Background: An Arg-Gly-Asp (RGD) motif in CagL anchors Helicobacter pylori to host cell integrins.
Results: Reduced pH causes a conformational change in CagL that buries the RGD motif.
Conclusion: A pH-induced conformational change regulates CagL RGD binding to host cells.
Significance: Regulation of the RGD-integrin interaction prevents premature host cell attachment.
Keywords: analytical ultracentrifugation, conformational change, Helicobacter pylori, integrin, x-ray crystallography
Abstract
Arginine-aspartate-glycine (RGD) motifs are recognized by integrins to bridge cells to one another and the extracellular matrix. RGD motifs typically reside in exposed loop conformations. X-ray crystal structures of the Helicobacter pylori protein CagL revealed that RGD motifs can also exist in helical regions of proteins. Interactions between CagL and host gastric epithelial cell via integrins are required for the translocation of the bacterial oncoprotein CagA. Here, we have investigated the molecular basis of the CagL-host cell interactions using structural, biophysical, and functional analyses. We solved an x-ray crystal structure of CagL that revealed conformational changes induced by low pH not present in previous structures. Using analytical ultracentrifugation, we found that pH-induced conformational changes in CagL occur in solution and not just in the crystalline environment. By designing numerous CagL mutants based on all available crystal structures, we probed the functional roles of CagL conformational changes on cell surface integrin engagement. Together, our data indicate that the helical RGD motif in CagL is buried by a neighboring helix at low pH to inhibit CagL binding to integrin, whereas at neutral pH the neighboring helix is displaced to allow integrin access to the CagL RGD motif. This novel molecular mechanism of regulating integrin-RGD motif interactions by changes in the chemical environment provides new insight to H. pylori-mediated oncogenesis.
Introduction
The Arg-Gly-Asp (RGD) motif is an important structural motif found in multiple proteins that allows binding to integrins (1, 2). The RGD motif is important for mediating cell-cell contact and adhesion to the extracellular matrix to form tissues and organs. Several structures of RGD containing proteins have been solved, resolving the RGD motif predominately in loops such as fibronectin (3–5). The RGD motif has also be found to be buried, as in collagen, where is only becomes exposed during denaturation when the collagen is damaged (6). Several pathogenic bacteria and viruses utilize RGD motifs as virulence factors including Group A Streptococcus, foot and mouth disease virus, Bordetella pertussis, and Helicobacter pylori (7–10).
H. pylori resides in the stomachs of 50% of the world's population, where it remains predominately asymptomatic for life (11). However, in ∼20% of infected individuals it causes peptic ulcers, symptomatic gastritis, and/or gastric adenocarcinomas (12, 13). H. pylori infection begins with colonization of the gastric epithelial lining, where the bacterium uses several adhesins, such as SabA or BabA, to anchor itself to a host gastric epithelial cell (14, 15). A type four secretion system (T4SS)2 comprised of ∼30 proteins is then used by the bacterium to inject its sole effector protein, the oncoprotein CagA, into the host cell cytoplasm (16, 17). The T4SS engages host cell integrins to bridge its pilus to the plasma membrane (8). Specifically, the T4SS protein CagL, which decorates the pilus exterior, interacts with α5β1, αVβ3,αvβ5, and αvβ6 integrins through an RGD motif (8, 18–21). Although weaker than the integrin binding affinity observed for fibronectin, CagL-integrin interactions have been shown to be essential for the translocation of CagA into host cells (8). X-ray crystal structures of CagL revealed that its RGD motif is located within the center of a long α helix, the first structure of a protein where the RGD sequence has been observed outside of an exposed loop conformation (22, 23). These structures also highlighted the dramatic conformational heterogeneity of CagL in that the length of the longest axis of the protein can effectively double, leading to the formation of a domain-swapped crystallographic dimer (22).
The root cause of these conformational changes in CagL, whether they exist in solution, and their potential role in host cell binding by the helical RGD motif have remained largely unexplored. Here, we have investigated the conformational changes and cell adherence properties of CagL using numerous complementary structural, biophysical, and functional analyses. Together, our data describe how CagL undergoes specific conformational changes in response to changes in pH to modulate its binding capacity to host cell integrins through exposure of its RGD motif. Our results describe a novel molecular mechanism by which integrin-RGD interactions can be regulated and provide mechanistic insight to the pathogenicity of a cancer-causing bacterium.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis of CagL
CagLWT (residues 23–237, H. pylori strain 26995) was cloned into an NcoI/XhoI cut pET21d vector with a C-terminal hexahistidine tag. This vector served as a template for whole plasmid mutagenesis to create CagLR76A, CagLΔ23–61, CagLΔ55, CagLΔ55–56, CagLΔ55–57, and CagLΔ55–58. Engineering of a disulfide bond across α5 and α6 to prevent the conformational change (CagLL156/F204C) was designed by submitting CagL3zci to the Disulfide by Design 2 server (24).
Protein Expression and Purification
CagLWT and all mutants were expressed in Escherichia coli BL21(DE3) pLysS cells and purified as described before (8). Briefly, cells were grown at 37 °C until an A600 nm of 0.6. Cells were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and grown for a further 2 h at 37 °C. Cells were resuspended in 50 mm KH2PO4-K2HPO4, 200 mm NaCl, pH 7.5 (ice-cold), and lysed by sonication. The soluble fraction was discarded and inclusion bodies were washed twice with 50 mm KH2PO4-K2HPO4, 200 mm NaCl, pH 7.5. Inclusion bodies (from 1 liter of LB) were solubilized in 5 ml of 50 mm KH2PO4-K2HPO4, 200 mm NaCl, 6 m guanidine hydrochloride, pH 7.5. CagL was refolded at 4 °C by diluting the re-dissolved inclusion bodies slowly (100 μl min−1) into 0.4 liters of 50 mm Tris, 20 mm NaCl, 0.1 mm KCl, 1 mm EDTA, 2 mm reduced glutathione, 0.2 mm oxidized glutathione, pH 8.3. Refolding was allowed to continue for 2.5 days at 4 °C. EDTA was quenched by the addition of MgCl2 to a final concentration of 2 mm. Protein precipitate was filtered and refolded CagL was concentrated by nickel affinity purification (HisPur Ni-NTA Resin, Thermo Scientific). CagL was further purified by size exclusion chromatography using a Superdex 200 column (GE Healthcare) equilibrated with 50 mm Tris, 200 mm NaCl, 1 mm EDTA, pH 7.5.
Thermal Stability
CagLWT, which was dialyzed against 50 mm Tris, 200 mm NaCl, pH 7.5 (1 mg ml−1), was mixed with ×5000 Sypro Orange (Sigma) to a final concentration of ×10. 12.5 μl was diluted 2-fold in a 96 White TempPlate with semi-skirt (USA Scientific) with 200 mm of various buffers spanning pH 3.5–9.5 in the absence and presence of 500 mm NaCl. Melting curves were measured on an iQ5 Multicolor Real Time PCR Detection System (Bio-Rad). Melting curves were measured from 25–95 °C with 1 °C intervals and 1-min dwell time at each temperature before measuring fluorescence. Data were analyzed as described by Niesen et al. (25).
V8 Protease Sensitivity
CagL variants were dialyzed against water. 190 pmol of each CagL construct was mixed 2-fold with either 200 mm ammonium bicarbonate, pH 7.8, or 200 mm ammonium acetate, pH 4.0, and 10 pmol of endoproteinase Glu-C from Staphylococcus aureus V8 (Sigma) was added. Samples were incubated at 37 °C for 3 h. Proteolytic degradation was analyzed by SDS-PAGE.
