Significance
A key step in protein synthesis is catalyzed by aminoacyl-tRNA synthetases (ARSs), which attach specific amino acids to tRNAs. Errors caused by ARS amino acid misactivation require proofreading activities encoded in editing domains. Approximately half of the ARSs possess intrinsic editing functions, and single-domain editing proteins also are encoded in some organisms. The present study used an in vivo screen coupled with in vitro assays to elucidate the substrate specificity of two free-standing editing domains, revealing that they are multifunctional Ser/Thr-tRNA deacylases with the ability to prevent errors caused by a number of ARSs. Trans-editing proteins with broad substrate specificities provide a growth advantage to microbes under conditions in which amino acid pools are altered and may represent novel targets.
Keywords: aminoacyl tRNA synthetase, protein synthesis, ProXp, quality control, trans-editing
Abstract
Aminoacyl-tRNA synthetases (ARSs) establish the rules of the genetic code, whereby each amino acid is attached to a cognate tRNA. Errors in this process lead to mistranslation, which can be toxic to cells. The selective forces exerted by species-specific requirements and environmental conditions potentially shape quality-control mechanisms that serve to prevent mistranslation. A family of editing factors that are homologous to the editing domain of bacterial prolyl-tRNA synthetase includes the previously characterized trans-editing factors ProXp-ala and YbaK, which clear Ala-tRNAPro and Cys-tRNAPro, respectively, and three additional homologs of unknown function, ProXp-x, ProXp-y, and ProXp-z. We performed an in vivo screen of 230 conditions in which an Escherichia coli proXp-y deletion strain was grown in the presence of elevated levels of amino acids and specific ARSs. This screen, together with the results of in vitro deacylation assays, revealed Ser- and Thr-tRNA deacylase function for this homolog. A similar activity was demonstrated for Bordetella parapertussis ProXp-z in vitro. These proteins, now renamed “ProXp-ST1” and “ProXp-ST2,” respectively, recognize multiple tRNAs as substrates. Taken together, our data suggest that these free-standing editing domains have the ability to prevent mistranslation errors caused by a number of ARSs, including lysyl-tRNA synthetase, threonyl-tRNA synthetase, seryl-tRNA synthetase, and alanyl-tRNA synthetase. The expression of these multifunctional enzymes is likely to provide a selective growth advantage to organisms subjected to environmental stresses and other conditions that alter the amino acid pool.
A high level of accuracy in protein synthesis is essential for normal cell function and proliferation. A critical step in this process is pairing the correct amino acid with the cognate tRNA species by aminoacyl-tRNA synthetases (ARSs). ARSs catalyze the aminoacyl-tRNA (aa-tRNA) formation in two steps involving amino acid activation (step 1) and transfer of the activated amino acid to tRNAs (step 2). Although ARSs have evolved to exhibit specific tRNA-recognition capabilities with an estimated error frequency of 10−6, a greater number of mistakes arise from the lack of discrimination of near-cognate amino acids, with an estimated error rate of 10−4 to 10−5 at this step (1). Misincorporation of amino acids into proteins can be harmful to both eukaryotic and prokaryotic cells (2, 3).
Quality control of aa-tRNA formation is achieved by hydrolysis of the aminoacyl-adenylate (“pre-transfer editing”) or deacylation of the mischarged aa-tRNA (“post-transfer editing”) (4, 5). Editing mechanisms are used by both classes of ARSs, and 7 of 22 ARSs possess posttransfer editing sites that are distinct from the aminoacylation active site. The connective polypeptide 1 editing domain is found in class I isoleucyl-tRNA synthetase (IleRS) (6), leucyl-tRNA synthetase (LeuRS) (7, 8), and valyl-tRNA synthetase (ValRS) (9). Editing domains of class II ARSs are more diverse and include the N-terminal domain of threonyl-tRNA synthetase (ThrRS) (10), the related editing domain of alanyl-tRNA synthetase (AlaRS) (11), the β3/ β4 domain of phenylalanine-tRNA synthetase (12), and the insertion domain (INS) of prolyl-tRNA synthetase (ProRS) (13, 14). Mutations in editing domains can have detrimental effects on cells. For example, a mutation in the editing site of AlaRS, which results in only a twofold increase in misacylation in vitro, results in a severe neurodegeneration phenotype in mice (15). Cellular degradation and apoptosis caused by a mutation in the editing domain of ValRS have been reported in murine cells (16). In addition, editing defects in bacteria often result in slower growth rates, delayed growth, or even death (17–22).
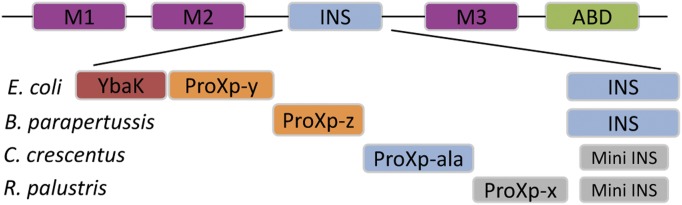
In addition to the cis-editing domain appended to ARSs, free-standing homologs of editing domains are distributed throughout organisms in all three kingdoms of life as additional checkpoints to maintain translational fidelity in trans. Autonomous trans-editing factors that are evolutionarily related to three class II ARSs, namely, AlaRS, ThrRS and ProRS, have been identified. A homolog of the AlaRS editing domain, AlaXp, is widely distributed and is shown to hydrolyze seryl-tRNA synthetase (Ser-tRNAAla) (23, 24). ThrRS-ed is an autonomous editing domain of Ser-tRNAThr in crenarchaeal genomes where thrS genes are truncated (25). Bacterial ProRS INS domain homologs include five proteins previously named “YbaK,” “ProXp-ala,” “ProXp-x,” “ProXp-y” (annotated YeaK), and “ProXp-z” (annotated PA2301) (Fig. 1) (26). These domains together with the INS domain of ProRS are collectively known as the “INS superfamily.” Although the INS domain edits mischarged Ala-tRNAPro (13, 14), YbaK deacylates Cys-tRNAPro (27), which is formed in the active site of ProRS because of the similar size of Cys and Pro (28). ProXp-ala is capable of clearing Ala-tRNAPro and compensates for the lack of an INS posttransfer editing domain in many bacteria (23, 26). Functions for ProXp-x, -y, and -z have not yet been reported. Here, for the first time to our knowledge, we explore the in vivo and in vitro substrate specificities of ProXp-y and ProXp-z. These two domains are phylogenetically distinct (26) and differ in length and in the identity of several highly conserved residues. Most notably, proXp-y encodes a functionally critical Lys residue that is present in all other INS superfamily members with the exception of proXp-z, which instead contains a strictly conserved Asn (26).
