Abstract
AIM: Gene expression profiling provides an unique opportunity to gain insight into the development of different types of gastric cancer. Tumor sample heterogeneity is thought to decrease the sensitivity and tumor specificity of microarray analysis. Thus, microdissection and preamp-lification of RNA is frequently performed. However, this technique may also induce considerable changes to the expression profile. To assess the effect of gastric tumor heterogeneity on expression profiling results, we measured the variation in gene expression within the same gastric cancer sample by performing a gene chip analysis with two RNA preparations extracted from the same tumor specimen.
METHODS: Tumor samples from six intestinal T2 gastric tumors were dissected under liquid nitrogen and RNA was prepared from two separate tumor fragments. Each extraction was individually processed and hybridized to an Affymetrix U133A gene chip covering approximately 18 000 human gene transcripts. Expression profiles were analyzed using Microarray Suite 5.0 (Affymetrix) and GeneSpring 6.0 (Silicon Genetics).
RESULTS: All gastric cancers showed little variance in expression profiles between different regions of the same tumor sample. In this case, gene chips displayed mean pair wise correlation coefficients of 0.94±0.02 (mean±SD), compared to values of 0.61±0.1 for different tumor samples. Expression of the variance between the two expression profiles as a percentage of "Total change "(Affymetrix) revealed a remarkably low average value of 1.18±0.78 for comparing fragments of the same tumor sample. In contrast, comparison of fragments from different tumors revealed a percentage of 24.4±4.5.
CONCLUSION: Our study indicates a low degree of expression profile variability within gastric tumor samples isolated from one patient. These data suggest that tumor tissue heterogeneity is not a dominant source of error for microarray analysis of larger tumor samples, making total RNA extraction an appropriate strategy for performing gene chip expression profiling of gastric cancer.
Keywords: Gastric cancer, Microarray analysis, Tissue heterogeneity
INTRODUCTION
Gastric cancer is one of the most common malignancies, accounting for almost 10% of new cancer cases diagnosed worldwide[1,2]. However, knowledge about the molecular mechanisms underlying tumor development and progression is limited and molecular features of gastric cancer are not commonly used for diagnosis and treatment
cDNA and oligonucleotide microarrays are capable of profiling gene expression patterns of tens of thousands of genes in a single experiment. They are an ideal tool to study cancer progression and development and to identify new molecular markers for tumor classification and prognosis. Microarrays have been successfully applied to study various tumors, including gastric cancers[3-13].
Like most solid tumors, gastric cancer consists of many different cell types including endothelium, different types of stromal cells and inflammatory cells. Such tissue heterogeneity is thought to decrease the sensitivity and tumor specificity of the microarray analysis[14-16]. One strategy to overcome the problem of tissue heterogeneity is the use of laser capture microdissection (LCM) for isolation of a defined cell population[17,18] followed by amplification of the RNA for subsequent microarray analysis[19,20]. However, amplification-associated bias may induce considerable changes to the expression profile[15,21]. Depending on both the degree of tissue heterogeneity and the robustness of the microarray analysis protocol, it may therefore sometimes be advantageous to avoid RNA amplification and instead extract total RNA directly from a larger tumor sample.
To assess the influence of gastric tumor heterogeneity on the outcome of the microarray analysis, we measured the variation between gene expression profiles derived from two RNA preparations extracted from the same gastric tumor specimen.
MATERIALS AND METHODS
Gastric tumor samples
Tumor samples from six patients who underwent surgery for gastric cancer at the University Hospital of Dresden were used for microarray analysis. Immediately after gastrectomy, a certified pathologist obtained representative samples of 1 cm3 from the tumor centers. Special care was taken to avoid tumor necrosis. Tumors were snap-frozen in liquid nitrogen and stored at -80°C. To ensure consistency of the study population, only tumors classified as intestinal and T2 by pathology were included in the study[22]. Clinical and histopathological tumor characteristics are summarized in Table 1.
Table 1.
