ABSTRACT
In preparation for embryo implantation and pregnancy, the uterine epithelium undergoes a genomic and biological transition that mediates adhesion and invasion of the blastocyst. These events resemble an inflammatory response, and the immune system likely takes an active role in the establishment and maintenance of pregnancy. Although glucocorticoids are primary mediators of the immune system, the functional role of glucocorticoid signaling in the uterine epithelium is not well defined. To investigate the dynamic relationship between glucocorticoids and reproductive hormones, we performed whole-genome microarray analysis in a human uterine endometrial cancer cell line (ECC1 cells) treated with the synthetic glucocorticoid dexamethasone (Dex) alone or in combination with estradiol (E2). Over 10 000 genes were significantly regulated in the presence of Dex and/or E2. Surprisingly, unique targets of Dex and E2 together represented the largest group of regulated genes. Ingenuity pathway analysis found both overlapping and independent regulated networks for each hormone. Several hundred genes were found to be coregulated by Dex and E2, including several that were antagonistically regulated. The effects of glucocorticoids and E2 are mediated primarily through the glucocorticoid receptor (NR3C1) and estrogen receptor (ESR1), respectively. In silico promoter analysis revealed that NR3C1 and ESR1 response elements are enriched in antagonistically regulated genes, and signaling through these receptors was required for antagonism. Glucocorticoid and E2 antagonism of target genes may represent a critical junction between the immune system and female reproductive system. Moreover, identification and ontology analysis of glucocorticoid-regulated genes in a uterine epithelial-like cell line suggests that glucocorticoid signaling regulates important biological functions, including immune cell trafficking and embryonic development.
Keywords: epithelial cells, ESR1, estrogen receptor, glucocorticoid receptor, microarray, NR3C1, uterus
INTRODUCTION
Despite advances in assisted reproductive technologies, current estimates indicate 72.4 million people worldwide are infertile, and the World Health Organization estimates female factor infertility accounts for 37% of infertility [1, 2]. Blastocyst adhesion and implantation into the uterus are decisive steps in mammalian reproduction and are dictated by an intricate series of signaling events and cellular interactions (reviewed in reference [3]). Uterine epithelial cells are the targets of many of these signaling events, coordinating the adhesion and invasion of the blastocyst during implantation. During this period, adhesion molecules, chemoattractant signals, and cells of the immune system dictate the relative success of this process [4–7].
The inflammatory response represents an important component of embryo implantation and mammalian reproduction. While the cells of the uterine epithelium represent the first line of defense against infection, the inflammatory response plays an important role in the development of immune tolerance to embryonic antigens and in subsequent embryonic development [8, 9]. Maternal immune tolerance is likely not a passive or suppressed response to pregnancy but rather a proactive participant in pregnancy progression [6, 10, 11]. Macrophages and dendritic cells recruited to the endometrium by cytokines and chemokines released from the luminal epithelium participate in tissue remodeling and vascular changes necessary to support the invading trophoblast [6, 12–14]. Differentiation and the activity of macrophages and dendritic cells are regulated by glucocorticoids, steroid hormones with well-characterized immunosuppressive and anti-inflammatory effects [15–18]. Such a link between the immune system and reproduction provides a potential mechanism for infertility in women with disorders of the adrenal gland, including Cushing syndrome and Addison disease [19, 20]. Further evidence for a link between glucocorticoids and hormones of the reproductive system is seen in rodent models of hormone exposure. Exogenous administration of the synthetic glucocorticoid dexamethasone (Dex) in rats and mice blocks estrogen-induced uterine growth and differentiation, leading to diminished rates of embryo implantation [21–23]. Similarly, the early inflammatory-like response to estradiol (E2), including edema of the stroma and myometrium, increased vascularization, infiltration of eosinophils, and potent antibacterial complement activity, is antagonized by the presence of Dex, suggesting glucocorticoids may act directly within the uterus to mediate the effects of E2 [23, 24].
In addition to their immune-modulatory properties, glucocorticoids play a pivotal role in many essential biological processes, including stress-induced activation of the hypothalamic-pituitary-adrenal axis [25–29]. The physiological actions of glucocorticoids are determined by binding and activating the glucocorticoid receptor (NR3C1), a member of the nuclear receptor superfamily of ligand-dependent transcription factors [30, 31]. Consistent with the broad effects of glucocorticoids, the glucocorticoid receptor is expressed in nearly all tissues and cell types and is necessary for life after birth [32]. Ligand-activated glucocorticoid receptor translocates to the nucleus and regulates the expression of target genes through multiple mechanisms: 1) binding directly to glucocorticoid response elements (GREs) in the promoter of target genes; (2) binding negative GREs to suppress gene expression; 3) in a composite manner where the glucocorticoid receptor directly binds a GRE and interacts with transcription factors bound to neighboring sites; or 4) tethering to other transcription factors apart from DNA binding [33–36]. Interestingly, in the human uterine ECC1 cell line, the glucocorticoid receptor and estrogen receptor-alpha were found to be recruited to the promoter of the immunomodulatory gene glucocorticoid-induced leucine zipper (GILZ) in response to Dex and E2 [37, 38]. Occupancy of the promoter by the glucocorticoid receptor at an upstream GRE and at the transcriptional start site mediated Dex-induced GILZ mRNA expression. Cotreatment with E2 antagonized Dex-induced expression and resulted in less glucocorticoid receptor and more estrogen receptor-alpha present at the GILZ promoter, suggesting glucocorticoids and E2 may directly regulate inflammatory response genes by receptor recruitment to promoter elements.
Human ECC1 cells, an endometrial cancer cell line, exhibit a luminal epithelial phenotype and maintain functional steroid receptors, making these cells an important model for the study of endocrine regulation of uterine epithelium in vitro [39]. To identify targets of endocrine regulation by glucocorticoids and E2, ECC1 cells were treated with Dex or E2 or both, and whole-genome microarray analysis was performed to evaluate the global transcriptional response and to identify targets of both of the hormones. Gene clustering and pathway analysis identified several genes antagonistically regulated in the presence of Dex and E2, including members of the inflammatory response pathway. Many of those genes that were identified contain GREs or estrogen response elements (EREs) within their promoters and may also represent additional targets of direct transcriptional regulation by the glucocorticoid and estrogen receptor-alpha. Interestingly, microarray analysis also identified a large group of genes that were regulated only in the presence of both Dex and E2. These genes represent many unique pathways and functions and could provide insight into the mechanism of glucocorticoid action in uterine epithelial cells. Collectively, these studies demonstrate that glucocorticoids regulate thousands of genes in a human uterine epithelial-like cell line, interacting with many targets of E2 to antagonize the expression of genes involved in significant biological processes, including embryonic development.
MATERIALS AND METHODS
Reagents
RPMI 1640 medium was purchased from Invitrogen (Gibco). Heat-inactivated fetal bovine serum (FBS) was purchased from Atlanta Biologicals. Charcoal stripped heat-inactivated FBS was purchased from Hyclone (Logan, UT). Dex (1,4-pregnadien-9α-fluoro-16α-methyl-11β,17,21-triol-3,20-dione), E2 (1,3,5[10] estratrien-3,17β-diol), and mifepristone (RU486; 4,9-estradien-17α-propynyl, 11β-[4-dimethynylamino] phenyl-17β-OL-3-One) were purchased from Steraloids, Inc.. Fulvestrant (ICI 182,780) was purchased from Sigma-Aldrich. TaqMan reverse transcription-PCR (RT-PCR) primer probes were purchased from Applied Biosystems. On-Target Plus Control Pool (nontargeting pool) and Smart Pool (human glucocorticoid receptor and estrogen receptor-alpha) were purchased from Thermo Scientific Dharmacon.
Culture of Human ECC1 Cells
An immortalized human uterine endometrial cancer cell line (ECC1) was obtained from American Type Culture Collection and grown in a standard tissue culture incubator at 37°C, with 95% humidity and 5% carbon dioxide atmosphere. ECC1 cells were maintained in RPMI 1640 medium supplemented with 5% FBS. Twenty-four hours prior to cell treatment, medium was changed to phenol red-free RPMI 1640 medium containing 5% charcoal dextran-treated (stripped) FBS. Cells were treated with 100 nM Dex prepared in phosphate-buffered saline (PBS) or 10 nM E2 prepared in EtOH for 6 h. For the glucocorticoid receptor or estrogen receptor-alpha antagonism experiments, 1 μM RU486 or 1 μM fulvestrant prepared in EtOH was added 1 h prior to hormone treatment.
RNA Isolation
Total RNA was extracted from ECC1 cells using the QIAshredder and RNeasy mini-kit (Qiagen) according to the manufacturer's protocol. DNase treatment was performed on-column using a ribonuclease-free DNase kit (Qiagen) according to the manufacturer's instruction. Samples were quantified with a spectrophotometer (Nanodrop ND-1000; Thermo Scientific), and purity was analyzed by the 260:280 nm absorbance ratio.
Quantitative RT-PCR
Quantitative real-time RT-PCR (QPCR) was performed with the 7900HT sequence detection system and predesigned primer-probe sets from Applied Biosystems according to the manufacturer's instructions. Real-time RT-PCR assays were performed with 100 ng of total RNA, using One-Step RT-PCR Universal Master Mix reagent. Standard curves were generated by serial dilution of a preparation of total RNA isolated from ECC1 cells. The signal obtained from each gene primer-probe set was normalized to that of the unregulated housekeeping gene peptidylprolyl isomerase B (PPIB) primer-probe set. Each primer-probe set was analyzed with at least three different sets of RNA.
Microarray Study
Gene expression analysis was conducted using whole Human Genome 4x44 multiplex format oligonucleotide arrays (product no. 014850; Agilent Technologies) following the Agilent one-color microarray-based gene expression analysis protocol. Starting with 500 ng of total RNA, we then produced Cy3-labeled cRNA according to the manufacturer's protocol. For each sample, 1.65 μg of Cy3-labeled cRNA was fragmented and hybridized for 17 h in a rotating hybridization oven. Slides were washed and scanned (Agilent). Data were obtained with Feature Extraction version 9.5 software (Agilent), using one-color defaults for all parameters. Feature Extraction software performed error modeling, adjusting for additive and multiplicative noise. Resulting data were processed using the Resolver version 7.2 software system (Rosetta Biosoftware). Principal component analysis (PCA) was performed with all samples and all probes to reduce dimensionality of data and to preserve variations in the data set. This allowed us to assess the similarities and differences of samples within a treatment group and between treatment groups. Error-weighted ratios that Resolver generated and associated P values were used to identify differentially expressed probes. The ratios used to determine these lists were E2-treated cells:vehicle-treated cells, Dex-treated cells:vehicle treated cells, Dex + E2-treated cells:vehicle-treated cells, Dex-treated cells:E2-treated cells, Dex + E2-treated cells:E2-treated cells, and Dex + E2-treated cells:Dex-treated cells. A heat map was generated using HeatPlus software (BioConductor). Normalized and sample replicate-averaged data for all significant probes were used for calculation of pairwise correlation. Dendrograms of samples (columns) and genes (rows) were generated by hierarchical clustering. The color scale is from 3-fold lower (log2-fold = −1.58) than mean (green) to 3-fold higher (log2-fold = 1.58) than the mean (red).
