Abstract
The highly conserved, multifunctional YB-1 is a powerful breast cancer prognostic indicator. We report on a pervasive role for YB-1 in which it associates with thousands of nonpolyadenylated short RNAs (shyRNAs) that are further processed into small RNAs (smyRNAs). Many of these RNAs have previously been identified as functional noncoding RNAs (http://www.johnlab.org/YB1). We identified a novel, abundant, 3′-modified short RNA antisense to Dicer1 (Shad1) that colocalizes with YB-1 to P-bodies and stress granules. The expression of Shad1 was shown to correlate with that of YB-1 and whose inhibition leads to an increase in cell proliferation. Additionally, Shad1 influences the expression of additional prognostic markers of cancer progression such as DLX2 and IGFBP2. We propose that the examination of these noncoding RNAs could lead to better understanding of prostate cancer progression.
Keywords: YB-1, noncoding RNAs, miRNAs, prostate cancer
INTRODUCTION
Human Y-box binding protein 1 (YB-1) is a multifunctional protein that belongs to the highly conserved superfamily of cold shock domain (CSD) proteins known to bind both DNA and RNA (Kohno et al. 2003). It is involved in many cellular functions including transcription/translation (Didier et al. 1988; Evdokimova et al. 2006), alternative splicing (Stickeler et al. 2001; Dutertre et al. 2010), and mRNA degradation/processing in P-bodies (Evdokimova et al. 2001, 2006; Stickeler et al. 2001). Inverse CAAT motifs, typically localized within promoter regions, serve as binding sites for YB-1. Occupancy of YB-1 at these sites can trigger either transcriptional activation through the recruitment of RNA polymerase II (Spitkovsky et al. 1992) or repression via displacement of bound activator proteins (Van de Putte et al. 2003). YB-1 has also been implicated as a protein partner in the splicing and transport of pre-mRNAs (Deckert et al. 2006; Agafonov et al. 2011). In the cytoplasm, YB-1 is also a component of the messenger ribonucleoprotein (mRNP) complexes formed by human Argonautes (AGO), which are central to small RNA-mediated silencing (Hammond et al. 2001; Meister et al. 2004; Girard et al. 2006; Höck et al. 2007). In addition to the link between YB-1 and mature microRNAs (miRNAs), YB-1's relatedness to lin-28, a pluripotency factor and a processor of the let-7 miRNA precursor (Moss and Tang 2003; Yu et al. 2007; Viswanathan et al. 2009) suggests that it may have a broader role in small RNA processing and function than previously recognized.
The role of YB-1 in human biology and disease has been most thoroughly examined in cancer. YB-1 expression levels have been shown to correlate with drug resistance and poor patient outcome in various cancers (Bargou et al. 1997). Furthermore, YB-1 may be a more powerful prognostic marker for relapse and survival in breast cancer patients than the commonly used markers Her-2 and estrogen receptor (Habibi et al. 2008). YB-1 has been shown to induce the expression of pro-proliferative genes such as Her-2, EGFR, MDR-1, cyclin A, and cyclin B (Bargou et al. 1997; Ise et al. 1999; Raffetseder et al. 2003; Wu et al. 2006). Specifically in breast cancer, YB-1 has been associated with BRCA1 mutations as identified by microarray (Sorlie et al. 2003). Finally, YB-1 expression levels have also been shown to correlate with malignant transformation and castrate resistant prostate cancer (Giménez-Bonafé et al. 2004). Although the mechanistic basis for YB-1's role in cancer has not been revealed, it is a partner of the p53 tumor suppressor protein and may therefore influence its many functions in tumor progression (Zhang et al. 2003).
Currently, 50 miRNAs have been associated with prostate cancer; however none of them have been shown to be directly related to disease pathogenesis (Pang et al. 2010). Instead, many miRNAs have been shown to correlate with different grades of prostate cancer and could potentially serve as biomarkers for diagnosis and treatment (Walter et al. 2013). A pilot project to predict RNA binding proteins important to small RNA pathways led us to test whether YB-1 associates with small RNAs in the androgen independent PC-3 prostate cancer cell line. In recent years, various classes of small and long noncoding RNAs have been identified in mammals (Gupta et al. 2010; Meiri et al. 2010; Lai et al. 2013). We found that YB-1 associates with a variety of noncoding RNAs in PC-3 cells. These RNAs correspond to thousands of distinct short YB-1–associated noncoding RNAs (shyRNAs) and their cognate processed YB-1–associated small RNAs (smyRNAs) that primarily match to the 5′ or 3′ terminal regions. Many of the shyRNAs that we identified are also identical to known regulatory RNAs such as Y RNAs, vault RNAs and snoRNAs. One of the shyRNAs derive from the antisense strand of the Dicer1 gene, termed Shad1 (short antisense to Dicer1), and found to regulate the proliferation of PC-3 cells. Furthermore, Shad1 RNA colocalizes with YB-1 in P-bodies and stress granules upon oxidative stress of PC-3 cells. Therefore, shyRNAs have the capacity to impact many fundamental biochemical pathways through the interaction with the global regulator, YB-1.
RESULTS
YB-1 associates with a variety of noncoding RNAs
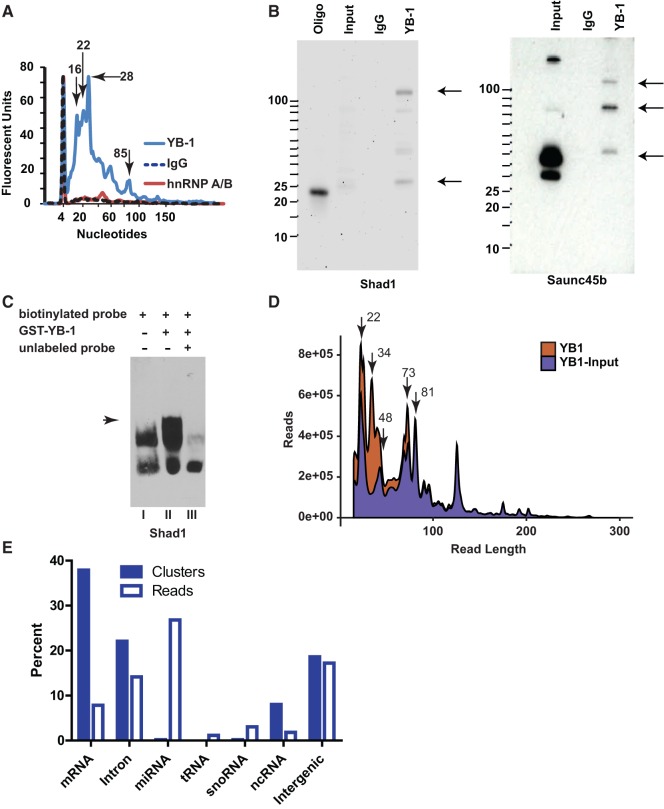
PC-3 cell lysates were subjected to immunoprecipitation with a YB-1 antibody under conditions expected to stabilize RNA–protein complexes. As shown in results of the Bioanalyzer analysis of RNA species enriched in YB-1 immunoprecipitates relative to nonimmune IgG complexes, the YB-1 RNA fraction is highly enriched in small RNAs with distinct and prominent peaks at 16, 22, and 28 nt (Fig. 1A). The profile of RNA species associated with another protein containing two RNA binding domains (i.e., hnRNP A/B) does not match that identified in YB-1 immunoprecipitates indicating that not all RNA binding proteins bind small RNAs. The YB-1–associated small RNAs (∼10–50 nt) were therefore size-fractionated and sequenced using the Illumina small RNA-sequencing protocol. A total of 185,105 distinct genomic loci (14,042,214 reads) were cloned with 156,006 locations (14,012,548 reads) representing >1 RPKM. One smyRNA-sequenced maps to a novel gene locus that is antisense to the first exon of the Dicer transcript, KI AA0928. Northern blot analysis validates that the Shad1 small RNA was indeed present in YB-1 immunoprecipitated samples as compared with input and IgG controls (Fig. 1B). Northern blotting also validated the presence of another highly sequenced smyRNAs selectively enriched in YB-1 immunoprecipitates (i.e., Saunc45b) (Fig. 1B). RNA EMSAs using purified GST-tagged YB-1 protein demonstrate a direct interaction between YB-1 and the DIG-labeled small Shad1 RNA, suggesting that the association between YB-1 and Shad1 in immunoprecipitates may be indicative of an interaction that occurs in vivo (Fig. 1C). Addition of excess unlabeled Shad1 eliminates the detectable shift due to the binding of the tagged protein. YB-1 is known to bind directly to the RNA transcribed from the fourth exon on the CD44 gene both in vivo and in vitro (Stickeler et al. 2001). To test the specificity of RNA binding, CD44v4 was used as a positive control in the EMSA not only to test the specificity with the addition of excess unlabeled probes, but also with increasing concentrations of either labeled probe or tagged YB-1 protein (Supplemental Fig. S1).
