Abstract
Programmed cell death, or apoptosis, is a highly conserved cellular process that is crucial for tissue homeostasis under normal development as well as environmental stress. Misregulation of apoptosis is linked to many developmental defects and diseases such as tumour formation, autoimmune diseases and neurological disorders. In this paper, we show a novel role for the exoribonuclease Pacman/Xrn1 in regulating apoptosis. Using Drosophila wing imaginal discs as a model system, we demonstrate that a null mutation in pacman results in small imaginal discs as well as lethality during pupation. Mutant wing discs show an increase in the number of cells undergoing apoptosis, especially in the wing pouch area. Compensatory proliferation also occurs in these mutant discs, but this is insufficient to compensate for the concurrent increase in apoptosis. The phenotypic effects of the pacman null mutation are rescued by a deletion that removes one copy of each of the pro-apoptotic genes reaper, hid and grim, demonstrating that pacman acts through this pathway. The null pacman mutation also results in a significant increase in the expression of the pro-apoptotic mRNAs, hid and reaper, with this increase mostly occurring at the post-transcriptional level, suggesting that Pacman normally targets these mRNAs for degradation. Our results uncover a novel function for the conserved exoribonuclease Pacman and suggest that this exoribonuclease is important in the regulation of apoptosis in other organisms.
Keywords: Apoptosis, Compensatory proliferation, RNA stability, Wing imaginal discs, XRN1
INTRODUCTION
Apoptosis, or programmed cell death, is crucial to normal embryonic development and metamorphosis of multicellular organisms, as well as being important in disease. Control of apoptosis is coordinated with that of proliferation, with many signalling pathways implicated in the normal control of tissue growth involved in both processes (Danial and Korsmeyer, 2004). The key components of apoptosis pathways are well known and highly conserved, and many of the signalling pathways that regulate apoptosis have been elucidated (Domingos and Steller, 2007; Fuchs and Steller, 2011; Hay and Guo, 2006; Salvesen and Abrams, 2004; Steller, 2008; Xu et al., 2009). Although post-transcriptional processes that work at the level of RNA stability are known to be important in a number of cellular processes [e.g. inflammation (Sanduja et al., 2011)] their contribution in the control of apoptosis are not well understood.
Drosophila provides an excellent model system for the study of apoptosis because of its genetic tractability and the similarities of its apoptosis pathways to that of other organisms. In Drosophila, the regulation of caspase activation is a major strategy by which apoptosis is regulated. Factors inducing apoptosis, such as developmental signals or irradiation, result in activation of pro-apoptotic proteins such as Reaper, Hid and Grim. These proteins then trigger ubiquitin-mediated degradation of DIAP1 (XIAP), releasing Dronc (Caspase-9-like) from DIAP1 inhibition. Together with the scaffolding protein Ark (Apaf-1), free Dronc proteolytically cleaves and activates the effector Caspase-3-like caspases DrICE (Drosophila interleukin-1-converting-enzyme) and Dcp-1 (death caspase-1). These caspases then trigger downstream events such as DNA fragmentation, inhibition of translation initiation and formation of apoptotic bodies (Domingos and Steller, 2007; Thomas and Lieberman, 2013).
This paper describes the discovery of a new player in the control of apoptosis, namely the Pacman/Xrn1 exoribonuclease. Pacman (Pcm) is a highly conserved processive 5′-3′ exoribonuclease that degrades mRNAs after they have been decapped (Garneau et al., 2007; Jones et al., 2012; Nagarajan et al., 2013). As well as being a key enzyme in RNA turnover, XRN1 homologues are also involved in nonsense-mediated decay (NMD) and degradation of mRNAs after they have been targeted by small interfering RNAs or miRNAs (Orban and Izaurralde, 2005). In Drosophila, mutations in pacman result in specific phenotypes such as defects in wound healing, epithelial closure and male fertility (Grima et al., 2008; Jones et al., 2013; Till et al., 1998; Zabolotskaya et al., 2008). The Pacman protein is maternally contributed and is differentially expressed during development (Grima et al., 2008).
In this study, we show that the 5′-3′ exoribonuclease Pacman/Xrn1 regulates apoptosis in Drosophila wing imaginal discs. We generated a null mutation in pacman (pcm14) and confirmed, using a novel assay, that it has no detectable 5′-3′ exoribonuclease activity in vivo. We found that the pcm14 mutation results in small imaginal discs and lethality at the pupation stage. Using mosaic analysis and immunocytochemistry, we show that the small wing imaginal discs result from increased apoptosis, even though compensatory proliferation is also occurring. Finally, we also demonstrate that Pacman acts through the caspase pathway and that the pcm14 mutation causes post-transcriptional upregulation of the pro-apoptotic genes hid and reaper. This is the first time that a 5′-3′ exoribonuclease has been found to specifically regulate apoptosis. The conservation of Pacman/Xrn1 throughout eukaryotes suggests that it may also regulate apoptosis in other organisms.
MATERIALS AND METHODS
Fly stocks and crosses
Fly stocks were cultivated on standard media at 25°C in uncrowded conditions. All the stocks used were from the Bloomington Stock Center unless otherwise stated.
Creation of the pcm5 allele was previously described (Grima et al., 2008); the same methodology was used to create the pcm14 mutant and its corresponding wild-type control (pcmWT). The pcm14 allele is a 3,501 bp deletion extending from the P-element insertion site towards pcm, deleting 3,068 bp into the 3′ of the gene, completely removing exons 7–11 and part of exon 6. The 5′ of the neighbouring non-coding RNA CR43260 is also deleted. Additionally, a new hypomorphic allele, pcm13, was created which is a 2,222 bp deletion extending in both directions from the P-element insertion site, deleting 590 bp from the 3′ of pacman (including exons 10 and 11), 529 bp from the 3′ of Nat1 and entirely deleting CR43260. Despite the additional deletions of CR43260 and the 3′ of Nat1, the phenotypes observed for pcm13 were weaker than those seen for the pcm5 allele, showing that deletion of CR43260 does not contribute to the pacman mutant phenotypes. For use as a wild-type control for pcm13 and pcm14, a line from which the P-element was excised without causing a deletion was selected (referred to as pcmWT). To ensure that the lethality of the pcm14 chromosome was due solely to the deletion at the pacman locus, chromosomal crossover was allowed to occur between the pcm14 chromosome (w1118 pcm14) and a chromosome containing multiple recessive markers (y1 cv1 v1 f1 car1, stock 1515). Phenotypes of males carrying recombinant chromosomes were examined and none were found without the f1 and car1 markers, indicating the lethality of the chromosome stemmed from this region, which contains the pacman locus. Additionally, a translocation from X to Y, T(1;Y)B92 [stock 101110 (In(1)FM7/T(1;Y)B92, y1 y+ BS) from the Kyoto Drosophila Genetics Research Center], which includes the pacman locus was able to rescue the lethality of the pcm14 chromosome (w1118 pcm14/T(1;Y)B92, y1 y+ BS males survived to adulthood).
GAL4 drivers used were nub-GAL4 (stock 25754; P{UAS-Dcr-2.D}1, w1118; P{GawB}nub-AC-62), 69B-GAL4 (stock 1774; w*;; P{GawB}69B) and en-GAL4 UAS-GFP-actin/CyO (kindly donated by Paul Martin, University of Bristol). w*; P{tubP-GAL80ts}20; TM2/TM6B, Tb1 was used to inhibit GAL4 activity. To prevent ectopic DIAP1 expression during embryogenesis the GAL80ts system was used to inhibit GAL4 activity until the larval stages of development. This was achieved by moving the larvae from 19°C to 29°C 48 hours AEL.
The UAS-pcmRNAi stock used was stock number 21677; w1118; P{GD10926}v21677 (Vienna Drosophila RNAi Centre). Construction of the UAS-pcmWT construct has been described previously (Grima et al., 2008). UAS-pcmND was created using the Stratagene QuikChange mutagenesis kit (cat. no. 200521) to mutate an A to a G residue (GAG→GGA) at the conserved position E177G. Alteration of the homologous residue (E178G) in yeast abolishes 99.9% of exonuclease activity (Page et al., 1998). The entire pacman construct was checked by sequencing. Germline transformation of w1118 flies was then carried out using standard methods (Grima et al., 2008).
The Df(1)ED7452 stock (full genotype Df(1)ED7452, w1118 P{3′.RS5+3.3′}ED7452/FM7i, P{ActGFP}JMR3) was created by the DrosDel method (Ryder et al., 2004) and was submitted to the Bloomington Stock Center (stock 38466). The source of the Adhfn6 allele was Adhfn6 cn1; ry506 (stock 1983).
For the mosaic analysis experiment the w1118 pcm14 chromosome was recombined with y1 w1118 P{neoFRT}19A (stock 1744) (as in Xu and Rubin, 1993) to give genotype w1118 pcm14 P{neoFRT}19A/FM7i, P{ActGFP}JMR3 which was then crossed to P{Ubi-mRFP.nls}1, w*, P{hsFLP}122 P{neoFRT}19A (stock 31418). By selecting against GFP and for RFP, the offspring from this cross used in the experiment were w1118 pcm14, P{neoFRT}19A/P{Ubi-mRFP.nls}1, w*, P{hsFLP}122 P{neoFRT}19A.
In order to inhibit apoptosis the following stocks were used; w*; P{lacW}Arkk11502 Ark82/CyO y+, Df(3L)H99 kniri-1 pp/TM3 Sb1 and w*;; P{UAS-DIAP1.H}3.
Measurement of wing and wing imaginal disc sizes
Wing imaginal discs were dissected from L3 larvae in Ringer's solution and photographed using a Nikon Digital Sight DS-Fi1 camera mounted on a Nikon SMZ800 dissecting microscope at a constant magnification. The area of each disc was then measured in arbitrary units using ImageJ (http://rsbweb.nih.gov/ij/) and normalised to wild type as a percentage. For images, L3 wing imaginal discs were dissected and mounted in Aqua-Poly/Mount (Polysciences, cat. no. 18606-20). Wings were mounted in DPX medium (Fisher Scientific, cat. no. 10050080) (weighted down overnight). Images were produced using Axiovision 4.7 on an Axioplan microscope (Carl Zeiss).