CagL Crystallization
CagLWT was dialyzed against 20 mm sodium acetate, 20 mm NaCl, pH 4.5. CagLWT was screened against Wizard Classic Screens (Rigaku) at 8 mg ml−1 with a protein:reservoir ratio of 100:100 nl at room temperature. Microcrystals grew in 40% (v/v) ethanol, 100 mm sodium phosphate:citric acid buffer, pH 4.2, 5% (w/v) PEG 1000 within a few days. Single crystals were grown with a protein:reservoir ratio of 600:600 nl at room temperature against a reservoir consisting of 30% (v/v) ethanol, 200 mm sodium phosphate:citric acid buffer, pH 4.2, 5% (w/v) PEG 1000, 1% (v/v) glycerol and after several rounds of microseeding. A single crystal was washed in the same condition containing 20% (v/v) ethylene glycol before being flash-cooled in liquid nitrogen. A single 360° data set (oscillations of 0.1°) was collected at 100 K on beamline 11-1 at SSRL, USA, using a Pilatus 6M PAD detector.
CagL Structure Determination
The data set was processed with XDS, and scaled and truncated with Aimless. The structure was solved by molecular replacement using CagL (PDB 3zci) as a search model in Phaser. The model was manually rebuild in Coot and refined with Refmac5 with three TLS groups. Data collection and refinement statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics for CagL
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P6522 |
| Unit cell parameters (Å) | a = b = 63.02, c = 243.8 |
| Resolution (Å) | 54.58–2.30 (2.39–2.30) |
| No. of observed reflections | 510,170 (58,592) |
| No. of unique reflections | 13,696 (1,484) |
| Multiplicity | 37.2 (39.5) |
| Completeness (%) | 100 (100) |
| Rmerge | 0.169 (5.247) |
| Rmeas | 0.173 (5.372) |
| Rp.i.m. | 0.037 (1.146) |
| 〈I〉/〈σ(I)〉 | 19.3 (1.3) |
| CC1/2 | 1.000 (0.731) |
| Wilson B factor (Å) | 54.6 |
| Refinement | |
| Resolution (Å) | 54.58–2.30 |
| Rcryst (%) | 25.5 |
| Rfree (%) | 31.3 |
| No. of reflections | |
| Work set | 12,955 |
| Test set | 801 |
| No. of molecules/atoms | |
| Protein | 1,647 |
| Water | 3 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.094 |
| Mean B factor (Å2) | 63.29 |
Analytical Ultracentrifugation
All sedimentation measurements were performed at 20 °C using a Beckman-Coulter XL-I analytical ultracentrifuge equipped with a 4-hole An-60Ti rotor. For both sedimentation equilibrium and velocity measurements, the protein was first dialyzed exhaustively against buffer containing either 50 mm Tris-HCl, pH 7.5, 150 mm NaCl or 50 mm sodium acetate, pH 4.5, 150 mm NaCl. The values of the protein partial specific volume and solvent density and viscosity used data analysis were calculated from the protein amino acid and the buffer composition using SedenTerp.
Sedimentation Equilibrium
The oligomerization properties of CagLWT were determined at pH 4.5 and 7.5 by equilibrium analytical ultracentrifugation. Protein samples were prepared in each of the buffers at three concentrations and loaded into cells equipped with 6-hole charcoal-filled epon centerpieces (1.2-cm path length) with sapphire windows. Centrifugation was carried out at 20,000, 23,000, and 26,000 rpm, and scans were acquired at 280 nm with a step size of 0.001 and 5 averages per step. The data were globally analyzed using a monomer-dimer model in WinNonLin (26).
Sedimentation Velocity
Protein samples dialyzed against the buffers described above were diluted to 10 μm concentration to ensure that the monomeric species was nearly exclusively populated and loaded into cells equipped with double sector, charcoal-filled epon centerpieces with sapphire windows. Cells were placed in a 4-hole An-60Ti rotor that had been pre-equilibrated at 20 °C for 1 h. The loaded rotor was placed in the microcentrifuge and temperature equilibration was continued for at least 2 h prior to starting centrifugation. Samples were centrifuged at a speed of 50,000 rpm and scans were collected in continuous mode at 239 nm to obtain a sufficiently high signal to noise ratio with the dilute samples. Data were analyzed to obtain the sedimentation coefficients using DCDT+ version 2.4.1 (27, 28).
Cell Adhesion
AGS cells (ATCC 1739) were grown and expanded at 37 °C with 5% (v/v) CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mm GlutaMAX, 100 units/ml of penicillin, and 100 units/ml of streptomycin (Life Technologies). Cells were initially grown in a T-25 flask for 2 days before expanding into a T-75 flask for 2 days before finally splitting into a T-175 flask for 2 days (Corning). MaxiSorp flat-bottom 96-well plates (Nunc) were coated at room temperature for 3 h with 50 μl of 500–2.5 nm CagL or 25–0.1 nm fibronectin (R&D Systems) diluted in PBS. 100 μl of 5% (w/v) nonfat dried milk powder (Foodhold) in PBS, pH 7.4, was added and incubated at room temperature for 2.5 h. Wells were emptied and 200 μl of fresh 5% (w/v) nonfat dried milk powder in PBS, pH 7.4, was added and left overnight at 4 °C. Harvested and combined nonattached cells and trypsinized detached cells were split and centrifuged at 200 × g for 10 min at room temperature. The supernatant was discarded and cells were resuspended in buffered RPMI 1640 medium at 5 × 105 cells/ml. Buffered RPMI 1640 was made by dissolving 4.16 g of RPMI 1640 medium with l-glutamine and without sodium bicarbonate (Sigma) in 300 ml of water. The medium was buffered to pH 4.5, 5.5, 5.75, 6.0, 6.25, 6.5 or 7.5 by the addition of buffer and acid/base to final concentrations of: pH 4.5, 14.7 mm sodium acetate, 8.4 mm HCl; pH 5.5, 32 mm MES, 7 mm NaOH; pH 5.75, 28.6 mm MES, 8.7 mm NaOH; pH 6.0, 24 mm MES, 11 mm NaOH; pH 6.5, 20.7 mm MES, 9.5 mm NaOH; pH 7.5, 23 mm HEPES, 11.5 mm NaOH. The buffered RPMI 1640 medium was then diluted to 400 ml with water and vacuum filtered through 0.22-μm filters (Denville Scientific) and afterward supplemented with 10% fetal calf serum, 2 mm GlutaMAX, 100 units/ml of penicillin, 100 units/ml of streptomycin (Life Technologies), and 10 μm CaCl2/MgCl2/MnCl2. The wells of the plate were emptied and 100 μl of cells were added. Plates were incubated at 37 °C with no CO2 for 2 h. Nonattached cells were removed and the plates were washed twice with 200 μl of PBS, pH 7.4. 60 μl of 1.9 mm p-nitrophenol-N-acetyl-β-d-glucosaminide (Sigma), 0.25% (v/v) Triton X-100, and 50 mm sodium citrate, pH 5.0, was added and incubated at room temperature for 75 min. Development of deprotonated p-nitrophenol was accomplished by the addition of 90 μl of 200 mm glycine, pH 10.4. The absorbance value of each plate was measured in a Victor2 1420 Multilabel counter (Wallac) at 405 nm. Background readings were subtracted from the data and were either normalized to 25 nm fibronectin, pH 7.5 (fibronectic samples), or 500 nm CagLWT, pH 7.5 (CagL variants). Cell viability measurements were made of the AGS cells immediately and 2–3 h after resuspension in buffered RPMI 1640 medium by tyrpan blue stain (Life Technologies). Viability was always greater than 90%.
CagA Translocation and IL-8 Induction
For generating an H. pylori strain that produces a disulfide-locked CagL variant (L156C/F204C), we used the recA integration plasmid pWS255 (pWS241-PcagA-cagL) as a template and transformed the resulting plasmid pPT30 into a cagL deletion mutant of H. pylori strain P12 (29). Functionality of the complemented strain was assessed using AGS cell infection, IL-8 ELISA, and tyrosine phosphorylation assays, as described previously (33). Briefly, AGS cells were grown in 6-well plates to 80% confluence, adjusted to different pH values by replacing the cell culture medium with buffered media as described above, and infected with H. pylori strains at a multiplicity of infection of 60 for 4 h at 37 °C, 5% CO2. Subsequently, co-culture supernatants were removed, centrifuged, and used for determination of IL-8 concentrations. The infected cells were washed twice with PBS and scraped into PBS containing 1 mm sodium orthovanadate, 1 mm PMSF, 10 μg/ml of leupeptin, and 10 μg/ml of pepstatin. Cells were collected by centrifugation, resuspended in SDS sample solution, and analyzed by immunoblotting.