The lists of probe sets generated using Resolver that were responsive to Dex, E2, or Dex + E2 were visually sorted by using a Venn diagram generator (http://www.pangloss.com/seidel/Protocols/venn.cgi) and further analyzed with the Pathway Analysis version 6.5 tool (Ingenuity Systems). The average expression value of duplicate identifiers for the same molecule was used in the analyses to eliminate redundancy. Functional pathway analysis identified pathways from the Ingenuity Pathway Analysis (IPA) library of functional pathways and ranked them by ratio (number of genes from the data set that mapped to the pathway divided by the total number of genes that mapped to the functional pathway). The highest ranked pathway that met a P value of <0.05 (Fischer exact test) is shown.
Promoter Analysis
The JASPAR CORE database, a curated, nonredundant set of profiles derived from published collections of experimentally defined transcription factor binding sites for eukaryotes, was used to identify predicted glucocorticoid receptor and estrogen receptor-alpha binding sites within 3500 bp upstream of the transcriptional start site and 500 bp downstream of the transcriptional start site [40]. Sequences were scanned at a profile score threshold of 80% and 85%.
Western Blotting Analysis
ECC1 cells grown in vitro were washed twice with ice-cold PBS, treated with 350 μl of radioimmunoprecipitation buffer with the addition of protease inhibitor cocktail tablets (cOmplete Mini, EDTA-free; Roche), scraped off the plate, snap frozen at −80°C, thawed, and rotated at 4°C for 30 min. Cellular debris was removed by centrifugation at 13 200 rpm (16 100 relative centrifugal force) for 20 min in a tabletop centrifuge (model 5415R; Eppendorf International) at 4°C, and supernatant was collected. Protein concentration was determined using a BCA protein quantitation kit (Pierce). Sample buffer (5×) was added to samples containing 40 μg of protein, heated to 95°C for 5 min, and separated on a 10% ReadyGel Tris-Gly gels (Bio-Rad). Proteins were then transferred onto a nitrocellulose membrane. Membranes were blocked for 1 h with 10% skim milk in Tris-buffered saline with 0.1% Tween 20 and subsequently probed with polyclonal antiglucocorticoid receptor antibodies (1:1000 dilution) [40]), monoclonal antiestrogen receptor-alpha antibodies (1:750 dilution; Immunotech), or monoclonal anti-ACTB antibodies (1:10 000 dilution; Millipore). Immunoreactivity was visualized by incubation with a mixture of goat anti-rabbit Alexa fluorophore 680-conjugated (Molecular Probes) and goat anti-mouse IRDye 800-conjugated (Rockland Immunochemicals) secondary antibodies for 1 h at room temperature and scanned (Odyssey imaging system).
RNA Interference
Human estrogen receptor-alpha, glucocorticoid receptor, and nontarget control (NTC) small interfering RNA (siRNA) SMARTpools were purchased from Thermo Scientific Dharmacon. For each biological replicate, cells were plated in 6-well plates at approximately 70% confluency 1 day prior to transfection. A 50 nM aliquot of each siRNA was transfected into cells with DharmaFECT 1 transfection reagent (Thermo Scientific Dharmacon) following the manufacturer's instructions. The next day, each transfected well was split into four wells of a six-well plate for RNA isolation following hormone treatment and one 10-cm dish for protein isolation. Forty-eight hours later, medium was changed to phenol-red free RPMI 1640 medium. After 72 h of siRNA treatment, cells were induced with 100 nM Dex, 10 nM E2, or Dex + E2, and RNA was harvested 6 h after the addition of hormone.
Statistical Analysis
Data are means ± SEM. Statistical significance was determined by ANOVA with Tukey post hoc analysis.
RESULTS
Microarray Analysis Identifies Dex- and Estradiol-Induced Changes to Global Gene Expression in Human ECC1 Cells
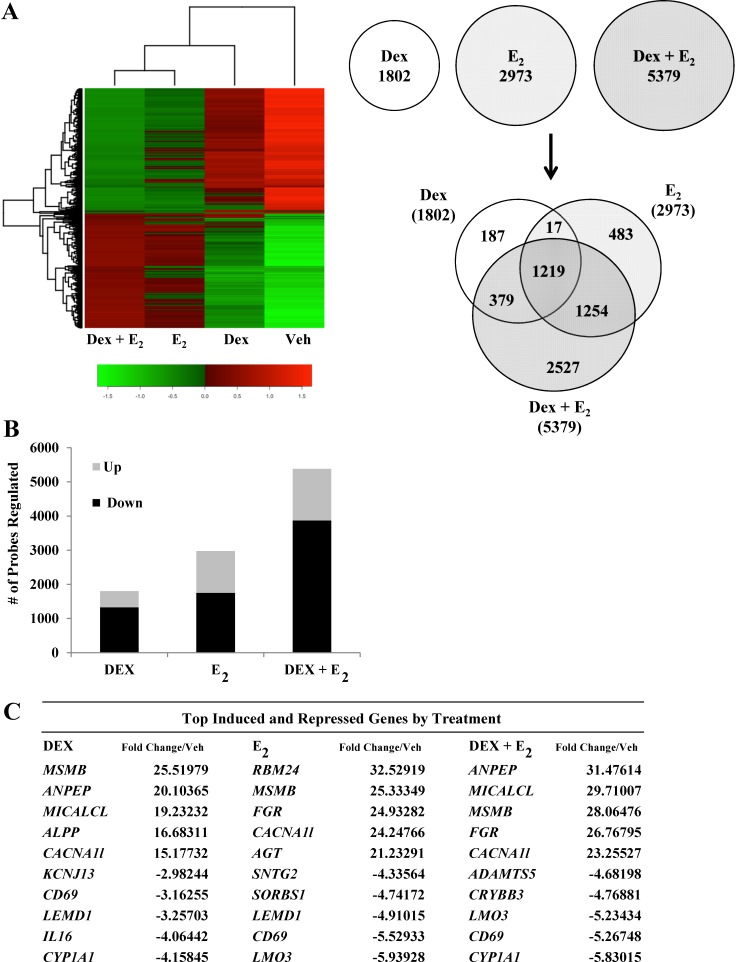
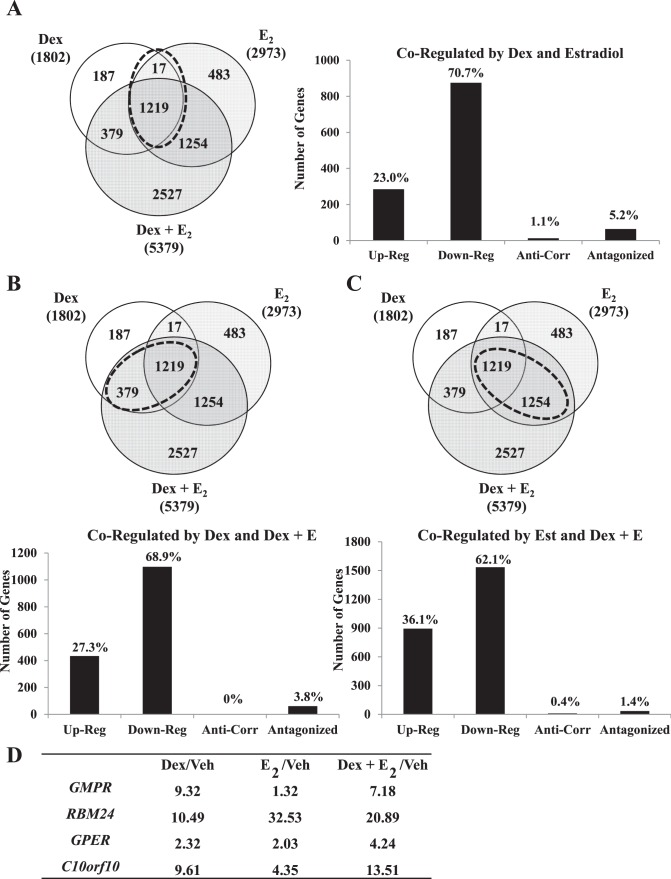
The human uterine epithelial-like cell line ECC1 represents the cell-type first contacted by the embryo during implantation and is a site for integrated transcriptional regulation by Dex and E2 [37]. To determine the global transcriptional response to Dex and E2 in human uterine epithelial cells, as well as to identify transcripts regulated by both hormones, we performed microarray analysis (whole Human Genome; Agilent) of ECC1 cells treated for 6 h to evaluate early response of genes to vehicle, 100 nM Dex, 10 nM E2, or Dex + E2. Gene lists were generated from the average of three biological replicates from each hormone treatment by selecting probes that were significant at a P value of <0.01 as determined by ANOVA. A heat map of averaged sample replicates within treatment groups and hierarchical clustering is shown to visualize global gene change (Fig. 1A). Comparison of significantly regulated hybridized probes identified 10 154 targets regulated by Dex, E2, or Dex + E2. Venn diagram analysis compared the gene lists to identify those genes that are unique and common among the three hormone paradigms (Fig. 1A). In ECC1 cells, 1802 gene probes were regulated after 6 h of treatment with Dex, 2973 gene probes were regulated in response to E2, and 5379 gene probes were regulated by Dex + E2. Interestingly, of the 5379 Dex + E2 probes, 2527 probes were uniquely regulated only by the combination of Dex and E2. Several genes were found to be coregulated by Dex and E2. A total of 1236 gene probes were commonly regulated by Dex and E2, 1598 were common to Dex and Dex + E2, and 2473 were common to E2 and Dex + E2. From the Dex and Dex + E2 treatment groups, most of the genes identified as regulated are repressed (Fig. 1B). Approximately three fourths of the genes regulated by Dex and Dex + E2 were down-regulated in ECC1 cells (74% and 72%, respectively). In E2-treated cells, gene expression was more evenly balanced between up- and down-regulation, with approximately 41% of genes induced and 59% repressed. The top five induced and repressed genes are listed (Fig. 1C). Several genes were common to all three treatments, although most differed in relative fold change (i.e., calcium channel, voltage dependent, T type, alpha 1l subunit [CACNA1l] and cluster of differentiation 69 [CD69]). IPA was performed to identify the networks significantly regulated in human uterine epithelial cells in response to Dex, E2, or a combination of Dex + E2 (Fig. 1D). Some of the top five networks were common among treatment groups (e.g., embryonic development, organ development, small molecule biochemistry, cellular movement, and connective tissue development and function). Embryonic and organ development is listed as the top network for Dex-regulated genes in ECC1 cells and represents the only network shared among all treatments. This network, given a colorimetric identity based on expression values from each treatment, illustrates how the genes of a common network are regulated in diverse manners by Dex, E2, or Dex + E2 (Fig. 1E). Antagonistic regulation of left-right determination factor 1 (LEFTY1) by Dex and E2 was validated on mRNA samples isolated independently of samples evaluated by microarray (Fig. 1F). Distinct functions were found for the significantly associated genes for all treatment groups. These data suggest that the genes common and uniquely regulated by Dex and E2 contribute to both shared and distinct functions in human uterine epithelial cells.