Measurement of larval size, development time and survival
Larval surface area was measured essentially as described in (Hou et al., 2012) using a Nikon Digital Sight DS-Fi1 camera mounted on a Nikon SMZ800 dissecting microscope. For survival experiments, larvae of the desired genotype were placed in fresh vials and the number of eclosing adults was counted. Larval development time was measured by placing L1 larvae into food vials and counting pupae as they pupated. For the larval weight experiment, larvae were staged by the addition of bromophenol blue to the food (0.05%) and selecting only larvae that had cleared the dyed food from their gut for weighing.
Immunocytochemistry
Immunocytochemistry was performed essentially as described in (Sullivan et al., 2000). Images were taken with a Zeiss Axiovert confocal microscope equipped with a LSM520 Meta. Primary antibodies used were anti-Pacman (Grima et al., 2008) (1:500), anti-Cleaved Caspase-3 (Asp175) (Cell Signaling, cat no. 9661;1:400), anti-phosphohistone H3 (Ser10) (Cell Signaling, cat. no. 9701; 1:400) and anti-Wingless (4D4) (Developmental Studies Hybridoma Bank; 1:400). Secondary antibodies used were Cy3-conjugated monoclonal goat anti-rabbit IgG (Jackson ImmunoResearch, cat. no. 711-165-152; 1:400), Cy3-conjugated monoclonal Donkey anti-mouse IgG (Jackson ImmunoResearch, cat. no.715-165-151; 1:400) and FITC-conjugated polyclonal goat anti-rabbit (Sigma, cat. no. F9887; 1:200).
BrdU incorporation
BrdU incorporation and labelling was performed essentially as previously described (http://theduroniolab.web.unc.edu/files/2013/10/Eye-disc-BrdU.pdf). 5′ bromodeoxyuridine was fed to 120-hour-old larvae at a concentration of 0.1 mg/ml. Wing discs were dissected, fixed in 4% formaldehyde and permeabilised for 45 min in PBS + 0.6% Triton X-100. Discs were incubated in 2N HCl for 30 min at room temperature and then neutralised in sodium borate. Discs were washed 3 times in PBS + 0.3% Triton X-100 before incubation in anti-BrdU overnight at 4°C (Developmental hybridoma bank (G3G4), 1:20). The secondary antibody was Cy3-conjugated monoclonal Donkey anti-mouse IgG (Jackson ImmunoResearch, cat. no.715-165-151; 1:350).
RNA extraction and qRT-PCR
RNA extractions were performed using a mirVana miRNA isolation kit (Life Technologies, cat. no. AM1560). Samples were treated with a DNA-free kit (Life Technologies, cat. no. AM1906) and their concentrations measured on a NanoDrop 1000 spectrophotometer (Thermo Scientific). For qRT-PCR, cDNA was prepared in duplicate from the RNA samples using a High Capacity cDNA Reverse Transcription Kit (Life Technologies, cat. no. 4368814) with random primers or oligo dT primers as appropriate. A “no RT” reaction was performed in parallel as a control to confirm that all genomic DNA had been degraded. qRT-PCR was performed on each cDNA replicate in duplicate (i.e. 4 technical replicates in all), using TaqMan Universal PCR Master Mix, No AmpErase UNG (Life Technologies, cat. no. 4324018) and an appropriate TaqMan mRNA/pre-mRNA assay (Life Technologies). All mRNA TaqMan assays used were pre-designed. For custom pre-mRNA assays, 100 nt of sequence of the desired target area was submitted to Life Technologies' web-based Custom TaqMan Assay Design Tool as in (Jones et al., 2013) (supplementary material Fig. S3). RpL32 (Rp49) was used for normalisation.
Western blotting
Western blotting was performed on samples containing 60 wing imaginal discs. Tubulin was used as an internal control. Mouse anti-Tubulin primary antibody (Sigma, cat. no. T9026) was used at a 1:2000 dilution with an anti-mouse-HRP conjugated secondary antibody (Sigma, cat. no. A2304) at 1:80,000. Rabbit anti-Pacman was used at 1:2,000 with an anti-rabbit-HRP conjugated secondary antibody (Sigma, cat. no. 1949) at 1:80,000. Antibody binding was detected using Amersham ECL detection reagents (GE Healthcare, cat. no. RPN2209). Relative quantification of bands was performed in ImageJ.
Mosaic analysis
48±4 hours old larvae of the desired genotype (see Fly stocks and crosses above) were subjected to heat shock at 37°C for 1 hour. The larvae were then placed at 25°C until L3 larvae had developed. The wing imaginal discs were then dissected in Ringer's solution and mounted on poly-l-lysine treated slides in Aqua-Poly/Mount. Images were taken with a Zeiss Axiovert confocal microscope equipped with a LSM520 Meta.
Calculating the mitotic/S phase index
To count the number of cells in M phase from the phosphohistone H3 staining, or S phase from the BrdU incorporation, the ImageJ plugin, DeadEasy MitoGlia was used (Forero et al., 2010). All discs were stained and photographed under the same conditions using the standard immunocytochemistry protocol above. All settings were kept as default except the minimum threshold was set to 60. The mitotic/S phase index was then calculated for each disc using the following calculation:
Statistical analyses
All statistical analyses were performed in GraphPad Prism 6. All data analysed were compatible with parametric tests. Two-sided two-sample t-tests were used to compare the means of single test groups to single control groups. If multiple comparisons were required, a one-way ANOVA was performed with a post-test to compare the means of each possible pair of samples.
RESULTS
A null mutation in pacman results in lethality
In our previous work, we analysed the phenotypic consequences of a hypomorphic allele of pcm, pcm5. This was found to result in a number of phenotypes including smaller wing imaginal discs and wings (Jones et al., 2013). Since this allele leaves intact a large portion of the Pacman protein (amino acids 1–1264 out of 1612) including the catalytic domain (amino acids 1–674), it was likely that there was still some exoribonuclease activity within the cells of the mutant flies. To fully understand the function of Pacman in development, it was necessary to generate a null allele of pcm, to eliminate all the activity of the Pacman enzyme. This was achieved using imprecise excision of the P-element P{EP}EP1526 to generate a new lethal allele (pcm14) (Fig. 1A). As expected, the pcm14 allele results in a stronger phenotype than the hypomorphic pcm5 allele in that hemizygous males and homozygous females die during pupation before any adult structures are formed (100% penetrance). To confirm that the lethality was directly caused by a lack of pacman expression we made use of the GAL4-UAS system to express wild-type pacman cDNA (UAS-pcmWT) in particular larval tissues, in an attempt to rescue the lethality. Using the 69B-GAL4 driver (which drives expression throughout the wing, eye, haltere and leg imaginal discs and in ectodermal tissue during stages 9–17 of embryogenesis) (Brand and Perrimon, 1993; Flybase Consortium, 1996) the lethality is completely rescued (supplementary material Fig. S1), showing that the lethality of pcm14 stems from the pacman locus. We also demonstrated that the pcm14 allele is genetically a null allele as it acts as a deficiency in combination with the pcm5 allele (Fig. 1C,D).
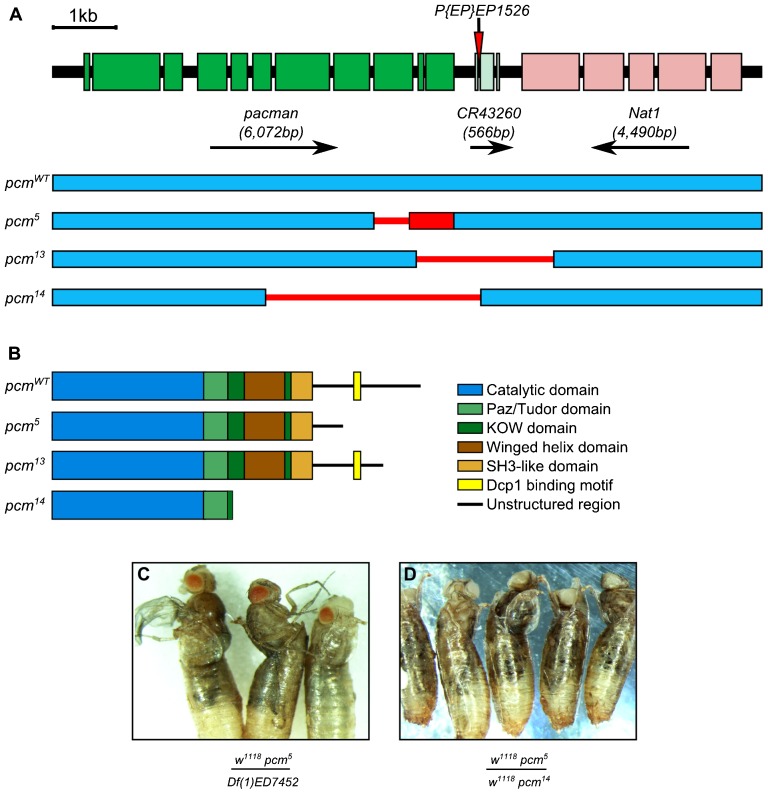
Fig. 1. pcm14 is a null allele of pcm.
(A) The genomic region of the pacman gene and the alleles pcm5, pcm13 and pcm14, created by imprecise excision of P{EP}EP1526. Green boxes represent exons of pacman, blue boxes represent wild-type genomic DNA sequences unaffected by the imprecise excisions and thin red lines indicate regions of the genomic DNA that are deleted in each allele. pcm5 is a hypomorphic allele (Grima et al., 2008; Jones et al., 2013) that consists of a 516 bp deletion causing the remainder of the coding region to be out of frame (red box). pcm13 is also a hypomorphic allele consisting of a 2,222 bp deletion extending in both directions from the P-element insertion site. pcm14 is a 3,501 bp deletion extending 3,068 bp into the 3′ of pacman, removing exons 7–11 and part of exon 6, as well as the 5′ region of CR43260. (B) The domains of Pacman proteins encoded by each allele. (C,D) When cultured at 19°C, pcm5/Df(1)ED7452 flies become stuck when eclosing from their pupal cases. Df(1)ED7452 is a 17,963 bp deficiency that removes four genes including pacman. Similar results are obtained for pcm5/Df(1)JA27 which removes at least 69 genes including pacman. The same phenotype is produced when pcm5/pcm14 flies are cultured at 19°C, showing that pcm14 is a null allele.
