Abstract

Drug development has a high attrition rate, with poor pharmacokinetic and safety properties a significant hurdle. Computational approaches may help minimize these risks. We have developed a novel approach (pkCSM) which uses graph-based signatures to develop predictive models of central ADMET properties for drug development. pkCSM performs as well or better than current methods. A freely accessible web server (http://structure.bioc.cam.ac.uk/pkcsm), which retains no information submitted to it, provides an integrated platform to rapidly evaluate pharmacokinetic and toxicity properties.
Introduction
Developing new drugs has become an increasingly challenging, costly, and risky endeavor with a low success rate. The vast majority of drugs evaluated in clinical trials do not reach the market due to either a lack of efficacy or unacceptable side effects.1 Drug development is a fine balance of optimizing drug like properties to maximize efficacy, safety, and pharmacokinetics. Many early stage drug discovery programs focus on identifying molecules that bind to a target of interest. While potency is a driving factor in these early stages, ultimately the pharmacokinetic and toxicity properties dictate whether it will ever advance its effectiveness and success therapeutically.
The interaction between pharmacokinetics, toxicity, and potency is crucial for effective drugs. The pharmacokinetic profile of a compound defines its absorption, distribution, metabolism, and excretion (ADME) properties. While optimal binding properties of a new drug to the therapeutic target are crucial, ensuring that it can reach the target site in sufficient concentrations to produce the physiological effect safely is essential for the introduction into the clinic. Appreciation of the importance of ADMET properties has led to their consideration in early stage drug development, leading to a significant reduction in the number of compounds that failed in clinical trials due to poor ADMET properties.2−6
One strategy that has been widely employed is the introduction of physicochemical filters, such as Lipinski’s “Rule of 5”7 or the PAINs filters8 as guidelines for what may constitute a successful drug. These try to identify broad chemical properties that may increase a molecules chances to reach the market, however, presenting the converse effect of limiting potential unexplored chemical space, from which successful drugs have been originated from.9 Even using the extensive data available within pharmaceutical companies can lead to conflicting rules,10,11 highlighting the difficulty associated with applying these filters. Ultimately, irrespective of filters, the early ADMET profiling of drug candidates is a crucial component in determining the potential success of a new compound and when integrated into the drug development process can hopefully mitigate the risk of attrition.
Experimental evaluation of small-molecule ADMET properties is both time-consuming and expensive and does not always scale between animal models and humans. The evolution of computational approaches to optimize pharmacokinetic and toxicity properties may enable the progression of discovery leads effectively and swiftly to drug candidates. The prediction of ADMET-associated properties of new chemicals, however, is a challenging task with only tenuous links between many physicochemical characteristics and pharmacokinetic and toxicity properties. This has led to a need for novel approaches to understand, explore, and predict ADMET properties of small molecules as a way to improve compound quality and success rate.12
Many in silico approaches for predicting pharmacokinetic and toxicity properties of compounds from their chemical structure have been developed,13 ranging from data-based approaches such as quantitative structure–activity relationship (QSAR),14,15 similarity searches,16,17 and 3-dimensional QSAR,18 to structure-based methods such as ligand–protein docking19 and pharmacophore modeling.20 Many of these are unfortunately not freely available, which limits their utility for the scientific community.
Numerous databases of experimentally measured ADMET properties have been compiled,21−30 some of which are freely available. Using these databases a number of QSAR models have been generated to predict some of these properties.22,31−36 The problem with these methods is that they tend to focus on recognition of certain substructure elements and are prone to be of limited use when exploring novel chemical entities beyond the scope of the experimental data used to generate the original models. Machine learning approaches, however, rely upon learning patterns between chemical composition, similarity, and pharmacokinetic and safety properties in order to build predictive models capable of generalization, i.e., discovering implicit patterns consistent and valid for unseen data.
Here we use the concept of graph-based structural signatures to study and predict a range of ADMET properties for novel chemical entities. We show that these signatures can be used successfully to train predictive models for a variety of ADMET properties. The approach, called pkCSM, also provides a platform for the analysis and optimization of pharmacokinetic and toxicity properties implemented in a user-friendly, freely available web interface (http://structure.bioc.cam.ac.uk/pkcsm), a valuable tool to help medicinal chemists find the balance between potency, safety, and pharmacokinetic properties. We have conducted a series of comparative experiments that indicate that pkCSM performs as well as or better than several other widely used methods.