Fig. 1.
Domain structure of E. coli and B. parapertussis ProRS and the INS superfamily. Conserved motifs 1, 2, and 3 (M1–M3, purple), the anticodon-binding domain (ABD, green), and the editing domain (INS domain, blue) are shown. Single-domain INS-like proteins YbaK, ProXp-ala, ProXp-x, ProXp-y, and ProXp-z encoded in the indicated species are shown together with the INS and a truncated mini-INS present in the corresponding ProRS. C. crescentus is Caulobacter crescentus, and R. palustris is Rhodopseudomonas palustris. The known activities of INS, YbaK, and ProXp-ala are color coded as follows: blue, Ala deacylation; red, Cys deacylation. The domains investigated in this work are in orange, and domains of unknown function are in gray.
Results
Effect of Escherichia coli proXp-y Deletion on Cell Growth in the Presence of Elevated Amino Acid Levels.
We first tested the in vitro editing activities of E. coli ProXp-y using Pro-, Ala-, and Cys-tRNAPro. However, no deacylation activity was detected for these substrates (Fig. S1).We next took advantage of the availability of an E. coli proXp-y–null strain (Keio collection) to screen for conditions under which growth defects in the null strain could be observed. We hypothesized that increased expression of certain error-prone synthetases together with elevated levels of a near-cognate amino acid may amplify any growth defect caused by mistranslation in the null strain. The WT and proXp-y–null strains were transformed with overexpression plasmids encoding each of the E. coli class II ARS genes. The E. coli genome encodes two class II lysyl-tRNA synthetase (LysRS) genes, constitutive lysS and stress-inducible lysU. The two corresponding enzymes share 88% identity but possess different affinities for Lys and different thermostabilities (29). The mischarging activities of these two LysRS isoforms have not been investigated extensively, and both were included in our screen. Among the 231 growth conditions screened, growth defects were observed in the null strain in the presence of Ser when AlaRS and LysU were overexpressed and with Thr when LysS and LysU were overexpressed (Fig. 2 and Table S1). Upon the addition of 10 mM Ser alone, a much smaller growth defect was observed in the proXp-y–null strain (Fig. S2A), whereas without the amino acid addition, overexpression of AlaRS, LysS, and LysU delayed growth in the WT and proXp-y–null strains to the same extent (Fig. S2 B–D).
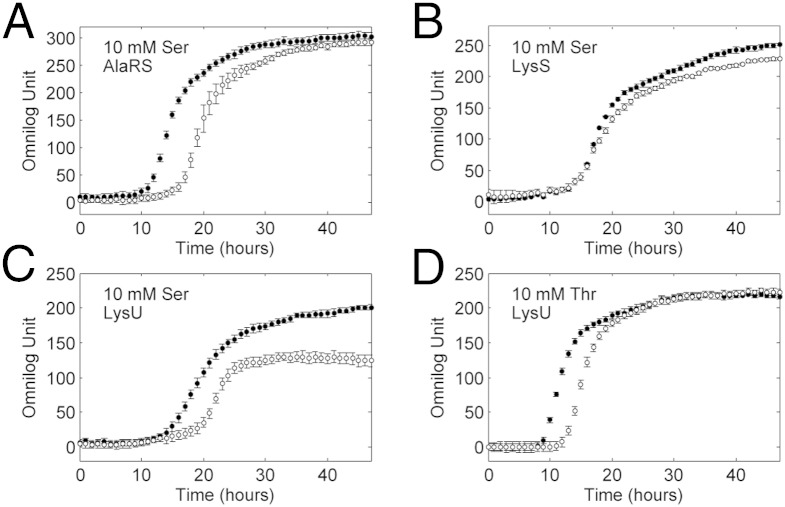
Fig. 2.
Comparison of growth curves of E. coli WT (closed circles) and proXp-y–null strains (open circles). E. coli WT BW25113 and proXp-y–null strains (Keio collection) were grown in M9 liquid minimal medium containing 10 mM Ser, AlaRS overexpressed (A); 10 mM Ser, LysS overexpressed (B); 10 mM Ser, LysU overexpressed (C); and 10 mM Thr, LysU overexpressed (D).
To confirm that intracellular amino acid concentrations were elevated when excess Ser and Thr were added to the extracellular medium, we used an LC-MS/MS approach (SI Experimental Procedures). The intracellular Ser concentration was elevated 3- to 10-fold, whereas the Thr concentration was increased ∼1.5-fold when 10 mM of each amino acid was added to the medium (Fig. S3). To confirm that the growth defects observed in Fig. 2 were caused by the lack of ProXp-y function, we performed a complementation assay. As shown in Fig. S2 E and F, the growth difference was abolished when both WT and null strains were grown in the presence of elevated amino acids and overexpressed ARS and ProXp-y. Taken together, these results indicate that growth defects were observed for the proXp-y–null strain in the presence of an altered intracellular amino acid pool and elevated ARS levels and suggest that ProXp-y is likely to function in aa-tRNA quality control. Interestingly, addition of homoserine (Hse) to the growth medium, which was tested because of its structural similarity to both Ser and Thr, also resulted in a phenotype when LysS, LysU, and seryl-tRNA synthetase (SerRS) were overexpressed (Table S1). In all cases, the main difference in growth observed in the WT and null strain was a difference in the lag phase before exponential growth. This effect is similar to growth defects observed with editing-deficient synthetases (22, 30).
