Abstract
The amino acid sequence of amyloid precursor protein (APP) is highly conserved, and age-related Aβ aggregates have been described in a variety of vertebrate animals, with the notable exception of mice and rats. Three amino acid substitutions distinguish mouse and human Aβ that might contribute to their differing properties in vivo. To examine the amyloidogenic potential of mouse Aβ, we studied several lines of transgenic mice overexpressing wild-type mouse amyloid precursor protein (moAPP) either alone or in conjunction with mutant PS1 (PS1dE9). Neither overexpression of moAPP alone nor co-expression with PS1dE9 caused mice to develop Alzheimer-type amyloid pathology by 24 months of age. We further tested whether mouse Aβ could accelerate the deposition of human Aβ by crossing the moAPP transgenic mice to a bigenic line expressing human APPswe with PS1dE9. The triple transgenic animals (moAPP × APPswe/PS1dE9) produced 20% more Aβ but formed amyloid deposits no faster and to no greater extent than APPswe/PS1dE9 siblings. Instead, the additional mouse Aβ increased the detergent solubility of accumulated amyloid and exacerbated amyloid deposition in the vasculature. These findings suggest that, although mouse Aβ does not influence the rate of amyloid formation, the incorporation of Aβ peptides with differing sequences alters the solubility and localization of the resulting aggregates.
Although genetically engineered mice have become a commonly used tool for Alzheimer disease research, wild-type rats and mice do not innately develop age-associated amyloid pathology (1). Many other animals, including dogs, bears, and primates, display amyloid lesions similar to those of Alzheimer disease in humans (2–6). The lesions are formed by the aggregation of a small peptide, Aβ, common to all of these species. The amyloid precursor protein (APP),3 from which Aβ is derived, is well conserved, and >96% of the amino acid sequence is identical between mouse, rat, monkey, and human (1, 7–9). The reason for rodents’ relative resistance to amyloid pathology remains unknown, although several hypotheses have been proposed, including the importance of amino acid sequence differences in the peptide (10), the short lifespan of rodents relative to other animals (11), and differences in processing of mouse APP by β-site APP-cleaving enzyme 1 (BACE1) (12).
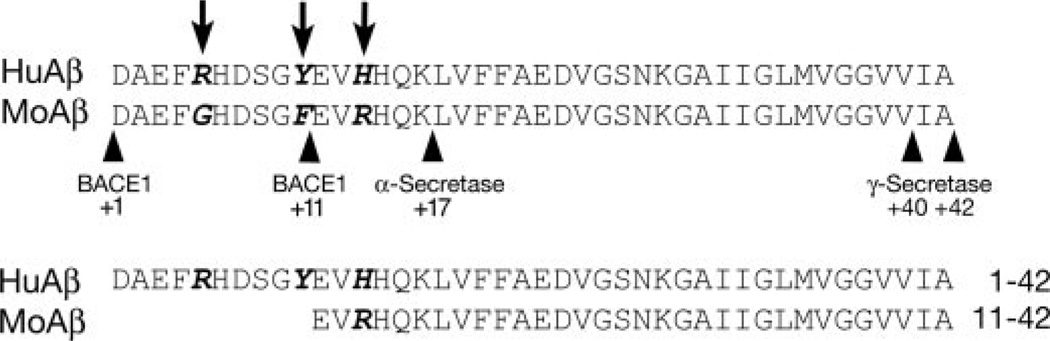
Only 17 amino acid differences distinguish moAPP from its human homolog; however, three of these differences are located within the N-terminal domain of the Aβ peptide (Fig. 1; Arg-5→Gly, Tyr-10→Phe, and His-13→Arg). This region is thought to be important for the specificity of interaction between Aβ peptides, whereas the hydrophobic C-terminal sequence governs the rate of Aβ aggregation (13–15). Despite these three amino acid substitutions, in vitro studies using synthetic Aβ peptides revealed no difference in the ability of rodent and human peptides to form congophilic filaments in aqueous buffers (16, 17). Peptides from the two species can also co-aggregate in solution to form mixed Aβ fibers (17). The sequence differences are not completely without consequence, however. The replacement of His-13 for Arg in rodent Aβ disrupts a metal coordination site, which renders the rodent peptide much less prone to zinc-induced aggregation in vitro (18–20).
FIGURE 1. Mouse and human Aβ differ in primary sequence and N-terminal processing.
Three amino acid differences at residues 5, 10, and 13 distinguish the rodent and human peptides. These substitutions influence the specificity of BACE1 cleavage. Human APP expressed in transgenic mice is preferentially cleaved at residue 1, producing peptides 38–43 amino acids in length. In contrast, endogenous mouse APP is preferentially cleaved by BACE1 at +11, generating peptides of 28–33 residues. Peptides ending at amino acid 42 are shown for comparison.
The amino acid sequence of Aβ also affects interactions with APP-processing enzymes (21). BACE1 is responsible for releasing the N terminus of Aβ from APP at either of two cleavage sites within the peptide sequence: +1 or +11. When murine BACE1 encounters human APP, the human protein is processed to generate Aβ starting predominantly at the +1 site. In contrast, murine BACE1 preferentially cleaves endogenous APP at the +11 site (12, 22–25). The effect of this truncation on the aggregation of Aβ is subject to debate: published reports describe both enhanced and reduced aggregation (oligomerization or sedimentation) in vitro of N-terminally truncated Aβ (26, 27).
Transgenic mice overexpressing human APP form amyloid deposits composed primarily of human Aβ. In addition, these mice harbor low levels of mouse Aβ beginning at both +1 and +11. Pype et al. (28) show that the amount of mouse Aβ extracted from the brains of transgenic mice increased dramatically with the formation of human Aβ deposits, suggesting that the mouse peptide was co-depositing in the amyloid. However, mouse peptide comprised only about 5% of the total Aβ recovered from the oldest animals tested. Moreover, Calhoun et al. (29) have shown that removing mouse APP expression in human APP transgenic mice had no impact on the extent or location of amyloid formation, indicating that the mouse peptide plays little or no role in the rate of amyloid formation in the presence of high levels of human Aβ.
To date, only one study has addressed the effects of elevating murine Aβ in vivo by overexpressing wild-type moAPP (30). This study examined animals only 3.5–4 months of age and, therefore, did not test whether overexpression of moAPP would induce late onset amyloid formation. A complicating factor in this initial study was the FVB background strain used to generate the transgenic lines. Transgenic mice of this strain are prone to premature death when human APP is expressed via the hamster prion protein promoter (PrP) vector (30). We have subsequently moved one of these original moAPP transgenic lines out of the FVB background by crossing onto a hybrid C3H/HeJ × C57BL/6J strain for more than five generations. This strategy eliminated the premature death of animals, allowing us to examine the potential for amyloid formation in aged moAPP transgenic mice. We report here that neither overexpression of moAPP alone, nor co-expression with the human presenilin-1 exon-9 deletion variant (PS1dE9 (11, 31)), resulted in amyloid composed only from mouse Aβ. We further demonstrate that mouse Aβ does not accelerate the deposition of human Aβ in transgenic mice overexpressing both peptides, but reveal that high levels of mouse peptide alter the solubility of the resulting Aβ aggregates and increase the prevalence of vascular deposits. We further find that Aβ11–40/42, predicted to be the predominant form of Aβ produced from mouse APP, does not appear to be a major co-depositing peptide. These findings provide insight into the potential role of specific Aβ sequences in modulating the solubility and distribution of amyloid deposits in rodent models.
EXPERIMENTAL PROCEDURES
Generation of Transgenic Mice
All transgenic mice used in this study have been described and fully characterized in earlier publications. All transgenes were expressed under control of the mouse prion protein promoter (MoPrP.Xho), which drives high protein expression in neurons and astrocytes of the central nervous system (31, 32). Line S-9, expressing human PS1 harboring the FAD exon-9 deletion (PS1dE9), is described in Lee et al. (33). Line 1874, expressing wild-type mouse APP (moAPPwt), is described in Hsiao et al. (30). Line 85, co-expressing human PS1dE9 and mouse/human (mo/hu) chimeric APP695 (humanized Aβ domain) harboring the Swedish (K594M/N595L) mutation, is described in Jankowsky et al. (11). Unlike lines S-9 and 1874, line 85 was created by co-injecting two transgenes, each driven by their own prion promoter element. The two transgenes co-integrated and co-segregate as a single locus (34). Lines 85 and S-9 have been deposited with Jackson Laboratories (Bar Harbor, ME) for distribution (Stock numbers 004462 and 005866, respectively). After these experiments were completed, line 1874 was lost through accidental mistyping of breeding stock.
Line 85 and line S-9 animals used in this study were maintained on a hybrid background by backcrossing to C3HeJ × C57BL/6J F1 animals obtained from Jackson Laboratories. Line 1874 was backcrossed to C57BL/6J for two generations after it was originally generated on the FVB background. After discovering premature lethality in the offspring, the line was crossed back to the hybrid C3HeJ × C57BL/6J F1 strain for two additional generations, which restored normal longevity to the line. Offspring from the second C3/B6 backcross were used as breeders to generate the cohorts described in this study.
Offspring were genotyped for the presence of the transgene by PCR amplification of genomic DNA extracted from 1-cm tail clippings as described previously (34). Reactions contained three primers, one antisense primer matching sequence within the vector that is also present in mouse genomic PrP (5′: GTG GAT ACC CCC TCC CCC AGC CTA GAC C), one sense primer specific for the transgene cDNA (PS1: 5′: CAG GTG GTG GAG CAA GAT G, huAPP: 5′: CCG AGA TCT CTG AAG TGA AGA TGG ATG, moAPP: 5′: CCT TCA GGA TTT GAA GTC CGC), and a second sense primer specific for the genomic PrP coding region, which has been removed from the MoPrP vector (5′: CCT CTT TGT GAC TAT GTG GAC TGA TGT CGG). All reactions give a 750-bp product from the endogenous PrP gene as a control for DNA integrity and successful amplification; transgene-positive samples have an additional band at 400 bp (huAPP), 350 bp (moAPP) or 1.3 kb (PS1).
Animals were housed in microisolator cages with free access to food and water. All procedures involving animals were approved by the Johns Hopkins University Institutional Animal Care and Use Committee.
Western Blotting
Mice of each genotype (NTg and 1874, n = 3–5; lines 85 and 1874 × 85, n = 10–15) were harvested at 8 months of age for assessment of amyloid pathology and APP/Aβ biochemistry. One-half of the brain was immersed in 4% paraformaldehyde and used for histology as described below. The remaining hemisphere was frozen on dry ice and prepared as a 20% homogenate that was used for Western blotting, filter trap assay, and ELISA.
Frozen hemi-brain samples were sonicated in 5 volumes of 1× PBS containing 5 mm EDTA and 1× protease inhibitor mixture (Mammalian cell mix, Sigma). Homogenates were further diluted 1:1 with additional PBS/EDTA/protease inhibitor and centrifuged at high speed for 10 min, and the supernatant was used for analysis. Approximately 50 µg of protein homogenate per sample (5 µg for 22C11) was loaded onto 4–12% BisTris Novex gels (Invitrogen) and electrophoresed at 175 V for 1.5–2 h in 1× MES buffer (Invitrogen). Proteins were transferred for 1 h at 100 V to 0.45-µm Optitran nitrocellulose (Schleicher and Schuell, Keene, NH) in 1× NuPAGE transfer buffer made with 10% methanol and 1% antioxidant solution (Invitrogen). Blots were blocked in PBS containing 5% nonfat dry milk powder for 30–60 min at room temperature. After blocking, blots were incubated with primary antibody for either 3 h at room temperature or overnight at 4 °C. The following primary antibodies and dilutions were used: 6E10 mouse anti-human Aβ monoclonal (Signet Laboratories, Dedham, MA) 1:2000; rabbit antirodent APP purified polyclonal antibody (AB5571P, Chemicon, Temecula, CA) 1:2000; 22C11 mouse anti-APP N terminus monoclonal, kind gift of Drs. Konrad Beyreuther and Andreas Weidemann, 1:2000 (35); m/hSOD1 rabbit anti-SOD1 polyclonal, 1:4000 (36). After incubation with primary antibody, the blots were washed several times with PBS containing 0.1% Tween 20, and then incubated with either goat anti-rabbit IgG or goat anti-mouse IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA) diluted 1:2500 to 1:5000 in blocking solution. After washing several times in PBS containing 0.1% Tween 20, blots were developed with enhanced chemiluminescence reagent (ECL Plus, Amersham Biosciences/GE Biosciences) and exposed to film. Intensity of immunostaining was quantified from digitally scanned films with ImageJ by first inverting to create a negative image and then measuring the integrated density of each band. Background values calculated from a blank portion of the gel were subtracted manually from each sample before assessing the average signal intensity for the genotype.
