Abstract
Resistance to the innate defenses of the intestine is crucial for the survival and carriage of Staphylococcus aureus, a common colonizer of the human gut. Bile salts produced by the liver and secreted into the intestines are one such group of molecules with potent antimicrobial activity. The mechanisms by which S. aureus is able to resist such defenses in order to colonize and survive in the human gut are unknown. Here we show that mnhF confers resistance to bile salts, which can be abrogated by efflux pump inhibitors. MnhF mediates the efflux of radiolabeled cholic acid both in S. aureus and when heterologously expressed in Escherichia coli, rendering them resistant. Deletion of mnhF attenuated the survival of S. aureus in an anaerobic three-stage continuous-culture model of the human colon (gut model), which represents different anatomical areas of the large intestine.
INTRODUCTION
Staphylococcus aureus is a ubiquitous and highly adaptable human pathogen responsible for a significant global burden of morbidity and mortality. The bacterium lives as a commensal in the nares of 20 to 25% of the population at any one time (1, 2). While nasal colonization is a well-established risk factor for most types of S. aureus infections, several recent studies have suggested that colonization of the intestine, which occurs in ca. 20% of individuals and which by and large has been overlooked, could have important clinical implications (3). Patients with S. aureus intestinal colonization can serve as an important source of transmission, as they often contaminate the adjacent environment (4). Similarly, such patients display an increased frequency of skin colonization (5). A study of intensive care and liver transplant units showed that patients with both rectal and naris colonization by methicillin-resistant S. aureus (MRSA) had a significantly higher risk of disease (40%) than did patients with nasal colonization alone (18%) (6). Furthermore, a study of hospitalized patients in the United States reported cocolonization by S. aureus and vancomycin-resistant enterococci in >50% of the individuals studied (7). Thus, it is likely that intestinal colonization by S. aureus provides the pathogen a potential opportunity to acquire new antibiotic resistance genes.
While the clinical implications of intestinal colonization by S. aureus are still relatively ill defined, it is assumed that carriage is a risk for intestinal infection; S. aureus can induce pseudomembranous colitis that is histologically distinct from that caused by Clostridium difficile (8). Multiple studies have demonstrated frequent intestinal colonization in infants, particularly those who were breast fed, and that there is a positive correlation with the development of allergies (9–13). While a role for S. aureus intestinal carriage in the development of systemic S. aureus disease has not been established, colonization of the intestinal lumen of mice can lead to the pathogen crossing the intestinal epithelial barrier and subsequently spreading to the mesenteric lymph nodes (14, 15).
As a common commensal and pathogen, S. aureus must resist the human host's innate defenses that have evolved to limit its in vivo growth and spread. In particular, bile represents a major challenge to bacteria that survive transit through the stomach and enter the intestines. Bile is a digestive secretion that plays an essential role in the emulsification and solubilization of lipids. We have previously demonstrated the survival of S. aureus in a human colonic model fed with physiological levels of bile (16). Resistance to bile salts has been demonstrated to be important for the intestinal survival of several enteric pathogens, but for S. aureus, such an understanding is lacking. The role of the S. aureus mnhABCDEFG locus in bile resistance was identified by using a Tn917 library screened for bile-sensitive mutants. MnhF is homologous to mammalian bile salt transporters; thus, we hypothesized that it was involved in bile resistance and, therefore, the survival of S. aureus under conditions modeling the human colon.
Here we provide molecular proof that a cause of bile salt resistance in S. aureus is efflux, catalyzed by MnhF. This represents the first description of an intestinal colonization factor in this pathogen.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The strains and plasmids used in this work are listed in Tables 1 and 2, respectively. Escherichia coli strains were grown on Luria-Bertani (LB) medium, using selection with the antibiotic ampicillin (100 μg/ml) where appropriate. S. aureus was grown on brain heart infusion (BHI) broth (Oxoid) at 37°C. Where appropriate, the following antibiotics were added: erythromycin at 5 μg/ml and lincomycin at 25 μg/ml. Phage transductions were performed as described previously (23).
TABLE 1.
Bacterial strains
| Strain | Description or genotype | Reference or source |
|---|---|---|
| S. aureus SH1000 | Wild type | 17 |
| S. aureus SH1001 | agr mutation in SH1000 | 17 |
| S. aureus RN4220 | Accepts E. coli DNA | 18 |
| S. aureus mnhA::Tn917 | Tn917 inserted into mnhA in SH1000 | This study |
| S. aureus ΔmnhF | ΔmnhF mutation in SH1000 | This study |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(Strr) endA1 λ− | Invitrogen |
| E. coli TG1 | F′ [traD36 proAB+ lacIq lacZΔM15] supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5 (rK− mK−) | Lucigen |
TABLE 2.
Plasmids
| Plasmid | Description | Antibiotic resistance(s) | Reference or source |
|---|---|---|---|
| pLTV1 | Carries Tn917 | Emr Tcr | 19 |
| pMAD | Temperature-sensitive (30°C) E. coli-S. aureus shuttle vector; pE194ts::pBR322 | Emr | 20 |
| pBAD/HisA | Expression vector containing the araBAD promoter | Apr | 21 |
| pRMC2 | S. aureus expression vector | Apr Cmr | 22 |
| pΔmnhF | Vector for ΔmnhF mutation | Emr | This study |
| pMnhF1 | pBAD/HisA containing the mnhF internal fragment | Apr | This study |
| pMnhF2 | pRMC2 containing the mnhF internal fragment | Apr Cmr | This study |
Determination of MIC.
The MICs of selected bile salts, sodium cholate (CA), sodium chenodeoxycholate (CDCA), sodium deoxycholate (DCA), sodium glycocholate (GCA), and sodium taurocholate (TCA), were determined by broth dilution. MICs were determined by doubling dilutions, and MICs were reproduced in 3 independent experiments.
Time course measurement of bacterial viability upon exposure to bile salts.
Cultures were grown overnight to mid-exponential phase in BHI broth at 37°C with shaking. After harvesting, cells were washed twice with sterile 5 mM HEPES buffer (pH 7.2) containing 10 mM glucose and then resuspended in the same buffer to an optical density at 600 nm (OD600) of 0.5. Cells were incubated with various concentrations of bile salt for 30 min at 37°C. At 10-min intervals, dilutions from each of the bile salt-treated groups were made with a sterile peptone saline diluent. Dilutions were plated onto tryptic soy agar plates and incubated overnight at 37°C. Colonies were counted, and percent viabilities were calculated based on the initial untreated cell suspension.
Generation of an in-frame mnhF mutant.
For the ΔmnhF mutant, DNA fragments corresponding to ca. 0.7 kb upstream and downstream of mnhF were amplified by using Pwo polymerase (Roche) with primer pairs ΔmnhFLFor/ΔmnhFLRev and ΔmnhFRFor/ΔmnhFRRev (Table 3). Following purification, PCR products were digested with BamHI/EcoRI and cloned into pMAD. The resulting plasmid was used to transform electrocompetent S. aureus RN4220 (24). Plasmids were transduced into SH1000 by using φ11 phage. The temperature-sensitive nature of plasmid replication was exploited to integrate the plasmid into the bacterial chromosome, by plating cells onto medium containing erythromycin and lincomycin at 42°C. After further rounds of plating, erythromycin- and lincomycin-sensitive colonies were isolated, and the loss of mnhF was confirmed by PCR.
