Abstract
Prostate cancer (PCa) progression is regulated by the androgen receptor (AR); however, patients undergoing androgen deprivation therapy (ADT) for disseminated PCa eventually develop castration resistant PCa (CRPC). Studies showed that AR, a transcription factor, occupies distinct genomic loci in CRPC compared to hormone-naïve PCa; however, the cause for this distinction was unknown. The E3 ubiquitin ligase Nrdp1 is a model AR target modulated by androgens in hormone-naïve PCa but not in CRPC. Using Nrdp1, we investigated how AR switches transcription programs during CRPC progression. The proximal Nrdp1 promoter contains an androgen response element (ARE); we demonstrated AR binding to this ARE in androgen-sensitive PCa. Analysis of hormone-naive human prostatectomy specimens revealed correlation between Nrdp1 and AR expression, supporting AR regulation of Nrdp1 levels in androgen-sensitive tissue. However, despite sustained AR levels, AR binding to the Nrdp1 promoter and Nrdp1 expression were suppressed in CRPC. Elucidation of the suppression mechanism demonstrated correlation of Nrdp1 levels with nuclear localization of the scaffolding protein Filamin A (FlnA) which, as we previously showed, is itself repressed following ADT in many CRPC tumors. Restoration of nuclear FlnA in CRPC stimulated AR binding to Nrdp1 ARE, increased its transcription, and augmented Nrdp1 protein expression and responsiveness to ADT, indicating that nuclear FlnA controls AR-mediated androgen-sensitive Nrdp1 transcription. Expressions of other AR-regulated genes lost in CRPC were also re-established by nuclear FlnA. Thus our data demonstrate that nuclear FlnA promotes androgen-dependent AR-regulated transcription in PCa, while loss of nuclear FlnA in CRPC alters the AR-regulated transcription program.
Keywords: Castration resistant prostate cancer, AR/androgen receptor, FLRF/RNF41/Nrdp1, ABP280/FilaminA, HER3/ErbB3
INTRODUCTION
Prostate cancer (PCa) development and progression is regulated by the androgen receptor (AR), a steroid nuclear receptor, both in early as well as in advanced stages of the disease (Yuan, et al. 2014). While localized PCa is mostly treated by surgery or radiation therapy, AR inhibition is a cornerstone of treatment for disseminated PCa. Although initially effective, patients undergoing androgen deprivation therapy (ADT) eventually fail this treatment due to the development of castration resistant PCa (CRPC) (Mitsiades 2013; Nelson 2012). Most CRPC patients develop relapsed tumors with high levels of AR-regulated activity, as evidenced by elevated serum levels of prostate specific antigen (PSA), an AR-dependent gene (Karantanos, et al. 2013; Yuan et al. 2014). With the advent of strong AR antagonists such as enzalutamide and androgen synthesis inhibitors such as abiraterone acetate, it was observed that 65–70% CRPC patients initially responded to these drugs (de Bono, et al. 2011; Scher, et al. 2012), confirming continued AR activity in CRPC.
One of the perplexing aspects of PCa progression observed using patient-derived tissues is that AR target genes identified in low grade localized PCa are often downregulated in high grade, high risk PCa and in metastasis, despite continued AR expression (Tomlins, et al. 2007). In addition, studies showed that the AR occupies a distinct set of genomic loci in CRPC compared to those occupied in androgen-dependent cells (Decker, et al. 2012; Hu, et al. 2012; Wang, et al. 2009b). AR binding sites in untreated PCa were lost upon ADT initiation, and although a proportion of these were regained with the emergence of CRPC and AR resurgence, others were not (Sharma, et al. 2013). AR mutations and alternately spliced AR variants that lack the ligand binding domain (LBD) (Guo, et al. 2009; Hu et al. 2012) offer partial explanation for the change in targets, but this discrepancy is observed even in tumors that do not harbor altered AR forms. A consequence of the altered AR transcriptome is that pathways not activated by AR in hormone naïve tumors are upregulated in CRPC, promoting tumor progression (Decker et al. 2012; Hu et al. 2012; Wang et al. 2009b). The overall goal of the present studies was to understand how the AR regulates a different transcription program in CRPC and whether this altered program can be reversed.
We previously showed that in androgen-dependent cells, the AR suppresses levels of the receptor tyrosine kinase ErbB3 by stimulating the E3 ubiquitin ligase Nrdp1 (Chen, et al. 2010b), which causes ErbB3 degradation (Cao, et al. 2007; Wu, et al. 2004; Yen, et al. 2006). However, in CRPC cells, Nrdp1 levels were downregulated despite continued AR expression (Chen et al. 2010b). Here, we identify Nrdp1 as an AR target gene in hormone-naive PCa but not in some CRPC tumors. Using Nrdp1 as a model, we investigated why the AR did not transcribe certain genes in CRPC cells although they were transcribed in hormone-naïve cells.
Transcriptional activity of the AR is tightly regulated via interaction with co-regulators (Parker, et al. 2013; van de Wijngaart, et al. 2012). The presence or absence of co-regulators determines transcriptional efficiency of the AR, independent of AR splicing or mutations. Here, we show that a scaffolding protein, Filamin A (FlnA), affects AR-regulated transcription of Nrdp1. FlnA is a 280kDa protein consisting of an actin binding domain (ABD) followed by 24 repeats of 96-amino acid units (Loy, et al. 2003). Upon proteolysis, FlnA cleaves to a 170kDa N-terminal and an 110kDa C-terminal fragment which further cleaves to a 90 kDa fragment (Loy et al. 2003). The 90kDa C-terminal fragment binds to AR and translocates to the nucleus (Ozanne, et al. 2000), whereas the N-terminal fragment remains cytoplasmic (Loy et al. 2003). Our previous studies showed that nuclear FlnA is observed in >75% of localized tumors but <45% of metastatic CRPC lesions (Bedolla, et al. 2009). We demonstrated that in the presence of nuclear FlnA, CRPC cells were sensitized to anti-androgens (Wang, et al. 2007), but ADT inhibited FlnA proteolysis, thereby preventing FlnA translocation to the nucleus, which persisted in CRPC (Mooso, et al. 2012). Thus, loss of FlnA nuclear localization is one characteristic of CRPC development, and in cells where resistance to anti-androgen therapy was FlnA-regulated, restoration of nuclear FlnA reinstated androgen-sensitive cell growth (Mooso et al. 2012; Wang et al. 2007).
In this study, we demonstrate that Nrdp1 is a direct AR transcriptional target, but only in the presence of nuclear FlnA, which is present in normal prostate and in hormone-naïve PCa but is reduced in most CRPC. Further, we observe that this influence of nuclear FlnA is also effective in the transcription of various other AR-regulated genes whose expression is reduced in CRPC, but is restored when nuclear FlnA levels are increased. In addition, our data show that nuclear FlnA-induced AR transcriptional activity is ligand-dependent, thus, expression of FlnA-upregulated genes can be suppressed by the use of anti-androgens, thereby restoring androgen-sensitivity to CRPC cells. In contrast, in the absence of nuclear FlnA, the expression of AR-transcribed genes, including PSA, are not suppressed by anti-androgens. These results indicate that loss of nuclear FlnA is one reason why in some CRPC cells, AR transcribes an altered transcriptional program, and that this program can be restored when FlnA is induced to re-enter the nucleus.
MATERIALS AND METHODS
Patient Characteristics
All data was collected with approval from the University of California Davis (UCD) or VA Northern California Health Care System (VANCHCS) Institutional Review Board. Sections from formalin fixed paraffin-embedded prostate tumors of 157 patients who underwent prostatectomy at UCD (79) or VANCHCS (78) were analyzed for these studies. Patient characteristics are described in Table 1. Tumor and non-tumor areas were identified by a pathologist and 60µm core samples were extracted. Specimens were arranged in triplicate in a tissue microarray (TMA) using a Beecher Instruments Manual Tissue Arrayer (Sun Prairie, WI). Hematoxylin-eosin staining was used as a reference for interpreting the additional sections of the TMA stained with antibodies to Nrdp1 and AR.
Table 1. Patient Characteristics.
Of the 157 patients included in this study, matching tumor and non-tumor tissue was available for 78, while for the remainder, only tumor tissue was available.
| NUMBER OF PATIENTS | 157 | |||
| RACE | Caucasian | 122 | ||
| African American | 22 | |||
| Mean BMI | 28.3 ± 4.69 | MAX: 45.5 | MIN: 19.7 | |
| Mean Pre-op PSA | 9.946 ± 8.114 | MAX: 57.8 | MIN: 1.0 | |
| GLEASON | Gleason 5–6 | 72 | ||
| Gleason 7 | 67 | |||
| Gleason 8–9 | 18 | |||
| STAGE | STAGE T1 | 43 | ||
| STAGE T2 | 88 | |||
| STAGE T3 | 18 | |||
| POSITIVE MARGINS | 34/114 | |||
| PSA FAILURE | 53/143 | |||
Cell culture and materials
LNCaP, CWR22Rv1 (ATCC, Manassas, VA), C4-2 (UroCor, Oklahoma City, OK), C4-2B (MDA Cancer Center, Houston, TX), CWR-R1 (Dr. Elizabeth Wilson, University of North Carolina), LNCaP-AI (Wang et al. 2007) and pRNS-1-1 (Dr. Johng Rhim, University of the Health Sciences, Bethesda, MD) cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic solutions. Stable transfectants of pRNS-1-1 cells expressing wild-type AR (WT-AR) could only be cultured in media containing 10% charcoal stripped serum (CSS) as they were growth-inhibited by the levels of hormones present in FBS. Stable transfectants of pRNS-1-1 expressing AR(T877A) and C4-2 cells expressing FlnA(16-24) were cultured in RPMI+10%FBS. All cell lines used here were investigated for the presence of contaminants and their cellular origins were verified prior to use. Cells were transiently transfected using Lipofectamine 2000 reagent (Invitrogen, Grand Island, NY) according to manufacturer’s specifications. Casodex (bicalutamide) was kindly provided by AstraZeneca, Cheshire, UK. Antibodies to the following proteins were employed: Nrdp1 (US Biologicals, San Antonio, TX), ErbB3, Lamin A, and α-Tubulin (Cell Signaling Technology, Beverly, MA), AR and β-Actin (Santa Cruz Biotechnology, Santa Cruz, CA), FlnA (Abcam, Cambridge, MA), and GAPDH (Millipore, Billerica, MA). Primers are described in Supplementary Table 1,Supplementary Table 2.
Plasmids
pCMV-FlnA, FlnA(16-24) and FlnA(1-15) plasmids were kindly provided by Dr. E.W. Yong, National University of Singapore, Singapore, and human PSA-luciferase construct (hPSA-luc) containing two AREs in the proximal PSA promoter was kindly provided by Dr. XuBao Shi, University of California Davis. Human Nrdp1-luciferase constructs pGl4.11 ARE3 and mutated ARE3 were constructed as follows: A 500bp fragment immediately upstream of the Nrdp1 transcriptional start site was amplified from LNCAP genomic DNA using primers CA TCA GAT GCGC GGT ACC GGT TAC GAA GCT CTG GGA TGC T and CA TCA GAT GCGC GCT AGC GAA GAC TCC TAC CAC TCG TCG C and then directionally cloned into the Kpn1 and Nhe1 cut pGL4.11 reporter construct (Promega). Mutagenesis was performed using the Stratagene QuikChangeII kit from Agilent technologies according to the manufacturer's instructions (Santa Clara, CA). Mutagenic primers were designed using the Agilent technologies QuikChange Primer Design program and were used to amplify nascent plasmid containing the desired mutation(s). All mutant plasmids described were fully sequenced for confirmation.
RNA inhibition
LNCaP cells were plated in 60 mm dishes and transfected with 50 pmoles of a pool of 3 duplexes sold as Filamin A siRNA (siRNA1, Santa Cruz Biotechnology, Santa Cruz, CA) with the following sequences: Strand #1: 5’-CCAUCACUGACAACAAAGA-3’, Strand #2: 5’-CUGCAGAGUUUAU-CAUUGA-3’, Strand #3: 5’-GCUACCUCAUCUCCAUCAA-3’. Control was a pool of 4 scrambled non-specific siRNA duplex (Santa Cruz Biotechnology, Santa Cruz, CA).
