Abstract
The negative global impact of the AIDS pandemic is well known. In this perspective article, the utility of magic angle spinning (MAS) NMR spectroscopy to answer pressing questions related to the structure and dynamics of HIV-1 protein assemblies is examined. In the recent years, MAS NMR has undergone major technological developments enabling studies of large viral assemblies. We discuss some of these evolving methods and technologies and provide a perspective on the current state of MAS NMR as applied to the investigations into structure and dynamics of HIV-1 assemblies of CA capsid protein and of Gag maturation intermediates.
Keywords: solid-state NMR, magic angle spinning, MAS, HIV-1, HIV-AIDS, Gag polyprotein, CA, capsid protein
Graphical abstract

1. Introduction
Since the discovery of the human immunodeficiency virus (HIV) as the causative agent for acquired immunodeficiency syndrome (AIDS) in 1983 [1, 2] three decades of intense research have followed. Current estimates reveal that 35 million people are living with HIV, a global prevalence rate of 0.8%. In some Sub-Saharan African nations, the HIV prevalence rate exceeds 20% [3]. Current treatment options for those infected with HIV are based on antiretroviral therapies from several different classes of drugs. While effective, they are often limiting due to prohibitive cost, debilitating short- and long-term side effects, and the ability to give rise to drug-resistant mutants (www.AIDS.gov). A better understanding of important atomic-level structural information of the various macromolecules and macromolecular assemblies comprising HIV and the molecular processes that govern the virus’ function could lead to enhanced therapies and hopefully an eventual cure.
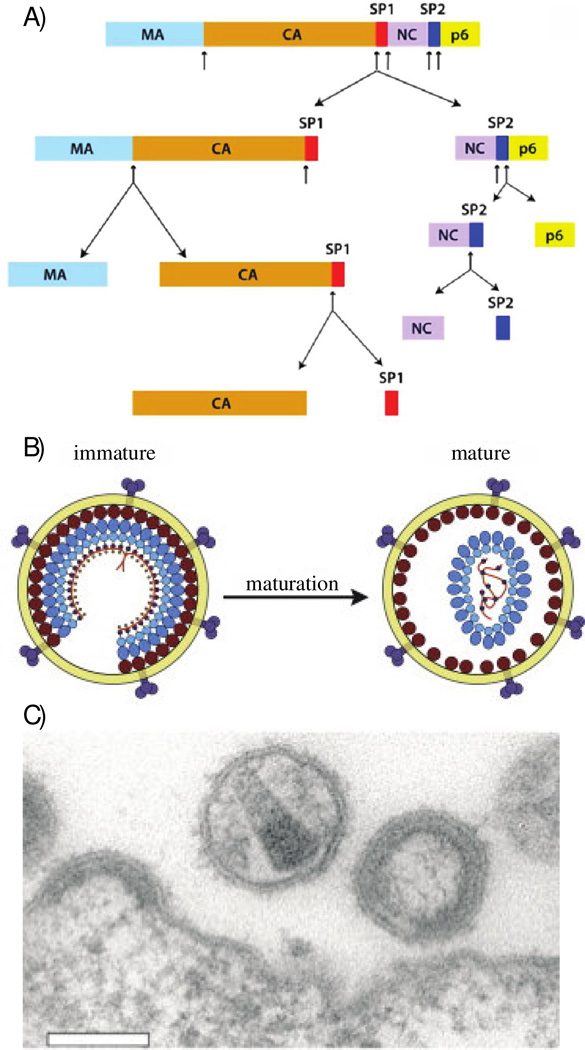
The three major genes comprising the HIV genome, gag, pol, and env, encode the main structural proteins (synthesized as polyproteins) and enzymes [4]. Six additional genes, tat, rev, nef, vpr, vif, and vpu, encode regulatory proteins (Tat and Rev) and auxillary proteins (Nef, Vpr, Vif, and Vpu). The HIV’s gag gene encodes the precursor Gag polyprotein, which contains the major structural proteins of the virus (Figure 1). These include matrix (MA), capsid (CA), spacer peptide 1 (SP1), nucleocapsid (NC), spacer peptide 2 (SP2), and P6 [5]. Immature, noninfectious viruses contain intact, uncleaved Gag polyprotein, see Figure 1. Proteolytic cleavage of the Gag polyprotein into its constituent protein domains results in an infectious virion through a process termed viral maturation [6]. Upon maturation, a characteristic capsid core is formed which encloses the viral RNA as well as complementary viral proteins, as shown in Figure 1 [7]. This Perspective article discusses on the recent structural and dynamics characterization of HIV-1 CA capsid protein assemblies, by magic angle spinning (MAS) NMR spectroscopy.
Figure 1.
A) Schematics of Gag polyprotein domain organization, including the cleavage pathway leading to viral maturation. Reprinted with permission from Han et al., J. Am. Chem. Soc., 2013, 135, 17793–17803. Copyright 2013 American Chemical Society. B) Artistic representation of structural differences between immature and mature HIV-1 virions. Reprinted with permission from Monroe et al., Structure, 2010, 18, 1483–1491 [111]. Copyright 2010 Cell Press. C) Electron micrograph image of HIV-1 virions. Both immature and mature virions are visualized here. Reprinted from open access V. Baumgartel et al., Viruses 4 2012 777–779 [112].
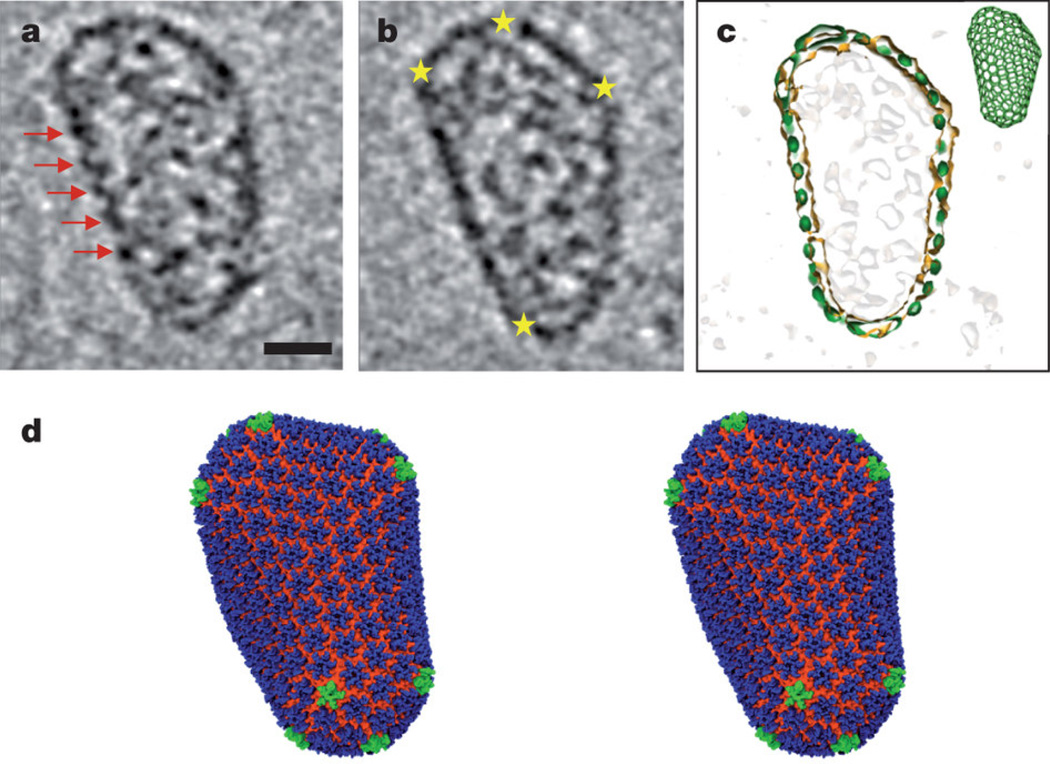
The molecular structure of mature HIV-1 capsid has been extensively studied at different levels of resolution using various methods, including X-ray crystallography [8–10], cryo-electron microscopy [7, 11–13], solution NMR spectroscopy [14, 15] and, most recently, MAS NMR spectroscopy [16–18] [19]. The HIV-1 capsid is assembled from 1200 copies of the 25.6 kDa CA protein, a cleavage product of the Gag polyprotein [7]. The capsid encloses two copies of viral RNA and a small complement of proteins for replication [20]. On the basis of the early cryo-EM and electron cryo-crystallography studies, an idealized “fullerene cone” model [11, 21] was developed to describe the structural organization of the mature capsid. In this model, 250 hexameric CA subunits are organized in a hexagonal lattice, with 12 pentameric CA subunits incorporated to close up the structure [7, 11, 21]. This model incorporated atomic-resolution NMR and X-ray structures of the individual N- and C-terminal CA domains (NTD and CTD, respectively), which were docked into the electron densities [8, 9, 14, 22–24]. Subsequently, high-resolution structures of capsid subunits were obtained by examining disulfide-stabilized CA hexamers and pentamers using X-ray crystallography, yielding more detailed structural information for the CA model [10, 13]. Most recently, a pseudoatomic capsid structure has been solved using a hybrid approach combining cryo-EM, solution NMR spectroscopy, and molecular dynamics simulations, as illustrated in Figure 2 [7].
Figure 2.
All-atom model of mature HIV-1 capsid. a, b. Representative slices of the three-dimensional cryo-ET data. Red arrows indicate arrays of CA hexamers, and yellow stars indicate locations of sharp curvature change. The scale bar is 20 nm. c. A overlay of the fullerene model (green) and the densities from the segmented capsid (orange). d. All-atom molecular dynamics equilibrated model of mature HIV-1 capsid comprising 216 CA hexamers (blue, NTD; orange, CTD) and 12 CA pentamers (green). Reprinted with permission from Zhao et al., Nature, 2013, 497, 643–646. Copyright 2013 Nature Publishing Group.
MAS NMR spectroscopy is an emerging technique for structural investigations of HIV protein assemblies. It is complementary to other structural biology techniques in that it is well suited for both structural and dynamic analysis of native-like protein assemblies, and possibly intact viral particles in the future, at atomic resolution. In contrast, solution NMR studies are limited to solubilized monomers or dimers of CA and other Gag maturation intermediates [25–28]. Higher order oligomers, such as cross-linked hexamers and pentamers (as well as smaller constructs) can be characterized with X-ray crystallography but the native-like assemblies are not amenable to X-ray analysis as they do not crystallize [9, 29, 30]. Cryo-electron microscopy (cryo-EM) is currently limited in resolution (~ 5Å), even though recent advances in instrumentation and data acquisition and analysis may push the resolution limits [31]. Furthermore, both X-ray crystallographic and cryo-EM microscopic structural analyses typically require non-physiological cryogenic temperatures, nor do they provide direct insights into internal dynamics of the HIV-1 protein assemblies, which have been found to be important for their biological function [7, 17, 25]. In contrast, MAS NMR experiments can be conducted at physiological or close to physiological temperatures [16–19, 25]. Using MAS NMR, many important features of CA and other HIV-1 proteins can be understood at atomic resolution, including the structure of the CA molecule in the assembled state, the interhexamer CTD-CTD contacts as well hexamer-forming NTD-NTD and CTD-NTD contacts. Tubular CA assemblies in particular appear to exhibit the same hexameric lattice and to mimic the curvature and intermolecular contacts as the native HIV-1 capsid core. Furthermore, MAS NMR experiments yield detailed insights into conformational dynamics on multiple timescales in HIV-1 protein assemblies, as we and others have demonstrated [17–19, 25]. Finally, interactions of HIV-1 protein assemblies with small molecules can be readily probed by MAS NMR, as has been demonstrated in other protein assemblies [32, 33].