TABLE 3.
Oligonucleotides
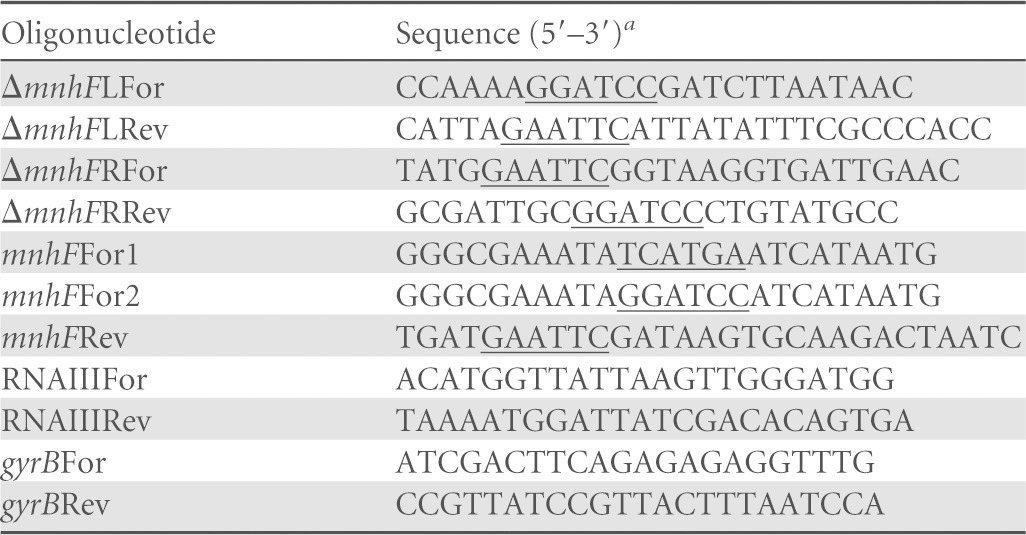
Restriction endonuclease sites are underlined.
Cloning and expression of mnhF.
The mnhF gene was amplified by PCR with S. aureus SH1000 DNA. For cloning into S. aureus, primers mnhFFor2 and mnhFRev (Table 3) were used. PCR products were digested with EcoRI and BamHI and ligated into similarly digested plasmid pRMC2. This created pMnhF2, where mnhF is fused to Pxyl/tetO, which is under the control of TetR and induced with anhydrotetracycline. For cloning into E. coli, oligonucleotides mnhFFor1 and mnhFRev (Table 3) were used. PCR products were digested with EcoRI and BspHI and ligated into similarly digested pBAD/HisA. This created pMnhF1, where mnhF is fused to PBAD, which is under the tight control of AraC.
Bile salt accumulation assay.
The accumulation of cholic acid in S. aureus was quantified by using a previously described method (25). Briefly, S. aureus and E. coli were grown in BHI and LB broth, respectively, at 37°C to an OD600 of ca. 0.6. Cells were centrifuged (5 min at 16,000 × g), washed twice in 25 mM potassium phosphate buffer (pH 7.0) containing 1 mM MgSO4, and resuspended in the same buffer to a concentration of 100 OD units/ml. A total of 1 μCi of 14C-labeled cholic acid (American Radiolabeled Chemicals) with a specific radioactivity of 55 mCi/mmol was added to a final concentration of 18 μM, and cells were incubated at 37°C for 2 h. Cells were then diluted to 10 OD units/ml in 25 mM potassium phosphate buffer (pH 7.0) containing 1 mM MgSO4, 20 mM glucose, and 0.2 mM nonradiolabeled cholic acid and incubated at 37°C. The incorporation of radiolabeled cholic acid was measured by scintillation counting. At the indicated times, 250 μl cells was centrifuged at 16,000 × g for 2 min, and the pellets were resuspended in 500 μl of sterile water and 3 ml of Ultima Gold scintillation cocktail (PerkinElmer). Counts per minute were determined with a Beckman Coulter LS 6500 liquid scintillation counter.
Quantitative real-time PCR.
mRNAs from mutant and wild-type strains were quantified by using quantitative real-time PCR (qRT-PCR). Cells were grown in triplicate as described above and then treated with RNA Protect (Qiagen), and RNA was isolated by using the Qiagen RNeasy minikit. DNA was removed by using Turbo DNase-Free (Life Technologies). Purified RNA was quantified by using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). A total of 0.5 μg of RNA was reverse transcribed by using the Tetro cDNA synthesis kit (Bioline). qRT-PCR was performed by using the Agilent qPCR system and iQ SYBR green supermix (Bio-Rad). The relative amounts of RNAIII mRNA in parental wild-type and mutant cells were determined by relative quantification using gyrB, based on consistent levels observed in previous studies (26–29). The oligonucleotides used for qRT-PCR are listed in Table 3.
Three-stage continuous-culture colonic model system (human gut model).
The three-stage continuous-culture model of the human colon was described previously (16, 30). The experiment was carried out in triplicate, using fecal samples from three different volunteers. After obtaining verbal informed consent, a standard questionnaire to collect information regarding health status, drug use, clinical anamnesis, and life-style was administered before the donor was asked to provide a fecal sample. No volunteers had received antibiotics, probiotics, steroids, or other drugs with a proven impact on gut microbiota for at least 3 months before sampling. None of them had any history of gastrointestinal disorder. All healthy fecal donors had the experimental procedure explained to them and were given the opportunity to ask questions. The University of Reading research ethics committee exempted this study from review because no donors were involved in any intervention and waived the need for written consent due to the fact that the samples received were not collected by means of intervention. All fecal samples were collected on site, kept in an anaerobic cabinet (10% H2, 10% CO2, and 80% N2), and used within a maximum of 15 min after collection. Samples were diluted 1/10 (wt/vol) in anaerobic PBS (0.1 mol/liter phosphate buffer solution, pH 7.4) and homogenized (Stomacher 400; Seward, West Sussex, United Kingdom) for 2 min at 460 paddle beats.
Samples were plated onto BHI agar containing 0.01% (wt/vol) potassium tellurite as a selective agent at different dilutions in PBS (from 102 to 109 CFU/ml) in triplicate for each time point to measure bacterial counts.
Statistical analysis.
All experiments were repeated three times, and data are presented as means ± standard errors of the means. Analysis was performed by using GraphPad Prism 5 software. Experimental data were analyzed by one-way analysis of variance (ANOVA) and two-way ANOVA methods, using Bonferroni posttest analysis.
RESULTS
Identification of a bile salt resistance locus.
Genes conferring resistance to bile were identified by replica plating of S. aureus SH1000 Tn917 insertion libraries onto BHI agar and onto BHI agar containing 18% (wt/vol) bile salts (Oxoid), which represented 0.8× MIC. Six colonies were unable to grow in the presence of bile salts but exhibited no growth defect on BHI agar in the absence of bile. Sequencing of the genomic DNA flanking the transposon insertion site of bile-sensitive strains was carried out in order to identify the DNA insertion sites of Tn917, revealing that all six strains were siblings containing the transposon inserted into the same gene, namely, the previously described mnhA gene, the first gene in the polycistronic mnhABCDEFG operon, which encodes a Na+/H+ antiporter (31). Bacillus subtilis contains the orthologous mrpABCDEFG operon, which has an identical function; however, mrpF and, by extension, mnhF are homologous to mammalian bile transporters, and mrpF mediates cholic acid efflux (32, 33).
