ABSTRACT
We describe here a novel, fast and inexpensive method for producing a 3D ‘heart’ structure that forms spontaneously, in vitro, from larval zebrafish (ZF). We have named these 3D ‘heart’ structures ‘zebrafish heart aggregate(s)’ (ZFHAs) and have characterised their basic morphology and structural composition using histology, immunohistochemistry, electron microscopy and mass spectrometry. After 2 days in culture, the ZFHA spontaneously form and become a stable contractile syncytium consisting of cardiac tissue derived by in vitro maturation, which beats rhythmically and consistently for more than 8 days. We propose this model as a platform technology, which can be developed further to study in vitro cardiac maturation, regeneration, tissue engineering and safety pharmacological/toxicology testing.
KEY WORDS: Drug screening, Ion channels, Tissue engineering, Cardiac tissue, Myocyte, Heart rate
Summary: Spontaneously beating 3D ‘heart’ structures can be developed in vitro from larval zebrafish using a novel, fast and inexpensive method that could be employed in ecotoxicology and biomedical safety testing.
INTRODUCTION
In vitro models are widely used in biomedical, toxicological and pharmacological research as they provide a simplified test system whilst aligning with the principles of reduction in animal use. In vitro systems routinely used for biomedical research draw on advances in developmental biology, biomaterials and stem cell technologies, and provide a mechanism for high throughput compound testing and safety pharmacology (Hirt et al., 2014; Amanfu and Saucerman, 2011). However, the biomedical applicability of many of these current in vitro heart models is limited because: (1) they are derived from embryonic or neonatal rodent tissue, which has electrophysiological properties that differ from those of the human heart (Shenje et al., 2008; Sauer et al., 1999), even when generated in a 3D scaffold-free system (Desroches et al., 2012), and (2) they are often 2D cell monolayers, which lack essential features of 3D hearts, including mature myocytes that couple as a functional syncytium (Ausma and Borgers, 2002). Assay systems using HEK cells (Thomas and Smart, 2005), or human embryonic and induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) avoid these issues, but are expensive to generate and also come with their own limitations, including ethical constraints in terms of the use of human embryos and the potential for tumorigenesis (see Novakovic et al., 2014; Mandel et al., 2012; de Boer et al., 2010). Here, we describe a method for generating an inexpensive, organisationally complex, stable, 3D and spontaneously active cell aggregate derived from larval zebrafish [ZF; Danio rerio (Hamilton 1822)] termed the zebrafish heart aggregate (ZFHA). This method builds on previous work by Grunow et al. (2011) and Mehnert et al. (2013) for salmonids but also offers new investigative potential by virtue of the role of ZF as a model organism for cardiac research. Work by us (Brette et al., 2008) and others (Nemtsas et al., 2010; Verkerk and Remme, 2012; Bovo et al., 2013) clearly shows that the ZF heart is a relevant model for human cardiac ion channel disorders, particularly those associated with cardiac repolarisation. ZF are also an established model species for environmental and (eco) toxicology testing (Strähle et al., 2011; Busch et al., 2011), with recent work focusing on the effect of pollutants on the ZF cardiovascular system (Parker et al., 2014). Thus, we believe this novel in vitro system has promise for toxicology, disease modelling, understanding in vitro maturation, and tissue engineering. The data presented herein outline the methodology required to generate the ZFHA model and provide basic structural and electrical characterisation. Further characterisation will be needed by the research community to fully exploit the potential of our ZFHA model for human and fish cardiac biology.
RESULTS AND DISCUSSION
Generation of ZFHAs
ZF from 1 to 4 days post-fertilisation (dpf) were initially used to develop ZFHAs. The greatest ZFHA formation ratio (i.e. number of ZFHAs developed/number larvae used) was achieved using 3–4 dpf larvae (formation ratio of 0.4) compared with earlier developmental stages (formation ratio of 0.2 or less) (Fig. 1A). All subsequent experiments were conducted on ZFHAs generated from 3–4 dpf larvae and a typical ZFHA from this age group is shown in Fig. 1B. These data are comparable with results from other fish species, where larvae just prior to hatching were optimal for formation of spontaneously beating cell aggregates (Grunow et al., 2012). We analysed ZFHA growth on days 2 and 7 in culture. The ZFHAs were ∼8000±900 µm2 on day 2 and ∼6000±600 µm2 after a week in culture, showing a 25% decrease in size (N=19, P<0.001, Wilcoxon). The reason for the decrease in size during culture was not identified, but may involve sub-optimal culture conditions, suggesting further optimisation of seeding density, survival factors and medium metabolites is required.
Fig. 1.