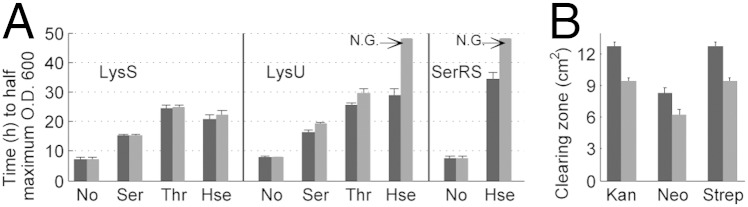
AlaRS is known to misactivate Ser (31), and LysRS has been reported to misactivate Ser, Thr, and Hse (32). Although E. coli SerRS has been reported not to misactivate Hse (33), we observed weak mischarging of tRNASer in the presence of 5 mM Hse (Fig. S4). The differences observed between the WT and proXp-y–null strains in Fig. 2 and Table S1 suggest roles for ProXp-y in editing tRNAs mischarged by LysRS, AlaRS, SerRS, and possibly ThrRS, which misactivates Ser (10). LysU overexpression appeared to result in a much larger growth defect than LysS overexpression. To explore these differences further, a dose-dependent analysis was carried out by varying the concentrations of added amino acids (Fig. S5 A–G). As summarized in Fig. 3A for the 5-mM concentrations of Ser, Thr, and Hse, the proXp-y–null strain expressing the stress-inducible lysU gene showed a dramatic increase in growth time to half-maximal OD600 compared with the WT strain. The largest effect was observed for Hse, where no growth was observed with LysU in the null strain. SerRS overexpression in the proXp-y–null strain showed a similar growth defect in the presence of Hse. Taken together, these results indicate that LysU may be more error prone than LysS, and ProXp-y may be required under physiological conditions where the lysU gene is induced. To test this hypothesis, we grew cells in 1 mM Leu in the absence of ARS overexpression, because increased Leu is known to induce LysU expression (34). We observe that the proXp-y–null strain displays a reduced growth phenotype even though ProXp-y does not edit Leu-tRNAs (Figs. S1 and S5H). This result is consistent with the requirement for ProXp-y under LysU overexpression conditions. If ProXp-y functions as a quality-control factor in vivo, we hypothesize that the editing-defective null strain would be hypermutagenic and therefore would develop antibiotic resistance more rapidly than the WT strain. To test this idea, halo assays were performed in the presence of three aminoglycoside antibiotics (neomycin, streptomycin, and kanamycin). Indeed, compared with the WT strain, the proXp-y–null strain showed a smaller clearing zone (Fig. 3B). The reduced antibiotic sensitivity of the null strain is consistent with an editing function for ProXp-y and with the notion that when cells are stressed by ribosome ambiguity (i.e., antibiotics), the regulated expression of trans editing domains provides adaptive variation to sustain growth (17).
Fig. 3.
Effect of amino acids and antibiotics on the growth of E. coli WT and proXp-y–null strains. (A) Effect of Ser, Thr, and Hse on the growth of WT (black bars) and proXp-y–null (gray bars) strains with LysS, LysU, or SerRS overexpression. E. coli WT and proXp-y–null strains were transformed with overexpression plasmids encoding lysS, lysU, or serS genes and were cultured at 37 °C overnight. Cells then were washed and inoculated into M9 minimal medium containing no amino acids (No) or 5 mM Ser, Thr, or Hse. Growth curves were monitored using the Omnilog system. The bars indicate the mean time to half-maximal OD600 of three independent growth curves. N.G., no growth of cells during the time (48 h) monitored. (B) Growth assays of WT (black bars) and proXp-y–null (gray bars) strains in the presence of kanamycin (Kan), neomycin (Neo), or streptomycin (Strep). The y axis indicates the area of the clearing zone, which was calculated by measuring the diameter of the circle in which no growth was observed. Error bars indicate the SD based on three independent experiments.
In Vitro Deacylation by E. coli ProXp-y and Bordetella parapertussis ProXp-z.
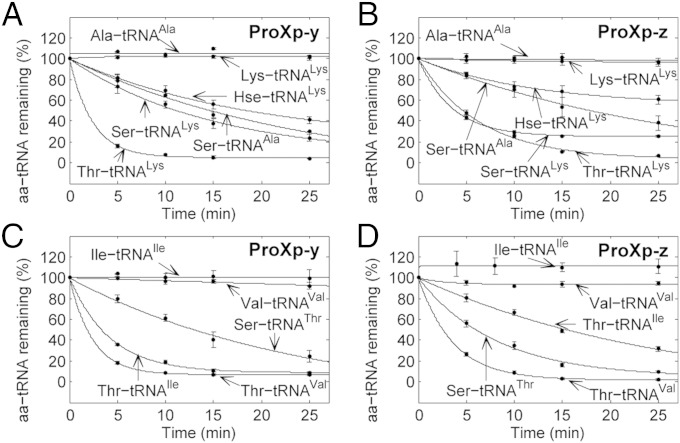
Based on the in vivo results using the proXp-y–null strain, we next examined the editing capability of both E. coli ProXp-y and the closely related ProXp-z in vitro. Because the later domain is not encoded in E. coli, we cloned and purified the homolog from B. parapertussis for these studies. For both proteins, moderate levels of deacylation were observed for mischarged Ser-tRNALys, Ser-tRNAAla, and Hse-tRNALys, and more robust activities were measured for Thr-tRNALys (Fig. 4 A and B). No deacylation of cognate Lys-tRNALys and Ala-tRNAAla was observed. In addition to the formation of mischarged Ser-tRNAAla, Ser-tRNALys, and Thr-tRNALys by AlaRS and LysRS, Thr and Ser are known to be misactivated by ValRS, IleRS, and ThrRS to form Thr-tRNAVal, Thr-tRNAIle, and Ser-tRNAThr, respectively. Thus, these substrates were prepared to test whether the activities of ProXp-y and ProXp-z extend beyond tRNAAla and tRNALys. Deacylation assays showed that ProXp-y and ProXp-z also edit Thr-tRNAVal, Thr-tRNAIle, and Ser-tRNAThr, whereas cognate Val-tRNAVal and Ile-tRNAIle species are not deacylated (Fig. 4 C and D). These results suggest the role of ProXp-y and ProXp-z as general deacylases with dual specificities for mischarged Ser/Thr-tRNAs. To ensure that the various levels of deacylation observed for the different substrates tested in Fig. 4 are caused by different amounts of uncharged tRNA in the aa-tRNA preparations, we performed deacylation assays for both Ser-tRNALys and Thr-tRNALys using ProXp-x and ProXp-y with various ratios of uncharged tRNA added to the reaction (Fig. S6 A and B). The results clearly show that the presence of high amounts of uncharged tRNA is not inhibitory and does not impact the deacylation activity of ProXp-y or -z.
