ABSTRACT
Reactive chlorine species (RCS) defense mechanisms are important for bacterial fitness in diverse environments. In addition to the anthropogenic use of RCS in the form of bleach, these compounds are also produced naturally through photochemical reactions of natural organic matter and in vivo by the mammalian immune system in response to invading microorganisms. To gain insight into bacterial RCS defense mechanisms, we investigated Azospira suillum strain PS, which produces periplasmic RCS as an intermediate of perchlorate respiration. Our studies identified an RCS response involving an RCS stress-sensing sigma/anti-sigma factor system (SigF/NrsF), a soluble hypochlorite-scavenging methionine-rich periplasmic protein (MrpX), and a putative periplasmic methionine sulfoxide reductase (YedY1). We investigated the underlying mechanism by phenotypic characterization of appropriate gene deletions, chemogenomic profiling of barcoded transposon pools, transcriptome sequencing, and biochemical assessment of methionine oxidation. Our results demonstrated that SigF was specifically activated by RCS and initiated the transcription of a small regulon centering around yedY1 and mrpX. A yedY1 paralog (yedY2) was found to have a similar fitness to yedY1 despite not being regulated by SigF. Markerless deletions of yedY2 confirmed its synergy with the SigF regulon. MrpX was strongly induced and rapidly oxidized by RCS, especially hypochlorite. Our results suggest a mechanism involving hypochlorite scavenging by sacrificial oxidation of the MrpX in the periplasm. Reduced MrpX is regenerated by the YedY methionine sulfoxide reductase activity. The phylogenomic distribution of this system revealed conservation in several Proteobacteria of clinical importance, including uropathogenic Escherichia coli and Brucella spp., implying a putative role in immune response evasion in vivo.
IMPORTANCE
Bacteria are often stressed in the environment by reactive chlorine species (RCS) of either anthropogenic or natural origin, but little is known of the defense mechanisms they have evolved. Using a microorganism that generates RCS internally as part of its respiratory process allowed us to uncover a novel defense mechanism based on RCS scavenging by reductive reaction with a sacrificial methionine-rich peptide and redox recycling through a methionine sulfoxide reductase. This system is conserved in a broad diversity of organisms, including some of clinical importance, invoking a possible important role in innate immune system evasion.
INTRODUCTION
Bacteria are often stressed in their natural environment by reactive chlorine species (RCS) of either anthropogenic or natural origin. RCS are often byproducts of antiseptic disinfecting agents of drinking water supplies, are used in the form of bleach in household products, and are produced naturally through photochemical reactions of organic and inorganic chlorine species in the environment (1). Furthermore, RCS production is a first-line defense of the innate immune system and mucosal epithelia of eukaryotes against invading microorganisms (2). A lesser known biological source of RCS is bacterial chlorate and perchlorate [collectively referred to as (per)chlorate] reduction, a respiratory process carried out by members of the Proteobacteria known as dissimilatory perchlorate-reducing bacteria (DPRB) (3). These bacteria are obligate respirers that use (per)chlorate as an electron acceptor, reducing it to chlorite in the periplasm with the heterodimer perchlorate reductase (PcrAB) enzyme (4). Chlorite is then converted to molecular oxygen and chloride by the chlorite dismutase (Cld) enzyme, which is conserved in both chlorate and perchlorate reducers (5, 6). Recent biochemical investigations (7) revealed the production of the RCS hypochlorite (OCl−) as an intermediate formed in micromolar quantities by Cld during its mediation of chlorite dismutation. This was responsible for irreversible enzyme inhibition through oxidative protein damage (7).
RCS have a unique profile of reactivity toward amino acids compared with that of common reactive oxygen species (ROS). One striking example of this is the rate of methionine oxidation by hypochlorite (3.8 × 107 M−1 ⋅ s−1) (8), which is significantly higher than that of hydrogen peroxide (~10−2 M−1 ⋅ s−1) (9). Methionine sulfoxide formation thus plays an important role in the mechanism of bacterial killing by hypochlorite produced in phagocytes by the myeloperoxidase enzyme (10). Methionine oxidation is also the activating mechanism of the hypochlorite-specific transcription factor HypT in Escherichia coli strain K-12 (11, 12). E. coli K-12 contains two other transcription factors that are responsive to hypochlorite (13). These transcription factors are conserved in other gammaproteobacteria, further suggesting that RCS defense systems are important for bacterial fitness in diverse environments.
During our previous investigations on perchlorate respiration in the model perchlorate reducer Azospira suillum strain PS, we found that perchlorate inhibited growth with nitrate by a Δcld mutant but had no effect on a ΔpcrA Δcld double mutant (14). We proposed that the epistatic interaction between these two genes was due to accumulation of RCS in the Δcld mutant via inadvertent reduction of perchlorate by basal synthesis of PcrA, which was eliminated in the ΔpcrA Δcld double mutant. DPRB have a horizontally transferred perchlorate reduction genomic island (PRI) that contains the genes responsible for perchlorate reduction, including pcrA and cld (15). Markerless deletions of each of the 17 genes in the PRI of PS demonstrated that 7 have no apparent role in perchlorate reduction yet are conserved to various extents in the PRIs of perchlorate reducers (14, 15). Two of these genes encode products similar to the ROS-responsive SigF-NrsF system described in two model alphaproteobacteria, Caulobacter crescentus and Bradyrhizobium japonicum (16, 17), and we hypothesized that SigF-NrsF in strain PS may also form part of an RCS response during perchlorate reduction. In this paper, we explore the role of the SigF-NrsF system and its regulon in RCS defense. In particular, we examined a methionine-rich peptide (MrpX) and a putative methionine sulfoxide reductase (YedY) that act in unison in the periplasm to scavenge hypochlorite.
RESULTS
SigF is required for Azospira suillum PS to respond to aerobic chlorite stress.
