To the Editor
Insulin receptor substrate 1 (IRS1) plays a key role in the insulin-stimulated signal transduction pathway [1]. Recently, a genome-wide association study showed that a variant (rs2943641) near the IRS1 gene was associated with insulin resistance, hyperinsulinaemia and the risk of type 2 diabetes [2].
In this study, we comprehensively examined the associations of the variants near the IRS1 gene with type 2 diabetes risk in two nested case–control studies from the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS). Because physical activity may modify the genetic effects on diabetes [3] and enhance insulin activation of IRS1-associated phosphatidylinositol 3-kinase activity [4], we further examined the gene–physical activity interactions in relation to the risk of diabetes.
Type 2 diabetes cases were identified by self-report methods that were confirmed with a validated supplementary questionnaire. The study was approved by the Human Research Committee at the Brigham and Women’s Hospital, Boston. General information was derived from the baseline questionnaire. Physical activity was expressed as metabolic equivalent task (MET) h of moderate to vigorous exercise per week in men, and as h/week in women because MET was not measured at baseline.
We used genotyping data from a genome-wide scan. We examined associations with type 2 diabetes for the single-nucleotide polymorphisms (SNPs) near the reported SNP rs2943641 [2] (electronic supplementary material [ESM] Fig. 1). We used logistic regression to estimate ORs after adjustment for potential risk factors. An additive genetic model was applied in the first instance. Tests of interaction between SNPs and physical activity were assessed by including the cross-product of the tested variables and the main effects in the model. All SNPs fitted Hardy–Weinberg equilibrium. The SAS statistical package was used (SAS, version 9.0 for UNIX; SAS Institute, Cary, NC, USA). The meta-analysis was carried out by using Mix 1.7 software [5].
Patients with type 2 diabetes had a significantly higher BMI, engaged in less physical activity and were more likely to smoke and have a family history of diabetes than control participants, among both men (HPFS) and women (NHS) (ESM Table 1).
The nominally associated SNPs fell into two major linkage disequilibrium (LD) blocks. In one block, the reported SNP, rs2943641, was significantly associated with type 2 diabetes (per T allele, OR 0.88, 95% CI 0.80–0.97; p=0.008) in the combined analysis. Subjects with the T allele had 15% and 9% decreased risk for men (HPFS) and women (NHS), respectively (Table 1).
Table 1.
Associations between SNPs rs1522813 and rs2943641 and type 2 diabetes in NHS and HPFS
| SNP | Genotype | NHS (women)
|
HPFS (men)
|
Meta-analysis
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n=1,467) | Controls (n=1,754) | Crude OR (95% CI) | p value | Adjusted OR (95% CI)a | p value | Cases (n=1,124) | Controls (n=1,298) | Crude OR (95% CI) | p value | Adjusted OR (95% CI)a | p value | OR (95% CI) | p value | ||
| rs1522813 | GG | 784 (53.4) | 1,003 (57.2) | 1.00 | – | 1.00 | – | 550 (48.9) | 691 (53.2) | 1.00 | – | 1.00 | – | – | – |
| GA | 581 (39.6) | 630 (35.9) | 1.18 (1.02–1.37) | 0.03 | 1.25 (1.03–1.51) | 0.02 | 470 (41.8) | 510 (39.3) | 1.16 (0.98–1.37) | 0.09 | 1.21 (1.00–1.46) | 0.06 | 1.23 (1.07–1.41) | 0.003 | |
| AA | 102 (7.0) | 121 (6.9) | 1.08 (0.82–1.43) | 0.60 | 1.21 (0.84–1.74) | 0.30 | 104 (9.3) | 97 (7.5) | 1.35 (1.00–1.82) | 0.05 | 1.38 (0.98–1.94) | 0.07 | 1.29 (1.01–1.66) | 0.042 | |
| Additive model | – | – | 1.10 (0.99–1.23) | 0.09 | 1.17 (1.01–1.35) | 0.03 | – | – | 1.16 (1.02–1.31) | 0.02 | 1.19 (1.03–1.37) | 0.019 | 1.18 (1.06–1.30) | 0.002 | |