Below, we discuss the recent methodological advances that have enabled atomic-resolution analysis of viral assemblies and provide a perspective on the current body of MAS NMR investigations into assemblies of HIV-1 CA protein and Gag maturation intermediate CA-SP1.
2. Sample Preparation and MAS NMR Methods for Structural Analysis of HIV-1 Protein Assemblies
2.1 Preparation of HIV-1 Protein Assemblies for MAS NMR
CA and CA-SP1 assembly conditions compatible with MAS NMR have been developed by us and by others [16–19, 25]. These conditions yield morphologically homogeneous assemblies of desired morphologies that exhibit high-resolution NMR spectra and are stable under magic angle spinning for extended periods of time [16–19]. Notably, the preparation of morphologically homogeneous assemblies typically requires high concentrations of NaCl, of the order of 1 – 2 M in the sample [17]. As we have demonstrated, the use of EFree or low-E probes enables multidimensional MAS NMR spectroscopy in these samples, with high-quality data readily attainable [17]. The typical 13C line widths in the HIV-1 CA and CA-SP1 assemblies of tubular morphologies are 40 – 100 Hz at the magnetic field strengths of 20.0–21.1 T, in fully protonated samples acquired at MAS frequencies of 14 – 20 kHz. It is important to corroborate the morphology of the samples prepared for the NMR experiments, and transmission electron microscopy (TEM) is used by us and by others for this purpose [16–19, 25].
It is interesting to note that there is some variability in the morphology of tubular assemblies of CA and CA-SP1 maturation intermediate, depending on the primary sequence and the preparation method. Tubular assemblies of CA and CA-SP1 have the same overall morphology, with diameters ~ 40–50 nm [17]. In another study, dark field transmission (tilted-beam) TEM has been used to determine the mass-per-length values of CA tubes, and indicates the presence of tubes formed from a single layer of CA, as well as multiwalled tubes [18]. Many studies utilize the A92E mutant of CA, which yields more homogeneous, single-walled tubes [15].
2.2 Through Space Correlations
Dipolar-based correlation experiments are extensively used in MAS NMR to obtain resonance assignments and distance restrains for protein structure determination. We have reviewed some recent methodological advances for through space correlation spectroscopy [34]. Here, we highlight methods that we found to be particularly useful in structural studies of HIV-1 CA protein assemblies.
Under moderate MAS conditions (10 – 30 kHz), conventional dipolar-based two-and three-dimensional DARR [35, 36], as well as NCA, NCO, NCACX, and NCOCX experiments [37] work efficiently for resonance assignments in HIV-1 CA protein assemblies, and yielded spectra with remarkably high resolution, including in samples containing 1 – 2 M NaCl [16, 17].
Under fast MAS conditions (40 – 110 kHz), the above experiments are no longer suitable, and our group has used the R2nv-symmetry based sequences [38–40] for 13C-13C correlation spectroscopy [41]. We have recently found that the combined supercycled R2nv-symmetry sequences, CORD, exhibit superior performance compared to the individual R2nv- blocks and deliver fully broadbanded excitation and high polarization transfer efficiencies at the MAS frequencies of 40 kHz [42]. It is worth nothing that R2nv and CORD experiments work well at moderate MAS frequencies, and the CORD sequence in particular yields strong aromatic correlations, which are typically weak or missing in the DARR and PDSD experiments [42].
2.3 Recoupling of Anisotropic Spin Interactions
Dipolar and CSA tensors contain valuable information on structure and dynamics. Their accurate site-specific measurement in macromolecules under MAS has been rather challenging, and multiple groups in the field have turned their attention to this problem, as discussed in recent reviews [34, 43, 44].
Our group has explored Levitt’s R-symmetry based sequences for the recoupling of heteronuclear (1H-13C/1H-15N) dipolar and CSA interactions in the context of 2D and 3D experiments, permitting site-resolved recording of dipolar [45] or CSA [46, 47] lineshapes in an indirect dimension. These RNnv-symmetry schemes work at both moderate and fast MAS frequencies. Since RNnv-symmetry sequences can efficiently suppress the homonuclear dipolar couplings and are selective for the first-order heteronuclear dipolar couplings, they are suitable for examining fully protonated samples.
We have employed RNnv-symmetry based dipolar recoupling experiments to characterize the backbone dynamics of HIV-1 CA protein assemblies using U-13C, 15N-Tyr enriched samples, and discovered that the hinge region between the N-terminal domain and C-terminal domain is rigid on the nano- to microsecond timescales and flexible on the millisecond timescales [25]. As an improvement to the dipolar recoupling experiments based on the individual RNnv-symmetry sequences, where 1H CSA persists and causes non-negligible errors for dipolar coupling measurements, we have recently reported Phase-Alternating R-Symmetry (PARS) sequences, which yield accurate heteronuclear dipolar couplings, due to the efficient suppression of 1H CSA [48].
Chemical shift anisotropy is a sensitive probe of structure, hydrogen bonding, and dynamics of proteins assemblies. We have established that γ-encoded RNnv-symmetry based CSA (RNCSA) sequences are very well suited for site-specific measurement of 13C and 15N CSA parameters in both U-13C,15N enriched and natural abundance 13C systems, in the context of 2D and 3D experiments. We have demonstrated that these experiments can be conducted under a broad range of MAS frequencies, including fast MAS [47], which is a significant improvement to the prior methods [49]. The development of these RNCSA sequences permitted us to examine backbone dynamics of CA in U-13C,15N-Tyr enriched HIV-1 CA protein assemblies [25, 47], as described in section 3 of this Perspective article.
The measurement of 1H CSA tensors has been challenging and, until recently, restricted to either solid systems with sparse proton sites or based on the indirect determination of anisotropic tensor components from solution cross-relaxation and liquid crystal NMR experiments [50, 51]. To overcome the technical challenges associated with the determination of accurate proton CSA tensors, our group has developed an MAS approach that permits site-resolved measurement of 1H CSA tensors in fully protonated systems with multiple sites [50, 51]. In this method, three 3D RN-symmetry based experiments are recorded, which are dominated by mainly 1H-15N dipolar, mainly 1H CSA, or combined 1H-15N dipolar and 1H CSA interactions. The 1H CSA tensors are extracted by a simultaneous triple fit of the data [51]. This approach yields accurate proton CSA tensors. 1H CSA, together with 15N and 13C CSA is expected to be a very useful reporter on local structure and dynamics of the proton sites, such as electrostatic interactions between the corresponding proton and neighboring molecules at the interfaces (being correlated with the length of hydrogen bonding) as well as structural disorder and dynamic flexibility [50–55]. 13C CSA tensors have been already used as restraints to improve protein structure quality determined by MAS NMR, and similar efforts are ongoing in our laboratory to utilize 1H CSA as a restraint in structure calculation. This approach is well suited for structural analysis of a broad range of biological systems, including HIV-1 protein assemblies.
2.4 Recent Advances for Enhancements of Sensitivity, Resolution, Data Collection and Analysis
Sensitivity and resolution remain major challenges in MAS NMR, particularly in analysis of large macromolecular assemblies, such as HIV-1 virus assemblies. High magnetic fields (17.6 T and higher) in conjunction with fast MAS probes capable of spinning at frequencies of 40 – 110 kHz have pushed the sensitivity and resolution boundaries permitting studies of large systems with small sample quantities, as we have reviewed recently [34, 56–63]. Proton detection gives rise to considerable sensitivity enhancements in deuterated [64–67] and fully protonated [62, 63, 68] macromolecules and appears to be a promising venue for the studies of HIV-1 based assemblies. A combination of fast MAS methods and the use of paramagnetic dopants for reducing the T1 relaxation time is another exciting approach for speeding up the data acquisition, and could be potentially useful in a wide range of systems [60, 69].
Dynamic nuclear polarization (DNP) is another highly active area of research, with large DNP-based enhancements making it very promising for analysis of viral assemblies [70, 71]. We note that DNP applications to structural analysis of proteins and protein assemblies are still at early stages of development, and extensive work is needed to establish experimental conditions that would permit rapid freezing of the samples to attain conformational homogeneity and prevent loss of resolution, as is typically observed in the current setup.
Alternative approaches to data collection have also been explored, including nonuniform sampling, where, in the regime of exponentially biased random sampling [72] schemes, time-domain sensitivity enhancements have been theoretically predicted and experimentally corroborated [73–75]. Other NUS approaches have been applied in MAS NMR as well, albeit, the sensitivity enhancements have been attained at the expense of resolution [76, 77]. Finally, computational protocols for streamlining and automation of resonance and distance restraint assignments are also exciting and active areas of research [78–84]. In the following sections, we highlight the recent progress in the above areas, not including DNP based research, which is beyond the scope of this article.
2.4.1 Proton detection at fast-MAS and high fields
Until recently, proton detection was not common in MAS NMR of fully protonated biological solids. The advent of fast MAS probes has enabled proton detection at MAS frequencies of 40 kHz and above. Zhou et al., Huber et al., Knight et al., Holland et al., and Asami et al. reported large sensitivity enhancements for 1H-detected experiments over their 13C or 15N detected counterparts, at MAS frequencies of 40 – 60 kHz [62, 63, 85–88]. It was demonstrated that the performance of proton-detected sequences increases sharply with the magnetic field strength, and at 23.5 T (the highest field strength attainable today), outstanding resolution and sensitivity have been observed in fully protonated samples [68]. Recently, Barbet-Massin et al. have established a set of 1H-detected triple-resonance experiments for facile resonance assignments in proteins [89]. This approach, consisting of six 3D experiments, is based on protocols employed in solution NMR. With this set of experiments and the use of an automatic resonance assignment protocol the authors were able to establish significant portions of assignments in five different proteins, with minimal manual input [89].
2.4.2 Nonuniform Sampling and Nonuniform Consecutive Data Acquisition Approaches
The use of nonuniform sampling (NUS) for solid-state NMR is attracting attention [69, 73, 75, 90–92]. In solution, NUS is typically applied to speed up the data collection, and this mode has also been explored in the solids NMR experiments. For example, in a recent report Xiang et al. demonstrated that a set of four-dimensional experiments, (H)CACO(CA)NH and (H)COCA(CO)NH, could be used to collect data with appropriate sensitivity and resolution in a reasonable amount of time using NUS. With 20% nonuniform sampling coverage, the combined experiment time could be reduced from 55 days to 11 days [92]. Another study by Lin and Opella demonstrated that with NUS coverage of 25–33%, two- and three-dimensional datasets could be collected and reconstructed using compressed sensing processing, thus saving experiment time [90].
In biosolids, many of the systems of interest, including HIV-1 protein assemblies, possess inherently low sensitivity. In our laboratory, we have explored NUS for attaining time-domain sensitivity enhancements without compromising the spectral resolution. Rovnyak established theoretically that random, exponentially biased sampling schedules can yield bona fide time-domain sensitivity enhancements on the order of two-fold in each dimension provided exponentially decaying signals (non-constant-time mode), without compromising on spectral line widths [74]. We have demonstrated experimentally that these predictions are valid, by performing various hetero- and homonuclear 2D and 3D MAS NMR experiments on different samples, including peptides, a U-15N,13C-LC8 protein, and a reassembled 1-73-(U-13C, 15N)/74-108-(U-15N) E. coli thioredoxin [69, 73, 75]. This approach is particularly advantageous for challenging samples possessing inherently low sensitivity, where with the appropriate design of NUS sampling schedules one can increase the number of scans, attaining significant sensitivity enhancements, which are compounded in each indirect dimension, without sacrificing the resolution [75].