Our recent genetic analysis of A. suillum strain PS demonstrated that 9 of the 17 genes that compose the core of the PRI are dispensable for perchlorate reduction (14). Of these genes, two are homologous to the sigF-nrsF sigma factor/anti-sigma factor system, which has been shown to be responsive to reactive oxygen species in Caulobacter crescentus (16, 18) and Bradyrhizobium japonicum, where it is referred to as ecfF-osrA (17). In this system, SigF is bound by the membrane protein NrsF until the oxidative signal is sensed. This results in the release of SigF and transcription of its stress response regulon (17, 18). The ecfF-osrA sigma factor/anti-sigma factor system is an example of the ECF (extracytoplasmic function) sigma factor family, in which an anti-sigma factor transduces environmental signals into transcriptional outputs (19). As chlorite is a known intermediate of perchlorate respiration, we hypothesized that the presence of the sigF-nrsF system in DPRB was related to amelioration of RCS stress. To test this, we initially assayed the ability of ΔsigF (Dsui_0155) and ΔnrsF (Dsui_0154) mutants to respond to chlorite treatment when grown under oxic conditions in the absence of perchlorate by adding two aliquots of chlorite during log phase (Fig. 1A) and one initial aliquot after inoculation during lag phase (Fig. 1B). The addition of chlorite arrested aerobic growth of the ΔsigF mutant, despite only slightly inhibiting the wild-type strain PS (Fig. 1A), indicating that sigF endowed the strain with a resistance to RCS even under oxic conditions. In contrast, the ΔnrsF strain challenged with chlorite during lag phase grew identically to unchallenged wild-type PS and better than the challenged wild-type PS (Fig. 1B).
FIG 1 .
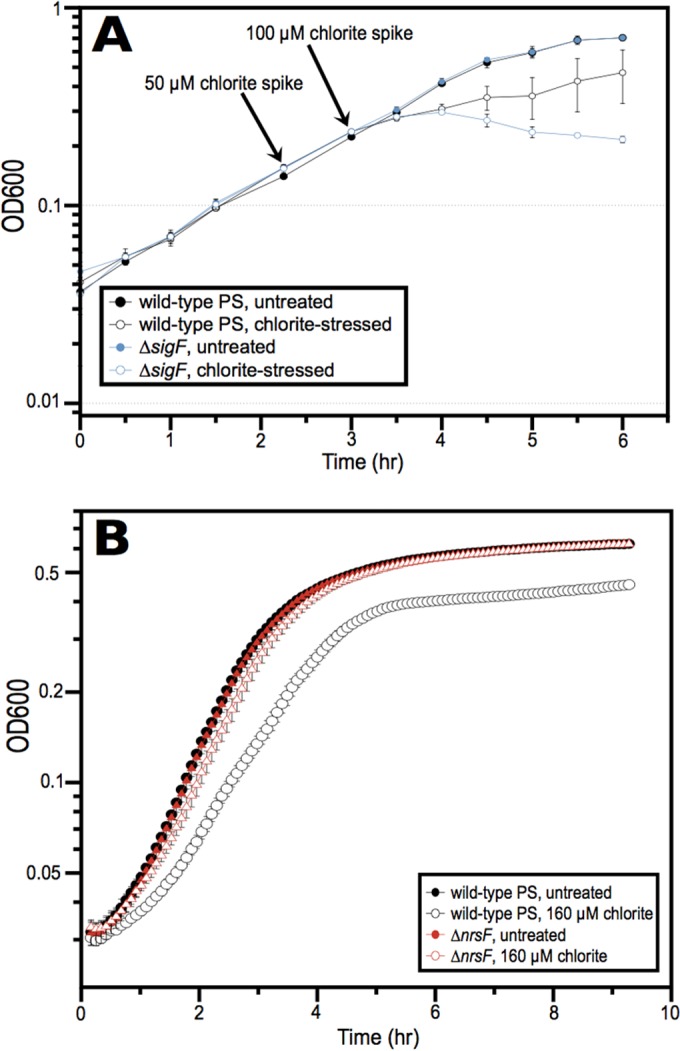
(A) Impact of chlorite treatment on wild-type and ΔsigF strains during log-phase growth in ALP medium. Chlorite was added in two successive spikes as indicated by the black arrows. (B) Difference in growth in ALP medium between wild-type and ΔnrsF cells when 160 µM chlorite is added immediately after inoculation during lag phase. OD600, optical density at 600 nm.
Although ΔsigF or ΔnrsF mutants did not have phenotypes during perchlorate reduction in previous work (14), this may have been due to the use of rich medium with an excess of electron donor. We surmised that oxidative stress may be more pronounced during growth in minimal medium with higher concentrations of electron acceptor, especially chlorate. Chlorate is reduced by PcrAB faster than perchlorate, potentially increasing the pool size of the RCS intermediates and resulting in increased oxidative stress. Using appropriately modified growth conditions to account for these aspects (see Materials and Methods), a reduction in the lag phase for the ΔnrsF mutant relative to the lag phase of the wild-type control was observed (Fig. 2).
FIG 2 .
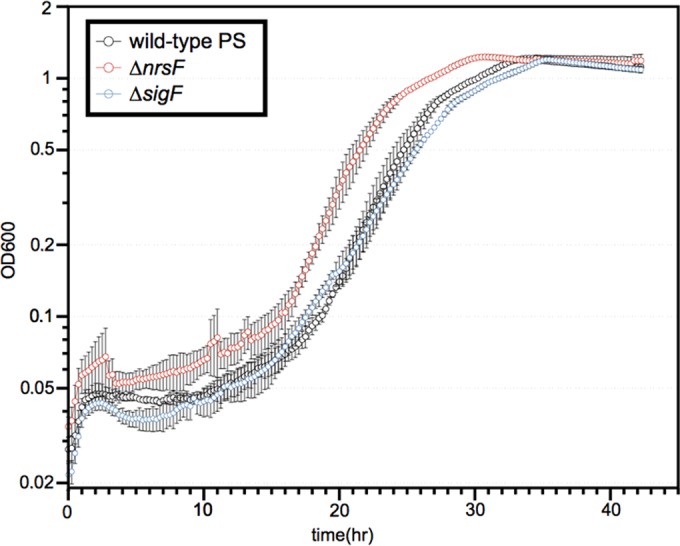
Anaerobic growth of wild-type, ΔsigF, and ΔnrsF strains in minimal medium containing 30 mM lactate and 20 mM chlorate.
An RNA-seq approach to identify the SigF regulon.