The Pacman protein comprises a N-terminal catalytic domain followed by PAZ/TUDOR, KOW, Winged helix and SH3 domains (Jinek et al., 2011) (Fig. 1B). The C-terminal domain is relatively unstructured and includes short sections of conserved amino acids which bind cofactors such as the decapping factor Dcp1(Braun et al., 2012). The lethal phenotype of Pacman could result from a lack of exoribonuclease activity or a lack of binding to decapping or other factors. To test this we used a nuclease-dead version of Pacman, where the critical glutamate in the conserved magnesium binding site has been mutated to a glycine (E177G). Expression of this nuclease-dead construct did not rescue the lethality when expressed with the 69B-GAL4 driver (supplementary material Fig. S1).
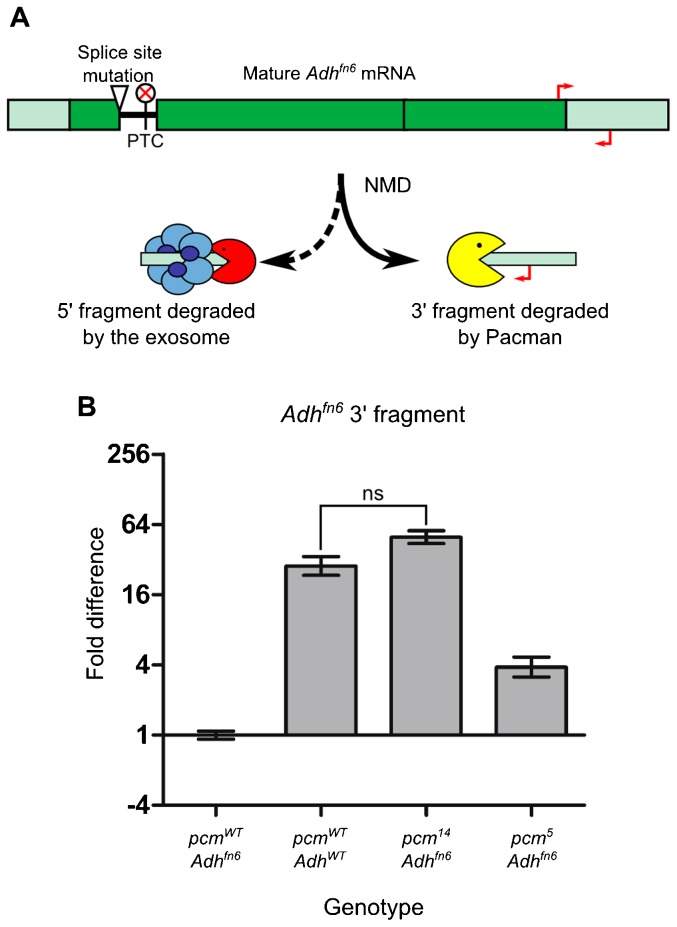
To confirm that the pcm14 allele had no enzymatic activity in vivo, we carried out a novel assay in whole larvae utilising the process of nonsense-mediated decay (NMD). NMD in Drosophila begins by endonucleolytic cleavage, creating two RNA fragments. It has been shown in cell culture that Pacman is required for degradation of the 3′ fragment (Gatfield and Izaurralde, 2004). Using an allele of Alcohol dehydrogenase, Adhfn6, which contains a premature termination codon (PTC) and is known to undergo NMD (Fig. 2A) (Benyajati et al., 1982; Brogna, 1999), we showed that the level of the 3′ fragment in pcm14; Adhfn6 double mutants is not significantly different from the level of the undegraded transcript in wild-type larvae (Fig. 2B). This is congruent with the genetic data demonstrating that pcm14 is a null allele (Fig. 1C,D). This also fits with previous findings that large C terminal deletions reduce the exoribonuclease activity to less than 10%, despite not affecting the catalytic domain (Page et al., 1998). The relative function of the pcm5 allele was calculated (using ΔΔCT values) as 66.6% [(6−2)/(6−0)*100 = 66.6%;], which demonstrates that the catalytic activity of the Pacman protein translated from the pcm5 allele is 66.6% functional. This fits in with previous findings that deletions removing the extreme C terminal of pacman, but not the SH3 domain, reduce the catalytic function to 65% that of wild type in S. cerevisiae (Page et al., 1998).
Fig. 2. Estimation of the relative function of Pacman protein produced from the pacman alleles in vivo.
(A) The Adhfn6 allele of Alcohol dehydrogenase contains a splice site mutation which retains an in-frame stop codon in the intron. This causes the Adhfn6 mRNA to undergo NMD, during which the 5′ fragment is degraded by the exosome and the 3′ fragment is degraded by Pacman. (B) To estimate the level of Pacman function in the pcm5 and pcm14 alleles, the level of the fragment degraded by Pacman was compared between lines containing the Adhfn6 allele and either pcm5 or pcm14 (n≥11. p<0.001 for all comparisons unless indicated, ns = not significant. Error bars represent standard error).
pcm14 mutant larvae have small L3 wing imaginal discs
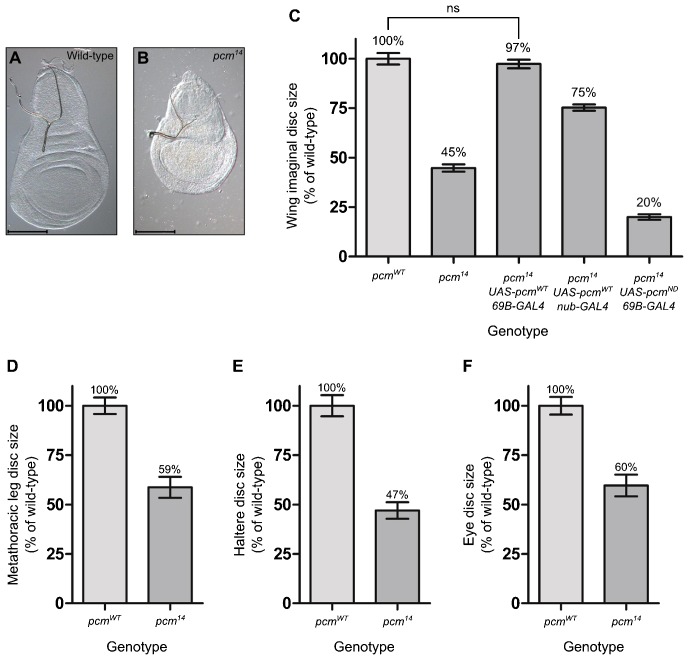
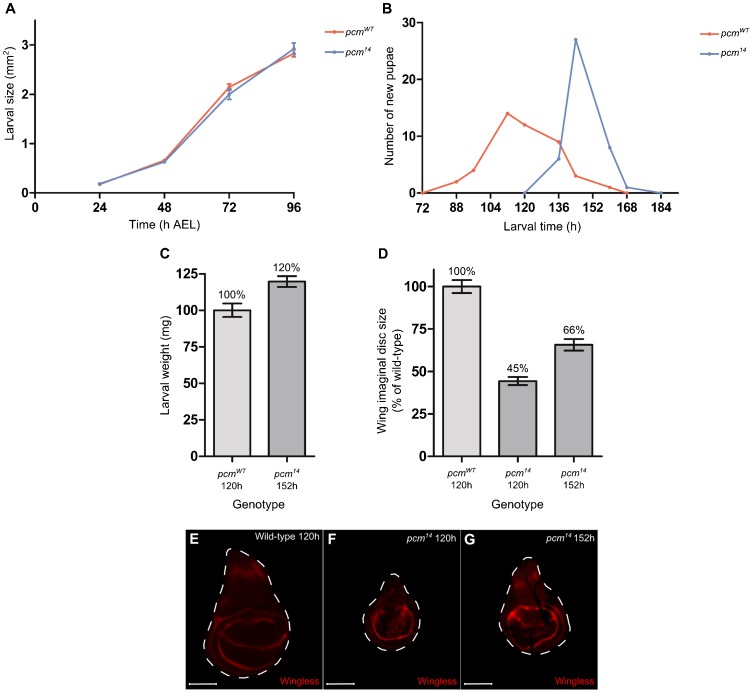
The pcm5 hypomorphic allele results in imaginal discs that are 18% smaller than those of wild type and also wings that are 16% smaller in viable adults (Jones et al., 2013). As expected, the pcm14 wing imaginal discs are substantially smaller than pcm5 discs, at 45% the size of wild type (Fig. 3A,B). We showed that the small wing disc size of the pcm14 mutants is a consequence of lack of expression of wild-type pacman in the wing discs by rescuing the phenotype using the 69B-GAL4 driver to drive expression of UAS-pcmWT over the entire wing disc (Fig. 3C). The nub-GAL4 driver, which drives expression only in the wing pouch, in combination with UAS-pcmWT rescues the wing imaginal disc size to 75% the size of wild type, demonstrating that Pacman is required throughout the wing disc (Fig. 3C). This rescue is dependent on the exoribonuclease activity of Pacman as no rescue was observed when the nuclease-dead pacman (UAS-pcmND) construct was expressed with the 69B-GAL4 driver and actually reduced the overall size significantly to 25% the size of wild type, suggesting a dominant negative effect (Fig. 3C). In order to determine whether this growth phenotype was specific to the wing imaginal discs or whether all imaginal discs were affected, we measured the size of the metathoracic leg, haltere and eye imaginal discs in wild-type and pcm14 L3 larvae (Fig. 3D–F). These other imaginal discs were smaller than those of wild type demonstrating that this phenotype is not specific to the wing imaginal discs. We chose to concentrate our investigations on the role of Pacman in the wing imaginal disc, as the development of this disc is better characterised than that of other discs and is also where the phenotypes are most apparent. The small size of the wing imaginal discs in pcm14 mutants is not due to a decrease in the overall size of the larvae as the growth rates of wild-type and mutant larvae are identical, suggesting that Pacman specifically affects the growth of the imaginal discs (Fig. 4A). We also observed that pcm14 larval development is significantly delayed, in that the majority of pupariation occurred at 136 hours after egg lay (AEL) in wild type compared to 168 hours AEL in pcm14 mutants (Fig. 4B). During this extra 32 hours of development the pcm14 larvae continue to feed and grow as pcm14 larvae weighed significantly more than wild-type larvae immediately prior to pupariation (Fig. 4C). In addition, the size of the wing imaginal discs also increased from 45% to 66% the size of wild-type during the extended 32 hours of pcm14 development (Fig. 4D). By staining the pcm14 discs for the Wingless protein, which is expressed in a distinct pattern in the mature wild-type L3 wing imaginal disc at 120 hours (Fig. 4E) (Couso et al., 1994) we showed that the wing discs at 120 hours had an incomplete pattern of Wingless expression (Fig. 4F) and were therefore immature, whereas at 152 hours, when the majority of the mutant larvae are about to pupate, the pattern of Wingless expression was similar to wild type (Fig. 4G). Therefore pcm14 wing discs are delayed in both growth and differentiation.
