Significance
Indole-3-acetic acid (IAA or auxin) is indispensable during the entire plant life cycle and regulates diverse processes and transitions via hierarchical gene expression. The hormone triggers rapid destruction of AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) repressors that control AUXIN RESPONSE FACTOR (ARF) activators in auxin-regulated gene transcription. AUX/IAA and ARF proteins interact physically via their C-terminal Phox and Bem1p (PB1) domain, which is shared between both families. This work reports the solution structure of a wild-type AUX/IAA PB1 domain and identifies amino acid residues that engage in directional interaction of monomers, largely via hydrogen bonds between conserved basic and acidic surface patches. Our study provides a framework for unraveling molecular determinants that confer specificity to complex interactions between AUX/IAA and ARF transcription factors.
Keywords: auxin signaling, transcription factors, AUX/IAA proteins, PB1 domain, NMR structure
Abstract
The plant hormone auxin activates primary response genes by facilitating proteolytic removal of AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA)-inducible repressors, which directly bind to transcriptional AUXIN RESPONSE FACTORS (ARF). Most AUX/IAA and ARF proteins share highly conserved C-termini mediating homotypic and heterotypic interactions within and between both protein families. The high-resolution NMR structure of C-terminal domains III and IV of the AUX/IAA protein PsIAA4 from pea (Pisum sativum) revealed a globular ubiquitin-like β-grasp fold with homologies to the Phox and Bem1p (PB1) domain. The PB1 domain of wild-type PsIAA4 features two distinct surface patches of oppositely charged amino acid residues, mediating front-to-back multimerization via electrostatic interactions. Mutations of conserved basic or acidic residues on either face suppressed PsIAA4 PB1 homo-oligomerization in vitro and confirmed directional interaction of full-length PsIAA4 in vivo (yeast two-hybrid system). Mixing of oppositely mutated PsIAA4 PB1 monomers enabled NMR mapping of the negatively charged interface of the reconstituted PsIAA4 PB1 homodimer variant, whose stoichiometry (1:1) and equilibrium binding constant (KD ∼6.4 μM) were determined by isothermal titration calorimetry. In silico protein–protein docking studies based on NMR and yeast interaction data derived a model of the PsIAA4 PB1 homodimer, which is comparable with other PB1 domain dimers, but indicated considerable differences between the homodimeric interfaces of AUX/IAA and ARF PB1 domains. Our study provides an impetus for elucidating the molecular determinants that confer specificity to complex protein–protein interaction circuits between members of the two central families of transcription factors important to the regulation of auxin-responsive gene expression.
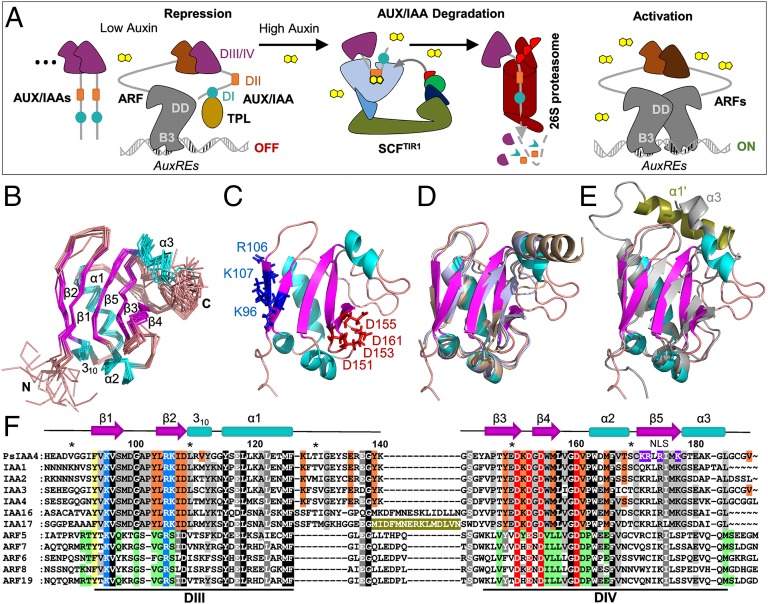
Indole-3-acetic acid (IAA or auxin) is an indispensable phytohormone that orchestrates numerous processes and developmental transitions during plant growth, largely via hierarchical control of gene expression (1). Dynamic perception of intracellular auxin differentials is accomplished by auxin-facilitated assembly of coreceptor complexes consisting of a F-box protein from the TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN SIGNALING F-BOX PROTEIN (AFB) family and a member of the AUXIN/IAA-INDUCIBLE (AUX/IAA) family of transcriptional repressors (2–4). Formation of ternary TIR1/AFB:auxin:AUX/IAA complexes recruit AUX/IAA repressors to Skp1–Cullin–F-box (SCF) E3 ubiquitin ligases for ubiquitylation and subsequent proteolysis (2, 5). AUX/IAA proteins interact physically with transcription factors of the AUXIN RESPONSE FACTOR (ARF) family (6, 7) bound to auxin-responsive elements (AuxREs) in many auxin-regulated genes (8). AUX/IAAs repress ARF function by sequestering histone deacetylase complexes via TOPLESS (TPL) or TPL-RELATED (TPR) corepressor proteins (9, 10). Thus, auxin-initiated AUX/IAA degradation relieves ARF repression and activates transcription of primary response genes (Fig. 1A).
Fig. 1.