FIGURE 1.
Characteristics of YB-1–associated small RNAs. (A) Length distribution of small RNAs immunoprecipitated with YB-1, hnRNP A/B (control), and rabbit IgG (control), based on Agilent Bioanalyzer (marker peak at 4 nt). (B) Northern blot validation of two abundant small RNAs, Shad1 and Saunc45b. A more abundant ∼100-nt long Shad1 isoform was also detected. (C) RNA EMSA using purified GST-YB-1 protein shows interaction with Shad1. (Lane I) DIG-labeled probe in the absence of YB-1. (Lane II) Labeled probed in the presence of YB-1. (Lane III) Labeled probe, YB-1 and an excess (20×) of unlabeled RNA probes as competitor. All reactions use unlabeled tRNA as a nonspecific competitor. (D) Size distribution of paired-end reads. The red distribution shows all read lengths from the YB-1 RNA immunoprecipitation. The blue distribution shows all read lengths from the YB-1 RNA immunoprecipitation twofold enriched over PC-3 input RNAs. (E) Analysis of distinct YB-1–associated small RNA sequences after removal of input RNAs.
In addition to validating the presence of sequenced smyRNAs, Northern blot analysis also identified additional higher molecular weight bands RNAs containing the same small RNA sequence, which could be precursors of Shad1 and Saunc45B RNAs (Fig. 1B). YB-1–associated RNAs were therefore size fractionated from ∼10 to 250 nt and paired-end sequenced to examine the different size populations. A total of 403,455 distinct genomic loci (32,916,224 reads) were mapped to the genome and of those, 281,686 loci (32,720,233 reads) represent locations >1 RPKM. The size distribution of each unique paired-end read shows two distinct populations of RNAs with an enrichment of RNAs in the range of 16–48 nt, similar to the distribution from the Bioanalyzer assays (Fig. 1D, red). To normalize for any artifacts related to ligation, amplification and nonspecific binding of abundant RNAs, the same size fraction was also sequenced from the PC-3 input samples. A total of 412,690 distinct genomic loci (67,175,643 reads) were mapped with 115,496 loci (66,688,207 reads) representing locations >1 RPKM. Only genomic locations (247,547 clusters; 22,938,551 reads) where there was at least a twofold enrichment over the input library and the locations with >1 RPKM were considered resulting in a conservative group of YB-1–associated RNAs. The size distribution of these YB-1–associated RNAs remains very similar to the nonnormalized data set (Fig. 1D, blue). Examination of the 188,061 smyRNA locations (7,594,709 reads) revealed that a small percentage of reads correspond to annotated tRNAs (6.5%) and snoRNAs (3%) (Fig. 1E). Because smyRNAs sequenced from the input samples were removed from the YB-1 IP samples, the reads attributed to tRNAs and snoRNAs are not due to nonspecific associations or ligation bias due to the sequencing technologies. The high percentage of snoRNAs sequenced suggests an enrichment of sdRNAs associated with YB-1 (Taft et al. 2009). On the other hand, miRNA reads accounted for 27% of total smyRNAs. The enrichment of known small noncoding RNAs associated with YB-1 suggests a functional role for YB-1 in the small RNA pathway.
A second population of RNAs, ∼63–86 nt in length, emerged upon sequencing a larger size distribution of RNAs associated with YB-1 (Fig. 1E). In total, we sequenced 67,132 distinct shyRNA loci (6,342,725 reads). Only a low percentage of all reads (7.7%) map to tRNA locations (Fig. 2A). In contrast, 52.5% of the reads within this shyRNA fraction still map to snoRNAs. Recurrent enrichment of snoRNAs in both the shy and small RNA fractions, over similarly sized RNAs, adds support to the notion that YB-1 preferentially associates with both snoRNAs and their processed, smaller sdRNAs. To analyze whether shyRNAs have polyadenylated isoforms, similar to that of exosome-dependent promoter associated RNAs (Preker et al. 2008), we identified the polyadenylation sites across the genome using Direct RNA sequencing (DRS) (Ozsolak et al. 2010) in PC-3 cells and found that the majority (83%) of shyRNA locations do not overlap with polyadenylation sites.
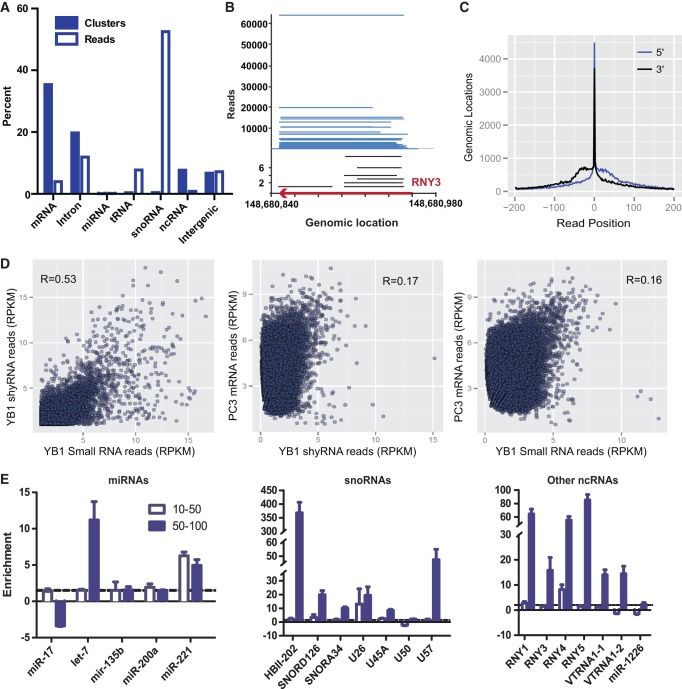
FIGURE 2.
