Summary
Macromolecular machines, such as the ribosome, undergo large-scale conformational changes during their functional cycles. While their mode of action is often compared to that of mechanical machines, a crucial difference is that at the molecular dimension, thermodynamic effects dominate functional cycles, with proteins fluctuating stochastically between functional states defined by energetic minima on an energy landscape. Here, we have used cryo-electron microscopy to image ex vivo-derived human polysomes as a source of actively translating ribosomes. Multiparticle refinement and three-dimensional variability analysis allowed us to visualize a variety of native translation intermediates. Significantly populated states include not only elongation cycle intermediates in pre- and post-translocational states, but also eEF1A-containing decoding and termination/recycling complexes. Focusing on the post-translocational state, we extended this assessment to the single-residue level, uncovering striking details of ribosome-ligand interactions and identifying both static and functionally important dynamic elements.
Introduction
At the heart of many biological processes are complex and dynamic macromolecular machines. Different from macroscopic machines, these operate intermittently rather than continuously. Because inertia is irrelevant at the nanometer scale, conformational changes are dominated by thermal forces (Frauenfelder et al, 1991; Purcell, 1977). Consequently, macromolecular machines randomly sample all conformational states available to them at a given temperature, instead of passing smoothly from one functional state to the other (Frauenfelder et al., 1991). Functional states represent local minima in their energy landscape, defined by energetically costly conformational changes required to transit to neighbouring minima.
The ribosome is an archetypical molecular machine, synthesizing proteins based on the primary sequence information encoded in messenger RNA (mRNA) templates (Frank and Spahn, 2006; Voorhees and Ramakrishnan, 2013). The ribosome consists of a large subunit (LSU; 60S in eukaryotes) containing the peptidyl transferase centre (PTC), and a small subunit (SSU; 40S in eukaryotes) containing the mRNA decoding centre (DC). Together, both subunits define three distinct transfer RNA-(tRNA-)binding sites in their intersubunit space, referred to as the aminoacyl-(A-)site responsible for binding and decoding incoming aminoacylated tRNAs, the peptidyl-(P-)site responsible for orienting the polypeptide-bearing P-site tRNA for efficient transamidation, and the exit-(E-)site responsible for subsequent release of deacylated tRNA.
Protein synthesis can be divided into the four phases initiation, elongation, termination and recycling (Melnikov et al., 2012). Each phase comprises numerous distinct functional states, and multiple large-scale intra- and inter-subunit rearrangements of the ribosome and its ligands drive the functional cycle (Dunkle and Cate, 2010; Korostelev et al., 2008). Dynamic single-molecule distance measurements show that these rearrangements are governed by a rugged energy landscape that is shaped by translation factors (Munro et al., 2009; Petrov et al., 2011). Many functional intermediates of translation have been structurally analysed employing both X-ray crystallography and cryo-electron microscopy (cryo-EM) (Moore, 2012; Voorhees and Ramakrishnan, 2013). The focus of these studies has been on bacterial complexes, while considerably less is known about the structures of functional states sampled by ribosomes from higher eukaryotes. Traditionally, such structural studies rely on in vitro assembled complexes and on the use of antibiotics, tRNA mimics, non-hydrolysable nucleotide analogues or genetic modifications in order to stall ribosomes in defined states. It is still largely unknown if or how in vitro assemblies differ from their in vivo counterparts that are assembled in the complex context of the living cell. Only by investigating samples in a native(-like) setting can these issues be addressed. While cryo-electron tomography allows the visualization of individual, active molecular machines inside cells (Brandt et al., 2010; Myasnikov et al., 2014), its resolution is limited.
Recognizing that in vitro systems are not able to account for the full complexity of in vivo environments, we considered studying native translation intermediates by imaging ex vivo-derived non-stalled and unmodified polysomes from human cell extracts using multiparticle cryo-EM. Polysomes are formed by actively translating ribosomes and are therefore expected to constitute a mixture of ribosomes in elongation states (Rich et al., 1963). Thus, polysomes bear the potential to study the process of translation using one single specimen and to obtain not only multiple structural snapshots of functional states from the same sample, but also to determine the native distribution of states to approximate the positions of minima on the energy landscape, if conformational and compositional heterogeneity can be overcome by particle image sorting procedures (Spahn and Penczek, 2009).
To further structural insights into the process of protein synthesis inside the living cell, we report here the structural analysis of ex vivo-derived human polysomes using multiparticle cryo-EM. We show that a variety of functional states are significantly populated, providing critical structural insights into minima of the energy landscape of the ribosomal elongation cycle and the rate-limiting steps close to the in vivo situation. We also demonstrate that subunit rolling (Budkevich et al., 2014) indeed constitutes a degree of freedom sampled in vivo. Focussing on a larger subset of particle images, we solve the structure of the human 80S ribosome in the post-translocational state at near-atomic resolution despite conformational and compositional heterogeneity. The high-resolution cryo-EM map shows details of native interactions of the ribosome with its ligands, revealing a striking difference in the binding mode between P- and E-site tRNA binding in the unrotated state, and allows identifying both static and functionally important dynamic elements.
Results
Distinct Functional States can be Reconstructed from Human Ex Vivo-derived Polysomes
To preserve the in vivo functional states of polysomes during purification, we switched from classical sucrose-gradient centrifugation to a considerably faster gel filtration-based enrichment strategy to isolate polysomes from the cytosol of human cells (Stephens and Nicchitta, 2007). Samples were vitrified in liquid ethane with minimal delay after cell lysis and enrichment, while they were still exhibiting hallmark features expected of a polysomal sample (Brandt et al., 2010; Rich et al., 1963), such as the distinct peak pattern in a sucrose gradient (Figure 1A) and clusters of ribosomes in the raw micrographs (Figure 1B). In order to sort particle images in silico, we employed unsupervised multiparticle analysis (Loerke et al., 2010) that was combined with three-dimensional (3D) variability analysis to identify regions of high conformational and/or compositional heterogeneity (Supplementary Experimental Procedures).
Figure 1. The Experimentally Observed Elongation Circle.
(A) Sucrose density-gradient analysis of the polysomal sample and (B) raw micrograph after size-exclusion gel-filtration. Scale bar represents 100 nm. (C) Overview of the cryo-EM maps in the framework of the elongation circle. Translocation and decoding-sampling/-recognition complexes (grayed out) were not observed experimentally and are represented by simulated maps based on published factor structures (PDB ID 4cxh, PDB ID 2p8w). For POST and classical PRE structures with different amounts of rolling were observed, represented by the blue-scale bar. All maps are filtered to 7.5 A. Insert: Relative occupancies are color-coded in grayscale. See also Figure S1.