RESULTS
CagL Stability Increases with Reduced pH
Translocation of CagA into gastric epithelial cells is dependent on the interaction of the pilus protein, CagL, with host cell integrins (8). To further investigate this interaction, we expressed full-length CagL without its signal peptide (residues 21–237) but including a C-terminal His6 tag. Our attempts to crystallize the CagL from a neutral pH storage buffer resulted almost exclusively in heavy precipitation. Thus, we used differential scanning fluorimetry to identify conditions that increased its thermal stability (Tm), a property shown to increase the success rate of protein crystallization (30, 31). We observed an increase in Tm of ∼10 K by lowering the pH from 7.5 to 4.0 (data not shown). We also assessed CagL stability by incubation with the V8 protease from S. aureus, as it retains the same specificity and activity (i.e. hydrolysis on the carboxylic side of glutamic acid residues) at both pH 4.0 and 7.8 (32). In agreement with our thermal shift assay results, we observed that CagL was more susceptible to proteolysis at pH 7.8 than at pH 4.0 (data not shown).
CagL Adopts an Elongated Conformation When Crystallized at Low pH
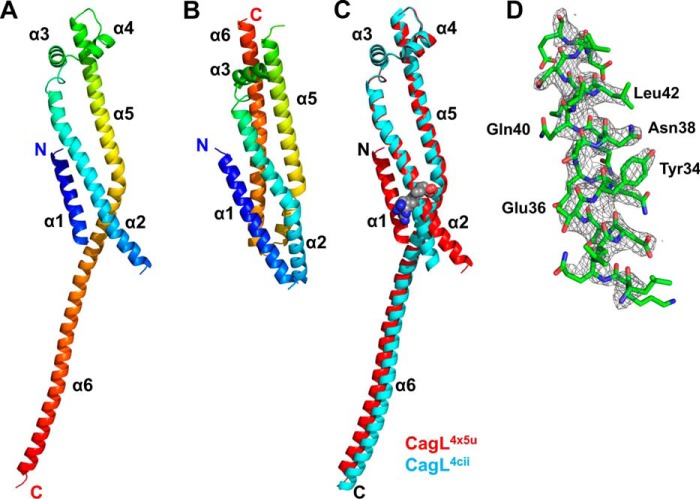
We exchanged purified CagL into a storage buffer at pH 4.5 and repeated our attempts to crystallize the protein. Under these conditions, we observed that CagL more readily achieved the metastable state required for crystallization and identified microcrystals growing in a pH 4.5 crystallization condition, which we optimized via microseeding to produce single crystals with a tetragonal bipyramidal morphology. We solved the CagL structure at 2.3-Å resolution by molecular replacement using the mutant CagLKKQEK crystal structure (PDB code 3zci) as a search model. Scaling and refinement statistics are shown in Table 1. Residues 27–237 of CagL are resolved in this structure (which has a PDB code of 4x5u and is hereafter referred to as CagL4x5u), with only the N-terminal 6 residues and residues 50–57 from the loop connecting the α1 and α2 helices lacking sufficient electron density to be modeled in the final structure (Fig. 1A). In the first two published structures of CagL (PDB entries 3zcj and 3zci), the protein adopts a compact 6-helix bundle structure as shown in Fig. 1B (23). A previously determined CagL structure (PDB entry 4cii) revealed that CagL could undergo a large conformational change (22); the loop residues 176–179 connecting the α5 and α6 helices convert to an α helix conformation, and together the α5 and α6 helices form a single long helix in this structure (Fig. 1C). This helical fusion/elongation results in a domain-swapped crystallographic dimer. In our structure, CagL also exists as a domain-swapped dimer in the crystal, very similar to CagL4cii, with a backbone root mean square deviation of 0.986 Å (Fig. 1C). However, the CagL structure that we report here exhibits two major differences relative to CagL4cii: 1) the N terminus of the α2 helix, which contains the RGD motif, is not kinked (Fig. 1C) and 2) the N-terminal helix, α1, is resolved (Fig. 1, C and D).
FIGURE 1.
Stability and structure of CagL. A, schematic ribbon of CagL structure solved at pH 4.5 showing the 6 helices from the N terminus (blue) to the C terminus (red). B, structure of the compact form of CagL3zci. C, superposition of CagL4cii (blue) onto the recent crystal structure, CagL4x5u (red). The presence of α1 in the new structure causes straightening of the N-terminal half of α2 below the RGD motif (gray spheres). D, representation of the 2Fo − Fc σA-weighted electron density map contoured at 1.5 σ showing α1 residues of CagL.
CagL Dimerization and Conformational Elongation Occur in Solution
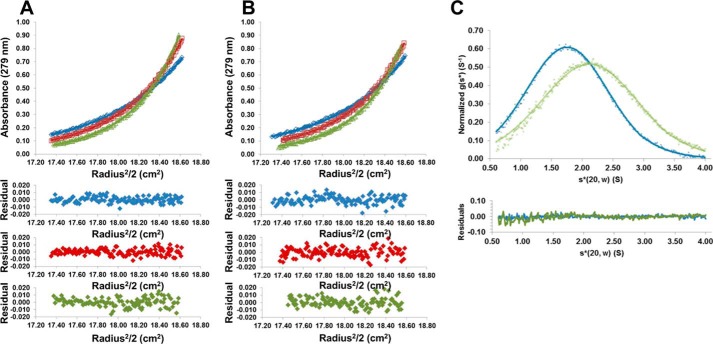
Although crystallization of CagL at low pH consistently results in an elongated structure that forms a crystallographic domain-swapped dimer (22), the oligomeric nature of CagL in solution has not been investigated. Therefore, we used analytical ultracentrifugation to measure the dimerization constant, Kdimerization, of CagL at pH 4.5 and 7.5 by sedimentation equilibrium analysis at concentrations ranging from 10 to 40 μm and rotor speeds of 20,000 to 26,000 rpm. Fig. 2, A and B, show typical sedimentation data of the highest concentration of CagL for three different rotor speeds at pH 4.5 and 7.5, respectively. By globally fitting these data to a monomer-dimer self-association model, we estimated a molecular mass of 29 ± 1 kDa and a Kdimerization of 400 ± 100 μm for CagL at pH 4.5. Fitting the same model to the pH 7.5 data, we observed a species characterized by a molecular mass of 31 ± 2 kDa and a Kdimerization of 170 ± 60 μm. These data indicate that weak dimerization occurs at both pH 4.5 and 7.5, and is approximately twice as strong at neutral pH.
FIGURE 2.
CagL conformational changes in solution. A, top, sedimentation equilibrium experiment of CagL (38 μm) in 50 mm sodium acetate, 150 mm sodium chloride, pH 4.5, with rotor speeds of 20,000, 23,000, and 26,000 rpm (blue, red, and green curves, respectively). Bottom, residuals of fitted data for each curve. B, same experiment as described in A except conducted in 50 mm Tris, 150 mm sodium chloride, pH 7.5. Dimerization constants were measured from these data as 380 ± 115 μm at pH 4.5 and 170 ± 55 μm at pH 7.5. C, sedimentation velocity experiment of CagL (10 μm) at 50,000 rpm in 50 mm sodium acetate, 150 mm sodium chloride, pH 4.5 (blue crosses), or 50 mm Tris, 150 mm sodium chloride, pH 7.5 (green crosses). Fitted data are shown as solid lines and the residuals (bottom) are plotted. CagL sediments with an s20, w* of 1.9 and 2.2 for pH 4.5 and 7.5, respectively.