Fig. 4.
Deacylation of 0.75 μM aa-tRNA by 0.5 μM E. coli ProXp-y or B. parapertussis ProXp-z. Ser-tRNALys, Ala, Thr-tRNALys, and Hse-tRNALys (A and B) and Ser-tRNAThr and Thr-tRNAVal, Ile (C and D) were deacylated by ProXp-y (A and C) or ProXp-z (B and D). Cognate aa-tRNALys, Ala, Val, Ile were tested also. All assays were carried out in triplicate as described in Experimental Methods; error bars indicate SD.
Deacylation Activity of ProXp-y and ProXp-z Toward Cognate Thr-tRNAThr.
Based on the robust deacylation of various Thr-tRNAs by ProXp-y and ProXp-z, we wondered whether cognate Thr-tRNAThr also was hydrolyzed by these two enzymes. Indeed, Thr-tRNAThr is effectively deacylated by ProXp-y and ProXp-z (Fig. S6 C and D). However, the presence of even a substoichiometric amount of ThrRS protects Thr-tRNAThr from hydrolysis, whereas excess ProRS does not. This result suggests that Thr-tRNAThr is not deacylated by these two free-standing editing domains in a physiological context.
Deacylation Activity of ProXp-y and ProXp-z Toward aa-tRNASer.
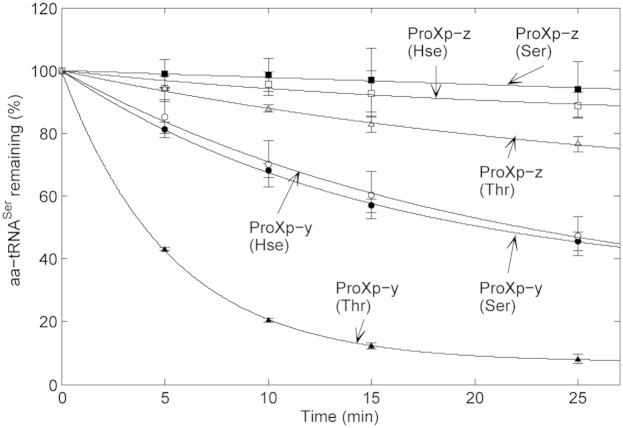
In contrast to the deacylation of Thr-tRNAThr by ProXp-y and ProXp-z, Ser-tRNASer is not cleared efficiently by ProXp-z, although it is a moderate substrate for ProXp-y (Fig. 5). Compared with the deacylation of Ser-tRNALys, Ser-tRNASer is deacylated with a threefold and 50-fold reduced rate by ProXp-y and ProXp-z, respectively. In both cases, despite the cell-based results showing a dramatic growth defect with added Hse (Fig. 3A), very similar deacylation rates were observed for Hse-tRNASer relative to Ser-tRNASer. In addition, Thr-tRNASer is a good substrate for ProXp-y, but not for ProXp-z, the only major difference in specificity observed for these two homologs.
Fig. 5.
Deacylation of 0.75 μM Ser-, Thr-, and Hse-tRNASer by 0.5 μM E. coli ProXp-y or B. parapertussis ProXp-z. All assays were carried out in triplicate as described in Experimental Methods; error bars indicate SD.
Distribution of AlaXp, ProXp-y, ProXp-z, and LysU.
The editing domain of AlaRS and the trans-editing domain AlaXp display similar capabilities for deacylating Ser-tRNAAla (23). Because ProXp-y and -z also deacylate Ser-tRNAAla, we investigated the distribution of the three trans-editing proteins. Of 1,597 genomes examined representing all kingdoms of life, 44% (37% when only bacteria are considered) encode at least one of these trans-editing domains. Because ProXp-y and -z are not encoded in archaea and are present in only a very few eukaryotes, only bacteria were considered further. No bacteria encode all three trans-editing domains, and there is very little overlap between ProXp-y and ProXp-z (10 species encode both) or between ProXp-z and AlaXp (11 species encode both). However, 51 species encode both AlaXp and ProXp-y (Fig. S7). An analysis of all bacterial genomes reveals 10 species (two Clostridium species and eight species of Gammaproteobacteria) that encode both lysS and lysU. Although the Clostridium species do not encode proXp-y or proXp-z, 100% of the lysU-containing genomes in Gammaproteobacteria also encode proXp-y.
Discussion
As a key step in maintaining high fidelity in protein synthesis, catalysis of aa-tRNA formation by ARSs is balanced between efficiency and specificity. Although the active sites of ARSs reject most noncognate amino acids in the “primary sieve,” their ability to differentiate proteinogenic and nongenetically encoded amino acids from cognate substrates is challenged by smaller and structurally related amino acids. Thus, “double-sieve” and “triple-sieve” editing mechanisms have evolved in all three kingdoms of life and become essential under certain growth conditions, including environmental stress (22, 35).
