Abstract
We present a novel parametric encoding scheme for efficiently recording white matter fiber bundle information obtained from diffusion tensor imaging. The coordinates of fiber tracts are parameterized using a cosine series expansion. For an arbitrary tract, a 19 degree expansion is found to be sufficient to reconstruct the tract with an average error of about 0.26 mm. Then each tract is fully parameterized with 60 parameters, which results in a substantial data reduction. Unlike traditional splines, the proposed method does not have internal knots and explicitly represents the tract as a linear combination of basis functions. This simplicity in the representation enables us to design statistical models, register tracts and perform subsequent analysis in a more streamlined mathematical framework. As an illustration, we apply the proposed method in characterizing abnormal tracts that pass through the splenium of the corpus callosum in autistic subjects.
I. Introduction
Diffusion tensor imaging (DTI) can be used to characterize the microstructure of biological tissues using measures of the magnitude, anisotropy and aniotropic orientation [2]. It is assumed that the direction of greatest diffusivity is most likely aligned to the local orientation of the white matter fibers. White matter tractography offers the unique opportunity to map out, segment and characterize the trajectories of white matter fiber bundles noninvasively in the brain. Most deterministic tractography algorithms use the local diffusion tensor orientation to estimate the local direction of propagation along the reconstructed pathway or fiber tract [3] [8] [13] [15]. Tractography has been used to visualize and map out major white matter pathways in individuals and brain atlases [6] [16] [23] [24], segment specific white matter areas for region of interest analyses [12], quantify white matter morphometry and connections [19] [22], and visualize the relationships between brain pathology and white matter anatomy for clinical applications like neurosurgical planning [1] [17] [18]. However, tractography data can be challenging to interpret and quantify. Whole brain tractography studies often generate many hundreds of thousand tracts. Recent efforts have attempted to cluster [20] and automatically segment white matter tracts [21] as well as characterize tract shape parameters [4]. Many of these techniques can be quite computationally demanding. Clearly efficient methods for representing tract shape, regional tract segmentation and clustering, tract registration and quantification would be of tremendous value to researchers.
In this paper, we present a novel approach for parameterizing tract shapes using Fourier descriptors. Fourier descriptors have been previously used to classify tracts [4]. The Fourier coefficients are computed by the Fourier transform that involves the both sine and cosine series expansion. Then the sum of the squared coefficients are obtained up to degree 30 for each tract and the k-means clustering is used to classify the fibers globally. Our approach differs from [4] in that we obtain local shape information employing cosine series only, a special case of Fourier series. Using the new representation, we demonstrate how to quantify abnormal pattern of white matter fibers passing through the splenium of the corpus callosum for autistic subjects.
II. Cosine Representation
We are interested in encoding a tract ℳ consisting of 𝔫 noisy control points 𝔭1, ⋯, 𝔭𝔫. Consider a mapping ζ−1 that maps the control point 𝔭𝔧 onto the unit interval [0, 1] as
| (1) |
This is the ratio of the arc-length from the point 𝔭1 to p𝔧, to 𝔭1 to 𝔭𝔫. We let this ratio to be 𝔱𝔧. We assume ζ−1(𝔭1) = 0. Then we parameterize the smooth inverse map
using the cosine basis functions:
The representation is first introduced in [7]. The constant is introduced to make the eigenfunctions orthonormal in [0, 1] so that
| (2) |
the Dirac-delta function. If we denote the coordinates of ζ as (ζ1, ζ2, ζ3), the 𝗄-th degree cosine series representation is given by
| (3) |
The Fourier coefficients 𝔠𝔩𝔬 are estimated by solving the system of equations obtained at the 𝔫 control points:
| (4) |
In the matrix notation, we write (4) as
where 𝖸 = (ζ𝔬(𝔱𝔧)), Ψ = (ψ𝔩(𝔱𝔧)) and 𝖢 = (𝔠𝔩𝔬). Then the least squares estimation of 𝖢 is given by
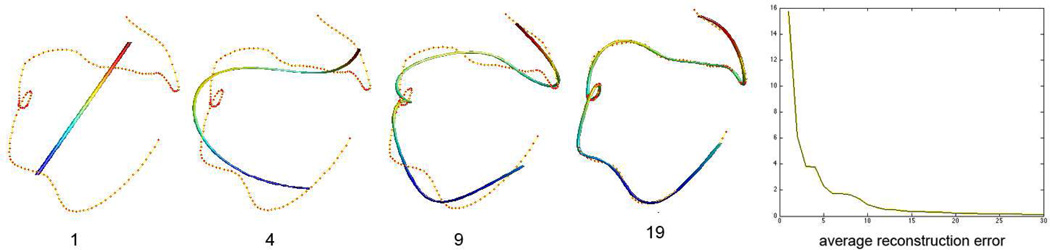
The proposed least squares estimation technique avoids using the Fourier transform (FT) [4] [5] [9]. The drawback of the FT is the need for a predefined regular grid system so some sort of interpolation is needed. After various experiments to obtain the optimal degree, we decided to use 𝗄 = 19 through out the paper (Figure 1). This gives the average error of 0.26mm along the tract. The plot of the average reconstruction error for other degrees is given in Figure 1.
