Abstract
Multidrug-resistant (MDR) enterococci are important nosocomial pathogens and a growing clinical challenge. These organisms have developed resistance to virtually all antimicrobials currently used in clinical practice using a diverse number of genetic strategies. Due to this ability to recruit antibiotic resistance determinants, MDR enterococci display a wide repertoire of antibiotic resistance mechanisms including modification of drug targets, inactivation of therapeutic agents, overexpression of efflux pumps and a sophisticated cell envelope adaptive response that promotes survival in the human host and the nosocomial environment. MDR enterococci are well adapted to survive in the gastrointestinal tract and can become the dominant flora under antibiotic pressure, predisposing the severely ill and immunocompromised patient to invasive infections. A thorough understanding of the mechanisms underlying antibiotic resistance in enterococci is the first step for devising strategies to control the spread of these organisms and potentially establish novel therapeutic approaches.
Keywords: antimicrobial resistance, enterococcus, mechanisms of resistance, vancomycin-resistant enterococci
Enterococci are gram-positive facultative anaerobes that live as commensals in the GI tract of a variety of organisms including humans [1]. Shaped by the selective pressures of their competitive environment, these bacteria have evolved a diverse array of responses and genetic plasticity allowing them to thrive in the modern healthcare environment. Advances in modern medicine have increased the ability to sustain life in critically ill patients increasing the susceptibility to infection and resulting in rapid patient turnover and widespread use of antibiotics. Armed with multiple antibiotic resistance determinants, enterococci ‘take advantage’ of this opportunity and expand within their ecologic niche (GI tract of hospitalized patients) to gain the upper hand and dominate the intestinal microbiota. From the GI tract, multidrug-resistant (MDR) enterococci disseminate rapidly in the hospital environment. Indeed, enterococci are leading causes of nosocomial infections and are second only to staphylococci as a cause of gram-positive nosocomial infections [2]. Here, we will discuss the historical context behind the rise of enterococci as MDR pathogens followed by a review of the current understanding of the molecular basis of enterococcal antimicrobial resistance. Finally, we will briefly address the need for the development of novel drug targets and combination therapies against these sturdy and recalcitrant organisms.
Evolution of antibiotic resistance in MDR enterococci
The advent of the antibiotic era spurred a new revolution in modern medicine. With the discovery of antimicrobial agents and the understanding of the microbiological basis of disease, infection became a treatable disease with remarkable results. However, clinicians rapidly realized that certain microorganisms appear to respond less well to specific antimicrobial agents. For example, with the introduction of penicillin, streptococcal infection was successfully treated with this agent. However, a subset of ‘streptococcal’ organisms (later known as enterococci) appeared to respond less well to penicillin due to an inherent tolerance to the killing action of these compounds [3]. It was later found that the addition of aminoglycosides (streptomycin was discovered in 1944 [4]) to penicillin produced synergistic activity improving the cure rates for enterococcal infective endocarditis from 40 to 88% [5]. This synergistic effect was seen despite the fact that enterococci are also inherently less susceptible to aminoglycosides compared to other gram-positive bacteria. Thus, the combination of a cell-wall active agent (i.e., ampicillin/penicillin) plus an aminoglycoside became the standard of care for deep-seated enterococcal infections and this combination is still used to the present day [6].
Though it was not known at that time, the seeds of the modern MDR enterococci were already being sown. Using comparative genomics, Lebreton et al. demonstrated that the modern-day MDR Enterococcus faecium belongs to a genetic clade that appears to have diverged from animal-adapted E. faecium isolates about 75 years ago, coinciding with the introduction of antibiotics in clinical practice [7]. Features of this clade (designated A1) include an increase in mobile genetic elements, alterations in metabolism and hypermutability, a skill set that provides E. faecium a malleable genome in the face of multiple selective pressures. The remarkable increase in the use of antimicrobials in clinical medicine in the latter half of the 20th century provided the selective environment for these microorganisms to evolve by recruiting a variety of antibiotic resistance determinants.
Among the most distinct examples of this adaptability is the acquisition of the genes encoding vancomycin resistance. Van-comycin use was associated with the emergence and spread of methicillin-resistant Staphylococcus aureus (MRSA) in the 1960s. However, unlike MRSA, enterococci have been able to recruit and maintain a variety of gene clusters encoding the biochemical machinery for vancomycin resistance. From the first recognition of vancomycin-resistant enterococci (VRE) in 1988 to late in the first decade of this century, the rates of van-comycin resistance in E. faecium in the USA have surpassed 80% [8]. Of concern, E. faecium is also increasingly identified in nosocomial infections, now occurring as frequently as isolates of Enterococcus faecalis (whose rates of vancomycin resistance are about 5%) [2]. More recently, enterococci have also served as donors of vancomycin resistance gene clusters to more pathogenic microorganisms such as MRSA, an event considered to be a serious public health threat [9,10].
Despite the availability of anti-gram-positive agents (e.g., line-zolid, quinupristin/dalfopristin [Q/D], daptomycin [DAP], tige-cycline), enterococci have rapidly adapted and emergence of resistance to all these newer agents have been well documented. This phenomenon makes the treatment of MDR enterococcal infections a daunting clinical challenge. Thus, in the following sections, we will review the molecular mechanisms of antibiotic resistance in enterococci with the objective to place such mechanisms in context within a clinical perspective and explore innovative strategies to combat these recalcitrant microorganisms.
Mechanisms of resistance to cell-wall active agents
Ampicillin/penicillin
Ampicillin and penicillin are the most active β-lactams against enterococci inhibiting the synthesis of peptidoglycan, the basic structure of the bacterial cell wall and a critical component needed for bacterial viability. As such, the peptidoglycan synthesis and maintenance machinery has long been a target for antimicrobial therapy. Furthermore, the lack of similar structures in eukaryotic cells decreases the toxicity profiles of many of these agents, making them an ideal target against bacteria. Penicillin-binding proteins (PBPs) are the workhorses of the cell wall synthesis machinery and they can be roughly divided into two groups: class A, which are bifunctional enzymes that possess both d,d-transpeptidase and transglycosylase activity, and class B, which possess only the transpeptidase domain and rely on the transglycosylase activity of other enzymes.
All enterococci produce at least five PBPs, which can be differentiated by migration pattern on gel electrophoresis and were originally named by convention on the order of migration [11]. Subsequent genomic analysis of both E. faecalis and E. faecium revealed six putative PBP genes, three of which are class A (ponA, pbpF, pbpZ) and three class B (pbp5, pbpA, pbpB) [12]. Intrinsic tolerance to the action of β-lactams is associated with the presence of a species-specific chromosomal gene, pbp5, which encodes a class B PBP with low binding affinity for ampicillin and the cephalosporins [13,14]. In E. faecium, pbp5 exists in an operon with two other genes implicated in cell wall synthesis; psr, for PBP synthesis repressor, as it was originally thought to repress transcription of pbp5 (this effect was later attributed to a deletion in the promoter region of the gene) and ftsW, a protein known to interact with Escherichia coli PBP3 during septum formation [15]. High-level resistance to ampicillin (MIC 128 μg/ml or more) was first associated with increased production of the enzyme, requiring a higher concentration of antibiotic to saturate the active site [16]. However, many resistant clinical isolates were found to have no difference in protein expression from their sensitive counterparts. Thus, resistance in these strains was attributed to mutations altering the protein sequence, specifically a Met485 → Ala substitution near the active serine residue and the insertion of a serine residue at position 466 in a loop structure predicted to interact with the active site complex [17]. Although the authors of this study observed a decrease in binding of β-lactams and a subsequent increase in the MIC, the relationship was not directly proportional. Additionally, copies of the pbp5 gene from highly resistant clinical isolates were unable to fully restore the resistant phenotype of a hypersensitive strain lacking a copy of pbp5 [14].
Sequence variation of PBP5 is sufficient to distinguish two groups of E. faecium, one possessing high-level ampicillin resistance (termed Pbp5-R) associated with the hospital environment and a community-associated variant (Pbp5-S) that results in lower MICs to ampicillin (usually <64 μg/ml), but is unable to completely explain the differing susceptibility profiles observed in clinical practice [18]. Thus, as yet unidentified factors play an important role in expressing resistance via Pbp5 in E. faecium. In E. faecalis, pbp5 appears to exist independent from the operon structure described in E. faecium, and although it also encodes a class B PBP, it has a lower intrinsic tolerance to ampicillin (typical MICs of E. faecalis isolates are 1–4 μg/ml). Both overexpression of the enzyme and mutations in amino acid sequence have been implicated in higher levels of resistance, though neither method produced changes in MIC as dramatic as seen in E. faecium [19,20].
Another form of ampicillin resistance, mediated by a β-lactamase that inactivates the antibiotic through the cleavage of the β-lactam ring, has been described in both E. faecalis and E. faecium [21,22]. Originally described in staphylococci, the gene blaZ encodes a β-lactamase as part of an operon with blaR1, a transmembrane sensor and signal transducer, and blaI, a repressor gene [23]. Sequence analysis of enterococci possessing the entire operon showed amino acid sequence identity of 97, 95 and 96% to the blaZ, blaI and blaR1 genes, respectively of S. aureus, reinforcing previous evidence that these genes likely originated in staphylococci [24]. In contrast to staphylococci, blaZ in enterococci is expressed constitutively and at a much lower level, resulting in a clinically significant inoculum effect. Thus, when inoculating bacteria at concentrations for routine suscepbility testing (generally 1 × 105 organisms per ml), the enterococci produce so little enzyme that they test susceptible, while at high inoculum, such as during infection, the presence of more enzyme leads to resistance. Once identified, however, the presence of a β-lactamase is of trivial clinical concern as the addition of the β-lactamase inhibitor sulbactam (i.e., ampicil-lin-sulbactam) is sufficient to restore the antibiotic efficacy.
Finally, the development of ampicillin resistance via an l,d-transpeptidase known as Ldtfm has also been described in vitro [25]. Constitutively expressed in E. faecium, Ldtfm utilizes a tetrapeptide substrate (unlike the d,d-transpeptidases which act on pentapeptides) and is thought to be involved in the maintenance of peptidoglycan in the stationary phase. Mutations in a cryptic locus (ddc), coding for a putative two-component signal transduction system (TCS) induced expression of a d,d-carboxypeptidase (DdcY), which shares similarities with the van resistance cluster (discussed below) and produced the necessary tetrapeptide substrate [26]. Of note, this pathway would allow the synthesis of a functional cell wall in the presence of glycopeptides [27].