MnhF mediates resistance to bile salts.
We hypothesized that MnhF was responsible for the observed bile salt resistance phenotype. To test this, an in-frame ΔmnhF strain was created in S. aureus SH1000. The mutant strain had no growth defect when grown on BHI solid or liquid medium in the absence of bile salts (results not shown). Compared to the parental wild-type strain, the ΔmnhF strain had a reduced MIC for unconjugated bile salts, in particular cholic acid (Table 4). Complementation of the mutation with mnhF under the control of an inducible promoter restored the bile resistance phenotype to that observed for the parent strain in the presence of anhydrotetracycline as an inducer (Table 4), whereas there was no such resistance in the absence of the inducer (results not shown). In killing assays, the ΔmnhF strain was significantly more sensitive than the parent. In the presence of 1 μg/ml anhydrotetracycline, the complemented strain exhibited a rate of cell death similar to that of the parental wild-type strain (Fig. 1). The increased sensitivity of the mutant strain was observed only with unconjugated bile salts. However, it should be noted that we were unable to determine the MIC of conjugated bile salts for S. aureus, as they were insoluble at concentrations of >200 mM.
TABLE 4.
MICs of bile salts for S. aureus SH1000 and the ΔmnhF mutanta
| Bile salt | MIC (mM) for strain |
|||
|---|---|---|---|---|
| Wild type | ΔmnhF | ΔmnhF(pMnhF2) | ΔmnhF(pRMC2) | |
| CA | 22 | 5 | 22 | 5 |
| DCA | 1.2 | 0.6 | 1.2 | 0.6 |
| CDCA | 1.2 | 0.6 | 1.2 | 0.6 |
| GCA | >200 | >200 | ND | ND |
| TCA | >200 | >200 | ND | ND |
CA, sodium cholate; DCA, sodium deoxycholate; CDCA, sodium chenodeoxycholate; GCA, sodium glycocholate; TCA, sodium taurocholate; ND, not determined.
FIG 1.
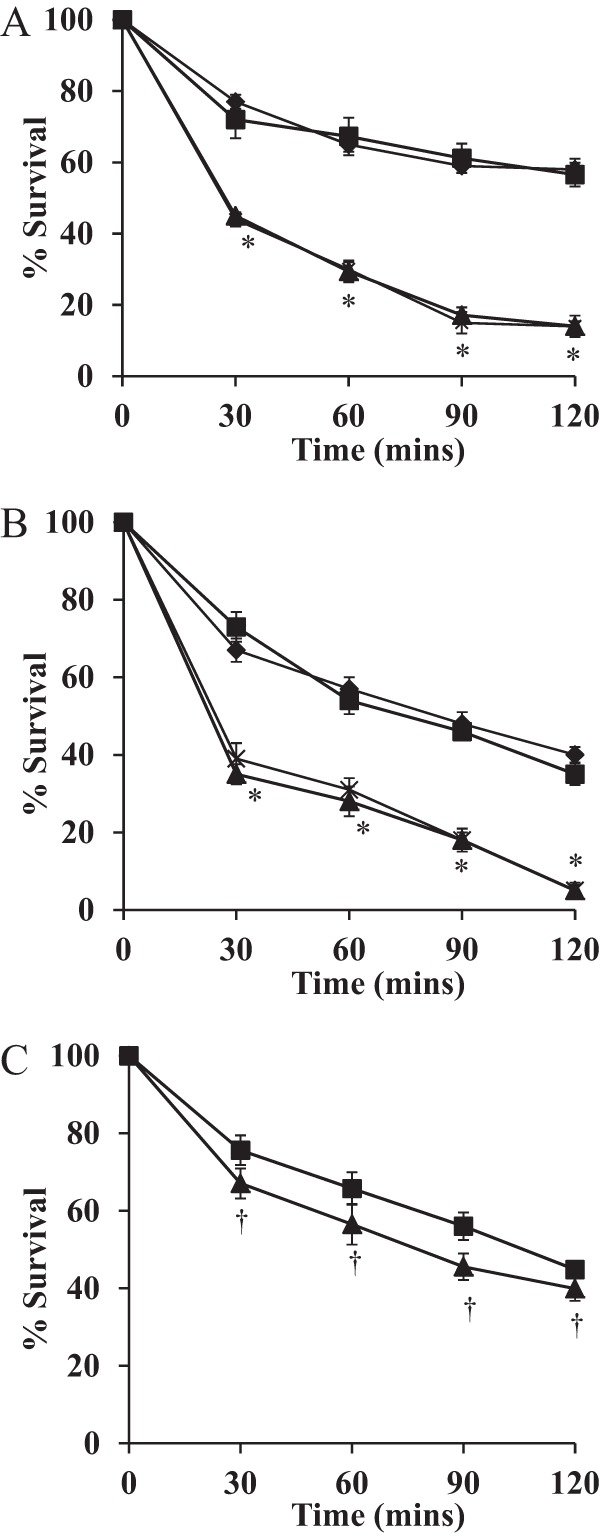
MnhF protects S. aureus against the bactericidal activity of bile salts. Shown are data on the viability of S. aureus SH1000 (■), the ΔmnhF strain (▲), the ΔmnhF(pMnhF2) strain (⧫), and the ΔmnhF(pRMC2) strain (×) treated with 2 mM CA (A), 0.25 mM DCA (B), and 20 mM GCA (C). Data represent means ± standard errors of the means from three independent experiments. *, P < 0.01; †, P > 0.05.
To confirm the role of mnhF in bile salt resistance, it was cloned under the control of the arabinose-inducible PBAD promoter of plasmid pBAD/HisA, which enabled arabinose-dose-dependent expression of MnhF in E. coli strains TG1 and TOP10. The expression of MnhF increased the MICs of both conjugated and unconjugated bile salts in both background strains, and in the case of cholic acid, the increased resistance was arabinose dose dependent (Table 5). Similarly, the expression of MnhF in E. coli decreased the bacteriostatic effects of bile salts on this bacterium (Fig. 2). Thus, MnhF was sufficient to enable bile salt resistance in the absence of the rest of the mnhABCDEFG operon.
TABLE 5.
MICs of bile salts for wild-type and recombinant E. coli strains expressing MnhF at different levels of arabinose induction
| Bile salt | MIC (mM) for strain |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type |
Vector control |
Recombinant |
||||||||
| TG1(pMnhF1) with: |
TOP10(pMnhF1) with: |
|||||||||
| TG1 | TOP10 | TG1(pBAD) | TOP10(pBAD) | 0% arabinose | 0.02% arabinose | 2% arabinose | 0% arabinose | 0.02% arabinose | 2% arabinose | |
| CA | 30 | 30 | 30 | 30 | 30 | 60 | 90 | 30 | 60 | 90 |
| DCA | 4 | 4 | 4 | 4 | 4 | >4 | >4 | 4 | >4 | >4 |
| CDCA | 4 | 4 | 4 | 4 | 4 | >4 | >4 | 4 | >4 | >4 |
| GCA | 50 | 50 | 50 | 50 | 50 | 100 | 100 | 50 | 100 | 100 |
| TCA | 50 | 50 | 50 | 50 | 50 | 100 | 100 | 50 | 100 | 100 |
FIG 2.