Results
pkCSM: Graph-Based Signatures
Graph modeling is an intuitive and well established mathematical representation of chemical entities, from which different descriptors encompassing both molecule structure and chemistry can be extracted. An intuitive graph representation of a compound can be achieved by representing atoms as nodes and their covalent bonds as edges. This simple representation can be decorated with labels denoting, for instance, physicochemical properties of atom and bonds, from which structural patterns could be prospected. Substructure matching, implemented for instance as a toxicophore search,37 frequent subgraph mining,38 and graph kernels,39 are examples of approaches for extracting patterns from these graphs. Together with experimental data on particular properties of interest (e.g., ADMET properties), these descriptors can then be used as evidence to train highly accurate predictive models via machine learning methods. Such a predictive capability may be an essential computational tool for property optimization and to guide screening initiatives.
An alternative way of extracting relevant patterns from molecular graphs is using the concept of structural signatures. In da Silveira et al.,40 we introduced the Cutoff Scanning algorithm to extract distance patterns from protein structure graphs and summarized them into a signature vector. These signatures have been shown to be a general, powerful, and scalable way to represent geometry and physicochemical properties of protein structures and have been successfully adapted and employed for different purposes, including protein structural classification and function prediction,41 receptor-based ligand prediction,42 and more recently, as a component of structure-based mutation analysis approaches.43−47
Here, we propose pkCSM, a novel method for predicting and optimizing small-molecule pharmacokinetic and toxicity properties which relies on distance-based graph signatures. We adapted the Cutoff Scanning concept to represent small-molecule structure and chemistry (expressed as atomic pharmacophores–node labels) in order to represent and predict their pharmacokinetic and toxicity properties, building 30 predictors divided into five major classes: absorption (seven predictors), distribution (four predictors), metabolism (seven predictors), excretion (two predictors), and toxicity (10 predictors).
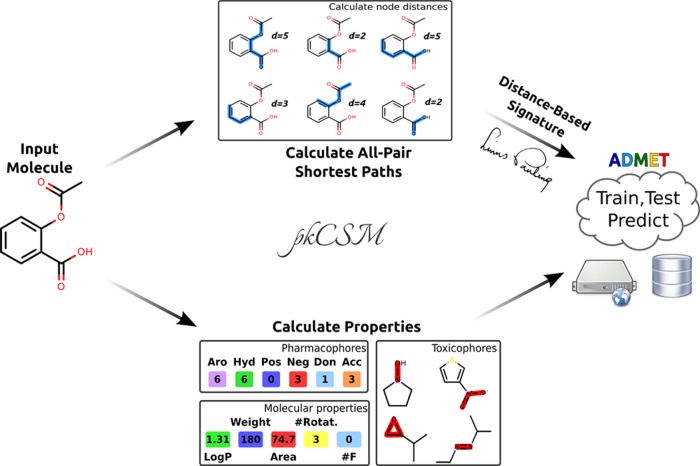
Figure 1 shows the pkCSM workflow. Given a set of input molecules, two main sets of descriptors are calculated and combined to be used in the subsequent machine learning step: general molecule properties and a distance-based graph signature.
Figure 1.
pkCSM workflow. Given an input molecule, two main sources of information are used to train and test machine learning-based predictors: compound general properties (including molecular properties, toxicophores and pharmacophore) and distance-based graph signatures.
The first major component of the pkCSM signature refers to molecular properties, which include:
-
•
A toxicophore fingerprint;
-
•
Atomic pharmacophore frequency count;
-
•
General molecular properties including lipophilicity (log P), molecular weight, surface area, number of rotatable bonds, among others.
The toxicophore fingerprint was calculated based on substructure matching from SMARTS queries proposed in ref (37) originally as potential indicators of AMES mutagenicity (available as Supporting Information). The toxicophore substructure matching, molecular properties, and pharmacophore calculations were obtained using the RDkit cheminformatics toolkit. A complete list of calculated properties can be found in the Supporting Information (Table S1). Six nonexclusive pharmacophore classes are considered (i.e., an atom can belong to more than one class): hydrophobic, aromatic, hydrogen acceptor, hydrogen donor, positive ionizable, and negative ionizable.
The second major component are distance-based patterns, represented as a cumulative distribution function, encoded in a small-molecule graph-based signature, which was adapted from the Cutoff Scanning algorithm.41,42 This way, each dimension of the signature denotes the number of atoms (categorized by pharmacophore type) within a certain distance in the molecular graph. The distance between any two nodes of the graph is given by the cost of their shortest path, calculated by Johnson’s algorithm.48 The cost of a shortest path is the sum of the weights of the edges on this path. We consider all the edges to have unitary weight. Thus, the cost of the shortest path is the number of edges in it.
Predicting Small-Molecule ADMET Properties
To build pkCSM, we performed a careful selection of data sets and recently published methods available in the literature. The validation methods chosen for each data set is consistent to the original work for comparison purposes and are available in Table S2 of Supporting Information.