Previously, two trans-editing domain homologs of the bacterial ProRS INS domain, YbaK and ProXp-ala, were shown to deacylate known ProRS editing errors, with specificity for Cys- and Ala-tRNAPro, respectively. YbaK gains tRNAPro specificity through interaction with ProRS (36), whereas ProXp-ala recognizes tRNAPro through interaction with the same acceptor stem identity elements as bacterial ProRS (26, 37). Here, we show that two other INS domain homologs, ProXp-y and ProXp-z, have specificity for Ser- and Thr-tRNAs and therefore are unlikely to clear ProRS mischarging errors. Instead, based on in vitro studies, these enzymes are capable of clearing mischarging errors caused by LysRS (Ser/Hse- and Thr-tRNALys), AlaRS (Ser-tRNAAla), and ThrRS (Ser-tRNAThr) as well as class I IleRS (Thr-tRNAIle) and ValRS (Thr-tRNAVal). Given their in vitro specificity, we renamed ProXp-y “ProXp-ST1” and ProXp-z “ProXp-ST2” (Fig. 6). ProXp-ST1 and -ST2 have very similar specificities in vitro and are more similar in sequence to each other than to other INS homologs (26), although they differ in a residue that is critical for activity in all INS homologs investigated to date (38). With the exception of ProXp-ST2, a Lys residue (K52 in E. coli ProXp-ST1) is strictly conserved in the active site of all INS-like domains and is proposed to play a role in A76 phosphate backbone recognition and substrate positioning (38, 39). In the case of ProXp-ST2, this residue is a strictly conserved Asn (see sequence alignment in Fig. S8A). B. parapertussis N66A ProXp-ST2 failed to deacylate Ser-tRNAThr unless very high concentrations (∼8 µM) of enzyme were added, as is consistent with a role for N66 in substrate binding (Fig. S8B).
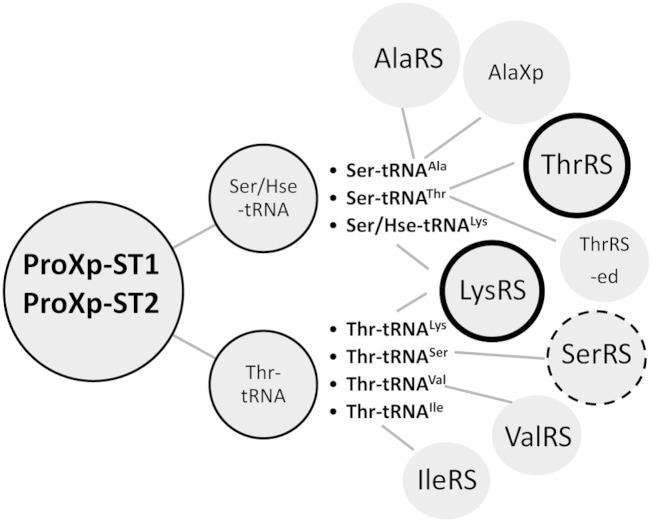
Fig. 6.
Summary of aa-tRNA deacylation capabilities of ProXp-ST1 (ProXp-y) and ProXp-ST2 (ProXp-z) and the ARSs that generate the mischarged substrates. The major substrates appear to be Ser- and Thr-tRNAs with weaker activity toward the nonprotein amino acid Hse. Based on the results of our analysis, the most likely candidates for substrates are (i) Ser-tRNAThr, which is formed by mischarging errors of ThrRS under oxidative stress (bold circle), (ii) Ser/Hse- and Thr-tRNALys, which are formed by LysRS (bold circle), and (iii) Thr-tRNASer, which is formed by SerRS (dotted circle indicating ProXp-ST1 only).
With the exception of LysRS and SerRS, all the other synthetases listed in Fig. 6 have known cis posttransfer editing capability, and in some cases trans-editing domains exist also. For example, redundant mechanisms exist for correcting errors made by AlaRS caused by the similar structure of Ser relative to cognate Ala (2). AlaXp, ProXp-ST1, and ProXp-ST2 can all deacylate Ser-tRNAAla, and their distribution is not always exclusive (Fig. S7). In the organisms where they do not overlap, differing environmental conditions may determine which enzyme is needed. In cases where there is overlap with AlaXp, the ProXp domains may have been retained for a function other than Ser-tRNAAla deacylation.
The ability to differentiate Ser and Thr from cognate substrates appears to be particularly challenging for ARSs, because at least 6 of the 20 synthetases misactivate these amino acids. ThrRS readily misactivates Ser, and in most species ThrRS encodes an N-terminal posttransfer editing domain to clear Ser-tRNAThr (10). In several crenarchaeal species, the editing domain ThrRS-ed is encoded as a separate trans-editing factor (25). Interestingly, in E. coli the ThrRS editing function is severely impaired under oxidative stress conditions, and this phenotype is attributed to an oxidized Cys residue in the editing domain active site (22). We hypothesize that trans-editing factors may be required to clear Ser-tRNAThr when ThrRS editing function is lost.
All trans-editing factors identified to date are related to class II synthetase editing domains (AlaRS, ThrRS, and ProRS) and are believed to assist in editing of tRNAs mischarged by these same synthetases. However, we show here that ProXp-type editing domains are able to deacylate a wide variety of tRNAs, including those potentially mischarged by class I IleRS and ValRS. Although Thr-tRNAIle and -tRNAVal were robustly deacylated by both ProXp-ST1 and -ST2, both these synthetases possess editing domains that clear these mischarged tRNAs under normal conditions. Although it is possible that the ProXp-type domains perform a redundant editing function under certain conditions, differences in aminoacylation kinetics between class I and class II ARSs suggest that trans editing may not be required for tRNAs charged by class I synthetases (40). Product release is rapid in class II ARSs, unlike class I enzymes in which this step is rate limiting (41). Thus, class I ARSs remain bound to their substrates long enough to edit any misacylated tRNA before release and normally would not require proofreading by a trans-editing factor (42). In the assay demonstrating ThrRS protection, we also expect that class II ThrRS releases the substrate rapidly; however, ThrRS competes effectively with ProXp-ST1 and -ST2 in rebinding the cognate Thr-tRNAThr substrate and therefore protects it from hydrolysis.
LysRS is a class II synthetase with no known posttransfer editing capability, despite reported mischarging of a variety of protein and nonprotein amino acids in vitro (32). Based on the results reported here, we propose that mischarging errors caused by LysRS are the primary substrates for ProXp-ST1 and -ST2 (Fig. 6). We hypothesize that these domains are required under conditions of cellular stress, which is when LysU levels are elevated. The overlapping distribution of lysU and proXp-y in Gammaproteobacteria supports a functional relationship between these two genes in this phylum. However, many species that encode ProXp-ST1 and/or ProXp-ST2 do not encode LysU (e.g., B. parapertussis). In these cases, LysS may be more error prone, especially under conditions of cellular stress, requiring the existence of a trans-editing factor.
