ABSTRACT
The molecular mechanisms of alginate polymerization/modification/secretion by a proposed envelope-spanning multiprotein complex are unknown. Here, bacterial two-hybrid assays and pulldown experiments showed that the catalytic subunit Alg8 directly interacts with the proposed copolymerase Alg44 while embedded in the cytoplasmic membrane. Alg44 additionally interacts with the lipoprotein AlgK bridging the periplasmic space. Site-specific mutagenesis of Alg44 showed that protein-protein interactions and stability were independent of conserved amino acid residues R17 and R21, which are involved in c-di-GMP binding, the N-terminal PilZ domain, and the C-terminal 26 amino acids. Site-specific mutagenesis was employed to investigate the c-di-GMP-mediated activation of alginate polymerization by the PilZAlg44 domain and Alg8. Activation was found to be different from the proposed activation mechanism for cellulose synthesis. The interactive role of Alg8, Alg44, AlgG (epimerase), and AlgX (acetyltransferase) on alginate polymerization and modification was studied by using site-specific deletion mutants, inactive variants, and overproduction of subunits. The compositions, molecular masses, and material properties of resulting novel alginates were analyzed. The molecular mass was reduced by epimerization, while it was increased by acetylation. Interestingly, when overproduced, Alg44, AlgG, and the nonepimerizing variant AlgG(D324A) increased the degree of acetylation, while epimerization was enhanced by AlgX and its nonacetylating variant AlgX(S269A). Biofilm architecture analysis showed that acetyl groups promoted cell aggregation while nonacetylated polymannuronate alginate promoted stigmergy. Overall, this study sheds new light on the arrangement of the multiprotein complex involved in alginate production. Furthermore, the activation mechanism and the interplay between polymerization and modification of alginate were elucidated.
IMPORTANCE
This study provides new insights into the molecular mechanisms of the synthesis of the unique polysaccharide, alginate, which not only is an important virulence factor of the opportunistic human pathogen Pseudomonas aeruginosa but also has, due to its material properties, many applications in medicine and industry. Unraveling the assembly and composition of the alginate-synthesizing and envelope-spanning multiprotein complex will be of tremendous significance for the scientific community. We identified a protein-protein interaction network inside the multiprotein complex and studied its relevance with respect to alginate polymerization/modification as well as the c-di-GMP-mediated activation mechanism. A relationship between alginate polymerization and modification was shown. Due to the role of alginate in pathogenesis as well as its unique material properties harnessed in numerous applications, results obtained in this study will aid the design and development of inhibitory drugs as well as the commercial bacterial production of tailor-made alginates.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic human pathogen which can become life-threatening in immunocompromised patients. It is the leading cause of morbidity and mortality in cystic fibrosis patients. This is due mainly to its ability to colonize lungs by forming structured biofilms which consist of bacterial cells embedded in a complex matrix predominantly composed of alginate. Bacterial cells in biofilms are protected against the immune system and antibiotics (1, 2). Alginates are anionic exopolysaccharides composed of variable proportions of 1,4-linked β-d-mannuronic acid (M) and its C-5 epimer α-l-guluronic acid (G). The alginate derived from P. aeruginosa is naturally acetylated and lacks consecutive G residues (GG-blocks) (3). Alginates exhibit unique gel-forming properties suitable for numerous medical and industrial applications (3, 4). The alginate structure strongly impacts its material properties. Hence, the development of bioengineering approaches to control the alginate structure will enable production of alginates with new material properties toward novel applications.
For many years, P. aeruginosa has been the model organism to study various aspects of alginate biosynthesis, such as polymerization, epimerization, acetylation, secretion, and regulation. Thirteen proteins are directly involved in the biosynthesis of alginate, and except for algC, their encoding genes are clustered in the alginate biosynthesis operon (algD, alg8, alg44, algK, algE, algG, algX, algL, algI, algJ, algF, algA) (5, 6). Except for soluble cytoplasmic proteins AlgA, AlgC, and AlgD, which are responsible for providing the activated nucleotide sugar precursor, GDP-mannuronic acid, proteins encoded by the operon are proposed to constitute an envelope-spanning multiprotein complex. Two cytoplasmic membrane-anchored proteins, the glycosyltransferase, Alg8, and the proposed copolymerase, Alg44, are necessary for alginate polymerization (7–9). The MucR sensor protein, a diguanylate cyclase (DGC)/phosphodiesterase (PDE) embedded in the cytoplasmic membrane, was proposed to provide c-di-GMP for binding to the cytoplasmic PilZ domain of Alg44 by which alginate polymerization is activated (10). Translocation of nascent alginate across the periplasm is coupled with modification processes, including O-acetylation and epimerization. O-Acetylation is independently catalyzed by AlgJ and AlgX (11), while the acetyl group donor is provided by AlgI and AlgF (12, 13). The AlgG epimerase converts M residues to G residues in the nascent alginate chain. AlgG, AlgX, and AlgK were suggested to form a periplasmic scaffold for guiding alginate through the periplasm for secretion via the outer membrane protein AlgE (14–19). It was also suggested that if alginate is misguided into the periplasm, then degradation would be mediated by the periplasmic AlgL lyase (20). Previous studies on protein-protein interactions and mutual stabilities of proposed subunits of the multiprotein biosynthesis machinery provided evidence of binary protein interactions, including AlgE-AlgK, AlgX-AlgK, AlgX-MucD (a serine protease), Alg44-AlgX, and Alg8-AlgG (21, 22). However, more experimental evidence is needed to map all protein-protein interactions within the multiprotein complex, in particular toward unraveling the molecular mechanisms of alginate polymerization, molecular mass control, and the relationship of modification events to polymerization.
In this study, protein-protein interactions within the multiprotein complex were investigated using the bacterial two-hybrid technique and pulldown assays. The proposed interacting protein surface of Alg44 was probed, and the molecular mechanism of c-di-GMP-mediated activation was studied (23, 24). The role of Alg8, Alg44, AlgG, and AlgX with respect to polymerization and modification was studied by analyzing the composition and material properties of alginates produced by various strains. We employed a constitutively alginate-producing strain of P. aeruginosa, PDO300, to generate isogenic single- and double-gene knockouts of alg8, alg44, algG, and algX. This allowed studying the role of the respective proteins in alginate polymerization and/or modifications by introducing additional copy numbers of subunits or their variants in trans. The impact of various alginate structures on motility, biofilm formation, and architecture was investigated.
RESULTS
Protein-protein interaction of membrane-anchored Alg8 and Alg44 toward constitution of an active alginate polymerase subunit.
The two cytoplasmic membrane-anchored proteins Alg8 and Alg44 were previously shown to be necessary for alginate polymerization. Alg8 is a glycosyltransferase catalyzing alginate polymerization, using the substrate GDP-mannuronic acid, while the c-di-GMP-binding PilZ domain containing Alg44 was proposed as copolymerase (9, 25). However, the functional and structural interactions of Alg8 and Alg44 had not been elucidated. Therefore, the marker-free isogenic double-gene-knockout PDO300Δalg8Δalg44 mutant was generated. This mutant lost the mucoid phenotype, while introduction of plasmid pBBR1MCS-5:alg44:alg8 restored alginate production and the mucoid phenotype.
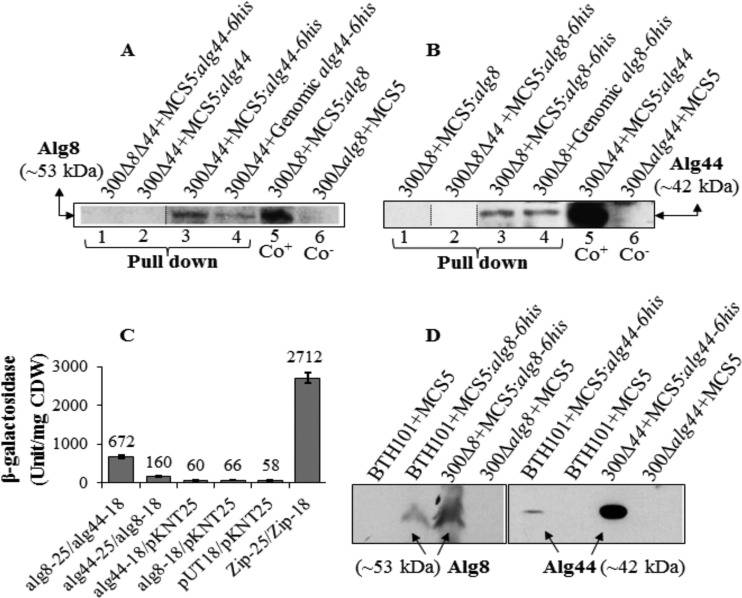
In order to investigate the proposed interaction of Alg8 and Alg44, functional His-tagged variants (Alg44-6His and Alg8-6His) were subjected to pulldown experiments under native conditions and to bacterial two-hybrid system assays. In pulldown experiments, wild-type Alg44 and Alg8 without His tag served as negative controls. To address possible stoichiometric effects, i.e., effects of increased copy numbers of individual subunits on the integrity of the multiprotein complex, single genes encoding Alg8-6His or Alg44-6His under the control of their native promoter were integrated into the genome. In contrast, in trans genes were present on plasmids in multiple copies under control of the strong constitutive lac promoter. Immunoblots showed that Alg44 with an apparent molecular mass of 41.8 kDa was copurified with Alg8-6His produced either from in trans or in cis encoding genes, and similarly Alg8 (~53 kDa) was copurified with Alg44-6His, while respective proteins were not detected for complemented mutants with native Alg44 and Alg8 as well as in double-knockout mutants with single Alg8-6His or Alg44-6His (Fig. 1A and B).
FIG 1 .
Alg8-Alg44 protein-protein interaction. (A and B) His tag-based pulldown assays (lanes 1 to 4) and immunoblotting using anti-Alg8 antibodies (A) and anti-Alg44 antibodies (B) show protein-protein interaction between Alg8 and Alg44. Alg8 and Alg44 interaction was detected by respective copurification of the non-His-tagged interacting protein partner encoded by genes either integrated into the genome or localized on a plasmid (lanes 3 and 4). Lanes 1 and 2 belong to negative controls (proteins without His tag or present individually in double-gene-knockout mutants) (see Materials and Methods). (C) In bacterial two-hybrid analysis, the appropriate pUT18 and pKNT25 derivatives were cotransferred into E. coli BTH101. The pUT18 and pKNT25 vectors were used as a negative control (background) in addition to transformants with one of the constructs and one empty vector, while the pUT18c-Zip and pKT25-Zip plasmid pair was used as a positive control. Upon induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 4-fold β-galactosidase activity higher than background was regarded as evidence for protein-protein interaction. (D) Immunoblots developed using anti-His tag antibodies show production and subcellular localization of Alg8 and Alg44 to the cytoplasmic membrane of E. coli BTH101 (see Materials and Methods). 300, PDO300; 8 and 44, Alg8 and Alg44; MCS5, pBBR1MCS-5; Co+/−, positive/negative control; CDW, cellular dry weight; −18, pUT18; −25, pKNT25. An irrelevant lane was deleted between lanes 2 and 3 in blot A, as indicated by a thin line. Lane 2 in blot B was inserted as a representative control blot lane, as it was obscured by artificial stains in the original bot. Insertion of lane 2 is indicated by thin lines. Between dividing lines in blot B, a representative control blot lane was inserted from a separate blot, because in the original blot, it was obscured by artificial dots/development artifacts.