Aβ Immunoprecipitation
50 µl of the 20% PBS homogenate described above was diluted 10-fold in radioimmunoprecipitation assay buffer (0.2% SDS, 0.5% Nonidet P-40, 0.5% deoxycholate, 5 mm EDTA, in 1× PBS) and boiled for 10 min. After cooling, protease inhibitors were added, and the solution was incubated overnight at 4 °C with 2 µl of purified 4G8 (Signet Laboratories). The antibody was recovered with protein A-agarose beads (1 h at 4 °C), and nonspecific binding was removed by several washes with additional radioimmunoprecipitation assay buffer at 4 °C. The beads were heated to 95 °C for 5 min in 2× Tricine-SDS sample buffer, and the entire reaction was loaded onto 10–20% Tricine gels (Bio-Rad). Gels were pre-run for 10 min prior to loading, and then run in 15-min voltage steps of 25, 50, and 100, before running the gel to completion at 150 V. Protein was transferred to 0.45-µm Optitran nitrocellulose (Schleicher and Schuell) in 1× Tris-glycine/20% methanol/0.1% SDS, after which the blot was boiled 5 min in 1× PBS and blocked in 2% ECL Advance blocking reagent (Amersham Biosciences/GE Biosciences)/1× TBS/0.1% Tween-20. Blots were incubated overnight at room temperature with 4G8 diluted 1:10,000 in Advance block with 0.1% sodium azide. After washing several times in blocking reagent, blots were incubated for 2–3 h at room temperature with peroxidase-conjugated anti-mouse IgG diluted 1:20,000 in block. Blots were washed thoroughly with TBS/0.1% Tween-20, developed with ECL Advance (Amersham Biosciences/GE Biosciences), and exposed to film ~1 h after developing with ECL. To demonstrate that 4G8 was capable of binding Aβ11–42, several reactions were run using 15 µl of 20% homogenate spiked with 10–50 ng of synthetic human Aβ11–42 (kindly provided by Dr. David Teplow, UCLA).
Aβ ELISA: Steady-state Levels (7–24 Weeks of Age)
Brain tissue used for ELISA was harvested from female mice prior to the onset of amyloid pathology (lines 1874, 85, and 1874 × 85: 7–10 weeks of age; line 1874 × S-9: 9–24 weeks of age). Frozen mouse hemi-brains (n = 4–6 per genotype) were extracted by sonication in 0.2% diethylamine (DEA)/50 mm NaCl at a concentration of 100 mg/ml. After centrifugation at 100,000 × g for 1 h at 4 °C, the supernatant was removed and saved as the DEA extract. The pellet was then sonicated in 70% formic acid (FA) diluted in water, using a volume equal to the original volume of DEA. After centrifugation at 100,000 × g for 1 h at 4 °C, the supernatant was removed and saved as the FA extract. The DEA extracts were neutralized by adding a 1/10 volume of 0.5 m Tris-HCl, pH 6.8. The FA extracts were neutralized and prepared for ELISA by diluting 1:20 in 1 m Tris-phosphate buffer, pH 11. The samples were then assayed by sandwich ELISA as described below.
Aβ ELISA: Amyloid Solubility (8 Months of Age)
An aliquot of PBS 20% homogenate generated for Western analysis described above was subjected to a three-step sequential extraction using PBS, 2% SDS, and 70% formic acid (NTg and line 1874, n = 4; lines 85 and 1874 × 85, n = 8). At each step, the sample was sonicated in appropriate buffer and centrifuged at 100,000 × g for 30 min at 4 °C. The supernatant was removed for analysis, and the pellet was sonicated in an equal volume of the next solution in sequence. The 2% SDS extracts were diluted at least 1:40 in EC buffer (0.02 m sodium phosphate buffer, pH 7.0, 2 mm EDTA, 400 mm NaCl, 0.2% bovine serum albumin, 0.05% CHAPS, 0.4% BlockAce (Dainippon Pharmaceuticals), 0.05% NaN3), prior to testing to bring the SDS concentration below 0.05%; the FA extracts were neutralized with 1 m Tris-phosphate buffer, pH 11, and then diluted with EC buffer prior to testing.
Brain extracts were measured by sandwich ELISA as described previously (37–39). Human Aβ was measured in each fraction using BAN50 for capture (epitope Aβ1–16) and BA27 and BC05 for detection (Aβ40 and Aβ42, respectively). Total Aβ (mouse plus human) was measured in each fraction using BNT77 for capture (epitope Aβ 11–28) and BA27 and BC05 for detection. Although BNT77 recognizes both mouse and human Aβ 1-x and 11-x, it does not bind α-secretase processed APP (40), and measurements with BNT77 therefore do not include p3. All values were calculated as picomoles per g based on the initial weight of brain tissue.
Filter Trap Assay
An aliquot of 20% PBS protein homogenate from each 8-month-old animal was partially solubilized by the addition of SDS to a final concentration of 1%. Serial 1:1 dilutions were made with 1× PBS/1% SDS, and 90 µl of each dilution was then vacuum-filtered through a pre-wet 0.22-µm cellulose acetate membrane (OE66, Schleicher and Schuell, Keene, NH). Each well was washed several times with PBS, after which blots were blocked for an hour in 1× TBS plus 5% nonfat dry milk powder. Blots were then incubated at 4 °C overnight with polyclonal anti-Aβ peptide antibody (71–5800, Zymed Laboratories) diluted 1:600 in blocking solution. After washing the blots several times in 1× TBS/0.1% Tween 20, the membrane was incubated for 1 h with an IRDye 800-conjugated goat anti-rabbit IgG secondary antibody (Rockland Immunochemicals, Gilbertsville, PA) diluted 1:5000 in blocking solution. The membranes were again washed three times with 1× TBS/0.1% Tween 20, given a final rinse in 1× TBS, and then imaged with an Odyssey fluorescence imager (LI-COR, Lincoln, NE). Staining intensity for each well was quantified using Odyssey analysis software, from which the linear range of the dilution series was determined and used for all genotype comparisons.
Histology
Brains from lines 1874, 85, and 1874 × 85 were harvested for histological analysis at 4 months (n = 3–4 per genotype) and at 8 months (n = 5–14 per genotype) of age. Mice were euthanized by ether inhalation, and the brain was removed for analysis. One half was used for biochemical analysis described above; the remaining hemisphere was used for histology. After immersion in 4% paraformaldehyde/1× PBS for 48 h at 4 °C, the fixed hemi-brains were transferred to PBS, dehydrated in an alcohol series, treated with cedar wood oil and methylsalicylate, and embedded in paraffin for sectioning.
Hirano Silver Stain
Silver impregnation histology was performed on 10-µm paraffin-embedded sections by Hirano’s modification of the Bielschowsky method (41, 42). Briefly, sections were deparaffinized through xylene and alcohols into tap water before being placed into fresh 20% silver nitrate solution for 20 min. After washing thoroughly with distilled water, slides were immersed in 20% silver nitrate solution titrated with fresh ammonium hydroxide. After 20 min, slides were washed with ammonia water before being individually developed with 100 µl of developer (20 ml of 37% formaldehyde, 100 ml of distilled water, 50 µl of concentrated nitric acid, and 0.5 g of citric acid) added to 50 ml of titrated silver nitrate solution. Slides were then rinsed in tap water, fixed in 5% sodium thiosulfate, and dehydrated through alcohols and xylene.
Campbell-Switzer Silver Stain
A detailed protocol for this stain was kindly provided by Dr. Bob Switzer of NeuroScience Associates.
Thioflavine-S Staining
Following deparaffinization of sections through xylene and alcohols, amyloid impregnation with thioflavine-S was performed according to the Guntern modification of the standard protocol. Slides were washed twice in distilled water, then immersed for 5 min in a 0.25% potassium permanganate solution, followed by 5 min in a 1% potassium metabisulfate/1% oxalic acid solution. After this preparation, slides were placed into a filtered aqueous 0.02% thioflavine-S solution (Chroma-Gesellschaft, Schmid GmbH and Co., Kongen, Germany) for 8 min. Excess stain was removed by two brief rinses in 80% ethanol, then two in distilled water, after which slides were finished in aqueous mounting medium for florescence photomicrography.
Aβ Immunohistochemistry
Prior to immunostaining, slides were deparaffinized by oven heating followed by immersion in xylene. After rehydration through graded alcohols into tap water, amyloid was partially denatured by immersing sections in 80% formic acid for 5 min, followed by rinsing in running tap water. Nonspecific staining was blocked for 1 h with 3% normal goat serum and 0.1% Triton-X 100 in TBS. Slides were then placed into primary antibody (rabbit anti-human Aβ peptide polyclonal antibody 71–5800 diluted 1:500, Zymed Laboratories; 6E10 mouse anti-human Aβ monoclonal antibody diluted 1:250, Signet Laboratories; or rabbit anti-rodent APP purified polyclonal antibody AB5571P diluted 1:250, Chemicon) in blocking solution and incubated overnight at room temperature. After washing with several changes of TBS, slides were incubated either with the Vectastain Elite antimouse secondary system according to the manufacturer’s directions (for diaminobenzidine-developed anti-Aβ immunostaining, Vector Laboratories, Burlingame, CA) or with Alexafluor-conjugated goat anti-rabbit (Alexa 568) and goat anti-mouse (Alexa 488) secondary antibodies diluted 1:100 in blocking solution (for double immunofluorescence immunostaining, Molecular Probes c/o Invitrogen). Slides were again rinsed several times in TBS and either mounted immediately in fluorescence mounting medium (6E10/roAPP) or developed with diaminobenzidine (Zymed Laboratories anti-Aβ), counterstained with hematoxylin, dehydrated and mounted.
Plaque Load Estimation
Amyloid burden was estimated using non-biased stereology or image-based threshold analysis. Three sagittal sections spaced at 200-µm intervals were analysed for each animal. Slides were analyzed by an investigator blind to the genotype of the samples.
Non-biased Stereology
StereoInvestigator software (MBF Biosciences, Colchester, VT) was used to estimate the surface area covered by plaques stained with Aβ immunohistochemistry (Zymed Laboratories, 71–5800 anti-Aβ polyclonal) using an area fraction fractionator grid (Cavalieri spacing: 500 × 500 µm, grid spacing: 10 µm, frame size: 85 × 110 µm, 40× magnification). Percent coverage within the cortex of each animal was averaged from three sections to obtain a final estimate of plaque burden.
ImageJ-based Threshold Analysis
Sections stained for Aβ immunohistochemistry or Campbell-Switzer silver were scanned using an Epson 4990 flatbed scanner set for film at 2400 dpi. Images were brought into ImageJ and converted to an 8-bit grayscale. The cortex was outlined manually using a Wacom Graphire tablet, the surrounding area was cleared, and the automatic threshold within the remaining image was determined (but not applied to the image). The percent surface area above threshold was then determined using the Analyze Particles command.
Vascular Amyloid Quantitation
Sections from 8-monthold mice that had been stained for Aβ immunohistochemistry were viewed at 40× under standard brightfield conditions. All amyloid deposits within one microscopic field of the dorsal cortical surface running from the frontal to occipital poles were examined. Blood vessels were identified by morphology with hematoxylin counterstain; all vessels positive for Aβ were manually counted by an investigator who was blind to the genotype of the sample. Only parenchymal vessels were considered; amyloid-positive vessels in the meninges at the pial surface were not counted. Three sections, spaced at 200-µm intervals, were averaged for each animal (line 85: n = 9; line 1874 × 85 n = 8).
Statistics
All data were analyzed for statistical significance by ANOVA followed with Tukey post-test using SigmaStat analysis software (Systat Software, Port Richmond, CA).