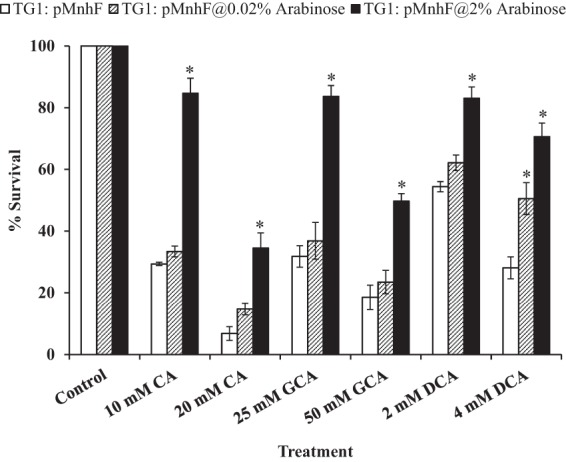
Heterologous expression of MnhF in E. coli protects against the bacteriostatic effects of bile salts. Shown are data on the viability of wild-type E. coli TG1 and E. coli TG1(pMnhF1) cells in LB medium containing CA (10 and 20 mM), DCA (2 and 4 mM), and GCA (25 and 50 mM) and then grown overnight at 37°C. Cell counts were then determined by viable plate counting. Data represent means ± standard errors of the means from three independent experiments. *, P < 0.001.
Effect of efflux pump inhibitors on bile salt resistance.
Given the ability of MnhF to confer bile salt resistance and its similarity to other known and putative bile efflux systems, its ability to mediate the removal of cholic acid from bacteria was tested. Both Phe-Arg-β-naphthylamide (PAβN), a synthetic dipeptide that inhibits bacterial efflux pumps, including bile salt efflux pumps of Gram-negative bacteria, and reserpine, a plant alkaloid which can inhibit multidrug efflux pumps in Gram-positive bacteria, were tested for their ability to reduce bile salt MICs in S. aureus. Both inhibitors caused reductions in the S. aureus MIC for cholic acid, and PAβN reduced the MIC for all three unconjugated bile salts (Table 6); however, the reduction was much smaller in the ΔmnhF strain than in the parental wild-type strain, possibly indicating the presence of other bile salt efflux systems in the pathogen. Similarly, in E. coli(pMnhF1), PAβN reduced bile salt MICs to levels lower than those in untreated E. coli(pBAD/HisA) (Table 7). Thus, in both S. aureus and E. coli, inhibitors of efflux pumps abrogated bile salt resistance in an MnhF-dependent manner.
TABLE 6.
Effect of efflux pump inhibitors on MICs of bile salts for S. aureusa
| Bile salt | MIC (mM) for strain |
|||||
|---|---|---|---|---|---|---|
|
S. aureus SH1000 |
S. aureus ΔmnhF |
|||||
| Control | PAβN | Reserpine | Control | PAβN | Reserpine | |
| CA | 22 | 2.5 | 10 | 5 | 2.5 | 2.5 |
| DCA | 1.2 | 0.3 | 1.2 | 0.6 | 0.3 | 0.3 |
| CDCA | 1.2 | 0.3 | 1.2 | 0.6 | 0.3 | 0.3 |
| GCA | >200 | 200 | >200 | >200 | 200 | >200 |
| TCA | >200 | 200 | >200 | >200 | 200 | >200 |
With PAβN at 20 μg/ml and reserpine at 40 μg/ml.
TABLE 7.
Effect of efflux pump inhibitors on MICs of bile salts for E. colia
| Bile salt | MIC (mM) |
|||||
|---|---|---|---|---|---|---|
|
E. coli TG1 |
E. coli TG1(pMnhF1) |
|||||
| Control | PAβN | Reserpine | Control | PAβN | Reserpine | |
| CA | 30 | 2.5 | 30 | 90 | 2.5 | 90 |
| DCA | 4 | 0.6 | >4 | >4 | 0.6 | >4 |
| CDCA | 4 | 0.6 | >4 | >4 | 0.6 | >4 |
| GCA | 50 | 10 | 50 | 100 | 10 | 100 |
| TCA | 50 | 10 | 50 | 100 | 10 | 100 |
With PAβN at 20 μg/ml and reserpine at 40 μg/ml.
MnhF transports cholic acid.
Given the ability of efflux pump inhibitors to reduce the MICs of certain bile salts in S. aureus, the capacity of MnhF to transport cholic acid was determined in vitro by using a 14C-radiolabeled cholic acid substrate, similarly to previous efflux assays (25, 34, 35). The S. aureus SH1000 and ΔmnhF strains were incubated with [14C]cholic acid (uptake period) and then diluted in buffer containing an excess of nonradiolabeled cholic acid (efflux period). The initial rate of [14C]cholic acid uptake was the same for both strains (10,962 ± 550 cpm for S. aureus SH1000 and 10,278 ± 278 cpm for the S. aureus ΔmnhF mutant), but throughout the efflux period, the S. aureus ΔmnhF strain retained significantly more of the radiolabel than did the parental wild-type strain (Fig. 3A). To further corroborate these findings, efflux assays were also carried out on E. coli expressing MnhF. E. coli TG1, E. coli TG1(pBAD), and E. coli TG1(pMnhF1) were grown overnight in LB supplemented with 1% arabinose at 37°C and then incubated with [14C]cholic acid. All the E. coli TG1 strains incorporated similar levels of [14C]cholic acid during the uptake period [20,774 ± 363 cpm for TG1, 23,274 ± 386 cpm for TG1(pBAD), and 22,435 ± 460 cpm for TG1(pMnhF1)]. At various points after the initial incorporation of radiolabeled cholic acid, cells were centrifuged, and cell-associated radioactivity was determined by the liquid scintillation method. E. coli TG1 cells expressing MnhF retained significantly (P < 0.05) lower levels of 14C-radiolabeled cholic acid than did parental TG1 cells and TG1 cells with the empty pBAD vector [TG1(pBAD)] (Fig. 3B). In both sets of experiments, the reason for the increasing cell-associated radiolabel during the efflux period, after which cells had been diluted in excess nonlabeled cholic acid, is unclear but has also been observed in previous studies on Listeria monocytogenes and may reflect continued incorporation of [14C]cholic acid during the efflux period after dilution (25).
FIG 3.