NMR structure of PsIAA4 DIII/IV reveals a type I/II PB1 domain. (A) Current model of auxin action. Under low-auxin condition, ARFs bound to AuxREs of early genes dimerize with AUX/IAAs via DIII/IV and are repressed by DI-mediated recruitment of TPL corepressor complexes. When auxin rises, ARF de-repression and gene activation is facilitated by recruitment of AUX/IAAs via DII to the SCFTIR1 E3 ubiquitin ligase coreceptor complex, leading to AUX/IAA degradation. (B) Backbone representation of the 10 lowest-energy structures. Structural elements are highlighted in color: helices (α1-α3, 310, cyan), β-strands (β1–β5, magenta), loops (salmon). (C) Ribbon representation of the lowest-energy structure (colored as in B). Conserved basic and acidic residues of the canonical type I/II PB1 features are presented as blue and red sticks, respectively. (D) Superimposition of PsIAA4 DIII/IV (colored as in B) with ARF5 PB1 (pale brown) and ARF7 PB1 (pale blue) reveals a similar PB1 domain architecture, although the three polypeptides share low sequence identity (∼26%). (E) Superimposition of PsIAA4 DIII/IV with IAA17 PB1 (pale gray; the insertion helix α1′ in olive green). (F) Multiple sequence alignment of the PB1 domains of PsIAA4, closely related Arabidopsis AUX/IAAs (IAA1 clade, IAA16 and IAA17), and ARF activators. The canonical features of type I/II PB1 are highlighted in blue (basic motif) and red (acidic OPCA motif). The insertion helix α1′ of IAA17 is highlighted in olive green. Additional residues of the homodimeric interface of PsIAA4 PB1 (HADDOCK-generated) and ARF5 PB1 (X-ray structure) are indicated in orange and green, respectively. The aromatic interface residue common to both groups is highlighted in yellow. Other conserved positions are highlighted in gray shades if not part of the interaction face. The conserved C-terminal SV40-type NLS of PsIAA4 PB1 is shown in purple.
The diversity of auxin signaling via the tripartite TIR1/AFB-AUX/IAA-ARF module is determined by tissue-specific expression and combinatorial interactions of its redundant components, which are encoded by 6 TIR1/AFB, 29 AUX/IAA, and 23 ARF genes in Arabidopsis thaliana (4). The AUX/IAA family plays a key role in these processes because its members largely determine auxin affinities of coreceptor pairs and engage in complex AUX/IAA-ARF interaction networks (3, 7). AUX/IAA proteins are often products of early auxin genes and may thus establish regulatory feedback loops (11, 12). Most AUX/IAA members contain four regions of amino acid sequence conservation, known as domains I–IV or DI–DIV (4). N-terminal DI recruits TPL/TPR corepressors and central DII interacts directly with TIR1/AFB proteins, which is facilitated by auxin presence. The C-terminal half comprising DIII and DIV mediates homodimerization as well as heterodimerization of AUX/IAA and ARF family members. Most ARF proteins share related C-terminal DIII/IV and contain an N-terminal B3-type DNA-binding domain that recognizes AuxREs. The B3 domain is followed by a separate dimerization domain promoting ARF homodimerization upon cooperative DNA binding (13) and by a variable middle region determining ARF activator or repressor function (8) (Fig. 1A).
Evolutionary reconstruction of the ubiquitin-like β-grasp fold revealed structural homologies with C-terminal DIII/IV of ARF proteins (14), which emerged from the more ancient Phox and Bem1p (PB1) protein–protein interaction domain. PB1 domains are composed of two helices and a mixed five-stranded β-sheet and are classified into three types containing a conserved acidic OPCA (octicosapeptide repeat, p40phox, Cdc24p, atypical PKC-interaction domain) motif (type I), an invariant lysine residue on the first β-strand (type II), or both characteristic features (type I/II) (15). Secondary structure predictions of ARF and AUX/IAA proteins suggested the presence of a type I/II PB1 domain in both families (16). The recently published tertiary DIII/DIV structures of Arabidopsis wild-type ARF5 as well as mutant ARF7 and IAA17 revealed a type I/II PB1 fold of opposite acidic and basic faces, which mediate via hydrogen bonds homotypic as well as heterotypic front-to-back interactions (17–19).
We here report the high-resolution NMR structure of wild-type DIII/IV of PsIAA4 from pea (Pisum sativum), a pioneering experimental system for exploring AUX/IAA function in auxin action (6, 20–30). We mapped interface residues of the homodimer in solution and performed in silico protein–protein docking studies based on NMR and biochemical data with protein variants. Our work provides the solution structure of a wild-type AUX/IAA PB1 domain and reveals molecular properties that determine and may predict complex interactions within and between AUX/IAA and ARF family members in auxin response networks.
Results
NMR Structure of the PsIAA4 Oligomerization Domain.
We determined the solution structure of the 13C,15N isotope-labeled C-terminal domain pair of PsIAA4, which comprises amino acid residues 86–189 (DIII/IV), by using an acidic buffer system to prevent aggregation of the affinity-purified, (His)6-tagged wild-type protein. Aggregation was reported over a broad pH range for other AUX/IAA proteins and truncated derivatives in vitro (6, 31). Compared to neutral pH conditions, sedimentation equilibrium analysis confirmed the monomeric and homogeneous state of PsIAA4 DIII/IV at low pH (Fig. S1). A pH scan (pH 2–7) revealed well-dispersed 1H-15N heteronuclear single quantum coherence (HSQC) spectra at pH 2.5, indicating that this condition is suitable for solution NMR experiments of the wild-type protein (Fig. S2A).
Standard 3D NMR spectra [i.e., amide proton-to-nitrogen-to-α-carbon correlation (HNCA), HNCACB, HNCO, HN(CO)CACB, H(C)CH-total correlation spectroscopy (TOCSY), and NOESY-HSQC] were analyzed for the complete assignment of backbone and side-chain nuclei of PsIAA4 DIII/IV. All NMR spectra showed a single set of resonances for the monomeric protein, which displayed the characteristics of a stably folded polypeptide chain. The 3D structure of the PsIAA4 DIII/IV monomer was calculated with the program ARIA (Version 2.3; ref. 32) by using NOE (Fig. S2B) and dihedral constraints. Experimental constraints and structure statistics of PsIAA4 DIII/IV are summarized in Table S1.