RESULTS
APP Expression and Steady-state Aβ Levels Are Elevated Severalfold by Transgenic Expression of moAPP
Two lines of transgenic mice were used for these studies. The first line, hereafter referred to as line 85, co-expressed a chimeric mouse/human APP 695 (human Aβ sequence) harboring the Swedish K594M/N595L mutation (using 695 numbering) alongside human PS1 harboring the exon-9 deletion mutation (PS1dE9). Each transgene is controlled by an independent mouse prion promoter. The two transgenes are co-integrated and segregate as a single locus, making all mice from this line transgenic for both proteins. The second line, hereafter referred to as line 1874, expressed wild-type mouse APP 695 under the control of the hamster prion promoter (30), which produces a similar pattern of transgene expression as the mouse prion promoter used for line 85 (32). Interbreeding of the two lines generated mice of four genotypes (non-transgenic (NTg), 1874 (single transgenic), 85 (double transgenic), and 1874 × 85 (triple transgenic)) that were analyzed to determine the impact of mouse Aβ on the timing and extent of human Aβ deposition.
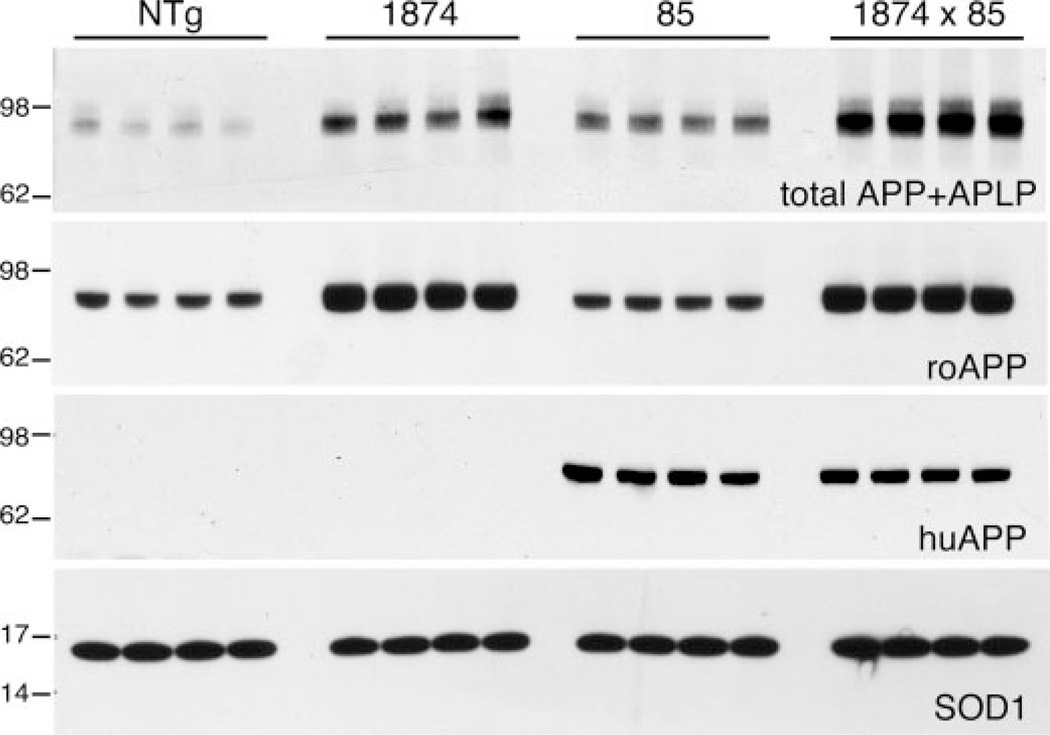
Both lines 85 and 1874 overexpressed transgenic APP at levels severalfold over the endogenous protein. Immunoblotting for the full-length protein was used to quantify how well the transgene expressed each line and to ensure that expression of one APP transgene was not diminished by co-expression of the second in 1874 × 85 triple transgenic (mo/huAPPswe/PS1dE9/moAPPwt) offspring. Three separate antibodies were used for this analysis: 22C11, which recognizes from both mouse and human the N terminus of mature and unprocessed APP as well as APP-like protein 2 (43); 6E10, which recognizes the N-terminal region of human Aβ; and a rodent-specific APP antibody, which recognizes the N-terminal region of mouse Aβ (Fig. 2). Quantitation with 22C11 and roAPP indicated that line 1874 expressed between 2.5- (2.46 ± 0.05; roAPP) and 4-fold (3.98 ± 0.36; 22C11) more APP than NTg. APP overexpression in line 85 appeared somewhat lower than in line 1874; blotting with 22C11 revealed ~3-fold more APP than NTg (2.89 ± 0.13). Expression levels in the triple transgenic line 1874 × 85 were roughly the sum of that in each parental line (7.28 ± 0.27; 22C11). Most importantly, comparison of mo/huAPPswe expression in lines 85 and 1874 × 85, and of moAPPwt in lines 1874 and 1874 × 85, indicated that co-expression of the second APP transgene did not diminish expression of the first. Levels of mo/huAPPswe measured by 6E10 immunoblotting were nearly identical in the line 85 and 1874 × 85 brains (2.24 ± 0.16 (line 85) versus 2.03 ± 0.06 (line 1874 × 85), in arbitrary units; ANOVA: F1,7 = 3.228, p = 0.122). Likewise, expression of mouse APP measured with roAPP was similar in lines 1874 and 1874 × 85 (5.64 ± 0.11 (line 85) versus 5.92 ± 0.06 (line 1874 × 85), in arbitrary units, ANOVA for all four genotypes F3,12 = 66.219, p = 0.001, Tukey post-hoc for pairwise comparison of 85 versus 1874 × 85 p = 0.900). These findings indicate that there is no change in the expression of one APP transgene caused by the introduction of a second.
FIGURE 2. Transgenic expression increases moAPP 3-fold without lowering co-expressed human APP.
Western blots compare mouse and human APP expression in brain homogenates from each of the four genotypes; four animals (2 male and 2 female) from each genotype are shown. Blots show immunodetection of total full-length APP and APP-like protein 2 (top panel, 22C11); rodent-specific APP (second panel, roAPP); human-specific APP (third panel, 6E10); and SOD1 (Cu/Zn superoxide dismutase 1) as an internal control (bottom panel). The 62- and 98-kDa markers in the upper two panels migrated more slowly than expected based on the known size of APP and were consistently positioned lower on the BisTris gels run in MES buffer than in previous studies using Tris-HCl gels in Tris-glycine-SDS buffer.
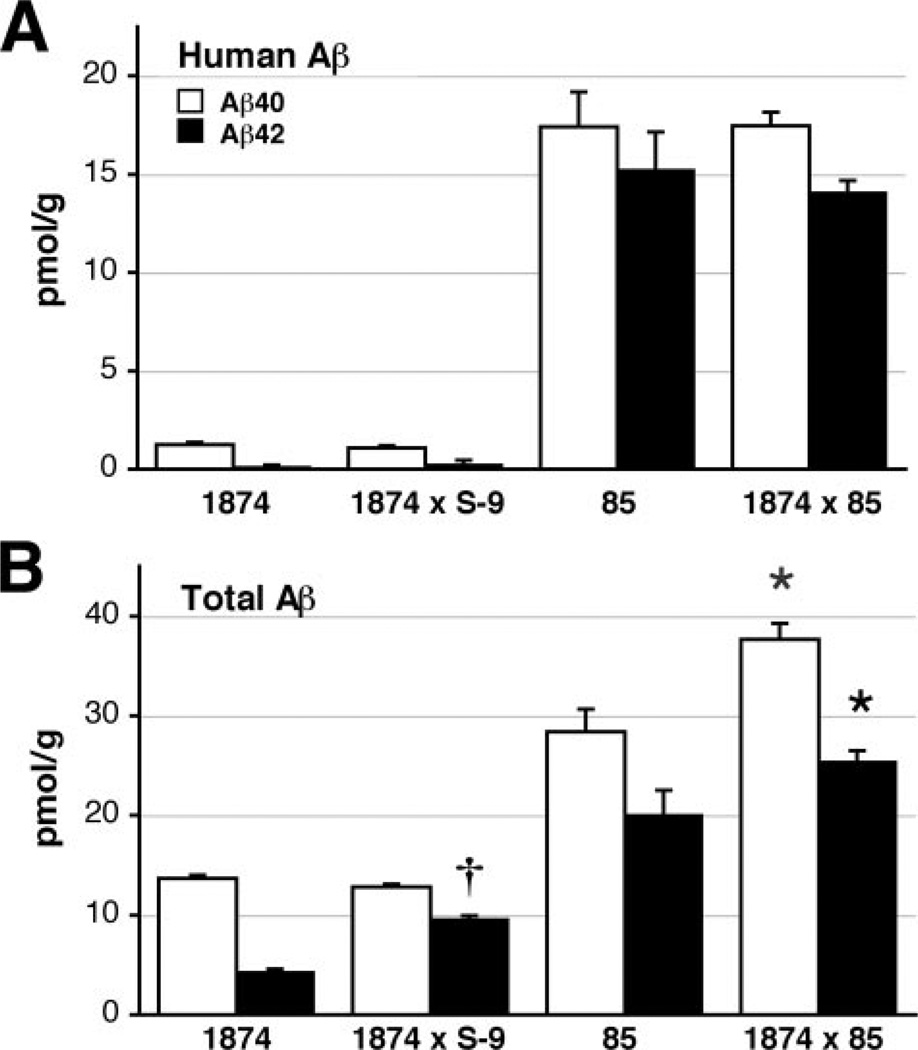
We next assessed steady-state levels of Aβ in the 1874, 85, and 1874 × 85 mice. Brain tissue was harvested from young mice prior to the formation of amyloid deposits. These analyses also included mice derived from the mating of line 1874 × line S-9, which overexpresses human PS1 encoding the exon 9 deletion. Both Aβ40 and Aβ42 levels were measured by ELISA using two different capture antibodies to distinguish human peptide from total Aβ (mouse plus human). The human-specific capture antibody BAN50 recognizes only full-length human Aβ1-x, whereas the BNT77 capture antibody that detects both mouse and human Aβ recognizes full-length Aβ in addition to Aβ11-x (but not α-secretase products 17-x). Therefore, when referring to ELISA results with BNT77, we designate the peptides as x-40 and x-42. Consistent with the immunoblot demonstrating equal expression of mo/huAPPswe, brain tissue from line 85 and 1874 × 85 mice contained nearly identical levels of human Aβ peptide (Table 1 and Fig. 3; Aβ40: 17.42 ± 3.53 (line 85) versus 17.47 ± 1.39 (line 1874 × 85) pmol/g tissue, ANOVA for all four genotypes: F3,15 = 139.75, p < 0.001, Tukey post-hoc for pairwise comparison of 85 versus 1874 × 85 p = 1.0; Aβ42: 15.24 ± 3.87 (line 85) versus 14.08 ± 1.17 (line 1874 × 85) pmol/g tissue, Tukey post-hoc p = 0.808). Similarly, levels of total Aβ (mouse plus human) paralleled the expression of total APP measured by 22C11. Line 1874 mice produced almost as much mouse Aβx-40 as line 85 mice produced of human Aβ1–40 (13.68 ± 0.76 pmol/g of tissue) but with much lower levels of Aβx-42 (4.31 ± 0.67 pmol/g). Introduction of the FAD-variant PS1dE9 in mice expressing moAPP more than doubled production of Aβx-42 without impacting Aβx-40 (line 1874 × S-9, Aβ40: 12.83 ± 0.69; Aβ42: 9.59 ± 0.81 pmol/g), consistent with our previous work on this PS1 variant (11). The amount of Aβ in the triple transgenic 1874 × 85 mice was roughly the sum of Aβ levels measured in each line separately (Aβ40: 37.66 ± 3.28, Aβ42: 25.41 ± 2.24 pmol/g). Steady-state levels of Aβ40 are significantly higher in the triple transgenic 1874 × 85 mice than in their double transgenic line 85 siblings (ANOVA: F3,15 = 98.94, p < 0.001, Tukey post-hoc p < 0.001); as are levels of Aβ42 (ANOVA: F3,15 = 66.47, p < 0.001, Tukey post-hoc p = 0.037). In sum, 1874 × 85 brain harbors significantly more total Aβ peptide than does line 85 (48.42 ± 4.76 (line 85) versus 63.08 ± 2.25 (line 1874 × 85) pmol/g). Based on our previous study of APP and APP/PS1 transgenic mice, these data predict an earlier onset of amyloid deposition, with a greater plaque burden at any given age in the triple transgenic mice compared with their double transgenic siblings (11).
TABLE 1. ELISA measurement of steady-state Aβ levels in 4-month-old mice.