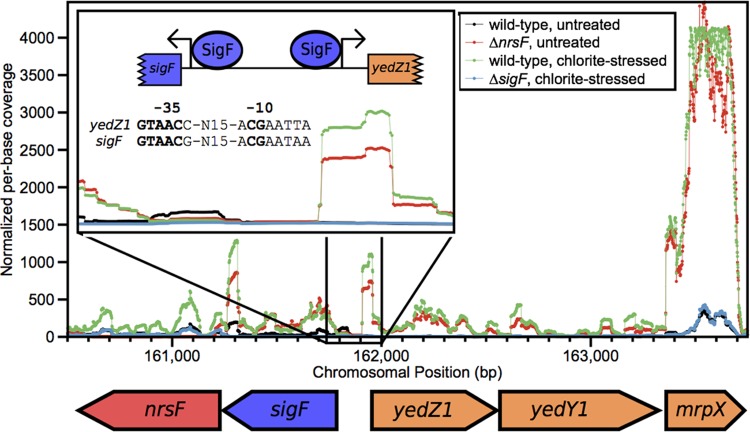
Because the SigF/NrsF system relies on a single anti-sigma factor that releases SigF in response to chlorite stress, ascertaining the regulon of SigF was possible with two experiments comparing RNA transcription in ΔsigF and ΔnrsF mutants relative to the RNA transcription in the wild type. We initially compared the global transcription of the chlorite-stressed ΔsigF mutant to that of chlorite-stressed wild-type PS (see Data Set S1 in the supplemental material) to identify genes specifically associated with chlorite stress. To delineate the subset of these genes whose transcription was a SigF-specific response, we also compared the transcriptomes of the unstressed ΔnrsF strain and wild-type PS (see Data Set S1). Comparative analysis revealed that only four genes were upregulated in the ΔnrsF mutant and downregulated in the ΔsigF mutant (Dsui_0156 to Dsui_0159 [Dsui_0156–Dsui_0159]). These genes compose an operon adjacent to the sigF-nrsF operon but on the opposite strand (Fig. 3). Many genes that were significantly regulated in one experiment but not the other were not deemed part of the SigF regulon (Data Set S1).
FIG 3 .
Average per-base coverage (y axis) across all RNA-seq replicates in the region of the sigF-nrsF and yedY1Z1-mrpX operons. The inset shows the transcriptional start sites that were identified and the SigF promoter found upstream from the start sites. The nucleotides also conserved in Caulobacter crescentus and Bradyrhizobium japonicum are indicated in bold.
Five genes carried in the PRI (Dsui_0154–Dsui_0158) were classified as the functional part of the SigF regulon (Fig. 3). Dsui_0156 and Dsui_0157 are annotated as yedY1 and yedZ1, which encode a molybdopterin-containing oxidoreductase complex that is conserved in many members of the Proteobacteria (20). YedZ is predicted to be an integral membrane protein, and YedY1 contains a signal for export via the Tat pathway. yedY homologs are also part of the SigF regulon in Bradyrhizobium japonicum and Caulobacter crescentus, but their role in the oxidative stress response is not known (17, 18). Dsui_0158 encodes a small protein composed almost entirely of methionines (20%) and charged residues (44%), which we have named MrpX (methionine-rich peptide X). MrpX contains a signal peptide, indicating that it is exported to the periplasm.
yedY1, yedZ1, and mrpX are carried on the same strand and likely form a single transcriptional unit. Likewise, the sigF and nrsF genes overlap and form another operon. We focused on the intergenic region between sigF and yedZ1 and identified two locations where the ΔnrsF and untreated wild-type transcriptome had increased coverage relative to the coverage in the ΔsigF mutant and the chlorite-stressed wild type (Fig. 3, inset). Upstream from these two putative transcriptional start sites, we found a conserved promoter similar to the SigF binding motifs reported for both Caulobacter crescentus and Bradyrhizobium japonicum (17, 18). The nucleotides that are conserved in the promoters in all three organisms are indicated in boldface in Fig. 3. We were unable to find a promoter with this structure anywhere else in the entire PRI.
We used a quantitative PCR (qPCR) assay to quantify the upregulation of mrpX after chlorite treatment, which also showed a 20- to 60-fold increase in transcription with respect to the level in an untreated control (see Fig. S1A in the supplemental material). This increase was on the same order of magnitude as observed in the RNA-seq experiments. Interestingly, mrpX was also upregulated to a similar degree by hypochlorite but not by hydrogen peroxide, demonstrating that SigF is an RCS-specific response and is not activated indiscriminately by all reactive oxygen species (see Fig. S1B).
BarSeq fitness profiling on different electron acceptors indicates that the SigF regulon is important during chlorate reduction.
Our previous genetic analysis showed that PRI genes are not important for aerobic growth or denitrification but that eight of them are essential for perchlorate reduction (14). To search for genes with more subtle phenotypes, including those outside the PRI, a saturated transposon mutant library containing unique TagModule bar codes in each insertion was generated (21). We then sequenced the library after growth under various conditions and used bar code abundance to calculate fitness values for individual genes (BarSeq).
The BarSeq data recapitulated previous clean deletion phenotypes of genes in the PRI; 8 of the 17 genes (pcrABCDPSR and cld) are essential for perchlorate reduction, and this is reflected by the significant fitness defect for all 8 genes under the perchlorate and chlorate conditions (Fig. 4A). The fitness defects for sigF, yedY, yedZ, and mrpX were milder than the fitness defects for the essential genes and seemed to be chlorate specific, as there was no measurable defect for the individual SigF regulon components during perchlorate reduction and only a slight defect for sigF itself (Fig. 4A and B). This is consistent with the previous observation that mutants with deletions of these genes were not defective when grown in rich medium with perchlorate as the electron acceptor (14).
FIG 4 .