Characteristics of YB-1–associated short RNAs. (A) Analysis of distinct YB-1–associated short RNAs (shyRNAs) after removal of input RNAs. (B) The RNY3 RNA is matched to YB-1–associated shyRNAs (blue) and low abundance small RNAs (black) that preferentially derive from either the 5′ or 3′ ends of RNY3. The y-axis represents the number of reads while the x-axis shows the genomic location of the reads. The most abundant read is nearly identical to RNY3. (C) Small RNAs preferentially match to the 5′ and 3′ ends of shyRNAs. (D) Correlation between shyRNAs, small RNAs, and mRNAs in PC-3 cells. (E) qPCR validation of different classes of small and shyRNAs.
The presence of longer RNA sequences in Northern blot analysis of YB-1–associated RNAs, the preferential association of snoRNAs and their derivatives with YB-1, and the evidence that long RNAs (>200 nt) are processed into short RNAs (<200 nt) raised the possibility that shyRNAs are processed into smaller RNAs (Kapranov et al. 2007). Indeed, among the 247,547 distinct shyRNA genomic locations, ∼62% generate YB-1–associated small RNAs that considerably overlap (≥80%) with their cognate short RNA sequences. This property is not seen with the small and short RNA fractions in the input controls with only 1340 of 84,981 locations (1.6%) reflective of a precursor/product relationship. An inspection of smyRNAs generated from longer shyRNAs, such as the case for RNY3, reveals a general preference for shyRNAs to generate small RNAs from either the 5′ or 3′ termini (Fig. 2B). The most abundant RNY3 small RNA is located toward the 5′ end of the short RNA. An additional small RNA that is seemingly less abundant is located close to the 3′ end. The lack of small RNAs deriving from other regions of RNY3 is reminiscent of other small RNAs that manifest terminal bias, such as tRNA-derived fragments (Lee et al. 2009) and snoRNAs-derived RNAs (Taft et al. 2009; Lin et al. 2012). A more comprehensive analysis of the locations of the small RNAs within shyRNAs indicates that shyRNAs are preferentially processed from their 5′ or 3′ ends (Fig. 2C). The correlation between the abundance of shyRNAs and their small RNAs is also high (R = 0.53) (Fig. 2D). To investigate whether mRNA expression correlates to their corresponding shyRNAs and the small RNAs, we quantified transcript levels of exons in PC-3 using Illumina RNA-seq data. Negligible correlation was seen between mRNAs and the shyRNAs, as well as mRNAs and the small RNAs (R = 0.17–0.16) (Fig. 2D). These results suggest that shyRNA sequences generally serve as precursors to YB-1–associated small RNAs. Quantitative RT-PCR (qPCR) validation was performed on size-fractionated YB-1–associated RNAs to examine the observed association of 26 RNAs (Fig. 2E). Several well characterized small/short noncoding RNAs such as mirtrons and vault RNAs are also present among the YB-1–associated RNA reads. For instance, the recently discovered MALAT1-associated small (∼60 nt) cytoplasmic RNA, mascRNA, generates a novel small RNA that is processed precisely from the 5′ end of the annotated mascRNA. Similarly, 77% (10/13) of a high-confidence set of cytoplasmic human mirtrons were detected (Sibley et al. 2011). Three of the four cytoplasmic vault RNAs also overlap with both shyRNAs and their terminal smyRNAs.
While the correlation between the shyRNAs and their equivalent smyRNA is high, there still exists the possibility that these YB-1–associated RNAs are simply a byproduct of some RNA degradation process within the cell. A recent study examined small noncoding RNAs (18–30 and 30–100 nt) and the effects of the processing of these RNAs in the absence of the endonuclease RRP40 or the exonucleases XRN1 and XRN2 (Valen et al. 2011). RNA categories identified in these analyses as either RRP40, XRN1, and XRN2-dependent or independent were then overlapped with the YB-1–associated RNAs in order to assess the potential contribution of these endo-/exonucleases to the processing these RNAs with the caveat that this comparison was being made with two distinct human cell lines. Nonetheless, only 3% of the small and 0.5%–0.7% of the shyRNAs could be considered dependent on RRP40 or XRN1/2 for processing. Similarly, 3% of the small and 0.4%–0.5% of the shyRNAs represents known degradation products of RRP40 and XRN1/2. Therefore, the vast majority of shyRNAs are unlikely to be generated by degradation by the major RNA processing endonuclease and exonucleases found in human cells.
Shad1 influences expression of prognostic markers of metastasis
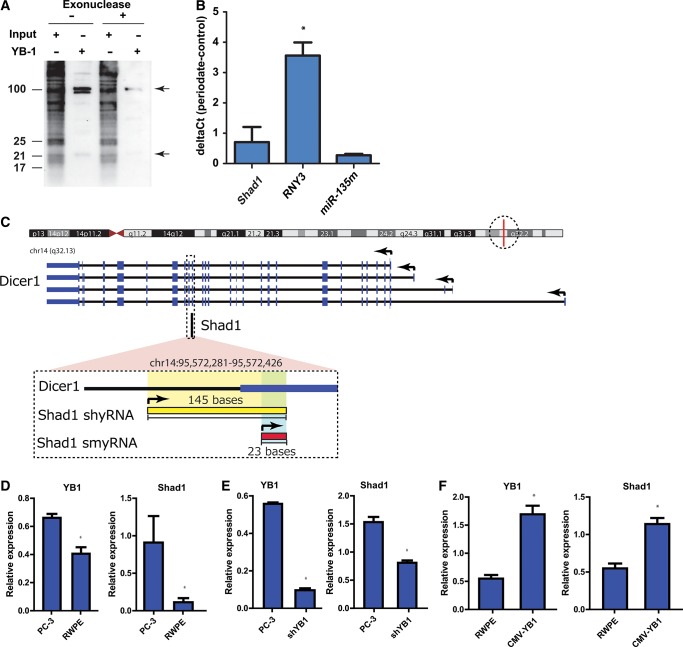
Surprisingly, while initial sequencing experiments recovered Shad1 small RNAs, it was not as abundantly present in the Illumina reads when the larger fraction was sequenced. This could result from either a 5′ or 3′ end modification in Shad1 RNA that would reduce the efficiency of adaptor ligation used to prep samples for Illumina sequencing. To test for 5′ terminal RNA modifications, YB-1 immunoprecipitated RNA was treated with a 5′ monophosphate-dependent exonuclease. For both the 25 and 100 nt forms of Shad1, the exonuclease treatment results in Shad1 degradation, ruling out the presence of a 5′ modification (Fig. 3A). To test for the presence of 3′ modifications, YB-1 immunoprecipitated RNA was subjected to periodate oxidation followed by polyadenylation and qPCR. qPCR of Shad1 with and without periodate oxidation reveals little change in amplification efficiency suggesting that a 3′ modification may exist (Fig. 3B). A synthetic miR-135 oligo containing a 2′-O-methylation at the 3′ end yields similar results, while the novel small RNA derived from the human Y RNA, RNY3, prominently sequenced in association with YB-1, manifests a clear difference. Therefore, this modification is likely responsible for limiting ligation of the 3′ sequencing adapter necessary for Illumina sequencing. Although the 3′ modification did limit the number of reads generated from Shad1, we inferred the genomic location of Shad1 through the combination of single-end and paired-end sequencing of YB-1–associated RNAs (Fig. 3C).
FIGURE 3.