Fig. 3. pcm14 larvae have significantly smaller imaginal discs than wild-type larvae.
(A,B) Representative wild-type and pcm14 wing imaginal discs. Scale bar represents 100 µm. (C) The mean size of pcm14 wing imaginal discs is 45% the size of wild type. This phenotype can be rescued by expressing a UAS-pcmWT construct throughout the wing imaginal disc cells using the 69B-GAL4 driver. Driving UAS-pcmWT expression with nub-GAL4 partially rescues this phenotype to 75% the size of wild type. Expressing a UAS-pcmND construct throughout the disc reduces the mean wing disc size to 20% of wild type (n≥31). (D) The mean size of pcm14 metathoracic leg discs is 59% the size of wild type (n≥21), pcm14 haltere discs (E) are 47% the size of wild-type (n≥21) and pcm14 eye discs (F) are 60% the size of wild type (n≥16). p<0.001 for all comparisons unless indicated, ns = not significant, error bars represent 95% confidence limits.
Fig. 4. pcm14 larvae are delayed in development.
(A) The growth rate of pcm14 larvae is not significantly different to wild type (n≥16). (B) Onset of pupariation of pcm14 larvae is delayed by around 32 hours compared to wild type (n≥42). (C) The mean weight of pcm14 larvae just prior to pupariation (152 hours) is 120% compared to the mean size of wild-type larvae just prior to pupariation (120 hours). (n≥35, p<0.0001). (D) During the extra 32 hours of development that pcm14 larvae undergo, the size of the wing imaginal discs increases from 45% to 66% the size of wild type. (n≥30, p<0.001 for all comparisons). Error bars represent 95% confidence limits. (E–G) Wing imaginal disc development in pcm14 larvae is morphologically delayed by 32 hours as determined by Wingless staining. (E) Wild-type wing imaginal disc at 120 hours displaying the correct pattern of Wingless. (F) pcm14 wing imaginal disc at 120 hours does not display the correct pattern of expression for this time point. The expression is more diffuse throughout the wing pouch and does not contain the two rings of expression surrounding the wing pouch. (G) pcm14 wing imaginal disc at 152 hours displaying the pattern of Wingless expression seen in wild type at 120 hours. Scale bars represent 100 µm.
We also tested whether the reduced growth of the wing imaginal discs in pcm14 mutants is due to a lack of functional Pacman protein in the wing imaginal disc cells themselves rather than a consequence of the whole larva failing to develop correctly and signalling to the wing imaginal discs to delay their development. Knockdown of pacman expression in specific domains of the wing disc using various GAL4 drivers resulted in loss of tissue in the corresponding domains of the adult wing (Fig. 5). Therefore Pacman would appear to be required cell autonomously.
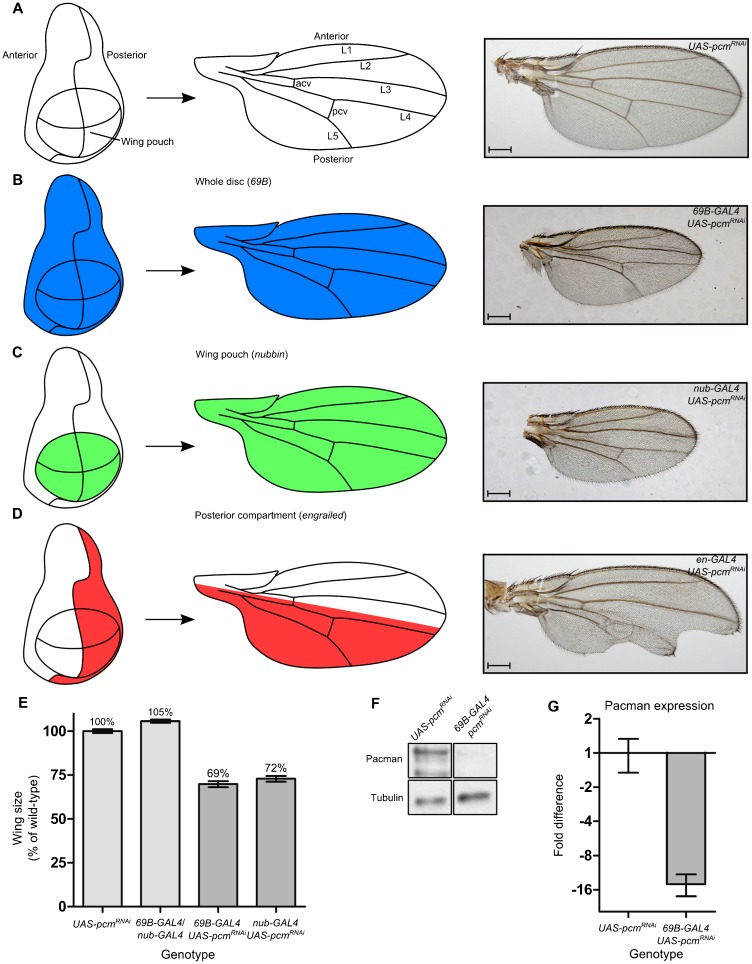
Fig. 5. Knockdown of pacman using RNAi specifically within the wing imaginal discs results in smaller wings and wing vein defects.
(A) Diagrammatic representation of a wing imaginal disc fate map forming the adult wing and a typical wild-type wing. (B) Knockdown of pacman throughout the wing imaginal discs using 69B-GAL4 leads to smaller wings and wing vein defects including loss of the anterior cross vein (94%), shortened L5 (8%) and shortened posterior cross vein (6%); (n = 49). (C) Knockdown of pacman specifically in the wing pouch using nub-GAL4 also leads to smaller wings and also wing vein abnormalities such as loss of the anterior cross vein (100%), and a shortened L5 vein (40%); (n = 33). (D) Knockdown of pacman specifically in the posterior compartment of the wing imaginal discs using en-GAL4 leads to defects in the posterior of the adult wing, The most common phenotypes recorded were blisters (66%), notches/loss of tissue (62%), and wing veins abnormalities, such as shortened L5 (19%) or branching of the posterior cross vein (13%) (n = 818). Scale bar represents 200 µm. (E) Wing sizes in 69B-GAL4/UAS-pcmRNAi and nub-GAL4/UAS-pcmRNAi wings compared with their parental controls. (n≥33, error bars represent 95% confidence limits. p<0.001 for all comparisons except between 69B-GAL4/UAS-pcmRNAi and nub-GAL4/UAS-pcmRNAi where p<0.05.). (F,G) Western blotting experiments to quantitate Pacman expression in 69B-GAL4/UAS-pcmRNAi wing imaginal discs, show that it is knocked down almost 16 fold when compared to the UAS-pcmRNAi parental control. (n = 3, error bars represent standard error.)
Populations of pcm14 wing imaginal disc cells are smaller than populations of wild-type wing imaginal disc cells
To confirm the cell autonomous requirement for Pacman in wing disc cells we used mosaic analysis where clones of cells with a mutant or wild-type homozygous genotype are induced in a heterozygous background by mitotic recombination. Populations of pcm14/pcm14 cells and pcm+/pcm+ cells were induced in a background of pcm14/pcm+ cells, using the FLP/FRT system. If the pcm14 mutation does result in a reduced growth rate, then a reduction in size of the mutant clones compared to their wild-type twin spots would be expected (Neto-Silva et al., 2009). When mitotic recombination was induced 48 hours AEL, mutant clones are clearly visible throughout the disc, alongside the wild-type twin spots, which were larger in size, showing that wild-type cells do indeed have a growth advantage over pcm14 cells (Fig. 6A). This result also demonstrates a cell autonomous requirement for Pacman for correct growth.
Fig. 6. Populations of pcm14 cells have reduced growth compared to populations of wild-type cells as a result of an increase in apoptosis.