To investigate whether CagL undergoes pH-dependent conformational changes in solution as suggested by the various crystal structures, we conducted sedimentation velocity experiments to measure the rate at which monomeric CagL sediments at both pH 4.5 and 7.5. At a concentration more than 1 log-fold lower than the Kdimerization, CagL is predominately monomeric regardless of pH, and if it undergoes elongation would sediment slower at pH 4.5. We found this to be the case, with CagL sedimenting as an ∼2.2 S particle at pH 7.5, whereas at pH 4.5 it sedimented as an ∼1.9 S particle (Fig. 2C). We calculated theoretical S values using the program HYDROPRO (33) for all four structures of CagL. For PDB entries 3zci and 3zcj, both at neutral pH, we obtained values of 2.1 and 2.0 S, respectively; for our structure, 4x5u, and PDB entry 4cii, both at acidic pH, we obtained a sedimentation coefficient of 1.7 S. Although the calculated values are lower than those that we measured experimentally, the differences between neutral and acidic pH are consistent with our observation that the S values are higher at pH 7.5 than at pH 4.5. These data indicate that CagL undergoes significant conformational changes in solution in response to pH that are equivalent to those observed in the crystal structures.
CagL Adherence to Host Cells and Functional Consequences Are pH-dependent
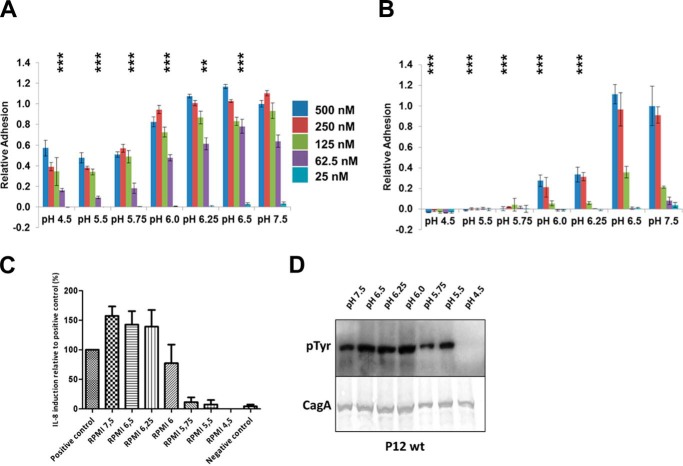
Numerous studies have shown that CagL binds to human gastric epithelial cells dependent on its RGD motif to cell surface integrins α5β1, αVβ5, and αVβ3 and that this interaction is an essential step for the delivery of the effector protein CagA into host cells (8, 19, 20). RGD motifs are common to many proteins that bind integrins and, prior to the originally published CagL structure (23), were found exclusively in loop sequences. CagL was the first, and remains the only, protein known to contain an RGD motif located within an α helix, specifically within the α2 helix (Fig. 1C). As pH has a pronounced effect on the structure of CagL, we investigated the role of pH on the adhesion of both fibronectin (in which the RGD motif is situated in a loop (34) and is likewise important for mediating cell-cell contact via integrin binding) and CagL to gastric epithelial cells. We grew AGS cells in RPMI 1640 buffered with sodium bicarbonate at 37 °C in the presence of 5% CO2 before splitting the cells and reconstituting them in buffered RPMI 1640 at pH values spanning pH 4.5 to 7.5 at concentrations that produce osmolarities identical to sodium bicarbonate to prevent cell shrinkage or lysis. We incubated AGS cells at distinct pH values with either immobilized fibronectin or CagL for 2 h and assessed adhesion using a hexosaminidase assay. We found that fibronectin adhered to AGS cells at all pH values investigated, although we observed reduced binding at pH 5.75 and lower (Fig. 3A), similar to the RDGLXX(L/I) motif within the prodomains of TGF-β1 and TGF-β3 binding to αVβ5 (35). CagL, in comparison, exhibited a substantially more pronounced pH dependence, with adhesion significantly reduced below pH 6.25 and undetectable below pH 6.0, even at the highest CagL concentration tested (Fig. 3B).
FIGURE 3.
pH dependence of fibronectin and CagL for host cell integrins. A, relative adhesion of AGS cells to fibronectin using a hexosaminidase enzyme assay to quantify adhesion at various pH values. Data were normalized to 25 nm fibronectin, pH 7.5, with standard deviations displayed. One-way analysis of variance statistics were compared with 25 nm fibronectin, pH 7.5; ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001. B, relative adhesion of AGS cells to CagLWT was measured using the same assay. Data were normalized to 500 nm CagLWT, pH 7.5, with standard deviations displayed. One-way analysis of variance statistics were compared with 500 nm CagLWT, pH 7.5. C, relative induction of IL-8 secretion as measured by ELISA at various pH values. Data were normalized to a positive control. D, translocation of CagA monitored by its phosphorylation at various pH values.
Previous studies have shown that CagL, as well as the entire T4SS machinery, induces IL-8 secretion from gastric epithelial cells through the mitogen-activated protein kinase, NF-κB, and NOD-1 pathways (36, 37). Thus, we tested the pH dependence of IL-8 production by AGS cells exposed to the wild type P12 strain of H. pylori at the same pH values used in the above described adhesion assays. We found that IL-8 induction is also pH-dependent, with secreted IL-8 levels decreasing below pH 6.0 and indistinguishable to background below pH 5.5 (Fig. 3C). Although two previous studies (38, 39) investigated the relationship of pH, H. pylori infection and IL-8 production with varying results, the pH of the medium had been adjusted by the direct addition of hydrochloric acid, instead of by resuspending the cells in a buffered solution, which potentially allowed the pH of the medium to change during the course of the experiment. By using chemically defined medium, we show here that IL-8 production by H. pylori infection is maximal above pH 6.25.
It has been shown that CagA translocation through the T4SS and into gastric epithelial cells is dependent on the expression of CagL and its presence in the pilus (8). We therefore also tested the ability of wild type P12 H. pylori to translocate CagA over the same pH range for which we examined CagL cell adhesion. By detecting and quantifying the presence of phosphorylated CagA in AGS cells, we found that CagA translocation was reduced at pH values below 6.0 and was undetectable at pH 4.5 (Fig. 3D), similar, although not identical, to the pH dependence of IL-8 secretion.
CagL Conformational Elongation Does Not Modulate AGS Cell Binding, IL-8 Secretion, or CagA Translocation
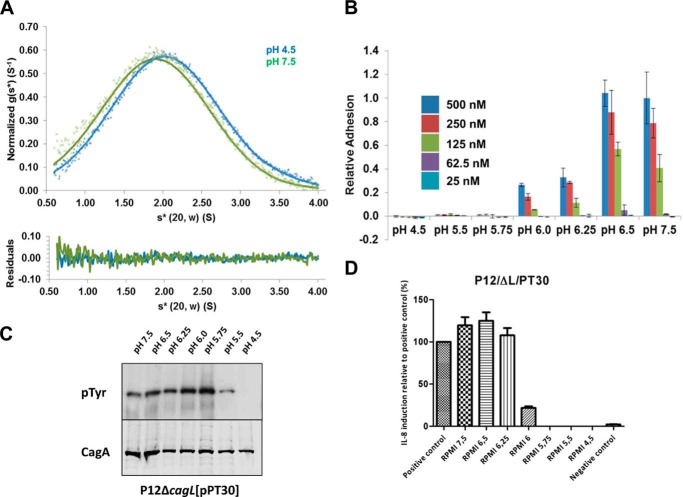
We hypothesized that the pH-mediated change in CagL binding to AGS cells was due to the large conformation change observed in the low pH crystal structures in which the α5 and α6 helices form a single long helix. To test this, we engineered a disulfide bond between residue Leu-156 in the α5 helix and residue Phe-204 in the α6 helix by mutating these residues both to cysteine (CagLL156C/F204C), to lock CagL in its compact, neutral pH conformation. We verified that CagLL156C/F204C maintained the compact conformation regardless of pH by: assaying with Ellman reagent, in which we observed no free cysteines; proteolytically degrading CagLL156C/F204C using V8 protease, for which we observed distinct cleavage patterns at pH 7.8 in the presence and absence of reducing reagent (data not shown); and sedimentation analysis at pH 4.5 and 7.5, in which we observed sedimenting species with similar S values of 2.1 and 2.0, respectively, slightly lower than for wild type CagL (Fig. 4A), likely due to other conformational changes that occur within the disulfide-locked variant. When we conducted cell binding assays with CagLL156C/F204C (Fig. 4B), we found it to adhere to AGS cells nearly identically to CagLWT (Fig. 3B) across the entire range of pH values that we tested. These data indicate that the large conformational change involving the fusion of the α5 and α6 helices and conformational elongation of the protein is not responsible for the pH-mediated cell adhesion properties of CagL.