Shad1 modification and correlation of expression with YB-1. (A) The Shad1 RNAs possess a 5′-phosphate as indicated by a 5′-phosphate-dependent exonuclease reaction. (B) Periodate oxidation followed by polyadenylation and qPCR reveals 3′ modifications in YB-1–associated RNAs and the positive control, miR-135m, a synthetic oligo with a 3′ end 2′-O-methyl modification. (C) Genomic location of Shad1 shyRNA and smyRNA as determined by sequencing of YB-1–associated RNAs. (D) In a normal prostate epithelial cell line (RWPE), YB-1 expression is less than that of the prostate cancer cell line (PC-3). Shad1 level is also down-regulated in RWPE compared with PC-3. (E) Upon lentiviral infection with shRNA for YB-1, Shad1 levels also decrease. (F) Upon overexpression of YB-1, Shad1 levels are also increased. (*) P-values <0.05; n = 3.
Although we show that Shad1 is directly bound by YB-1, we sought to examine if levels of Shad1 correlate with YB-1 expression. In order to differentiate between the Shad1 shyRNA and the Shad1 smyRNA, oligos were synthesized for complementary to the common short/small region (Shad1) or specifically to the short region (Shad1 [−50]). In a normal prostate epithelial cell line, RWPE-1, YB-1 is down-regulated compared with PC-3; in the same cell line, Shad1 is also significantly down-regulated (Fig. 3D). When YB-1 expression is knocked down with a lentiviral shRNA, Shad1 expression likewise decreases (Fig. 3E). Finally, when a CMV-YB-1 is stably transfected into the RWPE-1 cell line to increase the expression of YB-1, Shad1 expression increases with that of YB-1 (Fig. 3F). Under the same conditions, qPCR was performed using oligos specific to the Shad1 shyRNA only (Supplemental Fig. S2A–C). Similar to the Shad1 qPCR results, the expression of Shad1 shyRNA correlates with the expression of YB-1 in all cell lines. Examination of a gene not known to be regulated by YB-1 expression was also checked as a negative control, and there was no correlation of GusB expression with that of YB-1 or Shad1 (Supplemental Fig. S2D–F). These results in sum suggest that Shad1 expression is tightly linked to that of its protein partner, YB-1.
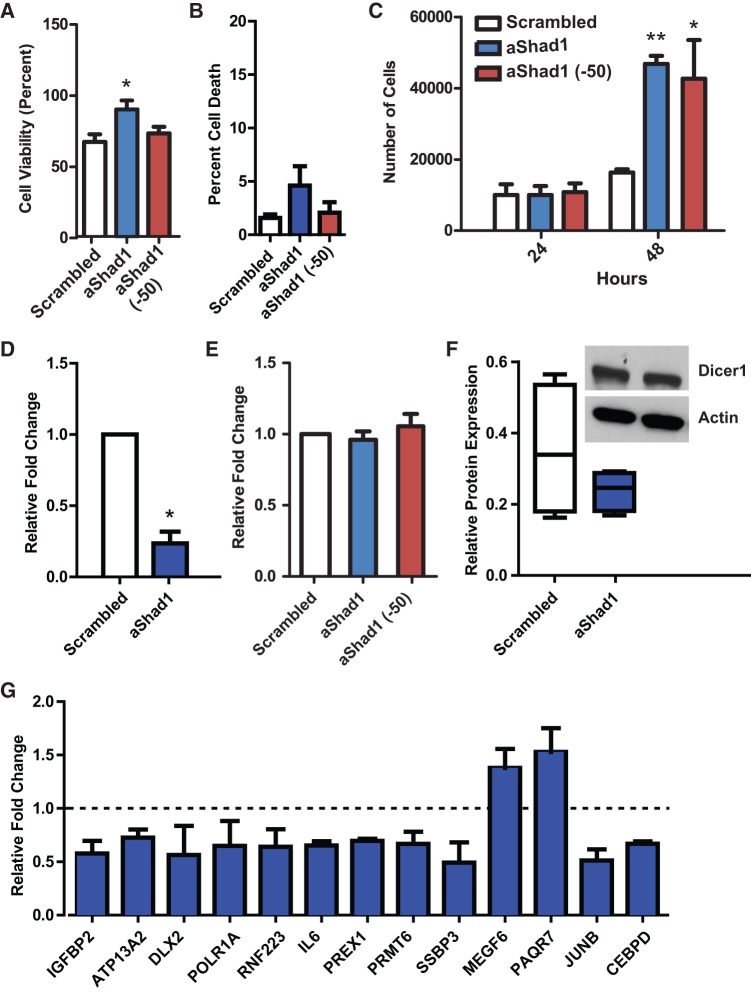
In order to investigate whether Shad1 shyRNA has a functional role in PC-3 cells, we designed LNA oligos antisense for both the short/small Shad1 isoform (aShad1) and the specific short Shad1 isoform (aShad1 (-50)). Transfections of aShad1 LNAs result in increase in metabolic activity compared with the scrambled control LNA oligo (Fig. 4A). Cytotoxicity assays of the transfections showed no increased cell death in the transfection reactions as opposed to negative (control untreated) and positive (fully lysed) controls (Fig. 4B). Cell counts also showed no cell death with increased proliferation in the cells transfected with both LNA oligos that are complementary to Shad1 (Fig. 4C). Taken together, Shad1 functions in PC-3 cells to repress proliferation. Since Shad1 is antisense to Dicer1, blocking Shad1 could remove its potential function as a regulator of Dicer1 expression. Shad1 inactivation was examined by qPCR between the scrambled (scr) control and the aShad1 LNA (Fig. 4D). When the aShad1 LNA was transfected, there is a statistically significant decrease in Shad1 expression. Further examination by qPCR of Dicer1 expression showed no significant difference in Dicer1 mRNA is PC-3 cells (Fig. 4E); Western blot showed no significant difference in Dicer1 protein (Fig. 4F). Since there was no significant change in Dicer1 expression or protein levels, Shad1 may be having a unique role in mRNA regulation independent of Dicer1. DRS was performed on both the scr control and the aShad1 LNA knockdown cells to examine gene expression changes, and there were 115 genes differentially expressed between the two conditions. A few of the most differentially expressed genes, DLX2, IGFBP2, and IL-6, were reconfirmed by qPCR and are known markers of cancer progression and poor patient outcome (Fig. 4G). Because of the closely linked nature of Shad1 and YB-1, we examined the Shad1-dependent genes in reference to YB-1 expression. Of the 13 genes examined in the Shad1 LNA knockdown cells, 10 genes remained significantly differentially regulated in response to YB-1 knockdown (Supplemental Fig. S3A). However, there were no consistent changes in gene expression when the genes were examined in normal prostate epithelial cells with or without YB-1 overexpression. Motif analysis of Shad1 regulated genes revealed a consensus motif shared between all disregulated genes, but this motif did not correspond to either the YB-1 binding site or Shad1 complementary sequence indicating that it is unlikely that YB-1 in conjunction with Shad1 binds directly to regulate the expression of these genes (Supplemental Fig. S3B). A TOMTOM scan of the Shad1 consensus motif revealed only 24 hits in 1329 total motifs in the vertebrate database (1.8%) (Gupta et al. 2007). Additionally, a FIMO search found no statistically significant enrichment of the Shad1 consensus motif in the human nucleotide database indicating that there is no enrichment of this motif among genes that are not Shad1 regulated (Grant et al. 2011). These results reveal that Shad1 and YB-1 expression is closely linked, and Shad1 likely plays an indirect role in the expression of other prognostic markers of cancer metastasis. These results also indicate that YB-1 and Shad1 expression alone is insufficient to disregulate genes associated with cancer metastasis in normal prostate epithelial cells.
FIGURE 4.