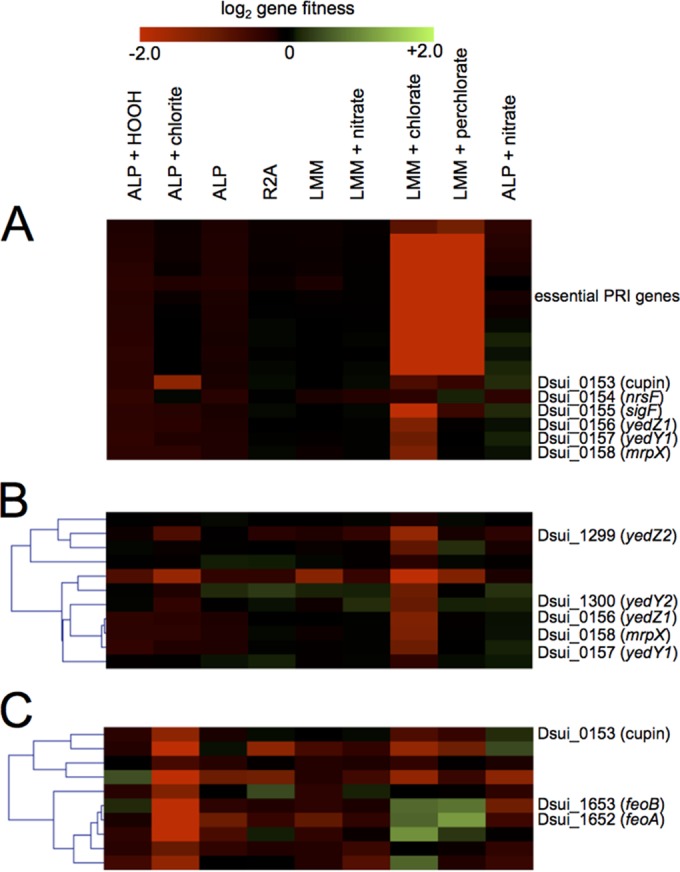
Heatmap generated using MeV to show the fitness values for all 17 genes that make up the core of the PRI (A), the cluster of genes that contains yedY1Z1 and yedY2Z2 (B), and the cluster of genes with the strongest defects specific to the chlorite stress condition (C). The gradient at the top indicates the magnitude of the growth defect; exact numbers and estimates of error can be found in Data Set S3 in the supplemental material.
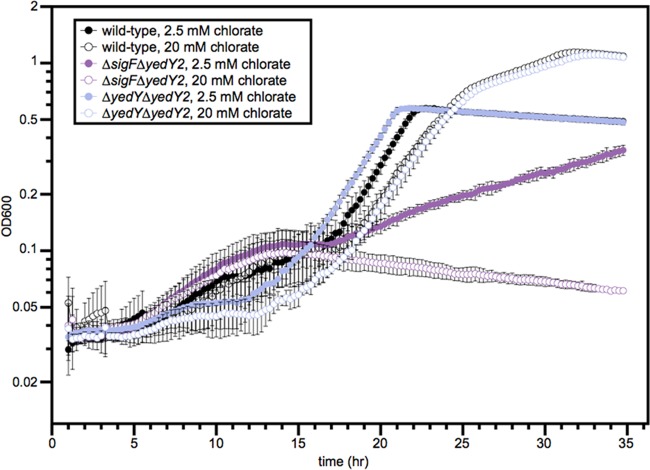
Because the sigF mutant only has a mild growth defect with chlorate (Fig. 2) despite the basal transcription of the SigF regulon, we hypothesized that there may be redundant mechanisms of chlorite resistance outside the SigF regulon. To find these genes, we used the hierarchical clustering (HCL) algorithm within the MeV software package to find genes with fitness profiles that clustered with the chlorate-specific defect seen in the members of the SigF regulon (22, 23). We extracted a cluster of 11 genes from the analysis that contained all 3 genes in the SigF regulon (Fig. 4B). Two of the other genes in this cluster were homologous to yedY1 and yedZ1 (Dsui_1300 and Dsui_1299, hereinafter called yedY2 and yedZ2), providing a possible redundant pathway for ameliorating RCS stress. We therefore created a ΔyedY2 deletion mutant, as well as a ΔsigF ΔyedY2 double knockout mutant. The ΔyedY2 single mutant had no obvious phenotype when grown with chlorate, but the ΔsigF ΔyedY2 mutant had a very strong defect with chlorate (Fig. 5). The severity of the defect was proportional to the chlorate concentration, with the double knockout mutant exhibiting no growth at 20 mM chlorate. The ΔsigF ΔyedY2 mutant was more defective than the ΔsigF ΔyedY1 mutant because yedY2 is not regulated by RCS or SigF and apparently plays a critical role in providing a redundant system for limiting RCS stress. Deleting yedY1 had no effect in the ΔsigF background because SigF is not present to upregulate yedY1.
FIG 5 .
Anaerobic growth curve showing the slight defect of the ΔyedY1 ΔyedY2 strain with 20 mM chlorate and severe defects of the ΔsigF ΔyedY2 strain with 2.5 mM or 20 mM chlorate. This experiment was carried out using minimal medium containing 30 mM lactate with a wild-type control.
Although individual SigF regulon mutants were not defective under acute chlorite stress, there were some genes that did have defects specific to the chlorite stress condition (Fig. 4C). These 10 genes were clustered together in the HCL analysis and included 1 gene from the PRI (Dsui_0153). This gene encodes a cupin domain protein that is not essential for (per)chlorate reduction. Mutants with mutations in a subset of genes (Dsui_1304, Dsui_1652, and Dsui_1653) in this cluster were sensitive to chlorite stress but had fitness benefits when grown with perchlorate and chlorate. Dsui_1304 encodes a c-type cytochrome, and Dsui_1652-Dsui_1653 encode FeoAB, a ferrous iron transporter (24). Control of cytoplasmic iron is one of the major outputs of one of the hypochlorite-sensitive transcription factors (HypT) and may represent a common strategy for dealing with RCS (11).
The methionine residues of MrpX are vulnerable to oxidation by chlorite.
We hypothesized that the physiological function of MrpX is to provide a source of methionine residues that can be sacrificially oxidized to remove RCS from the periplasm during (per)chlorate respiration. To monitor the oxidation state of MrpX resulting from chlorite treatment, we took advantage of the fact that methionine oxidation results in a disproportionate shift in protein migration in denaturing polyacrylamide gel electrophoresis (PAGE) (25). We used allelic replacement to add a C-terminal myc tag to the chromosomal mrpX in order to preserve the native transcriptional regulation. This replacement was also performed in the ΔnrsF background to generate a positive-control strain that constitutively overexpressed a tagged version of MrpX. We grew cultures of these two strains, as well as an untagged wild-type strain, and used a simple chloroform extraction to get periplasmic protein fractions from all three strains (26). We were able to detect MrpX::Myc via Western blotting against the Myc tag, and we could also see the MrpX::Myc protein in the ΔnrsF background via a simple total protein stain.