In addition, the bacterial two-hybrid system showed that the chimeric enzyme adenylate cyclase was reconstituted when its two complementary fragments (T18/T25) were brought together by Alg8 and Alg44 interaction. β-Galactosidase activity in those cells harboring two plasmids producing fusion proteins of Alg8 and Alg44 (pKNT25:alg8-pUT18:alg44) was on average 11-fold (672 U/mg [cellular dry weight]) greater than the negative controls without the fusion protein partner (background control) or compared with single fusion protein Alg8 or Alg44 (58 to 66 U/mg [cellular dry weight]) as well as the vice versa combination (pKNT25:alg44-pUT18:alg8), which showed a β-galactosidase activity of 160 U/mg (cellular dry weight) (Fig. 1C). Analysis of cytoplasmic membrane proteins by immunoblotting confirmed that both proteins were localized to the cytoplasmic membrane of Escherichia coli (Fig. 1D). These results provided the first experimental evidence for the direct interaction between the membrane-anchored proposed alginate polymerase (glycosyltransferase) Alg8 and the copolymerase Alg44.
Cytoplasmic membrane-anchored Alg44 interacts with outer membrane-anchored AlgK, while Alg44 is critical for structural integrity of the multiprotein alginate biosynthesis machinery.
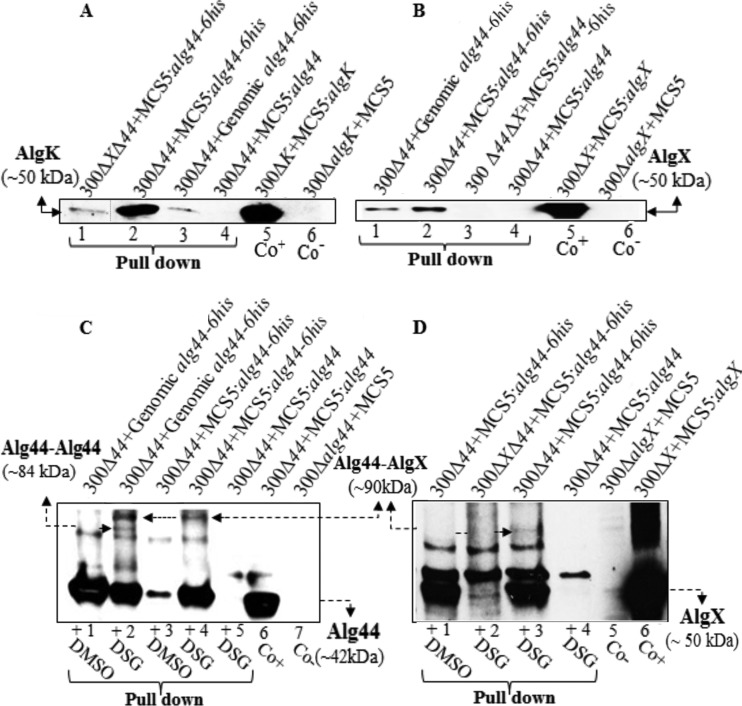
To assess whether Alg8 and Alg44 interact with other proposed subunits of the multiprotein complex, pulldown assays under native conditions were employed using Alg8-6His and Alg44-6His proteins followed by immunoblotting using anti-AlgX, -AlgK, and -AlgE antibodies. Additionally, to rule out indirect interactions, appropriate double-gene-knockout mutants harboring individual genes in trans [PDO300Δalg8Δalg44(pBBR1MCS5:alg8-6His), PDO300Δalg8Δalg44(pBBR1MCS5:alg44-6His), and PDO300Δalg44ΔalgX(pBBR1MCS5:alg44-6His) mutants] were included. To address stoichiometric effects, complemented PDO300Δalg8 and PDO300Δalg44 mutants, respectively, were used to generate single-gene-copy complementation strains by integrating alg8-6His and alg44-6His into the bacterial genome. The mutants producing native Alg8 and Alg44 proteins were used as negative controls. Resultant immunoblots (Fig. 2A and B) showed AlgK and AlgX but not AlgE were independently pulled down with Alg44-6His, both when alg44-6His was provided in trans or in cis. Hence, experimental evidence is provided, as previously proposed (21), that a protein-protein interaction network spanning the periplasm and constituted by Alg8-Alg44-AlgK-AlgE interactions exists.
FIG 2 .
Protein-protein interaction analysis indicates interaction of Alg44-AlgK, Alg44-AlgX, and probable Alg44-Alg44 (dimer). (A and B) His tag-based pulldown assays and immunoblotting using anti-AlgK (A) and anti-AlgX (B) antibodies show interactions of Alg44-AlgK and Alg44-AlgX (A and B, lanes 1 to 4). These interactions were detected in both cases of in trans and in cis complementation of mutants with alg44-6His (lanes 2 and 3 of blot A; lanes 1 and 2 of blot B). AlgK was pulled down using His-tagged Alg44 in the absence of AlgX (lane 1, blot A). The elution fraction derived from complemented mutants producing non-His-tagged protein lacked AlgK and AlgX (lane 4 in blots A and B). (C and D) Immunoblots suggesting in vivo Alg44 dimerization and Alg44-AlgX interaction using stabilization of interaction by chemical cross-linking combined with His-tagged Alg44-mediated pulldown assays under denaturing conditions (see Materials and Methods). A presumable Alg44 dimer band (~84 kDa) reacted only with anti-Alg44 antibody (blot C, lane 2) while the alg44-AlgX band (~90 kDa) reacted with Alg44 and AlgX antibodies in both blots (blot C, lanes 2 and 4; blot D, lane 3). These bands were not detected in the elution fraction derived from mutants with native Alg44 with DSG and Alg44-6His treated with DMSO as negative controls (blot C, lanes 1, 3, and 5; blot D, lanes 1 and 4). 300, PDO300; MCS5, BBR1MCS-5; Co+/−, positive/negative control. An irrelevant lane was deleted between lanes 1 and 2 in blot A, as indicated by a thin line.
Cross-linking experiments using a DSG cross-linking reagent with a spacer arm length of 7.7 Å followed by Alg44-6His pulldown under denaturing conditions showed a protein with an apparent molecular mass of ~84 kDa, which was detected only by the anti-Alg44 antibody. In addition, the previously shown Alg44-AlgX (21) interaction was confirmed by detecting a cross-linked protein with an apparent molecular mass of ~90 kDa binding both anti-Alg44 and anti-AlgX antibodies. The ~84-kDa protein was detected only in pulldown elution fractions obtained from genomic expression of alg44-6His but not from the plasmid-borne gene, while Alg44-AlgX interactions were found to be independent of the stoichiometry of the individual proteins (Fig. 2C and D). These proteins were not detected in elution fractions when the native protein Alg44 was present and in the negative control treated with dimethyl sulfoxide (DMSO).
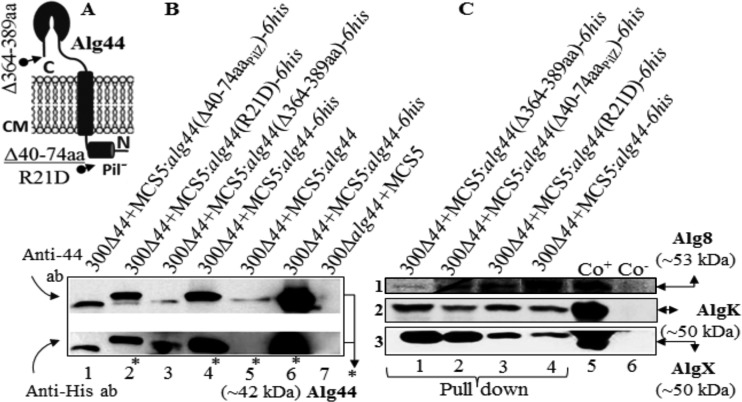
Alg44 variants with a truncated PilZ domain and C terminus were stable and maintained integrity of protein-protein interactions within the alginate biosynthesis multiprotein complex.
Previously, it was demonstrated that site-directed mutagenesis of the putative c-di-GMP-binding motifs (R17XXXR21) of the PilZ domain and a C-terminal truncation of Alg44 completely abolished alginate production (26). In comparison, it was shown that c-di-GMP binds directly to both PgaC and PgaD, the two cytoplasmic membrane components of the E. coli poly-β-1,6-N-acetylglucosamine synthesis machinery, which stimulated their glycosyltransferase activity by stabilizing their interaction (27). Here, it was investigated whether c-di-GMP binding to the PilZ domain of Alg44 and the C-terminal part itself impacts on protein-protein interactions and ultimately alginate polymerization.
His pulldown experiments under native conditions as described above were applied using His-tagged Alg44 variants [Alg44(R21D), Alg44(Δ40–74aaPilZ), and C-terminally truncated Alg44(Δ364–389aa)] (Fig. 3A). As shown in Fig. 3B, Alg44-6His variants were all found in the envelope fraction, which suggested their localization and stability were not affected. Interestingly, the abovementioned protein interaction network was confirmed, which signifies neither protein stability nor the interaction of Alg44 with AlgK and AlgX was disrupted by its defective PilZ domain or the C-terminal truncation. However, the C-terminal truncation of Alg44 impacted on the stability of Alg8 (Fig. 3C, lane 1), while the other variants of Alg44 did not (Fig. 3C).
FIG 3 .
Localization, stability, and protein-protein interaction of Alg44 variants. (A) Schematic view of the deleted or point-mutated region of Alg44. (B) Immunoblots of the envelope fraction developed using anti-Alg44 (upper) and anti-His tag (lower) antibodies showed with the various Alg44 variants localized to the envelope fraction (immunoblot B, lanes 1 to 3). Lanes 4 to 7 represent negative and positives controls (blot C, lanes 1 to 4). Immunoblots suggested protein-protein interaction of Alg44 variants with Alg8, AlgK, and AlgX by using pulldown experiments using His-tagged variants of Alg44. C-terminal deletion of Alg44 had a destabilizing effect on Alg8. Immunoblots were developed using anti-Alg8 (1), anti-AlgK (2), and anti-AlgX (3) antibodies, respectively. Asterisks indicate full-length Alg44. 300, PDO300; MCS5, pBBR1MCS-5; ab, antibody; Co+/−, positive/negative control.
C-di-GMP levels and growth mode impact on Alg44 stability.
Previous studies showed that introducing high copy numbers of mucR in the PDO300ΔmucR mutant resulted in greater production of alginate than found in the wild type, while increased copy numbers of rocR encoding a c-di-GMP-degrading phosphodiesterase (PDE) led to strongly reduced alginate production, presumably due to reduction in c-di-GMP levels. Therefore, it was suggested that MucR plays a specific role in the regulation of alginate biosynthesis by colocalizing with Alg44 and providing a localized c-di-GMP pool (10). Here, it was tested if Alg44 copy numbers in the envelope fraction might be affected by the presence or absence of MucR or RocR, respectively, i.e., by different c-di-GMP levels and within different physiological conditions, such as planktonic and biofilm growth modes. Immunoblotting analysis of envelope fractions of the various mutants showed the amount of Alg44 was not significantly affected by the absence of MucR in biofilm mode, while it was reduced in the envelope fraction of biofilm cells with a high copy number of rocR in the absence of MucR (see Fig. S1 in the supplemental material), indicating that the small amount of Alg44 corresponded with low c-di-GMP levels. However, the amount of Alg44 in the same mutants growing in planktonic mode did not significantly differ.
Is alginate polymerization controlled by an autoinhibition mechanism, as shown for the bacterial cellulose synthase?