FIG. 1.
Dex and E2 regulate global gene expression in human uterine epithelial cells. A) mRNA isolated from Dex-, E2-, and Dex + E2-treated ECC1 cells was analyzed using Whole Human Genome 4x44 multiplex format oligo array (Agilent) for gene expression. A heat map was generated with Heatplus software (BioConductor), using the averaged, normalized sample replicates of all significant probes. The number of probes that were statistically different (P < 0.01) between treatment groups were sorted by Venn diagram. B) The number of probes that were regulated are organized as either induced or repressed according to treatment group in Dex, E2, and Dex + E2 samples. C) The top five induced and repressed genes for each treatment group are listed. D) IPA of the top five regulated networks using the gene lists generated from Dex-, E2-, and Dex + E2-treated ECC1 cells. E) Embryonic development and organ development network is presented, with expression values of the Dex, E2, and Dex + E2 treatment groups overlaid, as a colorimetric indicator of up- or down-regulation. F) Independent validation of Left-Right Determination Factor 1 (LEFTY1), one of the antagonized genes identified in the embryonic development and organ development network. Values were normalized to the housekeeping gene Cyclophilin B. Bar graphs show means ± SEM. **Groups with statistically different means at P < 0.01, as determined by ANOVA.
FIG. 1.
Continued.
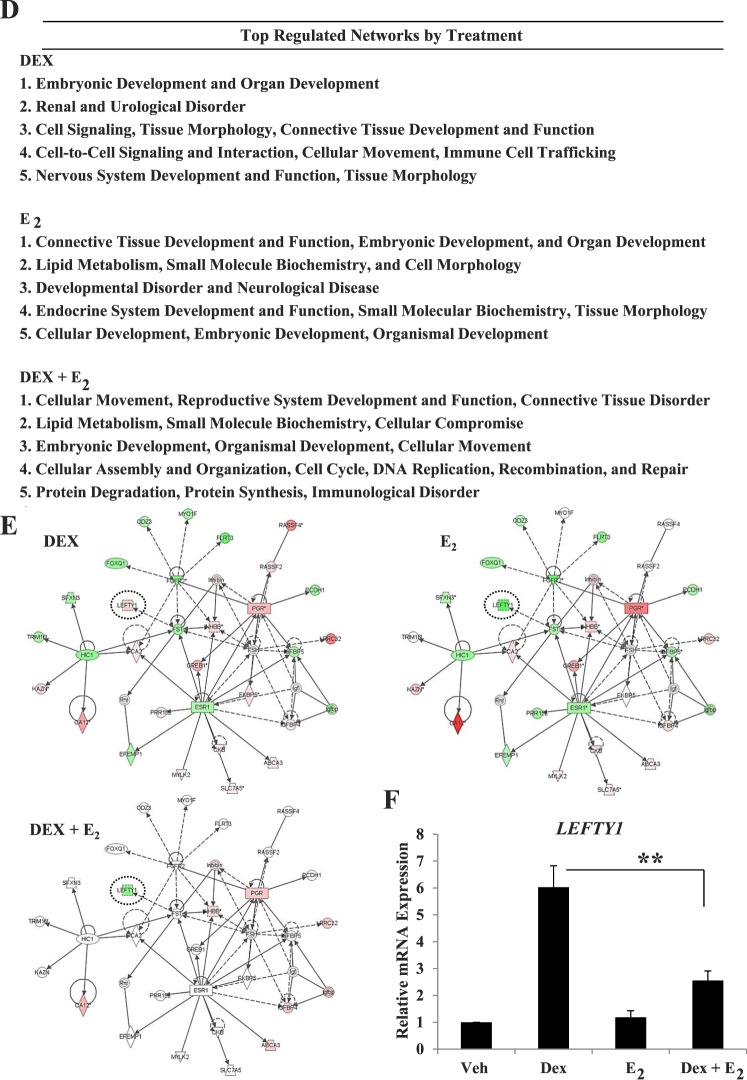
Immune cell trafficking and immunological disorder were highlighted by analysis of all Dex-regulated genes, and several inflammatory response genes were identified in the top five up- and down-regulated genes, which is consistent with actions of glucocorticoids in both innate and adaptive immune responses [18]. IPA software was used to create a network analysis of the inflammatory response pathway to visually represent changes in the expression levels of inflammatory response genes upon treatment with Dex, E2, or Dex + E2 (Fig. 2A). Several genes were identified as differentially regulated by Dex and E2. Tumor necrosis factor superfamily member 10 (TNFSF10), thrombopoietin (THPO), interleukin 23 alpha subunit (IL-23A), and IL-5, IL-6, and IL-16 were all down-regulated only in the presence of Dex. Interleukin 12 receptor beta 1 (IL-12RB1) was down-regulated by Dex, but Dex-mediated repression was antagonized by E2 in the Dex + E2-treatment group. Nuclear factor of kappa light chain inhibitor alpha (NFκB1A) was up-regulated by Dex in both the Dex and Dex + E2 treatment groups but down-regulated by E2 alone. Independent QPCR analysis of different samples validated the expression profiles of three of the inflammatory response genes determined to be differentially regulated by Dex and E2 (Fig. 2B). Expression of IL-6 mRNA duplicated the pattern of expression determined by microarray analysis. Dex and Dex + E2 significantly down-regulated IL-6 expression (P < 0.05). Microarray analysis suggested IL-6 was repressed by Dex and not regulated in the presence of E2. In independent QPCR validation, we discovered that E2 induced the expression of IL-6, and Dex-mediated repression was antagonized by E2 (P < 0.01). Independent validation of NFκBIA confirmed the anticorrelated regulation by Dex and E2 (P < 0.01).
FIG. 2.
Inflammatory response genes demonstrate differential response to Dex and E2. A) IPA identified regulated genes in the inflammatory response pathway. The expression values for each treatment are overlaid on the gene ID. Colors represent direction and degrees of regulation, red indicating up-regulation and green indicating down-regulation. B) mRNA expression was determined for Interleukin 6 (IL6), Interleukin 16 (IL16) and Nuclear Factor of Kappa Light Polypeptide Gene Enhancer in B-cells Inhibitor Alpha (NFκBIA) in four independent biological replicates. Values were normalized to those of the housekeeping gene Cyclophilin B. Bar graphs show means ± SEM, and the fold change value determined by microarray is listed below each treatment (N/A = no change compared to vehicle). Statistically significant at *P < 0.05 and ** P < 0.01, as determined by ANOVA.
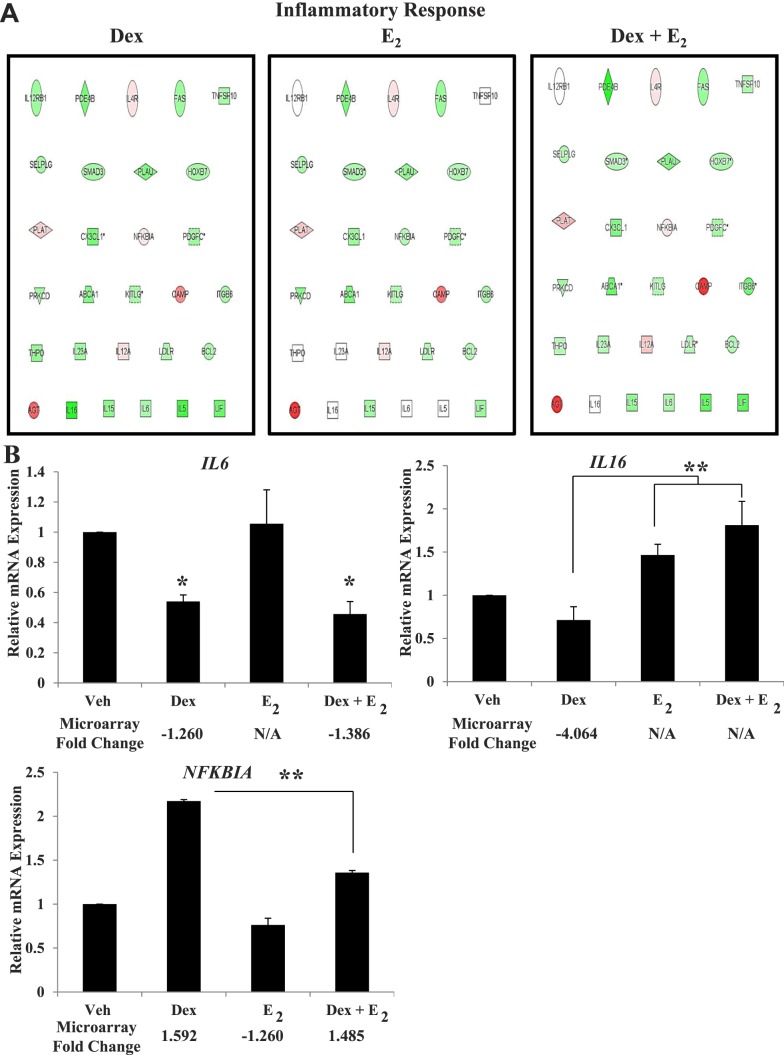
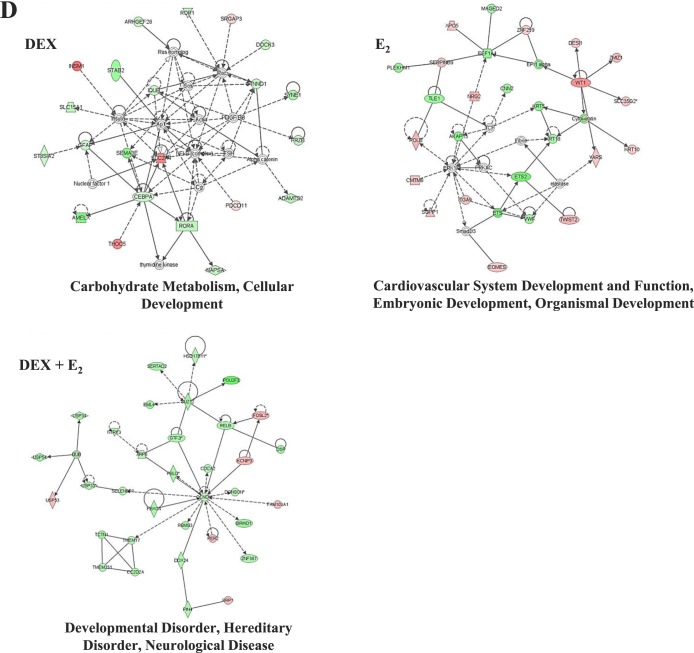
Venn diagram analysis also identified genes uniquely regulated in the presence of Dex, E2, or Dex + E2. Comparison of those gene lists showed 187 gene probes were unique to Dex treatment, 483 were E2 unique gene probes, and 2527 gene probes were regulated only in the presence of both hormones (Fig. 3A). The directional pattern of regulation for the uniquely regulated genes closely mirrors that of all regulated genes for each treatment (82.3% down-regulated for Dex, 39.7% down-regulated for E2, and 80.8% down-regulated for Dex + E2). The top five induced and repressed genes by treatment are listed (Fig. 3B). To determine if the uniquely regulated genes represent unique molecular functions, IPA analysis was performed with the unique gene lists, and the top five regulated networks are reported (Fig. 3C). Compared to those networks identified for all Dex-regulated genes, the genes unique regulated by Dex were from almost entirely different networks. Genes regulated only in the presence of E2 were also responsible for the regulation of several unique networks. The 2527 gene probes uniquely regulated only in the presence of Dex + E2 represent some distinct molecular and cellular functions (developmental disorder, hereditary disorder, neurological disease, gene expression, molecular transport, developmental disorder, and organismal injury and abnormalities), but there was also some overlap with networks from all Dex + E2-regulated genes. The top network with expression values overlaid from the Dex, E2, and Dex + E2 gene lists are presented to provide a reference for the unique molecular and cellular functions for this treatment group (Fig. 3D).