Fig. 1.
Cosine representation of a tract at various degrees. Red dots are control points. The degree 1 representation is a straight line that fits all the control points in a least squares fashion. The error plot displays the average reconstruction error in millimeter (vertical) vs. degree (horizontal).
The advantage of the cosine representation is that, instead of recording the coordinates of all control points, we only need to record 3 · (𝗄 + 1) number of parameters for all possible tract shape. This is a substantial data reduction considering that the average number of control points is 105 (315 parameters). We recommend readers to use 𝗄 ≤ 30 degrees for most applications.
III. Application to autism study
A. Image Acquisition and Preprocessing
DTI data were acquired on a Siemens Trio 3.0 Tesla Scanner with an 8-channel, receive-only head coil. Diffusion-weighted images were acquired in 12 non-collinear diffusion encoding directions with diffusion weighting factor 1000 s/mm2 in addition to a single reference image. Data acquisition parameters included the following: contiguous (no-gap) fifty 2.5mm thick axial slices with an acquisition matrix of 128×128 over a FOV of 256mm, 4 averages, repetition time (TR) = 7000 ms, and echo time (TE) = 84 ms.
Eddy current related distortion and head motion of each data set were corrected using AIR and distortions from field inhomogeneities were corrected using custom software algorithms based on [11]. Distortion-corrected DW images were interpolated to 2 × 2 × 2mm voxels and the six tensor elements were calculated using a multivariate log-linear regression method [2].
The images were isotropically resampled at 1mm3 resolution before applying the white matter tractography algorithm. The second order Runge-Kutta streamline algorithm with tensor deflection [13] was used. The trajectories were initiated at the center of the seed voxels and were terminated if they either reached regions with the factional anisotropy (FA) value smaller then 0.15 or if the angle between two consecutive steps along the trajectory was larger than π/4. Each tract consists of 105 ± 54 control points. The distance between control points is 1mm. Whole brain tracts are stored as a file of size approximately 600MB. Whole brain white matter tracts for 74 subjects are further aligned using the affine registration [10] of FA-maps to the average FA-map.
B. Autism Population Study
The representation provides 60 dimensional feature vectors (coefficients) for characterizing a single tract. We have investigated the utility of the proposed representation in the ability to discriminate the different clinical populations (42 autistic and 32 control subjects). We have focused our detailed anatomical study on the splenium of the corpus callosum, which is manually masked by J.E. Lee [14]. Then the tracts passing through a ball of radius 5mm at the spleninum are identified. Each subject have 1943 ± 1148 number of tracts passing through the ball. The within-subject tract averaging can be easily done within our representations by summing the coefficients of the same degree [7] (Figure 2). First two images in Figure 3 shows the 74 average within-subject tracts color coded according to autism (red) and controls (blue). The control subjects seem to show more clustering of fibers compared to autistic subjects. So we have tested the statistical significance of the clustering.