Mouse studies
All experiments were conducted as approved by UC Davis IACUC Committee. 4–5-week old nu/nu athymic male mice were obtained from Harlan Sprague Dawley, Inc. (Indianapolis, IN) and implanted subcutaneously with sustained release testosterone pellets (12.5 mg, 90-day release; Innovative Research of America, Sarasota, FL). Suspensions of CWR22 or CWR22Rv1 cells were made in 50% Matrigel solubilized basement membrane (BD Biosciences, Bedford, MA) and xenografts were established by subcutaneous injections of 2.5 × 106 cells/site. When palpable tumors were observed, the testosterone pellets were removed and animals were followed for approximately four weeks, after which the mice were euthanized; and the tumors were collected. Part of the tumors were processed for paraffin embedding for immunohistochemistry, while the rest were lysed and homogenized for western blotting in cell lysis buffer (50 mM Tris HCl, pH 7.4, 150mM NaCl, and 1% NP-40, and protease inhibitors: 0.1mM benzamidine, 1mM phenylmethylsulfonyl fluoride, 10mg/ml each of phenathroline, leupeptin, aprotinin, and pepstatin A) and phosphatase inhibitors: 20mM β-glycerol phosphate, 1mM Na-orthovanadate, and 10mM NaF. Proteins were quantitated using a BCA assay (Pierce, Rockford IL) and fractionated on 29:1 acrylamide-bis SDS-PAGE.
ChIP
Cells were treated using the Magna ChIP Chromatin Immunoprecipitation Kit (Millipore) according to the manufactures protocol. The lysates were sonicated using the Bioruptor UCD-200 (Diagenode, Denville, NJ) and immunoprecipitated using 3µg of ChIPAP+ Androgen Receptor (Millipore). After immunoprecipitation DNA was size selected by DNA electrophorisis between 100- and 300-bp and purified using the Gel Extraction Kit (Qiagen, Inc. CA). All experiments were conducted using a negative control –ARarfneg2, a region of the p14arf gene that the AR does not bind to, or ZNF333, a non-AR transcribed zinc-finger region, as per studies suggesting the use of an unresponsive region as negative control (Kidder, et al. 2011).
qPCR
Total RNA was prepared utilizing RNeasy mini kit (Qiagen, Inc. CA) based on the manufacturer’s protocol. cDNA was made using the Verso cDNA Synthesis kit (Thermo Scientific, Waltham, MA), according to manufacturer’s instructions. Expression levels were determined using the Luminaris Color HiGreen qPCR Master Mix (Thermo Scientific) and StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
Other Methods
Western blotting, MTT assays, immunofluorescence, immunohistochemistry and flow cytometry were performed as has been described earlier (Mooso et al. 2012). AR transcriptional activity was estimated in cells transfected with 2 µg of pGL3-hPSA-luc, pGL4.11-ARE3, or pGL4.11-mutated ARE3 and 1 µg β-galactosidase with or without co-transfection of 2 µg of the FlnA vectors as described earlier (Chen et al. 2010b). Subcellular Fractionation into cytoplasmic and nuclear fractions was conducted using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific) according to the manufacture’s protocol.
Statistical analysis
For immunohistochemical analysis of patient samples, median staining levels were compared between cancer and non-cancer cells from the same subject using Wilcoxon signed-rank tests. Median staining levels were classified as high or low and analyzed using chi-squared test for association, logistic regression or log-linear models and compared between levels of categorical demographic characteristics using Wilcoxon rank-sum tests for the case of demographic characteristics with two levels, or using Kruskal-Wallis tests for the case of demographic characteristics with more than 2 levels. The correlations between staining levels and continuous demographic characteristics were estimated using Spearmans' rho. Analyses were conducted using R, version 2.13.0 (R Development Core Team, 2011) or SAS version 9.3. Mice tumor data were analyzed by normalization of all measurements to pre-operation (sham or castration) measurements for each individual mouse, then mean and standard errors calculated for the aggregate group. For staining analysis, associations were based on Pearson’s product moment correlation coefficient. Similar statistical considerations had been reported earlier in more detail (Bedolla, et al. 2007; Kreisberg, et al. 2004).
RESULTS
Nrdp1 is a transcriptional target of the AR in hormone-sensitive PCa
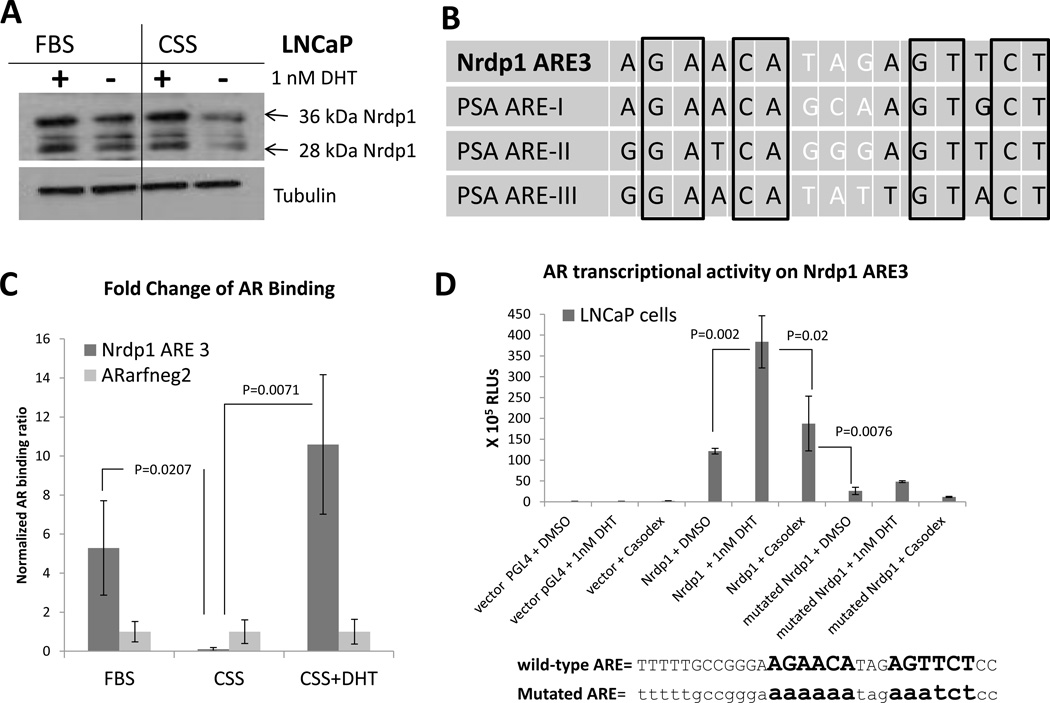
We previously demonstrated that expression of the E3 ubiquitin ligase Nrdp1 was androgen regulated in hormone-naïve PCa cells (Chen et al. 2010b). Prolonged culture of androgen-sensitive LNCaP cells in charcoal-stripped serum (CSS) (which was significantly stripped of various hormones including androgens) decreased expression of Nrdp1 (both 36kDa and 28kDa isoforms) compared to FBS (Figure 1A). Since CSS reduces a number of hormones and growth factors, to determine the specific effect of androgens, the cells were treated with the androgen dihydrotestosterone (DHT), a strong AR ligand, which significantly replenished Nrdp1 levels (Figure 1A). These results demonstrated hormone-sensitivity of Nrdp1 expression.
Figure 1. Nrdp1 is a transcriptional target of the AR in androgen-dependent LNCaP cells.
(A) LNCaP cells were cultured in complete media containing FBS or CSS with and without DHT (1 nM) for 72 hours. Cell lysates were blotted for Nrdp1 and two isoforms detected – 36 kDa and 28 kDa. Exposure to CSS caused a decrease in both 36 kDa and 28 kDa Nrdp1 but this effect was restored by DHT, showing that Nrdp1 is androgen regulated. (B) We identified an ARE in the proximal promoter of Nrdp1 gene. Comparison of PSA AREs and Nrdp1 ARE show that the PSA AREs and Nrdp1 ARE3 both contain a 15bp-palindromic ARE. (C) ChIP assay of AR binding in LNCaP cells to Nrdp1 ARE3. LNCaP cells were cultured in media containing CSS for 48 hours and then switched to complete media containing FBS or CSS with or without DHT. Chromatin samples were immunoprecipitated with an anti-AR antibody and analyzed by qPCR with primers flanking the Nrdp1 ARE3 region against a negative control (ARarfneg2). In LNCaP cells grown in normal FBS media the AR binds to the Nrdp1 ARE3 sequence, this binding no longer occurs in CSS media, but is restored in CSS media with the addition of DHT. In contrast the negative control is unaffected by these manipulations. (D) LNCaP cells were transfected with plasmids expressing luciferase under control of wild-type Nrdp1 ARE3 promoter, mutant Nrdp1 ARE3 promoter (as shown), or with the parental vector (pGL4), and assayed for luciferase activity in the presence of DMSO vehicle, or 1nM DHT, with or without 10µM bicalutamide (Casodex). The luciferase activity of Nrdp1 ARE3 in LNCaP cells was responsive to androgens but was abolished by the mutations.
We identified an androgen response element (ARE) 209-bp upstream of the transcriptional start site (ARE3), which is a 15-bp bipartite palindromic sequence very similar to ARE-I in the PSA proximal promoter (Cleutjens, et al. 1997; Cleutjens, et al. 1996) (Figure 1B). In LNCaP cells, chromatin immunoprecipitation (ChIP) analysis showed AR binding to Nrdp1 ARE3, but not to a negative control. Further, there was a decrease of AR binding to the Nrdp1 ARE3 in CSS compared to FBS and a restoration of AR binding in CSS with DHT (Figure 1C). A luciferase reporter containing Nrdp1 ARE3 was transfected into LNCaP cells, and further treated with vehicle, DHT or the anti-androgen bicalutamide. Significantly, AR transcriptional activity on the Nrdp1 promoter increased 3.158-fold following the addition of DHT (p=0.007), but was suppressed by addition of bicalutamide (p=0.02). In contrast, there was little to no luciferase activity when LNCaP cells were transfected with a construct containing a mutated ARE3, (fold change = 1.85, p=0.186) indicating that the site was required for AR-dependent transcription (Figure 1D).
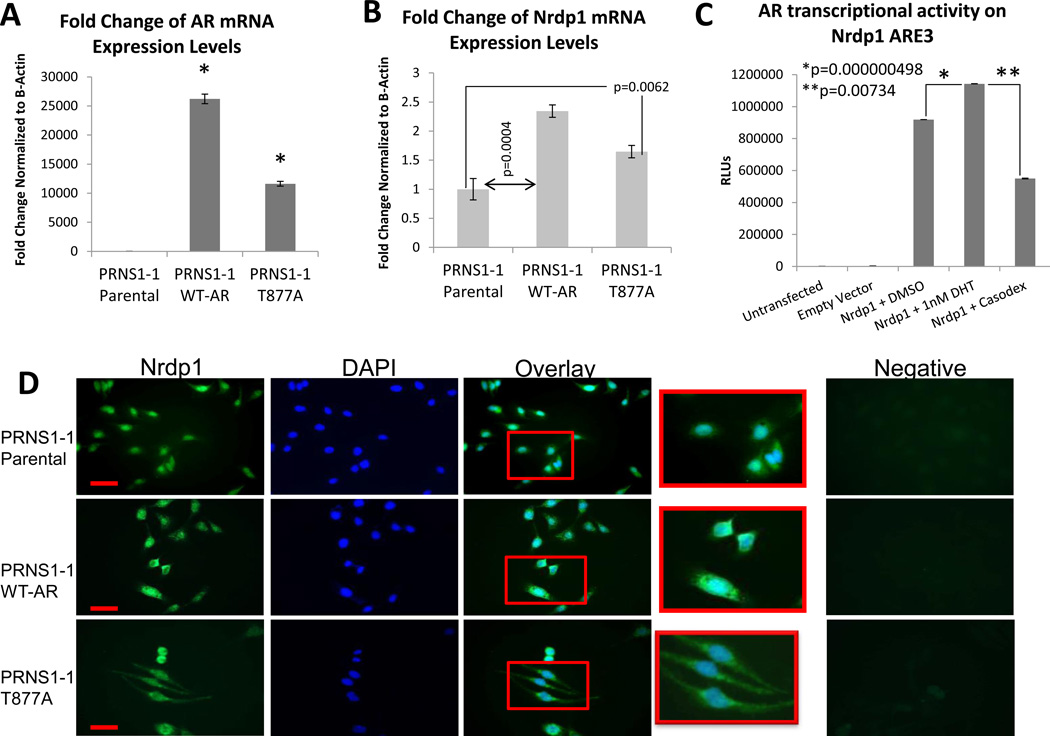
Since LNCaP cells carry a mutated AR(T877A), the androgen sensitivity of Nrdp1 transcription was tested in other cell lines as well. pRNS-1-1 is a cell line derived from a normal prostate (Shi, et al. 2007), which express low (normal) levels of wild-type AR. Stable pRNS-1-1 transfectants expressing an empty vector, wild-type AR or the AR(T877A) mutant (Figure 2A) showed that expression of wild-type or mutant AR significantly increased endogenous Nrdp1 levels (Figure 2B), thereby demonstrating that this effect is not due to the expression of the mutated AR alone. Luciferase assay in AR-expressing pRNS-1-1 cells showed that AR transactivation of the Nrdp1 promoter was androgen sensitive, similar to that observed in LNCaP (Figure 2C). (Similar experiments could not be conducted in pRNS-1-1 cells overexpressing WT-AR, since they have to be cultured in CSS and their activation with DHT induced cell death). Baseline levels of Nrdp1 protein was observed in all cells indicating the influence of other transcription factors in Nrdp1 expression, however, its levels increased when the cells were transfected with wild-type or mutant AR (Figure 2D), confirming AR dependence. Taken together, these results demonstrate that the Nrdp1 is a novel AR target gene regulated in a hormone-sensitive manner in androgen-dependent cells.