FIG. 3.
Unique genes represent distinct functions. A) Separation by Venn diagram identified uniquely regulated genes, which are listed by treatment type. The number of unique probes regulated is organized as either induced or repressed by treatment group in Dex, E2, and Dex + E2 samples. B) The top five induced and repressed genes for each treatment group are listed. C) IPA of the top regulated networks, using the gene lists generated from Dex-, E2-, and Dex + E2-treated ECC1 cells and isolating the uniquely regulated genes. D) The top network for each treatment is shown with expression values overlaid as a colorimetric indicator of up- or down-regulation.
FIG. 3.
Continued.
Analysis of Coregulated Genes Identifies Gene Targets of both Glucocorticoids and Estradiol
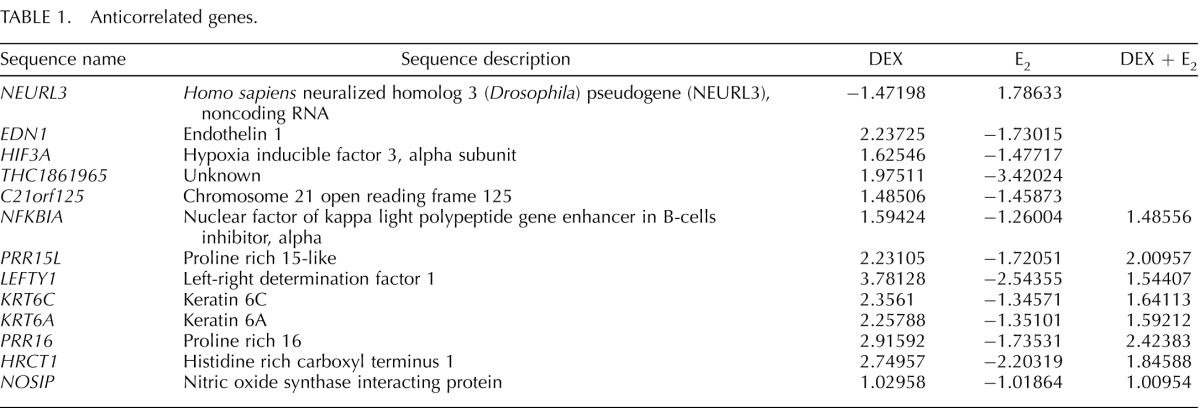
Venn diagram and network analyses determined several genes and pathways were targets of both glucocorticoids and E2. To further explore the extent of transcriptional cooperation and antagonism induced by glucocorticoid and E2 treatment, coregulated genes that displayed antagonistic patterns of expression were identified from the overlapping gene lists of Dex, E2, and D + E2 treatment groups (Fig. 4A). Interestingly, most of the coregulated genes were regulated by Dex and E2 in the same direction and repressed or induced by either treatment. Nonetheless, 13 genes were found to have anticorrelated regulation (opposing the direction of regulation, + vs −; 1.05% of all genes regulated) (Table 1), and 64 genes were found to be antagonistically regulated (greater than 2-fold difference between treatment types compared to Dex + E2 treatment group; 5.17% of all genes regulated). Analysis was expanded to include genes which might or might not have been regulated by E2 but in which the presence of E2 antagonized regulation by Dex (Fig. 4B). No anticorrelated genes were found, but 61 genes with antagonistic regulation were identified. Relative fold changes for guanosine monosphosphate reductase (GMPR) are shown as a representative of one of the antagonistically regulated genes identified (Fig. 4D). The reciprocal analysis was performed with the gene list that included targets regulated by E2 that might or might not have been regulated by Dex alone but were in the presence of both hormones (Fig. 4C). This analysis identified 10 anticorrelated genes and 34 antagonized genes (representing 1.37% of all genes regulated). RNA binding motif protein 24 (RBM24) is an example of one of the genes identified as antagonistically regulated by Dex and E2 (Fig. 4D). G protein-coupled estrogen receptor 1 (GPER) and chromosome 10 open reading frame 10 (C10orf10) illustrate the differentially regulated genes with additive expression values (the sum of D + E2 treatment is equal to the expression of Dex plus the expression of E2) (Fig. 4D).
FIG. 4.
Comparison of coregulated genes reveals several targets of Dex and E2 antagonism. A) Dex-, E2-, and Dex + E2-coregulated genes were separated by direction of regulation. Anticorrelated genes were classified as opposing directions of regulation (positive vs negative). Antagonized genes were classified as genes with greater than two-fold difference between treatments. B) Genes classified as regulated by Dex and Dex + E2 were separated by direction of regulation. Anticorrelated genes were classified as opposing directions of regulation (positive vs. negative). Antagonized genes were classified as genes with greater than 2-fold difference between treatment type compared to Dex + E2. C) Genes classified as regulated by E2 and Dex + E2 were separated by direction of regulation. Anticorrelated genes were classified as opposing directions of regulation (positive vs. negative). Antagonized genes were classified as genes with greater than two-fold difference between treatment type compared to Dex + E2. D) Guanosine monophosphate reductase (GMPR) represents the pattern of expression found for Dex and Dex + E2 antagonistically regulated genes. RNA Binding Motif Protein 24 (RBM24) represents one gene identified as antagonistically regulated by E2 and Dex + E2. G Protein-Coupled Estrogen Receptor 1 (GPER) and Chromosome 10 Open Reading Frame 10 (C10orf10) illustrate differentially regulated genes with additive expression values.
TABLE 1.
Anticorrelated genes.
Validation of Identified Target Genes
Analysis of coregulated genes generated several targets for glucocorticoid and E2 antagonism. Therefore, microarray results were confirmed by independent analysis of selected genes by QPCR in different samples (Supplemental Fig. S1, available online at www.biolreprod.org). Genes were selected as representatives of different patterns of expression. Relative mRNA expression is graphed and microarray fold change values are listed below for each gene validated. Expression levels of RBM24, alpha-2c-adrenergic receptor (ADRA2C), mucin 5B (MUC5B), natriuretic peptide type C (NPPC) were up-regulated by treatment with E2 but antagonized by Dex (P < 0.01). Period homolog 1 (PER1) and LEFTY1 were up-regulated in response to Dex, and E2 significantly antagonized this induction (P < 0.01). Validation of C10orf10 confirmed that for some genes, Dex and E2 act in an additive manner to regulate gene expression.
Predicted Glucocorticoid Receptor and Estrogen Receptor-Alpha Binding Sites Are Enriched in the Promoters of Antagonistically Regulated Genes
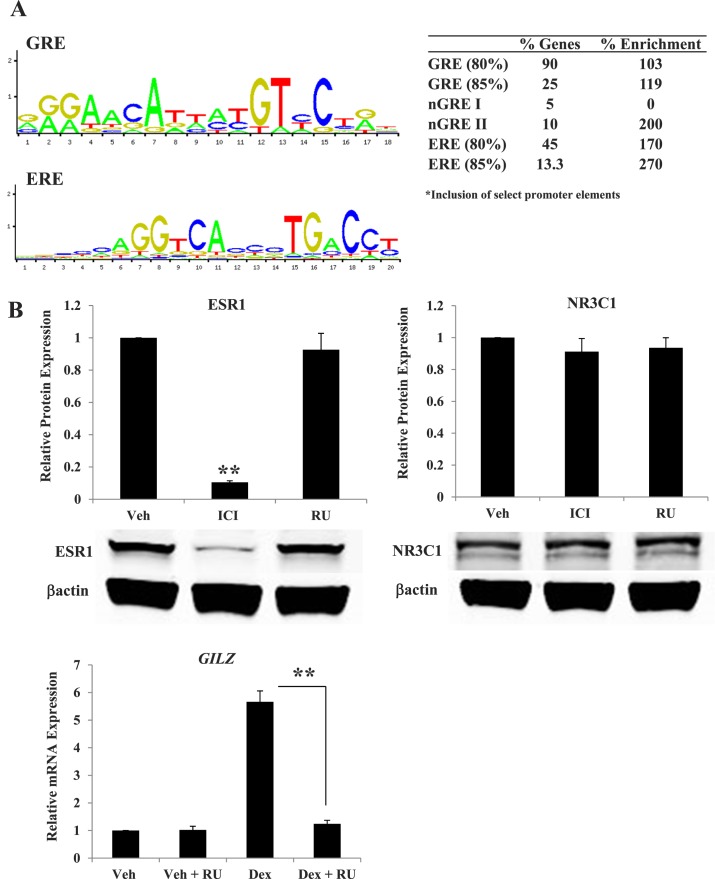
Direct DNA binding of steroid hormone receptors as transcription factors is one mechanism by which gene expression is regulated. Glucocorticoid receptor and estrogen receptor-alpha bind specific DNA sequences (GREs and EREs), respectively. Furthermore, the glucocorticoid receptor is able to mediate direct transcriptional repression through binding negative GRE sequences (nGREs), of which three nGRE sequences have been identified [41]. Although direct sequence binding represents only one mechanism by which steroid hormone receptors regulate transcription, glucocorticoid and E2 antagonism of the GILZ gene was found to involve glucocorticoid receptor and estrogen receptor-alpha binding at sites within the promoter containing their response elements [37]. A limited in silico promoter analysis was performed with genes identified as antagonistically regulated by Dex and E2 in the microarray analysis to determine whether the promoters of these genes contain GREs or EREs and could be direct targets of the glucocorticoid receptor or estrogen receptor-alpha binding. Promoter analysis was performed using the open-access database JASPAR to search for the inclusion of GREs and EREs 3500 bp upstream of the transcriptional start site and 500 bp downstream of the transcriptional start site [42]. Seventy-five genes were identified as antagonistically regulated and 60 of these possess an annotated promoter. Those genes possessing an annotated promoter were analyzed for the inclusion of a GRE, nGRE, or ERE sequence and compared to 100 genes not significantly regulated by Dex or E2. GREs were found in 90% of the promoters analyzed using an 80% threshold score (Fig. 5A). Although the presence of the GRE consensus sequence appears to be common among the genes analyzed, the inclusion of this sequence is enriched in the antagonized gene set compared to those genes included on the microarray gene platform not significantly regulated by Dex or E2. The presence of nGRE II (CTCCnnGGAGA; n indicates the inclusion of a nonspecified base pair) is enriched 200% in antagonistically regulated genes compared to those not regulated. Although not enriched, nGRE I (CTCCnGGAGE) is also present in 5% of regulated genes. The ERE sequence is present in 45% (80% threshold score) of Dex and E2 antagonistically regulated genes, which represents a 170% enrichment over genes not statistically regulated.