Characterization of probable Shad1 function. (A) MTT assay of cells 48 h post-transfection. Percent cell viability is based on nontransfected cells plated at the same initial density. (*) P-value <0.05; n = 3. (B) Cytotoxicity assay shows no significant cell death upon Shad1 knockdown. (C) Cell counts of transfected cells at 24 and 48 h post-transfection. (*) P-value <0.05; (**) P-value <0.001; n = 3. (D) Transfection with LNA antisense to Shad1 (aShad1) shows a decrease in Shad1 expression compared with scrambled control. (*) P-value <0.05; n = 3. (E) Shad1 knockdown shows no difference in Dicer1 mRNA levels (P-value = 0.5002; n = 3). (F) Shad1 knockdown shows no difference in Dicer1 protein levels (P-value = 0.2907; n = 4). Representative Western blot illustrates band intensities used for quantitation (inset). (G) qPCR validation of genes differentially expressed upon Shad1 knockdown. All genes are significantly differentially expressed with P-values <0.05; n = 3. Dashed line represents no relative change; anything above is up-regulated and below is down-regulated.
Shad1 is evolutionarily conserved and localizes to mammalian P-bodies/stress granules
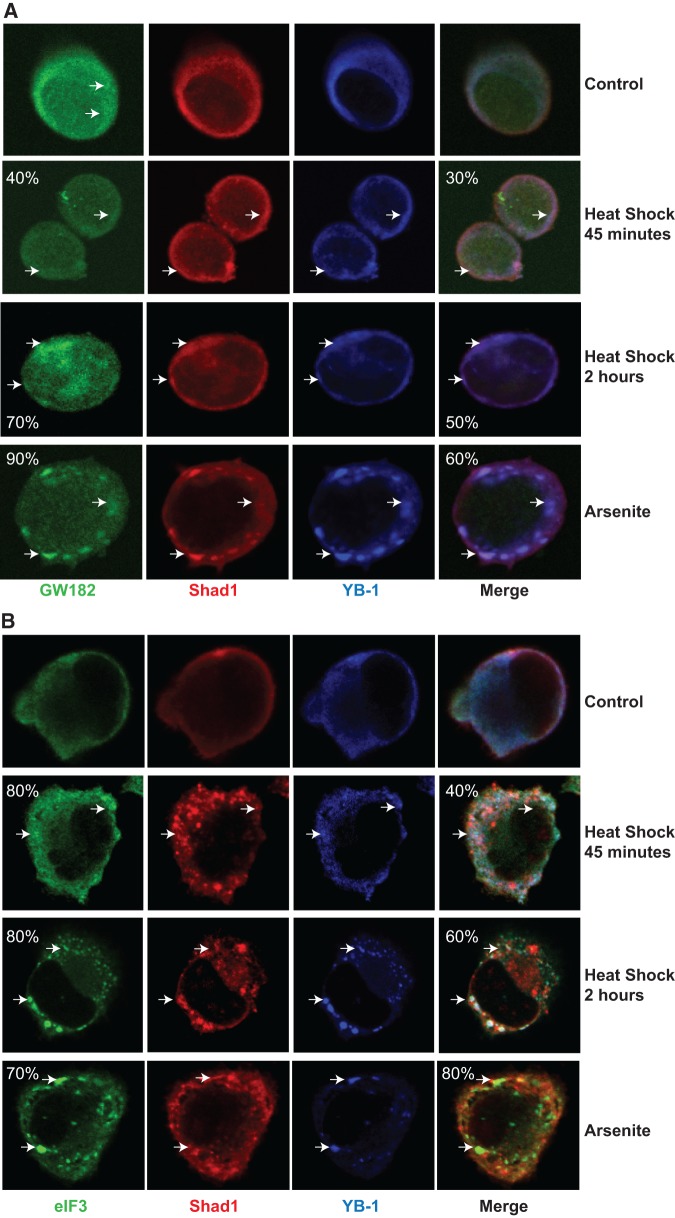
YB-1 is known to be highly conserved from bacteria to all vertebrate species examined and its subcellular localization can be influenced by cellular stress (Christov et al. 2006; Krude et al. 2009). For example, YB-1 associates with two distinct stress-responsive cellular structures, the mRNA degradation-related P-bodies (PB), and stress granules that are sites of mRNA processing (Kedersha and Anderson 2007). In six cell lines comprising human (PC-3, HeLa, HEK293, MCF-7), mouse (P19), and monkey (COS7), YB-1 consistently localizes to the cytoplasm of the cells under normal growth conditions (Fig. 5; Supplemental Figs. S4–S8). While Shad1 is also primarily localized within the cytoplasm, it is detectable in the nucleus of HeLa cells. When cells are exposed to stress conditions, YB-1 and Shad1 form punctate structures in the cytoplasm in all cell lines tested. For example, upon heat shock for 45 min, 30% of YB-1/Shad1 punctate structures colocalize with GW182, a marker for PBs but not SGs, which represents 40% of labeled PBs (Fig. 5A). Upon a 2 h heat shock, 50% of YB-1/Shad1 structures colocalize with PBs, representing 70% of PBs. When cells are treated with arsenite, an oxidative stress agent, 60% colocalize to 90% of PBs. Under the same arsenite exposure conditions, 80% of YB-1/Shad1 colocalizes in 70% of eIF3, a marker for SGs but not PBs (Fig. 5B). The colocalization of eIF3 with YB-1 and Shad1 is also visible upon heat shock (40% and 60% colocalization, respectively). In some instances, a minor fraction of Shad1 and YB-1 are also detected in the nucleus (Supplemental Fig. S4). To compare the localization differences between the Shad1 shyRNA and Shad1 smyRNA, we used another probe that targets the immediate upstream location of the Shad1-derived small RNA. Although the Shad1 shyRNA showed more punctate staining even under nonstress conditions in PC-3 cells, colocalization patterns with YB-1 and PBs or SGs remained the same when cells were subjected to heat shock and arsenite stress (Supplemental Fig. S9). To further confirm that the observed localization patterns are truly attributed to YB-1, we used a FLAG-tagged YB-1 in HeLa S3 cells to reconfirm the localization of Shad1 with YB-1 under stress conditions (Supplemental Fig. S10). To verify that the GW182 cytoplasmic structures were actually P-bodies, we also used Dcp1a as another maker of PBs (Supplemental Fig. S11). Finally, we tested whether Shad1 levels change after arsenite treatment. Northern blot analysis revealed that stress induced recruitment of Shad1 to PBs/SGs does not reflect an elevated level of Shad1 (Supplemental Fig. S12). Taken together, Shad1 is not only associated with YB-1 across a variety of cell types, but colocalizes to stress-responsive structures specifically with YB-1.
FIGURE 5.
YB-1 colocalizes with Shad1 to cytoplasmic bodies in PC-3 cells. (A) Upon heat shock or oxidative stress (arsenite), Shad1 localizes with YB-1 to P-bodies (GW182). (B) Under the same conditions, some of the Shad1 localizes with YB-1 to stress granules (eIF3). Percent colocalization of Shad1/YB-1 to cytoplasmic bodies are indicated on merged panels and the percent of cytoplasmic bodies with Shad1/YB-1 are indicated in the PB/SG panels.