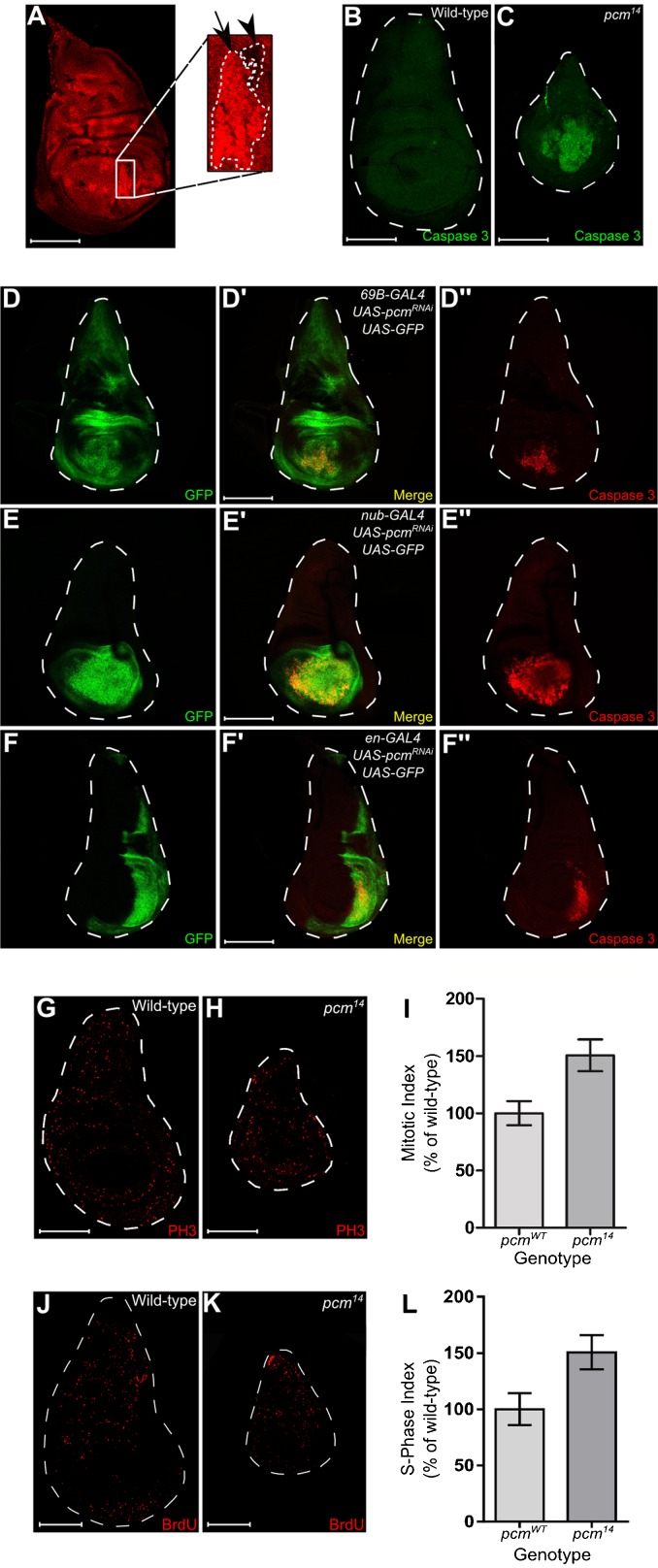
(A) Mosaic analysis was performed to directly compare growth rates between wild-type and pcm14 mutant cells. Background cells are pcmWT/pcm14, cells increased in fluorescence are pcmWT/pcmWT and cells with no fluorescence are pcm14/pcm14. Mutant clones (arrowhead) were significantly smaller than their wild-type twin spots (arrow) (n = 25). The boundaries of wild type and mutant clones are marked by a white dashed line. Wild-type and pcm14 wing imaginal discs were stained with an antibody for activated Caspase 3 which labels cells undergoing apoptosis. Cells throughout the wing pouch of the pcm14 discs (C) were undergoing apoptosis which is not seen in wild type (B) (n≥39). Knockdown of pacman using RNAi driven by (D–D″) 69B-GAL4, (E–E″) nub-GAL4 or (F–F″) en-GAL4 caused apoptosis to occur specifically within the wing pouch region of the disc. GFP fluorescence indicates regions of the disc expressing the UAS-pcmRNAi construct and Caspase 3 staining indicates regions of the disc where cells are undergoing apoptosis (n≥6). Note that these imaginal discs express UAS-GFP as well as UAS-pcmRNAi under the control of the relevant driver. (G,H) Wild-type and pcm14 wing imaginal discs were stained with an antibody for phosphohistone H3 which labels cells in M phase. (I) A mitotic index was calculated by dividing the number of cells in M phase by the area of the disc. The mitotic index was increased in pcm14 discs by 150% compared with wild type (n≥14, p<0.001. Error bars represent 95% confidence limits). BrdU incorporation of cells at S-phase within wild-type (J) and pcm14 wing imaginal discs (K) was visualised using an antibody for BrdU. (L) A S-phase index was calculated by dividing the number of cells in S-phase by the area of the disc. The S-phase index was increased in pcm14 discs by 154% compared with wild type (n≥8 p<0.01. Error bars represent 95% confidence limits). Scale bars represent 100 µm.
Loss of Pacman induces ectopic apoptosis in the wing pouch region of wing imaginal discs
The smaller size of the pcm14 wing imaginal discs could be due to an increase in apoptosis, a decrease in cell division, or a combination of both. To determine whether there was an increase in apoptosis in pcm14 wing imaginal discs compared to wild type, discs were stained with an anti-activated Caspase 3 antibody, which stains cells undergoing apoptosis. In the pcm14 wing imaginal discs, large groups of cells in the wing pouch undergo apoptosis (Fig. 6C). This does not occur in the wild-type discs (Fig. 6B). Therefore loss of pacman appears to induce apoptosis in the wing pouch which could account for the small size of the wing discs.
To determine whether loss of pacman induces apoptosis only in the wing pouch area of the disc we knocked down pacman in different regions of the disc using the GAL4-UAS system and monitored apoptosis activity by anti-activated Caspase 3 staining. As can be seen from Fig. 6D, knockdown of pacman over the entire disc using the 69B-GAL4 driver results in apoptosis only in the wing pouch (Fig. 6D″). In the case of the engrailed-GAL4 driver, which drives expression in the posterior half of the disc, apoptosis also occurs only in the posterior part of the wing pouch (Fig. 6F″) These results demonstrate that the ectopic apoptosis is specific to the wing pouch region of the disc during the L3 stage of development.
The pcm14 mutation results in compensatory proliferation of wing imaginal disc cells
The results of the mosaic analysis experiment above, where pcm14/pcm14 mutant clones were smaller than pcm+/pcm+ clones could also be explained by a decrease in cell division. To test this, the rate of cell division in the pcm14 imaginal discs, compared to wild type, was monitored by staining the discs with an anti-phosphohistone H3 antibody, which detects cells undergoing mitosis. The nuclei undergoing division were counted and the mitotic index (the number of cells in M phase/area of disc) was calculated. Surprisingly, these results suggest that the rate of cell division in mutant discs is 51% higher than in wild-type discs (Fig. 6G–I). To assess whether the cells stained using phosphohistone H3 are blocked in M phase, rather than undergoing proliferation, we also used bromodeoxyuridine (BrdU) incorporation to identify cells undergoing DNA synthesis (i.e. S-phase) (Schubiger and Palka, 1987). This data shows that cell division is 54% higher in mutant discs than the control discs (Fig. 6J–L), confirming the phosphohistone H3 staining results. Therefore, these results strongly suggest that cells within mutant discs are undergoing compensatory proliferation (Fan and Bergmann, 2008a; Martín et al., 2009) in an attempt to overcome the increased rate of apoptosis. Nevertheless, the smaller size of the mutant discs, even after the extended period of larval development, means that this compensatory proliferation is unable to counteract the increased levels of apoptosis.
Inhibition of apoptosis rescues the pcm14 wing imaginal disc phenotypes
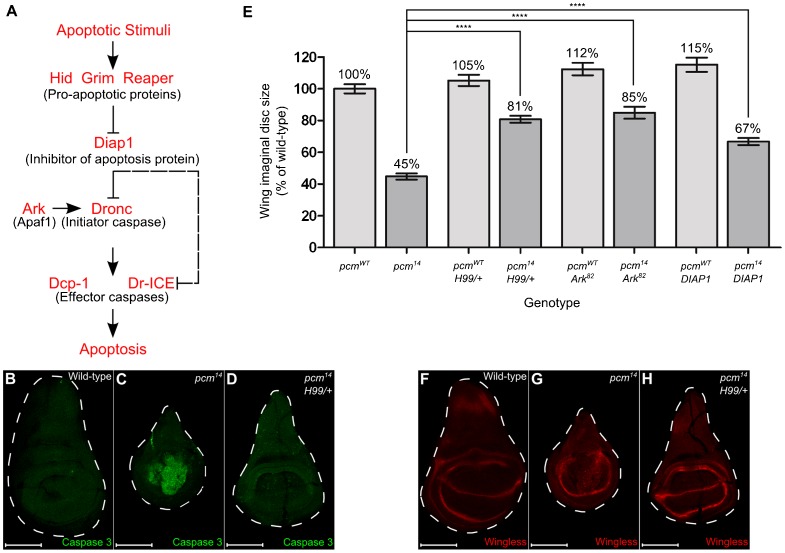
In Drosophila, the key activators of the caspase-induced apoptosis pathway are the Reaper, Hid and Grim proteins (Steller, 2008; Xu et al., 2009) (Fig. 7A). These are located adjacent to each other on the chromosome arm 3L and are removed by the deficiency Df(3L)H99 (Chen et al., 1996; Grether et al., 1995; White et al., 1994). To determine whether Pacman acts through this pathway, we crossed Df(3L)H99 into the pcm14 mutant as a heterozygote, and analysed the effects on wing imaginal discs. Fig. 7D shows that apoptosis in pcm14;;Df(3L)H99/+ imaginal discs is significantly reduced compared to pcm14 discs (Fig. 7C and Fig. 6C) and is similar to that of wild type (Fig. 7B and Fig. 6B). Furthermore, the size of the imaginal discs is partially rescued to 81% the size of wild-type discs compared to 45% the size for pcm14 discs (Fig. 7E). Staining pcm14;;Df(3L)H99/+ wing discs at 120 hours AEL shows the characteristic Wingless expression pattern (Fig. 7H) as seen in wild-type discs (Fig. 7F and Fig. 4E). Therefore the delay in development of pcm14 discs is also rescued by reduced expression of the caspase pathway genes.
Fig. 7. Inhibiting apoptosis partially rescues pcm14 phenotypes.
(A) A diagrammatic representation of the apoptosis pathway in Drosophila, indicating the main proteins involved in triggering apoptosis in response to an apoptotic stimuli. (B–D) Inhibiting apoptosis in pcm14 wing imaginal discs was achieved by crossing in the Df(3L)H99 deletion (which deletes pro-apoptotic genes hid, grim and reaper) as a heterozygote. The presence of Df(3L)H99 reduced the amount of Caspase 3 staining and increased the size of the pcm14 wing discs (compare C with D) (n = 13). (E) Inhibiting apoptosis partially rescues the size of pcm14 wing imaginal discs. Reducing the copy number of reaper, hid and grim from 2 to 1 using the Df(3L)H99 deletion as a heterozygote (pcm14;;Df(3L)H99/+) rescues the wing disc size from 45% to 81% (n≥31). Use of the Ark82 allele as a homozygote rescues wing disc size from 45% to 85% that of wild type, showing that the adaptor protein Ark is required for much of the pcm14 induced apoptosis (n≥19). Overexpression of the Inhibitor of apoptosis protein DIAP1 using the 69B-GAL4 driver (pcm14;GAL80ts/+;69B-GAL4/UAS-DIAP1) rescues the size of pcm14 discs from 45% to 67% that of wild type, confirming that Pacman is acting through the pathway in A (n≥19) ****p<0.0001. Inhibiting apoptosis in the pcmWT background increases wing disc size to 105% using the Df(3L)H99 deletion (p<0.05), 112% using the Ark82 homozygous mutation (p<0.0001) and 115% using the UAS-DIAP1 (p<0.0001). Error bars represent 95% confidence limits. (F–H) Inhibiting apoptosis rescues the delay in wing imaginal disc development as determined by Wingless staining at 120 hours (compare G with H) (n = 13). Scale bars represent 100 µm.