The ensemble of the 10 lowest-energy water-refined structures of PsIAA4 DIII/IV is well defined and converges with a backbone rmsd of 0.66 ± 0.07 Å (Fig. 1B). A ribbon representation of the lowest energy structure is shown in Fig. 1C. The well superimposing residues in the structure ensemble clearly reveal the canonical topology of a globular ubiquitin-like β-grasp fold, which is homologous to the recently solved PB1 domain of Arabidopsis ARF5, ARF7, and IAA17 (17–19). Accordingly, the NMR structure of PsIAA4 DIII/IV can be superimposed on the crystal structures of ARF5 PB1 and ARF7 PB1 (Fig. 1D) with rmsd values for the structured regions of 1.02 and 1.19 Å, respectively. Compared with PsIAA4, the loop connecting DIII and DIV of IAA17 contains an insertion of 13 amino acid residues (olive green in Fig. 1F), which folds into an additional α-helix (α1′) as revealed by NMR structure analysis of the IAA17 PB1 domain (19). Although the C-terminal α3 helix of IAA17 PB1 is oriented toward the α1′ helix, the typical β-grasp fold is not affected; thus, both AUX/IAA PB1 domains can be well superimposed (rmsd of 1.65 Å) (Fig. 1E). The structural similarity with other PB1 family members demonstrates that the NMR structure of the wild-type PsIAA4 DIII/IV monomer, solved at pH 2.5, represents the native architecture in solution, which is hereafter referred to as the PsIAA4 PB1 domain. Conserved DIII and DIV correspond to subdomain β1-α1 and β3-α3 of the PsIAA4 PB1 fold, respectively (Fig. 1F).
PsIAA4 PB1 Mutations Suppress Homotypic Interaction at Neutral pH.
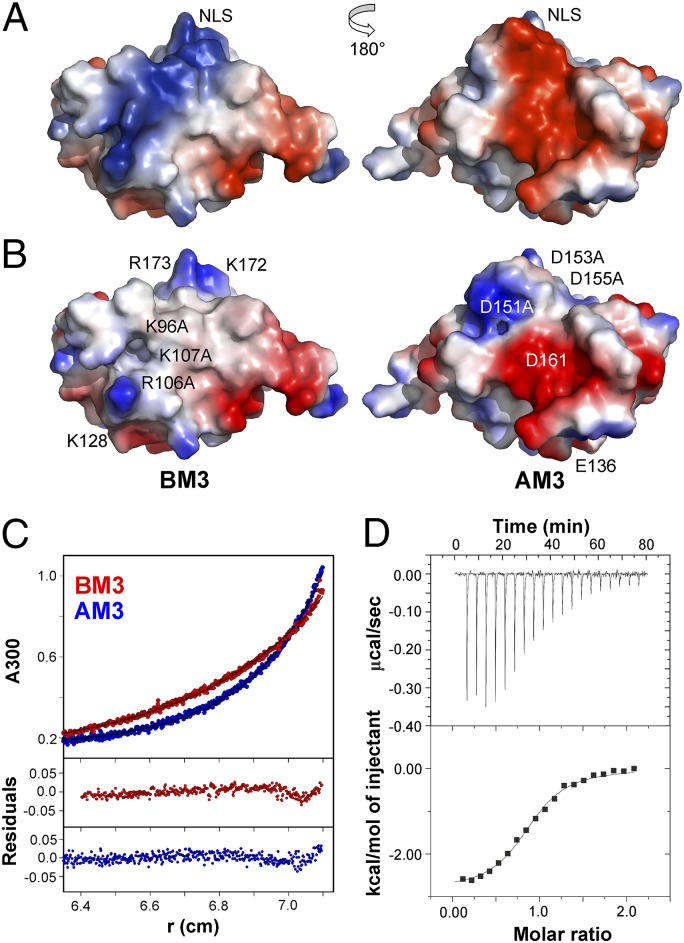
To determine the structural requirements for PsIAA4 dimerization or oligomerization (6), we systematically altered by site-directed mutagenesis basic and acidic surface residues on the type I/II PsIAA4 PB1 domain. Amino acid sequence alignment of the PB1 domains of PsIAA4, of closely related Arabidopsis AUX/IAA proteins (12), and of Arabidopsis ARF activators (8) identified an invariant and thus characteristic lysine residue (K96) on β1 and the canonical acidic OPCA motif (D151, D153, D155, D161) at both flanks of β4 (Fig. 1 C and F). The electrostatic surface potential of the PsIAA4 PB1 domain reveals a protracted, extended ridge of basic amino acid residues with K96 at its center, which is flanked by R106, K107, and K128, as well as by K172 and R173 (Fig. 2A). These basic residues are highly conserved in AUX/IAA proteins (Fig. 1E), and the latter two (K172 and R173) contribute to the functionally identified SV40-type nuclear localization signal (NLS) of PsIAA4 (33). Opposite to the NLS, the first three conserved aspartate residues of the OPCA motif form an acidic bulge that is part of an extended acidic patch complemented by conserved D161 and E136 (Fig. 2A).
Fig. 2.
Mutation of canonical features on PsIAA4 PB1 suppress homotypic interaction. (A) Electrostatic surface potentials of wild-type PsIAA4 PB1 reveal oppositely oriented basic (blue) and acidic (red) patches, containing (see B for residue labeling) invariant K96 and the OPCA motif (D151, D153, D155, and D161). K96 is flanked by R106, K107, and K128 and by K172 and R173 of the NLS. (B) Electrostatic surface potentials of in silico-generated PsIAA4 PB1BM3 and PsIAA4 PB1AM3 variants indicate disruption of each contiguous basic and acidic patch. (C) Sedimentation equilibrium analysis indicates homogeneous monomer species of PsIAA4 PB1BM3 (red curve) and PsIAA4 PB1AM3 (blue curve) samples at pH 6.25 (Upper, experimental data and fits; Lower, residuals). (D) Thermodynamic analysis of PsIAA4 PB1 homodimerization by ITC (pH 6.25 at 25 °C). PsIAA4 PB1AM3 was titrated with PsIAA4 PB1BM3 (Upper, ITC thermograms; Lower, binding isotherm). A single binding site model was fitted to the data.
