Abstract
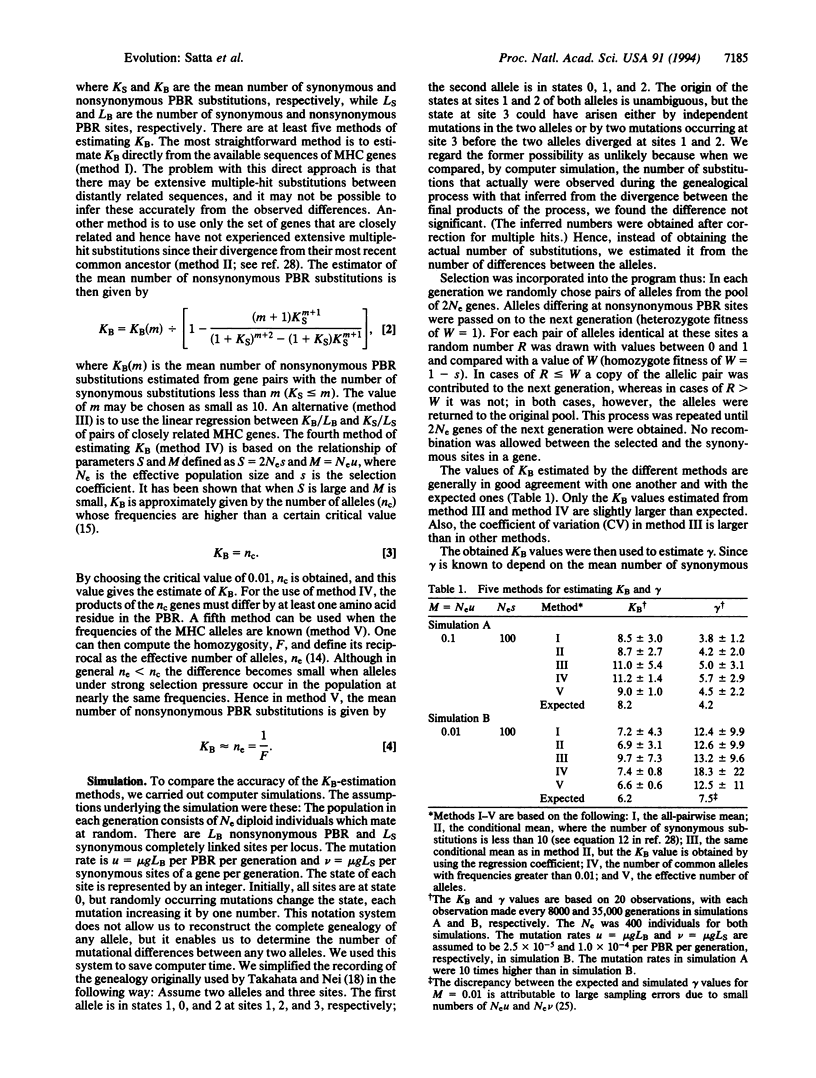
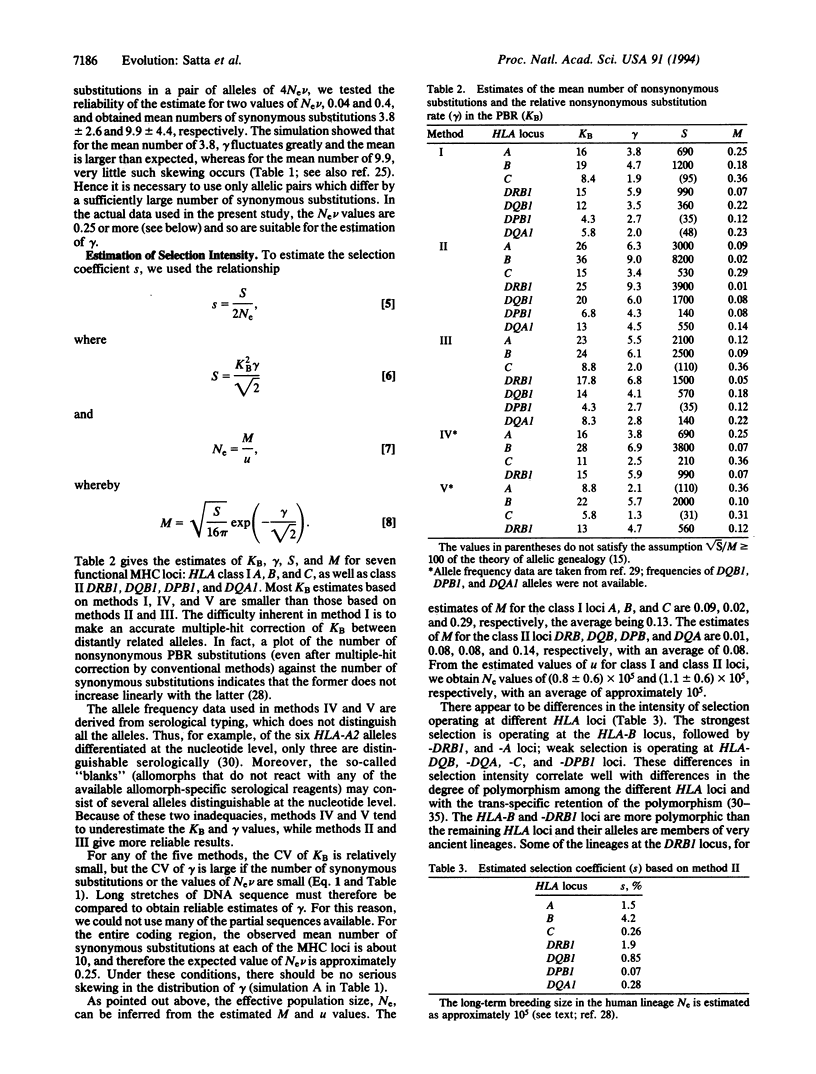
Long persistence of allelic lineages, prevalence of nonsynonymous over synonymous substitutions in the peptide-binding region (PBR), and deviation from neutrality of the expected gene identity parameter F all indicate indirectly that balancing selection is operating at functional major histocompatibility complex (MHC) loci. Direct demonstrations of the existence of balancing selection at MHC loci are, however, either lacking or not fully convincing. To define the conditions under which balancing selection could be demonstrated, we estimated its intensity from the mean number of nonsynonymous substitutions, KB, at the PBR and the mutation rate mu. We compared the five available methods for estimating KB by computer simulation and chose the most reliable ones for estimation of selection intensity. For the human MHC, the selection coefficients of the HLA-A, -B, -C, -DRB1, -DQB1, -DQA1, and -DPB1 loci are 0.015, 0.042, 0.0026, 0.019, 0.0085, 0.0028, and 0.0007, respectively. This low selection intensity places severe restrictions on the possibility of measuring selection directly in vertebrate populations.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bjorkman P. J., Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987 Oct 8;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Bontrop R. E. Nonhuman primate Mhc-DQA and -DQB second exon nucleotide sequences: a compilation. Immunogenetics. 1994;39(2):81–92. doi: 10.1007/BF00188610. [DOI] [PubMed] [Google Scholar]
- Briles W. E., Stone H. A., Cole R. K. Marek's disease: effects of B histocompatibility alloalleles in resistant and susceptible chicken lines. Science. 1977 Jan 14;195(4274):193–195. doi: 10.1126/science.831269. [DOI] [PubMed] [Google Scholar]
- Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993 Jul 1;364(6432):33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Christiansen F. B., Frydenberg O., Simonsen V. Genetics of Zoarces populations. IV. Selection component analysis of an esterase polymorphism using population samples including mother-offspring combinations. Hereditas. 1973;73(2):291–304. doi: 10.1111/j.1601-5223.1973.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Clark A. G., Feldman M. W. The estimation of epistasis in components of fitness in experimental populations of drosophila melanogaster II. Assessment of meiotic drive, viability, fecundity and sexual selection. Heredity (Edinb) 1981 Jun;46(3):347–377. doi: 10.1038/hdy.1981.45. [DOI] [PubMed] [Google Scholar]