Figure 2. Nrdp1 is transcribed by both wild-type and mutated AR in a normal prostate-derived cell line.
(A) Parental pRNS-1-1 cells derived from a normal prostate were stably transfected with an empty vector, wild-type AR (WT-AR), or mutant AR(T877A). qPCR to determine AR expression in parental pRNS1-1 cells, or those stably expressing AR expression goes up 120 fold in PRNS1-1 WT-AR cells (p ≤ 0.0001) and 53 fold in PRNS1-1 AR T877A (p ≤ 0.0001) compared to PRNS1-1 Parental cells. AR transcript levels were normalized to the corresponding values for β-Actin. (B) qPCR for Nrdp1 expression in parental pRNS1-1 cells, or those stably expressing WT-AR, or AR(T877A). Nrdp1 expression goes up 2.3 fold in pRNS1-1 WT-AR cells (p=0.0004) and 1.6 fold in pRNS1-1 AR T877A (p=0.0062) compared to parental pRNS1-1 cells. Nrdp1 transcript levels were normalized to the corresponding values for β-actin. (C) pRNS1-1 AR T877A cells were transfected with plasmids expressing luciferase under control of the Nrdp1 ARE3 promoter or with the parental vector (pGL4), and assayed for luciferase activity in the presence of DMSO (vehicle), 1 nM DHT, or 10µM bicalutamide (Casodex), and shows responsiveness to androgens. (D) Immunofluorescence of parental pRNS1-1 cells, or those stably expressing WT-AR, or AR(T877A). Cells were stained for Nrdp1 (green) or DAPI (blue) to demonstrate the location of the nucleus. (top) Note that while parental pRNS1-1 express very little Nrdp1, (middle) the expression of wt-AR or (bottom) AR(T877A) increased Nrdp1 in the cytoplasm. Negative control(s) denote staining with secondary antibody alone. Bar=20µm.
Nrdp1 expression is elevated in localized human PCa tissue compared to non-tumor prostate and correlates with active (nuclear) AR levels
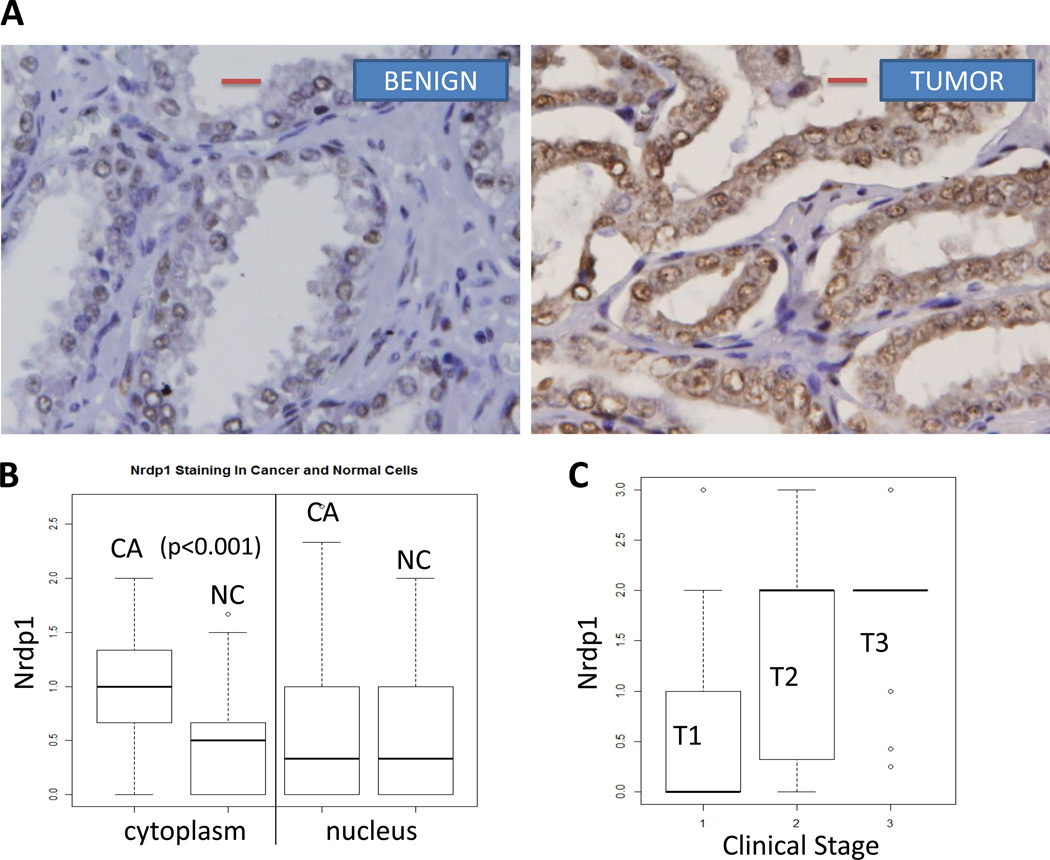
We next investigated the levels of Nrdp1 in primary prostate tissues from 157 individual patients. Of these, matched tumor and surrounding non-tumor tissues were available from 78 patients. Using a scoring system (0–3) based on immunohistochemistry (IHC), where 0 represents no staining and 3 represents 100% staining, we observed that Nrdp1 was strongly expressed in the epithelial cells of the prostate and could be observed in both the nucleus and the cytoplasm (Figure 3A) (specificity of the Nrdp1 antibody was verified with control and Nrdp1 shRNA (Supplemental Figure 1)). Cytoplasmic Nrdp1 was significantly increased in tumor vs non-tumor specimens (p<0.001); although low levels of nuclear Nrdp1 was observed in all samples (Figure 3B, Table 2). Within the tumor tissues, comparison with clinical stage in all 157 patients showed that Nrdp1 expression increased with the stage of the tumor (T1<T2,T3) (p<0.001) (Figure 3C; Table 3).
Figure 3. Nrdp1 is highly expressed in hormone naïve localized tumors from patients with PCa and correlate with intratumoral AR.
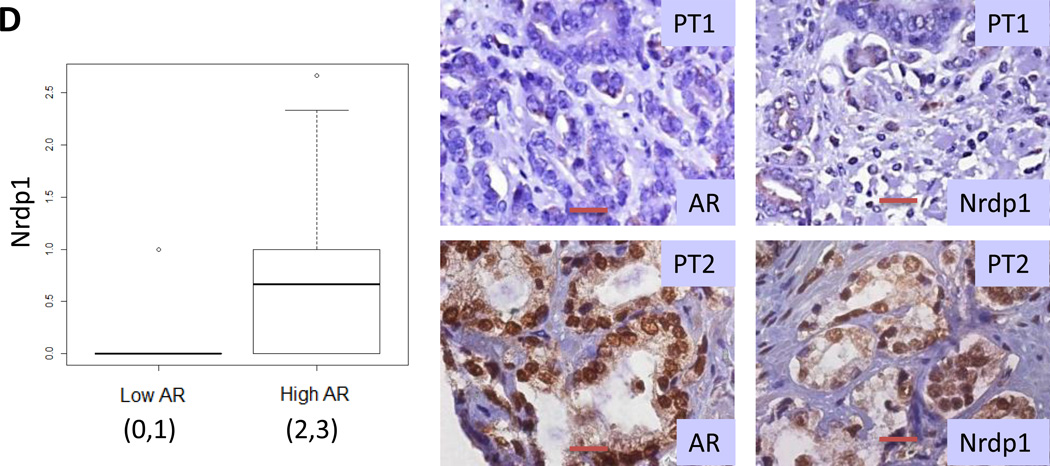
(A) Formalin fixed paraffin-embedded human localized prostate cancer specimens obtained by prostatectomy were arranged in a tissue microarray and stained with anti-Nrdp1 antibody. Nrdp1 expression (brown staining) was observed in the nucleus, cytoplasm or both and was scored on a 0 to 3 scale in both benign and cancerous prostate tissues. Shown are typical staining from a benign section (score 1) (left) and a section showing Gleason grade 3 tumor (score 3) (right) bar=20µm. (B) Boxplot depicting the distribution of Nrdp1 in the nucleus or cytoplasm of tumor compared to non-tumor tissue (n=78) demonstrate that the expression of nuclear Nrdp1 remains the same in both cancer and non-tumor tissues, whereas cytoplasmic expression of Nrdp1 increases in tumor compared to non-tumor tissue. (C) Nuclear Nrdp1 levels differed significantly by clinical stage (p < 0.001), with post-hoc testing showing significantly higher expression in Stage 2 (T2) and Stage 3 (T3) patients than in Stage 1 (T1) patients (p = 0.001). (D) (left) Boxplots showing the correlation between Nrdp1 and AR in tumor tissue (p<0.001). (right) IHC of AR and Nrdp1 in two patients with high and low AR vs Nrdp1. Note that the patient who expressed little AR also expressed little Nrdp1, whereas strong AR staining correlated with strong Nrdp1 expression. Bar=50µm.
Table 2. Statistical Analysis of Protein Staining in prostate tumors and non-tumor prostate.
Nuclear and cytoplasmic levels of both Nrdp1 and AR were analyzed individually. Cytoplasmic but not nuclear AR and Nrdp1 expression increase in human prostate tumor tissues compared to non-tumor prostate. P-value (p<0.05) were considered significant.
| Cytoplasm | Nucleus | |||||
|---|---|---|---|---|---|---|
| Protein | Median in Cancer Cells (n=78) |
Median in Non- Cancer Cells (n=78) |
P-Value (Wilcoxon Signed Rank Test) |
Median in Cancer Cells (n=78) |
Median in Non- Cancer Cells (n=78) |
P-Value (Wilcoxon Signed Rank Test) |
| Nrdp1 | 1.0 | 0.5 | <0.001 | 0.3 | 0.3 | 0.134 |
| AR | 1.0 | 0.8 | <0.001 | 2.0 | 2.0 | 0.071 |
Table 3.