Clinical data and histopathological features of gastric cancers included in the study
| Tumor number | Age/Sex | Site | Lauren | TNM |
| 1 | 72/Female | Antrum | Intestinal | T2 N2 G3 |
| 2 | 78/Male | Body | Intestinal | T2 N1 G2 |
| 3 | 81/Male | Antrum | Intestinal | T2 N1 G2 |
| 4 | 75/Female | Antrum | Intestinal | T2 N0 G3 |
| 5 | 76/Male | Cardia | Intestinal | T2 N2 G3 |
| 6 | 76/Male | Cardia | Intestinal | T2 N0 G3 |
RNA preparation
For RNA isolation, tumor samples were fragmented into smaller pieces under liquid nitrogen using a mortar and pestle. Subsequently, total RNA was extracted from two independent, randomly selected tumor fragments using RNAzol reagent according to the manufacturer's instructions (TelTest Inc., Friendswood, TX). Both extractions (extractions A and B) were processed individually until hybridization on two separate microarrays. To determine the influence of experimental variability, RNA was extracted only once from tumor no. 3 and aliquots of the same RNA preparation were used for further processing.
High-density oligonucleotide microarray analysis
Microarray analysis was performed according to the Affymetrix instructions for eukaryotic sample preparation[23]. In summary, double-stranded cDNA was synthesized from 5 mg of total RNA with oligo(dT)24 T7 primer (Affymetrix), followed byin vitro transcription of cRNA synthesis using Enzo BioArray High Yield RNA Transcript Labeling Kit (Affymetrix) and the biotinylated cRNA was hybridized to Affymetrix U133A gene chip arrays containing 22 253 probe sets (approximately 18 000 human gene transcripts). The hybridized probe array was stained with streptavidin-hycoerythrin conjugate and scanned by the Affymetrix gene chip scanner.
Statistical analysis
Expression profiles were analyzed using Microarray Suite 5.0 (Affymetrix) and GeneSpring 6.0 (Silicon Genetics). We used the Affymetrix software to identify changes in expression levels between fragments of the same tumor specimen using standard protocols recommended by Affymetrix[24]. For normalization, data from each expression array were scaled, so that the overall fluorescence intensity across each chip was equivalent (average target intensity set at 500). Only relative changes equal or greater than twofold level of expression were considered. For a given gene transcript in any chip-to-chip comparison, Microarray Suite Software generates a "change call" parameter ("Increase "or "Decrease") based on a consideration of signal specificity as well as intensity. In other words, the "change call "is based on an evaluation of the intensities of the signals generated from each gene transcript on one chip relative to the corresponding signal intensities on the other chip. Instances where the signal on the higher intensity chip is falsely elevated are called "Increased in a comparison between two chips derived from the same target preparation. "Decrease" calls represent instances where the signal on the lower intensity array has been falsely elevated. Consequently we define all "Increase "or "Decrease" calls in a comparison between arrays derived from the same target preparation as false positive. According to the Affymetrix specification, a percentage of "total change up to 2% between two chips derived from the same hybridization cocktail is acceptable. To confirm results derived by Microarray Suite, GeneSpring was used to perform multiple comparisons between arrays and to generate pairwise correlation coefficients (Spearman correlation) for all arrays.
RESULTS
RNA and hybridization quality control parameters
Extracted total RNA was evaluated on a 2% agarose gel for the presence of 28S and 18S rRNA bands, which were clearly visible in all RNA samples. RNA purity was assessed by UV spectrophotometry. An A260/280 ratio >1.9 was achieved in all samples. Total RNA was also used for RT-PCR of a housekeeping mRNA species to assess RNA quality. Human GAPDH (Applied Biosystems Inc., Foster City, CA, USA) was robustly amplified in all cases.
We followed the Affymetrix guidelines for efficient hybridization: Among the 22 253 probe sets, there was a consistent percentage of “present” calls for each of the 12 cRNA samples tested with a median of 50.68 (range 42.9-54.7). No array image showed grossly visible artifacts. The ratio of the 3 probe set to the 5 probe set of the GAPDH internal control gene, which acts as an indicator for overall RNA quality was below three in all cases. RNA and gene chip hybridization quality control parameters are summarized in Table 2.
Table 2.