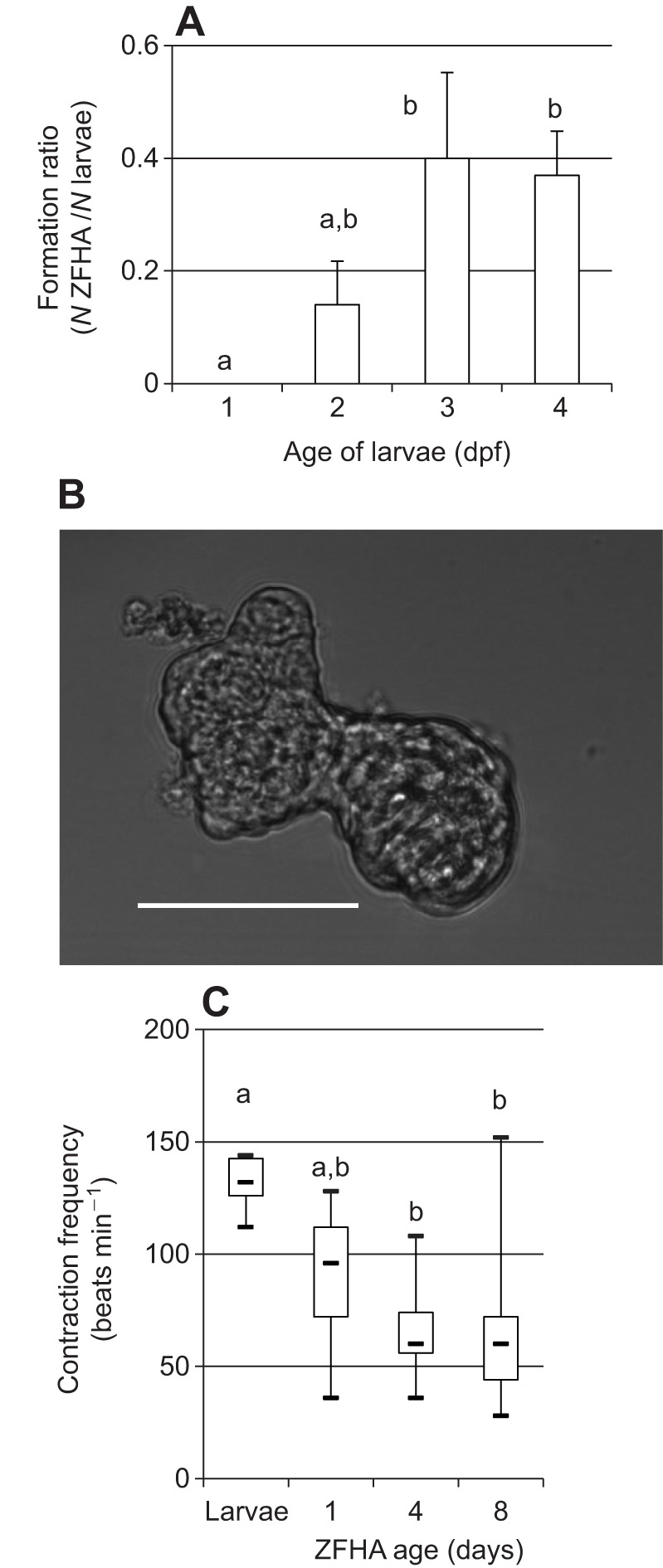
Formation and contraction frequency of zebrafish heart aggregates (ZFHAs) in vitro. (A) Formation ratio calculated as the number of ZFHA developed divided by the number of zebrafish (ZF) larvae dissociated. Number of dissected larvae was: 1 day post-fertilisation (dpf), N=57; 2 dpf, N=80; 3 dpf, N=30; and 4 dpf, N=38. (B) Phase contrast image of ZFHA after 4 days in culture generated using mortar/pestle from 3 dpf larval ZF. Scale bar, 100 µm. (C) Contraction frequency of larval hearts at 3 dpf (N=6) and ZFHA after day 1 (N=21), day 4 (N=19) and day 8 (N=17) in culture illustrated by box–whisker plots showing minimum and maximum contraction (crossbars at the end), median (central bar) and upper and lower quartile (borders of the box). Data represent means±s.e.m. Dissimilar letters indicate significance; one-way ANOVA or ANOVA on ranks, P<0.05 (SPSS22; IBM, USA).