A first tier of unsupervised multiparticle refinement (Figure S1) revealed tRNA-carrying ribosomes in either classical unrotated (66% of ribosomal particle images) or rotated (34% of ribosomal particle images) intersubunit arrangement. However, both rotated and unrotated complexes still featured localized 3D variability, indicating heterogeneity in the form of substoichiometric ligand binding. We therefore employed a second tier of unsupervised classification focussing on the heterogeneous areas to further split the data into defined functional states (Figures 1C, 2 and S2). The presence of density corresponding to the nascent chain (NC) in all complexes demonstrates that our ex vivo-derived polysomes are functional and contain predominantly active ribosomes. This is different from a recent microsomal sample, where only ∼13% of the ribosomes were found in an active state (Voorhees et al., 2014). Our approach indeed allows the structural analysis of functional ribosomal complexes derived from the native environment of the cell that all were assembled and isolated under identical conditions. The resulting maps can be regarded as snapshots of the ribosome “in midflight” (Moore, 2012) allowing key insights into in vivo protein synthesis.
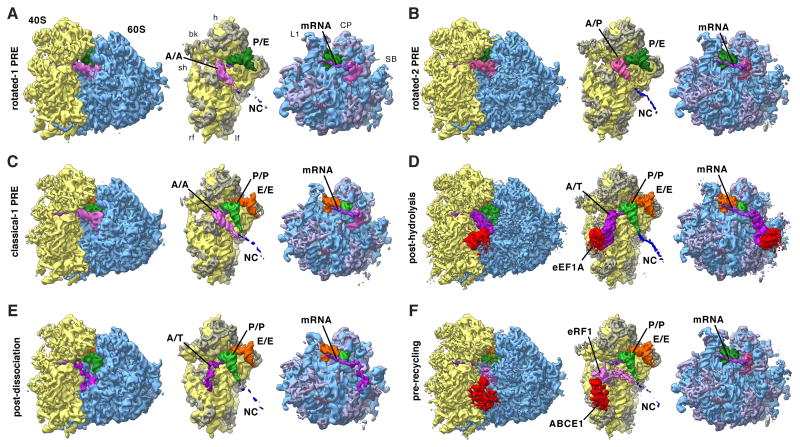
Figure 2. Functional States Reconstructed from Human Polysomes.
Cryo-EM maps filtered to their global resolution (Table S1) corresponding to (A) rotated-1 PRE, (B) rotated-2 PRE, (C) classical-1 PRE, (D) post-hydrolysis (E) post-dissociation and (F) pre-recycling states. For the POST state at high resolution see Figure 4. For the rotated PRE* state and states featuring intermediate amounts of rolling see Figure S2. Left: Ribosomal complexes with SSU depicted in yellow and LSU in blue. Right: Segmented cryo-EM maps, rotated by 80°: A/A-site tRNA (pink), A/T-site tRNA (dark pink), A/P-site tRNA (medium pink), eRF1 (pink), P-site tRNA (green), P/E-site tRNA (dark green), E-site tRNA (orange), mRNA (purple), eEF1 A (red), ABCE1 (red), NC (blue), 18S RNA (yellow), 40S r-proteins (gray-yellow), 28S, 5S, 5.8S RNA (blue), and 60S r-proteins (gray-blue). See also Figure S2 and Table S1.
For the rotated configuration, continued sorting revealed three distinct subpopulations. The first of theses contains an A/A- and a P/E-tRNA and thus represents a rotated-1 state (Figure 2A). This structure is almost identical to the in vitro rotated-1PRE state (Budkevich et al., 2011), with the A-tRNA contacting H89 and H69 and the CCA end being held in the A-site, but additionally shows a contact with the ASF (Figure 3A). The second rotated state contains A/P- and P/E-configured tRNAs (Figure 2B) similar to the in vitro rotated-2 PRE state (Budkevich et al., 2011) and the active fraction of microsomal porcine ribosomes (Voorhees et al., 2014). Intriguingly, the dominating rotated PRE in vivo corresponds to the rotated-2 PRE state with two hybrid tRNAs (Figure 1C, Insert, and Table S1), unlike the previous bacterial structures of the rotated 70S ribosome where only the P/E-tRNA is seen in a clear hybrid position (Agirrezabala et al., 2008; Julian et al., 2008). Unexpectedly, we observe a third rotated PRE conformation with three tRNAs in classical configurations (Figure S2A). Contacts of the A-tRNA with the LSU are similar to those of the rotated-1 state. Compared to POST, the SSU is rotated by ∼8°. We conclude that this rare sub-population may represent a short-lived intermediate PRE state (PRE*), however high flexibility of the tRNA and low resolution preclude a more detailed interpretation.
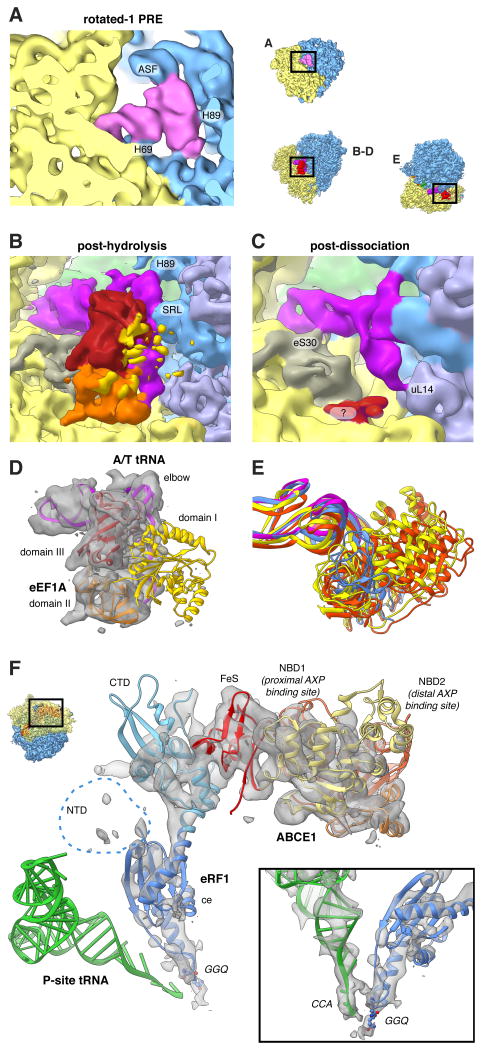
Figure 3. Imaging Ex vivo-derived Polysomes Allows the Visualisation of Transient States.
(A) Close-up view of the rotated-1 PRE state A-site tRNA and (B-E) of the post-decoding states. (B,D) Only domain III (red) and domain II (orange) of eEF1A feature strong density, while domain I (yellow) is fragmented. The A/T-tRNA (dark pink) elbow is connected to the SRL and H89. (C) For the post-dissociation state additional contacts with eS30 and uL14 are visible. A fragmented density of unclear origin is shown in red. (E) 18S RNA-based overlay of decoding-sampling (yellow), decoding-recognition (orange), post-decoding post-hydrolysis (blue) and post-decoding post-dissociation (pink) models for eEFIA and the A/T-tRNA elbow. (F) Close-up view of the pre-recycling state showing eRF1 (shades of blue) and ABCE1 (yellow to red). Atomic models are based on (Preis et al., 2014). See also Movie S1.