RNA and hybridization quality control parameters
| Extraction/Tumor number | A260/280 ratio | GAPDH CT | Present calls (%) | GAPDH 3’5’ ratio |
| A/1 | 2.18 | 15.86 | 52.5 | 1.08 |
| B/1 | 2.18 | 15.7 | 52.3 | 1.26 |
| A/2 | 2.09 | 15.8 | 49.2 | 0.89 |
| B/2 | 2.07 | 15.6 | 54.4 | 1.19 |
| A/3 | 2.1 | 15.2 | 53.5 | 1.09 |
| B/3 | 2.1 | 15.2 | 52.5 | 1.08 |
| A/4 | 2.07 | 15.5 | 48.8 | 0.99 |
| B/4 | 2.08 | 15.2 | 49.8 | 0.96 |
| A/5 | 2.15 | 16.07 | 51.7 | 1.95 |
| B/5 | 2.24 | 16.15 | 54.7 | 1.46 |
| A/6 | 2.19 | 16.08 | 42.9 | 1.58 |
| B/6 | 2.13 | 16.2 | 45.9 | 1.34 |
Intra-tumor variability
Expression profiles from five gastric tumors were used to determine the degree of intra-tumor variability. As shown in Table 3, all gastric cancers demonstrated very little variance in expression profiles between different regions of the same tumor sample. In this case, expression arrays displayed mean pairwise correlation coefficients of 0.95±0.02, in contrast to values of 0.61±0.1 when different tumor samples were compared to each other. In accordance, the variance between two expression profiles calculated with the Affymetrix software (percentage of "total change") revealed a remarkably low average value of 1.18±0.78 for comparing fragments of the same tumor sample. This result is even more striking in light of the Affymetrix guidelines, which suggest that a percentage of "total change" up to 2% is tolerable for comparing chips derived from the same hybridization cocktail. In contrast, when we compared tumor fragments from different tumors the percentage of "total change "was 24.4±4.5. As an example, Figure 1 illustrates different analyses results for two regions of the same gastric cancer sample (Figure 1A) and for two different gastric cancer samples (Figure 1B). In conclusion, there is a surprisingly low degree of expression profile variability within all gastric tumor samples studied.
Table 3.
Comparison results for all tumor samples analyzed
| Comparison | Pairwise correlation coefficient | Percentage of “total change" |
| Tumor 1, extraction A vs B | 0.96 | 1.07 |
| Tumor 2, extraction A vs B | 0.91 | 2.65 |
| Tumor 3, extraction A vs B | 0.99 | 0.08 |
| Tumor 4, extraction A vs B | 0.96 | 0.55 |
| Tumor 5, extraction A vs B | 0.94 | 1.28 |
| Tumor 6, extraction A vs B | 0.95 | 0.39 |
Tumor no. 3 was used to evaluate experimental variability.
Figure 1.
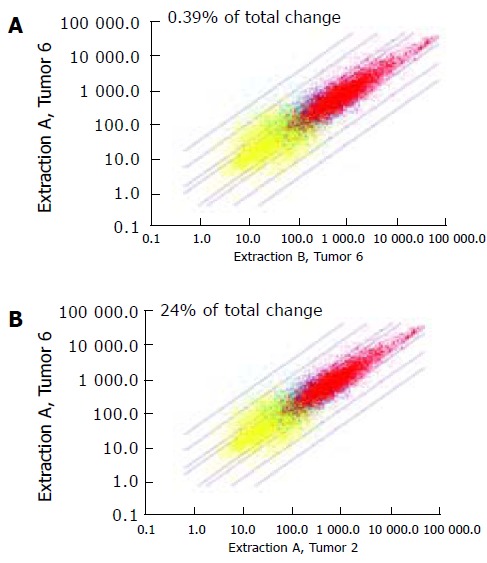
Scatter plots of the expression profiles derived from two different regions of the same gastric cancer sample (extraction A vs extraction B from tumor no. 6) (B) and from two different gastric cancer samples (extraction A from tumor no. 6 vs extraction A from tumor no. 2). The solid lines indicate 2-, 3-, 10-, and 30-fold expression level differences. Red dots represent gene transcripts detected in both arrays, yellow dots represent gene transcripts undetectable in both arrays, and blue dots represent transcripts detectable in one but undetectable in the other array. Gene transcripts detectable in both arrays (red dots) show a high correlation and thus a low percentage of "total change" for two different regions of the same tumor sample (B), whereas they show a poor correlation and thus a high percentage of "total change" for fragments of different tumors (A).
Experimental variability
During the whole procedure of the microarray experiment, so called experimental variability is generated by multiple factors including chip manufacture, preparation of cRNA, hybridization, washing steps, etc. To measure the degree of experimental variability, RNA was extracted only once from tumor no. 3. Subsequently, two aliquots from the same RNA preparation were used for further processing and hybridization on two separate Affymetrix U133A arrays. In this case, gene chip expression profiles differed in only 0.18% of all genes analyzed. Likewise the correlation coefficient was 0.99, respectively. Table 3 summarizes comparison of results from all samples.