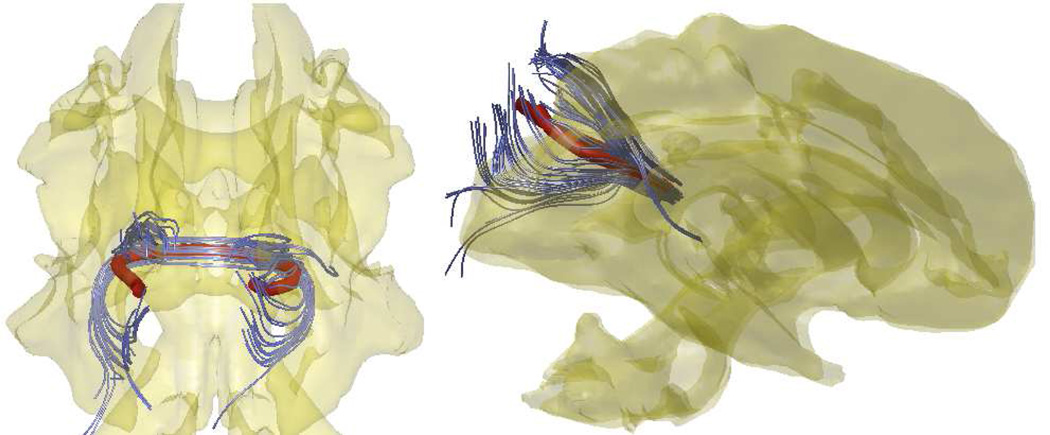
Fig. 2.
The within-subject average tract (red) of 2149 fibers. 2149 fiber tracts are subsampled to show few selective tracts (blue). The average tract is obtained by averaging the Fourier coefficients of 2149 cosine representations.
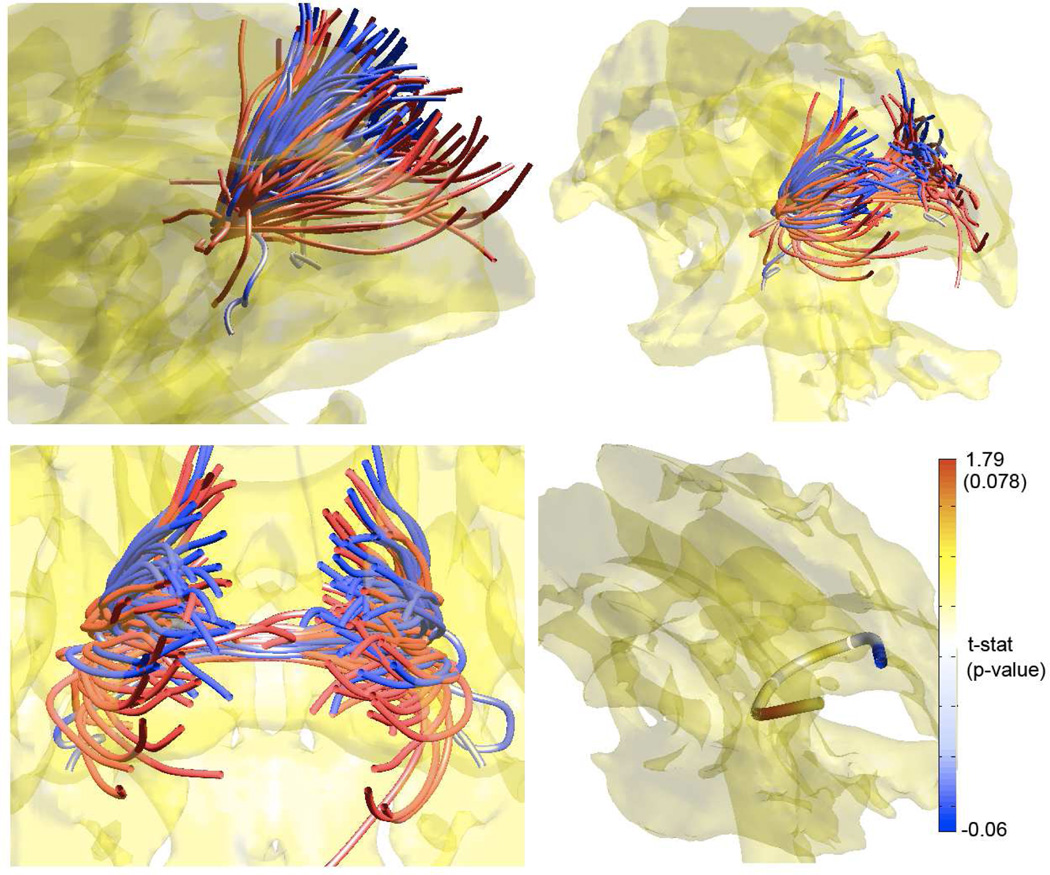
Fig. 3.