We next assessed the contraction frequency of ZFHAs over 8 days in culture (Fig. 1C). On day 1, ZFHAs had a mean contraction frequency of 100 beats min−1, which decreased to a mean of 67±4.3 beats min−1 and then stayed relatively stable for 8 days. We found no statistical relationship between ZFHA size and contraction frequency. The intrinsic contraction frequency of stable ZFHAs is closer to that of adult ZF hearts (60 beats min−1; Orfanidou et al., 2013) than larval hearts (∼130 beats min−1, Fig. 1C; Barrionuevo and Burggren, 1999) at the same temperature. The intrinsic contraction frequency of the ZFHAs is also akin to that of the adult human heart rate (60–90 beats min−1 at rest). The similarity in heart rate between ZF (at 20°C) and humans (at 37°C) is driven by an action potential with a very similar waveform; both have a prominent plateau phase, which reflects similarities in their underlying ion channel profiles (Brette et al., 2008; Nemtsas et al., 2010; Verkerk and Remme, 2012). Rodents, in contrast, have ∼10 times faster heart rates, and their action potential lacks a significant plateau phase, which is reflected in their ion channel expression (Li et al., 1999; Nerbonne et al., 2001). Grunow et al. (2011) and Mehnert et al. (2013) showed previously that aggregates generated from larval trout have beat rates and action potential characteristics that closely match those of humans and that two commonly used hERG blockers, which are known to be pro-arrhythmic in humans, prolong action potentials in fish aggregates (Mehnert et al., 2013). Whilst we have not confirmed these findings in ZFHAs, our previous work (Brette et al., 2008) suggests ZF have a similar complement of cardiac ion channels to those of adult mammalian myocytes (Nemtsas et al., 2010) and hiPSC-CM (van den Heuvel et al., 2014). Thus, electrical activity between fish and humans is more similar than that between human and mouse, and, as such, ZFHA may be suitable for high/medium throughput pharmacological screening for human safety pharmacology (Brette et al., 2008; Nemtsas et al., 2010; Mehnert et al., 2013).
Morphological and proteomic characterisation
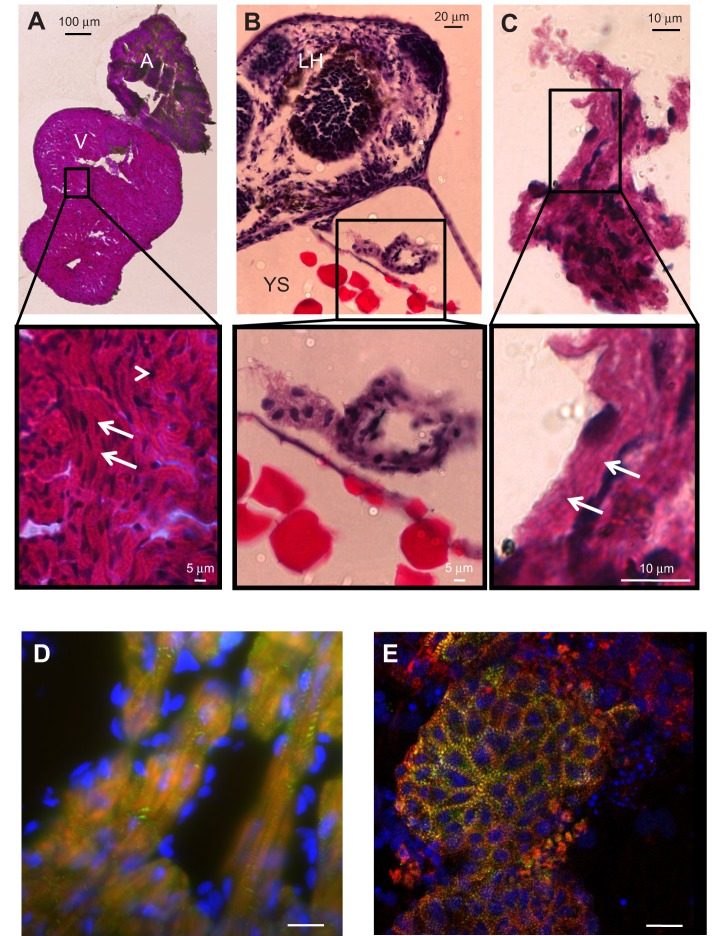
We next investigated the morphological phenotype of the ZFHAs compared with both adult and larval ZF hearts. H&E histological staining and bright field microscopy were used to compare the gross morphology of larval ZF hearts, ZFHAs after 4 days in culture and adult ZF heart (>1 year old) (Fig. 2). The adult ZF heart showed typical mature muscle organisation (Johnson et al., 2014) with a predominantly spongy atrial and ventricular myocardium and a small outer compact layer surrounding the ventricle (Fig. 2A). The myocytes were organised into bundles (Klaiman et al., 2011) with striations visible at higher resolution (see enlarged sections in Fig. 2A–C). In the larval heart, unorganised filamentous tissue was observed, reflecting immature cardiac tissue organisation (Zhang et al., 2013; Bakkers, 2011; Grunow et al., 2010). Importantly, ZFHAs exhibited well-organised gross myocardial structures after 4 days in culture. This suggests in vitro maturation from embryonic ZF progenitor myocardial cells to adult-like cardiac tissue. These findings corroborate those for aggregates derived from rainbow trout (Grunow et al., 2010) and support the idea that in fish, ex vivo maturation of a 3D cardiac phenotype is possible (Zhang et al., 2013). To provide greater resolution of structural organisation, we next used immunohistochemistry and confocal microscopy. Actin filaments were observed in both adult ZF heart (Fig. 2D) and ZFHAs (Fig. 2E). Well-organised, cross-striated, cardiac-specific α-actinin–actin complexes were evident in both ZFHAs and adult heart.
Fig. 2.