Cephalosporin resistance
Although the natural resistance of enterococci to cephalosporins is a well-known feature, the molecular basis of this phenotype is not completely understood. A common observation is that intrinsic resistance is associated with a decrease in binding affinity of cephalosporins for the enterococcal PBPs, specifically Pbp5 [28,29]. As a class B PBP possessing only transpeptidase enzymatic activity, Pbp5 must partner with a glycosyltransferase to synthesize peptidoglycan. In two studies performed by Arbeloa et al. [29], and Rice et al. [28], sequential deletion of the known class A PBPs (PbpF, PonA or PbpZ) in both E. faecalis and E. faecium was undertaken alone and in combination to infer their effect on the resistance phenotype. In both species, it was found that either PonA or PbpF was needed to express cephalosporin resistance; PbpZ alone was unable to provide the needed transglycosylase activity. Likewise, deletion of all three genes led to a susceptible phenotype in both species, however, the triple mutant was still viable, albeit at an impaired rate of growth. The enzyme providing transglycosylase activity in the absence of the class A PBPs was not identified, and genomic analysis looking for conserved sequences similar to known glyco-syltransferases did not suggest a suitable candidate. Interestingly, in E. faecium, it was noted that deletion of the class A PBPs showed a dissociation of cephalosporin and penicillin resistance, as MICs to ampicillin did not change. Further, an inducible resistance to cephalosporins in triple mutants (that is deletion of all class A PBPs) was unmasked by disk diffusion with penicillin, suggesting differential activation of regulatory pathways as a response to β-lactam antibiotics.
Several regulatory pathways controlled by bacterial two-component regulatory systems (TCS) have also been associated with the intrinsic resistance to cephalosporins. However, we are just beginning to scratch the surface of the action of their downstream effectors [30]. Among them, CroRS was shown to be important for cephalosporin resistance. CroS, a sensor with histidine kinase activity, phosphorylates the cognate response regulator CroR, which is postulated to alter transcription via a DNA binding domain [31]. Several genes under direct CroR regulation have been identified; however, none of them appears to alter pathways currently known to induce cephalosporin resistance [32,33]. Another TCS implicated in resistance consists of a serine/threonine kinase designated IreK and a phosphatase IreP [34]. E. faecalis with deletions of IreP were shown to exhibit increased resistance to ceftriaxone, but had a substantial reduction of fitness in the absence of antibiotics [35]. A third protein, IreB, was demonstrated to be the target of both IreK and IreP and to negatively regulate the expression of cephalo-sporin resistance [36]. Furthermore, the concomitant deletion of IreK and IreB, or the elimination of the threonine residue where phosphorylation occurs, was able to restore resistance via a release of inhibition of the pathway. This system seems to be specific to cephalosporin resistance, as MICs to ampicillin and other cell wall active agents were unchanged. One protein that may be involved in the downstream effects of the IreK signaling pathway is MurAA, one of a pair of homologs that catalyze the first committed step in peptidoglycan synthesis [37]. Deletion of the gene coding for this enzyme resulted in a loss of resistance to cephalosporins despite the ability of the homolog MurAB to carry out enzymatic activity as a uridine diphos-phate-N-acetylglucosamine 1-carboxyvinyl transferase. When expressed from a plasmid vector in ireK-deficient E. faecalis, but not in croR or pbp5 deletion mutants, MurAA was able to restore ceftriaxone resistance [37].
Glycopeptide resistance
Glycopeptides (vancomycin and teicoplanin) bind to the terminal d-alanine-d-alanine (d-Ala-d-Ala) moiety of peptidoglycan precursors, thus preventing cross-linking of peptidoglycan chains and inhibiting synthesis of the cell wall. Resistance to these agents, typified by vancomycin, can be described as high-level (MIC >64 μg/ml) or low-level (MIC between 4 and 32 μg/ml), both due to a change in the terminal amino acids of the peptido-glycan precursor from d-Ala-d-Ala to d-alanine-d-lactate (d-Ala-d-Lac) or, less frequently, d-alanine-d-serine (d-Ala-d-Ser) [38]. The type of amino acid change is relevant, as it determines the level of resistance. Thus, low-level resistance is conferred by the d-Ala-d-Ser-ending precursors, which decreases the binding affinity of the antibiotic about sevenfold. On the other hand, high-level resistance relies on the change of the terminal penta-peptide to d-Ala-d-Lac, a substitution that eliminates one of the five hydrogen bonds required for the binding of vancomycin to the peptidoglycan chain, reducing its affinity about 1000-fold.
The origin of vancomycin resistance gene clusters (designated van) likely lies in the day-to-day struggle for survival in the microbial world. Genes nearly identical to the vanA cluster found in enterococci have been described in the soil organisms Paenibacillus thiaminoluticus and P. apiaries [39]. To date, nine distinct vancomycin resistance clusters have been described in enterococci (vanA, vanB, vanC, vanD, vanE, vanG, vanL, vanM and vanN) [38,40–42]. In general, these clusters consist of three groups of genes encoding: TCS; enzymes necessary for the synthesis of new peptidoglycan precursors and enzymes that destroy the normal d-Ala-d-Ala-ending precursors. The different clusters can be differentiated by the identity at the amino acid level of the ligase enzyme (e.g., VanA vs VanB), by the degree of vancomycin resistance conferred, the ability of the system to be induced upon exposure to teicoplanin and the structural arrangement of the gene cluster.
The vanA cluster is the most common mediator of vancomycin resistance in enterococci and its expression is under the regulation of two promoters. The first is responsible for the transcription of VanRS, the TCS that regulates VanA expression and function. The sensor of the system is VanS, which is a transmembrane protein with a histidine kinase domain that responds to the presence of glycopeptides by phosphorylating the response regulator VanR [43]. Once activated, VanR binds to the second promoter region, located upstream of the resistance genes, activating their transcription. The first step in expressing vancomycin resistance is the transcription of vanH that encodes a dehydrogenase, which allows for the production of d-lactate from pyruvate. The next gene, vanA, produces a ligase that enables the addition of d-Lac to d-Ala before adding it to a tripeptide precursor. The resulting pentadepsipeptide is incorporated into the growing cell wall and allows for cross-linking of the peptidoglycan structure. VanX, a d,d-dipeptidase, and VanY, a DdcY, work to clear the usual d-Ala-d-Ala dipeptides (which will bind vancomycin if incorporated in the cell wall) and the normal d-Ala-ending pentapeptide chains from the pool of cell wall precursors, respectively. Thus, destruction of d-Ala-ending pentapeptide precursors is crucial for the mechanism of glyco-peptide resistance. A gene, designated vanZ, encodes for a putative protein whose function has not been completely elucidated, but that was shown to confer teicoplanin resistance when expressed independently in an E. faecium strain [44].
VanB is also commonly found in clinical enterococcal isolates and functions in a similar manner to the VanA cluster, with several important differences. The sensor kinase (VanSB) and response regulator (VanRB) share only a distant relationship (34 and 23% sequence identity, respectively) to their VanA homologs [43]. As such, VanSB is not activated in the presence of teicoplanin, allowing strains with the VanB pheno-type to remain susceptible to this antibiotic. Importantly, resistance to teicoplanin may arise from mutations in the sensor kinase, allowing the activation of the system in the presence of the antibiotic or impairment of the phosphatase regulatory ability of VanSB resulting in constitutive expression of the resistance genes [45]. VanB also lacks the vanZ gene and instead possesses vanW, whose function is also unknown.
An intrinsic feature of two species of enterococci, Enterococcus gallinarum and Enterococcus casseliflavus, is that they carry vanC, a chromosomally encoded gene cluster that confers (with other genes) low-level resistance to vancomycin through the change of the terminal dipeptide from d-Ala-d-Ala to d-Ala-d-Ser [46]. These species produce the altered d-Ala-d-Ser dipeptide via VanT, a serine racemase that catalyzes the change from l-Ser to d-Ser [47]. In addition, they also harbor a bifunctional d,d-pepti-dase/DdcY designated VanXYC that hydrolyzes the typical d-Ala-d-Ala precursors in the cytoplasm. Similar to VanC, VanE is also chromosomally encoded, expresses low-level resistance and is under the control of a single promoter [48]. Interestingly, Abadía Patiño et al. found that a functional VanE-resistant phenotype was fully expressed despite the inactivation of VanSE by a premature stop codon [49]. Activation and transcription of the operon by an undetermined endogenous TCS was proposed and underscores the robust nature of the enterococcal stress response.
The genes encoding the TCS of the van gene clusters are an elegant system that allows bacteria to balance the need of antimicrobial resistance and the cost in fitness they carry for the cell. Defects of this system, as evidenced in the VanD phenotype, can lead to an interesting series of compensatory mutations [50]. Enterococci expressing VanD lack an active signal TCS due to an inactivating mutation of VanSD or VanRD. As such, the resistant phenotype is constitutively expressed by the constant activation of the operon producing the d-Ala-d-Lac depsipeptide. This would be expected to saddle the bacteria with a substantial metabolic burden, as it is necessary to maintain a futile cycle of synthesis and destruction of d-Ala-d-Ala, despite the absence of the glycopeptide threat. However, these strains also harbor a second mutation in the gene ddl, encoding the ligase responsible for the production of normal peptidogly-can precursors. As a result, they express the resistant phenotype without a substantial reduction in fitness. In fact, loss of the ddl gene in other van cluster-containing enterococci (most commonly VanB) leads to vancomycin dependence as these bacteria are unable to synthesize normal peptidoglycan precursors and require the presence of vancomycin to activate the van operon in order to build a cell wall [51]. In this case, restoration of normal growth occurs with either a ‘gain-of-function’ mutation of ddl or constitutive expression of the van operon [52].
Mechanisms of DAP resistance
The bacterial cell membrane (CM) is a crucial structural and functional component that regulates a vast majority of cell processes by connecting the external milieu with the cytoplasm. This structure also serves to keep out toxic compounds and provides an anchor for vital membrane-based proteins. DAP is a lipopeptide antibiotic that targets the CM and is related to many cationic antimicrobial peptides (CAMPs) that are produced by the innate immune system of eukaryotic organisms. Insertion of DAP into the CM requires the presence of calcium ions and appears to bind preferentially at the division septum plane. Once inside the membrane, DAP oligomerizes in the outer leaflet and the DAP complexes reach the inner leaflet of the CM forming a ‘pore’ structure that disrupts the integrity and functionality of the CM resulting in a variety of processes (including leakage of ions) that lead to cell death [53]. Oligomerization of DAP in the outer leaflet appears to depend on the presence of the phospholipid phosphatidylglycerol [54], whereas another phospholipid, cardiolipin (CL), influences the ability of DAP oligomers to reach the inner leaflet of the CM [55]. In Bacillus subtilis, DAP has been shown to cause recruitment of a cell division protein (DivIVA) at the sites of CM damage, resulting in abnormal cell wall synthesis and defective cell division [56]. Of interest, enterococci are less susceptible to DAP than staphylococci, a fact that is reflected by the susceptibility breakpoints, which are four-times higher for enterococcal isolates than for staphylococci [57].