Values are in picomoles/g wet tissue weight (±S.E.).
| DEA |
FA |
n | |||
|---|---|---|---|---|---|
| 40 | 42 | 40 | 42 | ||
| pmol/g | |||||
| Human Aβ-BAN50 capture | |||||
| 1874a | 0.06 (0.035) | 0.02 (0.018) | 1.19 (0.103) | 0.10 (0.103) | 5 |
| 1874 × S-9a | 0.04 (0.037) | 0.02 (0.008) | 1.04 (0.147) | 0.23 (0.196) | 6 |
| 85 | 9.25 (1.19) | 6.71 (1.06) | 8.17 (0.56) | 8.53 (0.92) | 4 |
| 1874 × 85 | 8.85 (0.67) | 6.22 (0.46) | 8.62 (0.25) | 7.86 (0.38) | 4 |
| Total Aβ-BNT77 capture | |||||
| 1874 | 8.17 (0.33) | 1.61 (0.049) | 5.51 (0.35) | 2.70 (0.25) | 5 |
| 1874 × S-9 | 7.23 (0.33) | 3.98 (0.15) | 5.60 (0.31) | 5.61 (0.24) | 6 |
| 85 | 17.49 (1.78) | 9.81 (1.17) | 10.90 (0.57) | 10.23 (1.40) | 4 |
| 1874 × 85 | 23.30 (1.24) | 12.51 (0.28) | 14.37 (0.79) | 12.91 (0.87) | 4 |
The capture antibody for this assay (BAN50) is specific for human Aβ. Values obtained for 1874 and 1874 × S-9 represent background binding.
FIGURE 3. ELISA analysis confirms that transgenic expression of moAPP increases pre-deposit steady-state Aβ levels.
A, brain homogenates from each of the lines indicated were assayed at 2.5 (lines 1874, 85, and 1874 × 85) or 6 months of age (line 1874 × S-9) for human-specific Aβ40 (open bars) and Aβ42 (filled bars). As predicted, lines 85 and 1874 × 85 produce identical levels of human peptide. B, measurement of total Aβ (mouse plus human) using the same samples assayed for human Aβ in panel A. Note that Aβ levels in line 1874 × 85 are roughly the sum of Aβ levels in the separate lines. Data are shown ± S.E. *, p < 0.05 versus line 85. †, p < 0.05 versus line 1874.
Plaque Burden Is Not Increased or Accelerated by Overproduction of Mouse Aβ
Line 1874 produces 2- to 4-fold more APP than wild-type animals, which is in the range needed to produce amyloid pathology in transgenic lines expressing the mutant human protein (11). Despite high levels of transgene expression, long term study of line 1874 found no evidence for amyloid formation in mice up to 24 months of age (data not shown). Similarly, the 1874 × S-9 mice also failed to develop amyloid deposits by 2 years of age (data not shown). These findings suggest that either mouse Aβ is incapable of initiating amyloid deposits in vivo, or that there is too little Aβ42 produced in these lines to aggregate within the 24-month mouse lifespan.
We next addressed whether mouse Aβ could accelerate amyloid formation from human Aβ by crossing mice from line 1874 to line 85. Previous studies in line 85 indicated that amyloid formation begins at about 5–6 months of age in this line. We therefore chose two ages for analysis that bracketed this onset to test whether addition of extra mouse Aβ introduced in the triple transgenic 1874 (moAPPwt) × 85 (mo/huAPPswe/huPS1dE9) offspring would alter the rate of human Aβ deposition. The first set of mice was harvested at 4 months of age, with the expectation that raising the steady-state Aβ level in the triple transgenics would accelerate its aggregation into plaques. Instead we found no sign of amyloid formation in any of the mice examined at this age, regardless of genotype or gender (data not shown). Despite carrying significantly more total Aβ, the 1874 × 85 mice formed cored-amyloid deposits no faster than their line 85 siblings.
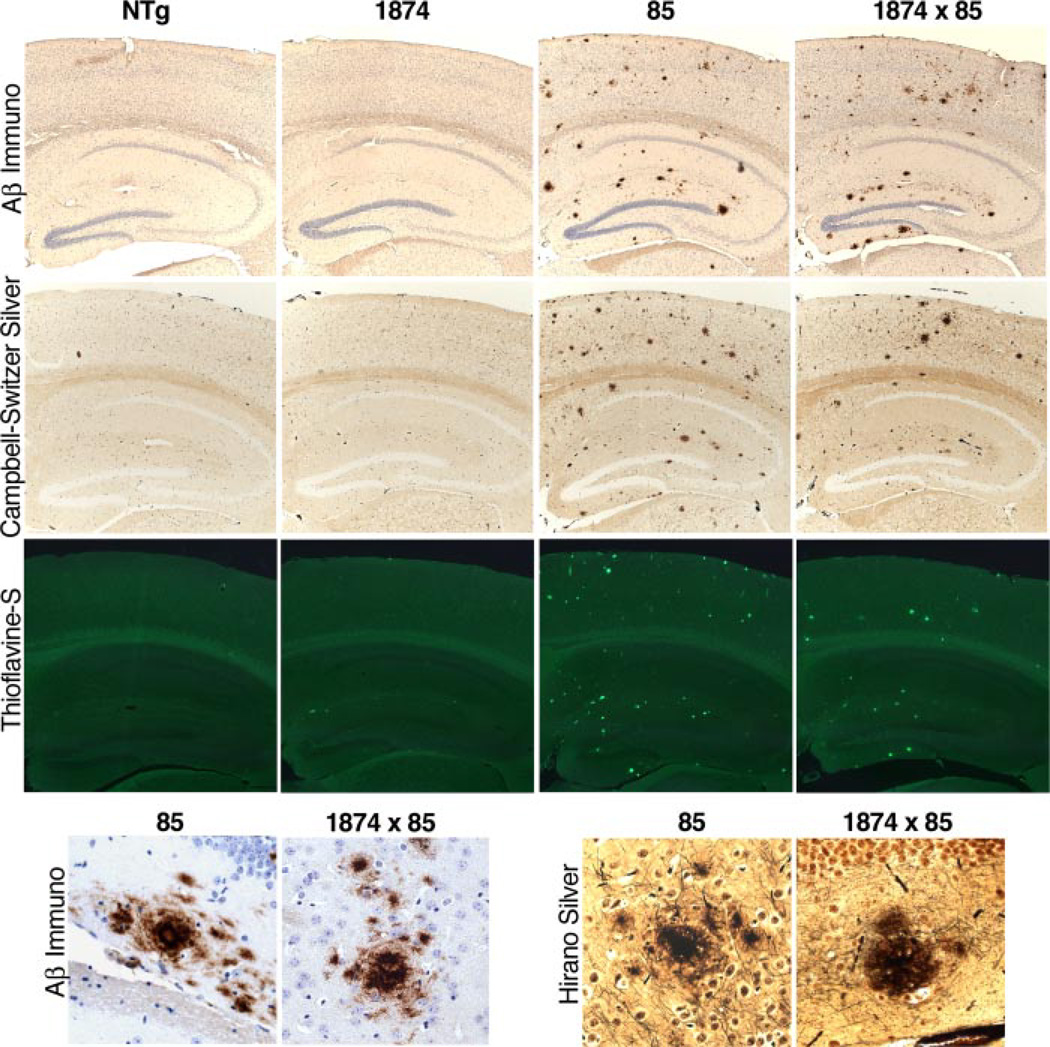
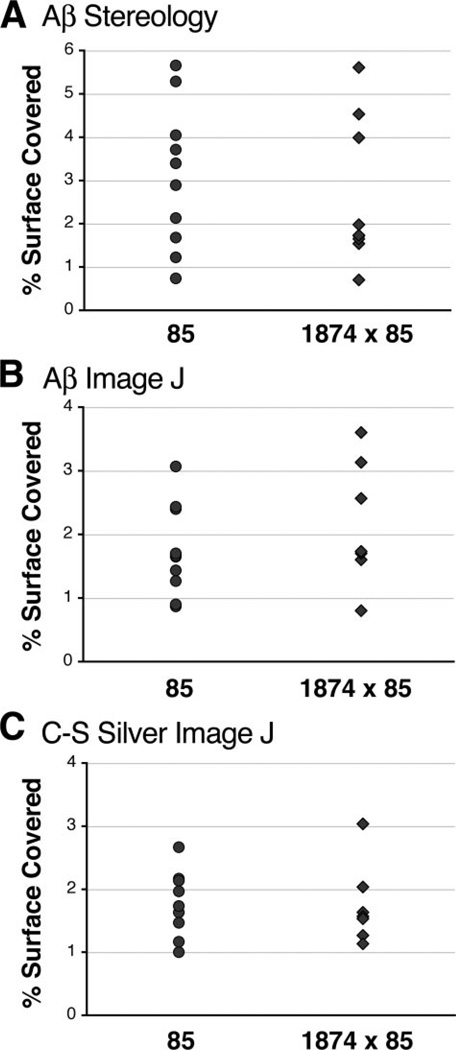
We next examined amyloid burden in 1874 × 85 offspring at a time point after deposits were known to appear in line 85 animals. We found that, by 8 months of age, amyloid deposits were apparent in brains from both the line 85 double transgenic animals and their 1874 × 85 triple transgenic siblings. Qualitatively, there was little to distinguish between genotypes: plaques were found throughout the cortex and hippocampus of most animals, with no obvious difference in the relative appearance of the deposits or the surrounding tissue (Fig. 4). The distribution of plaques appeared similar in line 85 and 1874 × 85 sections stained with silver or Aβ immunostaining, and a fraction of the deposits in both genotypes bound Thioflavine-S. Non-biased stereology supported this observation. Analysis of the percent surface area covered by plaques in the cortex of Aβ-immunostained sections revealed almost complete overlap of the two genotypes (Fig. 5; line 85: 3.08% ± 0.52; line 1874 × 85: 2.61% ± 0.56). This conclusion was confirmed by a second method of quantitation using ImageJ (see “Experimental Procedures”). Both silver and Aβ immunostained sections were analysed by digital imaging; both yielded the same outcome (Fig. 5; silver: 1.77% ± 0.17 (line 85) versus 1.71% ± 0.19 (line 1874 × 85); Aβ immunostaining: 1.74% ± 0.22 (line 85) versus 2.16% ± 0.40 (line 1874 × 85)). Overproduction of mouse Aβ did not alter the timing or the extent of amyloid formation in mice producing human Aβ.
FIGURE 4. Overexpression of mouse APP does not enhance senile plaque pathology in mice producing human Aβ.
Amyloid staining by Aβ immunohistochemistry (top row), Campbell-Switzer silver stain (second row), and thioflavine-S (third row) produce qualitatively similar images in 8-month-old mice overexpressing human Aβ (line 85) versus those overexpressing both mouse and human peptide (line 1874 × 85). High power (40×) images of plaques stained by Aβ immunohistochemistry (bottom row, left) and by Hirano silver stain (bottom row, right) suggest that the structure of amyloid aggregates in both lines is also similar. By itself, overexpression of mouse APP (line 1874) failed to produce amyloid plaques at any age examined.
FIGURE 5. Quantitation of senile plaque burden confirms that plaque formation is not increased by the addition of extra mouse Aβ.
The panels show scatter plots of the percent surface area covered by amyloid at 8 months of age within the cortex of individual mice for each genotype. Coverage was assessed by two independent methods: non-biased stereology with Stereo-Investigator software (A) or by digital threshold analysis with ImageJ (B and C). Sections used for analysis were stained for amyloid with an anti-Aβ antibody (A and B) or by silver impregnation using the Campbell-Switzer Alzheimer’s stain (C). The three analyses reached similar conclusions: overexpression of mouse Aβ does not change the extent of amyloid formation in the cortex of animals depositing human Aβ.