Histological and immunohistochemical imaging of ZF adult heart, larval heart and ZFHA. (A) Haematoxylin and Eosin (H&E) staining of atrium and ventricle of an adult ZF (>1 year old). Notice both chambers are composed of spongy myocardium with a small compact layer surrounding the ventricle. The expanded view shows the nuclei (dark stain, arrowhead) and the organisation of the muscle into myocyte bundles with striations visible (arrows). V, ventricle; A, atrium. (B) Section through the anterior of a 3 dpf larval ZF, with the yolk sac (YS) and larval head (LH) clearly visible. The larval heart is shown at higher magnification below. Nuclei are clearly visible, but the muscle structure lacks organisation. (C) ZFHA after 4 days in culture; areas of structural organisation are evident (arrows) and are greater than those of the larval hearts (tissue of origin for ZFHA). (D,E) Immunohistochemical imaging of (D) an adult ZF ventricle and (E) a ZFHA following 3 days in culture stained with α-actinin antibody (green) with actin filaments labelled with phalloidin (red). Nuclei were stained with DAPI (blue). The image in D was captured using epi-fluorescence (Zeiss Axioplan 2 upright microscope), which accounts for the out-of-focus light. The ZFHA image in E has greater resolution as it was captured with a Nikon C1 laser confocal microscope with 488 and 543 nm excitation. In both samples, the α-actinin–actin complex is visible throughout the tissue, highlighting the muscle cell striations. Scale bar, 20 µm for D and E.
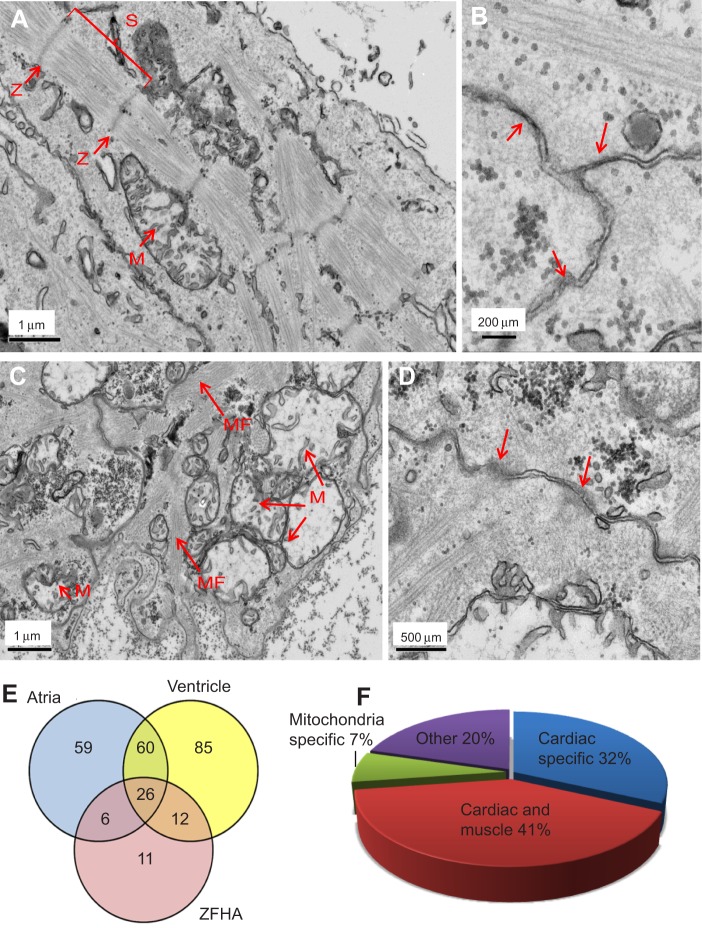
To assess ultrastructure, we employed transmission electron microscopy (TEM). TEM revealed well-developed myofilaments with highly organised sarcomeres in the ZFHAs (Fig. 3A,B). Sarcomere length ranged from 1.4 to 1.6 µm, which is typical of fixed adult ZF hearts (Jopling et al., 2010). In contrast, larval hearts exhibited an embryonic phenotype, with clear myofibrils, but without highly organised striations or Z-lines (Fig. 3C,D). Both preparations contained a high number of mitochondria, myofilaments and cell–cell contacts, indicative of a metabolically active contractile syncytium (Fig. 3B,D).
Fig. 3.
Ultrastructure and protein expression in ZFHA. (A) ZFHAs consist of muscle cells with sarcomeres (S) that are bordered by Z-lines (Z) and contain mitochondria (M). (B) Higher magnification showing cell–cell connections (arrows) indicating ZFHAs are a functional syncytium. (C) Larval heart shows less organised sarcomeric structures without clear Z-lines but containing myofilaments (MF). (D) Cell–cell connections are also visible in the larval heart. Mitochondria are present in all images. (E) Venn diagram showing the similarity between proteins in adult ZF atria, ventricle and ZFHA. The results showed that 44 known proteins (80% of total protein) were common to ZFHAs and/or atria and ventricle of the adult fish, with 59 proteins exclusively expressed in atria compared with 85 in ventricle alone. In ZFHA, 20% of total proteins were expressed in the adult ventricle and 21% in the atria. Only 11 proteins (20% of total known proteins) from ZFHA were not found in heart tissue from adult ZF. (F) Pie chart showing a representation of the ontology of proteins identified using mass spectroscopy in ZFHA. Minimal non-cardiac-related proteins (20%) were identified showing minimal contamination by unwanted tissue. Myocardial proteomic analysis was performed using LC-MS after extracting protein from ZFHA (N=20) following 3 days in culture, adult ZF ventricles (N=6) and adult ZF atria (N=5). A list of all proteins identified using LC-MS can be found in supplementary material Table S1.