Recent work has shed light on the genetic changes responsible for the DAP non-susceptible phenotype (hereafter referred to as DAP resistance [DAP-R] for clarity) in enterococci. Sequencing of a clinical strain pair of E. faecalis that developed DAP-R over the course of therapy revealed three genes responsible for the resistance phenotype [58]. The first is liaF that encodes a member of a three-component regulatory system (LiaFSR) that has been found to orchestrate the cell-envelope response to stress in gram-positive bacteria. In B. subtilis, LiaF negatively regulates LiaS, which is a histidine kinase that, upon cell-envelope stress, phosphorylates the response regulator LiaR [59]. In E. faecalis, a single deletion of an isoleucine at position 177 of LiaF increased the DAP MIC from 1 to 4 μg/ml (established breakpoint is 4 μg/ml) and, more importantly, abolished the bactericidal activity of DAP [60]. The other two genes implicated in DAP-R are involved in phospholipid metabolism: gpdD, which encodes a glycerol-phosphodiester phosphodiesterase, and cls, which encodes for a cardiolipinsynthase (Cls) [58,61].
In S. aureus, the actual mechanism of DAP resistance is related to the increase in cell envelope surface charge due to changes in MprF (a transmembrane protein that increases the content of the positive charged phospholipid lysyl-phosphatidylglycerol in the outer leaflet of the CM). Under this scenario, positively charged DAP is ‘repelled’ from the cell surface (also with positive charge due to high content of lysyl-phosphatidylglycerol) preventing its interaction with the CM [62]. In contrast, recent evidence in E. faecalis suggested that this microorganism uses a different strategy to withstand the DAP attack. Indeed, using a fluorescent derivative of DAP, Tran et al. showed that binding of DAP to the CM was similar between a DAP-susceptible E. faecalis and its DAP-resistant derivative [63], suggesting that repulsion of DAP from the cell surface was not relevant for the mechanism of DAP resistance. Instead, the resistance phenotype was associated with redistribution of CL microdomains in the CM which were relocated from the septum (the principal target of DAP) to non-septal areas. These CL microdomain remodeling was associated with changes in the LiaFSR system (see above). From these experiments, the authors hypothesized that a novel mechanism of resistance involved a stepwise process in which mutations in liaFSR resulted in mobilization of CL microdomains from the septum. As a result, DAP is diverted from the septum to other non-septal areas. The mechanism of resistance is completed by changing the content of CM phos-pholipids; the content of phosphatidylglycerol is decreased preventing DAP oligomerization and CL microdomains may ‘trap’ the diverted antibiotic away from the septum and prevent it from reaching the inner leaflet of the CM (Figure 1). Evidence for this ordered progression of resistance was recently provided by experimental evolution in vitro of E. faecalis [64]. Moreover, the important role of cls (encoding CL synthase) in mediating the DAP resistance phenotype was supported by experiments in which the overexpression of CL in a plasmid was sufficient to increase the DAP MICs.
Figure 1. Proposed mechanisms of daptomycin resistance in enterococci.
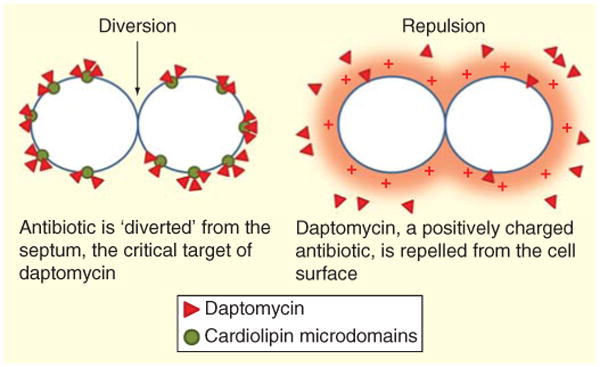
Two main mechanisms have been postulated to mediate DAP resistance in enterococci. The first is diversion of the antibiotic from the septum (black arrow) by redistribution of cardiolipin microdomains away from the division plane at the septum level. This mechanism, characterized in Enterococcus faecalis only, is initially mediated by substitutions in the LiaFSR signaling system that controls cell envelope homeostasis. Changes in phospholipid synthesis enzymes (cardiolipin synthase and possibly a glycerol-phosphodiester phosphodiesterase) complete the resistance phenotype. The second mechanism, seen in E. faecium, is electrostatic repulsion of the positively charged daptomycin/calcium complex from the cell membrane. Several genes may be involved in this mechanism. DAP: Daptomycin.
Development of DAP resistance in E. faecium seems to be more common than that observed in E. faecalis, since E. faecium isolates are commonly MDR and DAP is more frequently used against these organisms. As opposed to E. faecalis, DAP-R in E. faecium appears to be mainly dependent on repulsion of DAP from the cell surface rather than diversion from septal areas (Figure 1). The genetic basis for DAP resistance in E. faecium also appears to be different from E. faecalis, although genes encoding regulatory systems that are involved in cell envelope homeostasis and phospholipid metabolism also appear to play an important role. A recent comparative analysis of the genomes from a clinical strain-pair of E. faecium that developed resistance during therapy revealed changes in eight different genes, including a TCS capable of modulating the cell-wall stress response to DAP and designated YycFG [65]. Specific to the low G + C gram-positive bacteria, this system plays an important role in the homeostasis of the cell wall with high activity during cell division [66]. The mechanism by which changes in this system facilitate DAP-R in enterococci is not entirely clear and is currently part of active investigations. Another gene that was found to have changes and is hypothesized to be important is cfa, which encodes for a cyclopropane synthase involved in phospholipid metabolism. This enzyme adds a methylene group to the double bond of unsaturated fatty acids, which act to stabilize the CM to respond to a variety of cell envelope stressors [67]. Of note, changes in cls have also been commonly identified in DAP-R E. faecium isolates.
The LiaFSR system has also been implicated in DAP-R and tolerance in E. faecium. Recently, Diaz et al. analyzed the genomes of 19 E. faecium isolates with a wide range of DAP MICs (from 3 to 48 μg/ml) and compared them with all publicly available genomes, specifically searching for genetic changes previously associated with DAP-R (both in enterococci and staphylococci). Interestingly, the most frequent mutations identified were in liaFSR, followed by changes in the above-mentioned YycFG systems, supporting the hypothesis that changes in TCS are a pivotal step toward DAP-R in enterococci [68]. Furthermore, the majority (75%) of DAP-susceptible E. faecium bacteremia isolates whose MIC was in the higher range of susceptibility (i.e., between 3 and 4 μg/ml) harbored mutations in LiaFSR [69]. Conversely, none of the isolates of the same collection with DAP MIC ≤2 μg/ml exhibited changes in this system. More importantly, these changes were sufficient to abolish the in vitro bactericidal activity of DAP [60] and were associated with a clinical failure in a neutropenic patient with VRE bacteremia [70].
Interestingly, development of DAP resistance is inversely related to increased susceptibility to β-lactams. Indeed, combinations of DAP with ampicillin, ceftriaxone and ceftaroline have been noted to be synergistic in vitro and to improve clinical outcomes of patients failing DAP therapy [71–73]. Recent data showed that synergy with ampicillin was dependent on changes in LiaFSR but not in YycFG, suggesting the possibility that the synergism with β-lactams is influenced by the particular signaling pathway through which DAP resistance (or tolerance) is mediated [68]. Elucidation of the mechanism behind this interaction requires further study, as genetic information of DAP-resistance-associated mutations may eventually become relevant for clinicians as a useful tool to predict DAP failure and/or the need (and usefulness) of combination therapy.
Mechanisms of resistance to antibiotics that interfere with protein synthesis
Aminoglycosides
Enterococci display intrinsic tolerance (manifested by the lack of bactericidal activity) to the aminoglycosides. This phenomenon seems to be mediated by two main factors: poor uptake of the antibiotic requiring higher concentrations to promote entrance into the intracellular space and inactivation by covalent modification of the hydroxyl or amino groups of the aminoglycoside molecule carried out by naturally occurring enterococcal enzymes, creating a steric hindrance and decreasing the binding to the ribosomal target. Indeed, E. faecium possess a chromosomally encoded 6′-acetyltransferase enzyme (AAC(6′)-Ii) capable of modifying tobramycin, sisomicin, kanamycin and netilmicin [74]. In addition, many clinical isolates also possess the enzyme APH(3′)-IIIa, which results in resistance to kanamycin and amikacin through its phosphotransferase ability. Additionally, enterococci are capable of modifying the ribosomal target via a ribosomal RNA (rRNA) methyltransferase known as EfmM [75]. This enzyme recognizes a specific cytidine at position 1404 of the 16S rRNA in E. faecium, and methylation of this residue confers resistance to kanamycin and tobramycin.
Due to the above issues, only two aminoglycosides (gentamicin and streptomycin) are reliably used in clinical practice (for synergism with β-lactams) due to the fact that these compounds are not readily affected by intrinsic enzymes produced by enterococci. However, high-level resistance to aminoglycosides, defined as an MIC >2000 μg/ml for streptomycin and 500 μg/ml for gentamicin (agar dilution method), abolishes the synergistic effect of these compounds. Resistance to streptomycin occurs by one of two mechanisms. ‘Absolute’ inhibition at the level of the ribosome was demonstrated in clinical isolates that possessed MICs to streptomycin >128,000 μg/ml by precipitating the ribosomal complex and showing that they were able to translate polyU RNA (through the quantification of radiolabeled phenyl-alanine) in the presence of the drug [76]. Enzymatic inactivation due to acquisition of a streptomycin adenyltransferase confers high-level resistance and abolishes synergy [77]. Similarly, high-level resistance to gentamicin is primarily due to a bifunctional modifying enzyme AAC(6′)-Ie/APH(2′)-Ia that possesses both 6′-acetyltransferase and 2′-phosphotransferase activities and confers resistance to all aminoglycosides except streptomycin [78]. Three other acquired genes encoding phosphotransferases that may affect the activity of gentamicin have been identified: APH (2′)-Ic, which was originally isolated from E. gallinarum and has since been found in E. faecium and E. faecalis [79], has activity against gentamicin and tobramycin, but not amikacin or netilmicin; Aph(2′)-Id, which confers resistance to gentamicin but not amikacin and has been identified in E. casseliflavus and E. faecium [80] and aph(2′)-Ib is a gene that has been described in E. faecium and its presence results in resistance to all amino-glycosides except for streptomycin and amikacin [81].