For the unrotated configuration, a second tier of sorting resulted in five subpopulations. Comparison with structures from defined in vitro settings (Budkevich et al., 2011; 2014) identified these subpopulations as a classical-1 PRE state, two states with an A/T tRNA, a pre-recycling state and a POST state.
Further sorting of the classical PRE state, containing three classical A/A-, P/P- and E/E-tRNAs (Figures 2C), in a third tier of classification shows that it consists of two complexes with different amounts of rolling (Figures S2B,C). For the first state, the 40S subunit is rolled by ∼6° with respect to the unrotated POST configuration (Figure S2G), and the overall 80S configuration matches well that of a classical-1 configuration observed in vitro (Budkevich et al., 2014). A second state shows intermediate rolling of ∼1-2° with respect to the POST (Figure S2B) and may correspond to an accommodation intermediate (classical-i PRE), where the interaction of the A-site tRNA with the 80S ribosome is reminiscent of the classical-2 configuration.
Interestingly, both complexes that contain classical P/P- and E/E-tRNAs and an A/T configured tRNA (Figure 2D,E) are different from the decoding states observed in vitro where eEF1 A was trapped in the GTP state by the non-hydrolysable GTP analogue GMPPNP (Budkevich et al., 2014). It is thus likely that the present states correspond to later decoding intermediates after GTP hydrolysis. This is corroborated by the appearance of the factor density. For the first, higher populated complex, we observe clear density for both domain III and II of eEF1A in the factor-binding site, but density corresponding to the G-domain (domain I) is highly fragmented indicating flexibility (Figure 3B). The second complex lacks significant density in the factor-binding site, although there is some density present close to the surface of the SSU where domain II of eEF1A makes contact (Figure 3C). In addition to the contacts observed for the factor-bound state, we observe a contact of the acceptor stem of the A/T tRNA with uL14, potentially acting as a steric filter (Caulfield and Devkota, 2012), and a connection of the ASL region with the N-terminal region of eS30, which has previously been shown to also interact with eEF2 (Anger et al., 2013).
The fourth subpopulation of the unrotated states features density in the A-site that does not agree with an A-site tRNA, and density in the factor-binding site different from any expected for factors involved in the elongation cycle. Comparison with in vitro termination complexes (Georges et al., 2014; Preis et al., 2014) identifies this state as a pre-recycling state with bound eRF1 and ABCE1 (Figure 2F). eRF1 is in the extended conformation with the GGQ motif of domain ce facing towards the PTC (Figure 3F). As for the in vitro complex (Preis et al., 2014), its NTD is fragmented. Similarly, the distal nucleotide-binding domain (NBD2) of ABCE1 is fragmented.
A major fraction of particle images of our ex vivo-derived polysomes was assigned to the POST state. As POST state complexes appear to be stable in terms of conformation (Budkevich et al., 2014), we continued refinement of this subpopulation to improve its resolution. Although it has been demonstrated that near-atomic resolution maps for relative invariant parts of ribosome can be obtained, e.g. by focusing the refinement on the large ribosomal 60S subunit (Penczek et al., 2014), and composite near-complete atomic models of the eukaryotic ribosome can be constructed by combining the best resolved parts from different functional states (Voorhees et al., 2014), we tried to represent distinct complexes by a single cryo-EM map each. This was to ensure that we describe distinct functional states and are able to faithfully visualize structural links between remote functional sites (Agmon et al., 2005), e.g. tRNAs bridging the ribosomal subunits or the dynamic inter-subunit bridges (Gabashvili et al., 2000). Intriguingly, further sorting of the population representing POST state complexes revealed a degree of freedom with regard to the presence of subunit rolling (Figure S2D-F). While the majority of particles did not show any subunit rolling, and was refined to high resolution, we obtained two additional populations (POST-i2 and POST-i3) with ∼1° and ∼3° of subunit rolling, respectively (Figure S2D,E). Due to the limited resolution of these two states we cannot discern whether they constitute true energetic minima of subunit rolling, or encompass a continuous band of subunit rolling.
In total, our three-tier multiparticle refinement strategy enabled us to identify and visualize 11 distinct functional states of translating human ribosomes, the majority corresponding to elongation states. All are resolved to sub-nanometer resolution or better (Figure S3A-D and Table S1) and show robust ligand-densities (Figures 2 and S2).
Structure of the Native Human POST Complex at Near-Atomic Resolution
After refining the largest POST population of 313,321 particle images (16% of the total dataset) separately, we obtained a highly improved cryo-EM map for the POST state with a global resolution of 4.0 A using the 0.5 Fourier shell correlation (FSC) criterion, whereas the 0.143 FSC criterion suggests that the map is equivalent to an X-ray density map at 3.5 A resolution (Figure S3E). We corroborated this resolution estimate by a local resolution measurement that is independent of the FSC (Figure S3F). Visual inspection of the map agreed with the near-atomic resolution estimate, with the cryo-EM map (Figure 4) allowing direct observation of single-residue details for large parts of the map (Figures 4C-F and S4A-D). However, intrinsically flexible expansion segments remain less defined (Figure S3G,H), indicating that these structural elements are uncoupled from the functional state of the ribosome. Moreover, all elements endogenously present as mixtures remain less defined, with exception of the remarkably well-resolved P-site tRNA.
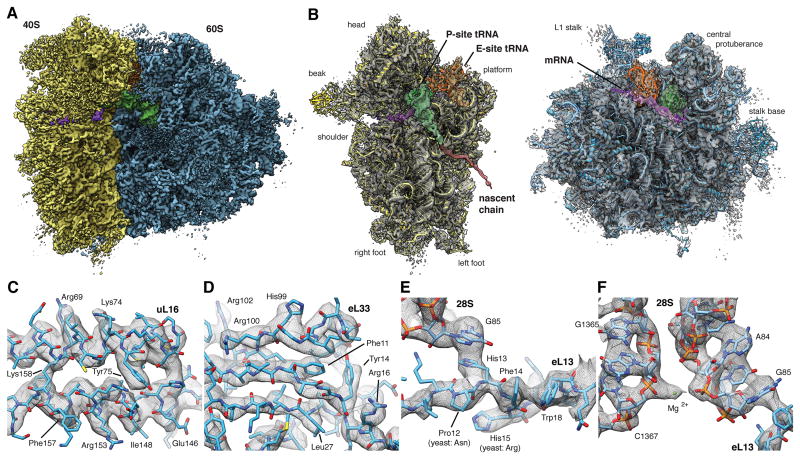
Figure 4. High-resolution Structure of the Human Ribosome in the POST State.
(A) Surface representation of the POST state cryo-EM map filtered to 3.5 A (blue: LSU, yellow: SSU, green: P-site tRNA, orange: E-site tRNA, purple: mRNA). (B) Individual subunit maps with the corresponding atomic models. Segmented density corresponding to the NC (red) is shown filtered to 7.0 A for clarity. Segmented maps are shown turned by 80°. (C-F) Enlarged regions of the cryo-EM map showing well-resolved (C) alpha-helices or (D) beta-strands with individual side-chains, (E,F) strong π -stacking interactions and (F) individual nucleotides with nearby ions. See also Figures S3 and S4. See also Tables S2, S3 and S4.