In order to determine whether the adapter protein Ark (Apaf-1) (Fig. 7A) is required for the ectopic apoptosis in pcm14 wing imaginal discs, Ark activity was completely removed using the null Ark82 mutation as a homozygote (Akdemir et al., 2006). This significantly rescued the size of pcm14 wing imaginal discs from 45% to 85% the size of wild type, suggesting that Ark is required for most but not all of the apoptosis occurring in these discs. This supports recent findings (Kang and Bashirullah, 2014) showing that apoptosis induced prior to late L3 is independent of apoptosome formation. In addition, overexpressing the inhibitor of apoptosis protein DIAP1 (Fig. 7A) in wing imaginal disc cells, rescued the size of pcm14 wing imaginal discs to 67% the size of wild type. This was achieved using the UAS-DIAP1 construct under 69B-GAL4 control. These results demonstrate that Pacman is indeed acting through the pathway depicted in Fig. 7A and that it is acting upstream of the Hid, Reaper and Grim proteins. Inhibiting apoptosis in a wild-type background increases wing imaginal disc size by 5% (Df(3L)H99/+), 12% (Ark82) or 15% (UAS-DIAP1), which is to be expected as it has been shown that there are low levels of apoptosis during normal wing imaginal disc development (Milán et al., 1997).
hid and reaper are post-transcriptionally upregulated in pcm14 wing imaginal discs
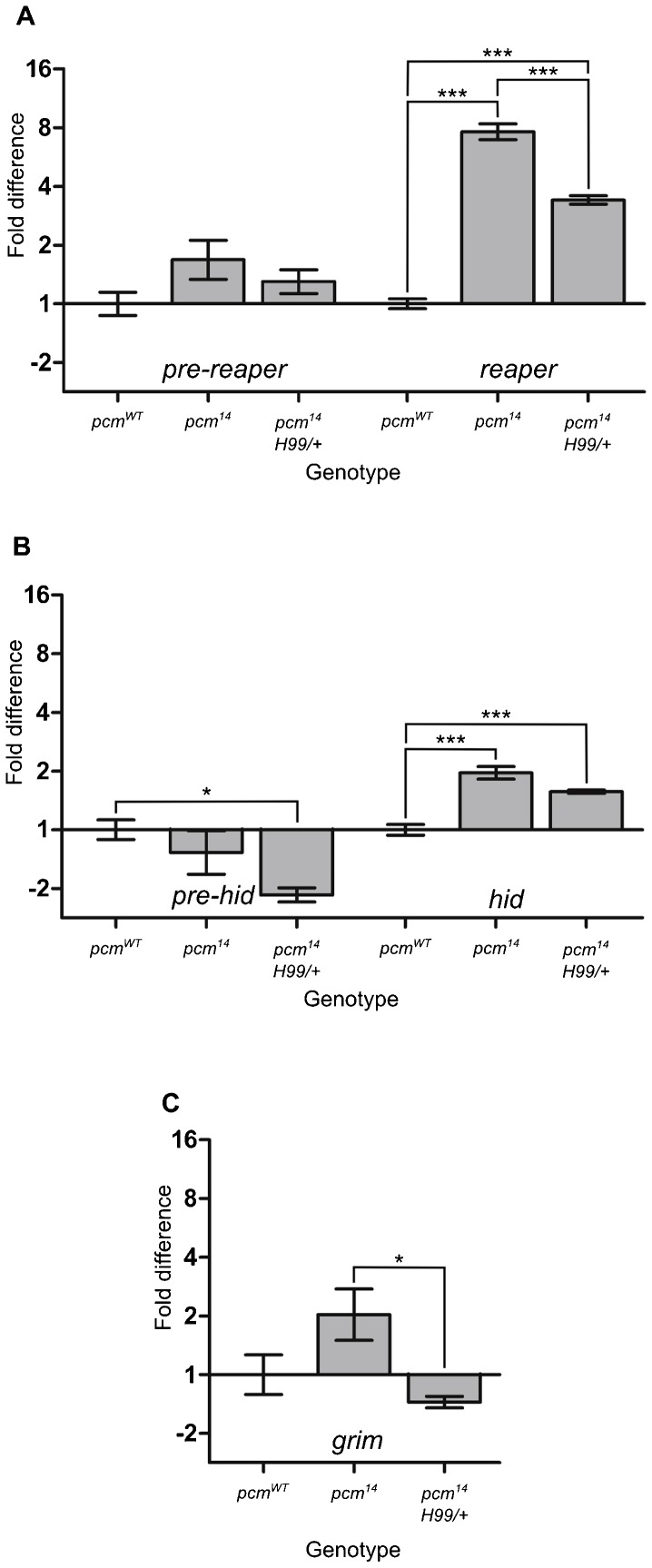
Pacman is an exoribonuclease that degrades mRNAs. We therefore hypothesised that Pacman could specifically target hid, grim and reaper mRNA with the consequence that the loss of Pacman would result in increased levels of these mRNAs. To test this, we used quantitative TaqMan qRT-PCR to measure the levels of hid, grim and reaper mRNAs in pcm14 mutant wing imaginal discs. As can be seen from Fig. 8, reaper mRNA is increased by 7.8-fold in pcm14 wing imaginal discs compared to wild type whereas hid mRNA is increased by 2-fold. The levels of grim mRNA were very low and variable and showed no significant difference to those of wild type. In the rescued wing imaginal discs (pcm14;;Df(3L)H99/+) reaper and hid mRNA levels were intermediate between pcm14 discs and wild-type discs, as expected.
Fig. 8. reaper and hid are post-transcriptionally upregulated in pcm14 wing imaginal discs as determined by qRT-PCR.
(A) Levels of reaper mRNA increase 7.8-fold in pcm14 mutant wing imaginal discs compared with wild type whereas levels of pre-reaper RNA are not significantly different. The increase in mature reaper RNA is halved to 3.4-fold in pcm14;;Df(3L)H99/+ mutant wing imaginal discs, where there is only one copy of the reaper, hid and grim genes. (n = 6 for wild type and pcm14 and n = 5 for pcm14;;Df(3L)H99. ***p<0.001. Error bars represent standard error). (B) Levels of hid mRNA increase 2-fold in pcm14 mutant wing imaginal discs compared with wild type, whereas levels of pre-hid RNA do not differ significantly. The levels of mature hid are reduced in pcm14;;Df(3L)H99/+ wing imaginal discs, although this difference only showed statistical significance if no correction for multiple comparisons was performed (i.e. if a t-test was used to compare hid levels in pcm14 and pcm14;;Df(3L)H99, rather than an ANOVA). (n = 6 for wild type and pcm14 and n = 5 for pcm14;;Df(3L)H99. ***p<0.001 and *p<0.05. Error bars represent standard error). (C) Levels of grim mRNA are not significantly different between wild-type and pcm14 wing imaginal discs. grim pre-mRNA could not be reliably detected. (n = 6 for wild type and pcm14 and n = 5 for pcm14;;Df(3L)H99. *p<0.05. Error bars represent standard error). RpL32 (Rp49) was used for normalisation.
If Pacman is involved in the degradation of reaper and hid mRNAs, then we would expect these mRNAs to increase at the post-transcriptional level but not at the transcriptional level in the pcm14 mutants. To test this, we used primer-probe assays designed to detect reaper and hid pre-mRNAs, but not their mRNAs (supplementary material Fig. S3). These experiments show that reaper and hid pre-mRNA levels in the pcm14 mutant are not significantly different from that of the wild-type control (p = 0.0808 and 0.3634 respectively; Fig. 8A,B). Therefore reaper and hid mRNAs show a significant increase at the post-transcriptional level but not the transcriptional level suggesting that Pacman normally targets these mRNAs for degradation.
To determine whether the upregulation of reaper and hid mRNA in pacman mutants is a specific effect, rather than a general effect on many RNAs, we also analysed the effect of the pacman null mutation on the levels of other RNAs. TaqMan qRT-PCR experiments using primer-probe assays for the cell cycle mRNAs string (cdc25), CyclinD and CyclinE mature mRNA showed no differences in expression levels between pcm14 and wild-type wing imaginal discs (supplementary material Fig. S4). This is consistent with a specific effect of Pacman on reaper and hid mRNAs in vivo.
An increase of reaper and hid mRNA in pcm14 mutants would normally mean that Reaper and Hid proteins are also increased. We were, however, unable to test this directly using western blotting as the antibodies available to us gave non-specific bands or no bands at all. Nevertheless, the phenotypic effects we see in pcm14 mutants, or as a result of pacman knockdown, and the genetic interactions of these mutants with other genes in the caspase pathway, are entirely consistent with protein being expressed from the increased reaper and hid mRNAs. Taken together, our results therefore show that Pacman can regulate apoptosis in Drosophila wing imaginal discs and that this regulation mainly takes place at the post-transcriptional level.
DISCUSSION
Apoptosis is a key process in developmental pathways and also in cancer. In the present study, we have generated a null mutation in pacman (pcm14) and used this to show that Pacman can control apoptosis in wing imaginal discs by regulating levels of hid and reaper mRNAs. Use of the Df(3L)H99 deletion, which removes one copy of the hid, grim and reaper genes, largely rescues the effect of the pcm14 mutation on growth of the wing imaginal discs and on developmental timing. However, the Df(3L)H99 deletion (Df(3L)H99/+) does not rescue the lethality of the pcm14 mutation. This suggests that there may be other targets of Pacman that are misregulated in pcm14 larvae or pupae.