Oxazolidinones
Linezolid is a bacteriostatic agent with broad activity against gram-positive bacteria. It binds to the 23S rRNA and disrupts the docking of the aminoacyl-tRNA in the A site of the ribosome, thus inhibiting the delivery of peptides and the subsequent elongation of the polypeptide chain [82,83]. Mutations in genes encoding the 23S rRNA, which is an important part of the drug-binding site at the ribosome, are the most common mechanisms of linezolid resistance. Of note, enterococci, as many other bacteria, carry multiple copies of the 23S rRNA gene and the number of mutated alleles correlates with the resistance phenotype [84]. Recombination between these alleles has been demonstrated to accelerate the increase in MIC in the E. faecalis JH2-2 compared to a recombination-deficient JH2-2 mutant [85]. Among these changes in the domain V of the 23S rRNA, substitution G2576T is the most common (see Table 1 for complete list of known mutations; E. coli numbering) and selection for mutations in rRNA is associated with longer duration of therapy [86]. Additionally, mutations in the ribosomal proteins L3 and L4, which border the peptidyl-transferase center where linezolid binds, are associated with an increase in the linezolid MIC. These mutations were originally described in linezolid-resistant staphylococci and have subsequently been identified in resistant enterococci as well [87,88]. Enzymatic modification of the 23S rRNA by methylation of an adenine in position 2503 has also been described in enterococci [89]. The responsible gene, cfr which encodes for a methylase (Cfr), is a plasmid-borne determinant of resistance that has been found in clinical isolates of E. faecalis as well as in other clinically relevant gram-positive organisms such as staphylococci [90,91]. The cfr gene has been associated with the mobile transposable element IS256, whose sequence is common in MDR staphylococci and enterococci, and this sequence has been found to mediate the transfer of antibiotic resistance genes, as well as altering the promoter sequence of regulatory proteins or activate the expression of existing resistance determinants [92]. This phenomenon could explain the ability of cfr to spread across species and portends the possibility of widespread dissemination in the clinical setting. Data from an in vivo mouse peritonitis model suggested that cfr resistance in staphylococci may be overcome by linezolid doses that mimic human pharmacokinetics; however, mutations in the 23S rRNA resulted in therapeutic failure [93].
Table 1. Mechanisms of antibiotic resistance in enterococci.
| Site of resistance | Strategy | Antibiotic | Gene(s) or gene product | Origin | Notes |
|---|---|---|---|---|---|
| Cell wall | Decreased affinity for PBPs | β-Lactams | pbp5 | Intrinsic | Low-affinity PBPs allow peptidoglycan synthesis in the presence of β-lactams |
| Drug inactivation | β-Lactams | blaZ | Acquired | β-Lactamase. Low-level constitutive production may be missed on routine laboratory screening | |
| Alternate pathway | β-Lactams/glycopeptides | LDTfm | Intrinsic | l,d-Transpeptidation only observed in vitro | |
| Cell signaling | Cephalosporins |
croRS ireK |
Intrinsic | Mutations in these pathways sensitize enterococci to cephalosporins | |
| Altered target | Glycopeptides | van operons | Acquired or intrinsic | Intrinsic low-level resistance in Enterococcus gallinarium and Enterococcus casseliflavus | |
|
| |||||
| Cell membrane | Altered target | Daptomycin | LiaFSR, YycFG Cls, GdpD | Acquired | Mutations in these genes alter membrane structure and composition leading to either repulsion or diversion of the antibiotic from the cell membrane target |
|
| |||||
| Ribosome | Decreased drug uptake | Aminoglycosides | – | Intrinsic | Polar molecules have difficulty penetrating to cytoplasm |
| Inactivation | Aminoglycosides (except gentamicin and streptomycin) | AAC(6′)-Ii APH(3′)-IIIa |
Intrinsic Intrinsic |
Chromosomally encoded AAC and APH | |
| Streptomycin | aadA/ANT(3′) | Acquired | Streptomycin adenyltransferase | ||
| Gentamicin | AAC(6′)-Ie/APH (2′)-Ia | Acquired | Inactivates all AG except streptomycin | ||
| Q/D | vatD and vatE | Acquired | Streptogramin A acetyltransferase | ||
| vgbA and vgbB | Acquired | Streptogramin B lactonase | |||
| Target modification | Aminoglycosides | efmM | Acquired | 16S rRNA methylase at C1404 | |
| Linezolid | rRNA | Acquired | Mutations in genes coding for 23S rRNA: G2576T, G2505A, U2500A, G2447U, C2534U, G2603U | ||
| Ribosomal protein | Acquired | Mutations in proteins L3 and L4 | |||
| cfr | Acquired | Methylation of A2503, associated with IS256 | |||
| Macrolides/Lincosamides/Streptogramins | ermB | Acquired | Methylation of A2508, MLSB phenotype | ||
| Efflux | Q/D | lsa | Intrinsic | Enterococcus faecalis only, putative ATP-binding protein. Not confirmed to function as an efflux protein | |
| msrC | Intrinsic | Enterococcus faecium | |||
| eatA | Acquired | E. faecium, C1349T mutation confers resistance | |||
| Tetracycline | tetK and tetL | Acquired | Resistance to tetracycline but not minocycline | ||
| Target protection | Tetracyclines | tetM, tetO and tetS | Acquired | Resistance to tetracycline and minocycline, but not tigecycline; transferable by Tn916 | |
| Nucleic acid metabolism | Altered target | Quinolones |
gyrA parC |
Acquired | Mutations in QRDR alter drug binding |
| Rifampicin | rpoB | Acquired | Mutations in β-subunit of RNA polymerase | ||
| Efflux | Quinolones | norA | Acquired | E. faecium | |
| Target protection | Quinolones | qnr | Acquired | Binds gyrase, described in E. faecalis | |
| Alternate pathway | TMP/SMX | – | Intrinsic | Use of exogenous folate | |
AAC: Acetyltransferase; AG: Aminoglycoside; ANT: Adenyltransferase; APH: Phosphotransferase; ATP: Adenosine triphosphate; LDT: l,d-Transpeptidase; MLSB: Macrolide/lincosamide/streptogramin B; PBP: Penicillin-binding protein; Q/D: Quinupristin/dalfopristin; QRDR: Quinolone resistance-determining regions; TMP/SMX: Trimethoprim/sulfamethoxazole.
Streptogramins/macrolides/lincosamides
Q/D is a mixture of pristinamycin derivatives, streptogramin A (dalfopristin) and B (quinupristin), which is effective against E. faecium, but not E. faecalis. Indeed, E. faecalis possess a chromosomal gene named lsa (for lincosamide and streptogramin A resistance), which encodes for a putative protein with an ATP-binding cassette motif of transporter proteins but not the trans-membrane region that would be expected for an efflux pump [94]. The exact molecular function and how it mediates resistance remains unknown, but its presence provides all E. faecalis with intrinsic resistance to streptogramin A and linco-samides, which explains the lack of action of Q/D against these microorganisms. Moreover, resistance to macrolides, lincosa-mides and streptogramin B (known as the MLSB phenotype) is prevalent in enterococci [95]. Cross-resistance with all macrolides arises from modification of the 23S rRNA target (A2508, as opposed to modification of A2503 by cfr in linezolid resistance) by a variety of methylase genes, most commonly ermB [96,97].
The mechanism of bactericidal action of Q/D results from a synergistic effect of both pristinamycin compounds. The binding of dalfopristin induces a conformational change in the ribo-some that unmasks a high-affinity binding site for quinupristin, leading to irreversible inhibition of the ribosome complex [98]. Resistance to Q/D in E. faecium is mediated by several mechanisms. First, modification of dalfopristin via the acetyltransferases VatD and VatE renders it ineffective, abolishing the synergy observed with quinupristin [99]. A second mechanism of resistance, originally described in staphylococci, involves the enzymatic cleavage of the ring structure of streptogramin B by the lactonases VgbA and VgbB [100]. Interestingly, the MLSB phenotype conferred by the erm genes modifies the target for quinupristin (streptogramin B); however, dalfopristin, as a streptogramin A, remains active. However, in vivo the presence of ermB may affect the efficacy of Q/D. Indeed, this phenomenon was demonstrated in a rat endocarditis model where Fantin et al. found that activity of Q/D was decreased in enterococci possessing MLSB due to the incomplete penetration of dalfopristin into the valvular vegetation, resulting in five treatment failures in the group with Q/D monotherapy, as compared with none in the amoxicillin group [101]. Finally, efflux pumps such as msrC [96] have also been implicated in playing a role in removing Q/D from the cell and, more recently, a mutation in the eatA gene (for Enterococcus ABC transporter) was shown to confer resistance to susceptible E. faecium strains [102].
Tetracyclines & glycylcyclines
Tetracyclines exert their antibacterial effect by binding to the ribosome and interfering with the docking of aminoacyl-tRNA. This occurs via association with several loops of the 16S rRNA and the ribosomal protein S7, however, this is a reversible process and these agents are bacteriostatic [103]. Resistance is mediated by multiple genes, but follows two general strategies, efflux of the antibiotic and ribosomal protection. Efflux pumps encoded by tetK and tetL are plasmid-borne determinants that encode proteins with 14 α-helices that make up the transmembrane domains and confer resistance to tetracycline but not minocycline [104]. Expression of resistance is regulated by translational attenuation in the absence of the antibiotic due to the formation of a stem loop structure in the mRNA, which masks the second of two ribosomal binding sites [105]. In the presence of tetracycline, the ribosome complex is unable to synthesize the normal leader peptide, an alternate loop structure forms in the mRNA and the second binding site becomes accessible, allowing for synthesis of the efflux pump. The genes tetM, tetO and tetS are chromosomal resistance determinants, which confer resistance to doxycycline and minocycline as well as tetracycline and can be transferred via the Tn916 transposon [106,107]. These genes code for a protein with a significant homology to bacterial elongation factors (EFs), and like EFs they are able to hydrolyze GTP in the presence of the ribosome, which alters ribosomal conformation and displaces bound tetracycline [104].
Tigecycline, a glycylcycline, is a synthetic derivative of minocycline with a broad spectrum of activity against gram-negative and gram-positive bacteria, including MRSA and VRE. This compound is US FDA-approved for the treatment of complicated skin and soft tissue infections and abdominal infections. Given the low achievable serum concentrations of the antibiotic, mono-therapy in serious infections is discouraged. Similar to all tetracy-clines, tigecycline binds to the 16S rRNA of the 30S subunit of the ribosome and inhibits the association of the aminoacyl-tRNA [108]. Unlike other tetracyclines, however, MICs are not affected by typical tetracycline resistance determinants [109]. To date, there have been two published reports of tigecycline resistance in enterococci, both related to intra-abdominal procedures [110,111]. The mechanism of resistance remains unknown.