Each streamtube is the average of tracts passing through a ball of 5mm radius around the splenium in a subject. White matter fibers in controls (blue) are more clustered together with smaller spreading compared to autism (red). Thick streamtube at the bottom right image is the population average tract of all 74 subjects. Based on the fiber concentration map, we constructed 𝔱-statistic and the corresponding 𝔭-value.
Given two tracts
the 𝖫2-distance between the two tracts is defined as
The metric ρ computes the Euclidean distance between corresponding points along the two tracts at each 𝔱. Given a fiber consisting of n tracts η1, ⋯, η𝔫 within a subject, the average tract η̄ is obtained by averaging the coefficients within the corresponding basis. Then we define the tract concentration map 𝒞 as
The value of 𝒞 increases as tracts get more clustered. The concentration map 𝒞 is a function of the parameter 𝔱 and can be projected along the average tract η̄. We can compute the average of the 74 average tracts by averaging the coefficients of the average tracts. We have constructed the two sample 𝔱-statistic (control - autism) using 74 𝒞-values and projected the statistic on the population average tract in the third image in Figure 3. We have detected the higher concentration of fibers in control subjects in the left hemisphere (𝔱-stat 1.79, 𝔭-value 0.078). Autistic subjects show abnormal brain lateralization effect in fibers passing through the splenium.
IV. Discussion
Although the cosine representation is efficient for normalizing and averaging tracts, unfortunately it is not translation, rotation and scale invariant. This might be a reason why the resulting signal is a bit weak (𝔭-value < 0.078). One simple way of obtaining translation, rotation and scale invariant representation is to project white matter fiber tracts onto a unit sphere. Consider directional vectors ν𝔦 = 𝔭𝔦 − 𝔭𝔦−1 with the convention ν1 = 𝔭1. The vectors ν𝔦 contain all the necessary information to reconstruct the original tract.
The advantage of using the spherical projection method is that it offers a translation, rotation and scale invariant tract representation. Two tracts with the identical shape but at different positions will be identically represented as the same spherical curve. The translation information is stored in ν1 value, which should be stored separately.
Since ν𝔦 are unit vectors (except ν1) in our tractography algorithm [13], they are all in 𝖲2. For a general case, which will likely happen for other tractography algorithms, we project ν𝔦 onto 𝖲2 via the spherical projection 𝖯:
𝗐𝔧 defines control points for a spherical curve. The spherical curves can be parameterized using the cosine representation
| (5) |
However, directly solving for each coordinate ζ𝔬 will violate the quadratic constraint that the spherical curve has to be embedded on 𝖲2, i.e.
| (6) |
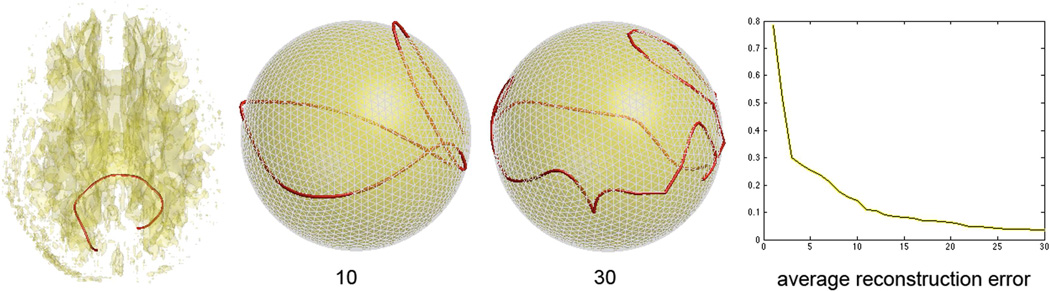
This is easily seen from Figure 4, in which the degree 10 representation is visibly not embedded in 𝖲2. The average absolute error for reconstruction is relatively large for low degree due to the fact that the representation is no longer embedded in 𝖲2. Note that at degree 30, the average absolute error is small enough, i.e. 0.0153mm, to be used for subsequent modeling.