We prepared mutant PsIAA4 PB1 protein samples containing single amino acid changes in either the basic (K96A, R106A, K107A) or the acidic (D151A, D153A, D155A) surface patch as well as double and triple mutations in either area (Fig. 2B), designated BM2 (K96A/R106A), AM2 (D151A/D153A), BM3 (K96A/R106A/K107A), and AM3 (D151A/D153A/D155A). Although each of the single mutations did not considerably improve PsIAA4 PB1 solubility at neutral pH, the double and triple amino acid substitutions prevented protein aggregation at pH 6.25, as indicated by sedimentation equilibrium analysis of PsIAA4 PB1BM3 and PsIAA4 PB1AM3 (Fig. 2C). Thus, suppression of protein aggregation and the resulting monomeric state of mutant PsIAA4 PB1 polypeptides with disrupted type I/II charged surface residues at near-neutral pH is consistent with homotypic front-to-back dimerization. Thermodynamic analysis of the PsIAA4 PB1BM3:PsIAA4 PB1AM3 interaction by isothermal titration calorimetry (ITC) indicated dimer formation (1:1 stoichiometry) with an equilibrium dissociation constant (KD) of 6.4 ± 0.9 μM (Fig. 2D). Formation of this mutant PsIAA4 PB1 dimer is driven by favorable enthalpy and entropy changes (Table S2). ITC experiments at different temperatures showed that the corresponding KD is largely independent of temperature upon dimer formation of the protein variants (Table S2).
Mapping the Acidic Interface of the PsIAA4 PB1 Dimer.
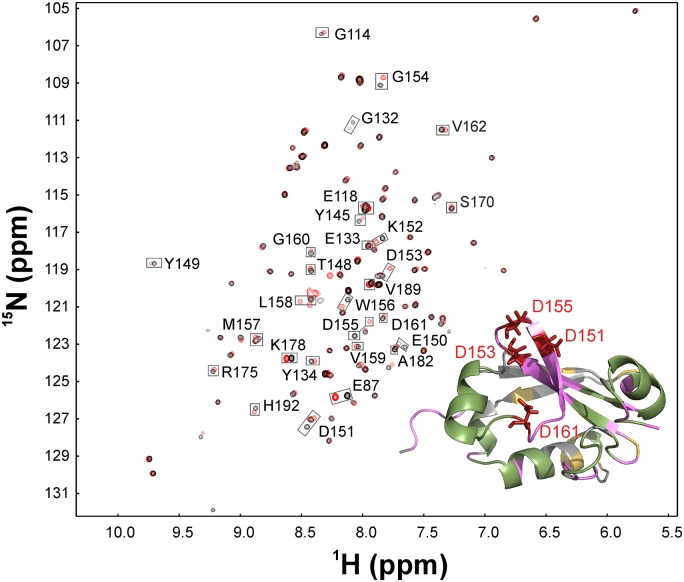
We took advantage of both mutated PsIAA4 PB1 type I/II motifs to further verify homotypic front-to-back interaction and to map by NMR spectrometry the acidic dimer face in solution. Isolated PsIAA4 PB1 samples with triple mutations in the basic or acidic surface patches, PB1BM3 and PB1AM3, remained monomeric at high protein concentration (c = 0.5 mM) under physiological conditions (pH 6.25) and were sufficiently stable in solution for NMR titration experiments. We expressed and purified 15N isotope-labeled PsIAA4 PB1BM3 and unlabeled PB1AM3 proteins. 1H-15N HSQC spectra of PsIAA4 PB1BM3 samples were initially assigned by using standard triple-resonance and pH scan NMR experiments (Fig. S3). Molecular interaction between PsIAA4 PB1BM3 and PB1AM3 mutant variants was probed by adding unlabeled PB1AM3 to labeled PB1BM3 protein sample (molar ratio of 1:4) and by recording 1H-15N HSQC spectra of the formed complex. Overlay of the 1H-15N HSQC spectra of free and bound PB1BM3 revealed major and minor chemical shift perturbations for specific cross peaks. When mapped onto the wild-type PsIAA4 PB1 structure, the residues corresponding to major chemical shift perturbations upon sample mixing and presumed dimer formation included amino acids of the conserved OPCA motif as well as additional residues surrounding the acidic surface patch (Fig. 3). As expected, amino acid residues at the opposite side of PsIAA4 PB1BM3, including the mutated basic patch, showed no or only minor perturbations upon titration (Fig. 3).
Fig. 3.
Acidic dimer interface of PsIAA4 PB1BM3 in solution. 1H-15N HSQC spectral superimposition of 15N-labeled PsIAA4 PB1BM3 monomer (black cross-peaks) and the complex of 15N-labeled PsIAA4 PB1BM3 with unlabeled PsIAA4 PB1AM3 (4:1 ratio; red cross-peaks), which revealed specific chemical-shift changes indicative of molecular interaction of both monomers. (Inset) Structural mapping of interacting residues of PsIAA4 PB1BM3 with PsIAA4 PB1AM3 as derived by backbone chemical shift perturbation (as indicated by gray boxes in the NMR spectra). Mapped residues include the OPCA motif (red) and additional interface residues (pink). Colored regions of the structure correspond to unambiguously assigned residues (red, pink, and green); residues with spectral overlap (yellow); and unassigned residues (gray).
In Vivo Validation of the Dimeric PsIAA4 Interface.