To further characterise ZFHAs, we compared protein expression profiles of ZFHAs after 3 days in culture with adult ZF ventricle and atrium using LC-MS mass spectrometry. We were able to successfully isolate enough protein (∼60 μg) from ZFHAs for detection. The results showed 44 proteins (80% of total proteins isolated) in ZFHAs were also present in ZF atria and/or ventricle (see areas of overlap on the Venn diagram in Fig. 3E). A complete list of identified proteins is provided in supplementary material Table S1). The majority of these peptides (73%) are cardiac specific/related and include: ventricle myosin heavy chain, myosin heavy chain 6 and tropomyosin 4a (Fig. 3F; supplementary material Table S1). ZFHAs express a relatively low number of proteins associated with non-cardiac tissues (20%) (Fig. 3F; supplementary material Table S1). This is important as ZFHAs were formed by culturing dissociated whole larval fish, not just larval myocardial tissue. These data indicate that our rapid, simple and inexpensive methodology enriches for cardiac cell populations spontaneously in vitro. Importantly, these data demonstrate that cells liberated from ZF embryonic progenitor populations mature ex vivo to form 3D, organised structures composed of typical cardiac proteins that are comparable to adult ZF hearts.
Conclusions
The ZF is an established model organism for cardiac research (Brette et al., 2008; Nemtsas et al., 2010; Bovo et al., 2013; Zhang et al., 2013). Here, we present a fast, simple and inexpensive method for generating spontaneously beating 3D ‘heart’ aggregates from larval ZF. This methodology builds on previous work in rainbow trout (Grunow et al., 2011), but also offers new and unique investigative opportunities. ZF breed continuously, with one fish generating >100 larvae per week. This provides inexpensive starting material from which ZFHAs can be rapidly generated. Furthermore, this method does not require the use of protected animals, thus supporting the tenets of the 3Rs of animal research (replacement, reduction and refinement). The similarity between ZF and human cardiac action potentials and the underlying ion channels have already been investigated in detail (Brette et al., 2008; Nemtsas et al., 2010; Verkerk and Remme, 2012; Bovo et al., 2013), as have the regenerative capabilities of the ventricle (Major and Poss, 2007; Kikuchi et al., 2010). These properties of ZF cardiology, together with relatively simple gene technology (e.g. Morpholino, TALENS) can be incorporated into the methodology we report here to create a stable culture tool for investigation without the complication of embryonic lethality. For example, mutant fish with fluorescent hearts [i.e. Tg(cmlc2:EGFP)] could be used for cell sorting and to visualise maturation. There are a number of well-characterised ZF cardiac mutants (e.g. breakdance/reggae/slomo) showing both morphological and functional phenotypes that could be used to generate disease-specific ZFHA, and morpholino oligonucleotide knock-downs could be used to target aspects of cardiac maturation including myocyte proliferation, regeneration, conductivity and ion channel function. Thus, we see ZFHAs complementing current in vitro investigative strategies (i.e. cardiac tissue engineering and hiPSC technology) by offering a stable, 3D, electrically and mechanically coupled spontaneously contracting syncytium that is simple and inexpensive to generate. The data presented here show the potential of this novel in vitro system. With further characterisation and refinement, we speculate that the ZFHA could form the basis of large-scale generation of cardiac tissue appropriate for human drug discovery, tissue engineering biology and (eco) toxicology safety testing.
MATERIALS AND METHODS
Generation and culture of ZFHA
All procedures adhere to the UK Home Office Animals (Scientific Procedures) Act 1986. ZF larvae (1–4 dpf) were collected and washed in cold Dulbecco's phosphate buffered saline (DPBS, Invitrogen, UK). Animals from each age group were aseptically dissected and cellular material liberated by mincing whole fish using springbow scissors, pestle/mortar or scalpel, followed by enzymatic digestion in 0.1% trypsin/EDTA for 1 min at room temperature (RT). Cell lysates were suspended in media comprising Dulbecco's modified Eagle’s medium (DMEM), 20% fetal calf serum (FCS; Hyclone, Thermo Scientific, UK), 1000 U ml−1 penicillin and 10 mg ml−1 streptomycin, and centrifuged for 5 min at 130 g. Isolated cells from 10 larvae were seeded per well of a 24-well plate (Greiner bio-one, UK) in 1 ml media. Prior to cell culture, tissue culture plates were coated with Matrigel (BD Biosciences, UK; 1:30 in DMEM) and incubated at 37°C for 1 h prior to use. Cells were propagated at 28°C and 2.5% CO2 and media replenished following the initial 24 h and every 48 h thereafter. Smaller tissue fragments and more single cells were observed 24 h post-seeding in preparations derived from grinding larvae with pestle/mortar and thus this method was used for the remainder of the study.