We grew the wild-type strain PS containing the tagged mrpX gene and then spiked in chlorite, harvesting samples for periplasmic protein extraction every 10 min. When the proteins were analyzed by Western blotting, we observed a gradual accumulation of MrpX, initially in the oxidized form, which shifted downwards over time, reaching the same level as the reduced protein control after 50 to 60 min (Fig. 6A). While the oxidation of the methionine residues could be the result of direct reaction with chlorite, we are unaware of definitive evidence supporting such reactivity. In contrast, hypochlorite is a potent oxidizer of methionine residues both in vivo and in vitro (10, 27), and previous studies have shown that hypochlorite is produced from redox-active proteins such as Cld during chlorite dismutation (7). To test which RCS species was responsible for methionine oxidation, we incubated purified MrpX with various ratios of chlorite and hypochlorite and measured the extent of oxidation via SDS-PAGE gel shift. Our results demonstrated that hypochlorite was much more reactive than chlorite toward MrpX (Fig. 6B).
FIG 6 .
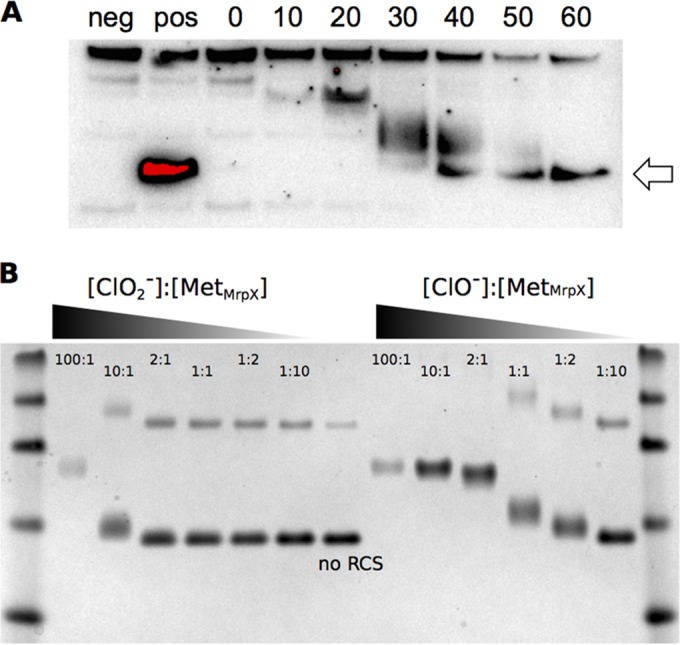
(A) Western blot of total protein extracted from the wild type using an anti-myc tag primary antibody and HRP-conjugated secondary antibody. The samples run were from strains with several genotypes: ΔmrpX (negative control), ΔnrsF mrpX::myc (positive control), and mrpX::myc (experiment). Aliquots of the mrpX::myc strain were withdrawn at various times following a chlorite treatment (from 0 to 60 min, indicated). The arrow shows the migration of MrpX in the untreated positive control. (B) Reactions of purified MrpX with various ratios of RCS. All ratios were calculated based on the methionine molarity of the purified protein. [MetMrpX], the concentration of methionine residues in MrpX, calculated by multiplying the number of predicted methionines found in MrpX by the total concentration of the MrpX peptide.
DISCUSSION
In this paper, we have characterized a novel mechanism for sensing and ameliorating RCS by redox cycling of sacrificial periplasmic methionine residues. The concept of methionine residues acting as antioxidants has been established previously (28). Thus, the physiological function of methionine sulfoxide reductases is 2-fold: not only do they repair nonfunctional proteins, but they also regenerate exposed methionines which act as sinks for oxidative stress (29). We propose that MrpX is primarily an oxidative stress sink; its lack of sequence conservation and high proportion of charged residues mean that it likely is natively unfolded, which may allow preferential oxidation and reduction of its methionine residues (30). A simple search of a large sequence database suggested that there are many small peptides in both prokaryotes and eukaryotes that contain many methionines and charged residues. One of these is the hydrophilin protein Sip18 from Saccharomyces cerevisiae, which has a role in desiccation and oxidative stress tolerance (31, 32).
The function of the YedY proteins was not initially clear, given that the only described member of this family does not have a known biological function. However, biochemical assays of this protein from E. coli showed that it is a reductase, not an oxidase, and is able to reduce free methionine sulfoxide in vitro (21, 33, 34). We propose that this is its physiological role in vivo in perchlorate reducers and other bacteria as well. A microarray study on E. coli O157:H7 showed upregulation of yedYZ in response to hypochlorite but not hydrogen peroxide (35). Second, fitness profiling of two Shewanella strains (MR-1 and SB2B) showed that yedYZ were dispensable for hundreds of conditions, with the single exception of chlorite stress, where insertions led to a strong growth defect (M. Price, unpublished data). Furthermore, a recent study from our laboratory demonstrated transcriptional upregulation of a yedYZ homolog under chlorate-respiring conditions in the chlorate-reducing Shewanella algae strain acdc (36). Future biochemical studies should bear out the precise function of YedY and answer crucial questions about its activity, such as questions regarding stereoselectivity and preference for free methionine sulfoxide versus peptide methionine sulfoxide (29).
We analyzed the distribution of YedY across more than 2,000 microbial genomes and found that it was widely distributed among the bacteria but was most prevalent in the Proteobacteria. It was frequently found adjacent to MrpX and SigF/NrsF but also linked to the canonical methionine sulfoxide reductases msrA and msrB (Fig. 7; see also Fig. S2 in the supplemental material). A certain kind of YedY seems to be enriched in host-associated organisms, such as uropathogenic E. coli (UPEC), Brucella spp., and members of the Rhizobiales (see Fig. S3 in the supplemental material), which suggests a putative role in pathogenesis. The full results of this analysis can be found in Text S1 in the supplemental material.
FIG 7 .