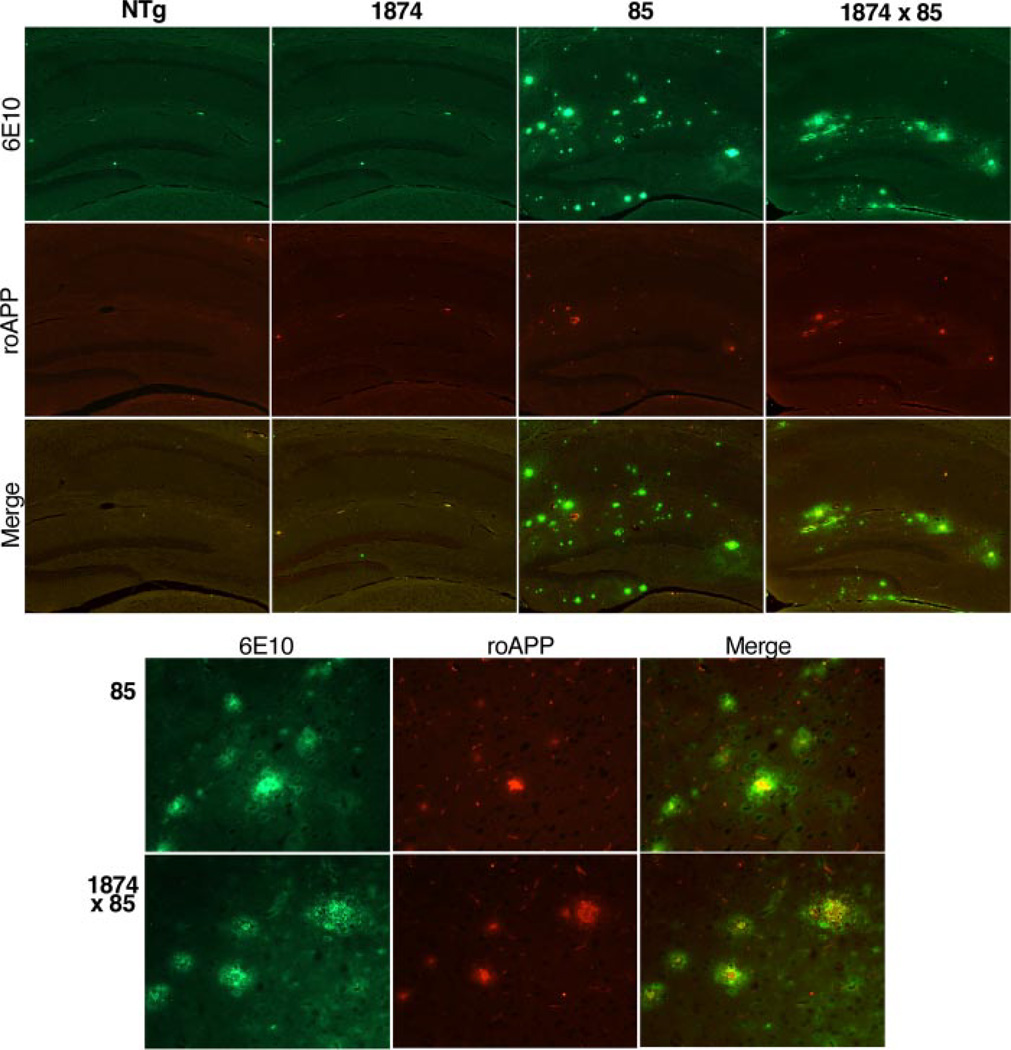
Although we found that amyloid burden in the brains of 1874 × 85 mice did not change as a result of overproducing mouse Aβ, we wanted to know whether the relative amount of mouse Aβ co-deposited with human peptide would increase as more mouse peptide was produced. We used double immunofluorescence to label mouse (roAPP) and human (6E10) APP/Aβ in 8-month-old mice of each genotype (Fig. 6). The predominant signal in the plaques of both 85 and 1874 × 85 mice is from the human peptide. Human immunostaining is present within all plaques observed in both genotypes. Mouse APP/Aβ immunostaining is present within most plaques but covers a much more restricted and focal area within the core of the deposits. Occasional plaques staining with the human antibody can be found that do not co-label for mouse peptide; these are predominantly either diffuse or very small deposits. The most notable finding of this experiment was the similarity of parenchymal amyloid staining in the 85 and 1874 × 85 mice. Although the triple transgenic mice produced much more mouse peptide than their double transgenic siblings, there was no change in the extent of immunostaining for mouse APP/Aβ in their plaques.
FIGURE 6. Participation of rodent Aβ in amyloid plaques does not increase with overexpression of mouse APP.
Double immunostaining for human (top row, 6E10, and green) and mouse Aβ (middle row, roAPP, and red) reveals that human peptide is the dominant species depositing in 8-month-old mice overproducing APP. A minimal core of rodent immunostaining is seen in most plaques; this signal does not increase even when mouse APP/Aβ levels are elevated severalfold (line 85 versus 1874 × 85). The top panels show immunostaining for each genotype at low power; the bottom panels highlight immunostaining in plaques at higher magnification (40×).
Vascular Amyloid Is Increased by Overexpression of Mouse Aβ
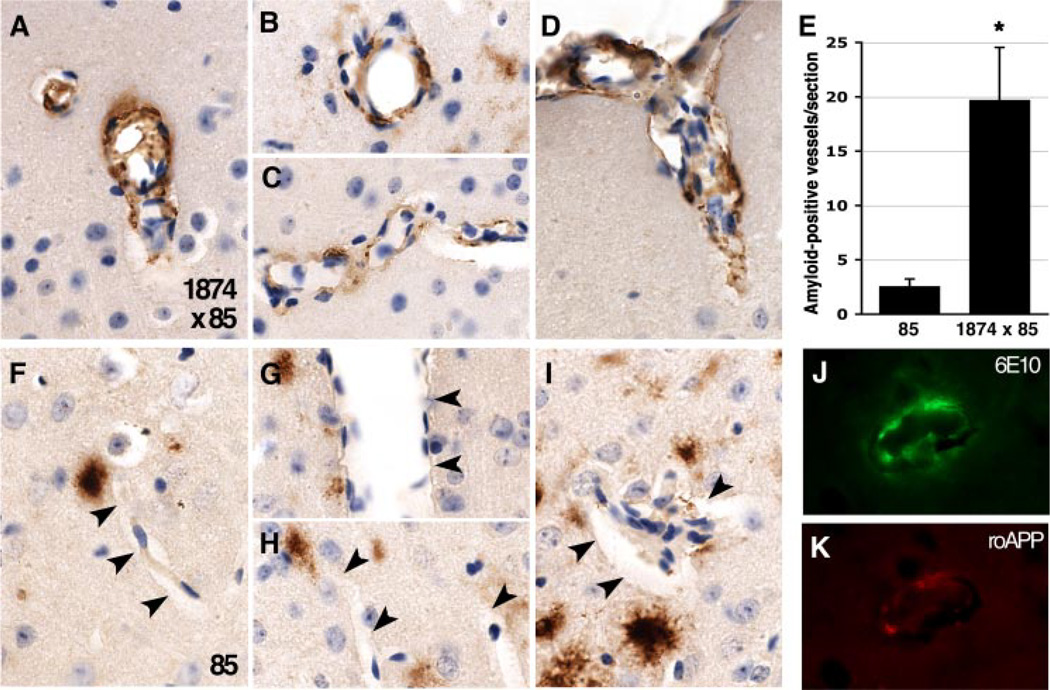
Careful examination of Aβ-immunostained sections revealed that vascular amyloid deposits were more common in the 1874 × 85 mice than in their double transgenic siblings. Although both genotypes displayed extensive accumulation in blood vessels at the pial surface, Aβ immunostaining was rare in vessels within the cortex for line 85. Small amyloid deposits were often seen within close proximity to blood vessels, but the vessel wall was usually clean. By comparison, the appearance of vascular amyloid reactive with human-specific 6E10 antibody was much more common in the triple transgenics (Fig. 7). Here, the entire circumference of some vessels stained for Aβ. However, amyloid-positive vessels still comprised only a small fraction of the total vasculature in each section. Nonetheless, the phenotype was consistent enough to allow a blinded investigator to correctly identify the genotype of tissues samples from 15 out of 19 animals based solely on the presence or absence of vascular deposits. This qualitative assessment was confirmed by quantitation: manual counting of amyloid-positive vessels within the cortex revealed the 1874 × 85 mice to have 7.8-fold more Aβ-positive vessels per section than their line 85 siblings (85: 2.53 ± 0.72 amyloid-positive vessels per section; 19.67 ± 4.89; ANOVA: F1,15 = 13.54, p < 0.005).
FIGURE 7. Overproduction of mouse Aβ increases deposition of human Aβ in cortical blood vessels.
A–D, immunostaining for Aβ is more often found within the vasculature of 8-month-old triple transgenic 1874 × 85 mice than in their double transgenic siblings. E, quantitation of amyloid-positive blood vessels in the cortex of Aβ-immunostained sections reveals a significant increase in cerebral amyloid angiopathy in 1874 × 85 mice compared with their line 85 siblings. F–I, amyloid deposits appear in close proximity to, but usually not within cortical blood vessels (indicated by arrowheads) of line 85 animals. J and K, like the parenchymal deposits, vascular amyloid in line 1874 × 85 mice contains both human(E; 6E10) and mouse (F; roAPP) APP/Aβ. Panels A–D and F–I show representative images from three animals for each genotype; all images were taken at 100×. *, p < 0.005 versus line 85.
Mouse Aβ Increases the Detergent Solubility of Human Peptide Aggregates
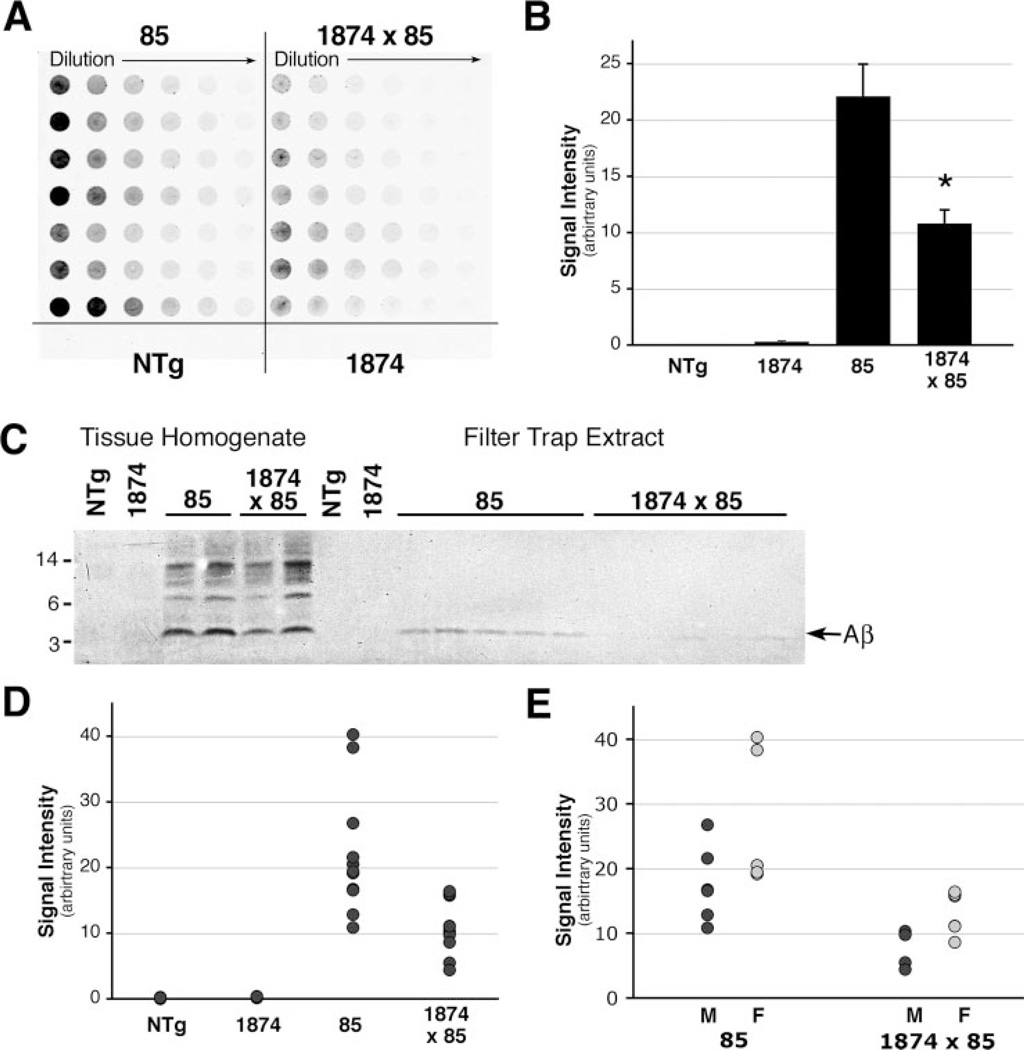
To extend our histological data, we used filter-trap and ELISA measurements to assess the solubility and composition of accumulated Aβ in mice of the various genotypes. The filter-trap assay uses a cellulose acetate filter to trap protein aggregates larger than the pore size within the membrane for detection by immunoblotting (Fig. 8). Serial dilutions are made in 1% SDS, which is harsh enough to partially solubilize even human AD amyloid (44). Unexpectedly, we found that protein extracts from the 1874 × 85 mice contained substantially less trapped Aβ than extracts from line 85. Quantitation of staining intensity within the linear range of the dilution series revealed that, on average, the triple transgenic animals harbored less than half the amount of filter-trapped Aβ than their double transgenic counterparts (22.10 ± 2.87 (line 85) versus 10.80 ± 1.20 (line 1874 × 85) in arbitrary units, ANOVA for all four genotypes: F3,24 = 14.60, p < 0.001, Tukey post-hoc for pairwise comparison of 85 versus 1874 × 85 p = 0.003). This outcome appeared to contradict our histological analyses showing similar amyloid burden in the brains of the two models. However, because the homogenates were prepared in 1% SDS, we thought it possible that the amyloid deposited in the 1874 × 85 mice might be more soluble in SDS than the amyloid accumulating in the line 85 mice.