The quality of the cryo-EM map in the well-ordered regions appears comparable to that of recent crystal structures of eukaryotic ribosomes (Ben-Shem et al., 2011; Klinge et al., 2011; Rabl et al., 2011), allowing us to resolve individual nucleotides with distinct densities for phosphates, bases and sugars as well as protein backbones with clearly visible bulky side-chains. Starting from our previous homology model (Figure S4E,F) of the human ribosome (Budkevich et al., 2014), we created an atomic model for the human ribosome (Tables S2 and S3) by iterating multiple rounds of (semi-)manual real-space fitting, energy minimization and geometric idealization (Supplementary Experimental Procedures, Table S4). The quality of the cryo-EM map allows rationalization of single point mutations compared to yeast (Figure 4E) and determination of correct residues for ambiguous protein sequences (Figure S4A). The high signal-to-noise-ratio of the ordered regions allows the visualization of individual charged ions (Figure 4F and S4B,C). We tentatively assigned these as either chelated or diffuse magnesia based on comparison to known magnesium binding-sites (Jenner et al., 2010) and -motifs (Klein et al., 2004). In total, the atomic model provides a detailed inventory of protein-protein, RNA-RNA and protein-RNA interactions that define the human ribosome in the native, unrotated POST state, while previous high-resolution structures of the 80S ribosome where all solved in rotated or partially rotated conformations (Ben-Shem et al., 2011; Voorhees et al., 2014).
Molecular Description of Eukaryotic-specific Bridges in the Unrotated Configuration
Our present map facilitates the assessment of interactions between the ribosomal subunits via eukaryotic bridges in the classical, unrotated subunit arrangement. As the dynamic nature of the intersubunit bridges is prerequisite to support large-scale conformational changes of the ribosome, like intersubunit rotation or 40S subunit rolling, molecular knowledge of the bridges in all relevant configurations is crucial. Our high-resolution structure now validates our initial assignment of intersubunit bridges (Budkevich et al., 2014) and reveals molecular details for most of the intersubunit interactions in the POST state (Table S5). Especially the lateral eukaryotic-specific bridges eB12 and eB13 are affected by intersubunit rearrangements. For example, the distal part of the C-terminal helix of eL19, forming bridge eB12, is displaced by up to 25 A (Figure 5A) in comparison to the yeast crystal structures (Ben-Shem et al., 2011). Remarkably, the interaction interface with the large groove of expansion segment es6E on the 40S side is hardly affected by this: e.g. the interaction between Argl63 of eL19 and U871 (yeast U813) of es6E is maintained irrespective of the intersubunit arrangement (Figure 5B). On the opposing side of the SSU, bridge eB13 acts akin to a tethered anchor, with a flexible linker of eL24 (residues 68-85) allowing for highly similar binding positions of the C-terminal kinked “anchor” regardless of the intersubunit arrangement (Figure 5C). Different from the lateral bridges, the central eukaryotic-specific bridge eB14 comprising the highly conserved peptide eL41 is largely unaffected by intersubunit rearrangements (Figure 5D). Interestingly, eL41 folds into a linear alpha-helix reminiscent of an axle that binds into a “socket” formed between several rRNA helices of the SSU. Potentially, eB14 could thus help defining the motion centre of 40S rolling and rotation.
Figure 5. Eukaryotic-specific Bridges eB12, eB13 and eB14 are Differentially Affected by Inter-subunit Rotation.
Comparison of yeast LSU crystal structures (“ribosome A”: orange, “ribosome B”: purple) (Ben-Shem et al., 2011) with the present unrotated human atomic model (blue). The orientation aid illustrates the orientation of the 80S in each panel. (A) In the unrotated state, the C-terminal helix of eL19 forming eB12 is bend compared to the yeast structures, however, (B) virtually identical interactions are observed between eL19 and es6E. (C) To visualize the flexible linker tethering the C-terminal kinked “anchor” of eL24 forming eB13, density is shown filtered to 7.0 Å. Despite a strong displacement of the “anchor”, its overall shape remains highly similar. (D) The central bridge eB14 is hardly affected by intersubunit rearrangements. See also Table S5.
Interactions of the Ribosome With a Classical P-site tRNA
Although ex vivo-derived polysomes contain a mixture of all endogenous tRNAs, the P-site tRNA density is almost completely defined to high resolution. Exceptions localize to regions with known structural variability, especially the variable loop and the D-stem loop (Figure 6A) (Giegé et al., 2012). The well-resolved density of the P-site tRNA implies that at least for the vast majority of endogenous tRNAs a single conformation is enforced by the P-site binding pocket. Comparison with crystal structures of Thermus thermophilus ribosomes (Selmer et al., 2006) demonstrates a striking level of structural conservation. Still, we note a direct interaction between the C-terminal Arg146 of uS9 and the tRNA at positions 33 and 35 (Figure 6B) different from bacterial structures (Selmer et al, 2006). It is apparently a swap of Lysl45 for a tyrosine compared to bacteria (Figures 6C and S5) that changes the electrostatic situation at the C-terminus, promoting the direct contact.
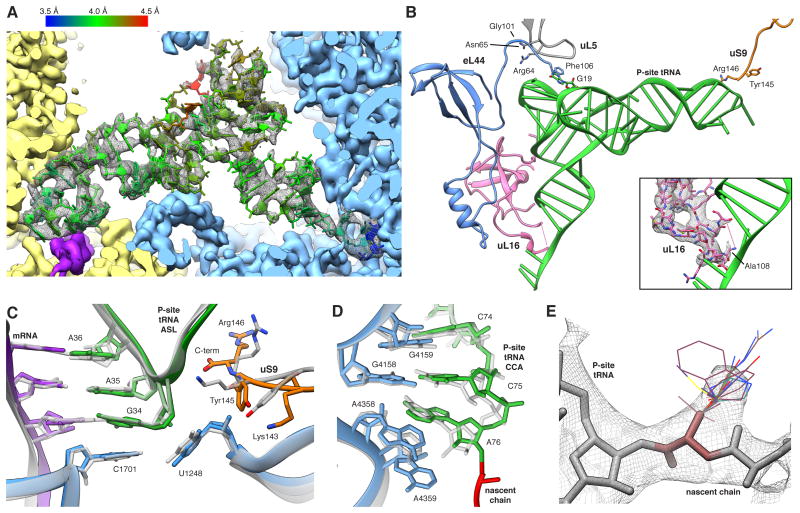
Figure 6. The P-site tRNA is Defined Despite Chemical Heterogeneity.