Traditional FFT protocols are not suitable for processing data collected with nonuniform sampling [93]. Several reconstruction protocols have been developed for NUS data processing, including: maximum entropy (MaxEnt), covariance transforms, G-matrix Fourier transform (GFT), spectroscopy by integration of frequency and time domain information (SIFT), multidimensional decomposition (MDD), and a variant of MaxEnt termed maximum entropy interpolation (MINT) [73, 90, 91, 93–96]. The choice of an alternative and appropriate reconstruction algorithm can be dictated by several considerations, such as whether the data were sampled on- vs. off-grid, the amount of computational time required for reconstruction, and the desired linearity of the reconstructed data.
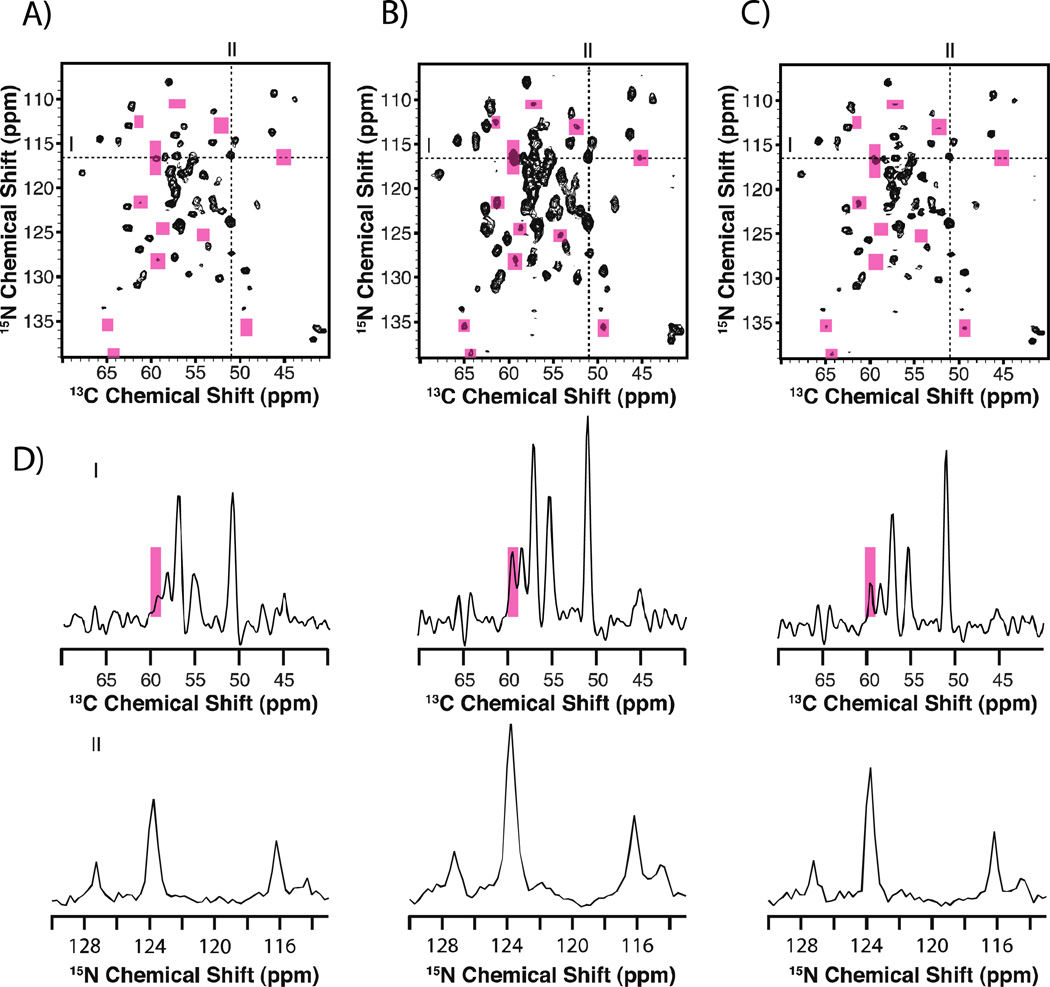
In our laboratory, we have adopted the use of maximum entropy interpolation (MINT) reconstruction that results in a highly linear transformation between the time-and frequency-domain [73]. This is accomplished by using entropy maximization to estimate the values of missing data samples, while tightly fitting the reconstructed spectra to match the experimentally measured data points. Under MINT conditions, high-fidelity spectral reconstructions are achieved for NUS data. Importantly, the linear behavior of MINT has been shown to extend to dynamic ranges of ~245x for 13C-13C homonuclear correlation experiments on model compounds and reassembled 1-73-(U-13C, 15N)/74-108-(U-15N) E. coli thioredoxin [75]. Figure 3 demonstrates the sensitivity gains and linearity attained with the NUS/MINT approach, as demonstrated on 1-73-(U-13C, 15N)/74-108-(U-15N) E. coli thioredoxin.
Figure 3.
Comparison of 2D NCA spectra for reassembled thioredoxin collected with US (A) and NUS (B and C). Panel A displays data collected with uniform sampling and processed with MINT. Panel B was collected with NUS and processed with MINT. Panel C shows the same dataset as panel B but processed with line width deconvolution, which improves resolution. The data are displayed at the same contour level. Pink boxes are used to highlight areas where new peaks are present in the NUS spectra. In D), line shapes extracted through the direct and indirect dimensions are displayed and show intensity enhancements for NUS data. Reprinted with permission from Paramasivam et al., J. Phys. Chem. B., 2012, 116, 7416–7427. Copyright American Chemical Society 2012
An alternative approach to nonuniform sampling is the use of nonuniform consecutive acquisition schemes (NUCA). NUCA, unlike NUS where data points are sampled nonlinearly, rely on sampling each point equidistantly but varying the number of scans per increment in the indirect dimensions. The collection of evenly spaced data points using the NUCA approach, allows for the use of FFT for data processing.
Qiang demonstrated the NUCA approach as applied to an Aβ fibril peptide [97]. In these 2D 13C-13C correlation experiments, the number of acquisitions per t1 point was reduced as a function of the evolution time. Different acquisition profiles including linear and Gaussian were created and used for data collection and comparison. This approach yielded enhancements on the order of 40–50% but also introduced spectral line broadening. Li et al., recently demonstrated a similar approach where NUCA was combined with covariance spectroscopy allowing for the total experiment time to be reduced by a factor of 3–6 without the loss of resolution or signal-to-noise ratio [98]. These 13C-13C experiments were performed on histidine, microcrystalline GB1, and lipid reconstituted proteorhodopsin.
2.4.3 Automated Resonance and Distance Restraint Assignment Techniques for MAS NMR
Resonance assignments is the cornerstone yet the most tedious and time-consuming part of the macromolecular structure determination protocol. In large biological systems like HIV-1 protein assemblies these challenges are amplified. For these cases, alternative resonance assignment protocols, which rely on computational algorithms, may need to be applied. Once the NMR spectra are automatically assigned, they can be reviewed and confirmed manually. In addition to automated resonance assignment, programs designed to assign distance restraints are also valuable. Some of these automated protocols, specifically designed for or applicable to MAS NMR data, are described below.
Tycko and colleagues described a Monte Carlo/simulated annealing algorithm that allowed for resonance assignments to be obtained computationally [83]. This approach requires the use of several 2D and/or 3D MAS NMR spectra [79]. The input for the original and the expanded program includes a list of 15N and 13C cross peaks, which are created through manual inspection of the two-dimensional spectra as well as the primary amino acid sequence. The list of cross peaks contains the chemical shifts, uncertainties in shift, degeneracies, and possible residue type, where multiple possibilities are allowed. The program then attempts to assign individual peaks to a given residue and determines a score. This approach was tested and demonstrated to work on several proteins, including α-spectrin’s SH3 domain, microcystalline GB1, and HET-s (218–289) [79]. In a later study by Bayro et al., this approach was used to assign tubular HIV-1 capsid assemblies. 50 independent runs were performed yielding 159 of 231 residues with 100% consistency [19].
An alternative recent approach by Schmidt and coworkers, only requires an input of the protein sequence and unassigned peak lists [81]. The program, ssFLYA, uses an algorithm based on the knowledge of through-bond or through-space polarization transfers and chemical shift statistics to generate expected peaks based on a set of rules for each spectrum. These assignment solutions are then scored and optimized based on their similarity to chemical shift statistics, peak alignment, completeness of assignment, and chemical shift degeneracy. The validity of this approach was tested using multiple runs on several protein systems. Compared to manual assignment, ssFLYA resulted in 77–90% agreement based on analysis from the individual runs.
Recently Nielsen et al., presented a different method termed GAMES_ASSIGN [80]. Similar to ssFLYA, this approach utilizes an input of only the unassigned peak list and the primary protein sequence. This approach begins by pairing peaks based on alignment to determine peak clusters and spin systems. These spin systems are linked with regions in the primary amino acid sequence through an iterative process. Finally, by combining the previous information spin systems are rebuilt, missing atoms are assigned, and resonance assignments are made. Using multiple independent runs can yield different results and a statistical approach can be used to determine the results. To validate the approach, three protein systems were chosen with different sizes and data quality. For GB1, ubiquitin, and CsmA, 91.8%, 74.2%, and 83.1% of assignments, respectively, were correctly made for all atoms.
Fossi and colleagues introduced the program SOLARIA for automatically assigning cross peaks arising from long-range 13C-13C and 15N-13C correlations [78]. These spectra are used to measure distance restraints that are critical for determining the three-dimensional structures of proteins. It is often the case that spectral overlap between signals leads to multiple assignment options per peak making manual assignments difficult and requiring the exclusion of ambiguous information that could be valuable. Peak lists derived by manual inspection of 2D 13C-13C and 3D 15N-13C-13C spectra, for α-spectrin’s SH3 domain, were used as input for SOLARIA. The program used these peak lists to perform automatic assignments of the distance restraints and structures were calculated in only 12 hours. Importantly, the automatic protocol allowed for the use of additional restraints, which were not previously assigned by manual inspection. The eleven lowest energy structures displayed an RMSD of 1.3 Å when compared to the X-ray reference.
Overall, automated resonance assignment and restraint protocols are anticipated to be an integral step in working with viral assemblies and other large biomolecular systems in the future.
3. HIV-1 CA and Gag Maturation Intermediate Protein Assemblies: Structural and Dynamics Studies by MAS NMR
To date, two laboratories have reported MAS NMR studies of HIV-1 capsid protein assemblies: our group at the University of Delaware and the Pittsburgh Center of HIV Protein Interactions (PCHPI), and the group of Dr. Robert Tycko at NIH. Investigations of assemblies of HIV-1 proteins are challenging because of the large size and the complex symmetry of the system, as well as the presence of high concentrations of NaCl typically required for the preparation of morphologically homogeneous samples. At the same time, CA and Gag maturation intermediates assemble into uniquely homogeneous and stable structures, yielding outstanding-resolution MAS NMR spectra, from which detailed atomic-resolution information on structure and dynamics of the assembled proteins can be inferred. As discussed below, the current body of parallel independent work from these two laboratories has revealed so far a consistent picture of the salient structural and dynamics features of CA in the assembled state, establishing MAS NMR spectroscopy as a reliable and information-rich method for the analysis of HIV-1 protein assemblies.