Fig. 4.
Left: a single white matter fiber tract passing through the splenium of the corpus callosum. Middle: the cosine representation of the spherical projection of tracts at degree 10 and 30. The error plot displays the average reconstruction error in millimeter (vertical) vs. degree (horizontal) in the spherical projection method.
The spherical projection based representation can not be obtained in a straightforward fashion and requires solving three least squares problem simultaneously with the quadratic constraint (6) that relates the three equations (5). We will not consider this issue in this paper and leave it for a future study.
Another possible reason for the weak signal might be the improper choice of the fiber concentration map 𝒞. Although 𝒞 increases as tracts get more clustered, it may not be a proper metric for separating the groups. Possibly a better metric would be to use the inverse of the sample variance, i.e.
This new metric is normalized by the total number of tracts accounting for variable number of tracts for different subjects.
Acknowledgements
The authors wish to thank Gary Pack at the university of Wisconsin-Madison for discussion on the cosine representation. This work was supported by NIH Mental Retardation/Developmental Disabilities Research Center (MRDDRC Waisman Center), NIMH 62015 (ALA), NIMH MH080826 (JEL) and NICHD HD35476 (University of Utah CPEA). N. Adluru is supported by Computational Informatics in Biology and Medicine (CIBM) program and Morgridge Institute for Research at the University of Wisconsin in Madison.
Contributor Information
Moo K. Chung, Waisman Laboratory for Brain Imaging and Behavior at the University of Wisconsin, Madison, USA. Department of Brain and Cognitive Science at the Seoul National University, Korea.
Nagesh Adluru, Waisman Laboratory for Brain Imaging and Behavior at the University of Wisconsin, Madison, USA..
Jee Eun Lee, Waisman Laboratory for Brain Imaging and Behavior at the University of Wisconsin, Madison, USA..
Mariana Lazar, Center for Biomedical Imaging at the New York university school of medicine, New York, USA..
Janet E. Lainhart, University of Utah at Salt Lake City.
Andrew L. Alexander, Waisman Laboratory for Brain Imaging and Behavior at the University of Wisconsin, Madison, USA.
References
- 1.Arfanakis K, Gui M, Lazar M. Optimization of white matter tractography for pre-surgical planning and image-guided surgery. Oncology Report. 2006;15:1061–1064. doi: 10.3892/or.15.4.1061. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo tractography using dt-mri data. Magnetic Resonance in Medicine. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 4.Batchelor PG, Calamante F, Tournier JD, Atkinson D, Hill DL, Connelly A. Quantification of the shape of fiber tracts. Magnetic Resonance in Medicine. 2006;55:894–903. doi: 10.1002/mrm.20858. [DOI] [PubMed] [Google Scholar]
- 5.Bulow T. Spherical diffusion for 3d surface smoothing. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2004;26:1650–1654. doi: 10.1109/TPAMI.2004.129. [DOI] [PubMed] [Google Scholar]
- 6.Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. neuroimage. NeuroImage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- 7.Chung MK, Pack G, Lazar M, Lange NT, Lainhart JE, Alexander AL, Lee JE. A unified parametric model of white matter fiber tracts. MICCAI 2008 Workshop on Computational Diffusion MRI. 2008 [Google Scholar]
- 8.Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. Tracking neuronal fiber pathways in the living human brain. Natl Acad Sci USA. 1999;96 doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu X, Wang YL, Chan TF, Thompson TM, Yau ST. Genus zero surface conformal mapping and its application to brain surface mapping. IEEE Transactions on Medical Imaging. 2004;23:1–10. doi: 10.1109/TMI.2004.831226. [DOI] [PubMed] [Google Scholar]