| rs2943641 | CC | 641 (43.7) | 722 (41.2) | 1.00 | – | 1.00 | – | 510 (45.4) | 539 (41.5) | 1.00 | – | 1.00 | – | – | – |
| CT | 636 (43.4) | 798 (45.5) | 0.90 (0.77–1.04) | 0.16 | 0.91 (0.75–1.11) | 0.36 | 489 (43.5) | 570 (43.9) | 0.91 (0.76–1.08) | 0.26 | 0.92 (0.76–1.12) | 0.41 | 0.92 (0.80–1.05) | 0.218 | |
| TT | 190 (13.0) | 234 (13.3) | 0.91 (0.74–1.14) | 0.42 | 0.83 (0.62–1.11) | 0.21 | 125 (11.1) | 189 (14.6) | 0.70 (0.54–0.90) | 0.006 | 0.67 (0.50–0.90) | 0.008 | 0.75 (0.61–0.92) | 0.006 | |
| Additive model | – | – | 0.94 (0.85–1.04) | 0.23 | 0.91 (0.80–1.04) | 0.18 | – | – | 0.86 (0.76–0.96) | 0.009 | 0.85 (0.74–0.97) | 0.016 | 0.88 (0.80–0.97) | 0.008 | |
Adjusted for age, BMI (five categories), family history of diabetes (yes, no), smoking (never, past or current), alcohol (five categories), menopausal status (pre- or post-menopausal [never, past or current hormone use]; women only), quintiles of physical activity (MET h/week for men in HPFS, h/week for women in NHS) and quintiles of energy-adjusted polyunsaturated fatty acid:saturated fatty acid ratio, and intakes of trans fat and cereal fibre
In another LD block, SNP rs1522813 showed the strongest association (ESM Figs. 1, 2), each A allele being associated with an 18% increased risk of diabetes (95% CI 1.06–1.30; p=0.002; Table 1). We further performed conditional analyses by simultaneously including SNPs from each LD block (the best-associated SNP rs1522813 and the reported SNP rs2943641); both SNPs remained significant in the conditional analyses (p=0.012 and 0.022, respectively). For the SNP rs1522813, each A allele was associated with an 11% increased risk of diabetes (95% CI 1.02–1.21); for the SNP rs2943641, each T allele was associated with a 9% decreased risk (95% CI 0.85–0.99). The population attributable risk of SNPs rs1522813 and rs2943641 was 9.28% and 6.58%, respectively. SNPs rs1522813 and rs2943641 showed weak LD in the study samples (r2=0.02). The previously reported missense polymorphism Gly972Arg (rs1801278) in IRS1 [6] was not associated with type 2 diabetes.
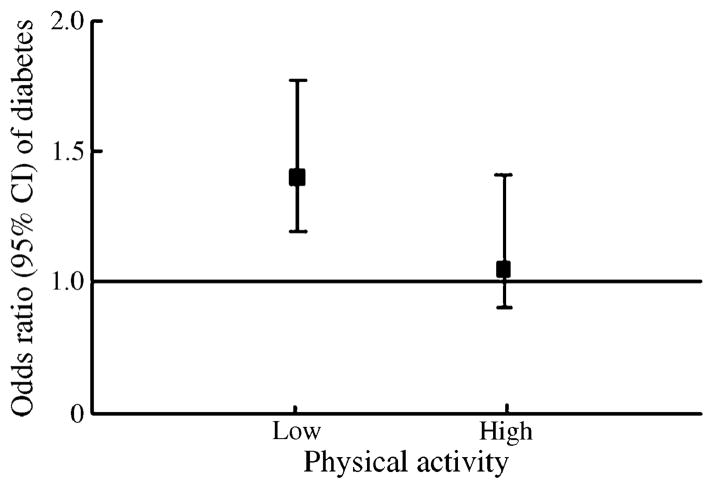
We further examined the associations of the two SNPs with diabetes risk stratified by physical activity. We observed a significant interaction (p=0.017; Fig. 1) between physical activity and rs1522813 in women (NHS). Among women with low levels of physical activity (low vs high levels, by median values), carriers of the A allele had a 39% increased risk of type 2 diabetes (p=0.007); among women with high levels of physical activity, carriers of the A allele had a 4% increased risk of diabetes (p=0.78). The test for interaction between rs1522813 and physical activity was not statistically significant in men (HPFS) (p=0.26). Physical activity did not interact with rs2943641 in either sex. In order to replicate our results, we also examined the associations between the two SNPs and the risk of type 2 diabetes in the Diabetes Genetics Replication and Meta-analysis Consortium (DIAGRAM). The ORs (95% CI) were 1.09 (1.04–1.13) for SNP rs2943641 and 1.01 (0.96–1.06) for rs1522813. In DIAGRAM, the associations for rs1522813 showed significant between-study heterogeneity. This may reflect the greater diversity in the DIAGRAM study samples, which were from a wide range of geographical regions (and therefore had more dissimilar LD structure). In the present study, with more homogeneous populations, SNP rs1522813 showed consistently significant associations with comparable effect size in two independent cohorts. The potential influence of environmental factors may also contribute to the difference.