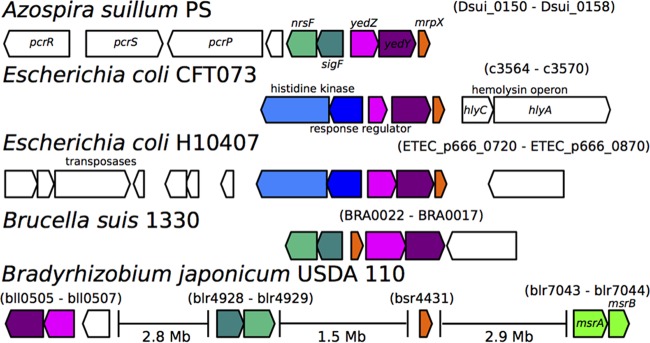
The genomic organization of genes of interest from several organisms, including perchlorate reducers and host-associated organisms.
The conservation of the yedYZ-mrpX operon in UPEC and Brucella spp. suggests that, similar to DPRB, methionine redox cycling plays a role in pathogenesis, perhaps in ameliorating hypochlorite stress caused by myeloperoxidase. In previous studies, virulence was greatly attenuated but growth was unimpaired in UPEC strain 536 when pathogenicity island II (PAI-II) containing yedYZ-mrpX was deleted (37). Many other genes are also part of this island, so future work is needed to ascertain the detailed role of this system in UPEC. However, the conservation of this system in Brucella spp. is also suggestive of its role in RCS defense and immune system evasion, as they are facultative intracellular pathogens that likely encounter reactive chlorine species at least periodically, despite their ability to attenuate the initial innate immune response in other ways (38).
In this paper, we set out to characterize an RCS defense mechanism in perchlorate-respiring organisms and determine whether this is a common strategy in general microbial species. The mechanism depends on SigF as a regulator of RCS oxidative stress and subsequent upregulation of a methionine-rich protein and a putative methionine sulfoxide reductase. The conservation of this system in many non-perchlorate-respiring species suggests that bacteria with diverse lifestyles require the ability to specifically respond to RCS. We present perchlorate-reducing bacteria and mammalian pathogens as two such groups but suspect that future work will uncover others.
MATERIALS AND METHODS
Bacterial culturing.
All strains of Azospira suillum PS were revived from freezer stocks by streaking for single colonies on agar plates containing ALP medium, a rich medium created for routine aerobic culturing of freshwater perchlorate reducers (14). ALP was also the medium used for several growth curves, but in other cases, a minimal medium was used. This minimal medium is composed of the same ingredients as ALP, with the exception of all electron donors (lactate, acetate, and pyruvate) and yeast extract. A detailed description of the growth conditions for various experiments can be found in Text S1 in the supplemental material.
Strain construction.
Gene deletions in Azospira suillum strain PS were created as previously described (14). Myc- and Strep-tagged versions of the mrpX gene were generated using two different methods, detailed descriptions of which can be found in Text S1 in the supplemental material.
Determining the SigF regulon using RNA-seq.
The cells for RNA-seq (described under “Bacterial culturing”) were removed from the incubator and placed on ice prior to centrifugation at 4,000 relative centrifugal force at 4°C for 15 min. The supernatant was removed, and the cells were resuspended in 1.5 ml TRIzol (Life Technologies). RNA was then extracted using the manufacturer’s method. DNA was removed using the Turbo DNA-free kit (Life Technologies) according to the manufacturer’s method. RNA quantity and purity were assessed using a NanoDrop ND-2000 (Thermo Scientific). Illumina library preparation and sequencing were carried out at the Vincent J. Coates Genomics Sequencing Laboratory. Ribosomal RNA was removed using the Ribo-Zero rRNA removal kit (Epicentre) prior to cDNA synthesis and library construction using the Apollo 324 (WaferGen Biosystems). Shearing of DNA and library quality checks were performed using a Covaris S2 and an Agilent 2100 Bioanalyzer. Samples were multiplexed on a single Illumina HiSeq 2000 lane for single-read sequencing of 50-bp reads.
Differential expression of genes was determined using DEseq (39), a complete description of which can be found in Text S1 in the supplemental material.
qPCR experiments.
For qPCR, RNA was extracted as described above for the RNA-seq sample prep. After quantification, 5 µl of total RNA (~250 ng) was used as the template for a SuperScript II reverse transcriptase reaction (Life Technologies) according to the manufacturer’s directions using random hexamer primers. rpoB was chosen as the housekeeping control gene based on its observed stable expression under all of the RNA-seq conditions tested. Primers for rpoB and mrpX were designed, and their primer efficiency was calculated against a genomic DNA dilution series, using 2× Maxima SYBR green master mix (Thermo Scientific) on the StepOnePlus (Life Technologies). The actual qPCR experiment was performed with both biological and technical triplicates and 1 µl cDNA as the template for all reactions (both rpoB and mrpX). Relative quantification was performed using the Pfaffl method, with the specific primer efficiencies for each primer pair being incorporated for normalization (40). All calculations were performed using StepOnePlus software.
Construction of the mutant library and fitness experiments.
A full description of the methods used to generate the fitness libraries and analyze the fitness data is expected to be published shortly in an upcoming paper (42). Briefly, the mutant library was made by electroporating a plasmid containing a barcoded transposon into electrocompetent strain PS cells prior to harvesting and recovery in ALP medium (41). The entire library was sequenced using a Tn-seq approach to identify the insertion site associated with each unique bar code.
The fitness of an individual strain is calculated as the log2 ratio of the abundance of a specific bar code before and after a treatment. Gene fitness is calculated by taking a weighted average of all strains carrying insertions in a given gene and then normalized such that genes with no phenotype have fitness values near zero. We performed two biological replicates for each condition, and the fitness values for the SigF regulon were consistent between replicates.
Overexpression and purification of Mrp.
Mrp was expressed in E. coli Tuner (DE3) competent cells (Novagen) and purified from cell homogenate using a two-column method. A detailed description of this process can be found in Text S1 in the supplemental material.
SDS-PAGE mobility shift assay.