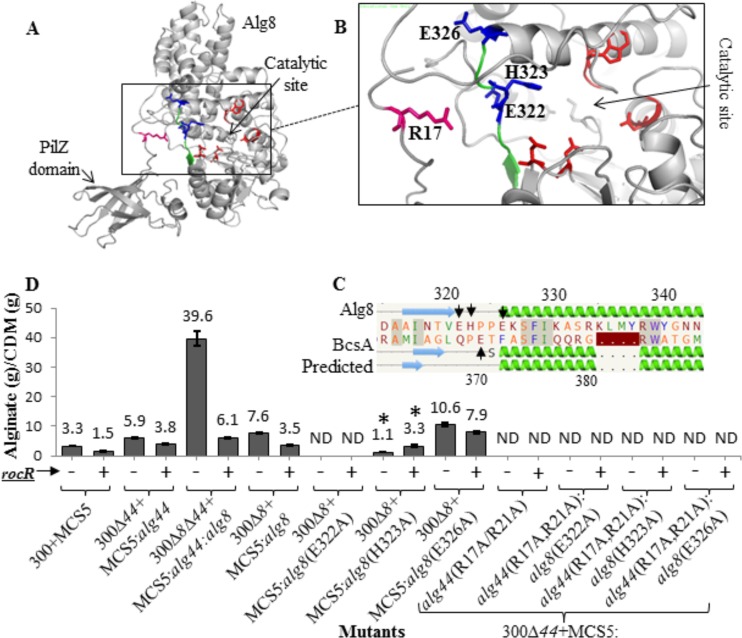
Alg8 and BcsA of bacterial cellulose synthase both belong to the glycosyltransferase family 2 (GT-2), and they share the same conserved signature motifs and residues experimentally known as critical for production of alginate and cellulose, respectively (28, 29). Recently, the structure of the bacterial cellulose synthase BcsA-BcsB complex was resolved. It was shown that the PilZ domain in this complex was in proximity to the catalytic site of BcsA (30). The first arginine of the PilZ domain’s R580XXXR584 motif formed a salt bridge with E371 preceding the RW motif (a signature of the glycosyltransferase family 2), consequently tethering the gating loop in the resting status and blocking the catalytic site. This steric hindrance was called the autoinhibiting mechanism, which was proposed to be eliminated upon c-di-GMP binding to R580, opening up the gate for precursors to enter into the catalytic site.
Accordingly, informed by the BcsA-BcsB structure and implementing bioinformatics analysis using the Phyre2 Protein Fold Recognition Server, an in silico structural model of Alg8 fused with the C-terminal PilZAlg44 domain was developed (31). A structural model homologous to BcsA (confidence, 100%; coverage, 93%) showed the PilZ domain in proximity to the catalytic site of Alg8 and close to the E322, H323, and E326 residues located on the BcsA-homologous loop preceding motif RW (residues 339 and 340), a site potentially involved in salt bridge formation (Fig. 4A to C). The impact of alanine substitutions of these residues, individually and in combination with R17 and/or R21 of Alg44’s RXXXR (residues 17 to 21) motif at different c-di-GMP levels (i.e., the presence or absence of overproduced c-di-GMP degrading RocR), on in vivo activity of respective Alg8 and Alg44 variants was assessed. The overproduction of RocR was confirmed to significantly reduce alginate production in the wild-type strain and complemented mutants. Substitution of R residues in the PilZAlg44 domain RXXXR motif (amino acids 17 to 21) and the E322 residue of Alg8 with alanine completely abolished alginate production. The mutagenesis of H323 [i.e., PDO300Δalg8(pBBR1MCS-5:alg8(H323A) mutant] lowered alginate production by 6.9-fold compared to that of the PDO300Δalg8(pBBR1MCS-5:alg8) mutant. Interestingly, RocR-mediated reduced intracellular c-di-GMP levels restored alginate production to PDO300Δalg8(pBBR1MCS-5:alg8:rocR) mutant levels (Fig. 4D, labeled with asterisk). Replacement of E326 by alanine in Alg8 increased alginate production by 1.3-fold compared with that of the PDO300Δalg8(pBBR1MCS-5:alg8) mutant. RocR production in this mutant background mediated decreased alginate production by about 2-fold compared to that of wild-type Alg8 (Fig. 4D). In summary, conserved R residues of Alg44 proposed to be involved in autoinhibition via salt bridge formation inactivated alginate polymerization. However, the replacement of H323 or E326 of Alg8 still mediated alginate production, while reduced levels of c-di-GMP did cause less or no reduction of alginate production compared to that of the reference strain [PDO300Δalg8(pBBR1MCS-5:alg8) mutant].
FIG 4 .
Bacterial cellulose synthase-associated autoinhibiting mechanism does not play a role in alginate polymerization. (A) In silico fusion of Alg8-PilZAlg44 was modeled using the Phyre2 server. (B) Highly conserved amino acids (blue sticks; E322, H323, and E326 are labeled with arrows in pairwise alignment of Alg8 with BcsA [C]) were identified in a similar position to that of BcsA and could form a salt bridge with R residues of the PilZAlg44 domain R17XXXR21 motif and were chosen to be replaced by alanine using site-specific mutagenesis. (D) Alginate quantification of PDO300Δalg8, PDO300Δalg44, and PDO300Δalg8Δalg44 transformants with plasmids harboring respective site-specific mutants of alg8 and alg44 with (+) and without (−) the rocR gene. Alg8’s mutated residue which positively responded to c-di-GMP level alteration by RocR is labeled with an asterisk. 300, PDO300; ND, not detectable; MCS5, pBBR1MCS-5.
Interplay of alginate polymerizing (Alg8-Alg44) and modifying (AlgG-AlgX) units on alginate composition and molecular mass.
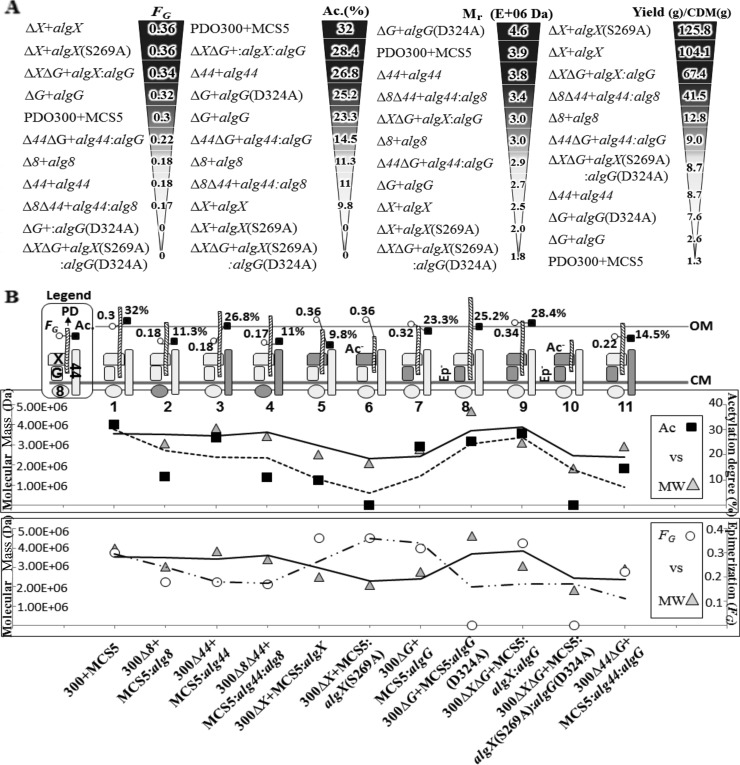
In order to investigate the relationship between alginate polymerization and modification, single- and double-gene-knockout mutants of P. aeruginosa PDO300 were generated and followed by individual and combinatorial in trans complementation using relevant genes, including alg8-alg44 (encoding alginate-polymerizing proteins), algX-algX(S269A) (encoding alginate-acetylating/nonacetylating AlgX), and algG-algG(D324A) (encoding alginate-epimerizing/nonepimerizing AlgG). Generated knockout mutants lost mucoidy, while mucoidy was restored upon in trans complementation with relevant genes. In order to shed light on the functional interaction between alginate-polymerizing and -modifying subunits, the polymerization degree, epimerization degree, and acetylation level of resulting alginates were assessed. The composition and the molecular masses of the respective alginates are summarized in Table 1. Figure 5A shows in descending order the values obtained in regard to epimerization, acetylation, and polymerization degree. 1H-nuclear magnetic resonance (NMR) spectra of compositional analysis of alginates are shown in Fig. S2 in the supplemental material. In order to investigate whether Alg8 and Alg44 are directly involved in polymannuronate synthesis, additional copies of both Alg8 and its interacting partner Alg44 were introduced into respective mutant backgrounds. Additional copies of Alg8 and/or Alg44 had a similar effect on alginate production, such as resulting in high molecular masses with reduced epimerization and acetylation compared to those of the wild-type control (Fig. 5A; Table 1). The same effect of Alg8 and Alg44 on alginate polymerization supported the hypothesis that both subunits constitute the alginate polymerase.
TABLE 1 .
Composition and molecular mass analyses of alginate produced by different mutantsa
| No. | Mutant description | FG | FM | FGM/MG | FMM | Ac. % | Wt avg molecular mass (kDa) | No. avg molecular mass (kDa) | PI | Alginate yield (g)/CDM (g) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 300+MCS5 | 0.3 | 0.7 | 0.29 | 0.41 | 32 | 3,927 (±0.864%) | 3,832 (±0.842%) | 1.025 (±1.2%) | 1.3 ± 0.03 |
| 2 | 300Δ8+MCS5:alg8 | 0.18 | 0.82 | 0.17 | 0.65 | 11.3 | 3,045 (±0.556%) | 3,037 (±0.551%) | 1.003 (±0.7%) | 12.8 ± 1.03 |
| 3 | 300Δ44+MCS5:alg44 | 0.18 | 0.82 | 0.18 | 0.64 | 26.8 | 3,831 (±0.963%) | 3,650 (±0.950%) | 1.05 (±1.3%) | 8.7 ± 0.53 |
| 4 | 300Δ44Δ8+MCS5:alg44:alg8 | 0.17 | 0.83 | 0.17 | 0.66 | 11 | 3,369 (±0.839%) | 3,352 (±0.821%) | 1.005 (±1.1%) | 41.5 ± 4.9 |
| 5 | 300ΔX+MCS5:algX | 0.36 | 0.64 | 0.36 | 0.28 | 9.8 | 2,460 (±0.932%) | 2,447 (±0.913%) | 1.005 (±1.3%) | 104.1 ± 5.5 |
| 6 | 300ΔX+MCS5:algX(S269A) | 0.36 | 0.64 | 0.36 | 0.28 | 0 | 2,086 (±0.960%) | 2,065 (±0.944%) | 1.010 (±1.3%) | 125.8 ± 9.9 |
| 7 | 300ΔG+MCS5:algG | 0.32 | 0.68 | 0.32 | 0.36 | 23.3 | 2,755 (±1.041%) | 2,726 (±0.986%) | 1.011 (±1.4%) | 2.6 ± 0.04 |
| 8 | 300ΔG+MCS5:algG(D324A) | 0 | 1 | 0 | 1 | 25.2 | 4,653 (±1.097%) | 4,575 (±1.117%) | 1.017 (±1.5%) | 7.6 ± 0.57 |
| 9 | 300ΔXΔG+MCS5:algX:algG | 0.34 | 0.66 | 0.34 | 0.32 | 28.4 | 3,076 (±1.051%) | 3,044 (±1.029%) | 1.011 (±1.4%) | 67.42 ± 4.8 |
| 10 | 300ΔXΔG+MCS5:algX(S269A):algG(D324A) | 0 | 1 | 0 | 1 | 0 | 1,811 (±0.884%) | 1,716 (±0.888%) | 1.055 (±1.2%) | 8.7 ± 0.42 |
| 11 | 300Δ44ΔG+MCS5:alg44+algG | 0.22 | 0.78 | 0.22 | 0.56 | 14.5 | 2,907 (±0.966%) | 2,861 (±0.944%) | 1.016 (±1.3%) | 9.0 ± 0.3 |
300, PDO300; MCS5, pBBR1MCS-5; FG, molar fraction of guluronate (G) residue; FM, molar fraction of mannuronate (M) residue; FGM/MG, molar fraction of two consecutive G and M residues; FMM, molar fraction of two consecutive M residues; Ac, acetylation; PI, polydispersity index; CDM, cell dry mass.