FIG. 5.
Promoters of antagonistically regulated genes contain glucocorticoid receptor and estrogen receptor-alpha response elements and require receptor for coregulation. A) JASPAR CORE Vertebrata server was used to search 3500 bp up-stream and 500 bp down-stream of the transcriptional start site of antagonistically regulated genes with annotated promoters. The search sequence for GRE or ERE is listed as provided by JASPAR. The percentage of genes that contain a GRE or ERE (at 80% and 85% profile score threshold) and nGRE I or II is listed. Antagonistically regulated genes were compared to 100 genes from the microarray platform that were not significantly regulated by Dex or E2. Enrichment was determined as the percentage of genes in the regulated list containing promoter element/percentage of genes in unregulated gene list. B) ECC1 cells treated for 7 h with the estrogen receptor antagonist
Glucocorticoid Receptor and Estrogen Receptor-Alpha Are Required for Transcriptional Antagonism Regulated by Dex and E2
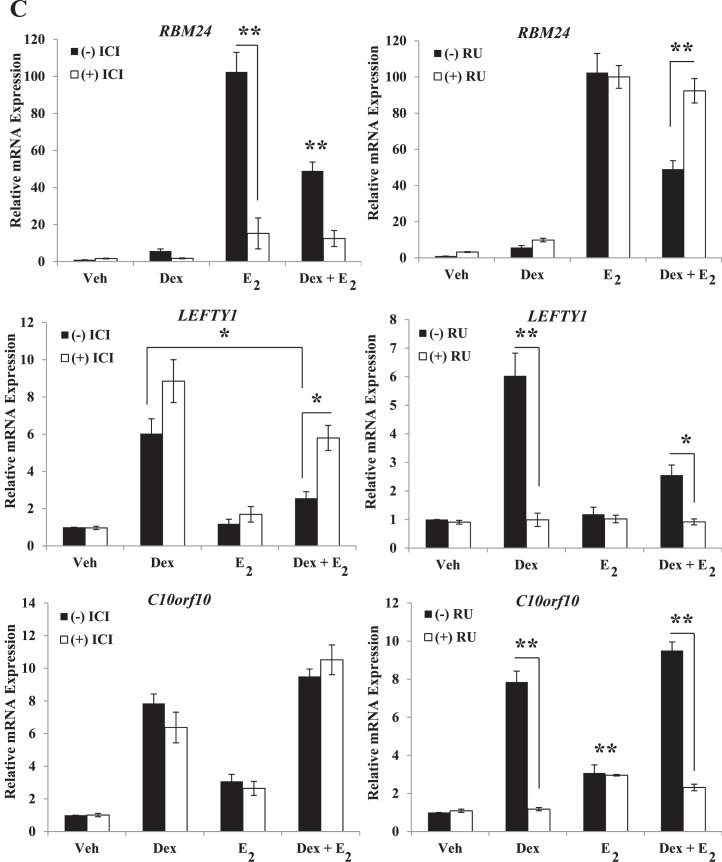
To understand whether each receptor is required for antagonistic gene regulation by Dex and E2 in genes containing glucocorticoid receptor or estrogen receptor-alpha response elements or both, ECC1 cells were pretreated with the glucocorticoid receptor antagonist RU486 or the estrogen receptor antagonist ICI 182,780, and gene expression levels of antagonistically regulated genes were analyzed 6 h after treatment with 100 nM Dex, 10 nM E2, or Dex + E2. Western blot analysis of total estrogen receptor-alpha protein levels demonstrated that 7 h of exposure to ICI 182,780 resulted in a significant decrease in estrogen receptor-alpha (∼90%) but not the glucocorticoid receptor, corresponding to its mechanism of inhibition (Fig. 5B) [43]. The glucocorticoid receptor antagonist RU486 did not alter estrogen receptor-alpha or glucocorticoid receptor protein levels but did significantly block Dex-induced expression of the GILZ gene, indicating the glucocorticoid receptor-mediated transcriptional regulation is blocked by RU486. The requirement of the glucocorticoid receptor and estrogen receptor for regulation of RBM24, LEFTY1, and C10orf10 expression was evaluated because of the significant coregulation of these genes by Dex and E2, as well as the inclusion of receptor response elements within the promoter region analyzed. Expression of RBM24 is induced by E2, and this induction is antagonized by the coadministration of Dex (Fig. 5C). Pretreatment with ICI 182,780 blocks E2-mediated induction, and the glucocorticoid antagonist RU486 blocks the antagonism by Dex, suggesting both of the receptors are required for coregulation of RBM24. Expression of LEFTY1 is up-regulated in response to Dex treatment, which is antagonized by coadministration of E2 with Dex. Induction of LEFTY1 expression by Dex is blocked by RU486, and antagonism of LEFTY1 expression by E2 is inhibited by pretreatment with ICI 182,780. Dex- and E2-mediated antagonism of LEFTY1 expression appears to require the activity of both the glucocorticoid receptor and estrogen receptor. Expression of C10orf10 is induced by both Dex and E2, and up-regulation is additive in the presence of both. Interestingly, estrogen receptor antagonism by ICI 182,780 did not block E2-mediated induction of C10orf10. Conversely, RU486 prevented Dex from up-regulating C10orf10 expression, suggesting the glucocorticoid receptor but not the estrogen receptor is required for Dex and E2 coregulation.
FIG. 5 cont.
ICI 182,780 or the glucocorticoid receptor antagonist RU486 were assayed for Estrogen Receptor α (ESR1) and Glucocorticoid Receptor (NR3C1) protein levels by Western blotting. Values were normalized to those of the housekeeping gene ACTB and quantified for four biological replicates. Bar graphs show means ± SEM. Statistically different means at **P < 0.01, as determined by ANOVA. mRNA expression of the Glucocorticoid-induced Leucine Zipper (GILZ) gene was determined following 1 h of pretreatment with ICI 182,780 or RU486 and 6 h of exposure to Dex, E2, or Dex and E2 in four biological replicates. Values were normalized to the housekeeping gene Cyclophilin B (PPIB) and graphed as means ± SEM. C) ECC1 cells were pretreated for 1 h with the estrogen receptor antagonist ICI 182,780 or the glucocorticoid receptor antagonist RU486. Dex, E2, or Dex + E2 was then added for 6 h, and mRNA was evaluated by QPCR. Gene expression was determined for RNA Binding Motif Protein 24 (RBM24), Left-Right Determination
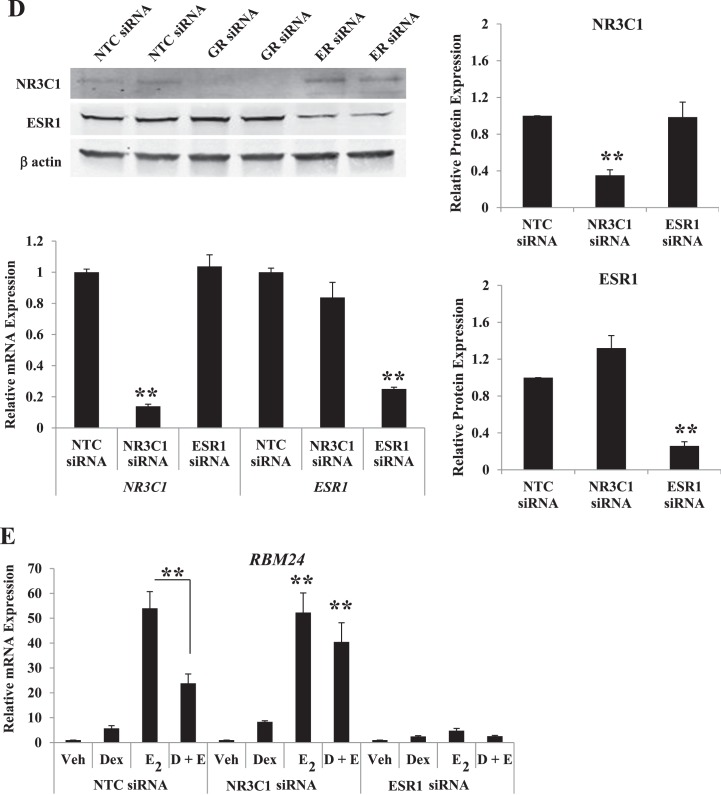
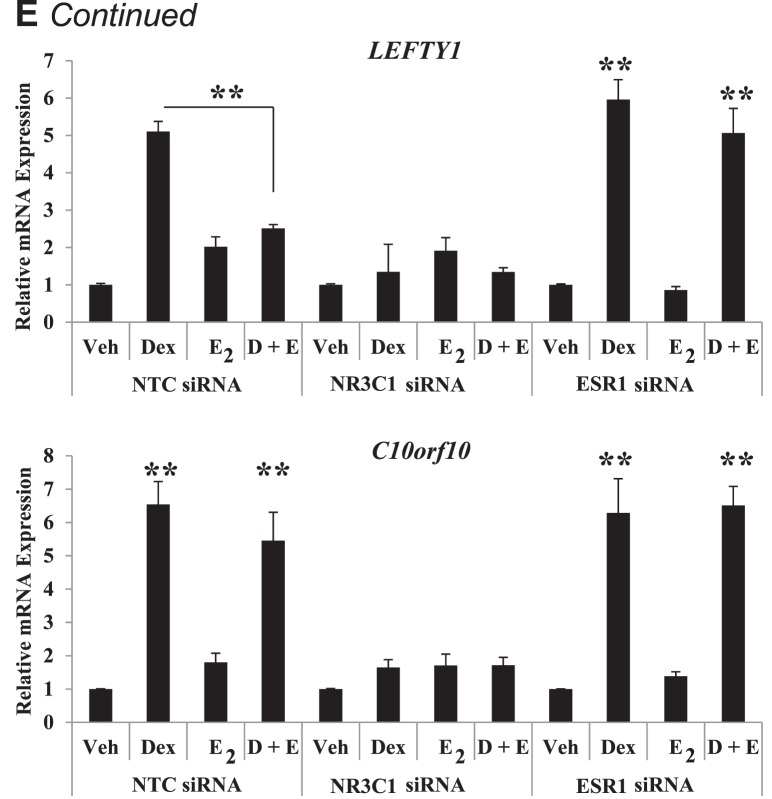
RU486 displays cross-reactivity to the progesterone receptor. To further isolate the role of the glucocorticoid receptor, specifically, and estrogen receptor-alpha in mediating glucocorticoid and E2 coregulation of gene expression, cells were transfected with siRNA against the glucocorticoid receptor, estrogen receptor-alpha, or a NTC pool 72 h prior to hormone treatment. Western blot analysis indicated a significant reduction in the protein expression of the glucocorticoid receptor (78%) and estrogen receptor-alpha (75%) in glucocorticoid receptor and estrogen receptor-alpha siRNA compared to NTC-transfected cells (Fig. 5D). Expression of the glucocorticoid receptor (86% reduced) and estrogen receptor-alpha (75% reduced) mRNA corresponded to knockdown of target genes. Evaluating mRNA expression of RBM24, LEFTY1, and C10orf10 following knockdown of the glucocorticoid receptor and estrogen receptor-alpha confirmed the requirement of these receptors for glucocorticoid and E2-mediated gene regulation (Fig. 5E). Expression of RBM24 was induced by E2 in NTC-transfected cells, and coadministration of Dex partially blocked this response. Antagonism by Dex is not present following transfection with siRNA targeted to the glucocorticoid receptor. Significantly reducing cellular levels of estrogen receptor-alpha with siRNA prevented RBM24 mRNA inductions by E2, indicating the glucocorticoid receptor and estrogen receptor-alpha are required for coregulation of RBM24 gene expression. LEFTY1 and C10orf10 are novel glucocorticoid-responsive genes and mRNA expression was induced by Dex treatment in NTC-transfected cells at levels comparable to those of nontransfected cells. Dex-induced expression of LEFTY1 was antagonized by coadministration of E2. In glucocorticoid receptor-diminished cells, induction of LEFTY1 is lost, and decreased levels of estrogen receptor-alpha block E2-mediated antagonism. LEFTY1 represents a glucocorticoid-responsive gene whose transcription can be regulated by both the glucocorticoid receptor and the estrogen receptor-alpha. Congruent to pretreatment with ICI 182,780 and RU486, C10orf10 mRNA expression appears to require the glucocorticoid receptor but not the estrogen receptor-alpha. Results from targeted knockdown of the glucocorticoid receptor and estrogen receptor-alpha further support the requirement of these receptors for glucocorticoid and E2-mediated regulation of common targets.
FIG. 5. cont.
Factor 1 (LEFTY1), and Chromosome 10 Open Reading Frame 10 (C10orf10). Values were normalized to those of PPIB. Bar graphs show means ± SEM of four biological replicates. Statistical significance at *P < 0.05 and **P < 0.01, as determined by ANOVA. D) Extent of receptor knockdown of ECC1 cells (transfected for 72 h with glucocorticoid receptor, estrogen receptor-alpha, or nontargeting control [NTC] siRNA) as assessed by QPCR was compared to that found by Western blotting. Values were normalized to those of the housekeeping genes PPIB (by QPCR) or ACTB (by Western blotting). Western blots present two representative biological replicates, and bar graphs show means ± SEM of four independent biological replicates. Statistical differences at **P < 0.01, as determined by ANOVA. E) ECC1 cells transfected with NTC, glucocorticoid receptor, or estrogen receptor-alpha siRNA were treated for 6 h with vehicle, Dex, E2, or Dex + E2, and mRNA expression levels of RBM24, LEFTY1, and C10orf10 were evaluated. Values were normalized to those of PPIB. Bar graphs show the means ± SEM of four biological replicates. The symbol Statistical differences at **P < 0.01, as determined by ANOVA.