Nrdp1 Levels by Clinical Stage.
| Stage T1 Median (Range) |
Stage T2 Median (Range) |
Stage T3 Median (Range) |
Kruskal- Wallis Test P-Value |
|
|---|---|---|---|---|
| No of patients | 43 | 88 | 18 | |
| Nrdp1 | 0 (0—3) |
2 (0—3) |
2 (0.3—3) |
<0.001 |
Comparison of AR and Nrdp1 protein levels revealed a significant correlation between Nrdp1 levels and nuclear (active) AR (cytoplasmic Nrdp1 vs nuclear AR: pairwise correlation coefficient: 0.42; p<0.001; nuclear Nrdp1 vs nuclear AR: pairwise correlation coefficient: 0.26; p=0.035) (Figure 3D). Examination of Oncomine datasets showed a similar correlation between Nrdp1 and AR mRNA levels in human patients comparing prostate tumor vs. non-tumor prostate as determined by various investigators (Supplementary Figure 2), supporting our observations. Taken together, these results indicate strong association between AR and Nrdp1 expression in localized human PCa, and support AR dependence of Nrdp1 expression.
Nrdp1 levels are reduced in CRPC compared to hormone-sensitive PCa
We also investigated whether correlation between Nrdp1 and AR is observed in CRPC. Well characterized CRPC sublines of LNCaP cells (Denmeade, et al. 2003; Vinall, et al. 2006; Wu, et al. 1994) were analyzed for comparative levels of Nrdp1. Although the AR in the CRPC sublines is known to be active (Ghosh, et al. 2005; Vinall et al. 2006), Nrdp1 expression was decreased in all three, indicating dissociation between AR and Nrdp1 in these CRPC lines (Figure 4A). Comparison of the AR binding site on the Nrdp1 promoter in LNCaP and C4-2 showed that they were identical (Supplementary Figure 3), however, AR binding to the Nrdp1 ARE was severely decreased in C4-2 cells compared to LNCaP (p<0.0001), despite the level of AR protein level in the C4-2 cells being comparable to that in LNCaP, as we have shown elsewhere (Ghosh et al. 2005; Wang, et al. 2008). Thus, the decrease in AR binding is not caused by a mutation in the AR binding site, and is not a result of a significantly different AR protein level. It may be noted that the Nrdp1 promoter is regulated in CRPC cells by other transcription factors that take over once the cell achieves a CRPC phenotype. As a result of the loss of AR binding to the Nrdp1 ARE, the levels of Nrdp1 mRNA and protein is significantly lower, but because it is now transcribed by other transcription factors, it is not completely eliminated.
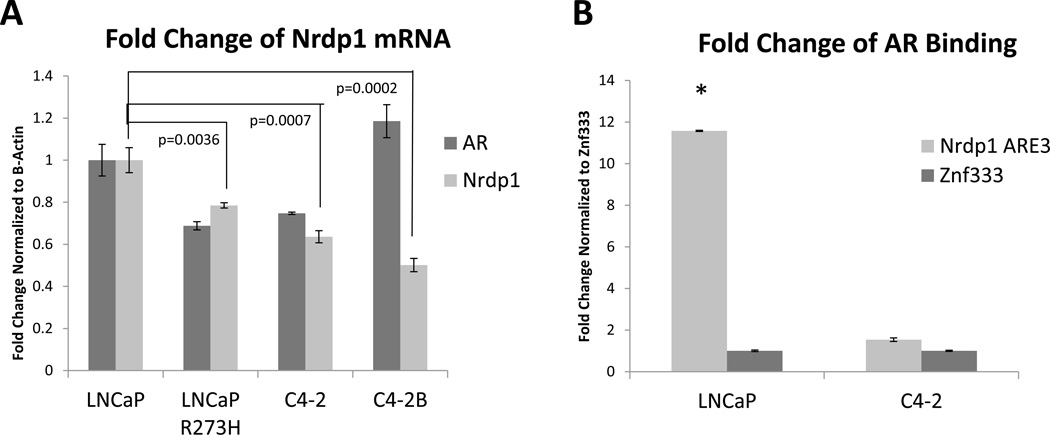
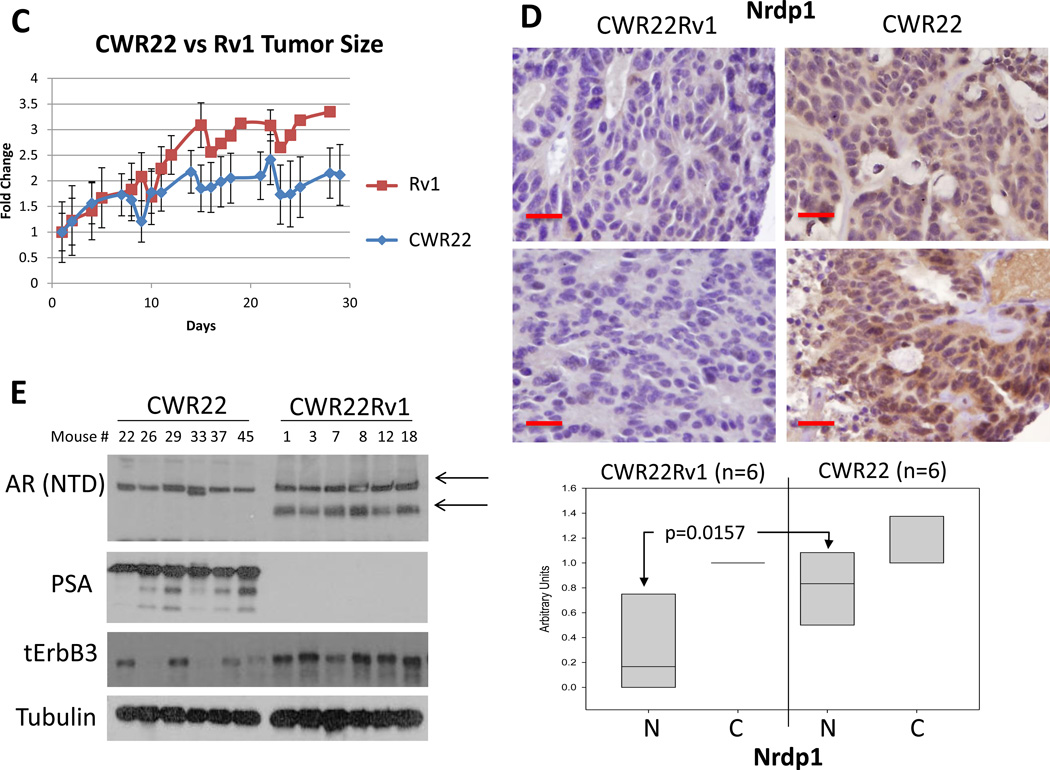
Figure 4. Loss of Nrdp1 expression and AR regulation of Nrdp1 transcription in CRPC compared to hormone naïve tumors.
(A) qPCR comparing Nrdp1 expression in LNCaP cells vs those in LNCaP R273H (p=0.0036), C4-2 (p=0.0007), and C4-2B (p=0.0002). Note the decrease in Nrdp1 levels in the latter three cell lines, which are all CRPC. Nrdp1 transcript levels were normalized to the corresponding values for β-Actin. (B) Comparison of AR binding to the Nrdp1 ARE3 in LNCaP vs C4-2 cells. Note the sharp decrease in AR binding in C4-2 compared to LNCaP (p<0.0001). Chromatin samples were immunoprecipitated with an anti-AR antibody and analyzed by qPCR with primers flanking the Nrdp1 ARE3 region with Znf333 as a negative control. (C) (Top) Nude mice were subcutaneously implanted with either CWR22 (right) or CWR22Rv1 (left) tumor and the tumors were allowed to grow for up to 29 days; the mice were euthanized and the tumors excised when tumors>150 cm3 or at the end of that period. Tumor size was measured as described and plotted over time. The Rv1 tumors were more aggressive compared to CWR22 (p=0.003). (D) Formalin fixed paraffin-embedded tumor specimens were stained with anti-Nrdp1 antibody. Note that the cells in CWR22 stained strongly for Nrdp1 (brown), while those in CWR22Rv1 did not (bar=30µm). (Bottom) Boxplot of Nrdp1 in CWR22 (n=6) vs CWR22Rv1 (n=6) in the nucleus (N) and cytoplasm (C). Primary CWR22 tumors expressed higher levels of nuclear Nrdp1 compared to recurrent CWR22Rv1 tumors (p=0.0157). (E) Whole cell lysates of xenografted tumors were run on a Western blotted and stained with AR, PSA and ErbB3, while tubulin levels were used as loading control. Results show that despite the sharp change in Nrdp1 between the two tumor types, there was no significant difference in AR levels (except that CWR22Rv1 tumors also expressed the alternately spliced forms). However, AR transcriptional activity in the CWR22Rv1 was significantly suppressed as shown by a decrease in PSA levels. In support of the decrease in Nrdp1 in CWR22Rv1 compared to CWR22, the former expressed higher levels of the Nrdp1 degradation target ErbB3.
We next investigated whether loss of Nrdp1 expression in CRPC is also observed in other models. A tumor line, CWR22, and its CRPC derivative CWR22Rv1 were implanted in nude mice and the tumors excised following volume ≥150cm3. The CWR22Rv1 tumors grew at a significantly rapid rate compared to CWR22 when normalized to day 1 (p=0.003) (Figure 4C). In addition, by immunohistochemistry (IHC), Nrdp1 was significantly lower in castration resistant CWR22Rv1 tumors (Median=0, n=6) compared to androgen sensitive CWR22 tumors (median=1, n=6) (p=0.0157) (Figure 4D) despite expression of full-length AR in both CWR22 and CWR22Rv1 (although CWR22Rv1 tumors in addition expressed AR splice variants) (Li, et al. 2013) (Figure 4E). We also determined whether Nrdp1 in the tumors (detected by IHC) was active by examining the levels of its ubiquitination target ErbB3. As expected, ErbB3 levels were higher in CWR22Rv1 compared to CWR22 (Figure 4E), thereby demonstrating negative correlation with Nrdp1. In all, these results demonstrate that despite AR dependence of Nrdp1 in hormone naïve PCa, this correlation is lost in advanced disease.
Expression of Nrdp1 and AR binding to the Nrdp1 promoter correlates with expression of the 90kDa C-terminal FlnA fragment
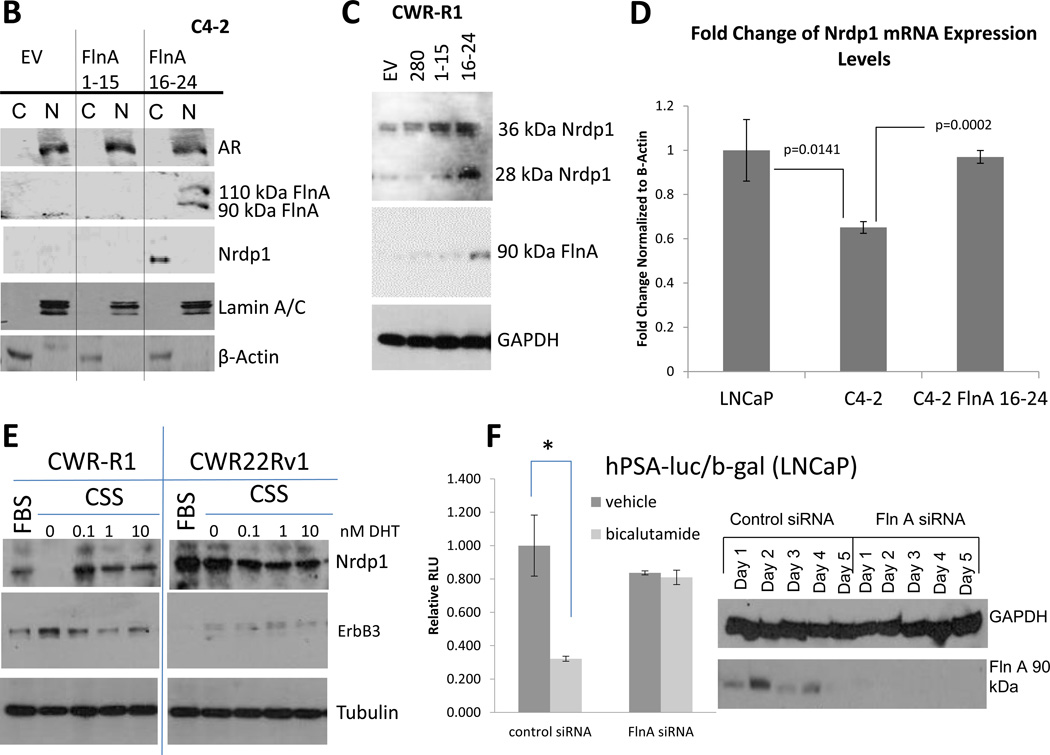
We now investigated the molecular mechanism leading to the reduction of Nrdp1 expression in advanced PCa. In support of lower AR binding to Nrdp1 ARE3 in Figure 4B, C4-2 cells expressed lower levels of Nrdp1 protein compared to LNCaP (Figure 5A). Since C4-2 cells express the same AR mutation as LNCaP, this difference cannot be attributed to an AR mutation. However, we previously demonstrated that FlnA expression was mostly nuclear in LNCaP cells, whereas in C4-2, it was mostly cytoplasmic (Supplementary Figure 4) (Bedolla et al. 2009; Wang et al. 2007). Comparison of the two lines showed that the levels of 90kDa FlnA correlated with Nrdp1 levels (Figure 5A). These results indicate a possible role for FlnA in determining the transcriptional activity of AR on Nrdp1.