FIGURE 8. Overexpression of mouse APP increases the solubility of human Aβ aggregates.
A, representative example of a filter-trap assay for aggregated Aβ in serial dilutions of brain homogenates from 8-month-old mice. Each row represents a separate mouse. Lacking aggregated peptide, NTg and 1874 homogenates showed no immunostaining for Aβ. Both lines 85 and 1874 × 85 have significant levels of aggregated Aβ, however, less peptide was retained from the 1% SDS homogenates in mice overexpressing mouse and human APP than in those overexpressing only the human protein. B, average intensity of immunostaining within the linear range of the filter-trap dilution series for each genotype reveals a dramatic reduction of aggregated Aβ in the SDS extracts of mice overexpressing mouse and human APP (line 1874 × 85) compared with animals overexpressing only the human protein (line 85). C, Western blot of protein homogenate (lanes 1–6) and extracted filter trap wells (lanes 7–18) probed with human-specific antibody 6E10. Consistent with the greater intensity of staining on the serial dilution filter trap shown in A, the extracted wells from line 85 mice contained more aggregated Aβ than those from 1874 × 85. D, filter-trap quantitation used to generate the genotype averages shown in C are plotted as individual data points. E, separation of genders within each genotype reveals that males carried the lowest, and females the highest, individual amyloid burdens within each group. This is consistent with previous work describing greater amyloid loads in female mice of several other APP transgenic lines (54, 55). *, p < 0.005 versus line 85.
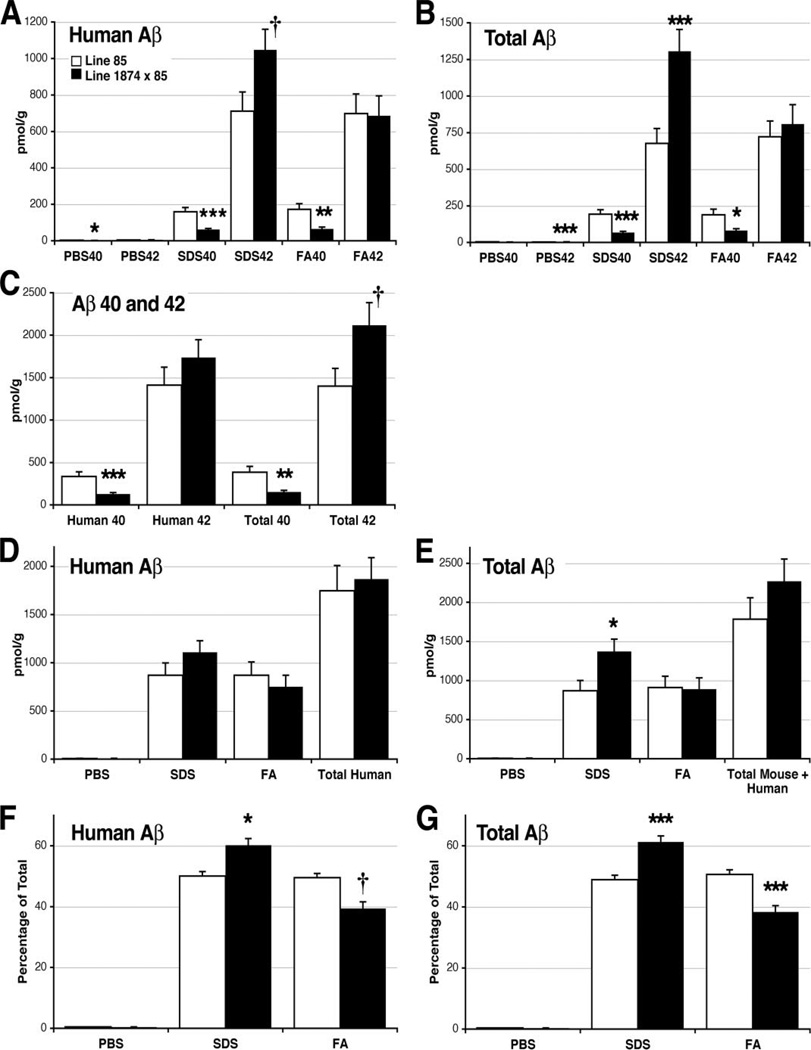
By contrast to the filter-trap assay, ELISA measurements of Aβ showed that the levels of total human Aβ in 8-month-old 1874 × 85 mice were similar to that of age-matched line 85 mice (Table 2). However, these ELISA experiments used a three-step sequential extraction and sedimentation protocol. Tissue homogenates were first extracted with PBS, followed by 2% SDS, and finally by 70% formic acid, revealing interesting differences in amyloid solubility at each step. As a fraction of the total human Aβ in each group, more of the accumulated Aβ was soluble in SDS in line 1874 × 85 mice than in line 85 mice, leaving less Aβ to recover in the final formic acid extraction (Table 2 and Fig. 9; SDS: 50.1 ± 1.4% (line 85) versus 60.2 ± 2.2% (line 1874 × 85), ANOVA for all four genotypes: F3,20 = 78.85, p < 0.001, Tukey post-hoc for pairwise comparison of line 85 versus 1874 × 85 p = 0.017; FA: 49.5 ± 1.5% (line 85) versus 39.4 ± 2.18% (line 1874 × 85), ANOVA: F3,20 = 45.53, p < 0.001, Tukey post-hoc p = 0.063, however direct comparison by Student’s t test yielded p < 0.001). Underlying this trend was a 47% increase in the amount of SDS-soluble human Aβ42 (710.6 pmol/g (line 85) versus 1047.3 pmol/g (line 1874 × 85); ANOVA: F3,20 = 22.68, p < 0.001, Tukey post-hoc p = 0.075, with direct comparison by Student’s t test, p = 0.047) along with a 62% decrease in the amount of FA-soluble human Aβ40 (172.2 pmol/g (line 85) versus 65.0 pmol/g (line 1874 × 85); ANOVA: F3,20 = 13.11, p < 0.001, Tukey post-hoc p = 0.005).
TABLE 2. ELISA measurement of human and total (mouse plus human) Aβ in brain tissue from 8-month-old mice.
Values are in picomoles/g wet tissue weight (±S.E.).
| PBS |
SDS |
FA |
n | ||||
|---|---|---|---|---|---|---|---|
| 40 | 42 | 40 | 42 | 40 | 42 | ||
| pmol/g | |||||||
| Human Aβ-BAN50 capture | |||||||
| 85 | 2.98 (0.33) | 4.13 (0.41) | 159.5 (23.4) | 710.6 (106.0) | 172.2 (31.3) | 697.6 (107.9) | 8 |
| 1874 × 85 | 2.15 (0.077) | 5.20 (0.34) | 61.9 (5.61) | 1047 (112.9) | 65.0 (10.1) | 685.8 (109.5) | 8 |
| Total Aβ-BNT77 capture | |||||||
| 85 | 3.56 (0.35) | 2.58 (0.25) | 192.9 (30.6) | 676.6 (102.1) | 118.4 (39.26) | 721.8 (107.8) | 8 |
| 1874 × 85 | 3.19 (0.10) | 3.79 (0.17) | 66.8 (8.61) | 1306 (148.6) | 81.7 (11.82) | 808.7 (132.1) | 8 |
FIGURE 9. ELISA analysis of aggregated Aβ in 8-month-old transgenic mice.
Brain homogenates were sequentially extracted with PBS, 2% SDS, and FA before each fraction was assayed for human-specific and total (mouse plus human) Aβ40 and Aβ42. Data from this experiment are tabulated in Table 2. A, accumulation of human Aβ40 was significantly reduced in all three fractions by the expression of exogenous mouse APP. Conversely, human Aβ42 levels increased in the SDS-fraction of triple transgenic mice. B, total Aβ levels (mouse plus human) mirror the differences found in human Aβ. SDS- and FA-soluble Aβ40 was significantly lower, whereas SDS-soluble Aβ42 was significantly higher, in line 1874 × 85 than in line 85. C, overexpression of mouse APP/Aβ significantly decreased the accumulation of both human and total (mouse plus human) Aβ40 summed across all three fractions. The accumulated sum of human Aβ42 is not significantly changed; however, the amount of total (mouse plus human) Aβ42 is substantially higher in the 1874 × 85 mice, suggesting that mouse Aβ42 may account for the extra peptide. D, the two genotypes harbor statistically indistinguishable amounts of human Aβ (40 plus 42) in each fraction and accumulate nearly identical amounts of total human peptide (PBS plus SDS plus FA). E, total mouse plus human Aβ (40 plus 42) differs between the two genotypes only in the SDS-soluble fraction. Despite this increase in SDS-soluble peptide, the overall amount of total Aβ (PBS plus SDS plus FA) in line 1874 × 85 is statistically indistinguishable from line 85. F, although the absolute amount of human Aβ extracted into each fraction is identical in each line, the relative levels differ substantially. A greater fraction of the accumulated Aβ is soluble in SDS in line 1874 × 85 than in line 85. The 1874 × 85 animals show an attendant decrease in the percentage of FA-soluble peptide. G, similar to the case for human Aβ, a greater fraction of total mouse plus human Aβ is soluble in SDS, and correspondingly less in FA, in the 1874 × 85 animals. Data are shown ± S.E. *, p < 0.05; **, p < 0.01; and ***, p < 0.005 versus line 85 by ANOVA/Tukey post-hoc; †, p < 0.05 versus line 85 by Student’s t test (but not by ANOVA).
The addition of excess mouse APP significantly decreased the accumulation of human Aβ40 in all three fractions (PBS: 28% decrease; 2.98 pmol/g (line 85) versus 2.15 pmol/g (line 1874 × 85), ANOVA for all four genotypes: F3,20 = 38.19, p < 0.001; Tukey post-hoc for pairwise comparison of 85 versus 1874 × 85 p = 0.041; SDS: 61% decrease, 159.5 pmol/g (line 85) versus 61.9 pmol/g (line 1874 × 85), ANOVA: F3,20 = 21.15, p < 0.001, Tukey post-hoc p < 0.001; FA: 62% decrease described above) causing a 61% overall decrease in total Aβ40 (334.7 pmol/g (line 85) versus 129.1 pmol/g (line 1874 × 85), ANOVA: F3,20 = 16.93, p < 0.001, Tukey post-hoc p = 0.002). By contrast, the amount of human Aβ42 did not differ significantly between the two groups.
In contrast to the marked decrease in Aβ 40, the amount of total Aβ42 (mouse plus human) was substantially higher in the 8-month-old 1874 × 85 mice than in their double transgenic siblings (Fig. 8; 1400.9 pmol/g (line 85) versus 2118.3 pmol/g (line 1874 × 85), ANOVA for all four genotypes: F3,20 = 19.55, p < 0.001; Tukey post-hoc for pairwise comparison of 85 versus 1874 × 85 p = 0.082; with direct comparison by Student’s t test, p = 0.05). The increase in Aβ42 was composed primarily from a 93% gain in SDS-soluble peptide (676.6 pmol/g (line 85) versus 1305.8 pmol/g (line 1874 × 85), ANOVA: F3,20 = 24.97, p < 0.001, Tukey post-hoc p = 0.003). The higher level of total Aβ42 in the 1874 × 85 mice suggests a substantial accumulation of mouse peptide.