The pkCSM platform for ADMET properties prediction can be divided in two groups of highly predictive models: (a) 14 regression models that aim to predict a numeric quantification of the pharmacokinetic or toxicity property and (b) 16 classification models, which categorize the outcome into two classes. A description of the models in pkCSM and how to interpret their predictions can be found in the Supporting Information.
Table 1 shows the comparative prediction performance for the regression models. Further information on the data sets used, number of data points, reference, and their validation procedure (i.e., cross-validation, external test set) can also be found in Supporting Information (Table S2). The performance for the classification models can be found in Table 2. pkCSM outperformed well established tools. For example, pkCSM AMES test achieved an accuracy of 83.8% compared to ToxTree49 (which achieved an accuracy of 75.8%).
Table 1. Comparative Regression Performance between pkCSM and Other Available Methods.
| previous methods |
pkCSM |
|||||
|---|---|---|---|---|---|---|
| data set | method | ref | std error | R2 | std error | R2 |
| water solubility | admetSAR | (22) | 0.823 | 0.810 | 0.692/0.497 | 0.943/0.967a |
| Caco2 permeability | admetSAR | (22) | 0.339 | 0.564 | 0.605/0.466 | 0.733/0.828a |
| intestinal absorption- human | Hou et al. | (50) | 10.28 | 0.890 | 12.80/9.51 | 0.846/0.902 |
| skin permeability | Alves et al. | (51) | 0.490 | 0.720d | 0.758/0.539 | 0.683/0.801 |
| steady state volume of distribution | Berellini et al. | (52) | 1.287 | 0.613 | 1.104/0.803 | 0.637/0.706 |
| fraction unbound- human (Fu) | Del Amo et al. | (53) | NA | 0.737 | 0.248/0.189 | 0.693/0.824 |
| blood–brain barrier permeability | Suenderhauf et al. | (54) | 0.580 | 0.900a | 0.379/0.287 | 0.807/0.862 |
| CNS Permeability | Suenderhauf et al. | (54) | NA | NAc | 0.825/0.665 | 0.690/0.794 |
| total clearance | Yap et al. | (55) | NA | 0.636 | 0.300/0.245 | 0.600/0.755 |
| maximum recommended tolerated dose (MRTD)-human | Liu et al. | (56) | 0.560 | 0.790a,b | 0.885/0.641 | 0.633/0.741 |
| oral rat accute toxicity (LD50) | admetSAR | (22) | 0.324 | 0.613 | 0.683/0.470 | 0.663/0.779a |
| oral rat chronic toxicity-lowest observed adverse effect (LOAEL) | Mazzatorta et al. | (57) | 0.727 | 0.500 | 0.744/0.591 | 0.683/0.776a |
| T. Pyriformis toxicity | admetSAR | (22) | 0.256 | 0.761 | 0.535/0.349 | 0.855/0.933a |
| flathead minnow toxicity (LC50) | admetSAR | (22) | 0.666 | 0.574 | 0.836/0.587 | 0.743/0.853a |
Denotes a statistically significant performance difference obtained via a Fisher r–to–z transformation, by calculating the z value, using a threshold of p ≤ 0.05 for significance. Two values are shown per column for pkCSM, denoting the performance on the entire data set and the performance after 10% outlier removal. NA: not available.
Results for 40-fold cross-validation.
Only classification methods were available.
Results reported for 0.77 data set coverage.
Table 2. Comparative Classification Performance between pkCSM and Related Methods.
| previous
method |
pkCSM |
|||||
|---|---|---|---|---|---|---|
| data set | method | ref | Q | AUC | Q | AUC |
| P-glycoprotein substrate | admetSAR | (22) | 0.735 | 0.768 | 0.780 | 0.814 |
| P-glycoprotein inhibitor I | admetSAR | (22) | 0.786 | 0.853 | 0.844 | 0.906a |
| P-glycoprotein inhibitor II | admetSAR | (22) | 0.866 | 0.922 | 0.898 | 0.948a |
| CYP450 1A2 inhibitor | admetSAR | (22) | 0.815 | 0.815 | 0.802 | 0.876a |
| CYP450 C19 inhibitor | admetSAR | (22) | 0.805 | 0.805 | 0.808 | 0.879a |
| CYP450 2C9 inhibitor | admetSAR | (22) | 0.802 | 0.802 | 0.807 | 0.868a |
| CYP450 2D6 inhibitor | admetSAR | (22) | 0.855 | 0.855a | 0.853 | 0.843 |
| CYP450 3A4 inhibitor | admetSAR | (22) | 0.645 | 0.848 | 0.780 | 0.847 |
| CYP450 2D6 substrate | admetSAR | (22) | 0.759 | 0.759 | 0.766 | 0.787 |
| CYP450 3A4 substrate | admetSAR | (22) | 0.638 | 0.638 | 0.656 | 0.676 |
| hERG I inhibitor | admetSAR | (22) | 0.870 | 0.820 | 0.853 | 0.881 |
| hERG II inhibitor | admetSAR | (22) | 0.784 | 0.849 | 0.813 | 0.876 |
| renal organic cation transporter | admetSAR | (22) | 0.795 | 0.807 | 0.797 | 0.810 |
| AMES toxicity | admetSAR | (22) | 0.851 | 0.908 | 0.838 | 0.909 |
| AMES toxicity | ToxTree | (49) | 0.758 | NA | 0.838 | 0.909 |
| hepatotoxicity | Fourches et al. | (58) | 0.639a | NA | 0.658 | 0.687 |
| skin sensitization | Alves et al. | (59) | NA | 0.820 | 0.810 | 0.850 |
Denotes a statistically significant performance difference calculated by nonparametric Wilcoxon statistic,60 using a threshold of ≤0.05 for significance.