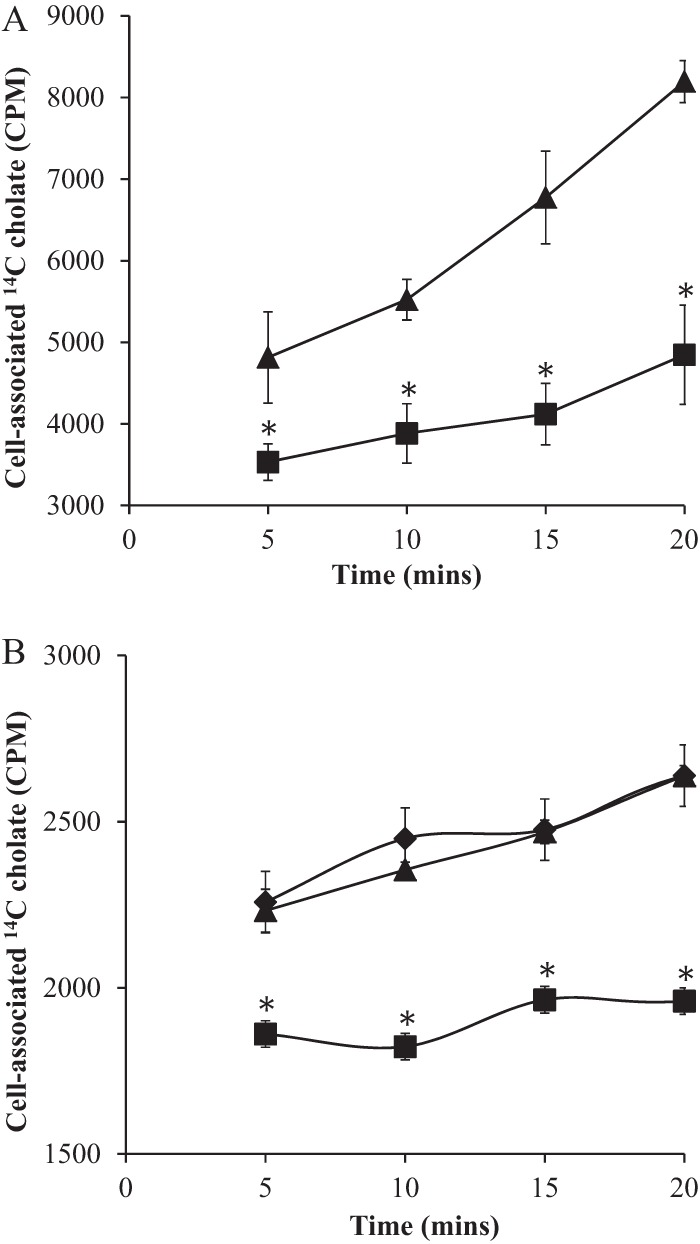
MnhF exports cholic acid. (A) S. aureus SH1000 wild-type (■) and ΔmnhF (▲) cells were loaded with 1 μCi of [14C]cholic acid and then diluted in a buffer containing an excess of nonradiolabeled cholic acid (0.2 mM). (B) E. coli TG1 parental cells (▲), E. coli TG1 cells expressing pBAD [TG1(pBAD)] (⧫), and E. coli TG1 cells expressing pMnhF1 [TG1(pMnhF1)] (■) grown overnight in LB medium under 1% arabinose induction were loaded with 1 μCi of [14C]cholic acid and then diluted in a buffer containing an excess of nonradiolabeled cholic acid (0.2 mM) and 1% arabinose. At the indicated times, the amounts of retained [14C]cholic acid in cell pellets were determined by liquid scintillation counting. Data represent means ± standard errors of the means of data from three independent experiments. *, P < 0.05.
Bile salt resistance is not affected by agr.
To examine whether the agr quorum sensing system is involved in bile salt resistance, the MICs for CA, DCA, and CDCA in S. aureus SH1001 (agr) were determined and found to be indistinguishable from those in the wild type (results not shown). Furthermore, the agr system is not inhibited by the mnhF mutation, as the RNAIII effector molecule is still produced (Fig. 4). Thus, we were unable to demonstrate a role for agr in bile resistance.
FIG 4.
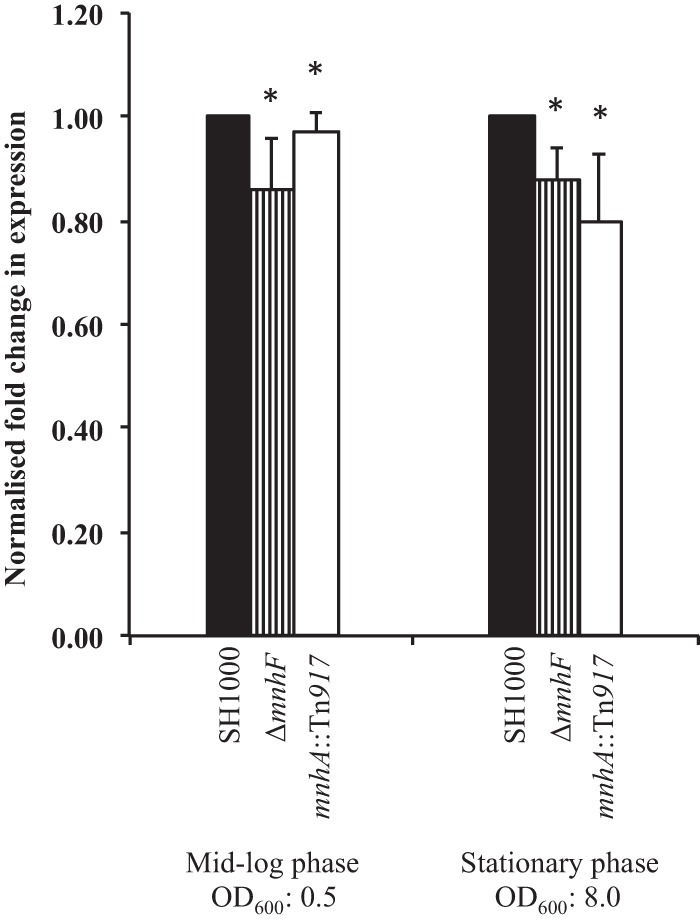
Mutation of mnhF does not affect agr. qRT-PCR was performed in order to quantify amounts of RNAIII in S. aureus strains during the exponential and stationary phases of growth. Data represent means ± standard errors of the means from three independent experiments. *, P > 0.05.
MnhF is required for survival of S. aureus in a human gut model.
To examine the role of MnhF in the survival of S. aureus under conditions found in the human colon, we used a three-stage continuous-culture gut model system designed to reproduce the spatial, temporal, nutritional, and physicochemical characteristics of the microbiota in the human colon. In vivo studies of colonic bacteria are hampered by the lack of suitable animal models, as these models do not correctly simulate the microbiota and physicochemical conditions of the human colon (36). We have previously used this in vitro model to study survival of S. aureus and the impact of infection on the host's intestinal microflora (16).
Mutational inactivation of the whole mnhABCDEFG operon does not affect the ability of S. aureus to grow at a range of pHs (37). In order to exclude the possibility that the normal pH range (5.5 to 7.5) found in the colon influenced the survival of the ΔmnhF mutant, we corroborated the previous observations at pH 5.5 to 8.5 using this strain (results not shown).
After inoculation of vessel 1 (which models the proximal colon) of the colonic models with S. aureus to a concentration of ca. 2 × 1010 CFU/ml as a single dose, the S. aureus populations stabilized at 6 to 7 log10 units over a period of up to 8 h. Survival of the S. aureus ΔmnhF strain was significantly attenuated compared to that of its parental strain in all three vessels (Fig. 5A to C).
FIG 5.