FIG 5 .
Impact of putative alginate polymerase subunits on alginate polymerase activity, alginate polymerization, and composition and correlation between polymerization and modification. (A) The values of molar fraction of G residue (FG), acetylation degrees (Ac. %), mean molecular masses, and alginate yield are aligned with the strains producing the respective alginates. (B) Correlation between degree of acetylation, epimerization, and molecular mass of alginate. Presumable features (no. 1 to 11) show protein complexes constituted by Alg8, Alg44, AlgG, and AlgX (see the legend at the top left corner of the plot). The subunit produced upon in trans complementation is shown as darker shape(s). Inactive AlgX(S269A) and AlgG(D324A) proteins are labeled as (Ac─) and (Ep─), respectively. The length of various alginates (PD) with respect to acetylation (Ac. %) and epimerization (FG) degrees are presented and proportionally illustrated for each feature. 300, PDO300; MCS5, pBBR1MCS-5; PD, polymerization degree; OM, outer membrane; CM, cytoplasmic membrane.
AlgF, AlgI, and AlgJ were proposed to form a protein complex constituting the alginate acetyltransferase/acetylesterase (12, 32, 33). Recently, AlgX was demonstrated to play an independent role in alginate acetylation. AlgX is a two-domain protein, including a domain with acetyltransferase activity and a carbohydrate-binding domain. Replacement of amino acid residues S269, H176, and D174, which were proposed to constitute the catalytic site, resulted in nonacetylated alginate (13). AlgG, the epimerase, contains a conserved DPHD motif (residues 324 to 327) (14, 19, 34, 35), the proposed active site involved in epimerization. Replacement of amino acid residues in this motif was shown to result in nonepimerized alginate, while modified AlgG retained its protective role on nascent alginate against degradation in the periplasm. Here, we used catalytically inactive variants of AlgX(S269A) and AlgG(D324A). When only the inactive AlgX variant was present, the resulting alginate was nonacetylated. Interestingly, additional copies of active AlgX or inactive AlgX resulted in the highest epimerization values of molar fraction of G residue (FG) = 0.36. Additional copies of both AlgG and AlgX or inactive variants increased the degree of epimerization of the resulting alginate (Fig. 5A). Additional copies of both AlgX and AlgG increased the degree of acetylation compared to additional copies of only AlgX (Fig. 5A). Interestingly, additional copies of Alg44 enhanced acetylation 2.7-fold compared to AlgX.
The correlation between the molecular mass of alginate and alginate modification, such as acetylation and epimerization, was assessed (Fig. 5B). The molecular mass of the various alginates was determined by size exclusion chromatography-multiangle laser light scattering (SEC-MALLS) (Table 1; see also Fig. S3 in the supplemental material). The highest molecular mass (4,653 ± 1.1% kDa, corresponding to about 22,876 uronic acid residues) was detected in alginates from strains with additional copies of the catalytically inactive epimerase variant AlgG(D324A). This was about a 70% increase in molecular mass compared to alginate produced from strains with additional copies of epimerizing AlgG. The lowest-molecular-weight alginates were produced by strains harboring additional copies of those subunits contributing the highest levels of epimerization, i.e., AlgX (FG = 0.36) and AlgX(S269A) (FG = 0.36), along with the lowest levels of acetylation (9.8% and 0). Nonacetylated and nonepimerized alginates (FG = 0, acetylation = 0%) showed the lowest molecular mass (1,811 ± 0.9% kDa). Since 1H-NMR spectra of the various alginates did not provide evidence for double bonds (between the C-4 and C-5 carbons, leading to 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid), which are introduced by alginate lyase-mediated degradation, the alginate lyase presumably did not influence the polymerization degree. Additional copies of Alg8 and Alg44 gave rise to increased molecular masses ranging from 3,000 to 3,800 kDa, supporting their direct involvement in alginate chain synthesis. These data suggested that the alginate molecular mass is inversely correlated with alginate epimerization but positively correlated with acetylation (Fig. 5B). Alginates produced by the various strains showed a narrow molecular mass distribution with a polydispersity index close to 1 (Table 1).
In vivo alginate polymerase activity.
Alginate produced by the various strains was isolated and quantified (Fig. 5A; Table 1). Although additional copies of the various proteins increased the amount of alginate produced compared to the PDO300(pBBR1MCS-5) reference strain, a significant variation of alginate productivity, i.e., alginate polymerase activity, was detected. Interestingly, additional copies of nonacetylating AlgX(S269A) and native AlgX mediated production of the largest amounts of alginate, while epimerizing and nonepimerizing AlgG mediated the lowest level of production (Fig. 5A). Pairwise comparison of these four strains showed that more alginate is produced in the absence of modification events. The enhancing role of AlgX in alginate production was further supported when additional copies of AlgX together with AlgG led to a strong production of alginate. However, the nonacetylating and nonepimerizing pair of them resulted in a much lower quantity (Fig. 5A). Furthermore, all attempts to restore alginate production in the PDO300Δalg44ΔalgX mutant with pBBR1MCS-5:alg44:algX failed. The mucoid phenotype of this double-gene-knockout mutant was restored only when one of the introduced complementing genes, either alg44 or algX, was integrated into the genome (in cis complementation using mini-CTX) and the other one presented in trans, resulting in alginate production of 1.9 g/cell dry mass (CDM) (g).
Microrheological analysis of various alginates.
Particle-tracking microrheology was applied to assess the viscoelastic properties of the various resulting alginates. All alginates showed viscoelastic properties in which the solid-like elastic modulus G' was greater than the liquid-like viscous modulus G'' (G' > G''). The plot of particle mean square displacement (MSD) versus correlation time showed MSD curves of the alginates are distributed in four distinct categories (see Fig. S4 in the supplemental material). In the first category, the alginates produced from the PDO300ΔalgG(pBBR1MCS-5:algG(D324A)) and PDO300Δalg8(pBBR1MCS-5:alg8) mutants, without G residues and with the highest molar fraction of MM-blocks, respectively, and both with very high molecular mass, showed the highest and quite similar viscoelastic properties (G' = 0.41, G'' = 0.3; G' = 0.40, G'' = 0.28, respectively). Interestingly, the alginates from the PDO300Δalg44(pBBR1MCS-5:alg44), PDO300ΔalgG(pBBR1MCS-5:algG), and PDO300(pBBR1MCS-5) strains dropped into the second category with lower viscoelastic property. In the third category, showing lower viscoelastic properties, those alginates with a molecular mass of ≤2,000 kDa produced by PDO300ΔalgX(pBBR1MCS-5:algX(S269A)) and PDO300ΔalgXΔalgG(pBBR1MCS-5:algX(S269A):algG(D324A)) mutants were found. Surprisingly, acetylated alginate from the PDO300ΔalgX(pBBR1MCS-5:algX) mutant was the only member of the fourth category, with the lowest viscoelastic property among all analyzed samples. These results suggested that viscoelasticity was positively impacted by the molecular mass combined with high M content, while the presence of G residues and acetyl groups in the alginate chain lowered viscoelasticity. All these polymers showed greater elasticity than viscosity (G' > G'').
The impact of various alginates on biofilm formation.
P. aeruginosa is capable of different modes of motility, such as twitching, swarming, and swimming, which are controlled by various regulatory pathways and environmental factors. These play an important role in biofilm formation and dispersal. Here, motility assays were conducted with strains capable of producing different alginates in order to assess the relationship between alginate composition/molecular mass, i.e., material properties, and motility, ultimately impacting on biofilm formation (see Fig. S5 in the supplemental material). All strains with alginate production greater than that of PDO300(pBBR1MCS-5) showed lower twitching motility, while non-alginate-producing knockout mutants showed greater twitching values. The lowest twitching motility among all strains was found for the PDO300Δalg44(pBBR1MCS-5:alg44) mutant. Twitching motility differences between the PDO300ΔalgX(pBBR1MCS-5:algX) and PDO300ΔalgX(pBBR1MCS-5:algX(S269A) mutants or between the PDO300ΔalgG(pBBR1MCS-5:algG) and PDO300ΔalgG(pBBR1MCS-5:algG(D324A)) mutants were insignificant (see Fig. S5 in the supplemental material).
Swarming motility, which occurs on semisolid surfaces and is regulated by quorum sensing, was assessed as being lower in alginate-producing strains than in their respective knockout mutants, except for the PDO300Δalg8(pBBR1MCS-5:alg8) mutant, which showed slightly greater swarming motility than the PDO300Δalg8(pBBR1MCS-5) mutant. Among the alginate-producing strains, the greatest value of swarming motility was found for the PDO300Δalg44(pBBR1MCS-5:alg44) mutant (11.33 mm) and the PDO300ΔalgX(pBBR1MCS-5:algX) mutant (11.0 mm). The PDO300ΔalgG(pBBR1MCS-5:algG(D324A)) mutant (6.83 mm) showed slightly greater swarming motility than the PDO300ΔalgG(pBBR1MCS-5:algG) mutant (6.0 mm).
Swimming, which occurs in aqueous environments, was tested, and the values obtained varied significantly among the strains. Generally, alginate-producing strains showed lower swimming capability than knockout mutants, except for the PDO300Δalg8(pBBR1MCS-5:alg8) and PDO300Δalg8(pBBR1MCS-5) mutants (see Fig. S5 in the supplemental material).