FIG. 5.
Continued.
DISCUSSION
Several microarray studies have defined altered gene expression in the uterus upon hormone treatment [44–46]. In contrast, relatively few studies have examined glucocorticoid-mediated gene expression patterns in the uterus in a cell-type-specific manner [47]. A previous study from our laboratory showed microarray data from whole-rat uterus following intraperitoneal injection with glucocorticoids or E2 or both [23]. However, it is difficult to interpret the results of that study in terms of the epithelial response due to the heterogeneous population of cell types in the uterus, where many epithelium-specific effects may be diluted. Therefore, the present study used a human uterine endometrial cancer cell line with uterine epithelial properties to isolate the pattern of gene expression in a uterine cell-type-specific manner. In the future, these in vitro experiments should be complementary to gene expression analysis following uterine epithelial cell isolation in mice treated with glucocorticoids and E2. Our microarray analysis identified over 10 000 gene probes that were significantly regulated, and large overlap exists for gene targets of Dex and E2 in ECC1 cells. This in itself is interesting to consider in light of the well-described antagonistic effects of Dex on the biological response to E2 in the uterus [23, 24, 37, 48]. These antagonistic effects could be linked to those unique functions identified in ECC1 cells treated with both Dex and E2, which likely are representative of the large number of genes regulated only in the presence of Dex and E2. Remarkably, the follicle-stimulating hormone receptor (FSHR) gene is one of the genes regulated only in the presence of Dex and E2 in ECC1 cells. Expression levels were 3.4-fold lower than those of the vehicle, which is supported by reports of glucocorticoid repression of FSHR in the male reproductive tract (epididymis and testicular tissue) [49]. FSHR is required for fertility and mRNA expression increases in the human endometrium following ovulation [50, 51]. Establishing an association between genes with known reproductive functions and novel glucocorticoid regulation provides further evidence of the glucocorticoid receptor's essential role in the uterus.
Several genes with immune-related functions were also identified in the top regulated gene list. Because of the significant regulation of immune or inflammatory genes and the division of Dex-regulated genes into immune networks, the inflammatory response network was evaluated. Interestingly, a considerable number of these genes were independently regulated by Dex and E2 in the same manner, suggesting that either hormone could elicit the same response in uterine epithelium. Although, most of the genes regulated in the inflammatory response network are down-regulated by both Dex and E2, NFκB1A exhibited anti-correlated regulation by Dex and E2. NFκB1A functions to inhibit the NFκB complex through cytoplasmic sequestration, a signaling pathway already linked to both glucocorticoids and estrogens [52, 53]. NFκB signaling in the uterus regulates the apoptotic activity of caspase 3 during pregnancy [54]. While little is known regarding the regulation of NFκB1A in the uterus, expression of NFκB1A increases in the late secretory phase, corresponding to increased rates of apoptosis in the uterus [55, 56]. Glucocorticoid and E2 regulation of NFκB1A may play a role in uterine cell death and survival decisions.
In the uterus, regulation of the inflammatory response requires bi-potential, where the epithelial cells act as both the first line of defense against pathogens and the regulators of immune tolerance in the female reproductive tract during implantation (reviewed in reference [57]). As part of the inflammatory response, cytokines provide signals between immune cells to coordinate their pro- and anti-inflammatory actions. In response to mating, cytokine expression is transiently induced in the uterus and has been implicated in conferring uterine receptivity [58, 59]. The uterine epithelium is a potent source of IL-6, a pleiotropic cytokine required for blastocyst attachment and embryo outgrowth [60, 61]. Repression by Dex parallels what has been described in human placental cytotrophoblasts, although here we show for the first time repression of IL-6 in Dex-treated human uterine epithelial cells [62]. Interestingly, expression of IL-16, a proinflammatory chemoattractant factor for CD4+ T lymphocytes, monocytes, and eosinophils, is up-regulated by E2, and repression by Dex is antagonized with coadministration of E2. Antagonistic gene regulation by glucocorticoids and E2 represents a unique opportunity for the immune and reproductive systems to regulate the same processes in an opposing manner with a few target genes. Furthermore, as a cell line derived from endometrial carcinoma, a pathology linked to the inflammatory response for both its initiation and progression, unique glucocorticoid gene targets involved in the inflammatory response may be implicated as both therapeutic targets and agents involved in the biology of uterine epithelium-like cells [63].
The analysis of coregulated genes identified 75 genes as antagonistically regulated in ECC1 cells, and several of these were independently validated. Enrichment of the glucocorticoid and estrogen response elements in those genes suggests they could be directly targeted by the glucocorticoid receptor and estrogen receptor-alpha, respectively. In fact, three of the genes validated for antagonistic regulation by Dex and E2 appear to require the glucocorticoid receptor and estrogen receptor-alpha for coregulation. Based on what is currently known regarding their function, all three genes are interesting targets of glucocorticoids and E2 in the uterus. We provide the first report indicating RBM24 is a target of the estrogen receptor-alpha in human uterine ECC1 cells. RBM24 plays a role in the post-transcriptional regulation of gene expression through regulating mRNA stability [64]. Dex antagonism of RBM24 expression indicates the scope of gene regulation by glucocorticoids in the uterus could include not only direct targets of the glucocorticoid receptor but also targets of estrogen signaling. C10orf10 encodes the decidual protein induced by progesterone (DE-PP), which is induced in the mid- and late-secretory phase and during the first trimester [65]. Progesterone-inducible DE-PP activates the ELK-1 transcription factor, modulating the effects of progesterone through regulation of gene expression. In ECC1 cells, Dex is a potent inducer of C10orf10 expression, which places the glucocorticoid receptor upstream of progesterone signaling. Furthermore, our data are the first to demonstrate glucocorticoid receptor regulation of LEFTY1 expression, a gene encoding a soluble cytokine of the TGF-β superfamily [66]. Transduction of LEFTY1 in the uterine horn of pregnant mice markedly attenuates uterine decidualization and results in decrease litter size [67]. Regulation of important fertility genes by the glucocorticoid receptor, such as LEFTY1, provides further indication that glucocorticoids may serve to integrate the immune system with the reproductive system in the uterus.