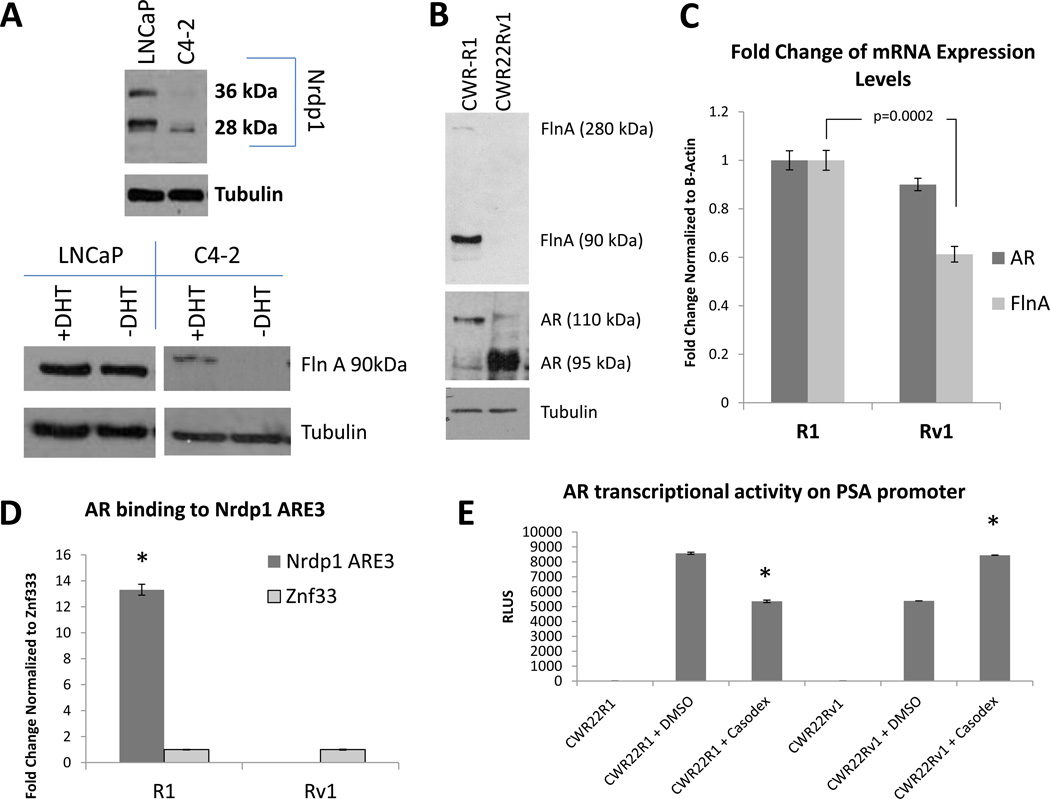
Figure 5. Correlation between Nrdp1 levels and expression of a 90kDa FlnA isoform.
(A) (upper) Comparison of Nrdp1 protein levels in LNCaP and C4-2 cell lines cultured in FBS. Cell lysates were immunoblotted with anti-Nrdp1, and anti-tubulin antibodies. Note that C4-2 cells expressed lower levels of this protein compared to LNCaP. (lower) Correspondingly, the levels of 90kDa FlnA was also assessed in the presence or absence of 1nM DHT. LNCaP cells cultured in FBS showed no difference in FlnA levels with DHT, however, in C4-2 cells these levels were much lower and change with DHT was immediately obvious. (B) Protein expression of FlnA in CWR-R1 and CWR22Rv1 cells cultured in FBS. Cell lysates were immunoblotted with anti-FlnA, anti-AR, and anti-tubulin antibodies. Whole cell lysates of CWR-R1 and CWR22Rv1 cells demonstrate the decreased expression of the 90 kDa fragment of FlnA in CWR22Rv1 cells. (C) qPCR for AR and FlnA expression in CWR-R1 and CWR22-Rv1. Higher expression of FlnA was observed in CWR-R1 cells (p=0.0002), although AR expression in the two cell lines was comparable (p>0.05). AR and FlnA transcript levels were normalized to the corresponding values for β-Actin. (D) ChIP assay of AR binding to ARE3 in CWR-R1 and CWR22Rv1 cells. AR binds to Nrdp1 ARE3 in CWR22R1 cells but not CWR22Rv1 cells. Chromatin samples were immunoprecipitated with an anti-AR antibody and analyzed by qPCR with primers flanking the Nrdp1 ARE3 region (p<0.0001), and Znf333 (p>0.05) as a negative control. (E) Androgen sensitivity in CWR-R1 vs. CWR22Rv1 cells. AR transcriptional activity was tested in untransfected cells, or cells transfected cells with luciferase driven by the PSA ARE. The decrease in luciferase activity in CWR-R1 cells but not in CWR22-Rv1 cells in the presence of 10µM bicalutamide indicates that CWR-R1 cells are more androgen sensitive compared to CWR22Rv1 cells, though both are considered castration resistant (bicalutamide decreased AR transcriptional activity on the PSA promoter in CWR-R1 cells by ~38%, p<0.0001).
To determine whether 90kDa FlnA indeed plays a role in AR transcription of Nrdp1, we compared two CRPC lines, CWR22Rv1 and CWR-R1, both derived from relapsed CWR22 tumors. Both lines expressed AR variants that lack the LBD and are essentially androgen-independent in phenotype (Chen, et al. 2010a). However, CWR-R1 cells express higher levels of FlnA compared to CWR22Rv1 (Figure 5B). Both lines express similar levels of total AR (detected using primers against the DNA binding domain of the AR, p>0.05); while CWR-R1 cells expressed higher levels of FlnA mRNA (p=0.0002) compared to CWR22Rv1 (Figure 5C). ChIP assay showed that in CWR-R1, but not in CWR22Rv1, the AR strongly bound to the Nrdp1 promoter, vs a negative control (Figure 5D). These results indicated correlation between Nrdp1 transcription by AR, and 90kDa FlnA. In support of a role for FlnA in androgen sensitivity, CWR-R1 which express high FlnA, but not CWR22Rv1 cells which express low FlnA, responded partially to treatment with the anti-androgen bicalutamide (Casodex), (Figure 5E). Therefore, despite castration-resistance in both lines, the response of the cells correlated with the expression of 90kDa FlnA.
Expression of 90kDa FlnA isoform restored expression of Nrdp1 in CRPC cells
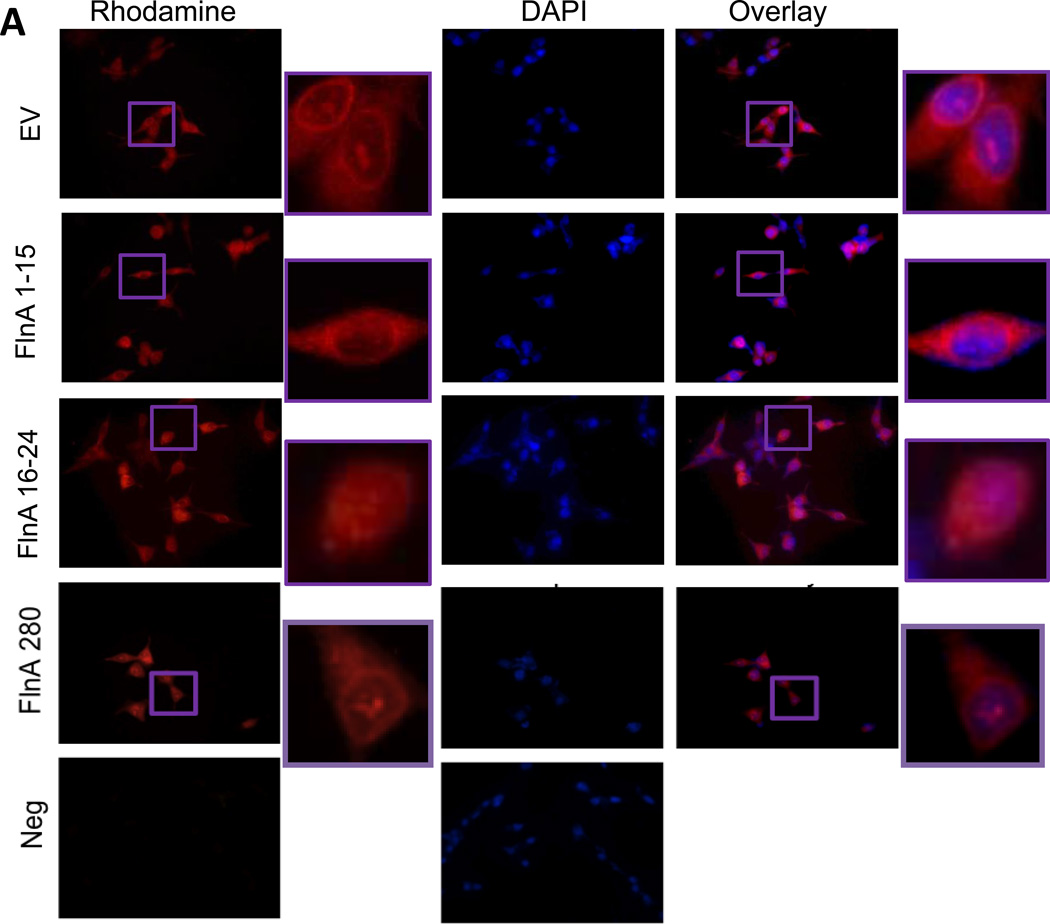
Since CWR22Rv1 cells expressed higher levels of the low molecular-weight AR variants compared to CWR-R1, to distinguish between the effects of the splice variants and 90kDa FlnA in regulating Nrdp1 transcription by AR, we utilized cells that did not express AR splice variants. Transfection of C4-2 cells with full-length FlnA (280kDa), N-terminal FlnA (FlnA1-15, 170kDa) or C-terminal FlnA (FlnA16-24, 90kDa) revealed that only FlnA16-24 localized to the nucleus (Figure 6A). These results were verified by isolation of C4-2 cytoplasmic and nuclear fractions, which also revealed that FlnA16-24, but not FlnA1-15, was expressed in the nuclear fragment (Figure 6B).
Figure 6. The 90kDa FlnA isoform localized to the nucleus and promoted apoptosis and growth arrest in a ligand dependent manner.