DISCUSSION
The YB-1 protein utilizes its DNA and RNA binding activity to influence various aspects of nucleic acid metabolism including DNA replication, transcription, mRNA splicing, and export. We have identified a unique regulatory mechanism for YB-1 that is mediated by a novel class of short RNAs termed shyRNAs and their processed small RNAs (smyRNAs) that specifically associate with YB-1, perhaps as shown in one case by a direct interaction. It seems likely that only a minor proportion of shyRNAs represent degradation products of longer noncoding RNAs or mRNAs since the overlap between shyRNAs and RNA-seq data from endo- and exonuclease knockdown cells are low. While the biosynthesis of these RNAs is unknown, the number of functional noncoding RNAs associated with YB-1 (i.e., RNY3, MALAT-1) suggests that YB-1 is important in the small RNA pathway. Previous studies have shown that YB-1 associates with AGO proteins so some of these shy- and smy-RNAs may associate with both YB-1 and AGO in a complex.
To examine the potential biological relevance of the unique YB-1–associated RNAs that we identified, functional assays were performed with validated shyRNAs such as one derived from the antisense strand of Dicer that we designated as Shad1. The increased proliferation of PC-3 cells upon Shad1 ablation seems contradictory since YB-1 overexpression reportedly plays a role in tumor progression and outcome. However, we show that blocking Shad1 decreases the expression of many other genes know to predict cancer progression and metastasis. The transcription factor DLX2 has been shown to inhibit apoptosis induced by TGFβ and the expression of DLX2 correlates with tumor progression and metastasis in melanoma, glioma, lung, and prostate cancers (Yilmaz et al. 2011). Another gene whose overexpression correlates with tumor progression and is down-regulated in the absence of Shad1 is IGFBP2. Previous studies have shown that IGFBP2 is overexpressed in glioma, prostate, and breast cancers (Kanety et al. 1993; Busund et al. 2005; Lin et al. 2009). In prostate cancer, serum levels of IGFBP2 can be measured and levels increase as the cancer progresses (Shariat et al. 2002; Bensalah et al. 2008). IL-6, a cytokine that can be both pro- and anti-inflammatory, plays an important role in the metastasis of prostate cancer (Shariat et al. 2002). Endothelial cells have been shown to excrete IL-6 resulting in a decrease in androgen receptor activity leading to increased invasion of the epithelial cells (Wang et al. 2013). In HER2 positive breast cancer, IL-6 expression is increased as well (Hartman et al. 2011). However, YB-1 overexpression in breast cancer cells actually decreases cell growth rather than promoting proliferation (Garand et al. 2011). Since one property of YB-1 overexpression is cisplatin resistance and cisplatin functions by forming DNA crosslinks during replication, the drop in proliferation could render treatment less efficacious. Instead, the overexpression of YB-1 in decreasing proliferation could be promoting enhanced migration and epithelial–mesenchymal transition (EMT) (Evdokimova et al. 2009a,b). Shad1 can be found not only in different mammalian cell lines we examined (mouse and monkey), but it is also expressed in normal cell lines at levels corresponding to YB-1 expression. The fact that Shad1 is antisense to Dicer1 means that any blockade of Shad1 requires a sequence that would not affect Dicer1 expression, making it a potential target that would specifically inhibit a handful of genes important to tumor progression instead of inhibiting a multifunctional protein like YB-1. Taken together with the observation that shyRNAs include many known regulatory short RNAs such as Y RNAs, pre-miRNAs, and snoRNAs, these observations support the notion that shyRNAs represent a group of RNAs that share a common pathway but have evolutionarily acquired diverse cellular roles.
MATERIALS AND METHODS
Cell culture and proliferation
PC-3, HeLa, HEK293, MCF-7, and COS7 cells were maintained in RPMI1640 supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. P19 cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. RWPE1 cells were maintained in Keratinocyte-serum free media supplemented with bovine pituitary extract and epidermal growth factor (EGF). Cell proliferation was determined with the Vybrant MTT cell proliferation assay (Invitrogen) and trypan blue cell count at 48 h post-transfection.
RNA immunoprecipitation
Cells were grown to 70% confluence in 15 cm plates and lysed with RNA immunoprecipitation lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.25% NP-40, 1.5 mM MgCl2, protease inhibitors, PMSF, and RNaseOUT). Protein A/G agarose beads (Santa Cruz) were prewashed with RNA wash buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% NP-40, 1.5 mM MgCl2). For antibody-immobilized beads, 2 μg of YB-1 (Abcam), normal rabbit IgG (Santa Cruz), or hnRNP A/B (Santa Cruz) antibody was added to the bead/wash buffer solution and incubated at 4°C for 90 min. Cell lysates were precleared with washed agarose beads for 60 min. Equal volumes of supernatant were either saved as input sample or applied to the antibody-immobilized beads and incubated at 4°C for 3 h. The antibody-immobilized beads–RNP complexes were washed three times in wash buffer and treated with proteinase K at 45°C for 30 min. After protein digestion, RNA was isolated using TRIzol (Invitrogen) and chloroform (Sigma).
RNA sequencing
RNA obtained from the immunoprecipitation experiments were size fractionated on a polyacrylamide-urea gel (National Diagnostics) and gel slices were crushed and resuspended in 0.4M NaCl, frozen, and incubated at 42°C overnight. The acrylamide was removed by passing the suspension through Nanosep 100K filters (Pall Corp.) and RNA was precipitated. 5′ phosphate-dependent cDNA libraries were prepared using the manufacturer TruSeq small RNA-sequencing protocol. Small RNA 3′ and 5′ adapters were ligated to the RNAs using T4 RNA Ligase 2 and cDNA synthesis was performed using the RNA RT primer (RTP) and SuperScript II reverse transcriptase. PCR amplification was performed with RNA PCR primer (RP1) and primer index (RPIX) and Phusion DNA polymerase. The cDNA library was then size fractionated on a 6%–12% PAGE gel and the library recovered for sequencing. The small RNA was single-end sequenced while the ∼10–250 nt library was paired-end sequenced (Tufts core facility).
Genomic analysis
All sequence mapping and filtering were performed in house. Illumina FASTQ files were processed to retrieve sequences with Phred quality score to ensure a base call accuracy of 99%. The 3% adapter sequences and reads shorter than 15 nucleotides were removed using cutadapt. The filtered RNA reads were mapped to the human genome (GRCh37) using Bowtie. The mapped locations were then further filtered to only retain those reads that matched to a single unique location in the genome and were >1 RPKM. Neighboring (distance <10 nt) reads from identical strands were considered as part of a single genomic cluster for annotations using UCSC Genome Browser and Bioconductor (Gentleman et al. 2004; Karolchik et al. 2009). For the paired-end sequencing reads, the libraries were normalized to the sequenced input RNA. Input RNAs that were greater than twofold enriched over YB-1 RNAs was considered to be the control RNA sequences; YB-1 RNAs that were greater than twofold enriched over input RNAs was considered to be the YB-1–associated RNA sequences.
Statistical analysis
Publicly available (embargo free) ChIP-seq data (ENCODE) of histone modifications (72 cell lines, 682 samples) and transcription factors (88 cell lines, 218 transcription factors, 1284 samples) from the UCSC genome browser portal were used to establish the preference of various genomic features to shyRNA locations. As a control group, the input RNA locations with at least twofold enrichment over YB-1 and >1 RPKM (96,335 locations) were defined. An equivalent number based on RPKM of YB-1–associated RNAs were selected. Bootstrapping analysis for YB-1 to histone modification/TF binding site enrichment was performed by randomly sampling (with replacement, n = 96,335) the described sets of both YB-1 and control samples to yield 10 different data sets for each sample (YB-1 or control). The distances to the closest histone modification or TF binding sites for each 5′ end of the YB-1–associated RNAs or its equivalent control 5′ locations were calculated and binned (size = 200 nt). P-values were computed using the two tailed t-test based on the probability distribution of the YB-1 and control bootstrapped data sets at position 0 (±100 nt).