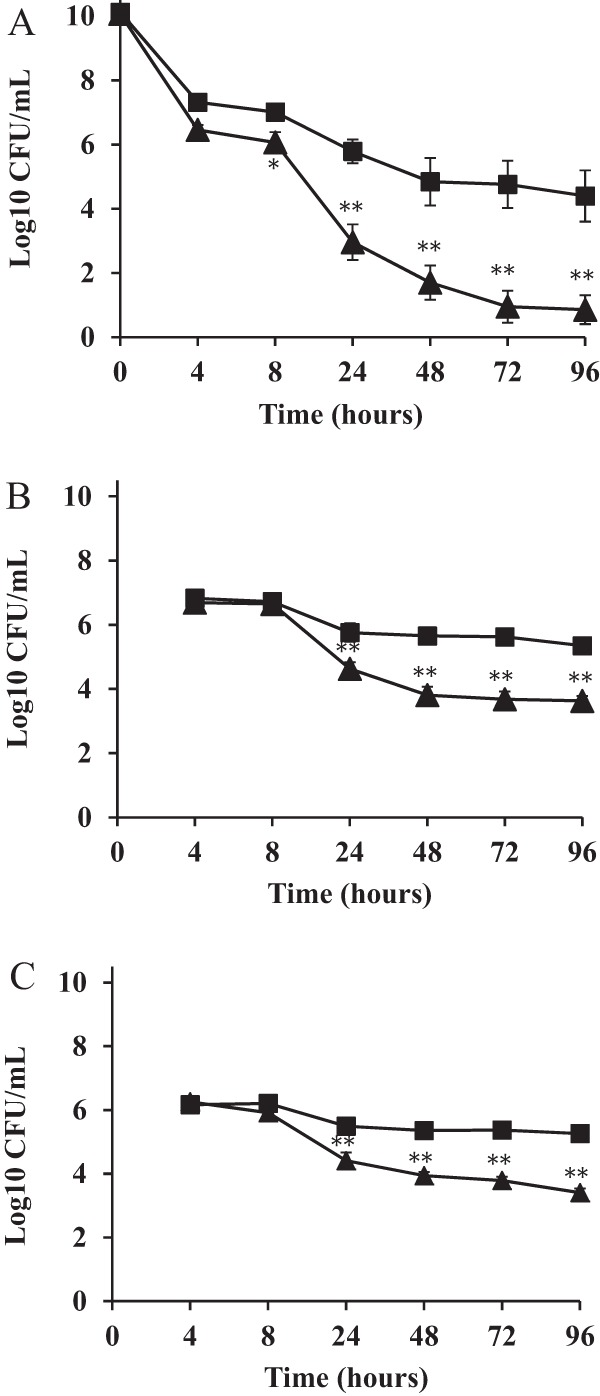
MnhF is required for S. aureus survival in the human colonic model. Survival of S. aureus SH1000 (■) and ΔmnhF (▲) cells in the human colonic model. (A) Model of the ascending colon (vessel 1); (B) model of the transverse colon (vessel 2); (C) model of the descending colon (vessel 3). Samples were taken at inoculation (0 h) and at 4, 8, 24, 48, 72, and 96 h postinfection. Results are reported as means (log10 CFU/ml) of the data from three colonic models ± standard errors of the means. *, P < 0.05; **, P < 0.001.
DISCUSSION
A complex set of interactions exists between S. aureus and its human host, as the bacterium is able to colonize several niches, as both an opportunistic pathogen of great medical importance and a common commensal. In order to defend against colonization by microorganisms, the host produces a range of antimicrobials, such as peptides, fatty acids, and bile. Bile represents a significant challenge to the gut microflora; in humans, the liver secretes up to 1 liter of bile per day into the intestines (38). Furthermore, molecules secreted by bacteria, including S. aureus, during infection are an important cause of metabolic cholestasis, an inability of hepatocytes to produce bile (39). Bile is a complex cocktail composed principally of bile salts, phospholipids, cholesterol, proteins, and bilirubin (40). Originally characterized as digestive molecules, bile salts have antimicrobial activity, which has been attributed to their ability to damage cell membranes (41). Additionally, they cause intracellular acidification and induce the formation of secondary structures in RNA, DNA damage, and misfolding and denaturation of proteins. Thus, bile salts represent a serious challenge to bacterial cells in the gastrointestinal tract, and bacteria that are able to colonize the gut should therefore be able to overcome their toxicity.
Bile salts that pass into the large intestine undergo modification by the normal microbiota (42). The major modifications include deconjugation; oxidation of hydroxyl groups at C-3, C-7, and C-12; and 7α/β-dehydroxylation (43, 44). Thus, the normal commensal inhabitants of the human gastrointestinal tract, such as Lactobacillus, Propionibacterium, and Bifidobacterium, are required by the host for maintenance of gut health and ecological balance by influencing the composition of the bile acids in the large intestine and, by extension, the gut microbiome (45, 46). Their ability to survive in the presence of bile salts indicates the existence of inherent bile resistance mechanisms. Indeed, colonic commensals deploy various different strategies for resisting bile. Lactobacillus plantarum produces a bile salt hydrolase, which detoxifies bile salts by deconjugating bile salts inside the cell, turning them into weaker acids and thus negating the drop in pH that they cause (47). Bifidobacteria possess a number of characterized bile salt resistance mechanisms. In addition to multiple efflux pumps, exposure to bile salts results in a modification of the cell envelope. Increased concentrations of membrane fatty acids and altered phospholipids increase membrane rigidity and reduce the permeation of lipophilic bile salts (48). Similarly, exposure of Bifidobacterium animalis subsp. lactis to bile salts induces increased expression of exopolysaccharides, which are proposed to form a protective layer around the bacterium (49).
Bile salts represent a physiological challenge for bacteria and an environmental cue; Salmonella enterica and Vibrio cholerae regulate intestinal colonization and virulence in response to bile (50, 51). However, pathogens that inhabit the human intestine are also exposed to the bactericidal nature of bile salts and hence must also exhibit resistance in order to survive. Generally, Gram-negative bacteria are more innately resistant than Gram-positive bacteria due to the presence of an outer membrane, which acts as a barrier (38). Indeed, maintenance of membrane integrity by lipopolysaccharide (LPS) in the cellular envelope of Gram-negative bacteria imparts protection against the actions of bile salts (52, 53). Salmonella enterica serovar Typhi and S. enterica serovar Typhimurium are able to grow at bile concentrations that are much higher than those encountered in vivo. This is due, at least in part, to the presence of outer membrane efflux pumps such as AcrAB (54). Similarly, HefC is an AcrB homologue that confers bile salt resistance to Helicobacter pylori (55). The multidrug efflux pump CmeABC of Campylobacter jejuni mediates bile salt resistance and is required for colonization of chickens (56). Gram-positive pathogens such as Enterococcus faecalis and L. monocytogenes also exhibit bile resistance. In addition to bile salt hydrolase activities, both bacteria possess multiple bile efflux systems. Exposure of E. faecalis to bile results in the upregulation of two open reading frames, EF0420 and EF1814, which are homologous to the QacA family of efflux pumps (57). L. monocytogenes OpuC, an osmolyte transporter, as well as the specialist bile transporters BilE and MdrT all confer bile salt resistance to the pathogen (58).