(A) Immunofluorescence of C4-2B cells transfected with empty vector (EV), FlnA 1-15, or FlnA 16-24. Cells were stained for C-terminal FlnA or DAPI to identify the location of the nucleus. (top panel) Control cells (transfected with empty vector) only express cytoplasmic FlnA, as demonstrated by unstained nuclear regions in FlnA stained cells, (2nd panel) and transfection of FlnA 1-15 did not affect the localization, (3rd panel) while those transfected with FlnA 16-24 express both cytoplasmic and nuclear FlnA. (4th panel) Transfection of full-length FlnA did not restore nuclear localization completely. (5th panel) Negative controls were treated the same but did not use the anti-FlnA antibody (bar=30µm). (B) Subcellular fractionation of C4-2 cells transfected with empty vector, FlnA 1-15, or FlnA 16-24. Fractionated cell lysates were immunoblotted with anti-Nrdp1, anti-AR, anti-FlnA (C-terminal), anti-β-Actin (to demonstrate specificity of cytoplasmic fraction), and anti-Lamin A/C (to demonstrate specificity of nuclear fraction) and demonstrates that transfection of FlnA 16-24 caused nuclear expression of FlnA and restores Nrdp1 protein in C4-2 cells, although AR levels were not altered. (C) Protein expression of Nrdp1 in CWR-R1 cells is regulated by the 90 kDa FlnA. Whole cell lysates of CWR-R1 cells that were transfected with empty vector, full length FlnA, FlnA 1-15, or FlnA 16-24 were immunoblotted with anti-FlnA, anti-Nrdp1, and anti-tubulin antibodies. Nrdp1 protein levels increased with the increased levels of the 90 kDa FlnA fragment. (D) FlnA restores Nrdp1 expression in CRPC cells. qPCR for Nrdp1 expression in LNCaP, C4-2, and stably transfected C4-2-FlnA(16-24) showed that Nrdp1 expression was reduced in C4-2 compared to LNCaP cells (p=0.0141), but expression of FlnA16-24 in C4-2 cells restored Nrdp1 expression to a level similar to LNCaP cells (p=0.0002 compared to C4-2). Nrdp1 transcript levels were normalized to the corresponding values for β-Actin. (E) Comparison of Nrdp1 response to changes in AR in CWR-R1 and CWR22-Rv1 cells cultured in FBS, CSS or CSS treated with increasing doses of DHT as indicated. Lysates were immunoblotted with anti-Nrdp1, anti-ErbB3, and anti-tubulin antibodies. While the levels of Nrdp1 in CWR22Rv1 cells were unaltered despite culture in CSS, in CWR-R1 these levels altered slightly. (F) (left) Reporter gene activity of AR on a luciferase-tagged PSA promoter section demonstrates that in control LNCaP cells, 10 µM bicalutamide is able to suppress AR activity whereas in cells where FlnA is downregulated by siRNA, bicalutamide failed to affect AR activity. (right) Western blotting demonstrating the efficacy of FlnA siRNA used.
Having established that FlnA16-24 transfection induces nuclear expression of this protein, we next investigated whether nuclear FlnA altered the expression of Nrdp1. Significantly, only FlnA16-24, but not FlnA1-15, restored Nrdp1 protein levels in C4-2 cells (Figure 6B), indicating a role for nuclear FlnA in this process. To demonstrate a role for FlnA in Nrdp1 expression independent of AR splice variants, we used CWR-R1 cells that expressed the splice variant. When CWR-R1 cells were transfected with full-length FlnA, FlnA1-15 and FlnA16-24, expression of the 90kDa FlnA band was seen only in the latter, and we observed the highest levels of Nrdp1 as determined by protein expression in cells expressing FlnA(16-24) (Figure 6C). FlnA(16-24) affected the rate of transcription as determined by the change in mRNA expression (Figure 6D) and by the extent of AR binding to the Nrdp1 promoter in the presence of FlnA 16-24 (Supplementary Figure 5A). Our data demonstrate that nuclear expression of 90kDa FlnA regulates Nrdp1 levels by modulating its transcription. In addition, Figure 6E shows that Nrdp1 and therefore ErbB3 levels are slightly androgen dependent in CWR-R1 cells (left), but not in CWR22Rv1 cells (right). These results support the observed AR binding to the Nrdp1 promoter in CWR-R1 but not CWR22Rv1 cells shown in the previous figure.
We next investigated whether other AR targets are also affected by the presence of FlnA. The best known AR target is PSA, which is known to be decreased upon bicalutamide treatment in androgen-dependent LNCaP cells (Ghosh et al. 2005). Downregulation of FlnA expression by siRNA revealed that bicalutamide failed to suppress AR activity on the PSA promoter in the absence of FlnA (Figure 6F). The above data did not specify which fragment of FlnA was needed to show a PSA response. Therefore, LNCaP cells were transfected with either FlnA 1–15 or FlnA 16-24. Similar to control cells (transfected with an empty vector), those transfected with FlnA16-24 responded to bicalutamide, whereas the cells transfected with FlnA 1–15 did not (Supplementary Figure 5B). Therefore, it is the C-terminal fragment that is needed for PSA response.
Nuclear FlnA regulates AR-mediated transcription of Nrdp1 in CRPC cells
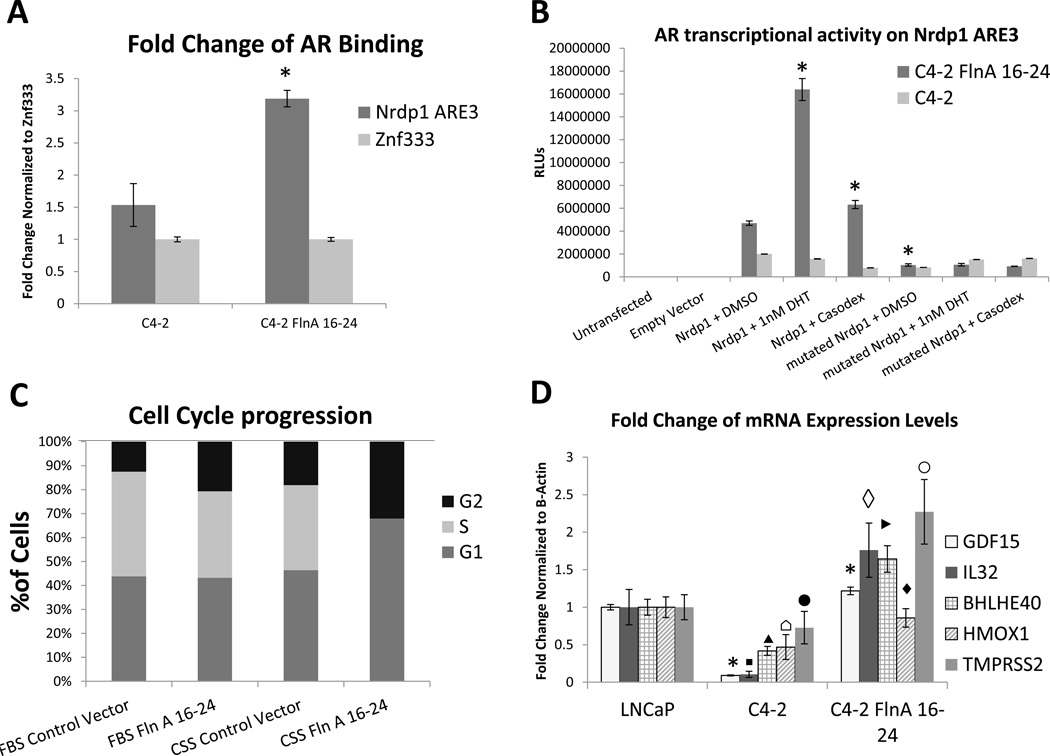
Since Nrdp1 is transcriptionally regulated by the AR, and FlnA(16-24) also regulates Nrdp1 transcription, we investigated whether AR-regulated Nrdp1 transcription is modulated by nuclear FlnA. Transfection of FlnA16-24 in C4-2 cells increased AR binding to Nrdp1 ARE3 (Figure 7A). Importantly, this re-establishment of AR binding restored androgen-sensitivity of AR-induced Nrdp1 transcription. C4-2 cells transfected with sham or FlnA16-24 and either wild-type or mutated Nrdp1 ARE3-luciferase constructs were treated with vehicle, DHT, or bicalutamide. Untransfected C4-2 cells showed very little AR transcriptional activity on Nrdp1 ARE3, and no response to either the androgen or the anti-androgen (p>0.05), whereas C4-2 cells transfected with FlnA16-24 responded to them. The mutant ARE3 construct showed little AR-dependent transactivation, but in C4-2 FlnA16-24 cells transfected with wild-type ARE3, transcription was increased in the presence of DHT and inhibited by bicalutamide, indicating a restoration of androgen-sensitivity (Figure 7B). In contrast, AR transcriptional activity stimulated by FlnA1-15 (cytoplasmic) was ligand-independent as indicated by the lack of response when cells were cultured in CSS (unlike nuclear FlnA16-24) (Supplementary Figure 5B), and is likely caused by increased actin cross-linking, which promotes AR transcriptional activity (de Vere White, et al. 1997; McGrath, et al. 2013). Thus, nuclear FlnA induced androgen-sensitive AR transcriptional activity while in the absence of nuclear FlnA, AR activity was ligand-independent.
Figure 7. Expression of the 90kDa FlnA isoform restored AR regulation of Nrdp1 transcription.
(A) AR binds to Nrdp1 ARE3 in the presence of FlnA 16-24 (90 kDa). Chromatin samples were immunoprecipitated with an anti-AR antibody and analyzed by qPCR with primers flanking the Nrdp1 ARE3 region with Znf333 as a negative control. Note that AR binding increased 2-fold upon FlnA16-24 transfection (p<0.0001). (B) AR transcriptional activity of Nrdp1 ARE3 is androgen regulated in the presence of FlnA 16-24. C4-2 cells transfected with empty vector or FlnA 16-24 were cultured in FBS medium and transfected with control vector, normal Nrdp1 ARE3, or mutant Nrdp1 ARE3. AR transcriptional activity was measured by luciferase assay. Cells were also treated with DMSO, 1nM DHT, or 10µM bicalutamide (Casodex). Luciferase was increased in the presence of FlnA 16-24, and regulated in an androgen dependent manner with normal Nrdp1 ARE3. (C) Flow cytometric analysis in PI-stained, ethanol fixed C4-2 cells to determine the effect of transfection with FlnA 16-24 on cell cycle. Cells were grown in FBS or CSS and transfected with either empty vector or FlnA 16-24. Tranfection with FlnA 16-24 has the same effect as growing the cells in CSS, however the combination of the two completely S phase. (D) FlnA acts as a regulator of mRNA expression levels in multiple genes. qPCR for GDF15, IL32, TMPRSS2, BHLHE40, and HMOX1 expression in LNCaP, C4-2, and C4-2 FlnA 16-24. The expression of 4 of 5 genes were significantly decreased in C4-2 compared to LNCaP cells [GDF15: p<0.0001 (*); IL-32: p=0.0029 (▪); BHLHE40: p=0.0012 (▲); HMOX1: p=0.0129 (⌂)] while that for TMPRSS2 was not significant (p>0.05; ●). The expression of FlnA 16-24 in C4-2 cells restored expression of these genes to a level similar or higher than LNCaP cells [GDF15: p<0.0001 (*); IL-32: p=0.0014 (◊); BHLHE40: p=0.0003 (►); HMOX1: p=0.0052 (♦); TMPRSS2: p=0.0312 (○)]. All transcript levels were normalized to the corresponding values for β-Actin.
Next, we investigated whether nuclear FlnA-regulated AR activity had any functional role on tumor response. Investigation of the effect of FlnA16-24 on cell proliferation by flow cytometric analysis revealed that in CRPC C4-2 cells, FlnA16-24 by itself had no significant effect, and neither did the removal of hormones (which includes androgens) by charcoal stripping; however, in the absence of hormones, FlnA16-24 induced severe growth arrest, causing cells to arrest in both G1 and G2 phases, with very few cells in S-phase (Figure 7C). FlnA16-24 also induced 3-fold increase in apoptosis (Supplementary Figure 6).
Finally, we determined whether other genes were also similarly regulated by FlnA. Five genes known to contain an ARE in the proximal promoter (IL-32, HMOX1, GDF15, BHLHE40, and TMPRSS2, see Supplemental Table 3) were examined to determine whether FlnA16-24 affected their transcription (Figure 7D). The expression of growth differentiation factor 15 (GDF15), a member of the transforming growth factor beta (TGFβ) superfamily, and interleukin-32 (IL-32), is known to induce apoptosis (Park, et al. 2012; Podar, et al. 2007; Wang, et al. 2012; Yun, et al. 2013), while TMPRSS2 is a known AR target suppressed in CRPC. Heme oxygenase 1 (HMOX1) counteracts oxidative and inflammatory damage and is implicated in the adhesive and morphological properties of tumor cells (Gueron, et al. 2014), while Basic loop-helix-loop E40 (BHLHE40) is a transcription factor is involved in the regulation of cell differentiation, response to hypoxia and carcinogenesis (Wu, et al. 2014). qPCR showed that each of these genes were under-expressed in C4-2 compared to LNCaP, while expression of nuclear FlnA restores their expression (Figure 7D). These results indicate that FlnA regulates a subset of genes associated with AR transcriptional activity in PCa.