Environmental stress and other factors may result in an imbalance of amino acid levels in the cell, and these imbalances can pose a difficult challenge for the fidelity of ARSs. In addition to Ser/Thr ambiguity, Hse is a structurally related nonprotein amino acid that is misactivated by LysRS (5) and is readily imported into the cell, resulting in toxic effects. Although the exact mechanism of Hse toxicity is unknown, Thr can rescue the effect, and it is proposed that the toxic effects may be caused by the misincorporation or misfolding of proteins (43). In addition to LysU, our results showed that Hse is detrimental to the proXp-y–null strain in the presence of overexpressed SerRS. However, our in vitro deacylation studies showed that Hse-tRNASer is not a very good substrate for ProXp-ST1 or -ST2 (Fig. 5). We hypothesize that overexpression of SerRS together with Hse may result in mischarging of noncognate tRNAs in vivo, as previously reported (44). Alternatively, because Hse is an intermediate in the biosynthetic pathway to Thr (43), elevated levels of Hse may result in increased Thr in the cell. Metabolomic studies to measure the levels of amino acids inside cells directly under a variety of conditions will be needed to address these open questions.
In summary, although ProXp-ST1 and -ST2 are homologs of the ProRS editing domain, they do not edit tRNAPro substrates. These multifunctional domains edit multiple amino acids (Ser, Hse, and Thr) and lack significant tRNA specificity, although ProXp-ST2 does not readily deacylate aa-tRNASer. This tRNA encodes a universally conserved G73 in bacteria (45), which may prevent deacylation by this factor. Indeed, mutation of E. coli tRNALys A73 to G73 abolished the deacylation activity of ProXp-ST2 but had little effect on the activity of ProXp-ST1 (Fig. S6 E and F). These results indicate either that A73 serves as an important positive recognition element for ProXp-ST2 or, alternatively, that the presence of other nucleotides such as G at this position acts as an anti-determinant to prevent deacylation. Both domains fail to deacylate Thr-tRNAThr in the presence of cognate ThrRS, and promiscuous editing of Hse, a nonprotein amino acid, would not be expected to be detrimental to the cell. Studies to explore the tRNA and protein-binding partners of E. coli ProXp-ST1 under different conditions are under way to help define the biological function of these proteins. It has been estimated that nearly 37% of E. coli enzymes are generalists with multiple substrates, shaped by the metabolic network under certain environmental circumstances (46). Trans-editing proteins such as ProXp-ST1 and ProXp-ST2 clearly fit into this category, with the ability to act on multiple aa-tRNA substrates depending on the pressures experienced by the cellular translational machinery.
Experimental Procedures
Materials and Strains.
All amino acids and chemicals were purchased from Sigma-Aldrich unless otherwise noted. [α-32P]ATP was from PerkinElmer Life Sciences. E. coli strain BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) and the proXp-y–null strain (Δyeak, JW1776) from the Keio collection were obtained from the Coli Genetic Stock Center (47). The plasmid pCP20, which carries the FLP recombinase, was used to remove the kanamycin cassette from JW1776 mutants (48). Plasmids encoding each ARS gene in vector pCA24N (-gfp) were from the ASKA collection (49).
Growth Curve Assays.
M9 minimal medium supplemented with 0.4% glucose and indicated concentrations of amino acids was used in all growth curve assays. E. coli cells were grown in LB medium overnight (for strains carrying ARS expression plasmids, 20 μg/mL chloramphenicol was added), collected, and washed twice with minimal medium. Cells were diluted to an OD600 of 0.01 and were grown at 37 °C in M9 minimal medium in the presence of Biolog Redox Dye Mix MA and 0.005 mM β-d-1-thiogalactopyranoside (IPTG) and in the absence or presence of the indicated concentration of specific amino acids. Cell growth was monitored at 37 °C for 48 h using the Omnilog system (Biolog Inc.).
Halo Assays.
Overnight cultures of WT and proXp-y–null strains grown in LB medium were diluted 1:100, and 1-mL aliquots were pipetted onto LB plates. Plates were dried, and a 200-μL plug was made in the middle of the plates. Solutions of kanamycin (33 mg/mL), neomycin (10 mg/mL), or streptomycin (50 mg/mL) (100 μL each) were allowed to absorb into the plates, which were incubated at 37 °C overnight followed by measurement of the diameter of the clearing zone. All conditions were tested in triplicate.
Enzyme Preparation.
The E. coli proXp-y gene was cloned into pET15b with an N-terminal His-tag and was overexpressed in E. coli BL21 (DE3) by the addition of 0.1 mM IPTG and incubation for 12 h at room temperature. The B. parapertussis pa2301 gene sequence encoding ProXp-z was codon-optimized for expression in E. coli and cloned into pET15b (Novagen) by Genewiz. ProXp-z overexpression was carried out in E. coli BL21 (DE3) as described for ProXp-y. To prepare ProXp-y and ProXp-z proteins, cells were pelleted by centrifugation at 4,300 × g for 20 min at 4 °C. Buffer containing 50 mM NaH2PO4 (pH 7), 300 mM NaCl, and 10 mM β-mercaptoethanol was used for cell lysis, followed by sonication. The His-tagged proteins were purified using HIS-select nickel resin and were eluted with an imidazole gradient. Fractions containing protein were concentrated using Amicon ultra centrifugal filters and were stored in 50 mM NaH2PO4 (pH 7), 150 mM NaCl, and 40% glycerol at −20 °C. WT E. coli ThrRS, E. coli C666A/Q584H AlaRS (11), E. coli K279A ProRS (14), E. coli T222P ValRS (50), E. coli LysRS, E. coli SerRS, and E. coli tRNA nucleotidyltransferase (51) were prepared as described above. Protein concentrations were determined by the Bradford assay (Bio-Rad).
aa-tRNA Substrate Preparation.