We demonstrated that the mnhABCDEFG operon in S. aureus confers bile salt resistance to the pathogen. Previous studies have shown that this operon encodes a multisubunit hetero-oligomeric antiporter system involved in the efflux of monovalent cations such as Na+, K+, and Li+ in exchange for H+ (59). Transposon insertion into mnhD (also called snoD) resulted in reduced susceptibility to platelet microbicidal protein 1 (37); thus, the operon also has the ability to sensitize the pathogen to other host innate antimicrobials. The function of individual components remains to be determined; however, mnhF is homologous to a hamster ileal bile salt transporter (60) and a rat liver organic anion transporter that was shown to efflux cholic acid (61). A transposon insertion at mnhA, which presumably had a polar effect on the rest of the operon, and an in-frame deletion of mnhF rendered the bacterium equally susceptible to bile salts. Together with our observation that cloning of mnhF in E. coli increased the bile salt MIC, we demonstrated that MnhF alone is sufficient to confer bile salt resistance. Furthermore, MnhF acted to exclude cholic acid from both S. aureus and E. coli.
In order to confirm that this increased sensitivity of S. aureus translated into a decreased ability of S. aureus to survive under conditions found in the human colon, we studied the survival of the mutant in a well-characterized in vitro three-stage system that models the microbial and physicochemical conditions of the proximal, transverse, and distal colon (30). The ΔmnhF strain was attenuated in its ability to survive in this model, compared to the parental wild-type strain. To date, no suitable in vivo models have been developed to study the carriage and survival of S. aureus in the human intestine. Laboratory mouse models of infection do not reproduce the human gut’s complex microbial ecosystem or physicochemical defenses (36).
The physiology of S. aureus in the human gut is very poorly understood, relative to other niches. A recent study to determine S. aureus genetic traits associated with observed higher rectal carriage rates was inconclusive (62); thus, this is the first report of an S. aureus intestinal colonization factor. Given the complex nature of the gut as a niche, it seems highly likely that other loci are similarly required. Indeed, it appears from our data that other bile resistance factors also exist. As such, much remains to be discovered about the behavior and survival of S. aureus in the human gut.
ACKNOWLEDGMENT
This work was funded by a Ph.D. Felix scholarship to T.H.S.
REFERENCES
- 1.Peacock SJ, de Silva I, Lowy FD. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol 9:605–610. doi: 10.1016/S0966-842X(01)02254-5. [DOI] [PubMed] [Google Scholar]
- 2.Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 3.Acton DS, Plat-Sinnige MJ, van Wamel W, de Groot N, van Belkum A. 2009. Intestinal carriage of Staphylococcus aureus: how does its frequency compare with that of nasal carriage and what is its clinical impact? Eur J Clin Microbiol Infect Dis 28:115–127. doi: 10.1007/s10096-008-0602-7. [DOI] [PubMed] [Google Scholar]
- 4.Masaki H, Asoh N, Watanabe H, Tao M, Watanabe K, Ikeda H, Matsumoto K, Oishi K, Nagatake T. 2003. Possible relationship between Staphylococcus aureus colonizing the respiratory tract and rectum and S. aureus isolated in a geriatric hospital environment. Intern Med 42:281–282. doi: 10.2169/internalmedicine.42.281. [DOI] [PubMed] [Google Scholar]
- 5.Bhalla A, Aron DC, Donskey CJ. 2007. Staphylococcus aureus intestinal colonization is associated with increased frequency of S. aureus on skin of hospitalized patients. BMC Infect Dis 7:105. doi: 10.1186/1471-2334-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Squier C, Rihs JD, Risa KJ, Sagnimeni A, Wagener MM, Stout J, Muder RR, Singh N. 2002. Staphylococcus aureus rectal carriage and its association with infections in patients in a surgical intensive care unit and a liver transplant unit. Infect Control Hosp Epidemiol 23:495–501. doi: 10.1086/502095. [DOI] [PubMed] [Google Scholar]
- 7.Ray AJ, Pultz NJ, Bhalla A, Aron DC, Donskey CJ. 2003. Coexistence of vancomycin-resistant enterococci and Staphylococcus aureus in the intestinal tracts of hospitalized patients. Clin Infect Dis 37:875–881. doi: 10.1086/377451. [DOI] [PubMed] [Google Scholar]
- 8.Froberg MK, Palavecino E, Dykoski R, Gerding DN, Peterson LR, Johnson S. 2004. Staphylococcus aureus and Clostridium difficile cause distinct pseudomembranous intestinal diseases. Clin Infect Dis 39:747–750. doi: 10.1086/423273. [DOI] [PubMed] [Google Scholar]
- 9.Björkstén B, Naaber P, Sepp E, Mikelsaar M. 1999. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy 29:342–346. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 10.Lindberg E, Nowrouzian F, Adlerberth I, Wold AE. 2000. Long-time persistence of superantigen-producing Staphylococcus aureus strains in the intestinal microflora of healthy infants. Pediatr Res 48:741–747. doi: 10.1203/00006450-200012000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Lindberg E, Adlerberth I, Hesselmar B, Saalman R, Strannegård I-L, Aberg N, Wold AE. 2004. High rate of transfer of Staphylococcus aureus from parental skin to infant gut flora. J Clin Microbiol 42:530–534. doi: 10.1128/JCM.42.2.530-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adlerberth I, Strachan DP, Matricardi PM, Ahrné S, Orfei L, Aberg N, Perkin MR, Tripodi S, Hasselmar B, Saalman R, Coates AR, Bonanno CL, Panetta V, Wold AE. 2007. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol 120:343–350. doi: 10.1016/j.jaci.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Lundell AC, Adlerberth I, Lindberg E, Karlsson H, Ekberg S, Aberg N, Saalman R, Hock B, Steinkasserer A, Hesselmar B, Wold AE, Rudin A. 2007. Increased levels of circulating soluble CD14 but not CD83 in infants are associated with early intestinal colonization with Staphylococcus aureus. Clin Exp Allergy 37:62–71. doi: 10.1111/j.1365-2222.2006.02625.x. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura Y, Aramaki Y, Kakiuchi T. 2001. A mouse model for postoperative fetal enteritis due to Staphylococcus infection. J Surg Res 96:35–43. doi: 10.1006/jsre.2000.6043. [DOI] [PubMed] [Google Scholar]
- 15.Hess DJ, Garni RM, Henry-Stanley MJ, Wells CL. 2005. Escherichia coli modulates extraintestinal spread of Staphylococcus aureus. Shock 24:376–381. doi: 10.1097/01.shk.0000180615.75822.fe. [DOI] [PubMed] [Google Scholar]
- 16.Sannasiddappa TH, Costabile A, Gibson GR, Clarke SR. 2011. The influence of Staphylococcus aureus on gut microbial ecology in an in vitro continuous culture human colonic model system. PLoS One 6:e23227. doi: 10.1371/journal.pone.0023227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 19.Camilli A, Portnoy A, Youngman P. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol 172:3738–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corrigan RM, Foster TJ. 2009. An improved tetracycline inducible expression vector for Staphylococcus aureus. Plasmid 61:126–129. doi: 10.1016/j.plasmid.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 24.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 73:133–138. [DOI] [PubMed] [Google Scholar]
- 25.Quillin SJ, Schwartz KT, Leber JH. 2011. The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol Microbiol 81:129–142. doi: 10.1111/j.1365-2958.2011.07683.x. [DOI] [PubMed] [Google Scholar]