(A) Cryo-EM map (mesh) and atomic model of the P-site tRNA (colored by local resolution as determined by ResMap). SSU yellow, LSU blue and mRNA density in purple. (B) Key interactions of the P-site tRNA with its binding pocket on LSU and SSU. The insert shows the fragmented density of the uL16 P-site loop (mesh). (C,D) Comparison between prokaryotic (transparent) and human (color) (C) ASL and (D) PTC. Atomic models of the prokaryotic (PDB ID 2J00) and the eukaryotic LSU were aligned based on the LSU rRNA. (E) Cryo-EM map (mesh) of A76 and the first residues of the NC. Stick representations depict the most abundant rotamers of each amino acid with the exception of phenylalanine and tyrosine, where less abundant rotamers are depicted, and proline, which is not shown. See also Figures S5 and S6.
At the tRNA elbow the P-site loop around Arg64 of uL5, monitoring P-site occupancy (Rhodin and Dinman, 2010), contacts the T-stem loop (Figure 6B). Moreover, the C-terminal Phe106 of eL44 establishes an additional tentative contact with the tRNA via aromatic stacking. Intriguingly, uL5 interacts through the neighbouring Asn65 side chain with the backbone carbonyl oxygen of Gly101 in eL44, suggesting a concerted action of both proteins at the P-site (Figure 6B). Contrary to our in vitro ribosomal complex (Budkevich et al., 2014) we do not observe a direct interaction of uL16 with position 1 of the P-site tRNA. Rather, at high resolution clear density for the residues comprising the tip of the loop is lacking, implying heterogeneity or flexibility (Figure 6B, insert). This agrees best with a transient interaction in the POST state, suggesting release after guiding peptidyl-tRNA from the A/P hybrid position to the classical P/P position in the POST state (Budkevich et al., 2011).
At the PTC, superimposing structures of T. thermophilus containing three tRNAs (Selmer et al., 2006) and Haloarcula marismortui LSU containing tRNA-mimics (Schmeing et al., 2003) with our model emphasizes the high conservation between the three domains of life. The backbone atoms of the PTC residues superimpose with a root-mean-square deviation of 0.77 Å and 0.84 Å, respectively. We observe identical interactions of the P-tRNA acceptor stem where residues C74, C75 and A76 stack (Figure 6D). C74 and C75 furthermore form Watson-Crick base pairs with residues G4159 (Escherichia coli numbering G2252; E. coli numbering will be given in brackets in the following) and G4158 (G2251) of the 28S RNA P-loop, respectively. The terminal A76 is stabilized by interaction with A4359 (A2451). The well-described A-minor interaction between A76 and the C3880 (C2063) and A4358 (A2450) pair (Selmer et al., 2006) is present in native human ribosomes.
Visualization of Chemically Heterogeneous NC and mRNA
Despite the chemical heterogeneity of the NC, we observe a continuous density extending from the P-site tRNA into the exit tunnel, most likely representing the first five to six residues of the NC. For the amino acid connected to the CCA-end of the P-tRNA, a smeared-out density bulge may represent a mixture of all endogenous side chains (Figure 6E). Limiting the resolution to 7 A allowed us to trace the path of the NC through the complete LSU (Figure S6A).
Similarly to the NC, the mRNA is expected to contain mixtures of nucleotides at each position. Although the mRNA density is largely fragmented, we are able to trace the path of approximately 28 nucleotides when limiting the resolution to 7 Å (Figure S6B). At the A-site on the SSU, only A1824 (A1492) and A1825 (A1493) of h44 are disordered (Figure S6C), most likely sampling flipped-in and -out positions as no A-site tRNA is present. At the P-site, despite the heterogeneity of codons, the mRNA density is well resolved at full resolution (Figure S6D), resembling the situation observed for the P-site tRNA. At the E-site, individual bases of the mRNA are still defined, especially at codon positions -1 and -2, but the wobble position -3 is partially fragmented (Figure S6D).
POST State Ribosomes Undergo Stable Interactions with E-site tRNA
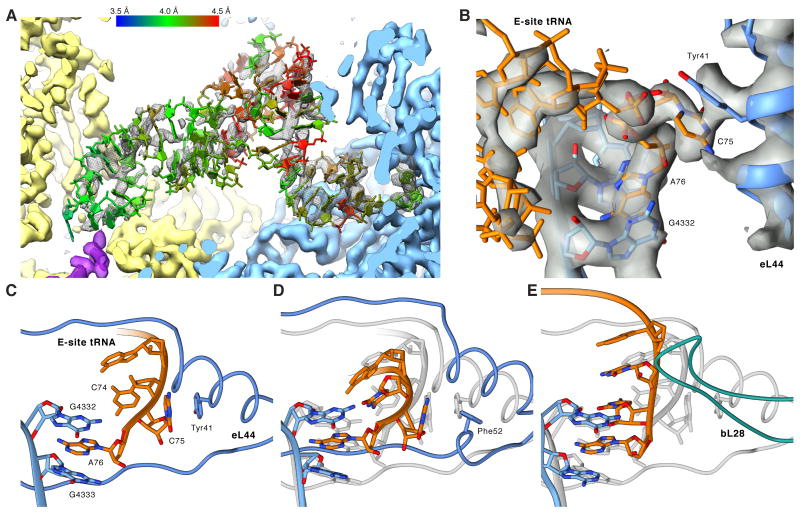
We observe distinct density corresponding to a tRNA bound to the E-site, demonstrating that the E-site tRNA in the POST state is at least stable enough to survive gel filtration. Different from the well-resolved P-site tRNA, the bulk of the E-site tRNA density is fragmented (Figure 7A), suggesting that the E-site allows for a more relaxed binding. While we discern no direct interactions with the SSU, on the LSU side we observe a delocalized interaction between the tRNA elbow and the L1 stalk (Figure S7A). The full definition of the CCA-end of the E-site tRNA (Figure 7B) implies that the fragmented appearance of the majority of the E-site density is not caused by sub stoichiometric occupancy but by conformational heterogeneity, which in turn may be caused by small differences among different tRNA species and/or flexibility/mobility. This is corroborated by the full presence of the whole E-site tRNA when the resolution of the map is limited to 7 Å (Figure S7B).
Figure 7. Native POST State Ribosomes Contain an E-site tRNA.
(A) Cryo-EM map (mesh) and atomic model of the E-site tRNA (colored by local resolution as determined by ResMap). (B) Close-up on the cryo-EM map (transparent gray) of the CCA-end of the E-site tRNA (orange) and surrounding LSU elements (blue) (C-E) Comparison between human, archaeal and bacterial E-site CCA-ends. Atomic models were aligned based on the LSU rRNA and are depicting (C) the human, (D) the H. marismortui and (E) the T. thermophilus CCA-end. In (D) and (E), the human model is shown in transparent gray for comparison. See also Figures S5, S6 and S7.