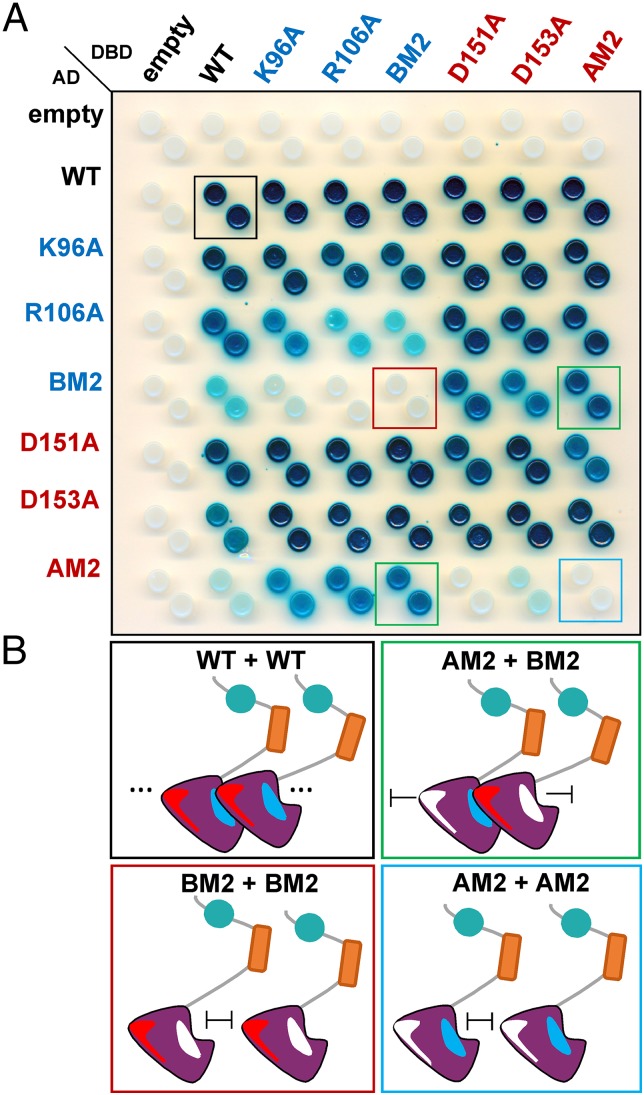
Homotypic interaction of PsIAA4 was previously demonstrated by using the yeast two-hybrid (Y2H) system, and deletion analyses identified C-terminal DIII/IV as the major region mediating PsIAA4 dimerization in vivo (6). To test whether the front-to-back mode of electrostatic interactions of type I/II PB1 domains also applies to full-length PsIAA4 in vivo, we replaced by site-directed mutagenesis one or two central amino acid residues on the basic (K96A, R106A, BM2) or acidic (D151A, D153A, AM2) face of the PsIAA4 PB1 domain and generated a Y2H protein–protein interaction matrix (Fig. 4). Immunoblot analysis confirmed expression of PsIAA4 protein fusions in yeast cells (Fig. S4). In either vector configuration of the Y2H system, full-length wild-type PsIAA4 interacted with itself or with each of the mutant PsIAA4 proteins tested, suggesting that a single electrostatic dimerization interface is sufficient for homotypic PsIAA4 interaction. Likewise, mutations of the basic PB1 face (PsIAA4K96A, PsIAA4R106A, or PsIAA4BM2) did not prevent PsIAA4 dimerization when paired with mutations of the acidic PB1 face (PsIAA4D151A, PsIAA4D153A, or PsIAA4AM2), and vice versa. However, both the PsIAA4AM2 and the PsIAA4BM2 mutant protein failed to interact with itself in Y2H assays, suggesting that the presence of both a negative and positive surface is necessary for homotypic PsIAA4 interaction in vivo. Thus, the Y2H study validates the mode of PsIAA4 dimerization via front-to-back interaction of its PB1 domain.
Fig. 4.
Y2H assays confirm type I/II PB1-mediated homotypic interaction of PsIAA4 in vivo. (A) Interaction matrix of full-length wild-type and mutant PsIAA4 variants with single or double amino acid changes on the basic or acidic PB1 interface. Diploids expressing wild-type and mutant DBD–PsIAA4 and AD–PsIAA4 protein fusions were generated and spotted on selective induction medium (Gal/Raf/–Ura/–Trp/–His/+X-Gal). β-galactosidase expression reporting protein–protein interaction (blue colonies) is shown 4 d after spotting. PsIAA4BM2 interacted with PsIAA4AM2 in either vector configuration (green boxes). Each double mutant failed to interact with itself, PsIAA4BM2 (red box), and PsIAA4AM2 (blue box). The wild-type PsIAA4 control corresponds to the black box. DBD, DNA-binding domain; AD, activation domain. (B) Scheme of wild-type and mutant PsIAA4 interactions from Y2H assays (colored boxes correspond to A).
Data-Driven Homodimer Docking of PsIAA4 PB1.