pkCSM regression models presented a range of Pearson correlation coefficients ranging from 0.6 to 0.9, using both cross-validation schemes and external validation data sets. In comparison with available methods, for most data sets, it presents a statistically significant improvement in predictive power.
Compounds were ranked based on the absolute prediction error, and the worst 10% were considered outliers for regression analysis purposes. It is interesting to note the increase in performance when 10% of the outliers are removed. For instance, pkCSM is able to achieve a correlation of R2 = 0.779 in 90% of the data for rat toxicity and R2 = 0.828 for Caco2 permeability, a significant improvement in comparison with the correlations for the whole data sets (R2 = 0.663 and R2 = 0.733, respectively). In cases where previous methods exhibit a better correlation coefficient than pkCSM, we observed that, after removing the outliers, pkCSM presented a comparable performance and/or a lower standard error, such as the case for the blood–brain barrier permeability data set (BBB). No distinguishable trends were identified in the analysis of physicochemical properties of outlier compounds in comparison with the remaining data set.
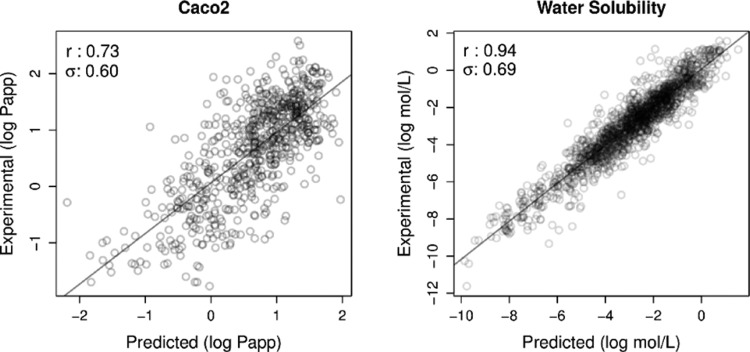
Figure 2 shows the plots between experimental and predicted values for regression absorption predictors. Figures S1 and S2 of Supporting Information, depict results for distribution and toxicity predictors, respectively. The pkCSM models were able to achieve good correlations despite the variability in data set sizes and distribution of experimental values.
Figure 2.
Regression analysis for absorption predictors considering cross-validation schemes. Pearson’s correlation coefficients and standard error are also shown at the top-left corner. The left graph shows the correlation between experimental and predicted values for Caco2 permeability, while the graph on the right for water solubility.
An external validation data set available for volume of distribution at steady state (VDss) presented a correlation of R2 = 0.637 (R2 = 0.706, after 10% outlier removal), performance compatible with the cross-validation results obtained, depicted in the left graph of Figure S1 of Supporting Information (R2 = 0.66).
Discussion and Conclusions
In summary, we have described here a novel approach to predicting pharmacokinetic and toxicology outcomes using graph-based signatures to represent small-molecule chemistry and topology. Using these signatures we have developed and implemented 14 quantitative regression models with actual numeric outputs and 16 predictive classification models with categorical outputs for predicting a wide arrange of ADMET properties for novel diverse molecules. We show pkCSM achieved a performance as good as or better than similar methods currently available, presenting a significant improve in performance for 11 data sets (water solubility, Caco2 permeability, rat, Tetrahymena pyriformis, and minnow toxicity, P-glycoprotein inhibitors, and CYP450 1A2, C19, and 2C9 inhibitors). While chemical modifications and drug carriers can improve a compounds ADMET properties,61−64 pkCSM provides a rapid and easy method to for early evaluation of compounds. In the Supporting Information, we apply these predictive models to understanding the pharmacokinetic and toxicity properties of diverse, challenging chemical sets, including macrocycles and antineoplastic drugs.