N-terminally Truncated Mouse Aβ Is Not a Major Component of Amyloid Formed in Transgenic Mice
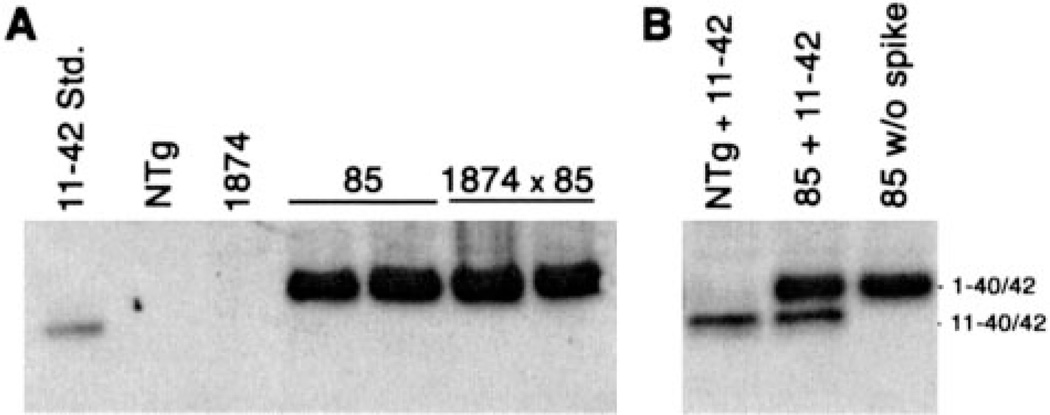
Prior studies of the interaction between BACE1 and APP suggested that the endogenous protease preferentially cleaves rodent APP at the +11 site within Aβ (12, 22–25). This cleavage generates predominantly N-terminally truncated Aβ 11-x from moAPP. To determine whether the alterations in amyloid solubility and distribution in the triple transgenic mice correlated with co-deposition of mouse Aβ11-x, we immunoprecipitated Aβ from brain homogenates of both line 85 and 1874 × 85 mice using an antibody that would detect both full-length and 11-x peptides (4G8). To our surprise, we found no evidence of Aβ11-40/42 in the brains of triple transgenic mice (Fig. 10). In contrast to the intense signal from full-length 1–40/42 peptides, Aβ11–40/42 was undetectable in the immunoprecipitates. Control experiments clearly demonstrated that 4G8 was capable of immunoprecipitating and detecting synthetic Aβ11-x added to homogenates prepared from line 85 mice with high amyloid burden (Fig 10). Moreover, these immunoprecipitations were highly sensitive: as little as 10 ng of exogenous Aβ11–42 could be recovered and detected from 1 mg of mouse brain protein by this method. Thus, we conclude that mouse Aβ11–42 is not a major component of the amyloid deposited in our triple transgenic animals and that factors other than peptide length must influence the solubility and localization of human Aβ in the line 1874 × 85 animals.
FIGURE 10. N-terminally truncated mouse Aβ is not a major component of amyloid aggregates.
A, immunoprecipitation of Aβ from brain homogenates of 85 and 1874 × 85 mice demonstrates the abundance of full-length Aβ, but fails to detect any sign of N-terminally truncated mouse Aβ 11-x. Peptide was immunoprecipitated and detected with purified 4G8, which binds a mid-region epitope common to both mouse and human Aβ. 5 ng of synthetic human Aβ11–42 was run alongside the IP samples as a positive control. B, immunoprecipitation of synthetic Aβ11–42 spiked into NTg or line 85 brain homogenates provides proof that 4G8 is capable of immunoprecipitating the N-terminally truncated peptide when present. 50 ng of added peptide is shown here, however, as little as 10 ng of exogenous Aβ11–42 could be recovered by immunoprecipitation under conditions identical to those shown in panel A.
DISCUSSION
We set out to test the amyloidogenic potential of mouse Aβ produced in its normal environment within the mouse brain and to understand how the presence of mouse Aβ influences the deposition of human Aβ in transgenic models for Alzheimer disease. Previous studies have shown that full-length mouse Aβ aggregates as readily as the human peptide in vitro and that the two species form co-polymers that are indistinguishable from pure human fibrils (16, 17). Our findings indicate a more complicated picture when these processes occur in vivo. We show that mouse Aβ on its own does not promote the formation of mature senile plaques as aggressively as the human peptide. We further show that mouse Aβ does not enhance the rate or severity of amyloid formed from human Aβ. Instead, the presence of excess mouse peptide had more subtle effects on human Aβ aggregation. The added mouse Aβ altered the detergent solubility of the human peptides and shifted the Aβ40:42 ratio of the aggregated human peptide; human Aβ40 levels in the triple transgenic 1874 × 85 mice were >50% lower than that of their age-matched line 85 siblings. Excess mouse Aβ also increased the relative burden of amyloid deposited around cortical blood vessels. Overall, we conclude that the addition of excess mouse APP/Aβ has multiple effects on the solubility, location, and composition of the amyloid deposited in mice producing human Aβ.
By itself, we found that overexpression of mouse Aβ did not lead to amyloid formation in moAPP transgenic mice. The lack of deposition could be due to inadequate production of the mouse peptide in our moAPP transgenic line. Transgenic mouse APP was expressed at levels 3- to 4-fold that of endogenous APP, but we had not included mutations that could have augmented overall Aβ production. However, increasing the relative production of Aβ42 by co-expressing PS1dE9 with the mouse APP transgene also failed to produce cored-amyloid deposits. Perhaps the most telling indicator of the minimal ability of mouse Aβ to induce deposition was its lack of effect on senile plaque formation in mice overproducing both mouse and human peptides. Despite its inability to influence the rate or extent of amyloid formation in the moAPP × APPswe/PS1dE9 triple transgenic animals, ELISA measurements of total Aβ levels suggest that mouse Aβ was indeed accumulating alongside the human peptide. Although the levels of human Aβ were nearly identical, 8-month-old triple transgenic animals harbored 30% more total Aβ (mouse plus human) than their age-matched APPswe/PS1dE9 siblings. The increase in total accumulated Aβ after the onset of amyloid deposition is roughly equivalent to the steady-state overproduction of Aβ in pre-deposit triple transgenic mice. Thus, mouse Aβ accumulation in these animals is equal to its overproduction, but the accumulation occurs without changing the rate or extent of amyloid burden. Intriguingly, the extra Aβ in the triple transgenic mice is composed almost entirely of SDS-soluble material.
The presence of exogenous mouse Aβ unexpectedly influenced the solubility of human Aβ accumulated in the brains of moAPP × APPswe/PS1dE9 mice. This effect suggests a close physical interaction between the two peptides. Previous studies comparing amyloid formed in several transgenic mouse models to that from human AD patients had shown that plaques formed in mice from the human peptide were more soluble in detergents than amyloid formed in the human brain (45, 46). Our data suggest that the higher solubility of amyloid formed in transgenic mice is due in large part to the presence of mouse peptide rather than differences in post-translational modification or local microenvironment. Three amino acids substitutions at positions 5, 10, and 13 may underlie this effect of mouse Aβ. In particular, the His to Arg substitution at position 13 disrupts a metal binding site in the mouse peptide (18–20, 47), which could influence the structure of the resulting mouse-human co-aggregates.
One of the more striking outcomes of co-expressing transgenic mouse APP with the mutant human protein was a dramatic increase in the appearance of vascular amyloid. A similar shift from parenchymal to vascular Aβ deposition has also been noted in several studies that have experimentally manipulated the ratio of Aβ40:42. However, depending on the mouse model studied, both elevated and reduced Aβ40:42 have been associated with increased cerebral amyloid angiopathy. Experimental manipulations such as co-expressing human apoE4 with APPswe (48), or introducing the E693Q Dutch mutation into APP transgenic mice (49), both elevated the ratio of Aβ40:42 and caused redistribution of amyloid deposits from the parenchyma to the vasculature. Conversely, several experiments designed to specifically lower the production of Aβ40 relative to Aβ42, such as co-expressing mutant PS1 with APP (50, 51) or expressing an Aβ42-exclusive transgene (52), are also reported to increase cerebral amyloid angiopathy in transgenic mice. Our current results are most consistent with the hypothesis that lowering Aβ40:42 increases the appearance of vascular amyloid. The triple transgenic animals accumulated considerably more Aβx-42 (and considerably less Aβx-40) than their double transgenic siblings and developed greater vascular pathology. Whether these two factors were causally related is unclear; it is most likely that the Aβ40:42 ratio is one of several factors that contributed to the increased amyloid angiopathy in our line 1874 × 85 mice.
Despite showing a significant effect on the solubility, composition, and location of human β-amyloid, the distribution of the accumulated mouse Aβ is unclear. Immunohistochemical analyses demonstrated that some mouse Aβ co-deposits with human peptide in cored senile plaques. In addition, the triple transgenic animals have considerably more amyloid in and around cortical blood vessels. The worsening of vascular amyloid is consistent with the overall increase in total Aβ levels in the triple transgenic mice. However, vascular deposits in these animals were reactive with both human-specific and rodent Aβ antibodies, and thus we cannot conclude that mouse Aβ initiates this pathology or concentrates in this location. That the majority of extra Aβ (presumably mouse Aβ) harbored in the triple transgenics is soluble in SDS could mean that this peptide accumulates as diffuse amyloid. Mouse Aβ may also be accumulating in oligomeric structures that could be difficult to detect histologically yet release substantial amounts of peptide into detergent extracts.
One motivation behind this study was to resolve a question that arose regarding our recent study of mice that express mutant APP via an inducible promoter (53). Induction of mutant APP expression for 6 months resulted in robust amyloid pathology that, although arrested by turning off the transgene, was not significantly diminished following long periods of suppression. One issue we could not resolve in these studies was whether mouse Aβ, which was not regulated by the transgene, could provide a source of peptide that would have maintained an equilibrium favoring amyloid stability. The experiments we present here suggest that this was likely not the case: mouse Aβ, even when present in excess, did not appear to promote senile plaque formation on its own or enhance the deposition of human Aβ in these structures. Instead, we find that mouse Aβ would decrease stability of aggregates formed by co-deposition of both peptides. These data make it unlikely that the presence of small amounts of endogenous mouse Aβ would have the ability to sustain amyloid in our inducible APP mice. We therefore conclude that the persistence of amyloid in our APP-inducible mice is due to inherent stability of the human amyloid.
Taken together, our studies demonstrate that mouse Aβ produced at 3- to 4-fold wild-type levels does not drive amyloid formation in vivo nor does it accelerate the deposition of human Aβ in mice overproducing both peptides. However, mouse Aβ does accumulate with the human peptide, where it increases the appearance of vascular deposits and alters the overall solubility of resulting amyloid. These effects appear to be mediated by full-length mouse Aβ 1–40/42; Aβ 11–40/42, the predominant BACE product described in vitro, does not accumulate to significant levels in vivo. That the solubility and location of human Aβ aggregates can be influenced by the presence of mouse peptide suggests that a better understanding of the effects of peptide sequence and species-specific processing may provide new insight into disease mechanisms.
Acknowledgments
We thank Gay Rudow for advice on stereology, Eduardo Marcora for help with the Odyssey imager, and Neil Segil for sharing his ultracentrifuge for this work. We are grateful to Takeda Chemical Industries Co., Ltd. for providing antibodies BAN50, BNT77, BA27, and BC05, and to Konrad Beyreuther and Andreas Weidemann for sharing the 22C11 antibody. We thank David Teplow for providing mouse and human Aβ peptide standards, and David Holtzman for sharing peptide electrophoresis protocols.
Footnotes
This work was supported in part by NIA, National Institutes of Health Grants 1 P50 AG-14-248 and 1 P01 AG-98-003 (to D. R. B.) and AG-06-656 (to S. G. Y.), the Robert H. and Clarice Smith and Abigail Van Buren Alzheimer’s Disease Research Program (to S. G. Y.).
Supported by the John Douglas French Alzheimer’s Foundation and by NIA, National Institutes of Health Grant KO1 AG-26144-01.