FIGURE 4.
The large conformational change of CagL is not responsible for integrin binding. A, top, sedimentation velocity experiment of CagLL156/F204C (10 μm) at 50,000 rpm in 50 mm sodium acetate, 150 mm sodium chloride, pH 4.5 (blue crosses), or 50 mm Tris, 150 mm sodium chloride, pH 7.5 (green crosses). Fitted data are shown as solid lines and the residuals (bottom) are plotted. CagLL156/F204C sediments with an s20,w* of 2.1 and 2.0 for pH 4.5 and 7.5, respectively. B, relative adhesion of AGS cells to CagLL156/F204C were measured using the same assay. Data were normalized to 500 nm CagLWT, pH 7.5, with standard deviations displayed. C, translocation of CagA monitored by its phosphorylation at various pH values by CagLL156/F204C. D, relative induction of IL-8 secretion as measured by ELISA at various pH values by CagLL156/F204C. Data were normalized to a positive control.
We also tested whether locking CagL into its compact 6-helix bundle conformation affected IL-8 secretion from, and CagA translocation into, AGS cells. To do so, we complemented a cagL deletion mutant of H. pylori strain P12 with the gene for CagLL156C/F204C and measured IL-8 secretion and CagA translocation as above over the same range of pH values. This CagLL156C/F204C mutant H. pylori strain exhibited a nearly indistinguishable pattern of IL-8 secretion (Fig. 4C) as wild type H. pylori (Fig. 3, C and D). In addition, CagA phosphorylation was substantially reduced for the CagLL156C/F204C mutant H. pylori strain at pH 5.5 and undetectable at pH 4.5, although it did appear to maximal at pH 5.5 to 6.0 (Fig. 4C). These data indicate that, like cell adhesion, conformational elongation of CagL is neither responsible for pH-dependent IL-8 secretion nor, most likely, CagA translocation.
AGS Cell Binding Is Regulated by pH-induced Movements of the CagL α1 Helix
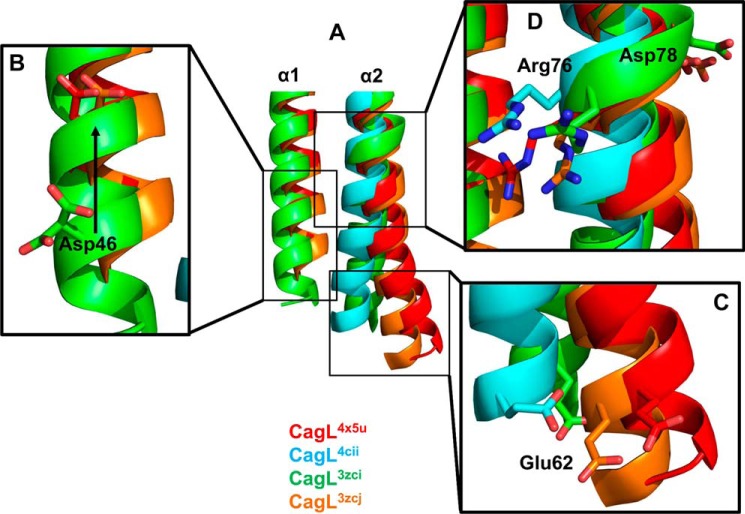
Having ruled out the large conformation change in CagL as the cause of pH-dependent cell adhesion, IL-8 secretion and CagA translocation, we evaluated additional conformation differences between our and previously determined CagL structures. By comparing all of the CagL structures, we observed differences in the α1 helix and the N-terminal portion of the α2 helix adjacent to the RGD motif (Fig. 5A). Residues comprising the α1 helix are resolved in only three of the four CagL crystal structures. In two of these structures, CagL4x5u and CagL3zcj, the α1 helix displays a registry shift relative to the third structure, CagL3zci, resulting in a 7.0-Å movement of the α1 helix by a full rotation, as highlighted by the relative movement of Asp46 in these structures (Fig. 5B). The registry shift of the α1 helix is correlated with the kinkiness of the α2 helix. In CagL structures in which the α1 helix is in the “up” position (CagL4x5u and CagL3zcj), the α2 helix adopts a relatively straight helical conformation with only a small difference, 2.5 Å, in positions of the Cα atom of residue Glu62 (Fig. 5C). In CagL structures in which the α1 helix is either missing or in the “down” position (CagL4cii and CagL3zci, respectively), the α2 helix is more substantially displaced, as monitored by relative movements of the Glu62 Cα atom, by 6.2 and 7.9 Å for PDB 3zci and 4cii structures, respectively (Fig. 5C).
FIGURE 5.
Structural changes of α1 and α2 in CagL in response to pH. A, superposition of all four CagL structures; CagL4x5u, pH 4.5 (red); CagL4cii, pH 4.2 (cyan); CagL3zci, pH 7.4 (green); and CagL3zcj, pH 6.5 (orange). B, lowering of the pH (CagL4x5u and CagL3zcj) causes a registry shift of α1 (CagL3zci) as monitored by Asp46 moving up by one turn of the helix. C, movement of α1 causes the N-terminal half of α2 below the RGD motif to move. With α1 in the up position (CagL4x5u and CagL3zcj), α2 is not kinked and remains straight. When α1 is in the down position or missing (CagL4cii and CagL3zci), α2 becomes kinked. Glu62 shows the degree of movement of α2. D, conformational changes in α1 and α2 result in small differences in the RGD motif in α2. In CagL4x5u, Arg76 forms a salt bridge to α1, whereas in the other structures, Arg76 is either free or the backbone is in a distorted conformation.
These differences in the relative positioning of the α1 and α2 helices correspond to structural changes in the RGD motif. The RGD motif of CagL3zci is distorted, with an α2 helical diameter wider than those observed in the RGD motifs of the other CagL structures. In two of the CagL structures, CagL3zci and CagL3zcj, the RGD motif does not engage any other residue, except for the peptide bond forming part of the α2 helix. In CagL4cii, where the α1 helix is absent, Asp78 hydrogen bonds to Asn-166, whereas in CagL4x5u, where the α1 helix is present, Arg76 forms a salt bridge to Asp46, whereas Asp78 hydrogen bonds to Thr70 (Fig. 5D). In all cases, the loop connecting the α1 and α2 helices lacks sufficient electron density to be resolved in the crystal structures. These data suggest that the α1 helix could control the affinity of the RGD motif for host cell integrins by adopting at least two different pH-dependent conformations that modulate its engagement with the RGD motif on the α2 helix.
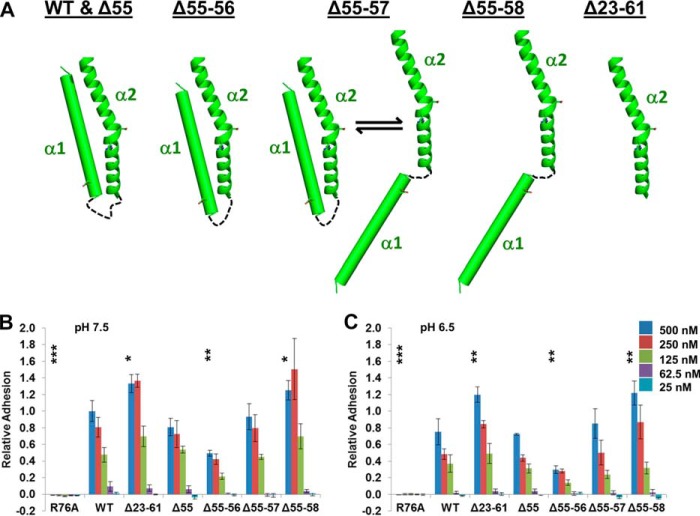
We tested this hypothesis by measuring the cell adhesive properties of CagL variants designed to modulate the ability of the α1 helix to adopt different registers relative to the α2 helix. These variants included one in which the α1 helix was deleted (CagLΔ23–61), as well as several in which the loop connecting the α1 and α2 helices was successively shortened by deletions of residues within the loop (CagLΔ55, CagLΔ55–56, CagLΔ55–57, and CagLΔ55–58). By measuring the distances between the end of the α1 helix and the beginning of the α2 helix in all of the CagL structures and calculating the maximum length of each truncated loop between these two helices, we modeled the following conformations for each variant (Fig. 6A): CagLΔ55, in which the loop is shortened by one residue, is still able to adopt the wild type α1 and α2 helix positions; CagLΔ55–56, in which the loop is shortened by two residues, is restrained to the down position for the α1 helix; CagLΔ55–57, in which three loop residues are deleted, is in equilibrium between the down position for the α1 helix and a conformation in which the α1 and α2 helices are unable to pack against one another; and CagLΔ55–58, missing four loop residues, invariably adopts a conformation in which the α1 and α2 helices are unable to pack against one another. Finally, we constructed an R76A mutation within the RGD motif (CagLR76A).