Another interesting aspect of pkCSM is its scalability, translated into an ability to handle large data sets, an important requirement for its application as a filter in screening initiatives. Over 10000 molecules compose the rat toxicity data set (prediction correlation depicted in the right graph of Figure S2 of Supporting Information) and up to 18000 compounds for the metabolism classifiers.
We have implemented a user-friendly web server that will enable researchers to freely predict ADMET properties for their molecules of interest, including in large batch formats. Considering the sensitive nature of many medicinal chemistry projects, the web server does not retain any information submitted to it. This will hopefully facilitate the drug development process by enabling the rapid design, evaluation, and prioritization of compounds.
Experimental Section
Available in the Supporting Information.
Acknowledgments
Newton Fund RCUK-CONFAP grant awarded by The Medical Research Council (MRC) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) [to D.E.V.P., T.L.B,. and D.B.A.]; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Centro de Pesquisas René Rachou (CPqRR/FIOCRUZ Minas), Brazil [to D.E.V.P.]; NHMRC CJ Martin Fellowship [APP1072476 to D.B.A.]; University of Cambridge and The Wellcome Trust for facilities and support [to T.L.B.]. Funding for open access charge: The Wellcome Trust.
Glossary
Abbreviations Used
- ADMET
absorption–distribution–metabolism–excretion–toxicity
- PAINs
pan-assay interference compounds
- QSAR
quantitative structure–activity relationship
- SMARTS
simplified molecular input line entry system arbitrary target specification
- VDss
volume of distribution at steady state
- BBB
blood–brain barrier
- SMILES
simplified molecular-input line-entry system
Supporting Information Available
Three additional tables describing implementation details and data sets used on pkCSM; four additional figures illustrating the performance of regression models, complete predictive models description, experimental methods, three case studies, and additional references. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
§ The manuscript was written through contributions of all authors. These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Kola I.; Landis J. Can the pharmaceutical industry reduce attrition rates?. Nature Rev. Drug Discovery 2004, 3, 711–715. [DOI] [PubMed] [Google Scholar]
- Merlot C. Computational toxicology—a tool for early safety evaluation. Drug Discovery Today 2010, 15, 16–22. [DOI] [PubMed] [Google Scholar]
- Eddershaw P. J.; Beresford A. P.; Bayliss M. K. ADME/PK as part of a rational approach to drug discovery. Drug Discovery Today 2000, 5, 409–414. [DOI] [PubMed] [Google Scholar]
- Li A. P. Screening for human ADME/Tox drug properties in drug discovery. Drug Discovery Today 2001, 6, 357–366. [DOI] [PubMed] [Google Scholar]
- Lin J.; Sahakian D. C.; de Morais S. M.; Xu J. J.; Polzer R. J.; Winter S. M. The role of absorption, distribution, metabolism, excretion and toxicity in drug discovery. Curr. Top. Med. Chem. 2003, 3, 1125–1154. [DOI] [PubMed] [Google Scholar]
- Thompson T. N. Early ADME in support of drug discovery: the role of metabolic stability studies. Curr. Drug Metab. 2000, 1, 215–241. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46, 3–26. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- Zhang M. Q.; Wilkinson B. Drug discovery beyond the ‘rule-of-five’. Curr. Opin. Biotechnol. 2007, 18, 478–488. [DOI] [PubMed] [Google Scholar]
- Muthas D.; Boyer S.; Hasselgren C. A critical assessment of modeling safety-related drug attrition. MedChemComm 2013, 4, 1058–1065. [Google Scholar]
- Hughes J. D.; Blagg J.; Price D. A.; Bailey S.; Decrescenzo G. A.; Devraj R. V.; Ellsworth E.; Fobian Y. M.; Gibbs M. E.; Gilles R. W.; Greene N.; Huang E.; Krieger-Burke T.; Loesel J.; Wager T.; Whiteley L.; Zhang Y. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg. Med. Chem. Lett. 2008, 18, 4872–4875. [DOI] [PubMed] [Google Scholar]
- Cumming J. G.; Davis A. M.; Muresan S.; Haeberlein M.; Chen H. Chemical predictive modelling to improve compound quality. Nature Rev. Drug Discovery 2013, 12, 948–962. [DOI] [PubMed] [Google Scholar]