Tissue processing
Adult ZF heart and whole larvae were fixed for 10 min in 4% paraformaldehyde (PFA) prior to wax processing using a Shandon Citadel 2000 Tissue Processor (Thermo Scientific). Because of the small size of ZFHA, they were first stained briefly with Haematoxylin (1 min) to help identify the cells during paraffin embedding. Samples were then processed as follows; 10 min in 70% ethanol, a series of alcohol washes (90%, 95% and 100%) for 15 min followed by xylene wash, then embedded in paraffin wax for 30 min. All samples were sectioned at 10 μm. Standard Haematoxylin and Eosin (H&E) staining was performed. Images were captured using a Zeiss Axiovert 200 inverted microscope with an AxioCamMR colour camera and processed using AxioVision software (Carl Zeiss AG).
Immunohistochemistry
Adult ZF hearts were cryo-embedded and stored at −80°C. Serial sections (10 μm) were taken using a cryostat (Cryotom Leica 350S) and fixed in 4% PFA for 30 min at RT prior to immunohistochemical analysis. Following DPBS washes, samples were permeabilised using 0.1% Triton-X, 1% bovine serum albumin (BSA) in DPBS for 20 min at RT. Non-specific binding was blocked by incubation with 10% normal goat serum for 1 h at RT. Samples were incubated with α-actinin (mouse monoclonal, 1:300) for 45 min at 37°C in a humid chamber and, following washes in DPBS, incubated for 30 min at 37°C with the secondary antibody (anti-mouse IgG Alexa Fluor 488, 1:200: Invitrogen, UK) and 0.1 μmol l−1 phalloidin dissolved in methanol. Tissues were mounted in Vectashield (Vector Labs, USA).
Electron microscopy
ZFHAs were washed in DPBS and transferred to a MATEK dish containing a gridded coverslip (P35G-2-14-CGRD, MatTek Corp., USA). These plates were coated with 1 mg ml−1 fibronectin prior to use (R&D Systems, UK). Samples were left to adhere overnight in the incubator prior to fixation 24 h later. ZF larvae were processed in parallel. ZFHAs were fixed for 12 h in 4% formaldehyde, 2.5% glutaraldehyde in 0.1 mol l−1 Hepes buffer (pH 7.2). Samples were post-fixed with 1% osmium tetroxide, 1.5% potassium ferrocynaide in 0.1 mol l−1 cacodylate buffer (pH 7.2) for 1 h, followed by incubation in 1% tannic acid in 0.1 mol l−1 cacodylate buffer (pH 7.2) for 1 h, and subsequently incubated in 1% uranyl acetate in water for a further 1 h. Samples were dehydrated in an ethanol series, infiltrated with TAAB 812 resin and polymerised for 24 h at 60°C. Sections were cut using a Reichert Ultramicrotome and visualised under an FEI Tecnai 12 Biotwin microscope at 100 kV accelerating voltage. Images were taken using a Gatan Orius SC1000 CCD camera.
Mass spectroscopy
Adult ZF ventricles (N=6), adult ZF atria (N=5) and ZFHAs following 3 days of culture (N=20) were frozen in liquid nitrogen. Tissues were solubilised with 0.005% Rapigest detergent (Waters), heated to 80°C for 10 min, reduced with 3 mmol l−1 DTT for 10 min at 60°C and alkylated with 9 mmol l−1 iodoacetamide for 30 min in the dark at RT before digestion with trypsin (1:50 ratio of trypsin to protein, ∼300 ng protein). The resultant peptides were cleaned and concentrated using reversed phase solid phase extraction on Poros R3 beads and analysed by LC-MSMS on a UltiMate® 3000 Rapid Separation LC (RSLC, Dionex Corporation, Sunnyvale, CA, USA) coupled to a LTQ Velos Pro (Thermo Fisher Scientific, Waltham, MA, USA) mass spectrometer. Peptides were selected for fragmentation automatically by data-dependent analysis. The data produced were converted to a peak list and searched against the SwissProt database with D. rerio species selected and the DANRE database using Mascot software (Matrix Science) and validated using Scaffold (Proteome software).
Supplementary Material
Acknowledgements
The authors are grateful to Robert Hallworth at the Biological Service Facility for the ZF. We thank Peter Walker from the Histology Facility, Aleksandr Mironov from the Electron Microscope Facility, David Knight and Ronan O’Cualain from the Biomolecular Analysis Facility at the University of Manchester. Thanks also to Alison Gurney for the use of her confocal microscope and to Catherine Merry for the Matrigel and many useful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
B.G., L.M. and H.A.S. designed the experiments. B.G. and L.M. carried out the experiments. B.G., L.M. and H.A.S. interpreted the data. All authors wrote the manuscript.