According to the data presented in Fig. 6C, the mutant wing discs are proportionately reduced in size, even though the majority of apoptosis occurs in the wing pouch. Our data also show that Pacman is expressed over the entire disc (supplementary material Fig. S2) and pcm14/pcm14 mutant clones are smaller than their wild-type twin spots throughout the disc (Fig. 6A). It is possible that apoptosis is occurring throughout the disc in earlier stages of development but is restricted to the wing pouch during L3. Our data is consistent with other studies reporting that cells within the wing pouch are particularly sensitive to apoptosis (Kang and Bashirullah, 2014), perhaps due to expression of particular apoptotic regulators in that region (Bejarano et al., 2010). The co-ordinate growth of the wing disc, even though apoptosis is occurring in a particular region of the disc, is likely to be due to long range signalling via morphogens which control overall patterning and growth of the wing disc. For example, Decapentaplegic (Dpp), a bone morphogenetic protein (BMP) functions as a long range morphogen to control patterning and growth (Vuilleumier et al., 2010). Furthermore, the Aegerter-Wilmsen model which explains how growth is constant throughout the disc suggests that growth of the peripheral cells within the disc is caused by stretching of the cells as a result of growth at the centre of the disc (Aegerter-Wilmsen et al., 2007; Aegerter-Wilmsen et al., 2012). Therefore, reduced growth at the centre of the disc, caused by apoptosis specifically in the pouch, is likely to cause reduced growth of the whole disc.
Our results also show that the pcm14 mutation induces cell proliferation as well as apoptosis. Apoptosis-induced compensatory proliferation is known to occur to maintain tissue homeostasis so that damaged tissues can be replaced allowing the organ to maintain its normal size (Fan and Bergmann, 2008a; Martín et al., 2009). In Drosophila, this occurs via the initiator caspase Dronc which induces compensatory proliferation as well as apoptosis (Fan and Bergmann, 2008b; Huh et al., 2004; Kondo et al., 2006; Wells et al., 2006). Since Dronc is activated by Hid and Reaper, the increase in hid and reaper mRNA in pcm14 cells is consistent with increased activity of Dronc. Nevertheless, the 51%–54% increase in cell division in the pcm14 wing imaginal discs is insufficient to compensate for the concurrent increase in apoptosis because the wing discs fail to develop and differentiate, leading to death of the pupa. This failure of the wing discs to regenerate could be explained by there being prolonged apoptosis in the pcm14 wing imaginal discs, whereas other experiments have induced a pulse of apoptosis, allowing time for the wing disc to recover (Milán et al., 1997; Pérez-Garijo et al., 2004).
The above results are consistent with reaper and hid being translated from the upregulated reaper and hid transcripts in pcm14 mutants. This would imply that these transcripts are both capped and polyadenylated. Biochemical analyses have shown that the less structured C-terminal domain of Pacman/Xrn1 includes short sections of conserved amino acids which bind co-factors such as the decapping protein Dcp1. Dcp1 associates with the decapping enzyme Dcp2, therefore coupling decapping to 5′-3′ degradation (Braun et al., 2012). In pcm14 cells where no Pacman is present, decapping would therefore be expected to be impaired, which is consistent with our data. The alternative and/or additional hypothesis is that reaper and hid are being translated in a cap independent manner. Indeed the 5′ UTRs of these genes have been shown to contain functional Internal Ribosome Entry Sites (IRES) and are still able to undergo translation in cells in which cap dependent translation is impeded (Hernández et al., 2004; Vazquez-Pianzola et al., 2007).
The above molecular mechanisms also are consistent with the “dominant negative” effect seen when we express the nuclease-dead version of Pacman in a pcm14 mutant background. In Drosophila tissue culture cells, over-expression of catalytically inactive Pacman inhibited both decapping and degradation of a reporter RNA leading to an accumulation of capped fragments (Braun et al., 2012). Therefore the dominant negative effect could result from the sequestration of the Decapping protein Dcp1 together with lack of exonuclease activity. Expressing a “nuclease dead” Pacman in pcm14 cells would not rescue any exoribonuclease activity but could impair decapping further. Our results therefore support the model (Jones et al., 2012) that Pacman/Xrn1 normally assembles a complex of 5′-3′ degradation factors including Dcp1 to provide a multicomponent complex which decaps and then degrades specific RNAs in a 5′-3′ direction.
Our data, using natural tissue rather than immortalised tissue culture cells, supports the idea that there is a network of RNA-protein interactions contributing to apoptosis and proliferation. This idea is supported by work on the deadenylases Ccr4a and Ccr4b which can affect cell survival in MCF7 human breast cancer cells (Mittal et al., 2011). Further, the RNA-binding protein HuR (homologue of Elav in Drosophila) has recently been shown to be cleaved in HeLa cells during caspase-mediated apoptosis with the two cleavage fragments binding to and stabilising caspase 9 mRNA, thus promoting apoptosis (von Roretz et al., 2013). Our data showing that the exoribonuclease Pacman is also involved in the control of apoptosis suggests a key role for the 5′-3′ degradation pathway in the regulation of apoptosis.
What are the mechanisms by which Pacman might be affecting the levels of mature hid and reaper mRNA? The simplest hypothesis is that Pacman is normally targeted to hid and reaper mRNA, resulting in degradation of these mRNAs. This targeting could be accomplished by specific RNA binding proteins and/or miRNAs binding to the 3′ UTRs of hid and reaper mRNAs and directing them to the 5′-3′ degradation machinery (Eulalio et al., 2007; Jones et al., 2012; Nishihara et al., 2013). The 3′ untranslated regions of hid and reaper contain many predicted and validated miRNA binding sites for miRNAs (Brennecke et al., 2003; Brennecke et al., 2005; Ge et al., 2012; Hilgers et al., 2010; Ruby et al., 2007; Xu et al., 2004). For example, the miRNA bantam has been shown to bind to the 3′ UTR of hid mRNA, thus regulating its expression (Brennecke et al., 2003). In addition, miR-2 is known to bind to the 3′ UTR of reaper, repressing its translation and directing it to P-body-like structures (Thermann and Hentze, 2007). A possible model to explain our results is that reaper and hid mRNAs are normally unstable because they are directed to the 5′-3′ degradation complex by miRNAs binding to their 3′ UTRs. In wild-type cells, these RNAs are rapidly decapped by decapping enzymes associated with Pacman and then degraded in a 5′-3′ direction. In the Pacman mutant, these mRNAs are not efficiently degraded because of the absence of Pacman. It is also possible that reaper and hid are particularly affected by loss of Pacman because the presence of IRES sequences within their 5′ UTRs (Hernández et al., 2004; Vazquez-Pianzola et al., 2007) means that these RNAs can be translated even if they are decapped. In a pacman mutant, these decapped RNAs may still be translated to produce Reaper and Hid protein. The exact mechanisms whereby Pacman regulates these mRNAs will require further research.
Supplementary Material
Acknowledgments
The authors would like to thank Clare Rizzo-Singh and Karen Scruby for technical help and Juan Pablo Couso, Claudio Alonso and Robert Ray for helpful discussions. We are grateful to Julie Aspden and Simon Morley for critical reading of the manuscript. The Wingless antibody developed by Stephen M. Cohen and the BrdU antibody developed by Stephen J. Kaufman were obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development (NICHD) and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. The fly stock engrailed-GAL4 UAS-GFP-actin/CyO was kindly donated by Paul Martin, University of Bristol, UK.
Footnotes
Author Contributions: J.A.W. designed the work and performed most of the Drosophila experiments; C.I.J. designed experiments, generated the null pcm14 mutation, and performed the in vivo activity assay; B.P.T. performed RNAi and BrdU experiments; A.L.P. carried out some of the TaqMan qRT-PCR experiments; D.P.G. generated the pcm5 mutation, characterised the nuclease-dead pacman mutant and provided advice on experiments; S.H. generated the nuclease-dead pacman mutant; S.H.C. carried out the growth experiments; M.V.Z. advised on confocal microscopy; S.F.N. designed experiments and wrote the manuscript.
Competing interests: The authors declare no competing or financial interests.
Funding
This work was funded by the Biotechnology and Biological Sciences Research Council [grant numbers BB/I021345/1 and BB/G002754/1]; a Medical Research Council Studentship and Medical Research Council Centenary Award [G1000409, MR/J500525/1 to J.A.W.]; a Nuffield Foundation Undergraduate Research bursary [URB/39931 to B.P.T.]; and a Biochemical Society Summer Vacation Studentship [to S.H.C.].