Unlike the body of the E-site tRNA, the acceptor stem features a well-defined density due to a contact with the 28S RNA at U3686 and G3711 and strong interactions of the CCA-end with the LSU (Figure 7B). As observed for prokaryotes and archaea, A76 is tightly packed in a sandwich between G4332 (G2421) and G4333 (C2422) forming a binding pocket excluding aminoacylated CCA-ends (Schmeing et al., 2003). In addition, C75 interacts by π-stacking with Tyr41 of eukaryote-specific eL44,while in prokaryotes C75 and C74 are stabilized by internal nucleotide stacking (Selmer et al., 2006). Converse to our findings, a preceding structure of the H. marismortu LSU in complex with a CCA tri-nucleotide suggested that instead of π-stacking Arg40 and Gly57 of eL44 provide additional stabilization of the E-site CCA-end (Schmeing et al., 2003). However, sequence alignment highlights that the eL44 interaction observed in our structure likely corresponds to the general case in eukaryotes as H. marismortui harbors an eL44 sequence unique to halobacter. Other archaea and eukarya feature a conserved Tyr or Phe at position 41 (Figure S5). Superimposing the H. marismortu crystal structure onto our model indicates that H. marismortui Phe52 occupies almost the same position as Tyr41 in human and could potentially rearrange under physiological conditions to interact with C75. Superposition also reveals that the loop extension of eL44 between Phe56 and Thr62, which widens upon E-site binding, is shifted up to 4 Å when compared to H. marismortui (Schmeing et al., 2003), most likely due to steric clashes between tRNA and the loop extension of eL44 (Figure 7C-E).
Discussion
The Multi-tiered Landscape of Translation Elongation Inside the Cell
Cytosolic polysomes comprise actively translating ribosomes sampling a large variety of functional states. Despite this heterogeneity, our data-driven in silico sorting scheme allowed us to uncover 11 distinct states and visualize them with at least sub-nanometer resolution (Figure 1). We cannot rule out that certain more transient states were lost during our purification procedure. However, the fact that the vast majority of particle images were assigned to bona fide functional states and the richness in functional states covering most of the ribosomal elongation cycle shows that the stable intermediates have been preserved. Comparing the relative SSU-LSU configurations among all states (Figure S2G-I), we note that in line with our preceding in vitro studies aimed at unveiling key transitory states of mammalian elongation (Budkevich et al., 2011; 2014) intersubunit rotation and eukaryotic-specific 40S subunit rolling are indeed conformational modes present in vivo.
However, we observe neither complexes with significant head swivelling nor complexes containing eEF2. Non-stalled eEF2 has been observed in a subpopulation of in vitro assembled complexes (Budkevich et al., 2014) and on the populations isolated from mammalian cells that showed inactive 80S complexes (Anger et al., 2013; Voorhees et al., 2014). Thus, eEF2 binding to actively translating 80S ribosomes appears less stable implying that translocation states are short-lived intermediates in vivo. Kinetic (Guo and Noller, 2012) and structural studies (Spahn et al., 2004, Ratje et al., 2010) have linked head swivelling to EF-G/eEF2 containing translocation intermediates. The simultaneous absence of eEF2 and significant head swivelling in our present elongation intermediates supports such a model.
While not sufficient to map the exact topology of the energy landscape of elongation, our structural description allows assessment of the distribution of the more stable functional states of active polysomes in a native setting (Figure 1C, Insert). This distribution correlates with the relative energetic stability of each state in comparison to the most stable one (Fischer et al., 2010; Frank, 2013). We note that neither is the unrolled configuration exclusive to the POST state nor the rolled configuration to the classical PRE states. Rather, rolling appears to be a spontaneous movement where the presence of an A-site tRNA shifts the equilibrium towards the rolled classical PRE-state.
From the local heterogeneity uncovered by our 3D variability analysis, it is moreover evident that the energetic minimum corresponding to each state can potentially be split into finer substates, given proper sorting of the particle images. While functionally defined, each of these states features localized heterogeneity due to structural elements that are more flexible - in line with the assumption of a multi-tiered hierarchical energy landscape governing elongation (Munro et al., 2009) and protein activity in general (Frauenfelder et al., 1991).
While after 3D variability analysis and focussed classification a major part of the POST complex can be resolved at single-residue resolution, there are still regions exhibiting a fragmented appearance. This localized disorder can be regarded as structural evidence for the finely split sub-valleys of the energy landscape. The most striking example is the highly localized disorder of the bases A1824 (A1492) and A1825 (A1493) involved in A-site decoding (Demeshkina et al., 2012; Ogle et al., 2003) in the otherwise well-ordered h44 (Figure S6C). Both bases are likely sampling both flipped-in and -out positions as no A-site tRNA is present. Such flexibility has been predicted from molecular dynamics simulations (Sanbonmatsu, 2006), but is in contrast to recent X-ray crystallization data from bacterial complexes with P-site tRNA and mRNA showing a partially preformed DC with A1493 stably flipped-out and A1492 stably flipped-in by stacking with A1913 (Demeshkina et al., 2012). Still, our findings can easily be reconciled with the known kinetics of decoding by assuming that the empty DC, behaving akin to a liquid unstructured region, rigidifies upon codon-anticodon interaction. In this “flow-fit” model, the mobile decoding bases allow fast sampling of the codon-anticodon duplex with the cognate tRNA inducing a stronger binding interface and thus rigidifying the DC with higher probability and higher speed. This matches well the experimental observation that both cognate and near-cognate tRNAs bind to the ribosome with the identical rates, but that cognate tRNAs dissociate not only more slowly from the DC compared to near-cognate tRNAs, but also exhibit faster rates for the forward reactions, i.e. GTPase activation (Pape et al., 1999; Geggier et al., 2010). Thus, the observed disorder of the bases A1824 (A1492) and A1825 (A1493) in an otherwise highly ordered environment exemplifies the potential biological importance of mobile, more “liquid” regions (Dyson, 2011), and demonstrates the potential of visualizing macromolecular machine at near-atomic resolution in a solution-like state under near-physiological conditions.
Native Proofreading Complexes Highlight Rate-Limiting Steps
The presence of ribosomal decoding complexes in polysomes is not immediately expected, as decoding complexes are believed to be short-lived intermediates. Accordingly, structural investigations of ribosome-bound ternary complexes rely on the use of non-hydrolysable GTP analogues or antibiotics to inhibit the transition of EF-Tu/eEF1 A from the GTP to the GDP conformation (Budkevich et al., 2014; Schmeing et al., 2009; Schuette et al., 2009; Villa et al., 2009; Voorhees et al., 2010). The visualization of a non-stalled ternary complex on the human ribosome, and of a second ribosomal decoding complex containing only A/T-tRNA but no factor, demonstrates the power of our approach that aims to derive structures of functional complexes from the on-going functional cycle instead of isolated functional complexes. Under steady-state/multi turnover conditions even shorter-lived states may be significantly populated, as they are constantly replenished.