It is difficult to ascribe an absolute function to glucocorticoids that is either pro- or anti-inflammatory during pregnancy, as it appears the actions of glucocorticoids are balanced between both positive and negative effects. True to the immunosuppressive actions of glucocorticoids, women with a history of recurrent miscarriages and elevated levels of uterine natural killer cells when given oral glucocorticoids report suppressed levels of natural killer cells and improved pregnancy outcomes [68–71]. In cell line models of human trophoblast invasion, synthetic glucocorticoids increase proliferation and enhance invasion [72]. However, an earlier study of first trimester human cytotrophoblasts found that Dex inhibited cytotrophoblast invasion in vitro [73]. In addition to conflicting results regarding trophoblast invasion, glucocorticoids may also negatively impact trophoblast growth [74]. Dex can induce both apoptosis and necrosis in primary cultures of human placental trophoblasts and in a cytotrophoblast-derived cell line [75]. In microarray data from the ECC1 cell line, the range of effects exerted by glucocorticoids in the uterus encompasses both induction and repression of members of the inflammatory response. Although we have focused on the antagonism of RBM24, LEFTY1, and C10orf10 by glucocorticoids and E2, global analysis of gene expression exposes a complex network of gene regulation in this uterine cell type. These findings have identified potential mediators of pregnancy that are under the control of glucocorticoids and E2 in the uterus, as well as, a potential mechanism by which antagonism by these hormones occurs.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Robert Oakley, Dr. Carl Bortner, Dr. Lindsay Buckley, and Dr. Mahita Kadmiel for critical reading of the manuscript.
Footnotes
Supported by the Intramural Research Program of the NIH National Institute of Environmental Health Sciences.
REFERENCES
- Boivin J, Bunting L, Collins JA, Nygren KG. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum Reprod. 2007;22:1506–1512. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- WHO Scientific Group on Recent Advances in Medically Assisted Conception. Recent Advances in Medically Assisted Conception. WHO Technical Report Series; p. 820. In. Geneva; 1992. [PubMed] [Google Scholar]
- Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006;7:185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- Shiokawa S, Yoshimura Y, Sawa H, Nagamatsu S, Hanashi H, Sakai K, Ando M, Nakamura Y. Functional role of arg-gly-asp (RGD)-binding sites on beta1 integrin in embryo implantation using mouse blastocysts and human decidua. Biol Reprod. 1999;60:1468–1474. doi: 10.1095/biolreprod60.6.1468. [DOI] [PubMed] [Google Scholar]
- Simon C, Valbuena D, Krussel J, Bernal A, Murphy CR, Shaw T, Pellicer A, Polan ML. Interleukin-1 receptor antagonist prevents embryonic implantation by a direct effect on the endometrial epithelium. Fertil Steril. 1998;70:896–906. doi: 10.1016/s0015-0282(98)00275-1. [DOI] [PubMed] [Google Scholar]
- Plaks V, Birnberg T, Berkutzki T, Sela S, BenYashar A, Kalchenko V, Mor G, Keshet E, Dekel N, Neeman M, Jung S. Uterine DCs are crucial for decidua formation during embryo implantation in mice. J Clin Invest. 2008;118:3954–3965. doi: 10.1172/JCI36682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard JW. Uterine DCs are essential for pregnancy. J Clin Invest. 2008;118:3832–3835. doi: 10.1172/JCI37733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SA. Seminal fluid signaling in the female reproductive tract: lessons from rodents and pigs. J Anim Sci. 2007;85:E36–44. doi: 10.2527/jas.2006-578. [DOI] [PubMed] [Google Scholar]
- Schuberth HJ, Taylor U, Zerbe H, Waberski D, Hunter R, Rath D. Immunological responses to semen in the female genital tract. Theriogenology. 2008;70:1174–1181. doi: 10.1016/j.theriogenology.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Saito S, Nakashima A, Myojo-Higuma S, Shiozaki A. The balance between cytotoxic NK cells and regulatory NK cells in human pregnancy. J Reprod Immunol. 2008;77:14–22. doi: 10.1016/j.jri.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Aluvihare VR, Kallikourdis M, Betz AG. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol. 2004;5:266–271. doi: 10.1038/ni1037. [DOI] [PubMed] [Google Scholar]
- Robertson SA, Mau VJ, Tremellen KP, Seamark RF. Role of high molecular weight seminal vesicle proteins in eliciting the uterine inflammatory response to semen in mice. J Reprod Fertil. 1996;107:265–277. doi: 10.1530/jrf.0.1070265. [DOI] [PubMed] [Google Scholar]
- Grant KS, Wira CR. Effect of mouse uterine stromal cells on epithelial cell transepithelial resistance (TER) and TNFalpha and TGFbeta release in culture. Biol Reprod. 2003;69:1091–1098. doi: 10.1095/biolreprod.103.015495. [DOI] [PubMed] [Google Scholar]
- Fahey JV, Schaefer TM, Channon JY, Wira CR. Secretion of cytokines and chemokines by polarized human epithelial cells from the female reproductive tract. Hum Reprod. 2005;20:1439–1446. doi: 10.1093/humrep/deh806. [DOI] [PubMed] [Google Scholar]
- Giles KM, Ross K, Rossi AG, Hotchin NA, Haslett C, Dransfield I. Glucocorticoid augmentation of macrophage capacity for phagocytosis of apoptotic cells is associated with reduced p130Cas expression, loss of paxillin/pyk2 phosphorylation, and high levels of active Rac. J Immunol. 2001;167:976–986. doi: 10.4049/jimmunol.167.2.976. [DOI] [PubMed] [Google Scholar]
- Lim HY, Muller N, Herold MJ, van den Brandt J, Reichardt HM. Glucocorticoids exert opposing effects on macrophage function dependent on their concentration. Immunology. 2007;122:47–53. doi: 10.1111/j.1365-2567.2007.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodrea J, Majai G, Doro Z, Zahuczky G, Pap A, Rajnavolgyi E, Fesus L. The glucocorticoid dexamethasone programs human dendritic cells for enhanced phagocytosis of apoptotic neutrophils and inflammatory response. J Leukoc Biol. 2012;91:127–136. doi: 10.1189/jlb.0511243. [DOI] [PubMed] [Google Scholar]
- Busillo JM, Cidlowski JA. The five Rs of glucocorticoid action during inflammation: ready, reinforce, repress, resolve, and restore. Trends Endocrinol Metab. 2013;24:109–119. doi: 10.1016/j.tem.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard J, Jochen AL, Sadur CN, Hofeldt FD. Cushing's syndrome in pregnancy. Obstet Gynecol Surv. 1990;45:87–93. doi: 10.1097/00006254-199002000-00002. [DOI] [PubMed] [Google Scholar]
- Erichsen MM, Husebye ES, Michelsen TM, Dahl AA, Lovas K. Sexuality and fertility in women with Addison's disease. J Clin Endocrinol Metab. 2010;95:4354–4360. doi: 10.1210/jc.2010-0445. [DOI] [PubMed] [Google Scholar]
- Bever AT, Hisaw FL, Velardo JT. Inhibitory action of desoxycorticosterone acetate, cortisone acetate, and testosterone on uterine growth induced by estradiol-17beta. Endocrinology. 1956;59:165–169. doi: 10.1210/endo-59-2-165. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dey SK. Role of histamine in implantation: dexamethasone inhibits estradiol-induced implantation in the rat. Biol Reprod. 1980;22:1136–1141. doi: 10.1093/biolreprod/22.5.1136. [DOI] [PubMed] [Google Scholar]
- Rhen T, Grissom S, Afshari C, Cidlowski JA. Dexamethasone blocks the rapid biological effects of 17beta-estradiol in the rat uterus without antagonizing its global genomic actions. FASEB J. 2003;17:1849–1870. doi: 10.1096/fj.02-1099com. [DOI] [PubMed] [Google Scholar]
- Rhen T, Cidlowski JA. Estrogens and glucocorticoids have opposing effects on the amount and latent activity of complement proteins in the rat uterus. Biol Reprod. 2006;74:265–274. doi: 10.1095/biolreprod.105.045336. [DOI] [PubMed] [Google Scholar]
- Ren R, Oakley RH, Cruz-Topete D, Cidlowski JA. Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology. 2012;153:5346–5360. doi: 10.1210/en.2012-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck A, Koritz SB. Studies on the mode of action of glucocorticoids in rats. I. Early effects of cortisol on blood glucose and on glucose entry into muscle, liver and adipose tissue. Biochim Biophys Acta. 1962;57:310–317. doi: 10.1016/0006-3002(62)91124-1. [DOI] [PubMed] [Google Scholar]
- Swingle WW, Davanzo JP, Glenister D, Wagle G, Osborne M, Rowen R. Effect of mineralo- and glucocorticoids on fasted adrenalectomized dogs subjected to electroshock. Proc Soc Exp Biol Med. 1960;104:184–188. doi: 10.3181/00379727-104-25773. [DOI] [PubMed] [Google Scholar]
- Ballard PL, Ballard RA. Glucocorticoid receptors and the role of glucocorticoids in fetal lung development. Proc Natl Acad Sci U S A. 1972;69:2668–2672. doi: 10.1073/pnas.69.9.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates FE, Leeman SE, Glenister DW, Dallman MF. Interaction between plasma corticosterone concentration and adrenocorticotropin-releasing stimuli in the rat: evidence for the reset of an endocrine feedback control. Endocrinology. 1961;69:67–80. doi: 10.1210/endo-69-1-67. [DOI] [PubMed] [Google Scholar]