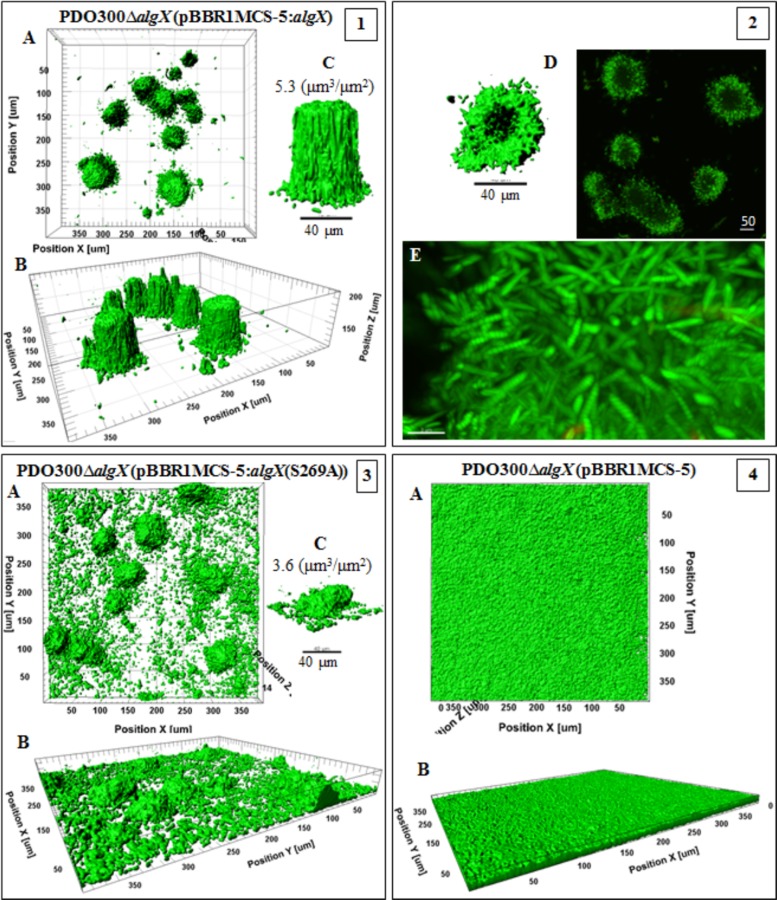
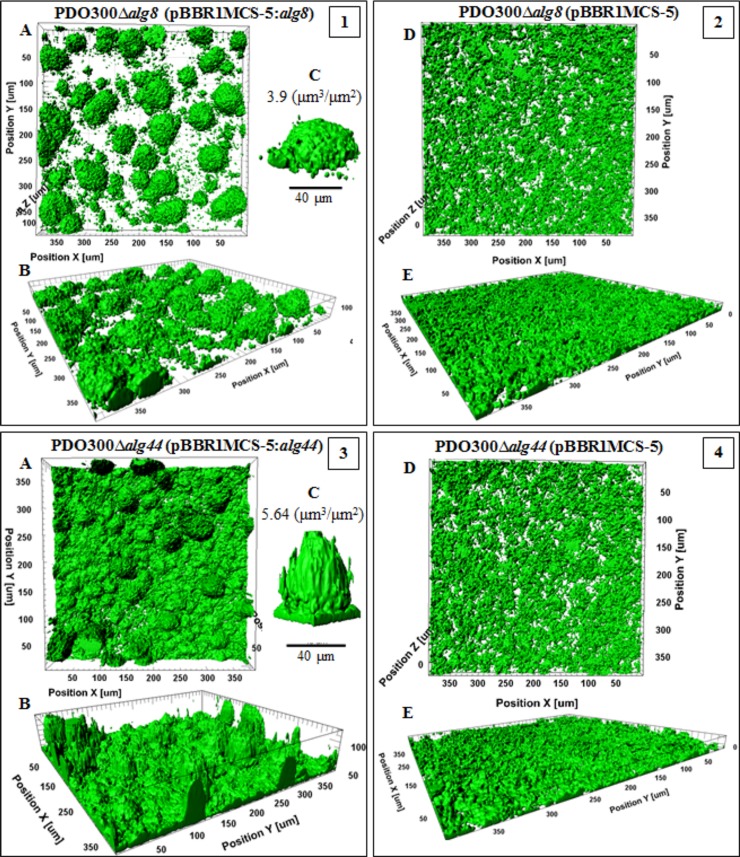
Confocal laser-scanning microscopy images of biofilms formed by the PDO300ΔalgX(pBBR1MCS-5:algX) mutant and its nonacetylating counterpart, the PDO300ΔalgX(pBBR1MCS-5:algX(S269A)) mutant, highlighted the crucial role of acetylation of alginate for developing biofilms and cellular arrangements (Fig. 6). Comparison of the two strains revealed significant differences in elevated structures and the distribution of microcolonies. For example, the structures formed by strains producing acetylated alginates were perfectly shaped and developed with a biovolume of 5.5 ± 1.26 µm3 ⋅ µm−2 and a maximum height of 83 µm, while those formed with nonacetylated alginate showed a smaller biovolume of 3.9 ± 0.2 µm3 ⋅ µm−2 and a reduced height of 26 µm, with irregular architecture. Interestingly, the strain producing acetylated alginate did not produce a multicellular base layer, and cells were organized in pillar-shaped architectures, similar to structures described for the architectures of the Psl-overproducing strain (i.e., P. aeruginosa WFPA801) (36, 37). In contrast, the strain producing nonacetylated alginate formed a biofilm with disordered and scattered microcolonies (Fig. 6, frame 3). Furthermore, the PDO300ΔalgX(pBBR1MCS-5:algX) mutant formed a biofilm with 1.5-fold more compactness and 31% more live cells than the biofilm formed by the PDO300ΔalgX(pBBR1MCS-5:algX(S269A)) mutant (Table 2). The PDO300ΔalgX(pBBR1MCS-5) mutant did not form a structured biofilm (Fig. 6, frame 4) but did form a multicellular layer with a thickness of 6 µm.
FIG 6 .
Biofilm architecture of mutants producing acetylated and nonacetylated alginates. This figure shows biofilm formation and architecture of the PDO300ΔalgX(pBBR1MCS-5:algX) (frames 1 and 2) and PDO300ΔalgX(pBBR1MCS-5:algX(S269A)) (frame 3) mutants, which produce acetylated and nonacetylated alginates, respectively, and the PDO300ΔalgX(pBBR1MCS-5) (frame 4) mutant with no alginate production. In all frames, A, B, and C show top views, side views, and a representative of typical highly structured cell community, respectively. The cell community dimensions are provided as µm3 ⋅ µm−2.
TABLE 2 .
Compactness and dead/live ratio calculated for analyzed biofilms
| Mutant descriptionc | Compactnessa | Dead/live ratiob |
|---|---|---|
| 300Δ8+MCS5:alg8 | 6.09E+02 | 1.43 ± 0.10 |
| 300Δ44+MCS5:alg44 | 4.43E+02 | 1.17 ± 0.04 |
| 300ΔG+MCS5:algG | 2.77E+02 | 0.70 ± 0.06 |
| 300ΔG+MCS5:algG(D324A) | 1.68E+02 | 0.97 ± 0.04 |
| 300ΔX+MCS5:algX | 3.00E+02 | 0.42 ± 0.02 |
| 300ΔX+MCS5:algX(S269A) | 2.20E+02 | 0.55 ± 0.02 |
| 300ΔGΔX+MCS5:algG(D324A):algX(S269A) | 1.20E+03 | 0.49 ± 0.03 |
| 300+MCS5 | 4.46E+02 | 2.40 ± 0.29 |
| 300Δ8+MCS5 | 1.39E+02 | 1.46 |
| 300Δ44+MCS5 | 1.42E+02 | 0.7 |
| 300ΔG+MCS5 | 8.90E+01 | 1.01 |
| 300ΔX+MCS5 | 1.16E+02 | 0.94 |
Total fluorescence per volume of biofilm.
Ratio between red and green fluorescence shown by each biofilm-forming mutant.
300, PDO300; MCS5, pBBR1MCS-5.
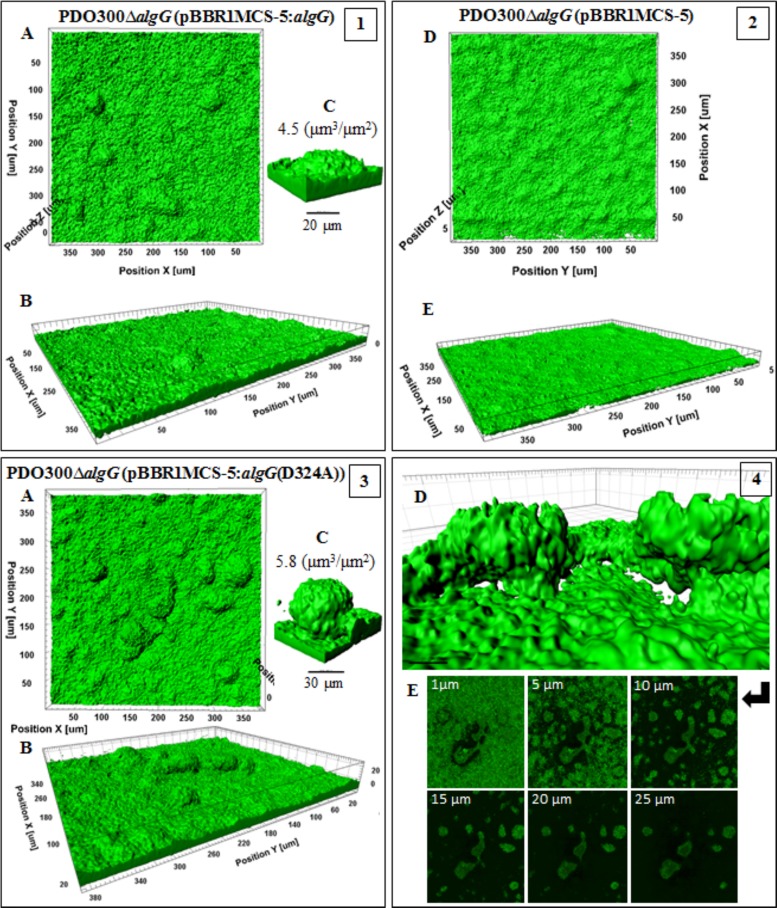
The PDO300ΔalgG(pBBR1MCS-5:algG(D324A)) mutant, which produced a high-molecular-mass, acetylated polymannuronate with strong viscoelasticity, acquired the largest biovolume of 6.0 ± 0.22 µm3 ⋅ µm−2 (Fig. 7, frame 3). Interestingly, adjacent structures were networked with horizontal appendages, with void spaces and channels formed underneath whole structures, likely to constitute water channels (Fig. 7, frame 4). On the other hand, the PDO300ΔalgG(pBBR1MCS-5:algG) mutant, which produced lower-molecular-mass, acetylated, and G residue-containing alginate, formed elevated but less developed structures with less biovolume, 4.8 ± 0.22 µm3 ⋅ µm−2 (Fig. 7, frame 1). The base layers formed by both strains were dense and covered the whole area of the surface. The biofilm of the PDO300ΔalgG(pBBR1MCS-5) mutant was a homogenous layer of cells (7-µm thickness) without elevated structures (Fig. 7, frame 2).
FIG 7 .
Biofilm architecture of mutants producing epimerized and nonepimerized alginates. This figure shows biofilm formation and architecture of the PDO300ΔalgG(pBBR1MCS-5:algG) (frame 1) and PDO300ΔalgG(pBBR1MCS-5:algG(D324A)) (frames 3 and 4) mutants, which produce, respectively, epimerized [poly(MG)] and nonepimerized [poly(M)] alginates, and the PDO300ΔalgG(pBBR1MCS-5) (frame 2) mutant with no alginate production. In all frames, A, B, and C show, respectively, top views, side views, and a representative of typical highly structured cell communities for that mutant with biovolume-per-area (µm3 ⋅ µm−2) ratio. In frame 3, poly(M) alginate-based biofilm is more highly developed than poly(MG) alginate-based biofilm in frame 1, presenting larger biovolume and biovolume-per-area ratios. Cells of both mutants covered the entire cover slide surface. Frames 4D and E represent the architecture of poly(M) alginate-based microcolonies in which two adjacent structures are connected with horizontal appendages and free-cell void cavities channeled underneath of microcolonies. Frame 4E shows 6 different slices of microcolonies with connected structures at the middle of the figures surrounded by free-cell and matrix areas. Frame 2 represents the homogenous cell community of a nonmucoid mutant without highly structured architecture.