Mechanisms of resistance to agents interfering with nucleic acid replication, transcription & synthesis
Quinolones
The introduction and relaxation of supercoils in DNA is important for transcription and the replication of the genome prior to cell division. The quinolones target two of the enzymes responsible for this process, DNA gyrase and topoisomerase IV. Both enzymes are tetramers composed of two different subunits: GyrA and GyrB form the DNA gyrase complex, while the topoisomerase IV is composed of ParC and ParE. The DNA gyrase introduces negative supercoils into the DNA strand, priming it for the initiation of replication and relaxing the strand in front of the advancing polymerase. On the other hand, topoisomerase IV separates the newly replicated interlocking DNA double helix allowing for segregation to occur before cell division. Both processes require double-stranded breaks in the DNA, and stabilization of the enzyme/DNA complex by quinolones results in a disruption of strand continuity and arrest of replication [112]. There is evidence that there may be differential inhibition of these two enzymes between gram-positive and gram-negative bacteria and varying grades of inhibition between the different types of quinolones [112,113].
Enterococci demonstrate low levels of intrinsic resistance to the quinolones, but can acquire high-level resistance through several mechanisms. Mutations in the target genes, specifically gyrA and parC, have been described in E. faecium and E. faecalis, but are absent from E. gallinarum and E. casseliflavus [114–116]. These changes affect the so-called ‘quinolone resistance-determining regions’, which presumably alter the binding affinity of the antibiotic. Externalization of the antibiotic through efflux pumps is another well-described mechanism of quinolone resistance. Among them, NorA and PmrA have been implicated in quinolone resistance in S. aureus and Streptococcus pneumoniae [117], and the former has also been described in E. faecium [112]. A third mechanism of resistance, found in E. faecalis [118], is mediated by qnr and encodes for a protein with a series of pentapeptide repeats similar to the plasmid-borne quinolone resistance genes described in Enterobacteriaceae. The presence of this protein is likely to protect DNA gyrase by decreasing DNA binding of the quinolone and the subsequent formation of the quinolone–gyrase complex [119].
Rifampicin
Rifampicin inhibits transcription of mRNA by binding to the β-subunit of the enterococcal DNA-dependent RNA polymerase. Resistance to these agents is widespread, occurring in 65.9% of E. faecium isolates from the USA and 67.5% of those from Europe [120]. Rifampicin resistance arises from a variety of mutations in the rpoB gene that encodes for the β-subunit of the RNA polymerase. Interestingly, a specific mutation in rpoB (H486Y) in both E. faecalis and E. faecium was shown to increase resistance to broad-spectrum cephalosporins, but did not affect other classes of cell-wall acting antibiotics (including ampicillin and vancomycin) [121]. The authors postulated that this was due to differential transcription of genes related to cephalosporin resistance by the mutated polymerase. In an independent study, assessing the fitness cost of rpoB mutations in E. faecium, it was noted that the same H486Y substitution showed minimal deleterious effect on growth both in vitro and in vivo [122]. Finally, Rand et al. described an isolate of E. faecium that developed rifampicin resistance without any evidence of mutations in the rpoB gene, enzymatic inactivation or efflux pumps. The exact mechanism of resistance is unclear, but interestingly, the resistant phenotype was reversible after incubation with subinhibitory concentrations of DAP [123].
Trimethoprim & sulfamethoxazole
Trimethoprim and sulfamethoxazole are inhibitors of bacterial enzymes involved in the folate synthesis pathway. Folic acid is necessary to carry out a variety of important cellular functions, including synthesis of nucleic acids, particularly thymidine. Most bacteria are unable to take up exogenous folate from the environment and instead must synthesize it from the p-amino benzoic acid precursor. Trimethoprim and sulfamethoxazole inhibit successive enzymes in this pathway, limiting the production of dihydrofolate and its subsequent conversion to tetrahy-drofolate. Enterococci show susceptibility to these compounds when tested in vitro; however, these compounds are ineffective in vivo due to the ability of enterococci to utilize exogenous sources of folate [124,125].
Alternative targets
In addition to the metabolic pathways targeted by traditional antimicrobial compounds, the rise of resistance has generated interest in adjunctive treatments to enhance host response and decrease the pathogenic potential of the enterococci. The presence of virulence factors, while not directly responsible for resistance, influence the ability of bacteria to persist in a hostile environment and resist host defenses. The widespread use of indwelling medical devices (e.g., central venous catheters, Foley catheters and endotracheal tubes) provide both a breach of host barriers and a surface amenable to supporting biofilm formation. A host of macromolecules associated with the enterococcal cell surface and secreted into the extracellular matrix act as passive barriers to prevent antimicrobial compounds from reaching their intended targets [126]. Moreover, at the interface of the host mucosa with external milieu, CAMPs of the innate immune system keep antimicrobial populations in check. These CAMPs possess a wide array of antimicrobial properties (including the ability to disrupt peptidoglycan synthesis, CM structure and activate autolysins) and individual proteins often display multiple modes of action, which may in part explain their effectiveness in the face of ever-evolving bacterial resistance [127]. In E. faecalis, Kandaswamy et al. demonstrated that human β-defensins are targeted to the division septum and specifically disrupt the translocation machinery responsible for export of virulence-associated macromolecules, even at subinhibitory concentrations [128]. Furthermore, it has been postulated that the long history of co-evolution between CAMPs and their bacterial targets could be fruitful grounds for developing therapies that temper the acquisition and dissemination of resistance determinants [129]. Host adaptive responses, specifically humoral immunity, have been exploited through vaccines to produce antibodies against pathogens. The lipoteichoic acids and diheteroglycans present in enterococcal cell walls are anti-genic motifs capable of inducing an antibody response that is protective against E. faecalis in a mouse bacteremia model [130]. Antibodies directed against these epitopes could be important adjuncts to antimicrobial therapy in the future; however, at present, they are limited by a lack of universal cross-reactivity and the presence of polysaccharides that protect the lipoteichoic acid motif from antibody binding to its corresponding target [131].
Expert commentary
Enterococci are prime examples of organisms with an impressive array of genetic versatility and unparalleled ability to recruit and express antimicrobial resistance determinants. These organisms have adapted through time to outcompete other bacteria in a specific biological niche such as the GI tract of eukaryotic organisms. From a simple commensal and tamed member of the intestinal microbiota, enterococci now have risen in importance and have become one of the leading causes of intra-hospital infections. This untaming of MDR enterococci has occurred with the massive increase in the use of antimicrobials in clinical medicine, which has played a significant role in the evolution and adaptation of these microorganisms. Indeed, selected by the use of broad-spectrum antimicrobials, their rugged durability enables them to persist and disseminate in the nosocomial environment. Furthermore, rising rates of obesity, diabetes and resulting comorbidities, advances in oncology and critical care and a demographic shift as the population ages are important events in modern medicine that increase the rates of hospital admissions and the prevalence of sicker patients.
Though enterococci lack the virulence armamentarium of S. aureus or pneumococci, they often pose a challenging problem to clinicians since antibiotic choices to treat these microorganisms are now extremely limited. Often, clinicians are faced with the dilemma of attempting to clear a deep-seated infection while balancing treatment-related toxicities. Such scenarios are not new to the treatment of enterococci. This is exemplified by a paper published in 1959 entitled ‘Deaf or Dead’ [132], in which the authors had to deal with extreme aminoglycoside toxicity to treat a patient with subacute endocarditis due to resistant E. faecalis.
In addition, enterococci is likely to function as a reservoir of drug resistance determinants and can serve as the springboard for the spread of these genes to other gram-positive pathogens. Indeed, it is now well documented that VRE are the source of vancomycin resistance genes that have been identified in MRSA [9,10]. Acquisition of vancomycin resistance in staphylococci with subsequent dissemination of such strains is deemed as one of the most pressing public health issues worldwide. Further exploration of the mechanisms of enterococcal resistance may therefore pay dividends in treating other infections and in preventing the widespread dissemination of resistance determinants in the future.
Five-year view
The dawn of the 21st century is seeing the advent of what many are calling the ‘post-antibiotic era’. As older therapies gave way to newer drugs, bacteria rapidly responded with a diverse array of defense tactics. We are fighting today's wars with yesterday's strategies, much like the massed troop assaults against fixed fortifications at the Somme and Verdun in World War I. Out of these battles, however, came new innovations, the rise of the airplane and the tank, and with them new strategies. Just two decades later, the idea of combined arms, manifested as Blitzkrieg, or ‘lightning war’ would change the battlefield of the 20th century.
Perhaps the idea of combined arms can be used to the clinician's advantage in the conflicts of the post-antibiotic era. Seemingly redundant therapy, such as the combination of the β-lactams, ampicillin and ceftriaxone, has been shown clinically to be as efficacious as more traditional combinations with aminoglycosides, with the benefit of less toxicity and the ability to bypass high-level resistance to aminoglycosides [133,134]. As more is known about the mechanisms by which enterococci subvert the assault of modern medicine, it will be possible to develop strategies that can be used to turn enterococcal biology against itself. The inverse relationship between DAP susceptibility and sensitivity to β-lactams [71], combinations of cephalosporins and fosfomycin [135] and the synergy between DAP and rifampicin [123] could provide a way to ‘hold the line’ against the rising tide of resistance. Once new agents are deployed, combination therapy could also assist in the prevention of resistance using the same rationale as combined antiretroviral therapy in human immunodeficiency virus infection.
Greater understanding of mechanisms of antibiotic action and resistance also offers the hope of new therapeutic targets to reload an empty antibiotic pipeline. Advances in the understanding of membrane physiology provided by research into DAP could provide new ways of attacking enterococcal phospholipid synthesis. Indeed, new agents need not kill on their own. By targeting the sensor and effector proteins of a stress response pathway, it may be possible to disable the defense network of a resistant microbe, leaving it vulnerable to decades old therapy. Moreover, monoclonal antibodies, which have been successfully used in cancer and auto-immune diseases, may be designed to specifically target enterococcal signal transduction pathways, leaving resistance gene clusters silent while the antibiotic pours in. Genomic medicine offers the promise of individually targeted treatments for every patient. There is no good reason to think antimicrobial therapy should be any different.
Key issues.
Enterococci are increasingly common nosocomial pathogens. The changing epidemiology of enterococcal infections with the rise of multidrug-resistant Enterococcus faecium in hospitals worldwide has important therapeutic implications.