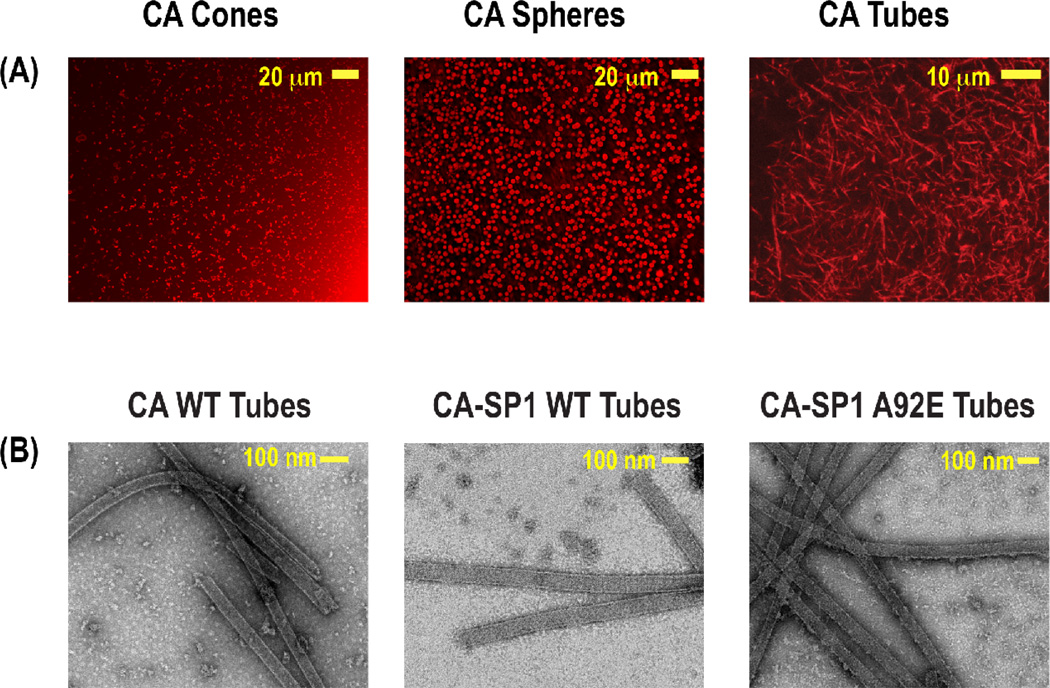
In our initial MAS NMR studies of the HIV-1 protein assemblies, we investigated conical and spherical assemblies of the 25.6 kDa CA capsid protein [16]. During this early work, we have established protocols for generating CA assemblies of different desired morphologies under conditions compatible with MAS NMR. Subsequently, we have established conditions for reliable preparation of tubular assemblies of various CA constructs, as well as of the corresponding CA-SP1 maturation intermediates of Gag polyprotein, see Figure 4 [17]. 13C-13C DARR spectra of U-13C,15N-labeled spherical and conical assemblies of CA exhibited relatively high resolution, and the analysis of chemical shifts indicated that the protein has the same secondary and tertiary structure in these assemblies possessing different morphologies. We were able to assign 63% of residues in CA on the basis of NCACB, NCACX, NCOCX, and CCC (DARR-DARR) 2D and 3D MAS NMR spectra of the conical assemblies of full-length CA and the solution NMR datasets of the CA’s individual CTD and NTD domains. Importantly, the high quality data that we obtained during this early work demonstrated that these HIV-1 protein assemblies are amenable to detailed characterization at atomic resolution by MAS NMR.
Figure 4.
A) Confocal images showing conical, spherical, and tubular morphologies of HIV-1 CA assemblies. B) TEM images of tubular assemblies of CA HXB2, CA-SP1 WT HXB2, and CA-SP1 NL4-3 A92E constructs. Reprinted with permission from Han et al., J. Am. Chem. Soc., 2010, 132, 1976–1987 and Han et al., J. Am. Chem. Soc., 2013, 135, 17793–17803. Copyright 2010 and 2013 American Chemical Society.
Structural plasticity of HIV-1 CA is a necessary prerequisite for the assembly of CA into pleiomorphic capsid cones [20, 99, 100]. Interestingly, structural polymorphism of the capsid core has been observed both in vivo and in vitro. The CA’s CTD forms intermolecular contacts critical for the assembly of the mature viral core. EM, X-ray, and solution NMR structures of the CTD dimerization domain indicated that this region can adopt a range of conformations [8, 10, 15, 24]. To examine the origin of conformational plasticity and establish the molecular mechanism of the CA assembly, we have addressed the conformational dynamics of CA by an integrated MAS and solution NMR approach. Using U-13C,15N-Tyr labeled protein, we performed a series of MAS NMR studies, including 13Cα-15N and 1H-15N dipolar coupling constant measurements, 15N CSA measurements, and 15N relaxation measurements to probe dynamics on the timescales spanning milliseconds through nanoseconds [25]. The samples used in these studies included conical assemblies of full length CA and CTD constructs of 2 different lengths containing various isotopic labels.
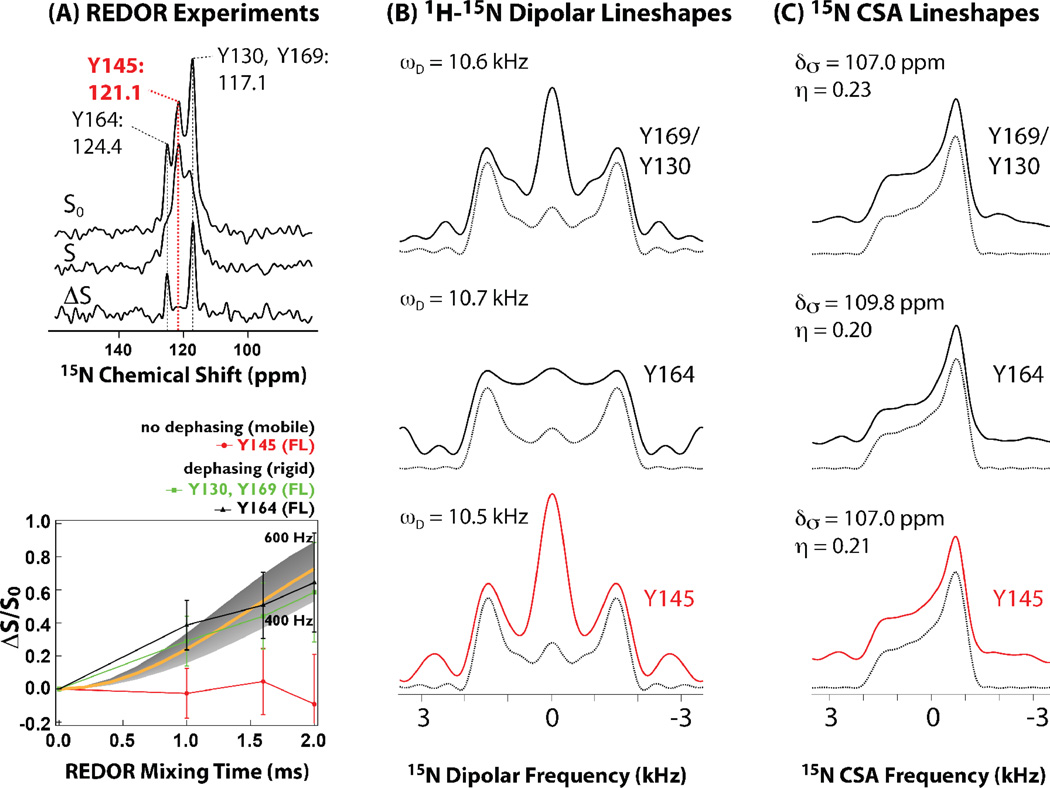
CA has four tyrosine residues. Y145 is in the hinge region between the CTD and NTD and has been shown to be critical for capsid assembly and viral infectivity. Y130 is located in the NTD while Y164 and Y169 are in the CTD. On the basis of multiple homo- and heteronuclear correlation spectra as well as 15N{13C} REDOR dephasing experiments in full-length assembled CA and the two CTD constructs we concluded that Y145 undergoes conformational dynamics on the timescales of ~ 10 ms, in contrast to the other tyrosine residues, which are rigid [25], as summarized in Figure 5. Interestingly, Y145 does not undergo motions on faster, micro- to nanosecond, timescales, as evidenced by their rigid-limit 1H-15N and 1H-13C dipolar lineshapes recorded by us in a series of RN-symmetry based experiments [25]. These MAS NMR data, in conjunction with solution NMR results led us to propose a molecular switch mechanism that appears to determine the ability of the CA protein to assemble into the conical capsids. The millisecond time scale motions of hinge region residues of CA open up the available conformational space. At the same time, the actual number of conformers accessible for the CA assembly into varied morphologies is finite, and their relative populations in solution appear associated with an intermolecular electrostatic interaction. This combined dynamic and electrostatic control appears to permit the formation of varied capsid morphologies in mature HIV virions.
Figure 5.
A) 15N{13C} REDOR spectra (top) and dephasing curves (bottom) for conical assemblies of full-length HIV-1 CA protein. The lack of dipolar dephasing for Y145 in all constructs indicates millisecond timescale dynamics, which average the 15N-13C dipolar coupling. B) 1H-15N dipolar experimental and best-fit line shapes for U-13C,15N-Tyr labeled full-length HIV-1 CA protein assemblies of conical morphology. Dipolar coupling constants that were extracted by numerical simulations show that each Tyr residue is rigid on the time scale measured experimentally. C) 15N CSA line shapes for U-13C,15N-Tyr labeled full-length HIV-1 CA protein assemblies of conical morphology. Experimental and best-fit line shapes for each residue are shown. Each Tyr residue exhibits rigidity on the time scale measured experimentally. Adapted with permission from Byeon et al., J. Am. Chem. Soc., 2012, 134, 6455–6466. Copyright 2012 American Chemical Society.
In parallel, we turned our attention to the investigations of the mechanism of HIV-1 maturation. The final step in the formation of the mature HIV-1 capsid is the cleavage of the SP1 peptide from the CA-SP1 maturation intermediate. This maturation step is of particular interest due to the recent discovery of antiretrovirals, such as Bevirimat [101], which abolish the SP1 peptide cleavage by binding to CA-SP1 [102]. One open question in the field of HIV research is what triggers SP1 cleavage and initiates the conical capsid assembly. One hypothesis proposes the change in CA lattice shape from spherical to conical upon the cleavage of the SP1 peptide [103]. The second one postulates that during the maturation, the immature lattice disassembles completely during the course of proteolytic cleavages of Gag followed by de novo reassembly of the mature core [5]; this hypothesis has been recently corroborated by several techniques, including cryoelectron tomography (cryo-ET) [104]. In the context of the maturation mechanism, the SP1 cleavage is thought to mediate disassembly of the immature CA during the final maturation step [105], and, therefore, its conformation in the various maturation intermediates has been a subject of interest and debate in the field. The SP1 conformation in the assembled HIV-1 CA-SP1 has remained unknown until our recent work. Another interesting question is how the primary CA sequence variability affects the conformation of assembled CA and CA-SP1 and the viral infectivity.