FIGURE 6.
Positioning of α1 is important for cell adhesion. A, schematic representation of α1 and α2 of wild type CagL and the deletion variants of CagL that were constructed. B, relative adhesion of AGS cells to CagLWT, CagLR76A, CagLΔ23–61, CagLΔ55, CagLΔ55–56, CagLΔ55–57, and CagLΔ55–58, at pH 7.5, using a hexosaminidase enzyme assay. Data were normalized to 500 nm CagLWT with standard deviations displayed. One-way analysis of variance statistics were compared with 500 nm CagLWT, pH 7.5; ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, as in B, except all experiments were conducted at pH 6.5.
For all of these CagL variants, we observed no adhesion below pH 5.75, and weak binding at pH 6.25 and 6.0 (data not shown). At higher pH values (7.5 and 6.5) we observed a similar pattern of binding at numerous concentrations for each of the mutants when compared with CagLWT, with binding being slightly weaker for each protein at pH 6.5 (Fig. 6, B and C). As expected, we observed no adhesion to gastric epithelial cells by CagLR76A, consistent with the RGD motif being the critical component of integrin binding for CagL. We detected stronger adhesion for CagLΔ23–61 relative to CagLWT, suggesting that removal of the entire α1 helix exposes the RGD motif such that it can make more productive interactions with integrins. In our series of variants in which the loop connecting the α1 and α2 helices was shortened by deleting residues, we observed the following: CagLΔ55 adhered to cells similarly to wild type CagL; CagLΔ55–56 adheres to cells less than CagLWT; CagLΔ55–57 adheres similarly to CagLΔ55 and CagLWT; and CagLΔ55–58 adheres more than CagLWT, in a manner indistinguishable from the variant with no α1 helix, CagLΔ23–61. These data suggest that the exposure of the RGD motif on the α2 helix as dictated by the relative position of the α1 helix controls the ability of CagL to bind integrin.
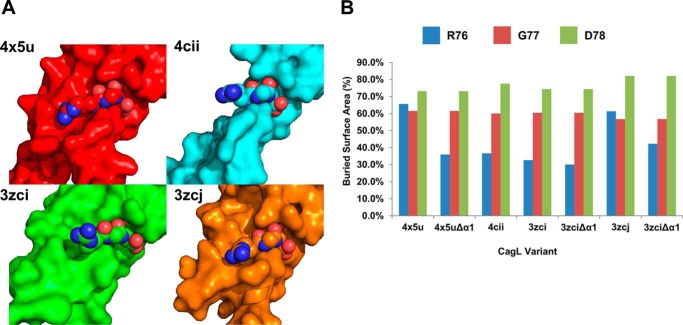
We also calculated the buried surface area of the RGD motif due to the presence and position of the α1 helix in all of the available CagL crystal structures (Fig. 7A). We found that the buried surface areas of the Gly77 and Asp78 residues of the RGD motif remain essentially constant across all structures, with an average percentage change of buried surface area for Gly77 of 60 ± 2% and Asp78 of 77 ± 4% (Fig. 7B). Notably, the α1 helix does not contact these two residues. However, the buried surface area of the Arg76 residue of the RGD motif is relatively more variable (44 ± 14%). When the α1 helix is in the down position, as found CagL3zci, only 32.7% of Arg76 is buried. Removal of the entire α1 helix decreased the burial of Arg76 to only 30.1%, demonstrating that this critical residue of the RGD motif is already essentially as solvent exposed as possible when the α1 helix adopts its neutral pH position. When the α1 helix is in the up position, as found in CagL3zcj and CagL4x5u, we observed that Arg76 becomes predominantly buried with 61.4 and 73.4% buried surface areas, respectively. Removal of the α1 helix in these structures clearly shows that this helix buries most of the surface area with Arg76 being buried only by 42.3 and 37.3% in CagL3zcj and CagL4x5u, respectively. This suggests that the relative movements of the α1 and α2 helices induced by pH result in exposure or burial of Arg76, which modulates the integrin binding capacity of CagL.
FIGURE 7.
Burial of the RGD by α1. A, surface projection of the four structures of CagL, PDB entries 3zci, 3zcj, and 4cii, and 4x5u with Arg76 of the RGD motif shown as spheres. B, a graph of the calculated burial percentage for each amino acid of the RGD motif for all four structures as well as when the α1 helix is removed in PDB entries 3zci, 3zcj, and 4x5u.
DISCUSSION
Integrins are transmembrane receptors that control cell adhesion to other cells and the extracellular matrix. When bound by their various ligands, integrins transduce signals that regulate cell morphology and motility, the cell cycle, and the expression of new receptors on the cell surface. Accordingly, integrins are critical components of the rapid and flexible responses by cells to their environment that allow for numerous biological functions including immune cell patrolling and cell migration. Although our understanding of integrin function and ligand binding at the molecular level has grown, the mechanisms that regulate integrin-ligand recognition remain unclear.
Our data describe a novel molecular mechanism by which integrin engagement by one type of integrin ligand, RGD motifs, can be regulated by pH. Only one previous study has investigated the role of pH dependence of RGD binding (35), in which a peptide of TGF-β3 containing an RGD motif was found to bind αVβ6 in a pH-dependent manner, exhibiting maximal binding above pH 7.0 and diminishing as the pH was reduced. Here, we investigated the effects of pH on the ability of the H. pylori protein CagL to engage integrin on host epithelial cells, an interaction that is important for the translocation of the oncoprotein CagA through the bacterial T4SS, of which CagL decorates the surface of the pilus. We found that the pH dependence of integrin binding by CagL is significantly more severe than for what had been observed previously by others for the TGF-β3 peptide or, by us, for another RGD motif-containing integrin ligand, fibronectin.
CagL undergoes numerous conformational changes in response to variations in pH. The most significant structural change is the formation of a single long α helix from the α5 and α6 helices in crystals at acidic pH, resulting in an approximate doubling of the long axis of the protein and a domain-swapped dimer. We showed that this conformational change and dimerization are not artifacts of crystallization, as they also occur in solution. However, by locking the α5 and α6 helices in their neutral pH positions, we found that this major conformational change was not responsible for reduced cell adhesion, IL-8 secretion, and CagA translocation that we observed at lower pH values.