RNA electromobility shift assay
GST-tagged YB-1 protein was constructed by subcloning the CDS of YB-1 into the vector pGEX-6P1. E. coli Rosetta was transformed and expression was induced at 18°C over night. Cells were lysed by sonication and protein was purified with Glutathione Sepharose 4B (GE Healthcare). Elution was done with Glutathione peptide, followed by dialysis over night against PBS. To generate synthetic RNAs for binding assays, DNA oligos were designed with a T7 promoter (underlined) AACCAATCTCAACCAGCCACTGCCCTATAGTGAGTCGTATTA (Shad1) and TCCTTGTGGTTGTCTGAAGTAGCACTTCCGGATTTGAATGGCTTGGGTTCCA CTGGGTCCAGTCCTGGTTCCTATAGTGAGTCGTATTA (CD44v4). T7 in vitro transcription was performed using the T7 RNA polymerase (NEB) and the RNA was purified with phenol/chloroform extraction and ethanol precipitation. Synthesized RNAs were then 3′-biotin labeled (Pierce) and RNA probes were purified with phenol/chloroform extraction and ethanol precipitation. The binding reaction was carried out in 20 μL of RNA binding buffer (100 mM HEPES pH 7.9, 100 mM KCl, 10 mM MgCl2, 10 mM DTT), 5% glycerol, 2 μg tRNA (nonspecific competitor RNA). Where indicated, a 20-fold excess of unlabeled RNA probe was added to the reaction as a control for specificity. The reactions were incubated at room temperature for an hour and resolved on a 4.5% native polyacrylamide gel in 0.5× TBE buffer. The samples were electrotransferred to positively charged nylon membranes and UV crosslinked at 120 mJ/cm2. Chemiluminescent detection of the biotin-labeled RNA probes was performed per manufacturer's instructions (Pierce).
Northern blots and qRT-PCR
RNA was separated by electrophoresis on 15% polyacrylamide-urea gels and electrotransferred to positively charged nylon membranes (Roche). Northern blots were performed using our previously described protocol utilizing LNA probes, EDC crosslinking and DIG-labeled probes (Kim et al. 2010). EDC crosslinking was performed at 60°C for 1–2 h to facilitate adequate RNA–membrane crosslinking. LNA or DNA oligonucleotide probes were 3′-end labeled with digoxigenin (DIG) using an End Tailing Kit (Roche). Probe sequences used for detection are ACAATCTCAACCAGCCACT (Shad1), AATTGAGATAACTCACTACCTTCGGA CCAGCC (RNY1), TAGTCAAGTGAAGCAGTGGGAGTGGAGA (RNY3), and CCTCCGCTGAC TAATATGCTTAAATTCAGCGGGTCGCCACGTCTGATCTGAGGTCGCG (Saunc45B) with LNA bases underlined. RNA−probe hybridizations were carried out overnight in Ultrahyb buffer (Ambion) at 42°C.
RNA obtained from the immunoprecipitation experiments were size fractionated and gel purified. The NCode VILO cDNA synthesis and qRT-PCR kit was used to polyadenylate the resulting RNA and oligo(DT) primers were used to synthesize the first strand cDNA (Invitrogen). For validation of small RNAs, one universal qPCR reverse primer was used to amplify the poly-dT region of the cDNA along with small RNA specific forward primers (Table 1). Gene-specific primers only were used to validate shyRNAs (Table 1). PCR was performed with input, IgG-associated, and YB-1–associated RNAs. The enrichment of the RNAs by IgG or YB-1 over the input RNA was calculated by subtracting the immunoprecipitated RNA values from the input values. The enrichment of the specific YB-1–associated RNAs over the nonspecific IgG controls were then calculated using the qPCR formula 2−(experimental − control), where the experimental would be the input normalized YB-1 and the control would be the input normalized IgG.
TABLE 1.
qRT-PCR primers for small RNA and shyRNA validation, and LNA oligos

Detection of 5′ phosphate and 3′ hydroxyl groups
RNAs from immunoprecipitation experiments were incubated with Terminator 5′-monophosphate-dependent exonuclease (Epicentre, manufacturer protocol) to probe for the presence of a 5′-monophosphate. After incubation, RNA was purified and used directly for Northern blot. For 3′ hydroxyl modification tests, RNAs were treated with sodium periodate (Alefelder et al. 1998). Briefly, RNA solution was diluted (1:10) in borax/boric acid buffer (pH 8.6). One-tenth volume of sodium periodate solution (250 mM in water) was added and the mixture was incubated at room temperature for 10 min. After incubation, one-tenth volume of glycerol was added, followed by 10 min incubation. The mixture was then diluted (1:10) in NaOH/borax/boric acid buffer (pH 9.5) and kept at 45°C for 90 min. Finally RNA was purified from the mixture using mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA). After purification, RNAs were polyadenylated and qRT-PCR was performed as described above.
LNA transfections and shRNA lentivirus infections
For transfections, cells were plated to 50% confluency in plates appropriate for downstream experiments. Transfections with 100 nM LNA oligo concentrations were performed for the Shad1 small and shyRNAs. Transfection for the YB-1 overexpression cell line was performed with 1 μg of plasmid DNA (Addgene). All transfections were done in media without antibiotics using Opti-MEM and Lipofectamine LTX with Plus Reagent (Invitrogen). For stable transfections, cells were treated with 200 μg/mL G418 selection 48 h after transfection. For lentivirus infections, 1 × 105 cells were plates per well in a 6-well plate in complete media for 24 h. Media was replaced with media containing 8μg/mL polybrene and lentivirus containing either shYB-1 or shScr control plasmids (Santa Cruz). Fresh media containing 2 μg/mL puromycin was added to cells 24 h post-infection.
Dual fluorescent immunocytochemistry and in situ hybridization
After heat shock or arsenite treatment, cells were fixed to glass coverslips at room temperature for 10 min with 4% paraformaldehyde in 0.1 M PIPES pH 6.9, 2 mM MgCl2, and 1.25 mM EGTA (Geiger and Neugebauer 2005). Coverslips were rinsed in PBS and permeabilized with 0.2% Triton X-100 and MgPBS (10 mM MgCl2 in PBS) for 10 min at room temperature. Primary and fluorescent secondary antibodies were diluted according to manufacturer's recommendations in protein block (3% BSA in PBS). All steps subsequent to secondary antibody incubation were performed in the dark to prevent photobleaching. Antibodies were fixed in 4% paraformaldehyde for 5 min and rinsed again with MgPBS. For in situ hybridization, coverslips were incubated in prehybridization buffer for 30 min. DIG-labeled LNA probes were diluted 1:100 in the prehybridization buffer and heated for 5 min at 80°C. Coverslips were incubated with LNA probes at 60°C overnight in a humidified chamber then incubated in 2× SSC with 50% formamide at 37°C for 30 min. After stringency washes in 2× and 1× SSC, coverslips were incubated with the anti-DIG-POD antibody diluted in FISH blocking buffer (0.25% BSA in 2× SSC). The antibody was removed with 3–10 min washes in PBST. Tyramide signal amplification (TSA) fluorescent detection was used to detect the DIG-labeled LNA probe. The TSA reagent was diluted 1:50 and applied directly to the coverslip and incubated for 10 min. Coverslips were mounted on slides using VectaShield reagent and slides were imaged using an LSM700 Confocal microscope equipped with a digital camera (Zeiss). Images were analyzed using the Zen 2009 Light Edition Software. Two percent colocalization values were calculated; one was the percentage of PB/SGs that contained both YB-1 and Shad1 staining and one was the percentage of dual labeled YB-1 and Shad1 localized to PB/SGs.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the laboratories of D.B.DeF. and B.J. for helpful discussions. We also thank Drs. Paula Grabowski and Mark Nichols for valuable advice. This work was primarily supported by the G. Harold and Leila Y. Mathers Foundation (B.J.), and partly by National Institutes of Health (GM079756 to B.J.) and American Cancer Society (RSG-09-054-01 to B.J.), Deutsche Forschungsgemeinschaft (SFB 960 to G.M.), the European Research Council (ERC grant “sRNAs” to G.M.), and the Bavarian Genome Research Network (BayGene to G.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045559.114.