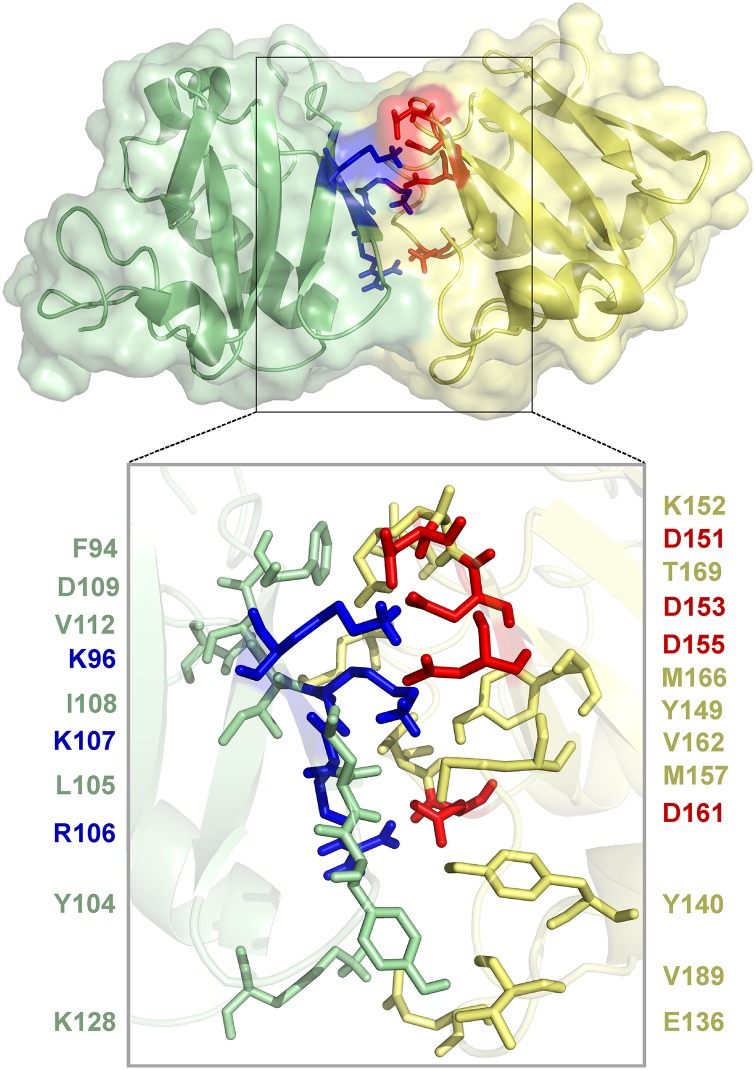
Based on NMR mapping of acidic interface residues (Fig. 3) and results of Y2H assays (Fig. 4), we performed data-driven protein–protein docking analyses by using the HADDOCK (High Ambiguity Driven protein–protein DOCKing) server (34) for refined modeling of the interaction interface. The experimentally identified residues of the acidic (D151, D153, D155, and D161) and basic (K96 and R106) interaction patch were specified, and passive surrounding residues were automatically selected for docking. The best PsIAA4 PB1 homodimer pose (Fig. 5) resembles the closest available PB1 dimeric protein structures, including the ARF5 PB1 homodimer despite different sets of interface residue. Further analysis pointed to an expanded interaction interface of the K96-centered (698 Å2) and OPCA (644 Å2) motifs. All together 23 interacting residues may engage in 9 hydrogen bonds and ∼70 nonbonded contacts (Figs. S5 and S6). Notably, the center of either electrostatic surface ridge is intersected at a similar angle (∼70°) by a stretch of hydrophobic and polar residues likely involved in nonbonded contacts, which thus contribute to complementary surfaces for asymmetric homodimer formation (Fig. S6).
Fig. 5.
HADDOCK-based data-driven model of the PsIAA4 PB1 homodimer. A protein–protein docked model of the PsIAA4 PB1 homodimer was generated by HADDOCK using data from NMR acidic interface mapping and Y2H analyses. The model highlights canonical electrostatic interactions between the monomers (box). Conserved basic (blue) and acidic (red) residues are shown as sticks. The box below shows all interface residues (sticks) that interact via hydrogen bonds (blue and red) and nonbonded contacts (light green and yellow).
Discussion
The primary auxin response gene PsIAA4 and its encoded protein have long been a conceptual model for exploring early auxin action (11, 35). However, structural insight into the pivotal interplay of AUX/IAA repressors and ARF activators in auxin-dependent gene transcription (Fig. 1A) has been impeded by low solubility of the recombinant proteins due to aggregation. Structural prediction of AUX/IAAs and ARFs suggested the presence of a C-terminal PB1 domain in both protein families (14, 16). After the NMR structure of PsIAA4 DIII/IV was released (Protein Data Bank ID code 2M1M), the tertiary structures of C-terminal DIII/IV of Arabidopsis ARF5, ARF7, and IAA17 were reported, which revealed canonical type I/II PB1 features for mediating electrostatic front-to-back interaction of monomers via oppositely charged and located surface patches (17–19). Whereas the ARF7 PB1 and IAA17 PB1 structures were solved after introducing charge-neutralizing mutations to prevent protein aggregation (18, 19), we determined the solution structure of the wild-type PsIAA4 DIII/IV monomer at pH 2.5, which suppresses homo-oligomerization in vitro. Even at this low pH, PsIAA4 DIII/IV displays the topology of a globular ubiquitin-like β-grasp fold that closely matches the structure of wild-type ARF5 PB1 solved by X-ray crystallography at physiological pH (Fig. 1D). The characteristic features of the well-defined type I/II PB1 domain of PsIAA4 are displayed on expanded but distinct acidic (OPCA motif) and basic (K96 motif) surface patches. These features mediate directional self-assembly in vitro, which is disrupted by protonation of OPCA residues (pH 2.5) or, at neutral pH, by site-directed mutagenesis of charged amino acid residues on either patch (Fig. 2 and Fig. S3). Their importance for front-to-back interaction of PsIAA4 in vivo is supported by Y2H assays of PsIAA4 variants with a mutated PB1 domain (Fig. 4).
Self-assembly of the PsIAA4 PB1 domain in vitro revealed a KD of 6.4 μM for the arrested dimer of two oppositely mutated monomer units that retain charge complementarity at their interface (Fig. 2D). The KD value remarkably matches the reported equilibrium-binding constants for homotypic interactions of various mutant IAA17 and IAA17 PB1 variants (KD = 5.5–6.6 μM), including an IAA17 deletion mutant that misses the extra α1′ helix inserted into the major loop of the PB1 domain (19). The latter KD determination implies that the insertion helix does not obstruct front-to-back interaction of monomers, which is consistent with the NMR structure and distant positioning of the α1′ helix (19) (Fig. 1E). Therefore, and because long insertions (∼15 residues) between DIII and DIV are not present in Arabidopsis ARF proteins as well as in more than half of the AUX/IAA family members (Fig. S7), the typical PB1 β-grasp fold is likely a general hallmark of both protein families.
Despite the similar equilibrium-binding constants, the thermodynamics of monomer interactions point to differences between both studied AUX/IAA proteins. Although mutant IAA17 homodimer formation is largely enthalpically driven (19), the interaction of the mutant PsIAA4 PB1 dimer is favored by both enthalpy and entropy changes (Fig. 2D and Table S2). This finding suggests that, in addition to electrostatic interactions between the acidic OPCA and basic K96 motifs, other contacts contribute to PsIAA4 PB1 homodimerization. Mapping of the PsIAA4 PB1 dimer interface by NMR titration followed by data-driven homodimer docking indicated that both interaction interfaces are possibly expanded by hydrophobic and polar residues (Figs. S5 and S6). Interestingly, the modeled dimeric PsIAA4 PB1 interface is similar in contact area, number of interacting side chains, and type of bonds formed compared with the experimentally determined interface of the wild-type ARF5 PB1 homodimer (17) (Fig. S6).