- 10.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 11.Jezzard P, Balaban RS. Correction for geometric distortion in echo planar images from b0 field variations. Magn. Reson. Med. 2007;34:65–73. doi: 10.1002/mrm.1910340111. [DOI] [PubMed] [Google Scholar]
- 12.Jones DK, Catani M, Pierpaoli C, Reeves SJ, Shergill SS, O’Sullivan M, Golesworthy P, McGuire P, Horsfield MA, Simmons A, Williams SC, Howard RJ. Age effects on diffusion tensor magnetic resonance imaging tractography measures of frontal cortex connections in schizophrenia. Human Brain Mapping. 2006;27:230–238. doi: 10.1002/hbm.20179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazar M, Weinstein DM, Tsuruda JS, Hasan KM, Arfanakis K, Meyerand ME, Badie B, Rowley H, Haughton V, Field A, Witwer B, Alexander AL. White matter tractography using tensor deflection. Human Brain Mapping. 2003;18:306–321. doi: 10.1002/hbm.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Hsu D, Alexander AL, Lazar M, Bigler D, Lainhart JE. A study of underconnectivity in autism using DTI: W-matrix tractography. Proceedings of ISMRM. 2008 [Google Scholar]
- 15.Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of Neurology. 1999;45:256–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Mori S, Kaufmann WE, Davatzikos C, Stieljes, Amodei L, Fredericksen K, Pearlson GD, Melhem ER, Solaiyappan M, Raymond GV, Moser HW, van Zijl PC. Imaging cortical association tracts in the human brain using diffusion-tensor-based axonal tracking. Magnetic Resonance in Medicine. 2002;47:215–223. doi: 10.1002/mrm.10074. [DOI] [PubMed] [Google Scholar]
- 17.Mller M, Frandsen J, Andersen G, Gjedde A, Vestergaard-Poulsen P, stergaard L. Dynamic changes in corticospinal tracts after stroke detected by fibretracking. Journal of Neurol. Neurosurg. Psychiatry. 2007;78:587–592. doi: 10.1136/jnnp.2006.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nimsky C, Ganslandt O, Hastreiter P, Wang R, Benner T, Sorensen AG, Fahlbusch R. Preoperative and intraoperative diffusion tensor imaging-based fiber tracking in glioma surgery. Neurosurgery. 2005;56:130–137. doi: 10.1227/01.neu.0000144842.18771.30. [DOI] [PubMed] [Google Scholar]
- 19.Nucifora PG, Verma R, Melhem ER, Gur RE, Gur RC. Leftward asymmetry in relative fiber density of the arcuate fasciculus. Neuroreport. 2005;16:791–794. doi: 10.1097/00001756-200505310-00002. [DOI] [PubMed] [Google Scholar]
- 20.O’Donnell LJ, Kubicki M, Shenton ME, Dreusicke MH, Grimson WE, Westin CF. A method for clustering white matter fiber tracts. American Journal of Neuroradiology. 2006;27:1032–1036. [PMC free article] [PubMed] [Google Scholar]
- 21.O’Donnell LJ, Westin CF. Automatic tractography segmentation using a high-dimensional white matter atlas. IEEE Transactions on Medical Imaging. 2007;26:1562–1575. doi: 10.1109/TMI.2007.906785. [DOI] [PubMed] [Google Scholar]
- 22.Roberts TP, Liu F, Kassner A, Mori S, Guha A. Fiber density index correlates with reduced fractional anisotropy in white matter of patients with glioblastoma. American Journal of Neuroradiology. 2005;26:2183–2186. [PMC free article] [PubMed] [Google Scholar]
- 23.Thottakara P, Lazar M, Johnson SC, Alexander AL. Probabilistic connectivity and segmentation of white matter using tractography and cortical templates. Neuroimage. 2006;29 doi: 10.1016/j.neuroimage.2005.08.051. [DOI] [PubMed] [Google Scholar]
- 24.Yushkevich PA, Zhang H, Simon TJ, Gee JC. Structure-specific statistical mapping of white matter tracts using the continuous medial representation; IEEE 11th International Conference on Computer Vision (ICCV); 2007. pp. 1–8. [Google Scholar]