- van de Waterbeemd H.; Gifford E. ADMET in silico modelling: towards prediction paradise?. Nature Rev. Drug Discovery 2003, 2, 192–204. [DOI] [PubMed] [Google Scholar]
- Obrezanova O.; Csanyi G.; Gola J. M.; Segall M. D. Gaussian processes: a method for automatic QSAR modeling of ADME properties. J. Chem. Inf. Model. 2007, 47, 1847–1857. [DOI] [PubMed] [Google Scholar]
- Khan M. T. Predictions of the ADMET properties of candidate drug molecules utilizing different QSAR/QSPR modelling approaches. Curr. Drug Metab. 2010, 11, 285–295. [DOI] [PubMed] [Google Scholar]
- Fröhlich H.; Wegner J. K.; Sieker F.; Zell A. Kernel Functions for Attributed Molecular Graphs—A New Similarity-Based Approach to ADME Prediction in Classification and Regression. QSAR Comb. Sci. 2006, 25, 317–326. [Google Scholar]
- Drwal M. N.; Banerjee P.; Dunkel M.; Wettig M. R.; Preissner R. ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014, 42, W53–W58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill M. A. Multi-dimensional QSAR in drug discovery. Drug Discovery Today 2007, 12, 1013–1017. [DOI] [PubMed] [Google Scholar]
- Moroy G.; Martiny V. Y.; Vayer P.; Villoutreix B. O.; Miteva M. A. Toward in silico structure-based ADMET prediction in drug discovery. Drug Discovery Today 2012, 17, 44–55. [DOI] [PubMed] [Google Scholar]
- Guner O. F.; Bowen J. P. Pharmacophore modeling for ADME. Curr. Top. Med. Chem. 2013, 13, 1327–1342. [DOI] [PubMed] [Google Scholar]
- Cao D.; Wang J.; Zhou R.; Li Y.; Yu H.; Hou T. ADMET evaluation in drug discovery. 11. PharmacoKinetics Knowledge Base (PKKB): a comprehensive database of pharmacokinetic and toxic properties for drugs. J. Chem. Inf. Model. 2012, 52, 1132–1137. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Li W.; Zhou Y.; Shen J.; Wu Z.; Liu G.; Lee P. W.; Tang Y. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [DOI] [PubMed] [Google Scholar]
- Matthews E. J.; Kruhlak N. L.; Benz R. D.; Contrera J. F. Assessment of the health effects of chemicals in humans: I. QSAR estimation of the maximum recommended therapeutic dose (MRTD) and no effect level (NOEL) of organic chemicals based on clinical trial data. Current Drug Discovery Technol. 2004, 1, 61–76. [DOI] [PubMed] [Google Scholar]
- Obach R. S.; Lombardo F.; Waters N. J. Trend analysis of a database of intravenous pharmacokinetic parameters in humans for 670 drug compounds. Drug Metab. Dispos. 2008, 36, 1385–1405. [DOI] [PubMed] [Google Scholar]
- Ahmed J.; Worth C. L.; Thaben P.; Matzig C.; Blasse C.; Dunkel M.; Preissner R. FragmentStore—a comprehensive database of fragments linking metabolites, toxic molecules and drugs. Nucleic Acids Res. 2011, 39, D1049–D1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U.; Struck S.; Gruening B.; Hossbach J.; Jaeger I. S.; Parol R.; Lindequist U.; Teuscher E.; Preissner R. SuperToxic: a comprehensive database of toxic compounds. Nucleic Acids Res. 2009, 37, D295–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miteva M. A.; Violas S.; Montes M.; Gomez D.; Tuffery P.; Villoutreix B. O. FAF-drugs: free ADME/tox filtering of compound collections. Nucleic Acids Res. 2006, 34, W738–W744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E.; Pon A.; Djoumbou Y.; Knox C.; Shrivastava S.; Guo A. C.; Neveu V.; Wishart D. S. T3DB: a comprehensively annotated database of common toxins and their targets. Nucleic Acids Res. 2010, 38, D781–D786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. Z.; Ji Z. L.; Chen X.; Wang J. F.; Chen Y. Z. ADME-AP: a database of ADME associated proteins. Bioinformatics 2002, 18, 1699–1700. [DOI] [PubMed] [Google Scholar]