Funding
This work was supported by the German Academic Exchange Service (DAAD). The Biomolecular Analysis Facility at the University of Manchester is supported by the Wellcome Trust [097820/Z/11/A]. Deposited in PMC for release after 6 months.
Supplementary material
Supplementary material available online at http://jeb.biologists.org/lookup/suppl/doi:10.1242/jeb.118224/-/DC1
References
- Amanfu R. K. and Saucerman J. J. (2011). Cardiac models in drug discovery and development: a review. Crit. Rev. Biomed. Eng. 39, 379-395 10.1615/CritRevBiomedEng.v39.i5.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausma J. and Borgers M. (2002). Dedifferentiation of atrial cardiomyocytes: from in vivo to in vitro. Cardiovasc. Res. 55, 9-12 10.1016/S0008-6363(02)00434-0 [DOI] [PubMed] [Google Scholar]
- Bakkers J. (2011). Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 91, 279-288 10.1093/cvr/cvr098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrionuevo W. R. and Burggren W. W. (1999). O2 consumption and heart rate in developing zebrafish (Danio rerio): influence of temperature and ambient O2. Am. J. Physiol. Regul. Integr. Comp. Physiol. 276, R505-R513. [DOI] [PubMed] [Google Scholar]
- Bovo E., Dvornikov A. V., Mazurek S. R., de Tombe P. P. and Zima A. V. (2013). Mechanisms of Ca2+ handling in zebrafish ventricular myocytes. Pflügers Arch. 465, 1775-1784 10.1007/s00424-013-1312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brette F., Luxan G., Cros C., Dixey H., Wilson C. and Shiels H. A. (2008). Characterization of isolated ventricular myocytes from adult zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 374, 143-146 10.1016/j.bbrc.2008.06.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W., Duis K., Fenske M., Maack G., Legler J., Padilla S., Strähle U., Witters H. and Scholz S. (2011). The zebrafish embryo model in toxicology and teratology, September 2–3, 2010, Karlsruhe, Germany. Reprod. Toxicol. 31, 585-588 10.1016/j.reprotox.2011.02.010 [DOI] [PubMed] [Google Scholar]
- de Boer T. P., van Veen T. A. B., Jonsson M. K. B., Kok B. G. J. M., Metz C. H. G., Sluijter J. P. G., Doevendans P. A., de Bakker J. M. T., Goumans M.-J. and van der Heyden M. A. G. (2010). Human cardiomyocyte progenitor cell-derived cardiomyocytes display a maturated electrical phenotype. J. Mol. Cell. Cardiol. 48, 254-260 10.1016/j.yjmcc.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Desroches B. R., Zhang P., Choi B.-R., King M. E., Maldonado A. E., Li W., Rago A., Liu G., Nath N., Hartmann K. M. et al. (2012). Functional scaffold-free 3-D cardiac microtissues: a novel model for the investigation of heart cells. Am. J. Physiol. Heart Circ. Physiol. 302, H2031-H2042 10.1152/ajpheart.00743.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunow B., Ciba P., Rakers S., Klinger M., Anders E. and Kruse C. (2010). In vitro expansion of autonomously contracting, cardiomyogenic structures from rainbow trout Oncorhynchus mykiss. J. Fish Biol. 76, 427-434 10.1111/j.1095-8649.2009.02535.x [DOI] [PubMed] [Google Scholar]
- Grunow B., Wenzel J., Terlau H., Langner S., Gebert M. and Kruse C. (2011). In vitro developed spontaneously contracting cardiomyocytes from rainbow trout as a model system for human heart research. Cell Physiol. Biochem. 27, 1-12. [DOI] [PubMed] [Google Scholar]
- Grunow B., Schmidt M., Klinger M. and Kruse C. (2012). Development of an in vitro cultivated, spontaneously and long-term contracting 3D heart model as a robust test system. J Cell Sci. Ther. 3, 1 10.4172/2157-7013.1000116 [DOI] [Google Scholar]
- Hirt M. N., Hansen A. and Eschenhagen T. (2014). Cardiac tissue engineering: state of the art. Circ. Res. 114, 354-367 10.1161/CIRCRESAHA.114.300522 [DOI] [PubMed] [Google Scholar]
- Johnson A. C., Turko A. J., Klaiman J. M., Johnston E. F. and Gillis T. E. (2014). Cold acclimation alters the connective tissue content of the zebrafish (Danio rerio) heart. J. Exp. Biol. 217, 1868-1875 10.1242/jeb.101196 [DOI] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A. and Izpisúa Belmonte J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Werdich A. A., Anderson R. M., Fang Y., Egnaczyk G. F., Evans T., MacRae C. A., Stainier D. Y. R. and Poss K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601-605 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaiman J. M., Fenna A. J., Shiels H. A., Macri J. and Gillis T. E. (2011). Cardiac remodeling in fish: strategies to maintain heart function during temperature change. PLoS ONE 6, e24464 10.1371/journal.pone.0024464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. R., Yang B., Feng J., Bosch R. F., Carrier M. and Nattel S. (1999). Transmembrane ICa contributes to rate-dependent changes of action potentials in human ventricular myocytes. Am. J. Physiol. 276, H98-H106. [DOI] [PubMed] [Google Scholar]