Although electrostatic contacts and hydrogen bonds are critical for PB1 oligomerization, additional (nonbonded) contacts are likely responsible for the reported high specificity and affinity of modular interactions of PB1 domain proteins (15), which may also confer specificity to intrafamily and interfamily interactions between AUX/IAA and ARF members. Indeed, compared with mutant IAA17 homodimer formation, ITC analysis measured considerably higher affinities of the ARF5 homodimer (KD ∼0.9 μM) and ARF5:IAA17 heterodimer (KD ∼0.075 μM), which was explained by differences in charge content and distribution at the respective interaction surfaces (19). A comparison of the basic and acidic PB1 surfaces of PsIAA4 and IAA17 reveals clear differences. Whereas the K96-centered motif of PsIAA4 forms a prominent ridge, the corresponding K114-centered patch on IAA17 is less compact, although it also includes part of the NLS conserved in AUX/IAAs (Fig. S8A). This topologic difference, and a possible contribution of nonbonded contacts, may explain why replacement of more than one basic residue in PsIAA4 PB1 is required to abolish homotypic monomer interactions in vitro (Fig. 2) and in vivo (Fig. 4). Conversely, the density and distribution of negative charges (OPCA motif) is similar for both AUX/IAA PB1 domains. However, for IAA17 the acidic surface is considerably expanded and incorporates residue D118 (Fig. S8B). Interestingly, the D118N mutation suppresses growth phenotypes caused by IAA17 protein stabilization, and Y2H analysis indicated that D118 is necessary for IAA17 homotypic and heterotypic (IAA3, ARF5, and ARF7) interactions (36, 37). As expected from the different PsIAA4 PB1 topology (Fig. S8B), the corresponding PsIAA4 mutation (D100N) does not prevent homodimerization in yeast (Fig. S8C), suggesting that the IAA17 interaction face differs from members of the IAA1/PsIAA4 clade (Fig. 1F). The interface residues conferring specificity likely vary between phylogenetic clades of both families (Fig. S7) and may thus determine combinatorial specificity of AUX/IAA-ARF interactions. For example, in Y2H assays, the 29 AUX/IAA proteins often homodimerize and heterodimerize and interact preferentially with the 5 ARF activators, whereas the 15 PB1 domain-containing ARF repressors show no or very limited interactions within this network (7).
The modular PB1 domain is well suited to assemble assorted protein complexes via directional (hetero)oligomerization (type I/II) and chain termination (type I or type II). These scaffold-like complexes may acquire additional subunits by noncanonical PB1 interactions for enabling specificity and fidelity of signal transduction pathways (15). It is of note that DIII/IV of certain AUX/IAA and ARF proteins has been reported to interact with other transcription factors (16) and that AUX/IAA multimerization with ARF activators is likely necessary for repressor function in planta, possibly by disrupting cooperative binding of ARF oligomers to AuxREs (13, 17, 18). Thus, the solution structure of wild-type PsIAA4 PB1, together with the recently solved ARF and AUX/IAA PB1 structures (17–19), provide a framework for elucidating the structural rules of intricate interactions between members of the two central families of transcription factors in early auxin action.
Experimental Procedures
Sample Preparation and Biochemical Analyses.
The pQE vector for expressing (His)6-tagged PsIAA4 DIII/IV was reported (30). Site-directed mutagenesis, protein labeling, sample preparation, and sample analyses by analytical ultracentrifugation and ITC are described in SI Experimental Procedures.
NMR Spectroscopy and Structure Calculation.
All NMR experiments for assignments of PsIAA4 PB1 were performed on a Bruker Avance III 600 MHz (QXI probe) or 800 MHz (cryogenic TCI probe) spectrometer by using 1.5 mM as protein concentration in 50 mM NaH2PO4, 2 mM MgCl2, 1 mM DTT, pH 2.5, 10% D2O (vol/vol) sample buffer at 25 °C. All 2D and 3D NMR datasets were processed with NMRPipe (38) and analyzed with NMRView (39). Backbone resonances of the 13C,15N-labeled sample were assigned by using the triple-resonance experiments HNCA, HNCACB, HNCO, and HN(CO)CACB. Side-chain information was obtained by recording H(C)CH-TOCSY. The 3D NOESY-edited HSQC experiments (120 ms mixing time each) for 15N and 13C aliphatic/aromatic nuclei confirmed and finalized the side-chain assignment. Four experiments were used to generate NOE restraints: 1H-1H NOESY, 1H-15N NOESY-HSQC, 1H-13C NOESY-HSQC (aliphatic and aromatic signal region were separated). ARIA (Version 2.3) runs and analysis (32) were performed by using ambiguous NOEs and TALOS (40) derived dihedral information (41) as structural restraints. An ensemble of the 10 lowest energy structures was used for further structural analysis. The Ramachandran analysis was performed by PROCHECK-NMR (42). The structure ensembles were aligned and illustrated by using PyMOL (Version 0.99rc6).
NMR Titration Experiments.