- Moda T. L.; Torres L. G.; Carrara A. E.; Andricopulo A. D. PK/DB: database for pharmacokinetic properties and predictive in silico ADME models. Bioinformatics 2008, 24, 2270–2271. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Ikenaga Y.; Zhou Y.; Yu Y.; Li W.; Shen J.; Du Z.; Chen L.; Xu C.; Liu G.; Lee P. W.; Tang Y. In silico assessment of chemical biodegradability. J. Chem. Inf. Model. 2012, 52, 655–669. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Shen J.; Yu Y.; Li W.; Liu G.; Lee P. W.; Tang Y. In silico prediction of Tetrahymena pyriformis toxicity for diverse industrial chemicals with substructure pattern recognition and machine learning methods. Chemosphere 2011, 82, 1636–1643. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Yu Y.; Shen J.; Yang L.; Li W.; Liu G.; Lee P. W.; Tang Y. Classification of cytochrome P450 inhibitors and noninhibitors using combined classifiers. J. Chem. Inf. Model. 2011, 51, 996–1011. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Yu Y.; Zhou Y.; Shen Z.; Xiao W.; Liu G.; Li W.; Lee P. W.; Tang Y. Insights into molecular basis of cytochrome p450 inhibitory promiscuity of compounds. J. Chem. Inf. Model. 2011, 51, 2482–2495. [DOI] [PubMed] [Google Scholar]
- Shen J.; Cheng F.; Xu Y.; Li W.; Tang Y. Estimation of ADME properties with substructure pattern recognition. J. Chem. Inf. Model. 2010, 50, 1034–1041. [DOI] [PubMed] [Google Scholar]
- Broccatelli F.; Carosati E.; Neri A.; Frosini M.; Goracci L.; Oprea T. I.; Cruciani G. A novel approach for predicting P-glycoprotein (ABCB1) inhibition using molecular interaction fields. J. Med. Chem. 2011, 54, 1740–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazius J.; McGuire R.; Bursi R. Derivation and validation of toxicophores for mutagenicity prediction. J. Med. Chem. 2005, 48, 312–320. [DOI] [PubMed] [Google Scholar]
- Takigawa I.; Mamitsuka H. Graph mining: procedure, application to drug discovery and recent advances. Drug Discovery Today 2013, 18, 50–57. [DOI] [PubMed] [Google Scholar]
- Rupp M.; Schneider G. Graph Kernels for Molecular Similarity. Mol. Inf. 2010, 29, 266–273. [DOI] [PubMed] [Google Scholar]
- da Silveira C. H.; Pires D. E.; Minardi R. C.; Ribeiro C.; Veloso C. J.; Lopes J. C.; Meira W. Jr.; Neshich G.; Ramos C. H.; Habesch R.; Santoro M. M. Protein cutoff scanning: a comparative analysis of cutoff dependent and cutoff free methods for prospecting contacts in proteins. Proteins 2009, 74, 727–743. [DOI] [PubMed] [Google Scholar]
- Pires D. E.; de Melo-Minardi R. C.; dos Santos M. A.; da Silveira C. H.; Santoro M. M.; Meira W. Jr. Cutoff Scanning Matrix (CSM): structural classification and function prediction by protein inter-residue distance patterns. BMC Genomics 2011, 12Suppl 4S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E.; de Melo-Minardi R. C.; da Silveira C. H.; Campos F. F.; Meira W. Jr. aCSM: noise-free graph-based signatures to large-scale receptor-based ligand prediction. Bioinformatics 2013, 29, 855–861. [DOI] [PubMed] [Google Scholar]
- Pires D. E.; Ascher D. B.; Blundell T. L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E.; Ascher D. B.; Blundell T. L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E.; Blundell T. L.; Ascher D. B. Platinum: a database of experimentally measured effects of mutations on structurally defined protein–ligand complexes. Nucleic Acids Res. 2015, 43, D387–D391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemethova M.; Radvanszky J.; Kadasi L.; Ascher D. B.; Pires D. E.; Blundell T. L.; Porfirio B.; Mannoni A.; Santucci A.; Milucci L.; Sestini S.; Biolcati G.; Sorge F.; Aurizi C.; Aquaron R.; Alsbou M.; Marques Lourenco C.; Ramadevi K.; Ranganath L. R.; Gallagher J. A.; van Kan C.; Hall A. K.; Olsson B.; Sireau N.; Ayoob H.; Timmis O. G.; Le Quan Sang K. H.; Genovese F.; Imrich R.; Rovensky J.; Srinivasaraghavan R.; Bharadwaj S. K.; Spiegel R.; Zatkova A. Twelve novel HGD gene variants identified in 99 alkaptonuria patients: focus on ‘black bone disease’ in Italy. Eur. J. Hum Genet 2015, 10.1038/ejhg.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher J. L.; Ascher D. B.; Pires D. E.; Milan A. M.; Blundell T. L.; Ranganath L. R. Analysis of HGD Gene Mutations in Patients with Alkaptonuria from the United Kingdom: Identification of Novel Mutations. J. Inherited Metab. Dis. Rep 2015, 10.1007/8904_2014_380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. B. Efficient Algorithms for Shortest Paths in Sparse Networks. J. ACM 1977, 24, 1–13. [Google Scholar]
- Patlewicz G.; Jeliazkova N.; Safford R. J.; Worth A. P.; Aleksiev B. An evaluation of the implementation of the Cramer classification scheme in the Toxtree software. SAR QSAR Environ. Res. 2008, 19, 495–524. [DOI] [PubMed] [Google Scholar]