DISCUSSION
Microarray studies on gastric cancer are currently based on two main methods of RNA preparation: the recently developed technique of LCM allows for the isolation of cancer cells and other subpopulations of interest from a heterogeneous piece of tumor tissue. The advantage of microdissection is that it directly focuses on the gastric cancer cells[5,10]. The disadvantage of LCM is at the level of resources and expertise. A certified pathologist is required to select the cells to be microdissected under the microscope, and microdissection itself requires an expensive technology and special training. Furthermore the amount of RNA obtained after LCM is not sufficient for microarray analysis and generally requires amplification. However, the amplification of small amounts of RNA from laser-captured samples is difficult and has been proved to be less reproducible than measurement of unamplified mRNA[15,21]. Even a minor degree of amplification bias might result in a substantial variation to the expression profile.
The approach we used here is based on total RNA extraction with RNA isolated directly from the gastric tumor tissue without the need for subsequent amplification. Hence, the microarray profile reflects all different tissue components present in the tumor. Most gastric cancer microarray studies are based on this method of RNA extraction[3,4,6-9,11-13]. This strategy is less laborious, because it does not require isolation of specific cell types. Moreover, there is usually a sufficient amount of high quality RNA obtained for microarray analysis. However, since the actual proportion of tumor cells in the gastric cancer tissue studied remains unknown, expression profiling based on this strategy may not reliably represent "true tumor changes. In other words, the level of intra-tumor variability is crucial for the sensitivity and tumor specificity of the microarray experiment. If there is a significant amount of intra-tumor variability, LCM becomes the strategy of choice.
In this study we found an unexpectedly low degree of expression profile variability within all gastric tumor samples studied. Randomly selected fragments from the same tumor sample displayed a striking similarity in expression profiles, while expression profiles derived from distinct tumor samples differed significantly. These data suggest that tissue heterogeneity is not a dominant source of error for microarray analysis of larger gastric tumor samples, making total RNA extraction an appropriate strategy for performing gene chip expression profiling of gastric cancer.
Our findings are in contrast to a recently published microarray study on human muscle biopsies[25], where tissue heterogeneity was a major source of expression profile variability. This indicates that our results may be exclusive for larger tumor samples and should not be generalized. It might also reflect the influence of study design and appropriate material selection on the outcome of any microarray experiment. We believe that an experienced pathologist who is able to carefully select an appropriate piece of gastric tumor for microarray experiments directly from a large tumor avoiding necrosis and infiltration by normal gastric mucosa is crucial for the quality of our results.
In our study, the degree of experimental variability was only minimal. In the early times of gene chip technology, Mills and Gordon[26] found a substantial level of experimental variability, with an average of 12% increase/decrease calls between the same RNA processed in parallel and hybridized on two Mu11k-A Affymetrix arrays. However, studies using newer, more robust Affymetrix array generations have achieved results consistent with our studies[25,27]. The low degree of experimental variability reflects the inherent advantage of the Affymetrix Gene Chip technology. Due to commercial mass production, they contain a robust series of controls designed to minimize chip-to-chip variation. The availability of standardized protocols together with the use of stringent laboratory quality control parameters also helps to reduce experimental variability.
In conclusion, we assume that gastric tumor expression profiling based on total RNA extraction reliably represents "true" tumor changes. Therefore, total RNA extraction may be the strategy of choice for microarray studies including large gastric cancer samples.