Comparing the ternary complex observed from polysomes to decoding-sampling and decoding-recognition complexes from our preceding in vitro study with GMPPNP-stalled eEF1 A (Budkevich et al., 2014), we note significant differences. Firstly, the elbow of the A/T-tRNA has already released the stalk base and appears more strongly bound to the sarcin-ricin loop (SRL, H95) instead (Figure 3B). Secondly, there is relevant disorder of domain I containing the GTP-binding pocket (Figure 3D). These differences can be readily reconciled by the notion that both sets of complexes represent different states along the pathway of tRNA selection. For the in vitro complexes eEF1 A was trapped in the GTP state by GMPPNP and accordingly the complexes were observed in the initial phase of decoding before GTP hydrolysis. (Budkevich et al., 2014). As in our present study the chemical step is not inhibited, and based on the trajectory of structural changes from decoding-sampling to decoding-recognition (Budkevich et al., 2014) to the present complex (Figure 3E and Movie S1), we infer that it corresponds to an aminoacyl(aa)-tRNA•eEF1 A•GDP ternary complex in the post-hydrolysis / proofreading state before the eEF1A dissociation and accommodation steps. This implies that the transition to the GDP-induced conformation of the factor and release of aa-tRNA from eEF1A•GDP do not occur immediately upon SRL-promoted GTP hydrolysis, but with a significant delay. This is in excellent agreement with kinetic studies in the bacterial system where GTP hydrolysis has been shown to be a very fast step, whereas tRNA accommodation and especially EF-Tu dissociation are rate-limiting during tRNA selection (Pape et al., 1998). Furthermore, our results rationalize recent studies on the formation and turnover of bacterial EF-Tu•GXP•EF-Ts•aa-tRNA quarternary complexes (Burnett et al., 2014; 2013), which proposed a novel role of EF-Ts in promoting release of aa-tRNA from EF-Tu•GDP. The presence of a second proofreading state containing A/T-tRNA, but lacking density for eEF1A, implies that also tRNA accommodation constitutes a second slow step after eEF1 A dissociation, in agreement with the hypothesis that necessary conformational changes in the tRNA elbow to allow A/T to A/A transition resemble a stochastic trial-and-error process and not a concerted pathway (Whitford et al., 2010; Geggier et al., 2010). We believe that from both post-hydrolysis states near-cognate tRNA can be rejected in line with the concept of kinetic proofreading (Hopfield, 1974).
A Stably Occupied E-site is an In Vivo Feature of Elongating Human Ribosomes
The properties of the E-site tRNA in the bacterial system have been controversially discussed for decades and it is still not generally agreed on whether the E-site is only transiently occupied directly after translocation, or whether the E-site tRNA is released at latter stages, i.e. during A-site occupation (Wilson and Nierhaus, 2006). Our maps now show an occupied E-site also in functional states subsequent to the POST state, with three tRNAs present in the classical PRE, post-decoding and PRE* complexes. Solely for the rotated-1 and -2 PRE complexes do we observe only two tRNAs. Remarkably, during human translation elongation the 60S E-site appears to be always occupied by the CCA-end of either an E/E- or a P/E-tRNA. While it can be argued that the presence of an E-site tRNA on in vitro-assembled complexes is due to the excess of deacylated tRNA used for technical reasons (Budkevich et al., 2011; 2014), this argument falls short for the present ex vivo-derived complexes. Thus, our findings strongly suggest that stable E-site occupation in all but the rotated-1 and rotated-2 PRE states is an in vivo feature of the human system.
Native PRE Complexes Pinpoint the Release of E-site tRNA
Given that our structures allow us to trace the transition from an unrotated PRE state with three classical tRNAs to the rotated PRE states with two tRNAs in either classical or hybrid configuration, and especially due to the observation of a rotated PRE* subpopulation with A/A-, P/P- and E/E-tRNAs, we can pinpoint the release of E-site tRNA during human translation elongation. Apparently, it is the rotation between the two subunits that critically destabilizes the E-site, leading to subsequent release of the E-site tRNA (Figure 1C). Still, while our structures definitely support the existence of the aforementioned pathway, it has to be noted that this observation does not preclude the possibility of parallel alternative pathways where either E-site tRNA is released concomitantly with intersubunit rotation, or even before rotation.
Conclusions
Our study and others have recently demonstrated that cryo-EM has opened up the way to study the structure of the ribosome at high resolution unconstrained by a crystal lattice. Furthermore, by relying on thorough in silico classification, we have demonstrated that defined structures corresponding to known and hitherto unknown intermediate states of translation can be obtained from ex vivo-derived elongating polysomes. Different from traditional “arrest and isolate” strategies, our approach has shed light on the preferred states of the human ribosome. This uncovered that while eEF2-mediated head rotation is paramount for translocation, corresponding functional states are only sparsely populated precluding visualization, corroborating the assumed short-lived nature of the translocation state. Analysing the full spectrum of significantly populated states of elongation has not only addressed the specific point of E-site tRNA release, but also uncovered that proofreading states of tRNA selection, after codon recognition and GTP hydrolysis, can be significantly populated, implying that tRNA accommodation is indeed a slower step. Importantly, despite the high degree of complexity of a native-like polysomal sample, we were able to overcome heterogeneity using a data-driven sorting scheme. This allowed us to resolve the native POST state to near-atomic resolution and thus highlight the divergent properties of P- and E-site and uncover dynamic elements in the ribosome, such as the decoding bases.
Experimental Procedures
Additional details can be found online in the Supplementary Experimental Procedures.
Polysome Isolation and Grid Preparation
Polysomes were prepared from the cytosolic fraction of digitonin permeabilized HEK293T cells (Hirashima and Kaji, 1970; Stephens and Nicchitta, 2007). The cytosolic fraction was further separated on a Sepharose 4B size-exclusion column, isolating polysomes as the first peak absorbing at 254 nm. Samples were immediately prepared for cryo-EM by vitrification.
Data Collection
Electron micrographs were collected automatically on an FEI Krios microscope equipped with a back-thinned Falcon II detector and on an FEI Tecnai G2 Polara equipped with a TC-F416 CMOS camera. The total data set comprised 51,282 micrographs yielding 1,823,338 particles (801,789 Krios, 1,121,549 Polara).
Data Processing
The dataset was processed using incremental K-means-like procedures (Loerke et al, 2010) in SPIDER (Frank et al, 1996). Initially, the two datasets split into subsets belonging to either rotated or unrotated ribosomal complexes or to artefactual particles. Particle images belonging to the rotated PRE and unrotated POST states were separated and artefactual particle images were removed. Particle images were refined and classified further using 3D variability analysis to guide sorting. The final map of the POST state, based on 313,321 particle images (130,953 Krios, 182,368 Polara), reached a resolution of 4.0/3.5 A. Cryo-EM density maps have been deposited with the EMDB (accession no. EMD-2875, EMD-2902, EMD-2903, EMD-2904, EMD-2905, EMD-2906, EMD-2907, EMD-2908, EMD-2909, EMD-2910 and EMD-2911) and coordinates for the POST state have been deposited with the Protein Data Bank (entry codes 5aj0 and 5aj5).
Model Building and Refinement
Initial atomic models of H. sapiens 40S and 60S subunits derive from our preceding study (Budkevich et al., 2014). Ligands were rebuilt based on crystal structures of prokaryotic or archaeal tRNAs. The NC poly-alanin model was built de novo. Overlapping stretches of the model were manually adjusted into the cryo-EM map as rigid bodies, followed by real space refinement and geometric idealisation for well-resolved densities. Structure models were further refined and validated using crystallography tools.