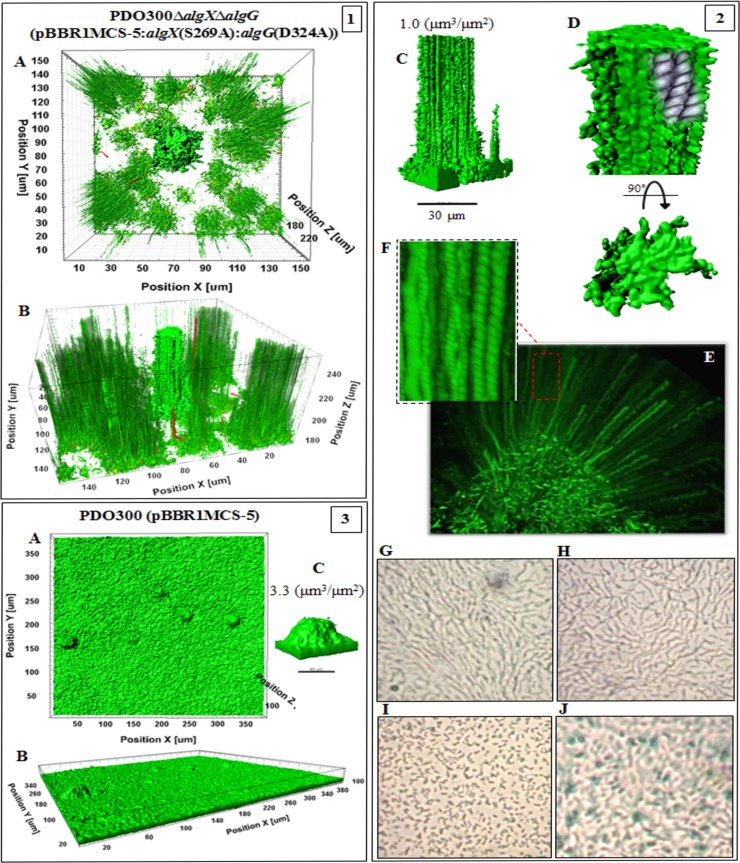
The PDO300ΔalgGΔalgX(pBBR1MCS-5:algX(S269A):algG(D324A)) mutant produced a nonacetylated polymannuronate with a low molecular mass, and the respective biofilm was composed of very long and narrowly elevated structures (Fig. 8, frames 1 and 2). The biovolume was 1.5 ± 0.2 µm3 ⋅ µm−2), which was less than for all the other investigated strains (Table 2).
FIG 8 .
Biofilm architecture of mutant-producing nonepimerized and nonacetylated alginates and the wild type. This figure shows biofilm formation and architecture of the PDO300ΔalgXΔalgG(pBBR1MCS-5:algX(S269A):algG(D324A)) (frames 1 and 2) and PDO300(pBBR1MCS-5) (frame 3) mutants. In all frames, A, B, and C show, respectively, top views, side views, and a representative of typical highly structured cell communities for that mutant with biovolume-per-area (µm3 ⋅ µm−2) ratio. The biofilm architecture visualized for the mutant producing nonacetylated poly(M) alginate (frames 1and 2) was remarkably different from that of other applied mutants. Affected by alginate properties, emerging biofilm consists of very narrow but long elevated microcolonies representing longitudinal cell trails or strips indicating stigmergic self-organization and adaptation of cells in weak matrices. Frames 2D to F represent close side and top views of one of the microcolonies and cell trails, and cell-cell interactions in each cell trail are depicted in sketches. Frames 2G to J show micrographs (×40 magnification) of the edge (H and I) and surface (G and J) of mucoid colonies of the PDO300ΔalgXΔalgG(pBBR1MCS-5:algX(S269A):algG(D324A)) (G and H) and PDO300(pBBR1MCS-5) (I and J) mutants forming a thin layer on PIA medium after incubation at 37°C for 18 h. Organization of cells of the PDO300ΔalgXΔalgG(pBBR1MCS-5:algX(S269A):algG(D324A)) mutant showed a linear filamentous aggregation pattern. Wild-type biofilm architecture is presented in frame 3.
The PDO300Δalg8(pBBR1MCS-5:alg8) and PDO300Δalg44(pBBR1MCS-5:alg44) strains established heterogeneous highly structured biofilms (Fig. 9). The former formed a biovolume of 3.95 ± 0.43 µm3 ⋅ µm−2 and compactness of 6.09E+02, while the latter generated very dense and large structures with a biovolume of 5.8 ± 0.43 µm3 ⋅ µm−2 but less compactness (4.43E+02). Both mutants showed higher numbers of dead cells than all applied mutants but fewer dead cells than the wild type (Table 2). Conversely, the PDO300Δalg8(pBBR1MCS-5) and PDO300Δalg44(pBBR1MCS-5) strains generated homogenous biofilm without elevated or highly structured architectures. Compactness values and dead/live ratios are summarized in Table 2.
FIG 9 .
Biofilm architecture of a mutant producing a high mannuronate molar fraction and M-block. This figure shows biofilm formation and architecture of the PDO300Δalg8(pBBR1MCS-5:alg8) (frame 1) and PDO300Δalg44(pBBR1MCS-5:alg44) (frame 3) mutants and nonmucoid mutants (frames 2 and 4). In all frames, A, B, and C show, respectively, top views, side views, and a representative of typical highly structured cell communities for that mutant with biovolume-per-area (µm3 ⋅ µm−2) ratio. Both mutants produce alginates with the highest degree of M-block occurrence but very different degrees of acetylation. The PDO300Δalg44(pBBR1MCS-5:alg44) (frame 3) mutant, which produces highly acetylated alginate, established very dense and highly developed and larger microcolonies than the PDO300Δalg8(pBBR1MCS-5:alg8) (frame 1) mutant. One explanation for this significance difference is the presence of an additional copy of Alg44 which senses c-di-GMP, which is a common secondary messenger in the cells governing the physiological condition of cells during colonization. However, nonmucoid mutants did not establish highly structured biofilm and microcolonies.
DISCUSSION
In this study, we investigated the relationship between alginate polymerization and modification, the functional role of the subunits Alg8, Alg44, AlgG, and AlgX, and their physical and functional interaction. A range of alginate compositions and molecular masses exhibiting various material properties were produced by engineered P. aeruginosa strains, and their impact on motility and biofilm formation was assessed. This study revealed protein-protein interaction between Alg8 and Alg44, proposed as alginate polymerase and copolymerize (Fig. 1). Alg44 was found to interact with AlgK (Fig. 2A), which is an outer membrane lipoprotein that aids the correct localization of the AlgE porin to the outer membrane. This study provides experimental evidence for the previously suggested presence of an Alg8-Alg44-AlgX-AlgK-AlgE multiprotein complex bridging the cell envelope forming the alginate polymerization/modification/secretion machinery (21). The absence of the previously observed AlgK-X interaction after DSG cross-linking and anti-AlgX immunoblotting (Fig. 2D) was presumable due to a lack of suitable cross-linking sites (K residues 7 Å apart from each other) (22). In addition, immunoblotting/cross-linking data suggested that Alg44 forms a dimer. However, this dimer was not observed when Alg44 was overproduced from a plasmid, indicating that Alg44 dimerization might be susceptible to changes in stoichiometry (Fig. 2C and D). Bioinformatics analysis of the periplasmic part of Alg44 suggests the presence of coil-coiled structures (Coil/Pcoils-based score of 0.4), which have been described for membrane fusion proteins (MFPs) such as MexA and polysaccharide copolymerases (PCPs) to contribute to oligomerization (38). Here, it was demonstrated that Alg44 localization, stability, and protein-protein interaction were not impacted by altering the c-di-GMP level or by non-c-di-GMP binding variants (Fig. 3; see also Fig. S1 in the supplemental material). Our results indicated that the c-di-GMP-mediated activation mechanism of alginate polymerization differs from the activation mechanism of cellulose synthase in which c-di-GMP releases an autoinhibited state by breaking a salt bridge (30). However, residue H323 of Alg8 might be involved in c-di-GMP-dependent activation of alginate polymerase, as reduced c-di-GMP levels did not impair in vivo Alg8 activity (Fig. 4).
The first experimental evidence was obtained that AlgX and AlgG exhibit a mutually auxiliary behavior, suggesting that the two modification events (acetylation and epimerization) are not competitive but linked (35). In addition, we propose a new auxiliary role for Alg44 in acetylation besides being necessary for c-di-GMP-dependent activation of alginate polymerization (Table 1; Fig. 5A). Failed attempts in complementing the PDO300Δalg44ΔalgX mutant with pBBR1MCS-5:alg44:algX, but successful complementation if one gene was present in cis and the other in trans, suggested that the stoichiometry of these two proteins is critical for proper performance of the multiprotein complex.
This study revealed how alginate polymerization (Alg8, Alg44) is aligned with alginate modification (AlgG, AlgX). As shown in Fig. 5A, additional copies of active or inactive AlgX acetyltransferase significantly increased the molar fraction of G residues as well as productivity, which appeared inversely correlated with the alginate molecular mass (Fig. 5A; Table 1). This suggests a new role of AlgX in epimerization and a periplasmic scaffold protein playing a key role in efficient translocation of the alginate chain across the periplasm. Recently, it was reported that AlgX binds to polymannuronic acid in a length-dependent manner and acts as a terminal acetyltransferase (11).
Interestingly, restoration of alginate production of the AlgG-negative mutant by an inactive variant of AlgG led to a significantly increased alginate molecular mass compared to that of active AlgG, suggesting that AlgG as a scaffold subunit is critical for processivity of alginate polymerization, while the actual epimerization event interferes with processivity (Fig. 5A and B). Furthermore, the role of AlgG-mediated epimerization on alginate length might be due to AlgG-mediated alginate degradation, as polysaccharide epimerases show a reaction mechanism similar to that of polysaccharide lyases (35). This finding might also explain why algal alginates with a high molar fraction of G residues introduced by epimerases have very low molecular masses (39). Acetylation was correlated with the molecular mass, suggesting that there was no impact on processivity of alginate polymerization (Fig. 5B).
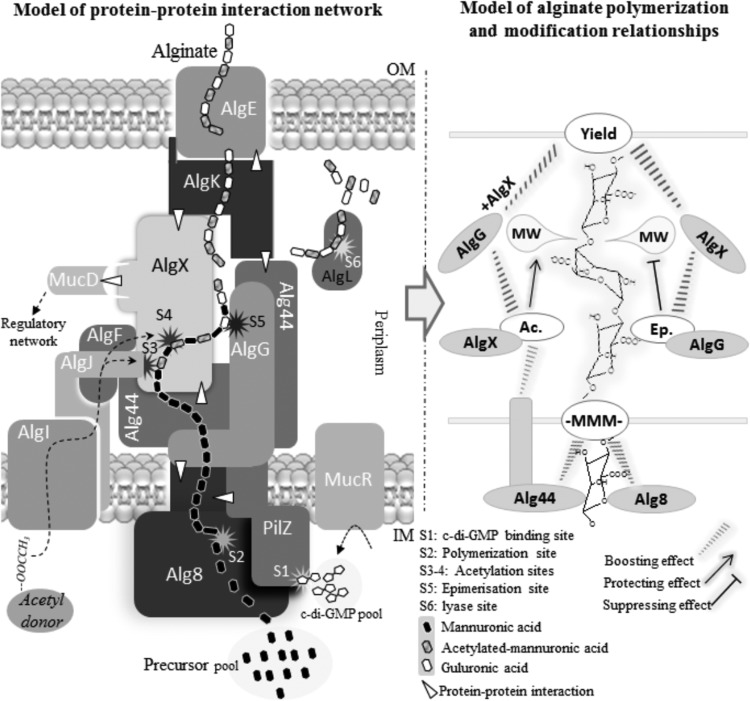
In general, additional copies of any subunit increased alginate production compared to that of the reference strain, indicating that the stoichiometry of the various subunits is less critical for the activity of the multiprotein complex (Table 1). Based on these results with regard to the roles of the investigated subunits in alginate synthesis and modification, a revised model of the alginate biosynthesis multiprotein complex was proposed (Fig. 10).