A seemingly more minor conformational change occurs at the N terminus of CagL. At acidic pH, the N-terminal α1 helix slides by one full rotation in the direction of the C terminus of the α2 helix, which contains the RGD motif. This register shift results in a relative increase in the surface burial of the Arg76 side chain at low pH as it becomes more protected from solvent. To probe whether this conformational change in CagL could affect its ability to engage integrin, we generated a series of CagL variants to rationally manipulate the relative positions of the α1 and α2 helices. When we consecutively shortened the loop connecting the α1 and α2 helices, we observed measurable effects on integrin adhesion. Removal of one residue had no effect, which would suggest the loop itself is slightly longer than needed to allow α1 to adopt the two different positions identified. Shortening the helix by four residues does not allow α1 to interact with α2, thereby causing it to disengage, remain free in solution, and expose the RGD motif fully, resulting in adhesion identical to a variant that contains no α1 helix, CagLΔ23–61. Removal of two loop residues caused a reduction in adhesion. This could be due to either α1 adopting predominately the up position as seen CagL3zcj and CagL4x5u, which is unlikely as we have shortened the loop, or a different conformation that can still interact with α2 but buries the RGD motif. Deletion of three residues causes adhesion similar to CagLWT, suggesting that α1 is interacting weakly with α2 and is thus a mixed state of two and four residues deleted. These data suggest a mechanism by which pH modulates the relative positions of the α1 and α2 helices to expose or protect the RGD motif and, thereby, regulate integrin engagement. Solvent accessibility may also be the molecular mechanism by which integrin binding by cryptic RGD motifs is regulated; in this case, an RGD motif could be liberated by proteolysis of blocking protein domains rather than by a pH-mediated helical sliding effect as for CagL.
Interestingly, the loop connecting the α1 and α2 helices in CagL has previously been shown to contain polymorphisms that can affect the severity of gastric cancer (40). As this loop would also have to move to allow the α1 helix to slide relative to the α2 helix, these polymorphisms may regulate the affinity of CagL for host cell integrins. As we have demonstrated that the conformational changes of α1 and α2 are caused by pH, control of the affinity of the RGD motif for host cell integrins could be modulated through these small movements via burial of Arg76. Indeed, we have shown that shortening of the loop connecting α1 and α2 can have a pronounced effect on integrin binding. In particular, the Tyr58/Glu59 polymorphisms were more likely to be found associated with gastric cancer (40). Furthermore, these polymorphisms were observed to result in a higher affinity for the α5β1 integrin than the Asp58/Lys59 polymorphisms and cause an increase in integrin expression through CagA translocation when using H. pylori strain 1033. A second study showed that the Tyr58/Glu59 polymorphisms prevent CagA translocation when expressed in H. pylori strain 26695 and also notes that the Tyr58/Glu59 polymorphisms are located at the N terminus of α2 and that this region is highly flexible (41). In light of our results, we propose that these polymorphisms could affect the properties of the loop between the α1 and α2 helices. This would alter the position of α1 and thus control integrin binding through burial or exposure of Arg76 of the CagL RGD motif, which is essential for integrin binding and translocation of CagA.
We propose that the function of the α1 helix in CagL is 2-fold. First, it serves as a pH sensor to allow selective adherence to gastric epithelial cells. Gastric epithelial cells have a rapid turnover, on the order of 2 to 3 days (42). This results in cells shedding and undergoing anoikis in the mucosa (43), or committing to apoptosis in the epithelial layer, where they form apoptotic bodies (44). These apoptopic bodies are either absorbed by neighboring cells or phagocytes and undergo destruction in the lysosome or disposed into a glandular lumen (44). Both gastric epithelial cells that are shed and apoptotic bodies could act as decoys to potentially recruit H. pylori and prevent colonization. During an acute infection of H. pylori, shed cells would be present in the gastric lumen, a low pH environment. Apoptotic bodies would experience this low pH during ingestion by lysosomes. The major habitat of H. pylori is adherent to gastric epithelial cells or within the mucus layer overlying the epithelium, where pH values are more or less neutral (pH 6 to 7). Bacteria in the gastric lumen (e.g. during acute infection, when adherent to epithelial cells shed into the gastric lumen, or under conditions of depletion or erosion of the protective mucus layer such as during ulcer development or progression) clearly encounter acidic conditions with pH values as low as 1 to 2 in vivo. In this regard, it also is logical that H. pylori activates its type IV secretion system only when adherent to intact, and not to shed, epithelial cells or apoptotic bodies. With CagL serving as a pH sensor, shed gastric cells that are released into the gastric juices or absorbed into phagocytes would cause release of the bacterium in response to lower pH. Indeed, the pH dependence of binding TGF-β3 for αVβ6 has been proposed to prevent TGF-β1 from binding the integrin during biosynthesis in the Golgi and transport in the endosomes, where the pH is below 7.0 (35, 45). Furthermore, although collagen contains an RGD motif, it is only accessible under partially denaturing conditions or by partial proteolysis (2). It is believed that the exposure of these cryptic RGD motifs might regulate tissue repair (6). Acid production in the stomach may be altered in the time course of a chronic infection and is dependent on the pathology, which may have an impact in CagL binding to host cell integrins (46). The activity of the T4SS of H. pylori does not generally correlate with ulcer risk but with gastric cancer risk (11), which is typically associated with low acid output. Therefore, it is not unreasonable to expect lower amounts of CagA being secreted at low pH, which could be related to the role of CagL as a pH sensor.
Second, the CagL α1 helix acts as a rheostat to finely tune the amount of CagA that is injected and to control inflammatory responses to H. pylori infection. We observed that fibronectin can still bind gastric epithelial cells well below pH 6.0, suggesting that the RGD motif of fibronectin does not need to be regulated as does CagL. Because fibronectin forms part of the extracellular matrix to adhere cells to form tissues and organs, there may have been little or no selective pressure to regulate its activity by pH, unlike CagL where it can experience a wide range of pH levels.
In summary, using structural, biophysical, and functional analyses, we have defined a novel molecular mechanism for regulating integrin engagement by an RGD motif. This mechanism relies on pH-mediated changes in protein structure that modulate the relative exposure of the RGD motif and, thus, its ability to bind integrin. We propose that this regulatory mechanism evolved in H. pylori to prevent premature host cell engagement and off-target delivery of its sole effector molecule, the oncoprotein CagA. We also suggest that cryptic RGD motifs are regulated by a similar mechanism, albeit where proteolysis provides the exposure trigger rather than changes in pH. Others have suggested that a less dramatic pH regulation prevents TGF-β1 binding to integrin inside cells.
This work was supported, in part, by an Alexander von Humboldt Foundation Experienced Researcher Fellowship (to E. J. S.) and Research Grants HA2697/16-1 and 17-1 from the Deutsche Forschungsgemeinschaft (to R. H.).
The atomic coordinates and structure factors (code 4x5u) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T4SS
- type four secretion system
- PDB
- Protein Data Bank.