- Figueroa F., O'hUigin C., Tichy H., Klein J. The origin of the primate Mhc-DRB genes and allelic lineages as deduced from the study of prosimians. J Immunol. 1994 May 1;152(9):4455–4465. [PubMed] [Google Scholar]
- Hill A. V., Allsopp C. E., Kwiatkowski D., Anstey N. M., Twumasi P., Rowe P. A., Bennett S., Brewster D., McMichael A. J., Greenwood B. M. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991 Aug 15;352(6336):595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Hughes M. K., Watkins D. I. Contrasting roles of interallelic recombination at the HLA-A and HLA-B loci. Genetics. 1993 Mar;133(3):669–680. doi: 10.1093/genetics/133.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988 Sep 8;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- KIMURA M., CROW J. F. THE NUMBER OF ALLELES THAT CAN BE MAINTAINED IN A FINITE POPULATION. Genetics. 1964 Apr;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968 Feb 17;217(5129):624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution: a review of recent evidence. Jpn J Genet. 1991 Aug;66(4):367–386. doi: 10.1266/jjg.66.367. [DOI] [PubMed] [Google Scholar]
- Klein J., Satta Y., O'hUigin C., Takahata N. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- Klitz W., Thomson G., Baur M. P. Contrasting evolutionary histories among tightly linked HLA loci. Am J Hum Genet. 1986 Sep;39(3):340–349. [PMC free article] [PubMed] [Google Scholar]
- Kondo R., Horai S., Satta Y., Takahata N. Evolution of hominoid mitochondrial DNA with special reference to the silent substitution rate over the genome. J Mol Evol. 1993 Jun;36(6):517–531. doi: 10.1007/BF00556356. [DOI] [PubMed] [Google Scholar]
- Lawlor D. A., Warren E., Taylor P., Parham P. Gorilla class I major histocompatibility complex alleles: comparison to human and chimpanzee class I. J Exp Med. 1991 Dec 1;174(6):1491–1509. doi: 10.1084/jem.174.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993 Jan;36(1):96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Marsh S. G., Bodmer J. G. HLA class II nucleotide sequences, 1992. Immunogenetics. 1993;37(2):79–94. doi: 10.1007/BF00216830. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Nei M. Genetic variability maintained by mutation and overdominant selection in finite populations. Genetics. 