Footnotes
Science Editor Guo SY Language Editor Elsevier HK
Supported by a MeDDrive grant From the University of Dresden 2003 and by a grant from the Dr. Mildred Scheel Stiftung No. 70-2923
References
- 1.Neugut AI, Hayek M, Howe G. Epidemiology of gastric cancer. Semin Oncol. 1996;23:281–291. [PubMed] [Google Scholar]
- 2.Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83:18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 3.Boussioutas A, Li H, Liu J, Waring P, Lade S, Holloway AJ, Taupin D, Gorringe K, Haviv I, Desmond PV, et al. Distinctive patterns of gene expression in premalignant gastric mucosa and gastric cancer. Cancer Res. 2003;63:2569–2577. [PubMed] [Google Scholar]
- 4.El-Rifai W, Frierson HF, Harper JC, Powell SM, Knuutila S. Expression profiling of gastric adenocarcinoma using cDNA array. Int J Cancer. 2001;92:832–838. doi: 10.1002/ijc.1264. [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012–7017. [PubMed] [Google Scholar]
- 6.Hippo Y, Yashiro M, Ishii M, Taniguchi H, Tsutsumi S, Hirakawa K, Kodama T, Aburatani H. Differential gene expression profiles of scirrhous gastric cancer cells with high metastatic potential to peritoneum or lymph nodes. Cancer Res. 2001;61:889–895. [PubMed] [Google Scholar]
- 7.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- 8.Inoue H, Matsuyama A, Mimori K, Ueo H, Mori M. Prognostic score of gastric cancer determined by cDNA microarray. Clin Cancer Res. 2002;8:3475–3479. [PubMed] [Google Scholar]
- 9.Kim B, Bang S, Lee S, Kim S, Jung Y, Lee C, Choi K, Lee SG, Lee K, Lee Y, et al. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63:8248–8255. [PubMed] [Google Scholar]
- 10.Mori M, Mimori K, Yoshikawa Y, Shibuta K, Utsunomiya T, Sadanaga N, Tanaka F, Matsuyama A, Inoue H, Sugimachi K. Analysis of the gene-expression profile regarding the progression of human gastric carcinoma. Surgery. 2002;131:S39–S47. doi: 10.1067/msy.2002.119292. [DOI] [PubMed] [Google Scholar]
- 11.Suganuma K, Kubota T, Saikawa Y, Abe S, Otani Y, Furukawa T, Kumai K, Hasegawa H, Watanabe M, Kitajima M, et al. Possible chemoresistance-related genes for gastric cancer detected by cDNA microarray. Cancer Sci. 2003;94:355–359. doi: 10.1111/j.1349-7006.2003.tb01446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tay ST, Leong SH, Yu K, Aggarwal A, Tan SY, Lee CH, Wong K, Visvanathan J, Lim D, Wong WK, et al. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 2003;63:3309–3316. [PubMed] [Google Scholar]
- 13.Liu LX, Liu ZH, Jiang HC, Qu X, Zhang WH, Wu LF, Zhu AL, Wang XQ, Wu M. Profiling of differentially expressed genes in human gastric carcinoma by cDNA expression array. World J Gastroenterol. 2002;8:580–585. doi: 10.3748/wjg.v8.i4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liotta L, Petricoin E. Molecular profiling of human cancer. Nat Rev Genet. 2000;1:48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- 15.Alizadeh AA, Ross DT, Perou CM, van de Rijn M. Towards a novel classification of human malignancies based on gene expression patterns. J Pathol. 2001;195:41–52. doi: 10.1002/path.889. [DOI] [PubMed] [Google Scholar]
- 16.Lakhani SR, Ashworth A. Microarray and histopathological analysis of tumours: the future and the past? Nat Rev Cancer. 2001;1:151–157. doi: 10.1038/35101087. [DOI] [PubMed] [Google Scholar]
- 17.Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 18.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 19.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 21.Nygaard V, Løland A, Holden M, Langaas M, Rue H, Liu F, Myklebost O, Fodstad Ø, Hovig E, Smith-Sørensen B. Effects of mRNA amplification on gene expression ratios in cDNA experiments estimated by analysis of variance. BMC Genomics. 2003;4:11. doi: 10.1186/1471-2164-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarbia M, Becker KF, Höfler H. Pathology of upper gastrointestinal malignancies. Semin Oncol. 2004;31:465–475. doi: 10.1053/j.seminoncol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Gene chip expression analysis technical manual Affymetrix Inc. 2003 [Google Scholar]
- 24.GeneChip Expression Analysis Data Analysis Fundamentals Affymetrix Inc. 2002 [Google Scholar]
- 25.Bakay M, Chen YW, Borup R, Zhao P, Nagaraju K, Hoffman EP. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics. 2002;3:4. doi: 10.1186/1471-2105-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills JC, Gordon JI. A new approach for filtering noise from high-density oligonucleotide microarray datasets. Nucleic Acids Res. 2001;29:E72–E72. doi: 10.1093/nar/29.15.e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unger MA, Rishi M, Clemmer VB, Hartman JL, Keiper EA, Greshock JD, Chodosh LA, Liebman MN, Weber BL. Characterization of adjacent breast tumors using oligonucleotide microarrays. Breast Cancer Res. 2001;3:336–341. doi: 10.1186/bcr317. [DOI] [PMC free article] [PubMed] [Google Scholar]