To address the above aspects of the HIV-1 maturation mechanism, we have examined assemblies of CA and CA-SP1 from two common viral strains, HXB2 and NL4-3. These two strains differ in sequence by 5 residues. The NL4-3 construct also contained an A92E mutation in the Cyclophilin A (CypA) loop. This A92E “escape” mutant possesses 10% of the viral infectivity of the wild type HIV-1 [106, 107]. The tubular assemblies of these four proteins yielded outstanding-quality MAS NMR data (Figures 6 and 7). From the multiple dipolar- and scalar-based homo- and heteronuclear correlation spectra we have unequivocally determined the conformation of SP1 in the assembled state of CA-SP1 and gained information on conformational and dynamic differences of the CA and CA-SP1 assemblies derived from the two HIV-1 strains, HXB2 and NL4-3 A92E. The salient conclusions of this study are discussed below.
Figure 6.
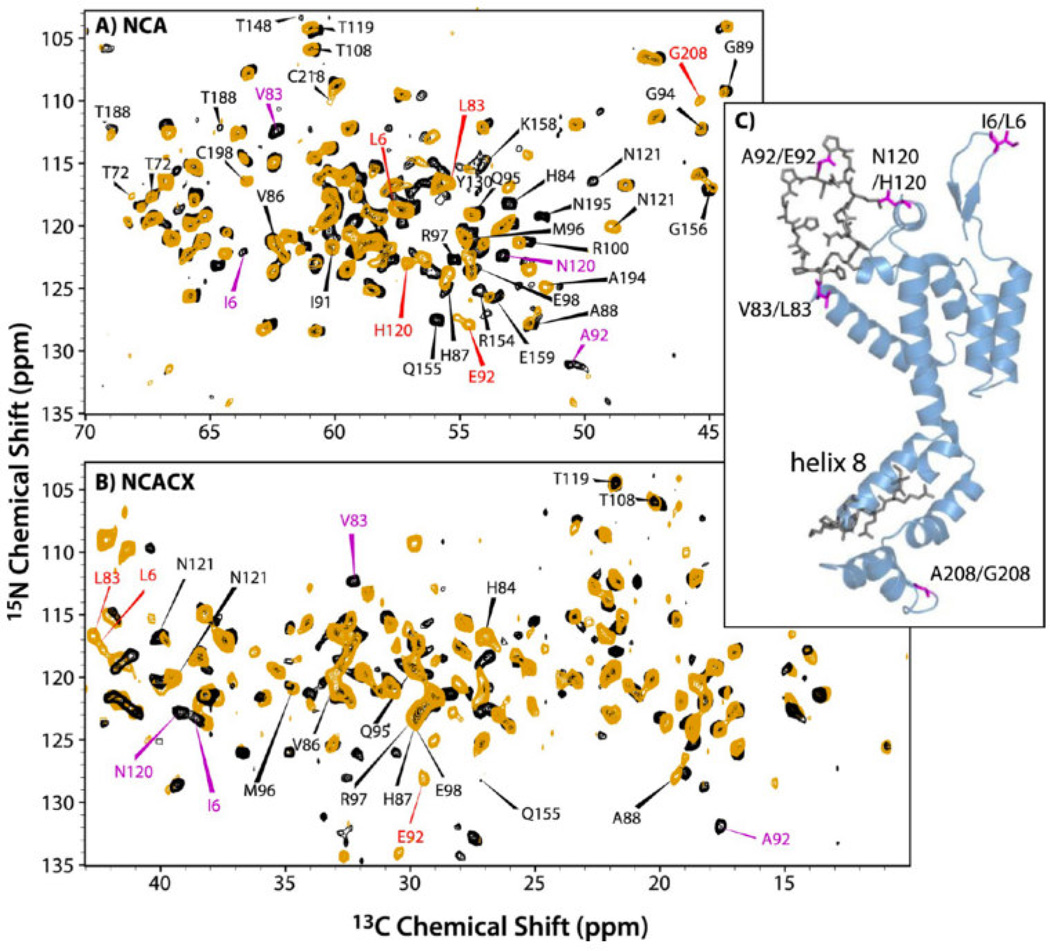
Overlay of (A) NCA and (B) NCACX spectra of CA NL4-3 A92E (orange) and CA HXB-2 (black). The NCACX spectrum is an expansion of the aliphatic sidechain region. Indicated with red and magenta arrows are distinct resonances observed for HXB2 and NL4-3, respectively, indicating differences in conformation or electronic environment. (C) A view of the 3D structure of CA (HXB2 variant, PDB file 3NTE). Residues highlighted in magenta vary between the two constructs. Residues shown in gray are conserved in the two strains, but exhibit chemical shift differences. Reprinted with permission from Han et al., J. Am. Chem. Soc., 2013, 17793–17803. Copyright 2013 American Chemical Society.
Figure 7.
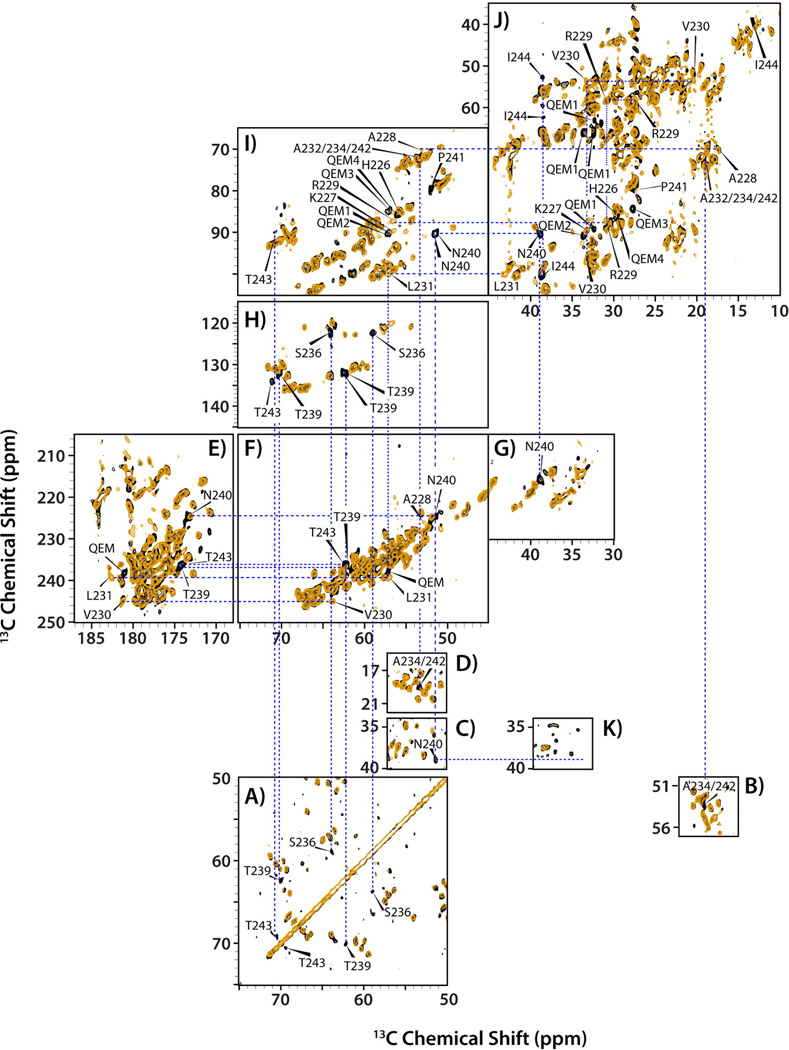
Overlay of spectra for tubular assemblies of CA NL4-3 A92E (orange) and CA-SP1 NL4-3 A92E (black). (A–D) Direct-DARR, (E–J) INADEQUATE, and (K) CP-DARR, acquired at 19.9 T and 4 °C. Labeled peaks indicate new resonances that appear in the CA-SP1 spectrum, but not the CA spectrum. The CP-DARR spectrum in (K) demonstrates the absence of signal from N240 of the SP1 peptide due to dynamics, while this resonance is present in the direct-DARR (C) and INADAQUATE (I) spectra. Reprinted with permission from Han et al., J. Am. Chem. Soc., 2013, 135, 17793–17803. Copyright 2013 American Chemical Society.
We discovered that the 6-residue difference in the primary sequence of HXB2 vs. NL4-3 A92E induced multiple conformational perturbations mapped to the CypA loop region (NTD) and to the loop preceding helix 8 (CTD), see Figure 6. The results indicate that the SP1 tail adopts a dynamic random coil, and the conformation of the peptide is similar in both constructs. Interestingly, the tail is more dynamic in the HXB2 CA-SP1. Furthermore, the presence of the SP1 tail enhances the mobility of several preceding residues and, possibly, of CypA loop as well. (These findings have been corroborated by our most recent dynamics studies using heteronuclear dipolar lineshapes, which will be reported in a forthcoming manuscript.) Thus, conformational plasticity in functionally important loop residues of CA and CA-SP1 is induced by primary sequence. Our results support the hypothesis that SP1 cleavage induces the disassembly of immature CA and subsequent re-assembly into the mature capsid core.
In parallel, Tycko and co-workers have also performed MAS NMR studies of CA tubular assemblies. In their early work, they have concluded on the basis of the various MAS NMR spectra of 1,3-13C-glycerol-labeled CA samples that CTD and NTD of CA are largely rigid and retain their general secondary and tertiary structure upon formation of tubes [18]. Using double quantum filtering for further spectral simplification, they were able to assign 111 cross peaks by comparison with solution NMR chemical shifts. INEPT and NOE-INEPT measurements also indicated that very few CA residues are mobile.
In the recent work from the same group, Bayro et al. performed de novo resonance assignments from solid-state shifts using the combination of 3D NCACX, and NCOCX spectra of U-13C,15N CA tubes; NCACX, NCOCX, and CONCA spectra of 1,3-13C glycerol/U-15N CA; and a CONCA spectrum of 2-13C glycerol/U-15N CA. Using this protocol, site-specific assignments were derived for 69% of the protein without comparison to solution NMR assignments [19]. Manual resonance assignment was supplemented by application of the automated assignment program MCASSIGN2 discussed in section 2.4.3 of this article.