The abbreviations used are: APP, amyloid precursor protein; Aβ, amyloid-β; BACE, β-APP cleaving enzyme; PrP, prion protein promoter; PS1, presenilin-1; dE9, PS1 encoding the exon-9 deletion mutation; DEA, diethylamine; ELISA, enzyme-linked immunosorbent assay; FA, formic acid; FAD, familial Alzheimer disease; hu, human; mo, mouse; ro, rodent; mo/hu, chimeric mouse/human; swe, APP encoding the Swedish mutation; wt, wild type; NTg, non-transgenic; PBS, phosphate-buffered saline; MES, 4-morpholineethanesulfonic acid; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; TBS, Tris-buffered saline; ANOVA, analysis of variance; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)-propane-1,3-diol; CHAPS, 3-[{3-cholamidopropyl)dimethylammonio}]-1-propanesulfonic acid.
REFERENCES
- 1.Shivers BD, Hilbich C, Multhaup G, Salbaum M, Beyreuther K, Seeburg PH. EMBO J. 1988;7:1365–1370. doi: 10.1002/j.1460-2075.1988.tb02952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selkoe DJ, Bell DS, Podlisny MB, Price DL, Cork LC. Science. 1987;235:873–877. doi: 10.1126/science.3544219. [DOI] [PubMed] [Google Scholar]
- 3.Struble RG, Price DL, Jr, Cork LC, Price DL. Brain Res. 1985;361:267–275. doi: 10.1016/0006-8993(85)91298-3. [DOI] [PubMed] [Google Scholar]
- 4.Wisniewski HM, Ghetti B, Terry RD. J. Neuropathol. Exp. Neurol. 1973;32:566–584. doi: 10.1097/00005072-197310000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Wisniewski H, Johnson AB, Raine CS, Kay WJ, Terry RD. Lab. Invest. 1970;23:287–296. [PubMed] [Google Scholar]
- 6.Johnstone EM, Chaney MO, Norris FH, Pascual R, Little SP. Brain Res. Mol. Brain Res. 1991;10:299–305. doi: 10.1016/0169-328x(91)90088-f. [DOI] [PubMed] [Google Scholar]
- 7.Yamada T, Sasaki H, Dohura K, Goto I, Sakaki Y. Biochem. Biophys. Res. Commun. 1989;158:906–912. doi: 10.1016/0006-291x(89)92808-8. [DOI] [PubMed] [Google Scholar]
- 8.Yamada T, Sasaki H, Furuya H, Miyata T, Goto I, Sakaki Y. Biochem. Biophys. Res. Commun. 1987;149:665–671. doi: 10.1016/0006-291x(87)90419-0. [DOI] [PubMed] [Google Scholar]
- 9.Kang J, Muller-Hill B. Nucleic Acids Res. 1989;17:2130. doi: 10.1093/nar/17.5.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selkoe DJ. Annu. Rev. Neurosci. 1989;12:463–490. doi: 10.1146/annurev.ne.12.030189.002335. [DOI] [PubMed] [Google Scholar]
- 11.Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR. Hum. Mol. Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 12.Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC. Nat. Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- 13.Hilbich C, Kisters-Woike B, Reed J, Masters CL, Beyreuther K. J. Mol. Biol. 1991;218:149–163. doi: 10.1016/0022-2836(91)90881-6. [DOI] [PubMed] [Google Scholar]
- 14.Jarrett JT, Berger EP, Lansbury PT., Jr Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 15.Jarrett JT, Lansbury PT., Jr Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 16.Hilbich C, Kisters-Woike B, Reed J, Masters CL, Beyreuther K. Eur. J. Biochem. 1991;201:61–69. doi: 10.1111/j.1432-1033.1991.tb16256.x. [DOI] [PubMed] [Google Scholar]
- 17.Fung J, Frost D, Chakrabartty A, McLaurin J. J. Neurochem. 2004;91:1398–1403. doi: 10.1111/j.1471-4159.2004.02828.x. [DOI] [PubMed] [Google Scholar]
- 18.Yang DS, McLaurin J, Qin K, Westaway D, Fraser PE. Eur. J. Biochem. 2000;267:6692–6698. doi: 10.1046/j.1432-1327.2000.01767.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu ST, Howlett G, Barrow CJ. Biochemistry. 1999;38:9373–9378. doi: 10.1021/bi990205o. [DOI] [PubMed] [Google Scholar]
- 20.Bush AI, Pettingell WH, Multhaup G, d Paradis M, Vonsattel JP, Gusella JF, Beyreuther K, Masters CL, Tanzi RE. Science. 1994;265:1464–1467. doi: 10.1126/science.8073293. [DOI] [PubMed] [Google Scholar]
- 21.De Strooper B, Simons M, Multhaup G, Van Leuven F, Beyreuther K, Dotti CG. EMBO J. 1995;14:4932–4938. doi: 10.1002/j.1460-2075.1995.tb00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R, Sweeney D, Gandy SE, Sisodia SS. J. Biol. Chem. 1996;271:31894–31902. doi: 10.1074/jbc.271.50.31894. [DOI] [PubMed] [Google Scholar]
- 23.Huse JT, Liu K, Pijak DS, Carlin D, Lee VM, Doms RW. J. Biol. Chem. 2002;277:16278–16284. doi: 10.1074/jbc.M111141200. [DOI] [PubMed] [Google Scholar]
- 24.Liu K, Doms RW, Lee VM. Biochemistry. 2002;41:3128–3136. doi: 10.1021/bi015800g. [DOI] [PubMed] [Google Scholar]
- 25.Gouras GK, Xu H, Jovanovic JN, Buxbaum JD, Wang R, Green-gard P, Relkin NR, Gandy S. J. Neurochem. 1998;71:1920–1925. doi: 10.1046/j.1471-4159.1998.71051920.x. [DOI] [PubMed] [Google Scholar]
- 26.Pike CJ, Overman MJ, Cotman CW. J. Biol. Chem. 1995;270:23895–23898. doi: 10.1074/jbc.270.41.23895. [DOI] [PubMed] [Google Scholar]
- 27.Bitan G, Vollers SS, Teplow DB. J. Biol. Chem. 2003;278:34882–34889. doi: 10.1074/jbc.M300825200. [DOI] [PubMed] [Google Scholar]
- 28.Pype S, Moechars D, Dillen L, Mercken M. J. Neurochem. 2003;84:602–609. doi: 10.1046/j.1471-4159.2003.01556.x. [DOI] [PubMed] [Google Scholar]
- 29.Calhoun ME, Burgermeister P, Phinney AL, Stalder M, Tolnay M, Wiederhold KH, Abramowski D, Sturchler-Pierrat C, Sommer B, Staufenbiel M, Jucker M. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14088–14093. doi: 10.1073/pnas.96.24.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsiao KK, Borchelt DR, Olson K, Johannsdottir R, Kitt C, Yunis W, Xu S, Eckman C, Younkin S, Price D, Iadecola C, Clark HB, Carlson G. Neuron. 1995;15:1203–1218. doi: 10.1016/0896-6273(95)90107-8. [DOI] [PubMed] [Google Scholar]
- 31.Lesuisse C, Xu G, Anderson J, Wong M, Jankowsky J, Holtz G, Gonzalez V, Wong PC, Price DL, Tang F, Wagner S, Borchelt DR. Hum. Mol. Genet. 2001;10:2525–2537. doi: 10.1093/hmg/10.22.2525. [DOI] [PubMed] [Google Scholar]
- 32.Borchelt DR, Davis J, Fischer M, Lee MK, Slunt HH, Ratovitsky T, Regard J, Copeland NG, Jenkins NA, Sisodia SS, Price DL. Genet Anal. 1996;13:159–163. doi: 10.1016/s1050-3862(96)00167-2. [DOI] [PubMed] [Google Scholar]
- 33.Lee MK, Borchelt DR, Kim G, Thinakaran G, Slunt HH, Ratovitski T, Martin LJ, Kittur A, Gandy S, Levey AI, Jenkins N, Copeland N, Price DL, Sisodia SS. Nat. Med. 1997;3:756–760. doi: 10.1038/nm0797-756. [DOI] [PubMed] [Google Scholar]
- 34.Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR. Biomol. Eng. 2001;17:157–165. doi: 10.1016/s1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- 35.Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 36.Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH, Jr, Price DL, Sisodia SS, Cleveland DW. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki N, Cheung TT, Cai XD, Odaka A, Otvos L, Jr, Eckman C, Golde TE, Younkin SG. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 38.Gravina SA, Ho L, Eckman CB, Long KE, Otvos L, Jr, Younkin LH, Suzuki N, Younkin SG. J. Biol. Chem. 1995;270:7013–7016. doi: 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- 39.Kawarabayashi T, Younkin LH, Saido TC, Shoji M, Ashe KH, Younkin SG. J. Neurosci. 2001;21:372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asami-Odaka A, Ishibashi Y, Kikuchi T, Kitada C, Suzuki N. Biochemistry. 1995;34:10272–10278. doi: 10.1021/bi00032a022. [DOI] [PubMed] [Google Scholar]
- 41.Hirano A, Zimmermann HM. Arch. Neurol. 1962;6:114–122. doi: 10.1001/archneur.1962.00450200028003. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto T, Hirano A. Neuropathol. Appl. Neurobiol. 1986;12:3–9. doi: 10.1111/j.1365-2990.1986.tb00677.x. [DOI] [PubMed] [Google Scholar]
- 43.Slunt HH, Thinakaran G, Von Koch C, Lo AC, Tanzi RE, Sisodia SS. J. Biol. Chem. 1994;269:2637–2644. [PubMed] [Google Scholar]
- 44.Xu G, Gonzales V, Borchelt DR. Alzheimer Dis. Assoc. Disord. 2002;16:191–195. doi: 10.1097/00002093-200207000-00010. [DOI] [PubMed] [Google Scholar]
- 45.Kalback W, Watson MD, Kokjohn TA, Kuo YM, Weiss N, Luehrs DC, Lopez J, Brune D, Sisodia SS, Staufenbiel M, Emmerling M, Roher AE. Biochemistry. 2002;41:922–928. doi: 10.1021/bi015685+. [DOI] [PubMed] [Google Scholar]
- 46.Kuo YM, Kokjohn TA, Beach TG, Sue LI, Brune D, Lopez JC, Kalback WM, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Roher AE. J. Biol. Chem. 2001;276:12991–12998. doi: 10.1074/jbc.M007859200. [DOI] [PubMed] [Google Scholar]
- 47.McLaurin J, Fraser PE. Eur. J. Biochem. 2000;267:6353–6361. doi: 10.1046/j.1432-1327.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 48.Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, Holtzman DM. J. Neurosci. 2005;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herzig MC, Winkler DT, Burgermeister P, Pfeifer M, Kohler E, Schmidt SD, Danner S, Abramowski D, Sturchler-Pierrat C, Burki K, van Duinen SG, Maat-Schieman ML, Staufenbiel M, Mathews PM, Jucker M. Nat. Neurosci. 2004;7:954–960. doi: 10.1038/nn1302. [DOI] [PubMed] [Google Scholar]
- 50.Van Dooren T, Muyllaert D, Borghgraef P, Cresens A, Devijver H, Van der Auwera I, Wera S, Dewachter I, Van Leuven F. Am. J. Pathol. 2006;168:245–260. doi: 10.2353/ajpath.2006.050752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Dorpe J, Smeijers L, Dewachter I, Nuyens D, Spittaels K, Van Den Haute C, Mercken M, Moechars D, Laenen I, Kuiperi C, Bruyn-seels K, Tesseur I, Loos R, Vanderstichele H, Checler F, Sciot R, Van Leuven F. Am. J. Pathol. 2000;157:1283–1298. doi: 10.1016/S0002-9440(10)64644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Onstead L, Randle S, Price R, Smithson L, Zwizinski C, Dickson DW, Golde T, McGowan E. J. Neurosci. 2007;27:627–633. doi: 10.1523/JNEUROSCI.4849-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jankowsky JL, Slunt HH, Gonzales V, Savonenko AV, Wen JC, Jenkins NA, Copeland NG, Younkin LH, Lester HA, Younkin SG, Borchelt DR. PLoS Med. 2005:e355. doi: 10.1371/journal.pmed.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Tanila H, Puolivali J, Kadish I, van Groen T. Neurobiol. Dis. 2003;14:318–327. doi: 10.1016/j.nbd.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Callahan MJ, Lipinski WJ, Bian F, Durham RA, Pack A, Walker LC. Am. J. Pathol. 2001;158:1173–1177. doi: 10.1016/s0002-9440(10)64064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]