FIG 10 .
A new proposed model of alginate biosynthesis machinery complex and interactive performances of protein functionality over alginate polymerization, acetylation, epimerization, and length determination. Based on experimental results obtained in the current study, functional and physical interactions of subunits (left side of figure) were modeled into a multiprotein complex. In this model, different binary interactions are marked with white triangles. Our finding of Alg8-Alg44 and Alg44-AlgK interactions together with the previously discovered interaction of AlgK-AlgE (21) constitute the periplasmic part of the multiprotein complex connecting the cytoplasmic membrane with the outer membrane. The model also includes the following interactions, Alg44-AlgX, Alg8-AlgG, and MucD-AlgX-AlgK (21, 22, 49), in support of the periplasmic scaffold guiding nascent alginate for translocation and secretion. This model shows alginate production is positively regulated by c-di-GMP binding to Alg44 (S1) which targets the catalytic site of Alg8 polymerase (S2) through an unknown mechanism. Then, translocation across the periplasmic scaffold is coupled with interactive functional performances of modification events and proteins (right side of model) for alginate length regulation and a series of associated modification events (S3 to S5). AlgL is responsible for degrading misguided alginate accumulating in periplasm (S6). MucD protein links the complex with the posttranslational alginate regulatory network via an interaction with AlgX.
To shed light on the structure-function relationship of the various alginates, their viscoelasticity was assessed. The presence of acetyl groups lowered viscoelasticity by possibly interfering with intermolecular alginate chain interactions (see Fig. S4 in the supplemental material). In contrast, increasing molar fractions of MM-blocks and higher molecular masses increased viscoelasticity (see Fig. S4 in the supplemental material). Acetylated alginates gave rise to well-developed and highly organized heterogeneous architectures and promoted cell aggregations (Fig. 6), which was consistent with previous studies (40), but these findings suggested viscoelasticity is not critical for biofilm architecture formation. Figure 7 shows that nonepimerized alginate (polymannuronate) with high molecular mass and the strong viscoelasticity supported the establishment of these biofilm features and that by controlling the molar fraction of G residue biofilm architecture, characteristics could be adapted to various environments. We showed that the lack of G residues and acetyl groups caused the formation of undeveloped and narrow microcolonies, which were supported by specific long trails or strips of cells emerging from stigmergic self-organization of cells affected by this particular alginate (Fig. 8) (41). This was further evidence for the role of alginate material properties on the formation of particular biofilm architectures and cellular aggregation patterns.
Motility mediated by twitching, swarming, and swimming was assessed using strains producing various alginates. These motilities are critical for biofilm development and dispersal (42). Here, we showed that overproduction of alginate interferes with motility. Previously, it was shown that high levels of c-di-GMP reduced motility while increasing production of various exopolysaccharides and biofilm formation (43). Moreover, our results showed these motilities are independent of the acetyl group and G residue content of the respective alginate (see Fig. S5 in the supplemental material). Overall, this study led to updating the protein-protein interaction network constituting the proposed alginate polymerization/modification/secretion multiprotein complex in P. aeruginosa and the development of strains producing a range of alginates, enabling the analysis of structure-function relationships from a material property and biological function perspective, such as demonstrating that viscoelasticity of alginate contributed to enhanced cell aggregation during biofilm formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in this study are summarized in Table S1 in the supplemental material. P. aeruginosa and Escherichia coli strains were grown at 37°C. For those assays designed for studying alginate production, bacterial phenotype, and protein-protein interaction and stability, Difco Pseudomonas isolation agar (PIA) medium was used. To rule out the effect of growth condition and media on alginate composition and quantity, all experiments were conducted under the same conditions and using the same batch culture at the same time (see the supplemental material).
Construction of isogenic mutants with single- and double-gene knockouts of alg8, alg44, algG, and algX genes.
The P. aeruginosa PDO300 strain was used to generate isogenic mutants with single- and double-gene knockouts of alg8, alg44, algG, and algX genes through two events of homologous recombination using suicide plasmid pEX100T containing knockout genes which were disrupted by the aacC1 gene (encoding gentamicin acetyltransferase) flanked by two FRT sites (see the supplemental material).
In trans complementation of single- and double-gene-knockout mutants and chromosomal integrations.
Relevant genes encoding Alg8, Alg44, AlgG, and AlgX (with and without the 6His tag) were individually or in combination transferred into generated mutants using the pBBR1MCS-5 plasmid and also incorporated into the genome using mini-CTX-lacZ plasmid (see the supplemental material). The pBBR1MCS-5 plasmid containing the various alginate genes was considered to study the impact on production of alginates and their characteristics in order to more sensitively detect changes in the alginates due to additional copy numbers of the respective alginate protein under investigation.
Site-specific mutations and deletions of alg44 and alg8.
Site-specific mutations and deletions of alg44 and alg8 genes were performed using the QuikChange II site-directed mutagenesis kit (Stratagene) or by DNA synthesis (GenScript), resulting in genes encoding the Alg8(E322A/H323/E326A) protein, Alg44(R17A/R21A/R21D) protein, and Alg44's N- and C-terminal truncation (see the supplemental material).
In vivo detection of the protein-protein interaction network.
In vivo detection of protein-protein interaction was performed by employing pulling down 6His-tagged proteins under native conditions, in vivo chemical cross-linking, and bacterial two-hybrid assay (see the supplemental material).
Assessment of the stability of Alg44 variants in the presence and absence of MucR (DGC/PDE)/RocR (PDE).
Previously generated mutants, including the PDO300ΔmucR, PDO300ΔmucR(pBBR1MCS-5:mucR), PDO300(pBBR1MCS-5:rocR), and PDO300ΔmucR(pBBR1MCS-5:rocR) mutants, were shown to positively or negatively regulate alginate production through c-di-GMP synthesis (by MucR) or degradation (by RocR) (24). Also to use presumably nonfunctional Alg44 in binding to c-di-GMP and/or in alginate polymerization (8, 26), the PDO300Δalg44(pBBR1MCS-5:alg44(R21D)-6His), PDO300Δalg44(pBBR1MCS-5:alg44-6His(Δ40-74aaPilZ)), and PDO300Δalg44(pBBR1MCS-5:alg44-6His(Δ364-389aa)) mutants were generated. Using these mutants, we examined the direct effect of c-di-GMP and defective variants of Alg44 on its localization and stability in planktonic and biofilm cells. Cells grown on solid media or in liquid cultures as described above were washed twice with saline. Enzymatic cell lysis was performed using the abovementioned lysis buffer prepared in buffer A followed by sonication and isolation of the cell envelope fraction by ultracentrifugation at 100,000 × g for 1 h at 4°C. Pellets were solubilized with buffer A for protein analysis.
Isolation of cytoplasmic membrane and general protein analysis.
To confirm the localization of Alg8 and Alg44 in the cytoplasmic membrane of E. coli BTH101, the cytoplasmic membrane fraction was isolated as described previously with some modifications (44) (see supplemental material).
Protein samples were generally analyzed utilizing SDS-PAGE (8% acrylamide gels) and immunoblotting (see the supplemental material).
Alginate purification and quantification.
Two milliliters of bacterial overnight culture grown in LB medium supplemented with the appropriate antibiotic was sedimented, and cells were washed twice with saline solution. Cells were suspended in 1 ml of saline solution, and 200 µl of cell suspension was plated onto PIA medium (in triplicates) containing 300 µg ⋅ ml−1 of gentamicin and then incubated at 37°C for 72 h. Cells of each agar plate were scraped off and suspended in saline solution until the biomass was completely suspended. Then, suspensions were pelleted and alginates in supernatants were precipitated with equal volume of ice-cold isopropanol. The alginate precipitants were freeze-dried and then redissolved in 50 mM Tris-HCl (pH 7.4) and 10 mM MgCl2 to a final concentration of 0.5% (wt/vol), followed by incubation with 15 µg ⋅ ml−1 DNase I and 15 µg ⋅ m ⋅ liter−1 RNase I at 37°C for 6 h. Then, pronase E was added to a final concentration of 20 µg ⋅ ml−1 and incubated for a further 18 h at 37°C. Alginate solutions were dialyzed (12- to 14-kDa molecular mass cutoff; ZelluTrans/Roth mini dialyzer; Carl Roth GmbH & Co.) against 5 liters of ultrapure H2O for 48 h. Finally, alginates were precipitated with an equal volume of ice-cold isopropanol and freeze-dried for uronic acid assay and biochemical analysis (see the supplemental material).
The analysis of molecular mass, composition, and viscoelastic properties of the alginates.
Various alginates produced by different complemented mutants were subjected to molecular mass analysis using size exclusion chromatography-multiangle laser light scattering (SEC-MALLS) and compositional analysis utilizing 1H-nuclear magnetic resonance (NMR) spectroscopy and Fourier transform infrared (FTIR) spectrometry (see Text S1 in the supplemental material. Microrheological analysis was used to measure the viscoelastic property of the alginates in which the mean square displacement (MSD) of probe particles embedded in the samples and in turn the viscoelastic moduli (G' [elastic] and G'' [viscose]) were measured (see the supplemental material).
Continuous-culture flow cell biofilms, quantitative analysis, and motility assays.
Biofilm architecture analysis was performed for those mutants producing alginates with very distinct composition and properties from each other, including the PDO300(pBBR1MCS-5), PDO300Δalg8(pBBR1MCS-5:alg8), PDO300Δalg44(pBBR1MCS-5:alg44), PDO300ΔalgG(pBBR1MCS-5:algG), PDO300ΔalgG(pBBR1MCS-5:algG(D324A)), PDO300ΔalgX(pBBR1MCS-5:algX), PDO300ΔalgX(pBBR1MCS-5:algX(S269A)), and PDO300ΔalgXΔalgG(pBBR1MCS-5:algX(S269A):algG(D324A)) mutants. Each mutant was grown in continuous-culture flow cells (channel dimensions of 4 mm by 40 mm by 1.5 mm) at 37°C (45). The flow cells were then incubated at 37°C for 24 h. Biofilms were stained utilizing the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes) and visualized using confocal laser scanning microscopy (Leica SP5 DM6000B). For quantitative analysis of biofilms, IMARIS image analysis software (Bitplane) was employed. Biofilm architecture and appearance, biovolume (µm3), the ratio of biovolume per unit area (µm3 ⋅ µm−2), dead-to-live ratio, compactness, and thickness of base layers were analyzed (46–48). Motilities, including twitching, swarming, and swimming, were assessed by the method explained by Pang et al. (42) with modification. Full experimental details are provided in the supplemental materials.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods. Download
Alg44 stability is independent of c-di-GMP. In envelope fractions of planktonic (Li, liquid) and biofilm (So, solid) cells and in the presence and/or absence of WspR (generating c-di-GMP) (lanes 1 and 2) and RocR (degrading c-di-GMP) (lanes 3 and 4) and MucR (alginate biosynthesis-associated c-di-GMP regulator) (lanes 5 and 6), Alg44 was detected using immunoblotting with anti-Alg44 antibody. Lanes 7 to 9 show negative and positive controls. 300, PDO300; MCS5, pBBR1MCS-5. Download
1H-NMR spectra of acetylated (A) and deacetylated (B) samples. (C) Representative 1H-NMR spectrum for M and G residue detection. H1-MM, H1-G, and H1-MG indicate the anomeric protons of M residues followed by another M residue, G residues along the alginate chain, and M residues followed by a G residue, respectively. (D) Representative 1H-NMR spectrum for O-acetylation indicates the methyl group of the acetyl residues along the alginate chain. H1-M indicates the anomeric proton of nonacetylated M residues. H1-MO-Ac, H2-MO-Ac, and H3-MO-Ac indicate the anomeric proton, hypotheses 2 and 3 of acetylated M residues. Download
Plots show molar mass of alginate samples versus time analyzed using SEC-MALLS. 300, PDO300; MCS5, pBBR1MCS-5. Download
Viscoelastic property of alginates was impacted by molecular mass and modifications. The plot of MSD versus time shows distribution of alginate viscoelastic properties in four distinct regions. Alginate composition and viscoelasticity moduli (G', elastic; G", viscous) are presented next to the plot. Bold and underlined letters indicate the predominance of mannuronate (M)/guluronate (G) molar fraction or acetylation (Ac.) degree among all analyzed alginates. 300, PDO300; MCS5, pBBR1MCS-5. Download
Impact of alginates on motility of P. aeruginosa. Twitching, swarming, and swimming were assessed in triplicates on 1%, 0.5%, and 0.3% agar plates, respectively, and after incubation at 30°C for 18 to 24 h. The means of colony diameter values ± standard deviation errors are graphed. Download
Strains and plasmids applied in this study.