Supplementary Material
Acknowledgments
We thank Helena Seibel and Brian Bauer for technical support during sample preparation and Alina Bretfeld and Mathias Brünner for initial structural work. The present work was supported by grants from the Deutsche Forschungsgemeinschaft DFG (SFB 740 to C.M.T.S., P.S. and T.M.; SFB 1078 to P.S), DFG Cluster of Excellence ‘Unifying Concepts in Catalysis’ (Research Field D3/E3-1) to P.S., HSFP and Senatsverwaltung fur Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum), Charite (Rahel-Hirsch stipend to T.V.B.) and by the National Institutes of Health (R01 GM60635) to P.A.P. E. B. holds a Freigeist-Fellowship from the Volkswagen Foundation. The authors acknowledge the North-German Supercomputing Alliance (HLRN) and the Texas Advanced Computing Center (TACC) at the University of Texas at Austin for providing high-performance computing resources that have contributed to the research results reported in this paper.
Footnotes
Author Contributions: T.V.B. and K.Y. prepared polysome samples. T.M. and M.R.V. supervised and coordinated cryo-EM data collection. J.B. performed data collection. J.L., E.B. and C.M.T.S. processed, refined and analysed cryo-EM data (based on refinement strategies designed by J.L., P.A.P. and C.M.T.S.). E.B., A.S. and P.S. refined atomic models. E.B., J.L. and C.M.T.S. interpreted electron densities and atomic models. E.B. and J.L. prepared figures. C.M.T.S. designed the study. E.B., J.L. and C.M.T.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon I, Bashan A, Zarivach R, Yonath A. Symmetry at the active site of the ribosome: structural and functional implications. Biol Chem. 2005;386:833–844. doi: 10.1515/BC.2005.098. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Brandt F, Carlson LA, Hartl FU, Baumeister W, Griinewald K. The Three-Dimensional Organization of Polyribosomes in Intact Human Cells. Mol Cell. 2010;39:560–569. doi: 10.1016/j.molcel.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Budkevich TV, Giesebrecht J, Behrmann E, Loerke J, Ramrath DJF, Mielke T, Ismer J, Hildebrand PW, Tung CS, Nierhaus KH, et al. Regulation of the Mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell. 2014;158:121–131. doi: 10.1016/j.cell.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich T, Giesebrecht J, Altaian RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CMT. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett BJ, Airman RB, Ferguson A, Wasserman MR, Zhou Z, Blanchard SC. Direct Evidence of an Elongation Factor-Tu/Ts-GTP-Aminoacyl-tRNA Quaternary Complex. J Biol Chem. 2014 doi: 10.1074/jbc.M114.583385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett BJ, Airman RB, Ferrao R, Alejo JL, Kaur N, Kanji I, Blanchard SC. Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor TuGTPaminoacyl-tRNA ternary complex. J Biol Chem. 2013;288:13917–13928. doi: 10.1074/jbc.M113.460014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield T, Devkota B. Motion of transfer RNA from the A/T state into the A-site using docking and simulations. Proteins. 2012;80:2489–2500. doi: 10.1002/prot.24131. [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- Dunkle JA, Cate JHD. Ribosome structure and dynamics during translocation and termination. Annu Rev Biophys. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- Dyson HJ. Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Frank J. Story in a sample-the potential (and limitations) of cryo-electron microscopy applied to molecular machines. Biopolymers. 2013;99:832–836. doi: 10.1002/bip.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Spahn CMT. The ribosome and the mechanism of protein synthesis. Rep Prog Phys. 2006;69:1383–1417. [Google Scholar]
- Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Geggier P, Dave R, Feldman MB, Terry DS, Altaian RB, Munro JB, Blanchard SC. Conformational sampling of aminoacy1-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;339:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges des A, Hashem Y, Unbehaun A, Grassucci RA, Taylor D, Hellen CUT, Pestova TV, Frank J. Structure of the mammalian ribosomal pre-termination complex associated with eRFl.eRF3.GDPNP. Nucleic Acids Research. 2014;42:3409–3418. doi: 10.1093/nar/gkt1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege R, Juhling F, Piitz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip Rev RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci USA. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima A, Kaji A. Factor dependent breakdown of polysomes. Biochem Biophys Res Commun. 1970;41:877–883. doi: 10.1016/0006-291x(70)90165-8. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. Proc Natl Acad Sci USA. 1974;71:4135–4139. [Google Scholar]
- Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- Julian P, Konevega AL, Scheres SHW, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin ChemBiol. 2008;72:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke J, Giesebrecht J, Spahn CMT. Multiparticle cryo-EM of ribosomes. Methods in Enzymology. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- Moore PB. How should we think about the ribosome? Annu Rev Biophys. 2012;41:1–19. doi: 10.1146/annurev-biophys-050511-102314. [DOI] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. Navigating the ribosome's metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myasnikov AG, Afonina ZA, Menetret JF, Shirokov VA, Spirin AS, Klaholz BP. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat Commun. 2014;5:5294. doi: 10.1038/ncomms6294. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacy1-tRNA on the ribosome. Embo J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. Embo J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek PA, Fang J, Li X, Cheng Y, Loerke J, Spahn CMT. CTER-Rapid estimation of CTF parameters with error assessment. Ultramicroscopy. 2014;140C:9–19. doi: 10.1016/j.ultramic.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Kornberg G, O'Leary S, Tsai A, Uemura S, Puglisi JD. Dynamics of the translational machinery. Curr Opin Struct Biol. 2011;21:137–145. doi: 10.1016/j.sbi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A, Heuer A, Barrio-Garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, Beckmann R. Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing eRFl-eRF3 or eRF1-ABCE1. CellReports. 2014:1–18. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Briinner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodin MHJ, Dinman JD. A flexible loop in yeast ribosomal protein L11 coordinates P-site tRNA binding. Nucleic Acids Research. 2010;38:8377–8389. doi: 10.1093/nar/gkq711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A, Warner JR, Goodman HM. The structure and function of polyribosomes. 1963;28:269–285. [Google Scholar]
- Sanbonmatsu KY. Energy landscape of the ribosomal decoding center. Biochimie. 2006;88:1053–1059. doi: 10.1016/j.biochi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FV, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. Embo J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Penczek PA. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr Opin Struct Biol. 2009;19:623–631. doi: 10.1016/j.sbi.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JPG, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. Embo J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SB, Nicchitta CV. In vitro and tissue culture methods for analysis of translation initiation on the endoplasmic reticulum. Biothermodynamics, Part D. 2007;431:47–60. doi: 10.1016/S0076-6879(07)31004-5. [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SHW, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Geggier P, Airman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY. Accommodation of aminoacy1-tRNA into the ribosome involves reversible excursions along multiple pathways. Rna. 2010;16:1196–1204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH. The E-site story: the importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell Mol Life Sci. 2006;63:2725–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.