E. coli tRNASer, tRNAAla, tRNAPro, tRNAIle, tRNAVal, tRNAThr, and tRNALys were prepared by in vitro transcription using T7 RNA polymerase as previously described (13). All tRNAs were 3′-[32P]–labeled by tRNA nucleotidyltransferase (51). Preparation of E. coli Ser-tRNASer, Ser-tRNAThr, Ser-tRNAPro, Leu-tRNAPro, and Ile-tRNAIle was carried out using biotinylated dinitro-flexizyme and Ser-3,5-dinitrobenzyl ester (Ser-DBE), Leu-DBE, or Ile-DBE as described (52). Thr-tRNASer and Thr-tRNAIle were prepared using enhanced-flexizyme and Thr-4-chlorobenzyl thioester (Thr-CBT) (52). E. coli C666A/Q584H AlaRS was used for preparation of E. coli Ala-, Gly-, and Ser-tRNAAla; E. coli K279A ProRS was used for preparation of Ala- and Pro-tRNAPro; E. coli WT ProRS was used for preparation of Cys-tRNAPro; E. coli T222P ValRS was used for preparation of Val- and Thr-tRNAVal; E. coli LysRS was used for preparation of Lys-, Ser-, and Thr-tRNALys; E. coli ThrRS was used for preparation of Thr-tRNAThr; and E. coli SerRS was used for preparation of Hse-tRNASer. Following aminoacylation reactions using standard protocols (26), aa-tRNAs were phenol chloroform-extracted followed by ethanol precipitation. Substrates for deacylation assays were stored as pellets at −80 °C. Mischarging assays of Hse-tRNASer were performed at 37 °C with WT E. coli SerRS (3 μM) in the presence of trace amounts of E. coli [32P]tRNASer, unlabeled E. coli tRNASer (8 μM), and Hse (5 mM). Reactions were initiated by addition of E. coli SerRS following standard protocols (24).
Deacylation Assays.
In vitro deacylation assays were performed at 20–25 °C in reactions containing ∼0.75 μM [32P]-labeled aa-tRNA, 50 mM Hepes (pH 7.5), 2 mM DTT, 20 mM KCl, 5 mM MgCl2, 0.1 mg/mL BSA, and 15 μg/mL inorganic pyrophosphatase. Reactions were initiated by addition of 0.5 μM E. coli ProXp-y or B. parapertussis ProXp-z and were quenched at the indicated time points by putting 2 μL of the reaction into 6 μL of a solution containing 200 mM NaOAc (pH 5.5) and 0.5 U/μL of P1 nuclease. The reaction products (aa-[32P]AMP and [32P]AMP) were analyzed on polyethyleneimine-cellulose TLC plates as previously described (51). Graphical analysis was performed using SigmaPlot (Systat Software) with error bars representing the SD of triplicate measurements. Reactions to measure spontaneous (no enzyme) hydrolysis were performed for each aa-tRNA substrate and were subtracted from deacylation reactions with enzyme. In addition, all aa-tRNA preparations were shown to be fully deacylated under basic conditions (0.1 mM NaOH).
Bioinformatic Analysis of the Distribution of ProXp-y, ProXp-z, AlaXp, and LysU in All Kingdoms of Life.
Complete annotated and curated reference proteomes were downloaded from UniProt (www.uniprot.org/). Selecting only one strain per species, we obtained a final dataset of 1,597 complete proteomes, which included 119 Archaea, 1143 Bacteria, and 335 Eukarya. Structure-guided multiple sequence alignments of experimentally validated AlaXp, ProXp-y, ProXp-z, and LysU sequences were built using T-coffee (53) and were converted into hidden Markov model (HMM) profiles using HMMER 3.0 (54). Each HMM profile then was used to query our dataset of reference proteomes using HMMER. To distinguish hits that belong to the functional group of interest (i.e., AlaXp, ProXp-y, ProXp-z, and LysU) from those that are homologous editing domains that belong to a functionally different group (e.g., YbaK), the hits obtained by the HMM profile searches were annotated manually and curated using phylogenetic analysis, using the HMM alignments of the hits as input for the PHYLIP package (55). Therefore, ProXp HMM hits were classified into ProXp-x, ProXp-y, ProXp-z, ProXp-ala, INS, and YbaK, and AlaXp HMM hits were classified into trans-editing AlaXp domains AlaRS, ThrRS, and TrpRS. In contrast, LysU sequences did not form an independent phylogenetic cluster in our analysis and thus could not be distinguished from LysS. Therefore, to identify LysU members, we first filtered the LysRS hits by size (removing LysX and PoxA members), leaving only LysU and LysS candidates. Among this set of filtered LysRS hits, we defined LysU-containing species as those for which we found two LysRS genes in the same species within our filtered dataset of LysRS hits.
Supplementary Material
Acknowledgments
We thank Drs. Michael Ibba, Margaret Saks, and Paul Schimmel for providing the plasmids encoding E. coli tRNAIle, E. coli tRNASer, and E. coli C666A/Q584H AlaRS, respectively; Drs. Jo Marie Bacusmo and Mom Das for preparation of E. coli tRNAIle and T222P ValRS, respectively; and Ms. Rachel Simari for technical support. This work was funded by National Institutes of Health Grants R01 GM049928 (to K.M.-F.) and SAF2012-32200 (to L.R.d.P.). E.M.N. was supported by a La Caixa/Institute for Research in Biomedicine International Ph.D. Program Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423664112/-/DCSupplemental.