- 26.Wolz C, Goerke C, Landmann R, Zimmerli W, Fluckiger U. 2002. Transcription of clumping factor A in attached and unattached Staphylococcus aureus in vitro and during device-related infection. Infect Immun 70:2758–2762. doi: 10.1128/IAI.70.6.2758-2762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valle J, Toledo-Arana A, Berasain C, Ghigo J-M, Amorena B, Penadés J, Lasa I. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 28.Kenny JG, Ward D, Josefsson E, Jonsson I-M, Hinds J, Rees HH, Lindsay JA, Tarkowski A, Horsburgh MJ. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. doi: 10.1371/journal.pone.0004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Shopsin B, Zhao Y, Smyth D, Wasserman GA, Fang C, Liu L, Kreiswirth BN. 2012. Real-time nucleic acid sequence-based amplification assay for rapid detection and quantification of agr functionality in clinical Staphylococcus aureus isolates. J Clin Microbiol 50:657–661. doi: 10.1128/JCM.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macfarlane GT, Macfarlane S, Gibson GR. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35:180–187. doi: 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
- 31.Hiramatsu T, Kodama K, Kuroda T, Mizushima T, Tsuchiya T. 1998. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J Bacteriol 180:6642–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito M, Guffanti AA, Oudega B, Krulwich TA. 1999. mnh, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J Bacteriol 181:2394–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito M, Guffanti AA, Wang W, Krulwich TA. 2000. Effects of nonpolar mutations in each of the seven Bacillus subtilis mnh genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J Bacteriol 182:5663–5670. doi: 10.1128/JB.182.20.5663-5670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sleator RD, Wemekamp-Kamphuis HH, Gahan CG, Abee T, Hill C. 2005. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol 55:1183–1195. doi: 10.1111/j.1365-2958.2004.04454.x. [DOI] [PubMed] [Google Scholar]
- 36.Hapfelmeier S, Hardt WD. 2005. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol 13:497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Bayer AS, McNamara P, Yeaman MR, Lucindo N, Jones T, Cheung AL, Sahl HG, Proctor RA. 2006. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. J Bacteriol 188:211–222. doi: 10.1128/JB.188.1.211-222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Begley M, Gahan CG, Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Minuk GY, Rascanin N, Sarjeant ES, Pai CH. 1986. Sepsis and cholestasis: the in vitro effects of bacterial products on 14C-taurocholate uptake by isolated rat hepatocytes. Liver 6:199–204. doi: 10.1055/s-2008-1040603. [DOI] [PubMed] [Google Scholar]
- 40.Esteller A. 2008. Physiology of bile secretion. World J Gastroenterol 14:5641–5649. doi: 10.3748/wjg.14.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholmerich J, Becher MS, Schmidt K, Schubert R, Kremer B, Feldhaus S, Gerok W. 1984. Influence of hydroxylation and conjugation of bile salts on their membrane-damaging properties—studies on isolated hepatocytes and lipid membrane vesicles. Hepatology 4:661–666. doi: 10.1002/hep.1840040416. [DOI] [PubMed] [Google Scholar]
- 42.Hoffman AF. 1999. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med 159:2647–2658. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 43.Ridlon J, Kang D-J, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Buffie CG, Bucci V, Stein RR, McKenny PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van der Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaing NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guarner F, Malagelada JR. 2003. Gut flora in health and disease. Lancet 361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 46.Pfeiler EA, Klaenhammer TR. 2009. Role of transporter proteins in bile tolerance of Lactobacillus acidophilus. Appl Environ Microbiol 75:6013–6016. doi: 10.1128/AEM.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Smet I, Van Hoorde L, Vande Woestyne M, Christiaens H, Verstraete W. 1995. Significance of bile salt hydrolytic activities of lactobacilli. J Appl Bacteriol 79:292–301. doi: 10.1111/j.1365-2672.1995.tb03140.x. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz L, Sanchez B, Ruas-Madiedo P, de Los Reyes-Gavilan CG, Margolles A. 2007. Cell envelope changes in Bifidobacterium animalis ssp. lactis as a response to bile. FEMS Microbiol Lett 274:316–322. doi: 10.1111/j.1574-6968.2007.00854.x. [DOI] [PubMed] [Google Scholar]
- 49.Ruas-Madiedo P, Gueimonde M, Arigoni F, de los Reyes-Gavilan CG, Margolles A. 2009. Bile affects the synthesis of exopolysaccharides by Bifidobacterium animalis. Appl Environ Microbiol 75:1204–1207. doi: 10.1128/AEM.00908-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prouty AM, Gunn JS. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect Immun 68:6763–6769. doi: 10.1128/IAI.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peterson KM. 2002. Expression of Vibrio cholerae virulence genes in response to environmental signals. Curr Issues Intest Microbiol 3:29–38. [PubMed] [Google Scholar]
- 52.Nesper J, Schild S, Lauriano CM, Kraiss A, Klose KE, Reidl J. 2002. Role of Vibrio cholerae O139 surface polysaccharides in intestinal colonization. Infect Immun 70:5990–5996. doi: 10.1128/IAI.70.11.5990-5996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crawford RW, Keestra AM, Winter SE, Xavier MN, Tsolis RM, Tolstikov V, Bäumler AJ. 2012. Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog 8:e1002918. doi: 10.1371/journal.ppat.1002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 55.Trainor EA, Horton KE, Savage PB, Testerman TL, McGee DJ. 2011. Role of the HefC efflux pump in Helicobacter pylori cholesterol-dependent resistance to ceragenins and bile salts. Infect Immun 79:88–97. doi: 10.1128/IAI.00974-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solheim M, Aakra A, Vebo H, Snipen L, Nes IF. 2007. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl Environ Microbiol 73:5767–5774. doi: 10.1128/AEM.00651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Begley M, Sleaton RD, Gahan GC, Hill C. 2005. Contribution of three bile-associated loci bsh, pva, and bltB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun 73:894–904. doi: 10.1128/IAI.73.2.894-904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swartz TH, Ito M, Ohira T, Natsui S, Hicks DB, Krulwich TA. 2007. Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J Bacteriol 189:3081–3090. doi: 10.1128/JB.00021-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong MH, Oelkers P, Craddock AL, Dawson PA. 1994. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J Biol Chem 269:1340–1347. [PubMed] [Google Scholar]
- 61.Jacquemin E, Hagenbuch B, Stieger B, Wolkoff AW, Meier PJ. 1994. Expression cloning of a rat liver Na(+)-independent organic anion transporter. Proc Natl Acad Sci U S A 91:133–137. doi: 10.1073/pnas.91.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lemmens N, van Wamel W, Snijders S, Lesse AJ, Faden H, van Belkum A. 2011. Genomic comparisons of USA300 Staphylococcus aureus colonizating the nose and rectum of children with skin abscesses. Microb Pathog 50:192–199. doi: 10.1016/j.micpath.2010.12.006. [DOI] [PubMed] [Google Scholar]