- Major R. J. and Poss K. D. (2007). Zebrafish heart regeneration as a model for cardiac tissue repair. Drug Discov. Today Dis. Models 4, 219-225 10.1016/j.ddmod.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel Y., Weissman A., Schick R., Barad L., Novak A., Meiry G., Goldberg S., Lorber A., Rosen M. R., Itskovitz-Eldor J. et al. (2012). Human embryonic and induced pluripotent stem cell-derived cardiomyocytes exhibit beat rate variability and power-law behavior. Circulation 125, 883-893 10.1161/CIRCULATIONAHA.111.045146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert J. M., Brandenburger M. and Grunow B. (2013). Electrophysiological characterization of spontaneously contracting cell aggregates obtained from rainbow trout larvae with multielectrode arrays. Cell Physiol. Biochem. 32, 1374-1385 10.1159/000356576 [DOI] [PubMed] [Google Scholar]
- Nemtsas P., Wettwer E., Christ T., Weidinger G. and Ravens U. (2010). Adult zebrafish heart as a model for human heart? An electrophysiological study. J. Mol. Cell. Cardiol. 48, 161-171 10.1016/j.yjmcc.2009.08.034 [DOI] [PubMed] [Google Scholar]
- Nerbonne J. M., Nichols C. G., Schwarz T. L. and Escande D. (2001). Genetic manipulation of cardiac K+ channel function in mice: what have we learned, and where do we go from here? Circ. Res. 89, 944-956 10.1161/hh2301.100349 [DOI] [PubMed] [Google Scholar]
- Novakovic G., Eschenhagen T. and Mummery C. (2014). Myocardial tissue engineering: in vitro models. Cold Spring Harb. Perspect. Med. 4, 1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orfanidou T., Papaefthimiou C., Antonopoulou E., Leonardos I. and Theophilidis G. (2013). The force of the spontaneously contracting zebrafish heart, in the assessment of cardiovascular toxicity: application on adriamycin. Toxicol. In Vitro 27, 1440-1444 10.1016/j.tiv.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Parker T., Libourel P.-A., Hetheridge M. J., Cumming R. I., Sutcliffe T. P., Goonesinghe A. C., Ball J. S., Owen S. F., Chomis Y. and Winter M. J. (2014). A multi-endpoint in vivo larval zebrafish (Danio rerio) model for the assessment of integrated cardiovascular function. J. Pharmacol. Toxicol. Methods 69, 30-38 10.1016/j.vascn.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Sauer H., Rahimi G., Hescheler J. and Wartenberg M. (1999). Effects of electrical fields on cardiomyocyte differentiation of embryonic stem cells. J. Cell Biochem. 75, 710-723 [DOI] [PubMed] [Google Scholar]
- Shenje L. T., Field L. J., Pritchard C. A., Guerin C. J., Rubart M., Soonpaa M. H., Ang K.-L. and Galiñanes M. (2008). Lineage tracing of cardiac explant derived cells. PLoS ONE 3, e1929 10.1371/journal.pone.0001929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strähle U., Scholz S., Geisler R., Greiner P., Hollert H., Rastegar S., Schumacher A., Selderslaghs I., Weiss C., Witters H. et al. (2011). Zebrafish embryos as an alternative to animal experiments-A commentary on the definition of the onset of protected life stages in animal welfare regulations. Reprod. Toxicol. 33, 128-132 10.1016/j.reprotox.2011.06.121 [DOI] [PubMed] [Google Scholar]
- Thomas P. and Smart T. G. (2005). HEK293 cell line: a vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods 51, 187-200 10.1016/j.vascn.2004.08.014 [DOI] [PubMed] [Google Scholar]
- van den Heuvel N. H. L., van Veen T. A. B., Lim B. and Jonsson M. K. B. (2014). Lessons from the heart: mirroring electrophysiological characteristics during cardiac development to in vitro differentiation of stem cell derived cardiomyocytes. J. Mol. Cell. Cardiol. 67, 12-25 10.1016/j.yjmcc.2013.12.011 [DOI] [PubMed] [Google Scholar]
- Verkerk A. O. and Remme C. A. (2012). Zebrafish: a novel research tool for cardiac (patho)electrophysiology and ion channel disorders. Front Physiol. 3, 255 10.3389/fphys.2012.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Han P., Yang H., Ouyang K., Lee D., Lin Y.-F., Ocorr K., Kang G., Chen J., Stainier D. Y. R. et al. (2013). In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497-501 10.1038/nature12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.