The extraction of periplasmic protein was performed based on a previously published method using chloroform (26). A detailed description of the sample preparation, electrophoresis, and Western blotting can be found in Text S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Boxplots indicating the levels of expression of mrpX as determined by RNA-seq (A) and qPCR (B). For the RNA-seq plot, the values are expressed in reads per kilobase per million mapped reads (RPKM), which is a value normalized by gene length and depth of coverage. The qPCR values are expressed as abundance relative to mrpX expression in the untreated control, and the whiskers indicate standard errors for the results from three biological replicates. The boxplots for the RNA-seq data illustrate the actual range of the data, with the central line indicating the median RPKM value and the whiskers indicating the maximum and minimum RPKM values from the three replicates under each condition. Download
Phylogeny of the HGT clade of YedY protein sequences with taxon labels showing both the SwissProt ID and the species of origin. Colored star markers indicate the presence of conserved accessory genes close to yedY in the genome. The gene diagrams at the left side of the tree show the striking difference in yedYZ synteny, which are labeled based on the PS genome (yedY1 yedZ2, paralogs in the PRI; yedY2 yedZ2, paralogs in the chromosome). Bootstrap support for a given node is illustrated by black (>90/100) or grey (>70/100) circles. Download
Taxonomic distribution of the two clades of YedY. (A) The distribution of YedY among the five major proteobacterial classes is detailed. (B) Distribution among several proteobacterial orders of interest, with arrows highlighting two cases explored in further detail in the text, uropathogenic E. coli and Brucella spp. Download
Strains, primers, and plasmids used in this study.
Differentially expressed genes from the RNA-seq experiments.
Replicate-averaged gene fitness scores and standard error.
Identifiers of genes and phylogenetic affiliations of members of the two YedY families.
ACKNOWLEDGMENTS
We thank all members of the Coates laboratory for discussions and feedback regarding this project. We would also like to acknowledge Andrew Hryckowian for discussions regarding uropathogenic E. coli physiology and phylogenomics.
Work on perchlorate and reactive chlorine species in the laboratory of J.D.C. is funded via the Energy Biosciences Institute. R.A.M. would also like to acknowledge financial support from a Philomathia graduate student fellowship. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation grants S10RR029668 and S10RR027303. Computational analyses were carried out at the Computational Genomics Resource Laboratory, which is part of the California Institute for Quantitative Biosciences.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Melnyk RA, Youngblut MD, Clark IC, Carlson HK, Wetmore KM, Price MN, Iavarone AT, Deutschbauer AM, Arkin AP, Coates JD. 2015. Novel mechanism for scavenging of hypochlorite involving a periplasmic methionine-rich peptide and methionine sulfoxide reductase. mBio 6(3):e00233-15. doi:10.1128/mBio.00233-15.
REFERENCES
- 1.Keene WC, Khalil MAK, Erickson DJ, McCulloch A, Graedel TE, Lobert JM, Aucott ML, Gong SL, Harper DB, Kleiman G, Midgley P, Moore RM, Seuzaret C, Sturges WT, Benkovitz CM, Koropalov V, Barrie LA, Li YF. 1999. Composite global emissions of reactive chlorine from anthropogenic and natural sources: reactive chlorine emissions inventory. J Geophys Res 104:8429–8440. doi: 10.1029/1998JD100084. [DOI] [Google Scholar]
- 2.Gray MJ, Wholey W-Y, Jakob U. 2013. Bacterial responses to reactive chlorine species. Annu Rev Microbiol 67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coates JD, Achenbach LA. 2004. Microbial perchlorate reduction: rocket-fueled metabolism. Nat Rev Microbiol 2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- 4.Kengen SW, Rikken GB, Hagen WR, van Ginkel CG, Stams AJ. 1999. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J Bacteriol 181:6706–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Ginkel CG, Rikken GB, Kroon AG, Kengen SW. 1996. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch Microbiol 166:321–326. doi: 10.1007/s002030050390. [DOI] [PubMed] [Google Scholar]
- 6.Clark IC, Melnyk RA, Engelbrektson A, Coates JD. 2013. Structure and evolution of chlorate reduction composite transposons. mBio 4(4):e00379-13. doi: 10.1128/mBio.00379-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofbauer S, Gruber C, Pirker KF, Sündermann A, Schaffner I, Jakopitsch C, Oostenbrink C, Furtmüller PG, Obinger C. 2014. Transiently produced hypochlorite is responsible for the irreversible inhibition of chlorite dismutase. Biochemistry 53:3145–3157. doi: 10.1021/bi500401k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattison DI, Davies MJ. 2001. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol 14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 9.Pan B, Abel J, Ricci MS, Brems DN, Wang DI, Trout BL. 2006. Comparative oxidation studies of methionine residues reflect a structural effect on chemical kinetics in rhG-CSF. Biochemistry 45:15430–15443. doi: 10.1021/bi061855c. [DOI] [PubMed] [Google Scholar]
- 10.Rosen H, Klebanoff SJ, Wang Y, Brot N, Heinecke JW, Fu X. 2009. Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc Natl Acad Sci U S A 106:18686–18691. doi: 10.1073/pnas.0909464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drazic A, Miura H, Peschek J, Le Y, Bach NC, Kriehuber T, Winter J. 2013. Methionine oxidation activates a transcription factor in response to oxidative stress. Proc Natl Acad Sci U S A 110:9493–9498. doi: 10.1073/pnas.1300578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebendorfer KM, Drazic A, Le Y, Gundlach J, Bepperling A, Kastenmüller A, Ganzinger KA, Braun N, Franzmann TM, Winter J. 2012. Identification of a hypochlorite-specific transcription factor from Escherichia coli. J Biol Chem 287:6892–6903. doi: 10.1074/jbc.M111.287219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker BW, Schwessinger EA, Jakob U, Gray MJ. 2013. The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J Biol Chem 288:32574–32584. doi: 10.1074/jbc.M113.503516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melnyk RA, Clark IC, Liao A, Coates JD. 2013. Transposon and deletion mutagenesis of genes involved in perchlorate reduction in Azospira suillum PS. mBio 5(1):e00769–13. doi: 10.1128/mBio.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melnyk RA, Engelbrektson A, Clark IC, Carlson HK, Byrne-Bailey K, Coates JD. 2011. Identification of a perchlorate reduction genomic island with novel regulatory and metabolic genes. Appl Environ Microbiol 77:7401–7404. doi: 10.1128/AEM.05758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Martinez CE, Baldini RL, Gomes SL. 2006. A Caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase. J Bacteriol 188:1835–1846. doi: 10.1128/JB.188.5.1835-1846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masloboeva N, Reutimann L, Stiefel P, Follador R, Leimer N, Hennecke H, Mesa S, Fischer HM. 2012. Reactive oxygen species-inducible ECF σ factors of Bradyrhizobium japonicum. PLoS One 7:e43421. doi: 10.1371/journal.pone.0043421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler C, Lourenço RF, Avelar GM, Gomes SL. 2012. Extracytoplasmic function (ECF) sigma factor σF is involved in Caulobacter crescentus response to heavy metal stress. BMC Microbiol 12:210. doi: 10.1186/1471-2180-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol 74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 20.Loschi L, Brokx SJ, Hills TL, Zhang G, Bertero MG, Lovering AL, Weiner JH, Strynadka NC. 2004. Structural and biochemical identification of a novel bacterial oxidoreductase. J Biol Chem 279:50391–50400. doi: 10.1074/jbc.M408876200. [DOI] [PubMed] [Google Scholar]