References
- Aegerter-Wilmsen T., Aegerter C. M., Hafen E., Basler K. (2007). Model for the regulation of size in the wing imaginal disc of Drosophila. Mech. Dev. 124, 318–326. 10.1016/j.mod.2006.12.005 [DOI] [PubMed] [Google Scholar]
- Aegerter-Wilmsen T., Heimlicher M. B., Smith A. C., de Reuille P. B., Smith R. S., Aegerter C. M., Basler K. (2012). Integrating force-sensing and signaling pathways in a model for the regulation of wing imaginal disc size. Development 139, 3221–3231. 10.1242/dev.082800 [DOI] [PubMed] [Google Scholar]
- Akdemir F., Farkas R., Chen P., Juhasz G., Medved'ová L., Sass M., Wang L., Wang X., Chittaranjan S., Gorski S. M. et al. (2006). Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development 133, 1457–1465. 10.1242/dev.02332 [DOI] [PubMed] [Google Scholar]
- Bejarano F., Smibert P., Lai E. C. (2010). miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 338, 63–73. 10.1016/j.ydbio.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Place A. R., Wang N., Pentz E., Sofer W. (1982). Deletions at intervening sequence splice sites in the alcohol dehydrogenase gene of Drosophila. Nucleic Acids Res. 10, 7261–7272. 10.1093/nar/10.22.7261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Braun J. E., Truffault V., Boland A., Huntzinger E., Chang C. T., Haas G., Weichenrieder O., Coles M., Izaurralde E. (2012). A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat. Struct. Mol. Biol. 19, 1324–1331. 10.1038/nsmb.2413 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M. (2003). bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36. 10.1016/S0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R. B., Cohen S. M. (2005). Principles of microRNA-target recognition. PLoS Biol. 3, e85 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S. (1999). Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA 5, 562–573. 10.1017/S1355838299981359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nordstrom W., Gish B., Abrams J. M. (1996). grim, a novel cell death gene in Drosophila. Genes Dev. 10, 1773–1782. 10.1101/gad.10.14.1773 [DOI] [PubMed] [Google Scholar]
- Couso J. P., Bishop S. A., Martinez Arias A. (1994). The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development 120, 621–636. [DOI] [PubMed] [Google Scholar]
- Danial N. N., Korsmeyer S. J. (2004). Cell death: critical control points. Cell 116, 205–219. 10.1016/S0092-8674(04)00046-7 [DOI] [PubMed] [Google Scholar]
- Domingos P. M., Steller H. (2007). Pathways regulating apoptosis during patterning and development. Curr. Opin. Genet. Dev. 17, 294–299. 10.1016/j.gde.2007.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Rehwinkel J., Stricker M., Huntzinger E., Yang S-F., Doerks T., Dorner S., Bork P., Boutros M., Izaurralde E. (2007). Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 21, 2558–2570. 10.1101/gad.443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Bergmann A. (2008a). Apoptosis-induced compensatory proliferation. The cell is dead. Long live the cell! Trends Cell Biol. 18, 467–473. 10.1016/j.tcb.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Bergmann A. (2008b). Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev. Cell 14, 399–410. 10.1016/j.devcel.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase Consortium(1996). FlyBase: the Drosophila database. Nucleic Acids Res. 24, 53–56. 10.1093/nar/24.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero M. G., Learte A. R., Cartwright S., Hidalgo A. (2010). DeadEasy Mito-Glia: automatic counting of mitotic cells and glial cells in Drosophila. PLoS ONE 5, e10557 10.1371/journal.pone.0010557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y., Steller H. (2011). Programmed cell death in animal development and disease. Cell 147, 742–758. 10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J. (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113–126. 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Gatfield D., Izaurralde E. (2004). Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429, 575–578. 10.1038/nature02559 [DOI] [PubMed] [Google Scholar]
- Ge W., Chen Y. W., Weng R., Lim S. F., Buescher M., Zhang R., Cohen S. M. (2012). Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell Death Differ. 19, 839–846. 10.1038/cdd.2011.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether M. E., Abrams J. M., Agapite J., White K., Steller H. (1995). The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9, 1694–1708. 10.1101/gad.9.14.1694 [DOI] [PubMed] [Google Scholar]
- Grima D. P., Sullivan M., Zabolotskaya M. V., Browne C., Seago J., Wan K. C., Okada Y., Newbury S. F. (2008). The 5′-3′ exoribonuclease pacman is required for epithelial sheet sealing in Drosophila and genetically interacts with the phosphatase puckered. Biol. Cell 100, 687–701. 10.1042/BC20080049 [DOI] [PubMed] [Google Scholar]
- Hay B. A., Guo M. (2006). Caspase-dependent cell death in Drosophila. Annu. Rev. Cell Dev. Biol. 22, 623–650. 10.1146/annurev.cellbio.21.012804.093845 [DOI] [PubMed] [Google Scholar]
- Hernández G., Vázquez-Pianzola P., Sierra J. M., Rivera-Pomar R. (2004). Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA 10, 1783–1797. 10.1261/rna.7154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Bushati N., Cohen S. M. (2010). Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 8, e1000396 10.1371/journal.pbio.1000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou D., Ruiz M., Andrulis E. D. (2012). The ribonuclease Dis3 is an essential regulator of the developmental transcriptome. BMC Genomics 13, 359 10.1186/1471-2164-13-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh J. R., Guo M., Hay B. A. (2004). Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 14, 1262–1266. 10.1016/j.cub.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Jinek M., Coyle S. M., Doudna J. A. (2011). Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41, 600–608. 10.1016/j.molcel.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. I., Zabolotskaya M. V., Newbury S. F. (2012). The 5′ → 3′ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip. Rev. RNA 3, 455–468. 10.1002/wrna.1109 [DOI] [PubMed] [Google Scholar]
- Jones C. I., Grima D. P., Waldron J. A., Jones S., Parker H. N., Newbury S. F. (2013). The 5′-3′ exoribonuclease Pacman (Xrn1) regulates expression of the heat shock protein Hsp67Bc and the microRNA miR-277-3p in Drosophila wing imaginal discs. RNA Biol. 10, 1345–1355. 10.4161/rna.25354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Bashirullah A. (2014). A steroid-controlled global switch in sensitivity to apoptosis during Drosophila development. Dev. Biol. 386, 34–41. 10.1016/j.ydbio.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Senoo-Matsuda N., Hiromi Y., Miura M. (2006). DRONC coordinates cell death and compensatory proliferation. Mol. Cell. Biol. 26, 7258–7268. 10.1128/MCB.00183-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín F. A., Peréz-Garijo A., Morata G. (2009). Apoptosis in Drosophila: compensatory proliferation and undead cells. Int. J. Dev. Biol. 53, 1341–1347. 10.1387/ijdb.072447fm [DOI] [PubMed] [Google Scholar]
- Milán M., Campuzano S., García-Bellido A. (1997). Developmental parameters of cell death in the wing disc of Drosophila. Proc. Natl. Acad. Sci. USA 94, 5691–5696. 10.1073/pnas.94.11.5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S., Aslam A., Doidge R., Medica R., Winkler G. S. (2011). The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol. Biol. Cell 22, 748–758. 10.1091/mbc.E10-11-0898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan V. K., Jones C. I., Newbury S. F., Green P. J. (2013). XRN 5′→3′ exoribonucleases: structure, mechanisms and functions. Biochim. Biophys. Acta 1829, 590–603. 10.1016/j.bbagrm.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva R. M., Wells B. S., Johnston L. A. (2009). Mechanisms of growth and homeostasis in the Drosophila wing. Annu. Rev. Cell Dev. Biol. 25, 197–220. 10.1146/annurev.cellbio.24.110707.175242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara T., Zekri L., Braun J. E., Izaurralde E. (2013). miRISC recruits decapping factors to miRNA targets to enhance their degradation. Nucleic Acids Res. 41, 8692–8705. 10.1093/nar/gkt619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban T. I., Izaurralde E. (2005). Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11, 459–469. 10.1261/rna.7231505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. M., Davis K., Molineux C., Kolodner R. D., Johnson A. W. (1998). Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucleic Acids Res. 26, 3707–3716. 10.1093/nar/26.16.3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Garijo A., Martín F. A., Morata G. (2004). Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development 131, 5591–5598. 10.1242/dev.01432 [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., Lai E. C. (2007). Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17, 1850–1864. 10.1101/gr.6597907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., Drummond J., Webster J., Gubb D., Gunton N., Johnson G. et al. (2004). The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167, 797–813. 10.1534/genetics.104.026658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen G. S., Abrams J. M. (2004). Caspase activation – stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene 23, 2774–2784. 10.1038/sj.onc.1207522 [DOI] [PubMed] [Google Scholar]
- Sanduja S., Blanco F. F., Dixon D. A. (2011). The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 2, 42–57. 10.1002/wrna.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M., Palka J. (1987). Changing spatial patterns of DNA replication in the developing wing of Drosophila. Dev. Biol. 123, 145–153. 10.1016/0012-1606(87)90436-2 [DOI] [PubMed] [Google Scholar]
- Steller H. (2008). Regulation of apoptosis in Drosophila. Cell Death Differ. 15, 1132–1138. 10.1038/cdd.2008.50 [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner A., Hawley R. S. (2000). Drosophila Protocols New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Thermann R., Hentze M. W. (2007). Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 447, 875–878. 10.1038/nature05878 [DOI] [PubMed] [Google Scholar]
- Thomas M. P., Lieberman J. (2013). Live or let die: posttranscriptional gene regulation in cell stress and cell death. Immunol. Rev. 253, 237–252. 10.1111/imr.12052 [DOI] [PubMed] [Google Scholar]
- Till D. D., Linz B., Seago J. E., Elgar S. J., Marujo P. E., Elias M. L., Arraiano C. M., McClellan J. A., McCarthy J. E. G., Newbury S. F. (1998). Identification and developmental expression of a 5′-3′ exoribonuclease from Drosophila melanogaster. Mech. Dev. 79, 51–55. 10.1016/S0925-4773(98)00173-7 [DOI] [PubMed] [Google Scholar]
- Vazquez-Pianzola P., Hernández G., Suter B., Rivera-Pomar R. (2007). Different modes of translation for hid, grim and sickle mRNAs in Drosophila. Cell Death Differ. 14, 286–295. 10.1038/sj.cdd.4401990 [DOI] [PubMed] [Google Scholar]
- von Roretz C., Lian X. J., Macri A. M., Punjani N., Clair E., Drouin O., Dormoy-Raclet V., Ma J. F., Gallouzi I. E. (2013). Apoptotic-induced cleavage shifts HuR from being a promoter of survival to an activator of caspase-mediated apoptosis. Cell Death Differ. 20, 154–168. 10.1038/cdd.2012.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier R., Springhorn A., Patterson L., Koidl S., Hammerschmidt M., Affolter M., Pyrowolakis G. (2010). Control of Dpp morphogen signalling by a secreted feedback regulator. Nat. Cell Biol. 12, 611–617. 10.1038/ncb2064 [DOI] [PubMed] [Google Scholar]
- Wells B. S., Yoshida E., Johnston L. A. (2006). Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr. Biol. 16, 1606–1615. 10.1016/j.cub.2006.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K., Grether M. E., Abrams J. M., Young L., Farrell K., Steller H. (1994). Genetic control of programmed cell death in Drosophila. Science 264, 677–683. 10.1126/science.8171319 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Xu P., Guo M., Hay B. A. (2004). MicroRNAs and the regulation of cell death. Trends Genet. 20, 617–624. 10.1016/j.tig.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Xu D., Woodfield S. E., Lee T. V., Fan Y., Antonio C., Bergmann A. (2009). Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin) 3, 78–90. 10.4161/fly.3.1.7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabolotskaya M. V., Grima D. P., Lin M. D., Chou T. B., Newbury S. F. (2008). The 5′-3′ exoribonuclease Pacman is required for normal male fertility and is dynamically localized in cytoplasmic particles in Drosophila testis cells. Biochem. J. 416, 327–335. 10.1042/BJ20071720 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.