1981 Jun;98(2):441–459. doi: 10.1093/genetics/98.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W. E., Jonker M., Klein D., Ivanyi P., van Seventer G., Klein J. Nucleotide sequences of chimpanzee MHC class I alleles: evidence for trans-species mode of evolution. EMBO J. 1988 Sep;7(9):2765–2774. doi: 10.1002/j.1460-2075.1988.tb03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'hUigin C., Bontrop R., Klein J. Nonhuman primate Mhc-DRB sequences: a compilation. Immunogenetics. 1993;38(3):165–183. doi: 10.1007/BF00211517. [DOI] [PubMed] [Google Scholar]
- Pamilo P., Bianchi N. O. Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol. 1993 Mar;10(2):271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- Prout T. The estimation of fitnesses from population data. Genetics. 1969 Dec;63(4):949–967. doi: 10.1093/genetics/63.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satta Y. How the ratio of nonsynonymous to synonymous pseudogene substitutions can be less than one. Immunogenetics. 1993;38(6):450–454. doi: 10.1007/BF00184527. [DOI] [PubMed] [Google Scholar]
- Satta Y., O'hUigin C., Takahata N., Klein J. The synonymous substitution rate of the major histocompatibility complex loci in primates. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7480–7484. doi: 10.1073/pnas.90.16.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2419–2423. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990 Apr;124(4):967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Satta Y., Klein J. Polymorphism and balancing selection at major histocompatibility complex loci. Genetics. 1992 Apr;130(4):925–938. doi: 10.1093/genetics/130.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins D. I., Zemmour J., Parham P. Non-human primate MHC class I sequences, 1992. Immunogenetics. 1993;37(5):317–330. doi: 10.1007/BF00216796. [DOI] [PubMed] [Google Scholar]
- Wright S. The Distribution of Self-Sterility Alleles in Populations. Genetics. 1939 Jun;24(4):538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour J., Parham P. HLA class I nucleotide sequences, 1992. Immunogenetics. 1993;37(4):239–250. doi: 10.1007/BF00187449. [DOI] [PubMed] [Google Scholar]
- de Vries R. R., Meera Khan P., Bernini L. F., van Loghem E., van Rood J. J. Genetic control of survival in epidemics. J Immunogenet. 1979 Aug;6(4):271–287. doi: 10.1111/j.1744-313x.1979.tb00684.x. [DOI] [PubMed] [Google Scholar]