- Ostrowski MC, Huang AL, Kessel M, Wolford RG, Hager GL. Modulation of enhancer activity by the hormone responsive regulatory element from mouse mammary tumor virus. EMBO J. 1984;3:1891–1899. doi: 10.1002/j.1460-2075.1984.tb02064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter JD, Tomkins GM. The relationship between glucocorticoid binding and tyrosine aminotransferase induction in hepatoma tissue culture cells. Proc Natl Acad Sci U S A. 1970;65:709–715. doi: 10.1073/pnas.65.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- Sakai DD, Helms S, Carlstedt-Duke J, Gustafsson JA, Rottman FM, Yamamoto KR. Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988;2:1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- Pearce D, Yamamoto KR. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 1993;259:1161–1165. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Whirledge S, Cidlowski JA. Estradiol antagonism of glucocorticoid-induced GILZ expression in human uterine epithelial cells and murine uterus. Endocrinology. 2013;154:499–510. doi: 10.1210/en.2012-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Adamio F, Zollo O, Moraca R, Ayroldi E, Bruscoli S, Bartoli A, Cannarile L, Migliorati G, Riccardi C. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7:803–812. doi: 10.1016/s1074-7613(00)80398-2. [DOI] [PubMed] [Google Scholar]
- Mo B, Vendrov AE, Palomino WA, DuPont BR, Apparao KB, Lessey BA. ECC-1 cells: a well-differentiated steroid-responsive endometrial cell line with characteristics of luminal epithelium. Biol Reprod. 2006;75:387–394. doi: 10.1095/biolreprod.106.051870. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR. the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeling AE, Dukes M, Bowler J. A potent specific pure antiestrogen with clinical potential. Cancer Res. 1991;51:3867–3873. [PubMed] [Google Scholar]
- Bethin KE, Nagai Y, Sladek R, Asada M, Sadovsky Y, Hudson TJ, Muglia LJ. Microarray analysis of uterine gene expression in mouse and human pregnancy. Mol Endocrinol. 2003;17:1454–1469. doi: 10.1210/me.2003-0007. [DOI] [PubMed] [Google Scholar]
- Hong SH, Nah HY, Lee JY, Gye MC, Kim CH, Kim MK. Analysis of estrogen-regulated genes in mouse uterus using cDNA microarray and laser capture microdissection. J Endocrinol. 2004;181:157–167. doi: 10.1677/joe.0.1810157. [DOI] [PubMed] [Google Scholar]
- Pan H, Zhu L, Deng Y, Pollard JW. Microarray analysis of uterine epithelial gene expression during the implantation window in the mouse. Endocrinology. 2006;147:4904–4916. doi: 10.1210/en.2006-0140. [DOI] [PubMed] [Google Scholar]
- Whirledge S, Dixon D, Cidlowski JA. Glucocorticoids regulate gene expression and repress cellular proliferation in human uterine leiomyoma cells. Horm Cancer. 2012;3:79–92. doi: 10.1007/s12672-012-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin DS, Johnson EO, Brandon DD, Liapi C, Chrousos GP. Glucocorticoids inhibit estradiol-mediated uterine growth: possible role of the uterine estradiol receptor. Biol Reprod. 1990;42:74–80. doi: 10.1095/biolreprod42.1.74. [DOI] [PubMed] [Google Scholar]
- Herrera-Luna CV, Budik S, Aurich C. Gene expression of ACTH, glucocorticoid receptors, 11betaHSD enzymes, LH-, FSH-, GH receptors and aromatase in equine epididymal and testicular tissue. Reprod Domest Anim. 2012;47:928–935. doi: 10.1111/j.1439-0531.2012.01993.x. [DOI] [PubMed] [Google Scholar]
- La Marca A. Carducci Artenisio A, Stabile G, Rivasi F, Volpe A. Evidence for cycle-dependent expression of follicle-stimulating hormone receptor in human endometrium. Gynecol Endocrinol. 2005;21:303–306. doi: 10.1080/09513590500402756. [DOI] [PubMed] [Google Scholar]
- Danilovich N, Babu PS, Xing W, Gerdes M, Krishnamurthy H, Sairam MR. Estrogen deficiency, obesity, and skeletal abnormalities in follicle-stimulating hormone receptor knockout (FORKO) female mice. Endocrinology. 2000;141:4295–4308. doi: 10.1210/endo.141.11.7765. [DOI] [PubMed] [Google Scholar]
- Rao NA, McCalman MT, Moulos P, Francoijs KJ, Chatziioannou A, Kolisis FN, Alexis MN, Mitsiou DJ, Stunnenberg HG. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011;21:1404–1416. doi: 10.1101/gr.118042.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vina J, Gambini J, Lopez-Grueso R, Abdelaziz KM, Jove M, Borras C. Females live longer than males: role of oxidative stress. Curr Pharm Des. 2011;17:3959–3965. doi: 10.2174/138161211798764942. [DOI] [PubMed] [Google Scholar]
- Jeyasuria P, Subedi K, Suresh A, Condon JC. Elevated levels of uterine anti-apoptotic signaling may activate NFKB and potentially confer resistance to caspase 3-mediated apoptotic cell death during pregnancy in mice. Biol Reprod. 2011;85:417–424. doi: 10.1095/biolreprod.111.091652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmowski WP, Ding J, Shen J, Rana N, Fernandez BB, Braun DP. Apoptosis in endometrial glandular and stromal cells in women with and without endometriosis. Hum Reprod. 2001;16:1802–1808. doi: 10.1093/humrep/16.9.1802. [DOI] [PubMed] [Google Scholar]
- Ponce C, Torres M, Galleguillos C, Sovino H, Boric MA, Fuentes A, Johnson MC. Nuclear factor kappaB pathway and interleukin-6 are affected in eutopic endometrium of women with endometriosis. Reproduction. 2009;137:727–737. doi: 10.1530/REP-08-0407. [DOI] [PubMed] [Google Scholar]
- Robertson SA. Immune regulation of conception and embryo implantation-all about quality control? J Reprod Immunol. 2010;85:51–57. doi: 10.1016/j.jri.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Sanford TR, De M, Wood GW. Expression of colony-stimulating factors and inflammatory cytokines in the uterus of CD1 mice during days 1 to 3 of pregnancy. J Reprod Fertil. 1992;94:213–220. doi: 10.1530/jrf.0.0940213. [DOI] [PubMed] [Google Scholar]
- Sherwin JR, Freeman TC, Stephens RJ, Kimber S, Smith AG, Chambers I, Smith SK, Sharkey AM. Identification of genes regulated by leukemia-inhibitory factor in the mouse uterus at the time of implantation. Mol Endocrinol. 2004;18:2185–2195. doi: 10.1210/me.2004-0110. [DOI] [PubMed] [Google Scholar]
- Robertson SA, Mayrhofer G, Seamark RF. Uterine epithelial cells synthesize granulocyte-macrophage colony-stimulating factor and interleukin-6 in pregnant and nonpregnant mice. Biol Reprod. 1992;46:1069–1079. doi: 10.1095/biolreprod46.6.1069. [DOI] [PubMed] [Google Scholar]
- Jacobs AL, Sehgal PB, Julian J, Carson DD. Secretion and hormonal regulation of interleukin-6 production by mouse uterine stromal and polarized epithelial cells cultured in vitro. Endocrinology. 1992;131:1037–1046. doi: 10.1210/endo.131.3.1505448. [DOI] [PubMed] [Google Scholar]
- Rosen T, Krikun G, Ma Y, Wang EY, Lockwood CJ, Guller S. Chronic antagonism of nuclear factor-kappaB activity in cytotrophoblasts by dexamethasone: a potential mechanism for antiinflammatory action of glucocorticoids in human placenta. J Clin Endocrinol Metab. 1998;83:3647–3652. doi: 10.1210/jcem.83.10.5151. [DOI] [PubMed] [Google Scholar]
- Wallace AE, Gibson DA, Saunders PT, Jabbour HN. Inflammatory events in endometrial adenocarcinoma. J Endocrinol. 2010;206:141–157. doi: 10.1677/JOE-10-0072. [DOI] [PubMed] [Google Scholar]
- Jin D, Hidaka K, Shirai M, Morisaki T. RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes Cells. 2010;15:1158–1167. doi: 10.1111/j.1365-2443.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Nonoguchi K, Sakurai T, Masuda T, Itoh K, Fujita J. A novel protein Depp, which is induced by progesterone in human endometrial stromal cells activates Elk-1 transcription factor. Mol Hum Reprod. 2005;11:471–476. doi: 10.1093/molehr/gah186. [DOI] [PubMed] [Google Scholar]
- Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature. 1996;381:151–155. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- Tang M, Naidu D, Hearing P, Handwerger S, Tabibzadeh S. LEFTY. a member of the transforming growth factor-beta superfamily, inhibits uterine stromal cell differentiation: a novel autocrine role. Endocrinology. 2010;151:1320–1330. doi: 10.1210/en.2009-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma CM, Keay SD, Macklon NS. Peri-implantation glucocorticoid administration for assisted reproductive technology cycles. Cochrane Database Syst Rev. 2007 doi: 10.1002/14651858.CD005996.pub2. CD005996. [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Aoki K. Successful uterine steroid therapy in a case with a history of ten miscarriages. Am J Reprod Immunol. 2000;44:253–255. doi: 10.1111/j.8755-8920.2000.440411.x. [DOI] [PubMed] [Google Scholar]
- Quenby S, Farquharson R, Young M, Vince G. Successful pregnancy outcome following 19 consecutive miscarriages: case report. Hum Reprod. 2003;18:2562–2564. doi: 10.1093/humrep/deg502. [DOI] [PubMed] [Google Scholar]
- Quenby S, Kalumbi C, Bates M, Farquharson R, Vince G. Prednisolone reduces preconceptual endometrial natural killer cells in women with recurrent miscarriage. Fertil Steril. 2005;84:980–984. doi: 10.1016/j.fertnstert.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Mandl M, Ghaffari-Tabrizi N, Haas J, Nohammer G, Desoye G. Differential glucocorticoid effects on proliferation and invasion of human trophoblast cell lines. Reproduction. 2006;132:159–167. doi: 10.1530/rep.1.00976. [DOI] [PubMed] [Google Scholar]
- Librach CL, Feigenbaum SL, Bass KE, Cui TY, Verastas N, Sadovsky Y, Quigley JP, French DL, Fisher SJ. Interleukin-1 beta regulates human cytotrophoblast metalloproteinase activity and invasion in vitro. J Biol Chem. 1994;269:17125–17131. [PubMed] [Google Scholar]
- Ma Y, Ryu JS, Dulay A, Segal M, Guller S. Regulation of plasminogen activator inhibitor (PAI)-1 expression in a human trophoblast cell line by glucocorticoid (GC) and transforming growth factor (TGF)-beta. Placenta. 2002;23:727–734. doi: 10.1016/s0143-4004(02)90863-5. [DOI] [PubMed] [Google Scholar]
- Crocker IP, Barratt S, Kaur M, Baker PN. The in-vitro characterization of induced apoptosis in placental cytotrophoblasts and syncytiotrophoblasts. Placenta. 2001;22:822–830. doi: 10.1053/plac.2001.0733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.