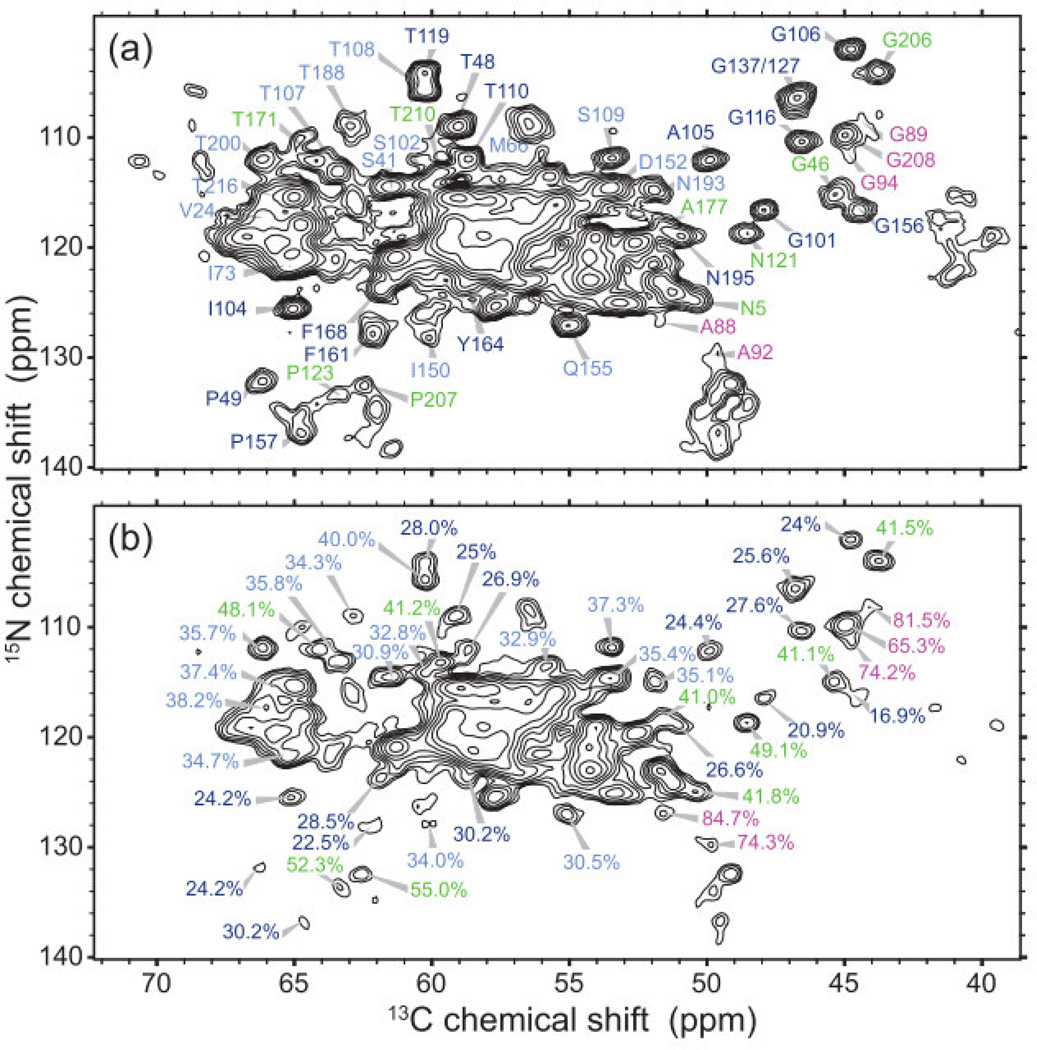
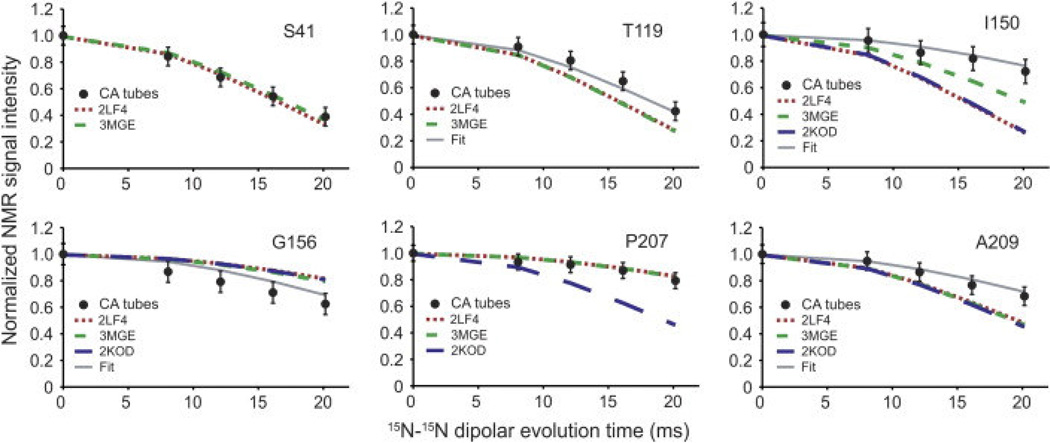
Another important conclusion of the above study is that while much of the CA protein is rigid and ordered in tubular assemblies, specific regions including the CypA loop, the N-terminal β hairpin, the hinge between NTD and CTD, as well as NTD-CTD intermolecular interaction sites, are disordered, indicated by the absence of resonances in spectra (Figure 8). 13C-13C TOBSY experiments were utilized to distinguish dynamically disordered regions from statically disordered regions and indicate that the CTD-NTD linker, as well as the segment between helices 8 and 9, is dynamically disordered. 1H T2-filtered NCA spectra demonstrated that different interhelical segments had different mobility, as indicated by greater loss in peak volume for rigid residues than partially mobile residues. The greatest mobility was observed in the CypA binding loop, consistent with our own results. 15N-15N backbone dipolar recoupling (15N-BARE) experiments were utilized for the measurement of the torsion angles ψi and ψi-1 (and φi to an extent). While most α-helical residues exhibited rapid dephasing due to relatively short interhelical distances, some residues displayed slower dephasing times, indicative of extended backbone conformations, illustrated in Figure 9. Using TALOS+ torsion angle predictions [108] and the 15N-BARE data as restraints [109], structure calculations of CA were run with Xplor-NIH [110] and revealed that the 310-helix near the CTD-NTD linker is in a more extended conformation than indicated in solution NMR studies, and also distinguished differences in the loops between H3 and H4 and H10 and H11, which may be caused by different intermolecular contacts than observed for dimers/monomers in solution or hexameric crystallography constructs. The findings by Bayro et al. suggest a curvature generation mechanism in this system: the curvature appears to be determined by the variations of the structure around the inter-domain linker region, the segments involved in intermolecular NTD-CTD interactions, and the C-terminal tail.
Figure 8.
Indications of mobility in tubular assemblies of CA. (A) Reference NCA spectrum with no T2 filter. (B) 1H T2-filtered NCA spectrum. Percentages represent the remaining peak volume as compared to the reference spectrum. Sites with a greater remaining peak volume are more dynamic, highlighted in green and magenta. Reprinted with permission from Bayro et al., J. Mol. Biol., 2014, 426, 1109–1127. Copyright 2014 Elsevier.
Figure 9.
15N-BARE decay curves demonstrated for 6 residues in tubular assemblies of CA. Experimental decay curves are indicated by filled circles. Broken and dotted lines are simulations based on 3 different PDB structures of CA monomers (green dashed line: 3MGE, red dotted line: 2LF4, blue dashed line: 2KOD). “Fit” indicates simulated curves based on the refined CA structure using the 15N-BARE data as a structural restraint. Reprinted with permission from Bayro et al., J. Mol. Biol., 2014, 426, 1109–1127. Copyright 2014 Elsevier.
It is worth noting that the above studies have paved the road for the analysis of other HIV-1 assemblies. With the current state of MAS NMR methodologies larger assemblies of HIV-1 proteins are becoming amenable to full 3D structural and dynamics characterization at atomic resolution. These investigations are ongoing in our laboratory.
4. Conclusions
The emergence of MAS NMR spectroscopy as an influential method to understand salient structural and dynamic features of HIV-1 assemblies is an important development in the field of HIV-1 structural biology. MAS NMR provides a wealth of information on the HIV-1 proteins in their assembled state and under conditions similar to those found in vivo, at atomic resolution. High-field magnets deliver the sensitivity and resolution necessary to work on these systems, which were considered intractable only several years ago. Fast MAS probe technologies have enabled proton detection in fully protonated samples, providing additional boosts in sensitivity and resolution and requiring several-fold less sample than used in the conventional probes. The developments of fast-MAS probes have also resulted in new efficient experiments for multidimensional correlation spectroscopy and for recording of various anisotropic NMR parameters that are key to structure and dynamics studies. EFree and low-E probes permitted spectroscopy of HIV-1 assemblies containing high salt concentration, opening doors to their detailed analysis. The use of nonuniform sampling and nonuniform consecutive acquisition methods allow for more efficient data collection and/or for bona fide time-domain sensitivity enhancements all of which is essential for the study of large HIV-1 and other viral assemblies. The ever increasing complexity of biomolecular systems has also necessitated the development of automated resonance and distance restraint assignment protocols, some of which have already been applied to the studies of HIV-1 protein assemblies. It is clearly an exciting time to be working in the field of MAS NMR spectroscopy. In our opinion, the potential for our discipline to make unique contributions to understanding structural biology of HIV-1 and other viral assemblies and even more complex systems is very high.
Highlights.
MAS NMR can yield unique atomic-level details about HIV protein assemblies.
Contemporary MAS methods enabling analysis of HIV protein assemblies are discussed.
Perspective is provided on recent MAS NMR studies of HIV-1 protein assemblies.
Acknowledgments
This work was supported by the National Institutes of Health (NIGMS Grant P50GM082251) and is a contribution from the Pittsburgh Center for HIV Protein Interactions. We acknowledge the support of the National Science Foundation (NSF Grant CHE0959496) and of the University of Delaware for the acquisition of the 850 MHz NMR spectrometer at the University of Delaware. We acknowledge the support of the core instrumentation facilities by the NIH-COBRE programs (5P30GM103519 and 5P20RR017716).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Christopher L. Suiter, Email: csuiter@udel.edu.
Caitlin Quinn, Email: cmquinn@udel.edu.
Manman Lu, Email: lumm@udel.edu.
Guangjin Hou, Email: hou@udel.edu.
Huilan Zhang, Email: zhang@udel.edu.
Tatyana Polenova, Email: tpolenov@udel.edu.