Ampicillin plus an aminoglycoside (gentamicin or streptomycin), the traditional combination for severe enterococcal infections, is increasingly ineffective due to emergence of resistance.
Resistance to ampicillin in E. faecium is mediated by PBP5, a transpeptidase that functions in the presence of high concentrations of β-lactams.
Resistance to cephalosporins is an intrinsic feature of enterococci and is mediated in part by CroRS, a two-component signaling pathway, and a system with competing kinase and phosphatase activity (IreK and IreP) that function to control expression of resistance while preserving fitness.
Glycopeptide resistance is mediated by the van operons, of which nine have currently been described in enterococci. In general, they consist of genes that encode two-component signal transduction systems, which activate the genes responsible for the synthesis of modified peptidoglycan precursors and destruction of ‘normal’ (d-alanine ending) precursors.
The vanA gene cluster, conferring resistance to vancomycin and teicoplanin, is the most commonly encountered cause of resistance to glycopeptides in the clinical setting and can be transferred between enterococci.
Enterococci are often intrinsically resistant to most aminoglycosides due to the presence of the 6′-acetyltransferase enzyme AAC(6′)-Ii. As such, only gentamicin or streptomycin should be used to achieve synergy with cell-wall agents.
Ampicillin plus ceftriaxone is a β-lactam combination against Enterococcus faecalis that appears to be as good as the ‘standard of care’ (ampicillin plus gentamicin) but with much less toxicity.
Daptomycin (DAP) resistance is associated with multiple mutations but usually involves genes encoding regulatory systems that control cell envelope homeostasis and enzymes that synthesize cell membrane phospholipids and/or are involved in phospholipid metabolism.
The combination of DAP with β-lactams may offer promise in the future to restore DAP susceptibility and prevent emergence of resistance.
Linezolid resistance in enterococci continues to be low, but increasing reports in enterococci have been associated with longer duration of therapy.
Quinupristin/dalfopristin retains bactericidal activity in vitro against in E. faecium (not E. faecalis), but the presence of Erm methylases (which are frequently found in clinical isolates) may decrease the bactericidal effect in vivo and reduce therapeutic efficacy as monotherapy.
Enterococci are often resistant to quinolones with several mechanisms of resistance including mutation of the quinolone targets, efflux pumps and a transferable plasmid containing qnr, a gene similar to plasmid-borne quinolone resistance in the Enterobacteriaceae.
Continued research focused on understanding the mechanisms of resistance in enterococci is important to develop novel combination therapies or new antimicrobial compounds.
Acknowledgments
JM Munita is supported in part by a grant from the Chilean Ministry of Education and by Clinical Alemana de Santiago and Universidad del Desarrollo School of Medicine, Chile. CA Arias is supported by NIH-NIAD grant R01 AI093749. CA Arias has received lecture fees, research support and consulting fees from Pfizer Inc. Lectures and consulting honoraria from Novartis, Cubist, Forest Pharmaceuticals, Astra-Zeneca and Bayer Pharmaceuticals. Research support to CA Arias has been provided by Forest Pharmaceuticals and Theravance Inc.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hidron AI, Edwards JR, Patel J, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 3.Williamson R, Calderwood SB, Moellering RC, et al. Studies on the mechanism of intrinsic resistance to beta-lactam antibiotics in group D streptococci. J Gen Microbiol. 1983;129:813–22. doi: 10.1099/00221287-129-3-813. [DOI] [PubMed] [Google Scholar]
- 4.Schatz A, Waksman S. Effect of streptomycin and other antibiotic substances upon Mycobacterium tuberculosis and related organisms. Proc Soc Exp Biol Med. 1944;57:244–8. [Google Scholar]
- 5.Robbins WC, Tompsett R. Treatment of enterococcal endocarditis and bacteremia; results of combined therapy with penicillin and streptomycin. Am J Med. 1951;10:278–99. doi: 10.1016/0002-9343(51)90273-2. [DOI] [PubMed] [Google Scholar]
- 6.Baddour LM, Wilson WR, Bayer AS, et al. Infective endocarditis: diagnosis, antimicrobial therapy, and management of complications: a statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils on Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association: endorsed by the Infectious Diseases Society of America. Circulation. 2005;111:e394–434. doi: 10.1161/CIRCULATIONAHA.105.165564. [DOI] [PubMed] [Google Scholar]
- 7.Lebreton F, van Schaik W, McGuire AM, et al. Emergence of Epidemic Multidrug-Resistant Enterococcus faecium from Animal and Commensal Strains. MBio. 2013;4:e00534–13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–78. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang S, Sievert D, Hageman J, et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:1342–7. doi: 10.1056/NEJMoa025025. [DOI] [PubMed] [Google Scholar]
- 10.Ray A, Pultz N, Bhalla A, et al. Coexistence of vancomycin-resistant enterococci and Staphylococcus aureus in the intestinal tracts of hospitalized patients. Clin Infect Dis. 2003;37:875–81. doi: 10.1086/377451. [DOI] [PubMed] [Google Scholar]
- 11.Williamson R, Gutmann L, Horaud T, et al. Use of Penicillin-binding Proteins for the identification of enterococci. J Gen Microbiol. 1986;132:1929–37. doi: 10.1099/00221287-132-7-1929. [DOI] [PubMed] [Google Scholar]
- 12.Duez C, Hallut S, Rhazi N, et al. The ponA gene of Enterococcus faecalis JH2-2 codes for a low-affinity class A penicillin-binding protein. J Bacteriol. 2004;186:4412–16. doi: 10.1128/JB.186.13.4412-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Signoretto C, Boaretti M, Canepari P. Cloning, sequencing and expression in Escherichia coli of the low-affinity penicillin binding protein of Enterococcus faecalis. FEMS Microbiol Lett. 1994;123:99–106. doi: 10.1111/j.1574-6968.1994.tb07207.x. [DOI] [PubMed] [Google Scholar]
- 14.Sifaoui F, Arthur M, Rice L, et al. Role of Penicillin-Binding Protein 5 in Expression of Ampicillin Resistance and Peptidoglycan Structure in Enterococcus faecium. Antimicrob Agents Chemother. 2001;45:2594–7. doi: 10.1128/AAC.45.9.2594-2597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice LB, Carias LL, Hutton-Thomas R, et al. Penicillin-Binding Protein 5 and Expression of Ampicillin Resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2001;45:1480–6. doi: 10.1128/AAC.45.5.1480-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontana R, Aldegheri M, Ligozzi M, et al. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1994;38:1980–3. doi: 10.1128/aac.38.9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice LB, Bellais S, Carias LL, et al. Impact of Specific pbp5 Mutations on Expression of Beta-Lactam Resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2004;48:3028–32. doi: 10.1128/AAC.48.8.3028-3032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galloway-Peña JR, Rice LB, Murray BE. Analysis of PBP5 of early U.S. isolates of Enterococcus faecium: sequence variation alone does not explain increasing ampicillin resistance over time. Antimicrob Agents Chemother. 2011;55:3272–7. doi: 10.1128/AAC.00099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duez C, Zorzi W, Sapunaric F, et al. The penicillin resistance of Enterococcus faecalis JH2-2r results from an overproduction of the low-affinity penicillin-binding protein PBP4 and does not involve a psr-like gene. Microbiology. 2001;147:2561–9. doi: 10.1099/00221287-147-9-2561. [DOI] [PubMed] [Google Scholar]
- 20.Ono S, Muratani T, Matsumoto T. Mechanisms of Resistance to Imipenem and Ampicillin in Enterococcus faecalis. Antimicrob Agents Chemother. 2005;49:2954–8. doi: 10.1128/AAC.49.7.2954-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray BE. Beta-lactamase-producing enterococci. Antimicrob Agents Chemother. 1992;36:2355–9. doi: 10.1128/aac.36.11.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coudron PE, Markowitz SM, Wong ES. Isolation of a betalactamase-producing, aminoglycoside-resistant strain of Enterococcus faecium. Antimicrob Agents Chemother. 1992;36:1125–6. doi: 10.1128/aac.36.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackbarth CJ, Chambers HF. blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–9. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarti M, Campanile F, Sabia C, et al. Polyclonal diffusion of beta-lactamase-producing Enterococcus faecium. J Clin Microbiol. 2012;50:169–72. doi: 10.1128/JCM.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mainardi JL, Legrand R, Arthur M, et al. Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J Biol Chem. 2000;275:16490–6. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- •26.Sacco E, Hugonnet JE, Josseaume N, et al. Activation of the L,D-transpeptidation peptidoglycan cross-linking pathway by a metallo-D,D-carboxypeptidase in Enterococcus faecium. Mol Microbiol. 2010;75:874–85. doi: 10.1111/j.1365-2958.2009.07014.x. Though only seen in vitro, this is a novel pathway of resistance to many cell-wall active agents. It demonstrates the resiliency of the enterococci. [DOI] [PubMed] [Google Scholar]
- 27.Cremniter J, Mainardi JL, Josseaume N, et al. Novel mechanism of resistance to glycopeptide antibiotics in Enterococcus faecium. J Biol Chem. 2006;281:32254–62. doi: 10.1074/jbc.M606920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •28.Rice LB, Carias LL, Rudin S, et al. Role of class A penicillin-binding proteins in the expression of beta-lactam resistance in Enterococcus faecium. J Bacteriol. 2009;191:3649–56. doi: 10.1128/JB.01834-08. Detailed investigations in both Enterococcus faecalis and Enterococcus faecium that outlines the importance of the partner glycosyltransferase to PBP5 in resistance to β-lactam antibiotics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Arbeloa A, Segal H, Hugonnet JE, et al. Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J Bacteriol. 2004;186:1221–8. doi: 10.1128/JB.186.5.1221-1228.2004. Detailed investigations in both E. faecalis and E. faecium that outlines the importance of the partner glycosyltransferase to PBP5 in resistance to β-lactam antibiotics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancock LE, Perego M. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J Bacteriol. 2004;186:7951–8. doi: 10.1128/JB.186.23.7951-7958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comenge Y, Quintiliani R, Jr, Li L, et al. The CroRS two-component regulatory system is required for intrinsic beta-lactam resistance in Enterococcus faecalis. J Bacteriol. 2003;185:7184–92. doi: 10.1128/JB.185.24.7184-7192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Breton Y, Muller C, Auffray Y, et al. New insights into the Enterococcus faecalis CroRS two-component system obtained using a differential-display random arbitrarily primed PCR approach. Appl Environ Microbiol. 2007;73:3738–41. doi: 10.1128/AEM.00390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller C, Le Breton Y, Morin T, et al. The response regulator CroR modulates expression of the secreted stress-induced SalB protein in Enterococcus faecalis. J Bacteriol. 2006;188:2636–45. doi: 10.1128/JB.188.7.2636-2645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci USA. 2007;104:3508–13. doi: 10.1073/pnas.0608742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristich CJ, Little JL, Hall CL, et al. Reciprocal regulation of cephalosporin resistance in Enterococcus faecalis. MBio. 2011;2:e00199–11. doi: 10.1128/mBio.00199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall CL, Tschannen M, Worthey EA, et al. IreB, a Ser/Thr kinase substrate, influences antimicrobial resistance in Enterococcus faecalis. Antimicrob Agents Chemother. 2013;57:6179–86. doi: 10.1128/AAC.01472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vesić D, Kristich CJ. MurAA is required for intrinsic cephalosporin resistance of Enterococcus faecalis. Antimicrob Agents Chemother. 2012;56:2443–51. doi: 10.1128/AAC.05984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •38.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42:S25–34. doi: 10.1086/491711. Informative review of vancomycin resistance in enterococci. [DOI] [PubMed] [Google Scholar]