Fig. 1.
Interaction between physical activity and SNP rs1522813 on type 2 diabetes risk in women. Physical activity was classified into two groups according to the median value in controls (cutoff point 3.0 h/week). The stratified analyses of rs1522813 (carries of A allele comparison with GG genotypes) with type 2 diabetes risk according to physical activity were adjusted for age, BMI (five categories), family history of diabetes (yes, no), smoking (never, past, current), alcohol (five categories), menopausal status (pre- or post-menopausal [never, past or current hormone use]; women only), and quintiles of energy-adjusted polyunsaturated fatty acid:saturated fatty acid ratio, and intakes of trans fat and cereal fibre. p value for interaction=0.017
We have demonstrated the associations of several previously reported genetic variants with the risk of type 2 diabetes in our cohorts [7]. In the present study, our data indicate that multiple variants near the IRS1 gene may independently affect the risk of diabetes. SNP rs1522813 was associated with diabetes risk even after conditioning for the reported SNP, rs2943641. Our data support the notion that allelic heterogeneity may exist in the genetic architecture of type 2 diabetes, in line with the observations for other metabolic traits, such as the independent associations of multiple SNPs in the MC4R locus with obesity [8]. In addition, we found that SNP rs1522813 interacted with physical activity in relation to type 2 diabetes risk in women but not in men. This may be partly attributable to a higher level of physical activity in men than in women (mean physical activity in men and women in 1986 was 21.1 and 14.2 MET h/week, respectively; p<0.001). Because of the replication nature of the study, we did not control for multiple testing, but our data are consistent with a previous genome-wide association study [2] and in line with evidence from experimental studies [4].
In summary, we confirmed that polymorphisms near IRS1 were significantly associated with the risk of type 2 diabetes, and we identified a new SNP independently associated with the disease. In addition, we report that physical activity might modify the association in women.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (DK58845, HL34594 and HL71981), an American Heart Association Scientist Development Award and the Boston Obesity Nutrition Research Center (DK46200 to L. Qi).
Abbreviations
- HPFS
Health Professionals Follow-up Study
- IRS1
Insulin receptor substrate 1
- LD
Linkage disequilibrium
- MET
Metabolic equivalent task
- NHS
Nurses’ Health Study
- SNP
Single nucleotide polymorphism
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00125-011-2123-7) contains supplementary material, which is available to authorised users.
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Contributor Information
M. A. He, Department of Nutrition, Harvard School of Public Health, Boston, MA 02115, USA. Institute of Occupational Medicine, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, People’s Republic of China. Department of Cardiology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, People’s Republic of China
T. Workalemahu, Department of Nutrition, Harvard School of Public Health, Boston, MA 02115, USA
M. C. Cornelis, Department of Nutrition, Harvard School of Public Health, Boston, MA 02115, USA
F. B. Hu, Department of Nutrition, Harvard School of Public Health, Boston, MA 02115, USA. Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA
L. Qi, Email: nhlqi@channing.harvard.edu, Department of Nutrition, Harvard School of Public Health, Boston, MA 02115, USA. Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA
References
- 1.Sun XJ, Rothenberg P, Kahn CR, et al. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 2.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 3.Qi L, Hu FB, Hu G. Genes, environment, and interactions in prevention of type 2 diabetes: a focus on physical activity and lifestyle changes. Curr Mol Med. 2008;8:519–532. doi: 10.2174/156652408785747915. [DOI] [PubMed] [Google Scholar]
- 4.Kirwan JP, del Aguila LF, Hernandez JM, et al. Regular exercise enhances insulin activation of IRS-1-associated PI3-kinase in human skeletal muscle. J Appl Physiol. 2000;88:797–803. doi: 10.1152/jappl.2000.88.2.797. [DOI] [PubMed] [Google Scholar]
- 5.Bax L, Yu LM, Ikeda N, Tsuruta H, Moons KG. Development and validation of MIX: comprehensive free software for meta-analysis of causal research data. BMC Med Res Methodol. 2006;6:50. doi: 10.1186/1471-2288-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jellema A, Zeegers MP, Feskens EJ, Dagnelie PC, Mensink RP. Gly972Arg variant in the insulin receptor substrate-1 gene and association with type 2 diabetes: a meta-analysis of 27 studies. Diabetologia. 2003;46:990–995. doi: 10.1007/s00125-003-1126-4. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis MC, Qi L, Zhang C, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in US men and women of European ancestry. Ann Intern Med. 2009;150:541–550. doi: 10.7326/0003-4819-150-8-200904210-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.