DISCUSSION
The AR regulates a very different transcription program in androgen-dependent PCa vs. CRPC, even in tumors that do not harbor AR mutations or alternately spliced forms (Decker et al. 2012; Hu et al. 2012; Wang et al. 2009b). To examine the cause of AR reprogramming, we used Nrdp1 as a model gene whose transcription is regulated by AR in hormone-naïve PCa, but not in CRPC. Nrdp1 was first identified as an E3 ubiquitin ligase that caused ErbB3 degradation in breast cancer cells (Cao et al. 2007; Wu et al. 2004; Yen et al. 2006). Since then, this RING finger containing protein has been found to regulate a number of other targets, including pro-inflammatory cytokines (Wang, et al. 2009a), type 1 cytokine receptor (Wauman, et al. 2011), inhibitor of apoptosis proteins (IAP) (Qiu, et al. 2004), parkin (Zhong, et al. 2005), and CCAAT/enhancer-binding protein β (C/EBPβ) (Ye, et al. 2012). Previous studies in breast cancer showed that Nrdp1 is lost during normal-to-tumor transition (Cao et al. 2007; Wu et al. 2004; Yen et al. 2006); however, our results showed that it increased in PCa compared to non-tumor prostate, indicating alternate pathways regulating its levels in various organs. Despite the initial increase in Nrdp1 with AR in hormone-naïve tumors, we observed reduced Nrdp1 expression in CRPC, although AR expression persisted in the latter. The current study makes three important observations. (i) In androgen-sensitive PCa, Nrdp1 is a direct transcription target of AR, and is increased in localized PCa, where AR levels increase compared to non-tumor prostate. (ii) In CRPC, despite further increase in AR activity, Nrdp1 levels decrease because the AR no longer regulates its transcription and (iii) this difference in AR-induced transcription is regulated by the availability of nuclear FlnA.
Full-length FlnA is a 280kDa actin-binding protein that acts as a scaffold to enable interaction of actin with other proteins to regulate diverse functions such as cell rigidity, adhesion and migration (Stossel, et al. 2001; van der Flier and Sonnenberg 2001). Full-length FlnA is important for proper embryonic development (Robertson, et al. 2003) but likely promotes metastasis if overexpressed in the cytoplasm of cancer cells (Castoria, et al. 2011; McGrath et al. 2013). Actin binding proteins are known to regulate AR transcriptional activity (de Vere White et al. 1997), and FlnA has been suggested to be a putative AR co-regulator (Parker et al. 2013). Androgen stimulation of quiescent NIH3T3 cells caused cytoplasmic FlnA binding to AR and co-localization of the FlnA/AR complex at intermediate actin filaments leading to extranuclear AR-mediated Rac1 activation and subsequent cell motility (Castoria et al. 2011). In normal epithelial cells of the adult prostate, however, FlnA cleaves to a 90kDa fragment which localizes to the nucleus and regulates AR transcriptional activity (Loy et al. 2003; Ozanne et al. 2000). Importantly, in the presence of nuclear FlnA, AR regulated its transcription only upon ligand-binding (Supplementary Figure 5B). FlnA is a scaffolding protein, and binds to a very large number of proteins of diverse functions. One way FlnA can promote AR binding to target genes is by regulating the interaction of the AR with other co-regulators. Both actin-binding proteins and DNA-repair proteins act as androgen receptor co-factors (Parker et al. 2013; van de Wijngaart et al. 2012) and FlnA is known to interact with both these classes of proteins (Stossel et al. 2001; van der Flier and Sonnenberg 2001) The significance of this observation is that in the presence of nuclear FlnA, treatment with anti-androgens such as bicalutamide or enzalutamide will prevent tumor growth or progression, whereas in the absence of nuclear FlnA, the anti-androgens are ineffective.
Basal levels of Nrdp1 are seen in all cells, as apart from AR, Nrdp1 promoter has binding sites for multiple other transcription factors. Thus in cells where Nrdp1 is transcribed by AR, its expression is androgen-dependent, while in other cells, its expression is regulated by other transcription factors in an androgen-independent manner. The effective finding of this paper is that AR binding and increased transcription in an androgen-dependent manner was seen only in the presence of nuclear FlnA. It may be remarked that both cytoplasmic and nuclear FlnA promoted AR transcriptional activity; however, AR transcriptional activity induced by cytoplasmic FlnA was ligand-independent whereas AR activity caused by nuclear FlnA was androgen-dependent. One mechanism by which nuclear FlnA can affect AR transcriptional activity is by its scaffolding action in regulating the interaction of the AR with other co-regulators. While we (Wang et al. 2007) and others (Loy et al. 2003) demonstrate direct interaction between AR and FlnA, the mechanism of AR interaction with FlnA in the nucleus by which androgen-sensitivity is maintained, is yet to be identified. AR binds to C-terminal FlnA (Ozanne et al. 2000); and although nuclear FlnA induced AR transcriptional activity, it prevented inappropriate activation of AR by non-specific ligands or by ligand-independent activation, thereby demonstrating anti-tumorigenic properties (Bedolla et al. 2009; Loy et al. 2003; Mooso et al. 2012; Sun, et al. 2014; Wang et al. 2007).
A relevant question is – how does FlnA 16-24 cause growth arrest and apoptosis in the absence of hormones? In this paper, we have not addressed this issue – but it is well known that in CRPC cells, AR regulates both cell cycle progression as well as cell survival, and anti-androgens are known to promote apoptosis and induce growth arrest. Now, full-length FlnA is required for cell cycle progression and cell survival as well; therefore, we believe that one way FlnA 16-24 can induce apoptosis and growth arrest is by preventing cell survival and cell cycle progression in cells where anti-androgens induce apoptosis and growth arrest, which would have been possible with full-length FlnA. On that note, FlnA is also known to be a major factor in DNA damage repair by interaction with BRCA1 (Velkova, et al. 2010) and BRCA2 (Yue, et al. 2009). Since anti-androgens also prevent DNA double strand break repair (Polkinghorn, et al. 2013), it is expected that FlnA16-24 will also promote ionizing radiation-induced apoptosis.
In a previous publication, we had shown that the expression of nuclear FlnA is also regulated by the AR (Mooso et al. 2012). FlnA proteolysis is prevented by its phosphorylation at S2152 (van der Flier and Sonnenberg 2001). Our data showed that in LNCaP cells, FlnA is not phosphorylated and undergoes proteolysis to the 90kDa fragment which then translocates to the nucleus. Prevention of FlnA phosphorylation appears to require AR activity. On the other hand, in C4-2 cells, which were developed by implantation of LNCaP cells in castrated mice, FlnA is phosphorylated, which likely resulted during its progression to castration resistant growth. Therefore, the phosphorylated FlnA in C4-2 cells does not undergo proteolysis and remains cytoplasm-bound (Mooso et al. 2012). It may be noted that FlnA has also been shown to localize to the nucleolus and associate with the RNA polymerase I (Pol I) transcription machinery to suppress rRNA gene transcription (Deng, et al. 2012). However, we found nucleolar FlnA to be present in C4-2 as well as LNCaP (Supplementary Figure 4), and when transfected with either the empty vector or a FlnA plasmid (Figure 6A), thus Nrdp1 expression seems to be independent of the FlnA fraction localizing to the nucleolus, although it is dependent on the fraction localizing to the nucleoplasm.
FlnA’s effect on broad gene expression is obvious from its effects on TMPRSS2, HMOX1, BHLHE40, GDF15 and IL-32, each of which was identified to have an ARE in the proximal promoter. Previous studies showed that TMPRSS2 mRNA expression, but not TMPRSS2-ERG gene fusion, is decreased in CRPC, since the gene fusion likely cause an increase in ERG expression instead of TMPRSS2 (Cai, et al. 2009). FlnA also upregulates two other genes, GDF15 and IL-32 that are also associated with increased apoptosis (Park et al. 2012; Wang et al. 2012; Yun et al. 2013). Therefore, it is likely that nuclear FlnA induces apoptosis by increasing GDF-15 and IL-32 levels. It is important to note that absence of BHLHE40 has been identified as a transcription factor is involved in the regulation of cell differentiation and prevention of carcinogenesis (Wu et al. 2014), which may be one way by which it has an effect on tumor cell progression.
In conclusion, in this paper, we demonstrate that FlnA16-24 regulates Nrdp1 transcription by the AR by acting as a co-activator of its transcriptional action. The regulation of Nrdp1 by AR in PCa cells that express 90 kDa FlnA, its loss in those that do not, and the adjustment upon re-introduction of FlnA into the nucleus, is an example of retooling of the transcriptional program regulated by the AR in PCa cells. We demonstrate that it is possible to restore the original androgen-sensitive program by re-introducing a key co-regulator that is frequently lost in CRPC (Bedolla et al. 2009). The AR is known to repress cell growth and induce differentiation in various tissues (Batch, et al. 1992; Govoroun, et al. 2001; Holdcraft and Braun 2004), but promote tumorigenesis in prostate cells (Berger, et al. 2004). Our results indicate that the availability of co-regulators may dictate which genes are transcribed by the AR.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. XuBao Shi, University of California, Davis, for stable pRNS-1-1 cell lines expressing an empty vector, wild-type androgen receptor and a mutated androgen receptor, Dr. Sheetal Singh, VA Northern California Health Care System, for comparing the nucleotide sequence of Nrdp1 ARE3 in LNCaP and C4-2 cell lines. We thank former interns Mohana Roy and Anisha Madhav for their valuable input in the preparation of the data. We also thank Ms. Stephanie Soares, University of California Davis, for the construction of the tissue microarrays used in this study, and Ms. Jessica Wald, Department of Biochemistry and Molecular Medicine, for technical assistance.
FUNDING
This work was supported by a Biomedical Laboratory Research & Development (BLRD) service Merit Award (I01BX000400, PMG) from the Department of Veterans Affairs, Office of Research and Development, Biomedical Laboratory Research Program, and by Awards R01CA133209 (PMG), R01CA185509 (PMG), and R01CA123541 (KLC) from the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest. The work reported here does not represent the views or opinions of the Department of Veteran Affairs or the United States Government.
AUTHOR CONTRIBUTIONS
Conception and design: Paramita M. Ghosh
Development of methodology: Paramita M. Ghosh, Maria Mudryj, Kermit L. Carraway, III
Acquisition of data: Rosalinda M. Savoy, Liqun Chen, Salma Siddiqui, Maitreyee K. Jathal, Thomas M. Steele, Swagata Bose, Benjamin A. Mooso, Leandro S. D’Abronzo, William H. Fry
Analysis and interpretation of data: Frank U. Melgoza, Blythe Durbin-Johnson, Christiana Drake, Paramita M. Ghosh.
Writing, review and/or revision of the manuscript: Rosalinda M. Savoy, Paramita M. Ghosh
Supplementary Information accompanies the paper.