REFERENCES
- Agafonov DE, Deckert J, Wolf E, Odenwälder P, Bessonov S, Will CL, Urlaub H, Lührmann R 2011. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol 31: 2667–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alefelder S, Patel BK, Eckstein F 1998. Incorporation of terminal phosphorothioates into oligonucleotides. Nucleic Acids Res 26: 4983–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargou RC, Jürchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dörken B, et al. 1997. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med 3: 447–450. [DOI] [PubMed] [Google Scholar]
- Bensalah K, Lotan Y, Karam JA, Shariat SF 2008. New circulating biomarkers for prostate cancer. Prostate Cancer Prostatic Dis 11: 112–120. [DOI] [PubMed] [Google Scholar]
- Busund LT, Richardsen E, Busund R, Ukkonen T, Bjørnsen T, Busch C, Stalsberg H 2005. Significant expression of IGFBP2 in breast cancer compared with benign lesions. J Clin Pathol 58: 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov CP, Gardiner TJ, Szüts D, Krude T 2006. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol 26: 6993–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 26: 5528–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD 1988. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci 85: 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D 2010. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol 17: 1358–1366. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N 2001. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J 20: 5491–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH 2006. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol 26: 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ, et al. 2009a. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15: 402–415. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Sorensen PH 2009b. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 8: 2901–2906. [DOI] [PubMed] [Google Scholar]
- Garand C, Guay D, Sereduk C, Chow D, Tsofack SP, Langlois M, Perreault E, Yin HH, Lebel M 2011. An integrative approach to identify YB-1-interacting proteins required for cisplatin resistance in MCF7 and MDA-MB-231 breast cancer cells. Cancer Sci 102: 1410–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger JA, Neugebauer KM 2005. Fluorescent detection of nascent transcripts and RNA-binding proteins in cell nuclei. In Handbook of RNA biochemistry (ed. Hartmann RK, et al. ), Vol. 2, pp. 729–736 Wiley-VCH Verlag GmbH, Weinheim, Germany. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Bonafé P, Fedoruk MN, Whitmore TG, Akbari M, Ralph JL, Ettinger S, Gleave ME, Nelson CC 2004. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate 59: 337–349. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS 2007. Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi G, Leung S, Law JH, Gelmon K, Masoudi H, Turbin D, Pollak M, Nielsen TO, Huntsman D, Dunn SE 2008. Redefining prognostic factors for breast cancer: YB-1 is a stronger predictor of relapse and disease-specific survival than estrogen receptor or HER-2 across all tumor subtypes. Breast Cancer Res 10: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hartman ZC, Yang XY, Glass O, Lei G, Osada T, Dave SS, Morse MA, Clay TM, Lyerly HK 2011. HER2 overexpression elicits a proinflammatory IL-6 autocrine signaling loop that is critical for tumorigenesis. Cancer Res 71: 4380–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G 2007. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep 8: 1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, Izumi H, Ohmori H, Okamoto T, Ohga T, et al. 1999. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res 59: 342–346. [PubMed] [Google Scholar]
- Kanety H, Madjar Y, Dagan Y, Levi J, Papa MZ, Pariente C, Goldwasser B, Karasik A 1993. Serum insulin-like growth factor-binding protein-2 (IGFBP-2) is increased and IGFBP-3 is decreased in patients with prostate cancer: correlation with serum prostate-specific antigen. J Clin Endocrinol Metab 77: 229–233. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Kent WJ 2009. The UCSC genome browser. Curr Protoc Bioinformatics Chapter 1: Unit 1.4. 10.1002/0471250953.bi0104s28. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P 2007. Mammalian stress granules and processing bodies. Methods Enzymol 431: 61–81. [DOI] [PubMed] [Google Scholar]
- Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B 2010. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res 38: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M 2003. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25: 691–698. [DOI] [PubMed] [Google Scholar]
- Krude T, Christov CP, Hyrien O, Marheineke K 2009. Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J Cell Sci 122: 2836–2845. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R 2013. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Jiang T, Zhou K, Xu L, Chen B, Li G, Qiu X, Zhang W, Song SW 2009. Plasma IGFBP-2 levels predict clinical outcomes of patients with high-grade gliomas. Neuro Oncol 11: 468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, et al. 2012. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res 40: 8460–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri E, Levy A, Benjamin H, Ben-David M, Cohen L, Dov A, Dromi N, Elyakim E, Yerushalmi N, Zion O, et al. 2010. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res 38: 6234–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10: 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Tang L 2003. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol 258: 432–442. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Young CY, Yuan H 2010. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin (Shanghai) 42: 363–369. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. [DOI] [PubMed] [Google Scholar]
- Raffetseder U, Frye B, Rauen T, Jürchott K, Royer HD, Jansen PL, Mertens PR 2003. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J Biol Chem 278: 18241–18248. [DOI] [PubMed] [Google Scholar]
- Shariat SF, Lamb DJ, Kattan MW, Nguyen C, Kim J, Beck J, Wheeler TM, Slawin KM 2002. Association of preoperative plasma levels of insulin-like growth factor I and insulin-like growth factor binding proteins-2 and -3 with prostate cancer invasion, progression, and metastasis. J Clin Oncol 20: 833–841. [DOI] [PubMed] [Google Scholar]
- Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ 2011. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res 40: 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci 100: 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitkovsky DD, Royer-Pokora B, Delius H, Kisseljov F, Jenkins NA, Gilbert DJ, Copeland NG, Royer HD 1992. Tissue restricted expression and chromosomal localization of the YB-1 gene encoding a 42 kD nuclear CCAAT binding protein. Nucleic Acids Res 20: 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J 20: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH 2011. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol 18: 1075–1082. [DOI] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y 2003. Mice lacking Zfhx1b, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease–mental retardation syndrome. Am J Hum Genet 72: 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. 2009. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 41: 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BA, Valera VA, Pinto PA, Merino MJ 2013. Comprehensive microRNA profiling of prostate cancer. J Cancer 4: 350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee SO, Xia S, Jiang Q, Luo J, Li L, Yeh S, Chang C 2013. Endothelial cells enhance prostate cancer metastasis via IL-6→androgen receptor→TGF-β→MMP-9 signals. Mol Cancer Ther 12: 1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci 103: 4034–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Maass D, Tiwari N, Waldmeier L, Schmidt P, Lehembre F, Christofori G 2011. Transcription factor Dlx2 protects from TGFβ-induced cell-cycle arrest and apoptosis. EMBO J 30: 4489–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Homer C, Edwards SJ, Hananeia L, Lasham A, Royds J, Sheard P, Braithwaite AW 2003. Nuclear localization of Y-box factor YB1 requires wild-type p53. Oncogene 22: 2782–2794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.