- 39.Guardabassi L, Agersø Y. Genes homologous to glycopeptide resistance vanA are widespread in soil microbial communities. FEMS Microbiol Lett. 2006;259:221–5. doi: 10.1111/j.1574-6968.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 40.Boyd DA, Willey BM, Fawcett D, et al. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob Agents Chemother. 2008;52:2667–72. doi: 10.1128/AAC.01516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, Lin D, Yan G, et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother. 2010;54:4643–7. doi: 10.1128/AAC.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebreton F, Depardieu F, Bourdon N, et al. D-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2011;55:4606–12. doi: 10.1128/AAC.00714-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••43.Depardieu F, Podglajen I, Leclercq R, et al. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev. 2007;20:79–114. doi: 10.1128/CMR.00015-06. Comprehensive review of the regulation of antibiotic resistance including two-component signaling, the role of insertion sequences and post-transcriptional regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arthur M, Depardieu F, Molinas C, et al. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995;154:87–92. doi: 10.1016/0378-1119(94)00851-i. [DOI] [PubMed] [Google Scholar]
- 45.Baptista M, Depardieu F, Reynolds P, et al. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol Microbiol. 1997;25:93–105. doi: 10.1046/j.1365-2958.1997.4401812.x. [DOI] [PubMed] [Google Scholar]
- 46.Arias CA, Courvalin P, Reynolds PE. vanC cluster of vancomycin-resistant Enterococcus gallinarum BM4174. Antimicrob Agents Chemother. 2000;44:1660–6. doi: 10.1128/aac.44.6.1660-1666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arias CA, Martín-Martinez M, Blundell TL, et al. Characterization and modelling of VanT: a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarum BM4174. Mol Microbiol. 1999;31:1653–64. doi: 10.1046/j.1365-2958.1999.01294.x. [DOI] [PubMed] [Google Scholar]
- 48.Fines M, Perichon B, Reynolds P, et al. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob Agents Chemother. 1999;43:2161–4. doi: 10.1128/aac.43.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abadía Patiño L, Courvalin P, Perichon B. vanE gene cluster of vancomycin-resistant Enterococcus faecalis BM4405. J Bacteriol. 2002;184:6457–64. doi: 10.1128/JB.184.23.6457-6464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Depardieu F, Kolbert M, Pruul H, et al. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2004;48:3892–904. doi: 10.1128/AAC.48.10.3892-3904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yowler CJ, Blinkhorn RJ, Fratianne RB. Vancomycin-dependent enterococcal strains: case report and review. J Trauma. 2000;48:783–5. doi: 10.1097/00005373-200004000-00035. [DOI] [PubMed] [Google Scholar]
- 52.San Millan A, Depardieu F, Godreuil S, et al. VanB-type Enterococcus faecium clinical isolate successively inducibly resistant to, dependent on, and constitutively resistant to vancomycin. Antimicrob Agents Chemother. 2009;53:1974–82. doi: 10.1128/AAC.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steenbergen JN, Alder J, Thorne GM, et al. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother. 2005;55:283–8. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]
- 54.Muraih JK, Harris J, Taylor SD, et al. Characterization of daptomycin oligomerization with perylene excimer fluorescence: stoichiometric binding of phosphatidylglycerol triggers oligomer formation. Biochim Biophys Acta. 2012;1818:673–8. doi: 10.1016/j.bbamem.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 55.Zhang T, Muraih JK, Tishbi N, et al. Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J Biol Chem. 2014;289:11584–91. doi: 10.1074/jbc.M114.554444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194:4494–504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cantón R, Ruiz-Garbajosa P, Chaves R, et al. A potential role for daptomycin in enterococcal infections: what is the evidence? J Antimicrob Chemother. 2010;65:1126–36. doi: 10.1093/jac/dkq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••58.Arias CA, Panesso D, McGrath DM, et al. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med. 2011;365:892–900. doi: 10.1056/NEJMoa1011138. Identification of multiple genes associated with daptomycin (DAP) resistance from a clinical strain pair of E. faecalis that developed resistance during therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jordan S, Junker A, Helmann JD, et al. Regulation of LiaRS-dependent gene expression in bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol. 2006;188:5153–66. doi: 10.1128/JB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munita JM, Tran TT, Diaz L, et al. A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 2013;57:2831–3. doi: 10.1128/AAC.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palmer KL, Daniel A, Hardy C, et al. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother. 2011;55:3345–56. doi: 10.1128/AAC.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones T, Yeaman MR, Sakoulas G, et al. Failures in clinical treatment of Staphylococcus aureus Infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother. 2008;52:269–78. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •63.Tran TT, Panesso D, Mishra NN, et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio. 2013;4:13. doi: 10.1128/mBio.00281-13. Investigation of the response to DAP in E. faecalis that demonstrated changes in membrane phospholipid content and subsequent relocalization of DAP from the dividing septum, suggesting a mechanism for DAP resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller C, Kong J, Tran TT, et al. Adaptation of enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother. 2013;57:5373–83. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••65.Tran TT, Panesso D, Gao H, et al. Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob Agents Chemother. 2013;57(1):261–8. doi: 10.1128/AAC.01454-12. Genomic analysis of the changes in E. faecium arising as a consequence of DAP therapy, including the newly implicated YycFG regulon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dubrac S, Bisicchia P, Devine KM, et al. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70:1307–22. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 67.Mishra NN, Bayer AS, Tran TT, et al. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One. 2012;7:e43958. doi: 10.1371/journal.pone.0043958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diaz L, Tran TT, Munita JM, et al. Whole-Genome Analyses of Enterococcus faecium Isolates with Diverse Daptomycin MICs. Antimicrob Agents Chemother. 2014;58:4527–34. doi: 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munita J, Panesso D, Diaz L, et al. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob Agents Chemother. 2012;56:4354–9. doi: 10.1128/AAC.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munita JM, Mishra NN, Alvarez D, et al. Failure of High-Dose Daptomycin for Bacteremia Caused by Daptomycin Susceptible Enterococcus faecium Harboring LiaSR Substitutions. Clin Infect Dis. 2014 doi: 10.1093/cid/ciu642. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakoulas G, Bayer AS, Pogliano J, et al. Ampicillin enhances daptomycin- and cationic host defense peptide-mediated killing of ampicillin- and vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother. 2012;56:838–44. doi: 10.1128/AAC.05551-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall Snyder A, Werth BJ, Barber KE, et al. Evaluation of the novel combination of daptomycin plus ceftriaxone against vancomycin-resistant enterococci in an in vitro pharmacokinetic/pharmacodynamic simulated endocardial vegetation model. J Antimicrob Chemother. 2014;69(8):2148–54. doi: 10.1093/jac/dku113. [DOI] [PubMed] [Google Scholar]
- 73.Sakoulas G, Rose W, Nonejuie P, et al. Ceftaroline restores daptomycin activity against daptomycin-nonsusceptible vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother. 2014;58:1494–500. doi: 10.1128/AAC.02274-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Costa Y, Galimand M, Leclercq R, et al. Characterization of the chromosomal aac (6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother. 1993;37:1896–903. doi: 10.1128/aac.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Galimand M, Schmitt E, Panvert M, et al. Intrinsic resistance to aminoglycosides in Enterococcus faecium is conferred by the 16S rRNA m5C1404-specific methyltransferase EfmM. RNA. 2011;17:251–62. doi: 10.1261/rna.2233511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eliopoulos GM, Farber BF, Murray BE, et al. Ribosomal resistance of clinical enterococcal to streptomycin isolates. Antimicrob Agents Chemother. 1984;25:398–9. doi: 10.1128/aac.25.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krogstad D, Korfhagen T, Moellering R, et al. Aminoglycoside-inactivating enzymes in clinical isolates of Streptococcus faecalis. An explanation for resistance to antibiotic synergism. J Clin Invest. 1978;62:480–6. doi: 10.1172/JCI109149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Courvalin P, Carlier C, Collatz E. Plasmid-mediated resistance to aminocyclitol antibiotics in group D streptococci. J Bacteriol. 1980;143:541–51. doi: 10.1128/jb.143.2.541-551.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chow J, Zervos M, Lerner S, et al. A novel gentamicin resistance gene in Enterococcus. Antimicrob Agents Chemother. 1997;41:511–14. doi: 10.1128/aac.41.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsai S, Zervos M, Clewell D, et al. A new high-level gentamicin resistance gene, aph (2″)-Id, in Enterococcus spp. Antimicrob Agents Chemother. 1998;42:1229–32. doi: 10.1128/aac.42.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chow J. Aminoglycoside resistance in enterococci. Clin Infect Dis. 2000;31:586–9. doi: 10.1086/313949. [DOI] [PubMed] [Google Scholar]