ACKNOWLEDGMENTS
We are thankful to Lynne Howell from the University of Toronto and Dennis E. Ohman from Virginia Commonwealth University for providing antibodies and Iain D. Hay for advice and technical assistance, the Manawatu Microscopy and Imaging Centre (MMIC) of Massey University, and especially Matthew Savoian for microscopic technical assistance and Martin A. K. Williams and Bradley W. Mansel for assisting us with microrheology analysis.
This work was supported by grants from the Massey University Research Fund and the MacDiarmid Institute of Advanced Materials and Nanotechnology (New Zealand) to B.H.A.R. M.F.M. is funded by a Massey University doctoral scholarship.
Footnotes
Citation Moradali MF, Donati I, Sims IM, Ghods S, Rehm BHA. 2015. Alginate polymerization and modification are linked in Pseudomonas aeruginosa. mBio 6(3):e00453-15. doi:10.1128/mBio.00453-15.
REFERENCES
- 1.Rehm BH, Grabert E, Hein J, Winkler UK. 1994. Antibody response of rabbits and cystic fibrosis patients to an alginate-specific outer membrane protein of a mucoid strain of Pseudomonas aeruginosa. Microb Pathog 16:43–51. doi: 10.1006/mpat.1994.1004. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen SS, Høiby N, Espersen F, Koch C. 1992. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 47:6–13. doi: 10.1136/thx.47.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hay ID, Ur Rehman Z, Ghafoor A, Rehm BHA. 2010. Bacterial biosynthesis of alginates. J Chem Technol Biotechnol 85:752–759. doi: 10.1002/jctb.2372. [DOI] [Google Scholar]
- 4.Hay ID, Ur Rehman Z, Moradali MF, Wang Y, Rehm BH. 2013. Microbial alginate production, modification and its applications. Microb Biotechnol 6:637–650. doi: 10.1111/1751-7915.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chitnis CE, Ohman DE. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol 8:583–593. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 7.Rehm BHA, Remminghorst U, Hay ID. 2009. Molecular characterization of Alg8, a putative glycosyltransferase, involved in alginate polymerisation. J Biotechnol 140:176–183. doi: 10.1016/j.jbiotec.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Lory S, Merighi M, Lee VT, Hyodo M, Hayakawa Y. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 9.Rehm BHA, Remminghorst U. 2006. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl Environ Microbiol 72:298–305. doi: 10.1128/AEM.72.1.298-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehm BHA, Hay ID, Remminghorst U. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker P, Ricer T, Moynihan PJ, Kitova EN, Walvoort MT, Little DJ, Whitney JC, Dawson K, Weadge JT, Robinson H, Ohman DE, Codée JD, Klassen JS, Clarke AJ, Howell PL. 2014. P. aeruginosa SGNH hydrolase-like proteins AlgJ and AlgX have similar topology but separate and distinct roles in alginate acetylation. PLoS Pathog 10:e1004334. doi: 10.1371/journal.ppat.1004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin MJ, Ohman DE. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J Bacteriol 184:3000–3007. doi: 10.1128/JB.184.11.3000-3007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riley LM, Weadge JT, Baker P, Robinson H, Codée JD, Tipton PA, Ohman DE, Howell PL. 2013. Structural and functional characterization of Pseudomonas aeruginosa AlgX: role of AlgX in alginate acetylation. J Biol Chem 288:22299–22314. doi: 10.1074/jbc.M113.484931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain S, Franklin MJ, Ertesvåg H, Valla S, Ohman DE. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol Microbiol 47:1123–1133. doi: 10.1046/j.1365-2958.2003.03361.x. [DOI] [PubMed] [Google Scholar]
- 15.Schiller NL, Robles-Price A, Wong TY, Sletta H, Valla S. 2004. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol 186:7369–7377. doi: 10.1128/JB.186.21.7369-7377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakkevig K, Sletta H, Gimmestad M, Aune R, Ertesvåg H, Degnes K, Christensen BE, Ellingsen TE, Valla S. 2005. Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J Bacteriol 187:8375–8384. doi: 10.1128/JB.187.24.8375-8384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keiski CL, Harwich M, Jain S, Neculai AM, Yip P, Robinson H, Whitney JC, Riley L, Burrows LL, Ohman DE, Howell PL. 2010. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18:265–273. doi: 10.1016/j.str.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehman ZU, Rehm BH. 2013. Dual roles of Pseudomonas aeruginosa AlgE in secretion of the virulence factor alginate and formation of the secretion complex. Appl Environ Microbiol 79:2002–2011. doi: 10.1128/AEM.03960-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douthit SA, Dlakic M, Ohman DE, Franklin MJ. 2005. Epimerase active domain of Pseudomonas aeruginosa AlgG, a protein that contains a right-handed beta-helix. J Bacteriol 187:4573–4583. doi: 10.1128/JB.187.13.4573-4583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albrecht MT, Schiller NL. 2005. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol 187:3869–3872. doi: 10.1128/JB.187.11.3869-3872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehman ZU, Wang Y, Moradali MF, Hay ID, Rehm BH. 2013. Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol 79:3264–3272. doi: 10.1128/AEM.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay ID, Schmidt O, Filitcheva J, Rehm BH. 2012. Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl Microbiol Biotechnol 93:215–227. doi: 10.1007/s00253-011-3430-0. [DOI] [PubMed] [Google Scholar]
- 23.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay ID, Remminghorst U, Rehm BH. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehm BHA, Remminghorst U. 2006. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett 580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 26.Ohman DE, Oglesby LL, Jain S. 2008. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steiner S, Lori C, Boehm A, Jenal U. 2013. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J 32:354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehm B. 2009. Microbial production of biopolymers and polymer precursors: applications and perspectives. Caister Academic, Wymondham, United Kingdom. [Google Scholar]
- 29.Morgan JL, Strumillo J, Zimmer J. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan JL, McNamara JT, Zimmer J. 2014. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. doi: 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 32.Franklin MJ, Ohman DE. 1993. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol 175:5057–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franklin MJ, Ohman DE. 1996. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J Bacteriol 178:2186–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, Ohman DE. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol 176:1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfram F, Kitova EN, Robinson H, Walvoort MT, Codée JD, Klassen JS, Howell PL. 2014. Catalytic mechanism and mode of action of the periplasmic alginate epimerase AlgG. J Biol Chem 289:6006–6019. doi: 10.1074/jbc.M113.533158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billings N, Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. 2013. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. 2009. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in Gram-negative bacteria. Microbiol Mol Biol Rev 73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galindo E, Peña C, Núñez C, Segura D, Espín G. 2007. Molecular and bioengineering strategies to improve alginate and polydydroxyalkanoate production by Azotobacter vinelandii. Microb Cell Fact 6:7. doi: 10.1186/1475-2859-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tielen P, Strathmann M, Jaeger KE, Flemming HC, Wingender J. 2005. Alginate acetylation influences initial surface colonization by mucoid Pseudomonas aeruginosa. Microbiol Res 160:165–176. doi: 10.1016/j.micres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Gloag ES, Javed MA, Wang H, Gee ML, Wade SA, Turnbull L, Whitchurch CB. 2013. Stigmergy: a key driver of self-organization in bacterial biofilms. Commun Integr Biol 6:e27331. doi: 10.4161/cib.27331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pang CM, Hong P, Guo H, Liu WT. 2005. Biofilm formation characteristics of bacterial isolates retrieved from a reverse osmosis membrane. Environ Sci Technol 39:7541–7550. doi: 10.1021/es050170h. [DOI] [PubMed] [Google Scholar]
- 43.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 44.Hancock RE, Nikaido H. 1978. Outer membranes of Gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol 136:381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campisano A, Schroeder C, Schemionek M, Overhage J, Rehm BH. 2006. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl Environ Microbiol 72:3066–3068. doi: 10.1128/AEM.72.4.3066-3068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghafoor A, Hay ID, Rehm BH. 2011. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuehn M, Hausner M, Bungartz HJ, Wagner M, Wilderer PA, Wuertz S. 1998. Automated confocal laser scanning microscopy and semiautomated image processing for analysis of biofilms. Appl Environ Microbiol 64:4115–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. [DOI] [PubMed] [Google Scholar]
- 49.Hay ID, Schmidt O, Filitcheva J, Rehm BH. 2012. Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl Microbiol Biotechnol 93:215–227. doi: 10.1007/s00253-011-3430-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods. Download
Alg44 stability is independent of c-di-GMP. In envelope fractions of planktonic (Li, liquid) and biofilm (So, solid) cells and in the presence and/or absence of WspR (generating c-di-GMP) (lanes 1 and 2) and RocR (degrading c-di-GMP) (lanes 3 and 4) and MucR (alginate biosynthesis-associated c-di-GMP regulator) (lanes 5 and 6), Alg44 was detected using immunoblotting with anti-Alg44 antibody. Lanes 7 to 9 show negative and positive controls. 300, PDO300; MCS5, pBBR1MCS-5. Download
1H-NMR spectra of acetylated (A) and deacetylated (B) samples. (C) Representative 1H-NMR spectrum for M and G residue detection. H1-MM, H1-G, and H1-MG indicate the anomeric protons of M residues followed by another M residue, G residues along the alginate chain, and M residues followed by a G residue, respectively. (D) Representative 1H-NMR spectrum for O-acetylation indicates the methyl group of the acetyl residues along the alginate chain. H1-M indicates the anomeric proton of nonacetylated M residues. H1-MO-Ac, H2-MO-Ac, and H3-MO-Ac indicate the anomeric proton, hypotheses 2 and 3 of acetylated M residues. Download
Plots show molar mass of alginate samples versus time analyzed using SEC-MALLS. 300, PDO300; MCS5, pBBR1MCS-5. Download
Viscoelastic property of alginates was impacted by molecular mass and modifications. The plot of MSD versus time shows distribution of alginate viscoelastic properties in four distinct regions. Alginate composition and viscoelasticity moduli (G', elastic; G", viscous) are presented next to the plot. Bold and underlined letters indicate the predominance of mannuronate (M)/guluronate (G) molar fraction or acetylation (Ac.) degree among all analyzed alginates. 300, PDO300; MCS5, pBBR1MCS-5. Download
Impact of alginates on motility of P. aeruginosa. Twitching, swarming, and swimming were assessed in triplicates on 1%, 0.5%, and 0.3% agar plates, respectively, and after incubation at 30°C for 18 to 24 h. The means of colony diameter values ± standard deviation errors are graphed. Download
Strains and plasmids applied in this study.