References
- 1.Barresinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axlerblin C, Vezinetbrun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune-deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo RC, Sarin PS, Gelmann EP, Robertguroff M, Richardson E, Kalyanaraman VS, Mann D, Sidhu GD, Stahl RE, Zollapazner S, Leibowitch J, Popovic M. Isolation of human T-cell leukemia-virus in acquired immune-deficiency syndrome (AIDS) Science. 1983;220:865–867. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- 3.UNAIDS report on the global AIDS epidemic. 2013:1–106. [Google Scholar]
- 4.Mushahwar IK. Human immunodeficiency viruses: molecular virology, pathogenesis, diagnosis and treatment. Persp. Med. V. 2007;13:75–87. [Google Scholar]
- 5.Briggs JAG, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 6.Freed EO. HIV-1 Gag proteins: Diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 7.Zhao GP, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning JY, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang PJ. Mature HIV-1 capsid structure by cryoelectron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamble TR, Yoo SH, Vajdos FF, vonSchwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 9.Worthylake DK, Wang H, Yoo SH, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 angstrom resolution. Acta Crystallogr. D. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 10.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua YZ, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 12.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469 doi: 10.1038/nature09640. 424-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 15.Byeon IJL, Meng X, Jung JW, Zhao GP, Yang RF, Ahn JW, Shi J, Concel J, Aiken C, Zhang PJ, Gronenborn AM. Structural convergence between cryo-EM and NMR reveals intersubunit Interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Y, Ahn J, Concel J, Byeon IJL, Gronenborn AM, Yang J, Polenova T. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Y, Hou GJ, Suiter CL, Ahn J, Byeon IJL, Lipton AS, Burton S, Hung I, Gor'kov PL, Gan ZH, Brey W, Rice D, Gronenborn AM, Polenova T. Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2013;135:17793–17803. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Tycko R. Structural and dynamical characterization of tubular HIV-1 capsid protein assemblies by solid state nuclear magnetic resonance and electron microscopy. Protein Sci. 2010;19:716–730. doi: 10.1002/pro.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayro MJ, Chen B, Yau WM, Tycko R. Site-specific structural variations accompanying tubular assembly of the HIV-1 capsid protein. J. Mol. Biol. 2014;426:1109–1127. doi: 10.1016/j.jmb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs JAG, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. Embo J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 22.Berthet-Colominas C, Monaco S, Novelli A, Sibai G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. Embo J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc. Natl. Acad. Sci. USA. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat. Struct. Mol. Biol. 2005;12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 25.Byeon IJL, Hou GJ, Han Y, Suiter CL, Ahn J, Jung J, Byeon CH, Gronenborn AM, Polenova T. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: implications for structural polymorphism of CA assemblies. J. Am. Chem. Soc. 2012;134:6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshmukh L, Schwieters CD, Grishaev A, Ghirlando R, Baber JL, Clore GM. Structure and dynamics of full-length HIV-1 capsid protein in solution. J. Am. Chem. Soc. 2013;135:16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol. Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 28.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Mol. Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 29.Gamble TR, Vajdos FF, Yoo SH, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 30.Yeager M. Design of in vitro symmetric complexes and analysis by hybrid methods reveal mechanisms of HIV capsid assembly. J. Mol. Biol. 2011;410:534–552. doi: 10.1016/j.jmb.2011.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. J. Struct. Biol. 2012;177:589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A, Heise H, Blommers MJJ, Krastel P, Schmitt E, Petersen F, Jeganathan S, Mandelkow E-M, Carlomagno T, Griesinger C, Baldus M. Interaction of Epothilone B (Patupilone) with Microtubules as Detected by Two-Dimensional Solid-State NMR Spectroscopy. Angewandte Chemie International Edition. 2010;49:7504–7507. doi: 10.1002/anie.201001946. [DOI] [PubMed] [Google Scholar]
- 33.Schütz AK, Soragni A, Hornemann S, Aguzzi A, Ernst M, Böckmann A, Meier BH. The Amyloid-Congo Red Interface at Atomic Resolution. Angewandte Chemie International Edition. 2011;50:5956–5960. doi: 10.1002/anie.201008276. [DOI] [PubMed] [Google Scholar]
- 34.Hou GJ, Suiter CL, Yan S, Zhang H, Polenova T. Magic angle spinning NMR studies of protein assemblies: Recent advances in methodology and applications. Ann. R. NMR S. 2013;80:293–257. [Google Scholar]
- 35.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical Physics Letters. 2001;344:631–637. doi: 10.1063/1.2364503. [DOI] [PubMed] [Google Scholar]
- 36.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 37.Luca S, Heise H, Baldus M. High-Resolution Solid-State NMR Applied to Polypeptides and Membrane Proteins. Accounts of Chemical Research. 2003;36:858–865. doi: 10.1021/ar020232y. [DOI] [PubMed] [Google Scholar]
- 38.Kristiansen PE, Carravetta M, van Beek JD, Lai WC, Levitt MH. Theory and applications of supercycled symmetry-based recoupling sequences in solid-state nuclear magnetic resonance. J. Chem. Phys. 2006;124 doi: 10.1063/1.2205857. [DOI] [PubMed] [Google Scholar]
- 39.Levitt MH. Symmetry in the design of NMR multiple-pulse sequences. J. Chem. Phys. 2008;128 doi: 10.1063/1.2831927. [DOI] [PubMed] [Google Scholar]
- 40.Zhao X, Eden M, Levitt MH. Recoupling of heteronuclear dipolar interactions in solid-state NMR using symmetry-based pulse sequences. Chem. Phys. Lett. 2001;342:353–361. [Google Scholar]
- 41.Hou GJ, Yan S, Sun SJ, Han Y, Byeon IJL, Ahn J, Concel J, Samoson A, Gronenborn AM, Polenova T. Spin diffusion driven by R-symmetry sequences: Applications to homonuclear correlation spectroscopy in MAS NMR of biological and organic solids. J. Am. Chem. Soc. 2011;133:3943–3953. doi: 10.1021/ja108650x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou GJ, Yan S, Trebosc J, Amoureux JP, Polenova T. Broadband homonuclear correlation spectroscopy driven by combined R2(n)(v) sequences under fast magic angle spinning for NMR structural analysis of organic and biological solids. J. Magn. Reson. 2013;232:18–30. doi: 10.1016/j.jmr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ladizhansky V. Dipolar-based torsion angle measurements for protein structure determination. In: Harris RK, Wasylishen RE, editors. Encyclopedia of NMR. Chichester: Wiley; 2009. [Google Scholar]
- 44.Ramamoorthy A. NMR spectroscopy of biological solides. CRC Press; 2005. [Google Scholar]
- 45.Hou GJ, Byeon IJL, Ahn J, Gronenborn AM, Polenova T. H-1-C-13/H-1-N-15 heteronuclear dipolar recoupling by R-symmetry sequences under fast magic angle spinning for dynamics analysis of biological and organic solids. J. Am. Chem. Soc. 2011;133:18646–18655. doi: 10.1021/ja203771a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou G, Paramasivam S, Byeon IJL, Gronenborn AM, Polenova T. Determination of relative tensor orientations by gamma-encoded chemical shift anisotropy/heteronuclear dipolar coupling 3D NMR spectroscopy in biological solids. Phys. Chem. Chem. Phys. 2010;12:14873–14883. doi: 10.1039/c0cp00795a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou GJ, Byeon IJL, Ahn J, Gronenborn AM, Polenova T. Recoupling of chemical shift anisotropy by R-symmetry sequences in magic angle spinning NMR spectroscopy. J. Chem. Phys. 2012;137 doi: 10.1063/1.4754149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou G, Lu X, Vega AJ, Polenova T. Accurate measurement of heteronuclear dipolar couplings by phase-alternating R-symmetry (PARS) sequences in magic angle spinning NMR spectroscopy. J. Chem. Phys. 2014;141:104202. doi: 10.1063/1.4894226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan JCC, Tycko R. Recoupling of chemical shift anisotropies in solid-state NMR under high-speed magic-angle spinning and in uniformly C-13-labeled systems. J. Chem. Phys. 2003;118:8378–8389. [Google Scholar]
- 50.Hou G, Gupta R, Polenova T, Vega AJ. A magic-angle-spinning NMR spectroscopy method for the site-specific measurement of proton chemical-shift-anisotropy in biological and organic solids. Isr. J. Chem. 2014;54:171–183. doi: 10.1002/ijch.201300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou GJ, Paramasivam S, Yan S, Polenova T, Vega AJ. Multidimensional magic angle spinning NMR spectroscopy for site-resolved measurement of proton chemical shift anisotropy in biological solids. J. Am. Chem. Soc. 2013;135:1358–1368. doi: 10.1021/ja3084972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma Y, Kwon OY, Brooks B, Tjandra N. An ab initio study of amide proton shift tensor dependence on local protein structure. J Am Chem Soc. 2002;124:327–335. doi: 10.1021/ja016859d. [DOI] [PubMed] [Google Scholar]
- 53.Tjandra N, Bax A. Solution NMR measurement of amide proton chemical shift anisotropy in N-15-enriched proteins. Correlation with hydrogen bond length. J Am Chem Soc. 1997;119:8076–8082. [Google Scholar]
- 54.Duma L, Abergel D, Tekely P, Bodenhausen G. Proton chemical shift anisotropy measurements of hydrogen-bonded functional groups by fast magic-angle spinning solid-state NMR spectroscopy. Chem Commun. 2008:2361–2363. doi: 10.1039/b801154k. [DOI] [PubMed] [Google Scholar]
- 55.Loth K, Pelupessy P, Bodenhausen G. Chemical shift anisotropy tensors of carbonyl, nitrogen, and amide proton nuclei in proteins through cross-correlated relaxation in NMR spectroscopy. J Am Chem Soc. 2005;127:6062–6068. doi: 10.1021/ja042863o. [DOI] [PubMed] [Google Scholar]
- 56.Agarwal V, Penzel S, Szekely K, Cadalbert R, Testori E, Oss A, Past J, Samoson A, Ernst M, Bockmann A, Meier BH. De novo 3D structure determination from sub-milligram protein samples by solid-state 100 kHz MAS NMR spectroscopy. Angew. Chem. Int. Edit. doi: 10.1002/anie.201405730. In Press. [DOI] [PubMed] [Google Scholar]
- 57.Bertini I, Emsley L, Felli IC, Laage S, Lesage A, Lewandowski JR, Marchetti A, Pierattelli R, Pintacuda G. High-resolution and sensitivity through-bond correlations in ultra-fast magic angle spinning (MAS) solid-state NMR. Chem. Sci. 2011;2:345–348. [Google Scholar]
- 58.Laage S, Sachleben JR, Steuernagel S, Pierattelli R, Pintacuda G, Emsley L. Fast acquisition of multi-dimensional spectra in solid-state NMR enabled by ultra-fast MAS. J. Magn. Reson. 2009;196:133–141. doi: 10.1016/j.jmr.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 59.Parthasarathy S, Nishiyama Y, Ishii Y. Sensitivity and resolution enhanced solid-state NMR for paramagnetic systems and biomolecules under very fast magic angle spinning. Acc. Chem. Res. 2013;46:2127–2135. doi: 10.1021/ar4000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickramasinghe NP, Parthasarathy S, Jones CR, Bhardwaj C, Long F, Kotecha M, Mehboob S, Fung LWM, Past J, Samoson A, Ishii Y. Nanomole-scale protein solid-state NMR by breaking intrinsic H-1 T-1 boundaries. Nat. Met. 2009;6:215–218. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan S, Suiter CL, Hou GJ, Zhang HL, Polenova T. Probing structure and dynamics of protein assemblies by magic angle spinning NMR spectroscopy. Acc. Chem. Res. 2013;46:2047–2058. doi: 10.1021/ar300309s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou DH, Shah G, Cormos M, Mullen C, Sandoz D, Rienstra CM. Proton-detected solid-state NMR Spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. J. Am. Chem. Soc. 2007;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- 63.Zhou DH, Shea JJ, Nieuwkoop AJ, Franks TW, Wylie BJ, Mullen C, Sandoz D, Rienstra CM. Solid-state protein-structure determination with proton-detected triple-resonance 3D magic-angle-spinning NMR spectroscopy. Angew. Chem. Int. Edit. 2007;46:8380–8383. doi: 10.1002/anie.200702905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asami S, Szekely K, Schanda P, Meier BH, Reif B. Optimal degree of protonation for H-1 detection of aliphatic sites in randomly deuterated proteins as a function of the MAS frequency. J. Biomol. NMR. 2012;54:155–168. doi: 10.1007/s10858-012-9659-9. [DOI] [PubMed] [Google Scholar]