- 21.Oh J, Fung E, Price MN, Dehal PS, Davis RW, Giaever G, Nislow C, Arkin AP, Deutschbauer A. 2010. A universal TagModule collection for parallel genetic analysis of microorganisms. Nucleic Acids Res 38:e146. doi: 10.1093/nar/gkq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
- 23.Eisen MB, Spellman PT, Po Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 25.Le DT, Liang X, Fomenko DE, Raza AS, Chong CK, Carlson BA, Hatfield DL, Gladyshev VN. 2008. Analysis of methionine/selenomethionine oxidation and methionine sulfoxide reductase function using methionine-rich proteins and antibodies against their oxidized forms. Biochemistry 47:6685–6694. doi: 10.1021/bi800422s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ames GF, Prody C, Kustu S. 1984. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J Bacteriol 160:1181–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawkins CL, Pattison DI, Davies MJ. 2003. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25:259–274. doi: 10.1007/s00726-003-0016-x. [DOI] [PubMed] [Google Scholar]
- 28.Levine RL, Mosoni L, Berlett BS, Stadtman ER. 1996. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci U S A 93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezraty B, Aussel L, Barras F. 2005. Methionine sulfoxide reductases in prokaryotes. Biochim Biophys Acta 1703:221–229. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Tarrago L, Kaya A, Weerapana E, Marino SM, Gladyshev VN. 2012. Methionine sulfoxide reductases preferentially reduce unfolded oxidized proteins and protect cells from oxidative protein unfolding. J Biol Chem 287:24448–24459. doi: 10.1074/jbc.M112.374520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Porrata B, Carmona-Gutierrez D, Reisenbichler A, Bauer M, Lopez G, Escoté X, Mas A, Madeo F, Cordero-Otero R. 2012. Sip18 hydrophilin prevents yeast cell death during desiccation stress. J Appl Microbiol 112:512–525. doi: 10.1111/j.1365-2672.2011.05219.x. [DOI] [PubMed] [Google Scholar]
- 32.López-Martínez G, Rodríguez-Porrata B, Margalef-Català M, Cordero-Otero R. 2012. The STF2p hydrophilin from Saccharomyces cerevisiae is required for dehydration stress tolerance. PLoS One 7:e33324. doi: 10.1371/journal.pone.0033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Havelius KG, Reschke S, Horn S, Döring A, Niks D, Hille R, Schulzke C, Leimkühler S, Haumann M. 2011. Structure of the molybdenum site in YedY, a sulfite oxidase homologue from Escherichia coli. Inorg Chem 50:741–748. doi: 10.1021/ic101291j. [DOI] [PubMed] [Google Scholar]
- 34.Brokx SJ, Rothery RA, Zhang G, Ng DP, Weiner JH. 2005. Characterization of an Escherichia coli sulfite oxidase homologue reveals the role of a conserved active site cysteine in assembly and function. Biochemistry 44:10339–10348. doi: 10.1021/bi050621a. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, Deng K, Zaremba S, Deng X, Lin C, Wang Q, Tortorello ML, Zhang W. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl Environ Microbiol 75:6110–6123. doi: 10.1128/AEM.00914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark IC, Melnyk RA, Iavarone AT, Novichkov PS, Coates JD. 2014. Chlorate reduction in Shewanella algae ACDC is a recently acquired metabolism characterized by gene loss, suboptimal regulation and oxidative stress. Mol Microbiol 94:107–125. doi: 10.1111/mmi.12746. [DOI] [PubMed] [Google Scholar]
- 37.Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Olschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A 103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roop RM, Gaines JM, Anderson ES, Caswell CC, Martin DW. 2009. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deutschbauer A, Price MN, Wetmore KM, Shao W, Baumohl JK, Xu Z, Nguyen M, Tamse R, Davis RW, Arkin AP. 2011. Evidence-based annotation of gene function in Shewanella oneidensis MR-1 using genome-wide fitness profiling across 121 conditions. PLoS Genet 7:e1002385. doi: 10.1371/journal.pgen.1002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. mBio, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Boxplots indicating the levels of expression of mrpX as determined by RNA-seq (A) and qPCR (B). For the RNA-seq plot, the values are expressed in reads per kilobase per million mapped reads (RPKM), which is a value normalized by gene length and depth of coverage. The qPCR values are expressed as abundance relative to mrpX expression in the untreated control, and the whiskers indicate standard errors for the results from three biological replicates. The boxplots for the RNA-seq data illustrate the actual range of the data, with the central line indicating the median RPKM value and the whiskers indicating the maximum and minimum RPKM values from the three replicates under each condition. Download
Phylogeny of the HGT clade of YedY protein sequences with taxon labels showing both the SwissProt ID and the species of origin. Colored star markers indicate the presence of conserved accessory genes close to yedY in the genome. The gene diagrams at the left side of the tree show the striking difference in yedYZ synteny, which are labeled based on the PS genome (yedY1 yedZ2, paralogs in the PRI; yedY2 yedZ2, paralogs in the chromosome). Bootstrap support for a given node is illustrated by black (>90/100) or grey (>70/100) circles. Download
Taxonomic distribution of the two clades of YedY. (A) The distribution of YedY among the five major proteobacterial classes is detailed. (B) Distribution among several proteobacterial orders of interest, with arrows highlighting two cases explored in further detail in the text, uropathogenic E. coli and Brucella spp. Download
Strains, primers, and plasmids used in this study.
Differentially expressed genes from the RNA-seq experiments.
Replicate-averaged gene fitness scores and standard error.
Identifiers of genes and phylogenetic affiliations of members of the two YedY families.