REFERENCES
- Batch JA, Williams DM, Davies HR, Brown BD, Evans BA, Hughes IA, Patterson MN. Role of the androgen receptor in male sexual differentiation. Horm Res. 1992;38:226–229. doi: 10.1159/000182548. [DOI] [PubMed] [Google Scholar]
- Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, Ghosh PM. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res. 2007;13:3860–3867. doi: 10.1158/1078-0432.CCR-07-0091. [DOI] [PubMed] [Google Scholar]
- Bedolla RG, Wang Y, Asuncion A, Chamie K, Siddiqui S, Mudryj MM, Prihoda TJ, Siddiqui J, Chinnaiyan AM, Mehra R, et al. Nuclear versus cytoplasmic localization of filamin A in prostate cancer: immunohistochemical correlation with metastases. Clin Cancer Res. 2009;15:788–796. doi: 10.1158/1078-0432.CCR-08-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger R, Febbo PG, Majumder PK, Zhao JJ, Mukherjee S, Signoretti S, Campbell KT, Sellers WR, Roberts TM, Loda M, et al. Androgen-induced differentiation and tumorigenicity of human prostate epithelial cells. Cancer Res. 2004;64:8867–8875. doi: 10.1158/0008-5472.CAN-04-2938. [DOI] [PubMed] [Google Scholar]
- Cai C, Wang H, Xu Y, Chen S, Balk SP. Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 2009;69:6027–6032. doi: 10.1158/0008-5472.CAN-09-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Wu X, Yen L, Sweeney C, Carraway KL. Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Molecular and Cellular Biology. 2007;27:2180–2188. doi: 10.1128/MCB.01245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoria G, D’Amato L, Ciociola A, Giovannelli P, Giraldi T, Sepe L, Paolella G, Barone MV, Migliaccio A, Auricchio F. Androgen-induced cell migration: role of androgen receptor/filamin A association. PLoS One. 2011;6:e17218. doi: 10.1371/journal.pone.0017218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Libertini SJ, George M, Dandekar S, Tepper CG, Al-Bataina B, Kung HJ, Ghosh PM, Mudryj M. Genome-wide analysis of androgen receptor binding and gene regulation in two CWR22-derived prostate cancer cell lines. Endocr Relat Cancer. 2010a;17:857–873. doi: 10.1677/ERC-10-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Siddiqui S, Bose S, Mooso B, Asuncion A, Bedolla RG, Vinall R, Tepper CG, Gandour-Edwards R, Shi X, et al. Nrdp1-mediated regulation of ErbB3 expression by the androgen receptor in androgen-dependent but not castrate-resistant prostate cancer cells. Cancer Res. 2010b;70:5994–6003. doi: 10.1158/0008-5472.CAN-09-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol Endocrinol. 1997;11:148–161. doi: 10.1210/mend.11.2.9883. [DOI] [PubMed] [Google Scholar]
- Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkmann AO, Trapman J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J Biol Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr, Saad F, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vere White R, Meyers F, Chi SG, Chamberlain S, Siders D, Lee F, Stewart S, Gumerlock PH. Human androgen receptor expression in prostate cancer following androgen ablation. Eur Urol. 1997;31:1–6. doi: 10.1159/000474409. [DOI] [PubMed] [Google Scholar]
- Decker KF, Zheng D, He Y, Bowman T, Edwards JR, Jia L. Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Res. 2012;40:10765–10779. doi: 10.1093/nar/gks888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lopez-Camacho C, Tang JY, Mendoza-Villanueva D, Maya-Mendoza A, Jackson DA, Shore P. Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription. Proc Natl Acad Sci U S A. 2012;109:1524–1529. doi: 10.1073/pnas.1107879109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denmeade SR, Sokoll LJ, Dalrymple S, Rosen DM, Gady AM, Bruzek D, Ricklis RM, Isaacs JT. Dissociation between androgen responsiveness for malignant growth vs. expression of prostate specific differentiation markers PSA, hK2, and PSMA in human prostate cancer models. Prostate. 2003;54:249–257. doi: 10.1002/pros.10199. [DOI] [PubMed] [Google Scholar]
- Ghosh PM, Malik SN, Bedolla RG, Wang Y, Mikhailova M, Prihoda TJ, Troyer DA, Kreisberg JI. Signal transduction pathways in androgen-dependent and -independent prostate cancer cell proliferation. Endocr Relat Cancer. 2005;12:119–134. doi: 10.1677/erc.1.00835. [DOI] [PubMed] [Google Scholar]
- Govoroun M, McMeel OM, D'Cotta H, Ricordel MJ, Smith T, Fostier A, Guiguen Y. Steroid enzyme gene expressions during natural and androgen-induced gonadal differentiation in the rainbow trout, Oncorhynchus mykiss. J Exp Zool. 2001;290:558–566. doi: 10.1002/jez.1106. [DOI] [PubMed] [Google Scholar]
- Gueron G, Giudice J, Valacco P, Paez A, Elguero B, Toscani M, Jaworski F, Leskow FC, Cotignola J, Marti M, et al. Heme-oxygenase-1 implications in cell morphology and the adhesive behavior of prostate cancer cells. Oncotarget. 2014;5:4087–4102. doi: 10.18632/oncotarget.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Kong X, Melamed J, Tepper CG, Kung HJ, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131:459–467. doi: 10.1242/dev.00957. [DOI] [PubMed] [Google Scholar]
- Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Hu G, Zhao K. ChIP-Seq: technical considerations for obtaining high-quality data. Nat Immunol. 2011;12:918–922. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisberg JI, Malik SN, Prihoda TJ, Bedolla RG, Troyer DA, Kreisberg S, Ghosh PM. Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res. 2004;64:5232–5236. doi: 10.1158/0008-5472.CAN-04-0272. [DOI] [PubMed] [Google Scholar]
- Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy CJ, Sim KS, Yong EL. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc Natl Acad Sci U S A. 2003;100:4562–4567. doi: 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath MJ, Binge LC, Sriratana A, Wang H, Robinson PA, Pook D, Fedele CG, Brown S, Dyson JM, Cottle DL, et al. Regulation of the transcriptional coactivator FHL2 licenses activation of the androgen receptor in castrate-resistant prostate cancer. Cancer Res. 2013;73:5066–5079. doi: 10.1158/0008-5472.CAN-12-4520. [DOI] [PubMed] [Google Scholar]
- Mitsiades N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res. 2013;73:4599–4605. doi: 10.1158/0008-5472.CAN-12-4414. [DOI] [PubMed] [Google Scholar]
- Mooso BA, Vinall RL, Tepper CG, Savoy RM, Cheung JP, Singh S, Siddiqui S, Wang Y, Bedolla RG, Martinez A, et al. Enhancing the effectiveness of androgen deprivation in prostate cancer by inducing Filamin A nuclear localization. Endocr Relat Cancer. 2012;19:759–777. doi: 10.1530/ERC-12-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PS. Molecular states underlying androgen receptor activation: a framework for therapeutics targeting androgen signaling in prostate cancer. J Clin Oncol. 2012;30:644–646. doi: 10.1200/JCO.2011.39.1300. [DOI] [PubMed] [Google Scholar]
- Ozanne DM, Brady ME, Cook S, Gaughan L, Neal DE, Robson CN. Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol Endocrinol. 2000;14:1618–1626. doi: 10.1210/mend.14.10.0541. [DOI] [PubMed] [Google Scholar]
- Park MH, Song MJ, Cho MC, Moon DC, Yoon do Y, Han SB, Hong JT. Interleukin-32 enhances cytotoxic effect of natural killer cells to cancer cells via activation of death receptor 3. Immunology. 2012;135:63–72. doi: 10.1111/j.1365-2567.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C, Nilsson S, Heinrich D, Helle SI, O'Sullivan JM, Fossa SD, Chodacki A, Wiechno P, Logue J, Seke M, et al. Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013;369:213–223. doi: 10.1056/NEJMoa1213755. [DOI] [PubMed] [Google Scholar]
- Podar K, Raab MS, Tonon G, Sattler M, Barila D, Zhang J, Tai YT, Yasui H, Raje N, DePinho RA, et al. Up-regulation of c-Jun inhibits proliferation and induces apoptosis via caspase-triggered c-Abl cleavage in human multiple myeloma. Cancer Res. 2007;67:1680–1688. doi: 10.1158/0008-5472.CAN-06-1863. [DOI] [PubMed] [Google Scholar]
- Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, Arora VK, Yen WF, Cai L, Zheng D, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–1253. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Markant SL, Yuan J, Goldberg AL. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 2004;23:800–810. doi: 10.1038/sj.emboj.7600075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, MacArthur S, Stark R, Warren AY, Mills IG, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Shi XB, Xue L, Tepper CG, Gandour-Edwards R, Ghosh P, Kung HJ, DeVere White RW. The oncogenic potential of a prostate cancer-derived androgen receptor mutant. Prostate. 2007;67:591–602. doi: 10.1002/pros.20544. [DOI] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Sun GG, Lu YF, Zhang J, Hu WN. Filamin A regulates MMP-9 expression and suppresses prostate cancer cell migration and invasion. Tumour Biol. 2014;35:3819–3826. doi: 10.1007/s13277-013-1504-6. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- van de Wijngaart DJ, Dubbink HJ, van Royen ME, Trapman J, Jenster G. Androgen receptor coregulators: recruitment via the coactivator binding groove. Mol Cell Endocrinol. 2012;352:57–69. doi: 10.1016/j.mce.2011.08.007. [DOI] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- Velkova A, Carvalho MA, Johnson JO, Tavtigian SV, Monteiro AN. Identification of Filamin A as a BRCA1-interacting protein required for efficient DNA repair. Cell Cycle. 2010;9:1421–1433. doi: 10.4161/cc.9.7.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinall RL, Tepper CG, Shi XB, Xue LA, Gandour-Edwards R, de Vere White RW. The R273H p53 mutation can facilitate the androgen-independent growth of LNCaP by a mechanism that involves H2 relaxin and its cognate receptor LGR7. Oncogene. 2006;25:2082–2093. doi: 10.1038/sj.onc.1209246. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat Immunol. 2009a;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009b;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chrysovergis K, Bienstock RJ, Shim M, Eling TE. The H6D variant of NAG-1/GDF15 inhibits prostate xenograft growth in vivo. Prostate. 2012;72:677–689. doi: 10.1002/pros.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kreisberg JI, Bedolla RG, Mikhailova M, deVere White RW, Ghosh PM. A 90 kDa fragment of filamin A promotes Casodex-induced growth inhibition in Casodex-resistant androgen receptor positive C4-2 prostate cancer cells. Oncogene. 2007;26:6061–6070. doi: 10.1038/sj.onc.1210435. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mikhailova M, Bose S, Pan CX, deVere White RW, Ghosh PM. Regulation of androgen receptor transcriptional activity by rapamycin in prostate cancer cell proliferation and survival. Oncogene. 2008;27:7106–7117. doi: 10.1038/onc.2008.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauman J, De Ceuninck L, Vanderroost N, Lievens S, Tavernier J. RNF41 (Nrdp1) controls type 1 cytokine receptor degradation and ectodomain shedding. J Cell Sci. 2011;124:921–932. doi: 10.1242/jcs.078055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer. 1994;57:406–412. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]
- Wu X, Yen L, Irwin L, Sweeney C, Carraway KL. Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Molecular and Cellular Biology. 2004;24:7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sato H, Suzuki T, Yoshizawa T, Morohashi S, Seino H, Kawamoto T, Fujimoto K, Kato Y, Kijima H. Involvement of c-Myc in the proliferation of MCF-7 human breast cancer cells induced by bHLH transcription factor DEC2. Int J Mol Med. 2014 doi: 10.3892/ijmm.2014.2042. [DOI] [PubMed] [Google Scholar]
- Ye S, Xu H, Jin J, Yang M, Wang C, Yu Y, Cao X. The E3 ubiquitin ligase neuregulin receptor degradation protein 1 (Nrdp1) promotes M2 macrophage polarization by ubiquitinating and activating transcription factor CCAAT/enhancer-binding Protein beta (C/EBPbeta) J Biol Chem. 2012;287:26740–26748. doi: 10.1074/jbc.M112.383265. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yen L, Cao Z, Wu X, Ingalla ERQ, Baron C, Young LJT, Gregg JP, Cardiff RD, Borowsky AD, Sweeney C, et al. Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Res. 2006;66:11279–11286. doi: 10.1158/0008-5472.CAN-06-2319. [DOI] [PubMed] [Google Scholar]
- Yuan X, Cai C, Chen S, Chen S, Yu Z, Balk SP. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33:2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Wang Q, Lu H, Brenneman M, Fan F, Shen Z. The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 2009;69:7978–7985. doi: 10.1158/0008-5472.CAN-09-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun HM, Oh JH, Shim JH, Ban JO, Park KR, Kim JH, Lee DH, Kang JW, Park YH, Yu D, et al. Antitumor activity of IL-32beta through the activation of lymphocytes, and the inactivation of NF-kappaB and STAT3 signals. Cell Death Dis. 2013;4:e640. doi: 10.1038/cddis.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Tan Y, Zhou A, Yu Q, Zhou J. RING finger ubiquitin-protein isopeptide ligase Nrdp1/FLRF regulates parkin stability and activity. J Biol Chem. 2005;280:9425–9430. doi: 10.1074/jbc.M408955200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.