REFERENCES
- 1. Barczyk M., Carracedo S., Gullberg D. (2010) Integrins. Cell Tissue Res. 339, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davis G. E. (1992) Affinity of integrins for damaged extracellular matrix: αvβ3 binds to denatured collagen type I through RGD sites. Biochem. Biophys. Res. Commun. 182, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 3. Leahy D. J., Aukhil I., Erickson H. P. (1996) 2.0-Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84, 155–164 [DOI] [PubMed] [Google Scholar]
- 4. Shi M., Zhu J., Wang R., Chen X., Mi L., Walz T., Springer T. A. (2011) Latent TGF-β structure and activation. Nature 474, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schürpf T., Chen Q., Liu J. H., Wang R., Springer T. A., Wang J. H. (2012) The RGD finger of Del-1 is a unique structural feature critical for integrin binding. FASEB J. 26, 3412–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis G. E., Bayless K. J., Davis M. J., Meininger G. A. (2000) Regulation of tissue injury responses by the exposure of matricryptic sites within extracellular matrix molecules. Am. J. Pathol. 156, 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mason P. W., Rieder E., Baxt B. (1994) RGD sequence of foot-and-mouth disease virus is essential for infecting cells via the natural receptor but can be bypassed by an antibody-dependent enhancement pathway. Proc. Natl. Acad. Sci. U.S.A. 91, 1932–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwok T., Zabler D., Urman S., Rohde M., Hartig R., Wessler S., Misselwitz R., Berger J., Sewald N., König W., Backert S. (2007) Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449, 862–866 [DOI] [PubMed] [Google Scholar]
- 9. Leininger E., Roberts M., Kenimer J. G., Charles I. G., Fairweather N., Novotny P., Brennan M. J. (1991) Pertactin, an Arg-Gly-Asp-containing Bordetella pertussis surface protein that promotes adherence of mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 88, 345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stockbauer K. E., Magoun L., Liu M., Burns E. H., Jr., Gubba S., Renish S., Pan X., Bodary S. C., Baker E., Coburn J., Leong J. M., Musser J. M. (1999) A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αvβ3 and αIIbβ3. Proc. Natl. Acad. Sci. U.S.A. 96, 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peek R. M., Jr., Blaser M. J. (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2, 28–37 [DOI] [PubMed] [Google Scholar]
- 12. Blaser M. J., Perez-Perez G. I., Kleanthous H., Cover T. L., Peek R. M., Chyou P. H., Stemmermann G. N., Nomura A. (1995) Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55, 2111–2115 [PubMed] [Google Scholar]
- 13. Parsonnet J., Friedman G. D., Orentreich N., Vogelman H. (1997) Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 40, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pang S. S., Nguyen S. T., Perry A. J., Day C. J., Panjikar S., Tiralongo J., Whisstock J. C., Kwok T. (2014) The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori. J. Biol. Chem. 289, 6332–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ilver D., Arnqvist A., Ogren J., Frick I. M., Kersulyte D., Incecik E. T., Berg D. E., Covacci A., Engstrand L., Borén T. (1998) Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279, 373–377 [DOI] [PubMed] [Google Scholar]
- 16. Censini S., Lange C., Xiang Z., Crabtree J. E., Ghiara P., Borodovsky M., Rappuoli R., Covacci A. (1996) cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. U.S.A. 93, 14648–14653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Odenbreit S., Gebert B., Püls J., Fischer W., Haas R. (2001) Interaction of Helicobacter pylori with professional phagocytes: role of the cag pathogenicity island and translocation, phosphorylation and processing of CagA. Cell Microbiol. 3, 21–31 [DOI] [PubMed] [Google Scholar]
- 18. Shaffer C. L., Gaddy J. A., Loh J. T., Johnson E. M., Hill S., Hennig E. E., McClain M. S., McDonald W. H., Cover T. L. (2011) Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 7, e1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Conradi J., Huber S., Gaus K., Mertink F., Royo Gracia S., Strijowski U., Backert S., Sewald N. (2012) Cyclic RGD peptides interfere with binding of the Helicobacter pylori protein CagL to integrins αVβ3 and α5β1. Amino Acids 43, 219–232 [DOI] [PubMed] [Google Scholar]
- 20. Wiedemann T., Hofbaur S., Tegtmeyer N., Huber S., Sewald N., Wessler S., Backert S., Rieder G. (2012) Helicobacter pylori CagL dependent induction of gastrin expression via a novel αvβ5-integrin-integrin linked kinase signalling complex. Gut 61, 986–996 [DOI] [PubMed] [Google Scholar]
- 21. Barden S., Niemann H. H. (2015) Adhesion of several cell lines to Helicobacter pylori CagL is mediated by integrin αVβ6 via an RGDLXXL motif. J. Mol. Biol. 427, 1304–1315 [DOI] [PubMed] [Google Scholar]
- 22. Barden S., Schomburg B., Conradi J., Backert S., Sewald N., Niemann H. H. (2014) Structure of a three-dimensional domain-swapped dimer of the Helicobacter pylori type IV secretion system pilus protein CagL. Acta Crystallogr. D Biol. Crystallogr. 70, 1391–1400 [DOI] [PubMed] [Google Scholar]
- 23. Barden S., Lange S., Tegtmeyer N., Conradi J., Sewald N., Backert S., Niemann H. H. (2013) A helical RGD motif promoting cell adhesion: crystal structures of the Helicobacter pylori type IV secretion system pilus protein CagL. Structure 21, 1931–1941 [DOI] [PubMed] [Google Scholar]
- 24. Dombkowski A. A. (2003) Disulfide by design: a computational method for the rational design of disulfide bonds in proteins. Bioinformatics 19, 1852–1853 [DOI] [PubMed] [Google Scholar]
- 25. Niesen F. H., Berglund H., Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 26. Johnson M. L., Correia J. J., Yphantis D. A., Halvorson H. R. (1981) Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 36, 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stafford W. F., 3rd. (1992) Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 203, 295–301 [DOI] [PubMed] [Google Scholar]
- 28. Philo J. S. (2006) Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 354, 238–246 [DOI] [PubMed] [Google Scholar]
- 29. Pham K. T., Weiss E., Jiménez Soto L. F., Breithaupt U., Haas R., Fischer W. (2012) CagI is an essential component of the Helicobacter pylori Cag type IV secretion system and forms a complex with CagL. PLoS One 7, e35341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 31. Dupeux F., Röwer M., Seroul G., Blot D., Márquez J. A. (2011) A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr. D Biol. Crystallogr. 67, 915–919 [DOI] [PubMed] [Google Scholar]
- 32. Houmard J., Drapeau G. R. (1972) Staphylococcal protease: a proteolytic enzyme specific for glutamoyl bonds. Proc. Natl. Acad. Sci. U.S.A. 69, 3506–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ortega A., Amorós D., García de la Torre J. (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101, 892–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Main A. L., Harvey T. S., Baron M., Boyd J., Campbell I. D. (1992) The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell 71, 671–678 [DOI] [PubMed] [Google Scholar]
- 35. Dong X., Hudson N. E., Lu C., Springer T. A. (2014) Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat. Struct. Mol. Biol. 21, 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gorrell R. J., Guan J., Xin Y., Tafreshi M. A., Hutton M. L., McGuckin M. A., Ferrero R. L., Kwok T. (2013) A novel NOD1- and CagA-independent pathway of interleukin-8 induction mediated by the Helicobacter pylori type IV secretion system. Cell Microbiol. 15, 554–570 [DOI] [PubMed] [Google Scholar]
- 37. Fischer W., Püls J., Buhrdorf R., Gebert B., Odenbreit S., Haas R. (2001) Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42, 1337–1348 [DOI] [PubMed] [Google Scholar]
- 38. Choi I. J., Fujimoto S., Yamauchi K., Graham D. Y., Yamaoka Y. (2007) Helicobacter pylori environmental interactions: effect of acidic conditions on H. pylori-induced gastric mucosal interleukin-8 production. Cell Microbiol. 9, 2457–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Takeuchi H., Nishioka M., Morimoto N., Kamioka M., Kumon Y., Sugiura T. (2009) Relationship of IL-8 production and the CagA status in AGS cells infected with Helicobacter pylori exposed to low pH and activating transcription factor 3 (ATF3). Microbiol. Res. 164, 180–190 [DOI] [PubMed] [Google Scholar]
- 40. Yeh Y. C., Chang W. L., Yang H. B., Cheng H. C., Wu J. J., Sheu B. S. (2011) H. pylori cagL amino acid sequence polymorphism Y58E59 induces a corpus shift of gastric integrin α5β1 related with gastric carcinogenesis. Mol. Carcinog. 50, 751–759 [DOI] [PubMed] [Google Scholar]
- 41. Tegtmeyer N., Lind J., Schmid B., Backert S. (2014) Helicobacter pylori CagL Y58/E59 mutation turns-off type IV secretion-dependent delivery of CagA into host cells. PLoS One 9, e97782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Belanger L. F., Leblond C. P. (1946) A method for locating radioactive elements in tissues by covering histological sections with a photographic emulsion. Endocrinology 39, 8–13 [DOI] [PubMed] [Google Scholar]
- 43. Frisch S. M., Francis H. (1994) Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124, 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stachura J., Tarnawski A., Dabroś W. (1993) Apoptosis: genetically programmed physiologic cell loss in normal gastric oxyntic mucosa and in mucosa of grossly healed gastric ulcer. J. Clin. Gastroenterol. 17, S70–77 [PubMed] [Google Scholar]
- 45. Paroutis P., Touret N., Grinstein S. (2004) The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 19, 207–215 [DOI] [PubMed] [Google Scholar]
- 46. Suerbaum S., Michetti P. (2002) Helicobacter pylori infection. New Engl. J. Med. 347, 1175–1186 [DOI] [PubMed] [Google Scholar]