- Hou T.; Wang J.; Zhang W.; Xu X. ADME evaluation in drug discovery. 7. Prediction of oral absorption by correlation and classification. J. Chem. Inf. Model. 2007, 47, 208–218. [DOI] [PubMed] [Google Scholar]
- Alves V. M.; Muratov E.; Fourches D.; Strickland J.; Kleinstreuer N.; Andrade C. H.; Tropsha A. Predicting chemically-induced skin reactions. Part II: QSAR models of skin permeability and the relationships between skin permeability and skin sensitization. Toxicol. Appl. Pharmacol. 2015, 10.1016/j.taap.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berellini G.; Springer C.; Waters N. J.; Lombardo F. In silico prediction of volume of distribution in human using linear and nonlinear models on a 669 compound data set. J. Med. Chem. 2009, 52, 4488–4495. [DOI] [PubMed] [Google Scholar]
- del Amo E. M.; Ghemtio L.; Xhaard H.; Yliperttula M.; Urtti A.; Kidron H. Applying linear and non-linear methods for parallel prediction of volume of distribution and fraction of unbound drug. PloS One 2013, 8, e74758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenderhauf C.; Hammann F.; Huwyler J. Computational prediction of blood–brain barrier permeability using decision tree induction. Molecules 2012, 17, 10429–10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap C. W.; Li Z. R.; Chen Y. Z. Quantitative structure–pharmacokinetic relationships for drug clearance by using statistical learning methods. J. Mol. Graphics Modell. 2006, 24, 383–395. [DOI] [PubMed] [Google Scholar]
- Liu R.; Tawa G.; Wallqvist A. Locally weighted learning methods for predicting dose-dependent toxicity with application to the human maximum recommended daily dose. Chem. Res. Toxicol. 2012, 25, 2216–2226. [DOI] [PubMed] [Google Scholar]
- Mazzatorta P.; Estevez M. D.; Coulet M.; Schilter B. Modeling oral rat chronic toxicity. J. Chem. Inf. Model. 2008, 48, 1949–1954. [DOI] [PubMed] [Google Scholar]
- Fourches D.; Barnes J. C.; Day N. C.; Bradley P.; Reed J. Z.; Tropsha A. Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species. Chem. Res. Toxicol. 2010, 23, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves V. M.; Muratov E.; Fourches D.; Strickland J.; Kleinstreuer N.; Andrade C. H.; Tropsha A. Predicting chemically-induced skin reactions. Part I: QSAR models of skin sensitization and their application to identify potentially hazardous compounds. Toxicol. Appl. Pharmacol. 2015, 10.1016/j.taap.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J. A.; McNeil B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [DOI] [PubMed] [Google Scholar]
- Chan L. J.; Bulitta J. B.; Ascher D. B.; Haynes J. M.; McLeod V. M.; Porter C. J.; Williams C. C.; Kaminskas L. M. PEGylation Does Not Significantly Change the Initial Intravenous or Subcutaneous Pharmacokinetics or Lymphatic Exposure of Trastuzumab in Rats but Increases Plasma Clearance after Subcutaneous Administration. Mol. Pharmaceutics 2015, 12, 794–809. [DOI] [PubMed] [Google Scholar]
- Kaminskas L. M.; Ascher D. B.; McLeod V. M.; Herold M. J.; Le C. P.; Sloan E. K.; Porter C. J. PEGylation of interferon alpha2 improves lymphatic exposure after subcutaneous and intravenous administration and improves antitumour efficacy against lymphatic breast cancer metastases. J. Controlled Release 2013, 168, 200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas L. M.; McLeod V. M.; Ascher D. B.; Ryan G. M.; Jones S.; Haynes J. M.; Trevaskis N. L.; Chan L. J.; Sloan E. K.; Finnin B. A.; Williamson M.; Velkov T.; Williams E. D.; Kelly B. D.; Owen D. J.; Porter C. J. Methotrexate-conjugated PEGylated dendrimers show differential patterns of deposition and activity in tumor-burdened lymph nodes after intravenous and subcutaneous administration in rats. Mol. Pharmaceutics 2015, 12, 432–443. [DOI] [PubMed] [Google Scholar]
- Landersdorfer C. B.; Caliph S. M.; Shackleford D. M.; Ascher D. B.; Kaminskas L. M. PEGylated Interferon Displays Differences in Plasma Clearance and Bioavailability Between Male and Female Mice and Between Female Immunocompetent C57Bl/6J. and Athymic Nude Mice. J. Pharm. Sci. 2015, 104, 1848–1855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.