A. M. (2014) Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology 147, 680–689 [DOI] [PubMed] [Google Scholar]
- 31. Patiño P. J., Rae J., Noack D., Erickson R., Ding J., de Olarte D. G., Curnutte J. T. (1999) Molecular characterization of autosomal recessive chronic granulomatous disease caused by a defect of the nicotinamide adenine dinucleotide phosphate (reduced form) oxidase component p67-phox. Blood 94, 2505–2514 [PubMed] [Google Scholar]
- 32. Lapouge K., Smith S. J., Walker P. A., Gamblin S. J., Smerdon S. J., Rittinger K. (2000) Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol. Cell 6, 899–907 [DOI] [PubMed] [Google Scholar]
- 33. De Ravin S. S., Naumann N., Cowen E. W., Friend J., Hilligoss D., Marquesen M., Balow J. E., Barron K. S., Turner M. L., Gallin J. I., Malech H. L. (2008) Chronic granulomatous disease as a risk factor for autoimmune disease. J. Allergy Clin. Immunol. 122, 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cale C. M., Morton L., Goldblatt D. (2007) Cutaneous and other lupus-like symptoms in carriers of X-linked chronic granulomatous disease: incidence and autoimmune serology. Clin. Exp. Immunol. 148, 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van den Berg J. M., van Koppen E., Ahlin A., Belohradsky B. H., Bernatowska E., Corbeel L., Español T., Fischer A., Kurenko-Deptuch M., Mouy R., Petropoulou T., Roesler J., Seger R., Stasia M. J., Valerius N. H., Weening R. S., Wolach B., Roos D., Kuijpers T. W. (2009) Chronic granulomatous disease: the European experience. PLoS One 4, e5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winkelstein J. A., Marino M. C., Johnston R. B., Boyle J., Curnutte J., Gallin J. I., Malech H. L., Holland S. M., Ochs H., Quie P., Buckley R. H., Foster C. B., Chanock S. J., Dickler H. (2000) Chronic granulomatous disease: report on a national registry of 368 patients. Medicine (Baltimore) 79, 155–169 [DOI] [PubMed] [Google Scholar]
- 37. Paclet M. H., Coleman A. W., Burritt J., Morel F. (2001) NADPH oxidase of Epstein-Barr virus immortalized B lymphocytes: effect of cytochrome b(558) glycosylation. Eur. J. Biochem. 268, 5197–5208 [DOI] [PubMed] [Google Scholar]
- 38. Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I. C., Lennon-Duménil A. M., Seabra M. C., Raposo G., Amigorena S. (2006) NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126, 205–218 [DOI] [PubMed] [Google Scholar]
- 39. Mantegazza A. R., Savina A., Vermeulen M., Pérez L., Geffner J., Hermine O., Rosenzweig S. D., Faure F., Amigorena S. (2008) NADPH oxidase controls phagosomal pH and antigen cross-presentation in human dendritic cells. Blood 112, 4712–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hultqvist M., Olsson L. M., Gelderman K. A., Holmdahl R. (2009) The protective role of ROS in autoimmune disease. Trends Immunol. 30, 201–208 [DOI] [PubMed] [Google Scholar]
- 41. Perl A. (2013) Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat. Rev. Rheumatol. 9, 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doherty E., Oaks Z., Perl A. (2014) Increased mitochondrial electron transport chain activity at complex I is regulated by N-acetylcysteine in lymphocytes of patients with systemic lupus erythematosus. Antioxid. Redox Signal. 21, 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vyshkina T., Sylvester A., Sadiq S., Bonilla E., Canter J. A., Perl A., Kalman B. (2008) Association of common mitochondrial DNA variants with multiple sclerosis and systemic lupus erythematosus. Clin. Immunol. 129, 31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.