- 82.Shinabarger D, Marotti K, Murray R, et al. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob Agents Chemother. 1997;41:2132–6. doi: 10.1128/aac.41.10.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leach K, Swaney S, Colca J, et al. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell. 2007;26:393–402. doi: 10.1016/j.molcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 84.Marshall S, Donskey C, Hutton-Thomas R, et al. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2002;46:3334–6. doi: 10.1128/AAC.46.10.3334-3336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boumghar-Bourtchaï L, Dhalluin A, Malbruny B, et al. Influence of recombination on development of mutational resistance to linezolid in Enterococcus faecalis JH2-2. Antimicrob Agents Chemother. 2009;53:4007–9. doi: 10.1128/AAC.01633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bourgeois-Nicolaos N, Massias L, Couson B, et al. Dose dependence of emergence of resistance to linezolid in Enterococcus faecalis in vivo. J Infect Dis. 2007;195:1480–8. doi: 10.1086/513876. [DOI] [PubMed] [Google Scholar]
- 87.Locke JB, Hilgers M, Shaw KJ. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother. 2009;53:5275–8. doi: 10.1128/AAC.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen H, Wu W, Ni M, et al. Linezolid-resistant clinical isolates of enterococci and Staphylococcus cohnii from a multicentre study in China: molecular epidemiology and resistance mechanisms. Int J Antimicrob Agents. 2013;42:317–21. doi: 10.1016/j.ijantimicag.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 89.Toh S, Xiong L, Arias C, et al. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol. 2007;64:1506–14. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mendes R, Deshpande L, Castanheira M, et al. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob Agents Chemother. 2008;52:2244–6. doi: 10.1128/AAC.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diaz L, Kiratisin P, Mendes R, et al. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob Agents Chemother. 2012;56:3917–22. doi: 10.1128/AAC.00419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hennig S, Ziebuhr W. Characterization of the transposase encoded by IS256, the prototype of a major family of bacterial insertion sequence elements. J Bacteriol. 2010;192:4153–63. doi: 10.1128/JB.00226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diaz L, Kontoyiannis DP, Panesso D, et al. Dissecting the mechanisms of linezolid resistance in a Drosophila melanogaster infection model of Staphylococcus aureus. J Infect Dis. 2013;208:83–91. doi: 10.1093/infdis/jit138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Singh K, Weinstock G, Murray B. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother. 2002;46:1845–50. doi: 10.1128/AAC.46.6.1845-1850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hershberger E, Donabedian S, Konstantinou K, et al. Quinupristin-dalfopristin resistance in gram-positive bacteria: mechanism of resistance and epidemiology. Clin Infect Dis. 2004;38:92–8. doi: 10.1086/380125. [DOI] [PubMed] [Google Scholar]
- 96.Portillo A, Ruiz-Larrea F, Zarazaga M, et al. Macrolide resistance genes in Enterococcus spp. Antimicrob Agents Chemother. 2000;44:967–71. doi: 10.1128/aac.44.4.967-971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–85. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Canu A, Leclercq R. Overcoming bacterial resistance by dual target inhibition: the case of streptogramins. Curr Drug Targets Infect Disord. 2001;1:215–25. doi: 10.2174/1568005014606152. [DOI] [PubMed] [Google Scholar]
- 99.Werner G, Klare I, Witte W. Molecular analysis of streptogramin resistance in enterococci. Int J Med Microbiol. 2002;292:81–94. doi: 10.1078/1438-4221-00194. [DOI] [PubMed] [Google Scholar]
- 100.Korczynska M, Mukhtar T, Wright G, et al. Structural basis for streptogramin B resistance in Staphylococcus aureus by virginiamycin B lyase. Proc Natl Acad Sci USA. 2007;104:10388–93. doi: 10.1073/pnas.0701809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fantin B, Leclercq R, Garry L, et al. Influence of inducible cross-resistance to macrolides, lincosamides, and streptogramin B-type antibiotics in Enterococcus faecium on activity of quinupristin-dalfopristin in vitro and in rabbits with experimental endocarditis. Antimicrob Agents Chemother. 1997;41:931–5. doi: 10.1128/aac.41.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Isnard C, Malbruny B, Leclercq R, et al. Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP phenotype) in Enterococcus faecium. Antimicrob Agents Chemother. 2013;57:4463–9. doi: 10.1128/AAC.01030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165:359–69. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 104.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–60. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schwarz S, Cardoso M, Wegener HC. Nucleotide sequence and phylogeny of the tet(L) tetracycline resistance determinant encoded by plasmid pSTE1 from Staphylococcus hyicus. Antimicrob Agents Chemother. 1992;36:580–8. doi: 10.1128/aac.36.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pepper K, Horaud T, Le Bouguénec C, et al. Location of antibiotic resistance markers in clinical isolates of Enterococcus faecalis with similar antibiotypes. Antimicrob Agents Chemother. 1987;31:1394–402. doi: 10.1128/aac.31.9.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bentorcha F, De Cespédès G, Horaud T. Tetracycline resistance heterogeneity in Enterococcus faecium. Antimicrob Agents Chemother. 1991;35:808–12. doi: 10.1128/aac.35.5.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bauer G, Berens C, Projan S, et al. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J Antimicrob Chemother. 2004;53:592–9. doi: 10.1093/jac/dkh125. [DOI] [PubMed] [Google Scholar]
- 109.Fluit A, Florijn A, Verhoef J, et al. Presence of tetracycline resistance determinants and susceptibility to tigecycline and minocycline. Antimicrob Agents Chemother. 2005;49:1636–8. doi: 10.1128/AAC.49.4.1636-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Werner G, Gfrörer S, Fleige C, et al. Tigecycline-resistant Enterococcus faecalis strain isolated from a German intensive care unit patient. J Antimicrob Chemother. 2008;61:1182–3. doi: 10.1093/jac/dkn065. [DOI] [PubMed] [Google Scholar]
- 111.Cordina C, Hill R, Deshpande A, et al. Tigecycline-resistant Enterococcus faecalis associated with omeprazole use in a surgical patient. J Antimicrob Chemother. 2012;67:1806–7. doi: 10.1093/jac/dks122. [DOI] [PubMed] [Google Scholar]
- 112.Hawkey P. Mechanisms of quinolone action and microbial response. J Antimicrob Chemother. 2003;51(Suppl 1):29–35. doi: 10.1093/jac/dkg207. [DOI] [PubMed] [Google Scholar]
- 113.Oyamada Y, Ito H, Fujimoto K, et al. Combination of known and unknown mechanisms confers high-level resistance to fluoroquinolones in Enterococcus faecium. J Med Microbiol. 2006;55:729–36. doi: 10.1099/jmm.0.46303-0. [DOI] [PubMed] [Google Scholar]
- 114.López M, Tenorio C, Del Campo R, et al. Characterization of the mechanisms of fluoroquinolone resistance in vancomycin-resistant enterococci of different origins. J Chemother. 2011;23:87–91. doi: 10.1179/joc.2011.23.2.87. [DOI] [PubMed] [Google Scholar]
- 115.Werner G, Fleige C, Ewert B, et al. High-level ciprofloxacin resistance among hospital-adapted Enterococcus faecium (CC17) Int J Antimicrob Agents. 2010;35:119–25. doi: 10.1016/j.ijantimicag.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 116.Yasufuku T, Shigemura K, Shirakawa T, et al. Mechanisms of and risk factors for fluoroquinolone resistance in clinical Enterococcus faecalis isolates from patients with urinary tract infections. J Clin Microbiol. 2011;49:3912–16. doi: 10.1128/JCM.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hooper D. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31(Suppl 2):S24–8. doi: 10.1086/314056. [DOI] [PubMed] [Google Scholar]
- 118.Arsène S, Leclercq R. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob Agents Chemother. 2007;51:3254–8. doi: 10.1128/AAC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tran J, Jacoby G, Hooper D. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005;49:118–25. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deshpande L, Fritsche T, Moet G, et al. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis. 2007;58:163–70. doi: 10.1016/j.diagmicrobio.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 121.Kristich C, Little J. Mutations in the β subunit of RNA polymerase alter intrinsic cephalosporin resistance in Enterococci. Antimicrob Agents Chemother. 2012;56:2022–7. doi: 10.1128/AAC.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Enne V, Delsol A, Roe J, et al. Rifampicin resistance and its fitness cost in Enterococcus faecium. J Antimicrob Chemother. 2004;53:203–7. doi: 10.1093/jac/dkh044. [DOI] [PubMed] [Google Scholar]
- 123.Rand K, Houck H, Silverman J, et al. Daptomycin-reversible rifampicin resistance in vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother. 2007;59:1017–20. doi: 10.1093/jac/dkm045. [DOI] [PubMed] [Google Scholar]
- 124.Chenoweth C, Robinson K, Schaberg D. Efficacy of ampicillin versus trimethoprim-sulfamethoxazole in a mouse model of lethal enterococcal peritonitis. Antimicrob Agents Chemother. 1990;34:1800–2. doi: 10.1128/aac.34.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grayson M, Thauvin-Eliopoulos C, Eliopoulos G, et al. Failure of trimethoprim-sulfamethoxazole therapy in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1990;34:1792–4. doi: 10.1128/aac.34.9.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Otto M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr Top Microbiol Immunol. 2006;306:251–8. doi: 10.1007/3-540-29916-5_10. [DOI] [PubMed] [Google Scholar]
- 127.Peschel A, Sahl H. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–36. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 128.Kandaswamy K, Liew T, Wang C, et al. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci USA. 2013;110:20230–5. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gilmore M, Lebreton F, Van Tyne D. Dual defensin strategy for targeting Enterococcus faecalis. Proc Natl Acad Sci USA. 2013;110:19980–1. doi: 10.1073/pnas.1319939110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Theilacker C, Kaczyński Z, Kropec A, et al. Serodiversity of opsonic antibodies against Enterococcus faecalis – glycans of the cell wall revisited. PLoS One. 2011;6:e17839. doi: 10.1371/journal.pone.0017839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Theilacker C, Kaczynski Z, Kropec A, et al. Opsonic antibodies to Enterococcus faecalis strain 12030 are directed against lipoteichoic acid. Infect Immun. 2006;74:5703–12. doi: 10.1128/IAI.00570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Havard CW, Garrod LP, Waterworth PM. Deaf or dead? Br Med J. 1959;1:688–9. doi: 10.1136/bmj.1.5123.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fernández-Hidalgo N, Almirante B, Gavaldà J, et al. Ampicillin plus ceftriaxone is as effective as ampicillin plus gentamicin for treating Enterococcus faecalis infective endocarditis. Clin Infect Dis. 2013;56:1261–8. doi: 10.1093/cid/cit052. [DOI] [PubMed] [Google Scholar]
- 134.Munita JM, Arias CA, Murray BE. Editorial commentary: Enterococcus faecalis infective endocarditis: is it time to abandon aminoglycosides? Clin Infect Dis. 2013;56:1269–72. doi: 10.1093/cid/cit050. [DOI] [PubMed] [Google Scholar]
- 135.Farina C, Russello G, Chinello P, et al. In vitro activity effects of twelve antibiotics alone and in association against twenty-seven Enterococcus faecalis strains isolated from Italian patients with infective endocarditis: high in vitro synergistic effect of the association ceftriaxone-fosfomycin. Chemotherapy. 2011;57:426–33. doi: 10.1159/000330458. [DOI] [PubMed] [Google Scholar]