- 65.Chevelkov V, Rehbein K, Diehl A, Reif B. Ultrahigh resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angew. Chem. Int. Edit. 2006;45:3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- 66.Hologne M, Chevelkov V, Reif B. Deuterated peptides and proteins in MAS solid-state NMR. Prog. Nucl. Magn. Reson. Spec. 2006;48:211–232. [Google Scholar]
- 67.Reif B. Ultra-high resolution in MAS solid-state NMR of perdeuterated proteins: Implications for structure and dynamics. J. Magn. Reson. 2012;216:1–12. doi: 10.1016/j.jmr.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 68.Marchetti A, Jehle S, Felletti M, Knight MJ, Wang Y, Xu ZQ, Park AY, Otting G, Lesage A, Emsley L, Dixon NE, Pintacuda G. Backbone assignment of fully protonated solid proteins by 1H detection and ultrafast magic-angle-spinning NMR spectroscopy. Angew. Chem. Int. Edit. 2012;51:10756–10759. doi: 10.1002/anie.201203124. [DOI] [PubMed] [Google Scholar]
- 69.Sun SJ, Yan S, Guo CM, Li MY, Hoch JC, Williams JC, Polenova T. A time-saving strategy for MAS NMR spectroscopy by combining nonuniform sampling and paramagnetic relaxation assisted condensed data collection. J. Phys. Chem. B. 2012;116:13585–13596. doi: 10.1021/jp3005794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andreas LB, Barnes AB, Corzilius B, Chou JJ, Miller EA, Caporini M, Rosay M, Griffin RG. Dynamic nuclear polarization study of inhibitor binding to the M2(18–60) proton transporter from influenza A. Biochemistry. 2013;52:2774–2782. doi: 10.1021/bi400150x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sergeyev IV, Day LA, Goldbourt A, McDermott AE. Chemical shifts for the unusual DNA structure in Pf1 bacteriophage from dynamic-nuclear-polarization-enhanced solid-state NMR spectroscopy. J. Am. Chem. Soc. 2011;133:20208–20217. doi: 10.1021/ja2043062. [DOI] [PubMed] [Google Scholar]
- 72.Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP. Exponential sampling, an alternative method for sampling in two-dimensional NMR experiments. Journal of Magnetic Resonance. 1987;73:69–77. [Google Scholar]
- 73.Paramasivam S, Suiter CL, Hou GJ, Sun SJ, Palmer M, Hoch JC, Rovnyak D, Polenova T. Enhanced sensitivity by nonuniform sampling enables multidimensional MAS NMR spectroscopy of protein assemblies. J. Phys. Chem. B. 2012;116:7416–7427. doi: 10.1021/jp3032786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rovnyak D, Sarcone M, Jiang Z. Sensitivity enhancement for maximally resolved two-dimensional NMR by nonuniform sampling. Magn. Reson. Chem. 2011;49:483–491. doi: 10.1002/mrc.2775. [DOI] [PubMed] [Google Scholar]
- 75.Suiter CL, Paramasivam S, Hou GJ, Sun SJ, Rice D, Hoch J, Rovnyak D, Polenova T. Sensitivity gains, linearity, and spectral reproducibility in nonuniformly sampled multidimensional MAS NMR spectra of high dynamic range. J. Biomol. NMR. 2014;59:57–73. doi: 10.1007/s10858-014-9824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiang W. Signal enhancement for the sensitivity-limited solid state NMR experiments using a continuous, non-uniform acquisition scheme. Journal of Magnetic Resonance. 2011;213:171–175. doi: 10.1016/j.jmr.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Wang Q, Zhang Z, Yang J, Hu B, Chen Q, Noda I, Deng F. Covariance spectroscopy with a non-uniform and consecutive acquisition scheme for signal enhancement of the NMR experiments. Journal of Magnetic Resonance. 2012;217:106–111. doi: 10.1016/j.jmr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 78.Fossi M, Castellani T, Nilges M, Oschkinat H, van Rossum BJ. SOLARIA: A protocol for automated cross-peak assignment and structure calculation for solid-state magic-angle spinning NMR spectroscopy. Angew. Chem. Int. Edit. 2005;44:6151–6154. doi: 10.1002/anie.200501884. [DOI] [PubMed] [Google Scholar]
- 79.Hu KN, Qiang W, Tycko R. A general Monte Carlo/simulated annealing algorithm for resonance assignment in NMR of uniformly labeled biopolymers. J. Biomol. NMR. 2011;50:267–276. doi: 10.1007/s10858-011-9517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nielsen JT, Kulminskaya N, Bjerring M, Nielsen NC. Automated robust and accurate assignment of protein resonances for solid state NMR. J. Biomol. NMR. 2014;59:119–134. doi: 10.1007/s10858-014-9835-1. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt E, Gath J, Habenstein B, Ravotti F, Szekely K, Huber M, Buchner L, Bockmann A, Meier BH, Guntert P. Automated solid-state NMR resonance assignment of protein microcrystals and amyloids. J. Biomol. NMR. 2013;56:243–254. doi: 10.1007/s10858-013-9742-x. [DOI] [PubMed] [Google Scholar]
- 82.Schmidt E, Guntert P. A new algorithm for reliable and general NMR resonance assignment. J. Am. Chem. Soc. 2012;134:12817–12829. doi: 10.1021/ja305091n. [DOI] [PubMed] [Google Scholar]
- 83.Tycko R, Hu KN. A Monte Carlo/simulated annealing algorithm for sequential resonance assignment in solid state NMR of uniformly labeled proteins with magic-angle spinning. J. Magn. Reson. 2010;205:304–314. doi: 10.1016/j.jmr.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Y, Fritzsching KJ, Hong M. Resonance assignment of the NMR spectra of disordered proteins using a multi-objective non-dominated sorting genetic algorithm. J. Biomol. NMR. 2013;57:281–296. doi: 10.1007/s10858-013-9788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holland GP, Cherry BR, Jenkins JE, Yarger JL. Proton-detected heteronuclear single quantum correlation NMR spectroscopy in rigid solids with ultra-fast MAS. J. Magn. Reson. 2010;202:64–71. doi: 10.1016/j.jmr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huber M, Hiller S, Schanda P, Ernst M, Bockmann A, Verel R, Meier BH. A proton-detected 4D solid-state NMR experiment for protein structure determination. Chemphyschem. 2011;12:915–918. doi: 10.1002/cphc.201100062. [DOI] [PubMed] [Google Scholar]
- 87.Knight MJ, Webber AL, Pell AJ, Guerry P, Barbet-Massin E, Bertini I, Felli IC, Gonnelli L, Pierattelli R, Emsley L, Lesage A, Herrmann T, Pintacuda G. Fast resonance assignment and fold determination of human superoxide dismutase by high-resolution proton-detected solid-state MAS NMR spectroscopy. Angew. Chem. Int. Edit. 2011;50:11697–11701. doi: 10.1002/anie.201106340. [DOI] [PubMed] [Google Scholar]
- 88.Asami S, Schmieder P, Reif B. High resolution 1H-detected solid-state NMR spectroscopy of protein aliphatic resonances: access to tertiary structure information. J Am Chem Soc. 2010;132:15133–15135. doi: 10.1021/ja106170h. [DOI] [PubMed] [Google Scholar]
- 89.Barbet-Massin E, Pell AJ, Retel JS, Andreas LB, Jaudzems K, Franks WT, Nieuwkoop AJ, Hiller M, Higman V, Guerry P, Bertarello A, Knight MJ, Felletti M, Le Marchand T, Kotelovica S, Akopjana I, Tars K, Stoppini M, Bellotti V, Bolognesi M, Ricagno S, Chou JJ, Griffin RG, Oschkinat H, Lesage A, Emsley L, Herrmann T, Pintacuda G. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J. Am. Chem. Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin EC, Opella SJ. Sampling scheme and compressed sensing applied to solid-state NMR spectroscopy. J. Magn. Reson. 2013;237:40–48. doi: 10.1016/j.jmr.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matsuki Y, Eddy MT, Griffin RG, Herzfeld J. Rapid three-dimensional MAS NMR spectroscopy at critical sensitivity. Angew. Chem. Int. Edit. 2010;49:9215–9218. doi: 10.1002/anie.201003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiang S, Chevelkov V, Becker S, Lange A. Towards automatic protein backbone assignment using proton-detected 4D solid-state NMR data. J. Biomol. NMR. 2014 doi: 10.1007/s10858-014-9859-6. [DOI] [PubMed] [Google Scholar]
- 93.Hoch J, Stern AS. NMR Data Processing. New York: Wiley; 1996. [Google Scholar]
- 94.Franks WT, Atreya HS, Szyperski T, Rienstra CM. GFT projection NMR spectroscopy for proteins in the solid state. J. Biomol. NMR. 2010;48:213–223. doi: 10.1007/s10858-010-9451-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoch J, Maciejewski MW, Mobli M, Schuyler AD, Stern AS. Nonuniform sampling in multidimensional NMR. In: Harris RK, Wasylishen RE, editors. Encyclopedia of NMR. Chichester: Wiley; 2012. [Google Scholar]
- 96.Orekhov VY, Ibraghimov I, Billeter M. Optimizing resolution in multidimensional NMR by three-way decomposition. J. Biomol. NMR. 2003;27:165–173. doi: 10.1023/a:1024944720653. [DOI] [PubMed] [Google Scholar]
- 97.Qiang W. Signal enhancement for the sensitivity-limited solid state NMR experiments using a continuous, non-uniform acquisition scheme. J. Magn. Reson. 2011;213:171–175. doi: 10.1016/j.jmr.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li YX, Wang Q, Zhang ZF, Yang J, Hu BW, Chen Q, Noda I, Deng F. Covariance spectroscopy with a non-uniform and consecutive acquisition scheme for signal enhancement of the NMR experiments. J. Magn. Reson. 2012;217:106–111. doi: 10.1016/j.jmr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 99.Cardone G, Purdy JG, Cheng NQ, Craven RC, Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–U693. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hyun JK, Radjainia M, Kingston RL, Mitra AK. Proton-driven assembly of the rous sarcoma virus capsid protein results in the formation of icosahedral particles. J. Biol. Chem. 2010;285:15056–15064. doi: 10.1074/jbc.M110.108209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci USA. 2003;100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen AT, Feasley CL, Jackson KW, Nitz TJ, Salzwedel K, Air GM, Sakalian M. The prototype HIV-1 maturation inhibitor, bevirimat, binds to the CA-SP1 cleavage site in immature Gag particles. Retrovirology. 2011;8 doi: 10.1186/1742-4690-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Datta SAK, Temeselew LG, Crist RM, Soheilian F, Kamata A, Mirro J, Harvin D, Nagashima K, Cachau RE, Rein A. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J. Virol. 2011;85:4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keller PW, Huang RK, England MR, Waki K, Cheng NQ, Heymann JB, Craven RC, Freed EO, Steven AC. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol. 2013;87:13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Krausslich HG. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aberham C, Weber S, Phares W. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 1996;70:3536–3544. doi: 10.1128/jvi.70.6.3536-3544.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qi ML, Yang RF, Aiken C. Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells. J. Virol. 2008;82:12001–12008. doi: 10.1128/JVI.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bax A, Shen Y, Delaglio F, Cornilescu G. TALOS plus : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular Nmr. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu KN, Qiang W, Bermejo GA, Schwieters CD, Tycko R. Restraints on backbone conformations in solid state NMR studies of uniformly labeled proteins from quantitative amide 15N-15N and carbonyl 13C-13C dipolar recoupling data. Journal of magnetic resonance (San Diego, Calif. : 1997) 2012;218:115–127. doi: 10.1016/j.jmr.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 111.Monroe EB, Kang S, Kyere SK, Li R, Prevelige PE. Hydrogen/deuterium exchange analysis of HIV-1 capsid assembly and maturation. Structure. 2010;18:1483–1491. doi: 10.1016/j.str.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baumgartel V, Muller B, Lamb DC. Quanitative live-cell imaging of human immunodeficiency virus (HIV-1) assembly. Viruses. 2012;4:777–799. doi: 10.3390/v4050777. [DOI] [PMC free article] [PubMed] [Google Scholar]