13C,15N-labeled PsIAA4 PB1BM3 and unlabeled PsIAA4 PB1AM3 were expressed and purified as described for wild-type PsIAA4 PB1, and dialyzed against 50 mM Na–citrate, 500 mM NaCl, 2 mM MgCl2, 3 mM Tris-(2-chloroxyethyl)-phosphine hydrochloride, pH 6.25. The concentrated labeled PsIAA4 PB1BM3 preparation showed a well-dispersed 1H-15N HSQC spectrum, indicating suitability for NMR experiments at pH 6.25. Molecular interaction studies were conducted by mixing the labeled and unlabeled mutant PsIAA4 PB1 samples. The 2D 1H-15N HSQC spectra were acquired for labeled PsIAA4 PB1BM3 (110 μM) prior to and after the addition of unlabeled PsIAA4 PB1AM3 (75 μM) in a molar ratio of 4:1. After NMR-based pH scanning of 15N-labeled PsIAA4 PB1BM3 (Fig. S4), the 1H-15N HSQC spectrum was assigned by using transverse relaxation-optimized spectroscopy-based 3D experiments (HNCO, HNCA, HNCACO, HNCACB, and HNCOCACB) at high protein concentration (450 μM) in low salt (150 mM NaCl) and in reference to the assigned spectrum of wild-type PsIAA4 PB1.
Y2H Assays.
Y2H assays were carried out with the LexA-based reporter system (3) as described in SI Experimental Procedures.
Data-Driven Protein–Protein Docking.
The HADDOCK Easy interface (haddock.science.uu.nl/services/HADDOCK) was used to analyze directional docking with two monomer structures as input (SI Experimental Procedures).
Supplementary Material
Acknowledgments
We thank A. Klamt, A. Kumar, and R. Patzschke for assistance with NMR data collection; and A. Khatri for helping in NMR data analysis. This research was supported by Deutsche Forschungsgemeinschaft Research Training School Grant GRK 1026 (to D.C.D., M.K., and M.G.); and by institutional core funding from the Federal Republic of Germany (to S.A.), the state of Saxony-Anhalt (to S.A., J.B., and H.L.), and the European Regional Development Fund (to J.B.). This work was inspired by and is dedicated to Athanasios (Sakis) Theologis on the occasion of his 70th birthday.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR, atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2M1M), and the Biological Magnetic Resonance Data Bank, www.bmrb.wisc.edu/ (BMRB accession no. 18870).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424077112/-/DCSupplemental.
References
- 1.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 2.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 3.Calderón Villalobos LI, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8(5):477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, Estelle M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94(22):11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319(5868):1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 10.Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012;139(22):4180–4190. doi: 10.1242/dev.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111(1):9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251(4):533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- 13.Boer DR, et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156(3):577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 14.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: The extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007(401):re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 16.Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 2012;190:82–88. doi: 10.1016/j.plantsci.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Nanao MH, et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014;5:3617–3624. doi: 10.1038/ncomms4617. [DOI] [PubMed] [Google Scholar]
- 18.Korasick DA, et al. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA. 2014;111(14):5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han M, et al. Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci USA. 2014;111(52):18613–18618. doi: 10.1073/pnas.1419525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abel S, Theologis A. Odyssey of auxin. Cold Spring Harb Perspect Biol. 2010;2(10):a004572. doi: 10.1101/cshperspect.a004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theologis A, Ray PM. Early auxin-regulated polyadenylylated mRNA sequences in pea stem tissue. Proc Natl Acad Sci USA. 1982;79(2):418–421. doi: 10.1073/pnas.79.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theologis A, Huynh TV, Davis RW. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol. 1985;183(1):53–68. doi: 10.1016/0022-2836(85)90280-3. [DOI] [PubMed] [Google Scholar]
- 23.Oeller PW, Keller JA, Parks JE, Silbert JE, Theologis A. Structural characterization of the early indoleacetic acid-inducible genes, PS-IAA4/5 and PS-IAA6, of pea (Pisum sativum L.) J Mol Biol. 1993;233(4):789–798. doi: 10.1006/jmbi.1993.1555. [DOI] [PubMed] [Google Scholar]
- 24.Ballas N, Wong LM, Theologis A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum) J Mol Biol. 1993;233(4):580–596. doi: 10.1006/jmbi.1993.1537. [DOI] [PubMed] [Google Scholar]
- 25.Abel S, Oeller PW, Theologis A. Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA. 1994;91(1):326–330. doi: 10.1073/pnas.91.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oeller PW, Theologis A. Induction kinetics of the nuclear proteins encoded by the early indoleacetic acid-inducible genes, PS-IAA4/5 and PS-IAA6, in pea (Pisum sativum L.) Plant J. 1995;7(1):37–48. doi: 10.1046/j.1365-313x.1995.07010037.x. [DOI] [PubMed] [Google Scholar]
- 27.Koshiba T, Ballas N, Wong LM, Theologis A. Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum) J Mol Biol. 1995;253(3):396–413. doi: 10.1006/jmbi.1995.0562. [DOI] [PubMed] [Google Scholar]
- 28.Ballas N, Wong LM, Ke M, Theologis A. Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid-inducible gene PS-IAA4/5. Proc Natl Acad Sci USA. 1995;92(8):3483–3487. doi: 10.1073/pnas.92.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong LM, et al. Differential activation of the primary auxin response genes, PS-IAA4/5 and PS-IAA6, during early plant development. Plant J. 1996;9(5):587–599. doi: 10.1046/j.1365-313x.1996.9050587.x. [DOI] [PubMed] [Google Scholar]
- 30.Colón-Carmona A, Chen DL, Yeh KC, Abel S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000;124(4):1728–1738. doi: 10.1104/pp.124.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan KE, Zarembinski TI, Theologis A, Abel S. Biochemical characterization of recombinant polypeptides corresponding to the predicted βαα fold in Aux/IAA proteins. FEBS Lett. 1999;454(3):283–287. doi: 10.1016/s0014-5793(99)00819-4. [DOI] [PubMed] [Google Scholar]
- 32.Rieping W, et al. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23(3):381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 33.Abel S, Theologis A. A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum) Plant J. 1995;8(1):87–96. doi: 10.1046/j.1365-313x.1995.08010087.x. [DOI] [PubMed] [Google Scholar]
- 34.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 35.Theologis A. Auxin-regulated gene expression in plants. Biotechnology. 1989;12:229–243. doi: 10.1016/b978-0-409-90068-2.50017-4. [DOI] [PubMed] [Google Scholar]
- 36.Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13(4):829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279(5355):1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- 38.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 39.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 40.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 41.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.