References
- 1.Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu Rev Genet. 2013;47(1):121–137. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schimmel P. Mistranslation and its control by tRNA synthetases. Philos Trans R Soc Lond B Biol Sci. 2011;366(1580):2965–2971. doi: 10.1098/rstb.2011.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perona JJ, Gruić-Sovulj I. Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top Curr Chem. 2014;344:1–41. doi: 10.1007/128_2013_456. [DOI] [PubMed] [Google Scholar]
- 4.Martinis SA, Boniecki MT. The balance between pre- and post-transfer editing in tRNA synthetases. FEBS Lett. 2010;584(2):455–459. doi: 10.1016/j.febslet.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakubowski H. Quality control in tRNA charging. Wiley Interdiscip Rev RNA. 2012;3(3):295–310. doi: 10.1002/wrna.122. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt E, Schimmel P. Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science. 1994;264(5156):265–267. doi: 10.1126/science.8146659. [DOI] [PubMed] [Google Scholar]
- 7.Chen JF, Guo NN, Li T, Wang ED, Wang YL. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry. 2000;39(22):6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 8.Mursinna RS, Martinis SA. Rational design to block amino acid editing of a tRNA synthetase. J Am Chem Soc. 2002;124(25):7286–7287. doi: 10.1021/ja025879s. [DOI] [PubMed] [Google Scholar]
- 9.Lin L, Hale SP, Schimmel P. Aminoacylation error correction. Nature. 1996;384(6604):33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 10.Dock-Bregeon A, et al. Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell. 2000;103(6):877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 11.Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22(3):668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23(23):4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beuning PJ, Musier-Forsyth K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 2000;97(16):8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong FC, Beuning PJ, Nagan M, Shiba K, Musier-Forsyth K. Functional role of the prokaryotic proline-tRNA synthetase insertion domain in amino acid editing. Biochemistry. 2002;41(22):7108–7115. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 15.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 16.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13(10):1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Bacher JM, de Crécy-Lagard V, Schimmel PR. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci USA. 2005;102(5):1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacher JM, Schimmel P. An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc Natl Acad Sci USA. 2007;104(6):1907–1912. doi: 10.1073/pnas.0610835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong YE, Yang XL, Schimmel P. Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J Biol Chem. 2008;283(44):30073–30078. doi: 10.1074/jbc.M805943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karkhanis VA, Mascarenhas AP, Martinis SA. Amino acid toxicities of Escherichia coli that are prevented by leucyl-tRNA synthetase amino acid editing. J Bacteriol. 2007;189(23):8765–8768. doi: 10.1128/JB.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cvetesic N, Palencia A, Halasz I, Cusack S, Gruić-Sovulj I. The physiological target for LeuRS translational quality control is norvaline. EMBO J. 2014;33(15):1639–1653. doi: 10.15252/embj.201488199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107(9):4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100(26):15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schimmel P, Ribas De Pouplana L. Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem Sci. 2000;25(5):207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 25.Korencic D, et al. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc Natl Acad Sci USA. 2004;101(28):10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vargas-Rodriguez O, Musier-Forsyth K. Exclusive use of trans-editing domains prevents proline mistranslation. J Biol Chem. 2013;288(20):14391–14399. doi: 10.1074/jbc.M113.467795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279(41):42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 28.Kamtekar S, et al. The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases. Proc Natl Acad Sci USA. 2003;100(4):1673–1678. doi: 10.1073/pnas.0437911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brevet A, Chen J, Lévêque F, Blanquet S, Plateau P. Comparison of the enzymatic properties of the two Escherichia coli lysyl-tRNA synthetase species. J Biol Chem. 1995;270(24):14439–14444. doi: 10.1074/jbc.270.24.14439. [DOI] [PubMed] [Google Scholar]
- 30.Pezo V, et al. Artificially ambiguous genetic code confers growth yield advantage. Proc Natl Acad Sci USA. 2004;101(23):8593–8597. doi: 10.1073/pnas.0402893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsui WC, Fersht AR. Probing the principles of amino acid selection using the alanyl-tRNA synthetase from Escherichia coli. Nucleic Acids Res. 1981;9(18):4627–4637. doi: 10.1093/nar/9.18.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakubowski H. Misacylation of tRNALys with noncognate amino acids by lysyl-tRNA synthetase. Biochemistry. 1999;38(25):8088–8093. doi: 10.1021/bi990629i. [DOI] [PubMed] [Google Scholar]
- 33.Gruić-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. Hydrolysis of non-cognate aminoacyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS Lett. 2007;581(26):5110–5114. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 34.Lévêque F, Gazeau M, Fromant M, Blanquet S, Plateau P. Control of Escherichia coli lysyl-tRNA synthetase expression by anaerobiosis. J Bacteriol. 1991;173(24):7903–7910. doi: 10.1128/jb.173.24.7903-7910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bullwinkle TJ, et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. eLife. 2014;3:e02501. doi: 10.7554/eLife.02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An S, Musier-Forsyth K. Cys-tRNA(Pro) editing by Haemophilus influenzae YbaK via a novel synthetase.YbaK.tRNA ternary complex. J Biol Chem. 2005;280(41):34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 37.Das M, Vargas-Rodriguez O, Goto Y, Suga H, Musier-Forsyth K. Distinct tRNA recognition strategies used by a homologous family of editing domains prevent mistranslation. Nucleic Acids Res. 2014;42(6):3943–3953. doi: 10.1093/nar/gkt1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartholow TG, et al. Strictly conserved lysine of prolyl-tRNA Synthetase editing domain facilitates binding and positioning of misacylated tRNA(Pro.) Biochemistry. 2014;53(6):1059–1068. doi: 10.1021/bi401279r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Das M, Hadad CM, Musier-Forsyth K. Aminoacyl-tRNA substrate and enzyme backbone atoms contribute to translational quality control by YbaK. J Phys Chem B. 2013;117(16):4521–4527. doi: 10.1021/jp308628y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadavalli SS, Ibba M. Quality control in aminoacyl-tRNA synthesis: Its role in translational fidelity. Adv Protein Chem Struct Biol. 2012;86:1–43. doi: 10.1016/B978-0-12-386497-0.00001-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct kinetic mechanisms of the two classes of Aminoacyl-tRNA synthetases. J Mol Biol. 2006;361(2):300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 42.Ling J, et al. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell. 2009;33(5):654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kingsbury JM, McCusker JH. Homoserine toxicity in Saccharomyces cerevisiae and Candida albicans homoserine kinase (thr1Δ) mutants. Eukaryot Cell. 2010;9(5):717–728. doi: 10.1128/EC.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson R, et al. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science. 1988;242(4885):1548–1551. doi: 10.1126/science.3144042. [DOI] [PubMed] [Google Scholar]
- 45.Jühling F, et al. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nam H, et al. Network context and selection in the evolution to enzyme specificity. Science. 2012;337(6098):1101–1104. doi: 10.1126/science.1216861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2(1):2006.0008.
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12(5):291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 50.Döring V, et al. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science. 2001;292(5516):501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 51.Ledoux S, Uhlenbeck OC. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008;44(2):74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods. 2006;3(5):357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 53.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 54.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23(1):205–211. [PubMed] [Google Scholar]
- 55.Felsenstein J. Phylogenies from molecular sequences: Inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.